Abstract
Since Aujeszky`s disease (pseudorabies), which is caused by Suid herpesvirus type 1 (SuHV-1), was first notified in Argentina in 1978, many SuHV-1 strains have been isolated from swine. However, this disease can affect other vertebrates, such as dogs (secondary hosts), and lead to fatal neurological disease. The objective of the current work is to report the first isolation and molecular characterization of SuHV-1 from a dead domestic dog from Santa Fe Province (Argentina), which had had nervous signs compatible with pseudorabies. Samples of brain and trigeminal ganglia from this dog were obtained and fixed in formol for histopathology, and virology studies were conducted after cell disruption. Supernatants of both samples were inoculated onto RK13 cells and, after 72 h, DNA was extracted with phenol-chloroform. Purified DNA was cut with a restriction enzyme and subjected to agarose gel and an aliquot was used to amplify the gD and gC genes by PCR. The gC sequence was compared with other public sequences. The strain isolated from the dog was similar to other Argentinean swine strains.
Keywords: Argentina, Domestic dog, Suid herpesvirus 1
Introduction
Pseudorabies, also known as Aujeszky’s disease, is caused by Suid herpesvirus type 1 (SuHV-1), a member of the Alphaherpesvirinae subfamily. In pigs, which are the natural hosts of pseudorabies, the symptoms include various degrees of respiratory distress, nervous and genital disorders, and mortality, according to the age of the pig and the virulence of the virus strain involved (Wittmann, 1986). In addition, infection of pregnant gilts or sows frequently results in resorption, abortion, or birth of mummified fetuses or stillborn neonates. In pigs surviving acute infection, SuHV-1 develops latency, primarily in neuronal tissues, but also in lymphoid tissues. The virus may be transmitted by nose-to-nose contact, coitus, artificial insemination, fomites, or transplacentally (Cramer et al., 2011). A wide range of mammals and other vertebrate species, such as carnivores, rodents and ungulates, are susceptible to SuHV-1 infection (Müller et al., 2011). In these secondary hosts, SuHV-1 infection leads to fatal neurological disease; animals succumb to massive neurological dysfunction within a few days of disease onset. In general, disease in secondary hosts is observed only sporadically (Steinrigl et al., 2012).
Pseudorabies is a notifiable disease that causes substantial economic losses to the swine industry and has a major economic impact due to trade implications and income losses for farmers. In Argentina, since SuHV-1 was first isolated in 1978 (Ambrogi et al., 1981), many outbreaks have occurred in different parts of the country (Davido, 1981; Sager et al., 1984; Echeverría et al., 1991, 1992). In 1996, the National Animal Health Service of Argentina (SENASA) established a control program based on serological detection of infected animals without vaccine usage, in which all seropositive animals are segregated and/or slaughtered. Although SuHV-1 seroprevalence in Argentina is relatively low (around 18%), in 1998, SENASA established a program that involved voluntary vaccination of animals with an inactivated glycoprotein E (gE)-deleted vaccine and an ELISA that differentiated infected from vaccinated animals. However, both the importation of the vaccine and the ELISA were discontinued in 2001 because of the economic crisis in Argentina. Later, in 2016, SENASA implemented compulsory vaccination with the same kind of vaccine to continue with the eradication program.
The pig population in Argentina is of around 5,200,000 pigs and is concentrated mainly in three Provinces: Buenos Aires, Córdoba and Santa Fe. Some pigs are also distributed in other Provinces such as Entre Ríos, Chaco and Salta. About 80% of the farms have about 10 animals each (small producers or subsistence production), whereas the other 20% of farms have about 100 to 500 animals each. The highest percentage of positive animals is found in small farms.
Herrmann et al. (1984) proposed the systematization of SuHV-1 genomic types based on genomic BamHI DNA restriction fragment length polymorphisms (RFLPs). The classification obtained was modified by Christensen (1995), who determined three genomic types (genotypes): type I (including seven subtypes and reported mainly in the United Kingdom, Sweden and New Zealand); type II (including two subtypes and reported in Japan and Central Europe); and type III (including two subtypes and reported in Denmark).
In our laboratory, eight SuHV-1 strains were isolated from 1988 to 2013 (Echeverría et al., 1992, 2000; Serena et al., 2010). Subtyping of the Argentinean field isolates of SuHV-1 (Serena et al., 2010) has clearly shown that all strains are genotype I, the same as that occurred in Central Europe (Herrmann et al., 1984), Northern Ireland (Todd and Mc Ferran, 1985), the United States (Ben-Porat et al., 1984; Pirtle et al., 1984), and New Zealand (Tisdall et al., 1988). Although only one strain isolated in Argentina and previously classified as genotype II has been isolated from animals imported from Holland (Echeverría et al., 1994), no new isolates belonging to genotype II have been reported since 1981. Previous reports mention that genotype II is common in Holland (Gielkens et al., 1985).
In Brazil, genotype II is predominant in the pig population (Piatti et al., 2001; Schaefer et al., 2006). As regards the SuHV-1 genotypes present in South America, no further information has been reported.
