Summary
In vivo studies of human brain cellular function face challenging ethical and practical difficulties. Animal models are typically used but display distinct cellular differences. One specific example is astrocytes, recently recognized for contribution to neurological diseases and a link to the genetic risk factor apolipoprotein E (APOE). Current astrocytic in vitro models are questioned for lack of biological characterization. Here, we report human induced pluripotent stem cell (hiPSC)-derived astroglia (NES-Astro) developed under defined conditions through long-term neuroepithelial-like stem (ltNES) cells. We characterized NES-Astro and astrocytic models from primary sources, astrocytoma (CCF-STTG1), and hiPSCs through transcriptomics, proteomics, glutamate uptake, inflammatory competence, calcium signaling response, and APOE secretion. Finally, we assess modulation of astrocyte biology using APOE-annotated compounds, confirming hits of the cholesterol biosynthesis pathway in adult and hiPSC-derived astrocytes. Our data show large diversity among astrocytic models and emphasize a cellular context when studying astrocyte biology.
Keywords: astrocytes, induced pluripotent stem cells, cell differentiation, glutamate plasma membrane transport proteins, neuroinflammation, calcium signaling, apolipoproteins E, high-throughput screening assays, neurodegenerative diseases, drug discovery
Graphical Abstract

Highlights
-
•
Expression and functional profiling display variation between astrocyte models
-
•
Development of NES-Astro showing functional astrocyte-associated glutamate receptor
-
•
NES-Astro is immune competent, displaying ATP and glutamate-driven calcium signaling
-
•
APOE HTS assay shows that compound hit finding depends on astrocytic model biology
Human studies can typically not be used to understand cellular functions of the brain. Astrocytes, important for neuronal circuit regulation and support, lack cellular model characterization and biological translation. Falk, Herland, and colleagues report striking differences in astrocyte models. A pilot screen of Alzheimer's disease-related drugs demonstrates dependence between compound hit finding and astrocytic model biology.
Introduction
Over the last decades the neuronal view of brain function has become more complex, recognizing the important role of astrocytes in the regulation of neuronal circuits, in addition to their previously identified support function (De Pittà et al., 2016). The identification of an increasingly diverse set of astrocytic functions has created a need for improved models of high relevance for astrocyte biology to better define their contribution to brain function, embryonic development, and neurological diseases. As human astrocytes display clear differences compared with mouse astrocytes in morphological complexity (Oberheim et al., 2009), transcriptomic profile (Zhang et al., 2016), and inflammatory response (Tarassishin et al., 2014), animal model-derived data may be of limited relevance. Astrocytes may also be important as part of preclinical pharmacological screening strategies for prevalent neurological disorders in which astrocyte function may be impaired. Historically, high failure rates of drug candidates in clinical trials for neurological diseases, such as Alzheimer's disease (AD) (Cummings et al., 2014), in combination with a recognized possible role of astrocytes in AD (Yu et al., 2014), creates a need for specific human cellular astrocyte models. A realistic astrocyte model should at least represent four key astrocytic features: the uptake of the neurotransmitter glutamate, essential for synapse dynamics; inflammatory response to trauma; calcium signaling response to neurotransmitters; and the secretion of apolipoprotein E (APOE), a lipid and cholesterol transporter in the brain (Khakh and Sofroniew, 2015, Bazargani and Attwell, 2016, Yu et al., 2014). In neurological diseases such as AD, the ɛ4 variant of the APOE-encoding gene, APOE ɛ4, has been shown to be the strongest genetic risk factor for non-familial AD (the common, sporadic form of the disease). The precise molecular mechanism underlying this association is currently unknown, but stimulating APOE secretion from astrocytes has been proposed as a potential therapeutic target (Yu et al., 2014).
Astrocytic functional studies in vitro use cells from various cell sources including the current “gold standard” primary fetal (Malik et al., 2014) or primary adult (Zhang et al., 2016) astrocytes. However, due to the ease of culture and greater availability, immortalized astrocytoma cell lines have been used in multiple screening applications (Fan et al., 2016, Finan et al., 2016). Moreover, since the development of human induced pluripotent stem cell (hiPSC) technology (Takahashi et al., 2007) there have also been an increasing number of published protocols for directed differentiation of hiPSCs toward astrocytes (Chen et al., 2014, Gupta et al., 2012, Kondo et al., 2013, Krencik et al., 2011, Palm et al., 2015, Santos et al., 2017, Serio et al., 2013, Shaltouki et al., 2013).
Cellular models for high-throughput screening (HTS) need high reproducibility, efficient upscaling, and short lead times, which can be fulfilled by glioma cell lines but whose biological relevance can be questioned (Auvergne et al., 2013). Primary neuronal cells are generally seen to have high biological relevance but do not normally meet other HTS criteria. hiPSC-derived astrocytes provide a promising source of astrocytes for screening models, but generally the protocols are extensive (Krencik et al., 2011) and use undefined conditions (Kondo et al., 2013), introducing variability. Additionally, besides technical and biological challenges there are stringent policy guidelines that need to be adhered to when pharmaceutical companies are to use cellular models for drug screening, limiting the use of fetal tissue and human embryonic stem cells (hESCs).
The aim of the present study was to evaluate the biological relevance and potential model diversity between commercial astrocytic models available for pharmaceutical industry in relation to hiPSC-derived models. We included primary human adult astrocytes (HMP202-4014, Neuromics), an astrocytoma cell line (CCF-SSTG1, ATCC), and a commercial hiPSC-derived astrocytic model (iCell Astrocytes, Cellular Dynamics International). However, none of these cellular systems were fully defined. In addition, we assessed whether long-term self-renewing hiPSC-derived neuroepithelial-like stem cells (ltNES) (Falk et al., 2012), previously used for undirected neuron and glia differentiation studies (Shahsavani et al., 2017, Tailor et al., 2013, Zhang et al., 2014), could be adapted for directed glia differentiation under fully defined conditions to generate a robust and functional astrocytic model applicable for HTS. To characterize model biology and identify key features of each model, we combined transcriptomic and proteomic profiling with astrocyte-associated functional assays. To assess the possibility of direct application into a pharmaceutical setting, we also performed a pilot screen of 10 compounds, modulating APOE secretion, in an HTS setting. The outcome will be of critical value to guide in the selection of appropriate astrocyte models for various biological questions and HTS.
