Abstract
Myocyte enhancer factor 2A (MEF2A) is widely distributed in various tissues or organs and plays crucial roles in multiple biological processes. To examine the potential effects of MEF2A on skeletal muscle myoblast, the functional role of MFE2A in myoblast proliferation and differentiation was investigated. In this study, we found that the mRNA expression level of Mef2a was dramatically increased during the myogenesis of bovine skeletal muscle primary myoblast. Overexpression of MEF2A significantly promoted myoblast proliferation, while knockdown of MEF2A inhibited the proliferation and differentiation of myoblast. RT-PCR and western blot analysis revealed that this positive effect of MEF2A on the proliferation of myoblast was carried out by triggering cell cycle progression by activating CDK2 protein expression. Besides, MEF2A was found to be an important transcription factor that bound to the myozenin 2 (MyoZ2) proximal promoter and performed upstream of MyoZ2 during myoblast differentiation. This study provides the first experimental evidence that MEF2A is a positive regulator in skeletal muscle myoblast proliferation and suggests that MEF2A regulates myoblast differentiation via regulating MyoZ2.
Introduction
Skeletal muscle is an important constituent in indicating muscle growth and livestock muscle quality [1]. Previous studies have demonstrated that the development of skeletal muscle is a complex process that determines the number of muscle fibers, mass, and fiber type [2–5]. At the genetic level, the maintenance of skeletal muscle function mainly depends on myocytes, the primary cellular component of skeletal muscle. Therefore, it is important to understand the molecular mechanisms that regulates skeletal muscle myogenesis.
MEF2A, an evolutionarily conserved transcription factor, is widely distributed in various tissues or organs and play crucial roles in multiple biological processes including cell fate determination, migration and shape [6–9]. Previous studies showed that MEF2A is a key regulator in myogenesis. However, the underlying mechanisms of regulation within the various stages of myogenesis, such as proliferation and differentiation of progenitor cells, has not been fully elucidated [10]. A limited number of studies has suggested that MEF2A may regulate cell proliferation. In mice, global deletion of MEF2A impaired regenerative myogenesis, and knockdown of MEF2A in C2C12 cells severely impaired myotube formation [11–13]. Only one study investigated the function of MEF2A in myoblast proliferation [14]. However, more robust evidence is needed to determine whether and how MEF2A regulates myoblast proliferation and differentiation.
MEF2A has a functional role in the differentiation of muscle cells [9, 13, 14]. Most of the present studies of MEF2A, however, have focused on rodents in C2C12 cell lines, which is not a suitable comparison to primary myocytes of other species because of the differences among organs and species as well as differences within the same species. For example, Snyder et al. (2013) reported that MEF2A knocked out in mice severely inhibited skeletal muscle regeneration [13]. Whereas, Liu et al. (2014) found that the deletion of MEF2A had no distinguishable effect on mice skeletal muscle histology [14]. Furthermore, the downstream genes that are regulated by MEF2A and the underlying mechanisms are far from well-studied [12], thus it is necessary to uncover the mechanisms of MEF2A in skeletal muscle myogenesis.
The aim of this study was to determine the function of MEF2A in myoblast proliferation and elucidate the regulatory mechanisms underlying the effects of MEF2A on the differentiation of myoblasts. We were able to demonstrate, for the first time, that MEF2A promoted the proliferation of skeletal muscle myoblasts. In addition, we found that MEF2A was an important transcription factor that bound to the MyoZ2 proximal promoter and performed upstream of MyoZ2 during myoblast differentiation. Our results reveal the roles of MEF2A in regulating bovine myoblast proliferation and differentiation in vitro, which can help inform theories in cattle skeletal muscle development and advance gene therapies.
Materials and methods
Ethics statement
A three-day old healthy Qinchuan beef cattle was used for myoblast isolation and cell culture. It was born and raised at the experimental farm of National Beef Cattle Improvement Center (Yangling, China) and slaughtered using mechanized slaughter line at Shaanxi Qinbao Animal Husbandry Development Co., Ltd. The experiments and animal care were performed according to the Regulations for the Administration of Affairs Concerning Experimental Animals (Ministry of Science and Technology, China, 2004) and approved by the Institutional Animal Care and Use Committee (College of Animal Science and Technology, Northwest A&F University, China).
Isolation and cell culture of bovine skeletal muscle primary myoblasts
Isolation of primary skeletal muscle myoblasts was performed as previously described by Springer et al. [15]. The muscle sample was obtained from slaughter house. The limb skin was rinsed with 75% ethanol and removed with sterile sharp curved surgical scissors to expose the muscle tissue. The hind limb muscle sample was then removed into 1×PBS supplemented with 10% penicillin/streptomycin and was immediately taken into the cell culture lab. Under a stereo dissecting microscope, the muscle sample was dissected away from the blood vessel and connective tissue with sterile forceps. The muscle sample was then minced and digested with 0.25% Collagenase Ⅱ (Sigma)/0.1% Diapase Ⅱ (Roche) solution at 37°C until the mixture is a fine slurry. The cell suspension was filtered through an 80-μm cell strainer and pelleted by centrifugation for 5 minutes at 350×g. The pellets were then resuspended and seeded in 60-mm collagen-coated culture plates. The primary skeletal muscle myoblasts were cultured in complete growth medium containing Dulbecco's Modified Eagle Medium/F-12 (DMEM/F-12, Gibco), 20% fetal bovine serum (Gibco) and 1% penicillin/streptomycin. Growth medium was changed every two days and cells were passaged at 70% confluence to avoid spontaneous differentiation. For induced myogenic differentiation, cells at 70% confluence were switched to differentiation medium containing DMEM/F-12, 2% horse serum (Gibco) and 1% penicillin/streptomycin. The differentiation medium was changed every two days.
MEF2A overexpression assay
Adenovirus carrying full length bovine Mef2a gene coding sequence (CDS) was generated as previously described [16]. Simply, the full length CDS sequence (Accession number: NM_001083638) was amplified and cloned into pAd-Track shuttle vector. The shutter vector was then linearized by Pme Ⅰ restriction enzyme (New England Biolabs) for recombination with pAdEasy-1 expression vector in BJ5183 competent cells. After digested with Pac Ⅰ restriction enzyme (New England Biolabs), the expression vector was purified and transfected into 293A cells to allow being packaged into adenovirus (OE-2A). Negative control (NC) adenovirus was generously provided by Changzhen Fu. (Dalian University) and Yaran Zhang. (Northwest A&F University). Viral particles were expanded in 293A cells and viral titer was determined using the end point dilution assay (Clontech).
MEF2A interference assay
Specific short hairpin RNA (shRNA) and NC oligonucleotides for Mef2a mRNA (NM_001083638) were designed by using the online software: BLOCK-iT adenoviral RNAi expression system (https://rnaidesigner.thermofisher.com/rnaiexpress/) [17]. The specific shRNA that had the highest interference efficiency was selected through psiCHECKTM-Ⅱ reporter system (Promega). The specific shRNA and negative control were cloned into the pENTRTM/U6 RNAi entry vector followed by recombined with the pAd/PL-DESTTM expression vector (Invitrogen). The expression vector was then transfected into 293A cells to allow to be packaged into adenoviruses (sh-2A/sh-NC). Viral particle expansion and viral titer detection were the same as OE-2A. Table 1 shows the shRNA sequences used in this study.
