Abstract
The phytohormones auxin and cytokinin control development and maintenance of plant meristems and stem cell systems. Fluorescent protein reporter lines that monitor phytohormone controlled gene expression programmes have been widely used to study development and differentiation in the model species Arabidopsis, but equivalent tools are still missing for the majority of crop species. Barley (Hordeum vulgare) is the fourth most abundant cereal crop plant, but knowledge on these important phytohormones in regard to the barley root and shoot stem cell niches is still negligible. We have now analysed the role of auxin and cytokinin in barley root meristem development, and present fluorescent protein reporter lines that allow to dissect auxin and cytokinin signalling outputs in vivo. We found that application of either auxin or cytokinin to barley seedlings negatively impacts root meristem growth. We further established a barley cytokinin reporter, TCSnew, which revealed significant cytokinin signalling in the stele cells proximal to the QC, and in the differentiated root cap cells. Application of exogenous cytokinin activated signalling in the root stem cell niche. Commonly employed auxin reporters DR5 or DR5v2 failed to respond to auxin in barley. However, analysis of putative auxin signalling targets barley PLETHORA1 (HvPLT1) is expressed in a similar pattern as its orthologue AtPLT1 from Arabidopsis, i.e. in the QC and the surrounding cells. Furthermore, the PINFORMED1 (HvPIN1) auxin efflux carrier was found to be expressed in root and shoot meristems, where it polarly localized to the plasma membrane. HvPIN1 expression is negatively regulated by cytokinin and its intracellular localisation is sensitive to brefeldinA (BFA). With this study, we provide the first fluorescent reporter lines as a tool to study auxin and cytokinin signalling and response pathways in barley.
Introduction
The phytohormones auxin and cytokinin are crucial regulators in plant development, for example in embryogenesis, phyllotaxis, gravitropism and root and shoot formation [1–5]. Auxin controlled gene expression is transcriptionally regulated by AUXIN RESPONSE FACTORS (ARFs), which bind to short DNA sequences termed auxin responsive elements (AuxRes) in the promoter of target genes. At low auxin concentrations, the co-repressor TOPLESS represses auxin-regulated transcription by mediating the binding of AUXIN/ INDOLE-3-ACETIC ACID (Aux/IAA) proteins to ARFs. Perception of auxin by the TRANSPORT INHIBITOR RESPONSE 1/AUXIN SIGNALING F-BOX (TIR1/AFB) family proteins, subunits of an SCF E3-ligase protein complex, target the Aux/IAA proteins for degradation via the ubiquitin-proteasome pathway, thereby leading to the activation of the ARFs and hence auxin responsive gene expression (reviewed in [6]). Cytokinins are perceived by histidine kinase receptors (AHKs) which carry an extracellular CHASE domain for hormone sensing. Cytokinin perception leads to autophosphorylation of the receptor kinase domain and subsequent transfer of the phosphoryl group onto a histidine phosphotransfer-protein (AHP). This enables AHP allocation to the nucleus and relay of the phosphoryl group to type-B response regulators (type-B ARRs), which in turn regulate transcription of cytokinin responsive genes. Among their targets are type-A ARRs which negatively influence cytokinin signalling, thereby creating a negative feedback loop (reviewed in [7]). Phytohormone distributions in a tissue can be visualized by the expression of reporter genes under the control of known auxin- or cytokinin-responsive elements, such as DR5 and DR5v2 for auxin signalling, and Two Component signalling Sensor (TCS) and TCSnew (TCSn) for cytokinin [8, 9]. A more direct way to analyse the presence of auxin is monitoring the degradation of reporter proteins fused to the highly conserved SCF-TIR1 complex recognition domain (DII) of Aux/IAA proteins [10, 11]. In the Arabidopsis shoot apical meristem (SAM), these reporters uncovered auxin maxima in the primordia, and high levels of cytokinins in the center of the meristem and the primordia. In the root apical meristems (RAM), an auxin maximum is formed in the Quiescent Center (QC), in the columella initials and in differentiated columella cells, while cytokinin maxima are observed in the differentiated columella and the stele [2, 9, 12–14]. Post-embryonic development of plant organs depends on the activity of meristems, and this specific phytohormone distribution was shown to be required for meristem patterning [1, 13, 15, 16]. At least for auxin, fine-tuned short-distance transport is essential to establish and maintain its distribution pattern. PINFORMED (PIN) proteins serve as auxin efflux carriers and establish a directional auxin flow to maintain the auxin maximum [1, 15, 17–21]. Different PINs are expressed in specific, partially overlapping domains within the root meristem and polarly localize to the cell membrane, thereby exporting auxin only in one specific direction [15, 20]. Among downstream targets of auxin signalling in Arabidopsis are the PLETHORA (PLT) genes, which are members of the AINTEGUMENTA-like (AIL) subclass of the APETALA2/ethylene-responsive element binding proteins (AP2/EREBP) family of transcription factors [12]. AtPLT1 and AtPLT2 are redundantly required for the embryonic specification of the QC and for the maintenance of root stem cells in Arabidopsis [12]. The AtPLTs are expressed in the stem cell niche forming a concentration gradient with a maximum in the QC and the distal stem cells (DSCs), therefore mirroring auxin distribution [12, 22, 23]. Auxin and cytokinin signalling are connected at multiple points: for example, auxin induces ARR expression [24, 25], while ARR1 promotes expression of the Aux/IAA gene SHORT HYPOCOTYL2 (SHY2). Furthermore, cytokinin influences auxin transport by regulating the expression of auxin influx (LIKE AUXIN RESISTANT 2) and efflux carriers (PINs), causing a relocation of auxin [26–28]. Homologues of PIN auxin efflux carriers, as well as auxin-responsive homologues of PLTs were also identified in monocots, indicating that the principles of auxin and cytokinin transport and signalling is conserved between monocots and dicots [17, 19–21, 29–31]. Even though barley (Hordeum vulgare) is the fourth most produced crop plant in the world (FAO statistics 2014; http://faostat.fao.org), highly salt tolerant in comparison to other cereal crops (Maas and Hoffman, 1977) and therefore a valuable model plant in regard to abiotic stresses, only very few studies about auxin and cytokinin exist for barley [32–35]. Initial studies indicate a connection between cytokinin signalling and drought stress resistance in barley, which so far has not been fully explored [35]. In this study, we analysed cytokinin signalling and downstream targets of auxin and cytokinin in the barley shoot and root meristem, utilizing phytohormone treatment, RNA in situ hybridisations and transgenic fluorescent reporter lines. Application of the hormones to barley seedlings impairs root growth and meristem maintenance. The expression pattern of the cytokinin signalling reporter TCSn reveals cytokinin signalling in the stele proximal to the QC and in the differentiated root cap cells. The homologue of the auxin-responsive gene AtPLT1, HvPLT1, is expressed in a pattern similar to AtPLT1 in Arabidopsis, in particular in and around the QC. Furthermore, the putative auxin transporter HvPIN1a is expressed and polarly localised in the root meristem, its expression is regulated by cytokinin and the intracellular localisation is affected by BFA, similar to Arabidopsis. With our study we provide the first fluorescent reporter lines for phytohormone transport, signalling or responses in barley which serve as valuable tools to analyse the role of auxin and cytokinin in barley development.
Material and methods
Plant growth
To monitor root growth and expression of reporter genes in the root, seedlings were grown on square plates as described in [36]. For all experiments either the cultivar (cv.) Morex or Golden Promise were used as indicated. SAMs were monitored in plants grown 8 DAG on agar plates (Waddington stage I, “transition apex”) or plants grown on soil under greenhouse conditions for around 3 weeks (Waddington stage II “double-ridge”) [37].
Cloning
The HvpPLT1:HvPLT1-mVENUS construct was built by PCR amplification of a 1929 bp fragment upstream of the start codon of HvPLT1 (MLOC_76811.2 on morex_contig_73008/ HORVU2Hr1G112280.5 [38, 39]) as the putative promoter region from Morex genomic DNA (gDNA) and cloned by restriction and ligation via a AscI site into a modified pMDC99 [40]. The whole HvPLT1 coding region without stop codon (3433 bp) was amplified from Morex gDNA and inserted downstream of the promoter in the pMDC99 vector by Gateway cloning (Invitrogen). A C-terminal mVENUS [41] was integrated downstream of the gateway site by restriction and ligation via PacI and SpeI. The HvpPIN1:HvPIN1-mV construct was produced the same way, using 3453 bp upstream of the start codon of HvPIN1 (AK357068/MLOC_64867 on morex_contig_101983/ HORVU6Hr1G076110.1 [38, 39]) as putative promoter region and the whole HvPIN1 coding region including the stop codon. The mVENUS sequence was inserted by restriction and ligation via a SmaI restriction site into the sequence coding for the central hydrophilic region of the HvPIN1 protein, as described for a PIN1 reporter construct in Arabidopsis [2]. The insertion of mVENUS is depicted in S7 Fig. For the TCSn:VENUS-H2B cytokinin reporter construct, the TCSn regulatory sequence [9] was obtained in the pDONR221 gateway vector from Invitrogen and subsequently inserted by Gateway cloning into the modified pMDC99 vector. The auxin reporter construct DR5v2:VENUS-H2B was built by amplifying the DR5v2 promoter from the pGIIK/DR5v2::NLS-tdTomato plasmid (kind gift of Dolf Weijers, [11]) and inserted by Gateway cloning into the modified pMDC99 vector. The pMDC99 modified for TCSn:VENUS-H2B and DR5v2:VENUS-H2B contained the gateway cassette, the coding sequence of VENUS [42] and a T3A terminator, which were inserted by restriction and ligation with AscI and SacI from pAB114 (described in [43]). Furthermore, it contains the coding sequence of Arabidopsis HISTONE H2B (AT5G22880) at the C terminus of the VENUS gene, inserted via restriction and ligation at a PacI restriction site. The DR5:ER-GFP contains the auxin-response promoter DR5 that consists of 9 inverted repeats of the 11 b-sequence 5′-CCTTTTGTCTC-3′, a 46-bp CaMV35S minimal promoter element, and a tobacco mosaic leader sequence as translational enhancer fused to endoplasmatic reticulum -targeted GFP [2, 44]. The plasmid was a kind gift from the Benková lab.
Barley transformation
The barley cv. Golden Promise was used for transformation as described previously [45], tested for hygromycin resistance by growth on medium containing hygromycin and via PCR to detect the hygromycin gene. For root expression analysis, the seeds of the plants recovered from the transformed scutella were used (T1) and again tested for the presence of the reporter construct by PCR with primers binding in the gene of interest and the downstream reporter gene.
Preparation of the reporter line samples
Clearing of the transgenic reporter lines was performed as described for pea root nodules with an altered fixation step [46]. Root samples were fixed with 4% para-formaldehyde in phosphate buffered saline (PBS) for 1 h with applied vacuum. Samples were incubated in the clearing solution for 1 week in darkness at 4 °C. The roots of plant lines with weak expression, or to be examined uncleared, were embedded in warm liquid 5% (w/v) agarose in dH2O for stabilization and sectioned longitudinally in the center by hand with a razor blade. SAM preparation was carried out by removing all leaves from the SAMs and the expression was directly monitored without clearing.
Cell wall and starch staining
Modified pseudo-Schiff propidium iodide (mPS-PI) staining and microscopy of the stained samples was performed as described previously [36].