Results
Efficient and Robust Generation of hiPSC-Derived Astroglia from a Neural Progenitor State
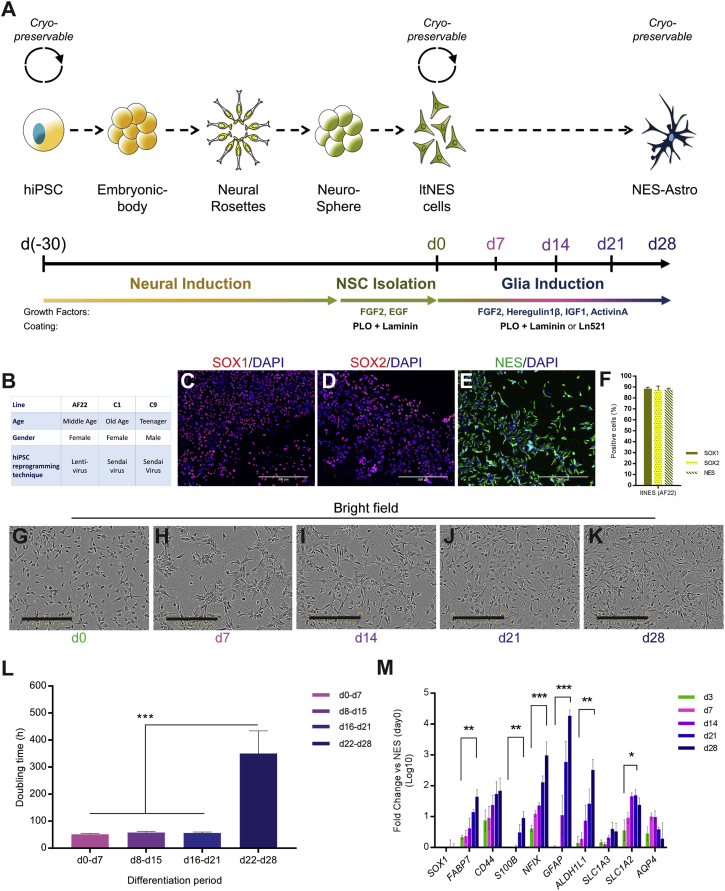
Since the majority of existing protocols for hiPSC-derived astrocytes are dependent on fetal bovine serum (FBS) (Kondo et al., 2013, Santos et al., 2017, Serio et al., 2013, Shaltouki et al., 2013), we adapted previously published protocols to try and meet the requirements of a fully defined cellular system (Figure 1A). Looking for a stable phenotypic and cryopreservable intermediate model we chose ltNES, which can be generated from multiple hESC and hiPSC lines (Falk et al., 2012), despite patient genomic background variation. ltNES can also be cultured up to 100 passages with maintained proliferation capacity, stable neuronal and glia differentiation propensity, and the characteristic ltNES transcription profile (Falk et al., 2012), enabling extensive qualitative upscaling. In this study we used three ltNES cell lines derived from three individuals differing in age and gender (Figure 1B). All ltNES lines were homogenously positive for neural stem cell markers SOX1, SOX2, and NES (Figures 1C–1F and S1A), and displayed similar proliferation characteristics (Figure S1B). To assess astroglia protocols we first focused on the well-established AF22 line, which resulted in the adaptation of a previously published protocol (Shaltouki et al., 2013). Astroglia differentiation was initiated by exposing ltNES (AF22) to a growth factor cocktail including fibroblast growth factor 2 (FGF2), heregulin-1β, insulin-like growth factor 1 (IGF1), and activin A, while culturing on a double extracellular matrix coating of poly-L-ornithine (PLO) and laminin (hereafter called the FHIA protocol) (Figure 1A). During the differentiation there was a significant morphological change from the triangular ltNES shape to a stellate morphology in combination with a non-overlapping cellular domain profile (Figures 1G–1K and Movie S1), similar to what is seen in vivo (Oberheim et al., 2009). Doubling time was unchanged during the first periods (day 0 [d0] to d7, d8–d15, d16–d21), but significantly shifted in the last period (d22–d28) (Figure 1L). A basal characterization of the glial development was performed during the differentiation process, showing increasing and significantly higher levels of glia- and astrocyte-associated markers at day 28 compared with day 3 (Figure 1M). Exploring the possibility for future clinical adaptation, we simultaneously assessed a complete animal-free component system (FHIA521 protocol) showing very similar developmental profiles (Figure S1C). We also assessed and show that NES-Astro (AF22) at day 28 can be cryopreserved with high post-thaw viability of 90.6% ± 1.9% (n = 3) (Figures S1D and S1E), facilitating instant post-thaw application.
Figure 1.
Differentiation of hiPSCs to Astroglia from a Long-Term Neuroepithelial-like Stem Cell Intermediate Stage
(A) Schematic overview of NES-Astro generation from hiPSCs.
(B) Age, gender, and reprogramming technique of cell lines AF22, C1, and C9.
(C–E) Immunocytochemical staining for SOX1 (C), SOX2 (D), and NES of ltNES (AF22) (E) culture.
(F) Image-based quantification of ltNES (AF22) population marker expression.
(G–K) Bright-field imaging over the time course of NES-Astro (AF22) differentiation time points day 0 (d0) (G), d7 (H), d24 (I), d21 (J), and d28 (K).
(L) Doubling time during the differentiation periods (d0–d7), (d8–d15), (d16–d21), and (d22–d28) (n = 6 for each period, p < 0.001).
(M) Real-time qPCR of mRNA expression for glia-associated genes at differentiation time points d3, d7, d14, d21, and d28.
Data shown from independent experiments, n = 3, mean ± SEM. Statistical analysis: one-way ANOVA (K and L); ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Scale bars, 300 μm.
In conclusion, we validated that ltNES show similar basic neural stem cell marker expression and equal proliferation profiles across hiPSC line origins. Applying a completely defined differentiation medium and coating system to ltNES (AF22) initiated glia differentiation with expression of common glia and astrocytic markers, developing into an NES-Astro (AF22) phenotype at day 28 of differentiation.
Transcriptomic Diversities between Astrocyte Models
To investigate astrocyte model diversity, we included three commercially available astrocytic models representing different cell sources: human primary adult astrocytes (phaAstro), astrocytoma cell line CCF-STTG1 (CCF), and hiPSC-derived astrocytes from Cellular Dynamics International (iCellAstro). Additionally we also included our NES-Astro model together with its parental cellular origin, ltNES, representing a non-astrocytic model. NES-Astro and ltNES were from three individual hiPSC lines, AF22, C1, and C9, respectively. Initially, to assess astrocytic marker expression and to identify possible differences between the cellular models, RNA sequencing (RNA-seq) was performed followed by subsequent differential analysis (Figure S2A).
Principal component analysis (PCA) (Figure 2A) showed five separate clustered groups reflecting the differences between the five main model types used in this study. Additionally, the PCA results showed a high homogeneity between ltNES lines generated from various donors, but more importantly a high reproducibility of our FHIA protocol by the tight clustering between the NES-Astro lines of different genotypes.
Figure 2.
Diversities in Transcriptomic Profiles between Neuronal Models
(A) Principal component analysis.
(B and C) Pearson correlation, comparing global gene expression (B), and (C) gene-to-gene correlation between ltNES, NES-Astro, primary human adult astrocytes (phaAstro), iCell astrocytes (iCellAstro), and CCF-STTG1 (CCF).
(D) Differential expressional analysis of neuronal models to (phaAstro) (data inclusion criteria; lfc > |3|, p = 0.05). Statistical analysis: Benjamini-Hochberg procedure.
(E) DESeq2 normalized RNA expression of 25 common astrocyte-associated genes. Red boxes highlight highest value among the models. Data shown from independent experiments, n = 3.
Moreover, Pearson correlation analysis (Figure 2B) between models using all coding genes confirms the results of the PCA analysis, but also shows a higher correlation between phaAstro and CCF compared with the hiPSC-derived models. These differences could potentially be attributed to the process of primary isolation and hiPSC derivation.
Gene-pair correlations (Figure 2C) extracted the top gene pairs representing the largest diversities across models. This resulted in a selection of 396 gene pairs out of 19,522 with a correlation of r = |0.000001| (Figure S2B; Tables S2 and S3).
We analyzed the gene counts with DESeq2 (Love et al., 2014) and the obtained fold changes, using phaAstro as a reference sample, were visualized in a heatmap (Figure 2D) to highlight the differences in gene expression levels between the models. To determine whether any of the models had a more profound astrocytic profile, we analyzed in more detail 25 genes commonly associated with mouse and human astrocytes (Zhang et al., 2016), fetal astrocytes (Malik et al., 2014), hpNPC (human primary neural progenitor cell)-derived astrocytes (Magistri et al., 2016), and hiPSC-derived astrocytes (Pei et al., 2016). However, none of the studied models had a uniform high expression of the 25 genes (Figure 2E). Comparing the selected list of 25 genes with single-cell transcriptomic astrocyte profiling (Darmanis et al., 2015, Spaethling et al., 2017) showed similar heterogeneity in gene expression across models (Table S4). However, a notable pattern was that functional astrocyte-related receptors such as SLC1A3, SLC1A2, AQP4, RYR3, and KCNJ10 were highly expressed in NES-Astro compared with the other models (Figure 2E).
In conclusion, transcriptomic analysis revealed clear differences between astrocytic models but that there was no single model that homogeneously correlate with the vast literature on astrocyte marker profiles.
