Abstract
Purpose
Colorectal cancer (CRC) is a common cause of death worldwide. Tumor-node-metastasis-system stage is currently used to guide therapy decisions but lacks precision. Prognostic biomarkers are needed to refine stratification of patients for chemotherapy but validated biomarkers are not yet available. Recently, a SNP in a lethal-7 (let-7) miRNA complementary site (LCS6) in the KRAS 3′ untranslated region was suggested to affect survival in metastatic CRC. Effects in early-stage CRC are however unknown. We studied KRAS-LCS6 genotype, hypothesizing that it might identify early-stage cases with a poor prognosis, and could potentially be used in therapy decision-making.
Experimental Design
We studied 409 early stage, 182 stage III, and 69 stage IV cases, and 1,886 subcohort members from the Netherlands Cohort Study. KRAS-LCS6 genotype was assessed with TaqMan PCR. Kaplan–Meier analyses or Cox regression were used to assess associations between genotype and CRC risk or cause-specific survival.
Results
Early-stage cases with the KRAS-LCS6 variant had a lower CRC risk (incidence-rate ratio 0.68; 95% CI: 0.49–0.94) and a better survival (log-rank P = 0.038; HR 0.46; 95% CI: 0.18–1.14). In patients with KRAS-mutated CRC carrying the KRAS-LCS6 variant, the better outcome was enhanced as no patients died of CRC (log-rank P = 0.017). In advanced patients, no clear association between genotype and CRC risk or survival was observed.
Conclusions
Our results indicate that early-stage CRC cases with the KRAS-LCS6 variant have a better outcome. In advanced disease, the better outcome no longer exists. For early-stage patients, KRAS-LCS6 genotype combined with KRAS mutations merits validation as a prognostic biomarker and consideration in therapy decision-making.
Introduction
Despite diagnostic and therapeutic innovations, colorectal cancer (CRC) remains the second cause of cancer death in the western world (1). The tumor-node-metastasis-system (TNM) is currently the main tool to provide prognostic information; it is highly predictive for prognosis at the extremes, but less predictive for intermediate stages (2, 3). According to current guidelines, adjuvant chemotherapy is not given to early-stage patients (T1-3-N0-M0 according to the International Union Against Cancer-TNM) as 5-year survival rates in this group are more than 70%. Nevertheless, 20% to 30% of early-stage patients (stage I and II) will die of CRC within 5 years, evoking the question whether these deaths could have been avoided if these patients were identified in advance and therapy was adapted accordingly. Previously, numerous studies have been published claiming a prognostic influence of molecular markers. Results however, are inconsistent and the question which molecular alterations influence prognosis remains unresolved (4).
Over the last years, a new class of gene regulators, micro-RNAs (miRNA), has been identified as important factors in cancer development and progression. Evidence suggests that one miRNA can regulate many mRNAs simultaneously (5) and miRNAs can act as both tumor suppressors and oncogenes (6). One of the first discovered miRNA families is the lethal-7 (let-7) family of miRNAs, and altered expression of these miRNAs has been described in many cancers (7). In lung cancer, let-7 is poorly expressed (8, 9), overexpression of let-7 inhibits cell growth in vitro (9) and in vivo (10, 11) suggesting that let-7 miRNAs may act as tumor suppressors (6). In colon cancer cells, let-7 expression was significantly decreased in tumor tissue as compared with adjacent noncancerous tissue (12). In addition, let-7 expression was increased and RAS expression was decreased in cell lines after transfection of a let-7a-1 miRNA precursor suggesting that let-7 is involved in regulating colon cancer cell growth (12). miRNAs usually control gene expression by binding to complementary elements in the 3′ untranslated region (UTR) of target mRNAs (6). It has been shown that let-7 induces RAS downregulation after binding to specific sites in the 3′-UTR KRAS mRNA (6, 7). Recently, a SNP in a let-7 complementary site (LCS) in the KRAS 3′-UTR (KRAS-LCS6) has been identified that affects let-7-mediated regulation of KRAS expression. The variant G-allele was observed to lead to higher KRAS levels and lower let-7 levels as compared with the wild type (13). G-allele carriers have been shown to have an increased lung cancer risk in moderate smokers (13), an increased ovarian cancer risk (14) although perhaps not for all women (15), an increased triple-negative breast cancer risk (16), a reduced survival in oral cancers (17) but not in lung cancer (18). Recently, in KRAS/BRAF-mutated CRC, G-allele carriers showed a reduced survival in late-stage CRC (19) and an altered response to cetuximab (19, 20), supporting the function of this variant in colon cancer. As the influence of KRAS-LCS6 genotype in early-stage CRC has not been determined, we assessed the influence on prognosis in 409 early-stage (TNM stage I and II; T1-4, N0, M0), 182 stage III (T1-4, N1, M0), and 69 stage IV (T1-4, N0-1, M1) CRC cases from a large prospective cohort study. We assessed the influence of KRAS-LCS6 genotype on CRC risk using data from 1,886 subcohort members from the Netherlands Cohort Study on diet and cancer (NLCS).
Translational Relevance.
We report for the first time that a SNP in a lethal-7 (let-7) complementary site (LCS) in the KRAS 3′-UTR (KRAS-LCS6) might be a prognostic biomarker in early-stage colorectal cancer (CRC). The KRAS-LCS6 variant is known to cause higher levels of the KRAS oncogenic protein and lower levels of the tumor suppressor let-7 miRNAs. We studied the influence of KRAS-LCS6 in 409 early-stage (stage I and II), 182 stage III, and 69 stage IV cases from the large, prospective Netherlands Cohort Study (NLCS). Early-stage patients with the KRAS-LCS6 variant had a better prognosis, especially those that also had KRAS mutations, and this was independent of microsatellite instability or other prognostic factors. In addition, we studied the influence of the KRAS-LCS6 variant on CRC risk using data from 1,886 subcohort members from the NLCS. The G-allele was associated with a decreased risk on early-stage CRC, but was not associated with advanced stage CRC risk, suggesting that the G-allele is not associated with the likelihood of advanced stage CRC. As our population is the only untreated population studied to date, our results give a first insight into the natural biology of CRC with the KRAS-LCS6 variant. The KRAS-LCS6 variant may become a new biomarker in CRC to guide treatment decisions in early-stage patients.
