Abstract
Sensitization to inhaled allergens is dependent on activation of conventional dendritic cells (cDCs) and on the adaptor molecule, MyD88. However, many cell types in the lung express Myd88, and it is unclear how signaling in these different cell types reprograms cDCs and leads to allergic inflammation of the airway. By combining ATAC-seq with RNA profiling, we found that MyD88 signaling in cDCs maintained open chromatin at select loci even at steady state, allowing genes to be rapidly induced during allergic sensitization. A distinct set of genes related to metabolism was indirectly controlled in cDCs through MyD88 signaling in airway epithelial cells (ECs). In mouse models of asthma, Myd88 expression in ECs was critical for eosinophilic inflammation, whereas Myd88 expression in cDCs was required for Th17 cell differentiation and consequent airway neutrophilia. Thus, both cell-intrinsic and cell-extrinsic MyD88 signaling controls gene expression in cDCs and orchestrates immune responses to inhaled allergens.
Asthma is a chronic disease of the airways characterized by reversible airway obstruction, airway hyperresponsiveness (AHR) and inflammation. This disease is now regarded as a heterogeneous set of lung pathologies that vary with regard to the provoking stimulus, age of onset, type of inflammation, mucus production, lung function and response to treatment.1 Whereas many patients display classic, type 2 cytokine-associated eosinophilic inflammation and are generally responsive to steroid-responsive inhaled corticosteroids, approximately half of patients with asthma have non-eosinophilic forms of the disease, often with neutrophilic inflammation of the airway.2–4 These patients are typically resistant to inhaled corticosteroids and can develop uncontrolled, life-threatening disease.5 Several lines of evidence suggest that IL-17-producing T helper (Th)17 cells, which are steroid-resistant,6,7 might drive the latter form of asthma by promoting recruitment of neutrophils to the airway.7–13
The initiation of adaptive immunity, including Th2 and Th17 responses to inhaled allergens, is dependent on Cd11c-expressing, conventional dendritic cells (cDCs).14 Multiple receptors on the surface of cDCs allow them to directly sense a variety of allergens and their associated microbial products, and to respond by increasing cytokine production and display of cell surface co-stimulatory molecules. This direct sensing of pathogen associated microbial patterns (PAMPs) is undoubtedly important for their function, but some features of cDCs might also be indirectly controlled by signals provided by other cells. To test this, bone marrow chimeric mice are frequently generated to compare gene function in radio-resistant and -sensitive cells, and this approach was used to show Toll-like receptor 4 (TLR4) signaling in airway structural cells is required for allergic sensitization to lipopolysaccharide (LPS)-containing preparations of house dust mite (HDM) allergen.15 Lung epithelial cells (ECs) are often presumed to be the major radio-resistant cell type that modifies the function of lung cDCs, but lung macrophages are also largely resistant to irradiation, which confounds interpretation of this type of experiment.16 Furthermore, radiation induces profound transcriptional changes in ECs that have unknown effects on their function.17 Thus, a major gap exists in our understanding of how direct and indirect recognition of PAMPs contribute to the epigenetic landscape, transcriptional profiles, and activities of cDCs in vivo.
Myeloid differentiation primary response gene 88 (MyD88) is an adaptor molecule used by all known TLRs, except TLR3. MyD88 is required for allergic sensitization through the airway in several models of asthma, including when LPS or flagellin (FLA) is used as an adjuvant to promote allergic responses to ovalbumin (OVA).18,19 To better understand cDC activation during allergic sensitization without the confounding aspects of irradiation, we used a genetic approach to selectively delete Myd88 in either ECs or Cd11c-expressing cells, and sensitized the animals through the airway with OVA/FLA, which is strictly dependent on MyD88.19 Myd88 expression in each of these cell types contributed to chromatin accessibility and gene expression in cDCs. Phenotypically, Myd88 expression in ECs was required for robust eosinophilic inflammation, whereas expression of this gene in Cd11c-expressing cells was required for Th17-associated neutrophilic inflammation. These findings reveal that reprogramming of lung cDCs and consequent induction of allergic pulmonary inflammation results from both cell-intrinsic and cell-extrinsic MyD88-dependent signaling.
RESULTS
Generation of mice lacking Myd88 in Cd11c-expressing cells
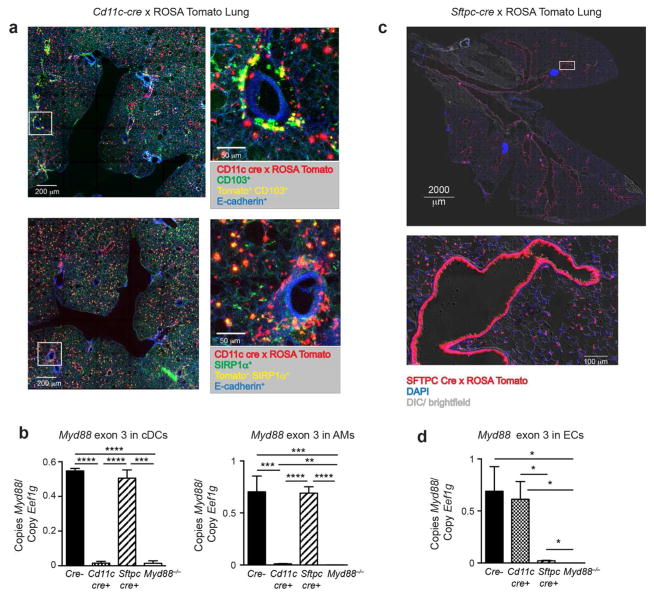
To understand how MyD88 signaling in cDCs affects their gene expression and thus allergic sensitization through the airway, we employed transgenic mice expressing Cre from the Cd11c promoter.20 We first confirmed cell specificity by crossing these animals to tdTomato reporter mice in which a LoxP-flanked transcriptional stop cassette (LoxP-Stop-LoxP) is positioned upstream of a tandem dimer (td)Tomato gene at the constitutively active Rosa26 locus. Precision cut lung slices (PCLS) from offspring of this cross confirmed the presence of tdTomato-expressing cells around the airways and within the lung parenchyma. Staining of the PCLS with antibodies against CD103 showed that one of the two major types of cDCs in the lung, CD103+ cDCs, are closely associated with the abluminal surface of the airway wall (Figure 1a, top panels). PCLS staining with antibodies to SIRP-1α, which identify CD11bhi cDCs21 and macrophages, showed that these cells reside throughout the lung parenchyma (Figure 1a, bottom panels). Flow cytometric analysis showed that >90% of CD103+ cDCs, CD11b+ cDCs, and alveolar macrophages (AMs) expressed tdTomato (Supplementary Figure 1a,b). Having confirmed the specificity of Cd11c-cre expression, we crossed Cd11c-cre mice to Myd88fx/fx mice, which bear a LoxP-flanked version of the third exon of Myd88.22 Using quantitative PCR, we confirmed that purified cDCs and AMs from Myd88fx/fxCD11c-cre mice (hereafter called DC-KO mice), had undergone deletion of the third exon, whereas it was retained in cDCs and AMs of Myd88fx/fx animals lacking Cre recombinase (hereafter called WT mice) (Figure 1b).
Figure 1. Cd11c drives Cre-mediated recombination in lung DCs and AMs while Sftpc drives Cre-mediated recombination in ECs.
(a) Fluorescent microscopic images of a PCLS prepared from progeny of Cd11c-cre x LoxP-Stop-LoxP-tdTomato cross. Top images show Tomato+ cells (red), CD103+ (green), Tomato+CD103+ DCs (yellow) and E-cadherin+ ECs (blue). Bottom images show Tomato+ cells (red), SIRP-1α+ (green), Tomato+SIRP-1α+ DCs (yellow) and E-cadherin+ ECs (blue). Low power images (left) include white squares marking an area also shown in higher power (right). (b) Real time PCR analysis of Myd88 exon 3 in cDCs and AMs sorted from Myd88fx/fx (Cre−), Myd88fx/fx Cd11c-cre (Cd11c-cre+), Myd88fx/fx Sftpc-cre (Sftpc-cre+), and Myd88−/− mice at 6h post-sensitization with OVA/FLA. Lung cDCs were gated on single cells that are CD11chi, nonautofluorescent, I-Ab+, Ly6C−, and CD88−. Data are representative of three to six separate cDC or AM sorts per genotype and are normalized to the Eef1g housekeeping gene. (c) Fluorescent microscopic images of a frozen lung section from progeny of Sftpc-cre x LoxP-Stop-LoxP-tdTomato cross. Shown are a low power image of the whole lung, including the bifurcation of the trachea (top), and a high power image of a representative small airway, showing fluorescent ECs in the airway and alveoli (bottom). tdTomato cells (red), DAPI (blue), and Differential Interference Contrast (DIC)/ brightfield (gray). (d) Real time PCR for the ‘floxed’ exon 3 region of the Myd88 gene in ECs sorted from Myd88fx/fx (Cre−), Myd88fx/fx Cd11c-cre (Cd11c-cre+), Myd88fx/fx Sftpc-cre (Sftpc-cre+), and Myd88−/− mice at 2h post-sensitization with OVA/FLA. Data is representative of three different EC sorts per genotype. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001
Generation of mice lacking Myd88 in ECs
To study how MyD88 signaling in ECs affects immune responses, including gene expression and function of lung cDCs, we used transgenic mice expressing Cre under control of the human surfactant protein C (Sftpc) promoter.23 Although Sftpc is not expressed in most mature ECs, it is expressed in their precursors, and genetic loci that undergo Cre-mediated recombination at that stage are reported to remain recombined in mature ECs.24 To confirm EC-specific, Cre-mediated recombination, we bred Sftpc-cre mice to the Cre-inducible tdTomato reporter strain. Tiled microscopic analysis of lung sections revealed fluorescent ECs throughout the entire respiratory tree of these animals, including the trachea, bronchi, bronchioles, and alveoli (Figure 1c). By comparing adjacent serial sections by fluorescent microscopy and hematoxylin and eosin staining, we confirmed that the tdTomato fluorescence was restricted to ECs and did not include endothelial cells lining the blood vessels (data not shown). Flow cytometric analysis confirmed that >90% of ECs expressed tdTomato (Supplementary Figure 1c,d), whereas less than 0.1% of Lin+ Epithelial cell adhesion molecule (EpCAM)− cells did. Having established that the Sftpc-cre transgene efficiently targets ECs, we bred Sftpc-cre mice to Myd88fx/fx mice. Sorted ECs from their Myd88fx/fx Sftpc-cre offspring (hereafter called EC-KO mice) had very little Myd88 mRNA compared with ECs from WT or DC-KO mice (Figure 1d).
