Abstract
Bacillus salis strain ES3T (= CSUR P1478 = DSM 100598) is the type strain of B. salis sp. nov. It is an aerobic, Gram-positive, moderately halophilic, motile and spore-forming bacterium. It was isolated from commercial table salt as part of a broad culturomics study aiming to maximize the culture conditions for the in-depth exploration of halophilic bacteria in salty food. Here we describe the phenotypic characteristics of this isolate, its complete genome sequence and annotation, together with a comparison with closely related bacteria. Phylogenetic analysis based on 16S rRNA gene sequences indicated 97.5% similarity with Bacillus aquimaris, the closest species. The 8 329 771 bp long genome (one chromosome, no plasmids) exhibits a G+C content of 39.19%. It is composed of 18 scaffolds with 29 contigs. Of the 8303 predicted genes, 8109 were protein-coding genes and 194 were RNAs. A total of 5778 genes (71.25%) were assigned a putative function.
Keywords: Bacillus salis, culturomics, genome, halophilic bacteria, human gut, taxonogenomics
Introduction
Halophiles are considered as microorganisms living in hypersaline environments which often require a high salt concentration for growth. They are involved in centuries-old processes, such as production of salt and fermentation of food consumed by humans [1], [2]. Today, with the emergence of new biologic technologies, these organisms have been isolated and described from many traditional foods [2] such as salt [3].
Despite recent technologic advances in molecular biology, pure culture is the only way to characterize the physiologic properties of bacteria and to evaluate their potential virulence [4]. Therefore, we tried to investigate the population of halophilic prokaryotes in the human gut and salty food by using a culturomics approach. This approach allowed us to isolate a new member of the Bacillus genus. This bacterium is Gram negative, strictly aerobic, moderately halophilic and motile. It was isolated from commercial table salt. This isolation was part of a culturomics study using high-salt culture conditions in order to cultivate halophilic bacteria from human faeces and environmental samples [5]. This isolate is described using a new and innovative method that we have implemented [6]. The old methods, based on 16S rRNA sequencing, phylogeny, G + C content and DNA-DNA hybridization (DDH), are fastidious and include many limitations [6], [7].
The emergence of new tools for DNA sequencing and technology, such as matrix-assisted desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), has allowed an increase in available genomic and proteomic data over the last few years [8], [9]. These technologic advances have allowed us to develop a new way of describing bacterial species that takes into account genomic and protonic information [10].
Here we present a summary classification and a set of features for B. salis strain ES3T (= CSUR P1478 = DSM 100598), together with the description of its complete genomic sequence and its annotation.
Materials and methods
Strain isolation and identification
Culture condition
Culture was realized in an aerobic atmosphere on a homemade culture medium consisting of a Columbia agar culture (Sigma-Aldrich, Saint-Quentin Fallavier, France) modified by adding (per liter): MgCl2 6H2O, 5 g; MgSO4 7H2O, 5 g; KCl, 2 g; CaCl2 2H2O, 1 g; NaBr, 0.5 g; NaHCO3, 0.5 g, glucose, 2 g and 100 g/L of NaCl. The pH was adjusted to 7.5 with 10 M NaOH before autoclaving [3].
MALDI-TOF MS identification
The identification of our strain was carried out by a MALDI-TOF MS analysis with a Microflex spectrometer (Bruker Daltonics, Leipzig, Germany) as previously described [11]. Obtained spectra were then compared by using MALDI Biotyper 3.0 software (Bruker) as well as the Unité des Maladies Infectieuses et Tropicales Emergentes's (URMITE) database, which is constantly updated. If no identification was possible at the genus or species level (score <1.7), sequencing of the 16S rRNA gene was performed to achieve a correct identification [12], [13].
Sequencing of 16S rRNA gene
DNA extraction was performed using the EZ1 DNA Tissue Kit and BioRobot EZ1 Advanced XL (Qiagen, Courtaboeuf, France). The 16S rRNA gene was amplified using PCR technology and universal primers fD1 and rP2 [12] (Eurogentec, Angers, France). The amplifications and sequencing of the amplified products were performed as previously described [14]. Then 16S rRNA gene sequences were assembled and corrected using Codoncode Aligner software (http://www.codoncode.com/) and compared with those available in GenBank (http://www.ncbi.nlm.nih.gov/genbank/). Identification at the species level was defined by a 16S rRNA gene sequence similarity of ≥99% with the sequence of the type strain in GenBank. When the percentage of identity was <98.7%, the studied strain was considered as a new species [15].
Phylogenetic classification
Phylogenetic analysis based on 16S rRNA of our isolate was performed to identify its phylogenetic affiliations with other close isolates, including other members of the genus Bacillus. MEGA 6 (Molecular Evolutionary Genetics Analysis) software allowed us to construct a phylogenetic tree [16]. Sequence alignment of the different species was performed using CLUSTAL W [17], and evolutionary distance matrices for the neighbour-joining method were calculated using the algorithm of the Kimura two-parameter model [18].
Physiologic and phenotypic characteristics
Phenotypic tests
The phenotypic characteristics of this strain were studied by testing different parameters. Regarding temperature, we studied growth at 25, 30, 37, 45 and 56°C. Growth at various NaCl concentrations (0.5, 5, 7.5, 10, 15, 200 and 250%) was also investigated. The optimal pH for growth was determined by testing different pHs: 5, 6, 6.5, 7, 7.5, 8, 9 and 10. Growth of strain ES3T was tested under aerobic atmosphere, in the presence of 5% CO2 and also under anaerobic and microaerophilic atmospheres, created using AnaeroGen (Thermo Fisher Scientific, Saint Aubin, France) and CampyGen (Thermo Fisher Scientific) respectively.
