Summary
Hypertrophic cardiomyopathy (HCM) is the most common cause of sudden cardiac death in young individuals. A potential role of mtDNA mutations in HCM is known. However, the underlying molecular mechanisms linking mtDNA mutations to HCM remain poorly understood due to lack of cell and animal models. Here, we generated induced pluripotent stem cell-derived cardiomyocytes (HCM-iPSC-CMs) from human patients in a maternally inherited HCM family who carry the m.2336T>C mutation in the mitochondrial 16S rRNA gene (MT-RNR2). The results showed that the m.2336T>C mutation resulted in mitochondrial dysfunctions and ultrastructure defects by decreasing the stability of 16S rRNA, which led to reduced levels of mitochondrial proteins. The ATP/ADP ratio and mitochondrial membrane potential were also reduced, thereby elevating the intracellular Ca2+ concentration, which was associated with numerous HCM-specific electrophysiological abnormalities. Our findings therefore provide an innovative insight into the pathogenesis of maternally inherited HCM.
Keywords: mitochondrion, hypertrophic cardiomyopathy, induced pluripotent stem cells, MT-RNR2, maternal inheritance
Highlights
-
•
Generation of HCM-specific iPSC-CMs carrying the m.2336T>C mutation in MT-RNR2
-
•
m.2336T>C mutation results in mitochondrial dysfunctions
-
•
Mitochondrial dysfunctions lead to increased [Ca2+]i and decreased ICaL
-
•
Abnormal Ca2+ homeostasis is associated with HCM-specific abnormalities
In this article, Yan Q, Liu Z, Huang W, and colleagues show that patient-specific iPSCs as well as their derived cardiomyocytes carrying the m.2336T>C mutation in MT-RNR2 were generated to understand the pathogenic mechanism of maternally inherited HCM. MT-RNR2 mutation resulted in mitochondrial dysfunctions and ultrastructure defects, which induced abnormal Ca2+ homeostasis, then HCM-specific cellular and electrophysiological characteristics in iPSC-CMs.
Introduction
Hypertrophic cardiomyopathy (HCM) is a primary myocardial disease characterized by left ventricle and asymmetric septal hypertrophy. It is the most common cause of sudden cardiac death in young individuals, affecting approximately 0.2% of the population (Hershberger et al., 2009, Maron et al., 2006). Familial HCM is mainly inherited as an autosomal dominant trait by mutations in sarcomeric genes (Alcalai et al., 2008, Morita et al., 2008). The maternal transmission of HCM has also been implicated in some pedigrees, suggesting that mtDNA mutations could contribute to this disorder (Bates et al., 2012, Song et al., 2011). However, the underlying molecular mechanisms by which mtDNA mutations contribute to HCM remain elusive due to lack of cell and animal models.
The mitochondrion is a semiautonomous organelle and contains its own genome (mtDNA), encoding for 13 subunits of complexes I, III, IV, and V of the electron transport chain (ETC) as well as 22 tRNAs and 2 rRNAs necessary for mitochondrial translational machinery. mtDNA mutations often lead to mitochondrial dysfunctions and contribute to mitochondrial disorders (Taylor and Tumbull, 2005, Wallace et al., 1988). Cardiomyopathy is one of most common clinical manifestations in mitochondrial diseases (Finsterer and Kothari, 2014). Since the m.3260A>G mutation in the MT-TL1 gene was identified to be associated with HCM (Zeviani et al., 1991), several other mutations have been reported in mitochondrial tRNA genes such as MT-TG (Merante et al., 1994), MT-TK (Santorelli et al., 1996), MT-TH (Shin et al., 2000), and MT-TI (Taylor et al., 2003) and in mitochondrial protein-encoding genes such as MT-CYB (Andreu et al., 2000), MT-ATP8 (Jonckheere et al., 2008), and MT-ATP6 (Ware et al., 2009). These mtDNA mutations result in some defects of mitochondrial functions and ultrastructures (Merante et al., 1994, Liu et al., 2014). Clinical and experimental studies have shown that abnormal energy metabolism is a common event in HCM pathogenesis (Pisano et al., 2016, Rosca et al., 2013, Luedde et al., 2009). The primary cardiomyocytes from patients are difficult to obtain due to ethical issues, biopsy inaccessibility, and culture span limitations. Previous studies were mainly performed using patient samples such as skin fibroblasts, lymphocytes, and transferring mitochondria cell lines (cybrids). However, these cells could not provide direct evidence of the impact of mtDNA mutations on the pathogenesis of HCM, because different tissues may show different susceptibilities to specific mtDNA mutations (Wu et al., 2016, Wallace and Fan, 2009, Zhang et al., 2014).
Recently, patient-specific induced pluripotent stem cells (iPSCs) have attracted great attention for their unique advantages in elucidating the mechanisms underlying mitochondrial disorders (Inoue et al., 2014, Yamanaka, 2007). Since each mammalian cell contains hundreds of double-membrane mitochondria and each mitochondrion contains multiple copies of mtDNA molecules, it is impossible to directly generate cellular and animal models with targeted mtDNA mutations by using plasmid transfecting or gene editing technology. Generation of patient-specific iPSCs circumvent the above technical hurdles and make reprogramming donor cells carrying mtDNA defects available. iPSCs from patients with mutations in the nuclear DNA (nDNA) have been widely established. iPSC-derived cardiomyocytes carrying mutations in sarcomeric genes possessed the characteristics of the HCM phenotype at the single-cell level (Lan et al., 2013, Birket et al., 2015). To date, only a few studies have been reported to reprogram iPSCs from patients with mtDNA mutations, such as Pearson marrow pancreas syndrome with mtDNA deletion (Cherry et al., 2013), diabetes mellitus with m.3243A>G mutation in tRNALeu(UUR) (Fujikura et al., 2012), and mitochondrial encephalomyopathy with lactic acidosis and stroke-like episodes with m.3243A>G mutation in tRNALeu(UUR) (Hamalainen et al., 2013). These studies indicated that the patient iPSCs contain a similar range of mtDNA heteroplasmy for disease-causing mutations (Cherry et al., 2013, Folmes et al., 2013), and iPSC-derived cells manifest cell-type-specific patterns of respiratory chain deficiency (Hamalainen et al., 2013). Genetically rescued patient iPSCs display normal metabolic function, compared with the impaired oxygen consumption and ATP production observed in mutant cells (Ma et al., 2015). However, studies on HCM-specific iPSCs with mtDNA mutations have not been reported yet.
We previously identified the m.2336T>C mutation in the MT-RNR2 gene from a Chinese maternally inherited HCM family (Liu et al., 2014). This mutation disturbed conserved 2336U-A2438 base pairing in the domain III stem-loop structure of mitochondrial ribosome 16S rRNA, resulting in mitochondrial dysfunctions in the immortalized lymphoblastoid cell lines (Liu et al., 2014). In this report, we generated patient-specific iPSCs as well as their derived cardiomyocytes as a disease model to understand the pathogenic mechanism of maternally inherited HCM.
Results
Establishment and Identification of HCM-iPSCs Carrying the m.2336T>C Mutation
We previously reported the clinical, genetic, and molecular characterization of a four-generation HCM family (Liu et al., 2014). Mutational analysis of the mitochondrial genome identified the m.2336T>C mutation, which was presented exclusively in all the maternal members. The clinical features of the HCM family members are summarized in Table S1. The proband of this family was affected with severe non-obstructive HCM, left ventricular remodeling, and implanted permanent pacemaker. His echocardiography parameters were as follows: maximum left ventricular wall thickness, 37.4 mm; interventricular septum thickness, 28.7 mm; and interventricular septum/left posterior wall ratio, 3.99.
Urine cells ([UCs] epithelial-like cells detached from tubules) from the proband (III-3) in the HCM family (HCM-UCs), and from healthy controls (Con-UCs) including the proband's son (IV-2) and two genetically unrelated individuals in the same region, were collected and cultured for approximately 2 weeks. The UCs were then infected with retroviral vectors containing Oct4, Sox2, Klf4, and c-Myc. After induction for 25–30 days, 20 clones of HCM-specific iPSCs (HCM-iPSCs) from the proband and 10 clones of iPSCs from each control individual (Con-iPSCs) were selected. These iPSCs were tightly packed and grown in colonies that were positively stained for alkaline phosphatase (Figure S1A). Immunofluorescence analysis of iPSCs confirmed positive signals for embryonic stem cell (ESC) marker proteins, including TRA-1-60, TRA-1-81, SSEA-3, SSEA-4, and NANOG (Figure S1A). Expression of endogenous pluripotency genes (OCT4, SOX2, NANOG, and REX-1) were similar to those in ESCs but significantly higher than those in UCs (Figure S2A). Expression of exogenous reprogramming factors (OCT4, SOX2, KLF4, and C-MYC) was silenced (Figure S2B) and integrated into iPSC genomes (Figure S2C). The methylation levels of pluripotency gene (OCT4 and NANOG) promoters were also similar to those in ESCs but significantly lower than those in primary UCs (Figure S2D). Karyotype analysis showed that iPSCs exhibited normal chromosomal morphology, number, and integrity (Figure S2E). Embryoid body and teratoma formation assays yielded cellular derivatives of ectodermal, mesodermal, and endodermal cells both in vitro (Figure S1B) and in vivo (Figure S1C), confirming the pluripotent nature of the generated iPSCs. We next analyzed the sequence of the complete mitochondrial genome from the HCM-iPSCs at the 10th, 23rd, and 33rd passages. The results confirmed the presence of the m.2336T>C mutation (Figure S3) and the absence of any other novel mtDNA mutations (Table S2). These results indicated that we successfully established patient-specific HCM-iPSCs.