Treatments
The cytokinins 6-benzylaminopurine (6-BA) (Duchefa) and trans-zeatin (t-Z) (Sigma-Aldrich) were used, as well as the auxins NAA (Duchefa) and 2,4D (Duchefa). For phytohormone treatments of wild type plants, the hormones were added to the growth medium at the concentrations indicated in the results section. The mock control was treated with water. For phytohormone treatment of the TCSn:VENUS-H2B and HvpPIN1:HvPIN1-mVENUS and DR5v2:VENUS-H2B reporter lines, the phytohormones were added to PBS and the plates with 7 day-old seedlings were flooded with the hormone solution or pure PBS as mock control and incubated for 2–3 h to allow phytohormone uptake. After removing the buffer, plates were placed back into the phytochamber at a 45 ° angle and examined 24 h later. Brefeldin-A (BFA) treatment of the HvpPIN1:HvPIN1-mVENUS reporter line was performed as described before [47]. Roots were cut around 1 cm above the tip, which was then placed in PBS as mock control or PBS containing 50 μM BFA. Pictures of the outer cortex cell layers of the roots and the epidermis of the SAMs were taken at the time of treatment (0 h) and 2 h later.
RNA in situ hybridisations
Probes for the HvPLT1 mRNA were prepared from genomic DNA of the barley cv. Morex from the HvPLT1 start to stop codon (3433 bp). The DNA was cloned into the pGGC000 entry vector of the GreenGate cloning system [48] and then amplified including the T7 and SP6 promoter sites by PCR. RNA probes were produced as described [49]. The RNA probes were hydrolysed by adding 50 μl carbonate buffer (0.08 M NaHCO3, 0.12 M Na2CO3) to 50 μl RNA probe and incubation at 60 °C for 58 min. On ice, 10 μl 10% acetic acid, 12 μl sodium acetate and 312 μl EtOH were added, the RNA was precipitated and dissolved in RNase-free dH2O. RNA in situ hybridisations were performed on roots of plants 8 days after germination (DAG) as described previously [36]. Polyvinyl alcohol was added to a final concentration of 10% to the NBT/BCIP staining buffer. Permanent specimens were created by washing the slides in 50% EtOH, 70% EtOH, 95% EtOH and 100% EtOH for 2 min each and for 10 s in xylol, and after drying, a few drops of Entellan (Merck) and a cover slip were added.
Microscopy
The transgenic reporter lines with mVENUS or VENUS fluorophores were examined with a 40x water objective with a numeric aperture (NA) of 1.20 using the Zeiss confocal laser scanning microscope (LSM) 780. Yellow fluorescence was excited using a 514 nm Argon laser and the emission was detected between 519 and 620 nm. The pinhole was set to 2,24 airy units. Transmitted light pictures were recorded with a transmitted light detector (T-PMT). Pictures were recorded with the tile scan function with 10% overlap, a threshold of 0.70 and automatically stitched using the microscope software. RNA in situ hybridizations were examined using a plan-neofluar 20x objective with a NA of 0.50 or a plan-neofluar 40x objective with a NA of 0.75 using the Zeiss Axioskop light microscope.
Phylogenetic trees
For the PLT phylogenetic tree, Arabidopsis PLT sequences were taken from TAIR12 (www.arabidopsis.org); rice PLT sequences were named according to Li and Xue (Li and Xue, 2011); maize PLT sequences were identified in a BLAST search with AtPLT1 as template (e-value below 5e-75) on the Phytozomev.12.0 website and named according to Zhang and colleagues [29]; barley homologue proteins were identified by BLAST-p search on http://webblast.ipk-gatersleben.de/barley/ with AtPLT1 as template (e-value below 4e-47 for high-confidence genes and 2e-11 for low-confidence genes) [38]. For the PIN phylogenetic tree, barley PINs were identified via http://webblast.ipk-gatersleben.de/barley/ with BLAST-p using HvPIN1a (MLOC_64867) as template (e-value below 1e-41 for high and low-confidence genes); rice sequences are taken from [18]; Arabidopsis PINs were found in TAIR12 (www.arabidopsis.org); maize PINs originate from Phytozome v12 (e-value below 4.3e-29) and named according to [50]. Selected SoPIN1 proteins from Brachypodium and tomato (Solanum lycopersicum) were taken from [31, 51] to define the SoPIN1 clade. Alignments and evolutionary analyses were performed using MEGA7.0 (Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets (Kumar, Stecher and Tamura 2015)) and a MUSCLE alignment; the evolutionary history was inferred using the Maximum Likelihood method based on the JTT matrix-based model. The bootstrap consensus tree inferred from 100 replicates is taken to represent the evolutionary history of the taxa analyzed. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (100 replicates) are shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value.
Analyses
Picture analyses were carried out using Fiji [52]. For root length measurements, the mean root length of all roots from a single plant were measured. For meristem length measurements, the border between meristem and elongation zone was defined by the first cell in the outermost cortex cell layer that doubled in cell length compared to its distal neighbour and analysis was carried out qualitatively from direct observation (as described in [53]). For analysing the DSC layers the starch-free cells of three columns in the center of the root cap below the QC were counted and the mean for one column was calculated. Only roots with mPS-PI stained starch-containing cells were used for analysis. The transmembrane domains of the PIN proteins were predicted using the TMHMM method [54]. Plots and statistics were created in R [55]. Significance was determined by a two-tailed Student's T-Test with the given p value. For image processing, Adobe Photoshop was used. Contrast and brightness were adjusted in the mPS-PI sample pictures manually to increase the cell wall and starch visibility. When the fluorescence brightness was compared, the same changes were performed equally for all samples. The surface of the SAMs was extracted in MorphoGraphX [56].
Results
Cytokinin inhibits root growth and root meristem maintenance
The effects of auxin and cytokinin signalling can easily be studied by manipulation of the hormone levels in the plant, for instance by externally adding an excess of the hormone, or by inhibiting biosynthesis or signal perception. Reduction of cytokinin levels by overexpression of degradation enzymes leads to enhanced root growth and longer meristems in Arabidopsis [57], whereas application of cytokinin reduces the root and meristem length [27, 53]. While the effect of a manipulation of the cytokinin signalling and biosynthesis pathway was already studied in barley roots on a whole-organ level, it has not yet been analysed on a cellular level [33, 35]. To test the effect of cytokinin, we used both trans-zeatin (t-Z), a naturally occurring isoprenoid-type cytokinin [58], and the synthetic cytokinin 6-benzylaminopurine (6-BA), both of which were shown to affect root and meristem length upon application in Arabidopsis [27, 53]. 6-BA had a negative effect on root length in both used concentrations (1 μM and 10 μM) measured 10 DAG, while t-Z did not affect root growth significantly (Fig 1A). Root longitudinal growth can primarily be attributed to cell divisions in the meristematic zone and cell elongation in the elongation zone. Interestingly, the effect on the root meristems was much stronger for both, 6-BA and t-Z, and resulted in a reduction in meristem size (Fig 1B and 1C). Both hormones seem to affect meristem size by changes in cell division and/or differentiation rate, since a reduced cell number was responsible for the difference in overall meristem size, rather than the mean length of the meristematic cells (S1 Fig). Furthermore, both cytokinins reduced the width of the root meristem (S2 Fig). In summary, cytokinin application causes a reduction in root growth in barley which is due to a reduced meristem size and thereby a lower production of new root cells for growth in length or diameter.
Fig 1. Root length and meristem size of the barley cv. Morex upon cytokinin treatment for 10 days.
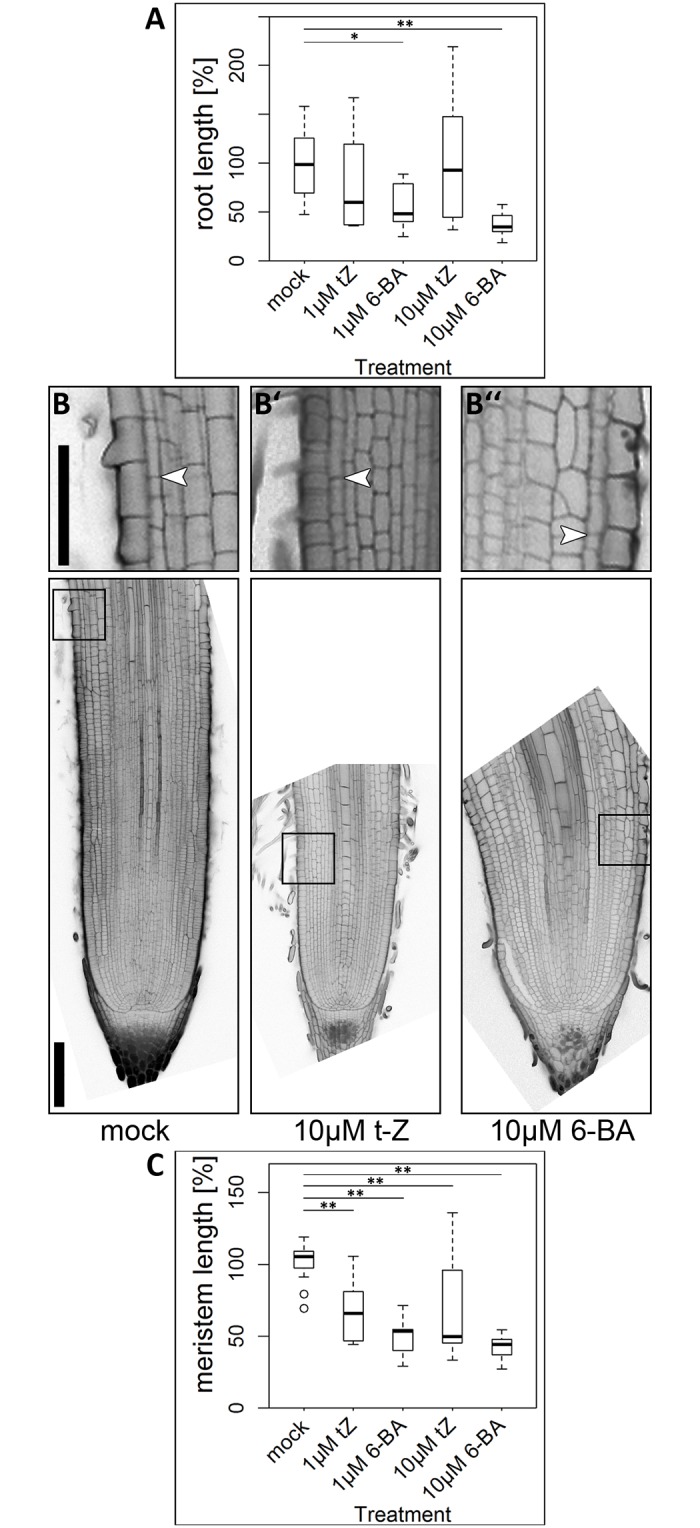
(A) Root length after 10 day-treatment with cytokinin; experiment was performed twice; values normalized to mock-treated plants; n = 7–18 plants per data point. (B)-(B”) Representative pictures of meristem phenotypes upon cytokinin treatment according to the captions; arrowheads mark the transition zones in the outer cortex layer; insets show magnifications of the transition zones; scale bars 200 μm (overviews) and 100 μm (magnified insets). (C) Meristem size after 10-day cytokinin treatment, measured by meristem length; experiment was performed twice; n = 11–16 roots per data point; significance was determined using the two-tailed Student’s t test, * = p<0.05, ** = p<0.001.
