Abstract
The effect of sulfur limitation on the partitioning of carbon, nitrogen, and sulfur was investigated in Dunaliella salina. D. salina was able to adapt to 6 μm sulfate; under these conditions, the cells showed reduced growth and photosynthetic rates. Whereas intracellular sulfate was depleted, phosphate, nitrate, and ammonium increased. Amino acids showed a general increase, and alanine became the most abundant amino acid. The activities of four key enzymes of carbon, sulfur, and nitrogen metabolism were differentially regulated: Adenosine 5′ triphosphate sulfurylase activity increased 4-fold, nitrate reductase and phosphoenolpyruvate (PEP) carboxylase activities decreased 4- and 11-fold, respectively, whereas carbonic anhydrase activity remained unchanged. Sulfur limitation elicited specific increase or decrease of the abundance of several proteins, such us Rubisco, PEP carboxylase, and a light harvesting complex protein. The accumulation of potentially toxic ammonium indicates an insufficient availability of carbon skeletons. Sulfur deficiency thus induces an imbalance between carbon and nitrogen. The dramatic reduction in PEP carboxylase activity suggests that carbon was diverted away from anaplerosis and possibly channeled into C3 metabolism. These results indicate that it is the coordination of key steps and components of carbon, nitrogen, and sulfur metabolism that allows D. salina to adapt to prolonged sulfur limitation.
Changes in environment require organisms to develop strategies of adaptations that allow them to optimize the use of available resources. Among the most frequent limitations of plant growth is the deficiency in macronutrients. General responses to carbon, nitrogen, and sulfur limitation are reduced growth and photosynthesis (Davies and Grossman, 1998). Since these compounds are integrated into major constituents of the cell, a converging sequence of events can be expected. The regulation of carbon and nitrogen metabolism has been extensively investigated and found to be intimately related on physiological and molecular levels (Huppe and Turpin, 1994; Kaiser, 1997). For example, the chemical form of nitrogen and its concentration modulate photosynthesis and anaplerosis in D. salinae due to the direct link existing between carbon skeleton availability and nitrogen fixation into amino acids (Huppe and Turpin, 1994; Giordano and Bowes, 1997). In turn, the availability of inorganic carbon strongly affects the ability to assimilate nitrate and ammonium (Larsson et al., 1985).
Little is known about the interaction of nitrogen metabolism with sulfur metabolism. Elements of this interaction may be the coregulation of nitrate and sulfate uptake, the down-regulation of nitrate reductase (NR) and changes in the level of O-acetyl-Ser, the intermediate in Cys biosynthesis under sulfur limitation (Clarkson et al., 1989; Brunold, 1993; Davies and Grossman, 1998). Even less information is available on the interaction of sulfur and carbon metabolism. So far, mostly isolated steps of the corresponding pathways have been investigated during limitation of sulfate as a nutrient, although sulfur is a key component of essential cell constituents (Hell, 1997).
Accessible forms of sulfur for assimilation can be limiting in both terrestrial and aquatic ecosystems. A decline in the yield of some crops due to decreased atmospheric sulfur input has been reported for northern Europe (Dämmgen et al., 1998). In the same region, the reduction in sulfur deposition has also been detected in aquatic environments (Tipping et al., 1998).
The structural, functional, and developmental complexity of vascular plants led us to select the wall-less green microalga (Dunaliella salina) as experimental system to investigate the mechanisms of resource partitioning between sulfur, nitrogen, and carbon metabolism during sulfur-limited growth. D. salina (Volvocales, Chlorophyta) inhabits hypersaline waters, where its halotolerance is predominantly mediated by glycerol as an osmoticum (Ginzburg, 1987). This alga is intensively cultured for the commercial production of both glycerol and the photoprotective pigment β-carotene (Ginzburg, 1987; Borowitzka and Borowitzka, 1988) and is therefore of great economic interest. Furthermore, Dunaliella is often present in the crystallizer ponds of saltworks, where the organic matter it releases negatively affects salt quality (Davis and Giordano, 1996).
In this study, instead of the dissection of the individual pathways, we analyzed key components and reactions of carbon, nitrogen, and sulfur metabolism with respect to resource allocation under sulfur limitation. To the best of our knowledge, this is the first report of a combined analysis of these nutrients applied to adaptive processes under nutrient deficiency.
RESULTS
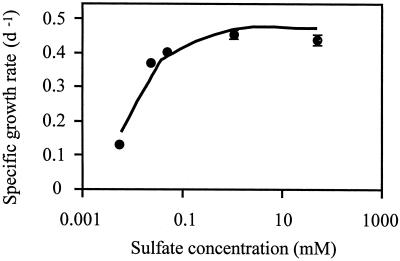
The sulfate requirement of D. salina was investigated in cultures acclimated for several generations to sulfate concentrations ranging from 6 μm to 48 mm as the sole sulfur source (Fig. 1). Specific growth rate of approximately 25% of the maximum rate was reached at 6 μm sulfate, whereas 90% of maximum rate was predicted at approximately 200 μm sulfate from the modeled growth rate. On the basis of these results, growth media containing 6 μm sulfate were used for sulfur-limited cultures, and growth media containing 48 mm sulfate were used for sulfur sufficient cultures.
