Abstract
Developmental Pluripotency-Associated-4 (DPPA4) is one of the few core pluripotency genes lacking clearly defined molecular and cellular functions. Here we used a proteomics screening approach of human embryonic stem cell (hESC) nuclear extract to determine DPPA4 molecular functions through identification of novel cofactors. Unexpectedly, the signaling molecule ERBB3-binding protein 1 (EBP1) was the strongest candidate binding partner for DPPA4 in hESC. EBP1 is a growth factor signaling mediator present in two isoforms, p48 and p42. The two isoforms generally have opposing functions, however their roles in pluripotent cells have not been established. We found that DPPA4 preferentially binds p48 in pluripotent and NTERA-2 cells, but this interaction is largely absent in non-pluripotent cells and is reduced with differentiation. The DPPA4-EBP1 interaction is mediated at least in part in DPPA4 by the highly conserved SAF-A/B, Acinus and PIAS (SAP) domain. Functionally, we found that DPPA4 transcriptional repressive function in reporter assays is significantly increased by specific p48 knockdown, an effect that was abolished with an interaction-deficient DPPA4 ΔSAP mutant. Thus, DPPA4 and EBP1 may cooperate in transcriptional functions through their physical association in a pluripotent cell specific context. Our study identifies EBP1 as a novel pluripotency cofactor and provides insight into potential mechanisms utilized by DPPA4 in regulating pluripotency through its association with EBP1.
Graphical abstract
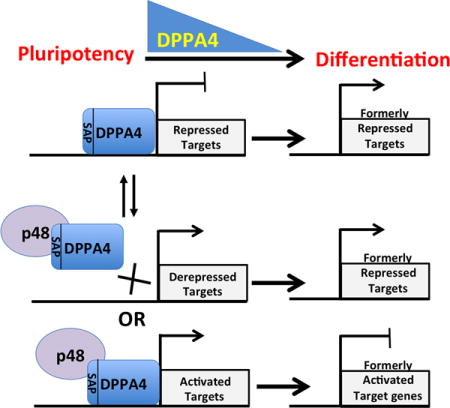
INTRODUCTION
Stem cells have strong potential in regenerative medicine; however, one of the main roadblocks remaining to clinical translation is the link between pluripotency factors and tumorigenesis [1]. A better understanding of the molecular machinery that both governs pluripotency and its link to cancer is essential for moving to the clinic. Several master pluripotency-related transcription factors, such as OCT-3/4 and SOX2 have been implicated in tumorigenesis [2,3], and epigenomic mechanisms play a role both in stem cells and cancer. For instance, Polycomb proteins are involved in regulating the biology of both pluripotent and cancer cells. In addition, specific histone modifying enzymes, DNA methyltransferases, and epigenetic states link cancer and pluripotency [4]. A number of these factors are also involved both in oncogenic transformation and reprogramming to an induced pluripotent state as well [5].
The closely related Developmental Pluripotency-Associated proteins, DPPA4 and DPPA2, function in both pluripotent stem and cancer cells. They were identified as embryonic stem cell (ESC)-specific transcripts that are downstream targets of the transcription factor OCT-4 [6–8] and are hypothesized to play an important role in maintaining the undifferentiated state of stem cells, but the mechanisms by which DPPA4 and DPPA2 function in stem cells remain unclear. Importantly, DPPA2 was also identified separately as a gene with the intriguing feature of only being expressed in embryonic and cancerous tissues. Hence it was given the name Embryo-cancer sequence A (ECSA) [7,9].
DPPA4 expression is normally restricted to early embryos and normally diminishes at E13.5 [10]. DPPA4-null mice displayed lung and skeletal defects along with perinatal death, suggesting that DPPA4 is essential for early embryogenesis and that defects due to DPPA4 depletion manifest substantially after its normal expression ceases [10–12] possibly due to a perturbation of epigenetic memory that impacts cell fate. DPPA4 and DPPA2 proteins are highly conserved [13] and their expression is routinely utilized as markers of pluripotency, although their actual functional impact on pluripotency remains unclear. Recent studies have shown that DPPA4 is a nuclear factor that associates with active chromatin [14,15], and DPPA4 and DPPA2 overexpression is evident in several types of cancer, including both germline and adult cancers [7,16]. Importantly, both DPPA4 and DPPA2 have recently been established as putative oncogenes with transformative capability similar to established oncogenes, such as Ras [17]. Aberrant expression of DPPA2 has been linked to poor prognosis and tumor metastasis of colorectal and gastric cancers [18]. A recent study concluded through genome-wide binding studies that DPPA2 functions outside the conventional pluripotency network in ESCs, [19] however, even less is known about DPPA4 function in pluripotent cells, and there is a particular gap on DPPA4 function in human cells. For instance, most existing data is on murine Dppa4 with very few reports on human DPPA4 function, and whether there are functional differences between murine and human DPPA4 proteins which are 56% identical is unclear.
As most studies to date have examined DPPA4 gene expression rather than DPPA4 protein function, here we pursued the identification of novel potential protein cofactors for human DPPA4 protein in hESCs with the goal of gaining insight into its molecular function in pluripotency and in cancer. Utilizing a proteomics screening approach, we identified putative endogenous DPPA4 cofactors from H9 hESC including most prominently ERBB3 Binding Protein 1 (EBP1), encoded by the PA2G4 gene. We validated that DPPA4 can directly interact with EBP1, a ubiquitous protein that is expressed in most cell lines examined, including germline cells [20,21]. However, to our knowledge there are no reports of PA2G4 or EBP1 expression or function in hESC or induced pluripotent stem cells (IPSC). Due to alternative splicing, EBP1 protein occurs as two isoforms, p48 and p42, that while differing by only 54 amino acids at the N-terminus, display largely opposing functions in non-stem cell types. While p48 promotes cell survival by suppressing apoptosis in an ERBB3-independent manner, p42 suppresses cell growth and promotes differentiation of cell lines upon stimulation of ERBB3 by heregulin [20–22]. We found that DPPA4 interacts specifically with p48 in pluripotent stem cells, but this interaction is much weaker or absent in normal, non-pluripotent cells. Additionally, the binding between DPPA4 and p48 is significantly reduced upon differentiation of pluripotent cells. DPPA4-EBP1 protein interaction is mediated in part by the highly conserved SAF-A/B, Acinus and PIAS (SAP) domain in DPPA4. Furthermore, p48 loss-of-function studies implicate it in attenuating DPPA4 transcription repression function in a SAP-domain dependent manner. Overall, our data indicate that EBP1 is a major cofactor of DPPA4 in pluripotent and cancer cells that impacts DPPA4 transcriptional function.
MATERIALS AND METHODS
Maltose Binding protein (MBP) Pulldown Assays and Mass spectrometry
Equal amounts of purified recombinant control MBP alone and MBP-fusion proteins (30ug) were bound to washed and BSA-blocked amylose resin and 100 uL nuclear or whole cell lysates (1 mg/ml) were added to amylose-bound MBP proteins. The mixture was rotated overnight at 4C. Following incubation, beads were washed ten times (for mass spectrometry) or four times for validation binding and Western blotting experiments with buffer containing 100 mM NaCl, 1M Tris-HCl, 0.5M EDTA, 1 mM DTT. Proteins bound to beads were submitted to the UC Davis Proteomics Core for LC-MS/MS analysis on an Orbitrap with Q-exactive Mass spectrometer. Peptide analysis was done using the Scaffold Proteome Software [23]. Analysis filters were set to 5% FDR and peptides that were present in the MBP-DPPA4 samples while absent in the MBP sample were chosen as potential candidates for validation. For validation interaction studies, proteins bound to beads were analyzed through Western blotting with antibodies to the candidate interacting protein. Proteins were quantified to ensure equal input of recombinant proteins into interaction assays.
Cell culture and transfections
H9 embryonic stem cells were cultured using feeder-free conditions utilizing Matrigel (BD Biosciences) coated plates and mTESR-1 medium (Stem Cell Technologies 0580). 293FT, NIH-3T3, mouse embryonic fibroblasts (MEFs), human dermal fibroblasts (HDF) and NTERA-2 clone D1 cells (supplied by Shiro Urayama) were cultured in DMEM supplemented with 10% FBS (Hyclone) and 1% L-glutamine. HEK 293FT were transfected with 5ug plasmid DNA (DPPA4 or p48 sequences in pcDNA3.1 or pCS2+-HA vector backbones) using the X-tremeGENE HP DNA transfection reagent (Roche, 06366236001) according to the manufacturer instructions. Media was changed 24-hours post transfection and cells were harvested for nuclear or whole cell lysates 48 hours post transfection. siRNA transfections were conducted as reverse transfections with 50 pmol siRNA utilizing the Lipofectamine RNAiMax Reagent (Thermo Fisher Scientific) according to the manufacturer instructions (https://tools.thermofisher.com/content/sfs/manuals/Lipofectamine_RNAiMAX_Reag_protocol.pdf). Briefly, siRNA and RNAiMax were diluted in OptiMEM (Thermo Fisher Scientific) and incubated for 25 minutes. The siRNA transfection complex was then plated onto empty cell culture plates on top of which trypsinized cells and media were plated. Cells were incubated for 24 hours to complete the reverse transfection prior to co-transfection with plasmid DNA. hIPSC were generated, validated in the Knoepfler laboratory and previously characterized [24]. The hIPSC lines tagged with mEGFP at the Tubulin-alpha 1b (TUBA1B) locus were obtained from the Allen Institute for Cell Science (cell line ID: AICS-0012). hIPSC were plated on Matrigel (Corning: 354230) in the presence of the 10uM Rock Inhibitor compound Y27632 (Reagents Direct: 53-B85 or StemRD: 50175997) and routinely cultured with mTeSR1 (Stem Cell Tech: 85850) as per the Allen Institute for Cell Science protocol [25].
