Summary
Cardiac hypertrophy accompanies many forms of cardiovascular diseases. The mechanisms behind the development and regulation of cardiac hypertrophy in the human setting are poorly understood, which can be partially attributed to the lack of a human cardiomyocyte-based preclinical test system recapitulating features of diseased myocardium. The objective of our study is to determine whether human embryonic stem cell-derived cardiomyocytes (hESC-CMs) subjected to mechanical stretch can be used as an adequate in vitro model for studying molecular mechanisms of cardiac hypertrophy. We show that hESC-CMs subjected to cyclic stretch, which mimics mechanical overload, exhibit essential features of a hypertrophic state on structural, functional, and gene expression levels. The presented hESC-CM stretch approach provides insight into molecular mechanisms behind mechanotransduction and cardiac hypertrophy and lays groundwork for the development of pharmacological approaches as well as for discovering potential circulating biomarkers of cardiac dysfunction.
Keywords: stem cells, human cardiomyocytes, hypertrophy, in vitro disease modeling, cardiomyocytes stretch response, mechanotransduction
Highlights
-
•
Cyclic stretch leads to the changes in hESC-CM size, contractility, and elasticity
-
•
hESC-CM stretch results in release of cardiac injury mediators
-
•
RNA-seq reveals signaling pathways associated with cardiac hypertrophy
-
•
Knockdown of FSTL3 and SP6 genes blunted the hypertrophic response
In this article, Berezikov, van der Meer, and colleagues used stem cell-derived cardiomyocytes to model human cardiac hypertrophy. Their approach provides novel insights into molecular mechanisms behind mechanotransduction and cardiac hypertrophy and lays groundwork for the development of new pharmacological approaches as well as for discovering new potential circulating biomarkers of cardiac dysfunction.
Introduction
Heart diseases are among the leading causes of death worldwide (Roth et al., 2017). Cardiac hypertrophy is a common manifestation of many forms of cardiac disease, including heart failure, myocardial infarction, and hypertension (Frey et al., 2004).
An important cause of cardiac hypertrophy is mechanical stretch (Frey et al., 2004, Ruwhof and van der Laarse, 2000). Due to volume or pressure overload, the heart starts sending stress signals activating a hypertrophic response program to compensate the wall stress. Persistent wall stress leads to progressive cardiac remodeling and eventually the heart goes into a failing state (Frey et al., 2004, Ruwhof and van der Laarse, 2000). Understanding the molecular mechanisms underlying development of cardiac hypertrophy is essential for advancing treatment of cardiac disease. Preclinical studies indicate that the hypertrophic response is detrimental from the start and therefore specific growth pathways responsible for hypertrophy may emerge as direct targets for therapeutic interventions (Schiattarella et al., 2017). Understanding the sequence of intracellular molecular events and mechanisms underlying the development of cardiac hypertrophy is essential for the treatment of cardiovascular disease. Adequate research models are needed for functional studies of molecular mechanisms of cardiac hypertrophy. Human in vitro models are limited by the amount of patient-derived primary cells of cardiovascular lineage and poor consistency, while existing animal models do not always accurately represent the mechanisms responsible for cardiac hypertrophy development due to significant inter-species differences in contractile features, stress response, and ion channel expression and distribution (Denning et al., 2016, Garbern et al., 2013, Sala et al., 2016). In addition, employment of stem cell-derived organ-specific cells in disease modeling would align with the 3Rs principles of refine, reduce, and replace the use of animals in research and answer to the incentives to limit the use of animal testing and help to replace it with alternatives, including the development of “human-on-a-chip” technology (report on a European Commission scientific conference, Cronin, 2017).
Cardiomyocytes (CMs) differentiated from human embryonic stem cells (hESCs) are a powerful tool for investigating cardiac development, function, and pathophysiology (Davis et al., 2012, Elliott et al., 2011). Subsequently, the availability of human embryonic stem cell-derived CMs (hESC-CMs) provided a relatively cheap platform for drug testing (Li et al., 2017, Mordwinkin et al., 2013). However, the use of hESC-CMs for studies of cardiac hypertrophy and downstream molecular effects has not been clearly demonstrated (Benam et al., 2015, Földes et al., 2011). The mechanic force overload, or stretch model, is based on induction of mechanical stress by physical stretching of CMs. This is a sustained in vitro model that mimics volume overload on the heart during cardiac hypertrophy development (Ruwhof and van der Laarse, 2000, Yamazaki et al., 1995, Yamazaki et al., 1998). An additional advantage of the stretch in vitro model compared with neurohumoral stimulation of CMs (i.e., phenylephrine stimulation) is that it allows researchers to distinguish direct effects of increased biomechanical load from secondary neurohumoral activation (Frank et al., 2008).
The aim of this study is to determine whether hESC-CMs subjected to mechanical stretch can be used as an informative in vitro model for the investigation of molecular mechanisms of CM hypertrophy and identification of potential targets involved in this process.
Results
Generation and Characterization of hESC-CMs
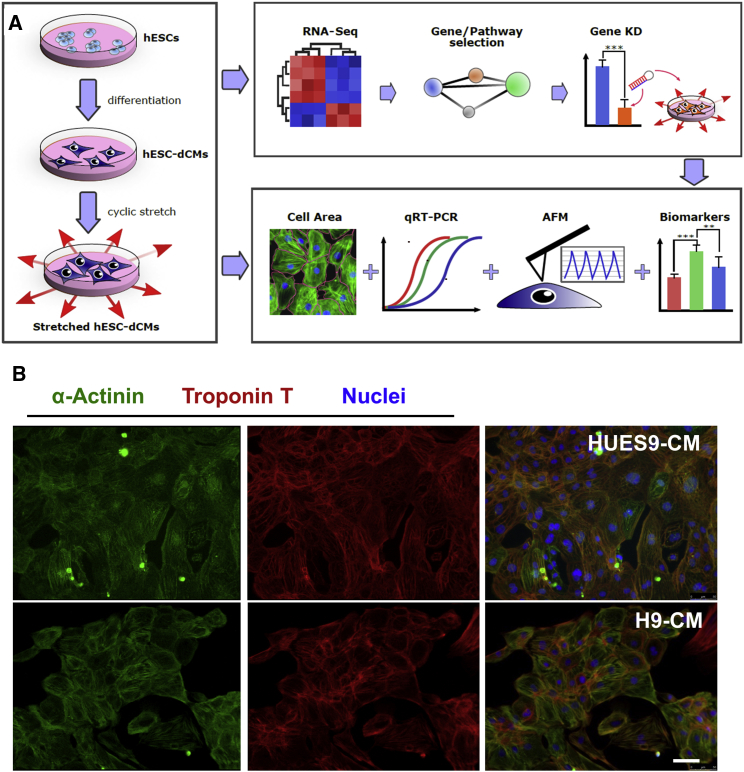
CMs were generated from two independent previously characterized hESC lines, HUES9 and H9, using small molecule-modulated differentiation and subsequent lactate purification (Burridge et al., 2014, Scott et al., 2009). Purified CMs were subjected to mechanical stretch (Figure S1A) and examined for cardiac hypertrophy-related alterations with several assays (Figure 1A). CM derived from both lines expressed standard CM-specific markers (Figure 1B) and exhibited spontaneous beating (Movies S1 and S2).
Figure 1.
Study Workflow
(A) CMs were subjected to mechanical stress for 48 hr and assessed for changes in cell size, gene expression, cell stiffness, reactive oxygen species (ROS) production, and stress marker release. AFM, atomic force microscopy; RNA-seq, RNA sequencing; hESC-dCMs, human embryonic stem cell-derived cardiomyocytes; KD, knockdown. ∗∗ p < 0.01, ∗∗∗ p < 0.001.
(B) Fluorescent microscopy images of differentiated hESC-CMs expressing cardiac-specific markers cardiac troponin T (TnT) and a-actinin (ACTN2). DAPI staining was used to label nuclei. Scale bar, 50 μm.
Mechanical Stretch Induces Hypertrophy, Reactive Oxygen Species Production, Cell Death, and Fetal Genes Expression in hESC-CMs
Heart cells are exposed to rhythmic contraction or regular cyclic stretch throughout adult life. However, chronic biomechanical stress is also an attribute of arterial hypertension or valvular heart disease, eventually leading to cardiac remodeling and cardiac hypertrophy development (Frey and Olson, 2003). There are several stretch parameters that could influence the hypertrophy response, including the duration of stretch and the amount of stretch. Before extensive phenotyping was started, we studied several conditions, including different durations of stretch (7, 24, and 48 hr) and different amounts of stretch (5% versus 15%). We applied 15% stretch to monitor the release of cardiac-specific biomarkers after 7, 24, and 48 hr of HUES9-CM stretch. Interestingly, the most robust release of the N-terminal prohormone of brain natriuretic peptide (NT-proBNP), a marker of cardiac stretch, as well as of troponin T (TnT), a marker of CM damage, appeared after 48 hr of 15% stretch (Figure S1B). In addition, mRNA expression level of TnT was not changed during the stretch at different time points, but the expression of NT-proBNP significantly increased after 7 hr of stretch and remained significantly upregulated (Figure S1C). We also looked at two different stretch regimes: 5% and 15%. In our setup, application of 5% to HUES9-CMs for 48 hr did not induce a clear phenotype. We did not record significant changes in TnT release, increase of CM size, or clear induction of stress genes expression (see Figure S2). Therefore, for further experiments, 15% cyclic stretch was applied for 48 hr.
