Abstract
Identifying the signals of artificial selection can contribute to further shaping economically important traits. Here, a chicken 600k SNP-array was employed to detect the signals of artificial selection using 331 individuals from 9 breeds, including Jingfen (JF), Jinghong (JH), Araucanas (AR), White Leghorn (WL), Pekin-Bantam (PB), Shamo (SH), Gallus-Gallus-Spadiceus (GA), Rheinlander (RH) and Vorwerkhuhn (VO). Per the population genetic structure, 9 breeds were combined into 5 breed-pools, and a ‘two-step’ strategy was used to reveal the signals of artificial selection. GA, which has little artificial selection, was defined as the reference population, and a total of 204, 155, 305 and 323 potential artificial selection signals were identified in AR_VO, PB, RH_WL and JH_JF, respectively. We also found signals derived from standing and de-novo genetic variations have contributed to adaptive evolution during artificial selection. Further enrichment analysis suggests that the genomic regions of artificial selection signals harbour genes, including THSR, PTHLH and PMCH, responsible for economic traits, such as fertility, growth and immunization. Overall, this study found a series of genes that contribute to the improvement of chicken breeds and revealed the genetic mechanisms of adaptive evolution, which can be used as fundamental information in future chicken functional genomics study.
Introduction
Chickens, like all other domestic animals, have a long history of artificial selection that dates back for centuries [1, 2]. Although the genetic information responsible for many notable changes in simple Mendelian traits have been successfully mapped, genetic signals for most economic traits that are modified during artificial selection are still unclear [3]. If successful, the detection of artificial selection signals, which is a central focus in the study of population genetics, can contribute to mapping the causal mechanisms related to economic traits in the genome and provide a clear understanding of how diversity evolved in various chicken breeds.
During domestication and subsequent commercial breeding, a variety of detectable selection signals were maintained in the genome [1, 4, 5]. Among them, signals shaped by artificial selection are consistently associated with some important economic traits during the process of breed formation. Given that genomic regions have a long-term divergent (bi-directional) selection, the extreme genetic differentiation in the respective regions will be generated and the corresponding allele frequency spectra will depart from what is expected under neutral conditions [6]. This is analogous to the diversity among domesticated, local and wild populations. In the twentieth century, domestic chickens have been refined to improve specialized economic traits for applications in modern breeding technologies based on quantitative genetics [1, 7]. Then, more and more commercial chicken lines, including both layer and broiler lines, have been bred with similar selection directions, which may share similar artificial selection evidence in specific genomic regions [2]. This genetic homogeneity was particularly promising in lines bred for similar purposes and selection pressure. Hence, they are excellent models for characterizing the genomic response to artificial selection of multiple populations.
With the recent implementation of high-throughput genotyping techniques for many populations, identifying selection signals at the genome level has become possible. In addition, there have also been many studies on statistical tests used for detecting selection signals based on different models [8–10]. The general model in which de-novo mutations arise and were selected to fixation within a short time frame is well known as hard sweep [7, 11–13]. In fact, a large number of identified signals with major effects can be categorized into this type of selection signal [7]. In addition to hard sweep, another selection signal model that signals for standing variation or reoccurring novel mutations at a locus has gradually gathered interest. These selection signals, known as soft sweeps, contrast with hard sweeps [7, 11–13]. So far, publications on soft sweep appear to be rare, but soft sweeps are likely to be prevalent in domestic animals, because they have almost no time to develop de novo mutations that mid-small effects that occur during the short process of artificial selection. In general, commercial traits due to human-driven selection are quantitative and have a polygenic basis, and they also respond rapidly under strong selection pressure [7]. Therefore, there is a good reason to believe that both soft sweep and hard sweep modes play an important role in the commercial breeding of economic traits.
In this study, we identified artificial selection signals using genome wide data from multiple chicken populations. In addition, we also tried to distinguish the potential artificial selection signals that are based on hard sweep from those resulting from soft sweep, and we found that the number of soft sweeps is greater than that of hard sweeps in the pool of artificial selection signals.
Materials and methods
Animals, genotyping data and quality control
All research involving animals was conducted under protocols (No. 5 proclaim of the Standing Committee of Hubei People’s Congress) approved by the Standing Committee of Hubei People’s Congress and the ethics committee of Huazhong Agricultural University in P. R. China. In addition, all experiments were performed in accordance with approved relevant guidelines and regulations. Genotypes obtained with the Affymetrix chicken 600 K Axiom-SNP-array were from a total of 331 individual chickens from nine different breeds, which included 110 Jingfen (JF), 82 Jinghong (JH), 12 Araucanas (AR), 16 White Leghorn (WL), 40 Pekin-Bantam (PB), 19 Shamo (SH), 16 Gallus-Gallus-Spadiceus (GA), 20 Rhinelander (RH) and 16 Vorwerkhuhn (VO). The breeds used have a wide geographical distribution and almost every major chicken category was represented, including layer, local breed, and chickens from wild population (Table 1). As typical representatives of the commercial line in this study, Jingfen and Jinghong chickens were bred in China to further improve egg-laying traits and have similar foundation stocks.
Table 1. Samples, origin, specialization and some characteristics of nine chicken populations used in this study.
| Breed (abbreviation) | No. of SNPs1 | Sample Size | Origin of samples | Specialization | Dwarf/ Normal | The other Breed Characteristics |
|---|---|---|---|---|---|---|
| Vorwerkhuhn (VO) | 392,280 | 16 | Germany | Local breed | Normal | Distinctive black and gold plumage; |
| Araucanas (AR) | 392,280 | 12 | Germany | Local breed | Normal | Blue eggs; |
| White Leghorn (WL) | 392,280 | 16 | Germany | Layer line | Normal | White shell; High production; |
| Pekin-Bantam (PB) | 392,280 | 40 | Germany | Local breed | Dwarf | Bantamized Cochin breed; |
| Rhinelander (RH) | 392,280 | 20 | Germany | Local breed | Normal | Medium size breed; mainly an exhibition breed today but originally used for egg-laying. |
| Shamo (SH) | 392,280 | 19 | Germany | Fancy breed | Normal | Game bird, heavy and upright posture; |
| Gallus Gallus Spadiceus (GA) | 392,280 | 16 | Thailand | Wild population | Normal | A subspecies of red junglefowl. |
| Jinghong (JH) | 366,571 | 82 | China | Layer line | Normal | Brown shell; High production; |
| Jingfen (JF) | 364,449 | 110 | China | Layer line | Normal | Brown shell; High production; |
1 A total of 286,564 high-quality SNPs are shared in 9 populations after quality control.
Quality control of the SNP data was determined by the following criteria: (i) individual call rate > 0.95; (ii) SNP call rate > 0.99; (iii) SNPs in Hardy–Weinberg equilibrium in each breed (p>10e–6); (iv) SNP minor allele frequency > 0.01; (v) only autosomal SNPs with known positions were used. After assessing the quality control, we imputed the missing genotypes and inferred haplotypes using BEAGLE [14]. The final dataset contained 286,564 common SNPs that were genotyped in 331 individuals from 9 chicken populations.
