Abstract
Latent transforming growth factor-β-binding protein (LTBP)2 is a member of the fibrillin/LTBP superfamily of extracellular matrix proteins, and has been demonstrated to exhibit tumor-promoting and tumor-suppressive functions in different types of cancer. However, the function of LTBP2 in gastric cancer (GC) remains unknown. The aim of the present study was to investigate the expression and molecular function of LTBP2 in GC, and to evaluate its prognostic value for patients with GC. The results revealed that the expression of LTBP2 was upregulated in GC tissues and cell lines. Increased LTBP2 expression was associated with poor overall survival in patients with early-stage [tumor-node-metastasis (TNM) I/II] and late-stage (TNM III/IV) GC. Furthermore, silencing of LTBP2 effectively suppressed the proliferation, migration, invasion and epithelial-mesenchymal transition in GC cells. These results suggested that LTBP2 may be considered as a potential therapeutic target and a promising prognostic biomarker for human GC.
Key words: gastric cancer, migration, invasion, latent transforming growth factor-β-binding protein 2, epithelial-mesenchymal transition
Introduction
Despite a generally decreasing trend in incidence, gastric cancer (GC) remains one of the most common types of cancer and the third leading cause of cancer-associated mortality worldwide (1,2). Although improved diagnostic and therapeutic strategies have improved early-stage GC detection and decreased patient mortality (3), the diagnosis of GC in >80% of patients is made at an advanced stage, which usually indicates unfavorable clinical outcomes (4). Invasion and metastasis are the primary reasons for the poor prognosis of GC. In patients with an initial diagnosis, lymph node metastasis has been identified in >50% of patients with GC, and peritoneum metastasis may be present in between 5 and 20% of patients undergoing curative surgery (5). As invasion and metastasis serve a critical function in the progression of GC (6), unveiling the molecular mechanisms of the invasion and metastasis of GC cells and identification of novel biomarkers remain of great importance for early diagnosis and effective treatment.
Latent transforming growth factor (TGF) β-binding protein (LTBP)2 belongs to the LTBP/fibrillin family of extracellular matrix (ECM) proteins, which includes LTBPs 1–4 and fibrillins 1–3 (7). LTBPs mainly consist of a repeated domain structure including calcium-binding epidermal growth factor-like domain repeats and 8-Cys TGF-β-binding domain (TB domain) repeats (8). LTBPs serve an important function in modulating the structural integrity of the ECM and targeting the latent TGF-β complex to the cell surface so that the latent TGF-β is efficiently activated (9). Unlike the other LTBPs, LTBP2 is unable to associate with small latent TGF-β (10). Previous studies have demonstrated that LTBP2 mutations serve a critical function in primary congenital glaucoma (11), microspherophakia (12), Weill-Marchesani syndrome (13) and acute dyspnea (14). In addition, previous studies have also indicated various functions of LTBP2 in different types of cancer. Increased expression of LTBP2 was observed in head and neck squamous cell carcinoma (15), and ovarian (16), cervical (17), thyroid (18) and liver (19) cancer, and was associated with unfavorable outcomes and tumor progression. In contrast, in esophageal squamous cell carcinoma (20) and nasopharyngeal carcinoma (21), LTBP2 was downregulated and served a tumor-suppressive function. However, the expression and function of LTBP2 in GC remains unknown.
In the present study, it was identified that LTBP2 is overexpressed in human GC tissues and cell lines, and its overexpression is associated with depth of tumor invasion, lymph node metastasis and tumor-node-metastasis (TNM) stage. Increased LTBP2 expression is associated with poor overall survival of patients with early-stage (TNM I/II) and late-stage (TNM III/IV) GC. LTBP2 knockdown impaired the proliferation, migration and invasion of GC cells, accompanied by alteration of the expression of epithelial-mesenchymal transition (EMT)-associated factors. The results of the present study suggested that LTBP2 acts as an oncogene, and that it serves an important function in promoting the invasion and migration of GC cells, and may be considered as a potential therapeutic target for GC.
Materials and methods
Patients and tissue specimens
GC and adjacent normal tissue samples were obtained from 174 patients with GC with histologically confirmed gastric adenocarcinoma between January 2007 and December 2010 at the First Hospital of Lanzhou University (Lanzhou, China). All cases were followed up for >5 years. All patients were treated with radical gastrectomy and D2 lymphadenectomy with postoperative chemotherapy based on fluorouracil regimens. None of the patients had been treated using chemotherapy or radiotherapy pre-operation. Each GC specimen was histologically graded by two pathologists in a double-blinded manner. Tumor stages were determined according to the 7th edition of the American Joint Committee on Cancer TNM classification system (22). A total of 16 paired tissue specimens from fresh GC and matched non-cancerous tissues were frozen at −80°C until being subjected to RNA extraction for reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis. The present study was approved by the Ethics Committee of the First Hospital of Lanzhou University and written informed consent was obtained from all enrolled patients. The clinical features of patients are summarized in Table I.
Table I.
Clinical features of patients with gastric cancer.
| Clinical characteristic | n (%) |
|---|---|
| Age, years | |
| ≥60 | 73 (41.95) |
| <60 | 101 (58.05) |
| Sex | |
| Male | 120 (68.97) |
| Female | 54 (31.03) |
| Histology grade | |
| G1 | 15 (8.62) |
| G2 | 45 (25.86) |
| G3 | 114 (65.52) |
| T stage | |
| T1 | 33 (18.97) |
| T2 | 25 (14.37) |
| T3 | 85 (48.85) |
| T4 | 31 (17.82) |
| N stage | |
| N0 | 71 (40.80) |
| N1 | 47 (27.01) |
| N2 | 44 (25.29) |
| N3 | 12 (6.90) |
| M stage | |
| M0 | 149 (85.63) |
| M1 | 25 (14.37) |
| TNM stage | |
| I | 46 (26.44) |
| II | 56 (32.18) |
| III | 47 (27.01) |
| IV | 25 (14.37) |
T, tumor; N, node; M, metastasis.
Cell culture
Human GC cell lines MGC803, SGC7901, BGC823, AGS and MKN-45 as well as the immortalized gastric epithelium cell line GES-1 were purchased from the Cell Bank of the Shanghai Institute of Cell Biology (Shanghai, China). Cells were cultured at 37°C in RPMI-1640 medium in a 100% humid atmosphere containing 5% CO2 medium supplemented with 10% fetal bovine serum (FBS) (both from HyClone; GE Healthcare, Logan, UT, USA).
Immunohistochemistry (IHC)
Immunohistochemical staining was performed to detect the expression of LTBP2 using a REAL™ EnVision™ Detection system (Dako; Agilent Technologies, Inc., Santa Clara, CA, USA), according to the manufacturer's protocol. Briefly, 4-µm-thick sections were cut and treated with xylene for deparaffinization and rehydration with a graded ethanol series. Tissue slides were incubated with rabbit anti-LTBP2 polyclonal antibody (1:300; ab121193; Abcam, Cambridge, UK) at 4°C overnight following antigen retrieval with EDTA (pH 9.0) in a pressure cooker for 15 min and blocking endogenous peroxidase with 3% H2O2 for 30 min at room temperature. Following washing with PBS three times to remove the primary antibody, tissue sections were incubated at 37°C for 30 min with a horseradish peroxidase-conjugated anti-mouse/anti-rabbit secondary antibody (cat. no. K5007; used undiluted; included in the REAL™ EnVision™ Detection system), according to the manufacturer's protocol. IHC staining was scored by multiplying staining intensity (0, no staining; 1, weak staining; 2, moderate staining; 3, strong staining) by the percentage of positive cancer cells (1, 1–25%; 2, 26–50%; 3, 51–75%; 4, >75%). The resulting IHC scores ranged between 0 and 12. An IHC score of 4 was determined as the optimal threshold value in the IHC cohort for all 174 patients with GC using X-tile software (version 3.6.1; Yale University School of Medicine, New Haven, CT, USA) (23) and SPSS (version 19.0; IBM Corporation, Armonk, NY, USA). Furthermore, an IHC score of 4 was determined in the same way as the threshold value for patients with TNM I/II and TNM III/IV tumors. Therefore, IHC scores ≥6 were classified as LTBP2high and scores ≤4 were classified as LTBP2low.
