Abstract
In tyrosinaemia type 1(HT1), a mosaic pattern of fumarylacetoacetase (FAH) immunopositive or immunonegative nodules in liver tissue has been reported in many patients. This aspect is generally explained by a spontaneous reversion of the mutation into a normal genotype. In one HT1 patient carrying the frequent FAH c.1062+5G>A mutation, a second somatic change (c.1061C>A) has been reported in the same allele, and found in immunopositive nodules. Here, we demonstrated that the c.1062+5G>A prevents usage of the exon 12 5’ splice site (ss), even when forced by an engineered U1snRNA specifically designed on the FAH 5’ss to strengthen its recognition. Noticeably the new somatic c.1061C>A change, in linkage with the c.1062+5G>A mutation, partially rescues the defective 5’ss and is associated to trace level (~5%) of correct transcripts. Interestingly, this combined genetic condition strongly favored the rescue by the engineered U1snRNA, with correct transcripts reaching up to 60%.
Altogether these findings elucidate the molecular basis of HT1 caused by the frequent FAH c.1062+5G>A mutation, and demonstrate the compensatory effect of the c.1061C>A change in promoting exon definition, thus unraveling a rare mechanism leading to FAH immune-reactive mosaicism.
Keywords: Hereditary tyrosinemia type I, splicing mutations, immuno-reactive reversion, FAH immune-reactive mosaicism, U1snRNA
Introduction
Hereditary tyrosinemia type I (HT1)(OMIM 276700) is an autosomal recessive disorder caused by genetic defects in the fumarylacetoacetate hydrolase (FAH), the last enzyme in the catabolic pathway of tyrosine1. The accumulation of toxic metabolites in liver is believed to cause liver failure, cirrhosis, hepatocellular carcinoma, and death.
Several patients (28% and up to 100% in the Saguenay-Lac-Saint-Jean region of Quebec)2 carry the FAH c.1062+5G>A mutation at the exon 12 5’splice site (ss). Investigations in patients3 and with minigenes4 demonstrated that this change induces exon 12 and exons 12-13 skipping as well as partial intron 12 inclusion with usage of an intronic cryptic 5’ss, thus explaining the FAH immune-negativity of liver sections from homozygotes. Differently, the presence of the new c.1061C>A (c.P354Q) somatic change with the c1062+5G>A mutation was found to be associated with hepatic FAH immune-positivity5. Here we demonstrated the compensatory effect of the c.1061C>A change in promoting FAH immune-positivity, which also renders the defective FAH 5’ss highly responsive to correction by a compensatory U1snRNA.
Material and Methods
To create the pFAHwt minigenes, the 804-bp genomic fragment spanning FAH intron 11 (from position c.960-357) trough intron 12 (until position c.1062+327) was amplified from genomic DNA of a normal subject using high-fidelity PfuI DNA-Polymerase (Transgenomic, Glasgow, UK) with primers 5’CATATGGACTGGAGGGTGTTCCCA3’ (forward) and 5’CATATGCCACCTCATCCTGGGAGGGT3’ (reverse), and cloned in the pTB expression vector by exploiting the NdeI restriction site within primers (underlined)6. The mutant pFAH constructs were generated by site-directed mutagenesis (QuickChange II XL Site directed Mutagenesis Kit; Agilent Technologies, Santa Clara, CA, USA) using primers 5’CATCAGCGGGCCGGTGAATATCTGGCTGCACTGAG3’ and 5’CTCAGTGCAGCCAGATATTCACCGGCCCGCTGATG3’, to introduce the c.1062+5G>A change, and primers 5’CCATCAGCGGGCDGGTGAGTATCTG3’ and 5’CAGATACTCACCHGCCCGCTGATGG3’, to introduce the c.1061C>A/T/G changes. The pU1FAH expression vector was created as previously described7.
