Summary
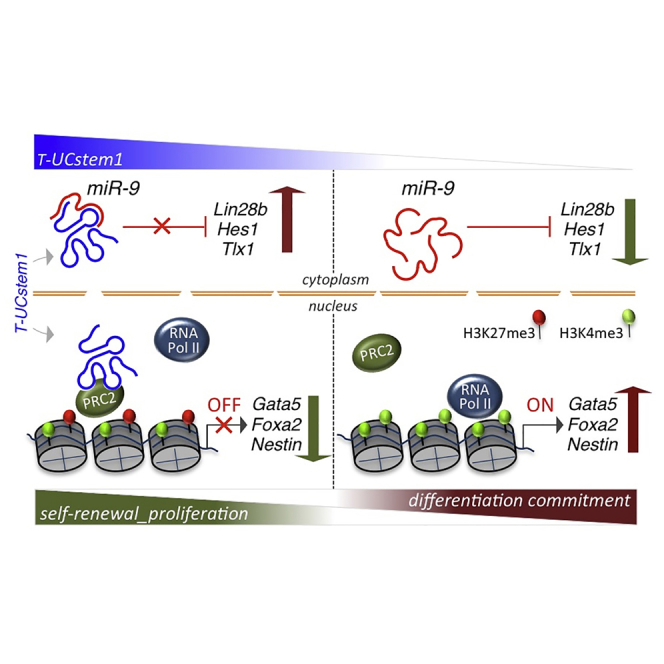
Ultraconserved elements (UCEs) show the peculiar feature to retain extended perfect sequence identity among human, mouse, and rat genomes. Most of them are transcribed and represent a new family of long non-coding RNAs (lncRNAs), the transcribed UCEs (T-UCEs). Despite their involvement in human cancer, the physiological role of T-UCEs is still unknown. Here, we identify a lncRNA containing the uc.170+, named T-UCstem1, and provide in vitro and in vivo evidence that it plays essential roles in embryonic stem cells (ESCs) by modulating cytoplasmic miRNA levels and preserving transcriptional dynamics. Specifically, while T-UCstem1::miR-9 cytoplasmic interplay regulates ESC proliferation by reducing miR-9 levels, nuclear T-UCstem1 maintains ESC self-renewal and transcriptional identity by stabilizing polycomb repressive complex 2 on bivalent domains. Altogether, our findings provide unprecedented evidence that T-UCEs regulate physiological cellular functions and point to an essential role of T-UCstem1 in preserving ESC identity.
Keywords: embryonic stem cells, self-renewal and differentiation, T-UCEs, non-coding RNAs, PRC2, bivalent genes
Graphical Abstract
Highlights
-
•
The transcribed ultraconserved element T-UCstem1 exerts a dual function in ESCs
-
•
T-UCstem1 controls ESC proliferation by regulating the miR-9/Lin28b molecular axis
-
•
T-UCstem1 maintains ESC self-renewal
-
•
T-UCstem1 preserves ESC transcriptional identity by stabilizing PRC2
In this article Fico, Minchiotti, and colleagues identify an ultraconserved element containing long non-coding RNA, named T-UCstem1, in embryonic stem cells (ESCs) and provide evidence that it regulates cell-cycle progression by modulating cytoplasmic miR-9 levels and preserves ESC identity and self-renewal by stabilizing polycomb repressive complex 2 (PRC2) on bivalent domains.
Introduction
Evolutionary conservation has become more and more a powerful tool to identify functionally important sequences in the genome (Dermitzakis et al., 2005). In this context, the ultraconserved elements (UCEs) are 481 genomic segments longer than 200 base pairs (bp), which are fully conserved (100% identity with no insertions or deletions) between human, mouse, and rat genomes (Bejerano et al., 2004). This complete conservation led to the hypothesis that UCEs likely have biological functions fundamental to mammal cells (Katzman et al., 2007). Despite extensive studies, our knowledge of UCEs is still limited. Indeed, increasing evidences indicate that UCEs play different functions in vertebrate genomes, acting as enhancer (Paparidis et al., 2007, Pennacchio et al., 2006), splicing (Ni et al., 2007), and epigenetic regulators (Bernstein et al., 2006, Lee et al., 2006), or functioning as transcriptional coactivators (Feng et al., 2006). In particular, many UCEs act as long-range enhancers during mouse development (Pennacchio et al., 2006), and it has been proposed that their removal in vivo would lead to a significant phenotypic impact. Nevertheless, knockout studies performed so far indicate that UCEs are dispensable for mice viability (Ahituv et al., 2007, Nobrega et al., 2004).
A large fraction of UCEs are transcribed (T-UCEs) in a tissue specific manner, and are deregulated in several human cancers (Calin et al., 2007, Fabbri et al., 2008, Fassan et al., 2014, Olivieri et al., 2016). Indeed, it has been shown that T-UCEs may also act as long non-coding RNAs (lncRNAs) regulating other RNAs (Calin et al., 2007, Liz et al., 2014). The main molecular mechanism of T-UCEs activity described so far is the “decoy” function. Indeed, T-UCEs sequester microRNAs (miRNAs) from the cytoplasm and eventually regulate cancer cell proliferation (Calin et al., 2007, Galasso et al., 2014, Olivieri et al., 2016). All together these findings provided robust evidence supporting the functional role of T-UCEs in the human genome, and highlighted a link between these genomic elements and human disease. Nevertheless, currently very little is known on the physiological role of this specific class of lncRNAs, as for instance in stem cell biology (Dinger et al., 2008, Feng et al., 2006, Mattick and Makunin, 2005).
Of note, several lncRNAs, including Hotair (Rinn et al., 2007), LincRNA-RoR (Loewer et al., 2010), Dali, MALAT1, Evf-2, and Nkx2.2AS (Chalei et al., 2014, Dinger et al., 2008, Guan et al., 2013, Ng et al., 2012, Ng and Stanton, 2013), are implicated in stemness and cell fate determination, even though their functional characterization is still incomplete. In this scenario, there is a lack of studies that directly investigate the potential role of T-UCE family members in molecular mechanisms orchestrating the balance between proliferation and differentiation of mouse embryonic stem cells (ESCs).
Here, we provide evidence of a functional role of T-UCEs in maintaining ESC self-renewal and get mechanistic insights into this process.
Results
Genome-wide Profiling Reveals T-UCEs Differentially Expressed during ESC Neural Differentiation
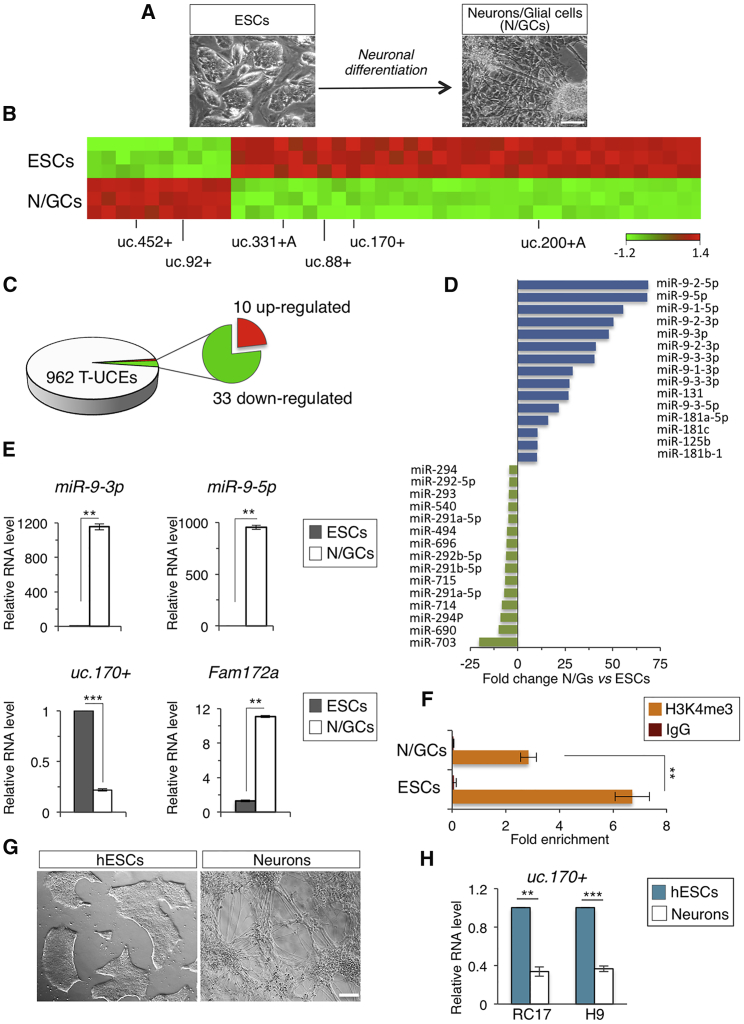
To investigate the role of UCEs in ESC self-renewal/differentiation, we first searched for T-UCEs differentially expressed in undifferentiated versus differentiated ESCs. To this end, we performed a genome-wide expression profile analysis of T-UCEs in ESC neural differentiation (Fico et al., 2008) by using a custom microarray designed to study the expression of both T-UCEs and miRNAs (Calin et al., 2007, Lujambio et al., 2010) and compared terminally differentiated cells (neurons and glia cells) with undifferentiated ESCs (Figure 1A). Interestingly, out of the 962 T-UCEs, only 43 were differentially expressed (p < 0.001), the large majority of which (77%) were downregulated (Figures 1B and 1C; Table S1), whereas ≈150 miRNAs resulted differentially expressed (p < 0.001) (Figure 1D; Table S1). Of note, miR-9 was the most upregulated miRNA on the array (Figure 1D), which was in line with the role of this miRNA family, both in the developing and adult vertebrate brain (Coolen et al., 2013). The microarray results were validated by qRT-PCR of randomly selected T-UCEs (uc.170+, uc.88+, uc.331+ A, uc.200+ A, uc.92+, and uc.452+) and miRNAs (miR-9-3p/5p, miR-714, miR-494, miR-181a, miR-411-5p, and miR-135b-5p) (Figures 1E, S1A, S1B, and S1C). Based on the T-UCE::miRNA functional interaction described in cancer cells (Calin et al., 2007, Olivieri et al., 2016), we hypothesized that such interaction may also occur in ESCs. Therefore, we focused our attention on the most upregulated miRNA family, and identified putative miR-9 target sites in the differentially expressed T-UCEs by using miRBase software. Specifically, we focused on the 33 T-UCEs that showed a negative correlation with miR-9, i.e., they were downregulated in ESC differentiation, and selected the uc.170+, which showed the lower minimum free energy of binding to both the mature forms of miR-9 (ΔG: −27 kcal/mol and −14.8 kcal/mol for miR-9-5p and miR-9-3p, respectively) (Rehmsmeier et al., 2004) (Figure S1D).
