Abstract
The microbiota is considered to be an important factor influencing the pathogenesis of autoimmunity at both barrier sites and internal organs. Impinging on innate and adaptive immunity, commensals exert protective or detrimental effects on various autoimmune animal models. Human microbiome studies of autoimmunity remain largely descriptive, but suggest a role for dysbiosis in autoimmune disease. Humanized gnotobiotic approaches have advanced our understanding of immune-commensal interactions, but little is known about the mechanisms in autoimmunity. We propose that, similarly to infectious agents, the microbiota mediates autoimmunity via bystander activation, epitope spread, and, particularly under homeostatic conditions, via crossreactivity. This review presents an overview of the current literature concluding with outstanding questions in this field.
Keywords: microbiome, commensal, bystander activation, epitope spread crossreactivity, molecular mimicry
Host-microbe interactions: from Pasteur to present
‘Messieurs, c’est les microbes qui auront le dernier mot.’ (Gentlemen, it is the microbes who will have the last word) – Louis Pasteur
Louis Pasteur was right in many ways. All animals are indeed ‘walking culture dishes’ and will be decomposed and metabolized by the collection of all commensals that colonize us: the microbiota (see Glossary). We are colonized from birth by a large diversity of microbes spanning all three domains of life that form evolutionary ties with the human host. These microorganisms have a range of relationships with their hosts, and exist as mutualists, symbionts, or pathobionts. To date, our understanding of commensal–immune interactions comes principally from studying the gut microbiota. However, the microbiota outside the gut is likely to be equally important, especially with regards to ‘barrier diseases’ that occur on mucosal or skin surfaces that are colonized with niche-specific microbiota [1].
Far beyond the crucial role of gut bacteria in nutrition and metabolism, it is well established that their influence reaches many host physiologic systems [2,3]. Gnotobiotic animal experiments have shed light on how the microbiota influences the metabolism of drugs, neurological function, immune development and homeostasis, and various chronic diseases of modern societies [3]. Commensals exert profound effects on the development and function of the immune system and therefore likely influence immune-mediated diseases [4]. Indeed, gut bacteria prevent, exacerbate, or induce numerous autoimmune, allergic, or inflammatory diseases and malignancies in animal models [5–26]. A causal role for multifactorial autoimmune diseases in humans is still outstanding, but such a role has been demonstrated in murine models of multiple sclerosis (MS) [7–9], rheumatoid arthritis (RA) [11–13], and type 1 diabetes (T1D) [14–18], and is likely to extend to systemic lupus erythematosus (SLE) [27]. Many autoimmune diseases have a female sex bias, and animal studies have provided insight into gut commensal effects on hormonal status and gender bias in autoimmunity [17,18]. The purpose of this review is to provide a basic overview of auto-immunity in the context of commensals, and to explore recent advances in the understanding of the complex interactions between commensal bacteria, the immune system, and the effects these interactions have on autoimmune disease.
Genetics, environment, and commensals: pieces of the autoimmunity puzzle
Genetic polymorphisms, in particular at the human leukocyte antigen (HLA) loci, play a key role in the predisposition to autoimmunity (Box 1) [28,29], but genetics alone cannot fully explain multifactorial autoimmunity. In addition to genetic factors, several environmental and dietary factors have been identified in epidemiologic studies and have been mechanistically linked to autoimmunity in vitro and in vivo. Exactly how these dietary or environmental factors influence autoimmunity in patients, however, is still largely unknown [30].
Box 1. Genetics of autoimmunity.
Autoimmunity is grossly broken down into monogenic and poly-genic multifactorial diseases. Monogenic autoimmune diseases represent rare and highly devastating diseases, typically arising from defects in either central or peripheral tolerance pathways such as FOXP3 and AIRE [119]. Polygenic multifactorial autoimmune diseases represent complex diseases that are caused by a combination of genetic and environmental factors. Diseases such as T1D, RA, MS, and SLE are multifactorial autoimmune diseases. Genetic influence over these diseases is becoming clearer in large part through advances in bioinformatic approaches and sharing of patient genomic data through large consortia [120,121]. How genetics influences the microbiota and thus autoimmunity is not well understood. Numerous genetically modified mouse models display an altered microbiota, although these findings could be biased by non-genetic factors [122].
Genome-wide association studies (GWAS) led to the identification of SNPs associated with disease such as those in genes within the HLA locus or cytokine receptor genes (e.g., IL2RA, IL7RA) implicated in Treg homeostasis and function [28,120]. Each SNP confers a small risk, but taken together the risk is thought to be additive [29]. Consistent with a shared autoimmune pathogenesis, particular SNPs impact upon immune pathways across autoimmune diseases such as HLA loci, co-stimulatory, and proinflammatory pathways [29]. Despite the wealth of information gained from GWAS, this approach has been criticized for lacking the power to identify causal variants. Only a few functional SNPs have been characterized to date, such as those in PTPN22 and TNFAIP3 [111,123]. Groups using novel bioinformatic techniques are addressing this critique [124]. Clearly, combining environmental/ commensal factors with genetics will lead to a more complete understanding of autoimmunity.
Because any dietary and environmental trigger needs to enter the host through one of the mucosal or skin barriers, its effects on autoimmunity are likely modulated by the colonizing microbiota, which is itself shaped by the environment and diet [31]. Diet and antibiotic use, for instance, are well established to profoundly alter the gut microbiota, leading to long-lasting changes in metabolic profiles [32,33]. These metabolites, such as short-chain fatty acids (SCFAs) or retinoic acid, can either dampen or ignite inflammation depending on their effects on host adaptive immune cells, in particular on regulatory T cells (Tregs) and T helper 17 (TH17) cells [34–37]. Environmental triggers of autoimmunity may thus enhance commensal-mediated inflammatory processes and thereby influence autoimmunity, but these links have yet to be demonstrated. However, independently of environmental factors, the gut microbiota is emerging as a key player in the development of autoimmunity.
The abundance and diversity of organisms colonizing the host (Box 2) represent an enormous challenge to the innate and adaptive immune system. On one hand, the immune system needs to recognize any potentially detrimental non-self antigen that enters the host, while on the other hand it cannot eliminate the commensal communities at barrier organs because it would negatively affects host fitness [38]. This conundrum was aptly termed ‘learning tolerance while fighting ignorance’ [39]. The interplay between pathogen and commensal recognition by the immune system is a delicate balance which, when altered, may lead to unintended consequences such as chronic inflammatory and autoimmune diseases.
Box 2. Commensals by the numbers.
It is estimated that human bacteria alone outnumber human cells by a factor of 10 with roughly 100 trillion bacteria colonizing an individual at any point in time [104]. The number of bacterial genes constituting the microbiome is even larger with recent reference catalogs of over 9.8 million non-redundant genes [105].
Improvements in 16S rDNA sequencing methods and dramatic decreases in sequencing costs are allowing a more complete and individualized picture of the microbial communities colonizing each person over time. It is believed that around 1000 species of gut bacteria are present across the human population, with each person harboring 100–160 species at a given moment in time [125,126]. Gut bacterial communities, in the absence of medical interventions (such as antibiotic use) or pathogen infection, tend to be stable over time [126,127]. It should, however, also be noted that the gut microbiota encodes 100-fold more proteins than the human genome, including over 2 million proteins that are found in less than 20% of individuals [128]. This immense number provides a major source for antigenic variation that could contribute to immune stimulation and crossreactivity in autoimmunity.
Dysbiosis: intestinal homeostasis and inflammatory bowel disease
Dietary changes, antibiotic use, and excessive hygiene disrupt commensal homeostasis leading to dysbiotic states that drive chronic inflammation (Figure 1). Chronic inflammation in the gut is the hallmark of inflammatory bowel disease (IBD), which encompasses a spectrum of diseases from ulcerative colitis (UC) to Crohn’s disease (CD). The gut microbiota is known to play an especially important role in IBD, which is not surprising given that the gut is a barrier organ that carries the largest microbiota, and an abnormal innate and adaptive immune response to multiple members of the gut microbiota is thought to drive inflammation [10,23,24,40].
