Abstract
Cushing's disease is primarily caused by pituitary adrenocorticotropin-secreting adenoma. However, its pathogenesis has remained obscure. In the present study, whole transcriptome analysis was performed by RNA sequencing (RNA-Seq) and expression of secreted frizzled-related protein 2 (SFRP2) was decreased in corticotroph tumors compared with normal pituitary glands. Furthermore, the RNA-Seq results were validated and the expression of SFRP2 in tumor tissues was analyzed by comparing another cohort of 23 patients with Cushing's disease and 3 normal human pituitary samples using reverse transcription-quantitative polymerase chain reaction, western blot and immunohistochemistry staining. Clinically, there was an association between lower SFRP2 expression and aggressive adenoma characteristics, including larger size and invasiveness. Conversely, SFRP2 overexpression reduced the ability of AtT20 cells to proliferate and migrate, and reduced production of the adrenocorticotrophic hormone in vitro. Mechanistically, overexpressed SFRP2 reduced the level of β-catenin in the cytoplasm and nucleus, and decreased Wnt signaling activity in AtT20 cells. Therefore, SFRP2 appears to act as a tumor suppressor in Cushing's disease by regulating the activity of the Wnt signaling pathway.
Keywords: Cushing's disease, corticotroph adenoma, secreted frizzled-related protein 2, Wnt signaling
Introduction
Cushing's disease (CD), which is characterized by excessive secretion of the adrenocorticotrophic hormone (ACTH), usually results from a pituitary adenoma or corticotroph hyperplasia (1). High levels of ACTH stimulate the adrenal glands to release excess glucocorticoids, resulting in hypercortisolism, which can induce a series of long-term complications, including cardiovascular disease, metabolic syndrome, muscle atrophy, osteoporosis, recurrent infections and psychiatric disorders that can cause substantial morbidity and mortality if not appropriately treated (2,3). To date, pituitary surgery remains the first line therapy, while pituitary radiotherapy or bilateral adrenalectomy have traditionally been used as adjuvant therapies for persistent hypercortisolism (4). However, even with combined therapy, the remission rate is low and the risk of recurrence is high (5). Thus, the development of novel therapeutic approaches for CD is required, which will depend closely on an improved understanding of the genetic pathogenesis of the disease.
To detect the intrinsic molecular defects of CD, RNA sequencing (RNA-Seq) was used to determine the gene expression profile of CD in the current study. Using RNA-Seq, the transcriptome of 13 cases of CD and 5 normal human pituitaries (NHPs) were analyzed. Marked downregulation of the gene secreted frizzled-related protein 2 (SFRP2) was identified in CD. SFRP2 belongs to the SFRP family and acts as a negative regulator of the canonical Wnt signaling pathway. It can competitively bind frizzled receptors that accelerate β-catenin degradation and reduce the nuclear translocation of β-catenin (6). Thus, decreased expression of SFRP2 results in upregulation of Wnt/β-catenin signaling, which regulates diverse developmental processes, such as differentiation, cell migration, cell adhesion and proliferation (7). In fact, it has been reported that decreased expression of SFRP2 activates Wnt/β-catenin signaling, which promotes the development of various tumors types, including colorectal cancer (8), esophageal carcinoma (9), medulloblastoma (10), hepatocellular carcinoma (11) and aggressive nonfunctioning pituitary adenoma (12).
The findings of the current study confirmed that SFRP2 expression is downregulated in human pituitary corticotroph tumors compared with normal pituitary tissue. In clinical analysis, a correlation between lower SFRP2 expression and larger tumor size and invasiveness was identified. Furthermore, it was demonstrated that SFRP2 is closely associated to corticotroph cell proliferation and invasiveness through downregulation of Wnt signaling in vitro. These results suggest that SFRP2 is a novel molecular marker underlying tumorigenesis of pituitary corticotroph adenoma, which may provide a rationale for SFRP2 as a potential therapeutic target for CD.
Materials and methods
Patients and human tissue samples
NHP glands (n=8; 3 males, 5 females; 22–67 years old) were obtained from fresh autopsy specimens between February, 2013 and October, 2015 at the Shanghai Jiaotong University School of Medicine (Shanghai, China). The 8 samples were rinsed in sterile saline and snap-frozen in liquid nitrogen. Suitable parts of each normal pituitary tissue were embedded in paraffin. All 36 human ACTH-secreting pituitary adenoma samples (30 females, 6 males; 24–59 years old) were obtained through endonasal transsphenoidal surgery between January, 2013 and September, 2015 at the Department of Neurosurgery of Rui-Jin Hospital, Shanghai Jiaotong University (Shanghai, China). Tumors were immediately 'flash-frozen' in liquid nitrogen until use. Suitable parts of each sample were embedded in paraffin. Clinical diagnoses were confirmed via endocrine assessment, clinical features, magnetic resonance imaging, histology and pituitary hormone immunohistochemistry. Clinical data were obtained from Rui-Jin Hospital (Shanghai, China), including plasma ACTH, urinary free cortisol and midnight serum cortisol. Informed consent was obtained from all participating patients, which was approved by the Ruijin Hospital Ethics Committee (Shanghai, China).
RNA extraction and purification
Total RNA was extracted from tissues and cells using TRIzol reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's instructions. The RNA integrity number was verified to inspect RNA integrity with an Agilent Bioanalyzer 2100 (Agilent Technologies, Inc., Santa Clara, CA, USA). The RNeasy Micro kit (cat. no. 74004) and the RNase-Free DNase Set (cat. no. 79254) (both from Qiagen GmbH, Hilden, Germany) were used to further purify total RNA.
RNA-Seq expression profiling
Preparation of the RNA libraries was performed according to the manufacturer's recommendations. All 18 libraries (5 NHP, 13 CD) were constructed using the TruSeq RNA Sample Prep Kit V2 (RS-122-2001; Illumina, Inc., San Diego, CA, USA). All sequencing was performed with the Illumina HiSeq 2500. HiSeq Control Software 2.0.12 (Illumina, Inc.) was used to analyze array images and extract the raw data. FASTX-Toolkit (version 0.0.13; hannonlab.cshl.edu/fastx_toolkit/) was used to filter the raw reads to acquire clean reads. TopHat (version 2.0.9; ccb.jhu.edu/software/tophat/downloads/) was employed to map the reads. Subsequently, cufflinks (version 2.1.1; ccb.jhu.edu/software/tophat/index.shtml) was used to quantify the mapping results. Differentially expressed genes (DEGs) were then identified through fold change and P-values calculated with a t-test. The threshold set for up and downregulated genes was fold change ≥2.0 and P≤0.05.
Functional enrichment analysis
Functional enrichment analysis was applied to investigate the potential functions of the DEGs. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (www.kegg.jp) was performed to reveal the physiological function of the DEGs in CD, and Fisher's exact test was applied to select significant pathways. The threshold of significance was defined according to the P-value and false discovery rate.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Following extraction of total RNA, 1 µg total RNA was converted into first-strand cDNA using an RT kit (Promega Corporation, Madison, WI, USA), with incubation at 42°C for 15 min, 95°C for 5 min and then 4°C for 5 min. The cDNA was the used as a template for qPCR using SYBR Green Supermix (Takara Bio, Inc., Otsu, Japan), and performed with the LightCycler 480 system (Roche Diagnostics, Basel, Switzerland) subjected to 40 cycles of amplification (95°C for 5 sec and 60°C for 1 min). The relative RNA levels were determined with the 2−ΔΔCq method (13) and normalized to the mRNA level of β-actin. Quantification was performed in quadruplicate, and the experiments were repeated in triplicate. The sequences of the primers are listed in Table I.
Table I.
