Abstract
Long-range enhancer interactions critically regulate gene expression, yet little is known about how their coordinated activities contribute to CNS development, or how this may in turn relate to disease states. By examining the regulation of the transcription factor NFIA in the developing spinal cord, we identified long-range enhancers that recapitulate NFIA expression across glial and neuronal lineages in vivo. Complementary genetic studies found that Sox9/Brn2 and Isl1/Lhx3 regulate enhancer activity and NFIA expression in glial and neuronal populations. Chromatin conformation analysis (3C) revealed that these enhancers and transcription factors form distinct architectures within these lineages in the spinal cord. In glioma models, the glial-specific architecture is present in tumors, and these enhancers are required for NFIA expression and contribute to glioma formation. By delineating three-dimensional mechanisms of gene expression regulation, our studies identify lineage specific chromatin architectures and associated enhancers that regulate cell fate and tumorigenesis in the CNS.
Introduction
Transcriptional control of gene expression is regulated at several levels, including site-specific transcription factors, epigenetic modifications of chromatin, and long-range enhancer interactions 1–3. Understanding how these diverse regulatory layers coordinate gene expression, in three-dimensions, across long-range enhancers to influence biological phenomena remains a fundamental question that has critical implications for development, physiology, and associated diseases. ES cells and the hematopoietic system have provided useful initial insights into how three-dimensional chromatin configurations are formed and regulate gene expression in vertebrates4–9. Despite these advances, how long-range enhancers and associated three-dimensional chromatin architecture contribute to the development of the central nervous system (CNS) and associated diseases remains unclear.
Decoding transcriptional regulation via three-dimensional architecture requires the identification and validation of distal enhancers that regulate long-range regulation of gene expression 10. This represents a major barrier as in silico approaches currently used can predict enhancer function, however these activities can vary across dynamic developmental states and cell lineage, ultimately requiring validation in appropriate in vivo models 11,12. These limitations highlight the need for tractable, in vivo systems to identify and validate enhancer elements, as these are key entry points for understanding how chromatin architecture influences gene expression and, more broadly, tissue development and disease.
The developing spinal cord offers an attractive system in which to identify enhancers and investigate these regulatory mechanisms in vivo, as it is an accessible embryonic tissue containing diverse CNS lineages that are generated in a well-defined spatial and temporal pattern 13,14. One of the key events that occurs during spinal cord development is a process termed the “gliogenic switch” 14–16. Initially, progenitor populations produce neurons between E3–E5 (E9–E11 in mouse), and subsequently produce glia beginning at E6 (E12 in mouse). Previously, we identified Nuclear Factor I-A (NFIA) as a key transcriptional determinant of the gliogenic switch, in which NFIA is induced in progenitor populations coincident with the onset of gliogenesis 17,18. Sox9 (Sex Determining Region Y-Box 9) directly regulates NFIA induction, however expression of NFIA is only mildly affected in the absence of Sox9 and NFIA is also expressed in motor neuron populations in the spinal cord, suggesting additional regulatory elements regulate NFIA expression across neuronal and glial lineages during development18.
To delineate these regulatory mechanisms, we performed an enhancer screen in the embryonic chick spinal cord and identified multiple, long-range enhancer elements that recapitulate NFIA expression in glial precursors and motor neurons. Chromatin conformation and genetic analysis revealed distinct chromatin architectures and associated transcriptional mechanisms regulating NFIA expression across glial and neuronal lineages. These chromatin architectures are present in glioma and regulate NFIA expression and tumorigenesis through the associated enhancers. Together, our multi-disciplinary approach, integrating in vivo enhancer discovery and long-range chromatin interactions with mouse genetics and associated disease models, defines a transcriptional mechanism that regulates CNS development and malignancy.
Results
NFIA enhancers demonstrate lineage specific activities and regulation
NFIA is specifically induced in glial precursors occupying the ventricular zone (VZ) and in motor neurons in mantel regions during the E4–E6 gliogenic switch interval in the embryonic chick spinal cord (Fig. 1A–B). That NFIA demonstrates expression across diverse CNS lineages raises the question of how it is selectively regulated within these cell populations. To identify the transcriptional mechanisms associated with its expression in the spinal cord, we performed an enhancer screen in the embryonic chick and identified an enhancer termed e161 that recapitulates the spatial and temporal patterns of NFIA induction in glial progenitors of the VZ (Fig. 1E–F, U; Supplemental Figures S1–S2). Subsequent overexpression and deletion mapping studies revealed that Brn2 (also known as Pou3f2) regulates the activity of e161 (Fig. 1I–N; V, Y–Z; Supplemental Figure S2). Our previous studies identified the e123 enhancer that is regulated by Sox9 and has a similar spatial/temporal pattern of activity as e161 (Fig. 1C–D, O; 18). Therefore we next investigated the specificity of the e123/Sox9 and e161/Brn2 regulatory relationships, finding that Sox9 and Brn2 do not cross-activate enhancers (Fig. 1L, P), suggesting that these are distinct regulatory nodes that collaboratively oversee NFIA induction during gliogenesis.
Figure 1. NFIA enhancers are regulated by lineage-specific mechanisms.
(A–B) NFIA expression in the chick spinal cord during the gliogenic switch. (C–H) Spatial and temporal activities of NFIA enhancers in the chick spinal cord, arrows denote GFP-reporter activity in VZ (D, F) and MNs (G–H). (I–T) Sox9-specifically induces e123 activity (L–N), Brn2 specifically induces e161 activity (O–Q), and Isl1/Lhx3 specifically induce e96 activity (R–T). (U) Map of the enhancer locations within the chick NFIA locus. (V) Location of Isl1/Lhx3 Mu5/6 binding sites within e96 and the Brn2 Mu4 binding site within e161; alignment of chick and mouse sequences reveals conservation across species. (W–X) Deletion of Mu5/6 sites within e96 eliminates MN activity. (Y–Z) Deletion of Mu4 site within e161 eliminates VZ activity at E6. Each chick electroporation experiment was performed at least 4 independent times/8 sections per spinal cord was analyzed; images are representative. “Con” is control/non-electroporated side of spinal cord; “Ep” is the electroporated side of spinal cord. Scale bars are 100um.
