Abstract
Mitochondrial genome integrity is fundamental to mammalian cell viability. Since mitochondrial DNA is constantly under attack from oxygen radicals released during ATP production, DNA repair is vital in removing oxidatively generated lesions in mitochondrial DNA, but the presence of a strong base excision repair system has not been demonstrated. Here, we addressed the presence of such a system in mammalian mitochondria involving the primary base lesion repair enzyme DNA polymerase (pol) β. Pol β was localized to mammalian mitochondria by electron microscopic-immunogold staining, immunofluorescence co-localization and biochemical experiments. Extracts from purified mitochondria exhibited base excision repair activity that was dependent on pol β. Mitochondria from pol β-deficient mouse fibroblasts had compromised DNA repair and showed elevated levels of superoxide radicals after hydrogen peroxide treatment. Mitochondria in pol β-deficient fibroblasts displayed altered morphology by electron microscopy. These results indicate that mammalian mitochondria contain an efficient base lesion repair system mediated in part by pol β and thus pol β plays a role in preserving mitochondrial genome stability.
Keywords: Mitochondria, DNA polymerase β, DNA polymerase γ, Base excision repair, Electron microscopy, Immunogold staining, Confocal immunofluorescence microscopy
1. Introduction
In higher eukaryotes, both nuclear and mitochondrial genomes encounter damage from a variety of physical and chemical agents, resulting in base loss and other DNA lesions. Failure to accurately repair such damage can lead to deleterious mutations and other forms of genomic instability. In an attempt to resolve DNA damage, cells employ multiple and overlapping DNA repair pathways that are instrumental in maintaining integrity of genomic DNA. Base excision repair (BER) is a highly-conserved pathway for DNA repair after single-base loss, singlestrand breaks and base damage, and these lesions arise from a wide variety of exogenous and endogenous sources including environmental genotoxicants, oxidizing agents, and other common exposures such as ionizing and UV irradiation [1–3].
Well-accepted sub-pathways for nuclear BER in mammalian cells include single strand break repair, single-nucleotide BER (SN BER) and long-patch BER (LP BER) that are differentiated by repair patch size and the enzymes involved [4,5]. For repair of base damage in both of the latter sub-pathways, the first step involves spontaneous base loss or recognition and removal of the base lesion by a DNA glycosylase that hydrolyzes the N-glycosylic bond linking the base to the sugar phosphate backbone [6,7]. The resulting apurinic/apyrimidinic (AP)-site is then incised 5′ to the sugar by AP endonuclease 1 (APE1), generating a 1-nt gapped repair intermediate with 3′-OH and 5′-deoxyribose phosphate (dRP) groups at the margins. Trimming of the 5′-dRP group generating a 5′-phosphate, and replacement of the damaged base are presumed to be mediated mainly by DNA polymerase β (pol β) in the nucleus and pol γ in mitochondria [8–11]. Finally, the repair intermediate is ligated by DNA ligases I and IIIα [12–14]. An alternate form of SN BER involves tailoring of the gap margins produced when oxidatively generated DNA base lesions are removed by bifunctional DNA glycosylases. Now polynucleotide kinase 3′-phosphatase (PNKP) and tyrosyl-DNA phosphodiesterase 1 (Tdp1) generate the 3′-OH and 5′-phosphate groups at the gap margins required for ligation [15–18].
The mammalian mitochondrial genome is a covalently closed circular DNA molecule (mtDNA) that is transmitted through the maternal lineage [19]. The mitochondrial genome encodes for tRNAs and rRNAs along with many protein components of the respiratory chain/oxidative phosphorylation system essential for normal mitochondrial function [20]. The cell contains hundreds of mitochondria, each with multiple copies of mtDNA. Replication of mtDNA is independent of cell cycle stage and occurs in dividing and non-dividing cells. Mitochondrial DNA has a higher mutation rate than nuclear DNA, and the human population exhibits a high level of mtDNA polymorphisms [21,22]. Many inherited diseases result from germline variations in the mitochondrial genome or in nuclear genes that encode for mitochondrial components [23,24]. In addition, somatic mutations in mtDNA are linked to common diseases, including age-related neurodegenerative disorders and cancers [25–28].
Mitochondria are known as the “energy powerhouse” of the cell generating ATP through oxidative phosphorylation via the electron transport chain (ETC). The ETC generates energy by passing electrons between its components creating an electrochemical gradient. The process produces free radicals as a natural byproduct during ATP production, and these radicals can cause DNA damage, including oxidized bases, AP-sites, and single- and double-strand breaks. Thus, the natural production of free radicals in mitochondria results in damaged DNA requiring repair [29]. While repair mechanisms of nuclear oxidatively damaged DNA have been extensively characterized, repair processes for the mitochondrial genome are less well understood. Initial studies of mtDNA repair focused on UV light-induced DNA damage and suggested mitochondria lack a functional DNA repair mechanism [30]. However, repair mechanisms thought to occur exclusively in the nucleus have been reported in mammalian mitochondria [31–35].
Until recently the mitochondrial replicative polymerase, pol γ, was considered the only DNA polymerase in mammalian mitochondria and the enzyme that participates in DNA repair [36]. Pol γ has dRP lyase activity capable of trimming the 5′-margin of gapped DNA in the BER intermediate, and enabling ligation during SN BER [11]. Additionally, BER can be reconstituted in vitro using purified BER factors along with purified pol γ. The enzyme is able to contribute the steps of SN gap filling and gap trimming of the 5′-dRP group in a manner similar to pol β in nuclear BER [11]. However, 5′-dRP removal by pol γ performed under steady-state enzymatic conditions was> 20-fold slower than that of pol β [11]. Considering the proximity of mtDNA to free radicals, the presence of a robust BER system in mitochondria seems likely. Yet, the existence of an efficient pol β-like enzyme in mammalian mitochondria has been ambiguous. An enzyme with pol β size and biochemical properties was observed in bovine heart mitochondria [37]. However, Loeb and coworkers [38] addressed the question of pol β in mammalian mitochondria and concluded the enzyme is not present. Their samples of purified mitochondria contained pol β, but they concluded this was due to contamination by nuclear and/or cytoplasmic pol β. Interestingly, a pol β-like mitochondrial enzyme(s) has been extensively characterize in trypanosomes [39,40], and pol β-like enzymes are well known in prokaryotes, including the most ancient organisms [41].
In light of uncertainty regarding the presence of pol β in mammalian cell mitochondria, we addressed the question using several biochemical and biophysical approaches. Mitochondrial localization of pol β was confirmed. Toward exploring a functional role of mitochondrial pol β, mouse fibroblasts were treated with hydrogen peroxide (H2O2) and repair of mtDNA damage was assessed by a quantitative PCR-based assay. Mitochondrial BER was stronger in pol β-expressing cells than in pol β-deficient cells. Further features of mitochondrial metabolism as a function of pol β expression also are described.
2. Materials and methods
2.1. Purification of mitochondria from cultured cells
Mitochondria from freshly grown HEK 293, HeLa S3, and pol β+/+ and pol β−/− mouse embryonic fibroblast (MEF) cells [9] in log phase were purified using a differential centrifugation protocol as previously reported [42]. Briefly, cell pellets were resuspended in 9 × of the packed cell volume (PCV) in hypotonic buffer (20 mM HEPES/KOH, pH 7.6, 5 mM KCl, 0.5 mM MgCl2, 0.5 mM DTT, 0.1 mM AEBSF, 1 mM benzamidine, 1 μg/ml pepstatin A, and 1 μg/ml leupeptin) and allowed to swell for 10–30 min on ice. Cells were homogenized with 20–40 strokes using a tight-fitting Dounce homogenizer. Disruption of cells and release of nuclei (over 90%) were confirmed by microscopy. Mitochondria were stabilized by addition of 2/3-volume 2.5 × mannitol-sucrose buffer (MSB) to bring the mixture to a final concentration of 1 × MSB (210 mM mannitol, 70 mM sucrose, 5 mM Tris-HCl, pH 7.5, 5 mM EDTA). Nuclei and unbroken cells were removed by centrifugation twice at 750 ×g for 10 min at 4 °C. Mitochondria were collected from the supernatant fractions (referred as the cytosolic fraction) by centrifugation at 12,000 × g for 10 min. The mitochondrial pellet was washed twice in 10 vols of 1× MSB and then resuspended in 1 × MSB. Mitochondria were further enriched and purified by discontinuous 1 M/1.5 M sucrose gradient centrifugation in SW28 tubes (25 × 89 mm). Centrifugation was performed at 22,000 rpm in a Beckman SW28 rotor for 30 min at 4 °C. Mitochondria sedimenting at the 1 M/1.5 M sucrose interface were carefully collected with a Pasteur pipette. The sample was diluted with 2–3 vols of 1 × MSB, and mitochondria were collected by centrifugation at 10,000 rpm for 10 min at 4 °C. The mitochondrial pellet was washed twice by resuspension in 10 vols 1 × MSB and centrifugation at 10,000 rpm for 10 min at 4 °C. Finally, the washed mitochondria were resuspended in a small volume of 1 × MSB, and the protein concentration was determined by the Bio-Rad assay. This sample is referred to here as “purified mitochondria.” Freshly purified mitochondria were used immediately in biochemical assays and Percoll gradient centrifugation or were flash frozen and stored at −80 °C until use.
2.2. Mitochondrial extract preparation
Frozen purified mitochondria were resuspended in a 2.5 × volume of lysis buffer (20 mM Tris-HCl, pH 8.0, 10% glycerol, 14 mM 2-mercaptoethanol, 300 mM NaCl, 1% Triton X-100, 0.1 mM AEBSF, 1 mM benzamidine, 1 μg/ml pepstatin A, and 1 μg/ml leupeptin). After incubation on ice for 5 min, the suspension was centrifuged, the clarified supernatant fraction was removed and the protein concentration was determined by a Bio-Rad assay. Mitochondrial lysates were then used in biochemical assays and/or were flash frozen in small aliquots and stored at −80 °C until use.
