Abstract
Salmonella infection causes morbidity and mortality throughout the world with the host immune response varying depending on whether the infection is acute and limited, or systemic and chronic. Additionally, Salmonella bacteria have evolved multiple mechanisms to avoid or subvert immunity to its own benefit and often the anatomical location of infection plays a role in both the immune response and bacterial fate. Here, we provide an overview of the interplay between the immune system and Salmonella, while discussing how different host and bacterial factors influence the outcome of infection.
Keywords: Salmonella, infectious disease, macrophage, neutrophil, dendritic cell, T cell, B cell, immunomodulation
1. Overview of Salmonella Infection
Organisms belonging to the Salmonella genus are flagellated rod-shaped Gram-negative facultative anaerobes. Within the Salmonella genus, Salmonella enterica is further subdivided into six-subspecies with at least 2500 serotypes that are distinguished by variations in O (somatic) and H (flagellar) antigens. Approximately 99% of the Salmonella strains that cause infection in humans or other mammals belong to the Salmonella enterica species. The three major diseases caused by Salmonella in humans are non-invasive non-typhoidal salmonellosis, invasive non-typhoidal salmonellosis, and typhoid fever, all of which are described in greater detail below.
1.1 Non-invasive, Non-Typhoidal Salmonellosis
Nontyphoidal salmonellosis (NTS) refers to any illnesses caused to humans by all serotypes of Salmonella, except for the distinct typhoidal serotypes: Typhi and Paratyphi A-C. Salmonellosis is an acute, gastroenteritis, typically acquired orally through contaminated water or comestibles. Annually, there are an estimated 1.3 billion cases of Salmonella gastroenteritis, leading to approximately 3 million deaths worldwide [1]. It is among the most commonly isolated foodborne pathogens associated with fresh fruits and vegetables such as apples, cantaloupes, alfalfa sprouts, mangos, lettuce, cilantro, tomatoes, melons, orange juice, celery and parsley [1]. The incidence of NTS gastroenteritis is highest in the developing world, but is also of considerable importance in developed countries [2].
Salmonellosis is characterized by acute enterocolitis, which is accompanied by inflammatory diarrhea, a symptom rarely observed in individuals infected with invasive serovars (i.e. S. Typhi). Infection occurs after ingestion of >50,000 bacteria in contaminated food or water, with symptoms typically occurring 6–72 hours after consumption. Onset of symptoms is marked by abdominal pain and diarrhea with or without blood, while nausea and vomiting are also common. Typically, the gastroenteritis will resolve itself in 5–7 days without need for treatment although symptoms are usually more severe and longer lasting in children [3]; however, in cases were fluid loss is substantial, oral or intravenous rehydration may be necessary. In adults, antibiotics are usually contraindicated unless there is evidence of invasive disease (i.e. bacteremia), as antibiotics are unlikely to lessen the duration of illness or decrease the severity of symptoms [2,4,5] and have the potential to increase bacterial antibiotic resistance. Notably, individuals can continue to shed bacteria and infect others even after they no longer exhibit symptoms. On average, non-typhoidal serotypes persist in the intestinal tract from 6 weeks to 3 months, depending on the serotype, but approximately one person in a thousand will continue to shed Salmonella in their feces for periods exceeding 1 year [6]. Despite the growing morbidity of NTS infections, mortality due to Salmonella gastroenteritis is predominantly restricted to the developing world. This may be due in part to lack of clean water supplies and adequate sanitation [7]. In addition, reduced healthcare infrastructure may play a role, as appropriate diagnosis could be delayed and antibiotic resistant strains might go unidentified. Lastly, there are no vaccines against non-typhoidal Salmonella strains, possibly due to the fact that there is a large variance between strains and an incomplete knowledge of protective antigens. This is of particular concern for invasive non-typhoidal strains of Salmonella, discussed below.
1.2 Invasive Non-Typhoidal Salmonellosis
In sub-Saharan Africa, an emerging Salmonella strain is evolving and has a unique pathogenesis, in comparison to its genetic counterparts. This emerging pathogen has been termed invasive non-typhoidal Salmonella (iNTS). Like non-invasive NTS, the Salmonella serotypes most commonly associated with iNTS are S. Typhimurium and S. Enteritidis; however, other serotypes such as Choleraesuis and Dublin are also known to cause invasive disease in humans [8,9]. Whole-genome sequencing of invasive isolates have identified dominant regional genotypes uniquely found in Africa. These isolates, from strain ST313, have several genetic differences compared with other strains and suggest distinct genotypes of Salmonella have emerged as new pathogenic clades in sub-Saharan Africa, and might have adapted to cause invasive disease in humans [10]. Notably, other invasive S. Typhimurium ST313 strains have been found elsewhere in the world, reflecting a potentially increasing problem with this disease spreading globally [11]. iNTS strains were described commonly as a cause of bloodstream infections in Africa children predating the HIV epidemic [12]; however, shortly after the discovery of AIDS in Africa, more reports surfaced of children and adults with invasive non-typhoidal Salmonella bacteremia and the first epidemiological link between invasive Salmonella infection and AIDS was made in New Jersey. By 1990, iNTS had been confirmed as a common HIV-related pathogen in sub-Saharan African adults, implicating a role for CD4 T cells in this disease as these cells are eliminated during HIV infection [10]. To this day, non-typhoidal Salmonella infections are the most common bacterial bloodstream infections isolated from both adults and children presenting with fever in sub-Saharan Africa [10].
iNTS typically presents as a febrile systemic illness where diarrhea is often absent (as compared to non-invasive NTS salmonellosis and acute gastroenteritis, where diarrhea is common). Diagnosis can be difficult without microbiological tests, because there is often clinical overlap with other bacterial or parasitic diseases, notably pneumonia and malaria [13]. Patients with iNTS frequently present with lower respiratory tract disease, commonly attributable to co-infections with other pathogens, such as Mycobacterium tuberculosis and Streptococcus pneumoniae [14,15]. Even when treated with the appropriate antimicrobial, iNTS has a case fatality rate of 22–47% in both African adults and children [16]. The main risk factor to adults for iNTS is undoubtedly advanced HIV infection, however only about 20% of children presenting with iNTS are infected with HIV. Other risk factors to children are thought to include malnutrition, malaria, sickle-cell anemia and schistosomiasis [10]. Due to the increasing number of iNTS infections, the mortality associated with iNTS and the increasing difficulty in treating iNTS (due to the emergence of antibiotic-resistant strains), there is a medical need for vaccines with broad serovar coverage and high efficacy against systemic salmonellosis. Unfortunately, it is unknown what antigens are most protective against non-typhoidal Salmonella strains; however, work is ongoing to define potential immunodominant antigens [17].
1.3 Typhoid Fever
Typhoid fever is caused by infection with Salmonella Typhi and is responsible for 21 million new cases each year leading to an estimated 200,000 deaths. The annual mortality from typhoid fever has increased by 39% between 1990 and 2010 [18]. Most cases occur in developing countries, or among travelers to these countries, and a recent analysis of global mortality data revealed that in endemic regions (such as sub-Saharan Africa and Asia), the relative years of life lost to enteric fever ranks similarly to those lost to breast cancer, prostate cancer, and leukemia in North America [19,20]. One particular difference between S. Typhi and NTS strains is the presence of the polysaccharide capsular antigen, Vi [21], which is thought to be a virulence factor of S. Typhi, allowing it to survive the acidic environment of the stomach early after infection, as acapsular S. Typhi is less virulent [22]. Unlike NTS broad host specificity, S. Typhi is restricted to humans only [23].
S. Typhi can survive and replicate within host cells, particularly phagocytes (i.e. macrophages, dendritic cells, neutrophils, etc.), and the bacteria uses these cells to translocate to systemic sites of the body, such as the liver, spleen and bone marrow. For reasons not fully understood, it is estimated 5% of infected individuals will fail to clear the infection within a year, and will progress instead to a chronic carrier state where the bacteria likely reside primarily the hepatobiliary tract and gallbladder in humans [19]. This observation is also supported by data collected from S. Typhi positive cadavers, showing S. Typhi present in 85.7% of liver tissues [24]. Furthermore, chronic carriage of Salmonella Typhi is a risk factor for gallbladder carcinomas [25]. A better understanding of the carrier state is needed, as these individuals remain the reservoir for future S. Typhi transmission. Cholecystectomy (removal of the gallbladder) in conjunction with antibiotic therapy is the most effective means of clearance but does not guarantee elimination of the carrier state, as additional foci of infection can persist elsewhere in the body [19], including the liver [26].
More concerning is the increasing prevalence of multidrug-resistant (MDR) strains of S. Typhi [27–31]. Antibiotic resistance is of particular concern in resource-limited countries, many of which are endemic for enteric fevers. As discussed above, NTS is rarely treated with antibiotics, because they can increase the duration of bacterial excretion; however, typhoid fever is immediately treated with antibiotics, due to the systemic nature of the infection. As early as the 1990s, physicians began moving away from first-line antibiotics (i.e. chloramphenicol, ampicillin, etc.), due to a general widespread resistance amongst many S. enterica serovars. Fluoroquinolones (i.e. ciprofloxacin) have become the primary treatment, however resistance to this class of antimicrobials is increasing, as evidenced in studies conducted in endemic regions [30]. Third generation cephalosporins are now often the second-line treatment for typhoid fever. MDR strains of both S. Typhi and iNTS are becoming increasingly more common, highlighting the need for better prophylactic therapeutics.