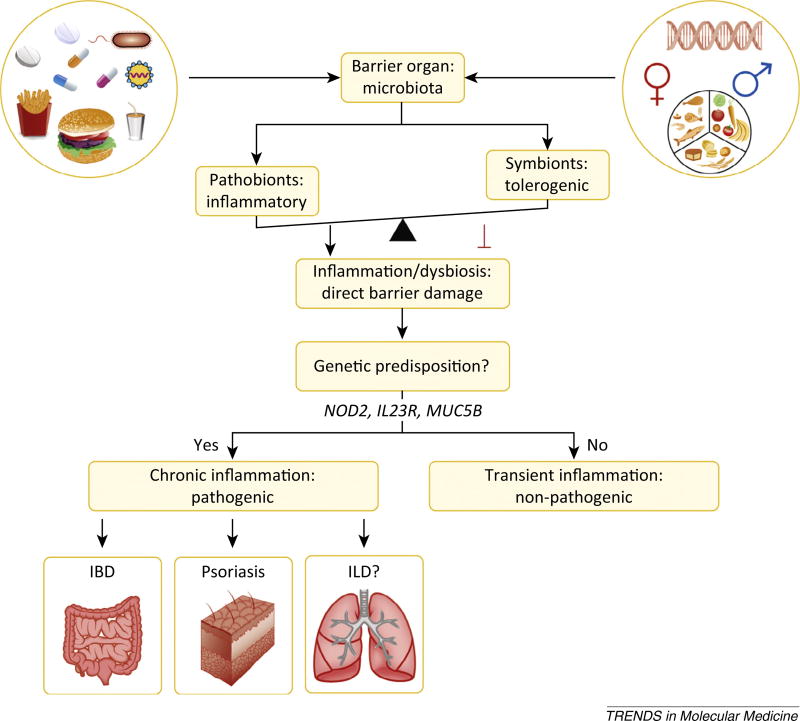
Figure 1.
Gut microbiota balance inflammation at barrier organs. Complex interactions between genetics, environment, and the microbiota shape the inflammatory status at barrier sites, and this impinges on autoinflammatory and autoimmune diseases. Diet, hygiene, antibiotics, pathogenic infections, and hormones shape the balance between pathobionts and symbionts at barrier organs. The barrier sites themselves, as well as the effects of each of these external factors on the barrier organs, are influenced by genetics. Increases in pathobionts or decreases in anti-inflammatory commensals favor aberrant immune interactions with these microbes, leading to dysbiosis and direct barrier damage. In genetically predisposed individuals, barrier damage is thought to instigate inflammatory bowel disease (IBD) in the gastrointestinal tract and psoriasis at cutaneous surfaces. The susceptible sites are determined by the colonization patterns of commensals. Other barrier sites might be equally affected by the microbiota, for example interstitial lung disease (ILD) in the lung. Individuals not genetically predisposed will have only transient or no inflammation with limited pathogenesis. Genes implicated in gut, skin, or lung barrier integrity are, for example, nucleotide-binding oligomerization domain-containing protein 2 (NOD2; polymorphic in IBD), interleukin-23 receptor (IL23R; polymorphic in psoriasis), and mucin 5B (MUC5B; polymorphic in ILD).
16S ribosomal DNA (rDNA) profiling studies revealed a decrease in the diversity of microbial communities in IBD compared to controls [41]. Despite efforts to understand these diseases, reports of the specific microbial communities associated with dysbiosis in IBD are variable [40,41]. Recent work attempts to overcome previous limitations by leveraging a standardized multicenter cohort of new-onset pediatric CD and healthy controls. Gevers and colleagues collected samples from 668 patients and controls from multiple sites within the intestinal tract before treatment. These efforts led to the identification of candidate organisms in microbial dysbiosis of pediatric CD. An increase in Enterobacteriaceae, Pasteurellaceae, Veillonellaceae, and Fusobacteriaceae, and a decrease in Erysipelotrichales, Bacteroidales, and Clostridiales, correlated with disease status [40]. While correlative studies in humans are proving useful to identify biomarkers and potential pathobionts, novel approaches (e.g., 16S rDNA sequencing of IgA-coated microbiota) and mechanistic studies in gnotobiotic models are key to better delineating the pathogenesis of IBD (Box 3) [24,42].
Box 3. IBD as a paradigm.
The contribution of the gut microbiota has been dissected in multiple IBD models. One well-studied animal model is the TRUC (T-bet−/−Rag2−/− ulcerative colitis) mouse model [129]. TRUC mice develop spontaneous ulcerative colitis due to genetic defects in innate (T-bet in dendritic cells) and adaptive immunity (Rag2 in lymphocytes). Interestingly, the altered microbial communities present in these mice were shown to transfer disease to genetically intact mice, supporting a dominant role of pathobionts over host genetics [129]. Recently, this model was shown to be important to decipher the effects of treatment modalities commonly used in human UC. Anti-TNF therapy, which is successfully used in human IBD patients, suppresses the pathobionts that mediate colitis [23]. Intriguingly, once anti-TNF therapy is discontinued, the pathobionts outgrow again, providing one plausible mechanism why biologic therapies currently used in autoimmune diseases do not cure patients and why patients usually flare after discontinuing immunosuppressive therapies. Furthermore, TRUC animals with active disease have a reduced potential in carbohydrate and energy metabolism, and an increased potential for flagellar assembly, tetrathionate respiration, and benzoate degradation [130]. These findings support the hypothesis that, during states of inflammation, particular bacteria can exploit host substrates, which leads to an increase in fitness. For example, the ability to respire tetrathionate improves the fitness of several Enterobacteriaceae and can also increase the bioavailability of ATP, which is linked to enhanced inflammatory TH17 skewing [131–133]. In addition to the human study of pediatric CD, increases in Enterobacteriaceae have also been shown in the TRUC model [23]. The specific disease trigger, however, may vary based on the maternally transmitted microbiome, the animal facility, or other external variables. Indeed, another group in Europe identified Helicobacter typhlonius as the causal pathobiont in the TRUC model [134]. It therefore remains to be seen if UC and CD are both instigated by similar communities within the gut microbiota, or if different communities drive each disease that may also vary in individual patients. In either case, these studies support that pathobionts exploit inflammation, which in turn drives additional proinflammatory feedback mechanisms. These processes could serve to promote IBD and also autoimmune diseases outside the intestine.
The microbiota shapes adaptive immunity and influences autoimmunity
Studies of gut microbial dysbiosis in IBD has greatly added to our understanding of host–microbiota interactions within the confines of the gut, but it is well established that gut commensals also affect peripheral immunity, even in the absence of dysbiosis [1,5]. Early observations of the profound effects of commensals on host immunity came from animals without natural microbial colonization at birth (germ-free animals). Germ-free mice have severely decreased numbers of CD4+ T cells in the periphery [43]. In addition, the balance of CD4+ TH cell populations are skewed towards a TH2 phenotype with a decrease in TH1 cells, supporting the hygiene hypothesis that the incidence of allergic and immune-mediated diseases rise with increased hygiene (Figure 2A) [19,20,44].
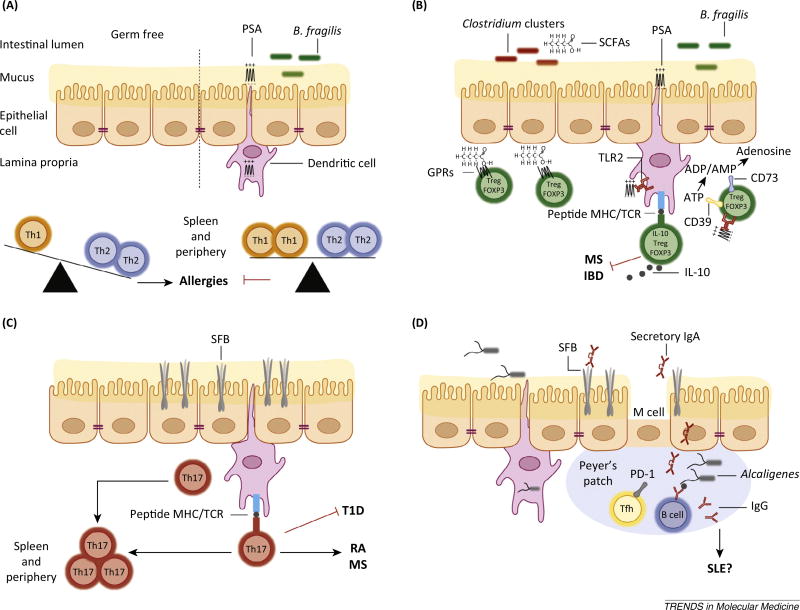
Figure 2.