2.3. Co-fractionation of pol β and pol γ during Percoll centrifugation
Purified mitochondria prepared as described above were subjected to Percoll gradient centrifugation, and the gradient was evaluated for migration of pol β and pol γ (as a marker for mitochondria). Purified mitochondria were suspended in 1 ml 1 × MSB containing 10% Percoll and layered onto 10 ml of 35% Percoll/1 × MSB in an OptiSeal tube (16 × 67 mm). Centrifugation was performed in a Beckman NVT65 vertical rotor at 23,000 rpm for 1 h at 4 °C, and a Percoll gradient is formed in situ during the centrifugation. After centrifugation, fractions (~400 μl each) were collected from the bottom of the tube. Proteins in the indicated fractions were separated by SDS-PAGE and analyzed for the presence of pol β and pol γ by immunoblotting as described below.
In an additional experiment described in the text, fractions containing pol β and pol γ were pooled and subjected to a second co-fractionation at a higher Percoll concentration. Experiments were similar to those described above except the Percoll concentration was 37%; this provided to have significantly different centrifugation properties than 35% Percoll.
2.4. Isolation and mass spectrometry analysis of the pol β affinity-capture fraction from purified mitochondrial extracts using anti-FLAG M2 affinity gel
Purified mitochondrial extracts were prepared from pol β−/− cells expressing FLAG-tagged pol β or pol β−/− cells (control), as above. The protein concentration of this extract was determined using the Bio-Rad protein assay, with bovine serum albumin as standard. For immunoprecipitation of FLAG-tagged pol β and its binding partners, 1 ml of each extract (~6 mg) of cell extract was diluted 1:1 with TBS (Tris-HCl, pH 7.5, 150 mM NaCl) without NaCl and then mixed with ~50 μl of EZview Red anti-FLAG M2 affinity gel (Sigma Aldrich), which was washed and pre-equilibrated with TBS according to the manufacturer’s instructions. The cell extract was mixed with anti-FLAG M2 affinity gel suspension and incubated with a gentle rotation for 2 h at 4 °C. After this incubation, the resin was centrifuged at 3000 rpm for 30 s at 4 °C. Supernatant fractions were carefully removed with a narrow-end pipette tip. The resin was washed 5 times with TBS, to remove non-specific proteins. Then, FLAG-tagged bound pol β and its interacting proteins were eluted with SDS-PAGE sample buffer and separated by 4–12% Nu-PAGE Bis-Tris gel. Subsequently, the gel was stained with Simply Blue SafeStain (Invitrogen) and analyzed by mass spectrometry, as described previously [18]. Mass spectrometry data were acquired and searched essentially as described previously [43]. The list in Table S1 was generated by including proteins that were identified with a Distinct Summed MSMS Score of 20 or higher, 4 or more spectra corresponding to 2 or more peptides, and having at least 5-fold more spectra in the pol β sample than control sample (pol β−/− MEF mitochondrial extract).
2.5. Trypsin digestion of mitochondria
Freshly purified mitochondria were subjected to trypsin digestion, since freeze/thawing compromised membrane integrity. Mitochondria (120 μg) were suspended in 100 μl 1 × MSB. An aliquot (20 μl) was withdrawn as the untreated sample and trypsin (Promega) was added to the remaining sample to a final concentration of 2 μg/ml. The reaction mixture was incubated at 30 °C. Equal aliquots were removed for analysis at the indicated times. Proteolysis was stopped by the addition of an equal volume of 20% trichloroacetic acid (TCA) with incubation on ice for 30 min. TCA precipitates were collected by centrifugation and resuspended in 10 μl of 1 M Tris-HCl, pH 7.5. SDS-PAGE buffer was added to each sample, and the sample was boiled for 5 min. Proteins were separated by Nu-PAGE 4–12% Bis-Tris mini-gel electrophoresis. Then, proteins were electro-transferred onto a nitrocellulose membrane, and the membrane was probed for pol β, pol γ, or VDAC1, as indicated in the figure legends. In some experiments purified mitochondria were supplemented with 10 ng purified pol β.
2.6. DNase I treatment of mitochondria
Purified mitochondria (240 μg) were suspended in 200 μl 1 × MSB containing 10 mM MgCl2. Equal volumes of the mitochondrial suspension (50 μl) were treated with DNase I (40, 100–200 μg/ml) or buffer alone and incubated for 30 min at 37 °C. Reaction mixtures were then centrifuged at 14,000 rpm for 10 min. The supernatant fraction was saved, and the pellet fraction was washed once and resuspended in an equivalent volume of 1 × MSB (50 μl). Equal volumes (30 μl) of supernatant and pellet fractions were analyzed for pol β and pol γ by immunoblotting as described below.
2.7. Alkali extraction of intact purified mitochondria
An alkali extraction assay of intact purified mitochondria was performed as previously described [44]. Briefly, purified mitochondria (37.5 μg) suspended in 1 × MSB were treated with 0.1 M Na2CO3 (pH 11.5) for 30 min on ice. Then, the mixture was centrifuged at 14,000 rpm for 15 min. The supernatant fraction was saved and the pellet fraction was washed twice with 1 × MSB. Untreated mitochondria and the supernatant and pellet fractions were separated by Nu-PAGE 4–12% Bis-Tris mini-gel electrophoresis, and proteins were transferred onto a nitrocellulose membrane. The membrane was probed for pol β, pol γ, SMAC, or VDAC1 using a specific antibody to each protein.
2.8. Immunoblotting
For immunoblotting analysis, proteins in mitochondrial samples were separated by SDS-PAGE using Nu-PAGE 4–12% Bis-Tris mini-gel electrophoresis and transferred onto nitrocellulose membranes. The membranes were incubated in 5% nonfat dry milk in Tris-buffered saline containing 0.1% (v/v) Tween 20 (TBS-T) and then probed with either monoclonal (18S) or affinity purified polyclonal antibody to pol β, monoclonal antibody to pol γ (Abcam), SMAC mouse monoclonal antibody (Cell Signaling), or VDAC1 antibody (Abcam), as indicted in the figure legends. Goat anti-rabbit or goat anti-mouse IgG conjugated to horseradish peroxidase (1:10,000 dilution) was used as secondary antibody, and the immobilized horseradish peroxidase activity was detected by enhanced chemiluminescence (ECL). These membranes were stripped by incubation in Restore Western Blot Stripping Buffer (Thermo Scientific) for 30 min at room temperature, followed by three washes with TBS-T. Then, the membranes were used for probing with another antibody, as above. For analysis of Percoll gradient fractions, an equal volume (100 μl) of each fraction was precipitated with 10% TCA on ice for 30 min, and the precipitates were collected by centrifugation. Precipitates were then suspended in 10 μl of 1 M Tris-HCl, pH 7.5, and 20 μl SDS-PAGE sample buffer.
2.9. Preparation of samples for electron microscopy and immunogold labeling
Pol β+/+ and pol β−/− MEF cells were fixed for 1 h at room temperature (RT) in freshly prepared 2.5% glutaraldehyde (Electron Microscopy Sciences, Hatfield, PA) and post-fixed with osmium tetroxide 1% (Electron Microscopy Sciences). Cells were then dehydrated in a graded series of ethanol solutions before embedding in EPON-812 Resin (Electron Microscopy Sciences) and polymerization for 48 h at 60 °C. A Leica EM UC-7 Ultramicrotome equipped with a diamond knife was used to cut ultra-thin sections of 50–60 μm which were collected on Nickel (Ni) grids. For immunostaining, thin sections were pretreated with 4% aqueous sodium metaperiodate and 1% aqueous periodic acid before overnight incubation at RT with rabbit anti-pol β primary antibody (1:50 dilution, ab26343, Abcam). After extensive washings, the samples were incubated with the secondary antibody, goat anti-rabbit IgG conjugated to 10-nm colloidal gold particles (1:20 dilution, ab27234, Abcam) for 1 h at RT and counterstained with 2% uranyl acetate and lead citrate.
For quantification, at least 100 images for each study group were captured using a Gatan Orius real time CCD camera (Pleasanton, CA) attached to an FEI Tecnai T12 TEM/STEM instrument (Hillsboro, OR) operated at 80 kV. EM subcellular analysis and immunogold particle localization were made using Gatan Digital Micrograph software.
Quantitative immunogold analysis was performed by the application of an overlay screen and by manually scoring the gold particles in the different subcellular compartments. The results were normalized to the nuclei, considered as 100%. Immunogold particle quantification in mitochondria was carried out by manually counting the gold particles in the outer-inner mitochondrial membrane as well as in the matrix of at least 40 mitochondria randomly chosen. Statistical analysis was performed using the non-parametric Mann-Whitney U test and data are presented as mean values. P-values<0.05 were considered statistically significant.
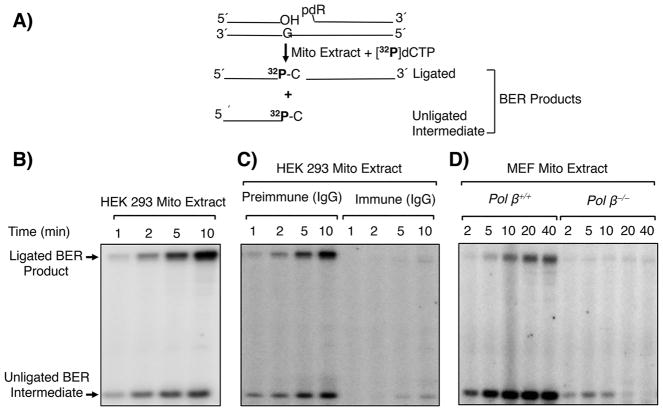
2.10. In vitro SN BER assay with mitochondrial extract
The BER assay was performed in a final reaction mixture volume of 20 μl, as described previously [18]. BER reaction mixtures contained 50 mM HEPES, pH 7.5, 0.5 mM EDTA, 2 mM DTT, 20 mM KCl, 5 mM MgCl2, 4 mM ATP, 5 mM phosphocreatine, 100 μg/ml phosphocreatine kinase, 0.5 mM NAD, 2.3 μM [α-32P]dCTP (specific activity, 1× 106 dpm/pmol), and a 35-base-pair oligonucleotide duplex (250 nM) pretreated with UDG and APE1, unless indicated otherwise. The repair reactions were then initiated by addition of 20 μg extract from purified mitochondria and incubated at 37 °C. In some experiments, the mitochondrial extract (20 μg) was pretreated with IgG from preimmune serum or immune serum specific to pol β for 30 min on ice prior to addition to the BER reaction mixture, as previously described [45]. Aliquots (4.5 μl) were removed for analysis at the indicated times. The reaction was terminated by addition of an equal volume of DNA gel loading buffer (95% formamide, 20 mM EDTA, 0.02% bromophenol blue, and 0.02% xylene cyanol). After incubation at 75 °C for 2 min, the reaction products were separated by electrophoresis in a 16% polyacrylamide gel containing 8 M urea in 89 mM Tris-HCl, pH 8.8, 89 mM boric acid, and 2 mM EDTA. A Typhoon PhosphorImager was used for gel scanning and imaging, and the data were analyzed by ImageQuant software.