In the course of screening our candidate enhancers, we identified another element, e96, which is specifically active in motor neurons (MNs) (Fig. 1G–H) and resembles NFIA expression in these populations (Fig. 1B). Deletion mapping and overexpression studies revealed that Isl1 and Lhx3, transcriptional partners that are required for MN development 19,20, regulate the activity of e96 (Fig. 1T, V–X; Supplemental Figure S3). Moreover, Sox9/Brn2 do not activate e96 and Isl1/Lhx3 do not active e123 or e161 (Fig. 1I–T), suggesting that regulation of the enhancers by these transcription factors is lineage specific. Put together, our enhancer studies suggest that distinct transcriptional mechanisms regulate NFIA expression across CNS lineages: Sox9/Brn2 in glial precursors and Isl1/Lhx3 in MNs.
Lineage-specific transcriptional mechanisms regulate NFIA expression
We next sought to determine whether the individual transcriptional mechanisms implicated by the enhancer studies regulate NFIA expression in glial precursors or motor neurons. The e123/e161 enhancer studies suggest that Sox9 and Brn2 collaboratively regulate NFIA induction during the gliogenic switch. In support of this model, we found that Sox9 and Brn2 are co-expressed in ventricular zone (VZ) populations at E12.5 in the mouse, concurrent with NFIA induction (Fig. 2A–B, E, I). To examine this putative relationship at the genetic level, we evaluated NFIA induction during the gliogenic switch in a series of Sox9 and Brn2 single and double mouse mutants 21,22 (Supplemental Figure S4). Analysis of Sox9 or Brn2 single mutant mice revealed a delay in NFIA induction at E11.5 (Fig. 2E–G) and reduced expression at E12.5 (Fig. 2I–K), suggesting compensation by other Sox- or Pou- family members. The Sox9;Brn2 double mutant demonstrated delayed induction at E11.5, which continued through E12.5 and was matched by a concordant reduction in the expression of the glial precursor marker Glast (Fig. 2H, L–P; Supplemental Figure S4). That the double Sox9;Brn2 mutants demonstrated a substantially greater reduction in NFIA expression compared to the single mutants (Fig. 2J,K v. L) indicates that genetic cooperation between these factors regulates NFIA induction during the gliogenic switch. To further understand the nature of the collaboration between Sox9 and Brn2, we examined whether there was also a biochemical relationship. Towards this, we performed co-immunoprecipitation (Co-IP) studies on mouse E12.5 spinal cord, finding that Sox9 and Brn2 associate in these tissues (Fig. 2C; Supplemental Figure S4), suggesting that cooperation between these factors is mediated at the biochemical level.
Figure 2. Brn2/Sox9 and Isl1 regulate NFIA expression in the spinal cord.
(A–C) Sox9 and Brn2 are co-expressed in VZ populations at 12.5 in the developing mouse spinal cord. (D) Immunoprecipitation of E12.5 spinal cord with Sox9 antibodies, reveals biochemical association between Sox9 and Brn2. (D–P) Single Sox9- (F, J, N) or Brn2 (G, K, O) mutants demonstrate delayed NFIA induction at E11.5 (F–G, arrows) and reduced expressed at E12.5 (J–K). Double Sox9;Brn2 mutants (G,K,O) demonstrate delayed induction at E11.5 (H, arrow), which continues through E12.5 (L, arrow). NFIA expression is detected via immunofluorescence. Glast expression, detected via in situ hybridization, also demonstrates a greater reduction in the Sox9;Brn2 double mutants at E12.5 (M–P). (Q–R) Isl1-mutants demonstrate reduced NFIA expression in MN populations at E11.5. Images from each mutant and timepoint are representative of at least three embryos per genetic condition and 8 sections analyzed per embryo. Statistics: (S) The percentage of Lhx3-expressing cells that co-express NFIA are presented as box-and-whisker plots where box limits represent first and third quartiles, center line represents median, and whiskers represent minimum and maximum values. *P<0.00001 represents results of unpaired two-tailed t-test. Scale bars are 100um. See Supplemental Figure S10 for uncropped gel images in D.
In parallel, we investigated the regulatory relationship between Isl1 and NFIA in MN populations, finding that they are co-expressed in MNs at E12.5 in the spinal cord (Supplemental Figure S4). Examination of their genetic relationship revealed that Isl1-mutants demonstrate a robust decrease in NFIA expression in the MN populations, where there is a drastic decrease in the number of Lhx3-cells that co-express NFIA (Fig. 2Q–S). These observations in MNs, coupled with our findings during the gliogenic switch, support a model whereby distinct transcriptional mechanisms regulate NFIA expression in MNs and glial precursors.
NFIA enhancers form distinct chromatin architectures in the spinal cord
Having established the genetic relationships that oversee NFIA induction in glial precursors and MNs, we next sought to determine the biochemical mechanism by which these regulatory relationships operate. Focusing first on the Sox9/Brn2 relationship in glial precursors, the relative locations of the associated e123 and e161 enhancers gave critical insight into the putative mechanism of action. In the mouse e123 is 90kb away from the transcriptional start site (TSS) and 120kb away from the e161 enhancer (Fig. 3A), while the e161 enhancer is 30kb away from the TSS and is located within the first intron (Fig. 3A). These data, coupled with our Co-IP data, suggest long-range DNA interactions between e123/e161 and the TSS are responsible for NFIA induction during the gliogenic switch. To test this possibility, we performed chromatin conformation capture (3C) 23,24 on E12.5 spinal cord to test the long-range DNA interactions between e123, e161, and the TSS/promoter across the encompassing 120kb interval within the NFIA locus (Fig. 3A). Using e123 as our anchor point for 3C, we identified strong associations with regions in close proximity to the TSS/promoter region (region6) and containing e161 (region 11) that are not present in other NFIA-expressing tissues, e.g. lung, (Fig. 3B). To further substantiate these interactions, we performed ChIP analysis on E12.5 spinal cord, finding that both Sox9 and Brn2 associate with their response sites within e123 and e161, respectively, as well as the TSS region, but not e96 (Fig. 3D; Supplemental Figure S5). Integration of these 3C and ChIP data, along with our Co-IP observations (Fig. 2C) suggests a double-looped conformation across the e123/TSS/e161 interval at the NFIA locus during the gliogenic switch in the spinal cord (E11.5–E12.5) (Fig. 3C). Importantly, given that NFIA induction is delayed in the single Sox9- and Brn2 mutants (Fig. 2I–L), it is likely other, compensatory Sox- and/or Pou-family members can also participate in this looped configuration under those specific genetic conditions.