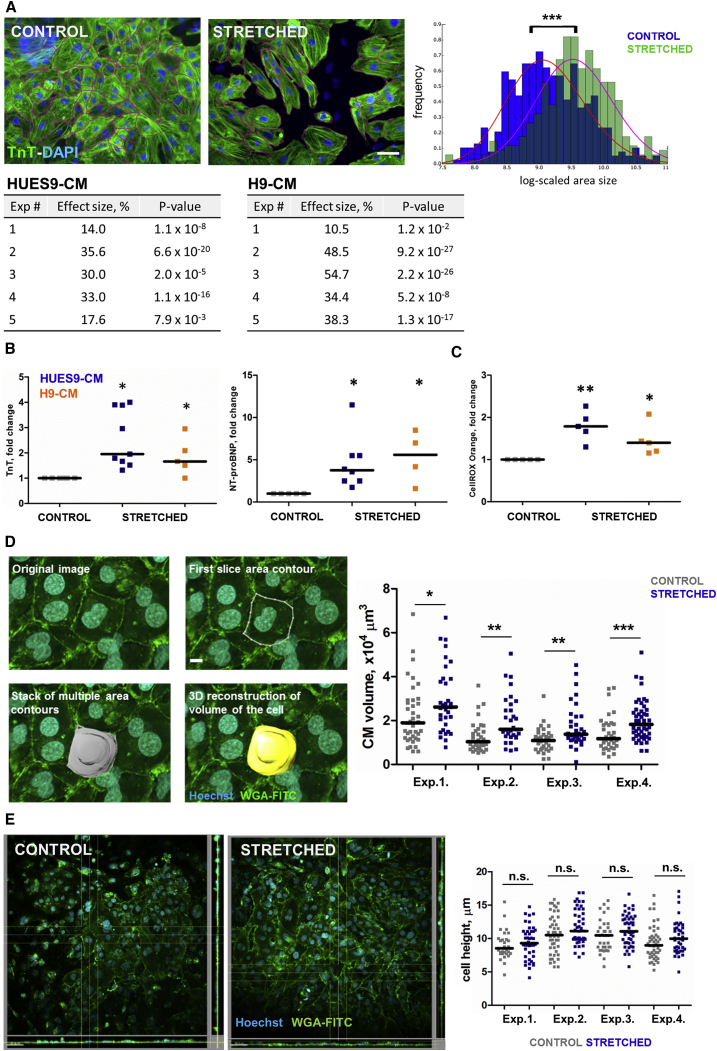
Exposure of hESC-CMs to mechanical stretch for 48 hr caused a significant increase in cell size in HUES9-CMs (26% ± 9.7%, p < 0.001) (Figures 2A and S3). In order to define whether inherited variabilities between hESC lines have an impact on the results, we validated the model using a different hESC line, H9. Cyclic stretch applied to H9-CM resulted in increase of CM size (37% ± 19.6%, p < 0.001), which indicated the robustness of the data (Figure 2A).
Figure 2.
Effects of Mechanical Stretch on hESC-CMs
(A) Changes in CM size. Areas of at least 200 cells per condition per experiment were measured; each experiment represents independent differentiation of hESCs into CMs. Two different hESC lines were used: HUES9 and H9. Scale bar, 50 μm. ∗∗∗p < 0.001 by Mann-Whitney U test.
(B) Levels of TnT and NT-proBNP after 48 hr stretch of HUES9- and H9-derived CMs. ∗p < 0.05.
(C) Levels of total ROS production in HUES9- and H9-derived CMs subjected to mechanical stress. Measured with CellROX Orange assay. For all graphs, each dot represents independent experiment and independent differentiation of hESCs into CMs. For all graphs, HUES9-CM is depicted in blue and H9-CM in orange. ∗p < 0.05, ∗∗p < 0.01.
(D) Changes in CM volume derived from 3D reconstruction of live confocal images of control and stretched CMs; cell membranes were stained with wheat germ agglutinin fluorescein isothiocyanate (WGA-FITC) and nuclei with Hoechst. Images show an example of how the volume reconstruction was performed. Volumes of at least 30 cells per condition per experiment were measured; each experiment represents independent differentiation of HUES9 into CM. Scale bar, 15 μm. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 by Mann-Whitney U test.
(E) Changes in CM height derived from orthogonal projections of live confocal images of control and stretched CMs. Heights of at least 30 cells per condition per experiment were measured; each experiment represents independent differentiation of HUES9 into CM. Scale bar, 50 μm. n.s., not significant by Mann-Whitney U test.
For both HUES9- and H9-derived CMs we observed a more than 2-fold increase of NT-pro-BNP and 1.5-fold increase of TnT release after 48 hr of stretch (Figure 2B). After 48 hr of stretch, a significant increase in reactive oxygen species (ROS) production in HUES9- and H9-derived CMs was observed (Figure 2C). In order to further substantiate our claim that cellular hypertrophy is present, we measured CM volume with the use of confocal laser scanning microscopy (Zeiss LSM 7MP) on live wheat germ agglutinin (WGA)/Hoechst stained cells. Confocal imaging was further combined with three-dimensional (3D) image processing software (Imaris 6.3.5) to generate a 3D structure of the CMs and determine cell volume. We chose not to fix CM and performed experiments on live cells, since fixation causes shrinkage of the cells in the z direction. Our results show that application of 15% stretch leads to a significant increase in CM volume compared with control (+41.7% ± 6.8%, p < 0.05, n = 4), Figure 2D. We performed an additional analysis of orthogonal projections of the confocal images in order to determine the height or thickness of the stretched and control cells. This analysis revealed that the height of stretched CMs was not changed compared with controls (10.78 ± 0.5 μm [stretched] versus 9.97 ± 0.56 μm [controls], p > 0.1, n = 4; Figures 2E and S4D).
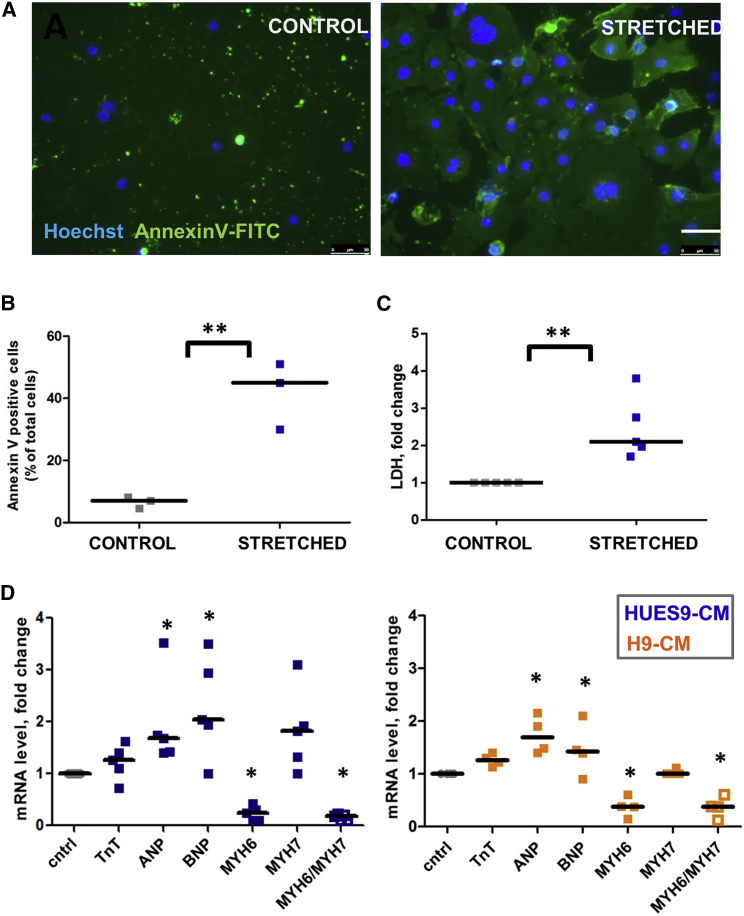
To assess CM apoptosis, an annexin V assay was used. Stretched CMs displayed a 4-fold (p < 0.01) increased staining of annexin V compared with control (Figures 3A and 3B). After 48 hr of stretching we observed a significant 2-fold increase of lactate dehydrogenase (LDH), a marker of necrosis, in the supernatant from stretched cells (Figure 3C).
Figure 3.
Effect of Mechanical Stretch on hESC-CM Viability
(A) Annexin V and Hoechst staining. Scale bar, 50 μm.
(B) The total number of annexin V-positive cells as a percentage of total cell number shown as median of three independent experiments.
