Abstract
MicroRNAs (miRNAs or miRs) are highly conserved small noncoding RNA molecules involved in gene regulation. An increasing number of studies have demonstrated that miRNAs act as oncogenes or antioncogenes in various types of cancer, including breast cancer (BC). However, the exact role of miR-671-3p in BC has not yet been reported. In the present study, in vitro experiments were implemented to explore the effects of miR-671-3p on the proliferation and apoptosis of BC cells, and reverse transcription-quantitative polymerase chain reaction was conducted using in-house clinical BC samples to address the expression level and clinical value of miR-671-3p in BC. Simultaneously, miR-671-3p target genes were collected, and subsequent bioinformatics analyses were executed to probe the potential signaling pathway through which miR-671-3p influenced the occurrence and progression of BC. According to the results, the expression level of miR-671-3p was lower in BC tissues compared with that in adjacent non-tumorous tissues (P=0.048), and the area under the curve was 0.697 (95% confidence interval=0.538-0.856), with a sensitivity and specificity of 0.818 and 0.579, respectively. Forced miR-671-3p expression in the BC cell line MDA-MB-231 evidently arrested cell proliferation and induced cell apoptosis. Furthermore, in silico enrichment analyses suggested that miR-671-3p may be involved in the initiation and progression of BC through the targeting of genes associated with the Wnt signaling pathway. In conclusion, the present study findings suggested that miR-671-3p may function as a tumor suppressor in BC by influencing the Wnt signaling cascade, which provides a prospective molecular target for the therapy of BC.
Key words: breast cancer, microRNA-671-3p, biological function, clinical value, molecular mechanism
Introduction
MicroRNAs (miRNAs or miRs) are a major type of small noncoding RNA molecules involved in gene regulation via binding to the 3′ untranslated region of their target mRNAs, and subsequently causing mRNA degradation or translation inhibition (1–4). An increasing volume of research has provided evidence that miRNAs act as oncogenes or tumor suppressors in various malignant tumors by influencing tumor growth, proliferation, apoptosis, invasion and metastasis (5–8). The close association between miRNAs and breast cancer (BC) has also been investigated. For instance, Ding et al (9) revealed that miR-145 is a tumor suppressor in BC and performs its anti-oncogenic role by inhibiting a cancer-associated gene, transforming growth factor-β1. Xiao et al (10) observed that miR-129 blocks BC regeneration by degrading estrogen receptor 1 mRNA at the posttranscriptional level, inhibiting the subsequent estrogen-induced NOTCH cascade and ultimately reducing the number of stem-like cells. A recent study conducted by Lu et al (11) demonstrated that miR-129-5p increases the sensitivity of human epidermal growth factor receptor 2 (Her2)-positive BC to trastuzumab by downregulating ribosomal protein S6. In addition, due to the ease of detecting miRNAs in tumor biopsies and body fluids, numerous researchers have highlighted the diagnostic and prognostic roles of miRNAs in cancer, including in BC. Jang et al (12) examined miR-9 and miR-155 expression levels in triple-negative BC and found that they functioned as biomarkers for prognosis prediction. Zhang et al (13) detected circulating miRNAs expression using a serum-direct multiplex detection assay based on reverse transcription-polymerase chain reaction (RT-PCR). The authors determined a 3-miRNA signature (miR-424, miR-199a and miR-29c) that exhibited the highest diagnostic accuracy for BC diagnosis (13). Our research group has also identified a 9-miRNA signature for BC diagnosis based on data from The Cancer Genome Atlas (TCGA) (14). Therefore, the investigation of miRNAs in cancer has provided promising results, and exploring miRNAs involved in the occurrence and development of BC is conducive to the diagnosis and treatment of patients with BC.
miR-671 precursors form two mature miRNAs, namely miR-671-5p and miR-671-3p. A recent study conducted by Tan et al (15) has demonstrated that miR-671-5p is an antioncogene in BC. The authors also observed that miR-671-5p was downregulated in BC and that forced miR-671-5p expression in the BC cell line MDA-MB-231 was able to inhibit cell proliferation and invasion, and sensitize cells to chemotherapy (15). Furthermore, Godfrey et al (16) compared global miRNA expression in 205 BC patients and 205 healthy volunteers using a microarray method, and observed a high expression of miR-671-3p in the serum samples of BC patients. However, a study concentrating on the biological function and molecular mechanism of miR-671-3p in BC has yet to be reported.
The present study focused on the expression level, biological function and potential molecular mechanism of miR-671-3p in BC. In vitro experiments were conducted to determine the biological effects of miR-671-3p on BC cells, and miR-671-3p expression was detected using RT-quantitative PCR (RT-qPCR). Simultaneously, all available microarray datasets were combined from the Gene Expression Omnibus (GEO) and ArrayExpress databases to verify the expression of miR-671-3p in BC. Furthermore, the predicted targets of miR-671-3p were examined, Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were conducted, and a protein-protein interaction (PPI) network was constructed to gain insight into the underlying molecular mechanism of miR-671-3p in BC.
Materials and methods
Patients and samples
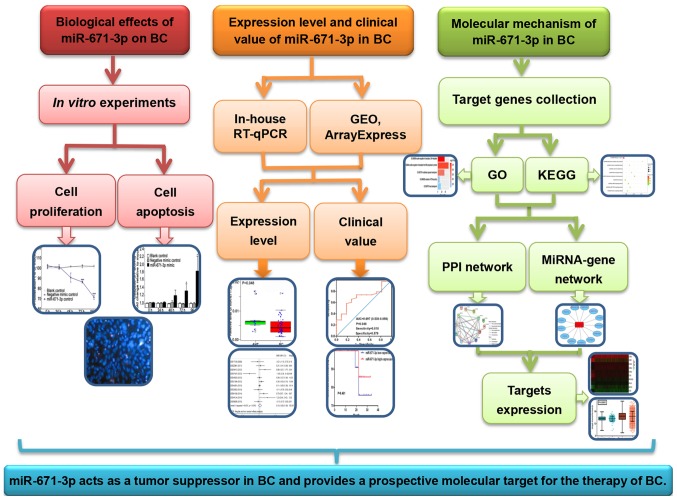
A total of 38 pairs of BC and adjacent non-tumorous tissues were obtained from patients who were pathologically diagnosed with BC (17). To ensure that the RT-qPCR results were not affected by the size of paraffin-embedded tissues, samples with a surface area that was too small or large were excluded. Finally, 38 BC tissues and 11 adjacent non-tumorous tissues with a surface area of ~1.5 cm2 were selected. All patients underwent initial surgery at the First Affiliated Hospital of Guangxi Medical University (Nanning, China) between January 2012 and December 2013, and had not received any preoperative radiotherapy or chemotherapy. Adjacent non-tumorous tissues were collected at >5 cm away from the tumorous node. Following excision, tissues were cryopreserved in liquid nitrogen and stored at −80°C prior to RNA extraction. All participants provided informed consent prior to sample collection. The Ethics Committee of the First Affiliated Hospital of Guangxi Medical University approved this investigation. The workflow of the present study is exhibited in Fig. 1.
Figure 1.
Workflow of the present study. miR, microRNA; BC, breast cancer; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; GEO, Gene Expression Omnibus; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, protein-protein interaction.
Cell culture and transfection with miR-671-3p mimic
The human BC-derived cell line MDA-MB-231 was provided by the American Type Culture Collection (Manassas, VA, USA) and cultured as described in previous studies (18,19). For transfection, MDA-MB-231 cells were seeded in a 96-well plate (2.5×103 cells per well), and subsequently incubated at 37°C for 24 h. Next, miR-671-3p mimic and negative mimic control (Ambion; Thermo Fisher Scientific, Inc., Waltham, MA, USA) were transfected into the cells at a concentration of 200 nmol/l with a CombiMag Magnetofection transfection kit (OZ Biosciences, Marseille, France) according to the manufacturer's protocol. A blank control was set with nothing transfected into cells. Following transfection, cell samples were collected at 0, 24, 48, 72 and 96 h for further analyses. All in vitro experiments were conducted in triplicate.