Materials and Methods
Study population
Until 1994, 925 incident CRC cases (ICD-O: 153.0–154.1) were identified within the NLCS which started in 1986 with 120,852 healthy persons between 55 and 69 years. Incident cancer cases were identified by linkage with the Netherlands Cancer Registry (NCR) and PALGA, a nationwide registry of histopathology and cytopathology (21). The NLCS has been described in detail elsewhere (22). A total of 815 patients could be linked to PALGA and paraffin-embedded tumor tissue was collected from 54 pathology registries throughout the Netherlands. We were able to extract sufficient, good quality DNA for 734 (90%) cases (23). At baseline, a subcohort of 5,000 healthy persons was randomly sampled from the entire cohort to estimate person-years at risk of the cohort through biennial follow-up of vital status. For 1,886 persons, DNA from buccal swabs was available for KRAS-LCS6 genotyping.
Data collection
Information on tumor localization, stage, differentiation grade, incidence date, and treatment in the 3 months after diagnosis, was available through the NCR. Vital status until May 2005 was retrieved from the Central Bureau of Genealogy and the municipal population registries and could be obtained for all 734 cases. Causes of death were retrieved through linkage with Statistics Netherlands. CRC-related deaths were defined as deaths as a result of a carcinoma in the colon, rectosigmoid, rectum, gastro-intestinal tract (nonspecific) or liver metastases. In the case of gastrointestinal (nonspecified) or liver metastases, we used the information from NCR and PALGA to eliminate the possibility of another primary cancer as cause of death.
DNA isolation and KRAS-LCS6 SNP determination
A 5-μm section of each tumor tissue block was stained with haematoxylin and eosin and revised by a pathologist. Five sections of 20 μm were deparaffinated and DNA was extracted using the Puregene DNA isolation kit (Gentra systems) according to the manufacturer’s instructions. In brief, cell lysis solution and proteinase K (20 mg/mL, Qiagen) were added to the tissue and incubated overnight at 55°C. DNA was extracted for 72 hours at 37°C, protein was removed, and DNA was precipitated using 100% 2-propanol. Finally, DNA was rehydrated in hydration buffer. Isolated DNA was amplified using TaqMan PCR assays designed specifically to identify the T or G allele of the KRAS-LSC6 SNP (Applied Biosciences). Although we used tumor DNA to assess genotype, it has previously been well documented that the genotype of normal and tumor tissue is the same in KRAS-LCS6 variant allele carriers (13).
KRAS and BRAF mutations were assessed by nested PCR and direct sequencing (KRAS), and restriction fragment length polymorphism (BRAF) as described previously (23, 24). Promoter methylation of RASSF1A, O6-MGMT, CHFR, and CIMP markers as proposed by Weisenberger (25) was assessed by chemical modification of genomic DNA with sodium bisulfite and methylation-specific PCR (MSP; refs. 24, 26, 27). Microsatellite instability (MSI) status was determined using BAT-26, BAT-25, NR-21, NR-22, and NR-24 as described previously (28). All assays were done and analyzed blinded to the main study endpoint, that is, CRC-related death.
Statistical analyses
Cause-specific survival was defined as time from cancer diagnosis until CRC-related death or end of follow-up. Kaplan–Meier curves and log-rank tests were used to estimate the influence of the KRAS variant on cause-specific survival. HR and corresponding 95% CI were assessed by use of Cox proportional hazard models adjusted for potential confounders. Factors were considered possible confounders if they were known prognostic factors for CRC and influenced the crude HR by more than 10%. Confounders that were included were age at diagnosis (continuous), sex, tumor differentiation grade (well, moderate, poor, and undifferentiated), and location (proximal, distal, rectosigmoid, and rectum). The proportional hazard assumption was tested using the Schoenfeld residuals and the log(−log) hazards plots. Survival analyses were restricted to 10 years after diagnosis as CRC-related cause of death was unlikely after that point. Incidence rate ratios (RR) and 95% CI were estimated using Cox proportional hazards models. Standard errors were estimated using the robust Huber–White sandwich estimator to account for additional variance introduced by sampling from the cohort. All analyses were done with the statistical package STATA10.0.
Results
CRC variables and the KRAS-LCS6 variant
Patients in this study were more often male (55.6%), diagnosed with an early-stage tumor (62.0%) or a proximal or distal tumor (65.3%; Table 1). During follow-up, 41.4% of the patients died of CRC. The KRAS-LCS6 variant was detected in 14.0% of early-stage (stage I and II), in 19.2% of stage III and 21.4% of stage IV patients (P = 0.160; Ptrend = 0.060). KRAS-LCS6 variant patients were more often diagnosed with advanced stage disease (47.5% vs. 36.9% in wild-type patients, P = 0.046). No other statistically significant differences were found between wild type and variant carriers for sex, age at diagnosis, differentiation grade, tumor location, MSI, or mutations in KRAS (Table 1), BRAF (P = 0.640), or RASSF1A promoter CpG island methylation (P = 0.423). As expected, patients with stage III or IV disease more often died from CRC (P < 0.001) and more often had a poorly differentiated tumor (P < 0.001). Advanced stage patients more often had a proximal (P = 0.036) or MSS tumor (P = 0.047) as compared with early-stage patients.
Table 1.
