Summary
Generation of hematopoietic stem cells (HSCs) from pluripotent stem cells, in vitro, holds great promise for regenerative therapies. Primarily, this has been achieved in mouse cells by overexpression of the homeotic selector protein HOXB4. The exact cellular stage at which HOXB4 promotes hematopoietic development, in vitro, is not yet known. However, its identification is a prerequisite to unambiguously identify the molecular circuits controlling hematopoiesis, since the activity of HOX proteins is highly cell and context dependent. To identify that stage, we retrovirally expressed HOXB4 in differentiating mouse embryonic stem cells (ESCs). Through the use of Runx1(−/−) ESCs containing a doxycycline-inducible Runx1 coding sequence, we uncovered that HOXB4 promoted the formation of hemogenic endothelium cells without altering endothelial cell development. Whole-transcriptome analysis revealed that its expression mediated the upregulation of transcription of core transcription factors necessary for hematopoiesis, culminating in the formation of blood progenitors upon initiation of Runx1 expression.
Keywords: HOXB4, hematopoietic stem cells, hemangioblast, hemogenic endothelium, hematopoietic specification, EHT, RUNX1, pluripotent stem cells
Highlights
-
•
HOXB4 induces hemogenic endothelium formation from differentiating ESCs
-
•
Hematopoiesis is not promoted at the cost of endothelial cell development
-
•
HOXB4 induces a transcriptional program closely resembling dorsal aorta HE cells
In this article, Klump and colleagues demonstrate that the human homeotic selector protein HOXB4 promotes ESC-derived hematopoiesis by inducing hemogenic endothelium formation, in vitro. It propels hematopoietic specification by upregulating the transcription of genes essential for hematopoietic development, such as those encoding members of the so-called heptad transcription factors.
Introduction
Patient-specific, induced pluripotent stem cells (iPSCs) present an attractive starting point for generation of autologous hematopoietic stem cells (HSCs) for treating certain hematologic diseases in the future (Klump et al., 2013, Wahlster and Daley, 2016). However, directed differentiation toward HSCs has remained inefficient, indicating that some key requirements necessary for full hematopoietic specification, in vitro, are still ill-defined, likely owing to our incomplete knowledge of HSC development, in vivo.
During embryonic development, definitive, multipotent HSCs are formed by a subset of cells lining the ventral floor of the dorsal aorta termed hemogenic endothelial (HE) cells (Bertrand et al., 2010, Jaffredo et al., 1998, Tavian et al., 2001, Zovein et al., 2010). Endothelial and blood cells share a similar gene expression pattern and emanate from common mesodermal progenitors (Kataoka et al., 2011, Liu et al., 2013, Wareing et al., 2012). When isolated from embryos or differentiating embryonic stem cell (ESC) cultures, these progenitors, so-called hemangioblasts, form blast colonies, in vitro, containing both endothelial and hematopoietic progenitors (Choi et al., 1998, Kennedy et al., 1997), and, in some reports, even smooth muscle cells (Ema et al., 2003). During blast colony formation, HE cells are generated as intermediate cells, which subsequently undergo endothelial-to-hematopoietic transition (EHT) (Lancrin et al., 2009).
Several transcription factors (TFs) are known to be important intrinsic regulators of endothelial as well as HSC development, such as SCL/TAL1, ETV2, GATA2, or MEIS1 (Azcoitia et al., 2005, Bloor et al., 2002, Liu et al., 2012, Liu et al., 2015). However, for the enforced generation of sufficient numbers of HSCs from differentiating pluripotent stem cells, in vitro, the instructive regulatory networks controlling separation of the endothelial and hematopoietic lineages and the critical time points of their action need to be identified. Single-cell transcriptional profiling during early mesoderm development has turned out to be a key approach for this purpose, allowing for an improved distinction of endothelial cells and hematopoietic progenitors during early mesodermal differentiation (Moignard et al., 2015, Scialdone et al., 2016, Swiers et al., 2013). Some of the identified hematopoiesis-associated genes are involved in a regulatory network controlling Runx1 transcription, which is essential for the formation of adult definitive HSCs by directly controlling EHT (Lacaud et al., 2002, Lancrin et al., 2009, North et al., 1999). Because only limited numbers of HSCs are present in embryos (Taoudi et al., 2008), the availability of HE cells may be a crucial bottleneck for the de novo generation of HSCs. Therefore, adequate numbers of HE cells likely need to be generated for the formation of sufficient numbers of HSCs, in vitro.
Except one report describing the development of hematopoietic stem and progenitor cells (HSPCs) from genetically unmanipulated ESCs (Pearson et al., 2015), significant and stable long-term repopulation of pluripotent stem cell-derived HSPCs has only been achieved using mouse cells, and only when the homeotic selector protein HOXB4 was ectopically expressed (Chan et al., 2008, Kyba et al., 2002, Lesinski et al., 2012, Lu et al., 2016, Pilat et al., 2005). However, differentiation of transplanted HSPCs was skewed toward myelopoiesis with a concomitant inhibition of lymphopoiesis and erythropoiesis in a dose-dependent manner, suggesting either an incomplete maturation of ESC-derived HSPCs and/or active interference with transcriptional circuitries controlling differentiation (Kyba et al., 2002, Pilat et al., 2005, Schiedlmeier et al., 2003). Although we, and others, have identified target genes of HOXB4 in differentiating mouse ESCs (Fan et al., 2012, Jackson et al., 2012, Oshima et al., 2011, Schiedlmeier et al., 2007), these findings have not yet led to a satisfying explanation on how HOXB4 promotes HSPC development, in vitro. The most likely reason is that the activities of HOX proteins, as modulators of transcription, are highly context dependent and influenced by the microenvironment of a given cell (Abate-Shen, 2002, Schiedlmeier et al., 2007, Will et al., 2006). So far, the cellular stage at which HOXB4 unfolds its activities during hematopoiesis, in vitro, is not yet known. Thus, its identification would be a prerequisite for unambiguously identifying the key molecular circuitries driving hematopoietic development and for the in vitro generation of HSPCs from pluripotent stem cells.
In this work, we demonstrate that HOXB4 promotes the generation of early hematopoietic progenitors from differentiating mouse ESCs, in vitro, by enforcing the development of HE cells poised for the induction of EHT by RUNX1. Molecularly, it does so, at least in part, by upregulating the expression of key TFs involved in hematopoietic development.
Results
Ectopic HOXB4 Expression Does Not Promote Mesoderm Specification
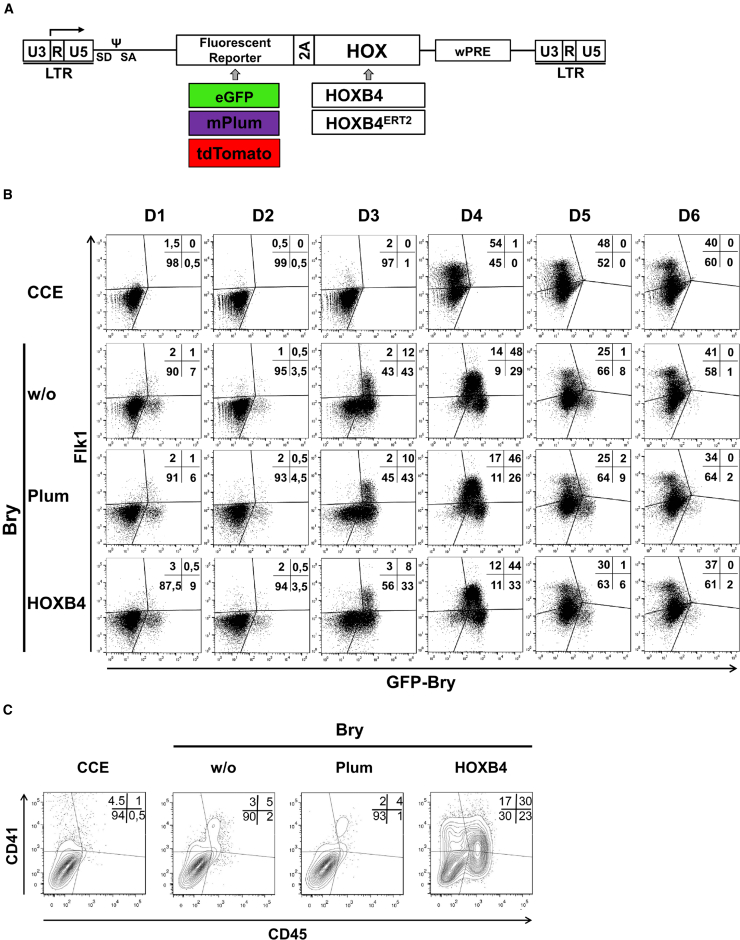
As hematopoietic cells originate from the mesoderm, we transduced Brachyury (Bry) reporter ESCs (GFP-Bry, kindly provided by J. Fehling, Ulm) (Fehling et al., 2003) with retroviral vectors co-expressing HOXB4 and the fluorescent protein mPlum (Figure 1A), and determined GFP as well as vascular endothelial growth factor receptor 2 (FLK-1) expression during differentiation (Nishikawa et al., 1998). The peak of BryGFP+FLK-1+ cells was detected between days 3 and 4 of embryoid body (EB) development, in vitro. At this stage of development, ectopic expression of HOXB4 did not promote specification of BryGFP+FLK-1+ mesodermal cells (Figure 1B). Later, however, HOXB4 strongly enhanced the formation of CD41+ and CD45+ hematopoietic progenitors after dissociation of EB day 6 GFP-Bry (EBd6) cells and subsequent co-culture on OP9 stromal cells, confirming our previous results with CCE ESCs (Figures 1C and S1; Movie S1a. Genesis of CD41+ Hematopoietic Cells, In Vitro, Promoted by HOXB4 (green: HOXB4, red: CD41), Movie S1b. Genesis of CD41+ Hematopoietic Cells, In Vitro, Promoted by HOXB4 (phase contrast)) (Chan et al., 2008, Lesinski et al., 2012). These results indicate that HOXB4 first acts downstream of early mesoderm specification and upstream of hematopoietic progenitor formation during differentiation of pluripotent stem cells.