RT-qPCR
Total RNA was isolated using the miRNeasy FFPE kit (Qiagen, Duesseldorf, German) following the manufacturer's protocol. The measurement of total RNA quality was conducted on NanoDrop-2000 Ultra Micro Spectrophotometer (Thermo Fisher Scientific, Inc.), and an RNA concentration >45 ng/µl was required based on the calculation by the instructions of miScript II RT kit and miScript SYBR-Green PCR kit. Then, total RNA was reversed into cDNA with miScript II RT kit (Qiagen). Subsequently, RT-qPCR was conducted with 7900HT Fast Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.) in accordance with the manufacturer's protocol. The thermal cycling conditions are shown in Table I. RNU44 was used as a reference gene for the normalization of miR-671-3p abundance. The primers for miR-671-3p (cat. no. 1000066; CCGGUUCUCAGGGCUCCACC) and RNU44 (cat. no. MS00033855) were synthesized by Qiagen. All the samples were examined in triplicate, and the mean of the three well values were used as the Cq value. The relative expression level of miR-671-3p was determined according to the 2−ΔCq method (ΔCq = CqmiR-671-3p-CqRNU44), as previously described (20,21).
Table I.
Thermal cycling conditions of RT-qPCR.
| Step | Time | Temperature (°C) | Cycle |
|---|---|---|---|
| Pre-denaturation | 15 min | 95 | 1 |
| Three steps of one cycle | 45 | ||
| Denaturation | 15 sec | 94 | |
| Annealing | 30 sec | 55 | |
| Elongation | 30 sec | 70 |
min, minute; sec, second.
Cell function detection
Cell viability and proliferation were examined by fluorometric detection with resorufin (CellTiter-Blue Cell Viability Assay; cat. no. G8080; Promega Corportation, Madison, WI, USA) and a colorimetric tetrazolium (MTS) assay (CellTiter96 AQUEOUS One Solution Cell Proliferation Assay; cat. no. G3580; Promega Corporation), respectively. Furthermore, cell apoptosis was assessed by Hoechst 33342 and propidium iodide (PI; both purchased from Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) double-fluorescent chromatin staining. Caspase-3/7 activity was also examined with a synthetic rhodamine-labeled caspase-3/7 substrate (Apo-ONE Homogeneous Caspase-3/7 Assay; cat. no. G7790; Promega Corporation). The protocols of these assays were performed as reported in previous studies (18,19,21–23).
Studies on GEO and ArrayExpress databases
To verify the expression level of miR-671-3p in BC, a meta-analysis was conducted based on data obtained from the GEO (www.ncbi.nlm.nih.gov/geo) and ArrayExpress (www.ebi.ac.uk/array-express) databases. Relevant records were retrieved with the following keywords: (cancer OR carcinoma OR tumor OR neoplas* OR malignan*) AND (breast OR mammary OR mastocarcinoma) AND (miRNA OR microRNA OR miR OR 'non-coding RNA' OR ncRNA). The last update time was October 25th, 2017. Datasets were included once they met the following inclusion criteria: i) The records provided expression data of miR-671-3p in BC; ii) the number of samples in each dataset in the cancerous and non-tumorous groups was >3; and iii) the subjects involved in the study were humans. The following datasets were excluded: i) Studies not associated with BC; ii) records with insufficient data to calculate the expression level of miR-671-3p; and iii) animal studies.
Two trained investigators independently screened all available datasets and extracted the following characteristics from all the enrolled studies: First author, publication year, country, data source, platform, sample size, and expression values of miR-671-3p in cancer and normal groups. Any disagreement was resolved through discussion with a third researcher.
The standard mean difference (SMD) with the 95% confi-dence interval (CI) was calculated to appraise miR-671-3p expression in BC. SMD <0 indicated that miR-671-3p was downregulated, and the result was consider as statistically significant if the corresponding 95% CI of SMD did not overlap zero. Heterogeneity among studies was evaluated by the χ2-based Q test and I2 statistic. If a statistically significant heterogeneity existed (I2 >50% or P<0.05), a random-effects model was applied. Otherwise, a fixed-effects model was selected (24). Furthermore, sensitivity analysis was conducted to evaluate whether the pooled result was stable. Begg's funnel plot and Egger's test were also used to determine the possible publication bias, with P>0.05 suggesting no publication bias. All the aforementioned calculations were performed with Stata software, version 12.0 (Stata Corporation, College Station, TX, USA).
Target prediction and bioinformatics analyses
The predicted target genes of miR-671-3p were determined using the miRWalk 2.0 database (zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2), which contains 12 online tools, namely Targetscan, RNAhybrid, RNA22, PITA, Pictar2, miRWalk, Microt4, miRNAMap, miRDB, mirbridge, miRanda and miRMap. Targets predicted by >3 algorithms were selected for GO functional annotation and KEGG pathway analyses using the DAVID online tool (david.ncifcrf.gov). Subsequently, the enriched results were visualized with the R package 'ggplot2' (cran.r-project.org/web/packages/ggplot2/index.html). To better understand the molecular mechanism of miR-671-3p in BC, target genes enriched in cancer-associated pathways were uploaded to the STRING database (string-db.org) for PPI network construction. Meanwhile, the regulatory network of the miRNA-gene was constructed and visualized with Cytoscape version 3.4.0 software (25). Furthermore, the expression of these target mRNAs was validated using data from TCGA (cancergenome.nih.gov). A heatmap was drawn with the R package 'pheatmap' (cran.r-project.org/web/packages/pheatmap/index.html), and a box-scatter plot was generated with GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA, USA).
Statistical analysis
Values are represented as the mean ± standard deviation. Mann-Whitney U test and Student's t-test were applied for comparison between continuous variables. Fisher's exact test was also conducted to evaluate the correlation between miR-671-3p expression and clinicopathological parameters of age, histological grade (26), T stage, N stage, M stage, TNM stage (27), molecular subtype, ER/PR status and Her2 status. A receiver operating characteristic (ROC) curve was generated to determine the ability of miR-671-3p to distinguish BC from non-tumorous breast tissues. A Kaplan-Meier survival curve with log-rank test was utilized to estimate the prognostic power of miR-671-3p in BC. All these analyses were conducted using SPSS version 22.0 software (IBM Corp., Armonk, NY, USA), and P<0.05 was considered to indicate a difference that was statistical significance.
Results
miR-671-3p mimic arrests the proliferation and induces the apoptosis of BC cells in vitro
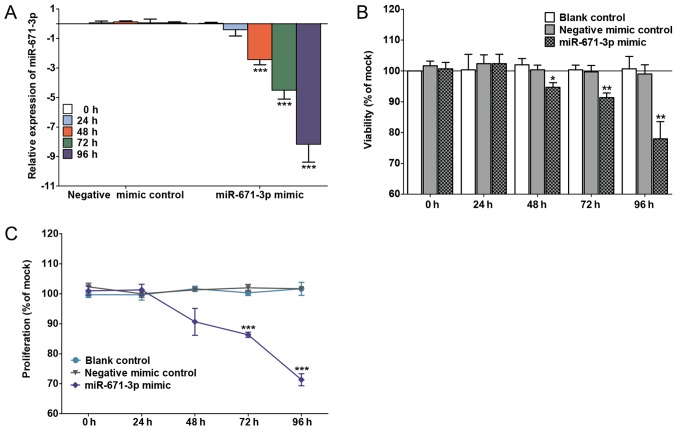
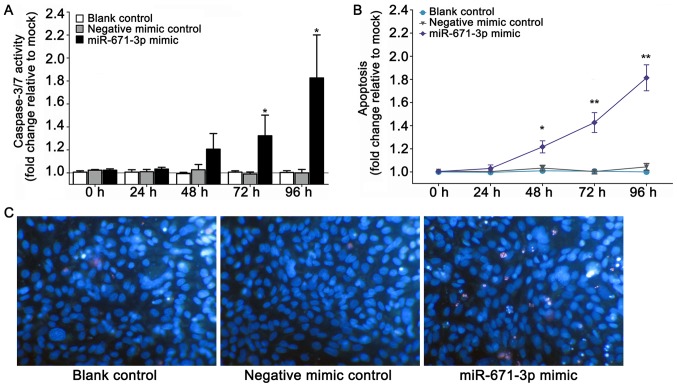
The transfection efficiency is shown in Fig. 2A. miR-671-3p was significantly upregulated in the miR-671-3p mimic group at 48 h (P<0.0001), 72 h (P<0.0001) and 96 h (P<0.0001) compared with the negative mimic control group. An unregulated miR-671-3p decreased cell viability and proliferation, as determined by the fluoro-metric resorufin and MTS assays, respectively (Fig. 2B and C). Furthermore, to explore the effects of the miR-671-3p mimic on cell apoptosis, CellTiter-Blue and fluorescent caspase-3/7 assays were conducted. The results indicated that the caspase-3/7 activity and apoptosis of cells transfected with miR-671-3p mimic increased compared the caspase-3/7 activity and apoptosis of cells in the blank and negative mimic control groups (Fig. 3A and B). In addition, viable and apoptotic cells were observed under a microscope subsequent to Hoechst 33342/PI double-fluorescent chromatin staining at 96 h. The results revealed that miR-671-3p overexpression evidently inhibited the viability and boosted the apoptosis of MDA-MB-231 cells in vitro (Fig. 3C).