Myd88 expression in Cd11c-expressing cells and ECs controls neutrophilia and eosinophilia, respectively, in mouse models of asthma
We next studied the role of cell-specific MyD88 in two different animal models of asthma. In the first model, we sensitized mice by allowing them to inhale OVA/FLA, then challenged the animals by exposing them to aerosolized OVA (Figure 2a). Following OVA challenge, DC-KO mice had markedly reduced neutrophilic inflammation compared to WT mice, whereas eosinophilic inflammation was similar in these two strains. Contrasting results were seen with EC-KO mice, which had markedly reduced eosinophilic inflammation compared to WT mice but only a modest reduction in neutrophils. Almost identical results were obtained in the second model of asthma in which an extract of house dust mites (HDM) was used to sensitize and challenge the animals (Figure 2b). Together, these results show that Myd88 expression in Cd11c-expressing cells is important for neutrophilic inflammation in these models of asthma, whereas Myd88 expression in ECs is critical for eosinophilic inflammation. While MyD88 is required for signaling responses to FLA, it is also required for responses to IL-33, which has been associated with some types of allergic responses.25 As expected, Myd88 null mice failed to become sensitized to OVA when IL-33 was used as an adjuvant, and they did not develop inflammation upon subsequent OVA challenge (Supplementary Figure 2a). By contrast, WT, DC-KO and EC-KO mice all displayed robust neutrophilic and eosinophilic inflammation. This suggests that in the OVA/FLA and HDM models of asthma, MyD88 is required in ECs and Cd11c-expressing cells for responsiveness to TLR ligands, not IL-33.
Figure 2. DC and EC expression of MyD88 control distinct facets of allergic pulmonary inflammation.
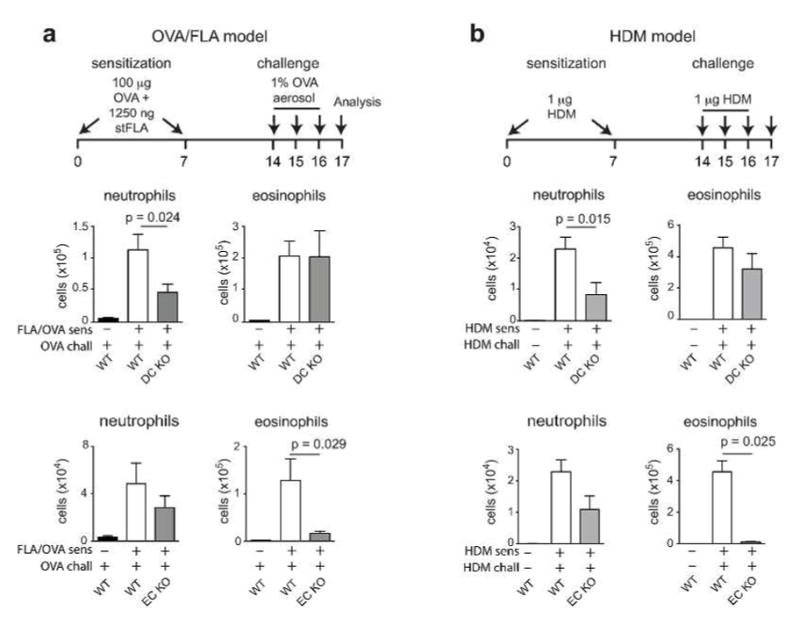
(a–b) Schematic of OVA/FLA (a) and HDM (b) mouse models of allergic asthma (top). Neutrophilic and eosinophilic inflammation of the airway in WT and DC-KO mice (middle) or WT and EC-KO mice (bottom) sensitized with OVA/FLA and challenged with OVA (a), or sensitized and challenged with HDM (b). Data shown are representative of two experiments, n = 9–12 mice per group (a,b).
Myd88 expression in ECs drives early immune responses during allergic sensitization
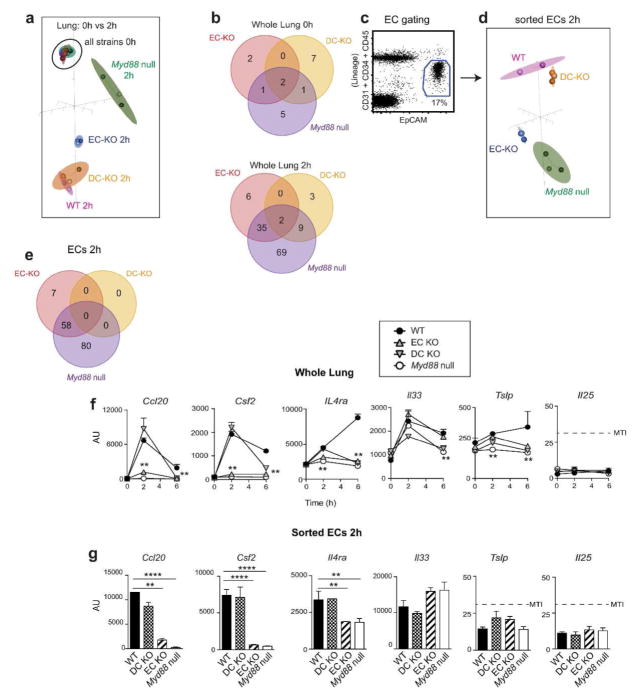
We next sought to obtain a molecular understanding of how Myd88 expression in Cd11c-expressing cells and ECs controls the expression of immune response genes during allergic sensitization through the airway. Following instillation of OVA/FLA, the expression of 547 known immune response genes was evaluated in whole lungs of the various recombinant mouse strains using the Nanostring Mouse Immunology Codeset (Supplementary Resource). Between group analyses (BGA) of the data revealed that at steady state, gene expression at the whole lung level was similar in WT, EC-KO, DC-KO and Myd88 null mice (Figure 3a), with only a small number of genes being affected by the absence of MyD88 (Figure 3b, top, and Supplementary Table 1). However, by 2h after allergic sensitization with OVA/FLA, genotype-specific differences in gene expression became apparent, with the greatest difference seen between WT and Myd88 null mice (Figure 3a and Supplementary Table 2, left). The expression pattern observed for EC-KO mice was intermediate between the patterns of WT mice and Myd88 null mice, whereas expression in DC-KO mice was similar to that of WT mice (Figure 3b, bottom, and Supplementary Table 3). ECs were also purified from the lung using flow cytometry-based sorting (Figure 3c) and analyzed. In agreement with data from the whole lung, ECs from EC-KO and Myd88 null mice displayed similar expression profiles, but were distinct from those of ECs from WT and DC-KO mice (Figure 3d,e and Supplementary Table 4). Ccl20, Csf2, and Il4ra were among the genes differentially expressed (above the minimum threshold intensity (MTI), ≥1.5 fold difference between groups, p <0.01; see Supplementary Methods) between EC-KO and WT mice at the whole lung level (Figure 3f) and in sorted ECs (Figure 3g). Although the epithelial-associated cytokines, Il25, Il33, and Tslp, have been linked to allergic pulmonary inflammation in some studies,25 none of these genes were differentially expressed among the various strains (Figure 3f,g). Loss of MyD88 in Cd11c-expressing cells did not affect gene expression in ECs. Together, these data suggest that shortly after inhalation of OVA/FLA, expression of select proinflammatory genes in ECs is increased in a manner dependent on MyD88 signaling in those cells.
Figure 3. Immune gene expression in whole lung and ECs at 2h post-sensitization.
(a) BGA plot of immune gene expression in whole lung at 0 and 2h after OVA/FLA-mediated sensitization of the indicated mouse strains. (b) Venn diagram showing the number of genes differentially expressed (≥1.5 fold; p≤0.01, MTI≥32 for at least one group) between lungs of WT mice (reference group) and the indicated mouse strains at 0 (top) and 2h (bottom) post-sensitization. (c) Gating strategy for purification of ECs. (d) BGA plot of immune gene expression in sorted ECs of the indicated mouse strains at 2h post-sensitization. (e) Venn diagrams showing the number of genes differentially expressed between sorted ECs of WT mice and the indicated strains (≥1.5 fold; p≤0.01, MTI≥32 for at least one group). (f–g) Expression of genes in whole lung (f) and ECs at 2h post-sensitization (g). n = 3 mice per group. MTI = minimum threshold intensity for gene detection, **p < 0.01, ****p < 0.0001.
