Abstract
Kinesin family member C1 (KIFC1, also known as HSET) is a minus end-directed motor protein, which is critical in centrosome clustering. The present study investigated the expression of KIFC1 in paired hepatocellular carcinoma (HCC) tissues and adjacent non-cancerous tissues from 91 patients by immunohistochemical analysis; clinical data were concomitantly collected. KIFC1 was expressed at high levels in HCC tissues, compared with that in peritumoral tissues (54.9 vs. 14.3%; P<0.01), and its expression correlated with tumor emboli, metastasis, recurrence and time of recurrence. Kaplan-Meier analysis showed that the expression of KIFC1 was significantly associated with tumor-free survival rates. In addition, multivariate analyses revealed that the overexpression of KIFC1was an independent predictive marker in patients with HCC. Consistently, data derived from GEPIA was in agreement with the results. In vitro, KIFC1 knockdown effectively decreased HCC cell viability, and induced apoptosis and cell death. KIFC1 knockdown also significantly suppressed tumor cell migration and invasion in vitro. Mechanistically, the apoptosis-related protein, B-cell lymphoma-2 (Bcl-2), was downregulated in KIFC1 small interfering RNA-treated groups, whereas thee levels of Bcl-2-associated X protein and p53 were upregulated. In addition, the expression levels of phosphorylated phosphoinositide 3-kinase and phosphorylated AKT were decreased significantly when KIFC1 was silenced. The epithelial-mesenchymal transition-related proteins, N-cadherin, matrix metalloproteinase-2 (MMP-2), β-catenin, Slug, and Zinc finger E-box-binding homeobox 1, were downregulated, whereas the expression of E-cadherin was upregulated. The overexpression of KIFC1 was correlated closely with the progression of HCC and poor prognosis, and suggested that the expression levels of KIFC1 are a potential prognostic biomarker and therapeutic target in HCC.
Keywords: kinesin family member C1, hepatocellular carcinoma, apoptosis, cell cycle, epithelial-mesenchymal transition
Introduction
Hepatocellular carcinoma (HCC) is a life-threatening malignancy that occurs worldwide; 250,000–1,000,000 new cases are diagnosed each year worldwide and the life expectancy from the time of diagnosis is ~6 months (1,2). As there are different major causative and risk factors, the incidence of HCC varies by geographical area. HCC is particularly common in Asia due to the high incidence of hepatitis B virus (HBV) and hepatitis C virus infection, and the intake of dietary aflatoxin. In China, HBV infection is the most common cause of HCC (3–5). Hepatectomy and liver transplantation are potentially curative approaches in HCC (6). However, as HCC tends to be diagnosed at the later stage, only a small proportion of patients are eligible for these treatments (6,7). Therefore, it is necessary to investigate the genes responsible for the development and progression of HCC, establish an improved prognostic model, and identify prognostic biomarkers with higher sensitivity, accuracy and specificity.
In normal somatic cells, centrosomes complete duplication precisely during cell division (8–10). Increased centrosome number or centrosome amplification causes chromosomal instability, aneuploidy and tumorigenesis, which affect the cell cycle and apoptosis, and are associated with tumor differentiation, metastasis and recurrence. However, cancer cells can evade these conditions and maintain survival by clustering the extra centrosomes (8–10). In previous studies, centrosome amplification has been recognized as a hallmark of cancer (9,11,12).
A C-type kinesin of the kinesin-14 family (13), kinesin family member C1 (KIFC1, also known as HSET), is a minus end-directed motor protein (14) with a critical role in centrosome clustering in cancer cells (15). KIFC1 is reported to exert its function in vesicular and organelle trafficking (16), oocyte development (17), spermatogenesis (18,19) and double-stranded DNA transportation (20). Previously, investigations have focused on KIFC1, as it is essential for the bipolar mitotic division of cancer cells but appears redundant in normal cells (15,21). KIFC1 is upregulated in ovarian cancer, breast cancer and non-small cell lung cancer, and it facilitates cancer initiation and progression (22–24). In addition, the overexpression of KIFC1 suppresses docetaxel-mediated apoptosis in breast cancer cells (25). However, the expression of KIFC1 in HCC has not been investigated. In addition, the effects of KIFC1 on apoptosis, the cell cycle and epithelial-mesenchymal transition (EMT) in HCC remain to be fully elucidated. Accordingly, the present study examined the expression and distribution of KIFC1 in HCC via immunohistochemistry, and assessed the expression of KIFC1 in HCC specimens, in terms of its association with HCC clinicopathological characteristics and tumor-free survival rates of patients. Furthermore, the effects of KIFC1 knockdown on the regulation of HCC cell malignant behaviors were examined in vitro.
Materials and methods
Patients and tissue samples
A total of 91 paired tumor tissues and corresponding normal liver tissues were obtained from patients pathologically diagnosed with HCC, who underwent surgical resection between 2009 and 2013 at the Second Affiliated Hospital of Nanchang University (Nanchang, China). These patients had not received chemotherapy or radiotherapy prior to surgery. The samples were cut into 4–5 mm3 sections, quick-frozen in liquid nitrogen and stored at −80°C. The clinicopathological data of the patients, including sex, age, tumor size and tumor stage, were collected concomitantly. Histologically, tumor stage was classified separately by two experienced pathologists according to the American Joint Committee on Cancer (26). The present study was approved by the Ethics Committee of the Second Affiliated Hospital of Nanchang University, written consent was obtained from each participant, and the study was performed in accordance with the ethical standards of the Declaration of Helsinki.
Immunohistochemistry
A two-step immunohistochemical method was used to perform the immunohistochemistry. Specifically, the 4-mm-thick paraffinized sections of the paired tumor and peritumoral tissues were dewaxed in a 65°C water bath for 90 min, deparaffinized twice in xylene for 15 min per deparaffinization, rehydrated in gradient alcohol for 5 min per concentration, and placed in a pressure cooker at 100°C for 15 min for antigen repair. The sections were incubated with 3% H2O2 at room temperature for 15 min to block endogenous peroxidase activity, washed with phosphate-buffered saline (PBS) three times (5 min per wash), and blocked with goat serum (Boster, Wuhan, China) for 30 min. Subsequently, the sections were incubated with rabbit anti-KIFC1 antibody (1:200; ABclonal, Wuhan, China) at 4°C overnight. The following day, the sections were incubated with the secondary antibody (PV-6000, 1:200; ZSGB-Bio, Beijing, China) for 30 min at 37°C. Following washing with PBS three times, diaminobenzidine and hematoxylin were used for the color reaction. Pathologists evaluated the immunostained tissue sections independently in a blinded manner using a microscope (Olympus, Tokyo, Japan). KIFC1 protein was localized in the nuclei. Staining was scored as 0 (no staining), 1 (weak staining), 2 (moderate staining) or 3 (strong staining) according to the staining intensity. Based on the percentage of staining, the extent was scored as 1 (<10% staining), 2 (10–40% staining), or 3 (>40% staining). The overall staining scores were then computed by multiplying the scores for staining intensity and extent of staining. The final score ranged between 0 and 9, with scores of 0–1 indicating negative expression of KIFC1 and scores of 2–9 indicating positive expression of KIFC1.