The molecular characterization of strains uncovers genetic variations but not necessarily epidemiological relationships, even if the strains analyzed come from geographically distant regions (Müller et al., 2010). By using RFLP and sequence analysis, in a previous study, we were able to distinguish the genotypes of the Argentinean SuHV-1 strains (Serena et al., 2010). The Argentinean genotype I strains were grouped mainly with isolates from North America, Brazilian genotype I strains, and the NIA-3 vaccine strain, whereas the Argentinean genotype II strains were grouped with the reference type II Yamagata S-81 strain and the Brazilian genotype II strains. Partial sequence analysis of this gC region allowed a clear differentiation between Argentinean isolates of genotype I and those of genotype II. All Argentinean genotype I strains and the NIA-3 gE-deleted vaccine strain had identical amino acid sequence (232 amino acids). This could indicate the introduction of SuHV-1 in Argentina from a common source. Although the Argentinean strains are not gE-deleted, they probably resulted from a recombination event (Serena et al., 2011).
Although, in many European countries, SuHV-1 has been eliminated from domestic pigs, it is being continuously reported in wild boar populations and in related hunting dogs (Cay and Letellier, 2009; Steinrigl et al., 2012). There has been an unpublished report of pseudorabies infection in Argentine dogs associated with an outbreak of SuHV-1 in La Pampa Province in 1986 (Moras et al., 1986).
The objective of the current work is to report the first isolation and molecular characterization of SuHV-1 from a dead domestic dog from Santa Fe Province, Argentina, which had had nervous signs compatible with pseudorabies.
Materials and Methods
Tissue sampling
Postmortem examination of the head from the domestic dog from Santa Fe Province was performed at the Virology Laboratory of the School of Veterinary Sciences of La Plata, Buenos Aires, Argentina. The map of geographical distribution of SuHV-1 in Caseros Department (Santa Fe, Argentina) was created using QGis (version 2.18.3). Samples collected at necropsy included samples of the brain and trigeminal ganglia. Half tissues were immersed in 10% neutral buffered formalin, and half stored at -20°C for virus isolation and DNA extraction.
Virus isolation and RFLP analysis
Frozen-stored samples were homogenized in minimal essential medium (MEM) with L-glutamine and centrifuged at 5000 rpm for 10 min. The supernatant was inoculated onto confluent monolayers of RK13 (rabbit kidney) cells grown on six-well culture plates. The growth medium consisted of MEM supplemented with 10% fetal bovine serum (FBS). The inocula were allowed to absorb for 1 h at 37°C in an atmosphere of 5% CO2 in air. After incubation, the inocula were removed and MEM with 2% FBS was added. Cell cultures were incubated at 37°C in an atmosphere of 5% CO2 and assessed daily for the occurrence of herpesvirus cytopathic effect (CPE). After exhibiting typical herpesvirus CPE, the supernatant was harvested, aliquoted and frozen (-80°C).
For DNA extraction, the infected cells were harvested and pelleted by centrifugation. The pellet cell was washed, suspended in phosphate buffered saline (PBS) and DNA was extracted using a commercial kit Wizard Genomic DNA Purification Kit (Promega) according to the manufacturer’s protocol. The quantity and quality of DNA was determined by measuring absorbance at an OD260/OD280 ratio in a spectrophotometer SmartSpec 3000 (Bio-Rad, USA).
For RFLP analysis, approximately 1.5 μg of DNA solution was digested overnight with 2UI of BamHI at 37 °C, and then separated by electrophoresis in 0.7% (w/v) agarose gel (140 x 150 x 5mm) in TAE buffer (40 mM Tris-acetate pH 7.8, 5 mM sodium acetate, and 1 mM EDTA) at 20 V for 16 h at room temperature, stained with ethidium bromide, visualized and photographed under a UV illuminator.
Detection of rabies virus
Viral RNA was extracted from a brain sample by using TRIzol® (Invitrogen, Carlsbad, CA, USA), following the manufacturer’s instructions. Reverse transcription and PCR amplification were achieved as previously described by Cisterna et al. (2005) and performed at the Virology Laboratory of the School of Veterinary Sciences of La Plata.
PCR amplification and sequencing
PCR was performed in a DNA Thermal Cycler (Perkin-Elmer Cetus, Norwalk, CT, USA). PCR reactions were carried out according to the manufacturer’s protocols using PCRMaster Mix (Promega, Madison, WI, USA). To amplify the glycoprotein D (gD) gene, the following primers were used: ForwgDm (5’-GTGCACGGAGGACGAGCTGGGGCT-3’) and RevgDm (5’-GACGTCCACGCCCCGCTTGAAGCT-3’). Denaturation, annealing and extension consisted of 35 cycles at 95°C for 45 s, 60°C for 45 s and 72°C for 45 s, respectively. The PCR products were visualized on 1% agarose stained with ethidium bromide.
Partial gC was amplified using the primers gC-2U GTTTCCTGATTCACGCCCACGC and gC-1L GAAGGGCTCACCGAAGAGGAC (Goldberg et al., 2001), which gave an expected product of 788 nucleotides. The reaction mixtures were cycled as follows: a denaturation step of 95°C for 5 min, 8 cycles of 95°C for 50 s, 67°C for 50 s, 72°C for 50 s, 27 cycles of 95°C for 50 s, 64°C for 50 s, 72°C for 50 s and an extension step of 72°C for 5 min (Fonseca et al., 2010). The PCR products were visualized on 1% agarose gels and purified according to the manufacturer’s protocols using Wizard SV Gel and PCR Clean-Up System (Promega, Madison, WI, USA), and sequenced on both strands of each product by using the Big Dye Terminator V 3.1 sequencing kit (Applied Biosystems, Germany) with the same primers used for amplification. The sequences were analyzed on an ABI3130XL genetic analyzer (Applied Biosystems, USA), at the Unidad de Genómica, INTA Castelar, Argentina.