The cytokinin signaling reporter TCSn is expressed in the stele and root cap and can be activated by cytokinin application
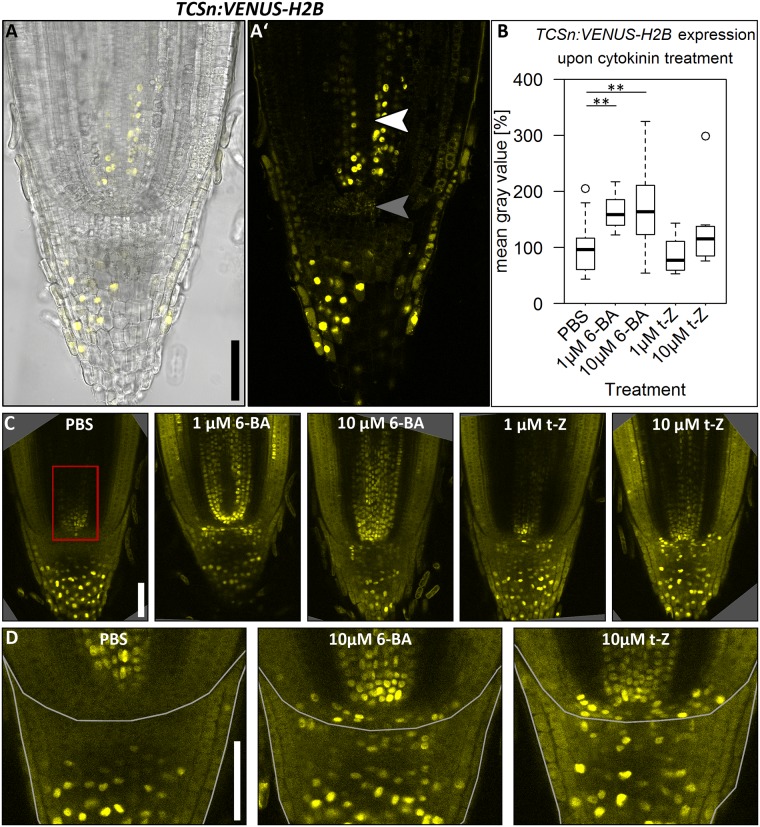
Since cytokinin application had an effect on root length and meristem size, we aimed to reveal the distribution of the phytohormone in the root. Therefore, we transformed barley cv. Golden Promise plants with a construct carrying a fluorescent VENUS reporter gene driven by the TCSn promoter within the vector backbone of pMDC99 [40]. This promoter carries concatemeric binding motifs for type-B ARRs combined with a minimal promoter, and responds to cytokinin signalling in Arabidopsis and maize protoplasts [9]. In mature barley root apical meristems (8 DAG), we observed expression of the cytokinin reporter TCSn:VENUS-H2B in the differentiated root cap and the stele (Fig 2A’), but not in the metaxylem (Fig 2A’, white arrow head), the QC or the surrounding initials (Fig 2A’, gray arrow head), resembling the expression pattern of the TCSn promoter in Arabidopsis [9]. 24 h treatment with 6-BA significantly increased TCSn:VENUS-H2B expression level in the stele, while there was only a weaker response to t-Z (Fig 2B and 2C). Cytokinins activated TCSn expression also in the DSCs, the cortex and endodermis initials, the epidermis initials and in a layer of the QC adjacent to the root cap (Fig 2D). In control plants, expression in the DSCs, the QC or the surrounding initials could never be observed (Fig 2D). However, we could not observe expression of the TCSn:VENUS-H2B in the SAM of any of the transgenic lines (S3 Fig).
Fig 2. Expression of the cytokinin reporter TCSn:VENUS-H2B in the root meristem of the barley cv. Golden Promise 8 DAG.
(A), (A’) TCSn:VENUS-H2B expression in untreated roots; transmitted light and VENUS emission (A)) and VENUS emission only (A’)); white arrow head in A’: metaxylem, gray arrow head in A’: QC; seven independent transgenic lines were examined and exhibit a similar expression pattern. (B) Quantification of TCSn:VENUS-H2B expression by the mean gray value of the region marked by red box in C); mean gray value normalized to the PBS control; significance was determined using the two-tailed Student’s t test, ** = p<0.001. (C) Representative pictures of TCSn:VENUS-H2B expression in root meristems upon 24 h of cytokinin treatment according to the captions; PBS without hormone was used as control; three independent transgenic lines were examined; the experiment was performed three times; n = 8–31 per treatment. (D) Magnification of the stem cell niche and root cap of roots upon treatments indicated by the captions; expression in the cortex/ endodermis initials, the DSCs, the QC layer adjacent to the root cap and the epidermis initials (PBS: 0/21 roots, 1 μM 6-BA: 1/9 roots, 10 μM 6-BA 8/18 roots, 1 μM t-Z 2/9 roots, 10 μM t-Z 5/8 roots); the root cap border is marked with a white frame; for a better comparison between samples, roots were cleared before microscopy (C), D), E)); scale bars 100 μm.
High auxin concentrations decrease root growth and negatively affect root meristem size
External application of auxin affects the root architecture of plants in regard to root length, meristem size and structure [20, 27, 59]. In barley, root growth was inhibited by all auxins tested [32]. To study barley root and meristem growth in more detail, we used the synthetic auxin NAA and the auxin analogue 2,4D, which cannot be exported from cells via auxin efflux carriers [60]. Low (10 nM) and high concentrations (1 μM and 10 μM) were used in comparison, as auxins are known to have opposite effects on meristem size at different concentrations [27]. Growing barley plants on medium containing either no phytohormone or the different auxins for 10 days revealed that neither root length nor root meristem length are affected by low concentrations (10 nM) of NAA or 2,4D, but both are decreased at high auxin concentrations (1μM 2,4D, 1μM and 10μM NAA) (Fig 3A, 3B and 3C). The reduction in meristem length went together with a reduction in meristematic cortex cell number (S1 Fig). Additionally, treatments with high concentrations (1 μM and 10 μM) increased meristem width (S2 Fig).
Fig 3. Root length, meristem size and DSC phenotype of the cv. Morex upon auxin treatment for 10 days.
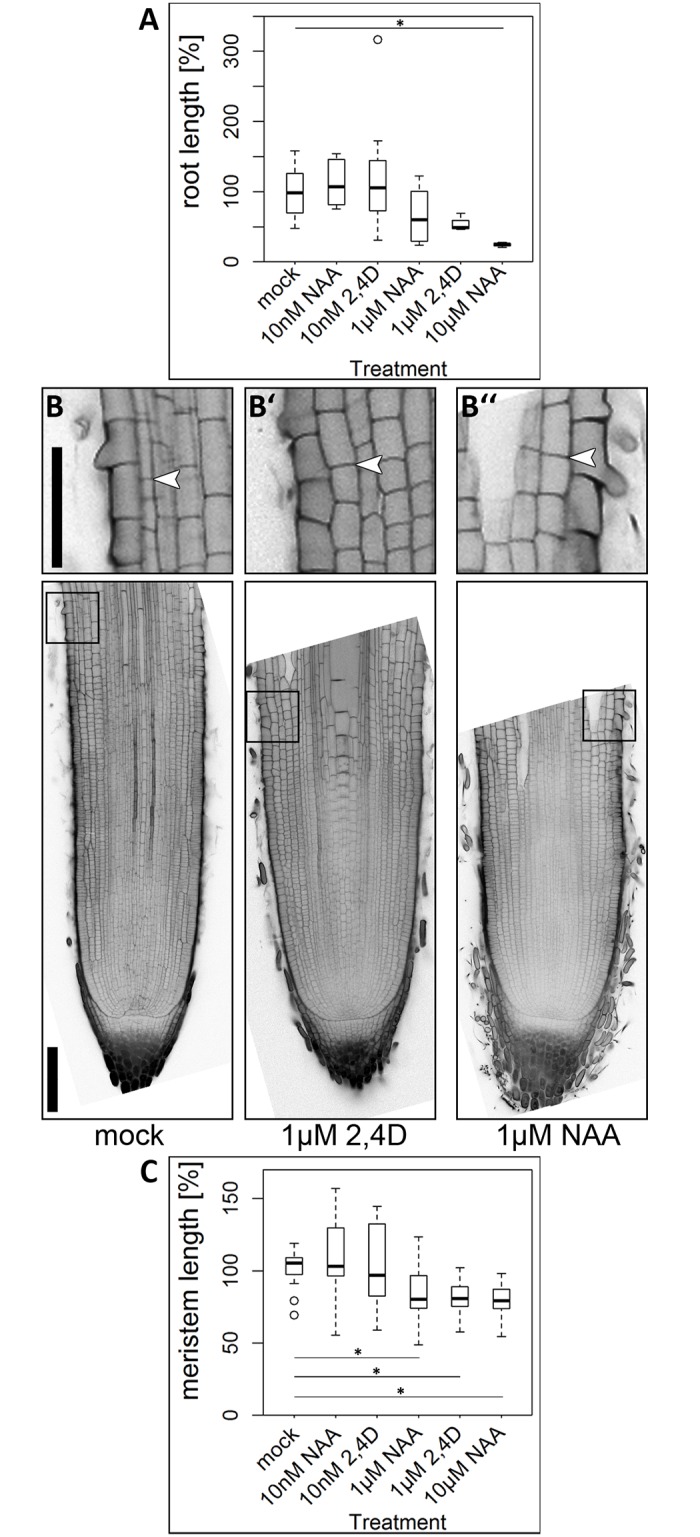
(A) Root length after 10 day-treatment with auxin; experiment was performed twice; for a better comparison between the experiments, all values were normalized to the mock-treated plants; n = 4–18 plants per data point. (B)–(B”) Representative pictures of the root meristem phenotype at 10 DAG upon hormone treatment according to the captions; arrow heads mark the transition zones; insets show magnifications of the transition zones; scale bars 200 μm and 100 μm in the magnification. (C) Meristem length upon hormone treatment, measured by meristem length; experiment was performed twice; all values are normalized to the mock-treated control; n = 7–17 roots per data point; significance was determined using the two-tailed Student’s t test, * = p<0.05, ** = p<0.001.
In Arabidopsis, auxin application results in differentiation of the DSCs, indicated by their accumulation of starch granules [61]. In barley, no significant difference in the number of DSC layers was detected after auxin application (S4 Fig).
The commonly used auxin signalling reporter DR5 and DR5v2 are not stably expressed in barley
As auxin reporters, two widely used regulatory sequences are the DR5 and the DR5v2, the former consisting of 9 inverted repeats of the auxin responsive element TGTCTC [8] and the latter of 9 repeats of the higher affinity ARF binding-site TGTCGG [11]. Here, the presence of auxin in a cell is indirectly determined through the activation of ARFs that bind to the synthetic promoters in an auxin-dependent manner, activating expression of the reporter genes. In Arabidopsis, the responsiveness of these reporters to auxin was confirmed by auxin application to the roots, leading to an enhanced expression of the reporter gene and a broadening of the expression domain [11]. The same reporters were successfully used in rice and maize to display the spatial domain of auxin signalling [14, 21], therefore, the same regulatory sequences might be usable to monitor auxin signalling in barley. Surprisingly, no expression of the DR5:GFP reporter could be detected in transgenic lines with this construct, and expression of the DR5v2:VENUS-H2B reporter lines was very weak and inconsistent between different roots and plant lines (S5 Fig). Furthermore, no increase of DR5v2:VENUS-H2B expression was detected even upon high auxin concentrations (10 μM 2,4D) (S5 Fig). Thus, the DR5 and DR5v2 reporters are not suitable to report on auxin signalling in barley.