Data mining
The data of expression of KIFC1 for liver HCC was obtained from the Gene Expression Profiling Interactive Analysis (GEPIA) online database (http://gepia.cancer-pku.cn/), a web server providing customizable functions (27). Tumors and normal samples in the GEPIA database were derived from The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) projects. The correlations of disease-free survival and overall survival rates with the expression of KIFC1 in HCC were also computed by the GEPIA database.
Cell lines and culture
The MHCC-97H, SMMC-7721 and HCC-LM3 human HCC cell lines, and the HL-7702 human liver cell line (also named LO2) were obtained from the Shanghai Institute for Biological Sciences, Chinese Academy of Sciences (Shanghai, China). The HL-7702 cells were used as the control. The cells were cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM; Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA) and maintained in a humidified incubator with 5% carbon dioxide at 37°C. Exponential growth-phase cells were used for the in vitro assays. The cells were used for in vitro assays or were subcultured to ~80% confluence when in the logarithmic growth phase.
Small interfering RNA (siRNA) depletion of KIFC1
Two human KIFC1 siRNA reagents were purchased from GenePharma (Shanghai, China); the sequences of siRNA1 (KIFC1-s1) and siRNA2 (KIFC1-s2) were 5′-UAACUGACCCUUUAAGUCCUU-3′ and 5′-UGGUCCAACGUUUGAGUCCUU-3′, respectively. A negative siRNA was used as the control. The HCC-LM3 and SMMC-7721 cells were transfected using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol for 48 h in 6-well plates. The transfected cells were used for protein extraction or other subsequent assays.
Protein extraction and western blot analysis
The cells in the 6-well plates were lysed using radioimmunoprecipitation assay buffer containing 1% phenylmethanesulfonyl fluoride to extract the total protein, and quantified using the Bradford method as previously described (28). Protein concentrations were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred onto polyvinylidene fluoride membranes. The membranes were blocked using 5% non-fat milk for 2 h at room temperature, and subsequently incubated overnight with anti-KIFC1 (ABclonal) or other primary antibodies at 4°C. The antibodies against BAX (#2772), Bcl-2 (#4223), p53 (#2527), E-cadherin (#3195), N-cadherin (#13116), β-catenin (#8480), MMP-2 (#13132), Slug (#9585), ZEB-1 (#3396), p-PI3K (#4228), PI3K (#4249), AKT (#4691) and p-AKT (#4060), were acquired from Cell Signaling Technology (Danvers, MA, USA) and the anti-GAPDH (ab8245) was acquired from Abcam (Cambridge, MA, USA). All these antibodies were used according to the manufacturers' instructions. Following three washes with Tris-buffered saline containing Tween-20 at 10 min per wash, the membranes were incubated at room temperature for 1 h with horseradish peroxidase (HRP)-conjugated secondary antibody (1:10,000; Abcam). The membranes were then washed three times, as previously, and the target protein immunoreactivity was detected using an enhanced chemiluminescence system (Clinx, Shanghai, China). As an endogenous protein, glyceraldehyde-3-phosphate dehydrogenase was used for normalization. The results of western blot was quantified by densitometric analysis using ImageJ software.
Cell viability assay
Cell viability was assayed using Cell Counting Kit-8 (CCK-8; Nanjing KeyGEN Biotech Co., Ltd., Nanjing, China). A total of 5,000 cells were seeded in each 96-well plate for 24 h following KIFC1 siRNA-transfection, and incubated for another 24, 48, 72 and 96 h. At the end of each incubation period, 10 µl CCK-8 reagent was added to each well and incubated for 1 h prior to measurement of the optical density of each well at 450 nm using a microplate reader.
Analysis of apoptosis
Apoptosis was detected using fluorescence-activated cell sorting (FACS) with an Annexin V Cell Apoptosis Analysis kit (Sungene Biotech, Tianjing, China). The cells (~1×105) were harvested for each assay, washed with cold PBS, and suspended twice with 1X binding buffer. Annexin V-fluorescein isothiocyanate was added (5 µl) and the cells were incubated for 10 min in the dark at room temperature. Subsequently, 5 µl propidium iodide (PI) solution was added and the samples were incubated for 5 min at room temperature in the dark. Finally, the samples were analyzed by flow cytometry (BD Biosciences, San Jose, CA, USA) within 1 h.
Cell cycle analysis
Cell cycle distribution was analyzed via FACS. The cells were harvested and the cell numbers adjusted to 1×106. The cells were then fixed overnight in cold 70% ethanol at 2–8°C, washed twice with cold PBS, resuspended with 0.5 ml PI/RNase (Sungene Biotech) staining solution, and incubated for 30 min at 37°C in dark prior to analysis by flow cytometry.
Transwell assay
The migration and invasive capabilities of the HCC-LM3 and SMMC-7721 cells were detected using Transwell assays. In the invasion assay, 60 µl diluted Matrigel (BD Biosciences) was added to Transwell chambers for 24 h at 37°C. The cells were resuspended in serum-free DMEM, the cell number was adjusted to 1×104, and the cells were added to the Transwell chambers. The chambers were placed in a 24-well plate and 600 µl DMEM containing 20% FBS was added to the lower surface of the chambers. Following incubation for 24 h, the non-invaded cells and Matrigel were wiped away carefully, and the invaded cells were fixed in 4% paraformaldehyde for 30 min, and stained with 0.1% crystal violet for 30 min. The number of invaded cells was calculated under a microscope (Olympus). The same steps were used in the cell migration assay; however, Matrigel was not used.
Statistical analysis
SPSS software (version 17.0) was used for all statistical analyses. The Mann-Whitney U test was used to compare the expression of KIFC1 between HCC tissues and paired normal tissues; the expression of KIFC1 and clinicopathological data were examined using the χ2 test. Tumor-free survival rates were evaluated using Kaplan-Meier curves, and the Cox proportional hazards regression model was used to evaluate the hazard ratio (HR) and to confirm independent prognostic predictor factors. The in vitro data are expressed as the means ± standard error and performed using one-way analysis of variance. Multiple comparison between the groups was performed using a Student-Newman-Keuls (S-N-K) test. P-values were based on a two-sided statistical analysis. All assays were performed independently three times. P≤0.05 was considered to indicate a statistically significant difference.