Glia-Associated Protein Profiles Specify Cellular Models
We next studied protein expression by immunocytochemistry (ICC) to assess both expression and cellular localization of cytoplasmic proteins, receptors, transcription factors, and intermediate filaments associated with a glial cell type. We also included HEK293 cells as a non-neuronal model. Bright-field images reveal distinct morphology differences (Figure S3A), which is also captured with staining for CD44, FABP7, S100B, GFAP (Figures 3A–3X), SOX9, NFIA, SOX2, NES, and VIM (Figures S3B–S3F). Image-based quantification highlighted the heterogeneity in marker expression (Figure 3Y and Table S5). CD44, a common glial marker, was expressed in all astrocyte models (Figures 3A–3D and S4A) but not in ltNES nor in HEK293 cells (Figures 3E and 3F), thus showing a discriminative expression profile. However, of the markers studied, CD44 was the only one that was able to make the distinction between all astrocytic- and non-astrocytic models. S100B was homogenously expressed in phaAstro, CCF, and NES-Astro, but not in iCellAstro (Figures 3M–3P and S4C). The common astrocyte classifier, GFAP (glial fibrillary acidic protein), was surprisingly only expressed at low level in iCellAstro (average 8%, Figure 3Y), and not detected in other models (Figures 3S–3X). This was consistent using multiple different GFAP antibodies (Figure S3G). The glia-associated transcription factor, NFIA (nuclear factor I-A) was expressed in 70% of the cell populations across astrocyte models (Figure 3Y). This was also seen for SOX9, except in iCellAstro that showed a lower expression (Figures 3Y, S3C, and S3D). Differential expressions of NES, VIM, and CD44 were small, but differences in signal intensity could be observed between models (Figure S3H). NES-Astro (AF22) shared similar protein expression profiles with NES-Astro (C1) and NES-Astro (C9) (Figures S4A–S4I), verifying the robustness and consistency in the FHIA protocol also at the protein level. Additionally, applying the complete animal-free FHIA521 protocol generated NES-521Astro with a protein profile similar to that of the regular FHIA protocol (Figures S4A–S4I). NES-Astro, consistently across multiple cell lines, was the model that displayed most of the glia-associated proteins, and also the only model expressing FABP7 (Figures 3J and S4B).
Figure 3.
Glia-Associated Protein Characterization of Cellular Models
(A–X) Immunocytochemical staining of phaAstro, CCF, iCellAstro, NES-Astro (AF22), ltNES (AF22), and HEK293 cells for glia-associated markers.
(Y) Image-based quantification of population expression level for glia-associated markers. See also Table S6 for DEseq2 expression of the proteins studied.
Data shown are as mean ± SEM. For images analyzed (n = 11–18), see Table S5 for additional details. Scale bars, 300 μm.
The expression of glial markers was heterogeneous across models. NES-Astro (AF22/C1/C9) models were the only ones that expressed all astrocytic markers, except for GFAP, which surprisingly was only detected in iCellAstro.
Astrocyte-Associated Glutamate Transporter Responsible for Uptake in NES-Astro
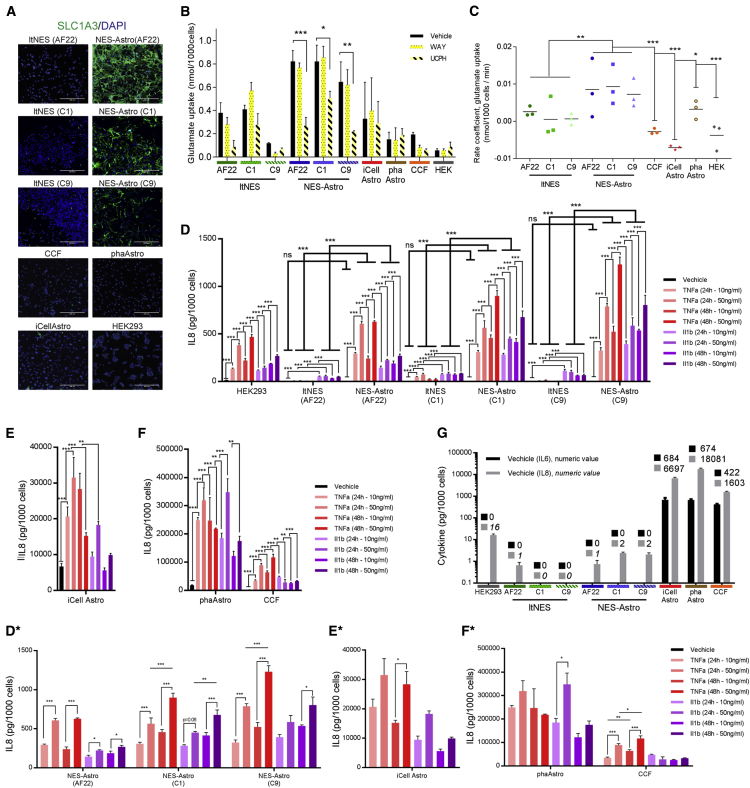
The lack of conclusive and consistent human astrocyte marker profiles might reflect the complexity of various astrocytic functions (Khakh and Sofroniew, 2015), hence making it difficult to validate model representation. Characterization of astrocytic models therefore needs to assess astrocytic function, such as uptake of extracellular glutamate, critical for maintaining synaptic signaling and avoiding excitotoxicity. The two excitatory amino acid transporters, SLC1A2 (EAAT2, Glt-1) and SLC1A3 (EAAT1, Glast) are considered the main astrocyte glutamate transporters (DeSilva et al., 2012, Sosunov et al., 2014). Evaluating the activity of these transporters showed that they were expressed on an RNA level in all neural samples (all cell lines except HEK293), while the translation into protein was only observed in the NES-Astro (AF22/C1/C9) models (Figures 4A and S5A). To validate whether either SLC1A2 or SLC1A3 were functionally active and contributed to the uptake of glutamate, we treated all cellular models with the SLC1A2 inhibitor WAY213613 or the SLC1A3 inhibitor UCPH101. The SLC1A3 inhibitor showed a significant effect on glutamate uptake in the NES-Astro (AF22/C1/C9) models (Figure 4B). To evaluate active uptake, we performed a time-course experiment including six time points over 60 min whereby we identified an acute recovery phase within the first 10 min for all cellular models. However, compared with the other models, NES-Astro (AF22/C1/C9) were the only models having continued uptake from 10 min onward as reflected by the significantly higher positive regression coefficient (Figure 4C). SLC1A2 contribution to glutamate uptake could not be demonstrated (Figure 4C), which could be explained by no or delocalized protein expression of SLC1A2 (NES-Astro C1/C9) (Figure S5B).
Figure 4.
Functional Characterization of Cellular Models
(A) Immunocytochemical staining of all models for SLC1A3.
(B) Glutamate uptake levels at 60 min during vehicle, WAY, and UCPH treatment.
(C) Glutamate uptake rate coefficient from linear regression was generated from time-course studies, from samples at 10, 20, 30, and 60 min after glutamate addition at time point 0.
(D–F) IL-8 secretion after 24 or 48 hr stimulation with either TNF-α or IL-1β at concentrations of 10 or 50 ng/mL of HEK293, ltNES-AF22/C1/C9, NES-Astro-AF22/C1/C9 (D), iCellAstro (E), and phaAstro and CCF (F). Highlighted are significant differences between treatment and vehicle, and between ltNES and NES-Astro for each cell line.
(D∗–F∗) Reuse of results in (D) to (F) highlighting the significant concentration-dependent responses of NES-Astro-AF22/C1/C9 (D∗), iCellAstro (E∗), and CCF and phaAstro (F∗).
(G) Basal level of IL-8 secretion during vehicle treatment, representing non-inflammatory condition.
Data shown from independent experiments, n = 3, mean ± SEM. Statistical analysis: one-way ANOVA (B, D, D∗, E, E∗, F, and F∗) and two-way ANOVA (C, and in D between ltNES and NES-Astro per line); ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Scale bars, 300 μm.
Glutamate transporter SLC1A3, which contributes to glutamate uptake, was detected at the RNA level across all models. However, NES-Astro (AF22/C1/C9) was the only model that displayed a strong protein expression at the cellular membrane, where it also contributed functionally to active glutamate uptake.
