Abstract
The oceans are a uniquely rich source of bioactive metabolites, of which sponges have been shown to be among the most prolific producers of diverse bioactive secondary metabolites with valuable therapeutic potential. Much attention has been focused on marine bioactive peptides due to their novel chemistry and diverse biological properties. As summarized in this review, marine peptides are known to exhibit various biological activities such as antiviral, anti-proliferative, antioxidant, anti-coagulant, anti-hypertensive, anti-cancer, antidiabetic, antiobesity, and calcium-binding activities. This review focuses on the chemistry and biology of peptides isolated from sponges, bacteria, cyanobacteria, fungi, ascidians, and other marine sources. The role of marine invertebrate microbiomes in natural products biosynthesis is discussed in this review along with the biosynthesis of modified peptides from different marine sources. The status of peptides in various phases of clinical trials is presented as well as the development of modified peptides including optimization of PK and bioavailability.
Keywords: Marine organisms, bioactive peptides, challenges, peptide isolation, biosynthesis, therapeutic peptides
I. INTRODUCTION
The oceans represent a vast resource for new bioactive natural products with utility in basic research, biomedical sciences, and the development of therapeutics. Marine organisms and in particular sponges produce an unprecedented variety of chemical classes with a wide range of biological activities and are considered the largest remaining reservoir of undiscovered natural molecules. Marine organisms produce unique molecules due to conditions that differ significantly from terrestrial environments which include aggressive, exigent, and competitive surroundings that lead to the production of potent active molecules [1].
Peptides are promising drug candidates based on the significance of the bioregulatory role of various endogenous peptides and the unique molecular mechanisms of action of bioactive peptides obtained from marine natural sources [2]. Complex cyclic and linear peptides discovered from marine sources have expanded our knowledge about ion-channels, antimicrobial agents, cytotoxic mechanisms of action, and other properties that has led to the introduction of marine peptides as novel and innovative therapeutics [1]. Trends in marine natural products chemistry has shifted toward the study of the microbiome that live in association with marine macroorganisms and the sustainable production of these compounds. In this review, we focus largely on the diversity of complex bioactive modified peptides isolated from various marine sources and the biological aspects of these molecules.
II. BIOACTIVE PEPTIDES FROM VARIOUS MARINE SOURCES
A. Sponges
Sponges belong to a large and diversified group of colonial organisms comprising the phylum Porifera. With thousands of various species distributed widely ranging from shallow estuaries to deep waters of the ocean, sponges are a source of varied and novel bioactive metabolites that include nucleoside derivatives, terpenoids, polyethers, alkaloids, and macrolides, in addition to modified peptides [3]. Sponges are an abundant source of bioactive and structurally diverse peptides that include linear peptides, depsipeptides, and cyclic peptides with the residue numbers spanning from two to forty eight. The diverse biological activities and the novel structural features of these metabolites have engendered considerable interest [4].
Most of the active peptides from sponges have unique structures which can either be linear or cyclic, possessing unusual amino acids that are rarely seen in terrestrial systems [1]. The tetradecapeptide discodermins were the first novel peptides isolated from sponges that showed cell growth inhibition [5]. Discodermins A [6], B, C, D [5], E [7], F, G, and H [8] isolated from the genus Discodermia are cytotoxic peptides possessing a chain of amino acids, some of which are common and others extremely rare, along with a macrocyclic ring. All of these molecules were found to be cytotoxic with IC50 values ranging from 0.02–20 μg/mL against P388 murine leukemia cells and A549 human lung cell-lines [1]. Kasumigamide is a tetrapeptide that was initially isolated from Microcystis aeruginosa, a freshwater cyanobacterium [9], but was later isolated from Discodermia calyx [10]. Kasumigamide is known to possess an N-terminal α-hydroxy acid and displayed antialgal activity at a minimum inhibitory concentration (MIC) of 2 μg/mL [9]. Previous studies reported the presence of a filamentous microorganism Entotheonella sp. in the lithistid sponge Discodermia species. Schmidt et al. [11] initially reported the genus Entotheonella in the δ-subdivision of ‘Candidatus Entotheonella palauensis,’ a Proteobacteria, isolated from the marine sponge Theonella swinhoei [12]. Exploration of Entotheonella symbionts in T. swinhoei revealed a large biosynthetic repertoire, including the potential for the production of polytheonamides, onnamide A, and theopederin A, along with cyclotheonamides, proteusins, nazumamide, and keramamides [13].
Phakellistatins are unique examples of proline-rich cycloheptapeptides that were isolated from Phakellia sp. where phakellistatin 1 was found to exhibit cell growth inhibition against P388 murine leukemia cell-line at ED50 of 7.5 μg/mL [14]. Phakellistatins 2 [15] and 14 [16] showed cell growth inhibitory activity against the murine P388 lymphocytic leukemia with ED50 values of 0.34 μg/mL and 5 μg/mL, respectively. Phakellistatin 3 and isophakellistatin 3 are two isomeric cyclo-heptapeptides of which phakellistatin 3 showed significant P388 inhibition at an ED50 of 0.33 μg/mL [17]. Phakellistatin 4 exhibited inhibition of L1210 leukemia, LNCAP lung carcinoma, KB human epidermoid carcinoma, and SK-OV-3 ovarian cancer cell-lines at concentrations of 31.6 μg/mL [18,19], while phakellistatin 5 showed GI50 value of 0.6 μM [20]. Phakellistatin 6 inhibited the growth of human cancer cell lines at GI50 values ranging between 0.1 to 0.01 μg/mL [21]. Phakellistatins 7–9 inhibited P388 cancer cell growth at ED50 values of 3.0, 2.9, and 4.1 μg/mL, respectively [22], while phakellistatins 10 and 11 exhibited inhibition at ED50 values of 2.1 and 0.2 μg/mL, respectively [23]. Phakellistatin 12 inhibited the P388 lymphocytic leukemia at an ED50 value of 2.8 μg/mL [24].
Phakellistatin 13 is a cyclic heptapeptide isolated from Phakellia fusca that showed significant cytotoxicity at an ED50 value of < 10−2 μg/mL against the BEL-7404 human hepatoma cell-line [25]. Phakellistatin 15 is a cyclic octapeptide with three proline moieties (in trans form) [26] that exhibited antitumor activity against P388 cell-line at an IC50 value of 8.5 μM, while phakellistatin 16 inhibited both P388 and BEL-7402 cell-lines at IC50 values of 5.4 and 14.3 μM, respectively [27]. Phakellistatins 17 and 18 exhibited no antitumor activity [27], while phakellistatin 19 showed antimitotic activity at IC50 values ranging between 84 to 420 nM [28].
Geodiamolides A-I are a group of cyclodepsipeptides isolated from Geodia sp., of which geodiamolides A and B showed antifungal activity against Candida albicans [29], while geodiamolide H exhibited in vitro cytotoxicity against a number of human cancer cell-lines [30]. Geodiamolides A-F are cytotoxic peptides isolated from Pseudaxinyssa sp., that showed cytotoxicity at 3.2, 2.6, 2.5, 39.0, 14.0, and 6.0 ng/mL, respectively [31]. Geodiamolides A, B, H, and I were anti-proliferative against breast-cancer cells through the disorganization of the actin filaments of T47D and MCF7 cancer cells [32]. Geodiamolides J-P, and R are cyclic depsipeptides isolated from the Cymbastela sp., while geodiamolide TA and neosiphoniamolide A were reported from Hemiastrella minor and Neosiphonia superstes, respectively. Geodiamolide G exhibited weak in vitro cytotoxicity against U373 human glioblastoma/astrocytoma and HEY human ovarian carcinoma cell-lines at IC50 values of 7.7 mg/mL and 8.6 mg/mL, respectively [33]. Jaspamide or jasplakinolide isolated from Jaspis johnstoni is a cyclic depsipeptide with a 15-carbon macrocyclic ring and three amino-acid residues. Jasplakinolide exhibited apoptosis in Jurkat T cells along with increased caspase-3 activity. The apoptosis induced by jaspamide is connected with reduced Bcl-2 protein expression and caspase-3 activation, along with enhanced Bax levels. Jaspamide is known to induce caspases-independent pathway of cell-death that is considered to be responsible for membrane and cytoplasmic changes in apoptosis cells, and a caspase-dependent cell death, responsible for PARP proteolysis [34]. Pipestelides A-C are cyclodepsipeptides isolated from Pipestela candelabra, and are non-ribosomal peptide synthetase - polyketide synthase (NRPS-PKS) hybrid macrolides that are biosynthetically related to jaspamide. Pipestelides AC hold a bromotyrosine [3-amino-3-(bromo-4-hydroxyphenyl)propanoic acid] unit, polypropionate with a double bond (Z), and 2-hydroxyquinolinone, respectively. Pipestelide A exhibited potent cytotoxicity at an IC50 value of 0.1 μM, while pipestelide B showed modest activity but these cytotoxicity’s were low compared to jaspamide [35].
Milnamide D, hemiasterlin, scleritodermin A, and diazonamide A are other marine peptides that exhibited potent tubulin-polymerization in various cancer-cells. Milnamides A and D isolated from the Cymbastela sp., were found to be potent against two colorectal cancer cell-lines: p53-deficient and HCT-116 cell-lines with IC50 values of 1.65 μM and 66.8 nM, respectively. Milnamide C was isolated from Auletta sp., and exhibited activity against MDA-MB-435 cancer cell-line at an IC50 value of 1.48 × 10−4 μg/mL along with microtubule cytoskeletal activity [36]. Milnamides A and D also inhibited tubulin polymerization at IC50 values of 6.02 μM and 16.90 μM, respectively [34,37,38]. Hemiasterlin, also known as milnamide B [36], is a natural tripeptide isolated from Hemiasterella minor, Cymbastela sp., Siphonochalina sp., and Auletta sp., and binds to the vinca peptide site in tubulin, resulting in the disruption of the normal microtubule dynamics and microtubule depolymerization. Hemiasterlin was also found to be potent against p53 and HCT-116 colorectal cancer cell-lines at an IC50 value of 6.8 nM [34,37]. Milnamides E-G and hemiasterlins A & D were isolated from Pipestela candelabra. Milnamides A-G exhibited cytotoxicity against prostate cancer (PC3) and human neonatal foreskin fibroblast non-cancer (NFF) cell-lines at IC50 values of 11.0 & 70.6 nM, 0.05 & 0.40 nM, 31.7 & 188 nM, 0.38 & 1.19 μM, 34.2 & 123 nM, 2.18 & 5.65 μM, and 2.87 & > 10 μM, respectively, while hemiasterlins A and D showed cytotoxicity against PC3 and NFF cancer cell-lines at IC50 values of 0.27 & 1.03 nM and 2.20 & 8.16 nM, respectively [39]. Scleritodermin A isolated from Scleritoderma nodosum, a lithistid sponge, is a cyclic peptide with potential in vitro cytotoxicity against human tumor cell-lines and an inhibitor of the tubulin-polymerization. Scleritodermin A was found to induce apoptosis at an IC50 of 1.3 μM, and was found to be cytotoxic at an IC50 value of < 2 μM [34,40,41].
Mirabamides isolated from Siliquariaspongia mirabilis showed potent inhibition against HIV-1 fusion. Among these, mirabamide A showed inhibition in HIV-1 fusion and neutralization assays with IC50 values of 140 and 40 nM, respectively, while mirabamides C and D exhibited lower potential with IC50 values between 1.3 μM & 140 nM and 3.9 μM & 190 nM, respectively. Mirabamides are known to inhibit HIV-1 at membrane fusion level, presumably from interactions with the HIV-1 envelope glycoproteins [42,43]. Mirabamides E-H are depsipeptides isolated from Stelletta clavosa and exhibited strong HIV-1 inhibition at IC50 values of 121, 62, 68, and 41 nM, respectively, in a neutralization assay [44].
Celebesides A-C and theopapuamides B-D were isolated from Siliquariaspongia mirabilis. Celebesides are exceptional cyclic depsipeptides that included a polyketide moiety along with five other amino-acid residues comprising a phosphoserine residue and an uncommon 3-carbamoyl threonine. Theopapuamides B-D are undecapeptides that possessed an N-terminal fatty acid moiety with two unreported amino acids: 4-amino-2,3-dihydroxy-5-methylhexanoic acid and 3-acetamido-2-aminopropanoic acid. Celebeside A inhibited HIV-1 in the neutralization assay at an IC50 value of 1.9 ± 0.4 μg/mL, while celebeside C was found to be inactive even at high concentrations (50 μg/mL). Theopapuamide A isolated from Theonella swinhoei and theopapuamides B-C isolated from S. mirabilis showed cytotoxicity against human colon HCT-116 carcinoma cell-line with IC50 values between 2.1 to 4.0 μg/mL along with strong antifungal activity against amphotericin-B resistant strains of Candida albicans at concentrations of 1–5 μg/disk [45].
Among the ten homophymines isolated from the Homophymia sp., homophymine A showed cytoprotective activity against HIV-1 infection with IC50 value of 75 nM [46]. Homophymines B-E and A1-E1 were other cyclodepsipeptides isolated from the same sponge and exhibited potent antiproliferative activity against a panel of human cancer cell-lines with IC50 values ranging between 2–100 nM [47].
Neamphamide A isolated from Neamphius huxleyi is a cyclic depsipeptide that included 11 amino-acid residues with an amide linked 3-hydroxy-2,4,6-trimethylheptanoic acid moiety. It showed potent cytotoxicity against HIV-1 infection at EC50 value of 28 nM [48,49]. Neamphamides B-D isolated from the same sponge exhibited cytotoxicity against human cancer cell-lines with IC50 values ranging between 88 to 370 nM, while neamphamide D displayed A549 cell-proliferation at sub-cytotoxic doses [50].
Callipeltins are cyclodepsipeptides isolated from the Callipelta sp. Callipeltin A is a decapeptide with three unusual amino-acid residues: (2R,3R,4S)-4-amino-7-guanidino-2,3-dihydroxyheptanoic acid, (3S,4R)-3,4-dimethyl-L-glutamine, and β-methoxytyrosine and was found to exhibit anti-HIV and antifungal activities. Callipeltin A is also a potent selective inhibitor of Na/Ca exchanger and a positive inotropic-agent when tested in guinea pig left-atria [51]. Callipeltin B showed weak inhibition activity at 4 μM, while callipeltins C and D exhibited no significant inhibitory activity on Na/Ca exchanger. All three callipeltins B-D did not induce any positive inotropic effect [51,52].
Callipeltin E [53] is a truncated linear peptide isolated from Latrunculia sp., with unique amino acids such as D-allothreonine, D-arginine, leucine, N-methylglutamine, N-methylalanine, and β-methoxytyrosine, while callipeltins F-I were also isolated from the same sponge. Callipeltins F-I exhibited antifungal activities against Candida at 10−4 M [54]. Callipeltins J-M are antifungal peptides isolated from Latrunculia sp., as well, where callipeltins K and J inhibited the growth of C. albicans at MIC value of 10−4 M [55]. Callipeltins N-Q are callipeltin derivatives isolated from the Asteropus sp., where callipeltins P and Q are acyclic callipeltins, while callipeltins N and O are cyclic callipeltins. Callipeltins N and O exhibited significant cytotoxicity against various cell-lines at an IC50 value of 0.16 μM explaining the significance of macrocyclisation as well as amino-acid composition in biological activity [56].
Microspinosamide is a cyclic depsipeptide (tridecapeptide) isolated from Sidonops microspinosa with numerous unusual amino acids and was the first naturally occurring peptide to comprise a β-hydroxy-ρ-bromophenylalanine residue. Microspinosamide exhibited cytopathic effect against HIV-1 infection with an EC50 value of 0.2 μg/mL in a XTT-based in vitro assay [57]. Carteritins A and B are cyclic heptapeptides isolated from Stylissa carteri. Carteritin A exhibited cytotoxicity against human cervical cancer HeLa, human colon cancer HCT116, and murine macrophage RAW264 cell-lines at IC50 values of 0.7, 1.3, and 1.5 μM, respectively [58].
Mycothiazole is a mixed polyketide/peptide derived compound isolated from Spongia mycofijiensis that exhibited in vitro anti-helminthic activity against Nippostrongylus braziliensis at 50 μg/mL. Mycothiazole was the first disubstituted thiazole that was known to inhibit the hypoxic HIF1 signal in tumor cells correlating with the HIF1 target gene suppression of VEGF expression. Mycothiazole is also known to selectively suppress the mitochondrial respiration at NADH-ubiquinone oxidoreductase [complex I] and can serve as a valuable molecular probe for HIF-mediated hypoxic signaling and mitochondrial biology. The only other examples of compounds with thiazole moiety are dysidenins [Ex. isodysidenin] isolated from Dysidea herbacea [34,59]. Dysideaprolines A-F and barbaleucamides A-B were isolated from Dysidea species. Dysideaprolines A-F are proline analogs of dysidenin, while barbaleucamides A-B are structural analogs of barbamide that were previously isolated from the marine cyanobacterium, Lyngbya majuscula. No biological activity was reported for either dysideaprolines A-F or barbaleucamides A-B [60]. Dysithiazolamide is a polychlorinated dipeptide which was also isolated from Dysidea species. Dysithiazolamide is a tetrachloro amino acid derivative with two leucine-like fragments with no reported biological activity [61].
Microcionamides A and B are linear peptides isolated from Clathria (Thalysias) abietina, a Philippine marine sponge. The C-terminus of these peptides is blocked by a 2-phenylethylenamine group with the peptides being cyclized via a cysteine moiety. Both these peptides showed inhibitory activity against Mycobacterium tuberculosis H37Ra and displayed potential cytotoxicity against human breast cancer cell-lines: SKBR-3 and MCF-7. Microcionamide A was active against SKBR-3 and MCF-7 cell-lines with IC50 values of 98 and 125 nM, respectively, while microcionamide B was active with IC50 values of 172 and 177 nM, respectively. Microcionamides A and B also displayed MIC values of 5.7 μM against M. tuberculosis H37Ra [34,62,63].
Halicylindramides A-C are tetradecapeptides isolated from Halichondria cylindrata, a Japanese marine sponge with their C-terminus lactonized with a threonine residue and the N-terminus blocked by a formyl group. Halicylindramides A-C were found to be cytotoxic against P388 murine leukemia cells with IC50 values of 0.54, 0.2, and 0.2 μg/mL, respectively, and exhibited antifungal activity against Mortierella ramanniana at 7.5 μg/disk. A seco-methyl ester generated from the halicylindramide B was found to be antifungal at 120 μg/disk, and was cytotoxic at 10 μg/mL [34,64].
Haligramides A and B are two cyclic hexapeptides isolated from Haliclona nigra along with a known peptide waiakeamide. Haligramide A was found to be cytotoxic against lung A-549, colon HCT-15, CNS SF-539, and CNS SNB-19 human cancer cell-lines at 5.17, 15.62, 9.00, and 9.08 μg/mL respectively, while haligramide B was cytotoxic at 3.89, 8.82, 5.01, and 6.56 μg/mL, respectively, towards the same cell-lines [34,65]. Waiakeamide was previously isolated from Ircinia dendroides and included three proline residues along with one thiazolylphenylalanine and two methionine sulfoxides. Waiakeamide exhibited potent activity against P388 cell-line at an IC50 value of 0.054 μg/mL [66]. A hexapeptide which is a sulfone derivative of waiakeamide was also isolated from Haliclona sp., along with waiakeamide, but did not exhibit any cytotoxicity [67].
Corticiamide A and cyclocinamide B are two halogenated cyclic peptides that were isolated from the Fijian sponge, Corticium sp. Corticiamide A is a member of a family with structural similarities to peptides including microspinosamide A, discodermins, polydiscamide A, and halicylindramides. Cyclocinamide A was isolated from the Psammocinia sp., [68] and exhibited potent in vitro selective cytotoxicity towards solid tumors while cyclocinamide B showed no cytotoxicity against HCT-116 cell-line [34,69]. Pembamide is an N-methylated linear peptide isolated from Cribrochalina species belonging to the family Niphatidae. Pembamide was found to exhibit cytotoxicity against human lung A-549 tumor, human colon HT-29, and human breast MDA-MB-231 cancer cell-lines at GI50 values of 2.46, 3.80, and 3.35 μM, respectively [70]. Kapakahines A-D are cyclic peptides that were isolated from a Pohnpei sponge, C. olemda. Kapakahines possessed a unique structural feature where the two tryptophan residues are linked with an N-C bond from the indole nitrogen of Trp-1, rather than an amide bond to the Trp-2 β-indole carbon. Kapakahines A, C, and D are octapeptides while kapakahine B is a hexapeptide. Kapakahines A-C exhibited moderate cytotoxicity against murine leukemia P388 cells at IC50 values of 5.4, 5.0, and 5.0 μg/mL, respectively, while kapakahine D did not show any cytotoxicity at 10 μg/mL. Kapakahine A was tested against several enzymes but possessed only 15% inhibition against protein phosphatase 2A (PP2A) at 30 μM [71]. Kapakahines E-G were also isolated from C. olemda where kapakahine E exhibited cytotoxicity against murine leukemia P388 cells at an IC50 value of 5.0 μg/mL, while kapakahines F and G showed weak cytotoxicity at this concentration [72].
Taumycins A and B are two related lipodepsipeptides that were isolated from the Madagascan sponge, Fascaplysinopsis sp., which possessed a 12-membered oxodepsipeptide ring-system. Both the taumycins were found to be toxic to brine-shrimp larvae at IC50 values of 10 μg/mL, but only taumycin A showed inhibition against human UT-7 leukemic cell-line at 1 μM [73].
Pipecolidepsins A and B are two cyclodepsipeptides isolated from Homophymia lamellosa. Of these, pipecolidepsin A exhibited cytotoxicity against A549 lung, HT29 colon, and MDA-MB-231 breast cancer cell-lines at GI50 values of 0.6, 1.12, and 0.7 μM, respectively, while pipecolidepsin B showed GI50 values of 0.04, 0.01, and 0.02 μM, respectively. The replacement of Asp2 residue in pipecolidepsin A with a HOAsp amino-acid gave rise to a 10-fold increase in bioactivity of pipecolidepsin B, revealing the key role of the hydrophilic nature and substitution at C-3 position of this amino-acid residue in the mode of action [74,75].
Halipeptins A-D [76,77] are cyclic depsipeptides isolated from the Haliclona sp. Halipeptin A is a 17-membered cyclic depsipeptide containing two alanines and three novel residues: 3-hydroxy-2,2,4-trimethyl-7-methoxydecanoic acid [HTMMD], N-methyl-δ-hydroxyisoleucine, and 1,2-oxazetidine-4-methyl-4-carboxylic acid. Halipeptin A exhibited potent in vivo anti-inflammatory activity at a dose of 300 μg/kg causing about 60% edema inhibition in mice [76]. Halipeptin D displayed strong in vitro inhibition against HCT-116 human colon cancer cell-line at IC50 value of 7 nM, and against BMS oncology diverse cell panel (ODCA) of tumor cell-lines at IC50 value of 420 nM [77].
Tausalarin C is a bioactive nitrogenous bismacrolide peptide isolated from the Madagascar sponge, Fascaplysinopsis sp. Tausalarin C was found to inhibit the proliferation of the K562 leukemia cell-line at 1 μM [78]. Arenastatin A is a cyclic didepsipeptide isolated from Dysidea arenaria, an Okinawan marine sponge that exhibited potent cytotoxicity at an IC50 value of 5 pg/mL against KB cell-line [79,80]. Axinastatins 1–3 are cycloheptapeptides isolated from Axinella species [81]. Pseudoaxinellin, also known as axinastatin 1 or malaysiatin [82] is a cyclic heptapeptide isolated from Pseudoaxinella massa [83]. Axinastatins-2 and -3 are cytostatic against six human cancer cell-lines at GI50 values ranging from 0.35 to 0.0072 μg/mL with axinastatin 3 being more potent against PS leukemia cell-line at an ED50 of 0.4 μg/mL [81].
Hymenamides A and B are proline rich cyclic heptapeptides having a prolylproline segment and were isolated from Hymeniacidon sp., an Okinawan marine sponge. Hymenamide A has an arginine residue, while hymenamide B possessed glutamic acid as hydrophilic moieties. Both hymenamides exhibited antifungal activity against C. albicans at MIC values of 33 and 66 μg/mL, respectively, and against Cryptococcus neoformans at > 133 and 33 μg/mL, respectively. Hymenamide B also showed cytotoxicity against in vitro human epidermoid KB carcinoma and murine lymphoma L1210 cell-lines with IC50 values of 6.0 and 3.2 μg/mL, respectively [84]. Hymenamides C-E are cyclic heptapeptides with two proline residues that were also isolated from the same sponge. Of these, hymenamides C and E exhibited antifungal activity against C. neoformans at MIC value of 133 μg/mL, but did not show any cytotoxicity against both human epidermoid KB carcinoma and murine lymphoma L1210 cell-lines at an IC50 value of > 10 μg/mL [85]. Hymenamide F was also isolated from Hymeniacidon sp., which is a cyclic heptapeptide with an arginine and prolylproline residues [86]. Hymenamides G, H, J, and K are cyclic octapeptides isolated from Hymeniacidon sp. Hymenamides G and K exhibited no cytotoxicity while hymenamide H showed cytotoxicity against L1210 cell-line at an IC50 value of 6.3 μg/mL. Hymenamide J showed cytotoxicity against human epidermoid KB carcinoma and murine leukemia L1210 cell-lines at IC50 values of 0.76 and 2.6 μg/mL, respectively. Hymenamides G and K also exhibited cytotoxicity against protein tyrosine-kinase c-erbB-214 at IC50 values of 63 and 73 μg/mL, respectively [87].
Wainunuamide is an unusual cyclic heptapeptide isolated from Stylotella aurantium, a Fijian marine sponge. This peptide included three proline residues along with a histidine residue, which is usually rare in cyclic peptides isolated from marine sponges. Wainunuamide was previously reported from the cyanobacterium, Oscillatoria agardhii, and was found to be weakly cytotoxic against K562 leukemia cancer and A2780 ovarian tumor cell-lines at ID50 values of 18.36 and 19.15 μg/mL, respectively [88]. Axinellins A and B are bioactive cyclopeptides that were isolated from Axinella carteri [89], while axinellins B and C are cyclic octapeptides that were isolated from Stylotella aurantium. Axinellins A and B exhibited moderate antitumor activity against NSCLC-N6 human bronchopulmonary non-small cell lung carcinoma cell-line at IC50 values of 3.0 and 7.3 μg/mL, respectively [89]. Axinellin C showed weak cytotoxicity against K562 leukemia cancer and A2780 ovarian tumor cell-lines at ID50 values of 4.46 and 13.17 μg/mL, respectively [90]. Cyclonellin is another cyclic octapeptide that was also isolated from A. carteri. Cyclonellin was found to be inactive when tested at 50 μg/mL against human colon COLO-205 and ovarian OVCAR-3 tumor cell-lines [91]. Stylopeptide 1 is a cycloheptapeptide that was also isolated from Stylotella sp. and Phakellia costata [92]. Stylopeptide 2 is a proline rich cyclodecapeptide isolated from the Stylotella sp., and exhibited inhibition against BT-549 and HS 578T breast cancer cell-lines at a dose of 10−5 M [93]. Stylostatin 1 is another cycloheptapeptide that has been isolated from S. aurantium which exhibited lymphocytic P388 leukemia cell-growth inhibition at an ED50 value of 0.8 μg/mL [94]. Stylostatin 2, also a cycloheptapeptide, was isolated from the Stylotella sp. and P. costata [95].
Fenestins A and B are cyclic peptides isolated from Leucophloeus fenestrata. Fenestin A is cyclo-[L-Pro-L-Pro-L-Leu-L-Ile], while fenestin B is cyclo-[L-Pro-L-Val-L-Pro-L-Leu-L-Ile]. Fenestins were tested against HT-29 and P388 cell-lines and were found to exhibit no activity at concentrations up to 20 μg/mL [96]. Hymenistatin 1 is an antineoplastic cyclic peptide isolated from Hymeniacidon sp., and exhibited cytotoxicity against NCI murine P388 lymphocytic leukemia cell-line at ED50 of 3.5 μg/mL [97].
Discobahamins A and B are bioactive peptides isolated from Discodermia and were found to exhibit weak antifungal activity against C. albicans [98]. Calyxamides A and B are thiazole and 5-hydroxytryptophan moieties containing cyclic peptides that were isolated from Discodermia calyx [99] with moderate cytotoxicity against murine leukemia P388 cell-line [100].
Microsclerodermins A-E [101,102] are cyclic hexapeptides isolated from the Microscleroderma sp. Microsclerodermins A and B exhibited antifungal activity against C. albicans at 2.5 μg/disk [101]. Microsclerodermins C-E and anhydromicrosclerodermin C were also isolated from Theonella sp., Microsclerodermins C-E and anhydromicrosclerodermin C were all found to exhibit antifungal activity against C. albicans with microsclerodermin C being the most active at 5 μg/disk, followed by microsclerodermin E at 10 μg/disk, anhydromicrosclerodermin C at 50 μg/disk, and microsclerodermin D at 100 μg/disk [102]. Microsclerodermins F-I are other cyclic peptides that were also isolated from Microscleroderma sp., and exhibited cytotoxicity against HCT-116 cell-lines with IC50 values of 1.8, 2.4, 1.0, and 1.1 μg/mL, respectively. Microsclerodermins F-I inhibited the growth of Candida albicans with microsclerodermin F being the most potent at 1.5 μg/disk, while microsclerodermins G-I were active at 3, 12, and 25 μg/disk, respectively [103]. Microsclerodermins J-K are cyclic hexapeptides with moderate antifungal activity [104].
Aciculitins A-C are cyclic peptides isolated from the lithistid sponge, Aciculites orientalis and were found to be cytotoxic against HCT-116 cell-line at an IC50 value of 0.5 μg/mL along with exhibiting antifungal activity against C. albicans at 2.5 μg/disk. Aciculitins included a bicyclic peptide with an unusual histidine-tyrosine bridge. To the bicyclic peptide, C13-C15 2,3-dihydroxy-4,6-dienoic acids bearing D-lyxose is attached at position 3. Aciculitamides A-B are artifacts obtained from the same sponge probably reacting to the methanol used in extraction, resulting in oxidation of the imidazole ring. Aciculitamide A did not show any cytotoxicity against HCT-116 and/or antifungal activity even at loadings of < 500 μg/disk [105].
Polydiscamide A is a depsipeptide comprising of 13 amino acids including the 3-methylisoleucine and was isolated from the Discodermia sp., Polydiscamide A inhibited proliferation of A549 human lung cancer cell-line in vitro at an IC50 value of 0.7 μg/mL along with inhibition of Bacillus subtilis growth at an MIC value of 3.1 μg/mL [106]. Polydiscamides B-D are potent human sensory neuron specific G protein coupled receptor (SNSR) agonists isolated from the Ircinia sp., at EC50 values of 1.26, 3.57, and 2.80 μM, respectively [107].
Criamides A and B are cytotoxic peptides isolated from the Cymbastela sp., and were found to be potent cytotoxins, both in vitro and in vivo. Criamide B exhibited cytotoxicity against A549 human lung, LOVO human colon, HEY human ovarian carcinoma, U373 human glioblastoma/astrocytoma, MCF7 human breast cancer, and P388 murine leukemia cell-lines at ED50 values of 0.29, 0.15, 0.19, 0.27, 6.8, and 0.0073 μg/mL, respectively [108]. Gombamide A is a cyclic thiopeptide isolated from Clathria gombawuiensis that displayed weak cytotoxicity against A549 and K562 cell-lines at LC50 values of 7.1 and 6.9 μM, respectively, along with moderate inhibition of Na+/K+-ATPase action at an LC50 value of 9.4 μM [109].
Euryjanicins A-D and dominicin are cyclic peptides isolated from Prosuberites laughlini. Euryjanicins A-D are proline-rich cycloheptapeptides while dominicin is a cyclooctapeptide. These peptides were marginally active to inactive when screened against NCI-60 tumor cell-line panel. The lost activity was reported due to the conformational changes of the cyclic peptides during the isolation or due to the binding ability of the peptides in low concentrations to the potent antineoplastic substances making them detectable only in biological screenings [110]. Euryjanicins E-G are cyclic heptapeptides with poly-phenylalanine and poly-proline residues, which were also isolated from P. laughlini with no anticancer activity when tested at 10 μM against NCI-60 tumor cell panel [111].
Neopetrosiamides A and B are diastereomeric (differ only in the configuration at the sulfoxide functionality) tricyclic peptides isolated from the Neopetrosia sp. Neopetrosiamides A and B were found to be active at 6 μg/mL in the amoeboid invasion assay with the potential to find drug-targets for the inhibition of amoeboid invasion of tumor-cells [112]. N-sulfoureidylated lipopeptides named sulfolipodiscamides A-C are cytotoxic peptides isolated from Discodermia kiiensis. Sulfolipodiscamides A-C possessed a unique feature of having an unprecedented N-sulfoureidyl group on the D-citrulline residue which is not found in other structurally similar lipodiscamides A-C. Of the three sulfolipodiscamides A-C, sulfolipodiscamide A exhibited a 2.3 fold increase in cytotoxicity against P388 murine leukemia cell-line compared to the parent compound [113]. Lipodiscamides A-C are lipodepsipeptides that were also isolated from D. kiiensis, and possessed an unprecedented dilactone macrocycle. Lipodiscamides A-C are probably the only lipopeptides that included a 4S-hydroxy-trans-2-enoate along with non-canonical amino acids: D-citrulline, E-dehydronorvaline, and L-3-ureidoalanine. Lipodiscamides A-C exhibited moderate cytotoxicity against murine P388 leukemia cells at IC50 values of 23, 20, and 31 μM, respectively, while showed weak to moderate cytotoxicity against HeLa cells at IC50 values of 18, 26, and 46 μM, respectively [114]. Jamaicensamide A is a cyclic peptide with a thiazole-homologated amino acid along with six other amino acids and was isolated from Plakina jamaicensis, a Bahamian sponge, with no known antifungal activity [115].
Stylissamides A-D are cyclic heptapeptides that were isolated from Stylissa caribica and included three proline residues in stylissamides A, C, & D and four proline residues in stylissamide B [116]. In addition, stylissamides E [117], F [117], G [118], and H [118] were also isolated from S. caribica. Stylissamide E included two proline residues while stylissamide F was a polar peptide with three proline residues [117]. Stylissamide H showed modest cytotoxicity against HCT-116 at an EC50 of 5.7 μM [118]. Stylissamide X is another proline-rich octapeptide isolated from the Stylissa sp. that showed inhibition of HeLa cell-migration at concentrations ranging between 0.1 to 10 μM and 75% cell viability [118]. Stylissatin A is another cyclic heptapeptide isolated from Stylissa massa and inhibited the nitric oxide production in LPS stimulated RAW264.7 murine macrophage cells at an IC50 value of 87 μM [119]. Stylissatins B-D were also isolated from S. massa, of which stylissatin B exhibited inhibition against a panel of human tumor cell-lines such as MCF7, HepG2, A2780, NCI-H1650, BGC-823, and HCT-116 at IC50 values ranging between 2.4 to 9.8 μM [120]. Apart from stylissamides, stylisins 1 and 2 were other cyclic heptapeptides that were isolated from S. caribica with no known anti-inflammatory, anti-microbial, anti-malarial, anti-cancer, anti-Mtb, and anti-HIV-1 activities [121].
Reniochalistatins A-E are cyclic peptides isolated from Reniochalina stalagmitis where reniochalistatins A-D are heptapeptides while reniochalistatin E is an octapeptide. Reniochalistatins C and D were closely related to phakellistatin 18 and stylissamide C with more than 70% similarity in their peptide sequences, while reniochalistatins A-E differed in more than 50% similarity within their peptide sequences. Reniochalistatin E exhibited cytotoxicity against RPMI-8226 myeloma and MGC-803 gastric cell-lines at IC50 values of 4.9 and 9.7 μM, respectively, with no activity against HeLa cervical, HepG2 hepatoma, and HL-60 leukemia cell-lines. Reniochalistins A-D did not possess any cytotoxicity [122]. Yaku’amides A and B are cytotoxic peptides that were isolated from Ceratopsion sp., which inhibited murine leukemia P388 cells at IC50 values of 14 and 4 ng/mL, respectively. When tested against a panel of 39 human cancer cell-lines, yaku’amide A was found to possess a unique mode of action in its growth-inhibition activity [123].
Chujamides A and B are cyclic cysteine-bridged peptides isolated from the Korean sponge, Suberites waedoensis. Chujamides A and B showed weak cytotoxicity against A549 cell-line at LC50 values of 10.1 and 26.4 μM, respectively, and against K562 cell-line at LC50 values of 37.0 and 55.6 μM, respectively. Chujamide B also showed moderate inhibition of Na+/K+-ATPase at an IC50 value of 17.2 μM [124]. Leucamide A is another bioactive cyclic heptapeptide that was isolated from Leucetta microraphis. Leucamide A included a distinct mixed 4,2-bisheterocycle tandem pair with a thiazole and methyloxazole subunit and exhibited moderate cytotoxicity against several tumor cell-lines such as Huh7, HepG2, and HM02 at GI50 values of 5.1, 5.9, and 5.2 μg/mL, respectively [125].
Azumamides A-E are cyclic tetrapeptides that were isolated from Mycale izuensis. Azumamides A-E were the first examples of marine cyclic peptides that exhibited histone deacetylase inhibition between the IC50 range of 0.045 – 1.3 μM. Azumamide A also exhibited moderate cytotoxicity against human leukemia K562 and human colon WiDr cancer cells at IC50 values of 4.5 and 5.8 μM, respectively [126]. Phoriospongins A-B are nematocidal depsipeptides that were isolated from Phoriospongia species and Callyspongia bilamellata. Phoriospongin A was structurally similar to cyclolithistide A and both the phoriospongins exhibited nematocidal activity with an LD99 of 8.3 μg/mL [127].
Callyaerins A-F and H are cytotoxic cyclic peptides that were isolated from Callyspongia aerizusa. Callyaerins included ring systems with 5–9 amino acids and side-chains of 2–5 amino acids in length. The ring closure has an unusual (Z)-2,3-diaminoacrylic acid unit template. All the peptides included three or more proline-residues with other hydrophobic residues where all the amino acids are in L form. Callyaerins E and H displayed strong activity against L5178Y cell-line with ED50 values of 0.39 and 0.48 μM, respectively, while the rest were less active with ED50 values between 2.92 to 4.14 μM. Callyaerin F was found to be inactive [128]. Callyaerin G was isolated from C. aerizusa and exhibited cytotoxicity against human cervix carcinoma HeLa, mouse lymphoma L5178Y, and rat brain tumor PC12 cell-lines at concentrations between 3–10 μg/mL [129].
Sponges are a generous source of compounds with unique chemical structures including peptides. The sponge Theonella swinhoei has been explored exhaustively for more than a decade yielding unprecedented peptide chemistry [4]. Motuporin, a cyclic pentapeptide possessed inhibitory activity towards protein phosphatase 1 at concentrations less than 1 nM [130]. It also showed cytotoxicity towards breast, brain, colon, ovarian, murine leukemia, and human lung cancer cell-lines at IC50 values of 12.4, 2.4, 2.3, 2.8, 6.0, and 2.4 μg/mL, respectively [131]. Theonellapeptolide Id showed moderate cytotoxicity towards L1210 at an IC50 of 2.4 μg/mL. It is also known to possess ion transport activities towards Na+, K+, and Ca2+ ions [132]. Nazumazoles A-F are cyclic pentapeptides that were isolated from T. swinhoei. Nazumazoles A-C displayed cytotoxicity against murine leukemia P388 cell-line at an IC50 value of 0.83 μM [133,134]. Nazumazoles D-F are protease inhibitors that cleaved amide-bonds adjacent to hydrophobic amino acid residues at IC50 values of 2, 3, and 10 μM, respectively, but did not show any inhibition against thrombin or trypsin nor exhibited P388 cytotoxicity at concentrations of 50 μM [134].
Orbiculamide A is a cyclic peptide that showed cytotoxicity at an IC50 of 4.7 μg/mL towards P388 murine leukemia cell-line [135]. Polytheonamides A, B, and C are cyclic peptides that showed cytotoxicity at IC50 values of 78, 68, and 68 pg/mL, respectively, against P388 leukemia cell-line. Polytheonamide A is an epimer of polytheonamide B which differ in the stereochemistry of the sulfoxide at the 44th residue [136,137].
There are a huge number of cyclic peptides with potent activity which included pseudotheonamides [138], and cyclotheonamides A and B [139], that act as serine protease inhibitors. Cyclotheonamide A was the first macrocyclic peptide to belong to the class of serine protease inhibitors. It is known to be a potent inhibitor of streptokinase and trypsin at IC50 values of 0.023 and 0.035 μM, respectively, and is also a moderate inhibitor of α-thrombin at an IC50 of 0.18 μM [140]. Cyclotheonamides E, E2, and E3 also showed inhibitory activities against thrombin at IC50 values of 2.9, 13.0, and 9.5 nM, respectively. They showed inhibition against trypsin at IC50 values of 30, 55, and 52 nM, respectively [139]. Nazumamide A is a linear tetrapeptide and is a thrombin inhibitor at an IC50 of 2.8 μg/mL. This was the first natural peptide with a N-2,5-dihydroxybenzoate terminus [141]. Pseudotheonamides A1, A2, B2, C, D, and dihydrocyclotheonamide A are all linear pentapeptides that have a rare piperidinoiminoimidazolone and piperazinone ring system. They inhibited thrombin at IC50 values of 1.0, 3.0, 1.3, 0.19, 1.4, and 0.33 μM, respectively. They also inhibited trypsin at 4.5, > 10, 6.2, 3.8, > 10, and 6.7 μM, respectively [138].
The common characteristics of the peptides isolated from Theonella spp. included high degree of isomerism and similarities represented in the peptide families of theonellamides, keramamides, and many more. Theonellamides are bicyclic peptides with bromine and carbohydrate substituents. Theonellamides A-E showed cytotoxicity towards P388 leukemia cell-line at IC50 values of 5.0, 1.7, 2.5, 1.7, and 0.9 μg/mL, respectively [142]. Theonellamide G was later isolated from T. swinhoei and was found to exhibit antifungal activity against wild and amphotericin-B resistant strains of C. albicans with IC50 values of 4.5 and 2.0 μM, respectively. Theonellamide G also displayed cytotoxicity against human colon adenocarcinoma HCT-16 cell-line at an IC50 value of 6.0 μM [143]. Keramamides are cyclic peptides containing oxazole or thiazole rings. Keramamide A is a cyclic hexapeptide with inhibitory activity against sarcoplasmic reticulum Ca2+-ATPase at an IC50 value of 3 × 10−4 moldm−3 [144]. Keramamides B-D showed inhibition towards human neutrophil superoxide generation at 5 × 10−8 M [145]. Keramamide F is a cytotoxic cyclic peptide containing unusual amino acids including an isoserine residue, a didehydrotryptophan, an α-ketoamide function as part of 3-amino-4-methyl-2-oxo-hexanoic acid, and an O-methylserylthiazole derivative [146]. Keramamides E, G, H, and J are thiazole or oxazole containing cyclic peptides where keramamide E showed cytotoxicity against human epidermoid KB carcinoma and murine leukemia L1210 cell-lines at IC50 values of 1.55 and 1.60 μg/mL, respectively. Keramamides G, H, and J exhibited weak cytotoxicity at IC50 value of ~ 10 μg/mL [147]. Keramamides K and L are cyclic peptides with an unusual tryptophan; keramamide K is a thiazole-containing cyclic peptide while keramamide L possessed a 6-chloro-N-methyltryptophan residue and an ureido bond. Keramamides K and L displayed cytotoxicity against murine leukemia L1210 cell-line at IC50 values of 0.72 and 0.46 μg/mL, respectively, and against epidermoid KB carcinoma cell-line at IC50 values of 0.42 and 0.9 μg/mL, respectively [148]. Keramamides M and N are cyclic peptides with a sulfate ester that were known to exhibit cytotoxicity against epidermoid KB carcinoma cell-line at IC50 values of 6.0 and 7.5 μg/mL, respectively, and against murine leukemia L1210 cell-line at IC50 values of 2.4 and 2.8 μg/mL, respectively [149].
Numerous peptides have been reported from Theonella spp. exhibiting promising activities such as anti-HIV-1, immunomodulatory, antifungal, antibacterial, enzyme inhibitory, and others. The diverse structures of peptides isolated from Theonella sp. range from linear to cyclic peptides, depsipeptides, and large bicyclic peptides. Koshikamides [150] and highly cytotoxic polytheonamides [136] are examples of linear peptides isolated from Theonella sp. Koshikamide A1 is a linear decapeptide with moderate cytotoxicity towards P388 leukemia cell-line at IC50 of 2.2 μg/mL [150]. Koshikamide A2 is a linear undecapeptide that showed moderate cytotoxicity towards P388 cell-line at IC50 of 6.7 μg/mL [151]. Koshikamide B is a cyclic peptide lactone and showed cytotoxicity towards human colon tumor and P388 leukemia cell-lines at IC50 values of 3.7 and 0.22 μM, respectively [152]. Koshikamides C-E, and G did not inhibit HIV entry [153], while koshikamides F and H showed entry inhibition at IC50 values of 2.3 and 5.5 μM, respectively. Koshikamide H exhibited cytotoxicity towards colon cancer at IC50 of 10 μM [152].
The depsipeptide nagahamide A exhibited antibacterial activity [154], while papuamides A-D are HIV-1 inhibitors [155]. Nagahamide A is also a weak antibacterial agent at 50 μg when applied to an inhibitory zone of 7 mm [154]. Papuamides A and B are cyclic depsipeptides that inhibited the human T-lymphoblastoid cell infection at EC50 of 4 ng/mL. Papuamide A also exhibited cytotoxicity towards human cancer cells at IC50 of 75 ng/mL [155]. Papuamides A and B showed 80% HIV viral entry inhibition at 710 nM, while papuamides C and D showed 30% and 55% entry inhibitions at approximate concentrations of 40- and 20-fold higher [156]. Two other depsipeptides, papuamides E and F were isolated from the marine sponge belonging to the genus Melophlus collected from the Solomon Islands along with known papuamides C and D. Papuamides E-F exhibited cytotoxicity against brine-shrimp with LD50 values of 92 and 106 μg/mL, respectively [157]. Bicyclic peptides theonellamide F and theonegramide were reported as antifungal agents [158]. Theonellamide F, a dodecapeptide exhibited cytotoxicity against P388 and L1210 leukemia cell-lines at IC50 values of 2.7 and 3.2 μg/mL, respectively [158]. Theonegramide is a glycopeptide combined with a bicyclic dodecapeptide that exhibited antifungal activity against Candida albicans at 10 μg/disk loading [159]. Cupolamide A is a cyclic heptapeptide isolated from Theonella cupola that included one D-Ser, one D-Leu, and two L-Val along with three uncommon amino-acid residues: L-2,4-diaminobutanoic acid [Dba], D-homoarginine [Har], and trans-4-hydroxy-L-proline [Hyp]. Cupolamide A exhibited cytotoxicity against murine leukemia P388 cells at an IC50 value of 7.5 μg/mL [160].
Theonella swinhoei collected from different regions of Indonesia is also a rich source of peptides. A series of cyclic peptides called barangamides were isolated from T. swinhoei collected at Barang Lompo Island, Indonesia. Barangamides A-D were reported as new cyclic undecapeptides with N-methylated amino acids and β-alanine [161]. Barangamides A, B, C, and D are cyclic peptides that were reported to lack cytotoxic and immunosuppressive activities [161,162]. In addition to barangamides, the sponge also included theonellapeptolides Ia, Id, Ie, IId, and IIe. Barangamide A showed sequence homology with the ring part of theonellapeptolide Id. Moreover, barangamides B, C, and D were derivatives of theonellapeptolides Ib, Ia, and Ic, respectively [162].
Theonellapeptolides possessed unique characteristics that were rich in β-, D, and N-methyl amino acids. Theonellapeptolides Ia, Ib, Ic, Id, and Ie were reported for the first time from the Okinawan sponge Theonella swinhoei. These are tridecapeptide lactones that showed moderate cytotoxicity against L1210 cell-line at IC50 values of 1.6, 1.3, 2.4, and 1.4 μg/mL respectively, except for theonellapeptolide Ia. These tridecapeptides inhibited the development of the fertilized eggs of the sea urchin, Hemicentrotus pulcherrimus [132], [163]. Theonellapeptolide Id [164] exhibited ion-transport activity for Na+, K+, and Ca2+ ions while theonellapeptolide Ie has activity for Na+ and K+ ions in erythrocyte membranes of humans [132]. Theonellapeptolide Ie also caused malformation of starfish oocytes Asterina pectinifera [165]. Theonellapeptolides IId and IIe showed cytotoxicity at 9.4 μM. Of these two, theonellapeptolide IId exhibited the strongest immunosuppressive activity [152]. Theonellapeptolide IIIe, a 36-membered cyclic peptolide exhibited moderate cytotoxicity towards P388 at 7.4 μg/mL [166].
The crystal structures of theonellapeptolides Id and IIIe have been reported [166,167,168]. Theonellapeptolide IIIe was reported as a cytotoxic constituent of the deep water sponge, Lamellomorpha strongylata. This finding suggested that the sponge-associated microbes may be responsible for the biosynthesis of the peptides [166].
Two congeners of theonellapeptolides have been isolated from the Okinawan sponge, Theonella sp [170]. Congener 1 replaced the methoxyacetyl of theonellapeptolide with a methylsulfinylacetyl group at the N-terminus, while congener 2 possessed an acetyl group. Congeners 1 and 2 exhibited antifungal activity against Trichophyton mentagrophytes (4.0 & 8.0 μg/mL), Aspergillus niger (> 66 & 8.0 μg/mL), and antibacterial activity against Staphylococcus aureus (8.0 & > 16 μg/mL), Micrococcus luteus (8.0 μg/mL), Bacillus subtilis (8.0 & 16 μg/mL), and Mycobacterium smegmatis (16 & 66 μg/mL), respectively. They exhibited cytotoxicity towards L1210 leukemia cell-line at IC50 values of 9.0 and 7.5 μg/mL, respectively [170]. Solomonamides A and B are two unprecedented cyclic peptides isolated from T. swinhoei where solomonamide A exhibited anti-inflammatory activity at a dose of 100 μg/kg [171].
Cyclolithistide A along with motuporin and theonellapeptolide Id were isolated from Theonella swinhoei collected from the Sangihe Island, Indonesia. Cyclolithistide A included the unique amino acids: 4-amino-3,5-dihydrohexanoic acid, formyl-leucine, and chloroleucine. Cyclolithistide A is a cyclic depsipeptide that exhibited antifungal activity at 20 μg/disk [172]. In addition, it has been reported that T. swinhoei collected from the same location also possessed a series of swinholides A-G, theonellamine B, and theopalauamide A. Theonellamine B is used as a synonym for theonellapeptolide Id [173]. Theopalauamide differs from theonegramide in the presence of D-galactose instead of D-arabinose [174]. Theopalauamide is a bicyclic glycopeptide that showed inhibition towards Candida albicans at 10 μg/disk, while isotheopalauamide exhibited inhibition at 50 μg/disk. Isotheopalauamide is a stable conformational isomer of theopalauamide [174]. Oriamide is another cyclic peptide with a novel 4-propenoyl-2-tyrosylthiazole amino acid [PTT] that was also isolated from T. swinhoei [175].
Miraziridine A, paltolides A-C, perthamides B-F, and mutremdamide A are other peptides isolated from Theonella. Miraziridine A is a linear peptide with a rare aziridine-2,3-dicarboxylic acid residue, and was found to be a cathepsin B inhibitor at an IC50 value of 2.1 μM. Paltolides A-C are anabaenopeptin-type peptides. Anabaenopeptin class compounds included an N-methylated amino acid prior and adjacent to the C-terminal residue that is cyclized to the ε-amine of lysine residue. Paltolide A was the first reported anabaenopeptin-type peptide missing an N-methyl group at this site. Due to the presence of a C-terminal tryptophan residue linkage to the ε-amine of the N-terminal lysine residue, paltolides belong to the rare sub-group of anabaenopeptins. Other compounds in this subgroup are carboxypeptidase U inhibitors. Paltolides A-B did not exhibit any biological activity in HCT-116 or HIV-1 entry assays, while the carboxypeptidase U inhibition has not been evaluated [152]. Perthamide B is a cyclic octapeptide with weak binding inhibition of [125I]IL-1β to the intact EL46.1 cells at an IC50 value of 27.6 μM [177]. Perthamides C-E displayed anti-inflammatory activity that is known to mediate via TNF-α up-regulation [178]. Perthamides C and D displayed 60% and 46% edema reductions at a dose of 0.3 mg/kg. Similar to perthamide C, perthamides H, I, and K also exhibited similar behavior at 0.3 mg/kg [179]. Perthamides inhibited TNF-α and IL-8 release in the human primary keratinocyte cells, and hence could be the potential leads for psoriasis treatment [178]. Mutremdamide A was isolated from T. swinhoei and T. cupola and differs from perthamide C in the mutual presence of sulfation and carbamoylation of β-OH and Nδ-amide groups of β-OHAsn, respectively. Perthamide C had a threo configuration at C-2/C-3, while mutremdamide A displayed an erythro configuration [153]. Mutremdamide A is a sulfated cyclic depsipeptide similar to perthamide B exhibiting 60% edema inhibition at 0.3 mg/kg and is at least 100 times more potent than naproxen which has an ED50 of 40 mg/kg [152].
From a biological point of view, Theonella swinhoei is unique because it is a host to various microorganisms. Histological and ultra-structural investigation of T. swinhoei from the Red Sea and Indian Ocean revealed that the sponge may have ingested filamentous bacteria. In addition, the polychaete Haplosyllis spongicola was present in the aquiferous system of the sponge [180].
The metabolites reported from the Lithistid sponge including Theonella sp., revealed a degree of similarity with metabolites from microorganisms [Figure 45] [181,182,183]. This led to the suggestion that the metabolites from these sponges may be produced by symbiotic microorganisms, specifically cyanobacteria. This suggestion is also supported by the fact that these marine peptides included D- and unusual amino acids as well as additional side chains which are characteristic of microbial metabolites [130]. Cellular localization of metabolites along with analysis of the types of symbionts also supported the hypothesis. The presence of filamentous microorganisms in the interior of T. swinhoei correlated with the presence of theopalauamide, while the presence of unicellular bacteria correlated with the biosynthesis of swinholide A [184].
Figure 45.

