Summary
MyoD and Myf5 are fundamental regulators of skeletal muscle lineage determination in the embryo, and their expression is induced in satellite cells following muscle injury. MyoD and Myf5 are also expressed by satellite cell precursors developmentally, although the relative contribution of historical and injury-induced expression to satellite cell function is unknown. We show that satellite cells lacking both MyoD and Myf5 (double knockout [dKO]) are maintained with aging in uninjured muscle. However, injured muscle fails to regenerate and dKO satellite cell progeny accumulate in damaged muscle but do not undergo muscle differentiation. dKO satellite cell progeny continue to express markers of myoblast identity, although their myogenic programming is labile, as demonstrated by dramatic morphological changes and increased propensity for non-myogenic differentiation. These data demonstrate an absolute requirement for either MyoD or Myf5 in muscle regeneration and indicate that their expression after injury stabilizes myogenic identity and confers the capacity for muscle differentiation.
Keywords: skeletal muscle regeneration, muscle stem cell programming, muscle differentiation, satellite cell, MyoD, Myf5, adipogenesis, fibrosis, conditional knockout, Cre/loxP
Highlights
-
•
MyoD or Myf5 expression in satellite cells is essential for muscle regeneration
-
•
Satellite cells lacking both regulatory genes exhibit labile myogenic programming
-
•
A single functional allele of either MyoD or Myf5 can support muscle regeneration
-
•
Satellite cells lacking both MyoD and Myf5 are maintained with aging
In this article, Goldhamer and colleagues show that loss of both MyoD and Myf5 in skeletal muscle satellite cells results in regenerative failure following injury. Satellite cell progeny accumulate in injured muscle and continue to express markers of myoblast identity, but do not undergo muscle differentiation, and exhibit a propensity for non-myogenic differentiation.
Introduction
The myogenic regulatory factors (MRFs) MYOD and MYF5 are fundamental regulators of skeletal muscle lineage determination in the embryo (Kassar-Duchossoy et al., 2004, Rudnicki et al., 1993) and are upregulated in activated muscle stem cells (satellite cells) as an early response to muscle injury (Yin et al., 2013). Although it is now well established that satellite cells are essential for muscle regeneration (McCarthy et al., 2011, Murphy et al., 2011, Sambasivan et al., 2011), the mechanisms by which myogenic programming is established and maintained, and the functions of Myod1 (MyoD) and Myf5 in satellite cell determination and differentiation, remain unknown. Satellite cells lacking either MyoD or Myf5 exhibit relatively mild differentiation and growth defects and remain stably committed to the myogenic fate (Gayraud-Morel et al., 2007, Megeney et al., 1996, Starkey et al., 2011, Ustanina et al., 2007, Yablonka-Reuveni et al., 1999). Determining whether this reflects genetic redundancy between these MRF family members, as in the embryo (Kassar-Duchossoy et al., 2004, Rudnicki et al., 1993), or the engagement of alternative or compensatory regulatory pathways in muscle regeneration, requires analyses of mice lacking both MRFs. To explore the regulatory relationship between MyoD and Myf5 in satellite cell function, we developed a conditional knockout allele of MyoD to circumvent the perinatal lethality exhibited by mice constitutively lacking both genes (Kassar-Duchossoy et al., 2004, Rudnicki et al., 1993).
Satellite cell precursors share a number of fundamental properties with fetal myoblasts or their progenitors, including their origin from the dermomyotome (Yin et al., 2013), sequential transcriptional activation of Pax3 and Pax7 (Relaix et al., 2004), and expression of MyoD (Kanisicak et al., 2009, Wood et al., 2013), Myf5 (Biressi et al., 2013), and the related myogenic regulatory gene, Myf6 (Mrf4) (Sambasivan et al., 2013). Given the dominant ability of each MRF to stably drive non-muscle cells into the myogenic program, historical MRF expression could be sufficient to program satellite cells for injury-induced myogenesis. Here, we explicitly test this hypothesis and determine the relevance of injury-induced MRF expression in satellite cell function.
Results
Generation of Mice Carrying a New Conditional MyoD Knockout Allele
We developed a Cre-dependent MyoD conditional knockout allele (MyoDL2G) by flanking the entire MyoD coding sequence and 3′ UTR with loxP sites, thereby ensuring production of an unambiguously null allele after Cre recombination (Figure S1). MyoDL2G was carried over the original constitutively null MyoD allele (Rudnicki et al., 1992) (referred to here as MyoDneo) such that production of homozygous-null cells required only a single recombination event. We utilized the constitutively null Myf5 allele, Myf5loxP, which does not affect transcription of the linked Mrf4 gene (Kassar-Duchossoy et al., 2004). All experimental mice carried the Pax7CreERT2 allele (Murphy et al., 2011) to selectively delete MyoD from quiescent satellite cells of adult muscle (Figure S1). Mice of the MRF genotype, MyoDL2G/neo;Myf5loxP/loxP were recovered at the expected Mendelian frequency at weaning (n = 239).
Satellite cells homozygous-null for both MyoD and Myf5 (double knockout [dKO]) were produced by administering tamoxifen to mice with the MRF genotype, MyoDL2G/neo;Myf5loxP/loxP. Mice also were generated that carried one wild-type allele of either MyoD or Myf5 after tamoxifen treatment to test for MRF dosage effects. The last dose of tamoxifen was administered at least 3 days prior to muscle injury or tissue collection. Using five daily doses of 10 mg tamoxifen by oral gavage, recombination efficiency of the Cre-dependent GFP reporter allele R26NG (Yamamoto et al., 2009) was ∼92% ± 4% of VCAM-1+; α7-integrin+ satellite cells (n = 10). The vast majority of GFP+ satellite cells were also recombined at the MyoD locus, as quantified in cultures of satellite cells fluorescence-activated cell sorting (FACS) isolated from total hindlimb muscles (93%; not shown) or on single extensor digitorum longus (EDL) fibers (∼98%) (Figure S1). No appreciable differences in tamoxifen-dependent recombination efficiencies or outcomes were noted with mice carrying either one or two copies of Pax7CreERT2 or R26NG and the results are presented together.
Satellite Cells Carrying a Single Functional Allele of MyoD or Myf5 Can Support Skeletal Muscle Regeneration
Regeneration in tamoxifen-treated MyoDL2G/+;Myf5loxP/loxP mice (resulting in satellite cells with a single functional MyoD allele [MyoDLG/+;Myf5loxP/loxP]; MyoD-SA) was comparable with that of double heterozygous controls (dHet), as demonstrated by similar kinetics of regeneration of the tibialis anterior (TA) muscle with no consistent occurrence of histopathological features such as hypercellularity or fibrosis (Figures 1E, 1F, and 1I). Although a single allele of Myf5 (MyoDLG/neo;Myf5loxP/+; Myf5-SA) supported regeneration (Figures 1J and 1P), signs of impairment were evident, particularly at early stages post-injury. For example, induction of the early differentiation marker myogenin was delayed in Myf5-SA muscle (Figures 2A–2H). In addition, there were fewer, disorganized, nascent fibers, and a greater number of mononuclear cells at 11 days post-injury (dpi) (Figure 1G). Further, whereas a modest and variable number of adipogenic cells was observed in both MyoD-SA and dHet muscle, fatty infiltrates were a more consistent and conspicuous feature of Myf5-SA muscle (Figures S2A–S2C and S2E–S2H; data not shown). Finally, average muscle weight and fiber diameter were reduced in regenerated Myf5-SA muscle (Figures 1I, 1J, and S3).