Figure 1.
HOXB4 Does Not Promote Early Mesoderm Specification, In Vitro
(A) Scheme depicting the gammaretroviral, FMEV-based expression vectors (Lesinski et al., 2012, Schiedlmeier et al., 2007). The vectors co-express a fluorescent protein (eGFP, mPlum, or tdTomato) together with HOXB4 or a 4-hydroxytamoxifen (Tam) inducible form, HOXB4ERT2. Co-translational separation of the proteins is mediated by the TAV-2A esterase. LTR, long terminal repeat; wPRE, woodchuck hepatitis virus posttranscriptional regulatory element.
(B) Vector-transduced GFP-Bry ESCs (mPlum +/− HOXB4) were differentiated as embryoid bodies (EBs). At the indicated days, eGFP fluorescence as well as FLK-1 expression were determined by flow cytometry. The percentages of eGFP+FLK-1+ cells are shown. CCE ESCs were used as eGFP-negative controls (top row).
(C) After 6 days of differentiation, GFP-Bry EBs were dissociated and 105 cells each co-cultured on OP9 stoma cells for further 8 days. Contour plots of flow cytometry analysis are shown, the percentages of CD41+ and CD45+ cells are indicated. For (B) and (C), representative results of n = 3 independent experiments are shown.
HOXB4 Promotes Formation of a Hemogenic Endothelium
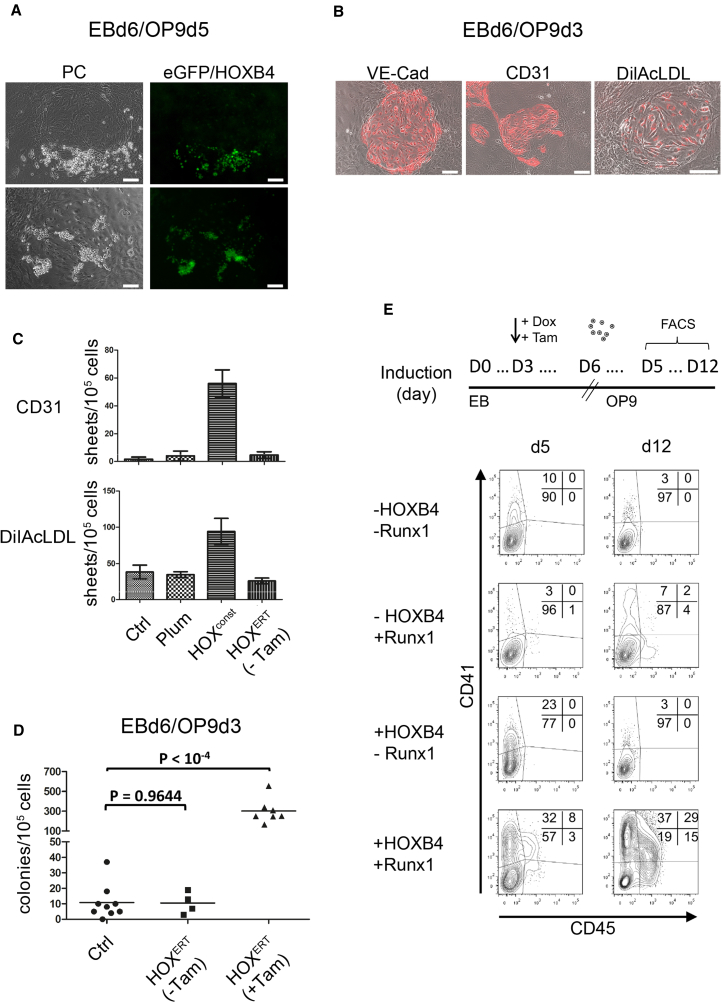
Concomitant to the marked formation of ESC-derived CD41+ and CD45+ progenitors in OP9 co-cultures, ectopic HOXB4 led to an increased number of adherent circular structures, which were associated with suspension cell clusters (Figure 2A), expressed VE-cadherin, CD31, and were capable of acetylated low-density lipoprotein uptake (Figures 2B and 2C). The suspension cells continued to proliferate only when HOXB4 was ectopically expressed and, after transplantation into immunodeficient NSG mice, mediated the typical pattern of myeloid-biased repopulation, as repeatedly reported by us and others (Figures S2A–S2D) (Kyba et al., 2002, Lesinski et al., 2012, Lu et al., 2016, McKinney-Freeman et al., 2009, Pilat et al., 2005, Wang et al., 2005). The adherent circular, cobblestone-like structures were reminiscent of previously described HE colonies (Eilken et al., 2009, Lancrin et al., 2009). To test this notion, we used a Runx1 knockout ESC line carrying a doxycycline-inducible Runx1 coding sequence stably integrated into the genome (iRunx cells) (Lancrin et al., 2009). These cells are blocked immediately prior to EHT due to the absence of Runx1 expression, which is essential for transition of the HE to hematopoietic cells. Importantly, the induction of its expression rescues the generation of blood cells (Lancrin et al., 2009). This system allowed us to answer the question of whether HOXB4 acts upstream of Runx1 in promoting the hematopoietic fate and to separate RUNX1-dependent from RUNX1-independent effects of HOXB4, as well. HOXB4 overexpression in the absence of RUNX1 led to a significant accumulation of endothelial colonies (Figure 2D). To test if these cells are truly hemogenic, we induced Runx1 expression by addition of doxycycline to the cultures (Movie S2a. Endothelial-to-Hematopoietic Transition of HOXB4+ Hemogenic Endothelium (without Runx induction), Movie S2b. Endothelial-to-Hematopoietic Transition of HOXB4+ Hemogenic Endothelium (after Runx1 induction)). After Runx1 induction, EHT of the endothelium cells initiated with a concomitant strong upregulation of CD41 expression, particularly when HOXB4 was activated (Figure 2E). Between day 5 and 12, a subpopulation of CD41+ cells also initiated CD45+ expression and continued to mature toward CD41−CD45+ cells. Without Runx1 induction, the proportion of cells expressing low levels of CD41 was also strongly increased by HOXB4. However, these cells did not undergo EHT, further upregulate CD41 expression, or even generate CD45+ cells. Instead, the proportion of CD41+ cells strongly diminished over time. Without ectopic human HOXB4, a much smaller proportion of cells became CD41+ or CD45+ after induction of Runx1, which was in line with the significantly reduced number of HE colonies present in the cultures. Moreover, transcription of the hemato-endothelial genes Scl/Tal1, Gata2, Lmo2, and Cdh5 (encoding VE-cadherin) and Pecam1 (Gritz and Hirschi, 2016) was upregulated by HOXB4 in the absence of Runx1 (Figure S3B). After induction of Runx1, expression of the key hematopoietic TF Pu.1 (Iwasaki et al., 2005) was induced, as well as Gfi1 and Gfi1b, two direct downstream effectors of RUNX1, which are necessary for the early and late phase of EHT, respectively (Lancrin et al., 2012, Thambyrajah et al., 2016). Noteworthy, induction of Runx1 alone without HOXB4 led to a transcriptional repression of the aforementioned hemato-endothelial genes, likely mediated by RUNX1 itself or GFI1 (Lancrin et al., 2012). Taken together, these results prove that the endothelial structures promoted by HOXB4 are indeed hemogenic.
Figure 2.
Formation of HE Colonies Is Promoted by HOXB4
(A) During co-culture on OP9 cells, circular sheet colonies were formed by the dissociated CCE-ESC-derived EBs (eGFP-HOXB4 transduced), which were commonly associated with hematopoietic suspension cell clusters. Left panel: phase contrast; right panel: eGFP-fluorescence. Scale bars, 100 μm.
(B) The observed endothelial colonies expressed VE-cadherin, CD31, and were capable of acetylated low-density lipoprotein (LDL) (DilAcLDL) uptake. Scale bars, 100 μm.
(C) The number of endothelial CD31+ and DilAcLDL+ colonies strongly increased when HOXB4 was ectopically expressed. Average colony numbers per 105 seeded cells are represented as columns, error bars represent SD of n = 3 independent experiments.
(D) iRunx-ESCs with and without a 4-hydroxytamoxifen (Tam) inducible form of HOXB4 (vector FMEV-tdTomato-2A-HOXB4ERT) were differentiated as EBs for 6 days, dissociated, and co-cultured on OP9 stroma cells for further 4 days without Runx1 induction (no addition of doxycycline); n = 9 and 4 independent experiments for controls, n = 7 for HOXB4. Without HOXB4 induction, the number of HE colonies per 105 seeded EB cells was comparable with unmanipulated controls. When HOXB4 was induced throughout differentiation, the number of HE colonies increased approximately 30-fold (p < 10−4). The p values were calculated using the two-sided, unpaired Student's t test with a significance level defined as 0.05.