Chemical structures of barangamides A-D, theonellapeptolides Ia-Ie, and IId-IIe
The structure of motuporin, an inhibitor of protein phosphatase 1 from T. swinhoei differs from nodularin isolated from the cyanobacteria, Nodularia pumigena only in the replacement of L-valine with L-arginine. Nodularin, a monocyclic pentapeptide exhibited inhibition towards protein phosphatases 1 and 2A at ED50 of 0.7 nM [185]. A cyclic peptide keramamide A isolated from the Theonella sp., [186] incorporated a 5-hydroxytryptophan residue and a ureido linkage, joining two amino acid residues that shared similarities with the main skeleton of ferintoic acids isolated from the fresh water cyanobacteria, Microcystis aeruginosa [187]. Keramamide A, a cyclic hexapeptide inhibited sarcoplasmic reticulum Ca2+-ATPase at an IC50 of 3 × 10−4 moldm−3 [144]. Ferintoic acids A and B are two cyclic hexapeptides that did not exhibit any chymotrypsin inhibition at concentrations up to 1.5 × 10−4 M [187]. The structure of mozamide A isolated from the Theonella sp., [144] is closely related to brunsvicamides A and B isolated from a cyanobacterial strain. The difference is in the presence of hydroxylated tryptophan and the absolute configuration of some amino acid residues [188]. Mozamides A and B are two cyclic peptides that did not exhibit any antimicrobial activity towards yeast or gram positive, and gram negative bacteria at low concentrations [189].
B. Marine Bacteria
Turnagainolides A and B isolated from Bacillus sp., are C-3 epimeric cyclic peptides where turnagainolide B stimulated the inositol 5-phosphatase SHIP1. Inositol 5-phosphatase SHIP1 is a negative regulator of PI3K (phosphatidylinositol-3-kinase) pathway, and its irregular functioning is associated with some cancers and inflammatory diseases [190]. Solonamides A and B are cyclic peptides isolated from a bacterial strain [tropical Pacific Ocean mussel] allied to Photobacterium halotolerans that hindered the agr quorum sensing system which controls the virulence gene expression in S. aureus [190]. Actinoramides A-C are modified peptides with unusual amino acids: 4-amino-3-hydroxy-2-methyl-5-phenylpentanoic acid and 2-amino-4-ureidobutanoic acid isolated from a bacterium closely associated to the genus Streptomyces [190]. Fijimycins A-C are three depsipeptides isolated from a Streptomyces strain that occurred as a complex conformational mixture and showed prominent activity against three MRSA strains [190]. Brunsvicamides A-C are cyclic hexapeptides isolated from the cyanobacterium of the Tychonema sp. Of these, brunsvicamides B and C showed selective inhibition against the protein tyrosine phosphatase B of Mycobacterium tuberculosis at IC50 values of 7.3 and 8.0 μM, respectively. However, brunsvicamide A exhibited weak inhibition at IC50 of 64.2 μM [191].
Malyngamide 2 isolated from a collection of Lyngbya sordida is a polyketide synthase-nonribosomal peptide synthetase [PKS-NRPS] derived metabolite that showed anti-inflammatory activity in LPS-induced [lipopolysaccharide] RAW macrophage cells with meek cytotoxicity within the cell-lines. Malyngamide 3 and cocosamides A and B, are three cyclic peptides isolated from L. majuscula with modest cytotoxicity against HT-29 colon and MCF7 breast cancer cell-lines [190,192]. Malyngamide 3 exhibited cytotoxicity against HT-29 and MCF7 cell-lines at IC50 values of 48 and 29 μM, respectively. Cocosamides A and B were cytotoxic against HT-29 at 24 and 11 μM, respectively, and against MCF7 at 30 and 39 μM, respectively [192]. Pitiprolamide is a proline-rich cyclic depsipeptide that was also isolated from L. majuscula and exhibited cytotoxicity against two HTCLs along with being weakly antibacterial against B. cereus and M. tuberculosis [190]. Pitipeptolides A and B isolated from L. majuscula and pitipeptolides C-F isolated from other collections were most active against M. tuberculosis and showed weak cytotoxicity against two HTCLs. Pitipeptolide C was formerly prepared as a hydrogenation artifact of pitipeptolides A and B [190,193].
Lagunamides A, B [194], and C [190] isolated from L. majuscula are cyclodepsipeptides with potent cytotoxicity towards HTCLs along with weak anti-swarming activity against Pseudomonas aeruginosa and significant anti-malarial activity against Plasmodium falciparum [190]. Wewakamide A and guineamide G are two cyclic depsipeptides [190] isolated from L. semiplena and L. majuscula, which showed potent toxicity to brine shrimp. Guineamide G was also found to be cytotoxic to mouse neuroblastoma cell lines [195]. Guineamides A-F are other cyclic depsipeptides isolated from L. majuscula [196].
Wewakazole is a cyclic dodecapeptide isolated from Lyngbya majuscula [197], while wewakazole B is a cytotoxic peptide isolated from the Red Sea Moorea producens (formerly L. majuscula). Wewakazole exhibited cytotoxicity against H460 human lung cancer cell-line at an IC50 value of 10 μM while wewakazole B displayed cytotoxicity against MCF7 human breast cancer and H460 human lung cancer cell-lines at IC50 values of 0.58 and 1.0 μM, respectively. Wewakazole and wewakazole B were the only isolated compounds from M. producens till date with both oxazole and methyloxazole moieties [198]. Wewakpeptins A-D are depsipeptides isolated from L. majuscula. Wewakpeptins have an unusual arrangement of hydroxy acid and amino subunits relative to well-known cyanobacterial peptides, along with a 2,2-dimethyl-3-hydroxyoctanoic acid or a 2,2-dimethyl-3-hydroxy-7-octynoic acid residue, a bis-ester, and a diprolyl group similar to dolastatin 15. Wewakpeptins A and B were the most cytotoxic peptides against human lung NCI-H460 tumor and neuro-2a mouse neuroblastoma cell-lines at an LC50 value of 0.4 μM [199].
Porpoisamides A and B isolated from Lyngbya sp., are two C-2 epimeric cyclic depsipeptides with weak cytotoxicity towards osteosarcoma U2OS and HCT-116 cell-lines [190]. Bisebromoamide [200,201,202] and its demethyl analog norbisebromoamide [202] isolated from the Okinawan Lyngbya sp., were found to be strongly anti-proliferative [202].
Somocystinamide A [ScA] and C-phycocyanin [C-PC] are other peptides isolated from the marine sources which exhibited potent caspases dependent anti-apoptotic activity in various cancer cell-lines. Somocystinamide A is a lipopeptide isolated from Lyngbya majuscula/Schizothrix sp., that stimulated apoptosis in angiogenic endothelial cells and various tumor cell-lines via both extrinsic and intrinsic pathways with the most effective mechanism being caspase 8 activation and its downstream-pathways [34,203]. C-phycocyanin is a major biliprotein (tetrapyrrole protein-complex) isolated from Spirulina platensis, Agmenellum quadruplicatum, and Mastigocladus laminosus that induced down regulation of anti-apoptotic gene-expression and pro-apoptotic gene activation facilitating the apoptosis signal-transduction resulting in in vitro HeLa cells apoptosis. C-PC also resulted in cytochrome-c release from mitochondria in to the cytosol when tested in C-PC treated HeLa cells [34].
Desmethoxymajusculamide C (DMMC) is a cyclic depsipeptide isolated from Lyngbya majuscula with selective and potent anti-solid tumor activity against HCT-116 human carcinoma cell-line, through the disruption of microfilament cellular networks at an IC50 value of 20 nM. There are significant differences in the anticancer activities of cyclic and linear DMMC where cyclic DMMC exhibited IC50 values of 0.02, 0.063, 0.22, and > 1.0 μM, and linear DMMC exhibited IC50 values of 0.016, 0.094, 0.23, and > 1.0 μM against HCT-116 human colon carcinoma, H-460 human large cell lung-carcinoma, MDA-MB-435 human carcinoma, and neuro-2A murine neuroblastoma cell-lines, respectively. The closely related compounds to DMMC include lyngbyastatins 1, 3, and dolastatin 12, which are considered to be mixtures of 4-amino-2,2-dimethyl-3-oxopentanoic acid unit [Ibu] epimers [S (minor) and R (major)], with majusculamide C being a single diastereomer with the S-Ibu unit [204]. Epilyngbyastatin 1 is a C-15 epimer of lyngbyastatin 1 which was also isolated from L. majuscula/Schizothrix calcicola. Epilyngbyastatin 1 displayed cytotoxicity against human nasopharyngeal KB carcinoma cell-line at MIC of 0.1 μg/mL, along with being a potent disrupter of cellular microfilament networks at concentrations of 2 and 0.2 μg/mL [205]. Majusculamides A and B were isolated from L. majuscula which at low concentrations were found to increase sea hare feeding, while at high concentrations inhibited feeding [206]. Majusculamide C, dolastatins 11, and 12 are other cyclic depsipeptides that showed significant cytotoxicity stimulating the microfilament hyperpolymerization by arresting cells in a time- and dose-dependent manner [34,207]. Majusculamide C exhibited strong antifungal activity, while majusculamide D and deoxymajusculamide D are two acyclic lipopentapeptides that showed moderate cytotoxicity at 0.2 μg/mL in CCRF-CEM cell culture [208]. Lyngbyastatin 3 was also isolated from L. majuscula and exhibited activity against LoVo and KB cell-lines at IC50 values of 400 and 32 nM, respectively [209].
Apratoxins A-C are cyclodepsipeptides isolated from Lyngbya majuscula with in vitro cytotoxicity against LoVo cell-line at 0.36–10.8 nM and against KB cell-line at 0.52–21.3 nM [210]. Apratoxin D is another peptide isolated from L. majuscula and L. sordida. Apratoxin D was found to be cytotoxic against H-460 human lung cancer cell-line with an IC50 value of 2.6 nM. Apratoxin D included the same macrocycle as apratoxins A-C with an additional unprecedented 3,7-dihydroxy-2,5,8,10,10-pentamethylundecanoic acid as a polyketide moiety [204]. Apratoxin E is another peptide isolated from L. bouillonii and exhibited better cytotoxicity compared to its closest analog the semi-synthetic E-dehydroapratoxin A against various cancer cell-lines derived from bone, colon, and cervix with values ranging between 21–72 nM, but less active compared to apratoxin A. This was speculated to be due to the conformational alteration in apratoxin E resulting from the dehydration of the polyketide chain, thereby reducing its activity [204].
Dragomabin along with carmabin A and dragonamides A-E are acyclic peptides isolated from Lyngbya majuscula and L. polychroa. These peptides included an 8 or 10-carbon long terminal alkynamide. Among these peptides dragomabin, carmabin A, and dragonamide A exhibited good antimalarial activities at IC50 values of 6.0, 4.3, and 7.7 μM, respectively. Dragonamides A and E showed activity against Leishmania donovani at IC50 values of 6.5 and 5.1 μM, respectively. Dragonamide B lacked activity suggesting that the aromatic amino-acid at the carboxyl terminus was necessary for the antiparasitic activity [204]. Carmabins A-B are linear lipotetrapeptides isolated from L. majuscula [211]. Carmabin A was found to be more cytotoxic against Vero cells at IC50 value of 9.8 μM compared to dragomabin or dragonamide A that showed IC50 values of 182.3 and 67.8 μM, respectively. The increased cytotoxicity of carmabin A over dragomabin was due to the long and more branched alkynamide chain in carmabin A [204]. Carmabin B lacked any antiproliferative activity [211]. Dragonamides C-D exhibited weak cytotoxicity in cancer cell viability assays against U2OS osteosarcoma cells at GI50 values of 56 and 59 μM, respectively, against IMR-32 neuroblastoma cells at 49 and 51 μM, respectively, and against HT29 colon adenocarcinoma cells at 22 and 32 μM, respectively. Dragonamides C-D lacked any antiparasitic activity [204]. Herbamide A was isolated from the marine sponge Dysidea herbacea [212], while herbamide B was isolated from L. majuscula [213]. Herbamides are modified linear peptides where herbamide A was found to be inactive in the NCI cytotoxicity screen [212] while herbamide B displayed antileishmanial activity at an IC50 value of 5.9 μM [213].
Almiramides A-C are lipopeptides isolated from L. majuscula, a cyanobacterium of the Caribbean coast of Panama. Almiramides B-C were found to have antileishmanial properties. Almiramides B and C possessed an extra Ala residue when compared to dragonamide A along with opposite configuration of the α-carbon of lipophilic side-chain and absence of a methyl group on Val1. Almiramides B and C exhibited antileishmanial activity at IC50 values of 2.4 and 1.9 μM, respectively, but lacked antimalarial properties at concentrations up to 13.5 μM. Almiramide A lacked antileishmanial activity due to the absence of an unsaturated terminus on lipophilic side-chain that played a significant role in other almiramides and dragonamides [204].
Grassystatins A-C are three statin unit [γ-amino-β-hydroxyacid] containing linear peptides isolated from Lyngbya confervoides. Grassystatins A and B exhibited similar selectivity and potency against cathepsin D with IC50 values of 7.27 and 26.5 nM, respectively, and against cathepsin E with IC50 values of 354 and 886 pM, respectively. Grassystatins showed higher affinity for cathepsin E over cathepsin D due to the interaction of their polar asparagine-residue with glutamine-303 of cathepsin E which corresponded to the non-polar residue of methionine-307 in cathepsin D. Grassystatin A also showed antigen reduction by dendritic-cells; a process that relies on cathepsin E. Grassystatin C is a truncated peptide analog that included two fewer residues compared to grassystatins A and B, and hence was less potent against cathepsins D and E. However, grassystatin C showed selectivity towards cathepsin E. Grassystatins A-C were also found to be metalloprotease tumor necrosis factor α converting enzyme [TACE] inhibitors with IC50 values of 1.23, 2.23, and 28.6 μM, respectively [204].
Obyanamide is a cyclic depsipeptide isolated from Lyngbya confervoides and exhibited moderate cytotoxicity against KB and LoVo cell-lines at IC50 values of 0.58 and 3.14 μg/mL, respectively [214]. Lobocyclamides A-C are lipopeptides isolated from L. confervoides. Lobocyclamides B and C were the first peptides with a unique amino acid 4-hydroxythreonine along with the rare long chain β-amino acids: 3-aminooctanoic acid and homologous 3-aminodecanoic acid, respectively. Lobocyclamides A-C displayed moderate antifungal activity in disk diffusion assays when tested against fluconazole-resistant fungi, Candida albicans and C. glabrata at 150 μg/disk. In microbroth dilution assay, lobocyclamide A exhibited antifungal activity against C. albicans at an MIC of 100 μg/disk, while lobocyclamide B showed similar activity between 30–100 μg/disk [215].
Hantupeptins A-C are cyclodepsipeptides isolated from Lyngbya majuscula. Hantupeptins A-C exhibited 100% brine-shrimp mortality between 100 and 10 ppm. This activity was found to be significantly higher compared to its closest analog trungapeptin A isolated from the same cyanobacterium with mild toxicity to brine shrimp. Hantupeptins A-C exhibited in vitro cytotoxicity against MOLT-4 leukemia cell-line at IC50 values of 32, 0.2, and 3.0 μM, respectively, and against MCF-7 breast cancer cell-line at IC50 values of 4.0, 0.5, and 1.0 μM, respectively [204].
Trungapeptins A-C are cyclodepsipeptides isolated from Lyngbya majuscula. These three peptides are closely related to the antanapeptins, which are a series of depsipeptides isolated from the same cyanobacterium. Trungapeptin A exhibited mild toxicity towards brine-shrimp at 10 ppm and ichthyotoxicity at 6.25 ppm, with no activity against LoVo colon carcinoma and KB cervical adenocarcinoma cell-lines at 10 μg/mL [216]. Antanapeptins A-D are other depsipeptides isolated from the same cyanobacterium with no antimicrobial activity at concentrations of 100 μg/disk [217].
Palmyramide A is an unusual cyclic depsipeptide isolated from Lyngbya majuscula which included three hydroxy acids and three amino acids. The 2,2-dimethyl-3-hydroxyhexanoic acid unit [Dmhha] found in palmyramide A was also found in guineamide F [196]. Palmyramide A is known to block the voltage-gated sodium channel in neuro-2a cells at an IC50 value of 17.2 μM, and exhibited mild cytotoxicity against human lung H-460 carcinoma cell-line at an IC50 value of 39.7 μM. Dudawalamides A-E are cyclic lipopeptides isolated from the Lyngbya sp. These depsipeptides are structurally similar to pitipeptolides A and B, antanapeptins, kulolides, and mantillamide A which were all isolated from the Lyngbya sp [218]. Dudawalamide A has a planar structure of 2,2-dimethyl-3-hydroxy-7-octynoic acid unit and was found to exhibit anti-parasitic activity [204]. Dudawalamides A-E displayed greatest biological activity when tested in Chagas, leishmania, anti-parasitic, and malarial assays. Dudawalamides A, B, D, and E displayed cytotoxicity against Plasmodium falciparum at IC50 values of 2.7, 7.6, 3.7, and 7.7 μM, respectively, and against Leishmania donovani at IC50 values of 25.9, 14.7, 2.6, and 2.6 μM, respectively. Dudawalamide E was cytotoxic against Trypanosoma cruzi at an IC50 value of 7.3 μM [218].
Grassypeptolide is a macrocyclic depsipeptide isolated from Lyngbya confervoides with one β-amino acid, an unusually high D-amino acid content, and two thiazolines. Grassypeptolide exhibited cytotoxicity against IMR-32 neuroblastoma, HeLa cervical carcinoma, HT29 colorectal adenocarcinoma, and U2OS human osteosarcoma cancer cell-lines with IC50 values ranging between 1.0–4.2 μM [204]. Carriebowmide is another cyclodepsipeptide isolated from L. polychroa and was previously isolated from L. majuscula along with two other depsipeptides: itralamides A and B. Carriebowmide included two rare amino acids: methionine sulfoxide and 3-amino-2-methylhexanoic acid. Carriebowmide was tested as a feeding deterrent whose effectiveness was not determined [204].
Hoiamide A is a bioactive cyclic depsipeptide isolated from Lyngbya majuscula. Hoiamide A possessed a 15 carbon subunit from C30 to C44 which was postulated to have been derived from the polyketide pathway. Hoiamide A is a partial agonist of the voltage-gated sodium channel α subunit at site-2 along with exhibiting modest cytotoxicity against cancer cells, and inhibiting batrachotoxin induced Na+ elevation in a concentration-dependent manner [204].
Tiglicamides A-C were isolated from Lyngbya confervoides along with largamides A-C. Tiglicamides and largamides differed by only one amino acid residue in the cyclic core. This difference could result from the unusual relaxed specificity arising from the adenylation domains of the NRPS assembly or from a separate biosynthetic pathway. Tiglicamides A-C and largamides A-C are serine-protease inhibitors with elastase selectivity over trypsin and chymotrypsin. The carboxylic-acid residue present in the compounds showed little effect on the elastase inhibitory activity which was confirmed from the semi-synthetic methyl ester analogs of largamides that exhibited low micromolar inhibitory activities [204].
Itralamides A and B are two depsipeptides isolated from Lyngbya majuscula. Itralamide B exhibited significant cytotoxicity in HEK-293 human embryonic kidney cell-line at an IC50 value of 6 ± 1 μM while itralamide A showed a ten-fold lower potency. The cytotoxic difference between the two itralamides showed that the biological activities could be dramatically altered from minor structural modifications [204].
Lyngbyastatins 4–6 were isolated from Lyngbya confervoides while lyngbyastatins 8–10 were isolated from L. semiplena. Other peptides including lyngbyastatin 7, kempopeptins A and B, and somamide B were isolated from another Lyngbya sp., collected from the mangrove channel in Florida at Summerland Key. This class of compounds was known as serine protease inhibitors along with exhibiting varied selectivity and wide potency range. Extensive studies on the structure-activity relationships and the crystal structures of a related depsipeptide scyptolin A bound to the elastase and the cyanopeptolin (A90720A) bound to trypsin exposed significant interactions in the enzyme binding site, providing an insight into the selectivity of this class of inhibitors. Lyngbyastatins 8–10 showed weak inhibition against porcine pancreatic elastase when compared to lyngbyastatin 7 [204]. Somamides A and B are depsipeptides isolated from L. majuscula and Schizothrix sp., whose structures are analogous to symplostatin 2 and dolastatin 13 structures. The biological activities of somamides A and B have not been reported [219].
Lyngbyastatins 7–10 shared the same depsipeptide core, but the reduced potency of lyngbyastatins 8–10 could have been associated with the differences in the side-chain residues. These compounds were known to include hydrophobic residues, exclusively in the pendant chain which are considered to be responsible for the hydrophobic bonding and electrostatic interactions with the enzyme. Lyngbyastatin 6, a O-methylated [Amp] derivative was able to retain the protease-inhibitory activity explaining the fact that the presence of a hydroxyl group in the Ahp unit is not significant for the inhibition of chymotrypsin or elastase [204].
Lyngbyazothrins A-D, schizotrin A, and pahayokolides A-B were known to show structural similarity. Lyngbyazothrins A-D and pahayokolides A-B were produced from the cultured Lyngbya sp., and from freshwater Lyngbya sp., respectively. Schizotrin A was isolated from Schizotrix sp., while tychonamides were isolated from Tchyonema sp. These cyclic undecapeptides included the [Val/Ile/Dhb]-Ser-Dhb-[Ser/Thr]-[homo-Phe/homo-Tyr]-Pro-X-Gln-Gly-Pro-[Pro/Phe] sequence where X is an unusual long chain: α, γ-hydroxy-β-amino acid. In lyngbyazothrins, schizotrin A, and pahayokolides, the α, γ-hydroxy-β-amino acid is a 3-amino-2,5,7,8-tetrahydroxy-10-methylundecanoic acid [Athmu], while in tychonamides it is a 3-amino-2,5,7-trihydroxy-8-phenyloctanoic acid moiety [Atpoa]. The γ-hydroxy group has not been reported to decorate the ester linkage on the N-acetyl-N-methyl tyrosine [lyngbyazothrins], an N-butyroyl-N-methyl alanine [schizotrin A] or an N-acetyl-N-methyl leucine [pahayokolides and tychonamides]. The mixture of lyngbyazothrins A and B exhibited low antimicrobial activity against Micrococcus flavus, while the mixture of lyngbyazothrins C and D was found to be active against Serratia marcescens, Bacillus subtilis, Pseudomonas aeruginosa, and Escherichia coli at concentrations between 25–200 μg/disk [204,220]. The acyl residue present at the C-5 position of Athmu appears to play a critical role in the antimicrobial activity which has been supported from the structure and activity of the pahayokolides A-B. Pahayokolide A showed acute toxicity towards zebrafish embryos at LC50 value of 2.15 μM, marginal toxicity against brine-shrimp at concentrations of 1 mg/mL, and inhibition against various cancer cell-lines at IC50 values ranging between 2.13 to 44.57 μM [204].
Grassypeptolides A-C are a group of closely related bis thiazoline containing cyclic depsipeptides [221] that were isolated from Lyngbya confervoides while grassypeptolides D-E and Ibu-epidemethoxylyngbyastatin 3 were isolated from Leptolyngbya collected off the Red Sea shipwreck, SS Thistlegorm. Grassypeptolide D exhibited cytotoxicity against neuro-2a mouse blastoma and HeLa cervical carcinoma cell-lines at IC50 values of 599 and 335 nM, respectively, while grassypeptolide E was cytotoxic at IC50 values of 407 and 192 nM, respectively. The cytotoxicity of grassypeptolide D was at least 1.5 times less compared to grassypeptolide E. Grassypeptolides D and E are threonine/N-methylleucine diastereomers while grassypeptolides A and C are N-methylphenylalanine epimers. Ibu-epidemethoxylyngbyastatin 3 showed low cytotoxicity to neuro-2a cell-line at an IC50 value of > 10 μM while grassypeptolides and dolastatin 12 were cytotoxic at an IC50 of > 1 μM [222].
Grassypeptolides F and G were isolated from Lyngbya majuscula. Grassypeptolides F and G are bis-thiazoline containing cyclic depsipeptides that included a rare β-amino acid, a large number of D-amino acids, and extensive N-methylation. Both grassypeptolides were found to exhibit moderate inhibition against the oncogenic AP-1 transcription factor at IC50 values of 5.2 and 6.0 μM, respectively [223].
Lyngbyapeptins A [224], B, C [225], and D [226] are tetrapeptides with a rare 3-methoxy-2-butenoyl moiety isolated from Lyngbya bouillonii [224,225]. All these peptides were found to be non-cytotoxic against LoVo and KB cell-lines at concentrations of < 5 μM [225]. Lyngbyabellin A [226], C [225], and J [226] are lipopeptides isolated from the same cyanobacterium. Lyngbyabellins A and C were found to be weakly cytotoxic against LoVo and KB cell-lines at IC50 values of 5.3 and 2.1 μM, respectively [225]. Lyngbyabellin B was isolated from L. majuscula along with lyngbyabellin A [227]. Lyngbyabellin B is a cyclic depsipeptide that was found to possess potent toxicity against the fungus Candida albicans at 100 μg/disk, and against brine shrimp at an LD50 value of 3.0 ppm [228]. Lyngbyabellins D-I were also isolated from L. majuscula where lyngbyabellins D, F, and H exhibited cytotoxicity against H460 and KB cancer cell-lines at LC50 or IC50 values ranging between 0.1 to 0.4 μM [229].
Lyngbyabellins K-N are lipopeptides isolated from two collections of extracts from marine cyanobacteria obtained from Palmyra Atoll. Lyngbyabellin L and its C-7 epimer, 7-epi-lyngbyabellin L, possessed a rare monochlorination on the 3-acyloxy-2-methyloctanoate residue while lyngbyabellin N included an unusual N,N-dimethylvaline terminus along with a leucine statin residue, showing strong cytotoxicity against HCT-116 colon cancer cell-line at an IC50 value of 40.9 ± 3.3 nM [230]. Alotamide A is another cyclic depsipeptide isolated from Lyngbya bouillonii which exhibited unique calcium-influx activation profile in the murine cyanobacterial metabolite at an EC50 value of 4.18 μM [231].
Barbamide is a chlorinated lipopeptide isolated from Lyngbya majuscula strain and is known for its potent molluscicidal activity [232]. Jamaicamides A-C are other lipopeptides isolated from L. majuscula. Jamaicamide A possessed a pyrrolinone ring, β-methoxy eneone system, alkynyl bromide, and vinyl chloride, and exhibited fish toxicity and sodium channel-blocking activity. All three jamaicamides exhibited cytotoxicity against neuro-2a mouse neuroblastoma and H-460 human lung cell-lines at an LC50 value of 15 μM along with sodium channel blocking activity at 5 μM. Jamaicamide C showed modest toxicity against brine shrimp at 10 ppm while jamaicamides A and B were inactive [233].
Hectochlorin is a lipopeptide isolated from Lyngbya majuscula. Hectochlorin exhibited potent antifungal activity against Candida albicans and antiproliferative activity in the NCI-60 cell-line assay. Hectochlorin also inhibited the human Burkitt lymphoma CA46 and actin cytoskeleton PtK2 cell-lines at IC50 values of 20 nM and 300 nM, respectively. Hectochlorin showed an 11 mm inhibition zone at 10 μg/disk against C. albicans and a 16 mm inhibition zone at 100 μg/disk but was inactive against P. aeruginosa, E. coli, S. aureus, B. subtilis, and Salmonella choleraesuis. Hectochlorin structurally resembled lyngbyabellins A and B, and dolabellin [234].
Apramides A-G are linear lipopeptides isolated from the lyngbyastatin-2 producing strain of Lyngbya majuscula. Apramides A-F were non-cytotoxic with no fungal, bacterial, or protease inhibiting activities but apramide A showed enhanced elastase-activity [235].
Aurilide is a potent cytotoxic cyclic depsipeptide isolated from Dolabella auricularia, a Japanese seahare [236], while aurilides B and C were isolated from Lyngbya majuscula. Aurilide induced apoptosis in human cells at picomolar to nanomolar range [236], while aurilides B and C exhibited cytotoxicity against neuro-2a mouse neuroblastoma and NCI-H460 human lung tumor cell-lines at LC50 values between 0.01 and 0.13 μM. Aurilide bound selectively to prohibitin-1 [PHB1] in mitochondria resulting in the activation of the proteolytic processing of optic-atrophy 1 [OPA1], thereby causing mitochondria induced apoptosis [236]. Aurilide B exhibited high level cytotoxicity against the NCI-60 cell-line with a GI50 concentration less than 10 nM, and was most active against prostate, leukemia, and renal cancer cell-lines [237]. Antillatoxin [238] is an ichthyotoxic cyclic lipopeptide isolated from L. majuscula at an LD50 value of 0.05 μg/mL, while barbaramide A [239] exhibited potent molluscicidal activity at an LD100 value of 10 μg/mL [240]. Kalkitoxin is another neurotoxic lipopeptide isolated from L. majuscula and was found to be a more potent ichthyotoxic towards common goldfish, Carassius auratus, at an LC50 value of 700 nM. Kalkitoxin also inhibited cell-division in fertilized sea-urchin embryo assay at an IC50 value of around 25 nM, and was potently toxic to brine shrimp at an LC50 value of 170 nM. Kalkitoxin was found to be a potent blocker of voltage sensitive sodium channel in mouse neuro-2a cell-line at an EC50 value of 1 nM [241].
Georgamide is a cyclic depsipeptide with an alkynoic acid residue and was isolated from the Australian cyanobacterium [242], while yanucamides A and B are two depsipeptides isolated from Lyngbya majuscula and Schizothrix sp. Both yanucamides possessed a unique 2,2-dimethyl-3-hydroxy-7-octynoic acid and exhibited strong brine-shrimp toxicity at an LD50 value of 5 ppm [243].
Dysidenamide, pseudodysidenin, and nordysidenin are lipopeptides isolated from Lyngbya majuscula while isodysidenin was isolated from the New Guinean and Australian sponge, Dysidea herbacea. Pseudodysidenin exhibited cytotoxicity against MEL-28, P-388, HT-29, and A-549 cell-lines at IC50 values of > 1 μg/mL [244].
Ulongamides A-F are cyclic depsipeptides isolated from Lyngbya majuscula. Ulongamides are β-amino acid containing depsipeptides where ulongamides A-E exhibited weak cytotoxicity against LoVo and KB cell-lines at IC50 values of 5 μM and 1 μM, respectively. Ulongamide F was inactive at < 10 μM against both the cell lines [245].
Malyngamides are a group of mixed polyketide-peptides isolated from Lyngbya majuscula. Malyngamides A and B, and isomalyngamides A and B were isolated from L. majuscula. Malyngamides A and B are chlorine-containing amides of 7(S)-methoxytetradec-4(E)-enoic acid. Isomalyngamides showed lethal toxicity towards crayfish [246]. Malyngamide C along with its stereoisomer 8-epi-malyngamide C were also isolated from L. majuscula. Both malyngamide C and 8-epi-malyngamide C exhibited cytotoxicity against HT29 colon cancer cell-line at IC50 values of 5.2 and 15.4 μM, respectively [247]. Malyngamides D and E are trans-7-methoxy-9-methylhexadec-4-enamides isolated from L. majuscula [248]. Malyngamide F was also isolated from L. majuscula. Malyngamide G is a chlorine containing 7(S)-methoxydodec-4(E)-enamide isolated from a blue-green alga, Cystoseira crinite [249] and from L. majuscula as well. Malyngamide F was found to be potent against NO assay at an IC50 value of 5.4 μM [250], while malyngamide G was found to be non-cytotoxic against KB cell-line but showed immunosuppressive activity at ED50 value of 6 μg/mL with lipopolysaccharide (LPS) and concanavaline K cells [251]. Malyngamides H-K were isolated from L. majuscula where malyngamides H, I, and K exhibited slight reduction of nitrite-production. Malyngamide I also exhibited brine shrimp toxicity at an LD50 value of 35 μg/mL, and goldfish toxicity at an LD50 of < 10μg/mL [252]. Malyngamides J and T did not show any anti-inflammatory activity due to their strong cytotoxicity of 60% and 9% cell-survival, respectively [250].
Malyngamides L and T were isolated from Lyngbya majuscula, while malyngamides M and N were isolated from Gracilaria coronopifolia, a Hawaiian red alga. Malyngamide N exhibited moderate cytotoxicity against mouse neuroblastoma (NB) cell-line at an IC50 value of 4.9 μg/mL, while malyngamide M showed weak cytotoxicity at an IC50 value of > 20 μM [253]. Malyngamides O and P were isolated from Stylocheilus longicauda, a marine sea hare. Malyngamide O exhibited moderate activity against human colon HT-29 carcinoma, lung A-549 carcinoma, and mouse P-388 lymphoma cell-lines at an IC50 value of 2 μg/mL [254]. Malyngamides Q and R were also isolated from L. majuscula where malyngamide Q did not show any biological activity due to its instability, while malyngamide R exhibited brine-shrimp toxicity at an LD50 value of 18 ppm [255]. Malyngamide S was isolated from Bursatella leachii, a New Zealand sea hare. Malyngamide S exhibited anti-proliferative activity against human leukemic HL60 cell-line at an IC50 value of ~ 6–8 μM and showed cytotoxicity towards P388 murine leukemia cell-line at an IC50 value of 29 μM and against NCI panel at LC50 value of 69.2 μM, GI50 value of 16.6 μM, and TGI value of 35.5 μM. Malyngamide S also exhibited cytotoxicity against BSC-1 cell-line at 120 μg/disk, and anti-tubercular activity at 6.25 μg/mL [256]. Malyngamides U-W were isolated from L. majuscula, and did not show any brine shrimp toxicity [257]. Malyngamide X was the first (7R)-lyngbic acid with a novel tripeptide backbone which was isolated from Bursatella leachii, a Thai sea hare [258]. Malyngamide X was found to possess antimalarial activity against P. falciparum, and was found to be active against M. tuberculosis as well [259]. Malyngamide Y was isolated from Moorea producens, and exhibited anticancer activity. Malyngamide Y possessed four amino acid units including alanine, valine, proline, and N-methyl phenylalanine along with an unique unit: 2,2-dimethyl-3-hydroxy-octanoic acid [260].
Hermitamides A-B are lipopeptides isolated from Lyngbya majuscula. Hermitamides A-B represented the aromatized malyngamide type compounds and exhibited brine shrimp toxicity at LD50 values of 5 and 18 μM, respectively. Hermitamide A also displayed weak goldfish ichthyotoxicity at an LD50 value of 19 μM while hermitamide B was inactive at 25 μM. Both hermitamides were inactive in molluscicidal assay at a concentration of 10 ppm while showed cytotoxicity against neuro-2a-neuroblastoma cell-line at IC50 values of 2.2 and 5.5 μM, respectively [261]. Laxaphycins A [262] and B [262,263] are cyclic lipopeptides isolated from L. majuscula. Laxaphycin A is a cyclic undecapeptide, while laxaphycin B is a cyclic dodecapeptide. Laxaphycin A was found to be inactive at concentrations of 20 μM while laxaphycin B showed cytotoxic activities against drug sensitive CCRF-CEM human leukemic lymphoblasts at IC50 value of 1.1 μM along with preserving equal cytotoxicity against altered DNA-topoisomerase II associated MDR and Pgp-MDR cells [264].
Tasiamide and tasiamide B are cytotoxic peptides isolated from Symploca sp. Tasiamide is an acyclic peptide that exhibited cytotoxicity against LoVo and KB cell-lines at IC50 values of 3.47 and 0.48 μg/mL, respectively [265], while tasiamide B was cytotoxic against KB cell-line with an IC50 value of 0.8 μM [266]. Tasiamides C-E are lipopeptides which were also isolated from Symploca sp., and were found to be inactive against HCT-116 colon cancer cell-line at 25 μM [267]. Tasiamide F is a peptide isolated from Lyngbya sp., with a structure similar to pepstatin A, a natural aspartic protease inhibitor produced by Actinomycetes. Tasiamide F included a Phe-derived statin core and inhibited cathepsins D and E at IC50 values of 57 and 23 nM, respectively [268]. Tasipeptins A and B are cytotoxic depsipeptides isolated from Symploca sp., and exhibited cytotoxicity against KB cell-line at IC50 values of 0.93 and 0.82 μM, respectively [269].
Symplocamide A was isolated from Symploca sp., and exhibited potent cytotoxicity towards neuro-2a neuroblastoma and H-460 lung cancer cell-lines at IC50 values of 29 nM and 40 nM, respectively [270]. Symplocin A is a N,N-dimethyl-terminated peptide which was also isolated from Symploca sp., and was found to be a potent inhibitor of cathepsin E at an IC50 value of 300 pM [271]. Veraguamides A-G were isolated from Symploca cf. hydnoides, and were found to be moderately cytotoxic towards HTCLs [272].
Symplostatin 1 has been isolated from Symploca hydnoides and is a methyl derivative of dolastatin 10, which was also isolated from S. hydnoides apart from being initially isolated from the marine mollusk Dolabella auricularia. Symplostatin 1 induced proapoptotic stimuli in cancer cells similar to dolastatin 10 [273]. Symplostatin 2 is a cyclic depsipeptide and an analog of dolastatin 13 that was isolated from S. hydnoides with no reported biological activity [274]. Symplostatin 3 is another analog of dolastatin 10 that differs only in the C-terminal unit where the 3-phenyllactic acid moiety replaces the dolaphenine. Symplostatin 3 exhibited IC50 values of 3.9 nM and 10.3 nM against KB and LoVo cell-lines respectively [275]. Symplostatin 4 [Sym4] is a cyanobacterial secondary metabolite isolated from Symploca sp. Sym4 was found to be a potent inhibitor of the malarial parasite, P. falciparum with IC50 values ranging between 36–100 nM. Symplostatin 4 was also known to cause a food vacuole phenotype in Plasmodium infected red blood cells and is known to inhibit the pathogen replication at an EC50 value of 0.7 μM [276].
Malevamides A-C are depsipeptides isolated from Symploca laete-viridis and were found to be inactive against human colon HT-29 carcinoma, mouse P388 lymphoma, and human lung A-549 carcinoma cell-lines at a concentration of 2 μg/mL. Malevamides A-C included structural features such as α-hydroxy acids, β-amino acids, and N-methylation [277]. Malevamide D and belamide A were isolated from S. hydnoides. Malevamide D is a highly cytotoxic peptide ester, while belamide A is a linear tetrapeptide belonging to similar compound class as dolastatin-10. Belamide A and malevamide D exhibited antiproliferative activity and subsequently, tubulin disrupting effects [273]. Largazole is another cytotoxic cyclodepsipeptide that was also isolated from Symploca sp. and included a substituted 4-methyl thiazoline, linearly fused to thiazole. Largazole exhibited cytotoxicity against human epithelial MDA-MB-231, murine mammary epithelial NMuMG, fibroblastic osteosarcoma U2OS, and nontransformed fibroblasts NIH3T3 cancer cell-lines at GI50 values of 7.7, 122, 55, and 480 nM, respectively [278].
Mitsoamide is a cytotoxic linear lipopeptide isolated from the Madagascar marine cyanobacterium, Geitlerinema sp., and included an unusual polyketide unit: 3,7-dimethoxy-5-methyl-nonanedioic acid [DMNA], along with a highly unusual piperidine aminal moiety, and a homolysine residue. Mitsoamide was found to be cytotoxic against NCI-H460 human lung tumor cell-line at an IC50 value of 460 nM [279]. Gallinamide A is an antimalarial peptide isolated from Schizothrix sp. Gallinamide A exhibited potent antimalarial activity against W2 chloroquine-resistant strain of Plasmodium falciparum at an IC50 value of 8.4 μM [280].
Nostocyclamide is a macrocyclic thiazole containing allelochemical isolated from the cyanobacteria belonging to the Nostoc sp. Nostocyclamide is cyclic peptide and an antialgal and an anticyanobacterial secondary metabolite with toxicity against Brachionus calyciflorus. Nostocyclamide also inhibited the growth of Anabaena P-9 at a concentration of 0.1 μM [281]. Tenuecyclamides A-D are cyclic hexapeptides isolated from Nostoc spongiaeforme var. tenue. Tenuecyclamide A inhibited sea-urchin embryos at an ED100 value of 10.8 μM, while tenuecyclamides C and D exhibited inhibition at ED100 values of 9.0 μM and 19.1 μM, respectively [282]. Cryptophycin was also isolated from Nostoc genus and is a cytotoxic dioxadiazacyclohexadecenetetrone exhibiting antifungal activity with unknown mechanism. Cryptophycin was a novel antimicrotubule compound and was a poor substrate for P-glycoprotein compared to Vinca alkaloids [283].
Microcystins are monocyclic heptapeptide liver-toxins from cyanobacteria of both marine and/or freshwater belonging to the genera Nostoc, Oscillatoria, Anabaena, and Microcystis. Over 50 different microcystins have been isolated so far that differ primarily in two L-amino acids plus demethylation and methylation on the two unusual amino acids. All microcystins included an Adda [(2S,3S,8S,9S)-3-amino-9-methoxy-2,6,8-trimethyl-10-phenyldeca-4,6-dienoic acid] that is essential for the biological activity. Microcystins were found to be potent protein phosphatase types 1 and 2A inhibitors as well as tumor-promoters [284]. Raocyclamides A and B are two thiazole- and oxazole-containing cyclic hexapeptides isolated from Oscillatoria raoi where raocyclamide A exhibited moderate cytotoxicity against sea-urchin embryos [285]. Venturamides A-B are cyclic hexapeptides that were isolated from marine Oscillatoria species. Venturamides A-B exhibited antimalarial activity against W2 chloroquine-resistant strain of malaria, Plasmodium falciparum at IC50 values of 8.2 and 5.6 μM, respectively, while also exhibiting mild cytotoxicity against mammalian Vero cells at IC50 values of 86 and 56 μM, respectively [286].
Anabaenopeptins A and B are cyclic peptides isolated from Anabaena flos-aquae, and produced concentration dependent relaxations in rat aortic preparations with endothelium at concentrations of 10–400 μg/mL [287]. Anabaenopeptins C and D were isolated from the hepatotoxic cyanobacteria, Anabaena sp. [288], while anabaenopeptins E and F are ureido bond containing cyclic peptides isolated from Oscillatoria agardhii with no reported biological activities [289]. Anabaenopeptins G and H were also isolated from O. agardhii as potent carboxypeptidase-A [CPA] inhibitors at IC50 values of 0.0018 and 3.4 μg/mL, respectively, with no activity against proteases [290]. Pompanopeptins A and B were isolated from L. confervoides. Pompanopeptin A is a 3-amino-6-hydroxy-2-piperidone containing peptolide, while pompanopeptin B is a cyclic pentapeptide. Pompanopeptin B possessed N-methyl-2-amino-6-(4′-hydroxyphenyl) hexanoic acid [N-Me-Ahpha] and was structurally related to the carboxypeptidase A inhibitors: anabaenopeptins I and J, which were also isolated from the cyanobacterium Aphanizomenon flos-aquae. The N-methyl-L-Ahpha and L-Htyr in pompanopeptin B were replaced by the N-methyl-L-alanine and L-leucine/L-phenylalanine residues of anabaenopeptins J and I, respectively. Pompanopeptin A was found to selectively inhibit trypsin over chymotrypsin and elastase at an IC50 value of 2.4 μM which was due to the presence of the arginine residue in the cyclic core. There was no biological data reported for pompanopeptin B [204].
(+)-Floridamide is a cyclic depsipeptide isolated from Moorea producens and exhibited cancer cell cytotoxicity. The cyclic depsipeptide possessed four amino acid residues including proline, alanine, valine, and N-methyl phenylalanine [260]. Coibamide A [291] is a potent antiproliferative depsipeptide isolated from Leptolyngbya sp. Coibamide A exhibited potent cytotoxicity against mouse neuro-2a and NCI-H460 lung cancer cell-lines at LC50 values of < 23 nM but did not interfere with actin or tubulin in the cytoskeletal assays. Coibamide A was known to dose dependently increase the number of cells in G1 phase of cell cycle with a little change in the G2/M phase and a complete loss of cells in the S phase. Coibamide A was also evaluated against the in vitro panel of NCI 60 cancer cell-lines with high potency against MDA-MB-231, HL-60 (TB), SNB-75, and LOX IMVI at GI50 values of 2.8, 7.4, 7.6, and 7.4 nM, respectively, and showed good selectivity for CNS, ovarian, breast, and colon cancer cell-lines. Coibamide A could be a promising lead in the drug discovery of cancer with potentially new mechanism of action [291]. Hormothamnin A is a lipophilic cyclic peptide isolated from Hormothamnion enteromorphoides, a tropical marine cyanobacterium. Hormothamnin A was an undecapeptide that exhibited antimicrobial and cytotoxic properties and resembled laxaphycin A with minute differences in the stereochemistry of the DHHA unit [292]. Trichamide is a small cyclic peptide isolated from a bloom-forming Trichodesmium erythraeum with no significant activity [293]. Scytonemin is a small molecule polo-like kinase 1 inhibitor isolated from Stigonema sp., and possessed a unique chemical structure with a potential scaffold, for further chemical modification that could be used for the development of therapeutically useful compounds in the treatment of hyperproliferative disorders due to its nontoxic and antiproliferative activity. Scytonemin is a natural product thought to stem from the condensation of tryptophan and phenylpropanoid derived subunits. Scytonemin inhibited the capability of GST-polo-like kinase 1 to phosphorylate GST-cdc25C at an IC50 value of 2.3 μM ± 3.6 in a concentration dependent manner [294].
Arenamides A-C are cyclohexadepsipeptides isolated from the marine bacterial strain, Salinispora arenicola. Arenamides A and B were known to block the TNF-induced activation in a time- and dose-dependent manner at IC50 values of 3.7 and 1.7 μM, respectively. These peptides also inhibited the PGE2 and nitric oxide (NO) production with LPS-induced RAW 264.7 macrophages, but exhibited moderate cytotoxicity with HCT-116 human colon carcinoma cell-line [295]. Caldoramide is a linear pentapeptide isolated from Caldora penicillata. Caldoramide showed cytotoxicity against HCT116 KRAS cell-line at IC50 values of 3.9 μM to 5.2 μM [296].
Viequeamides A-F belonging to the kulolide superfamily of cyclic depsipeptides were isolated from the Rivularia sp., where viequeamide A was found to be extremely toxic to the human lung cancer H460 cell-line at an IC50 value of 60 ± 10 nM, while viequeamides B-F were found to be inactive. Viequeamides belonged to the 2,2-dimethyl-3-hydroxy-7-octynoic acid [Dhoya] comprising cyclic depsipeptides [297]. Companeramides A-B were cyclic depsipeptides with a 3-amino-2-methyl-7-octynoic acid [Amoya] and hydroxy isovaleric acid along with eight other α-amino acid units. Companeramides A-B were isolated from the same cyanobacteria as coibamide A and exhibited nanomolar in vitro anti-plasmodial activity, but were inactive against cancer cell-lines at concentrations of 1 μM [298].
Nodulapeptins A-B and spumigins A-C were isolated from the toxic Nodularia spumigena. Nodulapeptins A-B are cyclic peptides similar to anabaenopeptins that were isolated from Anabaena species, while spumigins A-C are linear peptides which were considered to be the precursors of aeruginosins containing agmatin, argininal, and argol isolated from the freshwater cyanobacteria, Microcystis aeruginosa [299]. Spumigins D-H were also isolated from N. spumigena. Spumigins are linear tetrapeptides with proteinogenic amino acids which included a C-terminal alcohol derivative of arginine [300]. Spumigin E and aeruginosins were also isolated from N. spumigena. Aeruginosins are a family of linear peptides that are known to be serine protease inhibitors [301].
Kailuins A-D are cyclic acyldepsipeptides that were isolated from the liquid cultures of BH-107, a Gram-negative bacterium collected from the Kailua beach. Kailuins A-D showed mild cytotoxicity against A-549 lung cancer cell-lines at GI50 values of 3, 2, 3, and 2 μg/mL, respectively, MCF-7 breast cancer cell-lines at GI50 values of 3, 2, 4, and 3 μg/mL, respectively, and HT-29 colon cancer cell-lines at GI50 values of 3, 3, 3, and 2 μg/mL, respectively [302]. Kailuins E-H were also isolated from Photobacterium halotolerans similar to kailuins A-D. Kailuins B, C, D, E, G, and H exhibited cytotoxicity against HCT-116 cancer cell-line at IC50 values of 22, 50, 28, 18, 32, and 17 μM, respectively, while kailuin D showed cytotoxicity against MCF-7 breast cancer cell-line at IC50 value of 39 μM [303].
Ngercheumicins A-I [304,305] are other depsipeptides that were isolated from the Photobacterium strains. Ngercheumicins were found to be active against Pseudovibrio denitrificans. Of these, ngercheumicins A-B included a depsipeptide macrocycle with one Phe and two Leu residues with various fatty-acid tails while ngercheumicins C-E included a macrocycle with three Leu, two Thr, and one Ser with no fatty-acid tail [304]. Ngercheumicins F-I inhibited the regulatory rnaIII transcription in S. aureus which is an effector molecule of agr QS system, making these a novel class of peptide-signaling molecules in the marine environment [305]. Unnarmicins A and C are antibiotic depsipeptides that were isolated from the fermentation broth of Photobacterium species [306]. Unnarmicins A and C inhibited rhodamine 6G efflux at IC50 values of 3.61 and 5.65 μM, respectively, along with inhibition of the CaCdr1p ATPase activity (in vitro) with IC50 values of 0.495 and 0.688 μM, respectively. Unnarmicins A and C could be potential candidates as adjuvants for antifungal chemotherapy [307].
Ariakemicins A and B are linear hybrid polyketide nonribosomal peptide antibiotics that were isolated from the fermentation extract of Rapidithrix species. Ariakemicins A and B are positional isomers with a double bond and included a threonine, δ-isovanilloylbutyric acid, and two ω-amino-(ω−3)-methyl carboxylic acids with triene or diene units. Ariakemicins exhibited cytotoxicity against baby hamster BHK kidney and human lung A549 cancer cells at IC50 values of 15 and 25 μg/mL, respectively [308]. Mollemycin A is a glyco hexadepsipeptide-polyketide that was isolated from a marine derived Streptomyces species. Mollemycin A exhibited extremely selective and potent growth-inhibition against gram-negative and gram-positive bacteria at IC50 values of 10–50 nM and multi-drug resistant and drug-sensitive clones of Plasmodium falciparum at IC50 values of 9 and 7 nM, respectively [309]. Thiocoraline is a depsipeptide that was isolated from the marine actinomycete strain, L-13-ACM2-092 and exhibited potent cytotoxicity against MEL-28, P-388, and A-549 cell-lines at an IC50 value of 0.002 μg/mL along with strong anti-microbial activity against gram-positive microorganisms by binding to the super-coiled DNA and inhibiting the RNA-synthesis [310]. Cyclomarins A-C are cyclic heptapeptides that were isolated from Streptomyces sp. CNB-982. Cyclomarin A exhibited potent anti-inflammatory activity with 92% inhibition at a dose of 50 μg/ear [311]. Cyclomarin D along with cyclomarazines A and B are other cyclic peptides that were isolated from Salinispora arenicola CNS-205. Cyclomarazines A-B are diketopiperazine dipeptides [312].
Surugamides A-E are cyclic octapeptides that were isolated from the broth of a marine-derived Streptomyces species. Surugamides A-E included four D-amino acid residues and exhibited inhibition against bovine cathepsin-B at IC50 values of 21, 27, 36, 18, and 16 μM, respectively [313]. Surugamide F is a linear decapeptide with a 3-amino-2-methylpropionic acid residue that was isolated from the mycelium extract of Streptomyces sp. JAMM992 [314]. Champacyclin is another cyclic octapeptide that was also isolated from the marine sediments of Streptomyces champavatii and exhibited antimicrobial activity against Erwinia amylovora, a phytopathogen, responsible for the fire-blight disease in plants. Champacyclin showed 40% inhibition of E. amylovora at 25 μM concentration [315].
Tumescenamides A and B are cyclic depsipeptides isolated from the marine bacterium Streptomyces tumescens YM23-260. The two tumescenamides differed in the replacement of 2,4-dimethylheptanoyl residue at α-NH2 position in tumescenamide A with a 2,4,6-trimethylnonanoyl residue in tumescenamide B. Tumescenamide A induced reporter gene-expression under controlled insulin degrading enzyme (IDE) promoter and hence could be a promising candidate for the treatment of Alzheimer’s disease [316]. Tumescenamide C is another cyclic lipodepsipeptide that was isolated from the broth of Streptomyces sp. KUSC_F05. Tumescenamide C is a congener of tumescenamides A and B and exhibited selective antimicrobial activity against Streptomyces strains with a growth inhibitory zone at 3.0 μg/disk [317]. Streptocidins A-D are cyclic decapeptide antibiotics that were isolated from the culture of Streptomyces sp. Tu 6071. Streptocidins possessed a distinct feature in that they included L-Leu at position 6 unlike loloatins and tyrocidines which possessed L-Trp or L-Phe [318,319]. Streptocidins C and D showed antimicrobial activity against B. subtilis, S. aureus, S. viridochromogenes, and S. sp. Tu 6071 at MIC values between 1–3 μg/mL [320].
Salinamides A-E are anti-inflammatory depsipeptides that were isolated from Streptomyces sp. CNB-091. Salinamides A and B exhibited 84% and 83% inhibition of edema at a testing dose of 50 μg/ear, respectively, and also showed moderate antibiotic activity against gram-positive bacteria Streptomyces pyrogenes and S. pneumoniae at MIC values of 4 μg/mL for salinamide A and 2 μg/mL for salinamide B [321]. Salinamide F was later isolated from the same Streptomyces strain on further exploration. Similar to salinamide A, salinamide F also exhibited significant antibacterial and RNA-polymerase (RNAP) inhibitory activities. Salinamide F displayed significant antibacterial activity against E. coli at MIC value of 0.20 μg/mL, Enterococcus faecalis and Haemophilus influenzae at MIC values of 12.5 μg/mL, Neisseria gonorrhoeae with MIC value of 25 μg/mL, Enterobacter cloacae with MIC value of 50 μg/mL, and S. aureus at MIC value of 100 μg/mL. Salinamide F exhibited potent gram-negative and gram-positive bacterial RNAP inhibition against S. aureus with an IC50 value of 4 μM and against E. coli RNAP with IC50 value of 2 μM [322].
Tauramamide is an antibiotic lipopeptide that was isolated from the marine bacterial cultures of Brevibacillus laterosporus PNG276. Tauramamide is acetylated at the N-terminus and included two D amino acids; the hallmarks indicating the non-ribosomal peptide synthase biosynthetic origin. Tauramamide exhibited selective and potent inhibition of the pathogenic Enterococcus sp. at minimum inhibitory concentration of 0.1 μg/mL [323]. Tupuseleiamides A and B are acyldipeptides that were also isolated from B. laterosporus PNG 276. Tupuseleiamides A and B had a nonprotein D configuration and differed only in the presence of a methyloctanoyl fragment [324]. Bogorols A-E are linear cationic peptides that were also isolated from B. laterosporus PNG276. Bogorols were the first known linear cationic peptides with both N-terminal α-hydroxy acid and C-terminal aminol modifications. Bogorols A-E exhibited potent and selective activity against methicillin-resistant Staphylococcus aureus [MRSA] at MIC of 1–8 μg/mL range and also against vancomycin-resistant Enterococcus spp. [VRE] at MIC of 9–75 μg/mL range [325].
Loloatins A-D are cyclic decapeptide antibiotics that were isolated from a tropical marine bacterium MK-PNG-276A. Loloatins A-D showed potent antibiotic activity against MRSA, VRE, and the drug-resistant Streptococcus pneumoniae, but only loloatin C exhibited antibacterial activity against the gram-negative E. coli. Loloatin D was four fold less active against gram-positive bacteria compared to other loloatins [326]. Marthiapeptide A is a tristhiazole-thiazoline containing cyclic peptide that was isolated from Marinactinospora thermotolerans SCSIO 00652. Marthiapeptide A showed antibacterial activity against gram-positive bacteria between MIC values of 2.0 to 8.0 μg/mL, and exhibited anticancer activity between IC50 values of 0.38 to 0.52 μM [327].
Loihichelins A-F are amphiphilic peptide siderophores that were isolated from Halomonas sp. LOB-5 which is a heterotrophic Mn(II)-oxidizing bacterium [328]. Siderophores are high affinity iron (III) ligands with low molecular-weight, which are produced by bacteria in order to solubilize and promote the uptake of iron under low iron-conditions [329]. Loihichelins included a hydrophilic head group with an octapeptide comprising of a series of amino acids, appended by fatty acids ranging from tetradecanoic acid to decanoic acid [328]. Marinobactins are another class of marine bacterial siderophores with a distinct amphiphilic structure that were isolated from a Marinobacter sp. DS40M6. Marinobactins included a unique peptidic head-group which coordinates with iron (III) along with one series of fatty acid appendages. The membrane affinity of marinobactins and the changes due to iron-binding could help aid in the receptor-assisted iron acquisition process [330].
Aquachelins are other amphiphilic peptide siderophores that were isolated from Halomonas aquamarina and are characterized with a 7-aa peptide with N-terminal appendages that range from C-12 to C-14 fatty acids. The fatty acids present in both aquachelins and marinobactins included both unsaturated and saturated alkyl chains with one double bond in cis configuration [331]. Petrobactin is a photoreactive citrate based catecholate-type siderophore that was isolated from the culture of Marinobacter hydrocarbonoclasticus, an oil-degrading marine bacterium. Petrobactin is a bis-catecholate α-hydroxy acid siderophore which included a unique 3,4-dihydroxybenzoyl chelating subunit rather than the usual 2,3-dihydroxybenzoyl moiety [332]. Amphibactins are amphiphilic siderophores that were isolated from Vibrio sp. R-10, a γ-Proteobacterium. All amphibactins possessed similar Tris-hydroxamate containing peptidic headgroup with one serine residue and three ornithine residues but differed in the C-14 to C-18 acyl appendage, thereby varying in the degree of hydroxylation and saturation [331]. Aerobactin is a hydroxamate-siderophore that was isolated from the marine Vibrio sp. DS40M5 which was previously isolated from other organisms [333]. Amphi-enterobactin is another siderophore that was isolated from V. harveyi BAA-1116. Amphi-enterobactins are fatty acid derivatives with a fourth serine incorporated into the trilactone backbone to form a tetralactone ring. The presence of an acyl moiety increased the hydrophobicity of amphi-enterobactins causing them to associate with the cell [334].
Alterobactins A and B are other siderophores that were isolated from Alteromonas luteoviolacea. Alterobactins were the first natural products that included 4,8-diamino-3-hydroxyoctanoic acid known for its implication in peptide protease inhibition [335]. Pseudoalterobactins A and B are siderophores that were isolated from Pseudoalteromonas sp. KP20-4 collected from the marine sponge, Cinachyrella australiensis. Alterobactins and pseudoalterobactins resembled in the possession of a catechol and two β-hydroxy-Asp residues [336].
Trivanchrobactin, divanchrobactin, and vanchrobactin are catecholamide compounds isolated from Vibrio sp. DS40M4, a marine bacterium. Vanchrobactin included a L-serine, D-serine, and 2,3-dihydroxybenzoic acid, while trivanchrobactin is a trimer of vanchrobactin joined with two serine ester-linkages [337]. Anguibactin is another siderophore which belonged to the phenolate category isolated from Vibrio anguillarum 775 [338]. Anguibactin exhibited cytotoxicity against murine P388 leukemia cell-line at an IC50 value of 15 μM [337]. Vibrioferrin is another siderophore that was isolated from V. parahaemolyticus with the structure: 1-{2-[2[(5-carboxy-5-hydroxy-2-oxo-1-pyrrolidinyl)propionamide]ethyl} citrate existing in two epimeric forms that resulted from the cyclization of the keto group of 2-ketoglutaric acid residue and the amidic-nitrogen of the alanine residue. Vibrioferrin exhibited iron-uptake at Vmax and Km values of 54 pmol and 67 nM Fe/mg cell protein/min, respectively, showing that this was receptor-mediated [339].
Ochrobactins A-C are citrate-derived amphiphilic siderophores that were isolated from Ochrobactrum sp. SP18. Ochrobactins included a citric acid backbone amide that is linked to two lysine residues. Each ochrobactin had two appendages of which one is an acylated appendage: (E)-2-decenoic acid. The other acylated appendage is (E)-2-octenoic acid, octanoic acid, and (E)-2-decenoic acid for ochrobactins A, B, and C, respectively [340]. Synechobactins are other amphiphilic siderophores that were isolated from Synechococcus sp. PCC 7002. Synechobactins also possessed a citric acid backbone that is linked to two 1,3-diaminopropane units. Synechobactins A, B, and C differ in the terminal fatty-acid residues which were dodecanoic acid, decanoic acid, and octanoic acid, respectively [341].
C. Marine Fungi
Unguisins A, B [342], C [343], and D [344] are four cyclic heptapeptides isolated from a marine derived strain of Emericella unguis. Unguisin E is a γ-aminobutyric acid containing cyclic heptapeptide that was isolated from Aspergillus sp., [345,190] and unguisin F is another cyclic heptapeptide that was obtained from the endophytic fungus, Mucor irregularis, isolated from a medicinal plant Moringa stenopetala [346].
An inseparable mixture of two cyclic pentapeptides [2:1]: versicotides A-B [347], and the cyclopentapeptides: versicoloritides A-C were isolated from Aspergillus versicolor. Fellutamides A and B are cytotoxic peptides that were isolated from the cultured fungus, Penicillium fellutanum obtained from the gastro-intestine of the marine fish Apogon endekataenia [348]. Fellutamides C and D are two aldehyde peptides that were isolated from an undescribed species of Metulocladosporiella (Chaetothyriales), which are potent inhibitors of the fungal proteasome activity along with exhibiting substantial potency against human PC-3 tumor cell-line [349]. Fellutamide F is a lipopeptide that was isolated from A. versicolor and exhibited cytotoxicity against a panel of HTCLs [190]. Asperterrestide A is another antiviral and cytotoxic cyclic tetrapeptide that was isolated from Aspergillus terreus. Asperterrestide A exhibited cytotoxicity against MOLT4 and U937 cell-lines at IC50 values of 6.2 and 6.4 μM, respectively, while displaying antiviral activity against H1N1 and H3N2 influenza strains at IC50 values of 15 and 8.1 μM, respectively [350].
Sansalvamide A is a cyclic depsipeptide isolated from the mycelium of the fungus belonging to Fusarium genus obtained from the surface of the sea-grass Halodule wrightii, and exhibited potent anticancer activity. An analog of sansalvamide A caused G-phase cell cycle arrest in CD18 and AsPC-1 human pancreatic cancer cell-lines. Sansalvamide A also caused topoisomerase I inhibition resulting in cell-death with some apoptotic characteristics in certain cancer cells. Sansalvamide A showed selective in vitro cytotoxicity towards SK-MEL-2 melanoma and COLO 205 colon cancer cell-lines at IC50 values of 5.9 μg/mL and 3.5 μg/mL, respectively [34,351]. Scopularides A and B are two cyclodepsipeptides isolated from Scopulariopsis brevicaulis obtained from the marine sponge Tethya aurantium. Scopularides showed weak activity against gram-positive bacteria and no activity against gram-negative bacteria, but exhibited significant activity against several tumor cell-lines at 10 μg/mL [352,353]. Exumolides A-B are cyclic depsipeptides that were isolated from the marine fungus belonging to Scytalidium genus and were found to display anti-microalgal activity at 20 μg/mL against the marine chlorophyte alga belonging to Dunaliella species [354].
Chemical analysis of Penicillium bilaii, an Australian marine derived fungi, yielded cyclo-(L-Phe-L-Pro), cyclo-(L-Pro-L-Tyr), cyclo-(L-Pro-L-Val), and cis-bis(methylthio)silvatin. cis-bis(methylthio)silvatin was found to be weakly cytotoxic at an LD99 value of 0.15 μM [355].
Penilumamide is a lumazine peptide which was the first and the only peptide isolated from a red algal derived Penicillium sp., (CNL-388 strain) fungus. Penilumamide is a linear peptide with an unusual 1,3 dimethyllumazine-6-carboxylic acid starter unit and did not exhibit any cytotoxicity [356]. Penilumamides B-D are other lumazine peptides isolated from a gorgonian-derived marine fungus Aspergillus sp., (XS-20090B15) which was obtained from the inner part of the gorgonian Muricella abnormaliz along with a cyclic pentapeptide asperpeptide A. Penilumamides A-D did not exhibit any antiviral and/or acetylcholinesterase inhibitory activities, while asperpeptide A displayed antibacterial activity against Staphylococcus epidermidis and Bacillus cereus at an MIC value of 12.5 μM [357].
Emericellamides A and B are cyclic depsipeptides isolated from the co-culture of the fungus belonging to Emericella sp., and Salinispora arenicola, an actinomycete. Emericellamide A exhibited antimicrobial activity against MRSA (methicillin-resistant Staphylococcus aureus) at an MIC of 3.8 μM, while also displaying cytotoxicity against human colon carcinoma HCT-116 cell-line at an IC50 value of 23 μM. Emericellamide B showed moderate cytotoxicity against MRSA at an IC50 value of 6.0 μM [358,359]. Guangomides A and B are cyclic depsipeptides isolated from an unidentifiable sponge-derived fungus cultured from salt-water. Along with guangomides A and B, homodestcardin which was the first example of a destruxin derivative was also isolated. Guangomides A and B exhibited weak antibacterial activity against Enterococcus durans and Staphylococcus epidermidis at an MIC value of 100 μg/mL [360]. Azonazine is an unusual hexacyclic dipeptide isolated from Aspergillus insulicola which was a marine sediment-derived fungus. Azonazine displayed anti-inflammatory activity through NF-κB luciferase inhibition at an IC50 value of 8.37 μM and nitrite production at an IC50 value of 13.70 μM [361].
Endolides A-D and hirsutide were isolated from the sponge derived fungus Stachylidium sp. Hirsutide was previously isolated from the entomopathogenic fungus belonging to Hirsutella species. Endolides A and B were N-methylated peptides, while endolides C and D were tetrapeptide analogs. Endolides A and B showed affinity for vasopressin receptor 1A and serotonin receptor 5HT2B, respectively [362].
Dictyonamides A and B are peptides derived from the mycelium of the fungus isolated from the red alga Ceratodictyon spongiosum with no reported biological activity [363]. Simplicilliumtides A-H were linear tetrapeptides isolated from a deep-sea derived fungus Simplicillium obclavatum. Simplicilliumtides A-B are linear tetrapeptides with a 2-aminobenzoic acid residue, while simplicilliumtides C-H are acetylated linear di- or tri-peptides. Simplicilliumtides were evaluated for cytotoxic, acetylcholinesterase inhibition, antibacterial, and antifouling activities. Simplicilliumtides A and G exhibited weak cytotoxicity against human HL-60 leukemia cell-line at IC50 values of 64.7 and 100 μM, respectively, while simplicilliumtides E and H displayed cytotoxicity against K562 cell-line at IC50 values of 39.4 and 73.5 μM, respectively. Simplicilliumtide D displayed potent antifouling activity against Bugula neritina larvae settlement indicating its potential as a natural antifouling candidate [364].
Halolitoralin A is a cyclic hexapeptide while halolitoralins B-C are cyclic tetrapeptides isolated from the marine sediment derived Halobacillus litoralis along with three cyclic dipeptides: cyclo(Ile-Val), cyclo (Pro-Leu), and cyclo(Pro-Val). Halolitoralins A-C exhibited moderate antifungal activity along with weak anti-tumor activities in vitro, against human gastric BGC cell-line [365]. Sclerotides A-B were isolated from the marine derived halotolerant Aspergillus sclerotiorum. Sclerotides A-B are cyclic hexapeptides with dehydroamino acid and anthranilic acid units. Sclerotides A-B exhibited moderate antifungal activity against Candida albicans at MIC values of 7.0 and 3.5 μM, respectively, while sclerotide B also displayed weak cytotoxicity against HL-60 cell-line at an IC50 value of 56.1 μM and antibacterial activity against Pseudomonas aeruginosa at an MIC of 35.3 μM [366].
RHM1 and RHM2 were N-methylated linear octapeptides which were isolated from the fungus belonging to Acremonium species cultured from a marine sponge along with efrapeptin G. Efrapeptin G exhibited potent cytotoxicity while RHM1 and RHM2 displayed weak cytotoxicity against murine cancer cell-lines [367]. Efrapeptins are (peptides) potent inhibitors of mitochondrial ATPase isolated from the entomopathogenic fungus Tolypocladium niveum [368].
Cordyheptapeptides C-E were cycloheptapeptides that were isolated from Acremonium persicinum, a marine fungus. Cordyheptapeptides C and E exhibited cytotoxicity against NCI-H460, SF-268, and MCF-7 tumor cell-lines with IC50 values between 2.5 to 12.1 μM [369]. Cordyheptapeptides A and B were isolated from an insect pathogenic fungus Cordyceps sp. BCC 16173. Cordyheptapeptide A possessed an N-methyl-L-tyrosine while cordyheptapeptide B has an N-methyl-L-phenylalanine residue. Cordyheptapeptide A showed antimalarial activity at an IC50 value of 3.8 μM, while cordyheptapeptide B was inactive. Cordyheptapeptides A and B also exhibited moderate cytotoxicity against NCI-H187 cells with IC50 values of 0.18 and 3.1 μM, BC cells with IC50 values of 0.20 and 0.66 μM, Vero cells with IC50 values of 14 and 1.6 μM, and KB cells with IC50 values of 0.78 and 2.0 μM, respectively [370]. Oryzamides A-E were cyclohexadepsipeptides that were isolated from the Nigrospora oryzae PF18, a marine fungus obtained from the sponge Phakellia fusca. Oryzamides did not exhibit any antiparasitic, NF-κB inhibition, cytotoxic, or antibacterial activities [371].
D. Ascidians
Cycloxazoline was a cyclic hexapeptide isolated from Lissoclinum bistratum and showed cytokinetic inhibition and HL-60 leukemia cell accumulation in G2/M. Cycloxazoline exhibited cytotoxicity against T24 and MRC5CV1 cell-lines at an IC50 value of 0.5 μg/mL [34,372]. Diazonamide A was a cytotoxic peptide isolated from Diazona angulata and possessed a unique binding-site on tubulin, differing from the dolastatin 10 and Vinca alkaloid binding-sites. Diazonamide A and its analog were also shown to strongly bind to the microtubule ends and weakly bind to the unpolymerized tubulin. Diazonamide A showed potent in vitro activity against B-16 murine melanoma and HCT-116 human colon carcinoma cell-lines with IC50 values of < 15 ng/mL while diazonamide B isolated from D. chinensis was found to be less active [34,373,374]. The structure of diazonamide A has been revised with the altered spectroscopic and physical characteristics relative to those of the original structure [375]. Diazonamides C-E were macrocyclic peptides isolated from Diazona sp., and displayed moderate cytotoxicity against human tumor cell-lines [376].
Vitilevuamide was a bicyclic 13 amino-acid residue isolated from Didemnum cuculiferum and Polysyncranton lithostrotum. Vitilevuamide exhibited inhibition of tubulin-polymerization at an IC50 value of 2 μM possibly through interaction at a unique site. Vitilevuamide showed in vitro activity against P388 lymphocytic leukemia at 30 μg/kg along with stabilizing colchicine binding to tubulin at 9 μg/mL. Vitilevuamide was also found to be cytotoxic against various human tumor cell-lines at LC50 values of 6–311 nM [34,377].
Thiazoline-, thiazole-, and oxazoline-based cyclic octa- and heptapeptides were isolated from Lissoclinum patella. Examples include lissoclinamides, patellamides, ulithiacyclamide A which included a disulfide-bridge, and ascidiacyclamide which incorporated a C2-symmetry. Examples of compounds including oxazoline-based cyclic hexapeptides were bistratamides isolated from the Australian Great Barrier Reef and the Philippine ascidian L. bistratum. These hexapeptides were associated with the C3-symmetric tris-oxazoline westiellamide isolated from L. bistratum [378,379]. Bistratamides A and B exhibited cytotoxicity against MRC5CV1 fibroblasts and T24 bladder carcinoma cell-lines at IC50 values of 50 and > 100 μg/mL, respectively. This toxicity was similar to certain cyclic hepta- and octapeptides isolated from L. patella: lissoclinamide 5 and patellamide D [380]. Bistratamides C and D were cyclic hexapeptides isolated from Lissoclinum sp., where bistratamide D exhibited depressant effects in mice at a dose of 65 μg [381]. Bistratamides E-J were moderate cytotoxic hexapeptides isolated from L. bistratum. Bistratamides E-J exhibited cytotoxicity against HCT-116 human colon tumor cell-line at IC50 values of 7.9, 28.0, 5.0, 1.7, 9.0, and 1.0 μg/mL, respectively [382]. Didmolamides A and B were bis-thiazole based compounds isolated from Didemnum molle collected from Madagascar [378].
Lissoclinamides isolated from the aplousobranch ascidian Lissoclinum patella showed anti-neoplastic properties against human bladder carcinoma and fibroblast cell-lines, and normal lymphocytes. Lissoclinamide 7 was the most potent compound with two thiazoline rings that rivaled in vitro didemnin B in cytotoxicity. Patellamides also displayed potent cytotoxicity [34,378,379].
N-methylamino acids are commonly found in marine peptides such as jaspamide, didemnins, and others. Didemnins A-E, M, N, X, Y, nordidemnin N, epididemnin A1, and acyclodidemnin A were cyclic depsipeptides that were isolated from a Caribbean tunicate Trididemnum solidum [383,384]. Didemnin B was a branched N-methylated cyclic peptolide and was shown to induce cell-death with apoptotic morphology, DNA fragmentation, and DNA ladder generation in a variety of transformed cells with no known mechanism. Many analogs of didemnin B were known to possess cytotoxicity, antiviral, and immunosuppressive properties [385]. Didemnin H was a cyclodepsipeptide isolated from Trididemnum cyanophorum [386]. Didemnins M, N, X, Y, A, and B exhibited cytotoxicity against P388 cell-line at IC50 values of 2.0, 50, 2.0, 2.0, 30, and 0.5 ng/mL, respectively. Epididemnin A1 and acyclodidemnin A also showed weak cytotoxicity at IC50 values of 2.0 and 0.2 μg/mL, respectively. Didemnin M expressed potent immunosuppressive activity in both graft vs host and lymphocyte reaction assays [384].
Styelin D was a C-terminal amidated 32-residue antimicrobial peptide isolated from the hemocytes of Styela clava, a solitary ascidian. Styelin D included two novel amino acids: dihydroxylysine and dihydroxyarginine, and two distinct unusual ones: 3,4-dihydroxyphenylalanine and 6-bromotryptophan. Styelin D was found to be cytotoxic and hemolytic to eukaryotic cells [34,387]. Eusynstyelamide was a modified dimer peptide isolated from Eusynstyela misakiensis and exhibited weak cytotoxicity at IC50 values of 100 μg/mL against human colon HCT-116 tumor cell-line [34,388]. Eudistomides A and B were five residue cysteine-linked cyclic peptides that were isolated from Eudistoma sp. Eudistomides were flanked with a 12-hydroxy- or 12-oxo-tetradecanoyl moiety and a C-terminal methyl ester and were reported to be the first ascidian derived peptides cyclized with a disulfide bridge [389].
Mollamide was a cytotoxic cyclic heptapeptide isolated from Didemnum molle. Mollamide was found to be cytotoxic against CV1 monkey kidney fibroblasts, A549 human lung carcinoma, and HT29 human colon carcinoma cell-lines at an IC50 value of 2.5 μg/mL, and against P388 murine leukemia cell-line at an IC50 value of 1 μg/mL [390]. Mollamide was also an RNA synthesis inhibitor at an IC50 value of 1 μg/mL. Mollamides -B and -C were two cyclic hexapeptides that were isolated from D. molle along with keenamide A. These peptides were previously reported from a notaspidean mollusk Pleurobranchus forskalii, and shared peculiar reverse prenylated ethers of threonine and serine amino acids. Mollamide B and keenamide A were found to be cytotoxic against various cancer cell-lines. Mollamide B showed moderate anti-malarial activity against W2 and D6 clones of Plasmodium falciparum with IC50 values of 2.1 and 2.0 μg/mL, respectively, while marginal activity was seen with Leishmania donovani with IC90 and IC50 values of 35 and 18 μg/mL, respectively. Mollamide B also exhibited in vitro activity against HIV-1 with an EC50 value of 48.7 μM [34,391]. Keenamide A exhibited significant activity against MEL-20, P-388, and A-549 cell-lines at an IC50 value of 2.5 μg/mL, and showed activity against HT-29 cell-line at an IC50 value of 5.0 μg/mL [392]. Cycloforskamide was another macrocyclic peptide isolated from P. forskalii which exhibited cytotoxicity against murine P388 leukemia cell-line at an IC50 value of 5.8 μM [393].
Other marine peptides such as virenamides A-C, sansalvamide A, and cycloxazoline exhibited potent anti-apoptotic activity in various cancer-cells, but the exact targets of these compounds are unknown. Virenamides A-C were linear cytotoxic tripeptides isolated from a didemnid ascidian Diplosoma virens, and possessed modest cytotoxicity against a panel of cultured cells. Virenamide A was potent at an IC50 value of 10 μg/mL against CV1, A549, and HT29 cell-lines and against P388 cell-line at an IC50 value of 2.5 μg/mL. Virenamide A also exhibited topoisomerase II inhibition. Both virenamides B and C were potent at an IC50 value of 2.5 μg/mL against CV1, A549, HT29, and P388 cell-lines [34,394].
Cyclodidemnamide was a cyclic heptapeptide isolated from Didemnum molle, and was found to be weakly cytotoxic against human colon HCT-116 tumor cell-line at an ED50 value of 16 μg/mL [395]. Along with cyclodidemnamide, D. molle also yielded two cyclic hexapeptides: comoramides A and B, and two cyclic heptapeptides: mayotamides A and B [396]. Comoramides and mayotamide peptides were screened against various tumor cell-lines including MEL-28, A549, and HT29 and were found to be cytotoxic at IC50 values of 5–10 μg/mL [396]. Didmolamides A and B were isolated from D. molle along with prepatellamide A, and tamandarins A and B. Didmolamides A and B exhibited cytotoxicity against HT29, MEL28, and A549 at IC50 values of 10–20 μg/mL [397]. Prepatellamide A was initially isolated from Lissoclinum patella along with three cyclic peptides: patellamides A-C. Prepatellamide A and patellamides A-C exhibited cytotoxicity at an IC50 value of 5 μg/mL against murine leukemia P388 cell-line [398]. Tamandarin A inhibited protein-biosynthesis in the reticulocyte cell-lysates of rabbit at an IC50 value of 1.3 μM along with being more potent against various human cancer cell-lines compared to didemnin B [399].
Patellins 1–6 and trunkamide A were isolated from Lissoclinum patella. Patellins 1–2 were cyclic hexapeptides while patellins 3–6 were cyclic octapeptides; trunkamide A is a cyclic heptapeptide. Patellins 1–5 and trunkamide A did not exhibit any in vitro cytotoxicity, but patellin 6 displayed cytotoxicity against CV1, A549, HT29, and P388 cell-lines at an IC50 value of 2 μg/mL, and topoisomerase II inhibition at an IC50 value of 2.5 μg/mL [400].
Kulolides 1–3, kulokainalide-1, and kulomo’opunalides 1–2 were depsipeptides isolated from the marine mollusk Philinopsis speciosa along with the linear peptide, pupukeamide. Kulolide-1 exhibited cytotoxicity at a concentration of 50 μM in rat fibroblast cells. Similar activity was also seen with other kulolides with the strongest activity seen with kulokainalide-1 at a concentration of 5 μM [401].
Ulicyclamide, preulithiacyclamide [402], ulithiacyclamide, and ulithiacyclamide B were small peptides that were isolated from Lissoclinum patella [403]. Ulithiacyclamide B exhibited cytotoxicity against solid tumor cells at an IC50 value of 17 ng/mL. Ulithiacyclamide and ulithiacyclamide B exhibited cytotoxicity against KB cell-line at IC50 values of 35 ng/mL and 17 ng/mL, respectively [404]. Ulithiacyclamide was also a potent inhibitor of Macrophage Scavenger Receptor (MSR) at an IC50 value of 98 nM, while ulicyclamide was inhibitor of MSR at 51 μM [402].
E. Other Marine Sources
A family of peptidic metabolites namely nobilamides A-E were isolated from the Streptomyces bacterial strain [Chicoreus nobilis] while nobilamides A and F-H were isolated from another Streptomyces strain [Conus tribblei] separated from molluscs [190]. A-3302-A and A-3302-B are related peptides which were previously obtained from the terrestrial B. subtilis and were later isolated from a marine source [190]. N-acetyl-L-phenylalanyl-L-leucinamide [405], a known synthetic peptide, was initially isolated from a natural source. Nobilamide B and A-3302-B are potent, long-acting antagonists of human and mouse transient receptor potential vanilloid-1 channels (TRPV1) as inflammation and pain mediators [406].
Aplidine is also known with several names such as aplidin, dehydrodidemnin B, and DDB, and was isolated from a Mediterranean tunicate Alpidium albicans. Low concentrations of aplidine were found to be sensitive against melanoma, non-small cell lung, and breast cancer cell-lines. Several mechanisms of action were involved such as inhibition of protein-synthesis and cell-cycle arrest. Aplidine resulted in a persistent and rapid activation of p38 MAPK phosphorylation and JNK activation resulting in the downstream release of cytochrome-c along with inducing early oxidative-stress. Aplidine also activated PARP cleavage and caspases-3 and -9 representing the mitochondrial apoptotic-pathway mediation during the process. Aplidine induced apoptosis in MDA-MB-231 breast cancer cell-line along with sustained activation of the serine/threonine kinases p38 MAPK and JNK, epidermal growth factor receptor [EGFR], and the non-receptor protein tyrosine kinase Src. Two proposed mechanisms regarding the JNK activation of aplidine included MKP-1 phosphatase down-regulation and Rac1 small GTPase activation. Aplidine is also known as plitidepsin, and following the completion of phase I clinical trials, has proceeded to phase II clinical trials well-tolerating the minor toxicity [34].