Currently, there are three licensed vaccines against S. Typhi: a killed whole cell vaccine, a live attenuated vaccine (Ty21a), and a Vi capsular polysaccharide vaccine. The killed whole cell vaccine is no longer manufactured due to its high rate of reactogenicity, where it causes profound inflammation-driven systemic and local reactions [32–34]; however, it was successfully used to control typhoid fever in Thailand [35]. After years of use and post-licensure data, the Ty21a vaccine is known to be well tolerated and safe, but only has a moderate efficacy of around 50% [36]. Furthermore, the vaccine is not licensed in children under the age of six years old, a population that is highly susceptible to typhoid fever [37]. The Vi capsular vaccine, while well tolerated and 60% effective, has several limitations. First is that the vaccine is administered parenterally, which requires specialized training, making it less useful in developing countries [36]. Second, being comprised exclusively of a bacterial polysaccharide, the vaccine does not generate potent immunological memory (efficacy lasts about 2–3 years) and cannot be boosted with repeated vaccination, due to its T cell independent nature [38]. Thus, the vaccine is unlikely to be efficacious in children under 18 months of age, owing to the immaturity of their splenic marginal zones, which are required for T cell independent antibody responses [39]. It should also be noted that the Vi capsular vaccine cannot protect against disease caused by S. Paratyphi A, which is increasing in Asia, as this serovar doesn’t express Vi capsule and so immunity directed against this antigen is rendered ineffective [40,41]. No vaccines currently exist against iNTS. Clearly a need for new therapies to treat salmonellosis is desired [7] and a preventative, broad-spectrum vaccine would be especially useful in reducing the need for antibiotics.
1.4 Animal Models of Salmonella Infection
To better understand the host immune responses against NTS and to aid in the development of new vaccines and therapies, animal models of salmonellosis are necessary. Unfortunately, as with most animal modeling, mimicking human disease can be challenging. The best models for studying human gastroenteritis caused by NTS are non-human primates (NHP) and bovines, both of which mirror human pathology [42]. Additionally, NHP rhesus macaques can be used as a model of iNTS infection, where co-infection with simian immunodeficiency virus results in immune responses comparable to those seen in HIV-infected individuals [43]; however, both the bovine and NHP models are restricted by their financial costs and the inability to genetically manipulate the host genome. Furthermore, the use of primates generates numerous ethical concerns. Therefore, due to their low cost, ease of housing/handling, and the possibility for genetic manipulation, inbred mice are the most widely used model organisms to study salmonellosis [7]. Expectedly, these models have their own disadvantages. One of these is that mice do not typically mimic the gastroenteritis seen in human NTS patients, but rather develop a systemic typhoid-like illness. This issue can be circumvented by pretreating mice with antibiotics, usually streptomycin [44,45], which eliminates the host gut microbiota, permitting S. Typhimurium to overcome the natural intestinal colonization resistance posed by the normal flora and allowing for efficient colonization of the cecum and colon, without disseminating and killing the mouse [42].
Further challenges arise when utilizing animals to model typhoid fever. Because S. Typhi is human restricted, animal models are often inadequate and do not accurately reflect the human disease state. Historically, chimpanzees have been infected with S. Typhi, which develop a mild form of disease resembling typhoid fever, however, the infectious dose is high (~1×1011 CFUs), compared to the dose needed to cause typhoid fever in humans (~500 CFUs) [23]. Notably, mice infected with S. Typhimurium become systemically infected, with minimal intestinal pathology, akin to S. Typhi infections in humans [3]. Both the acute and chronic stages can be studied in mice; however, establishment of long-term infection is dependent on mouse strain. Many common inbred mouse strains have a single nucleotide polymorphism (SNP) in the natural resistance-associated protein 1 (Nramp1, also known as Slc11a1), a divalent transition metal (iron and manganese) transporter involved in iron metabolism and host resistance to certain pathogens. This SNP causes a single amino acid change (glycine to asparagine), which renders the Nramp1 protein non-functional. As such, phagocytic cells are incapable of depriving Salmonella containing vacuoles (SCVs) of metal ions, allowing the bacteria to proliferate uncontrollably. Susceptible mice (Nramp1−/− i.e C57Bl/6, Balb/c and others) are less able to control infection with virulent, non-attenuated Salmonella strains and often succumb to infection within two to three weeks, while resistant mice (Nramp1+/+ i.e 129Sv, DBA or Nramp1+/− C57Bl/6×129x1/sv (F1 hybrids) commonly survive acute infection, develop a carriage state, and can transmit the bacteria through their feces akin to what is seen during human typhoid fever [46,47]. More recently, two other mouse typhoid models have been developed. One uses a Toll-like receptor 11-knockout mouse strain (TLR11−/−), as TLR11 is present in mice and recognizes S. Typhi flagellin, but is absent in humans. TLR11−/− mice have reduced intestinal responses and exhibit a systemic infection when orally infected with S. Typhi [48]; however, these mice do not appear to develop chronic infection and succumb to infection within a few weeks. Notably, multiple groups, including the group that originally characterized the model, have recently reported that the original S. Typhi infection model in the TLR11−/− is not reproducible and that these mice may not actually be susceptible to infection with S. Typhi [49,50]. As such TLR11−/− mice are likely not a suitable model for the study of S. Typhi infection. The other recently described typhoid mouse model makes use of bone marrow humanized mice allowing for S. Typhi replication in the spleen, livers, and gallbladders [51]. Notably, however, these mice succumb to S. Typhi infection, and do not persist into the carriage state seen with the human infection [52,53]. Additionally, humanized mice can be expensive and labor intensive to generate, not to mention subject to considerable variability given the genetic heterogeneity of human bone marrow donors and the variable degree of engraftment [54]. Importantly, no small animal model of iNTS currently exists and such a model has the potential to provide valuable information about this emerging infection. Lastly, while invaluable information has been gained from the use of animal models in understanding Salmonella virulence factors, host inflammatory responses, dissemination and transmission, any relevance to human disease must be carefully inferred, as is true with any animal model of human disease.
2. Immunity to Salmonella infection
2.1 innate immune response to Salmonella infection
Because Salmonella is transmitted via the fecal-oral route, recognition by the immune system is initiated during invasion of intestinal epithelial cells, which can identify the pathogenic bacteria and initiate an inflammatory response through the recruitment of various phagocytic cell lineages. Recognition of bacterial LPS by TLR4 causes these cells to release cytokines and chemokines that serve as the initial signal for phagocytic cell recruitment [55]. The early innate immune responses initiated in the Peyer’s patches and mesenteric lymph nodes (MLNs) involve the recruitment of neutrophils and inflammatory monocytes, which help slow the spread of bacteria to systemic tissues [56,57].
Research indicates that a variety of innate immune cells are responsible for the early, acute control of Salmonella. It has been previously reported that neutrophil depletion leads to increased bacterial loads of Salmonella in the liver suggesting neutrophils play an important role in the prevention of bacterial dissemination to systemic sites [58]. The significance of neutrophils in early Salmonella infections has been further highlighted in a recent study, which demonstrated neutrophils are a key source of cellular IFN-γ during the acute phase of S. Typhimurium infection [59]. However, neutrophils are not the only key producers of IFN-γ during this early phase of Salmonella infection. Natural Killer (NK) cells have also been shown to make IFN-γ and may contribute to early resistance [60].
During initial Salmonella infection, inflammatory monocytes rapidly accumulate in Peyer’s patches and mesenteric lymph nodes of infected mice, where they produce a number of anti-microbial factors, including iNOS, TNF-α and IL-1β [56]. Additionally resident macrophages within infected tissues are capable of phagocytosing Salmonella and subsequently producing pro-inflammatory cytokines such as IL-1β and IL-18 through the recognition of cytosolic flagellin via the NLRC4 inflammasome complex [61,62]. Resident dendritic cells (DC) can also recognize Salmonella LPS and flagellin, which induces DC maturation, enhancing antigen presentation and inducing their migration to T cell areas of various lymphoid tissues to initiate the adaptive phase of the immune response [63].
The type of cytokine response to infection is essential to control bacterial persistence and replication. IFN-γ, IL-12 and TNF-α have all been demonstrated to be important for Salmonella clearance [64], while IL-10 and IL-4 have been shown to be permissive of infection [65–67]. Cytokines detected in patient serum consists of high levels of the inflammatory cytokines IFN-γ, IL-18, IL-12, IL-15, TNF-α as well as the anti-inflammatory cytokine IL-10 [68,69]. Likewise, PBMCs from volunteers orally immunized with the live-attenuated Ty21a vaccine secrete Th1 cytokines including IFN-γ, TNF-α, but also IL-10 [70]. Various animal studies have corroborated the cytokine profiles seen in human patients [71,72].
2.2 Adaptive immune response to Salmonella infection
Normally, Salmonella infection is controlled and cleared after the generation of both T and B cell immunity specific for the pathogen. Additionally, these types of immune responses are important for protecting the host against a secondary infection. Notably, the exact role of the humoral immune response during Salmonella infection has been contested [73]. In humans, most individuals who survive typhoid fever generally acquire protective immunity to future infections, with recurrence rates around 2–3%, and recurrence occurs only if an individual is exposed to a high secondary inoculum of organisms or received early antibiotic therapy for the initial infection (thus prohibiting a robust adaptive immune response from occurring) [74]. Volunteers immunized with the live-attenuated Ty21a vaccine develop significant levels of IgA and IgG against S. Typhi LPS; however, it is unclear whether these antibody responses contribute to protective immunity [75]. Similarly, volunteers receiving the purified Vi antigen injected vaccine, show infection rates of 4.1%, compared to 16.2% for an unvaccinated group and these volunteers also show significant increases of serum antibodies toward the Vi antigen [76]. Conversely chronic carriers, where S. Typhi persists for years, also have high levels of circulating anti-Vi antibodies, as well as antibodies against the O- (LPS) and H-antigens (flagella), yet they never clear the infection, making it unclear what role these antibodies play during infection [77]. One possibility is that anti-S. Typhi antibodies only protect the host from invading bacteria, but are not sufficient to clear the bacteria after establishment of an intracellular infection. In animal models, the role of antibody-mediated protection against challenge is even more contentious. For instance, in experiments where serum from Salmonella infected mice was transferred to naïve animals, which were then subsequently challenged with Salmonella, some investigators have observed protection [78,79], while others have not [80]. Nonetheless, B cells appear to be important in controlling bacterial replication, as mice deficient in B cells were able to eventually clear Salmonella, yet had higher bacterial burdens during both primary and secondary infections, compared to wild type mice [81]. Nanton et al. showed the susceptibility of B cell–deficient mice correlated with reduced IFN-γ production from CD4 T cells; however, the B cell role may be antibody-independent, since mice harboring B cells that were unable to produce antibodies did not show an increased susceptibility to Salmonella infection, nor was there a deficit in IFN-γ production from CD4 T cells [82]. Therefore, the cross-talk between humoral and cell-mediated immunity in salmonellosis seems crucial for the clearance of Salmonella, although a correlation between the presence of antibodies alone and resistance to reinfection appears complex [83]. One possible explanation for these findings is that, in addition to antibody production, B cells perform alternative functions, including antigen presentation and cytokine secretion, which could have an important function in anti-Salmonella immune responses [84,85].