Table 1. shRNA sense strand sequences for Mef2a mRNA and negative control in this study.
| Name | Sense strand (5’-3’) | Loop |
|---|---|---|
| shRNA-571 | GCAGAACCAACUCGGAUAUUG | UCAAGAG |
| shRNA-1193 | GCCUCCACUGAAUACCCAAAG | UCAAGAG |
| shRNA-1219 | GCAGUUCUCAAGCCACUCAAC | UCAAGAG |
| shRNA-1418 | GCAGCACCAUUUAGGACAAGC | UCAAGAG |
| shRNA-NC | GUUCCACGACCAAAUCAGCUC | UCAAGAG |
Note: shRNA: short hairpin RNA; “571”, “1193”, “1219”, “1418” means the position of the shRNA sequence in the mRNA region of Mef2a.
Cell flow cytometry assay
Skeletal muscle myoblasts were grown in normal growth medium and passaged at 70% confluence. When grown to 60% confluence, cells in 6-well plates were infected with OE-2A or sh-2A in triplicate. 48 hours after infection, cells were washed, harvested and permeabilized by using One Step Cell Cycle Straining Kit (MultiSciences Biotech). After the nuclei was strained with propidium iodide (PI) for 30 minutes, cell cycle was detected by measuring DNA content using Flow Cytometry (FACS Calibur, BD, USA) through counting 20000 cells.
EdU labeling assay
To detect cell proliferation, 5-ethynyl-2-deoxyuridine (EdU) assays were performed using Click-iT® EdU Imaging Kit (Invitrogen) according to the manufacturer’s instructions. Cells were plated on coverslips at 60% density and treated with OE-2A or OE-NC. 48 hours after infection, cells were treated with 10μM EdU solution in normal growth medium for 1 hour. Subsequently, cells were fixed with 3.7% paraformaldehyde and permeabilized with 0.5% Triton® X-100 (Sigma, USA). After incubation for 20 minutes, cells were treated with Alexa Fluor® 594 azide and DAPI to stain the nuclei. Immunofluorescence images were taken by Olympus IX71 microscope (OLYMPUS).
Vectors and plasmids
To select the specific shRNA, the full length CDS of Mef2a gene was cloned into psiCHECKTM-Ⅱ vector and the shRNA was cloned into pENTR/CMV-GFP/U6 vector. For luciferase reporter assay, the bovine MyoZ2 proximal promoter (190bp) containing the MFE2 binding site was cloned into pGL3-Basic vector (Promega). The mutant MyoZ2 promoter sequence was chemically synthesized (Sangon Biotech) by mutating -33bp MEF2 site TATATA to GGGGGG and was also cloned into pGL3-Basic vector. pRL-TK vector was used as internal control.
Luciferase activity assay
The relative luciferase activity was tested 40 hours after transfection of pGL3-Basic or psiCHECKTM-Ⅱ vector by using Dual-Luciferase® Reporter Assay System (Promega). In brief, cells were seeded in 12-well plates at a density of 1×106 cells per well. Cells were treated according to the experimental design and when the cells grown to 70% confluence, cells were lysed by 1×Passive Lysis Buffer for 15 minutes at room temperature. To measure the firefly luciferase activity, 50μl Luciferase Assay Buffer Ⅱ was mixed with 20μl cell lysate followed by absorbance detection. The Renilla luciferase activity was determined by mixing 50μl 1×Stop & Glo® reagent with the previous mixture. Absorbance was detected on microplate reader (TECAN, Infinite® 200 PRO NanoQuant). All the luciferase assays were performed in triplicate wells and the experiment was performed 3 times.
Cell culture immunofluorescence
The myoblasts were cultured in 6-well culture plates, fixed with 4% paraformaldehyde for 15 minutes, washed with PBS, permeabilized with 0.2% Triton X-100 for 15 minutes and then incubated in 10% (vol/vol) normal donkey serum/1% BSA (Sigma) /0.3 M glycine (Sigma) for 1h to block non-specific protein-protein interactions. For immunofluorescence, the cells were incubated with the primary antibody (diluted in 10% normal donkey serum/1% BSA/0.3 M glycine) overnight at 4°C. The cells were then washed with PBS and incubated with secondary antibody protected from light at 37°C for 1h. The nuclei were stained protected from light with DAPI (Sigma) at room temperature for 15 minutes. The antibodies were used as follows: anti-α-actinin (H-300) (1:200, Santa), and donkey anti-rabbit IgG H&L (Alexa Fluor® 555) (1:1000, Abcam). DAPI was used at the final concentration of 1μg/ml. Immunofluorescence images were taken by Olympus IX71 microscope (OLYMPUS).
siRNA transfection
Cells were seeded in 6-well plates and when cells grown to 70% confluence, siRNAs were transfected according to the standard protocol at a final concentration of 25nM. Briefly, 1.32μl X-tremeGENE HP DNA transfection reagent (Roche) and 0.66μg siRNA were diluted in Opti-MEM (Gibco) respectively for 10 minutes. The two mixtures were then mixed for another 15 minutes at room temperature to allow to form transfection reagent-siRNA complexes. The complex mixture was then added to the cell culture medium. Cells were replaced with fresh complete growth medium 8 hours later. The siRNA transfection was performed in triplicate wells and the experiment was performed with 3 repeats.
Quantitative real time-PCR
Total RNA from myoblasts (n = 3) were isolated by Trizol reagent (Takara) according to the manufacturer’s instructions. The RNA was then applied to synthesize cDNA using PrimeScriptTM RT reagent Kit with gDNA Eraser (Takara). The reverse transcript reaction was performed at 37°C for 15 minutes followed by 85°C for 5s. The cDNA was then used for quantitative real time-PCR (qRT-PCR) in triplicate wells using GoTaq® qPCR Master Mix (Promega) in 7500 Real Time PCR System (Applied Biosystems). The relative mRNA expression level was normalized to GAPDH. The detection of qRT-PCR was performed with 3 repeats. The experiment data were analyzed by using 2-ΔΔCT method [18]. Summary information of the genes used for qRT-PCR in this study were listed in S1 Table.