Dolastatin 3 was a cyclic peptide initially isolated from Dolabella auricularia and later from Lyngbya majuscula. Dolastatin 3 exhibited a GI50 value of < 1 μM against murine leukemia P388 cell-line [393]. Homodolastatin 3 and kororamide were other peptides isolated from L. majuscula with no known biological activity [407]. Dolastatin 10–15 were peptides isolated from a marine mollusk D. auricularia and included several unique amino-acid subunits. Dolastatin 10 was a linear pentapeptide known to inhibit the growth of L1210 murine leukemia-cell-line along with causing aggregation of cold-stable tubulin formation. Other properties of dolastatin 10 included tubulin-binding of Vinca alkaloids, inhibition of microtubule assembly, and tubulin-dependent GTP hydrolysis. Dolastatin 10 was also known to prevent the loss of stabilizing effects on colchicine binding-activity of tubulin. A segment [tripeptide] of dolastatin 10 inhibited the GTP-hydrolysis and tubulin-polymerization, but did not show any significant inhibition towards nucleotide exchange or vincristine binding [34]. Dolastatin 11 exhibited a mean GI50 value of ~ 0.07 μM in the NCI-60 cell-line panel, while dolastatin 12 showed GI50 values ranging from ~ 1 nM in human NCI-H460 non-small cell lung cancer [NSCLC] to ~ 30 nM in human SF-295 CNS cancer when assayed in five different cancer cell-lines. Moreover, it showed GI50 values of > 1 μM in P388 leukemia cell-line and ~ 0.1 μM in mouse neuro-2a neuroblastoma cell-line. Dolastatins 13 and 14 displayed GI50 values of 14 nM and 20 nM in murine P388 leukemia cell-line, respectively [393]. Dolastatins 14 and 15 exhibited antineoplastic activity, with dolastatin 15 being the most potent [408]. Epidolastatin 12 was a C-15 epimer of dolastatin 12 that was isolated from L. majuscula/Schizotrix calcicola. Epidolastatin 12 exhibited cytotoxicity against human nasopharyngeal KB carcinoma cell-line at an MIC of < 0.05 μg/mL and disrupted the microfilament network at concentrations of 0.2 and 2 μg/mL [205].
Dolastatin 16 was another cyclic depsipeptide that was isolated from Dolabella auricularia, Lyngbya majuscula, and Symploca cf. hydnoides. Dolastatin 16 included two amino acids: dolaphenvaline and dolamethylleucine and displayed GI50 values in low nanomolar ranges in a panel of five leukemia cell-lines and four human solid cancer cell-lines with a mean GI50 value of ~ 0.3 μM [393]. Homodolastatin 16 was a cyclic depsipeptide which was also isolated from L. majuscula and was a higher homologue of dolastatin 16. Homodolastatin 16 exhibited moderate cytotoxicity against two oesophageal WHCO1 and WHCO6 cell-lines at IC50 values of 4.3 and 10.1 μg/mL, respectively, and against cervical ME180 cancer cell-line at an IC50 value of 8.3 μg/mL [409]. Dolastatin 17 was a cyclodepsipeptide that included dolayne (Doy), a novel acetylenic β-amino acid similar to onchidin. It showed submicromolar range of GI50 values when assayed in four cancer cell-lines. Dolastatin 18 was unique in the presence of a thiazole ring in its structure, and has submicromolar range GI50 values in the human NCI-H460 NSCLC and the mouse P388 lymphocytic leukemia cell-lines [393].
Dolastatin C was a depsipeptide structurally related to dolastatins 10 and 15 and was isolated from Dolabella auricularia. Dolastatin C exhibited weak cytotoxicity against HeLa S3 cell-line at an IC50 value of 17 μg/mL [410]. Dolastatin D was another cyclic depsipeptide that exhibited a GI50 value of ~ 4 μM in human cancer HeLa S3 cell-line, while dolastatin G and nordolastatin G were cyclic depsipeptides with GI50 values of ~ 1 and ~ 5 μM, respectively, against the same cancer cell-lines [393]. Dolastatin E was a cyclic hexapeptide with three five-membered heterocycles: thiazolone, thiazole, and oxazole isolated from D. auricularia. Dolastatin E exhibited cytotoxicity against HeLa S3 cell-line at IC50 values ranging between 22–40 μg/mL [411]. Lyngbyastatin 2 and norlyngbyastatin 2 were cytotoxic analogs of dolastatin G and nordolastatin G which were isolated from L. majuscula. Lyngbyastatin 2 exhibited toxicity at LD100 of 3 mg/kg in mice and this was hypothesized due to the presence of an enol ether functionality but was inactive at sub-lethal doses against in vivo C38 murine colon adenocarcinoma. Both lyngbyastatin 2 and norlyngbyastatin 2 were not inhibitors of microfilament assembly or microtubule, nor were topoisomerase I inhibitors [412]. Dolastatin H and isodolastatin H were linear peptides isolated from D. auricularia and were closely related to dolastatin 10. Isodolastatin H exhibited in vivo antitumor activity against murine P388 leukemia and was slightly weaker than dolastatin 10, while synthetic dolastatin H displayed a GI50 value of 2 nM against human HeLa S3 cancer cell-line [393]. Dolastatin I was a cyclic hexapeptide with three five-membered heterocycles: oxazole, oxazoline, and thiazole isolated from D. auricularia. Dolastatin I displayed cytotoxicity against HeLa S3 cell-line at an IC50 value of 12 μg/mL [413].
Dolabellin was another novel cytotoxic metabolite isolated from the Japanese sea hare Dolabella auricularia. Dolabellin included a novel dechlorinated β-hydroxy acid along with two thiazole hydroxy-acids. Dolabellin exhibited cytotoxicity against HeLa S3 cell-line with an IC50 value of 6.1 μg/mL [414]. Doliculide was a cytotoxic cyclodepsipeptide that was isolated from the Japanese sea hare D. auricularia. Doliculide was a mixed peptide-polyketide having a 15-carbon polyketide unit, a unique D-amino acid, and glycine and showed potent cytotoxicity against HeLa S3 cell-line at an IC50 value of 0.001 μg/mL [415].
Kulokekahilide-1 was a cytotoxic depsipeptide that was isolated from a cephalaspidean mollusk Philinopsis speciosa and included two unusual amino acids: 3-amino-3-methylhexanoic acid and 4-phenylvaline. Kulokekahilide-1 was cytotoxic against P388 murine leukemia cell-line at an IC50 value of 2.1 μg/mL [416]. Kulokekahilide-2 was another cytotoxic depsipeptide that was isolated from the same cephalaspidean mollusk and was active against various cancer cell-lines such as A-10, P388, MDA-MB-435, and SK-OV-3 with IC50 values of 59.1, 4.2, 14.6, and 7.5 nM, respectively [417]. Aurilide was a cyclodepsipeptide that was isolated from a Japanese sea hare Dolabella auricularia and was similar to kulokekahilide-2. Aurilide was a potent cytotoxic peptide that may serve as a small molecule tool for studies of mitochondria-induced apoptosis [236]. Aurilide was a 26-membered cyclodepsipeptide that showed cytotoxicity against HeLa S3 cell-line at an IC50 value of 0.011 μg/mL [418].
Several bioactive peptides were isolated from the marine coral reef belonging to the Okeania genus. These included viridamides A-B [419] which were isolated from Okeania comitata; microcolins A-B [420,421] from O. erythroflocculosa; grassypeptolides A-C [221] and grassystatins A-B from O. lorea; tumonoic acids A-C, lyngbyastatin 1, and dolastatin 12 [205] from O. plumata; and malyngamide C, malyngamide C acetate [422], and lyngbic acid from O. hirsuta [423]. Microcolin A [Mic-1] induced apoptosis in murine-thymocytes at 10–100 nM [424], while microcolin B was found to be a potent immunosuppressant [421]. Other tumonoic acids D-I were isolated from the marine cyanobacterium Blennothrix cantharidosmum where tumonoic acid I exhibited moderate antimalarial activity at an IC50 value of 2 μM. Most of the tumonoic acids exhibited moderate bioluminescence inhibition with tumonoic acid F being the most active with an IC50 value of 62 μM. All the tumonoic acids were initially isolated from L. majuscula and Schizothrix calcicola [425]. Janadolide was a cyclic polyketide-peptide hybrid isolated from Okeania sp., and exhibited anti-trypanosomal activity at an IC50 value of 47 nM with no cytotoxicity towards human cells at 10 μM. Janadolide structure has a tert-butyl group similar to antillatoxins and apratoxins [426].
Onchidin and onchidin B were cyclic depsipeptides that were isolated from Onchidium sp., a pulmonate mollusk. The structure of onchidin included two identical halves of MeVal-Amo-Val-Hiv-Hiv with a novel β-amino acid: 3-amino-2-methyl-oct-7-ynoic acid (Amo). Onchidin was a cytotoxic depsipeptide that was found to be a rich source of γ-pyrone polypropionates [427]. Onchidin B included four α-amino acids: two units of proline and two units of N-methyl valine, two units of β-hydroxy acid: 3-hydroxy-2-methyloct-7-ynoic acid [Hymo], and four α-hydroxy acids: two 2-hydroxy-3-methylpentanoic acid moieties [Hmp] and two 2-hydroxyisovaleric acids [Hiv]. Onchidin B showed 97% inhibition of Kb cells at 10 μg/mL [428]. Norcardiamides A-B were cyclic hexapeptides that were isolated from the marine derived actinomycete Nocardiopsis species. Norcardiamides possessed a triple-Val subunit, and exhibited negligible antimicrobial activities with no cytotoxicity against human colon carcinoma HCT-116 cell-line [429].
Kahalalides were cyclic depsipeptides isolated from a sacoglossan mollusk Elysia rufescens. Kahalalides ranged from a C31 tripeptide to a C75 tridecapeptide. Kahalalides F and G included a rare dehydroaminobutyric acid that separated these from the other kahalalides, while the others included the same amino acids. Kahalalide A (KA) exhibited moderate antimalarial activity against Plasmodium falciparum while kahalalide E (KE) showed selectivity against HSV II (Herpes simplex virus II). Kahalalide F displayed selective activity against few AIDS opportunistic infections (OI) and solid tumors [430]. Kahalalide F (KF) also exhibited antitumor activity against human breast and prostate cancer cell-lines at IC50 values ranging from 0.07 μM to 0.28 μM while being less sensitive to non-tumor human cells at IC50 values ranging from 1.6–3.1 μM [431]. Of the different kahalalides, only kahalalides A, E, F, isoKF, R1, isoKA, and 5OH-KF showed significant biological activities. Kahalalides R1 and F exhibited cytotoxicity against MCF-7 cell-line at IC50 values of 0.14 and 0.22 μM, respectively, while kahalalide R1 displayed cytotoxicity against L1578 Y cell-line at an IC50 value of 4.3 nM [432]. KA exhibited antimicrobial activity with 83% growth inhibition against Mycobacterium tuberculosis at 12.5 μg/mL. KF showed antifungal activity against Aspergillus fumigatus at IC50 value of 3.2 μM, Cryptococcus neoformans at 1.5 μM, and Candida albicans at 3.0 μM, along with antiviral activity against HSV II at 0.5 μg/mL, immunosuppressive activity at an IC50 value of 3 μg/mL, and antileishmanial activity. Kahalalide R1 displayed antifungal activity with inhibition zones of 16 and 24 mm [432].
Two isomers of bioactive thiazole containing cyclic heptapeptides: cis,cis-ceratospongamide and trans,trans-ceratospongamide were isolated from the marine red alga, Rhodophyta: Ceratodictyon spongiosum with the symbiotic sponge Sigmadocia symbiotica. trans,trans-Ceratospongamide exhibited anti-inflammatory activity with potent sPLA2 expression inhibition at an ED50 value of 32 nM while the cis,cis isomer was inactive. The trans, trans isomer also exhibited the human-sPLA2 promoter based inhibition by 90% [433]. Mebamamides A-B were lipopeptides isolated from the green alga Derbesia marina. Mebamamides included a 3,8-dihydroxy-9-methyldecanoic acid residue along with four D-amino acid residues. Mebamamide A did not display any growth-inhibition against HL60 and HeLa cells at 10 μM while mebamamide B did not exhibit any growth inhibition at 100 μM. Mebamamide B also persuaded the differentiation of HL60 cells into macrophage like cells at 100 μM [434]. Galaxamide was a cyclic pentapeptide that was isolated from Galaxaura filamentosa, a marine alga, and exhibited antiproliferative activity. Galaxamide included two N-methyl leucines and three leucines [435]. Galaxamide showed cytotoxicity against human cancer U87 and MCF-7 cell-lines with IC50 values of 10.6 and 14.9 μg/mL, respectively [436].
Conantokin G and conotoxin GV were isolated from a fish eating snail Conus geographus [437], while conantokin T was isolated from the venom of a fish hunting cone snail Conus tulipa [438]. Conantokin G was a 17 amino acid and possessed N-methyl-D-aspartate (NMDA) antagonist activity blocking the NMDA induced cGMP elevation at an IC50 value of 171 nM [437]. Conantokin-T was a 21 amino acid peptide that included 4 residues of a modified amino acid: γ-carboxyglutamate. Conantokin T exhibited NMDA antagonist activity [438]. Conotoxin GV was a sleeper heptadecapeptide whose structure was determined to be Gly-Glu-Gla-Gla-Leu-Gln-Gla-Asn-Gln-Gla-Leu-Ile-Arg-Gla-Lys-Ser-Asn-NH2. Low doses of this peptide induced a sleep-like state in mice under 2 weeks old at doses of 4–30 pmol/g while older mice tended to be more hyperactive [439]. Actinia equine equinatoxin II [EqT-II] represented a family of basic, pore-forming, polypeptide toxins isolated from the sea-anemones which are now called as actinoporins. Actinoporins are 15–20 kDa cytolytic polypeptides that were isolated from the sea-anemones Actiniaria and exhibited pore forming activity in model and natural lipid membranes. Of the three isotoxins: actinoporins, sphingomyelin, and equinatoxin II, EqT-II was found to be highly lethal in mammals at an LD50 value of 35 μg/kg and also strongly hemolytic [440]. A mitogenic hexapeptide named SECMA 1 isolated from the alga belonging to Ulva sp., with the sequence Glu-Asp-Arg-Leu-Lys-Pro was found to be active against human foreskin fibroblasts at 4 μg/mL [441,442].
Arasin-1 was a proline-arginine rich antimicrobial peptide that was isolated from the hemocyte extracts of Hyas araneus, a spider crab. Arasin-1 included 37 amino acids with a C-terminal domain having two disulfide linkages and an N-terminal domain rich in arginine and proline. The amino acid sequence of arasin-1 is SRWPSPGRPRPFPGRPKPIFRPRPCNCYAPPCPCDRW with an average mass of 4345.10 Da and a calculated monoisotopic mass of 4342.06 Da. Aracin-1 exhibited activity against Corynebacterium glutamicum at an MIC value of 0.8 μM, while showing antibacterial activity at higher concentrations [443]. Arenicins 1 and 2 were antimicrobial peptides with 21-residues that were isolated from the coelomocytes of Arenicola marina, a marine polychaeta. Arenicin-1 has a molecular mass of 2758.3 Da with the amino acid sequence: RWCVYAYVRVRGVLVRYRRCW, while arenicin-2 has a molecular mass of 2772.3 Da with the amino acid sequence: RWCVYAYVRIRGVLVRYRRCW. Arenicins 1 and 2 exhibited antimicrobial activity at 25 μg/mL. Arenicins showed activity against Listeria monocytogenes at an MIC value of 0.6 μg/mL at low salt conditions and were less potent against E. coli and C. albicans at MIC values of 4.0 and 4.5 μg/mL, respectively [444].
III. CHALLENGES INVOLVED WITH PEPTIDE ISOLATION
Peptides are an interesting class of therapeutics that shows growing interest. Peptides are used in various therapeutic areas like anti-infectives, arthritis, allergy, cardiovascular, oncology, obesity, and diabetes besides others. Peptides also possess various advantages such as specificity, potency, minimized drug-drug interactions, high activity, less tissue accumulation; thereby low toxicity, and also offer a significant chemical and biological diversity. Along with advantages, peptides do have some disadvantages limiting their success. The major disadvantage is the stability of peptides in the body that could result in lesser bioavailability and shorter half-life along with other limitations such as formulation challenges and expensive synthesis. Solubility and pH signify important characteristics that should be considered for drug-formulation. During synthesis, one can encounter problems such as aggregation and solubility hurdles. There is only an 11 per cent success rate for peptides to reach from phase I clinical trials to drug approval, which could be higher with cardiovascular peptide-drugs and lower with anticancer peptide-drugs [445]. A total of 19 peptides were approved between 2001 and 2012, while in 2012 alone five of the 40 drugs approved were peptides [446].
One of the major challenges in peptide-based drug development is the optimization between the required peptide length and the pharmacologically useful levels for receptor-activation. The numerous variables that are considered include: (1) size and accessibility of ligand binding surfaces; (2) possible induced fit; (3) receptor residency time and ligand-stability. Another drawback with peptide drugs is their increased proteolytic instability compared to other small-molecule drugs which can be overcome by altering both the side-chains and amide-bonds making the peptidomimetics resistant to proteolytic degradation. Serum stability assay helps measure the peptide stability along with other important pharmacokinetic behavior of drugs [446].
In order to overcome the challenges of lower bioavailability and shorter half-life, the peptide backbone can be modified by introducing D-amino acids or unnatural amino acids or the peptide bonds can be altered with reduced amide bonds or β-amino acids in order to constrain the backbone by introducing cyclization or using constrained amino acids to induce less flexibility and enzyme-digestion. With some peptides, a fatty-acid attachment might result in increased bioavailability and half-life along with more target-specific binding, thereby resulting in fewer side-effects [445].
There are drug-delivery technologies (DDT) to improve the pharmacokinetics of the peptides and recent advancements to modulate their in vivo stability. Most peptides work by exerting their action on the cell-surface by binding to the membrane-proteins and hence various techniques were developed for intracellular delivery of peptides. One of the techniques is to fuse the peptides with nanoparticles or liposomes in order to penetrate the cell-membrane for the release of their contents in to the intracellular-domain, while the other technique involves the use of protein transduction domains (PTD) which are short peptide sequences, in order to allow the peptide-delivery into the cell. However, the challenge involved with liposomal formulation involves the inefficiency of encapsulation of the active pharmaceutical ingredient (API) into the liposomes; however this technology is still one of the best solutions with certain drug-formulations. Some of the achievements with this technology are Exubera® and inhaled insulin, opening the doors for further market-growth [445].
The process development of peptide synthesis is costly. This depends on the specifications or requirements and targeted volumes, which in turn affect the manufacturing costs. Delivering expected purities and yields is another problem associated with peptides along with identifying critical process parameters and proven acceptable ranges. The costs can also increase during drug-development. Peptide synthesis involves either solid-phase or liquid-phase or a combination of both. Liquid phase peptide synthesis (LPPS) is suitable for shorter peptide sequences (< 15 residues) and gives higher purity compared to solid-phase peptide synthesis (SPPS). LPPS process is often longer than an SPPS process. SPPS is used for longer peptide sequences (> 20 residues). The major drawbacks with SPPS involve waste volumes, solvents, and high raw-material costs. Along with these, the coupling reagents, choice of the resins, etc. should also be optimized [445].
Another challenge involved in peptide synthesis is aggregation. Some peptide sequences tend to self-associate due to aggregation [β-sheet formations]. This problem could be resolved by using pseudoprolines that tend to break the structures since the amide bond adopts a cis conformation (similar to proline residues in peptide sequences), which further leads to better solvation and chain-accessibility for further couplings. But the use of cysteine pseudoprolines shows limitations in peptide synthesis due to the high thiazolidine ring-stability which could require stronger acidic-conditions for deprotection. Hence, the deprotection conditions depend on the peptide sequences present and so require a case to case study [445].
Chemical biology comes into play to increase the sensitivity of the quantitative analysis of peptides. Another limitation is the oral bioavailability of peptides. The chemical and physical modifications that improve the oral bioavailability of peptides include conjugation to active and passive transport enhancers. Chemical biology has twofold function: to improve or solve suboptimal parameters such as poor pharmacokinetics of peptides or lack of oral activity, along with educating biotechnology investors the misconceptions of peptide-based therapeutics, which in reality are highly active and safe therapy options [446].
One noteworthy example of a peptide which was led from discovery to clinical development is caspofungin acetate, a parenteral antifungal agent. Caspofungin was a semisynthetic derivative of a natural, lipophilic cyclic peptide, pneumocandin B0, isolated from the fungus Glarea lozoyensis. There were many challenges faced by caspofungin to get approved for clinical use, starting with its discovery to final approval. These included the discovery of pneumocandin B0, fermentation development of pneumocandin B0, semisynthetic modifications of pneumocandin B0 using medicinal chemistry approaches to improve its properties, purification, formulation-development, commercial synthesis, and clinical development for the final approval of caspofungin as an antifungal drug [447].
Initially echinocandin B, a cyclic lipopeptide, was reported by Sandoz in 1974. The screening efforts at Merck’s natural products led to the discovery of pneumocandin series with two structural differences to echinocandins. In 1987, the structural characterization and biological evaluation of pneumocandin A0 as an antifungal agent was established. During the same year, pneumocandin B0, a desmethyl congener of pneumocandin A0, was identified. The actual structure of pneumocandin B0 was not established due to insufficient amounts of natural product and also based on the fact that the pneumocandin A0 lead was discontinued due to the lack of antifungal activity, poor water solubility, and oral bioavailability. When pneumocandin A0 was tested in an immunosuppressed rat model of Pneumocystis carinii pneumonia, it exhibited superior activity compared to the existing trimethoprim/sulfamethoxazole therapy. This led to reinstate pneumocandin B0 as a lead compound in order to improve its properties using medicinal chemistry program and make it a successful drug [447].
The complete structure of pneumocandin B0 was established, followed by its relative stereochemistry by X-ray crystallography and the absolute stereochemistry from acid hydrolysis and HPLC analysis. The challenge that was faced with the determination of relative stereochemistry was the presence of a large portion of fatty side chain that was disordered, making the dataset acquisition process slow. This also prevented the establishment of relative stereochemistry of two methyl groups in the side chain. Following this, the next major challenge was with the purification of pneumocandin B0. The purification of pneumocandin B0 was a challenge due to its structural similarity to not only pneumocandin A0, but also to its regioisomer, pneumocandin C0. Pneumocandin B0 was purified in a silica gel thin layer chromatography using a combination of dichloromethane, methanol, and water. For commercial scale production of pneumocandin B0, a normal phase silica-gel based HPLC method is being used [447].
Since pneumocandin A0 was the dominant product, efforts were made to improve the fermentation development of pneumocandin B0 to selectively increase the production and purity of pneumocandin B0. This was done using environmental approach which included optimizing the medium composition, pH, temperature, oxygen transfer, and carbondioxide concentrations. The other approach used was the genetic approach where the selective mutant for pneumocandin B0 was isolated. The optimization of the physicochemical and pharmacological properties of pneumocandin B0 followed the discovery, isolation, and fermentation of pneumocandins. These included stability, solubility, pharmacokinetic properties, and antifungal spectrum of pneumocandin B0. This is where the medicinal chemistry approaches come into play. In order to enhance the water solubility, pneumocandin B0 prodrugs with charged groups were developed. At a pH range of 5 to 8, pneumocandin B0 underwent accelerated ionization or ring-opening at hemiaminal. Under acidic conditions, both the benzylic hydroxyl groups and hemiaminal were ionized and hydride was delivered with improved acid and base stability. This chemistry was used for further discovery of caspofungin analogs to improve the stability. Various structural modifications on pneumocandin B0 to improve the pharmacokinetic parameters along with pharmacology, drug metabolism studies, and minimizing the toxicity led to MK-0991, which was later named as caspofungin [447].
Caspofungin synthesis involved modifications at two sites on the pneumocandin B0 peptide core: reduction of the primary amide of 3-hydroxyglutamine to an amine, and the condensation of the hemiaminal moiety with ethylenediamine. These transformations led to significant synthetic challenges where the need for the control of the region-, chemo-, and stereoselectivity during the peptide core modification on an industrial scale was required. Though the starting material pneumocandin B0 was purified to 90%, it still retained close to 20 analogs that were difficult to remove. Standard workups were impossible due to the detergent-like physical properties of the compounds with a lipophilic side chain and a polar peptide core. Non-conventional workups and purification processes were required. Finally, the compound was unstable without any crystalline intermediates or final product availability when the compound entered development [447].
As mentioned before, medicinal chemistry approaches came into play during the peptide core modification, to an analog with improved water solubility, half-life, and potency. Following the identification of the lead candidate, the process chemistry group helped in the development of a scalable and practical process, defining a route that was economically viable, green, and robust. The synthesis of the final compound was a 5-step route with an overall yield of 6–8% and poor α:β selectivity at the hemiaminal center. Even after the development of caspofungin acetate, there were significant challenges faced by the pharmaceutical scientists with regards to its instability. Caspofungin acetate decomposed by oxidation and hydrolysis even at neutral pH and also underwent dimerization. Hence caspofungin acetate is commercialized as a lyophilized powder as it is the only viable option. It took more than a decade from the start of the discovery to the entry of caspofungin into clinical development. Caspofungin serves as an example to understand the various challenges of getting a peptide to be approved for therapeutic use [447].
IV. BIOSYNTHESES OF MARINE PEPTIDES
Marine invertebrate derived natural products have biosynthetic origins that are not always clear, and increasing evidence shows the involvement of associated microorganisms. The following highlights the marine source biosyntheses based on the cellular localization studies [182]. Feeding experiments with isotope-labelled precursors were used to evaluate the biosynthetic pathways. For deeper understanding of complex marine microbial pathways, modern molecular techniques concerning recombinant technology and bioinformatics were used. Most biosynthetic pathways were initially elucidated from marine bacteria and cyanobacteria to complex peptides, polyketides, and hybrids, opening the door for novel research opportunities engaging metabolic biocatalysis and engineering [448].
Nonribosomal peptides and polyketides are a diverse group of natural products possessing complex chemical structures and vast pharmaceutical potential. These are synthesized by a conserved thiotemplate mechanism on the modular non-ribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) enzyme complexes. NRPS and PKS are known for their involvement across three domains of life from the discovery of 3,339 gene clusters from 991 organisms with a total of 2,699 genomes. A total of 1,147 hybrid NRPS/PKS clusters were discovered, explaining their widespread occurrence. Sequence analysis indicated NRPS evolution from a combination of common descent and horizontal gene transfer. Related siderophore NRPS gene clusters have been identified, which were known to encode modular and non-modular NRPS enzyme organization in a gradient. Higher frequencies of NRPS and PKS gene clusters were detected from bacteria in comparison to archaea or eukarya along with common occurrence in the phyla of Actinobacteria, Cyanobacteria, Firmicutes, and Proteobacteria in bacteria and the phylum of Ascomycota in fungi [449]. Most of these NRPS and PKS gene clusters have unknown end products (referred to as orphan gene clusters) [450], emphasizing the significance of genome mining in the identification of novel genetic machinery for the biosynthesis of secondary metabolites [449].
A. Marine Cyanobacteria
Cyanobacteria include slow-growing photosynthetic bacteria [451] and hence produce diverse natural products, some of which possess potential pharmaceutical activities while others are toxic [450]. Most of the biosynthetic mechanisms are unique to cyanobacteria and are rarely described from other organisms [451]. Genome mining is one way of enabling the characterization and identification of natural product gene clusters; yet the present number of cyanobacterial genomes is low compared to other phyla. This issue has been overcome by increasing the sequenced cyanobacterial genomes, enabling the identification of biosynthetic gene clusters for structurally distinct metabolites including ribosomal peptides, non-ribosomal peptides, alkaloids, fatty acids, terpenes, and UV-absorbing compounds. Various bioinformatic tools have been used for the prediction of the orphan gene clusters; however the prediction could be problematic in cases of complexity of the cyanobacterial pathways. Use of mass spectrometry guided natural product genome mining and/or heterologous expression helped solve this issue exploring the cyanobacterial natural product gene cluster [450]. The above studies demonstrate the role of cyanobacteria in encoding huge diverse cryptic gene clusters for natural product production, and the known chemical diversity is only a fraction of the true biosynthetic capabilities of this ancient group of organisms [451].
Marine blue-green algae/cyanobacteria are considered to be the prolific producers of bioactive natural products, most of which originate from the mixed polyketide-peptide biosynthetic pathways. The biosynthesis of microcystin LR, a cyclic heptapeptide, isolated from the alga Microcystis aeruginosa originated from the hybrid PKS-NRPS. The first cluster that represented the complete characterization of a complex cyanobacterial secondary metabolic-pathway belonged to M. aeruginosa PCC7806, which was about 55-kb and included 10 bi-directionally transcribed open reading frames (ORFs) arranged in two operons [mcyABC and mcyDEFGHIJ]. The microcystin synthetase included a loading di-domain, adenylation-peptidyl carrier protein (A-PCP) at PKS McyG, which was the N-terminus that was known to activate and load phenylacetate, the starter unit for consequent extension by six amino-acids and four malonate residues to McyC-bound linear microcystin-precursor. C-terminal thioesterase (TE) of NRPS McyC catalyzed the hydrolysis and cyclization of microcystin-LR. McyF was the tailoring racemase enzyme that reported the preliminary probing of microcystin pathway. Characterizing the microcystin synthetase genes allowed for the study of pathway regulations, elucidation of genetic-variants correlating different microcystin isoforms, and detection of microcystin producing cyanophytes from various environmental samples. The cyclic pentapeptide nodularin is structurally related to microcystins, and the nodularin biosynthetic gene-cluster identified from N. spumigena was found to proceed through a similar pathway to that of microcystin-LR [448].
In general, Lyngbya majuscula is a prolific producer of structurally diverse natural-products with broad biological activities. The biosynthesis of barbamide was chemically probed with labeled precursors as well as genetically probed via sequence analysis of the biosynthetic gene cluster. Previous studies revealed that barbamide was derived from one molecule each of acetate, L-cysteine, L-phenylalanine, and L-leucine along with two methyl-groups from S-adenosyl-L-methionine [SAM]. The direct precursor of this polyketide-peptide molecule was 5,5,5-trichloroleucine formed from the chlorination of the inactivated pro-R methyl-group of leucine. Sequence analysis of the 12 ORF cluster, which extended to 26-kb, exposed a co-linear genetic arrangement of barbamide cluster. L-leucine undergoes chlorination on the PCP [BarA]-bound leucine molecule by BarB1 and BarB2 halogenases before the transfer of 5,5,5-trichloroleucine to the PCP domain of NRPS BarE. An uncharacterized one carbon truncation of 5,5,5-trichloroleucine into a trichloroisovaleryl moiety precedes a malonate extension on BarE-BarF complex with an E-double bond to a diketide. The pathway was completed when cysteine and phenylalanine were added to PCP-bound diketide-dipeptide intermediate on the bimodular NRPS BarG undergoing the formation of a novel thiazole ring catalyzed by BarG C-terminal TE. The intermediacy of L-leucine, 5,5,5-trichloroleucine, and L-cysteine amino acids was supported by the biochemical characterization of the adenylation domains of BarD, BarE, and BarG, respectively. The related barbaleucamide supported the presumption that many Dysidea natural products are of cyanobacterial origin [448].
Jamaicamide A was a mixed polyketide-peptide with a number of unique structural features including a vinyl chloride, pyrrolinone ring, and alkynyl bromide. Jamaicamide A was known to be derived from ten acetate units, L-alanine, β-alanine, and two methyl groups from SAM. Jamaicamide A was a ~ 58-kb biosynthetic gene-cluster that revealed a high integrated PKS-NRPS system on 17 ORFs that were organized co-linearly with regards to its biosynthetic utilization. A β-keto diketide intermediate was formed from the ligation of 5-hexynoic acid to ACP JamC by acyl ACP synthetase JamA, followed by the extension of malonate on hexadomain PKS JamE. The N-terminal region of jamJ product and jamFGHI led to a “β-keto modifying gene cassette,” which was involved in the formation of pendent carbon-atom having vinyl chloride functionality. The addition of acetate to β-keto intermediate was catalyzed by the HMG-CoA synthase like homolog: JamH. The nascent polyketide underwent two rounds of NRPS and five rounds of PKS extension after the formation of vinyl (chloride). JamQ, an NRPS condensation (C)-related protein, facilitated the cyclization reaction, resulting in ACP-bound linear-intermediate on PKS JamP and the formation of pyrrolinone ring residue [448].
The biosynthesis of lyngbyatoxin was established from the molecular cloning of its biosynthetic gene-cluster of 11.3-kb with four ORFs [448]. A copy control fosmid vector pCC1FOS has been used to include a genomic fosmid library made from a lyngbyatoxin producing strain of L. majuscula. (–)-Indolactam V was the core structure of lyngbyatoxins A, B, and C, which was partially assembled using an NRPS system with a C-terminal reductase domain. Based on the Red1 motif of NRPS-associated Red-domains and the A3 motif of the adenylation (A)-domains, a highly specific probe was isolated by PCR from the genomic DNA of Lyngbya strain using degenerate primers. The probe was 1.4 kilobase (kb) and was used to isolate the fosmid clones that were analyzed by Southern blot and restriction mapping experiments. For DNA sequencing, a single fosmid clone fos-DE3-86 was chosen. The ltx gene cluster was then sequenced using fragments that were subcloned into pBluescript II SK(+) and a 17.1 kb region [452].
The ltx gene cluster was an 11.3 kb region and had four open reading frames (ORFs) that were all transcribed in a similar direction. ltxA was the first ORF that encoded for a two module NRPS protein. The first module included an A-domain specific for L-Val based on the traditional A-domain binding pocket designations, an N-methylation domain (NM), and a peptidyl carrier protein (PCP). There was a second module that included a condensation (C) domain, A-domain specific for L-Trp, a PCP, and a Red domain responsible for NADPH-dependent reductive release of N-Me-L-Val-L-Trp from NRPS for the generation of 5. ltxB was the second ORF that encoded for the cytochrome P450 monooxygenase that included an N-terminal with 80 amino acids, which was similar to MbtH, an unknown domain seen in numerous diverse NRPS containing pathways. LtxB was known for its involvement in the oxidation of the indole-ring of 5 and in subsequent cyclization for the formation of 4. ltxC was the third ORF that encoded proteins with little resemblance to other known enzymes. ltxD was the fourth ORF that encoded for proteins related to diverse families of oxidase or reductase-type proteins. LtxD might be responsible for the conversion of lyngbyatoxin A into minor metabolites: lyngbyatoxins B and C [452].
The dolastatins are hybrid polyketide-peptides produced by cyanobacteria and isolated from the sea-hares. Due to the low isolation yields of dolastatins from molluscs, cyanobacterial fermentation and subsequent bioengineering afforded new opportunities for the drug discovery and development of these promising anti-cancer agents [448].
Recently, molecular networking was employed, especially in industrial settings, to help expedite the discovery of innovative small molecule therapeutic leads from complex marine sources. Sirenas LLC has integrated chemometrics-based bioactivity predictions with molecular networking in a drug discovery platform that has been successful over the years in identifying new classes of molecules, especially with the well-studied dolastatin 10 structural class. A new member belonging to the dolastatin 10 structural class has been discovered with better physicochemical profile and increased potency using molecular networking. This was done by screening the marine fraction library against several cancer cell-lines, followed by the LC-MS/MS analysis. Following this, activity scores were assigned to all the metabolites found to exhibit cytotoxic activity that were generated from the relative abundance of each metabolite. Sometimes a single metabolite could be present across multiple fractions in varying abundances, resulting in rank-order correlations between the strength of the biological activity and the relative abundance of the metabolite. An activity score or a correlative value was yielded by the global analysis of the fraction that falls between 0 and 1, where 1 represents a perfect correlation between the biological activity of the fraction and the abundance of the metabolite. Mapping the scores to their corresponding metabolites in a molecular network provides visualization between the bioactivity and the structural relationship between molecules. Interrogation of the above network resulted in revealing a molecular family with strong activity scores, and numerous member nodes were de-replicated as dolastatin 10 along with the related symplostatins 1 and 3, malevamide D family members, and a novel analog SMD5041 [453].
Anabaenopeptins are assembled on a NRP synthetase enzyme complex coded by a 32 kb apt gene cluster encoded with two alternative starter modules arranged in separate bimodular proteins. The starter modules exhibited strong substrate specificity for L-Arg/L-Lys and L-Tyr, respectively, allowing specific biosynthesis of various anabaenopeptin variants. NRPSs are huge enzymes with modular structures where in each module catalyzed the activation and incorporation of an amino acid or in some instances a carboxylic acid to the growing peptide in a stepwise fashion. The modules included catalytic domains for amino acid condensation, activation, and thiolation. Additional modifications on the amino acids included N-methylation, epimerization, and heterocyclization. The collinearity rule is applied to the NRPSs where the substrate specificity and the module number reflect the product composition. In the NRPS enzyme complexes, the adenylation domains determined the amino acid selection while the condensation domains played a selective role in the non-ribosomal peptide synthesis. Discrete NRPS enzyme complexes are capable of synthesizing sets of peptides simultaneously. Incomplete modification and lack of substrate specificity are a few reasons for natural variation in the non-ribosomal peptide chemical structures [454].
Anabaena species included a 32.5 kb gene cluster with five ORFs: aptA1, aptA2, aptB, aptC, and aptD with the amino acid sizes of 2195, 2174, 1069, 2565, and 1403, respectively. Each AptA1 and AptA2 was comprised of two NRPS modules, both of which had an adenylation and thiolation domains, one epimerization, and one condensation domain. AptB and AptD included a single module containing an adenylation, thiolation, and condensation, while AptD also included a thioesterase domain. AptC included two modules; each with an adenylation, thiolation, and condensation domain. The latter module also included an N-methyltransferase domain embedded in the adenylation domain. The gene cluster also included two genes: aptE and aptF, which did not code for NRPS. The catalytic domain order and the modular structure matched the NRPS organization for the assembly of anabaenopeptins from Anabaena species [454].
The genome of Nodularia spumigena included a 17.6 kb gene cluster for aeruginosin (aer) as a single contig that encoded a peptide synthetase with reductive release mechanism as reactive peptide aldehydes. Aeruginosins and spumigins are structurally similar families with linear peptide aldehydes utilizing separate peptide synthetases. The spumigin gene cluster was located at 11 kb and 133 kb apart from the nodulapeptin gene cluster and aer gene cluster, respectively. Eight proteins were encoded by the aer gene cluster that was organized in a single operon. The substrate specificities predicted for AerB, AerG, and AerM peptide synthetases were L-Tyr, Choi, and L-Arg from comparison with other biosynthetic pathways. Similar to other non-ribosomal biosynthetic pathways, aeruginosin biosynthesis started from loading short chain fatty acids by the AerB condensation domain. The condensation domain resulted in lipidation of aeruginosins. Genes that encoded AerD, AerE, and AerF enzymes have been located on the aer gene cluster along with AerI, which encoded a putative glycosyltransferase. The reductase domain AerM resulted in the release of the C-terminal arginine as reactive aldehyde [301].
Spumigins are linear peptides with an N-terminal hydroxyphenyl lactic acid exclusively with D-homotyrosine, 4-methylproline (mPro) or proline, and an arginine derivative or a C-terminal lysine. Spumigins were assembled on the NRPS enzyme complex, offloading the peptides as reactive aldehydes. Necessary enzymatic steps for the unusual mPro were encoded in the 21-kb gene cluster along with the peptide synthetases [301].
The biosynthesis of thiocoraline involved at least one initiation module and four modules for amino-acid incorporation. The starter unit can be activated in two ways. When the starter unit was an amino-acid, the first module included an adenylation (A) and a peptidyl carrier protein (P) domain. When the starter unit was an aromatic compound (not an amino-acid), an adenosine monophosphate (AMP) ligase and an aryl-carrier protein were found. An AMP-ligase containing module was expected since the starter unit of thiocoraline was 3HQA. TioJ and TioO proteins formed the initiation module. There was no candidate for an aryl-carrier protein in the thiocoraline cluster while TioJ was expected to be the 3HQA AMP-ligase. TioO was expected to play the role of 3HQA-carrier protein. TioK was an NRP related enzyme formed by P8 and A8 domains, which were also known to involve in 3HQA activation. The P domain present was unusual and independent. TioX was involved in the methylation of sulfhydryl groups in two cysteine residues in thiocoraline [455].
The biosynthetic gene cluster of thiocoraline was a 64.6 kbp DNA region sequence with 36 complete ORFs and two incomplete ones. The NRPS that was involved in the biosynthesis of thiocoraline backbone constituted TioR and TioS. Thiocoraline included two moieties of 3-hydroxy-quinaldic acid [3HQA], which might have acted as the starter unit during biosynthesis and was derived from tryptophan. tioF, tioG, tioH, and tioI were the genes involved in the biosynthesis of 3HQA [455]. The role of TioN, which was initially thought to be inactive, was identified in the formation of 3HQA [456]. TioG was similar to kynurenine transaminases and aspartate aminotransferases. TioH was the only oxidoreductase in the cluster that had similarity with several NADP+ and NAD+ oxidoreductases. TioI was similar to cytochrome P450s and acted as quinaldate-3-hydroxylase for the conversion of quinaldic acid to 3HQA, which was the starter unit in biosynthesis of thiocoraline. TioF was involved in the conversion of L-tryptophan to N-formyl-L-kynurenine. tioL or tioM encoded formamidase resulting in the generation of L-kynurenine that was later transformed to kynurenic acid by TioG. TioH and TioI were responsible for 3HQA formation with an oxido-reduction step by eliminating the 4-hydroxy group in kynurenic acid followed by a hydroxylation step for the introduction of 3-hydroxy group [455].
The biosynthetic gene cluster of thiocoraline also included TioN. TioN was the A domain and a potential candidate for S-Me-L-Cys formation. An A domain included 10 core signature sequences, and in a variety of natural products, it is usually interrupted between a8 and a9 core signature sequences. Contrary to this, the interruption of A domain by M domain occurred between a2 and a3 core signature sequences in TioN. A domain could be involved in the S-methylation of L-Cys. TioN was also unique in that it was the only A domain that was interrupted. Initially TioT was found to be inactive, but later it was found that TioT was essential for TioN enzymatic activity. TioT was not required for the TioN expression in its soluble form. TioN was involved in the methylation of both L-Cys-AMP and L-Cys-S-TioS. TioN was a bifunctional [adenylating and methylating] stand-alone A domain that was interrupted in an NRPS complex [456].
There were four modules responsible for the biosynthesis of the thiocoraline backbone with four A domains that activated L-cysteine, glycine, L-cysteine, and L-cysteine. An epimerization (E) domain should be present in the first module as the first amino-acid in the peptide backbone was a D-cysteine. The last two modules must include a proper N-methyltransferase (M) domain since the last two L-cysteines were N-methylated. The last module ends up with a thioesterase (TE) domain for the peptide chain release and cyclization [455].
Cyclomarin A was a marine microbial peptide that was largely derived non-ribosomally from the nonproteinogenic amino-acid residues. Salinispora arenicola has a 5.8 Mb long DNA sequence analysis with a circular genome and a 47 kb biosynthetic gene-cluster (cym) with 23 open reading frames. The cym locus is controlled by the largest ORF with 23,358 bp cymA, which encoded the 7-module (heptamodular) NRPS and led to the assembly of the full length cyclomarin heptapeptides and also truncated cyclomarazine dipeptides. The megasynthetase CymA possessed an unprecedented biosynthetic feature that helped synthesize different sized peptides in vivo, which was triggered by the β-oxidation level of the priming tryptophan-residue. This tryptophan residue was unoxidized in cyclomarazines and oxidized in the cyclomarin series. Prior to the release of the cyclic peptide from CymA, the tryptophan residue was reverse prenylated by CymD while the cytochrome P450 CymV introduced the epoxide group on the cyclomarin C isoprene post NRPS assembly. Lastly, the 2-amino-3,5-dimethylhex-4-enoic acid in the cyclomarin series was derived from the condensation of the isobutyraldehyde and pyruvate followed by the S-adenosylmethionine methylation. Hence, the biosynthesis of cyclomarins started with the tryptophan derivative and ended with ADH yielding a linear heptapeptide intermediate that was bound to the thiolation (T) domain. The C-terminal thioesterase (TE) then helped in subsequent release and macrocyclization. In case of cyclomarazines, the diketopiperazines (DKPs) were yielded from module-2 bound diketide that was cleaved from CymA, which in turn was facilitated by type II TE CymQ [312].
The cym cluster included four oxygenases: cymO, cymS, cymV, and cymW along with an O-MT (cymP), a putative PTase (cymD), a four gene operon (cymE-H) involved with ADH biosynthesis, four other genes putatively involved in resistance and regulation, and two in phosphoenolpyruvate metabolic flux. The cym locus was flanked with pseudogenes including fragments of transposases and integrases suggesting this cluster to be horizontal [312].
The putative gene cluster of the surugamides included four NRPS genes: surA, surB, surC, and surD. Surugamides A-E were produced by the surA and surD NRPS genes while surugamide F was produced by surB and surC NRPS genes. The modular compositions of SurA and SurB were similar, and vice versa with SurC and SurD. While SurA and SurB started with a A-domain and ended with a peptidyl carrier protein, SurC and SurD started with a condensation (C) domain and ended with an epimerization (E) domain [314].
Four contiguous NRPS genes encoding 18 A-domains had the biosynthetic genes for SA-SE present at their head and tail. The enzymes responsible for SA-SE and SF production lacked domains such as condensation, thioesterase, or terminal reductase for chain termination. Hence, the biosynthesis was expected to be terminated by a putative α/β-hydrolase (Orf2) or a putative β-lactamase (Orf9) that encoded upstream of surA – surD [314].
The gene cluster for the biosynthesis of salinamides was 47.6 kbp encoding 14 sequences. This is a NRPS-PKS hybrid gene cluster designated in the sln locus. The sln cluster included four genes coding the NRPS proteins and one gene coding the PKS/NRPS hybrid along with enzymes involved in the precursor biosynthesis and post-NRPS tailoring enzymes. The genes supporting the biosynthesis of ρ-HPG, which is a non-proteinogenic amino-acid residue, were sub-clustered in the sln1-3 gene cassette. The sln cluster also included two genes: sln6 and sln9, which encoded the tetra-domain NRPS modules comprising the C-terminal TE domain. This suggested that one of the type I TE domains played a significant role in the addition of (4-methylhexa-2,4,-dienoyl)-glycine moiety to the Ser-OH [457].
Sln9 was a nonribosomal peptide synthetase that played a central role in the construction and installation of a distinctive acyl-glycine “basket” handle to salinamides. Sln9 was a thioesterase domain that resulted in the transesterification of the serine residue in desmethylsalinamide E with acylated glycyl thioesters in order to yield the desmethylsalinamide C. The construction of salinamides involved ATP-dependent activation by multifunctional assembly-line proteins, and their amino-acid precursors were transferred to the arrayed carrier protein (CP) domains. NRPS assembled the mature peptide intermediate and then the peptide product was released by a myriad of protein off-loading reactions. The terminal reaction was catalyzed by the thioesterase (TE) proteins that occupied the NRPS terminal domain, and the product was released as linear or macrocyclized peptides [457].
The biosynthesis of amphibactins involved a genome with two putative NRPSs: ABO_2093 and ABO_2092, which in turn possessed four classical NRPS modules. The M1 of ABO_2093 started with an N-terminal condensation domain, suggesting the presence of an external ligase that activated the fatty acid in order to be incorporated into the peptidic headgroup. There was no ACP domain present in ABO_2092 to tether and transport this activated fatty acid. The biosynthesis then proceeded in a linear fashion, ending with a thioesterase domain which was preceded by a domain with unknown function [458].
There are two putative NRPSs involved in the biosynthesis of marinobactins: ENO16763 and ENO16762. ENO16763 was the first module that encoded a unique domain with high homology to fatty acyl-AMP ligases (FAALs). This module activated the fatty acid to get it incorporated into the peptidic headgroup. The biosynthesis of marinobactins also occurred in a linear fashion and was terminated by a thioesterase domain. Hence the biosynthesis of marinobactins started with an N-terminal acyl AMP-ligase starter domain that activated the fatty acid, thereby initiating the biosynthesis [458].
The biosynthetic pathway of amphi-enterobactin involved six genes: aebA-F, to encode the homologous proteins similar to enterobactin biosynthetic machinery. EntA, EntB (N-terminal domain), and EntC converted the chorismate to 2,3-dihydroxybenzoic acid (DHBA). EntB (C-terminal domain), EntD, EntE, and EntF were four NRPS proteins that catalyzed the formation of the DHBA L-Ser unit, polymerized three units, and cyclized the trimer to form the lactone-backbone. In Vibrio harveyi, the presence of a proximal aebG gene was a distinct feature that encoded a long-chain fatty acid CoA ligase (FACL). The FACL enzymes activated the fatty acids to fatty acyl-CoA thioesters prior to their incorporation on to the acylated non-ribosomal peptides. Hence, it was predicted that the gene cluster aebA-F was responsible for the formation of the amphi-enterobactin core-structure, and the aebG gene was involved in acylation of the serine residue. AebB and AebE were necessary for the DHBA activation, AebE for DHBA-AMP ligase, and AebB as DHBA carrier protein [334].
B. Marine Fungi
The biosynthesis of scopularide A isolated from the marine fungi Scopulariopsis brevicaulis is discussed here. Scopularide A is a lipopeptide. Lipopeptides are usually linear or cyclic compounds having a fatty acid attached to the N-terminal amino-acid of the peptide portion with hydroxyl acids or amino acids. This peptide core was synthesized by the NRPSs consisting of modules that enabled the sequential synthesis of the growing peptide chain. The NRPS module usually included an amino acid substrate recognizing adenylation [A] domain and a peptide acyl carrier [T or PCR] domain that transferred amino acid to condensation [C] domain where peptide-bond formation occurred. The lipid portion attached to the peptide core could be derived from various sources, including the PKSs or the primary lipid metabolism. Similar to NRPSs, PKSs were multi-domain enzymes with three core domains: β-ketosynthase [KS], acyltransferase [AT], and acyl-carrier protein [ACP]. The carbon chain in lipopeptides provided by the PKSs included additional reducing domains such as ketoreductase [KR], enoylreductase [ER], dehydratase [DH], and an optional methylation domain [MET]. During biosynthesis, the reduced carbon chain was merged into the peptidyl backbone by a process called lipoinitiation. Scopularide A is structurally similar to emericellamide A produced by a marine Aspergillus sp [353].
The gene clusters in both S. brevicaulis and A. nidulans included a CoA-ligase and an acyltransferase. The two enzymes were considered to be responsible for the activation and loading of the reduced polyketide to the NRPS. There was an additional gene between the NRPS and PKS in A. nidulans with unknown function, and was not involved in the biosynthesis of emericellamide. There were five additional genes between NRPS1 and PKS2 in S. brevicaulis. These genes were expected to encode a transporter, a copper amide oxidase, a mannosyl transferase, transcription factor, and a gene with unknown function [353].
The proteins that are involved in the biosynthesis of a specific secondary metabolite are frequently encoded by genes located in close proximity, forming a cluster. The responsible gene clusters for emericellamide A have been identified. The carbon chains in emericellamide A were provided by PKSs that included the reducing and methylation domains. The resultant reduced polyketides were then transformed to CoA thioesters by acyl-CoA ligases and were loaded on to acyltransferases that shuttled the polyketide intermediates to first thiolation [T] domain of NRPSs [353].
The most probable biosynthetic gene cluster in the production of scopularide A was NRPS1/PKS2 based on the in silico analyses of the S. brevicaulis genome. This was indirectly proven from the enhanced production of scopularides in two transformants TF1-5-1 and TF5-2, which included the gene that encoded the putative transcription factor from a gene cluster under a constitutive promoter. Based on the emericellamide model, the following biosynthesis of scopularide A was proposed where the reduced polyketide that was produced by the PKS2 was loaded to the NRPS1 via the CoA ligase and the acyl transferase [353].
The biosynthetic origin of the N-methyl-3-(3-furyl)alanine moiety of the cyclic peptide endolide A was described by Kӧnig et al in 2016. The initial hypothesis of the 3-(3-furyl)alanine biosynthesis was based on its structural relationship to a phenylalanine skeleton. Both erythrose-4-phosphate and phosphoenolpyruvate were the precursor molecules, and the shikimate pathway was established to be the biosynthetic route for the 3-(3-furyl)alanine moiety in endolide A, where the N-methyl group was known to originate from methionine [362].
C. Sponges
Entotheonella sp., was found to be responsible for the production of kasumigamide from Discodermia calyx when a single-cell analysis was conducted in conjunction with PCR. This bacterial phylotype was reported to be a symbiotic producer of multiple natural products in Theonellidae sponges. Four different kasumigamide gene clusters were detected in four different bacterial species, namely Entotheonella [a marine sponge symbiont], Microcystis aeruginosa [a free-living cyanobacterium], Delftia acidovorans [a human oral bacterium], and Herbaspirillum sp. [the endosphere of Populus deltoids tree] [10].
The biosynthetic gene cluster included a hybrid PKS-NRPS cluster with 9 ORFs, of which the kasA-C formed the PKS-NRPS core. KasA had an adenylation (A) domain [KasA-A1], a ketoreductase (KR) domain, and a peptidyl carrier protein (PCP) domain. KasB included three modules [2–4], where the first two coded for NRPSs and the latter encoded a PKS. Modules 5–6 were encoded in kasC. From the NRPS codes, the substrates for five A domains were predicted, which were KasA-A1, KasB-A1, A2, KasC-A1, and A2, with the exception of KasA-A1, which was annotated to A domain employing phenyl pyruvic acid [PP]. Kasumigamide that was initially isolated from a fresh water cyanobacterium M. aeruginosa was predicted to be the chemical structure of PKS and NRPS hybrid compound [10].
There were some peculiar features in the module or domain organization of the kas family gene clusters. One of the noteworthy points was the shift of PKS module in the M. aeruginosa kas-cluster [makasA-D] while in other kas genes the PKS module was located on module 4. D-erythro-PS was the C-terminal residue of kasumigamide. L-Phe was initially hydroxylated by MakasD for the generation of L-threo-PS. Subsequently the L-threo-PS was loaded onto the PCP domain by MakasC-A2. Finally MakasC-E1 epimerized L-threo-PS to D-erythro-PS that was encoded between modules 4 and 5 [10].
Other unusual features of kas family genes included the likely chain termination by a condensation (C) domain and the absence of a thioesterase (TE) domain. Due to the fact that some C domains functioned as a TE domain, it was expected that the C-terminal C domain encoded in kasC, hkasC, and dakasC served as a thioesterase. The release mechanism of makas pathway remained uncertain because the C-terminal C domain was absent in makasA-D [10].
Polytheonamides are the largest among the known NRPS-synthesized secondary metabolites. A ribosomal pathway was also necessary to introduce multiple D-configured and C-methylated residues. The DNA region included 11 additional genes that were clustered around the ORF. poy genes poyA-I were nine ORFs that formed the operon that was known from the short or absent intergenic regions. The 3ʹ terminus of the poyA included 48 codons, which matched with the complete polytheonamide precursor. The three 3-hydroxyvaline units originated from two residues: C-methylation of residue 16 from threonine (Thr) and hydroxylation of residues 23 and 31 from valine (Val). There was also a long 5ʹ sequence in poyA exhibiting homology to nitrile-hydratases. The production of polytheonamides from proteinogenic residues involved four hydroxylations, 18 epimerizations, and at least 21 methylations. PoyB, PoyC, and PoyD were homologous to the radical S-adenosylmethionine (rSAM) superfamily members; PoyE was homologous to the SAM-dependent methyltransferases; PoyF was homologous to dehydratase domain of lantibiotic LanM-type synthetases; and PoyI was homologous to the Fe(II)/α-ketoglutarate oxidoreductases. The cluster also encoded other proteins on the basis of homologies that were involved in transport, regulation, and proteolytic removal of the leader region: PoyJGH. The homology of PoyK is unknown. Individual enzymes converted structurally and positionally diverse residues [459].
The biosynthetic genes of keramamides isolated from Theonella swinhoei Y (ker) were homologous to the keramamide (krm) genes isolated from T. swinhoei WA. The keramamide gene-cluster krm included the unusual 5-hydroxytryptophan 6-halogenase gene. The notable differences between the two clusters included krmR and krmI. An O-methylserine extension similar to keramamide H was exhibited by the keramamides identified from T. swinhoei WA while krmR encoded the NRPS module that activated and incorporated O-methylserine. The krmI C-terminal region resembled the flavin-dependent tryptophan halogenases [FDTHs] and functionalized position-6 of indole. FDHs usually encoded in the natural product biosynthetic pathway gene-cluster and halogenated phenol, pyrrole, and indole motifs. A second N-terminal domain was also present in krmI with a distinct homology to ThiF enzyme family members. These were the enzymes that utilized ATP by adenylation to activate the carboxylic acids [460].
The key difference between the ker and krm genes was the presence of krmI. krmI was a flavin-dependent halogenase that generated the 6-halo containing keramamides and was the first halogenase to use the L-5-hydroxytryptophan as its preferred substrate. KrmI also exhibited promising broad substrate-specificity and accommodated a series of indole-derived substrates. Following protein engineering, krmI can also be a promising starting point for the generation of brominated and chlorinated products because of its inherent substrate-flexibility [460].
The biosynthesis of microsclerodermins involved a multi-modular PKS/NRPS system with the involvement of a set of enzymes for the biosynthesis of side-chains and post-assembly line modification. The enzymes that were involved in the side-chain biosynthesis were responsible for modifications such as oxidation or halogenation of the pyrrolidone ring. The gene cluster was about 60 kbp in length. The gene mscK encoded a major facilitator superfamily transporter followed by mscJ, a type-II thioesterase located upstream to mscA. The core biosynthetic assembly covered three PKS modules and five NRPS modules encoded on genes mscA to mscI. mscL encoded an additional halogenase near the downstream boundary [461].
The biosynthesis of microsclerodermin was initiated with the build-up of a phenyl group in conjugation to a double-bond at the side-chain. In such a case, benzoyl-CoA or trans-cinnamoyl-CoA was the activated starter unit that was recruited by the enzyme. But the observed biosynthesis suggested otherwise, and so phenylacetyl-CoA was found to be the starter unit in the biosynthesis of microsclerodermin. MscA and MscC modules helped in the elongation of the phenylacetate unit, using two times 3-hydroxymalonate and three times malonate as extender units. The modules 2 (MscB) and 4 (MscD) exhibited a combination of functional KS domain attached to an inactive AT domain. The PKS derived unit was then forwarded to the module MscF: the first PCP domain. MscF module harbored two additional uncommon domains that showed high homology to the amino-transferase (AMT) and monooxygenase families located downstream to the PCP domain. This domain resulted in the oxidation of the β-hydroxyl group of bound intermediate to β-keto functionality, followed by conversion to the β-amino moiety, which underwent macrocyclization [461].
The biosynthesis continued with a set of PKS- and NRPS-based reaction cycles. The A domain of the MscF module was specific for asparagine activation. Such an asparagine-derived pyrrolidone system was only found in koshikamides and microsclerodermins. The protein MscE was responsible for the cyclization step and was similar to the amidohydrolase class. Tryptophan halogenation as well as pyrrolidone ring oxidation was catalyzed by the tailoring enzymes. MscL was a halogenase located downstream and was responsible for tryptophan chlorination. MscN belonged to the SAM-dependent methyl transferase family, while MscM showed homology with Fe(II)/α-ketoglutarate dependent dioxygenases. Hence, MscM and MscN were responsible for the oxidation and methylation of the pyrrolidone ring, respectively [461].
D. Ascidians
A single coding sequence (CDS) was identified that was found to include the required sequence necessary for patellamide C. This gene was found to be a candidate for patE: a patellamide precursor peptide. The presence of two peptide products on a single CDS proposed the importance of synergy in the mechanism of action of patellamides. There were several other CDSs surrounding patE which included patA – patG genes in a 11-kbp cluster. patE was surrounded by patA: a protease enzyme, patD: an adenylating enzyme-hydrolase hybrid, and patG: an oxidoreductase-protease hybrid. This cluster also included patB, patC, and patF, which were other CDSs with low or no similarity to other proteins; and the cluster ended with a gene that was assigned to primary metabolism, which was a DNA-photolyase homolog and a putative structural gene on the other side extending to approximately 1 kbp which was upstream of patA [462].
patE encoded a peptide of 71 aa of which the first 37 served as a leader sequence for processing. Of the remaining 34 aa, 16 encoded patellamide C and A sequences while 18 make up motifs directed the cyclization of patellamides. The patellamide C was 8 aa upstream of the patellamide A sequence. Prior to the two peptides, there was a 5 aa conserved region that included G(L/V)E(A/P)S. The patellamide A sequence was terminated by AYDGE sequence and preceded the stop codon directly. An 8 aa sequence AYDGVEPS encoded between the two patellamides for both start and stop cyclization sequences with the consensus stop sequence being AYDG(E/V). The start consensus motif in PatE was G(L/)E(A/P)S, which resembled the consensus sequence (GAEPR) in some lantibiotics [462].
The pat cluster included 7 genes: patA – patG which were all transcribed in the same direction and included an operon. patA was involved in the cleavage of the PatE precursor peptide. Similar to PatA, PatD also included two domains: an N-terminal and a C-terminal domain. There were two proposed roles for PatD: the involvement of PatD2 in the cyclization of threonine and cysteine residues of PatE that resulted in oxazoline and thiazoline ring formation, and activation of cleaved patellamide precursors as adenylated by PatD1, which were then cyclized to final patellamide structures. PatG was a large multidomain protein involved in the oxidation and maturation of PatE. PatB, PatC, and PatF have no obvious roles in the biosynthesis of patellamides while epimerization was possible in tandem with heterocycle oxidation [462].
PEPTIDES IN THE DEVELOPMENT PIPELINE
The marine pharmaceutical pipelines include few FDA [Food and Drug Administration] approved drugs along with some natural products in various phases of clinical trials. Several hundred marine derived compounds with potential value moved into the preclinical pipeline. Hence, the marine pharmaceutical pipeline remains active and now has enough momentum to deliver novel compounds to the near future [463].
The initial marine derived natural products that entered into the pharmacopeia include cytarabine [Ara C] and vidarabine [Ara A] in 1974, following which it took at least 30 years before more marine derived natural products gained approval and became part of the pharmacopeia. The following marine compounds were either accepted or are in various phases of clinical trials [464,465].
Ziconotide was the first marine drug isolated from Conus magus, a fish eating cone snail, which has been approved by the FDA. Ziconotide is a neuroactive peptide that is synthetically equivalent to ω-MVIIA [ω-conotoxin], a component of marine snail venom. Ziconotide is known to selectively block the neuronal N-type voltage sensitive calcium channels [NVSCCs], thereby inhibiting the activity of a subset of pain sensing primary nociceptor neurons and providing a unique mechanism of action [466,467].
Leconotide is a 27-residue, similar to ziconotide, which is currently in Phase I clinical trials sponsored by Relevare Pharmaceuticals, previously known as CNSBio. Leconotide includes three internal CYS-CYS bonds and was developed for the treatment of cancer related pain. Leconotide is a ω-conotoxin that blocks the neuronal voltage sensitive calcium channels [468] and is used via systemic administration [469].
Xen-2174 is a naturally occurring χ-conotoxin isolated from the venom of a marine cone snail, Conus marmoreus. Xen-2174 is a modified 13-residue chi-conopeptide that selectively inhibits the neuronal norepinephrine transporter. Xen-2174 was developed for the treatment of chronic pain and postoperative pain including cancer pain and is currently in phase-II clinical trials [470,469].
Brentuximab vedotin (adcetris®) originated from a mollusk associated cyanobacteria and is known to treat Hodgkin’s lymphoma and patients with systemic anaplastic large cell lymphoma [467, 471]. Brentuximab vedotin is an immunoconjugate derived from dolastatin 10, monomethylauristatin E (MMAE), which is a secondary metabolite from Symploca species and is approved for the treatment of CD30 positive lymphoproliferative disorders [469].
Soblidotin and tasidotin [ILX-651] are synthetic analogs of dolastatin 10 [472]. Soblidotin differs from dolastatin 10 in the replacement of the thiazole moiety of dolaphenine in dolastatin 10 to a phenylalanine methyl ester. Soblidotin was found to possess in vivo anticancer activity in murine P388 leukemia, Lewis Lung carcinoma, M5076 sarcoma, B16 melanoma, human MX-1 breast cancer, and Colon26 colon cancer cell-lines and SBC-3 SCLC and LX-1 xenografts, but was found to be ineffective as a single agent and hence was discontinued from Phase II clinical trials [393]. Cemadotin is a synthetic water-soluble analog of dolastatin 15 and is known to inhibit the cell-proliferation and the growth of human tumor xenografts. Cemadotin is also in clinical trials as a potential cancer chemotherapeutic agent. Cemadotin binds to tubulin by strongly suppressing the microtubule dynamics and thereby blocking mitosis. Cemadotin binds to tubulin with affinity to two binding sites with Kd values of 19.4 and 136 μM [473].
Elisidepsin or isokahalalide F is a cyclic peptide that belongs to the kahalalide family of compounds, which was previously in clinical trials. Elisidepsin was found to be cytotoxic against a panel of human NSCLC [non-small cell lung cancer] cell-lines [474] along with inducing rapid oncosis in ErbB3 expressing cells [475].
HTI-286 is a synthetic analog of the tripeptide hemiasterlin and was found to inhibit tubulin polymerization along with the disruption of microtubule organization in cells and induced mitotic arrest and apoptosis. HTI-286 was a potent inhibitor of human tumor cell proliferation at an IC50 value of 1.16 μM, with less P-glycoprotein interaction compared to the current anti-microtubule agents [476].
Glembatumumab vedotin [CR-011-vc-MMAE] is a monoclonal antibody (mAb)-drug conjugate developed by Celldex Therapeutics Inc., for treatment of glycoprotein non-metastatic melanoma protein-B [GPNMB] expressing cancers. MMAE is a full human mAb directed against extracellular GPNMB domain expressed in human breast melanomas and cancers. Glembatumumab is conjugated to the monomethylauristatin E, a potent microtubule inhibitor, with a cathepsin cleavable valine-citrulline [vc] dipeptide linker. While glembatumumab did possess potent antitumor activity, the most common toxicity associated is skin rash due to GPNMB expression in healthy skin [477].
ABT-414 is an antibody drug conjugate (ADC) linked to the anti-Epidermal Growth Factor Receptor (EGFR) ABT-806 antibody, where monomethylauristatin F (MMAF) is used. Abbvie, renamed Abbott Pharmaceutical Division, developed this ADC, which is currently extended to Phase II clinical trials and was developed as an anti-EGFR mAb [469].
There are many monoclonal antibodies coupled to either MMAE or MMAF that are currently in various phases of clinical trials with different therapeutic applications. PSMA-ADC is a human mAb against prostate specific antigen coupled via the valine-citrulline dipeptide linker to MMAE, which was designed to release via proteolysis by human cathepsin B and is currently in Phase II clinical trials. DCDT-2980S is an IgG1 antibody against CD22 epitope with MMAE, a potent microtubule disrupting agent, linked to the antibody on the sulfhydryl groups (reduced cysteines) via a protease cleavable linker, maleimidocaproyl valine-citrulline-ρ-aminobenzoyloxycarbonyl (MC-vc-PAB). DCDT-2980S is currently in Phase II clinical trials. DCDS-4501A is another ADC with MMAE linked to anti-CD79b monoclonal for the treatment of follicular B cell lymphoma. DCDS-4501A was developed by Genentech/Roche pharmaceuticals and is currently in Phase II clinical trials. Enfortumab vedotin is another human IgG1k antibody linked to MMAE with a valine-citrulline linker and is also known as AGS-22MSE and AGS-22ME [469]. Enfortumab vedotin was developed to target Nectin-4, which is found in many cancers including pancreatic, breast, urothelial, and breast [478]. Enfortumab vedotin was developed by Astellas Pharma and Agensys, Seattle Genetics and is currently in Phase I clinical trials [469].
Vorsetuzumab Mafdotin is an ADC developed by Seattle Genetics, which is a MMAF linked to anti-CD70 mAb 1F6 through a non-cleavable maleimidocaproyl linker, where the release relies on invagination followed by proteolytic digestion. This is used against relapsed and refractory non-Hodgkin’s lymphoma and metastatic renal cancer expressing CD70 epitope and is currently in Phase I trials. SGN-19A is another ADC developed by Seattle Genetics with an anti-CD19 antibody linked to MMAF via a maleimidocaproyl valine-citrulline linker. SGN-19A is used for the treatment of lymphomas including Burkitt’s lymphoma and is currently in Phase I clinical trials. BAY 79-4620 is an ADC with a MMAE linked to an antibody against human carbonic anhydrase IX. BAY 79-4620 was developed by Seattle Genetics for the determination of maximum tolerated dose (MTD) with advanced solid tumors and is currently in Phase I clinical trials. AGS-16C3F is IgG2k mAb against AGS-16 antigen, conjugated to MMAF with a non-cleavable maleimido caproyl linker. AGS-16C3F is used against liver and renal carcinomas from AGS-16 antigen and has currently completed Phase I clinical trials [469].
DMUC-5754A is a mAb against MUC16 epitope linked to MMAE, which is used for the treatment against ovarian carcinomas and is currently under Phase I clinical trials. DNIB-0600A is an IgG1 mAb against NaPi2b epitope linked to MMAE. DNIB-0600A is currently in Phase I trials for treatment against non-small cell lung cancer (NSCLC) and platinum resistant ovarian cancer. A1-mcMMAF is an ADC with MMAF linked to the maleimidocaproyl to a mAb against 5T4 tumor antigen. A1-mcMMAF exhibited potent antitumor activity. DMOT-4039A is a mAb identified as MMOT-0530A against antigen overexpressed in ovarian and pancreatic cancers, which is conjugated to MMAE. DMOT-4039A is currently in Phase I clinical trials. RG-7600 is an ADC developed by Genentech and is currently in Phase I trials for the treatment of ovarian cancers. DEDN-6526A is an ADC currently in Phase I trials for unresectable melanoma. The warhead is expected to be a auristatin derivative with the antibody directed against endothelin ETB receptors [469].
DSTP-3086S is an ADC developed by Genentech/Roche with an antibody against anti-STEAP1 IgG1 antibody modified with reactive thiols modification and coupled to MMAE. The antibody is directed against a six transmembrane epithelial antigen of prostate 1 (STEAP1) and is currently in Phase I clinical trials for its use in metastatic castration resistant prostate cancer. MLN-0264 is an ADC with monoclonal IgG antibody [5F9] against guanylyl cyclase (GCC), conjugated to MMAE with a cleavable linker. MLN-0264 is currently in Phase I clinical trials for its use in gastrointestinal tumors expressing GCC. RG-7598 is an ADC currently in Phase I trials for its use in multiple myeloma with a auristatin warhead. SGN-LIV1A is an anti-LIV-1 mAb linked to MMAE, which was developed by Seattle Genetics. The LIV-1 epitope is also known as ZIP6 or SLC39A6 and is a member of zinc transporter family. SGN-LIV1A is currently in Phase I trials for its use that leads to significant delay of tumor growth [469].
ASG-15E is also known as ASG-15ME and is an ADC with IgG2 mAb [AGS15 which targets SLITRK6]. ASG-15E is conjugated to MMAE via a maleimidocaproyl valine-citrulline linker. This antibody targets SLITRK6, a member of SLITRK family of neuronal transmembrane proteins. ASG-15E was developed by Seattle Genetics and is currently in Phase I clinical trials for its use in human bladder transitional cell carcinomas and advanced primary and metastatic tumors [469]. AGS67E is an anti-CD37 antibody, AGS67C, which has been linked through reduced cysteine to MMAE, with maleimidocaproyl valine-citrulline ρ-aminobenzyloxycarbonyl, a protease linker. AGS67E is known to induce cell-cycle arrest and apoptosis in various cell-lines [479].
ABBV-399 is another ADC that has been generated with c-Met targeting ABT-700 antibody. This led in delivering a potent cytotoxin to the c-Met overexpressing tumor cells, resulting in cell-death without relying on MET signaling. ABBV-399 is currently in Phase I clinical trials where it is well tolerated and producing positive responses in c-Met expression in non-small cell lung cancer [NSCLC] patients [480]. GSK2857916 is an IgG1 anti-BCMA antibody that has been conjugated to MMAF, a microtubule disrupter, via a maleimidocaproyl linker that is stable and protease resistant. GSK2857916 exhibited enhanced antibody dependent cell mediated cytotoxicity, and this is the result of afucosylation of the FC domain that is known to increase the FCγRIIIa affinity expressed on the immune effector cells. GSK2857916 is currently in Phase I clinical trials for its use in relapsed/refractory multiple myeloma [481].
There are few ADC’s currently in advanced preclinical trials with minimal information. CDX-014 [CR-014-vcMMAE] is hypothesized to be a valine-citrulline linked MMAE to an anti-TIM1 mAB CR-014 based on the code name. This antibody targets a type I transmembrane (TIM 1) protein that is known to be expressed on the surface of the renal and ovarian carcinoma cells. HuMax-CD74 is an auristatin linked ADC with HuMax-CD74 antibody known to target the HLA class II histocompatibility antigen gamma chain [CD74] and is used in solid tumors and hematological malignancies. HuMab-TF-011-vcMMAE is an ADC developed for its use in solid tumors. It includes a human tissue factor [TF] specific antibody [TF-011] linked to valine-citrulline cleavable linker and MMAE [469].
V. CONCLUSION
Marine natural products provide tremendous opportunities for the study of unique and diverse secondary metabolites including modified peptides, providing opportunities for expansion of the pharmaceutical pipeline. Marine natural products have escalated beyond their original role in the identification of novel prototype drug-leads to studies involving the design of unique molecules and utilizing biosynthesis in association with synthesis. As mentioned in this review, there are several marine peptides that have been placed in the market as therapeutic agents contributing to improved human health. Enhanced methods in fermentation technologies to grow bacteria/fungi in addition to biosynthesis and syntheses have opened new doors to both generate and supply drug-leads from various marine sources. Examples include ziconotide, the first FDA approved marine peptide for pain treatment while dolastatin-derived agent (adcetris®) [484] is a treatment for cancer which was approved by the FDA as well, suggesting the importance of marine derived peptides in the field of science. Hence, the potential for marine based compounds in various diseases is immense, and there are approximately 25,000 defined chemical compounds that have been reported thus far [484] supporting the significance of drug-discovery from marine sources.
Figure 1.