Figure 2.
miR-671-3p mimic inhibited the proliferation and viability of MDA-MB-231 cells. (A) Transfection efficiency of miR-671-3p. Effect of miR-671-3p mimic on (B) cell proliferation determined by MTS Assay, and (C) cell viability determined by CellTiter-Blue Cell Viability Assay. *P<0.05, **P<0.01 and ***P<0.001 vs. negative control at 0 h. miR, microRNA.
Figure 3.
miR-671-3p mimic accelerated the apoptosis of MDA-MB-231 cells. (A) Effect of miR-671-3p mimic on cell apoptosis in terms of the caspase-3/7 activity, as determined by an Apo-ONE Homogeneous Caspase-3/7 assay. (B) Effect of miR-671-3p mimic on cell apoptosis, determined by fluorescence microscopy following Hoechst 33342/PI double-fluorescent chromatin staining. (C) Viable and apoptotic cells were observed under a microscope following Hoechst 33342/PI staining (magnification, x200) at 96 h. Viable cells exhibited Hoechst 33342-positive and PI-negative staining; early apoptotic cells exhibited Hoechst 33342-positive and PI-negative staining, with blue fragmentation in the cells; late apoptotic cells exhibited Hoechst 33342-positive and PI-positive staining, with red fragmentation in the cells; and necrotic cells exhibited PI-positive staining with debris signals. *P<0.05 and **P<0.01 vs. negative control at 0 h. miR, microRNA; PI, propidium iodide.
Expression level and clinical value of miR-671-3p in BC via in-house RT-qPCR
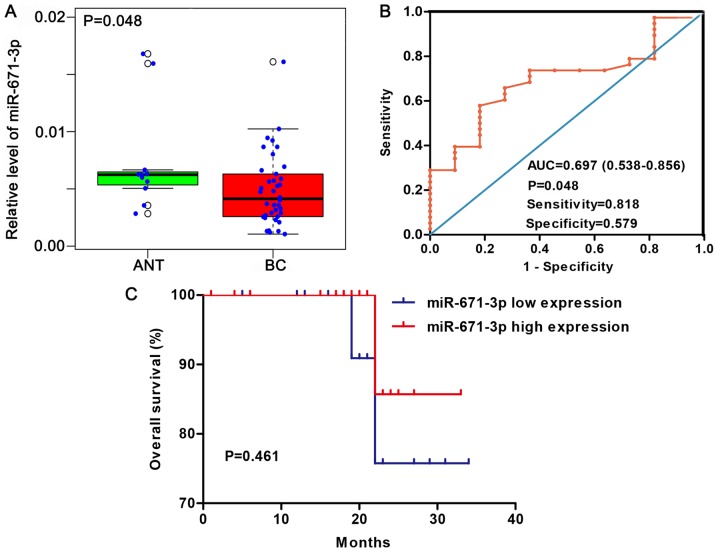
According to the median expression of miR-671-3p in 38 BC tissues and 11 adjacent non-tumorous tissues, samples were divided into high- and low-miR-671-3p group. A total of 17 tumor samples (44.7%) exhibited high miR-671-3p expression, while 9 non-tumorous samples (81.8%) presented high miR-671-3p expression (81.8%). The expression pattern of miR-671-3p was visualized as a box-scatter plot and ROC curve. According to the results, the miR-671-3p expression was significantly lower in cancer tissues when compared with that in adjacent non-tumorous tissues (P=0.048; Fig. 4A). The area under the ROC curve was 0.697 (95% CI=0.538-0.856; P=0.048; Fig. 4B), with a sensitivity and specificity of 0.818 and 0.579, respectively.
Figure 4.
Expression level and clinical value of miR-671-3p in BC based on in-house reverse transcription-quantitative polymerase chain reaction. (A) Expression levels of miR-671-3p in ANT and BC tissues. (B) Receiver operating characteristic curve and (C) prognostic value of miR-671-3p in BC. miR, microRNA; BC, breast cancer; ANT, adjacent non-tumorous tissues; AUC, area under the curve.
The association of miR-671-3p expression with the clinicopathological characteristics of patients, including the age, histological grade, T stage, N stage, M stage, TNM stage, molecular subtype, ER/PR status and Her2 status, was investigated. As shown in Table II, no statistically significant differences were detected. Furthermore, the association between miR-671-3p expression and prognosis was examined. The patients were divided into high and low miR-671-3p expression groups according to the median value of miR-671-3p expression. Patients with highly expressed miR-671-3p had a mean survival time of 31.429 months, while patients with low miR-671-3p expression had a mean survival time of 30.818 months. However, no statistically significant difference was observed between the two groups (P=0.461; Fig. 4C).
Table II.
Association between miR-671-3p expression and clinicopathological parameters of patients (n=38).
| Characteristic | Group | Count | miR-671-3p expression
|
P-value | |
|---|---|---|---|---|---|
| Low (%) | High (%) | ||||
| Age (years) | ≤50 | 18 | 11 (61.1) | 7 (38.9) | 0.532 |
| >50 | 20 | 10 (50) | 10 (50) | ||
| Tumor size | ≤2.5 cm | 11 | 7 (63.6) | 4 (36.4) | 0.721 |
| >2.5 cm | 27 | 14 (51.9) | 13 (48.1) | ||
| Pathological grade | IDC I | 5 | 3 (60) | 2 (40) | 0.396 |
| IDC II | 24 | 15 (62.5) | 9 (37.5) | ||
| IDC III | 9 | 3 (33.3) | 6 (66.7) | ||
| T stage | T1-T2 | 30 | 17 (56.7) | 13 (43.3) | 1 |
| T3-T4 | 8 | 4 (50) | 4 (50) | ||
| N stage | N0 | 18 | 10 (55.6) | 8 (44.4) | 1 |
| N1-N3 | 20 | 11 (55) | 9 (45) | ||
| M stage | M0 | 38 | 21 (55.3) | 17 (44.7) | – |
| M1 | 0 | – | – | ||
| TNM stage | I | 6 | 4 (66.7) | 2 (33.3) | 0.91 |
| II | 19 | 10 (52.6) | 9 (47.4) | ||
| III | 13 | 7 (46.2) | 6 (53.8) | ||
| Molecular subtype | Luminal | 10 | 6 (60) | 4 (40) | 0.315 |
| Her2-positive | 18 | 7 (38.9) | 11 (61.1) | ||
| Triple negative | 10 | 7 (70) | 3 (30) | ||
| ER/PR | Negative | 28 | 15 (53.6) | 13 (46.4) | 1 |
| Positive | 10 | 6 (60) | 4 (40) | ||
| Her2 | Negative | 20 | 14 (70) | 6 (30) | 0.101 |
| Positive | 18 | 7 (38.9) | 11 (61.1) | ||
miR, microRNA; IDC, invasive ductal carcinoma; ER/PR, estrogen receptors/progesterone receptors; Her2, human epidermal growth factor receptor 2.