Inflammatory Activation
Neuropathologies may cause astrocyte reactivity specific to the type and degree of the insult, highlighting the context dependency (Sofroniew, 2015). Inflammatory responses have been studied in hPSC-derived astrocytes (Santos et al., 2017, Tcw et al., 2017) as well as in human fetal primary astrocytes (Santos et al., 2017). To assess whether the models in this study could be used for studying inflammatory responses, we used tumor necrosis factor α (TNF-α) and interleukin-1β (IL-1β) as pro-inflammatory stimulation to evaluate IL-6 and IL-8 secretion. All models, including non-astrocytic models, responded to treatment with the inflammatory cytokines TNF-α and IL-1β by secretion of IL-8 (Figures 4D–4F). The responses were different in treatment dosing and exposure time, but also showed differences in secretion concentration levels of 4 orders of magnitude. Importantly, the basal level of IL-8 and IL-6 secretion during vehicle treatment, representing non-stimulated conditions, displayed the same concentration difference. While HEK, ltNES (AF22/C1/C9), and NES-Astro (AF22/C1/C9) had close to zero secretion, iCellAstro, CCF, and phaAstro secreted between 400 and 18,000 pg/1,000 cells during non-stimulating conditions (Figure 4G). NES-Astro (AF22/C1/C9) displayed a significantly higher immune activation compared with their source cells during both TNF-α and IL-1β treatment. However, there was no significant difference between NES-Astro and ltNES during vehicle conditions (Figure 4D), demonstrating a complete inflammatory inactive state. NES-Astro also showed a dose-dependent response to TNF-α and IL-1β, not observed in ltNES (Figure 4D∗). IL-8 dose-dependent secretion in response to IL-1β treatment was only observed in NES-Astro (Figure 4D∗) and phaAstro models (Figure 4F∗), while dose dependency to TNF-α treatment could also be observed in CCF (Figure 4F∗), iCellAstro (Figure 4E∗), and HEK (Figure S5C). In contrast to the IL-8 response, significantly higher IL-6 levels in relation to exposure time and cytokine treatment concentration were observed to be less frequent. Across models a significant IL-6 response was more frequently observed for higher stimulant concentration at 48 hr than for lower stimulant concentration (Figure S5D).
Together, these data show that the NES-Astro (AF22/C1/C9) model is immune competent and consistently shows a dose-dependent response of IL-8 to TNF-α and IL-1β treatment, which was only observed sporadically in the other astrocytic models. There are very large cytokine concentration differences between models, in both inflammatory activated states but also in non-stimulated conditions, whereby NES-Astro is shown to possess unmeasurable cytokine secretion in contrast to the other astrocytic models.
Calcium Signaling Response
Astrocytes are known to respond to synaptic release of glutamate and ATP through increased intracellular calcium levels (Bazargani and Attwell, 2016, Zhang et al., 2016). Slow calcium signal activation in human astrocytes has previously been reported using hPSC-derived and primary astrocytes in response to mechanical (Krencik et al., 2011, Santos et al., 2017, Serio et al., 2013), ATP (Serio et al., 2013), and glutamate (Tcw et al., 2017) stimulation. We used FLIPR Calcium 5, a calcium-sensitive dye, to study whether the astrocyte models were calcium signaling responsive to injection control (shear stimulation) or 3 μM ATP or 3 μM glutamate. Stimulation was followed by time-lapse acquisition of calcium-induced fluorescence (Figure 5A). We found that NES-Astro (AF22/C1/C9) and phaAstro responded with a significant response to ATP compared with the injection control, whereas only the NES-Astro (C1/C9) responded to glutamate stimulation (Figures 5B and 5C). NES-Astro (AF22) and phaAstro had a fraction of approximately 60% ATP-responding cells, and NES-Astro (C1/C9) had up to 90% ATP-responding cells. Additionally, NES-Astro (C1/C9) had 83% and 66% calcium glutamate-responsive cell populations, respectively (Figure 5D). NES-Astro (C1) showed a homogeneous functional diversity, having the same fraction of ATP- and glutamate-responsive cells (Figure 5D), making it possible to study treatment response patterns as previously reported for human primary adult and fetal astrocytes (Zhang et al., 2016). Single-cell analysis revealed that compared with glutamate, ATP induced an increased number of cellular responses for a longer period (Figures 5E and 5F), which corroborates previous findings (Zhang et al., 2016).
Figure 5.
Astrocytic Calcium Signaling Response
(A) Representative 45-s time-lapse images of calcium response to shear stimulation (top panels), 3 μM ATP (middle panels), and 3 μM glutamate (bottom panels) of NES-Astro (C1).
(B) Average well signal intensity over time (s) for injection control (shear stimulation) (left), 3 μM ATP (middle), and 3 μM glutamate (right) of each astrocytic model.
(C) Average signal intensity over the experimental period for injection control, ATP, and glutamate.
(D) Percentage of responsive cells (cells were classified as responsive if signal intensity >2-fold the average well intensity of injection control set per cell model).
(E) Rate of signal change during ATP and glutamate stimulation for NES-Astro (C1). Each trace represents a single cell.
(F) Number of cellular responses per cell over 125 s.
Data shown from independent experiments, n = 3, mean ± SEM. Statistical analysis: two-way ANOVA (C and D) and Student's t test (F); ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Scale bars, 300 μm.
In summary, the ATP- and glutamate-induced calcium responses recapitulates previous patterns observed following ATP and glutamate stimulation (Zhang et al., 2016). We could, however, also conclude that there is a large variability in calcium responsiveness between models.
Models Display Large Variability in Activity of APOE-Modulating Compounds
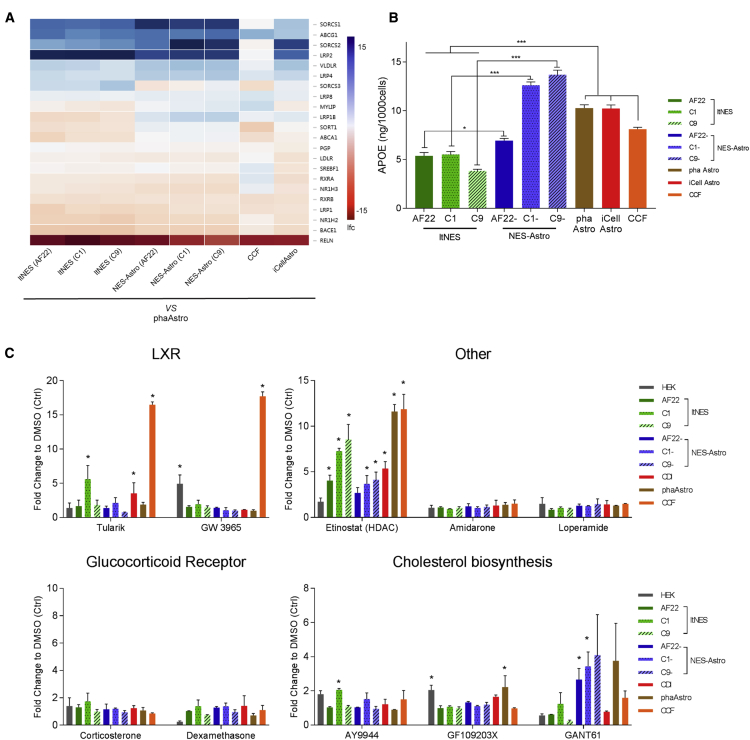
Given the importance of APOE to astrocyte biology and neurological disease, we analyzed the transcriptomic differences between the models of APOE-associated genes (Figure S6A) enriched in the brain (Figure 6A). Additionally, by assessing basal APOE levels in cell media we observed that astrocyte models have a significantly higher secretion compared with the neural stem cells (Figure 6B). Next, we wanted to extend the transcriptomic, proteomic, and functional characterization to also study basic modulation of astrocyte biology in relation to APOE secretion. A pilot screen was performed (Figure S6B) to find out whether previous findings of APOE-modulating compounds could be captured in any of our astrocyte models (Fan et al., 2016, Finan et al., 2016). Compounds with a known APOE-modulating effect were evaluated together with two glucocorticoid receptor modulators serving as negative controls (Figure 6C and Table 1). The astrocytoma cell line CCF, used as a positive control and for Z′ calculations (Figure S6), showed high fold changes in response to liver X receptor (LXR) agonists Tularik and GW3965. The histone deacetylase (HDAC) annotated compound, MS-275, increased APOE secretion in most of the models, including ltNES, without showing any astrocyte specificity. Interestingly, NES-Astro showed a significant increase of APOE secretion in response to the cholesterol biosynthesis annotated compound, GANT61, in accordance with previous findings for primary astrocytes (Finan et al., 2016). A tendency toward increased APOE secretion for GANT61 could also be observed for phaAstro.