Sequences of the primers for reverse transcription-quantitative polymerase chain reaction.
| Species Gene name | Primer sequences (5′–3′) |
|---|---|
| Human SFRP2 | F: ACGGCATCGAATACCAGAACA |
| R: CTCGTCTAGGTCATCGAGGCA | |
| Human GADD45B | F: CAGATCCATTTTACGCTGATCCA |
| R: TCCTCGCAAAACAGGCTGAG | |
| Human JUNB | F: ACAAACTCCTGAAACCGAGCC |
| R: CGAGCCCTGACCAGAAAAGTA | |
| Human FOSL2 | F: CAGAAATTCCGGGTAGATATGCC |
| R: GGTATGGGTTGGACATGGAGG | |
| Human TFPI2 | F: TCCTGCCCCTAGACTACGG |
| R: CTCCCAGGTGTAGAAATTGTTGG | |
| Human ACTB | F: CATGTACGTTGCTATCCAGGC |
| R: CTCCTTAATGTCACGCACGAT | |
| Human TBX19 | F: ACGGACCAATCAATCCAGGG |
| R: TACACCCCATCCTGGTCTCA | |
| Human CYR61 | F: GCATTCCTCTGTGTCCCCAA |
| R: TTAGTGTCCATCCGCACCAG | |
| Mouse Sfrp2 | F: CGTGGGCTCTTCCTCTTCG |
| R: ATGTTCTGGTACTCGATGCCG | |
| Mouse Pomc | F: ATGCCGAGATTCTGCTACAGT |
| R: TCCAGCGAGAGGTCGAGTTT | |
| Mouse c-myc | F: ATGCCCCTCAACGTGAACTTC |
| R: GTCGCAGATGAAATAGGGCTG | |
| Mouse Pttg1 | F: TCTGATCCGCTGTACTCTCCT |
| R: AGGCGGCAATTCAACATCCA | |
| Mouse MMP2 | F: CTGCCACTGTCCCAGGAAG |
| R: CTCGCGGCAAGTCTTCAGAG | |
| Mouse MMP7 | F: CTTACCTCGGATCGTAGTGGA |
| R: CCCCAACTAACCCTCTTGAAGT | |
| Mouse MMP9 | F: GCAGAGGCATACTTGTACCG |
| R: TGATGTTATGATGGTCCCACTTG | |
| Mouse Ctnnb1 | F: ATGGAGCCGGACAGAAAAGC |
| R: CTTGCCACTCAGGGAAGGA | |
| Mouse Actb | F: GGCTGTATTCCCCTCCATCG |
| R: CCAGTTGGTAACAATGCCATGT |
F, forward; R, reverse; SFRP2, secreted frizzled related protein 2; GADD45B, growth arrest and DNA damage inducible β; JUNB, JunB proto-oncogene AP-1 transcription factor subunit; FOSL2, FOS like 2 AP-1 transcription factor subunit; TFPI2, tissue factor pathway inhibitor 2; ACTB, actin β; TBX19, T-box 19; CYR61, cysteine rich angiogenic inducer 61; Sfrp2, secreted frizzled-related protein 2; Pomc, pro-opiomelanocortin-α; c-Myc, myelocytomatosis oncogene; Pttg1, pituitary tumor-transforming gene 1; MMP, matrix metallopeptidase; Ctnnb1, catenin (cadherin associated protein) β1.
Protein extraction and western blot analysis
NHP (n=3) and CD (n=23) fresh-frozen tumor tissues were used for western blotting. Total protein of each tissue was extracted using in radioimmunoprecipitation assay (RIPA) buffer containing Protease Inhibitor Cocktail (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 15 min at 0°C, and then supernatant was collected. For cells, total cell lysate was also prepared by incubating in RIPA buffer containing protease inhibitor cocktail on ice. Protein concentrations were measured using a Bio-Rad protein assay reagent (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Nuclear and cytoplasmic protein extracts were prepared using the Nuclear Extract Kit (Active Motif, Carlsbad, CA, USA) according to the manufacturer's instructions. Equal amounts of protein (20 µg per sample) were boiled for 5 min and resolved by SDS-PAGE on 8 or 10% gels. Following transfer, membranes were blocked for 1 h at room temperature in TBS-Tween-20 containing 5% non-fat dried milk. The membranes were incubated with the following primary antibodies: Anti-SFRP2 (dilution 1:1,000; cat. no. ab86379), anti-β-catenin (dilution, 1:5,000; cat. no. ab32572; all from Abcam, Cambridge, UK); anti-α-tubulin (dilution 1:1,000; cat. no. T8328; Sigma-Aldrich; Merck KGaA); anti-cyclin E (dilution 1:1,000; cat. no. 4129), anti-proliferating cell nuclear antigen (PCNA; dilution 1:2,000; cat. no. 2586), anti-phospho-extracellular signal-regulated kinase (p-Erk; dilution 1:2,000; cat. no. 9101), anti-Erk (dilution 1:1,000; cat. no. 9102), anti-c-myc (dilution 1:1,000; cat. no. 9402; all from Cell Signaling Technology, Inc. Danvers, MA, USA); anti-β-actin (dilution 1:10,000; cat. no. 60008-1-Ig), anti-lamin (dilution 1:5,000; cat. no. 66095-1-Ig), anti-GAPDH (dilution 1:10,000; cat. no. 10494-1-AP; all from ProteinTech Group, Inc., Chicago, IL, USA). Following washing, membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (dilution 1:4,000; cat. nos. 7076 and 7074; Cell Signaling Technology, Inc.) for 1 h at room temperature. The signal was detected using enhanced chemiluminescence (NEL104001EA; PerkinElmer, Inc., Waltham, MA, USA) on the 5200 Chemiluminescence Imager (Tanon Science and Technology Co., Ltd., Shanghai, China). The densitometry analysis was performed using ImageJ software (version 1.47v; National Institutes of Health, Bethesda, MD, USA).
Immunohistochemistry staining (IHC)
The 5 µm thick sections were prepared from the formalin-fixed and paraffin embedded tissues (3 NHP and 23 CD). IHC staining was performed as previously reported (12). Specific primary antibody against SFRP2 (dilution 1:200; cat. no. ab86379; Abcam) was used. All the IHC slides were evaluated by two pathologists (Department of Pathology, Rui-Jin Hospital, Shanghai, China). The staining intensity was graded as follows: No staining, 0; weakly positive, 1; moderately positive, 2; and strongly positive, 3. The staining percentage was graded as follows: 0–25% staining, 1; 26–50% staining, 2; 51–75% staining, 3; and 76–100% staining, 4. The immunoreactive score was calculated as intensity of the staining multiplied by the percentage of positive cells, which was then categorized as low (0–6) and high (7–12) expression.
Cell culture and transfection
The AtT20 cell line and 293T cells were obtained from the American Type Tissue Collection (Manassas, VA, USA). The cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (FBS) and 2 mM L-glutamine and 100 µg/ml penicillin/streptomycin (all from Gibco; Thermo Fisher Scientific, Inc.) in a humidified incubator under 5% CO2 conditions at 37°C. The murine SFRP2 coding sequence was cloned into the retroviral pMSCVpuro vector (Clontech Laboratories, Inc., Mountainview, CA, USA) to generate SFRP2 expression plasmids. The retroviral vector pMSCVpuro vector was used as the negative control. 293T cells (2.5×105 in 2 ml media per well) were seeded in a 6-well plate. After 24 h, the recombinant construct was co-transfected into 293T cells together with two packaging vectors psPAX2 and pMD2.G (plasmid cat. nos. 12260 and 12259; obtained from Addgene, Inc., Cambridge, MA, USA) in a ratio of 3:2.25:0.75 µg of pMSCVpuro, psPAX2 and pMD2.G, respectively, per well. Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific, Inc.) was used according to the manufacturer's instructions. The supernatant was collected at 48 h post-transfection, and filtered through 0.45-µm pore size filter. Viral titers were determined by QuickTiter Quantitation Kit (cat. no. VPK-120; Cell Biolabs, Inc., San Diego, CA, USA) following manufacturer's instructions. AtT20 cells were infected with pMSCVpuro-SFRP2 vector and pMSCVpuro vector at the multiplicity of infection of 10. Stably overexpressed SFRP2 AtT20 and control cells were established following puromycin selection (100 ng/ml) for 14 days. To determine whether recombinant SFRP2 protein can effect ACTH secretion, proliferation and clonogenic ability of AtT20 cells, the cells were treated with recombinant mouse SFRP2 protein (cat. no. 1169-FR-025; R&D Systems, Inc., Minneapolis, MN, USA) at different doses (0–100 nM) and time points (24–72 h).
