Abstract
The RNA-guided CRISPR-Cas9 nuclease from Streptococcus pyogenes (SpCas9) has been widely repurposed for genome editing1–4. High-fidelity (SpCas9-HF1) and enhanced specificity (eSpCas9(1.1)) variants exhibit substantially reduced off-target cleavage in human cells, but the mechanism of target discrimination and the potential to further improve fidelity were unknown5–9. Using single-molecule Förster resonance energy transfer (smFRET) experiments, we show that both SpCas9-HF1 and eSpCas9(1.1) are trapped in an inactive state10 when bound to mismatched targets. We find that a non-catalytic domain within Cas9, REC3, recognizes target complementarity and governs the HNH nuclease to regulate overall catalytic competence. Exploiting this observation, we designed a new hyper-accurate Cas9 variant (HypaCas9) that demonstrates high genome-wide specificity without compromising on-target activity in human cells. These results offer a more comprehensive model to rationalize and modify the balance between target recognition and nuclease activation for precision genome editing.
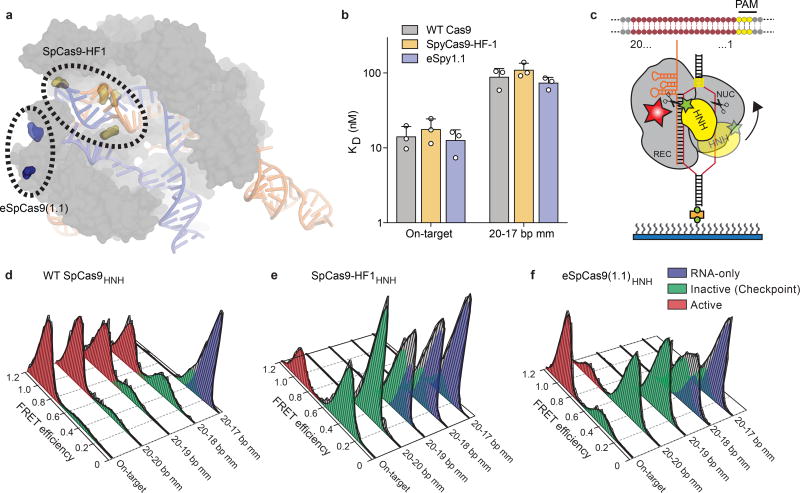
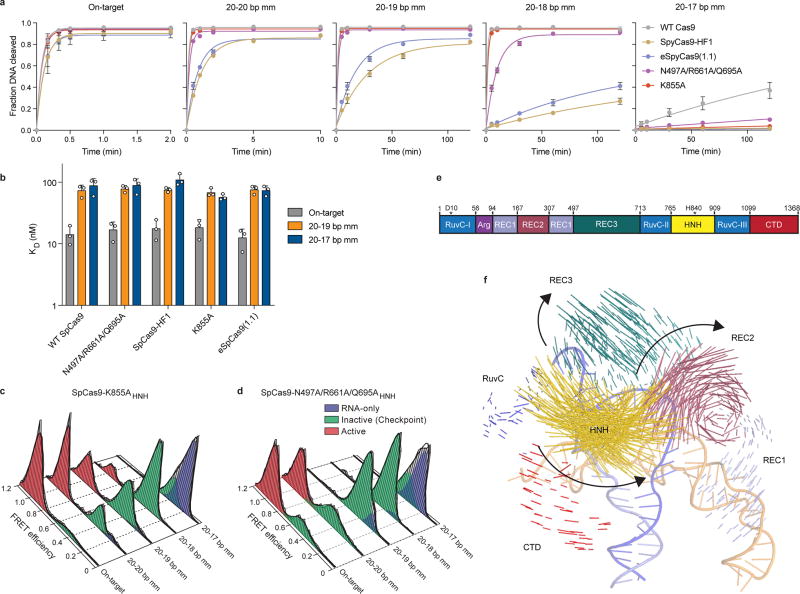
Efforts to minimize off-target cleavage by CRISPR-Cas9 have motivated the development of SpCas9-HF1 and eSpCas9(1.1) variants that contain amino acid substitutions predicted to weaken the energetics of target site recognition and cleavage8,9 (Figure 1a). Biochemically, we found that these Cas9 variants cleaved the on-target DNA with rates similar to that of wild-type (WT) SpCas9, whereas their cleavage activity was significantly reduced on substrates bearing mismatches (Extended Data Figures 1a, 2a). To test the hypothesis that SpCas9 with its single-guide RNA (sgRNA) might exhibit a greater affinity for its target than is required for effective recognition9,11, we measured DNA binding affinity and cleavage of SpCas9-HF1 and eSpCas9(1.1) variants. Contrary to a potential hypothesis that mutating these charged residues to alanine weakens target binding11, the affinities of these variants for on-target and PAM-distal mismatched substrates were similar to WT SpCas9 (Figure 1b, Extended Data Figures 1a, 2b), indicating that cleavage specificity is improved through a mechanism distinct from a reduction of target binding affinity11.
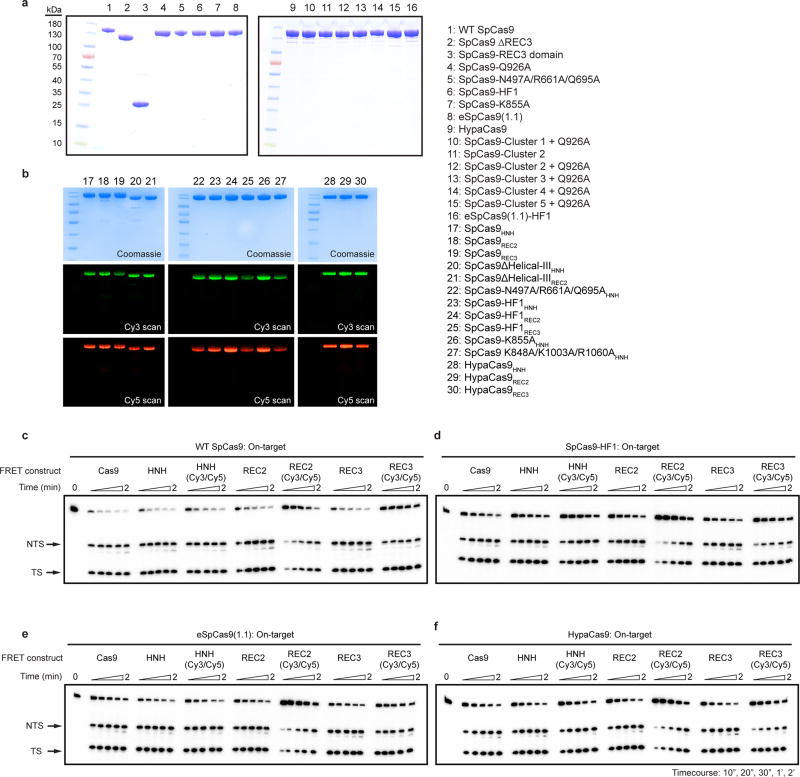
Figure 1. High-fidelity Cas9 variants enhance cleavage specificity through HNH conformational control.
a, Locations of amino acid alterations in existing high-fidelity SpCas9 variants mapped onto the dsDNA-bound SpCas9 crystal structure (PDB ID: 5F9R); HNH domain is omitted for clarity. b, Dissociation constants with mean and s.d. shown; n = 3 independent experiments (overlaid as white circles). c, Cartoon of DNA-immobilized SpCas9 for measuring HNH conformation by smFRET, with DNA target numbering scheme. d–f, smFRET histograms showing HNH conformation with indicated Cas9 variants bound to on-target and mismatched targets using nucleotide numbers diagramed in panel c. Black curves represent a fit to multiple Gaussian peaks.