Online database analyses
The mRNA expression microarray data of patients with GC were downloaded from the publicly available National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database (www-ncbi.nlm.nih.gov/gds; accession nos. GSE27342, GSE26272 and GSE54129). Expression data were log2-transformed and quantile-normalized using R software (version 3.2.5; R Foundation for Statistical Computing, Vienna, Austria; cran.r-project.org) with packages of the BioConductor project (www.bioconductor.org) as described previously (24). For The Cancer Genome Atlas (TCGA) dataset analysis, the mRNA levels of LTBP2 in 408 GC tissues and 211 normal tissues [TCGA-Stomach Adenocarcinoma (STAD)] were compared using the Gene Expression Profiling Interactive Analysis online database (gepia.cancer-pku.cn), as described previously (25).
For the survival analysis of LTBP2, the KM Plotter database (www.kmplot.com/gastric) was used to validate the relevance of LTBP2 mRNA expression to overall survival in patients with GC (26–28). Among the 876 enrolled patients with GC in the KM Plotter database, there were 380 cases with surgical treatment. The prognostic value of LTBP2 was analyzed in all 876 patients as well as in the 380 surgical patients. The autoselected best threshold value was used to identify the high and low groups. The hazard ratio (HR) with 95% confidence interval (CI) and P-value were calculated, and analyzed data were then downloaded from the website. Kaplan-Meier estimator survival curves were plotted with the downloaded data using GraphPad Prism software (version 6.01; GraphPad Software, Inc., La Jolla, CA, USA).
Western blotting
Total protein was extracted using radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology, Haimen, China) with phenylmethylsulfonyl fluoride proteinase inhibitor and the concentration was determined using the bicinchoninic acid protein assay (both from Thermo Fisher Scientific, Inc., Waltham, MA, USA). Equal amounts of total protein (30 µg) were separated by PAGE (10% gel) and transferred onto polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA). The membranes were blocked with 5% non-fat milk in Tris-buffered saline containing 0.3% Tween-20 (TBST) at room temperature for 2 h and incubated with the primary antibodies at 4°C overnight. The primary antibodies were as follows: Anti-LTBP2 (cat. no. sc-166199; 1:1,000; Santa Cruz Biotechnology, Dallas, TX, USA), anti-β-actin (cat. no. AF0003; 1:1,000; Beyotime Institute of Biotechnology), anti-epithelial (E-)cadherin (cat. no. ab1416; 1:400), anti-vimentin (cat. no. ab8979; 1:400) and anti-neuronal (N-) cadherin (cat. no. ab98952; 1:400) (all from Abcam). Following washing of the membranes with TBST, the membranes were then incubated with corresponding horseradish peroxidase-conjugated anti-mouse immunoglobulin G secondary antibody (cat. no. A0216; 1:5,000; Beyotime Institute of Biotechnology) at room temperature for 1 h. Reactive bands were detected using the enhanced chemiluminescence detection system (SuperSignal West Femto Maximum Sensitivity Substrate; Thermo Fisher Scientific, Inc.).
RNA extraction and RT-qPCR
Total RNA was extracted from cells and the 16 pairs of frozen tissue specimens using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. RT was performed using the Prime Script RT reagent kit with gDNA Eraser (Takara Biotechnology Co., Ltd., Dalian, China), according to the manufacturer's protocol. The resulting cDNA was subjected to qPCR using a CFX96 Real-Time System (Bio-Rad Laboratories, Inc., Hercules, CA, USA) with SYBR Premix Ex Taq II (Takara Biotechnology Co., Ltd.), according to the manufacturer's protocol. The relative mRNA expression levels were normalized to those of β-actin using the 2−ΔΔCq method (29). Experiments were repeated in triplicate. The primer sequences used are presented in Table II.
Table II.
Sequences of primers used for the quantitative polymerase chain reaction.
| Gene | Sequence |
|---|---|
| LTBP2 | F: 5′-TTACAAGCAGAGACTCACT-3′ |
| R: 5′-ACAACAGAAGAGACCAGAT-3′ | |
| E-cadherin | F: 5′-TGCCTGAGAACGAGGCTAAC-3′ |
| R: 5′-TGGGGGCTTCATTCACATCC-3′ | |
| N-cadherin | F: 5′-TCTGGGTCTGTTTTATTACTCCTGG-3′ |
| R: 5′-CGAGCTGATGACAAATAGCGG-3′ | |
| Vimentin | F: 5′-CCAGGCAAAGCAGGAGTC-3′ |
| R: 5′-GGGTATCAACCAGAGGGAGT-3′ | |
| β-actin | F: 5′-TTGCGTTACACCCTTTCTTG-3′ |
| R: 5′-CACCTTCACCGTTCCAGTTT-3′ |
LTBP2, latent transforming growth factor-β-binding protein 2; E-cadherin, epithelial cadherin; N-cadherin, neuronal cadherin; F, forward; R, reverse.
Plasmid construction and stable short hairpin (sh)RNA transfection
shRNAs targeting LTBP2 (shLTBP2) and non-targeting control (mock) (Table III) were designed and synthesized by Shanghai GeneChem Co., Ltd. (Shanghai, China). The transfection of lentiviral vectors containing target sequence or control non-target sequence and packaging vectors into 293T cells were carried out using Lipofectamine® 2000 transfection reagent (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The viral supernatant was collected 24 h after transfection. SGC7901 and BGC823 cells were infected with purified lentiviruses containing shRNA sequence targeting LTBP2 or control sequence lentivirus for 24 h without FBS. Stably transfected cells were isolated using 2 µg/ml puromycin (Sigma; Merck KGaA, Darmstadt, Germany).
Table III.
Sequences of LTBP2 knockdown and mock shRNAs.
| Name | Sequence |
|---|---|
| shLTBP2-1 | F: 5′-CACCGTAGCAGAAGCAGGAGTAGGACCGAAGTCCTACTCCTGCTTCTGCTA-3′ |
| R: 5′-AAAATAGCAGAAGCAGGAGTAGGACTTCGGTCCTACTCCTGCTTCTGCTAC-3′ | |
| shLTBP2-2 | F: 5′-CACCGTTGGTGCACTTTCCATGGGCACGAATGCCCATGGAAAGTGCACCAA-3′ |
| R: 5′-AAAATTGGTGCACTTTCCATGGGCATTCGTGCCCATGGAAAGTGCACCAAC-3′ | |
| shLTBP2-3 | F: 5′-CACCGAAGGAGCCTTCTAGGTTGGTGCGAACACCAACCTAGAAGGCTCC-3′ |
| R: 5′-AAAAGGAGCCTTCTAGGTTGGTGTTCGCACCAACCTAGAAGGCTCCTTC-3′ | |
| Mock | F: 5′-CCGGTTACGCGTAGCGTAATACGCTCGAGCGTATTACGCTACGCGTAATTTTTG-3′ |
| R: 5′-AATTCAAAAATTACGCGTAGCGTAATACGCTCGAGCGTATTACGCTACGCGTAA-3′ |
LTBP2, latent transforming growth factor-β-binding protein 2; sh, short hairpin; F, forward; R, reverse.
Cell proliferation assay
Cell proliferation was measured using the Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan), according to the manufacturer's protocol. GC cells with different treatments (mock, shLTBP2-1 and shLTBP2-3) were seeded into 96-well plates (~5×103 cells per well) and cultured for 24, 48, 72 or 96 h. The medium was removed from each well, and 10 µl CCK-8 solution was added to each well and incubated at 37°C for 1 h. The optical density at 450 nm (OD450) was determined using an auto microplate reader. The OD450 reading was proportional to the degree of cell proliferation. The assays were performed in triplicate and repeated three times.