Hormone assays
The culture media was collected from each well, which was incubated for a specified time interval, and the level of ACTH was measured using the ACTH ELISA kit (cat. no. EK-001-21; Phoenix Pharmaceuticals, Inc., Burlingame, CA, USA). Each experimental group was analyzed five times and the experiments were repeated independently three times.
Cell proliferation and colony formation assay
Cell viability was tested using the Cell Counting Kit-8 assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) at the indicated time points. In the colony formation test, AtT20 cells were plated in a 6-well plate at a density of 103 cells/well. After 3 weeks, colonies were fixed, stained and counted as previously described (14).
Flow cytometry
The cells were digested with 0.5% trypsin-EDTA, transferred into flow tubes and centrifuged at 800 × g for 5 min. Then cells were washed twice with cold PBS and resuspended cells in binding buffer, and incubated in the dark at room temperature for 5 min with 5 µl Annexin V-fluorescein isothiocyanate (FITC) and 10 µl propidium iodide prior to detection (FITC Annexin V Apoptosis Detection kit I; BD Pharmingen, San Jose, CA, USA). The apoptotic results were analyzed using FlowJo software (Tree Star, Inc., Ashland, OR, USA) The results of the flow cytometry experiments were displayed as four quadrants, in which the upper right quadrant indicated advanced apoptosis rate, the lower right quadrant represents the apoptosis rate during the early stages, the upper left quadrant represents dead cells, and the lower left quadrant represents living cells. This experiment was repeated six times.
Scratch assay
Cells were cultured to 90% confluence in 6-well plates. A small scratching area was then removed with a 100 µl plastic pipette tip, following which the cells were cultured in low serum medium (containing 2% FBS). Cells were inspected microscopically over 30 h and ImageJ software (version 1.47v; National Institutes of Health, Bethesda, MD, USA) was applied to measure the relative scratch width.
Statistical analysis
Statistical analysis was performed using SPSS version 19.0 software (IBM Corporation, Armonk, NY, USA). All experimental data were presented as the mean ± standard deviation. The correlation of expression of the DEGs between CD and NHP samples, and the correlation between SFRP2 expression and hormone parameters were analyzed using Spearman's correlation. Association between clinicopathological parameters and SFRP2 expression were evaluated by χ2 tests. When sample numbers in some categorical cells were less than five, Fisher's exact test was employed. Unpaired Student's t-test or the Mann-Whitney U test was applied to analyze significant differences. One-way analysis of variance was used to compare the differences among subgroups with post hoc contrasts by Student-Newman-Keuls tests. P<0.05 was considered to indicate a statistically significant difference.
Results
RNA-Seq reveals decreased expression of SFRP2 in human pituitary corticotroph adenoma
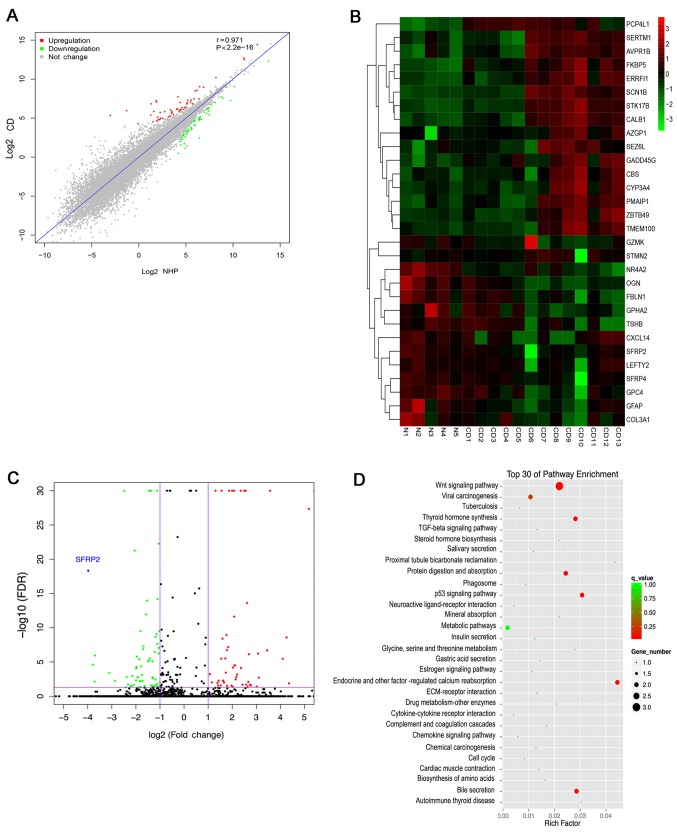
To determine the differences in mRNA expression levels between CD and NHP samples, the transcriptomes of 13 CD and 5 NHP samples were analyzed using RNA-Seq. Stringent statistical filters with P<0.05 and fold change >2 were applied. The correlation analysis revealed that the expression of genes between CD and NHP samples were highly correlated, with a correlation coefficient r of 0.97 (Fig. 1A). Based on the RNA-Seq data, 122 differentially expressed genes were identified in tumor tissues; 57 genes were upregulated and 65 genes were downregulated in CD samples compared with the NHP samples. The 30 most significant DEGs are listed in Table II. α-2-glycoprotein 1 zinc-binding (fold change 331.2874) exhibited the greatest upregulation, while SFRP2 (fold change 15.9491) exhibited the greatest downregulation. Hierarchical clustering analysis was also used to show the mRNA expression levels of all specimens (Fig. 1B). As shown in the heat map and volcano plot (Fig. 1C), SFRP2 was the most downregulated gene of all the identified DEGs. Furthermore, the top 30 pathways associated with these genes were determined using KEGG pathway enrichment analysis. This analysis indicted that the greatest number of genes were classified as associated with the Wnt signaling pathway, which was significantly enriched (Fig. 1D). These results suggest that the decreased expression of SFRP2 may result in aberrant Wnt signaling in ACTH-secreting pituitary adenomas.
Figure 1.
Transcriptome profile analysis between CD tumor and NHP samples. (A) Heat-map and hierarchical clustering analysis of differentially expressed mRNAs. Rows and columns represent each mRNA and tissue sample, respectively. The relative gene expression is depicted according to the color scale. Red and green indicate upregulation and downregulation, respectively. (B) Expression analysis of CD and NHP. Spearman's correlation coefficient r=0.97. (C) Volcano plot of DEGs. The red and green points represent the up and downregulated genes, respectively. The most downregulated gene SFRP2 is indicated. (D) Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis of the DEGs. The top 30 pathways are shown, in the Wnt signaling pathway was significantly enriched. DEG, differentially expressed gene; CD, Cushing's disease; NHP, normal human pituitary; SFRP2, secreted frizzled-related protein 2.
Table II.