2.11. Immunofluorescence and confocal microscopy
Wild-type MEF cells (from Dr. Shigemi Matsuyama, Cleveland, OH) were grown at 37 °C in a 10% CO2 incubator in Dulbecco’s modified Eagle’s medium (DMEM; Hyclone, Logan, UT) supplemented with glutamine, 10% fetal bovine serum (FBS), 1% non-essential amino acids, and 1% sodium pyruvate [46,47]. For pol β knock down, a lentiviral plasmid construct, pol β shRNA-468 (TRCN0000348053) was obtained from Sigma-Aldrich to obtain stable depletion of mouse pol β mRNA, as previously described [48]. Wild-type MEF cells were transduced with lentiviral particles at multiplicity of infection (MOI) of 5 per 50,000 cells, and stable cell lines were recovered after puromycin selection (6 μg/ml, Life Technologies). Single cell clones were isolated and characterized, and the cell line with strongest pol β knock down (K/D) was confirmed by western blotting (Fig. S6).
The protocol described below does not involve the SDS permeabilization step required for nuclear staining by the anti-pol β antibody [49,50]. Thus, the protocol enabled localization analysis of cytoplasmic pol β, but not nuclear pol β. Use of SDS was found to result in degradation of MitoTracker staining. Wild-type and pol β K/D MEF cells were seeded in 35-mm glass-bottomed petri dishes (MatTek Corporation) at 2× 105 cells/dish and incubated in growth medium for 24 h. Then, medium was removed and 100 nM MitoTracker Deep Red FM (Life Technologies) in phosphate buffered saline (PBS; Hyclone) was added for mitochondrial staining. The cells were returned to the incubator for 30 min at 37 °C and 5% CO2. The mitochondrial stain was then removed, and cells fixed in acetone at −20 °C for 5 min. After fixation, cells were washed three times with PBS and then permeabilized by adding 0.25% Triton X-100 for 10 min. After washing three times with PBS, the cells were blocked in PBS plus 1% BSA (blocking buffer) for 30 min. Cells were then incubated with anti-pol β antibody (1:200 dilution, ab26343, Abcam) in blocking buffer for 1 h, washed three times with PBS and incubated in Alexa 546 conjugated anti-rabbit antibody (1ː2000; Life Technologies) in blocking buffer for 1 h. Stained cells were mounted with ProLong Gold anti-fade with DAPI (Life Technologies) and cured overnight in the dark at room temperature. Fluorescence images were acquired with a Nikon A1rsi laser scanning confocal microscope with a Plan Apo λ 100X/NA 1.45 oil immersion objective. Single plane images only were used to create Figs. 4 and 5.
Fig. 4.
Localizing pol β in mitochondria by immunofluorescence imaging. Representative immunofluorescence images of co-localization of pol β and the mitochondrial marker MitoTracker in wild-type MEF cells and after pol β siRNA knockdown (K/D). Pol β (red), mitochondria (green) and co-localized mitochondria and pol β (yellow) are seen in merged images of wild-type cells (middle panels). The boxed region in the merged image along with the corresponding regions in the pol β (red) and mitochondrial marker (green) stained images were magnified to illustrate examples for precise co-localization of pol β and the mitochondrial marker (bottom panels). Similar co-localization was not observed in the pol β K/D cells (top panels). Nuclei were stained with DAPI (blue). Scale bars: 10 μm for the top and middle panels; 1 μm for the bottom panels.
Fig. 5.
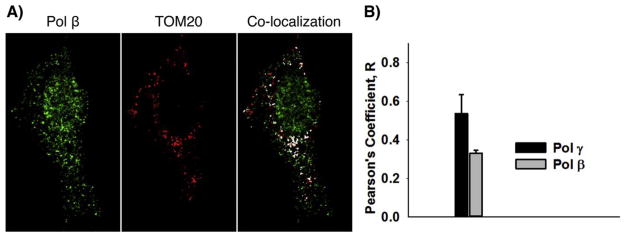
Immunofluorescence confocal microscopy of pol β in mitochondria. A) Mitochondria in MEF cells expressing FLAG-tagged pol β were stained with anti-TOM20 (red) and pol β was stained with anti-FLAG (green). Pol γ was stained with anti-pol γ antibody (not shown) as a control. Co-localization is illustrated in white pixels. B) Co-localization data were subjected to Pearson’s Coefficient (R) statistical analysis; for pol β: R = 0.330 ± 0.016 9 (n = 12); and for pol γ: R = 0.536 ± −0.0.098 (n = 29). Scale bar: 10 μm.
2.12. Modified method for immunofluorescence
In these experiments, SDS permeabilization was not required to obtain nuclear staining by anti-FLAG conjugated antibody. Pol β−/− cells expressing FLAG-tagged pol β [9] were seeded in 35-mm glassbottomed petri dishes (MatTek Corporation) at 2 ×105 cells/dish and incubated in growth medium for 24 h at 37 °C and 10% CO2. The cells were fixed with 100% methanol for 5 min for FLAG (pol β staining) or in 4% paraformaldehyde for 10 min for pol γ staining. Cells were then washed three times with PBS and permeabilized by adding 0.25% Triton X-100 for 5 min. After again washing three times with PBS, the cells were blocked with 1% BSA/10% normal goat serum/0.3 M glycine in 0.1% PBS-Tween for 1 h. The cells were then incubated with anti-TOM20-Alexa Fluor 647 conjugated antibody (1:4000 dilution, Abcam ab209606) and either anti-FLAG M2-FITC conjugated antibody (1:100 dilution, Sigma F4049) or anti-DNA pol γ-Alexa Fluor 488 conjugate (1:200 dilution, Bioss bs-13017R-A488) in blocking buffer for 1 h. Stained cells were mounted with ProLong Gold anti-fade with DAPI (Life Technologies) and cured overnight in the dark at room temperature.
Fluorescence images were acquired with 512× 512 pixels, bidirectional mode averaging of 4 lines, zoom 2.0 and pixel dwell time 6.3 μs with an EC Plan-Neofluor 40×/1.30 Oil DIC M27 objective on a Zeiss LSM780 microscope controlled by Zen 2012 SP2 software. Excitation lasers’ power were kept low, under 4% of 25 mW for 488 nm laser and under 8% of 5 mW for 633 nm laser. To avoid bleed through between fluorescence emission channels and for accurate co-localization assessment, we carefully selected single labeled spots and tested the correct range of emission. Emission ranges of 490 nm–550 nm and 640 nm–700 nm were used for two channel detection. For crisp images, the GaAsP detector was set on the most sensitive digital photon counting mode.
To quantify the degree of co-localization between β-and γ-polymerases and TOM20-labeled mitochondria in confocal images, a commonly accepted method, Pearson’s Correlation Coefficient (degree of synchrony), was used. Pearson’s coefficients were recorded above thresholds. Fiji open source imaging software with coloc 2 plug-in was used for calculation [51]. Statistics were determined using SigmaPlot (SysStat Software Inc.). Co-localization indices were evaluated using only the cytoplasmic area of each cell.
2.13. Hydrogen peroxide (H2O2) treatment
The final conditions for the experiments described were chosen after a series of preliminary experiments. These involved surveys of H2O2 treatment conditions to induce DNA damage, measurement of cell viability after treatment, and measurement of the time required for full repair of mtDNA. Cells were seeded in a 12-well plate (Corning) at a density of 1.5–2×105 cells per well in DMEM with 10% FBS and incubated overnight in a 10% CO2 incubator at 37 °C. The next day, culture medium was removed and cells were washed once with DMEM before the treatment with 1.5 mM H2O2 (prepared in DMEM medium with serum). Cells were incubated with H2O2 at 37 °C for 30 min. After washing with PBS, cells were either harvested immediately or allowed to recover in the complete culture medium for up to 2 h under usual growth conditions. An alternate protocol for H2O2 treatment conditions was evaluated also. The cells were washed once with DMEM before the treatment with 140 μM H2O2 (prepared in DMEM medium without serum). Cells were incubated with H2O2 at 37 °C for 30 min and processed as above.
2.14. Mitochondrial DNA damage and repair analyses using quantitative PCR
Genomic and mtDNA were simultaneously isolated using a DNeasy kit (Qiagen) and quantified with a Quant-iT Picogreen dsDNA kit (Invitrogen) according to the protocols supplied. The mtDNA damage and repair was assayed using the long-range quantitative polymerase chain reaction (QPCR) method as described previously with minor modification [52]. In brief, 15 ng of total DNA was the starting material for large fragment (10.1 kb) amplification in 50 μl total reaction volume, using published primer pairs (sense primer 5′-GCCAGCCTGACCCATAGCCATAATAT-3′ and antisense primer 5′-GAGAGATTTTATGGGTGTAATGCGG-3′). With an Elongase Enzyme Mix (Invitrogen), PCR amplification was performed at 94 °C for 30 s followed by 20 cycles at 94 °C for 30 s, 60 °C for 30 s, 68 °C for 10 min and a final elongation for 10 min. To normalize the input mtDNA template in each sample, a short fragment (117 bp) of mtDNA was amplified using the following primer pairs: sense primer 5′-CCCAGCTACTACCATCATTCAAGT-3′ and anti-sense primer 5′-GATGGTTTGGGAGATTGGTTGATG-3′. The PCR reaction condition was as follows: 94 °C for 3 min followed by 23 cycles at 94 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s and a final elongation for 5 min. All of the reactions were conducted in triplicate and two independent experiments were performed. The PCR products were quantified using Quant-iT Picogreen dsDNA assay kit (Invitrogen) and analyzed according to the previously reported method [52]. The same PCR samples also were analyzed by agarose gel electrophoresis, to verify the results obtained by Quant-iT Picogreen dsDNA assay kit. The relative QPCR product of the 10 kb mtDNA fragment was normalized to mtDNA copy number that was determined by PCR amplification of a 117 bp mtDNA fragment. Statistical analysis and graphing were performed using GraphPad Prism 7.0 software.