Differentiation of HCM-iPSCs into HCM-iPSC-CMs
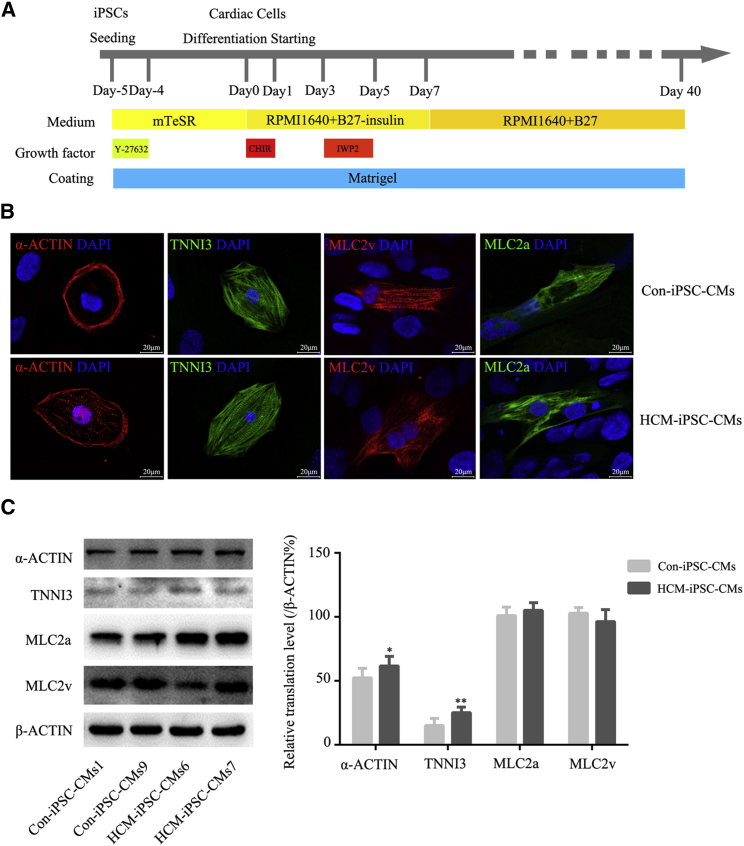
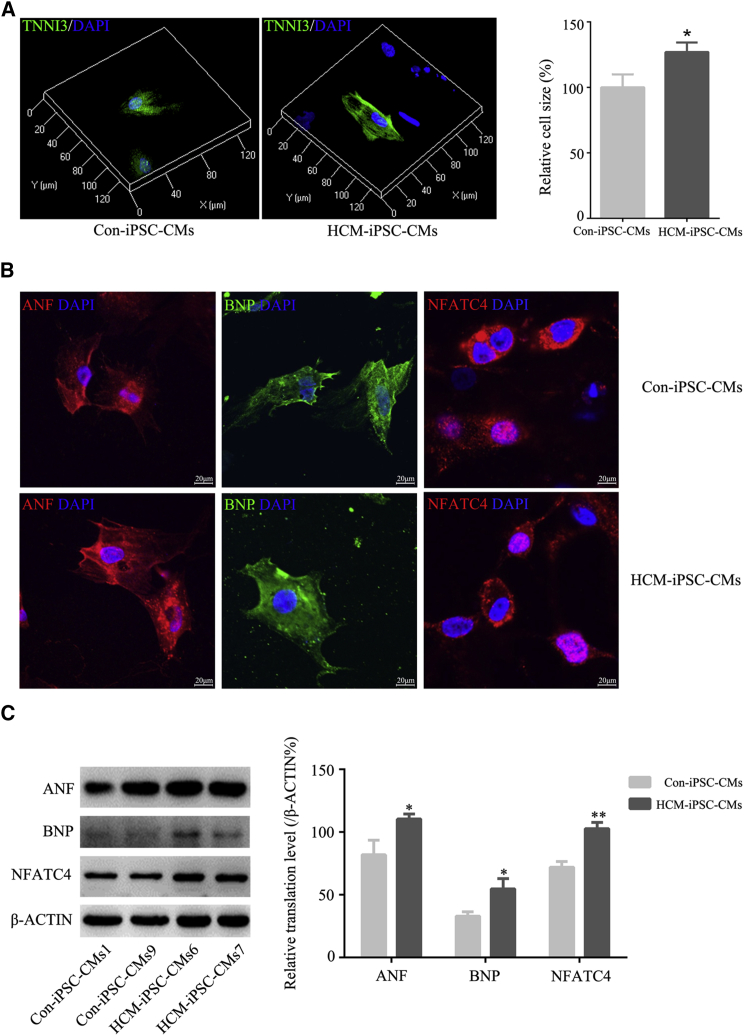
We employed a modified monolayer myocardial differentiation protocol (Figure 1A) to generate cardiomyocytes (iPSC-CMs). Spontaneously beating cardiomyocytes were observed on days 8–12 after the induction of differentiation. By day 40, mature cardiomyocytes were obtained for functional analysis. These cardiomyocytes were maintained as spontaneously beating cells in culture for more than 4 months. The differentiation efficiency of cardiomyocytes exceeded 90%. HCM-iPSC-CMs displayed a disorder of rhythmic beating (Movie S1), whereas Con-iPSC-CMs displayed persistent and spontaneous rhythmic beating (Movie S2). Based on cardiac troponin I (TNNI3) immunostaining, the purity of HCM-iPSC-CMs and Con-iPSC-CMs was approximately 78.8% and 84.1%, respectively. Immunofluorescence analysis indicated that HCM-iPSC-CMs exhibited positive staining for the regular arrangement of sarcomeric proteins such as TNNI3, α-ACTIN, MLC2v, and MLC2a (Figure 1B). Such immunostaining results were also confirmed by western blotting analysis of these sarcomeric proteins (Figure 1C). The double immunostaining of MLC2v and MLC2a and quantification of single- and double-positive cells showed that there was no significant difference in the ventricular and atrial differentiation between HCM-iPSC-CMs and Con-iPSC-CMs (Figure S4). Both expression of MLC2v and MLC2a indicated that those iPSC-CMs would be immaturities. Next, cell size was measured using the ImageJ 3D Objects Counter plugin under a laser scanning confocal microscope. Based on TNNI3 immunostaining, the average size of HCM-iPSC-CMs was approximately 27% larger than that of the Con-iPSC-CMs (Figure 2A). Immunostaining and western blotting showed that the expression of other HCM marker proteins such as ANF, BNP, and NFATC4 in HCM-iPSC-CMs was significantly higher than in Con-iPSC-CMs (Figures 2B and 2C). In addition, cardiomyocytes with positive nuclear NFATC4 staining were also significantly higher in HCM-iPSC-CMs (43.1%, n = 51) than in Con-iPSC-CMs (26.1%, n = 46) (p < 0.05) (Figure 2B). These results indicated that the m.2336T>C mutation did not alter the directional differentiation from HCM-iPSCs into HCM-iPSC-CMs but that HCM-iPSC-CMs might possess some major histopathological features of hypertrophied cardiomyocytes and provide a model to understand the pathological mechanism of maternally inherited HCM.
Figure 1.
Differentiation and Identification of iPSC-CMs
(A) Schematic of cardiomyocyte differentiation protocol.
(B) Representative immunostaining for α-ACTIN, cardiac troponin I (TNNI3), MLC2v, and MLC2a in HCM-iPSC-CMs and Con-iPSC-CMs. Nuclei are stained in blue with DAPI. Scale bar, 100 μm.
(C) Western blotting and quantification analysis of α-ACTIN, cardiac troponin I (TNNI3), MLC2v, and MLC2a. β-ACTIN is shown as a loading control. Data are represented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01.
Figure 2.
Identification of Hypertrophied Characteristics of HCM-iPSC-CMs
(A) 3D fluorescent images showing the cell size of HCM-iPSC-CMs and Ctrl-iPSC-CMs by the ImageJ 3D Objects Counter plugin under laser scanning confocal microscope. Quantification of cell size in HCM-iPSC-CMs (n = 52) compared with that in Con-iPSC-CMs (n = 63).
(B) Representative immunostaining for ANF, BNP, and NFATC4 in HCM-iPSC-CMs and Con-iPSC-CMs. Nuclei are stained in blue with DAPI.
(C) Western blotting and quantification analysis of ANF, BNP, and NFATC4. β-ACTIN is shown as a loading control.
Data are represented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01.