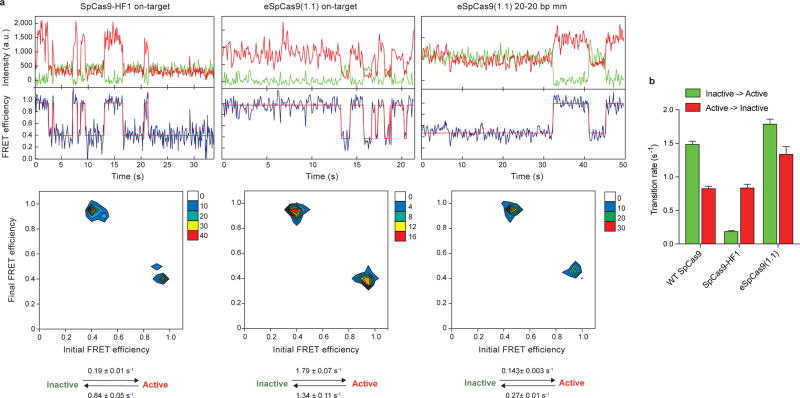
The HNH nuclease domain of SpCas9 undergoes a substantial conformational rearrangement upon target binding12–15, which activates the RuvC nuclease for concerted cleavage of both strands of the DNA12,16. It was previously shown that the HNH domain stably docks in its active state with an on-target substrate, but becomes loosely trapped in a catalytically-inactive conformational checkpoint when bound to mismatched targets10,12. We therefore hypothesized that SpCas9-HF1 and eSpCas9(1.1) variants may employ a more sensitive threshold for HNH domain activation to promote off-target discrimination. To test this possibility, we labeled catalytically active WT SpCas9 (SpCas9HNH), SpCas9-HF1 (SpCas9-HF1HNH) and eSpCas9(1.1) (eSpCas9(1.1)HNH) with Cy3/Cy5 FRET pairs at positions S355C (within the “stationary” REC1 domain) and S867C (within the “mobile” HNH domain) to measure HNH conformational states upon dsDNA binding (Figure 1c–f, Extended Data Figure 1c–e)12. Whereas SpCas9HNH stably populated the active state with on-target and mismatched substrates as observed by steady-state smFRET (Figure 1d), only ~32% of SpCas9-HF1HNH molecules occupied the HNH active state (EFRET = 0.97) with an on-target substrate, with the remaining ~68% trapped in the inactive intermediate state (EFRET = 0.45) (Figure 1e). Of the dynamic molecules (~36% of all smFRET traces) observed for SpCas9-HF1HNH, kinetics analysis further revealed that the HNH transition rate from the inactive to active states was ~8-fold slower compared to that of WT SpCas9HNH (~3% dynamic molecules10) (Extended Data Figure 3). However, when SpCas9-HF1HNH was bound to a substrate with a single mismatch at the PAM-distal end (20-20 bp mm), stable docking of the HNH nuclease was entirely ablated (Figure 1e). In addition, eSpCas9(1.1)HNH and other high fidelity variants8,9 reduced the HNH active state in the presence of mismatches (Figure 1f, Extended Data Figure 2c–d). We therefore propose that high fidelity variants of Cas9 reduce off-target cleavage by raising the threshold for HNH conformational activation when bound to DNA substrates.
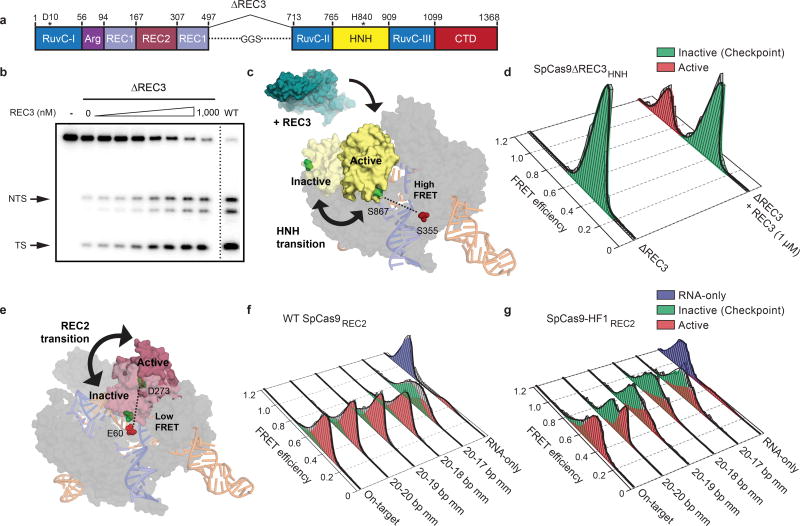
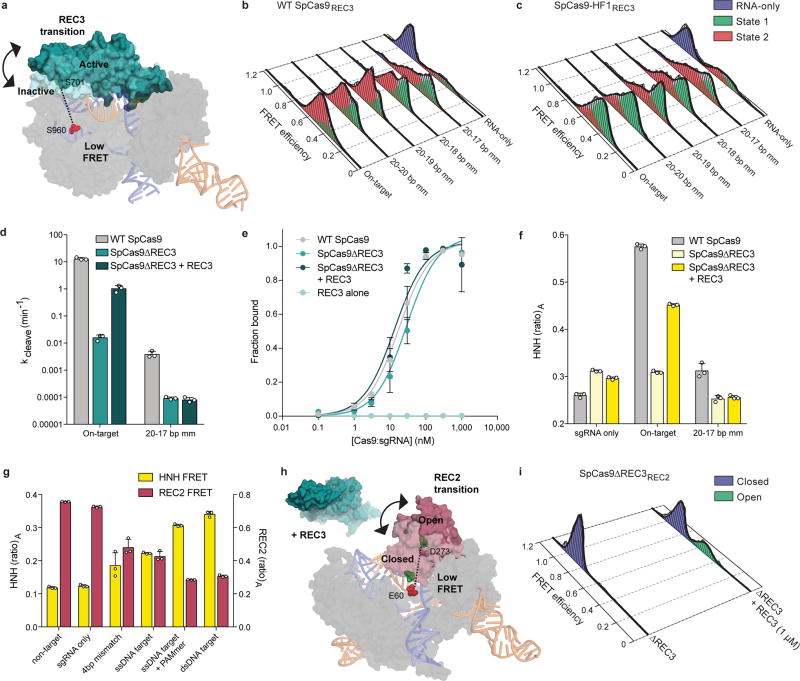
Since the HNH domain does not directly contact nucleic acids at the PAM-distal end13,17–19, it is likely that a separate domain of Cas9 senses target complementarity to govern HNH domain mobility. Structural studies suggested that a domain within the Cas9 recognition (REC) lobe (REC3) interacts with the RNA/DNA heteroduplex and undergoes conformational changes upon target binding (Extended Data Figure 2e–f)13,14,17–19. Because the function of this non-catalytic domain was previously unknown, we labeled SpCas9 with Cy3/Cy5 dyes at positions S701C (within the “mobile” REC3 domain) and S960C (within the “stationary” RuvC domain) to generate SpCas9REC3 and observed that the conformational states of REC3 become more heterogeneous as PAM-distal mismatches increase (Extended Data Figure 4a–c). To determine whether PAM-distal sensing precedes HNH activation, we deleted REC3 from WT Cas9 (SpCas9ΔREC3) (Figure 2a). Deletion of REC3 decreased the cleavage rate by ~1000-fold compared to WT Cas9, despite retaining near-WT affinity for the on-target (Extended Data Figure 4d–e). Unexpectedly, in vitro complementation of REC3 rescued the on-target cleavage rate by ~100-fold in a concentration-dependent manner (Figure 2b, Extended Data Figure 4d). Furthermore, the HNH domain in SpCas9ΔREC3 (SpCas9Δ REC3HNH) occupied the active state only when REC3 was supplemented in trans (Figure 2c–d, Extended Data Figure 4f). We therefore propose that REC3 acts as an allosteric effector that recognizes RNA/DNA heteroduplex to allow for HNH nuclease activation.
Figure 2. The alpha-helical lobe regulates HNH domain activation.
a, Domain organization of SpCas9ΔREC3. b, On-target DNA cleavage assay using SpCas9ΔREC3 with increasing concentrations of the REC3 domain supplied in trans, resolved by denaturing PAGE; repeated three independent times with similar results. c, Schematic of SpCas9ΔREC3HNH, with REC3 added in trans. Inactive to active structures represent HNH in the sgRNA-bound (PDB ID: 4ZT0) to dsDNA-bound (PDB ID: 5F9R) forms, respectively. d, smFRET histograms showing HNH conformation with SpCas9ΔREC3HNH bound to an on-target substrate, with and without REC3. e, Schematic of SpCas9REC2; HNH domain is omitted for clarity. Inactive to active structures represent REC2 in the sgRNA- (PDB ID: 4ZT0) to dsDNA-bound (PDB ID: 5F9R) forms, respectively. f–g, smFRET histograms showing REC2 conformation with f, WT SpCas9REC2 and g, SpCas9-HF1REC2 bound to on-target and mismatched targets. For panels d, f and g, black curves represent a fit to multiple Gaussian peaks.