Expression pattern of HvPLT1
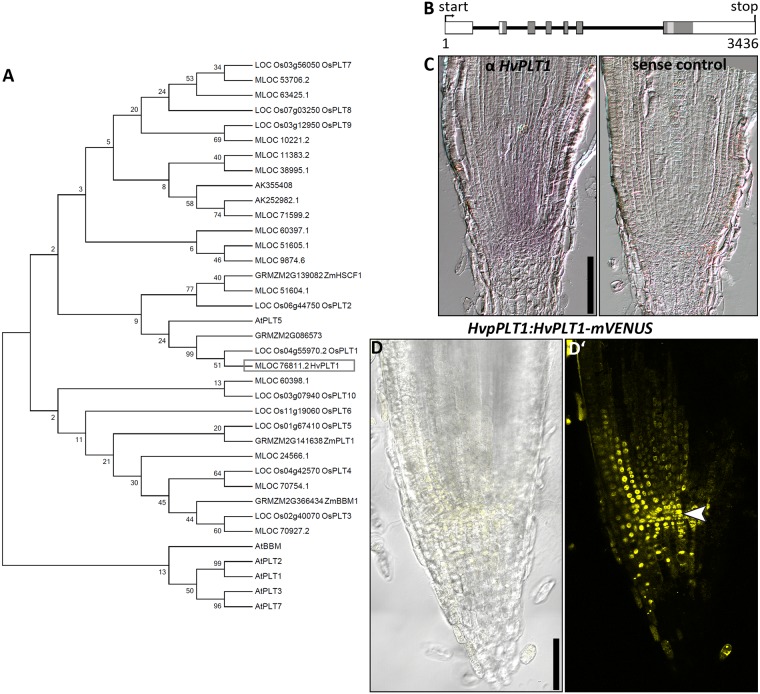
We therefore identified further genes known to be involved in auxin signalling, among them the PLT transcription factors [12, 22, 30]. In Arabidopsis, the expression of the AtPLTs is dependent on auxin signaling, and the expression of AtPLT1 and AtPLT2 spatially reflects the auxin distribution in the root tip, observed by the expression of the DR5 auxin reporters [12, 22, 23]. We searched the barley proteome [38] and created an unrooted tree of PLT family proteins from barley, rice, Arabidopsis and maize (Fig 4A). The rice OsPLT1 protein grouped together with AtPLT1-3 and AtBBM (AtPLT4) and therefore might have a similar function in the stem cell niche maintenance [30]. Consequently, we focused on MLOC_76811 as the closest homologue of OsPLT1 (Fig 4A), which we named HvPLT1 accordingly. HvPLT1 consists of two repeats of the conserved AP2 DNA binding domain and a conserved linker region (Fig 4B, http://pgsb.helmholtz-muenchen.de/plant/barley/) similar to AtPLT1 and AtPLT2 from Arabidopsis. We created transgenic reporter lines that expressed HvPLT1 fused to mVENUS under the control of 1929 bp of putative HvPLT1 promoter sequences. The reporter lines showed expression of HvPLT1 with the maximum in the QC and the surrounding cells, which gradually decreased towards the root cap, the proximal meristems and the outer root layers (Fig 4D and 4D’). Non-transgenic control plants did not show any expression (S6 Fig). RNA in situ hybridisations with a probe for HvPLT1 confirmed this expression pattern (Fig 4C).
Fig 4. PLT phylogenetic tree, HvPLT1 gene structure, promoter activity and protein localization in the root meristem of the barley cv. Golden Promise 8 DAG.
(A) Phylogenetic tree of PLT homologue proteins. (B) Genomic structure of the HvPLT1 coding sequence; boxes represent exons, black horizontal lines represent introns; dark gray boxes indicate coding sequence for AP2 domains, light gray boxes indicate coding sequence for the linkers between AP2 domains. (C) Representative picture of RNA in situ hybridizations with a probe for HvPLT1 (purple staining, C)) or the respective sense probe (C’)). (D) Representative picture of the HvpPLT1:HvPLT1-mVENUS emission in the root meristem; transmitted light and mVENUS emission (D)), mVENUS emission only (D’)); arrow head in D’) points to the QC; hand sections; seven independent transgenic lines were examined and exhibited similar expression patterns; scale bars 100 μm.
Identification of a PIN1 homologue in barley
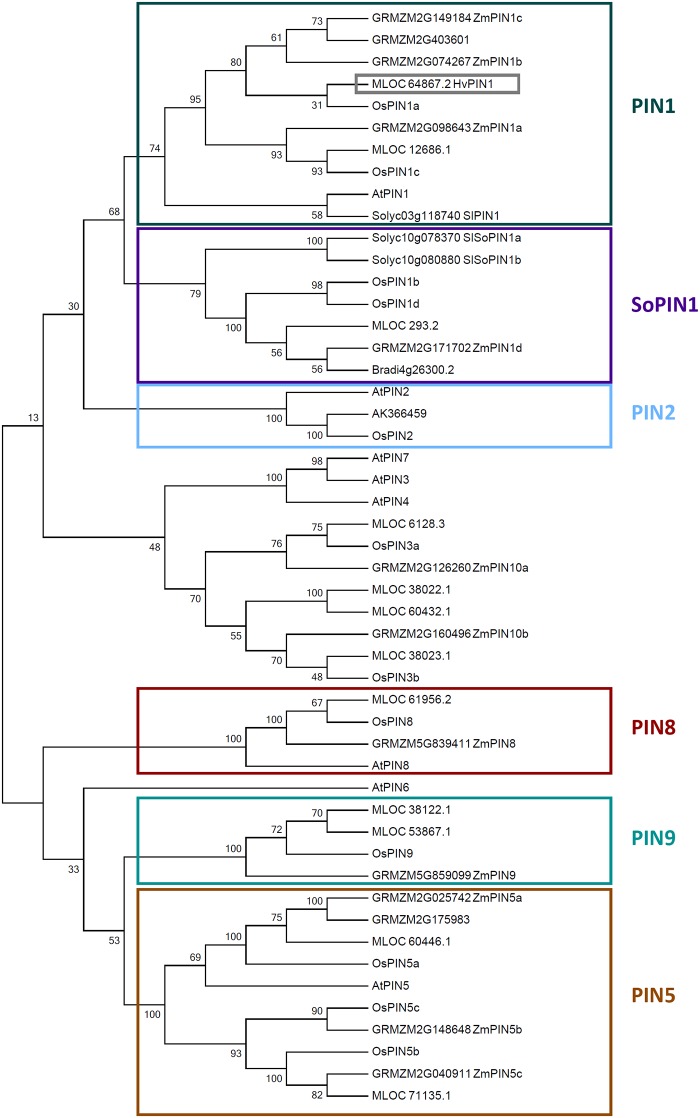
The HvPLT1 expression pattern suggests the presence of an auxin maximum in the QC and the root stem cell niche also in barley. A major source of auxin are young aerial tissues, from where the hormone is transported towards the root via the phloem [6] and subsequent cell-to-cell transport is facilitated by PIN proteins [15, 17, 20]. A search in the barley protein database for homologues of AtPINs discovered 13 putative HvPIN protein sequences that we used to build a phylogenetic tree and analyze their topology [38] (Fig 5 and S7 Fig). Based on the structure of the phylogenetic tree, we identified PIN1 (MLOC_64867.2, MLOC_12686.1), PIN2 (AK366549), PIN5 (MLOC_60446.1, MLOC_71135.1), PIN8 (MLOC_61956.2), PIN9 (MLOC_38112.1, MLOC_53867.1), PIN3 (MLOC_6128.3, MLOC_38023.1) and PIN10 (MLOC_38022.1, MLOC_60432.1) homologues and named them accordingly (Fig 5). Like maize and rice, also barley does not encode PIN4 and PIN7 homologues [17, 50]. For maize it was hypothesized that in the root apex, the three ZmPIN1s could take over the roles of PIN3, PIN4 and PIN7 efflux carriers [50]. The same distribution of functions could hold true for barley. Additionally, the barley genome encodes one member of the SISTER OF PIN1 (SoPIN1) clade (MLOC_293.2, Fig 5), which is conserved in flowering plants, but lost in Arabidopsis [31]. A transmembrane helices prediction analysis revealed that the HvPIN1 and HvPIN2 carry 4–5 transmembrane domains that group around a central hydrophilic region, similar to AtPIN1 (S7 Fig). HvPIN5, HvPIN8 and HvPIN9 in contrast exhibit only a short central hydrophilic region (S7 Fig). HvPIN3s and HvPIN10s did not show any typical structure of PIN proteins, and either lack a large hydrophilic region (HvPIN10a, HvPIN10b and HvPIN3b) or their hydrophilic region is not central (HvPIN3a) (S7 Fig). For the subsequent work on PIN protein localization in barley, we focussed on PIN1, as this is the best studied PIN protein in other model plants. Both in maize and rice, the two maize PIN1-like proteins and OsPIN1 show a similar transmembrane helices prediction profile, with two hydrophobic domains at the N and C termini and a central hydrophilic region [17, 19, 20]. From the HvPINs that grouped together with the other PIN1s, HvPIN1a is the one with the transmembrane helices prediction profile most similar to AtPIN1 (S7 Fig).
Fig 5. Phylogenetic tree of maize, Arabidopsis, rice and barley PINs.
Protein subfamilies are framed with the same colour; gray frame marks HvPIN1a.