Myd88 expression in both DCs and ECs controls gene transcription in lung DCs at 6h post-sensitization
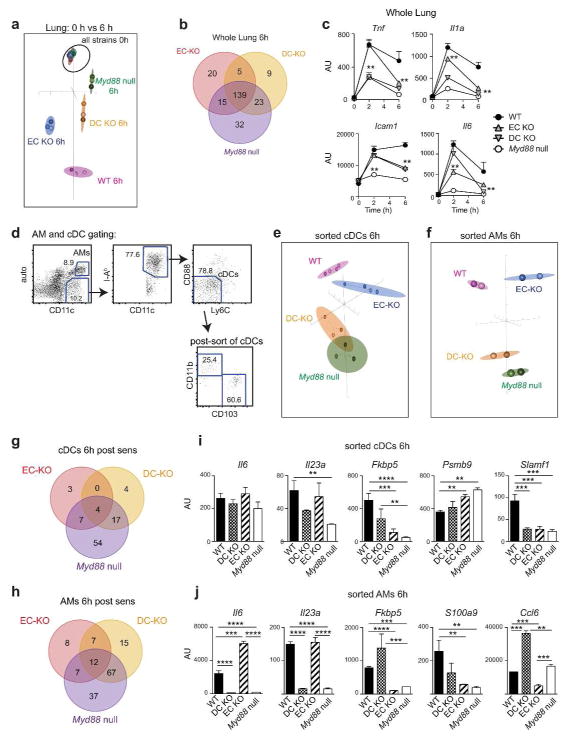
To determine if MyD88 signaling in Cd11c-expressing cells was important for gene expression in the lung at later time points during sensitization, we studied RNA prepared from WT, DC-KO, EC-KO, and Myd88 null mice at 6h post-sensitization (Supplementary Table 2, right). Many of the genes expressed at 6h were primarily dependent on MyD88 signaling in Cd11c-expressing cells (Figure 4a,b and Supplementary Table 5). These genes included Tnf and Il1a (Figure 4c), the latter being important for Th17 differentiation.26 In addition, several genes were identified whose expression in whole lung was affected by MyD88 deletion in both ECs and Cd11c-expressing cells, including Il6, which is also important for Th17 differentiation27, and the cell adhesion molecule, Icam1. Because the Cd11c-cre transgene is expressed in AMs as well as DCs, we separately purified these cells types from mice 6h after OVA/FLA-mediated allergic sensitization (Figure 4d). cDCs from DC-KO mice and Myd88 null mice had similar gene expression profiles, but differed from the profiles of cDCs from WT and EC-KO mice (Figure 4e,g and Supplementary Table 6). Similar results were obtained when expression in AMs was studied (Figure 4f,h and Supplementary Table 7). These results confirm that OVA/FLA-induced immune gene expression in cDCs and AMs is largely controlled by cell-intrinsic MyD88 signaling. Several cytokine encoding genes, including Il1a, Il6 and Il23a, were much more highly expressed in AMs than in cDCs or ECs (Figure 4i,j and Supplementary Figure 2b), suggesting that through their production of IL-1α, IL-6 and IL-23, AMs may cooperate with cDCs to promote Th17 differentiation.
Figure 4. Immune gene expression in whole lung, cDCs and AMs at 6h post-sensitization.
(a) BGA plot of immune gene expression in whole lung at 0 and 6h after OVA/FLA-mediated allergic sensitization of the indicated mouse strains. (b) Venn diagram showing the number of genes differentially expressed between lungs of WT mice (reference group) and lungs of the indicated mouse strains at 6h post-sensitization (≥1.5 fold; p≤0.01, MTI≥32 for at least one group). (c) Time course of lung gene expression in the indicated strains. **p ≤ 0.01 as compared to WT reference strain (d) Gating strategy for purification of AMs and cDCs. (e–f) BGA plot of immune gene expression in cDCs (e) and AMs (f) sorted from lungs of the indicated mouse strains 6h after OVA/FLA-mediated sensitization. (g–h) Venn diagrams showing the number of immune genes differentially expressed (≥1.5 fold; p≤0.01, MTI≥32 for at least one group) in cDCs (g) and AMs (h) of the indicated strains of mice compared to reference WT mice. (i–j) Relative expression of the indicated genes in purified cDCs (i) and AMs (j) of the indicated mouse strains at 6h post-sensitization. For purified AMs and cDCs, lungs from 3 mice were pooled prior to sorting each cell sample. n = 3–4 (cDCs) and n = 2 (AMs). **p <0.01, ***p < 0.001, ****p < 0.0001.
Our observation that Myd88 expression in ECs contributes to Th2 development and eosinophilic inflammation suggested that there might be crosstalk between ECs and DCs. To test this, we compared gene expression in cDCs of WT and EC-KO mice. We identified multiple immune genes whose expression in cDCs was partly controlled by MyD88 signaling in ECs, including Fkbp5, Psmb9, and Slamf1 (Figure 4i and Supplementary Table 6). Expression of some genes in AMs was also controlled by MyD88 signaling in ECs. For example, Fkbp5, S100a9, and Ccl6 were all decreased in AMs of EC-KO mice, whereas Il6 was increased in these cells (Figure 4j and Supplementary Table 7). Together, these data show that during allergic sensitization, expression of some immune genes in cDCs and AMs is controlled in part by signals they receive from ECs.
MyD88 controls chromatin accessibility at select promoters in cDCs
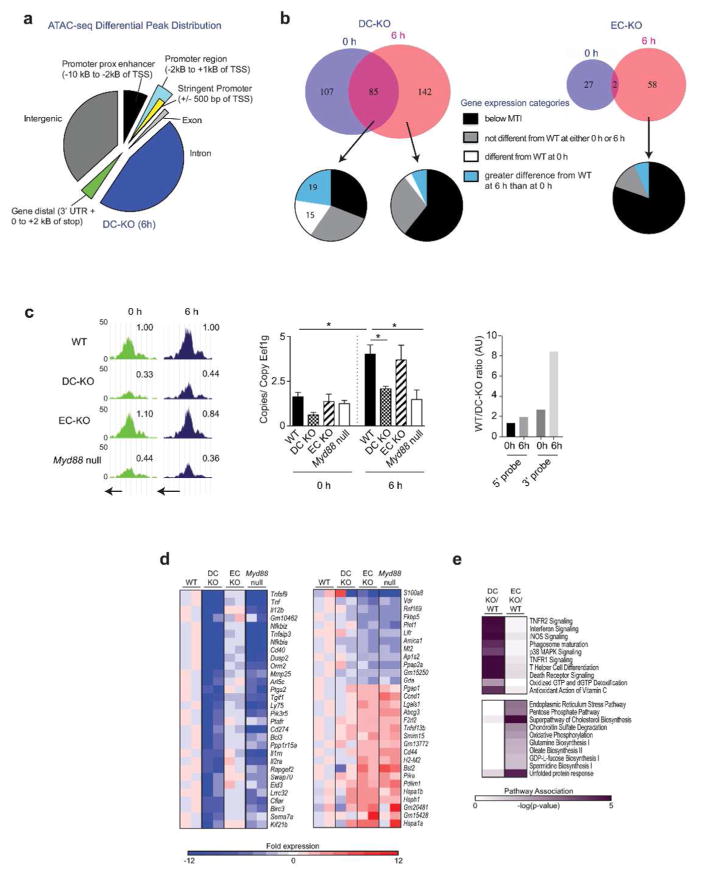
To gain additional insight into how MyD88 controls gene expression in lung cDCs, we examined the impact of cell-specific MyD88 signaling on chromatin accessibility in cDCs using ATAC-seq, an assay based on the ability of transposases to introduce DNA sequencing adaptors into open chromatin.28 As expected, ‘peaks’ corresponding to multiple overlapping sequencing reads at open chromatin mapped to transcription start sites (TSS) and enhancer regions, as well as intergenic and intronic regions (Figure 5a and Supplementary Table 8). Patterns of peak distribution among these categories were similar for all genotypes examined (data not shown). Restricting our analyses to chromatin at promoter regions defined as being within 500 base pairs (bp) of known TSS, we identified 227 genes whose chromatin accessibility differed (≥1.3-fold, p≤0.05, at least one peak ≥100 reads) between WT and DC-KO mice 6h after allergic sensitization (Figure 5b, top left). Of these, 85 genes were also different between these same genotypes at steady state (0h), whereas 142 loci were different only after sensitization (6h). We identified an additional 60 genes whose chromatin accessibility in lung cDCs at 6h was different between WT and EC-KO mice (Figure 5b, top right), suggesting that signals from ECs can also affect chromatin accessibility at some loci in cDCs.
Figure 5. ATAC-seq and whole transcriptome analysis of lung cDCs.
(a) Pie chart showing genomic distribution of ATAC-seq peaks that significantly differ (p≤0.05) in size between cDCs of WT mice (reference group) and DC-KO mice at 6h post-OVA/FLA. (b) Venn diagram showing the number of genes whose ATAC-seq peak sizes at the promoter differ by at least 1.3-fold (p≤0.05, at least one peak ≥100 reads) between cDCs of WT mice (reference group) and either DC-KO mice (top left), or EC-KO mice (top right). Pie charts below Venn diagrams show percentages of genes assigned to the indicated categories of gene expression (see Supplementary Methods). Data are from two biological replicates of cDCs sorted from 3 mice each. (c) ATAC-seq peaks and gene expression of Ccr7 in cDCs at 0 and 6h post-OVA/FLA. Left panel shows ATAC-seq plots, with the numbers above the plots representing peak areas for each genotype normalized to WT peaks. Arrows below plots show transcriptional start sites. Data are from 2 separate biological replicates with cDCs from 3 mice pooled and sorted for each replicate. Middle panel shows Ccr7 RNA levels in cDCs, as measured by qPCR and normalized to the Eef1g housekeeping gene. Data represent 2 biological replicates at the 0h time point and 3–4 biological replicates at the 6h time point, with cDCs from 3 mice per sorted group. Right panel compares Ccr7 expression in cDCs of WT and DC-KO mice, as measured with microarray probes corresponding to the 5′ or 3′ region of the mRNA. (d) Heat map showing the 30 genes most differentially expressed in cDCs of DC-KO (left) or EC-KO (right) mice, as determined by whole transcriptome microarray analysis. Individual fold-change values were normalized to the average WT reference. Data shown indicate 2 separate biological replicates with 3 mice per sorted replicate. (e) Heat map showing top 10 Ingenuity Pathways affected primarily in DC-KO (top) or EC-KO (bottom).