Aberrantly expressed mRNAs and six random verified genes in microarray for Cushing's disease pituitaries and normal human pituitaries.
| Gene name | GenBank accession no. | Fold change (absorbance) | P-value | Regulation |
|---|---|---|---|---|
| AZGP1 | NM_001185.3 | 331.2874 | 1.38e-10 | Up |
| CYP3A4 | NM_017460.5 | 150.9702 | 2.24e-16 | Up |
| PMAIP1 | NM_021127.2 | 45.64176 | 2.08e-27 | Up |
| CALB1 | NM_004929.3 | 36.33607 | 3.56e-31 | Up |
| GZMK | NM_002104.2 | 20.56787 | 6.93e-05 | Up |
| PCP4L1 | NM_001102566.1 | 17.1511 | 8.66e-09 | Up |
| ERRFI1 | NM_018948.3 | 11.91691 | 3.42e-38 | Up |
| STK17B | XM_011512169.1 | 10.96635 | 4.21e-10 | Up |
| AVPR1B | NM_000707.3 | 8.601595 | 8.06e-06 | Up |
| SERTM1 | NM_203451.2 | 8.40439 | 1.37e-09 | Up |
| TMEM100 | NM_001099640.1 | 8.094779 | 0.0002328 | Up |
| SCN1B | NM_199037.4 | 6.870989 | 0.000160445 | Up |
| CBS | XM_017028490.1 | 6.652037 | 0.000109376 | Up |
| GADD45G | NM_006705.3 | 6.484037 | 1.70e-09 | Up |
| SEZ6L | NM_021115.4 | 6.369464 | 3.95e-05 | Up |
| ZBTB49 | NM_001330625.1 | 6.233656 | 0.00026919 | Up |
| FKBP5 | NM_001145775.2 | 6.123551 | 2.88e-17 | Up |
| STMN2 | NM_001199214.1 | 6.000352 | 0.000109376 | Up |
| SFRP2 | NM_003013.2 | 15.77754 | 4.56e-22 | Down |
| SFRP4 | NM_003014.3 | 13.79746 | 2.44e-05 | Down |
| GPHA2 | XM_011544776.2 | 13.12417 | 3.61e-05 | Down |
| GFAP | NM_002055.4 | 13.00376 | 2.65e-09 | Down |
| LEFTY2 | NM_001172425.2 | 8.61589 | 1.38e-06 | Down |
| OGN | NM_024416.4 | 7.314456 | 2.44e-05 | Down |
| TSHB | XM_011542065.2 | 5.587824 | 8.25e-140 | Down |
| NR4A2 | NM_006186.3 | 4.645393 | 0.000184 | Down |
| COL3A1 | NM_000090.3 | 4.219145 | 0.000112 | Down |
| CXCL14 | NM_004887.4 | 4.147009 | 5.25e-25 | Down |
| GPC4 | NM_001448.2 | 3.987747 | 2.14e-05 | Down |
| FBLN1 | NM_006487.2 | 3.951956 | 2.88e-07 | Down |
| FOSL2 | NM_005253.3 | 1.956347 | 3.48e-06 | Up |
| GADD45B | NM_015675.3 | 3.139243 | 4.41e-05 | Up |
| TBX19 | NM_005149.2 | 4.090872 | 2.02e-07 | Up |
| CYR61 | NM_001554.4 | 5.372227 | 0.00032136 | Down |
| JUNB | NM_002229.3 | 2.079143 | 1.33e-09 | Down |
| TFPI2 | NM_006528.3 | 1.893254 | 4.26e-07 | Down |
Validation of the expression of SFRP2 in CD
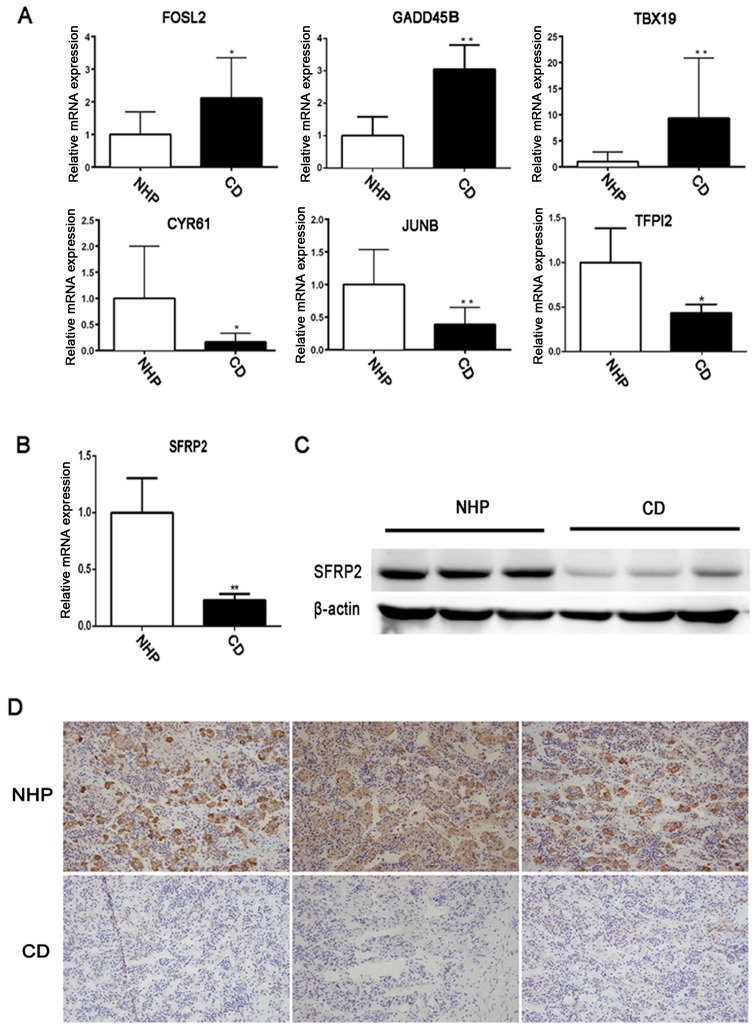
To verify the RNA-Seq data, further analysis was performed for six differentially expressed mRNAs, which were included in the 122 DEGs, in another cohort of 23 CD patients and three NHP samples by RT-qPCR. The DEGs, namely FOS like 2 AP-1 transcription factor subunit, growth arrest and DNA damage inducible β, T-box 19, cysteine rich angiogenic inducer 61, JunB proto-oncogene AP-1 transcription factor subunit and tissue factor pathway inhibitor 2, were randomly selected (Table II). The qPCR results of the six mRNAs were consistent with the RNA-Seq data, indicating the validity of the RNA-Seq data (Fig. 2A). Furthermore, the RNA-Seq results were validated with respect to SFRP2 by measuring its mRNA levels in samples procured from 23 patients diagnosed with CD and three NHPs. There was a significant decrease in SFRP2 mRNA expression in CD compared with the NHP samples (Fig. 2B). IHC and western blot analysis also revealed lower levels of SFRP2 expression in tumor tissues compared with normal controls (Fig. 2C and D). These results further confirmed the decreased expression of SFRP2 in corticotroph adenomas. The analysis of SFRP2 expression and association with clinicopathological features of patients with CD is shown in Table IV.
Figure 2.
Validation of the expression of SFRP2 in human pituitary corticotroph adenoma. (A) Expression of six randomly selected differentially expressed genes in CD tumor and NHP samples were tested by RT-qPCR. (B) RT-qPCR and (C) western blot analysis were used to detect SFRP2 expression in CD and NHP samples. (D) Immunohistochemical staining of SFRP2 in a representative NPH and CD corticotroph adenoma. *P<0.05 and **P<0.01. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; NHP, normal human pituitary; CD, Cushing's disease; FOSL2, FOS like 2 AP-1 transcription factor subunit; GADD45B, growth arrest and DNA damage inducible β; TBX19, T-box 19; CYR61, cysteine rich angiogenic inducer 61; JUNB, JunB proto-oncogene AP-1 transcription factor subunit; TFPI2, tissue factor pathway inhibitor 2; SFRP2, secreted frizzled-related protein 2.
Table IV.