Figure 1.
Genome-wide Expression Profiling of T-UCEs and miRNAs in ESC Differentiation
(A) Schematic representation and representative photomicrographs of the ESC neural differentiation. Scale bar, 100 μm.
(B) Heatmap diagram of differentially expressed sense (+) and antisense (+A) T-UCEs, in ESC-derived neurons/glial cells (N/GCs) versus undifferentiated cells (ESCs) (p < 0.001; two-sample t test). Randomly selected T-UCEs are indicated among all the deregulated.
(C) Microarray-based pie chart representing global distribution of differentially expressed T-UCEs in N/GCs versus ESCs.
(D) Bar plot showing the differentially expressed miRNAs in N/GCs versus ESCs with the largest change in expression (p < 0.001; two-sample t test).
(E) qRT-PCR analysis of miR-9-5p/3p, uc.170+, and Fam172a in ESCs and N/GCs. Relative RNA level was normalized to either Gapdh or U6 for coding/non-coding genes, respectively. Data are mean ± SEM (n = 3 independent experiments); ∗∗p < 0.005, ∗∗∗p < 0.001.
(F) ChIP-qPCR of H3K4me3 at uc.170+ (ultraconserved region) locus in ESCs and N/GCs. Data are mean ± SEM (n = 3 independent experiments); ∗∗p < 0.01.
(G) Representative photomicrographs of the human ESC neural differentiation. Scale bar, 200 μm.
(H) qRT-PCR analysis of uc.170+ in human ESCs and human neurons (RC17 and H9 are two different human ESC lines). Relative RNA level was normalized to U6. Data are mean ± SEM (n = 3 independent experiments); ∗∗p < 0.005, ∗∗∗p < 0.001. See also Figure S1.
In mouse, uc.170+ is localized on chromosome 13 within intron 6 of the Fam172a host gene on the opposite strand (Figure S1E); uc.170+ and Fam172a transcripts showed opposite expression profiles. Indeed, while uc.170+ expression was downregulated in ESC neuronal differentiation, Fam172a strongly increased (Figure 1E). We also examined the chromatin status at the uc.170+ locus by performing chromatin immunoprecipitation analysis (ChIP)-qPCR. Consistent with the expression profile of uc.170+, we found a specific enrichment of H3K4me3 in undifferentiated ESCs that was reduced in differentiated cells (Figure 1F).
Bioinformatic analysis of the genomic region 5′ upstream of the uc.170+ predicted a promoter region located at about 1.5 kb upstream of the uc.170+ that was not described so far, based on the presence of a TATA box, a transcriptional initiator (Inr), and/or a transcription start site (Figure S1E). The predicted transcript containing the uc.170+ (T-UCstem1) was validated by northern blot analysis (Figure S2A). Furthermore, by using rapid amplification of cDNA ends-PCR analysis followed by sequencing, we identified the 5′ and 3′ extremities of the unspliced transcript and determined that its length is 1813 bp (Chr13: 78031716–78033528, strand –; Figure S2B). Analysis of secondary structure prediction using two different algorithms showed that T-UCstem1 is able to form a complex secondary structure with several highly stable stem-loops, thus explaining the size of the transcript under native conditions (Figures S2C and S2A).
Finally, we found that T-UCstem1 was expressed also in hESCs, and it was downregulated upon neuronal differentiation (Figures 1G and 1H), suggesting that it may similarly regulate human ESCs.
Direct and Functional Interaction of T-UCstem1 and miR-9
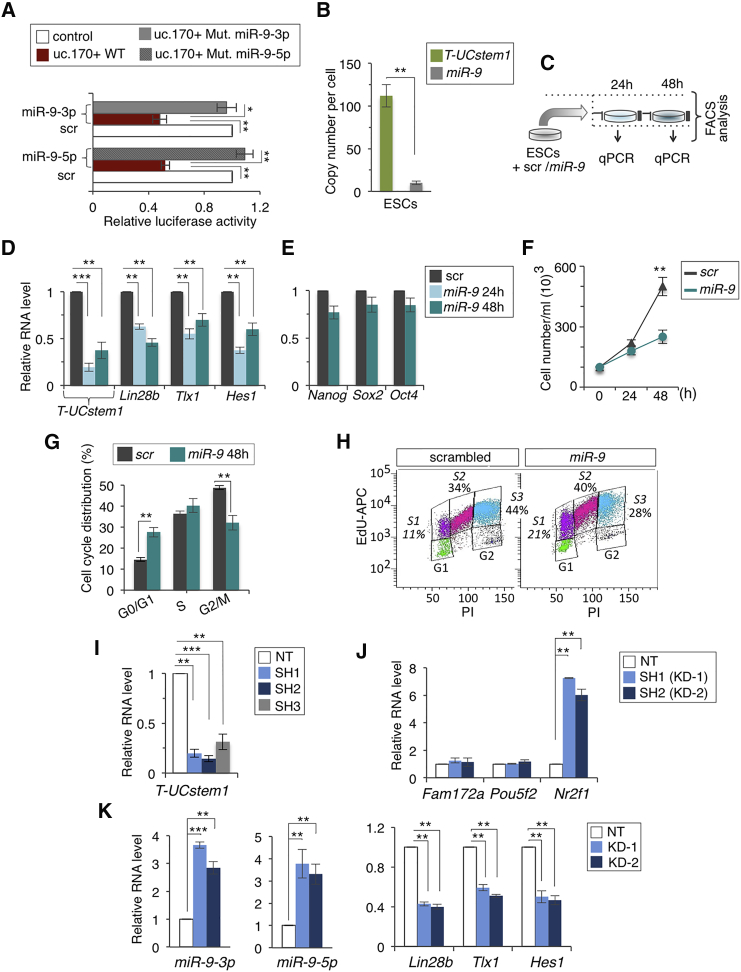
To assess whether there is a functional interaction between T-UCstem1 and miR-9, we performed a luciferase assay in 293FT cells. To this end, the uc.170+ was cloned in a luciferase reporter vector and co-transfected either with mimics of the mature forms of miR-9(3p/5p) or with a scrambled control (Figure 2A). The luciferase activity was significantly reduced in the presence of miR-9(3p/5p), which was consistent with the computationally predicted T-UCstem1::miR-9 interaction (Figure 2A). Furthermore, site directed mutagenesis of the miR-9-3p and miR-9-5p seed sequences confirmed the specificity of the T-UCstem1::miR-9 interaction (Figure 2A). To get further insights into the T-UCstem1::miR-9-3p/5p interaction, we extended the analysis to ESCs. First, we quantified the exact copy numbers of T-UCstem1 and miR-9 per cells in self-renewing ESCs by qRT-PCR. The mature miR-9 was present at a copy number of 9.6 ± 2.1 molecules/cell, which was significantly lower than T-UCstem1 (112 ± 13.8 copies/cell; Figure 2B), thus supporting the idea that T-UCstem1 may be able to function as a sponge for miR-9 (Wang et al., 2013). We therefore transfected ESCs with miR-9-3p/5p and assessed the expression of both T-UCstem1 and the miR-9 targets Lin28b, Tlx1, and Hes1 (Zhao et al., 2009, Zhong et al., 2010, Coolen et al., 2013) (Figure 2C). T-UCstem1 expression was strongly reduced already at 24 hr after transfection (Figure 2D), thus providing further evidence of a functional interaction between T-UCstem1 and miR-9 also in ESCs. Interestingly, while the expression of pluripotency genes (Nanog, Sox2, and Oct4) was comparable (Figure 2E), proliferation was reduced in miR-9 compared with scramble-transfected ESCs (Figure 2F). This observation prompted us to further investigate this phenotype. Consistently, cell-cycle distribution analysis of miR-9-transfected ESCs showed a significant G1-phase accumulation before S-phase progression compared with control, which was accompanied by a robust reduction in G2/M phase (Figure 2G). This was also confirmed by 5-ethynyl-2′-deoxyuridine/propidium iodide (PI) double staining, which showed an enlargement of the S1 subphase in miR-9-transfected ESCs (21% miR-9-transfected ESCs versus 11% Control) (Figure 2H).