Expression pattern and polar localization of HvPIN1a
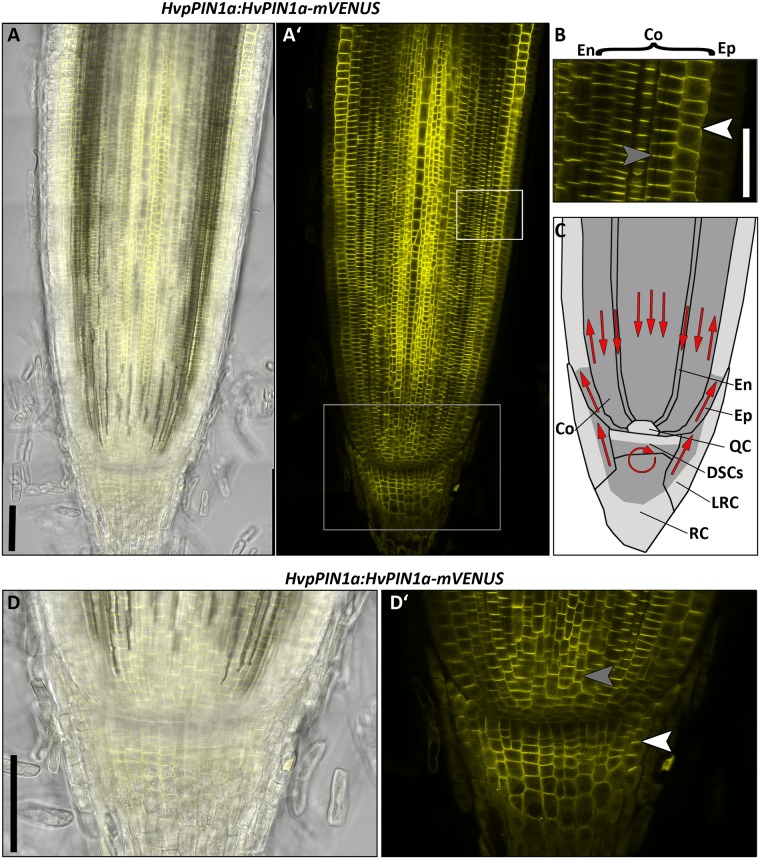
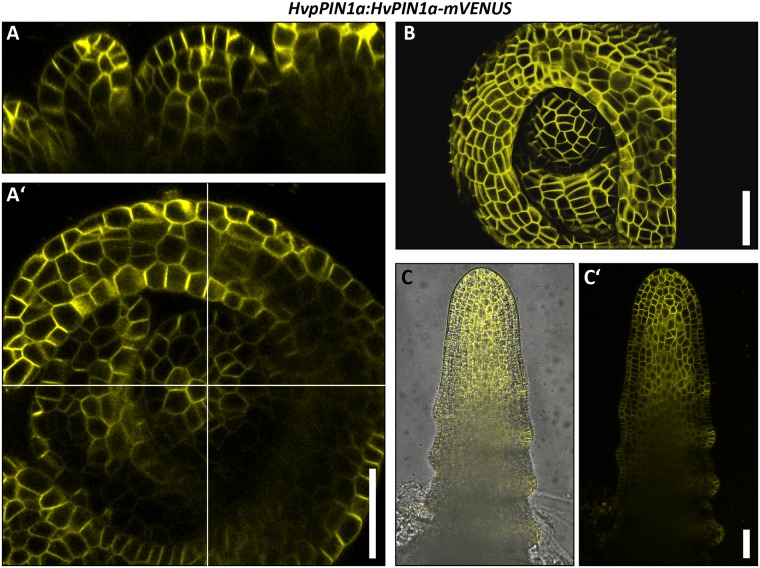
In the Arabidopsis root, PIN1 is expressed in the root meristem, in particular in the vasculature and endodermis, and weaker in the epidermis and cortex [15]. PIN1 homologues of maize and rice are expressed in the root meristem, and also in the root cap [17, 50]. We generated transgenic barley reporter lines using the genomic HvPIN1a sequence under control of the putative endogenous HvPIN1a regulatory sequences, consisting of 3453 bp upstream of the start codon. The sequence of the fluorophore mVENUS was inserted into the part of the HvPIN1a gene sequence that encodes the intracellular hydrophilic region of the protein, as it was described for the AtPIN1-GFP reporter [2] (S7 Fig). Strong expression was detected in the whole root meristem, except for the area of the presumed QC, where expression was weaker compared to surrounding tissues (Fig 6A’ and 6D’). High expression was observed in the stele, the endodermis, the cortex and DSCs, and the differentiated root cap (Fig 6D’). In the two analysed stages of the SAM, HvPIN1a was expressed throughout the whole meristem (Fig 7A and 7C). The PIN1s from Arabidopsis, maize and rice were shown to be mostly polarly localised to the plasma membranes at defined sides of the cells. In barley, a basal plasma membrane localisation was detected in the stele, endodermis and the inner cortex cell layers (gray arrow heads in Fig 6B and 6D’), but apical localisation was observed in the outermost cortex cell layer and the lateral root cap (white arrow head in Fig 6B and 6D’). In the central region of the root cap, polar localisation was not detectable, and HvPIN1a appeared evenly distributed at the plasma membrane (Fig 6D’). In the SAM, HvPIN1a was expressed everywhere, with expression peaks at the tips of the developing primordia (Fig 7). Here, we did not observe any polar localization of HvPIN1a (Fig 7).
Fig 6. HvPIN1a expression in the root meristem of the barley cv. Golden Promise 8 DAG.
(A) Representative picture of HvpPIN1a:HvPIN1a-mVENUS expression; transmitted light and mVENUS emission (A)), mVENUS emission only (A’)); six independent transgenic lines were examined which vary only in expression level but not in localisation or pattern; white box in A’) marks magnification in B); gray box in A’) marks magnification in D). (B) Magnification of the epidermal, cortical and endodermal cell layers depicted with white frame in A’). (C) Schematic illustration of HvPIN1a expression in the root meristem, high = dark gray, low = gray; red arrows indicate possible auxin flow created by localisation of PIN1 auxin transporters; En = endodermis, Co = cortex, Ep = epidermis, LRC = lateral root cap, RC = root cap. (D) Magnification of the stem cell niche depicted with gray frame in A’); transmitted light and mVENUS emission (D)), mVENUS emission only (D’)); white arrow heads mark apically localised PIN1, gray arrow heads mark basally localised PIN1; scale bars 100 μm in A), D); 50 μm in B).
Fig 7. HvPIN1a expression in the shoot meristem of the barley cv. Golden Promise.
(A), (A’) Representative picture of HvpPIN1a:HvPIN1a-mVENUS expression in the barley SAM in Waddington stage I; longitudinal view (A’)) and top view (A’)) of the same SAM. (B) Surface projection of HvpPIN1a:HvPIN1a-mVENUS of the same SAM as in A), created with MorphoGraphX [56]; (C), (C’) Representative picture of HvpPIN1a:HvPIN1a-mVENUS expression in the barley SAM in Waddington stage II; transmitted light and mVENUS emission (C)), mVENUS emission only (C’)); four independent transgenic lines were examined and vary only in expression strength but not in localisation and pattern; scale bars 50 μm.
HvPIN1a-mVENUS accumulates in vesicles upon Brefeldin-A (BFA) treatment
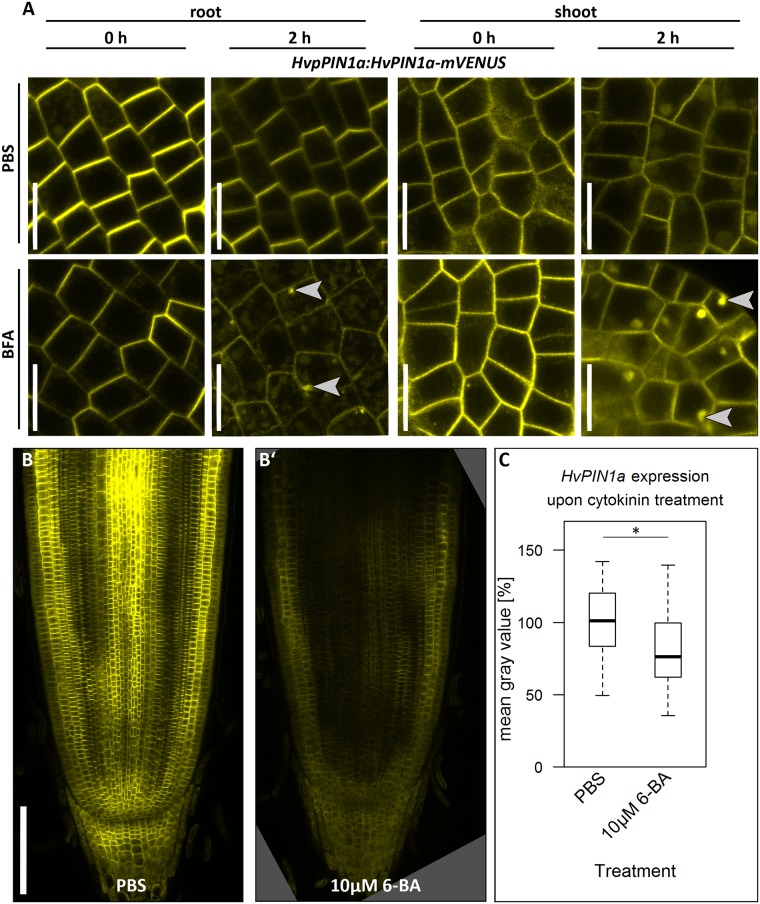
PINs are continuously recycled from the cell membrane to endosomes. Trafficking of basally localised PINs in Arabidopsis requires the GDP/GTP exchange factor for small G proteins of the ADP-ribosylation factor class (ARF-GEFs), which contain Sec7 domains [62–64]. The inhibitor of protein secretion BFA stabilizes an intermediate of the reaction of the ARF-GEF Sec7 domain with GDP, thereby blocking the cycle of activation of the ARF-GEFs and the recycling pathways [65]. Therefore, BFA can be used to reveal the involvement of BFA-sensitive ARF-GEFs in the PIN recycling pathways. Treatment with BFA induces intracellular accumulation of AtPIN1 by blocking the exocytosis of PIN1, which normally cycles rapidly between plasma membrane and endosomal compartments [47, 64]. To test if the PIN1 recycling mechanism is conserved in barley, we treated HvPIN1a-mVENUS expressing roots and SAMs with 50 μM BFA and monitored the HvPIN1a-mVENUS localisation after 2 h in the outer cortex cell file and the epidermis in root and shoot meristem, respectively. While HvPIN1a-mVENUS was exclusively localised at the apical cell membranes before the BFA treatment and upon mock controls, the formation of vesicles within the cells could be observed after 2 h of BFA treatment (gray arrow heads in Fig 8A). This indicates the existence of a conserved mechanism of PIN1 recycling between endosomal compartments and the plasma membrane in barley.
Fig 8. HvPIN1a localisation is influenced by BFA and its expression is decreased by cytokinin.
(A) Representative pictures of the HvpPIN1a:HvPIN1a-mVENUS expression in the outer cortex cell layer of the root meristem or in the epidermis of the SAM immediately after (0 h) or 2 h after mock (PBS) or 50 μM BFA treatment; gray arrow heads point to vesicles; scale bars 20 μm; three independent transgenic lines were examined; experiments were performed twice; n = 4–6; one transgenic line was used (in case of the root meristem); experiments were performed twice; n = 3–5; two transgenic lines were used (in case of shoot meristem). (B) Representative pictures of HvpPIN1a:HvPIN1a-mVENUS expression upon either mock (PBS, B)) or cytokinin (B’)) treatment as indicated in the captions; scale bar 200 μm. (C) Quantitative analysis of the HvpPIN1a:HvPIN1a-mVENUS expression upon cytokinin expression in B), measured by the mean gray value of the whole root meristem and the root cap; values are normalized to the PBS-control; five different independent transgenic lines were used; experiment was performed twice; n = 24 per treatment; significance was determined using the two-tailed Student’s t test, * = p<0.05.
HvPIN1a expression is regulated by cytokinin
As the recycling of the HvPIN1a protein is similarly affected by BFA as it is in Arabidopsis, we examined if gene expression of HvPIN1a is regulated by the same factors as in Arabidopsis. Dello Ioio and colleagues showed that AtPIN1 expression is downregulated by cytokinin [26, 27]. In barley, treatment with the cytokinin 6-BA for 24 h reduced HvPIN1a-mVENUS expression as well (Fig 8B and 8C). The downregulation was mostly visible in the stele, the place of cytokinin signaling (Fig 8B). However, if this downregulation occurs at the level of transcription like in Arabidopsis, or on protein level, we cannot distinguish here [26].