Results
KIFC1 is overexpressed in HCC clinical samples
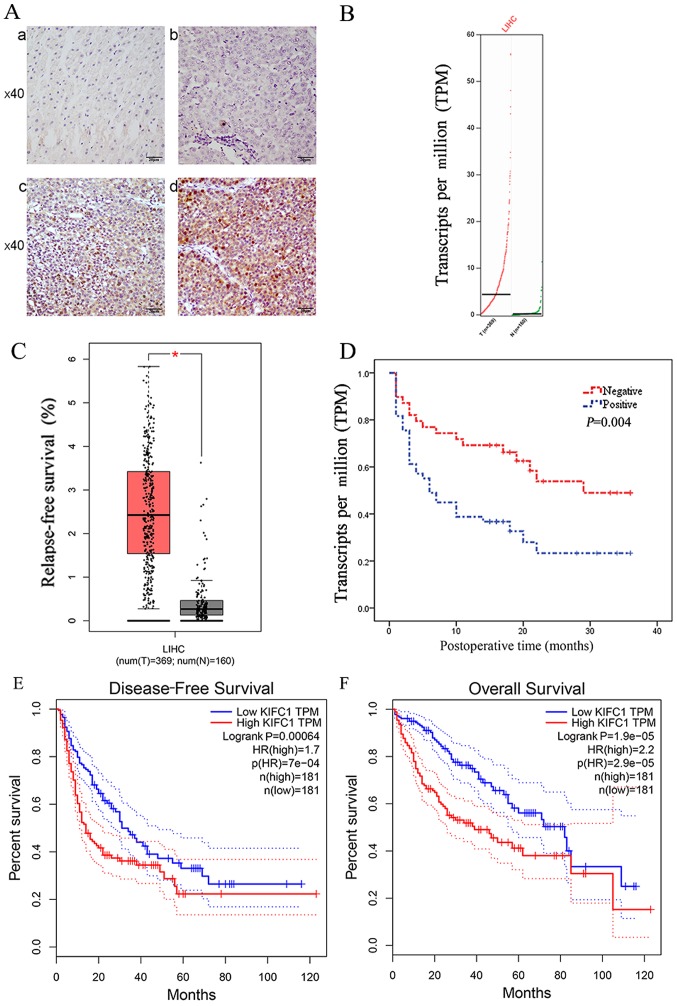
Using immunohistochemistry, the present study evaluated the protein expression of KIFC1 in 91 paired HCC clinical samples and corresponding normal tissues for the first time, to the best of our knowledge. 78/91 normal tissue samples (Fig. 1A-a) and 41/91 HCC tissue samples (Fig. 1A-b) had negligible expression of KIFC1. There was positive KIFC1 expression in 50/91 samples (Fig. 1A-c and d), and KIFC1 was mainly localized to the nuclei in the HCC samples. Therefore, KIFC1 was expressed at high levels in HCC tissues, compared with the peritumoral tissues (54.9 vs. 14.3%, P<0.01). Furthermore, it was found that the mRNA level of KIFC1 was significantly upregulated in HCC clinical samples, compared with that in the normal tissues via the online GEPIA database, the samples of which were from TCGA and the GTEx projects. (P<0.05) (Fig. 1B and C).
Figure 1.
Expression of KIFC1. (A) Immunohistochemical staining of KIFC1 in HCC and adjacent normal tissues: (a) Negative KIFC1 staining in paired normal tissues; (b) Negative KIFC1 staining in tumor tissues; (c) Weak positive KIFC1 staining in tumor tissues; (d) Strong positive KIFC1 staining in tumor tissues. (B) From the GEPIA database, KIFC1 gene expression profile was shown. (C) From the GEPIA database, KIFC1 gene expression was significantly upregulated in HCC (n=369) compared with corresponding normal tissues (n=160). *P<0.05. (D) Kaplan-Meier curves stratified by the expression level of KIFC1 in HCC showed a significantly correlation between the expression level of KIFC1 and tumor-free survival rates. Kaplan-Meier curves of KIFC1 for (E) disease-free and (F) overall survival were computed by GEPIA. HCC, hepatocellular carcinoma; KIFC1, kinesin family member C1; GEPIA, Gene Expression Profiling Interactive Analysis.
Overexpression of KIFC1 is associated with HCC clinicopathological characteristics and tumor-free survival rates
The association between the protein expression of KIFC1 and clinicopathological data in 91 HCC clinical samples is shown in Table I. Specifically, a high level of KIFC1 was significantly associated with tumor emboli (P=0.033), metastasis (P=0.001), recurrence (P=0.015) and time of recurrence (P=0.015). However, the expression of KIFC1 was not associated with sex, age, HBV, cirrhosis, α-fetoprotein (AFP), tumor size, tumor amount, differentiation, tumor capsule, tumor-node-metastasis (TNM) stage or Barcelona Clinic Liver Cancer (BCLC) stage. The Kaplan-Meier analysis showed that the expression of KIFC1 was significantly associated with tumor-free survival rate (P=0.004) (Fig. 1D). Furthermore, univariate analysis showed a significant association between tumor-free survival rate and tumor differentiation [HR=0.453; 95% confidence interval (95% CI)=0.210–0.974; P=0.043), tumor emboli (HR=0.365; 95% CI=0.188–0.709; P=0.003), TNM stage (HR=0.375; 95% CI=0.207–1.680; P=0.001) and expression of KIFC1 (HR=0.438; 95% CI=0.242–0.792; P=0.006). However, there was no statistical significance between tumor-free survival rate and sex, age, HBV, cirrhosis, AFP, tumor size, amount, tumor capsule or BCLC stage (Table II). The multivariate analysis also showed that the expression of KIFC1 (HR=0.518; 95% CI=0.281–0.953; P=0.035) was an independent predictive marker in patients with HCC (Table II). To confirm the above results, it was also found that the mRNA expression of KIFC1 was significantly associated with disease-free survival rate (P=0.00064) (Fig. 1E) and overall survival rate (P=1.9e-0) (Fig. 1F) by GEPIA.
Table I.
