Summary
Quiescent neural stem cells (qNSCs) with radial morphology are the only proven source of new neurons in the adult mammalian brain. Our understanding of the roles of newly generated neurons depends on the ability to target and manipulate adult qNSCs. Although various strategies have been developed to target and manipulate adult hippocampal qNSCs, they often suffer from prolonged breeding, low recombination efficiency, and non-specific labeling. Therefore, developing a readily manufactured viral vector that allows flexible packaging and robust expression of various transgenes in qNSCs is a pressing need. Here, we report a recombinant adeno-associated virus serotype 4 (rAAV4)-based toolkit that preferentially targets hippocampal qNSCs and allows for lineage tracing, functional analyses, and activity manipulation of adult qNSCs. Importantly, targeting qNSCs in a non-Cre-dependent fashion opens the possibility for studying qNSCs in less genetically tractable animal species and may have translational impact in gene therapy by preferentially targeting qNSCs.
Keywords: adult hippocampal neural stem cells, rAAV4, quiescence, radial, toolkit
Highlights
-
•
rAAV4 vectors preferentially target quiescent NSCs in the adult hippocampus
-
•
rAAV4 vectors with distinct promoters reveal differential selectivity for radial NSCs
-
•
rAAV4 allows for genetic manipulation and lineage tracing of quiescent NSCs
-
•
rAAV4 allows for calcium imaging and activity manipulation of quiescent NSCs
In this article, Song and colleagues report a recombinant adeno-associated virus serotype 4-based toolkit comprising several distinct rAAV4 vectors that preferentially target qNSCs within the hippocampus. They demonstrate that rAAV4-mediated transgene expression is robust in qNSCs and allows for lineage tracing, genetic manipulation using the Cre-Lox system, functional analyses using electrophysiology and calcium imaging, and activity manipulation using chemogenetics.
Introduction
Adult hippocampal neurogenesis, a developmental process of generating functionally integrated neurons, occurs throughout life in the dentate gyrus (DG) of the mammalian brain, highlighting the plastic nature of the mature CNS. Quiescent neural stem cells (qNSCs) with radial morphology in the DG are thought to be the most primitive NSCs in the adult brain, essential substrates for continuous neurogenesis throughout life. Currently our understanding of the cellular, molecular, and circuitry mechanisms regulating qNSCs and the process of hippocampal neurogenesis is limited, largely due to the lack of efficient and robust tools for qNSC manipulation.
To date, various viral and transgenic strategies have been developed to genetically manipulate NSCs and their progeny (Enikolopov et al., 2015). However, a number of limitations inherent to these approaches prevent efficient analysis of qNSCs and their progeny. For instance, classical onco-retroviral-mediated labeling approaches require active cell proliferation, thus preferentially labeling proliferating neural progenitors, not qNSCs (Breunig et al., 2007, Ming and Song, 2011, Suh et al., 2007). Recent transgenic approaches using indelible labeling of qNSCs and their progeny require the bigenic combination of neural stem/progenitor cell-specific Cre promoter (such as NestinCreER or Gli1CreER) and a floxed reporter gene. This approach is the most commonly used strategy to target qNSCs for lineage tracing, but often suffers from prolonged breeding and low recombination efficiency (Sun et al., 2014). This issue is further complicated by cell-autonomous manipulation of genes regulating qNSCs, which requires the generation of triple transgenic mice comprising stem cell-specific Cre recombinase, floxed reporter gene, and floxed gene(s) of interest. Additionally, generating triple transgenic combination could become extremely difficult if the Cre promoter and floxed gene(s) of interest are located at similar regions of the same chromosome. Furthermore, investigations of functional properties unique to qNSCs in response to extrinsic stimuli using calcium imaging and the contributions of qNSCs and their progeny to the hippocampal circuit using circuit-based approaches have been limited, largely due to low-level transgene expression or non-specificity associated with the bigenic combination of Cre promoter and floxed transgenes such as GCaMP6 or designer receptors exclusively activated by designer drugs (DREADDs). To overcome these technical caveats, developing a readily manufactured viral vector that allows flexible packaging of different transgenes and supports high-level transgene expression is a pressing need in the field.
Various types of viral vectors, particularly recombinant adeno-associated virus (rAAV)-, lentivirus (LV)-, and retrovirus (RV)-based vectors, have been designed for gene delivery. Among these vectors, rAAV vectors mediate long-term stable expression and induce minimal immune responses as compared with LV and RV (Kaplitt et al., 2007), thus becoming an effective platform for in vivo gene delivery. Ample evidence showed that rAAVs with different capsid structures (also termed “serotypes”) have altered cellular tropism when introduced into the adult mammalian brain (Murlidharan et al., 2014, Schultz and Chamberlain, 2008). rAAV vectors of various serotypes have demonstrated the capacity for efficient gene delivery to neurons and glia in the adult CNS (Berry and Asokan, 2016), but are inefficient in transducing adult NSCs in vivo. The recent development of AAVr3.45 variant provided a promising venue for targeting adult NSCs in vivo (Kotterman et al., 2015). However, further applications of this AAV variant in lineage tracing and activity manipulation of adult NSCs are limited, because a significant portion of rAAVr3.45-transduced cells are mature neurons.
AAV serotype 4, the African green monkey isolate, is one of the evolutionarily and structurally most distinct serotypes known to date, and has previously been shown to transduce type-B astrocytes in the subventricular zone (SVZ) and glial cells overlying the neural tube of the rostral migratory stream (Liu et al., 2005, Murlidharan et al., 2015). Since the slowly dividing type-B astrocytes are thought to be NSCs in the SVZ and the SGZ neurogenic niche lacks ependymal cells, we asked whether rAAV4 vectors selectively transduce qNSCs in the adult DG. Here we report an rAAV4-based toolkit comprising several distinct rAAV4 vectors that preferentially target qNSCs when introduced into the adult DG. We believe that this rAAV4-based approach is a timely addition to the ever growing need in the adult neurogenesis field for rapid targeting, robust gene expression, functional analyses, and activity manipulation of the qNSCs.
Results
rAAV4 Vectors Target Quiescent NSCs in the Adult Hippocampus
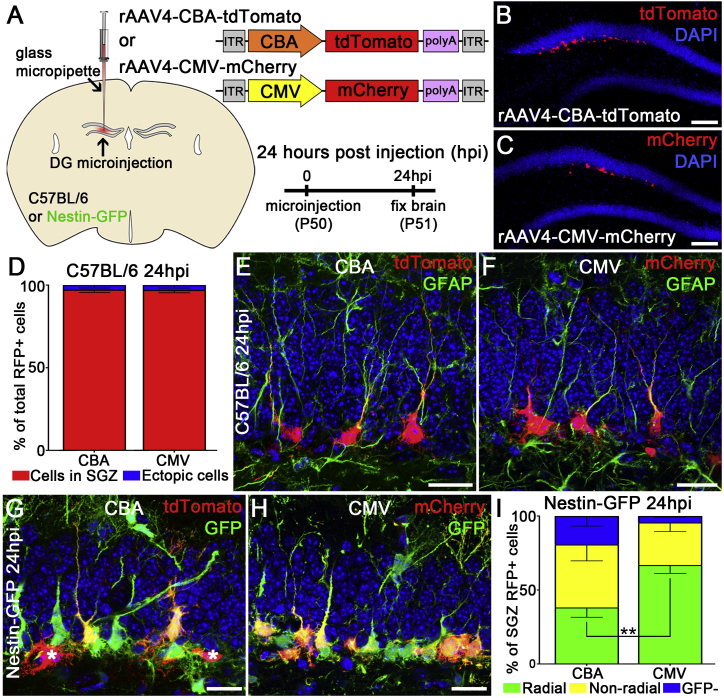
We characterized the selectivity of two rAAV4 vectors in targeting qNSCs with distinct promoters in the adult DG: one vector with mCherry reporter gene driven by human cytomegalovirus (CMV) promoter (rAAV4-CMV-mCherry), and the other with tdTomato (tdTom) reporter gene driven by human CMV enhancer/chicken β-actin promoter (CBA) (rAAV4-CBA-tdTom). First, we titrated each rAAV4 virus to ensure viral labeling is restricted within the SGZ (for details see Experimental Procedures). We injected two rAAV4 vectors (CBA-tdTom: 1.3 × 1010 particles/mL; CMV-mCherry: 9.7 × 109 particles/mL) into the DG of adult C57BL/6 mice (Figure 1A). To minimize tissue injuries associated with microinjection, we used a thin-tip glass pipette pulled from an electrode puller to deliver the viruses to the DG. We chose the brain sections away from the injection site for analyses to ensure intact tissue integrity and neurogenic capacity (Figure S1A). Twenty-four hours after viral injection, we stained the injected brain tissues with anti-RFP to amplify both tdTomato and mCherry signals. Strikingly, we observed that the vast majority of rAAV4-labeled RFP+ cells are located along SGZ (CBA-tdTom: 96.3% ± 1.7%; CMV-mCherry: 97.1% ± 1.7%) (Figures 1B–1D). Furthermore, many of these RFP+ cells have radial processes and are co-localized with glial fibrillary acidic protein (GFAP) (Figures 1E and 1F), suggesting that rAAV4 labels radial NSCs (rNSCs). Besides the vast majority of rAAV4-labeled cells along SGZ, we observed a small percentage of ectopic cells (CBA-tdTom: 2.8% ± 1.7%; CMV-mCherry: 2.9% ± 1.7%) located in the middle/outer granule cell layer and the molecular layer, which is likely associated with the tropism of rAAV4. We further characterized the identity of these rAAV4-labeled ectopic cells and found that some of them express S100β (Figure S1B), suggesting that they are astrocytes.