Sequence analysis and alignment of nucleotide sequences
The sequence was edited using BioEdit Sequence Alignment Editor. Homology analyses were performed with the BLASTN program (National Center for Biotechnology Information [http://www.ncbi.nlm.nih.gov/BLAST/]).
The sequence alignments of the partial gC gene were performed with the ClustalW method, using the MEGA 7.0 program. The phylogenetic analysis was carried out with the same program, using the neighbor-joining (NJ) method with the Kimura two-parameter model, and bootstrap analyses were conducted using 1000 replicates.
The dataset included 68 strains from different countries available through GenBank, including the Argentinean SuHV-1 swine strains (Table 1).
Table 1.
Origin, species and accession numbers of the SuHV-1 strains used in this study.
| GenBank Accession N° | Origin | Species | GenBank Accession N° | Origin | Species |
|---|---|---|---|---|---|
| AF176495 | USA | Swine | JF460034.1 | Argentina | Swine |
| AF176484.1 | USA | Dog | JF460035 | Argentina | Swine |
| AF176488 | USA | Swine | JF767011.1 | USA | Dog |
| AF176489 | USA | Swine | JQ768109 | Italy | Dog |
| AF176489.1 | USA | Swine | JQ768122 | Italy | Dog |
| AF176491 | USA | Swine | JQ768125 | Italy | Dog |
| AF403051 | China | Cow | JQ768151 | Italy | Swine |
| AF158090 | China | Swine | JQ768152 | Italy | Swine |
| D49435.1 | Japan | Swine | JQ768154 | Italy | Dog |
| D49436.1 | USA | Swine | JQ768156 | Italy | Dog |
| D49437.1 | Ireland | Swine | JQ081285.1 | Austria | Hunting dog |
| EU622054.1 | Brazil | Swine | JQ081286.1 | Austria | Hunting dog |
| EU622055 | Brazil | Swine | JQ081289.1 | Austria | Hunting dog |
| EU622056 | Brazil | Swine | JQ081291.1 | Austria | Hunting dog |
| EU622057.1 | Brazil | Swine | JQ081292.1 | Austria | Hunting dog |
| EU622058.1 | Brazil | Swine | JQ081293.1 | Austria | Hunting dog |
| EU622059.1 | Brazil | Swine | KC865672 | Croatia | Swine |
| EU622069.1 | Brazil | Swine | KC865680.1 | Croatia | Dog |
| EU622071.1 | Brazil | Swine | KF779458 | Belgium | Hunting dog |
| EU622079 | Brazil | Cow | KF779463 | Belgium | Swine |
| GQ862778.1 | Germany | Hunting dog | KF779468 | Belgium | Swine |
| GQ259098 | France | Hunting dog | KP780805.1 | Italy | Dog |
| GQ259099.1 | France | Hunting dog | KP780806.1 | Italy | Dog |
| GQ259100 | France | Hunting dog | KP862611.1 | Italy | Hunting dog |
| GQ259105 | Germany | Hunting dog | KP862612.1 | Italy | Hunting dog |
| GQ259106 | Germany | Hunting dog | KP862613.1 | Italy | Hunting dog |
| GQ259115 | France | Hunting dog | KP862614.1 | Italy | Hunting dog |
| GQ259116 | Germany | Hunting dog | KP862615.1 | Italy | Hunting dog |
| JF460027 | Argentina | Swine | KP862616.1 | Italy | Dog |
| JF460029 | Argentina | Swine | KP862617.1 | Italy | Dog |
| JF460030.1 | Argentina | Swine | KP862618.1 | Italy | Dog |
| JF460031.1 | Argentina | Swine | KP862619.1 | Italy | Hunting dog |
| JF460032.1 | Argentina | Swine | KP862620.1 | Italy | Hunting dog |
| JF460033.1 | Argentina | Swine | KP862621.1 | Italy | Hunting dog |
Virus neutralization test
Sera from pigs living in a farm near the place where the dog lived were analyzed by the virus neutralization test. The serum samples were heat-inactivated at 56°C for 30 min and serial two-fold dilutions were prepared in serum-free medium in 96-well flat-bottom tissue culture plates (Nunc, Rochester, NY, USA).
Virus suspension with a titer of 100 TCID50 in 25 μl was added to each serum dilution well and the mixture was incubated for 1 h at 37º and 5% CO2 and then 100 μl of an RK13 cell line suspension (3 × 105 cells/ml) was added to each well and incubated for 72 h. Appropriate serum, virus and cell controls were included in this test. The plates were observed under a microscope for CPE.
Histopathological analysis
Samples of the lateral area of a cerebral hemisphere (including cranial, media and caudal zones) were embedded in paraffin wax for histological examination, sectioned at 3-4 µm and stained with hematoxylin and eosin. Neither cerebellum nor brain stem was available.
Results
History and origin of the dog samples
The brain tissue and trigeminal ganglia analyzed were from a dog that lived close (about 1500 mts) to the SuHV-1-positive pig farm located in Chañar Ladeado, Santa Fe Province.
The map with georeferencing of the sample is shown in Figure 1.
Fig. 1.