Discussion
Creation and validation of auxin and cytokinin reporter lines in barley
With this study, we started to address the roles of auxin and cytokinin in barley root and shoot meristem development at cellular resolution. We found that the DR5 and DR5v2 regulatory elements that are successfully used in many other plant species to determine auxin signalling were not consistently expressed in our transgenic barley lines [11, 14, 50] (S5 Fig). In Arabidopsis, the two auxin reporters DR5 and DR5v2 showed differences in their expression patterns, indicating that ARFs have different binding affinity towards the respective auxin response elements TGTCTC (DR5) and TGTCGG (DR5v2) [11]. Moreover, it was shown that the spacing between the auxin responsive elements, their flanking sequences and the number of repeats are important for the reactivity of the reporter to auxin [8]. We have to conclude from the data presented here that in barley, different auxin responsive elements and/or a different composition of the reporters are necessary to create transcriptional reporter lines for auxin signalling. We could, however, successfully monitor the expression of auxin-related genes. We found that the expression pattern of HvPLT1 in root meristems is similar to that of AtPLT1 in Arabidopsis and the root-specific rice PLTs, with an expression maximum around the QC [22, 30], both on RNA and protein level (Fig 4). This suggests that the expression of PLTs is conserved between these plant species, indicating that the well described auxin- and PLT-mediated cell specification mechanism in the root meristem is conserved between Arabidopsis and barley [12, 22, 23]. The expression of PIN1 in the root is conserved between species, but expression in individual tissues differs (Fig 6A’) [15, 17, 50]. Furthermore, the intracellular localisation of PINs differs depending on their domain topology. Křeček and colleagues sorted the eight Arabidopsis PINs into two subfamilies, namely the “long” and the “short” PINs according to the length of their hydrophilic region [66]. The “long” PIN subfamily is characterised by its central hydrophilic loop, separating two hydrophilic domains, each consisting of five trans-membrane regions. They are primarily localised to the plasma membrane in the cell [2, 3, 15]. The "short" PINs, however, possess a short central hydrophilic region and localise to internal cell membranes [67]. Accordingly, HvPIN1a localised to the plasma membrane (Fig 6B and 6D’). We furthermore discovered PINs that cannot be assigned as "long" or "short" PIN because their transmembrane topology does not follow either structure (S7 Fig). This divergence in transmembrane topology has not been reported for PINs in Arabidopsis, rice and maize and therefore, the localisation and function of these PINs should be subjected to a closer examination. The expression pattern and polar localization of HvPIN1a indicate that also in barley, an auxin flow is created that is directed towards the QC, the stem cell niche and the root cap and also a flow from the stem cell niche to the proximal meristem via the outer cortex cell layers (Fig 6C), as it was proposed for the Arabidopsis PINs [15]. In the SAM, AtPIN1 is expressed in the epidermis and in subepidermal cells, and only below the primordia, the expression becomes restricted to the presumed provascular tissues [68]. In maize, ZmPIN1a is also predominantly expressed in the epidermis of axillary meristems and the inflorescence meristem, but also in underlying tissues [21]. Like in the root, HvPIN1a is more broadly expressed than PIN1 in Arabidopsis, not only in the epidermis but evenly in all tissue layers of the SAM (Fig 7). Like in the root, it is possible that close PIN1 homologues take over the role of a tissue-layer specific auxin transporter in barley shoots (S7 Fig).
Genetic analysis and BFA-treatment experiments revealed that in Arabidopsis, the basal localisation of PINs is dependent on the ARF-GEF GNOM [64]. The kinases D6 PROTEIN KINASE and PINOID regulate PIN localization by phosphorylating the PINs at the plasma membrane, making them less affine to the GNOM-dependent basal recycling pathway. The phosphorylated PIN proteins are then recruited to the apical GNOM-independent trafficking pathways [62–64]. For the apically localised AtPIN2 in the Arabidopsis epidermis, however, it was shown that its vacuolar trafficking is independent of GNOM and involves an additional, BFA-sensitive ARF-GEF [69]. In the outer cortex cell layer, where HvPIN1a is localised apically, and in the SAM epidermis, where we could not observe a polar localisation, BFA caused the accumulation of HvPIN1a in vesicles (Fig 8A), indicating that in barley, too, BFA-sensitive components are involved in PIN1 trafficking. Besides intracellular localisation of the PIN1 protein, also the expression of PIN1 is subject to regulation by other factors. Dello Ioio and colleagues showed that in Arabidopsis, AtPIN1 expression is downregulated by cytokinin [26]. In rice, the PIN1 homologues OsPIN1a, OsPIN1b and OsPIN1c, however, are not transcriptionally regulated by cytokinin [17]. In barley, HvPIN1a expression is downregulated by cytokinin application (Fig 8B and 8C). Thus, HvPIN1a expression is similarly regulated in barley as it is in Arabidopsis, but different from rice, where OsPIN1 expression is cytokinin independent [17, 26].
In contrast to the auxin reporters DR5 and DR5v2, the cytokinin signaling sensor TCSn was functional in barley, since its expression pattern is conserved between Arabidopsis and barley, and, more importantly, the expression pattern changes upon cytokinin application (Fig 2) [9]. However, although the TCSn reporter confers expression in the shoot meristem in Arabidopsis [9], we could not detect any expression of TCSn:VENUS-H2B in barley SAMs. Since the TCSn reporter consists of concatemeric repeats of the DNA-binding motif of the Arabidopsis type-B ARR [9], it is possible that in barley, only some tissues express proteins that induce the TCSn regulatory sequence, so that despite ongoing cytokinin signalling in the cell, the reporter gene is not activated.
Role of auxin and cytokinin for barley root meristems
Previous studies on hormone activities in barley had addressed how manipulation of endogenous cytokinin levels and auxin affects root growth, at the whole organ level [32, 33, 35]. We have extended these analyses to the meristem and cellular level. The reduction in root growth as well as a reduction in meristem size upon 6-BA application in barley is similar to that observed for Arabidopsis (Fig 1) [27, 53]. In Arabidopsis, components of cytokinin signalling in the root stem cell niche control the differentiation at the transition zone [25, 53]. Expression of the cytokinin reporter TCSn:VENUS-H2B in barley was enhanced in the stem cell niche upon cytokinin treatment, indicating that the reduction of meristem size upon cytokinin treatment might also depend on enhanced cytokinin signalling in the QC, as observed for Arabidopsis [25]. Interestingly, all effects on root growth and root meristem size were less pronounced upon treatment with the cytokinin t-Z compared to 6-BA. In Arabidopsis, CYTOKININ OXIDASES/DEHYDROGENASES (AtCKXs), which are involved in the degradation of cytokinins, preferentially cleave isoprenoid cytokinins including t-Z, but not 6-BA; similarly, CKX1 from maize predominately cleaves free cytokinin bases including t-Z [70, 71]. In barley, thirteen putative members of the HvCKX family were identified [34]. Their presence could lead to an enhanced degradation of the externally added t-Z, thereby leading to a reduced influence on root growth and meristem maintenance in comparison to 6-BA.
Besides cytokinin, we analysed the influence of the synthetic auxin NAA and the non-transportable synthetic auxin analogue 2,4D on barley root growth. In contrast to studies in Arabidopsis, where low concentrations of auxins were shown to increase the root growth rate [72, 73], we observed no enhancement of root growth rates upon applications of low concentrations of NAA and 2,4D (10 nM) in barley (B). Likewise, positive effects on root growth rate have not been reported in monocots before [14, 59]. The auxin concentrations used in Arabidopsis that increased the root growth rate, however, ranged from 0.001 to 10 nM [72, 73], whereas the applied auxin concentrations in maize, rice and our study were much higher [14, 59]. Thus, either lower auxin concentrations would indeed lead to an enhancement of root growth in monocots, or auxin levels are already saturated in monocots and therefore, no positive effect on root growth can be achieved.
In accordance to studies on Arabidopsis, maize and rice, we observed a reduced root length and meristem size upon treatment with high concentrations of auxin, both in regard to meristem cell number and meristem length (Fig 3, S1 Fig) [14, 27, 59, 73]. Besides the effect of auxin application on longitudinal root growth and meristem size in Arabidopsis, the phytohormone also influences the DSCs that give rise to the columella cells. Auxin application leads to a differentiation of these stem cells, marked by accumulation of starch granules [61]. In barley, however, we could not observe any starch granule accumulations in additional cell files (S4 Fig). Previously, we have published similar observations for the application of a CLE peptide. CLE peptides were shown to cause both a differentiation of the proximal root meristem and the DSCs in Arabidopsis. Application of the CLE peptide in barley, however, did only affect the proximal root meristem but not the DSC differentiation [36, 74]. This indicates that DSC maintenance, in contrast to root meristem maintenance, is regulated differently in barley than in Arabidopsis.
In summary, we have shown here important similiarities and differences in root meristem development and the role of phytohormones between barley, other crop species and the model organism Arabidopsis. We have also characterised a first set of fluorescent reporter lines for barley at cellular resolution, which will be useful for further in-depth studies of the poorly understood development of one of the major crop plants worldwide.
Supporting information
A) Meristem cell number upon 10-day cytokinin treatment; experiment was performed twice; n = 11–16 roots per data point. B) Meristem length upon auxin treatment; experiment was performed twice; all values are normalized to the mock-treated control; n = 7–17 roots per data point; significance was determined using the two-tailed Student’s t test, * = p<0.05, ** = p<0.001.
(TIF)
Meristem width measured at the transition zone from root meristems exemplarily shown in Figs 1B and 3B. A) Roots were treated with cytokinin for 10 days; experiments were performed twice; n = 15–25 roots per data point. B) Roots were treated with auxin for 10 days; experiment was performed twice, n = 12–25 roots per data point; significance was determined using the two-tailed Student’s t test, * = p<0.05, ** = p<0.001.
(TIF)
A), A’) Undectectable TCSn:VENUS-H2B expression in SAMs in waddington stage I; transmitted light and VENUS emission (A)) and VENUS emission only (A’)). B), B’) Undectectable TCSn:VENUS-H2B expression in SAMs in waddington stage II; transmitted light and VENUS emission (B)) and VENUS emission only (B’)). Seven independent transgenic lines were examined and show no expression in the SAM; scale bars 100 μm; insets in A’) and B’) show respective pictures with tonal correction to show autofluorescence.
(TIF)
A) Exemplary pictures of the root stem cell niche upon mock or auxin treatment as indicated; scale bar 100 μm. B) Number of DSC layers upon 10-day treatment with auxin; no significant difference to mock-treated plants; experiment was performed twice; n = 5–20 per data point. Significance was determined using the two-tailed Student’s t test, * = p<0.05, ** = p<0.001.
(TIF)
A) Exemplary picture of DR5:GFP root; transmitted light and (undetectable) GFP emission (A)), undetectable GFP emission only (A’)); four independent transgenic lines were examined and show no GFP epression; scale bar 200 μm. B) DR5v2:VENUS-H2B lines show only variable or no expression and do not show a consistent reaction on 2,4D treatment; number of expressing DR5v2:VENUS-H2B lines (B)); number of plants that show the respective expression change upon treatment with 10 μM 2, 4D for 24 h (B’)).
(TIF)
A) Representative picture of the root meristem of a non-transgenic Golden Promise seedling 8 DAG; transmitted light and mVENUS emission (A)), mVENUS emission only (A’)), same settings as in Fig 4B and 4B’; hand-sections as described in Material and Methods; only background signal with mVENUS excitation. B) Representative picture of the root meristem of a non-transgenic Golden Promise seedling 8 DAG; transmitted light and mVENUS emission (B)), mVENUS emission only (B’)), same settings as in Fig 5A’; cleared as described in Material and Methods; only background signal with mVENUS excitation; scale bars 100 μm; inset in A’) shows respective pictures with tonal correction to show autofluorescence.
(TIF)
Topology of the transmembrane barley PIN proteins in comparison to AtPIN1; domains predicted to the inside of the cell are shown in light-gray, transmembrane domains are shown in dark-gray and domains outside the cell are depicted in black according to the legend; in the protein topology of MLOC_64867—HvPIN1a the asterisk marks the site where mVENUS is inserted for the reporter line shown in Fig 5; newly identified HvPINs are named according to their topology and the cluster of the Arabidopsis, maize and rice PIN family to which they belong.
(TIF)
Acknowledgments
We thank Carin Theres for technical assistance in the lab, Marc Somssich and Ikram Blilou for valuable discussions about the manuscript and Dolf Weijers for kindly providing the DR5v2 plasmid. We would furthermore like to acknowledge the Center of Advanced Imaging (CAi) at the Heinrich Heine University Düsseldorf for help with microscopy and MorphographX. Work in RS and MvKS labs was supported by the DFG through CEPLAS (EXC1028).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Work in Rüdiger Simon's lab has been supported by the Deutsche Forschungsgemeinschaft through CEPLAS (EXC1028) (www.ceplas.eu).