SFRP2 expression and clinicopathological features in patients with Cushing's disease.
| Variable | SFRP2 expression [n (%)]
|
P-value | |
|---|---|---|---|
| High expression | Low expression | ||
| Sex | 1.000 | ||
| Male | 1 (4.35%) | 2 (8.70%) | |
| Female | 9 (39.13%) | 11 (47.82%) | |
| Age (years) | 45±10.88 | 44.69±11.58 | 0.949 |
| Tumor size | 0.104 | ||
| Microadenoma | 10 (43.48%) | 9 (39.13%) | |
| Macroadenoma | 0 (0%) | 4 (17.39%) | |
| Invasiveness | 0.339 | ||
| Yes | 1 (4.35%) | 4 (17.39%) | |
| No | 9 (39.13%) | 9 (39.13%) | |
| Midnight serum cortisol (µg/24 h) | 21.08±5.81 | 20.74±8.08 | 0.912 |
| Urinary free cortisol (µg/24 h) | 524.09±239.90 | 710.53±369.07 | 0.181 |
| Plasma ACTH (pg/ml) | 148.01±173.53 | 309.79±508.49 | 0.348 |
SFRP2, secreted frizzled-related protein 2; ATCH, adrenocorticotro-phic hormone.
Lower expression of SFRP2 in corticotroph adenoma is associated with larger size and increased invasiveness
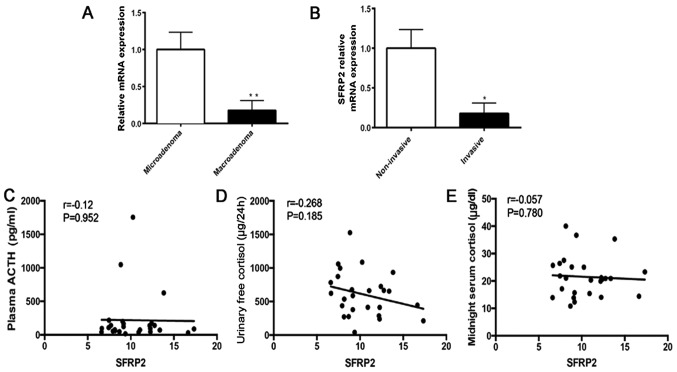
To further analyze the association between decreased SFRP2 expression and clinical phenotype, the SFRP2 expression in 23 patients with CD and 3 NHP samples was evaluated (Table III). Analysis of relative RNA expression indicated that the expression of SFRP2 was significantly decreased in macroadenoma compared with mircroadenoma of CD (Fig. 3A). Significant reduction of SFRP2 was observed in invasive adenoma compared with non-invasive adenoma of CD (Fig. 3B). These results indicated that lower expression of SFRP2 in tumors is associated with a more aggressive character of pituitary ACTH-secreting adenomas. In addition, the correlation between SFRP2 expression and hormone parameters in patients with CD was analyzed, including plasma ACTH, urinary free cortisol and midnight serum cortisol (data from the Department of Clinical Laboratory, Ruijin Hospital, Shanghai, China). There was no significant correlation between SFRP2 expression and these parameters (Fig. 3C–E). However, the results of the correlation analysis may not be indicative of the true correlation, because of the limited sample number.
Table III.
Clinical characteristics of the 26 patients with Cushing's disease.
| Case | Sex/age (years) | Midnight serum cortisol (µg/dl) | Urinary free cortisol (µg/24 h) | Plasma ACTH (pg/ml) | Tumor size | Invasiveness |
|---|---|---|---|---|---|---|
| 1 | F/51 | 13.95 | 239.02 | 94.29 | Microadenoma | No |
| 2 | F/52 | 15.39 | 414.46 | 37.575 | Negative | No |
| 3 | F/53 | 20.96 | 535.32 | 63.31 | Microadenoma | No |
| 4 | F/54 | 10.8 | 275.1 | 48.04 | Microadenoma | No |
| 5 | F/55 | 20.3 | 662.88 | 74.355 | Microadenoma | No |
| 6 | F/56 | 23.3 | 213.01 | 87.875 | Microadenoma | No |
| 7 | F/57 | 35.28 | 934.65 | 625.835 | Microadenoma | No |
| 8 | F/58 | 13.79 | 675.64 | 198.79 | Microadenoma | No |
| 9 | F/59 | 40.02 | 273.03 | 76.23 | Microadenoma | No |
| 10 | M/41 | 25.03 | 1085.83 | 1754 | Microadenoma | No |
| 11 | F/34 | 14.4 | 445.83 | 30.13 | Microadenoma | No |
| 12 | F/35 | 21.77 | 1059.03 | 217.56 | Huge adenoma | Invasive |
| 13 | F/36 | 19.85 | 287.36 | 139.055 | Microadenoma | No |
| 14 | F/37 | 12.34 | 379.49 | 122.12 | Macroadenoma | Invasive |
| 15 | F/38 | 25.7 | 623.1 | 95.09 | Microadenoma | No |
| 16 | F/39 | 15.69 | 587.5 | 166.37 | Microadenoma | No |
| 17 | F/40 | 20.91 | 654.26 | 71.19 | Microadenoma | No |
| 18 | M/24 | 25.08 | 1525.94 | 1048.19 | Macroadenoma | Invasive |
| 19 | F/31 | 21.2 | 412.62 | 46.29 | Microadenoma | No |
| 20 | F/32 | 17.09 | 996.96 | 140.595 | Huge adenoma | Invasive |
| 21 | F/33 | 26.39 | 872.34 | 107.58 | Negative | No |
| 22 | M/34 | 20.67 | 724.75 | 170.045 | Microadenoma | No |
| 23 | F/54 | 13.86 | 782.85 | 40.515 | Microadenoma | No |
| 24 | F/55 | 36.65 | 40.28 | 16.47 | Negative | No |
| 25 | F/56 | 20.93 | 666.54 | 141 | Microadenoma | Invasive |
| 26 | F/57 | 27.51 | 437.1 | 56.52 | Microadenoma | No |
M, male; F, female.
Figure 3.
Correlation analysis between SFRP2 expression and clinical phenotype in CD. Relative expression of SFRP2 in (A) microadenoma (n=19) or macroadenoma (n=4) of CD, and (B) in non-invasive (n=21) or invasive (n=5) corticotroph adenoma. Correlation of SFRP2 expression and hormone parameters in CD; (C) plasma ACTH, (D) urinary free cortisol, (E) midnight serum cortisol. *P<0.05 and **P<0.01. CD, Cushing's disease; SFRP2, secreted frizzled-related protein 2; ACTH, adrenocorticotrophic hormone.