Defects in Mitochondrial Ultrastructure
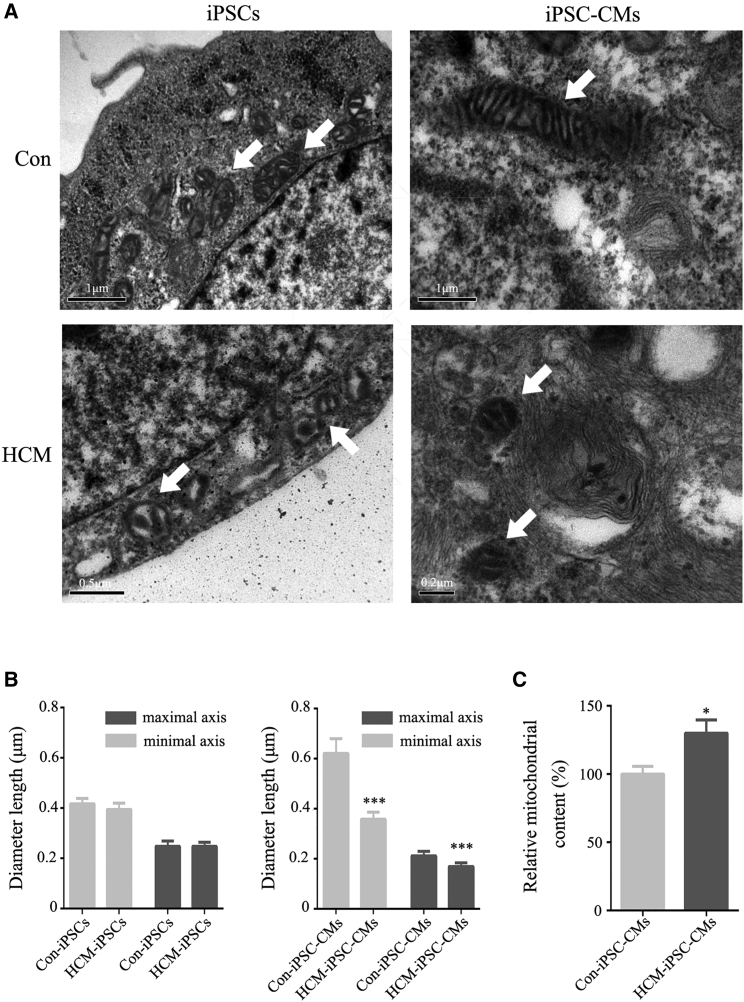
To determine whether the m.2336T>C mutation affects mitochondrial morphology, we used transmission electron microscopy (TEM) to examine the mitochondrial ultrastructure. As expected, we observed some changes in the mitochondrial structure within undifferentiated and differentiated cells. For control samples, the reprogrammed Con-iPSCs exhibited immature rounded mitochondria with rare cristae as well as an expanded matrix. The differentiated Con-iPSC-CMs showed few mature elongated mitochondria with many well-defined cristae and a compacted matrix (Figure 3A). However, for patient samples, HCM-iPSCs exhibited immature rounded mitochondria similar to Con-iPSCs (Figure 3A). HCM-iPSC-CMs displayed mainly round-shaped mitochondria with underdeveloped cristae and few elongated mitochondria with developed cristae (Figure 3A), while mitochondrial number was significantly increased in comparison with that in controls (Figure 3C). The length of the mitochondrial maximal axis significantly decreased during reprogramming and then markedly increased upon differentiation in control cells, whereas no similar changes were detected in patient cells (Figure 3B). These findings indicated that the m.2336T>C mutation impaired the mitochondrial morphology during cardiomyocyte differentiation.
Figure 3.
Mitochondrial Morphology in iPSCs and iPSC-CMs
(A) Transmission electron microscopy (TEM) was used to investigate the mitochondrial morphology changes during reprogramming and differentiation. The arrows show mitochondria.
(B) Morphometric analysis of mitochondrial length in HCM-iPSCs (n = 55), Con-iPSCs (n = 50), HCM-iPSC-CMs (n = 40), and Con-iPSC-CMs (n = 40).
(C) Relative mitochondrial content in HCM-iPSC-CMs (n = 40) compared with Con-iPSC-CMs (n = 40).
Data are represented as mean ± SEM. ∗p < 0.05, ∗∗∗p < 0.001.
Reduction in the Stability of 16S rRNA and Mitochondrial Translation
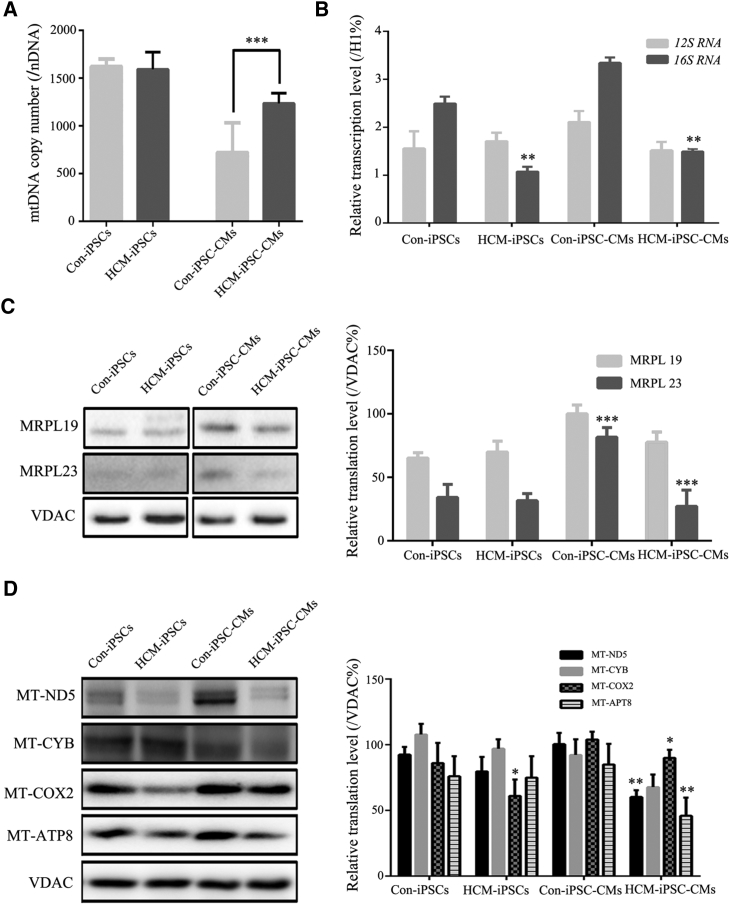
To determine whether the m.2336T>C mutation has effects on mitochondrial gene expression, we examined the mtDNA copy number, levels of 16S rRNA, and mitochondrial translation from three cell lines for HCM-iPSCs, Con-iPSCs, HCM-iPSC-CMs, and Con-iPSC-CMs. The average mtDNA copy number in HCM-iPSC-CMs was 1,236 ± 107 compared with 722 ± 51 in Con-iPSC-CMs. Consistent with the mitochondrial content via TEM observations, mtDNA copy number in HCM-iPSC-CMs was significantly increased by 71.2% in comparison with Con-iPSC-CMs (p < 0.05) (Figure 4A). It was also consistent with the situation of primary samples, such as UCs, from HCM patients and normal controls. The steady-state levels of mitochondrial 16S rRNA were measured by qPCR and normalized to the average levels in the same cell lines for the reference VDAC (a nuclear-encoded outer mitochondrial membrane protein). The average level of 16S rRNA in HCM-iPSC-CMs was significantly decreased by 55% (p < 0.01) compared with that in Con-iPSC-CMs (Figure 4B). A western blot analysis was performed to examine the steady-state levels of mitochondrial 16S rRNA binding proteins and some ETC complex subunits with VDAC as a loading control. The relative levels of mitochondrial 16S rRNA binding proteins were significantly decreased in the mutant cell lines compared with those of the controls: the steady-state levels of MRPL19 and MRPL23 in HCM-iPSC-CMs were 51.1% (p < 0.01) and 23.8% (p < 0.01) of those in Con-iPSC-CMs, respectively (Figure 4C). The average levels of mtDNA-encoded ETC complex subunits, such as p.MT-ND5 (subunit 5 of NADH dehydrogenase), p.MT-CYB (cytochrome b), p.MT-CO2 (subunit 2 of cytochrome c oxidase), and p.MT-ATP8 (subunit 8 of H+-ATPase) in HCM-iPSC-CMs were 59% (p < 0.01), 73% (p > 0.05), 84% (p < 0.05), and 54% (p < 0.01) of those in Con-iPSC-CMs. However, there were no significant differences in the mtDNA copy number and average levels of mitochondrial translation between HCM-iPSCs and Con-iPSCs (Figure 4D). These results indicated that the m.2336T>C mutation decreased the stability of 16S rRNA and the steady-state levels of its binding proteins, impaired mitochondrial ribosomal assembly, and led to reduced levels of mitochondrial proteins in HCM-iPSC-CMs.
Figure 4.
Mitochondrial Gene Expression in iPSCs and iPSC-CMs
(A) mtDNA copy number was performed by qPCR.
(B) qPCR analysis of 16S rRNA from HCM-iPSC-CMs compared with Con-iPSC-CMs.
(C) Western blotting and quantification analysis of mitochondrial large ribosomal subunit 16S rRNA binding proteins (MRPL19 and MRPL23). VDAC is shown as a loading control.
(D) Western blotting and quantification analysis of respiratory electron transport chain complex subunits, including MT-ND5, MT-CYB, MT-COX2, and MT-ATP8. VDAC is shown as a loading control.
Data are represented as mean ± SEM. ∗∗p < 0.01, ∗∗∗p < 0.001.
Reduction in the Mitochondrial Membrane Potential and ATP/ADP Ratio
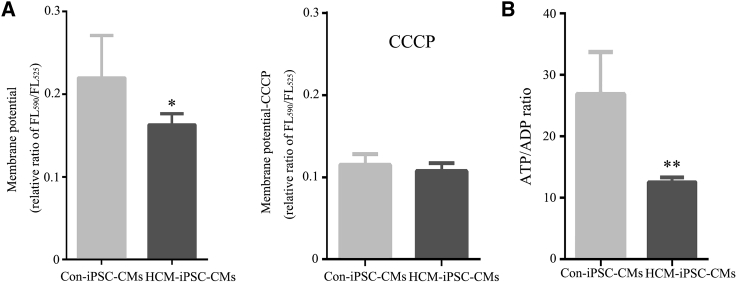
The mitochondrial membrane potential (Δψm) is a key indicator of the integrality of mitochondrial structure and function. The relative ratios of FL590/FL525 geometric mean between mutant and control cell lines were calculated to show the level of Δψm. Carbonyl cyanide 3-chlorophenylhydrazone (CCCP) was used as a protonophore uncoupler to allow for maximum proton (H+) flux through the ETC. The Δψm drove complex V of the ETC to synthesize ATP. In the absence of CCCP, the level of Δψm in HCM-iPSC-CMs was 75% (p < 0.05) of that in Con-iPSC-CMs (Figure 4A). In the presence of CCCP, the levels of Δψm in mutant cells were comparable with those in the control cell lines (p > 0.05) (Figure 5A). The decreased Δψm could reduce the ATP generation capacity, which was consistent with the observation of a 47% (p < 0.01) ATP/ADP ratio in HCM-iPSC-CMs relative to that in Con-iPSC-CMs (Figure 5B). These results indicate that the defective ETC complexes by the m.2336T>C mutation led to mitochondrial dysfunctions, including reduction of mitochondrial membrane potential and the ATP/ADP ratio.
