Abstract
Maternal hexachlorophene (HCP) exposure causes transient disruption of hippocampal neurogenesis in mouse offspring. We examined epigenetically hypermethylated and downregulated genes related to this HCP-induced disrupted neurogenesis. Mated female mice were dietary exposed to 0 or 100 ppm HCP from gestational day 6 to postnatal day (PND) 21 on weaning. The hippocampal dentate gyrus of male offspring was subjected to methyl-capture sequencing and real-time reverse transcription-polymerase chain reaction analyses on PND 21. Validation analyses on methylation identified three genes, Dlx4, Dmrt1, and Plcb4, showing promoter-region hypermethylation. Immunohistochemically, DLX4+, DMRT1+, and PLCB4+ cells in the dentate hilus co-expressed GAD67, a γ-aminobutyric acid (GABA)ergic neuron marker. HCP decreased all of three subpopulations as well as GAD67+ cells on PND 21. PLCB4+ cells also co-expressed the metabotropic glutamate receptor, GRM1. HCP also decreased transcript level of synaptic plasticity-related genes in the dentate gyrus and immunoreactive granule cells for synaptic plasticity-related ARC. On PND 77, all immunohistochemical cellular density changes were reversed, whereas the transcript expression of the synaptic plasticity-related genes fluctuated. Thus, HCP-exposed offspring transiently reduced the number of GABAergic interneurons. Among them, subpopulations expressing DLX4, DMRT1, or PLCB4 were transiently reduced in number through an epigenetic mechanism. Considering the role of the Dlx gene family in GABAergic interneuron migration and differentiation, the decreased number of DLX4+ cells may be responsible for reducing those GABAergic interneurons regulating neurogenesis. The effect on granule cell synaptic plasticity was sustained until the adult stage, and reduced GABAergic interneurons active in GRM1–PLCB4 signaling may be responsible for the suppression on weaning.
Keywords: epigenetic gene regulation, GABAergic interneuron, hexachlorophene, hippocampal dentate gyrus, hypermethylation, neurogenesis
The hippocampal dentate gyrus of the mammalian brain is crucial for higher brain functions, such as learning and memory that are correlated with adult hippocampal neurogenesis and activity-dependent synaptic plasticity (Vivar et al., 2013). During postnatal life, adult neurogenesis continues in the subgranular zone (SGZ) of the dentate gyrus (Zhao et al., 2008). The γ-aminobutyric acid (GABA)ergic interneurons in the hilus of the dentate gyrus innervate granule cell lineage populations to control neurogenesis in the SGZ (Masiulis et al., 2011). In addition to GABAergic neuronal inputs, various neurons outside the SGZ create synaptic connections with neurons in the dentate gyrus, such as cholinergic neurons and glutamatergic neurons (Fonnum et al., 1979; Zhu et al., 2008). Both cholinergic and glutamatergic inputs to the SGZ are important for maintaining proper proliferation and differentiation of granule cell lineages (Cameron et al., 1995; Zhu et al., 2008).
Hexachlorophene (HCP), used as an antimicrobial agent in soaps, liquid detergents, and cosmetics during the 1960s, had been widely used in agriculture as a plant fungicide and pesticide (Kennedy et al., 1976). HCP, now established as a typical neurotoxin, induces myelin vacuolation corresponding to the splitting of the intraperiod line of the myelin sheath in the cerebral white matter of rodents (Lampert et al., 1973). In chronically exposed rats, segmental demyelination and remyelination progress, and a few fibers undergo axonal degeneration (Maxwell and Le Quesne, 1979). Experimentally, HCP can be transferred into offspring through placenta and milk (Kennedy et al., 1977). In our previous study, developmental exposure of mice to HCP also induced myelin vacuolation in the brains of the offspring (Kato et al., 2016).
Because adult neurogenesis includes all processes from neuronal production to maturation, it has been thought that hippocampal neurogenesis is a sensitive target of both developmental and adult neurotoxicants. In particular, chemical toxicity could affect self-renewal of stem cells, proliferation and migration of progenitor cells, neuritogenesis, synaptogenesis, and myelinogenesis. It is possible that developmental neurotoxicant exposure causes abnormalities in the formation of the dentate gyrus and the regulatory system of adult neurogenesis to result in alterations in neurogenesis. We have recently shown that developmental exposure to glycidol, which targets axon terminals, impairs late-stage differentiation of the neurogenesis process involving density changes of interneuron subpopulations in the dentate hilus of rat offspring (Akane et al., 2013a). Furthermore, developmental exposure to HCP impairs intermediate-stage progenitor cells in the SGZ in rat or mouse offspring, probably by reducing nerve conduction velocity of regulatory neurons of neurogenesis (Itahashi et al., 2015; Kato et al., 2016). In our mouse HCP developmental exposure study, however, we observed no fluctuations in the density of GABAergic interneuron subpopulations in the dentate hilus, whereas transcript downregulations of cholinergic receptors and glutamate receptors were observed in the dentate gyrus, likely related to the disruption of hippocampal neurogenesis (Supplementary Figs. 1–3; Kato et al., 2016).
Recent studies indicate that various epigenetic mechanisms, including DNA methylation, are involved in regulating different aspects of adult neurogenesis (Sun et al., 2011). The understanding of DNA methylation as a long-lasting cellular memory necessary to maintain a cellular phenotype has recently been challenged by discoveries of its dynamic nature (Covic et al., 2010). This relationship is particularly common in CpG sites at promoter regions, where DNA methylation may directly interfere with transcription factor binding to DNA or indirectly suppress transcription through methylated DNA-binding proteins that recruit histone deacetylases, leading to chromatin condensation and subsequent gene silencing (Jones et al., 1998). Although the results of epigenetic changes on neurogenesis have remained unexplored, environmentally induced disruption of DNA methylation warrants further study (Ceccatelli et al., 2013), given the clear importance of DNA methylation to neuronal development. In fact, exposure to stress (Mueller and Bale, 2008), toxicants (Kundakovic et al., 2013), and maternal neglect (Weaver et al., 2004) in early life has shown to disrupt epigenetic programming involving DNA methylation in the brain, with lasting consequences for brain gene expression and behavior.
The present study was performed to clarify the potential effects of maternal HCP exposure on epigenetic regulation in the development of the hippocampal dentate gyrus that may affect adult neurogenesis in mouse offspring. For this purpose, the hippocampal dentate gyrus of male offspring was first subjected to a search for genes with downregulated transcripts induced by promoter region hypermethylation. We then examined the reversibility of the methylation status, transcript levels, and cellular distribution of the corresponding proteins in the dentate gyrus.
MATERIALS AND METHODS
Chemicals and animals
HCP (purity: >99%) was purchased from MP Biomedicals, LLC (Santa Ana, California). Mated female Slc: ICR mice were purchased from Japan SLC Inc. (Hamamatsu, Japan) at gestational day (GD) 1 (appearance of vaginal plugs was designated GD 0) and individually housed with their offspring in plastic cages containing paper chip bedding until postnatal day (PND) 21. Animals were maintained in an air-conditioned animal room (temperature: 23 ± 2°C, relative humidity: 55% ± 15%) with a 12-h light/dark cycle and provided MF basal diet purchased from Oriental Yeast Co., Ltd. (Tokyo, Japan) mixed with HCP and tap water from GD 6 to PND 21. Beginning on PND 21, male and female offspring were separated and reared with three or four animals per cage and provided MF basal diet and tap water ad libitum.
Animal experiment
The animal experimental protocol and samples used were identical to that previously reported (Kato et al., 2016), and exposure period and number of animals per group were in accordance with the OECD test guideline for the testing chemicals (Test No. 426: Developmental Neurotoxicity Study; OECD, 2007). In brief, mated female mice were randomly divided into three groups of 12 animals each and were treated with 0, 33, or 100 ppm HCP, which was mixed with their food, from GD 6 to day 21 after delivery, which was the day of weaning (Figure 1). On PND 3, the litters were randomly culled to preserve eight males per litter. If dams had fewer than eight male pups, female pups were added to maintain a total of eight pups per litter. Because neurogenesis is influenced by circulating levels of steroid hormones during the estrous cycle (Pawluski et al., 2009), only male offspring were used for all analyses in the present study. On PND 21, 10–11 male animals per group (one male offspring per dam) were randomly selected for use in brain immunohistochemistry assays and subjected to perfusion fixation through the left cardiac ventricle with cold 4% (w/v) paraformaldehyde (PFA) after deep anesthesia with CO2/O2. For perfusion, a Masterflex peristaltic pump (EW-7553-70/71; Cole-Parmer, Vernon Hills, Illinois) was used to apply a flow rate of 10 mL/min. On PND 77, 10–11 male offspring per group (one male offspring per dam) were also randomly selected for immunohistochemistry analysis and perfused with cold 4% PFA at a flow rate of 10 mL/min.
