Humans are walking culture dishes but we tend to forget what important tasks the trillions of microbes that colonize us are performing on a daily basis. With the advent of high-throughput sequencing and culturing techniques, rapid progress has been made in the enumeration, characterization and classification of the human microbiota. Using gnotobiotic models, several groups have also gained deep insights into host-microbe interactions that occur at steady state, and how perturbation of the microbial community composition can contribute to disease. The Special Issue ‘The Microbiome in Immune Diseases’ is focused on summarizing several advances in autoimmune, rheumatic, and allergic diseases that are mediated by host-microbiota interactions.
It has long been appreciated that immune cells are profoundly affected by the gut microbiota. Studies in the 1990’s, for instance, on gut epithelium-adhering commensals called ‘segmented filamentous bacteria’ or SFB demonstrated that gut lymphocytes and IgA are potently induced by monocolonization [1–3]. Earlier and subsequent studies have collectively shown that the microbiota is instrumental in immune development and maturation. Besides intimate interactions between innate immunity and the microbiota colonizing barrier organs like the gut, skin, or lung, several recent studies have revealed that adaptive immunity, in particular B cell development, IgA repertoire selection, and CD4 helper T cell differentiation are orchestrated by the microbiota [4]. Most notably, it is now well established that Th17 cells are induced by SFB and other commensals that are yet to be determined. Similarly, certain Clostridia species are capable of inducing regulatory T cell (Treg) subsets and thereby promoting immunologic tolerance and protection from inflammatory and allergic diseases [5]. Interestingly, colonic Tregs recognize specifically antigens from the gut microbiota, suggesting that a longstanding co-evolutionary path has led to acceptance of multiple gut commensals as an ‘extended self’. Disruptions in the homeostasis between the host and the microbiota are likely key steps in the multifactorial pathogenesis of immune-mediated diseases. Barrier disruption was recently shown to generate immunologic memory to gut commensals [6], which may have important implications for autoimmune diseases that could be influenced by this phenomenon [7]. This Special Issue aims to summarize several disease-relevant aspects in this intricate interplay between the immune cells at barrier sites and the colonizing microbiota.
First, Flavell and colleagues provide a broad overview of host immune–microbiota interactions, summarizing not only how innate immunity is crucial in maintaining a non-inflammatory state, but also how IgA sensing of pathobionts could be used to identify key candidates in inflammatory bowel disease (IBD) [8]. Zeissig and colleagues expand on the role of adaptive immunity in IBD with a special focus on lipid-reactive NKT cells and their impact on early host-microbiota development, exerting long-ranging effects on immune tolerance in adulthood consistent with the hygiene hypothesis [9]. Stoll provides a succinct overview of the current state of the gut microbiota in ankylosing spondylitis, a rheumatic disease closely linked with IBD but little explored in humans even though one of the earliest evidence of a role for the gut microbiome in rheumatic diseases stems from murine studies using germfree HLA-B27-transgenic animals [10]. In general, mechanistic insight is best gained using gnotobiotic models, that is, germfree animals colonized with a single species or a defined microbial community. Remarkable progress has been made in understanding the protective role of the gut microbiota in models of type 1 diabetes. Wen and colleagues summarize the latest studies in this autoimmune disease and explain also how diet and environmental factors can influence the gut microbiota and thereby autoimmunity [11]. Furthermore, sex hormones can shape the gut microbial community composition. A bidirectional interaction between the microbiota and sex hormones was first shown in gnotobiotic studies of type 1 diabetes. A gender-selective microbiome is also implicated in rheumatoid arthritis and Taneja and colleagues review studies on the interactions between gender, hormones, and the microbiota [12]. Following the emerging paradigm of immune–hormone–microbiota interactions, Aliprantis and colleagues present elegantly how the gut microbiota could impact systemic bone disease [13]. They provide a refreshing look at an underappreciated topic that connects bone homeostasis with host immune–microbiota cross-talk. McCoy and colleagues go beyond autoimmune and metabolic diseases and summarize the role of the microbiota in allergies and asthma [14]. They review recent studies that strongly support a fundamental role of the microbiota in the hygiene hypothesis and discuss potential interactions of the microbiota with the virome. They also introduce the lung microbiome as a plausible susceptibility factor in allergic diseases. Huffnagle and colleagues then summarize extensively the current state of the lung microbiome in health and disease, in particular in chronic lung disease [15]. Finally, Barin and colleagues state the latest findings presented at a recent symposium on the role of the microbiota in autoimmunity [16], covering several of the disease topics listed above as well as multiple sclerosis that was recently reviewed [17].
Much still remains to be learned about the host-microbiota interactions that influence apparently so many disease states (Fig. 1). It will be particularly important to bridge the highly insightful but artificial monocolonization models with the purely descriptive phylotyping studies in humans. This Special Issue gives a first glimpse at how complex the gene–environment–microbiota interactions are in immune-mediated diseases. No matter how complicated the microbiome is, there is no question that manipulations of the microbiota will have tremendous therapeutic opportunities for immunologic diseases.
Figure 1.