Baseline characteristics for the total population, KRAS-LCS6 variant and wild-type carriers and early-stage and advanced stage CRC cases within the NLCS on diet and cancer, 1986–1994
| Overall | KRAS-LCS6 wild-type TT | KRAS-LCS6 variant G-allele (He+Ho) | P | Early-stage (stage I and II) CRC | Stage III | Stage IV | P | ||
|---|---|---|---|---|---|---|---|---|---|
| Total population, n (%) | 734 (100) | 567 (83.6) | 111 (16.4) | 409 (62.0) | 182 (27.6) | 69 (10.5) | |||
| Sex [male, n (%)] | Male | 406 (55.6) | 308 (54.3) | 66 (59.5) | 0.320 | 219(53.6) | 102 (56.0) | 33(47.8) | 0.506 |
| Age at diagnosis (mean, SD) | 67.9 (4.3) | 67.9 (4.3) | 67.9 (4.4) | 0.885 | 68.0 (4.4) | 67.5 (4.1) | 68.5 (3.8) | 0.203 | |
| CRC-related death (yes, n (%)) | Yes | 302 (41.4) | 230 (40.6) | 46 (42.2) | 0.761 | 95 (23.3) | 107 (8.8) | 65(95.6) | <0.001 |
| Cancer stage, n (%) | Early stage (I and II) | 409 (62.0) | 326 (63.1) | 53 (52.5) | |||||
| III | 182 (27.6) | 137 (26.5) | 33 (32.7) | ||||||
| IV | 69 (10.5) | 54 (10.4) | 15 (14.9) | 0.124 | |||||
| Differentiation, n (%) | Well | 74 (11.5) | 58 (11.8) | 9 (8.7) | 46 (12.7) | 13 (7.8) | 3 (5.0) | ||
| Moderate | 457 (71.0) | 354 (71.8) | 72 (69.9) | 277(76.5) | 109 (65.3) | 36 (60.0) | |||
| Poor | 106 (16.5) | 75 (15.2) | 21 (20.4) | 37 (10.2) | 41 (24.6) | 20 (33.3) | |||
| Undifferentiated | 7 (1.1) | 6 (1.2) | 1 (1.0) | 0.532 | 2 (0.6) | 4 (2.4) | 1 (1.7) | <0.001 | |
| Location, n (%) | Proximal | 239 (33.2) | 196 (35.4) | 34 (31.2) | 128 (31.5) | 63 (34.8) | 33 (49.3) | ||
| Distal | 231 (32.1) | 177 (32.0) | 37 (33.9) | 125 (30.7) | 61 (33.7) | 22 (32.8) | |||
| Rectosigmoid | 80 (11.1) | 59 (10.6) | 11 (10.1) | 53 (13.0) | 17 (9.4) | 5 (7.5) | |||
| Rectum | 169 (23.5) | 122 (22.0) | 27 (24.8) | 0.824 | 101 (24.8) | 40 (22.1) | 7 (10.5) | 0.036 | |
| Molecular characteristics, n (%) | MSS | 578 (87.3) | 463 (87.5) | 88 (84.6) | 314 (84.9) | 149 (88.7) | 63 (95.5) | ||
| MSI | 84 (12.7) | 66 (12.5) | 16 (15.4) | 0.420 | 56 (15.1) | 19 (11.3) | 3 (4.5) | 0.047 | |
| CIMP+ | 167 (27.7) | 127 (24.5) | 34 (35.4) | ||||||
| CIMP− | 436 (72.3) | 352 (73.5) | 62 (64.6) | 0.076 | 0.121 | ||||
| KRAS mutations, n (%) | Wild type | 464 (63.2) | 362 (63.8) | 69 (62.2) | 263 (64.3) | 121 (66.5) | 39 (56.5) | ||
| KRAS mutated | 270 (36.8) | 205 (36.2) | 42 (37.8) | 0.736 | 146 (35.7) | 61 (33.5) | 30 (43.5) | 0.336 | |
| KRAS variant | Wild type | 567 (83.6) | 326 (86.0) | 137 (80.6) | 54 (78.3) | ||||
| Variant He | 107 (15.8) | 51 (13.5) | 32 (18.8) | 15 (21.7) | |||||
| Variant Ho | 4 (0.6) | 2 (0.5) | 1 (0.6) | – | 0.298 |
Stage IV G-allele KRAS-LCS6 carriers were more likely to be female (66.7%; P = 0.097), and to present with a proximal tumor (71.4%; P = 0.004) as compared with G-allele carriers in other stages (Table 2).
Table 2.
Baseline and molecular characteristics for early stage, stage III and IV patients according to KRAS-LCS6 status
| KRAS-LCS6 wild-type TT | KRAS-LCS6 variant G-allele (He+Ho) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Stage I and II | Stage III | Stage IV | P | Stage I and II | Stage III | Stage IV | P | ||
| Total population, n (%) | 326 (63.1) | 137 (26.5) | 54 (10.4) | 53 (52.5) | 33 (32.7) | 15 (14.9) | |||
| T1 | 43 (13.2) | 1 (0.7) | – | 8 (15.1) | – | – | |||
| T2 | 95 (29.1) | 13 (9.6) | 3 (5.6) | 17 (32.1) | 5 (15.6) | – | |||
| T3 | 174 (53.4) | 113 (83.1) | 41 (75.9) | 27 (50.9) | 24 (75.0) | 12 (80.0) | |||
| T4 | 14 (4.3) | 9 (6.6) | 10 (185) | <0.001 | 1 (1.9) | 3 (9.4) | 3 (20.0) | 0.001 | |
| Sex [male, n (%)] | Male | 173 (53.1) | 75 (54.7) | 28 (51.9) | 0.920 | 29 (54.7) | 22 (66.7) | 5 (33.3) | 0.097 |