To better understand how MyD88-dependent changes in chromatin accessibility affect gene expression, we used transcriptome wide arrays to study expression of genes that mapped to loci whose ATAC-seq peak sizes were dependent on Myd88 expression. Almost half of the genes whose chromatin differed between cDCs of WT and DC-KO mice at both 0h and 6h were expressed ie. above the minimum threshold intensity (MTI) (see Supplementary Methods). Of these genes, 15 were differentially expressed (≥ 1.5-fold) between cDCs of WT and DC-KO mice at baseline but not 6h. Another 19 genes were induced, ie. differentially expressed at 6h (Figure 5b, bottom left, and Supplementary Table 9a). By contrast, the majority of genes whose genotype-specific differences in chromatin accessibility were only apparent at 6h, but not 0h, did not yet display transcriptional differences between cDCs of WT and DC-KO mice (Supplementary Table 9b). Very few genes whose chromatin accessibility in cDCs was different between WT and EC-KO mice were expressed above the MTI (Figure 5b, bottom right, and Supplementary Table 9c).
Cell-intrinsic MyD88 controls gene expression through multiple mechanisms
Transcriptome-wide arrays have multiple probes for each gene, allowing simultaneous quantification of the 5′ and 3′ regions of RNA. Using this platform to measure RNAs in cDCs from the various strains (see Supplementary Methods), we identified two major types of MyD88-dependent gene induction that occurred during allergic sensitization. In the first, the ratio of gene expression between WT and DC-KO mice measured using 5′ probes was similar to ratios obtained using 3′ probes, consistent with regulatory control at the level of transcriptional initiation (see Supplementary Methods). In the second, the ratio of gene expression in WT and DC-KO mice was higher when measured using 3′ probes than with 5′ probes. The latter pattern is consistent with an indirect role for MyD88 in RNA elongation. Approximately one third of the genes induced in cDCs by allergic sensitization met the criteria to be included in this category (Supplementary Table 9a). As an illustrative example, chromatin at the Ccr7 locus was found to be more accesible in cDCs of WT mice than in those of either DC-KO or Myd88 null mice, both at 0 and 6h (Figure 5c, left). Despite these differences in chromatin accessibility, at 0h there were no apparent differences in Ccr7 mRNA levels among the various genotypes (Figure 5c, middle). However, after allergic sensitization, Ccr7 mRNA levels were markedly higher in cDCs of WT and EC-KO mice than in those of DC-KO and Myd88 null animals. Furthermore, the ratio of Ccr7 expression in DCs of WT mice compared to those of DC-KO mice was greater when expression was measured using a 3′ probe than with a 5′ probe (Figure 5c, right), suggestive of transcriptional control at the level of RNA elongation. Thus, MyD88 contols gene expression through multiple, distinct mechanisms. For Ccr7 and several other genes, MyD88 maintains open chromatin without affecting baseline gene expression, and during allergic sensitization promotes efficient RNA elongation.
MyD88 signaling in DCs and ECs control distinct cellular processes in cDCs
Analysis of gene expression in cDCs across the genome, without regard to ATAC-seq status, identified approximately 250 genes whose expression in cDCs was dependent on cell-intrinsic MyD88 signaling (Supplementary Table 10). Many of these genes have known roles in immunity and include Tnf, Il12b, and Cd40, as well as genes that regulate canonical NF-κb signaling, such as Tnfaip3 (which encodes A20), Nfkbiz, and Nfkbia (Figure 5d, left). Ingenuity Pathway Analysis (IPA) confirmed that these genes contribute to immune signaling pathways (Figure 5e, top). We also identified genes whose expression in lung cDCs was controlled indirectly, through MyD88 signaling in ECs. Approximately half of these genes were negatively regulated by MyD88 signaling in ECs (Supplementary Table 10), and included H2-m2; Cd44; the heat shock protein genes, Hspa1a, Hspa1b, and Hsph1; the cell cycle and DNA damage sensing proteins, Ccnd1 and Ddit3. A different set of genes in cDCs was positively regulated by MyD88 signaling in ECs, including S100a8, S100a9, Fkbp5 and Amica1 (Figure 5d, right). Amica1 encodes junctional adhesion molecule-like protein (JAML)29, binding partner of the epithelial tight junction protein, coxsackie-adenovirus receptor (CAR). Interestingly, IPA analysis revealed that MyD88 signaling in ECs affected pathways in cDCs that are associated with endoplasmic reticulum stress, oxidative phosphorylation, and cholesterol biosynthesis (Figure 5e, bottom). This suggests that ECs may indirectly affect the ability of cDCs to sense cellular stress and thereby modulate the downstream immune response.
Effects of cell-specific Myd88 expression on DC phenotype and function
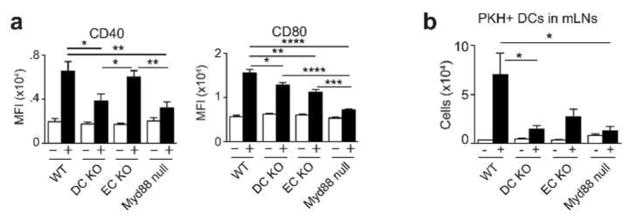
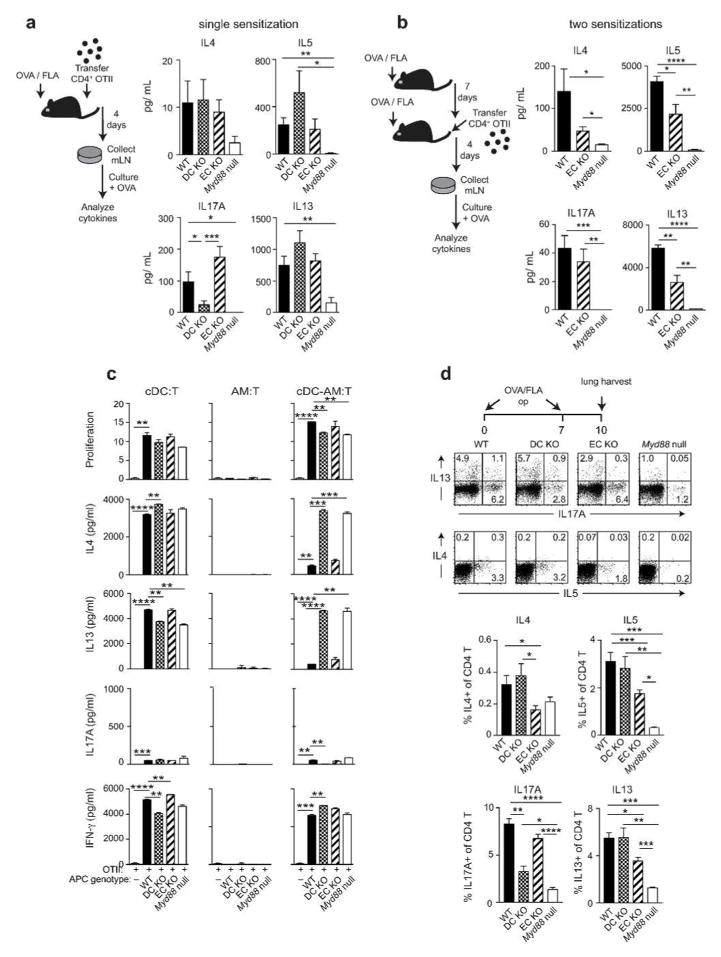
We next examined the consequences of cell-intrinsic and extrinsic MyD88 signaling on DC phenotype and function. Following allergic sensitization, allergen-bearing cDCs increase their display of co-stimulatory molecules and migrate from the lung to the regional mediastinal (m)LNs. At 16h post-sensitization with OVA/FLA, we noted that CD40 was markedly reduced on DCs of DC-KO mice, and CD80 was modestly reduced on DCs of both DC-KO mice and EC-KO mice (Figure 6a), in agreement with the observed Cd40 and Cd80 transcriptional differences at 6h (Supplementary Resource). We next studied the effect of cell-specific expression of Myd88 on DC trafficking by labelling the DCs with the fluorescent dye, PKH and inducing their migration with OVA/FLA. Analysis of mLNs 24h after OVA/FLA instillation revealed fewer PKH+ migratory DCs in mLNs of DC-KO mice than in those of WT mice (Figure 6b). There was also a trend towards fewer migratory DCs in EC-KO mice, but it was not statistically significant. To investigate how these and other changes related to Myd88 expression in Cd11c-expressing cells and ECs affects early T helper cell development in regional LNs, we adoptively transferred OVA-specific, naïve CD4+ T cells to recipient animals and sensitized them on a single occasion with OVA/FLA. mLNs of mice were excised from the sensitized mice and these mLN cells were cultured with OVA to stimulate cytokine production. As expected, Myd88 null mice had markedly lower amounts of type 2 cytokines and of IL-17A than did their WT counterparts (Figure 7a). IL-17A was also reduced in mLNs from DC-KO mice, but not in mLNs of EC-KO mice. Type 2 cytokines were similar in WT, DC-KO and EC-KO mice after a single sensitization, but after two sensitizations, mLNs of EC-KO mice had significantly lower amounts of IL-5 and IL-13 than did mLNs of WT mice (Figure 7b). These data suggest that Cd11c-MyD88 mediated effects on Th17 differentiation are direct, and that EC-MyD88 dependent effects on Th2 polarization are indirect, possibly requiring another cell type. To investigate this, we separately purified Cd11c-expressing AMs and cDCs from the lung at 6h post sensitization with OVA/FLA, and tested their abilities to promote proliferation of naïve OT-II T cells and stimulate their production of cytokines. We found that cDCs, but not AMs, could promote antigen-specific T cell proliferation and production of type 2 cytokines and IFNγ, whereas only very small amounts of IL-17 were produced. Surprisingly, purified cDCs from each of the strains displayed similar capacities to stimulate T cells (Figure 7c). Although AMs on their own were incapable of stimulating T proliferation or cytokine production, we nonetheless investigated the impact of adding them to the cDC: T cell co-cultures. No effect on T cell proliferation was seen, but AMs from WT mice and EC-KO mice markedly reduced type 2 cytokine production by T cells, whereas AMs from Myd88 null mice DC-KO mice did not. AMs did not affect Th1 differentiation, as inferred from IFNγ production. These data suggest that at least in this ex vivo experimental system, AMs produce one or more factors that selectively block Th2 differentiation in a manner dependent on MyD88 signaling in Cd11c-expressing cells.