Association between protein expression of KIFC1 and clinicopathological parameters in patients with hepatocellular carcinoma.
| Characteristic | Patients (n) | KIFC1 status
|
χ2 | P-value | |
|---|---|---|---|---|---|
| Negative | Positive | ||||
| Sex | |||||
| Male | 76 | 35 | 41 | 0.185 | 0.667 |
| Female | 15 | 6 | 9 | ||
| Age (years) | |||||
| <60 | 78 | 34 | 44 | 0.473 | 0.491 |
| ≥60 | 13 | 7 | 6 | ||
| Hepatitis B status | |||||
| Negative | 13 | 8 | 5 | 1.665 | 0.197 |
| Positive | 78 | 33 | 45 | ||
| Cirrhosis | |||||
| No | 33 | 14 | 19 | 0.145 | 0.704 |
| Yes | 58 | 33 | 45 | ||
| AFP (µg/l) | |||||
| <400 | 60 | 29 | 31 | 0.765 | 0.382 |
| ≥400 | 31 | 12 | 19 | ||
| Tumor size (cm) | |||||
| ≤3 | 16 | 5 | 11 | 1.495 | 0.222 |
| >3 | 75 | 36 | 39 | ||
| Tumor number | |||||
| 1 | 75 | 34 | 41 | 0.013 | 0.908 |
| >1 | 16 | 7 | 9 | ||
| Tumor capsule | |||||
| No | 49 | 22 | 27 | 0.001 | 0.974 |
| Yes | 42 | 19 | 23 | ||
| Differentiation | |||||
| High | 21 | 13 | 8 | 3.131 | 0.077 |
| Low to medium | 70 | 28 | 42 | ||
| Tumor emboli | |||||
| No | 76 | 38 | 38 | 4.554 | 0.033b |
| Yes | 15 | 3 | 12 | ||
| Metastasis | |||||
| No | 51 | 31 | 20 11.596 | 0.001b | |
| Yes | 40 | 10 | 30 | ||
| TNM stage | |||||
| T1-2 | 69 | 34 | 35 | 2.054 | 0.152 |
| T3-4 | 22 | 7 | 15 | ||
| BCLC stage | |||||
| A | 62 | 30 | 32 | 1.138 | 0.566 |
| B | 14 | 6 | 8 | ||
| C | 15 | 5 | 10 | ||
| Recurrencea | |||||
| No | 37 | 22 | 15 | 5.931 | 0.015b |
| Yes | 51 | 17 | 34 | ||
| Recurrence time (months)a | |||||
| <6 | 34 | 9 | 25 | 8.393 | 0.015b |
| 6≤ to <12 | 8 | 3 | 5 | ||
| ≥12 | 46 | 27 | 19 | ||
Metastasis was defined as patients whose recurrence time was <6 months or with lymph node invasion or cancer emboli, including portal vein tumor thrombus, biliary tract tumor thrombus and hepatic vein tumor thrombus.
Data of three patients missing;
P<0.05 was considered statistically significant. KIFC1, kinesin family member C1; AFP, α-fetoprotein; TNM, tumor-node-metastasis; BCLC, Barcelona Clinic Liver Cancer.
Table II.
Univariate and multivariate analyses of relapse-free survival rates in hepatocellular carcinoma.
| Variable | Univariate analysis
|
Multivariate analysis
|
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | |
| Sex (male/female) | 2.098 | 0.893–4.930 | 0.089 | |||
| Age (<60/≥60 years) | 1.468 | 0.583–3.700 | 0.415 | |||
| Hepatitis B (no/yes) | 0.746 | 0.336–1.661 | 0.474 | |||
| Cirrhosis (no/yes) | 1.458 | 0.834–2.549 | 0.186 | |||
| AFP (<400/≥400 µg/l) | 0.757 | 0.428–1.336 | 0.337 | |||
| Tumor size | 0.615 | 0.277–1.367 | 0.233 | |||
| Tumor number | 0.651 | 0.333–1.271 | 0.209 | |||
| Tumor capsule (no/yes) | 0.801 | 0.458–1.400 | 0.436 | |||
| Differentiation (high/low to medium) | 0.453 | 0.210–0.974 | 0.043a | 0.463 | 0.205–1.045 | 0.064 |
| Tumor emboli (no/yes) | 0.365 | 0.188–0.709 | 0.003a | 0.597 | 0.234–1.524 | 0.281 |
| TNM stage (T1-2/T3-4) | 0.375 | 0.207–1.680 | 0.001a | 0.367 | 0.193–0.696 | 0.002a |
| BCLC stage A (A/C) | 0.577 | 0.283–1.176 | 0.130 | |||
| B (B/C) | 0.908 | 0.358–2.304 | 0.839 | |||
| KIFC1 status (negative/positive) | 0.438 | 0.242–0.792 | 0.006a | 0.518 | 0.281–0.953 | 0.035a |
P<0.05 was considered statistically significant. HR, hazard ratio; CI, confidence interval; AFP, α-fetoprotein; TNM, tumor-node-metastasis; BCLC, Barcelona Clinic Liver Cancer; KIFC1, kinesin family member C1.
Expression of KIFC1 in HCC cell lines
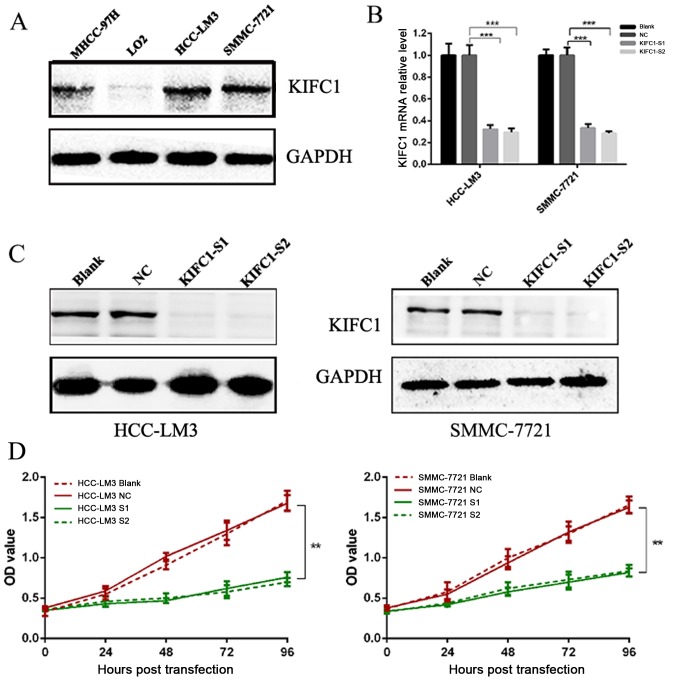
To further validate the role of KIFC1 in HCC, western blot analysis was performed to evaluate the expression of KIFC1 in human HCC cell lines and human normal liver epithelial cells. The expression of KIFC1 was high in MHCC-97H, SMMC-7721 and HCC-LM3 cells, but was absent in LO2 cells (Fig. 2A). As the HCC-LM3 and SMMC-7721 cell lines had high protein levels of KIFC1, these two cell lines were selected for the subsequent cell function investigation.
Figure 2.
Knockdown of KIFC1. (A) Western blot analysis of KIFC1 protein in hepatocellular carcinoma cell lines and liver epithelial cells. KIFC1 was expressed at high levels in MHCC-97H, SMMC-7721 and HCC-LM3 cells, but was absent in LO2 cells. (B) Both siRNAs effectively knocked down the mRNA expression of KIFC1 in HCC-LM3 and SMMC-7721 cells. ***P<0.001. (C) Both siRNAs effectively knocked down the protein expression of KIFC1 in HCC-LM3 and SMMC-7721 cells. (D) Effects of KIFC1 siRNAs on HCC-LM3 and SMMC-7721 cell growth over 96 h. **P<0.01. KIFC1, kinesin family member C1; siRNA, small interfering RNA; S1, siRNA 1; S2, siRNA 2; NC, negative control.