Like B cells, the role of Salmonella-specific cytotoxic (CD8) T cells is not well defined. Generally, CD8 T cells are not thought to contribute to the primary clearance of Salmonella [86] and depletion of CD8 T cells during the persistent stages of infection does not lead to increased bacterial burdens in systemic organs [87,88]. Conversely, studies using β2M-deficient mice lacking surface MHC Class I expression demonstrate these mice are capable of resolving infection with attenuated Salmonella [89], however, these mice are also deficient in the expression of non-classical MHC molecules and CD1, confounding the interpretation of the role of MHC Class I restricted antigens in protective Salmonella immune responses [90]. Mice that specifically lack only the MHC class I molecule or CD8+ T cell cytotoxic granules have demonstrated only a mildly protective role for CD8 T cells in the resolution of primary Salmonella infection [91]. Interestingly this same study demonstrated that memory CD8 T cells are dispensable during secondary infection with Salmonella [91]. Another report showed that the magnitude of the CD8 T cell response correlates directly with the intracellular proliferation of Salmonella, however the CD8 T cells in this study were inferior due to reduced proliferation, cytotoxic functionality, and cytokine production [92]. It should be noted that while many studies investigating CD8 T cell involvement have used attenuated Salmonella strains, a recent study showed an important role for MHC Class I-dependent CD8 T cells in protection against virulent Salmonella [93]. Taken altogether, it may be too early to assign a functional role to CD8 T cells during salmonellosis, or to dismiss their participation in anti-Salmonella immunity. More research is clearly needed to fully elucidate their role during both acute and persistent infection.
The importance of helper T cell immunity during Salmonella infection is well established. One indication that CD4 T cells are essential mediators of immunity against NTS infection in humans comes from the finding that normally non-invasive NTS can cause a severe, invasive, systemic infection (iNTS) in HIV-infected individuals, where CD4 T cells are naturally depleted [94]. In fact, antiretroviral HIV therapy has been shown to decrease the incidence of iNTS infection in humans, demonstrating that CD4 T cells are among the most important immune cells mediating immunity against salmonellosis [95]. One mechanism used by CD4 T cells during Salmonella infection is the activation of innate immune responses against intracellular bacteria, particularly through the production of IFN-γ. IFN-γ helps control intracellular bacterial replication by signaling through the IFN-γR on phagocytes, which activates JAK/STAT signaling to induce the expression of iNOS, which reacts with the cellular substrates L-arginine and oxygen to produce nitric oxide, a free radical that can cause bacterial DNA damage [96]. Th1 cells, which require the transcription factor T-bet, are essential producers of IFN-γ during Salmonella infection and can activate macrophages [97,98]. Studies using susceptible mice lacking Th1 cells, due to a deficiency in T-bet, have confirmed Th1 cells are necessary to resolve a primary, acute Salmonella infection [99]. It was also shown in this study that Salmonella-specific cells were unable to produce IFN-γ, and mice exhibited increased levels of IL-10. Direct depletion of CD4 T cells in both resistant [88] and susceptible [87] mice causes a significant increase in bacterial burdens in multiple organs further demonstrating the importance of maintaining CD4 T cell immunity during Salmonella infection. This increase is likely due to a loss in the production of IFN-γ by CD4 T cells, as neutralization of IFN-γ causes bacterial burdens to increase in persistently infected organs [47]. Likewise, mice lacking IL-12, IFN-γ or iNOS all have deficiencies in their ability to clear Salmonella infection [100]. Notably, Th17 T cells have been shown to be important for neutrophil recruitment to the gut where those neutrophils prevent systemic Salmonella dissemination of [43]; however, Th17 T cells may be dispensable for controlling systemic Salmonella infection [101]. CD4 T cells have also been shown to be important for Salmonella-specific antibody class switching. Mice deficient in Th1 cells had significant decreases in the production of Salmonella-specific IgG2a, leading to an increased susceptibility to infection [99]. It is also likely CD4 T cells are needed for optimal IFN-γ expression by CD8 T cells during chronic infection, as mice depleted of CD4 T cells have a reduction in IFN-γ producing CD8 effector cells, a phenomenon also observed during persistent M. tuberculosis infection [102]. It is clear that CD4 T cells play multiple, essential roles during Salmonella infection and targeting these cells with future vaccines would likely provide the greatest benefit. In fact, it has been shown that a single CD4 T cell peptide epitope derived from a Salmonella secreted effector protein (SseI) is capable of offering a significant level of protection against acute infection [103].
2.3 Organ-specific infection and immunity
While much attention has been paid to the various roles of individual cell types during Salmonella infection, less is known about how these cells might contribute or potentially differ functionally in the distinct anatomical tissues where Salmonella infection occurs. It has been shown Salmonella attacks the intestinal epithelial layer at the antigen-sampling microfold (M) cells. Subsequently, Salmonella encounter dendritic cells (DCs) and macrophages, followed by an influx of neutrophils, monocytes and more macrophages [57]. These phagocytic cells serve as the portals of entry and bacterial dissemination to more distant anatomical sites [104–107]. Some studies have shown that Salmonella infection of the intestine can occur independently of M cells and that this may be attributable to a direct luminal bacterial interaction with phagocytes such as DCs [108,109]. This allows for a rapid, systemic dissemination and may bypass M cell-induced inflammation. Less is known regarding the role for intestinal mucosal associated invariant T (MAIT) cells although it was recently shown that human B cells infected with S. Typhi can activate MR1-restricted CD8+ MAIT cells in a primary human cell culture system [110]. Notably, a more recent study showed that human volunteers orally infected with S. Typhi who were susceptible to the development of typhoid fever, had a drop in activated, peripheral blood MAIT cells, and these cells expressed the intestinal homing marker CCR9 [111]. This contrasted with volunteers who weren’t susceptible to disease where MAIT cell numbers fluctuated around normal and did not appear as activated. This details a possible contribution of these cells in combatting Salmonella infection in humans. In another study, Salmonella secreted effectors drove inflammasome production of IL-18, which lead to NK cell recruitment and perforin production, inducing potent, early inflammation at the intestinal site of infection [112]. Intriguingly, this is a new potential role for gut-homing NK cells which may contribute to the inflammation-induced diarrhea seen during the early stages of infection.
Following intestinal infection in mice, Salmonella are quickly trafficked to the MLN [113]. It has been demonstrated that the MLN are a major site of persistent infection in the chronic mouse model and that control of bacterial dissemination is dependent on IFN-γ production in the MLN [47]. Likewise, removal of the MLNs has been shown to correlate with increased bacterial burdens and severe pathology in systemic organs of infected mice [105,114]. Control of bacterial replication in the MLN during persistent infection is dependent on CD4 T cell maintenance and further, long-term preservation of these Salmonella-specific CD4 T cells requires continuous peptide:MHCII stimulation with Salmonella antigens [98]. The spleen is the other major lymphoid organ involved during the systemic phase of infection, which may be due to the presence of multiple subsets of splenic phagocytic cells, where Salmonella would likely reside. While the spleen is probably infected temporally after the MLNs, the immune response is phenotypically similar, with Salmonella-specific CD4 T cells producing anti-microbial cytokines, likely contributing to the control of the bacteria [99]. Why bacteria are not cleared by the potent immune response during these persistent infections is not clear and is worthy of further study.
While multiple mouse models of infection have shown that the MLNs are the most likely site of persistent infection, the story is less clear for the human disease. Indeed, it seems that the hepatobiliary tract and gallbladder are survival niches in humans [19]. This observation is supported by data collected from S. Typhi positive cadavers, showing S. Typhi present in 85.7% of liver tissues and may contribute to chronic carriage of bacteria [24,115]. Furthermore, chronic carriage of Salmonella Typhi is a risk factor for gallbladder carcinomas [25]. It is also clear that Salmonella does disseminate to the liver in mice, where they reside in multiple cell types, particularly Kupffer cells, liver sinusoidal endothelial cells and hepatocytes [116–118]. Kupffer cells (liver-resident macrophages) are numerically the largest distinct population of macrophages in the body. In the liver, they combine scavenger functions with immune homeostasis. They can also inhibit T cell activation through the direct production of PGE2 and IL-10, and by triggering regulatory T cells to produce IL-10 [119,120]. Kupffer cells have likewise been shown to have a role in attenuating infection-induced liver immunopathology during viral infection [121]; however, this phenomenon has not been demonstrated for bacterial infections. Furthermore, inflammatory Th1 effector CD4 T cells, known to be important in lymphoid tissues for controlling Salmonella infection, lose the capacity to produce cytokines, but do not deviate from their Th1 programming upon entering the liver [122]. While not known currently, it’s possible the immunosuppressive nature of the liver prevents Salmonella clearance during infection by attenuating essential CD4 T cell responses. The gallbladder also provides a potential niche for Salmonella infection; however, the immune response to infection in this organ is almost completely unknown and understanding what type of immunity occurs here will be essential to understanding persistent Salmonella pathogenesis.