Figure 6.
Characterization and Regulation of Endogenous APOE Secretion through APOE-Modulating Compounds
(A) Differential expressional analysis of neuronal models to phaAstro including genes linked to brain-associated APOE.
(B) Basal APOE secretion levels of neuronal models.
(C) APOE secretion levels after compound stimulation. Compound concentration resulting in the highest APOE secretion increase is shown as fold change compared with DMSO control.
Data shown from independent experiments, n = 3, mean ± SEM. Statistical analysis: one-way ANOVA (B) and t test (C); ∗p < 0.05, ∗∗∗p < 0.001.
Table 1.
Change in APOE Secretion in Response to CompoundStimulation
| Compound | Annotation | CID | ltNES |
NES-Astro |
iCellAstro | CCF | phaAstro | HEK | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C1 | C9 | AF22 | C1 | C9 | AF22 | |||||||
| Corticosterone | glucocorticoid receptor (GR) agonist (used as negative control) | 5753 | 1.7 ± 0.5 | 1.0 ± 0.1 | 1.3 ± 0.2 | 1.2 ± 0.0 | 1.0 ± 0.1 | 1.2 ± 0.3 | 1.2 ± 0.2 | 0.9 ± 0.0 | 1.1 ± 0.2 | 1.4 ± 0.5 |
| Dexamethasone | GR agonist (used as negative control) | 5743 | 1.4 ± 0.4 | 0.8 ± 0.0 | 1.0 ± 0.0 | 1.4 ± 0.2 | 1.0 ± 0.1 | 1.3 ± 0.1 | 1.4 ± 0.6 | 1.1 ± 0.3 | 0.7 ± 0.1 | 0.2 ± 0.1 |
| Tularik | liver X receptor (LXR) agonist | 447912 | 5.6 ± 1.6 | 1.8 ± 0.6 | 1.7 ± 0.7 | 2.1 ± 0.6 | 0.8 ± 0.0 | 1.4 ± 0.2 | 3.5 ± 1.3 | 16.5 ± 0.3 | 1.9 ± 0.3 | 1.4 ± 0.6 |
| GW3965 | LXR agonist | 447905 | 1.9 ± 0.5 | 1.5 ± 0.2 | 1.6 ± 0.1 | 1.1 ± 0.3 | 1.0 ± 0.1 | 1.4 ± 0.1 | 1.1 ± 0.0 | 17.7 ± 0.5 | 1.0 ± 0.1 | 4.9 ± 1.0 |
| Etinostat | histone deacetylase (HDAC) inhibitor | 4261 | 7.3 ± 0.3 | 8.6 ± 1.3 | 4.0 ± 0.5 | 3.7 ± 0.7 | 4.1 ± 0.7 | 2.7 ± 0.5 | 5.4 ± 0.6 | 11.9 ± 1.3 | 11.6 ± 0.6 | 1.7 ± 0.3 |
| Amidarone | calcium channel blocker | 2157 | 0.9 ± 0.0 | 1.1 ± 0.1 | 1.1 ± 0.1 | 1.0 ± 0.1 | 1.2 ± 0.2 | 1.2 ± 0.2 | 1.3 ± 0.5 | 1.5 ± 0.3 | 1.4 ± 0.2 | 1.1 ± 0.2 |
| Loperamide | μ-opioid receptor agonist | 3955 | 1.0 ± 0.1 | 0.9 ± 0.1 | 0.8 ± 0.1 | 1.2 ± 0.0 | 1.5 ± 0.5 | 1.3 ± 0.2 | 1.4 ± 0.3 | 1.5 ± 0.0 | 1.3 ± 0.0 | 1.5 ± 0.5 |
| AY9944 | cholesterol biosynthesis DHCR7 inhibitor | 9704 | 2.1 ± 0.1 | 1.1 ± 0.0 | 1.0 ± 0.0 | 1.5 ± 0.3 | 1.0 ± 0.1 | 1.0 ± 0.0 | 1.2 ± 0.2 | 1.5 ± 0.4 | 0.9 ± 0.0 | 1.8 ± 0.2 |
| GF109203X | cholesterol biosynthesis DHCR24 inhibitor | 2396 | 1.1 ± 0.1 | 1.0 ± 0.1 | 1.0 ± 0.1 | 1.1 ± 0.0 | 1.2 ± 0.1 | 1.3 ± 0.1 | 1.7 ± 0.1 | 1.0 ± 0.0 | 2.2 ± 0.5 | 2.1 ± 0.2 |
| GANT61 | cholesterol biosynthesis DHCR7 agonist | 421610 | 1.2 ± 0.5 | 0.2 ± 0.0 | 0.6 ± 0.0 | 3.4 ± 0.7 | 4.1 ± 1.9 | 2.7 ± 0.5 | 0.8 ± 0.0 | 1.6 ± 0.3 | 3.8 ± 1.8 | 0.6 ± 0.1 |
Fold change values from simulated APOE secretion compared with DMSO control are specified for each compound and cell type (fold change ± SEM). Data shown from independent experiments, n = 3. Significant increases compared with DMSO control are highlighted in boldface. See also Figure S6 for experimental procedure.
CID, PubChem CID; ltNES, long-term self-renewing neuroepithelial-like stem cells; NES-Astro, ltNES-derived astroglia; iCellAstro, iCell astrocytes; CCF, CCF-STTG1; phaAstro, primary human adult astrocytes; APOE, apolipoprotein E.
In conclusion, no clear pattern of general astrocytic APOE-modulating compounds could be observed from this pilot screen. Our results clearly indicate the relevance of cellular context during modulation of astrocyte APOE biology.
Discussion
LtNES cells have mainly been used for studying the undirected differentiation process toward a neuronal fate (Falk et al., 2012, Tailor et al., 2013, Zhang et al., 2014). Using ltNES in this study we show (1) the application of ltNES in directed glia generation using the FHIA protocol, with (2) high reproducibility across hiPSC lines, and (3) proven astrocyte functions including SLC1A3-driven glutamate uptake, immune competence, and calcium responsiveness to neurotransmitters. We also show (4) the use of an animal-free culture system applicable for good manufacturing practice and clinical adaptation, and (5) NES-Astro as a cryopreservable cell model for direct application in pharmacological HTS. For astrocyte modeling, we demonstrate that (1) there is a high variability between models and (2) the selection of the model is dependent on the functionality to be studied.
Global transcriptomic analysis has been applied to profile human primary astrocytes (Malik et al., 2014, Zhang et al., 2016), human primary NPC-derived astrocytes (Magistri et al., 2016), hiPSC-derived astrocytes (Pei et al., 2016), and glioma (Auvergne et al., 2013). However, a cross-comparison has not previously been performed between human adult primary astrocytes, astrocytoma, and hiPSC-derived astrocytes. Our comprehensive analysis of transcriptomic profiling shows clear differences between these astrocyte models, and also the developmental stage of neural stem cell (ltNES) and astroglia (NES-Astro). Protein evaluation corroborated the developmental transition, displaying expression of key glial lineage-associated transcription factors SOX9 and NFIA (Wilczynska et al., 2009), surface receptor CD44 (Sosunov et al., 2014), and cytoplasmic proteins FABP7 (Howard et al., 2006) and S100B (Steiner et al., 2007). NES-Astro also showed a strong and homogeneous expression of SLC1A3, a strong indication of astrocyte identity (DeSilva et al., 2012). Additionally, protein expression patterns confirm the transcriptional diversity that is indicative of differences in model representation. The expression of GFAP, the most common marker used to claim astrocyte lineage, was predominantly absent in the models. However, GFAP is not expressed by all human adult astrocytes including protoplasmic astrocytes (DeSilva et al., 2012), highlighting the importance of studying the role of GFAP in astrocytes in relation to their developmental stage and cellular subtypes.