Chemical structures of discodermins A-H and kasumigamide
Figure 2.

Chemical structures of phakellistatins 1–12 and isophakellistatin 3
Figure 3.

Chemical structures of phakellistatins 13–19
Figure 4.

Chemical structures of geodiamolides A-R, geodiamolide-TA, neosiphoniamolide, jasplakinolide, and pipestelides A-C
Figure 5.

Chemical structures of milnamides A-G, hemiasterlins A, D, and scleritodermin A
Figure 6.

Chemical structures of mirabamides A-H
Figure 7.

Chemical structures of celebesides A-C and theopapuamides A-D
Figure 8.

Chemical structures of homophymines A-E and A1-E1
Figure 9.

Chemical structures of neamphamides A-D
Figure 10.

Chemical structures of callipeltins A-D
Figure 11.

Chemical structures of callipeltins E-K
Figure 12.

Chemical structures of callipeltins L-Q
Figure 13.

Chemical structures of microspinosamide and carteritins A-B
Figure 14.

Chemical structures of mycothiazole, dysidenin, isodysidenin, dysideaprolines A-F, barbaleucamides A-B, and dysithiazolamide
Figure 15.

Chemical structures of microcionamides A-B
Figure 16.

Chemical structures of halicylindramides A-C and seco-methyl ester of halicylindramide B
Figure 17.

Chemical structures of haligramides A-B, waiakeamide, and sulfone derivative of waiakeamide
Figure 18.
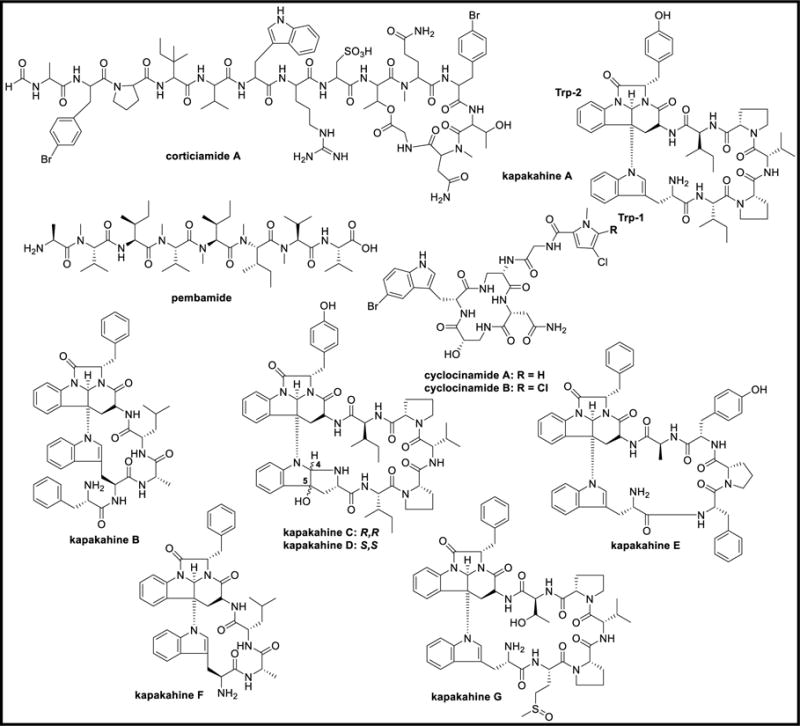
Chemical structures of corticiamide A, pembamide, cyclocinamides A-B, and kapakahines A-G
Figure 19.

Chemical structures of taumycins A-B
Figure 20.

Chemical structures of pipecolidepsins A-B
Figure 21.
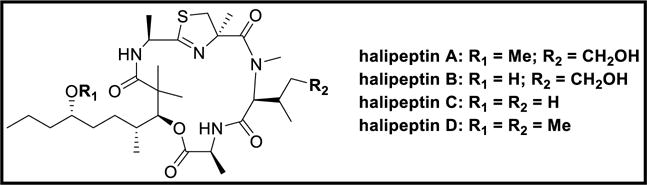
Chemical structures of halipeptins A-D
Figure 22.

Chemical structures of tausalarin C, arenastatin A, and axinastatins 1–3
Figure 23.

Chemical structures of hymenamides A-H and J-K
Figure 24.

Chemical structures of wainunuamide, axinellins A-C, stylopeptides 1–2, stylostatins 1–2, and cyclonellin
Figure 25.

Chemical structures of fenestins A-B and hymenistatin 1
Figure 26.

Chemical structures of discobahamins A-B and calyxamides A-B
Figure 27.

Chemical structures of microsclerodermins A-K and anhydromicrosclerodermin C
Figure 28.
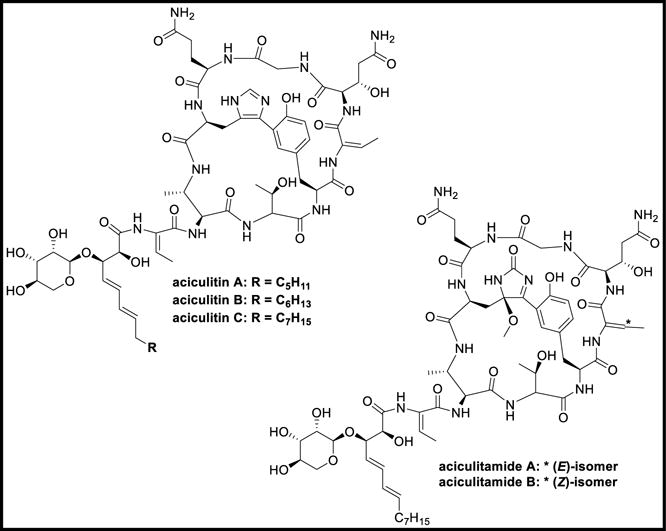
Chemical structures of aciculitins A-C and aciculitamides A-B
Figure 29.

Chemical structures of polydiscamides A-D
Figure 30.

Chemical structures of criamides A-B and gombamide A
Figure 31.
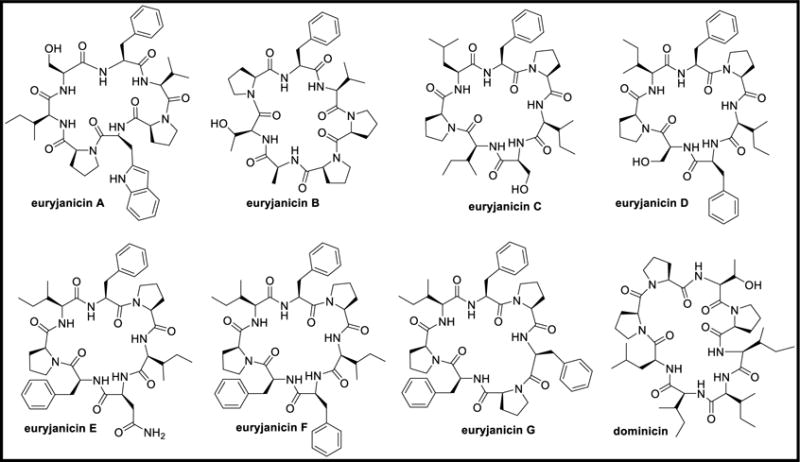
Chemical structures of euryjanicins A-G and dominicin
Figure 32.

Chemical structures of neopetrosiamides A-B, jamaicensamide A, sulfolipodiscamides A-C, and lipodiscamides A-C
Figure 33.

Structures of stylissamides A-H, X, stylissatins A-D, and stylisins 1–2
Figure 34.

Chemical structures of reniochalistatins A-E and yaku’amides A-B
Figure 35.

Chemical structures of chujamides A-B and leucamide A
Figure 36.

Chemical structures of azumamides A-E and phoriospongins A-B
Figure 37.

Chemical structures of callyaerins A-H
Figure 38.

Chemical structures of motuporin, theonellapeptolide Id, and nazumazoles A-F
Figure 39.

Chemical structures of polytheonamides A-C
Figure 40.

Chemical structures of pseudotheonamides A1-A2, B2, C-D, dihydrocyclotheonamide A, nazumamide A, cyclotheonamides A-B, and orbiculamide A
Figure 41.

Chemical structures of cyclotheonamides E, E2, and E3
Figure 42.

Chemical structures of theonellamides A-G, keramamides A-H, and J-N
Figure 43.

Chemical structures of koshikamides A1-A2 and B-H
Figure 44.

Chemical structures of papuamides A-F, theonegramide, nagahamide A, and cupolamide A
Figure 46.

Chemical structure of theonellapeptolide IIIe [169]
Figure 47.

Chemical structures of congeners 1–2 and solomonamides A-B
Figure 48.

Chemical structures of cyclolithistide A, revised structure of cyclolithistide A [176], theopalauamide A, isotheopalauamide, and oriamide
Figure 49.

Chemical structures of miraziridine A, paltolides A-C, perthamides B-K, and mutremdamide A
Figure 50.

Structural similarity of peptides isolated from Theonella sp., and microorganisms
Figure 51.

Chemical structures of turnagainolides A-B, solonamides A-B, actinoramides A-C, fijimycins A-C, and brunsvicamides A-C
Figure 52.

Chemical structures of malyngamides 2–3, cocosamides A-B, pitiprolamide, and pitipeptolides A-F
Figure 53.

Chemical structures of lagunamides A-C, wewakamide A, and guineamides A-G
Figure 54.

Chemical structures of wewakazole, wewakazole B, and wewakpeptins A-D
Figure 55.

Chemical structures of porpoisamides A-B, bisebromoamide, and norbisebromoamide
Figure 56.

Chemical structure of somocystinamide A
Figure 57.

Chemical structures of cyclic desmethoxymajusculamide C, linear desmethoxymajusculamide C, majusculamides A-D, dolastatins 11–12, 57-normajusculamide C, lyngbyastatins 1, 3, epilyngbyastatin 1, and deoxymajusculamide D
Figure 58.

Chemical structures of apratoxins A-E and dehydroapratoxin A
Figure 59.

Chemical structures of dragonamides A-E, herbamides A-B, dragomabin, and carmabins A-B
Figure 60.
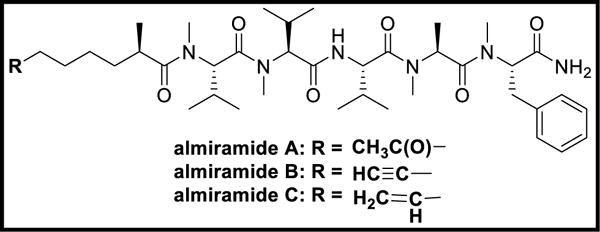
Chemical structures of almiramides A-C
Figure 61.

Chemical structures of grassystatins A-C
Figure 62.

Chemical structures of lobocyclamides A-C and obyanamide
Figure 63.

Chemical structures of hantupeptins A-C
Figure 64.

Chemical structures of trungapeptins A-C and antanapeptins A-D
Figure 65.

Chemical structures of palmyramide A, dudawalamides A-E, and mantillamide
Figure 66.

Chemical structures of grassypeptolide and carriebowmide
Figure 67.