Other tissues appear to be involved in some capacity during Salmonella infection. It is known that the bone marrow harbors S. Typhi during infection [123]. Other studies have shown that systemic Salmonella infection alters the phenotype and function of hematopoietic progenitors, possibly contributing to a dysregulated immune response to persistent or recurrent infection [124,125]. Some infections, particularly viral, infect the thymus and affect thymus output. As the thymus is the primary site of T cell maturation, this is likely an evolutionary adaptation that attenuates the T cell response against the thymus-infecting pathogen. Interestingly, while Salmonella also infects the thymus and induces thymic atrophy, mature CD4 T cell output is largely unaffected [126]. Conversely, double negative (DN; CD4-CD8-) thymocyte populations do appear to be reduced during both acute and chronic thymic infection by Salmonella [127]. In this same study, single positive (SP; CD4+ or CD8+) numbers were largely unaffected; however, it did appear these cells were somewhat more immature, possibly because they had only recently committed to the SP lineage to compensate for the loss of DN cells. While these findings are potentially conflicting, it demonstrates that the thymus has a remarkable capacity to adjust to infection induced atrophy to maintain thymic output. The implication of these findings is that T cells output is essential for control of Salmonella infection; however, this remains to be explored and would be an important finding for the role of T cells recently released from the thymus during this infection.
3. Salmonella exploitation and subversion of the immune response
3.1 Type III secretion system modulation of the anti-Salmonella immune response
To invade and disseminate, Salmonella makes use of Type-III secretion systems (T3SS) encoded on two pathogenicity islands (hereafter SPI-1 and SPI-2), which serve different functions. Once the bacteria have reached sufficient numbers in the gut, by outcompeting the gut flora, they utilize their first T3SS to facilitate entry into the M cells. Effectors secreted by T3SS/SPI-1 and T3SS/SPI-2 induce changes in host cells, such as rearrangement of the cytoskeleton and cell membrane and disconnection of epithelial cell junctions, which enable Salmonella invasion, particularly in macrophages [128,129]. Once phagocytosed, Salmonella survives acidification of the SCV and utilizes the second T3SS to inject SPI-2 effector proteins into the cell cytoplasm where these effectors mature the vacuole to allow for intracellular replication [130–132]. Some T3SS products directly modify immune cell function from within. For example, the T3SS/SPI-2 Salmonella secreted effector I (SseI) inhibits DC migration to the spleen which in turn impedes bacterial clearance [133]. Interestingly, a recent strain of iNTS, which is endemic to sub-Sarahan Africa, was recently shown to hyperdisseminate into systemic organs of mice, correlating with the natural pseudogenization of SseI, rendering this effector untranslatable during infection [134]. This ascribes a direct effect of the T3SS on bacterial dissemination through immune cell modulation. Notably, S. Typhi, which also causes systemic disease in humans lacks expression of SseI [135]. Perhaps evolutionary pressure has induced this effector loss in S. Typhi to allow for greater organ distribution via infected DCs. Further comparison of iNTS and typhoidal bacterial serovars may provide further insight into what effectors play a role in this type of disseminated disease.
3.2 Other Salmonella adaptations for avoidance or modulation of immunity
In addition to the T3SS, Salmonella has evolved other mechanisms to survive and create a niche during infection. While it is known Salmonella infects phagocytic cells, recent work has revealed that during a persistent infection, Salmonella can invade a specialized subset of cells known as a hemophagocytic macrophages, which engulf erythrocytes during infection [136]. Salmonella can influence these macrophages to increase erythrocyte engulfment as infection progresses, allowing the bacteria to acquire ferrous iron from the engulfed red blood cells [137,138]. This is a remarkable adaptation to the intracellular niche where the very cell that would be expected to destroy the invading organism instead provides essential nutrients. In a twist on the Salmonella/macrophage interaction, a recent study demonstrated that macrophages lacking TLRs led to, paradoxically, reduced bacterial virulence and mice lacking multiple TLRs were less susceptible to infection [139]. It appears Salmonella requires cues from the innate immune response normally induced through TLR ligation, such as acidification of the SCV, to allow the bacteria to recognize that they have entered the intracellular compartment and should begin replication. Other work has shown that Salmonella in the SCV is a heterogeneous population and acidification of the vacuole induces the formation of a group of non-replicating persister bacteria that may provide a potential pool of Salmonella for long term persistence in host macrophages [140].
As previously discussed, Salmonella seems well suited for gallbladder colonization in both humans and mice. In support of this, 3.5% of cholecystectomy (gallbladder removal) patients in S. Typhi endemic areas harbored antibiotic resistant Salmonella in the bile or gallbladder [141]. It is possible that some of these bacteria reside in gallbladder epithelial tissues, as has been shown in mice [142]; however, nearly 90% of people persistently infected with S. Typhi also have gallstones, indicating that these may provide a protected niche for Salmonella persistence [143]. Indeed, in a mouse model of persistent Salmonella infection fed a lithogenic diet where cholesterol gallstones develop, the bacteria form stable biofilms on these gallstones which potentially protects them against antibiotic treatment and the host immune response [144]. More strikingly, in a typhoid endemic region, some gallstones removed from people displayed S. Typhi biofilms, while gallstones removed from patients with E. coli infection had no biofilm formation [144]. Biofilms are notoriously difficult to eradicate with antibiotics and so these findings reveal that Salmonella may have found the perfect niche for persistence [145].
4. Concluding remarks
Salmonella infection and the resulting immune response is multifaceted, especially given the systemic nature of some infections, where different tissues are likely to display unique immunity to infection. This is made more complex by the fact that different Salmonella infections can vary from self-limiting gastroenteritis to invasive systemic disease to a systemic, but persistently infected asymptomatic carrier state. A growing recognition of these different types of infections and the resulting immune responses has revealed intriguing mechanisms employed by the immune system to control or eliminate infection and equally interesting means used by Salmonella to avoid immune recognition or effector functions. As this review highlights, the field has made great strides in our understanding of these host pathogen interactions. Future studies will hopefully further explore the interplay between immunity and bacteria in different infected organs, such as the liver, gallbladder, and gut. Such studies may have the potential to reveal novel treatments for hard to clear infections and may provide insights into how other systemic bacterial infections (i.e M. tuberculosis or H. pylori) interact with the immune system.
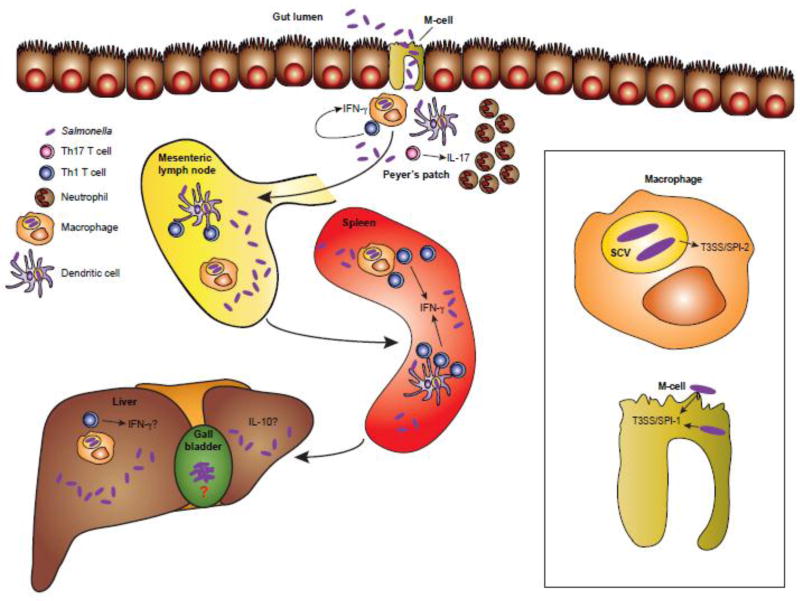
Figure 1. Overview of the immune response to Salmonella infection.
Salmonella bacteria enter via the intestine and use the T3SS/SPI-1 to induce uptake by the specialized M cells of the gut. Following translocation into the Peyer’s patches, Salmonella are engulfed by phagocytic cells such as macrophages, neutrophils monocytes, and DCs. Bacterial antigens are transported by DCs to the gut-draining MLNs where Salmonella-specific T cells are activated and traffic back to the intestine. These IFN-γ producing Th1 cells further activate macrophages while IL-17 producing Th17 cells recruit large numbers of neutrophils to combat infection. Salmonella use the T3SS/SPI-2 to inject effector proteins from within the SCV to modulate the immune response, such as preventing DC migration to lymph nodes. Th1 cells in the MLNs and spleen continue to activate antimicrobial macrophages to combat systemic infection in these organs. The liver is also colonized with bacteria and while it is possible that Th1 cells are important here, far less is known about the liver immune response to Salmonella infection; however, it is known that the liver tends towards an immunosuppressive, tolerant phenotype. Salmonella also infects the gallbladder where bacteria are known to persist as biofilms attached to gallstones. Almost nothing is known about the anti-Salmonella response in the gallbladder. Abbreviations: T3SS, type three secretion system; SPI, Salmonella pathogenicity island; DC, dendritic cell; MLN, mesenteric lymph node; Th, T helper; IFN, interferon; IL, interleukin; SCV, Salmonella containing vacuole.
Highlights.
Salmonella bacteria causes a different array of diseases and infection can be acute or chronic and can be limited to the intestine or distributed systemically
The immune response to systemic Salmonella infection is potent diverse and includes both innate and adaptive immune aspects
Salmonella uses multiple mechanisms to subvert or modulate the immune response directed against it
Acknowledgments
This work was supported by NIH grants R01 AI103343 and U01 AI124289 (to J.B.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interests.