Colony-formation assay
GC cells with different treatments (mock, shLTBP2-1 and shLTBP2-3) were plated into 6-well tissue culture plates at a density of 500 cells/well and cultured at 37°C for 14 days in RPMI-1640 medium containing 10% FBS. Following washing the cells twice with PBS, clones were stained using 0.1% crystal violet (Beyotime Institute of Biotechnology) for 10 min after fixation with 4% paraformaldehyde. The colonies containing >50 cells were counted by eye under light microscopy at ×100 magnification. Experiments were performed in triplicate.
Transwell chamber migration and invasion assay
Cell migration and invasion assays were performed with modified Boyden chambers (Corning Incorporated, Corning, NY, USA) with 8-µm pore filter inserts in 24-well plates (BD Biosciences, San Jose, CA, USA). GC cells were serum-starved overnight prior to being used for Transwell migration and invasion assays. For the invasion assay, the insert membranes were coated with Matrigel (BD Biosciences) on the upper side for 30 min. A total of 2×104 cells were seeded in the upper chamber in 200 µl RPMI-1640 medium without FBS, whereas 500 µl RPMI-1640 medium supplemented with 10% FBS was added to the lower chamber. Following incubation at 37°C for 24 h, cells on the upper side of the membranes were removed with a cotton swab. Cells that had passed through the filter were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet solution. The number of infiltrating cells was counted in five randomly selected fields using an inverted light microscope (magnification, ×200) and images were captured.
Wound-healing assay
Cell migration ability was determined using a wound-healing assay following silencing of LTBP2. GC cells were incubated in 6-well plates until reaching 90% confluence prior to wounding. A wound was made by dragging 10-µl pipette tips along the center of the plate. Following washing the cells twice with PBS, serum-free medium was added. Following an additional 8 and 24 h of culture, images were captured and cell migration was determined. The migration rate was quantified as the cell coverage compared with the initial cell-free gap.
Statistical analysis
Statistical analysis was performed using SPSS software (version 19.0; IBM Corp, Armonk, NY, USA) and GraphPad Prism software. Results are presented as the mean ± standard deviation. Student's t-test was used for comparisons between two groups. The comparison of the IHC scores of LTBP2 and the mRNA levels of LTBP2 in fresh tissue samples were achieved using a paired Student's t-test. The comparisons of GSE27342 and GSE29272 were performed by single Student's t-test. Comparisons of three or more groups were performed using one-way analysis of variance followed by a least significant difference test as a post hoc test. The association between clinical variables and LTBP2 expression was analyzed using Pearson χ2 test. The Kaplan-Meier estimator method was used for survival analysis, and differences in survival were examined using the log-rank test. Cox's proportional hazards regression model was used for univariate and multivariate analyses. The threshold value of the LTBP2 IHC score was calculated using the statistical software X-tile (23) and receiver operating characteristic analysis was performed using SPSS software. P<0.05 was considered to indicate a statistically significant difference.
Results
LTBP2 is highly expressed in human GC tissues and cell lines
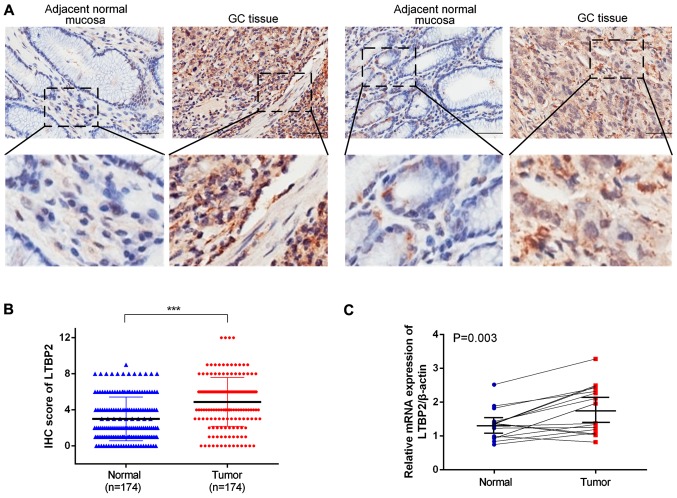
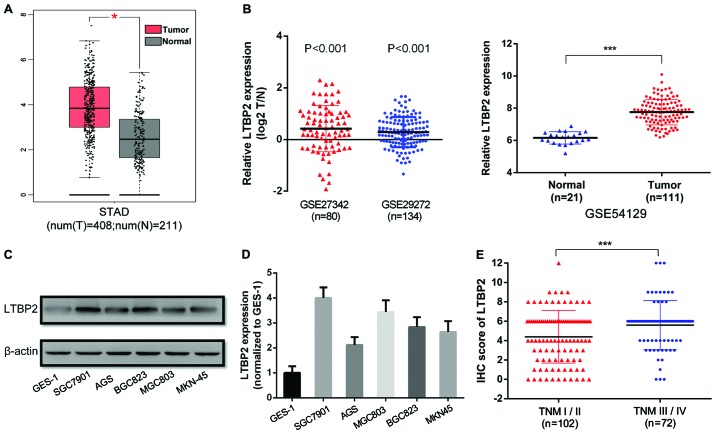
To investigate LTBP2 expression in GC tissues, IHC staining was performed in 174 GC tissues and paired adjacent normal tissues (representative images are presented in Fig. 1A). The proportion of LTBP2high specimens in GC tissues (54.60%, 95/174) was significantly increased compared with that in the paired adjacent normal tissues (23.56%, 41/174) (P<0.001; Table IV). Furthermore, the IHC score of LTBP2 in GC tissues was significantly increased compared with that of the paired normal tissues (P<0.001; Fig. 1B). The results indicated that LTBP2 was highly expressed in GC tissues compared with adjacent normal tissues. The mRNA levels of LTBP2 were determined in GC tissues using RT-qPCR analysis. A total of 16 pairs of GC and corresponding adjacent normal tissues were analyzed and the results indicated that LTBP2 mRNA levels were significantly upregulated in GC tissues compared with that of the adjacent normal specimens (P=0.003; Fig. 1C). Furthermore, the analyses of TCGA-STAD (P<0.05, Fig. 2A), GSE27342, GSE26272 and GSE54129 datasets (P<0.001; Fig. 2B) demonstrated that the mRNA levels of LTBP2 were significantly increased in GC tissues compared with in adjacent normal gastric mucosa. The results indicated that LTBP2 protein and mRNA levels were increased in GC tissues compared with in adjacent normal tissues.
Figure 1.
LTBP2 is highly expressed in GC tissues. (A) Representative IHC images of LTBP2 in adjacent normal mucosa and GC tissue. Scale bar, 50 µm. (B) The IHC score of LTBP2 was significantly higher in GC tissues compared with in adjacent normal tissues. ***P<0.001. (C) LTBP2 expression in 16 paired fresh surgical gastric tumor specimens and corresponding adjacent normal tissues detected using the reverse transcription-quantitative polymerase chain reaction. LTBP2, latent transforming growth factor-β-binding protein 2; GC, gastric cancer; IHC, immunohistochemistry.
Table IV.
LTBP2 expression in adjacent normal mucosa and gastric cancer tissues.
| Group | LTBP2low | LTBP2high | χ2 | P-value |
|---|---|---|---|---|
| Carcinoma | 79 | 95 | 35.196 | <0.001 |
| Adjacent normal mucosa | 133 | 41 |
LTBP2, latent transforming growth factor-β-binding protein 2.
Figure 2.