Figure 1.
Specific growth rate of D. salina cells as a function of the sulfate concentration in the growth medium. The error bars show the sd obtained from the analysis of three different cultures. The line represents the growth rate response modeled by the function μ = ([SO42−] × μmax × a)/([a × {SO42−}] + μmax), where μ is the specific growth rate and a is the initial slope of the curve.
Total protein as well as chlorophyll content were found to be unaffected by the sulfur concentration (Table I). The levels of β-carotene in sulfur-limited cultures were higher by approximately 20% on a per cell basis, as compared with the levels in sulfur sufficient cultures. Despite the lower growth rate, D. salina cells acclimated to growth at 6 μm sulfate had a cell volume 2.4-fold smaller than the cells cultured at 48 mm sulfate (Table I). The cell shape was unaffected by the sulfate concentration, and the ratio between the major and minor axes was 1.3 regardless of the growth conditions (data not shown).
Table I.
Measurements of cell volume, protein, chlorophyll, and β-carotene in D. salina cells cultured at either sufficient or limiting sulfate concentration
| Cellular Parameter | Sulfate in the Growth Medium
|
|
|---|---|---|
| 48 mm | 6 μm | |
| Cell volume (μm3) | 1,028 (387) | 427 (42) |
| Total protein (μg/106 cells) | 22.04 (6.05) | 22.5 (7.21) |
| Chlorophyll (μg/106 cells) | 2.30 (0.83) | 2.22 (1.44) |
| β-Carotene (μg/106 cells) | 0.77 (0.05) | 0.92 (0.05) |
The values in parentheses indicate the sd obtained from analysis of three cultures.
Growth of D. salina under sulfur-limited conditions profoundly affected the accumulation of macronutrients (Table II). Cells cultured in the presence of 6 μm sulfate contained about 6 times less intracellular sulfate than their high sulfate-grown counterparts, although the sulfate concentration of the sulfur-limited medium was almost 4 orders of magnitude lower than that of the sulfate-sufficient medium. Cells acclimated to growth in the presence of 6 μm sulfate accumulated about 11 times more nitrate and 7 times more phosphate than cells grown in the presence of 48 mm sulfate, despite equal concentrations of these ions in both growth media. Even though ammonium was not supplied to the cultures, it was detected in cells grown under both conditions but accumulated to 4- to 5-fold higher levels in the low-sulfate grown cells. When the contents of the nutrients are based on cell volume, as given in Table I, instead of a per cell basis, cells cultured at 6 μm sulfate still contain approximately 2 to 3 times less sulfate than high-sulfate-grown cells, whereas the increase of levels of nitrate, ammonium, and phosphate is enhanced between 2- and 4-fold.
Table II.
Determination of macronutrients in D. salina cells grown at either sufficient or limiting sulfate concentrations
| Sulfate in the Growth Medium
|
||
|---|---|---|
| 48 mm | 6 μm | |
| nmol/106 cells | ||
| Sulfate | 0.82 (0.11) | 0.14 (0.05) |
| Phosphate | 0.06 (0.02) | 0.42 (0.12) |
| Nitrate | 2.36 (0.43) | 25.6 (2.59) |
| Ammonium | 2.76 (1.10) | 12.7 (2.01) |
The values in parentheses indicate the sd.
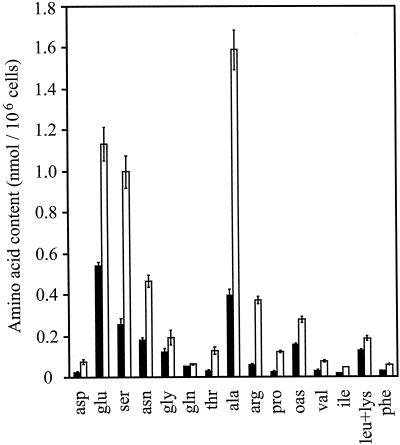
Total amino acid contents of low-sulfate cultures were almost 3 times higher than those of high-sulfate cultures due to an increase of almost all detected amino acids (Fig. 2). The three most abundant amino acids under both culture conditions were Glu, Ala, and Ser, whereas Gln was among the least abundant amino acids found. Ala increased 4-fold and became the most abundant amino acid under sulfur limitation, contributing 27% of the total. Arg, Pro, and Thr increased 6.5-, 4.9-, and 4.2-fold, respectively, but together contributed only 11% to the total amino acid content. Cys and Met contents were close to or below the detection limit of 5 pmol/106 cells given by the analysis method used, even under sulfur sufficient conditions. Cys pools were determined to 7 ± 1 pmol/106 cells and only trace amounts of Met could be detected, thus no quantification between high- and low-sulfate cultures was carried out.
Figure 2.
Contents of amino acids of D. salina cells grown at either sufficient (48 mm, black bars) or limiting sulfate (6 μm, white bars) concentration. OAS, O-Acetyl-Ser. The values shown are the means ± sd of five extractions with two replicates each.