Figure 2.
Functional Interaction of T-UCstem1 and miR-9 in ESCs
(A) Luciferase reporter assay with Renilla luciferase under control of uc.170+ sequence wild-type (WT) or mutant (Mut) co-transfected with miR-9-5p/3p or scrambled (scr) miRNA (100 nmol) in 293FT cells. The luciferase activity of Firefly was used as internal control. Data are mean ± SEM (n = 3 independent experiments); ∗p < 0.01, ∗∗p < 0.005.
(B) T-UCstem1 and miR-9 copy number per cell quantified with qRT-PCR in ESCs. Data are mean ± SEM (n = 3 independent experiments); ∗∗p < 0.005.
(C) Schematic representation of the experimental procedure.
(D and E) qRT-PCR analysis of (D) T-UCstem1, miR-9 targets (Lin28b, Tlx1, and Hes1), and (E) pluripotency-associated genes (Nanog, Sox2, and Oct4) in ESCs transfected with miR-9 or scr (100 nmol) for 24 and/or 48 hr. Data are mean ± SEM (n = 3 independent experiments); ∗∗p < 0.005, ∗∗∗p < 0.001.
(F) Automated cell counting of miR-9 and Control (scr) transfected ESCs at 24 and/or 48 hr. Data are mean ± SEM (n = 3 independent experiments); ∗∗p < 0.005.
(G and H) FACS-based cell-cycle distribution analysis of ESCs transfected with miR-9 or miRNA scr for 48 hr (G) after PI or (H) after double 5-ethynyl-2′-deoxyuridine (EdU)/propidium iodide (PI) staining (representative FACS plots of biological duplicates are shown). ∗∗p < 0.005.
(I and J) qRT-PCR analysis of (I) T-UCstem1, (J) host and neighbor genes (Fam172a, Pou5f2, and Nr2f1) in NT and independent T-UCstem1 KD ESC clones. Relative RNA level was normalized to either Gapdh or U6 for coding/non-coding genes, respectively. Data are mean ± SEM (n = 3 independent experiments); ∗∗p < 0.005, ∗∗∗p < 0.001.
(K) qRT-PCR analysis of miR-9-3p/5p and its target genes, Lin28b, Tlx1, and Hes1. Relative RNA level was normalized to either Gapdh or U6 for coding/non-coding genes, respectively. Data are mean ± SEM (n = 3 independent experiments); ∗∗p < 0.005, ∗∗∗p < 0.001. See also Figures S1 and S2.
To further analyze this phenotype and to evaluate the role of T-UCstem1 without altering the enhancer activity of genomic uc.170 (Pennacchio et al., 2006), we generated stable T-UCstem1 knockdown (KD) ESC clones using custom-designed short hairpin RNAs (shRNAs) targeting non-overlapping regions of the transcript (SH1-3; Figure S1E) that markedly reduced (≥70% of reduction) T-UCstem1 expression (Figures 2I and S2A). We first evaluated the effect of T-UCstem1 KD on the expression of the host gene Fam172a and two neighbor genes, Pou5f2 and Nr2f1. Fam172a was expressed at comparable levels in TUCstem1 KD (KD-1 and KD-2) and in Control ESCs, showing that T-UCstem1 does not regulate the host gene mRNA levels (Figure 2J). Of note, Nr2f1, but not Pou5f2, was significantly overexpressed in T-UCstem1 silenced ESCs (Figure 2J). Furthermore, we found a significant and consistent increase of both miR-9 mature forms upon T-UCstem1 KD (Figure 2K) and a consequential downregulation of the miR-9 targets Lin28b, Tlx1, and Hes1 (Figure 2K).
Altogether, these findings provide evidence of a functional interplay between T-UCstem1 and miR-9 in ESCs, and show that increased miR-9 cellular levels affect ESC proliferation.
T-UCstem1 Controls ESC Proliferation by Modulating miR-9 Intracellular Levels
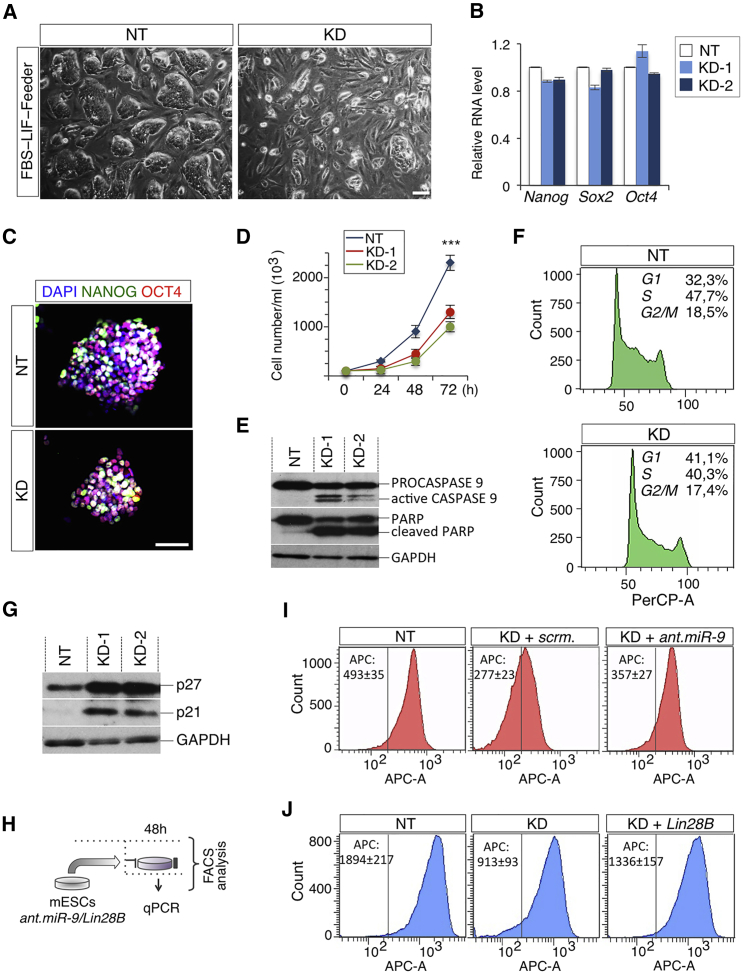
To further investigate the functional role of T-UCstem1 in ESCs, we analyzed the molecular and cellular features of T-UCstem1 KD ESCs. Under FBS/Lif/Feeders culture conditions, T-UCstem1 KD ESC colonies appeared flat, disorganized, and smaller in size compared with Control, which conversely showed the expected domed and tightly packed phenotype (Figure 3A). Expression levels of the pluripotency triad Oct4-Sox2-Nanog was comparable in KD and non-targeted (NT) ESCs (Figures 3B and 3C); however, T-UCstem1 KD ESCs showed reduced proliferation rate (Figure 3D), which was accompanied by reduced cell viability (Figure S3A) and induced apoptosis, as shown by CASPASE9 activation and PARP cleavage (Figure 3E). Furthermore, PI staining showed accumulation of T-UCstem1 KD ESCs in G1 phase, which eventually resulted in S-phase pauperization (Figure 3F). Accordingly, the expression of cell-cycle inhibitors p27 and p21 was strongly upregulated in T-UCstem1 KD cells compared with NT Control (Figure 3G). This mutant phenotype was further supported by carboxyfluorescein succinimidyl ester (CFSE) staining, which tracks cell division in living cells over the time (Ramírez et al., 2011) (Figures S3B–S3D). Interestingly, small interfering RNA-based knockdown of T-UCstem1 in hESCs (Figure S3F) resulted in decreased colony size and reduced cell proliferation rate (Figures S3G and S3H), suggesting a conserved role of T-UCstem1 in hESCs.
Figure 3.