Microbiota influence CD4+ helper T (TH) cell subsets. (A) Germ-free animals have decreased numbers of CD4+ T cells in the periphery. The splenic CD4+ T cell population of germ-free animals is skewed towards TH2 cells over TH1 cells, favoring allergic diseases. This phenotype is reversed by monocolonization with the zwitterionic polysaccharide-A (PSA)-producing Bacteroides fragilis (B. fragilis) or oral administration of PSA alone. This process is mediated through the recognition and presentation of PSA by dendritic cells (DCs) sampling the intestinal lumen. (B) In the gut, PSA signals through the innate toll-like receptor 2 (TLR2) mediating the expansion of CD39+ FOXP3+ interleukin-10+ (IL-10) Tregs. The ectonucleotidases CD39 in conjunction with CD73 promote degradation of ATP. PSA-induced Tregs have been shown to inhibit multiple sclerosis (MS) and colitis in animal models. Additional anti-inflammatory effects are elicited by bacterial production of short-chain fatty acids (SCFAs) that also dampen colitis. SCFAs promote the stabilization of Tregs by inhibiting histone deacetylases and triggering chemotactic G protein-coupled receptors (GPRs) on FOXP3+ Tregs. (C) Segmented filamentous bacteria (SFB) induce the differentiation of inflammatory CD4+ TH17 cells through direct interactions with intestinal epithelial cells and MHC class II-dependent antigen presentation of SFB antigens to CD4+ T cells by intestinal DCs. The induction of intestinal TH17 cells by SFB has been linked to inhibition of type 1 diabetes (T1D) and the induction of rheumatoid arthritis (RA) and MS in animal models. (D) SFB and Alcaligenes species promote the antigen-specific and antigen-independent production of immunoglobulin A (IgA) in the intestinal lumen and in the periphery. The gut microbiota influences the production of T cell dependent immunoglobulin (Ig) production likely through PD-1+ follicular TH cells (TFH) that are known to instruct B cells to produce antigen-specific Ig. This process could influence systemic lupus erythematosus (SLE) or other autoantibody-mediated autoimmune diseases.
Bacteroides fragilis regulates TH cell balance
How the microbiota influences adaptive immune responses was largely unknown until a groundbreaking study by Mazmanian et al. demonstrated that B. fragilis corrects several immune defects seen in germ-free mice through the production of the zwitterionic polysaccharide A (PSA) [45]. PSA is now considered a symbiosis factor because orally administered PSA alone replicates the effect of B. fragilis monocolonization on TH1/TH2 balance, whereas a PSA-deficient strain of B. fragilis failed to do so (Figure 2A) [45].
Remarkably, PSA exerted protective effects in a mouse model of MS known as experimental autoimmune encephalomyelitis (EAE). PSA was shown to act through the innate immunity toll-like receptor 2 (TLR2) and to mediate the expansion of tissue-specific CD39+ FOXP3+ interleukin-10+ Tregs (Figure 2B) [46,47]. It has recently been shown that plasmacytoid dendritic cells interacting with CD4+ T cells are necessary for the protective effect of PSA in EAE and colitis [48]. The anti-inflammatory effects of PSA may be beneficial in other autoimmune models via several mechanisms. In addition to the induction of the anti-inflammatory cytokine IL-10, upregulation of the ectonucleotidase CD39 in conjunction with CD73 on the cell surface of Tregs promotes degradation of ATP, ADP, and AMP to adenosine, which is thought to dampen the inflammatory effects of excess ATP (Figure 2B) [49].
It should be noted that not all strains of B. fragilis are anti-inflammatory and they may have immunoregulatory roles that are independent of PSA [50,51]. Particular strains of B. fragilis are highly pathogenic, producing the enterotoxin B. fragilis toxin (BFT). BFT-producing B. fragilis are known to induce pathology leading to inflammation-mediated diarrhea, and recent studies suggest it may also play a role in gastrointestinal tumorigenesis [50]. Furthermore, B. fragilis was shown to protect from autism-like disease in an animal model that was linked to immune dysregulation [51]. These studies illustrate how the same commensal species can exert pleiotropic effects depending on the model studied, and on the molecular details of how commensals influence the immune system.
Clostridium clusters and SCFAs: ‘quenching the fire’
Clostridium clusters IV, XIVa, and XVIII have been identified as inducers of FOXP3+ CD4+ Tregs in mice [34]. Unlike the individual commensal effects described above, this work highlights the importance of community composition on immune modulation because individual species of Clostridia were unable to replicate the effect of the collection of 46 strains of Clostridia on Treg induction [34]. Follow-up work showed that 17 strains of Clostridia isolated from a human fecal sample could also induce Tregs in germ-free mice colonized with this mix [52]. Similarly to murine Clostridia, a minimum number of strains was necessary to promote human Treg numbers and function [53]. The induction of Tregs by these bacteria appears to be mediated through the production of SCFAs, in particular butyrate (Figure 2B) [54].
Several groups have recently characterized the molecular mechanisms of SCFA-induced Tregs. It has been shown that butyrate inhibits histone deacetylases that in turn promote Treg induction and maintenance as well as intestinal macrophage function [55,56]. In addition, SCFAs signal through G protein-coupled receptors eliciting gut homing of cells expressing these receptors. Depending on which immune cells are recruited and in what context, SCFA-mediated homing contributes to both pro- and anti-inflammatory responses; for instance, recruitment of FOXP3+ Tregs by SCFAs creates an anti-inflammatory environment [36,37,57–59]. The effect of SCFAs on Tregs is not dependent on the presence of bacteria because orally administered SCFAs induce changes in Tregs similar to those observed in animals colonized with SCFA-producing bacteria [36,55]. Transcriptional regulation of Tregs is also partially mediated by signals from Clostridia. A recent study demonstrated that the DNA methylation adaptor Uhrf1 is rapidly upregulated in germ-free mice upon colonization with these Clostridium clusters, and that this functions to stabilize and promote Tregs [60]. Collectively, these studies represent a framework to understand some of the intricate molecular pathways affected by the production of SCFAs in the gastrointestinal tract.
Clostridium clusters are a heterogeneous group containing pathogenic and commensal bacteria [61]. As discussed above, clusters IV, XIVa, and XVIII create an anti-inflammatory milieu, but species within other clusters are implicated in colitis and autoimmunity. For instance, members of the Allobaculum genus (cluster XVI) were increased compared to controls in an HLA-transgenic model of RA [62]. Further, Clostridium difficile (cluster XI), which can be part of the normal human microbiota in about 3% of healthy subjects, is a frequent causative agent of antibiotic-associated colitis, with differences in pathogenesis being accounted for by strain differences [63]. The Clostridium clusters thus highlight the complexity of microbial interactions with the host immune system, ranging from anti-inflammatory responses to fulminant colitis depending on the clusters, genera, species, and strains.
Commensals are potent inducers of TH17 cells: segmented filamentous bacteria and beyond
Seminal work by Ivanov et al. showed that the murine commensal segmented filamentous bacterium (SFB), a non-culturable Clostridium-related species, induces proinflammatory CD4+ TH17 cells in the small intestinal lamina propria [64]. This effect is likely related to the adherent colonization of the intestinal epithelium unique to SFB. SFB penetrate the mucosal layer and adhere tightly to the surface of the epithelial cells in the terminal ileum (Figure 2C) [64]. Interestingly, the complete absence of SFB from particular commercial suppliers of laboratory mice was linked to a dearth of lamina propria TH17 cells in these animals [16,64]. This fact likely contributes to the variation in disease phenotypes observed within and across animal facilities [16].
Given the importance of TH17 cells in inflammation and TH17-mediated autoimmune diseases, it is plausible that SFB would have an effect on autoimmune pathogenesis. Indeed, several groups have shown that SFB colonization status modulates the outcomes in autoimmune models [9,12,16,18]. It is notable that SFB colonization can both promote the pathogenesis in animals models of autoimmunity, such as RA and MS, and have protective effects in others, such as the non-obese diabetic (NOD) animal model of T1D [9,12,16,18]. The differences are likely dependent on genetic background and on which CD4+ TH cell phenotype drives a particular type of autoimmune disease.
Germ-free rederivation of the K/B × N mouse model of RA led to protection from disease with a decrease in systemic TH17 cells, follicular TH cells (TFH), and germinal centers [12]. Upon monocolonization with SFB these mice again developed disease, but were protected by elimination of SFB via antibiotics [12]. Similar results were found in EAE, with colonization of SFB driving not only TH17 responses in the gut but also IL-17A-secreting TH17 cells in the central nervous system (CNS) [9].