Western blot analysis
Cells were rinsed with PBS, digested with 0.25% trypsin (Gibco), and harvested into centrifuge tubes. Proteins were extracted using cell lysis buffer for western blot. Protein concentration was measured with BCA method (Takara). Cellular proteins were then mixed with protein loading buffer and denatured at 100°C for 10 minutes. 20μg protein samples were then subjected to 12% SDS-PAGE gel and transferred to PVDF membrane. For immune blot assay, the membrane was blocked with 5% skim milk (BD) for 2h and incubated with primary antibody diluted in blocking buffer at 4°C overnight. The membrane was then incubated with the secondary antibody protected from light at room temperature for 2h. Chemiluminescent HRP substrate (MILLIPORE) was used for taking immune blot images on BIO-RAD Molecular Imager. The images were analyzed using Image Lab software. All the immune blots were analyzed 3 times. The antibodies were used as follows: Anti-CDK1 antibody [EPR165] (Rabbit monoclonal primary antibody, 1:1000, Abcom), Anti-CDK2 antibody [E304] (Rabbit monoclonal primary antibody, 1:1000, Abcom), Anti-PCNA antibody [EPR3821] (Rabbit monoclonal primary antibody, 1:5000, Abcom), Anti-MEF2A antibody[EP1706Y] (Rabbit monoclonal primary antibody, 1:1000, Abcom), Anti-GAPDH antibody[EPR16884] (Rabbit monoclonal primary antibody, 1:10000, Abcom), Anti-β-actin antibody (Rabbit Polyclonal primary antibody, 1:10000, Novus), Anti-α-actinin Antibody (H-300) (Rabbit Polyclonal primary antibody, 1:200, Santa Cruz Biotechnology), Goat anti-IgG H&L (HRP) (1:2000, Abcom), Donkey Anti-Rabbit IgG H&L (Alexa Fluor® 555, 1:2000, Abcom).
Statistical analysis
All data are presented as mean ± SEM. Statistically significant differences between two groups were analyzed using Independent-samples t-test, and among three or more groups were analyzed by one-way analysis of variance (ANOVA). P < 0.05 was statistically significant [13].
Results
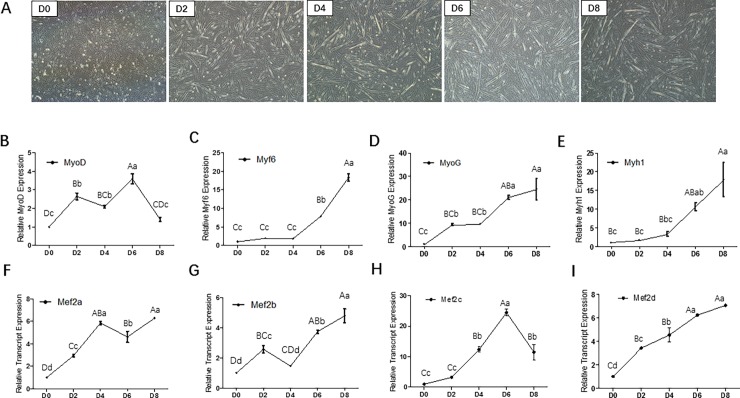
The Mef2 genes expression patterns during myoblast differentiation
To perform this study, primary bovine myoblasts were isolated from hind limb muscle and induced to myogenic differentiation. Forty-eight hours after myogenic induction, myoblasts began to form short myotubes (Fig 1A). As the culture continued, cells gradually elongated and most of the cultured cells fused into myotubes. The mRNA expression levels of the key myogenic transcription factors including MyoD, Mrf4, MyoG and Myh1 were measured at 0, 2, 4, 6 and 8 days after induction. The results showed that the mRNA levels of MyoD was higher in the early stage of differentiation and then gradually decreased (Fig 1B). In contrast, the mRNA levels of Mrf4, MyoG and Myh1 were continuously elevated during myoblast differentiation (Fig 1C–1E). These results indicated that the isolated primary myoblasts were satisfactory for the subsequent experiments.
Fig 1. Mef2 mRNA expression patterns in bovine myocytes.
(A) Cell culture of isolated bovine skeletal muscle primary myoblast and induction of myogenesis in vitro (OLYMPUS IX71 40×). (B~E) Relative mRNA expression of MyoD, Mrf4, MyoG and Myh1. (F~I) Relative mRNA expression of Mef2a, Mef2b, Mef2c and Mef2d. Error bars represent s.e.m. Different lowercases among different columns represent P < 0.05. Different uppercases among different columns represent P < 0.01.
To investigate the relationships between Mef2 and myoblast differentiation, the mRNA levels of Mef2a, Mef2b, Mef2c and Mef2d were also measured at 0, 2, 4, 6 and 8 days after induction. The mRNA levels of the four genes were all up-regulated during myoblast differentiation compared to that of the non-differentiated myoblasts (Fig 1F–1I). Moreover, the expression patterns of the four genes differed from each other. Mef2a was one of the first genes to exhibit differential expression in the time-course analysis. These differences suggest that the four genes of the MEF2 family perform distinct roles in skeletal muscle development. The time dependent expression pattern may reflect a specific role for MEF2A in bovine skeletal muscle myogenesis.
Construction of recombinant adenovirus to overexpress or interfere MEF2A
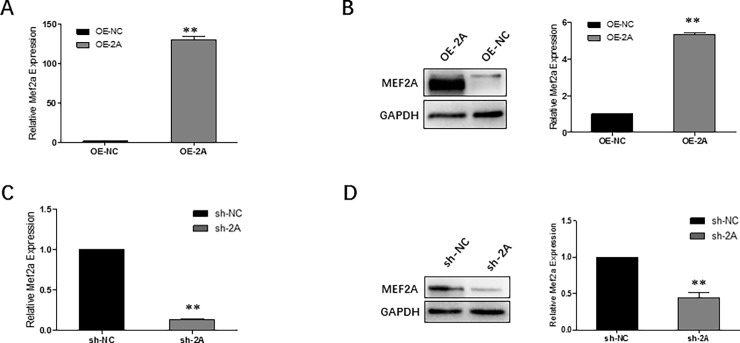
Specific adenovirus either to overexpress or knockdown MEF2A were generated in 293A cells. Adenovirus carrying full length bovine Mef2a CDS was successfully packaged within 8 days (S1A Fig). Cells were infected at a multiplicity of infection (MOI) of 50. The expression efficiency of OE-2A was examined through infection of skeletal muscle myoblasts. The relative mRNA expression level of Mef2a increased nearly 150 times in OE-2A infected cells compared to the control group (Fig 2A). The MEF2A protein expression level also significantly up-regulated in OE-2A infected myoblasts (Fig 2B).
Fig 2. Construction of recombinant adenovirus to overexpress or interfere MEF2A.
(A~B) OE-2A efficiently elevated mRNA expression level (A) and protein expression level (B)of Mef2a in myoblasts. (C~D) sh-2A efficiently interfered MEF2A mRNA expression level (C) and protein expression level (D) in myoblasts. Error bars represent s.e.m. *P < 0.05; **P < 0.01.