Figure 1.
rAAV4 Preferentially Transduces Quiescent NSCs in the Adult DG
(A) Schematic of viral microinjection and viral constructs.
(B) Low-magnification confocal image of the DG injected with AAV4-CBA-tdTomato (CBA) at 24 hr post injection (hpi).
(C) Low-magnification confocal image of the DG injected with AAV4-CMV-mCherry (CMV) at 24 hpi.
(D) Quantification of the distribution of RFP+ cells in the SGZ versus elsewhere in the DG (n = 3 mice).
(E and F) Representative confocal image of RFP+ cells transduced with rAAV4-CBA-tdTomato (E) or rAAV4-CMV-mCherry (F) at 24 hpi in C57BL/6 mice. Note the RFP+ cells expressing GFAP marker and showing radial morphology.
(G and H) Representative confocal image of RFP+ cells transduced with rAAV4-CBA-tdTomato (G) or rAAV4-CMV-mCherry (H) at 24 hpi in Nestin-GFP mice. RFP+ cells were either co-labeled with tdTomato and GFP (yellow color) or RFP only (indicated by asterisk).
(I) Quantification of co-localized RFP+ GFP+ rNSCs in the SGZ of Nestin-GFP mice injected with rAAV4-CBA or rAAV4-CMV. RFP+ cells were classified as (1) radial: rNSCs with a radial process and GFP+ (green); (2) non-radial: neural precursors without radial processes and GFP+ (yellow); and (3) GFP− cells (blue). The percentage of rAAV4-CMV-labeled rNSCs was significantly higher (∗∗p < 0.005, n = 3 mice) than that of rAAV4-CBA-labeled rNSCs.
Scale bars, 100 μm (B and C) and 20 μm (E–H). Values represent mean ± SD. See also Figure S1.
Next we characterized the identity of rAAV4-transduced cells in greater detail by injecting these rAAV4 vectors into the DG of Nestin-GFP mice. The Nestin-GFP transgenic line used in this study has previously been validated to represent rNSCs and early neural progenitor cells (NPCs) in the adult DG (Encinas and Enikolopov, 2008). Using the established titer and injection volume, rAAV4-CMV-mCherry labeled ∼30% of the Nestin-GFP+ cell population (27.21% ± 1.7%). We then quantified the percentage of rAAV4-transduced cells co-localized with Nestin-GFP expression out of the total number of rAAV4-labeled cells along the SGZ 24 hr after viral injection. Our results showed that 80.9% ± 6.5% of CBA-tdTom-labeled cells and 95.3% ± 0.8% CMV-mCherry-labeled cells are co-localized with Nestin-GFP cells (Figures 1G and 1H). Interestingly, although both rAAV4 vectors exhibited prominent targeting of Nestin-GFP cells, the distribution of radial and non-radial Nestin-GFP populations labeled by these two rAAV4 vectors are significantly different from each other (p < 0.005, t(4) = 5.68) (Figure 1I): rAAV4-CBA-tdTom-transduced cells display 38.3% ± 6.7% radial GFP+ NSCs and 42.6% ± 10.9% non-radial GFP+ cells, while rAAV4-CMV-mCherry-labeled cells display 66.7% ± 6.0% radial GFP+ NSCs and 28.6% ± 5.9% non-radial GFP+ cells. These data suggest that both rAAV4-CBA-tdTom and rAAV4-CMV-mCherry vectors are capable of targeting qNSCs, but rAAV4-CMV-mCherry is significantly more selective for rNSCs than rAAV4-CBA-tdTom.
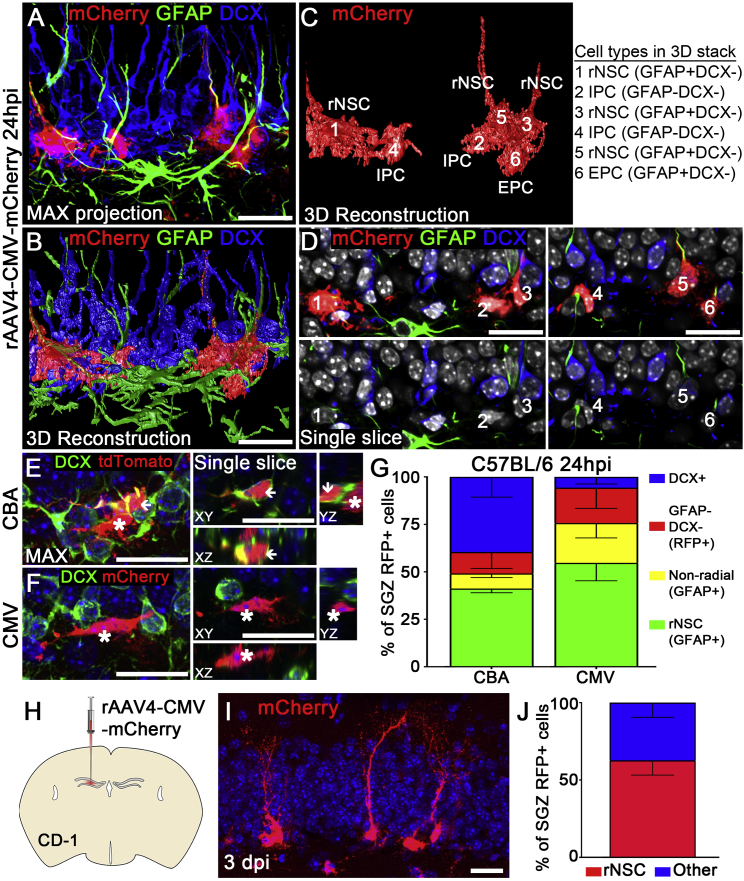
Since both rAAV4 vectors target a small population of cells negative for Nestin-GFP expression, we further classified rAAV4-labeled cells along SGZ into the following categories with validated cellular markers and established morphology at 24 hr after viral injection (Figures 2A–2D and Movie S1): NSCs with radial morphology (rNSCs: GFAP+ DCX−), early progenitor cells without radial morphology (EPCs: GFAP+DCX−), intermediate progenitor cells (IPCs: GFAP−DCX−), and neuroblasts (NB: GFAP−DCX+). Our results showed that the majority of CMV-mCherry-labeled cells are GFAP+DCX− rNSCs (54.7% ± 9.5%), compared with a moderate portion of CBA-tdTom-labeled GFAP+DCX− rNSCs (41.2% ± 2.2%) (Figure 2G). These results aligned with the data obtained from Nestin-GFP mice. Interestingly, a significant portion of CBA-tdTom-labeled cells are GFAP−DCX+ neuroblasts (39.6% ± 10.7%) compared with CMV-mCherry-labeled GFAP−DCX+ neuroblasts (5.7% ± 3.7%) (Figures 2E–2G), suggesting that rAAV4-CBA-tdTom targets both undifferentiated rNSCs and neuroblasts compared with rAAV4-CMV-mCherry that preferentially targets undifferentiated rNSCs. Taken together, these data suggest that the tropism of two distinct promoter-serotype combinations is different: rAAV4-CMV-mCherry transduction is more selective for GFAP+DCX− rNSCs, while rAAV4-CBA-tdTom transduction is equally selective for both GFAP+DCX− rNSCs and GFAP−DCX+ neuroblasts. We then chose the promoter-serotype combination of AAV4-CMV for further analysis and lineage tracing studies to achieve more specific targeting of rNSCs.
Figure 2.
rAAV4 Vectors with Distinct Promoters Reveal Differential Selectivity for rNSCs
(A) Confocal imaging of AAV4-CMV-mCherry-transduced cells stained for RFP, GFAP, and DCX. z-Stack confocal data were maximum-intensity projected in ImageJ.
(B) 3D reconstruction of a deconvoluted confocal image in (A) using Huygens Professional software.