(E) Flow cytometric analysis showing the proportion of CD41+ and CD45+ cells in OP9 co-cultures after 5 and 12 days. Dissociated iRunx EBd6 were co-cultured on OP9 cells with or without addition of doxycycline (0.1 μg/mL) to induce Runx1 expression and with or without addition of 100 nM Tam for induction of HOXB4ERT (FMEV-tdTomato-2A-HOXB4ERT). Induction of Runx1 and HOXB4ERT started from day 3 of EB development on until day 5 of OP9 co-culture. Cells were harvested after 5 or 12 days of OP9 co-culture, and the proportion of CD41- and CD45-expressing cells determined by flow cytometry. OP9 cells were removed with an anti-CD140b antibody.
HOXB4 Primes Hemangioblast Progeny toward the Hemogenic Endothelium
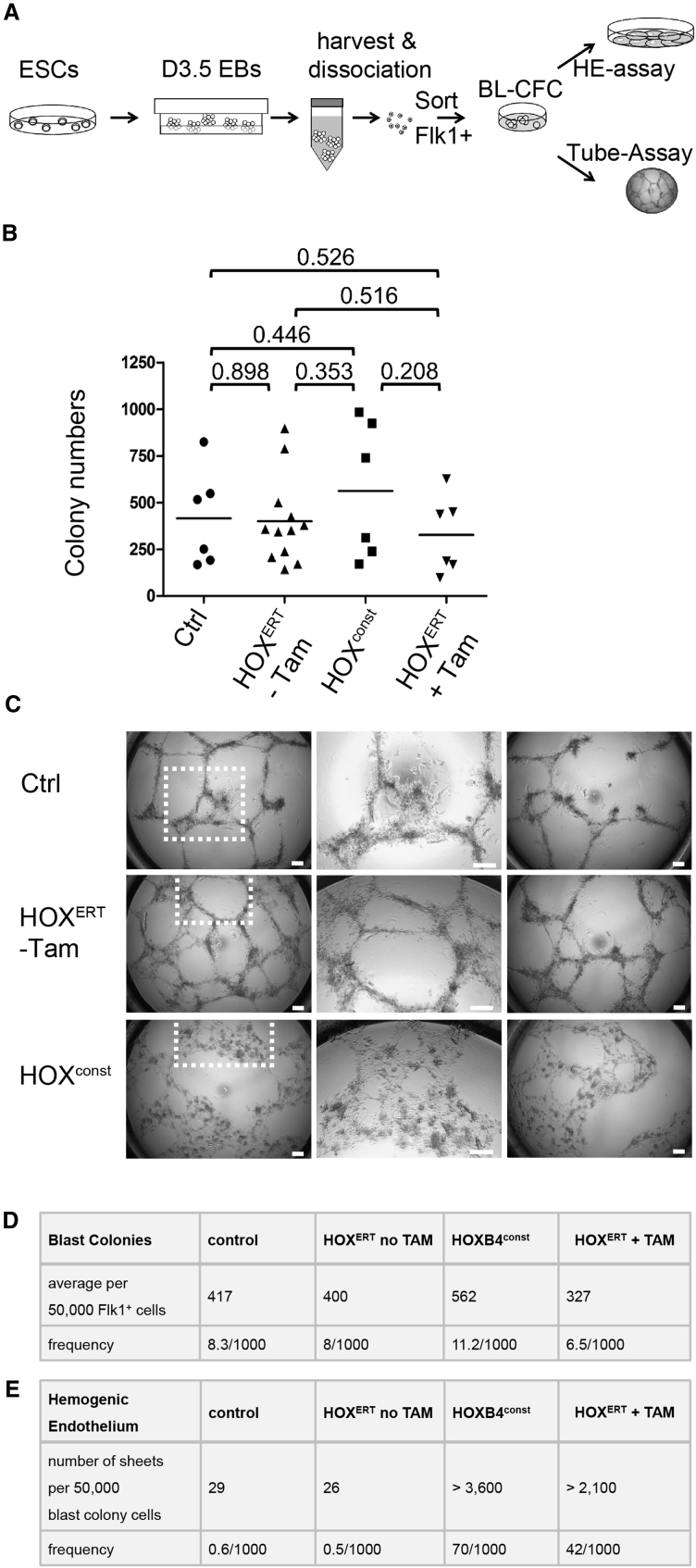
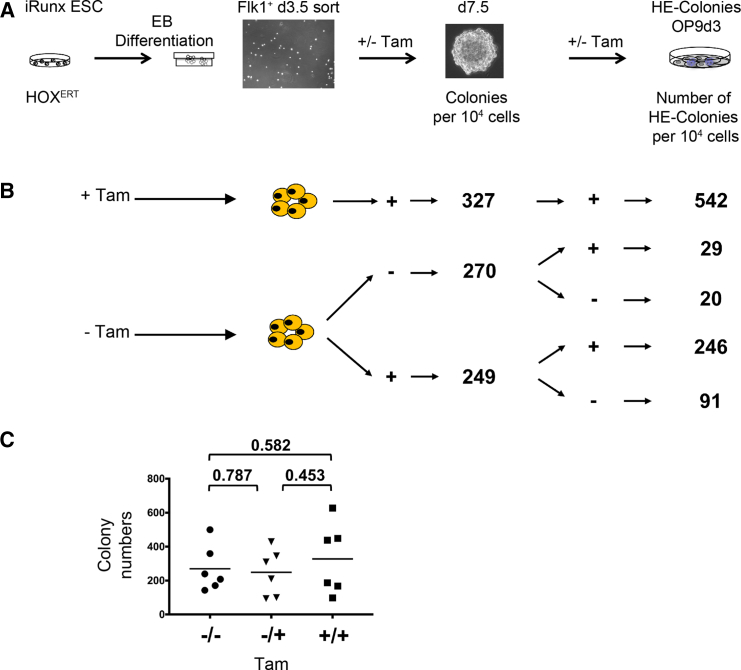
Because the number of HE colony-forming cells was increased by HOXB4, we asked whether it expands its upstream progenitor, the hemangioblast (Lancrin et al., 2009). To determine its frequencies, we performed blast colony-forming assays (Choi et al., 1998). As the frequency of FLK-1+ hemangioblasts (blast colony-forming cells [BL-CFCs]) was highest around day 3.5 of EB development, we isolated FLK-1+ iRunx cells at that time point and performed blast colony-forming assays with or without HOXB4 expressed constitutively (HOXconst) or in an inducible form, HOXERT (4-hydroxytamoxifen [Tam]-inducible HOXB4-ERT2) (Figure 3A). Two morphologically distinct types of colonies can be detected in this assay, smooth colonies and blast colonies (Lancrin et al., 2009). However, the average numbers of both types of colonies did not significantly change by ectopic HOXB4 expression. The colony frequencies were all at approximately 1%, which is the commonly reported range (Figures 3B and 3D) (Faloon et al., 2000). Thus, HOXB4 did not promote the formation of BL-CFCs. We therefore asked whether HOXB4 alters their fate as an explanation for the observed increase in the number of HE cells. For this purpose, we evaluated the developmental fate of pooled colonies by subjecting those cells into an angiogenic tube formation assay in Matrigel (Arnaoutova and Kleinman, 2010). While control iRunx cells and non-activated HOXB4ERT cells (without Tam) formed meshworks with cords of tubules in Matrigel, cells expressing HOXB4 formed a morphologically distinct mesh of flattened cells without a tubular network (Figure 3C). An aliquot was also placed onto OP9 cells to quantify HE colonies. The number of HE colonies was again over 100-fold higher when HOXB4 was constitutively expressed or induced (Figure 3E), therefore suggesting that HOXB4 may alter the developmental fate of either hemangioblasts or their progeny contained within the blast/smooth colonies. To examine this more closely, we induced HOXB4ERT either from the beginning of EB differentiation on, or at different time points prior to and after the blast colony-forming stage in differentiating iRunx cells. Again, the numbers of blast and smooth colonies did not change whether or not HOXB4 was induced in FLK-1+ cells (Figures 4A and 4C). When HOXB4 was first induced after their dissociation, the number of HE structures subsequently formed on OP9 stroma did not increase (Figure 4B, −/−/+ Tam). In contrast, when HOXB4 was already induced during the formation of blast/smooth colonies, the number of subsequent HE colonies markedly increased (Figure 4B, −/+/− Tam). Continuous presence throughout blast and HE colony formation even led to a further increase of HE colonies on OP9 cells (Figure 4B, −/+/+ Tam). These results indicate that HOXB4 primarily acts during the blast culture phase, in cells downstream of the hemangioblast, which may present immediate colony-forming precursors of the HE.
Figure 3.
HOXB4 Does Not Alter Blast Colony-Forming Cell (Hemangioblast) Frequencies
(A) Depicted is an overview of FLK-1+ hemangioblast frequency determination, subsequent HE quantitation, and evaluation of tube formation propensities. iRunx cells were differentiated as EBs for 3.5 days, FLK-1+ cells with and without ectopically expressed HOXB4 were sorted, and 50,000 cells each subjected to blast colony-forming assays to retrospectively quantify the number of hemangioblasts 4 days later. All colonies were harvested, dissociated, and 50,000 cells each placed onto OP9 stroma cells to determine the number of HE colony-forming progenitors 3 days later. A total of 40,000 cells each were placed into a Matrigel-based tube formation assays. All assays were performed at least in triplicate, without Runx1 induction.
(B) Ectopic HOXB4 expression did not significantly alter the total number of colonies. For statistical analysis, p values were calculated based on the two-sided, unpaired Student's t test, n = 6–12; the significance level was defined as p < 0.05. The individual colony numbers are shown as symbols, the arithmetic means depicted as lines.
(C) HOXB4 altered the ability of blast colony cells to form tubes. Instead of the small, thin tubular network observed in the controls (Ctrl and HOXB4ERT without Tam), flat, adherent structures were formed with differing morphologies. The mid-panels show magnifications of the areas indicated in the left pictures. Scale bars, 50 μm.