We next considered allosteric interactions that could couple the discontinuous REC3 and HNH domains. Structural studies suggested that REC2 occludes the HNH domain from the scissile phosphate in the sgRNA-bound state19, and undergoes a large outward rotation upon binding to double-stranded DNA (dsDNA)13,14 (Figure 2e). To test whether the REC2 domain regulates access of HNH to the target strand scissile phosphate, we labeled SpCas9 with Cy3/Cy5 dyes at positions E60C (within the “stationary” Arginine-rich helix) and D273C (within the “mobile” REC2 domain) to generate SpCas9REC2 in order to detect REC2 conformational changes (Extended Data Figure 1b–c). We observed reciprocal changes in bulk FRET values ((ratio)A)20 between SpCas9HNH and SpCas9REC2 across multiple DNA substrates (Extended Data Figure 4g), which suggest that the REC2 and HNH domains are tightly coupled to ensure catalytic competence. smFRET experiments further confirmed a large opening of REC2 during the transition from the sgRNA-bound state (EFRET = 0.96) to the target-bound state (EFRET = 0.43) (Figure 2e–f). In contrast to WT SpCas9REC2, SpCas9-HF1REC2 occupies an intermediate state (EFRET = 0.63) when bound to a target with a single PAM-distal mismatch (Figure 2g). Together with the observation that the HNH domain of SpCas9-HF1 does not occupy the active state with PAM-distal mismatches, these experiments suggest that REC2 sterically occludes HNH in the conformational checkpoint when SpCas9 is bound to off-target substrates.
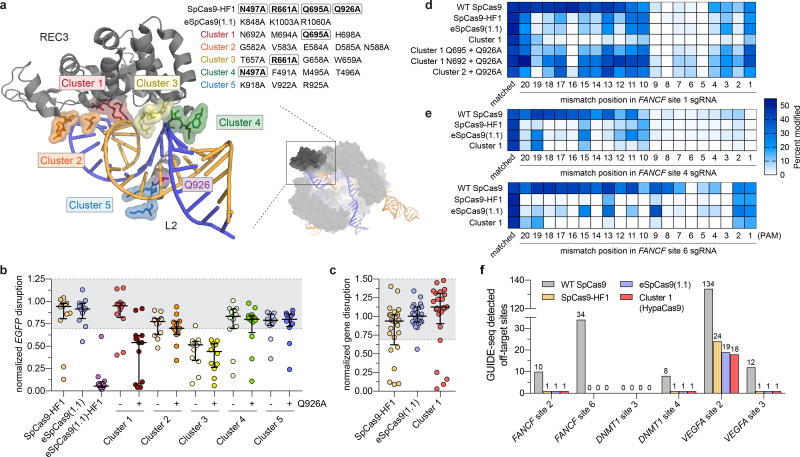
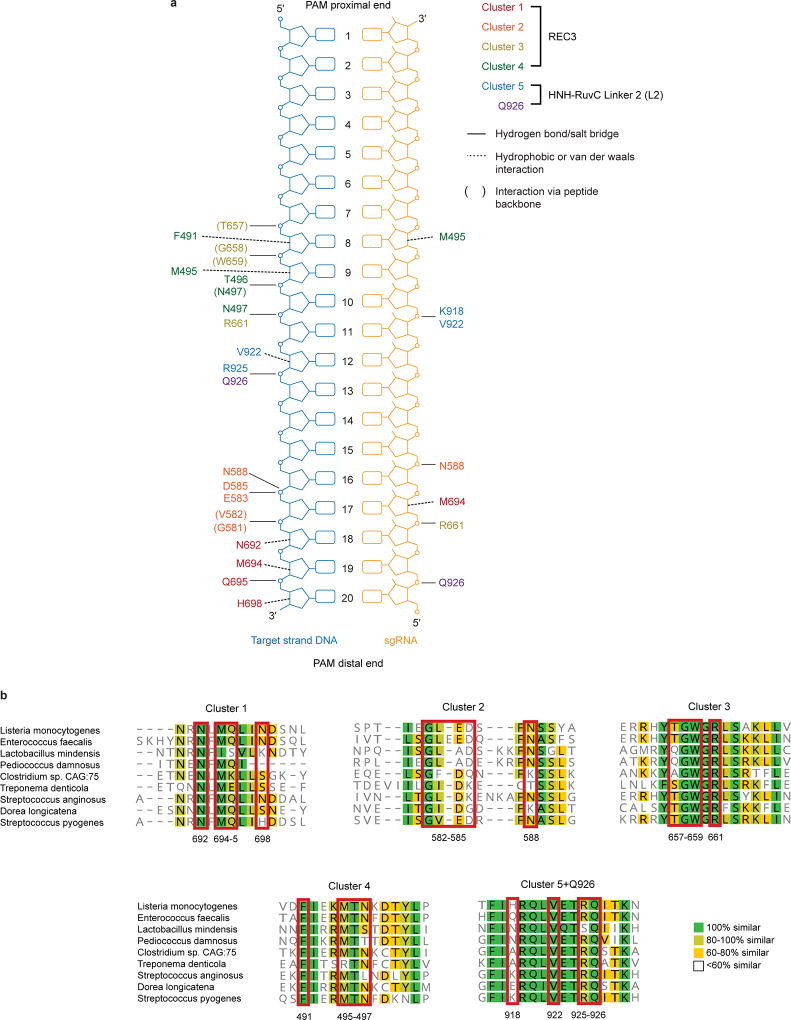
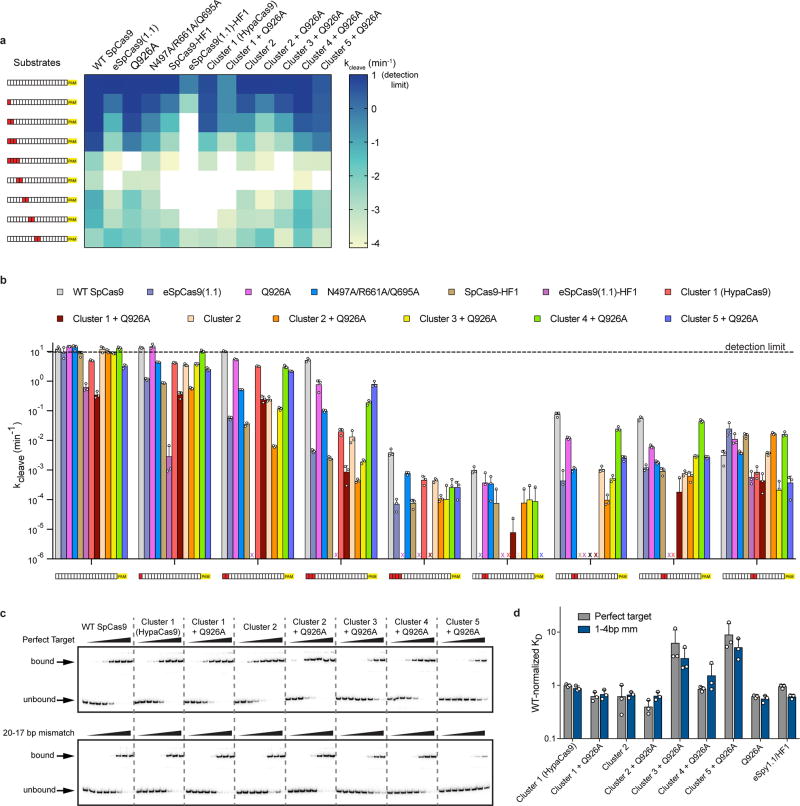
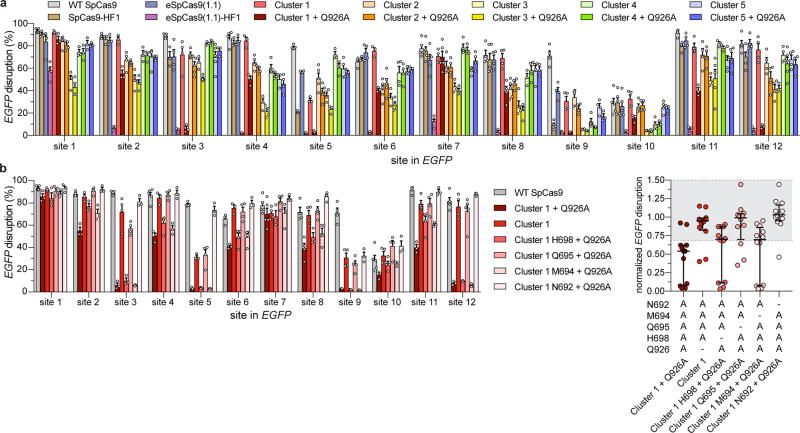
Next, we investigated if this conformational proofreading mechanism could be rationally exploited to design novel hyper-accurate Cas9 variants. We identified five clusters of residues containing conserved amino acids within 5 Å of the RNA/DNA interface, four of which are located within REC3 and one in the HNH-RuvC Linker 2 (L2) (Figure 3a, Extended Data Figure 5). Alone or in combination with Q926A, a substitution within L2 that confers higher specificity9, we generated alanine substitutions for each residue within the five different clusters of amino acids (Clusters 1–5 ± Q926A) (Figure 3a). We tested whether these cluster mutations affect cleavage accuracy and equilibrium binding in vitro, and found that Cluster 1 alone and Cluster 2 + Q926A suppressed off-target cleavage while retaining target binding affinities comparable to WT (Extended Data Figure 6). We next screened all cluster variants in human cells using an enhanced GFP (EGFP) disruption assay5. On-target activity for Cluster 1 was comparable to that of SpCas9-HF1 or eSpCas9(1.1), whereas Cluster 2 variants displayed generally lower activity (Figure 3b, Extended Data Figure 7a). Furthermore, Cluster 1 retained high on-target activity (> 70% of WT) at 19/24 endogenous gene sites tested, compared to 18/24 for SpCas9-HF1 and 23/24 for eSpCas9(1.1) (Figure 3c, Extended Data Figure 8a).