2.15. Mitochondrial superoxide analysis by flow cytometry
Cells (pol β+/+ and pol β−/−) were seeded in 100 mm dishes at a density of 1 ×106. The following day, cells were treated with 50 μM H2O2 for 1 h, then washed once and further incubated with control medium for 3 h. Cells were harvested by trypsinization, then centrifuged, and washed with PBS. The cell pellet was prepared for flow cytometry using the MitoSOX Red™ Superoxide Indicator for Live-Cell Imaging (Thermo Fisher Scientific). Cells were resuspended in 500 μl of PBS and 2.5 μl of a 1 mM MitoSOX Red stock was added to each tube (5 μM final). Cells were incubated at 37 °C for 30 min. To distinguish dead cells, 1 μl of Sytox Blue was added to each tube just before analysis. Samples were read on an LSR II flow cytometer (BD Biosciences) and analyzed using FACSDiva software (BD Biosciences).
3. Results
Pol β is critical for BER of nuclear DNA, whereas pol γ is considered the major DNA polymerase responsible for both mtDNA replication and repair. In exploratory experiments for this work, extracts from purified mitochondria exhibited 5′-dRP lyase activity that was dependent on pol β rather than pol γ, indicating pol β was either a mitochondrial component or contaminant in the purified mitochondrial sample. In light of the significance of this topic and conflicting reports of pol β presence in mitochondria, we further examined purified mitochondria from human and mouse cultured cell lines using a number of biochemical and physical approaches.
3.1. Biochemical evidence for pol β in mitochondria
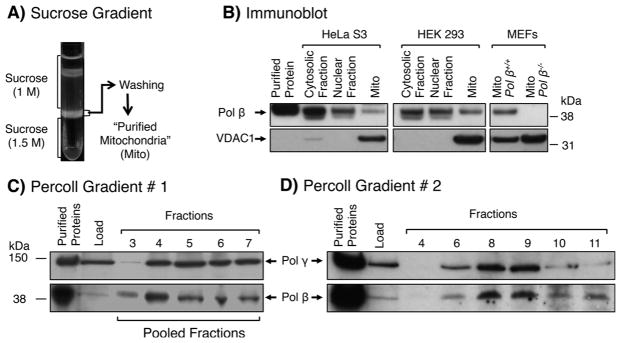
The presence of pol β was examined in sucrose gradient purified mitochondria from two human cell lines, i.e., HEK 293, HeLa S3, and also from MEF cells (Fig. 1A and B). Purified mitochondria from the pol β-expressing cell lines contained pol β, whereas mitochondria from pol β null MEF cells did not (Fig. 1B). The mitochondrial membrane protein, VDAC1 (voltage-dependent anion-selective channel 1), was used in these experiments as the mitochondrial marker. The purified mitochondrial samples from HEK 293 cells also were evaluated for cytoplasmic and nuclear contaminating proteins, including Importin β, β-actin, PCNA, and GAPDH; these proteins were not detected in the purified mitochondria (Fig. S1).
Fig. 1.
Purification of mitochondria from HeLa S3, HEK 293 and MEFs by discontinuous sucrose gradient centrifugation and co-fractionation of pol β and mitochondria in Percoll gradients. A) Mitochondria from cultured cells were prepared as described under “Materials and Methods.” Mitochondria were enriched and purified by centrifugation in a discontinuous sucrose gradient. The sample obtained at the 1.5 M sucrose layer was washed and is referred to as “purified mitochondria.” B) Immunoblotting of purified mitochondria (Mito) prepared from HeLa S3, HEK 293 and pol β+/+ and pol β−/− MEFs, along with cytosolic fraction (after removing nuclei) and nuclear fraction. Migration positions of protein markers, pol β and VDAC1, are indicated. C) Percoll gradient #1 (10–35%). After centrifugation, proteins in the indicated fractions were analyzed by immunoblotting for the presence of pol β and pol γ. D) Percoll gradient #2 (10–37%). Pooled fractions in gradient #1 containing pol β and pol γ (fractions 3–7) were subjected to a second co-fractionation using a higher concentration Percoll gradient. Proteins in the indicated fractions were analyzed as in (C). Fraction numbers, relative positions of protein markers, and pol β and pol γ positions are indicated.
The presence of pol β in purified mitochondria was confirmed using an alternate mitochondrial purification procedure (Fig. S3). This procedure makes use of magnetic microbeads conjugated to a TOM22 (mitochondrial translocase of the outer membrane 22) antibody and provides for rapid affinity isolation of mitochondria. Mitochondria purified by this method from HeLa S3 and HEK 293 cells were found to contain abundant pol β (Fig. S2).
We further analyzed purified mitochondrial samples to determine if pol β and a mitochondrial marker (pol γ) would co-fractionate during Percoll gradient centrifugation. After centrifugation, fractions were collected from the bottom of the tube, and immunoblotting analysis was performed to track pol β and pol γ. The results revealed pol β in fractions containing pol γ, corresponding to a visual band near the bottom of the gradient (Fig. 1C). To verify co-fractionation of pol β and mitochondria, pooled fractions containing these enzymes were analyzed using a second gradient centrifugation with a higher concentration of Percoll. The results revealed that pol γ and pol β co-sedimented in this gradient (Fig. 1D).
The presence of pol β in purified mitochondria was verified by mass spectrometry analysis. Purified mitochondria from MEF cells were subjected to an antibody affinity-capture procedure, along with purified mitochondria from pol β null cells as a negative control. Multiple pol β peptides were found in the mitochondrial sample, verifying that pol β was present in the mitochondrial extract prepared from pol β expressing cells (Table S1). A number of mitochondrial proteins also were found in the pol β affinity-capture fraction (Table S1). However, none of the BER factors found in the nuclear pol β affinity-capture studies were present [18].
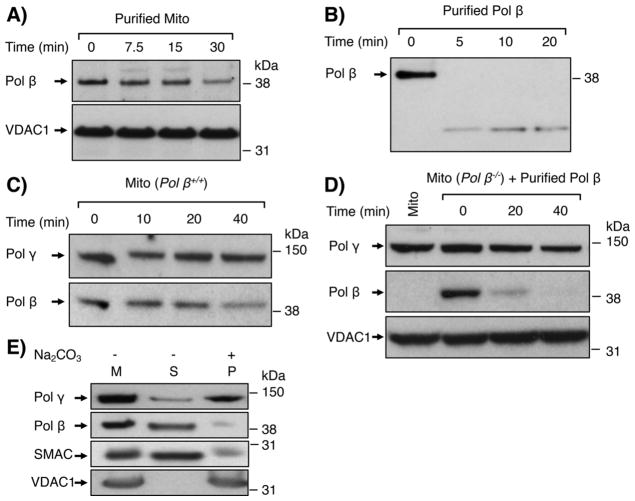
We further evaluated the possibility of nuclear or cytoplasmic pol β contamination of the purified mitochondrial samples. First, a “protease digestion” assay was applied to distinguish contaminating proteins from proteins that are internal to mitochondria. Contaminating proteins are susceptible to trypsin or proteinase K digestion, whereas proteins inside mitochondria are relatively resistant, depending on their sub-mitochondrial location. Trypsin digestion assays were conducted with purified mitochondria from HEK 293 cells (Fig. 2A) and with purified pol β as a control (Fig. 2B). The mitochondrial-associated pol β was partially resistant to digestion over a 30 min incubation, whereas purified pol β was completely digested within the first 5 min. Under identical reaction conditions, both outer membrane protein VDAC1 and matrix protein pol γ were completely resistant to trypsin digestion (Fig. 2A and C). Similar results were obtained with proteinase K digestion (not shown). Second, to rule out a concern that the trypsin resistance of mitochondrial-associated pol β could have been due to inhibition of protease activity in the purified mitochondria, rather than the internal location of pol β, further proteolysis experiments were conducted. Trypsin digestion was performed with purified mitochondria from pol β+/+ MEF cells (Fig. 2C), and pol β digestion was compared with that of purified pol β added to mitochondria from pol β−/− cells (Fig. 2D). The results revealed the endogenous mitochondrial-associated pol β was resistant to digestion for up to 20 min (Fig. 2C), whereas the exogenous pol β was almost completely digested within the 20 min of incubation (Fig. 2D). Thus, the observed trypsin resistance of mitochondrial-associated pol β did not appear to be due to protease inhibition by factors in the mitochondrial extract.
Fig. 2.
Trypsin digestion of mitochondrial-associated pol β. A) Purified mitochondria were treated with trypsin (2 μg/ml) at 30 °C for 7.5, 15 and 30 min. Immunoblotting was performed to probe for pol β and VDAC1. VDAC1 served as a mitochondrial outer membrane protein marker and a loading control. B) Purified pol β (10 ng) was treated with trypsin as in (A) for 5, 10 and 20 min and the immunoblot was processed for detection of pol β as in panel (A). C and D) Trypsin digestion was performed for the indicated times under similar reaction conditions as in panel (A), with either (C) mitochondria from pol β+/+ MEF cells or (D) mitochondria from pol β−/− MEF cells mixed with purified pol β (10 ng). Membranes were immunoblotted with antibodies against pol β, pol γ and VDAC1. Relative positions of protein markers (on the right) and of pol β, pol γ and VDAC1, are indicated. E) Sub-mitochondrial localization of pol β by Na2CO3 treatment of purified mitochondria. Purified mitochondria (37.5 μg) were treated with 0.1 M Na2CO3 (pH 11.5) for 30 min on ice, as described under “Material and Methods.” Untreated mitochondria (M), supernatant (S) and pellet (P) fractions were probed for pol β, pol γ, SMAC or VDAC1 proteins using specific antibodies. SMAC and VDAC1 served as an inner membrane space protein marker and outer membrane protein marker, respectively. Representative phosphorimages are at least of two repeats. Relative positions of protein markers (on the right) are indicated.