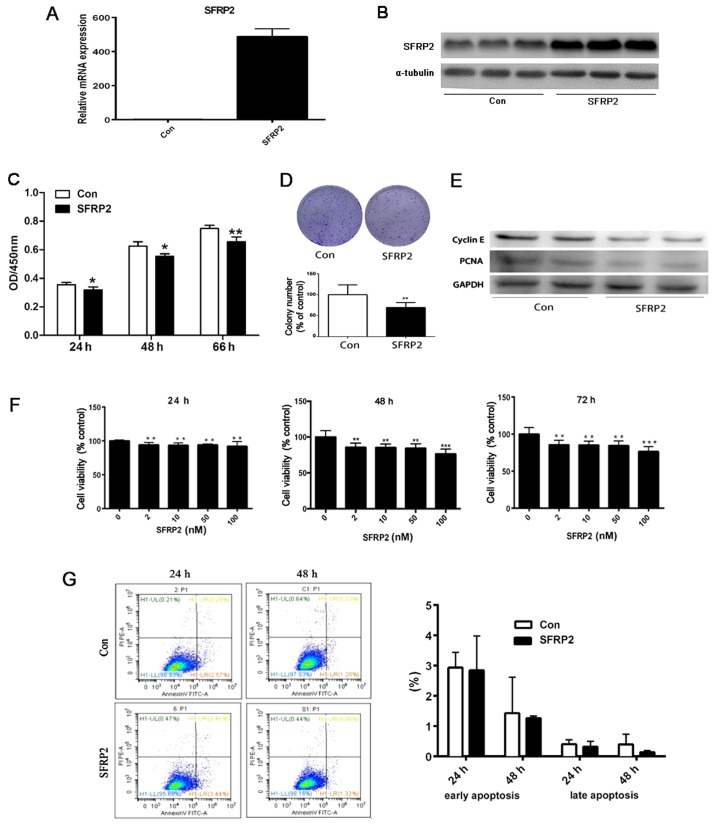
SFRP2 decreased the proliferation and clonogenic ability of AtT20 cells in vitro
Since the expression of SFRP2 is downregulated in CD and associated with larger tumor size, whether the overexpression of SFRP2 reversed the biological characteristics of murine pituitary corticotroph AtT20 cells was evaluated. A retroviral vector containing wild type SFRP2 cDNA or empty vector (EV) to generate stably transfected AtT20 cells. RT-qPCR and immunoblotting were performed to evaluate the efficiency of overexpression. The results demonstrated abundant SFRP2 mRNA and protein expression in SFRP2-transfected AtT20 cells, but not in EV transfected cells (Fig. 4A and B). Subsequently, it was determined that cell proliferation was partly inhibited in a time-dependent manner in AtT20 cells overexpressing SFRP2 with a decrease in proliferation observed from 24 to 66 h (P<0.05) (Fig. 4C). In addition, SFRP2 overexpression resulted in a reduction in the number AtT-20 cell colonies compared with the vector control (P<0.01) (Fig. 4D). This decrease in cell growth in AtT-20 cells overexpressing SFRP2 was confirmed by the reduction in the levels of the proteins, PCNA and cyclin E, which are indicative of proliferation, as determined by western blot analysis (Fig. 4E). In addition, AtT20 cells were treated with recombinant SFRP2 protein and the absorbance was detected at 450 nm. There was a decrease in cell proliferation in a dose-dependent manner from 24–72 h (Fig. 4F). However, there was no marked difference in the percentage of apoptotic cells between the SFRP2-transfected or vector-transfected AtT20 cells when analyzed by flow cytometry (Fig. 4G).
Figure 4.
SFRP2 decreases the proliferation and clonogenic ability of AtT20 cells in vitro. (A) Reverse transcription-quantitative polymerase chain reaction and (B) western blot analysis confirmed increased expression of SFRP2 in stably transfected AtT20 cells. (C) Cell proliferation was measured with the CCK-8 assay from 24 to 66 h. (D) Colony formation and the quantitative representation. (E) Expression of PCNA and cyclin E determined by western blot analysis in SFRP2-transfected AtT20 cells compared with controls. (F) Following SFRP2 treatment, AtT20 cell viability was detected from 24 to 72 h. (G) Flow cytometric analysis and the ratio of apoptosis (upper right quadrant, advanced apoptosis rate; the lower right quadrant, early apoptosis) between SFRP2-transfected and vector-transfected AtT20 cells. *P<0.05, **P<0.01 and ***P<0.005 vs. Con. Con, control; SFRP2, secreted frizzled-related protein 2; PCNA, proliferating cell nuclear antigen.
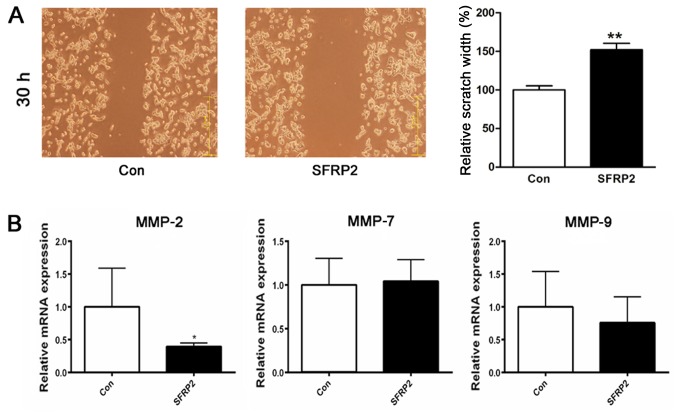
SFRP2 attenuates the migration of AtT20 cells in vitro
Previously, analysis of the clinical phenotype indicated that the decreased expression of SFRP2 in corticotroph adenoma was associated with higher invasiveness. Thus, it was sought to determine whether SFRP2 expression also affected the cellular migration of AtT20 cells. Migration was assayed in SFRP2-transfected and vector-transfected AtT20 cells using a wound-healing assay; the results revealed an evident delay in the wound closure rate of the SFRP2-transfected cells at 30 h compared with vector-transfected AtT20 cells (Fig. 5A). For further validation, the expression of the matrix metallo-proteinases MMP-2, MMP-7 and MMP-9, which are known to be associated with tumor invasion, were analyzed RT-qPCR revealed that SFRP2 overexpression attenuated the expression of MMP-2 in AtT20 cells (Fig. 5B).
Figure 5.
SFRP2 inhibits AtT20 cell migration. (A) Representative images of the wound healing assay in the SFRP2-transfected or vector-transfected AtT20 cells at 30 h. (B) Relative expression of MMP-2, MMP-7 and MMP-9 in two groups. *P<0.05 and **P<0.01. Con, control; SFRP2, secreted frizzled-related protein 2; MMP, matrix metallopeptidase.
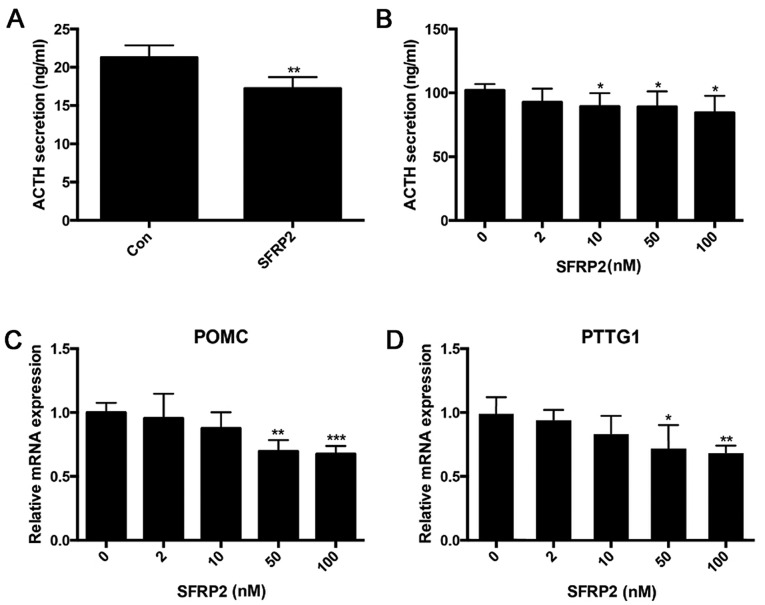
Overexpression of SFRP2 inhibits ACTH secretion in AtT20 cells
Given the obvious decrease in SFRP2 expression in pituitary corticotroph tumors, it was determined whether the level of SFRP2 affected the secretion of ATCH. ACTH secretion was measured in SFRP2-transfected and vector-transfected AtT20 cells, the results demonstrated that overexpression of SFRP2 suppressed the secretion of ACTH (Fig. 6A). Treating AtT20 cells with a different dose of recombinant SFRP2 protein demonstrated that SFRP2 inhibited the secretion of ACTH in a dose-dependent manner (Fig. 6B). Additionally, SFRP2 suppressed the transcription of POMC and PTTG (Fig. 6C and D).
Figure 6.
Overexpression of SFRP2 attenuates ACTH secretion in vitro. (A) Measurement of ACTH secretion in SFRP2-transfected cells and controls. (B) Variation in ACTH secretion following treatment with SFRP2 at different concentrations. Reverse transcription-quantitative polymerase chain reaction confirmed decreased levels of (C) POMC and (D) PTTG1 mRNA in stable SFRP2 transfected cells. *P<0.05, **P<0.01 and ***P<0.005 vs. Con or 0 nM. ATCH, adrenocorticotrophic hormone; Con, control; SFRP2, secreted frizzled-related protein 2; POMC, pro-opiomelanocortin-α; Pttg1, pituitary tumor-transforming gene 1.