The effects of the microbiota on the NOD mouse model of T1D were theorized based on the variance of diabetes incidence in NOD mice housed at different institutions, with complete penetrance in germ-free NOD mice [14]. However, natural segregation of SFB colonization in NOD mice (i.e., tracking physiologic spread of commensals within a colony) was correlated with protection against diabetes, although only in the more-susceptible female mice [16]. Monocolonization of NOD mice with SFB modestly protected male mice, likely due to substrain differences or the combined effects of SFB with associated commensals that are missing in the monocolonized setting [18]. The mechanistic basis for these gender-specific protective effects of SFB has yet to be elucidated in detail, but it is remarkable that SFB increased testosterone levels in this setting, which is known to protect against female-biased autoimmunity [18].
In addition to the effects SFB has on immune cells, the influence of SFB on mouse models of autoimmunity appears to be highly dependent on genetic background. Using a spontaneous mouse model of relapsing–remitting MS, the SJL/J mouse model, a recent study found that the phenotype was dependent on the microbiota, but was only partially affected by SFB colonization [8]. In this model, the gut microbiota was necessary to activate autoantigen-specific T cells in the gut-associated lymphoid tissue, and to recruit autoantibody-producing B cells to the CNS [8]. Additional work demonstrated that B10.S mice, which are genetically resistant to EAE induction, harbor significantly higher populations of TH17 cells in the intestine compared to SJL/J EAE-susceptible mice. Blockade of the gut-homing integrin α4β7 abrogated resistance in B10.S mice [65]. The link between intestinal immune cells and autoimmunity outside the gut remains to be fully elucidated. These studies nevertheless provide intriguing clues to better understand microbiota effects in the heterogeneous human population.
The link between genetics, gut microbiota, and target organ autoimmunity may not be limited to SFB and EAE because altered gut microbial communities and increased gut permeability have been demonstrated in an HLA-DRB1*0401-susceptible mouse model of RA [62]. Together, these studies serve as an important reminder that observations in one animal model of autoimmunity may not hold in another model of the same disease, and that one must consider the complex interplay between the genetic background of the host and the composition of the microbiota.
Furthermore, one must also take into account the host specificity of the microbiota [66]. SFB have yet to be readily identified in humans, although one study suggests that colonization is age-dependent and is mainly found within the first 2 years of life [67]. It is unknown if these SFB, identified by comparative studies, induce lamina propria TH17 in humans. In adults, it is likely that there are other gut commensals capable of inducing TH17 cells. Interestingly, a chloroform-sensitive fraction of the human adult microbiota is capable of inducing TH17 cells in gnotobiotic mice, suggesting that such microbial communities exist [52]. Certainly, identifying human commensals that could promote similar TH cell phenotypes will be of great importance.
Commensal bacteria shape the IgA repertoire at mucosal surfaces
Also blurring the distinction between host and microbe, Alcaligenes species were shown to inhabit Peyer’s patches (PP) in both mice and humans, and to induce murine antigen-specific IgA production [68]. SFB, and particularly Alcaligenes, are potent inducers of not only antigen-specific but also total IgA (Figure 2D) [69,70]. The intimate contact with host tissue (epithelial cells for SFB and PP for Alcaligenes) is likely crucial for this process, but the mechanisms must be distinct as SFB expand in the absence of IgA, whereas Alcaligenes decline [68,71], suggesting an evolutionary dependence of this unusual commensal on IgA induction.
IgA synthesis can be T cell dependent or independent [72]. TFH and follicular regulatory T cells (TFR) control T cell dependent immunoglobulin production and are also implicated in autoimmunity [73,74]. The co-inhibitory receptor PD-1 on TFH cells selects IgA plasma cell repertoires and gut microbiota composition [75]. Whether commensal IgA dynamics are involved in autoimmunity is not clear, but it is notable that IgA deficiency predisposes to multiple autoimmune diseases [76]. Also, TFR-induced IgA selection enhances the diversification of the gut microbiota through selection of the antibody repertoire [77].
A diverse microbiota is generally a sign of gut health, and restricted diversity emerges as a signature of a variety of chronic disease states including autoimmunity [6]. It is therefore likely that IgA–microbiota interactions are key in the pathogenesis of autoimmunity, as supported by recent studies on the IgA-bound fraction of fecal microbiota from IBD patients [24]. Interestingly, TH17 cells have also been shown to convert to TFH cells in the PP of the gut and aid IgA production in mice [78]. Finally, mucosal IgA production can be shifted to systemic IgG, especially in humans that are regularly exposed to environmental gut irritants and pathogens [79]. This was illustrated by a recent study showing that HIV-specific B cell clones from patients crossreacted with commensal bacteria and were detected in the gut as well as peripheral blood [79]. Taken together, there are multiple ways that the microbiota influences B cell responses, which in turn could impact on autoimmunity.
Sex, drugs, and bugs
Host immune–microbiota interactions in autoimmunity become even more complex when considering also the effects of hormones, medications, and non-bacterial microbiomes. Most autoimmune diseases are female-biased, including RA, MS, and SLE, and commensals also appear to contribute to this gender bias [31]. Studies have shown that the gut microbiota influences the gender bias seen in the NOD mouse model of T1D [17,18]. While T1D is not female-biased in humans, the NOD mouse develops other endocrine autoimmunity (thyroiditis, adrenalitis) resembling the female-biased autoimmune polyglandular syndrome type II, and also serves as a model for the highly female-biased Sjo¨gren’s syndrome [80]. This work thus highlights the importance of exploring not only sex hormones and X-linked genes but also the microbiome in autoimmune gender bias [31].
Further, the commensal metabolome is vast, and directs not only aspects of adaptive immunity but also of host metabolism and drug metabolism [81]. The exact mechanisms of drug metabolism by commensals are not yet well characterized for most drugs. The cardiac drug digoxin, however, was recently shown to be inactivated by the specific strain Eggerthella lenta DSM2243, whereas other strains were incapable of inactivating digoxin [82]. Strain-specific effects thus appear to be particularly important in understanding drug metabolism.
Finally, it is important to consider non-bacterial commensals in host immunity and disease. While the roles of endogenous retroviruses, viral elements, bacteriophages, and commensal fungi on autoimmunity are poorly studied, it is an area of intense investigation and is probably also an important aspect of autoimmunity and human disease [27,83,84]. In addition, chronic infestation with helminths, which is largely absent in industrialized nations, is linked to protection from various immune-mediated diseases, including allergies, IBD, T1D, and MS, by creating an anti-inflammatory environment in the host [85]. Taken together, the influence of viral, bacterial, and eukaryotic microbiomes on autoimmunity is likely much larger than currently appreciated, and future studies on the precise mechanisms of the host–commensal interactions will be crucial to fully understanding autoimmune diseases.
Commensal-specific T cells: are antigens the issue?
Commensal antigen-specific T cells are detectable distant from the gut and are generated following mucosal breaches in the gut barrier [86]. Parallel studies from two groups have demonstrated that TH17 cells induced by SFB colonization are SFB-specific and dependent on antigen presentation via MHC class II on intestinal dendritic cells [87,88]. Using an SFB-specific T cell receptor (TCR)transgenic model, it was shown that most TH17 cells in the small intestinal lamina propria of mice colonized with SFB were antigen-specific for this commensal [87]. In addition, this work explored the role of competing bacteria on the induction of SFB TCR transgenic helper subsets. Co-colonization with Listeria monocytogenes, a potent inducer of TH1 populations, had no effect on the expression of RORγt in SFB antigen-specific T cells. TH17 cells that are SFB-specific are thus robustly induced and not easily skewed [87]. More importantly, they can exit the mucosal environment and circulate in the periphery, potentially affecting the biology at distant sites [87]. How SFB promote non-SFB antigen-specific TH17 cells in the periphery, and how these cells affect autoimmune pathogenesis, are crucial questions in the field that remain to be elucidated.
In addition to commensal-specific TH1 and TH17 cells, commensal-specific Tregs have also been identified [89]. These particular cells likely evolved to maintain tolerance to commensal bacteria that are beneficial to the human host [39,90,91]. There is evidence that both thymic selection and peripheral education influences commensal-specific Treg populations [92,93]. These commensal-specific Tregs are not fixed and can switch to effector phenotypes during homeostatic disruptions – as evidenced during infections [86]. This switch from regulatory to effector phenotypes may have profound implications for the development of autoimmunity both at barrier organs (i.e., skin, gut, and lung) and systemic target organs such as the brain in MS, or joints in RA.