Figure 6. DC costimulatory molecule expression and DC migration to mLNs are regulated by MyD88.
(a) Display of the costimulatory molecules, CD40 and CD80 on lung DCs (autofluorescence− CD11c+ I-Ab hi) from untreated mice (−) and 16h after OVA/FLA instillation (+). Mean fluorescence intensity; (MFI). Data are representative of 4 experiments with 4–6 mice per group. (b) Migration of PKH-labeled DCs from the lung to mLNs at baseline (−) and 24h after OVA/FLA instillation (+). Data are representative of 3 experiments with 3–6 mice per group. * p < 0.05, ** p < 0.01, ***p < 0.001, ****p < 0.0001.
Figure 7. Differential control of Th2 and Th17 cytokine production by EC and DC expression of Myd88.
(a–b) Cytokines in mLNs after a single sensitization (a) or two sensitizations (b) with 100 μg OVA and 1.25 μg FLA. (c) T cell proliferation and cytokine production in co-cultures of OT-II T cells mixed with the indicated APC(s) sorted from lungs of mice 6h after sensitization with OVA/FLA. (d) Intracellular staining for IL-17 and type 2 cytokines in CD4+ T cells of mice sensitized with OVA/FLA op on days 0 and 7 and harvested on day 10. Lung cells were stimulated with PMA and ionomycin prior to staining. Data shown are (a) representative of 2 experiments with 8–10 mice per group; (b) from a single experiment, representative of two, 4–5 mice per group; (c) representative of a single experiment performed with duplicate samples; and (d) from a single experiment with 3–4 mice per genotype. * p < 0.05, ** p < 0.01, ***p < 0.001, ****p < 0.0001.
Effects of cell-specific Myd88 expression on T helper subsets in the lung
The reduced amounts of IL-17 in mLNs of DC-KO mice, as well as their decrements in neutrophilic inflammation, suggested that Th17 cells in the lungs of these animals might be reduced. The reduction in type 2 cytokines in mLNs of EC-KO mice following a second sensitization with OVA/FLA and the decreased eosinophilia in lungs of these mice suggested that Th2 cells in the lung might be similarly reduced. To test this, we performed intracellular staining for IL-4, IL-5, IL-13 and IL-17A in CD4+ T cells sorted from lungs 3 days after the second OVA/FLA sensitization (day 10). Compared with WT mice, DC-KO mice and Myd88 null mice had fewer Th17 cells in the lung, whereas no reduction was seen in EC-KO mice (Figure 7d). Conversely, intracellular staining for IL-4, -5 and -13 revealed that EC-KO mice had fewer Th2 cells compared with WT mice, whereas no reduction in Th2 cells was seen with DC-KO mice. Together, these data suggest that MyD88 signaling in Cd11c-expressing cells promotes Th17 development and consequent neutrophilic inflammation, whereas MyD88 signaling in ECs favors Th2 development and eosinophilic inflammation.
DISCUSSION
It is well established that MyD88 signaling is critical for innate immune responses to a wide variety of stimuli, including inhaled allergens. However, the extent to which MyD88 signaling in different cell types in the lung contributes to asthma is poorly understood. Here, we found using two different models of asthma that MyD88 expression in ECs impacts gene expression in cDCs and also profoundly affects immune responses. Specifically, MyD88 expression in ECs was found to be required for robust eosinophilia, whereas MyD88 expression in Cd11c-expressing cells was required for strong neutrophilic responses. This suggests that inhibiting MyD88 function in only one of these cell types might ameliorate disease features in individuals that display predominantly neutrophilic or eosinophilic asthma. Our current findings are consistent with studies showing that Tlr4 expression in non-hematopoietic cells contributes to eosinophilia15,30 and that Tlr4 expression in hematopoietic cells promotes airway neutrophilia in mouse models of asthma30 and further identifies ECs and Cd11c-expressing cells as requiring Myd88 expression for the development of eosinophilic and neutrophilic inflammation, respectively.
MyD88 signaling in Cd11c-expressing cells was required for robust expression of Ccr7, and in agreement with this, DC migration from the lung to regional LNs was reduced in DC-KO mice. This likely contributed to the reduced amounts of IL-17 in lung-draining LNs of DC-KO mice, and consequently reduced numbers of Th17 cells and neutrophils in their lungs after allergen challenge. Although Ccr7 expression was not reduced in EC-KO mice, they had a trend towards fewer migratory DCs in regional LNs. Ccr7 is required for DC migration, but other genes can also contribute31,32. It is possible that altered gene expression in ECs can directly impact DC migration by affecting DC: EC adhesion, or indirectly affect migration by modifying gene expression in DCs.
MyD88 signaling in Cd11c-expressing cells was required for robust expression in cDCs of the Th17-promoting cytokines, Il6 and Il23a, and also for their display of CD40 which contributes to Th17 development33, 34. While these changes are consistent with reduced numbers of Th17 cells and airway neutrophis in DC-KO mice, our finding that AMs have higher expression of Il1a, Il6 and Il23a than do cDCs suggested that AMs might also have a role in directing Th17 differentiation. On their own, AMs failed to activate T cells, and when added to DC: T cell cultures had a minimal effect on IL-17 production, possibly because the cultures lacked a cell type or cytokine necessary for robust Th17 differentiation. The apparent discrepancy between this result and our observation that Myd88 null mice do not produce detectable amounts of cytokines in LNs might be partly due to the fact that equivalent numbers of cDCs were added to the ex vivo co-cultures, which would circumvent the DC migration defect in Myd88 null mice. Also, cDCs harvested from the lung at 6h post-sensitization have not undergone the MyD88-dependent increases in co-stimulatory molecules to the extent that are seen in DCs arriving in regional LNs. Interestingly, when added to the cDC: T cell cultures, AMs selectively inhibited type 2 cytokine production in a manner dependent on Myd88. Whether this effect of AMs on T cells in vitro are relevant to T cell differentiation in vivo is unclear as AMs are non-migratory and therefore unable to traffic to regional LNs where most effector T cell differentiation is thought to occur. Naïve T cells primarily reside in the blood and lymphatic system, but some of them also enter non-lymphoid organs, including the lung,33,35 and mice whose DCs cannot migrate to regional LNs can nonetheless develop strong responses to allergens.33,36 Thus, it is possible that naïve T cells can undergo differentiation to Th17 cells in the lung itself, where their differentiation may be influenced by AM-derived cytokines.
Due to their position on the luminal surface of the entire respiratory tree, ECs are exposed to a wide variety of antigens and potentially damaging environmental insults. We have shown that the expression of many genes in ECs are dependent on MyD88 signaling in those cells, including genes expected to affect APC function. These genes included Csf2, which encodes GM-CSF, a potent activator of alveolar macrophages and DCs,33,37 and s100a8/a9, which encodes proteins that bind TLR4 and activate DCs.33,38 Previous studies have suggested roles for other EC-derived factors in shaping responses to inhaled allergens, including ATP, uric acid, IL-25, IL-33 and TSLP.33,39. We did not detect mRNA for Il25 or Tslp in sorted ECs, although low amounts of Tslp were seen in whole lung, in agreement with a previous report that ECs are not a major source of this cytokine.40 By contrast, Il33 mRNA was detected at relatively high levels in sorted ECs, consistent with the previous observation that alveolar type II epithelial cells are the major pulmonary source of IL-33.33,41 It is also possible that allergen inhalation triggers release of pre-formed IL-33. Whether that cytokine directly activates lung DCs is unclear, however, because Myd88 expression in DCs was not required for the adjuvant activity of IL-33.
RNA profiling and analysis of chromatin accessibility by ATAC-seq revealed control of gene expression in cDCs by both cell-intrinsic and -extrinsic MyD88 signaling. We unexpectedly found that chromatin accessibility at many promoter regions is established in a MyD88-dependent manner even prior to allergic sensitization. The ligand/receptor interactions that engage MyD88 to maintain an open chromatin at these loci during steady state are unknown. It is possible that low amounts of microbial products in the lung might signal through one or more TLRs to maintain a basal level of MyD88 signaling. Alternatively, constitutive production of endogenous cytokines, such as IL-1 or IL-33, which also signal through MyD88, might sustain open chromatin at these loci. How this MyD88 signaling leads to chromatin opening is also unknown, but might involve the induction of pioneer transcription factors (TFs), so named because of their ability to bind and open closed chromatin.42 Additional studies will be required to test this hypothesis and to determine whether cell-intrinsic and -extrinsic MyD88 signaling trigger chromatin binding by different pioneer TFs.
