Abstract
X-linked severe combined immunodeficiency (SCID-X1) is caused by mutations in the interleukin-2 receptor γ chain gene (IL2RG), and it is characterized by profound defects in T, B, and natural killer (NK) cell functions. Transplantation of hematopoietic stem/progenitor cells (HSPCs) genetically corrected with early murine leukemia retrovirus (MLV)-derived gammaretroviral vectors showed restoration of T cell immunity in patients, but it resulted in vector-induced insertional oncogenesis. We developed a self-inactivating (SIN) lentiviral vector carrying a codon-optimized human IL2RG cDNA driven by the EF1α short promoter (EFS-IL2RG), and we tested its efficacy and safety in vivo by transplanting transduced Il2rg-deficient Lin− HSPCs in an Il2rg−/−/Rag2−/− mouse model. The study showed restoration of T, B, and NK cell counts in bone marrow and peripheral blood and normalization of thymus and spleen cellularity and architecture. High-definition insertion site analysis defined the EFS-IL2RG genomic integration profile, and it showed no sign of vector-induced clonal selection or skewing in primarily and secondarily transplanted animals. The study enables a phase I/II clinical trial aimed at restoring both T and B cell immunity in SCID-X1 children upon non-myeloablative conditioning.
Keywords: gene therapy, immunodeficiency, SCID-X1, lentiviral vector, integration, genotoxicity
Introduction
Transplantation of autologous hematopoietic stem/progenitor cells (HSPCs) genetically corrected with a retroviral vector has been clinically tested as a treatment for four primary immunodeficiencies (PIDs), i.e., adenosine deaminase-deficient severe combined immunodeficiency (ADA-SCID), X-linked SCID (SCID-X1), Wiskott-Aldrich syndrome (WAS), and X-linked chronic granulomatous disease (X-CGD), each presenting different challenges in terms of achieving optimal correction and clinical efficacy.1 The first clinical trials were based on vectors derived from the Moloney murine leukemia retrovirus (MLV), carrying a therapeutic gene under the control of the MLV long terminal repeat (LTR) promoter/enhancer.2, 3, 4, 5, 6, 7 Although this form of gene therapy has been beneficial for most patients, all trials except those for ADA-SCID were characterized by severe adverse events, i.e., the occurrence of leukemia or myelodysplasia promoted by the insertional activation of proto-oncogenes (LMO2, CCND2, and MECOM) by the MLV vector.1 The molecular basis of these events is still ill defined, though it is likely that insertional deregulation of one or more proto-oncogenes led to clonal expansion and eventually transformation of HSPCs, a major genotoxic consequence of the genetic modification intended to correct the immunodeficiency.8, 9
The recognition of the MLV LTR as a major component contributing to the insertional gene activation led to the design of self-inactivating (SIN) gammaretroviral vectors, which do not contain the LTR promoter/enhancer sequences and instead incorporate mammalian promoters devoid of long-range acting enhancers to drive the expression of the therapeutic gene. This vector design proved safe and efficacious in one clinical trial of gene therapy for SCID-X1.10 However, LTR modifications do not change the overall retroviral vector target site selection,11 which is directed by the interaction of the MLV pre-integration complex with acetylated, active transcriptional control elements in the host cell genome.12
To overcome the risk of insertional oncogenesis, a new generation of vectors, derived from the human immunodeficiency lentivirus HIV-1, has largely replaced MLV-derived vectors in clinical trials involving genetically modified HSPCs. The integration pattern of lentiviral vectors (LVs) is inherently safer than that of MLV vectors, as indicated by in vitro as well as in vivo studies.13, 14 LVs preferentially integrate throughout the transcribed portion of active genes, targeting at high-frequency genes located in the outer portion of the nucleus in proximity to the nuclear pore,15 with no preference for regulatory elements or specific gene categories.8, 12, 16 LVs have been used to transduce HSPCs in clinical trials of gene therapy for WAS,17, 18 adrenoleukodystrophy (ALD),19 metachromatic leukodystrophy (MLD),20 and hemoglobinopathies,21, 22 providing strong evidence of clinical efficacy in the absence of treatment-related adverse events. Early data indicate the potential of LVs also for gene therapy of SCID-X1.23
Here we report a pre-clinical study addressing the efficacy and safety of gene therapy for SCID-X1 by transplantation of HSPCs transduced by a lentiviral vector. SCID-X1 is caused by mutations in the gene encoding the interleukin-2 receptor γ chain (IL2RG), and it is characterized by a lack of response to common γ chain-dependent cytokines and profound defects in T, B, and natural killer (NK) cell functions.24 Early clinical studies based on HSPCs genetically corrected with MLV or SIN-MLV vectors expressing IL2RG in non-conditioned patients showed long-lasting restoration of T cell immunity, though not of B or NK cells, despite the high frequency of severe adverse events observed with the use of MLV vectors carrying wild-type LTRs.25, 26 To develop a safer alternative to MLV vectors, we developed a third-generation lentiviral vector carrying a codon-optimized human IL2RG cDNA27 under the control of the human short elongation factor 1α (EFS) promoter. The performance of the vector was demonstrated in vitro by the restoration of a normal level of IL2RG mRNA or protein in a human IL2RG-deficient T cell line and by high-efficiency transduction of human-mobilized CD34+ HSPCs. The in vivo safety and efficacy were tested in a preclinical model of SCID-X1 gene therapy based on transplantation of genetically corrected Lin− cells from IL2rg−/− donor mice into sub-lethally irradiated IL2rg−/−/Rag2−/− recipients. The vector genotoxic profile was investigated by an in vitro immortalization assay (IVIM)28 and in vivo by insertion site analysis, in pre-transplant Lin− cells and in bone marrow (BM), thymus, and peripheral blood (PB) of transplanted mice, to detect the presence of any clonal skewing or any deviation from a normal lentiviral integration pattern. These studies enable a phase I/II clinical trial aimed at establishing the safety of lentiviral vector-mediated gene therapy for SCID-X1 after non-myeloablative marrow conditioning and its efficacy in achieving sustained restoration of T, B, and NK cell immunity.
Results
Design of a SIN Lentiviral Vector for SCID-X1 Gene Therapy
A SIN lentiviral vector was constructed by cloning a codon-optimized IL2RG cDNA sequence (IL2RGco), encoding the interleukin-2 receptor (IL2R) common γ chain under the transcriptional control of the EFS promoter and the mutated WPRE* in the CCL-SIN-18 LV vector backbone (EFS-IL2RG; Figure S1A). The efficiency of the VSV-G-pseudotyped EFS-IL2RG vector in driving IL2RG mRNA and protein expression was tested in a human leukemic T cell line lacking endogenous γ chain expression (ED7R cells) and compared to an identical vector containing the cDNA for the native (wild-type [WT]) sequence of the human IL2RG. ED7R cells were transduced with both vectors at increasing MOIs (0.1–100) and maintained in liquid culture for 12 days before evaluating IL2RG mRNA and protein expression. IL2RGco mRNA level in transduced cells was measured by qRT-PCR, normalized to a housekeeping gene (TFIID), and expressed as fold change with respect to the constitutively expressed, endogenous IL2RG mRNA in the Jurkat T cell line. IL2RG protein expression was evaluated by flow cytometry after staining with an anti-human γ chain-specific antibody (anti-CD132). The EFS-IL2RG vector drove significantly higher expression (t test, p < 0.05) of IL2RG mRNA and protein with respect to the vector containing the WT IL2RG sequence (Figures S1B and S1C). Transduction efficiency of EFS-IL2RG in human HSPCs was determined by transducing cord blood CD34+ cells derived from 5 different donors. HSPCs were transduced at increasing MOIs (10, 50, and 100) and maintained in liquid culture for 8 days or cultured as individual progenitors (colony-forming cells [CFCs]) in semi-solid medium for 2 weeks. The average vector copy number (VCN) in the bulk population of transduced CD34+ cells was 0.6 ± 0.1, 1.8 ± 0.5, and 2.8 ± 0.7 (mean ± SD), respectively, while the percentage of transduced individual CFCs (myeloid + erythroid + mixed colonies) was 54% ± 34%, 83% ± 10%, and 81% ± 7%, respectively.
Correction of the SCID-X1 Phenotype by Transplantation of EFS-IL2RG-Transduced Hematopoietic Stem Cells in Il2rg-Deficient Mice
The efficacy of the EFS-IL2RG vector in correcting the SCID-X1 phenotype was tested in an Il2rg-deficient murine model of the disease. BM-derived lineage-negative (Lin−) Il2rg−/− cells from male donors of three different age groups (2–4, 5–10, and 16 weeks) were separately transduced with one or two rounds of infection with the EFS-IL2RG vector and transplanted into sub-lethally irradiated Rag2−/−/Il2rg−/− female recipients (Figure 1A). The rationale for transplanting cells from mice of different ages was to show potential age-related differences in engrafting or reconstitution capacity of stem cells with this disease background. Rag2−/−/Il2rg−/− mice were used as recipients since they completely lack T, B, and NK cells, and they represent a better, less leaky background with respect to the Il2rg−/− single knockout to study immune cell reconstitution. As control arms, Rag2−/−/Il2rg−/− mice were transplanted with WT C57BL/6J or Il2rg−/− BM Lin− cells transduced with a control lentiviral vector (PGK-GFP) expressing the GFP gene under the human phosphoglycerate kinase (PGK) promoter (groups WT + PGK-GFP and knockout [KO] + PGK-GFP in Figure 1A).