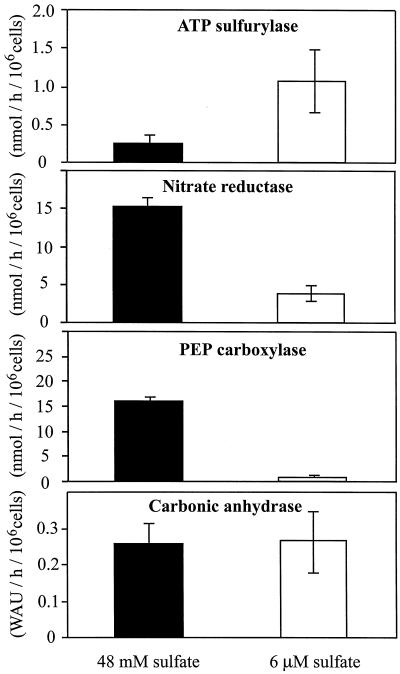
The activities of key enzymes of sulfur, nitrogen, and carbon metabolism were determined for high and low sulfur-grown D. salina cells (Fig. 3). Growth at low sulfur concentration caused ATP sulfurylase activity to increase by more than 4-fold. The same growth regime repressed NR activity by a factor of 4. An even larger decrease of activity resulted for PEP carboxylase (11-fold) as a consequence of adaptation to growth under low-sulfur condition. In contrast to the changes observed for these enzymes, the activity of periplasmic carbonic anhydrase, generally considered one of the major components of CO2 concentrating mechanisms, was unaffected by the sulfur concentration in the growth medium.
Figure 3.
Enzymatic activities of ATP sulfurylase, PEP carboxylase, NR, and carbonic anhydrase from D. salina cells grown at either sufficient (48 mm) or limiting sulfate (6 μm) concentration. The data represent the means ± sd of four independent measurements. WAU refers to Wilbur-Anderson units of carbonic anhydrase activity.
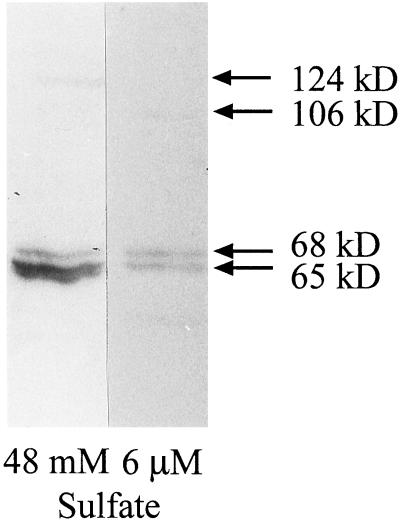
The strong decrease in PEP carboxylase activity prompted us to investigate the variation in protein levels using an anti-PEP carboxylase antibody (Fig. 4). Three bands were visible under both growth conditions. Two bands were present regardless of the sulfate concentration in the growth medium; the band of 65 kD, however, was more abundant in high sulfur-grown cells and was substantially reduced under sulfur limitation. The optical density of the upper band (68 kD), on the contrary, did not change when cells were adapted to low sulfur. For each growth regime, one additional band was detected, which had an apparent molecular mass of 124 kD in sulfur sufficient cells and of 106 kD in sulfur-limited cells. These higher Mr bands were much less abundant than the bands of 68 and 65 kD.
Figure 4.
Immunoblot of D. salina protein extracts obtained from high and low sulfur-grown cells using an anti-PEP carboxylase antibody. The experiment was repeated four times, and the image shows a representative result.
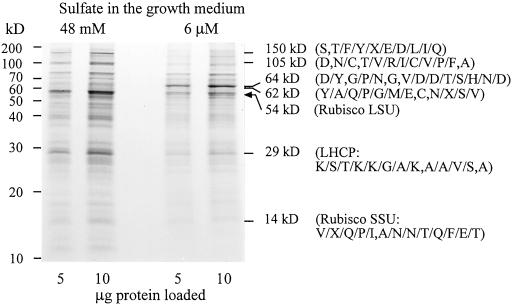
To investigate whether additional proteins changed in their abundance as a result of acclimation, total protein extracts were separated by denaturing gel electrophoresis (Fig. 5). Rubisco protein was strongly reduced by adaptation to low-sulfur concentration as indicated by the change of large-subunit abundance. A similar response was observed when cells were transferred from high- to low-sulfur medium (data not shown). Using high-resolution gels with varying acrylamide concentrations, several differentially expressed protein bands were detected; this finding is summarized in Figure 5, where a gel of intermediate polyacrylamide concentration is shown. Under low sulfur, proteins of 150, 110, 29, and 15 kD declined, whereas the abundance of proteins of 64 and 62 kD increased. These proteins were isolated from gels and partially sequenced to reveal their identities. Internal sequences of 8 to 12 amino acids in length were obtained but revealed no match in the SwissProt database for the peptides obtained from the bands of 150, 110, 64, and 62 kD. The 29-kD protein was identified as chlorophyll a/b binding protein of light harvesting complex II on the basis of homology with D. salina light harvesting complex (EMBL accession no. M23531). A second identification was obtained for the peptide derived from the 14-kD band that matched with the Rubisco small subunit of Chlamydomonas moewusii (SwissProt accession no. P17537) by 75% amino acid identity, independently confirming the observed decrease of the large subunit of Rubisco.