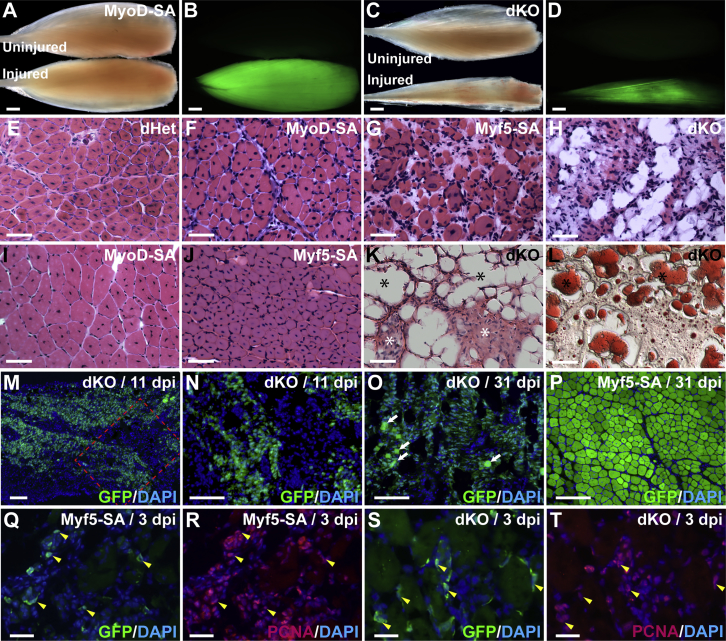
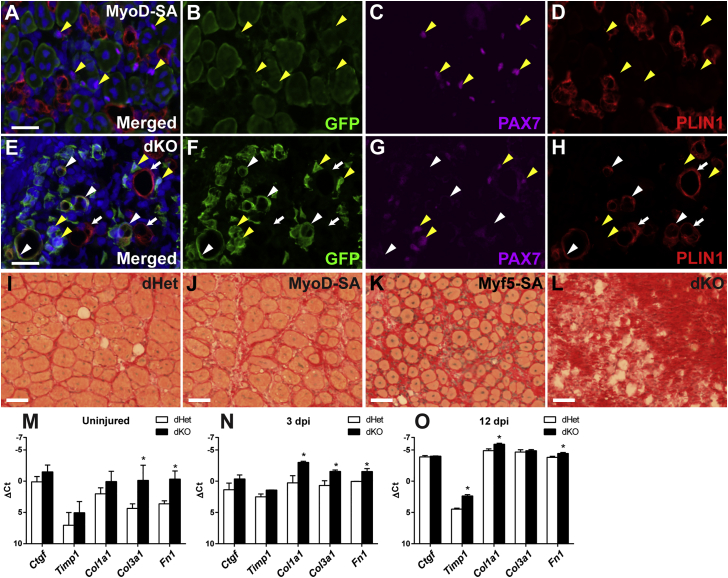
Figure 1.
Whole-Mount and Histological Analyses of Skeletal Muscle Regeneration Directed by Varying Gene Dosages of MyoD and Myf5 in Satellite Cells
(A–D) Whole-mount images of uninjured and injured (11 dpi) TA muscles from MyoD-SA (A and B) and dKO (C and D) mice. Mice carried the Cre-dependent GFP reporter, R26NG. Fluorescence images in (A) and (C) are shown in (B) and (D), respectively. GFP+ satellite cells in uninjured muscles are not visible at this magnification and exposure (B and D); top. The exposure times to capture the GFP images were 126 ms (B) and 892 ms (D). Scale bars represent 1 mm (A–D).
(E–L) Histological analyses at 11 dpi (E–H) and 31 dpi (I–L). Sections were stained with H&E (E–K) or oil red O (L). The same section is shown in (K and L). dKO muscle was filled with adipocytes (black asterisks) (K and L) and apparent fibrotic tissue (white asterisks) (K). Scale bars represent 50 μm.
(M–O) Lineage-labeled dKO cells populated the injured TA muscle. The boxed area in (M) is magnified in (N). Occasional, minute fibers are evident in (O) (arrows).
(P) Myf5-SA mice exhibited the typical architecture of regenerated muscle at 31 dpi.
(Q–T) PCNA immunofluorescence of Myf5-SA and dKO satellite cells at 3 dpi. Yellow arrowheads indicate representative PCNA+ GFP+ cells. Scale bars represent 100 μm (M–P) and 25 μm (Q–T).
See also Figures S2 and S3.
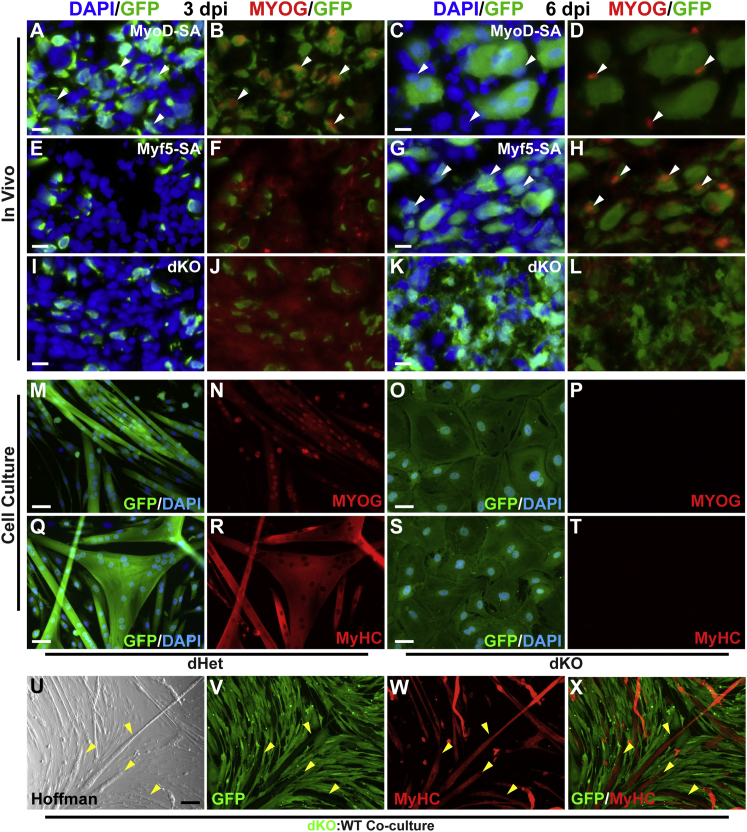
Figure 2.
dKO Cells Exhibit a Block in Muscle Differentiation
(A–L) Expression of myogenin (MYOG) in injured muscle is dependent on MyoD or Myf5. MYOG expression (arrowheads) was delayed or undetectable in Myf5-SA (F and H) and dKO (J and L) satellite cells, respectively.
(M–T) Cultured dKO satellite cells fail to fuse or express MYOG or MyHC after 21 days in DM.
(U–X) Co-culturing of equivalent numbers of wild-type and GFP+ dKO cells for 14 days in DM did not rescue fusion or differentiation capacity of dKO cells. Arrowheads identify representative fibers, which are GFP– and MyHC+. Scale bars represent 10 μm (A–L), 50 μm (M–T), or 100 μm (U–X).
MyoD or Myf5 Function in Satellite Cells Is Essential for Muscle Regeneration
Histological analyses through 72 dpi (n = 16) revealed an absolute requirement for MyoD or Myf5 in muscle regeneration. The dKO TA muscle was small and misshapen and did not recover in size (Figures 1A–1D) or weight (Figure S3). Myogenin protein was undetectable in dKO satellite cells at 3 and 6 dpi (Figures 2I–2L). At 11 dpi, the TA muscle was mostly comprised of lipid-filled adipocytes and fibrotic tissue (Figure 1H), histopathological features that are characteristic of late-stage muscle degenerative diseases such as Duchenne muscular dystrophy. That loss of MyoD and Myf5 resulted in regenerative failure rather than delayed regeneration was shown by analyses up to 10 weeks after injury (Figures 1K and 1L; data not shown). Although profound histopathology was common to all dKO mice, considerable mouse-to-mouse and regional variation was evident; a fibrotic response predominated in some mice or muscle regions, whereas massive adipocyte accumulations characterized others (Figures 1K, 1L, S2D, and S2I–S2L).
Lineage-Labeled dKO Satellite Cell Progeny Persist and Accumulate in Injured Muscle
Although dKO satellite cells cultured in growth medium (GM) exhibited reduced 5-ethynyl-2′-deoxyuridine (EdU) incorporation (Figure S4), dKO satellite cells in vivo were abundant at all post-injury times and mutant cells “filled” the damaged muscle, resulting in disorganized accumulations of GFP+ cells (Figures 1D and 1M–1O). The percentage of GFP+ dKO cells expressing proliferating cell nuclear antigen (PCNA) at 3 dpi (58.4%) was comparable with that of MyoD-SA (60.3%) and Myf5-SA (60.9%) satellite cells (Figures 1Q–1T), and apoptotic TUNEL+ dKO cells were rarely observed (data not shown). The percentage of PCNA+ dKO cells remained high at 6 dpi (46.7%), a post-injury time point when the number of wild-type satellite cell-derived myoblasts is already substantially reduced due to cell fusion. dKO cells were far more numerous than satellite cells of other genotypic classes at 31 dpi (Figure 1O; n = 4) and remained abundant through 72 dpi, the latest time tested. These data demonstrate that muscle regenerative failure cannot be explained by satellite cell loss.