To further study astrocyte biology in vitro it is important that the model represents main astrocytic functional features, including maintaining glutamate homeostasis, as observed around the synaptic cleft (Schousboe et al., 2013). NES-Astro showed a significantly higher active uptake of glutamate compared with the other astrocytic models, which did not show any difference compared with non-astrocytic nor non-neuronal models. Astrocyte-associated SLC1A3 was only expressed in NES-Astro (AF22/C1/C9), and significantly contributed to the glutamate uptake. Linking functional glutamate uptake to a specific astrocyte-associated glutamate transporter has not previously been shown using hiPSC-derived astrocytes (Chen et al., 2014, Gupta et al., 2012, Krencik et al., 2011, Palm et al., 2015, Santos et al., 2017, Serio et al., 2013, Shaltouki et al., 2013).
Astrocyte reactivity is a process to limit tissue damage and cell stress and restore normal tissue homeostasis. The reactivity is gradual and circumstantial, linked to specific types of neuropathology, and is characterized by brain region, niche, severity, and delimiting features (Sofroniew, 2015). The inflammatory competence of astrocytes is a key biological function. The high basal secretion of inflammatory cytokines observed in CCF, iCellAstro, and phaAstro during non-stimulated conditions correlates with results found in previous studies (Santos et al., 2017, Tcw et al., 2017). A common factor between these models is that they are cultured with FBS that has been shown to induce an inflammatory active state also during “non-stimulated” conditions (Magistri et al., 2016, Zhang et al., 2016). NES-Astro cells, which are cultured without FBS, had almost no secretion of inflammatory cytokines under non-stimulated conditions. NES-Astro also responded to pro-inflammatory stimulation in a dose-dependent manner, demonstrating robust immune competence. Additionally, NES-Astro morphologically display a stellate phenotype similar to acutely isolated primary astrocytes, a phenotype shown to be lost upon FBS treatment (Zhang et al., 2016).
Astrocytic calcium signaling and subsequent response effects have been intensely discussed (Bazargani and Attwell, 2016), suggesting that improved in vitro modeling to accompany in vivo findings would be beneficial. Mechanical stimulation has been previously proven to induce calcium response in astrocytes (Krencik et al., 2011, Santos et al., 2017, Serio et al., 2013), as also indicated by the injection controls for all models in this study. However, only NES-Astro AF22/C1/C9 and phaAstro responded to ATP, whereas NES-Astro C1/C9 was the only glutamate-responsive astrocyte model. Calcium responses have previously been described following ATP (Serio et al., 2013) and glutamate (Tcw et al., 2017, Zhang et al., 2016) treatments. ATP and glutamate responsiveness can discriminate between a fetal and adult stage, respectively, and it has been suggested that the latter is acquired by astrocytes after mid-gestation (Zhang et al., 2016). In comparison with acutely isolated human primary adult astrocytes (Zhang et al., 2016), NES-Astro (C1) display similar calcium signaling patterns with heterogeneous ATP and homogeneous glutamate responses. It would be interesting to speculate whether NES-Astro calcium signaling responsiveness could be linked to its high level of human astrocyte-specific RYR3 expression (Zhang et al., 2016), but this needs further extensive studies. The diversity in glutamate-responsive cell populations of 0%, 66%, and 83% between hiPSC-derived NES-Astro lines AF22, C1, and C9, respectively, could indicate different developmental stages and level of maturation, which could provide a future tool to study the functional development of astrocytes.
Screening for APOE-modulating compounds in astrocytes have been performed using CCF and human primary fetal astrocytes (Fan et al., 2016, Finan et al., 2016). To our knowledge the application of hiPSC-derived astrocyte in comparison with human primary adult astrocytes in an APOE screening assay has not previously been shown.
In agreement with previous reports (Fan et al., 2016, Finan et al., 2016), the responses in APOE secretion are much greater in CCF than in any of the other models, which might be reflected by its transcriptomic profile. The study by Finan et al. reported high responses in CCF and low responses in primary astrocytes to the LXR agonist Tularik (Finan et al., 2016) and GW3965 (Fan et al., 2016), which is confirmed in our study. The HDAC inhibitor MS-275 has been shown to modulate APOE expression (Sylvain et al., 2013), a response observed in almost all models, indicating that the compound is not astrocyte specific. The three cholesterol biosynthesis annotated compounds were selected to differentially target cholesterol-associated proteins DHCR24 and DHCR7 (Finan et al., 2016). GF109203X, a DHCR24 inhibitor, significantly increases APOE secretion in phaAstro (Figure 5C), which also have been observed in human fetal astrocytes (Finan et al., 2016). In contrast to AY9944 (a DHCR7 inhibitor) the DHCR7 agonist, GANT61, showed a significant effect and increased tendency of APOE secretion in NES-Astro (AF22/C1) and phaAstro, respectively. Together with previously published data on human fetal astrocyte response to cholesterol biosynthesis modulators (Finan et al., 2016), we can confirm these hits in human adult primary (phaAstro) and hiPSC-derived astrocytes (NES-Astro) and highlight this pathway as a future potential target of interest.
Taken together, based on this pilot screen there is a lack of consistency regarding astrocyte-specific APOE modulators. A possible explanation could be the large model diversities observed in the transcriptomic, protein, and functional characterization. These results emphasize the importance of comparing several astrocyte models to understand the biological relevance of screening data. However, the exciting results presented in this study introduce iPSC-derived astroglia as an additional valuable tool for screening.
Experimental Procedures
Long-Term Neural Epithelial Stem Cell Culture
We previously showed that ltNES could be generated from several hiPSCs and hESCs (Falk et al., 2012). Generation of ltNES lines have been performed previously (iPS Core at Karolinska Institutet) from human hiPSC lines AF22 (Falk et al., 2012), C1 (Shahsavani et al., 2017), and C9 (Uhlin et al., 2017), which represent a variation in gender, age, and reprogramming technique.
ltNES-Derived Astroglia Differentiation
ltNES cells were plated at 60,000 cells/cm2 on 2 μg/cm2 poly-L-ornithine and 0.2 μg/cm2 laminin (PLO-Laminin) (Sigma, St. Louis, MO) double-coated culture vessels in FHIA-differentiation medium; DMEM/F12, N2 supplement (1:100; Invitrogen, Carlsbad, CA), B27 (1:100; Invitrogen), FGF2 (8 ng/mL; PeproTech, Rocky Hill, NJ), heregulin 1β (10 ng/mL; Sigma), IGF1 (200 ng/mL; Sigma), activin A (10 ng/mL; PeproTech). The medium was changed every other day and cells were passaged once they reached 80% confluence; 7–9 passages during the differentiation protocol of 28 days. This was performed for ltNES lines AF22, C1, and C9 generating the NES-Astro phenotype for each line. For a complete animal free system the regular B27 (0080085SA, 1:100, Invitrogen) was exchanged for Xenofree B27 (A1486701; 1:100; Invitrogen) and the double coating of PLO-Laminin (both from Sigma) for Laminin 521, 1 μg/cm2 (Biolamina, Sweden). Gene expression was assessed by applying real-time qPCR using TaqMan Assays (Applied Biosystems, Foster City, CA). Proliferation during differentiation was assessed by estimation of doubling time via cell count using a Cedex HiRes Analyzer (Roche, Switzerland).
Cells
Human brain astrocytes (HBA) were purchased from Neuromics (Edina, MN), and iCell astrocytes from Cellular Dynamics International (Madison, WI), while the astrocytoma cell line CCF-STTG1 and HEK293 cells were both purchased from ATCC (Manassas, VA). A summary of cell lines included in the different experiments is specified in Table S7.