Since pol β could potentially bind to and be carried into the purified mitochondrial sample by contaminating nuclear DNA, we tested the effect of DNase I treatment on the presence of pol β. Purified mitochondria were incubated with increasing amounts of DNase I. After the incubation, each reaction mixture was separated into supernatant and mitochondrial pellet fractions by centrifugation, and these fractions were analyzed for pol β and pol γ by immunoblotting. (Fig. S3). The results revealed pol β, like pol γ, was not released into in the supernatant fraction by DNase I treatment, and there was no decrease in mitochondrial pellet pol β as a function of increasing concentrations of DNase I (Fig. S3). These results indicated the presence of pol β in the purified mitochondrial sample was not due to binding to contaminating nuclear DNA.
Finally, to test the internal location of pol β in mitochondria relative to that of pol γ, we used an “alkali extraction” assay [44]. Purified mitochondria were incubated with sodium carbonate (Na2CO3) and then subjected to centrifugation (Fig. 2E). As expected, the membrane protein VDAC1 was recovered in the pellet fraction, whereas the intermembrane space marker protein SMAC (second mitochondria-derived activator of caspases) was recovered in the supernatant fraction [31,33]. More pol β was recovered in the supernatant fraction as compared to the pellet fraction. As expected, pol γ was recovered in the pellet fraction (Fig. 2E). These results illustrate that pol β and pol γ do not have identical alkali extraction properties.
3.2. Morphological evidence for pol β in mitochondria
i) Electron Microscopic Analysis of Immunogold Staining of Pol β
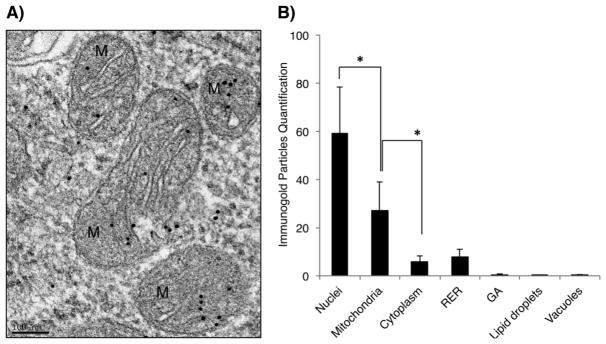
Localization of pol β to mitochondria was confirmed by immunogold EM analysis using pol β+/+ MEF cells. Cells were collected, prepared for EPON resin embedding, and after resin polymerization, multiple ultrathin sections were taken and labeled with an antibody against pol β (Fig. 3A). Although the protein was mainly distributed in the nuclei of these cells (Figs. 3A, B and S4), as expected from earlier work (Kirby et al., 2016), a considerable portion was observed in mitochondria (Fig. 3B). To verify the immuno-labeling specificity, embedded ultrathin sections were incubated with the secondary antibody only, anti-rabbit IgG conjugated to 10-nm colloidal gold particles (Fig. S5); appreciable gold particle signal was not detected.
Fig. 3.
Immunogold detection and quantification of pol β in mitochondria. A) EM representative micrograph of mitochondria in pol β+/+ MEFs cells. High magnification of pol β+/+ MEF cells revealed specific immunogold labeling of pol β in the mitochondria. Black dots represent immunogold particle. Scale bar: 100 nm. B) Subcellular quantification in pol β+/+ MEFs cells. *P- value ≤0.0001. RER: Rough Endoplasmic Reticulum; GA: Golgi Apparatus.
Immunofluorescence
Finally, confocal immunofluorescence microscopy images of MEF cells were analyzed after co-staining with an antibody against pol β (red) and the mitochondria-specific MitoTracker dye (green) (Fig. 4). Isogenic cell lines were used that were either wild-type for pol β expression or siRNA knockdown for pol β (Fig. S6). Immunofluorescence staining revealed pol β co-localization with the mitochondrial marker (Fig. 4, middle panels). Pol β staining was not observed in the pol β siRNA knockdown cells, as expected (Fig. 4, top panels).
During these initial immunostaining experiments, a concern arose that staining with MitoTracker dye was not entirely mitochondrial specific, and additionally we wished to obtain results with an alternate method of assessing pol β co-localization with a mitochondrial marker. A FLAG-tagged pol β expressing MEF cell line and anti-FLAG antibody were selected for staining pol β (Fig. 5), and an anti-TOM20 antibody (staining mitochondrial outer membrane), was selected for mitochondria. Pol γ mitochondrial localization also was established in these experiments as a reference and to validate the presence of pol β in mitochondria. We found that similar results were obtained for mitochondrial co-localization of pol β as observed in Fig. 4. Co-localization was verified using Pearson’s Coefficient analysis (Fig. 5B). The Pearson’s Coefficient value for pol β was similar to other mitochondrial proteins previously reported [53]. Taken together, these morphological results further corroborate the biochemical data in Figs. 1 and 2.
3.3. Biological roles of mitochondrial pol β
To examined whether pol β in purified mitochondria could be proficient in supporting in vitro BER, mitochondrial extract-mediated BER assays were conducted (Fig. 6A). Mitochondrial extract from HEK 293 cells showed single-nucleotide BER activity (Fig. 6B); endogenous UDG and APE1 activities also were observed (Fig. S7). To clarify the DNA polymerase(s) responsible for the repair activity, a neutralizing anti-pol β antibody was utilized. This pol β antibody is specific to pol β and does not recognize other polymerases, including pol γ [8]. Mitochondrial extracts were preincubated either with preimmune IgG or anti-pol β IgG and then repair activity was measured. Almost all of the repair activity was lost in reaction mixtures containing this antibody, whereas there was little effect in reaction mixtures with the preimmune IgG (Fig. 6C). The role for pol β in mitochondrial extract-mediated BER was further confirmed by comparing repair activity with mitochondria purified from pol β+/+ and pol β−/− MEF cells. BER activity was observed in the pol β+/+ mitochondrial extract, but activity was negligible in the pol β−/− extract (Fig. 6D). These results indicate pol β was responsible for the majority of the BER polymerase function in these purified mitochondrial extracts.
Fig. 6.
In vitro BER in mitochondrial extracts. A) Schematic representation of the in vitro BER substrate and the expected BER products after gap-filling (insertion of [32P]dCMP) and ligation steps. B) Representative phosphorimage of a BER time course showing repair activity of HEK 293 mitochondrial extract. C) Use of pol β-neutralizing antibody to eliminate pol β-dependent repair. Phosphorimage of a BER time course showing the repair capacity of the mitochondrial extract after incubation with preimmune IgG or anti-pol β immune IgG specific to pol β. D) Phosphorimage of a BER time course showing the repair capacity of mitochondrial extracts from pol β+/+ and pol β−/− MEF cells. The migration positions of the ligated BER product and unligated BER intermediate are indicated. A representative phosphorimage of two repeats is shown.
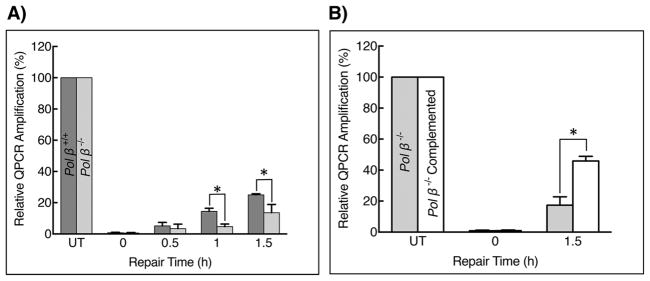
Next, we used MEF cells in culture to assess a potential role of pol β in protection against oxidatively induced DNA damage in mitochondria. Cells in mid- to late-log phase were treated briefly with H2O2 and then allowed to undergo DNA repair for up to 1.5 h in complete medium. The experiments were designed to compare mtDNA repair in wild-type MEF cells, isogenic pol β−/− fibroblasts, and pol β−/− fibroblasts stably expressing wild-type pol β. Repair of mtDNA damage was assessed by quantitative PCR analysis [52], and results are shown in Fig. 7A and B. The repair of H2O2-induced DNA damage was 2-fold higher in pol β wild-type than in pol β null cells (Fig. 7A). The deficiency in repair was complemented in pol β−/− cells expressing wildtype pol β (Fig. 7B). In these experiments, the copy number of mtDNA was found to be identical in all three cell types. In addition, similar results were obtained under alternate H2O2 treatment conditions (Fig. S8). These results suggest pol β is capable of contributing to the repair of oxidatively damaged DNA in MEF mitochondria.
Fig. 7.
Repair of oxidatively induced damage in mtDNA. A) Pol β+/+ and pol β−/− fibroblasts were treated with H2O2 (1.5 mM for 30 min) and then allowed to undergo DNA repair for 0.5, 1.0 and 1.5 h. Repair of mtDNA was assessed by QPCR analysis using total cellular DNA and the relative QPCR products were plotted. The PCR product in untreated (UT) cells was set at 100%. Error bars represent mean ± SD. The results were from triplicate measurements in two independent experiments. The repair capacity of pol β+/+ and pol β−/− cells was statistically significant, * p ≤ 0.05. B) Pol β−/− and pol β complemented pol β−/− MEF cells were treated with 1.5 mM H2O2 for 30 min and then allowed to undergo DNA repair for 1.5 h. Repair of mtDNA was assessed by QPCR analysis using total cellular DNA and the relative QPCR products were plotted. The PCR product in untreated (UT) cells was set at 100%. Error bars represent mean ± SD. Results were from extensive pilot experiments and the results shown were from triplicate measurements in two independent experiments; the difference in repair capacity of the pol β−/− and pol β complemented cells was statistically significant, * p ≤ 0.05.