Chemical structure of hoiamide A
Figure 68.

Chemical structures of tiglicamides A-C, largamides A-C, and methyl esters of largamides A-C
Figure 69.

Chemical structures of itralamides A-B
Figure 70.

Chemical structures of lyngbyastatins 4–6 and 8–10
Figure 71.

Chemical structures of somamides A-B, lyngbyastatin 7, kempopeptins A-B, and scyptolin A
Figure 72.

Chemical structures of lyngbyazothrins A-D, pahayokolides A-B, schizotrin A, and tychonamides A-B
Figure 73.

Chemical structures of grassypeptolides A-E and ibu-epidemethoxylyngbyastatin
Figure 74.

Chemical structures of grassypeptolides F-G
Figure 75.

Chemical structures of lyngbyapeptins A-D and lyngbyabellins A-J
Figure 76.

Chemical structures of alotamide A, lyngbyabellins K-N, and 7-epi-lyngbyabellin L
Figure 77.

Chemical structures of barbamide and jamaicamides A-C
Figure 78.

Chemical structure of hectochlorin
Figure 79.

Chemical structures of apramides A-G
Figure 80.

Chemical structures of antillatoxin, barbaramide A, aurilide, aurilides B-C, and kalkitoxin
Figure 81.

Chemical structures of georgamide and yanucamides A-B
Figure 82.

Chemical structures of dysidenamide, pseudodysidenin, nordysidenin, and isodysidenin
Figure 83.

Chemical structures of ulongamides A-F
Figure 84.

Chemical structures of isomalyngamides A-B, malyngamides A-K, and 8-epi-malyngamide C
Figure 85.

Chemical structures of malyngamides L-Y
Figure 86.

Chemical structures of hermitamides A-B and laxaphycins A-B
Figure 87.

Chemical structures of tasiamide, tasiamides B-F, pepstatin A, and tasipeptins A-B
Figure 88.

Chemical structures of symplocamide A, symplocin A, veraguamides A-G, and semisynthetic veraguamide
Figure 89.

Chemical structures of symplostatins 1–4
Figure 90.

Chemical structures of malevamides A-D, belamide A, and largazole
Figure 91.

Chemical structures of mitsoamide and gallinamide
Figure 92.

Chemical structures of nostocyclamide, tenuecyclamides A-D, and cryptophycin
Figure 93.

Chemical structures of microcystin, raocyclamides A-B, and venturamides A-B
Figure 94.

Chemical structures of anabaenopeptins A-J and pompanopeptins A-B
Figure 95.

Chemical structures of floridamide, coibamide A, scytonemin, hormothamnin A, and trichamide,
Figure 96.

Chemical structures of arenamides A-C and caldoramide
Figure 97.

Chemical structures of viequeamides A-F and companeramides A-B
Figure 98.

Chemical structures of nodulapeptins A-B, aeruginosins NAL2, NOL6, spumigins A, C-H, and spumigins B1-B2
Figure 99.

Chemical structures of kailuins A-H
Figure 100.

Chemical structures of ngercheumicins A-I, unnarmicins A and C
Figure 101.

Chemical structures of ariakemicins A-B, mollemycin A, thiocoraline, cyclomarins A-D, and cyclomarazines A-B
Figure 102.

Chemical structures of surugamides A-F and champacyclin
Figure 103.

Chemical structures of tumescenamides A-C and streptocidins A-D
Figure 104.

Chemical structures of salinamides A-F
Figure 105.

Chemical structures of tauramamide, tupuseleiamides A-B, and bogorols A-E
Figure 106.

Chemical structures of loloatins A-D and marthiapeptide A
Figure 107.

Chemical structures of loihichelins A-F, marinobactins A-C, D1, D2, and E
Figure 108.

Chemical structures of aquachelins A-D, amphibactins B-I, petrobactin, petrobactin sulfate, aerobactin, and amphi-enterobactin
Figure 109.

Chemical structures of alterobactins A-B and pseudoalterobactins A-B
Figure 110.
Chemical structures of trivanchrobactin, divanchrobactin, vanchrobactin, anguibactin, and vibrioferrin
Figure 111.

Chemical structures of ochrobactins A-C and synechobactins A-C
Figure 112.

Chemical structures of unguisins A-F
Figure 113.

Chemical structures of versicotides A-B, versicoloritides A-C, fellutamides A-D, F, and asperterrestide A
Figure 114.

Chemical structures of sansalvamide A, scopularides A-B, and exumolides A-B
Figure 115.

Chemical structures of cyclo-(L-Pro-L-Tyr), cyclo-(L-Pro-L-Val), cyclo-(L-Phe-L-Pro), and cis-bis(methylthio)silvatin
Figure 116.

Chemical structures of penilumamide, penilumamides B-D, and asperpeptide A
Figure 117.

Chemical structures of emericellamides A-B, guangomides A-B, homodestcardin, and azonazine
Figure 118.

Chemical structures of endolides A-D and hirsutide
Figure 119.

Chemical structures of simplicilliumtides A-H and dictyonamides A-B
Figure 120.

Chemical structures of halolitoralins A-C, sclerotides A-B, cyclo(L-Pro-L-Val), cyclo(L-Pro-L-Leu), and cyclo(L-Ile-L-Val)
Figure 121.

Chemical structures of RHM1, RHM2, and efrapeptin G
Figure 122.

Chemical structures of cordyheptapeptides A-E and oryzamides A-E
Figure 123.
Chemical structures of cycloxazoline, diazonamides A-E, revised structure of diazonamide A, and diazonamide A analog
Figure 124.

Chemical structure of vitilevuamide
Figure 125.

Chemical structures of bistratamides A-J, westiellamide, and didmolamides A-B
Figure 126.

Chemical structures of lissoclinamides 1–8, patellamides A-E, ulithiacyclamide A, and ascidiacyclamide
Figure 127.

Chemical structures of didemnins A-E, G-H, M-N, X-Y, epididemnin A1, nordidemnin N, and acyclodidemnin A
Figure 128.

Chemical structures of eusynstyelamide and eudistomides A-B
Figure 129.

Chemical structures of mollamide, mollamides B-C, keenamide A, and cycloforskamide
Figure 130.

Chemical structures of virenamides A-C
Figure 131.

Chemical structures of cyclodidemnamide, comoramides A-B, mayotamides AB, didmolamides A-B, prepatellamide A, patellamides A-C, and tamandarins A-B
Figure 132.

Chemical structures of patellins 1–6 and trunkamide A
Figure 133.

Chemical structures of kulolides 1–3, kulokainalide-1, kulomo’opunalides 1–2, and pupukeamide
Figure 134.

Chemical structures of ulicyclamide, preulithiacyclamide, ulithiacyclamide, and ulithiacyclamide B
Figure 135.

Chemical structures of nobilamides A-H, A-3302 A-B, and N-acetyl-L-phenylalanyl-L-leucinamide
Figure 136.

Chemical structure of aplidine
Figure 137.

Chemical structures of dolastatins 3, 10–15, homodolastatin 3, kororamide, and epidolastatin 12
Figure 138.

Chemical structures of dolastatins 16–18 and homodolastatin 16
Figure 139.

Chemical structures of dolastatins C-E, G-I, nordolastatin G, isodolastatin H, lyngbyastatin 2, and norlyngbyastatin 2
Figure 140.

Chemical structures of dolabellin and doliculide
Figure 141.

Chemical structures of kulokekahilides 1–2 and aurilide
Figure 142.

Chemical structures of viridamides A-B, dolastatin 12, lyngbyastatin 1, malyngamide C, malyngamide C acetate, lyngbic acid, janadolide, and microcolins A-B
Figure 143.

Chemical structures of tumonoic acids A-B, D-I, and epi-tumonoic acid
Figure 144.

Chemical structures of onchidins A-B and nocardiamides A-B
Figure 145.

Chemical structures of kahalalides A-H, J, W, norkahalalide A, isokahalalide F, and 5OH-kahalalide F
Figure 146.

Chemical structures of kahalalides K, O-Q, R1-R2, S1-S2, V,X, and Y
Figure 147.

Chemical structures of cis,cis-ceratospongamide, trans,trans-ceratospongamide, galaxamide, and mebamamides A-B
Figure 148.
Chemical structures of echinocandin B, pneumocandins A0, B0, C0, and caspofungin acetate
Figure 149.

Biosynthesis of nodularin
Figure 150.

Biosynthesis of barbamide & barbaleucamide
Figure 151.

Biosynthesis of jamaicamide A
Figure 152.

Biosynthesis of lyngbyatoxins [452]
Figure 153.

Molecular networking of dolastatin 10 tetrapeptide molecular family
Figure 154.

Biosynthesis of anabaenopeptins
Figure 155.

Biosyntheses of aeruginosin and spumigin
Figure 156.

Biosynthesis of thiocoraline
Figure 157.

Biosynthesis of cyclomarin A and cyclomarazine A
Figure 158.

Biosynthesis of surugamides
Figure 159.

Biosynthesis of salinamides
Figure 160.

Biosyntheses of amphibactins and marinobactins
Figure 161.

Biosynthesis of amphi-enterobactin
Figure 162.

Biosynthetic gene clusters of scopularide and emericellamide
Figure 163.

Biosynthesis of Scopularide in Scopulariopsis brevicaulis
Figure 164.

Biosynthesis of emericellamide in Aspergillus nidulans
Figure 165.

Biosynthesis of N-methyl-3-(3-furyl)alanine
Figure 166.

Biosynthesis of kasumigamide
Figure 167.

Biosynthesis of polytheonamides
Figure 168.

Biosynthesis of keramamides
Figure 169.

Biosynthesis of microsclerodermins
Figure 170.

Biosynthesis of patellamides
Figure 171.

Chemical structure of ziconotide
Figure 172.

Chemical structure of leconotide
Figure 173.

Chemical structure of Xen-2174
Figure 174.

Chemical structures of brentuximab vedotin and monomethylauristatin E
Figure 175.

Chemical structures of soblidotin, tasidotin, and cemadotin
Figure 176.

Chemical structure of elisidepsin
Figure 177.

Chemical structure of HTI-286
Figure 178.

Chemical structure of glembatumumab vedotin
Figure 179.