Cellular Differentiation
NTERA-2 cells
NTERA-2 cells were plated in T75 flasks and differentiated by addition of 10uM all-trans retinoic acid (at-RA) as previously described [26,27]. Media containing at-RA was changed every 2-3 days for 8 or 18 days after which cells were lysed with lysis buffer for whole cell lysates. Untreated cells were used as a control.
Embryoid Body Formation
Embryoid bodies (EBs) were formed based on protocol established by Lancaster and Knoblich [28]. Briefly, hiPSC were washed with PBS and incubated with Accutase (Stem Cell Technologies: 07920) until cells lifted. Cells were washed further with PBS, counted and resuspended in low bFGF media (20% KOSR, 3% FBS, 1% Glutamax, 1% NEAA, 0.0007% B-me and 4 ng/ml bFGF) plus Y27632. Either 9,000 (EB-1) or 18,000 (EB-2 and EB-3) were plated per well of a round-bottom 96-well plate (Corning: 3799) in 150ul low bFGF/Y27632 media and incubated at 37 degrees Celsius with 5% CO2. Half of the media was replaced every other day and EBs were harvested for nuclear lysates at 7-15 days.
Neural Induction
hiPSC were differentiated into neural lineages using the Gibco neural induction kit and recommended protocol (Gibco: A1647801). Cells were split as mentioned before, and 3×105 cells were plated into each well of a matrigel-coated 6 well plate. 24 hours later, mTeSR1 media was aspirated and replaced with 2.5ml of the neural induction media. Media was changed every other day for 7 days at which point cells were split and propagated onto a matrigel coated plate and switched to the neural expansion media. All cell passaging was conducted using 10uM Y27632.
Dual Luciferase Reporter Assays
Reporter assays were conducted in 293FT cells seeded at 400,000 cells/well. Briefly, cells were reverse transfected with 50pmol pooled (4 individual siRNAs) of either non-targeting siRNA or EBP1 siRNA (Dharmacon), and transiently transfected with GAL4 reporter plasmids (pGL3-basic, Promega) and GAL4-DBD containing constructs (pCMX-Gal4) as previously described [17]. Renilla reporter plasmids (pRL-CMV, 20ng) was co-transfected to account for transfection variability. Luciferase activity was detected at 48-hours post plasmid transfection using a Dual Luciferase Reporter assay kit (Promega) measured on a Spectramax I.3 machine (Molecular Devices). Experiments were performed as at least three independent biological replicates.
Additional Methods are described in Supplemental Methods.
RESULTS
A proteomics screen for DPPA4 cofactors in hESC identifies candidate cofactors including EBP1
We conducted a proteomics screen with the goal of identifying novel DPPA4 protein cofactors in human pluripotent stem cells. The screen used purified recombinant human DPPA4 fused at its amino-terminal end to maltose binding protein (MBP-DPPA4) as a bait to identify proteins in H9 hESC nuclear extracts that bind to DPPA4 by pulldown (Figure 1A, Supplemental Figures S1A–C). This kind of screen can identify both directly interacting proteins as well as proteins together found in multi-protein complexes. The screen here was repeated as two independent biological replicates and 74 candidate DPPA4-binding proteins were identified. The strongest candidate DPPA4 cofactor was ERBB3-binding protein 1 (EBP1), which was present in both replicate experiments when using MBP-DPPA4 as bait but was entirely absent in the MBP alone control samples (Figure 1B). EBP1 was present with 100% probability of identification at 48% and 36% coverage in each experiment, respectively, and FDR <0.05. Regarding the two EBP1 isoforms present in cells, both the unique N-terminal region of p48 and shared regions of p42 and p48 were identified in the 16 and 10 unique peptides pulled out with the MBP-DPPA4 samples in the two biological replicate experiments, respectively, indicating that DPPA4 either bound only the p48 isoform or both isoforms, but not p42 alone (Figure 1C).
FIGURE 1. A DPPA4 proteomics screen identifies candidate interacting proteins, including EBP1.
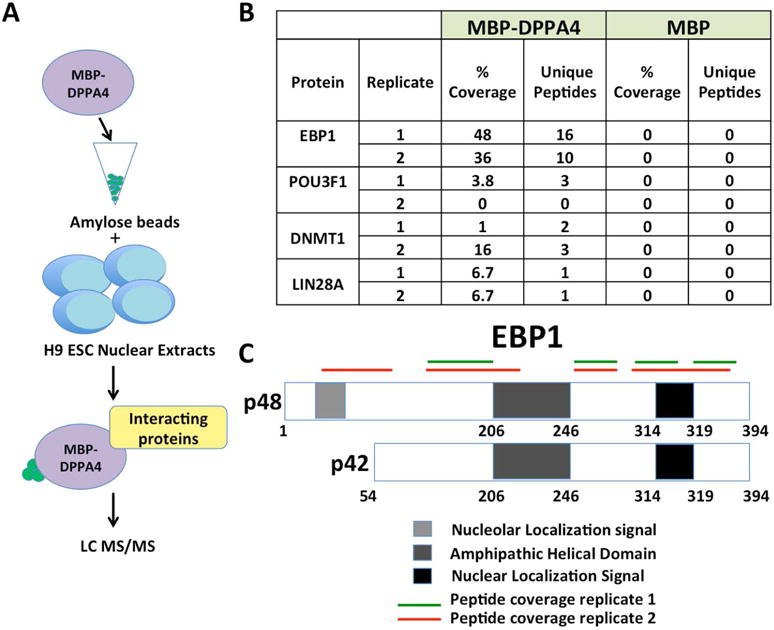
(A) MBP or MBP-DPPA4 was bound to amylose beads and incubated with nuclear extracts from H9 ESCs. Unbound proteins were washed off and interacting proteins were identified through LC MS/MS. Two biological replicates were performed. (B) Table of representative proteins identified through LC-MS/MS to be bound to DPPA4-MBP at 0.05 FDR. (C) Mass spectrometry peptide coverage of EBP1 from two biological replicates shows coverage of both isoforms of EBP1. Both the unique region of p48, within the first 54 amino acids, and regions shared by p42 and p48 isoforms were detected.
Several additional putative DPPA4 binding proteins were also identified. The DNA methyltransferase DNMT1 was present in both experimental replicates. Other candidate cofactors including some that are known to be specific to stem cells or even pluripotent cells, such as LIN28A, were also identified. Some members of the POU-domain family of transcription factors were also present, including OCT4 and OCT6. However, in each case these appeared in the screen results with much lower scores including relatively low peptide counts present in both biological replicates, or only present in one biological replicate alone (Supplemental Table S1 and Supplemental Table S2).
Proteins that were identified with at least one unique peptide in both of the MBP-DPPA4 samples while absent in the MBP control in both biological replicates, or with at least two unique peptides in one of the MBP-DPPA4 samples while absent in the MBP control were included for STRING database analysis of predicted protein-protein interactions [29]. As almost nothing is known of the DPPA4 protein network especially in human cells, we utilized this approach to identify several predicted interconnected protein networks in human ESCs. Interestingly, the STRING protein network included a majority of proteins that are not strictly associated with pluripotency or not reported to have pluripotent-specific functions, including EBP1 (PA2G4) as centrally linked (Figure 2A). Predicted molecular functions based on gene ontology generated by the STRING database implicated RNA, DNA and macromolecular complex binding as potential activities for these networks (Fig 2B). Gene ontology analysis of biological processes related to the predicted protein networks also highlighted several roles involving RNA binding including RNA splicing and RNA processing (Figure 2C), a notable finding given that DPPA4 contains a SAP domain, a domain type implicated in both RNA and DNA binding [11]. EBP1 (PA2G4) was linked to all predicted molecular functions shown except for macromolecular complex binding.
FIGURE 2. A DPPA4-interacting protein network.
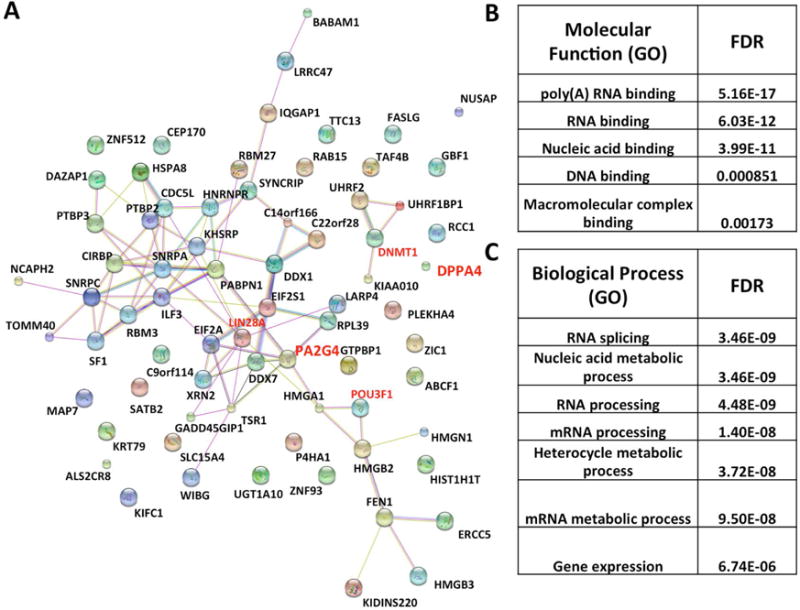
(A) STRING database analysis of proteins identified as DPPA4-interacting proteins through mass spectrometry. Proteins were chosen for analysis based on the following criteria: proteins that had at least one unique peptide in MBP-DPPA4 sample while absent in MBP sample and were present in both biological replicates, or proteins that had at least two unique peptides in MBP-DPPA4 sample while absent in MBP sample that were present in at least one biological replicate were chosen for analysis. (B) Predicted Molecular Function for the protein networks by STRING. (C) Predicted Biological processes for the protein networks by STRING.