(D and E) Frequencies of BL-CFCs (arithmetic means, as described in B) (D) and of HE colonies (arithmetic means of n = 3 independent replicates) without HOXB4 induction (HOXB4ERT no Tam) or with constitutively expressed (HOXB4 const) or induced HOXB4 (HOXB4ERT + Tam) (E).
Figure 4.
The Progeny of Hemangioblasts Is the Prime Target of HOXB4 Activity
(A) Ectopic HOXB4 activity was induced by addition of Tam at the indicated stages of iRunx-ESC differentiation. BL-CFC assays were performed with 10,000 FLK-1+ iRunx EBd3.5 cells each, +/− Tam.
(B) The total number of colonies was counted 4 days later, the average numbers of n = 3 independent experiments are indicated. +,+,+, Tam (HOXB4) was continuously present; −,−,−, without Tam (no HOXB4 induction); −/−/+, HOXB4 was first induced after dissociation of colonies; −/+/+, HOXB4 was first induced in EBd3.5 FLK-1-sorted cells; −/+/−, HOXB4 was only transiently induced during blast colony formation by FLK-1+ hemangioblasts.
(C) Individual colony numbers and arithmetic means (bars): −/−, no Tam; −/+, induction of HOXB4 in FLK-1-sorted cells; +/+, continuously induced HOXB4. For statistical analysis, an unpaired, two-sided t test with Welch's correction (n = 6) was performed. The p values of individual comparisons are indicated with a significance level defined as p < 0.05. To evaluate HE formation, 10,000 cells of pooled, dissociated colonies were placed onto OP9 stroma cells +/− Tam. The numbers of HE colonies grown on OP9 cells after 3 days are shown.
HOXB4 Induces Hemogenic Endothelial Cells without Affecting Endothelial Cell Development
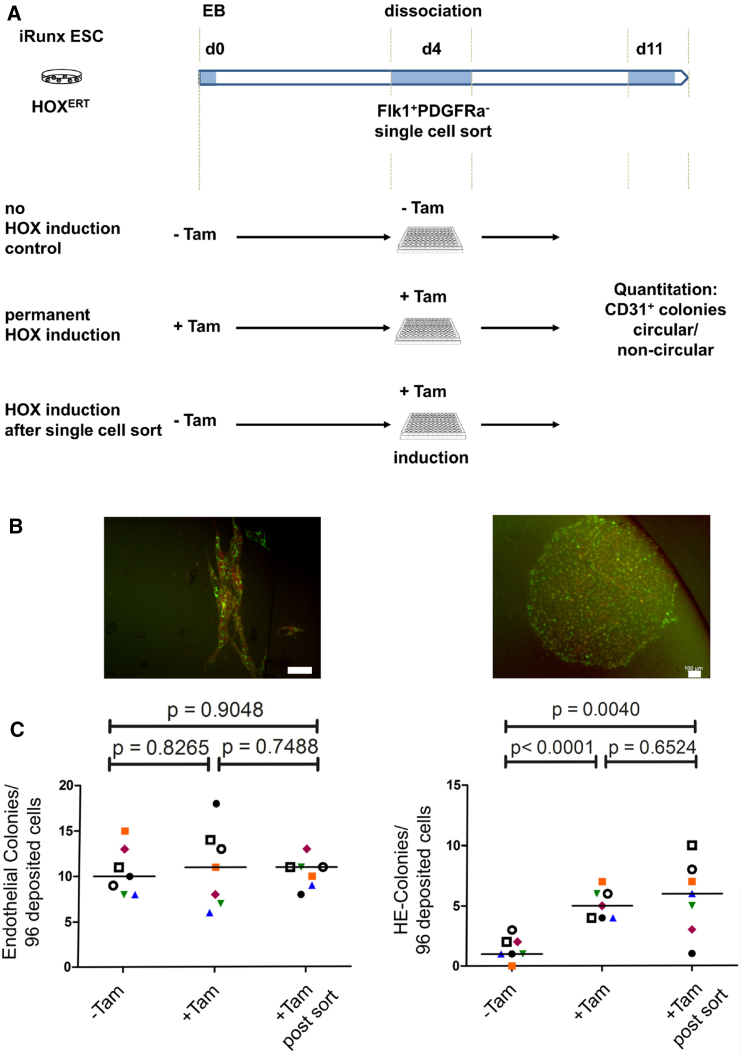
Fate mapping experiments in embryos have suggested that hematopoietic and endothelial cells specify independently from each other or, alternatively, separate early during development (Padron-Barthe et al., 2014, Ueno and Weissman, 2006). Both populations are contained within an FLK-1+PDGFRα− population, in vivo and in vitro, which derives from Etv2+(ER71)+ mesodermal progenitors (Kataoka et al., 1997, Liu et al., 2012, Sakurai et al., 2006, Schatteman et al., 1992, Wareing et al., 2012). To test whether HOXB4 re-specifies endothelial cells toward the hematopoietic fate during early Runx1-independent stages, we deposited single FLK-1+ PDGFRα− cells from d4 iRunx EBs on OP9 stromal cells and determined the frequencies of circular, cobblestone-like CD31+ (Pecam1) hemogenic and of elongated endothelial colonies formed after further 4 days. We then asked whether the frequencies change after induction of HOXB4 (Figure 5A). Irrespective if HOXB4 was induced or not, the frequencies of CD31+ endothelial, strand-like colonies were unchanged (Figures 5B and 5C). In contrast, when HOXB4 was induced, either throughout differentiation or immediately after single-cell deposition, the number of circular HE colonies significantly increased compared with the non-induced control. Possible explanations are that HOXB4 either mediated enhanced proliferation of HE precursors or, alternatively, protected them from apoptosis. To test these possibilities we induced HOXB4 in FLK-1+ cells purified from day 3.5 EBs and subjected them to blast culture conditions. After 24 and 48 hr, we either pulse-labeled replicating DNA with EdU for 2.5 hr or determined the proportion of apoptotic cells by annexin V and DAPI staining. Neither proliferation nor apoptosis of FLK-1+ progenitor cells was altered after induction of HOXB4 (Figure S4).
Figure 5.
HOXB4 Does Not Re-specify Endothelial Cells to a Hematopoietic Fate
(A) iRunx ESCs expressing tamoxifen-inducible HOXB4 (tdTomato-2A-HOXB4ERT) were differentiated as EBs for 4 days, dissociated, and single FLK-1+PDGFRα− cells deposited and co-cultured with OP9 cells in 96-well plates. After a further 4 days, CD31+ tdTomato/HOXB4+ colonies were counted and qualified either as "endothelial colony" or "HE colony."
(B) Typical colony morphologies. Scale bars, 100 μm.
(C) Quantification of endothelial and HE colonies formed by single deposited cells +/− HOXB4 induction. –Tam, without HOXB4ERT induction; +Tam, permanent induction of HOXB4ERT. +Tam post sort, induction after single-cell deposition. The p values as calculated by Student's t test, paired and two-sided, with a significance level defined as 0.05, are indicated above. Forms and colors indicate endothelial and HE colonies grown on the same 96-well plate.
Taken together, the results indicate the following: (1) HOXB4 does not re-specify FLK-1+PDGFRα− endothelial cells toward a hematopoietic fate because, otherwise, the endothelial colony numbers would have decreased after HOXB4 induction. (2) Induction of HOXB4 in single mesodermal FLK-1+ cells does not enhance their proliferative activity or inhibit apoptosis, but instead activates an endogenous program allowing them to form HE colonies and realize their hematopoietic potential as soon as Runx1 is switched on.