Immunocytochemistry
Immunocytochemistry was performed according to standard methods applying blocking procedures followed by primary and secondary antibody staining and ending with nuclei staining. Details of primary, secondary, and iso-type control antibodies are presented in Table S1.
Transcriptomic Analysis
Total RNA was isolated for all samples and run using Illumina products and an Illumina NextSeq 500 Sequencer (Illumina, San Diego, CA). The raw count data were processed using DESeq2 according to previously published methods (Love et al., 2014) followed by more in-depth bioinformatics analysis including principal component analysis, Pearson correlation analysis of samples and gene pairs, and differential gene expression analysis.
Glutamate Clearance Assay
Glutamate reduction was measured on medium samples using a Glutamine/Glutamate Determination Kit (Sigma) at sequential time points for 60 min. To modulate specific glutamate transporters, we used UCPH 101 (Abcam, UK) and WAY 213613 (Tocris, UK) to inhibit SLC1A3 and SLC1A2, respectively.
Inflammatory Assay
Inflammatory response was assessed by incubating each cell model for 24 or 48 hr with 10 or 50 ng/mL recombinant human TNF-α, or 10 or 50 ng/mL recombinant human IL-1β (both from R&D systems). IL-6 and IL-8 cytokines in the samples were quantified using the MultiCyt QBeads assay according to the manufacturer's instructions with an assay miniaturization modification (Intellicyt, Albuquerque, NM, USA).
Calcium Signaling Response
Calcium signaling responses were measured using Calcium5 (Molecular Devices) following stimulation with 3 μM ATP, 3 μM L-glutamic acid (both from Sigma), or 200 μM DHPG (Tocris). Images were taken with 20× magnification at 0.5-s intervals for 2 min. Image analysis, to extract single cell responses, was performed using Fiji (Schindelin et al., 2012). Subsequent data analysis was performed using TIBCO Spotfire (TIBCO Software, CA, USA).
APOE Stimulation and Quantification
APOE secretion was measured on medium samples using a sandwich ELISA, which were adapted to a screening setting through semi-automation. To modulate APOE secretion we treated all cellular models with 10 different compounds, respectively. All compounds are available in the public domain and are presented in Table 1 with PubChem CID.
Statistical Analysis
Student's t test and one- and two-way ANOVA statistical analysis were used. Specifically, for RNA-seq analysis DEseq2, principal component analysis, and Pearson correlation analysis were applied.
Author Contributions
A.L., L.D., G.B., R.H., A.H., and A.F., conception and design. A.L., L.D., M.C., A.S., and M.D., collection and/or assembly of data. A.L., L.D., P.R., J. Sanchez, A.S., R.H., A.H., and A.F., data analysis and interpretation. A.L., L.D, P.R., J. Sanchez, A.S., M.D., J. Synnergren, H.Z., G.B., R.H., A.H., and A.F., manuscript writing. A.L., R.H., A.H., and A.F., final approval of manuscript. A.F.: provision of study material or patients.
Acknowledgments
We thank Ola Engqvist and Johan Muller for sharing scientific experience and helpful discussion in regard to the APOE assay. We thank the iPS Core at Karolinska Institutet for derivation of iPSCs and ltNES cells. This work was supported by The Swedish Research Council (grant number 2013-5731), SSF (grant number IB13-0074), VINNOVA, VINNMER Marie Curie National qualification grant (number 2009-04085), VINNOVA, VINNMER Marie Curie National qualification grant (number 2010-01013), the Wallenberg Foundation (grant number 2015.0178), and the Swedish Knowledge Foundation (grant number 2014-0298). A.L., L.D., M.C., P.R., J.S., A.S., M.D., G.B., and R.H. are employees of AstraZeneca.
Published: February 15, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, seven tables, and one movie and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.01.021.
Contributor Information
Anna Herland, Email: aherland@kth.se.
Anna Falk, Email: anna.falk@ki.se.
Accession Numbers
The accession number for the RNA-seq data reported in this paper is GEO: GSE109001.
Supplemental Information
During the differentiation of ltNES to NES-Astro one bright-field image was taken each day from differentiation day 0 to day 28. Scale bars, 300 μm. ltNES, long-term self-renewing neuroepithelial-like stem cells; NES-Astro, ltNES-derived astroglia.
References
- Auvergne R.M., Sim F.J., Wang S., Chandler-Militello D., Burch J., Al Fanek Y., Davis D., Benraiss A., Walter K., Achanta P. Transcriptional differences between normal and glioma-derived glial progenitor cells identify a core set of dysregulated genes. Cell Rep. 2013;3:2127–2141. doi: 10.1016/j.celrep.2013.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazargani N., Attwell D. Astrocyte calcium signaling: the third wave. Nat. Neurosci. 2016;19:182–189. doi: 10.1038/nn.4201. [DOI] [PubMed] [Google Scholar]
- Chen C., Jiang P., Xue H., Peterson S.E., Tran H.T., McCann A.E., Parast M.M., Li S., Pleasure D.E., Laurent L.C. Role of astroglia in Down's syndrome revealed by patient-derived human-induced pluripotent stem cells. Nat. Commun. 2014;5:4430. doi: 10.1038/ncomms5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings J.L., Morstorf T., Zhong K. Alzheimer’s disease drug-development pipeline: few candidates, frequent failures. Alzheimer's Res. Ther. 2014;6:37. doi: 10.1186/alzrt269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmanis S., Sloan S.A., Zhang Y., Enge M., Caneda C., Shuer L.M., Hayden Gephart M.G., Barres B.A., Quake S.R. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. USA. 2015;112:7285–7290. doi: 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pittà M., Brunel N., Volterra A. Astrocytes: orchestrating synaptic plasticity? Neuroscience. 2016;323:43–61. doi: 10.1016/j.neuroscience.2015.04.001. [DOI] [PubMed] [Google Scholar]
- DeSilva T.M., Borenstein N.S., Volpe J.J., Kinney H.C., Rosenberg P.A. Expression of EAAT2 in neurons and protoplasmic astrocytes during human cortical development. J. Comp. Neurol. 2012;520:3912–3932. doi: 10.1002/cne.23130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk A., Koch P., Kesavan J., Takashima Y., Ladewig J., Alexander M., Wiskow O., Tailor J., Trotter M., Pollard S. Capture of neuroepithelial-like stem cells from pluripotent stem cells provides a versatile system for in vitro production of human neurons. PLoS One. 2012;7:e29597. doi: 10.1371/journal.pone.0029597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Zareyan S., Zhao W., Shimizu Y., Pfeifer T.A., Tak J.H., Isman M.B., Van den Hoven B., Duggan M.E., Wood M.W. Identification of a chrysanthemic ester as an apolipoprotein E Inducer in astrocytes. PLoS One. 2016;11:e0162384. doi: 10.1371/journal.pone.0162384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan G.M., Realubit R., Chung S., Lutjohann D., Wang N., Cirrito J.R., Karan C., Kim T.W. Bioactive compound screen for pharmacological enhancers of apolipoprotein E in primary human astrocytes. Cell Chem. Biol. 2016;23:1526–1538. doi: 10.1016/j.chembiol.2016.10.015. [DOI] [PubMed] [Google Scholar]
- Gupta K., Patani R., Baxter P., Serio A., Story D., Tsujita T., Hayes J.D., Pedersen R.A., Hardingham G.E., Chandran S. Human embryonic stem cell derived astrocytes mediate non-cell-autonomous neuroprotection through endogenous and drug-induced mechanisms. Cell Death Differ. 2012;19:779–787. doi: 10.1038/cdd.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard B., Chen Y., Zecevic N. Cortical progenitor cells in the developing human telencephalon. Glia. 2006;53:57–66. doi: 10.1002/glia.20259. [DOI] [PubMed] [Google Scholar]