Specific shRNA used to inhibit MEF2A expression was subsequently designed (S1B Fig). To select the specific shRNA with the highest interference efficiency, psiCHECKTM-Ⅱ luciferase reporter assay was carried out in 293A cells. As shown in S1C Fig, sh-1219 was selected as the specific sh-RNA to interfere MFE2A expression. Adenovirus carrying the selected shRNA were successfully packaged within 15 days (sh-2A and sh-NC) and viral particles were expanded in 293A cells (S1D Fig). Primary skeletal muscle myoblasts were infected with sh-2A or sh-NC at MOI of 50. As shown in Fig 2C, the shRNA had efficiently reduced mRNA expression level Mef2a gene nearly up to 87%. Although sh-NC still had nearly 20% knockdown efficiency, the final Mef2a transcript expression level showed no differences between sh-NC and control group (data not shown). Western blot analysis showed that MEF2A protein expression also significantly decreased in sh-2A infected myoblasts (Fig 2D).
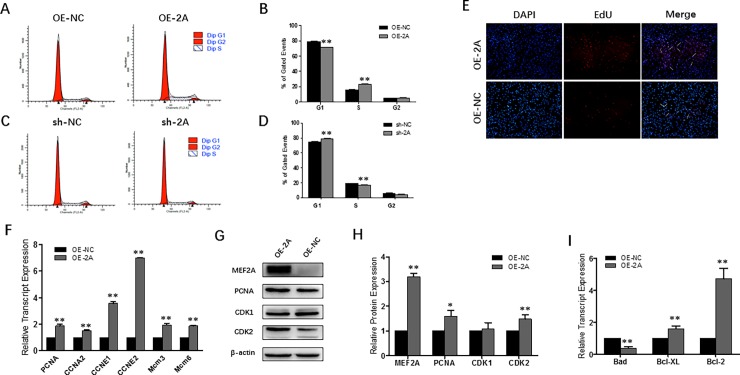
MEF2A promotes myoblast proliferation by triggering cell cycle progression
Overexpression of MEF2A in myoblasts induced a noticeable decrease in G1 phase cell counts and increased S phage cell counts (Fig 3A and 3B). While suppressing MEF2A expression, it showed the opposite effects (Fig 3C and 3D) in that cells were blocked in the G1 phase in sh-2A infected myoblasts. The apparent transition from the G1 phase to S phase in OE-2A infected myoblasts suggested a positive effect of MEF2A on myoblast proliferation. To confirm whether increased cell cycle activity was associated with increased DNA synthesis, EdU incorporation assay was performed. Results showed that overexpression of MEF2A induced a noticeable increase in EdU+ myoblasts (Fig 3E).
Fig 3. MEF2A promotes myoblast proliferation through triggering cell cycle progression.
(A~B) Flow cytometric measurement of DNA content using propidium iodide (PI) staining in OE-2A/OE-NC treated proliferating myoblast. (C~D) Flow cytometric measurement of DNA content using propidium iodide (PI) staining in sh-2A/sh-NC treated proliferating myoblast. (E) Images of the EdU assay: DAPI staining is shown in blue and EdU staining is in red (OLYMPUS IX71 100×). (F) Relative mRNA expression of cell cycle genes: PCNA, CCNA2, CCNE1, CCNE2, Mcm3 and Mcm6. (G~H) Western blot and protein expression analysis of PCNA, CDK1 and CDK2 showed that MFE2A activated CDK2 expression but not CDK1 expression. (I) Relative mRNA expression of pro-apoptotic gene (Bad) and pro-survival genes (Bcl-2 and Bcl-XL) at early apoptotic stage. Error bars represent s.e.m. *P < 0.05; **P < 0.01.
Proliferating cell nuclear antigen (PCNA) is an essential cofactor in DNA replication. RT-PCR and Western blot analysis showed that PCNA was up-regulated upon overexpression of MEF2A (Fig 3F–3H). We also found that the mRNA levels of cell cycle genes, including CCNA2, CCNE1, CCNE2, Mcm3 and Mcm6, were all up-regulated in OE-2A treated myoblasts (Fig 3F). In vertebrates, cyclin E binds to and activates CDK2 and then promotes the G1 to S phase transition. Cyclin A binds to CDK1 and activates G2 phase [19]. In this study, we found that overexpression of MEF2A increased CDK2 but not CDK1 protein expression (Fig 3G and 3H), resulting in promoted G1 to S phase transition. Bad is a pro-apoptotic gene and Bcl-XL, Bcl-2 are pro-survival genes that functions at early apoptotic stage in mitochondria. In this study, mRNA expression of these genes was also examined in OE-2A infected myoblasts. Theas results showed that MEF2A overexpression didn’t induce myoblast apoptosis or cell death.
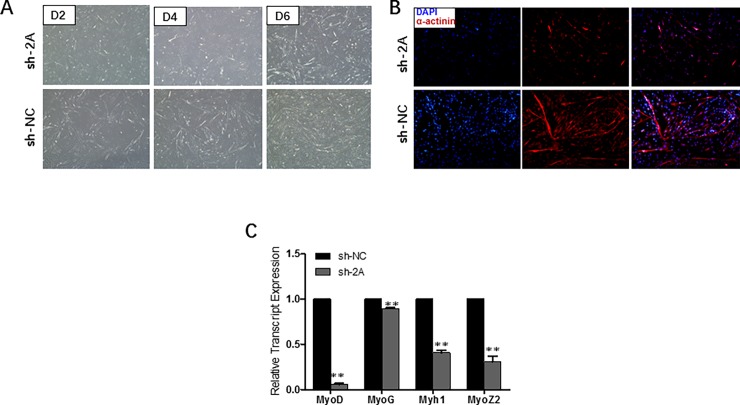
MEF2A knockdown inhibits myoblast differentiation
To investigate the function of MEF2A in regulating myoblast differentiation, MEF2A interference assay was carried out by using primary myoblasts. As shown in Fig 4A, MEF2A knocked down in differentiating myoblasts resulted in severely impaired myotube formation. During myogenic differentiation, sh-2A infected myocytes exhibited poor differentiation potential from D0 to D6. The amount of α-actinin+ myocytes at D6 of differentiation decreased significantly in MEF2A interfered myoblasts (Fig 4B). Upon determining the roles of MEF2A in myoblast differentiation, the relative transcript expression levels of MRFs and myosin were also examined because MRFs play essential roles in myogenic determination and terminal differentiation process [20, 21]. The mRNA expression levels of MyoD, Mrf4, MyoG, Myh1 and MyoZ2 were significantly down-regulated in MEF2A interfered myoblasts. In this process, MEF2A affects both the early myogenic determination and the terminal differentiation stages.
Fig 4. MEF2A knockdown inhibits myoblast differentiation.
(A) Morphological changes of differentiating bovine skeletal primary myoblasts at 2-day (D2), 4-day (D4) and 6-day (D6) after infection of sh-2A and sh-NC (OLYMPUS IX71 40×) (B) Images of immunofluorescence assay at differentiation day 6 treated with sh-2A or sh-NC (OLYMPUS IX71 100×). DAPI staining is shown in blue and α-actinin is shown in red. (C) Relative mRNA expression levels of myoblast differentiation marker genes (MyoD, Mrf4, MyoG and Myh1) significantly reduced in sh-2A treated cells. Error bars represent s.e.m. *P < 0.05; **P < 0.01.