The Transcriptional Signature of ESC-Derived Mesodermal Cells Ectopically Expressing HOXB4 Indicates the Acquisition of a Hemogenic Endothelial Fate
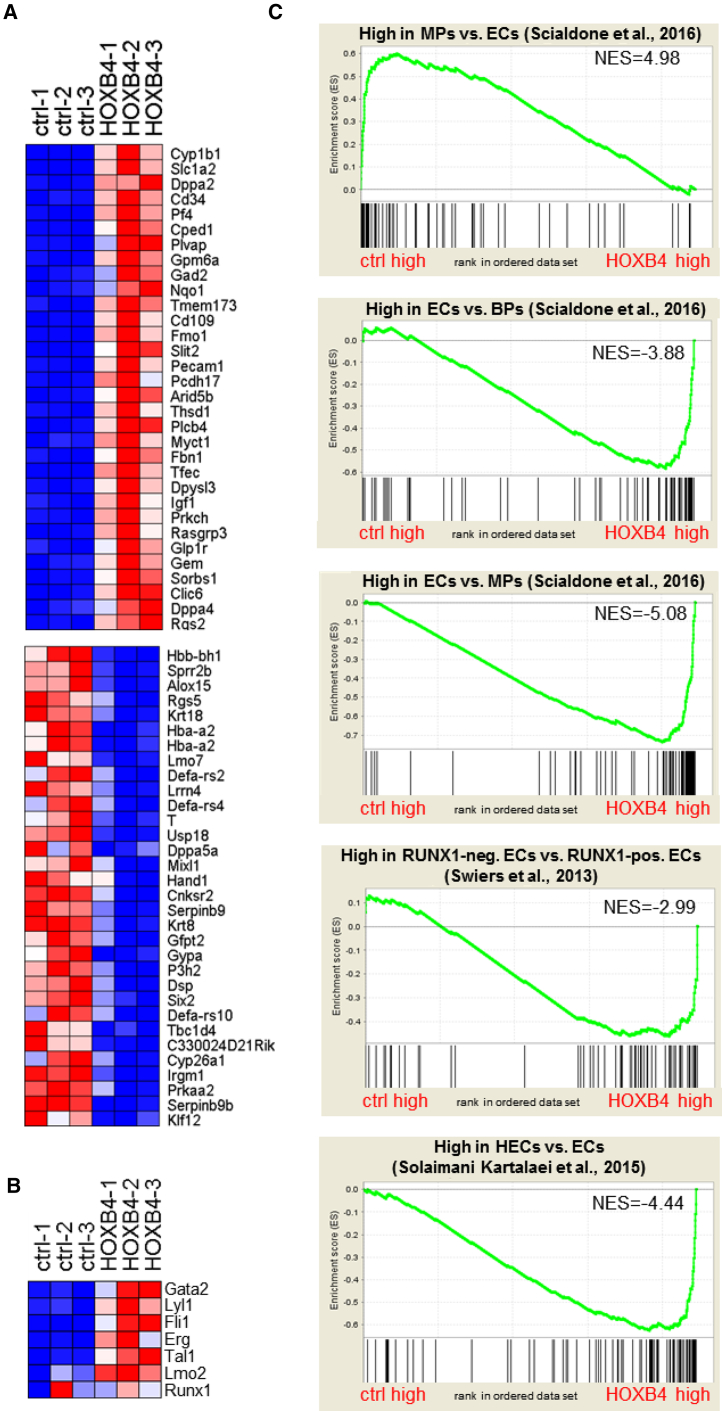
As HOXB4 does not appear to re-specify endothelial cells but, instead, activates a hematogenic program in single FLK-1+PDGFRα− cells, we asked how HOXB4 rewires the transcriptional regulatory circuits during a RUNX1-independent stage of hematopoietic development. To answer that question, we compared the transcriptional profiles of FLK-1+PDGFRα− cell populations purified from iRunx day 4 EBs with and without ectopic HOXB4 expression. Among mRNAs that were upregulated (321), we found transcripts that are necessary for hematopoietic specification, such as Scl/Tal1, Gata2, Fli1, Tie2, Erg, Kit, Egfl7, Lyl1, Myb, Sox7, or Hhex, as well as transcripts encoding hemato-endothelial surface molecules Cdh5 (VE-cadherin), CD34, or CD109 (Figures 6A and 6B; Table S1). Notably, most of the so-called heptad TF-encoding genes, which co-operate to promote hematopoiesis, were significantly upregulated in the HOXB4-iRunx cells (Tal1, Lyl1, Fli1, Gata2, Erg, and Lmo2, except Runx1) (Figure 6B) (Wilson et al., 2010). Downregulated mRNAs (178) comprised transcripts that encode proteins associated with mesodermal and cardiovascular development, such as Hand1, Mixl1, Gata4, Foxf1, Foxh1, Lef1, Etv2, Fgf15, or Tgfb2 (Figure 6A, lower panel; Table S1).
Figure 6.
Gene Expression Profiling of FLK-1+PDGFRα− Cells
(A) Heatmap showing the top 32 genes (fold-change ranked) either up- or downregulated in FLK-1+PDGFRα− cell populations from iRunx day 4 EBs without (ctrl) or with ectopically expressed HOXB4. Genes were considered differentially expressed if they met two criteria: q ≤ 0.2 and fold-change ≥ 2.0 (499 genes) (for full list see Table S1).
(B) Heatmap for the expression of heptad transcription factors.
(C) GSEA using gene sets specific for mesodermal progenitors ([MPs] top panel) compared with endothelial cells (ECs), ECs compared with blood progenitors ([BPs] second panel) and endothelial cells compared with mesodermal progenitors (third panel) (Scialdone et al., 2016); E8.5 RUNX− endothelial cells (RUNX1-neg. ECs) compared with RUNX1+ endothelial cells (RUNX1-pos. ECs, fourth panel) (Swiers et al., 2013) and E10 hemogenic endothelial cells (HECs) compared with endothelial cells (ECs, fifth panel) (Solaimani Kartalaei et al., 2015). Genes were drawn according to their rank from left (high expression in control) to right (high expression in HOXB4-expressing cells) and gene sets plotted on top with each black bar representing a gene. The enrichment score is plotted in on the vertical axis.
To test if these cells displayed a gene expression signature corresponding to a hemato-endothelial precursor, in vivo, we first asked which mesodermal cell type of the gastrulating embryo the iRunx-derived cells +/− HOXB4 most closely relate to molecularly (Scialdone et al., 2016). Gene set enrichment analysis (GSEA) (Subramanian et al., 2005) uncovered that FLK-1+PDGFRα−RUNX1− cells without ectopic HOXB4 most closely resembled a transcriptional signature of early mesodermal progenitors of the neural plate stage (E7.5). In contrast, cells expressing activated HOXB4 displayed a significant enrichment of genes expressed in hemato-endothelial progenitors of the head-fold stage (E7.75) (Figure 6C; Table S2). To further refine the analysis we asked how closely these iRunx-derived cells resembled hemato-endothelial cells of E8.5 transgenic Runx1-reporter (“+23GFP”) mouse embryos (Swiers et al., 2013). GSEA indicated a closer relationship of FLK-1+PDGFRα− EBd4 iRunx cells to 23GFP−/RUNX1− hemato-endothelial cells then to GFP+/RUNX1+ HE cells (Figure 6C; Table S2). This was not surprising, as Runx1 was not induced in the iRunx cells. However, the results point out that ectopic HOXB4 cannot initiate those transcriptional programs critical for subsequent steps of hematopoietic development without the activity of RUNX1. We finally asked whether the transcriptome of HOXB4 cells rather displayed an endothelial or HE signature using gene sets from dorsal aorta cells of E10 transgenic Sca1-reporter (Ly6aGFP) mice (Solaimani Kartalaei et al., 2015). Although Runx1 was not induced in the iRunx cells, GSEA clearly indicated similarity of HOXB4-expressing EBd4 iRunx cells to dorsal aorta-resident HE cells (Figure 6C; Table S2).
Discussion
Two main concepts exist with regard to how the HE is formed during embryonic development. One is based on a dichotomic model in which binary fate decisions happen during cell divisions throughout development. The FLK-1+ hemangioblast, a progenitor cell thought to generate all vascular endothelial and hematopoietic cells fits to the idea (Choi et al., 1998, Lancrin et al., 2009, Nishikawa, 2012). The other possibility is that endothelial and hematopoietic lineages separate during an earlier developmental stage and develop independently of each other. This concept is supported by cell tracing analysis in embryos (Padron-Barthe et al., 2014, Ueno and Weissman, 2006). In that case, the hemangioblast could present a specialized, transiently existing endothelial cell with the competence to acquire a hematopoietic fate upon environmental cues mediated, for example, by Notch or Wnt signaling (Richard et al., 2013).
When ectopically expressing HOXB4 in differentiating iRunx cells, we did not observe a significant change in the numbers of blast colony-forming hemangioblasts. However, when we tested the developmental competence of the progeny of these cells to form HE colonies, we observed a strong increase in their numbers when HOXB4 was induced during blast colony formation of FLK-1+ cells. Hence, HOXB4 appeared to promote the hematopoietic fate in HE precursors downstream of the hemangioblast. Supporting this idea, deposition of single FLK-1+PDGFRα− cells and induction of HOXB4 significantly promoted the formation of circular, cobblestone-like HE colonies, which underwent EHT after induction of Runx1. Interestingly, the number of cord-formed endothelial colonies was unchanged, irrespective if HOXB4 was activated or not. If HOXB4 would directly act as a fate determinant in hemangioblasts, the increase of HE colonies must have been accompanied by a concomitant decrease in the number of endothelial colonies. However, that was not the case, thus supporting the interpretation that HOXB4 acts after the hemangioblast stage in precursors of the HE.
Genome-wide transcriptional profiling revealed that HOXB4 induced the expression of a plethora of genes essential for hematopoiesis. Particularly, most of the heptad-TF genes were upregulated during early differentiation of iRunx-ESCs (except Runx1). The encoded TFs are known to co-operatively regulate genes essential for hematopoietic development (Wilson et al., 2010). Importantly, HOXB4 has previously been shown to physically bind upstream of all of these seven genes (Fan et al., 2012). For example, it directly binds to the conserved +85 kb enhancer element of Erg (Moignard et al., 2015, Wilson et al., 2009), which is also bound by most of the heptad TFs (Schutte et al., 2016). Furthermore, HOXB4 binds to DNA in close proximity to the TFs FLI-1, MEIS1, RUNX1, and SCL (Fan et al., 2012), strongly implying its participation in a TF complex regulating common hematopoietic target genes. GSEA with gene sets derived from hematopoietic tissues that had been isolated by different means and different stages (E7.5–E10) of embryonic development (Scialdone et al., 2016, Solaimani Kartalaei et al., 2015, Swiers et al., 2013) indicated that HOXB4+FLK-1+PDGFRα− cells most closely resembled HE cells of the dorsal aorta in E10 embryos. Given that endogenous mouse HoxB4 is also expressed at low levels around this stage, in vivo (Moignard et al., 2015), ectopic overexpression of human HOXB4 in differentiating ESCs may simply represent a fortification of embryonic hematopoiesis.