References
- 1.Pui CF, Wong WC, Chai LC, Robin T, Ponniah J, Mohd Sahroni NH, et al. Salmonella: a foodborne pathogen. International Food Research Journal. 2011;18:465–473. [Google Scholar]
- 2.Angulo FJ, Mølbak K. Human Health Consequences of Antimicrobial Drug—Resistant Salmonella and Other Foodborne Pathogens. Clin. Infect. Dis. 2005;41:1613–1620. doi: 10.1086/497599. [DOI] [PubMed] [Google Scholar]
- 3.Coburn B, Grassl GA, Finlay BB. Salmonella, the host and disease: a brief review. Immunol Cell Biol. 2007;85:112–118. doi: 10.1038/sj.icb.7100007. [DOI] [PubMed] [Google Scholar]
- 4.McGovern VJ, Slavutin LJ. Pathology of salmonella colitis. Am. J. Surg. Pathol. 1979;3:483–490. doi: 10.1097/00000478-197912000-00001. [DOI] [PubMed] [Google Scholar]
- 5.Boyd JF. Pathology of the alimentary tract in Salmonella typhimurium food poisoning. Gut. 1985;26:935–944. doi: 10.1136/gut.26.9.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crum-Cianflone NF. Salmonellosis and the gastrointestinal tract: More than just peanut butter. Curr Gastroenterol Rep. 2008;10:424–431. doi: 10.1007/s11894-008-0079-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gal-Mor O, Boyle EC, Grassl GA. Same species, different diseases: how and why typhoidal and non-typhoidal Salmonella enterica serovars differ. Front Microbiol. 2014;5:391. doi: 10.3389/fmicb.2014.00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jean S-S, Wang J-Y, Hsueh P-R. Bacteremia caused by Salmonella enterica serotype Choleraesuis in Taiwan. J Microbiol Immunol Infect. 2006;39:358–365. [PubMed] [Google Scholar]
- 9.Fang FC, Fierer J. Human infection with Salmonella dublin. Medicine (Baltimore) 1991;70:198–207. doi: 10.1097/00005792-199105000-00004. [DOI] [PubMed] [Google Scholar]
- 10.Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA. Invasive non-typhoidal salmonella disease: an emerging and neglected tropical disease in Africa. Lancet. 2012;379:2489–2499. doi: 10.1016/S0140-6736(11)61752-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Almeida F, Seribelli AA, da Silva P, Medeiros MIC, Dos Prazeres Rodrigues D, Moreira CG, et al. Multilocus sequence typing of Salmonella Typhimurium reveals the presence of the highly invasive ST313 in Brazil. Infect. Genet. Evol. 2017;51:41–44. doi: 10.1016/j.meegid.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 12.Duggan MB, Beyer L. Enteric fever in young Yoruba children. Arch. Dis. Child. 1975;50:67–71. doi: 10.1136/adc.50.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morpeth SC, Ramadhani HO, Crump JA. Invasive non-Typhi Salmonella disease in Africa. Clin. Infect. Dis. 2009;49:606–611. doi: 10.1086/603553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinson NA, Karstaedt A, Venter WDF, Omar T, King P, Mbengo T, et al. Causes of death in hospitalized adults with a premortem diagnosis of tuberculosis: an autopsy study. Aids. 2007;21:2043–2050. doi: 10.1097/QAD.0b013e3282eea47f. [DOI] [PubMed] [Google Scholar]
- 15.Gordon MA, Banda HT, Gondwe M, Gordon SB, Boeree MJ, Walsh AL, et al. Non-typhoidal salmonella bacteraemia among HIV-infected Malawian adults: high mortality and frequent recrudescence. Aids. 2002;16:1633–1641. doi: 10.1097/00002030-200208160-00009. [DOI] [PubMed] [Google Scholar]
- 16.Gordon MA, Kankwatira AMK, Mwafulirwa G, Walsh AL, Hopkins MJ, Parry CM, et al. Invasive non-typhoid salmonellae establish systemic intracellular infection in HIV-infected adults: an emerging disease pathogenesis. Clin. Infect. Dis. 2010;50:953–962. doi: 10.1086/651080. [DOI] [PubMed] [Google Scholar]
- 17.Bumann D. Identification of Protective Antigens for Vaccination against Systemic Salmonellosis. Front Immunol. 2014;5:381. doi: 10.3389/fimmu.2014.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crump JA, Luby SP, Mintz ED. The global burden of typhoid fever. Bull. World Health Organ. 2004;82:346–353. [PMC free article] [PubMed] [Google Scholar]
- 19.Gunn JS, Marshall JM, Baker S, Dongol S, Charles RC, Ryan ET. Salmonella chronic carriage: epidemiology, diagnosis, and gallbladder persistence. Trends Microbiol. 2014;22:648–655. doi: 10.1016/j.tim.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parry CM, Dougan G. Typhoid fever. N Engl J Med. 2002;347:1770–1782. doi: 10.1056/NEJMra020201. [DOI] [PubMed] [Google Scholar]
- 22.Waddington CS, Darton TC, Pollard AJ. The challenge of enteric fever. J. Infect. 2014;68(Suppl 1):S38–50. doi: 10.1016/j.jinf.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 23.Edsall G, Gaines S, Landy M, Tigertt WD, Sprinz H, Trapani RJ, et al. Studies on infection and immunity in experimental typhoid fever. I. Typhoid fever in chimpanzees orally infected with Salmonella typhosa. J Exp Med. 1960;112:143–166. doi: 10.1084/jem.112.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nath G, Singh YK, Maurya P, Gulati AK, Srivastava RC, Tripathi SK. Does Salmonella Typhi primarily reside in the liver of chronic typhoid carriers? J Infect Dev Ctries. 2010;4:259–261. doi: 10.3855/jidc.820. [DOI] [PubMed] [Google Scholar]
- 25.Nagaraja V, Eslick GD. Systematic review with meta-analysis: the relationship between chronic Salmonella typhi carrier status and gall-bladder cancer. Aliment. Pharmacol. Ther. 2014;39:745–750. doi: 10.1111/apt.12655. [DOI] [PubMed] [Google Scholar]
- 26.Nath G, Pratap CB, Patel SK, Gulati AK, Tripathi SK. Isolation of Salmonella typhi from apparently healthy liver. Infect. Genet. Evol. 2011;11:2103–2105. doi: 10.1016/j.meegid.2011.08.029. [DOI] [PubMed] [Google Scholar]
- 27.Rowe B, Ward LR, Threlfall EJ. Multidrug-resistant Salmonella typhi: a worldwide epidemic. Clin. Infect. Dis. 1997;24(Suppl 1):S106–9. doi: 10.1093/clinids/24.supplement_1.s106. [DOI] [PubMed] [Google Scholar]
- 28.Chiou C-S, Alam M, Kuo J-C, Liu Y-Y, Wang P-J. Chromosome-mediated multidrug resistance in Salmonella enterica serovar Typhi. Antimicrob. Agents Chemother. 2015;59:721–723. doi: 10.1128/AAC.04081-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tatavarthy A, Luna VA, Amuso PT. How multidrug resistance in typhoid fever affects treatment options. Annals of the New York Academy of Sciences. 2014;1323:76–90. doi: 10.1111/nyas.12490. [DOI] [PubMed] [Google Scholar]
- 30.Qamar FN, Azmatullah A, Kazi AM, Khan E, Zaidi AKM. A three-year review of antimicrobial resistance of Salmonella enterica serovars Typhi and Paratyphi A in Pakistan. J Infect Dev Ctries. 2014;8:981–986. doi: 10.3855/jidc.3817. [DOI] [PubMed] [Google Scholar]
- 31.Rahman BA, Wasfy MO, Maksoud MA, Hanna N, Dueger E, House B. Multi-drug resistance and reduced susceptibility to ciprofloxacin among Salmonella enterica serovar Typhi isolates from the Middle East and Central Asia. New Microbes New Infect. 2014;2:88–92. doi: 10.1002/nmi2.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang S, Kingsley RA, Santos RL, Andrews-Polymenis H, Raffatellu M, Figueiredo J, et al. Molecular pathogenesis of Salmonella enterica serotype typhimurium-induced diarrhea. Infect Immun. 2003;71:1–12. doi: 10.1128/IAI.71.1.1-12.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee CA, Silva M, Siber AM, Kelly AJ, Galyov E, McCormick BA. A secreted Salmonella protein induces a proinflammatory response in epithelial cells, which promotes neutrophil migration. Proc Natl Acad Sci USA. 2000;97:12283–12288. doi: 10.1073/pnas.97.22.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loetscher Y, Wieser A, Lengefeld J, Kaiser P, Schubert S, Heikenwalder M, et al. Salmonella transiently reside in luminal neutrophils in the inflamed gut. PLoS ONE. 2012;7:e34812. doi: 10.1371/journal.pone.0034812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bodhidatta L, Taylor DN, Thisyakorn U, Echeverria P. Control of typhoid fever in Bangkok, Thailand, by annual immunization of schoolchildren with parenteral typhoid vaccine. Rev. Infect. Dis. 1987;9:841–845. doi: 10.1093/clinids/9.4.841. [DOI] [PubMed] [Google Scholar]
- 36.Anwar E, Goldberg E, Fraser A, Acosta CJ. Vaccines for preventing typhoid fever - Cochrane Database of Systematic Reviews - Anwar - Wiley Online Library. The Cochrane. 2014 doi: 10.1002/14651858.CD001261.pub3/full. [DOI] [PubMed] [Google Scholar]
- 37.Waddington CS, Darton TC, Woodward WE, Angus B, Levine MM, Pollard AJ. Advancing the management and control of typhoid fever: a review of the historical role of human challenge studies. J. Infect. 2014;68:405–418. doi: 10.1016/j.jinf.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 38.Guzman CA, Borsutzky S, Griot-Wenk M, Metcalfe IC, Pearman J, Collioud A, et al. Vaccines against typhoid fever. Vaccine. 2006;24:3804–3811. doi: 10.1016/j.vaccine.2005.07.111. [DOI] [PubMed] [Google Scholar]
- 39.Lin FY, Ho VA, Khiem HB, Trach DD, Bay PV, Thanh TC, et al. The efficacy of a Salmonella typhi Vi conjugate vaccine in two-to-five-year-old children. N. Engl. J. Med. 2001;344:1263–1269. doi: 10.1056/NEJM200104263441701. [DOI] [PubMed] [Google Scholar]
- 40.Hu X, Chen Z, Xiong K, Wang J, Rao X, Cong Y. Vi capsular polysaccharide: Synthesis, virulence, and application. Crit. Rev. Microbiol. 2017;43:440–452. doi: 10.1080/1040841X.2016.1249335. [DOI] [PubMed] [Google Scholar]