(C) Level of LDH after 48 hr stretch of hESC-CMs, shown as a fold change between control and stretched samples obtained from four independent experiments.
(D) qPCR analysis of the mRNA levels of ANP, BNP, TnT, and α- and β-myosin heavy chain (MHC) isoforms in HUES9- and H9-derived CMs after 48 hr stretch. Each dot represents the mean Ct (cycle threshold) values of triplicate measurements normalized against the values obtained for the 36b4 gene for the same sample.
For all graphs, each dot represents an independent experiment and independent differentiation of hESCs into CMs. Median values for each condition are shown. ∗p < 0.05, ∗∗p < 0.01.
Cardiac hypertrophy is often accompanied by reactivation of fetal genes; i.e., genes that are active during fetal cardiac development and quiescent in adult hearts (Chien et al., 1993, Felkin et al., 2011, van der Pol et al., 2017). We tested several such genes and showed that expression of atrial natriuretic peptide (ANP), B-type natriuretic peptide (BNP), but not TnT was altered by mechanical stretch in HUES9- and H9-derived CMs (Figure 3D). In addition, 48 hr of stretch shifted the balance between α- and β-cardiac myosin heavy chain (MYH6 and MYH7, respectively) expression (Figure 3D), which is a common response to cardiac injury and a hallmark of cardiac hypertrophy (Hamdani et al., 2008, Izumo et al., 1987).
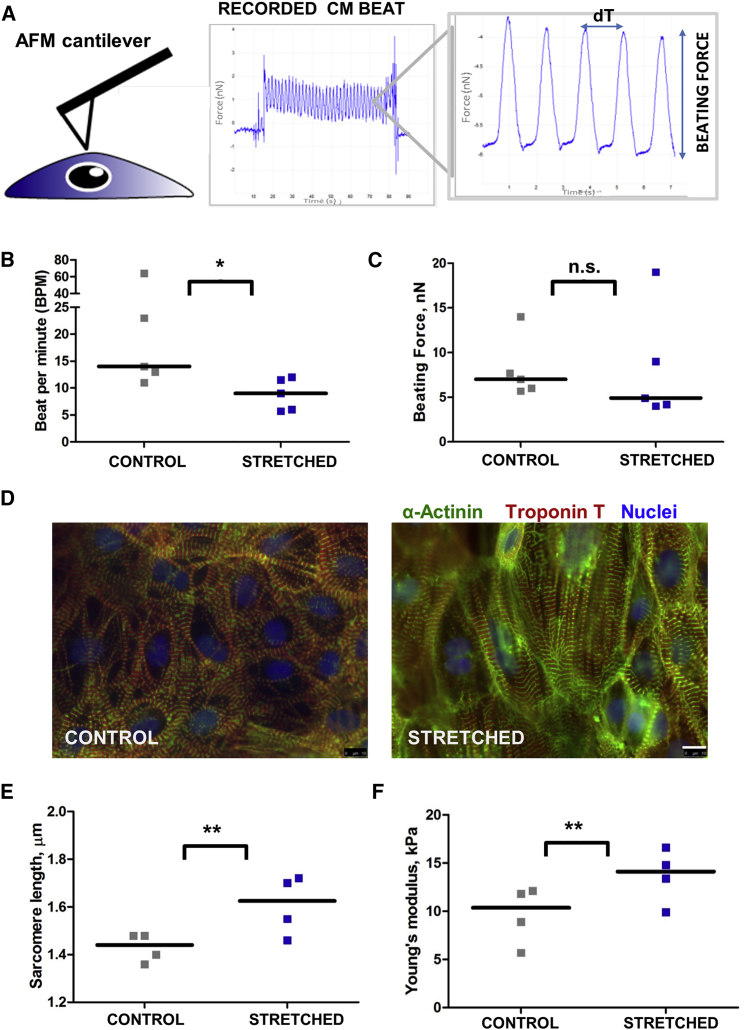
Mechanical Stretch Leads to Contractility Dysfunction and Increases Stiffness of hESC-CMs
In order to determine whether stretch leads to the development of functional defects (e.g., contractile dysfunction), we assessed CM contraction force with atomic force microscopy (AFM) (Figure 4A). CMs subjected to mechanical stress showed a 1.6-fold slower beating frequency but not significantly affected beating force amplitude compared with control (Figures 4B and 4C). However, AFM observations of CM beating behavior have some limitations, since all the measurements were performed at room temperature. We also noticed changes in myofibril structure and sarcomere length after 48 hr of stretch (Figures 4D and 4E). The size of the sarcomeres was significantly increased after stretch (Figures 4D and 4E). We found that myofibrils in stretched hESC-CMs were distributed in parallel and appeared wider, whereas in control CMs myofibrils exhibited a more branched pattern and appeared more spatially separated (Figure 4D). In addition, the elastic modulus, measured with AFM, was significantly higher in stretched CMs compared with untreated control cells, demonstrating that mechanical stretch leads to an increase of CM stiffness (Figure 4F). It is important to note that thinner cells potentially lead to inaccurate measurements of the elastic modulus. However, as described above, stretching did not result in flatter cells (Figures 2D and 2E). Moreover, we were able to image CMs by contact-mode atomic force scan using colloidal probe cantilever and show that there is no significant difference in CM height of control versus stretched CMs (6.1 ± 0.5 versus 6.4 ± 1, p > 0.1, Figure S4). In addition, as was shown before, substrate contributions to the cell's elastic modulus measurement can be neglected if the AFM tip indents less than 10%–20% of the cell thickness (Kuznetsova et al., 2007, Gavara, 2017).
Figure 4.
Effect of Mechanical Stretch on HUES9-CM Contraction and Sarcomere Length
(A) AFM cantilever brought into contact with cardiac cell. The gray box shows typical beating force trajectory. Peaks represent contraction of the CM: height of the peaks characterizes force of the CM beat (nN) and beat-to-beat distance represents beating rate (dT, seconds).
(B) Effect of 48 hr mechanical stretch on CM beating rate, beats per minute (BPM).
(C) Effect of 48 hr mechanical stretch on CM beating force.
(D) Myofibrillar organization in control and stretched HUES9-CM. CMs were stained for troponin T and α-actinin with specific antibodies (red and green, respectively). Nuclei were stained with DAPI. Magnification, 63× (oil). Scale bar, 10 μm.
(E) Effect of mechanical stretch on CM sarcomere length measured as a distance between z-discs.
(F) Young's modulus of the HUES9-CMs subjected to mechanical stretch for 48 hr. The plots show the difference in elasticity for the untreated control cells and stretched CMs.
All graphs show results of at least four independent experiments. Median values for each condition are shown. ∗p < 0.05, ∗∗p < 0.01; n.s., not significant.