Results of meta-analyses based on GEO and ArrayExpress databases
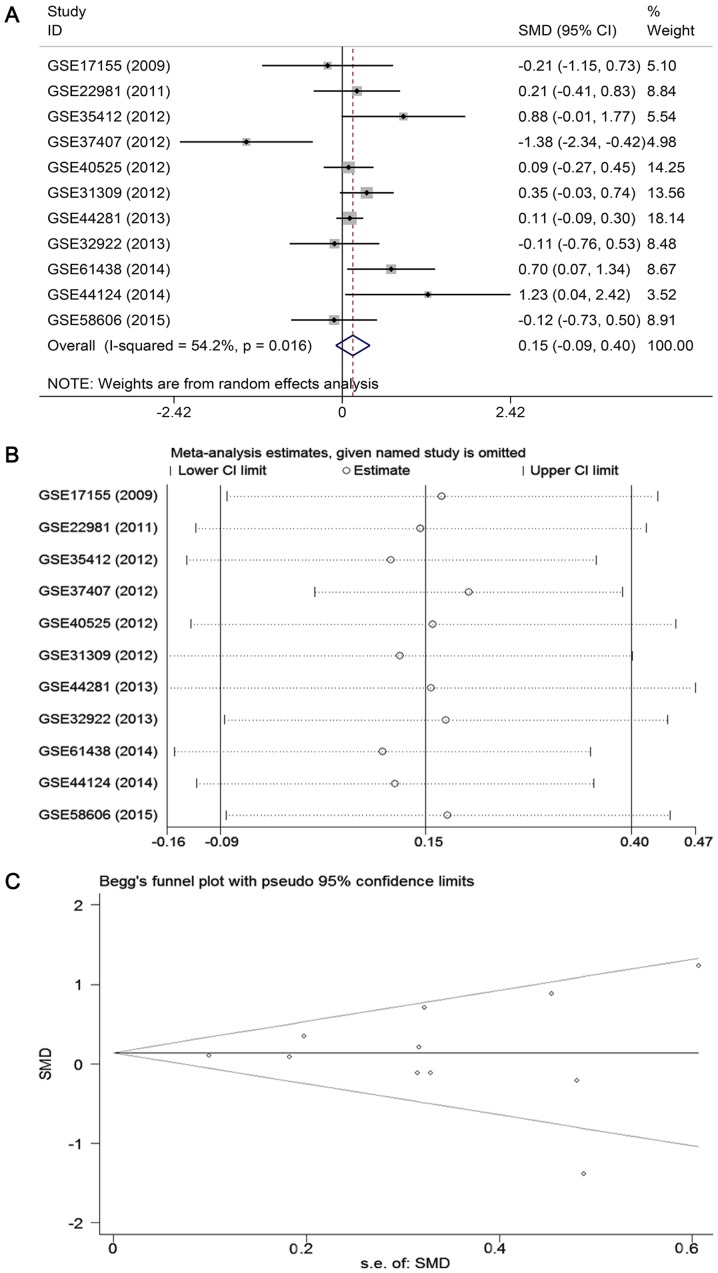
To further verify the expression level of miR-671-3p in BC, a meta-analysis based on microarray datasets was conducted. A total of 11 studies with 688 BC patients and 400 healthy subjects were included (16,28-36). The basic characteristics of the 11 available datasets are displayed in Table III. The combined SMD from a random-effects model demonstrated that the expression of miR-671-3p in BC samples was similar to that in normal controls (SMD: 0.15, 95% CI: -0.09 to 0.40, P=0.222; heterogeneity: I2=54.2%, P=0.016; Fig. 5A). Sensitivity analysis revealed that the pooled result was stable (Fig. 5B). Begg's and Egger's tests indicated that no publication bias was observed among the 11 records (Begg's test, P=0.755; Egger's test, P=0.564; Fig. 5C).
Table III.
Main characteristics of the 11 included microarray datasets obtained from the GEO and ArrayExpress databases.
| First author (year) | Country | Data source | Platform | Sample size (T/N) | miR-671-3p expressiona
|
Ref. | |
|---|---|---|---|---|---|---|---|
| T | N | ||||||
| Fassan et al (2009) | Italy | GEO: GSE17155 | GPL8871 | 33/5 | 7.98±0.95 | 8.17±0.32 | (28) |
| Zhao et al (2010) | USA | GEO: GSE22981 | GPL8179 | 20/20 | 9.39±1.95 | 9.00±1.75 | (29) |
| Romero-Cordoba et al (2012) | Mexico | GEO: GSE35412 | GPL9731 | 34/6 | 4.41±0.30 | 4.13±0.42 | (30) |
| Gravgaard et al (2012) | Sweden | GEO: GSE37407 | GPL13703 | 50/5 | 6.23±0.03 | 6.27±0.01 | (31) |
| Biagioni et al (2012) | Israel | GEO: GSE40525 | GPL8227 | 61/59 | 2.45±0.33 | 2.42±0.33 | (32) |
| Schrauder et al (2012) | Germany | GEO: GSE31309 | GPL14132 | 48/57 | 5.81±0.79 | 5.55±0.69 | (33) |
| Godfrey (2013) | USA | GEO: GSE44281 | GPL14613 | 205/205 | 2.24±0.70 | 2.17±0.59 | (16) |
| Tanic (2013) | Spain | GEO: GSE32922 | GPL7723 | 22/16 | 6.08±0.09 | 6.09±0.09 | No ref. available |
| Yan (2015) | Australia | GEO: GSE61438 | GPL8179 | 44/13 | 7.96±0.68 | 7.43±0.97 | (34) |
| Feliciano (2013) | Spain | GEO: GSE44124 | GPL14767 | 50/3 | 5.42±0.09 | 5.31±0.07 | (35) |
| Matamala (2015) | Spain | GEO: GSE58606 | GPL18838 | 121/11 | 6.43±0.71 | 6.51±0.04 | (36) |
Represented as the mean ± standard deviation. T, breast tumor; N, normal control; GEO, Gene Expression Omnibus.
Figure 5.
Expression level of miR-671-3p in BC based on data obtained from the GEO and ArrayExpress databases. (A) Forest plot of the 11 datasets evaluating miR-671-3p expression in BC (random-effects model). SMD <0 indicates that miR-671-3p is downregulated in BC. (B) Sensitivity analysis evaluating the stability of the meta-analysis. (C) Funnel plot assessing the publication bias in the 11 datasets. miR, microRNA; BC, breast cancer; GEO, Gene Expression Omnibus; SMD, standard mean difference; 95% CI, 95% confidence interval; s.e., standard error.
Prospective molecular mechanism of miR-671-3p in BC, as determined by enrichment analyses
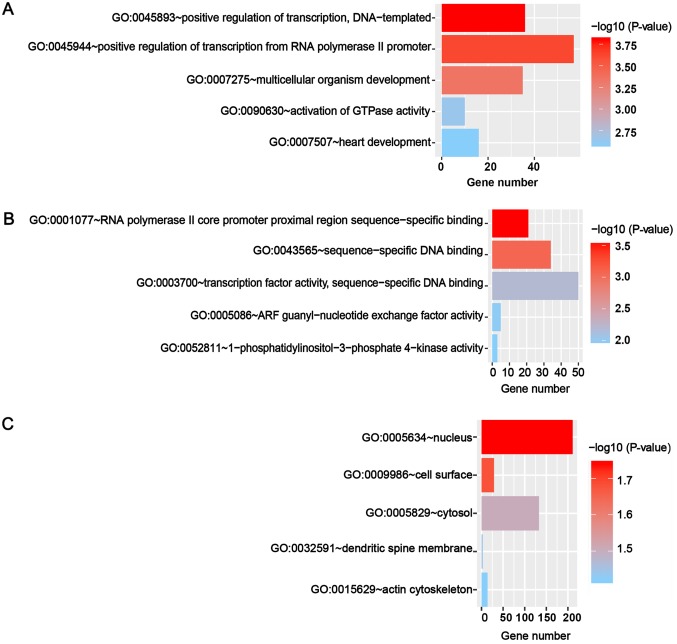
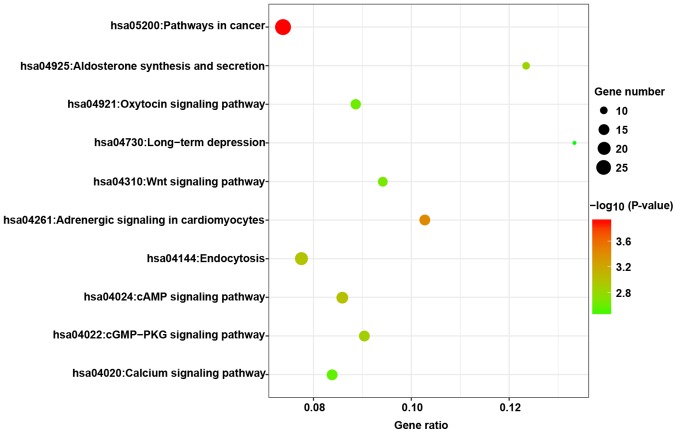
The putative targets of miR-671-3p were predicted in 12 online prediction databases (Targetscan, RNAhybrid, RNA22, PITA, Pictar2, miRWalk, Microt4, miRNAMap, miRDB, mirbridge, miRanda and miRMap), and targets confirmed by >3 algorithms were selected for further analyses. A total of 1,470 targets combined were obtained by GO and KEGG analyses. The GO analyses included three categories, namely biological process, molecular function and cellular component. The top five GO annotations are shown in Fig. 6, and the results suggested that the targets of miR-671-3p were markedly enriched in the cell nucleus and participated in transcription regulation. The top 10 KEGG pathways are presented in Fig. 7, and it was observed that these predicted targets may be involved in several tumor-associated pathways, including pathways in cancer and the Wnt signaling pathway.