- 41.Arndt MB, Mosites EM, Tian M, Forouzanfar MH, Mokhdad AH, Meller M, et al. Estimating the burden of paratyphoid a in Asia and Africa. PLoS Negl Trop Dis. 2014;8:e2925. doi: 10.1371/journal.pntd.0002925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santos RL, Zhang S, Tsolis RM, Kingsley RA, Adams LG, Baumler AJ. Animal models of Salmonella infections: enteritis versus typhoid fever. Microbes Infect. 2001;3:1335–1344. doi: 10.1016/s1286-4579(01)01495-2. [DOI] [PubMed] [Google Scholar]
- 43.Raffatellu M, Santos RL, Verhoeven DE, George MD, Wilson RP, Winter SE, et al. Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat Med. 2008;14:421–428. doi: 10.1038/nm1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barthel M, Hapfelmeier S, Quintanilla-Martínez L, Kremer M, Rohde M, Hogardt M, et al. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun. 2003;71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rivera-Chávez F, Zhang LF, Faber F, Lopez CA, Byndloss MX, Olsan EE, et al. Depletion of Butyrate-Producing Clostridia from the Gut Microbiota Drives an Aerobic Luminal Expansion of Salmonella. Cell Host Microbe. 2016;19:443–454. doi: 10.1016/j.chom.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Govoni G, Vidal S, Gauthier S, Skamene E, Malo D, Gros P. The Bcg/Ity/Lsh locus: genetic transfer of resistance to infections in C57BL/6J mice transgenic for the Nramp1 Gly169 allele. Infect Immun. 1996;64:2923–2929. doi: 10.1128/iai.64.8.2923-2929.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Monack DM, Bouley DM, Falkow S. Salmonella typhimurium persists within macrophages in the mesenteric lymph nodes of chronically infected Nramp1+/+ mice and can be reactivated by IFNgamma neutralization. J Exp Med. 2004;199:231–241. doi: 10.1084/jem.20031319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mathur R, Oh H, Zhang D, Park S-G, Seo J, Koblansky A, et al. A mouse model of salmonella typhi infection. Cell. 2012;151:590–602. doi: 10.1016/j.cell.2012.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song J, Wilhelm CL, Wangdi T, Maira-Litran T, Lee S-J, Raetz M, et al. Absence of TLR11 in Mice Does Not Confer Susceptibility to Salmonella Typhi. Cell. 2016;164:827–828. doi: 10.1016/j.cell.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mathur R, Zeng W, Hayden MS, Ghosh S. Mice Lacking TLR11 Exhibit Variable Salmonella typhi Susceptibility. Cell. 2016;164:829–830. doi: 10.1016/j.cell.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 51.Song J, Willinger T, Rongvaux A, Eynon EE, Stevens S, Manz MG, et al. A mouse model for the human pathogen Salmonella typhi. Cell Host Microbe. 2010;8:369–376. doi: 10.1016/j.chom.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Firoz Mian M, Pek EA, Chenoweth MJ, Ashkar AA. Humanized mice are susceptible to Salmonella typhi infection. Cell. Mol. Immunol. 2011;8:83–87. doi: 10.1038/cmi.2010.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Libby SJ, Brehm MA, Greiner DL, Shultz LD, McClelland M, Smith KD, et al. Humanized nonobese diabetic-scid IL2rgammanull mice are susceptible to lethal Salmonella Typhi infection. Proc. Natl. Acad. Sci. U.S.a. 2010;107:15589–15594. doi: 10.1073/pnas.1005566107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mian MF, Pek EA, Chenoweth MJ, Coombes BK, Ashkar AA. Humanized mice for Salmonella typhi infection: new tools for an old problem. Virulence. 2011;2:248–252. doi: 10.4161/viru.2.3.16133. [DOI] [PubMed] [Google Scholar]
- 55.Kaiser P, Hardt W-D. Salmonella typhimurium diarrhea: switching the mucosal epithelium from homeostasis to defense. Curr Opin Immunol. 2011;23:456–463. doi: 10.1016/j.coi.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 56.Rydström A, Wick MJ. Monocyte recruitment, activation, and function in the gut-associated lymphoid tissue during oral Salmonella infection. J Immunol. 2007;178:5789–5801. doi: 10.4049/jimmunol.178.9.5789. [DOI] [PubMed] [Google Scholar]
- 57.Tam MA, Rydström A, Sundquist M, Wick MJ. Early cellular responses to Salmonella infection: dendritic cells, monocytes, and more. Immunol Rev. 2008;225:140–162. doi: 10.1111/j.1600-065X.2008.00679.x. [DOI] [PubMed] [Google Scholar]
- 58.Conlan JW. Neutrophils prevent extracellular colonization of the liver microvasculature by Salmonella typhimurium. Infect Immun. 1996;64:1043–1047. doi: 10.1128/iai.64.3.1043-1047.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spees AM, Kingsbury DD, Wangdi T, Xavier MN, Tsolis RM, Bäumler AJ. Neutrophils are a source of gamma interferon during acute Salmonella enterica serovar Typhimurium colitis. Infect Immun. 2014;82:1692–1697. doi: 10.1128/IAI.01508-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kupz A, Scott TA, Belz GT, Andrews DM, Greyer M, Lew AM, et al. Contribution of Thy1+ NK cells to protective IFN-γ production during Salmonella typhimurium infections. Proc. Natl. Acad. Sci. U.S.a. 2013;110:2252–2257. doi: 10.1073/pnas.1222047110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winter SE, Thiennimitr P, Nuccio SP, Haneda T, Winter MG, Wilson RP, et al. Contribution of flagellin pattern recognition to intestinal inflammation during Salmonella enterica serotype typhimurium infection. Infect Immun. 2009;77:1904–1916. doi: 10.1128/IAI.01341-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Broz P, Newton K, Lamkanfi M, Mariathasan S, Dixit VM, Monack DM. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. Journal of Experimental Medicine. 2010;207:1745–1755. doi: 10.1084/jem.20100257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sierro F, Dubois B, Coste A, Kaiserlian D, Kraehenbuhl JP, Sirard JC. Flagellin stimulation of intestinal epithelial cells triggers CCL20-mediated migration of dendritic cells. Proc Natl Acad Sci USA. 2001;98:13722–13727. doi: 10.1073/pnas.241308598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nauciel C, Espinasse-Maes F. Role of gamma interferon and tumor necrosis factor alpha in resistance to Salmonella typhimurium infection. Infect Immun. 1992;60:450–454. doi: 10.1128/iai.60.2.450-454.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arai T, Hiromatsu K, Nishimura H, Kimura Y, Kobayashi N, Ishida H, et al. Effects of in vivo administration of anti-IL-10 monoclonal antibody on the host defence mechanism against murine Salmonella infection. Immunology. 1995;85:381–388. [PMC free article] [PubMed] [Google Scholar]
- 66.Lee K-S, Jeong E-S, Heo S-H, Seo J-H, Jeong D-G, Choi Y-K. IL-10 suppresses bactericidal response of macrophages against Salmonella Typhimurium. J. Microbiol. 2011;49:1050–1053. doi: 10.1007/s12275-011-1043-z. [DOI] [PubMed] [Google Scholar]
- 67.Everest P, Allen J, Papakonstantinopoulou A, Mastroeni P, Roberts M, Dougan G. Salmonella typhimurium infections in mice deficient in interleukin-4 production: role of IL-4 in infection-associated pathology. J Immunol. 1997;159:1820–1827. [PubMed] [Google Scholar]
- 68.Mizuno Y, Takada H, Nomura A, Jin C-H, Hattori H, Ihara K, et al. Th1 and Th1-inducing cytokines in Salmonella infection. Clinical and Experimental Immunology. 2003;131:111–117. doi: 10.1046/j.1365-2249.2003.02060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stoycheva M, Murdjeva M. Serum levels of interferon-gamma, interleukin-12, tumour necrosis factor-alpha, and interleukin-10, and bacterial clearance in patients with gastroenteric Salmonella infection. Scand. J. Infect. Dis. 2005;37:11–14. doi: 10.1080/00365540410026068. [DOI] [PubMed] [Google Scholar]
- 70.Wahid R, Salerno-Gonçalves R, Tacket CO, Levine MM, Sztein MB. Cell-mediated immune responses in humans after immunization with one or two doses of oral live attenuated typhoid vaccine CVD 909. Vaccine. 2007;25:1416–1425. doi: 10.1016/j.vaccine.2006.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mastroeni P, Ugrinovic S, Chandra A, MacLennan C, Doffinger R, Kumararatne D. Resistance and susceptibility to Salmonella infections: lessons from mice and patients with immunodeficiencies. Reviews in Medical Microbiology. 2003;14 [Google Scholar]
- 72.Eckmann L, Kagnoff MF. Cytokines in host defense against Salmonella. Microbes Infect. 2001;3:1191–1200. doi: 10.1016/s1286-4579(01)01479-4. [DOI] [PubMed] [Google Scholar]
- 73.Mastroeni P, Chabalgoity JA, Dunstan SJ, Maskell DJ, Dougan G. Salmonella: immune responses and vaccines. Vet. J. 2001;161:132–164. doi: 10.1053/tvjl.2000.0502. [DOI] [PubMed] [Google Scholar]
- 74.Marmion DE, Naylor GR, Stewart IO. Second attacks of typhoid fever. J Hyg (Lond) 1953;51:260–267. doi: 10.1017/s0022172400015680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Forrest BD, LaBrooy JT, Beyer L, Dearlove CE, Shearman DJ. The human humoral immune response to Salmonella typhi Ty21a. J. Infect. Dis. 1991;163:336–345. doi: 10.1093/infdis/163.2.336. [DOI] [PubMed] [Google Scholar]
- 76.Acharya IL, Lowe CU, Thapa R, Gurubacharya VL, Shrestha MB, Cadoz M, et al. Prevention of typhoid fever in Nepal with the Vi capsular polysaccharide of Salmonella typhi. A preliminary report. N. Engl. J. Med. 1987;317:1101–1104. doi: 10.1056/NEJM198710293171801. [DOI] [PubMed] [Google Scholar]