Effects of Mechanical Stretch on Gene Expression
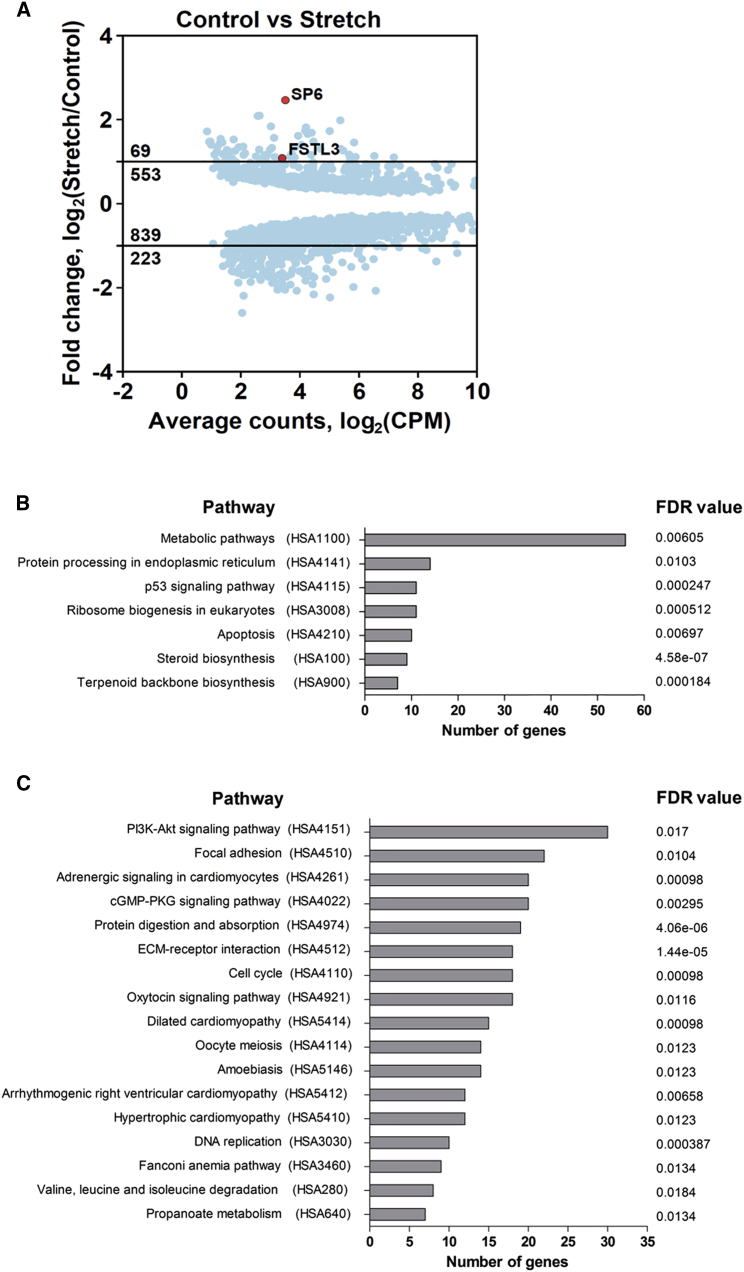
In order to determine genome-wide gene expression changes induced by mechanical stretch we performed RNA sequencing (RNA-seq) of seven independently derived pairs of control and stretched HUES9-CMs. Analysis of RNA-seq data identified 622 upregulated genes and 1022 downregulated genes with a false discovery rate (FDR) below 0.01 (Figure 5A, Tables S1 and S2, a Web interface for data search and visualization is available at http://cardio.genomes.nl). Gene Ontology (GO) term enrichment analysis revealed that downregulated genes were mainly enriched in GOs such as cell cycle, chromosome organization, DNA replication, and animal organ development (Table 1, Figure S4). Interestingly, animal organ development includes such GO terms as regulation of heart contraction, muscle structure development, and muscle system process (Figure S5). Upregulated GO terms were mainly enriched in regulation of apoptotic process, sterol biosynthetic process, and cytoskeleton organization (Table 1, Figure S5). In addition, KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis identified seven pathways that were upregulated (FDR <0.02, Figure 5B, Table S3) and 17 pathways that were downregulated by stretch (FDR < 0.02, Figure 5C, Table S4). Several of the downregulated pathways were enriched for a common set of regulatory (i.e., channels and mediators of Ca2+ concentration across the cell and contractility modulators) and structural genes (i.e., dystrophin, sarcoglycan alpha, members of integrin and collagen families) related to various cardiomyopathies (Table S4). KEGG pathway analysis also pointed toward significant upregulation of apoptosis and of p53 signaling pathways, as well as upregulation of steroid biosynthesis (Figure 5B, Table S3). The KEGG pathway maps (obtained via KEGG Mapper v3.1 and available at http://cardio.genomes.nl) allow further detailed exploration of the networks and the effects of stretch on gene expression.
Figure 5.
Changes in CM Gene Expression upon Mechanical Stretch Identified by RNA-Seq
(A) Mean (average) plots showing statistically significant (FDR < 0.01) differentially expressed genes in HUES9-CMs after 48 hr of stretch versus control. Average gene expression level is shown in counts per million (CPM). Position of SP6 and FSTL3 genes used in subsequent functional studies is indicated in red. See also Tables S1 and S2.
(B) KEGG pathway analysis for upregulated genes in stretched CM. See also Table S3.
(C) KEGG pathway analysis for downregulated genes in stretched CM. See also Table S4.
Table 1.
Significantly Enriched GO Categories for Genes Differentially Expressed upon Stretch
| GO Term | Description | Number of Sub-terms | p Value | FDR q Value |
| UP | ||||
| GO:0042981 | regulation of apoptotic process | 23 | 9.2 × 10−8 | 6.9 × 10−5 |
| GO:0016126 | sterol biosynthesis process | 20 | 4.5 × 10−16 | 6.8 × 10−12 |
| GO:0042221 | response to chemical | 20 | 7.0 × 10−9 | 1.0 × 10−5 |
| GO:0007010 | cytoskeleton organization | 8 | 2.5 × 10−6 | 8.8 × 10−4 |
| GO:0018126 | protein hydroxylation | 3 | 7.6 × 10−8 | 5.9 × 10−5 |
| GO:0031323 | cellular metabolism | 3 | 8.0 × 10−5 | 1.2 × 10−2 |
| DOWN | ||||
| GO:0022402 | cell-cycle process | 26 | 5.6 × 10−20 | 8.4 × 10−16 |
| GO:0071495 | cellular response to endogenous stimulus | 20 | 8.0 × 10−9 | 3.2 × 10−6 |
| GO:0051276 | chromosome organization | 16 | 1.6 × 10−14 | 5.9 × 10−11 |
| GO:0048513 | animal organ development | 14 | 9.8 × 10−12 | 1.3 × 10−8 |
| GO:0006260 | DNA replication | 12 | 8.5 × 10−11 | 1.6 × 10−7 |
(Upper panel) Significantly enriched GO terms in genes upregulated upon stretch. (Lower panel) Significantly enriched GO terms in genes downregulated upon stretch. See also Figure S5.
Dysregulation of SP6 and FSTL3 Expressions Plays a Role in Stretch-Induced Hypertrophy Development in hESC-CMs
We chose to further explore two genes of particular interest. The first gene, specificity protein 6 (SP6), was the most upregulated gene and transcription factor identified in our RNA-seq dataset (Figure 5A). SP6 is a member of the SP family of transcription factors, which contains zinc finger DNA-binding domains in its structure and localizes in the nucleus (Scohy et al., 2000, Talamillo et al., 2010). We tested the hypothesis that knockdown of SP6 expression in human HUES9-CM would prevent the development of stretch-induced hypertrophy. The second gene, follistatin-like 3 (FSTL3), was chosen to validate already known molecular targets of cardiac hypertrophy development. Follistatin (FST) and follistatin-like genes (FSTL1, FSTL3) play an important role in heart failure development, and were linked to both disease severity and mechanisms underlying recovery (Lara-Pezzi et al., 2008, Maruyama et al., 2016, Oshima et al., 2009, Panse et al., 2012, Shimano et al., 2011). Interestingly, all of these genes were dysregulated upon stretch (Tables S1 and S2), with FSTL3 being the most dysregulated among them at ∼2-fold upregulation upon stretch (Figure 5A). Changes in expression of FSTL3 and SP6 upon stretch were also validated on H9-CM (Figure S6A).
HUES9-CMs were transduced by lentiviral vector expressing anti-FSTL3 short hairpin RNA (shRNA), anti-SP6 shRNA, and scrambled control (SCR) shRNA. qRT-PCR analysis confirmed that transduction resulted in more than 70% reduction in FSTL3 and SP6 gene expression compared with the SCR (Figure S6B) and SCR subjected to mechanical stretch (Figure S6C).
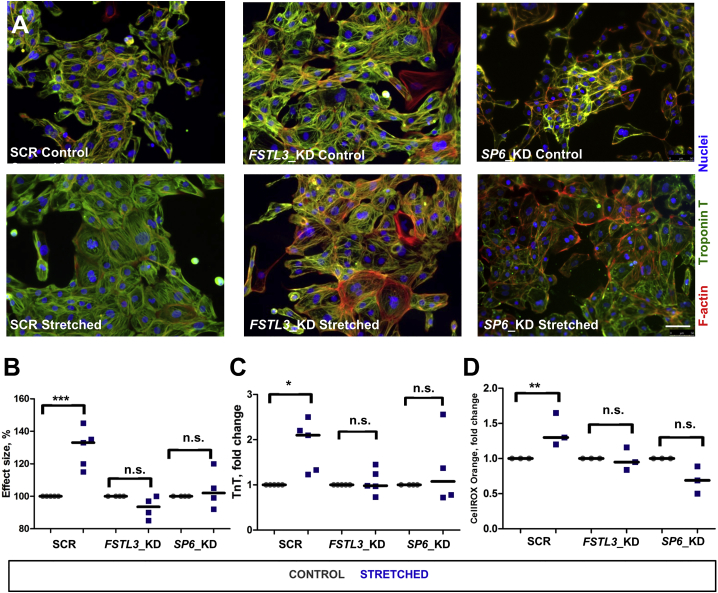
hESC-CMs expressing SCR-, FSTL3-, and SP6-specific shRNAs were stretched for 48 hr and assessed for cell size changes (Figure 6A). In CMs expressing SCR-shRNA, exposure to 15% mechanical stretch led, as expected, to a significant increase of cell size area (Figure 6B), TnT release (Figure 6C), and ROS production (Figure 6D). Knockdown of FSTL3 and SP6 in human CMs resulted in resilience to stretch-induced cardiac hypertrophy and resilience to oxidative stress (Figures 6B–6D).
Figure 6.
Knockdown of FSTL3 and SP6 in HUES9-CMs Decreases Level of Stretch-Induced Hypertrophy
(A) Immunostaining of HUES9-CMs expressing SCR-, FSTL3-, or SP6-specific shRNAs with specific antibodies against cardiac troponin T (green) and phalloidin. Images were taken on the control samples and samples subjected to mechanical stress for 48 hr. Scale bar, 50 μm. See also Figures S3 and S4.
(B) Changes of cell size after stretch in CMs expressing SCR-, FSTL3-, or SP6-specific shRNAs were measured. Measurements were performed in at least four independent experiments. ∗∗∗p < 0.001; n.s., not significant by Mann-Whitney U test.