| Age at diagnosis (mean, SD) | 68.0 (0.3) | 67.5 (0.4) | 68.5 (0.5) | 0.283 | 68.0 (0.6) | 67.5 (0.7) | 68.5 (1.2) | 0.756 | |
| CRC-related death [yes, n (%)] | Yes | 79 (24.2) | 79 (57.7) | 51 (96.2) | <0.001 | 8 (15.7) | 19 (57.6) | 14 (93.3) | <0.001 |
| Differentiation, n (%) | Well | 38 (13.2) | 9 (7.3) | 2 (4.4) | 5 (10.6) | 2 (6.3) | 1 (7.1) | ||
| Moderate | 224 (77.8) | 77 (62.1) | 31 (67.4) | 35 (74.5) | 24 (75.0) | 5 (35.7) | |||
| Poor | 25 (8.7) | 34 (27.4) | 12 (26.1) | 6 (12.8) | 6 (18.8) | 8 (57.1) | |||
| Undifferentiated | 1 (0.4) | 4 (3.2) | 1 (2.2) | <0.001 | 1 (2.1) | – | – | 0.028 | |
| Location, n (%) | Proximal | 107 (33.0) | 55 (40.2) | 23 (43.4) | 15 (28.3) | 6 (18.8) | 10 (71.4) | ||
| Distal | 100 (30.9) | 42 (30.7) | 20 (37.7) | 14 (26.4) | 17 (53.1) | 2 (14.3) | |||
| Rectosigmoid | 40 (12.4) | 12 (8.8) | 4 (7.6) | 8 (15.1) | 2 (6.3) | 1 (7.1) | |||
| Rectum | 77 (23.8) | 28 (20.4) | 6 (11.3) | 0.230 | 16 (30.2) | 7 (21.9) | 1 (7.1) | 0.004 | |
| Molecular characteristics, n (%) | MSS | 258 (85.2) | 113 (86.9) | 51 (100) | 0.013 | 40 (81.6) | 30 (93.8) | 12 (80.0) | 0.259 |
| CIMP+ | 72 (26.5) | 34 (28.3) | 16 (34.8) | 0.504 | 14 (30.4) | 8 (27.6) | 8 (57.1) | 0.126 | |
| KRAS mutations, n (%) | KRAS mutated | 115 (35.3) | 44 (32.1) | 23 (42.6) | 0.393 | 20 (37.7) | 11 (33.3) | 7 (46.7) | 0.676 |
The KRAS-LCS6 variant is associated with better survival in early-stage CRC
No statistically significant difference was observed in Kaplan–Meier analyses for the KRAS-LCS6 variant and cause-specific survival in the total population (log-rank test, P = 0.864) (Supplementary Fig. S1).
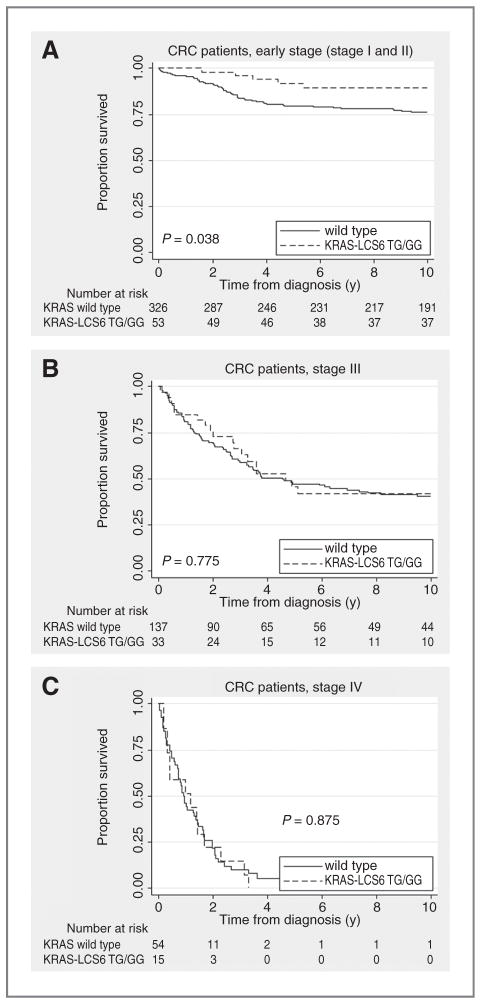
As survival depends on cancer stage, we conducted analyses stratified for stage. Early-stage G-allele carriers showed a statistically significantly better survival as compared with wild-type cases (log-rank test, P = 0.038; Fig. 1A). This difference was not seen for advanced stage cases (Fig. 1B and C; log rank, P = 0.775 and 0.875 for stage III and IV cases, respectively).
Figure 1.
A, Kaplan–Meier curve for the KRAS-LCS6 variant and cause-specific survival in early-stage (stage I and II) CRC. B, Kaplan–Meier curve for the KRAS-LCS6 variant and cause-specific survival in stage III CRC. C, Kaplan–Meier curve for the KRAS-LCS6 variant and cause-specific survival in stage IV CRC.
KRAS/BRAF mutation status enhances the association between the KRAS-LCS6 variant and survival
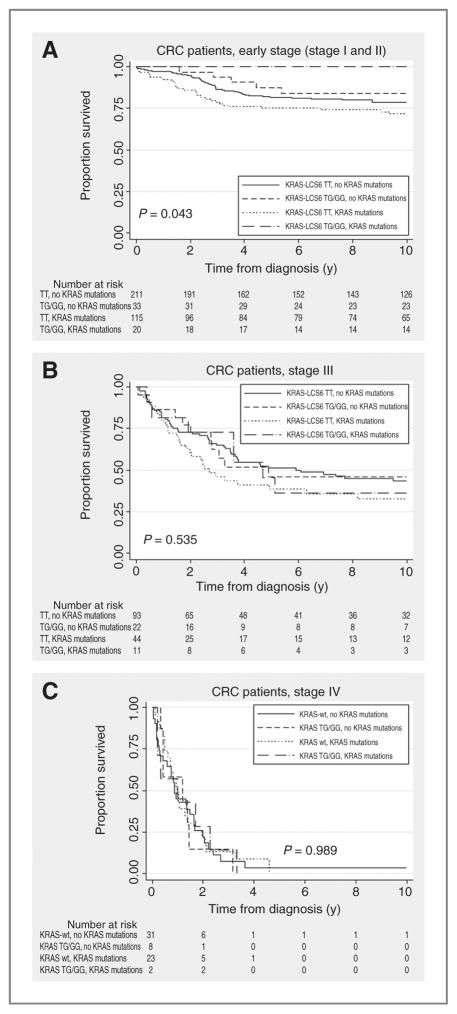
Figure 2A shows Kaplan–Meier analyses for early-stage (stage I and II) CRC cases with the KRAS-LCS6 variant and KRAS mutations. In our population, none of the 20 G-allele carriers with KRAS mutations died due to CRC. KRAS-LCS6 wild-type patients had a poorer survival, especially if they had KRAS mutations (log-rank test, P = 0.043; log-rank test KRAS-LCS6 G-allele carriers with KRAS mutations compared with KRAS-LCS6 G-allele carriers without KRAS mutations P = 0.017). This observation was independent of T stage; among 115 KRAS-LCS6 wild-type cases with KRAS mutations, only 5 (4%) were diagnosed as high-risk stage IIb (T4N0M0). Among G-allele carriers, no patients were diagnosed as stage IIb. For advanced stage patients, no survival difference was seen (Fig. 2B and C, log-rank test, P = 0.535 for stage III and P = 0.989 for stage IV)) although results for stage III patients suggest that KRAS-LCS6 wild-type patients with KRAS mutations have the worst prognosis. Subgroup analysis showed that the better outcome for early-stage KRAS-LCS6 variant carriers was mainly caused by stage II cases. Analyses stratified for T stage were not possible due to limited patient numbers (data not shown).