Figure 3. Enhancers demonstrate differential architecture at the NFIA locus.
(A) Map of the enhancer locations within the mouse NFIA locus. (B) Chromatin conformation capture (3C) assay performed on E12.5 spinal cord and lung. Red text denotes e123 as the anchor point from which long-range DNA interactions across the e123 – e161 interval were measured. (C) Schematic representation of the prospective long-range DNA interactions and transcription factor associations across the e123 – e161 interval during the gliogenic switch. The associated transcription factors in this model reflect what normally occurs during the gliogenic switch at E11.5 and does not account for potential compensation by other Pou- and Sox-family members in the Sox9 and/or Brn2 mutants (see figure 2I–L) (D) Chromatin immunoprecipitation (ChIP) assays on E12.5 spinal cord reveal that Sox9 and Brn2 associate with each of the gliogenic regulatory regions across the NFIA locus: e123, promoter, e161. (E) Chromatin conformation capture (3C) assay performed on E12.5 spinal cord and lung. Red text denotes promoter region as the anchor point from which long-range DNA interactions across the e96 - promoter interval were measured. (F) Schematic representation of the prospective long-range DNA interactions across the e96 – promoter interval. (G) Chromatin immunoprecipitation (ChIP) assays on E12.5 spinal cord reveal that Isl1 and Lhx3 associate with their regulatory regions across the NFIA locus: e96 and promoter. Three independent libraries were assayed per experiment. Note that in each 3C experiment a no ligase control was performed to assess the level of ligase-independent non-specific PCR products. Control regions for ChIP in D and G correspond to intronic sequences within NFIA devoid of predicted Sox9, Brn2, Lhx3, or Isl1 binding sites (see supplemental methods). Statistics: In (B) and (E) error bars indicate SD, and results of two-tailed t-tests are shown. *P<0.002. See Supplemental Figure S10 for uncropped gel images in D and G.
Next we addressed the mechanism by which Isl1/Lhx3 regulate the expression of NFIA in MNs. Similar to the gliogenic enhancers, e96 is also a long-range enhancer element, as it lies approximately 135kb away from the TSS (Fig. 3A). Using the TSS as the anchor point, we performed 3C assays on E12.5 spinal cord, finding a strong association between the TSS and regions containing e96 (region 14) and that the associations between the TSS/promoter and e96 are not present in other NFIA-expressing tissues, e.g. lung (Fig. 3E). ChIP assays on E12.5 spinal cord confirmed association of both Isl1 and Lhx3 with e96 and the TSS, but not e123/e161 (Fig. 3G; Supplemental figure S5). These data, coupled with our 3C data, suggest a single looped conformation across the e96/TSS interval at the NFIA locus during the gliogenic switch in the spinal cord. (Fig. 3F). These data, combined with our observations from the gliogenic enhancers, provide genetic, biochemical, and three-dimensional conformational evidence that differential chromatin looping at the NFIA locus drives its expression across diverse CNS lineages in the developing spinal cord (Fig. 3C, F).
Gliogenic chromatin architecture forms prior to NFIA induction
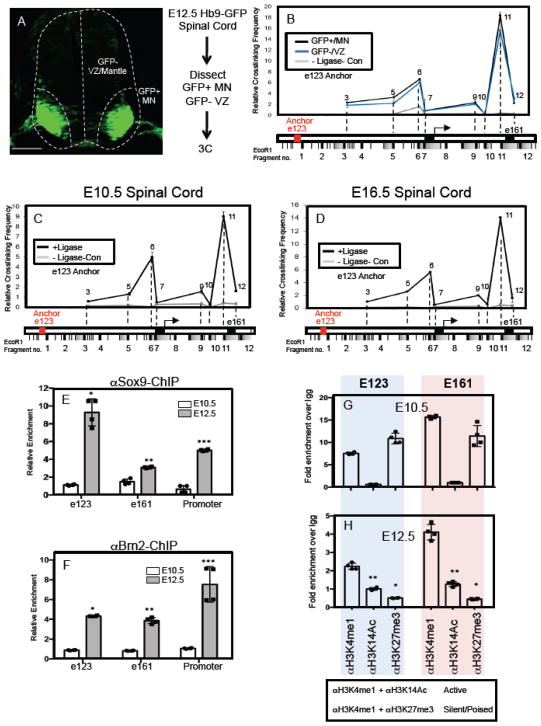
To further understand the properties of these NFIA-associated chromatin loops, we focused on the gliogenic chromatin loop and investigated whether this chromatin architecture is regulated by the associated transcription factors, Sox9 and Brn2. Given the difficulty of breeding a sufficient number of Sox9;Brn2 double mutant embryos in which to perform 3C, coupled with possible redundancy with other Sox- and Pou-family members, we addressed this question by using the Hb9-GFP reporter mouse. This mouse line allows us to use GFP expression in MNs to physically dissect MN populations from VZ populations in the E12.5 spinal cord, such that the GFP-negative, VZ populations contains Sox9/Brn2 and the GFP-positive, MN populations do not contain Sox9/Brn2 (Fig. 4A). In this way we can assess gliogenic loop formation in cell populations that express Sox9/Brn2 or those that do not. As shown in Figure 4B, the e123/e161 chromatin configuration is present in both the GFP-negative and the GFP-positive cell populations in the E12.5 spinal cord. The presence of the gliogenic loop in the GFP-positive populations that lack Sox9/Brn2 expression, suggests that its formation occurs independent of these factors.