Figure 6.
Top 5 enriched GO annotations of the putative target genes of miR-671-3p. The top 5 enriched (A) biological processes, (B) molecular functions, and (C) cellular components are shown. miR, microRNA; GO, Gene Ontology.
Figure 7.
Top 10 enriched KEGG pathways of the putative target genes of miR-671-3p. miR, microRNA; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Construction of PPI network and miRNA-gene regulatory network
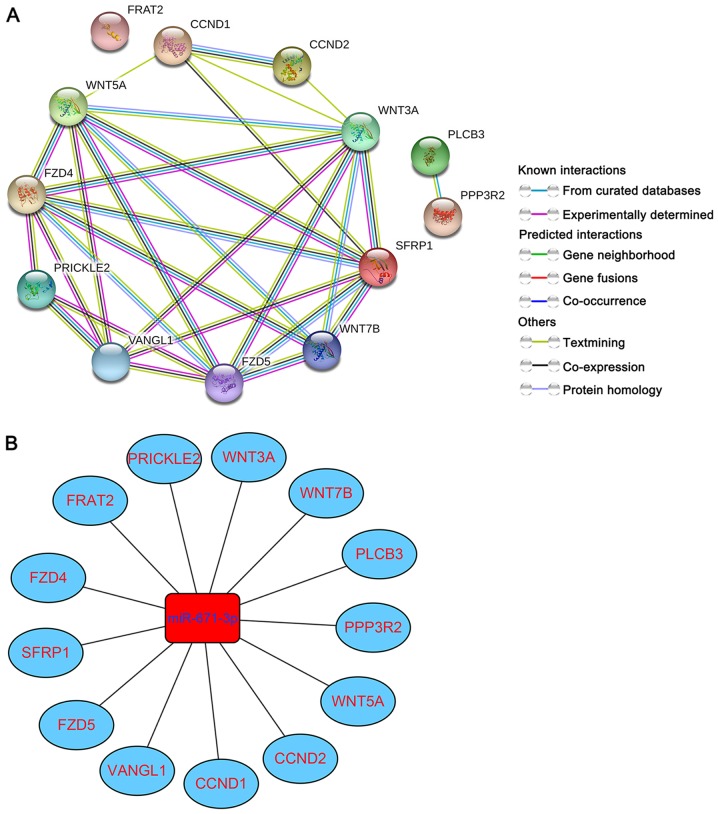
To further delve into the potential molecular mechanism of miR-671-3p in BC, targets enriched in the Wnt signaling pathway were selected for PPI network construction. The following 13 genes were selected: WNT5A, WNT7B, WNT3A, PLCB3, CCND1, CCND2, VANGL1, SFRP1, PRICKLE2, PPP3R2, FRAT2, FZD5 and FZD4. As shown in Fig. 8A, a total of 13 nodes and 29 edges were involved in the PPI network. Simultaneously, the miRNA-gene regulatory network was formed based on the 13 miRNA-gene pairs (Fig. 8B).
Figure 8.
PPI and miRNA-gene regulatory networks. (A) PPI network of 13 miR-671-3p target genes involved in the Wnt signaling pathway. (B) miRNA-gene regulatory network based on 13 miR-671-3p-gene pairs. miR, microRNA; PPI, protein-protein interaction.
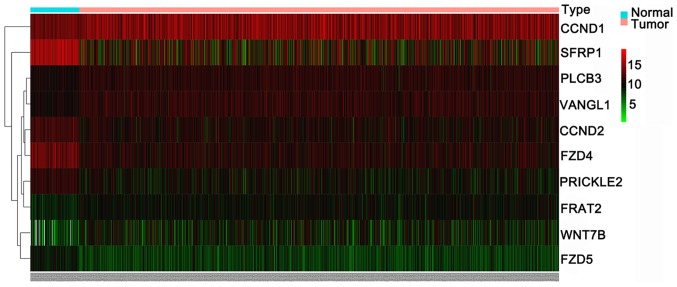
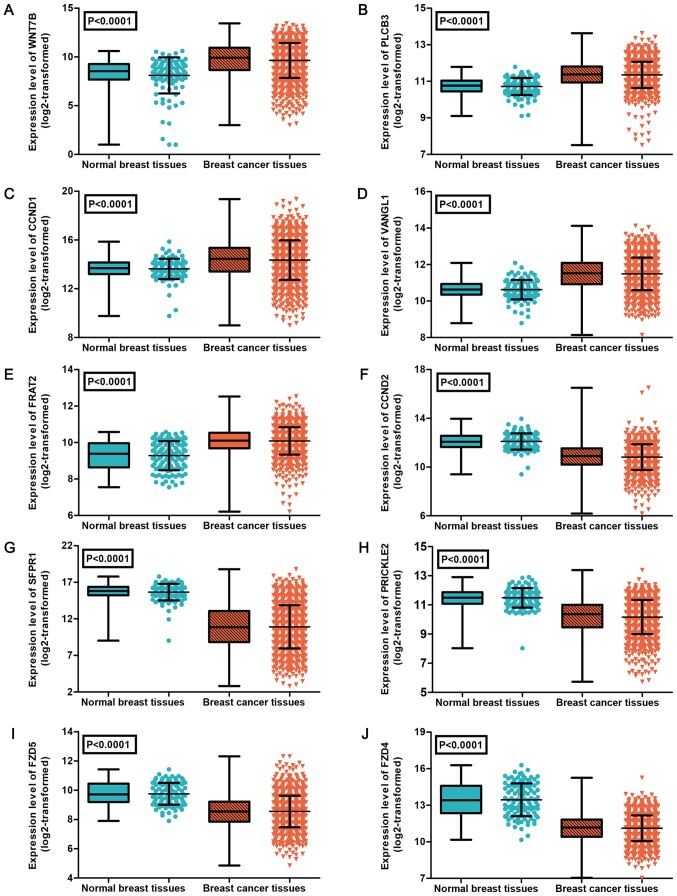
Subsequently, the expression levels of the 13 genes were verified using data from TCGA. Due to the expression of genes WNT3A and PPP3R2 being zero in >10% of samples, those genes were removed from the expression analysis (37). As shown in Table IV, the results identified 10 differently expressed target genes in BC, including 5 upregulated genes (WNT7B, PLCB3, CCND1, VANGL1 and FRAT2) and 5 downregulated genes (CCND2, SFRP1, PRICKLE2, FZD5 and FZD4). The expression patterns of the 10 differently expressed genes were visualized as a heatmap (Fig. 9) and box-scatter plots (Fig. 10).
Table IV.