Cultured dKO Satellite Cells Adopt a Fibroblast-like Morphology and Exhibit an Absolute Block in Myogenic Differentiation
We next cultured dKO satellite cells that were freshly isolated by FACS (Figure S5) to address whether they retained an intrinsic capacity for myogenic differentiation, which could be inhibited by cell non-autonomous effects of an abnormal muscle environment. Satellite cells were positively selected based on GFP fluorescence without the use of cell surface markers (Figure S5) in case marker expression was altered in MRF-deficient cells. Essentially 100% of GFP+ dHet and dKO satellite cells expressed PAX7 when assayed 24 hr after plating (data not shown), validating the specificity of the sorting method. Strikingly, dKO satellite cells cultured for up to 4 weeks in differentiation medium (DM) failed to fuse or express early (myogenin) or late (myosin heavy chain [MyHC]) muscle differentiation markers (Figures 2M–2T). The occasional muscle fibers observed in these cultures were invariably MYOD+, and, therefore, resulted from differentiation of “contaminating” cells that retained the unrecombined MyoD allele. Co-culturing of dKO and wild-type satellite cells did not rescue fusion capacity of the former (Figures 2U–2X). Surprisingly, despite the substantial myogenic activity of Myf5-SA satellite cells in vivo (Figures 1J and 1P), the fusion and differentiation capacity of cultured Myf5-SA cells was severely impaired (Figure S4A).
Cell morphology was dramatically altered in dKO satellite cells, which eventually adopted a greatly enlarged cell profile and a morphology often exhibited by senescing cells (Figures 2O, 2S, 3K, 3O, 3W, and 3A’). Whereas dKO cells displayed a typical satellite cell morphology for the first few days after plating in GM (Figures 3C and 3G), dKO cells adopted a fibroblast-like morphology and substantially increased in size between culture days 4 and 7 (Figure 3D).
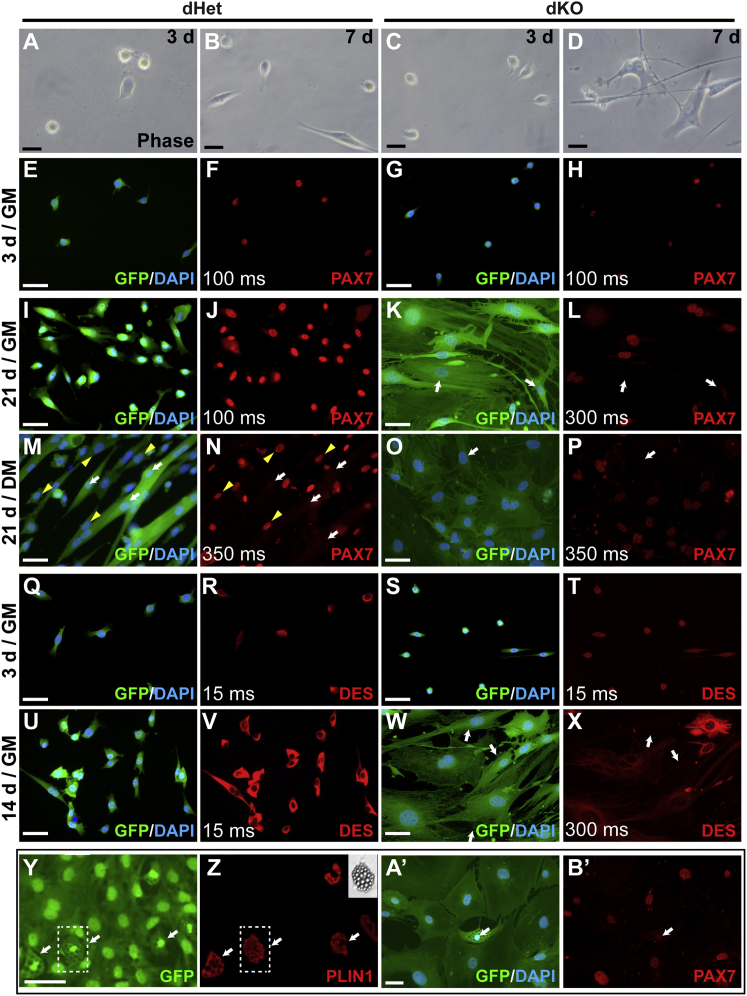
Figure 3.
Cultured dKO Cells Show Evidence of Altered Myogenic Programming
(A–X) Cultured dKO satellite cells adopt a fibroblast-like appearance but continue to express PAX7 and desmin (DES). (A–D) Phase images of dKO and dHet satellite cells after 3 or 7 days in culture. (E–L) Immunofluorescence analysis of PAX7 expression in dHet (E, F, I, and J) and dKO (G, H, K, and L) satellite cells after culturing for 3 (E–H) or 21 (I–L) days in GM. Occasional PAX7– cells were observed at day 21 (arrows) (K and L). (M–P) dKO cells maintained PAX7 expression after 21 days in DM. In dHet cells (M and N), PAX7 was downregulated in myotubes (arrows), but maintained in mononuclear cells (examples at arrowheads). Arrows in (O) and (P), example of an occasional PAX7– dKO cell. (Q–X) Comparison of DES expression in dHet (Q, R, U, and V) and dKO (S, T, W, and X) satellite cells 3 or 14 days in GM. dKO cells lacking detectable DES were common by day 14 (X), arrows.
(Y–B′) dKO satellite cells show a propensity for adipogenic differentiation. dKO cells were cultured in GM for 7 days and switched to DM for 13 (Y and Z) or 21 (A’ and B’) days. Approximately 10% of cells accumulated lipid droplets and were PLIN1+; examples at arrows in (Y and Z). Inset in (Z), phase contrast image of boxed lipid-filled cell in (Y) and (Z). PAX7 is downregulated in adipogenic cells; arrows in (A’) and (B’). Exposure times (ms) are shown in some panels to allow comparisons of apparent expression levels between genotypes. Scale bars represent 50 μm.
dKO Satellite Cells Retain Aspects of Myogenic Programming but Exhibit a Propensity for Adipogenic and Fibrogenic Differentiation
The large majority of dKO satellite cells isolated from uninjured muscle and cultured in GM for up to 3 weeks maintained PAX7 expression (Figures 3G, 3H, 3K, and 3L). The percentage of PAX7+ cells declined subsequently, although precise quantification was difficult due to the presence of unrecombined cells. The PAX7 immunofluorescence signal was consistently reduced in “fibroblastic” dKO cells compared with dHet control cells (Figures 3J and 3L). Interestingly, unlike wild-type satellite cells, which downregulate PAX7 during muscle differentiation (Olguin and Olwin, 2004, Zammit et al., 2004), dKO satellite cells cultured in DM retained PAX7 expression in virtually all fibroblast-like cells for at least 4 weeks (Figures 3O and 3P). In this regard, dKO cells responded to differentiation conditions like mononuclear “reserve cells,” which retain PAX7 expression and downregulate MYOD expression under differentiation conditions (Yoshida et al., 1998, Zammit et al., 2004).
We next addressed whether dKO cells that accumulate in injured muscle express markers of quiescent satellite cells or their daughters. The calcitonin receptor, which is expressed by quiescent satellite cells and their proliferative progeny (Fukada et al., 2007, Gnocchi et al., 2009), was not detected in dKO satellite cell progeny at 31 dpi (data not shown). In contrast, dKO satellite cells expressed PAX7 through 31 dpi (latest time tested), albeit at apparently reduced levels compared with control cells (Figures 4A–4F). Pax7 expression was confirmed by qRT-PCR of freshly isolated dKO cells from uninjured and injured skeletal muscle (Figures 4G–4I). Importantly, transcripts from the knocked out Myf5 allele (see Kassar-Duchossoy et al., 2004) were expressed at control levels and persisted at least through 12 dpi (Figures 4G–4I), indicating that dKO cells retain important aspects of myogenic programming. In addition, the myoblast marker desmin was expressed in both cultured and freshly isolated dKO cells, although with time in culture apparent desmin levels were greatly reduced and highly variable relative to control cells (Figures 3Q–3X).