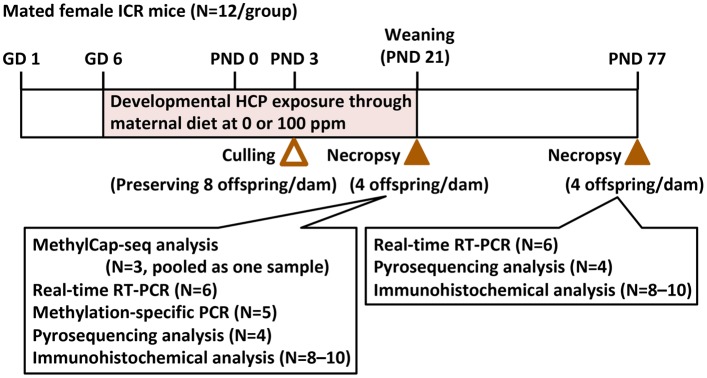
Figure 1.
Experimental design for the developmental exposure study of hexachlorophene (HCP) using mated female mice. Eight offspring were preserved in each dam after culling on postnatal day (PND) 3, and offspring were subjected to molecular and immunohistochemical analyses in the hippocampal dentate gyrus. One offspring per dam was used in each analysis.
All procedures in this study were conducted in compliance with the Guidelines for Proper Conduct of Animal Experiments (Science Council of Japan, June 1, 2006) and according to the protocol approved by the Animal Care and Use Committee of Tokyo University of Agriculture and Technology. All efforts were made to minimize animal suffering.
DNA and RNA extraction
For DNA and RNA extraction, male offspring brains were removed under CO2/O2 anesthesia on PND 21 and PND 77, fixed with methacarn solution for 5 h at 4°C, and then dehydrated in ice-cold absolute ethanol overnight at 4°C, as described previously (Akane et al., 2013b). A coronal brain slice obtained from a position at −2.2 mm from the bregma was prepared. Portions of the hippocampal dentate gyrus were collected using a biopsy punch (Ф1.0 mm; Kai Industries Co. Ltd., Seki, Japan) and stored in ethanol at −80°C until used for extraction.
For methylation analysis, genomic DNA and total RNA were extracted from tissue samples of the 0 ppm controls and the 100-ppm HCP-exposed group on PND 21 and PND 77 using an All-prep DNA/RNA mini kit (Qiagen, Hilden, Germany). As shown in Figure 1, extracted DNA was used for methyl-capture sequencing (MethylCap-seq) analysis (n = 3/group, pooled as one sample), quantitative methylation-specific PCR analysis (n = 5/group), and pyrosequencing assays (n = 4/group). Extracted total RNA was used for real-time RT-PCR analysis (n = 6/group).
MethylCap-seq analysis
MethylCap-seq analysis was performed in accordance with the manufacturers’ provided protocols using pooled genomic DNA sample of the 0 ppm controls and the 100-ppm HCP-exposed group on PND 21. In brief, genomic DNA was fragmented using a Bioruptor UCD-250 sonicator (Cosmo Bio Co. Ltd., Tokyo, Japan), and methylated DNA was enriched with methyl-CpG-binding domain 2 (MBD2) protein using an EpiXploreTM Methylated DNA Enrichment kit (Clontech Laboratories, Inc., Mountain View, Canada). Subsequently, enriched methylated DNA was used to construct libraries for sequencing using a DNA NEB® Next ChIP-Seq Library Prep Master Mix Set for Illumina® (New England Biolabs, Inc., Ipswich, Massachussets). The libraries, one from 0 ppm controls and the other from 100-ppm HCP-exposed group, were sequenced using a Miseq Sequencing System (Illumina, Inc., San Diego, California), and then data analysis was performed using Strand NGS (next-generation sequencing) analysis software (Strand Genomics, Inc., San Francisco, California). The genomic regions showing hypermethylation of CpG sites in the promoter region up to 1000 bp upstream from the transcription start site of the genes were selected using an enriched region detection algorithm with the criterion that the enrichment factor (ratio of HCP-exposed sample read counts/control sample read counts) was greater than 5.
Transcript expression analysis of candidate genes
Real-time RT-PCR quantification of mRNA was performed for genes selected as being hypermethylated using the MethylCap-seq data analysis with the RNA samples isolated from the 0 ppm controls and the 100-ppm HCP-exposed group on PND 21. The genes showing transcript downregulation on PND 21 were also analyzed for expression on PND 77. First-strand complementary DNA was synthesized using SuperScript® III Reverse Transcriptase (Thermo Fisher Scientific, Waltham, Massachussets) in a total reaction mixture of 20 μl from 1.0 μg of total RNA. Analysis of the transcript levels for candidate genes listed in Table 1 was performed using the PCR primers designed with Primer Express software Ver. 3.0 (Supplementary Table 1; Thermo Fisher Scientific). Real-time RT-PCR with Power SYBR® Green PCR Master Mix (Thermo Fisher Scientific) was conducted using a StepOnePlusTM Real-time PCR System (Thermo Fisher Scientific). The relative differences in gene expression between the 0 ppm controls and the 100-ppm HCP-exposed group were calculated using threshold cycle (CT) values that were first normalized to those of Hprt or Gapdh, which served as endogenous controls in the same sample, and then relative to a control CT value using the 2−ΔΔCT method (Livak and Schmittgen, 2001).
Table 1.
Genes Downstream of Hypermethylated CpG Sites in the Hippocampal Dentate Gyrus of Mouse Offspring Developmentally Exposed to HCP
| Genome Location | Gene ID | Gene Symbol | Description | Read Counts |
|
|---|---|---|---|---|---|
| 100 ppm HCP | 0 ppm HCP (Control) | ||||
| Chr 1 | MGI: 2443881 | Rasal2 | RAS protein activator like 2 | 19 | 3 |
| Chr 2 | MGI: 1890647 | Fign | Fidgetin | 26 | 2 |
| Chr 2 | MGI: 2675669 | Nusap1 | Nucleolar and spindle associated protein 1 | 26 | 0 |
| Chr 2 | MGI: 107464 | Plcb4 | Phospholipase C, beta 4 | 24 | 1 |
| Chr 4 | MGI: 1921896 | Spsb1 | SplA/ryanodine receptor domain and SOCS box containing 1 | 19 | 3 |
| Chr 4 | MGI: 1914775 | Trim62 | Tripartite motif-containing 62 | 24 | 4 |
| Chr 5 | MGI: 2441950 | Adgrl3 | Adhesion G protein-coupled receptor L3 | 28 | 3 |
| Chr 7 | MGI: 1338823 | Maz | MYC-associated zinc finger protein (purine-binding transcription factor) | 18 | 3 |
| Chr 7 | MGI: 3612342 | Vmn1r90 | Vomeronasal 1 receptor 90 | 24 | 2 |
| Chr 8 | MGI: 1261835 | Vps37a | Vacuolar protein sorting 37A | 28 | 5 |
| Chr 10 | MGI: 1196373 | Reps1 | RalBP1 associated Eps domain containing protein | 26 | 1 |
| Chr 11 | MGI: 94904 | Dlx4 | Distal-less homeobox 4 | 21 | 1 |
| Chr 11 | MGI: 3044668 | Gsdma3 | Gasdermin A3 | 22 | 4 |
| Chr 11 | MGI: 96911 | Mafg | v-Maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) | 23 | 4 |
| Chr 11 | MGI: 1933227 | Tex14 | Testis expressed gene 14 | 27 | 3 |
| Chr 13 | MGI: 1923387 | Mcur1 | Mitochondrial calcium uniporter regulator 1 | 23 | 2 |
| Chr 17 | MGI: 3587025 | Cdkl4 | Cyclin-dependent kinase-like 4 | 26 | 4 |
| Chr 19 | MGI: 1354733 | Dmrt1 | Doublesex and mab-3 related transcription factor 1 | 20 | 0 |
| Chr 19 | MGI: 1919449 | Mms19 | MMS19 (MET18 S. cerevisiae) | 24 | 3 |
Abbreviation: HCP, hexachlorophene.
Quantitative methylation-specific PCR analysis of candidate genes
Eight genes (Dlx4, Dmrt1, Fign, Gsdma3, Maz, Mms19, Plcb4, and Reps1) were selected for quantitative methylation-specific PCR analysis with the DNA samples isolated from the 0 ppm controls and the 100-ppm HCP-exposed group on PND 21. The isolated genomic DNA was sonicated using a Bioruptor UCD-250 sonicator (Cosmo Bio Co. Ltd.), mixed with incubation buffer, and then denatured with heat. Twenty percent of the mixture was stored as input DNA at −20°C until use. The remaining mixture was incubated with MBD2/magnetic bead complexes and then eluted. The methylation-enriched DNA was purified using an EpiXplore Methylated DNA Enrichment kit (Clontech Laboratories, Inc.). Input and methylation-enriched DNA samples were used as templates for quantitative measurement of methylation at target CpG sites by real-time PCR using Power SYBR® Green PCR Master Mix (Thermo Fisher Scientific) and a StepOnePlus Real-time PCR System (Thermo Fisher Scientific). The PCR primers for the target gene CpG sites were designed using Methyl Primer Express software Ver. 1.0 (Thermo Fisher Scientific) and Primer Express software Ver. 3.0 (Supplementary Table 2; Thermo Fisher Scientific). The quantification was based on the comparative CT method and involved a comparison of the CT values of the methylation-enriched DNA to the CT values of the input DNA.