References
- 1.Reinhardt D, Pesce E-R, Stieger P, Mandel T, Baltensperger K, Bennett M, et al. Regulation of phyllotaxis by polar auxin transport. Nature. 2003;426: 255–260. doi: 10.1038/nature02081 [DOI] [PubMed] [Google Scholar]
- 2.Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, et al. Local, Efflux-Dependent Auxin Gradients as a Common Module for Plant Organ Formation. Cell. 2003;115: 591–602. doi: 10.1016/S0092-8674(03)00924-3 [DOI] [PubMed] [Google Scholar]
- 3.Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, et al. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature. 2003;426: 147–53. doi: 10.1038/nature02085 [DOI] [PubMed] [Google Scholar]
- 4.Gordon SP, Chickarmane VS, Ohno C, Meyerowitz EM. Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc Natl Acad Sci U S A. 2009;106: 16529–34. doi: 10.1073/pnas.0908122106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marchant A, Kargul J, May ST, Muller P, Delbarre A, Perrot-Rechenmann C, et al. AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J. 1999;18: 2066–73. doi: 10.1093/emboj/18.8.2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saini S, Sharma I, Kaur N, Pati PK. Auxin: A master regulator in plant root development. Plant Cell Rep. 2013;32: 741–757. doi: 10.1007/s00299-013-1430-5 [DOI] [PubMed] [Google Scholar]
- 7.Bishopp A, Benková E, Helariutta Y. Sending mixed messages: auxin-cytokinin crosstalk in roots. Curr Opin Plant Biol. 2011;14: 10–6. doi: 10.1016/j.pbi.2010.08.014 [DOI] [PubMed] [Google Scholar]
- 8.Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA Proteins Repress Expression of Reporter Genes Containing Natural and Highly Active Synthetic Auxin Response Elements. Plant Cell. 1997;9: 1963–1971. doi: 10.1105/tpc.9.11.1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zurcher E, Tavor-Deslex D, Lituiev D, Enkerli K, Tarr PT, Muller B. A Robust and Sensitive Synthetic Sensor to Monitor the Transcriptional Output of the Cytokinin Signaling Network in Planta. PLANT Physiol. 2013;161: 1066–1075. doi: 10.1104/pp.112.211763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunoud G, Wells DM, Oliva M, Larrieu A, Mirabet V, Burrow AH, et al. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature. 2012;482: 103–106. doi: 10.1038/nature10791 [DOI] [PubMed] [Google Scholar]
- 11.Liao C, Smet W, Brunoud G, Yoshida S, Vernoux T, Weijers D. Reporters for sensitive and quantitative measurement of auxin response. Nat Methods. Nature Publishing Group; 2015;12: 207–210. doi: 10.1038/nmeth.3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, et al. The PLETHORA Genes Mediate Patterning of the Arabidopsis Root Stem Cell Niche. Cell. 2004;119: 109–120. doi: 10.1016/j.cell.2004.09.018 [DOI] [PubMed] [Google Scholar]
- 13.Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, et al. An Auxin-Dependent Distal Organizer of Pattern and Polarity in the Arabidopsis Root. Cell. 1999;99: 463–72. http://www.ncbi.nlm.nih.gov/pubmed/10589675 [DOI] [PubMed] [Google Scholar]
- 14.Yang J, Yuan Z, Meng Q, Huang G, Périn C, Bureau C, et al. Dynamic Regulation of Auxin Response during Rice Development Revealed by Newly Established Hormone Biosensor Markers. Front Plant Sci. 2017;8: 256 doi: 10.3389/fpls.2017.00256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, et al. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature. 2005;433: 39–44. doi: 10.1038/nature03184 [DOI] [PubMed] [Google Scholar]
- 16.Stahl Y, Simon R. Plant stem cell niches. Int J Dev Biol. 2005;49: 479–89. doi: 10.1387/ijdb.041929ys [DOI] [PubMed] [Google Scholar]
- 17.Wang JR, Hu H, Wang GH, Li J, Chen JY, Wu P. Expression of PIN genes in rice (Oryza sativa L.): Tissue specificity and regulation by hormones. Mol Plant. The Authors. All rights reserved.; 2009;2: 823–831. doi: 10.1093/mp/ssp023 [DOI] [PubMed] [Google Scholar]
- 18.Miyashita Y, Takasugi T, Ito Y. Identification and expression analysis of PIN genes in rice. Plant Sci. Elsevier Ireland Ltd; 2010;178: 424–428. doi: 10.1016/j.plantsci.2010.02.018 [Google Scholar]
- 19.Xu M, Zhu L, Shou H, Wu P. A PIN1 Family Gene, OsPIN1, involved in Auxin-dependent Adventitious Root Emergence and Tillering in Rice. Plant Cell Physiol. 2005;46: 1674–1681. doi: 10.1093/pcp/pci183 [DOI] [PubMed] [Google Scholar]
- 20.Carraro N, Forestan C, Canova S, Traas J, Varotto S. ZmPIN1a and ZmPIN1b Encode Two Novel Putative Candidates for Polar Auxin Transport and Plant Architecture Determination of Maize. Plant Physiol. 2006;142: 254–64. doi: 10.1104/pp.106.080119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallavotti A, Yang Y, Schmidt RJ, Jackson D. The Relationship between Auxin Transport and Maize Branching. Plant Physiol. 2008;147: 1913–1923. doi: 10.1104/pp.108.121541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, et al. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature. 2007;449: 1053–7. doi: 10.1038/nature06206 [DOI] [PubMed] [Google Scholar]
- 23.Mähönen AP, ten Tusscher K, Siligato R, Smetana O, Díaz-Triviño S, Salojärvi J, et al. PLETHORA gradient formation mechanism separates auxin responses. Nature. 2014;515: 125–129. doi: 10.1038/nature13663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Müller B, Sheen J. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature. 2008;453: 1094–7. doi: 10.1038/nature06943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moubayidin L, Di Mambro R, Sozzani R, Pacifici E, Salvi E, Terpstra I, et al. Spatial coordination between stem cell activity and cell differentiation in the root meristem. Dev Cell. Elsevier Inc.; 2013;26: 405–15. doi: 10.1016/j.devcel.2013.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, et al. A Genetic Framework for the Control of Cell Division and Differentiation in the Root Meristem. Science (80-). 2008;322: 1380–1384. doi: 10.1126/science.1164147 [DOI] [PubMed] [Google Scholar]
- 27.Ruzicka K, Simásková M, Duclercq J, Petrásek J, Zazímalová E, Simon S, et al. Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc Natl Acad Sci U S A. 2009;106: 4284–9. doi: 10.1073/pnas.0900060106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang W, Swarup R, Bennett M, Schaller GE, Kieber JJ. Cytokinin induces cell division in the quiescent center of the Arabidopsis root apical meristem. Curr Biol. Elsevier Ltd; 2013;23: 1979–89. doi: 10.1016/j.cub.2013.08.008 [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Paschold A, Marcon C, Liu S, Tai H, Nestler J, et al. The Aux/IAA gene rum1 involved in seminal and lateral root formation controls vascular patterning in maize (Zea mays L.) primary roots. J Exp Bot. 2014;65: 4919–4930. doi: 10.1093/jxb/eru249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li P, Xue H. Structural characterization and expression pattern analysis of the rice PLT gene family. ActaBiochim Biophys Sin. 2011;43: 688–697. [DOI] [PubMed] [Google Scholar]
- 31.O’Connor DL, Runions A, Sluis A, Bragg J, Vogel JP, Prusinkiewicz P, et al. A Division in PIN-Mediated Auxin Patterning during Organ Initiation in Grasses. PLoS Comput Biol. 2014;10: 21–24. doi: 10.1371/journal.pcbi.1003447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tagliani L, Nissen S, Blake TK. Comparison of Growth, Exogenous Auxin Sensitivity, and Endogenous Indole-3-Acetic Acid Content in Roots of Hordeum vulgare L. and an Agravitropic Mutant. Biochem Genet. 1986;24: 839–848. doi: 10.1007/BF00554523 [DOI] [PubMed] [Google Scholar]
- 33.Zalewski W, Galuszka P, Gasparis S, Orczyk W, Nadolska-Orczyk A. Silencing of the HvCKX1 gene decreases the cytokinin oxidase/dehydrogenase level in barley and leads to higher plant productivity. J Exp Bot. 2010;61: 1839–51. doi: 10.1093/jxb/erq052 [DOI] [PubMed] [Google Scholar]
- 34.Zalewski W, Gasparis S, Boczkowska M, Rajchel IK, Kała M, Orczyk W, et al. Expression Patterns of HvCKX Genes Indicate Their Role in Growth and Reproductive Development of Barley. Zhang J-S, editor. PLoS One. 2014;9: e115729 doi: 10.1371/journal.pone.0115729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pospíšilová H, Jiskrová E, Vojta P, Mrízová K, Kokáš F, Čudejková MM, et al. Transgenic barley overexpressing a cytokinin dehydrogenase gene shows greater tolerance to drought stress. N Biotechnol. 2016;33: 692–705. doi: 10.1016/j.nbt.2015.12.005 [DOI] [PubMed] [Google Scholar]
- 36.Kirschner GK, Stahl Y, Von Korff M, Simon R. Unique and Conserved Features of the Barley Root Meristem. Front Plant Sci. 2017;8 doi: 10.3389/fpls.2017.01240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waddington SR, Cartwright PM, Wall PC. A quantitative scale of spike initial and pistil development in barley and wheat. Ann Bot. 1983;51: 119–130. http://aob.oxfordjournals.org/content/51/1/119.short [Google Scholar]
- 38.Mayer KFX, Waugh R, Langridge P, Close TJ, Wise RP, Graner A, et al. A physical, genetic and functional sequence assembly of the barley genome. Nature. Nature Publishing Group; 2012;491: 711–716. doi: 10.1038/nature11543 [DOI] [PubMed] [Google Scholar]
- 39.Mascher M, Gundlach H, Himmelbach A, Beier S, Twardziok SO, Wicker T, et al. A chromosome conformation capture ordered sequence of the barley genome. Nature. Nature Publishing Group; 2017;544: 427–433. doi: 10.1038/nature22043 [DOI] [PubMed] [Google Scholar]
- 40.Curtis MD, Grossniklaus U. A Gateway Cloning Vector Set for High-Throughput Functional Analysis of Genes in Planta. Plant Physiol. 2003;133: 462–469. doi: 10.1104/pp.103.027979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koushik S V, Chen H, Thaler C, Puhl HL, Vogel SS. Cerulean, Venus, and VenusY67C FRET Reference Standards. Biophys J. Elsevier; 2006;91: L99–L101. doi: 10.1529/biophysj.106.096206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20: 87–90. doi: 10.1038/nbt0102-87 [DOI] [PubMed] [Google Scholar]
- 43.Bleckmann A, Weidtkamp-Peters S, Seidel CAM, Simon R. Stem Cell Signaling in Arabidopsis Requires CRN to Localize CLV2 to the Plasma Membrane. Plant Physiol. 2010;152: 166–76. doi: 10.1104/pp.109.149930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Friml J, Benkova E, Blilou I, Wisniewska J, Hamann T, Ljung K, et al. AtPIN4 Mediates Sink-Driven Auxin Gradients and Root Patterning in Arabidopsis. Cell. 2002;108: 661–673. [DOI] [PubMed] [Google Scholar]
- 45.Imani J, Li L, Schäfer P, Kogel K-H. STARTS—A stable root transformation system for rapid functional analyses of proteins of the monocot model plant barley. Plant J. 2011;67: 726–735. doi: 10.1111/j.1365-313X.2011.04620.x [DOI] [PubMed] [Google Scholar]