Figure 2.
A, Kaplan–Meier curve for the KRAS-LCS6 variant, KRAS mutations and cause-specific survival in early-stage (stage I and II) CRC, P = 0.875. B, Kaplan–Meier curve for the KRAS-LCS6 variant, KRAS mutations and cause-specific survival in stage III CRC. C, Kaplan–Meier curve for the KRAS-LCS6 variant, KRAS mutations and cause-specific survival in stage IV CRC.
BRAF mutated CRCs carrying the G-allele showed a similar better outcome, although this was not statistically significant (log-rank test, P = 0.166) possibly due to small number of patients carrying both events (9 patients). Similarly, G-allele carriers with aberrant RASSF1A promoter hypermethylation, another gene involved in the Ras pathway, had a better prognosis, although only borderline statistically significant, as compared with wild-type carriers without RASSF1A hypermethylation (log-rank test, P = 0.062). Analyses combining KRAS, BRAF, and RASSF1A status showed that early-stage G-allele carriers with additional alterations in KRAS, BRAF, or RASSF1A have a better prognosis (log-rank test, P = 0.026). In contrast, when adding methylation status of genes not involved in the Ras pathway such as MGMT or CHFR, no survival differences were observed (MGMT: log-rank test, P = 0.220; CHFR: log-rank test, P = 0.118; data not shown).
The survival impact of the KRAS-LCS6 variant combined with KRAS mutation status is independent of other prognostic factors
In multivariate analyses, no statistically significant differences in cause-specific survival were found for early-stage (HR 0.46; 95% CI: 0.18–1.14), stage III (HR 0.98, 95% CI: 0.55–1.74) or stage IV cases (HR 0.42; 95% CI: 0.17–1.06) with the G-allele variant as compared with wild types, although early-stage and stage IV G-allele carriers seemed to have a slightly better survival (Table 3).
Table 3.
HRs and 95% CI for cause-specific mortality and clinicopathologic and the KRAS-LCS6 variant in 734 CRC cases from the Netherlands Cohort Study on diet and cancer
| Early stage (stage I and II) CRC | Stage III CRC | Stage IV CRC | ||
|---|---|---|---|---|
| KRAS-LCS6 variant | 0.46 (0.18–1.14) | 0.98 (0.55–1.74) | 0.42 (0.17–1.06) | |
| KRAS-LCS6 variant without KRAS mutations | 0.77 (0.30–1.97) | 0.95 (0.44–2.05) | 0.35 (0.11–1.13) | |
| KRAS-LCS6 variant with KRAS mutations | No CRC-related deaths | 1.52 (0.66–3.54) | 0.60 (0.19–1.91) | |
| Sex (male) | 0.97 (0.60–1.57) | 0.92 (0.59–1.45) | 0.85 (0.44–1.64) | |
| Age at diagnosis | 0.99 (0.94–1.05) | 1.01 (0.96–1.06) | 1.02 (0.93–1.10) | |
| Grade | 1 | 1 (reference) | 1 (reference) | 1 (reference) |
| 2 | 1.40 (0.51–5.70) | 0.91 (0.34–2.45) | 2.14 (0.28–16.38) | |
| 3 | 0.77 (0.09–6.72) | 1.90 (0.52–6.94) | 14.47 (1.25–167.07) | |
| 4 | – | 4.17 (0.72–24.05) | 62.36 (2.11–1837.24) | |
| Sublocation of the tumor | Proximal | 1 (reference) | 1 (reference) | 1 (reference) |
| Distal | 0.76 (0.41–1.43) | 0.67 (0.37–1.19) | 0.55 (0.24–1.24) | |
| Rectosigmoid | 0.32 (0.11–0.94) | 0.60 (0.24–1.48) | 0.95 (0.27–3.35) | |
| Rectum | 0.49 (0.18–1.36) | 0.24 (0.08–0.69) | 0.35 (0.06–1.87) |
Early-stage G-allele carriers with KRAS mutations seemed to have a good prognosis; none of these patients died due to CRC. In contrast, no statistically significant differences in survival were found between KRAS nonmutated early-stage (HR 0.77; 95% CI: 0.30–1.97), stage III (HR 0.95; 95% CI: 0.44–2.05) or stage IV cases (HR 0.35; 95% CI: 0.11–1.13) with the KRAS-LCS6 variant. However, stage III G-allele carriers with KRAS mutations seemed to have a poor prognosis (HR 1.52; 95% CI: 0.66–3.54) although not statistically significant.
As Dutch guidelines did not advise adjuvant treatment at the time patients were diagnosed with CRC in the NLCS, the proportion of patients that received adjuvant treatment was very low. Within the early-stage cases, only 9% received adjuvant chemotherapy, for the advanced stage patients this was 31% for stage III and 19% for stage IV. Exclusion of treated patients did not alter our conclusions (data not shown) although it even enhanced the difference between early-stage and stage III G-allele carriers with KRAS mutations (early stage: no CRC-related deaths; stage III: HR 2.36 95% CI: 0.99–5.67) implying that stage III G-allele carriers might indeed have a worse natural course of the disease. However, this analysis is based on small patient numbers.