Population structure analysis
We employed the program ADMIXTURE v1.23 [15], which uses a block relaxation algorithm, to investigate the population structure with default parameters. The number of populations considered was from 2 to 12. To infer the phylogenetic tree, PLINK v1.07 [16] was used to generate the IBS distance for all pairs of individuals and then a neighbour-joining tree was constructed with FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/) based on the distance matrix. Finally, principal component analysis (PCA) was performed using the flashpcaR package[17] based on the whole genome genotype data that remained after quality control was assessed. After filtering SNPs to have a Pearson product-moment correlation of allele frequency (r2) less than 0.8 in 50 SNP windows, we ran ADMIXTURE and flashpcaR on all 231,545 remaining SNPs again.
Linkage disequilibrium analysis
We calculated the correlation coefficient (r2) for every pair of SNPs to measure the LD level in each population using PLINK v1.07 [16]. The parameters were set as follows:—r2—ld-window-kb 1000—ld-window 99999—ld-window-r2 0. To visualize the LD decay in this analysis, the r2 values for 1000-bp distance bins were averaged and the corresponding figure was drawn by R script.
Identifying signals under artificial selection
In accordance with the population genetic structure, five breed pools (or breed) of genotype data were used to detect potential selection signals in this analysis: Jinghong and Jingfen (JH_JF), Pekin-Bantam (PB), Rheinlander and White Leghorn (RH_WL), Araucanas and Vorwerkhuhn (AR_VO), Gallus-Gallus-Spadiceus (GA). To identify the signals under artificial selection, GA was defined as the common reference population because it was associated with limited amounts of artificial selection for commercial traits.
Multiple elementary selection signal statistics were employed to search for the evidence of artificial selection in two steps (hereafter termed the ‘two-step’ strategy). The first step is positive selection detection, which was performed using the composite likelihood ratio (CLR) [18] and the integrated haplotype score (iHS) [19] test. Among them, the CLR method calculates the likelihood ratio of selection signals by comparing the spatial distribution of allele frequencies in an observed window to the frequency spectrum of the whole genome. In this analysis, SweepFinder [18] software was employed to calculate the CLR with a grid size of 10 kb. To explore ongoing selection signals, the iHS method was performed, which searches for haplotype structures that have ancestral and derived alleles. Single marker scores for unstandardized iHS were calculated using the rehh R package [20] and then the |iHS| scores averaged across a non-overlapping 10 kb window across the genome. In the second step, we identified the underlying artificial selection signals from the above detected positive selection signals by calculating the FST statistic for pairwise sites between the observed populations and the common reference population. The unbiased FST estimate proposed by Weir and Cockerham was used to measure the population differentiation, with values ranging from 0 (no differentiation) to 1 (complete differentiation) [21]. To produce comparable CLR and iHS test results, the 10 kb grid size was also used for determining FST statistics. Finally, the selection signals were detected by using the |iHS|, CLR and FST scores in the genome level. The outliers for all methods were defined as signals with statistics surpassing the significance threshold (P<0.05) determined using 1,000 permutation tests. Accordingly, the chromosomal regions within a 200 kb window surrounding the outliers were defined as potential selection regions (PSR). This window size in this analysis was determined by the linkage disequilibrium decay in real data (S1 Fig). In addition, we further calculated the ‘observed heterozygosity’ (Het), which should be decreased in regions that are affected by a selection signal. As a by-product, the major (minor) allele frequency was also calculated by PLINK v1.07 [16]. Note that the artificial selection signals in this study were defined in the genomic region in which both positive selection signals and the FST statistical value were greater than the cut-off value at the genome level.
Haplotype analysis
To characterize the core haplotype of underlying selection signals, haplotype blocks in the detected artificial selection regions were inferred using Haploview v4.2 [22] software based on the solid spin algorithm. The parameters were set as follows: -blockoutput SPI -blockSpineDP 0.8. Core haplotypes were defined as haplotype blocks that have a remarkable signal, and a high frequency in an observed population suggests that the presence of a core haplotype in the potential selection region may be abnormal in neutral scenarios. In general, in sweep analysis, the remarkable signals should be significant and the frequency should be greater than 0.8, according to previous reports [4].
Functional annotation and gene ontology (GO) term enrichment analysis
In this analysis, bioinformatics analyses were performed to explore the potential biological functions of genes located in putative artificial selection regions. This analysis involved all the selected genes in the 200 kb window surrounding the significant signals, which was determined by the linkage disequilibrium decay in chicken populations. Detailed biological functions were matched for all highlighted candidate genes using the NCBI database (https://www.ncbi.nlm.nih.gov/gene). The gene annotation files for chickens that were downloaded from the database from ensemble (http://www.ensembl.org/info/data/ftp/index.html) were used to identify the exact position of markers in selected regions and highlight those that fell into selected regions using the textual mining R script. Gene-based annotations of Single nucleotide polymorphisms (SNPs) located in selected regions were performed using ANNOVAR [23]. Using ENSEMBL genes, we investigated where the SNPs are located in the regions of gene components. In addition, genes located in putatively selected regions were identified using the BioMart program (http://www.biomart.org/, Kasprzyk, 2011), and then an enrichment analysis, which included the terms cellular component, molecular function, and biological process, was performed for the identified genes using DAVID 6.7 (http://david.abcc.ncifcrf.gov/).
Results and discussion
Phylogenetic and population structure
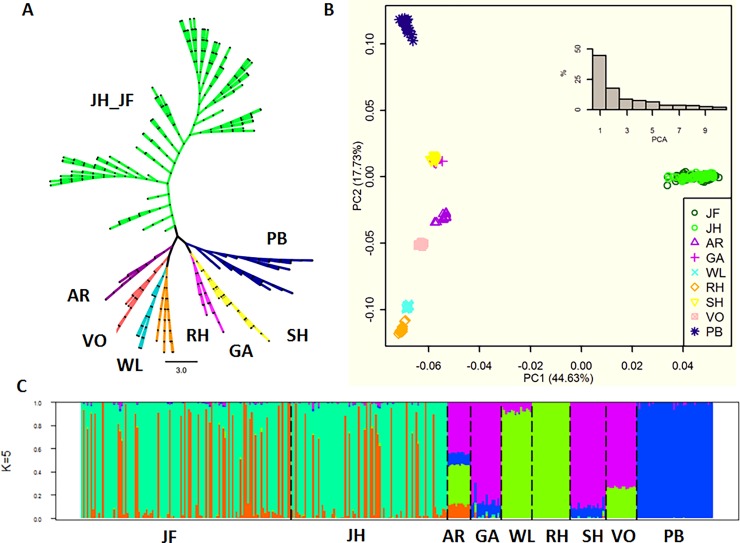
After quality control assessment, a total of 286,564 high-quality SNPs were used to explore the genetic population structure and relatedness among 9 chicken breeds (Table 1). The neighbour-joining tree, based on IBD distance, showed that the Chinese commercial lines (JH_JF) were aggregated as a genetic group with a few mixed clades. In addition, the other 7 populations can be divided into 2 geographic groups as Asian breeds (PB, GA and SH) and Western breeds (AR, VO, WL and RH) (Fig 1A). These findings indicated that two Chinese commercial lines may be derived from the same ancestral population and that the geographic locations contribute to the genetic clusters.