Figure 3. Targeted mutagenesis within the REC3 domain reveals a SpCas9 variant with hyper-accurate behavior in human cells.
a, Zoomed image of the REC3 domain and Linker 2 (L2) with amino acids of Cluster variants indicated (PDB ID: 5F9R). Boxed residues indicate amino acids also present in SpCas9-HF1. b, WT-normalized activity of Cas9 variants, using sgRNAs targeting 12 different sites within EGFP. c, WT-normalized endogenous gene disruption activity measured by T7 endonuclease 1 (T7E1) assay across 24 sites. For panels b and c, error bars represent median and interquartile ranges for n = 12 or 24 biologically independent samples, respectively; the interval with > 70% of wild-type activity is highlighted in light grey. d, Activities of WT and high-fidelity Cas9 variants when programmed with singly mismatched sgRNAs against FANCF site 1. e, Activities of Cas9 variants when programmed with singly mismatched sgRNAs against FANCF site 4 and FANCF site 6. f, Histogram of the total number of GUIDE-seq detected off-target sites for Cas9 variants with six different sgRNAs.
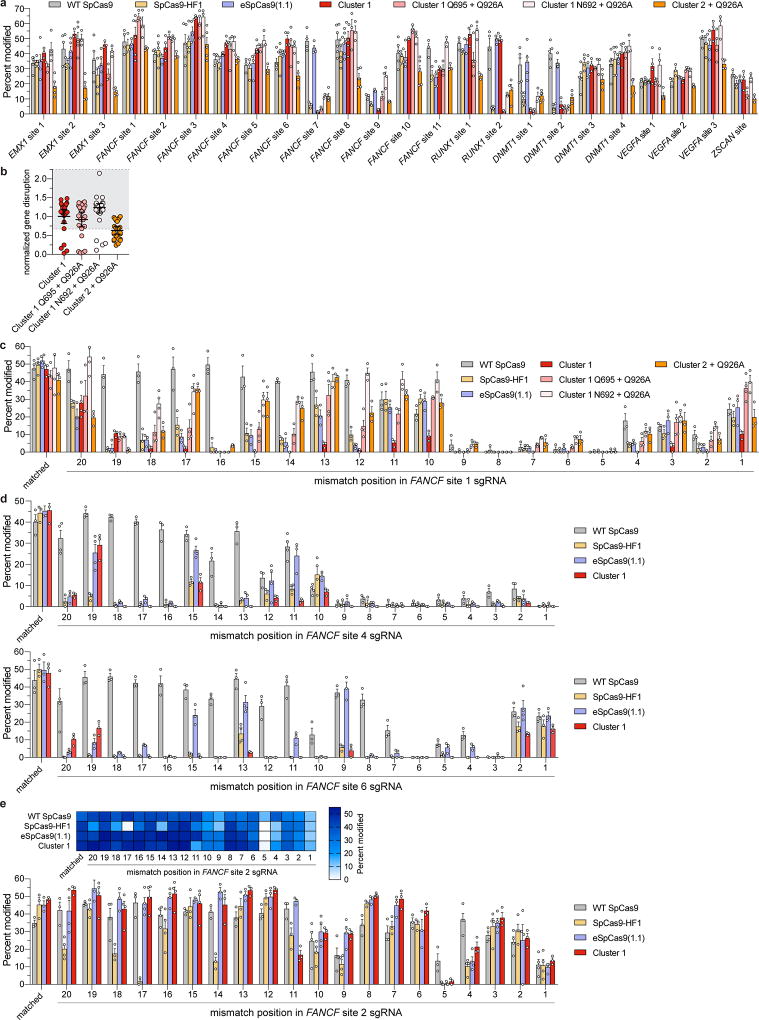
We then focused on the specific contributions of mutations within Cluster 1 by restoring each individual mutated residue to its wild-type identity, along with the Q926A mutation, and tested the resulting variants for on-target editing efficiency in human cells. On-target activity was significantly compromised when N692A/Q695A/Q926A mutations occurred together, but restoring either N692 (Cluster 1 N692 + Q926A) or Q695 (Cluster 1 Q695 + Q926A) alone led to robust on-target efficiency comparable to Cluster 1, signifying differential contributions from these mutations to activity and specificity (Extended Data Figure 7b–c, 8a–c). Using sgRNAs with single mismatches against the endogenous human gene target FANCF site 1, we found that Cluster 1 exhibited greater specificity than both SpCas9-HF1 and eSpCas9(1.1) in the middle and PAM proximal regions of the spacer (Figure 3d, Extended Data Figure 8c). Additional single mismatch tolerance assays on FANCF sites 4 and 6 further corroborated the superior accuracy of Cluster 1 (N692A/M694A/Q695A/H698A, hereafter referred to as HypaCas9) against mismatches at positions 1 through 18; however, single mismatches along FANCF site 2 were still tolerated across all SpCas9 variants tested (Figure 3e, Extended Data Figure 8d, e).
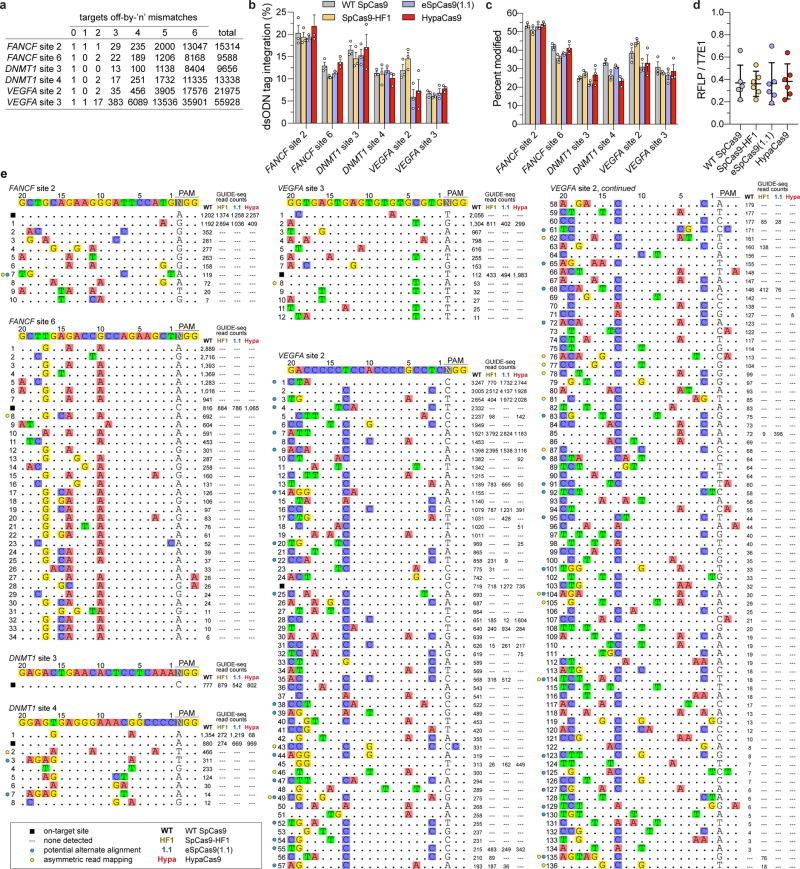
Next, we performed GUIDE-seq 6 to compare the genome-wide specificities of WT SpCas9, SpCas9-HF1, eSpCas9(1.1), and HypaCas9 using three sgRNAs previously shown to exhibit substantial off-target effects (FANCF site 2, VEGFA sites 2 and 3)6,9, and three previously uncharacterized sgRNAs with a moderate number of in silico predicted off-target sites (FANCF site 6, DNMT1 sites 3 and 4; Extended Data Figure 9a). We assessed GUIDE-seq tag integration and on-target editing and observed comparable efficiencies among the four nucleases for all six sgRNAs (Extended Data Figures 9b–d). Our GUIDE-seq analysis revealed that HypaCas9 exhibits dramatically improved genome-wide specificity compared to WT SpCas9, and shows equivalent or better genome-wide specificity relative to both SpCas9-HF1 and eSpCas9(1.1) for all sgRNAs examined (Figure 3f, Extended Data Figures 9e and 10). These results corroborate the enhanced mismatch intolerance of HypaCas9 and demonstrate that its specificity improvements may extend beyond the PAM-proximal and middle regions of the spacer sequence.