DPPA4 binds both EBP1 isoforms in vitro, but only p48 when expressed in cells
To validate DPPA4-EBP1 binding, we conducted direct in vitro pulldown assays to test for DPPA4 and EBP1 interaction utilizing MBP-DPPA4 and MBP with in vitro transcribed and translated (TNT) hemagglutinin (HA)-tagged EBP1 (Figure 3A). To account for variations in protein expression and antibodies used, all pulldown binding assays were normalized to expression of input sample. The inclusion of a beads alone sample also verified that the EBP1 detected in MBP pulldown assays was not bound to the amylose beads alone. HA-EBP1 bound specifically to DPPA4 and not the MBP alone control. Because of homology between DPPA4 and DPPA2 as well as potential DPPA4-DPPA2 dimerization [17], (Supplemental Figure S2), we also conducted in vitro pulldown assays utilizing MBP-DPPA2 and TNT HA-EBP1 (protein purification and validation shown in Supplemental Figure S1D). We observed binding of DPPA2 with EBP1 (Figure 3B). Since the HA-EBP1 plasmid utilized in these tests did not differentiate between its isoforms as it contains the entire open reading frame that if spliced can also make p42, we next conducted a binding assay utilizing TNT hemagglutinin (HA) tagged p48 or p42 with MBP-DPPA4 and MBP-DPPA2. Both EBP1 isoforms bound DPPA4 and DPPA2 in vitro (Figures 3C and 3D) with a stronger interaction observed between p42 and DPPA2 versus p48 (Supplemental Figure S3). The in vitro findings using recombinant MBP fusion proteins with TNT-produced EBP1 proteins are most likely indicative of direct DPPA4 (or DPPA2) binding to EBP1, but it is formally possible that there are other proteins in TNT reticulocyte lysate that can bridge the proteins together in complexes.
FIGURE 3. p48 and p42 can bind to DPPA4/2 in vitro, but only p48 binds DPPA4 in cells.
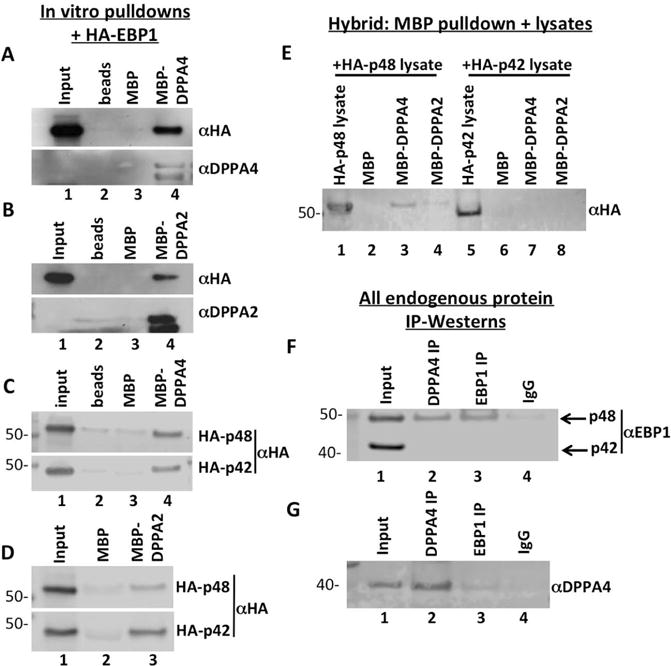
(A, B) MBP-DPPA4 or MBP-DPPA2 was bound with amylose beads and incubated with in vitro transcribed and translated HA-tagged EBP1, beads were washed, and bound protein was detected through Western blotting with anti-HA antibody. (C, D) EBP1 isoform binding to DPPA4 or DPPA2 was tested by binding MBP-DPPA4 or MBP-DPPA2 to amylose beads and incubated with in vitro transcribed and translated HA-tagged p48 or p42. Beads were washed and bound protein was detected through Western blotting with anti-HA antibody. Both p48 and p42 were found to bind to either DPPA4 or DPPA2 in vitro. (E) HA-tagged p48 or HA-tagged p42 plasmid constructs were transiently transfected into 293FT cells and cells were harvested 48-hour post transfection. Whole cell lysates were incubated with MBP-DPPA4 or MBP-DPPA2 bound to amylose beads, and bound protein was detected through western blotting with anti-HA antibody. Only p48-HA expressed in 293FT cells bound to MBP-DPPA4 and MBP-DPPA2. (F) Co-immunoprecipitation was performed utilizing nuclear lysates of NTERA-2 cells expressing completely endogenous levels of EBP1 and DPPA4. p48, but not p42, co-purified with DPPA4 immunoprecipitates. (G) Co-immunoprecipitation was performed utilizing whole cell lysates of NTERA-2 cells expressing completely endogenous levels of EBP1 and DPPA4.
To examine the nature of the DPPA4-EBP1 interaction within cellular lysates, HA-p48 and HA-p42 were expressed in HEK 293FT cells and lysates from those were incubated with MBP-DPPA4 or MBP-DPPA2 for pulldown assays. To distinguish between binding of endogenous EBP1 and exogenously expressed factors after the MBP pulldown, Western blots were probed with HA antibody so as to be specific for the exogenous factors. Only HA-p48 and not HA-p42 from cell lysates was able to bind to DPPA4 and DPPA2 (Figure 3E). Both EBP1 isoforms appeared to be expressed roughly equally, suggesting selective binding by DPPA4 of the p48 isoform when expressed in cells.
We used NTERA-2 (NT2) embryonic carcinoma cells (ECCs), a pluripotent cell line reported to have even higher endogenous expression of both DPPA4 and EBP1 [30] to analyze whether entirely endogenous DPPA4 and EBP1 proteins could interact, and if the interaction was isoform-specific. Additionally, as NT2s are a pluripotent cancer cell line, their use allowed us to investigate the link between DPPA4 function in stem cells and tumorigenesis, which is particularly important given the tumorigenic potential of pluripotent stem cells to form teratoma and more rarely malignant tumors. While both p48 and p42 were robustly expressed in NT2, only endogenous p48 co-immunoprecipitated (co-IP) with DPPA4 in nuclear lysates of NT2s, indicating that under completely endogenous conditions, DPPA4 preferably interacts with p48 (Figure 3F). Additionally, a co-IP of EBP1 in NT2 whole cell lysates demonstrated a weak, but detectable, interaction between endogenous DPPA4 and p48 (Figure 3G). We did not detect any interaction of endogenous p42 with DPPA4 in these experiments; however, since the anti-EBP1 antibody utilized did not appear to immunoprecipitate p42, binding between DPPA4 and p42 under endogenous conditions was not ruled out.
DPPA4 strongly binds p48 in pluripotent stem cells, but only weakly or not at all in fibroblasts
Since, to our knowledge, there have been no direct studies of differential expression of EBP1 isoforms in pluripotent stem cells, we examined the expression of EBP1 in H9 hESCs, human IPSCs previously generated in our lab (see Methods), and F1A mouse ESCs (mESCs; Figure 4). Both EBP1 isoforms were endogenously expressed in human pluripotent cells (Figure 4A). Consistent with previous reports that only one form of EBP1 protein is expressed in mouse cells [31], we observed only one isoform in mESCs that migrated as p48 (Figure 4B). Pulldown assays utilizing MBP-DPPA4 or MBP-DPPA2 incubated with total cellular protein lysates from NT2 cells were conducted, and while both isoforms were expressed in NT2s, only p48 was detectably bound to DPPA4 and DPPA2 (Figure 4C) with approximately equal loading of MBP-tagged proteins in interaction assays (Supplemental Figure S4A-B). Similarly, preferential interaction of DPPA4 and DPPA2 with p48 was observed with endogenous nuclear protein lysates from H9 ESCs and IPSCs (Figure 4D). The isoform ratio of input p48 and p42 in human pluripotent stem cells varied to some degree depending on cell type and experiment, however while the ratios were generally similar across pluripotent cells, non-pluripotent HDF cells exhibited a much higher p48/p42 ratio (Supplemental Table S3).
FIGURE 4. EBP1 p48 in ECC and ESCs, but not fibroblasts, binds to DPPA4.