Chemical structure of monomethylauristatin F
Table 1.
Peptides from Marine Sponges
| Compound | Biological Source | Biological Activity | Inhibitory Concentration | Ref |
|---|---|---|---|---|
| Discodermins A–H | Discodermia | Cytotoxic | IC50 0.01 – 11.9 μM | 5–8 |
| Kasumigamide | D. calyx | Antialgal | MIC 2.54 μM | 9–10 |
| Phakellistatins 1–3 | Phakellia sp. | Cytotoxic | ED50 0.4 – 9.1 μM | 14–15, 17 |
| Phakellistatin 4 | Phakellia sp. | Cytotoxic | ED50 40 μM | 18–19 |
| Phakellistatin 5 | Phakellia sp. | Cytotoxic | GI50 0.6 μM | 20 |
| Phakellistatin 6 | Phakellia sp. | Cytotoxic | GI50 11.8 – 117.5 nM | 21 |
| Phakellistatins 7–14 | Phakellia sp. | Cytotoxic | ED50 0.01 – 6.4 μM | 16, 22–25 |
| Phakellistatin 15 | Phakellia sp. | Antitumor | IC50 8.5 μM | 26–27 |
| Phakellistatin 16 | Phakellia sp. | P388 & BEL-7402 cytotoxicity | IC50 5.4 & 14.3 μM | 27 |
| Phakellistatin 19 | Phakellia sp. | Antimitotic | IC50 420 to 84 nM | 28 |
| Geodiamolides A–F | Pseudaxinyssa | Cytotoxic | 4.99, 4.37, 4.54, 62.15, 24.11, & 11.19 nM | 31 |
| Geodiamolide G | Geodia | Human glioblastoma/astrocyto ma U373 cytotoxicity HEY human ovarian carcinoma cytotoxicity |
IC50 11.75 mM IC50 13.12 mM |
33 |
| Jasplakinolide | Jaspis johnstoni | Apoptosis | – | 34 |
| Pipestelide A | Pipestela candelabra | Cytotoxic | IC50 0.1 μM | 35 |
| Milnamide A | Cymbastela | Cytotoxic Tubulin polymerization PC3 & NFF cytotoxic |
IC50 1.65 μM IC50 6.02 μM IC50 11.0 & 70.6 nM |
34, 36–39 |
| Milnamide B/Hemiasterlin | Hemiasterella, Cymbastela, Siphonochalina & Auletta | Cytotoxic PC3 & NFF cytotoxic |
IC50 6.8 nM IC50 0.05 & 0.40 nM |
34, 37, 39 |
| Milnamide C | Auletta | Anticancer PC3 & NFF cytotoxic | IC50 267.77 pM IC50 31.7 & 188 nM |
36, 39 |
| Milnamide D | Cymbastela | Cytotoxic Tubulin polymerization PC3 & NFF cytotoxic |
IC50 66.8 nM IC50 16.9 μM IC50 0.38 & 1.19 μM |
34, 36–39 |
| Milnamide E | Pipestela candelabra | PC3 & NFF cytotoxic | IC50 34.2 & 123 nM | 39 |
| Milnamide F | P. candelabra | PC3 & NFF cytotoxic | IC50 2.18 & 5.65 μM | 39 |
| Milnamide G | P. candelabra | PC3 cytotoxic | IC50 2.87 μM | 39 |
| Hemiasterlin A | P. candelabra | PC3 & NFF cytotoxic | IC50 0.27 & 1.03 nM | 39 |
| Hemiasterlin D | P. candelabra | PC3 & NFF cytotoxic | IC50 2.20 & 8.16 nM | 39 |
| Scleritodermin A | Scleritoderma nodosum | Apoptosis Cytotoxic |
IC50 1.3 μM IC50 < 2 μM |
34, 40–41 |
| Mirabamide A | Siliquariaspongi a mirabilis | HIV-1 fusion & neutralization | IC50 140 & 40 nM | 42–43 |
| Mirabamide C | S. mirabilis | HIV-1 fusion & neutralization | IC50 1.3 μM & 140 nM | 42–43 |
| Mirabamide D | S. mirabilis | HIV-1 fusion & neutralization | IC50 3.9 μM −190 nM | 42–43 |
| Mirabamides E–H | Stelletta clavosa | HIV-1 inhibition | IC50 121, 62, 68, & 41 nM | 44 |
| Celebeside A | S. mirabilis | HIV-1 neutralization assay | IC50 2.1 μM | 45 |
| Theopapuamides A–C | T. swinhoei & S. mirabilis | Cytotoxic Antifungal |
IC50 1.3 – 2.6 μM 1-5 μg/disk | 45 |
| Homophymine A | Homophymia sp. | HIV-1 cytoprotective activity | IC50 75 nM | 46 |
| Homophymines B–E | Homophymia sp. | Antiproliferative | IC50 2–100 nM | 47 |
| Homophymines A1–E1 | Homophymia sp. | Antiproliferative | IC50 2-100 nM | 47 |
| Neamphamide A | Neamphius huxleyi | HIV-1 cytotoxicity | EC50 28 nM | 48–49 |
| Neamphamides B–D | N. huxleyi | Anticancer | IC50 88–370 nM | 50 |
| Callipeltin A | Callipelta | Anti-HIV, antifungal, inhibition of Na/Ca exchanger | – | 51 |
| Callipeltin B | Callipelta | Inhibition of Na/Ca exchanger | 4 μM | 51–52 |
| Callipeltins F–I | Callipelta | Antifungal activity | MIC 10−4 M | 54 |
| Callipeltin K | Callipelta | Antifungal activity | MIC 10−4 M | 55 |
| Callipeltin J | Callipelta | Antifungal activity | MIC 10−4 M | 55 |
| Callipeltins N–O | Asteropus | Cytotoxic | IC50 0.16 μM | 56 |
| Microspinosamide | Sidonops microspinosa | HIV-1 cytopathic effect | EC50 115.8 nM | 57 |
| Carteritin A | Stylissa carteri | HeLa, HCT116, & RAW264 cytotoxicity | IC50 0.7, 1.3, & 1.5 μM | 58 |
| Mycothiazole | Spongia mycofijiensis | Anti-helminthic | 123.59 μM | 34, 59 |
| Microcionamide A | Clathria abietina | SKBR-3 & MCF-7 activity Anti-tubercular |
IC50 98 & 125 nM MIC 5.7 μM |
34, 62–63 |
| Microcionamide B | C. abietina | SKBR-3 & MCF-7 activity Anti-tubercular |
IC50 172 & 177 nM MIC 5.7 μM |
34, 62–63 |
| Halicylindramides A–C | Halichondria cylindrata | Cytotoxic Anti-fungal |
IC50 0.3, 0.1, & 0.1 μM MIC 7.5 μg/disk |
34, 64 |
| Halicylindramide B methyl ester | H. cylindrata | Antifungal Cytotoxic |
120 μg/disk 5.5 μM |
34, 64 |
| Haligramide A | Haliclona nigra | A549, HCT-15, SF-539, SNB-19 cytotoxicity | 6.6, 19.9, 11.5, & 11.6 μM | 34, 65 |
| Haligramide B | H. nigra | A549, HCT-15, SF-539, SNB-19 cytotoxicity | 4.9, 11.0, 6.3, & 8.2 μM | 34, 65 |
| Waiakeamide | Ircinia dendroides | Cytotoxic | IC50 66.2 nM | 66 |
| Cyclocinamide A | Psammocinia sp. | Cytotoxic | – | 34, 68–69 |
| Pembamide | Cribrochalina | A-549, HT-29, MDA-MB-231 cytotoxicity | GI50 2.46, 3.80, & 3.35 μM | 70 |
| Kapakahines A–C | C. olemda | Cytotoxic | IC50 5.1, 5.9, & 4.7 μM | 71 |
| Kapakahine E | C. olemda | Cytotoxic | IC50 5.0 μM | 72 |
| Taumycin A | Fascaplysinopsis | Brine shrimp toxicity Anti-leukemic |
IC50 17.9 μM 1 μM |
73 |
| Taumycin B | Fascaplysinopsis | Brine shrimp toxicity | IC50 17.9 μM | 73 |
| Pipecolidepsin A | Homophymia lamellosa | A549, HT29, MDA-MB-231 cytotoxicity | GI50 0.6, 1.12, & 0.7 μM | 74–75 |
| Pipecolidepsin B | H. lamellosa | A549, HT29, MDA-MB-231 cytotoxicity | GI50 0.04, 0.01, 0.02 μM | 74–75 |
| Halipeptin A | Haliclona sp. | Anti-inflammatory | 0.3 mg/kg | 76–77 |
| Halipeptin D | Haliclona sp. | Cytotoxic Anti-tumor |
IC50 7 nM IC50 420 nM |
77 |
| Tausalarin C | Fascaplysinopsis | Anti-leukemic | 1 μM | 78 |
| Arenastatin A | Dysidea arenaria | Cytotoxic | IC50 8.2 pM | 79–80 |
| Axinastatin 2 | Axinella sp. | Cytostatic | GI50 9.4 – 456.4 nM | 81 |
| Axinastatin 3 | Axinella sp. | Cytostatic Anti-leukemic |
GI50 9.2 – 448.2 nM ED50 512.2 nM |
81 |
| Hymenamide A | Hymeniacidon | Anti-fungal | MIC 37.5 μM | 84 |
| Hymenamide B | Hymeniacidon | Anti-fungal Cytotoxic to KB cells and murine lymphoma L1210 cells |
MIC 79.4 μM IC50 7.2 & 3.8 μM |
84 |
| Hymenamides C & E | Hymeniacidon | Antifungal | MIC 160.7 & 155.7 μM | 85 |
| Hymenamide H | Hymeniacidon | Cytotoxic | IC50 6.96 μM | 87 |
| Hymenamide J | Hymeniacidon | KB & L1210 cytotoxicity | IC50 691.2 nM & 2.4 μM | 87 |
| Hymenamides G & K | Hymeniacidon | Cytotoxic | IC50 70.5 & 72.5 μM | 87 |
| Wainunuamide | Stylotella aurantium | Cytotoxic against K562 & A2780 | ID50 24.6 & 25.7 μM | 88 |
| Axinellins A–B | Axinella carteri | Antitumor | IC50 3.7 & 7.8 μM | 89 |
| Axinellin C | A. carteri | K562 & A2780 cytotoxicity | ID50 4.8 & 14.0 μM | 90 |
| Stylopeptide 2 | Stylotella sp. & | Anticancer | 10−5 M | 93 |
| Stylostatin 1 | S. aurantium | Anti-leukemic | ED50 1.1 μM | 94 |
| Hymenistatin 1 | Hymeniacidon | Cytotoxic | ED50 3.9 μM | 97 |
| Discobahamins A–B | Discodermia | Antifungal | – | 98 |
| Calyxamides A–B | D. calyx | Cytotoxic | – | 99–100 |
| Microsclerodermins A. A–B | Microscleroderma | Antifungal | 2.5 μg/disk | 101 |
| Microsclerodermin C | Theonella | Antifungal | 5 μg/disk | 102 |
| Microsclerodermin D | Theonella | Antifungal | 100 μg/disk | 102 |
| Microsclerodermin E | Theonella | Antifungal | 10 μg/disk | 102 |
| Microsclerodermins F–I | Microscleroderma | Cytotoxic Antifungal | 1.9, 2.6, 1.1, & 1.2 μM 1.5, 3.0, 12.0, & 25.0 μg/disk | 103 |
| Microsclerodermins J–K | Microscleroderma | Antifungal | – | 104 |
| Anhydromicrosclerode rmin C | Theonella | Antifungal | 50 μg/disk | 102 |
| Aciculitins A–C | Aciculites orientalis | Cytotoxic Antifungal | IC50 0.4 μM 2.5 μg/disk |
105 |
| Polydiscamide A | Discodermia sp. | Anticancer Antimicrobial | IC50 403.8 nM MIC 1.8 μM |
106 |
| Polydiscamides B–D | Ircinia | SNSR agonists | EC50 1.26, 3.57, & 2.80 μM | 107 |
| Criamide B | Cymbastela | A549, LOVO, HEY, U373, & P388 cytotoxicity MCF7 cytotoxicity |
ED50 424.3, 219.5, 277.9, 395.0, & 10.2 nM ED50 9.9 μM |
108 |
| Gombamide A | Clathria gombawuiensis | A549 & K562 cytotoxicity Na+/K+-ATPase inhibition |
LC50 7.1 & 6.9 μM LC50 9.4 μM |
109 |
| Neopetrosiamides A & B | Neopetrosia sp. | Inhibition of amoeboid invasion of tumor cells | 1.95 μM | 112 |
| Sulfolipodiscamides A–C | Discodermia kiiensis | Cytotoxicity | – | 113 |
| Lipodiscamides A–C | D. kiiensis | P388 cytotoxic HeLa cytotoxic |
IC50 23, 20, & 31 μM IC50 18, 26, & 46 μM |
114 |
| Stylissamide H | Stylissa caribica | Cytotoxic | EC50 5.7 μM | 118 |
| Stylissamide X | Stylissa sp. | Inhibition of HeLa cell-migration | 0.1– 10 μM | 118 |
| Stylissatin A | S. massa | NO Inhibition | IC50 87 μM | 119 |
| Stylissatin B | S. massa | Antitumor | IC50 2.4 – 9.8 μM | 120 |
| Reniochalistatin E | Reniochalina stalagmitis | RPMI-8226 & MGC- 803 cytotoxicity | IC50 4.9 & 9.7 μM | 122 |
| Yaku’amides A–B | Ceratopsion sp. | Cytotoxic | IC50 8.5 & 2.4 nM | 123 |
| Chujamide A | Suberites waedoensis | A549 & K562 cytotoxicity | LC50 10.1 & 37.0 μM | 124 |
| Chujamide B | S. waedoensis | A549 & K562 cytotoxicity Na+/K+-ATPase inhibition |
LC50 26.4 & 55.6 μM IC50 17.2 μM |
124 |
| Leucamide A | Leucetta microraphis | Huh7, HepG2, & HM02 cytotoxic | GI50 8.3, 9.6, & 8.5 μM | 125 |
| Azumamide A | Mycale izuensis | Histone deacetylase inhibition K562 & WiDr cytotoxic |
IC50 0.045 μM IC50 4.5 & 5.8 μM |
126 |
| Azumamides B-E | M. izuensis | Histone deacetylase inhibition | IC50 0.11, 0.11, 1.3 & 0.064 μM | 126 |
| Phoriospongins A-B | Phoriospongia sp. & Callyspongia bilamellata | Nematocidal | LD99 7.2 μM | 127 |
| Callyaerins E | C. aerizusa | Cytotoxic | ED50 0.39 μM | 128 |
| Callyaerin H | C. aerizusa | Cytotoxic | ED50 0.48 μM | 128 |
| Callyaerin G | C. aerizusa | Cytotoxic | 2.3 – 7.7 μM | 129 |
| Motuporin | T. swinhoei | Protein phosphatase-1 inhibitor & Cytotoxic | IC50 < 1.0 nM IC50 3.1 – 16.2 μM |
130–131 |
| Theonellapeptolide Id | T. swinhoei | Cytotoxic | IC50 1.7 μM | 132 |
| Nazumazoles A-C | T. swinhoei | Cytotoxic | IC50 0.8 μM | 133–134 |
| Nazumazoles D-F | T. swinhoei | Protease inhibitors | IC50 2.0, 3.0, & 10.0 μM | 133–134 |
| Orbiculamide A | Theonella sp. | Cytotoxic | IC50 4.8 μM | 135 |
| Polytheonamides A–C | T. swinhoei | Cytotoxic | IC50 15.5, 13.5, & 13.5 pM | 136–137 |
| Cyclotheonamide A | T. swinhoei | Serine protease inhibitor | Streptokinase: IC50 0.02 μM Trypsin: IC50 0.04 μM α-thrombin: IC50 0.18 μM |
140 |
| Cyclotheonamides E, E2 & E3 | T. swinhoei | Thrombin inhibitor Trypsin inhibitor | IC 50 2.9, 13.0 & 9.5 nM IC50 30.0, 55.0 & 52.0 nM |
139 |
| Nazumamide A | T. swinhoei | Thrombin inhibitor | IC50 4.6 μM | 141 |
| Pseudotheonamides A1, A2, B2, C, D | T. swinhoei | Thrombin inhibitors Trypsin inhibitors | IC50 1.0, 3.0, 1.3, 0.2, & 1.4 μM IC50 4.5, > 10, 6.2, 3.8, & > 10 μM |
138 |
| Dihydrocyclotheonami de A | T. swinhoei | Thrombin inhibitor Trypsin inhibitor | IC50 0.33 μM IC50 6.7 μM |
138 |
| Theonellamides A–C | Theonella sp. | Cytotoxic | IC50 2.8, 1.1, 1.6 μM | 142 |
| Theonellamides D-E | Theonella sp. | Cytotoxic | IC50 953.2, & 496.3 nM | 142 |
| Theonellamide G | T. swinhoei | Antifungal Cytotoxic |
IC50 4.5 & 2.0 μM IC50 6.0 μM |
143 |
| Keramamide A | T. swinhoei | Ca2+-ATPase inhibitor | IC50 3 × 10−4 moldm−3 | 144 |
| Keramamides B–D | T. swinhoei | Inhibition of human neutrophil superoxide generation | 5 × 10−8 M | 145 |
| Keramamide E | T. swinhoei | KB & L1210 cytotoxicity | IC50 1.37 & 1.42 μM | 147 |
| Keramamide F | T. swinhoei | Cytotoxic | – | 146 |
| Keramamide G–H | T. swinhoei | Cytotoxic | ~ 10 μM | 147 |
| Keramamide J | T. swinhoei | Cytotoxic | ~ 10 μM | 147 |
| Keramamides K–L | T. swinhoei | L1210 cytotoxicity KB cytotoxicity |
IC50 768.1 & 495.9 nM IC50 448.0 & 970.4 nM |
148 |
| Keramamides M–N | T. swinhoei | KB cytotoxicity L1210 cytotoxicity |
IC50 5.1 & 6.2 μM IC50 2.0 & 2.3 μM |
149 |
| Koshikamide A1 | T. swinhoei | Cytotoxic | IC50 1.7 μM | 150 |
| Koshikamide A2 | T. swinhoei | Cytotoxic | IC50 4.6 μM | 151 |
| Koshikamide B | T. swinhoei | Human colon tumor & P388 cytotoxicity | IC50 3.7& 0.2 μM | 152 |
| Koshikamide F | T. swinhoei | HIV entry inhibition | IC50 2.3 μM | 152 |
| Koshikamide H | T. swinhoei | HIV entry inhibition Cytotoxic |
IC50 5.5 μM IC50 10 μM |
152 |
| Nagahamide A | Theonella sp. | Antibacterial | 50 μg | 154 |
| Papuamide A | Theonella sp. | Anti-infective Cytotoxic HIV entry |
EC50 2.8 nM IC50 52.9 nM 710 nM |
155–156 |
| Papuamide B | Theonella sp. | Anti-infective HIV-entry inhibition |
EC50 2.8 nM 710 nM | 155–156 |
| Papuamides C–D | Theonella sp. | HIV entry | 28.4 & 14.2 μM | 156 |
| Papuamides E–F | Melophlus | Brine shrimp toxicity | LD50 66.5 & 77.4 μM | 157 |
| Theonellamide F | Theonella sp. | P388 & L1210 cytotoxicity | IC50 1.6 & 1.9 μM | 158 |
| Theonegramide | Theonella sp. | Antifungal | 10 μg/disk | 159 |
| Cupolamide A | T. cupola | Cytotoxic | IC50 7.2 μM | 160 |
| Theonellapeptolide Ib–Ie | T. swinhoei | Cytotoxic | IC50 1.2, 0.9, 1.7, & 0.9 μM | 163 |
| Theonellapeptolide IId–IIe | T. swinhoei | Cytotoxic | 9.4 μM | 152 |
| Theonellapeptolide IIIe | T. swinhoei | Cytotoxic | 4.8 μM | 166 |
| Congeners 1–2 | Theonella sp. | Cytotoxic | IC50 6.3 & 5.4 μM | 170 |
| Solomonamide A | T. swinhoei | Anti-inflammatory | 0.1 mg/kg | 171 |
| Cyclolithistide A | T. swinhoei | Antifungal | 20 μg/disk | 172 |
| Theopalauamide | T. swinhoei | Antifungal | 10 μg/disk | 174 |
| Isotheopalauamide | T. swinhoei | Antifungal | 50 μg/disk | 174 |
| Miraziridine A | Theonella | Cathepsin B inhibitor | IC50 2.1 μM | 152 |
| Perthamide B | Theonella | [125I]IL-1β inhibition | IC50 27.6 μM | 177 |
| Perthamide C–D | Theonella | Anti-inflammatory Edema reduction |
– 0.3 mg/kg |
178–179 |
| Perthamide E | Theonella | Anti-inflammatory | – | 178 |
| Perthamides H–I | Theonella | Edema reduction | 0.3 mg/kg | 179 |
| Perthamide K | Theonella | Edema reduction | 0.3 mg/kg | 179 |
| Mutremdamide A | Theonella | Edema inhibition | 0.3 mg/kg | 152 |
Table 2.
Peptides isolated from Marine Cyanobacteria
| Compound | Biological Source | Biological Activity | Inhibitory Concentration | Ref |
|---|---|---|---|---|
| Nodularin | Nodularia pumigena | Protein phosphatases 1 & 2A inhibitor | ED50 0.7 nM | 185 |
| Turnagainolide B | Bacillus sp. | Inositol 5-phosphatase stimulator | – | 190 |
| Solonamides A-B | Photobacterium halotolerans | Viral gene expression controller | – | 190 |
| Fijimycins A-C | Streptomyces | Antibacterial | – | 190 |
| Brunsvicamide A | Tychonema sp. | Antitubercular | IC50 64.2 μM | 191 |
| Brunsvicamides B-C | Tychonema sp. | Antitubercular | IC50 7.3 & 8.0 μM | 191 |
| Malyngamide 2 | L. sordida | Anti-inflammatory | – | 190, 192 |
| Malyngamide 3 | L. majuscula | HT-29 & MCF7 cytotoxicity | IC50 48 & 29 μM | 192 |
| Cocosamides A-B | L. majuscula | HT-29 cytotoxicity MCF7 cytotoxicity |
IC50 24 & 11 μM IC50 30 & 39 μM |
192 |
| Pitiprolamide | L. majuscula | Cytotoxic & antibacterial | – | 190 |
| Pitipeptolides A-F | L. majuscula | Anti-tubercular & cytotoxicity | – | 190, 193 |
| Lagunamides A-C | L. majuscula | Cytotoxic, anti-swarming & antimalarial | – | 190, 194 |
| Wewakamide A | L. semiplena | Brine shrimp toxicity | – | 190, 195 |
| Guineamide G | L. majuscula | Brine shrimp toxicity & cytotoxic | – | 190, 195 |
| Wewakazole | L. majuscula | Cytotoxic | IC50 10 μM | 197–198 |
| Wewakazole B | Moorea producens | MCF7 & H460 cytotoxicity | IC50 0.58 & 1.0 μM | 197–198 |
| Wewakpeptins A-B | L. majuscula | Cytotoxic | LC50 0.4 μM | 199 |
| Porpoisamides A-B | Lyngbya sp. | Cytotoxic | – | 190 |
| Bisebromoamide | Lyngbya sp. | Anti-proliferative | – | 200–202 |
| Norbisebromoamide | Lyngbya sp. | Anti-proliferative | – | 200–202 |
| Somocystinamide A | L. majuscula/Schizothrix sp. | Apoptosis | – | 34, 203 |
| C-phycocyanin | S. platensis, A. quadruplicatum & M. laminosus | Apoptosis | – | 34 |
| Desmethoxymajusc ulamide C | L. majuscula | Antitumor | IC50 20 nM | 204 |
| Epilyngbyastatin 1 | L. majuscula/Schizothr ix calcicola | Cytotoxic Cellular microfilament disrupter |
MIC 0.1 μM 2.0 & 0.2 μM |
205 |
| Majusculamide C | L. majuscula | Cytotoxic & antifungal | – | 34, 207–208 |
| Majusculamide D | L. majuscula | Cytotoxicity | 0.25 μM | 208 |
| Deoxymajusculamid e D | L. majuscula | Cytotoxicity | 0.3 μM | 208 |
| Lyngbyastatin 3 | L. majuscula | LoVo & KB cytotoxicity | IC50 400 & 32 nM | 209 |
| Apratoxins A-C | L. majuscula | LoVo cytotoxic KB cytotoxic |
IC50 0.36–10.8 nM IC50 0.52–21.3 nM |
210 |
| Apratoxin D | L. majuscula & L. sordida | Cytotoxic | IC50 2.6 nM | 204 |
| Apratoxin E | L. bouillonii | Cytotoxic | 21–72 nM | 204 |
| Dragomabin | L. majuscula/L. polychroa | Antimalarial Cytotoxic |
IC50 6.0 μM IC50 182.3 μM |
204 |
| Dragonamide A | L. majuscula/L. polychroa | Antimalarial Antileishmanial Cytotoxic |
IC50 7.7 μM IC50 6.5 μM IC50 67.8 μM |
204 |
| Dragonamide C | L. majuscula/L. polychroa | U2OS, IMR-32, & HT29 cytotoxicity | GI50 56, 49, 22 μM | 204 |
| Dragonamide D | L. majuscula/L. polychroa | U2OS, IMR-32, & HT29 cytotoxicity | GI50 59, 51, 32 μM | 204 |
| Dragonamide E | L. majuscula/L. polychroa | Antileishmanial | IC50 5.1 μM | 204 |
| Carmabin A | L. majuscula/L. polychroa | Antimalarial Cytotoxic |
IC50 4.3 μM IC50 9.8 μM |
204 |
| Herbamide B | L. majuscula | Antileishmanial | IC50 5.9 μM | 213 |
| Almiramides B-C | L. majuscula | Antileishmanial | IC50 2.4 & 1.9 μM | 204 |
| Grassystatins A-B | L. confervoides | Cathepsin D inhibitor Cathepsin E inhibitor TACE inhibitor |
IC50 7.3 & 26.5 nM IC50 354 & 886 pM IC50 1.2 & 2.2 μM |
204 |
| Grassystatin C | L. confervoides | TACE inhibitor | IC50 28.6 μM | 204 |
| Obyanamide | L. confervoides | KB & LoVo cytotoxicity | IC50 0.97 & 5.20 μM | 214 |
| Lobocyclamide A | L. confervoides | Antifungal | MIC 100 μg/disk | 215 |
| Lobocyclamide B | L. confervoides | Antifungal | MIC 30–100 μg/disk | 215 |
| Lobocyclamide C | L. confervoides | Antifungal | MIC 150 μg/disk | 215 |
| Hantupeptins A-C | L. majuscula | Brine shrimp toxicity MOLT-4 cytotoxicity MCF-7 cytotoxicity |
100–10 ppm IC50 32.0, 0.2, & 3.0 μM IC50 4.0, 0.5, & 1.0 μM |
204 |
| Trungapeptin A | L. majuscula | Brine shrimp toxicity Ichthyotoxicity | 10 ppm 6.25 ppm | 216 |
| Palmyramide A | L. majuscula | Sodium-channel blocker Cytotoxic |
IC50 17.2 μM IC50 39.7 μM |
196, 218 |
| Dudawalamide A | Lyngbya | Anti-parasitic Antimalarial Antileishmanial |
– IC50 2.7 μM IC50 25.9 μM |
204, 218 |
| Dudawalamide B | Lyngbya | Antimalarial Antileishmanial |
IC50 7.6 μM IC50 14.7 μM |
218 |
| Dudawalamide D | Lyngbya | Antimalarial Antileishmanial |
IC50 3.7 μM IC 2.6 μM 50 |
218 |
| Dudawalamide E | Lyngbya | Antimalarial Antileishmanial Antitrypanosomal |
IC50 7.7 μM IC50 2.6 μM IC50 7.3 μM |
218 |
| Grassypeptolide | L. confervoides | Cytotoxic | IC50 1.0–4.2 μM | 204 |
| Carriebowmide | L. majuscula/L. polychroa | Feeding deterrent | – | 204 |
| Hoiamide A | L. majuscula | Sodium channel agonist & cytotoxic | – | 204 |
| Tiglicamides A-C | L. confervoides | Serine protease inhibitors | – | 204 |
| Largamides A-C | L. confervoides | Serine protease inhibitors | – | 204 |
| Itralamide B | L. majuscula | Cytotoxic | IC50 6.0±1.0 μM | 204 |
| Lyngbyastatins 4–6 | L. confervoides | Serine protease inhibitors | – | 204 |
| Lyngbyastatin 7 | Lyngbya sp. | Serine protease inhibitors | – | 204 |
| Lyngbyastatins 8–10 | L. semiplena | Serine protease inhibitors | – | 204 |
| Kempopeptins A-B | Lyngbya sp. | Serine protease inhibitors | – | 204 |
| Lyngbyazothrins AB | Lyngbya sp. | Antimicrobial | – | 204, 220 |
| Lyngbyazothrins C-D | Lyngbya sp. | Antimicrobial | 25–200 μg/disk | 204, 220 |
| Pahayokolide A | Lyngbya sp. | Zebra fish toxicity Brine shrimp toxicity Anticancer |
LC50 2.2 μM 679.0 μM IC50 2.1–44.6 μM |
204 |
| Grassypeptolides D-E | Leptolyngbya | Neuro-2a mouse blastoma cytotoxic HeLa cervical carcinoma cytotoxic |
IC50 599 & 407 nM IC50 335 & 192 nM |
222 |
| Grassypeptolides F-G | L. majuscula | AP-1 inhibitor | IC50 5.2 & 6.0 μM | 223 |
| Ibuepidemethoxylyn gbyastatin 3 | Leptolyngbya | Cytotoxic | IC50 > 10 μM | 223 |
| Lyngbyabellin A | L. bouillonii | Cytotoxic | IC50 5.3 μM | 225–226 |
| Lyngbyabellin B | L. majuscula | Antifungal Brine shrimp toxicity |
100 μg/disk LD50 3.0 ppm |
227–228 |
| Lyngbyabellin C | L. bouillonii | Cytotoxic | IC50 2.1 μM | 225 |
| Lyngbyabellin D | L. bouillonii, L. majuscula | Cytotoxic | IC50 0.1–0.4 μM | 229 |
| Lyngbyabellin F | L. majuscula | Cytotoxic | IC50 0.1–0.4 μM | 229 |
| Lyngbyabellin H | L. majuscula | Cytotoxic | IC50 0.1–0.4 μM | 229 |
| Lyngbyabellin N | L. majuscula | Cytotoxic | IC50 40.9±3.3 nM | 230 |
| Alotamide A | L. bouillonii | Calcium influx activator | EC50 4.18 μM | 231 |
| Barbamide | L. majuscula | Molluscicidal | – | 232 |
| Jamaicamide A | L. majuscula | Cytotoxic Sodium channel blocker |
LC50 15 μM 5 μM |
233 |
| Jamaicamide B | L. majuscula | Cytotoxic Sodium channel blocker |
LC50 15 μM 5 μM |
233 |
| Jamaicamide C | L. majuscula | Cytotoxic Sodium channel blocker Brine shrimp toxicity |
LC50 15 μM 5 μM 10 ppm |
233 |
| Hectochlorin | L. majuscula | Antifungal CA46 & PtK2 cytotoxicity |
10–100 μg/disk IC50 20 & 300 nM |
234 |
| Apramide A | L. majuscula | Elastase activity | – | 235 |
| Aurilide | Dolabella auricularia | Apoptosis | – | 236 |
| Aurilides B-C | L. majuscula | Cytotoxic | LC50 0.01–0.13 μM | 236 |
| Antillatoxin | L. majuscula | Ichthyotoxic | LD50 99.3 nM | 238 |
| Barbaramide A | L. majuscula | Molluscicidal activity | LD100 21.6 μM | 239–240 |
| Kalkitoxin | L. majuscula | Ichthyotoxic Cell division inhibitor Brine shrimp toxicity Sodium channel blocker |
LC50 700 nM IC50 25 nM LC50 170 nM EC50 1 nM |
241 |
| Yanucamides A-B | L. majuscula/Schizothrix sp. | Brine shrimp toxicity | LD50 5 ppm | 243 |
| Pseudodysidenin | L. majuscula | Cytotoxic | IC50 > 1.8 μM | 244 |
| Ulongamides A-E | L. majuscula | LoVo & KB cytotoxic | IC50 5.0 & 1.0 μM | 245 |
| Isomalyngamides | L. majuscula | Crayfish toxicity | – | 246 |
| Malyngamide C | L. majuscula | Cytotoxic | IC50 5.2 μM | 247 |
| 8-epi-malyngamide C | L. majuscula | Cytotoxic | IC50 15.4 μM | 247 |
| Malyngamide F | L. majuscula | NO inhibition | IC50 5.4 μM | 250 |
| Malyngamide G | L. majuscula | Immunosuppressive | ED50 14.2 μM | 251 |
| Malyngamides H-I | L. majuscula | Reduction of nitrite production | – | 250–251 |
| Malyngamide I | L. majuscula | Brine shrimp toxicity Goldfish toxicity |
LD50 72.3 μM LD < 20.7 μM 50 |
252 |
| Malyngamide K | L. majuscula | Reduction of nitrite production | – | 251 |
| Malyngamide R | L. majuscula | Brine shrimp toxicity | LD50 18 ppm | 255 |
| Malyngamide Y | Moorea producens | Anticancer | – | 260 |
| Hermitamide A | L. majuscula | Brine shrimp toxicity Ichthyotoxicity Cytotoxic |
LD50 5 μM LD50 19 μM IC50 2.2 μM |
261 |
| Hermitamide B | L. majuscula | Brine shrimp toxicity Cytotoxic |
LD50 18 μM IC50 5.5 μM |
261 |
| Laxaphycin B | L. majuscula | Cytotoxic | IC50 1.1 μM | 262–264 |
| Tasiamide | Symploca sp. | LoVo & KB cytotoxic | IC50 4.2 & 0.6 μM | 265 |
| Tasiamide B | Symploca sp. | Cytotoxic | IC50 0.8 μM | 266 |
| Tasiamide F | Lyngbya sp. | Cathepsin D & E inhibitor | IC50 57 & 23 nM | 268 |
| Tasipeptins A-B | Symploca sp. | Cytotoxic | IC50 0.9 & 0.8 μM | 269 |
| Symplocamide A | Symploca sp. | Neuro-2a neuroblastoma & H-460 cytotoxicity | IC50 29 & 40 nM | 270 |
| Symplocin A | Symploca sp. | Cathepsin E inhibitor | IC50 300 pM | 271 |
| Veraguamides A-G | Symploca cf. hydnoides | Cytotoxic | – | 272 |
| Symplostatin 1 | Symploca hydnoides | Proapoptosis | – | 273 |
| Symplostatin 3 | S. hydnoides | KB & LoVo cytotoxicity | IC50 3.9 & 10.3 nM | 275 |
| Symplostatin 4 | Symploca sp. | Antimalarial Pathogen replication inhibitor |
IC50 36–100 nM EC50 0.7 μM |
276 |
| Malevamide D | S. hydnoides | Cytotoxic, antiproliferative & tubulin disruption | – | 273 |
| Belamide A | S. hydnoides | Antiproliferative & tubulin disruption | – | 273 |
| Largazole | Symploca sp. | MDA-MB-231, NMuMG, U2OS, & N1H3T3 cytotoxic | GI50 7.7, 122.0, 55.0, & 480.0 nM | 278 |
| Mitsoamide | Geitlerinema sp. | Cytotoxic | IC50 460 nM | 279 |
| Gallinamide A | Schizothrix sp. | Antimalarial | IC50 8.4 μM | 280 |
| Nostocyclamide | Nostoc sp. | Antialgal & anticyanobacterial Anabaena P-9 growth inhibitor |
– 0.1 μM |
281 |
| Tenuecyclamide A | Nostoc spongiaeforme | Sea-urchin embryo inhibitor | ED100 10.8 μM | 282 |
| Tenuecyclamides C-D | N. spongiaeforme | Sea-urchin embryo inhibitors | ED100 9.0 & 19.1 μM | 282 |
| Cryptophycin | Nostoc | Antifungal Antimicrotubule |
– | 283 |
| Microcystins | Nostoc | Protein phosphatase 1 & 2A inhibitors Tumor promoters |
– | 284 |
| Raocyclamide A | Oscillatoria raoi | Cytotoxic | – | 285 |
| Venturamides A-B | Oscillatoria sp. | Antimalarial Cytotoxic |
IC50 8.2 & 5.6 μM IC50 86 & 56 μM |
286 |
| Anabaenopeptins A-B | Anabaena flos-aquae | Rat aortic relaxations | 11.8 – 477.3 μM | 287 |
| Anabaenopeptins G-H | O. agardhii | Carboxypeptidase A-inhibitors | IC50 2.2 nM & 3.7 μM | 290 |
| Anabaenopeptins I-J | Aphanizomenon flos-aquae | Carboxypeptidase-A inhibitors | – | 205 |
| Pompanopeptin A | L. confervoides | Trypsin inhibitor | IC50 2.4 μM | 205 |
| (+)-floridamide | Moorea producens | Anticancer | – | 260 |
| Coibamide A | Leptolyngbya sp. | Cytotoxic MDA-MB-231, HL-60, SNB-75 & LOX IMVI cytotoxicity |
LC50 < 23 nM GI50 2.8, 7.4, 7.6, & 7.4 nM |
291 |
| Hormothamnin A | Hormothamnion enteromorphoides | Antimicrobial & cytotoxic | – | 292 |
| Scytonemin | Stigonema sp. | GST-polo-like kinase 1 inhibitor | IC50 2.3 μM ± 3.6 | 294 |
| Arenamides A-B | Salinispora arenicola | TNF-induced activation blockers PGE2 inhibitors NO production inhibitors Anticancer |
IC50 3.7 & 1.7 μM | 295 |
| Caldoramide | Caldora penicillata | Cytotoxic | IC50 3.9 – 5.2 μM | 296 |
| Viequeamide A | Rivularia sp. | Anticancer | IC50 60 ± 10 nM | 297 |
| Companeramides A-B | Leptolyngbya sp. | Anti-plasmodial | – | 298 |
| Aeruginosins | Nodularia spumigena | Serine protease inhibitors | – | 301 |
| Kailuin A | BH-107 | A-549, MCF-7, & HT-29 cytotoxicity | GI50 4.3 μM | 302 |
| Kailuin B | BH-107 | A-549, MCF-7, & HT-29 cytotoxicity HCT-116 cytotoxicity |
GI50 2.8, 2.8, & 4.1 μM IC50 22 μM |
302–303 |
| Kailuin C | BH-107 | A-549, MCF-7, & HT-29 cytotoxicity HCT-116 cytotoxicity |
GI50 4.1, 5.5, & 4.1 μM IC50 50 μM |
302–303 |
| Kailuin D | BH-107 | A-549, MCF-7, & HT-29 cytotoxicity HCT-116 cytotoxicity |
GI50 2.7, 3.9, & 2.7 μM IC50 28 μM |
302–303 |
| Kailuin E | Photobacterium halotolerans | HCT-116 cytotoxicity | IC50 18 μM | 303 |
| Kailuins G & H | Photobacterium halotolerans | HCT-116 cytotoxicity | IC50 32 & 17 μM | 303 |
| Ngercheumicins F-I | Photobacterium sp. | Inhibition of rnaIII transcription | – | 305 |
| Unnarmicin A | Photobacterium sp. | Rhodamine 6G efflux inhibition CaCdr1p ATPase inhibition |
IC50 3.6 μM IC50 0.5 μM |
307 |
| Unnarmicin C | Photobacterium sp. | Rhodamine 6G efflux inhibition CaCdr1p ATPase inhibition | IC50 5.6 μM IC50 0.7 μM | 307 |
| Ariakemicins A-B | Rapidithrix sp. | Cytotoxic | IC50 25.4 & 42.4 μM | 308 |
| Mollemycin A | Streptomyces sp. | Antibacterial Antimalarial |
IC50 10–50 nM IC50 7–9 nM |
309 |
| Thiocoraline | L-13-ACM2-092 | Cytotoxic | IC50 1.7 nM | 310 |
| Cyclomarin A | Streptomyces sp. | Anti-inflammatory | 50 μg/ear | 311 |
| Surugamides A-E | Streptomyces sp. | Cathepsin-B inhibitors | IC50 21, 27, 36, 18, & 16 μM | 313 |
| Champacyclin | S. champavatii | Antimicrobial | 25 μM | 315 |
| Tumescenamide A | S. tumescens | Alzheimer’s disease | – | 316 |
| Tumescenamide C | S. tumescens | Antimicrobial | 3.0 μg/disk | 317 |
| Streptocidins C-D | Streptomyces sp. Tu 6071 | Antimicrobial | MIC 0.8 – 2.4 μM | 320 |
| Salinamide A | Streptomyces sp. CNB-091 | Edema Antibiotic |
50 μg/ear MIC 3.9 μM |
321 |
| Salinamide B | Streptomyces sp. CNB-091 | Edema Antibiotic |
50 μg/ear MIC 1.9 μM |
321 |
| Salinamide F | Streptomyces sp. | Antibacterial RNAP inhibiton |
MIC 0.2 – 96.3μM IC50 2 – 4 μM |
322 |
| Tauramamide | Brevibacillus laterosporus PNG276 | Antibacterial | MIC 0.12 μM | 323 |
| Bogorols A-E | B. laterosporus | MRSA inhibition VRE inhibition |
MIC 0.6 – 5.1 μM MIC 5.6 – 48.2 μM |
325 |
| Loloatins A-B | MK-PNG-276A | Antibiotics | – | 326 |
| Loloatin C-D | MK-PNG-276A | Antibiotic & antibacterial | – | 326 |
| Marthiapeptide A | Marinactinospora thermotolerans | Antibacterial Anticancer |
MIC 3 – 12 μM IC50 0.4–0.5 μM |
327 |
| Anguibactin | Vibrio anguillarum | Cytotoxic | IC50 15 μM | 337–338 |
Table 3.
Peptides isolated from Marine Fungi
| Compound | Biological Source | Biological Activity | Inhibitory Concentration | Ref |
|---|---|---|---|---|
| Fellutamides A-B | Penicillium fellutanum | Cytotoxic | – | 348 |
| Fellutamides C-D | Metulocladosporiella | Fungal proteasome inhibitors Antitumor |
– | 349 |
| Fellutamide F | Aspergillus versicolor | Cytotoxic | – | 190 |
| Asperterrestide A | Aspergillus terreus | MOLT4 & U937 cytotoxicity H1N1 & H3N2 antiviral |
IC50 6.2 & 6.4 μM IC50 15.0 & 8.1 μM |
350 |
| Sansalvamide A | Fusarium | Anticancer, Topoisomerase I inhibition SK-MEL-2 & COLO 205 colon cancer cells cytotoxicity |
– IC50 10.1 & 5.9 μM |
34, 351 |
| Scopularides A-B | Scopulariopsis brevicaulis | Antitumor | 15.0 – 15.5 μM | 352–353 |
| Exumolides A-B | Scytalidium | Anti-microalgal | 28 μM | 354 |
| Cis-bis(methylthio)silvatin | Penicillium bilaii | Cytotoxic | LD99 0.15 μM | 355 |
| Asperpeptide A | Aspergillus sp. | Antibacterial | MIC 12.5 μM | 357 |
| Emericellamide A | Emericella sp. & Salinispora arenicola | Antimicrobial Cytotoxic |
MIC 3.8 μM IC50 23 μM |
358–359 |
| Emericellamide B | Emericella sp. & S. arenicola | Cytotoxic | IC50 6.0 μM | 358–359 |
| Guangomides A-B | Unidentifiable fungus | Antibacterial | MIC 166 μM | 360 |
| Azonazine | Aspergillus insulicola | Anti-inflammatory Nitrite production |
IC50 8.4 μM IC50 13.7 μM |
361 |
| Endolides A-B | Stachylidium sp. | Affinity for vasopressin receptor 1A & serotonin receptor 5HT2B | – | 362 |
| Simplicilliumtide A | Simplicillium obclavatum | Cytotoxic | IC50 64.7 μM | 364 |
| Simplicilliumtide D | S. obclavatum | Antifouling | – | 364 |
| Simplicilliumtide E | S. obclavatum | Cytotoxic | IC50 39.4 μM | 364 |
| Simplicilliumtide G | S. obclavatum | Cytotoxic | IC50 100 μM | 364 |
| Simplicilliumtide H | S. obclavatum | Cytotoxic | IC50 73.5 μM | 364 |
| Halolitoralins A-C | Halobacillus litoralis | Antifungal | – | 365 |
| Sclerotide A | Aspergillus sclerotiorum | Antifungal | MIC 7.0 μM | 366 |
| Sclerotide B | A. sclerotiorum | Antifungal Cytotoxic Antibacterial |
MIC 3.5 μM IC50 56.1 μM MIC 35.3 μM |
366 |
| RHM1 | Acremonium | Cytotoxic | – | 367 |
| RHM2 | Acremonium | Cytotoxic | – | 367 |
| Efrapeptin G | Acremonium | Cytotoxic Mitochondrial ATPase inhibitor |
– | 367–368 |
| Cordyheptapeptide A | Cordyceps sp. | Antimalarial Cytotoxic |
IC50 3.8 μM IC50 0.2–14.0 μM |
370 |
| Cordyheptapeptide B | Cordyceps sp. | Cytotoxic | IC50 0.6–3.1 μM | 370 |
| Cordyheptapeptides C-E | A. persicinum | Cytotoxic | IC50 2.5–12.1 μM | 369 |
Table 4.
Peptides from Marine Ascidians
| Compound | Biological Source | Biological Activity | Inhibitory Concentration | Ref |
|---|---|---|---|---|
| Cycloxazoline | Lissoclinum bistratum | Cytotoxic | IC50 0.9 μM | 34, 372 |
| Diazonamide A | Diazona angulata | Cytotoxic | IC50 < 19.6 nM | 34, 373–374 |
| Diazonamides C-E | Diazona sp. | Cytotoxic | – | 376 |
| Vitilevuamide | Didemnum cuculiferum & Polysyncranton lithostrotum | Tubulin polymerization inhibition Lymphocytic leukemia inhibition Colchicine binding stabilization Cytotoxic |
IC50 2 μM IC50 30 μg/kg IC50 5.6 μM LC50 6–311 nM |
34, 377 |
| Bistratamides A-B | Lissoclinum bistratum | Cytotoxic | IC50 87.6 & > 175.8 μM | 380 |
| Bistratamide D | Lissoclinum sp. | Depressant | 65 μg | 381 |
| Bistratamides E-J | L. bistratum | Cytotoxic | IC50 14.4, 51.4, 9.5, 3.1, 16.4, & 1.8 μM | 382 |
| Lissoclinamide 7 | L. patella | Anti-neoplastic | – | 34, 378–379 |
| Patellamides | L. patella | Cytotoxic | – | 34, 378–379 |
| Didemnin A | Trididemnum solidum | Cytotoxic | IC50 0.03 μM | 383–384 |
| Didemnin B | T. solidum | Cytotoxic Apoptosis |
IC50 0.5 nM – |
383–385 |
| Didemnin M | T. solidum | Cytotoxic Immunosuppressive |
IC50 1.5 nM – |
383–384 |
| Didemnin N | T. solidum | Cytotoxic | IC50 46.1 nM | 383–384 |
| Didemnins X-Y | T. solidum | Cytotoxic | IC50 1.2 & 1.1 nM | 383–384 |
| Epididemnin A1 | T. solidum | Cytotoxic | IC50 2.1 μM | 383–384 |
| Acyclodidemnin A | T. solidum | Cytotoxic | IC50 0.2 μM | 383–384 |
| Styelin D | Styela clava | Cytotoxic & hemolytic | – | 34, 387 |
| Eusynstyelamide | Eusynstyela misakiensis | Cytotoxic | IC50 126.6 μM | 34, 388 |
| Mollamide | Didemnum molle | A549 & HT29 cytotoxicity P388 cytotoxicity RNA synthesis inhibitor |
IC50 3.1 μM IC50 1.2 μM IC50 1.2 μM |
34, 390–391 |
| Mollamide B | D. molle & Pleurobranchus forskalii | Anti-malarial against W2 and D6 of P. falciparum Antileishmanial HIV-1 inhibition |
IC50 3.0 & 2.9 μM IC90 50.2 μM & IC50 25.8 μM EC50 48.7 μM |
34, 391 |
| Keenamide A | D. molle & P. forskalii | MEL-20, P-388 & A-549 cytotoxicity HT-29 cytotoxicity |
IC50 4.0 μM IC50 8.0 μM |
392 |
| Cycloforskamide | P. forskalii | Cytotoxic | IC50 5.8 μM | 393 |
| Virenamide A | Diplosoma virens | CV1, A549, & HT29 cytotoxicity P388 cytotoxicity Topoisomerase-II inhibition |
IC50 18.6 μM IC50 4.6 μM – |
34, 394 |
| Virenamides B-C | D. virens | Cytotoxic | IC50 4.8 μM | 34, 394 |
| Cyclodidemnamide | Didemnum molle | Cytotoxic | ED50 23.1 μM | 395 |
| Comoramides A-B | D. molle | Cytotoxic | IC50 7.5 – 14.9 μM | 396 |
| Mayotamides A-B | D. molle | Cytotoxic | IC50 7.4 – 14.7 μM | 396 |
| Didmolamides A-B | D. molle | Cytotoxic | IC50 18.0 – 37.1 μM | 397 |
| Prepatellamide A | Lissoclinum patella & D. molle | Cytotoxic | IC50 6.6 μM | 398 |
| Patellamides A-C | L. patella & D. molle | Cytotoxic | IC50 6.4 – 6.6 μM | 398 |
| Tamandarin A | Didemnidae Family | Reticulocyte cell-lysate protein biosynthesis inhibition | IC50 1.3 μM | 399 |
| Patellin 6 | L. patella | Cytotoxic Topoisomerase II inhibition |
IC50 2.1 μM IC50 2.6 μM |
400 |
| Kulolide-1 | Philinopsis speciosa | Cytotoxic | 50 μM | 401 |
| Kulokainalide-1 | P. speciosa | Cytotoxic | 5 μM | 401 |
| Ulithiacyclamide | L. patella | Cytotoxic Macrophage Scavenger Receptor (MSR) inhibitor |
IC50 0.04 μM IC50 98 nM |
402–404 |
| Ulithiacyclamide B | L. patella | Cytotoxic | IC50 21.3 nM | 403–404 |
| Ulicyclamide | L. patella | Macrophage Scavenger Receptor (MSR) inhibitor | IC50 51 μM | 402–403 |
Table 5.
Peptides from Other Marine Sources
| Compound | Biological Source | Source | Biological Activity | Inhibitory Concentration | Ref |
|---|---|---|---|---|---|
| Malyngamide M | Gracilaria coronopifolia | Red alga | Cytotoxic | IC50 > 20 μM | 253 |
| Malyngamide N | G. coronopifolia | Red alga | Cytotoxic | IC50 10.5 μM | 253 |
| Malyngamide O | Stylocheilus longicauda | Sea hare | Cytotoxic | IC50 4.2 μM | 254 |
| Malyngamide S | Bursatella leachii | Sea hare | Antiproliferative Cytotoxic Antitubercular |
IC50 ~ 6–8 μM IC50 29 μM IC50 12.9 μM |
256 |
| Malyngamide X | B. leachii | Sea hare | Antimalarial & antitubercular | – | 258–259 |
| Nobilamide B | Chicoreus nobilis | Mollusk/cyanobacteria | Anti-inflammatory & pain mediator | – | 190, 406 |
| A-3302-B | Unknown | – | Anti-inflammatory & pain mediator | – | 190, 406 |
| Aplidine | Alpidium albicans | Tunicate | Anticancer | – | 34 |
| Dolastatin 3 | Dolabella auricularia/Lyngbya majuscula | Mollusk/cyanobacteria | Cytotoxic | GI50 < 1 μM | 393 |
| Dolastatin 10 | D. auricularia | Mollusk | Growth inhibition of L1210 murine leukemia cells, inhibition of microtubule assembly, & tubulin-dependent GTP hydrolysis | – | 34 |
| Dolastatin 11 | D. auricularia | Mollusk | Anticancer | GI50 ~ 0.07 μM | 34, 393 |
| Dolastatin 12 | D. auricularia | Mollusk | Anticancer | GI50 ~1 to ~30 nM | 34, 393 |
| Epidolastatin 12 | L. majuscula/S. calcicola | Cyanobacteria | Cytotoxic Microfilament disruption |
MIC < 0.05 μM 0.2 – 2.1 μM |
206 |
| Dolastatin 13 | D. auricularia | Mollusk | Anticancer | GI50 14 nM | 34, 393 |
| Dolastatin 14 | D. auricularia | Mollusk | Anticancer Antineoplastic |
GI50 20 nM – |
34, 393, 408 |
| Dolastatin 15 | D. auricularia | Mollusk | Antineoplastic | – | 34, 408 |
| Dolastatin 16 | D. auricularia/L. majuscula/Symploca | Mollusk/Cyanobacteria | Antileukemic & anticancer | GI50 ~ 0.3 μM | 393 |
| Homodolastatin 16 | L. majuscula | Cyanobacteria | WHCO1 & WHCO6 cytotoxicity ME180 cytotoxicity |
IC50 4.8 & 11.3 μM IC50 9.3 μM |
409 |
| Dolastatin 17 | D. auricularia | Mollusk | Anticancer | – | 393 |
| Dolastatin 18 | D. auricularia | Mollusk | Antileukemic | – | 393 |
| Dolastatin C | D. auricularia | Mollusk | Cytotoxic | IC50 26.4 μM | 410 |
| Dolastatin D | D. auricularia | Mollusk | Anticancer | GI50 ~ 4.0 μM | 393 |
| Dolastatin E | D. auricularia | Mollusk | Cytotoxic | IC50 44.8 – 81.5 μM | 411 |
| Dolastatin G | D. auricularia | Mollusk | Anticancer | GI50 ~ 1.0 μM | 393 |
| Nordolastatin G | D. auricularia | Mollusk | Anticancer | GI50 ~ 5.0 μM | 393 |
| Lyngbyastatin 2 | L. majuscula | Cyanobacteria | Cytotoxic | LD100 3 mg/kg | 412 |
| Norlyngbyastati n 2 | L. majuscula | Cyanobacteria | Cytotoxic | – | 412 |
| Isodolastatin H | D. auricularia | Mollusk | Antitumor | – | 393 |
| Dolastatin H | D. auricularia | Mollusk | Anticancer | GI50 2 nM | 393 |
| Dolastatin I | D. auricularia | Mollusk | Cytotoxic | IC50 23.2 μM | 413 |
| Dolabellin | D. auricularia | Mollusk | Cytotoxic | IC50 9.97 μM | 414 |
| Doliculide | D. auricularia | Mollusk | Cytotoxic | IC50 1.6 nM | 415 |
| Kulokekahilide-1 | Philinopsis speciosa | Mollusk | Cytotoxic | IC50 2.2 μM | 416 |
| Kulokekahilide-2 | P. speciosa | Mollusk | A-10, P388, MDA-MB-435, & SK-OV-3 cytotoxicity | IC50 59.1, 4.2, 14.6, & 7.5 nM | 417 |
| Aurilide | Dolabella auricularia | Japanese sea hare | Cytotoxic | IC50 13.2 nM | 237, 418 |
| Microcolin A | Okeania erythroflocculosa | Coral reef | Apoptosis in murine thymocytes | 10–100 nM | 420–421, 424 |
| Microcolin B | O. erythroflocculosa | Coral reef | Immunosuppressant | – | 420–421 |
| Tumonoic acid F | L. majuscula/S. calcicola | Cyanobacteria | Bioluminescence inhibition | IC50 62 μM | 425 |
| Tumonoic acid I | Blennothrix cantharidosmum | Cyanobacteria | Antimalarial | IC50 2.0 μM | 425 |
| Janadolide | Okeania sp. | Cyanobacteria | Anti-trypanosomal | IC50 47 nM | 426 |
| Onchidin | Onchidium sp. | Pulmonate mollusk | Cytotoxic | – | 427 |
| Onchidin B | Onchidium sp. | Pulmonate mollusk | Kb cells inhibition | IC97 8.7 μM | 427–428 |
| Norcardiamides A-B | Nocardiopsis | Actinomycete | Antimicrobial | – | 429 |
| Kahalalide A | Elysia rufescens | Sacoglossan mollusk | Antimalarial activity Antimicrobial |
– 13.98 μM |
430, 432 |
| Kahalalide E | E. rufescens | Sacoglossan mollusk | HSV-II inhibition | – | 430 |
| Kahalalide F | E. rufescens | Sacoglossan mollusk | Antitumor activity Cytotoxic HIV OI inhibitor Antifungal HSV-II inhibition Immunosuppressive Antileishmanial |
IC50 0.07 – 0.28 μM IC50 0.22 μM – IC50 1.5 – 3.2 μM IC50 0.3 μM IC50 2.03 μM – |
430–432 |
| Kahalalides R1 | E. rufescens | Sacoglossan mollusk | MCF-7 cytotoxicity L1578 cytotoxicity Antifungal |
IC50 0.14 μM IC50 4.3 nM – |
432 |
| Trans,trans-ceratospongami de | Ceratodictyon spongiosum | Red alga/sponge | Anti-inflammatory Inhibition of human-sPLA2 promoter | ED50 32 nM – |
433 |
| Mebamamide B | Derbesia marina | Green alga | HL60 differentiation | 100 μM | 434 |
| Galaxamide | Galaxaura filamentosa | Alga | U87 & MCF-7 cytotoxicity | IC50 17.8 & 25.1 μM | 436 |
| Conantokin G | Conus geographus | Cone snail | NMDA antagonist | IC50 171 nM | 437 |
| Conantokin T | C. tulipa | Cone snail | NMDA antagonist | – | 438 |
| Conantokin GV | C. geographus | Cone snail | Sleep-inducer | 4–30 pmol/g | 437, 439 |
| Actinoporins | Actiniaria | Sea-anemone | Pore forming | – | 440 |
| Equinatoxin II | Actinia equine | Sea anemone | Lethal | LD50 35 μg/kg | 440 |
| SECMA 1 | Ulva sp. | Alga | Human foreskin fibroblast inhibitor | 5.3 μM | 441–442 |
| Arasin-1 | Hyas araneus | Spider crab | Antibacterial | MIC 0.8 μM | 443 |
| Arenicins 1-2 | Arenicola marina | Polychaeta | Antimicrobial | 9.1 μM | 444 |
Table 6.
| Clinical Status | Compound Name | Chemical Class | Source | Disease area |
|---|---|---|---|---|
| Approved | Ziconotide | Peptode | Cone snail | Pain |
| Brentuximab vedotin [SGN 35] | ADC [MMAE] | Mollusk/cyanobacterium | Hodgkin lymphoma & systemic anaplastic large cell lymphoma | |
| Phase III | Plitidepsin/aplidine | Depsipeptide | Tunicate | Relapsed/refractory multiple myeloma |
| Phase II | Soblidotin [TZT-1027] | Dolastatin 10 derivative | Mollusk/cyanobacterium | Sarcoma & lung cancer |
| Xen-2174 | chi-conopeptide | Cone snail | Pain | |
| Elisidepsin; PM02734; erlotinib | Cyclic depsipeptide | Mollusk | Advanced malignant solid tumors | |
| Tasidotin/synthadotin [ILX-651] | Dolastatin 15 derivative | Mollusk/cyanobacterium | Melanoma, non-small cell lung carcinoma, & hormone refractory prostate cancer | |
| ABT-414 | ADC [MMAF] | Mollusk/cyanobacterium | Glioblastoma, gliosarcoma, & glioblastoma multiforme | |
| Glembatumumab vedotin | ADC [MMAE] | Mollusk/cyanobacterium | Squamous cell carcinoma of lung, recurrent osteosarcoma, recurrent uveal melanoma, stage IV uveal melanoma, melanoma, metastatic gpNMB over-expressing triple negative breast cancer, breast cancer, & unresectable stage III or stage IV melanoma | |
| AGS-16C3F | ADC [MMAF] | Mollusk/cyanobacterium | Metastatic renal cell carcinoma | |
| PSMA-ADC | ADC [MMAE] | Mollusk/cyanobacterium | Prostate cancer, glioblastoma multiforme, & gliosarcoma | |
| Phase I/II | Pinatuzumab vedotin [DCDT-2980S] | ADC [MMAE] | Mollusk/cyanobacterium | Non-Hodgkin’s lymphoma, chronic lymphocytic leukemia, lymphoma, follicular, & diffuse large B-cell lymphoma |
| Polatuzumab vedotin [DCDS-4501A] | ADC [MMAE] | Mollusk/cyanobacterium | Non-Hodgkin’s lymphoma, chronic lymphocytic leukemia, lymphoma, follicular, & diffuse large B-cell lymphoma | |
| Tisotumab vedotin [HuMax-TF-ADC or HuMab-TF-011-vcMMAE] | ADC [MMAE] | Mollusk/cyanobacterium | Ovary cancer, cervix cancer, endometrium cancer, bladder cancer, prostate cancer, esophagus cancer, lung cancer, & squamous cell carcinoma of the head and neck | |
| Lifastuzumab vedotin [DNIB0600A; RG-7599] | ADC [MMAE] | Mollusk/cyanobacterium | Ovarian cancer, epithelial tumors, malignant, fallopian tube cancer, peritoneal neoplasms, & non-squamous non-small cell lung cancer | |
| CDX-014 | ADC [MMAE] | Mollusk/cyanobacterium | Renal cell carcinoma, clear-cell renal cell carcinoma, papillary renal cell carcinoma, kidney diseases, kidney neoplasms, & metastatic renal cell carcinoma | |
| Phase I | Leconotide [CNSB004; AM336] | Peptode | Cone snail | Calcium channel blocker |
| Taltobulin [HTI-286; SPA-110] | Hemiasterlin analog | Sponge | Cancer | |
| Enfortumab vedotin [ASG-22CE; AGS-22M6E] | ADC [MMAE] | Mollusk/cyanobacterium | Metastatic urothelial cancer, other malignant solid tumors, tumors, medical oncology, & neoplasms | |
| Vorsetuzumab Mafdotin [SGN-75] | ADC [MMAF] | Mollusk/cyanobacterium | Non-Hodgkin’s lymphoma & renal cell carcinoma | |
| Telisotuzumab vedotin [ABBV-399] | ADC [MMAE] | Mollusk/cyanobacterium | Advanced solid tumor & cancer | |
| GSK2857916 | ADC [MMAF] | Mollusk/cyanobacterium | Multiple myeloma | |
| ASG-15ME; AGS15E | ADC [MMAE] | Mollusk/cyanobacterium | Metastatic urothelial cancer | |
| AGS-67E | ADC [MMAE] | Mollusk/cyanobacterium | Relapsed/refractory lymphoid malignancy, acute myeloid leukemia | |
| MLN-0264 | ADC [MMAE] | Mollusk/cyanobacterium | Gastrointestinal malignancies | |
| SGN-19A; SGN-CD19A | ADC [MMAF] | Mollusk/cyanobacterium | Burkitt’s lymphoma & follicular lymphoma | |
| SGN-LIV1A | ADC [MMAE] | Mollusk/cyanobacterium | Breast cancer | |
| BAY79-4620 | ADC [MMAE] | Mollusk/cyanobacterium | Neoplasms | |
| DMUC-5754A | ADC [MMAE] | Mollusk/cyanobacterium | Ovarian & pancreatic cancers | |
| DMOT4039A; RG-7600 | ADC [MMAE] | Mollusk/cyanobacterium | Ovarian cancer | |
| DEDN6526A; RG-7636 | ADC [MMAE] | Mollusk/cyanobacterium | Malignant melanoma | |
| Vandortuzumab vedotin; DSTP3086S; RG7450; MSTP2109A | ADC [MMAE] | Mollusk/cyanobacterium | Prostate cancer | |
| DFRF4539A; RG-7598 | ADC [MMAE] | Mollusk/cyanobacterium | Multiple myeloma | |
| Preclinical Trials | HuMax-CD74 | ADC [MMAE/MMAF] | Mollusk/cyanobacterium | Solid tumors & hematological malignancies |
| Discontinued | Cemadotin | Dolastatin 15 analog | Mollusk/cyanobacterium | Cancer |
| Kahalalide F | Depsipeptide | Alga | Cancer | |
| PF-06263507; A1-mcMMAF | ADC [MMAF] | Mollusk/cyanobacterium | Neoplasms, carcinoma, non-small cell lung, breast neoplasms & ovarian neoplasms |
Highlights.
Peptides from marine sources
Challenges involved with peptide isolation
Biosynthesis of peptides from marine cyanobacteria, sponges, fungi, and ascidians
Therapeutic peptides approved by FDA or in clinical trials
Acknowledgments
Mark T. Hamann is supported in part by an NIH NCCIH grant 1R01AT007318, KraftHeinz, Biosortia, An Endowment by Charles and Carol Cooper and SmartState Program and by the Drug Discovery and Biomedical Sciences, Medical University of South Carolina. The authors would like to acknowledge Noer Kasanah from the Department of Fisheries, Universitas Gadjah Mada, Indonesia for her assistance with the preparation of the manuscript and Xiaojuan Wang from the Medical University of South Carolina, for her help in revising the chemical structures of this review. The authors especially wish to thank Drs. Prabhakar Reddy Polepally and Francisco León from the University of Mississippi for their assistance in carefully reviewing the manuscript.
Footnotes
Notes
The authors declare no competing financial interest.
References
- 1.Aneiros A, Garateix A. Bioactive peptides from marine sources: Pharmacological properties and isolation procedures. J Chromatogr B: Anal Technol Biomed Life Sci. 2004;803:41–53. doi: 10.1016/j.jchromb.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 2.Miijanich GP. Venom peptides as human pharmaceuticals. Sci Med. 1997;4:6–15. [Google Scholar]
- 3.Cragg GM, Newman DJ, Weiss RB. Coral reefs, forests, and thermal vents: The worldwide exploration of nature for novel antitumor agents. Semin Oncol. 1997;24:156–163. [PubMed] [Google Scholar]
- 4.Matsunaga S, Fusetani N. Nonribosomal peptides from marine sponges. Curr Org Chem. 2003;7:945–966. [Google Scholar]
- 5.Matsunaga S, Fusetani N, Konosu S. Bioactive marine metabolites VII. Structures of discodermins B, C, and D, antimicrobial peptides from the marine sponge Discodermia kiiensis. Tetrahedron Lett. 1985;26:855–856. doi: 10.1021/np50038a006. [DOI] [PubMed] [Google Scholar]
- 6.Matsunaga S, Fusetani N, Konosu S. Bioactive marine metabolites VI. Structure elucidation of discodermin A, an antimicrobial peptide from the marine sponge Discodermia kiiensis. Tetrahedron Lett. 1984;25:5165–5168. doi: 10.1021/np50038a006. [DOI] [PubMed] [Google Scholar]
- 7.Ryu G, Matsunaga S, Fusetani N. Discodermin E, a cytotoxic and antimicrobial tetradecapeptide, from the marine sponge Discodermia kiiensis. Tetrahedron Lett. 1994;35:8251–8254. [Google Scholar]
- 8.Ryu G, Matsunaga S, Fusetani N. Discodermins F-H, cytotoxic and antimicrobial tetradecapeptides from the marine sponge Discodermia kiiensis: Structure revision of discodermins A-D. Tetrahedron. 1994;50:13409–13416. [Google Scholar]
- 9.Ishida K, Murakami M. Kasumigamide, an antialgal peptide from the cyanobacterium Microcystis aeruginosa. J Org Chem. 2000;65:5898–5900. doi: 10.1021/jo991918f. [DOI] [PubMed] [Google Scholar]
- 10.Nakashima Y, Egami Y, Kimura M, Wakimoto T, Abe I. Metagenomic analysis of the sponge Discodermia reveals the production of the cyanobacterial natural product kasumigamide by ‘Entotheonella’. PLoS One. 2016;11:e0164468. doi: 10.1371/journal.pone.0164468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt EW, Obraztsova AY, Davidson SK, Faulkner DJ, Haygood MG. Identification of the antifungal peptide-containing symbiont of the marine sponge Theonella swinhoei as a novel δ-proteobacterium,“Candidatus Entotheonella palauensis”. Mar Biol. 2000;136:969–977. [Google Scholar]
- 12.Brück WM, Sennett SH, Pomponi SA, Willenz P, McCarthy PJ. Identification of the bacterial symbiont Entotheonella sp. in the mesohyl of the marine sponge Discodermia sp. The ISME Journal. 2008;2:335–339. doi: 10.1038/ismej.2007.91. [DOI] [PubMed] [Google Scholar]
- 13.Flórez LV, Biedermann PHW, Engl T, Kaltenpoth M. Defensive symbioses of animals with prokaryotic and eukaryotic microorganisms. Nat Prod Rep. 2015;32:904–936. doi: 10.1039/c5np00010f. [DOI] [PubMed] [Google Scholar]
- 14.Pettit GR, Cichacz Z, Barkoczy J, Dorsaz AC, Herald DL, Williams MD, Doubek DL, Schmidt JM, Tackett LP, Brune DC, Cerny RL, Hooper JNA, Bakus GJ. Isolation and structure of the marine sponge cell growth inhibitory cyclic peptide phakellistatin 1. J Nat Prod. 1993;56:260–267. doi: 10.1021/np50092a011. [DOI] [PubMed] [Google Scholar]
- 15.Pettit GR, Rhodes MR, Tan R. Antineoplastic Agents. 400. Synthesis of the Indian Ocean Marine Sponge Cyclic Heptapeptide Phakellistatin 2. J Nat Prod. 1999;62:409–414. doi: 10.1021/np980168m. [DOI] [PubMed] [Google Scholar]
- 16.Pettit GR, Tan R. Isolation and Structure of Phakellistatin 14 from the Western Pacific Marine Sponge Phakellia sp. J Nat Prod. 2005;68:60–63. doi: 10.1021/np040092w. [DOI] [PubMed] [Google Scholar]
- 17.Pettit GR, Tan R, Herald DL, Williams MD, Cerny RL. Antineoplastic agents. 277. Isolation and structure of phakellistatin 3 and isophakellistatin 3 from a Republic of Comoros marine sponge. J Org Chem. 1994;59:1593–1595. [Google Scholar]
- 18.Pettit GR, Xu JP, Cichacz Z, Schmidt JM, Dorsaz AC, Boyd MR, Cerny RL. Isolation and structure of the human cancer cell growth inhibitory phakellistatin 4 from the Western Pacific sponge Phakellia costata. Heterocycles. 1995;40:501–506. [Google Scholar]
- 19.Mechnich O, Kessler H. What are the structures of phakellistatin 2 and phakellistatin 4? Lett Pept Sci. 1997;4:21–28. [Google Scholar]
- 20.Pettit GR, Xu JP, Cichacz ZA, Williams MD, Dorsaz AC, Brune DC, Boyd MR, Cerny RL. Antineoplastic agents 315. Isolation and structure of the marine sponge cancer cell growth inhibitor phakellistatin 5. Bioorg Med Chem Lett. 1994;4:2091–2096. [Google Scholar]
- 21.Pettit GR, Xu JP, Cichacz ZA, Williams MD, Chapuis JC, Cerny RL. Antineoplastic agents 323. Isolation and structure of phakellistatin 6 from a Chuuk archipelago marine sponge. Bioorg Med Chem Lett. 1994;4:2677–2682. doi: 10.1016/s0960-894x(02)01054-5. [DOI] [PubMed] [Google Scholar]
- 22.Pettit GR, Xu JP, Dorsaz AC, Williams MD, Boyd MR, Cerny RL. Isolation and structure of the human cancer cell growth inhibitory cyclic decapeptides phakellistatins 7, 8 and 9. Bioorg Med Chem Lett. 1995;5:1339–1344. [Google Scholar]
- 23.Pettit GR, Tan R, Ichihara Y, Williams MD, Doubek DL, Tackett LP, Schmidt JM, Cerny RL, Boyd MR, Hooper JNA. Antineoplastic agents, 325. Isolation and structure of the human cancer cell growth inhibitory cyclic octapeptides phakellistatin 10 and 11 from Phakellia sp. J Nat Prod. 1995;58:961–965. doi: 10.1021/np50120a025. [DOI] [PubMed] [Google Scholar]
- 24.Pettit GR, Tan R. Antineoplastic agents 390. Isolation and structure of phakellistatin 12 from a Chuuk archipelago marine sponge. Bioorg Med Chem Lett. 2003;13:685–688. doi: 10.1016/s0960-894x(02)01054-5. [DOI] [PubMed] [Google Scholar]
- 25.Li WL, Yi YH, Wu HM, Xu QZ, Tang HF, Zhou DZ, Lin HW, Wang ZH. Isolation and structure of the cytotoxic cycloheptapeptide phakellistatin 13. J Nat Prod. 2003;66:146–148. doi: 10.1021/np020223y. [DOI] [PubMed] [Google Scholar]
- 26.Shaheen F, Ziaee MA, Ali SA, Simjee SU, Ahmed A, Choudhary MI. The first solid-phase synthesis and structural studies on phakellistatin 15. Rec Nat Prod. 2016;10:397–406. [Google Scholar]
- 27.Zhang HJ, Yi YH, Yang GJ, Hu MY, Cao GD, Yang F, Lin HW. Proline-containing cyclopeptides from the marine sponge Phakellia fusca. J Nat Prod. 2010;73:650–655. doi: 10.1021/np9008267. [DOI] [PubMed] [Google Scholar]
- 28.Pelay-Gimeno M, Meli A, Tulla-Puche J, Albericio F. Rescuing biological activity from synthetic phakellistatin 19. J Med Chem. 2013;56:9780–9788. doi: 10.1021/jm401520x. [DOI] [PubMed] [Google Scholar]
- 29.Chan WR, Tinto WF, Manchand PS, Todaro LJ. Stereostructures of geodiamolides A and B, novel cyclodepsipeptides from the marine sponge Geodia sp. J Org Chem. 1987;52:3091–3093. [Google Scholar]
- 30.Tinto WF, Lough AJ, McLean S, Reynolds WF, Yu M, Chan WR. Geodiamolides H and I, further cyclodepsipeptides from the marine sponge Geodia sp. Tetrahedron. 1998;54:4451–4458. [Google Scholar]
- 31.de Silva ED, Andersen RJ, Allen TM. Geodiamolides C to F, new cytotoxic cyclodepsipeptides from the marine sponge Pseudaxinyssa sp. Tetrahedron Lett. 1990;31:489–492. [Google Scholar]
- 32.Rangel M, Prado MP, Konno K, Naoki H, Freitas JC, Machado-Santelli GM. Cytoskeleton alterations induced by Geodia corticostylifera depsipeptides in breast cancer cells. Peptides. 2006;27:2047–2057. doi: 10.1016/j.peptides.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 33.Coleman JE, Van Soest R, Andersen RJ. New geodiamolides from the sponge Cymbastela sp. collected in Papua New Guinea. J Nat Prod. 1999;62:1137–1141. doi: 10.1021/np990155o. [DOI] [PubMed] [Google Scholar]
- 34.Zheng LH, Wang YJ, Sheng J, Wang F, Zheng Y, Lin XK, Sun M. Antitumor peptides from marine organisms. Mar Drugs. 2011;9:1840–1859. doi: 10.3390/md9101840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sorres J, Martin MT, Petek S, Levaique H, Cresteil T, Ramos S, Thoison O, Debitus C, Al-Mourabit A. Pipestelides A–C: Cyclodepsipeptides from the Pacific marine sponge Pipestela candelabra. J Nat Prod. 2012;75:759–763. doi: 10.1021/np200714m. [DOI] [PubMed] [Google Scholar]
- 36.Sonnenschein RN, Farias JJ, Tenney K, Mooberry SL, Lobkovsky E, Clardy J, Crews P. A further study of the cytotoxic constituents of a milnamide-producing sponge. Org Lett. 2004;6:779–782. doi: 10.1021/ol036446c. [DOI] [PubMed] [Google Scholar]
- 37.Chevallier C, Richardson AD, Edler MC, Hamel E, Harper MK, Ireland CM. A new cytotoxic and tubulin-interactive milnamide derivative from a marine sponge Cymbastela sp. Org Lett. 2003;5:3737–3739. doi: 10.1021/ol035476c. [DOI] [PubMed] [Google Scholar]
- 38.Liu C, Masuno MN, MacMillan JB, Molinski TF. Enantioselective total synthesis of (+)- milnamide A and evidence of its autoxidation to (+)- milnamide D. Angew Chem, Int Ed. 2004;43:5951–5954. doi: 10.1002/anie.200461245. [DOI] [PubMed] [Google Scholar]