LTBP2 is overexpressed in GC tissues and cell lines, and is associated with the TNM stage of patients with GC. (A) Analysis of The Cancer Genome Atlas-STAD dataset exhibiting increased LTBP2 expression in GC tissues compared with adjacent normal tissues. (B) Analyses of the National Center for Biotechnology Information Gene Expression Omnibus datasets GSE27342, GSE29272 and GSE54129 indicating increased LTBP2 expression in GC tissues compared with adjacent normal tissues. Data in GES27342 and GSE29272 are paired data that was log2-transformed, whereas data in GES54129 are non-paired. (C) Western blotting and (D) RT-qPCR analyses of LTBP2 expression in five GC cell lines (AGC, SGC7901, MGC803, BGC823 and MKN-45) and GES-1 cells. (E) IHC score of LTBP2 in early-stage (TNM stage I/II) and late-stage (TNM stage III/IV) GC. *P<0.05; ***P<0.001. LTBP2, latent transforming growth factor-β-binding protein 2; GC, gastric cancer; TNM, tumor-node-metastasis; STAD, Stomach Adenocarcinoma; IHC, immunohistochemistry; T, tumor; N, normal.
To further confirm these results, the expression of LTBP2 was analyzed in five GC cell lines (SGC7901, MGC803, BGC823, AGS and MKN-45) and the immortalized gastric epithelium cell line GES-1 using western blotting and RT-qPCR assays. The results demonstrated that protein and mRNA levels of LTBP2 were increased in the five GC cell lines compared with in GES-1 cells (Fig. 2C and D, respectively). Among the five GC cell lines, SGC7901 and BGC823 expressed relatively increased levels of LTBP2. As a result, SGC7901 and BGC823 were selected for further study.
Increased expression of LTBP2 is associated with clinicopathological features and predicted poor outcomes of patients with GC
The association between the LTBP2 expression and the clinicopathological features of patients with GC was analyzed. The detailed information of patients and the association between LTBP2 expression and clinicopathological parameters are presented in Table V. The LTBP2 expression level was significantly associated with T stage (P=0.005), N stage (P=0.036), M stage (P=0.018) and TNM stage (P=0.017) of patients with GC. However, LTBP2 expression was not associated with the histological grade (P=0.691), age (P=0.332) or sex (P=0.140) (Table V). To further confirm that LTBP2 expression indicated an advanced clinical stage of patients with GC, the IHC score of LTBP2 was analyzed in early-stage (TNM I/II) and late-stage (TNM III/IV) GC (30,31). The results indicated that the IHC scores of LTBP2 were significantly increased in late-stage GC compared with in early-stage GC (P<0.001; Fig. 2E).
Table V.
Association between LTBP2 expression and clinicopathological features of patients with gastric cancer.
| Clinicopathological features | n | LTBP2 expression
|
χ2 | P-value | |
|---|---|---|---|---|---|
| Low | High | ||||
| Age, years | 0.941 | 0.332 | |||
| ≥60 | 73 | 30 | 43 | ||
| <60 | 101 | 49 | 52 | ||
| Sex | 2.177 | 0.140 | |||
| Male | 120 | 50 | 70 | ||
| Female | 54 | 29 | 25 | ||
| Histological grade | 0.158 | 0.691 | |||
| G1+G2 | 60 | 26 | 34 | ||
| G3 | 114 | 53 | 61 | ||
| T stage | 7.836 | 0.005 | |||
| T1–T2 | 58 | 35 | 23 | ||
| T3–T4 | 116 | 44 | 72 | ||
| N stage | 4.392 | 0.036 | |||
| N0 | 71 | 39 | 32 | ||
| N1–N3 | 103 | 40 | 63 | ||
| M stage | 7.600 | 0.006 | |||
| M0 | 149 | 74 | 75 | ||
| M1 | 25 | 5 | 20 | ||
| TNM stage | 5.652 | 0.017 | |||
| I–II | 102 | 54 | 48 | ||
| III–IV | 72 | 25 | 47 | ||
LTBP2, latent transforming growth factor-β-binding protein 2; T, tumor; N, node; M, metastasis.
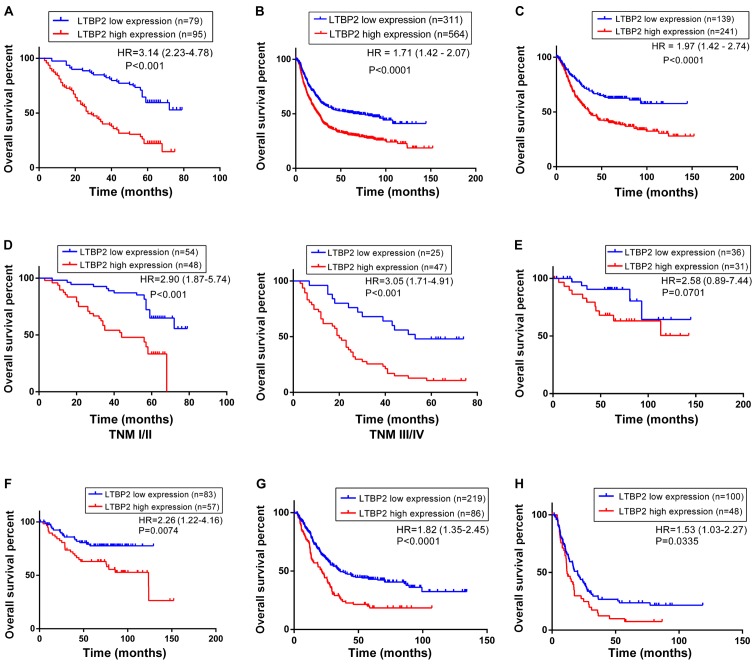
To determine the prognostic significance of LTBP2 in GC, the association between LTBP2 expression and the overall survival rates of 174 patients with GC was investigated. Kaplan-Meier estimator survival analysis indicated that the overall survival time for patients with LTBP2low (n=79) tumors was significantly increased compared with those with LTBP2high (n=95) tumors (HR, 3.14; 95% CI, 2.23–4.78; P<0.001; Fig. 3A). Multivariate analysis identified that high LTBP2 expression was an independent prognostic indicator for patients with GC (Table VI). To confirm the prognostic significance of LTBP2 expression in a large-scale cohort, the KM Plotter database, which includes a total of 875 GC samples (including 380 surgical samples) from published microarray data from the NCBI GEO database, was used. The prognostic significance in all 875 patients with GC as well as in the 380 surgical cases was analyzed. The results suggested that high levels of LTBP2 were significantly associated with poor prognosis in the cohort of 875 patients with GC (HR, 1.71; 95% CI, 1.42–2.07; P<0.001; Fig. 3B) and the cohort of 380 surgical patients (HR, 1.97; 95% CI, 1.42–2.74; P<0.001; Fig. 3C).
Figure 3.
Increased LTBP2 expression is associated with poor overall survival of patients with GC. (A) Kaplan-Meier estimator analysis of 174 patients with GC indicating decreased overall survival time of patients with LTBP2high GC. (B) Kaplan-Meier estimator survival curve for overall survival of 875 patients with GC from the NCBI GEO database generated using the KM Plotter database. (C) Kaplan-Meier estimator survival curve for overall survival of 380 surgical patients with GC from the NCBI GEO database generated using the KM Plotter database. (D) Kaplan-Meier estimator analyses of overall survival of patients with GC in early-stage (TNM stage I/II) and late-stage (TNM stage III/IV) GC according to LTBP2 expression. (E) Kaplan-Meier estimator analyses of overall survival rate of patients with GC at TNM stage I (F) II, (G) III and (H) IV stage generated using KM Plotter database. LTBP2, latent transforming growth factor-β-binding protein 2; GC, gastric cancer; NCBI, National Center for Biotechnology Information; GEO, Gene Expression Omnibus; TNM, tumor-node-metastasis.
Table VI.