(C) Six cells were classified for cell identity based on morphology and cell stage markers.
(D) Confocal images showing marker expression of cells (from A) at a single focal plane. Three RFP+ cells (cells 1, 3, 5) with radial processes expressed GFAP but not DCX. These cells were classified as rNSCs. One non-radial precursor cell (cell 6) expressed GFAP but not DCX. Cells of this stage were classified as early progenitor cells (EPCs). Two RFP+ cells (cells 2 and 4) did not express GFAP and DCX. These cells were classified as intermediate progenitor cells (IPCs).
(E and F) Confocal images showing the DCX expression in RFP+ cells transduced by rAAV4-CBA-tdTomato (E) and rAAV4-CMV-mCherry (F). Note some rAAV4-CBA-tdTomato-transduced RFP+ cells were DCX+ (arrowhead), and some were DCX− (asterisks); in contrast, most rAAV4-CMV-mCherry-transduced RFP+ cells were DCX−.
(G) Quantification of the cell types found in CBA-injected and CMV-injected mice at 24 hpi (n = 3 mice).
(H–J) rAAV4-CMV-mCherry displayed similar tropism for rNSCs in CD-1 mice compared with C57BL/6 mice at 3 days post injection (dpi) (n = 3 mice).
Scale bars, 20 μm. Values represent mean ± SD. See also Figures S2 and S3.
As rNSCs are largely quiescent, we sought to confirm the quiescence of rAAV4-transduced NSCs. First, we injected the thymidine analog 5-ethynyl-2′-deoxyuridine (EdU) into mice 3 days after rAAV4-CMV-mCherry injection to label proliferating cells. We quantified EdU incorporation in rAAV4-transduced mCherry+ cells, and our results showed that a small percentage of mCherry+ radial NSCs and non-radial precursors incorporated EdU (radial: 1.7% ± 0.5%; non-radial: 1.46% ± 1.4%) (Figure S2A). Next, we used a well-established cell proliferation marker, MCM2, to confirm the quiescence of rAAV4-transduced NSCs. We then quantified the co-localization of MCM2 and mCherry in rNSCs and non-radial precursors labeled by rAAV4-CMV-mCherry, and again observed a small percentage of NSCs labeled with MCM2 (radial: 3.99% ± 2.2%; non-radial: 3.75% ± 2.6%) (Figure S2B). These data together suggest that rAAV4 targets quiescent radial and non-radial precursors.
To address whether rAAV4 selectivity is dependent on the genetic background of different mouse strains, we injected rAAV4-CMV-mCherry into the DG of CD1 mice (Figure 2H), and observed similar rAAV4 selectivity in rNSCs compared with that in C57BL/6 mice (62.7% ± 9.5%) (Figures 2I and 2J), suggesting that rAAV4 selectivity is independent of the mouse strain. Furthermore, we evaluated the generality of rAAV4 for targeting NSCs in other rodent species by injecting the virus into the DG of the Sprague-Dawley rat (Figure S3A). Consistent with the selectivity of rAAV4 in NSCs observed in mice, our results revealed that rAAV4 is capable of targeting rNSCs in rats (Figures S3B and S3C).
rAAV4 Allows for Genetic Manipulation of Quiescent NSCs in the Adult Hippocampus
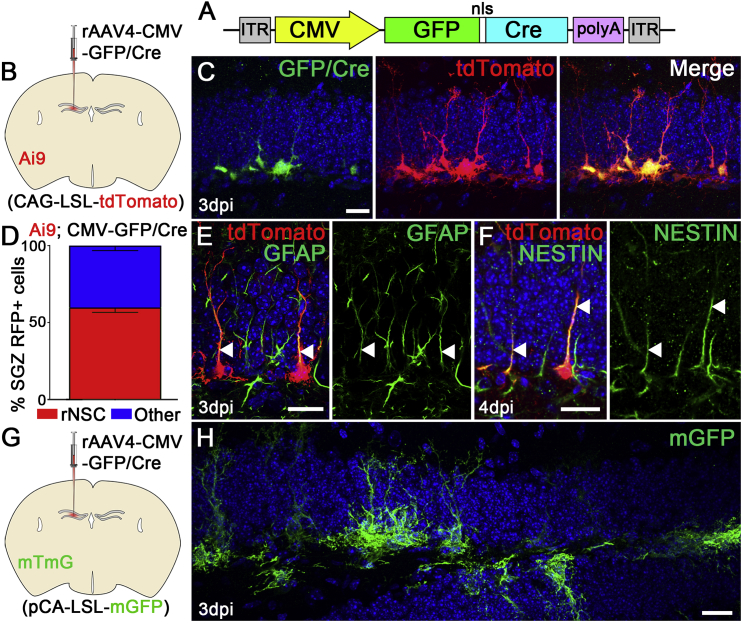
We next examined whether the rAAV4 system can be used with a floxed mouse line to achieve Cre-dependent recombination in the rNSCs. We packaged a CMV-GFP/Cre plasmid to the rAAV4 capsid to generate rAAV4-CMV-GFP/Cre (Figure 3A), and injected rAAV4-CMV-GFP/Cre (1.1 × 1011 particles/mL) into the DG of the Ai9 reporter mice harboring a loxP-flanked STOP cassette that prevents transcription of the downstream RFP variant (tdTomato) in the absence of Cre recombinase (Madisen et al., 2010) (Figure 3B). Three days after viral injection, we examined the expression of GFP (for Cre expression) and tdTomato (for Cre-dependent recombination) in AAV4-labeled cells, and observed that both GFP and tdTomato signals are robust and all of the tdTomato+ cells are co-localized with GFP (Figure 3C), suggesting that Cre-mediated recombination using CMV-GFP/Cre is highly efficient. Importantly, we observed that the majority of rAAV4-labeled tdTomato+ cells are rNSCs (59.7% ± 3.2%, Figure 3D) and co-localized with NESTIN and GFAP markers (Figures 3E and 3F). Similar to the tropism of rAAV4-CBA-tdTom and rAAV4-CMV-mCherry, we observed a small percentage of rAAV4-labeled ectopic cells (3.4% ± 1.5%) located in the middle/outer granule cell layer and the molecular layer that appear to be astrocytes expressing S100β (Figure S4A) but not γ-aminobutyric acid (GABA) (Figure S4B). Thus, the rAAV4-Cre system allows for genetic manipulation of qNSCs in a Cre-Lox-dependent fashion.
Figure 3.
rAAV4 Allows for Genetic Manipulation of Quiescent NSCs in the Adult Hippocampus
(A) Schematic of the AAV-CMV-GFP/Cre construct. The CMV promoter drives expression of a GFP-Cre fusion gene.
(B) AAV4-CMV-Cre was injected into adult Ai9 mice, which harbor a lox-STOP-lox-tdTomato allele in the ROSA locus. Cre-mediated recombination in these mice leads to constitutive cellular expression of tdTomato.
(C) Co-localization of GFP/Cre and tdTomato 3 days after rAAV4-CMV-GFP/Cre injection into the Ai9 mice.
(D) Quantification of the percentage of tdTom+ rNSCs in the SGZ at 3 dpi (n = 3 mice).
(E and F) tdTom+ rNSCs expressed NSC markers GFAP (E) and NESTIN (F). Note the co-localization of GFAP or NESTIN in their radial processes (arrowheads).
(G) rAAV4-CMV-GFP/Cre was injected into the DG of MTMG mice.
(H) mGFP+ rNSCs are labeled throughout the DG at 3 dpi.
Scale bars, 20 μm. Values represent mean ± SD. See also Figure S4.