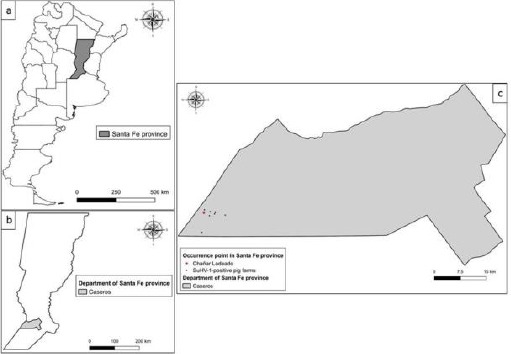
Map of Argentina (a) showing the location of Santa Fe Province (b) and Chañar Ladeado City (c) (QGis (version 2.18.3).
Epidemiological investigations were conducted by a veterinary doctor, who declared that the animal had direct contact with the pigs or had been fed with SuHV-1-infected meat. Clinical signs in the dog included neurological signs, such tremors, trismus, spasms of muscles of the larynx and pharynx, dyspnea, vomiting and pruritus. Death occurred within 24-48 h.
Virus isolation and identification
SuHV-1 was isolated from the brain and trigeminal ganglia in tissue culture in a confluent monolayer of RK13 cells. CPE was observed on day 4 of the first passage. Typical herpesvirus CPE, including lysis and syncytium formation, was observed in the two samples analyzed. Two more passages were done to increase the virus titer, but the magnitude of the CPE remained constant (Fig. 2).
Fig. 2.
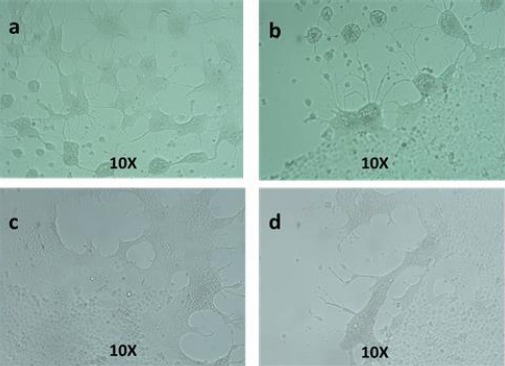
Typical herpesvirus cytopathic effect characterized by lysis and syncytium formation, detected in RK13 cells. Brain (a and b), trigeminal ganglia (c and d) after 72 h pi.
SuHV-1 genotyping of the virus isolated from the brain of the dog analyzed was performed by RFLP. The strain analyzed exhibited the classical restriction pattern present in SuHV-1 genotype I (Herrmann et al., 1984). In addition, it showed no alterations involving the gain or loss of BamHI cleavage sites, as also seen in all Argentinean strains reported previously, which present almost the same pattern, with small differences and a main variation in the 5+14’, 10 and 12 fragments (Serena et al., 2010) (Fig. 3).
Fig. 3.
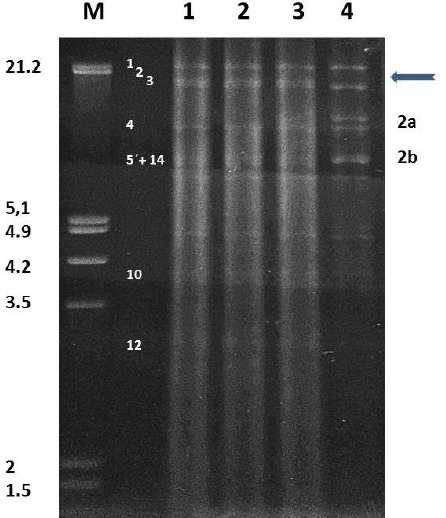
Restriction fragments obtained with the BamHI enzyme. M: Lambda DNA cleaved with EcoRI + HindIII (size expressed in Kbp). Lane 1: DNA ARG-Dog 2015 isolated from brain. Lane 2: DNA ARG-Dog 2015 isolated from trigeminal ganglion. Lane 3: Indiana-S Type I reference strain. Lane 4: Yamagata S-81 Type II reference strain. Representative Type fragments are numbered according to Hermann et al. (1984) for type I (left numbers) and type II (right numbers). The arrow indicates absence of fragment 2 in type II.
The DNA extracted from the infected cells was found to be SuHV-1-positive by PCR detecting gD (217 bp). The gC gene was then specifically amplified and the product was the expected size.
The PCR assay generated a product yielding a sharp, visible band around 750 bp on an ethidium bromide gel. An identical band was obtained from the positive control and no band was observed in the negative control used in the assay. The partial sequence of the gC gene of the dog strain analyzed was confirmed by BLAST analysis, which revealed 100 % of similarity with the SuHV-1 strain NIA-3 (complete genome KU900059.1).
No rabies virus was detected in the brain sample by using the RT-PCR technique. All the serum samples from the pigs from the neighboring farm analyzed by the virus neutralization test were SuHV-1-positive, with titers between 1:8 and 1:32.
Histopathological analysis
In all examinated areas of brain, the histopathological study revealed sparse mononuclear cell infiltration in meninges, and mild diffuse gliosis and neuronal satellitosis in gray matter.
Most neurons appeared unaffected, although a few evidenced shrinkage with condensed hypereosinophilic cytoplasm and pyknotic nuclei. These findings were compatible with mild non-specific encephalitis (Fig. 4).
Fig. 4.