The survival impact of the KRAS-LCS6 variant is independent of MSI
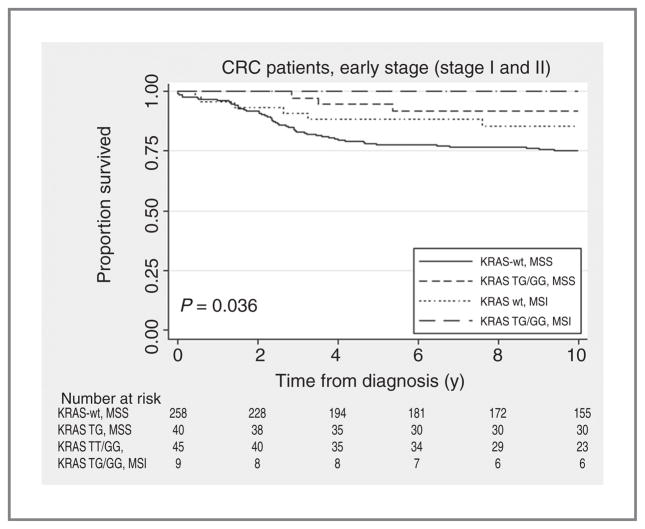
As MSI is currently the only established molecular prognostic marker in CRC, we studied the effect of KRAS-LCS6 genotype stratified for MSI. Exclusion of patients that had an MSI tumor, which is associated with a good prognosis, did not alter our conclusions; both MSI and MSS cases with the KRAS-LCS6 G-variant had a good prognosis. In contrast, patients with the KRAS-LCS6 wild type had a poor prognosis, even if they had an MSI tumor (log-rank test, P = 0.036) (Fig. 3). Additional analyses stratified for sex, tumor sublocation or differentiation grade within MSI patients were not possible due to limited patient numbers.
Figure 3.
Kaplan–Meier curve for the KRAS-LCS6 variant, MSI status and cause-specific survival in early-stage (stage I and II) CRC.
The risk of advanced stage CRC is not associated with the KRAS-LCS6 variant
To study the possibility that the KRAS-LSC6 G-allele predisposes for advanced stage CRC, we studied the association between KRAS-LSC6 genotype and CRC risk. The KRAS-LSC6 G-allele was found in 18% of the subcohort members. For CRC, we observed a decreased risk of developing early-stage (stage I or II) CRC when carrying the KRAS-LSC6 G-allele (RR 0.68, 95% CI: 0.49–0.94). The risk of developing advanced stage CRC (stage III or IV) was however not influenced by the KRAS-LSC6 genotype (RR stage III: 1.02, 95% CI: 0.68–1.53; RR stage IV: 1.15, 95% CI: 0.63–2.09).
Discussion
In this study, we tested the hypothesis that a T>G variant in the LCS6 in the 3′ UTR region of KRAS affects prognosis in early-stage (stage I and II) CRC. The KRAS-LCS6 G-variant was present in 16.4% of the cases whereas it is only found in 6% of world populations (13), and 12% to 15% in persons from European descent (14). We found an increased frequency of the KRAS-LCS6 G-allele in advanced cases (early-stage 14%, 19.2%, and 21.4% in stage III and IV patients, respectively), which is comparable with previously reported frequencies in stage III (19). The G-allele was found in 18% of the subcohort members. We found a statistically significant association between the KRAS-variant and an increased presentation with advanced colon cancer, perhaps giving some insight into the natural biology of colon cancer in KRAS-LCS6 variant carriers. Furthermore, we found a statistically significant increase in survival for early-stage CRC cases with the KRAS-LCS6 G-variant; among KRAS-mutated patients none of the early-stage patients carrying the G-allele died from CRC. This effect was independent of other prognostic factors such as tumor differentiation or sublocation. As T4 tumors were rare in our group of early-stage cases, a higher frequency of stage IIb cases among KRAS-LCS6 wild types is not the cause of the observed worse outcome. No statistically significant effect was seen in stage III or IV, although results suggested a slightly worse prognosis for stage III cases with the G-variant and KRAS mutations. In addition, we studied the effect of the KRAS-LSC6 G-allele on CRC risk and observed a slightly decreased risk of early-stage CRC, but no effect on the risk of advanced stage CRC, suggesting that the G-allele is not associated with a higher likelihood of advanced stage CRC.
In a number of previous studies, mutations in KRAS have been associated with a poorer prognosis. However, we and others have recently described that results on this topic are inconsistent and the clinical relevance of these findings are unclear (4). Acquired KRAS mutations are however not the same as the KRAS-LCS6 variant, which is congenital and could therefore have a different effect on tumor development, biology, and thus prognosis.
The unexpected finding that the KRAS-LCS6 variant is associated with an increased survival in early-stage CRC is intriguing. Previous research has suggested that cellular senescence can be triggered by overexpression of oncogenic Ras and might contribute to growth cessation in premalignant or benign neoplasms (29). Tumor cell senescence has been reported in human cancers, and premalignant colon adenomas display features of senescence as well (30). Previous studies have often suggested that oncogene-induced senescence plays a role in premalignant lesions only. Nevertheless, recent evidence suggests that physiologic levels of KRAS can induce senescence in the absence of the transcription factor Wilms tumor 1 (WT1) and lung cancer patients with a high KRAS gene expression had a good prognosis if they had decreased expression of WT1 related genes (31). These results imply that other molecular factors can be involved in the determination of cell fate, and that oncogene-induced senescence can occur after an altered expression of other genetic or epigenetic targets. Hypothetically, this could also play a role in CRC, and the KRAS-LCS6 genotype could either lead to an advanced stage tumor, or an early-stage tumor with a better prognosis based on the other (epi)genetic markers that are affected. However, evidence on this concept is scarce and more research is needed to elucidate whether our findings can be explained by oncogene-induced senescence.