Next we asked whether the selectivity of the AAV4-Cre system in targeting NSCs can be recapitulated in other Cre reporter lines, as non-specific targeting of NSCs using the combination of NestinCreERT2 inducible line with different Cre reporter lines was observed (Bonaguidi et al., 2011, Sun et al., 2014). We injected rAAV4-CMV-GFP/Cre into another reporter line called mTmG, a double-fluorescent Cre reporter mouse that expresses membrane-targeted tdTomato (mT) prior to Cre-mediated excision and membrane-targeted GFP (mG) after excision (Muzumdar et al., 2007) (Figure 3G). Because GFP/Cre is localized in the nucleus, the membrane-targeted GFP signal around the soma and radial processes of NSCs in the mTmG reporter mice indicates the occurrence of recombination. We observed that rAAV4-GFP/Cre targets rNSCs with high efficiency (Figure 3H), suggesting that the rAAV4-Cre system can target rNSCs using different Cre-inducible reporter lines.
rAAV4 Allows for Lineage Tracing of Quiescent NSCs in the Adult Hippocampus at both Population and Clonal Levels
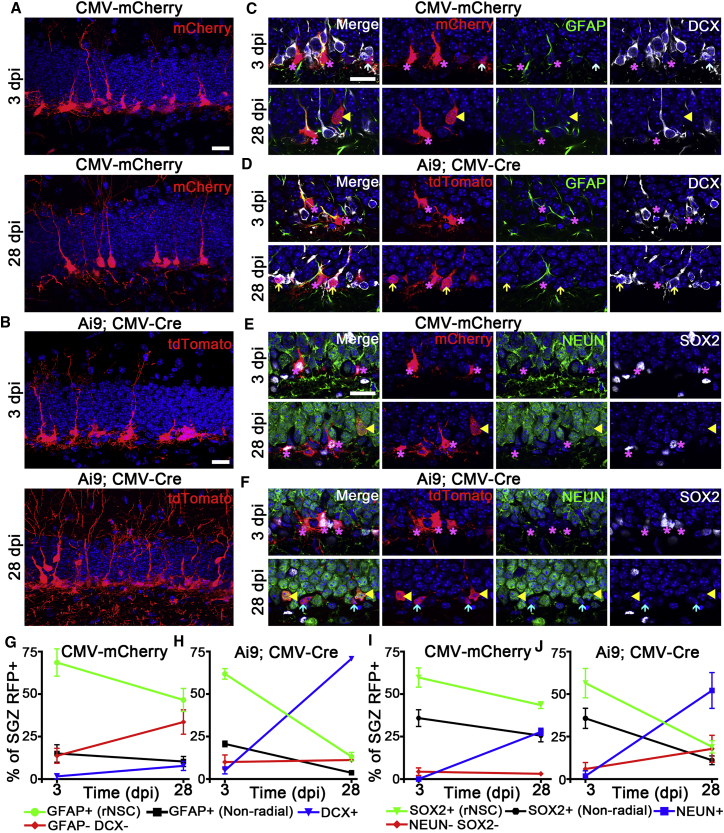
To evaluate the potential applications of Cre- and non-Cre rAAV4 systems in lineage tracing and fate mapping of rAAV4-labeled NSCs, we examined cellular components labeled by rAAV4-CMV-mCherry in C57BL/6 mice and rAAV4-CMV-GFP/Cre in Ai9 mice at 3 and 28 days after viral injection (Figures 4A and 4B). We used two sets of validated cell-stage-specific markers including GFAP and DCX, and SOX2 and NeuN to compare the cellular components between Cre- and non-Cre rAAV4 systems at 3 and 28 days after viral injection (Figures 4C–4F). The majority of adult-born hippocampal cells are thought to become neurons derived from qNSCs via transit amplifying intermediate progenitors (Bonaguidi et al., 2011, Doetsch and Hen, 2005, Dranovsky et al., 2011, Encinas et al., 2011). We thus expected to see an increase in the proportion of mature neurons with a corresponding decline in the proportion of rNSCs within the rAAV4-labeled lineage over time. Indeed, our lineage tracing of rAAV4-CMV-mCherry and rAAV4-CMV-GFP/Cre;Ai9-labeled qNSCs revealed that the percentage of GFAP+DCX− and SOX2+NeuN− rNSCs significantly decreased and the percentage of SOX2−NeuN+ mature neurons significantly increased from 3 to 28 days post injection (dpi) in both systems (Figures 4G–4J). Additionally we observed a small percentage (less than 2%) of GFAP+ stellate astrocytes in the SGZ labeled by both rAAV4-CMV-mCherry and rAAV4-CMV-GFP/Cre at both 3 and 28 dpi, which remain static over time (Figures S5A and S5B). We believe that these astrocytes represent the astroglia progeny derived from qNSCs, because rAAV4 appears to have a low propensity in labeling astrocytes in the SGZ (Figure S5C). These results are consistent with the observations in the bigenic mice harboring NestinCreERT2 and floxed reporter in which GFAP+ rNSCs decrease and NeuN+ mature neurons increase over time (Bonaguidi et al., 2011, Dranovsky et al., 2011).
Figure 4.
rAAV4 Allows for Lineage Tracing of Quiescent NSCs in the Adult Hippocampus
(A) Maximum projection of rAAV4-CMV-mCherry-injected mice stained for mCherry using RFP antibody at 3 and 28 dpi. RFP+ rNSCs were found at 3 and 28 dpi, and RFP+ mature neurons were found at 28 dpi.
(B) Maximum projection of Ai9; rAAV4-CMV-Cre/GFP-injected mice stained for tdTomato using RFP antibody at 3 and 28 dpi. RFP+ rNSCs were found at 3 and 28 dpi, and RFP+ mature neurons were observed at 28 dpi.
(C–F) Lineage tracing was performed using two sets of cell stage markers, including GFAP/DCX and SOX2/NeuN, and representative confocal images shown at 3 and 28 dpi from the two viral strategies. In (C), at 3 dpi RFP+ cells were GFAP+ rNSCs (purple asterisks) and GFAP− and DCX− IPCs (light-blue arrow); at 28 dpi RFP+ cells were GFAP+ rNSCs (purple asterisks) and GFAP−, DCX− mature neurons (yellow arrowheads). In (D), at 3 dpi RFP+ cells in Ai9 mice were GFAP+ rNSCs (purple asterisks); at 28 dpi RFP+ cells were GFAP+ rNSCs (purple asterisks) and DCX+, GFAP− neuroblasts (light-yellow arrows). In (E), at 3 dpi RFP+ cells were SOX2+, NeuN− rNSCs (purple asterisks); at 28 dpi RFP+ cells were SOX2+, NeuN− rNSCs (purple asterisks) and NeuN+, SOX2− neurons (yellow arrowheads). In (F), at 3 dpi RFP+ cells in Ai9 mice were SOX2+, NeuN− rNSCs (purple asterisks); at 28 dpi RFP+ cells were NeuN+, SOX2− neurons (yellow arrowheads) and NeuN−, SOX2− neuroblast-like cells (light-blue arrows).
(G–J) Quantification of stage marker co-expression across time using either GFAP/DCX (G and H) or SOX2/NeuN (I and J) for CMV-mCherry (G and I) and Ai9; CMV-Cre (H and J) (n = 3 mice).
Scale bars, 20 μm. Values represent mean ± SD. See also Figures S5–S7.
It bears noting that the lineage relationship of rAAV4-CMV-mCherry-labeled cells appears to be different from rAAV4-CMV-Cre-labeled cells. For instance, the decrease of the radial qNSC pool and increase of the DCX+ and NeuN+ populations using the Cre-rAAV4 system is more prominent than using the non-Cre rAAV4 system (Figures 4G–4J). There are two proposed possibilities: (1) different radial NSC populations may be targeted by the Cre and non-Cre AAV4 systems; and (2) episomal dilution of rAAVs (Schultz and Chamberlain, 2008) associated with multiple rounds of cell divisions in NPCs using the non-Cre rAAV4 system may lead to loss of or low-level fluorescence in the newborn progeny. To test the first possibility, we injected AAV4-CMV-GFP/Cre and AAV4-CMV-mCherry vectors into the DG of C57BL/6 mice. We found a significant percentage of mCherry+ NSCs co-localized with GFP+ (72.4% ± 10.4%) (Figures S6A–S6C), suggesting that these two viruses label the same NSC population. To support the episomal dilution hypothesis associated with the non-Cre AAV4 vector, we examined the florescence intensity associated with NeuN+ mature neurons transduced by the non-Cre vector (rAAV4-CMV-mCherry). We observed low-level mCherry expression often associated with NeuN+ mature neurons, but not rNSCs (Figure S6D). In addition, we compared the density of rAAV4-transduced cells between Cre and non-Cre rAAV4 vectors at 3 and 28 dpi, and observed similar radial NSC density in both Cre and non-Cre AAV4 systems (Figure S6E) but reduced DCX+ immature neuron density in the non-Cre rAAV4 system compared with the Cre rAAV4 system (Figure S6F). Importantly, with similar numbers of the starting population transduced by Cre and non-Cre rAAV4 vectors at 3 dpi, the number of rAAV4-CMV-mCherry-labeled RFP+ cells was significantly lower than that of AAV4-CMV-Cre-labeled RFP+ cells (Figure S6G). These data together support that episomal dilution associated with cell division may account for the observed difference in neuronal differentiation of the rNSCs labeled by Cre and non-Cre rAAV4 vectors. The episomal dilution issue is completely circumvented in the rAAV4-Cre system, as rAAV4-Cre-labeled cells exhibited lineage relationship similar to the previous characterization of NestinCreERT2:Rosa-YFP double transgenic mice at the population level (Dranovsky et al., 2011). Together, these data suggest that both Cre and non-Cre rAAV4 vectors can be used for lineage tracing of qNSCs, however, caution should be exercised when using the non-Cre rAAV4 system because the potential episomal dilution could lead to an underestimate of the production of neuronal progeny derived from qNSCs.