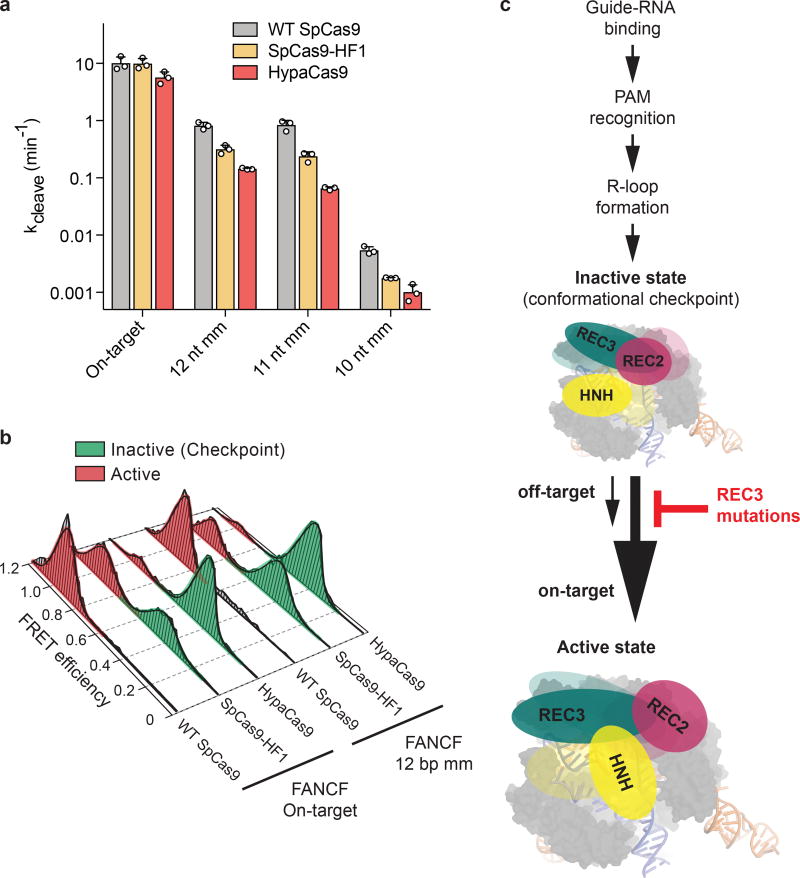
To biochemically validate cleavage specificity in the middle region of the spacer with HypaCas9, we measured cleavage rates against the FANCF site 1 sequence with or without internal mismatches. Although HypaCas9 retained on-target activity comparable to WT and SpCas9-HF1 in human cells, its in vitro cleavage rate was slightly reduced for the one target site examined (Figure 4a). However, the cleavage rate with internally mismatched substrates was considerably slower compared to WT and SpCas9-HF1, which may be explained by the altered threshold of HNH activation (Figure 4a, b).
Figure 4. Mutating residues involved in proofreading increases the threshold for conformational activation to ensure targeting accuracy.
a, DNA cleavage kinetics of SpCas9 variants with the FANCF site 1 on-target and internally mismatched substrates; mean and s.d. shown; n = 3 independent experiments (overlaid as white circles). b, smFRET histograms showing HNH conformation for indicated SpCas9 variants with a FANCF site 1 on-target and mismatched substrate at the 12th position; black curves represent a fit to multiple Gaussian peaks. c, Model for alpha-helical lobe sensing and regulation of the RNA/DNA heteroduplex for HNH activation and cleavage.
Our findings provide direct evidence to support previous speculation that Cas9 relies on protospacer sensing to enable accurate targeting21,22. In particular, we propose that REC3 binding to the RNA-DNA duplex is necessary for re-orienting REC2, which enables HNH docking at the active site (Extended Data Figure 4h–i). Mutation of residues within REC3 that are involved in RNA/DNA heteroduplex recognition, such as those mutated in HypaCas9 or SpCas9-HF1, prevents transitions by the REC2 domain, which more stringently traps the HNH domain in the conformational checkpoint in the presence of mismatches (Figure 4c, Extended Data Figure 10). Curiously, nearly all of the amino acids within the cluster variants were strongly conserved (Extended Data Figure 5), suggesting that these residues may also be involved in protospacer sensing and HNH nuclease activation across Cas9 orthologues. Furthermore, this observation may address how nature apparently has not selected for a highly precise Cas9 protein, whose native balance between mismatch tolerance and specificity may be optimized for host immunity. Our study delineates a general strategy for improving Cas9 specificity by tuning the natural conformational threshold, and offers opportunities for rational design of hyper-accurate Cas9 variants that do not compromise efficiency.
METHODS
Protein purification and dye labelling
S. pyogenes Cas9 and truncation derivatives were cloned into a custom pET-based expression vector containing an N-terminal His6-tag, maltose-binding protein (MBP) and TEV protease cleavage site. Point mutations were introduced by Gibson assembly or around-the-horn PCR and verified by DNA sequencing. Proteins were purified as described23, with the following modifications: after Ni-NTA affinity purification and overnight TEV cleavage at 4 °C, proteins were purified over an MBPTrap HP column connected to a HiTrap Heparin HP column for cation exchange chromatography. The final gel filtration step (Superdex 200) was carried out in elution buffer containing 20 mM Tris-HCl, pH 7.5, 200 mM NaCl, 5% (v/v) glycerol and 1 mM TCEP. For FRET experiments, Cy3/Cy5-dye positions were selected within a cysteine-free Cas9 protein based on a structural alignment of the sgRNA-bound (4ZT0) to dsDNA-bound (5F9R) structures. Each FRET pair consisted of one cysteine substitution within the “mobile” domain (HNH, REC2 or REC3) and another within the relatively “stationary” domain (REC1, Arginine-rich helix or RuvC), such that the inter-residue distance change from the sgRNA-bound to dsDNA-bound states was between 10–90 Å (Extended Data Figure 10). Dye-labeled Cas9 samples were subsequently prepared as described12. A list of all protein variants and truncations is in Supplementary Table 2.
Nucleic acid preparation
sgRNA templates were PCR amplified from a pUC19 vector containing a T7 promoter, 20 nt target sequence and optimized sgRNA scaffold. The amplified PCR product was extracted with phenol:chloroform:isoamyl alcohol and served as the DNA template for sgRNA transcription reactions, which were performed as described24. DNA oligonucleotides and 5′end biotinylated DNAs (Supplementary Table 3) were synthesized commercially (Integrated DNA Technologies), and DNA duplexes were prepared and purified by native PAGE as described23.
DNA cleavage and binding assays
DNA duplex substrates were 5′-[32P]-radiolabeled on both strands. For cleavage experiments, Cas9 and sgRNA were pre-incubated at room temperature for at least 10 min in 1X binding buffer (20 mM Tris-HCl, pH 7.5, 100 mM KCl, 5 mM MgCl2, 1 mM DTT, 5% glycerol, 50 µg ml−1 heparin) before initiating the cleavage reaction by addition of DNA duplexes. For REC3 in vitro complementation experiments, SpCas9ΔREC3 and sgRNA were pre-incubated with 10-fold molar excess of REC3 for at least 10 min at room temperature before addition of radiolabeled substrate. DNA cleavage experiments were performed and analyzed as previously described12. DNA binding assays were conducted in 1X binding buffer without MgCl2 + 1 mM EDTA at room temperature for 2 hr. DNA-bound complexes were resolved on 8% native PAGE (0.5X TBE + 1 mM EDTA, without MgCl2) at 4 °C, as previously described10. Experiments were replicated at least three times, and presented gels are representative results.
Bulk FRET experiments
All bulk FRET assays were performed at room temperature in 1X binding buffer, containing 50 nM SpCas9HNH (C80S/S355C/C574S/S867C labeled with Cy3/Cy5), SpCas9ΔREC3HNH(M1–N497,GGS,V713–D1368 + C80S/S355C/C574S/S867C) or SpCas9REC2 (E60C/C80S/D273C/C574S labeled with Cy3/Cy5) with 200 nM sgRNA and DNA substrate where indicated. Fluorescence measurements were collected and analyzed as described12. For REC3 in vitro complementation FRET experiments, SpCas9ΔREC3HNH and sgRNA were pre-incubated with 10-fold molar excess of REC3 for at least 10 min at room temperature before measuring bulk fluorescence.