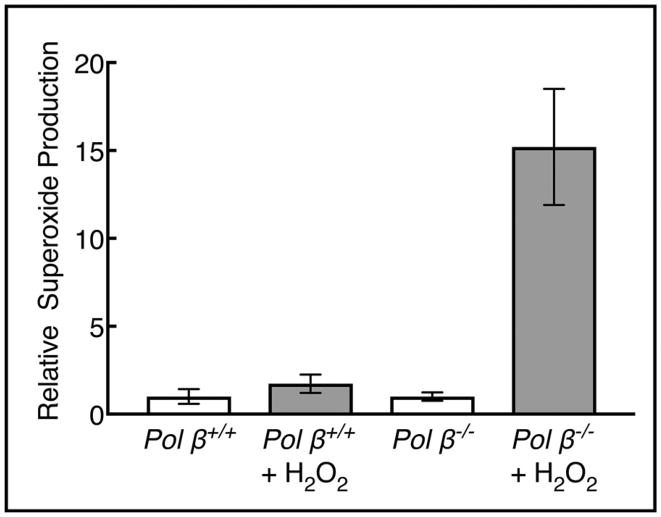
In light of the results described above, we compared mitochondrial metabolism in pol β+/+ and pol β−/− fibroblasts. It is well established that oxidative stress is characterized by the overproduction of various reactive oxygen species, and this can be used as an indicator of reduced mitochondrial function [54]. Using MitoSOX™ Red staining after cells were treated with H2O2, an increase in superoxide production was observed in pol β−/− cells compared with pol β+/+ cells (Fig. 8). These results indicate that the ability to handle H2O2-induced oxidative stress was compromised in the pol β null cells.
Fig. 8.
Superoxide radicals in H2O2-treated cells. MEF cells were treated with 50 μMH2O2 for 1 h, and then harvested after a further 3 h incubation. Increased production of superoxide radicals was observed in pol β−/− compared with pol β+/+ cells. The results represent three independent experiments and values for the untreated cells were set to 1.0. Error bars represent mean ± SD.
Finally, we evaluated the morphology of mitochondria by transmission electron microscopy of ultrathin sections of pol β+/+ and pol β−/− MEF cells. Wild-type cells showed normal mitochondria (average size 1.3 μm), with defined outer-inner mitochondrial membranes and cristae that maintained lamellar structures (Fig. 9A). The dense matrix, containing most of the mitochondrial protein, appeared dark, whereas the intermembranes and crista lumen were less-electron dense. Surprisingly, mitochondria in pol β−/− MEF cells were aberrant in shape and bigger in size (average size 1.7 μm). They appeared as swollen elongated spheres, had abnormal and fragmented cristae, contained vacuoles and less defined intermembranes and had an electron poor matrix, indicative of a loss of matrix content (Fig. 9B). The majority of mitochondria in pol β−/− MEF cells redistributed in the cytoplasm around the nuclear area and accumulation of lipid droplets was also observed. Interestingly, these morphological findings in the absence of pol β are reminiscent the morphological differences observed in polg2−/− 8.5 day embryos [55].
Fig. 9.
EM morphological analysis of mitochondria in pol β+/+ and pol β−/− MEFs. Representative EM images of pol β+/+ and pol β−/− MEF cells are shown. A) EM image of pol β+/+ MEF cells revealed normal nuclei (N) and mitochondria (M) with distinct outer and inner mitochondrial membranes and parallel cristae. B) Mitochondria in pol β−/− MEF were aberrant in shape with abnormal cristae and with many lipid droplets (L). Scale bar: 0.2 μm.
4. Discussion
Maintenance of the mammalian mitochondrial genome against DNA reactive agents is fundamental to normal cellular function, but little is known about DNA repair systems in mitochondria. Oxidatively damaged DNA is constantly formed in mtDNA because of its proximity to the electron transport system, and this is considered to contribute to the higher levels of oxidatively induced lesions found in mtDNA as compared to nuclear genomic DNA [56,57]. In mitochondria, it seems likely that the BER pathway would be used to repair oxidatively induced DNA base damage, and mitochondria are known to possess many of the core BER enzymes [58–61]. However, a DNA polymerase that could perform robust single-nucleotide gap-filling and dRP removal in mitochondrial BER has not been identified. Until recently, pol γ was considered the sole mitochondrial polymerase capable of performing these steps in the SN BER sub-pathway, but pol γ has relatively weak lyase activity in vitro [11]. Therefore, a question has remained as to whether pol β might be present in mitochondria where efficient BER is essential. Despite the presence of pol β-like enzymes in bacteria [41] and trypanosome mitochondria [39,40], conflicting reports and skepticism existed toward the presence of pol β in mammalian mitochondria [37,38].
Pol β is the nuclear enzyme considered most well equipped to handle the gap filling and lyase requirements of BER [8]. In the present study, we identified pol β in purified mitochondria from several mammalian cell lines and obtained evidence ruling out the possibility that the pol β in these samples was due to contamination by nuclear or cytoplasmic pol β. The mitochondrial-associated pol β was localized to mitochondria by immunogold EM, immunofluorescence microscopy and biochemical experiments. More work will be required to assign the sub-mitochondrial location of pol β. Importantly, pol β−/− MEFs were shown to be less proficient in repair of H2O2-induced mtDNA damage than isogenic cells expressing pol β. In addition, pol β−/− fibroblasts had increased levels of respiratory reactive oxygen species compared with wild-type cells after oxidative stress treatment. Finally, extracts from purified mitochondria exhibited strong in vitro BER that was dependent on pol β as revealed by antibody neutralization and genetic experiments.
It had been proposed that repair of oxidatively generated damaged bases in mtDNA is mediated by the SN BER sub-pathway [62,63], but the in vitro repair activity of pol γ, although present, was not striking [11]. Yet, LP BER and its strand incision factors, FEN1 and DNA2, were recently found in mitochondrial extracts, and along with pol γ activity, extracts were shown to be capable of conducting LP BER [23,64–67]. In addition, many of the core nuclear BER factors have been identified in mitochondria including DNA glycosylases, APE1, PNKP, Tdp1 and DNA ligases [32,36,68,69]. In some cases, mitochondrial targeting sequences in these repair enzymes are generated through alternative splicing of mRNAs of nuclear genes [68], and in others, the repair enzymes are imported into mitochondria through protein-protein interactions. For example, APE1 uses the mitochondrial import and assembly protein Mia40 for its translocation into mitochondria [70]. In the present study, we failed to identify a classical mitochondrial-targeting sequence for pol β, and further investigation will be necessary to understand how pol β is transported into the mitochondria. Although mitochondria import many different proteins from the cytosol, more than 50% of these proteins do not have a classical mitochondrial-targeting sequence, but instead have alternate mechanisms of mitochondrial targeting [71]. Thus, the mechanism of pol β mitochondrial transport is unknown, but this also is true for a number of other mitochondrial DNA enzymes, including Tdp1, MSH5, XPD, MSH5, PARP-1 and pol θ [31,33,53,70,72,73].
In closing, it is not clear why Loeb and coworkers failed to find pol β in their samples of mitochondria [38]. In those studies, pol β was found in samples of sucrose gradient purified mitochondria, but was not observed in mitochondria after Percoll gradient centrifugation. Yet, there had been earlier indications that pol β could be a mammalian mitochondrial component. First, pol β is a phylogenetically conserved enzyme and could have been integral in an ancient mitochondrial precursor. Second, pol β has been well characterized as a mitochondrial component in the trypanosomatid Crithidia fasciculata [40]. The Crithidia fasciculata mitochondrial pol β is capable of both of the BER enzymatic activities found in mammalian nuclear pol β, i.e., gap-filling and lyase. Third, Low and associates [37] identified pol β in bovine heart mitochondria in a series of biochemical experiments that are consistent with the results reported here. Also, a cross-linked protein consistent with pol β was observed in Percoll gradient purified mitochondria, but was considered a contaminant [65]. Fourth, Bohr and his associates recently found mitochondrial metabolism deficiency phenotypes in a mouse model with reduced expression of pol β due to haploinsufficiency of the pol β gene [74,75]. Interestingly, after the current manuscript was prepared Bohr and his associates confirmed the absence of pol β in mitochondria isolated from mouse liver and also detected pol β in mitochondria from brain tissue as well as from cultured mammalian cells [76]. The presence of pol β in mitochondria suggest that repair of oxidatively generated base lesions could be stronger than with pol γ alone. Additional experiments are expected to reveal further pol β phenotypes in mitochondrial DNA integrity, and this could involve disease susceptibility phenotypes in the presence of genetic deficiencies in mitochondrial factors. An example of this may be the case revealed in the pol β haploinsufficiency mouse model for aging and Alzheimer’s Disease [74].
Supplementary Material
Acknowledgments
Funding
This work was supported in part by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences, Project Numbers Z01ES050158 and Z01ES050159 (to SHW), Z01ES065078 (to WCC), R00ES023813 (to NRG) and NIH grants GM31819 and ES13773 (to JDG).
We thank: William Beard, Yesenia Rodriguez and Janine Santos for critical reading of the manuscript and Janine Santos for advice regarding conditions for the QPCR experiments; Robert Petrovich, the NIEHS Protein Expression Core Laboratory, for cultured cells; and Jason Williams, the NIEHS Mass Spectrometry Research and Support Group for assistance with mass spectrometry analyses.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.dnarep.2017.10.011.
Footnotes
Conflict of interest statement
The authors declare that there are no conflicts of interest.