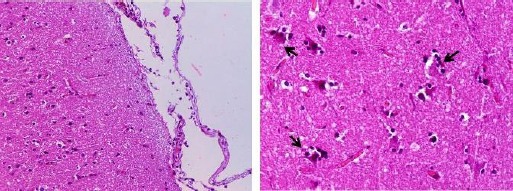
The histopathological study revealed sparse mononuclear cell infiltration in meninges, H&E (left 20X) and few neurons evidence acute eosinphilic degeneration with shrunken eosinophilic soma and pyknotic nuclei in brain (gray matter). Phagocytes around the cell body (satellitosis) are seen, H&E (right 40X).
Phylogenetic analysis based on gC
The partial nucleotide sequence data for the partial gC gene reported here have been previously submitted to GenBank under accession number MF101748.
Phylogenetic analysis of the gene encoding the gC protein demonstrated that the dog strain analyzed, named ARG-Dog 2015, showed no differences with swine Argentinean SuHV-1 strains and all together grouped in Cluster A, together with the NIA-3 reference strain. This cluster showed three groups: group a1, represented by swine Argentinean strains including the new Dog strain, swine Brazilian strains, one canine US strain and two reference strains (NIA-3 and Indiana-S); group a2, represented by one canine US strain and various swine US strains; and group a3, represented by four canine Italian strains.
Cluster B was formed by two defined groups, b1 and b2: b1 is represented by three hunting dog Austrian strains, whereas group b2 is represented by two swine Belgian strains, one swine Croatian strain, one canine Croatian strain, two hunting dog strains from Germany and France, two swine Italian strains, five canine Italian strains and two swine Brazilian strains, all of them with a high degree of homology to the reference strain Yamagata S-81. Cluster C was represented by three different groups: group c1, which included hunting dog strains from Germany, France and Belgium; group c2, which included three strains from Austria; and group c3, which included nine Italian strains. Groups c2 and c3 were closely related to the Shope reference strain. The Asian strains Fa and Ea were clustered separately and formed a new cluster named Cluster D, which was genetically divergent from all strains analyzed (Fig. 5).
Fig. 5.
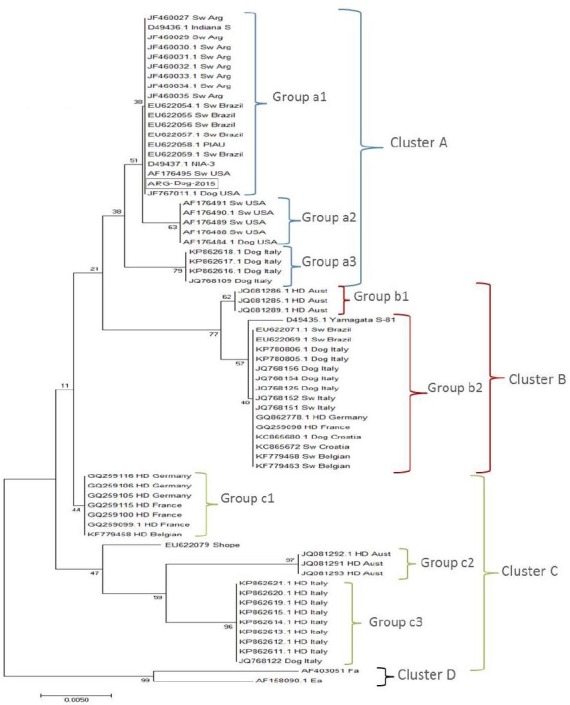
Phylogenetic tree obtained by the neighbor-joining (NJ) method with the Kimura two-parameter model and bootstrap analyses using 1000 replicates from the analysis of the partial gC gene amplified by polymerase chain reaction of Argentinean and international SuHV-1 strains.
An alignment of the deduced amino acid sequences of the gC protein showed several insertions and/or deletions as described previously in positions 24, 25, 38, 39, 181 and 182. Among the strains of group a1, the new Dog strain, the swine Argentinean strains, NIA-3 and one canine US strain (JF767011.1) showed a gap in positions 181 and 182. The rest of the strains of group a1, except the Indiana-S reference strain, also presented another gap in position 39. The Indiana-S strain showed the same variation inside the group because it presented one gap in position 181 and another gap in positions 24 and 25, but did not present the gap in position 39. The strains belonging to group a2 represented by US isolates showed only one gap in position 181. All strains of group a3 formed by canine Italian isolates presented all the gaps described. In Cluster B, none of the strains that formed group b1 and three strains of group b2 (GQ862778.1, KC865680.1 and KC865672) presented no gaps. The rest of the strains of group b2, except the Yamagata S-81 reference strain, presented two gaps: one in positions 24 and 25, and another in position 39. The Yamagata S-81 strain was the only strain of group b2 that had only one gap in positions 24 and 25. Analysis of Cluster C showed that all the strains of group c1 showed gaps in positions 181 and 182. One strain of this group (GQ259115) also presented one gap in positions 24 and 25, while the rest of the strains belonging to the group and all the strains from group c2 presented two gaps: one in the same position and another in positions 38 and 39. The strains of group c3 contained two deletions at positions 24 and 181. Cluster D was represented by Asian strains characterized by a long insertion (eight amino acids) from position 62 to position 69, present only in these strains. The Ea and Fa reference strains also had an insertion of two amino acids in positions 181 and 182. The phylogenetic tree from the amino acid sequence analysis maintained the same distribution of groups, differing only in the branch positions (data not shown).