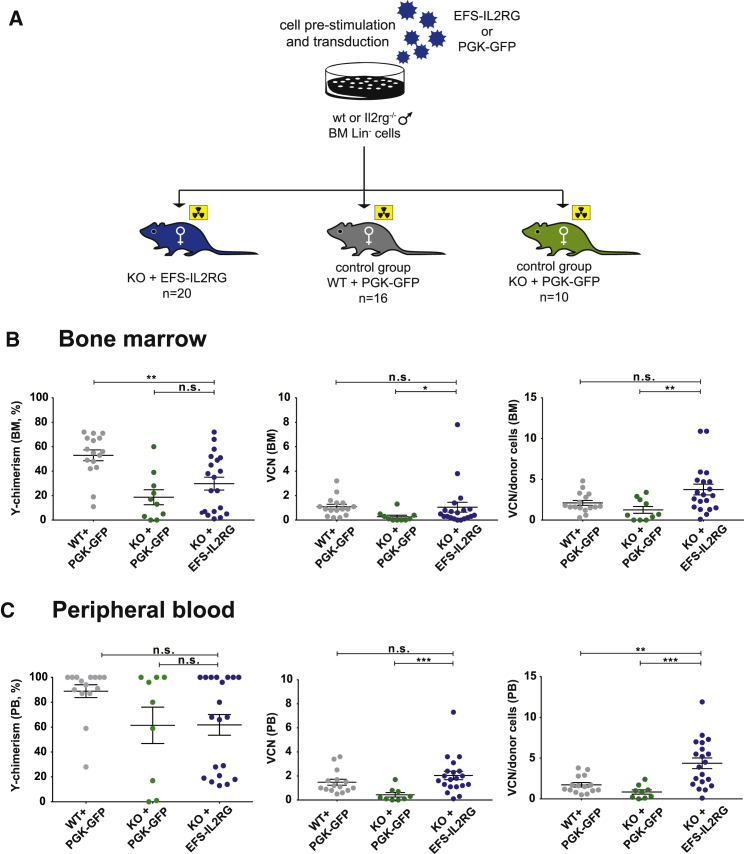
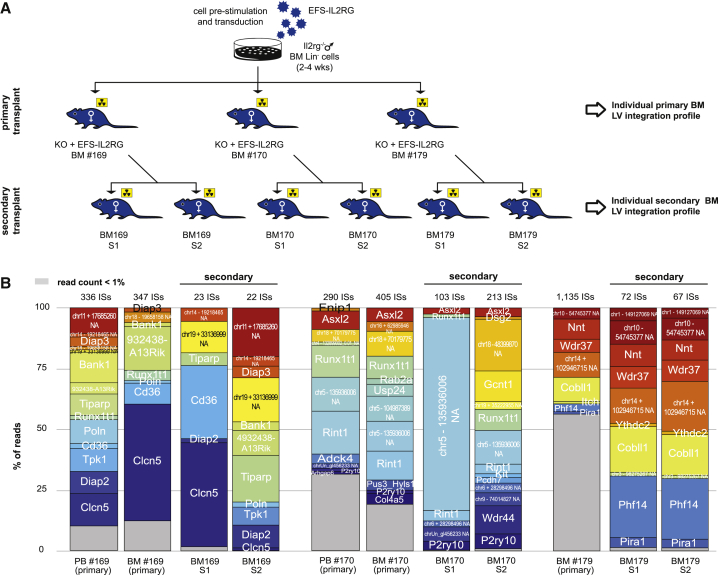
Figure 1.
Transplantation of HSPCs Transduced with the EFS-IL2RG in Il2rg-Deficient Mice
(A) Scheme of the in vivo study. (B) Chimerism, VCN, and VCN/donor cell in the bone marrow of Rag2−/−/Il2rg−/− mice transplanted with wild-type C56Bl6 Lin− cells transduced with a PGK-GFP vector (WT + PGK-GFP), Il2rg−/− Lin− cells transduced with the PGK-GFP vector (KO + PGK-GFP), and Il2rg−/− Lin− cells transduced with the EFS-IL2RG vector (KO + EFS-IL2RG), 6 months after transplantation. (C) Chimerism, VCN, and VCN/donor cell in the peripheral blood of the same animals. Data are presented as individual animals and as means ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001.
Lin− cells were pre-activated overnight with a cytokine cocktail and transduced once or twice with EFS-IL2RG or PGK-GFP at a vector concentration of 1 × 108 TU/mL (MOI 400) and transplanted by retro-orbital venous injection. An aliquot of the transduced cells was maintained in liquid culture for a week for VCN evaluation and vector integration analysis, or it was cultured as individual progenitors in semi-solid medium for 2 weeks. In these cells, the VCN averaged 2.5 and 4.5 after one or two rounds of transduction with EFS-IL2RG, respectively, with 78% and 92% of transduced individual progenitors. Vector-driven IL2RG mRNA expression ranged from 0.5- to 1.2-fold with respect to the endogenous Il2rg mRNA in WT cells (Table 1). Overall, eight mice died or were sacrificed in the first 4 weeks after transplantation because of loss of weight or poor health, with no obvious difference in the test (KO + IL2RG) versus control groups. VCN, blood cell count, and immune cell phenotype were evaluated in PB at 3 and 6 months and in the BM at 6 months. At sacrifice, mice were also analyzed for cell engraftment and biodistribution.
Table 1.
Analysis of Transduced Murine Lin− Cells Maintained in Bulk or Clonogenic Culture
| VCN (Bulk) | IL2RG mRNA (Bulk) | GFP+ Cells (%) | LV+ CFCs (%) | |
|---|---|---|---|---|
| WT + GFP | 3.5 ± 0.4 | 1 | 80.5 ± 0.7 | 88 |
| KO + GFP | 2.1 ± 0.5 | – | 64 ± 9 | 78 |
| KO + IL2RG 1 hit | 2.5 ± 1.1 | 0.46 ± 0.22 | – | 78 |
| KO + IL2RG 2 hits | 4.5 ± 1.0 | 1.17 ± 0.47 | – | 92 |
At the end of the study, donor chimerism was estimated in the BM by qPCR analysis with donor-specific primers for the Y chromosome, and it showed a significantly lower engraftment of Il2rg−/− stem cells, transduced by either the IL2RG or the GFP vector, with respect to mice engrafted with WT cells (Mann-Whitney test, p value < 0.01) (Figure 1B). No significant difference in chimerism was observed in the PB from the same mice (Figure 1C). In the test group, the average VCN in both the BM and PB correlated with values measured in pre-transplant cells, and it showed efficient transduction of repopulating stem cells by the EFS-IL2RG vector (Figures 1B and 1C). In these mice, VCNs were on average higher in PB than in the BM (2.04 ± 0.35 versus 1.05 ± 0.40), although the difference was borderline significant (p = 0.07), suggesting a selective advantage in the PB of cells expressing higher levels of IL2RG (Figure 1). Mice transplanted with Lin− cells from donors of different age showed non-significant differences in engraftment and average VCN, in both the BM and PB (data not shown). Likewise, no significant difference in either chimerism or VCN was observed in mice transplanted with cells transduced by one or two hits of either the test or the control vector (Figure S2). Therefore, all transplanted mice were subsequently analyzed as a single group.
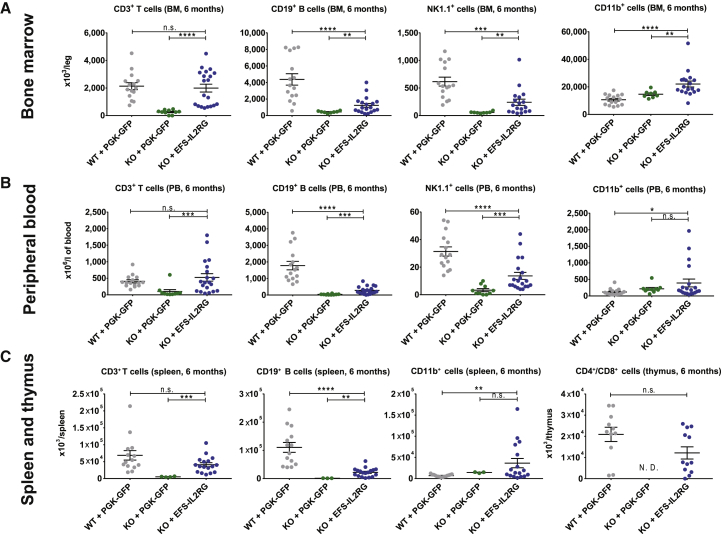
Overall, white blood cell counts in the test group (KO + EFS-IL2RG) were comparable to those of mice transplanted with WT cells (WT + PGK-GFP) and significantly higher (Mann-Whitney test, p value < 0.001) than those of mice transplanted with non-corrected Il2rg−/− cells (KO + PGK-GFP). Red blood cell (RBC) and platelet counts, Hb levels, and hematocrit showed no difference among groups (Figure S3). Immune reconstitution was evaluated by immunophenotyping of the BM, PB, spleen, and thymus cells at the end of the study (Figure 2). Cells isolated were stained with antibodies against specific surface antigens of T cells (CD3+), thymocytes (CD4+/CD8+), B cells (CD19+), NK cells (NK1.1+), and myeloid cells (CD11b+), and they were analyzed by flow cytometry. In the BM, PD, spleen, and thymus of mice transplanted with corrected cells, T cell reconstitution was comparable to that of mice receiving WT cells and significantly higher than mice receiving non-corrected cells (Mann-Whitney test, p value < 0.0001; Figure 2A), indicating that the EFS-IL2RG vector drives robust T cell development and repopulation of lymphoid organs. In the B and NK cell compartment, cell reconstitution was less efficient compared to mice receiving WT cells, but it was still significantly higher than in mice receiving non-corrected cells (Mann-Whitney test, p value < 0.01) in all organs (Figure 2). The restoration of a functional B cell compartment was confirmed by the presence of immunoglobulins (IgM and IgG) in the serum of the treated mice, at levels comparable with those of mice receiving normal cells (Figure S4). In the thymus, the number of CD4+/CD8+ thymocytes was comparable in mice transplanted with corrected or WT cells, while in mice receiving non-corrected cells this population was undetectable (Figure 2C). Overall, histopathological analysis of spleen and thymus revealed comparable size, cellularity, and organ architecture in mice treated with either corrected or WT cells (Figure S5). No sign of malignancy was observed in mice transplanted with corrected Il2rg−/− cells, while a thymic disseminated lymphoma was observed in a single mouse transplanted with WT cells transduced by PGK-GFP.