T-UCstem1 Depletion Affects ESC Cell Proliferation and Induces Apoptosis
(A) Representative photomicrographs of FBS/LIF/Feeders non-targeted (NT) and T-UCstem1 KD (KD) ESCs. Scale bar, 100 μm.
(B) qRT-PCR of pluripotency genes (Nanog, Sox2, and Oct4) in Control and two independent KD ESC clones. Relative RNA level was normalized to Gapdh. Data are mean ± SEM (n = 3 independent experiments).
(C) Representative pictures of OCT4/NANOG double immunostaining of NT and KD ESCs. Nuclei were stained with DAPI. Scale bars, 75 μm.
(D) Time course analysis of automated cell counting of FBS/LIF/Feeders Control (NT) and KD ESCs. Data are mean ± SEM (n = 3 independent experiments); ∗∗∗p < 0.001.
(E) Western blot analysis of PROCASPASE/active CASPASE9 and PARP full-length form and cleaved fragment in Control (NT) and two independent T-UCstem1 KD ESC clones. GAPDH was used as loading controls.
(F) Cell-cycle distribution by PI incorporation in Control and KD ESCs. Representative FACS plots of biological triplicates are shown.
(G) Western blot analysis of cell-cycle inhibitors (p27and p21) in Control (NT) and two independent T-UCstem1 KD ESC clones. GAPDH was used as a loading control.
(H) Schematic representation of the experimental procedure.
(I and J) FACS-based analysis of cell proliferation quantification by EdU incorporation in NT and in KD ESCs upon antagomiR-9 5p/3p/scr (100 nmol) (I) or Lin28B-Flag/empty vector at 48 hr after transfection (J). Representative FACS plots of biological triplicates are shown. Data are mean ± SEM. See also Figure S3.
We then asked if uc.170+ overexpression was able to rescue the molecular and functional phenotype of T-UCstem1 KD ESCs. Interestingly, uc.170+ overexpression fully rescued mir-9 levels and expression of the target genes (Lin28b and Tlx1; Figures S3I and S3J), as well as T-UCstem1 KD colony size and proliferation rate (Figures S3K and S3L). Of note, mir-9 overexpression (Figures 2D–2I) and T-UCstem1 depletion (Figures 3D, 3F, and 3G) similarly affected ESC proliferation, thus suggesting that T-UCstem1 KD phenotype could be due to increased mir-9 levels. To test this hypothesis, T-UCstem1 KD ESCs were transfected with antagomiR-9 or scrambled control (Figure 3H). AntagomiR-9 reduced miR-9 levels and restored Lin28b and Tlx1 expression in T-UCstem1 KD ESC (Figures S4A and S4B), and rescued proliferation (Figures 3I and S4C). Furthermore, overexpression of the miR-9 target gene Lin28b (Figures S4D and S4E) was also able to fully rescue T-UCstem1 KD ESC proliferation (Figures 3H and 3J).
All together, these data demonstrate that a T-UCstem1/miR-9/Lin28b axis controls cell-cycle progression in ESCs.
T-UCstem1 Preserves ESC Self-Renewal Properties In Vitro and In Vivo
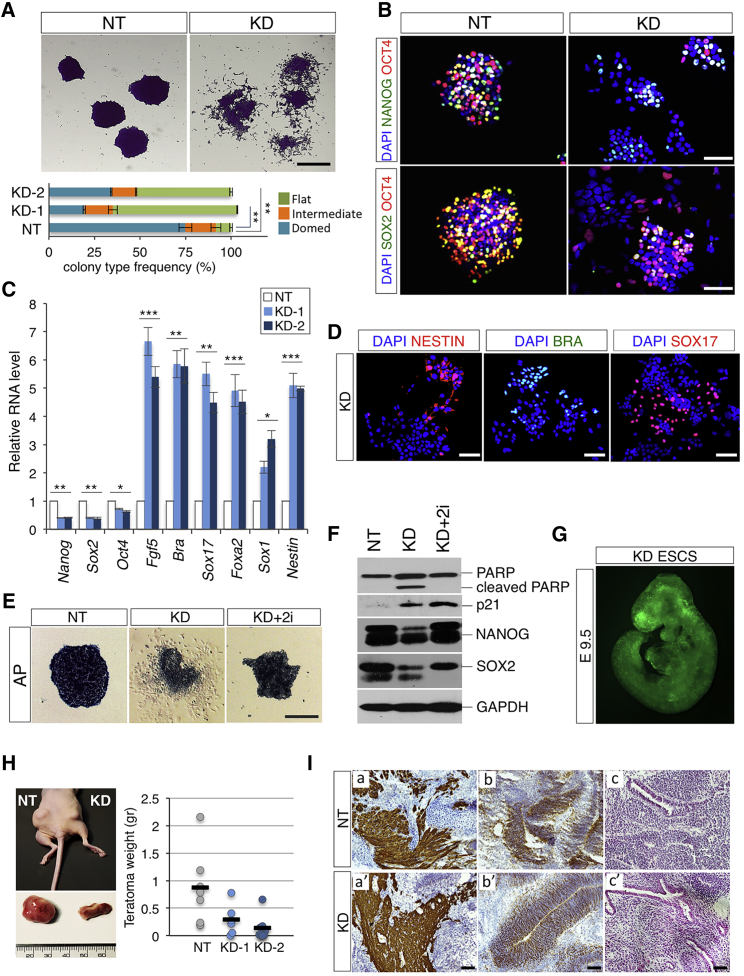
Despite their morphology and altered proliferation rate, FBS/Lif/Feeders T-UCstem1 KD ESCs retained a proper expression of pluripotency markers (Figures 3B and 3C). Nevertheless, when T-UCstem1 KD ESCs were plated at low density in colony-formation assay, and in the absence of feeders (Paling et al., 2004), colonies with a flat morphology massively increased at the expense of the classical domed colonies (Figure 4A), and this effect was exacerbated already after one passage in culture (Figures S4F and S4G). In line with these observations, expression of pluripotency factors was strongly reduced in T-UCstem1 KD ESC colonies in these culture conditions (FBS/Lif, low density) (Figures 4B, 4C, and S4H–S4J). Interestingly, this correlated with a significant increase in the expression of cell lineage commitment genes (i.e., Fgf5, Brachyury, Sox17, Foxa2, Sox1, and Nestin) at both RNA (Figure 4C) and protein (Figure 4D) levels.
Figure 4.
T-UCstem1 Sustains ESC Self-Renewal In Vitro and In Vivo
(A) Representative pictures of crystal violet-stained and colony-type frequency (∼100 colonies scored/condition) of colonies generated from Control (NT) and T-UCstem1 KD ESCs, at day 6 after plating. Data are mean ± SEM (n = 3 independent experiments); ∗∗p < 0.005. Scale bar, 100 μm.
(B) Representative immunofluorescence of NANOG/OCT4 and SOX2/OCT4 of Control (NT) and KD ESC colonies at day 6 after plating. Nuclei were stained with DAPI. Scale bars, 75 μm.
(C) qRT-PCR analysis of pluripotency (Nanog, Sox2, and Oct4), mesodermal (Fgf5 and Brachyury [Bra]), neural (Sox1 and Nestin), and endodermal-associated genes (Foxa2 and Sox17) in Control (NT) and two independent T-UCstem1 KD ESC clones. Relative RNA level was normalized to Gapdh expression. Data are mean ± SEM (n = 3 independent experiments); ∗p < 0.01, ∗∗p < 0.005, ∗∗∗p < 0.001.
(D) Representative pictures of NESTIN, BRA, and SOX17 immunostaining in KD ESC colonies. Nuclei were stained with DAPI. Scale bars, 75 μm.
(E) Representative pictures of alkaline phosphatase (AP)-stained colonies generated from Control (NT), KD, and KD + 2i (CHIR99021 + PD0325901) ESCs, at day 6 after plating. Scale bar, 100 μm.
(F) Western blot analysis of PARP full-length form and cleaved fragment, p21, NANOG, and SOX2 in Control (NT), KD, and KD + 2i (CHIR99021 + PD0325901) ESCs, at day 6 after plating. GAPDH was used as a loading control.
(G) Representative photomicrograph by Axio Zoom.V16 Zeiss microscopy (original magnification ×10) of chimeric embryos from EGFP-labelled T-UCstem1 KD ESCs injected into morula and dissected at embryonic day 9.5 (E9.5).
(H) Effect of T-UCstem1 depletion on teratoma formation. Representative picture of an immunodeficient mouse injected with Control (NT) and T-UCstem1KD ESCs, and the dissected ectopic tissues (left panel). Quantitative analysis of tissue weights (right panel). Data are mean ± SEM (seven mice/group).
(I) Immunohistochemistry analysis showing tissues deriving from ectoderm (nestin; a, a'), mesoderm (MF-20; b, b'). Scale bars, 75 μm., H&E staining displaying endoderm derivatives (glandular epithelial structures; c, c'). Scale bar, 150 μm. See also Figures S4 and S5.