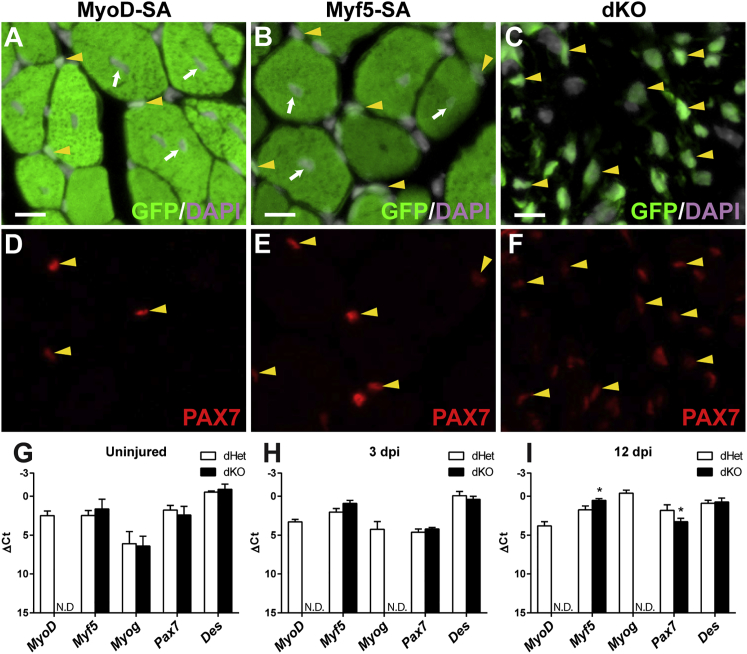
Figure 4.
dKO Satellite Cells Express Markers of Myogenic Commitment after Muscle Injury
(A–F) PAX7 immunofluorescence of transverse cryosections of the TA muscle 31 dpi. PAX7+ cells (examples at yellow arrowheads) carrying a wild-type allele of MyoD (A and D) or Myf5 (B and E) were associated with regenerated fibers, identified by their central nucleation (examples at arrows). PAX7+ dKO cells were numerous at 31 dpi (C and F). Apparent PAX7 levels were reduced in dKO satellite cells. Exposure times were identical between genotypes. Scale bars represent 10 μm.
(G–I) qRT-PCR analysis of myogenic gene expression of FACS-isolated satellite cells. ΔCt values were calculated using the average Ct values of the internal controls, Gapdh and EiF1a, which were highly consistent between genotypes within a given stage. Average Ct values were as follows: uninjured, 23.75; 3 dpi, 20.92; 12 dpi, 24.54. Each data point represents the average value of three biological samples and three technical replicates for each sample. Error bars represent standard deviations. p < 0.05 was considered significant (asterisks). Ct value >30 were considered N.D., not detected.
Approximately 10% of cultured dKO satellite cells adopted an adipogenic fate in standard muscle DM (Figures 3Y and 3Z), as assessed by morphology and perilipin 1 (PLIN1) staining, and these cells downregulated the expression of PAX7 (Figures 3A’ and 3B’). Adipogenic differentiation was not observed in MyoD-SA satellite cells, whereas Myf5-SA cells exhibited an intermediate level of adipogenic differentiation (data not shown). To further address the developmental state of cultured dKO cells, we quantified expression of a panel of 96 genes, which included markers of stem cells and several mesodermal lineages (Tables 1 and S1). In line with the results above, most muscle differentiation genes were significantly downregulated or undetected in dKO cells. Notably, expression of Mrf4 and Myf5 was comparable in cultured dKO and dHet cells.
Table 1.
qRT-PCR Analysis of Genes that Were Up- or Downregulated in Cultured dKO Satellite Cells
| Growth Medium |
Differentiation Medium |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Upregulated Genesa |
Downregulated Genesb |
Upregulated Genes |
Downregulated Genes |
||||||||
| Genec | Fold Changed (dKO/dHet) | Celle Type | Gene | Fold Change (dHet/dKO) | Cell Type | Gene | Fold Change (dKO/dHet) | Cell Type | Gene | Fold Change (dHet/dKO) | Cell Type |
| Anpep (Cd13) | 5.0 | many | Cdh15 (M-Cad) | NA | SC SkM | Bmp4 | NA | many | Cd44 | 7.1 | many |
| Bmp4 | NA | many | Cxcr4 | NA | S/P | Cebpa | 11.0 | A | Cdh15 (M-Cad) | NA | SC SkM |
| Cd34 | NA | S/P SC | Mef2c | NA | SkM | Cebpb | NA | A | Cxcr4 | NA | S/P |
| Cebpa | 4.3 | A | Met (c-Met) | 2.1 | SC | Col1a1 | 46.0 | F/M | Id1 | 6.5 | many |
| Cebpb | NA | A | Mstn | NA | SkM | Fabp4 (Ap2) | NA | A | Id2 | NA | many |
| Col1a1 | 18.3 | F/M | Myh4 | NA | SkM | Il4ra | 2.5 | many | Itga7 | 10.8 | SC |
| Fabp4 (Ap2) | NA | A | Myh7 | NA | SkM | Lpl | 6.5 | A | Lbr | 3.2 | many |
| Grem1 | 91.4 | many | Myod1 | NA | SkM | Ly6a (Sca1) | 43.5 | S/P | Mef2c | NA | SkM |
| Il4ra | 16.5 | many | Myog | NA | SkM | Pdgfra | NA | F/M | Met (c-Met) | NA | SC |
| Klf4 | 5.2 | many | Peg3 (Pw1) | 16.8 | S/P | Pdgfrb | NA | F/M | Mstn | NA | SkM |
| Ly6a (Sca1) | 125.8 | S/P | Tagln (Sm22a) | 9.2 | SM | Myh7 | NA | SkM | |||
| Numb | 3.0 | many | Twist1 | NA | many | Myod1 | NA | SkM | |||
| Pdgfra | NA | F/M | Myog | 36.3 | SkM | ||||||
| Pdgfrb | 19.2 | F/M | Peg3 (Pw1) | 21.1 | S/P | ||||||
| Pparg | 5.1 | A | |||||||||
| Sdc4 | 3.9 | SC | |||||||||
| Tagln (Sm22a) | 3.7 | SM | |||||||||
| Thy1 | NA | S/P | |||||||||
| Twist1 | NA | many | |||||||||
Genes upregulated in dKO cells compared with dHet cells.
Genes downregulated in dKO cells compared with dHet cells.
Only genes showing significant changes in gene expression (p < 0.05), or whose expression was detected in only one genotype, are shown. All genes assayed, with corresponding ΔCt values, are listed in Table S1.
Gene expression was quantified by qRT-PCR using a custom TaqMan low-density array. Fold changes were calculated using Bmpr2 as the reference gene, as it yielded highly consistent Ct values between genotypes. NA, fold change could not be calculated for genes expressed in cells of only one genotype (genes with Ct values ≤30 were considered to be expressed).
Cell types are listed for genes whose expression is cell type specific or restricted, or which are expressed by cells of a specific functional class. Satellite cells are listed for genes/proteins that are commonly used for their identification or isolation, even if more broadly expressed. A, adipogenic; F/M, fibroblast or mesenchyme; SC, satellite cell; SkM, skeletal muscle; SM, smooth muscle; S/P, stem/progenitor cell; many, genes that are broadly expressed. See also Table S1.
A total of 21 genes were significantly upregulated in dKO cells under growth or differentiation conditions (Table 1). Although most assayed genes are not unique to a specific cell type or developmental process, we note that at least three classes of genes are represented in this group: stem/progenitor cell markers, fibroblast/mesenchyme markers, and adipogenic markers. Brown adipogenic markers were not detected (Table S1; Cidea, Elovl3, Ucp1, and Prdm16), indicating that dKO satellite cells adopted a white adipocyte fate.
Injured muscles contained variable amounts of adipogenic cells and fibrotic tissue, with the extent of fibrosis and fatty infiltration correlating with satellite cell genotype, following the allelic series: dKO >>> Myf5-SA > MyoD-SA ≈ dHet (Figures 5A–5L and S2). MyoD-SA (Figures 5A–5D) and Myf5-SA (data not shown) satellite cells did not detectably adopt a PLIN1+ adipogenic fate. In contrast, PLIN1+ adipocytes derived from lineage-labeled dKO cells were readily observed. At 6 dpi, for example, approximately 7% of GFP+ dKO cells in highly adipogenic regions expressed PLIN1 (Figures 5E–5H; n = 2; 434 GFP+ cells counted). The remaining GFP+ cells in adipogenic regions, and all GFP+ dKO cells in apparent fibrogenic regions, assumed a mesenchymal appearance. As in culture, GFP+ adipocytes did not retain the expression of PAX7 protein, whereas the great majority of GFP+ mesenchymal cells expressed PAX7 (Figures 4C, 4F, and 5E–5H).