MEF2A transcriptionally regulates the MyoZ2 proximal promoter in bovine myoblasts
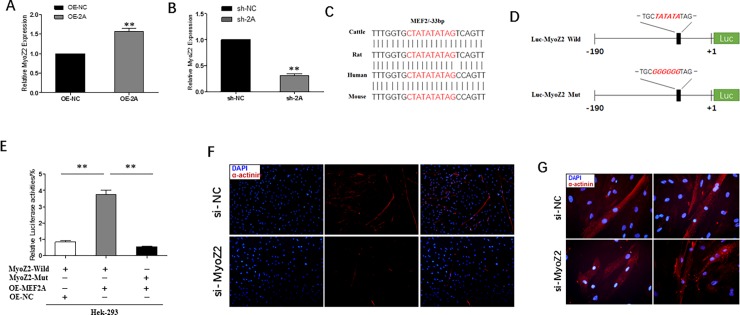
MYOZ2 is a sarcomeric calcineurin-binding protein residing in the Z-disk that plays a crucial role in myofiber formation and human hypertrophic cardiomyopathy [22–24]. Previous studies from our laboratory predicted that MEF2A was an important transcription factor that binds to the MyoZ2 proximal promoter and might regulate MyoZ2 transcriptional activity. In the present study, we found that MyoZ2 closely co-expressed with Mef2a (Fig 5A and 5B). Sequence alignment of the bovine MyoZ2 promoter revealed that the only -33bp MEF2 binding site, C/T TA(A/T)4TA G/A [25, 26], and its flanking sequences were nearly completely conserved among human, mouse, rat and bovine genomes (Fig 5C). To investigate whether MEF2A could regulate MyoZ2 transcriptional activity, MyoZ2 luciferase reporter vector with wild or mutant -33bp MEF2 binding site was generated and transfected into 293A cells (Fig 5D). The results showed that MEF2A could activate the MyoZ2 proximal promoter that harbored the MEF2 transcription factor binding site. 293A cells co-transfected with OE-2A and the MyoZ2 promoter resulted in two-fold higher levels of luciferase activity compared to the control group (Fig 5E). On the other hand, mutation of MEF2 binding site significantly reduced MyoZ2 transcriptional activity (Fig 5E). These results indicated that MEF2A promoted MyoZ2 expression by activating the promoter region.
Fig 5. MyoZ2 transcriptional activity is regulated by MEF2A and silencing MyoZ2 inhibited myoblast differentiation.
(A~B) Relative mRNA expression analysis of MyoZ2 showed that MyoZ2 was closely co-expressed with Mef2a. (C) Sequence alignments of MEF2 transcription binding site in the MyoZ2 promoter among cattle, rat, human and mouse was highly conserved. (D) Structure of MyoZ2 luciferase reporter vector with wild or mutant -33 bp MEF2 binding site. The mutant sequence was marked in italic red. (E) Luciferase analysis showed that MEF2A could efficiently promote MyoZ2 transcriptional activity. (F~G) Images of immunofluorescence assay at differentiation day 6 transfected with MyoZ2 specific siRNA or negative control siRNA (F: OLYMPUS IX71 100×; G: OLYMPUS IX71 400×). Error bars represent s.e.m. *P < 0.05; **P < 0.01.
Silencing MyoZ2 in myoblasts results in impaired myotube formation
To investigate the role of MyoZ2 in myoblast differentiation, MyoZ2 silencing assay were performed by using specific siRNA. Consistent with the inhibitory effect of MEF2A, interference of MyoZ2 also resulted in severely impaired myotube formation (Fig 5F). In detail, we could see the deficiency effects more morphologically from Fig 5G. In the control group, there were more myofibers and myotubes were much longer. Fusion of myotubes was much more better with more nuclei in each myofiber. However, in siRNA treated myoblasts, myotubes were shorter and fragmented. The majority of the myoblasts did not fuse into myotubes and only few myoblasts were poorly fused. In addition, expression of α-actinin was significantly lower due to the abnormal distribution of α-actinin protein and myotubes compared to that of the control. These results indicated that MyoZ2 was also likely involved in myotube formation and performed downstream of MEF2A in regulating myoblast differentiation.
Discussion
Skeletal muscle myoblast proliferation and differentiation are very important in determining muscle growth and muscle quality. In the present study, we investigated the roles of Mef2a in regulating the proliferation and differentiation of bovine skeletal muscle primary myoblast. There were two major findings in our study. First, we found that MEF2A expression promoted myoblast proliferation by triggering cell cycle progression through up-regulation of CDK2 expression. This is the first evidence that MEF2A is required for skeletal muscle myoblast proliferation. Second, interference of MEF2A expression in myoblasts blocked myogenic differentiation by down-regulation of MyoZ2 transcriptional activity.
Multiple lines of evidence have suggested that the genes of the MEF2 family play pivotal roles in embryonic development [7, 27], skeletal muscle fiber formation [28, 29], and muscle or cardiac disease [4, 30–32]. To some extent, gene expression patterns of MEF2 genes can reflect their functions in the relevant tissue or cells. In rodents, the four MEF2 genes are all up-regulated in myogenesis [13]. In skeletal muscle development, Mef2a, Mef2c and Mef2d play more important roles than Mef2b. During embryonic development, Mef2a expresses later than Mef2c [27]. During skeletal muscle regeneration, Mef2a is one of the earliest genes that can be detected, whereas Mef2c and Mef2d expression occurs much later [13]. In our study, we also observed a general trend of the gradual expression of the four MEF2 genes. However, expression of Mef2a occurs much earlier than expression of Mef2c although Mef2c exhibits obviously differential expression pattern. This notion suggests that Mef2a may play different roles compared to other genes of the MEF2 family in skeletal muscle.
MEF2A has been known to regulate proliferation, survival, and apoptotic pathways in a variety of specialized cell types, such as neurons, cardiomyocytes, immune cells, vascular smooth muscle cells and endothelial cells [7, 9, 33–35]. Until now, it was unknown whether MEF2A could regulate skeletal muscle myoblast proliferation. Generally, the prototypical eukaryotic cell cycle is divided into four phases that are tightly controlled by CDK-Cyclin complexes [36]. During mammalian cell cycle, CDK4 and CDK6 together with D-type cyclins promote the transition from the gap 0 (G0) phase to gap 1 (G1) phase. Subsequently, CDK2 controls entry into the S phase in complex with Cyclin E and Cyclin A. Cdk1, in conjunction with Cyclin A and Cyclin B, then controls the entry and progression through the M phase [37]. The major checkpoints are G1-S, G2-M and progression into anaphase. In our study, by detecting cell cycle with flow cytometry, we found that MEF2A expression promoted transition of bovine skeletal myoblasts from the G1 to S phase, while MEF2A inhibition arrests the cell cycle in the G1 phase. This positive effect activates CDK2 but not CDK1 protein expression. These results differ from what Liu (2014) reported that MEF2 is dispensable in mouse satellite cell proliferation [14]. However, we believe their result is not very precise because the methods used were too subjective. In addition, their EdU labeling time was too long and likely allowed some cells to have finished one cell cycle and entered into another. Although further investigations on how MEF2A activates myoblast proliferation is needed, we conclude that MEF2A is likely a positive regulator in the process of bovine myoblast cell cycle.