Microscopy
Gram staining and motility were observed with a DM1000 light microscope (Leica Microsystems, Nanterre, France). Cell morphology was studied using a Tecnai G20 Cryo (FEI Company, Limeil-Brévannes, France) transmission electron microscope operated at 200 keV after negative staining of bacteria. Cells were first fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer for at least 1 hour at 4°C. A drop of cell suspension was deposited for approximately 5 minutes on glow-discharged formvar carbon film on 400 mesh nickel grids (FCF400-Ni; Electron Microscopy Sciences (EMS), Hatfield, PA, USA). The grids were dried on blotting paper, and cells were negatively stained for 10 seconds with 1% ammonium molybdate solution in filtered water at room temperature. Formation of spores was determined after thermal shock and observed under a microscope.
Biochemical test
Acid production from carbohydrates was determined by using the API 50CHB system (bioMérieux, Marcy l’Etoile, France). Other physiologic tests were performed with the API 20NE system (bioMérieux) and API ZYM (bioMérieux), according to the manufacturer's instructions.
Antibiotic susceptibility test
Antibiotic susceptibility was determined on Mueller-Hinton agar in a petri dish using the disc diffusion method according to European Committee on Antimicrobial Susceptibility Testing recommendations (bioMérieux) [19]. The following antibiotics were tested: doxycycline, rifampicin, vancomycin, nitrofurantoin, amoxicillin, erythromycin, ampicillin, ceftriaxone, ciprofloxacin, gentamicin, penicillin, trimethoprim/sulfamethoxazole, imipenem and metronidazole.
Fatty acid analysis
Cellular fatty acid methyl ester (FAME) analysis was performed by gas chromatography/mass spectrometry (GC/MS). Two samples were prepared with approximately 85 mg of bacterial biomass per tube collected from several culture plates. FAMEs were prepared as described by Sasser [20]. GC/MS analyses were carried out as previously described [21]. Briefly, FAMEs were separated using an Elite 5-MS column and monitored by mass spectrometry (Clarus 500–SQ 8 S; Perkin Elmer, Courtaboeuf, France). Spectral database search was performed using MS Search 2.0 operated with the Standard Reference Database 1A (National Institute of Standards and Technology, Gaithersburg, MD, USA) and the FAME mass spectral database (Wiley, Chichester, UK).
Genome sequencing
Genomic DNA (gDNA) of Bacillus salis was extracted in two steps. A mechanical treatment was first performed by acid-washed glass beads (G4649-500g; Sigma-Aldrich, St. Louis, MO, USA) using a FastPrep BIO 101 instrument (Qbiogene, Strasbourg, France) at maximum speed (6.5 m/s) for 90 seconds. Then after a 2-hour lysozyme incubation at 37°C, DNA was extracted on the EZ1 biorobot (Qiagen) with an EZ1 DNA tissue kit. The elution volume was 50 μL. gDNA was quantified by a Qubit assay with the high-sensitivity kit (Life Technologies, Carlsbad, CA, USA) to 120 ng/μL.
gDNA was sequenced with MiSeq Technology (Illumina, San Diego, CA, USA) with the mate-pair strategy. The gDNA was barcoded to be mixed with 11 other projects with the Nextera Mate Pair sample prep kit (Illumina). The mate-pair library was prepared with 1.5 μg gDNA using the Nextera mate-pair Illumina guide. The gDNA sample was simultaneously fragmented and tagged with a mate-pair junction adapter. The pattern of the fragmentation was validated on an Agilent 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA, USA) with a DNA 7500 labchip. The DNA fragments ranged in size from 1.5 to 11 kb, with an optimal size of 6.859 kb. No size selection was performed, and 600 ng of tagmented fragments were circularized. The circularized DNA was mechanically sheared to small fragments with an optimal at 921 bp on the Covaris S2 device in T6 tubes (Covaris, Woburn, MA, USA). The library profile was visualized on a High Sensitivity Bioanalyzer LabChip (Agilent Technologies), and the final concentration library was measured at 39.94 nmol/L. The libraries were normalized at 2 nM, and this library was added as two spots and all were pooled. After a denaturation step and dilution at 15 pM, the pool of libraries was loaded onto the reagent cartridge and then onto the instrument along with the flow cell. Automated cluster generation and a sequencing run were performed in a single 39-hour run with a 2 × 251 bp read length. Total information of 5.5 Gb was obtained from a 572K/mm2 cluster density, with a cluster passing quality control filters of 96.33% (11 740 000 passing filter paired reads). Within this run, the index representation for Bacillus salis was determined to be 14.60%. The 1 662 573 paired reads were trimmed and then assembled.
Genome annotation and comparison
The genome's assembly was performed with a pipeline that enabled us to create an assembly with different software (Velvet [22], Spades [23] and Soap Denovo [24]) on trimmed (MiSeq and Trimmomatic softwares) [25] or untrimmed data (only MiSeq software). For each of the six assemblies performed, GapCloser [24] was used to reduce gaps. Then contamination with Phage Phix was identified (BLASTn against Phage Phix174 DNA sequence) and eliminated. Finally, scaffolds of size <800 bp were removed, and scaffolds with a depth value of <25% of the mean depth were removed (identified as possible contaminants). The best assembly was selected by using different criteria (number of scaffolds, N50, number of N). For the studied strain, Spades gave the best assembly, with a depth coverage of 99×.