Our results with HOXB4 may be paradigmatic for the activities of all HOX4 paralogs, as ectopic expression of the other orthologs HOXA4, C4, and D4 similarly enhanced hematopoietic development of differentiating ESCs (Iacovino et al., 2009). HOXB4 directly binds to a region between the proximal (P2) and distal (P1) promoters of Runx1, which correlates with demethylation and activation of the P1 promoter (Webber et al., 2013). This may probably explain our observation of Runx1 upregulation in a small subpopulation of ESC-derived HE cells, in vitro (Teichweyde et al., 2017). How homeotic selector proteins mechanistically influence cell fate decisions during development is not well understood yet. It appears that their activities as activators or repressors of transcription depend on their expression levels, the cell type they are expressed in, and probably also the microenvironment of the cells (Klump et al., 2005, Pilat et al., 2005, Schiedlmeier et al., 2003, Schiedlmeier et al., 2007, Will et al., 2006). It could be envisioned that these master switches of development integrate signaling events allowing for the realization of certain genetic programs by epigenetically promoting enhancer activation or decommission.
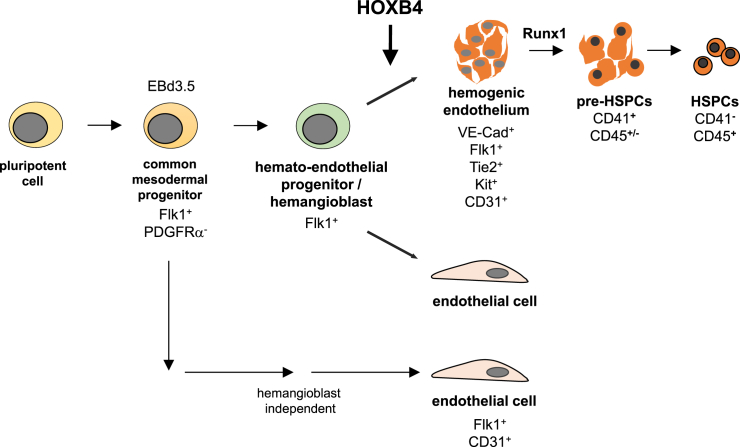
In the present work we have shown that HOXB4 first unfolds its hematopoiesis-promoting activity in HE precursors (summarized in Figure 7). By inducing a hematopoietic program in those cells, HOXB4 promotes the acquisition of an HE identity and, thus, enforces early hematopoietic development. It remains to be shown if this also holds true for differentiating human pluripotent stem cells, as this would open the possibility to enhance the generation of patient-specific HSPCs. In fact, a recent report strongly suggests that ectopic HOXB4 expression also promotes hematopoietic development of human ESCs (Jackson et al., 2016), therefore supporting the idea of what is true for mouse hematopoietic development also holds true for humans.
Figure 7.
Model of HOXB4 Activity during Pluripotent Stem Cell Differentiation
During mouse ESC differentiation, HOXB4 appears to promote hematopoietic commitment after the hemangioblast stage in VE-Cad+FLK-1+TIE2+KIT+CD31+ precursors of HE cells by upregulating the expression of key hematopoietic transcription factors, in a RUNX1-independent fashion. Importantly, our results indicate that this does not happen at the expense of endothelial cells. These results are also compatible with the notion that (at least a part of) endothelial cells develop hemangioblast-independently (Padron-Barthe et al., 2014, Ueno and Weissman, 2006).
Experimental Procedures
For production of retroviral vectors, ESC culture, hematopoietic differentiation, mouse transplantation, colony-forming assays, gene expression analysis, and time-lapse microscopy, see Supplemental Information. All animal experiments were approved by the Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen (LANUV) (reference number 84-02.04.2013.A350) and were performed according to official guidelines and regulations.
Blast Colony-Forming Assays
Blast colony assays were performed as described (Kennedy and Keller, 2003). In brief, 5 × 105 ESCs were differentiated as EBs for 3.5 days, dissociated and FLK-1+ cells sorted by flow cytometry (FACSAria III, BD Biosciences). Cells were subsequently cultured at a concentration of 5 × 104 cells/mL for 4–5 days in IMDM containing 10% (v/v) fetal calf serum (GE Healthcare), 2 mM L-glutamine, 20% (v/v) D4T conditioned medium, 1% (v/v) penicillin/streptomycin, 4.5 × 10−4 M MTG, 25 ng/mL ascorbic acid, 300 μg/mL holotransferrin (Sigma-Aldrich), 10 ng/mL rhIL-6, 5 ng/mL rhVEGF, 25% methylcellulose solution, under hypoxic conditions (3% O2) at 37°C in a H2O saturated atmosphere.
Tube Formation Assay
To assess the angiogenic activity of cells contained within blast colonies, 4 × 104 cells were cultured in μ-Slide Angiogenesis chambers (ibidi) filled with 10 μL Matrigel matrix (Corning Life Sciences) for 19 hr at 37°C in H2O saturated atmosphere, 3% O2, 5% CO2.
Flow Cytometry Analysis and Cell Sorting
Differentiating ESCs, bone marrow and peripheral blood of transplanted mice were analyzed by flow cytometry using fluorochrome-conjugated monoclonal antibodies. Details regarding employed antibodies (specificities, manufacturer, catalog numbers) are listed in the Supplemental Experimental Procedures section.
For FACS analysis, dead cells were excluded by DAPI staining. Gates were set based on control samples stained according to the Fluorescence Minus One method. Flow cytometry measurements and single-cell sorting were performed on a FACSAria III instrument (BD Biosciences) using FACSDiva software, datasets were analyzed using FlowJo software (Tree Star).
Magnet-Activated Cell Sorting
For removal of stroma cells, magnet-activated cell sorting (MACS) technology was used (Miltenyi). Harvested cells were incubated either with Feeder Removal MicroBeads (for removal of CF1-MEFs) or with an anti-CD140b PE-labeled mAB (OP9) and anti-PE microbeads, and subsequently depleted with MACS LS Columns, according to the manufacturer's recommendations.
Transcriptome Analysis
iRunx EBd4 were dissociated and FLK-1+PDFGRα− cells with or without retroviral expression of tdTomato/HOXB4 were sorted by FACS. Total RNA was extracted using TRIzol and subsequently purified with RNeasy columns (QIAGEN). Each sample (500 ng) was converted into fragmented, biotinylated cDNA hybridized to a microarray chip (Clariome D MTA 1.0) and fluorescently labeled according to the standard protocol (Affymetrix, Santa Clara, CA). Raw data were processed in Expression Console (Affymetrix) using RMA normalization and expression values calculated for each Refseq-annotated gene (23,781 total). The expression data were processed in Gene Pattern (Broad Institute, Cambridge, MA). Non-expressed genes were filtered out, and the resulting expression matrix was analyzed with the comparative marker module. GSEA (Broad Institute) was performed as described previously (Subramanian et al., 2005). Genes sets (Table S2) were derived from associated publications as indicated in the text.
Statistical Analysis
Significance testing of differences between mean values was performed by calculating p values using the two-sided unpaired, Student's t test. Significance levels were defined as p < 0.05. For all calculations, GraphPad Prism6 software (GraphPad, CA, USA) was used.
Author Contributions
Conceptualization, H.K.; Methodology, N.T., L.K., S.H., S.C., G.L., V.K., and H.K.; Formal Analysis, N.T., L.K., and S.H.; Investigation, N.T. and L.K.; Resources, G.L. and V.K.; Data Curation, N.T., L.K., S.H., and H.K.; Writing – Original Draft, H.K.; Writing – Review & Editing, N.T., L.K., S.H., S.C., G.L., V.K., P.A.H., and H.K.; Visualization, N.T., L.K., S.H., and H.K.; Supervision, H.K.; Project Administration, H.K.; Funding Acquisition, H.K. and P.A.H.
Acknowledgments
We wish to thank Jörg Fehling (Institute for Immunology, University of Ulm, Germany) for providing the GFP-Bry mouse reporter ESCs, Samantha Langer and Martina Cremanns for their help during mouse transplantation experiments, and Antje Kleinbielen and Susanne Skibbe for excellent technical support. This work was supported by the Deutsche Forschungsgemeinschaft (DFG grants KL1311/5-1 and KL1311/8-1) and the Else Kröner-Fresenius-Stiftung (support of ELAN graduate program student L.K.). Research in G.L.’s laboratory is supported by the Biotechnology and Biological Sciences Research Council (BB/I001794/1), Bloodwise (12037), the European Union’s Horizon 2020 (GA6586250), and Cancer Research UK (C5759/A20971). Research in V.K.’s laboratory is supported by the Medical Research Council (MR/P000673/1) and the Biotechnology and Biological Sciences Research Council (BB/I001794/1).
Published: February 15, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, two tables, and two movies and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.01.009.
Accession Numbers
The accession number for the microarray data reported in this paper is GEO: GSE103627.