Figure 5.
Mitochondrial Functional Assays in iPSC-CMs
(A) Mitochondrial membrane potential analysis was measured using a fluorescence probe JC-10 assay system. The relative ratio of JC-10 fluorescence intensity at FL590/FL525 in the absence and presence of CCCP.
(B) ATP/ADP analysis was measured using a bioluminescent assay system.
Data are represented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01.
Abnormal Ca2+ Homeostasis and L-type Ca2+ Current
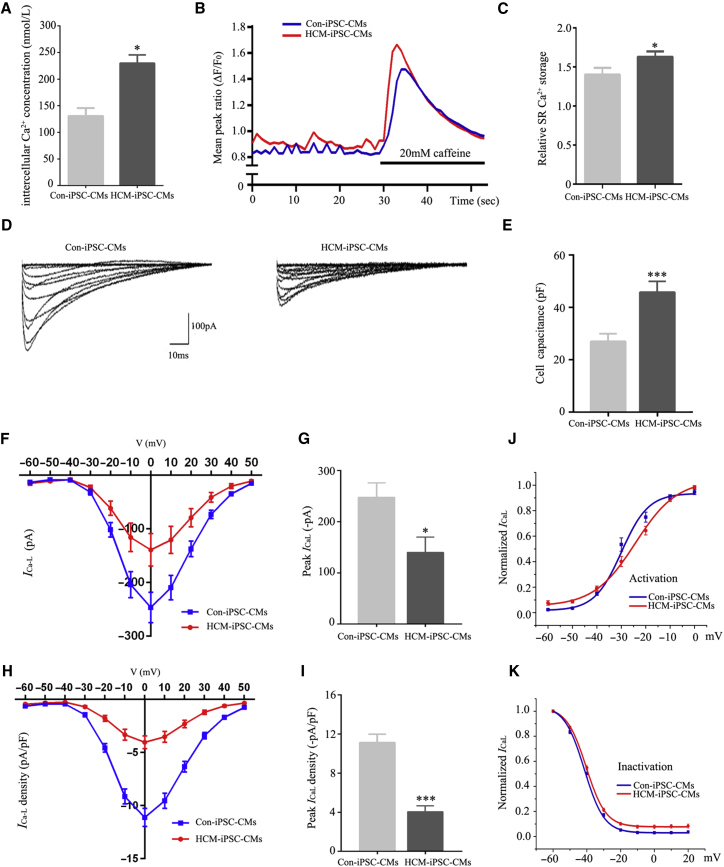
Calcium (Ca2+) plays a fundamental role in the regulation of electrophysiological signaling in cardiomyocyte (Bers, 2008). To determine whether the m.2336T>C mutation affects intracellular Ca2+ homeostasis, we compared intracellular Ca2+ concentration ([Ca2+]i) and L-type Ca2+ current (ICaL) between HCM-iPSC-CMs and Con-iPSC-CMs. [Ca2+]i in HCM-iPSC-CMs was 213 ± 14 nmol/L, significantly higher than 131 ± 14 nmol/L in Con-iPSC-CMs (p < 0.05) (Figure 6A). Representative Ca2+ transient traces were performed using fura-2-acetoxymethyl ester (fura-2AM) and exposed to caffeine, which induces release of sarcoplasmic reticulum (SR) Ca2+ stores into the cytoplasm (Figure 6B). Different from previous reports (Lan et al., 2013), HCM-iPSC-CMs exhibited significantly elevated SR Ca2+ release compared with Con-iPSC-CMs (Figure 6C). The steady-state level of mitochondrial calcium uniporter (MCU) in HCM-iPSC-CMs was decreased by 40% (p < 0.01) compared with that in Con-iPSC-CMs (Figure S5). The expression analysis of some Ca2+ transport marker proteins, such as SERCA, NCX, and PLB, showed no significant difference between Con-iPSC-CMs and HCM-iPSC-CMs (Figure S6A). The PLB-p/PLB ratio in Con-iPSC-CMs and HCM-iPSC-CMs also showed no significant difference (Figure S6B). These results also confirmed that the increment of [Ca2+]i was not due to reduced uptake of Ca2+ by the SR. Next, we measured ICaL of cardiomyocytes by whole-cell patch clamping at −60 to +50 mV (Figure 6D). The average cell capacitance (Cm) of HCM-iPSC-CMs (46.1 ± 3.9 pF) was significantly larger than that of Con-iPSC-CMs (28.7 ± 2.1 pF) (Figure 6E). The maximal ICaL was −138 ± 56 pA in HCM-iPSC-CMs at 0 mV, which was a 42.9% (p < 0.01) reduction compared with −242 ± 60 pA in the Con-iPSC-CMs (Figures 5F and 6G). Accordingly, normalized by Cm, the maximal density of ICaL was −4.52 ± 0.58 pA/pF in HCM-iPSC-CMs at 0 mV, which was a 57.2% (p < 0.01) reduction compared with −10.55 ± 0.73 pA/pF in the Con-iPSC-CMs (Figures 6H and 6I). Moreover, steady-state activation and inactivation of the ICaL were tested and investigated. The results showed that there was a remarkable voltage-positive shift for the activation curve, with a V1/2 elevation from −28.6 ± 1.5 mV in Con-iPSC-CMs to −23.6 ± 1.6 mV in HCM-iPSC-CMs (Figure 6J and Table S3), while for the inactivation curve, no marked difference was observed (Figure 6K and Table S3). These results indicated that the m.2336T>C mutation caused the reduction of mitochondrial membrane potential and then led to the increased [Ca2+]i and decreased ICaL density in HCM-iPSC-CMs.
Figure 6.
Ca2+ Handling Properties of HCM-iPSC-CMs
(A) Quantification of [Ca2+]i was measured on a fluorescence spectrometer by staining with fura-2 AM fluorescence dye.
(B) Representative Ca2+ transient traces from HCM-iPSC-CMs (n = 65) and Con-iPSC-CMs (n = 58) followed by caffeine exposure.
(C) Mean peak amplitudes of ΔF/F0 ratios after caffeine administration representing release of SR Ca2+ load for HCM-iPSC-CMs (n = 65) and Con-iPSC-CMs (n = 58).
(D) Representative traces for ICaL using whole-cell patch clamp from HCM-iPSC-CMs (n = 36) and Con-iPSC-CMs (n = 38).
(E) Cell capacitance measurements of cell size using patch clamp for HCM-iPSC-CMs (n = 34) and Con-iPSC-CMs (n = 39).
(F) Current-voltage relationship of ICaL was determined from −60 to +50 mV in 10-mV increments from HCM-iPSC-CMs (n = 36) and Con-iPSC-CMs (n = 38).
(G) Mean peak of ICaL in HCM-iPSC-CMs (n = 36) and Con-iPSC-CMs (n = 38). Peak ICaL was determined as the difference between the peak inward current and baseline current at the end of depolarization.
(H) ICaL density-voltage relationship was determined from −60 to +50 mV in 10-mV increments from HCM-iPSC-CMs (n = 36) and Con-iPSC-CMs (n = 38).
(I) Peak ICaL density in HCM-iPSC-CMs (n = 36) and Con-iPSC-CMs (n = 38). Current density (pA/pF) was obtained by normalizing currents to membrane capacitance.
(J) Average steady-state activation curves for HCM-iPSC-CMs (n = 36) and Con-iPSC-CMs (n = 38).
(K) Average steady-state inactivation curves for HCM-iPSC-CMs (n = 36) and Con-iPSC-CMs (n = 38).
Data are represented as mean ± SEM. ∗p < 0.05, ∗∗∗p < 0.001.