Figure 5.
Differential abundance of proteins from cultures adapted to 48 mm and 6 μm sulfate in the growth medium. The image shows a representative polyacrylamide gel (10%). To allow quantitative comparisons, 5 and 10 μg of total proteins from each extract were loaded. Internal peptide sequences derived from the isolated bands indicated by their molecular mass are shown in brackets. Alternative amino acids for the same position are included.
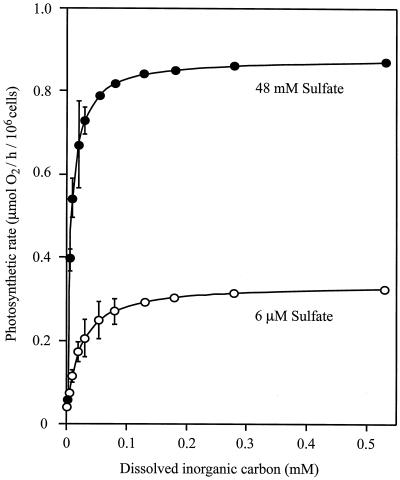
To assess the influence that the observed changes in Rubisco protein exert on photosynthesis and to ascertain whether a down-regulation occurred in the fixation of inorganic carbon to meet the reduced metabolic demands associated with the adaptation to low sulfate, the photosynthetic response of D. salina cultures adapted to high and low sulfur was investigated (Fig. 6). The maximal photosynthetic rate (Pmax) under sulfur-limited conditions was found to be approximately 3 times lower as compared with the Pmax of high sulfate grown cells. The Pmax was not affected by the sulfate concentration used during the assay, but depended exclusively on the sulfate concentration during growth (data not shown). In addition, the photosynthesis of low sulfate grown cells showed lower affinity for dissolved inorganic carbon (DIC) than that of the cells cultured at 48 mm sulfate. When the DIC affinity is estimated from the initial slope (α) of the DIC response curve, the photosynthesis of low-sulfate-grown cells appears to have a 7-fold lower affinity; the difference between the photosynthetic affinity of cells cultured in the presence of high and low sulfate remains substantial even when K½(DIC) is used as an indicator of affinity, although the difference is reduced to 3.4-fold. Dark respiration rates were independent of the sulfate concentration used for growth and had values of 0.1 μmol O2 (106 cells)−1 h−1.
Figure 6.
Photosynthetic response to dissolved inorganic carbon of D. salina cells cultured in the presence of high (●) and low (○) sulfate concentrations. The error bars show the sd of three replicates.
DISCUSSION
From the results presented, it is concluded that D. salina is able to adapt to sulfate concentrations of 6 μm in its growth medium. This nutrient limitation results in a reduced growth rate, cell volume, and photosynthetic activity but allows the organism to maintain protein and chlorophyll levels similar to sulfate sufficient grown cells. This adaptation is mediated by an array of physiological responses that strongly suggest regulatory interactions between the assimilatory pathways of sulfur, nitrogen, and carbon.
The observed strong reduction of photosynthesis under sulfur-limited growth correlates with a substantial decline of Rubisco and chlorophyll a/b-binding proteins. Yet the lowered photosynthetic response to carbon cannot be attributed to periplasmic carbonic anhydrase, whose activity was independent of sulfur concentration in the growth media. This decline in photosynthesis could therefore be the consequence of a combined reduction of major components of photosynthetic light and dark reaction. At the same time, due to its high abundance, Rubisco constitutes a large reservoir of reduced sulfur that has been estimated to reach 50 mm in the chloroplast (Ferreira and Teixeira, 1992). Therefore, lower ratios of Rubisco protein to total protein could facilitate the allocation of reduced sulfur and carbon skeletons to proteins specifically induced by sulfur limitation without an aggravation in sulfur and carbon consumption, as has been suggested for acclimation of wheat leaves to sulfate deprivation (Gilbert et al., 1997). The difference in cell volume between sulfur-limited and sulfur-sufficient cells enhances this process: On a volume basis, in fact, the amount of resources made available for sulfur-limited cells by the decrease of Rubisco would be even larger. At the onset of sulfur limitation, Rubisco may act as a source of fixed sulfur and carbon; in acclimated cells, the reduced synthesis of Rubisco would allow to shift resources to other more needed proteins.
The reduction of intracellular sulfate was several orders of magnitude smaller than the difference of concentrations in the high- and low-sulfate media, suggesting that the cells adapt by an induction of plasmalemma sulfate transporters as found for Chlamydomonas (Yildiz et al., 1994). When D. salina cells were transferred from high- to low-sulfate medium in short-term experiments, an increase of sulfate uptake capacity was observed (V. Pezzoni and M. Giordano, unpublished data). The increase in the activity of ATP sulfurylase, the first enzyme of the sulfate reduction pathway, is a further indication of a stimulation of sulfate assimilation. In contrast to sulfate, phosphate accumulates in low sulfur grown cells; this could be due to the reduced photosynthesis and growth, which result in a decline of sinks for phosphate such as Calvin cycle intermediates and nucleic acids.