Figure 4. Gliogenic chromatin loop forms prior to NFIA induction.
(A) GFP expression in MN populations in the E12.5 spinal cord of the Hb9-GFP mouse and the associated workflow for 3C experiments in GFP-positive MN populations and GFP-negative VZ/mantle populations. (B) Chromatin conformation capture (3C) assay performed on E12.5 spinal cord from Hb9-GFP mice, where the GFP+/MN and GFP-/VZ regions were manually dissected. Red text denotes e123 as the anchor point from which long-range DNA interactions across the e123 – e161 interval were measured. (C–D) Chromatin conformation capture (3C) assay performed on E10.5 (C) and E16.5 (D) spinal cord from wild type mice. Red text denotes e123 as the anchor point from which long-range DNA interactions across the e123 – e161 interval were measured. For each 3C experiment (B–D), three independent libraries were assayed per experiment. Note that in each 3C experiment a no ligase control was performed to assess the level of ligase-independent non-specific PCR products. (E–F) Chromatin immunoprecipitation assay (ChIP) assays on E10.5 and E12.5 spinal cord, comparing the relative enrichment of Sox9 (E) and Brn2 (F) associations with e123, e161, and the core promoter. (G–H) ChIP assays from spinal cord comparing the methylation and acetylation status of the E123 and E161 enhancers before (E10.5) and during the gliogenic switch (E12.5). Blue shaded panels represent data from e123 at both E10.5 and E12.5, while the pink shaded panels represent data from e161 at both E10.5 and E12.5. Statistics: (E) *p=0.03, **p=0.04, and ***0.007. In (F) *p=9x10−5, **p=0.004, ***p=0.05. In (H) *p= 0.01, **p=0.04. All error bars are SD. Scale bars are 100um.
One feature of many chromatin loops that regulate gene expression during development is that they form prior to the induction of the target gene and are, therefore, considered “pre-formed” loops25. These pre-formed structures are thought to provide a permissive configuration through which tissue specific transcription factors provide timely responses to developmental stimuli26. That the NFIA gliogenic loop forms independent of Sox9/Brn2 suggests that it may also be a pre-formed loop. To evaluate if indeed this is the case we determined whether the NFIA gliogenic loop is present prior to NFIA-induction at E10.5 and after its induction at E16.5. As shown in Figure 4C–D, the gliogenic loop is found at both E10.5 and E16.5, indicating that it is “pre-formed” at E10.5 and stable across early glial development.
That the gliogenic chromatin loop is pre-formed prior to NFIA induction at E10.5 raises the question of how NFIA is induced at E11.5. Given that formation of the loop does not require Sox9 or Brn2, yet these factors are required for NFIA expression (see figure 2), one possibility is that these transcription factors are not able to bind their respective enhancers at E10.5. To test this possibility, we performed ChIP assays on E10.5 spinal cord, assessing the association between Sox9/Brn2 and e123/e161/promoter regions. These experiments revealed that Sox9 and Brn2 do not efficiently ChIP with the e123/e161/promoter regions at E10.5, yet these factors efficiently associate with these regulatory elements at E12.5 (Figure 4E–F). One potential explanation for this shift in enhancer occupancy by Sox9 and Brn2 is changes in chromatin state, which we assessed via ChIP-qPCR on E10.5 and E12.5 spinal cord using antibodies for poised/silent enhancers (H3K4me1 + H3K27me3) and active enhancers (H3K4me1 + H3K14Ac). We found that both e123 and e161 enhancers contain chromatin marks consistent with closed/poised chromatin at E10.5 and that these marks (H3K27me3) are dramatically reduced at E12.5 (Figure 4G–H). These changes likely reflect the acquisition of a more permissive chromatin state at E12.5, which allows for the binding of Sox9/Brn2 to their respective enhancers. In parallel, we also found a concordant increase in “active” chromatin, reflected by an increase in H3K14Ac marks across the E10.5 – E12.5 interval. Together, these data indicate that the gliogenic loop is formed prior to NFIA induction and that the precise timing of NFIA induction is further coordinated by the specific association of Sox9/Brn2 with their enhancers during the gliogenic switch.
Glial-specific enhancers regulate NFIA expression and tumorigenesis
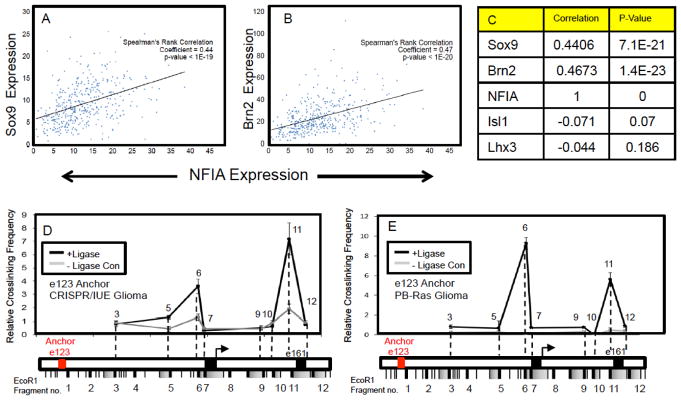
Given that developmental processes are often reutilized during tumorigenesis, we sought to determine whether similar regulatory mechanisms are associated with NFIA expression in glioma 27,28. Taking a bioinformatics approach, we assessed the co-expression of NFIA with its associated transcriptional regulators across a cohort of >400 high-grade human glioma expression datasets29. Using Spearman’s correlation co-efficiency analysis, we found that NFIA expression is very highly correlated with both Sox9 and Brn2, but not Lhx3 and Isl1 (Fig. 5A–C; Supplemental Figure S6). These relationships were corroborated in additional glioma datasets comprised of both high-and low-grade glioma (Supplemental Figure S6) 30, and together suggest that the transcriptional mechanisms controlling NFIA expression during gliogenesis similarly regulate its expression in glioma. To test this hypothesis, we used two different mouse models of glioma that harness in utero electroporation (IUE) of the embryonic cortex to facilitate gene manipulation. One model utilizes CRISPR/Cas9 (herein CRISPR/IUE)31 gene editing of NF1, PTEN, and p53, tumor suppressors that are commonly mutated in human glioma32–34, while the other utilizes PiggyBac (PB) targeting of glial lineages with oncogenic Ras to produce malignant glioma (herein PB-Ras/IUE) (Supplemental Figure S7; 28,35). Mutations in Ras are rare in glioma, however EGFR, PDGFR, and NF1 are frequently mutated and all of these pathways signal through Ras. Therefore, while Ras itself is not directly mutated, the pathway is frequently hyperactivated in glioma, and can be used as a surrogate for mutation of these key glioma-associated pathways.