Sample preparation for smFRET assay
99% PEG and 1% biotinylated-PEG coated quartz slides were received from MicroSurfaces, Inc. Sample preparation was performed as previously described10. Briefly, the glass surface was pre-blocked with casein (10 mg/ml) for 10 min. The sample chamber was washed with 1X binding buffer, then incubated with 20 µL streptavidin (1 mg ml−1) for 10 min. Unbound streptavidin was washed away with 40 µL of 1X binding buffer. To immobilize SpCas9 on its DNA substrate, 2.5 nM biotinylated DNA substrate was introduced and incubated in sample chamber for 5 min. Excess DNA was washed with 1X binding buffer. SpCas9-sgRNA complexes were prepared by mixing 50 nM Cas9 and 50 nM sgRNA in 1X binding buffer and incubated for 10 min at room temperature. SpCas9-sgRNA was diluted to 100 pM, introduced to sample chamber and incubated for 10 min. Before data acquisition, 20 µL imaging buffer (1 mg ml−1 glucose oxidase, 0.04 mg ml−1 catalase, 0.8% dextrose (w/v) and 2 mM Trolox in 1X binding buffer) was flown into chamber. The REC3 in vitro complementation assay was performed similar to steady-state FRET experiments: 2.5nM biotinylated DNA substrate (on-target) was immobilized on surface, and excess DNA was washed with 1X binding buffer. SpCas9-sgRNA complexes were prepared by mixing 50 nM SpCas9ΔREC3 and 50 nM sgRNA in 1X binding buffer and incubated for 10 min at room temperature. SpCas9-sgRNA was diluted to 100 pM, introduced to the sample chamber and incubated for 10 min. Before data acquisition, 20 µL imaging buffer was flowed into the chamber. After data acquisition, the sample chamber was washed with 1X binding buffer. 20 µL imaging buffer supplemented with 1 µM REC3 was flowed into the sample chamber and incubated for 10 min. After incubation, data for REC3 complementation was collected.
Microscopy and data analysis
A prism-type TIRF microscope was setup using a Nikon Ti-E Eclipse inverted fluorescent microscope equipped with a 60× 1.20 N.A. Plan Apo water objective and the perfect focusing system (Nikon). A 532-nm solid state laser (Coherent Compass) and a 633-nm HeNe laser (JDSU) were used for Cy3 and Cy5 excitation, respectively. Cy3 and Cy5 fluorescence were split into two channels using an Optosplit II image splitter (Cairn Instruments) and imaged separately on the same electron-multiplied charged-coupled device (EM-CCD) camera (512×512 pixels, Andor Ixon EM+). Effective pixel size of the camera was set to 267 nm after magnification. Movies for steady-state FRET measurements were acquired at 10 Hz under 0.3 kW cm−2 532-nm excitation. Steady-state and dynamic FRET data analysis was performed as described previously10. Briefly, for steady-state FRET analysis, two fluorescent channels were registered with each other using fiducial markers (20 nm diameter Nile Red Beads, Life Technologies) to determine the Cy3/Cy5 FRET pairs. Cy3/Cy5 pairs that photobleached in one step and showed anti-correlated signal changes were used to build histograms. FRET values were corrected for donor leakage and the histograms were normalized to determine the percentage of distinct FRET populations. Only samples showing greater than 3% of molecules with active transitions were subjected to dynamic FRET analysis.
Human cell culture and transfection
Descriptions of nuclease and guide RNA plasmids used for human cell culture are available in Supplementary Tables 2 and 3. Nuclease variants were generated by isothermal assembly into JDS246 (Addgene #43861)5, and guide RNAs were cloned into BsmBI-digested BPK1520 (Addgene #65777)25. Both U2OS cells (a gift from T. Cathomen, Freiburg) and U2OS-EGFP cells (encoding a single integrated copy of a pCMV-EGFP–PEST cassette)26 were cultured at 37 °C with 5% CO2 in advanced DMEM containing 10% heat-inactivated fetal bovine serum, 2 mM GlutaMax, penicillin/streptomycin, and 400 µg ml−1 Geneticin (for U2OS-EGFP cells only). Cell culture reagents were purchased from Thermo Fisher Scientific, cell line identities were validated by STR profiling (ATCC) and deep-sequencing, and cell culture supernatant was tested bi-weekly for mycoplasma. Transfections were performed using a Lonza 4-D Nucleofector with the SE Kit and the DN-100 program on ~200k cells with 750 ng of nuclease and 250 ng of guide RNA plasmids.
Human cell EGFP disruption assay
EGFP disruption experiments were performed as previously described5,26. Briefly, transfected cells were analyzed ~52 hr post-transfection for loss of EGFP fluorescence using a Fortessa flow cytometer (BD Biosciences). Background loss was determined by gating a negative control transfection (containing nuclease and empty guide RNA plasmid) at ~2.5% for all experiments.
T7 endonuclease I assay
Roughly 72 hr post-transfection, genomic DNA was extracted from U2OS cells using the Agencourt DNAdvance Genomic DNA Isolation Kit (Beckman Coulter Genomics), and T7 endonuclease I (T7E1) assays were performed as previously described26. Briefly, 600–800 nt amplicons surrounding on-target sites were amplified from ~100 ng of genomic DNA using Phusion Hot-Start Flex DNA Polymerase (New England Biolabs, NEB) using the primers listed in Supplementary Table 3. PCR products were visualized (using a QIAxcel capillary electrophoresis instrument, Qiagen), and purified (Agencourt Ampure XP cleanup, Beckman Coulter Genomics), Denaturation and annealing of ~200 ng of the PCR product was followed by digestion with T7EI (NEB). Digestion products were purified (Ampure) and quantified (QIAxcel) to approximate the mutagenesis frequencies induced by Cas9-sgRNA complexes.
GUIDE-seq
GUIDE-seq experiments were performed with WT SpCas9, SpCas9-HF1, eSpCas9(1.1), and HypaCas9 for six different sgRNAs, essentially as previously described6. Briefly, U2OS cells were transfected as described above with the addition of 100 pmol of an end-protected double-stranded oligo (dsODN) GUIDE-seq tag. Approximately 72 hr post-nucleofection, genomic DNA was extracted and gene disruption was quantified via T7E1 assay (as described above). GUIDE-seq tag-integration efficiencies were assessed using restriction fragment length polymorphism (RFLP) assays as previously described9. Briefly, PCR reactions amplified from ~100 ng of genomic DNA from GUIDE-seq treated samples, using Phusion Hot-Start Flex DNA Polymerase (NEB), were treated with 20 U NdeI (NEB) for 3 hr. Digested products were purified (Ampure) and quantified (QIAxcel) to approximate GUIDE-seq tag integration efficiencies. To perform GUIDE-seq, sample libraries were assembled as previously described6 and sequenced on an Illumina MiSeq machine. Data was analyzed using open-source guideseq software (version 1.1)27. GUIDE-seq data can be found in Supplementary Table 1, and are deposited with the NCBI Sequence Read Archive. Potential alternate alignments shown in Supplementary Table 1, resulting from RNA or DNA bulges28, depict one of many possible alternate alignments.
Data Availability Statement
Plasmids encoding the high-fidelity SpCas9 variants described in this manuscript have been deposited with the non-profit plasmid distribution service Addgene (http://www.addgene.org/). All sequencing data from this study are available through the NCBI Sequence Read Archive (SRA) under accession number SRP116962. The authors declare that all other data supporting the findings of this study are available within the paper and its supplementary information files.
Extended Data
Extended Data Figure 1. Dually-labeled SpCas9 variants are fully functional for DNA cleavage.
a, Sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) analysis of unlabeled Cas9 variants. b, SDS-PAGE analysis of Cy3/Cy5-labeled Cas9 variants. The gel was scanned for Cy3/Cy5 fluorescence (middle, bottom) before staining with Coomassie blue (top). c–f, DNA cleavage time courses of Cas9 FRET constructs and their dually-labeled counterparts for c, WT SpCas9, d, SpCas9-HF1, e, eSpCas9(1.1) and f, HypaCas9. For panels a–f, experiments were repeated three independent times with similar results.
Extended Data Figure 2. HNH domain in eSpCas9 variants still populate the docked state in the presence of PAM-distal mismatches.
a, Quantification of DNA cleavage time courses comparing WT SpCas9, SpCas9-HF and eSpCas9(1.1) variants with perfect and PAM-distal mismatched targets. b, Dissociation constants comparing WT SpCas9, SpCas9-HF and eSpCas9(1.1) variants with perfect and PAM-distal mismatched targets, as measured by electrophoretic mobility shift assays. For panels a–b, mean and s.d. shown; n = 3 independent experiments (overlaid as white circles in panel b). c–d, smFRET histograms for c, SpCas9-K855A and d, SpCas9-N497A/R661A/Q695A. For panels c and d, black curves represent a fit to multiple Gaussian peaks. e, Schematic of SpCas9 domain structure with color coding for separate domains. f, Vector map of global SpCas9 conformational changes from the sgRNA- (PDB ID: 4ZT0) to dsDNA-bound structures (PDB ID: 5F9R), domains colored as in panel e.