Nitrate dramatically increased in sulfur-limited cells, probably as a consequence of the lowered activity of NR. For higher plants, it has been suggested that the down-regulation of NR is mediated by products of nitrogen assimilation like Gln, Asn, and Arg (Brunold, 1993; Migge et al., 2000). In sulfur-limited D. salina, however, the cell contents of these amino acids undergo only very limited changes. Since it has been reported that the NR of green algae is extremely susceptible to ammonium rather than to its metabolic products (Franco et al., 1984), it seems likely that the observed increase in cellular ammonium of sulfur-limited D. salina was responsible for the drop in the NR activity.
The accumulation of nitrate, ammonium, and amino acids indicates that an imbalance exists between sulfur and nitrogen in sulfur-limited cells. The difference in ammonium concentration between high- and low-sulfate-grown cells is 10.0 nmol (106 cells)−1, compared with a difference of 2.8 nmol (106 cells)−1 determined for the total amounts of soluble amino acids. This suggests that cells are lacking carbon skeletons for nitrogen assimilation. Sulfate-limited growth caused a predominant accumulation of C3 amino acids, most notably Ala and Ser, whereas Gln was unaffected and Glu, Asn, and Arg showed only moderate increases as compared with higher plants (Brunold, 1993; Migge et al., 2000). The preferential accumulation of C3 amino acids might serve the purpose of saving carbon especially because in D. salina a conspicuous and unavoidable sink of carbon is constituted by glycerol, the main osmoticum of this alga in its hypersaline environment (Avron and Ben-Amotz, 1992). Under the conditions used for this study, the cellular concentration of glycerol is in the order of 3 m (Giordano and Bowes, 1997).
Thus the observed dramatic reduction in PEP carboxylase activity under sulfur deficiency could be an even more efficient way to prevent allocation of reduced carbon into C4 and longer carbon skeletons. Instead, PEP could be funneled into glycerol metabolism via inverse glycolysis. Since Glu levels were increased under sulfur limitation, this pathway could also be favored by the inhibition of pyruvate kinase by Glu, as reported for the green alga Selenastrum (Lin et al., 1989). The decreased activity was flanked by a reduction of PEP carboxylase protein in sulfur-limited cells. Immunodetection using an antibody originally directed against PEP carboxylase from Selenastrum (Rivoal et al., 1996) revealed strong cross-reaction with two bands of similar molecular mass of which the more abundant 65-kD band declined at low sulfur. Catalytic subunits of PEP carboxylase of most sources have molecular masses of approximately 100 kD, a range in which only very weak cross-reaction was observed for Dunaliella extracts; however, subunit sizes of 60 kD to 70 kD have also been reported (Sako et al., 1996; Rivoal et al., 1998). At present it cannot be distinguished whether several genetically defined isoforms exist in D. salina or whether PEP carboxylase isoforms are post-translationally modified.
The specific alterations in enzymatic activities and protein levels prompted us to compare protein profiles of high- and low-sulfate adapted cultures. Several differentially expressed bands were detected in crude extracts using Coomassie staining, indicating a high abundance of these proteins. Sulfur does not simply cause a general decline in proteins, since two proteins were found to increase as part of the adaptation response of D. salina. The higher molecular mass proteins (150, 105, 64, and 62 kD) could not be attributed to accessions in the protein databases. They either represent new polypeptides or the internal peptide sequences obtained were not suited to allow homology-based identification between D. salina and other plant species.
In conclusion, D. salina can adapt to sulfur limitation by reduction of growth and photosynthesis. The basis for this general stress response is represented by specific up-regulation of sulfur assimilation, down-regulation of nitrogen assimilation, and a partial redirection of carbon toward C3 metabolites to optimize carbon usage. Therefore, Dunaliella is a suitable organism to investigate the interplay of the reductive assimilation pathways of sulfur, nitrogen, and carbon.
MATERIALS AND METHODS
Cell Culture
Dunaliella salina was cultured at constant temperature (30°C) and photon flux density (50 μmol m−2 s−1). The growth medium contained 1.5 m NaCl, 24 mm MgSO4, 24 mm Na2SO4, 20 mm MgCl2, 10 mm KCl, 10 mm CaCl2, 5 mm NaNO3, 185 μm H3BO3, 100 μm NaH2PO4, 7 μm MnCl2, 1.5 μm FeEDTA, 0.8 μm ZnCl2, 0.02 μm CoCl, and 0.002 μm CuCl2. The medium was buffered with 20 mm Tris (tris[hydroxymethyl]aminomethane)-HCl, pH 7.6. Algae acclimated to different sulfate concentrations were cultured in the same media, using MgSO4 at 6, 25, or 50 μm as the sole sulfur source. The reduction in Na+ and Mg2+ concentrations consequent to these changes in the media composition did not affect per se any of the parameters measured. Growth was recorded on three different cultures with a Burker hemocytometer. All experiments were carried out on cells in the midexponential growth phase. The responses of growth rates to changing sulfate concentration was modeled with the equation μ = ([SO42−] × μmax × a)/([a × {SO42−}] + μmax), where μ is the specific growth rate and a is the initial slope of the curve.