The hunt for new immunomodulatory commensals
As it becomes clear that commensals exert immunomodulatory functions that have profound effects on human autoimmunity, it will be necessary to identify both individual commensal species and commensal communities that exhibit these capacities. The identification and study of immunomodulatory commensals is expedited by sophisticated techniques such as the colonization of germ-free mice using combinatorial communities combined with high-throughput screening [94,95]. Similarly, profiling the IgA-coated fraction of human fecal microbiota of IBD patients revealed communities that drive IBD in humanized, gnotobiotic models [24]. Identifying these species is the first step to being able to perform mechanistic studies that will ultimately lead to new diagnostic and therapeutic approaches in humans. While studies using germ-free mice are artificial, and are associated with an abnormal neonatal immune development that could bias autoimmune studies in adult animals [96], the use of advanced techniques, such as those mentioned above, are crucial to better understand the influence of the microbiota on autoimmunity in vivo.
Commensals: mediators of autoimmunity?
In the absence of a breach of mucosal barriers, commensal-specific T and B cells are generally restricted to mucosal compartments and are tolerogenic [89,97]. As discussed earlier, commensal-specific Tregs are capable of switching to effector phenotypes during disruption of mucosal barriers, as evidenced during infections [86]. This switch from regulatory to effector phenotypes would allow one to draw an analogy to how infectious agents breach immunologic tolerance in autoimmunity, chiefly via three mechanisms: bystander activation, epitope spreading, and molecular mimicry (Figure 3) [98].
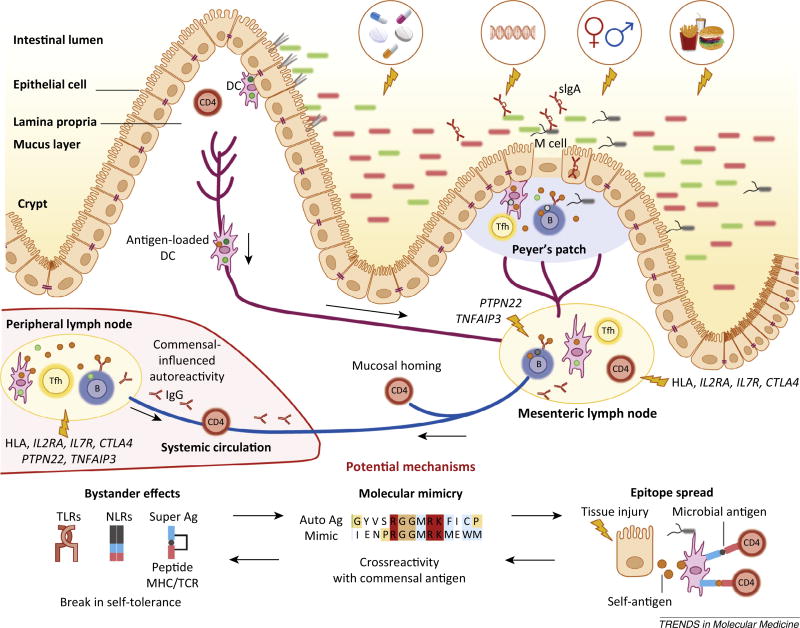
Figure 3.
The gut microbiota impinges on non-gut autoimmunity. The gut microbiota balances the development of organ-specific and systemic autoimmunity in animal models. Dietary, antibiotic, genetic, hormonal, and hygienic factors affect the composition of the intestinal microbiota. Altered microbial community composition or function (dysbiosis) influences immune homeostasis locally and systemically. Genetic susceptibility (particularly via HLA haplotypes and genes regulating inflammation), combined with barrier disruption and dysbiosis, will increase the propensity of antigen-presenting cells [e.g., dendritic cells (DCs) or B cells] to take up antigen at different sites (lamina propria and Peyer’s patches), become activated, and present antigen to cognate T cells in secondary lymphoid organs. Follicular TH cells (TFH; shown as Tfh) cells help B cells in the germinal centers of lymph nodes or Peyer’s patches to produce different classes of antibodies, such as secretory IgA (sIgA) in the gut and IgG and IgA in the periphery. Activated immune cells traffic from the intestine to the mesenteric lymph nodes where they become imprinted with intestinal-homing markers. Some cells will also circulate systemically entering peripheral lymph nodes and target tissues. The development of autoreactive lymphocytes is proposed to occur due to three mechanisms: bystander effects, molecular mimicry, and epitope spreading. These mechanisms are not mutually exclusive and can also affect barrier organ autoimmunity. Each of these three mechanisms applied to commensals could be implicated in the development of non-barrier organ autoimmune diseases, for example type 1 diabetes (T1D), rheumatoid arthritis (RA), multiple sclerosis (MS), or systemic lupus erythematosus (SLE), similarly to transient autoimmune syndromes induced by pathogenic infections affecting for instance the joints (rheumatic fever) or nervous system (Guillan–Barré syndrome). Without genetic susceptibility, the proposed anti-commensal responses would be limited and lead to no overt autoimmunity. In a genetically predisposed host, several scenarios can occur depending on the genetics and pathobiont/ symbiont balance. Dysbiosis could be sustained by a genetic predisposition that leads to innate or B cell hyper-responsiveness, for example via polymorphisms in protein tyrosine phosphatase 22 (PTPN22) or tumor necrosis factor α-induced protein 3 (TNFAIP3). Without overt dysbiosis, antigen-specific recognition of commensals could lead to autoimmunity via crossreactivity if HLA polymorphisms or genetically encoded defects in Tregs or T cell homeostasis are present, for example in interleukin-2 receptor a (IL2RA), interleukin-7 receptor (IL7R), or cytotoxic T lymphocyte-associated protein 4 (CTLA4).
Bystander activation refers to concomitant presentation of a self- and a microbial antigen (including superantigen) to antigen-specific T cells during tissue damage, with triggering of pattern-recognition receptors, such as toll-like receptors (TLRs) and nod-like receptors (NLRs), that override tolerogenic signals [99]. Commensals, either during a breach in mucosal barriers or under homeostatic conditions in a genetically predisposed individual, may trigger a break in tolerance that overrides tolerogenic signals of auto-antigen specific T and B cells.
Epitope spreading occurs during an immune response to microbial antigens that leads to tissue destruction and apoptosis, release of self-antigens, and concomitant presentation of self- and microbial-antigens. This generates a break in tolerance to multiple self-antigens that spreads from epitope to epitope [100]. Similarly to bystander activation, dysbiotic conditions may lead to disruptions of the mucosal barriers, and commensal-specific immune recognition would switch from tolerogenic to effector responses, and this would drive tissue destruction and possible epitope spreading.
These latter two mechanisms occur mainly in the setting of overt inflammation and tissue damage as evoked by invading pathogens or a dysbiotic commensal environment. The vast majority of gut commensals, however, co-exist in the host without eliciting an inflammatory response [39]. In this case, molecular mimicry or crossreactivity with commensal antigens would be most plausible.
The microbiota: a crossreactive universe?
The idea that molecular mimicry is a driver in autoimmunity was proposed long ago. Although it is difficult to prove definitively, this mechanism has been supported by several in vitro and in vivo experiments in cases of human autoimmunity [101]. Some of the best-studied examples are rheumatic fever (triggered by Streptococci) and Guillain– Barré syndrome (induced by Campylobacter jejuni infections) [102,103], both of which are transient autoimmune syndromes. While sequelae from the autoimmune attack (carditis or paralysis) can be long-lasting, autoreactivity typically wanes over time. It is tempting to speculate that chronic autoimmune diseases are instead sustained by crossreactivity with persistently colonizing commensals, especially when considering the enormous number of commensals and their proteomes (Box 2) [104,105].