Figure 5. Gliogenic chromatin architecture is present in mouse models of glioma.
(A–C) NFIA and Sox9, Brn2 expression is strongly correlated in human glioma. Spearman’s correlation coefficient plots are derived from human glioma RNA-Seq expression profiles from TCGA datasets representing a total of 403 high-grade glioma samples. Parallel analysis was performed on additional datasets that include low-grade glioma and showed a similar relationship (see Supplemental Figure S6). (D–E) Chromatin conformation capture (3C) assay performed on CRISPR/IUE generated tumors at P70 (D) and PB-Ras generated glioma tumors at P14 (E). In both cases, Red text denotes e123 as the anchor point from which long-range DNA interactions across the e123 – e161 interval in these tumors were measured.
Analysis of tumors from our mouse models revealed extensive co-expression of NFIA with Sox9 and Brn2, with nominal expression of Isl1 and Lhx3, indicating that these models reflect these features of human glioma (Supplemental Figure S8). Next, to determine whether NFIA is similarly regulated in glioma we interrogated the three-dimensional conformation of the e123/TSS/e161 interval within the NFIA locus in both the CRISPR/IUE and PB-Ras/IUE glioma models using 3C. As indicated in Figures 5C–D, the e123 anchor demonstrates a strong association with regions in close proximity to the TSS/promoter (region 6) and those containing e161 (region 11), indicating that the NFIA locus in both glioma models demonstrates a three-dimensional conformation that parallels developmental gliogenesis (see Fig. 3B).
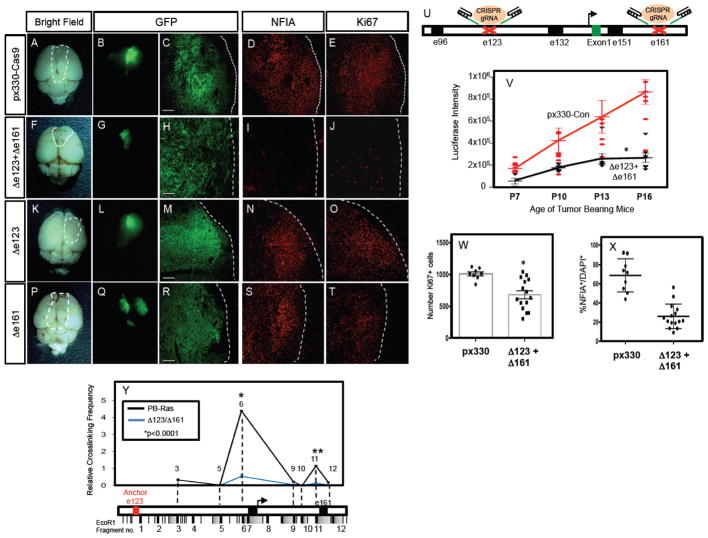
The forging observations suggest that NFIA expression in glioma is governed by the e123/Sox9 and e161/Brn2 regulatory axis and the long-range interactions between these enhancer elements. To directly test whether these interactions and the resulting three dimensional conformation of the NFIA locus is required for its expression in our glioma model, we turned to CRISPR-mediated gene editing to delete the e123 and e161 enhancers 36,37. Towards this, we generated guide RNAs that efficiently target regions surrounding the conserved Sox9 and Brn2 response sites in the mouse e123 and e161 enhancers (Fig. 6U; Supplemental Figure S9; see methods). To test the contributions of these enhancers, we used the PB-Ras/IUE model because tumors are produced 2–3 weeks after birth, making it a tractable model system in which to determine whether deletions of these enhancers influence NFIA expression and tumor growth. As indicated in Figure 6A–Y, introduction of the guide RNAs/CAS9 to both e123 and e161 resulted in decreased rate of gross tumor growth (Fig. 6V), cell proliferation (Fig. 6W), and loss of NFIA expression within the tumor (Fig. 6X) (also see Supplemental Figure S8); efficient deletion of the targeted e123 and e161 regions within the tumor was confirmed via deep sequencing (Supplemental Figure S9 and methods). These effects on NFIA expression and tumor growth are not secondary to reduced expression of Sox9 and Brn2 within the tumor, as their expression remained unchanged in the presence of e123 and e161 guide RNAs (Supplemental Figure S8). Finally, introduction of single e123 or e161 guide RNAs did not impact tumor growth or NFIA expression (Fig. 6K–T; Supplemental Figure S8).
Figure 6. Glial enhancers are required for NFIA expression and tumorigenesis.