References
- 1.Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, Lindahl T, Barnes DE. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc Natl Acad Sci. 1999;96:13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindahl T. DNA repair enzymes. Annu Rev Biochem. 1982;51:61–87. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- 3.Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 4.Fortini P, Pascucci B, Parlanti E, Sobol RW, Wilson SH, Dogliotti E. Different DNA polymerases are involved in the short- and long-patch base excision repair in mammalian cells. Biochemistry. 1998;37:3575–3580. doi: 10.1021/bi972999h. [DOI] [PubMed] [Google Scholar]
- 5.Frosina G, Fortini P, Rossi O, Carrozzino F, Raspaglio G, Cox LS, Lane DP, Abbondandolo A, Dogliotti E. Two pathways for base excision repair in mammalian cells. J Biol Chem. 1996;271:9573–9578. doi: 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- 6.Mosbaugh DW, Bennett SE. Uracil-excision DNA repair. Prog Nucleic Acid Res Mol Biol. 1994;48:315–370. doi: 10.1016/s0079-6603(08)60859-4. [DOI] [PubMed] [Google Scholar]
- 7.Slupphaug G, Eftedal I, Kavli B, Bharati S, Helle NM, Haug T, Levine DW, Krokan HE. Properties of a recombinant human uracil-DNA glycosylase from the UNG gene and evidence that UNG encodes the major uracil-DNA glycosylase. Biochemistry. 1995;34:128–138. doi: 10.1021/bi00001a016. [DOI] [PubMed] [Google Scholar]
- 8.Singhal RK, Prasad R, Wilson SH. DNA polymerase β conducts the gap-filling step in uracil-initiated base excision repair in a bovine testis nuclear extract. J Biol Chem. 1995;270:949–957. doi: 10.1074/jbc.270.2.949. [DOI] [PubMed] [Google Scholar]
- 9.Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH. Requirement of mammalian DNA polymerase-β in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 10.Sobol RW, Prasad R, Evenski A, Baker A, Yang XP, Horton JK, Wilson SH. The lyase activity of the DNA repair protein β-polymerase protects from DNA-damage-induced cytotoxicity. Nature. 2000;405:807–810. doi: 10.1038/35015598. [DOI] [PubMed] [Google Scholar]
- 11.Longley MJ, Prasad R, Srivastava DK, Wilson SH, Copeland WC. Identification of 5′-deoxyribose phosphate lyase activity in human DNA polymerase γ and its role in mitochondrial base excision repair in vitro. Proc Natl Acad Sci. 1998;95:12244–12248. doi: 10.1073/pnas.95.21.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caldecott KW, Tucker JD, Stanker LH, Thompson LH. Characterization of the XRCC1-DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res. 1995;23:4836–4843. doi: 10.1093/nar/23.23.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prasad R, Singhal RK, Srivastava DK, Molina JT, Tomkinson AE, Wilson SH. Specific interaction of DNA polymerase β and DNA ligase I in a multiprotein base excision repair complex from bovine testis. J Biol Chem. 1996;271:16000–16007. doi: 10.1074/jbc.271.27.16000. [DOI] [PubMed] [Google Scholar]
- 14.Tomkinson AE, Chen L, Dong Z, Leppard JB, Levin DS, Mackey ZB, Motycka TA. Completion of base excision repair by mammalian DNA ligases. Prog Nucleic Acid Res Mol Biol. 2001;68:151–164. doi: 10.1016/s0079-6603(01)68097-8. [DOI] [PubMed] [Google Scholar]
- 15.Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, Mitra S, Hazra TK. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 16.Weinfeld M, Mani RS, Abdou I, Aceytuno RD, Glover JN. Tidying up loose ends: the role of polynucleotide kinase/phosphatase in DNA strand break repair. Trends Biochem Sci. 2011;36:262–271. doi: 10.1016/j.tibs.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Debethune L, Kohlhagen G, Grandas A, Pommier Y. Processing of nucleopeptides mimicking the topoisomerase I-DNA covalent complex by tyrosyl-DNA phosphodiesterase. Nucleic Acids Res. 2002;30:1198–1204. doi: 10.1093/nar/30.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prasad R, Williams JG, Hou EW, Wilson SH. Pol β associated complex and base excision repair factors in mouse fibroblasts. Nucleic Acids Res. 2012;40:11571–11582. doi: 10.1093/nar/gks898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giles RE, Blanc H, Cann HM, Wallace DC. Maternal inheritance of human mitochondrial DNA. Proc Natl Acad Sci. 1980;77:6715–6719. doi: 10.1073/pnas.77.11.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 21.Barja G, Herrero A. Oxidative damage to mitochondrial DNA is inversely related to maximum life span in the heart and brain of mammals. FASEB J. 2000;14:312–318. doi: 10.1096/fasebj.14.2.312. [DOI] [PubMed] [Google Scholar]
- 22.Wallace DC, Brown MD, Lott MT. Mitochondrial DNA variation in human evolution and disease. Gene. 1999;238:211–230. doi: 10.1016/s0378-1119(99)00295-4. [DOI] [PubMed] [Google Scholar]
- 23.Chan SS, Copeland WC. DNA polymerase γ and mitochondrial disease: understanding the consequence of POLG mutations. Biochim Biophys Acta. 2009;1787:312–319. doi: 10.1016/j.bbabio.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tuppen HA, Blakely EL, Turnbull DM, Taylor RW. Mitochondrial DNA mutations and human disease. Biochim Biophys Acta. 2010;1797:113–128. doi: 10.1016/j.bbabio.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 25.Polyak K, Li Y, Zhu H, Lengauer C, Willson JK, Markowitz SD, Trush MA, Kinzler KW, Vogelstein B. Somatic mutations of the mitochondrial genome in human colorectal tumours. Nat Genet. 1998;20:291–293. doi: 10.1038/3108. [DOI] [PubMed] [Google Scholar]
- 26.Radpour R, Fan AX, Kohler C, Holzgreve W, Zhong XY. Current understanding of mitochondrial DNA in breast cancer. Breast J. 2009;15:505–509. doi: 10.1111/j.1524-4741.2009.00767.x. [DOI] [PubMed] [Google Scholar]
- 27.Dasgupta S, Hoque MO, Upadhyay S, Sidransky D. Mitochondrial cytochrome B gene mutation promotes tumor growth in bladder cancer. Cancer Res. 2008;68:700–706. doi: 10.1158/0008-5472.CAN-07-5532. [DOI] [PubMed] [Google Scholar]
- 28.Jin X, Zhang J, Gao Y, Ding K, Wang N, Zhou D, Jen J, Cheng S. Relationship between mitochondrial DNA mutations and clinical characteristics in human lung cancer. Mitochondrion. 2007;7:347–353. doi: 10.1016/j.mito.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Lan L, Nakajima S, Oohata Y, Takao M, Okano S, Masutani M, Wilson SH, Yasui A. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc Natl Acad Sci U S A. 2004;101:13738–13743. doi: 10.1073/pnas.0406048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clayton DA, Doda JN, Friedberg EC. The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. Proc Natl Acad Sci. 1974;71:2777–2781. doi: 10.1073/pnas.71.7.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J, Fang H, Chi Z, Wu Z, Wei D, Mo D, Niu K, Balajee AS, Hei TK, Nie L, Zhao Y. XPD localizes in mitochondria and protects the mitochondrial genome from oxidative DNA damage. Nucleic Acids Res. 2015;43:5476–5488. doi: 10.1093/nar/gkv472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prakash A, Doublie S. Base excision repair in the mitochondria. J Cell Biochem. 2015;116:1490–1499. doi: 10.1002/jcb.25103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bannwarth S, Figueroa A, Fragaki K, Destroismaisons L, Lacas-Gervais S, Lespinasse F, Vandenbos F, Pradelli LA, Ricci JE, Rotig A, Michiels JF, Vande Velde C, Paquis-Flucklinger V. The human MSH5 (MutSHomolog 5) protein localizes to mitochondria and protects the mitochondrial genome from oxidative damage. Mitochondrion. 2012;12:654–665. doi: 10.1016/j.mito.2012.07.111. [DOI] [PubMed] [Google Scholar]
- 34.Martin SA, McCabe N, Mullarkey M, Cummins R, Burgess DJ, Nakabeppu Y, Oka S, Kay E, Lord CJ, Ashworth A. DNA polymerases as potential therapeutic targets for cancers deficient in the DNA mismatch repair proteins MSH2 or MLH1. Cancer Cell. 2010;17:235–248. doi: 10.1016/j.ccr.2009.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mason PA, Matheson EC, Hall AG, Lightowlers RN. Mismatch repair activity in mammalian mitochondria. Nucleic Acids Res. 2003;31:1052–1058. doi: 10.1093/nar/gkg167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krasich R, Copeland WC. DNA polymerases in the mitochondria: a critical review of the evidence. Front Biosci. 2017;22:692–709. doi: 10.2741/4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nielsen-Preiss SM, Low RL. Identification of a β-like DNA polymerase activity in bovine heart mitochondria. Arch Biochem Biophys. 2000;374:229–240. doi: 10.1006/abbi.1999.1590. [DOI] [PubMed] [Google Scholar]
- 38.Hansen AB, Griner NB, Anderson JP, Kujoth GC, Prolla TA, Loeb LA, Glick E. Mitochondrial DNA integrity is not dependent on DNA polymerase-β activity. DNA Repair. 2006;5:71–79. doi: 10.1016/j.dnarep.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 39.Torri AF, Kunkel TA, Englund PT. A β-like DNA polymerase from the mitochondrion of the trypanosomatid Crithidia fasciculata. J Biol Chem. 1994;269:8165–8171. [PubMed] [Google Scholar]
- 40.Saxowsky TT, Matsumoto Y, Englund PT. The mitochondrial DNA polymerase β from Crithidia fasciculata has 5′-deoxyribose phosphate (dRP) lyase activity but is deficient in the release of dRP. J Biol Chem. 2002;277:37201–37206. doi: 10.1074/jbc.M206654200. [DOI] [PubMed] [Google Scholar]