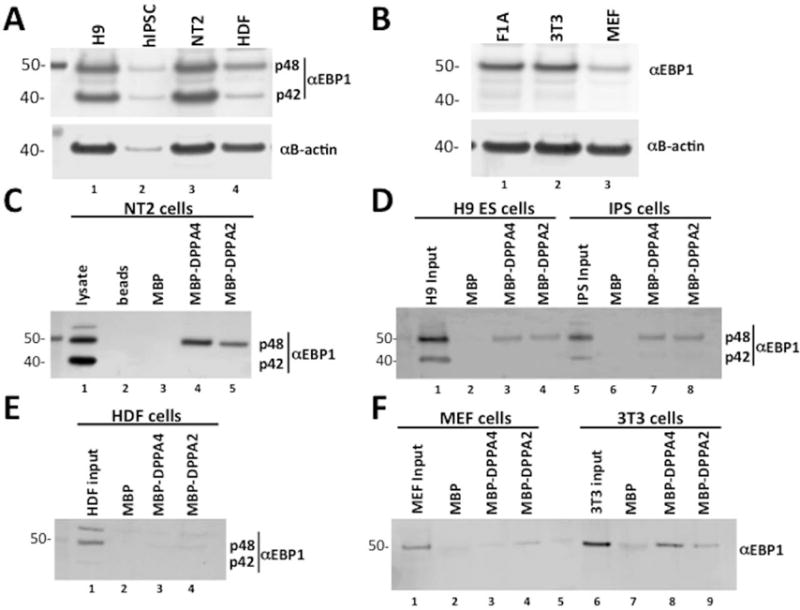
(A) Western blot analysis of EBP1 protein expression in human pluripotent cell lines. Both p48 and p42 are endogenously expressed in H9 ESCs, IPSC, and NTERA-2 (NT2) ECCs. (B) Western blot analysis of EBP1 expression in mouse pluripotent cell lines. Only one isoform of EBP1 is expressed in F1A mESCs. (C) Interaction of EBP1 p48 with DPPA4 and DPPA2 was detected through binding of MBP-DPPA4 or MBP-DPPA2 to amylose beads, incubating the beads with NT2 whole cell lysates, removal of unbound proteins and detection through Western blotting with anti-EBP1 antibody. (D) Interaction of EBP1 p48 with DPPA4 and DPPA2 was detected in H9 ESCs and IPSCs through binding of MBP-DPPA4 or MBP-DPPA2 to amylose beads, incubating the beads with H9 or IPS nuclear extracts and detection through western blotting with anti-EBP1 antibody. (E) EBP1 interaction with DPPA4 or DPPA2 was not detected through incubation of amylose beads bound MBP-DPPA4 or MBP-DPPA2 with whole cell lysates from human dermal fibroblasts followed by western blotting with anti-EBP1 antibody. (F) Interaction of mEBP1 with DPPA4 was not detected through incubation of whole cell lysates of mouse embryonic fibroblasts with amylose beads bound MBP-mDPPA4 followed by western blotting with anti-EBP1 antibody, while weak interaction was detected with MBP-mDPPA2. Binding of mEBP1 in 3T3 cells was detected through incubation of whole cell lysates with MBP-mDPPA4 or MBP-mDPPA2 followed by western blotting with anti-EBP1. Lane 5 is a molecular weight marker.
Given the consistent interaction of DPPA4 with p48 from ESCs and ECC, we further tested the ability of EBP1 from human dermal fibroblasts (HDF) to bind to DPPA4. We found very weak to no interaction of HDF EBP1 with DPPA4 and DPPA2 relative to MBP control (Figure 4E, Supplemental Figure S4C). Similarly, a test of interaction of EBP1 in mouse embryonic fibroblasts (MEF) with DPPA4 or DPPA2 showed at most a slight interaction and only with DPPA2 above background, while EBP1 from immortalized NIH-3T3 cells demonstrated moderate binding to DPPA4 and DPPA2 (Figure 4F). Collectively, these data indicate that EBP1 (p48) binds to DPPA4 most strongly in cells that share characteristics between pluripotent and cancer cells. Notably, in addition to p48, we found weak interaction of p42 with DPPA4 and DPPA2 in IPS cells compared to what was observed for p42 in H9 or NT2 cells (Supplemental Figure S4D-S4E).
Differentiation of NT2s significantly reduces binding of p48 to DPPA4, with more variable results with differentiation of hIPSCs
Since we found the DPPA4-EBP1 interaction to be specific to pluripotent or cancer cells, we tested the hypothesis that differentiation of NT2s would reduce this interaction. NT2 is an established cellular model of induced differentiation via all-trans retinoic acid [26]. MBP binding assays with extracts from NT2 cells differentiated for 8 days showed a significant reduction of approximately 40% in the ability of p48 to bind to MBP-DPPA4 compared to control undifferentiated lysates (Figure 5A-B). We observed some binding to DPPA2, but the overall DPPA2-p48 interaction was relatively weak. OCT4 and DPPA4 levels were examined by Western blotting to validate the loss of pluripotency with differentiation and EBP1 protein levels were moderately decreased (Figure 5C). Additionally, we tested interaction of EBP1 with DPPA4 via the same approach but at 18 days of differentiation of NT2 cells and found no additional decrease of binding between the proteins at day 18 compared to day 8, while expression of p42 and p48 remained similar to Day 8 (data not shown). These data support a pluripotent-specific role of DPPA4-EBP1 interaction. We conducted similar experiments differentiating hIPSCs to examine the impact on DPPA4-EBP1 interaction, but found more variable influence of differentiation in that context, ranging from reduced binding to no effect depending on the experiment (Supplemental Figure S5).
FIGURE 5. EBP1 displays reduced binding to DPPA4 upon differentiation of NT2 cells.
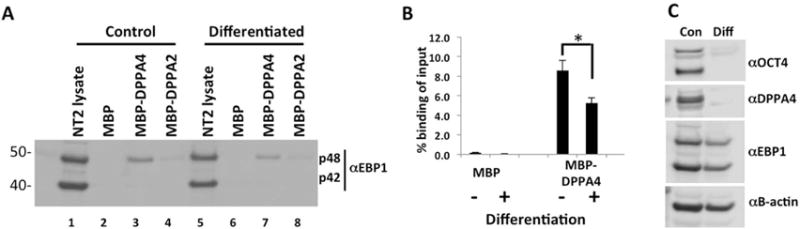
(A) Whole cell lysates of NT2 cells differentiated for 8 days with all-trans retinoic acid (at-RA) were incubated with amylose-bound MBP-DPPA4 or MBP-DPPA2, unbound proteins were washed, and interacting protein was detected through western blotting with anti-EBP1. Undifferentiated whole cell lysates were included as a control. Western blot is representative of three independent biological replicates. (B) Quantitation of panel A. Western blot analysis demonstrated reduced binding of EBP1 p48 to DPPA4, normalized to p48 input, in differentiated NT2s. (n=3, *p<0.05). Error bars are standard error of the mean (S.E.M). (C) Downregulation of OCT4, DPPA4 and EBP1 protein expression by western blot analysis to verify differentiation of NT2s with at-RA.
The DNA/RNA binding SAP domain of DPPA4 is necessary for interaction with EBP1
The only putative domain within DPPA4 is a SAP (SAF-A/B, Acinus, and PIAS) domain in the N-terminal region that is conserved between mouse and human species [11,13] (Figure 6A). SAP domains are predicted DNA/RNA binding motifs [32]. In order to test the hypothesis that the SAP domain is necessary for DPPA4 interaction with EBP1, we conducted in vitro pulldown assays with a fusion protein of MBP-p48 or MBP incubated with full length DPPA4 or DPPA4 with an internal SAP domain deletion (DPPA4ΔSAP). Deletion of the SAP domain abolished binding to p48 compared to the full-length protein (Figure 6B) with approximately equal loading of MBP and MBP-fused proteins in the assay. Additionally, using the reverse approach, we detected weak but reproducible interaction of endogenous EBP1 from 3T3 cell lysates with the DPPA4 SAP domain alone fused to MBP (Figure 6C). Notably, we also detected stronger interaction of EBP1 with the N115 domain of DPPA4 that contains the SAP domain (N81-N115) and other sequences (Figures 6A,C) even relative to full-length DPPA4. Together, these findings indicate that the SAP domain of DPPA4 can bind EBP1 and is necessary for its interaction, but other interaction surfaces could be present as well.
FIGURE 6. The DPPA4 SAP domain is necessary for its interaction with p48 and p48 knockdown enhances the transcriptional repressive function of DPPA4.