Fig 1. Analysis of the phylogenetic relationship and population structure of 9 chicken breeds.
(A) Neighbor-joining tree constructed using SNP data. (B) Principal component analysis. (C) Population structure analysis of the 331 chicken individuals, showing the distribution of the K = 5 genetic clusters. See material and method for the abbreviations of the breeds.
In principal components analysis, two Chinese commercial lines formed a tight cluster, which further confirmed the findings from phylogenetic analysis. Western breeds and Asian breeds could be further subdivided into AR_VO (AR and VO) and RH_WL (RH and WL), and PB and GA_SH (GA and SH), respectively (Fig 1B). Among them, three layer groups, including JH_JF, PB and RH_WL, formed a congruent equilateral triangle around a centre wide group (GA_SH) in this analysis. Similar results were obtained when the markers that had a high degree of linkage disequilibrium after filtering the SNPs were used (S2 Fig). It may suggest that diversity is caused by a special geographic environment, commercial purpose or other unknown elements, which formed layers from the wide population with similar distances (Fig 1B).
In addition, the ADMIXTURE program was employed to investigate population structure and introgression, with the number of given populations (K) varying from 2 to 12 using all 286,564 SNPs and the 231,545 SNPs that remained after filtering for a high degree of linkage disequilibrium (Fig 1C, S3 and S4 Figs). As K increased, individuals were classified into the expected subpopulations based on the geographic position or the introgression event. However, no matter how the number of given populations changed, the two commercial lines were indistinguishable, which is consistent with the analysis above. It indicated that Jinghong and Jingfen are two commercial chicken lines from the same ancestral population, rather than two chicken breeds with an independent breeding history. Instead, the other populations can be clearly divided, especially when K is large enough, which suggests that those seven breeds have a long, independent breeding history compared with the two Chinese commercial lines. Note that the population structure analysis using PCA and the neighbour-joining tree method presented a similar result when K = 5 (or 4). This phenomenon indicated that some populations in this study may share a similar genetic basis. Therefore, Jinghong and Jingfen, Rheinlander and White Leghorn, Araucanas and Vorwerkhuhn were merged as JH_JF, RH_WL and AR_VO, respectively. Gallus-Gallus-Spadiceus (GA) was treated as the common reference population for identifying the artificial selection signals. It is possible that this combination strategy causes some positive selection signals for each breed to be lost. However, the drift can also mimic selection signals, especially if the effective population size is/has been small. Therefore, in this study, the pool population strategy based on the above analysis can reduce the impact of random drift with a little cost.
Genome-wide artificial selection signals
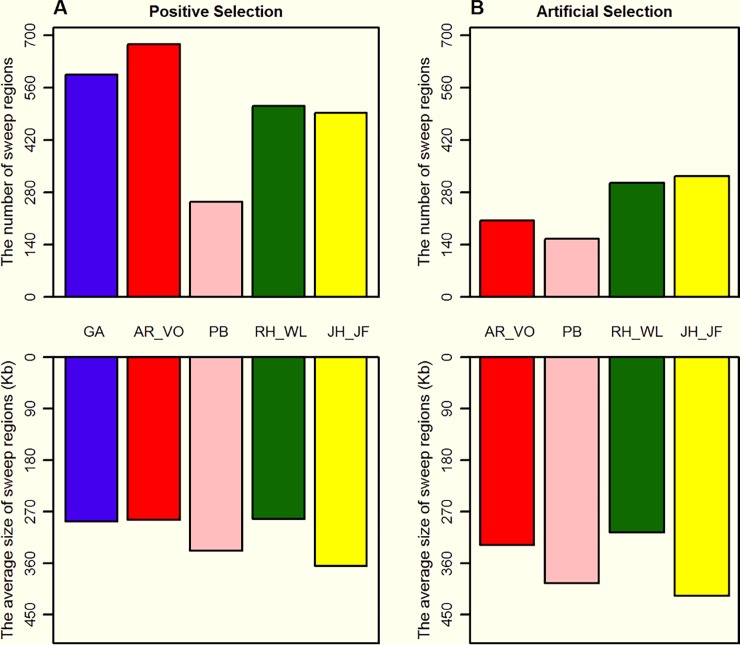
To detect positive selection, unstandardized iHS scores were calculated per site and the absolute values were averaged across non-overlapping sliding 10 kb windows across the whole genome. Correspondingly, the selection signals fell in 575, 626, 214, 365 and 194 potential selection regions in GA, AR_VO, PB, RH_WL and JH_JF, respectively. Similarly, a total of 26, 81, 79, 186 and 311 potential selection regions were also identified by the CLR test. In general, iHS is known to be good for detecting ongoing selection signals and CLR is most sensitive for detecting fixed selection signals. Therefore, the potential selection regions detected by iHS and CLR tests for each population were further merged when their interval distance was within a 200 kb window (hereafter termed iHS-CLR). Finally, a total of 595, 675, 254, 510 and 492 potential selection regions, spanning lengths of 170.93 Mb, 191.72 Mb, 85.90 Mb, 144.12 Mb and 179.26 Mb, were identified in GA, AR_VO, PB, RH_WL and JH_JF, respectively (Fig 2A).
Fig 2. Visualization of the trend of sweep regions in all observed populations.
(A) The potential positive selection regions identified by the iHS-CLR test. (B) The potential artificial selection regions identified by the ‘two-step method’, GA was defined as reference population in this analysis.
To further identify the artificial selection signals, FST statistics were used to evaluate the differentiation between domestic and wild populations. When GA was treated as a common reference population, a total of 720, 1151, 1205 and 1057 selection regions containing the signals at the 5% significant level were found in AR_VO, PB, RH_WL and JH_JF, respectively. The overlapping selection regions detected by iHS-CLR and FST were defined as the artificial selection signals for further analysis. Collectively, 204, 155, 305 and 323 potential artificial selection signals spanning lengths of 66.90 Mb, 61.16 Mb, 93.28 Mb and 134.65 Mb were separately identified in AR_VO, PB, RH_WL and JH_JF compared with the wild population (Figs 2B and 3, S5 and S7 Figs). Remarkably, the average size of sweep regions underlying artificial selection is longer than all underlying positive selection (Fig 2). Although only about 54% (data ranged from 30.22% to 65.65%) of positive selection signals were shaped by artificial selection, the length of genomic fragments under artificial selection accounted for about 61.5% (ranged from 34.89% to 75.11%) of all positive selection regions. The results indicated that artificial selection is more powerful than natural selection in shaping the genome, resulting in rapid improvement of goal-directed economic traits in domestic populations[24].