Univariate and multivariate analysis of prognostic parameters for survival in patients with gastric cancer.
| Prognostic variable | Univariate analysis
|
Multivariate analysis
|
||
|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Age, years | 1.415 (0.968–2.068) | 0.073 | 1.187 (0.783–1.798) | 0.419 |
| Sex | 1.041 (0.698–1.554) | 0.844 | 1.296 (0.853–1.971) | 0.225 |
| Histological grade | 0.861 (0.581–1.275) | 0.454 | 0.792 (0.528–1.186) | 0.257 |
| T stage | 2.130 (1.382–3.282) | 0.001 | 1.219 (0.702–2.116) | 0.483 |
| N stage | 2.063 (1.376–3.093) | <0.001 | 1.219 (0.723–2.056) | 0.457 |
| M stage | 2.792 (1.721–4.531) | <0.001 | 1.331 (0.747–2.372) | 0.332 |
| TNM stage | 2.400 (1.638–3.514) | <0.001 | 1.647 (0.924–2.938) | 0.091 |
| High LTBP2 expression | 3.249 (2.146–4.920) | <0.001 | 2.909 (1.896–4.464) | <0.001 |
HR, hazard ratio; CI, confidence interval; T, tumor; N, node; M, metastasis; LTBP2, latent transforming growth factor-β-binding protein 2.
The prognostic value of LTBP2 was investigated following further stratification by TNM stage. The results from the GC cohort of 174 patients indicated that high LTBP2 expression was associated with a lower survival rate in early-stage (TNM I/II) and late-stage (TNM III/IV) GC (P<0.001; Fig. 3D). The prognostic significance of LTBP2 was further analyzed in different TNM stages using the KM Plotter database. Although Kaplan-Meier estimator analysis of LTBP2 expression in patients with TNM I stage GC did not exhibit a significant difference (HR, 2.58; 95% CI, 0.89–7.44; P=0.0701; Fig. 3E), there appeared to be a general trend that expression of LTBP2 was associated with unfavorable outcome in these patients. This may be due to a limited number of cases with TNM I stage GC. However, survival analyses of patients with TNM II stage (HR, 2.26; 95% CI, 1.22–4.16; P=0.0074; Fig. 3F), TNM III stage (HR, 1.82; 95% CI, 1.35–2.45; P<0.0001; Fig. 3G) and TNM IV stage (HR, 1.53; 95% CI, 1.03–2.27; P=0.0335; Fig. 3H) GC demonstrated that the overall survival rates for patients with LTBP2high GC were significantly lower compared with patients with LTBP2low tumors. Survival analyses demonstrated that high LTBP2 expression was associated with poor overall survival in early- and late-stage GC.
LTBP2 depletion impairs the proliferative ability of GC cells in vitro
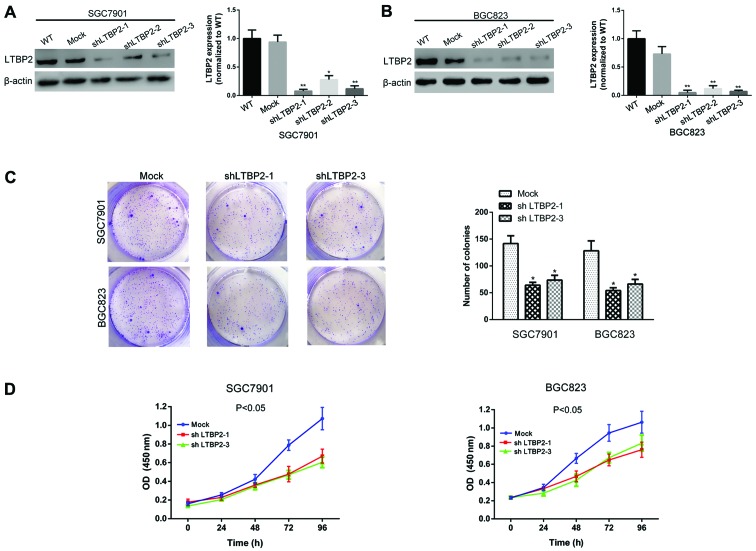
To investigate the effect of LTBP2 on the proliferation of GC cells, knockdown of LTBP2 was performed in SGC7901 and BGC823 cells. The knockdown of LTBP2 was verified using western blotting and RT-qPCR analyses (Fig. 4A and B). Colony-formation and CCK-8 assays were performed to investigate the potential effect of LTBP2 on the proliferative ability of the GC cells. Results of the colony-formation assay indicated that knockdown of LTBP2 abrogated the colony-formation ability in SGC7901 and BGC823 cells (P<0.05; Fig. 4C). The CCK-8 assays demonstrated that the absorbance of LTBP2-knockdown SGC7901 and BGC823 cells was significantly decreased compared with that of mock cells, suggesting that knockdown of LTBP2 abrogated the proliferative ability of GC cells in vitro (P<0.05; Fig. 4D). These results indicated a promotive function of LTBP2 in the proliferation of GC cells.
Figure 4.
Silencing LTBP2 decreases the proliferative ability of GC cells. The knockdown efficiency of LTBP2 in (A) SGC7901 and (B) BGC823 cells was investigated using western blotting and reverse transcription-quantitative chain reaction analyses. *P<0.05 and **P<0.01 vs. WT (C) Representative images of the colony-formation assay and quantification of the results indicating decreased colony-formation ability of GC cells with knockdown of LTBP2. *P<0.05 vs. Mock (D) CCK-8 assays indicating impaired proliferative ability of GC cells with knockdown of LTBP2. LTBP2, latent transforming growth factor-β-binding protein 2; GC, gastric cancer; WT, wild-type; Mock, cells infected with mock lentivirus; OD, optical density; sh, short hairpin.
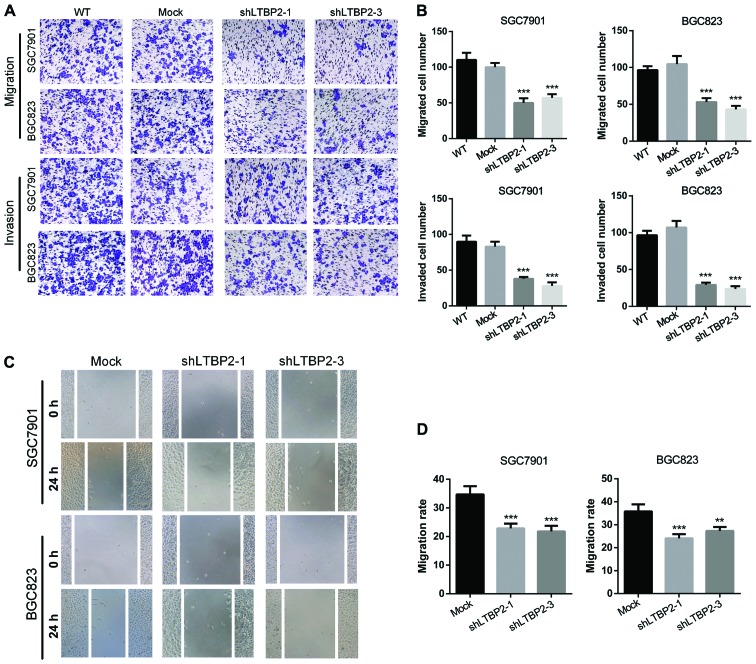
Knockdown of LTBP2 inhibits the migratory and invasive abilities of GC cells
To investigate the functions of LTBP2 in cell migration and invasion, Transwell migration and invasion assays were performed using LTBP2-knockdown and mock GC cells (SGC7901 and BGC823). The results demonstrated that knockdown of LTBP2 decreased the number of migratory and invasive cells in SGC7901 and BGC823 cells (Fig. 5A), which was identified to be significant (P<0.001; Fig. 5B). Furthermore, wound-healing assays were performed to validate the observations that LTBP2 knockdown abrogated the migratory ability of GC cells. The results indicated that silencing LTBP2 in GC cells (SGC7901 and BGC823) decreased the migratory capabilities compared with the mock cells (Fig. 5C), which was identified to be significant (P<0.05; Fig. 5D). These results suggested that LTBP2 promotes the migratory and invasive capabilities of GC cells in vitro.