Lineage tracing of qNSCs at the population level provides valuable information on the fate decisions of NSCs as a whole. However, to address the heterogeneity of NSCs in their fate decisions, we require clonal analysis of individual NSCs. To evaluate the feasibility for clonal labeling of NSCs by rAAV4, we injected serially diluted rAAV4-CMV-Cre-GFP into the DG of the Ai9 mice to titrate the viral titer that could produce ∼10 clusters per hemisphere to minimize the mixture of progeny from different NSCs (Figure S7A). Glass micropipettes with very thin tips were used to deliver the viruses, so no significant injuries were observed at the injection sites (Figure S7A, yellow circled areas). However, we noticed more NSC clusters at or close to injection sites compared with the sections further away from the injection sites. Therefore, we excluded those sections at or close to injection sites from the analysis to avoid ambiguity in assigning radial NSC clones. We monitored fate choices of individual rAAV4-labeled radial NSC clusters at 7 days after viral infection and observed three modes of fate decisions: (1) quiescent NSCs (Figure S7B); (2) asymmetric neurogenic division that produced one radial NSC and one progenitor (Figure S7C); and (3) symmetric division yielding two rNSCs (Figure S7D). Together, these data suggest that the rAAV4 system is amenable for fate mapping of NSCs at the clonal level.
rAAV4-Transduced Quiescent NSCs Preserve Normal Functional Properties of NSCs
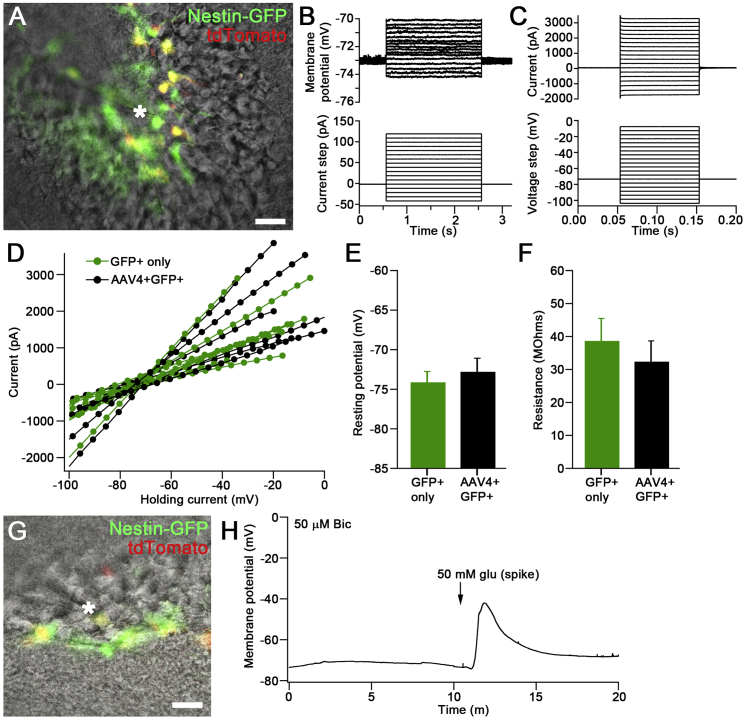
To address whether rAAV4-transduced cells preserve the functional properties of qNSCs, we performed electrophysiological recording in rAAV4-labeled NSCs. We injected rAAV4-CBA-tdTom into the DG of the Nestin-GFP mice, and 7 days after viral injection we were able to detect bright fluorescence associated with tdTomato expression in rNSCs under the acute slice preparation (Figure 5A). We performed whole-cell recordings from radial GFP+tdTom+ NSCs and compared their functional properties with GFP+ rNSCs. We observed passive membrane properties and low input resistance typical of astrocytes in both GFP+tdTom+ and GFP+ rNSCs (Figures 5B–5D). Additionally, their membrane resistance and membrane potential are indistinguishable from each other (Figures 5E and 5F), suggesting that rAAV4 transduction does not affect physiological properties of rNSCs. Next we tested whether rAAV4-labeled cells respond to extrinsic stimulation. As our recent study using single-cell RNA sequencing in Nestin-GFP mice has shown that rNSCs express glutamate receptors (Shin et al., 2015), including both AMPA and NMDA receptors, we recorded GFP+ tdTom+ rNSCs under the current-clamp mode upon bath application of glutamate and observed robust depolarization of rNSCs (Figures 5G and 5H). Together, these data suggest that rAAV4-transduced rNSCs preserved the functional properties of non-viral labeled rNSCs and were capable of responding to extrinsic stimulation with a change of the membrane potential.
Figure 5.
rAAV4-Transduced Quiescent NSCs Exhibit Normal Electrophysiological Properties
(A) Live differential interference contrast (DIC) and fluorescence image from whole-cell electrophysiology recording showing the labeled rAAV4-CBA-tdTom cells in the DG of the Nestin-GFP. Also visible is the patch electrode, indicated by an asterisk. Scale bar, 20 μm.
(B and C) Passive electrical properties of tdTom+ cells characteristic of NSCs. (B) Top: overlaid series of membrane potential changes due to a series of whole-cell current injections (bottom). (C) Top: overlaid holding currents in response to a series of hyperpolarizing or depolarizing voltage steps (bottom).
(D–F) Overlapping distributions of current/voltage relationships in several tdTom+ and tdTom− Nestin-GFP-labeled cells showing linear responses. Electrophysiological determinations of (E) resting membrane potentials and (F) membrane resistances were unchanged in rAAV4-transduced cells (n = 6 cells from 3 mice for each group).
(G) Sample DIC and fluorescent image during a whole-cell recording from a double-positive radial NSC in current-clamp mode. The tip of the recording electrode is labeled with an asterisk next to the recorded cell (yellow central cell). Scale bar, 20 μm.
(H) Large depolarizing response of the rAAV4-labeled NSC in (G) following spike application of 50 mM glutamate in the presence of 50 μM bicuculline.
Values represent mean ± SEM.
rAAV4 Allows for Calcium Imaging of Quiescent NSCs
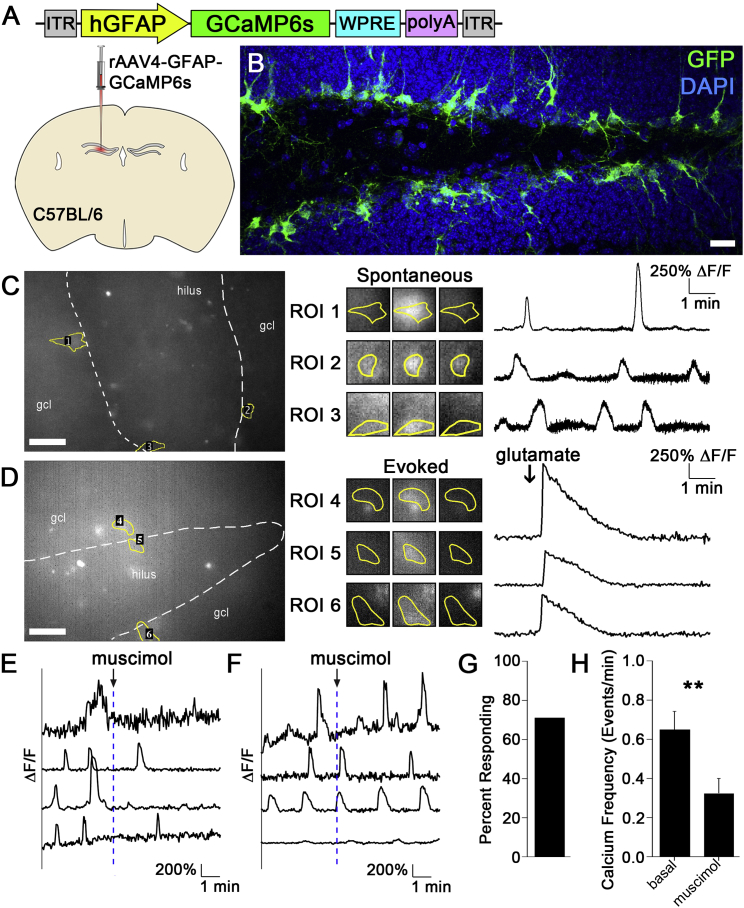
The utility of rAAV4 for studying activity-dependent regulation of adult NSCs will depend on its ability to mediate high-level expression of genetically encoded indicators and effectors. We first assessed the ability to monitor activity in qNSCs through the rAAV4 system. We packaged a GFAP-GCaMP6s construct into the rAAV4 capsid to generate rAAV4-GFAP-GCaMP6s (Figure 6A). GFAP is also known as a marker for astrocytes; however, we did not observe extensive labeling of astrocytes within the DG and the vast majority of rAAV4-GCaMP6s-labeled cells (∼77%, 58/74) were rNSCs (Figure 6B). This is probably due to the low propensity of rAAV4 to label astrocytes (Figure S5). To test whether the level of GCaMP6s expression in NSCs is sufficient to allow their activity to be monitored, we injected rAAV4-GFAP-GCaMP6s (4.1 × 1010 particles/mL) into the DG of adult C57BL/6 mice and performed calcium imaging in acute brain slices prepared from viral injected mice at 10 dpi. Consistent with their high-level expression, robust spontaneous calcium transients were detected in GCaMP6s+ NSCs (Figure 6C). Additionally, we observed increased calcium transients in GCaMP6s+ NSCs upon spike application of glutamate (Figure 6D), consistent with the depolarizing effect observed in rNSCs upon glutamate application (Figures 5G and 5H). Together, these data demonstrate that the GFAP promoter is specific to restricting GCaMP6 expression in the NSCs and the GCaMP6 expression is high, allowing for detection of both spontaneous and evoked calcium activity of qNSCs.