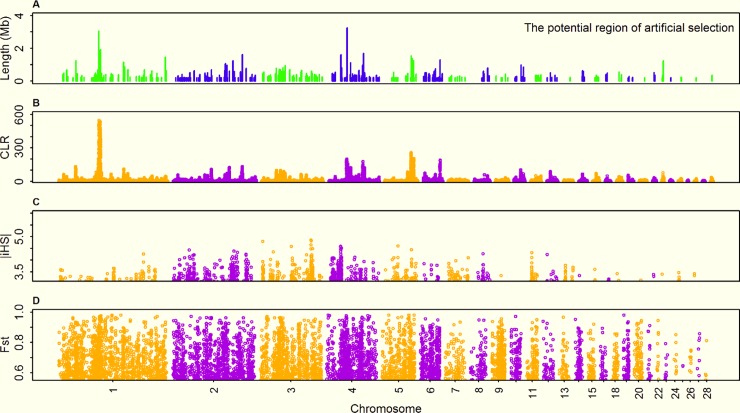
Fig 3. Summary of the genomic regions underlying artificial selection in JH_JF.
(A) The lines illustrate the positions and lengths of genomic regions underlying artificial selection. (B, C, D) Manhattan plots based on CLR, iHS and FST tests, the y axis values are statistics scores, and the x axis shows positions along each chromosome.
To investigate the effects of selection on different gene components, we annotated where the selected SNPs are located in the genome. Out of 286,563 SNPs with an average density of 3.2 SNPs/kb on the genome, 37,737, 14,514, 15,425 and 21,671 SNPs fall into the artificial selection regions in JH_JF, PB, AR_VO and RH_WL, respectively. Taken JH_JF as an example, 31,520 were located in intergenic regions, 5,305 were in intronic regions and 350 were in exonic regions. Similar proportions of the SNPs that were located in different gene components were also observed in the genomic regions without artificial selection (S1 Table). Simultaneously, it seems that there is no difference among the effects of selection on regulatory region, coding sequence and the other gene components. We explain this phenomenon by the hitchhiking effect of selection that would shape the pattern of diversity around the selected locus and thus leaded to the difficulty of the causal selected locus mapping. However, the detailed annotation of SNPs in selected region is still helpful in facilitating chicken functional genomics study in the future (S2 Table).
Goal-directed economic traits in chicken were improved by human-driven selection in recent decades[1, 4]. Correspondingly, the genomic signals, such as long range haplotype homozygosity and skewed allele frequency spectra, would be generated under the great selection pressure[25]. From a statistical perspective, it’s difficult to distinguish the artificial selection signals from the positive selection signals only using a single method. By combining multiple methods, it is possible to identify artificial selection signals by utilizing both the wild and domestic populations. Our previous simulation study indicated that iHS and CLR statistics performed best and can be complementary in detecting positive selection signals [26]. The divergent signals that derived from wild population and further shaped by artificial selection can be detected by FST statistics. Accordingly, we used multiple methods to detect artificial selection signals to benefit from advantageous complementarities across methods in hope of improving the reliability of selection signals. In addition, the previous simulation studies also suggested that combining several methods can greatly increase the power to pinpoint the selected region [26, 27].
Hard sweep and soft sweep
As described by Hermisson and Pennings [11], the signals underlying adaptive responses generally fall into two categories: hard sweep signals arise from newly arisen beneficial mutations and soft sweep signals arise from standing genetic variations. Chickens are an important animal economically, and most of their economic traits show quantitative heredity. In addition, the high polygenic basis for the important identified economic traits have been proven in recent genome-wide association studies [28, 29]. Therefore, we argue that selection of beneficial alleles already present in the population is the main reason for a rapid adaptation of economic traits in chickens.
To evaluate the contributions from standing genetic variations to artificial selection in chickens, we assumed the beneficial alleles for the important economic traits are still neutral or nearly neutral in Gallus-Gallus-Spadiceus. It may be closer to the ancestral population, but they have been prevalent in domestic populations because of artificial selection. Therefore, artificial selection regions with a haplotype allele frequency greater than 0.8 in domestic populations were reliable, and the corresponding signals based on emerging alleles (frequency is 0 in GA) and existing alleles (frequency is greater than 0.125, at least 4 gametes with the beneficial allele, in GA) were treated as a hard sweep and a soft sweep, respectively, in this study. In total, 10, 33, 43 and 113 potential hard sweeps of artificial selection (PAHSR) and 47, 60, 63 and 122 potential soft sweeps of artificial selection (PASSR) were identified in AR_VO, PB, RH_WL and JH_JF, respectively. It is generally known that artificial selection in chickens has yielded rapid changes in commercial traits that only have a history of approximately 100 years [1]. This means that commercial traits are likely to experience rapid improvement as a result of selection from standing genetic variation, especially when the trait is controlled by many mid-small effect loci [7]. However, there are also some mutations that have major effects that have been fixed or nearly fixed in domestic animals, such as mutations for double muscling in some beef cattle [30] and an IGF2 mutation in pigs [31]. Therefore, the results may suggest that both standing and de-novo genetic variations have taken an active role in the process of artificial selection, probably at different time scales and with different selection coefficients. Here, we tried to disentangle hard sweep and soft sweep using wild populations with no strong artificial selection as a reference. However, there are several subspecies of red jungle fowl that have contributed to domestic chickens [32]. Therefore, the detected sweeps based on standing and de-novo genetic variations here are just one small part of them.