(A–J) CRISPR-mediated deletion of the response regions within both e123 and e161 results in a loss of NFIA expression (G v. L), attenuated tumor growth (E v. J, Z), and reduced tumor proliferation (H v. M, AA). Single deletions of e123 (K–O) and e161 (P–T) are also shown for comparison. (U) Schematic of CRISPR-mediated gene editing of e123 and e161 within the NFIA locus. (V) Quantification of bioluminescence imaging of PB-Ras tumors containing CRISPR mediated deletions in e123/e161 across the P7-P16 tumor development timecourse. The deletion of both e123/e161 significantly reduces the rate of tumor growth across this timecourse. Values for each timepoint are derived from at least 6 mice per condition, two-way ANOVA *p<0.0001 and error bars are SEM. (W–X) Quantification of Ki-67 (W) and NFIA (X) in P14 PB-Ras tumors with e123/e161 deletions and controls. (Y) Chromatin conformation capture (3C) assay performed on PB-Ras generated glioma tumors that have been subjected to CRISPR-mediated gene editing of the response regions within e123 and e161. Red text denotes e123 as the anchor point from which long-range DNA interactions were measured. Three independent libraries were assayed per experiment.: In W–Y error bars indicate SD, and results of two-tailed t-tests are shown. In Y, *p=0.0001272 and **p=0.0001962. In Z * anova=0.0062, ttest=0.005. In AA *p=0.0000181. Scale bars are 100um.
That CRISPR-mediated deletion of response sites within e123 and e161 resulted in decreased NFIA expression and tumor growth, in the presence of normal levels of Sox9 and Brn2, suggests that the three-dimensional conformation of the e123/TSS/e161 interval within the NFIA locus is disrupted. To test this, we performed 3C on tumors generated in the presence of the e123 and e161 guide RNAs, finding that the long range interactions between the e123 anchor point and the TSS/promoter (region 6) were disrupted, while the interaction with e161 (region 11) was significantly reduced (Fig. 6Y). These data, in conjunction with our phenotypic analysis of glioma formation, indicate that the three dimensional conformation of the e123/TSS/e161 interval within the NFIA locus is essential for NFIA expression in glioma and tumorigenesis.
Discussion
Leveraging insight gained from the in vivo validation of enhancer elements during the gliogenic switch in the chick, we used a combination of genetic, biochemical, and 3C approaches to identify lineage-specific chromatin looping mechanisms that regulate expression of NFIA in the developing CNS. These studies delineate a comprehensive three-dimensional chromatin regulatory mechanism in the developing CNS and demonstrate that the chromatin architecture and associated regulatory mechanisms for the same gene can vary between cell lineages in vivo (Fig. 3).
Dissection of these lineage specific processes identified Brn2 and Isl1/Lhx3 as novel transcription factors for NFIA induction in glial precursors and MNs, respectively. Brn2 is a member of the POU-family of transcription factors and has been implicated in the migration of cortical neurons and iPS reprogramming to the neuronal lineage 22,38–40, however its role in glial lineage development has not been characterized. Our genetic and biochemical studies implicate Brn2 expression and its association with Sox9 as key events during the initiation of gliogenesis in the spinal cord. Indeed, cooperative gene regulation between Sox- and POU-family members has been observed in a wide range of tissues and model systems, suggesting that it is a more general transcriptional mechanism41–43. While expression and function of NFIA in glial progenitors has been well-studied, its role in motor neurons has remained undefined. Our studies reveal that key regulators of motor neuron development, Isl1/Lhx3 directly regulate NFIA expression, suggesting that NFIA may participate in the well-described transcriptional networks regulating motor neuron development and physiology44. While motor neurons do not demonstrate any overt developmental defects in the absence of NFIA (not shown), additional genetic, biochemical, physiological analysis in these populations will be required to explore this in more detail.
Our genetic studies indicate that selective NFIA expression in glial precursors and MNs is regulated by the expression of these cell fate determinants in the associated populations, Sox9/Brn2 regulate NFIA expression in the VZ, while Isl1/Lhx3 regulate NFIA expression in MNs. Further examination of this phenomenon revealed that these factors and associated enhancers form specific chromatin configurations across the NFIA genomic locus. Moreover, we found that the gliogenic chromatin conformation forms prior to NFIA induction and its formation does not require Sox9/Brn2. These “pre-formed” loops are thought to create a permissive configuration that facilitates transcription factor associations, enabling timely gene induction during dynamic developmental states. In this case, the pre-formed gliogenic loop serves to facilitate the association between Sox9 and Brn2, which drives gliogenic expression of NFIA. Given that Sox- and Pou- family members associate and cooperate to drive gene expression across numerous systems, it’s possible that pre-formed chromatin loops are a conserved mechanism that engenders cooperation between these transcription factor families in other tissues.
Additional studies on the timing of NFIA induction revealed that Sox9/Brn2 do not ChIP with the e123/e161 regulatory elements at E10.5, further reinforcing our observation that the gliogenic loop is pre-formed. These data lead to a model in which the gliogenic loop forms, but cannot activate gene transcription until timely association of Sox9/Brn2 with the e123/e161 elements after E10.5. Association of Sox9/Brn2 with their enhancers is likely mediated by changes in chromatin state, as both e123 and e161 demonstrate a drastic reduction in silent/poised chromatin marks across the E10.5 – E12.5 interval that are correlated with Sox9/Brn2 binding (Fig. 4E–H). These data suggest that additional epigenetic regulators are likely to be involved in the induction of NFIA. Moreover, that both e123 and e161 demonstrate coordinated changes in epigenetic states, further reinforces the utility of the pre-formed chromatin loop at E10.5 as a key facilitator of these inductive events across long distances.
In addition to epigenetic mechanisms, these observations also suggest specific interactions between Sox9/Brn2 and the transcriptional mechanisms that oversee chromatin looping. Recent studies using 3C in ES cells identified Med12 as a key regulator of chromatin looping that operates at the core promoter and functions to maintain ES cells in an undifferentiated state8. Interestingly, in other tissues and model systems, Sox9 associates with Med12 and functions as its co-activator 45–47. Moreover, Sox10 has been shown to also interact with Med12 and facilitate the recruitment of the mediator complex in Schwann cells 48, suggesting that Sox-proteins may serve as transcriptional interfaces with factors regulating chromatin architecture. It will be important to further dissect how these factors selectively interface with established components of the chromatin looping machinery (i.e. CTCF, Mediator, Cohesion)8.