Figure 2.
Correction of the SCID-X1 Phenotype in Il2rg-Deficient Mice
(A–C) Reconstitution of T, B, NK, and myeloid cells in the bone marrow (A), peripheral blood (B), and spleen and thymus (C) of Rag2−/−/Il2rg−/− mice transplanted with wild-type C56Bl6 Lin− cells transduced with a PGK-GFP vector (WT + PGK-GFP), Il2rg−/− Lin− cells transduced with the PGK-GFP vector (KO + PGK-GFP), and Il2rg−/− Lin− cells transduced with the EFS-IL2RG vector (KO + EFS-IL2RG), 6 months after transplantation. Data are presented as individual animals and as means ± SEM. Statistical differences are expressed as follows: n.s., non-significant; **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Two individual mice in the KO + IL2RG group (169 and 170), transplanted with cells from 2- to 4-week-old donors, showed a population of double-positive CD4+/CD8+ cells in the PB (∼25% of the total mononuclear cells; Figure S6), but no sign of malignancy. Mouse 169 had virtually no thymus, while mouse 170 showed a thymus with normal architecture (data not shown) and T cell counts and phenotype indistinguishable from those of a mouse transplanted with WT cells (Figure S6). The BMs of these animals, together with that of a mouse from the same group that had no double-positive population in the periphery (179), were serially transplanted in Il2rg−/− recipients (two for each donor), which were analyzed for cell phenotype 3 months after transplantation. None of the animals showed signs of a double-positive T cell population in their PB (Figure S6).
Analysis of the In Vitro Genotoxic Potential of the EFS-IL2RG Vector
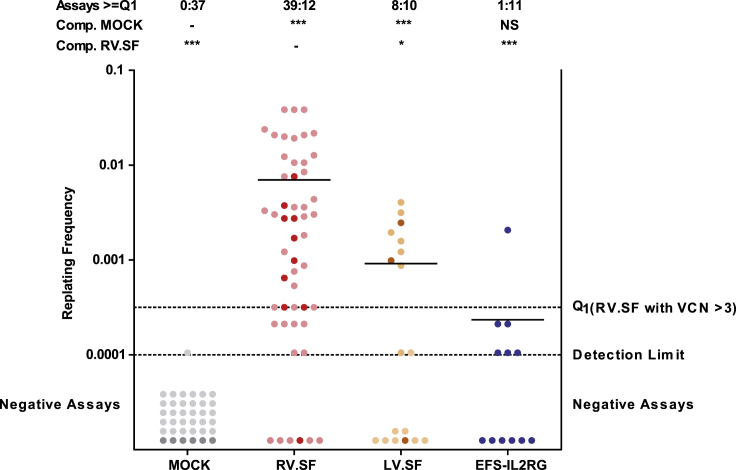
We used the IVIM assay28, 29 to estimate the insertional mutagenesis potential of the EFS-IL2RG vector in vitro. Mutagenic vectors with strong enhancer/promoter sequences can activate proto-oncogenes near the insertion sites. In the IVIM assay, these mutants show a proliferation advantage when cells are seeded at very low density. While non-immortalized cells stop growing, insertional mutants can be quantified by their replating phenotype on a 96-well plate. The EFS-IL2RG vector was tested with non-transduced (MOCK) cells as a negative control and two vectors with known mutagenic potential as positive controls, i.e., a gammaretroviral vector with a spleen focus-forming virus (SFFV) promoter/enhancer element in the intact LTRs (RV.SF) and a SIN lentiviral vector with the same SFFV elements in an internal position (LV.SF). For RV.SF, we observed insertional mutants in 9 of 10 assays. Vector LV.SF triggered immortalization in 2 of 3 assays. In contrast, EFS-IL2RG had a significantly lower incidence and fitness of insertional mutants (Figure 3): only 1 of 12 assays showed cells with a replating potential. As this phenotype is most often elicited by an overexpression of Mecom genes, we expanded these positive wells and performed a qPCR-based gene expression assay. We observed no upregulation of Mecom in the EFS-IL2RG-transduced, re-plated cells, unlike the RV.SF-transduced control clone that was expanded and measured in parallel (Table S1).
Figure 3.
In Vitro Immortalization Assay
Insertional mutants are identified by clonal outgrowth on a replating assay, where non-immortalized cells do not grow (negative, below detection limit). The number of positive wells is used to calculate the replating frequency (RF) according to Poisson statistics. Positive assays above the Q1 level (replating frequency [RF] of 3.17 × 10−4) are counted as positive. Each dot represents one assay. Control data (MOCK, RV.SF, and LV.SF) from previous experiments conducted under the same standard operation procedure are included. Darker colors mark the actual assays and lighter colors indicate the meta data. Bars show the mean RF. Above the graph, the ratio of assays above and below the Q1 level are given together with a statistical analysis on the incidence of positive and negative plates. EFS-IL2RG had a significantly lower mutagenic potential compared to RV.SF and was indistinguishable from MOCK. (NS, not significant; ***p < 0.001 and *p < 0.05, Fisher’s exact test with Benjamini-Hochberg multiple comparison correction).
Analysis of the Integration Profile of the EFS-IL2RG Vector in Murine Il2rg−/− Lin− Progenitors
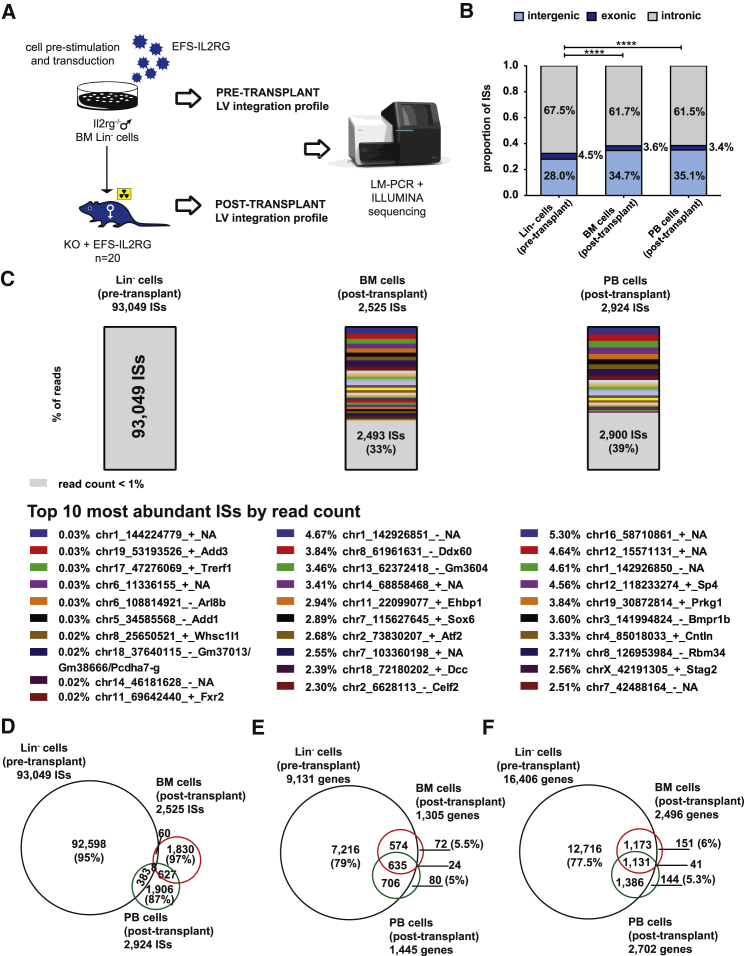
The global integration pattern of the EFS-IL2RG vector was determined on an aliquot of the transduced Il2rg−/− Lin− cells. Integration sites (ISs) were recovered by ligation-mediated PCR (LM-PCR) followed by Illumina sequencing, and they were mapped to the mouse genome (mm10 assembly) by a custom-designed bioinformatic pipeline (Figure 4A). Sequencing of the three LM-PCR libraries obtained by transduction of Il2rg−/− Lin− cells of the three different age groups generated a total of 26 million reads and 93,049 ISs univocally mapped on the murine genome. The EFS-IL2RG vector showed the canonical LV integration profile, with a prevalence of intragenic ISs (72%), either intronic (67%) or exonic (5%) (Figure 4B). Virtually all the ISs were represented by less than 1% of the total sequencing reads, indicating a highly polyclonal composition of the original pool of transduced Lin− cells (Figure 4C). The 66,532 intragenic ISs targeted 9,131 genes, defined as spanning from the transcription start site (TSS) to the end of the gene. By applying a less stringent gene definition, which includes 50 kb upstream of the TSS, we identified a pool of 16,406 targeted genes enriched in 35 gene ontology functional categories (Table S2) and 20 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (Table S3) by Database for Annotation, Visualization and Integrated Discovery (DAVID) analysis (p < 0.05 after Bonferroni correction for false discovery rate). Enriched pathways included metabolism and transport of nucleic acids and proteins, chromatin modifications, regulation of gene expression and mitosis, ubiquitination, splicing, and endocytosis.