A particularly interesting candidate selection region
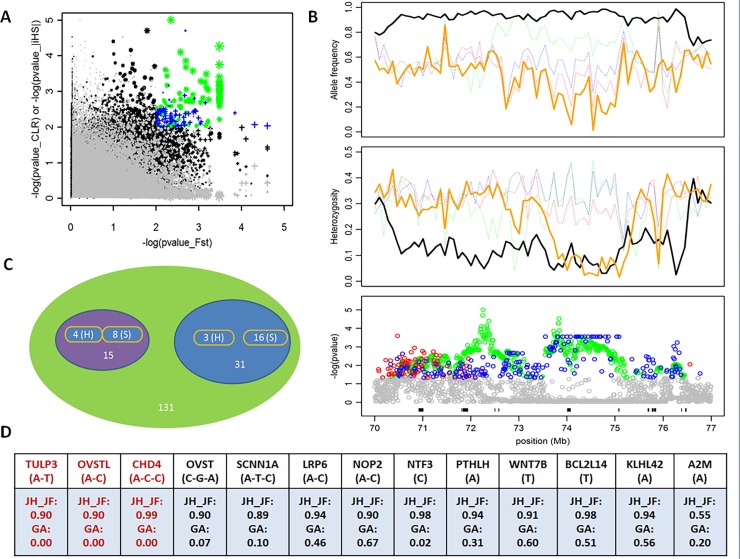
We highlighted the genomic region in JH_JF with the strongest artificial selection signal (Fig 4A), which is located between positions 70–77 Mb on GGA1 and includes a total of 131 genes. As shown in Fig 4B, a series of remarkable positive selection signals in this region were identified by CLR analyses. The differentiation of loci between JH_JF and GA were detected by the FST test, which is suggestive of artificial selection during improvement of this commercial line. Notably, we also observed lower heterozygosity in this candidate selection region compared with background levels in GA from JH_JF. Correspondingly, the extreme differences in allele frequency between JH_JF and GA further show a strong indication of artificial selection in this region. The gradient variation of allele frequency in five populations indicated that divergent selection has further enriched the inter-population genetic diversity of chickens in this region. Out of 131 genes embedded in this candidate region, only 46 were overlapped with SNP markers in exon regions in this analysis. Among them, 31 genes were covered by a single SNP marker and the other 15 genes were covered by multiple SNP markers. In accordance with the above criteria, a total of 7 and 24 genes are believed to bind to hard sweep and soft sweep regions, respectively (Fig 4C). Finally, we further analysed the biological functions of the 46 genes located in this region using the available database. For JH_JF, at least 13 genes harboured in this region were related to egg quality [33–36], immune response [37], bone development [33], neuron functions [38, 39] and fertility [40] (Fig 4D). Notably, most of them correspond with the beneficial alleles in the reference population, with the exception of three genes that were selected from the novel present alleles when haplotype analysis was performed using the markers in the exon region. In conclusion, these results demonstrated that this genomic region was not only under strong artificial selection, but it also harboured soft and hard sweep, simultaneously.
Fig 4. Highlight of a candidate selection region on GGA 1 in JH_JF.
(A) Distribution of log10 (PCLR or P|iHS|) and log10 (PFST) values calculated in sliding windows. The dots of color (corresponding to signals surpassing the 5% significance threshold of the empirical log10 (PCLR or P|iHS|) and signals surpassing the 5% significance threshold of the empirical log10 (PFST) distribution) are genomic regions underlying artificial selection in JH_JF. The dots in green represent the significant signals located in this candidate selection region. (B) The bottom Manhattan plot shows all selection signals identified by CLR (the dots in green) and FST (the dots in blue) in the 70–77 Mb region of GGA1, respectively. The positions of 131 gene models in this region are displayed by the black segments. The middle plot shows the pattern of heterozygosity of four domestic populations in this region. The top plot shows the major allele frequencies in JH_JF and their corresponding frequencies in three other domestic populations in this region. The lines in black, orange, green, red and blue separately represents JH_JF, GA, PB, RH_WL and AR_VO. (C) Classification of 15 genes with multiple makers and 31 genes with single marker. H and S indicate hard sweep and soft sweep, respectively. (D) 13 genes and their core haplotype frequencies (or major allele) in JH_JF (Obs.) and GA (Ref.), See Supplementary S4 Table for the exact nucleotide positions.
Underlying selected genes and Go terms for artificial selection
Based on the signals identified by the ‘two-step’ strategy, a total of 663, 668, 975 and 1394 genes were harboured in all artificial selection regions in AR_VO, PB, RH_WL and JH_JF, respectively. To further investigate the changes in biological process during artificial selection, those orthologous genes with human were used to perform an enrichment analysis using DAVID 6.7 (S3 Table). Most of terms did not give any intuitive information on artificial selection, such as some genes specific to PB and AR_VO were separate over-represented in the ‘ion transport’ (49 genes) and ‘low-density lipoprotein particle remodeling’(4 genes). However, Genes identified in JH_JF were enriched for the reproductive process, which mainly included ‘hsa04914: Progesterone-mediated oocyte maturation’ (14 genes), ‘hsa04114: Oocyte meiosis’ (14 genes) and ‘urogenital system development’ (15 genes), growth, which mainly included ‘regulation of growth’ (35 genes), and ‘growth’ (19 genes). These findings may reflect that artificial selection for JH_JF, a layer population, did improve the fertility and growth in recent decades. Similarly, genes found in RH_WL were mainly enriched for ‘embryonic development ending in birth or egg hatching’ (27 genes) and ‘in utero embryonic development’ (16 genes), which may contribute to fertility production. In addition, Gene Ontology (GO) analysis in four populations also revealed a series of biological process, including ‘behavioral fear response’ (3 genes in AR_VO), ‘behavior’ (42 genes in JH_JF), and ‘neuron development’ (32 genes in RH+WL), that are consistent with signal-related genes in this study. The findings may suggest that the signal-related genes and their corresponding biological processes may play an important role in changes of behaviour, habits, and commercial traits, such as modest temperament and the ability to grow fast.
Although there were no overall significant GO terms after Bonferroni correction, we found that genes related with a number of terms previously implicated in domestication-related changes are present within or close to these sweep regions. Among them, some candidate genes were observed that are related to immunity [37], neural system development [38], growth [41], feed intake [42] and fertility [40] (Table 2). We note that the established selective sweep around the TSHR gene in domestic chickens was also identified in our four populations. In general, this gene was considered as proof of principle demonstrating that the identification of selection signals should be reliable [1, 4]. Another interesting selection signal is located on GGA3 (105.92–106.09 Mb) and harbours the NCOA1 gene, which is associated with total egg production [43]. For most layer history, egg production has consistently been treated as an objective trait of breeding. Correspondingly, the quality of the eggshell would also be considered and designed in the breeding programme. In this study, three putative genes (PTHLH, OVST and SCNN1A) overlapping with the strongest selection signals are related to eggshell characteristics. Among them, the PTHLH gene, located in the 72.51–72.52 Mb region of GGA1, plays an important role in calcium regulation[33]. As an estrogenic-stimulated gene involved in the process of oviduct development, the OVST gene is also associated with the formation of eggshells by regulating eggshell matrix protein secretion [35]. These results indicated that high levels of egg productivity would promote the evolution of calcium transportation system in layers. Note that the SCNN1A gene is precisely responsible for calcium transportation [34]. In contrast to egg production, another apparent characteristic of layer chickens is low weight. Here, a series of artificial selection signals at the FGF6, TGFB2 and IGF2 loci across different populations were detected. In accordance to previous reports [41, 44, 45], all of them are important candidate genes for growth traits. Remarkably, we also found three significant sweep regions that separately overlapped with the PMCH gene, which contributes to appetite [1], as well as the KLHL42 and LPCAT3 genes, which are related to feed intake [42, 46]. These findings indicated that the artificial selection of economic traits could have a cascading effect on phenotypes with genetic correlation, such as egg production, eggshell, growth, and feed intake. In addition, we also found a few genes with significant signals that are functionally plausible for neurodevelopment and immune response, which is consistent with previous studies of genes associated with chicken domestication [4].