Effects of the knockdown of KIFC1 on cell viability, apoptosis and cell cycle in HCC-LM3 and SMMC-7721 cells
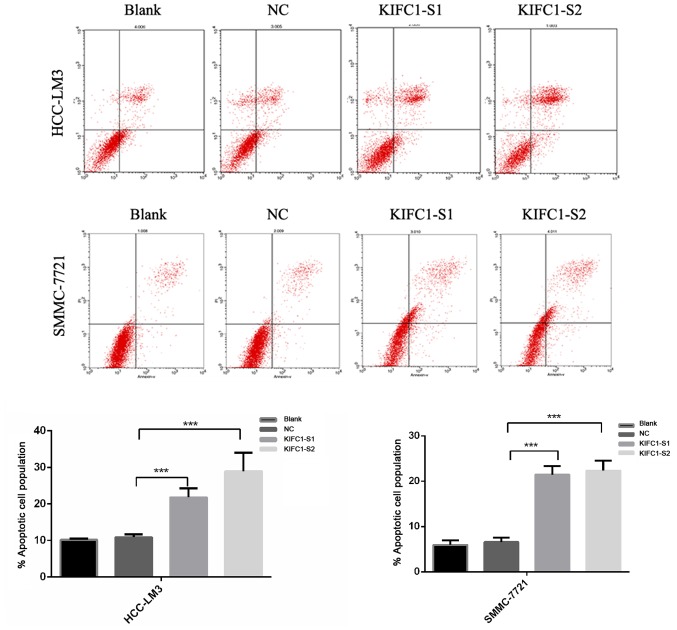
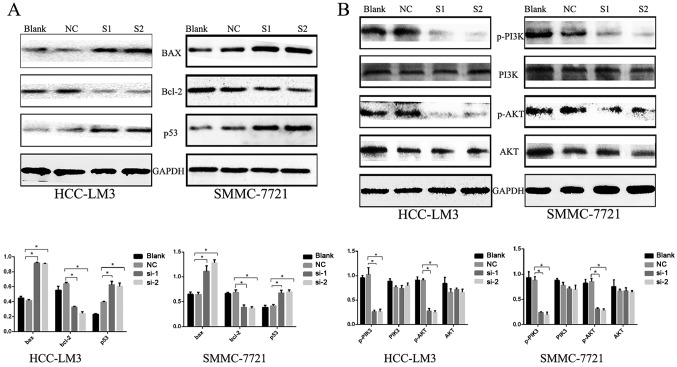
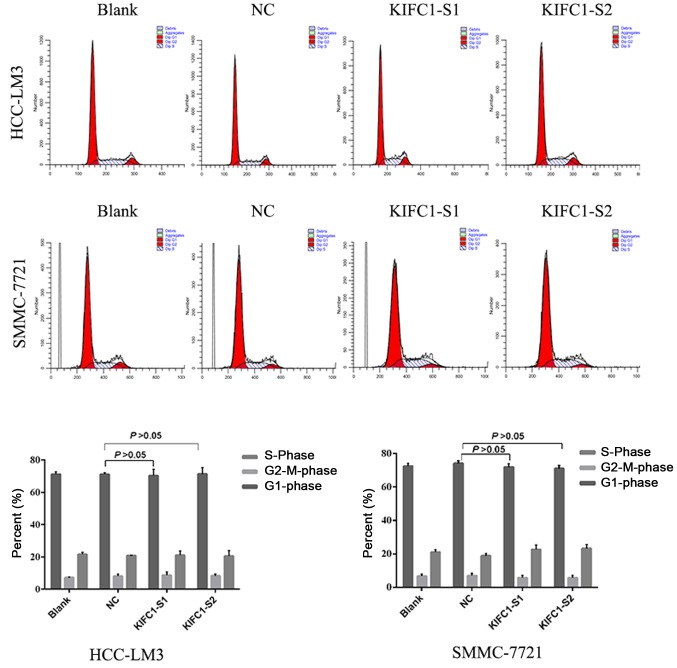
Two independent siRNAs targeting KIFC1 were used to examine the role of KIFC1 in the development and progression of HCC, with that the mRNA and protein expression of KIFC1 shown to be significantly silenced by RT-qPCR and western blot analyses at 48 h post-siRNA transfection (Fig. 2B and C). The cell viability of the two cell lines was significantly suppressed following KIFC1 knockdown (Fig. 2D). In addition, the KIFC1 siRNA-treated cells showed a marked increase in the rate of apoptosis, compared to the control (P<0.001) (Fig. 3). In confirming these results, it was demonstrated that the apoptosis-related protein Bcl-2 was downregulated, whereas Bax and p53 were upregulated (Fig. 4A). Furthermore, it was found that the expression levels of phosphorylated (p-)phosphoinositide 3-kinase (PI3K) and p-AKT were decreased significantly when KIFC1 was silenced (Fig. 4B). However, when DNA content was analyzed to assess cell cycle distribution, there was minimal change in the KIFC1 siRNA-treated group, compared with the control (P=0.254) (Fig. 5). These results demonstrated that the knockdown of KIFC1 be an effective strategy for inhibiting HCC growth.
Figure 3.
Induction of apoptosis by siRNA. Fluorescence-activated cell sorting measurement of KIFC1 siRNA induction of apoptosis in HCC-LM3 and SMMC-7721 cells. ***P<0.001. KIFC1, kinesin family member C1; siRNA, small interfering RNA; S1, siRNA 1; S2, siRNA 2; NC, negative control; PI, propidium iodide.
Figure 4.
Expression of apoptosis-related proteins. (A) Western blots showing downregulated Bcl-2, and upregulated Bax and p53 following KIFC1 knockdown in HCC-LM3 and SMMC-7721 cells. *P<0.05. (B) Western blot analysis was used to determine the PI3K/AKT pathway proteins. *P<0.05. KIFC1, kinesin family member C1; Bcl-2, B-cell lymphoma-2; Bax, Bcl-2-associated X protein; PI3K, phosphoinositide 3-kinase, p-, phosphorylated; NC, negative control; si-1/S1, small interfering RNA 1; si-2/S2, small interfering RNA 2.
Figure 5.
Cell cycle distribution. Minimal change in cell cycle distribution was observed following KIFC1 knockdown in HCC-LM3 and SMMC-7721 cells. P>0.05. KIFC1, kinesin family member C1; NC, negative control; S1, small interfering RNA 1; S2, small interfering RNA 2.