- 41.Bienstock RJ, Beard WA, Wilson SH. Phylogenetic analysis and evolutionary origins of DNA polymerase X-family members. DNA Repair. 2014;22:77–88. doi: 10.1016/j.dnarep.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bogenhagen DF. Purification of mitochondria for enzymes involved in nucleic acid transactions. Methods Mol Biol. 2002;197:199–210. doi: 10.1385/1-59259-284-8:199. [DOI] [PubMed] [Google Scholar]
- 43.Choi JH, Williams J, Cho J, Falck JR, Shears SB. Purification, sequencing, and molecular identification of a mammalian PP-InsP5 kinase that is activated when cells are exposed to hyperosmotic stress. J Biol Chem. 2007;282:30763–30775. doi: 10.1074/jbc.M704655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vande Velde C, Miller TM, Cashman NR, Cleveland DW. Selective association of misfolded ALS-linked mutant SOD1 with the cytoplasmic face of mitochondria. Proc Natl Acad Sci. 2008;105:4022–4027. doi: 10.1073/pnas.0712209105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Braithwaite EK, Prasad R, Shock DD, Hou EW, Beard WA, Wilson SH. DNA polymerase λ mediates a back-up base excision repair activity in extracts of mouse embryonic fibroblasts. J Biol Chem. 2005;280:18469–18475. doi: 10.1074/jbc.M411864200. [DOI] [PubMed] [Google Scholar]
- 46.Gama V, Gomez JA, Mayo LD, Jackson MW, Danielpour D, Song K, Haas AL, Laughlin MJ, Matsuyama S. Hdm2 is a ubiquitin ligase of Ku70-Akt promotes cell survival by inhibiting Hdm2-dependent Ku70 destabilization. Cell Death Differ. 2009;16:758–769. doi: 10.1038/cdd.2009.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gassman NR, Coskun E, Stefanick DF, Horton JK, Jaruga P, Dizdaroglu M, Wilson SH. Bisphenol a promotes cell survival following oxidative DNA damage in mouse fibroblasts. PLoS One. 2015;10:e0118819. doi: 10.1371/journal.pone.0118819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Horton JK, Gassman NR, Dunigan BD, Stefanick DF, Wilson SH. DNA polymerase β-dependent cell survival independent of XRCC1 expression. DNA Repair. 2015;26:23–29. doi: 10.1016/j.dnarep.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gassman NR, Stefanick DF, Kedar PS, Horton JK, Wilson SH. Hyperactivation of PARP triggers nonhomologous end-joining in repair-deficient mouse fibroblasts. PLoS One. 2012;7:e49301. doi: 10.1371/journal.pone.0049301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson DM, 3rd, Bianchi C. Improved immunodetection of nuclear antigens after sodium dodecyl sulfate treatment of formaldehyde-fixed cells. J Histochem Cytochem. 1999;47:1095–1100. doi: 10.1177/002215549904700814. [DOI] [PubMed] [Google Scholar]
- 51.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Santos JH, Meyer JN, Mandavilli BS, Van Houten B. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol Biol. 2006;314:183–199. doi: 10.1385/1-59259-973-7:183. [DOI] [PubMed] [Google Scholar]
- 53.Wisnovsky S, Jean SR, Kelley SO. Mitochondrial DNA repair and replication proteins revealed by targeted chemical probes. Nat Chem Biol. 2016;12:567–573. doi: 10.1038/nchembio.2102. [DOI] [PubMed] [Google Scholar]
- 54.Guo C, Sun L, Chen X, Zhang D. Oxidative stress, mitochondrial damage and neurodegenerative diseases. Neural Regen Res. 2013;8:2003–2014. doi: 10.3969/j.issn.1673-5374.2013.21.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Humble MM, Young MJ, Foley JF, Pandiri AR, Travlos GS, Copeland WC. Polg2 is essential for mammalian embryogenesis and is required for mtDNA maintenance. Hum Mol Genet. 2013;22:1017–1025. doi: 10.1093/hmg/dds506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hamilton ML, Guo Z, Fuller CD, Van Remmen H, Ward WF, Austad SN, Troyer DA, Thompson I, Richardson A. A reliable assessment of 8-oxo-2-deoxyguanosine levels in nuclear and mitochondrial DNA using the sodium iodide method to isolate DNA. Nucleic Acids Res. 2001;29:2117–2126. doi: 10.1093/nar/29.10.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hamilton ML, Van Remmen H, Drake JA, Yang H, Guo ZM, Kewitt K, Walter CA, Richardson A. Does oxidative damage to DNA increase with age? Proc Natl Acad Sci. 2001;98:10469–10474. doi: 10.1073/pnas.171202698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Souza-Pinto NC, Eide L, Hogue BA, Thybo T, Stevnsner T, Seeberg E, Klungland A, Bohr VA. Repair of 8-oxodeoxyguanosine lesions in mitochondrial DNA depends on the oxoguanine DNA glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial DNA of OGG1-defective mice. Cancer Res. 2001;61:5378–5381. [PubMed] [Google Scholar]
- 59.Englander EW, Hu Z, Sharma A, Lee HM, Wu ZH, Greeley GH. Rat MYH, a glycosylase for repair of oxidatively damaged DNA, has brain-specific isoforms that localize to neuronal mitochondria. J Neurochem. 2002;83:1471–1480. doi: 10.1046/j.1471-4159.2002.01259.x. [DOI] [PubMed] [Google Scholar]
- 60.Tell G, Damante G, Caldwell D, Kelley MR. The intracellular localization of APE1/Ref-1: more than a passive phenomenon? Antioxid Redox Signal. 2005;7:367–384. doi: 10.1089/ars.2005.7.367. [DOI] [PubMed] [Google Scholar]
- 61.Lakshmipathy U, Campbell C. The human DNA ligase III gene encodes nuclear and mitochondrial proteins. Mol Cell Biol. 1999;19:3869–3876. doi: 10.1128/mcb.19.5.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stierum RH, Dianov GL, Bohr VA. Single-nucleotide patch base excision repair of uracil in DNA by mitochondrial protein extracts. Nucleic Acids Res. 1999;27:3712–3719. doi: 10.1093/nar/27.18.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bogenhagen DF, Pinz KG, Perez-Jannotti RM. Enzymology of mitochondrial base excision repair. Prog Nucleic Acid Res Mol Biol. 2001;68:257–271. doi: 10.1016/s0079-6603(01)68105-4. [DOI] [PubMed] [Google Scholar]
- 64.Kalifa L, Beutner G, Phadnis N, Sheu SS, Sia EA. Evidence for a role of FEN1 in maintaining mitochondrial DNA integrity. DNA Repair. 2009;8:1242–1249. doi: 10.1016/j.dnarep.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu P, Qian L, Sung JS, de Souza-Pinto NC, Zheng L, Bogenhagen DF, Bohr VA, Wilson DM, Shen B, Dernple B. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol Cell Biol. 2008;28:4975–4987. doi: 10.1128/MCB.00457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kazak L, Reyes A, He J, Wood SR, Brea-Calvo G, Holen TT, Holt IJ. A cryptic targeting signal creates a mitochondrial FEN1 isoform with tailed R-Loop binding properties. PLoS One. 2013;8:e62340. doi: 10.1371/journal.pone.0062340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Copeland WC, Longley MJ. DNA2 resolves expanding flap in mitochondrial base excision repair. Mol Cell. 2008;32:457–458. doi: 10.1016/j.molcel.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kang D, Hamasaki N. Maintenance of mitochondrial DNA integrity: repair and degradation. Curr Genet. 2002;41:311–322. doi: 10.1007/s00294-002-0312-0. [DOI] [PubMed] [Google Scholar]
- 69.Mandal SM, Hegde ML, Chatterjee A, Hegde PM, Szczesny B, Banerjee D, Boldogh I, Gao R, Falkenberg M, Gustafsson CM, Sarkar PS, Hazra TK. Role of human DNA glycosylase Nei-like 2 (NEIL2) and single strand break repair protein polynucleotide kinase 3′-phosphatase in maintenance of mitochondrial genome. J Biol Chem. 2012;287:2819–2829. doi: 10.1074/jbc.M111.272179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barchiesi A, Wasilewski M, Chacinska A, Tell G, Vascotto C. Mitochondrial translocation of APE1 relies on the MIA pathway. Nucleic Acids Res. 2015;43:5451–5464. doi: 10.1093/nar/gkv433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bolender N, Sickmann A, Wagner R, Meisinger C, Pfanner N. Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep. 2008;9:42–49. doi: 10.1038/sj.embor.7401126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Das BB, Dexheimer TS, Maddali K, Pommier Y. Role of tyrosyl-DNA phosphodiesterase (TDP1) in mitochondria. Proc Natl Acad Sci. 2010;107:19790–19795. doi: 10.1073/pnas.1009814107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rossi MN, Carbone M, Mostocotto C, Mancone C, Tripodi M, Maione R, Amati P. Mitochondrial localization of PARP-1 requires interaction with mitofilin and is involved in the maintenance of mitochondrial DNA integrity. J Biol Chem. 2009;284:31616–31624. doi: 10.1074/jbc.M109.025882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sykora P, Misiak M, Wang Y, Ghosh S, Leandro GS, Liu D, Tian J, Baptiste BA, Cong WN, Brenerman BM, Fang E, Becker KG, Hamilton RJ, Chigurupati S, Zhang Y, Egan JM, Croteau DL, Wilson DM, 3rd, Mattson MP, Bohr VA. DNA polymerase β deficiency leads to neurodegeneration and exacerbates Alzheimer disease phenotypes. Nucleic Acids Res. 2015;43:943–959. doi: 10.1093/nar/gku1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Misiak M, Vergara Greeno R, Baptiste BA, Sykora P, Liu D, Cordonnier S, Fang EF, Croteau DL, Mattson MP, Bohr VA. DNA polymerase beta decrement triggers death of olfactory bulb cells and impairs olfaction in a mouse model of Alzheimer’s disease. Aging Cell. 2017;16:162–172. doi: 10.1111/acel.12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sykora P, Kanno S, Akbari M, Kulikowicz T, Baptiste BA, Leandro GS, Lu H, Tian J, May A, Becker KA, Croteau DL, Wilson DM, 3rd, Sobol RW, Yasui A, Bohr VA. DNA polymerase beta participates in mitochondrial DNA repair. Mol Cell Biol. 2017 doi: 10.1128/MCB.00237-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.