Figure 4.
Integration Profile of the EFS-IL2RG Vector
(A) Scheme of the vector integration analysis study. ISs were retrieved by LM-PCR and Illumina high-throughput sequencing in pre-transplant Lin− cells and post-transplant BM and PB of Rag2−/−/Il2rg−/− mice transplanted with and Il2rg−/− Lin− cells transduced with the EFS-IL2RG vector (see Figure 1). (B) Genomic distribution of the vector ISs in pre- and post-transplant cells. The statistical significance of the difference in the percentage of intragenic (intronic + exonic) ISs in pre- versus post-transplant cells is calculated by a χ2 test. ****p < 0.0001. (C) Histograms of the percentage of sequence reads associated to each IS in the pre- and post-transplant cells. In each bar, the fraction of ISs with read count <1% is shown in gray, while ISs with read counts >1% are shown in different colors. The top 10 most abundant ISs in each sample are identified by their genomic coordinates. (D) Venn diagram showing the number of ISs identified in pre- and post-transplant cells. (E) Venn diagram showing the number of target genes identified in pre- and post-transplant cells by applying a stringent gene definition (from the +1 nt to the gene end). (F) Venn diagram showing the number of target genes identified in pre- and post-transplant cells by applying an enlarged gene definition, which includes 50 kb upstream of the +1 nt.
Analysis of the Integration Profile of the EFS-IL2RG Vector in the BM and PB of Transplanted Mice
The aim of the analysis was to compare pre- and post-transplantation integration profiles in terms of frequency and function of the targeted gene pool that might suggest positive or negative selection of cells harboring specific integration events. The ex vivo, post-transplantation EFS-IL2RG integration profile was determined on 14/20 mice surviving at 6 months. The integration profile was determined in the BM and PB of mice belonging to each transduction group as pools (2–4, 5–10, and 16 weeks). Mice 169 and 170, showing the abnormal CD4+/CD8+ cell population in the PB (see above), were analyzed individually, as well as one control mouse (179) belonging to the same transduction group (2–4 weeks), and they were excluded from the pooled post-transplant IS datasets. Overall, we retrieved 2,525 and 2,924 post-transplant ISs from the BM and PB, respectively (merged pool of all transduction groups). A large fraction of ISs accounted for <1% of the total read counts in both tissues (in gray in Figure 4C), and no IS accounted for >4.7% and >5.3% of the total reads in the BM and PB, respectively, indicating a largely polyclonal hematopoietic reconstitution in the transplanted mice, although we cannot exclude a more prominent oligoclonality in individual mice engrafted with a lower number of transduced stem cells. A polyclonal pattern was observed also in the three transduction groups analyzed as individual pools, despite the necessarily lower number of IS per sample (Figure S7). Notably, the top 10 most abundant ISs by read count were different in the BM and PB, and they included both intragenic and intergenic events (Figure 4C).
Interestingly, the proportion of intragenic (intronic + exonic) ISs was significantly decreased in the post-transplant samples with respect to the pre-transplant Lin− sample (65% in both the BM and PB versus 72% in Lin− cells, χ2 test p value < 0.0001) (Figure 4B), suggesting a negative selection of cells carrying an intragenic LV insertion in vivo. The difference was significant also when considering only the integrations in exons (3.6% in the BM and 3.4% in the PB versus 4.5% in Lin− cells, p = 0.002) (Figure 4B). When analyzed in individual transduction groups, the difference remained significant except for the 16-week group (69.1% in the BM and 68.7% in the PB versus 71.3% in pre-transplant cells, p = 0.14), where the relatively low number of post-transplant integrations may have affected the significance of the test. Most of the post-transplant ISs were not found in the large pool of pre-transplant ISs (98% and 87% in the BM and PB samples, respectively; Figure 4D), while, as expected, a significant fraction of ISs was shared between the post-transplant BM and PB pools (635 of 2,525 BM and 2,924 PB ISs or 25% and 22%, respectively; Figure 4D).
Overall, the intragenic ISs targeted 1,305 and 1,445 genes in the post-transplant BM and PB samples, respectively, 93% of which were in common with those targeted in the pre-transplant, Lin− cells (Figure 4E). Uniquely targeted genes were 96 in the BM and 104 in the PB, all hit by one IS, except the Sspn gene that was targeted by 2 ISs. By applying the gene definition that includes 50 kb upstream of the TSS, we identified 2,496 and 2,702 target genes in the BM and PB, respectively, 93% of which were again in common with those targeted in pre-transplant cells using the same definition (Figure 4F). To determine whether any gene was targeted at a different frequency in post- versus pre-transplant cells, we calculated an expected targeting frequency for each gene by multiple (1,000) random sampling of a comparable number of integrations from the corresponding pre-transplant IS collection. We observed no significant difference between observed and expected targeting frequency for any gene in the post- versus pre-transplant samples (p > 0.05) after Bonferroni correction for false discovery rate, regardless of the gene definition. Similarly, we observed no significant enrichment of gene ontology (GO) categories or KEGG pathways in post-transplant samples by running a DAVID analysis using the corresponding pre-transplant gene lists as background. These results indicate no positive or negative selection in vivo for cells carrying integrations in specific genes or class of genes.
IS Analysis in Individual Animals
Mice 169 and 170, showing the CD4+/CD8+ double-positive cell population in the PB, and a control mouse (179) belonging to the same 2- to 4-week transduction group, were analyzed individually. The IS analysis revealed a polyclonal hematopoietic reconstitution, with 347 and 405 ISs retrieved from the BM of mice 169 and 170 and 1,135 ISs from mouse 179 (Figure 5). None of the most abundant ISs (>1% of the total reads) was shared between mice 169 and 170, although two different ISs targeted the same gene (Runx1t1) 20,283 bp apart in opposite orientation (Figure 5). Eight and 12 ISs represented >80% of the reads in the BM of mice 169 and 170, respectively, indicating substantial clonal skewing, while in mouse 179 the 8 most abundant ISs accounted for only 30% of the total reads, indicating a more polyclonal and more balanced clonal composition in this animal. None of the most abundant ISs in the PB was in common between mice 169 (14 ISs) and 170 (11 ISs), arguing against the hypothesis that the CD4+/CD8+ double-positive cell populations originated from an integration-driven clonal expansion. We were unable to separately analyze the two cell populations to formally prove this point or to show whether they originated from donor, transduced cells.
Figure 5.
Integration Site Analysis in Individual Mice
(A) Scheme of the vector integration study in three individual Rag2−/−/Il2rg−/− mice transplanted with and Il2rg−/− Lin− cells transduced with the EFS-IL2RG vector (169, 170, and 179) and the corresponding, secondarily transplanted Il2rg-deficient mice. (B) Clonal composition in the BM of secondarily transplanted mice compared to that of the parental mice. The total number of ISs retrieved in each sample is shown on the top of the histograms, while the percentage of sequence reads associated to each ISs is indicated in gray (<1% of the total) or in different colors (>1% of the total). For the intragenic ISs, the name of the targeted gene is reported in the colored portion of the bar. Intergenic ISs are identified by NA followed by the genomic coordinates.
To further analyze the clonal composition in the hematopoiesis of mice 169, 170, and 179, we analyzed the ISs in the BM of secondarily transplanted Il2rg−/− mice. As described above, each primary BM was transplanted in two recipient mice, termed S1 and S2 (Figure 5A). As expected, we retrieved substantially less ISs in the secondary BMs (22–213) compared to the primarily transplanted animals (347–1,135). In all recipient mice, however, the most abundant ISs were essentially the same observed in the corresponding donor BM, although with a somewhat different relative proportion, with mice receiving the BM 179 having a practically overlapping IS profile (Figure 5B). These results argue against any integration-driven clonal expansion in mice 169 and 170, even in the condition of reduced stem cell numbers and stressed hematopoiesis. Of note, the proportion of intragenic integrations further decreased in BM cells from secondarily transplanted animals (from 67% of 1,919 ISs in the three primary recipients to 42% of 500 ISs in the six secondary recipients, p = 0), indicating a pronounced counterselection of cells harboring an intragenic provirus upon serial transplantation.