Table 2. Some candidate genes overlap with the potential regions of artificial selection in four domestic chicken populations.
| Chr. | Pos. (Mb)1 | P-value. (method)2 | Gene | Gene function |
|---|---|---|---|---|
| 1 | 55.42–55.43 | Fst0.039,iHS0.025;Fst0.005,CLR0.002; Fst0.008; Fst0.035; | PMCH | appetite, food intake[1] |
| 1 | 70.84–70.94 | Fst0.027,iHS0.027;Fst0.005;Fst0.005; Fst <0.001,Clr<0.001; | WNT7B | the developing central nervous system[38] |
| 1 | 71.72–71.74 | Fst0.025,iHS0.017;Fst0.002;Fst0.005,iHS0.035;Fst<0.001, Clr<0.001; | BCL2L14 | immune[47] |
| 1 | 72.42–72.44 | Fst0.025,iHS0.039;Fst<0.001;Fst0.005,iHS0.035;Fst<0.001, Clr<0.001; | KLHL42 | feed intake[42] |
| 1 | 72.51–72.53 | Fst0.025,iHS0.039;Fst<0.001;Fst0.005,iHS0.035;Fst<0.001, Clr<0.001; | PTHLH | bone development, eggshell[33] |
| 1 | 73.16–73.17 | Fst0.026,iHS0.018;Fst<0.001;Fst0.001;Fst<0.001,Clr<0.001; | FGF6 | growth[41] |
| 1 | 76.34–76.44 | Fst0.025,iHS0.004;Fst0.001,iHS0.006; Fst0.031,iHS0.004,CLR0.01; Fst<0.001,Clr<0.001; | OVST | eggshell[35] |
| 1 | 76.90–76.92 | Fst0.025,iHS0.003;Fst0.007,iHS0.008;Fst0.031,iHS0.004,CLR0.01;Fst0.013,CLR<0.001; | SCNN1A | cation transportation, eggshell[34] |
| 1 | 77.35–77.38 | Fst0.025,iHS0.003;iHS0.008;Fst0.015,iHS0.004;Fst<0.001,CLR<0.001; | LPCAT3 | feed intake[46] |
| 2 | 14.04–14.88 | Fst0.042,iHS0.036;Fst0.036,CLR0.004;Fst0.007,iHS0.005,CLR0.014; Fst0.037,iHS 0.041,CLR0.014; | ITGB1 | skeletal myogenesis[48] |
| 3 | 19.37–19.43 | Fst0.038,iHS0.048; Fst0.031; Fst0.004;Fst 0.006, Clr 0.006; | TGFB2 | growth traits[44] |
| 3 | 105.92–106.1 | Fst 0.019,iHS0.013; Fst 0.022; Fst 0.001; Fst 0.049; | NCOA1 | total egg production[43] |
| 5 | 13.77–13.78 | Fst0.009,iHS0.026;Fst0.031;Fst0.001,CLR0.017;Fst0.004,iHS0.008; | IGF2 | growth and carcass traits[45] |
| 5 | 40.81–40.86 | Fst0.005,iHS0.004,CLR0.016;Fst<0.001;Fst<0.001,CLR0.007; Fst <0.001, CLR 0.004; | TSHR | the reproductive machinery[1, 4] |
| 5 | 49.02–49.03 | Fst0.006,iHS0.031;Fst0.005;Fst0.036,iHS0.047;Fst<0.001,CLR<0.001; | DLK1 | muscle hypertrophy[4] |
| 6 | 14.82–14.84 | Fst0.032,iHS0.003;Fst0.001;Fst0.036,iHS0.023,CLR0.008;Fst0.013,iHS0.021,CLR0.008; | COMTD1 | Pigmentation[49] |
| 7 | 22.54–22.55 | Fst0.038,iHS0.018;Fst0.022,CLR0.004;Fst0.004,iHS0.014;Fst0.033; | PRKAG3 | Meat quality[3] |
| 11 | 16.18–16.63 | Fst0.037,iHS0.012;Fst0.005;Fst0.021;Fst<0.001,CLR0.007; | CDH13 | Disease-resistant[50] |
| 18 | 4.47–4.51 | Fst0.006;Fst0.007;Fst0.004,CLR0.007;Fst0.004,CLR0.015; | PRPSAP1 | Abdominal fatness deposition[51] |
1 This column presents the position of candidate genes which overlap with or close to the potential regions of artificial selection.
2 This column presents the genome-wide P-values of three statistics, corresponding to AR_VO, PB, RH_WL and JH_JF with semicolons.
Conclusions
In this study, we identified the artificial selection signals based on nine chicken breeds using multiple statistical methods and further discussed the possible classifications of those signals. The successful application of the ‘two-step’ method suggested that artificial selection signals can be detected by combining multiple statistical methods and populations (or breeds). In some special cases, the pool population strategy based on the genetic relationship of multiple populations can reduce the impact from random drift in sweep analysis with the small cost of losing some signals for unique populations. According to the results, we found that both standing and de-novo genetic variations have contributed to adaptive evolution during the process of artificial selection. As a promising approach to studying population genomics, sweep analysis revealed a series of genes that contribute to the improvement of chicken breeds, which can be used as fundamental information in future chicken functional genomics study.
Supporting information
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(A) The lines illustrate the positions and lengths of genomic regions underlying artificial selection. (B, C, D) Manhattan plots based on CLR, iHS and FST tests, the y axis values are–log(P-value), and the x axis shows positions along each chromosome.
(TIFF)
(A) The lines illustrate the positions and lengths of genomic regions underlying artificial selection. (B, C, D) Manhattan plots based on CLR, iHS and FST tests, the y axis values are–log(P-value), and the x axis shows positions along each chromosome.
(TIFF)
(A) The lines illustrate the positions and lengths of genomic regions underlying artificial selection. (B, C, D) Manhattan plots based on CLR, iHS and FST tests, the y axis values are–log(P-value), and the x axis shows positions along each chromosome.
(TIFF)
(DOC)
(XLSX)
(XLSX)
(DOCX)
Acknowledgments
We thank Chunyan Mu for her helpful comments. We also thank Steffen Weigend for kindly providing the detailed information of seven chicken breeds.
Data Availability
The data are all contained within the paper and Supporting Information files. Please contact lishijun@mail.hzau.edu.cn for additional information.
Funding Statement
This research was financially supported by the National Natural Science Foundation of China (31772585, 31601916) and the Fundamental Research Funds for the Central Universities (0900202930, 2662015QD018). The funders did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the “author contributions” section.