Figure 5.
dKO Satellite Cells Can Adopt an Adipogenic or Fibroblast-like Fate Following Muscle Injury
(A–H) Immunofluorescence localization of PAX7 and PLIN1 on transverse cryosections of TA muscles collected 6 dpi. Some dKO satellite cells gave rise to GFP+ PLIN1+ adipocytes (E, F, and H); examples at white arrowheads, whereas MyoD-SA satellite cells did not adopt an adipogenic fate (A, B, and D). Representative PAX7+ cells are shown with yellow arrowheads. Arrows in (E, F, and H), represent examples of GFP–, PLIN1+ adipocytes.
(I–L) Picrosirius red staining of TA cross-sections at 11 dpi. Scale bars represent 25 μm (A and E) and 50 μm (I–L).
(M–O) qRT-PCR analysis of fibrogenic gene expression of freshly isolated satellite cells. Internal controls and methods used to calculate ΔCt values are described in the legend to Figure 4. Each data point represents the average value of three biological samples and three technical replicates for each sample. Error bars represent standard deviations. p < 0.05 was considered significant (asterisks). See also Figure S2.
By qRT-PCR, dKO cells freshly isolated from skeletal muscle at 3 and 12 dpi showed increased expression of several fibroblast markers (Figures 5N and 5O). Fibrogenic gene expression was dynamic, and statistically significant increases in mRNA abundance in the dKO population were modest (2- to 4-fold) and both stage and gene specific (Figures 5N and 5O). Unexpectedly, the most marked increase in expression of fibroblast markers in dKO cells was observed in satellite cells isolated from uninjured muscle (Figure 5M). We note that a substantial minority of fibroblast-like cells, and the majority of PLIN1+ adipocytes (706 of 883 PLIN1+ cells scored; n = 4) in injured muscle, were not derived from dKO satellite cells (Figures 5E–5H and S2I–S2L).
dKO Satellite Cells Are Stably Maintained in Uninjured Aging Muscle
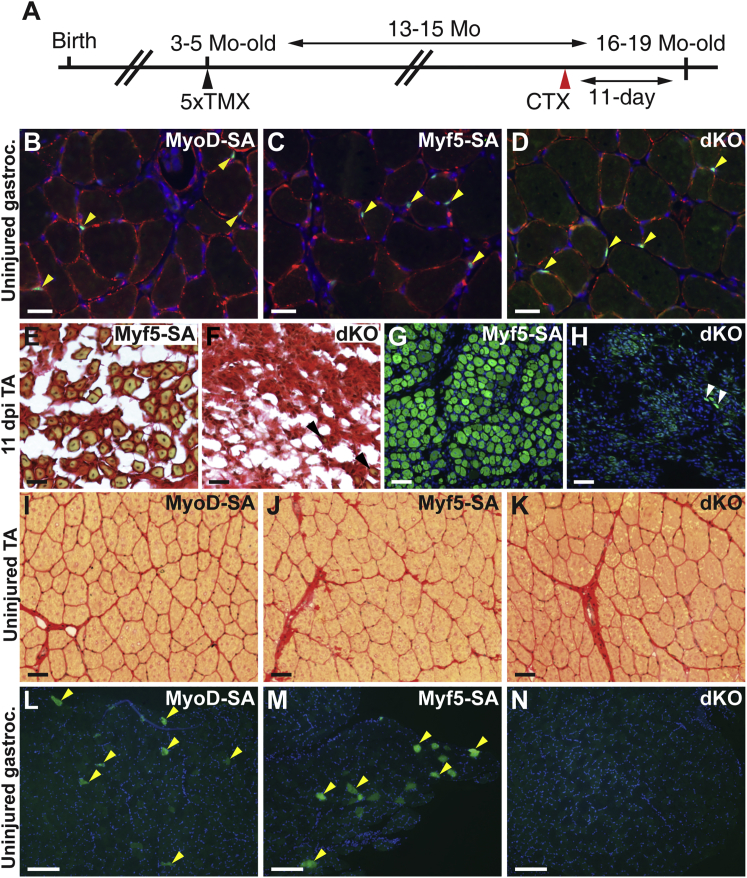
The upregulation of fibroblast genes in dKO satellite cells derived from uninjured muscle raised the question of whether mutant satellite cells adopt a fibroblast-like phenotype with aging, possibly resulting in increased fibrosis. To test this possibility, Pax7CreERT2/+;R26NG/+ mice of different MRF genotypes were administered tamoxifen at 3 to 5 months of age, and long-term maintenance of satellite cells and accumulation of fibrotic tissue was assessed at 16 to 19 months of age (Figure 6A). Satellite cell numbers were comparable in the gastrocnemius muscle of dKO, MyoD-SA, and Myf5-SA mice, as assessed by lineage labeling and PAX7 immunofluorescence (Figures 6B–6D and S6). That PAX7+ satellite cells in dKO muscle represented the maintenance of dKO cells and not the accretion of MyoD-unrecombined, functional, satellite cells was demonstrated by the failure of aged dKO muscle to regenerate after injury (Figures 6F and 6H). As evaluated by Picrosirius red staining, however, no increase in intramuscular fibrotic tissue was evident in uninjured dKO muscle (Figures 6I–6K).
Figure 6.
dKO Satellite Cells Are Maintained with Aging
(A) Schematic representation of tamoxifen/cardiotoxin administration schedule.
(B–D) Merged images of gastrocnemius cryosections stained for PAX7 (green; arrowheads), laminin (red), and DAPI (blue).
(E–H) Cross-sections of injured Myf5-SA (E and G) or dKO (F and H) TA muscles 11 dpi. Sections were stained with Picrosirius red to detect collagen accumulations (E and F) or represent merged GFP/DAPI images (G and H). The exposure time to capture the GFP signal was three times longer in (H) than in (G). Rare nascent fibers are labeled with arrowheads in (F) and (H).
(I–K) Cryosections of uninjured TA muscles stained with Picrosirius red.
(L–N) Low-magnification micrographs of uninjured gastrocnemius muscles imaged for GFP and DAPI. Examples of lineage-labeled GFP+ myofibers are shown (L and M, arrowheads).
Scale bars represent 25 µm (B–F), 50 μm (G–K), and 200 µm (L–N). See also Figure S6.
Recent data have shown that satellite cell fusion to uninjured muscle fibers is a general phenomenon characteristic of anatomically and physiologically diverse muscles (Pawlikowski et al., 2015). Consistent with the behavior of wild-type satellite cells, MyoD-SA and Myf5-SA satellite cells were capable of fusing with fibers of the TA and gastrocnemius muscles in the absence of injury, resulting in GFP+ lineage-labeled muscle fibers (Figures 6L and 6M). Importantly, however, GFP+ muscle fibers were rarely observed in muscles of dKO mice (Figure 6N), demonstrating that nuclear addition to uninjured muscle fibers by satellite cell fusion requires MyoD or Myf5.
Discussion
Using conditional mutagenesis, we demonstrated an absolute requirement for either MyoD or Myf5 in skeletal muscle regeneration. Whereas mutations in either MyoD or Myf5 cause comparatively modest regeneration and stem cell deficits (Asakura et al., 2007, Cornelison et al., 2000, Gayraud-Morel et al., 2007, Megeney et al., 1996, Sabourin et al., 1999, Schuierer et al., 2005, Ustanina et al., 2007, Yablonka-Reuveni et al., 1999), satellite cells homozygous-null for both MRFs exhibited profound stem cell dysfunction, resulting in the complete failure of injured skeletal muscle to regenerate. As the great majority of activated satellite cells express both MyoD and Myf5 (Cornelison and Wold, 1997, Zammit et al., 2002), genetic redundancy between these factors likely reflects functional compensatory activities within individual satellite cells rather than regulative behavior of distinct MyoD- or Myf5-dependent populations. The loss of regenerative capacity was not due to satellite cell loss, as lineage-marked dKO satellite cell daughters persisted in the damaged muscle. Notably, culture cells, removed from influences of the abnormal muscle environment, also exhibited a complete blockade in differentiation and a propensity for adipogenic differentiation, indicating severe, intrinsic stem cell dysfunction resulting from the loss of MyoD and Myf5. Further, since conditional MyoD deletion was restricted to satellite cells (i.e., cells that expressed Pax7 prior to injury), the failure of muscle to regenerate argues against a significant myogenic role of other stem/progenitor cell populations in muscle regeneration, consistent with results of satellite cell ablation studies (McCarthy et al., 2011, Murphy et al., 2011, Sambasivan et al., 2011).