Average cell volume was determined from the analysis of 60 to 100 cells from each of three different cultures as described by Giordano and Bowes (1997).
Cell Constituents
Chlorophyll and β-carotene were extracted in 90% (v/v) acetone. Chlorophyll was determined according to Jeffrey and Humphrey (1975); β-carotene content was estimated according to Jensen (1978). Total proteins were measured following the procedure described by Peterson (1977). The measurements were effected in four replicates.
Internal sulfate and nitrate determinations were done on algae grown in the presence of either 48 mm or 6 μm sulfate. The cells were carefully washed with an isotonic NaCl solution. A deproteinized extract was obtained from approximately 20 × 106 cells, diluted 10 times with ultrapure water, and assayed for sulfate and nitrate with an ion chromatograph Dionex 4000i that was equipped with a 4-mm anion-exchange column Dionex IonPAc AS9-SC. Anions were eluted with a solution of 7 mm NaHCO3/Na2CO3 and determined with a suppressed conductivity detector (Dionex, Sunnyvale, CA). The measurements were performed on samples from three different cultures.
The cell content of ammonium was estimated using the Spectroquant kit (Merck, Rahway, NJ). The samples were extracted by sonication in a medium containing 50 mm HEPES (4-[2-hydroxyethyl]-1-piperazineethanesulfonic acid), pH 7.8, 5 mm EDTA, 5 mm dithiothreitol, 5 mm phenylmethylsulfonyl fluoride, 2% (w/v) polyvinylpyrrolidone-40, 10 mm MgSO4. The influence on the measurements of amino group release from protein was checked by assaying bovine serum albumin solutions and found to be negligible.
Amino acids were determined from supernatants of cell extracts in 0.5 mL of 0.1 n HCl, 1 mm EDTA, 4% (w/v) polyvinylpyrrolidone-40, after ultrasonication and centrifugation (15 min at 13,000g). Fluorescence derivatization with the Accq-Tag system was carried out according to the supplier (Waters, Milford, MA). The derivatized samples were analyzed with a Waters HPLC system equipped with Novapak C18 column (3.9 × 150 mm) with a precolumn. The separation of the amino acids was achieved using a gradient of eluent A (140 mm sodium acetate, 7 mm triethanolamine, pH 5.8) and eluent B (100% [v/v] acetonitrile). The gradient proceeded with 0% to 5% B in 27 min, 5% to 9% B in 1.5 min, and 9% to 18% B in 16 min at a flow rate of 1 mL min−1. Detection, identification, and quantification of fluorescence signals were carried out using the Waters Millenium software and individual standards for each amino acid.
Gas Exchange Measurements
Photosynthesis was measured as net O2 evolution in a Clark-type Hansatech DW2 oxygen electrode. Cells were harvested by centrifugation at 1,000g for 10 min and washed twice in growth medium free of DIC. All experiments were carried out at a temperature of 30°C, a cell concentration of 5 × 105 cells mL−1, and a saturating light of 420 μmol photons m−2 s−1 supplied by a red LED source (Hansatech, King's Lynn, UK). The measurements were started at 10% of O2 saturation and ended at approximately 50% of O2 saturation. Prior to the beginning of the experiments, cells were let photosynthesize to deplete the residual DIC in the medium and to reduce the internal DIC pool. Cells were then primed with 5 μm DIC. When net oxygen evolution was zero, sequential addition of DIC were effected to obtain a DIC response curve of O2 evolution. The results were modeled to the equation y = (Pmax × α × DIC)/(Pmax + [α × DIC]) describing a rectangular hyperbola (Smith, 1936), where Pmax is the carbon- and light-saturated rate and α is the initial slope of the curve. Dark respiration was measured as O2 consumption using the same Hansatech system and the same conditions used for the photosynthetic rate determinations. The values reported are those measured after incubation of the algae in the dark for 20 min.
Enzyme Activities
ATP-sulfurylase (ATP-S), PEPC, and NR activities were measured on crude extracts obtained from cells harvested at 1,000g for 10 min and washed with an isosmotic NaCl solution. The cells were broken on ice with a Ten-Broeck homogenizer in an extraction medium containing the following: for ATP-S, 50 mm Tris-HCl, pH 8.0, 1 mm EDTA, 10 mm MgCl2; for PEPC, 25 mm HEPES, pH 7.5, 20% (v/v) glycerol, 1 mm Na2EDTA, 10 mm MgCl2, 0.1% (v/v) Triton X-100, 5 mm malate, 0.05% (v/v) protease inhibitors (Sigma P-2714); and for NR, 50 mm HEPES, pH 7.8, 10 mm MgSO4, 5 mm EDTA, 5 mm dithiothreitol, 5 mm phenylmethylsulfonyl fluoride, 2% (w/v) polyvinylpyrrolidone-40. The homogenate was spun down at 7,900g for 15 min at 4°C; the supernatant was used for the activity assays. ATP-S activity was measured spectrophotometrically at 30°C according to Burnel (1984). PEP carboxylase activity was determined spectrophotometrically at 40°C following the method by Holdsworth and Bruck (1977). NR activity was determined in an assay mix that contained 50 mm phosphate buffer, pH 7.5, 0.35 mm NADH, 10 mm KNO3, measuring the decrease in the absorbence of NADH at 340 nm at 30°C. Periplasmic carbonic anhydrase activity was measured on whole cells using the electrometric method by Wilbur and Anderson (1948) as modified by Miyachi et al. (1983). The activity of this enzyme was expressed in arbitrary units according to Wilbur and Anderson (1948).