(C) HUES9-CMs expressing SCR-, Fstl3-, or SP6-specific shRNAs were stretched for 48 hr and troponin T level in the growth media was measured. ∗p < 0.05; n.s., not significant.
(D) HUES9-CM expressing SCR-, Fstl3-, or SP6-specific shRNAs were stretched for 48 hr and levels of total ROS production were measured. ∗∗p < 0.01; n.s., not significant.
Discussion
In the present study, we employ mechanical stretching of hESC-CMs as a human disease model for cardiac hypertrophy. We show dysregulation at the cellular, functional, and genomic levels upon stretching. All specific hypertrophic hallmarks, such as increase of cell size, elevated levels of stress biomarkers, and shift in fetal genes expression, are observed. In addition, we notice an increase in cellular stiffness and decreased contractility accompanied by changes of the sarcomeric structure. Intervention via knockdown of genes of interest resulted in resilience to the hypertrophic phenotype upon stretch.
Several proteins are involved in sensing and responding to stretch in the heart, the so-called mechanosensors (Lyon et al., 2015, Sequeira et al., 2014). Upon mechanical load, alterations in these proteins result in mechanical signaling leading to hypertrophy (Knoll et al., 2011, Lyon et al., 2015). These changes can take place at various sites in the CM, including the sarcomere, sarcolemma, and intercalated disc (Frank and Frey, 2011, Lyon et al., 2015). To determine the global gene expression changes involved in the hypertrophic response to mechanical stress, we used an RNA-seq approach. For instance, the ANKRD1 gene, which is 1.5-fold upregulated in our dataset (Table S1), is a transcription factor that interacts with the sarcomeric protein titin and plays a role in the myofibrillar stretch-sensor system. Increased levels of this protein have been detected in heart tissue of patients suffering from ischemic cardiomyopathy and dilated cardiomyopathy (Herrer et al., 2014, Zheng et al., 2010). It was also demonstrated that ANKRD1 upregulation is associated with altered systolic/diastolic function and that mutations in this gene result in a differential stretch-induced gene expression pattern (Herrer et al., 2014, Moulik et al., 2009). FHL1 is another titin-binding protein that has been shown to be upregulated in mouse models in response to pressure-overload-induced hypertrophy and in the hearts of human patients exhibiting hypertrophic cardiomyopathy (Chu et al., 2000, Lim et al., 2001). This is in line with our RNA-seq data that show a significant 1.5-fold upregulation of FHL1 in stretched hESC-CMs (Table S1). Also other members of the LIM domain family that have been implicated in mechanical strain signaling, such as PDLIM3 and LDB3, were significantly dysregulated upon stretch (2-fold up and 1.5-fold down, respectively). LDB3 and PDLIM3 are known to interact with the α-actinin rod domain within z-disc and ablation of these proteins in mice resulted in abnormal cardiac function and severe myopathy (Gautel, 2008, Zheng et al., 2010).
Our AFM data showed that hESC-CMs subjected to mechanical stretch were significantly stiffer. Increased CM stiffness is one of the hallmarks of myocardial remodeling (Mohamed et al., 2016, Paulus and Tschope, 2013). There are several factors that influence cardiac stiffness, such as changes in sarcomere length, myofibril density, shift in N2A to N2B titin's isoforms expression, and titin phosphorylation (Hutchinson et al., 2015, Mohamed et al., 2016, Paulus and Tschope, 2013). Even though titin expression was not changed upon stretch, KEGG pathway analysis revealed downregulation of cGMP-PKG signaling pathway. This downregulation further contributes to the deficit of titin phosphorylation and overall increase of CM stiffness (Kovacs et al., 2016). Also stiffness increases under oxidative stress conditions due to disulfide-bridge formation, which can occur in titin filaments (Linke and Kruger, 2010). We believe that changes in sarcomeric organization observed upon CM stretch could also contribute to the changes in cardiac stiffness. There is a significant dysregulation in the expression of MYOM2, MYPN, FHL1, SQSTM1, CRYAB, OBSCN, PBK, and MDM2. All these genes encode proteins, interact with titin, and affect myofibril formation (Linke and Kruger, 2010, Sequeira et al., 2014).
In addition to changes in myofibril architecture, regulation of cardiac muscle contraction was one of the significantly downregulated processes according to the GO term enrichment analysis. Our RNA-seq data showed significant deregulation of ion homeostasis, one of the essential regulators of cardiac contraction (Rosati and McKinnon, 2004). We observed significant downregulation of Ca2+-channels, such as RYR2, CACNA1D, CACNG6, CACNA1H, and CACNB2 (Table S2). We also found a downregulation of potassium (K+) and sodium (Na+) channels encoding genes such as SCN9A, SCN8A, ATP1A2, KCNQ1, HCN1, KCNN2, KCNAB2, and KCNH7 (Table S2). Multiple studies demonstrated that alterations in Ca2+ release and disturbance of K+ and Na+ ion channels cause abnormalities in CM beating behavior (Communal et al., 2002, Gorski et al., 2015, Zhang et al., 2013). These observations are also in line with our AFM results showing that stretched CMs need more time to generate beats of the same force amplitude compared with control.
The GO term enrichment analysis of differentially expressed genes showed that mechanical stretch upregulates biological processes such as apoptosis, sterol biosynthesis, and cytoskeleton organization. This is in line with our phenotyping data, which also demonstrate that mechanical stretch causes increase of ROS production, necrosis, as well as apoptosis in CMs, and could be responsible for cell loss. Similar findings have been reported for rat CM and human biopsy studies, in which cardiac hypertrophy development was associated with induction of significant apoptotic and necrotic responses (Condorelli et al., 1999, Fujita and Ishikawa, 2011, Mohamed et al., 2016, Okada et al., 2004).
As the heart begins to fail, disruption of its homeostasis leads to the release of stress-related cytokines, proteins, or peptides, into the circulation. In our in vitro model system NT-pro-BNP and TnT are released in the supernatant upon stretching. Another well-known cardiac biomarker is GDF15. It has been shown to be upregulated in patients with various forms of heart failure and is associated with an impaired prognosis (Dewey et al., 2016, Lok et al., 2013). Interestingly, GDF-15 was among the top ten upregulated genes identified in our RNA-seq dataset. These findings suggest that our in vitro model combined with the RNA-seq approach can be used for identification and validation of circulating biomarkers involved in cardiac failure.
We used obtained RNA-seq data not only for studying potential candidates, such as SP6, the most induced transcription factor in the dataset, but also for validation of previously identified candidate genes such as FSTL3. This gene has been reported as an important player in cardiac remodeling. Expression of FSTL3 was shown to be elevated in tissue biopsies of patients with heart failure and levels correlated with α-skeletal actin and BNP, both markers of disease development (Lara-Pezzi et al., 2008, Oshima et al., 2009, Shimano et al., 2011). As well as BNP, plasma levels of FSTL3 were shown to be increased in heart failure patients and associated with the severity of the disease, suggesting its potential role as a circulating biomarker (Assadi-Khansari et al., 2016). In addition to being secreted, FSTL3 can be found in both the nucleus and cytoplasm, which implies its involvement in transcriptional regulation of cardiac remodeling (Lara-Pezzi et al., 2008, Shimano et al., 2011). Knockdown of FSTL3, as well as SP6, caused significant desensitization of CMs toward stretch-induced stress. This is in line with previously obtained in vivo and in vitro studies that show FSTL3 induction under myocardial stress and its negative effect on CM survival. A cardiac-specific FSTL3 knockout mouse model showed significantly smaller infarct sizes and lower number of apoptotic myocytes after ischemia/reperfusion injury. Knockdown of FSTL3 in cultured rat CMs inhibited phenylephrine-induced cardiac hypertrophy (Oshima et al., 2009, Panse et al., 2012, Shimano et al., 2011). Similarly, in our study, FSTL3 ablation in human CMs subjected to mechanical stretch led to the reduced level of cardiac hypertrophy and less cardiac damage assessed by level of TnT release and ROS production. Transcription factor SP6 has never been studied in relation to cardiac hypertrophy development. However, two other members of the SP family, SP1 and SP3, are both significantly overexpressed in hypertrophied and fetal murine hearts and are believed to be involved in cardiac hypertrophy development (Sack et al., 1997).
Current advances in tissue engineering techniques made 3D cardiac tissue cultures feasible and potentially useful for pharmacological and clinical applications (Feric and Radisic, 2016, Kimbrel and Lanza, 2015, Li et al., 2017, Shinozawa et al., 2017, Weinberger et al., 2016). In addition, two-dimensional (2D) and 3D stem cell-derived CM cultures were used in a number of studies in order to address the immaturity of stem cell-derived CMs by employing a variety of biochemical and biophysical signals (Kosmidis et al., 2015, Jackman et al., 2016, Huebsch et al., 2016, Shen et al., 2017). Furthermore, there are several studies that applied 3D tissue culturing for studying stretch- or phenylephrine-induced cardiac hypertrophy (Rupert et al., 2017, Yang et al., 2016).