Similar to KRAS mutations, we observed a better outcome for early-stage (stage I and II) cases with the KRAS-LCS6 G-variant and BRAF mutations or RASSF1A hypermethylation, 2 other genes involved in the Ras signaling pathway. BRAF-associated senescence has previously been reported to occur in melanoma (32) but a possible role of RASSF1A in oncogence-induced senescence is currently unknown. As in our population both events are less common, statistical significance was not reached. When combining these (epi) genetic events, the better outcome of patients with the variant G-allele and KRAS, BRAF, or RASSF1A alterations was even more enhanced. Along this line, we hypothesize that Ras overexpression due to the KRAS-LCS6 G-allele, in combination with (epi)genetic alterations in genes from the Ras pathway, could induce senescence in early-stage CRC thereby influencing survival. For advanced cases on the other hand, an increasing number of molecular pathways are affected that all have a role in prognosis. Although intriguing, this is only speculative; more research is needed to determine whether the KRAS-LCS6 G-variant and (epi) genetic alterations in Ras-associated genes can lead to oncogene-induced senescence in early-stage CRC.
Several studies have previously shown a tumor growth suppression effect of the let-7 miRNA (10–12, 33–35), and lower let-7 expression and higher KRAS levels in the presence of the KRAS-LCS6 G-variant (13). Following this, it would be expected that patients with the KRAS-LCS6 variant have a worse prognosis and this has been shown for oral cancer (17). For CRC, there are 2 published reports studying the effect of KRAS-LCS6 genotype (19, 20). One reports a poor survival among a small population of irinotecan-refractory metastatic patients with the KRAS-LCS6 G-variant, and an association with KRAS mutations and the absence of BRAF mutations (19), these findings could not be replicated in our study. The second reports a better response to cetuximab in meta-static CRC and a longer survival in patients with the G-variant without KRAS mutations, but not statistically significant (20). We also observe a slightly better prognosis in stage IV G-allele carriers, although not statistically significant, but our group of stage IV patients is small rendering instable results. Although previous studies used germline tissue to assess the KRAS-LCS6 genotype, we used tumor DNA to assess genotype. However, it has previously been well documented that genotype of normal and tumor tissue is the same for the KRAS-LCS6 variant (13).
The conflicting results in early and advanced stage CRC also raises questions on the origin and progression of tumors in different cancer stages, and whether early-stage CRC might develop through a molecular distinct pathway as compared with advanced stage. Our results indicate that the KRAS-LCS6 variant is more common among cases with advanced stage disease, however, those patient that are diagnosed early with the KRAS-LCS6 variant seem to have a more advantageous outcome. This might imply a different biology in early-stage as compared with advanced stage cases. In addition, our finding that early-stage KRAS-LCS6 wild-type patients have a poor prognosis, even if they have a MSI tumor, might indicate that these patients are possibly in need of (additional) adjuvant treatment. However, further research including randomized clinical trials, is needed to assess whether these early-stage patients with a poor prognosis would benefit from additional adjuvant treatment. Up until now, MSI has been considered to be a marker for good prognosis (36) however, our data suggest a better outcome for KRAS-LCS6 G-allele carriers independent of MSI status. Even though our study is the largest study on the KRAS-LCS6 genotype in CRC up until now, patient numbers in specific subgroups are still small. Larger, prospective studies and randomized clinical trials are needed to validate the potential role of KRAS-LCS6 genotype as a prognostic biomarker in therapy decision-making.
In conclusion, our assessment of the influence of the KRAS-LCS6 G-variant in early-stage CRC cases showed a better outcome for early-stage G-allele carriers with KRAS mutations. Although the population used in this study is among the largest studies that have been done on the KRAS-LCS6 variant, subgroups were small. However, this population is the only group studied to date that is generally untreated, and for the first time gives insight into the natural biology of CRC with the LCCS6 variant. Future studies validating our results are however needed. Nevertheless, our data should be regarded as hypothesis generating providing a first indication that the KRAS-LCS6 genotype is a possible prognostic biomarker for early-stage CRC that can be used to identify CRC patients with a good prognosis.
Acknowledgments
Grant Support
This study was funded by Maastricht University Medical Center through a personal grant for K. Smits from the Centre for Research Innovation and Policy (CRISP).