Abnormal Electrophysiological Properties
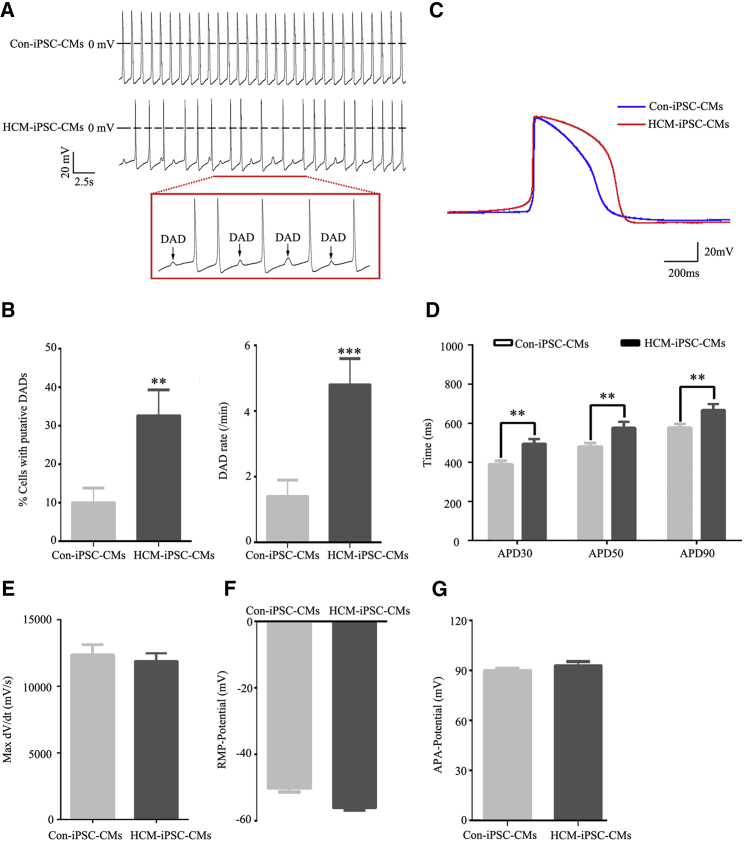
The electrophysiological properties of the cardiomyocytes were examined by whole-cell patch clamping (Table S4). The HCM-iPSC-CMs exhibited delayed afterdepolarizations (DADs) more frequently (Figure 7A), which failed to trigger action potentials (APs) and clustered beats. Cells with putative DADs were 32.6% ± 6.7% in HCM-iPSC-CMs and 10.0% ± 3.8% in Con-iPSC-CMs, whereas DAD rates were 4.8 ± 0.8 in HCM-iPSC-CMs and 1.4 ± 0.5 in Con-iPSC-CMs (Figure 7B). Representative APs were recorded from HCM-iPSC-CMs and Con-iPSC-CMs (Figure 7C). Statistical analysis indicated that the action potential duration (APD) of the HCM-iPSC-CMs was approximately 21% (p < 0.01), 17% (p < 0.01), and 14% (p < 0.01) longer than that of Con-iPSC-CMs at the 30% repolarization (APD30), 50% repolarization (APD50), and 90% repolarization (APD90) stage, respectively (Figure 7D). There was no significant difference in the maximum (Max) velocity of phase 0 depolarization (dV/dt Max) (Figure 7E), resting membrane potential (Figure 7F), and AP amplitude (Figure 7G) between HCM-iPSC-CMs and HCM-iPSC-CMs. The potassium currents in both control and HCM-iPSC-CMs were also measured via whole-cell patch clamping (Figure S7A). Although different from maximal ICaL, the maximal IK showed no significant difference between HCM-iPSC-CMs (151.6 ± 29.0 pA) and Con-iPSC-CMs (119.4 ± 12.9 pA) (Figures S7B and 7C). Similar to maximal density of ICaL, the maximal density of IK was 2.3 ± 0.4 pA/pF in HCM-iPSC-CMs, which was a 58.9% (p < 0.01) reduction compared with 5.6 ± 0.7 pA/pF in the Con-iPSC-CMs (Figures S7D and 7E). The prolongation of the APD at 30%, 50%, and 90% of the repolarization phase might result from multi-components of the AP contour, including ICaL and IK. These results suggest that HCM-iPSC-CMs with the m.2336T>C mutation exhibited some cell-specific features of HCMs, such as arrhythmias, DADs, and prolonged AP duration.
Figure 7.
Electrophysiological Properties of iPSC-CMs
(A) Electrophysiological measurements of spontaneous action potential were performed by whole-cell patch clamp in HCM-iPSC-CMs compared with Con-iPSC-CMs. The boxes indicate the underlined portion of the HCM-iPSC-CM waveform, demonstrating delayed afterdepolarization (DAD)-like arrhythmia.
(B) Quantification of cells with putative DADs, and DAD rate (total DADs/total beats) in HCM-iPSC-CMs (n = 43) and Con-iPSC-CMs (n = 40).
(C) Representative action potential recordings from HCM-iPSC-CMs (n = 43) and Con-iPSC-CMs (n = 40).
(D) Quantification of action potential duration (APD) at 30% (APD30), 50% (APD50), and 90% (APD90) repolarization in HCM-iPSC-CMs (n = 43) and Con-iPSC-CMs (n = 40) from (A).
(E) Quantification of the maximal rate of depolarization (max dV/dt) in HCM-iPSC-CMs (n = 43) and Con-iPSC-CMs (n = 40) from (A).
(F) Quantification of resting membrane potential (RMP) in HCM-iPSC-CMs (n = 43) and Con-iPSC-CMs (n = 40) from (A).
(G) Quantification of action potential amplitude (APA) in HCM-iPSC-CMs (n = 43) and Con-iPSC-CMs (n = 40) from (A).
Data are represented as mean ± SEM. ∗∗p < 0.01, ∗∗∗p < 0.001.
Discussion
In the present study, we investigated the molecular and pathological mechanisms underlying the HCM-associated m.2336T>C mutation by generating a patient-specific iPSC-derived cardiomyocytes model. The HCM-iPSC-CMs not only maintained the original m.2336T>C mutation but also possessed the major properties of hypertrophied cardiomyocytes. We observed a significant reduction in the steady-state level of 16S rRNA and its binding proteins in HCM-iPSC-CMs compared with that in Ctrl-iPSC-CMs, suggesting that the m.2336T>C mutation impaired the stability of 16S rRNA and assembly of mitochondrial ribosomes. Interestingly, some mutations in genes encoding mitochondrial ribosome binding proteins were identified from HCM families (Galmiche et al., 2011, Smits et al., 2011, Carroll et al., 2013). The L156R mutation in MRPL44 affected the assembly of the large ribosomal subunit and stability of 16S rRNA, thereby leading to complex IV deficiency (Carroll et al., 2013). The Leu215Pro mutation in MRPS22 caused a decrease in 12S rRNA (Smits et al., 2011). Furthermore, the m.2336T>C mutation caused the defect of mitochondrial ETC complexes to transfer less H+ from the matrix into the intermembrane space, which formed the lower H+ potential difference and concentration gradient (Δψm) across the mitochondrial inner membrane. This lower Δψm resulted in less proton motive force to generate ATP via complex V of the ETC. The reduction of the ATP/ADP ratio means a low ATP synthesis efficiency (Maldonado and Lemasters, 2014). We also observed more immature mitochondria with less cristae in HCM-iPSC-CMs than in Con-iPSC-CMs, similar to those in lymphoblastoid cell lines (Liu et al., 2014). Mitochondrial morphology has been shown to regulate cardiomyocyte differentiation and embryonic heart development (Kasahara et al., 2013, Hom et al., 2011). The defect of mitochondrial ultrastructure contributes to the pathogenesis of congestive heart failure complications (Arbustini et al., 1998, Wanet et al., 2015, Xu et al., 2013).
The mitochondrion is a major storage organism of Ca2+ and regulates the intracellular Ca2+ homeostasis with the SR (Santo-Domingo and Demaurex, 2010). Evidence of elevation in [Ca2+]i was provided as an initiating factor in iPSC-CMs derived from HCM patients with the Arg663His mutation in MYH7 (Lan et al., 2013). Here, we observed a reduction of MCU and an elevation of [Ca2+]i and SR Ca2+ stores, along with a significant decrease of Δψm in HCM-iPSC-CMs. The uptake of Ca2+ into the mitochondrial matrix is mediated by MCU, which is powered by the mitochondrial membrane potential (Ben-Hail et al., 2014, Kamer and Mootha, 2015). Functional exclusion of the mitochondrial compartment by MCU blockade caused a shift of Ca2+ to the SR (Ronchi et al., 2017). The lower Δψm caused by the m.2336T>C mutation forced less cytoplasmic Ca2+ into the mitochondrial matrix and resulted in increased [Ca2+]i. These results indicated that the mitochondrial dysfunctions might disrupt the Ca2+ homeostasis and induce the downstream Ca2+-dependent cardiac hypertrophy signaling pathways in HCM-iPSC-CMs. For example, elevated [Ca2+]i can activate Ca2+-sensitive calcineurin via Ca2+/calmodulin (Hudson and Price, 2013) or mediate the activation of myocyte enhancer factor 2 (Molkentin et al., 1998) and thereby can cause HCM. Furthermore, HCM-iPSC-CMs exhibited some electrophysiological abnormalities (such as prolonged APD and increased DADs) and cellular arrhythmias. Generation of APs results from sequential activation and inactivation of ion (Na+, Ca2+, and K+) channels. The large, rapid influx of Na+ (INa) results in fast membrane depolarization in the AP upstroke. The inward flow of Ca2+ (ICaL) through L-type calcium channels (LCCs) leads to the plateau phase (Morotti et al., 2012, Veerman et al., 2015). In this study, we identified a more voltage-positive shift of ICaL activation but little shift of ICaL inactivation. The more potential difference between steady-state activation and inactivation, increasing the total amount of Ca2+ entry through the LCC channel during the plateau phase, would prolong the APD. IK is the primary determinant of AP repolarization, such as the efflux of K+ (Ito) in a brief repolarization, and the rapid and slow delayed rectifier K+ channels (IKr and IKs) in membrane repolarization to its original state (Veerman et al., 2015). Here, the reduced IK would slow the repolarization to contribute to the prolonged APD in HCM-iPSC-CMs. In addition, Ca2+ influx during the plateau phase triggers the massive release of SR Ca2+ and influences the kinetic properties of ICaL. The higher SR Ca2+ content may promote diastolic Ca2+ leakage with potential effects on APD. The HCM-iPSC-CMs with the MYH7 Arg663His mutation displayed evidence of DAD, but not altered APD status (Lan et al., 2013), whereas the HCM-iPSC-CMs with the MYH7 Arg442Gly mutation displayed prolonged APD, but not DAD (Han et al., 2014). We observed that HCM-iPSC-CMs with the m.2336T>C mutation exhibited both DAD and prolonged APD. The decreased ICaL together with IK might result in the prolongation of the APD at 30%, 50%, and 90% of the repolarization phase. These results might suggest the existence of multiple manifestations among these HCM-iPSC-CMs due to different pathogenic mutations. Ca2+ homeostasis played an important role in the regulation of excitation-contraction coupling and development of cardiac arrhythmias (Berlin et al., 1989, Bers, 2008, Deferrari et al., 1995).