- 46.Warner CA, Biedrzycki ML, Jacobs SS, Wisser RJ, Caplan JL, Sherrier DJ. An Optical Clearing Technique for Plant Tissues Allowing Deep Imaging and Compatible with Fluorescence Microscopy. Plant Physiol. 2014;166: 1684–1687. doi: 10.1104/pp.114.244673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geldner N, Friml J, Stierhof YD, Jürgens G, Palme K. Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature. 2001;413: 425–428. doi: 10.1038/35096571 [DOI] [PubMed] [Google Scholar]
- 48.Lampropoulos A, Sutikovic Z, Wenzl C, Maegele I, Lohmann JU, Forner J. GreenGate—A Novel, Versatile, and Efficient Cloning System for Plant Transgenesis. Janssen PJ, editor. PLoS One. 2013;8: e83043 doi: 10.1371/journal.pone.0083043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hejátko J, Blilou I, Brewer PB, Friml J, Scheres B, Benková E. In situ hybridization technique for mRNA detection in whole mount Arabidopsis samples. Nat Protoc. 2006;1: 1939–1946. doi: 10.1038/nprot.2006.333 [DOI] [PubMed] [Google Scholar]
- 50.Forestan C, Farinati S, Varotto S. The Maize PIN Gene Family of Auxin Transporters. Front Plant Sci. 2012;3: 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martinez CC, Koenig D, Chitwood DH, Sinha NR. A sister of PIN1 gene in tomato (Solanum lycopersicum) defines leaf and flower organ initiation patterns by maintaining epidermal auxin flux. Dev Biol. Elsevier; 2016;419: 85–98. doi: 10.1016/j.ydbio.2016.08.011 [DOI] [PubMed] [Google Scholar]
- 52.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9: 676–682. doi: 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, et al. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol. 2007;17: 678–682. doi: 10.1016/j.cub.2007.02.047 [DOI] [PubMed] [Google Scholar]
- 54.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J Mol Biol. 2001;305: 567–580. doi: 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 55.R Core Team, R Development Core Team R. R: A Language and Environment for Statistical Computing. Team RDC, editor. R Found. Stat. Comput. R Foundation for Statistical Computing; 2014. p. 409. [Google Scholar]
- 56.de Reuille PB, Routier-Kierzkowska AL, Kierzkowski D, Bassel GW, Schüpbach T, Tauriello G, et al. MorphoGraphX: A platform for quantifying morphogenesis in 4D. Elife. 2015;4: 1–20. doi: 10.7554/eLife.05864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Werner T, Nehnevajova E, Köllmer I, Novák O, Strnad M, Krämer U, et al. Root-Specific Reduction of Cytokinin Causes Enhanced Root Growth, Drought Tolerance, and Leaf Mineral Enrichment in Arabidopsis and Tobacco. Plant Cell. 2010;22: 3905–3920. doi: 10.1105/tpc.109.072694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Podlešáková K, Zalabák D, Čudejková M, Plíhal O, Szüčová L, Doležal K, et al. Novel Cytokinin Derivatives Do Not Show Negative Effects on Root Growth and Proliferation in Submicromolar Range. Rahman A, editor. PLoS One. 2012;7: e39293 doi: 10.1371/journal.pone.0039293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martínez-de la Cruz E, García-Ramírez E, Vázquez-Ramos JM, Reyes de la Cruz H, López-Bucio J. Auxins differentially regulate root system architecture and cell cycle protein levels in maize seedlings. J Plant Physiol. Elsevier GmbH.; 2015;176: 147–156. doi: 10.1016/j.jplph.2014.11.012 [DOI] [PubMed] [Google Scholar]
- 60.Delbarre A, Muller P, Imhoff V, Guern J. Comparison of mechanisms controlling uptake and accumulation of 2,4-dichlorophenoxy acetic acid, naphthalene-1-acetic acid, and indole-3-acetic acid in suspension-cultured tobacco cells. Planta. 1996;198: 532–541. doi: 10.1007/BF00262639 [DOI] [PubMed] [Google Scholar]
- 61.Ding Z, Friml J. Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc Natl Acad Sci U S A. 2010;107: 12046–12051. DCSupplemental. www.pnas.org/cgi/doi/10.1073/pnas.1000672107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kleine-Vehn J, Huang F, Naramoto S, Zhang J, Michniewicz M, Offringa R, et al. PIN Auxin Efflux Carrier Polarity Is Regulated by PINOID Kinase-Mediated Recruitment into GNOM-Independent Trafficking in Arabidopsis. Plant Cell. 2009;21: 3839–3849. doi: 10.1105/tpc.109.071639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Steinmann T, Geldner N, Grebe M, Mangold S, Jackson CL, Paris S, et al. Coordinated Polar Localization of Auxin Efflux Carrier PIN1 by GNOM ARF GEF. Science (80-). 1999;286: 316–18. doi: 10.1126/science.286.5438.316 [DOI] [PubMed] [Google Scholar]
- 64.Geldner N, Anders N, Wolters H, Keicher J, Kornberger W, Muller P, et al. The Arabidopsis GNOM ARF-GEF Mediates Endosomal Recycling, Auxin Transport, and Auxin-Dependent Plant Growth. Cell. 2003;112: 219–230. doi: 10.1016/S0092-8674(03)00003-5 [DOI] [PubMed] [Google Scholar]
- 65.Peyroche A, Antonny B, Robineau S, Acker J, Cherfils J, Jackson CL. Brefeldin A Acts to Stabilize an Abortive ARF–GDP–Sec7 Domain Protein Complex. Mol Cell. 1999;3: 275–285. doi: 10.1016/S1097-2765(00)80455-4 [DOI] [PubMed] [Google Scholar]
- 66.Křeček P, Skůpa P, Libus J, Naramoto S, Tejos R, Friml J, et al. The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol. 2009;10: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ganguly A, Lee SH, Cho M, Lee OR, Yoo H, Cho H-T. Differential Auxin-Transporting Activities of PIN-FORMED Proteins in Arabidopsis Root Hair Cells. PLANT Physiol. 2010;153: 1046–1061. doi: 10.1104/pp.110.156505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heisler MG, Ohno C, Das P, Sieber P, Reddy G V., Long JA, et al. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol. 2005;15: 1899–1911. doi: 10.1016/j.cub.2005.09.052 [DOI] [PubMed] [Google Scholar]
- 69.Kleine-Vehn J, Leitner J, Zwiewka M, Sauer M, Abas L, Luschnig C, et al. Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc Natl Acad Sci U S A. 2008;105: 17812–17817. doi: 10.1073/pnas.0808073105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Galuszka P, Popelková H, Werner T, Frébortová J, Pospíšilová H, Mik V, et al. Biochemical Characterization of Cytokinin Oxidases/Dehydrogenases from Arabidopsis thaliana Expressed in Nicotiana tabacum L. J Plant Growth Regul. 2007;26: 255–267. doi: 10.1007/s00344-007-9008-5 [Google Scholar]
- 71.Mrízová K, Jiskrová E, Vyroubalová S, Novák O, Ohnoutková L, Pospíšilová H, et al. Overexpression of Cytokinin Dehydrogenase Genes in Barley (Hordeum vulgare cv. Golden Promise) Fundamentally Affects Morphology and Fertility. PLoS One. 2013;8: e79029 doi: 10.1371/journal.pone.0079029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Evans ML, Ishikawa H, Estelle MA. Responses of Arabidopsis roots to auxin studied with high temporal resolution: Comparison of wild type and auxin-response mutants. Planta. 1994;194: 215–222. doi: 10.1007/BF00196390 [Google Scholar]
- 73.Müssig C, Shin G-H, Altmann T. Brassinosteroids promote root growth in Arabidopsis. Plant Physiol. 2003;133: 1261–1271. doi: 10.1104/pp.103.028662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stahl Y, Wink RH, Ingram GC, Simon R. A Signaling Module Controlling the Stem Cell Niche in Arabidopsis Root Meristems. Curr Biol. 2009;19: 909–914. doi: 10.1016/j.cub.2009.03.060 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Meristem cell number upon 10-day cytokinin treatment; experiment was performed twice; n = 11–16 roots per data point. B) Meristem length upon auxin treatment; experiment was performed twice; all values are normalized to the mock-treated control; n = 7–17 roots per data point; significance was determined using the two-tailed Student’s t test, * = p<0.05, ** = p<0.001.
(TIF)
Meristem width measured at the transition zone from root meristems exemplarily shown in Figs 1B and 3B. A) Roots were treated with cytokinin for 10 days; experiments were performed twice; n = 15–25 roots per data point. B) Roots were treated with auxin for 10 days; experiment was performed twice, n = 12–25 roots per data point; significance was determined using the two-tailed Student’s t test, * = p<0.05, ** = p<0.001.
(TIF)
A), A’) Undectectable TCSn:VENUS-H2B expression in SAMs in waddington stage I; transmitted light and VENUS emission (A)) and VENUS emission only (A’)). B), B’) Undectectable TCSn:VENUS-H2B expression in SAMs in waddington stage II; transmitted light and VENUS emission (B)) and VENUS emission only (B’)). Seven independent transgenic lines were examined and show no expression in the SAM; scale bars 100 μm; insets in A’) and B’) show respective pictures with tonal correction to show autofluorescence.
(TIF)
A) Exemplary pictures of the root stem cell niche upon mock or auxin treatment as indicated; scale bar 100 μm. B) Number of DSC layers upon 10-day treatment with auxin; no significant difference to mock-treated plants; experiment was performed twice; n = 5–20 per data point. Significance was determined using the two-tailed Student’s t test, * = p<0.05, ** = p<0.001.
(TIF)
A) Exemplary picture of DR5:GFP root; transmitted light and (undetectable) GFP emission (A)), undetectable GFP emission only (A’)); four independent transgenic lines were examined and show no GFP epression; scale bar 200 μm. B) DR5v2:VENUS-H2B lines show only variable or no expression and do not show a consistent reaction on 2,4D treatment; number of expressing DR5v2:VENUS-H2B lines (B)); number of plants that show the respective expression change upon treatment with 10 μM 2, 4D for 24 h (B’)).
(TIF)
A) Representative picture of the root meristem of a non-transgenic Golden Promise seedling 8 DAG; transmitted light and mVENUS emission (A)), mVENUS emission only (A’)), same settings as in Fig 4B and 4B’; hand-sections as described in Material and Methods; only background signal with mVENUS excitation. B) Representative picture of the root meristem of a non-transgenic Golden Promise seedling 8 DAG; transmitted light and mVENUS emission (B)), mVENUS emission only (B’)), same settings as in Fig 5A’; cleared as described in Material and Methods; only background signal with mVENUS excitation; scale bars 100 μm; inset in A’) shows respective pictures with tonal correction to show autofluorescence.
(TIF)
Topology of the transmembrane barley PIN proteins in comparison to AtPIN1; domains predicted to the inside of the cell are shown in light-gray, transmembrane domains are shown in dark-gray and domains outside the cell are depicted in black according to the legend; in the protein topology of MLOC_64867—HvPIN1a the asterisk marks the site where mVENUS is inserted for the reporter line shown in Fig 5; newly identified HvPINs are named according to their topology and the cluster of the Arabidopsis, maize and rice PIN family to which they belong.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.