Figure 5.
Silencing LTBP2 results in decreased migratory and invasive abilities of GC cells. (A) Representative images of Transwell migration and invasion assays, indicating decreased invasive and migratory capability of GC cells with knockdown of LTBP2. (B) Quantification of the Transwell migration and invasion assay results. ***P<0.001 vs. WT. (C) Wound-healing assays indicating impaired migratory capability of GC cells with knockdown of LTBP2. (D) Quantification of the wound-healing assay results. **P<0.01 and ***P<0.001 vs. Mock. LTBP2, latent transforming growth factor-β-binding protein 2; GC, gastric cancer; WT, wild-type; Mock, cells infected with mock lentivirus; sh, short hairpin.
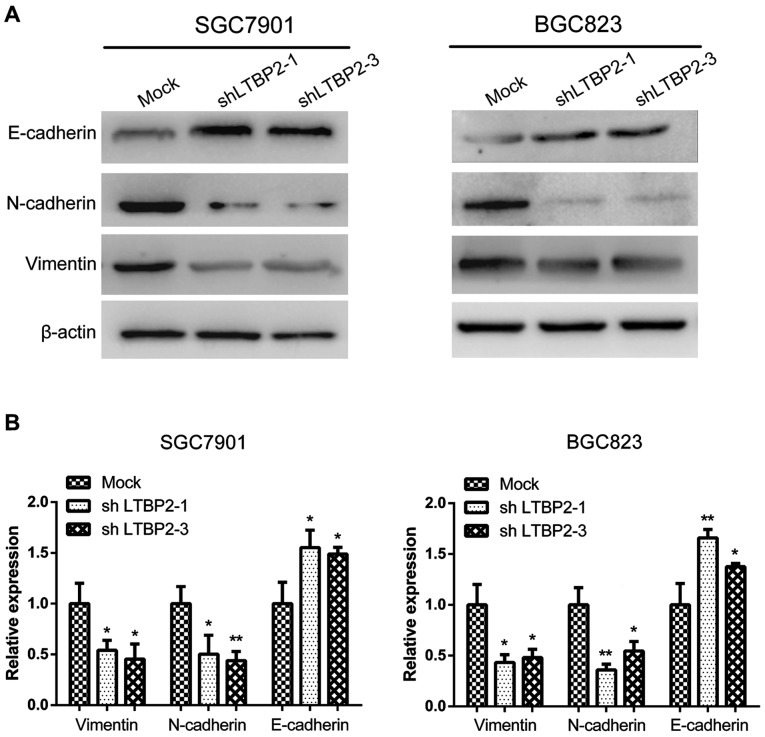
Knockdown of LTBP2 alters the expression of EMT-associated markers
It has been demonstrated previously that EMT serves an essential function in tumor progression and metastasis (32). Cancer cells may undergo EMT prior to metastasis, during which the cells lose cell-cell adhesion, apical-basolateral polarity and epithelial markers, and acquire motility, a spindle-cell shape and mesenchymal markers (33). Thus, EMT is thought to facilitate cancer cell motility, invasion and metastasis. On the basis of this knowledge and the results of the present study that knockdown of LTBP2 inhibited the invasion and migration of GC cells, it was investigated whether LTBP2 knockdown altered the expression of the EMT-associated markers E-cadherin, N-cadherin and vimentin. The results of western blotting analyses indicated a decrease in the expression of N-cadherin and vimentin, and an increase in the expression of E-cadherin following LTBP2 knockdown in SGC7901 and BGC823 cells (Fig. 6A). Furthermore, RT-qPCR assays demonstrated that the mRNA levels of E-cadherin were significantly upregulated following LTBP2 knockdown, whereas N-cadherin and vimentin were significantly downregulated (Fig. 6B). These results indicated that LTBP2 knockdown led to inhibition of EMT.
Figure 6.
Silencing LTBP2 leads to inhibition of the EMT phenotype in SGC7901 and BGC823 GC cells. (A) Protein levels of EMT-associated markers (E-cadherin, N-cadherin and vimentin) in LTBP2-knockdown and mock cells. (B) RT-qPCR analysis of E-cadherin, N-cadherin and vimentin in LTBP2-knockdown and mock cells. *P<0.05; **P<0.01 vs. Mock. LTBP2, latent transforming growth factor-β-binding protein 2; EMT, epithelial-mesenchymal transition; GC, gastric cancer; E-cadherin, epithelial cadherin; N-cadherin, neuronal cadherin; Mock, cells infected with mock lentivirus; sh, short hairpin.
Discussion
The TGF-β signaling pathway regulates various cellular processes including proliferation, apoptosis, differentiation, ECM modification, cytokine secretion and migration, in normal and malignant cells (34,35). Nevertheless, TGF-β has contrasting functions in cancer, acting as a tumor suppressor in the development of pre-malignant tumors and as an oncogene in the progression of advanced-stage tumors, particularly during tumor cell invasion and metastasis (36,37). The majority of cells secrete TGF-β in a latent form, non-covalently bound to TGF-β propeptide [latency-associated peptide (LAP)] (38). LAP associates with LTBPs, which may assist with the proper folding of TGF-β precursor protein, aiding its secretion as well as directing its association with the ECM (9). LTBP1 promotes TGF-β activation by anchoring to the ECM and creating traction when LAP is bound by cell-surface integrins. This traction facilitates LAP deformation and aids in the release of TGF-β from LAP-LTBP complexes, which is essential for the activation of TGF-β (39). However, LTBP2 is different from other LTBPs as it is the only member that does not bind to latent TGF-β (40). It has been demonstrated that LTBP1 and LTBP2 exhibited comparable binding affinities for fibrillin-1 because they compete for the same binding site (41). This led to the hypothesis that LTBP2 may indirectly modulate the TGF-β activation by releasing LTBP1 from microfibrils. However, the definite function of LTBP2 in regulating TGF-β remains unclear, although tumor-promoting and tumor-suppressive functions have been proposed for LTBP2 (15,21).
In head and neck squamous cell carcinoma, LTBP2 expression was upregulated and associated with lymph node metastasis and poor overall survival (15). Knockdown of LTBP2 attenuated the proliferation, invasion and EMT phenotype through the inhibition of the phosphoinositide 3-kinase/protein kinase B signaling pathway in thyroid carcinoma cells (18). LTBP2 was ubiquitously expressed in colorectal carcinoma (CRC) liver metastases, and was absent from normal colon tissues and inflammatory liver diseases, suggesting that LTBP2 served a critical function during CRC cell metastasis (42). In pancreatic, cervical and serous ovarian cancers, overexpression of LTBP2 was observed in cancer tissues and was associated with poor prognosis (16,17,43). However, tumor-suppressive functions of LTBP2 were also identified in several types of cancer. LTBP2 expression was significantly decreased in nasopharyngeal carcinoma tissues and cell lines, and re-expression of LTBP2 suppressed migration, angiogenesis, vascular endothelial growth factor secretion and tumorigenicity accompanied by the inactivation of the nuclear factor-κB p65 signaling pathway (21,44). LTBP2 was epigenetically downregulated through promoter hypermethylation and served a tumor-suppressive function in esophageal squamous cell carcinoma (ESCC), and decreased LTBP2 expression predicted improved overall survival rates in patients with ESCC (20). In the present study, it was identified that LTBP2 acts as an oncogene in GC. It was identified that upregulation of LTBP2 was associated with tumor invasion depth, lymph node metastasis and TNM stage of patients with GC. Furthermore, it was identified that increased LTBP2 expression predicted a lower overall survival rate for patients with early- or late-stage GC. To the best of our knowledge, the present study is the first to investigate the prognostic value and molecular function of LTBP2 in GC.