Despite their open chromatin at baseline, many genes were not strongly expressed until after OVA/FLA-mediated allergic sensitization. This suggests that for these genes, MyD88 is not only required to open the chromatin at steady state, but also required for responses to a second, activating signal. For many genes, the ratio of expression of WT to DC-KO mice after sensitization was higher when measured using 3′ probes than with 5′ probes, suggesting that this second signal can act by promoting RNA elongation. This hypothesis is consistent with the known ability of RNA polymerase II (Pol II) to initially associate with an incomplete transcriptional complex and generate only short (20-65 nucleotide) RNA segments until receiving an activation signal required to generate full-length mRNA.43 This second step can be triggered by the binding of additional TFs and is associated with release of negative elongation factors from the complex through the positive transcription elongation factor b (P-TEFb kinase). It remains to be determined whether different TFs are involved in this putative elongation step than those that direct chromatin opening, and whether these TFs are differentially induced by cell-intrinsic and -extrinsic MyD88 signaling.
IPA software revealed that MyD88 signaling in Cd11c-expressing cDCs primarily affected genes associated with the innate immune response and cDC effector function. Most of these genes are likely controlled by MyD88 signaling in the cDCs themselves, but because Cd11c-cre is also expressed in AMs, it is also possible that MyD88 deficiency in those cells indirectly affects gene expression in cDCs. Nonetheless, the genetic approach used here avoids the confounding problem of inefficient and variable depletion of AMs by irradiation in bone marrow chimeric mice16 and provides a good model to evaluate the role of Myd88 in these cells. Although IL-1α can be produced by bronchial epithelial cells,44 our data suggests that the primary MyD88-dependent cell source in the lung is the alveolar macrophage. Interestingly, the loss of MyD88 in ECs was associated with increased transcription in cDCs of genes involved in cellular stress and lipid metabolism, including several genes in the superpathway of cholesterol synthesis. Several intermediates in the cholesterol biosynthetic pathway function as ligands of RORγt, the nuclear receptor critical for the development of Th17 cells,45 and blocking cholesterol synthesis impairs the differentiation of these cells.46 Furthermore, accumulation of intracellular cholesterol leads to increased Th17 responses, but decreased Th2 responses, in the lung.47 Whether transfer of lipids from DCs to T cells contributes to Th17 differentiation is not known. However, lipids and other material can be transferred from DCs to T cells by secreted exosomes,48 or through direct contact at the immunological synapse.49 It will be of interest to identify the MyD88-dependent signals from ECs that act on lung cDCs to control their cholesterol metabolism, and to determine if the altered cholesterol pathways in cDCs contributes to the increased Th17 cell differentiation seen in EC-KO mice.
Our finding that both cell-intrinsic and -extrinsic MyD88 signaling contributes to cDC reprogramming provides important insights into the cellular and molecular basis of allergic sensitization. Given the heterogeneity seen in clinical asthma and the differential responsiveness of patients to current therapies, these insights will be useful in the design of new therapies specifically tailored to target asthma that is predominantly eosinophilic or neutrophilic in nature.
MATERIALS AND METHODS
Mice
The following strains were purchased from Jackson Laboratory, Bar Harbor, ME: C57BL/6, OTII (C57BL/6-Tg(TcraTcrb)425Cbn/J), Myd88−/− (B6.129P2(SJL)-Myd88tm1.1Defr/J), Myd88fx/fx (B6.129P2(SJL)-Myd88tm1Defr/J), Cd11c-cre (B6.Cg-Tg(Itgax-cre)1-1Reiz/J) and B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J. Brigid Hogan (Duke University) provided B6;D2-Tg(Sftpc-cre)1Blh (Sftpc-cre) mice.23 EC-KO mice and DC-KO mice were generated by crossing conditionally mutant Myd88fx/fx mice22 to Sftpc-cre mice and Itgax-cre mice, respectively. Mice were housed in specific pathogen-free conditions at the NIEHS and used between 6 and 12 weeks of age in accordance with guidelines provided by the Institutional Animal Care and Use Committees.
Processing frozen sections
Lungs were inflated with 4% paraformaldehyde, serially transferred into 15%, 20%, and then 30% sucrose, and finally into a 1:1 mixture of 30% sucrose in PBS:OCT. The lungs were then frozen on dry ice and sectioned using a cryostat.
Images of frozen sections
Tiled images of frozen sections were acquired at 400x magnification on an Automated Epifluorescence Microscope (Zeiss, Oberkochen, Germany) and assembled using Zen Blue software (Zeiss).
Images of precision cut lung slices
Lungs were inflated with 2% low-melt agarose at 40°C, and the upper right apical lung lobe was sliced into 150 μm sections using a VF-300 Compresstome (Precisionary Instruments, Greenville, NC). Each tiled image was taken at 200x magnification on a 710 Confocal Microscope (Zeiss) and assembled using Zen Black software (Zeiss).
Mouse models of asthma
For the OVA/FLA model of asthma, mice were sensitized by oropharyngeal (o.p.) instillations of 100 μg endotoxin-free OVA together with 1.25 μg standard FLA from Salmonella typhimurium (InvivoGen, San Diego, CA) on days 0 and 7, then challenged on days 14, 15, and 16 with 1% OVA aerosol. (This FLA preparation signals through both TLR4 and TLR5 because it contains 103–104 endotoxin units/mg protein). In the HDM model of asthma, mice were sensitized with 1 μg HDM (Greer, Lenoir, NC) by o.p. instillation on days 0 and 7 and challenged by the same route on days 14, 15, and 16. For both models, mice were harvested at 4h post-challenge to examine cytokines and chemokines, and at 24h post-challenge to assess leukocyte recruitment to the airway. For the OVA/IL33 model of asthma, mice were sensitized by o.p. instillations of 100 μg endotoxin-free OVA together with 500 ng IL33 (Biolegend, San Diego, CA) on days 0 and 7, then challenged on days 14, 15, and 16 with 1% OVA aerosol.
Flow cytometry
Cells were blocked with anti-mouse CD16/CD32 (2.4G2) and normal mouse and rat serum (Jackson ImmunoResearch, West Grove, PA). For staining of surface antigens, cells were incubated with one or more of the following antibodies: CD3 (145-2C11), CD8α (53-6.7), CD8β (53-5.8), CD19 (1D3), CD44 (IM7), IL-4 (11B11), IL-5 (TRFK5), IL-17A (TC11-18H10), Ly6C (AL-21), Ter119, (BD Biosciences, San Jose, CA); CD34 (MEC14.7), CD45 (104), CD88 (20/70), EpCAM/CD326 (G8.8) (Biolegend, San Diego, CA); B220 (RA3-6B2), CD25 (PC61.5), CD4 (GK1.5), CD11b (M1/70), CD11c (N418), CD31 (390), CD40 (1C10), CD49b (DX5), CD80 (16-10A1), CD103 (2E7), eF780 viability dye, I-Ab (AF6-120.1), IL-13 (eBio13A), Ly6C/G (Gr-1) (RB6-8C5), SIRP-1α/CD172a (P84), streptavidin eFluor450, 7-AAD (eBioscience, San Diego, CA). Lung DCs were examined for costimulatory molecules at baseline and 16h after o.p. inhalation of OVA/FLA.
Preparation of sorted ECs
Lungs were perfused, inflated and digested with 4.5 U/mL elastase for 45 minutes at 37°C. Minced lung was then digested with DNase for another 15 minutes. Cells were strained through a 70 μm filter, depleted for non-epithelial lineage markers (CD31, CD34, CD45) using AutoMACS column (Miltenyi Biotec, San Diego, CA). ECs were FACS sorted (7-AAD− CD31− CD34− CD45− EpCAM+) with purity >99%.
Preparation of sorted cDCs and AMs
Lungs were perfused, minced and digested in collagenase, DNAse, hyaluronidase, and Liberase TM for 1h. Single cells were isolated using a 70 μm filter, and a 14.5% Nycodenz gradient was used to separate AMs and cDCs from other lung leukocytes. AMs were sorted for CD11c+ autofluorescence+, while cDCs were sorted for CD11c+ autofluorescence− I-Ab hi Ly6C− CD88− by flow cytometry. Ly6C and CD88 were used to exclude inflammatory DCs and monocyte-derived (mo)DCs respectively.50 Purity was estimated at >94% for AMs and cDCs.
RNA isolation from whole lung and sorted cells
Lungs were harvested from mice at various times after oropharyngeal administration of 100 μg OVA mixed with 1.25 μg stFLA, and RNA was isolated using RNeasy kit (Qiagen). RNA from sorted cells was isolated by Trizol, followed by Qiagen RNeasy MinElute Cleanup Kit to remove any organic solvents.
Nanostring Mouse Immunology Gene Expression
RNA from whole lung and sorted ECs, cDCs, and AMs was hybridized to the Nanostring nCounter Mouse Immunology Gene Expression Codeset according to manufacturer’s instructions and quantified on the Nanostring nCounter machine (Nanostring, Seattle, WA). Only genes having a group mean intensity ≥32 (log2 ≥5) for at least one sample were analyzed (see Supplementary Materials and Methods).
Whole Transcriptome Microarray Gene Expression
RNA from sorted cDCs was processed and analyzed on the Affymetrix Mouse Whole Transcriptome Array 1.0 according to manufacturer’s instructions (Affymetrix, Santa Clara, CA). Only genes having a maximum mean bi-weight log2 probe signal of ≥ 6.0 were considered above the MTI.
Pathway analysis
Pathway analysis was performed on whole transcriptome microarray data using the Ingenuity Pathway Analysis software (Redwood City, CA).