We then asked whether this phenotype could be rescued in 2i (CHIR99021 + PD0325901) culture conditions (Guo et al., 2009). Intriguingly, while T-UCstem1 KD ESC self-renewal was fully rescued in 2i, as indicated by alkaline phosphatase staining (Figure 4E) and expression of pluripotency-associated genes (Figures S4K and 4F), the reduced proliferation rate (Figure S4L), as well as the expression of cell-cycle inhibitor p21 persisted (Figure 4F), thus suggesting that different mechanisms control T-UCstem1-dependent regulation of ESC proliferation and self-renewal.
In line with our in vitro findings that T-UCstem1 silencing affected ESC proliferation and self-renewal, but not pluripotency, T-UCstem1 KD ESCs generated teratomas significantly smaller in size compared with Control, but not different in histological composition (Figures 4H and 4I). Furthermore, EGFP-labeled T-UCstem1 KD ESCs efficiently contributed to chimeric embryos upon injection into morula (Figures 4G and S5A; eight out of nine embryos analyzed).
Altogether, these findings provide evidence of a crucial role of T-UCstem1 in preserving ESC self-renewal and proliferation both in vitro and in vivo, without affecting pluripotency.
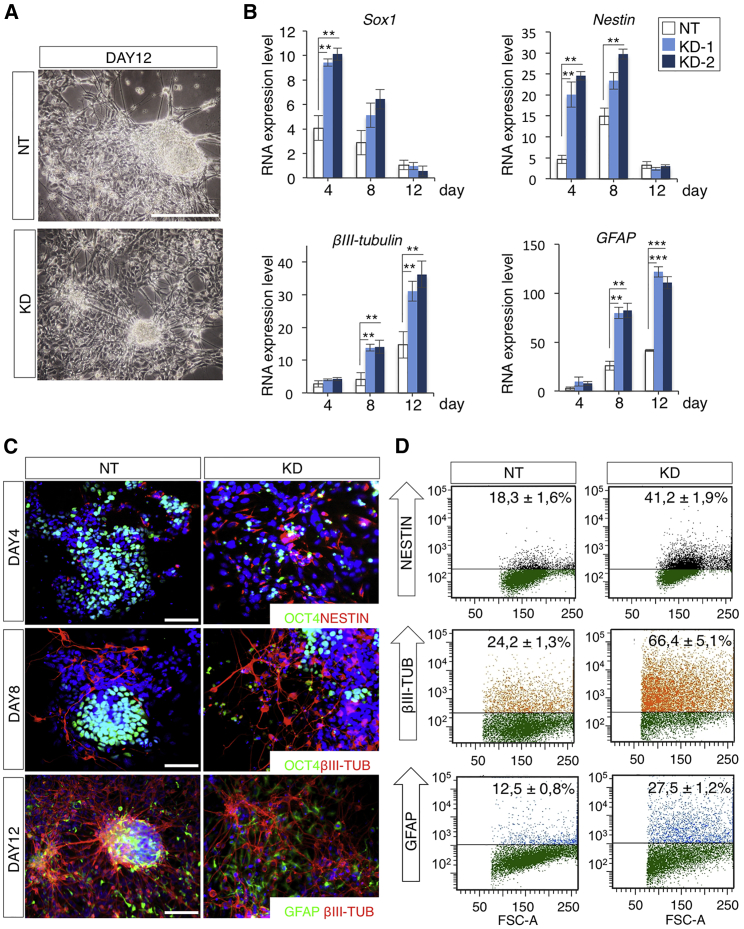
T-UCstem1 Silencing Accelerates and Enhances ESC Differentiation
To further assess the functional role of T-UCstem1 in ESCs, we evaluated the impact of T-UCstem1 KD on ESC differentiation. A time course analysis of ESC neural differentiation showed a significant difference in the differentiation kinetics of T-UCstem1 KD and Control ESCs. Specifically, neural and glial differentiation strongly increased in T-UCstem1 KD ESCs compared with Control (Figure 5A). Indeed, markers of neural precursors (Sox1 and Nestin), fully differentiated neurons (βIII-tubulin), and glial cells (glial fibrillary acidic protein) were all upregulated in T-UCstem1 KD ESCs already at earlier time points of differentiation (Figures 5B and 5C), suggesting that differentiation was accelerated in T-UCstem1 KD cells. Consistently, while Oct4 was downregulated in T-UCstem1 KD culture, its expression persisted in Control ESCs (Figure 5C). Finally, fluorescence-activated cell sorting (FACS) analysis showed increased neural and glial cells in T-UCstem1 KD culture, thus further supporting the idea that ESC differentiation was accelerated and was more efficient upon T-UCstem1 downregulation (Figure 5D). To further evaluate this phenotype, we evaluated the effect of T-UCstem1 KD on ESC cardiac differentiation. Time course expression analysis showed a transient upregulation (day 6) of the pan-mesodermal marker Brachyury in T-UCstem1 KD cultures, which progressively decreased (Figure S5B), and increased expression of both early (Nkx2.5) and late (α-myosin heavy chain [αMHC]) cardiac markers throughout differentiation (days 6–10; Figure S5B), suggesting that cardiac specification and differentiation was enhanced and accelerated. Accordingly, FACS analysis showed that Brachyury-positive cells were strongly reduced (10.8% ± 2.2% KD versus 21.1% ± 3.7% Control) at day 8, while αMHC (MF20)-positive cells almost doubled at day 10 (27% ± 3.7% KD versus 12.3% ± 3.1% Control) in T-UCstem1 KD cell culture compared with Control (Figure S5C), which is in line with accelerated differentiation.
Figure 5.
T-UCstem1 Silencing Promotes ESC Neural Differentiation
(A) Representative photomicrographs of Control (NT) and T-UCstem1 KD (KD) ESCs differentiated in neurons. Scale bar, 100 μm.
(B) Time course expression profiles of neural (Sox1, Nestin, and βIII-tubulin) and glial (GFAP) markers in Control (NT) and two independent T-UCstem1 KD clones. RNA expression level was normalized to Gapdh expression. Data are mean ± SEM (n = 3 independent experiments); ∗∗p < 0.005, ∗∗∗p < 0.001.
(C) Representative pictures of OCT4/NESTIN, OCT4/βIII-TUBULIN, and GFAP/βIII-TUBULIN double immunostaining in Control (NT) and KD ESC neural differentiation at the indicated time points. Nuclei were stained with DAPI. Scale bars, 75 μm.
(D) FACS-based quantification of NESTIN (day 4), βIII-TUBULIN (day 12), and GFAP (day 12) positive cells in Control (NT) and KD ESC neural differentiation. Data are mean ± SEM (n = 3 independent experiments). See also Figure S5.
All together, these results indicate that T-UCstem1 is required to regulate ESC differentiation.