Abnormal energy metabolism is a common feature of HCM (Rosca et al., 2013). The mutant hearts in transgenic mice expressing the HCM-associated Lys104Glu mutation in the myosin regulatory light chain displayed significantly higher mitochondrial content with preserved overall architecture (Huang et al., 2014). Measurement of transgenic cardiomyocyte sections in transgenic rats overexpressing a truncated cardiac troponin T molecule revealed a significant increase in the mitochondrial fraction (Luedde et al., 2009). Cardiomyocytes from hypertrophic hearts showed a markedly increased number and reduced size of mitochondria (Pisano et al., 2016). These results indicated that the inefficient cellular ATP utilization and the consequently increased energy demand contributed to the pathogenesis of HCM with mutations in nDNA encoding sarcomeric genes (Ashrafian et al., 2003). Here, we further reported changes in mitochondrial biogenesis with a primary mtDNA defect. The m.2336T>C mutation led to a reduction of the ATP/ADP ratio, implying a lower ATP synthesis efficiency and fewer ATP products in each mitochondrion. The increased mtDNA copy number per nDNA and mitochondrial content in each cardiomyocyte were detected by qPCR and TEM. It can be considered as energy compensatory in HCM due to lower ATP synthesis efficiency by mtDNA mutations, similar to inefficient cellular ATP utilization by sarcomeric gene mutations.
Limitations to our study should be also noted. First, although Yamanaka factor-based reprogramming remains the most convenient way of generating iPSCs, the integration of retroviral vectors into the host genome potentially caused immunogenicity and tumorigenesis and greatly hindered its application in clinic (Takahashi and Yamanaka, 2016). Second, human iPSC-derived cardiomyocytes exhibit high phenotypic immaturity and variability, which presents some differences compared with adult cardiomyocytes (Sala et al., 2016). More optimal differentiation approaches and culture conditions will be tried in the future to improve the maturation of iPSC-CMs. Third, the AP was recorded from spontaneously beating cells and not from cells stimulated at a fixed frequency. APD values might then be a bit biased by the different baseline beating frequency.
In summary, our results demonstrate that the MT-RNR2 mutation results in mitochondrial dysfunctions and ultrastructure defects, which induced HCM-specific cellular and electrophysiological characteristics in iPSC-CMs. Furthermore, the MT-RNR2 mutation induced an increase of mitochondrial content to compensate for the energy-generating shortage. Our findings thus provide insight into the pathogenesis of HCM contributed by mtDNA mutations.
Experimental Procedures
Ethical Statement
The study was approved by the Ethics Committee of the First Affiliated Hospital, Zhejiang University School of Medicine, China, and written informed consent was obtained from each participant before enrollment.
Generation and Characterization of HCM-iPSCs
The generation of patient-specific iPSCs from urine samples was performed (Zhou et al., 2012, Zhang et al., 2016). Characterization of iPSCs was performed as reported previously, including alkaline phosphatase staining, expression of endogenous pluripotent genes, silencing of exogenous transgenes, transgene integration, immunocytochemistry, karyotyping, DNA methylation, and in vitro and in vivo differentiation.
Differentiation of iPSCs into Cardiomyocytes
The established iPSC lines from HCM patients and the control group were differentiated directionally into cardiomyocyte lineages (iPSC-CMs) by monolayer myocardial differentiation protocols (Lian et al., 2012). The beating cardiomyocytes were picked up, dissociated, and seeded on coverslips for functional assays, such as calcium imaging, patch clamp, immunostaining, and more.
Measurement of Cell Size
Cell size measurements were performed by TNNI3 immunostaining as described previously, with modifications (Parra et al., 2014). Confocal image stacks were captured with a Zeiss LSM710 laser scanning confocal microscope, and the analysis software was Zen2011. Cell size was analyzed using the ImageJ 3D Objects Counter plugin.
ATP/ADP Measurements
The ATP/ADP level in cells was measured using bioluminescent detection (ADP/ATP Ratio Assay Kit, Abcam) according to the manufacturer's instructions. The cells were plated in 96-well microplates with RPMI 1640 medium containing 20% fetal bovine serum for 72 hr. The bioluminescent intensities were measured on a multi-mode microplate reader (Synergy H1 Hybrid, BioTek).
Mitochondrial Membrane Potential Assay
The mitochondrial membrane potential was measured using fluorescence detection (JC-10 Assay Kit, Abcam) according to the manufacturer's instructions. The cells were cultured on 96-well, black-walled, clear-bottom plates and dyed with 50 μL of JC-10 solution. The fluorescence intensities (excitation/emission [Ex/Em] = 485/525 nm and Ex/Em = 540/590 nm) were measured on a multi-mode microplate reader (Synergy H1 Hybrid, BioTek).
Measurement of Intracellular Ca2+
[Ca2+]i was measured as described previously (Liao et al., 2016). iPSC-CMs were dissociated and loaded in culture medium containing fura-2AM (Dojindo Laboratories). The calcium flux was measured using excitation at 340 nm and 380 nm in a fluorescence spectrometer (LS55, PerkinElmer Life Sciences). The intracellular Ca2+ concentrations were calculated using a fluorescence spectrometer measurement program.
Calcium Imaging
Changes in intracellular Ca2+ concentration were assessed with ratiometric calcium measurement using fura-2AM as described previously (Zou et al., 2017). iPSC-CMs were dissociated and seeded in gelatin-coated chambers (SPL, Korea). Cells were loaded with 5 μmol/L fura-2 AM in a bath solution at room temperature. Imaging was acquired in a BX51WI microscope (Olympus) with a 40× objective lens on an Andor DL-604M EMCCD camera. Data were collected using Macro-manager software.
Patch Clamping
The electrophysiological analysis, including single-cell AP, ICaL, and potassium currents, was performed by the whole-cell patch-clamp configuration (Axopatch-700A and MultiClamp 700B amplifier, Axon Instruments, Sunnyvale, CA) (Hamill et al., 1981). pClamp10.2 software (Axon Instruments, Molecular Devices) was used for data analysis.
Western Blotting Analysis
Twenty micrograms of various proteins were loaded on 12% SDS-PAGE, then electro-transferred to a polyvinylidene difluoride membrane and subjected to western blotting. The primary antibodies used for this experiment are listed in Table S5. Signals were detected using the CLINX ChemiScope and ECLsystem (CWBIO).
Quantitative Real-Time PCR
Total RNA preparations were obtained by using Trizol reagent (Invitrogen) from 1 × 107 cells. Quantitative real-time PCR, for mtDNA copy number, expression of mtDNA, endogenous pluripotent genes, and exogenous transgenes and more, was performed on an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). The forward and reverse primers for PCR amplification are shown in Table S6.
Statistical Analysis
Statistical analysis was carried out using Student's unpaired, two-tailed t test in Microsoft Excel. All data represent three control individuals (three clones) and one patient (three clones), with at least three independent experiments. Several replicates for each clone were performed in every independent experiment. n is the total replicates of each group. Data are represented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Author Contributions
Q.Y. conceived and designed the research. S.L., H.P., C.T., Y. Sun, Y. Song, X.Z., W.Y., X.W., D.L., Y.D., Q.M., C.X., X.Z., L.K., Y.F., X.X., and J.S. performed the experiments. Q.Y., W.H., and Z.L. analyzed the data and wrote the manuscript. F.H., N.Z., and D.Q. helped analyze the data and discuss the results.
Acknowledgments
This work was supported by the National Basic Research Program of China (2014CB943001, 2012CB966804), the National Natural Science Foundation of China (31771398, 31571299, 81570216, 81470686), Zhejiang Provincial Natural Science Foundation of China (LY14C060004, LY14H140004), and the NIH (NCI 2R01CA139158). We thank Professor Min-Xin Guan at Zhejiang University School of Medicine for mentorship and support and Professor Xiaohang Yang at Zhejiang University School of Medicine for discussions. We also thank Dr. Nancy Linford at City of Hope National Medical Center for editing.
Published: February 15, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, seven tables, and two movies and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.01.013.
Contributor Information
Wendong Huang, Email: whuang@coh.org.
Zhong Liu, Email: liuzhong_68@hotmail.com.
Qingfeng Yan, Email: qfyan@zju.edu.cn.