Satellite cells have the capacity for brown adipogenic differentiation, and recent studies have identified MyoD and Myf5 as negative regulators of prdm16, a master regulator of the brown adipocyte fate (An et al., 2017, Pasut et al., 2016, Seale et al., 2008, Seale et al., 2007, Wang et al., 2017). In this context, the conversion of dKO satellite cells to an apparent white adipocyte fate was unexpected. In injured muscle, the minority of lineage-labeled dKO cells that adopted a PLIN1+ adipogenic fate were characterized by a single, large, fat droplet, a morphological feature that typifies white adipocytes. Gene expression analysis of cultured dKO satellite cells also indicated a white adipocyte fate, as definitive markers of brown adipogenesis, including the thermogenic gene, Ucp1, were not detected. Although population-level gene expression analyses cannot exclude the possibility that a small number of dKO cells exhibited a brown fat phenotype, the results suggest that conversion to white adipocytes is the preferred adipogenic fate under the conditions employed. Importantly, dKO cells in culture adopted an adipocyte fate “spontaneously,” without the use of adipogenesis-inducing medium. This is in contrast to previous studies, in which acquisition of the brown adipocyte fate was apparently dependent on the use of adipogenic inducers. Further, expression of Prdm16, which was not detected in cultured dKO cells and is both necessary and sufficient for acquisition and maintenance of the brown adipocyte fate, is induced in satellite cell-derived myoblasts cultured in pro-adipogenic media (An et al., 2017, Pasut et al., 2016, Seale et al., 2008, Wang et al., 2017). Collectively, these data suggest that the MRFs are negative regulators of both white and brown adipogenic fates, and that acquisition of the brown fat phenotype requires additional molecular inputs, including activation of Prdm16 expression.
Although either MyoD or Myf5 is sufficient to support regeneration, satellite cell defects are much more pronounced in MyoD mutant mice (Asakura et al., 2007, Cornelison et al., 2000, Gayraud-Morel et al., 2007, Megeney et al., 1996, Sabourin et al., 1999, Schuierer et al., 2005, Ustanina et al., 2007, Yablonka-Reuveni et al., 1999). The differential capacity of these MRFs to support regeneration was also shown here under conditions of reduced gene dosage in which satellite cells carried only a single functional allele of either MyoD or Myf5. Whereas a single allele of MyoD supported apparently normal regeneration, mice with a single Myf5 allele showed delayed regeneration, reduced fiber diameter at endpoint, and increased fatty infiltration. In addition, satellite cells with a single Myf5 allele exhibited greatly reduced fusion and differentiation capacity and a propensity for adipogenic differentiation. These differences are probably attributable to MRF-specific transcriptional activities; although MYOD and MYF5 bind the same genomic sites, show similar chromatin remodeling activities, and activate a similar set of target genes, only MYOD efficiently recruits Pol II and robustly activates transcription (Conerly et al., 2016). In this context, the degree to which satellite cells with a single Myf5 allele were able to support regeneration is perhaps surprising, and it will be important to define MYF5 transcriptional functions in satellite cells and further explore the regulatory relationships between members of the MRF family.
In contrast to the absolute requirement for either MyoD or Myf5 in muscle regeneration, Mrf4 alone can drive early myogenic determination and differentiation in the embryo (Kassar-Duchossoy et al., 2004). A priori, this difference could indicate that distinct regulatory mechanisms control embryonic and adult myogenesis, or might be attributable to the timing of Mrf4 expression relative to the myogenic program. In this latter regard, western blot analysis showed MRF4 protein expression in satellite cell cultures only after the onset of differentiation, arguing against a role in satellite cells or their proliferative daughter cells (Gayraud-Morel et al., 2007). In the present study, Mrf4 transcripts were readily detected in cultured dKO satellite cells grown in GM, which may indicate that MRF4 is not sufficient to activate functional levels of myogenin gene expression or execute the myogenic differentiation program in the absence of MYOD and MYF5. This interpretation should be viewed with caution, however, as we could not assess the presence or levels of MRF4 protein by immunofluorescence due to the unavailability of suitable anti-MRF4 antibodies (our unpublished data).
Given that MyoD (Kanisicak et al., 2009, Wood et al., 2013), Myf5 (Biressi et al., 2013), and Mrf4 (Sambasivan et al., 2013) are historically expressed in satellite cell precursors, we speculate that initial myogenic programming of satellite cells is established prenatally through epigenetic mechanisms (Conerly et al., 2016, Sartorelli and Juan, 2011), which may account for the expression of markers of myoblast identity in dKO satellite cells. Yet, the present data clearly show that myogenic gene expression in the embryo, and prior to injury in quiescent satellite cells (Gayraud-Morel et al., 2012, Kuang et al., 2007), is not sufficient for stable commitment to a functional muscle stem cell fate; ongoing regulatory inputs from either MYOD or MYF5 are required for successful regeneration. Without such inputs, stem cell myogenic programming shows greater lability and muscle differentiation competency is lost. It will be important to determine the relative contribution of transcriptional activating functions and chromatin remodeling activities to the maintenance of a functional muscle stem cell state.
Experimental Procedures
Generation of the MyoDL2G Conditional Knockout Mice
Animal procedures were reviewed and approved by the University of Connecticut’s Institutional Animal Care and Use Committee.
loxP sites were inserted immediately upstream of the MyoD first ATG and downstream of the third exon. The 3′ loxP site was followed by an EGFP-bGH polyadenylation sequence and an FRT-flanked PGKNeo cassette. Standard subcloning and recombineering (Liu et al., 2003) methods were used for the targeting vector construction (Figure S1). Embryonic stem cell (ESC) electroporation and production of chimeras by aggregation with CD1 embryo was performed by the University of Connecticut Gene Targeting and Transgenic Facility (GTTF) using 129S6/C57BL6 hybrid ESCs (D1: established by GTTF). Nested PCR was used to screen for homologous recombination in ESCs and transmission of the targeted allele (Supplemental Experimental Procedures). One established targeted line was crossed to Rosa26FLP/FLP mice (Farley et al., 2000) (Jackson Laboratories) to remove the PGKNeo cassette, resulting in the MyoDL2G allele.
Mouse Breeding and Genotyping
Experimental mice were maintained on a mixed background predominantly comprised of FVB, CD1, and C57BL/6 strains. To generate experimental mice, Myf5loxP (Kassar-Duchossoy et al., 2004), MyoDm1 (Rudnicki et al., 1992), Pax7CreERT2 (Murphy et al., 2011), and the Cre-dependent reporters, R26NG (Yamamoto et al., 2009) or R26mTmG (Muzumdar et al., 2007) were introduced into MyoDL2G mice by sequential breeding. PCR genotyping of tail DNA was performed by standard methods (Supplemental Experimental Procedures).
Tamoxifen and Cardiotoxin Treatment
Tamoxifen (Toronto Research Chemical, T006000) was dissolved in corn oil at a concentration of 50 mg/mL. Unless otherwise noted, all experimental mice were between 8 and 13 weeks of age and were administered tamoxifen at a dose of 10 mg via oral gavage for 4 or 5 consecutive days. Muscle injury was induced at least 3 days after the last tamoxifen dose by injecting 100 μL of 10 μM Naja mossambica mossambica cardiotoxin (Sigma-Aldrich, C9759) in PBS into the left TA or gastrocnemius muscle.
Tissue Collection and Histology
To prevent shrinkage, TA, EDL, and gastrocnemius muscles were dissected together with the connected tendons and bones, fixed in 4% paraformaldehyde (PFA)/PBS at 4°C, and washed in PBS several times. TA muscles were weighed after isolation from the connected tendon and bone. Muscles were incubated in 30% sucrose/PBS and embedded in O.C.T. compound (Tissue-Tek). For PAX7 immunodetection in uninjured aging muscles and calcitonin receptor immunohistochemistry, freshly dissected muscles isolated from the connected tendon and bone were directly embedded in O.C.T. compound. Cryosections were collected on Superfrost Plus slides or on pre-coated adhesive slides using the CryoJane tape transfer system (Leica). To detect lipid-filled adipose tissue, cryosections were incubated in 0.3% oil red O (ORO)/0.1 μg/μL DAPI/60% isopropanol briefly and rinsed in water. ORO and DAPI images were captured without coverslipping to avoid smearing and loss of lipid droplets. H&E staining used routine methods. Picrosirius red staining was performed according to the manufacturer's protocol (American MasterTech).
Myofiber Cultures
Myofibers were isolated from the uninjured EDL muscle as described previously (Starkey et al., 2011). Isolated myofibers were cultured for 1 day in DMEM (Life Technologies, 11995065) supplemented with 10% horse serum (Life Technologies, 26050), 0.5% chicken embryo extract (Accurate Chemical, CE-650-TL) and 1× penicillin-streptomycin (Pen/Strep; Caisson Laboratories, PSL01). Myofibers were fixed in 4% PFA/PBS for 5 min, washed in PBS/0.5% Triton X-100 (PBT) and processed for immunofluorescence localization of PAX7 and MYOD.