Expression levels of the 11 target genes of miR-671-3p based on data obtained from TCGA.
| Gene | Sample size (T/N) | miR-671-3p expressiona
|
t-test
|
||
|---|---|---|---|---|---|
| Tumor | Normal | t-value | P-value | ||
| WNT5A | 1109/113 | 10.03±1.62 | 10.16±1.03 | 1.234 | 0.219 |
| WNT7B | 1109/108 | 9.64±1.79 | 7.61±2.66 | 7.975 | <0.0001 |
| PLCB3 | 1109/113 | 11.35±0.72 | 10.71±0.47 | 12.995 | <0.0001 |
| CCND1 | 1109/113 | 14.36±1.62 | 13.63±0.84 | 7.832 | <0.0001 |
| VANGL1 | 1109/113 | 11.49±0.89 | 10.62±0.54 | 15.203 | <0.0001 |
| FRAT2 | 1109/113 | 10.09±0.75 | 9.29±0.79 | 10.238 | <0.0001 |
| CCND2 | 1109/113 | 10.81±1.06 | 12.10±0.67 | 18.331 | <0.0001 |
| SFRP1 | 1109/113 | 10.89±2.96 | 15.66±1.13 | 34.327 | <0.0001 |
| PRICKLE2 | 1109/113 | 10.16±1.17 | 11.48±0.67 | 18.326 | <0.0001 |
| FZD5 | 1109/113 | 8.55±1.08 | 9.76±0.74 | 15.64 | <0.0001 |
| FZD4 | 1109/113 | 11.11±1.06 | 13.45±1.34 | 17.961 | <0.0001 |
Represented as the mean ± standard deviation. miR, microRNA; TCGA, The Cancer Genome Atlas.
Figure 9.
Heatmap of 10 differently expressed targets of miR-671-3p in BC and normal breast tissues based on data obtained from TCGA. miR, microRNA; BC, breast cancer; TCGA, The Cancer Genome Atlas.
Figure 10.
Expression levels of 10 differently expressed targets of miR-671-3p in BC and normal breast tissues based on data obtained from TCGA. (A) WNT7B, (B) PLCB3, (C) CCND1, (D) VANGL1 and (E) FRAT2 were upregulated in BC. (F) CCND2, (G) SFPR1, (H) PRICKLE2, (I) FZD5 and (J) FZD4 were downregulated in BC. miR, microRNA; BC, breast cancer; TCGA, The Cancer Genome Atlas.
Discussion
BC is an aggressive cancer that threatens the health of women. It is the most prevalent tumor and the main cause of oncogenic mortality in women worldwide, with an estimated 1,676,600 newly diagnosed cases and 521,900 associated mortalities annually (38). Although specific molecular biomarkers for optimizing clinical management have been explored (39–41), their clinical application remains limited due to the heterogeneity and complexity of BC. Therefore, probing effective biomarkers to better understand the molecular mechanism of BC and to provide appropriate target treatments for BC patients is essential. The aim of the present study was to address the expression level, clinical value, biological function and molecular mechanism of miR-671-3p in BC and, further, to explore the potential clinical usability of miR-671-3p for therapeutic decisions in BC.
In the present study, in vitro experiments were conducted to examine the biological effects of miR-671-3p on the proliferation and apoptosis of BC cells by transfecting a miR-671-3p mimic into MDA-MB-231 cells. The results revealed that the miR-671-3p mimic evidently attenuated cell proliferation and induced cell apoptosis, indicating that miR-671-3p was a tumor suppressor in BC.
Only one previous study conducted by Godfrey et al (16) has reported the expression pattern of miR-671-3p in BC. The authors detected miRNA expression in serum of BC patients and healthy participants using array Affymetrix arrays and demonstrated that 16 miRNAs, including miR-671-3p, were overexpressed in the serum of BC patients. The authors validated the expression levels of the three miRNAs with the highest expression using RT-qPCR; however, no statistically significant differences were detected. Given the low accuracy of gene chip technology, large-scale studies based on RT-qPCR are imperative for verifying the serum level of miR-671-3p in BC patients. In the present study, miR-671-3p expression was examined in BC and non-tumorous tissues using RT-qPCR, and the results indicated that miR-671-3p was downregulated in cancer tissues. All available relevant microarray datasets were also collected in order to further confirm the expression level of miR-671-3p in BC. However, the result revealed that the expression of miR-671-3p in BC tissues was similar to that in the normal controls, which was not consistent with the results of RT-qPCR. Differences in sample sources and detection methods may explain this inconsistency in the findings. For instance, the expression of miR-671-3p may vary across individuals. In addition, the accuracy of RT-qPCR is higher compared with that of microarray analysis. Although a total of 11 microarray datasets were included to assess the expression level of miR-671-3p in BC and the pooled result was stable, the results should be interpreted with caution, since these datasets were obtained from different platforms, resulting in a significant heterogeneity in the current study. Additionally, the confounders induced by different RNA extraction methods and diverse RNA detection platforms may also limit the validity of the present meta-analysis results. Thus, further and larger-scale RT-qPCR-based investigations will be indispensable in deciphering the exact expression of miR-671-3p in BC.
The association of miR-671-3p expression with the clinicopathological parameters and overall survival of patients was also analyzed to determine whether miR-671-3p can function as a predictive indicator in BC. However, no association between miR-671-3p expression and the clinicopathological factors was detected. Patients with high miR-671-3p expression survived longer in comparison with patients with low miR-671-3p expression, however, no statistically significant difference was observed. Considering the relatively small sample size of 38 cases in the present study, further investigation including more samples is required to determine the prognostic value of miR-671-3p in patients with BC.
Finally, the in silico GO and KEGG analyses in the present study revealed the potential molecular mechanism of miR-6713p in the tumorigenesis of BC. It was observed that the predicted targets of miR-671-3p were involved in several tumor-associated pathways, including the Wnt signaling pathway. The Wnt signaling pathway is known to be closely linked with the initiation and progression of various malignant tumors, including BC, by driving of the epithelial-mesenchymal transition and metastasis of tumor cells (42-47). Accumulating evidence has suggested that the Wnt signaling cascade is regulated by miRNAs (46,48-51). A study performed by Cai et al (46) indicated that miR-374a activates the Wnt signaling pathway in BC by inhibiting negative modulators of the Wnt signaling cascade, including WNT5A, WIF1 and PTEN. A study conducted by Yi et al (51) demonstrated that miR-214 negatively regulates the Wnt signaling cascade and, consequently, inhibits cell proliferation in BC. However, the regulatory effects of miR-671-3p on the Wnt signaling pathway have yet to be determined. In the current study, it was observed that 13 target genes (WNT5A, WNT7B, WNT3A, PLCB3, CCND1, CCND2, VANGL1, SFRP1, PRICKLE2, PPP3R2, FRAT2, FZD5 and FZD4) of miR-671-3p were significantly enriched in the Wnt signaling pathway. To further determine the roles of the 13 genes in BC, their expression patterns were investigated using data from TCGA. The results revealed that the genes WNT7B, PLCB3, CCND1, VANGL1 and FRAT2 were evidently upregulated in BC, indicating that they may act as oncogenes, while WNT3A, CCND2, SFRP1, PRICKLE2, FZD5 and FZD4 were markedly downregulated in BC, suggesting that they may function as tumor suppressor genes in this tumor. Furthermore, it is inferred that miR-671-3p may suppress the tumorigenesis and development of BC by interacting with these genes, and further inhibiting the Wnt signaling pathway. However, further rigorous investigation is requited to confirm the targeting effects of miR-671-3p on these genes.
In conclusion, the present study provided evidence that miR-671-3p functions as a tumor suppressor to inhibit the initiation and development of BC by influencing the Wnt signaling pathway, as observed by in vitro experiments, in-house RT-qPCR, microarray datasets and bioinformatics analyses. The findings also demonstrated a possible molecular mechanism of miR-671-3p in BC, indicating that miR-671-3p may be a prospective molecular target for the therapy of BC. Further investigation is required to verify these conclusions.
Acknowledgments
Not applicable.