T-UCstem1 Preserves the Transcriptional Dynamics of ESCs by Stabilizing PRC2 Complex
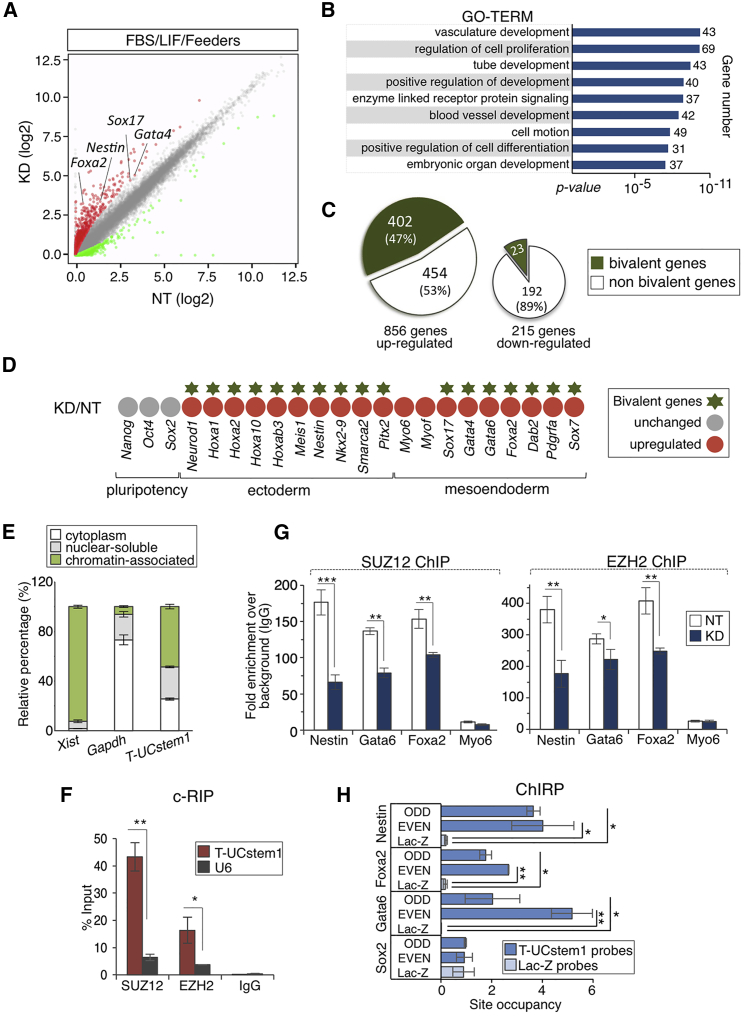
To get mechanistic insight into the role of T-UCstem1 in ESC self-renewal and differentiation, we compared RNA sequencing (RNA-seq) transcriptome profiling of FBS/Lif/Feeders T-UCstem1 KD and Control ESCs, and identified more than 1,000 deregulated genes (fold change ≥2; p < 0.05; Figure 6A; Table S2). Remarkably, gene ontology (GO) analysis showed striking enrichment in genes involved in regulation of cell proliferation, positive regulation of development, and positive regulation of cell differentiation (Figure 6B), which are in line with the functional role of T-UCstem1 (Figures 3, 4, and 5). The large majority of the deregulated genes were indeed upregulated (∼70%) in T-UCstem1 KD ESCs compared with Control and, remarkably, ∼50% were associated with bivalent chromatin domains (Figures 6C and 6D). Bivalent domains are characterized by the co-presence of the activating H3K4me3 and the repressive H3K27me3 histone marks, and are associated with silencing of developmental genes that would activate cell differentiation, while keeping these genes poised and ready to be induced (Bernstein et al., 2006). We thus first analyzed the status of H3K4me3 and H3K27me3 at the bivalent domains of Nestin, Foxa2, and Gata6 genes in T-UCstem1 KD and Control ESCs by ChIP analysis. In line with the expression data (Figures 6A and S6A), the H3K4me3/H3K27me3 ratio (De Gobbi et al., 2011) significantly increased in T-UCstem1 KD ESCs at the promoter of these representative genes of the three germ layers (Figure S6C). Of note, we found derepression of Nestin, Foxa2, and Gata6 genes also in T-UCstem1 KD hESCs (Figure S6B), suggesting that T-UCstem1 function may be conserved in humans. Given the very well-documented role of polycomb repressive complex 2 (PRC2) in maintaining the bivalent domains in ESCs and the involvement of several lncRNAs in this mechanism (Margueron and Reinberg, 2011), we questioned whether T-UCstem1 could directly interact with the PRC2, and eventually regulate bivalent gene expression. To address this issue, we first analyzed the subcellular localization of T-UCstem1 by RNA subcellular fractionation. According to its interaction with miR-9 in ESCs, T-UCstem1 was detected in the cytoplasmic fraction (Figure 6E). However, T-UCstem1 was also present both in the nuclear soluble and in the chromatin-associated fractions (Figure 6E), supporting the idea of a potential role of this lncRNA also in the nucleus. We then assessed whether T-UCstem1 might physically interact with components of PRC2, and carried out a crosslinked RNA immunoprecipitation using SUZ12 and EZH2 specific antibodies. We found that T-UCstem1 was able to specifically bind both SUZ12 and EZH2 proteins of PRC2 (Figure 6F). Native RIP carried out on cell lysate revealed that SUZ12 shows presumably higher affinity in this interaction than EZH2 (Figure S6D). These findings led us to hypothesize a role of T-UCstem1 in stabilizing the PRC2 complex on its target genes. To address this question, we analyzed SUZ12 and EZH2 binding on Nestin, Foxa2, and Gata6 promoters by ChIP experiments and found that T-UCstem1 downregulation significantly reduced PRC2 occupancy on all the target sites analyzed (Figures 6G and S6E). Of note, SUZ12 and EZH2 protein levels were comparable in T-UCstem1 KD and Control ESCs (Figure S6F). To prove that T-UCstem1 interacts with Nestin, Gata6, and Foxa2 genes showing changes in H3K4me3/H3K27me3 ratio, we performed chromatin isolation by RNA purification (ChIRP). We found a significant enrichment of T-UCstem1 at the target sites of PRC2 occupancy, while this enrichment was not detected at Sox2 promoter (Figure 6H).
Figure 6.
T-UCstem1 Depletion Remodels the Epigenomic Signature of ESCs
(A) Scatterplot of RNA-seq data shows differentially expressed genes in T-UCstem1 KD (KD) versus Control (NT) ESCs. The gray circles delineate the boundaries of less than 2-fold difference in gene expression levels. Genes showing a significant (p < 0.05) higher or lower expression level in KD versus NT ESCs are indicated as red and green circles, respectively. Three independent KD ESC clones were analyzed and the average values were reported in the plot.
(B) Gene ontology (GO) (http://david.abcc.ncifcrf.gov; setting as background the genes expressed in ESCs) of protein coding genes deregulated in KD versus Control (NT) ESCs.
(C) Chart showing a significant overlap between bivalent domain-associated genes and upregulated genes in KD versus Control (Hypergeometric test; p <10−16).
(D) Heatmap of upregulated developmental genes in KD cells versus Control (NT).
(E) Total RNA from ESCs was separated into cytoplasmic, nuclear-soluble, and chromatin-bound fractions. The relative abundance of T-UCstem1 in the different fractions was measured by qRT-PCR. Gapdh and Xist were analyzed as control. Data are mean ± SEM (n = 3 independent experiments).
(F) Crosslinked RNA immunoprecipitation (c-RIP) of T-UCstem1 in ESCs, using antibodies against SUZ12, EZH2, or immunoglobulin (IgG) as control. U6 was analyzed as negative control. Data are mean ± SEM (n = 3 independent experiments); ∗p < 0.05, ∗∗p < 0.005.
(G) ChIP-qPCR of SUZ12 and EZH2 binding at selected genes in NT and KD cells. Myo6 promoter has been reported as control. Data are mean ± SEM (n = 3 independent experiments); ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005.
(H) ChIRP-qPCR for T-UCstem1 in ESCs. Enrichments of Nestin, Gata6, Foxa2, and Sox2 promoter regions were quantified in the chromatin fraction precipitated using biotinylated DNA probes complementary to T-UCstem1 and LacZ as negative control. Sox2 promoter has been reported as negative control. Data are mean ± SEM (n = 3 independent experiments); ∗p < 0.05, ∗∗p < 0.005. See also Figure S6.
Overall, these data point to a key role of T-UCstem1 in maintaining ESC transcriptional identity by protecting the epigenetic status of key developmental regulatory genes, stabilizing PRC2 on their bivalent domains.
Discussion
This work provides evidence of a functional role of T-UCEs in regulating the finely tuned balance between pluripotency and differentiation in mouse ESCs. So far, the T-UCEs have been mostly linked to cancer, whereas their physiological role is still poorly understood. Based on the hypothesis that the T-UCE::miR interaction described in cancer cells can similarly occur in ESCs, we focused on uc.170+::mir9 since (1) uc.170+ carries the seed sequence for miR-9, and (2) uc.170+ and mir-9 expression inversely correlate in ESC neural differentiation. Here, we demonstrate that T-UCstem1 and miR-9 functionally interact and show that T-UCstem1::miR-9 interplay regulates ESC proliferation. According to our findings, recent data showed that miR-9 inhibits neural precursor cell and ESC proliferation by targeting Tlx1 (Qu et al., 2010, Zhao et al., 2009) and Lin28b (Xu et al., 2009, Zhong et al., 2010), respectively. Our results that Lin28b overexpression rescues the proliferation defects of T-UCstem1 KD ESCs support the conclusion that a T-UCstem1/miR-9/Tlx1-Lin28b axis controls cell-cycle progression in ESCs.
Besides its pro-proliferative activity, T-UCstem1 also acts as a brake for ESC differentiation. Indeed, genome-wide and targeted analysis indicate that, upon T-UCstem1 silencing, FBS/Lif/Feeders ESCs retain expression of key pluripotency factors but concomitantly induce the expression of a large set of developmental genes of the three germ layers (ectoderm, mesoderm, and endoderm). In line with this peculiar molecular signature, FBS/Lif/Feeders T-UCstem1 KD ESCs keep pluripotency features and are able to differentiate in vitro and contribute to chimeric embryos in vivo. On the other hand, in less-permissive culture conditions (low density without feeders) FBS/Lif T-UCstem1 KD ESCs rapidly exit pluripotency and undergo differentiation, pointing to a key role of T-UCstem1 in preserving ESC self-renewal rather than pluripotency. Of note, since T-UCstem1 expression is not fully abrogated in T-UCstem1 KD ESCs, we cannot rule out the possibility that a complete loss of T-UCstem1 expression could give a more dramatic effect.