Discussion
SuHV-1 was identified in a dog with history of exposure to a serologically positive swine farm, clinical signs and histopathological lesions. The virus was isolated and the infection was confirmed by RFLP, PCR and sequence analysis. Besides, the presence of virus antibodies in the neighboring swine farm was analyzed by the virus neutralization test from serum samples and positive results were detected. In the countries where SuHV-1 has been eradicated, researchers investigated the presence of the virus in wild boars and in dogs related to the swine farm or feral swine as well as in hunting dogs (Cramer et al., 2011; Steinrigl et al., 2012; Moreno et al., 2015).
Argentina has a valid eradication program of pseudorabies based on a serological test in the swine farm population and feral swine where we, as a reference laboratory, collaborate with SENASA. This program has shown a high prevalence of SuHV-1 in Santa Fe Province, where we detected the canine clinical case.
Pseudorabies is one of the most economically important diseases of farmed pigs. Both farmed pigs and wild boars can act as reservoirs and might represent a potential threat for occasional animals as dogs. Several reports have described the situation of SuHV-1 in dogs and their association and contact with live and/or dead feral pigs. It is not known whether the dogs become infected by consuming carcasses of dead feral pigs or whether they are exposed to the virus during hunting or fighting with live pigs (Cramer et al., 2011).
The dog analyzed presented a variety of clinical signs compatible with SuHV-1 infection, including the most classical clinical sign: facial pruritus (Pejsak and Truszczynski, 2006). Differential diagnosis for dogs exhibiting these clinical signs includes rabies infection which was discarded by the negative RT-PCR result.
The RFLP pattern of the ARG-Dog 2015 isolate suggests that it belongs to genotype I, as all the swine Argentinean strains previously isolated. The swine Argentinean viruses have been classified as type I, suggesting that no major variation has occurred in SuHV-1 spreading in Argentina since the first outbreak was noticed. The new isolate from the dog analyzed here supports this appreciation. The Argentinean type I strains had a high degree of BamHI pattern identity to the Indiana-S reference strain, especially in fragment 5+14’ (Serena et al., 2010). It is well known that the most noticeable change in the vaccine strain profile is the absence of BamHI fragment #7 (Petrovskis et al., 1986). No deletion of BamHI fragment #7 was observed in the ARG-Dog 2015 isolate or the Argentinean strains, indicating that they are wild-type viruses. The RFLP analysis, together with the phylogenetic analysis, allowed differentiating groups of strains relating genotype and origin of isolates.
In contrast to RFLP analysis, phylogenetic analysis is faster, avoids the need for virus isolation, and has the advantage that reference sequences are easily available. Goldberg et al. (2001) reported significant correlation between RFLP and gC sequence distance.
The histological pattern typical of canine SuHV-1 is characterized by nonsuppurative inflammation showing mononuclear cell perivascular cuffing, gliosis, neuronal degeneration, neuronophagia and intranuclear inclusion bodies in neurons and astrocytes (Cramer et al., 2011).
While the histopathological study revealed a lesion compatible with non-specific encephalitis, it must be pointed out that the sample of brain analyzed was partial. As described by Cramer et al. (2011), the most common lesion in dogs is observed in the brainstem, including the cranial nerves, but these samples were not available in this study. The phylogenetic tree based on the analysis of the gC gene revealed four different clusters. There was a clear distinction between the viral strains isolated from the Americas and those isolated from Europe, as well as between the strains isolated from hunting dogs and the strains isolated from working farm dogs and/or domestic pigs. Similar results were obtained by Moreno et al. (2015), who detected that the Italian strains were distributed into three different clusters and that there was a clear distinction between the wild boar strains (and those isolated from dogs that were used for hunting and subsequently traced back to wild boars) and the strains isolated from working farm dogs (and found to be closely related to strains in domestic pigs). The new Dog strain was located in a separate group (a1) inside Cluster A, together with swine Argentinean and Brazilian strains belonging to genotype I, with a US strain isolated from a dog (JF767011.1) and with the reference strains NIA-3 and Indiana-S. A similar distribution was observed by Cramer et al. (2011), who found that the dog strain isolate was grouped separately from other US strains and demonstrated notable divergence from the Indiana-S strain. In our study, including Argentinean strains, they formed a separate group, even with Indiana-S. Group a3 was represented by dog strains from Italy previously described as related to those found in domestic pigs clustered in clade 2 by Moreno et al. (2015).
SuHV-1 isolates representative of genomic type I are found in the USA and Central Europe, while isolates showing genomic type II are found predominantly in Central Europe and Japan (Sozzi et al., 2014). Genomic characterization based on sequencing of the gC gene showed a good correlation between genotype I and Cluster A as well as between genotype II and Cluster B. Inside Cluster B, there was a clear distinction between swine strains and canine strains originating from working dogs in pig farms (group b2), as described by Sozzi et al. (2014) and classified mainly as genotype II by Müller et al. (2010), and those originating from hunting dogs from Austria (group b1), represented as lineage 2 in a previous study and described as genotype I (Steinrigl et al., 2012).
Cluster C separated strains from hunting dogs related to wild boar strains from Europe. We obtained a distribution of the groups similar to that obtained by other authors, where cluster C in Sozzi’s report and clade 1 in Moreno’s report were represented in group c3 in our study. The differentiation of the groups in the tree based on the amino acid sequences revealed insertions and deletions and the results were similar to those of other studies (Müller et al., 2010; Steinrigl et al., 2012; Sozzi et al., 2014; Moreno et al., 2015).