Funding
This study was supported by grants from the Natural Science Foundation of Guangxi, China (grant nos. 2017GXNSFAA198067 and 2015GXNSFAA139187).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
DDX collected and analyzed the data from the GEO and Oncomine databases, participated in the statistical analyses and was a major contributor in the writing of the manuscript. HC performed in-house RT-qPCR and in vitro experiments. RQH, AHL and JCZ conducted the bioinformatics analyses. GC examined all of the data presented in the manuscript. ZBF and KLW guided the design of all of the experiments and the writing of the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
All participants provided informed consent prior to sample collection. The Ethical Committee of the First Affiliated Hospital of Guangxi Medical University approved this investigation.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91:827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 3.Fan FY, Deng R, Yi H, Sun HP, Zeng Y, He GC, Su Y. The inhibitory effect of MEG3/miR-214/AIFM2 axis on the growth of T-cell lymphoblastic lymphoma. Int J Oncol. 2017;51:316–326. doi: 10.3892/ijo.2017.4006. [DOI] [PubMed] [Google Scholar]
- 4.Yu M, Xue H, Wang Y, Shen Q, Jiang Q, Zhang X, Li K, Jia M, Jia J, Xu J, et al. miR-345 inhibits tumor metastasis and EMT by targeting IRF1-mediated mTOR/STAT3/AKT pathway in hepatocellular carcinoma. Int J Oncol. 2017;50:975–983. doi: 10.3892/ijo.2017.3852. [DOI] [PubMed] [Google Scholar]
- 5.He R, Yang L, Lin X, Chen X, Lin X, Wei F, Liang X, Luo Y, Wu Y, Gan T, et al. MiR-30a-5p suppresses cell growth and enhances apoptosis of hepatocellular carcinoma cells via targeting AEG-1. Int J Clin Exp Pathol. 2015;8:15632–15641. [PMC free article] [PubMed] [Google Scholar]
- 6.Xie Z, Chen W, Chen Y, Wang X, Gao W, Liu Y. miR-768-3p is involved in the proliferation, invasion and migration of non-small cell lung carcinomas. Int J Oncol. 2017;51:1574–1582. doi: 10.3892/ijo.2017.4133. [DOI] [PubMed] [Google Scholar]
- 7.Shang A, Yang M, Shen F, Wang J, Wei J, Wang W, Lu W, Wang C, Wang C. MiR-1-3p suppresses the proliferation, invasion and migration of bladder cancer cells by up-regulating SFRP1 expression. Cell Physiol Biochem. 2017;41:1179–1188. doi: 10.1159/000464379. [DOI] [PubMed] [Google Scholar]
- 8.Chen G, He M, Yin Y, Yan T, Cheng W, Huang Z, Zhang L, Zhang H, Liu P, Zhu W, et al. miR-1296-5p decreases ERBB2 expression to inhibit the cell proliferation in ERBB2-positive breast cancer. Cancer Cell Int. 2017;17:95. doi: 10.1186/s12935-017-0466-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding Y, Zhang C, Zhang J, Zhang N, Li T, Fang J, Zhang Y, Zuo F, Tao Z, Tang S, et al. miR-145 inhibits proliferation and migration of breast cancer cells by directly or indirectly regulating TGF-β1 expression. Int J Oncol. 2017;50:1701–1710. doi: 10.3892/ijo.2017.3945. [DOI] [PubMed] [Google Scholar]
- 10.Xiao G, Li X, Li G, Zhang B, Xu C, Qin S, Du N, Wang J, Tang SC, Zhang J, et al. MiR-129 blocks estrogen induction of NOTCH signaling activity in breast cancer stem-like cells. Oncotarget. 2017;8:103261–103273. doi: 10.18632/oncotarget.21143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu X, Ma J, Chu J, Shao Q, Zhang Y, Lu G, Li J, Huang X, Li W, Li Y, et al. MiR-129-5p sensitizes the response of Her-2 positive breast cancer to trastuzumab by reducing Rps6. Cell Physiol Biochem. 2017;44:2346–2356. doi: 10.1159/000486122. [DOI] [PubMed] [Google Scholar]
- 12.Jang MH, Kim HJ, Gwak JM, Chung YR, Park SY. Prognostic value of microRNA-9 and microRNA-155 expression in triple-negative breast cancer. Hum Pathol. 2017;68:69–78. doi: 10.1016/j.humpath.2017.08.026. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L, Xu Y, Jin X, Wang Z, Wu Y, Zhao D, Chen G, Li D, Wang X, Cao H, et al. A circulating miRNA signature as a diagnostic biomarker for non-invasive early detection of breast cancer. Breast Cancer Res Treat. 2015;154:423–434. doi: 10.1007/s10549-015-3591-0. [DOI] [PubMed] [Google Scholar]
- 14.Xiong DD, Lv J, Wei KL, Feng ZB, Chen JT, Liu KC, Chen G, Luo DZ. A nine-miRNA signature as a potential diagnostic marker for breast carcinoma: An integrated study of 1,110 cases. Oncol Rep. 2017;37:3297–3304. doi: 10.3892/or.2017.5600. [DOI] [PubMed] [Google Scholar]
- 15.Tan X, Fu Y, Chen L, Lee W, Lai Y, Rezaei K, Tabbara S, Latham P, Teal CB, Man YG, et al. miR-671-5p inhibits epithelial-to-mesenchymal transition by downregulating FOXM1 expression in breast cancer. Oncotarget. 2016;7:293–307. doi: 10.18632/oncotarget.6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Godfrey AC, Xu Z, Weinberg CR, Getts RC, Wade PA, DeRoo LA, Sandler DP, Taylor JA. Serum microRNA expression as an early marker for breast cancer risk in prospectively collected samples from the Sister Study cohort. Breast Cancer Res. 2013;15:R42. doi: 10.1186/bcr3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gradishar WJ, Anderson BO, Balassanian R, Blair SL, Burstein HJ, Cyr A, Elias AD, Farrar WB, Forero A, Giordano SH, et al. Invasive Breast Cancer Version 1.2016, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2016;14:324–354. doi: 10.6004/jnccn.2016.0037. [DOI] [PubMed] [Google Scholar]
- 18.Chen G, Rong M, Luo D. TNFRSF6B neutralization antibody inhibits proliferation and induces apoptosis in hepatocellular carcinoma cell. Pathol Res Pract. 2010;206:631–641. doi: 10.1016/j.prp.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 19.Rong M, Chen G, Dang Y. Increased miR-221 expression in hepatocellular carcinoma tissues and its role in enhancing cell growth and inhibiting apoptosis in vitro. BMC Cancer. 2013;13:21. doi: 10.1186/1471-2407-13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rong M, He R, Dang Y, Chen G. Expression and clinicopathological significance of miR-146a in hepatocellular carcinoma tissues. Ups J Med Sci. 2014;119:19–24. doi: 10.3109/03009734.2013.856970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen G, Kronenberger P, Teugels E, De Grève J. Influence of RT-qPCR primer position on EGFR interference efficacy in lung cancer cells. Biol Proced Online. 2010;13:1. doi: 10.1186/1480-9222-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen G, Umelo IA, Lv S, Teugels E, Fostier K, Kronenberger P, Dewaele A, Sadones J, Geers C, De Grève J. miR-146a inhibits cell growth, cell migration and induces apoptosis in non-small cell lung cancer cells. PLoS One. 2013;8:e60317. doi: 10.1371/journal.pone.0060317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen G, Kronenberger P, Teugels E, Umelo IA, De Grève J. Effect of siRNAs targeting the EGFR T790M mutation in a non-small cell lung cancer cell line resistant to EGFR tyrosine kinase inhibitors and combination with various agents. Biochem Biophys Res Commun. 2013;431:623–629. doi: 10.1016/j.bbrc.2012.12.070. [DOI] [PubMed] [Google Scholar]
- 24.Xiao F, Lan A, Lin Z, Song J, Zhang Y, Li J, Gu K, Lv B, Zhao D, Zeng S, et al. Impact of CAG repeat length in the androgen receptor gene on male infertility - a meta-analysis. Reprod Biomed Online. 2016;33:39–49. doi: 10.1016/j.rbmo.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 25.Kohl M, Wiese S, Warscheid B. Cytoscape: Software for visualization and analysis of biological networks. Methods Mol Biol. 2011;696:291–303. doi: 10.1007/978-1-60761-987-1_18. [DOI] [PubMed] [Google Scholar]