Figure 6.
rAAV4 Allows for Calcium Imaging of Quiescent NSCs
(A and B) The viral construct (A) of hGFAP-GCaMP6 was packaged into AAV4 and microinjected into the DG of C57BL/6 mice. (B) Sample expression of GCaMP6s in the DG visualized by immunostaining against GFP. Scale bar, 20 μm.
(C) Live snapshot (left) of GCaMP6+ rNSCs in an acute brain slice. Three spontaneously active cells were indicated with regions of interest (ROI) 1–3. Zoomed-in time-series snapshots (center) of the three indicated cells before, during, and after spontaneous calcium events. Quantifications (right) of ΔF/F calcium signals for these cells reveal multiple calcium events in each cell with varying intensities. Scale bar, 50 μm.
(D) Evoked calcium event of a GCaMP6+ rNSC in response to a spike application of 50 mM glutamate in the presence of 50 μM bicuculline. Scale bar, 50 μm.
(E and F) Sample ΔF/F measurements of rNSCs responsive to GABAA agonist muscimol (spike application at 100 mM) (E) and rNSCs non-responsive to muscimol application (F).
(G) Quantification of the percentage of rNSCs responsive to muscimol (n = 14 cells).
(H) Quantification of the frequencies of calcium events under basal conditions (4 min), and following muscimol (4 min) from the responding rNSCs (n = 10 cells; ∗∗p < 0.001).
Values represent mean ± SEM.
Using this system, we can start to address the fundamental question on functional heterogeneity of rNSCs, a question that has not been extensively investigated thus far largely due to low-level transgene expression of genetically encoded indicators or effectors in rNSCs. Our previous data showed that GABA signaling plays a role in inhibiting qNSC activation (Song et al., 2012, Bao et al., 2017), suggesting that GABA may play an inhibitory role in regulating rNSC activity. As our calcium imaging showed that rNSCs exhibit high-level spontaneous activity at the baseline (Figure 6C), we examined the effects of muscimol on the spontaneous calcium activity in rNSCs labeled with rAAV4-GFAP-GCaMP6. Interestingly, we found that 71% of GCaMP6-labeled rNSCs that exhibit spontaneous calcium activity respond to muscimol application (Figures 6E–6G and Movie S2). Consistent with an inhibitory effect on NSC activity, muscimol application led to reduced spontaneous calcium events in rNSCs (Figure 6H). These results highlight the functional heterogeneity of the rNSCs in response to GABA.
rAAV4 Allows for Chemogenetic Modulation of Quiescent NSC Activity
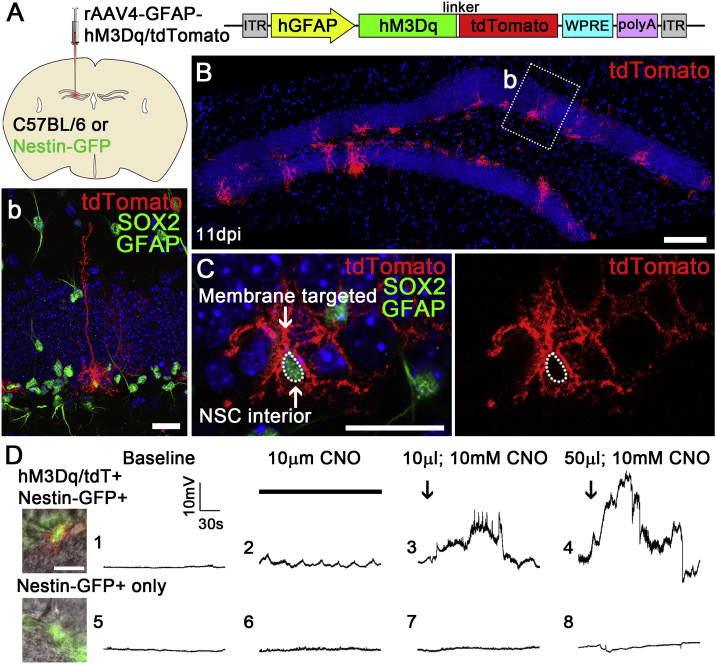
Having established that our rAAV4 system enables robust and preferential expression of fluorescent reporters and the activity indicator GCaMP6s in qNSCs, we next sought to examine whether chemogenetic approaches could be used to manipulate qNSC activity. DREADDs are modified human muscarinic receptors that can be activated by clozapine-N-oxide (CNO), a pharmacologically inert and orally bioavailable drug (Armbruster et al., 2007). To date their utilization in qNSCs for activity manipulation has not been extensively reported, likely due to the low recombination efficiency associated with the transgenic approaches. To test whether we could use the rAAV4 system to gain chemogenetic control of qNSCs, we generated an rAAV4-hM3Dq-tdTomato vector under the control of the GFAP promoter (Figure 7A). Similar to rAAV4-GFAP-GCaMP6s, AAV4-GFAP-hM3Dq-tdTomato efficiently targets rNSCs expressing GFAP and SOX2 (∼80%, 21/26) (Figure 7B). In addition, consistent with the expected expression of a functional receptor, the Gq-DREADD is located at the membrane of the infected cells (Figure 7C). Next, we tested the functionality of the Gq-DREADD within rNSCs. We injected rAAV4-hM3Dq-tdTomato (7.3 × 109 particles/mL) into adult Nestin-GFP mice in order to achieve better visualization of the tdTom+ NSCs for electrophysiology recording, because hM3Dq-tdTomato is localized on the membrane. We recorded tdTom+ GFP+ rNSCs upon low-concentration bath CNO perfusion (10 μM) and high-concentration spike application of CNO (10 mM) to achieve dose-dependent responses from tdTom+ rNSCs expressing Gq-DREADD. Interestingly, upon low-concentration bath CNO perfusion (10 μM), tdTom+GFP+ rNSCs exhibited small rhythmic membrane depolarization (Figure 7D2), which is absent at the baseline without CNO (Figure 7D1). In contrast, high-concentration spike application of CNO (10 mM) induced more robust membrane depolarization of tdTom+GFP+ rNSCs (Figure 7D3 and 4). Furthermore, such CNO-induced depolarization effects are specific to tdTom+GFP+ rNSCs, because low-concentration bath CNO perfusion (10 μM) and high-concentration bath spike application of CNO (10 mM) do not have any effects on tdTom−GFP+ rNSCs (Figure 7D5–8). Together, these experiments demonstrate that rAAV4-GFAP-hM3Dq allows specific, functional, and restricted expression of Gq-DREADD and that CNO treatment effectively and selectively increases the activity of qNSCs.
Figure 7.
rAAV4 Allows for Chemogenetic Modulation of Quiescent NSCs
(A) rAAV4-hGFAP-hM3Dq/tdTomato was microinjected into the DG of C57BL/6 mice or Nestin-GFP mice.
(B) Sample confocal imaging showing expression of hM3Dq/tdTomato in the DG at 11 dpi. Scale bar, 100 μm. Inset (b): a transduced cell displaying a radial morphology and expressing GFAP and SOX2. Scale bar, 20 μm.
(C) Confocal image showing membrane localization of tdTomato. The NSC nucleus was labeled by SOX2 expression. Scale bar, 20 μm.
(D) Dose-dependent depolarizing responses to CNO in tdTom+ cells by patch-clamp electrophysiology. Baseline membrane potentials (traces 1 and 5) from tdTom+ and tdTom− cells at approximately −77 to −80 mV. Low-concentration CNO applied via bath perfusion evoked small voltage oscillations in transduced cells (trace 2), but not in control cells (trace 6). Direct application of CNO at higher concentrations to the recording chamber evoked progressively larger depolarizing responses (traces 3 and 4). No equivalent responses were detected in tdTom− cells (traces 7 and 8). Scale bar, 20 μm.