At present, the role of MEF2A in skeletal muscle cell differentiation has been extensively studied, and it has been found that MEF2A nearly had no effect on skeletal muscle development [14, 38]. However, contradictory results have also been reported. Estrella et al. and Synder et al. revealed that MEF2A was quite necessary for mice skeletal muscle differentiation and regeneration [12, 13]. They explained that the phenotypic difference was due to radically different MEF2 temporal expression in C2C12 cells and developmental stages [13]. In our study, we found that expression of MEF2A was necessary for bovine skeletal muscle myotube formation. What should be noticed was that all these studies mentioned above were performed under different developmental stages and experimental conditions (in vitro or in vivo) in different animal models even though on the same organ: skeletal muscle. Thus, the functions of MEF2A in skeletal muscle is time-, dose-, environment- and species-dependent. MyoD is a myogenic determination gene that is expressed prior to differentiation. Mrf4 acts as both a determination and a differentiation factor, whereas MyoG is a terminal differentiation gene that expresses later [39]. Meanwhile, myosin is a superfamily that constructs myofibers. Myosin disfunction can result in severe muscular dystrophy and muscle diseases [40, 41]. Overall, the impairment differentiation effect of interfering MEF2A in proliferating myoblasts occurred from the determination stage to the terminal differentiation stage.
MyoZ2 is an important Z-disk gene in the sarcomere structure that is required for normal myotube formation as well as muscle development [42]. It is also an inhibitor of calcineurin (also called protein phosphatase 2B, PP2B) that regulates myofiber type and muscle development through the calcineurin-NFAT signaling pathway [43]. Meanwhile, MEF2A is an important transcription factor in CaMK-HDACs, calcineurin and MAPK pathway [8, 9]. To data, there is nearly no evidence that clarifies whether MEF2A interacts with MyoZ2 in myoblasts. However, in our study, we noticed a tight co-expression of MyoZ2 and MEF2A in bovine skeletal muscle myoblasts. We also found that MyoZ2 transcriptional activity was directly regulated by MEF2A. Moreover, a ChIP-seq study found that MyoZ2 was immune precipitated by MEF2A antibody [44]. However, further investigation is needed to conclusively determine whether MEF2A directly interacts with MyoZ2. Cardiac hypertrophy is the abnormal muscle development. Current evidence suggests that overexpression of MyoZ2 can inhibit calcineurin-dependent signaling and result in overload-induced cardiac hypertrophy as well as arterial hypertension [45, 46]. In the present study, MyoZ2 may function through the similar mechanisms in skeletal myoblast differentiation. Notably, we found that MyoZ2 inhibition also reduced α-actinin expression which caused the abnormal protein distribution in myotubes. Therefore, while α-actinin forms the major components of the contractile apparatus at the Z-disk and regulates dystrophin [47, 48], it is likely that MyoZ2 inhibition induced myoblast differentiation defect is closely related to Z-disk protein activity.
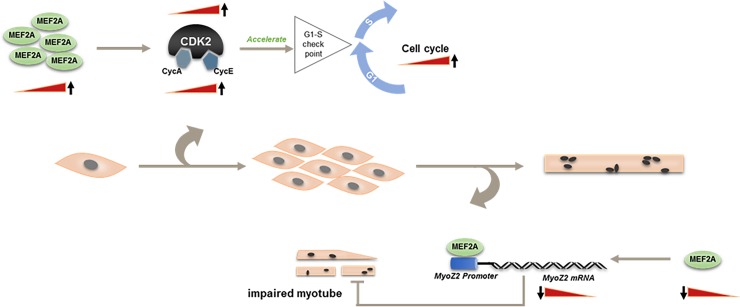
In conclusion, we report that MEF2A is a positive regulator in the proliferation and differentiation of bovine skeletal muscle primary myoblasts. MEF2A promotes myoblast proliferation by triggering cell cycle progression by activating CDK2 expression and regulates myoblast differentiation through transcriptional regulation of MyoZ2 (Fig 6). This study sheds some light on the roles of MEF2A in regulating bovine myoblast proliferation and differentiation in vitro and can help inform theories in cattle skeletal muscle development and improve gene therapies.
Fig 6. Mechanisms of MEF2A in regulating myogenesis of bovine skeletal muscle primary myoblasts.
MEF2A acts as a positive regulator in myoblast proliferation. It can promote cell cycle transition from G1 to S phase by activating CDK2. In addition, knockdown of MEF2A in myoblasts inhibits myogenic differentiation via transcriptionally regulating MyoZ2 expression.
Supporting information
(A) Recombinant adenovirus carrying full length bovine Mef2a CDS (OE-2A) was packaged within 8 days in 293A cells (40×). (B) Locations of sh-RNA that are specific for MEF2A. The four sh-RNAs are separated from each other vary from exon 4 to exon 10. (C) All the four specific sh-RNAs could significantly reduce MEF2A transcription efficiency. (D) Recombinant adenovirus carrying specific shRNA (sh-2A) and negative control shRNA (sh-NC) were packaged within 15 days in 293A cells (40×). Error bars represent s.e.m. Different lowercases among different columns represent P < 0.05. Different uppercases among different columns represent P < 0.01.