References
- 1.Rubin C-J, Zody MC, Eriksson J, Meadows JR, Sherwood E, Webster MT, et al. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature. 2010;464(7288):587–91. doi: 10.1038/nature08832 [DOI] [PubMed] [Google Scholar]
- 2.Perrins CM. Firefly encyclopedia of birds: Firefly Books Limited; 2003. [Google Scholar]
- 3.Rubin CJ, Megens HJ, Barrio AM, Maqbool K, Sayyab S, Schwochow D, et al. Strong signatures of selection in the domestic pig genome. P Natl Acad Sci USA. 2012;109(48):19529–36. doi: 10.1073/pnas.1217149109 ISI:000312313900015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qanbari S, Seidel M, Strom T-M, Mayer KF, Preisinger R, Simianer H. Parallel selection revealed by population sequencing in chicken. Genome biology and evolution. 2015;7(12):3299–306. doi: 10.1093/gbe/evv222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang H, Wang S-Z, Wang Z-P, Da Y, Wang N, Hu X-X, et al. A genome-wide scan of selective sweeps in two broiler chicken lines divergently selected for abdominal fat content. BMC genomics. 2012;13(1):704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheng Z, Pettersson ME, Honaker CF, Siegel PB, Carlborg Ö. Standing genetic variation as a major contributor to adaptation in the Virginia chicken lines selection experiment. Genome biology. 2015;16(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pritchard JK, Pickrell JK, Coop G. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Current biology. 2010;20(4):R208–R15. doi: 10.1016/j.cub.2009.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ai H, Fang X, Yang B, Huang Z, Chen H, Mao L, et al. Adaptation and possible ancient interspecies introgression in pigs identified by whole-genome sequencing. Nature genetics. 2015;47(3):217–25. doi: 10.1038/ng.3199 [DOI] [PubMed] [Google Scholar]
- 9.Qanbari S, Pausch H, Jansen S, Somel M, Strom TM, Fries R, et al. Classic selective sweeps revealed by massive sequencing in cattle. PLoS Genet. 2014;10(2):e1004148 doi: 10.1371/journal.pgen.1004148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki Y. Statistical methods for detecting natural selection from genomic data. Genes & genetic systems. 2010;85(6):359–76. [DOI] [PubMed] [Google Scholar]
- 11.Hermisson J, Pennings PS. Soft sweeps: Molecular Population Genetics of Adaptation From Standing Genetic Variation. Genetics. 2005;169(4):2335–52. doi: 10.1534/genetics.104.036947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pennings PS, Hermisson J. Soft sweeps II—molecular population genetics of adaptation from recurrent mutation or migration. Molecular biology and evolution. 2006;23(5):1076–84. doi: 10.1093/molbev/msj117 [DOI] [PubMed] [Google Scholar]
- 13.Pennings PS, Hermisson J. Soft sweeps III: the signature of positive selection from recurrent mutation. PLoS Genet. 2006;2(12):e186 doi: 10.1371/journal.pgen.0020186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Browning BL, Browning SR. Genotype imputation with millions of reference samples. The American Journal of Human Genetics. 2016;98(1):116–26. doi: 10.1016/j.ajhg.2015.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome research. 2009;19(9):1655–64. doi: 10.1101/gr.094052.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics. 2007;81(3):559–75. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abraham G, Inouye M. Fast principal component analysis of large-scale genome-wide data. PloS one. 2014;9(4):e93766 doi: 10.1371/journal.pone.0093766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nielsen R, Williamson S, Kim Y, Hubisz MJ, Clark AG, Bustamante C. Genomic scans for selective sweeps using SNP data. Genome research. 2005;15(11):1566–75. doi: 10.1101/gr.4252305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4(3):e72 doi: 10.1371/journal.pbio.0040072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gautier M, Vitalis R. rehh: an R package to detect footprints of selection in genome-wide SNP data from haplotype structure. Bioinformatics. 2012;28(8):1176–7. doi: 10.1093/bioinformatics/bts115 [DOI] [PubMed] [Google Scholar]
- 21.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. evolution. 1984;38(6)(6):1358–70. doi: 10.1111/j.1558-5646.1984.tb05657.x [DOI] [PubMed] [Google Scholar]
- 22.Barrett J, Fry B, Maller J, Daly M. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- 23.Wang K, Li MY, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16). ARTN e16410.1093/nar/gkq603. PubMed PMID: ISI:000281720500004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li M, Tian S, Jin L, Zhou G, Li Y, Zhang Y, et al. Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nature genetics. 2013;45(12):1431–8. doi: 10.1038/ng.2811 [DOI] [PubMed] [Google Scholar]
- 25.Qanbari S, Simianer H. Mapping signatures of positive selection in the genome of livestock. Livestock Science. 2014;166:133–43. [Google Scholar]
- 26.Ma Y, Ding X, Qanbari S, Weigend S, Zhang Q, Simianer H. Properties of different selection signature statistics and a new strategy for combining them. Heredity. 2015;115(5):426–36. doi: 10.1038/hdy.2015.42 ISI:000362824300005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vatsiou AI, Bazin E, Gaggiotti OE. Detection of selective sweeps in structured populations: a comparison of recent methods. Mol Ecol. 2016;25(1):89–103. doi: 10.1111/mec.13360 ISI:000367908800007. [DOI] [PubMed] [Google Scholar]
- 28.Yuan J, Wang K, Yi G, Ma M, Dou T, Sun C, et al. Genome-wide association studies for feed intake and efficiency in two laying periods of chickens. Genetics Selection Evolution. 2015;47(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Psifidi A, Banos G, Matika O, Desta TT, Bettridge J, Hume DA, et al. Genome-wide association studies of immune, disease and production traits in indigenous chicken ecotypes. Genetics Selection Evolution. 2016;48(1):74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savarese MC, Marchitelli C, Crisa A, Filippini F, Valentini A, Nardone A. Dating the onset of some mutations in myostatin gene determining the double muscled phenotype in beef cattle. Ital J Anim Sci. 2003;2:64–6. PubMed PMID: ISI:000208276800020. [Google Scholar]
- 31.Van Laere AS, Nguyen M, Braunschweig M, Nezer C, Collette C, Moreau L, et al. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature. 2003;425(6960):832–6. ISI:000186118500044. doi: 10.1038/nature02064 [DOI] [PubMed] [Google Scholar]