- 26.Phukan JP, Sinha A, Deka JP. Cytological grading of breast carcinoma on fine needle aspirates and its relation with histological grading. South Asian J Cancer. 2015;4:32–34. doi: 10.4103/2278-330X.149948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chavez-MacGregor M, Mittendorf EA, Clarke CA, Lichtensztajn DY, Hunt KK, Giordano SH. Incorporating tumor characteristics to the American Joint Committee on Cancer Breast Cancer Staging System. Oncologist. 2017;22:1292–1300. doi: 10.1634/theoncologist.2017-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fassan M, Baffa R, Palazzo JP, Lloyd J, Crosariol M, Liu CG, Volinia S, Alder H, Rugge M, Croce CM, et al. MicroRNA expression profiling of male breast cancer. Breast Cancer Res. 2009;11:R58. doi: 10.1186/bcr2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao H, Shen J, Medico L, Wang D, Ambrosone CB, Liu S. A pilot study of circulating miRNAs as potential biomarkers of early stage breast cancer. PLoS One. 2010;5:e13735. doi: 10.1371/journal.pone.0013735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romero-Cordoba S, Rodriguez-Cuevas S, Rebollar-Vega R, Quintanar-Jurado V, Maffuz-Aziz A, Jimenez-Sanchez G, Bautista-Piña V, Arellano-Llamas R, Hidalgo-Miranda A. Identification and pathway analysis of microRNAs with no previous involvement in breast cancer. PLoS One. 2012;7:e31904. doi: 10.1371/journal.pone.0031904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gravgaard KH, Lyng MB, Laenkholm AV, Søkilde R, Nielsen BS, Litman T, Ditzel HJ. The miRNA-200 family and miRNA-9 exhibit differential expression in primary versus corresponding metastatic tissue in breast cancer. Breast Cancer Res Treat. 2012;134:207–217. doi: 10.1007/s10549-012-1969-9. [DOI] [PubMed] [Google Scholar]
- 32.Biagioni F, Bossel Ben-Moshe N, Fontemaggi G, Canu V, Mori F, Antoniani B, Di Benedetto A, Santoro R, Germoni S, De Angelis F, et al. miR-10b*, a master inhibitor of the cell cycle, is down-regulated in human breast tumours. EMBO Mol Med. 2012;4:1214–1229. doi: 10.1002/emmm.201201483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schrauder MG, Strick R, Schulz-Wendtland R, Strissel PL, Kahmann L, Loehberg CR, Lux MP, Jud SM, Hartmann A, Hein A, et al. Circulating micro-RNAs as potential blood-based markers for early stage breast cancer detection. PLoS One. 2012;7:e29770. doi: 10.1371/journal.pone.0029770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan M, Shield-Artin K, Byrne D, Deb S, Waddell N, Haviv I, Fox SB, kConFab Investigators, kConFab Comparative microRNA profiling of sporadic and BRCA1 associated basal- like breast cancers. BMC Cancer. 2015;15:506. doi: 10.1186/s12885-015-1522-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feliciano A, Castellvi J, Artero-Castro A, Leal JA, Romagosa C, Hernández-Losa J, Peg V, Fabra A, Vidal F, Kondoh H, et al. miR-125b acts as a tumor suppressor in breast tumorigenesis via its novel direct targets ENPEP, CK2-α, CCNJ, and MEGF9. PLoS One. 2013;8:e76247. doi: 10.1371/journal.pone.0076247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matamala N, Vargas MT, González-Cámpora R, Miñambres R, Arias JI, Menéndez P, Andrés-León E, Gómez-López G, Yanowsky K, Calvete-Candenas J, et al. Tumor microRNA expression profiling identifies circulating microRNAs for early breast cancer detection. Clin Chem. 2015;61:1098–1106. doi: 10.1373/clinchem.2015.238691. [DOI] [PubMed] [Google Scholar]
- 37.Zeng JH, Liang L, He RQ, Tang RX, Cai XY, Chen JQ, Luo DZ, Chen G. Comprehensive investigation of a novel differentially expressed lncRNA expression profile signature to assess the survival of patients with colorectal adenocarcinoma. Oncotarget. 2017;8:16811–16828. doi: 10.18632/oncotarget.15161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 39.Schnitt SJ. Classification and prognosis of invasive breast cancer: From morphology to molecular taxonomy. Mod Pathol. 2010;23(Suppl 2):S60–S64. doi: 10.1038/modpathol.2010.33. [DOI] [PubMed] [Google Scholar]
- 40.Song C, Zhang L, Wang J, Huang Z, Li X, Wu M, Li S, Tang H, Xie X. High expression of microRNA-183/182/96 cluster as a prognostic biomarker for breast cancer. Sci Rep. 2016;6:24502. doi: 10.1038/srep24502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marino AL, Evangelista AF, Vieira RA, Macedo T, Kerr LM, Abrahão-Machado LF, Longatto-Filho A, Silveira HC, Marques MM. MicroRNA expression as risk biomarker of breast cancer metastasis: A pilot retrospective case-cohort study. BMC Cancer. 2014;14:739. doi: 10.1186/1471-2407-14-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun L, Liu T, Zhang S, Guo K, Liu Y. Oct4 induces EMT through LEF1/β-catenin dependent WNT signaling pathway in hepatocellular carcinoma. Oncol Lett. 2017;13:2599–2606. doi: 10.3892/ol.2017.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yanaka Y, Muramatsu T, Uetake H, Kozaki K, Inazawa J. miR-544a induces epithelial-mesenchymal transition through the activation of WNT signaling pathway in gastric cancer. Carcinogenesis. 2015;36:1363–1371. doi: 10.1093/carcin/bgv106. [DOI] [PubMed] [Google Scholar]
- 44.Jin B, Wang W, Meng XX, Du G, Li J, Zhang SZ, Zhou BH, Fu ZH. Let-7 inhibits self-renewal of hepatocellular cancer stem-like cells through regulating the epithelial-mesenchymal transition and the Wnt signaling pathway. BMC Cancer. 2016;16:863. doi: 10.1186/s12885-016-2904-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu ZJ, Liu HL, Zhou HC, Wang GC. TIPE2 Inhibits Hypoxia-induced Wnt/β-catenin pathway activation and EMT in glioma cells. Oncol Res. 2016;24:255–261. doi: 10.3727/096504016X14666990347356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cai J, Guan H, Fang L, Yang Y, Zhu X, Yuan J, Wu J, Li M. MicroRNA-374a activates Wnt/β-catenin signaling to promote breast cancer metastasis. J Clin Invest. 2013;123:566–579. doi: 10.1172/JCI65871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao C, Wu CH, Hu HZ. LncRNA UCA1 promotes epithelial-mesenchymal transition (EMT) of breast cancer cells via enhancing Wnt/beta-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2016;20:2819–2824. [PubMed] [Google Scholar]
- 48.Xiao G, Zhang B, Meng J, Wang J, Xu C, Tang SC, Li X, Zhang J, Liang R, Ren H, et al. miR-367 stimulates Wnt cascade activation through degrading FBXW7 in NSCLC stem cells. Cell Cycle. 2017;16:2374–2385. doi: 10.1080/15384101.2017.1380136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fasihi A, M Soltani B, Atashi A, Nasiri S. Introduction of hsa-miR-103a and hsa-miR-1827 and hsa-miR-137 as new regulators of Wnt signaling pathway and their relation to colorectal carcinoma. J Cell Biochem. 2017 Aug 17; doi: 10.1002/jcb.26357. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 50.Zhang W, Shen C, Li C, Yang G, Liu H, Chen X, Zhu D, Zou H, Zhen Y, Zhang D, et al. miR-577 inhibits glioblastoma tumor growth via the Wnt signaling pathway. Mol Carcinog. 2016;55:575–585. doi: 10.1002/mc.22304. [DOI] [PubMed] [Google Scholar]
- 51.Yi SJ, Li LL, Tu WB. MiR-214 negatively regulates proliferation and WNT/β-catenin signaling in breast cancer. Eur Rev Med Pharmacol Sci. 2016;20:5148–5154. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.