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
J.B. Weidhaas has a consultant and advisory role in MiraDX, her husband is a board member of MiraDX (uncompensated), and she has stock ownership of MiraDX. The other authors disclosed no potential conflicts of interest.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. GLO-BOCAN 2008, Cancer incidence and mortality worldwide: IARC cancer base no. 10. Lyon, France: International Agency for Research on Cancer; 2010. [Google Scholar]
- 2.Klump B, Nehls O, Okech T, Hsieh CJ, Gaco V, Gittinger FS, et al. Molecular lesions in colorectal cancer: impact on prognosis? Original data and review of the literature. Int J Colorectal Dis. 2004;19:23–42. doi: 10.1007/s00384-003-0499-7. [DOI] [PubMed] [Google Scholar]
- 3.Wallner M, Herbst A, Behrens A, Crispin A, Stieber P, Goke B, et al. Methylation of serum DNA is an independent prognostic marker in colorectal cancer. Clin Cancer Res. 2006;12:7347–52. doi: 10.1158/1078-0432.CCR-06-1264. [DOI] [PubMed] [Google Scholar]
- 4.Smits KM, Cleven AH, Weijenberg MP, Hughes LA, Herman JG, de Bruine AP, et al. Pharmacoepigenomics in colorectal cancer: a step forward in predicting prognosis and treatment response. Pharmacogenomics. 2008;9:1903–16. doi: 10.2217/14622416.9.12.1903. [DOI] [PubMed] [Google Scholar]
- 5.Paranjape T, Slack FJ, Weidhaas JB. MicroRNAs: tools for cancer diagnostics. Gut. 2009;58:1546–54. doi: 10.1136/gut.2009.179531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 7.Jerome T, Laurie P, Louis B, Pierre C. Enjoy the silence: The story of let-7 microRNA and cancer. Curr Genomics. 2007;8:229–33. doi: 10.2174/138920207781386933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–6. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 10.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105:3903–8. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–64. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 12.Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903–6. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 13.Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–40. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ratner E, Lu L, Boeke M, Barnett R, Nallur S, Chin LJ, et al. A KRAS-variant in ovarian cancer acts as a genetic marker of cancer risk. Cancer Res. 2010;70:6509–15. doi: 10.1158/0008-5472.CAN-10-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pharoah PD, Palmieri RT, Ramus SJ, Gayther SA, Andrulis IL, Anton-Culver HA, et al. The role of KRAS rs61764370 in invasive epithelial ovarian cancer: implications for clinical testing. Clin Cancer Res. 2011;17:3742–50. doi: 10.1158/1078-0432.CCR-10-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paranjape T, Heneghan H, Lindner R, Keane FK, Hoffman A, Hollestelle A, et al. A 3′-untranslated region KRAS variant and triple-negative breast cancer: a case-control and genetic analysis. Lancet Oncol. 2011;12:377–86. doi: 10.1016/S1470-2045(11)70044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christensen BC, Moyer BJ, Avissar M, Ouellet LG, Plaza SL, McClean MD, et al. A let-7 microRNA-binding site polymorphism in the KRAS 3′ UTR is associated with reduced survival in oral cancers. Carcinogenesis. 2009;30:1003–7. doi: 10.1093/carcin/bgp099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson HH, Christensen BC, Plaza SL, Wiencke JK, Marsit CJ, Kelsey KT. KRAS mutation, KRAS-LCS6 polymorphism, and non-small cell lung cancer. Lung Cancer. 2010;69:51–3. doi: 10.1016/j.lungcan.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graziano F, Canestrari E, Loupakis F, Ruzzo A, Galluccio N, Santini D, et al. Genetic modulation of the Let-7 microRNA binding to KRAS 3′-untranslated region and survival of metastatic colorectal cancer patients treated with salvage cetuximab-irinotecan. Pharmacoge-nomics J. 2010;10:458–64. doi: 10.1038/tpj.2010.9. [DOI] [PubMed] [Google Scholar]
- 20.Zhang W, Winder T, Ning Y, Pohl A, Yang D, Kahn M, et al. A let-7 microRNA-binding site polymorphism in 3′-untranslated region of KRAS gene predicts response in wild-type KRAS patients with metastatic colorectal cancer treated with cetuximab monotherapy. Ann Oncol. 2011;22:104–9. doi: 10.1093/annonc/mdq315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van den Brandt PA, Schouten LJ, Goldbohm RA, Dorant E, Hunen PM. Development of a record linkage protocol for use in the Dutch Cancer Registry for Epidemiological Research. Int J Epidemiol. 1990;19:553–8. doi: 10.1093/ije/19.3.553. [DOI] [PubMed] [Google Scholar]
- 22.van den Brandt PA, Goldbohm RA, van’t Veer P, Volovics A, Hermus RJ, Sturmans F. A large-scale prospective cohort study on diet and cancer in The Netherlands. J Clin Epidemiol. 1990;43:285–95. doi: 10.1016/0895-4356(90)90009-e. [DOI] [PubMed] [Google Scholar]
- 23.Brink M, de Goeij AF, Weijenberg MP, Roemen GM, Lentjes MH, Pachen MM, et al. K-ras oncogene mutations in sporadic colorectal cancer in The Netherlands Cohort Study. Carcinogenesis. 2003;24:703–10. doi: 10.1093/carcin/bgg009. [DOI] [PubMed] [Google Scholar]
- 24.de Vogel S, Bongaerts BW, Wouters KA, Kester AD, Schouten LJ, de Goeij AF, et al. Associations of dietary methyl donor intake with MLH1 promoter hypermethylation and related molecular phenotypes in sporadic colorectal cancer. Carcinogenesis. 2008;29:1765–73. doi: 10.1093/carcin/bgn074. [DOI] [PubMed] [Google Scholar]
- 25.Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, et al. CpG island methylator phenotype underlies sporadic micro-satellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–93. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 26.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Derks S, Lentjes MH, Hellebrekers DM, de Bruine AP, Herman JG, van Engeland M. Methylation-specific PCR unraveled. Cell Oncol. 2004;26:291–9. doi: 10.1155/2004/370301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suraweera N, Duval A, Reperant M, Vaury C, Furlan D, Leroy K, et al. Evaluation of tumor microsatellite instability using five quasimonomorphic mononucleotide repeats and pentaplex PCR. Gastroenterology. 2002;123:1804–11. doi: 10.1053/gast.2002.37070. [DOI] [PubMed] [Google Scholar]
- 29.Mooi WJ, Peeper DS. Oncogene-induced cell senescence–halting on the road to cancer. N Engl J Med. 2006;355:1037–46. doi: 10.1056/NEJMra062285. [DOI] [PubMed] [Google Scholar]
- 30.Collado M, Serrano M. Senescence in tumours: evidence from mice and humans. Nat Rev Cancer. 2010;10:51–7. doi: 10.1038/nrc2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vicent S, Chen R, Sayles LC, Lin C, Walker RG, Gillespie AK, et al. Wilms tumor 1 (WT1) regulates KRAS-driven oncogenesis and senescence in mouse and human models. J Clin Invest. 2010;120:3940–52. doi: 10.1172/JCI44165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–4. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 33.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–22. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 34.Torrisani J, Bournet B, du Rieu MC, Bouisson M, Souque A, Escourrou J, et al. let-7 MicroRNA transfer in pancreatic cancer-derived cells inhibits in vitro cell proliferation but fails to alter tumor progression. Hum Gene Ther. 2009;20:831–44. doi: 10.1089/hum.2008.134. [DOI] [PubMed] [Google Scholar]
- 35.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, et al. let-7 regulates self renewalandtumorigenicityofbreastcancercells. Cell. 2007;131:1109–23. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 36.Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–87. e3. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]