HepG2 cells were transfected with Lipofectamine 2000 (ThermoFisher SCIENTIFIC, Carlsbad, CA, USA) in 12-well plate with pFAH minigenes (1 μg) alone or with a molar excess (1.5X) of pU1FAH. Total RNA extraction was performed 24 hours post-transfection using TRIreagent (ThermoFisher SCIENTIFIC) and reverse transcribed using the M-MLV (ThermoFisher SCIENTIFIC). The primer couples α-2,3 (5’CAACTTCAAGCTCCTAAGCCA CTGC3’) and Bra (5’TAGGATCCGGTCACCAGGAAGTTGGTTAAATCA3’) in the neighboring exons of the hybrid minigene, or FAH12F in exon 12 (5’ACATGTACTGGACG ATGCTGCA3’) and Bra, indicated by arrows in figure 1, were used for the RT-PCR that was run for 40 cycles at the following conditions: 95°C for 30 s, 56 °C for 30 s, 72°C for 40 s. The densitometric analysis of bands was performed using ImageJ software.
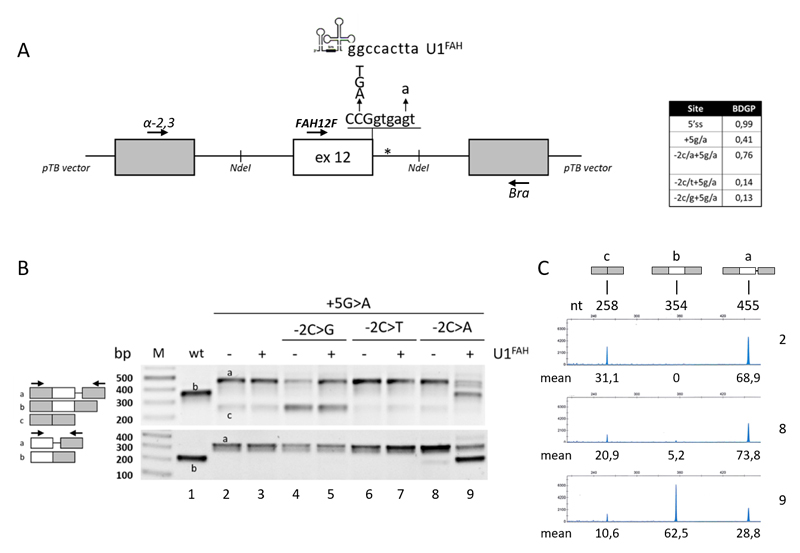
Figure 1. Splicing patterns of FAH minigenes in HepG2.
A) Schematic representation of the FAH minigene, cloned within the NdeI sites (indicated) in the pTB vector, with the sequence of the exon 12 5’ss and of the investigated changes as well as of the 5’ tail of the compensatory U1FAH. The asterisk indicates the crypric 5’ss.
The table reports the predicted score of the wild type or mutated 5’ss (http://www.fruitfly.org/seq_tools/splice.html).
B) FAH splicing patterns in HepG2 cells transiently transfected with pFAH minigenes (1 μg) alone or with a molar excess (1.5X) of pU1FAH. RT-PCR was conducted with primers α-2,3 and Bra (upper panel) or primers FAH12F and Bra (lower panel), and amplicons were separated on 2% agarose gel. The scheme of amplicons is reported on the left together with primers used (arrows).
C) FAH splicing patterns as in the upper panel of section B (lanes 2, 8 and 9) evaluated by fluorescently-labelled RT-PCR with primers α-2,3 and BraFAM, followed by denaturing capillary electrophoresis. The scheme of amplicons and the length (nt) are reported on top. Numbers below peaks indicate the relative amount (%, mean from three independent experiments) of each transcript.
For the denaturing capillary electrophoresis8 on the automated ABI-3100 instrument, the RT-PCR has been performed with primer α-2,3 and Bra, the latter labelled with FAM dye (BraFAM).
Results and Discussion
To dissect the mechanisms triggered by the HT1-causing FAH c.1062+5G>A mutation alone or in combination with the somatic FAH c.1061C>A change we performed expression studies with FAH minigenes (Figure 1A). Since the splicing process is cell specific, the experiments were conducted into human hepatoma cell lines (HepG2), being liver the major physiologic site of FAH synthesis.