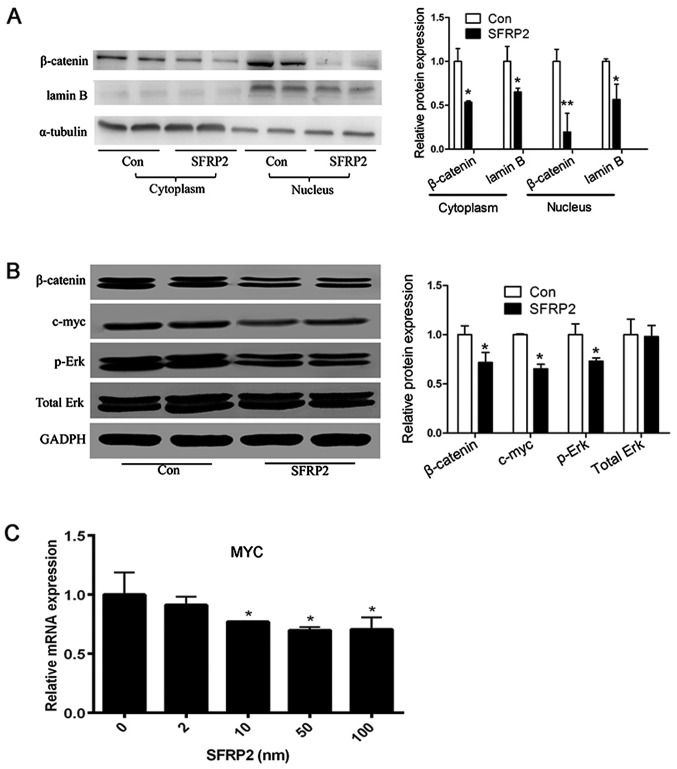
SFRP2 leads to decreased Wnt signaling activity in AtT20 cells
Given that SFRP2 is an antagonist of the Wnt signaling pathway, whether SFRP2 influences the activity of Wnt signaling was investigated in vitro. In SFRP2-transfected AtT20 cells, over-expression of SFRP2 reduced the amount of β-catenin in the cytoplasm and nucleus (Fig. 7A). Furthermore, the expression level of c-myc was detected, which is a downstream target of Wnt signaling. The level of c-myc was markedly decreased in SFRP2-transfected AtT20 cells, and the level of Erk phosphorylation was also decreased compared with the vector-transfected AtT20 cells (Fig. 7B). In addition, the expression level of c-myc was decreased in AtT20 cells treated with recombinant SFRP2 (Fig. 7C). These data demonstrated that SFRP2 could attenuated the activity of Wnt signaling in corticotroph adenoma AtT20 cells.
Figure 7.
SFRP2 leads to decreased activity of Wnt signaling in AtT20 cells. (A) Overexpression of SFRP2 reduced the levels of β-catenin both in the cytoplasm and nucleus. (B) SFRP2 overexpression reduced the expression of c-myc and β-catenin, and also decreased the phosphorylation but not effect on total of Erk in AtT20 cells. (C) SFRP2 treatment decreased the relative mRNA level of c-myc. *P<0.05 and **P<0.01 vs. Con or 0 nM. Con, control; SFRP2, secreted frizzled-related protein 2; p-, phospho-; Erk, extracellular signal-regulated kinase.
Discussion
As a typical type of next-generation sequencing technology, high-throughput RNA-Seq has been widely used in the recent studies of oncology due to its high sensitivity and efficiency. Numerous valuable studies have reported on the underlying molecular etiology or therapeutic targets in diverse malignancies, including gallbladder carcinoma (15), prostate cancer (16) and liver cancer (17). This is the first report, however, of the use of RNA-Seq in the study of CD.
In the present study, RNA-Seq was used to sequence the transcriptome of human pituitary corticotroph adenoma, which detected decreased expression of SFRP2. In addition, IHC and western blot analysis of CD and NHP samples also confirmed the decreased expression of SFRP2 in tumor tissues. Clinically, lower expression of SFRP2 was associated with larger tumor sizes and increased invasiveness. Thus, SFRP2 may be a useful marker for predicting characteristics of pituitary ACTH adenoma. In fact, researchers have already reported that SFRP2 expression is associated with the degree of tumor malignancy and invasive ability in various human cancers (12,18). However, this is the first description of the aberrant expression of SFRP2 in the tumorigenesis of CD.
The effects of SFRP2 overexpression on tumor cell biology were analyzed in vitro. The data suggested that overexpression of SFRP2 decreased the proliferation of AtT20 cells, and reduce clonal ability and migration. In addition, increased SFRP2 attenuated the secretion of ACTH in AtT20 cells. These data are consistent with the clinical analysis of CD.
KEGG pathway enrichment analysis revealed that genes classified in the Wnt signaling pathway were significantly enriched in the CD samples. Indeed, several lines of evidence suggested that SFRP2 has a pivotal role in the Wnt pathway, where it primarily functions as an antagonist of Wnt signaling (19,20). The Wnt signaling pathway has been reported to be involved in the tumorigenesis of several cell types (21–25). It has been reported that decreased expression of SFRP2 in aggressive nonfunctioning pituitary adenoma results in nuclear accumulation of β-catenin, and inhibitors of the Wnt pathway are strongly downregulated in pituitary tumors (12,26). To investigate whether SFRP2 influences Wnt signaling in CD, it was confirmed that SFRP2 treatment reduced β-catenin and c-myc expression. Similarly, AtT20 cells treated with SFRP2 exhibited reduced expression of c-myc. These data demonstrate that SFRP2 can antagonize Wnt signaling in AtT20 cells.
The major limitation of the current study is that all the effects of SFRP2 were only verified in AtT20 cells. The reason the AtT20 cell line was selected for the study is that it is a very typical and representative cell line in CD research (27). Currently, experiments are being performed involving SFRP2 expression, its functional verification and associated mechanism in other cell lines and mouse models.
In conclusion, the findings of the present study demonstrated that SFRP2 was downregulated in pituitary corticotroph adenomas. In addition, lower expression of SFRP2 resulted in a dysregulation of Wnt signaling. Thus, SFRP2 represents a promising biomarker and therapeutic target for pituitary ACTH adenoma. A pituitary tissue-specific SFRP2 knockdown mouse model will be established in future research. Using transgenic mice and primary cells, the phenotypes will be determined following SFRP2 overexpression and knockdown, and the underlying mechanism of SFRP2 in CD will be investigated.
Acknowledgments
Not applicable.
Funding
This study was supported by grants from the National Natural Science Foundation of China (nos. 81770779 and 81700692).
Availability of data and materials
All data generated or analyzed during this study are available from the corresponding author on reasonable request.
Authors' contributions
JR, FJ, HJ collected the data and wrote the manuscript. YS and SP analyzed the correlation between SFRP2 expression and clinical characteristics. CG and XC were involved in data analysis and interpretation. As endocrinologists, WW and GN made substantial contribution to the diagnosis of Cushing's disease, provided technical assistance and revised the manuscript. LB and QS were involved in the conception and design of the study, and QS was the leader of this study. The final version of the manuscript was read and approved by all authors.