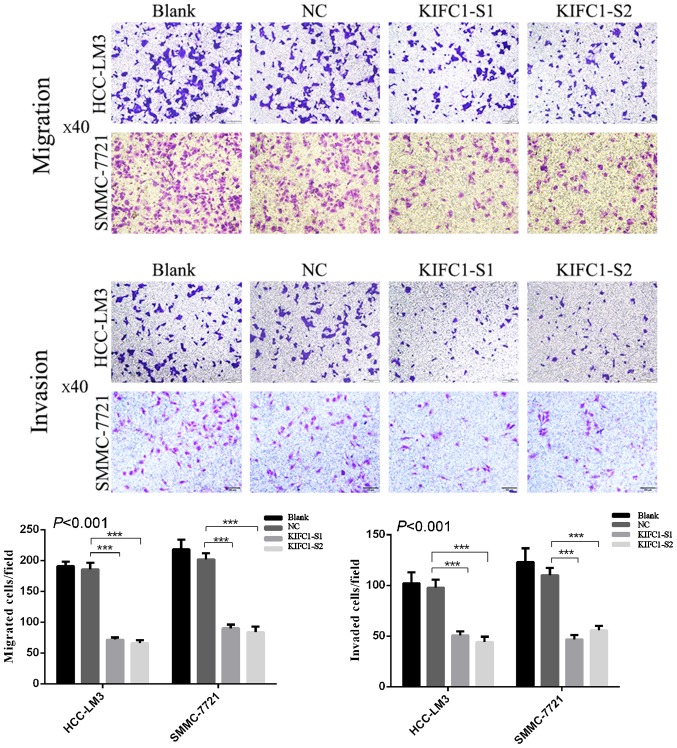
Effects of KIFC1 knockdown on inhibition of HCC-LM3 and SMMC-7721 cell migration and invasion
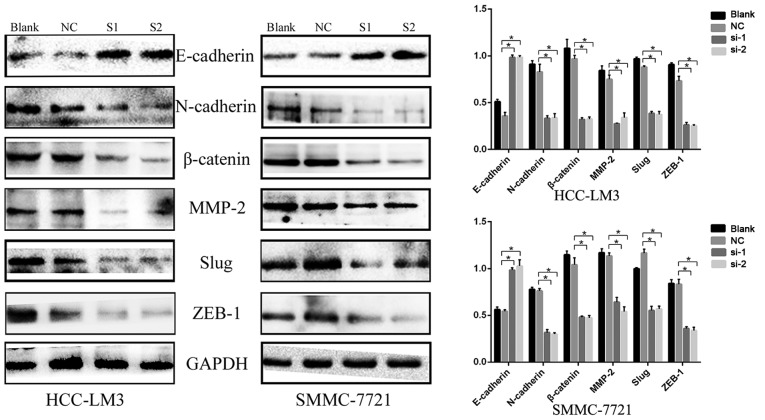
To investigate the effects of the inhibition of KIFC1 on HCC cell migration and invasion, Transwell assays were performed following transfection of KIFC1 siRNA or control siRNA into the two HCC cell lines. KIFC1 siRNA-treated cells had markedly impaired migration ability, compared with the blank and control groups (P<0.001) (Fig. 6), and there was a parallel downward trend in invasive ability (P<0.001) (Fig. 6). In addition, the expression of EMT-related proteins, including N-cadherin, matrix metalloproteinase-2 (MMP-2), β-catenin, Slug, Zinc finger E-box-binding homeobox 1 (ZEB1) and E-cadherin, were examined using western blot analysis. Compared with the negative control group, N-cadherin, MMP-2, β-catenin, Slug and ZEB1 were downregulated, whereas E-cadherin was upregulated (Fig. 7). These results indicated that the expression of KIFC1 may be associated with tumor invasion and metastasis in HCC.
Figure 6.
Effect of KIFC1 on cell migration and invasion. Knockdown of KIFC1 markedly suppresses HCC-LM3 and SMMC-7721 cell migration and invasion. ***P<0.001. KIFC1, kinesin family member C1; NC, negative control; S1, small interfering RNA 1; S2, small interfering RNA 2.
Figure 7.
Expression of proteins involved in epithelial-mesencchymal transition. Western blots show downregulated N-cadherin, MMP-2, β-catenin, Slug and ZEB1 and upregulated E-cadherin following KIFC1 knockdown. *P<0.05. KIFC1, kinesin family member C1; MMP2, matrix metalloproteinase 2; ZEB1, Zinc finger E-box-binding homeobox 1; NC, negative control; si-1/S1, small interfering RNA 1; si-2/S2, small interfering RNA 2.
Discussion
HCC is a life-threatening malignancy worldwide (1). Although traditional TNM classification systems provide a basic prognostic model, difficulty remains in differentiating outcomes considering the asymptomatic nature of early-stage HCC (29). Therefore, it is imperative to clarify the molecular mechanism underlying the progression of HCC and identify a novel HCC marker and therapeutic target.
KIFC1, a minus end-directed motor protein (14), is critical in centrosome clustering in cancer cells and is essential for cancer cell survival (15). KIFC1 predicts poor prognosis and shorter overall survival rate, and may serve as a potential marker of metastasis in ovarian cancer (22). In non-small cell lung cancer, together with Alzheimer antigen and N-cadherin, the overexpression of KIFC1 predicts brain metastasis in patients with early and advanced disease (23). KIFC1 is also overexpressed in breast cancer tissues and cell lines, and KIFC1 knockdown reduces breast cancer cell viability (24). However, the correlation between the expression of KIFC1 and HCC has not been reported. KIFC1 is essential for bipolar mitotic division of cancer cells, but appears redundant in normal cells (15,21), suggesting that KIFC1 is a promising thera peutic target in investigations on highly selective anticancer agents. The present study reported for the first time, to the best of our knowledge, that KIFC1 was expressed at a high level in HCC tissues, compared with that in peritumoral tissues, and its expression was correlated with tumor emboli, metastasis, recurrence and time of recurrence. Kaplan-Meier analysis showed that the expression of KIFC1 was significantly associated with tumor-free survival rates. In addition, multivariate analyses revealed that the overexpression of KIFC1 was an independent predictive marker in patients with HCC. Consistently, data derived from the TCGA and the GTEx projects via the online GEPIA database confirmed that the expression of KIFC1 was upregulated and significantly associated with disease-free survival and overall survival rates in HCC. These results indicated that a high expression of KIFC1 may be a crucial prognostic indicator for HCC.