Electrophoretic Patterns, Western Blots, and Peptide Sequences
Cell extracts were prepared as described for amino acids analysis. Proteins were loaded on 7.5%, 10%, or 15% (v/v) SDS gels, depending on the molecular size of the relevant proteins. Gels were stained with Coomassie Brilliant Blue R-250.
Western blots were carried out according to Towbin et al. (1979). The immunodetection was effected using rabbit anti-PEP carboxylase antibody raised against PEPC1 of Selenastrum minutum (Rivoal et al., 1996; kindly donated to M.G. by Dr. Jean Rivoal).
Proteins for sequencing of internal peptides were separated by high-resolution SDS-PAGE and extracted from gel slices. The isolated proteins were treated with trypsin and the peptides obtained were isolated by reverse phase HPLC prior to Edman degradation as described by Hellmann et al. (1994). For N-terminal sequencing, proteins were blotted on polyvinylidene difluoride membranes according to Eckerskorn and Lottspeich (1993).
ACKNOWLEDGMENTS
We wish to thank Giancarlo Gobbi (Dipartimento dei Materiali e della Terra, Università di Ancona) for his assistance in the determination of intracellular anion contents. We are also indebted to Claudia Krüger, Christian Horstmann, and Udo W. Stephan (IPK Gatersleben) for peptide sequencing and amino acids analysis.
LITERATURE CITED
- Avron M, Ben-Amotz A. Dunaliella: Physiology, Biochemistry and Biotechnology. London: CRC Press; 1992. [Google Scholar]
- Borowitzka MA, Borowitzka LJ. Dunaliella. In: Borowitzka MA, Borowitzka LJ, editors. Microalgal Biotechnology. Cambridge, UK: Cambridge University Press; 1988. pp. 27–58. [Google Scholar]
- Brunold C. Regulatory interactions between sulfate and nitrate assimilation. In: De Kok LJ, Stulen I, Rennenberg H, Brunold C, Rauser WE, editors. Sulfur Nutrition and Assimilation in Higher Plants. The Hague, The Netherlands: SPB Academic Publishing; 1993. pp. 61–75. [Google Scholar]
- Burnel JN. Sulfate assimilation in C4 plants. Plant Physiol. 1984;75:873–875. doi: 10.1104/pp.75.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson DT, Saker LR, Purves JV. Depression of nitrate and ammonium influx in barley plants with diminished sulfate-status: evidence for co-regulation of nitrogen and sulfate intake. J Exp Bot. 1989;40:953–963. [Google Scholar]
- Dämmgen U, Walker K, Grünhage L, Jäger H-M. The atmospheric sulphur cycle. In: Schnug E, editor. Sulphur in Agroecosystems. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 75–114. [Google Scholar]
- Davies JP, Grossman AR. Responses to deficiencies in macronutrients. In: Goldschmidt-Clermont M, Merchant S, editors. The Molecular Biology of Chloroplast and Mitochondria in Chlamydomonas. Amsterdam, The Netherlands: Kluwer Academic Publishers; 1998. pp. 613–633. [Google Scholar]
- Davis JS, Giordano M. Biological and physical events involved in the origin, effects and control of organic matter in solar saltwork. Int J Salt Lake Res. 1996;4:335–347. [Google Scholar]
- Eckerskorn C, Lottspeich F. Structural characterization of blotting membranes and the influence of membrane parameters for electroblotting and subsequent amino acid sequence analysis of proteins. Electrophoresis. 1993;14:831–838. doi: 10.1002/elps.11501401133. [DOI] [PubMed] [Google Scholar]
- Ferreira RMB, Teixeira ARN. Sulfur starvation in Lemna leads to degradation of ribulose/bisphosphate carboxylase without plant death. J Biol Chem. 1992;267:6253–6257. [PubMed] [Google Scholar]
- Franco AR, Cardenas J, Fernandez E. Ammonium (methylammonium) is the co-repressor of nitrate reductase in Chlamydomonas reinhardtii. FEBS Lett. 1984;176:453–456. [Google Scholar]
- Gilbert SM, Clarkson DT, Cambridge M, Lambers H, Hawkesford MJ. SO42− deprivation has an early effect on the content of ribulose-1,5-bisphosphate carboxylase/oxygenase and photosynthesis in young leaves of wheat. Plant Physiol. 1997;115:1231–1239. doi: 10.1104/pp.115.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginzburg M. Dunaliella: a green alga adapted to salt. Adv Bot Res. 1987;14:93–183. [Google Scholar]