Supplemental Information
References
- Abate-Shen C. Deregulated homeobox gene expression in cancer: cause or consequence? Nat. Rev. Cancer. 2002;2:777–785. doi: 10.1038/nrc907. [DOI] [PubMed] [Google Scholar]
- Arnaoutova I., Kleinman H.K. In vitro angiogenesis: endothelial cell tube formation on gelled basement membrane extract. Nat. Protoc. 2010;5:628–635. doi: 10.1038/nprot.2010.6. [DOI] [PubMed] [Google Scholar]
- Azcoitia V., Aracil M., Martínez A.C., Torres M. The homeodomain protein Meis1 is essential for definitive hematopoiesis and vascular patterning in the mouse embryo. Dev. Biol. 2005;280:307–320. doi: 10.1016/j.ydbio.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Bertrand J.Y., Chi N.C., Santoso B., Teng S., Stainier D.Y., Traver D. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 2010;464:108–111. doi: 10.1038/nature08738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloor A.J., Sanchez M.J., Green A.R., Gottgens B. The role of the stem cell leukemia (SCL) gene in hematopoietic and endothelial lineage specification. J. Hematother. Stem Cell Res. 2002;11:195–206. doi: 10.1089/152581602753658402. [DOI] [PubMed] [Google Scholar]
- Chan K.M., Bonde S., Klump H., Zavazava N. Hematopoiesis and immunity of HOXB4-transduced embryonic stem cell-derived hematopoietic progenitor cells. Blood. 2008;111:2953–2961. doi: 10.1182/blood-2007-10-117366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K., Kennedy M., Kazarov A., Papadimitriou J.C., Keller G. A common precursor for hematopoietic and endothelial cells. Development. 1998;125:725–732. doi: 10.1242/dev.125.4.725. [DOI] [PubMed] [Google Scholar]
- Eilken H.M., Nishikawa S., Schroeder T. Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature. 2009;457:896–900. doi: 10.1038/nature07760. [DOI] [PubMed] [Google Scholar]
- Ema M., Faloon P., Zhang W.J., Hirashima M., Reid T., Stanford W.L., Orkin S., Choi K., Rossant J. Combinatorial effects of Flk1 and Tal1 on vascular and hematopoietic development in the mouse. Genes Dev. 2003;17:380–393. doi: 10.1101/gad.1049803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faloon P., Arentson E., Kazarov A., Deng C.X., Porcher C., Orkin S., Choi K. Basic fibroblast growth factor positively regulates hematopoietic development. Development. 2000;127:1931–1941. doi: 10.1242/dev.127.9.1931. [DOI] [PubMed] [Google Scholar]
- Fan R., Bonde S., Gao P., Sotomayor B., Chen C., Mouw T., Zavazava N., Tan K. Dynamic HoxB4-regulatory network during embryonic stem cell differentiation to hematopoietic cells. Blood. 2012;119:e139–147. doi: 10.1182/blood-2011-12-396754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehling H.J., Lacaud G., Kubo A., Kennedy M., Robertson S., Keller G., Kouskoff V. Tracking mesoderm induction and its specification to the hemangioblast during embryonic stem cell differentiation. Development. 2003;130:4217–4227. doi: 10.1242/dev.00589. [DOI] [PubMed] [Google Scholar]
- Gritz E., Hirschi K.K. Specification and function of hemogenic endothelium during embryogenesis. Cell. Mol. Life Sci. 2016;73:1547–1567. doi: 10.1007/s00018-016-2134-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacovino M., Hernandez C., Xu Z., Bajwa G., Prather M., Kyba M. A conserved role for Hox paralog group 4 in regulation of hematopoietic progenitors. Stem Cells Dev. 2009;18:783–792. doi: 10.1089/scd.2008.0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H., Somoza C., Shigematsu H., Duprez E.A., Iwasaki-Arai J., Mizuno S., Arinobu Y., Geary K., Zhang P., Dayaram T. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood. 2005;106:1590–1600. doi: 10.1182/blood-2005-03-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M., Axton R.A., Taylor A.H., Wilson J.A., Gordon-Keylock S.A., Kokkaliaris K.D., Brickman J.M., Schulz H., Hummel O., Hubner N. HOXB4 can enhance the differentiation of embryonic stem cells by modulating the hematopoietic niche. Stem Cells. 2012;30:150–160. doi: 10.1002/stem.782. [DOI] [PubMed] [Google Scholar]
- Jackson M., Ma R., Taylor A.H., Axton R.A., Easterbrook J., Kydonaki M., Olivier E., Marenah L., Stanley E.G., Elefanty A.G. Enforced expression of HOXB4 in human embryonic stem cells enhances the production of hematopoietic progenitors but has no effect on the maturation of red blood cells. Stem Cells Transl. Med. 2016;5:981–990. doi: 10.5966/sctm.2015-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffredo T., Gautier R., Eichmann A., Dieterlen-Lievre F. Intraaortic hemopoietic cells are derived from endothelial cells during ontogeny. Development. 1998;125:4575–4583. doi: 10.1242/dev.125.22.4575. [DOI] [PubMed] [Google Scholar]
- Kataoka H., Hayashi M., Nakagawa R., Tanaka Y., Izumi N., Nishikawa S., Jakt M.L., Tarui H., Nishikawa S. Etv2/ER71 induces vascular mesoderm from Flk1+PDGFRalpha+ primitive mesoderm. Blood. 2011;118:6975–6986. doi: 10.1182/blood-2011-05-352658. [DOI] [PubMed] [Google Scholar]
- Kataoka H., Takakura N., Nishikawa S., Tsuchida K., Kodama H., Kunisada T., Risau W., Kita T., Nishikawa S.I. Expressions of PDGF receptor alpha, c-Kit and Flk1 genes clustering in mouse chromosome 5 define distinct subsets of nascent mesodermal cells. Dev. Growth Differ. 1997;39:729–740. doi: 10.1046/j.1440-169x.1997.t01-5-00009.x. [DOI] [PubMed] [Google Scholar]
- Kennedy M., Firpo M., Choi K., Wall C., Robertson S., Kabrun N., Keller G. A common precursor for primitive erythropoiesis and definitive haematopoiesis. Nature. 1997;386:488–493. doi: 10.1038/386488a0. [DOI] [PubMed] [Google Scholar]
- Kennedy M., Keller G.M. Hematopoietic commitment of ES cells in culture. Methods Enzymol. 2003;365:39–59. doi: 10.1016/s0076-6879(03)65003-2. [DOI] [PubMed] [Google Scholar]
- Klump H., Schiedlmeier B., Baum C. Control of self-renewal and differentiation of hematopoietic stem cells: HOXB4 on the threshold. Ann. N. Y. Acad. Sci. 2005;1044:6–15. doi: 10.1196/annals.1349.002. [DOI] [PubMed] [Google Scholar]
- Klump H., Teichweyde N., Meyer C., Horn P.A. Development of patient-specific hematopoietic stem and progenitor cell grafts from pluripotent stem cells, in vitro. Curr. Mol. Med. 2013;13:815–820. doi: 10.2174/1566524011313050012. [DOI] [PubMed] [Google Scholar]
- Kyba M., Perlingeiro R.C., Daley G.Q. HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell. 2002;109:29–37. doi: 10.1016/s0092-8674(02)00680-3. [DOI] [PubMed] [Google Scholar]
- Lacaud G., Gore L., Kennedy M., Kouskoff V., Kingsley P., Hogan C., Carlsson L., Speck N., Palis J., Keller G. Runx1 is essential for hematopoietic commitment at the hemangioblast stage of development in vitro. Blood. 2002;100:458–466. doi: 10.1182/blood-2001-12-0321. [DOI] [PubMed] [Google Scholar]
- Lancrin C., Mazan M., Stefanska M., Patel R., Lichtinger M., Costa G., Vargel O., Wilson N.K., Moroy T., Bonifer C. GFI1 and GFI1B control the loss of endothelial identity of hemogenic endothelium during hematopoietic commitment. Blood. 2012;120:314–322. doi: 10.1182/blood-2011-10-386094. [DOI] [PubMed] [Google Scholar]
- Lancrin C., Sroczynska P., Stephenson C., Allen T., Kouskoff V., Lacaud G. The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature. 2009;457:892–895. doi: 10.1038/nature07679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesinski D.A., Heinz N., Pilat-Carotta S., Rudolph C., Jacobs R., Schlegelberger B., Klump H., Schiedlmeier B. Serum- and stromal cell-free hypoxic generation of embryonic stem cell-derived hematopoietic cells in vitro, capable of multilineage repopulation of immunocompetent mice. Stem Cells Transl. Med. 2012;1:581–591. doi: 10.5966/sctm.2012-0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Bhang S.H., Arentson E., Sawada A., Kim C.K., Kang I., Yu J., Sakurai N., Kim S.H., Yoo J.J. Enhanced hemangioblast generation and improved vascular repair and regeneration from embryonic stem cells by defined transcription factors. Stem Cell Reports. 2013;1:166–182. doi: 10.1016/j.stemcr.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Kang I., Park C., Chang L.W., Wang W., Lee D., Lim D.S., Vittet D., Nerbonne J.M., Choi K. ER71 specifies Flk-1+ hemangiogenic mesoderm by inhibiting cardiac mesoderm and Wnt signaling. Blood. 2012;119:3295–3305. doi: 10.1182/blood-2012-01-403766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Li D., Yu Y.Y., Kang I., Cha M.J., Kim J.Y., Park C., Watson D.K., Wang T., Choi K. Induction of hematopoietic and endothelial cell program orchestrated by ETS transcription factor ER71/ETV2. EMBO Rep. 2015;16:654–669. doi: 10.15252/embr.201439939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y.F., Cahan P., Ross S., Sahalie J., Sousa P.M., Hadland B.K., Cai W., Serrao E., Engelman A.N., Bernstein I.D. Engineered murine HSCs reconstitute multi-lineage hematopoiesis and adaptive immunity. Cell Rep. 2016;17:3178–3192. doi: 10.1016/j.celrep.2016.11.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney-Freeman S.L., Naveiras O., Yates F., Loewer S., Philitas M., Curran M., Park P.J., Daley G.Q. Surface antigen phenotypes of hematopoietic stem cells from embryos and murine embryonic stem cells. Blood. 2009;114:268–278. doi: 10.1182/blood-2008-12-193888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moignard V., Woodhouse S., Haghverdi L., Lilly A.J., Tanaka Y., Wilkinson A.C., Buettner F., Macaulay I.C., Jawaid W., Diamanti E. Decoding the regulatory network of early blood development from single-cell gene expression measurements. Nat. Biotechnol. 2015;33:269–276. doi: 10.1038/nbt.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S. Hemangioblast: an in vitro phantom. Wiley Interdiscip. Rev. Dev. Biol. 2012;1:603–608. doi: 10.1002/wdev.38. [DOI] [PubMed] [Google Scholar]