The observation that T-UCstem1 KD ESC self-renewal, but not the proliferation defects, were rescued in 2i culture conditions, suggests different mechanisms of action of T-UCstem1-dependent control of ESC proliferation and self-renewal. Indeed, a large number (∼50%) of all the developmental regulatory genes that are upregulated in T-UCstem1 KD ESCs are bivalent domains-associated genes, which are characterized by a distinctive histone modification signature that combines the activating H3K4me3 and the repressive H3K27me3 marks. These bivalent domains are considered to poise expression of developmental genes, allowing timely activation, while maintaining repression in the absence of differentiation signals (Voigt et al., 2013). Increasing evidence indicates that PRC2 plays a crucial role in maintaining the bivalent domains in ESCs (Aranda et al., 2015) by ensuring a proper and robust differentiation. Withdrawal of PRC2 activity from ESCs results in global gene derepression of bivalent-associated genes (Azuara et al., 2006, Boyer et al., 2006, Lee et al., 2006) and spontaneous differentiation (Boyer et al., 2006, Endoh et al., 2008). PRC2 interacts with many lncRNAs in ESCs (e.g., HOTAIR, Malat1, and Gtl2), and these facilitate its recruitment to chromatin (Zhao et al., 2010). Furthermore, recent findings indicate that non-coding RNAs recruit PRC2 complex to chromatin either in cis or in trans, thereby causing changes in chromatin composition (Holoch and Moazed, 2015). Our findings indicate that T-UCstem1 is a new player in this complex scenario and provide evidence of a direct involvement of T-UCstem1 in switching the balance of these histone modifications in ESCs. Indeed, we show that T-UCstem1 directly interacts both with PRC2 complex and the bivalent domain-associated genes Nestin, Gata6, and Foxa2, and that this interaction may stabilize/guide PRC2 activity in determining the typical histone modifications at these bivalent domains. Thus, we propose a model wherein PRC2 is displaced in the absence of T-UCstem1, and this results in increased H3K4me3/H3K27me3 ratio on bivalent promoters of differentiation genes, which eventually induces their expression. Notably, we demonstrate that, besides the regulation on bivalent genes localized on different chromosomes (i.e., Nestin, Gata6, and Foxa2), T-UCstem1 also controls the expression of the neighbor bivalent gene Nr2f1 (Laursen et al., 2013), thus suggesting that the T-UCstem1 tethers PRC2 both in cis and in trans. Of note, considering the low abundance of T-UCstem1 and the ∼400 bivalent genes that are deregulated in the T-UCstem1 KD ESCs, we speculate that they might represent both direct and indirect targets of T-UCstem1.
In summary, we provide unprecedented evidence that a UCE-containing lncRNA is a key regulator of ESCs and get mechanistic insights into the mode of action. Indeed, we propose that T-UCstem1 exerts a dual function in ESCs; specifically, it controls ESC proliferation by regulating miR-9/Lin28b cellular levels in the cytoplasm, and maintains ESC transcriptional dynamics and self-renewal, at least in part through PRC2 stabilization in the nucleus. Overall, our study points to a functional role of T-UCEs in ESC biology, and pave the way for a better understanding of the complex molecular machinery controlling ESC pluripotency and lineage specification.
Experimental Procedures
T-UCstem1 KD Mouse ESC Generation
Animal experiments were done in accordance with the law on animal experimentation (article 7; D.L. 116/92) under the Animal Protocol approved by the Italian Ministry of Health.
Stable T-UCstem1 KD ESC clones (designated as KD-1 and KD-2) were generated by using custom-designed shRNAs targeting non-overlapping regions of the transcript (different shRNAs were used in order to limit the off-target effects). For this purpose, we used the BLOCK-iT Inducible H1 RNAi Entry Vector Kit (Invitrogen, cat. no. K4920-00 and K4925-00), a Gateway-adapted entry vector for regulated expression of shRNA in mammalian cells, following the manufacturer's instructions. In brief, we designed custom shRNA taking advantage of Invitrogen's RNAi designer and we cloned them into pENTR/H1/TO vector. Next, plasmid constructs that direct shRNA expression were introduced into ESCs (TBV2 (129/SvP)) by electroporation and drug selection (Zeocin). Then, we isolated ESC constitutively expressing shRNAs clones, we propagated them, and the degree of knockdown was assessed by qRT-PCR. Selected ESC clones were used for this study.
In particular, to silence T-UCstem1, we used three different shRNAs reported in Supplemental Information. Moreover, we generated NT ESCs, to use as Control, by transfecting the cells with shRNA targeting LacZ gene and supplied in the kit.
Whole-Genome Expression Analysis
RNA was extracted from NT and KD ESCs using TRIzol reagent (Thermo Fisher Scientific). To identify the expression profile of T-UCEs and miRNAs, the total RNA was hybridized to a custom ncRNA microarray (OSU-CCC 4.0, Ohio State University Comprehensive Cancer Center), which included sense and antisense probes, one corresponding to the sense genomic sequence (+) and the other to the complementary sequence (+A) for all 481 human ultraconserved sequences reported by Bejerano et al. (in total there are probes for 962 possible T-UCEs). The GEO describes the OSU-CCC 4.0 platform under accession number GPL14184. T-UCEs were retained when present in at least 20% of samples and when at least 20% of them had a fold change of more than 1.0 from the gene median. Absent calls were thresholded prior to normalization and statistical analysis. Normalization was performed using quantiles (Table S1).
RNA-seq was performed at the Institute for Applied Genomics using the Illumina HiSeq 2500 platform (http://www.igatechnology.com/). The data were analyzed by aligning the reads to a reference genome (Mus musculus mm9) using Tophat (Trapnell et al., 2009), which is also able to align sequences that span exon-exon junctions. Then, we performed a differential expression analysis using Cufflinks (Trapnell et al., 2010), which is able to calculate transcript abundance and abundance of different gene isoforms. Finally, this analysis showed the most deregulated transcripts when comparing them with the different groups (Table S2). Actually, in order to avoid clonal effects, we analyzed three independent KD ESC clones generated as described above and the average values were reported. A custom R-script was used to create the plot in Figure 6A.
T-UCstem1 ChIRP
ChIRP assay was performed using the Magna ChIRPTM RNA Interactome Kits (Millipore) according to the manufacturer's instructions. Biotin-labeled antisense T-UCstem1 DNA probe sequences are reported in Supplemental Information. Isolated DNA was used for qPCR analyses to estimate the site occupancy of T-UCstem1 on the Nestin, Gata6, and Foxa2 promoter. The site occupancy was calculated as ratio between the percent input of each specific target qPCR and the average of the percent input of the internal negative control (Sox2). Data are shown as the site occupancy mean.
Author Contributions
A. Fico and G.M. conceived and designed the study. M.R.M. contributed to the concept and design of the epigenetic study. A. Fiorenzano, E.P., M. Gagliardi, M.R.M., A. Fico, and G.M. planned and designed the experiments and analyzed the data. A. Fiorenzano, E.P., M. Gagliardi, S.T., M.P., G.A., and A. Fico performed the experiments. M. Gagliardi, M. Galasso, G.M.T., and C.T. performed bioinformatics analysis. E.J.P., A.C., and M.R.M. gave conceptual advice and edited the manuscript. G.M and A. Fico wrote the manuscript. All authors reviewed and approved the manuscript.
Acknowledgments
We thank the Mouse Modeling, Integrated Microscopy, and FACS Facilities of IGB-CNR, Naples. Dr. Laura Pisapia is acknowledged for flow cytometry analyses. We are grateful to Prof. Chris Ponting and George A. Calin for insightful scientific discussion. We thank Dr. Monica Autiero for helpful discussion and careful reading of the manuscript. We are indebted to Prof. Malin Parmar and her lab for support with human ESC experiments. Dr Silvia Parisi is acknowledged for providing Lin28 cDNA plasmid and Dr Valeria Tarallo for helpful discussion and experimental support in setting the northern blot conditions. This work is supported by Epigenomics Flagship Project (EPIGEN) MIUR-CNR; TRANSCAN-2 Project BeFIT; the Italian Ministry of Education-University-Research (grant CTN01_00177); AIRC (IG 20736); and Telethon grant GP15209. Emilia Pascale was supported by a PhD fellowship financed by IGB, CNR.
Published: February 15, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, and four tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.01.014.
Contributor Information
Gabriella Minchiotti, Email: gabriella.minchiotti@igb.cnr.it.
Annalisa Fico, Email: annalisa.fico@igb.cnr.it.
Accession Numbers
The accession numbers for the ncRNA (T-UCE and miRNAs) expression profile and RNA-seq data are ArrayExpress: E-MTAB-6391 and GEO: GSE108662, respectively.