(TIF)
(DOCX)
Acknowledgments
The study is supported by National Modern Agricultural Industry Special Program (No. CARS-38), National 863 Program of China (No.2013AA102505), National Science and Technology Support Projects (No. 2015BAD03B04), Shaanxi Technological Innovation Engineering Program (No. 2014KTZB02-02-01) and the National Natural Science Foundation of China (Grant No. 31501937).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by 1. National Modern Agricultural Industry Special Program (Grant No. CARS-38) is funded by Ministry of Agriculture of the People’s Republic of China (URL: http://www.moa.gov.cn/) and Ministry of Finance of the People’s Republic of China (URL: http://www.mof.gov.cn/index.htm), Lin-Sen Zan received the funding; 2. National 863 Program of China (Grant No. 2013AA102505) is funded by Ministry of Science and Technology of the People’s Republic of China (URL: http://www.most.gov.cn/), Lin-Sen Zan received the funding; 3. National Science and Technology Support Projects (Grant No. 2015BAD03B04) is funded by Ministry of Science and Technology of the People’s Republic of China (URL: http://www.most.gov.cn/), Lin-Sen Zan received the funding; 4. Shaanxi Technological Innovation Engineering Program (Grant No. 2014KTZB02-02-01) is funded by Department of Science and Technology of Shaanxi Province (URL: http://www.sninfo.gov.cn), Lin-Sen Zan received the funding; 5. the National Natural Science Foundation of China (Grant No. 31501937) is funded by National Natural Science Foundation of China (URL: http://www.nsfc.gov.cn/), Ying-Ying Zhang received the funding; The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Almada AE, Wagers AJ. Molecular circuitry of stem cell fate in skeletal muscle regeneration, ageing and disease. Nat Rev Mol Cell Biol. 2016; 17(5): 267–279. doi: 10.1038/nrm.2016.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braun T, Gautel M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat Rev Mol Cell Biol. 2011; 12(6): 349–361. doi: 10.1038/nrm3118 [DOI] [PubMed] [Google Scholar]
- 3.Bryson-Richardson RJ, Currie PD. The genetics of vertebrate myogenesis. Nat Rev Genet. 2008; 9(8): 632–646. doi: 10.1038/nrg2369 [DOI] [PubMed] [Google Scholar]
- 4.Cohen S, Nathan JA, Goldberg AL. Muscle wasting in disease: molecular mechanisms and promising therapies. Nat Rev Drug Discov. 2015; 14(1): 58–74. doi: 10.1038/nrd4467 [DOI] [PubMed] [Google Scholar]
- 5.Wang YX, Rudnicki MA. Satellite cells, the engines of muscle repair. Nat Rev Mol Cell Biol. 2012; 13(2): 127–133. [DOI] [PubMed] [Google Scholar]
- 6.Pon JR, Marra MA. MEF2 transcription factors: developmental regulators and emerging cancer genes. Oncotarget. 2016; 7(3): 2297–2312. doi: 10.18632/oncotarget.6223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007; 134(23): 4131–4140. doi: 10.1242/dev.008367 [DOI] [PubMed] [Google Scholar]
- 8.Schiaffino S, Serrano A. Calcineurin signaling and neural control of skeletal muscle fiber type and size. Trends Pharmacol Sc. 2002; 23(12): 569–575. [DOI] [PubMed] [Google Scholar]
- 9.Timothy AM, Zhang CL, Olson EN. MEF2: a calcium-dependent regulator of cell division, differentiation and death. TRENDS in Biochemical Sciences. 2002; 27(1):40–47. [DOI] [PubMed] [Google Scholar]
- 10.Buckingham M, Rigby PW. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev Cell. 2014; 28(3): 225–238. doi: 10.1016/j.devcel.2013.12.020 [DOI] [PubMed] [Google Scholar]
- 11.Clark AL, Naya FJ. MicroRNAs in the Myocyte Enhancer Factor 2 (MEF2)-regulated Gtl2-Dio3 Noncoding RNA Locus Promote Cardiomyocyte Proliferation by Targeting the Transcriptional Coactivator Cited2. J Biol Chem. 2015; 290(38): 23162–23172. doi: 10.1074/jbc.M115.672659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Estrella NL, Desjardins CA, Nocco SE, Clark AL, Maksimenko Y, Naya FJ. MEF2 transcription factors regulate distinct gene programs in mammalian skeletal muscle differentiation. J Biol Chem. 2015; 290(2): 1256–1268. doi: 10.1074/jbc.M114.589838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Snyder CM, Rice AL, Estrella NL, Held A, Kandarian SC, Naya FJ. MEF2A regulates the Gtl2-Dio3 microRNA mega-cluster to modulate WNT signaling in skeletal muscle regeneration. Development. 2013; 140(1): 31–42. doi: 10.1242/dev.081851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu N, Nelson BR, Bezprozvannaya S, Shelton JM, Richardson JA, Bassel-Duby R, et al. Requirement of MEF2A, C, and D for skeletal muscle regeneration. Proc Natl Acad Sci U S A. 2014; 111(11): 4109–4114. doi: 10.1073/pnas.1401732111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Springer ML, Rando T, Blau H M. Gene Delivery to Muscle. 2001. [DOI] [PubMed]
- 16.Wang H, Cheng G, Fu CZ, Wang HB, Yang WC, Wang HC. Sequence analysis of bovine C/EBPdelta gene and its adipogenic effects on fibroblasts. Mol Biol Rep. 2014; 41(1): 251–257. doi: 10.1007/s11033-013-2858-y [DOI] [PubMed] [Google Scholar]
- 17.Estrella NL, Clark AL, Desjardins CA, Nocco SE, Naya FJ. MEF2D deficiency in neonatal cardiomyocytes triggers cell cycle re-entry and programmed cell death in vitro. J Biol Chem. 2015; 290(40): 24367–24380. doi: 10.1074/jbc.M115.666461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu S, Sun GJ, Yuan B, Zhang LJ, Gao Y, Jiang H, et al. miR-375 negatively regulates porcine preadipocyte differentiation by targeting BMPR2. FEBS Lett. 2016; 590(10): 1417–1427. doi: 10.1002/1873-3468.12169 [DOI] [PubMed] [Google Scholar]
- 19.Kaushik Tiwari M, Adaku N, Peart N, Rogers FA. Triplex structures induce DNA double strand breaks via replication fork collapse in NER deficient cells. Nucleic Acids Res. 2016; 44(16): 7742–7754. doi: 10.1093/nar/gkw515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol. 2005; 16(4–5): 585–595. doi: 10.1016/j.semcdb.2005.07.006 [DOI] [PubMed] [Google Scholar]
- 21.Moncaut N, Rigby PW, Carvajal JJ. Dial M(RF) for myogenesis. FEBS J. 2013; 280(17): \3980–3990. doi: 10.1111/febs.12379 [DOI] [PubMed] [Google Scholar]
- 22.Frey N, Richardson JA, Olson EN. Calsarcins, a novel family of sarcomeric calcineurin-binding proteins. Proc Natl Acad Sci U S A. 2000; 97(26): 14632–14637. doi: 10.1073/pnas.260501097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol. 2006; 7(8): 589–600. doi: 10.1038/nrm1983 [DOI] [PubMed] [Google Scholar]
- 24.Osio A, Tan L, Chen SN, Lombardi R, Nagueh SF, Shete S. Myozenin 2 is a novel gene for human hypertrophic cardiomyopathy. Circ Res. 2007; 100(6): 766–768. doi: 10.1161/01.RES.0000263008.66799.aa [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andres V, Cervera M, Mahdavi V. Determination of the consensus binding site for MEF2 expressed in muscle and brain reveals tissue-specific sequence constraints. J Biol Chem. 1995; 270(40): 23246–23269. [DOI] [PubMed] [Google Scholar]