Consistently with previous data3–5 the c.1062+5G>A mutation, as revealed by both densitometric analysis of bands or fluorescent RT-PCR followed by denaturing capillary electrophoresis, induced exon 12 skipping (~31% of total transcripts) and partial intron retention (~69%), with no traces of correct transcripts (Figure 1B, upper panel and Figure 1C), a splicing profile that validated our experimental approach. Interestingly, the introduction of the c.1061C>A substitution in the c.1062+5G>A background partially rescued the defective 5’ss recognition and led to residual levels of correct transcripts (5,2±0,9%)(Figure 1C), which was further demonstrated by a RT-PCR focused on exon 12 (Figure 1B, lower panel).
Computational analysis of 5’ss scores (Figure 1A, Table), an estimate of the complementarity between the 5’ss sequence and the 5’ tail of the key spliceosomal U1 small nuclear RNA (U1snRNA)9, predicts that the c.1061C>A change, but not the other c.1061C>T or c.1061C>G substitutions, strengthen the affected 5’ss. This would favor the utilization of the mutated 5’ss and the production of correct transcripts. Consistently, the impact of the other changes (c.1061C>T or c.1061C>G) on the splicing profile of the c.1062+5G>A mutation was negligible (Figure 1B).
Altogether these observations demonstrated that in the c.1062+5G>A background the c.1061C>A somatic change accounts for trace levels of correct transcripts. In turn, this would explain the residual FAH protein expression and immune-positivity in HT1 liver nodules with the above mentioned genetic profile, thus providing an additional mechanism leading to FAH immune-reactive mosaicism. It is worth noting that the FAH immune-reactive mosaicism has been so far attributed to the reversion of the mutated allele into the wild type10, as also reported in other diseases11. Both mechanisms would be favored by the high mutation rate of HT1 hepatocytes,11 followed by positive selection of FAH-expressing hepatocites, which would lead to the formation of FAH-immunopositive nodules.
Splicing mutations, relatively frequent in metabolic disorders (http://www.hgmd.cf.ac.uk) as well as in others human diseases12, represent potential targets for RNA-based therapies. Several studies have indicated that splicing mutations can be counteracted by engineered U1snRNAs with increased complementarity with the 5’ss of the defective exon13–15. We therefore created a U1snRNA variant specifically designed on the FAH exon 12 5’ss (U1FAH). However, in co-expression experiment, the compensatory U1FAH was ineffective on the c.1062+5G>A mutation, which demonstrated the severe impairment of the 5’ss recognition. Notably, and consistently with a slightly improved 5’ss, the combination of the HT1-causative mutation with the c.1061C>A change resulted in remarkable responsiveness to the U1FAH that promoted exon 12 inclusion, as witnessed by the robust synthesis of correct transcripts (62,5±1,2%)(Figure 1B,C).
Conclusion
Altogether these findings elucidate the molecular basis of HT1 caused by the frequent FAH c.1062+5G>A mutation, and demonstrate the compensatory effect of the c.1061C>A change in promoting exon definition, thus unraveling a rare mechanism leading to FAH immune-reactive mosaicism.
Acknowledgments
The study was supported by grants from Telethon Foundation (GGP14190 to M.P and D.B), the AMC Foundation (to SFJvdG) and the University of Ferrara.
Footnotes
Conflict of Interest
D.S., C.S., S.F.J.G, and D.B. have no competing interests to declare. M.P. and F.B. are cofounders of the start-up company Raresplice.