Supplemental Information
References
- Ahituv N., Zhu Y., Visel A., Holt A., Afzal V., Pennacchio L.A., Rubin E.M. Deletion of ultraconserved elements yields viable mice. PLoS Biol. 2007;5:e234. doi: 10.1371/journal.pbio.0050234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda S., Mas G., Di Croce L. Regulation of gene transcription by Polycomb proteins. Sci. Adv. 2015;1:e1500737. doi: 10.1126/sciadv.1500737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V., Perry P., Sauer S., Spivakov M., Jørgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Bejerano G., Pheasant M., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Calin G.A., Liu C.G., Ferracin M., Hyslop T., Spizzo R., Sevignani C., Fabbri M., Cimmino A., Lee E.J., Wojcik S.E. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- Chalei V., Sansom S.N., Kong L., Lee S., Montiel J.F., Vance K.W., Ponting C.P. The long non-coding RNA Dali is an epigenetic regulator of neural differentiation. Elife. 2014;3:e04530. doi: 10.7554/eLife.04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolen M., Katz S., Bally-Cuif L. miR-9: a versatile regulator of neurogenesis. Front. Cell. Neurosci. 2013;7:220. doi: 10.3389/fncel.2013.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gobbi M., Garrick D., Lynch M., Vernimmen D., Hughes J.R., Goardon N., Luc S., Lower K.M., Sloane-Stanley J.A., Pina C. Generation of bivalent chromatin domains during cell fate decisions. Epigenetics Chromatin. 2011;4:9. doi: 10.1186/1756-8935-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermitzakis E.T., Reymond A., Antonarakis S.E. Conserved non-genic sequences - an unexpected feature of mammalian genomes. Nat. Rev. Genet. 2005;6:151–157. doi: 10.1038/nrg1527. [DOI] [PubMed] [Google Scholar]
- Dinger M.E., Amaral P.P., Mercer T.R., Pang K.C., Bruce S.J., Gardiner B.B., Askarian-Amiri M.E., Ru K., Soldà G., Simons C. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh M., Endo T.A., Endoh T., Fujimura Y., Ohara O., Toyoda T., Otte A.P., Okano M., Brockdorff N., Vidal M. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development. 2008;135:1513–1524. doi: 10.1242/dev.014340. [DOI] [PubMed] [Google Scholar]
- Fabbri M., Garzon R., Andreeff M., Kantarjian H.M., Garcia-Manero G., Calin G.A. MicroRNAs and noncoding RNAs in hematological malignancies: molecular, clinical and therapeutic implications. Leukemia. 2008;22:1095–1105. doi: 10.1038/leu.2008.30. [DOI] [PubMed] [Google Scholar]
- Fassan M., Dall'Olmo L., Galasso M., Braconi C., Pizzi M., Realdon S., Volinia S., Valeri N., Gasparini P., Baffa R. Transcribed ultraconserved noncoding RNAs (T-UCR) are involved in Barrett's esophagus carcinogenesis. Oncotarget. 2014;5:7162–7171. doi: 10.18632/oncotarget.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Bi C., Clark B.S., Mady R., Shah P., Kohtz J.D. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fico A., Manganelli G., Simeone M., Guido S., Minchiotti G., Filosa S. High-throughput screening-compatible single-step protocol to differentiate embryonic stem cells in neurons. Stem Cells Dev. 2008;17:573–584. doi: 10.1089/scd.2007.0130. [DOI] [PubMed] [Google Scholar]
- Galasso M., Dama P., Previati M., Sandhu S., Palatini J., Coppola V., Warner S., Sana M.E., Zanella R., Abujarour R. A large scale expression study associates uc.283-plus lncRNA with pluripotent stem cells and human glioma. Genome Med. 2014;6:76. doi: 10.1186/s13073-014-0076-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan D., Zhang W., Zhang W., Liu G.H., Belmonte J.C. Switching cell fate, ncRNAs coming to play. Cell Death Dis. 2013;4:e464. doi: 10.1038/cddis.2012.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., Yang J., Nichols J., Hall J.S., Eyres I., Mansfield W., Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holoch D., Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 2015;16:71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzman S., Kern A.D., Bejerano G., Fewell G., Fulton L., Wilson R.K., Salama S.R., Haussler D. Human genome ultraconserved elements are ultraselected. Science. 2007;317:915. doi: 10.1126/science.1142430. [DOI] [PubMed] [Google Scholar]
- Laursen K.B., Mongan N.P., Zhuang Y., Ng M.M., Benoit Y.D., Gudas L.J. Polycomb recruitment attenuates retinoic acid-induced transcription of the bivalent NR2F1 gene. Nucleic Acids Res. 2013;41:6430–6443. doi: 10.1093/nar/gkt367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.I., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liz J., Portela A., Soler M., Gomez A., Ling H., Michlewski G., Calin G.A., Guil S., Esteller M. Regulation of pri-miRNA processing by a long noncoding RNA transcribed from an ultraconserved region. Mol. Cell. 2014;55:138–147. doi: 10.1016/j.molcel.2014.05.005. [DOI] [PubMed] [Google Scholar]
- Loewer S., Cabili M.N., Guttman M., Loh Y.H., Thomas K., Park I.H., Garber M., Curran M., Onder T., Agarwal S. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat. Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A., Portela A., Liz J., Melo S.A., Rossi S., Spizzo R., Croce C.M., Calin G.A., Esteller M. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene. 2010;29:6390–6401. doi: 10.1038/onc.2010.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R., Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick J.S., Makunin I.V. Small regulatory RNAs in mammals. Hum. Mol. Genet. 2005;14 Spec No 1:R121–R132. doi: 10.1093/hmg/ddi101. [DOI] [PubMed] [Google Scholar]
- Ng S.Y., Johnson R., Stanton L.W. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31:522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S.Y., Stanton L.W. Long non-coding RNAs in stem cell pluripotency. Wiley Interdiscip. Rev. RNA. 2013;4:121–128. doi: 10.1002/wrna.1146. [DOI] [PubMed] [Google Scholar]
- Ni J.Z., Grate L., Donohue J.P., Preston C., Nobida N., O'Brien G., Shiue L., Clark T.A., Blume J.E., Ares M., Jr. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega M.A., Zhu Y., Plajzer-Frick I., Afzal V., Rubin E.M. Megabase deletions of gene deserts result in viable mice. Nature. 2004;431:988–993. doi: 10.1038/nature03022. [DOI] [PubMed] [Google Scholar]
- Olivieri M., Ferro M., Terreri S., Durso M., Romanelli A., Avitabile C., De Cobelli O., Messere A., Bruzzese D., Vannini I. Long non-coding RNA containing ultraconserved genomic region 8 promotes bladder cancer tumorigenesis. Oncotarget. 2016;7:20636–20654. doi: 10.18632/oncotarget.7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paling N.R., Wheadon H., Bone H.K., Welham M.J. Regulation of embryonic stem cell self-renewal by phosphoinositide 3-kinase-dependent signaling. J. Biol. Chem. 2004;279:48063–48070. doi: 10.1074/jbc.M406467200. [DOI] [PubMed] [Google Scholar]
- Paparidis Z., Abbasi A.A., Malik S., Goode D.K., Callaway H., Elgar G., deGraaff E., Lopez-Rios J., Zeller R., Grzeschik K.H. Ultraconserved non-coding sequence element controls a subset of spatiotemporal GLI3 expression. Dev. Growth Differ. 2007;49:543–553. doi: 10.1111/j.1440-169X.2007.00954.x. [DOI] [PubMed] [Google Scholar]
- Pennacchio L.A., Ahituv N., Moses A.M., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Qu Q., Sun G., Li W., Yang S., Ye P., Zhao C., Yu R.T., Gage F.H., Evans R.M., Shi Y. Orphan nuclear receptor TLX activates Wnt/beta-catenin signalling to stimulate neural stem cell proliferation and self-renewal. Nat. Cell Biol. 2010;12:31–40. doi: 10.1038/ncb2001. sup pp 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez M.Á., Pericuesta E., Yáñez-Mó M., Palasz A., Gutiérrez-Adán A. Effect of long-term culture of mouse embryonic stem cells under low oxygen concentration as well as on glycosaminoglycan hyaluronan on cell proliferation and differentiation. Cell Prolif. 2011;44:75–85. doi: 10.1111/j.1365-2184.2010.00732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt P., Tee W.W., Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Xu Z., Jiang J., Xu C., Kang J., Xiao L., Wu M., Xiong J., Guo X., Liu H. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev. Cell. 2013;25:69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Xu B., Zhang K., Huang Y. Lin28 modulates cell growth and associates with a subset of cell cycle regulator mRNAs in mouse embryonic stem cells. RNA. 2009;15:357–361. doi: 10.1261/rna.1368009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Sun G., Li S., Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat. Struct. Mol. Biol. 2009;16:365–371. doi: 10.1038/nsmb.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Ohsumi T.K., Kung J.T., Ogawa Y., Grau D.J., Sarma K., Song J.J., Kingston R.E., Borowsky M., Lee J.T. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Li N., Liang S., Huang Q., Coukos G., Zhang L. Identification of microRNAs regulating reprogramming factor LIN28 in embryonic stem cells and cancer cells. J. Biol. Chem. 2010;285:41961–41971. doi: 10.1074/jbc.M110.169607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.