Pyrosequencing analysis of candidate genes
The percentages of methylated CpG sites in the target sequences of Dlx4, Dmrt1, Plcb4, and Reps1 were measured with bisulfite-converted DNA using the PyroMark Q24 pyrosequencing system (Qiagen) with the DNA samples isolated from the 0 ppm controls and the 100-ppm HCP-exposed group on PND 21 and PND 77. The isolated genomic DNA was bisulfite converted with an EpiTect® Plus DNA Bisulfite kit (Qiagen) and then used as a template (10 ng) for biotin PCR reactions utilizing a Qiagen PyroMark PCR kit under the following conditions: 95°C for 15 min, (94°C for 30 s, 56°C for 30 s, and 72°C for 30 s) × 45 cycles, and 72°C for 10 min. The sequencing reactions were performed using PyroMark Gold Q24 reagents (Qiagen). Specific pyrosequencing primers were designed to amplify CpG sites using Pyrosequencing Assay Design software Ver. 2.0 (Supplementary Table 3; Qiagen).
Immunohistochemistry
For immunohistochemical analyses, perfusion-fixed brains were additionally fixed by permeation with 4% PFA overnight. Two millimeter coronal slices were prepared at −2.2 mm from bregma in PND 21 offspring brains and at −2.8 mm from bregma in PND 77 offspring brains (n = 8–10/group). Brain slices from offspring on PND 21 and PND 77 were further permeation-fixed with 4% PFA overnight at 4°C. Brain slices were processed using a standard protocol for paraffin embedding and were sectioned to a thickness of 3 μm. For immunohistochemistry using “mirror-image” pairs of paraffin sections, the first section of each pair was inverted before floating onto the water bath and picked up on the slide upside-down, and the second section was collected in the conventional manner.
Immunohistochemistry was performed by incubating 3-μm-thick brain tissue sections overnight at 4°C with the antibodies listed in Supplementary Table 4. To quench endogenous peroxidase, the sections were first incubated in 0.3% (v/v) hydrogen peroxide in absolute methanol for 30 min. Immunodetection was performed using a VECTASTAIN®Elite ABC kit (Vector Laboratories Inc., Burlingame, California) with 3, 3′-diaminobenzidine/H2O2 as the chromogen. The sections were then counterstained with hematoxylin and coverslipped for microscopic examination.
The following three proteins were selected for evaluation of immunohistochemical distribution after confirming their hypermethylation and transcript downregulation: distal-less homeobox 4 (DLX4), which is encoded by Dlx4, doublesex and mab-3-related transcription factor 1 (DMRT1), which is encoded by Dmrt1, and phospholipase C beta 4 (PLCB4), which is encoded by Plcb4.
For confirming the identity of DLX4+, DMRT1+, and PLCB4+ cells in the dentate hilus as GABAergic interneurons, immunohistochemistry of glutamic acid decarboxylase 67 (GAD67), a GABA-producing enzyme (Houser, 2007), was performed with DLX4, DMRT1, or PLCB4 using mirror sections in a pairwise fashion from an 0 ppm control brain. In addition, immunohistochemistry was performed examining GAD67 and GAD65 for evaluation of any change in the density of GABAergic interneurons between the 0 ppm controls and those exposed to 100 ppm HCP. To examine co-localization of PLCB4 with glutamate metabotropic receptor 1 (GRM1), a glutamate receptor subtype (Maejima et al., 2005), immunohistochemistry was performed examining PLCB4 and GRM1 using serial mirror sections from an 0 ppm control brain. In addition, immunohistochemistry of activity-regulated cytoskeleton (ARC)-associated protein, a member of the immediate-early genes involved in neuronal plasticity (Guzowski, 2002), was performed to investigate the functional relationship of PLCB4 and GRM1.
Morphometry of immunolocalized cells
The DLX4+, DMRT1+, PLCB4+, and GAD67+ cells distributed in the dentate hilus were bilaterally counted in an operator-blinded manner and normalized to the number per unit area of the hilar area (Supplementary Figure 4). ARC+ cells distributed in the granule cell layer (GCL) were also bilaterally counted in an operator-blinded manner and normalized to the length of the SGZ (Supplementary Figure 4). For quantitative measurement of each immunoreactive cellular component, digital photomicrographs at 400-fold magnification were captured using a BX53 microscope (Olympus Corp., Tokyo, Japan) attached to a DP72 Digital Camera System (Olympus Corp.), and quantitative measurements were performed using the WinROOF image analysis software package (version 5.7, Mitani Corp., Fukui, Japan).
Transcript expression analysis of synaptic plasticity-related genes
To investigate synaptic plasticity changes in the dentate gyrus, transcript levels of synaptic plasticity-related genes, including Dlg4, Gabbr1, Gabbr2, Gabra1, Gabrb2, Homer1, Stx4a, and Syp, were analyzed by real-time RT-PCR on PND 21 and PND 77 using the PCR primers listed in Supplementary Table 1. Gabbr1, Gabbr2, Gabra1, and Gabrb2 are GABA receptor family genes and are related to GCL long-term potentiation (Bramham and Sarvey, 1996). Dlg4 and Homer1 encode postsynaptic density proteins that play a role in stabilizing expression of the glutamate receptor on the postsynaptic membrane (Kempf et al., 2014; Tu et al., 1999). Stx4a and Syp encode synaptic vesicle proteins that play a role in synaptic vesicle exocytosis (Mohanasundaram and Shanmugam, 2010; Yong et al., 2013).
Statistical analysis
Numerical data are presented as mean ± SD. Immunoreactive cell counts for each antigen were analyzed using the litter as the experimental unit. Significant differences between the 0 ppm controls and the 100-ppm HCP-exposed group were evaluated as follows. Comparisons of numerical data between 0 ppm controls and 100-ppm HCP-exposed group were made using the F test for homogeneity of variance, and Student’s t test was applied when the variance was homogeneous between the groups as assessed using a test for equal variance. If a significant difference in variance was observed, the Aspin–Welch’s t test was performed. All analyses were conducted using an Excel Statistics 2010 software package (Social Survey Research Information Co. Ltd., Tokyo, Japan).
RESULTS
In Life Data
With regard to the parameters of dams, no significant difference was observed in reproductive parameters, body weight and food consumption during experiment, and body and brain weights at necropsy on weaning at PND 21 between the 0 ppm controls and 100-ppm HCP-exposed group (Supplementary Tables 5–8; Kato et al., 2016). With regard to the parameters of male offspring, body weight was significantly decreased from PND 13 to PND 21, and brain weight was also significantly decreased on PND 21 in 100-ppm HCP-exposed group compared with 0 ppm controls (Supplementary Tables 9 and 10; Kato et al., 2016).
Hypermethylated Genes Detected by MethylCap-seq Analysis
Nineteen genes with hypermethylated CpG sites located at the promoter region up to 1000 bp upstream from the transcription start site of the gene sequence showed ≥5-fold increase in methylation signals in the 100-ppm HCP-exposed group compared with the 0 ppm controls on PND 21 (Table 1).
Transcript Expression Changes of Candidate Genes in the Dentate Gyrus
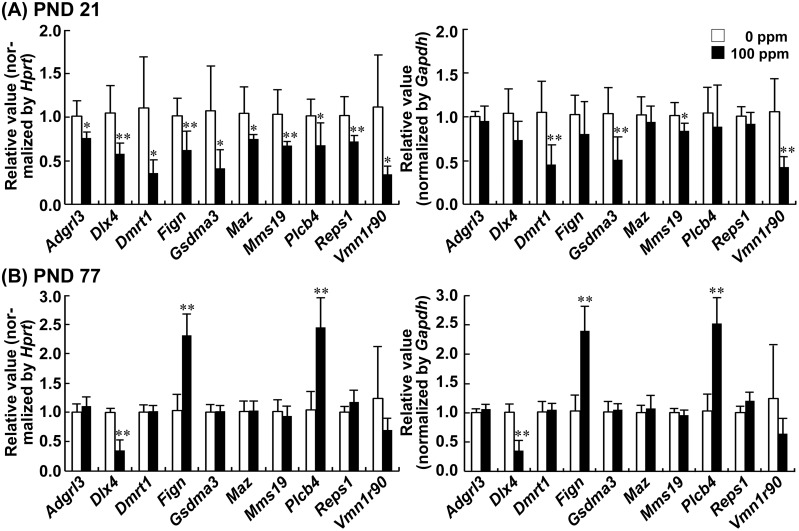
Among the 19 genes selected as showing ≥5-fold increase in methylation signals compared with the 0 ppm controls on PND 21, the transcript levels of Dmrt1, Gsdma3, Mms19, and Vmn1r90 were decreased after normalization with Hprt and Gapdh, and the transcript levels of Adgrl3, Dlx4, Fign, Maz, Plcb4, and Reps1 were decreased after normalization with Hprt in the 100-ppm HCP-exposed group compared with the 0 ppm controls on PND 21 (Figure 2A). By contrast, no 100-ppm HCP-exposure induced change was detected in transcript levels for Cdkl4, Mafg, Mcur1, Nusap1, Rasal2, Spsb1, Tex14, Trim62, and Vps37a (Supplementary Table 11).