References
- 1).Lindblad B, Lindstedt S, Steen G. On the enzymic defects in hereditary tyrosinemia. Proc Natl Acad Sci U S A. 1977;74(10):4641–5. doi: 10.1073/pnas.74.10.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2).Grompe M, St-Louis M, Demers SI, al-Dhalimy M, Leclerc B, Tanguay RM. A single mutation of the fumarylacetoacetate hydrolase gene in French Canadians with hereditary tyrosinemia type I. N Engl J Med. 1994;331(6):353–7. doi: 10.1056/NEJM199408113310603. [DOI] [PubMed] [Google Scholar]
- 3).Rootwelt H, Kristensen T, Berger R, Høie K, Kvittingen EA. Tyrosinemia type 1--complex splicing defects and a missense mutation in the fumarylacetoacetase gene. Hum Genet. 1994;94(3):235–9. doi: 10.1007/BF00208276. [DOI] [PubMed] [Google Scholar]
- 4).Pérez-Carro R, Sánchez-Alcudia R, Pérez B, Navarrete R, Pérez-Cerdá C, Ugarte M, Desviat LR. Functional analysis and in vitro correction of splicing FAH mutations causing tyrosinemia type I. Clin Genet. 2014;86(2):167–71. doi: 10.1111/cge.12243. [DOI] [PubMed] [Google Scholar]
- 5).Bliksrud YT, Brodtkorb E, Andresen PA, van den Berg IE, Kvittingen EA. Tyrosinaemia type I--de novo mutation in liver tissue suppressing an inborn splicing defect. J Mol Med (Berl) 2005;83(5):406–10. doi: 10.1007/s00109-005-0648-2. [DOI] [PubMed] [Google Scholar]
- 6).Tajnik M, Rogalska ME, Bussani E, Barbon E, Balestra D, Pinotti M, Pagani F. Molecular Basis and Therapeutic Strategies to Rescue Factor IX Variants That Affect Splicing and Protein Function. PLoS Genet. 2016;12(5) doi: 10.1371/journal.pgen.1006082. e1006082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Balestra D, Barbon E, Scalet D, Cavallari N, Perrone D, Zanibellato S, Bernardi F, Pinotti M. Regulation of a strong F9 cryptic 5'ss by intrinsic elements and by combination of tailored U1snRNAs with antisense oligonucleotides. Hum Mol Genet. 2015;24(17):4809–16. doi: 10.1093/hmg/ddv205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).Cavallari N, Balestra D, Branchini A, Maestri I, Chuamsunrit A, Sasanakul W, Mariani G, Pagani F, Bernardi F, Pinotti M. Activation of a cryptic splice site in a potentially lethal coagulation defect accounts for a functional protein variant. Biochim Biophys Acta. 2012;1822(7):1109–13. doi: 10.1016/j.bbadis.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9).Roca X, Krainer AR, Eperon IC. Pick one, but be quick: 5' splice sites and the problems of too many choices. Genes Dev. 2013;27(2):129–44. doi: 10.1101/gad.209759.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Kvittingen EA, Rootwelt H, Berger R, Brandtzaeg P. Self-induced correction of the genetic defect in tyrosinemia type I. J Clin Invest. 1994;94(4):1657–61. doi: 10.1172/JCI117509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Hirschhorn R. In vivo reversion to normal of inherited mutations in humans. J Med Genet. 2003;40(10):721–8. doi: 10.1136/jmg.40.10.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Sterne-Weiler T, Howard J, Mort M, Cooper DN, Sanford JR. Loss of exon identity is a common mechanism of human inherited disease. Genome Res. 2011;21(10):1563–71. doi: 10.1101/gr.118638.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13).Fernandez Alanis E, Pinotti M, Dal Mas A, Balestra D, Cavallari N, Rogalska ME, Bernardi F, Pagani F. An exon-specific U1 small nuclear RNA (snRNA) strategy to correct splicing defects. Hum Mol Genet. 2012;21(11):2389–98. doi: 10.1093/hmg/dds045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14).Scalet D, Balestra D, Rohban S, Bovolenta M, Perrone D, Bernardi F, Campaner S, Pinotti M. Exploring Splicing-Switching Molecules For Seckel Syndrome Therapy. Biochim Biophys Acta. 2017;1863(1):15–20. doi: 10.1016/j.bbadis.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 15).Balestra D, Scalet D, Pagani F, Rogalska ME, Mari R, Bernardi F, Pinotti M. An Exon-Specific U1snRNA Induces a Robust Factor IX Activity in Mice Expressing Multiple Human FIX Splicing Mutants. Mol Ther Nucleic Acids. 2016;5(10):e370. doi: 10.1038/mtna.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]