Ethics approval and consent to participate
Informed consent was obtained from all participating patients. For the use of human samples, the protocol for this study was approved by the Ruijin Hospital Ethics Committee (Shanghai, China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Nieman LK. Update in the medical therapy of Cushing's disease. Curr Opin Endocrinol Diabetes Obes. 2013;20:330–334. doi: 10.1097/MED.0b013e3283631809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prague JK, May S, Whitelaw BC. Cushing's syndrome. BMJ. 2013;346:f945. doi: 10.1136/bmj.f945. [DOI] [PubMed] [Google Scholar]
- 3.Gadelha MR, Vieira Neto L. Efficacy of medical treatment in Cushing's disease: A systematic review. Clin Endocrinol (Oxf) 2014;80:1–12. doi: 10.1111/cen.12345. [DOI] [PubMed] [Google Scholar]
- 4.Tritos NA, Biller BM, Swearingen B. Management of Cushing disease. Nat Rev Endocrinol. 2011;7:279–289. doi: 10.1038/nrendo.2011.12. [DOI] [PubMed] [Google Scholar]
- 5.Wagenmakers MA, Netea-Maier RT, van Lindert EJ, Timmers HJ, Grotenhuis JA, Hermus AR. Repeated transsphenoidal pituitary surgery (TS) via the endoscopic technique: A good therapeutic option for recurrent or persistent Cushing's disease (CD) Clin Endocrinol (Oxf) 2009;70:274–280. doi: 10.1111/j.1365-2265.2008.03334.x. [DOI] [PubMed] [Google Scholar]
- 6.Saini S, Majid S, Dahiya R. The complex roles of Wnt antagonists in RCC. Nat Rev Urol. 2011;8:690–699. doi: 10.1038/nrurol.2011.146. [DOI] [PubMed] [Google Scholar]
- 7.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, Van Engeland M, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 9.Saito T, Mitomi H, Imamhasan A, Hayashi T, Mitani K, Takahashi M, Kajiyama Y, Yao T. Downregulation of sFRP-2 by epigenetic silencing activates the β-catenin/Wnt signaling pathway in esophageal basaloid squamous cell carcinoma. Virchows Arch. 2014;464:135–143. doi: 10.1007/s00428-014-1538-1. [DOI] [PubMed] [Google Scholar]
- 10.Kongkham PN, Northcott PA, Croul SE, Smith CA, Taylor MD, Rutka JT. The SFRP family of WNT inhibitors function as novel tumor suppressor genes epigenetically silenced in medulloblastoma. Oncogene. 2010;29:3017–3024. doi: 10.1038/onc.2010.32. [DOI] [PubMed] [Google Scholar]
- 11.Takagi H, Sasaki S, Suzuki H, Toyota M, Maruyama R, Nojima M, Yamamoto H, Omata M, Tokino T, Imai K, et al. Frequent epigenetic inactivation of SFRP genes in hepatocellular carcinoma. J Gastroenterol. 2008;43:378–389. doi: 10.1007/s00535-008-2170-0. [DOI] [PubMed] [Google Scholar]
- 12.Wu Y, Bai J, Hong L, Liu C, Yu S, Yu G, Zhang Y. Low expression of secreted frizzled-related protein 2 and nuclear accumulation of β-catenin in aggressive nonfunctioning pituitary adenoma. Oncol Lett. 2016;12:199–206. doi: 10.3892/ol.2016.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Jian FF, Li YF, Chen YF, Jiang H, Chen X, Zheng LL, Zhao Y, Wang WQ, Ning G, Bian LG, et al. Inhibition of ubiquitin-specific peptidase 8 suppresses adrenocorticotropic hormone production and tumorous corticotroph cell growth in AtT20 cells. Chin Med J (Engl) 2016;129:2102–2108. doi: 10.4103/0366-6999.189047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu X, Li B, Jiang M, Fang M, Ji J, Wang A, Wang M, Jiang X, Gao C. RNA sequencing reveals differentially expressed genes as potential diagnostic and prognostic indicators of gallbladder carcinoma. Oncotarget. 2015;6:20661–20671. doi: 10.18632/oncotarget.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pflueger D, Terry S, Sboner A, Habegger L, Esgueva R, Lin PC, Svensson MA, Kitabayashi N, Moss BJ, MacDonald TY, et al. Discovery of non-ETS gene fusions in human prostate cancer using next-generation RNA sequencing. Genome Res. 2011;21:56–67. doi: 10.1101/gr.110684.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho DW, Yang ZF, Yi K, Lam CT, Ng MN, Yu WC, Lau J, Wan T, Wang X, Yan Z, et al. Gene expression profiling of liver cancer stem cells by RNA-sequencing. PLoS One. 2012;7:e37159. doi: 10.1371/journal.pone.0037159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marsit CJ, Karagas MR, Andrew A, Liu M, Danaee H, Schned AR, Nelson HH, Kelsey KT. Epigenetic inactivation of SFRP genes and TP53 alteration act jointly as markers of invasive bladder cancer. Cancer Res. 2005;65:7081–7085. doi: 10.1158/0008-5472.CAN-05-0267. [DOI] [PubMed] [Google Scholar]
- 19.Chung MT, Lai HC, Sytwu HK, Yan MD, Shih YL, Chang CC, Yu MH, Liu HS, Chu DW, Lin YW. SFRP1 and SFRP2 suppress the transformation and invasion abilities of cervical cancer cells through Wnt signal pathway. Gynecol Oncol. 2009;112:646–653. doi: 10.1016/j.ygyno.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Chen H. Genistein attenuates WNT signaling by up-regulating sFRP2 in a human colon cancer cell line. Exp Biol Med (Maywood) 2011;236:714–722. doi: 10.1258/ebm.2011.010347. [DOI] [PubMed] [Google Scholar]
- 21.Sano M, Driscoll DR, De Jesus-Monge WE, Klimstra DS, Lewis BC. Activated wnt signaling in stroma contributes to development of pancreatic mucinous cystic neoplasms. Gastroenterology. 2014;146:257–267. doi: 10.1053/j.gastro.2013.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, IV, Morris JP, IV, Yan W, Schofield HK, Gurney A, Simeone DM, Millar SE, Hoey T, Hebrok M, Pasca di Magliano M. Canonical wnt signaling is required for pancreatic carcinogenesis. Cancer Res. 2013;73:4909–4922. doi: 10.1158/0008-5472.CAN-12-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chambers TJ, Giles A, Brabant G, Davis JR. Wnt signalling in pituitary development and tumorigenesis. Endocr Relat Cancer. 2013;20:R101–R111. doi: 10.1530/ERC-13-0005. [DOI] [PubMed] [Google Scholar]
- 24.Li Q, Shen K, Zhao Y, He X, Ma C, Wang L, Wang B, Liu J, Ma J. MicroRNA-222 promotes tumorigenesis via targeting DKK2 and activating the Wnt/β-catenin signaling pathway. FEBS Lett. 2013;587:1742–1748. doi: 10.1016/j.febslet.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 25.Nojima M, Suzuki H, Toyota M, Watanabe Y, Maruyama R, Sasaki S, Sasaki Y, Mita H, Nishikawa N, Yamaguchi K, et al. Frequent epigenetic inactivation of SFRP genes and constitutive activation of Wnt signaling in gastric cancer. Oncogene. 2007;26:4699–4713. doi: 10.1038/sj.onc.1210259. [DOI] [PubMed] [Google Scholar]
- 26.Elston MS, Gill AJ, Conaglen JV, Clarkson A, Shaw JM, Law AJ, Cook RJ, Little NS, Clifton-Bligh RJ, Robinson BG, et al. Wnt pathway inhibitors are strongly down-regulated in pituitary tumors. Endocrinology. 2008;149:1235–1242. doi: 10.1210/en.2007-0542. [DOI] [PubMed] [Google Scholar]
- 27.Liu N-A, Araki T, Cuevas-Ramos D, Hong J, Ben-Shlomo A, Tone Y, Tone M, Melmed S. Cyclin E-mediated human proopiomelanocortin regulation as a therapeutic target for cushing disease. J Clin Endocrinol Metab. 2015;100:2557–2564. doi: 10.1210/jc.2015-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are available from the corresponding author on reasonable request.