Open reading frames (ORFs) were predicted using Prodigal [26] with default parameters, but the predicted ORFs were excluded if they were spanning a sequencing gap region (contained N). The predicted bacterial protein sequences were searched against the Clusters of Orthologous Groups (COGs) database using BLASTP (E value 1e-03, coverage 0.7 and identity percentage 30%). If no hit was found, sequences were searched against the NR database using BLASTP with a E value of 1e-03, coverage 0.7 and identity percentage 30%. If the sequence length was smaller than 80 aa, we used an E value of 1e-05. The tRNAScanSE tool [27] was used to find transfer RNA genes, whereas ribosomal RNAs were found using RNAmmer [28]. Lipoprotein signal peptides and the number of transmembrane helices were predicted using Phobius [29]. ORFans were identified if the BLASTP performed did not give positive results (E value was lower than 1e-03 for ORFs with sequence size >80 aa; if alignment lengths were <80 aa, we used an E value of 1e-05). Such parameter thresholds have been used in previous work to define ORFans. The annotation process was performed in DAGOBAH [30], which includes Figenix [31] libraries that provided pipeline analysis.
Artemis was used for data management and DNAPlotter [32] for visualization of genomic features. The Mauve alignment tool (version 2.3.1) was used for multiple genomic sequence alignment [33]. To estimate the mean level of nucleotide sequence similarity at the genome level, we used MAGI homemade software to calculate the average genomic identity of orthologous gene sequences (AGIOS) among compared genomes. Briefly, this software is combined with the Proteinortho software [34] for detecting orthologous proteins in pairwise genomic comparisons; it then retrieves the corresponding genes and determines the mean percentage of nucleotide sequence identity among orthologous ORFs using the Needleman-Wunsch global alignment algorithm. Genomes from the genus Bacillus and closely related genera were used for the calculation of AGIOS values. The genomic similarity was evaluated among studied species close to the isolate by digital DNA-DNA hybridization (http://ggdc.dsmz.de/distcalc2.php).
Results and discussion
Strain identification and phylogenetic analyses
Strain ES3T was first isolated in May 2014 (Table 1) after 30 days of preincubation in aerobic culture on our homemade culture medium at 37°C. No significant MALDI-TOF MS score was obtained for strain ES3T against the Bruker and URMITE databases, suggesting that our isolate was not a member of a known species [9]. An almost complete 16S rRNA gene sequence of strain ES3T (accession no. LN827530) comprising 1505 nt was analysed. Comparative 16S rRNA gene sequences analyses showed that strain ES3T is phylogenetically affiliated with the Bacillus genus (Fig. 1). The phylogenetic distinctiveness (16S rRNA gene sequence similarity of <97%) confirms that strain ES3T represents a distinct species from the recognized species belonging to Bacillus genus [35]. In fact, strain ES3T exhibited 97.5% nucleotide sequence similarity with Bacillus aquimaris, the phylogenetically closest species with a validly published name [36]. The reference spectrum for strain ES3T was thus incremented in our database (Fig. 2), then compared to other known species of the genus Bacillus. The differences exhibited are shown in Fig. 3 in the obtained gel view.
Table 1.
Classification and general features of Bacillus salis strain ES3T
| Property | Term |
|---|---|
| Current classification | Domain: Bacteria |
| Phylum: Firmicutes | |
| Class: Bacilli | |
| Order: Bacillales | |
| Family: Bacillaceae | |
| Genus: Bacillus | |
| Species: Bacillus salis | |
| Type strain: ES3T | |
| Gram stain | Positive |
| Cell shape | Rod shaped |
| Motility | Motile |
| Sporulation | Endospore forming |
| Temperature range | Mesophile |
| Optimum temperature | 37°C |
| Optimum pH | 7.5 |
| Salinity | 5.0–200 g/L |
| Optimum salinity | 100 g/L |
| Oxygen requirement | Aerobic |
Fig. 1.
Phylogenetic tree highlighting position of Bacillus salis strain ES3T relative to other close species. GenBank accession numbers are indicated in parentheses. Sequences were aligned using CLUSTAL W, and phylogenetic inferences were obtained by Kimura two-parameter model within MEGA 6 software. Bacteroides thetaiotaomicron was used as outgroup. Scale bar represents 0.05% nucleotide sequence divergence.
Fig. 2.
Reference mass spectrum from Bacillus salis strain ES3T. Spectra from 12 individual colonies were compared and reference spectrum generated.
Fig. 3.
Gel view comparing Bacillus salis strain ES3T to members of genera Bacillus and Paenibacillus. Gel view displays raw spectra of all loaded spectrum files arranged in pseudo–gel-like look. X-axis records m/z value. Left y-axis displays running spectrum number originating from subsequent spectra loading. Peak intensity is expressed by greyscale scheme code. Colour bar and right y-axis indicate relation between colour peak; peak intensity is expressed in arbitrary units. Displayed species are indicated at left.