- Nishikawa S.I., Nishikawa S., Hirashima M., Matsuyoshi N., Kodama H. Progressive lineage analysis by cell sorting and culture identifies FLK1+VE-cadherin+ cells at a diverging point of endothelial and hemopoietic lineages. Development. 1998;125:1747–1757. doi: 10.1242/dev.125.9.1747. [DOI] [PubMed] [Google Scholar]
- North T., Gu T.L., Stacy T., Wang Q., Howard L., Binder M., Marin-Padilla M., Speck N.A. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development. 1999;126:2563–2575. doi: 10.1242/dev.126.11.2563. [DOI] [PubMed] [Google Scholar]
- Oshima M., Endoh M., Endo T.A., Toyoda T., Nakajima-Takagi Y., Sugiyama F., Koseki H., Kyba M., Iwama A., Osawa M. Genome-wide analysis of target genes regulated by HoxB4 in hematopoietic stem and progenitor cells developing from embryonic stem cells. Blood. 2011;117:e142–e150. doi: 10.1182/blood-2010-12-323212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padron-Barthe L., Temino S., Villa del Campo C., Carramolino L., Isern J., Torres M. Clonal analysis identifies hemogenic endothelium as the source of the blood-endothelial common lineage in the mouse embryo. Blood. 2014;124:2523–2532. doi: 10.1182/blood-2013-12-545939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson S., Cuvertino S., Fleury M., Lacaud G., Kouskoff V. In vivo repopulating activity emerges at the onset of hematopoietic specification during embryonic stem cell differentiation. Stem Cell Reports. 2015;4:431–444. doi: 10.1016/j.stemcr.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilat S., Carotta S., Schiedlmeier B., Kamino K., Mairhofer A., Will E., Modlich U., Steinlein P., Ostertag W., Baum C. HOXB4 enforces equivalent fates of ES-cell-derived and adult hematopoietic cells. Proc. Natl. Acad. Sci. USA. 2005;102:12101–12106. doi: 10.1073/pnas.0505624102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard C., Drevon C., Canto P.Y., Villain G., Bollérot K., Lempereur A., Teillet M.A., Vincent C., Rosselló Castillo C., Torres M. Endothelio-mesenchymal interaction controls runx1 expression and modulates the notch pathway to initiate aortic hematopoiesis. Dev. Cell. 2013;24:600–611. doi: 10.1016/j.devcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai H., Era T., Jakt L.M., Okada M., Nakai S., Nishikawa S., Nishikawa S. In vitro modeling of paraxial and lateral mesoderm differentiation reveals early reversibility. Stem Cells. 2006;24:575–586. doi: 10.1634/stemcells.2005-0256. [DOI] [PubMed] [Google Scholar]
- Schatteman G.C., Morrison-Graham K., van Koppen A., Weston J.A., Bowen-Pope D.F. Regulation and role of PDGF receptor alpha-subunit expression during embryogenesis. Development. 1992;115:123–131. doi: 10.1242/dev.115.1.123. [DOI] [PubMed] [Google Scholar]
- Schiedlmeier B., Klump H., Will E., Arman-Kalcek G., Li Z., Wang Z., Rimek A., Friel J., Baum C., Ostertag W. High-level ectopic HOXB4 expression confers a profound in vivo competitive growth advantage on human cord blood CD34+ cells, but impairs lymphomyeloid differentiation. Blood. 2003;101:1759–1768. doi: 10.1182/blood-2002-03-0767. [DOI] [PubMed] [Google Scholar]
- Schiedlmeier B., Santos A.C., Ribeiro A., Moncaut N., Lesinski D., Auer H., Kornacker K., Ostertag W., Baum C., Mallo M. HOXB4's road map to stem cell expansion. Proc. Natl. Acad. Sci. USA. 2007;104:16952–16957. doi: 10.1073/pnas.0703082104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutte J., Wang H., Antoniou S., Jarratt A., Wilson N.K., Riepsaame J., Calero-Nieto F.J., Moignard V., Basilico S., Kinston S.J. An experimentally validated network of nine haematopoietic transcription factors reveals mechanisms of cell state stability. Elife. 2016;5:e11469. doi: 10.7554/eLife.11469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scialdone A., Tanaka Y., Jawaid W., Moignard V., Wilson N.K., Macaulay I.C., Marioni J.C., Gottgens B. Resolving early mesoderm diversification through single-cell expression profiling. Nature. 2016;535:289–293. doi: 10.1038/nature18633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solaimani Kartalaei P., Yamada-Inagawa T., Vink C.S., de Pater E., van der Linden R., Marks-Bluth J., van der Sloot A., van den Hout M., Yokomizo T., van Schaick-Solerno M.L. Whole-transcriptome analysis of endothelial to hematopoietic stem cell transition reveals a requirement for Gpr56 in HSC generation. J. Exp. Med. 2015;212:93–106. doi: 10.1084/jem.20140767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiers G., Baumann C., O'Rourke J., Giannoulatou E., Taylor S., Joshi A., Moignard V., Pina C., Bee T., Kokkaliaris K.D. Early dynamic fate changes in haemogenic endothelium characterized at the single-cell level. Nat. Commun. 2013;4:2924. doi: 10.1038/ncomms3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoudi S., Gonneau C., Moore K., Sheridan J.M., Blackburn C.C., Taylor E., Medvinsky A. Extensive hematopoietic stem cell generation in the AGM region via maturation of VE-cadherin+CD45+ pre-definitive HSCs. Cell Stem Cell. 2008;3:99–108. doi: 10.1016/j.stem.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Tavian M., Robin C., Coulombel L., Péault B. The human embryo, but not its yolk sac, generates lympho-myeloid stem cells: mapping multipotent hematopoietic cell fate in intraembryonic mesoderm. Immunity. 2001;15:487–495. doi: 10.1016/s1074-7613(01)00193-5. [DOI] [PubMed] [Google Scholar]
- Teichweyde N., Horn P.A., Klump H. HOXB4 increases Runx1 expression to promote the de novo formation of multipotent hematopoietic cells. Transfus. Med. Hemother. 2017;44:128–134. doi: 10.1159/000477130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thambyrajah R., Mazan M., Patel R., Moignard V., Stefanska M., Marinopoulou E., Li Y., Lancrin C., Clapes T., Moroy T. GFI1 proteins orchestrate the emergence of haematopoietic stem cells through recruitment of LSD1. Nat. Cell Biol. 2016;18:21–32. doi: 10.1038/ncb3276. [DOI] [PubMed] [Google Scholar]
- Ueno H., Weissman I.L. Clonal analysis of mouse development reveals a polyclonal origin for yolk sac blood islands. Dev. Cell. 2006;11:519–533. doi: 10.1016/j.devcel.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Wahlster L., Daley G.Q. Progress towards generation of human haematopoietic stem cells. Nat. Cell Biol. 2016;18:1111–1117. doi: 10.1038/ncb3419. [DOI] [PubMed] [Google Scholar]
- Wang Y., Yates F., Naveiras O., Ernst P., Daley G.Q. Embryonic stem cell-derived hematopoietic stem cells. Proc. Natl. Acad. Sci. USA. 2005;102:19081–19086. doi: 10.1073/pnas.0506127102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wareing S., Mazan A., Pearson S., Gottgens B., Lacaud G., Kouskoff V. The Flk1-Cre-mediated deletion of ETV2 defines its narrow temporal requirement during embryonic hematopoietic development. Stem Cells. 2012;30:1521–1531. doi: 10.1002/stem.1115. [DOI] [PubMed] [Google Scholar]
- Webber B.R., Iacovino M., Choi S.H., Tolar J., Kyba M., Blazar B.R. DNA methylation of Runx1 regulatory regions correlates with transition from primitive to definitive hematopoietic potential in vitro and in vivo. Blood. 2013;122:2978–2986. doi: 10.1182/blood-2013-03-489369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will E., Speidel D., Wang Z., Ghiaur G., Rimek A., Schiedlmeier B., Williams D.A., Baum C., Ostertag W., Klump H. HOXB4 inhibits cell growth in a dose-dependent manner and sensitizes cells towards extrinsic cues. Cell Cycle. 2006;5:14–22. doi: 10.4161/cc.5.1.2304. [DOI] [PubMed] [Google Scholar]
- Wilson N.K., Foster S.D., Wang X., Knezevic K., Schutte J., Kaimakis P., Chilarska P.M., Kinston S., Ouwehand W.H., Dzierzak E. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Wilson N.K., Miranda-Saavedra D., Kinston S., Bonadies N., Foster S.D., Calero-Nieto F., Dawson M.A., Donaldson I.J., Dumon S., Frampton J. The transcriptional program controlled by the stem cell leukemia gene Scl/Tal1 during early embryonic hematopoietic development. Blood. 2009;113:5456–5465. doi: 10.1182/blood-2009-01-200048. [DOI] [PubMed] [Google Scholar]
- Zovein A.C., Turlo K.A., Ponec R.M., Lynch M.R., Chen K.C., Hofmann J.J., Cox T.C., Gasson J.C., Iruela-Arispe M.L. Vascular remodeling of the vitelline artery initiates extravascular emergence of hematopoietic clusters. Blood. 2010;116:3435–3444. doi: 10.1182/blood-2010-04-279497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.