Figure 2.
Transcript expression results obtained from MethylCap-seq analysis of hypermethylated genes in the hippocampal dentate gyrus of mice on postnatal day (PND) 21 and PND 77. A, PND 21. B, PND 77. Values are normalized to Hprt (left) or Gapdh (right) and expressed as the mean + SD; n = 6/group. *p < .05, **p < .01, significantly different from 0 ppm controls by Student’s or Aspin-Welch’s t test.
On PND 77, Dlx4 transcript levels were decreased, whereas Fign and Plcb4 transcript levels were increased in the 100-ppm HCP-exposed group compared with the 0 ppm controls after normalization with Hprt and Gapdh (Figure 2B). By contrast, no HCP-exposure induced change was detected in transcript levels for Adgrl3, Dmrt1, Gsdma3, Maz, Mms19, Reps1, and Vmn1r90.
Validation of Hypermethylation Status by Quantitative Methylation-Specific PCR
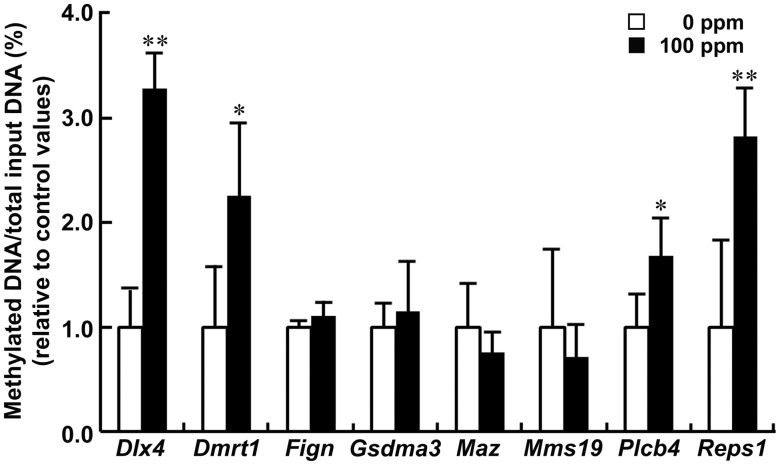
The methylation status of Dlx4, Dmrt1, Plcb4, and Reps1 significantly increased in the 100-ppm HCP-exposed group compared with 0 ppm controls, whereas Fign, Gsdma3, Maz, and Mms19 displayed no change in methylation status, on PND 21 (Figure 3). Although Adgrl3 and Vmn1r90 transcript levels were decreased on PND 21, both genes were excluded from quantitative methylation-specific PCR analysis because we were unable to construct appropriate primers.
Figure 3.
Quantitative methylation-specific PCR data of selected genes in the hippocampal dentate gyrus on postnatal day (PND) 21. Values are expressed as the mean + SD; n = 5/group. *p < .05, **p < .01, significantly different from 0 ppm controls by Student’s or Aspin-Welch’s t test.
Evaluation of DNA Methylation Status by Pyrosequencing
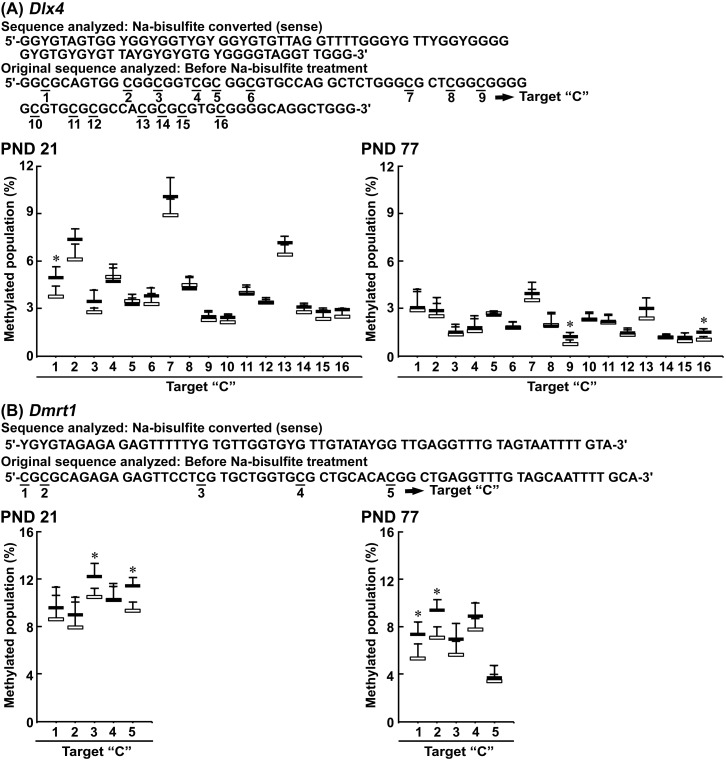
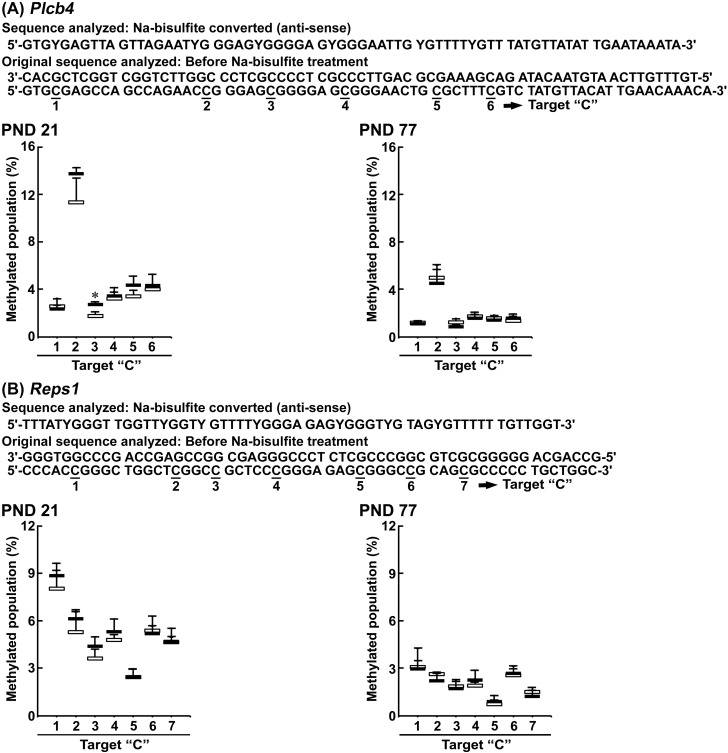
In Dlx4, Dmrt1, Plcb4, and Reps1, pyrosequencing of nucleotides 368–451, 160–222, 455–524, and 444–500 from the transcription start site, respectively, have CpG sites numbered as 1–16 in Dlx4, 1–5 in Dmrt1, 1–6 in Plcb4, and 1–7 in Reps1 (Figs. 4 and 5).
Figure 4.
Pyrosequencing results for Dlx4 and Dmrt1 in the hippocampal dentate gyrus on postnatal day (PND) 21. A, Dlx4. B, Dmrt1. All cytosine bases within CpG site are numbered as 1–16 or 1–5. White columns, 0 ppm controls; black columns, 100 ppm hexachlorophene (HCP). Values are expressed as the mean + SD.; n = 4/group. *p < .05, significantly different from 0 ppm controls by Student’s or Aspin-Welch’s t test.
Figure 5.
Pyrosequencing results for Plcb4 and Reps1 in the hippocampal dentate gyrus on postnatal day (PND) 21. A, Plcb4. B, Reps1. All cytosine bases within CpG site are numbered as 1–6 or 1–7. White columns, 0 ppm controls; black columns, 100 ppm hexachlorophene (HCP). Values are expressed as the mean + SD; n = 4/group. *p < .05, significantly different from 0 ppm controls by Student’s or Aspin-Welch’s t test.
On PND 21, cytosine bases in site numbers 1 of Dlx4, 3 and 5 of Dmrt1, and 3 of Plcb4 carried greater levels of methylation in the 100-ppm HCP-exposed group than in the 0 ppm controls. Methylation levels of cytosine bases in Reps1 were unchanged between the two groups.
On PND 77, cytosine bases in site numbers 9 and 16 of Dlx4, and 1 and 2 of Dmrt1 carried greater levels of methylation in the 100-ppm HCP-exposed group than in the 0 ppm controls. Methylation levels of cytosine bases in Plcb4 and Reps1 were unchanged between the two groups.