We elected to use 2D CM cultures as a basis for our study for several reasons. While 3D tissues provide a more physiologically relevant structure, the major incentive for using this model is the requirement of non-CMs to be incorporated in 3D tissues for optimal function and tissue functionality (Hussain et al., 2013). This would impede the attribution of stretch-induced effects specifically to CM. Moreover, constructed 3D tissues are based on various compositions of extracellular matrix (ECM), which undergoes cell-mediated remodeling at all times. Consequently, changes in ECM characteristics might alter the strain on specific parts of a tissue, thereby making the tissue unstable and prone to break, and reproducibility is rendered difficult. 3D tissues also have different diffusion rates of soluble factors depending on the location of individual cells within the tissue, resulting in heterogeneous cell behavior and therefore matrix remodeling. Combined, these traits of 3D tissues make characterization (of tissue as a whole) more complicated than with 2D cultures (Riehl et al., 2012).
To summarize, in the present study we show that mechanical stretching of hESC-CMs can serve as an in vitro disease model for studying human CM hypertrophy. This model broadens our understanding of the molecular mechanisms behind mechanotransduction and cardiac hypertrophy and can serve as a launch platform for development of pharmacological approaches as well as for discovering potential circulating biomarkers of cardiac dysfunction.
Experimental Procedures
Full experimental procedures are provided in the Supplemental Information section.
Cardiac Differentiation of hESCs
To differentiate hESCs into CMs, a small molecule-based protocol utilizing modulation of Wnt/β-catenin signaling was employed. hESCs were treated with 6 μM GSK3-β inhibitor CHIR99021 (Cayman Chemicals) for 2 days, followed by inhibition of WNT signaling using 2 μM Wnt-C59 (Tocris Bioscience). To further increase CM population purity, metabolic differences between CMs and non-CMs were exploited and cells were subjected to glucose starvation in presence of 5 mM DL-lactate for 6 days. For a more detailed protocol, see the Supplemental Information.
RNA-Seq Data Accessibility
A Web interface that provides search and visualization capabilities for the generated datasets is available at cardio.genomes.nl.
Statistical Analysis
Values are displayed as medians of at least three independently performed experiments. For all graphs, each dot represents results of independent experiments performed on the independently derived CM, and each dot is a mean of at least two technical replicates. Unless stated otherwise, statistical comparisons were performed using two-tailed, unpaired Student's t test (Prism, GraphPad Software). For qRT-PCR, analysis was performed using GenEx software (MultiD Analyses AB). The following indications of significance were used throughout the manuscript: ∗p < 0.05, ∗∗p < 0.01 ∗∗∗p < 0.001.
Author Contributions
E.O., M.H., E.B., and P.v.d.M. designed the experiments. E.O., M.H., K.U., and E.B. performed experiments and collected and analyzed the data. N.B. and T.V.d.J. provided technical support. H.v.d.M. gave conceptual advice. E.O., M.H., E.B., and P.v.d.M. wrote the manuscript.
Acknowledgments
We thank Joop de Vries for assistance with implementing the AFM protocol for studying hESC-CMs and help with analysis of the data. We also thank Klaas Sjolemma for technical assistance and help with obtaining and analyzing confocal images. Confocal imaging was performed at UMCG Microscopy and Imaging Center (UMIC), sponsored by NOW grant 175-010-2009-023. This research was supported by the Dutch Heart Foundation (grant 2012T047 to P.v.d.M.).
Published: February 15, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, seven tables, and two movies and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.01.016.
Contributor Information
Eugene Berezikov, Email: e.berezikov@umcg.nl.
Peter van der Meer, Email: p.van.der.meer@umcg.nl.
Accession Numbers
The accession numbers for the RNA-seq data reported in this paper are GenBank: SRR5875410–SRR5875423.
Supplemental Information
The table represents fold change of gene expression, significance (p value and FDR), and gene description.
The table represents fold change of gene expression, significance (p value and FDR), and gene description.
References
- Assadi-Khansari B., Liu S., Ajero C., Chua S., Horowitz J., Sverdlov A., Ngo D. Follistatin-like 3 is elevated in acute heart failure patients. Heart Lung Circ. 2016;25:S109–S110. [Google Scholar]
- Benam K.H., Dauth S., Hassell B., Herland A., Jain A., Jang K.J., Karalis K., Kim H.J., MacQueen L., Mahmoodian R. Engineered in vitro disease models. Annu. Rev. Pathol. 2015;10:195–262. doi: 10.1146/annurev-pathol-012414-040418. [DOI] [PubMed] [Google Scholar]
- Burridge P.W., Matsa E., Shukla P., Lin Z.C., Churko J.M., Ebert A.D., Lan F., Diecke S., Huber B., Mordwinkin N.M. Chemically defined generation of human cardiomyocytes. Nat. Methods. 2014;11:855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien K.R., Zhu H., Knowlton K.U., Miller-Hance W., van-Bilsen M., O’Brien T.X., Evans S.M. Transcriptional regulation during cardiac growth and development. Annu. Rev. Physiol. 1993;55:77–95. doi: 10.1146/annurev.ph.55.030193.000453. [DOI] [PubMed] [Google Scholar]
- Chu P.H., Ruiz-Lozano P., Zhou Q., Cai C., Chen J. Expression patterns of FHL/SLIM family members suggest important functional roles in skeletal muscle and cardiovascular system. Mech. Dev. 2000;95:259–265. doi: 10.1016/s0925-4773(00)00341-5. [DOI] [PubMed] [Google Scholar]
- Communal C., Sumandea M., de Tombe P., Narula J., Solaro R.J., Hajjar R.J. Functional consequences of caspase activation in cardiac myocytes. Proc. Natl. Acad. Sci. USA. 2002;99:6252–6256. doi: 10.1073/pnas.092022999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condorelli G., Morisco C., Stassi G., Notte A., Farina F., Sgaramella G., de Rienzo A., Roncarati R., Trimarco B., Lembo G. Increased cardiomyocyte apoptosis and changes in proapoptotic and antiapoptotic genes bax and bcl-2 during left ventricular adaptations to chronic pressure overload in the rat. Circulation. 1999;99:3071–3078. doi: 10.1161/01.cir.99.23.3071. [DOI] [PubMed] [Google Scholar]
- Cronin, M. (2017). Non-animal approaches - the way forward. Report on a European Commission scientific conference held on 6-7 December, 2016, at the Egg, Brussels, Belgium. doi:10.2779/373944.