EMT in cancer cells is a cellular plasticity process characterized by the loss of cell-cell junctions and acquisition of spindle morphology, which confers on tumor cells increased motility to establish a motile and invasive phenotype (45). Although the function and mechanism of LTBP2 in modulating the function of TGF-β remain ambiguous, previous studies have demonstrated that activation of TGF-β signaling pathway promotes EMT induction in numerous types of cancer (46–50). In the present study, it was investigated whether LTBP2 had an effect on EMT in GC cells. It was identified that knockdown of LTBP2 abrogated the mRNA and protein levels of EMT-associated markers in SGC7901 and BGC823 GC cells. The results of the present study suggested that EMT may be involved in the LTBP2-promoted migration and invasion of GC cells.
In summary, the results of the present study identified the critical function of LTBP2 in the malignant features of GC. It was identified that expression of LTBP2 was increased in GC tissues and cell lines; its overexpression was associated with poor prognosis in patients with early- and late-stage GC. Furthermore, LTBP2 promotes the proliferation, migration, invasion and EMT phenotypes of GC cells. Considering the contrasting function that LTBP2 serves in different types of cancer, the elucidation of the function of LTBP2 in GC is expected to aid in our understanding of the function of LTBP2 and TGF-β in tumor progression. Thus, LTBP2 may be considered as a potential therapeutic target and a promising prognostic biomarker for human GC.
Acknowledgments
Not applicable.
Abbreviations
- GC
gastric cancer
- LTBP2
latent transforming growth factor-β-binding protein 2
- TNM
tumor-node-metastasis
- EMT
epithelial-mesenchymal transition
Funding
The present study was supported by the Natural Science Foundation of Gansu Province, China (grant no. 1208RJZA222).
Availability of data and materials
Not applicable.
Authors' contributions
JW, WJL, GTM, HPW and WC performed all the experiments and drafted the manuscript. JW, GTM, HPW and NY participated in study design, data analysis and interpretation of results. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of The First Hospital of Lanzhou University and written informed consent was obtained from all enrolled patients.
Consent for publication
Not applicable.
Conflicts of interest
The authors declare that they have no competing interests.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Zong L, Abe M, Seto Y, Ji J. The challenge of screening for early gastric cancer in China. Lancet. 2016;388:2606. doi: 10.1016/S0140-6736(16)32226-7. [DOI] [PubMed] [Google Scholar]
- 4.Cervantes A, Roda D, Tarazona N, Roselló S, Pérez-Fidalgo JA. Current questions for the treatment of advanced gastric cancer. Cancer Treat Rev. 2013;39:60–67. doi: 10.1016/j.ctrv.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 5.Glockzin G, Piso P. Current status and future directions in gastric cancer with peritoneal dissemination. Surg Oncol Clin N Am. 2012;21:625–633. doi: 10.1016/j.soc.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Liu JJ, Liu JY, Chen J, Wu YX, Yan P, Ji CD, Wang YX, Xiang DF, Zhang X, Zhang P, et al. Scinderin promotes the invasion and metastasis of gastric cancer cells and predicts the outcome of patients. Cancer Lett. 2016;376:110–117. doi: 10.1016/j.canlet.2016.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morén A, Olofsson A, Stenman G, Sahlin P, Kanzaki T, Claesson-Welsh L, ten Dijke P, Miyazono K, Heldin CH. Identification and characterization of LTBP-2, a novel latent transforming growth factor-beta-binding protein. J Biol Chem. 1994;269:32469–32478. [PubMed] [Google Scholar]
- 8.Jensen SA, Robertson IB, Handford PA. Dissecting the fibrillin microfibril: Structural insights into organization and function. Structure. 2012;20:215–225. doi: 10.1016/j.str.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 9.Miyazono K, Olofsson A, Colosetti P, Heldin CH. A role of the latent TGF-beta 1-binding protein in the assembly and secretion of TGF-beta 1. EMBO J. 1991;10:1091–1101. doi: 10.1002/j.1460-2075.1991.tb08049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson SB, Goldberg AL, Whitman M. Identification of a novel pool of extracellular pro-myostatin in skeletal muscle. J Biol Chem. 2008;283:7027–7035. doi: 10.1074/jbc.M706678200. [DOI] [PubMed] [Google Scholar]
- 11.Jelodari-Mamaghani S, Haji-Seyed-Javadi R, Suri F, Nilforushan N, Yazdani S, Kamyab K, Elahi E. Contribution of the latent transforming growth factor-β binding protein 2 gene to etiology of primary open angle glaucoma and pseudoexfoliation syndrome. Mol Vis. 2013;19:333–347. [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar A, Duvvari MR, Prabhakaran VC, Shetty JS, Murthy GJ, Blanton SH. A homozygous mutation in LTBP2 causes isolated microspherophakia. Hum Genet. 2010;128:365–371. doi: 10.1007/s00439-010-0858-8. [DOI] [PubMed] [Google Scholar]
- 13.Haji-Seyed-Javadi R, Jelodari-Mamaghani S, Paylakhi SH, Yazdani S, Nilforushan N, Fan JB, Klotzle B, Mahmoudi MJ, Ebrahimian MJ, Chelich N, et al. LTBP2 mutations cause Weill-Marchesani and Weill-Marchesani-like syndrome and affect disruptions in the extracellular matrix. Hum Mutat. 2012;33:1182–1187. doi: 10.1002/humu.22105. [DOI] [PubMed] [Google Scholar]
- 14.Breidthardt T, Vanpoucke G, Potocki M, Mosimann T, Ziller R, Thomas G, Laroy W, Moerman P, Socrates T, Drexler B, et al. The novel marker LTBP2 predicts all-cause and pulmonary death in patients with acute dyspnoea. Clin Sci (Lond) 2012;123:557–566. doi: 10.1042/CS20120058. [DOI] [PubMed] [Google Scholar]
- 15.Han L, Tang MM, Xu X, Jiang B, Huang J, Feng X, Qiang J. LTBP2 is a prognostic marker in head and neck squamous cell carcinoma. Oncotarget. 2016;7:45052–45059. doi: 10.18632/oncotarget.8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshihara K, Tajima A, Komata D, Yamamoto T, Kodama S, Fujiwara H, Suzuki M, Onishi Y, Hatae M, Sueyoshi K, et al. Gene expression profiling of advanced-stage serous ovarian cancers distinguishes novel subclasses and implicates ZEB2 in tumor progression and prognosis. Cancer Sci. 2009;100:1421–1428. doi: 10.1111/j.1349-7006.2009.01204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren Y, Lu H, Zhao D, Ou Y, Yu K, Gu J, Wang L, Jiang S, Chen M, Wang J, et al. LTPB2 acts as a prognostic factor and promotes progression of cervical adenocarcinoma. Am J Transl Res. 2015;7:1095–1105. [PMC free article] [PubMed] [Google Scholar]
- 18.Wan F, Peng L, Zhu C, Zhang X, Chen F, Liu T. Knockdown of latent transforming growth factor-β (TGF-β)-binding protein 2 (LTBP2) inhibits invasion and tumorigenesis in thyroid carcinoma cells. Oncol Res. 2017;25:503–510. doi: 10.3727/096504016X14755368915591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.da Costa AN, Plymoth A, Santos-Silva D, Ortiz-Cuaran S, Camey S, Guilloreau P, Sangrajrang S, Khuhaprema T, Mendy M, Lesi OA, et al. Osteopontin and latent-TGF β binding-protein 2 as potential diagnostic markers for HBV-related hepatocellular carcinoma. Int J Cancer. 2015:172–181. doi: 10.1002/ijc.28953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan SH, Yee Ko JM, Chan KW, Chan YP, Tao Q, Hyytiainen M, Keski-Oja J, Law S, Srivastava G, Tang J, et al. The ECM protein LTBP-2 is a suppressor of esophageal squamous cell carcinoma tumor formation but higher tumor expression associates with poor patient outcome. Int J Cancer. 2011;129:565–573. doi: 10.1002/ijc.25698. [DOI] [PubMed] [Google Scholar]