Distribution of Immunolocalized Cells
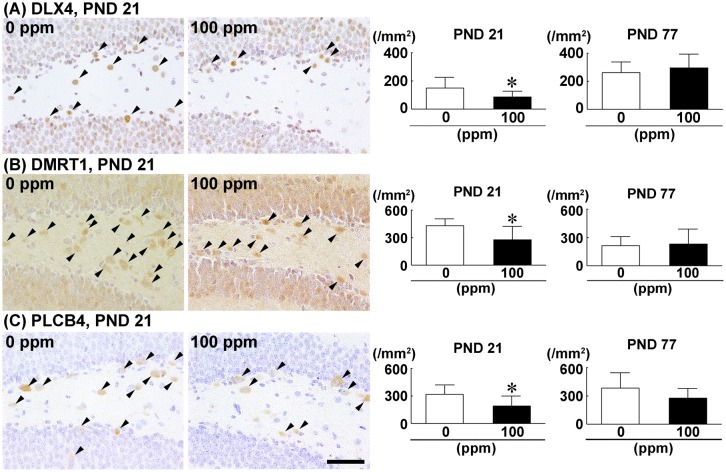
Using immunohistochemical analysis, we found cells in the hilus of the dentate gyrus expressing protein in their cytoplasm encoded by the hypermethylated genes (i.e., Dlx4, Dmrt1, and Plcb4). Densities of the DLX4+, DMRT1+, and PLCB4+ cells in the 100-ppm HCP-exposed group were significantly decreased compared with the 0 ppm controls on PND 21 (Figs. 6A–C). On PND 77, no significant difference was observed in the densities of DLX4+, DMRT1+, and PLCB4+ cells between the two groups (Figs. 6A–C;Supplementary Figs. 5A–C).
Figure 6.
Density of cells immunoreactive for DLX4, DMRT1, and PLCB4 in the hippocampal dentate hilus of male offspring on postnatal day (PND) 21 and PND 77 after maternal exposure to hexachlorophene (HCP) from gestational day 6 to PND 21. A, DLX4. B, DMRT1. C, PLCB4. Representative images from mice exposed to 0 ppm controls (left) and 100 ppm HCP (right) on PND 21. Arrowheads indicate immunoreactive cells. Magnification, ×400; scale bar, 50 µm. Graphs show the density of cells immunoreactive for the indicated molecule in the dentate hilus. n = 10/group. *p < .05, significantly different from 0 ppm controls by Student’s or Aspin-Welch’s t test.
An analysis of serial mirror sections showed that all subpopulations of cells in the dentate hilus expressing DLX4, DMRT1, or PLCB4 also expressed GAD67 (Figs. 7A–C). The density of GAD67+ cells in the dentate hilus was significantly decreased in the 100-ppm HCP-exposed group compared with the 0 ppm controls on PND 21 (Figure 7D). On PND 77, no significant difference was observed in the density of GAD67+ cells between the two groups (Supplementary Figure 5D). GAD65 immunoreactivity was only observed in the neuropil, and there were no countable GAD65+ cells showing cytoplasmic immunoreactivity in the dentate hilus.
Figure 7.
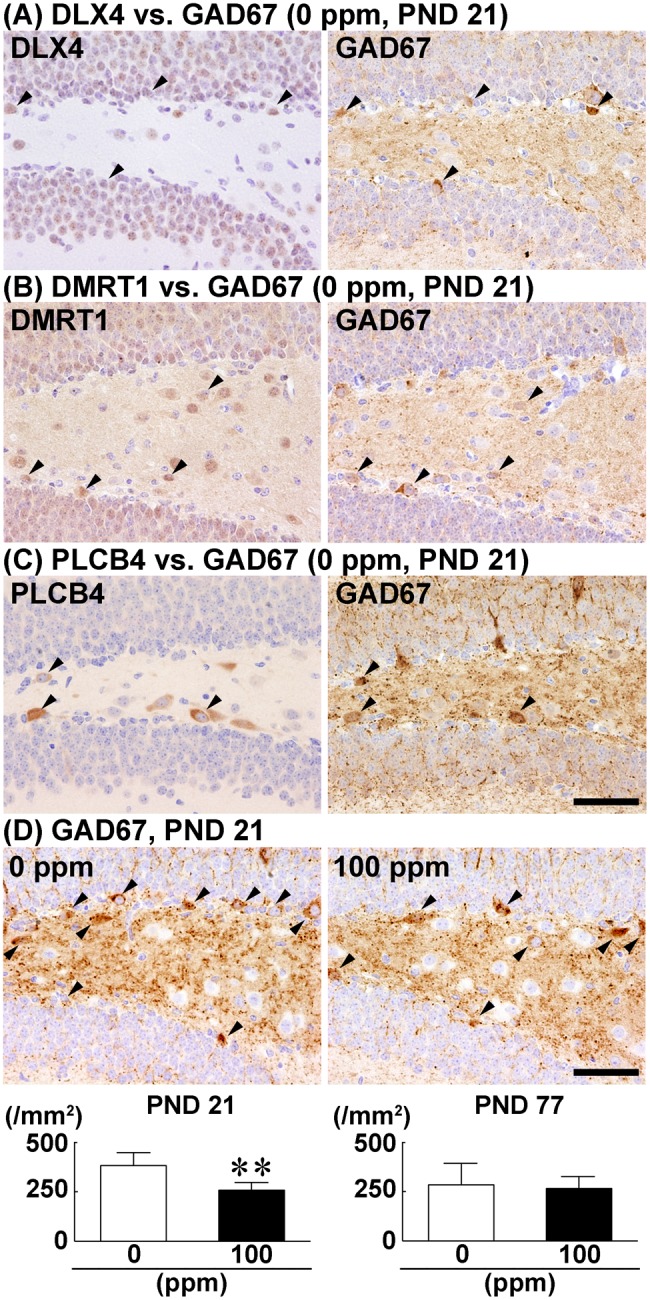
Density of cells immunoreactive for DLX4, DMRT1, and PLCB4 in the dentate hilus in relation to GAD67+ cells. A–C, Cellular identity of DLX4, DMRT1, and PLCB4 with GAD67+ cells analyzed using serial mirror sections. A, DLX4 vs. GAD67. B, DMRT1 vs. GAD67. C, PLCB4 vs. GAD67. Arrowheads indicate cells positive for both GAD67 and the indicated molecule. Magnification, ×400; scale bar, 50 µm. D, Density of GAD67+ cells in the dentate hilus of male offspring on postnatal day (PND) 21 and PND 77 after maternal exposure to hexachlorophene (HCP) from gestational day 6 to PND 21. Representative images from mice exposed to 0 ppm controls (left) and 100 ppm HCP (right) on PND 21. Arrowheads indicate GAD67+ cells. Magnification, ×400; scale bar, 50 µm. Graphs show the density of GAD67+ cells in the dentate hilus. n = 10/group. **p < .01, significantly different from 0 ppm controls by Student’s or Aspin–Welch’s t test.
With regard to PLCB4, an analysis of serial mirror sections showed that the subpopulation of cells in the dentate hilus expressing PLCB4 also expressed GRM1 (Figure 8A). The number of ARC+ cells in the GCL was decreased in 100-ppm HCP-exposed group compared the 0 ppm controls on PND 21 (Figure 8B). However, on PND 77, no significant difference was observed in the number of ARC+ cells between the two groups (Supplementary Figure 5E).
Figure 8.
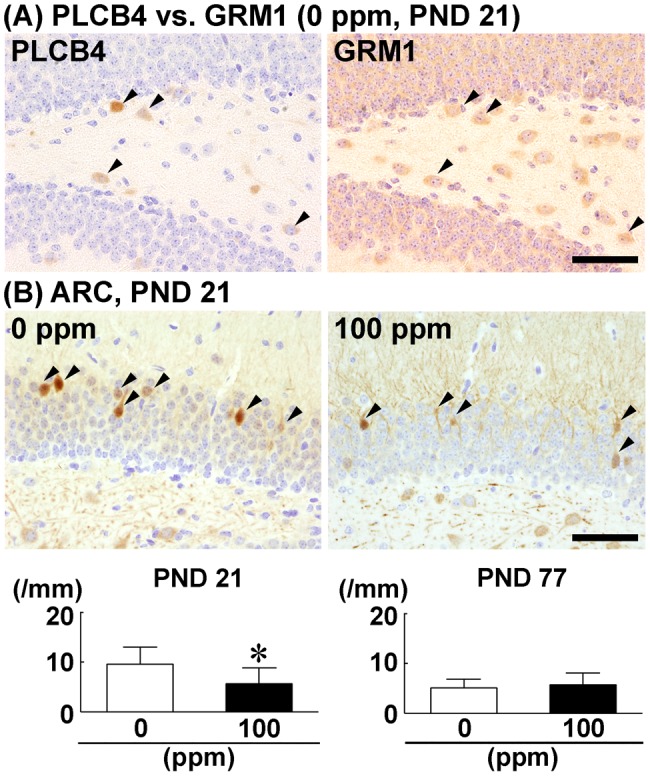
Distribution of GRM1+ cells in the dentate hilus in relation to PLCB4+ cells, and alteration in the number of ARC+ granule cells after maternal exposure to hexachlorophene (HCP). A, Identity of PLCB4+ cells as GRM1+ cells analyzed using serial mirror sections. Arrowheads indicate cells positive for both PLCB4 and GRM1. Magnification, ×400; scale bar, 50 µm. B, Number of ARC+ granule cells in male offspring on postnatal day (PND) 21 and PND 77 after maternal exposure to HCP from gestational day 6 to PND 21. Representative images from mice exposed to 0 ppm controls (left) and 100 ppm HCP (right) on PND 21. Magnification, ×400; scale bar, 50 µm. Graphs show the number of ARC+ cells in the granule cell layer. n = 8–10/group. *p < .05, significantly different from 0 ppm controls by Student’s or Aspin–Welch’s t test.