Extended Data Figure 3. Kinetic analysis of transitions between active and inactive states of the HNH domain.
a, Representative time traces (top), transition density plots (TDPs, middle) and rates of the transitions in TDPs (bottom) for SpCas9-HF1 with on-target DNA (left), eSpCas9(1.1) with on-target DNA (middle) and eSpCas9(1.1) with 20-20 bp mm DNA (right); mean and s.e.m. shown; n = 107, 24, and 74 individual molecules, respectively. The percentage of molecules that show at least one such transitions was 36%, 7% and 29% for SpCas9-HF1 with on-target, eSpCas9(1.1) with on-target and eSpCas9(1.1) with 20-20 bp mm DNA, respectively. Kinetics analysis of other cases (SpCas9-HF1 and eSpCas9(1.1) bound to other off-target substrates, and HypaCas9 bound to on- and off-target substrates) is not shown, because the percentage of molecules that show at least one such transitions was less than 3%. b, Comparison of on-target transition rates for WT SpCas9, SpCas9-HF1, and eSpCas9(1.1); mean and s.e.m. shown; n = 51, 107 and 24 individual molecules, respectively. Transition rates for WT SpCas9 collected from ref. 10.
Extended Data Figure 4. Nucleic acid sensing requires engagement with the REC3 domain and outward rotation of the REC2 domain.
a, Schematic of SpCas9REC3 with FRET dyes at positions S701C and S960C, with HNH domain omitted for clarity. Inactive to active structures represent REC3 in the sgRNA-bound (PDB ID: 4ZT0) to dsDNA-bound (PDB ID: 5F9R) forms, respectively. b–c, smFRET histograms showing HNH conformational activation with black curves representing a fit to multiple Gaussian peaks for b, WT SpCas9REC3 and c, SpCas9-HF1REC3 bound to perfect and PAM-distal mismatched targets. The purple peak denotes the sgRNA-only bound state, while the red and green peaks represent two states of REC3 with conformational flexibility upon binding to DNA substrates. d, REC3 in vitro complementation assay with SpCas9ΔREC3 by measuring cleavage rate constants. e, On-target DNA binding assay in the presence or absence of the REC3 domain; mean and s.d. shown. f, REC3 in vitro complementation assay with SpCas9ΔREC3 by measuring HNH activation with (ratio)A values. g, (Ratio)A data with SpCas9REC2 and SpCas9HNH showing reciprocal FRET states with the indicated substrates. For panels d–g, mean and s.d. shown; n = 3 independent experiments (overlaid as white circles in panels d, f, and g). h, Schematic of SpCas9ΔREC3REC2 with FRET dyes at positions E60C and D273C, with the REC3 domain added in trans. Inactive to active structures represent REC2 in the sgRNA-bound (PDB ID: 4ZT0) to dsDNA-bound (PDB ID: 5F9R) forms, respectively. i, smFRET histograms measuring REC2 conformational states with SpCas9ΔREC3REC2 in the absence and presence of the REC3 domain when bound to an on-target substrate.
Extended Data Figure 5. Identification of Cluster variants based on nucleic acid proximity and multiple sequence alignment of residues within Clusters 1–5.
a, Schematic depicting interactions of WT SpCas9 residues within Clusters 1–5 with the RNA/DNA heteroduplex, based on PDB accession 5F9R (adapted from ref 9). b, Alignment of selected Cas9 orthologues using MAFFT and visualized in Geneious 10.0, with red boxes outlining residues mutated to alanine within each cluster variant.
Extended Data Figure 6. Mutation clusters in the REC3 domain along the RNA/DNA heteroduplex demonstrate localized sensitivity to mismatches along the target sequence.
a–b, Quantified DNA cleavage rates (dotted line indicates detection limit for kcleave set at 10 min−1) displayed as a a, heatmap and b, bar graph. c–d, Target DNA binding assay c, resolved by native polyacrylamide gel electrophoresis (PAGE) mobility shift assays; repeated three independent times with similar results and d, quantification with WT-normalized dissociation constants. For panels b and d, mean and s.d. shown; n = 3 independent experiments (overlaid as white circles).
Extended Data Figure 7. On-target activities of altered specificity variants using a human cell EGFP disruption assay.
a, Summary of EGFP disruption activities for SpCas9-HF1, eSpCas9(1.1), eSpCas9(1.1)-HF1 and Cluster variants ± Q926A with mean and s.e.m., where n = at least 3 biologically independent samples (overlaid as white circles). b, Summary of EGFP disruption activities for the series of Cluster 1 variants with each substituted residue restored to the canonical amino acid; mean and s.e.m. where n = at least 3 biologically independent samples (overlaid as white circles); WT, Cluster 1 (HypaCas9), and Cluster 1 + Q926A data from panel a is re-plotted for comparison. c, WT-normalized plot of data in panel b; error bars represent median and interquartile range for n = 12 biologically independent samples; the interval with >70% of WT activity is highlighted in light grey.
Extended Data Figure 8. Activities and specificities of high-fidelity SpCas9 variants targeted to endogenous human cell sites.
a, On-target activities of WT SpCas9, SpCas9-HF1, Cluster 1 and Cluster 2 variants across 24 endogenous human genes, assessed by T7E1 assay; mean and s.e.m. shown; n = at least 3 biologically independent samples (overlaid as white circles). b, WT-normalized endogenous gene disruption data from panel a, for Cluster 1 and 2 variants. Error bars represent median and interquartile ranges of 24 biologically independent samples with the >70% interval of WT activity highlighted in light grey; Cluster 1 (HypaCas9) data from Fig. 3b is replotted for comparison. c–e, Summary of single mismatch tolerance of WT SpCas9, SpCas9-HF1, eSpCas9(1.1), and Cluster 1 and Cluster 2 variants on c, FANCF site 1 d, FANCF sites 4 and 6, and e, FANCF site 2. Percent modification in panels c–e assessed by T7E1 assay; mean and s.e.m. shown for n = at least 3 biologically independent samples (overlaid as white circles).
Extended Data Figure 9. Genome-wide specificity profiles of high fidelity SpCas9 variants defined using GUIDE-seq.
a, Number of in silico predicted target sites mismatched by ‘n’ positions for six sgRNAs against the reference human genome (hg38) via Cas-OFFinder29. b, Assessment of GUIDE-seq dsODN tag integration at the on-target site for each nuclease and guide combination, detected by RFLP assay. c, On-target editing, determined by T7E1 assay; mean and s.e.m.; n = 3 biologically independent samples (overlaid as white circles) for panels b and c. d, dsODN tag-integration efficiency ratios (integration:mutagenesis, from panels b and c) for each nuclease and guide combination, with means and 95% confidence intervals shown for n = 6 biologically independent samples. e, GUIDE-seq genome-wide specificity profiles for WT SpCas9, SpCas9-HF1, eSpCas9(1.1), and HypaCas9 each paired with six different sgRNAs. Mismatched positions in off-target sites are highlighted in color; GUIDE-seq read counts shown to the right of the sequences, which correlate with approximate cleavage efficiency at a given site; blue circles indicate sites with potential alternate alignments due to RNA or DNA bulges30 (see Supplementary Table 1); yellow circles indicate off-target sites that are only supported by asymmetric GUIDE-seq reads.
Extended Data Figure 10. Conformational gating drives targeting accuracy for SpCas9 variants.
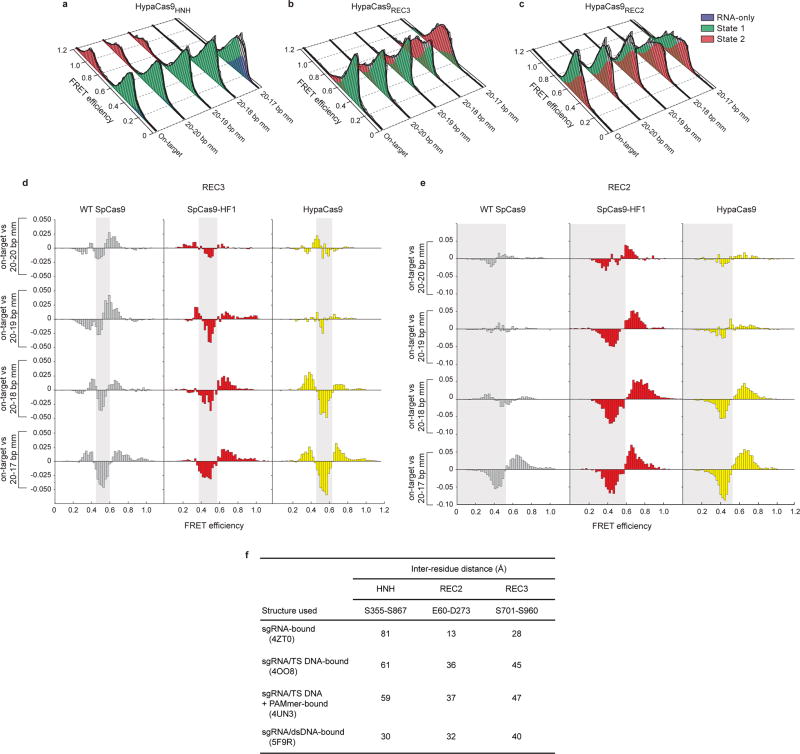
a–c, Steady state smFRET histograms measuring a, HNH, b, REC2 and c, REC3 conformational states for HypaCas9 bound to on-target and PAM-distal mismatched substrates. Black curves represent a fit to multiple Gaussian peaks. d–e, Steady state smFRET histograms of Cas9 variants bound to PAM distal mismatched substrates were normalized to and subtracted from that of on-target smFRET histograms. This analysis reveals transitions from one FRET population (negative peak, shaded region) to another population (positive peak, unshaded regions) for d, REC3 and e, REC2. f, Measured distances between residues labelled with Cy3/Cy5 FRET dyes for different substrate-bound Cas9 structures. Residue pairs were designed to report conformational changes of the specified domain (HNH, REC2 or REC3). The distances were measured between Cα atoms of the indicated residues for the associated PDB structures.