- 26.Pollock R, Treisman R. Human SRF-related proteins: DNA-binding properties and potential regulatory targets. Genes Dev. 1991; 5(12A): 2327–2341. [DOI] [PubMed] [Google Scholar]
- 27.Edmondson DG, Lyons GE, Martin JF, Olson EN. Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development. 1994; 120(5): 1251–1263. [DOI] [PubMed] [Google Scholar]
- 28.Estrella NL, Naya FJ. Transcriptional networks regulating the costamere, sarcomere, and other cytoskeletal structures in striated muscle. Cell Mol Life Sci. 2014; 71(9): 1641–1656. doi: 10.1007/s00018-013-1512-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu H, Naya FJ, McKinsey TA, Mercer B, Shelton JM, Chin ER. MEF2 responds to multiple calcium-regulated signals in the control of skeletal muscle fiber type. Embo Journal. 2000; 19(9): 1963–1973. doi: 10.1093/emboj/19.9.1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cante-Barrett K, Pieters R, Meijerink JP. Myocyte enhancer factor 2C in hematopoiesis and leukemia. Oncogene. 2014; 33(4): 403–410. doi: 10.1038/onc.2013.56 [DOI] [PubMed] [Google Scholar]
- 31.Chen XY, Liu GL, Zhang W, Zhang JN, Yan YG, Dong WQ. Inhibition of MEF2A prevents hyperglycemia-induced extracellular matrix accumulation by blocking Akt and TGF-beta1/Smad activation in cardiac fibroblasts. Int J Biochem Cell Biol. 2015; 69: 52–61. doi: 10.1016/j.biocel.2015.10.012 [DOI] [PubMed] [Google Scholar]
- 32.Konno T, Chen D, Wang L, Wakimoto H, Teekakirikul P, Nayor M. Heterogeneous myocyte enhancer factor-2 (Mef2) activation in myocytes predicts focal scarring in hypertrophic cardiomyopathy. Proc Natl Acad Sci U S A. 2010; 107(42): 18097–18102. doi: 10.1073/pnas.1012826107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolodziejczyk SM, Wang L, Balazsi K, Derepentigny Y, Kothary R, Megeney LA. MEF2 is upregulated during cardiac hypertrophy and is required for normal post-natal growth of the myocardium. Current Biology. 1999; 9(20): 1203–1206. doi: 10.1016/S0960-9822(00)80027-5 [DOI] [PubMed] [Google Scholar]
- 34.Passier R, Zeng H, Frey N, Naya FJ, Nicol RL, McKinsey TA. CaM kinase signaling induces cardiac hypertrophy and activates the MEF2 transcription factor in vivo. J Clin Invest. 2000; 105(10): 1395–1406. doi: 10.1172/JCI8551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Welten SMJ, de Vries MR, Peters EAB, Agrawal S, Quax PHA, Nossent AY. Inhibition of Mef2a Enhances Neovascularization via Post-transcriptional Regulation of 14q32 MicroRNAs miR-329 and miR-494. Mol Ther Nucleic Acids. 2017; 7: 61–70. doi: 10.1016/j.omtn.2017.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cai L, Tu BP. Driving the cell cycle through metabolism. Annu Rev Cell Dev Biol. 2012; 28: 59–87. doi: 10.1146/annurev-cellbio-092910-154010 [DOI] [PubMed] [Google Scholar]
- 37.Harashima H, Dissmeyer N, Schnittger A. Cell cycle control across the eukaryotic kingdom. Trends Cell Biol. 2013; 23(7): 345–356. doi: 10.1016/j.tcb.2013.03.002 [DOI] [PubMed] [Google Scholar]
- 38.Potthoff MJ, Arnold MA, McAnally J, Richardson JA, Bassel-Duby R, Olson EN. Regulation of skeletal muscle sarcomere integrity and postnatal muscle function by Mef2c. Mol Cell Biol. 2007; 27(23): 8143–8151. doi: 10.1128/MCB.01187-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kassar-Duchossoy L, Gayraud-Morel B, Gomes D, Rocancourt D, Buckingham M, Shinin V, et al. Mrf4 determines skeletal muscle identity in Myf5: Myod double-mutant mice. Nature. 2004; 431(7007): 466–471. doi: 10.1038/nature02876 [DOI] [PubMed] [Google Scholar]
- 40.Heissler SM, Sellers JR. Various Themes of Myosin Regulation. J Mol Biol. 2016; 428(9 Pt B): 1927–1946. doi: 10.1016/j.jmb.2016.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Talbot J, Maves L. Skeletal muscle fiber type: using insights from muscle developmental biology to dissect targets for susceptibility and resistance to muscle disease. Wiley Interdiscip Rev Dev Biol. 2016; 5(4): 518–534. doi: 10.1002/wdev.230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takada F, Vander Woude DL, Tong HQ, Thompson TG, Watkins SC, Kunkel LM, et al. Myozenin: an alpha-actinin- and gamma-filamin-binding protein of skeletal muscle Z lines. Proc Natl Acad Sci U S A. 2001; 98(4): 1595–1600. doi: 10.1073/pnas.041609698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruggiero A, Chen SN, Lombardi R, Rodriguez G, Marian AJ. Pathogenesis of hypertrophic cardiomyopathy caused by myozenin 2 mutations is independent of calcineurin activity. Cardiovasc Res. 2013; 97(1): 44–54. doi: 10.1093/cvr/cvs294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wales S, Hashemi S, Blais A, McDermott JC. Global MEF2 target gene analysis in cardiac and skeletal muscle reveals novel regulation of DUSP6 by p38MAPK-MEF2 signaling. Nucleic Acids Res. 2014; 42(18): 11349–11362 doi: 10.1093/nar/gku813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frank D, Kuhn C, van Eickels M, Gehring D, Hanselmann C, Lippl S. Calsarcin-1 protects against angiotensin-II induced cardiac hypertrophy. Circulation. 2007; 116(22): 2587–2596. doi: 10.1161/CIRCULATIONAHA.107.711317 [DOI] [PubMed] [Google Scholar]
- 46.Paulsson AK, Franklin S, Mitchell-Jordan SA, Ren S, Wang Y, Vondriska TM. Post-translational regulation of calsarcin-1 during pressure overload-induced cardiac hypertrophy. J Mol Cell Cardiol. 2010; 48(6): 1206–12014. doi: 10.1016/j.yjmcc.2010.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beggs AH, Byers TJ, Knoll JH, Boyce FM, Bruns GA, Kunkel LM. Cloning and characterization of two human skeletal muscle alpha-actinin genes located on chromosomes 1 and 11. J Biol Chem. 1992; 267(13): 9281–9288. [PubMed] [Google Scholar]
- 48.Lee FX, Houweling PJ, North KN, Quinlan KJ. How does alpha-actinin-3 deficiency alter muscle function? Mechanistic insights into ACTN3, the 'gene for speed'. Biochim Biophys Acta. 2016; 1863(4): 686–693. doi: 10.1016/j.bbamcr.2016.01.013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Recombinant adenovirus carrying full length bovine Mef2a CDS (OE-2A) was packaged within 8 days in 293A cells (40×). (B) Locations of sh-RNA that are specific for MEF2A. The four sh-RNAs are separated from each other vary from exon 4 to exon 10. (C) All the four specific sh-RNAs could significantly reduce MEF2A transcription efficiency. (D) Recombinant adenovirus carrying specific shRNA (sh-2A) and negative control shRNA (sh-NC) were packaged within 15 days in 293A cells (40×). Error bars represent s.e.m. Different lowercases among different columns represent P < 0.05. Different uppercases among different columns represent P < 0.01.
(TIF)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.