In the present study, KIFC1 knockdown significantly suppressed HCC cell viability and markedly increased the rate of apoptosis. In addition, Bcl-2 was downregulated, whereas Bax and p53 were upregulated; these proteins act as regulatory proteins in the process of apoptosis (30). Apoptosis is one of the most characteristic hallmarks of the cell death process (31). Dysregulation of the balance between cell death and proliferation has been identified as a protumorigenic process during hepatic tumorigenesis (32). These results provide novel clues to the development and progression of HCC, and indicate that silencing KIFC1 can be critical in the anti-apoptotic process and correlate closely with apoptosis-related proteins. In addition, a previous study indicated that the PI3K/Akt signal pathway is involved in cell proliferation and apoptosis (33). In the present study, KIFC1 knockdown inhibited the expression of p-PI3K and p-PAKT, which indicated that the downregulation of KIFC1 in HCC cells induced apoptosis via the PI3K/AKT pathway. Investigations in the future aim to elucidate the mechanisms more directly.
In cancer, metastasis is considered the leading cause of mortality (34). The poor prognosis of patients with HCC is mainly due to intrahepatic and distant metastasis. Metastasis involves a series of interdependent events, including cancer cell proliferation, migration and invasion (35). EMT promotes HCC cell migration and invasive ability (36). Evidence suggests that EMT confers tumor cells with various malignant characteristics, including increased mobility, aggressiveness, evasion from apoptosis and a stem-like phenotype (37). Accordingly, the present study investigated the effects of the inhibition of KIFC1 on HCC cell migration and invasion via a Transwell assay, and the resulting data indicated that KIFC1 was involved in tumor metastasis. In addition, the present study examined the expression of the EMT-related proteins, N-cadherin, MMP-2, β-catenin, Slug, ZEB1 and E-cadherin, using western blot analysis. The results of the western blot analysis were in agreement with those of the Transwell assay, and also showed that KIFC1 mediated HCC metastasis by modulating EMT. Therefore, KIFC1 knockdown may be an ideal therapeutic strategy for metastatic HCC.
In conclusion, the present study is the first, to the best of our knowledge, to show that KIFC1 was overexpressed in HCC tissues compared with peritumoral tissues. Based on the above results, it is possible to conclude that the overexpression of KIFC1 is correlated closely with the progression of HCC and poor prognosis, and the expression level of KIFC1 may be a potential prognostic biomarker and therapeutic target for HCC.
Acknowledgments
Not applicable.
Funding
This study was supported in part by grants from the Key Research Project of Jiangxi Province, China (grant no. 20171BBG70063), the Natural Science Foundation of Jiangxi Province, China (grant no. 20171BAB215038), the Health Department of Jiangxi Province, China (grant no. 20131077), the Natural Science Foundation of nation, China (grant no. 81760438) and the Innovation Research Foundation of Graduate School of Nanchang University (grant no. cx2016330).
Availability of data and materials
The analyzed datasets generated during the study are available from the corresponding author on reasonable request.
Authors' contributions
XF, BL and LF were involved in the conception and design of the study; XF, YZh, BZ, YZo, CW, PW and JW were involved in data acquisition; XF, YZh and BL were involved in data analysis and interpretation; HC and PD were invovled in patient follow-up; XF, YZh and BL performed the statistical analysis; XF and LF drafted and edited the manuscript; XF, YZh, BL and LF revised and reviewed the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the Second Affiliated Hospital of Nanchang University, written consent was obtained from each participant, and the study was performed in accordance with the ethical standards of the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Llovet JM, Bruix J. Molecular targeted therapies in hepatocellular carcinoma. Hepatology. 2008;48:1312–1327. doi: 10.1002/hep.22506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feo F, Pascale RM, Simile MM, De Miglio MR, Muroni MR, Calvisi D. Genetic alterations in liver carcinogenesis: Implications for new preventive and therapeutic strategies. Crit Rev Oncog. 2000;11:19–62. doi: 10.1615/CritRevOncog.v11.i1.20. [DOI] [PubMed] [Google Scholar]
- 3.Michielsen PP, Francque SM, van Dongen JL. Viral hepatitis and hepatocellular carcinoma. World J Surg Oncol. 2005;3:27. doi: 10.1186/1477-7819-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun W, Xing B, Sun Y, Du X, Lu M, Hao C, Lu Z, Mi W, Wu S, Wei H, et al. Proteome analysis of hepatocellular carcinoma by two-dimensional difference gel electrophoresis: Novel protein markers in hepatocellular carcinoma tissues. Mol Cell Proteomics. 2007;6:1798–1808. doi: 10.1074/mcp.M600449-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Tang ZY. Hepatocellular carcinoma - cause, treatment and metastasis. World J Gastroenterol. 2001;7:445–454. doi: 10.3748/wjg.v7.i4.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sherman M. Modern approach to hepatocellular carcinoma. Curr Gastroenterol Rep. 2011;13:49–55. doi: 10.1007/s11894-010-0163-7. [DOI] [PubMed] [Google Scholar]
- 7.Calvisi DF, Pascale RM, Feo F. Dissection of signal transduction pathways as a tool for the development of targeted therapies of hepatocellular carcinoma. Rev Recent Clin Trials. 2007;2:217–236. doi: 10.2174/157488707781662715. [DOI] [PubMed] [Google Scholar]
- 8.Fukasawa K. Oncogenes and tumour suppressors take on centrosomes. Nat Rev Cancer. 2007;7:911–924. doi: 10.1038/nrc2249. [DOI] [PubMed] [Google Scholar]
- 9.Ogden A, Rida PC, Aneja R. Let's huddle to prevent a muddle: Centrosome declustering as an attractive anticancer strategy. Cell Death Differ. 2012;19:1255–1267. doi: 10.1038/cdd.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderhub SJ, Krämer A, Maier B. Centrosome amplification in tumorigenesis. Cancer Lett. 2012;322:8–17. doi: 10.1016/j.canlet.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Ogden A, Rida PC, Aneja R. Heading off with the herd: How cancer cells might maneuver supernumerary centrosomes for directional migration. Cancer Metastasis Rev. 2013;32:269–287. doi: 10.1007/s10555-012-9413-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pannu V, Rida PC, Ogden A, Clewley R, Cheng A, Karna P, Lopus M, Mishra RC, Zhou J, Aneja R. Induction of robust de novo centrosome amplification, high-grade spindle multipolarity and metaphase catastrophe: A novel chemotherapeutic approach. Cell Death Dis. 2012;3:e346. doi: 10.1038/cddis.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ando A, Kikuti YY, Kawata H, Okamoto N, Imai T, Eki T, Yokoyama K, Soeda E, Ikemura T, Abe K, et al. Cloning of a new kinesin-related gene located at the centromeric end of the human MHC region. Immunogenetics. 1994;39:194–200. doi: 10.1007/BF00241260. [DOI] [PubMed] [Google Scholar]