Phenotypic description
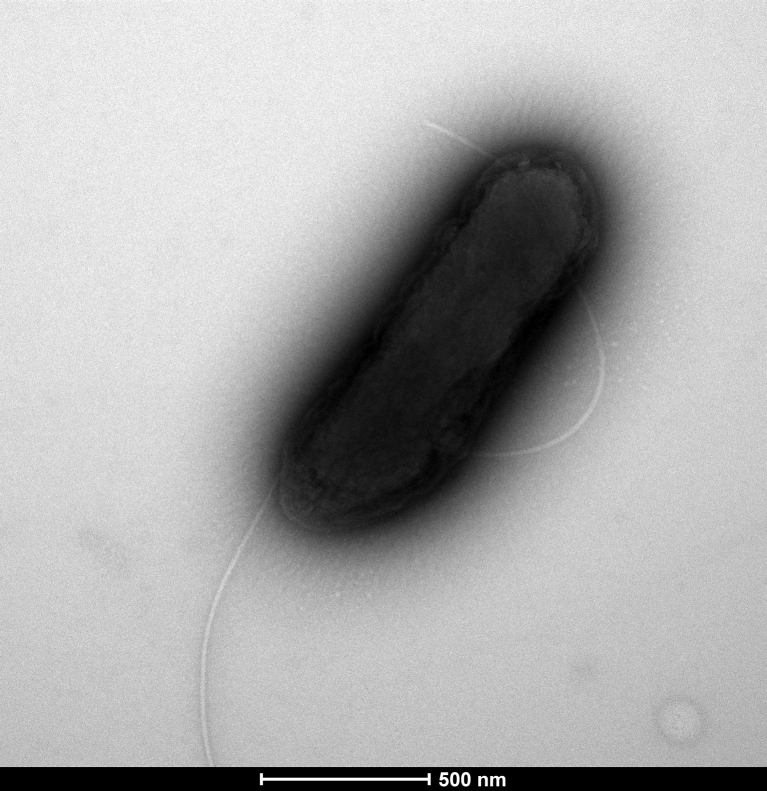
Strain ES3T formed creamy, smooth, circular and slightly irregular colonies 5 to 8 mm in diameter after incubation at 37°C for 2 days on our halophilic medium under an aerobic atmosphere. Growth occurred between 25 and 40°C, but not at 55°C. No growth was observed without NaCl, and the strain grew at salt concentrations ranging from 1% to 25% (w/v) NaCl, with optimum growth occurring at 10% (w/v) NaCl. Growth occurred between pH 6 and 10, with an optimum at pH 7.5. Cells were motile and spore forming. Gram staining (Fig. 4) showed Gram-positive rods. Strain ES3T exhibited catalase activity but no oxidase. Measured by electron microscopy, the rods had a mean diameter of 1.8 μm and a length of 5.9 μm (Fig. 5).
Fig. 4.
Gram staining of Bacillus salis strain ES3T.
Fig. 5.
Transmission electron microscopy of Bacillus salis strain ES3T. Cells were observed with Tecnai G20 transmission electron microscope operated at 200 keV. Scale bar = 500 nm.
Biochemical test
Using API 50CH strip, positive reactions was observed for d-glucose, d-fructose, d-mannose, arbutin, esculin ferric citrate, salicin, d-maltose, d-saccharose, d-trehalose, melezitose, d-raffinose and amidon; and negative reactions were recorded for glycerol, erythritol, d-arabinose, l-arabinose, d-ribose, d-xylose, l-xylose, d-adonitol, methyl-βd-xylopyranoside, d-galactose, l-sorbose, l-rhamnose, dulcitol, inositol, d-mannitol, d-sorbitol, methyl-αd-mannopyranoside, methyl-αd-glucopyranoside, N-acetyl-glucosamine, d-cellobiose, inulin, glycogen, xylitol, gentiobiose, d-turanose, d-lyxose, d-tagatose, d-fucose, l-fucose, d-arabitol, l-arabitol, potassium gluconate, potassium 2-ketogluconate and potassium 5-ketogluconate.
Using API 20NE, positive reactions were obtained for esculin ferric citrate, potassium nitrate, l-tryptophane, d-glucose (fermentation), l-arginine and urea. Glucose was assimilated. Nitrophenyl-βd-galactopyranoside, l-arabinose, d-mannose, d-mannitol, N-acetyl-glucosamine, d-maltose, potassium gluconate, capric acid, adipic acid, malic acid, trisodium citrate and phenylacetic acid were not assimilated.
When assayed with the API ZYM system, alkaline phosphatase, esterase (C4), esterase lipase (C8), acid phosphatase and naphthol-AS-BI-phosphohydrolase had an enzymatic activity, but lipase (C14), leucine arylamidase, valine arylamidase, cystine arylamidase, trypsin, α-chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase and α-fucosidase had no activity. Table 2 compares these features with closely related species.
Table 2.
Differential characteristics of Bacillus salis strain ES3T and Bacillus marisflavi strain TF-11T[36], Bacillus endophyticus strain 2DTT[37], Halobacillus halophilus strain Sl-4T[38], Paenibacillus terrae strain AM141T[39] and Paenibacillus sabinae strain T27T[40]
| Characteristic | B. salis | B. marisflavi | B. endophyticus | H. halophilus | P. terrae | P. sabinae |
|---|---|---|---|---|---|---|
| Cell diameter (μm) | 1.8 | 0.6–0.8 | 0.5–1.5 | 0.6–0.8 | 0.8–1.1 | 0.7–3.2 |
| Oxygen requirement | Aerobic | Aerobic | Aerobic | Aerobic | Aerobic | Aerobic |
| Gram stain | + | + to v | + to v | + | v | + |
| Motility | + | + | − | + | + | + |
| Endospore formation | + | + | − | + | + | + |
| Production of: | ||||||
| Catalase | + | + | − | + | + | + |
| Oxidase | − | − | + | + | − | − |
| Nitrate reductase | + | NA | − | − | + | + |
| Urease | + | − | − | − | − | NA |
| β-Galactosidase | − | NA | NA | NA | − | NA |
| N-acetyl-β-glucosaminidase | − | NA | NA | NA | + | NA |
| Acid from: | ||||||
| l-Arabinose | − | − | + | NA | − | − |
| d-Ribose | + | + | + | NA | − | + |
| d-Mannose | + | + | + | + | + | NA |
| d-Mannitol | − | + | − | − | + | NA |
| d-Sucrose | − | − | + | − | + | − |
| d-Glucose | + | + | + | − | + | + |
| d-Fructose | + | + | − | − | − | − |
| d-Maltose | − | − | − | NA | + | + |
| d-Lactose | − | − | − | NA | − | − |
| Starch | + | + | + | NA | NA | NA |
| Gelatin | + | + | + | NA | NA | NA |
| Habitat | Table salt | Seawater | Soil sediment | Soil | Soil | Salt lake |
+, positive result; −, negative result; v, variable result; NA, data not available.