- 32.Eriksson J, Larson G, Gunnarsson U, Bed'hom B, Tixier-Boichard M, Stromstedt L, et al. Identification of the Yellow skin gene reveals a hybrid origin of the domestic chicken. Plos Genetics. 2008;4(2):e1000010 ISI:000255386100011. doi: 10.1371/journal.pgen.1000010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dale MD, Mortimer EM, Kolli S, Achramowicz E, Borchert G, Juliano SA, et al. Bone-Remodeling Transcript Levels Are Independent of Perching in End-of-Lay White Leghorn Chickens. International journal of molecular sciences. 2015;16(2):2663–77. doi: 10.3390/ijms16022663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan Y-F, Hou Z-C, Yi G-Q, Xu G-Y, Yang N. The sodium channel gene family is specifically expressed in hen uterus and associated with eggshell quality traits. BMC genetics. 2013;14(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marie P, Labas V, Brionne A, Harichaux G, Hennequet-Antier C, Rodriguez-Navarro AB, et al. Data set for the proteomic inventory and quantitative analysis of chicken eggshell matrix proteins during the primary events of eggshell mineralization and the active growth phase of calcification. Data in brief. 2015;4:430–6. doi: 10.1016/j.dib.2015.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dunn I, Wilson P, Lu Z, Bain M, Crossan C, Talbot R, et al. New hypotheses on the function of the avian shell gland derived from microarray analysis comparing tissue from juvenile and sexually mature hens. General and comparative endocrinology. 2009;163(1):225–32. [DOI] [PubMed] [Google Scholar]
- 37.Reemers SS, van Haarlem DA, Koerkamp MJG, Vervelde L. Differential gene-expression and host-response profiles against avian influenza virus within the chicken lung due to anatomy and airflow. Journal of general virology. 2009;90(9):2134–46. [DOI] [PubMed] [Google Scholar]
- 38.Schubert M, Holland LZ, Holland ND. Characterization of two amphioxus Wnt genes (AmphiWnt4 and AmphiWnt7b) with early expression in the developing central nervous system. Developmental Dynamics. 2000;217(2)(2):205–15. doi: 10.1002/(SICI)1097-0177(200002)217:2<205::AID-DVDY7>3.0.CO;2-F [DOI] [PubMed] [Google Scholar]
- 39.Okamoto R, Uchikawa M, Kondoh H. Sixteen additional enhancers associated with the chicken Sox2 locus outside the central 50‐kb region. Development, growth & differentiation. 2015;57(1)(1):24–39. [DOI] [PubMed] [Google Scholar]
- 40.Berti F, Nogueira JM, Wöhrle S, Sobreira DR, Hawrot K, Dietrich S. Time course and side‐by‐side analysis of mesodermal, pre‐myogenic, myogenic and differentiated cell markers in the chicken model for skeletal muscle formation. Journal of anatomy. 2015;227(3):361–82. doi: 10.1111/joa.12353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Kaam J, Van Arendonk J, Groenen M, Bovenhuis H, Vereijken A, Crooijmans R, et al. Whole genome scan for quantitative trait loci affecting body weight in chickens using a three generation design. Livestock Production Science. 1998;54(2):133–50. [Google Scholar]
- 42.Lindholm-Perry AK, Butler AR, Kern RJ, Hill R, Kuehn LA, Wells JE, et al. Differential gene expression in the duodenum, jejunum and ileum among crossbred beef steers with divergent gain and feed intake phenotypes. Anim Genet. 2016;47(4):408–27. doi: 10.1111/age.12440 [DOI] [PubMed] [Google Scholar]
- 43.LI S-f, ZHAO Z-h, HUANG H-y, ZHANG J, DING Y-r. Association of Polymorphisms of the NCOA1 Gene with Egg Production Traits in Female Line of Shaobo Chicken [J]. Journal of Yunnan Agricultural University (Natural Science). 2011;2:014. [Google Scholar]
- 44.Zhang Y, Li H, Lian Z, Li N. Normal fibroblasts promote myodifferentiation of myoblasts from sex‐linked dwarf chicken via up‐regulation of β1 integrin. Cell biology international. 2010;34(11):1119–27. doi: 10.1042/CBI20090351 [DOI] [PubMed] [Google Scholar]
- 45.Tang S, Sun D, Ou J, Zhang Y, Xu G, Zhang Y. Evaluation of the IGFs (IGF1 and IGF2) genes as candidates for growth, body measurement, carcass, and reproduction traits in Beijing You and Silkie chickens. Animal biotechnology. 2010;21(2):104–13. doi: 10.1080/10495390903328090 [DOI] [PubMed] [Google Scholar]
- 46.Lee J, Karnuah AB, Rekaya R, Anthony NB, Aggrey SE. Transcriptomic analysis to elucidate the molecular mechanisms that underlie feed efficiency in meat-type chickens. Molecular Genetics & Genomics. 2015;290(5):1673–82. [DOI] [PubMed] [Google Scholar]
- 47.Reemers SS, van Haarlem DA, Koerkamp MJG, Vervelde L. Differential gene-expression and host-response profiles against avian influenza virus within the chicken lung due to anatomy and airflow. Journal of General Virology. 2009;90:2134–46. ISI:000269676300010. doi: 10.1099/vir.0.012401-0 [DOI] [PubMed] [Google Scholar]
- 48.Zhang Y, Li H, Lian ZX, Li N. Normal fibroblasts promote myodifferentiation of myoblasts from sex-linked dwarf chicken via up-regulation of beta 1 integrin. Cell Biology International. 2010;34(11):1119–27. ISI:000284134800011. doi: 10.1042/CBI20090351 [DOI] [PubMed] [Google Scholar]
- 49.Eriksson J. Genetic and Genomic Studies in Chicken: Assigning Function to Vertebrate Genes. Acta Universitatis Upsaliensis; 2012. [Google Scholar]
- 50.Connell S, Meade KG, Allan B, Lloyd AT, Downing T, O’Farrelly C, et al. Genome-Wide Association Analysis of Avian Resistance to Campylobacter jejuni Colonization Identifies Risk Locus Spanning the CDH13 Gene. G3 (Bethesda, Md). 2013;3(5):881–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang H, Wang SZ, Wang ZP, Da Y, Wang N, Hu XX, et al. A genome-wide scan of selective sweeps in two broiler chicken lines divergently selected for abdominal fat content. Bmc Genomics. 2012;13 Artn 704 doi: 10.1186/1471-2164-13-704. ISI:000314654600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(A) The lines illustrate the positions and lengths of genomic regions underlying artificial selection. (B, C, D) Manhattan plots based on CLR, iHS and FST tests, the y axis values are–log(P-value), and the x axis shows positions along each chromosome.
(TIFF)
(A) The lines illustrate the positions and lengths of genomic regions underlying artificial selection. (B, C, D) Manhattan plots based on CLR, iHS and FST tests, the y axis values are–log(P-value), and the x axis shows positions along each chromosome.
(TIFF)
(A) The lines illustrate the positions and lengths of genomic regions underlying artificial selection. (B, C, D) Manhattan plots based on CLR, iHS and FST tests, the y axis values are–log(P-value), and the x axis shows positions along each chromosome.
(TIFF)
(DOC)
(XLSX)
(XLSX)
(DOCX)
Data Availability Statement
The data are all contained within the paper and Supporting Information files. Please contact lishijun@mail.hzau.edu.cn for additional information.