Discussion
Transgenic- and retroviral-based approaches have revolutionized our understanding of developmental processes during adult hippocampal neurogenesis. However, this success highlights the limitations posed by our present inability to efficiently identify and manipulate qNSCs and their progeny. Here we report a recombinant rAAV4-based toolkit that preferentially targets hippocampal qNSCs and allows for lineage tracing, functional analyses, and activity manipulation of adult qNSCs.
The transduction mechanism underlying the tropism of rAAV4 vector in NSCs in the adult DG remains unknown. A key step in viral entry into the CNS is the recognition of specific cell surface membrane glycoproteins as receptors. One of the most versatile host glycans that have been used as viral attachment factors are the family of sialic acids (Neu et al., 2011). In the brain, two families of sialoglycans are of particular interest: gangliosides and polysialic acid (PSA). Of these two families, PSA can selectively bind to neural cell adhesion molecule (NCAM) (Schnaar et al., 2014) and plays an important role in regulating AAV4 transduction and tropism (Murlidharan et al., 2015). Interestingly, our recent study using single-cell RNA sequencing in Nestin-GFP mice has showed that NSCs express high-level NCAM, including NCAM1 and NCAM2 (Shin et al., 2015), thus providing a plausible explanation for the AAV4 selectivity in NSCs in the adult brain. An intriguing finding from this work is also the differential tropism of the CMV and CBA promoters when combined with the AAV4 serotype. Although both rAAV4-CMV-mCherry and rAAV4-CBA-tdTom target rNSCs, rAAV4-CMV-mCherry is more selective for rNSCs than rAAV4-CBA-tdTom. These results suggest that the tropism of rAAV4 vectors is determined by the combination of promoter and serotype. We speculate that the CMV promoter may be more selective for undifferentiated cells with glial properties (such as NSCs) than the CBA promoter. Supporting this, the CMV promoter has been reported to be transcriptionally active in multiple undifferentiated mouse embryonic stem cell lines (Ward and Stern, 2002). In contrast, the CBA promoter has been extensively used in conjunction with rAAVs to increase transduction efficiency in neurons (Fitzsimons et al., 2002), suggesting that the CBA promoter may preferentially transduce neurons. Although rAAV4 vectors preferentially target qNSCs, they also target a significant portion of non-radial precursors, similar to the genetic system using bitransgenic combination of NestinCreERT2 and floxed Rosa-YFP reporter (Dranovsky et al., 2011). This is a challenge for the field, as currently no genetic or viral systems can exclusively target adult rNSCs. Future efforts in identifying qNSC-specific molecular markers and creating functional promoters based on these qNSC-unique markers will help improve the targeting specificity of current rAAV4 vectors in qNSCs.
Our rAAV4-based approach provides a toolbox for studying the most primitive NSCs in the adult hippocampus. We demonstrated that the rAAV4-Cre delivery system is amenable for lineage tracing and fate mapping. Using the Cre reporter lines, rAAV4-Cre-labeled cells showed lineage relationships similar to the previous characterization of Nestin-CreERT2:Rosa-YFP double transgenic mice (Dranovsky et al., 2011). Importantly, this system is amenable for clonal analysis by tracing individual rNSCs; therefore, a fundamentally important question on the heterogeneity of qNSCs can be addressed using this system. Since ∼30% of rAAV4-transduced cells are non-radial precursors, one would expect to observe some rAAV4-labeled non-radial precursor clones. However, this will not pose major concerns for the overall data interpretation, as only NSC clones containing rNSCs can be chosen for data analysis to assess the heterogeneity. The non-Cre rAAV4 system is also amenable for lineage tracing of qNSCs, but the concern is the potential underestimate of the production of neuronal progeny derived from qNSCs due to episomal dilution associated with NPC division. Future studies of incorporating the CRISPR/Cas9 gene editing system into the non-Cre rAAV4 delivery system will circumvent this issue through permanent genome editing.
One hallmark of adult neurogenesis is the dynamic regulation by neuronal network activity. However, the cellular and circuit mechanisms underlying activity-dependent regulation of qNSCs remain largely unexplored, in part due to the technical limitations associated with simultaneous manipulation of specific neural circuit components and qNSCs. The development of rAAV4 vectors expressing fluorescence and activity indicator GCaMP6 will allow simultaneous manipulation of specific neural circuits and qNSC recording using the combination of cell-type-specific Cre (or Flp) lines (for activity manipulation) and the rAAV4 system (for electrophysiology or calcium imaging of qNSCs). Furthermore, neural circuit activity-dependent fate mapping of qNSCs can be performed using a double-recombinase system in which qNSCs can be labeled by the rAAV4-Cre system along with a Cre-inducible reporter line, and specific circuits of interest can be labeled via the Flp-FRT recombination system. With an increasing list of Flp mouse lines becoming publicly available (He et al., 2016), this approach thus opens the possibility to explore distinct neural circuits regulating in vivo behaviors of qNSCs. Furthermore, recent success in rAAV-mediated targeting of GABAergic neurons through mDlx/hDlx enhancer (Dimidschstein et al., 2016) holds the promise that many neuronal types can be potentially targeted using a viral-based approach, and circuit-based regulation of adult neurogenesis can be studied using a dual-viral system in non-transgenic wild-type mice.
Experimental Procedures
Animals
The following adult mice (7–9 weeks old) were used in this study: wild-type C57BL6 (JAX), CD1 (Charles River), Nestin-GFP (Encinas and Enikolopov, 2008), Ai9 (JAX stock #007909), and MTMG mice (JAX stock # 007676). Adult Sprague-Dawley rats (Charles River) were also used to examine the tropism of rAAV4 in the DG. The general procedure of stereotactic delivery of the virus to the DG was followed as previously described (Bao et al., 2017, Song et al., 2012, Song et al., 2013). All procedures were conducted in accordance with the guide for the Care and Use of Laboratory Animals, as adopted by the NIH, and with approval of the institutional Animal Care and Use Committee at the University of North Carolina at Chapel Hill.
Recombinant AAV4 Vector Production
rAAV4 was generated using a triple plasmid transfection protocol (Murlidharan et al., 2015). In brief, viral vectors were purified using iodixanol density gradient ultracentrifugation. Vectors packaging were subsequently subjected to buffer exchange and concentration using Sartorius vivaspin2 100-kDa molecular weight cutoff centrifugation columns. Following purification, viral genome titers were determined by qPCR.
In Vivo Viral Titration Experiments
Viral titration experiments were performed for rAAV4 vectors based on the original titer to determine the viral titer for microinjection in order to label an appropriate number of cells in the SGZ for analyses. For all the experiments we performed in this study, the viral titers ranging from 2.3 × 109 particles/mL to 1 × 1011 particles/mL were used to achieve specific labeling of an appropriate number of qNSCs for various experimental purposes and analysis.
Slice Electrophysiology and Ca2+ Imaging
Adult mice were used at 7–14 days post rAAV4 injection for slice preparation. Acute brain slice preparation and conditions for electrophysiological recording and calcium imaging were described previously (Bao et al., 2017). For stimulation and recording of hM3Dq+ NSCs, CNO was either added by dilution into the continuously supplied ACSF or spiked directly into the recording chamber at higher concentrations for transient activation. GABA and glutamate were either bath applied or directly spiked to the recording chamber to record GCaMP6-labeled NSCs.
Immunohistochemistry, Imaging, and Quantification
Sections without signs of tissue damage were chosen for immunostaining (see also Figure S1A). The immunohistology protocol was followed as previously described (Bao et al., 2017). The following primary antibodies were used: mouse anti-GFAP (Millipore #MAB360, 1:1,000), mouse anti-NeuN (Millipore #MAB377, 1:1,000), rabbit anti-GABA (Sigma #A2052, 1:500), goat anti-MCM2 (R&D Systems #AF5778, 1:500), chicken anti-Nestin (Aves Labs #NES, 1:1,000), chicken anti-GFP (Aves Labs #GFP-1020, 1:1,000), rabbit anti-RFP (Rockland #600-401-379, 1:1,000), rabbit anti-S100B (Abcam #ab52642, 1:1,000), rat anti-mCherry (Invitrogen #M11217, 1:500), goat anti-DCX (Santa Cruz Biotechnology #sc-8066, 1:100), goat anti-GFP (Rockland #600-101-215, 1:500), and goat anti-SOX2 (Santa Cruz #sc-17320, 1:250). All fluorescent secondary antibodies were obtained from Life Technologies and diluted to 1:500.
All images for quantification were acquired as a z stack, ≤1 μm axial step, then loaded into ImageJ (Fiji) software as a composite image and counted using the Cell Counter plugin. For 3D reconstruction, images were deconvolved using Huygens Professional software (SVI, the Netherlands). 3D reconstruction data were displayed using Surface Renderer in Huygens.