- 14.DeLuca JG, Newton CN, Himes RH, Jordan MA, Wilson L. Purification and characterization of native conventional kinesin, HSET, and CENP-E from mitotic hela cells. J Biol Chem. 2001;276:28014–28021. doi: 10.1074/jbc.M102801200. [DOI] [PubMed] [Google Scholar]
- 15.Kleylein-Sohn J, Pöllinger B, Ohmer M, Hofmann F, Nigg EA, Hemmings BA, Wartmann M. Acentrosomal spindle organization renders cancer cells dependent on the kinesin HSET. J Cell Sci. 2012;125:5391–5402. doi: 10.1242/jcs.107474. [DOI] [PubMed] [Google Scholar]
- 16.Nath S, Bananis E, Sarkar S, Stockert RJ, Sperry AO, Murray JW, Wolkoff AW. Kif5B and Kifc1 interact and are required for motility and fission of early endocytic vesicles in mouse liver. Mol Biol Cell. 2007;18:1839–1849. doi: 10.1091/mbc.E06-06-0524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall VJ, Compton D, Stojkovic P, Nesbitt M, Herbert M, Murdoch A, Stojkovic M. Developmental competence of human in vitro aged oocytes as host cells for nuclear transfer. Hum Reprod. 2007;22:52–62. doi: 10.1093/humrep/del345. [DOI] [PubMed] [Google Scholar]
- 18.Yang WX, Sperry AO. C-terminal kinesin motor KIFC1 participates in acrosome biogenesis and vesicle transport. Biol Reprod. 2003;69:1719–1729. doi: 10.1095/biolreprod.102.014878. [DOI] [PubMed] [Google Scholar]
- 19.Yang WX, Jefferson H, Sperry AO. The molecular motor KIFC1 associates with a complex containing nucleoporin NUP62 that is regulated during development and by the small GTPase RAN. Biol Reprod. 2006;74:684–690. doi: 10.1095/biolreprod.105.049312. [DOI] [PubMed] [Google Scholar]
- 20.Farina F, Pierobon P, Delevoye C, Monnet J, Dingli F, Loew D, Quanz M, Dutreix M, Cappello G. Kinesin KIFC1 actively transports bare double-stranded DNA. Nucleic Acids Res. 2013;41:4926–4937. doi: 10.1093/nar/gkt204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwon M, Godinho SA, Chandhok NS, Ganem NJ, Azioune A, Thery M, Pellman D. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–2203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pawar S, Donthamsetty S, Pannu V, Rida P, Ogden A, Bowen N, Osan R, Cantuaria G, Aneja R. KIFCI, a novel putative prognostic biomarker for ovarian adenocarcinomas: Delineating protein interaction networks and signaling circuitries. J Ovarian Res. 2014;7:53. doi: 10.1186/1757-2215-7-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grinberg-Rashi H, Ofek E, Perelman M, Skarda J, Yaron P, Hajdúch M, Jacob-Hirsch J, Amariglio N, Krupsky M, Simansky DA, et al. The expression of three genes in primary non-small cell lung cancer is associated with metastatic spread to the brain. Clin Cancer Res. 2009;15:1755–1761. doi: 10.1158/1078-0432.CCR-08-2124. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, Lu W, Chen D, Boohaker RJ, Zhai L, Padmalayam I, Wennerberg K, Xu B, Zhang W. KIFC1 is a novel potential therapeutic target for breast cancer. Cancer Biol Ther. 2015;16:1316–1322. doi: 10.1080/15384047.2015.1070980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De S, Cipriano R, Jackson MW, Stark GR. Overexpression of kinesins mediates docetaxel resistance in breast cancer cells. Cancer Res. 2009;69:8035–8042. doi: 10.1158/0008-5472.CAN-09-1224. [DOI] [PubMed] [Google Scholar]
- 26.Chun YH, Kim SU, Park JY, Kim DY, Han KH, Chon CY, Kim BK, Choi GH, Kim KS, Choi JS, et al. Prognostic value of the 7th edition of the AJCC staging system as a clinical staging system in patients with hepatocellular carcinoma. Eur J Cancer. 2011;47:2568–2575. doi: 10.1016/j.ejca.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang B, Zheng W, Fang L, Wu L, Zhou F, Yin X, Yu X, Zou Z. Overexpressed targeting protein for Xklp2 (TPX2) serves as a promising prognostic marker and therapeutic target for gastric cancer. Cancer Biol Ther. 2016;17:824–832. doi: 10.1080/15384047.2016.1195046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huitzil-Melendez FD, Capanu M, O'Reilly EM, Duffy A, Gansukh B, Saltz LL, Abou-Alfa GK. Advanced hepatocellular carcinoma: Which staging systems best predict prognosis? J Clin Oncol. 2010;28:2889–2895. doi: 10.1200/JCO.2009.25.9895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hao J, Yang Z, Wang L, Zhang Y, Shu Y, Jiang L, Hu Y, Lv W, Dong P, Liu Y. Downregulation of BRD4 inhibits gallbladder cancer proliferation and metastasis and induces apoptosis via PI3K/AKT pathway. Int J Oncol. 2017;51:823–831. doi: 10.3892/ijo.2017.4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Estaquier J, Vallette F, Vayssiere JL, Mignotte B. The mitochondrial pathways of apoptosis. Adv Exp Med Biol. 2012;942:157–183. doi: 10.1007/978-94-007-2869-1_7. [DOI] [PubMed] [Google Scholar]
- 32.Fabregat I. Dysregulation of apoptosis in hepatocellular carcinoma cells. World J Gastroenterol. 2009;15:513–520. doi: 10.3748/wjg.15.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin HL, Yang MH, Wu CW, Chen PM, Yang YP, Chu YR, Kao CL, Ku HH, Lo JF, Liou JP, et al. 2-Methoxyestradiol attenuates phosphatidylinositol 3-kinase/Akt pathway-mediated metastasis of gastric cancer. Int J Cancer. 2007;121:2547–2555. doi: 10.1002/ijc.22963. [DOI] [PubMed] [Google Scholar]
- 34.Rudmik LR, Magliocco AM. Molecular mechanisms of hepatic metastasis in colorectal cancer. J Surg Oncol. 2005;92:347–359. doi: 10.1002/jso.20393. [DOI] [PubMed] [Google Scholar]
- 35.Kumar S, Weaver VM. Mechanics, malignancy, and metastasis: The force journey of a tumor cell. Cancer Metastasis Rev. 2009;28:113–127. doi: 10.1007/s10555-008-9173-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El-Nahas AM. Plasticity of kidney cells: Role in kidney remodeling and scarring. Kidney Int. 2003;64:1553–1563. doi: 10.1046/j.1523-1755.2003.00255.x. [DOI] [PubMed] [Google Scholar]
- 37.Sato M, Shames DS, Hasegawa Y. Emerging evidence of epithelial-to-mesenchymal transition in lung carcinogenesis. Respirology. 2012;17:1048–1059. doi: 10.1111/j.1440-1843.2012.02173.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed datasets generated during the study are available from the corresponding author on reasonable request.