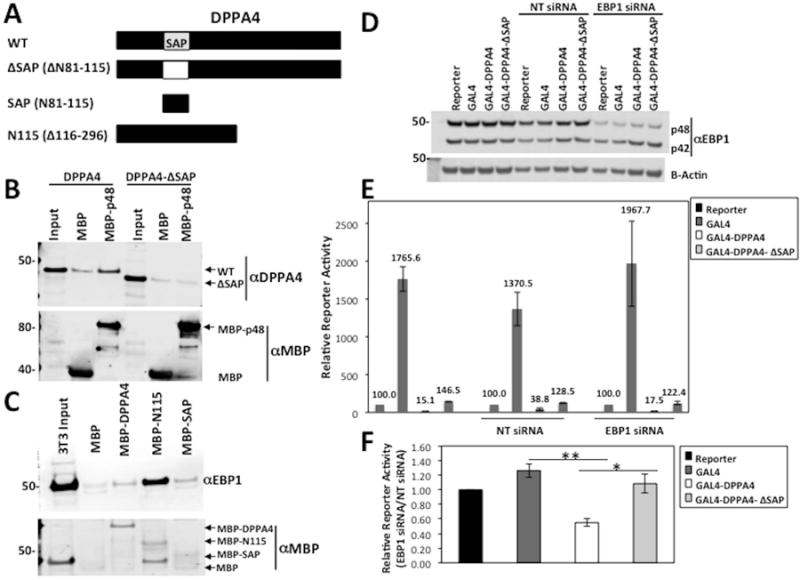
(A) Schematic of MBP-fusion protein truncations of DPPA4 for interaction mapping. Deleted amino acid (AA) region in each truncation is specified in brackets. (B) MBP or MBP-p48 was incubated with in vitro TNT DPPA4 or DPPA4-ΔSAP and bound proteins were analyzed through Western blot and probed for DPPA4 binding. Image is representative of three independent biological replicates. (C) MBP, MBP-DPPA4, MBP-N115, and MBP-SAP were incubated with 3T3 whole cell lysates, and bound proteins were analyzed through Western blot and probed for EBP1 binding. Image is representative of three independent assays. (D) Western blot analysis of pooled siRNA targeting EBP1 results in specific knockdown of p48 in HEK 293FT cells. Image is representative of three independent biological replicates. (NT= Non-targeting) (E) 293FT cells were reverse transfected with NT or EBP1 siRNA and the indicated plasmids. Relative reporter activity was calculated after 48 hours by normalization of luciferase signal to Renilla signal and represented relative to Reporter background. Data is representative of four or three independent biological replicates for GAL4 and GAL4-DPPA4 or GAL4-DPPA4-ΔSAP respectively. Error bars represent standard error of the mean (S.E.M). (F) Relative Reporter activity of Reporter, GAL4 GAL4-DPPA4, or GAL4-DPPA4-ΔSAP in the presence of EBP1 siRNA were normalized to each in NT siRNA control background. Values shown are the mean of at least three independent biological replicates. Error bars are S.E.M. * p< 0.05 ** p<0.001.
DPPA4 transcriptional repressive function is enhanced with p48 knockdown
A fusion of the GAL4-DNA binding domain with DPPA4 exhibits a transcriptional repressive function in cells [17]. GAL4-fusion based reporter assays are used as there is no known DPPA4 consensus DNA binding sequence. Given the predicted molecular functions for the DPPA4-EBP1 protein network (Figure 2), we asked whether EBP1 can affect the transcriptional repression of DPPA4. We performed dual luciferase reporter assays in 293FT control cells, a cell line we chose because it is routinely utilized for reporter assays, it is the one we used for our original characterization of DPPA4 repressive function [17], and one in which we demonstrated consistent DPPA4-EBP1 interaction (Figure 3E). We were also able to substantially lower p48 protein levels through siRNA-mediated knockdown in 293FT cells. The knockdown was specific to p48, with little to no change to p42 (Figure 6D, Supplemental Figures S6A-S6B) allowing us to measure the specific effect of p48 on the transcriptional repression by DPPA4 in this assay. As previously described [17], GAL4-DPPA4 exhibited greater than 100-fold transcriptional repression compared to the GAL4 alone in 293FT cells (Figure 6E). We found that p48 knockdown (relative to cells transfected with control non-targeting (NT) siRNA) increased repression by GAL4-DPPA4 by 2-fold, while reporter activity of GAL4 itself was not reduced (Figure 6E-F, p=0.0005). Therefore, DPPA4 is a better repressor in the context of lower p48 levels in reporter assays, suggesting p48 may normally impair DPPA4 repressive function.
To examine whether the increase in DPPA4 transcriptional repression in the context of reduced p48 levels we observed was specifically due to reduced interaction of DPPA4 and p48, we tested the relative effect of EBP1 siRNA on the transcriptional repression activity of GAL4 fused to DPPA4ΔSAP containing an internal deletion of the SAP domain (GAL4-DPPA4ΔSAP) rendering it EBP1-interaction deficient. The expression of GAL4-DPPA4 and GAL4-DPPA4ΔSAP plasmids was verified through Western blot (Supplemental Figure S6C). The GAL4-DPPA4ΔSAP exhibited transcriptional repression in WT cells as previously described [17], although substantially weaker than the WT DPPA4 protein. However, the GAL4-DPPA4ΔSAP still has some repressive effects indicating that while the SAP domain influences transcriptional repression by DPPA4, there are other mechanisms of DPPA4 repression. Notably, in contrast to GAL4-DPPA4, the transcriptional repression by GAL4-DPPA4ΔSAP was unaffected by p48 knockdown (p=0.03 versus GAL4-DPPA4, Figure 6E-F) suggesting the effect of p48 knockdown on repression by GAL4-DPPA4 is at largely SAP domain-dependent as in the absence of the SAP domain, DPPA4 becomes relatively insensitive to EBP1 status. Together, these results indicate (1) the transcriptional repression of DPPA4 is decreased by interaction with p48 demonstrating that DPPA4 may be a stronger repressor without p48, (2) p48 may impede the transcriptional repression of DPPA4 via the SAP-domain, most likely through mediating protein-protein interaction, and (3) the SAP domain overall plays an important even if not all-encompassing functional role in DPPA4 transcriptional repression.
DISCUSSION
DPPA4 function in stem cell biology has not been well understood. Here, through probing for molecular partners of DPPA4, we have identified several putative protein cofactors for DPPA4, with validation of EBP1 as a major interacting-partner of DPPA4, providing new insights into DPPA4 function. Collectively, our data support a model in which DPPA4 specifically can bind to the EBP1 p48 isoform in a pluripotent context. In this model, DPPA4 can mediate transcriptional repression, but upon binding to p48, the DPPA4-p48 complex may either (a) transcriptionally activate target genes or (b) p48 may inhibit the ability of DPPA4 to transcriptionally repress target genes. We theorize that activation or repression of specific target genes by DPPA4 either with or without EBP1 impacts cell biology. However, at this point relatively little is known about the direct gene targets that DPPA4 can regulate, with only a few published studies supporting transcriptional modulation by DPPA4 including our past report of DPPA4 being a strong repressor when fused to the GAL4 DNA-binding domain [17]. In contrast, DPPA4 has been found associated with transcriptionally active chromatin through its co-localization with both phosphorylated RNA polymerase II and the H3K4Me3 histone mark also associated with actively transcribed chromatin in mouse ESCs [14]. While we consistently find that DPPA4 has robust transcriptional repression activity through reporter assays, the increase in transcriptional repression observed in the absence of p48 suggests that p48 may inhibit the transcriptional repression of DPPA4 thereby facilitating stem cell-related transcriptional regulation at specific genomic loci, or they may cooperate in active transcription at native targets. EBP1 itself has been identified in a repressive complex including YY1, NuRD, HP1 and Trim28 in mouse ESCs although the functional targets remain unclear [33].
A recent report of novel interacting proteins of the variant polycomb repressive complex (vPRC) component, Polycomb Group Ring Finger 1 (PCGF1), identified DPPA4 as a PCGF1-interacting protein among several other pluripotency specific proteins, through an IP-Mass spectrometry screen of protein partners for PCGF1 in NT2 cells [34]. Utilizing molecular mass and stoichiometry analysis in that study, the authors concluded that DPPA4 and PCGF1 form a complex independent of the vPRC1. Their stoichiometry data also indicated that in addition to the participation of DPPA4 in several high mass protein complexes, DPPA4 elutes as a 60kDA protein complex, which could indicate a DPPA4 dimer. Taking into account our data, key remaining questions include whether DPPA4 can form multiple distinct dimers that consist of DPPA4 homodimers or DPPA4-DPPA2 heterodimers, and does EBP1 interact with DPPA4 dimers or a monomer. DPPA4 and DPPA2 were also both identified through a screen to pinpoint interacting partners of the es-BAF chromatin remodeling complex in mESCs [35]. This complex is essential for self-renewal and pluripotency of mESCs. Similar to our findings of DNMT1 as a potential DPPA4-interacting protein, the authors of the es-BAF study also identified DNMT-family members as potential interacting proteins of the es-BAF complex along with DPPA4 and DPPA2.
A previous study demonstrated the binding of DPPA4 to core histone H3, both in vitro and in vivo, via its c-terminal region. Additionally, it was shown that DPPA4 can bind to plasmid DNA directly [15]. However, to our knowledge there are no reports describing DPPA4 genomic binding specificity or properties, and in our hands in an in vitro screen for DPPA4 DNA binding specificity no clear consensus sequence emerged (Knoepfler and Segal, unpublished). The association of DPPA4 with histones and chromatin modifying proteins as well as DNA together has suggested possible transcriptional or epigenetic roles for DPPA4. These studies, along with candidate proteins identified in our proteomics screen, highlight the potentially versatile role for DPPA4 in associating with other pluripotent and non-pluripotent-specific proteins. Together, DPPA4 with such partners or within such complexes likely functions to carry out different molecular functions that may ultimately aid in maintaining potency and directing appropriate cell fate during differentiation, but when perturbed may contribute to tumorigenesis.