SuHV-1 has virtually disappeared from domestic pigs in many parts of the world, but is still a problem in some South American countries. The genomic characterization of SuHV-1 strains originating from swine and other mammals might help to better understand the population diversity and facilitate tracing the infection chain back to its origin (Sozzi et al., 2014). The results of this study provide an update of the knowledge of pseudorabies in dogs and the role of these animals in the viral infection in relation with their contact with pigs.
Acknowledgements
We thank to Mr. Claudio Leguizamón, Ms. Adriana Conde and Ms. Mariela Vazzano for their technical assistance.
Conflict of interest
The authors declare that there is no conflict of interests.
References
- Ambrogi A, Giraudo J, Busso J, Bianco B, Bagnat E, Segura de Aramburu M, Ramos B, Ceriatti S. Primer diagnóstico de la Enfermedad de Aujeszky en cerdos en la República Argentina. Gac.Vet. 1981;43:58–64. [Google Scholar]
- Ben-Porat T, Deatly A, Easterday B, Galloway D, Kaplan A, McGregor S. Latency of pseudorabies virus. In: Wittmann G, Gaskell RM, Rziha H.J, editors. Latent Herpes Virus Infections in Veterinary Medicine, Current Topics in Veterinary Medicine and Animal Sciences. Vol. 27. Boston, USA: MartinusNijhoff Publishers; 1984. pp. 365–383. [Google Scholar]
- Cay A, Letellier C. Isolation of Aujeszky's disease virus from two hunting dogs in Belgium after hunting wild boars. VlaamsDiergeneeskundigTijdschrift. 2009;78:194–195. [Google Scholar]
- Christensen L. The population biology of Suid Herpes virus 1. APMIS. 1995;(Suppl. 48):1–48. [PubMed] [Google Scholar]
- Cisterna D, Bonaventura R, Cailloub S, Pozoc O, Andreaud M, Dalla Fontana L, Echegoyen C, de Mattos C, Russo S, Novaroh L, Elbergerh D, Freire MC. Antigenic and molecular characterization of rabies virus in Argentina. Virus Res. 2005;109:139–147. doi: 10.1016/j.virusres.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Cramer S, Campbell G, Morgan S, Smith S, McLin W, Brodersen B, Wise A, Scherba G, Langohr I, Maes R. Pseudorabies virus infection in Oklahoma hunting dogs. J. Vet. Diag. Invest. 2011;23:915–923. doi: 10.1177/1040638711416628. [DOI] [PubMed] [Google Scholar]
- Davido M. Enfermedad de Aujeszky en el sur de la Provincia de Córdoba. Gac. Vet. 1981;44:291–296. [Google Scholar]
- Echeverría M, Nosetto E, Petruccelli M, Gimeno E, Etcheverrigaray M. Ocurrencia y diagnóstico de la Enfermedad de Aujeszky en las zonas de Chañar Ladeado y Saladillo. Vet. Argentina. 1991;8:252–257. [Google Scholar]
- Echeverría M, Nosetto E, Etcheverrigaray M, Galosi C, Fonrouge R, Pereyra N, Belak K, Gimeno E. Pseudorabies (Aujeszky's disease) in Argentina. Rev. Sci. Tech. Off. Int. Epiz. 1992;11:819–827. doi: 10.20506/rst.11.3.614. [DOI] [PubMed] [Google Scholar]
- Echeverría M, Norimine J, Galosi C, Oliva G, Etcheverrigaray M, Nosetto E. The genotype of Aujeszky's Disease viruses isolated in Argentina. J. Vet. Med. Sci. 1994;56:985–987. doi: 10.1292/jvms.56.985. [DOI] [PubMed] [Google Scholar]
- Echeverría M, Pecoraro M, Pereyra N, Pidone C, Galosi C, Etcheverriagaray M, Nosetto E. Rapid diagnosis of pseudorabies virus in swine tissues using a polymerase chain reaction method. Rev. Argent. Microbiol. 2000;32:109–115. [PubMed] [Google Scholar]
- Fonseca A, FernandezCamargos M, Macedo de Oliveira A, Ciacci-Zanella J, Patricio M, Braga A, Cunha E, D'Ambros R, Heinemann M, CerqueiraLeite R, Pimenta dos Reis J. Molecular epidemiology of Brazilianpseudorabies viral isolates. Vet. Microbiol. 2010;141:238–245. doi: 10.1016/j.vetmic.2009.09.018. [DOI] [PubMed] [Google Scholar]
- Gielkens A, Van Oirschot J, Berns A. Genome differences among field isolates and vaccine strains of pseudorabies virus. J. Gen. Virol. 1985;66:69–82. doi: 10.1099/0022-1317-66-1-69. [DOI] [PubMed] [Google Scholar]
- Goldberg T, Weigel R, Hahn E, Scherba G. Comparative utility of restriction fragment length polymorphism analysis and gene sequencing to molecular epidemiological investigation of a viral outbreak. Epidemiol. Infect. 2001;126:415–424. doi: 10.1017/s0950268801005489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann S, Heppner B, Ludwig H. Wittmann G, Gaskell R, Rziha H. Latent herpesvirus infections in veterinary medicine, current topics in veterinary medicine and animal science. Boston, USA: MartinusNujhoff Pub; 1984. Pseudorabies virus from clinical outbreaks and latent infections grouped into four major genome types; pp. 387–401. [Google Scholar]