Library preparation for ATAC-seq
Following their isolation, cDCs were subjected to ATAC-seq as previously described28 with the following modifications; 25,000 cDCs were lysed in cold lysis buffer (10 mM Tris-HCl pH 7.5, 10 mM NaCl, 3 mM MgCl2, 0.1% NP-40) for 5 minutes, and nuclei were incubated with 2.5 μl of Tn5 Transposase in 25 μl of reaction buffer. Purified DNA fragments were amplified with 9 cycles of PCR, the PCR products purified by using AMPure XP beads (1:3 ratio of sample to beads, Beckman Coulter), and the libraries sequenced on a NextSeq 500 (Illumina) in 50 bp paired-end format.
PKH+ DC migration from lung to mLN
Mice were given 50 μL of 10 μM PKH26 oropharyngeally to label cells in vivo in the lung. 24h later, mice were either not sensitized (baseline) or given OVA/FLA to sensitize the mice. After another 24h, the draining mediastinal lymph node was harvested. Cells were isolated and stained for flow cytometry. PKH+ DCs were identified based on staining for PKH as well as autofluorescent− CD11c+ I-Ab + staining.
OT-II cell transfer and draining lymph node cytokine measurement
Recipient mice were injected with 0.5–3 × 106 CD4+ T cells from OT-II mice, and sensitized 2–4h later with o.p. instillation(s) of OVA/FLA. mLNs were harvested 4 days later, and 5×106 cells/mL were cultured in cRPMI with 20 μg/mL OVA. 48 hours later, supernatants were analyzed by ELISA for IL-4, IL-5, IL-13, IL-17A and IFN-γ according to the manufacturer’s instructions.
DC-AM co-culture with naïve T cells
Mice were sensitized by instillation of OVA/FLA as described above, and 6h later AMs and cDCs from 3 pooled mice were sorted separately for each genotype. Naïve CD4+ OTII cells were isolated from the spleens and lymph nodes of OTII mice and depleted using an antibody cocktail including CD8α, CD8β, CD11b, CD11c, CD16/32, CD19, CD25, B220, CD49b, I-Ab Ly6C/G, TER119, and CD44 on the AutoMACS Pro (Miltenyi Biotec, Auburn, CA). Naïve OTII cells (5×104/well) were cultured together with lung cDCs and/or AMs (5 × 103/well) in a 5% CO2 incubator in a U-bottom plate (BD Biosciences). After 5 days, T cells were counted to measure proliferation. T cells (105/well) were then transferred to a 96-well flat bottom plate coated with 1 μg/mL anti CD3 and 5 μg/mL anti-CD28 antibodies (eBioscience). 24h later, supernatants were analyzed by ELISA for IL-4, IL-13, IL-17A and IFN-γ according to the manufacturer’s instructions
Intracellular cytokine staining of lung T cells following 2 sensitizations
Mice were sensitized oropharyngeally with OVA/FLA on days 0 and 7. On day 10, lungs were harvested, perfused, minced and digested in collagenase, DNAse, hyaluronidase, and Liberase TM for 30 minutes. Single cells were isolated using a 70 μm filter, and a Histopaque-1083 gradient was used to separate the lymphocyte fraction. Cells were stimulated with PMA and ionomycin for 6h and then cells were stained for extracellular markers, fixed and permeabilized, and stained for intracellular cytokines.
Statistical analysis
Unless stated otherwise, statistical significance in animal models of asthma were determined by Student’s unpaired t-test. Statistical analysis of Nanostring, whole transcriptome array and ATAC-seq data are described in detail in the Supplementary methods.
Supplementary Material
Acknowledgments
We thank Maria Sifre and Carl Bortner for help with flow cytometry, Kevin Gerrish and Liwen Liu for assistance with microarray processing and data analysis advice, Rick Fannin and Esther Hou for help with Nanostring processing and preliminary data analysis, Ligon Perrow for support with animal experiments, Charles Tucker for assistance with confocal microscopy, Otis Lyght and Natasha Clayton for frozen tissue processing and sectioning, Gordon Flake for histological comparison of H&E and fluorescence serial sections, Michael Sanderson and Jun Chen for guidance in processing and analyzing lung slices, John House for advice on pathway analysis, and Michael Fessler and Jennifer Martinez (NIEHS) for critical reading of the manuscript.
FUNDING
This work was supported by the Intramural Research Program of the NIEHS ZIA ES102025-09 to D.N.C. and ES101965 to P.A.W.
Footnotes
AUTHOR CONTRIBUTIONS
S.Y.T, G.S.W, M.T, J.M.W, XX, K.N., M.L-C., H.N., K.M.G., P.A.W. performed experiments and/or analyzed the data. D.N.C. conceived of the project, and S.Y.T. and D.N.C. designed the experiments and wrote the paper.
DISCLOSURE
The authors declare that they have no competing interests.
GEO ACCESSION NUMBER
All Nanostring, microarray and ATAC-seq data have been deposited at the National Center for Biotechnology Information in the Gene Expression Omnibus repository under accession number: GSE79615.
Gene expression data for whole lung at various timepoints, as well as sorted ECs at 2h, cDCs at 6h, and AMs at 6h are included in an online interactive display: https://jmw86069.github.io/myd88-asthma/
References
- 1.Moore WC, Fitzpatrick AM, Li X, Hastie AT, Li H, Meyers DA, et al. Clinical heterogeneity in the severe asthma research program. Annals of the American Thoracic Society. 2013;10(Suppl):S118–124. doi: 10.1513/AnnalsATS.201309-307AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hastie AT, Moore WC, Meyers DA, Vestal PL, Li H, Peters SP, et al. Analyses of asthma severity phenotypes and inflammatory proteins in subjects stratified by sputum granulocytes. The Journal of allergy and clinical immunology. 2010;125(5):1028–1036. e1013. doi: 10.1016/j.jaci.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poon AH, Eidelman DH, Martin JG, Laprise C, Hamid Q. Pathogenesis of severe asthma. Clinical and experimental allergy: journal of the British Society for Allergy and Clinical Immunology. 2012;42(5):625–637. doi: 10.1111/j.1365-2222.2012.03983.x. [DOI] [PubMed] [Google Scholar]
- 4.Moore WC, Hastie AT, Li X, Li H, Busse WW, Jarjour NN, et al. Sputum neutrophil counts are associated with more severe asthma phenotypes using cluster analysis. The Journal of allergy and clinical immunology. 2014;133(6):1557–1563. e1555. doi: 10.1016/j.jaci.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung KF, Wenzel SE, Brozek JL, Bush A, Castro M, Sterk PJ, et al. International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma. Eur Respir J. 2014;43(2):343–373. doi: 10.1183/09031936.00202013. [DOI] [PubMed] [Google Scholar]
- 6.Green RH, Brightling CE, Woltmann G, Parker D, Wardlaw AJ, Pavord ID. Analysis of induced sputum in adults with asthma: identification of subgroup with isolated sputum neutrophilia and poor response to inhaled corticosteroids. Thorax. 2002;57(10):875–879. doi: 10.1136/thorax.57.10.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McKinley L, Alcorn JF, Peterson A, Dupont RB, Kapadia S, Logar A, et al. TH17 cells mediate steroid-resistant airway inflammation and airway hyperresponsiveness in mice. Journal of immunology. 2008;181(6):4089–4097. doi: 10.4049/jimmunol.181.6.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barczyk A, Pierzchala W, Sozanska E. Interleukin-17 in sputum correlates with airway hyperresponsiveness to methacholine. Respir Med. 2003;97(6):726–733. doi: 10.1053/rmed.2003.1507. [DOI] [PubMed] [Google Scholar]
- 9.Hellings PW, Kasran A, Liu Z, Vandekerckhove P, Wuyts A, Overbergh L, et al. Interleukin-17 orchestrates the granulocyte influx into airways after allergen inhalation in a mouse model of allergic asthma. Am J Respir Cell Mol Biol. 2003;28(1):42–50. doi: 10.1165/rcmb.4832. [DOI] [PubMed] [Google Scholar]
- 10.Kaminska M, Foley S, Maghni K, Storness-Bliss C, Coxson H, Ghezzo H, et al. Airway remodeling in subjects with severe asthma with or without chronic persistent airflow obstruction. The Journal of allergy and clinical immunology. 2009;124(1):45–51. e41–44. doi: 10.1016/j.jaci.2009.03.049. [DOI] [PubMed] [Google Scholar]
- 11.Molet S, Hamid Q, Davoine F, Nutku E, Taha R, Page N, et al. IL-17 is increased in asthmatic airways and induces human bronchial fibroblasts to produce cytokines. The Journal of allergy and clinical immunology. 2001;108(3):430–438. doi: 10.1067/mai.2001.117929. [DOI] [PubMed] [Google Scholar]
- 12.Wilson RH, Whitehead GS, Nakano H, Free ME, Kolls JK, Cook DN. Allergic sensitization through the airway primes Th17-dependent neutrophilia and airway hyperresponsiveness. American journal of respiratory and critical care medicine. 2009;180(8):720–730. doi: 10.1164/rccm.200904-0573OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chesne J, Braza F, Mahay G, Brouard S, Aronica M, Magnan A. IL-17 in severe asthma. Where do we stand? American journal of respiratory and critical care medicine. 2014;190(10):1094–1101. doi: 10.1164/rccm.201405-0859PP. [DOI] [PubMed] [Google Scholar]
- 14.Lambrecht BN, Hammad H. Lung dendritic cells in respiratory viral infection and asthma: from protection to immunopathology. Annual review of immunology. 2012;30:243–270. doi: 10.1146/annurev-immunol-020711-075021. [DOI] [PubMed] [Google Scholar]