While crossreactivity to commensals has not been proven to drive autoimmunity, recent work has established that commensals can induce crossreactive lymphocytes, which may have evolved to increase host fitness [106]. Healthy individuals contain memory T cells specific to viruses to which they have never been exposed [106]. The virus-specific memory cells were shown to cross-react with both exogenous and commensal antigens [106]. Despite this crossreactivity, commensal/self-reactive T cells should be of a regulatory phenotype under normal conditions. However, it may suffice that Treg responses to gut commensals are genetically or environmentally skewed to a pathogenic TH subset (TH1, TH17, or TFH cells), for instance in the setting of barrier disruption or genetic predisposition with single-nucleotide polymorphisms in IL2RA or IL7R as in MS [107]. Support for TH cell skewing during barrier disruption comes from a study that demonstrated a break in T cell tolerance to commensals during pathogen-induced breaches of the intestinal barrier [86]. Therefore, the physiologic adaptive immune responses that are generated under homeostasis to contain the gut microbiota within the mucosal barrier might suffice to elicit a crossreactive response under conditions of physiologic stress.
Crossreactivity to commensals is not restricted to T cell recognition because antibody responses in the gut are directed to commensal and self-antigens, with up to 25% being polyreactive [108]. Increased B cell polyreactivity is caused by defects in central and peripheral B cell tolerance pathways, and is an important factor in autoimmunity [109,110]. Defects in B cell tolerance checkpoints are well established in patients with T1D, RA, or MS [111–113].
Recent work in mice suggests that B cell lineages can develop in the lamina propria and that the B cell receptor light-chain repertoire is influenced by the microbiota [114]. Supporting a role for commensal crossreactivity to increase host fitness, HIV-1 envelope protein gp41-specific antibodies crossreact with commensal bacteria (specifically an intracellular E. coli RNA polymerase) [79].
In any of these situations, such as homeostatic responses to gut commensals at steady-state, inflammatory breaches of the gut barrier, or genetic lesions predisposing to aberrant immune system responses to commensals, crossreactive T and B lymphocytes could be generated that sustain an immune response to self-antigen in chronic autoimmune diseases. This hypothesis is testable in vitro with human samples and in vivo in animal models, especially if a genetic deletion of the autoantigen is available. However, predicting crossreactive antigens remains a challenge.
A recent study attempting to overcome these limitations combined high-throughput in vitro screens with predictive algorithms to determine crossreactive candidates [115]. These methods successfully predicted several candidate antigens and showed that the degree of crossreactivity in MS autoantigen-specific T cell clones is broader than previously anticipated [115]. While this study only focused on limited dominant epitopes from MS T cell clones, it opens the possibility to explore crossreactivity between commensal antigens and autoreactive T cells in a variety of autoimmune diseases.
Concluding remarks and future perspectives
Antigen-specific recognition of self is the hallmark of autoimmune diseases. Commensals exert remarkable effects on the pathogenesis of autoimmunity in a variety of models, but many questions remain unanswered (Box 4). Because recent work supports the generation of commensal antigen-specific TH cell subsets, we propose bystander activation, epitope spread, and crossreactivity as potential mechanisms of how immune recognition of the microbiota could mediate disease (Figure 3). Given the ‘antigenic universe’ that is encoded in the microbiome, we suspect that crossreactive commensal immune responses are common and underappreciated. We have focused in this review primarily on CD4+ T cell responses but would like to extend this hypothesis equally to cytotoxic T cell and B cell responses. Crossreactive follicular TH cells that support B cell production of autoantibodies could theoretically fuel antibody-mediated autoimmune diseases. In addition, conformational or linear crossreactive B cell epitopes could similarly be derived from gut commensals.
Box 4. Outstanding questions.
Does the neonatal microbial community composition (and thus the mode of delivery and nursing) affect autoimmune predisposition later in life?
Do HLA and other genetic risk loci select for a pathogenic microbiota in autoimmune patients?
Do common gut microbial or metagenomic signatures exist across autoimmune diseases similarly to shared genetic risk loci?
What is the role of the microbiota in the pathogenesis of systemic autoimmunity?
Do commensal antigens crossreact with autoantigens in genetically predisposed individuals, thereby driving chronic autoreactivity at steady-state?
Is IgA coating of pathobionts consistently identifiable in autoimmune patients, and could it serve as a novel biomarker?
Can dietary interventions favor a balanced microbiota in autoimmune patients with dysbiosis?
Do bacteriophages, viral, or eukaryotic microbiomes influence autoimmunity by interacting with the bacterial microbiome?
Finally, we propose that the mechanisms of how autoimmunity is induced and sustained by commensal bacteria are different in diseases that affect the barrier organs versus those targeting distant organs. As briefly discussed in this review, IBD is likely instigated by a large pool of gut bacteria with antigen-specific responses being secondary to innate inflammation at the barrier site. Similar mechanisms are probably involved in psoriasis or interstitial lung disease [116,117]. Gut, lung, or skin inflammation might therefore be propagated by many commensals provided that they induce a local inflammatory response, whereas RA, lupus, or organ-specific autoimmune diseases that affect distant or immune-privileged sites (e.g., T1D, MS, uveitis, autoimmune hepatitis) might be driven by the antigen-specific effects laid out in this review (Figure 3). Much work still needs to be done to dissect the contribution of the microbiota in innate and adaptive immune responses involved at barrier sites and beyond, but the journey is worthwhile if one considers the therapeutic opportunities that lie within the microbiota [118]. To close again with Louis Pasteur: ‘The role of the infinitely small in nature is infinitely great.’
Acknowledgments
We would like to thank all members of the laboratory of M.A.K. for constructive discussions on this topic and Drs Amar Manvar and Teri Greiling for critical review of the manuscript. This work was supported by grants from the National Institutes of Health (NIH) (K08 AI095318), the Yale Rheumatic Diseases Research Core (NIH P30 AR053495), the Women’s Health Research at Yale, the O’Brien Center at Yale (NIH P30DK079310), the Arthritis National Research Foundation (all to M.A.K.), and the Yale Interdisciplinary Immunology Training Program (NIH T32AI07019) (to W.E.R.).
Glossary
- Bacteroides fragilis
B. fragilis is a Gram-negative, human gut commensal with unique immunoregulatory functions. Strains containing polysaccharide A (PSA) induce gut Treg and splenic TH1 cell responses. Other strains contain toxins implicated in chronic colon inflammation and tumorigenesis that are not covered in this review
- Clostridia
Gram-positive, spore-forming bacteria that include pathogens, soil bacteria, and several commensals with immunomodulatory functions. Strains within Clostridium clusters IV, XIVa, and XVIII are able to induce Tregs
- Gnotobiotic
a state describing germ-free animals or animals colonized with a defined microbiota, usually with a small community (e.g., altered Schaedler flora – ASF) or single species (monocolonization)
- Microbiome
the genetic material within a microbiota, typically defined by 16S rDNA sequencing. Often used synonymously with microbiota although strictly speaking not identical. The full gene content and composition of microbiomes is termed the metagenome, which can be assessed by whole-genome shotgun sequencing
- Microbiota
the sum of all commensals within a niche (gut, skin, mouth, lung, vagina, etc.). Preferentially used compared to outdated terms such as flora or microflora that describe the same
- Molecular mimicry
a longstanding theory that T and B cells specific for pathogen-derived antigens can crossreact with a self-antigen, leading to the development of autoimmunity
- Pathobiont
a commensal that promotes non-infectious disease and is detrimental to health and homeostasis under particular circumstances (e.g., genetic predisposition to over-react to a commensal, inflammation and translocation due to barrier disruption, suppression of beneficial commensals within the same niche). A pathobiont should be differentiated from a pathogen or opportunistic pathogen that causes an infectious disease
- Segmented filamentous bacteria (SFB)
until recently non-culturable commensal bacteria that can act as pathobionts or symbionts in autoimmunity depending on the host or model. SFB are capable of inducing IgA, intraepithelial lymphocytes, and antigen-specific TH17 cell responses in the small intestine, which is their physiologic niche in mice
- Short-chain fatty acids (SCFAs)
acetate, butyrate, and propionate are produced by the gut microbiota, particularly after a high-fiber diet. SCFAs act via multiple mechanisms including histone modification and G protein-coupled receptor signaling. SCFAs have multiple biological effects including the induction and recruitment of Tregs
- Symbiont
a commensal beneficial for the host. Both the commensal and the host need each other, and thus are living in symbiosis by providing factors that improve fitness
References