- 77.Sztein MB, Salerno-Gonçalves R, McArthur MA. Complex adaptive immunity to enteric fevers in humans: lessons learned and the path forward. Front Immunol. 2014;5:516. doi: 10.3389/fimmu.2014.00516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saxén H. Mechanism of the protective action of anti-Salmonella IgM in experimental mouse salmonellosis. J. Gen. Microbiol. 1984;130:2277–2283. doi: 10.1099/00221287-130-9-2277. [DOI] [PubMed] [Google Scholar]
- 79.Marecki NM, Hsu HS, Mayo DR. Cellular and humoral aspects of host resistance in murine salmonellosis. Br J Exp Pathol. 1975;56:231–243. [PMC free article] [PubMed] [Google Scholar]
- 80.Mastroeni P, Villarreal-Ramos B, Hormaeche CE. Adoptive transfer of immunity to oral challenge with virulent salmonellae in innately susceptible BALB/c mice requires both immune serum and T cells. Infect Immun. 1993;61:3981–3984. doi: 10.1128/iai.61.9.3981-3984.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mittrücker HW, Raupach B, Köhler A, Kaufmann SH. Cutting edge: role of B lymphocytes in protective immunity against Salmonella typhimurium infection. J Immunol. 2000;164:1648–1652. doi: 10.4049/jimmunol.164.4.1648. [DOI] [PubMed] [Google Scholar]
- 82.Nanton MR, Way SS, Shlomchik MJ, McSorley SJ. Cutting edge: B cells are essential for protective immunity against Salmonella independent of antibody secretion. J Immunol. 2012;189:5503–5507. doi: 10.4049/jimmunol.1201413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jones BD, Falkow S. Salmonellosis: host immune responses and bacterial virulence determinants. Annu Rev Immunol. 1996;14:533–561. doi: 10.1146/annurev.immunol.14.1.533. [DOI] [PubMed] [Google Scholar]
- 84.Janeway CA, Ron J, Katz ME. The B cell is the initiating antigen-presenting cell in peripheral lymph nodes. J Immunol. 1987;138:1051–1055. [PubMed] [Google Scholar]
- 85.Matsuzaki G, Vordermeier HM, Hashimoto A, Nomoto K, Ivanyi J. The role of B cells in the establishment of T cell response in mice infected with an intracellular bacteria, Listeria monocytogenes. Cell Immunol. 1999;194:178–185. doi: 10.1006/cimm.1999.1503. [DOI] [PubMed] [Google Scholar]
- 86.McSorley SJ. Immunity to intestinal pathogens: lessons learned from Salmonella. Immunol Rev. 2014;260:168–182. doi: 10.1111/imr.12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nauciel C. Role of CD4+ T cells and T-independent mechanisms in acquired resistance to Salmonella typhimurium infection. J Immunol. 1990;145:1265–1269. [PubMed] [Google Scholar]
- 88.Johanns TM, Ertelt JM, Rowe JH, Way SS. Regulatory T cell suppressive potency dictates the balance between bacterial proliferation and clearance during persistent Salmonella infection. PLoS Pathog. 2010;6:e1001043. doi: 10.1371/journal.ppat.1001043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hess J, Ladel C, Miko D, Kaufmann SH. Salmonella typhimurium aroA-infection in gene-targeted immunodeficient mice: major role of CD4+ TCR-alpha beta cells and IFN-gamma in bacterial clearance independent of intracellular location. J Immunol. 1996;156:3321–3326. [PubMed] [Google Scholar]
- 90.Pham OH, McSorley SJ. Protective host immune responses to Salmonella infection. Future Microbiol. 2015;10:101–110. doi: 10.2217/fmb.14.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee S-J, Dunmire S, McSorley SJ. MHC class-I-restricted CD8 T cells play a protective role during primary Salmonella infection. Immunol Lett. 2012;148:138–143. doi: 10.1016/j.imlet.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sad S, Dudani R, Gurnani K, Russell M, van Faassen H, Finlay B, et al. Pathogen proliferation governs the magnitude but compromises the function of CD8 T cells. J Immunol. 2008;180:5853–5861. doi: 10.4049/jimmunol.180.9.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Patel R, Sad S. Transcription factor Batf3 is important for development of CD8+ T-cell response against a phagosomal bacterium regardless of the location of antigen. Immunol Cell Biol. 2016;94:378–387. doi: 10.1038/icb.2015.98. [DOI] [PubMed] [Google Scholar]
- 94.Uche IV, MacLennan CA, Saul A. A Systematic Review of the Incidence, Risk Factors and Case Fatality Rates of Invasive Nontyphoidal Salmonella (iNTS) Disease in Africa (1966 to 2014) PLoS Negl Trop Dis. 2017;11:e0005118. doi: 10.1371/journal.pntd.0005118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Keddy KH, Takuva S, Musekiwa A, Puren AJ, Sooka A, Karstaedt A, et al. An association between decreasing incidence of invasive non-typhoidal salmonellosis and increased use of antiretroviral therapy, Gauteng Province, South Africa, 2003–2013. PLoS ONE. 2017;12:e0173091. doi: 10.1371/journal.pone.0173091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Blanchette J, Jaramillo M, Olivier M. Signalling events involved in interferon-gamma-inducible macrophage nitric oxide generation. Immunology. 2003;108:513–522. doi: 10.1046/j.1365-2567.2003.01620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee S-J, McLachlan JB, Kurtz JR, Fan D, Winter SE, Bäumler AJ, et al. Temporal expression of bacterial proteins instructs host CD4 T cell expansion and th17 development. PLoS Pathog. 2012;8:e1002499. doi: 10.1371/journal.ppat.1002499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nelson RW, McLachlan JB, Kurtz JR, Jenkins MK. CD4+ T cell persistence and function after infection are maintained by low-level peptide:MHC class II presentation. J Immunol. 2013;190:2828–2834. doi: 10.4049/jimmunol.1202183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ravindran R, Foley J, Stoklasek T, Glimcher LH, McSorley SJ. Expression of T-bet by CD4 T cells is essential for resistance to Salmonella infection. J Immunol. 2005;175:4603–4610. doi: 10.4049/jimmunol.175.7.4603. [DOI] [PubMed] [Google Scholar]
- 100.VanCott JL, Chatfield SN, Roberts M, Hone DM, Hohmann EL, Pascual DW, et al. Regulation of host immune responses by modification of Salmonella virulence genes. Nat Med. 1998;4:1247–1252. doi: 10.1038/3227. [DOI] [PubMed] [Google Scholar]
- 101.Schulz SM, Köhler G, Schütze N, Knauer J, Straubinger RK, Chackerian AA, et al. Protective immunity to systemic infection with attenuated Salmonella enterica serovar enteritidis in the absence of IL-12 is associated with IL-23-dependent IL-22, but not IL-17. J Immunol. 2008;181:7891–7901. doi: 10.4049/jimmunol.181.11.7891. [DOI] [PubMed] [Google Scholar]
- 102.Bold TD, Ernst JD. CD4+ T cell-dependent IFN-γ production by CD8+ effector T cells in Mycobacterium tuberculosis infection. J Immunol. 2012;189:2530–2536. doi: 10.4049/jimmunol.1200994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kurtz JR, Petersen HE, Frederick DR, Morici LA, McLachlan JB. Vaccination with a Single CD4 T Cell Peptide Epitope from a Salmonella Type III-Secreted Effector Protein Provides Protection against Lethal Infection. Infect Immun. 2014;82:2424–2433. doi: 10.1128/IAI.00052-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vazquez-Torres A, Jones-Carson J, Baumler AJ, Falkow S, Valdivia R, Brown W, et al. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature. 1999;401:804–808. doi: 10.1038/44593. [DOI] [PubMed] [Google Scholar]
- 105.Griffin AJ, Li L-X, Voedisch S, Pabst O, McSorley SJ. Dissemination of persistent intestinal bacteria via the mesenteric lymph nodes causes typhoid relapse. Infect Immun. 2011;79:1479–1488. doi: 10.1128/IAI.01033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Worley MJ, Nieman GS, Geddes K, Heffron F. Salmonella typhimurium disseminates within its host by manipulating the motility of infected cells. Proc Natl Acad Sci USA. 2006;103:17915–17920. doi: 10.1073/pnas.0604054103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Watson KG, Holden DW. Dynamics of growth and dissemination of Salmonella in vivo. Cell Microbiol. 2010;12:1389–1397. doi: 10.1111/j.1462-5822.2010.01511.x. [DOI] [PubMed] [Google Scholar]
- 108.Martinoli C, Chiavelli A, Rescigno M. Entry route of Salmonella typhimurium directs the type of induced immune response. Immunity. 2007;27:975–984. doi: 10.1016/j.immuni.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 109.Niess JH, Brand S, Gu X, Landsman L, Jung S, McCormick BA, et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307:254–258. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- 110.Salerno-Gonçalves R, Rezwan T, Sztein MB. B cells modulate mucosal associated invariant T cell immune responses. Front Immunol. 2014;4:511. doi: 10.3389/fimmu.2013.00511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Salerno-Gonçalves R, Luo D, Fresnay S, Magder L, Darton TC, Jones C, et al. Challenge of Humans with Wild-type Salmonella enterica Serovar Typhi Elicits Changes in the Activation and Homing Characteristics of Mucosal-Associated Invariant T Cells. Front Immunol. 2017;8:398. doi: 10.3389/fimmu.2017.00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Müller AA, Dolowschiak T, Sellin ME, Felmy B, Verbree C, Gadient S, et al. An NK Cell Perforin Response Elicited via IL-18 Controls Mucosal Inflammation Kinetics during Salmonella Gut Infection. PLoS Pathog. 2016;12:e1005723. doi: 10.1371/journal.ppat.1005723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.McSorley SJ, Asch S, Costalonga M, Reinhardt RL, Jenkins MK. Tracking salmonella-specific CD4 T cells in vivo reveals a local mucosal response to a disseminated infection. Immunity. 2002;16:365–377. doi: 10.1016/s1074-7613(02)00289-3. [DOI] [PubMed] [Google Scholar]