Satellite Cell Cultures and Quantification of Proliferation and Differentiation
Cells were isolated by FACS (Supplemental Experimental Procedures) and used at passage number 4 or earlier. Freshly isolated cells were plated in GM (20% FBS [HyClone, SH30071.03], 2.5 ng/mL basic-FGF [Life Technologies, PHG0264], and 1× Pen/Strep in DMEM) on 35-mm tissue culture plates coated with 1 mg/mL Matrigel (Corning, 356231). To minimize differentiation in GM, cells were plated at a low density, fed every 1 to 2 days, and were passed at a confluency of <50%.
To assess differentiation capacity, cells were maintained in GM until reaching approximately 70% confluency (5–6 days) and then passed into Matrigel-coated 8-well chamber slides (Ibidi, 80826) in GM until reaching >70% confluency (2–3 days). Cells were then switched to DM (10% horse serum and 1× Pen/Strep in DMEM) and fed every 3 days. Cultured cells were fixed for 5 min in 4% PFA/PBS after 1, 3, 7, 14, 21, or 28 days in GM, or after 3, 7, 14, 21, and 28 days in DM, rinsed in PBS and PBS-T (0.1% Triton X-100 in PBS), and processed for immunofluorescence.
For proliferation assays, cells were grown in GM and passed after 5 days at a density of ∼2.0 × 103 cells/cm2. Cells were then grown an additional 2 days in GM, and EdU incorporation quantified after a 4-hr exposure to 10 μM EdU using the Click-iT Plus EdU Alexa Fluor 647 Imaging kit (Thermo Fisher, C10640). All samples were done in biological triplicate.
For RNA analyses, GM samples were passed 1–2 times after initial plating and kept in GM for a total of 7 days prior to RNA isolation. DM samples were passed once in GM and grown until ∼70% confluency, after which DM was added and cells cultured for 7 days, for a total time in culture of 12–13 days.
Cells used to calculate fusion and differentiation indices were enriched via magnetic-activated cell sorting with Miltenyi's manual cell separator (Supplemental Experimental Procedures). Cells were directly plated into Matrigel-coated 8-well chamber slides and grown for 2–3 days until reaching >70% confluency. They were then switched to DM for 3 days and fixed and stained as described above. All samples were done in biological triplicate. Fusion index = (no. of nuclei in multinucleated MyHC+ cells/total no. of nuclei in GFP+ cells). Differentiation index = (no. of nuclei in MyHC+ cells/total no. of nuclei in GFP+ cells).
Protein Localization by Immunofluorescence
Immunofluorescence methods are detailed in the Supplemental Experimental Procedures. The following primary antibodies were used: chicken anti-GFP (Abcam, 13970); rabbit anti-PLIN1 (Sigma-Aldrich, P1817); mouse anti-chicken PAX7 (Developmental Studies Hybridoma Bank [DSHB]); mouse anti-rat myogenin (mAb F5D; DSHB); rabbit anti-PCNA (Santa Cruz, FL-261); mouse anti-human MYOD (mAb 5.8A, BD Biosciences, 554130); rabbit anti-desmin (Sigma, P1873); rabbit anti-rat calcitonin receptor (Bio-Rad, AHP635); and mouse anti-chicken MyHC (mAb MF20; DSHB).
Photography and Images
Fluorescence images of sections were captured on a Nikon Eclipse E600 (Spot RT3 digital camera) microscope, whole-mount fluorescence images were captured on a Leica MZFLIII stereomicroscope (Spot RT3 digital camera), and images of cultured cells were captured on either a Nikon TMS (phase: Nikon Coolpix 950 digital camera) or a Nikon Eclipse TE2000-U inverted microscope (fluorescence: QImaging Retiga EXi digital camera). Minor adjustments in brightness, midtone, color, and contrast, which were applied to the entire image, were made in Photoshop.
Measurement of Fiber Cross-Sectional Area
H&E-stained muscle cross-sections were digitally captured on the Nikon Eclipse E600 with the Spot RT3 digital camera. Choosing multiple areas enriched with muscle fibers with centrally localized myonuclei, the circumference of each fiber was outlined using Image Studio Lite (LI-COR) to generate fiber cross-sectional area (CSA). More than 300 fibers were analyzed for each mouse. Microsoft Excel was used to process the CSA data and to generate graphs. Adobe Illustrator was used to format the graphs.
Tissue Dissociation for Satellite Cell Isolation
Satellite cells were isolated as described previously (Pasut et al., 2012), with some modifications. Total mouse hindlimb muscles were minced for 7 min and digested with 0.1% (w/v) collagenase type II (Gibco, 17101-015) and 2.5 units/mL Dispase II (Gibco, 17105-041) in DMEM for 1 hr with gentle trituration every 15 min. The digestion was terminated by the addition of 20% FBS in DMEM and processed for FACS analysis.
RNA Isolation and qRT-PCR
RNA isolation and qRT-PCR are described in Supplemental Experimental Procedures.
Author Contributions
D.J.G., M.Y., and N.P.L. designed the study. M.Y., N.P.L., S.Y., A.A.B., and A.L. performed the experiments. M.Y. and N.P.L. compiled the data. D.J.G., M.Y., and N.P.L. analyzed and interpreted the data. G.K. and S.T. characterized and provided mouse lines. D.J.G. wrote the manuscript with input from all authors.
Acknowledgments
We thank members of the Goldhamer lab for helpful comments throughout the course of this work, Patrick Monico for technical assistance, and Youfen Sun for assistance with mouse maintenance and genotyping. We also thank FACS facility scientists Dr. Carol Norris and Robert Smith for expert technical assistance. This work was supported by grants from the Muscular Dystrophy Association (no. 186605) and the Connecticut Regenerative Medicine Research Fund (12-SCB-UCON-01) to D.J.G.
Published: February 22, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, and one table and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.01.027.