- Giordano M, Bowes G. Gas exchange and allocation in Dunaliella salina cells in response to the N source and CO2 concentration used for growth. Plant Physiol. 1997;115:1049–1056. doi: 10.1104/pp.115.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell R. Molecular physiology of plant sulfur metabolism. Planta. 1997;202:138–148. doi: 10.1007/s004250050112. [DOI] [PubMed] [Google Scholar]
- Hellmann U, Wernstedt C, Gonez J, Heldin CH. Improvement of an “in-gel” digestion procedure for the micropreparation of internal protein fragments for amino acid sequencing. Anal Biochem. 1994;224:451–455. doi: 10.1006/abio.1995.1070. [DOI] [PubMed] [Google Scholar]
- Holdsworth ES, Bruck K. Enzymes concerned with β-carboxylation in marine phytoplankton: purification and properties of phosphoenolpyruvate carboxykinase. Arch Biochem Biophys. 1977;182:87–94. doi: 10.1016/0003-9861(77)90286-7. [DOI] [PubMed] [Google Scholar]
- Huppe HC, Turpin DH. Integration of carbon and nitrogen metabolism in plants and algal cells. Annu Rev Plant Physiol Mol Biol. 1994;45:577–607. [Google Scholar]
- Jeffrey SW, Humphrey GF. New spectrophotometric equations for determining chlorophylls a, b, c1, c2 in higher plants, algae and natural phytoplankton. Biochem Physiol Pflanz. 1975;167:191–194. [Google Scholar]
- Jensen A. Chlorophylls and carotenoids. In: Hellebust JH, Craigie JS, editors. Handbook of Phycological Methods. Physiological and Biochemical Methods. Cambridge, UK: Cambridge University Press; 1978. pp. 59–70. [Google Scholar]
- Kaiser WM. Regulatory interaction of carbon- and nitrogen metabolism. Prog Bot. 1997;58:150–163. [Google Scholar]
- Larsson M, Olsson T, Larsson C-M. Distribution of reducing power between photosynthetic carbon and nitrogen assimilation in Scenedesmus. Planta. 1985;164:246–253. doi: 10.1007/BF00396088. [DOI] [PubMed] [Google Scholar]
- Lin M, Turpin DH, Plaxton W. Pyruvate kinase isozymes from the green alga Selenastrum minutum: II. Kinetic and regulatory properties. Arch Biochem Biophys. 1989;269:228–238. doi: 10.1016/0003-9861(89)90104-5. [DOI] [PubMed] [Google Scholar]
- Migge A, Bork C, Hell R, Becker TW (2000) Negative regulation of nitrate reductase gene expression by glutamate or asparagine accumulation in leaves of sulfur-deprived tobacco. Planta (in press) [DOI] [PubMed]
- Miyachi S, Tsuzuki M, Avramova T. Utilization modes of inorganic carbon for photosynthesis in various species of Chlorella. Plant Cell Physiol. 1983;24:441–451. [Google Scholar]
- Peterson GL. Simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal Biochem. 1977;83:346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Rivoal J, Dunford R, Plaxton WC, Turpin DH. Purification and properties of four phosphoenolpyruvate carboxylase isoforms from the green alga Selenastrum minutum: evidence that association of the 102-kDa catalytic subunit with unrelated polypeptides may modify the physical and kinetic properties of the enzyme. Arch Biochem Biophys. 1996;332:47–57. doi: 10.1006/abbi.1996.0315. [DOI] [PubMed] [Google Scholar]
- Rivoal J, Plaxton WC, Turpin DH. Purification and characterization of high- and low-molecular-mass isoforms of phosphoenolpyruvate carboxylase from Chlamydomonas reinhardtii. Biochem J. 1998;331:201–209. doi: 10.1042/bj3310201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sako Y, Takai K, Uchida A, Ishida Y. Purification and characterization of phosphoenolpyruvate carboxylase from the hyperthermophilic archeon Methanothermus sociabilis. FEBS Lett. 1996;392:148–152. doi: 10.1016/0014-5793(96)00805-8. [DOI] [PubMed] [Google Scholar]
- Smith EL. Photosynthesis in relation to light and carbon dioxide. Proc Natl Acad Sci USA. 1936;22:504–511. doi: 10.1073/pnas.22.8.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipping E, Carrick TR, Hurley MA, James JB, Lawlor AJ, Lofts S, Rigg E, Sutcliffe DW, Woof C. Reversal of acidification in upland waters of the English lake district. Environ Pollut. 1998;103:143–151. [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur KM, Anderson NG. Electrometric and colorimetric determination of carbonic anhydrase. J Biol Chem. 1948;176:147–154. [PubMed] [Google Scholar]
- Yildiz FH, Davies JP, Grossman AR. Characterization of sulfate transport in Chlamidomonas reinhardtii during sulfur-limited and sulfur-sufficient growth. Plant Physiol. 1994;104:981–987. doi: 10.1104/pp.104.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]