Here we focused primarily on the DPPA4-EBP1 interaction in NT2 ECCs due to the links of DPPA4 with both oncogenesis and pluripotency. NT2 cells are a pluripotent testicular embryonic carcinoma cell line with gene expression and chromatin profiles that resemble human ESCs when undifferentiated. Therefore, NT2s are an ideal model in which to study DPPA4 since they are both stem and cancer cells. This is particularly important given our past studies defining the oncogenic functions of DPPA4 and the ability of pluripotent cells to form tumors. NT2s are widely described as a neoplastic pluripotent line as characterized by their expression of pluripotent factors such as OCT-4, SSEA-3 and their pattern of differentiation strongly resembles human ES cells [36]. They are an effective model of differentiation, as administration of retinoic acid downregulates the pluripotency factors and promotes cellular differentiation into the neuroectodermal lineage. As NT2 cells differentiate in this neuronal lineage and DPPA4 levels decline, its binding with p48 decreases both due to reduced levels and potentially other differentiation-associated mechanisms. This reduced binding with differentiation is most apparent in NT2 cells and less consistently so in hIPSC. This variability and difference between NT2s and hIPSC may be due to differences in the cells or in the molecular process of retinoic-acid induced differentiation of NT2s versus embryoid body or neural induction induced differentiation of hIPSCs.
NT2s are most widely used for neuronal differentiation, but other protocols to differentiate NT2s into distinct lineages do exist [37]. We utilized retinoic-acid induced differentiation of NT2s in our study as it is straightforward and the most standard method of differentiation with these cells. As the number of studies differentiating them into distinct lineages is few, a moderate drawback of using NT2s is that there is some uncertainty about the full extent of their potency relative to fully validated pluripotent stem cells such as hESCs and hIPSCs. For that reason, in this study we did not analyze whether differences in EBP1 and DPPA4 binding exists in pluripotent cells that are differentiating towards other lineages. Even so, the decrease in DPPA4-p48 binding in differentiated NT2s is very consistent and may be due to either a post-translational modification to p48 that disrupts binding to DPPA4, or the association of p48 with other factors in the cell upon differentiation that renders it less available to bind to DPPA4.
There is a growing appreciation of the complex links between stem cell potency and cancer, including specifically roles for DPPA4. Perturbed DPPA4-p48 complex function may also contribute to cancer. A study examining the expression of self-renewal genes in several types of cancer found an association between higher grade prostate, bladder, and colon cancer and increased DPPA4 transcription levels in these tissues where DPPA4 is not normally expressed [16]. Stable expression of DPPA4 caused the formation of tumors in both in vitro transformation assays and in in vivo experiments in immunodeficient mice [17]. In that study, we also found that DPPA4 increases cell proliferation through upregulation of pro-cell cycle genes associated with the bypassing the G1/S transition. The two isoforms of EBP1 itself have also been recognized for their roles in oncogenesis. While p42 is considered to function similar to a tumor suppressor [38,39], p48 has characteristics similar to an oncogene with the ability to promote cell growth and invasion of several human cancer cells such as glioblastomas, oral squamous cell carcinomas, and breast cancer cells [40–42]. Several mechanisms have been suggested for p48’s ability to promote aggressive tumor behavior. In glioblastomas, p48 can promote the poly-ubiquitination of the tumor suppressor p53, leading to its degradation [40]. Interestingly, ChIP assays with DPPA4 showed that DPPA4 can bind to the promoter region of TRp53 [17].
What is already known about EBP1 function including in stem cells? A previous proteomics expression study identified EBP1, along with DPPA4, in a long list of proteins that are highly expressed in ECCs versus ESCs, including in NT2s [30]. Otherwise, we could find no other published reports on EBP1 specifically in pluripotent stem cells. The one notable published EBP1 functional study in stem cells reported that EBP1’s p48 isoform was found to have a role in controlling the proliferation and differentiation of adult muscle stem cells [43]. In that study, while overexpression of EBP1 did not have a major effect on quiescent satellite cells, siRNA mediated knockdown of EBP1 greatly inhibited the differentiation and proliferation of satellite cells. The role of EBP1 in muscle stem cell growth was found to involve the p48 isoform only and the authors reported that this function occurs independent of the ERBB3 receptor. Given that we found DPPA4 interacts with endogenous p48 but not p42 under the cellular conditions tested in our study, future studies investigating the direct involvement of ERBB3 signaling through growth factor stimulation will be of interest. Since DPPA4 and p42 can directly bind in vitro, it is possible that they bind in vivo, but only in certain contexts such as in specific types of cells.
We find that deletion of the conserved SAP domain renders DPPA4 unable to interact with EBP1. While other domains of DPPA4 may also be important for its interaction with EBP1, here we focused on the SAP domain as it is implicated in transcriptional and chromatin binding functions [32,44–46]. To our knowledge, the SAP domain has not been previously implicated in protein-protein interactions highlighting a potentially novel role for this conserved protein domain. As DPPA4 dimerizes with DPPA2 and here we find that EBP1 can directly bind to DPPA2, although to a lesser extent than DPPA4, the possibility of a trimeric complex involving these three proteins is of interest. Analysis of the spatial and temporal expression of the mouse Dppa4 and Dppa2 genes have suggested that they may be controlled by different regulatory mechanisms [11] making it possible that DPPA4 and/or DPPA2 form complexes independently with EBP1 in some circumstances to carry out various molecular functions.
Analysis of our proteomics interaction data through the STRING database has pointed to an entirely novel, but as yet hypothetical RNA-binding function for DPPA4. The possibility of DPPA4 regulation pluripotency through RNA binding suggested by STRING analysis is an exciting potential future direction. Although DPPA4 has been shown to bind to plasmid DNA in vitro through its SAP domain and it has been linked to several epigenetic roles through its putative ability to modulate chromatin [11], it is still unknown whether it can bind RNA and function as an RNA regulatory protein, but SAP domains are also putative RNA binding domains. In addition, importantly our STRING data suggest that DPPA4 could be involved in the regulation of translation, in which EBP1 itself is also known to be involved [47]. EBP1 was demonstrated to be a DNA or double stranded-RNA binding protein associated with ribosomes that can mediate the phosphorylation of translation factor, EIF2A, another candidate protein-interaction partner for DPPA4 that was present in our interaction data [48]. Thus, DPPA4-EBP1 complexes could impact RNA functions and translation. STRING database analysis of our proteomics data also implicate other molecular functions for DPPA4, both shared with EBP1 and independent of EBP1 that include other pluripotent specific proteins such as LIN28A.
Overall, our data support a previously uncharacterized function for DPPA4 in pluripotent cells through identifying its interaction with EBP1, hereto not reported to function in ESCs, in reprogramming to make IPSCs, or in other pluripotent cell types. The connected role of both these proteins in potentially modulating transcription may point towards the contributions they bring as a novel mechanism in maintaining pluripotency or directing later differentiation. Delineating links of pluripotency factors such as DPPA4 and DPPA2 to oncogenesis will also be a major factor in the development of safer, more effective stem cell and hIPSC-mediated applications that can be potentially translated to the clinic [49].
Supplementary Material
Supplemental Figure S1. Expression of MBP-fusion recombinant proteins. (A) Coomassie Brilliant Blue stained acrylamide gel of different fractions of the purification process for MBP-DPPA4. (B) Coomassie Brilliant Blue stained acrylamide gel of different fractions of the purification process for MBP. (C) Western Blot of purified MBP-DPPA4 probed with anti-DPPA4 to show specificity of purified MBP-DPPA4. (D) Western Blot of purified MBP-DPPA2 probed with anti-DPPA2 to show specificity of purified MBP-DPPA2.
Supplemental Figure S2. DPPA2 binds to DPPA4 directly in vitro. MBP or MBP-DPPA2 was bound with amylose beads and incubated with in vitro transcribed and translated DPPA2 protein, beads were washed, and bound protein was detected through Western blotting with anti-DPPA2 antibody.
Supplemental Figure S3. p42 demonstrates higher percentage binding to DPPA2. (A) Quantitation of Figure 3C. Western blot analysis of p48-HA or p42-HA binding to DPPA4 show that both can bind to DPPA4 in vitro. (B) Quantitation of Figure 3D. Western blot analysis of p48-HA or p42-HA shows that p42 can interact with DPPA2 to a higher percentage over input compared to p48, in vitro.
Supplemental Figure S4. Endogenous p48, but not p42, binds to DPPA4. (A) Western blot of Figure 4C probed with anti-MBP to demonstrate presence of purified MBP proteins in hybrid pulldown assays. (B) Quantitation of Figure 4C. Western blot analysis of p48 or p42 in NT2 cells bound to MBP-DPPA4 or MBP-DPPA2. (C) Quantitation of panel 4E. MBP-tagged proteins normalized to MBP show quantity of protein in HDF-hybrid pulldown assays. (D) Quantitation of Figure 4D. Western blot analysis of p48 or p42 binding in H9 ESCs to MBP-DPPA4 or MBP-DPPA2. (E) Quantitation of Figure 4D. Western blot analysis of p48 or p42 in IPS cells bound to MBP-DPPA4 and MBP-DPPA2.