Supplementary Material
Supplemental Table 1: This file contains GUIDE-seq data.
Supplemental Table 2: This file contains DNA plasmids and proteins used in this study. All enhanced specificity, high-fidelity, cluster and hyper-accurate SpCas9 variants tested in this study, with Addgene ID numbers for deposited plasmids. The HNH, REC2 or REC3 subscript designation with an enhanced specificity, high-fidelity or cluster SpCas9 variant denotes combination of residue substitutions with indicated FRET construct.
Supplemental Table 3: This file contains a list of nucleic acids used in the study.
Acknowledgments
We thank A.V. Wright, S.N. Floor, J.C. Cofsky, D. Burstein, C. Fellman, B.L. Oakes and O. Mavrothalassitis for discussions and critical reading of the manuscript, M.S. Prew for technical assistance, and J.M. Lopez for assistance with GUIDE-seq data processing. J.S.C. and L.B.H. are supported by National Science Foundation Graduate Research Fellowships, and B.P.K. from Banting (Natural Sciences and Engineering Research Council of Canada) and Charles A. King Trust Postdoctoral Fellowships. J.A.D. is an Investigator of the Howard Hughes Medical Institute. This work has been supported by NIH (GM094522 and GM118773 (A.Y.), R35 GM118158 (J.K.J.)), NSF (MCB-1617028 (A.Y.) and MCB-1244557 (J.A.D.)), and the Desmond and Ann Heathwood MGH Research Scholar Award (J.K.J.).
Footnotes
SUPPLEMENTARY INFORMATION is available in the online version of the paper.
AUTHOR CONTRIBUTIONS
J.S.C., Y.S.D. and B.P.K. contributed equally to the work, conceived of and designed experiments with input from L.B.H., S.H.S, J.K.J., A.Y. and J.A.D. J.S.C. performed protein expression, labeling and biochemical experiments. Y.S.D. performed single-molecule fluorescence assays and related data analysis. B.P.K. and M.M.W. performed human cell-based assays, and B.P.K. and A.A.S. performed and analyzed GUIDE-seq experiments. J.S.C., Y.S.D., B.P.K., J.K.J., A.Y. and J.A.D. wrote the manuscript.
The authors declare competing financial interests: details are available in the online version of the paper.
COMPETING FINANCIAL INTERESTS (to be included in online version only)
J.K.J. has financial interests in Beacon Genomics, Beam Therapeutics, Editas Medicine, Pairwise Plants, Poseida Therapeutics, and Transposagen Biopharmaceuticals. J.K.J.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies. J.A.D. is a co-founder of Caribou Biosciences, Editas Medicine, and Intellia Therapeutics; a scientific advisor to Caribou, Intellia, eFFECTOR Therapeutics and Driver; and executive director of the Innovative Genomics Institute at UC Berkeley and UCSF. S.H.S. is an employee of Caribou Biosciences, Inc. S.H.S., J.S.C. and J.A.D. are inventors on a patent application entitled “Reporter Cas9 variants and methods of use thereof” (PCT/US2016/036754), filed by The Regents of the University of California. B.P.K. and J.K.J. are inventors on a patent application entitled “Engineered CRISPR-Cas9 nucleases” (US 15/060,424), filed by The General Hospital Corporation. J.S.C, Y.S.D., B.P.K, A.Y., J.K.J. and J.A.D. have filed a patent application related to this work through The General Hospital Corporation and The Regents of the University of California.
References
- 1.Doudna JA, Charpentier E Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 2.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat. Methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrangou R, Horvath P. A decade of discovery: CRISPR functions and applications. Nat. Microbiol. 2017;2:17092. doi: 10.1038/nmicrobiol.2017.92. [DOI] [PubMed] [Google Scholar]
- 5.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsai SQ, Joung JK. Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat. Rev. Genet. 2016;17:300–312. doi: 10.1038/nrg.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slaymaker IM, et al. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleinstiver BP, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dagdas YS, Chen JS, Sternberg SH, Doudna JA, Yildiz A. A conformational checkpoint between DNA binding and cleavage by CRISPR-Cas9. Sci. Adv. 2017;3:eaao0027. doi: 10.1126/sciadv.aao0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bisaria N, Jarmoskaite I, Herschlag D. Lessons from Enzyme Kinetics Reveal Specificity Principles for RNA-Guided Nucleases in RNA Interference and CRISPR-Based Genome Editing. Cell Syst. 2017;4:21–29. doi: 10.1016/j.cels.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sternberg SH, LaFrance B, Kaplan M, Doudna JA. Conformational control of DNA target cleavage by CRISPR-Cas9. Nature. 2015;527:110–113. doi: 10.1038/nature15544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang F, et al. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science. 2016;351:867–871. doi: 10.1126/science.aad8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palermo G, Miao Y, Walker RC, Jinek M, McCammon JA. Striking Plasticity of CRISPR-Cas9 and Key Role of Non-target DNA, as Revealed by Molecular Simulations. ACS Cent. Sci. 2016;2:756–763. doi: 10.1021/acscentsci.6b00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palermo G, Miao Y, Walker RC, Jinek M, McCammon JA. CRISPR-Cas9 conformational activation as elucidated from enhanced molecular simulations. Proc. Natl Acad. Sci. USA. 2017 doi: 10.1073/pnas.1707645114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishimasu H, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang F, Zhou K, Ma L, Gressel S, Doudna JA. A Cas9-guide RNA complex preorganized for target DNA recognition. Science. 2015;348:1477–1481. doi: 10.1126/science.aab1452. [DOI] [PubMed] [Google Scholar]
- 20.Majumdar ZK, Hickerson R, Noller HF, Clegg RM. Measurements of internal distance changes of the 30S ribosome using FRET with multiple donor-acceptor pairs: quantitative spectroscopic methods. J. Mol. Biol. 2005;351:1123–1145. doi: 10.1016/j.jmb.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 21.Szczelkun MD, et al. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc. Natl Acad. Sci. USA. 2014;111:9798–9803. doi: 10.1073/pnas.1402597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cencic R, et al. Protospacer adjacent motif (PAM)-distal sequences engage CRISPR Cas9 DNA target cleavage. PLoS One. 2014;9:e109213. doi: 10.1371/journal.pone.0109213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jinek M, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright AV, et al. Rational design of a split-Cas9 enzyme complex. Proc. Natl Acad. Sci. USA. 2015;112:2984–2989. doi: 10.1073/pnas.1501698112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleinstiver BP, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reyon D, et al. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai SQ, Topkar VV, Joung JK, Aryee MJ. Open-source guideseq software for analysis of GUIDE-seq data. Nat. Biotechnol. 2016;34:483. doi: 10.1038/nbt.3534. [DOI] [PubMed] [Google Scholar]
- 28.Lin Y, et al. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014;42:7473–7485. doi: 10.1093/nar/gku402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bae S, Park J, Kim JS. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin Y, et al. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014;42:7473–7485. doi: 10.1093/nar/gku402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1: This file contains GUIDE-seq data.
Supplemental Table 2: This file contains DNA plasmids and proteins used in this study. All enhanced specificity, high-fidelity, cluster and hyper-accurate SpCas9 variants tested in this study, with Addgene ID numbers for deposited plasmids. The HNH, REC2 or REC3 subscript designation with an enhanced specificity, high-fidelity or cluster SpCas9 variant denotes combination of residue substitutions with indicated FRET construct.
Supplemental Table 3: This file contains a list of nucleic acids used in the study.
Data Availability Statement
Plasmids encoding the high-fidelity SpCas9 variants described in this manuscript have been deposited with the non-profit plasmid distribution service Addgene (http://www.addgene.org/). All sequencing data from this study are available through the NCBI Sequence Read Archive (SRA) under accession number SRP116962. The authors declare that all other data supporting the findings of this study are available within the paper and its supplementary information files.