- 39.Tran TD, Pham NB, Fechner GA, Hooper JNA, Quinn RJ. Potent cytotoxic peptides from the Australian marine sponge Pipestela candelabra. Mar Drugs. 2014;12:3399–3415. doi: 10.3390/md12063399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmidt EW, Raventos-Suarez C, Bifano M, Menendez AT, Fairchild CR, Faulkner DJ. Scleritodermin A, a cytotoxic cyclic peptide from the Lithistid sponge Scleritoderma nodosum. J Nat Prod. 2004;67:475–478. doi: 10.1021/np034035z. [DOI] [PubMed] [Google Scholar]
- 41.Liu S, Cui YM, Nan FJ. Total synthesis of the originally proposed and revised structures of scleritodermin A. Org Lett. 2008;10:3765–3768. doi: 10.1021/ol801419m. [DOI] [PubMed] [Google Scholar]
- 42.Gogineni V, Schinazi RF, Hamann MT. Role of marine natural products in the genesis of antiviral agents. Chem Rev. 2015;115:9655–9706. doi: 10.1021/cr4006318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plaza A, Gustchina E, Baker HL, Kelly M, Bewley CA. Mirabamides A-D, depsipeptides from the sponge Siliquariaspongia mirabilis that inhibit HIV-1 fusion. J Nat Prod. 2007;70:1753–1760. doi: 10.1021/np070306k. [DOI] [PubMed] [Google Scholar]
- 44.Lu Z, Van Wagoner RM, Harper MK, Baker HL, Hooper JNA, Bewley CA, Ireland CM. Mirabamides E−H, HIV-inhibitory depsipeptides from the sponge Stelletta clavosa. J Nat Prod. 2011;74:185–193. doi: 10.1021/np100613p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plaza A, Bifulco G, Keffer JL, Lloyd JR, Baker HL, Bewley CA. Celebesides A−C and theopapuamides B−D, depsipeptides from an Indonesian sponge that inhibit HIV-1 entry. J Org Chem. 2009;74:504–512. doi: 10.1021/jo802232u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zampella A, Sepe V, Luciano P, Bellotta F, Monti MC, D’Auria MV, Jepsen T, Petek S, Adeline M-T, Laprevote O, Aubertin AM, Debitus C, Poupat C, Ahond A. Homophymine A, an anti-HIV cyclodepsipeptide from the sponge Homophymia sp. J Org Chem. 2008;73:5319–5327. doi: 10.1021/jo800583b. [DOI] [PubMed] [Google Scholar]
- 47.Zampella A, Sepe V, Bellotta F, Luciano P, D’Auria MV, Cresteil T, Debitus C, Petek S, Poupat C, Ahond A. Homophymines B–E and A1–E1, a family of bioactive cyclodepsipeptides from the sponge Homophymia sp. Org Biomol Chem. 2009;7:4037–4044. doi: 10.1039/b910015f. [DOI] [PubMed] [Google Scholar]
- 48.Oku N, Gustafson KR, Cartner LK, Wilson JA, Shigematsu N, Hess S, Pannell LK, Boyd MR, McMahon JB. Neamphamide A, a new HIV-inhibitory depsipeptide from the Papua New Guinea marine sponge Neamphius huxleyi. J Nat Prod. 2004;67:1407–1411. doi: 10.1021/np040003f. [DOI] [PubMed] [Google Scholar]
- 49.Oku N, Krishnamoorthy R, Benson AG, Ferguson RL, Lipton MA, Phillips LR, Gustafson KR, McMahon JB. Complete stereochemistry of neamphamide A and absolute configuration of the β-methoxytyrosine residue in papuamide B. J Org Chem. 2005;70:6842–6847. doi: 10.1021/jo0508853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tran TD, Pham NB, Fechner G, Zencak D, Vu HT, Hooper JNA, Quinn RJ. Cytotoxic cyclic depsipeptides from the Australian marine sponge Neamphius huxleyi. J Nat Prod. 2012;75:2200–2208. doi: 10.1021/np3006474. [DOI] [PubMed] [Google Scholar]
- 51.Zampella A, Randazzo A, Borbone N, Luciani S, Trevisi L, Debitus C, D’Auria MV. Isolation of callipeltins A–C and of two new open-chain derivatives of callipeltin A from the marine sponge Latrunculia sp. A revision of the stereostructure of callipeltins. Tetrahedron Lett. 2002;43:6163–6166. [Google Scholar]
- 52.D’Auria MV, Zampella A, Paloma LG, Minale L, Debitus C, Roussakis C, Le Bert V. Callipeltins B and C; bioactive peptides from a marine Lithistida sponge Callipelta sp. Tetrahedron. 1996;52:9589–9596. [Google Scholar]
- 53.Kikuchi M, Nosaka K, Akaji K, Konno H. Solid phase total synthesis of callipeltin E isolated from marine sponge Latrunculia sp. Tetrahedron Lett. 2011;52:3872–3875. [Google Scholar]
- 54.Sepe V, D’Orsi R, Borbone N, D’Auria MV, Bifulco G, Monti MC, Catania A, Zampella A. Callipeltins F–I: new antifungal peptides from the marine sponge Latrunculia sp. Tetrahedron. 2006;62:833–840. [Google Scholar]
- 55.D’Auria MV, Sepe V, D’Orsi R, Bellotta F, Debitus C, Zampella A. Isolation and structural elucidation of callipeltins J–M: Antifungal peptides from the marine sponge Latrunculia sp. Tetrahedron. 2007;63:131–140. [Google Scholar]
- 56.Stierhof M, Hansen KØ, Sharma M, Feussner K, Subko K, Díaz-Rullo FF, Isaksson J, Pérez-Victoria I, Clarke D, Hansen E, Jaspars M, Tabudravu JN. New cytotoxic callipeltins from the Solomon Island marine sponge Asteropus sp. Tetrahedron. 2016;72:6929–6934. [Google Scholar]
- 57.Rashid MA, Gustafson KR, Cartner LK, Shigematsu N, Pannell LK, Boyd MR. Microspinosamide, a new HIV-inhibitory cyclic depsipeptide from the marine sponge Sidonops microspinosa. J Nat Prod. 2001;64:117–121. doi: 10.1021/np0002379. [DOI] [PubMed] [Google Scholar]
- 58.Afifi AH, El-Desoky AH, Kato H, Mangindaan REP, De Voogd NJ, Ammar NM, Hifnawy MS, Tsukamoto S. Carteritins A and B, cyclic heptapeptides from the marine sponge Stylissa carteri. Tetrahedron Lett. 2016;57:1285–1288. [Google Scholar]
- 59.Crews P, Kakou Y, Quinoa E. Mycothiazole, a polyketide heterocycle from a marine sponge. J Am Chem Soc. 1988;110:4365–4368. [Google Scholar]
- 60.Harrigan GG, Goetz GH, Luesch H, Yang S, Likos J. Dysideaprolines A−F and barbaleucamides A−B, novel polychlorinated compounds from a Dysidea Species. J Nat Prod. 2001;64:1133–1138. doi: 10.1021/np0101999. [DOI] [PubMed] [Google Scholar]
- 61.Ardá A, Rodríguez J, Nieto RM, Bassarello C, Gomez-Paloma L, Bifulco G, Jiménez C. NMR J-based analysis of nitrogen-containing moieties and application to dysithiazolamide, a new polychlorinated dipeptide from Dysidea sp. Tetrahedron. 2005;61:10093–10098. [Google Scholar]
- 62.Davis RA, Mangalindan GC, Bojo ZP, Antemano RR, Rodriguez NO, Concepcion GP, Samson SC, De Guzman D, Cruz LJ, Tasdemir D, Harper MK, Feng X, Carter GT, Ireland CM. Microcionamides A and B, bioactive peptides from the Philippine sponge Clathria (Thalysias) abietina. J Org Chem. 2004;69:4170–4176. doi: 10.1021/jo040129h. [DOI] [PubMed] [Google Scholar]
- 63.Hill RA. 5 Marine natural products. Annu Rep Prog Chem, Sect B: Org Chem. 2005;101:124–136. [Google Scholar]
- 64.Li HY, Matsunaga S, Fusetani N. Halicylindramides A−C, antifungal and cytotoxic depsipeptides from the marine sponge Halichondria cylindrata. J Med Chem. 1995;38:338–343. doi: 10.1021/jm00002a015. [DOI] [PubMed] [Google Scholar]
- 65.Rashid MA, Gustafson KR, Boswell JL, Boyd MR. Haligramides A and B, two new cytotoxic hexapeptides from the marine sponge Haliclona nigra. J Nat Prod. 2000;63:956–959. doi: 10.1021/np000051+. [DOI] [PubMed] [Google Scholar]
- 66.Mau CMS, Nakao Y, Yoshida WY, Scheuer PJ, Kelly-Borges M. Waiakeamide, a cyclic hexapeptide from the sponge Ircinia dendroides. J Org Chem. 1996;61:6302–6304. doi: 10.1021/jo960771e. [DOI] [PubMed] [Google Scholar]
- 67.Sera Y, Adachi K, Fujii K, Shizuri Y. A new antifouling hexapeptide from a Palauan sponge, Haliclona sp. J Nat Prod. 2003;66:719–721. doi: 10.1021/np020271i. [DOI] [PubMed] [Google Scholar]
- 68.Clark WD, Corbett T, Valeriote F, Crews P. Cyclocinamide A. An unusual cytotoxic halogenated hexapeptide from the marine sponge Psammocinia. J Am Chem Soc. 1997;119:9285–9286. [Google Scholar]
- 69.Laird DW, LaBarbera DV, Feng X, Bugni TS, Harper MK, Ireland CM. Halogenated cyclic peptides isolated from the sponge Corticium sp. J Nat Prod. 2007;70:741–746. doi: 10.1021/np060489v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Urda C, Pérez M, Rodriguez J, Jiménez C, Cuevas C, Fernandez R. Pembamide, a N-methylated linear peptide from a sponge Cribrochalina sp. Tetrahedron Lett. 2016;57:3239–3242. [Google Scholar]
- 71.Yeung BKS, Nakao Y, Kinnel RB, Carney JR, Yoshida WY, Scheuer PJ, Kelly-Borges M. The kapakahines, cyclic peptides from the marine sponge Cribrochalina olemda. J Org Chem. 1996;61:7168–7173. doi: 10.1021/jo960725e. [DOI] [PubMed] [Google Scholar]
- 72.Nakao Y, Kuo J, Yoshida WY, Kelly M, Scheuer PJ. More kapakahines from the marine sponge Cribrochalina olemda. Org Lett. 2003;5:1387–1390. doi: 10.1021/ol026830u. [DOI] [PubMed] [Google Scholar]
- 73.Bishara A, Rudi A, Aknin M, Neumann D, Ben-Califa N, Kashman Y. Taumycins A and B, two bioactive lipodepsipeptides from the Madagascar sponge Fascaplysinopsis sp. Org Lett. 2008;10:4307–4309. doi: 10.1021/ol801750y. [DOI] [PubMed] [Google Scholar]
- 74.Pelay-Gimeno M, García-Ramos Y, Martin MJ, Spengler J, Molina-Guijarro JM, Munt S, Francesch AM, Cuevas C, Tulla-Puche J, Albericio F. The first total synthesis of the cyclodepsipeptide pipecolidepsin A. Nat Commun. 2013;4:2352–2361. doi: 10.1038/ncomms3352. [DOI] [PubMed] [Google Scholar]
- 75.Coello L, Reyes F, Martín MJ, Cuevas C, Fernández R. Isolation and structures of pipecolidepsins A and B, cytotoxic cyclic depsipeptides from the Madagascan sponge Homophymia lamellosa. J Nat Prod. 2014;77:298–303. doi: 10.1021/np400888e. [DOI] [PubMed] [Google Scholar]
- 76.Randazzo A, Bifulco G, Giannini C, Bucci M, Debitus C, Cirino G, Gomez-Paloma L. Halipeptins A and B: Two novel potent anti-inflammatory cyclic depsipeptides from the Vanuatu marine sponge Haliclona species. J Am Chem Soc. 2001;123:10870–10876. doi: 10.1021/ja010015c. [DOI] [PubMed] [Google Scholar]
- 77.Yu S, Pan X, Ma D. Asymmetric total syntheses of marine cyclic depsipeptide halipeptins A–D. Chem -Eur J. 2006;12:6572–6584. doi: 10.1002/chem.200600383. [DOI] [PubMed] [Google Scholar]
- 78.Bishara A, Rudi A, Goldberg I, Aknin M, Neumann D, Ben-Califa N, Kashman Y. Tausalarin C: a new bioactive marine sponge-derived nitrogenous bismacrolide. Org Lett. 2009;11:3538–3541. doi: 10.1021/ol9011019. [DOI] [PubMed] [Google Scholar]
- 79.Kobayashi M, Aoki S, Ohyabu N, Kurosu M, Wang W, Kitagawa I. Arenastatin A, a potent cytotoxic depsipeptide from the Okinawan marine sponge Dysidea arenaria. Tetrahedron Lett. 1994;35:7969–7972. [Google Scholar]
- 80.Yadav JS, Purnima KV, Reddy BVS, Nagaiah K, Ghamdi AK. Total synthesis of cryptophycin-24 (arenastatin A) via Prins cyclization. Tetrahedron Lett. 2011;52:6709–6712. [Google Scholar]
- 81.Pettit GR, Gao F, Cerny RL, Doubek DL, Tackett LP, Schmidt JM, Chapuis JC. Antineoplastic agents. 278. Isolation and structure of axinastatins 2 and 3 from a Western Caroline Island marine sponge. J Med Chem. 1994;37:1165–1168. doi: 10.1021/jm00034a014. [DOI] [PubMed] [Google Scholar]
- 82.Konat RK, Mathä B, Winkler J, Kessler H. Axinastatin 1 or malaysiatin? proof of constitution and 3D structure in solution of a cyclic heptapeptide with cytostatic properties. Eur J Org Chem. 1995;1995:765–774. [Google Scholar]
- 83.Kong F, Burgoyne DL, Andersen J, Allen TM. Pseudoaxinellin, a cyclic heptapeptide isolated from the Papua New Guinea sponge Pseudoaxinella massa. Tetrahedron Lett. 1992;33:3269–3272. [Google Scholar]
- 84.Kobayashi JI, Tsuda M, Nakamura T, Mikami Y, Shigemori H. Hymenamides A and B, new proline-rich cyclic heptapeptides from the Okinawan marine sponge Hymeniacidon sp. Tetrahedron. 1993;49:2391–2402. [Google Scholar]
- 85.Tsuda M, Shigemori H, Mikami Y, Kobayashi JI. Hymenamides C~E, new cyclic heptapeptides with two proline residues from the Okinawan marine sponge Hymeniacidon sp. Tetrahedron. 1993;49:6785–6796. [Google Scholar]
- 86.Kobayashi JI, Nakamura T, Tsuda M. Hymenamide F, new cyclic heptapeptide from marine sponge Hymeniacidon sp. Tetrahedron. 1996;52:6355–6360. [Google Scholar]
- 87.Tsuda M, Sasaki T, Kobayashi JI. Hymenamides G, H, J, and K, four new cyclic octapeptides from the Okinawan marine sponge Hymeniacidon sp. Tetrahedron. 1994;50:4667–4680. [Google Scholar]
- 88.Tabudravu J, Morris LA, Kettenes-van den Bosch JJ, Jaspars M. Wainunuamide, a histidine-containing proline-rich cyclic heptapeptide isolated from the Fijian marine sponge Stylotella aurantium. Tetrahedron Lett. 2001;42:9273–9276. [Google Scholar]
- 89.Randazzo A, Dal Piaz F, Orrù S, Debitus C, Roussakis C, Pucci P, Gomez-Paloma L. Axinellins A and B: New proline-containing antiproliferative cyclopeptides from the Vanuatu sponge Axinella carteri. Eur J Org Chem. 1998;1998:2659–2665. [Google Scholar]
- 90.Tabudravu JN, Morris LA, Kettenes-van den Bosch JJ, Jaspars M. Axinellin C, a proline-rich cyclic octapeptide isolated from the Fijian marine sponge Stylotella aurantium. Tetrahedron. 2002;58:7863–7868. [Google Scholar]
- 91.Milanowski DJ, Rashid MA, Gustafson KR, O’Keefe BR, Nawrocki JP, Pannell LK, Boyd MR. Cyclonellin, a new cyclic octapeptide from the marine sponge Axinella carteri. J Nat Prod. 2004;67:441–444. doi: 10.1021/np030336x. [DOI] [PubMed] [Google Scholar]
- 92.Pettit GR, Srirangam JK, Herald DL, Xu JP, Boyd MR, Cichacz Z, Kamano Y, Schmidt JM, Erickson KL. Isolation and crystal structure of stylopeptide 1, a new marine porifera cycloheptapeptide. J Org Chem. 1995;60:8257–8261. [Google Scholar]
- 93.Brennan MR, Costello CE, Maleknia SD, Pettit GR, Erickson KL. Stylopeptide 2, a proline-rich cyclodecapeptide from the sponge Stylotella sp. J Nat Prod. 2008;71:453–456. doi: 10.1021/np0704856. [DOI] [PubMed] [Google Scholar]
- 94.Pettit GR, Srirangam JK, Herald DL, Erickson KL, Doubek DL, Schmidt JM, Tackett LP, Bakus GJ. Isolation and structure of stylostatin 1 from the Papua New Guinea marine sponge Stylotella aurantium. J Org Chem. 1992;57:7217–7220. [Google Scholar]
- 95.Pettit GR, Srirangam JK. Stylostatin 2, US5494893A. 1996 [Google Scholar]
- 96.Omar S, Tenenbaum L, Manes LV, Crews P. Novel marine sponge derived amino acids 7. The fenestins. Tetrahedron Lett. 1988;29:5489–5492. [Google Scholar]
- 97.Pettit GR, Clewlow PJ, Dufresne C, Doubek DL, Cerny RL, Rützler K. Antineoplastic agents. 193. Isolation and structure of the cyclic peptide hymenistatin 1. Can J Chem. 1990;68:708–711. [Google Scholar]
- 98.Gunasekera SP, Pomponi SA, McCarthy PJ. Discobahamins A and B, new peptides from the Bahamian deep water marine sponge Discodermia sp. J Nat Prod. 1994;57:79–83. doi: 10.1021/np50103a011. [DOI] [PubMed] [Google Scholar]
- 99.Kimura M, Wakimoto T, Egami Y, Tan KC, Ise Y, Abe I. Calyxamides A and B, cytotoxic cyclic peptides from the marine sponge Discodermia calyx. J Nat Prod. 2012;75:290–294. doi: 10.1021/np2009187. [DOI] [PubMed] [Google Scholar]
- 100.Rouf A, Tanyeli C. Bioactive thiazole and benzothiazole derivatives. Eur J Med Chem. 2015;97:911–927. doi: 10.1016/j.ejmech.2014.10.058. [DOI] [PubMed] [Google Scholar]
- 101.Bewley CA, Debitus C, Faulkner DJ. Microsclerodermins A and B. Antifungal cyclic peptides from the Lithistid sponge Microscleroderma sp. J Am Chem Soc. 1994;116:7631–7636. [Google Scholar]
- 102.Schmidt EW, Faulkner DJ. Microsclerodermins C–E, antifungal cyclic peptides from the Lithistid marine sponges Theonella sp. and Microscleroderma sp. Tetrahedron. 1998;54:3043–3056. [Google Scholar]
- 103.Qureshi A, Colin PL, Faulkner DJ. Microsclerodermins F–I, Antitumor and antifungal cyclic peptides from the Lithistid sponge Microscleroderma sp. Tetrahedron. 2000;56:3679–3685. [Google Scholar]
- 104.Zhang X, Jacob MR, Rao RR, Wang YH, Agarwal AK, Newman DJ, Khan IA, Clark AM, Li XC. Antifungal cyclic peptides from the marine sponge Microscleroderma herdmani. Res Rep Med Chem. 2012;2:7–14. doi: 10.2147/RRMC.S30895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bewley CA, He H, Williams DH, Faulkner DJ. Aciculitins A–C: Cytotoxic and antifungal cyclic peptides from the lithistid sponge Aciculites orientalis. J Am Chem Soc. 1996;118:4314–4321. [Google Scholar]
- 106.Gulavita NK, Gunasekera SP, Pomponi SA, Robinson EV. Polydiscamide A: A new bioactive depsipeptide from the marine sponge Discodermia sp. J Org Chem. 1992;57:1767–1772. [Google Scholar]
- 107.Feng Y, Carroll AR, Pass DM, Archbold JK, Avery VM, Quinn RJ. Polydiscamides B−D from a marine sponge Ircinia sp. as potent human sensory neuron-specific G protein coupled receptor agonists. J Nat Prod. 2008;71:8–11. doi: 10.1021/np070094r. [DOI] [PubMed] [Google Scholar]
- 108.Coleman JE, de Silva ED, Kong F, Andersen RJ, Allen TM. Cytotoxic peptides from the marine sponge Cymbastela sp. Tetrahedron. 1995;51:10653–10662. [Google Scholar]
- 109.Woo JK, Jeon JE, Kim CK, Sim CJ, Oh DC, Oh KB, Shin J. Gombamide A, a cyclic thiopeptide from the sponge Clathria gombawuiensis. J Nat Prod. 2013;76:1380–1383. doi: 10.1021/np4003367. [DOI] [PubMed] [Google Scholar]
- 110.Vera B, Vicente J, Rodríguez AD. Isolation and structural elucidation of euryjanicins B−D, proline-containing cycloheptapeptides from the Caribbean marine sponge Prosuberites laughlini. J Nat Prod. 2009;72:1555–1562. doi: 10.1021/np9004135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Avilés E, Rodríguez AD. Euryjanicins E–G, poly-phenylalanine, and poly-proline cyclic heptapeptides from the Caribbean sponge Prosuberites laughlini. Tetrahedron. 2013;69:10797–10804. doi: 10.1016/j.tet.2013.10.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Williams DE, Austin P, Diaz-Marrero AR, Soest RV, Matainaho T, Roskelley CD, Roberge M, Andersen RJ. Neopetrosiamides, peptides from the marine sponge Neopetrosia sp. that inhibit amoeboid invasion by human tumor cells. Org Lett. 2005;7:4173–4176. doi: 10.1021/ol051524c. [DOI] [PubMed] [Google Scholar]
- 113.Tan KC, Wakimoto T, Abe I. Sulfoureido lipopeptides from the marine sponge Discodermia kiiensis. J Nat Prod. 2016;79:2418–2422. doi: 10.1021/acs.jnatprod.6b00586. [DOI] [PubMed] [Google Scholar]
- 114.Tan KC, Wakimoto T, Abe I. Lipodiscamides A–C, new cytotoxic lipopeptides from Discodermia kiiensis. Org Lett. 2014;16:3256–3259. doi: 10.1021/ol501271v. [DOI] [PubMed] [Google Scholar]
- 115.Jamison MT, Molinski TF. Jamaicensamide A, a peptide containing β-Amino-α-keto and thiazole-homologated η-amino acid residues from the sponge Plakina jamaicensis. J Nat Prod. 2016;79:2243–2249. doi: 10.1021/acs.jnatprod.6b00336. [DOI] [PubMed] [Google Scholar]
- 116.Schmidt G, Grube A, Köck M. Stylissamides A–D – New proline-containing cyclic heptapeptides from the marine sponge Stylissa caribica. Eur J Org Chem. 2007;2007:4103–4110. [Google Scholar]
- 117.Cychon C, Köck M. Stylissamides E and F, cyclic heptapeptides from the Caribbean sponge Stylissa caribica. J Nat Prod. 2010;73:738–742. doi: 10.1021/np900664f. [DOI] [PubMed] [Google Scholar]
- 118.Wang X, Morinaka BI, Molinski TF. Structures and solution conformational dynamics of stylissamides G and H from the Bahamian sponge Stylissa caribica. J Nat Prod. 2014;77:625–630. doi: 10.1021/np400891s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kita M, Gise B, Kawamura A, Kigoshi H. Stylissatin A, a cyclic peptide that inhibits nitric oxide production from the marine sponge Stylissa massa. Tetrahedron Lett. 2013;54:6826–6828. [Google Scholar]
- 120.Sun J, Cheng W, de Voogd NJ, Proksch P, Lin W. Stylissatins B–D, cycloheptapeptides from the marine sponge Stylissa massa. Tetrahedron Lett. 2016;57:4288–4292. [Google Scholar]
- 121.Mohammed R, Peng J, Kelly M, Hamann MT. Cyclic heptapeptides from the Jamaican sponge Stylissa caribica. J Nat Prod. 2006;69:1739–1744. doi: 10.1021/np060006n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhan KX, Jiao WH, Yang F, Li J, Wang SP, Li YS, Han BN, Lin HW. Reniochalistatins A–E, cyclic peptides from the marine sponge Reniochalina stalagmitis. J Nat Prod. 2014;77:2678–2684. doi: 10.1021/np5006778. [DOI] [PubMed] [Google Scholar]
- 123.Ueoka R, Ise Y, Ohtsuka S, Okada S, Yamori T, Matsunaga S. Yaku’amides A and B, cytotoxic linear peptides rich in dehydroamino acids from the marine sponge Ceratopsion sp. J Am Chem Soc. 2010;132:17692–17694. doi: 10.1021/ja109275z. [DOI] [PubMed] [Google Scholar]
- 124.Song J, Jeon JE, Won TH, Sim CJ, Oh DC, Oh KB, Shin J. New cyclic cysteine bridged peptides from the sponge Suberites waedoensis. Mar Drugs. 2014;12:2760–2770. doi: 10.3390/md12052760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kehraus S, König GM, Wright AD, Woerheide G. Leucamide A: A new cytotoxic heptapeptide from the Australian sponge Leucetta microraphis. J Org Chem. 2002;67:4989–4992. doi: 10.1021/jo020058r. [DOI] [PubMed] [Google Scholar]
- 126.Nakao Y, Yoshida S, Matsunaga S, Shindoh N, Terada Y, Nagai K, Yamashita JK, Ganesan A, van Soest RWM, Fusetani N. Azumamides A–E: Histone deacetylase inhibitory cyclic tetrapeptides from the marine sponge Mycale izuensis. Angew Chem, Int Ed. 2006;45:7553–7557. doi: 10.1002/anie.200602047. [DOI] [PubMed] [Google Scholar]
- 127.Capon RJ, Ford J, Lacey E, Gill JH, Heiland K, Friedel T. Phoriospongin A and B: Two new nematocidal depsipeptides from the Australian marine sponges Phoriospongia sp. and Callyspongia bilamellata. J Nat Prod. 2002;65:358–363. doi: 10.1021/np010329d. [DOI] [PubMed] [Google Scholar]
- 128.Ibrahim SRM, Min CC, Teuscher F, Ebel R, Kakoschke C, Lin W, Wray V, Edrada-Ebel R, Proksch P. Callyaerins A–F and H, new cytotoxic cyclic peptides from the Indonesian marine sponge Callyspongia aerizusa. Bioorg Med Chem. 2010;18:4947–4956. doi: 10.1016/j.bmc.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 129.Ibrahim SRM, Edrada-Ebel R, Mohamed GA, Youssef DTA, Wray V, Proksch P. Callyaerin G, a new cytotoxic cyclic peptide from the marine sponge Callyspongia aerizusa. Arkivoc. 2008;12:164–171. [Google Scholar]
- 130.Fusetani N, Matsunaga S. Bioactive sponge peptides. Chem Rev. 1993;93:1793–1806. [Google Scholar]
- 131.de Silva ED, Williams DE, Andersen RJ, Klix H, Holmes CFB, Allen TM. Motuporin, a potent protein phosphatase inhibitor isolated from the Papua New Guinea sponge Theonella swinhoei Gray. Tetrahedron Lett. 1992;33:1561–1564. [Google Scholar]
- 132.Kobayashi M, Lee NK, Shibuya H, Momose T, Kitagawa I. Marine natural products. XXVI. Biologically active tridecapeptide lactones from the Okinawan marine sponge Theonella swinhoei (Theonellidae). (2). Structures of theonellapeptolides Ia, Ib, Ic, and Ie. Chem Pharm Bull. 1991;39:1177–1184. doi: 10.1248/cpb.39.1177. [DOI] [PubMed] [Google Scholar]
- 133.Fukuhara K, Takada K, Okada S, Matsunaga S. Nazumazoles A–C, cyclic pentapeptides dimerized through a disulfide bond from the marine sponge Theonella swinhoei. Org Lett. 2015;17:2646–2648. doi: 10.1021/acs.orglett.5b01020. [DOI] [PubMed] [Google Scholar]
- 134.Fukuhara K, Takada K, Okada S, Matsunaga S. Nazumazoles D–F, cyclic pentapeptides that inhibit chymotrypsin, from the marine sponge Theonella swinhoei. J Nat Prod. 2016;79:1694–1697. doi: 10.1021/acs.jnatprod.6b00261. [DOI] [PubMed] [Google Scholar]
- 135.Fusetani N, Sugawara T, Matsunaga S, Hirota H. Orbiculamide A: A novel cytotoxic cyclic peptide from a marine sponge Theonella sp. J Am Chem Soc. 1991;113:7811–7812. [Google Scholar]
- 136.Hamada T, Matsunaga S, Yano G, Fusetani N. Polytheonamides A and B, highly cytotoxic, linear polypeptides with unprecedented structural features, from the marine sponge, Theonella swinhoei. J Am Chem Soc. 2005;127:110–118. doi: 10.1021/ja045749e. [DOI] [PubMed] [Google Scholar]
- 137.Inoue M, Shinohara N, Tanabe S, Takahashi T, Okura K, Itoh H, Mizoguchi Y, Iida M, Lee N, Matsuoka S. Total synthesis of the large non-ribosomal peptide polytheonamide B. Nat Chem. 2010;2:280–285. doi: 10.1038/nchem.554. [DOI] [PubMed] [Google Scholar]
- 138.Nakao Y, Masuda A, Matsunaga S, Fusetani N. Pseudotheonamides, serine protease inhibitors from the marine sponge Theonella swinhoei. J Am Chem Soc. 1999;121:2425–2431. [Google Scholar]
- 139.Nakao Y, Oku N, Matsunaga S, Fusetani N. Cyclotheonamides E2 and E3, new potent serine protease inhibitors from the marine sponge of the genus Theonella. J Nat Prod. 1998;61:667–670. doi: 10.1021/np970544n. [DOI] [PubMed] [Google Scholar]
- 140.Maryanoff BE, Qiu X, Padmanabhan KP, Tulinsky A, Almond HR, Jr, Andrade-Gordon P, Greco MN, Kauffman JA, Nicolaou K, Liu A. Molecular basis for the inhibition of human alpha-thrombin by the macrocyclic peptide cyclotheonamide A. Proc Natl Acad Sci U S A. 1993;90:8048–8052. doi: 10.1073/pnas.90.17.8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fusetani N, Nakao Y, Matsunaga S. Nazumamide A, a thrombin-inhibitory tetrapeptide, from a marine sponge, Theonella sp. Tetrahedron Lett. 1991;32:7073–7074. [Google Scholar]
- 142.Matsunaga S, Fusetani N. Theonellamides A–E, cytotoxic bicyclic peptides, from a marine sponge Theonella sp. J Org Chem. 1995;60:1177–1181. [Google Scholar]
- 143.Youssef DTA, Shaala LA, Mohamed GA, Badr JM, Bamanie FH, Ibrahim SRM. Theonellamide G, a potent antifungal and cytotoxic bicyclic glycopeptide from the Red Sea marine sponge Theonella swinhoei. Mar Drugs. 2014;12:1911–1923. doi: 10.3390/md12041911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhang P, Liu R, Cook JM. An enantiospecific synthesis of L(−)- and D(+)-6-Chloro-5-hydroxytryptophan: An unusual amino acid residue from the cyclic hexapeptide keramamide A. Tetrahedron Lett. 1995;36:7411–7414. [Google Scholar]
- 145.Kobayashi J, Itagaki F, Shigemori H, Ishibashi M, Takahashi K, Ogura M, Nagasawa S, Nakamura T, Hirota H. Keramamides B−D: Novel peptides from the Okinawan marine sponge Theonella sp. J Am Chem Soc. 1991;113:7812–7813. [Google Scholar]
- 146.Sowinski JA, Toogood PL. Synthetic studies towards keramamide F. Tetrahedron Lett. 1995;36:67–70. [Google Scholar]
- 147.Kobayashi JI, Itagaki F, Shigemori I, Takao T, Shimonishi Y. Keramamides E, G, H, and J, new cyclic peptides containing an oxazole or a thiazole ring from a Theonella sponge. Tetrahedron. 1995;51:2525–2532. [Google Scholar]
- 148.Uemoto H, Yahiro Y, Shigemori H, Tsuda M, Takao T, Shimonishi Y, Kobayashi JI. Keramamides K and L, new cyclic peptides containing unusual tryptophan residue from Theonella sponge. Tetrahedron. 1998;54:6719–6724. [Google Scholar]
- 149.Tsuda M, Ishiyama H, Masuko K, Takao T, Shimonishi Y, Kobayashi JI. Keramamides M and N, two new cyclic peptides with a sulfate ester from Theonella sponge. Tetrahedron. 1999;55:12543–12548. [Google Scholar]
- 150.Fusetani N, Warabi K, Nogata Y, Nakao Y, Matsunaga S, van Soest RRM. Koshikamide A1, a new cytotoxic linear peptide isolated from a marine sponge, Theonella sp. Tetrahedron Lett. 1999;40:4687–4690. [Google Scholar]
- 151.Araki T, Matsunaga S, Fusetani N. Koshikamide A2, a cytotoxic linear undecapeptide isolated from a marine sponge of Theonella sp. Biosci, Biotechnol, Biochem. 2005;69:1318–1322. doi: 10.1271/bbb.69.1318. [DOI] [PubMed] [Google Scholar]
- 152.Winder PL, Pomponi SA, Wright AE. Natural products from the Lithistida: A review of the literature since 2000. Mar Drugs. 2011;9:2643–2682. doi: 10.3390/md9122643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Plaza A, Bifulco G, Masullo M, Lloyd JR, Keffer JL, Colin PL, Hooper JNA, Bell LJ, Bewley CA. Mutremdamide A and koshikamides C−H, peptide inhibitors of HIV-1 entry from different Theonella species. J Org Chem. 2010;75:4344–4355. doi: 10.1021/jo100076g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Okada Y, Matsunaga S, van Soest RWM, Fusetani N. Nagahamide A, an antibacterial depsipeptide from the marine sponge Theonella swinhoei. Org Lett. 2002;4:3039–3042. doi: 10.1021/ol0262791. [DOI] [PubMed] [Google Scholar]
- 155.Ford PW, Gustafson KR, McKee TC, Shigematsu N, Maurizi LK, Pannell LK, Williams DE, de Silva ED, Lassota P, Allen TM, Van Soest R, Andersen RJ, Boyd MR. Papuamides A−D, HIV-inhibitory and cytotoxic depsipeptides from the sponges Theonella mirabilis and Theonella swinhoei collected in Papua New Guinea. J Am Chem Soc. 1999;121:5899–5909. [Google Scholar]
- 156.Andjelic CD, Planelles V, Barrows LR. Characterizing the anti-HIV activity of papuamide A. Mar Drugs. 2008;6:528–549. doi: 10.3390/md20080027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Prasad P, Aalbersberg W, Feussner KD, Van Wagoner RM. Papuamides E and F, cytotoxic depsipeptides from the marine sponge Melophlus sp. Tetrahedron. 2011;67:8529–8531. doi: 10.1016/j.tet.2011.08.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Matsunaga S, Fusetani N, Hashimoto K, Walchli M. Theonellamide F. A novel antifungal bicyclic peptide from a marine sponge Theonella sp. J Am Chem Soc. 1989;111:2582–2588. [Google Scholar]
- 159.Bewley CA, Faulkner DJ. Theonegramide, an antifungal glycopeptide from the Philippine lithistid sponge Theonella swinhoei. J Org Chem. 1994;59:4849–4852. [Google Scholar]
- 160.Bonnington LS, Tanaka J, Higa T, Kimura J, Yoshimura Y, Nakao Y, Yoshida WY, Scheuer PJ. Cupolamide A: A cytotoxic cyclic heptapeptide from two samples of the sponge Theonella cupola. J Org Chem. 1997;62:7765–7767. [Google Scholar]
- 161.Roy MC, Ohtani II, Tanaka J, Higa T, Satari R. Barangamide A, a new cyclic peptide from the Indonesian sponge Theonella swinhoei. Tetrahedron Lett. 1999;40:5373–5376. [Google Scholar]
- 162.Roy MC, Ohtani II, Ichiba T, Tanaka J, Satari R, Higa T. New cyclic peptides from the Indonesian sponge Theonella swinhoei. Tetrahedron. 2000;56:9079–9092. [Google Scholar]
- 163.Kitagawa I, Lee NK, Kobayashi M, Shibuya H. Structure of theonellapeptolide Ie, a new tridecapeptide lactone from an Okinawan marine sponge, Theonella sp.(Theonellidae) Chem Pharm Bull. 1987;35:2129–2132. doi: 10.1248/cpb.35.2129. [DOI] [PubMed] [Google Scholar]
- 164.Kitagawa I, Lee NK, Kobayashi M, Shibuya H. Marine natural products. XXV. Biologically active tridecapeptide lactones from the Okinawan marine sponge Theonella swinhoei (Theonellidae). Structure of theonellapeptolide Id. Tetrahedron. 1991;47:2169–2180. doi: 10.1248/cpb.39.1177. [DOI] [PubMed] [Google Scholar]
- 165.Ohta E, Okada H, Ohta S, Kobayashi M, Kitagawa I, Horiike S, Takahashi T, Hosoya H, Yamamoto K, Ikegami S. Malformation of immature starfish oocytes by theonellapeptolide Ie, a tridecapeptide lactone from a marine sponge Petrosia species, through disturbance of cortical F-actin distribution. Biosci, Biotechnol, Biochem. 2003;67:1908–1915. doi: 10.1271/bbb.67.1908. [DOI] [PubMed] [Google Scholar]
- 166.Li S, Dumdei EJ, Blunt JW, Munro MHG, Robinson WT, Pannell LK. Theonellapeptolide IIIe, a new cyclic peptolide from the New Zealand deep water sponge, Lamellomorpha strongylata. J Nat Prod. 1998;61:724–728. doi: 10.1021/np970417r. [DOI] [PubMed] [Google Scholar]
- 167.Doi M, Ishida T, Kobayashi M, Deschamps JR, Flippen-Anderson JL. The highly solvated structure of theonellapeptolide Id, a tridecapeptide lactone from the Okinawa marine sponge Theonella swinhoei. Acta Crystallogr, Sect C: Cryst Struct Commun. 1999;55:796–798. [Google Scholar]
- 168.Doi M, Ishida T, Kobayashi M, Katsuya Y, Mezaki Y, Sasaki M, Terashima A, Taniguchi T, Tanaka C. Amphipathic structure of theonellapeptolide-Id, a hydrophobic tridecapeptide lactone from the Okinawa marine sponge Theonella swinhoei. Biopolymers. 2000;54:27–34. doi: 10.1002/(SICI)1097-0282(200007)54:1<27::AID-BIP30>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 169.Faulkner DJ. Marine natural products. Nat Prod Rep. 2000;17:7–55. doi: 10.1039/a809395d. [DOI] [PubMed] [Google Scholar]
- 170.Tsuda M, Shimbo K, Kubota T, Mikami Y, Kobayashi JI. Two theonellapeptolide congeners from marine sponge Theonella sp. Tetrahedron. 1999;55:10305–10314. [Google Scholar]
- 171.Festa C, De Marino S, Sepe V, D’Auria MV, Bifulco G, Débitus C, Bucci M, Vellecco V, Zampella A. Solomonamides A and B, new anti-inflammatory peptides from Theonella swinhoei. Org Lett. 2011;13:1532–1535. doi: 10.1021/ol200221n. [DOI] [PubMed] [Google Scholar]
- 172.Clark DP, Carroll J, Naylor S, Crews P. An antifungal cyclodepsipeptide, cyclolithistide A, from the sponge Theonella swinhoei. J Org Chem. 1998;63:8757–8764. [Google Scholar]
- 173.Nakamura H, Kobayashi JI, Nakamura Y, Ohizumi Y, Kondo T, Hirata Y. Theonellamine B, a novel peptidal Na, K-atpase inhibitor, from an Okinawan marine sponge of the genus Theonella. Tetrahedron Lett. 1986;27:4319–4322. [Google Scholar]
- 174.Schmidt EW, Bewley CA, Faulkner DJ. Theopalauamide, a bicyclic glycopeptide from filamentous bacterial symbionts of the Lithistid sponge Theonella swinhoei from Palau and Mozambique. J Org Chem. 1998;63:1254–1258. [Google Scholar]
- 175.Chill L, Kashman Y, Schleyer M. Oriamide, a new cytotoxic cyclic peptide containing a novel amino acid from the marine sponge Theonella sp. Tetrahedron. 1997;53:16147–16152. [Google Scholar]
- 176.Tajima H, Wakimoto T, Takada K, Ise Y, Abe I. Revised structure of cyclolithistide A, a cyclic depsipeptide from the marine sponge Discodermia japonica. J Nat Prod. 2014;77:154–158. doi: 10.1021/np400668k. [DOI] [PubMed] [Google Scholar]
- 177.Gulavita NK, Pomponi SA, Wright AE, Yarwood D, Sills MA. Isolation and structure elucidation of perthamide B, a novel peptide from the sponge Theonella sp. Tetrahedron Lett. 1994;35:6815–6818. [Google Scholar]
- 178.Festa C, De Marino S, Sepe V, D’Auria MV, Bifulco G, Andrés R, Terencio MC, Payá M, Debitus C, Zampella A. Perthamides C–F, potent human antipsoriatic cyclopeptides. Tetrahedron. 2011;67:7780–7786. [Google Scholar]
- 179.Festa C, De Marino S, D’Auria MV, Monti MC, Bucci M, Vellecco V, Debitus C, Zampella A. Anti-inflammatory cyclopeptides from the marine sponge Theonella swinhoei. Tetrahedron. 2012;68:2851–2857. [Google Scholar]
- 180.Magnino G, Sarà A, Lancioni T, Gaino E. Endobionts of the coral reef sponge Theonella swinhoei (Porifera, Demospongiae) Invertebr Biol. 1999;118:213–220. [Google Scholar]
- 181.Bewley CA, Faulkner DJ. Lithistid sponges: Star performers or hosts to the stars. Angew Chem, Int Ed. 1998;37:2162–2178. doi: 10.1002/(SICI)1521-3773(19980904)37:16<2162::AID-ANIE2162>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 182.Moore BS. Biosynthesis of marine natural products: Microorganisms and macroalgae. Nat Prod Rep. 1999;16:653–674. doi: 10.1039/a805873c. [DOI] [PubMed] [Google Scholar]
- 183.Piel J. Metabolites from symbiotic bacteria. Nat Prod Rep. 2004;21:519–538. doi: 10.1039/b310175b. [DOI] [PubMed] [Google Scholar]
- 184.Bewley CA, Holland ND, Faulkner DJ. Two classes of metabolites from Theonella swinhoei are localized in distinct populations of bacterial symbionts. Cell Mol Life Sci. 1996;52:716–722. doi: 10.1007/BF01925581. [DOI] [PubMed] [Google Scholar]
- 185.Yoshizawa S, Matsushima R, Watanabe MF, Harada KI, Ichihara A, Carmichael WW, Fujiki H. Inhibition of protein phosphatases by microcystis and nodularin associated with hepatotoxicity. J Cancer Res Clin Oncol. 1990;116:609–614. doi: 10.1007/BF01637082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kobayashi JI, Sato M, Ishibashi M, Shigemori H, Nakamura T, Ohizumi Y. Keramamide A, a novel peptide from the Okinawan marine sponge Theonella sp. J Chem Soc, Perkin Trans. 1991;1:2609–2611. [Google Scholar]
- 187.Williams DE, Craig M, Holmes CFB, Andersen RJ. Ferintoic acids A and B, new cyclic hexapeptides from the freshwater cyanobacterium Microcystis aeruginosa. J Nat Prod. 1996;59:570–575. [Google Scholar]
- 188.König GM, Kehraus S, Seibert SF, Abdel-Lateff A, Müller D. Natural products from marine organisms and their associated microbes. ChemBioChem. 2006;7:229–238. doi: 10.1002/cbic.200500087. [DOI] [PubMed] [Google Scholar]
- 189.Schmidt EW, Harper MK, Faulkner DJ. Mozamides A and B, cyclic peptides from a theonellid sponge from Mozambique. J Nat Prod. 1997;60:779–782. [Google Scholar]
- 190.Blunt JW, Copp BR, Keyzers RA, Munro MH, Prinsep MR. Marine natural products. Nat Prod Rep. 2013;30:237–323. doi: 10.1039/c2np20112g. [DOI] [PubMed] [Google Scholar]
- 191.Müller D, Krick A, Kehraus S, Mehner C, Hart M, Küpper FC, Saxena K, Prinz H, Schwalbe H, Janning P, Waldmann H, Kӧnig GM. Brunsvicamides A–C: Sponge-related cyanobacterial peptides with Mycobacterium tuberculosis protein tyrosine phosphatase inhibitory activity. J Med Chem. 2006;49:4871–4878. doi: 10.1021/jm060327w. [DOI] [PubMed] [Google Scholar]
- 192.Gunasekera SP, Owle CS, Montaser R, Luesch H, Paul VJ. Malyngamide 3 and cocosamides A and B from the marine cyanobacterium Lyngbya majuscula from Cocos Lagoon, Guam. J Nat Prod. 2011;74:871–876. doi: 10.1021/np1008015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Montaser R, Paul VJ, Luesch H. Pitipeptolides C–F, antimycobacterial cyclodepsipeptides from the marine cyanobacterium Lyngbya majuscula from Guam. Phytochemistry. 2011;72:2068–2074. doi: 10.1016/j.phytochem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Tripathi A, Puddick J, Prinsep MR, Rottmann M, Tan LT. Lagunamides A and B: Cytotoxic and antimalarial cyclodepsipeptides from the marine cyanobacterium Lyngbya majuscula. J Nat Prod. 2010;73:1810–1814. doi: 10.1021/np100442x. [DOI] [PubMed] [Google Scholar]
- 195.Nan HB, Gross H, Mcphail KL, Goeger D, Maier CS, Gerwick WH. Wewakamide A and guineamide G, cyclic depsipeptides from the marine cyanobacteria Lyngbya semiplena and Lyngbya majuscula. J Microbiol Biotechnol. 2011;21:930–936. doi: 10.4014/jmb.1105.05011. [DOI] [PubMed] [Google Scholar]
- 196.Tan LT, Sitachitta N, Gerwick WH. The guineamides, novel cyclic depsipeptides from a Papua New Guinea collection of the marine cyanobacterium Lyngbya majuscula. J Nat Prod. 2003;66:764–771. doi: 10.1021/np020492o. [DOI] [PubMed] [Google Scholar]
- 197.Nogle LM, Marquez BL, Gerwick WH. Wewakazole, a novel cyclic dodecapeptide from a Papua New Guinea Lyngbya majuscula. Org Lett. 2003;5:3–6. doi: 10.1021/ol026811k. [DOI] [PubMed] [Google Scholar]
- 198.Lopez JAV, Al-Lihaibi SS, Alarif WM, Abdel-Lateff A, Nogata Y, Washio K, Morikawa M, Okino T. Wewakazole B, a cytotoxic cyanobactin from the cyanobacterium Moorea producens collected in the Red Sea. J Nat Prod. 2016;79:1213–1218. doi: 10.1021/acs.jnatprod.6b00051. [DOI] [PubMed] [Google Scholar]
- 199.Han B, Goeger D, Maier CS, Gerwick WH. The wewakpeptins, cyclic depsipeptides from a Papua New Guinea collection of the marine cyanobacterium Lyngbya semiplena. J Org Chem. 2005;70:3133–3139. doi: 10.1021/jo0478858. [DOI] [PubMed] [Google Scholar]
- 200.Teruya T, Sasaki H, Fukazawa H, Suenaga K. Bisebromoamide, a potent cytotoxic peptide from the marine cyanobacterium Lyngbya sp.: Isolation, stereostructure, and biological activity. Org Lett. 2009;11:5062–5065. doi: 10.1021/ol9020546. [DOI] [PubMed] [Google Scholar]
- 201.Gao X, Liu Y, Kwong S, Xu Z, Ye T. Total synthesis and stereochemical reassignment of bisebromoamide. Org Lett. 2010;12:3018–3021. doi: 10.1021/ol101021v. [DOI] [PubMed] [Google Scholar]
- 202.Sasaki H, Teruya T, Fukazawa H, Suenaga K. Revised structure and structure– activity relationship of bisebromoamide and structure of norbisebromoamide from the marine cyanobacterium Lyngbya sp. Tetrahedron. 2011;67:990–994. [Google Scholar]
- 203.Wrasidlo W, Mielgo A, Torres VA, Barbero S, Stoletov K, Suyama TL, Klemke RL, Gerwick WH, Carson DA, Stupack DG. The marine lipopeptide somocystinamide A triggers apoptosis via caspase 8. Proc Natl Acad Sci U S A. 2008;105:2313–2318. doi: 10.1073/pnas.0712198105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Liu L, Rein KS. New peptides isolated from Lyngbya species: A review. Mar Drugs. 2010;8:1817–1837. doi: 10.3390/md8061817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Harrigan GG, Yoshida WY, Moore RE, Nagle DG, Park PU, Biggs J, Paul VJ, Mooberry SL, Corbett TH, Valeriote FA. Isolation, structure determination, and biological activity of dolastatin 12 and lyngbyastatin 1 from Lyngbya majuscula/Schizothrix calcicola cyanobacterial assemblages. J Nat Prod. 1998;61:1221–1225. doi: 10.1021/np9801211. [DOI] [PubMed] [Google Scholar]
- 206.Nagle DG, Paul VJ. Production of secondary metabolites by filamentous tropical marine cyanobacteria: Ecological functions of the compounds. J Phycol. 1999;35:1412–1421. [Google Scholar]
- 207.Simmons TL, Nogle LM, Media J, Valeriote FA, Mooberry SL, Gerwick WH. Desmethoxymajusculamide C, a cyanobacterial depsipeptide with potent cytotoxicity in both cyclic and ring-opened forms. J Nat Prod. 2009;72:1011–1016. doi: 10.1021/np9001674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Moore RE, Entzeroth M. Majusculamide D and deoxymajusculamide D, two cytotoxins from Lyngbya majuscula. Phytochemistry. 1988;27:3101–3103. [Google Scholar]
- 209.Williams PG, Moore RE, Paul VJ. Isolation and structure determination of lyngbyastatin 3, a lyngbyastatin 1 homologue from the marine cyanobacterium Lyngbya majuscula. Determination of the configuration of the 4-amino-2, 2-dimethyl-3-oxopentanoic acid unit in majusculamide C, dolastatin 12, lyngbyastatin 1, and lyngbyastatin 3 from cyanobacteria. J Nat Prod. 2003;66:1356–1363. doi: 10.1021/np0302145. [DOI] [PubMed] [Google Scholar]
- 210.Gutiérrez M, Suyama TL, Engene N, Wingerd JS, Matainaho T, Gerwick WH. Apratoxin D, a potent cytotoxic cyclodepsipeptide from Papua New Guinea collections of the marine cyanobacteria Lyngbya majuscula and Lyngbya sordida. J Nat Prod. 2008;71:1099–1103. doi: 10.1021/np800121a. [DOI] [PubMed] [Google Scholar]
- 211.Hooper GJ, Orjala J, Schatzman RC, Gerwick WH. Carmabins A and B, new lipopeptides from the Caribbean cyanobacterium Lyngbya majuscula. J Nat Prod. 1998;61:529–533. doi: 10.1021/np970443p. [DOI] [PubMed] [Google Scholar]
- 212.Clark WD, Crews P. A novel chlorinated ketide amino acid, herbamide A, from the marine sponge Dysidea herbacea. Tetrahedron Lett. 1995;36:1185–1188. [Google Scholar]
- 213.Balunas MJ, Linington RG, Tidgewell K, Fenner AM, Ureña LD, Togna GD, Kyle DE, Gerwick WH. Dragonamide E, a modified linear lipopeptide from Lyngbya majuscula with antileishmanial activity. J Nat Prod. 2010;73:60–66. doi: 10.1021/np900622m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Williams PG, Yoshida WY, Moore RE, Paul VJ. Isolation and structure determination of obyanamide, a novel cytotoxic cyclic depsipeptide from the marine cyanobacterium Lyngbya confervoides. J Nat Prod. 2002;65:29–31. doi: 10.1021/np0102253. [DOI] [PubMed] [Google Scholar]
- 215.MacMillan JB, Ernst-Russell MA, de Ropp JS, Molinski TF. Lobocyclamides A−C, lipopeptides from a cryptic cyanobacterial Mat containing Lyngbya confervoides. J Org Chem. 2002;67:8210–8215. doi: 10.1021/jo0261909. [DOI] [PubMed] [Google Scholar]
- 216.Bunyajetpong S, Yoshida WY, Sitachitta N, Kaya K. Trungapeptins A−C, cyclodepsipeptides from the marine cyanobacterium Lyngbya majuscula. J Nat Prod. 2006;69:1539–1542. doi: 10.1021/np050485a. [DOI] [PubMed] [Google Scholar]
- 217.Nogle LM, Gerwick WH. Isolation of four new cyclic depsipeptides, antanapeptins A−D, and dolastatin 16 from a Madagascan collection of Lyngbya majuscula. J Nat Prod. 2002;65:21–24. doi: 10.1021/np010348n. [DOI] [PubMed] [Google Scholar]
- 218.Malloy KL. Structure elucidation of biomedically relevant marine cyanobacterial natural products. Diss University of California, San Diego. 2011 https://escholarship.org/uc/item/2v22c1c5. 07/15/2016.
- 219.Nogle LM, Williamson RT, Gerwick WH. Somamides A and B, two new depsipeptide analogues of dolastatin 13 from a Fijian cyanobacterial assemblage of Lyngbya majuscula and Schizothrix Species. J Nat Prod. 2001;64:716–719. doi: 10.1021/np000634j. [DOI] [PubMed] [Google Scholar]
- 220.Zainuddin EN, Jansen R, Nimtz M, Wray V, Preisitsch M, Lalk M, Mundt S. Lyngbyazothrins A−D, antimicrobial cyclic undecapeptides from the cultured cyanobacterium Lyngbya sp. J Nat Prod. 2009;72:1373–1378. doi: 10.1021/np8007792. [DOI] [PubMed] [Google Scholar]
- 221.Kwan JC, Ratnayake R, Abboud KA, Paul VJ, Luesch H. Grassypeptolides A−C, cytotoxic bis-thiazoline containing marine cyclodepsipeptides. J Org Chem. 2010;75:8012–8023. doi: 10.1021/jo1013564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Thornburg CC, Thimmaiah M, Shaala LA, Hau AM, Malmo JM, Ishmael JE, Youssef DTA, McPhail KL. Cyclic depsipeptides, grassypeptolides D and E and Ibu-epidemethoxylyngbyastatin 3, from a Red Sea Leptolyngbya cyanobacterium. J Nat Prod. 2011;74:1677–1685. doi: 10.1021/np200270d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Popplewell WL, Ratnayake R, Wilson JA, Beutler JA, Colburn NH, Henrich CJ, McMahon JB, McKee TC. Grassypeptolides F and G, cyanobacterial peptides from Lyngbya majuscula. J Nat Prod. 2011;74:1686–1691. doi: 10.1021/np2005083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Klein D, Braekman JC, Daloze D, Hoffmann L, Castillo G, Demoulin V. Lyngbyapeptin A, a modified tetrapeptide from Lyngbya bouillonii (Cyanophyceae) Tetrahedron Lett. 1999;40:695–696. [Google Scholar]
- 225.Luesch H, Yoshida WY, Moore RE, Paul VJ. Structurally diverse new alkaloids from Palauan collections of the apratoxin-producing marine cyanobacterium Lyngbya sp. Tetrahedron. 2002;58:7959–7966. [Google Scholar]
- 226.Matthew S, Salvador LA, Schupp PJ, Paul VJ, Luesch H. Cytotoxic halogenated macrolides and modified peptides from the apratoxin-producing marine cyanobacterium Lyngbya bouillonii from Guam. J Nat Prod. 2010;73:1544–1552. doi: 10.1021/np1004032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Luesch H, Yoshida WY, Moore RE, Paul VJ. Isolation and structure of the cytotoxin lyngbyabellin B and absolute configuration of lyngbyapeptin A from the marine cyanobacterium Lyngbya majuscula. J Nat Prod. 2000;63:1437–1439. doi: 10.1021/np000104n. [DOI] [PubMed] [Google Scholar]
- 228.Milligan KE, Marquez BL, Williamson RT, Gerwick WH. Lyngbyabellin B, a toxic and antifungal secondary metabolite from the marine cyanobacterium Lyngbya majuscula. J Nat Prod. 2000;63:1440–1443. doi: 10.1021/np000133y. [DOI] [PubMed] [Google Scholar]
- 229.Tan LT. Bioactive natural products from marine cyanobacteria for drug discovery. Phytochemistry. 2007;68:954–979. doi: 10.1016/j.phytochem.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 230.Choi H, Mevers E, Byrum T, Valeriote FA, Gerwick WH. Lyngbyabellins K–N from two Palmyra Atoll collections of the marine cyanobacterium Moorea bouillonii. Eur J Org Chem. 2012;2012:5141–5150. doi: 10.1002/ejoc.201200691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Soria-Mercado IE, Pereira A, Cao Z, Murray TF, Gerwick WH. Alotamide A, a novel neuropharmacological agent from the marine cyanobacterium Lyngbya bouillonii. Org Lett. 2009;11:4704–4707. doi: 10.1021/ol901438b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Chang Z, Flatt P, Gerwick WH, Nguyen VA, Willis CL, Sherman DH. The barbamide biosynthetic gene cluster: A novel marine cyanobacterial system of mixed polyketide synthase (PKS)-non-ribosomal peptide synthetase (NRPS) origin involving an unusual trichloroleucyl starter unit. Gene. 2002;296:235–247. doi: 10.1016/s0378-1119(02)00860-0. [DOI] [PubMed] [Google Scholar]
- 233.Edwards DJ, Marquez BL, Nogle LM, McPhail K, Goeger DE, Roberts MA, Gerwick WH. Structure and biosynthesis of the jamaicamides, new mixed polyketide-peptide neurotoxins from the marine cyanobacterium Lyngbya majuscula. Chem Biol. 2004;11:817–833. doi: 10.1016/j.chembiol.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 234.Marquez BL, Watts KS, Yokochi A, Roberts MA, Verdier-Pinard P, Jimenez JI, Hamel E, Scheuer PJ, Gerwick WH. Structure and absolute stereochemistry of hectochlorin, a potent stimulator of actin assembly. J Nat Prod. 2002;65:866–871. doi: 10.1021/np0106283. [DOI] [PubMed] [Google Scholar]
- 235.Luesch H, Yoshida WY, Moore RE, Paul VJ. Apramides A−G, novel lipopeptides from the marine cyanobacterium Lyngbya majuscula. J Nat Prod. 2000;63:1106–1112. doi: 10.1021/np000078t. [DOI] [PubMed] [Google Scholar]
- 236.Sato SI, Murata A, Orihara T, Shirakawa T, Suenaga K, Kigoshi H, Uesugi M. Marine natural product aurilide activates the OPA1-mediated apoptosis by binding to prohibitin. Chem Biol. 2011;18:131–139. doi: 10.1016/j.chembiol.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 237.Han B, Gross H, Goeger DE, Mooberry SL, Gerwick WH. Aurilides B and C, cancer cell toxins from a Papua New Guinea collection of the marine cyanobacterium Lyngbya majuscula. J Nat Prod. 2006;69:572–575. doi: 10.1021/np0503911. [DOI] [PubMed] [Google Scholar]
- 238.Yokokawa F, Fujiwara H, Shioiri T. Total synthesis and revision of absolute configuration of antillatoxin, an ichthyotoxic cyclic lipopeptide of marine origin. Tetrahedron Lett. 1999;40:1915–1916. [Google Scholar]
- 239.Orsini MA, Pannell LK, Erickson KL. Polychlorinated acetamides from the cyanobacterium Microcoleus lyngbyaceus. J Nat Prod. 2001;64:572–577. doi: 10.1021/np000452p. [DOI] [PubMed] [Google Scholar]
- 240.Orjala J, Nagle DG, Hsu V, Gerwick WH. Antillatoxin: An exceptionally ichthyotoxic cyclic lipopeptide from the tropical cyanobacterium Lyngbya majuscula. J Am Chem Soc. 1995;117:8281–8282. [Google Scholar]
- 241.Wu M, Okino T, Nogle LM, Marquez BL, Williamson RT, Sitachitta N, Berman FW, Murray TF, McGough K, Jacobs R, Colsen K, Asano T, Yokokawa F, Shioiri T, Gerwick WH. Structure, synthesis, and biological properties of kalkitoxin, a novel neurotoxin from the marine cyanobacterium Lyngbya majuscula. J Am Chem Soc. 2000;122:12041–12042. [Google Scholar]
- 242.Wan F, Erickson KL. Georgamide, a new cyclic depsipeptide with an alkynoic acid residue from an Australian cyanobacterium. J Nat Prod. 2001;64:143–146. doi: 10.1021/np0003802. [DOI] [PubMed] [Google Scholar]
- 243.Sitachitta N, Williamson RT, Gerwick WH. Yanucamides A and B, two new depsipeptides from an assemblage of the marine cyanobacteria Lyngbya majuscula and Schizothrix Species. J Nat Prod. 2000;63:197–200. doi: 10.1021/np990466z. [DOI] [PubMed] [Google Scholar]
- 244.Jiménez JI, Scheuer PJ. New lipopeptides from the Caribbean cyanobacterium Lyngbya majuscula. J Nat Prod. 2001;64:200–203. doi: 10.1021/np000462q. [DOI] [PubMed] [Google Scholar]
- 245.Luesch H, Williams PG, Yoshida WY, Moore RE, Paul VJ. Ulongamides A−F, new β-amino acid-containing cyclodepsipeptides from Palauan collections of the marine cyanobacterium Lyngbya sp. J Nat Prod. 2002;65:996–1000. doi: 10.1021/np0200461. [DOI] [PubMed] [Google Scholar]
- 246.Kan Y, Sakamoto B, Fujita T, Nagai H. New malyngamides from the Hawaiian cyanobacterium Lyngbya majuscula. J Nat Prod. 2000;63:1599–1602. doi: 10.1021/np000250t. [DOI] [PubMed] [Google Scholar]
- 247.Kwan JC, Teplitski M, Gunasekera SP, Paul VJ, Luesch H. Isolation and biological evaluation of 8-epi-malyngamide C from the Floridian marine cyanobacterium Lyngbya majuscula. J Nat Prod. 2010;73:463–466. doi: 10.1021/np900614n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Mynderse JS, Moore RE. Malyngamides D and E, two trans-7-methoxy-9-methylhexadec-4-enamides from a deep water variety of the marine cyanophyte Lyngbya majuscula. J Org Chem. 1978;43:4359–4363. [Google Scholar]
- 249.Praud A, Valls R, Piovetti L, Banaigs B. Malyngamide G: Proposition de structure pour un nouvel amide chloré d’une algue bleu-verte epiphyte de Cystoseira crinita. Tetrahedron Lett. 1993;34:5437–5440. [Google Scholar]
- 250.Villa FA, Lieske K, Gerwick L. Selective MyD88-dependent pathway inhibition by the cyanobacterial natural product malyngamide F acetate. Eur J Pharmacol. 2010;629:140–146. doi: 10.1016/j.ejphar.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Mesguiche V, Valls R, Piovetti L, Peiffer G. Characterization and synthesis of (−)-7-methoxydodec-4(E)-enoic acid, a novel fatty acid isolated from Lyngbya majuscula. Tetrahedron Lett. 1999;40:7473–7476. [Google Scholar]
- 252.Todd JS, Gerwick WH. Malyngamide I from the tropical marine cyanobacterium Lyngbya majuscula and the probable structure revision of stylocheilamide. Tetrahedron Lett. 1995;36:7837–7840. [Google Scholar]
- 253.Kan Y, Fujita T, Nagai H, Sakamoto B, Hokama Y. Malyngamides M and N from the Hawaiian Red Alga Gracilaria coronopifolia. J Nat Prod. 1998;61:152–155. doi: 10.1021/np970423n. [DOI] [PubMed] [Google Scholar]
- 254.Gallimore WA, Scheuer PJ. Malyngamides O and P from the sea hare Stylocheilus longicauda. J Nat Prod. 2000;63:1422–1424. doi: 10.1021/np0000365. [DOI] [PubMed] [Google Scholar]
- 255.Chen J, Fu XG, Zhou L, Zhang JT, Qi XL, Cao XP. A convergent route for the total synthesis of malyngamides O, P, Q, and R. J Org Chem. 2009;74:4149–4157. doi: 10.1021/jo9003103. [DOI] [PubMed] [Google Scholar]
- 256.Appleton DR, Sewell MA, Berridge MV, Copp BR. A new biologically active malyngamide from a New Zealand collection of the sea hare Bursatella leachii. J Nat Prod. 2002;65:630–631. doi: 10.1021/np010511e. [DOI] [PubMed] [Google Scholar]
- 257.McPhail KL, Gerwick WH. Three new malyngamides from a Papua New Guinea collection of the marine cyanobacterium Lyngbya majuscula. J Nat Prod. 2003;66:132–135. doi: 10.1021/np0204186. [DOI] [PubMed] [Google Scholar]
- 258.Suntornchashwej S, Suwanborirux K, Koga K, Isobe M. Malyngamide X: The first (7R)- lyngbic acid that connects to a new tripeptide backbone from the Thai sea hare Bursatella leachii. Chemistry–An Asian Journal. 2007;2:114–122. doi: 10.1002/asia.200600219. [DOI] [PubMed] [Google Scholar]
- 259.Suntornchashwej S, Suwanborirux K, Isobe M. Total synthesis of malyngamide X and its 7′S-epi isomer. Tetrahedron. 2007;63:3217–3226. [Google Scholar]
- 260.Sabry OM, Goeger DE, Gerwick WH. Biologically active new metabolites from a Florida collection of Moorea producens. Nat Prod Res. 2016;31:555–561. doi: 10.1080/14786419.2016.1207074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Tan LT, Okino T, Gerwick WH. Hermitamides A and B, toxic malyngamide-type natural products from the marine cyanobacterium Lyngbya majuscula. J Nat Prod. 2000;63:952–955. doi: 10.1021/np000037x. [DOI] [PubMed] [Google Scholar]
- 262.Bonnard I, Rolland M, Salmon JM, Debiton E, Barthomeuf C, Banaigs B. Total structure and inhibition of tumor cell proliferation of laxaphycins. J Med Chem. 2007;50:1266–1279. doi: 10.1021/jm061307x. [DOI] [PubMed] [Google Scholar]
- 263.Boyaud F, Mahiout Z, Lenoir C, Tang S, Wdzieczak-Bakala J, Witczak A, Bonnard I, Banaigs B, Ye T, Inguimbert N. First total synthesis and stereochemical revision of laxaphycin B and its extension to lyngbyacyclamide A. Org Lett. 2013;15:3898–3901. doi: 10.1021/ol401645m. [DOI] [PubMed] [Google Scholar]
- 264.Bonnard I, Rolland M, Francisco C, Banaigs B. Total structure and biological properties of laxaphycins A and B, cyclic lipopeptides from the marine cyanobacterium Lyngbya majuscula. Lett Pept Sci. 1997;4:289–292. [Google Scholar]
- 265.Williams PG, Yoshida WY, Moore RE, Paul VJ. Tasiamide, a cytotoxic peptide from the marine cyanobacterium Symploca sp. J Nat Prod. 2002;65:1336–1339. doi: 10.1021/np020184q. [DOI] [PubMed] [Google Scholar]
- 266.Williams PG, Yoshida WY, Moore RE, Paul VJ. The isolation and structure elucidation of tasiamide B, a 4-amino-3-hydroxy-5-phenylpentanoic acid containing peptide from the marine cyanobacterium Symploca sp. J Nat Prod. 2003;66:1006–1009. doi: 10.1021/np030114z. [DOI] [PubMed] [Google Scholar]
- 267.Mevers E, Haeckl FPJ, Boudreau PD, Byrum T, Dorrestein PC, Valeriote FA, Gerwick WH. Lipopeptides from the tropical marine cyanobacterium Symploca sp. J Nat Prod. 2014;77:969–975. doi: 10.1021/np401051z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Al-Awadhi FH, Ratnayake R, Paul VJ, Luesch H. Tasiamide F, a potent inhibitor of cathepsins D and E from a marine cyanobacterium. Bioorg Med Chem. 2016;24:3276–3282. doi: 10.1016/j.bmc.2016.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Williams PG, Yoshida WY, Moore RE, Paul VJ. Tasipeptins A and B: New cytotoxic depsipeptides from the marine cyanobacterium Symploca sp. J Nat Prod. 2003;66:620–624. doi: 10.1021/np020582t. [DOI] [PubMed] [Google Scholar]