Supplemental Information
References
- Alcalai R., Seidman J.G., Seidman C.E. Genetic basis of hypertrophic cardiomyopathy: from bench to the clinics. J. Cardiovasc. Electrophysiol. 2008;19:104–110. doi: 10.1111/j.1540-8167.2007.00965.x. [DOI] [PubMed] [Google Scholar]
- Andreu A.L., Checcarelli N., Iwata S., Shanske S., DiMauro S. A missense mutation in the mitochondrial cytochrome b gene in a revisited case with histiocytoid cardiomyopathy. Pedia. Res. 2000;48:311–314. doi: 10.1203/00006450-200009000-00008. [DOI] [PubMed] [Google Scholar]
- Arbustini E., Diegoli M., Fasani R., Grasso M., Morbini P., Banchieri N., Bellini O., Dal Bello B., Pilotto A., Magrini G. Mitochondrial DNA mutations and mitochondrial abnormalities in dilated cardiomyopathy. Am. J. Pathol. 1998;153:1501–1510. doi: 10.1016/S0002-9440(10)65738-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafian H., Redwood C., Blair E., Watkins H. Hypertrophic cardiomyopathy: a paradigm for myocardial energy depletion. Trends Genet. 2003;19:263–268. doi: 10.1016/S0168-9525(03)00081-7. [DOI] [PubMed] [Google Scholar]
- Bates M.D., Bourke J.P., Giordano C., d’Amati G., Turnbull D.M., Taylor R.W. Cardiac involvement in mitochondrial DNA disease: clinical spectrum, diagnosis, and management. Eur. Heart J. 2012;33:3023–3033. doi: 10.1093/eurheartj/ehs275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Hail D., Palty R., Shoshan-Barmatz V. Measurement of mitochondrial Ca2+ transport mediated by three transport proteins: VDAC1, the Na+/Ca2+ exchanger, and the Ca2+ uniporter. Cold Spring Harb. Protoc. 2014;2014:161–166. doi: 10.1101/pdb.top066241. [DOI] [PubMed] [Google Scholar]
- Berlin J.R., Cannell M.B., Lederer W.J. Cellular-origins of the transient inward current in cardiac myocytes-role of fluctuations and waves of elevated intracellular calcium. Circ. Res. 1989;65:115–126. doi: 10.1161/01.res.65.1.115. [DOI] [PubMed] [Google Scholar]
- Bers D.M. Calcium cycling and signaling in cardiac myocytes. Annu. Rev. Physiol. 2008;70:23–49. doi: 10.1146/annurev.physiol.70.113006.100455. [DOI] [PubMed] [Google Scholar]
- Birket M.J., Ribeiro M.C., Kosmidis G., Ward D., Leitoguinho A.R., van de Pol V., Dambrot C., Devalla H.D., Davis R.P., Mastroberardino P.G. Contractile defect caused by mutation in MYBPC3 revealed under conditions optimized for human PSC-cardiomyocyte function. Cell Rep. 2015;13:733–745. doi: 10.1016/j.celrep.2015.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C.J., Isohanni P., Pöyhönen R., Euro L., Richter U., Brilhante V., Götz A., Lahtinen T., Paetau A., Pihko H. Whole-exome sequencing identifies a mutation in the mitochondrial ribosome protein MRPL44 to underlie mitochondrial infantile cardiomyopathy. J. Med. Genet. 2013;50:151–159. doi: 10.1136/jmedgenet-2012-101375. [DOI] [PubMed] [Google Scholar]
- Cherry A.B., Gagne K.E., McLoughlin E.M., Baccei A., Gorman B., Hartung O., Miller J.D., Zhang J., Zon R.L., Ince T.A. Induced pluripotent stem cells with a pathological mitochondrial DNA deletion. Stem Cells. 2013;31:1287–1297. doi: 10.1002/stem.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deferrari G.M., Viola M.C., D'Amato E., Antolini R., Forti S. Distinct patterns of calcium transients during early and delayed afterdepolarizations induced by isoproterenol in ventricular myocytes. Circulation. 1995;91:2510–2515. doi: 10.1161/01.cir.91.10.2510. [DOI] [PubMed] [Google Scholar]
- Finsterer J., Kothari S. Cardiac manifestations of primary mitochondrial disorders. Int. J. Cardiol. 2014;177:754–763. doi: 10.1016/j.ijcard.2014.11.014. [DOI] [PubMed] [Google Scholar]
- Folmes C.D., Martinez-Fernandez A., Perales-Clemente E., Li X., McDonald A., Oglesbee D., Hrstka S.C., Perez-Terzic C., Terzic A., Nelson T.J. Disease-causing mitochondrial heteroplasmy segregated within induced pluripotent stem cell clones derived from a patient with MELAS. Stem Cells. 2013;31:1298–1308. doi: 10.1002/stem.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikura J., Nakao K., Sone M., Noguchi M., Mori E., Naito M., Taura D., Harada-Shiba M., Kishimoto I., Watanabe A. Induced pluripotent stem cells generated from diabetic patients with mitochondrial DNA A3243G mutation. Diabetologia. 2012;55:1689–1698. doi: 10.1007/s00125-012-2508-2. [DOI] [PubMed] [Google Scholar]
- Galmiche L., Serre V., Beinat M., Assouline Z., Lebre A.S., Chretien D., Nietschke P., Benes V., Boddaert N., Sidi D. Exome sequencing identifies MRPL3 mutation in mitochondrial cardiomyopathy. Hum. Mutat. 2011;32:1225–1231. doi: 10.1002/humu.21562. [DOI] [PubMed] [Google Scholar]
- Hamalainen R.H., Manninen T., Koivumaki H., Kislin M., Otonkoski T., Suomalainen A. Tissue- and cell-type-specific manifestations of heteroplasmic mtDNA 3243A>G mutation in human induced pluripotent stem cell-derived disease model. Proc. Natl. Acad. Sci. USA. 2013;110:E3622–E3630. doi: 10.1073/pnas.1311660110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamill O.P., Marty A., Neher E., Sakmann B., Sigworth F.J. Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflugers Arch. 1981;391:85–100. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]
- Han L., Li Y., Tchao J., Kaplan A.D., Lin B., Mich-Basso J., Lis A., Hassan N., London B., Bett G.C. Study familial hypertrophic cardiomyopathy using patient-specific induced pluripotent stem cells. Cardiovasc. Res. 2014;104:258–269. doi: 10.1093/cvr/cvu205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberger R.E., Cowan J., Morales A., Siegfried J.D. Progress with genetic cardiomyopathies: screening, counseling, and testing in dilated, hypertrophic, and arrhythmogenic right ventricular dysplasia/cardiomyopathy. Circ. Heart Fail. 2009;2:253–261. doi: 10.1161/CIRCHEARTFAILURE.108.817346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hom J.R., Quintanilla R.A., Hoffman D.L., Karen M.B., Molkentin J.D., Sheu S.S., Porter G.A., Jr. The permeability transition pore controls cardiac mitochondrial maturation and myocyte differentiation. Dev. Cell. 2011;21:469–478. doi: 10.1016/j.devcel.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Liang J., Kazmierczak K., Muthu P., Duggal D., Farman G.P., Sorensen L., Pozios I., Abraham T.P., Moore J.R. Hypertrophic cardiomyopathy associated Lys104Glu mutation in the myosin regulatory light chain causes diastolic disturbance in mice. J. Mol. Cell. Cardiol. 2014;74:318–329. doi: 10.1016/j.yjmcc.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson M.B., Price S.R. Calcineurin: a poorly understood regulator of muscle mass. Int. J. Biochem. Cell Biol. 2013;45:2173–2178. doi: 10.1016/j.biocel.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H., Nagata N., Kurokawa H., Yamanaka S. iPS cells: a game changer for future medicine. EMBO J. 2014;33:409–417. doi: 10.1002/embj.201387098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonckheere A.I., Hogeveen M., Nijtmans L.G., van den Brand M.A., Janssen A.J., Diepstra J.H., van den Brandt F.C., van den Heuvel L.P., Hol F.A., Hofste T.G. A novel mitochondrial ATP8 gene mutation in a patient with apical hypertrophic cardiomyopathy and neuropathy. J. Med. Genet. 2008;45:129–133. doi: 10.1136/jmg.2007.052084. [DOI] [PubMed] [Google Scholar]
- Kamer K.J., Mootha V.K. The molecular era of the mitochondrial calcium uniporter. Nat. Rev. Mol. Cell Biol. 2015;16:545–553. doi: 10.1038/nrm4039. [DOI] [PubMed] [Google Scholar]
- Kasahara A., Cipolat S., Chen Y., Dorn G.W., 2nd, Scorrano L. Mitochondrial fusion directs cardiomyocyte differentiation via calcineurin and Notch signaling. Science. 2013;342:734–737. doi: 10.1126/science.1241359. [DOI] [PubMed] [Google Scholar]
- Lan F., Lee A.S., Liang P., Sanchez-Freire V., Nguyen P.K., Wang L., Han L., Yen M., Wang Y., Sun N. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell. 2013;12:101–113. doi: 10.1016/j.stem.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X., Hsiao C., Wilson G., Zhu K., Hazeltine L.B., Azarin S.M., Raval K.K., Zhang J., Kamp T.J., Palecek S.P. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. USA. 2012;109:E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Lu B., Ma Q., Wu G., Lai X., Zang J., Shi Y., Liu D., Han F., Zhou N. Human neuropeptide S receptor is activated via a Gαq protein-biased signaling cascade by a human neuropeptide S analog lacking the C-terminal 10 residues. J. Biol. Chem. 2016;291:7505–7516. doi: 10.1074/jbc.M115.704122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Song Y., Li D., He X., Li S., Wu B., Wang W., Gu S., Zhu X., Wang X. The novel mitochondrial 16S rRNA 2336T>C mutation is associated with hypertrophic cardiomyopathy. J. Med. Genet. 2014;51:176–184. doi: 10.1136/jmedgenet-2013-101818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedde M., Flögel U., Knorr M., Grundt C., Hippe H.J., Brors B., Frank D., Haselmann U., Antony C., Voelkers M. Decreased contractility due to energy deprivation in a transgenic rat model of hypertrophic cardiomyopathy. J. Mol. Med. 2009;87:411–422. doi: 10.1007/s00109-008-0436-x. [DOI] [PubMed] [Google Scholar]