Transcript Expression Changes of Synaptic Plasticity-Related Genes in the Dentate Gyrus
On PND 21, the transcript level of Stx4a was decreased after normalization with Hprt and Gapdh, and transcript levels of Dlg4, Gabbr1, and Gabra1 were decreased after normalization with Hprt in the 100-ppm HCP-exposed group compared with the 0 ppm controls (Table 2). By contrast, transcript levels of Gabbr2, Gabrb2, Homer1, and Syp did not differ between the two groups. On PND 77, transcript levels of Gabbr2, Gabra1, and Gabrb2 were increased, whereas those for Homer1 and Stx4a were decreased after normalization with Hprt and Gapdh in the 100-ppm HCP-exposed group compared with the 0 ppm controls. Transcript levels of Dlg4, Gabbr1, and Syp did not differ between the two groups.
Table 2.
Transcript Expression of Synaptic Plasticity-Related Genes in the Hippocampal Dentate Gyrus
| 0 ppm HCP (Control) |
100 ppm HCP |
|||
|---|---|---|---|---|
| Relative Transcript Level Normalized to |
Relative Transcript Level Normalized to |
|||
| Gapdh | Hprt | Gapdh | Hprt | |
| No. of Animals Examined | 6 | 6 | 6 | 6 |
| PND 21 | ||||
| Dlg4 | 1.06±0.39 | 1.07±0.45 | 0.79±0.32 | 0.61±0.22* |
| Gabbr1 | 1.05±0.39 | 1.03±0.29 | 0.77±0.15 | 0.65±0.12* |
| Gabbr2 | 1.06±0.37 | 1.06±0.40 | 0.74±0.18 | 0.62±0.11 |
| Gabra1 | 1.03±0.26 | 1.04±0.30 | 0.76±0.18 | 0.64±0.13* |
| Gabrb2 | 1.01±0.17 | 1.02±0.26 | 1.10±0.31 | 0.96±0.38 |
| Homer1 | 1.02±0.20 | 1.03±0.26 | 1.30±0.52 | 1.10±0.61 |
| Stx4a | 1.02±0.19 | 1.03±0.26 | 0.74±0.14* | 0.63±0.12* |
| Syp | 1.02±0.21 | 1.04±0.35 | 0.88±0.15 | 0.75±0.16 |
| PND 77 | ||||
| Dlg4 | 1.02±0.22 | 1.04±0.32 | 0.92±0.07 | 0.95±0.14 |
| Gabbr1 | 1.00±0.08 | 1.04±0.30 | 1.13±0.22 | 1.17±0.21 |
| Gabbr2 | 1.03±0.23 | 1.03±0.29 | 1.53±0.39* | 1.59±0.42* |
| Gabra1 | 1.02±0.21 | 1.03±0.26 | 1.53±0.34* | 1.56±0.28** |
| Gabrb2 | 1.02±0.21 | 1.02±0.25 | 1.53±0.30** | 1.58±0.32** |
| Homer1 | 1.02±0.21 | 1.04±0.32 | 0.52±0.11** | 0.54±0.09* |
| Stx4a | 1.03±0.24 | 1.04±0.28 | 0.71±0.18* | 0.72±0.10* |
| Syp | 1.01±0.17 | 1.02±0.24 | 1.16±0.18 | 1.21±0.21 |
Data are expressed as the mean ± SD.
Abbreviations: Dlg4, discs large MAGUK scaffold protein 4; Gabbr1, gamma-aminobutyric acid (GABA) B receptor, 1; Gabbr2, gamma-aminobutyric acid (GABA) B receptor, 2; Gabra1, gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1; Gabrb2, gamma-aminobutyric acid (GABA) A receptor, subunit beta 2; Gapdh, glyceraldehyde 3-phosphate dehydrogenase; HCP, hexachlorophene; Homer1, homer scaffolding protein 1; Hprt, hypoxanthine guanine phosphoribosyl transferase; PND, postnatal day; Stx4a, syntaxin 4 A (placental); Syp, synaptophysin.
p < .05, and **p < .01, significantly different from 0 ppm controls by Student’s or Aspin–Welch’s t test.
DISCUSSION
In the present study, we found 19 genes in the hippocampal dentate gyrus of mice exposed to 100 ppm HCP using MethylCap-seq analysis to show hypermethylation of the gene promoter regions up to 1000 bp upstream from the transcription start site sequences. Among them, transcript levels of 10 genes decreased, and 3 of these, Dlx4, Dmrt1, and Plcb4, were confirmed to be hypermethylated by both quantitative methylation-specific PCR and pyrosequencing analyses. Immunohistochemically, the gene products of the three hypermethylated genes DLX4, DMRT1, and PLCB4 were expressed in variable populations of neuron in the dentate hilus and co-expressed GAD67 in variable population in an analysis by mirror section method. It is well-known that not all GABAergic interneurons express GAD67 (Houser, 2007). Therefore, even though not every DLX4+, DMRT1+, or PLCB4+ cells co-expressed GAD67, they are still likely to be GABAergic interneurons. Interestingly, the densities of DLX4+, DMRT1+, and PLCB4+ cells in the dentate hilus were decreased on weaning by developmental HCP exposure, in contrast to the unchanged density of hilar GABAergic interneurons expressing reelin, parvalbumin or calbindin D-28K as demonstrated in our previous study (Kato et al., 2016).
We previously showed that developmental HCP exposure of mice causes myelin vacuolation in the offspring brains (Kato et al., 2016). Although all dentate granule cells have unmyelinated axons (Kress et al., 2008), GABAergic interneurons, as well as other neurons regulating hippocampal neurogenesis from outside the SGZ, have myelinated axons (Jinno et al., 2007). Therefore, developmental HCP exposure may primarily affect the myelin sheath of neurons regulating neurogenesis, including GABAergic interneurons, during development to cause their dysfunction. We have previously discussed the possibility that such HCP-induced myelin vacuolation may reduce the nerve conduction velocity of the cholinergic inputs and GABAergic interneurons (Kato et al., 2016). We also previously found dysfunctional glutamatergic signaling in the dentate gyrus by developmental HCP exposure (Kato et al., 2016). Therefore, the dysfunction of the neurons regulating neurogenesis by developmental HCP exposure in mice may directly or indirectly influence the epigenetic gene expression of Dlx4, Dmrt1, and Plcb4 in GABAergic interneurons toward downregulation.
Dlx4 is a member of the distal-less homeobox genes (Panganiban and Rubenstein, 2002). Dlx1/Dlx2 double mutant mice display marked reduction in the numbers of GABAergic interneurons in multiple brain regions (Panganiban and Rubenstein, 2002). This reduction is thought to be due to the lack of the tangential migration of immature GABAergic interneurons from the subcortical telencephalon into the cerebral cortex (Anderson et al., 1997). Therefore, HCP-induced epigenetic downregulation of Dlx4 observed here may affect the migration of immature GABAergic interneurons to the dentate gyrus. Conversely, it is reported that forced expression of DLX4 in embryonic stem cells induces differentiation to GABAergic neuron-like cells expressing GAD67 (Teratani-Ota et al., 2016). In the present study, developmental HCP exposure induced reduction of GAD67+ GABAergic interneurons in the dentate hilus, suggestive of a suppressed differentiation of a progenitor cell population to a GABAergic interneuron subpopulation by epigenetically downregulating Dlx4. Because hilar GABAergic interneurons receive cholinergic signals for control of hippocampal neurogenesis (Zhu et al., 2008), the developmental HCP exposure-induced cholinergic receptor downregulation in the dentate gyrus that we have previously shown may be a reflection of reduced GABAergic interneuron subpopulations. Regarding the possibility of the functional involvement of Dlx family genes on neurogenesis, Dlx1/Dlx2 double mutant mice or Dlx5 or Dlx6 single mutant mice show aberrant differentiation of progenitor cells in subventricular zone neurogenesis (Panganiban and Rubenstein, 2002). Although the functional involvement of DLX4 on neurogenesis has not been previously shown, there is a possibility that the reduced density of DLX4+ GABAergic interneurons suppresses neurogenesis. Importantly, hilar GABAergic interneuron subpopulations regulate proliferation and differentiation of type-2 SGZ progenitor cells (Tozuka et al., 2005). Thus, decline of intermediate progenitor cells by developmental HCP exposure as revealed in our previous study may be caused by the reduction in hilar GABAergic interneurons in relation with the reduction of DLX4.