- Moras E, Ierace A, Barboni de Stella A, Iribarren F, Barcos O, Menchaca E. II Congreso Argentino de Virología, 20-24 de octubre 1986. Argentina: Córdoba; 1986. Segundo aislamiento de Herpes suis en un brote de enfermedad de Aujeszky en caninos de la provincia de La Pampa. [Google Scholar]
- Moreno A, Sozzi E, Grilli G, Gibelli L, Gelmetti D, Lelli D, Chiari M, Prati P, Alborali G, Boniotti M, Lavazza A, Cordioli P. Detection and molecular analysis of Pseudorabies virus strains isolated from dogs and a wild boar in Italy. Vet. Microbiol. 2015;177:359–365. doi: 10.1016/j.vetmic.2015.04.001. [DOI] [PubMed] [Google Scholar]
- Müller T, Klupp B, Freuling C, Hoffmann B, Mojcicz M, Capua I, Palfi V, Toma B, Lutz W, Ruiz-Fon F, Gortarzar C, Hlinak A, Schaarschmidt U, Zimmer K, Conraths F, Hahn E, Mettenleiter T. Characterization of pseudorabies virus of wild boar origin from Europe. Epidemiol. Infect. 2010;138:1590–1600. doi: 10.1017/S0950268810000361. [DOI] [PubMed] [Google Scholar]
- Müller T, Hahn E, Tottewitz F, Kramer M, Klupp B, Mettenleiter T, Freuling C. Pseudorabies virus in wild swine: a global perspective. Arch. Virol. 2011;156:1691–1705. doi: 10.1007/s00705-011-1080-2. [DOI] [PubMed] [Google Scholar]
- Pejsak Z, Truszczynski M. Aujeszky's disease (pseudorabies) In: Straw BE, Zimmerman JJ, D'Allaire S, Taylor DJ, editors. In: Diseases of swineed. 9th ed. Blackwell Ames IA: 2006. pp. 419–433. [Google Scholar]
- Petrovskis E, Timmins J, Gierman T, Post L. Deletions in vaccine strains of pseudorabies virus and their effect on synthesis of glycoprotein gp63. J. Virol. 1986;60:1166–1169. doi: 10.1128/jvi.60.3.1166-1169.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti R, Ikuno A, Cunha E, D'Ambros R, Gregori F, Soares R, Cortez A, Richtzenhain L. Characterization of Aujeszky's disease virus isolates from South and Southeast Brazil by RFLP analysis. Braz. J. Microbiol. 2001;32:144–146. [Google Scholar]
- Pirtle E, Wathen M, Mengeling W, Sacks J. Evaluation of field isolates of pseudorabies virus (Aujeszky's disease) as determined by restriction endonuclease analysis and hybridisation. Am. J. Vet. Res. 1984;45:1906–1912. [PubMed] [Google Scholar]
- Sager R, Rossanigo C, Vázquez R, Avila J, Fondevila N. Enfermedad de Aujeszky en cerdos en Villa Mercedes (San Luis) Argentina. Revista de Medicina Veterinaria. 1984;65:86–89. [Google Scholar]
- Schaefer R, Ciacci-Zanella J, Mores N, Kleitton A, Dambros R, Schiochet M, Coldebella M. Characterization of Aujeszky's disease virus isolated from South Brazil in the last twenty years by restriction enzyme analysis. Braz. J. Microbiol. 2006;37:390–394. [Google Scholar]
- Serena M, Metz G, Martín Ocampos G, Gambaro S, Mórtola E, Echeverría M. Characterization of Suidherpesvirus 1 field isolates from Argentina. Rev. Argent. Microbiol. 2010;42:179–183. [PubMed] [Google Scholar]
- Serena M, Metz G, Mórtola E, Echeverría M. Phylogenetic analysis of SuidHerpesvirus 1 isolates from Argentina. Vet. Microbiol. 2011;154:78–85. doi: 10.1016/j.vetmic.2011.06.028. [DOI] [PubMed] [Google Scholar]
- Sozzi E, Moreno A, Lelli D, Cinotti S, Alborali GL, Nigrelli A, Luppi A, Bresaola M, Catella A, Cordioli P. Genomic Characterization of Pseudorabies Virus Strains Isolated in Italy. Transbound. Emerg. Dis. 2014;61:334–340. doi: 10.1111/tbed.12038. [DOI] [PubMed] [Google Scholar]
- Steinrigl A, Revilla-Fernández S, Kolodziejek J, Wodak E, Bagó Z, Nowotny N, Schmoll F, Köfer J. Detection and molecular characterization of Suidherpesvirus type 1 in Austrian wild boar and hunting dogs. Vet. Microbiol. 2012;157:276–284. doi: 10.1016/j.vetmic.2011.12.033. [DOI] [PubMed] [Google Scholar]
- Tisdall D, Bentley C, Horner G. Genetic variation between New Zealand and Western Samoan isolates of Aujeszky's disease virus. Vet. Microbiol. 1988;17:335–343. doi: 10.1016/0378-1135(88)90047-8. [DOI] [PubMed] [Google Scholar]
- Todd D, McFerran J. Restriction endonuclease analysis of Aujeszky's disease (pseudorabies) virus DNA comparison of Northern Ireland isolates and isolates from other countries. Arch. Virol. 1985;86:167–176. doi: 10.1007/BF01309822. [DOI] [PubMed] [Google Scholar]
- Wittmann G. La enfermedad de Aujeszky. Rev. Sci. Tech. Off. Int. Epiz. 1986;5:995–1009. [Google Scholar]