Antibiotic susceptibility test
Cells were resistant to metronidazole but susceptible to imipenem, doxycycline, rifampicin, vancomycin, amoxicillin, ceftriaxone, gentamicin, trimethoprim/sulfamethoxazole, erythromycin, ciprofloxacin, nitrofurantoin, ampicillin and penicillin.
Fatty acids analysis
The major fatty acids found for this strain were branched: 12-methyl-tetradecanoic acid (60%), 14-methyl-hexadecanoic acid (17%) and 13-methyl-tetradecanoic acid (10%). The most abundant fatty acids were saturated ones (99%) (Table 3).
Table 3.
Cellular fatty acid composition (%)
| Fatty acid | IUPAC Name | Mean relative %a |
|---|---|---|
| 15:0 anteiso | 12-methyl-Tetradecanoic acid | 59.6 ± 1.1 |
| 17:0 anteiso | 14-methyl-Hexadecanoic acid | 17.3 ± 1.0 |
| 15:0 iso | 13-methyl-Tetradecanoic acid | 10.1 ± 1.6 |
| 16:0 | Hexadecanoic acid | 3.7 ± 0.2 |
| 14:0 | Tetradecanoic acid | 2.7 ± 0.4 |
| 16:0 iso | 14-methyl-Pentadecanoic acid | 2.1 ± 0.3 |
| 17:0 iso | 15-methyl-Hexadecanoic acid | 1.5 ± 0.1 |
| 16:1n9 | 7-Hexadecenoic acid | TR |
| 5:0 anteiso | 2-methyl-Butanoic acid | TR |
| 14:0iso | 12-methyl-Tridecanoic acid | TR |
| 13:0 anteiso | 10-methyl-Dodecanoic acid | TR |
| 17:1 iso | 15-methyl-Hexadecenoic acid | TR |
| 19:0 anteiso | 16-methyl-Octadecanoic acid | TR |
| 18:0 | Octadecanoic acid | TR |
| 16:1 iso | 14-methyl-Pentadecenoic acid | TR |
| 13:0 iso | 11-methyl-Dodecanoic acid | TR |
| 12:0 | Dodecanoic acid | TR |
IUPAC, International Union of Pure and Applied Chemistry; TR, trace amounts < 1%.
Mean peak area percentage.
Genome properties
The draft genome of strain ES3T is 8 329 771 bp long with 39.19% G+C content (Table 4, Fig. 6). It is composed of 18 scaffolds with 29 contigs. Of the 8303 predicted genes, 8109 were protein-coding genes and 194 were RNAs (20 genes 5S rRNA, two genes 16S rRNA, two genes 23S rRNA and 170 genes tRNA). A total of 5778 genes (71.25%) were assigned a putative function (by COGs or by NR BLAST). A total of 180 genes (2.22%) were identified as ORFans. The remaining genes were annotated as hypothetical proteins (1748 genes, 21.569%). Table 4 summarizes the genome's properties. Table 5 presents the distribution of genes into COGs functional categories.
Table 4.
Nucleotide content and gene count levels of genome
| Attribute | Value | % of totala |
|---|---|---|
| Size (bp) | 8 329 771 | 100 |
| G+C content (bp) | 3 263 777 | 39.18 |
| Coding region (bp) | 6 920 184 | 83.07 |
| Total genes | 8303 | 100 |
| RNA genes | 194 | 2.33 |
| Protein-coding genes | 8109 | 97.66 |
| Genes with function prediction | 5778 | 71.25 |
| Genes assigned to COGs | 5277 | 65.07 |
| Genes with peptide signals | 869 | 10.71 |
| Genes with transmembrane helices | 2032 | 25.05 |
COGs, Clusters of Orthologous Groups database.
The total is based on either the size of the genome in base pairs or the total number of protein coding genes in the annotated genome.
Fig. 6.
Circular map of Bacillus salis strain ES3T chromosome. From outside to centre: outer two circles show open reading frames oriented in forward (coloured by COGs categories) and reverse (coloured by COGs categories) directions, respectively. Third circle marks tRNA genes (green). Fourth circle shows G+C% content plot. Innermost circle shows GC skew, with purple indicating negative values and olive positive values. COGs, Clusters of Orthologous Groups database.
Table 5.
Number of genes associated with 25 general COGs functional categories
| Code | Value | % value | Description |
|---|---|---|---|
| J | 475 | 5.85 | Translation |
| 0 | 0 | RNA processing and modification | |
| K | 400 | 4.93 | Transcription |
| L | 215 | 2.65 | Replication, recombination and repair |
| B | 2 | 0.02 | Chromatin structure and dynamics |
| D | 102 | 1.25 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0 | Nuclear structure |
| V | 130 | 1.60 | Defense mechanisms |
| T | 288 | 3.55 | Signal transduction mechanisms |
| M | 260 | 3.20 | Cell wall/membrane biogenesis |
| N | 118 | 1.45 | Cell motility |
| Z | 0 | 0 | Cytoskeleton |
| W | 15 | 0.18 | Extracellular structures |
| U | 66 | 0.81 | Intracellular trafficking and secretion |
| O | 234 | 2.88 | Posttranslational modification, protein turnover, chaperones |
| X | 56 | 0.69 | Mobilome: prophages, transposons |
| C | 358 | 4.41 | Energy production and conversion |
| G | 431 | 5.31 | Carbohydrate transport and metabolism |
| E | 571 | 7.04 | Amino acid transport and metabolism |
| F | 208 | 2.56 | Nucleotide transport and metabolism |
| H | 318 | 3.92 | Coenzyme transport and metabolism |
| I | 333 | 4.10 | Lipid transport and metabolism |
| P | 323 | 3.98 | Inorganic ion transport and metabolism |
| Q | 176 | 2.17 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 560 | 6.90 | General function prediction only |
| S | 403 | 4.96 | Function unknown |
| — | 2832 | 34.92 | Not in COGs |
COGs, Clusters of Orthologous Groups database.