- 21.Kan R, Shuen WH, Lung HL, Cheung AK, Dai W, Kwong DL, Ng WT, Lee AW, Yau CC, Ngan RK, et al. NF-κB p65 subunit is modulated by latent transforming growth factor-β binding protein 2 (LTBP2) in nasopharyngeal carcinoma HONE1 and HK1 cells. PLoS One. 2015;10:e0127239. doi: 10.1371/journal.pone.0127239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Washington K. 7th edition of the AJCC cancer staging manual: Stomach. Ann Surg Oncol. 2010;17:3077–3079. doi: 10.1245/s10434-010-1362-z. [DOI] [PubMed] [Google Scholar]
- 23.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: A new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 24.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Förster S, Gretschel S, Jöns T, Yashiro M, Kemmner W. THBS4, a novel stromal molecule of diffuse-type gastric adenocarcinomas, identified by transcriptome-wide expression profiling. Mod Pathol. 2011;24:1390–1403. doi: 10.1038/modpathol.2011.99. [DOI] [PubMed] [Google Scholar]
- 27.Kim HK, Choi IJ, Kim CG, Kim HS, Oshima A, Yamada Y, Arao T, Nishio K, Michalowski A, Green JE. Three-gene predictor of clinical outcome for gastric cancer patients treated with chemotherapy. Pharmacogenomics J. 2012;12:119–127. doi: 10.1038/tpj.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szász AM, Lánczky A, Nagy Á, Förster S, Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A, Győrffy B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Wang S, Cheng Y, Du W, Lu L, Zhou L, Wang H, Kang W, Li X, Tao Q, Sung JJ, et al. Zinc-finger protein 545 is a novel tumour suppressor that acts by inhibiting ribosomal RNA transcription in gastric cancer. Gut. 2013;62:833–841. doi: 10.1136/gutjnl-2011-301776. [DOI] [PubMed] [Google Scholar]
- 31.Yie SM, Ye SR, Ma XL, Xie K, Zhang JB, Cao M, He X, Hu ZB, Yang CL, Zhang J, et al. A protein fragment derived from DNA-topoisomerase I as a novel tumour-associated antigen for the detection of early stage carcinoma. Br J Cancer. 2016;115:1555–1564. doi: 10.1038/bjc.2016.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prieto-García E, Díaz-García CV, García-Ruiz I, Agulló-Ortuño MT. Epithelial-to-mesenchymal transition in tumor progression. Med Oncol. 2017;34:122. doi: 10.1007/s12032-017-0980-8. [DOI] [PubMed] [Google Scholar]
- 33.Sun J, Sun B, Zhu D, Zhao X, Zhang Y, Dong X, Che N, Li J, Liu F, Zhao N, et al. HMGA2 regulates CD44 expression to promote gastric cancer cell motility and sphere formation. Am J Cancer Res. 2017;7:260–274. [PMC free article] [PubMed] [Google Scholar]
- 34.Massagué J. TGFβ signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heldin CH, Vanlandewijck M, Moustakas A. Regulation of EMT by TGFβ in cancer. FEBS Lett. 2012;586:1959–1970. doi: 10.1016/j.febslet.2012.02.037. [DOI] [PubMed] [Google Scholar]
- 36.Ikushima H, Miyazono K. TGF-β signal transduction spreading to a wider field: A broad variety of mechanisms for context-dependent effects of TGF-β. Cell Tissue Res. 2012;347:37–49. doi: 10.1007/s00441-011-1179-5. [DOI] [PubMed] [Google Scholar]
- 37.Massagué J. TGFbeta in cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brunner AM, Marquardt H, Malacko AR, Lioubin MN, Purchio AF. Site-directed mutagenesis of cysteine residues in the pro region of the transforming growth factor beta 1 precursor. Expression and characterization of mutant proteins. J Biol Chem. 1989;264:13660–13664. [PubMed] [Google Scholar]
- 39.Ono RN, Sengle G, Charbonneau NL, Carlberg V, Bächinger HP, Sasaki T, Lee-Arteaga S, Zilberberg L, Rifkin DB, Ramirez F, et al. Latent transforming growth factor beta-binding proteins and fibulins compete for fibrillin-1 and exhibit exquisite specificities in binding sites. J Biol Chem. 2009;284:16872–16881. doi: 10.1074/jbc.M809348200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saharinen J, Keski-Oja J. Specific sequence motif of 8-Cys repeats of TGF-beta binding proteins, LTBPs, creates a hydrophobic interaction surface for binding of small latent TGF-beta. Mol Biol Cell. 2000;11:2691–2704. doi: 10.1091/mbc.11.8.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hirani R, Hanssen E, Gibson MA. LTBP-2 specifically interacts with the amino-terminal region of fibrillin-1 and competes with LTBP-1 for binding to this microfibrillar protein. Matrix Biol. 2007;26:213–223. doi: 10.1016/j.matbio.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 42.Turtoi A, Blomme A, Debois D, Somja J, Delvaux D, Patsos G, Di Valentin E, Peulen O, Mutijima EN, De Pauw E, et al. Organized proteomic heterogeneity in colorectal cancer liver metastases and implications for therapies. Hepatology. 2014;59:924–934. doi: 10.1002/hep.26608. [DOI] [PubMed] [Google Scholar]
- 43.Turtoi A, Musmeci D, Wang Y, Dumont B, Somja J, Bevilacqua G, De Pauw E, Delvenne P, Castronovo V. Identification of novel accessible proteins bearing diagnostic and therapeutic potential in human pancreatic ductal adenocarcinoma. J Proteome Res. 2011;10:4302–4313. doi: 10.1021/pr200527z. [DOI] [PubMed] [Google Scholar]
- 44.Chen H, Ko JM, Wong VC, Hyytiainen M, Keski-Oja J, Chua D, Nicholls JM, Cheung FM, Lee AW, Kwong DL, et al. LTBP-2 confers pleiotropic suppression and promotes dormancy in a growth factor permissive microenvironment in nasopharyngeal carcinoma. Cancer Lett. 2012;325:89–98. doi: 10.1016/j.canlet.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 45.Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 46.Kitamura K, Kasuya K, Tsuchida A, Mimuro A, Inoue K, Aoki T, Aoki T, Koyanagi Y. Immunohistochemical analysis of transforming growth factor beta in gallbladder cancer. Oncol Rep. 2003;10:327–332. [PubMed] [Google Scholar]
- 47.Fukai Y, Fukuchi M, Masuda N, Osawa H, Kato H, Nakajima T, Kuwano H. Reduced expression of transforming growth factor-beta receptors is an unfavorable prognostic factor in human esophageal squamous cell carcinoma. Int J Cancer. 2003;104:161–166. doi: 10.1002/ijc.10929. [DOI] [PubMed] [Google Scholar]
- 48.Sato Y, Harada K, Itatsu K, Ikeda H, Kakuda Y, Shimomura S, Shan Ren X, Yoneda N, Sasaki M, Nakanuma Y. Epithelial-mesenchymal transition induced by transforming growth factor-{beta}1/Snail activation aggravates invasive growth of cholangiocarcinoma. Am J Pathol. 2010;177:141–152. doi: 10.2353/ajpath.2010.090747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma GF, Miao Q, Zeng XQ, Luo TC, Ma LL, Liu YM, Lian JJ, Gao H, Chen SY. Transforming growth factor-β1 and -β2 in gastric precancer and cancer and roles in tumor-cell interactions with peripheral blood mononuclear cells in vitro. PLoS One. 2013;8:e54249. doi: 10.1371/journal.pone.0054249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voon DC, Wang H, Koo JK, Chai JH, Hor YT, Tan TZ, Chu YS, Mori S, Ito Y. EMT-induced stemness and tumorigenicity are fueled by the EGFR/Ras pathway. PLoS One. 2013;8:e70427. doi: 10.1371/journal.pone.0070427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.