- Davis R.P., Casini S., van den Berg C.W., Hoekstra M., Remme C.A., Dambrot C., Salvatori D., Oostwaard D.W., Wilde A.A.M., Bezzina C.R. Cardiomyocytes derived from pluripotent stem cells recapitulate electrophysiological characteristics of an overlap syndrome of cardiac sodium channel disease. Circulation. 2012;125:3079–3091. doi: 10.1161/CIRCULATIONAHA.111.066092. [DOI] [PubMed] [Google Scholar]
- Denning C., Borgdorff V., Crutchley J., Firth K.S.A., George V., Kalra S., Kondrashov A., Hoang M.D., Mosqueira D., Patel A. Cardiomyocytes from human pluripotent stem cells: from laboratory curiosity to industrial biomedical platform. Biochim. Biophys. Acta. 2016;1863:1728–1748. doi: 10.1016/j.bbamcr.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey C.M., Spitler K.M., Ponce J.M., Hall D.D., Grueter C.E. Cardiac-secreted factors as peripheral metabolic regulators and potential disease biomarkers. J. Am. Heart Assoc. 2016;5 doi: 10.1161/JAHA.115.003101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott D.A., Braam S.R., Koutsis K., Ng E.S., Jenny R., Lagerqvist E.L., Biben C., Hatzistavrou T., Hirst C.E., Yu Q.C. NKX2-5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat. Methods. 2011;8:1037–1040. doi: 10.1038/nmeth.1740. [DOI] [PubMed] [Google Scholar]
- Felkin L.E., Lara-Pezzi E.A., Hall J.L., Birks E.J., Barton P.J. Reverse remodelling and recovery from heart failure are associated with complex patterns of gene expression. J. Cardiovasc. Transl. Res. 2011;4:321–331. doi: 10.1007/s12265-011-9267-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric N.T., Radisic M. Strategies and challenges to myocardial replacement therapy. Stem Cells Transl. Med. 2016;5:410–416. doi: 10.5966/sctm.2015-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Földes G., Mioulane M., Wright J.S., Liu A.Q., Novak P., Merkely B., Gorelik J., Schneider M.D., Ali N.N., Harding S.E. Modulation of human embryonic stem cell-derived cardiomyocyte growth: a testbed for studying human cardiac hypertrophy? J. Mol. Cell. Cardiol. 2011;50:367–376. doi: 10.1016/j.yjmcc.2010.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D., Frey N. Cardiac Z-disc signaling network. J. Biol. Chem. 2011;286:9897–9904. doi: 10.1074/jbc.R110.174268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D., Kuhn C., Brors B., Hanselmann C., Lüdde M., Katus H.A., Frey N. Gene expression pattern in biomechanically stretched cardiomyocytes: evidence for a stretch-specific gene program. Hypertension. 2008;51:309–318. doi: 10.1161/HYPERTENSIONAHA.107.098046. [DOI] [PubMed] [Google Scholar]
- Frey N., Olson E.N. Cardiac hypertrophy: the good, the bad, and the ugly. Annu. Rev. Physiol. 2003;65:45–79. doi: 10.1146/annurev.physiol.65.092101.142243. [DOI] [PubMed] [Google Scholar]
- Frey N., Katus H.A., Olson E.N., Hill J.A. Hypertrophy of the heart: a new therapeutic target? Circulation. 2004;109:1580–1589. doi: 10.1161/01.CIR.0000120390.68287.BB. [DOI] [PubMed] [Google Scholar]
- Fujita T., Ishikawa Y. Apoptosis in heart failure. -The role of the beta-adrenergic receptor-mediated signaling pathway and p53-mediated signaling pathway in the apoptosis of cardiomyocytes- Circ. J. 2011;75:1811–1818. doi: 10.1253/circj.cj-11-0025. [DOI] [PubMed] [Google Scholar]
- Garbern J.C., Mummery C.L., Lee R.T. Model systems for cardiovascular regenerative biology. Cold Spring Harb. Perspect. Med. 2013;3:a014019. doi: 10.1101/cshperspect.a014019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautel M. The sarcomere and the nucleus: functional links to hypertrophy, atrophy and sarcopenia. Adv. Exp. Med. Biol. 2008;642:176–191. doi: 10.1007/978-0-387-84847-1_13. [DOI] [PubMed] [Google Scholar]
- Gavara N. A beginner’s guide to atomic force microscopy probing for cell mechanics. Microsc. Res. Tech. 2017;80:75–84. doi: 10.1002/jemt.22776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski P.A., Ceholski D.K., Hajjar R.J. Altered myocardial calcium cycling and energetics in heart failure–a rational approach for disease treatment. Cell Metab. 2015;21:183–194. doi: 10.1016/j.cmet.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdani N., Kooij V., van Dijk S., Merkus D., Paulus W.J., Remedios C.D., Duncker D.J., Stienen G.J., van der Velden J. Sarcomeric dysfunction in heart failure. Cardiovasc. Res. 2008;77:649–658. doi: 10.1093/cvr/cvm079. [DOI] [PubMed] [Google Scholar]
- Herrer I., Rosello-Lleti E., Rivera M., Molina-Navarro M.M., Tarazon E., Ortega A., Martinez-Dolz L., Trivino J.C., Lago F., Gonzalez-Juanatey J.R. RNA-sequencing analysis reveals new alterations in cardiomyocyte cytoskeletal genes in patients with heart failure. Lab. Invest. 2014;94:645–653. doi: 10.1038/labinvest.2014.54. [DOI] [PubMed] [Google Scholar]
- Hussain A., Collins G., Yip D., Cho C.H. Functional 3-D cardiac co-culture model using bioactive chitosan nanofiber scaffolds. Biotechnol. Bioeng. 2013;110:637–647. doi: 10.1002/bit.24727. [DOI] [PubMed] [Google Scholar]
- Huebsch N., Loskill P., Deveshwar N., Spencer C.I., Judge L.M., Mandegar M.A., Fox C.B., Mohamed T.M.A., Ma Z., Mathur A. Miniaturized iPS-cell-derived cardiac muscles for physiologically relevant drug response analyses. Sci. Rep. 2016;6:24726. doi: 10.1038/srep24726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson K.R., Saripalli C., Chung C.S., Granzier H. Increased myocardial stiffness due to cardiac titin isoform switching in a mouse model of volume overload limits eccentric remodeling. J. Mol. Cell. Cardiol. 2015;79:104–114. doi: 10.1016/j.yjmcc.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumo S., Lompré A.M., Matsuoka R., Koren G., Schwartz K., Nadal-Ginard B., Mahdavi V. Myosin heavy chain messenger RNA and protein isoform transitions during cardiac hypertrophy. Interaction between hemodynamic and thyroid hormone-induced signals. J. Clin. Invest. 1987;79:970–977. doi: 10.1172/JCI112908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman C.P., Carlson A.L., Bursac N. Dynamic culture yields engineered myocardium with near-adult functional output. Biomaterials. 2016;111:66–79. doi: 10.1016/j.biomaterials.2016.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimbrel E.A., Lanza R. Current status of pluripotent stem cells: moving the first therapies to the clinic. Nat. Rev. Drug Discov. 2015;14:681. doi: 10.1038/nrd4738. [DOI] [PubMed] [Google Scholar]
- Knoll R., Buyandelger B., Lab M. The sarcomeric Z-disc and Z-discopathies. J. Biomed. Biotechnol. 2011;2011:569628. doi: 10.1155/2011/569628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmidis G., Bellin M., Ribeiro M.C., Van Meer B., Ward-Van Oostwaard D., Passier R., Tertoolen L.G.J., Mummery C.L., Casini S. Altered calcium handling and increased contraction force in human embryonic stem cell derived cardiomyocytes following short term dexamethasone exposure. Biochem. Biophys. Res. Commun. 2015;4:998–1005. doi: 10.1016/j.bbrc.2015.10.026. [DOI] [PubMed] [Google Scholar]
- Kovacs A., Alogna A., Post H., Hamdani N. Is enhancing cGMP-PKG signalling a promising therapeutic target for heart failure with preserved ejection fraction? Neth. Heart J. 2016;24:268–274. doi: 10.1007/s12471-016-0814-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsova T.G., Starodubtseva M.N., Yegorenkov N.I., Chizhik S.A., Zhdanov R.I. Atomic force microscopy probing of cell elasticity. Micron. 2007;38:824–833. doi: 10.1016/j.micron.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Lara-Pezzi E., Felkin L.E., Birks E.J., Sarathchandra P., Panse K.D., George R., Hall J.L., Yacoub M.H., Rosenthal N., Barton P.J. Expression of follistatin-related genes is altered in heart failure. Endocrinology. 2008;149:5822–5827. doi: 10.1210/en.2008-0151. [DOI] [PubMed] [Google Scholar]
- Li J., Minami I., Shiozaki M., Yu L., Yajima S., Miyagawa S., Shiba Y., Morone N., Fukushima S., Yoshioka M. Human pluripotent stem cell-derived cardiac tissue-like constructs for repairing the infarcted myocardium. Stem Cell Reports. 2017;9:1546–1559. doi: 10.1016/j.stemcr.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim D.S., Roberts R., Marian A.J. Expression profiling of cardiac genes in human hypertrophic cardiomyopathy: insight into the pathogenesis of phenotypes. J. Am. Coll. Cardiol. 2001;38:1175–1180. doi: 10.1016/s0735-1097(01)01509-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke W.A., Kruger M. The giant protein titin as an integrator of myocyte signaling pathways. Physiology (Bethesda) 2010;25:186–198. doi: 10.1152/physiol.00005.2010. [DOI] [PubMed] [Google Scholar]
- Lok D.J., Klip I.T., Lok S.I., de la Porte P.W.B.-A., Badings E., van Wijngaarden J., Voors A.A., de Boer R.A., van Veldhuisen D.J., van der Meer P. Incremental prognostic power of novel biomarkers (growth-differentiation factor-15, high-sensitivity C-reactive protein, galectin-3, and high-sensitivity troponin-T) in patients with advanced chronic heart failure. Am. J. Cardiol. 2013;112:831–837. doi: 10.1016/j.amjcard.2013.05.013. [DOI] [PubMed] [Google Scholar]