Genome comparison
We compared the genome sequence of strain ES3T (accession no. FNMN00000000) with that of halophilic bacteria close to our strain: Halobacillus halophilus strain DSM 2266 (HE717023), Bacillus endophyticus Hbe603 (NZ_CP011974), Bacillus marisflavi JCM 11544 (LGUE00000000), Paenibacillus sabinae T27 (CP004078) and Paenibacillus terrae HPl-003 (CP003107). The draft genome of strain ES3T (8.32 Mb) was larger than that of B. endophyticus, B. marisflavi, H. halophilus, P. sabinae and P. terrae (4.86, 4.31, 4.17, 5.27 and 6.08 Mb respectively). Its G+C content (39.19%) was smaller than that of B. marisflavi, H. halophilus, P. sabinae and P. terrae (48.60, 41.82, 52.6 and 46.80% respectively) but larger than that of B. endophyticus (36.60%). The gene content of strain ES3T (8303) was larger than that of B. endophyticus, B. marisflavi, H. halophilus, P. sabinae and P. terrae (4816, 4319, 4857 and 5396 respectively). However, the distribution of genes into COGs categories was similar in all compared genomes (Fig. 7). In addition, strain ES3T shared more orthologous genes with species belonging to the same genus (B. endophyticus, B. marisflavi, 1153 and 1151 genes respectively) than with other species belonging to other genus (H. halophilus, P. sabinae and P. terrae respectively shared 997, 701 and 725 orthologous genes) (Table 6). The average percentage of nucleotide sequence identity ranged from 65.34% to 65.84% at the intraspecies level between strain ES3T and the two Bacillus species, but it ranged from 57.74% to 60.05% between strain ES3T and the two other Paenibacillus species. Similar results were obtained for the analysis of DDH using Genome-to-Genome Distance Calculator (GGDC) software (Table 7).
Fig. 7.
Distribution of functional classes of predicted genes according to Clusters of Orthologous Groups of proteins.
Table 6.
Number of orthologous proteins shared between genomes (upper right) and AGIOS values obtained (lower left)
| BS | BE | BM | PS | PT | HH | |
|---|---|---|---|---|---|---|
| BS | 8118 | 1153 | 1151 | 701 | 725 | 997 |
| BE | 65.34% | 4846 | 1036 | 657 | 717 | 818 |
| BM | 65.84% | 62.01% | 4356 | 639 | 678 | 822 |
| PS | 57.74% | 57.64% | 60.32% | 4866 | 735 | 518 |
| PT | 60.05% | 60.41% | 60.35% | 67.59% | 5446 | 528 |
| HH | 66.03% | 62.50% | 61.65% | 57.85% | 59.29% | 4055 |
The bold represents the total number of orthologous proteins for each species.
AGIOS, average genomic identity of orthologous gene sequences; BE, Bacillus endophyticus strain Hbe603; BM, Bacillus marisflavi strain JCM 11544; BS, Bacillus salis strain ES3T; HH, Halobacillus halophilus strain DSM 2266; PS, Paenibacillus sabinae strain T27T; PT, Paenibacillus terrae strain HPl-003.
Table 7.
Pairwise comparison of strain ES3T with other species using GGDC, formula 2 (DDH estimates based on identities/HSP length)
| BE | BM | PS | PT | HH | |
|---|---|---|---|---|---|
| BS | 23.20 ± 2.38% | 19.0 ± 2.30% | 30.50 ± 2.45% | 22.00 ± 2.39% | 20.40 ± 2.32% |
| BE | 26.50 ± 2.42% | 29.20 ± 2.44% | 28.50 ± 2.44% | 29.80 ± 2.45% | |
| BM | 28.90 ± 2.44% | 28.50 ± 2.44% | 22.70 ± 2.37% | ||
| PS | 26.00 ± 2.41% | 29.40 ± 2.44% | |||
| PT | 28.70 ± 2.44% |
Confidence intervals indicate inherent uncertainty in estimating DDH values from intergenomic distances based on models derived from empirical test data sets (which are always limited in size). These results are in accordance with 16S rRNA (Fig. 1) and phylogenomic analyses as well as GGDC results.
BE, Bacillus endophyticus strain Hbe603; BM, Bacillus marisflavi strain JCM 11544; BS, Bacillus salis strain ES3T; DDH, DNA-DNA hybridization; GGDC, Genome-to-Genome Distance Calculator; HH, Halobacillus halophilus strain DSM 2266; HSP, high-scoring segment pairs; PS, Paenibacillus sabinae strain T27; PT, Paenibacillus terrae strain HPl-003.
Conclusion
On the basis of the phenotypic properties (Table 2), phylogenetic tree (Fig. 1), MALDI-TOF MS analyses (Fig. 3), genomic comparison via taxonogenomics (Table 6, Table 7) and GGDC results, we propose the creation of Bacillus salis sp. nov., represented by the type strain ES3T.
Description of Bacillus salis sp. nov.
Bacillus salis (sa'lis, L. gen. n., salis, from ‘salt,’ in which the strain was first identified)
Colonies which grew after 48 hours' incubation at 37°C on our homemade culture medium were creamy, smooth, circular and slightly irregular, and measured 5 to 8 mm in diameter. Cells were Gram-positive rods and had a mean diameter of 1.8 μm and a length of 5.9 μm. The strain was able to form subterminal ellipsoidal spores and was motile with a single polar flagella. Growth occurred optimally at 37°C, pH 7.5 and 10% NaCl.