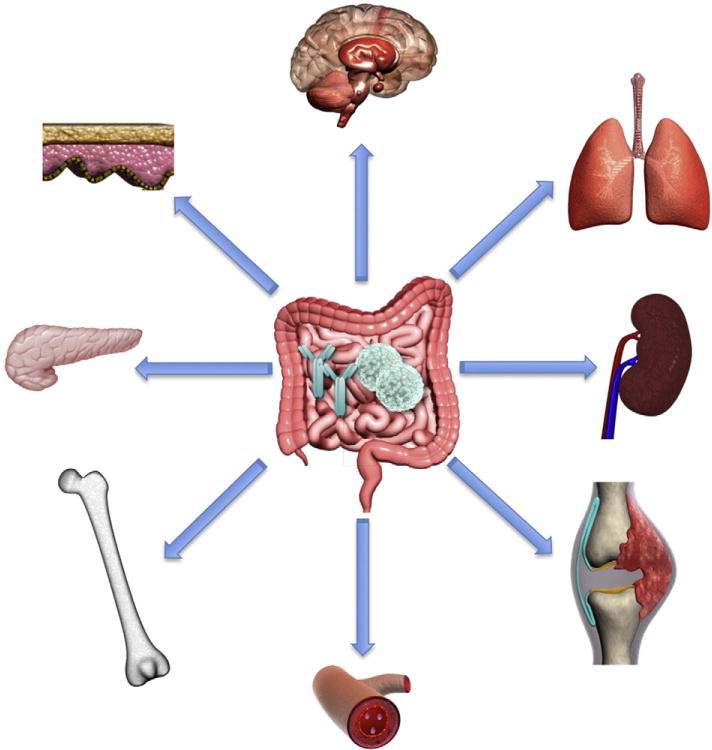
This cartoon summarizes the various organ systems and tissues affected by the gut microbiota and its interactions with the host immune system (depicted as lymphocytes and antibodies in the center of the gastrointestinal tract). These include the brain, lung, kidneys, joints, blood system, bones, endocrine organs (e.g. pancreas), and skin. It should be noted that the skin and lung as barrier organs host also a microbiota themselves that influences disease at these sites.
Acknowledgments
This work was supported by the National Institutes of Health (NIH K08 AI095318), the Arthritis National Research Foundation (ANRF #14-003679), the Lupus Research Institute (LRI 2015), the Yale Rheumatic Diseases Research Core (NIH P30 AR053495), the Women’s Health Research at Yale (WHRY #661), and the George M. O’Brien Kidney Center at Yale (NIH P30 DK079310).
Footnotes
Conflict of interest statement
The author declares no conflicts of interest.
References
- 1.Klaasen HL, Van der Heijden PJ, Stok W, Poelma FG, Koopman JP, Van den Brink ME, Bakker MH, Eling WM, Beynen AC. Apathogenic, intestinal, segmented, filamentous bacteria stimulate the mucosal immune system of mice. Infect Immun. 1993 Jan;61(1):303–306. doi: 10.1128/iai.61.1.303-306.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Umesaki Y, et al. Segmented filamentous bacteria are indigenous intestinal bacteria that activate intraepithelial lymphocytes and induce MHC class II molecules and fucosyl asialo GM1 glycolipids on the small intestinal epithelial cells in the exgerm-free mouse. Microbiol Immunol. 1995;39:555–562. doi: 10.1111/j.1348-0421.1995.tb02242.x. [DOI] [PubMed] [Google Scholar]
- 3.Talham GL, et al. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immun. 1999;67:1992–2000. doi: 10.1128/iai.67.4.1992-2000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maynard CL, et al. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489:231–241. doi: 10.1038/nature11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hand TW, et al. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science. 2012;337:1553–1556. doi: 10.1126/science.1220961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruff WE, Kriegel MA. Autoimmune host-microbiota interactions at barrier sites and beyond. Trends Mol Med. 2015;21:233–244. doi: 10.1016/j.molmed.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palm NW, de Zoete MR, Flavell RA. Immune-microbiota interactions in health and disease. Clin Immunol. 2015;159:122–127. doi: 10.1016/j.clim.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dowds CM, Blumberg RS, Zeissig S. Control of intestinal homeostasis through crosstalk between natural killer T cells and the intestinal microbiota. Clin Immunol. 2015;159:128–133. doi: 10.1016/j.clim.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stoll M. Gut microbes, immunity, and spondyloarthritis. Clin Immunol. 2015;159:134–142. doi: 10.1016/j.clim.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 11.Gülden E, Wong FS, Wen L. The gut microbiota and type 1 diabetes. Clin Immunol. 2015;159:143–153. doi: 10.1016/j.clim.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomez A, Luckey D, Taneja V. The gut microbiome in autoimmunity: sex matters. Clin Immunol. 2015;159:154–162. doi: 10.1016/j.clim.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charles JF, Ermann J, Aliprantis A. The intestinal microbiome and skeletal fitness: connecting bugs and bones. Clin Immunol. 2015;159:163–169. doi: 10.1016/j.clim.2015.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCoy K, Köller Y. New developments providing mechanistic insight into the impact of the microbiota on allergic diseases. Clin Immunol. 2015;159:170–176. doi: 10.1016/j.clim.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huffnagle GB. The bacterial microbiota in inflammatory lung diseases. Clin Immunol. 2015;159:177–182. doi: 10.1016/j.clim.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barin J, Tobias LD, Peterson DA. The microbiome and autoimmune disease: report from a Noel R. Rose Colloquium. Clin Immunol. 2015;159:183–188. doi: 10.1016/j.clim.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Mielcarz DW, Kasper LH. The gut microbiome in multiple sclerosis. Curr Treat Options Neurol. 2015;17:344. doi: 10.1007/s11940-015-0344-7. [DOI] [PubMed] [Google Scholar]