- 1.Belkaid Y, Naik S. Compartmentalized and systemic control of tissue immunity by commensals. Nat. Immunol. 2013;14:646–653. doi: 10.1038/ni.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat. Immunol. 2013;14:676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sommer F, Backhed F. The gut microbiota - masters of host development and physiology. Nat. Rev. Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 4.Maynard CL, et al. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489:231–241. doi: 10.1038/nature11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ochoa-Reparaz J, et al. Role of gut commensal microflora in the development of experimental autoimmune encephalomyelitis. J. Immunol. 2009;183:6041–6050. doi: 10.4049/jimmunol.0900747. [DOI] [PubMed] [Google Scholar]
- 8.Berer K, et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479:538–541. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- 9.Lee YK, et al. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. U.S.A. 2011;108(Suppl 1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rath HC, et al. Normal luminal bacteria, especially Bacteroides species, mediate chronic colitis, gastritis, and arthritis in HLA-B27/human beta2 microglobulin transgenic rats. J. Clin. Invest. 1996;98:945–953. doi: 10.1172/JCI118878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdollahi-Roodsaz S, et al. Stimulation of TLR2 and TLR4 differentially skews the balance of T cells in a mouse model of arthritis. J. Clin. Invest. 2008;118:205–216. doi: 10.1172/JCI32639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu HJ, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scher JU, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pozzilli P, et al. NOD mouse colonies around the world – recent facts and figures. Immunol. Today. 1993;14:193–196. doi: 10.1016/0167-5699(93)90160-M. [DOI] [PubMed] [Google Scholar]
- 15.Wen L, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kriegel MA, et al. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc. Natl. Acad. Sci. U.S.A. 2011;108:11548–11553. doi: 10.1073/pnas.1108924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Markle JG, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 18.Yurkovetskiy L, et al. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013;39:400–412. doi: 10.1016/j.immuni.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sudo N, et al. The requirement of intestinal bacterial flora for the development of an IgE production system fully susceptible to oral tolerance induction. J. Immunol. 1997;159:1739–1745. [PubMed] [Google Scholar]
- 20.von Mutius E, Vercelli D. Farm living: effects on childhood asthma and allergy. Nat. Rev. Immunol. 2010;10:861–868. doi: 10.1038/nri2871. [DOI] [PubMed] [Google Scholar]
- 21.Olszak T, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cahenzli J, et al. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe. 2013;14:559–570. doi: 10.1016/j.chom.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garrett WS, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palm NW, et al. Immunoglobulin a coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kostic AD, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rubinstein MR, et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vieira SM, et al. Diet, microbiota and autoimmune diseases. Lupus. 2014;23:518–526. doi: 10.1177/0961203313501401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho JH, Gregersen PK. Genomics and the multifactorial nature of human autoimmune disease. N. Engl. J. Med. 2011;365:1612–1623. doi: 10.1056/NEJMra1100030. [DOI] [PubMed] [Google Scholar]
- 29.Cotsapas C, Hafler DA. Immune-mediated disease genetics: the shared basis of pathogenesis. Trends Immunol. 2013;34:22–26. doi: 10.1016/j.it.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 30.Selmi C, et al. Heritability versus the role of the environment in autoimmunity. J. Autoimmun. 2012;39:249–252. doi: 10.1016/j.jaut.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 31.Tiniakou E, et al. Sex-specific environmental influences on the development of autoimmune diseases. Clin. Immunol. 2013;149:182–191. doi: 10.1016/j.clim.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turnbaugh PJ, et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cox LM, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–721. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cha HR, et al. Downregulation of Th17 cells in the small intestine by disruption of gut flora in the absence of retinoic acid. J. Immunol. 2010;184:6799–6806. doi: 10.4049/jimmunol.0902944. [DOI] [PubMed] [Google Scholar]
- 36.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maslowski KM, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rakoff-Nahoum S, et al. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 39.Sansonetti PJ, Medzhitov R. Learning tolerance while fighting ignorance. Cell. 2009;138:416–420. doi: 10.1016/j.cell.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 40.Gevers D, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manichanh C, et al. The gut microbiota in IBD. Nat. Rev. Gastroenterol. Hepatol. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 42.Huttenhower C, et al. Inflammatory bowel disease as a model for translating the microbiome. Immunity. 2014;40:843–854. doi: 10.1016/j.immuni.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dobber R, et al. The involvement of the intestinal microflora in the expansion of CD4+ T cells with a naive phenotype in the periphery. Dev. Immunol. 1992;2:141–150. doi: 10.1155/1992/57057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989;299:1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mazmanian SK, et al. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 46.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. U.S.A. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, et al. An intestinal commensal symbiosis factor controls neuroinflammation via TLR2-mediated CD39 signalling. Nat. Commun. 2014;5:4432. doi: 10.1038/ncomms5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dasgupta S, et al. Plasmacytoid dendritic cells mediate anti-inflammatory responses to a gut commensal molecule via both innate and adaptive mechanisms. Cell Host Microbe. 2014;15:413–423. doi: 10.1016/j.chom.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Antonioli L, et al. CD39 and CD73 in immunity and inflammation. Trends Mol. Med. 2013;19:355–367. doi: 10.1016/j.molmed.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sears CL, et al. Bacteroides fragilis subverts mucosal biology: from symbiont to colon carcinogenesis. J. Clin. Invest. 2014;124:4166–4172. doi: 10.1172/JCI72334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hsiao EY, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Atarashi K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 53.Narushima S, et al. Characterization of the 17 strains of regulatory T cell-inducing human-derived Clostridia. Gut Microbes. 2014;5:333–339. doi: 10.4161/gmic.28572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 55.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chang PV, et al. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. U.S.A. 2014;111:2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim SV, et al. GPR15-mediated homing controls immune homeostasis in the large intestine mucosa. Science. 2013;340:1456–1459. doi: 10.1126/science.1237013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim MH, et al. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology. 2013;145:396–406. doi: 10.1053/j.gastro.2013.04.056. [DOI] [PubMed] [Google Scholar]
- 59.Singh N, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40:128–139. doi: 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Obata Y, et al. The epigenetic regulator Uhrf1 facilitates the proliferation and maturation of colonic regulatory T cells. Nat. Immunol. 2014;15:571–579. doi: 10.1038/ni.2886. [DOI] [PubMed] [Google Scholar]
- 61.Lopetuso LR, et al. Commensal Clostridium: leading players in the maintenance of gut homeostasis. Gut Pathog. 2013;5:23. doi: 10.1186/1757-4749-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gomez A, et al. Loss of sex and age driven differences in the gut microbiome characterize arthritis-susceptible 0401 mice but not arthritis-resistant 0402 mice. PLoS ONE. 2012;7:e36095. doi: 10.1371/journal.pone.0036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Loo VG, et al. Host and pathogen factors for Clostridium difficile infection and colonization. N. Engl. J. Med. 2011;365:1693–1703. doi: 10.1056/NEJMoa1012413. [DOI] [PubMed] [Google Scholar]
- 64.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Berer K, et al. Selective accumulation of pro-inflammatory T cells in the intestine contributes to the resistance to autoimmune demyelinating disease. PLoS ONE. 2014;9:e87876. doi: 10.1371/journal.pone.0087876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chung H, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yin Y, et al. Comparative analysis of the distribution of segmented filamentous bacteria in humans, mice and chickens. ISME J. 2013;7:615–621. doi: 10.1038/ismej.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Obata T, et al. Indigenous opportunistic bacteria inhabit mammalian gut-associated lymphoid tissues and share a mucosal antibody-mediated symbiosis. Proc. Natl. Acad. Sci. U.S.A. 2010;107:7419–7424. doi: 10.1073/pnas.1001061107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reading NC, Kasper DL. The starting lineup: key microbial players in intestinal immunity and homeostasis. Front. Microbiol. 2011;2:148. doi: 10.3389/fmicb.2011.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kunisawa J, Kiyono H. Alcaligenes is commensal bacteria habituating in the gut-associated lymphoid tissue for the regulation of intestinal IgA responses. Front. Immunol. 2012;3:65. doi: 10.3389/fimmu.2012.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suzuki K, et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc. Natl. Acad. Sci. U.S.A. 2004;101:1981–1986. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fagarasan S, et al. Adaptive immune regulation in the gut: T cell-dependent and T cell-independent IgA synthesis. Annu. Rev. Immunol. 2010;28:243–273. doi: 10.1146/annurev-immunol-030409-101314. [DOI] [PubMed] [Google Scholar]
- 73.Craft JE. Follicular helper T cells in immunity and systemic autoimmunity. Nat. Rev. Rheumatol. 2012;8:337–347. doi: 10.1038/nrrheum.2012.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kato LM, et al. The role of the adaptive immune system in regulation of gut microbiota. Immunol. Rev. 2014;260:67–75. doi: 10.1111/imr.12185. [DOI] [PubMed] [Google Scholar]