Using these developmental processes to gain insight into malignant glioma, we found that the NFIA locus in glioma demonstrates a similar chromatin conformation as in glial precursors (Fig. 4). Strikingly, CRISPR-mediated deletion of both the e123 and e161 enhancers resulted in decreased tumor growth and NFIA expression, and disrupted chromatin conformation. That tumor growth is attenuated when NFIA expression is reduced in the absence of both e123/e161 is consistent with other studies demonstrating that it plays a multiple key role in glioma proliferation and tumorigenesis27,28,49. In mouse glioma stem cell and human cell line xenograft models, NFIA regulates cell proliferation via repression of p21, whereas in an IUE-based model of oligodendroglioma, NFIA influences the generation of glioma “fates” or sub-types29,30,52. Consistent with a role for NFIA in glioma tumor cell proliferation, we found a significant decrease in cell proliferation when both e123 and e161 were deleted (Fig. 6V–W). Importantly, while our findings further implicate NFIA in glioma tumorigenesis additional genetic studies are necessary to definitely prove that NFIA itself is required for glioma tumorigenesis, as our experiments indirectly manipulate NFIA expression via its associated enhancer elements.
Nevertheless, these results do provide direct evidence that the e123/e161 enhancers are essential for NFIA expression and more broadly, implicate the architecture of the NFIA locus as a key regulator of gliogenesis and glioma tumorigenesis. That CRISPR-targeted deletion of 123/e161 did not result in a complete elimination of NFIA expression is likely due to incomplete deletion of the enhancers (Figures 6X and S9). Other possibilities include infiltrating non-tumor derived reactive astrocytes that express NFIA and the presence of other, undefined long-range enhancers that oversee its expression. Interestingly, deletion of individual e123 or e161 enhancers did not affect NFIA expression, suggesting that Sox9 and Brn2 are able to associate in the presence of just one enhancer and this association is sufficient for the synergistic activities of this complex to drive NFIA expression. Alternatively, in the context of tumorigenesis, individual expression of Sox9 or Brn2 may be sufficient to drive NFIA expression.
Despite these possibilities, our chromatin looping studies strongly suggest a cooperative mechanism of action between Sox9 and Brn2 in glioma. Moreover, given our previous findings that Sox9 and NFIA cooperate18, its possible that NFIA itself is a part of this transcriptional regulatory node for glioma tumorigenesis. Along these same lines, previous studies found that Brn2, Sox2, Olig2, and Sall2 comprise a key transcriptional node in glioma stem cell systems50. Given that NFIA and Sox9 interact and modulate Olig2 function28, it’s likely that they are also a part of this broader developmental transcriptional regulatory node operating in glioma stem cells. Alternatively, given that Sox9 and NFIA both antagonize Olig2 function, it may be that they function in glioma cell lineages downstream of stem cells to promote differentiation into pathological glia, which importantly, comprise the vast majority of the bulk tumor. Indeed our recent studies have identified several astrocyte-like subpopulations of cells within mouse and human glioma31, and it may be that the NFIA, Sox9, Brn2 node functions to promote the differentiation of these populations during tumorigenesis. Future studies will be aimed at understanding the role of Sox9, NFIA, and Brn2 in glioma stem cells, how these new transcriptional nodes influence the production of diverse glioma cell populations, and the nature of their relationship with the glioma stem cell transcriptional node.
In many respects, tumorigenesis is in part of recapitulation of development. Our study has identified a transcriptional mechanism that oversees the induction of a key glial fate determinant, NFIA. Critically, this mechanism is reutilized during glioma tumorigenesis and plays a critical role in tumor formation. The broad implications from these findings are that along with raw expression profiles and epigenetic characteristics, the antecedent 3D-chromatin architectures of key regulatory factors are also conserved across developmental lineages implicated in malignancy.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (NS071153 to BD and K01CA190235 and 5-T32HL092332-08 to SG), Cancer Prevention Research Institute of Texas (RP510334 and RP160192 to BD and CC), and Sontag Foundation (BD).