A Different Set of Genes Is Targeted by Lentiviral Integration in Human and Murine HSPCs
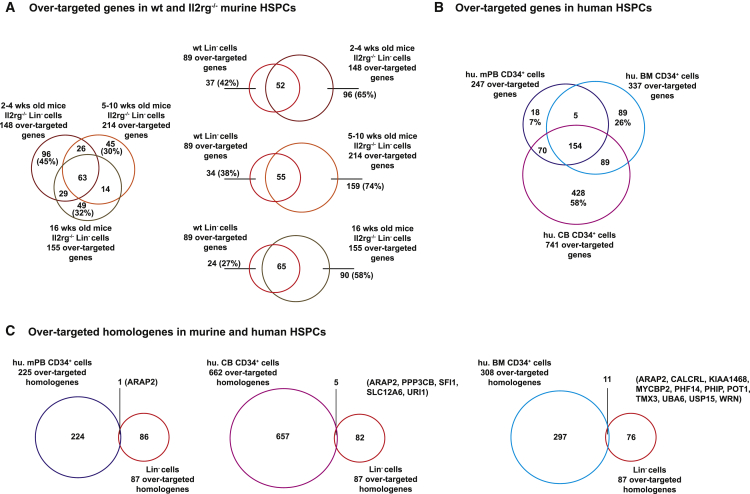
To gain insight into the differences in integration profiles in murine Lin− cells coming from mice of different age groups or between WT and Il2rg-deficient Lin− cells, we defined the sets of genes preferentially targeted by the vector in each cell type by comparing the actual targeting frequency of each gene with an expected targeting frequency obtained by multiple sampling of a set of random computational ISs, setting the threshold p value at < 0.001. By this analysis, we identified 148, 214, and 155 over-targeted genes of the 5,997, 6,599, and 5,723 genes targeted by the vector in the 2- to 4-, 5- to 10-, and 16-week Il2rg−/− Lin− cells, respectively. Most of the over-targeted genes were shared among the three age groups, with 63 genes common to all groups (Figure 6A). We then generated a set of 22,570 EFS-IL2RG ISs in Lin− cells obtained from the BM of 8-week-old, WT C57BL/6J mice, which we compared with the datasets obtained in the three groups of pre-transplant Il2rg−/− cells (28,886 ISs in 2- to 4-week, 37,084 in 5- to 10-week, and 27,947 in 16-week cells). The statistical analysis identified a set of 89 over-targeted genes in WT cells, the majority of which (58%–73%) were shared with the set of genes over-targeted in Il2rg−/− cells (Figure 6A), indicating that the vector identifies essentially the same preferential target genes in WT and Il2rg-deficient hematopoietic progenitors.
Figure 6.
Genes Over-Targeted by the EFS-IL2RG in Human and Murine HSPCs
(A) Venn diagrams showing the number of genes over-targeted by EFS-IL2RG in Lin− cells obtained from the BM of both wild-type and Il2rg-deficient mice of different age groups. (B) Over-targeted genes in common between human CD34+ HSPCs obtained from adult bone marrow, umbilical cord blood, and G-CSF-mobilized peripheral blood. (C) Over-targeted homologous genes in common between murine Lin− and human-mobilized CD34+ HSPCs. Only one gene (ARAP2) is over-targeted by EFS-IL2RG in both populations.
To compare the integration characteristics of LV vectors in murine versus human cells, we compared the integration profile of the EFS-IL2RG vector in murine Lin− cells with that obtained by transducing with the same vector granulocyte colony-stimulating factor (G-CSF)-mobilized PB CD34+ HSPCs. From these cells, we retrieved 25,682 ISs targeting 6,498 genes, 247 of which were significantly over-targeted. We also analyzed larger datasets previously obtained from BM- or umbilical cord blood-derived derived HSPCs transduced with an LV expressing the human WAS gene18 or a GFP gene30 in the same CCL-SIN-18-WPRE backbone, which identified 337 and 741 over-targeted genes, respectively (Table 2). While a substantial number of over-targeted genes was in common between the two adult human cell samples, almost 60% of the genes over-targeted in CB-derived HSPCs were specific for this cell type (Figure 6B). Interestingly, when we compared the genes preferentially targeted in murine Lin− cells with the human orthologs (NCBI HomoloGene definition) over-targeted in human mobilized CD34+ cells, we found only one gene in common (ARAP2). The comparison with the other two human datasets showed again no similarity (Figure 6C). Table S4 lists the top 50 over-targeted genes in murine Lin− cells and human-mobilized CD34+ cells ranked by number of ISs. The human list contains most of the genes found frequently targeted in pre- and post- transplantation samples in recent LV-based human gene therapy trials (e.g., KDM2A, PACS1, NPLOC4, GPATCH8, ASH1L, and FCHSD2),17, 18, 20, 31 none of which is targeted at any significant frequency in murine progenitors. These data show substantial differences in the pool of genes targeted at high frequency by LVs in murine versus human cells, which may potentially impact on the predictivity of LV-driven perturbation of specific genes in mouse studies.
Table 2.
Summary of Integration Data in Human CD34+ HSPCs from Different Sources
| HSPCs | ISs | Intergenic ISs (%) | Intronic ISs (%) | Exonic ISs (%) | Targeted Genes | Over-targeted Genes (p < 0.001) | ISs (Range) |
|---|---|---|---|---|---|---|---|
| BM CD34+ | 37,423 | 22.60 | 72.50 | 4.80 | 7,425 | 337 | 1–70 |
| CB CD34+ | 58,294 | 19.40 | 75.60 | 5.00 | 8,911 | 741 | 1–207 |
| PB CD34+ | 25,682 | 21.20 | 74.30 | 4.50 | 6,498 | 247 | 1–61 |
BM, bone marrow; CB, umbilical cord blood; PB, mobilized peripheral blood.
Discussion
The current therapy for SCID-X1 is transplantation of CD34+ HSPCs from histocompatible donors. The efficacy of the therapy depends on age, availability of a suitable donor, and absence of infections at the time of treatment.32 Selective advantage allows donor progenitor cells to repopulate the patient’s lymphopoietic organs even in the absence of marrow conditioning, which is traditionally avoided to reduce treatment toxicity. However, non-T immune cell reconstitution is variable in the absence of myeloablation, reducing the overall efficacy of the therapy. Gene therapy, i.e., transplantation of genetically corrected, autologous HSPCs, reduces the risk associated with allogeneic transplantation and is theoretically accessible to all patients irrespective of donor availability. The first trials proved the efficacy of the therapy in reconstituting T cell immunity in pediatric patients, and they showed that the absence of marrow conditioning impairs reconstitution of B and NK cell immunity also in an autologous setting.3, 6, 10 Gene therapy was only partially efficacious in older patients, probably due to involution of niches for new T cell development.33
To improve the efficacy of gene therapy, several groups developed LV vectors expressing IL2RG, and they attempted to introduce non-myeloablative conditioning to allow for a better clinical response.23, 27, 34 We developed a SIN lentiviral vector, EFS-IL2RG, expressing a codon-optimized cDNA driven by the constitutive EFS promoter, which showed no tendency for insertional gene activation in preclinical models35, 36, 37 and in clinical trials for ADA-SCID. We show reconstitution of T, B, and NK cells in sub-lethally irradiated IL2rg−/−/Rag2−/− mouse transplanted with IL2rg−/− Lin− HSPCs transduced with the EFS-IL2RG vector. IL2rg−/−/Rag2−/− mice completely lack T, B, and NK cells, and they provide a better, less leaky background to analyze immune reconstitution with respect to Il2rg−/− single-knockout mice. Gene therapy showed an efficacy comparable to transplantation with syngeneic wild-type HSPCs, in the absence of treatment-related adverse events. Reconstitution of B and NK cells was lower in animals receiving gene-corrected cells compared to controls, although serum immunoglobulin levels (IgM and IgG) were comparable. The good performance of the vector was confirmed by the high proportion (>80%) of gene-marked clonogenic progenitors grown in culture from human CD34+ HSPCs transduced by the EFS-IL2RG at an MOI of 100. Overall, the study shows the potential of the EFS-IL2RG vector in correcting SCID-X1 after non-myeloablative conditioning.
Vector IS analysis carried out at the end of the study showed that BM repopulation was sustained by a largely polyclonal repertoire of hematopoietic stem cells (HSCs), with no signs of vector-driven clonal dominance and no significant selection of cells carrying integration in specific genes, as evaluated after pooling BM and PB cells from animals receiving the same transduced cell batches. This type of analysis averages engraftment efficiency and estimates clonal representation, although it cannot exclude oligoclonality in individual mice with very low chimerism. Interestingly, we observed a reduction in intragenic integrations in BM and PB cells in primary and even more in secondary transplant recipients with respect to pre-transplant Lin− cells, indicating either a different integration bias in repopulating HSCs with respect to a progenitor cell pool or a negative in vivo selection of cells carrying integrations in exons and introns. Integrations in exons cause mono-allelic gene knockout, while the presence of LV proviruses in introns may induce post-transcriptional alterations of gene expression through the insertion of splicing and polyadenylation signals in primary transcripts, as previously shown in human hematopoietic cells.38, 39 Our data suggest that these integration events may reduce cell fitness in vivo and be counterselected when occurring in repopulating HSCs. However, we did not observe significant differences in the categories of genes targeted by LV integration before and after transplantation, indicating that, if intragenic integrations cause a reduction of cell fitness, they do so in a relatively random fashion.