Supplemental Information
References
- An Y., Wang G., Diao Y., Long Y., Fu X., Weng M., Zhou L., Sun K., Cheung T.H., Ip N.Y. A molecular switch regulating cell fate choice between muscle progenitor cells and Brown adipocytes. Dev. Cell. 2017;41:382–391.e5. doi: 10.1016/j.devcel.2017.04.012. [DOI] [PubMed] [Google Scholar]
- Asakura A., Hirai H., Kablar B., Morita S., Ishibashi J., Piras B.A., Christ A.J., Verma M., Vineretsky K.A., Rudnicki M.A. Increased survival of muscle stem cells lacking the MyoD gene after transplantation into regenerating skeletal muscle. Proc. Natl. Acad. Sci. USA. 2007;104:16552–16557. doi: 10.1073/pnas.0708145104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biressi S., Bjornson C.R.R., Carlig P.M.M., Nishijo K., Keller C., Rando T.A. Myf5 expression during fetal myogenesis defines the developmental progenitors of adult satellite cells. Dev. Biol. 2013;379:195–207. doi: 10.1016/j.ydbio.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conerly M.L., Yao Z., Zhong J.W., Groudine M., Tapscott S.J. Distinct activities of Myf5 and MyoD indicate separate roles in skeletal muscle lineage specification and differentiation. Dev. Cell. 2016;36:375–385. doi: 10.1016/j.devcel.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelison D.D., Olwin B.B., Rudnicki M.A., Wold B.J. MyoD(–/–) satellite cells in single-fiber culture are differentiation defective and MRF4 deficient. Dev. Biol. 2000;224:122–137. doi: 10.1006/dbio.2000.9682. [DOI] [PubMed] [Google Scholar]
- Cornelison D.D., Wold B.J. Single-cell analysis of regulatory gene expression in quiescent and activated mouse skeletal muscle satellite cells. Dev. Biol. 1997;191:270–283. doi: 10.1006/dbio.1997.8721. [DOI] [PubMed] [Google Scholar]
- Farley F.W., Soriano P., Steffen L.S., Dymecki S.M. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. [PubMed] [Google Scholar]
- Fukada S.-I., Uezumi A., Ikemoto M., Masuda S., Segawa M., Tanimura N., Yamamoto H., Miyagoe-Suzuki Y., Takeda S. Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells. 2007;25:2448–2459. doi: 10.1634/stemcells.2007-0019. [DOI] [PubMed] [Google Scholar]
- Gayraud-Morel B., Chrétien F., Flamant P., Gomès D., Zammit P.S., Tajbakhsh S. A role for the myogenic determination gene Myf5 in adult regenerative myogenesis. Dev. Biol. 2007;312:13–28. doi: 10.1016/j.ydbio.2007.08.059. [DOI] [PubMed] [Google Scholar]
- Gayraud-Morel B., Chrétien F., Jory A., Sambasivan R., Negroni E., Flamant P., Soubigou G., Coppée J.-Y., Di Santo J., Cumano A. Myf5 haploinsufficiency reveals distinct cell fate potentials for adult skeletal muscle stem cells. J. Cell Sci. 2012;125:1738–1749. doi: 10.1242/jcs.097006. [DOI] [PubMed] [Google Scholar]
- Gnocchi V.F., White R.B., Ono Y., Ellis J.A., Zammit P.S. Further characterisation of the molecular signature of quiescent and activated mouse muscle satellite cells. PLoS One. 2009;4:e5205. doi: 10.1371/journal.pone.0005205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanisicak O., Mendez J.J., Yamamoto S., Yamamoto M., Goldhamer D.J. Progenitors of skeletal muscle satellite cells express the muscle determination gene, MyoD. Dev. Biol. 2009;332:131–141. doi: 10.1016/j.ydbio.2009.05.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassar-Duchossoy L., Gayraud-Morel B., Gomès D., Rocancourt D., Buckingham M., Shinin V., Tajbakhsh S. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature. 2004;431:466–471. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- Kuang S., Kuroda K., Le Grand F., Rudnicki M.A. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Jenkins N.A., Copeland N.G. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy J.J., Mula J., Miyazaki M., Erfani R., Garrison K., Farooqui A.B., Srikuea R., Lawson B.A., Grimes B., Keller C. Effective fiber hypertrophy in satellite cell-depleted skeletal muscle. Development. 2011;138:3657–3666. doi: 10.1242/dev.068858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megeney L.A., Kablar B., Garrett K., Anderson J.E., Rudnicki M.A. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 1996;10:1173–1183. doi: 10.1101/gad.10.10.1173. [DOI] [PubMed] [Google Scholar]
- Murphy M.M., Lawson J.A., Mathew S.J., Hutcheson D.A., Kardon G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development. 2011;138:3625–3637. doi: 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar M.D., Tasic B., Miyamichi K., Li L., Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Olguin H.C., Olwin B.B. Pax-7 up-regulation inhibits myogenesis and cell cycle progression in satellite cells: a potential mechanism for self-renewal. Dev. Biol. 2004;275:375–388. doi: 10.1016/j.ydbio.2004.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasut A., Oleynik P., Rudnicki M.A. Isolation of muscle stem cells by fluorescence activated cell sorting cytometry. Methods Mol. Biol. 2012;798:53–64. doi: 10.1007/978-1-61779-343-1_3. [DOI] [PubMed] [Google Scholar]
- Pasut A., Chang N.C., Rodriguez U.G., Faulkes S., Yin H., Lacaria M., Ming H., Rudnicki M.A. Notch signaling rescues loss of satellite cells lacking Pax7 and promotes Brown adipogenic differentiation. Cell Rep. 2016;16:333–343. doi: 10.1016/j.celrep.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlikowski B., Pulliam C., Betta N.D., Kardon G., Olwin B.B. Pervasive satellite cell contribution to uninjured adult muscle fibers. Skelet. Muscle. 2015;5:42. doi: 10.1186/s13395-015-0067-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F., Rocancourt D., Mansouri A., Buckingham M. Divergent functions of murine Pax3 and Pax7 in limb muscle development. Genes Dev. 2004;18:1088–1105. doi: 10.1101/gad.301004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnicki M.A., Braun T., Hinuma S., Jaenisch R. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- Rudnicki M.A., Schnegelsberg P.N., Stead R.H., Braun T., Arnold H.H., Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- Sabourin L.A., Girgis-Gabardo A., Seale P., Asakura A., Rudnicki M.A. Reduced differentiation potential of primary MyoD–/– myogenic cells derived from adult skeletal muscle. J. Cell Biol. 1999;144:631–643. doi: 10.1083/jcb.144.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambasivan R., Comai G., Le Roux I., Gomès D., Konge J., Dumas G., Cimper C., Tajbakhsh S. Embryonic founders of adult muscle stem cells are primed by the determination gene Mrf4. Dev. Biol. 2013;381:241–255. doi: 10.1016/j.ydbio.2013.04.018. [DOI] [PubMed] [Google Scholar]
- Sambasivan R., Yao R., Kissenpfennig A., Van Wittenberghe L., Paldi A., Gayraud-Morel B., Guenou H., Malissen B., Tajbakhsh S., Galy A. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development. 2011;138:3647–3656. doi: 10.1242/dev.067587. [DOI] [PubMed] [Google Scholar]
- Sartorelli V., Juan A.H. Sculpting chromatin beyond the double helix: epigenetic control of skeletal myogenesis. Curr. Top. Dev. Biol. 2011;96:57–83. doi: 10.1016/B978-0-12-385940-2.00003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuierer M.M., Mann C.J., Bildsoe H., Huxley C., Hughes S.M. Analyses of the differentiation potential of satellite cells from myoD–/–, mdx, and PMP22 C22 mice. BMC Musculoskelet. Disord. 2005;6:15. doi: 10.1186/1471-2474-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P., Bjork B., Yang W., Kajimura S., Chin S., Kuang S., Scimè A., Devarakonda S., Conroe H.M., Erdjument-Bromage H. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P., Kajimura S., Yang W., Chin S., Rohas L.M., Uldry M., Tavernier G., Langin D., Spiegelman B.M. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkey J.D., Yamamoto M., Yamamoto S., Goldhamer D.J. Skeletal muscle satellite cells are committed to myogenesis and do not spontaneously adopt nonmyogenic fates. J. Histochem. Cytochem. 2011;59:33–46. doi: 10.1369/jhc.2010.956995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustanina S., Carvajal J., Rigby P., Braun T. The myogenic factor Myf5 supports efficient skeletal muscle regeneration by enabling transient myoblast amplification. Stem Cells. 2007;25:2006–2016. doi: 10.1634/stemcells.2006-0736. [DOI] [PubMed] [Google Scholar]
- Wang C., Liu W., Nie Y., Qaher M., Horton H.E., Yue F., Asakura A., Kuang S. Loss of MyoD promotes fate transdifferentiation of myoblasts into Brown adipocytes. EBioMedicine. 2017;16:212–223. doi: 10.1016/j.ebiom.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W.M., Etemad S., Yamamoto M., Goldhamer D.J. MyoD-expressing progenitors are essential for skeletal myogenesis and satellite cell development. Dev. Biol. 2013;384:114–127. doi: 10.1016/j.ydbio.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yablonka-Reuveni Z., Rudnicki M.A., Rivera A.J., Primig M., Anderson J.E., Natanson P. The transition from proliferation to differentiation is delayed in satellite cells from mice lacking MyoD. Dev. Biol. 1999;210:440–455. doi: 10.1006/dbio.1999.9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Shook N.A., Kanisicak O., Yamamoto S., Wosczyna M.N., Camp J.R., Goldhamer D.J. A multifunctional reporter mouse line for Cre- and FLP-dependent lineage analysis. Genesis. 2009;47:107–114. doi: 10.1002/dvg.20474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Price F., Rudnicki M.A. Satellite cells and the muscle stem cell niche. Physiol. Rev. 2013;93:23–67. doi: 10.1152/physrev.00043.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida N., Yoshida S., Koishi K., Masuda K., Nabeshima Y. Cell heterogeneity upon myogenic differentiation: down-regulation of MyoD and Myf-5 generates 'reserve cells'. J. Cell Sci. 1998;111(Pt 6):769–779. doi: 10.1242/jcs.111.6.769. [DOI] [PubMed] [Google Scholar]
- Zammit P.S., Heslop L., Hudon V., Rosenblatt J.D., Tajbakhsh S., Buckingham M.E., Beauchamp J.R., Partridge T.A. Kinetics of myoblast proliferation show that resident satellite cells are competent to fully regenerate skeletal muscle fibers. Exp. Cell Res. 2002;281:39–49. doi: 10.1006/excr.2002.5653. [DOI] [PubMed] [Google Scholar]
- Zammit P.S., Golding J.P., Nagata Y., Hudon V., Partridge T.A., Beauchamp J.R. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J. Cell Biol. 2004;166:347–357. doi: 10.1083/jcb.200312007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.