Footnotes
On-Line Methods and Supplemental Information
See Attached Documents; Please see Life Sciences Reporting Summary for statistical information. The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.Li G, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phillips-Cremins JE, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palstra RJ, Grosveld F. Transcription factor binding at enhancers: shaping a genomic regulatory landscape in flux. Front Genet. 2012;3:195. doi: 10.3389/fgene.2012.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 5.de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 2012;26:11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donohoe ME, Silva SS, Pinter SF, Xu N, Lee JT. The pluripotency factor Oct4 interacts with Ctcf and also controls X-chromosome pairing and counting. Nature. 2009;460:128–132. doi: 10.1038/nature08098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim YJ, Cecchini KR, Kim TH. Conserved, developmentally regulated mechanism couples chromosomal looping and heterochromatin barrier activity at the homeobox gene A locus. Proc Natl Acad Sci U S A. 2011;108:7391–7396. doi: 10.1073/pnas.1018279108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Handoko L, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pombo A, Dillon N. Three-dimensional genome architecture: players and mechanisms. Nat Rev Mol Cell Biol. 2015;16:245–257. doi: 10.1038/nrm3965. [DOI] [PubMed] [Google Scholar]
- 11.Buecker C, Wysocka J. Enhancers as information integration hubs in development: lessons from genomics. Trends Genet. 2012;28:276–284. doi: 10.1016/j.tig.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 13.Tanabe Y, Jessell TM. Diversity and pattern in the developing spinal cord. Science. 1996;274:1115–1123. doi: 10.1126/science.274.5290.1115. [DOI] [PubMed] [Google Scholar]
- 14.Rowitch DH, Kriegstein AR. Developmental genetics of vertebrate glial-cell specification. Nature. 2010;468:214–222. doi: 10.1038/nature09611. [DOI] [PubMed] [Google Scholar]
- 15.Morrison SJ, Shah NM, Anderson DJ. Regulatory mechanisms in stem cell biology. Cell. 1997;88:287–298. doi: 10.1016/s0092-8674(00)81867-x. [DOI] [PubMed] [Google Scholar]
- 16.Morrison SJ, et al. Transient Notch activation initiates an irreversible switch from neurogenesis to gliogenesis by neural crest stem cells. Cell. 2000;101:499–510. doi: 10.1016/s0092-8674(00)80860-0. [DOI] [PubMed] [Google Scholar]
- 17.Deneen B, et al. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron. 2006;52:953–968. doi: 10.1016/j.neuron.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 18.Kang P, et al. Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron. 2012;74:79–94. doi: 10.1016/j.neuron.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfaff SL, Mendelsohn M, Stewart CL, Edlund T, Jessell TM. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell. 1996;84:309–320. doi: 10.1016/s0092-8674(00)80985-x. [DOI] [PubMed] [Google Scholar]
- 20.Thaler JP, Lee SK, Jurata LW, Gill GN, Pfaff SL. LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein-protein interactions. Cell. 2002;110:237–249. doi: 10.1016/s0092-8674(02)00823-1. [DOI] [PubMed] [Google Scholar]
- 21.Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16:2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McEvilly RJ, de Diaz MO, Schonemann MD, Hooshmand F, Rosenfeld MG. Transcriptional regulation of cortical neuron migration by POU domain factors. Science. 2002;295:1528–1532. doi: 10.1126/science.1067132. [DOI] [PubMed] [Google Scholar]
- 23.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 24.Hagege H, et al. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat Protoc. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- 25.de Laat W, Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 2013;502:499–506. doi: 10.1038/nature12753. [DOI] [PubMed] [Google Scholar]
- 26.Bouwman BA, de Laat W. Getting the genome in shape: the formation of loops, domains and compartments. Genome Biol. 2015;16:154. doi: 10.1186/s13059-015-0730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glasgow SM, et al. The miR-223/Nuclear Factor I-A Axis Regulates Glial Precursor Proliferation and Tumorigenesis in the CNS. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:13560–13568. doi: 10.1523/JNEUROSCI.0321-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glasgow SM, et al. Mutual antagonism between Sox10 and NFIA regulates diversification of glial lineages and glioma subtypes. Nat Neurosci. 2014;17:1322–1329. doi: 10.1038/nn.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brennan CW, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ceccarelli M, et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell. 2016;164:550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin JCC, et al. Identification of diverse astrocyte populations and their malignant analogs. Nat Neurosci. 2017;20:396–405. doi: 10.1038/nn.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gundry MC, et al. Highly Efficient Genome Editing of Murine and Human Hematopoietic Progenitor Cells by CRISPR/Cas9. Cell Rep. 2016;17:1453–1461. doi: 10.1016/j.celrep.2016.09.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de la Iglesia N, et al. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev. 2008;22:449–462. doi: 10.1101/gad.1606508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xue W, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514:380–384. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen F, LoTurco J. A method for stable transgenesis of radial glia lineage in rat neocortex by piggyBac mediated transposition. Journal of neuroscience methods. 2012;207:172–180. doi: 10.1016/j.jneumeth.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sugitani Y, et al. Brn-1 and Brn-2 share crucial roles in the production and positioning of mouse neocortical neurons. Genes Dev. 2002;16:1760–1765. doi: 10.1101/gad.978002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vierbuchen T, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pang ZP, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wegner M. Secrets to a healthy Sox life: lessons for melanocytes. Pigment Cell Res. 2005;18:74–85. doi: 10.1111/j.1600-0749.2005.00218.x. [DOI] [PubMed] [Google Scholar]
- 42.Ma Y, et al. Functional interactions between Drosophila bHLH/PAS, Sox, and POU transcription factors regulate CNS midline expression of the slit gene. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2000;20:4596–4605. doi: 10.1523/JNEUROSCI.20-12-04596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dailey L, Basilico C. Coevolution of HMG domains and homeodomains and the generation of transcriptional regulation by Sox/POU complexes. J Cell Physiol. 2001;186:315–328. doi: 10.1002/1097-4652(2001)9999:9999<000::AID-JCP1046>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 44.Davis-Dusenbery BN, Williams LA, Klim JR, Eggan K. How to make spinal motor neurons. Development. 2014;141:491–501. doi: 10.1242/dev.097410. [DOI] [PubMed] [Google Scholar]
- 45.Rau MJ, Fischer S, Neumann CJ. Zebrafish Trap230/Med12 is required as a coactivator for Sox9-dependent neural crest, cartilage and ear development. Dev Biol. 2006;296:83–93. doi: 10.1016/j.ydbio.2006.04.437. [DOI] [PubMed] [Google Scholar]
- 46.Wang X, Yang N, Uno E, Roeder RG, Guo S. A subunit of the mediator complex regulates vertebrate neuronal development. Proc Natl Acad Sci U S A. 2006;103:17284–17289. doi: 10.1073/pnas.0605414103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou R, et al. SOX9 interacts with a component of the human thyroid hormone receptor-associated protein complex. Nucleic Acids Res. 2002;30:3245–3252. doi: 10.1093/nar/gkf443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vogl MR, et al. Sox10 cooperates with the mediator subunit 12 during terminal differentiation of myelinating glia. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:6679–6690. doi: 10.1523/JNEUROSCI.5178-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song HR, et al. Nuclear factor IA is expressed in astrocytomas and is associated with improved survival. Neuro-oncology. 2010;12:122–132. doi: 10.1093/neuonc/nop044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suva ML. Genetics and epigenetics of gliomas. Swiss Med Wkly. 2014;144:w14018. doi: 10.4414/smw.2014.14018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.