We observed an immature, CD4+/CD8+ double-positive T cell population in the peripheral circulation of two transplanted mice, which disappeared upon secondary transplantation, indicating their non-malignant nature and/or lack of repopulating capacity. Vector IS analysis showed that the two animals, as well as a control animal transplanted with the same batch of transduced cells at the same age, were engrafted by a polyclonal HSC cell population that further engrafted secondary recipients in a similar relative proportion, with no evidence of expansion of clones carrying integration in specific genes. On the contrary, cells carrying integrations in potential proto-oncogenes, such as Runx1t1, Clcn5, or Tpk1, either disappeared or were less prevalent in secondary versus primary recipients, indicating a random fluctuation of stem cell clones independently from the provirus insertion site. Together with the results of the IVIM, the in vivo vector integration analysis demonstrates the favorable genotoxic profile of the EFS-IL2RG vector.
The EFS-IL2RG vector showed a statistically significant over-targeting of a set of genes with no obvious characteristics in terms of size, gene expression activity, or function. Integration into hotspots is a typical feature of LV vectors reported in pre- and post-transplant hematopoietic cells in several clinical studies,17, 18, 19, 20, 21, 22 which depends on the topological distribution of euchromatin in the membrane-proximal compartment of the cell nucleus, favored by lentiviral integration.15 Interestingly, the set of genes targeted by EFS-IL2RG in murine Lin− cells was completely different from that targeted in human CD34+ cells obtained from BM, mobilized PB, or umbilical cord blood, suggesting a very different topological organization of homologous genes in HSPCs, and most likely in repopulating stem cells, between mice and humans. This evidence suggests that mouse models may not be entirely predictive of the genotoxic consequences of lentiviral integration when analyzing events happening in specific genes or gene clusters, due to the exceedingly different targeting frequency between the two species.
Overall, this study paves the way for application of an improved lentiviral gene therapy for SCID-X1. Combined with state-of-the-art cell processing and increased automation, the vector described in the study offers a clear potential for commercial drug development.
Materials and Methods
Vector Production and Titration
Lentiviral vectors pCCL_pEF1a_IL2RGWT_WPRE* and pCCL_pEF1a_IL2RGcoWPRE* (EFS-IL2RG) were designed and generated at Hannover Medical School, and they were packaged by triple transfection of HEK293T cells with the CMV-GAG/POL, CMV-REV, and CMV-VSV-G plasmids in 225-cm2 tissue culture flasks. The supernatants were collected 48 hr post-transfection, concentrated by ultracentrifugation, and resuspended in X-VIVO 20 medium (Lonza). For the animal study, EFS-IL2RG was produced by transfecting HEK293T cells at a 50-l scale in CF10 cell factories, purified by ion-exchange chromatography, concentrated by tangential-flow filtration, and formulated in X-VIVO 20, as previously described.40 The viral titer was calculated by transduction of the human colorectal carcinoma cell line (HCT116) with serial dilution of the viral preparation followed by qPCR, as previously described.41
Transduction of Human Cells
The ED7R cell line was derived from an adult human T cell leukemia lacking IL2RG expression.42, 43 ED7R cells were grown in RPMI-1640 medium supplemented with 10% fetal calf serum, penicillin/streptomycin, and Glutamax (all from Gibco). 5 × 105 ED7R cells were transduced at an MOI of 0.1–100 for 6 hr with polybrene (6 μg/mL, Sigma-Aldrich) and maintained in culture for 12 days. IL2RG expression was analyzed by flow cytometry.
CD34+ cells were isolated from cord blood or G-CSF-mobilized PB samples provided by the Centre Hospitalier Sud Francilien (Evry, France) or the Institut Gustave Roussy (Paris, France), under full informed consent. CD34+ cells were immunoselected with the CliniMACS system (Miltenyi Biotec), following the manufacturer’s instructions; pre-activated by overnight culture in X-vivo 20 (Lonza) containing 25 ng/mL human stem cell factor (SCF; CellGenix), 50 ng/mL human Flt-3 ligand (CellGenix), 25 ng/mL human thrombopoietin (TPO; CellGenix) ,and 10 ng/mL human IL-3 (CellGenix); and transduced at different MOIs for 6 hr in the same medium in the presence of 4 μg/mL protamine sulfate (Sigma-Aldrich). After transduction, cells were washed and maintained in liquid culture in the activation medium or grown as individual progenitors in semi-solid Methocult medium (H3434, STEMCELL Technologies) for 2 weeks. Individual colonies were counted and harvested for VCN determination after 2 weeks of culture.
Transduction of Murine Lin− Cells
Lin− cells were purified from the BM of Il2rg−/− male mice by magnetic bead cell sorting (Miltenyi Biotec) and activated overnight in X-vivo 20 supplemented with 50 μg/mL penicillin, 50 ng/mL streptomycin, 20 ng/mL murine interleukin-3 (Miltenyi Biotec), 100 ng/mL human TPO (CellGenix), 100 ng/mL human Flt-3 ligand (CellGenix), and 100 ng/mL murine SCF (Miltenyi Biotec). Cells were transduced with the CCL-EF1α-coILRG-WPRE or the CCL-PGK-GFP-WPRE vector at an MOI of 100 for 6 hr in the presence of 6 μg/mL protamine sulfate (Sigma-Aldrich). Cells receiving two rounds of infection were washed and re-exposed overnight to the vector under the same conditions.
Transplantation of Il2rg-Deficient Mice
Mice were housed at the Genethon animal facility in accordance with national and European ethical guidelines, following the animal study protocol DAP 2014-005-A. BM cells were flushed from femurs and tibias of C57Bl6 mice (from Charles River Laboratories), Il2rg−/− mice (B6.129S4-Il2rgtm1Wjl/J, from Charles River Laboratories), and Rag2−/−/Il2rg−/− mice (B6-Rag2/tm1Fwa/Il2rg/tm1Wjl, from Taconic). Transduced Lin− cells (5 × 105 cells/mouse) were administered intravenously by retro-orbital injection to Rag2−/−/Il2rg−/− female mice that were given 6-Gy total body irradiation as a single exposure 3 hr before transplantation.
Analysis of VCN, Chimerism, and IL2RG Expression
Genomic DNA was extracted from transduced cells expanded in liquid culture by the Wizard genomic DNA purification kit (Promega). DNA from colonies was extracted after lysis with proteinase K (Thermo Fisher Scientific) under standard conditions. The average VCN per cell (VCN/cell) was analyzed by duplex Taqman qPCR with primers and probes annealing to the HIV psi sequence and a reference gene, the human ALB or the murine Ttn gene. Results were calculated based on a standard curve of a plasmid containing the two sequences. Donor cell chimerism was measured by Taqman qPCR with primers specific for the murine Y chromosome and Ttn as a normalizer gene. Results were expressed as percentage of male cells in the total cells, based on a reference titration curve consisting of different proportions of male/female blood cells. All PCR measurements were performed at least in duplicates in an ABI PRISM 7700 system (Thermo Fisher Scientific). Total RNA was extracted with the SV Total RNA Isolation System (Promega), reverse transcribed into cDNA with the Super-Script II RT (Gibco) using random hexamer primers, and amplified with the TaqMan Universal PCR Master Mix (Thermo Fisher Scientific) on an ABI PRISM 7700 sequence detector (Thermo Fisher Scientific). Vector-derived IL2RG mRNA was measured by qRT-PCR in duplicates with primers annealing to WPRE and the human or murine TFIID gene as a normalizer. All primers and probes are listed in the Supplemental Materials and Methods.
Flow Cytometry
Cells were resuspended in PBS and incubated for 30 min at 4°C with antibodies for specific antigens or with isotype control, following the manufacturer’s instructions. Stained cells were washed and measured on a BD LSR II flow cytometer (BD Biosciences). Analyses were performed with the FlowJo software version (v.)7.6.5. The complete list of antibodies used in the study is in the Supplemental Materials and Methods.
Statistical Analysis
Statistical analyses of the results of the animal study were performed using GraphPad Prism v.7.0 for Windows. Results are reported as mean ± SEM. Statistical differences between means were evaluated by Mann-Whitney or χ2 test as appropriate. Differences were considered significant at a p value < 0.05.
IVIM Assay
The IVIM assay was performed as previously described.28, 29, 44 Briefly, 1 × 105 Lin− BM cells from C57BL/6J mice were transduced with gammaretroviral or lentiviral vectors, expanded for 15 days, and seeded on 96-well plates (100 cells/well). After a further incubation for 2 weeks, insertional mutants were detected by microscopic evaluation or absorbance measurement of a reduced formazan (MTT assay) in a microplate reader.45 Positive wells were counted and the replating frequencies (RFs) calculated by the R package limdil.46 Plates with an RF ≥ 3.17 × 10−4 were defined as positive assays. The incidence of positive assays was compared by a Fisher’s exact test with Benjamini-Hochberg correction for multiple comparisons. For selected clones, Mecom gene expression was analyzed as described in the Supplemental Materials and Methods.