- 270.Linington RG, Edwards DJ, Shuman CF, McPhail KL, Matainaho T, Gerwick WH. Symplocamide A, a potent cytotoxin and chymotrypsin inhibitor from the marine cyanobacterium Symploca sp. J Nat Prod. 2008;71:22–27. doi: 10.1021/np070280x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Molinski TF, Reynolds KA, Morinaka BI. Symplocin A, a linear peptide from the Bahamian cyanobacterium Symploca sp. configurational analysis of N,N-dimethylamino acids by chiral-phase HPLC of naphthacyl esters. J Nat Prod. 2012;75:425–431. doi: 10.1021/np200861n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Salvador LA, Biggs JS, Paul VJ, Luesch H. Veraguamides A−G, cyclic hexadepsipeptides from a dolastatin 16-producing cyanobacterium Symploca cf. hydnoides from Guam. J Nat Prod. 2011;74:917–927. doi: 10.1021/np200076t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Salvador-Reyes LA, Luesch H. Biological targets and mechanisms of action of natural products from marine cyanobacteria. Nat Prod Rep. 2015;32:478–503. doi: 10.1039/c4np00104d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Harrigan GG, Luesch H, Yoshida WY, Moore RE, Nagle DG, Paul VJ. Symplostatin 2: A dolastatin 13 analogue from the marine cyanobacterium Symploca hydnoides. J Nat Prod. 1999;62:655–658. doi: 10.1021/np980553b. [DOI] [PubMed] [Google Scholar]
- 275.Ramaswamy AV, Flatt PM, Edwards DJ, Simmons TL, Han B, Gerwick WH. The secondary metabolites and biosynthetic gene clusters of marine cyanobacteria. Applications in biotechnology. Front Mar Biotechnol: Horizon Biosci. 2006:175–224. [Google Scholar]
- 276.Stolze SC, Deu E, Kaschani F, Li N, Florea BI, Richau KH, Colby T, van der Hoorn RAL, Overkleeft HS, Bogyo M, Kaiser M. The antimalarial natural product symplostatin 4 is a nanomolar inhibitor of the food vacuole falcipains. Chem Biol. 2012;19:1546–1555. doi: 10.1016/j.chembiol.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Horgen FD, Yoshida WY, Scheuer PJ. Malevamides A−C, new depsipeptides from the marine cyanobacterium Symploca laete-viridis. J Nat Prod. 2000;63:461–467. doi: 10.1021/np990449+. [DOI] [PubMed] [Google Scholar]
- 278.Taori K, Paul VJ, Luesch H. Structure and activity of largazole, a potent antiproliferative agent from the Floridian marine cyanobacterium Symploca sp. J Am Chem Soc. 2008;130:1806–1807. doi: 10.1021/ja7110064. [DOI] [PubMed] [Google Scholar]
- 279.Andrianasolo EH, Goeger D, Gerwick WH. Mitsoamide: A cytotoxic linear lipopeptide from the Madagascar marine cyanobacterium Geitlerinema sp. Pure Appl Chem. 2007;79:593–602. [Google Scholar]
- 280.Linington RG, Clark BR, Trimble EE, Almanza A, Ureña LD, Kyle DE, Gerwick WH. Antimalarial peptides from marine cyanobacteria: Isolation and structural elucidation of gallinamide A. J Nat Prod. 2009;72:14–17. doi: 10.1021/np8003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Todorova AK, Juettner F, Linden A, Pluess T, von Philipsborn W. Nostocyclamide: A new macrocyclic, thiazole-containing allelochemical from Nostoc sp. 31 (cyanobacteria) J Org Chem. 1995;60:7891–7895. [Google Scholar]
- 282.Banker R, Carmeli S. Tenuecyclamides A−D, cyclic hexapeptides from the cyanobacterium Nostoc spongiaeforme var. tenue. J Nat Prod. 1998;61:1248–1251. doi: 10.1021/np980138j. [DOI] [PubMed] [Google Scholar]
- 283.Smith CD, Zhang X, Mooberry SL, Patterson GML, Moore RE. Cryptophycin: A new antimicrotubule agent active against drug-resistant cells. Cancer Res. 1994;54:3779–3784. [PubMed] [Google Scholar]
- 284.An J, Carmichael WW. Use of a colorimetric protein phosphatase inhibition assay and enzyme linked immunosorbent assay for the study of microcystins and nodularins. Toxicon. 1994;32:1495–1507. doi: 10.1016/0041-0101(94)90308-5. [DOI] [PubMed] [Google Scholar]
- 285.Admi V, Afek U, Carmeli S. Raocyclamides A and B, novel cyclic hexapeptides isolated from the cyanobacterium Oscillatoria raoi. J Nat Prod. 1996;59:396–399. [Google Scholar]
- 286.Linington RG, González J, Ureña LD, Romero LI, Ortega-Barría E, Gerwick WH. Venturamides A and B: Antimalarial constituents of the Panamanian marine cyanobacterium Oscillatoria sp. J Nat Prod. 2007;70:397–401. doi: 10.1021/np0605790. [DOI] [PubMed] [Google Scholar]
- 287.Harada KI, Fujii K, Shimada T, Suzuki M, Sano H, Adachi K, Carmichael WW. Two cyclic peptides, anabaenopeptins, a third group of bioactive compounds from the cyanobacterium Anabaena flos-aquae NRC 525-17. Tetrahedron Lett. 1995;36:1511–1514. [Google Scholar]
- 288.Fujii K, Sivonen K, Nakano T, Harada KI. Structural elucidation of cyanobacterial peptides encoded by peptide synthetase gene in Anabaena species. Tetrahedron. 2002;58:6863–6871. [Google Scholar]
- 289.Shin HJ, Matsuda H, Murakami M, Yamaguchi K. Anabaenopeptins E and F, two new cyclic peptides from the cyanobacterium Oscillatoria agardhii (NIES-204) J Nat Prod. 1997;60:139–141. [Google Scholar]
- 290.Itou Y, Suzuki S, Ishida K, Murakami M. Anabaenopeptins G and H, potent carboxypeptidase A inhibitors from the cyanobacterium Oscillatoria agardhii (NIES-595) Bioorg Med Chem Lett. 1999;9:1243–1246. doi: 10.1016/s0960-894x(99)00191-2. [DOI] [PubMed] [Google Scholar]
- 291.Medina RA, Goeger DE, Hills P, Mooberry SL, Huang N, Romero LI, Ortega-Barría E, Gerwick WH, McPhail KL. Coibamide A, a potent antiproliferative cyclic depsipeptide from the Panamanian marine cyanobacterium Leptolyngbya sp. J Am Chem Soc. 2008;130:6324–6325. doi: 10.1021/ja801383f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Gerwick WH, Jiang ZD, Agarwal SK, Farmer BT. Total structure of hormothamnin A, a toxic cyclic undecapeptide from the tropical marine cyanobacterium Hormothamnion enteromorphoides. Tetrahedron. 1992;48:2313–2324. [Google Scholar]
- 293.Sudek S, Haygood MG, Youssef DTA, Schmidt EW. Structure of trichamide, a cyclic peptide from the bloom-forming cyanobacterium Trichodesmium erythraeum, predicted from the genome sequence. Appl Environ Microbiol. 2006;72:4382–4387. doi: 10.1128/AEM.00380-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Stevenson CS, Capper EA, Roshak AK, Marquez B, Eichman C, Jackson JR, Mattern M, Gerwick WH, Jacobs RS, Marshall LA. The identification and characterization of the marine natural product scytonemin as a novel antiproliferative pharmacophore. J Pharmacol Exp Ther. 2002;303:858–866. doi: 10.1124/jpet.102.036350. [DOI] [PubMed] [Google Scholar]
- 295.Asolkar RN, Freel KC, Jensen PR, Fenical W, Kondratyuk TP, Park EJ, Pezzuto JM. Arenamides A−C, cytotoxic NFκB inhibitors from the marine actinomycete Salinispora arenicola. J Nat Prod. 2009;72:396–402. doi: 10.1021/np800617a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Gunasekera SP, Imperial L, Garst C, Ratnayake R, Dang LH, Paul VJ, Luesch H. Caldoramide, a modified pentapeptide from the marine cyanobacterium Caldora penicillata. J Nat Prod. 2016;79:1867–1871. doi: 10.1021/acs.jnatprod.6b00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Boudreau PD, Byrum T, Liu WT, Dorrestein PC, Gerwick WH. Viequeamide A, a cytotoxic member of the kulolide superfamily of cyclic depsipeptides from a marine button cyanobacterium. J Nat Prod. 2012;75:1560–1570. doi: 10.1021/np300321b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Vining OB, Medina RA, Mitchell EA, Videau P, Li D, Serrill JD, Kelly JX, Gerwick WH, Proteau PJ, Ishmael JE, McPhail KL. Depsipeptide companeramides from a Panamanian marine cyanobacterium associated with the coibamide producer. J Nat Prod. 2015;78:413–420. doi: 10.1021/np5007907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Fujii K, Sivonen K, Adachi K, Noguchi K, Sano H, Hirayama K, Suzuki M, Harada KI. Comparative study of toxic and non-toxic cyanobacterial products: Novel peptides from toxic Nodularia spumigena AV1. Tetrahedron Lett. 1997;38:5525–5528. [Google Scholar]
- 300.Fewer DP, Jokela J, Rouhiainen L, Wahlsten M, Koskenniemi K, Stal LJ, Sivonen K. The non-ribosomal assembly and frequent occurrence of the protease inhibitors spumigins in the bloom-forming cyanobacterium Nodularia spumigena. Mol Microbiol. 2009;73:924–937. doi: 10.1111/j.1365-2958.2009.06816.x. [DOI] [PubMed] [Google Scholar]
- 301.Fewer DP, Jokela J, Paukku E, Österholm J, Wahlsten M, Permi P, Aitio O, Rouhiainen L, Gomez-Saez GV, Sivonen K. New structural variants of aeruginosin produced by the toxic bloom forming cyanobacterium Nodularia spumigena. PLoS One. 2013;8:e73618. doi: 10.1371/journal.pone.0073618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Harrigan GG, Harrigan BL, Davidson BS. Kailuins A–D, new cyclic acyldepsipeptides from cultures of a marine-derived bacterium. Tetrahedron. 1997;53:1577–1582. [Google Scholar]
- 303.Theodore CM, Lorig-Roach N, Still PC, Johnson TA, Drašković M, Schwochert JA, Naphen CN, Crews MS, Barker SA, Valeriote FA, Lokey RS, Crews P. Biosynthetic products from a nearshore-derived gram-negative bacterium enable reassessment of the kailuin depsipeptides. J Nat Prod. 2015;78:441–452. doi: 10.1021/np500840n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Desriac F, Jégou C, Balnois E, Brillet B, Chevalier PL, Fleury Y. Antimicrobial peptides from marine proteobacteria. Mar Drugs. 2013;11:3632–3660. doi: 10.3390/md11103632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Kjaerulff L, Nielsen A, Mansson M, Gram L, Larsen TO, Ingmer H, Gotfredsen CH. Identification of four new agr quorum sensing-interfering cyclodepsipeptides from a marine Photobacterium. Mar Drugs. 2013;11:5051–5062. doi: 10.3390/md11125051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Oku N, Kawabata K, Adachi K, Katsuta A, Shizuri Y. Unnarmicins A and C, new antibacterial depsipeptides produced by marine bacterium Photobacterium sp. MBIC06485. J Antibiot. 2008;61:11–17. doi: 10.1038/ja.2008.103. [DOI] [PubMed] [Google Scholar]
- 307.Tanabe K, Lamping E, Adachi K, Takano Y, Kawabata K, Shizuri Y, Niimi M, Uehara Y. Inhibition of fungal ABC transporters by unnarmicin A and unnarmicin C, novel cyclic peptides from marine bacterium. Biochem Biophys Res Commun. 2007;364:990–995. doi: 10.1016/j.bbrc.2007.10.110. [DOI] [PubMed] [Google Scholar]
- 308.Oku N, Adachi K, Matsuda S, Kasai H, Takatsuki A, Shizuri Y. Ariakemicins A and B, novel polyketide-peptide antibiotics from a marine gliding bacterium of the genus Rapidithrix. Org Lett. 2008;10:2481–2484. doi: 10.1021/ol8007292. [DOI] [PubMed] [Google Scholar]
- 309.Raju R, Khalil ZG, Piggott AM, Blumenthal A, Gardiner DL, Skinner-Adams TS, Capon RJ. Mollemycin A: An antimalarial and antibacterial glyco-hexadepsipeptide-polyketide from an Australian marine-derived Streptomyces sp. (CMB-M0244) Org Lett. 2014;16:1716–1719. doi: 10.1021/ol5003913. [DOI] [PubMed] [Google Scholar]
- 310.Romero F, Espliego F, Baz JP, De Quesada TG, Grávalos D, De La Calle F, Fernandez-Puentes JL. Thiocoraline, a new depsipeptide with antitumor activity produced by a marine Micromonospora. J Antibiot. 1997;50:734–737. doi: 10.7164/antibiotics.50.734. [DOI] [PubMed] [Google Scholar]
- 311.Renner MK, Shen YC, Cheng XC, Jensen PR, Frankmoelle W, Kauffman CA, Fenical W, Lobkovsky E, Clardy J. Cyclomarins A−C, new antiinflammatory cyclic peptides produced by a marine bacterium (Streptomyces sp.) J Am Chem Soc. 1999;121:11273–11276. [Google Scholar]
- 312.Schultz AW, Oh DC, Carney JR, Williamson RT, Udwary DW, Jensen PR, Gould SJ, Fenical W, Moore BS. Biosynthesis and structures of cyclomarins and cyclomarazines, prenylated cyclic peptides of marine actinobacterial origin. J Am Chem Soc. 2008;130:4507–4516. doi: 10.1021/ja711188x. [DOI] [PubMed] [Google Scholar]
- 313.Takada K, Ninomiya A, Naruse M, Sun Y, Miyazaki M, Nogi Y, Okada S, Matsunaga S. Surugamides A–E, cyclic octapeptides with four D-amino acid residues, from a marine Streptomyces sp.: LC–MS-aided inspection of partial hydrolysates for the distinction of D-and L-amino acid residues in the sequence. J Org Chem. 2013;78:6746–6750. doi: 10.1021/jo400708u. [DOI] [PubMed] [Google Scholar]
- 314.Ninomiya A, Katsuyama Y, Kuranaga T, Miyazaki M, Nogi Y, Okada S, Wakimoto T, Ohnishi Y, Matsunaga S, Takada K. Biosynthetic gene cluster for surugamide A encompasses an unrelated decapeptide, surugamide F. ChemBioChem. 2016;17:1709–1712. doi: 10.1002/cbic.201600350. [DOI] [PubMed] [Google Scholar]
- 315.Pesic A, Baumann HI, Kleinschmidt K, Ensle P, Wiese J, Süssmuth RD, Imhoff JF. Champacyclin, a new cyclic octapeptide from Streptomyces strain C42 isolated from the Baltic Sea. Mar Drugs. 2013;11:4834–4857. doi: 10.3390/md11124834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Motohashi K, Toda T, Sue M, Furihata K, Shizuri Y, Matsuo Y, Kasai H, Shin-Ya K, Takagi M, Izumikawa M, Horikawa Y, Seto H. Isolation and structure elucidation of tumescenamides A and B, two peptides produced by Streptomyces tumescens YM23-260. J Antibiot. 2010;63:549–552. doi: 10.1038/ja.2010.73. [DOI] [PubMed] [Google Scholar]
- 317.Kishimoto S, Tsunematsu Y, Nishimura S, Hayashi Y, Hattori A, Kakeya H. Tumescenamide C, an antimicrobial cyclic lipodepsipeptide from Streptomyces sp. Tetrahedron. 2012;68:5572–5578. [Google Scholar]
- 318.Höltzel A, Jack RW, Nicholson GJ, Jung G, Gebhardt K, Fiedler HP, Süssmuth RD. Streptocidins A–D, novel cyclic decapeptide antibiotics produced by Streptomyces sp. Tü 6071. J Antibiot. 2001;54:434–440. doi: 10.7164/antibiotics.54.434. [DOI] [PubMed] [Google Scholar]
- 319.Qin C, Zhong X, Ng NL, Bu X, Chan WS, Guo Z. Facile solid-phase synthesis of cyclic decapeptide antibiotic streptocidins A–D. Tetrahedron Lett. 2004;45:217–220. [Google Scholar]
- 320.Gebhardt K, Pukall R, Fiedler HP. Streptocidins A–D, novel cyclic decapeptide antibiotics produced by Streptomyces sp. Tü 6071. I. Taxonomy, fermentation, isolation and biological activities. J Antibiot. 2001;54:428–433. doi: 10.7164/antibiotics.54.428. [DOI] [PubMed] [Google Scholar]
- 321.Moore BS, Trischman JA, Seng D, Kho D, Jensen PR, Fenical W. Salinamides, antiinflammatory depsipeptides from a marine streptomycete. J Org Chem. 1999;64:1145–1150. [Google Scholar]
- 322.Hassan HM, Degen D, Jang KH, Ebright RH, Fenical W. Salinamide F, new depsipeptide antibiotic and inhibitor of bacterial RNA polymerase from a marine-derived Streptomyces sp. J Antibiot. 2015;68:206–209. doi: 10.1038/ja.2014.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Desjardine K, Pereira A, Wright H, Matainaho T, Kelly M, Andersen RJ. Tauramamide, a lipopeptide antibiotic produced in culture by Brevibacillus laterosporus isolated from a marine habitat: Structure elucidation and synthesis. J Nat Prod. 2007;70:1850–1853. doi: 10.1021/np070209r. [DOI] [PubMed] [Google Scholar]
- 324.Barsby T, Kelly MT, Andersen RJ. Tupuseleiamides and basiliskamides, new acyldipeptides and antifungal polyketides produced in culture by a Bacillus laterosporus isolate obtained from a tropical marine habitat. J Nat Prod. 2002;65:1447–1451. doi: 10.1021/np0201321. [DOI] [PubMed] [Google Scholar]
- 325.Barsby T, Warabi K, Sørensen D, Zimmerman WT, Kelly MT, Andersen RJ. The bogorol family of antibiotics: Template-based structure elucidation and a new approach to positioning enantiomeric pairs of amino acids. J Org Chem. 2006;71:6031–6037. doi: 10.1021/jo060667p. [DOI] [PubMed] [Google Scholar]
- 326.Gerard JM, Haden P, Kelly MT, Andersen RJ. Loloatins A−D, cyclic decapeptide antibiotics produced in culture by a tropical marine bacterium. J Nat Prod. 1999;62:80–85. doi: 10.1021/np980219f. [DOI] [PubMed] [Google Scholar]
- 327.Zhou X, Huang H, Chen Y, Tan J, Song Y, Zou J, Tian X, Hua Y, Ju J. Marthiapeptide A, an anti-infective and cytotoxic polythiazole cyclopeptide from a 60 L scale fermentation of the deep sea-derived Marinactinospora thermotolerans SCSIO 00652. J Nat Prod. 2012;75:2251–2255. doi: 10.1021/np300554f. [DOI] [PubMed] [Google Scholar]
- 328.Homann VV, Sandy M, Tincu JA, Templeton AS, Tebo BM, Butler A. Loihichelins A–F, a suite of amphiphilic siderophores produced by the marine bacterium Halomonas LOB-5. J Nat Prod. 2009;72:884–888. doi: 10.1021/np800640h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Homann VV, Edwards KJ, Webb EA, Butler A. Siderophores of Marinobacter aquaeolei: Petrobactin and its sulfonated derivatives. Biometals. 2009;22:565–571. doi: 10.1007/s10534-009-9237-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Xu G, Martinez JS, Groves JT, Butler A. Membrane affinity of the amphiphilic marinobactin siderophores. J Am Chem Soc. 2002;124:13408–13415. doi: 10.1021/ja026768w. [DOI] [PubMed] [Google Scholar]
- 331.Martinez JS, Carter-Franklin JN, Mann EL, Martin JD, Haygood MG, Butler A. Structure and membrane affinity of a suite of amphiphilic siderophores produced by a marine bacterium. Proc Natl Acad Sci. 2003;100:3754–3759. doi: 10.1073/pnas.0637444100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Barbeau K, Zhang G, Live DH, Butler A. Petrobactin, a photoreactive siderophore produced by the oil-degrading marine bacterium Marinobacter hydrocarbonoclasticus. J Am Chem Soc. 2002;124:378–379. doi: 10.1021/ja0119088. [DOI] [PubMed] [Google Scholar]
- 333.Haygood MG, Holt PD, Butler A. Aerobactin production by a planktonic marine Vibrio sp. Limnol Oceanogr. 1993;38:1091–1097. [Google Scholar]
- 334.Zane HK, Naka H, Rosconi F, Sandy M, Haygood MG, Butler A. Biosynthesis of amphi-enterobactin siderophores by Vibrio harveyi BAA-1116: Identification of a bifunctional nonribosomal peptide synthetase condensation domain. J Am Chem Soc. 2014;136:5615–5618. doi: 10.1021/ja5019942. [DOI] [PubMed] [Google Scholar]
- 335.Holt PD, Reid RR, Lewis BL, Luther GW, III, Butler A. Iron(III) coordination chemistry of alterobactin A: A siderophore from the marine bacterium Alteromonas luteoviolacea. Inorg Chem. 2005;44:7671–7677. doi: 10.1021/ic0512072. [DOI] [PubMed] [Google Scholar]
- 336.Kanoh K, Kamino K, Leleo G, Adachi K, Shizuri Y. Pseudoalterobactin A and B, new siderophores excreted by marine bacterium Pseudoalteromonas sp. KP20-4. J Antibiot. 2003;56:871–875. doi: 10.7164/antibiotics.56.871. [DOI] [PubMed] [Google Scholar]
- 337.Sandy M, Han A, Blunt J, Munro M, Haygood M, Butler A. Vanchrobactin and anguibactin siderophores produced by Vibrio sp. DS40M4. J Nat Prod. 2010;73:1038–1043. doi: 10.1021/np900750g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Actis LA, Fish W, Crosa JH, Kellerman K, Ellenberger SR, Hauser FM, Sanders-Loehr J. Characterization of anguibactin, a novel siderophore from Vibrio anguillarum 775(pJM1) J Bacteriol. 1986;167:57–65. doi: 10.1128/jb.167.1.57-65.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Yamamoto S, Okujo N, Yoshida T, Matsuura S, Shinoda S. Structure and iron transport activity of vibrioferrin, a new siderophore of Vibrio parahaemolyticus. J Biochem. 1994;115:868–874. doi: 10.1093/oxfordjournals.jbchem.a124432. [DOI] [PubMed] [Google Scholar]
- 340.Martin JD, Ito Y, Homann VV, Haygood MG, Butler A. Structure and membrane affinity of new amphiphilic siderophores produced by Ochrobactrum sp. SP18, JBIC. J Biol Inorg Chem. 2006;11:633–641. doi: 10.1007/s00775-006-0112-y. [DOI] [PubMed] [Google Scholar]
- 341.Ito Y, Butler A. Structure of synechobactins, new siderophores of the marine cyanobacterium Synechococcus sp. PCC 7002. Limnol Oceanogr. 2005;50:1918–1923. [Google Scholar]
- 342.Malmstrøm J. Unguisins A and B: New cyclic peptides from the marine-derived fungus Emericella unguis. J Nat Prod. 1999;62:787–789. doi: 10.1021/np980539z. [DOI] [PubMed] [Google Scholar]
- 343.Hunter L, Chung JH. Total synthesis of unguisin A. J Org Chem. 2011;76:5502–5505. doi: 10.1021/jo200813a. [DOI] [PubMed] [Google Scholar]
- 344.Malmstrøm J, Ryager A, Anthoni U, Nielsen PH. Unguisin C, a GABA-containing cyclic peptide from the fungus Emericella unguis. Phytochemistry. 2002;60:869–872. doi: 10.1016/s0031-9422(02)00150-4. [DOI] [PubMed] [Google Scholar]
- 345.Liu S, Shen Y. A new cyclic peptide from the marine fungal strain Aspergillus sp. AF119. Chem Nat Compd. 2011;47:786–788. [Google Scholar]
- 346.Akone SH, Daletos G, Lin W, Proksch P. Unguisin F, a new cyclic peptide from the endophytic fungus Mucor irregularis. Zeitschrift für Naturforschung C. 2016;71:15–19. doi: 10.1515/znc-2015-0137. [DOI] [PubMed] [Google Scholar]
- 347.Peng J, Gao H, Zhang X, Wang S, Wu C, Gu Q, Guo P, Zhu T, Li D. Psychrophilins E–H and versicotide C, cyclic peptides from the marine-derived fungus Aspergillus versicolor ZLN-60. J Nat Prod. 2014;77:2218–2223. doi: 10.1021/np500469b. [DOI] [PubMed] [Google Scholar]
- 348.Shigemori H, Wakuri S, Yazawa K, Nakamura T, Sasaki T, Kobayashi JI. Fellutamides A and B, cytotoxic peptides from a marine fish-possessing fungus Penicillium fellutanum. Tetrahedron. 1991;47:8529–8534. [Google Scholar]
- 349.Xu D, Ondeyka J, Harris GH, Zink D, Kahn JN, Wang H, Bills G, Platas G, Wang W, Szewczak AA, Liberator P, Roemer T, Singh SB. Isolation, structure, and biological activities of fellutamides C and D from an undescribed Metulocladosporiella (Chaetothyriales) using the genome-wide Candida albicans fitness test. J Nat Prod. 2011;74:1721–1730. doi: 10.1021/np2001573. [DOI] [PubMed] [Google Scholar]
- 350.He F, Bao J, Zhang XY, Tu ZC, Shi YM, Qi SH. Asperterrestide A, a cytotoxic cyclic tetrapeptide from the marine-derived fungus Aspergillus terreus SCSGAF0162. J Nat Prod. 2013;76:1182–1186. doi: 10.1021/np300897v. [DOI] [PubMed] [Google Scholar]
- 351.Belofsky GN, Jensen PR, Fenical W. Sansalvamide: A new cytotoxic cyclic depsipeptide produced by a marine fungus of the genus Fusarium. Tetrahedron Lett. 1999;40:2913–2916. [Google Scholar]
- 352.Yu Z, Lang G, Kajahn I, Schmaljohann R, Imhoff JF. Scopularides A and B, cyclodepsipeptides from a marine sponge-derived fungus, Scopulariopsis brevicaulis. J Nat Prod. 2008;71:1052–1054. doi: 10.1021/np070580e. [DOI] [PubMed] [Google Scholar]
- 353.Lukassen MB, Saei W, Sondergaard TE, Tamminen A, Kumar A, Kempken F, Wiebe MG, Sørensen JL. Identification of the scopularide biosynthetic gene cluster in Scopulariopsis brevicaulis. Mar Drugs. 2015;13:4331–4343. doi: 10.3390/md13074331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Jenkins KM, Renner MK, Jensen PR, Fenical W. Exumolides A and B: Antimicroalgal cyclic depsipeptides produced by a marine fungus of the genus Scytalidium. Tetrahedron Lett. 1998;39:2463–2466. [Google Scholar]
- 355.Capon RJ, Stewart M, Ratnayake R, Lacey E, Gill JH. Citromycetins and bilains A–C: New aromatic polyketides and diketopiperazines from Australian marine-derived and terrestrial Penicillium spp. J Nat Prod. 2007;70:1746–1752. doi: 10.1021/np0702483. [DOI] [PubMed] [Google Scholar]
- 356.Meyer SW, Mordhorst TF, Lee C, Jensen PR, Fenical W, Köck M. Penilumamide, a novel lumazine peptide isolated from the marine-derived fungus, Penicillium sp. CNL-338. Org Biomol Chem. 2010;8:2158–2163. doi: 10.1039/b910629d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Chen M, Shao CL, Fu XM, Kong CJ, She ZG, Wang CY. Lumazine peptides penilumamides B–D and the cyclic pentapeptide asperpeptide A from a Gorgonian-derived Aspergillus sp. fungus. J Nat Prod. 2014;77:1601–1606. doi: 10.1021/np5001686. [DOI] [PubMed] [Google Scholar]
- 358.Oh DC, Kauffman CA, Jensen PR, Fenical W. Induced production of emericellamides A and B from the marine-derived fungus Emericella sp. in competing co-culture. J Nat Prod. 2007;70:515–520. doi: 10.1021/np060381f. [DOI] [PubMed] [Google Scholar]
- 359.Ma JY, Xu LF, Huang WF, Wei BG, Lin GQ. Total synthesis of emericellamide A: A secondary metabolite of marine cyclic depsipeptide with antimicrobial properties. Synlett. 2009;8:1307–1310. [Google Scholar]
- 360.Amagata T, Morinaka BI, Amagata A, Tenney K, Valeriote FA, Lobkovsky E, Clardy J, Crews P. A chemical study of cyclic depsipeptides produced by a sponge-derived fungus. J Nat Prod. 2006;69:1560–1565. doi: 10.1021/np060178k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Wu QX, Crews MS, Draskovic M, Sohn J, Johnson TA, Tenney K, Valeriote FA, Yao XJ, Bjeldanes LF, Crews P. Azonazine, a novel dipeptide from a Hawaiian marine sediment-derived fungus, Aspergillus insulicola. Org Lett. 2010;12:4458–4461. doi: 10.1021/ol101396n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.El Maddah F, Kehraus S, Nazir M, Almeida C, König GM. Insights into the biosynthetic origin of 3-(3-furyl) alanine in Stachylidium sp. 293 K04 tetrapeptides. J Nat Prod. 2016;79:2838–2845. doi: 10.1021/acs.jnatprod.6b00601. [DOI] [PubMed] [Google Scholar]
- 363.Komatsu K, Shigemori H, Kobayashi JI. Dictyonamides A and B, new peptides from marine-derived fungus. J Org Chem. 2001;66:6189–6192. doi: 10.1021/jo0156767. [DOI] [PubMed] [Google Scholar]
- 364.Liang X, Zhang XY, Nong XH, Wang J, Huang ZH, Qi SH. Eight linear peptides from the deep-sea-derived fungus Simplicillium obclavatum EIODSF 020. Tetrahedron. 2016;72:3092–3097. [Google Scholar]
- 365.Yang L, Tan RX, Wang Q, Huang WY, Yin YX. Antifungal cyclopeptides from Halobacillus litoralis YS3106 of marine origin. Tetrahedron Lett. 2002;43:6545–6548. [Google Scholar]
- 366.Zheng J, Zhu H, Hong K, Wang Y, Liu P, Wang X, Peng X, Zhu W. Novel cyclic hexapeptides from marine-derived fungus, Aspergillus sclerotiorum PT06-1. Org Lett. 2009;11:5262–5265. doi: 10.1021/ol902197z. [DOI] [PubMed] [Google Scholar]
- 367.Boot CM, Tenney K, Valeriote FA, Crews P. Highly N-methylated linear peptides produced by an atypical sponge-derived Acremonium sp. J Nat Prod. 2006;69:83–92. doi: 10.1021/np0503653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Gupta S, Krasnoff SB, Roberts DW, Renwick JAA, Brinen LS, Clardy J. Structures of the efrapeptins: Potent inhibitors of mitochondrial ATPase from the fungus Tolypocladium niveum. J Am Chem Soc. 1991;113:707–709. [Google Scholar]
- 369.Chen Z, Song Y, Chen Y, Huang H, Zhang W, Ju J. Cyclic heptapeptides, cordyheptapeptides C–E, from the marine-derived fungus Acremonium persicinum SCSIO 115 and their cytotoxic activities. J Nat Prod. 2012;75:1215–1219. doi: 10.1021/np300152d. [DOI] [PubMed] [Google Scholar]
- 370.Isaka M, Srisanoh U, Lartpornmatulee N, Boonruangprapa T. ES-242 derivatives and cycloheptapeptides from Cordyceps sp. strains BCC 16173 and BCC 16176. J Nat Prod. 2007;70:1601–1604. doi: 10.1021/np070357h. [DOI] [PubMed] [Google Scholar]
- 371.Ding LJ, Yuan W, Liao XJ, Han BN, Wang SP, Li ZY, Xu SH, Zhang W, Lin HW. Oryzamides A–E, cyclodepsipeptides from the sponge-derived fungus Nigrospora oryzae PF18. J Nat Prod. 2016;79:2045–2052. doi: 10.1021/acs.jnatprod.6b00349. [DOI] [PubMed] [Google Scholar]
- 372.Hambley TW, Hawkins CJ, Lavin MF, van den Brenk A, Watters DJ. Cycloxazoline: A cytotoxic cyclic hexapeptide from the ascidian Lissoclinum bistratum. Tetrahedron. 1992;48:341–348. [Google Scholar]
- 373.Lindquist N, Fenical W, Van Duyne GD, Clardy J. Isolation and structure determination of diazonamides A and B, unusual cytotoxic metabolites from the marine ascidian Diazona chinensis. J Am Chem Soc. 1991;113:2303–2304. [Google Scholar]
- 374.Cruz-Monserrate Z, Vervoort HC, Bai R, Newman DJ, Howell SB, Los G, Mullaney JT, Williams MD, Pettit GR, Fenical W, Hamel E. Diazonamide A and a synthetic structural analog: Disruptive effects on mitosis and cellular microtubules and analysis of their interactions with tubulin. Mol Pharmacol. 2003;63:1273–1280. doi: 10.1124/mol.63.6.1273. [DOI] [PubMed] [Google Scholar]
- 375.Li J, Burgett AWG, Esser L, Amezcua C, Harran PG. Total synthesis of nominal diazonamides—Part 2: On the true structure and origin of natural isolates. Angew Chem, Int Ed. 2001;40:4770–4773. doi: 10.1002/1521-3773(20011217)40:24<4770::aid-anie4770>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 376.Fernández R, Martín MJ, Rodríguez-Acebes R, Reyes F, Francesch A, Cuevas C. Diazonamides C–E, new cytotoxic metabolites from the ascidian Diazona sp. Tetrahedron Lett. 2008;49:2283–2285. [Google Scholar]
- 377.Edler MC, Fernandez AM, Lassota P, Ireland CM, Barrows LR. Inhibition of tubulin polymerization by vitilevuamide, a bicyclic marine peptide, at a site distinct from colchicine, the vinca alkaloids, and dolastatin 10. Biochem Pharmacol. 2002;63:707–715. doi: 10.1016/s0006-2952(01)00898-x. [DOI] [PubMed] [Google Scholar]
- 378.Bertram A, Pattenden G. Marine metabolites: Metal binding and metal complexes of azole-based cyclic peptides of marine origin. Nat Prod Rep. 2007;24:18–30. doi: 10.1039/b612600f. [DOI] [PubMed] [Google Scholar]
- 379.Wipf P, Fritch PC, Geib SJ, Sefler AM. Conformational studies and structure-activity analysis of lissoclinamide 7 and related cyclopeptide alkaloids. J Am Chem Soc. 1998;120:4105–4112. [Google Scholar]
- 380.Degnan BM, Hawkins CJ, Lavin MF, McCaffrey EJ, Parry DL, Watters DJ. Novel cytotoxic compounds from the ascidian Lissoclinum bistratum. J Med Chem. 1989;32:1354–1359. doi: 10.1021/jm00126a035. [DOI] [PubMed] [Google Scholar]
- 381.Foster MP, Concepcion GP, Caraan GB, Ireland CM. Bistratamides C and D. Two new oxazole-containing cyclic hexapeptides isolated from a Philippine Lissoclinum bistratum ascidian. J Org Chem. 1992;57:6671–6675. [Google Scholar]
- 382.Perez LJ, Faulkner DJ. Bistratamides E−J, modified cyclic hexapeptides from the Philippines ascidian Lissoclinum bistratum. J Nat Prod. 2003;66:247–250. doi: 10.1021/np0204601. [DOI] [PubMed] [Google Scholar]
- 383.Rinehart KL, Kishore V, Bible KC, Sakai R, Sullins DW, Li KM. Didemnins and tunichlorin: Novel natural products from the marine tunicate Trididemnum solidum. J Nat Prod. 1988;51:1–21. doi: 10.1021/np50055a001. [DOI] [PubMed] [Google Scholar]
- 384.Sakai R, Stroh JG, Sullins DW, Rinehart KL. Seven new didemnins from the marine tunicate Trididemnum solidum. J Am Chem Soc. 1995;117:3734–3748. [Google Scholar]
- 385.Grubb DR, Wolvetang EJ, Lawen A. Didemnin B induces cell death by apoptosis: The fastest induction of apoptosis ever described. Biochem Biophys Res Commun. 1995;215:1130–1136. doi: 10.1006/bbrc.1995.2580. [DOI] [PubMed] [Google Scholar]
- 386.Boulanger A, Abou-Mansour E, Badre A, Banaigs B, Combaut G, Francisco C. The complete spectral assignment of didemnin H a new constituent of the tunicate Trididemnum cyanophorum. Tetrahedron Lett. 1994;35:4345–4348. [Google Scholar]
- 387.Taylor SW, Craig AG, Fischer WH, Park M, Lehrer RI. Styelin D, an extensively modified antimicrobial peptide from ascidian hemocytes. J Biol Chem. 2000;275:38417–38426. doi: 10.1074/jbc.M006762200. [DOI] [PubMed] [Google Scholar]
- 388.Swersey JC, Ireland CM, Cornell LM, Peterson RW. Eusynstyelamide, a highly modified dimer peptide from the ascidian Eusynstyela misakiensis. J Nat Prod. 1994;57:842–845. doi: 10.1021/np50108a027. [DOI] [PubMed] [Google Scholar]
- 389.Whitson EL, Ratnayake AS, Bugni TS, Harper MK, Ireland CM. Isolation, structure elucidation, and synthesis of eudistomides A and B, lipopeptides from a Fijian ascidian Eudistoma sp. J Org Chem. 2009;74:1156–1162. doi: 10.1021/jo8022582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Carroll AR, Bowden BF, Coll JC, Hockless DCR, Skelton BW, White AH. Studies of Australian ascidians. IV. Mollamide, a cytotoxic cyclic heptapeptide from the compound ascidian Didemnum molle. Aust J Chem. 1994;47:61–69. [Google Scholar]
- 391.Donia MS, Wang B, Dunbar DC, Desai PV, Patny A, Avery M, Hamann MT. Mollamides B and C, cyclic hexapeptides from the Indonesian tunicate Didemnum molle. J Nat Prod. 2008;71:941–945. doi: 10.1021/np700718p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Wesson KJ, Hamann MT. Keenamide A, a bioactive cyclic peptide from the marine mollusk Pleurobranchus forskalii. J Nat Prod. 1996;59:629–631. doi: 10.1021/np960153t. [DOI] [PubMed] [Google Scholar]
- 393.Ciavatta ML, Lefranc F, Carbone M, Mollo E, Gavagnin M, Betancourt T, Dasari R, Kornienko A, Kiss R. Marine mollusk-derived agents with antiproliferative activity as promising anticancer agents to overcome chemotherapy resistance. Med Res Rev. 2017;37:702–801. doi: 10.1002/med.21423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Carroll AR, Feng Y, Bowden BF, Coll JC. Studies of Australian ascidians. 5. Virenamides A–C, new cytotoxic linear peptides from the colonial didemnid ascidian Diplosoma virens. J Org Chem. 1996;61:4059–4061. doi: 10.1021/jo951379o. [DOI] [PubMed] [Google Scholar]
- 395.Toske SG, Fenical W. Cyclodidemnamide: A new cyclic heptapeptide from the marine ascidian Didemnum molle. Tetrahedron Lett. 1995;36:8355–8358. [Google Scholar]
- 396.Rudi A, Aknin M, Gaydou EM, Kashman Y. Four new cytotoxic cyclic hexa- and heptapeptides from the marine ascidian Didemnum molle. Tetrahedron. 1998;54:13203–13210. [Google Scholar]
- 397.Rudi A, Chill L, Aknin M, Kashman Y. Didmolamide A and B, two new cyclic hexapeptides from the marine ascidian Didemnum molle. J Nat Prod. 2003;66:575–577. doi: 10.1021/np020531w. [DOI] [PubMed] [Google Scholar]
- 398.Fu X, Su J, Zeng L. Prepatellamide A, a new cyclic peptide from the ascidian Lissoclinum patella. Sci China, Ser B: Chem. 2000;43:643–648. [Google Scholar]
- 399.Vervoort H, Fenical W, Epifanio RDA. Tamandarins A and B: New cytotoxic depsipeptides from a Brazilian ascidian of the family Didemnidae. J Org Chem. 2000;65:782–792. doi: 10.1021/jo991425a. [DOI] [PubMed] [Google Scholar]
- 400.Carroll AR, Coll JC, Bourne DJ, MacLeod JK, Zabriskie TM, Ireland CM, Bowden BF. Patellins 1-6 and trunkamide A: Novel cyclic hexa-, hepta- and octa-peptides from colonial ascidians, Lissoclinum sp. Aust J Chem. 1996;49:659–667. [Google Scholar]
- 401.Nakao Y, Yoshida WY, Szabo CM, Baker BJ, Scheuer PJ. More peptides and other diverse constituents of the marine mollusk Philinopsis speciosa. J Org Chem. 1998;63:3272–3280. [Google Scholar]
- 402.Patil AD, Freyer AJ, Killmer L, Chambers-Myers C, Johnson RK. Preulithiacyclamide, a new cyclic peptide from the ascidian Lissoclinum patella. Nat Prod Lett. 1997;9:181–187. [Google Scholar]
- 403.Ireland C, Scheuer PJ. Ulicyclamide and ulithiacyclamide, two new small peptides from a marine tunicate. J Am Chem Soc. 1980;102:5688–5691. [Google Scholar]
- 404.Williams DE, Moore RE, Paul VJ. The structure of ulithiacyclamide B. Antitumor evaluation of cyclic peptides and macrolides from Lissoclinum patella. J Nat Prod. 1989;52:732–739. doi: 10.1021/np50064a011. [DOI] [PubMed] [Google Scholar]
- 405.Gorlero M, Wieczorek R, Adamala K, Giorgi A, Schininà ME, Stano P, Luisi PL. Ser- His catalyses the formation of peptides and PNAs. FEBS Lett. 2009;583:153–156. doi: 10.1016/j.febslet.2008.11.052. [DOI] [PubMed] [Google Scholar]
- 406.Lin Z, Reilly CA, Antemano R, Hughen RW, Marett L, Concepcion GP, Haygood MG, Olivera BM, Light A, Schmidt EW. Nobilamides A–H, long-acting transient receptor potential vanilloid-1 (TRPV1) antagonists from mollusk-associated bacteria. J Med Chem. 2011;54:3746–3755. doi: 10.1021/jm101621u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Mitchell SS, Faulkner DJ, Rubins K, Bushman FD. Dolastatin 3 and two novel cyclic peptides from a Palauan collection of Lyngbya majuscula. J Nat Prod. 2000;63:279–282. doi: 10.1021/np990353f. [DOI] [PubMed] [Google Scholar]
- 408.Pettit GR, Kamano Y, Herald CL, Fujii Y, Kizu H, Boyd MR, Boettner FE, Doubek DL, Schmidt JM, Chapuis JC, Michel C. Isolation of dolastatins 10–15 from the marine mollusc Dolabella auricularia. Tetrahedron. 1993;49:9151–9170. [Google Scholar]
- 409.Davies-Coleman MT, Dzeha TM, Gray CA, Hess S, Pannell LK, Hendricks DT, Arendse CE. Isolation of homodolastatin 16, a new cyclic depsipeptide from a Kenyan collection of Lyngbya majuscula. J Nat Prod. 2003;66:712–715. doi: 10.1021/np030014t. [DOI] [PubMed] [Google Scholar]
- 410.Sone H, Nemoto T, Ojika M, Yamada K. Isolation, structure, and synthesis of dolastatin C, a new depsipeptide from the sea hare Dolabella auricularia. Tetrahedron Lett. 1993;34:8445–8448. [Google Scholar]
- 411.Ojika M, Nemoto T, Nakamura M, Yamada K. Dolastatin E, a new cyclic hexapeptide isolated from the sea hare Dolabella auricularia. Tetrahedron Lett. 1995;36:5057–5058. [Google Scholar]
- 412.Luesch H, Yoshida WY, Moore RE, Paul VJ. Lyngbyastatin 2 and norlyngbyastatin 2, analogues of dolastatin G and nordolastatin G from the marine cyanobacterium Lyngbya majuscula. J Nat Prod. 1999;62:1702–1706. doi: 10.1021/np990310z. [DOI] [PubMed] [Google Scholar]
- 413.Sone H, Kigoshi H, Yamada K. Isolation and stereostructure of dolastatin I, a cytotoxic cyclic hexapeptide from the Japanese sea hare Dolabella auricularia. Tetrahedron. 1997;53:8149–8154. [Google Scholar]
- 414.Sone H, Kondo T, Kiryu M, Ishiwata H, Ojika M, Yamada K. Dolabellin, a cytotoxic bisthiazole metabolite from the sea hare Dolabella auricularia: Structural determination and synthesis. J Org Chem. 1995;60:4774–4781. [Google Scholar]
- 415.Ishiwata H, Nemoto T, Ojika M, Yamada K. Isolation and stereostructure of doliculide, a cytotoxic cyclodepsipeptide from the Japanese sea hare Dolabella auricularia. J Org Chem. 1994;59:4710–4711. [Google Scholar]
- 416.Kimura J, Takada Y, Inayoshi T, Nakao Y, Goetz G, Yoshida WY, Scheuer PJ. Kulokekahilide-1, a cytotoxic depsipeptide from the cephalaspidean mollusk Philinopsis speciosa. J Org Chem. 2002;67:1760–1767. doi: 10.1021/jo010176z. [DOI] [PubMed] [Google Scholar]
- 417.Nakao Y, Yoshida WY, Takada Y, Kimura J, Yang L, Mooberry SL, Scheuer PJ. Kulokekahilide-2, a cytotoxic depsipeptide from a cephalaspidean mollusk Philinopsis speciosa. J Nat Prod. 2004;67:1332–1340. doi: 10.1021/np049949f. [DOI] [PubMed] [Google Scholar]
- 418.Suenaga K, Mutou T, Shibata T, Itoh T, Fujita T, Takada N, Hayamizu K, Takagi M, Irifune T, Kigoshi H, Yamada K. Aurilide, a cytotoxic depsipeptide from the sea hare Dolabella auricularia: Isolation, structure determination, synthesis, and biological activity. Tetrahedron. 2004;60:8509–8527. [Google Scholar]
- 419.Simmons TL, Engene N, Ureña LD, Romero LI, Ortega-Barría E, Gerwick L, Gerwick WH. Viridamides A and B, lipodepsipeptides with antiprotozoal activity from the marine cyanobacterium Oscillatoria nigro-viridis. J Nat Prod. 2008;71:1544–1550. doi: 10.1021/np800110e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Koehn FE, Longley RE, Reed JK. Microcolins A and B, new immunosuppressive peptides from the blue-green alga Lyngbya majuscula. J Nat Prod. 1992;55:613–619. doi: 10.1021/np50083a009. [DOI] [PubMed] [Google Scholar]
- 421.Andrus MB, Li W, Keyes RF. Synthesis of microcolin B, a potent new immunosuppressant using an efficient mixed imide formation reaction. J Org Chem. 1997;62:5542–5549. [Google Scholar]
- 422.Ainslie RD, Barchi JJ, Jr, Kuniyoshi M, Moore RE, Mynderse JS. Structure of malyngamide C. J Org Chem. 1985;50:2859–2862. [Google Scholar]
- 423.Engene N, Paul VJ, Byrum T, Gerwick WH, Thor A, Ellisman MH. Five chemically rich species of tropical marine cyanobacteria of the genus Okeania gen. nov. (Oscillatoriales, Cyanoprokaryota) J Phycol. 2013;49:1095–1106. doi: 10.1111/jpy.12115. [DOI] [PubMed] [Google Scholar]
- 424.Zhang LH, Longley RE. Induction of apoptosis in mouse thymocytes by microcolin A and its synthetic analog. Life Sci. 1999;64:1013–1028. doi: 10.1016/s0024-3205(99)00028-4. [DOI] [PubMed] [Google Scholar]
- 425.Clark BR, Engene N, Teasdale ME, Rowley DC, Matainaho T, Valeriote FA, Gerwick WH. Natural products chemistry and taxonomy of the marine cyanobacterium Blennothrix cantharidosmum. J Nat Prod. 2008;71:1530–1537. doi: 10.1021/np800088a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Ogawa H, Iwasaki A, Sumimoto S, Kanamori Y, Ohno O, Iwatsuki M, Ishiyama A, Hokari R, Otoguro K, O̅mura S, Suenaga K. Janadolide, a cyclic polyketide– peptide hybrid possessing a tert-butyl group from an Okeania sp. marine cyanobacterium. J Nat Prod. 2016;79:1862–1866. doi: 10.1021/acs.jnatprod.6b00171. [DOI] [PubMed] [Google Scholar]
- 427.Rodríguez J, Fernández R, Quiñoá E, Riguera R, Debitus C, Bouchet P. Onchidin: A cytotoxic depsipeptide with C2 symmetry from a marine mollusc. Tetrahedron Lett. 1994;35:9239–9242. [Google Scholar]
- 428.Fernández R, Rodríguez J, Quiñoá E, Riguera R, Muñoz L, Fernández-Suárez M, Debitus C. Onchidin B: A new cyclodepsipeptide from the mollusc Onchidium sp. J Am Chem Soc. 1996;118:11635–11643. [Google Scholar]
- 429.Wu ZC, Li S, Nam SJ, Liu Z, Zhang C. Nocardiamides A and B, two cyclohexapeptides from the marine-derived actinomycete Nocardiopsis sp. CNX037. J Nat Prod. 2013;76:694–701. doi: 10.1021/np400009a. [DOI] [PubMed] [Google Scholar]
- 430.Hamann MT, Otto CS, Scheuer PJ, Dunbar DC. Kahalalides: Bioactive peptides from a marine mollusk Elysia rufescens and its algal diet Bryopsis sp. J Org Chem. 1996;61:6594–6600. doi: 10.1021/jo960877+. [DOI] [PubMed] [Google Scholar]
- 431.Suárez Y, González L, Cuadrado A, Berciano M, Lafarga M, Muñoz A. Kahalalide F, a new marine-derived compound, induces oncosis in human prostate and breast cancer cells. Mol Cancer Ther. 2003;2:863–872. [PubMed] [Google Scholar]
- 432.Gao J, Hamann MT. Chemistry and biology of kahalalides. Chem Rev. 2011;111:3208–3235. doi: 10.1021/cr100187n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Tan LT, Williamson RT, Gerwick WH, Watts KS, McGough K, Jacobs R. cis, cis- and trans,trans-Ceratospongamide, new bioactive cyclic heptapeptides from the Indonesian red alga Ceratodictyon spongiosum and symbiotic sponge Sigmadocia symbiotica. J Org Chem. 2000;65:419–425. doi: 10.1021/jo991165x. [DOI] [PubMed] [Google Scholar]
- 434.Iwasaki A, Ohno O, Sumimoto S, Matsubara T, Shimada S, Sato T, Suenaga K. Mebamamides A and B, cyclic lipopeptides isolated from the green alga Derbesia marina. J Nat Prod. 2015;78:901–908. doi: 10.1021/acs.jnatprod.5b00168. [DOI] [PubMed] [Google Scholar]
- 435.Xu WJ, Liao XJ, Xu SH, Diao JZ, Du B, Zhou XL, Pan SS. Isolation, structure determination, and synthesis of galaxamide, a rare cytotoxic cyclic pentapeptide from a marine algae Galaxaura filamentosa. Org Lett. 2008;10:4569–4572. doi: 10.1021/ol801799d. [DOI] [PubMed] [Google Scholar]
- 436.Xiao X, Liao X, Qiu S, Liu Z, Du B, Xu S. Synthesis, cytotoxicity and apoptosis induction in human tumor cells by galaxamide and its analogues. Mar Drugs. 2014;12:4521–4538. doi: 10.3390/md12084521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Mena EE, Gullak MF, Pagnozzi MJ, Richter KE, Rivier J, Cruz LJ, Olivera BM. Conantokin-G: A novel peptide antagonist to the N-methyl-D-aspartic acid (NMDA) receptor. Neurosci Lett. 1990;118:241–244. doi: 10.1016/0304-3940(90)90637-o. [DOI] [PubMed] [Google Scholar]
- 438.Haack JA, Rivier J, Parks TN, Mena EE, Cruz LJ, Olivera BM. Conantokin-T. A gamma-carboxyglutamate containing peptide with N-methyl-d-aspartate antagonist activity. J Biol Chem. 1990;265:6025–6029. [PubMed] [Google Scholar]
- 439.Rivier J, Galyean R, Simon L, Cruz LJ, Olivera BM, Gray WR. Total synthesis and further characterization of the γ-carboxyglutamate-containing “sleeper” peptide from Conus geographus venom. Biochemistry. 1987;26:8508–8512. doi: 10.1021/bi00400a002. [DOI] [PubMed] [Google Scholar]
- 440.Maček P, Belmonte G, Pederzolli C, Menestrina G. Mechanism of action of equinatoxin II, a cytolysin from the sea anemone Actinia equina L. belonging to the family of actinoporins. Toxicology. 1994;87:205–227. doi: 10.1016/0300-483x(94)90252-6. [DOI] [PubMed] [Google Scholar]
- 441.Anand TP, Chellaram C, Kuberan G, Archana H. Bioactive peptides from marine sources — A review. Indian J Innov Dev. 2012;1:61–64. [Google Scholar]
- 442.Ennamany R, Saboureau D, Mekideche N, Creppy EE. SECMA 1®, a mitogenic hexapeptide from Ulva algeae modulates the production of proteoglycans and glycosaminoglycans in human foreskin fibroblast. Hum Exp Toxicol. 1998;17:18–22. doi: 10.1177/096032719801700103. [DOI] [PubMed] [Google Scholar]
- 443.Stensvåg K, Haug T, Sperstad SV, Rekdal Ø, Indrevoll B, Styrvold OB. Arasin 1, a proline–arginine-rich antimicrobial peptide isolated from the spider crab, Hyas araneus. Dev Comp Immunol. 2008;32:275–285. doi: 10.1016/j.dci.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 444.Ovchinnikova TV, Aleshina GM, Balandin SV, Krasnosdembskaya AD, Markelov ML, Frolova EI, Leonova YF, Tagaev AA, Krasnodembsky EG, Kokryakov VN. Purification and primary structure of two isoforms of arenicin, a novel antimicrobial peptide from marine polychaeta Arenicola marina. FEBS Lett. 2004;577:209–214. doi: 10.1016/j.febslet.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 445.Ayoub M, Scheidegger D. Peptide drugs, overcoming the challenges, a growing business. Chim Oggi. 2006;24:46–48. [Google Scholar]
- 446.Otvos L, Jr, Wade JD. Current challenges in peptide-based drug discovery. Front Chem. 2014;2:62. doi: 10.3389/fchem.2014.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Balkovec JM, Hughes DL, Masurekar PS, Sable CA, Schwartz RE, Singh SB. Discovery and development of first in class antifungal caspofungin (CANCIDAS®) — A case study. Nat Prod Rep. 2014;31:15–34. doi: 10.1039/c3np70070d. [DOI] [PubMed] [Google Scholar]
- 448.Moore BS. Biosynthesis of marine natural products: Microorganisms (Part A) Nat Prod Rep. 2005;22:580–593. doi: 10.1039/b404737k. [DOI] [PubMed] [Google Scholar]
- 449.Wang H, Fewer DP, Holm L, Rouhiainen L, Sivonen K. Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc Natl Acad Sci U S A. 2014;111:9259–9264. doi: 10.1073/pnas.1401734111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Micallef ML, D’Agostino PM, Al-Sinawi B, Neilan BA, Moffitt MC. Exploring cyanobacterial genomes for natural product biosynthesis pathways. Mar Genomics. 2015;21:1–12. doi: 10.1016/j.margen.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 451.Dittmann E, Gugger M, Sivonen K, Fewer DP. Natural product biosynthetic diversity and comparative genomics of the cyanobacteria. Trends Microbiol. 2015;23:642–652. doi: 10.1016/j.tim.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 452.Edwards DJ, Gerwick WH. Lyngbyatoxin biosynthesis: Sequence of biosynthetic gene cluster and identification of a novel aromatic prenyltransferase. J Am Chem Soc. 2004;126:11432–11433. doi: 10.1021/ja047876g. [DOI] [PubMed] [Google Scholar]
- 453.Quinn RA, Nothias LF, Vining O, Meehan M, Esquenazi E, Dorrestein PC. Molecular networking as a drug discovery, drug metabolism, and precision medicine strategy. Trends Pharmacol Sci. 2017;38:143–154. doi: 10.1016/j.tips.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 454.Rouhiainen L, Jokela J, Fewer DP, Urmann M, Sivonen K. Two alternative starter modules for the non-ribosomal biosynthesis of specific anabaenopeptin variants in Anabaena (Cyanobacteria) Chem Biol. 2010;17:265–273. doi: 10.1016/j.chembiol.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 455.Lombó F, Velasco A, Castro A, De la Calle F, Braña AF, Sánchez-Puelles JM, Méndez C, Salas JA. Deciphering the biosynthesis pathway of the antitumor thiocoraline from a marine actinomycete and its expression in two Streptomyces species. ChemBioChem. 2006;7:366–376. doi: 10.1002/cbic.200500325. [DOI] [PubMed] [Google Scholar]
- 456.Al-Mestarihi AH, Villamizar G, Fernández J, Zolova OE, Lombó F, Garneau-Tsodikova S. Adenylation and S-methylation of cysteine by the bifunctional enzyme TioN in thiocoraline biosynthesis. J Am Chem Soc. 2014;136:17350–17354. doi: 10.1021/ja510489j. [DOI] [PubMed] [Google Scholar]
- 457.Ray L, Yamanaka K, Moore BS. A peptidyl-transesterifying type I thioesterase in salinamide biosynthesis. Angew Chem. 2016;128:372–375. doi: 10.1002/anie.201508576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Kem MP, Butler A. Acyl peptidic siderophores: Structures, biosyntheses and post-assembly modifications. Biometals. 2015;28:445–459. doi: 10.1007/s10534-015-9827-y. [DOI] [PubMed] [Google Scholar]
- 459.Freeman MF, Gurgui C, Helf MJ, Morinaka BI, Uria AR, Oldham NJ, Sahl HG, Matsunaga S, Piel J. Metagenome mining reveals polytheonamides as posttranslationally modified ribosomal peptides. Science. 2012;338:387–390. doi: 10.1126/science.1226121. [DOI] [PubMed] [Google Scholar]
- 460.Smith DRM, Uria AR, Helfrich EJN, Milbredt D, van Pée KH, Piel J, Goss RJM. An unusual flavin-dependent halogenase from the metagenome of the marine sponge Theonella swinhoei WA. ACS Chem Biol. 2017;12:1281–1287. doi: 10.1021/acschembio.6b01115. [DOI] [PubMed] [Google Scholar]
- 461.Hoffmann T, Müller S, Nadmid S, Garcia R, Müller R. Microsclerodermins from terrestrial myxobacteria: An intriguing biosynthesis likely connected to a sponge symbiont. J Am Chem Soc. 2013;135:16904–16911. doi: 10.1021/ja4054509. [DOI] [PubMed] [Google Scholar]
- 462.Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG, Ravel J. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc Natl Acad Sci U S A. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Mayer AMS, Glaser KB, Cuevas C, Jacobs RS, Kem W, Little RD, McIntosh JM, Newman DJ, Potts BC, Shuster DE. The odyssey of marine pharmaceuticals: A current pipeline perspective. Trends Pharmacol Sci. 2010;31:255–265. doi: 10.1016/j.tips.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 464.Haefner B. Drugs from the deep: Marine natural products as drug candidates. Drug Discovery Today. 2003;8:536–544. doi: 10.1016/s1359-6446(03)02713-2. [DOI] [PubMed] [Google Scholar]
- 465.Cheung RCF, Ng TB, Wong JH. Marine peptides: Bioactivities and applications. Mar Drugs. 2015;13:4006–4043. doi: 10.3390/md13074006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Miljanich GP. Ziconotide: Neuronal calcium channel blocker for treating severe chronic pain. Curr Med Chem. 2004;11:3029–3040. doi: 10.2174/0929867043363884. [DOI] [PubMed] [Google Scholar]
- 467.Lee JY, Orlikova B, Diederich M. Signal transducers and activators of transcription (STAT) regulatory networks in marine organisms: From physiological observations towards marine drug discovery. Mar Drugs. 2015;13:4967–4984. doi: 10.3390/md13084967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Kolosov A, Goodchild CS, Cooke I. CNSB004 (Leconotide) causes antihyperalgesia without side effects when given intravenously: A comparison with ziconotide in a rat model of diabetic neuropathic pain. Pain Med. 2010;11:262–273. doi: 10.1111/j.1526-4637.2009.00741.x. [DOI] [PubMed] [Google Scholar]
- 469.Newman DJ, Cragg GM. Marine-sourced anti-cancer and cancer pain control agents in clinical and late preclinical development. Mar Drugs. 2014;12:255–278. doi: 10.3390/md12010255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Nielsen CK, Lewis RJ, Alewood D, Drinkwater R, Palant E, Patterson M, Yaksh TL, McCumber D, Smith MT. Anti-allodynic efficacy of the χ-conopeptide, Xen2174, in rats with neuropathic pain. Pain. 2005;118:112–124. doi: 10.1016/j.pain.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 471.Younes A, Yasothan U, Kirkpatrick P. Brentuximab vedotin. Nat Rev Drug Discovery. 2012;11:19–20. doi: 10.1038/nrd3629. [DOI] [PubMed] [Google Scholar]
- 472.Waters AL, Hill RT, Place AR, Hamann MT. The expanding role of marine microbes in pharmaceutical development. Curr Opin Biotechnol. 2010;21:780–786. doi: 10.1016/j.copbio.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Jordan MA, Walker D, de Arruda M, Barlozzari T, Panda D. Suppression of microtubule dynamics by binding of cemadotin to tubulin: Possible mechanism for its antitumor action. Biochemistry. 1998;37:17571–17578. doi: 10.1021/bi9817414. [DOI] [PubMed] [Google Scholar]
- 474.Ling YH, Aracil M, Jimeno J, Perez-Soler R, Zou Y. Molecular pharmacodynamics of PM02734 (elisidepsin) as single agent and in combination with erlotinib; Synergistic activity in human non-small cell lung cancer cell lines and xenograft models. Eur J Cancer. 2009;45:1855–1864. doi: 10.1016/j.ejca.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Serova M, de Gramont A, Bieche I, Riveiro ME, Galmarini CM, Aracil M, Jimeno J, Faivre S, Raymond E. Predictive factors of sensitivity to elisidepsin, a novel kahalalide F-derived marine compound. Mar Drugs. 2013;11:944–959. doi: 10.3390/md11030944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Loganzo F, Discafani CM, Annable T, Beyer C, Musto S, Hari M, Tan X, Hardy C, Hernandez R, Baxter M, Singanallore T, Khafizova G, Poruchynsky MS, Fojo T, Nieman JA, Ayral-Kaloustian S, Zask A, Andersen RJ, Greenberger LM. HTI-286, a synthetic analogue of the tripeptide hemiasterlin, is a potent antimicrotubule agent that circumvents P-glycoprotein-mediated resistance in vitro and in vivo. Cancer Res. 2003;63:1838–1845. [PubMed] [Google Scholar]
- 477.Naumovski L, Junutula JR. Glembatumumab vedotin, a conjugate of an anti-glycoprotein non-metastatic melanoma protein B mAb and monomethyl auristatin E for the treatment of melanoma and breast cancer. Curr Opin Mol Ther. 2010;12:248–257. [PubMed] [Google Scholar]
- 478.Challita-Eid PM, Satpayev D, Yang P, An Z, Morrison K, Shostak Y, Raitano A, Nadell R, Liu W, Lortie DR, Capo L, Verlinsky A, Leavitt M, Malik F, Aviña H, Guevara CI, Dinh N, Karki S, Anand BS, Pereira DS, Joseph IBJ, Doñate F, Morrison K, Stover DR. Enfortumab vedotin antibody–drug conjugate targeting nectin-4 is a highly potent therapeutic agent in multiple preclinical cancer models. Cancer Res. 2016;76:3003–3013. doi: 10.1158/0008-5472.CAN-15-1313. [DOI] [PubMed] [Google Scholar]
- 479.Jagadeesh D, Smith MR. Antibody drug conjugates (ADCs): Changing the treatment landscape of lymphoma. Curr Treat Options Oncol. 2016;17:55. doi: 10.1007/s11864-016-0428-y. [DOI] [PubMed] [Google Scholar]
- 480.Wang J, Anderson MG, Oleksijew A, Vaidya KS, Erwin B, Tucker L, Zhang Q, Han EK, Palma JP, Naumovski L, Reilly EB. ABBV-399, a c-Met antibody–drug conjugate that targets both MET–amplified and c-Met–overexpressing tumors, irrespective of MET pathway dependence. Clin Cancer Res. 2017;23:992–1000. doi: 10.1158/1078-0432.CCR-16-1568. [DOI] [PubMed] [Google Scholar]
- 481.Cohen AD, Popat R, Trudel S, Richardson PG, Libby EN, III, Lendvai N, Anderson LD, Jr, Sutherland HJ, DeWall S, Ellis CE, He Z, Mazumdar J, Wang C, Opalinska JB, Voorhees PM. First in human study with GSK2857916, an antibody drug conjugated to microtubule-disrupting agent directed against B-cell maturation antigen (BCMA) in patients with relapsed/refractory multiple myeloma (MM): Results from study BMA117159 Part 1 dose escalation. Blood. 2016;128:1148. [Google Scholar]
- 482.Mayer AMS. Marine Pharmaceuticals: The Clinical Pipeline. http://marinepharmacology.midwestern.edu/clinPipeline.htm (accessed 02/02/2017)
- 483.U.S. National Institutes of Health. ClinicalTrials.gov. https://clinicaltrials.gov/ (accessed 02/03/2017)
- 484.Newman DJ, Cragg GM. Drugs and drug candidates from marine sources: An assessment of the current “State of Play”. Planta Med. 2016;82:775–789. doi: 10.1055/s-0042-101353. [DOI] [PubMed] [Google Scholar]