Supplemental Figure S5. Variable effects of differentiation on EBP1-DPPA4 binding in hIPSC. (A) Whole cell lysates of hIPSC differentiated for 7 days with Gibco Neural Induction media (catalog A1647801) produced neurally induced (NI) cells. Alternatively, we cultured hIPSC in suspension with low bFGF media (20% KOSR, 3% FBS, 1% Glutamax, 1% NEAA, 0.0007% B-mercaptoethanol and 2 mg/ml bFGF) for 7-15 days to form embryoid bodies (EBs). Western blot of whole cell lysate probed with EBP1, OCT4, DPPA4 and B-actin shows strong to complete elimination of pluripotency factors in both NI cells and EBs. (B) (Top) Differentiated or hIPSC lysates were incubated with amylose-bound MBP-DPPA4, or MBP as a control, and were washed to remove non-specific binding, and interacting protein was detected through Western blotting with anti-EBP1. Two independent NI cultures were used (NI-1 and NI-2) and three separate cultures of EBs. Undifferentiated whole hIPSC lysates were included as a control. Western blot shown is representative of replicates. Quantitation of the mean values from seven pulldowns for each of the four cell lysate types is shown under the blot for the signal of MBP-DPPA4 binding of EBP1’s p48 isoform, with binding using undifferentiated hIPSC lysate set as 1.0. In each case signal was normalized to percent input. An overexposed (OE) blot for the EB pulldown is shown at right to show that some faint binding to EBP1 is apparent and quantifiable. Only NI-2 exhibited a statistically significant change in binding, with a two-fold decrease relative to hIPSC. (Bottom) Western blot analysis of the amount of MBP or MBP-DPPA4 protein loaded in each pulldown.
Supplemental Figure S6. EBP1 siRNA results in p48 knockdown specifically. (A) Quantitation of p48 protein levels of Western blot in Figure 6D. (B) Quantitation of p42 protein levels of Western blot in Figure 6D. (C) Expression of GAL4-DPPA4 plasmids in luciferase assay experiments. Arrow indicates the expected band for GAL4-DPPA4 or GAL4-DPPA4-ΔSAP.
Acknowledgments
The authors wish to thank Po-Yuan Tung for reagents. We thank Kuang-Yui Chen and Rachel Klein for helpful reading of the manuscript, and Colleen Sweeney for feedback on the manuscript, reagents, and helpful advice on experimental design. We thank Enoch Baldwin for purification of mouse recombinant proteins. We thank Shiro Urayama for the NT2 cells. This work was supported by funding from NIH Grants 1R01GM100782 and 1R01GM116919 (PK) and CIRM Grant RN2-00922-1 (to PK).
Footnotes
Author Contributions
PS conducted all experiments with the exception of the truncation interaction mapping which was done by KMB. PK conceived of the project and both PK and PS did the experimental design, data interpretation, and manuscript writing with PS writing the initial draft.
References
- 1.Knoepfler PS. Deconstructing Stem Cell Tumorigenicity : A Roadmap to Safe Regenerative Medicine. Stem Cells. 2009;27:1050–1056. doi: 10.1002/stem.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boumahdi S, Driessens G, Lapouge G, et al. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature. 2014;511:246–253. doi: 10.1038/nature13305. [DOI] [PubMed] [Google Scholar]
- 3.Kumar SM, Liu S, Lu H, et al. Acquired cancer stem cell phenotypes through Oct4-mediated dedifferentiation. Oncogene. 2012;31:4898–4911. doi: 10.1038/onc.2011.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watanabe A, Yamada Y, Yamanaka S. Epigenetic regulation in pluripotent stem cells: a key to breaking the epigenetic barrier. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120292. doi: 10.1098/rstb.2012.0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riggs JW, Barrilleaux BL, Varlakhanova N, et al. Induced Pluripotency and Oncogenic Transformation. Stem Cells Dev. 2013;22(1):37–50. doi: 10.1089/scd.2012.0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakravarthy H, Boer B, Desler M, et al. Identification of DPPA4 and other genes as putative Sox2:Oct-3/4 target genes using a combination of in silico analysis and transcription-based assays. J Cell Physiol. 2008;216:651–662. doi: 10.1002/jcp.21440. [DOI] [PubMed] [Google Scholar]
- 7.Monk M, Hitchins M, Hawes S. Differential expression of the embryo/cancer gene ECSA(DPPA2), the cancer/testis gene BORIS and the pluripotency structural gene OCT4, in human preimplantation development. Mol Hum Reprod. 2008;14:347–355. doi: 10.1093/molehr/gan025. [DOI] [PubMed] [Google Scholar]
- 8.Mitsui K, Tokuzawa Y, Itoh H, et al. The homeoprotein nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 9.John T, Caballero OL, Svobodov SJ, et al. ECSA/DPPA2 is an embryo-cancer antigen that Is coexpressed with cancer-testis antigens in non-small cell lung cancer. Clin Cancer Res. 2008;14:3291–3298. doi: 10.1158/1078-0432.CCR-07-1322. [DOI] [PubMed] [Google Scholar]
- 10.Madan B, Madan V, Weber O, et al. The pluripotency-associated gene Dppa4 is dispensable for embryonic stem cell identity and germ cell development but essential for embryogenesis. Mol Cell Biol. 2009;29:3186–3203. doi: 10.1128/MCB.01970-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maldonado-Saldivia J, van den Bergen J, Krouskos M, et al. Dppa2 and Dppa4 are closely linked SAP motif genes restricted to pluripotent cells and the germ line. Stem Cells. 2007;25:19–28. doi: 10.1634/stemcells.2006-0269. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura T, Nakagawa M, Ichisaka T, et al. Essential roles of ECAT15-2/Dppa2 in functional lung development. Mol Cell Biol. 2011;31:4366–4378. doi: 10.1128/MCB.05701-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siegel D, Schuff M, Oswald F, et al. Functional dissection of XDppa2/4 structural domains in Xenopus development. Mech Dev. 2009;126:974–989. doi: 10.1016/j.mod.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 14.Masaki H, Nishida T, Kitajima S, et al. Developmental pluripotency-associated 4 (DPPA4) localized in active chromatin inhibits mouse embryonic stem cell differentiation into a primitive ectoderm lineage. J Biol Chem. 2007;282:33034–33042. doi: 10.1074/jbc.M703245200. [DOI] [PubMed] [Google Scholar]
- 15.Masaki H, Nishida T, Sakasai R, et al. DPPA4 modulates chromatin structure via association with DNA and core histone H3 in mouse embryonic stem cells. Genes Cells. 2010;15:327–337. doi: 10.1111/j.1365-2443.2010.01382.x. [DOI] [PubMed] [Google Scholar]
- 16.Amini S, Fathi F, Mobalegi J, et al. The expressions of stem cell markers: Oct4, Nanog, Sox2, nucleostemin, Bmi, Zfx, Tcl1, Tbx3, Dppa4, and Esrrb in bladder, colon, and prostate cancer, and certain cancer cell lines. Anat Cell Biol. 2014;47:1–11. doi: 10.5115/acb.2014.47.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tung PY, Varlakhanova NV, Knoepfler PS. Identification of DPPA4 and DPPA2 as a novel family of pluripotency-related oncogenes. Stem Cells. 2013;31:2330–2342. doi: 10.1002/stem.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raeisossadati R, Abbaszadegan MR, Moghbeli M, et al. Aberrant expression of DPPA2 and HIWI genes in colorectal cancer and their impacts on poor prognosis. Tumor Biol. 2014;35:5299–5305. doi: 10.1007/s13277-014-1690-x. [DOI] [PubMed] [Google Scholar]
- 19.Engelen E, Brandsma JH, Moen MJ, et al. Proteins that bind regulatory regions identified by histone modification chromatin immunoprecipitations and mass spectrometry. Nat Commun. 2015;6:7155. doi: 10.1038/ncomms8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia X, Lessor TJ, Zhang Y, et al. Analysis of the expression pattern of Ebp1, an ErbB-3-binding protein. Biochem Biophys Res Commun. 2001;289:240–244. doi: 10.1006/bbrc.2001.5942. [DOI] [PubMed] [Google Scholar]
- 21.Liu Z, Ahn JY, Liu X, et al. Ebp1 isoforms distinctively regulate cell survival and differentiation. Proc Natl Acad Sci U S A. 2006;103:10917–10922. doi: 10.1073/pnas.0602923103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lessor TJ, Yoo JY, Xia X, et al. Ectopic expression of the ErbB-3 binding protein ebp1 inhibits growth and induces differentiation of human breast cancer cell lines. J Cell Physiol. 2000;183:321–329. doi: 10.1002/(SICI)1097-4652(200006)183:3<321::AID-JCP4>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 23.Searle BC. Scaffold: A bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics. 2010;10:1265–1269. doi: 10.1002/pmic.200900437. [DOI] [PubMed] [Google Scholar]
- 24.Cerdeño VM, Barrilleaux BL, Mcdonough A, et al. Behavior of xeno-transplanted undifferentiated human induced pluripotent stem cells is impacted by microenvironment without evidence of tumors Stem Cells Dev. 2017;26(19):1409–1423. doi: 10.1089/scd.2017.0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts B, Haupt A, Tucker A, et al. Systematic gene tagging using CRISPR/Cas9 in human stem cells to illuminate cell organization. 2017:1–44. doi: 10.1091/mbc.E17-03-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee VM, Andrews PW. Differentiation of NTERA-2 clonal human embryonal carcinoma cells into neurons involves the induction of all three neurofilament proteins. J Neurosci. 1986;6:514–521. doi: 10.1523/JNEUROSCI.06-02-00514.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baldassarre G, Boccia A, Bruni P, et al. Retinoic acid induces neuronal differentiation of embryonal carcinoma cells by reducing proteasome-dependent proteolysis of the cyclin-dependent inhibitor p27. Cell Growth Differ. 2000;11:517–526. [PubMed] [Google Scholar]