Vector IS Analysis
3′ LTR vector-genome junctions were amplified by LM-PCR. Briefly, 0.3–2 μg genomic DNA was digested 6 hr at 37°C with the Tru91 restriction enzyme (Roche), purified by NucleoSpin Gel and PCR Clean-up kit (MACHEREY-NAGEL), and ligated overnight to a TA-protruding double-stranded DNA linker by T4 DNA Ligase (New England Biolabs). Ligated DNA was purified, digested overnight at 37°C with SacI (Roche), and purified again. Multiple nested PCRs (10–20) were performed with specific primers annealing to the linker and the 3′ vector LTR, adapted to Illumina sequencing and containing a 4-bp sample-specific bar code for sample identification. The pre-transplant Lin− cell library was derived from a pool of DNAs collected from cells transduced by 1 or 2 hits (1 μg each). The post-transplant libraries were derived from pools of DNAs from individual mice in each transduction group or from DNAs of individual primarily or secondarily transplanted mice. Libraries were quantified on a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific), and they were loaded onto a 1% agarose gel for amplicon size selection. Amplicons ranging from 200 to 500 kb were manually extracted from the gel and purified by NucleoSpin Gel and PCR Clean-up kit. About 1 μg of the final libraries was subsequently processed with MiSeq Reagent Kit v3 (2 × 300-bp pair-end sequencing), following the manufacturer’s instructions adding a 6-bp sample-specific index, and sequenced to saturation on an Illumina MySeq machine at IGA Technology Services (Udine, Italy). ISs in human cord blood-derived and adult BM CD34+ cells were determined by re-analyzing raw reads from previously published studies.18, 30
Raw reads from Illumina paired-end sequencing were bioinformatically trimmed to recover the host genome sequences adjacent to the 3′ proviral LTR by the Skewer software (mismatch rate of 0.12, minimal length of 20 bp after trimming, and a minimal match length equal to the pre-trimming sequence), and they were mapped on the reference genome (human GRCh37.75/hg19 and murine GRCm38/mm10) by the Bowtie2 software (>95% identity). The sequences are available at GenBank Sequence Read Archive (SRA): SRP135780. The genomic coordinates of the first nucleotide in the host genome adjacent to the viral LTR were indicated as IS. ISs originated by different reads mapping in the same genomic position were collapsed, recovering the number of corresponding reads (read count). Unique ISs were annotated on the RefSeq gene database as intergenic or intragenic (exonic or intronic). Genes (defined by the most upstream TSS to the most downstream 3′ end) hosting at least one IS were identified as target genes. Genes with targeting frequency significantly higher than random, defined by 200 random sampling of virtual ISs corresponding to 5,330,124 and 5,812,911 Tru91 sites in the human and murine genome, respectively, were defined as over-targeted after Bonferroni correction of the p values for false discovery rate. Genes with at least 3 ISs and a Bonferroni-corrected p value of < 0.001 were defined as over-targeted. To compare over-targeted genes in the human and murine genomes, we retrieved the orthologs genes by the HomoloGene tool on the NCBI webpage https://www.ncbi.nlm.nih.gov/homologene/statistics. Additional information about the bioinformatics processing of the sequencing reads can be found in the Supplemental Materials and Methods.
Author Contributions
Study Design, S.C., V.P., A.J.T., A.S., and F.M.; Study Execution, S.C., B.G., A.V., V.P., G.C., F.Z., M.R., and A.S.; Data Analysis, S.C., B.G., V.P., G.C., M.R., A.S., and F.M.; Manuscript Drafting, V.P., S.C., and M.R.; Manuscript Review, Discussion, and Finalization, H.B.G., A.J.T., and F.M.
Conflicts of Interest
A.J.T. is a founder and consultant of Orchard Therapeutics, and H.B.G. is Chief Scientific Officer of Orchard Therapeutics.
Acknowledgments
This work was funded by grants from the European Commission (H2020 Program SCIDNET and FP7 program CELL-PID), the Wellcome Trust (104807/Z/14/Z), the DFG (REBIRTH Cluster of Excellence), and the NIH Research Biomedical Research Centre at Great Ormond Street Hospital for Children, NHS Foundation Trust and University College London. F.Z. was supported by the EC FP7 grants CELL-PID (261387) and SUPERSIST (HEALTH-F4-2013-601958).
Footnotes
Supplemental Information includes Supplemental Materials and Methods, seven figures, and four tables and can be found with this article online at https://doi.org/10.1016/j.omtm.2018.03.002.
Supplemental Information
References
- 1.Booth C., Gaspar H.B., Thrasher A.J. Treating Immunodeficiency through HSC Gene Therapy. Trends Mol. Med. 2016;22:317–327. doi: 10.1016/j.molmed.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Aiuti A., Slavin S., Aker M., Ficara F., Deola S., Mortellaro A., Morecki S., Andolfi G., Tabucchi A., Carlucci F. Correction of ADA-SCID by stem cell gene therapy combined with nonmyeloablative conditioning. Science. 2002;296:2410–2413. doi: 10.1126/science.1070104. [DOI] [PubMed] [Google Scholar]
- 3.Hacein-Bey-Abina S., Le Deist F., Carlier F., Bouneaud C., Hue C., De Villartay J.P., Thrasher A.J., Wulffraat N., Sorensen R., Dupuis-Girod S. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N. Engl. J. Med. 2002;346:1185–1193. doi: 10.1056/NEJMoa012616. [DOI] [PubMed] [Google Scholar]
- 4.Ott M.G., Schmidt M., Schwarzwaelder K., Stein S., Siler U., Koehl U., Glimm H., Kühlcke K., Schilz A., Kunkel H. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat. Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 5.Aiuti A., Cattaneo F., Galimberti S., Benninghoff U., Cassani B., Callegaro L., Scaramuzza S., Andolfi G., Mirolo M., Brigida I. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N. Engl. J. Med. 2009;360:447–458. doi: 10.1056/NEJMoa0805817. [DOI] [PubMed] [Google Scholar]
- 6.Hacein-Bey-Abina S., Hauer J., Lim A., Picard C., Wang G.P., Berry C.C., Martinache C., Rieux-Laucat F., Latour S., Belohradsky B.H. Efficacy of gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 2010;363:355–364. doi: 10.1056/NEJMoa1000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boztug K., Schmidt M., Schwarzer A., Banerjee P.P., Díez I.A., Dewey R.A., Böhm M., Nowrouzi A., Ball C.R., Glimm H. Stem-cell gene therapy for the Wiskott-Aldrich syndrome. N. Engl. J. Med. 2010;363:1918–1927. doi: 10.1056/NEJMoa1003548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biasco L., Baricordi C., Aiuti A. Retroviral integrations in gene therapy trials. Mol. Ther. 2012;20:709–716. doi: 10.1038/mt.2011.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavazza A., Moiani A., Mavilio F. Mechanisms of retroviral integration and mutagenesis. Hum. Gene Ther. 2013;24:119–131. doi: 10.1089/hum.2012.203. [DOI] [PubMed] [Google Scholar]
- 10.Hacein-Bey-Abina S., Pai S.Y., Gaspar H.B., Armant M., Berry C.C., Blanche S., Bleesing J., Blondeau J., de Boer H., Buckland K.F. A modified γ-retrovirus vector for X-linked severe combined immunodeficiency. N. Engl. J. Med. 2014;371:1407–1417. doi: 10.1056/NEJMoa1404588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moiani A., Miccio A., Rizzi E., Severgnini M., Pellin D., Suerth J.D., Baum C., De Bellis G., Mavilio F. Deletion of the LTR enhancer/promoter has no impact on the integration profile of MLV vectors in human hematopoietic progenitors. PLoS ONE. 2013;8:e55721. doi: 10.1371/journal.pone.0055721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demeulemeester J., De Rijck J., Gijsbers R., Debyser Z. Retroviral integration: Site matters: Mechanisms and consequences of retroviral integration site selection. BioEssays. 2015;37:1202–1214. doi: 10.1002/bies.201500051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naldini L. Ex vivo gene transfer and correction for cell-based therapies. Nat. Rev. Genet. 2011;12:301–315. doi: 10.1038/nrg2985. [DOI] [PubMed] [Google Scholar]
- 14.Naldini L. Gene therapy returns to centre stage. Nature. 2015;526:351–360. doi: 10.1038/nature15818. [DOI] [PubMed] [Google Scholar]
- 15.Marini B., Kertesz-Farkas A., Ali H., Lucic B., Lisek K., Manganaro L., Pongor S., Luzzati R., Recchia A., Mavilio F. Nuclear architecture dictates HIV-1 integration site selection. Nature. 2015;521:227–231. doi: 10.1038/nature14226. [DOI] [PubMed] [Google Scholar]