API 50CH strip testing showed positive reactions for d-glucose, d-fructose, d-mannose, arbutin, esculin ferric citrate, salicin, d-maltose, d-saccharose, d-trehalose, melezitose, d-raffinose and amidon. Negative reactions were recorded for glycerol, erythritol, d-arabinose, l-arabinose, d-ribose, d-xylose, l-xylose, d-adonitol, methyl-βd-xylopyranoside, d-galactose, l-sorbose, l-rhamnose, dulcitol, inositol, d-mannitol, d-sorbitol, methyl-αd-mannopyranoside, methyl-αd-glucopyranoside, N-acetyl-glucosamine, d-cellobiose, inulin, glycogen, xylitol, gentiobiose, d-turanose, d-lyxose, d-tagatose, d-fucose, l-fucose, d-arabitol, l-arabitol, potassium gluconate, potassium 2-ketogluconate and potassium 5-ketogluconate, potassium gluconate, potassium 2-ketogluconate and potassium 5-ketogluconate.
API 20NE testing showed positive reactions for esculin ferric citrate, potassium nitrate, l-tryptophane, d-glucose (fermentation), l-arginine and urea. Glucose was assimilated. Nitrophenyl-βd-galactopyranoside, l-arabinose, d-mannose, d-mannitol, N-acetyl-glucosamine, d-maltose, potassium gluconate, capric acid, adipic acid, malic acid, trisodium citrate and phenylacetic acid were not assimilated.
When assayed with the API ZYM system, alkaline phosphatase, esterase (C4), esterase lipase (C8), acid phosphatase and naphthol-AS-BI-phosphohydrolase had an enzymatic activity, but lipase (C14), leucine arylamidase, valine arylamidase, cystine arylamidase, trypsin, α-chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase and α-fucosidase had no activity.
The type strain was sensitive to imipenem, doxycycline, rifampicin, vancomycin, amoxicillin, ceftriaxone, gentamicin (500 μg), trimethoprim/sulfamethoxazole, erythromycin, ciprofloxacin, nitrofurantoin, ampicillin, penicillin and gentamicin (15 μg) but resistant to metronidazole (500 μg).
The major fatty acids found for this strain were branched: 12-methyl-tetradecanoic acid (60%), 14-methyl-hexadecanoic acid (17%) and 13-methyl-tetradecanoic acid (10%). The most abundant fatty acids were saturated ones (99%). The G+C content of the genome was 39.19%. The 16S rRNA gene sequence and whole-genome shotgun sequence of B. salis strain ES3T were deposited in GenBank under accession numbers LN827530 and FNMN00000000, respectively. The type strain of Bacillus salis is strain ES3T (= CSUR P1478 = DSM 100598) and was isolated from salt.
Acknowledgements
The authors thank the Xegen Company (www.xegen.fr) for automating the genomic annotation process, and M. Lardière for English-language editorial work. This study was funded by the Fondation Méditerranée Infection.
Conflict of interest
None declared.
References
- 1.Kivistö A.T., Karp M.T. Halophilic anaerobic fermentative bacteria. J Biotechnol. 2011;152:114–124. doi: 10.1016/j.jbiotec.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 2.Lee H.S. Diversity of halophilic archaea in fermented foods and human intestines and their application. J Microbiol Biotechnol. 2013;23:1645–1653. doi: 10.4014/jmb.1308.08015. [DOI] [PubMed] [Google Scholar]
- 3.Diop A., Khelaifia S., Armstrong N., Labas N., Fournier P.-E., Raoult D. Microbial culturomics unravels the halophilic microbiota repertoire of table salt: description of Gracilibacillus massiliensis sp. nov. Microb Ecol Health Dis. 2016;27:32049. doi: 10.3402/mehd.v27.32049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vartoukian S.R., Palmer R.M., Wade W.G. Strategies for culture of ‘unculturable’ bacteria. FEMS Microbiol Lett. 2010;309:1–7. doi: 10.1111/j.1574-6968.2010.02000.x. [DOI] [PubMed] [Google Scholar]
- 5.Lagier J.C., Armougom F., Million M., Hugon P., Pagnier I., Robert C. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 6.Ramasamy D., Mishra A.K., Lagier J.C., Padhmanabhan R., Rossi M., Sentausa E. A polyphasic strategy incorporating genomic data for the taxonomic description of novel bacterial species. Int J Syst Evol Microbiol. 2014;64:384–391. doi: 10.1099/ijs.0.057091-0. [DOI] [PubMed] [Google Scholar]
- 7.Auch A.F., von Jan M., Klenk H.P., Göker M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci. 2010;2:117–134. doi: 10.4056/sigs.531120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., Manichanh C. A human gut microbial gene catalog established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seng P., Abat C., Rolain J.M., Colson P., Lagier J.C., Gouriet F. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol. 2013;51:2182–2194. doi: 10.1128/JCM.00492-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bouvet P., Ferraris L., Dauphin B., Popoffa M.-R., Butelb M.J., Julio Aires J. 16S rRNA gene sequencing, multilocus sequence analysis, and mass spectrometry identification of the proposed new species ‘Clostridium neonatale’. J Clin Microbiol. 2014;52:4129–4136. doi: 10.1128/JCM.00477-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lo C.I., Fall B., Ba S., Diawara S., Gueye M.W., Mediannikov O. MALDI-TOF mass spectrometry: a powerful tool for clinical microbiology at Hôpital principal de Dakar, Senegal (West Africa) PLoS One. 2015;10:e0145889. doi: 10.1371/journal.pone.0145889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weisburg W.G., Barns S.M., Pelletier D.A., Lane D.J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drancourt M., Bollet C., Carlioz A., Martelin R., Gayral J.P., Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol. 2000;38:3623–3630. doi: 10.1128/jcm.38.10.3623-3630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morel A.S., Dubourg G., Prudent E., Edouard S., Gouriet F., Casalta J.-P. Complementarity between targeted real-time specific PCR and conventional broad-range 16S rDNA PCR in the syndrome-driven diagnosis of infectious diseases. Eur J Clin Microbiol Infect Dis. 2015;34:561–570. doi: 10.1007/s10096-014-2263-z. [DOI] [PubMed] [Google Scholar]