- Lyon R.C., Zanella F., Omens J.H., Sheikh F. Mechanotransduction in cardiac hypertrophy and failure. Circ. Res. 2015;116:1462–1476. doi: 10.1161/CIRCRESAHA.116.304937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama S., Nakamura K., Papanicolaou K.N., Sano S., Shimizu I., Asaumi Y., van den Hoff M.J., Ouchi N., Recchia F.A., Walsh K. Follistatin-like 1 promotes cardiac fibroblast activation and protects the heart from rupture. EMBO Mol. Med. 2016;8:949–966. doi: 10.15252/emmm.201506151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed B.A., Schnelle M., Khadjeh S., Lbik D., Herwig M., Linke W.A., Hasenfuss G., Toischer K. Molecular and structural transition mechanisms in long-term volume overload. Eur. J. Heart Fail. 2016;18:362–371. doi: 10.1002/ejhf.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordwinkin N.M., Burridge P.W., Wu J.C. A review of human pluripotent stem cell-derived cardiomyocytes for high-throughput drug discovery, cardiotoxicity screening, and publication standards. J. Cardiovasc. Transl. Res. 2013;6:22–30. doi: 10.1007/s12265-012-9423-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulik M., Vatta M., Witt S.H., Arola A.M., Murphy R.T., McKenna W.J., Boriek A.M., Oka K., Labeit S., Bowles N.E. ANKRD1, the gene encoding cardiac ankyrin repeat protein, is a novel dilated cardiomyopathy gene. J. Am. Coll. Cardiol. 2009;54:325–333. doi: 10.1016/j.jacc.2009.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K., Minamino T., Tsukamoto Y., Liao Y., Tsukamoto O., Takashima S., Hirata A., Fujita M., Nagamachi Y., Nakatani T. Prolonged endoplasmic reticulum stress in hypertrophic and failing heart after aortic constriction: possible contribution of endoplasmic reticulum stress to cardiac myocyte apoptosis. Circulation. 2004;110:705–712. doi: 10.1161/01.CIR.0000137836.95625.D4. [DOI] [PubMed] [Google Scholar]
- Oshima Y., Ouchi N., Shimano M., Pimentel D.R., Papanicolaou K.N., Panse K.D., Tsuchida K., Lara-Pezzi E., Lee S.J., Walsh K. Activin A and follistatin-like 3 determine the susceptibility of heart to ischemic injury. Circulation. 2009;120:1606–1615. doi: 10.1161/CIRCULATIONAHA.109.872200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panse K.D., Felkin L.E., Lopez-Olaneta M.M., Gomez-Salinero J., Villalba M., Munoz L., Nakamura K., Shimano M., Walsh K., Barton P.J. Follistatin-like 3 mediates paracrine fibroblast activation by cardiomyocytes. J. Cardiovasc. Transl. Res. 2012;5:814–826. doi: 10.1007/s12265-012-9400-9. [DOI] [PubMed] [Google Scholar]
- Paulus W.J., Tschope C. A novel paradigm for heart failure with preserved ejection fraction: comorbidities drive myocardial dysfunction and remodeling through coronary microvascular endothelial inflammation. J. Am. Coll. Cardiol. 2013;62:263–271. doi: 10.1016/j.jacc.2013.02.092. [DOI] [PubMed] [Google Scholar]
- Riehl B.D., Park J.H., Kwon I.K., Lim J.Y. Mechanical stretching for tissue engineering: two-dimensional and three-dimensional constructs. Tissue Eng. Part B Rev. 2012;18:288–300. doi: 10.1089/ten.teb.2011.0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosati B., McKinnon D. Regulation of ion channel expression. Circ. Res. 2004;94:874–883. doi: 10.1161/01.RES.0000124921.81025.1F. [DOI] [PubMed] [Google Scholar]
- Roth G.A., Johnson C., Abajobir A., Abd-Allah F., Abera S.F., Abyu G., Ahmed M., Aksut B., Alam T., Alam K. Global, regional, and national burden of cardiovascular diseases for 10 causes, 1990 to 2015. J. Am. Coll. Cardiol. 2017;70:1–25. doi: 10.1016/j.jacc.2017.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupert C.E., Chang H.H., Coulombe K.L.K. Hypertrophy changes 3D shape of hiPSC-cardiomyocytes: implications for cellular maturation in regenerative medicine. Cell. Mol. Bioeng. 2017;10:54–62. doi: 10.1007/s12195-016-0462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruwhof C., van der Laarse A. Mechanical stress-induced cardiac hypertrophy: mechanisms and signal transduction pathways. Cardiovasc. Res. 2000;47:23–37. doi: 10.1016/s0008-6363(00)00076-6. [DOI] [PubMed] [Google Scholar]
- Sack M.N., Disch D.L., Rockman H.A., Kelly D.P. A role for Sp and nuclear receptor transcription factors in a cardiac hypertrophic growth program. Proc. Natl. Acad. Sci. USA. 1997;94:6438–6443. doi: 10.1073/pnas.94.12.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala L., Bellin M., Mummery C.L. Integrating cardiomyocytes from human pluripotent stem cells in safety pharmacology: has the time come? Br. J. Pharmacol. 2016 doi: 10.1111/bph.13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiattarella G.G., Hill T.M., Hill J.A. Is load-induced ventricular hypertrophy ever compensatory? Circulation. 2017;136:1273–1275. doi: 10.1161/CIRCULATIONAHA.117.030730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scohy S., Gabant P., Van Reeth T., Hertveldt V., Drèze P.L., Van Vooren P., Rivière M., Szpirer J., Szpirer C. Identification of KLF13 and KLF14 (SP6), novel members of the SP/XKLF transcription factor family. Genomics. 2000;70:93–101. doi: 10.1006/geno.2000.6362. [DOI] [PubMed] [Google Scholar]
- Scott C.T., McCormick J.B., Owen-Smith J. And then there were two: use of hESC lines. Nat. Biotechnol. 2009;27:696–697. doi: 10.1038/nbt0809-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira V., Nijenkamp L.L., Regan J.A., van der Velden J. The physiological role of cardiac cytoskeleton and its alterations in heart failure. Biochim. Biophys. Acta. 2014;1838:700–722. doi: 10.1016/j.bbamem.2013.07.011. [DOI] [PubMed] [Google Scholar]
- Shen N., Knopf A., Westendorf C., Kraushaar U., Riedl J., Bauer H., Pöschel S., Layland S.L., Holeiter M., Knolle S. Steps toward maturation of embryonic stem cell-derived cardiomyocytes by defined physical signals. Stem Cell Reports. 2017;9:122–135. doi: 10.1016/j.stemcr.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimano M., Ouchi N., Nakamura K., Oshima Y., Higuchi A., Pimentel D.R., Panse K.D., Lara-Pezzi E., Lee S.J., Sam F. Cardiac myocyte-specific ablation of follistatin-like 3 attenuates stress-induced myocardial hypertrophy. J. Biol. Chem. 2011;286:9840–9848. doi: 10.1074/jbc.M110.197079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozawa T., Nakamura K., Shoji M., Morita M., Kimura M., Furukawa H., Ueda H., Shiramoto M., Matsuguma K., Kaji Y. Recapitulation of clinical individual susceptibility to drug-induced QT prolongation in healthy subjects using iPSC-derived cardiomyocytes. Stem Cell Reports. 2017;8:226–234. doi: 10.1016/j.stemcr.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talamillo A., Delgado I., Nakamura T., de-Vega S., Yoshitomi Y., Unda F., Birchmeier W., Yamada Y., Ros M.A. Role of Epiprofin, a zinc-finger transcription factor, in limb development. Dev. Biol. 2010;337:363–374. doi: 10.1016/j.ydbio.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Pol A., Gil A., Silljé H.H.W., Tromp J., Ovchinnikova E.S., Vreeswijk-Baudoin I., Hoes M., Domian I.J., van de Sluis B., van Deursen J.M. Accumulation of 5-oxoproline in myocardial dysfunction and the protective effects of OPLAH. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aam8574. [DOI] [PubMed] [Google Scholar]
- Weinberger F., Breckwoldt K., Pecha S., Kelly A., Geertz B., Starbatty J., Yorgan T., Cheng K.-H., Lessmann K., Stolen T. Cardiac repair in Guinea pigs with human engineered heart tissue from induced pluripotent stem cells. Sci. Transl. Med. 2016;8:363ra148. doi: 10.1126/scitranslmed.aaf8781. [DOI] [PubMed] [Google Scholar]
- Yamazaki T., Komuro I., Kudoh S., Zou Y., Shiojima I., Mizuno T., Takano H., Hiroi Y., Ueki K., Tobe K. Angiotensin II partly mediates mechanical stress-induced cardiac hypertrophy. Circ. Res. 1995;77:258–265. doi: 10.1161/01.res.77.2.258. [DOI] [PubMed] [Google Scholar]
- Yamazaki T., Komuro I., Kudoh S., Zou Y., Nagai R., Aikawa R., Uozumi H., Yazaki Y. Role of ion channels and exchangers in mechanical stretch-induced cardiomyocyte hypertrophy. Circ. Res. 1998;82:430–437. doi: 10.1161/01.res.82.4.430. [DOI] [PubMed] [Google Scholar]
- Yang H., Schmidt L.P., Wang Z., Yang X., Shao Y., Borg T.K., Markwald R., Runyan R., Gao B.Z. Dynamic myofibrillar remodeling in live cardiomyocytes under static stretch. Sci. Rep. 2016;6:20674. doi: 10.1038/srep20674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Gomez A.M., Wang X., Yan Y., Zheng M., Cheng H. ROS regulation of microdomain Ca(2+) signalling at the dyads. Cardiovasc. Res. 2013;98:248–258. doi: 10.1093/cvr/cvt050. [DOI] [PubMed] [Google Scholar]
- Zheng M., Cheng H., Banerjee I., Chen J. ALP/Enigma PDZ-LIM domain proteins in the heart. J. Mol. Cell Biol. 2010;2:96–102. doi: 10.1093/jmcb/mjp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The table represents fold change of gene expression, significance (p value and FDR), and gene description.
The table represents fold change of gene expression, significance (p value and FDR), and gene description.
Data Availability Statement
A Web interface that provides search and visualization capabilities for the generated datasets is available at cardio.genomes.nl.