- 15.Hammad H, Chieppa M, Perros F, Willart MA, Germain RN, Lambrecht BN. House dust mite allergen induces asthma via Toll-like receptor 4 triggering of airway structural cells. Nature medicine. 2009;15(4):410–416. doi: 10.1038/nm.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janssen WJ, Barthel L, Muldrow A, Oberley-Deegan RE, Kearns MT, Jakubzick C, et al. Fas determines differential fates of resident and recruited macrophages during resolution of acute lung injury. American journal of respiratory and critical care medicine. 2011;184(5):547–560. doi: 10.1164/rccm.201011-1891OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Citrin DE, Shankavaram U, Horton JA, Shield W, 3rd, Zhao S, Asano H, et al. Role of type II pneumocyte senescence in radiation-induced lung fibrosis. J Natl Cancer Inst. 2013;105(19):1474–1484. doi: 10.1093/jnci/djt212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piggott DA, Eisenbarth SC, Xu L, Constant SL, Huleatt JW, Herrick CA, et al. MyD88-dependent induction of allergic Th2 responses to intranasal antigen. The Journal of clinical investigation. 2005;115(2):459–467. doi: 10.1172/JCI22462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson RH, Maruoka S, Whitehead GS, Foley JF, Flake GP, Sever ML, et al. The Toll-like receptor 5 ligand flagellin promotes asthma by priming allergic responses to indoor allergens. Nature medicine. 2012;18(11):1705–1710. doi: 10.1038/nm.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caton ML, Smith-Raska MR, Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. The Journal of experimental medicine. 2007;204(7):1653–1664. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlitzer A, Sivakamasundari V, Chen J, Sumatoh HR, Schreuder J, Lum J, et al. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nature immunology. 2015;16(7):718–728. doi: 10.1038/ni.3200. [DOI] [PubMed] [Google Scholar]
- 22.Hou B, Reizis B, DeFranco AL. Toll-like receptors activate innate and adaptive immunity by using dendritic cell-intrinsic and -extrinsic mechanisms. Immunity. 2008;29(2):272–282. doi: 10.1016/j.immuni.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okubo T, Knoepfler PS, Eisenman RN, Hogan BL. Nmyc plays an essential role during lung development as a dosage-sensitive regulator of progenitor cell proliferation and differentiation. Development. 2005;132(6):1363–1374. doi: 10.1242/dev.01678. [DOI] [PubMed] [Google Scholar]
- 24.Eblaghie MC, Reedy M, Oliver T, Mishina Y, Hogan BL. Evidence that autocrine signaling through Bmpr1a regulates the proliferation, survival and morphogenetic behavior of distal lung epithelial cells. Developmental biology. 2006;291(1):67–82. doi: 10.1016/j.ydbio.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 25.Lloyd CM, Saglani S. Epithelial cytokines and pulmonary allergic inflammation. Current opinion in immunology. 2015;34:52–58. doi: 10.1016/j.coi.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Chung Y, Chang SH, Martinez GJ, Yang XO, Nurieva R, Kang HS, et al. Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity. 2009;30(4):576–587. doi: 10.1016/j.immuni.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441(7090):235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 28.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature methods. 2013;10(12):1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moog-Lutz C, Cave-Riant F, Guibal FC, Breau MA, Di Gioia Y, Couraud PO, et al. JAML, a novel protein with characteristics of a junctional adhesion molecule, is induced during differentiation of myeloid leukemia cells. Blood. 2003;102(9):3371–3378. doi: 10.1182/blood-2002-11-3462. [DOI] [PubMed] [Google Scholar]
- 30.McAlees JW, Whitehead GS, Harley IT, Cappelletti M, Rewerts CL, Holdcroft AM, et al. Distinct Tlr4-expressing cell compartments control neutrophilic and eosinophilic airway inflammation. Mucosal immunology. 2015;8(4):863–873. doi: 10.1038/mi.2014.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halim TY, Steer CA, Matha L, Gold MJ, Martinez-Gonzalez I, McNagny KM, et al. Group 2 innate lymphoid cells are critical for the initiation of adaptive T helper 2 cell-mediated allergic lung inflammation. Immunity. 2014;40(3):425–435. doi: 10.1016/j.immuni.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krishnaswamy JK, Singh A, Gowthaman U, Wu R, Gorrepati P, Sales Nascimento M, et al. Coincidental loss of DOCK8 function in NLRP10-deficient and C3H/HeJ mice results in defective dendritic cell migration. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(10):3056–3061. doi: 10.1073/pnas.1501554112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mann J, Oakley F, Johnson PW, Mann DA. CD40 induces interleukin-6 gene transcription in dendritic cells: regulation by TRAF2, AP-1, NF-kappa B, AND CBF1. The Journal of biological chemistry. 2002;277(19):17125–17138. doi: 10.1074/jbc.M109250200. [DOI] [PubMed] [Google Scholar]
- 34.Perona-Wright G, Jenkins SJ, O’Connor RA, Zienkiewicz D, McSorley HJ, Maizels RM, et al. A pivotal role for CD40-mediated IL-6 production by dendritic cells during IL-17 induction in vivo. Journal of immunology. 2009;182(5):2808–2815. doi: 10.4049/jimmunol.0803553. [DOI] [PubMed] [Google Scholar]
- 35.Cose S, Brammer C, Khanna KM, Masopust D, Lefrancois L. Evidence that a significant number of naive T cells enter non-lymphoid organs as part of a normal migratory pathway. European journal of immunology. 2006;36(6):1423–1433. doi: 10.1002/eji.200535539. [DOI] [PubMed] [Google Scholar]
- 36.Grinnan D, Sung SS, Dougherty JA, Knowles AR, Allen MB, Rose CE, 3rd, et al. Enhanced allergen-induced airway inflammation in paucity of lymph node T cell (plt) mutant mice. The Journal of allergy and clinical immunology. 2006;118(6):1234–1241. doi: 10.1016/j.jaci.2006.07.036. [DOI] [PubMed] [Google Scholar]
- 37.Nambiar JK, Ryan AA, Kong CU, Britton WJ, Triccas JA. Modulation of pulmonary DC function by vaccine-encoded GM-CSF enhances protective immunity against Mycobacterium tuberculosis infection. European journal of immunology. 2010;40(1):153–161. doi: 10.1002/eji.200939665. [DOI] [PubMed] [Google Scholar]
- 38.Averill MM, Barnhart S, Becker L, Li X, Heinecke JW, Leboeuf RC, et al. S100A9 differentially modifies phenotypic states of neutrophils, macrophages, and dendritic cells: implications for atherosclerosis and adipose tissue inflammation. Circulation. 2011;123(11):1216–1226. doi: 10.1161/CIRCULATIONAHA.110.985523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lambrecht BN, Hammad H. Allergens and the airway epithelium response: gateway to allergic sensitization. The Journal of allergy and clinical immunology. 2014;134(3):499–507. doi: 10.1016/j.jaci.2014.06.036. [DOI] [PubMed] [Google Scholar]
- 40.Dewas C, Chen X, Honda T, Junttila I, Linton J, Udey MC, et al. TSLP expression: analysis with a ZsGreen TSLP reporter mouse. Journal of immunology. 2015;194(3):1372–1380. doi: 10.4049/jimmunol.1400519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hardman CS, Panova V, McKenzie AN. IL-33 citrine reporter mice reveal the temporal and spatial expression of IL-33 during allergic lung inflammation. European journal of immunology. 2013;43(2):488–498. doi: 10.1002/eji.201242863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes & development. 2011;25(21):2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henriques T, Gilchrist DA, Nechaev S, Bern M, Muse GW, Burkholder A, et al. Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Molecular cell. 2013;52(4):517–528. doi: 10.1016/j.molcel.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willart MA, Deswarte K, Pouliot P, Braun H, Beyaert R, Lambrecht BN, et al. Interleukin-1alpha controls allergic sensitization to inhaled house dust mite via the epithelial release of GM-CSF and IL-33. The Journal of experimental medicine. 2012;209(8):1505–1517. doi: 10.1084/jem.20112691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santori FR, Huang P, van de Pavert SA, Douglass EF, Jr, Leaver DJ, Haubrich BA, et al. Identification of natural RORgamma ligands that regulate the development of lymphoid cells. Cell metabolism. 2015;21(2):286–297. doi: 10.1016/j.cmet.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu X, Wang Y, Hao LY, Liu X, Lesch CA, Sanchez BM, et al. Sterol metabolism controls T(H)17 differentiation by generating endogenous RORgamma agonists. Nature chemical biology. 2015;11(2):141–147. doi: 10.1038/nchembio.1714. [DOI] [PubMed] [Google Scholar]
- 47.Draper DW, Gowdy KM, Madenspacher JH, Wilson RH, Whitehead GS, Nakano H, et al. ATP binding cassette transporter G1 deletion induces IL-17-dependent dysregulation of pulmonary adaptive immunity. Journal of immunology. 2012;188(11):5327–5336. doi: 10.4049/jimmunol.1101605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nature reviews Immunology. 2002;2(8):569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 49.Villarroya-Beltri C, Gutierrez-Vazquez C, Sanchez-Madrid F, Mittelbrunn M. Analysis of microRNA and protein transfer by exosomes during an immune synapse. Methods in molecular biology. 2013;1024:41–51. doi: 10.1007/978-1-62703-453-1_4. [DOI] [PubMed] [Google Scholar]
- 50.Nakano H, Moran TP, Nakano K, Gerrish KE, Bortner CD, Cook DN. Complement receptor C5aR1/CD88 and dipeptidyl peptidase-4/CD26 define distinct hematopoietic lineages of dendritic cells. Journal of immunology. 2015;194(8):3808–3819. doi: 10.4049/jimmunol.1402195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.