PLCB4 catalyzes the formation of inositol 1,4,5-trisphosphate and diacylglycerol to function as a second messenger of G protein-coupled receptors, such as the glutamate metabotropic receptor GRM1 (Maejima et al., 2005). We have previously reported that developmental HCP exposure decreases the Grm1 transcript level in the dentate gyrus in association with disrupted hippocampal neurogenesis (Kato et al., 2016). In the present study, we revealed that a subpopulation of PLCB4+ cells was identical to GRM1+ cells, and that hilar PLCB4+ cells were reduced by developmental HCP exposure. These results suggest that developmental HCP exposure targets the GRM1–PLCB4 signaling cascade in this GABAergic interneuron subpopulation. Pharmacological blockade of perisynaptically distributed GRM1 or GRM5 in interneurons abolishes the long-term potentiation of granule cell mossy fibers through suppression of synaptic plasticity (Hainmüller et al., 2014). In the present study, developmental HCP exposure reduced the number of ARC+ cells in the GCL on weaning. ARC is known to play a role in the axonal and synaptic plasticity (Guzowski, 2002). We also demonstrated here the developmental HCP exposure-induced transcript downregulation of the synaptic plasticity-related genes, i.e., Dlg4, Gabbr1, Gabra1, and Stx4a (Bramham and Sarvey, 1996; Kempf et al., 2014; Mohanasundaram and Shanmugam, 2010; Tu et al., 1999; Yong et al., 2013), in the dentate gyrus on weaning, suggesting confirmation of suppressed synaptic plasticity on weaning. The transcript downregulation of Stx4a was sustained on PND 77, whereas that of Homer1 first appeared on PND 77, suggestive of sustained suppressed synaptic plasticity until the adult stage. By contrast, transcript upregulation was observed for Gabbr2, Gabra1, and Gabrb2 on PND 77, suggestive of an operating compensatory mechanism for suppressed synaptic plasticity. Therefore, it is plausible that developmental HCP exposure causes decreased GCL neuronal plasticity through the reduction of GABAergic interneuron subpopulations active in the GRM1-PLCB4 signaling cascade, and this reduction may involve epigenetic downregulation of Plcb4 as one mechanism.
DMRT1, a member of a protein family that may share a novel DNA-binding motif called the DM domain, is a transcription factor involved in sexual differentiation, meiosis, and pluripotency in male germline stem cells (Takashima et al., 2013). Overexpression of Dmrt1 in embryonic stem cells induces differentiation toward neuron-like cells in vitro (Yamamizu et al., 2013). However, the role of DMRT1 in neuronal development is not reported. Our current finding that the hilar DMRT1+ cells are GABAergic interneurons suggests an epigenetic downregulation of Dmrt1 in relation with the suppression of GABAergic interneuron differentiation after developmental HCP exposure.
In the present study, Dlx4 and Dmrt1 showed promoter region hypermethylation on PND 77 as well as on PND 21 after developmental HCP exposure; however, the hypermethylated CpG sites differed in both genes between the two time points. Developmental bisphenol A exposure reportedly induces sustained transcript downregulation of Bdnf through promoter region hypermethylation in the hippocampus of mouse offspring until the adult stage, with different CpG sites hypermethylated on weaning and adult stages (Kundakovic et al., 2015). However, transcript downregulation of Dmrt1 was not sustained on PND 77 in the present study. The previous detection of methylation-sensitive and methylation-insensitive regulatory sequences for transcription in the gene promoter region (Kumar et al., 2016) suggests the hypermethylation at methylation-insensitive CpG sites of Dmrt1 on PND 77 after developmental HCP exposure. With regard to Dlx4, promoter region hypermethylation and transcript downregulation were sustained through PND 77, although these HCP-induced changes were not reflected in the hilar density of the DLX4+ cells. The reason for the unchanged density of DLX4+ cells was unclear, but compensatory mechanisms, such as those involving translational or posttranslational gene control, might have played a role. The epigenetic downregulation of Plcb4 observed in the present study on weaning was also reversed on PND 77, in parallel with the recovery from aberrant neurogenesis. We have previously shown that a reversibility in hypermethylation and expression downregulation in genes expressed in GABAergic interneurons, in contrast to a sustained downregulation in genes expressed in granule cell lineages, after developmental manganese exposure to cause sustained disruption of hippocampal neurogenesis in mice (Wang et al., 2012, 2013). It may be reasonable to suggest that hypermethylated neural stem cells continue production of hypermethylated granule cell lineage subpopulations to cause sustained hypermethylation through the adult stage. By contrast, methylation in nonproliferative postmitotic GABAergic interneurons may not be increased at the adult stage, and the number of hypermethylated cell populations that can be subjected to removal of excessive methyl bases by demethylation are limited, which differs from granule cell lineages.
In conclusion, our results suggested that the developmental HCP exposure that transiently affected hippocampal neurogenesis caused a transient reduction in GABAergic interneurons expressing DLX4, DMRT1, or PLCB4 in the hilus of the dentate gyrus through epigenetic gene expression downregulation. Developmental HCP exposure also reduced GAD67+ GABAergic interneurons. Considering the role of the Dlx gene family in GABAergic interneuron migration and differentiation, the reduction in DLX4+ cells might have decreased the GABAergic interneuron regulating neurogenesis. Developmental HCP exposure also induced influence on granule cell synaptic plasticity to be sustained through the adult stage, and the reduction in GABAergic interneurons active in GRM1–PLCB4 signaling might have been responsible for the suppressed plasticity observed on weaning.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Shigeko Suzuki for her technical assistance in preparing the histological specimens.
FUNDING
Grant-in-Aid for Scientific Research (B) from the Japan Society for the Promotion of Science (JSPS; 25292170); Research Fund from the Institute of Global Innovation Research, Tokyo University of Agriculture and Technology.
REFERENCES
- Akane H., Shiraki A., Imatanaka N., Akahori Y., Itahashi M., Ohishi T., Mitsumori K., Shibutani M. (2013a). Glycidol induces axonopathy by adult-stage exposure and aberration of hippocampal neurogenesis affecting late-stage differentiation by developmental exposure in rats. Toxicol. Sci. 134, 140–154. [DOI] [PubMed] [Google Scholar]
- Akane H., Saito F., Yamanaka H., Shiraki A., Imatanaka N., Akahori Y., Morita R., Mitsumori K., Shibutani M. (2013b). Methacarn as a whole brain fixative for gene and protein expression analyses of specific brain regions in rats. J. Toxicol. Sci. 38, 431–443. [DOI] [PubMed] [Google Scholar]
- Anderson S., Eisenstat D., Shi L., Rubenstein J. (1997). Interneuron migration from basal forebrain to neocortex: Dependence on Dlx genes. Science 278, 474–476. [DOI] [PubMed] [Google Scholar]
- Bramham C. R., Sarvey J. M. (1996). Endogenous activation of μ and δ-1 opioid receptors is required for long-term potentiation induction in the lateral perforant path: Dependence on GABAergic inhibition. J. Neurosci. 16, 8123–8131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron H. A., McEwen B. S., Gould E. (1995). Regulation of adult neurogenesis by excitatory input and NMDA receptor activation in the dentate gyrus. J. Neurosci. 15, 4687–4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccatelli S., Bose R., Edoff K., Onishchenko N., Spulber S. (2013). Long-lasting neurotoxic effects of exposure to methylmercury during development. J. Intern. Med. 273, 490–497. [DOI] [PubMed] [Google Scholar]
- Covic M., Karaca E., Lie D. C. (2010). Epigenetic regulation of neurogenesis in the adult hippocampus. Heredity (Edinb.) 105, 122–134.http://dx.doi.org/10.1038/hdy.2010.27 [DOI] [PubMed] [Google Scholar]
- Fonnum F., Karlsen R. L., Malthe-Sørenssen D., Skrede K. K., Walaas I. (1979). Localization of neurotransmitters, particularly glutamate, in hippocampus, septum, nucleus accumbens and superior colliculus. Prog. Brain Res. 51, 167–191. [DOI] [PubMed] [Google Scholar]
- Guzowski J. F. (2002). Insights into immediate-early gene function in hippocampal memory consolidation using antisense oligonucleotide and fluorescent imaging approaches. Hippocampus 12, 86–104.http://dx.doi.org/10.1002/hipo.10010 [DOI] [PubMed] [Google Scholar]