- 114.Voedisch S, Koenecke C, David S, Herbrand H, Förster R, Rhen M, et al. Mesenteric lymph nodes confine dendritic cell-mediated dissemination of Salmonella enterica serovar Typhimurium and limit systemic disease in mice. Infect Immun. 2009;77:3170–3180. doi: 10.1128/IAI.00272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nath G, Mauryal P, Gulati AK, Singh TB, Srivastava R, Kumar K, et al. Comparison of Vi serology and nested PCR in diagnosis of chronic typhoid carriers in two different study populations in typhoid endemic area of India. Southeast Asian J. Trop. Med. Public Health. 2010;41:636–640. [PubMed] [Google Scholar]
- 116.Nnalue NA, Shnyra A, Hultenby K, Lindberg AA. Salmonella choleraesuis and Salmonella typhimurium associated with liver cells after intravenous inoculation of rats are localized mainly in Kupffer cells and multiply intracellularly. Infect Immun. 1992;60:2758–2768. doi: 10.1128/iai.60.7.2758-2768.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vazquez-Torres A, Vallance BA, Bergman MA, Finlay BB, Cookson BT, Jones-Carson J, et al. Toll-like receptor 4 dependence of innate and adaptive immunity to Salmonella: importance of the Kupffer cell network. J Immunol. 2004;172:6202–6208. doi: 10.4049/jimmunol.172.10.6202. [DOI] [PubMed] [Google Scholar]
- 118.Richter-Dahlfors A, Buchan AM, Finlay BB. Murine salmonellosis studied by confocal microscopy: Salmonella typhimurium resides intracellularly inside macrophages and exerts a cytotoxic effect on phagocytes in vivo. J Exp Med. 1997;186:569–580. doi: 10.1084/jem.186.4.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.You Q, Cheng L, Kedl RM, Ju C. Mechanism of T cell tolerance induction by murine hepatic Kupffer cells. Hepatology. 2008;48:978–990. doi: 10.1002/hep.22395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Breous E, Somanathan S, Vandenberghe LH, Wilson JM. Hepatic regulatory T cells and Kupffer cells are crucial mediators of systemic T cell tolerance to antigens targeting murine liver. Hepatology. 2009;50:612–621. doi: 10.1002/hep.23043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sitia G, Iannacone M, Aiolfi R, Isogawa M, van Rooijen N, Scozzesi C, et al. Kupffer cells hasten resolution of liver immunopathology in mouse models of viral hepatitis. PLoS Pathog. 2011;7:e1002061. doi: 10.1371/journal.ppat.1002061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Klugewitz K, Blumenthal-Barby F, Schrage A, Knolle PA, Hamann A, Crispe IN. Immunomodulatory effects of the liver: deletion of activated CD4+ effector cells and suppression of IFN-gamma-producing cells after intravenous protein immunization. J Immunol. 2002;169:2407–2413. doi: 10.4049/jimmunol.169.5.2407. [DOI] [PubMed] [Google Scholar]
- 123.Gasem MH, Keuter M, Dolmans WMV, Van Der Ven-Jongekrijg J, Djokomoeljanto R, van der Meer JWM. Persistence of Salmonellae in blood and bone marrow: randomized controlled trial comparing ciprofloxacin and chloramphenicol treatments against enteric fever. Antimicrob. Agents Chemother. 2003;47:1727–1731. doi: 10.1128/AAC.47.5.1727-1731.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hyland L, Villarreal-Ramos B, Clarke B, Baaten B, Hou S. Bone marrow immunosuppression in Salmonella-infected mice is prolonged following influenza virus infection. Exp. Hematol. 2005;33:1477–1485. doi: 10.1016/j.exphem.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 125.Ross EA, Flores-Langarica A, Bobat S, Coughlan RE, Marshall JL, Hitchcock JR, et al. Resolving Salmonella infection reveals dynamic and persisting changes in murine bone marrow progenitor cell phenotype and function. Eur J Immunol. 2014;44:2318–2330. doi: 10.1002/eji.201344350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ross EA, Coughlan RE, Flores-Langarica A, Lax S, Nicholson J, Desanti GE, et al. Thymic Function Is Maintained during Salmonella-Induced Atrophy and Recovery. J Immunol. 2012;189:4266–4274. doi: 10.4049/jimmunol.1200070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Majumdar S, Deobagkar-Lele M, Adiga V, Raghavan A, Wadhwa N, Ahmed SM, et al. Differential susceptibility and maturation of thymocyte subsets during Salmonella Typhimurium infection: insights on the roles of glucocorticoids and Interferon-gamma. Sci Rep. 2017;7:40793. doi: 10.1038/srep40793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.House D, Bishop A, Parry C, Dougan G, Wain J. Typhoid fever: pathogenesis and disease. Curr. Opin. Infect. Dis. 2001;14:573–578. doi: 10.1097/00001432-200110000-00011. [DOI] [PubMed] [Google Scholar]
- 129.Hensel M, Shea JE, Raupach B, Monack D, Falkow S, Gleeson C, et al. Functional analysis of ssaJ and the ssaK/U operon, 13 genes encoding components of the type III secretion apparatus of Salmonella Pathogenicity Island 2. Mol Microbiol. 1997;24:155–167. doi: 10.1046/j.1365-2958.1997.3271699.x. [DOI] [PubMed] [Google Scholar]
- 130.Steele-Mortimer O. The Salmonella-containing vacuole: moving with the times. Curr Opin Microbiol. 2008;11:38–45. doi: 10.1016/j.mib.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gorvel JP, Méresse S. Maturation steps of the Salmonella-containing vacuole. Microbes Infect. 2001;3:1299–1303. doi: 10.1016/s1286-4579(01)01490-3. [DOI] [PubMed] [Google Scholar]
- 132.Bakowski MA, Braun V, Brumell JH. Salmonella-containing vacuoles: directing traffic and nesting to grow. Traffic. 2008;9:2022–2031. doi: 10.1111/j.1600-0854.2008.00827.x. [DOI] [PubMed] [Google Scholar]
- 133.McLaughlin LM, Govoni GR, Gerke C, Gopinath S, Peng K, Laidlaw G, et al. The Salmonella SPI2 effector SseI mediates long-term systemic infection by modulating host cell migration. PLoS Pathog. 2009;5:e1000671. doi: 10.1371/journal.ppat.1000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Carden SE, Walker GT, Honeycutt J, Lugo K, Pham T, Jacobson A, et al. Pseudogenization of the Secreted Effector Gene sseI Confers Rapid Systemic Dissemination of S. Typhimurium ST313 within Migratory Dendritic Cells. Cell Host Microbe. 2017;21:182–194. doi: 10.1016/j.chom.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Miao EA, Miller SI. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc Natl Acad Sci USA. 2000;97:7539–7544. doi: 10.1073/pnas.97.13.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nix RN, Altschuler SE, Henson PM, Detweiler CS. Hemophagocytic macrophages harbor Salmonella enterica during persistent infection. PLoS Pathog. 2007;3:e193. doi: 10.1371/journal.ppat.0030193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pilonieta MC, Moreland SM, English CN, Detweiler CS. Salmonella enterica infection stimulates macrophages to hemophagocytose. MBio. 2014;5:e02211. doi: 10.1128/mBio.02211-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nagy TA, Moreland SM, Detweiler CS. Salmonella acquires ferrous iron from haemophagocytic macrophages. Mol Microbiol. 2014;93:1314–1326. doi: 10.1111/mmi.12739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Arpaia N, Godec J, Lau L, Sivick KE, McLaughlin LM, Jones MB, et al. TLR Signaling Is Required for Salmonella typhimurium Virulence. Cell. 2011;144:675–688. doi: 10.1016/j.cell.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Helaine S, Cheverton AM, Watson KG, Faure LM, Matthews SA, Holden DW. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science. 2014;343:204–208. doi: 10.1126/science.1244705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dongol S, Thompson CN, Clare S, Nga TVT, Duy PT, Karkey A, et al. The Microbiological and Clinical Characteristics of Invasive Salmonella in Gallbladders from Cholecystectomy Patients in Kathmandu. Nepal, PLoS ONE. 2012;7:e47342. doi: 10.1371/journal.pone.0047342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gonzalez-Escobedo G, Gunn JS. Gallbladder Epithelium as a Niche for Chronic Salmonella Carriage. Infect Immun. 2013;81:2920–2930. doi: 10.1128/IAI.00258-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Schiøler H, Christiansen ED, Høybye G, Rasmussen SN, Greibe J. Biliary calculi in chronic Salmonella carriers and healthy controls: a controlled study. Scand. J. Infect. Dis. 1983;15:17–19. doi: 10.3109/inf.1983.15.issue-1.04. [DOI] [PubMed] [Google Scholar]
- 144.Crawford RW, Rosales-Reyes R, de L.L. Ramírez-Aguilar M, Chapa-Azuela O, Alpuche-Aranda C, Gunn JS. Gallstones play a significant role in Salmonella spp. gallbladder colonization and carriage. Proc. Natl. Acad. Sci. U.S.a. 2010;107:4353–4358. doi: 10.1073/pnas.1000862107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Parsek MR, Singh PK. Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 2003;57:677–701. doi: 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]