- 75.Kawamoto S, et al. The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science. 2012;336:485–489. doi: 10.1126/science.1217718. [DOI] [PubMed] [Google Scholar]
- 76.Wang N, et al. Selective IgA deficiency in autoimmune diseases. Mol. Med. 2011;17:1383–1396. doi: 10.2119/molmed.2011.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kawamoto S, et al. Foxp3+ T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity. 2014;41:152–165. doi: 10.1016/j.immuni.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 78.Hirota K, et al. Plasticity of Th17 cells in Peyer’s patches is responsible for the induction of T cell-dependent IgA responses. Nat. Immunol. 2013;14:372–379. doi: 10.1038/ni.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Trama AM, et al. HIV-1 Envelope gp41 antibodies can originate from terminal ileum B cells that share cross-reactivity with commensal bacteria. Cell Host Microbe. 2014;16:215–226. doi: 10.1016/j.chom.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hunger RE, et al. Male gonadal environment paradoxically promotes dacryoadenitis in nonobese diabetic mice. J. Clin. Invest. 1998;101:1300–1309. doi: 10.1172/JCI1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nicholson JK, et al. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 82.Haiser HJ, et al. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science. 2013;341:295–298. doi: 10.1126/science.1235872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Norman JM, et al. Kingdom-agnostic metagenomics and the importance of complete characterization of enteric microbial communities. Gastroenterology. 2014;146:1459–1469. doi: 10.1053/j.gastro.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stetson DB, et al. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weinstock JV, Elliott DE. Helminth infections decrease host susceptibility to immune-mediated diseases. J. Immunol. 2014;193:3239–3247. doi: 10.4049/jimmunol.1400927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hand TW, et al. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science. 2012;337:1553–1556. doi: 10.1126/science.1220961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang Y, et al. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature. 2014;510:152–156. doi: 10.1038/nature13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Goto Y, et al. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity. 2014;40:594–607. doi: 10.1016/j.immuni.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kuhn KA, Stappenbeck TS. Peripheral education of the immune system by the colonic microbiota. Semin. Immunol. 2013;25:364–369. doi: 10.1016/j.smim.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cong Y, et al. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc. Natl. Acad. Sci. U.S.A. 2009;106:19256–19261. doi: 10.1073/pnas.0812681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Alexander KL, et al. Microbiota activation and regulation of innate and adaptive immunity. Immunol. Rev. 2014;260:206–220. doi: 10.1111/imr.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lathrop SK, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cebula A, et al. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature. 2013;497:258–262. doi: 10.1038/nature12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Faith JJ, et al. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci. Transl. Med. 2014;6:220ra211. doi: 10.1126/scitranslmed.3008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ahern PP, et al. Mining the human gut microbiota for effector strains that shape the immune system. Immunity. 2014;40:815–823. doi: 10.1016/j.immuni.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Smith K, et al. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol. 2007;19:59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 97.Konrad A, et al. Tight mucosal compartmentation of the murine immune response to antigens of the enteric microbiota. Gastroenterology. 2006;130:2050–2059. doi: 10.1053/j.gastro.2006.02.055. [DOI] [PubMed] [Google Scholar]
- 98.Munz C, et al. Antiviral immune responses: triggers of or triggered by autoimmunity? Nat. Rev. Immunol. 2009;9:246–258. doi: 10.1038/nri2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Horwitz MS, et al. Diabetes induced by Coxsackie virus: initiation by bystander damage and not molecular mimicry. Nat. Med. 1998;4:781–785. doi: 10.1038/nm0798-781. [DOI] [PubMed] [Google Scholar]
- 100.Miller SD, et al. Persistent infection with Theiler’s virus leads to CNS autoimmunity via epitope spreading. Nat. Med. 1997;3:1133–1136. doi: 10.1038/nm1097-1133. [DOI] [PubMed] [Google Scholar]
- 101.Oldstone MB. Molecular mimicry, microbial infection, and autoimmune disease: evolution of the concept. Curr. Top. Microbiol. Immunol. 2005;296:1–17. doi: 10.1007/3-540-30791-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fae KC, et al. Mimicry in recognition of cardiac myosin peptides by heart-intralesional T cell clones from rheumatic heart disease. J. Immunol. 2006;176:5662–5670. doi: 10.4049/jimmunol.176.9.5662. [DOI] [PubMed] [Google Scholar]
- 103.Ang CW, et al. The Guillain-Barre syndrome: a true case of molecular mimicry. Trends Immunol. 2004;25:61–66. doi: 10.1016/j.it.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 104.Ley RE, et al. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 105.Li J, Jia H. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 2014;32:834–841. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- 106.Su LF, et al. Virus-specific CD4+ memory-phenotype T cells are abundant in unexposed adults. Immunity. 2013;38:373–383. doi: 10.1016/j.immuni.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nylander A, Hafler DA. Multiple sclerosis. J. Clin. Invest. 2012;122:1180–1188. doi: 10.1172/JCI58649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Benckert J, et al. The majority of intestinal IgA+ and IgG+ plasmablasts in the human gut are antigen-specific. J. Clin. Invest. 2011;121:1946–1955. doi: 10.1172/JCI44447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tiller T, et al. Autoreactivity in human IgG+ memory B cells. Immunity. 2007;26:205–213. doi: 10.1016/j.immuni.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wardemann H, et al. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 111.Menard L, et al. The PTPN22 allele encoding an R620W variant interferes with the removal of developing autoreactive B cells in humans. J. Clin. Invest. 2011;121:3635–3644. doi: 10.1172/JCI45790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Samuels J, et al. Impaired early B cell tolerance in patients with rheumatoid arthritis. J. Exp. Med. 2005;201:1659–1667. doi: 10.1084/jem.20042321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kinnunen T, et al. Specific peripheral B cell tolerance defects in patients with multiple sclerosis. J. Clin. Invest. 2013;123:2737–2741. doi: 10.1172/JCI68775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wesemann DR, et al. Microbial colonization influences early B-lineage development in the gut lamina propria. Nature. 2013;501:112–115. doi: 10.1038/nature12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Birnbaum ME, et al. Deconstructing the peptide-MHC specificity of T cell recognition. Cell. 2014;157:1073–1087. doi: 10.1016/j.cell.2014.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Alekseyenko AV, et al. Community differentiation of the cutaneous microbiota in psoriasis. Microbiome. 2013;1:31. doi: 10.1186/2049-2618-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dickson RP, et al. The role of the microbiome in exacerbations of chronic lung diseases. Lancet. 2014;384:691–702. doi: 10.1016/S0140-6736(14)61136-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sonnenburg JL, Fischbach MA. Community health care: therapeutic opportunities in the human microbiome. Sci. Transl. Med. 2011;3 doi: 10.1126/scitranslmed.3001626. 78ps12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Husebye ES, Anderson MS. Autoimmune polyendocrine syndromes: clues to type 1 diabetes pathogenesis. Immunity. 2010;32:479–487. doi: 10.1016/j.immuni.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hafler DA, et al. Risk alleles for multiple sclerosis identified by a genomewide study. N. Engl. J. Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 121.Sawcer S, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ubeda C, et al. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J. Exp. Med. 2012;209:1445–1456. doi: 10.1084/jem.20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Adrianto I, et al. Association of a functional variant downstream of TNFAIP3 with systemic lupus erythematosus. Nat. Genet. 2011;43:253–258. doi: 10.1038/ng.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Farh KK, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Topol EJ. Individualized medicine from prewomb to tomb. Cell. 2014;157:241–253. doi: 10.1016/j.cell.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Faith JJ, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ding T, Schloss PD. Dynamics and associations of microbial community types across the human body. Nature. 2014;509:357–360. doi: 10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Garrett WS, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rooks MG, et al. Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. ISME J. 2014;8:1403–1417. doi: 10.1038/ismej.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Winter SE, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Winter SE, et al. The dynamics of gut-associated microbial communities during inflammation. EMBO Rep. 2013;14:319–327. doi: 10.1038/embor.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Atarashi K, et al. ATP drives lamina propria TH17 cell differentiation. Nature. 2008;455:808–812. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- 134.Powell N, et al. The transcription factor T-bet regulates intestinal inflammation mediated by interleukin-7 receptor+ innate lymphoid cells. Immunity. 2012;37:674–684. doi: 10.1016/j.immuni.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]