- Khakh B.S., Sofroniew M.V. Diversity of astrocyte functions and phenotypes in neural circuits. Nat. Neurosci. 2015;18:942–952. doi: 10.1038/nn.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T., Asai M., Tsukita K., Kutoku Y., Ohsawa Y., Sunada Y., Imamura K., Egawa N., Yahata N., Okita K. Modeling Alzheimer's disease with iPSCs reveals stress phenotypes associated with intracellular Abeta and differential drug responsiveness. Cell Stem Cell. 2013;12:487–496. doi: 10.1016/j.stem.2013.01.009. [DOI] [PubMed] [Google Scholar]
- Krencik R., Weick J.P., Liu Y., Zhang Z.J., Zhang S.C. Specification of transplantable astroglial subtypes from human pluripotent stem cells. Nat. Biotechnol. 2011;29:528–534. doi: 10.1038/nbt.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magistri M., Khoury N., Mazza E.M., Velmeshev D., Lee J.K., Bicciato S., Tsoulfas P., Faghihi M.A. A comparative transcriptomic analysis of astrocytes differentiation from human neural progenitor cells. Eur. J. Neurosci. 2016;44:2858–2870. doi: 10.1111/ejn.13382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik N., Wang X., Shah S., Efthymiou A.G., Yan B., Heman-Ackah S., Zhan M., Rao M. Comparison of the gene expression profiles of human fetal cortical astrocytes with pluripotent stem cell derived neural stem cells identifies human astrocyte markers and signaling pathways and transcription factors active in human astrocytes. PLoS One. 2014;9:e96139. doi: 10.1371/journal.pone.0096139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberheim N.A., Takano T., Han X., He W., Lin J.H., Wang F., Xu Q., Wyatt J.D., Pilcher W., Ojemann J.G. Uniquely hominid features of adult human astrocytes. J. Neurosci. 2009;29:3276–3287. doi: 10.1523/JNEUROSCI.4707-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm T., Bolognin S., Meiser J., Nickels S., Träger C., Meilenbrock R.-L., Brockhaus J., Schreitmüller M., Missler M., Schwamborn J.C. Rapid and robust generation of long-term self-renewing human neural stem cells with the ability to generate mature astroglia. Scientific Rep. 2015;5:16321. doi: 10.1038/srep16321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Y., Peng J., Behl M., Sipes N.S., Shockley K.R., Rao M.S., Tice R.R., Zeng X. Comparative neurotoxicity screening in human iPSC-derived neural stem cells, neurons and astrocytes. Brain Res. 2016;1638:57–73. doi: 10.1016/j.brainres.2015.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos R., Vadodaria K.C., Jaeger B.N., Mei A., Lefcochilos-Fogelquist S., Mendes A.P.D., Erikson G., Shokhirev M., Randolph-Moore L., Fredlender C. Differentiation of inflammation-responsive astrocytes from glial progenitors generated from human induced pluripotent stem cells. Stem Cell Reports. 2017;8:1757–1769. doi: 10.1016/j.stemcr.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schousboe A., Bak L.K., Waagepetersen H.S. Astrocytic control of biosynthesis and turnover of the neurotransmitters glutamate and GABA. Front. Endocrinol. 2013;4:102. doi: 10.3389/fendo.2013.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio A., Bilican B., Barmada S.J., Ando D.M., Zhao C., Siller R., Burr K., Haghi G., Story D., Nishimura A.L. Astrocyte pathology and the absence of non-cell autonomy in an induced pluripotent stem cell model of TDP-43 proteinopathy. Proc. Natl. Acad. Sci. USA. 2013;110:4697–4702. doi: 10.1073/pnas.1300398110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahsavani M., Pronk R.J., Falk R., Lam M., Moslem M., Linker S.B., Salma J., Day K., Schuster J., Anderlid B.M. An in vitro model of lissencephaly: expanding the role of DCX during neurogenesis. Mol. Psychiatry. 2017 doi: 10.1038/mp.2017.175. [DOI] [PubMed] [Google Scholar]
- Shaltouki A., Peng J., Liu Q., Rao M.S., Zeng X. Efficient generation of astrocytes from human pluripotent stem cells in defined conditions. Stem Cells. 2013;31:941–952. doi: 10.1002/stem.1334. [DOI] [PubMed] [Google Scholar]
- Sofroniew M.V. Astrogliosis. Cold Spring Harb. Perspect. Biol. 2015;7:a020420. doi: 10.1101/cshperspect.a020420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosunov A.A., Wu X., Tsankova N.M., Guilfoyle E., McKhann G.M., 2nd, Goldman J.E. Phenotypic heterogeneity and plasticity of isocortical and hippocampal astrocytes in the human brain. J. Neurosci. 2014;34:2285–2298. doi: 10.1523/JNEUROSCI.4037-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaethling J.M., Na Y.J., Lee J., Ulyanova A.V., Baltuch G.H., Bell T.J., Brem S., Chen H.I., Dueck H., Fisher S.A. Primary cell culture of live neurosurgically resected aged adult human brain cells and single cell transcriptomics. Cell Rep. 2017;18:791–803. doi: 10.1016/j.celrep.2016.12.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner J., Bernstein H.-G., Bielau H., Berndt A., Brisch R., Mawrin C., Keilhoff G., Bogerts B. Evidence for a wide extra-astrocytic distribution of S100B in human brain. BMC Neurosci. 2007;8:2. doi: 10.1186/1471-2202-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvain E., Ramaswamy G., Beyna M., Duerr J., Lanyon L., LaChapelle E., Pettersson M., Steppan C., Bales K. A novel LXRα- or RX-independent mechanism for increasing brain APOE levels. Alzheimer's & Dementia. 2013;9:P855. [Google Scholar]
- Tailor J., Kittappa R., Leto K., Gates M., Borel M., Paulsen O., Spitzer S., Karadottir R.T., Rossi F., Falk A. Stem cells expanded from the human embryonic hindbrain stably retain regional specification and high neurogenic potency. J. Neurosci. 2013;33:12407–12422. doi: 10.1523/JNEUROSCI.0130-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tarassishin L., Suh H.S., Lee S.C. LPS and IL-1 differentially activate mouse and human astrocytes: role of CD14. Glia. 2014;62:999–1013. doi: 10.1002/glia.22657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcw J., Wang M., Pimenova A.A., Bowles K.R., Hartley B.J., Lacin E., Machlovi S.I., Abdelaal R., Karch C.M., Phatnani H. An efficient platform for astrocyte differentiation from human induced pluripotent stem cells. Stem Cell Reports. 2017;9:600–614. doi: 10.1016/j.stemcr.2017.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlin E., Ronnholm H., Day K., Kele M., Tammimies K., Bolte S., Falk A. Derivation of human iPS cell lines from monozygotic twins in defined and xeno free conditions. Stem Cell Res. 2017;18:22–25. doi: 10.1016/j.scr.2016.12.006. [DOI] [PubMed] [Google Scholar]
- Wilczynska K.M., Singh S.K., Adams B., Bryan L., Rao R.R., Valerie K., Wright S., Griswold-Prenner I., Kordula T. Nuclear factor I isoforms regulate gene expression during the differentiation of human neural progenitors to astrocytes. Stem Cells. 2009;27:1173–1181. doi: 10.1002/stem.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.T., Tan L., Hardy J. Apolipoprotein E in Alzheimer's disease: an update. Annu. Rev. Neurosci. 2014;37:79–100. doi: 10.1146/annurev-neuro-071013-014300. [DOI] [PubMed] [Google Scholar]
- Zhang D., Pekkanen-Mattila M., Shahsavani M., Falk A., Teixeira A.I., Herland A. A 3D Alzheimer's disease culture model and the induction of P21-activated kinase mediated sensing in iPSC derived neurons. Biomaterials. 2014;35:1420–1428. doi: 10.1016/j.biomaterials.2013.11.028. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Sloan S.A., Clarke L.E., Caneda C., Plaza C.A., Blumenthal P.D., Vogel H., Steinberg G.K., Edwards M.S., Li G. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
During the differentiation of ltNES to NES-Astro one bright-field image was taken each day from differentiation day 0 to day 28. Scale bars, 300 μm. ltNES, long-term self-renewing neuroepithelial-like stem cells; NES-Astro, ltNES-derived astroglia.