- 16.Poletti V., Mavilio F. Interactions between Retroviruses and the Host Cell Genome. Mol. Ther. Methods Clin. Dev. 2017;8:31–41. doi: 10.1016/j.omtm.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aiuti A., Biasco L., Scaramuzza S., Ferrua F., Cicalese M.P., Baricordi C., Dionisio F., Calabria A., Giannelli S., Castiello M.C. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 2013;341:1233151. doi: 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hacein-Bey Abina S., Gaspar H.B., Blondeau J., Caccavelli L., Charrier S., Buckland K., Picard C., Six E., Himoudi N., Gilmour K. Outcomes following gene therapy in patients with severe Wiskott-Aldrich syndrome. JAMA. 2015;313:1550–1563. doi: 10.1001/jama.2015.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eichler F., Duncan C., Musolino P.L., Orchard P.J., De Oliveira S., Thrasher A.J., Armant M., Dansereau C., Lund T.C., Miller W.P. Hematopoietic Stem-Cell Gene Therapy for Cerebral Adrenoleukodystrophy. N. Engl. J. Med. 2017;377:1630–1638. doi: 10.1056/NEJMoa1700554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biffi A., Montini E., Lorioli L., Cesani M., Fumagalli F., Plati T., Baldoli C., Martino S., Calabria A., Canale S. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- 21.Cavazzana-Calvo M., Payen E., Negre O., Wang G., Hehir K., Fusil F., Down J., Denaro M., Brady T., Westerman K. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ribeil J.A., Hacein-Bey-Abina S., Payen E., Magnani A., Semeraro M., Magrin E., Caccavelli L., Neven B., Bourget P., El Nemer W. Gene Therapy in a Patient with Sickle Cell Disease. N. Engl. J. Med. 2017;376:848–855. doi: 10.1056/NEJMoa1609677. [DOI] [PubMed] [Google Scholar]
- 23.De Ravin S.S., Wu X., Moir S., Anaya-O’Brien S., Kwatemaa N., Littel P., Theobald N., Choi U., Su L., Marquesen M. Lentiviral hematopoietic stem cell gene therapy for X-linked severe combined immunodeficiency. Sci. Transl. Med. 2016;8:335ra57. doi: 10.1126/scitranslmed.aad8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buckley R.H. Primary immunodeficiency diseases: dissectors of the immune system. Immunol. Rev. 2002;185:206–219. doi: 10.1034/j.1600-065x.2002.18517.x. [DOI] [PubMed] [Google Scholar]
- 25.Hacein-Bey-Abina S., Garrigue A., Wang G.P., Soulier J., Lim A., Morillon E., Clappier E., Caccavelli L., Delabesse E., Beldjord K. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howe S.J., Mansour M.R., Schwarzwaelder K., Bartholomae C., Hubank M., Kempski H., Brugman M.H., Pike-Overzet K., Chatters S.J., de Ridder D. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J. Clin. Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huston M.W., van Til N.P., Visser T.P., Arshad S., Brugman M.H., Cattoglio C., Nowrouzi A., Li Y., Schambach A., Schmidt M. Correction of murine SCID-X1 by lentiviral gene therapy using a codon-optimized IL2RG gene and minimal pretransplant conditioning. Mol. Ther. 2011;19:1867–1877. doi: 10.1038/mt.2011.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Modlich U., Bohne J., Schmidt M., von Kalle C., Knöss S., Schambach A., Baum C. Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity. Blood. 2006;108:2545–2553. doi: 10.1182/blood-2005-08-024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zychlinski D., Schambach A., Modlich U., Maetzig T., Meyer J., Grassman E., Mishra A., Baum C. Physiological promoters reduce the genotoxic risk of integrating gene vectors. Mol. Ther. 2008;16:718–725. doi: 10.1038/mt.2008.5. [DOI] [PubMed] [Google Scholar]
- 30.Cattoglio C., Pellin D., Rizzi E., Maruggi G., Corti G., Miselli F., Sartori D., Guffanti A., Di Serio C., Ambrosi A. High-definition mapping of retroviral integration sites identifies active regulatory elements in human multipotent hematopoietic progenitors. Blood. 2010;116:5507–5517. doi: 10.1182/blood-2010-05-283523. [DOI] [PubMed] [Google Scholar]
- 31.Cartier N., Hacein-Bey-Abina S., Bartholomae C.C., Veres G., Schmidt M., Kutschera I., Vidaud M., Abel U., Dal-Cortivo L., Caccavelli L. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- 32.Pai S.Y., Logan B.R., Griffith L.M., Buckley R.H., Parrott R.E., Dvorak C.C., Kapoor N., Hanson I.C., Filipovich A.H., Jyonouchi S. Transplantation outcomes for severe combined immunodeficiency, 2000-2009. N. Engl. J. Med. 2014;371:434–446. doi: 10.1056/NEJMoa1401177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thrasher A.J., Hacein-Bey-Abina S., Gaspar H.B., Blanche S., Davies E.G., Parsley K., Gilmour K., King D., Howe S., Sinclair J. Failure of SCID-X1 gene therapy in older patients. Blood. 2005;105:4255–4257. doi: 10.1182/blood-2004-12-4837. [DOI] [PubMed] [Google Scholar]
- 34.Zhou S., Mody D., DeRavin S.S., Hauer J., Lu T., Ma Z., Hacein-Bey Abina S., Gray J.T., Greene M.R., Cavazzana-Calvo M. A self-inactivating lentiviral vector for SCID-X1 gene therapy that does not activate LMO2 expression in human T cells. Blood. 2010;116:900–908. doi: 10.1182/blood-2009-10-250209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cesana D., Ranzani M., Volpin M., Bartholomae C., Duros C., Artus A., Merella S., Benedicenti F., Sergi Sergi L., Sanvito F. Uncovering and dissecting the genotoxicity of self-inactivating lentiviral vectors in vivo. Mol. Ther. 2014;22:774–785. doi: 10.1038/mt.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montini E., Cesana D., Schmidt M., Sanvito F., Ponzoni M., Bartholomae C., Sergi Sergi L., Benedicenti F., Ambrosi A., Di Serio C. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat. Biotechnol. 2006;24:687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- 37.Carbonaro D.A., Zhang L., Jin X., Montiel-Equihua C., Geiger S., Carmo M., Cooper A., Fairbanks L., Kaufman M.L., Sebire N.J. Preclinical demonstration of lentiviral vector-mediated correction of immunological and metabolic abnormalities in models of adenosine deaminase deficiency. Mol. Ther. 2014;22:607–622. doi: 10.1038/mt.2013.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cesana D., Sgualdino J., Rudilosso L., Merella S., Naldini L., Montini E. Whole transcriptome characterization of aberrant splicing events induced by lentiviral vector integrations. J. Clin. Invest. 2012;122:1667–1676. doi: 10.1172/JCI62189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moiani A., Paleari Y., Sartori D., Mezzadra R., Miccio A., Cattoglio C., Cocchiarella F., Lidonnici M.R., Ferrari G., Mavilio F. Lentiviral vector integration in the human genome induces alternative splicing and generates aberrant transcripts. J. Clin. Invest. 2012;122:1653–1666. doi: 10.1172/JCI61852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merten O.W., Charrier S., Laroudie N., Fauchille S., Dugué C., Jenny C., Audit M., Zanta-Boussif M.A., Chautard H., Radrizzani M. Large-scale manufacture and characterization of a lentiviral vector produced for clinical ex vivo gene therapy application. Hum. Gene Ther. 2011;22:343–356. doi: 10.1089/hum.2010.060. [DOI] [PubMed] [Google Scholar]
- 41.Charrier S., Ferrand M., Zerbato M., Précigout G., Viornery A., Bucher-Laurent S., Benkhelifa-Ziyyat S., Merten O.W., Perea J., Galy A. Quantification of lentiviral vector copy numbers in individual hematopoietic colony-forming cells shows vector dose-dependent effects on the frequency and level of transduction. Gene Ther. 2011;18:479–487. doi: 10.1038/gt.2010.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ishii N., Asao H., Kimura Y., Takeshita T., Nakamura M., Tsuchiya S., Konno T., Maeda M., Uchiyama T., Sugamura K. Impairment of ligand binding and growth signaling of mutant IL-2 receptor gamma-chains in patients with X-linked severe combined immunodeficiency. J. Immunol. 1994;153:1310–1317. [PubMed] [Google Scholar]
- 43.Kumaki S., Ishii N., Minegishi M., Tsuchiya S., Cosman D., Sugamura K., Konno T. Functional role of interleukin-4 (IL-4) and IL-7 in the development of X-linked severe combined immunodeficiency. Blood. 1999;93:607–612. [PubMed] [Google Scholar]
- 44.Huang J., Liu Y., Au B.C., Barber D.L., Arruda A., Schambach A., Rothe M., Minden M.D., Paige C.J., Medin J.A. Preclinical validation: LV/IL-12 transduction of patient leukemia cells for immunotherapy of AML. Mol. Ther. Methods Clin. Dev. 2016;3:16074. doi: 10.1038/mtm.2016.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 46.Bonnefoix T., Bonnefoix P., Verdiel P., Sotto J.J. Fitting limiting dilution experiments with generalized linear models results in a test of the single-hit Poisson assumption. J. Immunol. Methods. 1996;194:113–119. doi: 10.1016/0022-1759(96)00077-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.