- 15.Tindall B.J. The designated type strain of Pseudomonas halophila Fendrich 1989 is DSM 3051, the designated type strain of Halovibrio variabilis Fendrich 1989 is DSM 3050, the new name Halomonas utahensis (Fendrich 1989) Sorokin and Tindall 2006 is created for the species represented by DSM 3051 when treated as a member of the genus Halomonas, the combination Halomonas variabilis (Fendrich 1989) Dobson and Franzmann 1996 is rejected, and the combination Halovibrio denitrificans Sorokin et al. 2006 is validly published with an emendation of the description of the genus Halovibrio Fendrich 1989 emend. Sorokin et al. 2006. Opinion 93. Judicial Commission of the International Committee on Systematics of Prokaryotes. Int J Syst Evol Microbiol. 2014;64:3588–3589. doi: 10.1099/ijs.0.069195-0. [DOI] [PubMed] [Google Scholar]
- 16.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 19.Matuschek E., Brown D.F.J., Kahlmeter G. Development of the EUCAST disk diffusion antimicrobial susceptibility testing method and its implementation in routine microbiology laboratories. Clin Microbiol Infect. 2014;20:O255–O266. doi: 10.1111/1469-0691.12373. [DOI] [PubMed] [Google Scholar]
- 20.Sasser M. Microbial ID; Newark, NY: 2006. Bacterial identification by gas chromatographic analysis of fatty acids methyl esters (GC-FAME) [Google Scholar]
- 21.Dione N., Sankar S.A., Lagier J.C., Khelaifia S., Michele C., Armstrong N. Genome sequence and description of Anaerosalibacter massiliensis sp. nov. New Microbe New Infect. 2016;11(10):66–76. doi: 10.1016/j.nmni.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bankevich A., Nurk S., Antipov D., Edouard S., Gouriet F., Casalta J.P. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hyatt D., Chen G.L., LoCascio P.F., Land M.L., Larimer F.W., Hauser L.J. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:1. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lowe T.M., Eddy S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lagesen K., Hallin P., Rodland E.A., Staerfeldt H.H., Rognes T., Ussery D.W. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Käll L., Krogh A., Sonnhammer E.L. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 30.Gouret P., Paganini J., Dainat J., Louati D., Darbo E., Pontarotti P. Integration of evolutionary biology concepts for functional annotation and automation of complex research in evolution: the multi-agent software system DAGOBAH. In: Pontarotti P., editor. Evolutionary biology: concepts, biodiversity, macroevolution and genome evolution. Springer Verlag; Berlin: 2011. pp. 71–87. [Google Scholar]
- 31.Gouret P., Vitiello V., Balandraud N., Gilles A., Pontarotti P., Danchin E.G. FIGENIX: intelligent automation of genomic annotation: expertise integration in a new software platform. BMC Bioinform. 2005;6:198. doi: 10.1186/1471-2105-6-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carver T., Thomson N., Bleasby A., Berriman M., Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25:119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darling A.C., Mau B., Blattner F.R., Perna N.T. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lechner M., Findeiss S., Steiner L., Marz M., Stadler P.F., Prohaska S.J. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinform. 2011;12:124. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coorevits A., Logan N.A., Dinsdale A.E., Halket G., Scheldeman P., Heyndrickx M. Bacillus thermolactis sp. nov., isolated from dairy farms, and emended description of Bacillus thermoamylovorans. Int J Syst Evol Microbiol. 2011;61:1954–1961. doi: 10.1099/ijs.0.024240-0. [DOI] [PubMed] [Google Scholar]
- 36.Yoon J.H., Kim I.G., Kang K.H., Oh T.K., Park Y.H. Bacillus marisflavi sp. nov. and Bacillus aquimaris sp. nov., isolated from sea water of a tidal flat of the Yellow Sea in Korea. Int J Syst Evol Microbiol. 2003;53:1297–1303. doi: 10.1099/ijs.0.02365-0. [DOI] [PubMed] [Google Scholar]
- 37.Reva O.N., Smirnov V.V., Pettersson B., Priest F.G. Bacillus endophyticus sp. nov., isolated from the inner tissues of cotton plants (Gossypium sp.) Int J Syst Evol Microbiol. 2002;52:101–107. doi: 10.1099/00207713-52-1-101. [DOI] [PubMed] [Google Scholar]
- 38.Spring S., Lidwing W., Marquez M.C., Ventosa A., Schleifer K.-H. Halobacillus gen. nov., with descriptions of Halobacillus litoralis sp. nov., and Halobacillus trueperi sp. nov., and transfer of Sporosarcina halophila to Halobacillus halophilus comb. nov. Int J Syst Evol Microbiol. 1996;46:492–496. [Google Scholar]
- 39.Yoon J.H., Oh H.M., Yoon B.D., Kang K.H., Park Y.H. Paenibacillus kribbensis sp. nov. and Paenibacillus terrae sp. nov., bioflocculants for efficient harvesting of algal cells. Int J Syst Evol Microbiol. 2003;53:295–301. doi: 10.1099/ijs.0.02108-0. [DOI] [PubMed] [Google Scholar]
- 40.Ma Y., Xia Z., Liu X., Chen S. Paenibacillus sabinae sp. nov., a nitrogen-fixing species isolated from the rhizosphere soils of shrubs. Int J Syst Evol Microbiol. 2007;57:6–11. doi: 10.1099/ijs.0.64519-0. [DOI] [PubMed] [Google Scholar]