- Ma H., Folmes C.D.L., Wu J., Morey R., Mora-Castilla S., Ocampo A., Ma L., Poulton J., Wang X., Ahmed R. Metabolic rescue in pluripotent cells from patients with mtDNA disease. Nature. 2015;524:234–238. doi: 10.1038/nature14546. [DOI] [PubMed] [Google Scholar]
- Maldonado E.N., Lemasters J.J. ATP/ADP ratio, the missed connection between mitochondria and the Warburg effect. Mitochondrion. 2014;19(Pt A):78–84. doi: 10.1016/j.mito.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maron B.J., Towbin J.A., Thiene G., Antzelevitch C., Corrado D., Arnett D., Moss A.J., Seidman C.E., Young J.B., American Heart Association Council on Clinical Cardiology, Heart Failure and Transplantation Committee. Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups. Council on Epidemiology and Prevention Contemporary definitions and classification of the cardiomyopathies: an American heart association scientific statement from the council on clinical cardiology, heart failure and transplantation committee; quality of care and outcomes research and functional genomics and translational biology interdisciplinary working groups; and council on epidemiology and prevention. Circulation. 2006;113:1807–1816. doi: 10.1161/CIRCULATIONAHA.106.174287. [DOI] [PubMed] [Google Scholar]
- Merante F., Tein I., Benson L., Robinson B.H. Maternally inherited hypertrophic cardiomyopathy due to a novel T-to-C transition at nucleotide 9997 in the mitochondrial tRNA-glycine gene. Am. J. Hum. Genet. 1994;55:437–446. [PMC free article] [PubMed] [Google Scholar]
- Molkentin J.D., Lu J.R., Antos C.L., Markham B., Richardson J., Robbins J., Grant S.R., Olson E.N. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita H., Rehm H.L., Menesses A., McDonough B., Roberts A.E., Kucherlapati R., Towbin J.A., Seidman J.G., Seidman C.E. Shared genetic causes of cardiac hypertrophy in children and adults. N. Engl. J. Med. 2008;358:1899–1908. doi: 10.1056/NEJMoa075463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morotti S., Grandi E., Summa A., Ginsburg K.S., Bers D.M. Theoretical study of L-type Ca2+ current inactivation kinetics during action potential repolarization and early afterdepolarizations. J. Physiol. 2012;590:4465–4481. doi: 10.1113/jphysiol.2012.231886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra V., Verdejo H.E., Iglewski M., Del Campo A., Troncoso R., Jones D., Zhu Y., Kuzmicic J., Pennanen C., Lopez-Crisosto C. Insulin stimulates mitochondrial fusion and function in cardiomyocytes via the Akt-mTOR-NFkB-Opa-1 signaling pathway. Diabetes. 2014;63:75–88. doi: 10.2337/db13-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisano A., Cerbelli B., Perli E., Pelullo M., Bargelli V., Preziuso C., Mancini M., He L., Bates M.G., Lucena J.R. Impaired mitochondrial biogenesis is a common feature to myocardial hypertrophy and end-stage ischemic heart failure. Cardiovasc. Pathol. 2016;25:103–112. doi: 10.1016/j.carpath.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronchi C., Torre E., Rizzetto R., Bernardi J., Rocchetti M., Zaza A. Late sodium current and intracellular ionic homeostasis in acute ischemia. Basic Res. Cardiol. 2017;112:12. doi: 10.1007/s00395-017-0602-9. [DOI] [PubMed] [Google Scholar]
- Rosca M.G., Tandle B., Hoppel C.L. Mitochondria in cardiac hypertrophy and heart failure. J. Mol. Cell. Cardiol. 2013;55:31–41. doi: 10.1016/j.yjmcc.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala L., Bellin M., Mummery C.L. Integrating cardiomyocytes from human pluripotent stem cells in safety pharmacology: has the time come? Br. J. Pharmacol. 2016;174:3749–3765. doi: 10.1111/bph.13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santo-Domingo J., Demaurex N. Calcium uptake mechanisms of mitochondria. Biochim. Biophys. Acta. 2010;1797:907–912. doi: 10.1016/j.bbabio.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Santorelli F.M., Mak S.C., El-Schahawi M., Casali C., Shanske S., Baram T.Z., Madrid R.E., DiMauro S. Maternally inherited cardiomyopathy and hearing loss associated with a novel mutation in the mitochondrial tRNA(lys) gene (G8363A) Am. J. Hum. Genet. 1996;58:933–939. [PMC free article] [PubMed] [Google Scholar]
- Shin W.S., Tanaka M., Suzuki J., Hemmi C., Toyo-oka T. A novel homoplasmic mutation in mtDNA with a single evolutionary origin as a risk factor for cardiomyopathy. Am. J. Hum. Genet. 2000;67:1617–1620. doi: 10.1086/316896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits P., Saada A., Wortmann S.B., Heister A.J., Brink M., Pfundt R., Miller C., Haas D., Hantschmann R., Rodenburg R.J. Mutation in mitochondrial ribosomal protein MRPS22 leads to Cornelia de Lange-like phenotype, brain abnormalities and hypertrophic cardiomyopathy. Eur. J. Hum. Genet. 2011;19:394–399. doi: 10.1038/ejhg.2010.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.R., Liu Z., Gu S.L., Qian L.J., Yan Q.F. Advances in the molecular pathogenesis of hypertrophic cardiomyopathy. Yi Chuan. 2011;33:549–557. doi: 10.3724/sp.j.1005.2011.00549. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat. Rev. Mol. Cell Biol. 2016;17:183–193. doi: 10.1038/nrm.2016.8. [DOI] [PubMed] [Google Scholar]
- Taylor R.W., Tumbull D.M. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R.W., Giordano C., Davidson M.M., d'Amati G., Bain H., Hayes C.M., Leonard H., Barron M.J., Casali C., Santorelli F.M. A homoplasmic mitochondrial transfer ribonucleic acid mutation as a cause of maternally inherited hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 2003;41:1786–1796. doi: 10.1016/s0735-1097(03)00300-0. [DOI] [PubMed] [Google Scholar]
- Veerman C.C., Kosmidis G., Mummery C.L., Casini S., Verkerk A.O., Bellin M. Immaturity of human stem-cell-derived cardiomyocytes in culture: fatal flaw or soluble problem? Stem Cells Dev. 2015;24:1035–1052. doi: 10.1089/scd.2014.0533. [DOI] [PubMed] [Google Scholar]
- Wallace D.C., Singh G., Lott M.T., Hodge J.A., Schurr T.G., Lezza A.M., Elsas L.J., 2nd, Nikoskelainen E.K. Mitochondrial DNA mutation associated with Leber's hereditary optic neuropathy. Science. 1988;242:1427–1430. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- Wallace D.C., Fan W. The pathophysiology of mitochondrial disease as modeled in the mouse. Genes Dev. 2009;23:1714–1736. doi: 10.1101/gad.1784909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanet A., Arnould T., Najimi M., Renard P. Connecting mitochondria, metabolism and stem cell fate. Stem Cells Dev. 2015;24:1957–1971. doi: 10.1089/scd.2015.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware S.M., El-Hassan N., Kahler S.G., Zhang Q., Ma Y.W., Miller E., Wong B., Spicer R.L., Craigen W.J., Kozel B.A. Infantile cardiomyopathy caused by a mutation in the overlapping region of mitochondrial ATPase 6 and 8 genes. J. Med. Genet. 2009;46:308–314. doi: 10.1136/jmg.2008.063149. [DOI] [PubMed] [Google Scholar]
- Wu J., Ocampo A., Belmonte J.C. Cellular metabolism and induced pluripotency. Cell. 2016;166:1371–1385. doi: 10.1016/j.cell.2016.08.008. [DOI] [PubMed] [Google Scholar]
- Xu X., Duan S., Yi F., Ocampo A., Liu G.H., Belmonte J.C. Mitochondrial regulation in pluripotent stem cells. Cell Metab. 2013;18:325–332. doi: 10.1016/j.cmet.2013.06.005. [DOI] [PubMed] [Google Scholar]
- Yamanaka S. Strategies and new developments in the generation of patient-specific pluripotent stem cells. Cell Stem Cell. 2007;1:39–49. doi: 10.1016/j.stem.2007.05.012. [DOI] [PubMed] [Google Scholar]
- Zeviani M., Gellera C., Antozzi C., Rimoldi M., Morandi L., Villani F., Tiranti V., DiDonato S. Maternally inherited myopathy and cardiomyopathy- association with mutation in mitochondrial-DNA transfer RNALeu(UUR) Lancet. 1991;338:143–147. doi: 10.1016/0140-6736(91)90136-d. [DOI] [PubMed] [Google Scholar]
- Zhang X., Li S., Yang W., Pan H., Qin D., Zhu X., Yan Q. Mitochondrial disease-specific induced pluripotent stem cell models: generation and Characterization. Methods Mol. Biol. 2016;1353:322–342. doi: 10.1007/7651_2014_195. [DOI] [PubMed] [Google Scholar]
- Zhang X., Li S., Yang W., Qin D., Yu L., Yan Q. Patient-specific induced pluripotent stem cell models in mitochondrial diseases. Curr. Stem Cell Res. Ther. 2014;9:134–140. doi: 10.2174/1574888x09666131230142018. [DOI] [PubMed] [Google Scholar]
- Zhou T., Benda C., Dunzinger S., Huang Y., Ho J.C., Yang J., Wang Y., Zhang Y., Zhuang Q., Li Y. Generation of human induced pluripotent stem cells from urine samples. Nat. Protoc. 2012;7:2080–2089. doi: 10.1038/nprot.2012.115. [DOI] [PubMed] [Google Scholar]
- Zou W., Cheng H., Li S., Yue X., Xue Y., Chen S., Kang L. Polymodal responses in C. elegans phasmid neurons rely on multiple intracellular and intercellular signaling pathways. Sci. Rep. 2017;7:42295. doi: 10.1038/srep42295. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.