- 28.Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nature Protoc. 2014;9:2329–2340. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaerkady R, Kerr CL, Kandasamy K, et al. Comparative proteomics of human embryonic stem cells and embryonal carcinoma cells. Proteomics. 2010;10:1359–1373. doi: 10.1002/pmic.200900483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Radomski N, Jost E. Molecular cloning of a murine cDNA encoding a novel protein, p38-2G4, which varies with the cell cycle. Exp Cell Res. 1995;220:434–445. doi: 10.1006/excr.1995.1335. [DOI] [PubMed] [Google Scholar]
- 32.Aravind L, Koonin EV. SAP - A putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 2000;25:112–114. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- 33.Schlesinger S, Goff SP. Retroviral transcriptional regulation and embryonic stem cells: war and peace. Mol Cell Biol. 2015;35:770–777. doi: 10.1128/MCB.01293-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oliviero G, Munawar N, Watson A, et al. The variant Polycomb Repressor Complex 1 component PCGF1 interacts with a pluripotency sub-network that includes DPPA4, a regulator of embryogenesis. Sci Rep. 2015;5:18388. doi: 10.1038/srep18388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ho L, Ronan JL, Wu J, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A. 2009;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Draper JS, Pigott C, Thomson JA, et al. Surface antigens of human embryonic stem cells : changes upon differentiation in culture. J Anat. 2002:249–258. doi: 10.1046/j.1469-7580.2002.00030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simões PD, Ramos T. Human pluripotent embryonal carcinoma NTERA2 cl.D1 cells maintain their typical morphology in an angiomyogenic medium. J Negat Results Biomed. 2007;6:5. doi: 10.1186/1477-5751-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ko HR, Nguyen TL, Kim CH, et al. P42 Ebp1 functions as a tumor suppressor in non-small cell lung cancer. BMB Rep. 2014;48:159–165. doi: 10.5483/BMBRep.2015.48.3.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Linn D, Liu Z, et al. EBP1, an ErbB3-binding protein, is decreased in prostate cancer and implicated in hormone resistance. Mol Cancer Ther. 2008;7:3176–3186. doi: 10.1158/1535-7163.MCT-08-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim CK, Nguyen TL, Joo KM, et al. Negative regulation of p53 by the long isoform of ErbB3 binding protein Ebp1 in brain tumors. Cancer Res. 2010;70:9730–9741. doi: 10.1158/0008-5472.CAN-10-1882. [DOI] [PubMed] [Google Scholar]
- 41.Ko HR, Kim CK, Ahn JY. Phosphorylation of the N-terminal domain of p48 Ebp1 by CDK2 is required for tumorigenic function of p48. Mol Carcinog. 2015;54:1283–1291. doi: 10.1002/mc.22203. [DOI] [PubMed] [Google Scholar]
- 42.Mei Y, Zhang P, Zuo H, et al. Ebp1 activates podoplanin expression and contributes to oral tumorigenesis. Oncogene. 2014;33:3839–3850. doi: 10.1038/onc.2013.354. [DOI] [PubMed] [Google Scholar]
- 43.Figeac N, Serralbo O, Marcelle C, et al. ErbB3 binding protein-1 (Ebp1) controls proliferation and myogenic differentiation of muscle stem cells. Dev Biol. 2014;386:135–151. doi: 10.1016/j.ydbio.2013.11.017. [DOI] [PubMed] [Google Scholar]
- 44.Sook Ahn J, Whitby MC. The role of the SAP motif in promoting holliday junction binding and resolution by SpCCE1. J Biol Chem. 2003;278:29121–29129. doi: 10.1074/jbc.M302314200. [DOI] [PubMed] [Google Scholar]
- 45.Kipp M, Göhring F, Ostendorp T, et al. SAF-Box, a conserved protein domain that specifically recognizes scaffold attachment region DNA. Mol Cell Biol. 2000;20:7480–7489. doi: 10.1128/mcb.20.20.7480-7489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kelsey JS, Blumberg DD. A SAP domain-containing protein shuttles between the nucleus and cell membranes and plays a role in adhesion and migration in D. discoideum. Biol Open. 2013;2:396–406. doi: 10.1242/bio.20133889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Squatrito M, Mancino M, Donzelli M, et al. EBP1 is a nucleolar growth-regulating protein that is part of pre-ribosomal ribonucleoprotein complexes. Oncogene. 2004;23:4454–4465. doi: 10.1038/sj.onc.1207579. [DOI] [PubMed] [Google Scholar]
- 48.Squatrito M, Mancino M, Sala L, et al. Ebp1 is a dsRNA-binding protein associated with ribosomes that modulates eIF2alpha phosphorylation. Biochem Biophys Res Commun. 2006;344:859–868. doi: 10.1016/j.bbrc.2006.03.205. [DOI] [PubMed] [Google Scholar]
- 49.Barrilleaux B, Knoepfler PS. Inducing iPSCs to Escape the Dish. Cell Stem Cell. 2012;9:103–111. doi: 10.1016/j.stem.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. Expression of MBP-fusion recombinant proteins. (A) Coomassie Brilliant Blue stained acrylamide gel of different fractions of the purification process for MBP-DPPA4. (B) Coomassie Brilliant Blue stained acrylamide gel of different fractions of the purification process for MBP. (C) Western Blot of purified MBP-DPPA4 probed with anti-DPPA4 to show specificity of purified MBP-DPPA4. (D) Western Blot of purified MBP-DPPA2 probed with anti-DPPA2 to show specificity of purified MBP-DPPA2.
Supplemental Figure S2. DPPA2 binds to DPPA4 directly in vitro. MBP or MBP-DPPA2 was bound with amylose beads and incubated with in vitro transcribed and translated DPPA2 protein, beads were washed, and bound protein was detected through Western blotting with anti-DPPA2 antibody.
Supplemental Figure S3. p42 demonstrates higher percentage binding to DPPA2. (A) Quantitation of Figure 3C. Western blot analysis of p48-HA or p42-HA binding to DPPA4 show that both can bind to DPPA4 in vitro. (B) Quantitation of Figure 3D. Western blot analysis of p48-HA or p42-HA shows that p42 can interact with DPPA2 to a higher percentage over input compared to p48, in vitro.
Supplemental Figure S4. Endogenous p48, but not p42, binds to DPPA4. (A) Western blot of Figure 4C probed with anti-MBP to demonstrate presence of purified MBP proteins in hybrid pulldown assays. (B) Quantitation of Figure 4C. Western blot analysis of p48 or p42 in NT2 cells bound to MBP-DPPA4 or MBP-DPPA2. (C) Quantitation of panel 4E. MBP-tagged proteins normalized to MBP show quantity of protein in HDF-hybrid pulldown assays. (D) Quantitation of Figure 4D. Western blot analysis of p48 or p42 binding in H9 ESCs to MBP-DPPA4 or MBP-DPPA2. (E) Quantitation of Figure 4D. Western blot analysis of p48 or p42 in IPS cells bound to MBP-DPPA4 and MBP-DPPA2.
Supplemental Figure S5. Variable effects of differentiation on EBP1-DPPA4 binding in hIPSC. (A) Whole cell lysates of hIPSC differentiated for 7 days with Gibco Neural Induction media (catalog A1647801) produced neurally induced (NI) cells. Alternatively, we cultured hIPSC in suspension with low bFGF media (20% KOSR, 3% FBS, 1% Glutamax, 1% NEAA, 0.0007% B-mercaptoethanol and 2 mg/ml bFGF) for 7-15 days to form embryoid bodies (EBs). Western blot of whole cell lysate probed with EBP1, OCT4, DPPA4 and B-actin shows strong to complete elimination of pluripotency factors in both NI cells and EBs. (B) (Top) Differentiated or hIPSC lysates were incubated with amylose-bound MBP-DPPA4, or MBP as a control, and were washed to remove non-specific binding, and interacting protein was detected through Western blotting with anti-EBP1. Two independent NI cultures were used (NI-1 and NI-2) and three separate cultures of EBs. Undifferentiated whole hIPSC lysates were included as a control. Western blot shown is representative of replicates. Quantitation of the mean values from seven pulldowns for each of the four cell lysate types is shown under the blot for the signal of MBP-DPPA4 binding of EBP1’s p48 isoform, with binding using undifferentiated hIPSC lysate set as 1.0. In each case signal was normalized to percent input. An overexposed (OE) blot for the EB pulldown is shown at right to show that some faint binding to EBP1 is apparent and quantifiable. Only NI-2 exhibited a statistically significant change in binding, with a two-fold decrease relative to hIPSC. (Bottom) Western blot analysis of the amount of MBP or MBP-DPPA4 protein loaded in each pulldown.
Supplemental Figure S6. EBP1 siRNA results in p48 knockdown specifically. (A) Quantitation of p48 protein levels of Western blot in Figure 6D. (B) Quantitation of p42 protein levels of Western blot in Figure 6D. (C) Expression of GAL4-DPPA4 plasmids in luciferase assay experiments. Arrow indicates the expected band for GAL4-DPPA4 or GAL4-DPPA4-ΔSAP.