Statistics
Data graphs were plotted using GraphPad Prism 7.01. Statistical analysis in Figure 1 was performed using the two-tailed, unpaired Student's t test to determine statistical significance.
Author Contributions
J.S. and A.J.C. conceived the project and wrote the manuscript; A.J.C. and S.-A.L. performed surgeries, tissue processing, immunostaining, and confocal imaging; B. Asrican performed electrophysiology and calcium imaging; B. Albright and A.A. packaged rAAV4. D.H.C. and Y.-Y.I.S. assisted with rat experiments. J.W., C.-Y.Y., H.B., and J.H. assisted with experiments.
Acknowledgments
We thank members of the Song lab for comments and discussions. We thank Ken McCarthy's lab for providing the GFAP-GCAMP6s and GFAP-hM3Dq-TdTomato plasmids and Hannah Verrilli for technical support. This work was supported by grants to J.S. from the American Heart Association (14SDG20440029), the Whitehall Foundation, and the NIH (R01MH111773, R21MH106939) and F31MH110146 to A.J.C.
Published: February 22, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and two movies and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.01.018.
Supplemental Information
References
- Armbruster B.N., Li X., Pausch M.H., Herlitze S., Roth B.L. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc. Natl. Acad. Sci. USA. 2007;104:5163–5168. doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao H., Asrican B., Li W., Gu B., Wen Z., Lim S.A., Haniff I., Ramakrishnan C., Deisseroth K., Philpot B. Long-range GABAergic inputs regulate neural stem cell quiescence and control adult hippocampal neurogenesis. Cell Stem Cell. 2017;21:604–617 e5. doi: 10.1016/j.stem.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry G.E., Asokan A. Cellular transduction mechanisms of adeno-associated viral vectors. Curr. Opin. Virol. 2016;21:54–60. doi: 10.1016/j.coviro.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaguidi M.A., Wheeler M.A., Shapiro J.S., Stadel R.P., Sun G.J., Ming G.L., Song H. In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell. 2011;145:1142–1155. doi: 10.1016/j.cell.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breunig J.J., Arellano J.I., Macklis J.D., Rakic P. Everything that glitters isn't gold: a critical review of postnatal neural precursor analyses. Cell Stem Cell. 2007;1:612–627. doi: 10.1016/j.stem.2007.11.008. [DOI] [PubMed] [Google Scholar]
- Dimidschstein J., Chen Q., Tremblay R., Rogers S.L., Saldi G.A., Guo L., Xu Q., Liu R., Lu C., Chu J. A viral strategy for targeting and manipulating interneurons across vertebrate species. Nat. Neurosci. 2016;19:1743–1749. doi: 10.1038/nn.4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch F., Hen R. Young and excitable: the function of new neurons in the adult mammalian brain. Curr. Opin. Neurobiol. 2005;15:121–128. doi: 10.1016/j.conb.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Dranovsky A., Picchini A.M., Moadel T., Sisti A.C., Yamada A., Kimura S., Leonardo E.D., Hen R. Experience dictates stem cell fate in the adult hippocampus. Neuron. 2011;70:908–923. doi: 10.1016/j.neuron.2011.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encinas J.M., Enikolopov G. Identifying and quantitating neural stem and progenitor cells in the adult brain. Methods Cell Biol. 2008;85:243–272. doi: 10.1016/S0091-679X(08)85011-X. [DOI] [PubMed] [Google Scholar]
- Encinas J.M., Michurina T.V., Peunova N., Park J.H., Tordo J., Peterson D.A., Fishell G., Koulakov A., Enikolopov G. Division-coupled astrocytic differentiation and age-related depletion of neural stem cells in the adult hippocampus. Cell Stem Cell. 2011;8:566–579. doi: 10.1016/j.stem.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enikolopov G., Overstreet-Wadiche L., Ge S. Viral and transgenic reporters and genetic analysis of adult neurogenesis. Cold Spring Harb. Perspect. Biol. 2015;7:a018804. doi: 10.1101/cshperspect.a018804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzsimons H.L., Bland R.J., During M.J. Promoters and regulatory elements that improve adeno-associated virus transgene expression in the brain. Methods. 2002;28:227–236. doi: 10.1016/s1046-2023(02)00227-x. [DOI] [PubMed] [Google Scholar]
- He M., Tucciarone J., Lee S., Nigro M.J., Kim Y., Levine J.M., Kelly S.M., Krugikov I., Wu P., Chen Y. Strategies and tools for combinatorial targeting of GABAergic neurons in mouse cerebral cortex. Neuron. 2016;92:555. doi: 10.1016/j.neuron.2016.10.009. [DOI] [PubMed] [Google Scholar]
- Kaplitt M.G., Feigin A., Tang C., Fitzsimons H.L., Mattis P., Lawlor P.A., Bland R.J., Young D., Strybing K., Eidelberg D. Safety and tolerability of gene therapy with an adeno-associated virus (AAV) borne GAD gene for Parkinson's disease: an open label, phase I trial. Lancet. 2007;369:2097–2105. doi: 10.1016/S0140-6736(07)60982-9. [DOI] [PubMed] [Google Scholar]
- Kotterman M.A., Vazin T., Schaffer D.V. Enhanced selective gene delivery to neural stem cells in vivo by an adeno-associated viral variant. Development. 2015;142:1885–1892. doi: 10.1242/dev.115253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Martins I.H., Chiorini J.A., Davidson B.L. Adeno-associated virus type 4 (AAV4) targets ependyma and astrocytes in the subventricular zone and RMS. Gene Ther. 2005;12:1503–1508. doi: 10.1038/sj.gt.3302554. [DOI] [PubMed] [Google Scholar]
- Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming G.L., Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron. 2011;70:687–702. doi: 10.1016/j.neuron.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murlidharan G., Corriher T., Ghashghaei H.T., Asokan A. Unique glycan signatures regulate adeno-associated virus tropism in the developing brain. J. Virol. 2015;89:3976–3987. doi: 10.1128/JVI.02951-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murlidharan G., Samulski R.J., Asokan A. Biology of adeno-associated viral vectors in the central nervous system. Front. Mol. Neurosci. 2014;7:76. doi: 10.3389/fnmol.2014.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar M.D., Tasic B., Miyamichi K., Li L., Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Neu U., Bauer J., Stehle T. Viruses and sialic acids: rules of engagement. Curr. Opin. Struct. Biol. 2011;21:610–618. doi: 10.1016/j.sbi.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaar R.L., Gerardy-Schahn R., Hildebrandt H. Sialic acids in the brain: gangliosides and polysialic acid in nervous system development, stability, disease, and regeneration. Physiol. Rev. 2014;94:461–518. doi: 10.1152/physrev.00033.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz B.R., Chamberlain J.S. Recombinant adeno-associated virus transduction and integration. Mol. Ther. 2008;16:1189–1199. doi: 10.1038/mt.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Berg D.A., Zhu Y., Shin J.Y., Song J., Bonaguidi M.A., Enikolopov G., Nauen D.W., Christian K.M., Ming G.L. Single-cell RNA-seq with waterfall reveals molecular cascades underlying adult neurogenesis. Cell Stem Cell. 2015;17:360–372. doi: 10.1016/j.stem.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Sun J., Moss J., Wen Z., Sun G.J., Hsu D., Zhong C., Davoudi H., Christian K.M., Toni N. Parvalbumin interneurons mediate neuronal circuitry-neurogenesis coupling in the adult hippocampus. Nat. Neurosci. 2013;16:1728–1730. doi: 10.1038/nn.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Zhong C., Bonaguidi M.A., Sun G.J., Hsu D., Gu Y., Meletis K., Huang Z.J., Ge S., Enikolopov G. Neuronal circuitry mechanism regulating adult quiescent neural stem-cell fate decision. Nature. 2012;489:150–154. doi: 10.1038/nature11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh H., Consiglio A., Ray J., Sawai T., D'Amour K.A., Gage F.H. In vivo fate analysis reveals the multipotent and self-renewal capacities of Sox2+ neural stem cells in the adult hippocampus. Cell Stem Cell. 2007;1:515–528. doi: 10.1016/j.stem.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M.Y., Yetman M.J., Lee T.C., Chen Y., Jankowsky J.L. Specificity and efficiency of reporter expression in adult neural progenitors vary substantially among nestin-CreER(T2) lines. J. Comp. Neurol. 2014;522:1191–1208. doi: 10.1002/cne.23497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward C.M., Stern P.L. The human cytomegalovirus immediate-early promoter is transcriptionally active in undifferentiated mouse embryonic stem cells. Stem Cells. 2002;20:472–475. doi: 10.1634/stemcells.20-5-472. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.