- Hainmüller T., Krieglstein K., Kulik A., Bartos M. (2014). Joint CP-AMPA and group I mGlu receptor activation is required for synaptic plasticity in dentate gyrus fast-spiking interneurons. Proc. Natl. Acad. Sci. USA 111, 13211–13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houser C. R. (2007). Interneurons of the dentate gyrus: An overview of cell types, terminal fields and neurochemical identity. Prog. Brain Res. 163, 217–232. [DOI] [PubMed] [Google Scholar]
- Itahashi M., Abe H., Tanaka T., Mizukami S., Kimura M., Yoshida T., Shibutani M. (2015). Maternal exposure to hexachlorophene targets intermediate-stage progenitor cells of the hippocampal neurogenesis in rat offspring via dysfunction of cholinergic inputs by myelin vacuolation. Toxicology 328, 123–134. [DOI] [PubMed] [Google Scholar]
- Jinno S., Klausberger T., Marton L. F., Dalezios Y., Roberts J. D., Fuentealba P., Bushong E. A., Henze D., Buzsáki G., Somogyi P. (2007). Neuronal diversity in GABAergic long-range projections from the hippocampus. J. Neurosci. 27, 8790–8804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. L., Veenstra G. J., Wade P. A., Vermaak D., Kass S. U., Landsberger N., Strouboulis J., Wolffe A. P. (1998). Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19, 187–191. [DOI] [PubMed] [Google Scholar]
- Kato M., Abe H., Itahashi M., Kikuchihara Y., Kimura M., Mizukami S., Yoshida T., Shibutani M. (2016). Maternal exposure to hexachlorophene targets intermediate-stage progenitor cells in the hippocampal neurogenesis involving myelin vacuolation of cholinergic and glutamatergic inputs in mice. J. Appl. Toxicol. 36, 211–222. [DOI] [PubMed] [Google Scholar]
- Kempf S. J., Casciati A., Buratovic S., Janik D., Toerne C., Ueffing M., Neff F., Moertl S., Stenerlöw B., Saran A. (2014). The cognitive defects of neonatally irradiated mice are accompanied by changed synaptic plasticity, adult neurogenesis and neuroinflammation. Mol. Neurodegener 9, 57.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy G. L., Dressler I. A., Richter W. R., Keplinger M. L., Calandra J. C. (1976). Effects of hexachlorophene in the rat and their reversibility. Toxicol. Appl. Pharmacol. 35, 137–145. [DOI] [PubMed] [Google Scholar]
- Kennedy G. L. Jr, Dressler I. A., Keplinger M. L., Calandra J. C. (1977). Placental and milk transfer of hexachlorophene in the rat. Toxicol. Appl. Pharmacol. 40, 571–576. [DOI] [PubMed] [Google Scholar]
- Kress G. J., Dowling M. J., Meeks J. P., Mennerick S. (2008). High threshold, proximal initiation, and slow conduction velocity of action potentials in dentate granule neuron mossy fibers. J. Neurophysiol. 100, 281–291.http://dx.doi.org/10.1152/jn.90295.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D. L., Kumar P. L., James P. F. (2016). Methylation-dependent and independent regulatory regions in the Na, K-ATPase alpha4 (Atp1a4) gene may impact its testis-specific expression. Gene 575, 339–352.http://dx.doi.org/10.1016/j.gene.2015.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundakovic M., Gudsnuk K., Franks B., Madrid J., Miller R. L., Perera F. P., Champagne F. A. (2013). Sex-specific epigenetic disruption and behavioral changes following low-dose in utero bisphenol A exposure. Proc. Natl. Acad. Sci. USA 110, 9956–9961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundakovic M., Gudsnuk K., Herbstman J. B., Tang D., Perera F. P., Champagne F. A. (2015). DNA methylation of BDNF as a biomarker of early-life adversity. Proc. Natl. Acad. Sci. USA 112, 6807–6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert P., O’Brien J., Garrett R. (1973). Hexachlorophene encephalopathy. Acta Neuropathol. 23, 326–333. [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Maejima T., Oka S., Hashimotodani Y., Ohno-Shosaku T., Aiba A., Wu D., Waku K., Sugiura T., Kano M. (2005). Synaptically driven endocannabinoid release requires Ca2+-assisted metabotropic glutamate receptor subtype 1 to phospholipase C β4 signaling cascade in the cerebellum. J. Neurosci. 25, 6826–6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masiulis I., Yun S., Eisch A. J. (2011). The interesting interplay between interneurons and adult hippocampal neurogenesis. Mol. Neurobiol. 44, 287–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell I. C., Le Quesne P. M. (1979). Conduction velocity in hexachlorophene neuropathy: Correlation between electrophysiological and histological findings. J. Neurol. Sci. 43, 95–110.http://dx.doi.org/10.1016/0022-510X(79)90075-3 [DOI] [PubMed] [Google Scholar]
- Mohanasundaram P., Shanmugam M. M. (2010). Role of syntaxin 4 in activity-dependent exocytosis and synaptic plasticity in hippocampal neurons. Sci. Signal. 3, jc7..http://dx.doi.org/10.1126/scisignal.3144jc7 [DOI] [PubMed] [Google Scholar]
- Mueller B. R., Bale T. L. (2008). Sex-specific programming of offspring emotionality after stress early in pregnancy. J. Neurosci. 28, 9055–9065.http://dx.doi.org/10.1523/JNEUROSCI.1424-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [OECD] Organisation for Economic Co-operation and Development (2007). Test No. 426: Developmental Neurotoxicity Study OECD Guidelines for the Testing of Chemicals, Section 4. OECD Publishing, Paris, France. [Google Scholar]
- Panganiban G., Rubenstein J. L. (2002). Developmental functions of the Distal-less/Dlx homeobox genes. Development 129, 4371–4386. [DOI] [PubMed] [Google Scholar]
- Pawluski J. L., Brummelte S., Barha C. K., Crozier T. M., Galea L. A. (2009). Effects of steroid hormones on neurogenesis in the hippocampus of the adult female rodent during the estrous cycle, pregnancy, lactation and aging. Front. Neuroendocrinol. 30, 343–357. [DOI] [PubMed] [Google Scholar]
- Sun J., Sun J., Ming G. L., Song H. (2011). Epigenetic regulation of neurogenesis in the adult mammalian brain. Eur. J. Neurosci. 33, 1087–1093.http://dx.doi.org/10.1111/j.1460-9568.2011.07607.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima S., Hirose M., Ogonuki N., Ebisuya M., Inoue K., Kanatsu-Shinohara M., Tanaka T., Nishida E., Ogura A., Shinohara T. (2013). Regulation of pluripotency in male germline stem cells by Dmrt1. Genes Dev. 27, 1949–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teratani-Ota Y., Yamamizu K., Piao Y., Sharova L., Amano M., Yu H., Schlessinger D., Ko M. S., Sharov A. A. (2016). Induction of specific neuron types by overexpression of single transcription factors. In Vitro Cell Dev. Biol. Anim. 52, 961–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tozuka Y., Fukuda S., Namba T., Seki T., Hisatsune T. (2005). GABAergic excitation promotes neuronal differentiation in adult hippocampal progenitor cells. Neuron 47, 803–815. [DOI] [PubMed] [Google Scholar]
- Tu J. C., Xiao B., Naisbitt S., Yuan J. P., Petralia R. S., Brakeman P., Doan A., Aakalu V. K., Lanahan A. A., Sheng M. et al. , (1999). Coupling of mGluR/Homer and PSD-95 complexes by the Shank family of postsynaptic density proteins. Neuron 23, 583–592. [DOI] [PubMed] [Google Scholar]
- Vivar C., Potter M. C., van Praag H. (2013). All about running: Synaptic plasticity, growth factors and adult hippocampal neurogenesis. Curr. Top. Behav. Neurosci. 15, 189–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Ohishi T., Shiraki A., Morita R., Akane H., Ikarashi Y., Mitsumori K., Shibutani M. (2012). Developmental exposure to manganese chloride induces sustained aberration of neurogenesis in the hippocampal dentate gyrus of mice. Toxicol. Sci. 127, 508–521. [DOI] [PubMed] [Google Scholar]
- Wang L., Shiraki A., Itahashi M., Akane H., Abe H., Mitsumori K., Shibutani M. (2013). Aberration in epigenetic gene regulation in hippocampal neurogenesis by developmental exposure to manganese chloride in mice. Toxicol. Sci. 136, 154–165. [DOI] [PubMed] [Google Scholar]
- Weaver I. C., Cervoni N., Champagne F. A., D’Alessio A. C., Sharma S., Seckl J. R., Dymov S., Szyf M., Meaney M. J. (2004). Epigenetic programming by maternal behavior. Nat. Neurosci. 7, 847–854. [DOI] [PubMed] [Google Scholar]
- Yamamizu K., Piao Y., Sharov A. A., Zsiros V., Yu H., Nakazawa K., Schlessinger D., Ko M. S. (2013). Identification of transcription factors for lineage-specific ESC differentiation. Stem Cell Rep. 1, 545–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong Z., Yan L., Dong Z., Wang X., Su R., Gong Z. (2013). The effect of chronic thienorphine administration on long-term potentiation and synaptic structure in rat hippocampus. Synapse 67, 779–785.http://dx.doi.org/10.1002/syn.21682 [DOI] [PubMed] [Google Scholar]
- Zhao C., Deng W., Gage F. H. (2008). Mechanisms and functional implications of adult neurogenesis. Cell. 132, 645–660. [DOI] [PubMed] [Google Scholar]
- Zhu G., Okada M., Yoshida S., Ueno S., Mori F., Takahara T., Saito R., Miura Y., Kishi A., Tomiyama M. et al. , (2008). Rats harboring S284L Chrna4 mutation show attenuation of synaptic and extrasynaptic GABAergic transmission and exhibit the nocturnal frontal lobe epilepsy phenotype. J. Neurosci. 28, 12465–12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.