Abstract
The volume that the root system can occupy is associated with the efficiency of water and nutrient uptake from soil. Genetic improvement of root length, which is a limiting factor for root distribution, is necessary for increasing crop production. In this report, we describe identification of two quantitative trait loci (QTLs) for maximal root length, QUICK ROOTING 1 (QRO1) on chromosome 2 and QRO2 on chromosome 6, in cultivated rice (Oryza sativa L.). We measured the maximal root length in 26 lines carrying chromosome segments from the long-rooted upland rice cultivar Kinandang Patong in the genetic background of the short-rooted lowland cultivar IR64. Five lines had longer roots than IR64. By rough mapping of the target regions in BC4F2 populations, we detected putative QTLs for maximal root length on chromosomes 2, 6, and 8. To fine-map these QTLs, we used BC4F3 recombinant homozygous lines. QRO1 was mapped between markers RM5651 and RM6107, which delimit a 1.7-Mb interval on chromosome 2, and QRO2 was mapped between markers RM20495 and RM3430-1, which delimit an 884-kb interval on chromosome 6. Both QTLs may be promising gene resources for improving root system architecture in rice.
Keywords: CSSLs, Oryza sativa, root elongation, root system architecture, QTLs
For terrestrial plants, the root is an essential organ capturing water and nutrients from the surrounding soil. Many wild plants have evolved root systems that are able to adapt to various environmental stresses (Canadell et al. 1996). For example, under drought conditions, drought-resistant plants tend to have deep root systems and can thus avoid drought stress by absorbing water from subsoil. In crops, root system adaptations to a range of environments, which differ from region to region, may be beneficial for improving productivity (de Dorlodot et al. 2007; Uga et al. 2015a). In maize (Zea mays L.), a deep root system would be favorable for rapid acquisition of water and nitrogen from subsoil, because water and the nitrogen dissolved in it move downward owing to gravity (Lynch 2013). Saengwilai et al. (2014) reported that maize accessions with few crown roots take up more nitrogen from low-nitrogen soils than accessions with many crown roots. In rice (Oryza sativa L.), high-yielding cultivars often have higher root biomass in the deep soil layers of paddy fields in comparison with low-yielding varieties (Kawata et al. 1978; Morita et al. 1986, 1988). However, breeding for root traits has progressed slowly because phenotypic selection is more laborious and time consuming for root traits than for aboveground traits. Therefore, marker-assisted selection for quantitative trait loci (QTLs) without phenotypic selection may be a promising breeding strategy for improving root system architecture (de Dorlodot et al. 2007; Yamamoto et al. 2014).
The vertical distribution of root systems in monocotyledonous crops such as maize, wheat, and rice is determined mainly by root growth angle and the length of seminal and crown roots (Abe and Morita 1994; Araki et al. 2002). In rice, several QTLs for root growth angle have been detected or fine-mapped (Uga et al. 2011, 2012, 2013b, 2015b; Kitomi et al. 2015; Lou et al. 2015). Among them, DEEPER ROOTING 1 (DRO1) on chromosome 9 has been cloned (Uga et al. 2013a). The mapping population was derived from a cross between the shallow-rooted lowland cultivar IR64 and the deep-rooted upland cultivar Kinandang Patong (Uga et al. 2013a). Kinandang Patong has a functional allele of DRO1, whereas IR64 has a nonfunctional allele. Field experiments using a near-isogenic line (Dro1-NIL), which carried the Kinandang Patong allele of DRO1 in the IR64 genetic background, demonstrated that alteration of root system architecture from shallow to deep enhances drought avoidance and increases yields under drought stress (Uga et al. 2013a).
Several genes that affect root length have been identified in rice through mutant analyses. Most mutants have short roots as a result of repressed cell division, cell elongation, or both, and have abnormalities of genes involved in cell wall construction [OsDGL1 (Qin et al. 2013), OsMOGS (Wang et al. 2014), ROOT GROWTH INHIBITING/OsGLU3 (Inukai et al. 2012; Zhang et al. 2012)] and auxin signaling [OsARF12 (Qi et al. 2012)]. Because all the reported mutant alleles reduce root length, they cannot be used for improving the efficiency of water and nutrient acquisition or early seedling establishment. On the other hand, favorable alleles of >100 QTLs for root length, as summarized by Courtois et al. (2009), might improve root system architecture in rice. Among these, a few QTLs have been narrowed down to small chromosome regions, although no QTLs for root length have been isolated in rice so far. Under hydroponic conditions, qRL6.1, a QTLs for root length detected at the seedling stage, was delimited to a 337-kb interval on chromosome 6 (Obara et al. 2010), and qRL7, a QTLs for root length detected at the heading stage, was mapped to a 657-kb interval on chromosome 7 (Wang et al. 2013). Thus, only a few genes can be considered promising at this stage for increasing root length and root growth angle in rice.
DRO1 affects root growth angle but has no marked effect on root length or number, shoot length, or tiller number (Uga et al. 2013a). Therefore, QTLs for root length are needed to further improve root system architecture in rice; the efficiency of water and nutrient acquisition could be improved by combining such QTLs with DRO1. In this study, we measured maximal root length of rice chromosome segment substitution lines (CSSLs) grown in hydroponic culture to detect chromosomal regions increasing maximal root length. Next, we conducted rough mapping of five such regions. Finally, we fine-mapped two candidate QTLs.
Materials and Methods
Plant materials
For identification of genomic regions associated with maximal root length in rice, we used 26 CSSLs derived from a cross between IR64 and Kinandang Patong (IK-CSSLs in the IR64 genetic background; Uga et al. 2015b). IR64 is a modern lowland cultivar (indica; IRGC 66970) developed by the International Rice Research Institute in the Philippines, which is widely grown in South and Southeast Asia. Kinandang Patong is a traditional upland cultivar (tropical japonica; IRGC 23364) that originated in the Philippines. In the IK-CSSLs, each chromosome was covered by one to four lines that carried overlapping chromosomal segments, although small regions on chromosomes 2, 5, 7, 8, and 10 were not covered (Uga et al. 2015b).
For the QTLs analyses in five candidate regions associated with maximal root length in IK-CSSLs, we used five BC4F2 populations derived from self-pollinated BC4F1 plants with heterozygous target chromosomal regions, which had been selected during IK-CSSL development (Uga et al. 2015b). The BC4F2 for SL1006, SL1011, SL1015, SL1016, and SL1019 were #09-6642-34 (n = 144), #09-6647-13 (n = 79), #15-6456-7 (n = 119), #09-6651-10 (n = 74), and #09-S19-1 (n = 80), respectively.
For fine mapping of QTLs for maximal root length on chromosomes 2, 6, and 8, we developed homozygous BC4F3 lines in which recombination had occurred within the region containing the target QTLs. The number of BC4F3 line pairs for mapping of QTLs on chromosomes 2, 6, and 8 were 6, 16, and 7, respectively. Their BC4F4 progenies were used to determine the genotype of each QTLs in these lines.
Measurement of maximal root length in hydroponic culture
We grew IR64, Kinandang Patong, and IK-CSSLs in hydroponic culture. Seeds were washed in sterilized water three times and sterilized in 1% PPM (plant preservative mixture; Plant Cell Technology, Inc., Washington, DC) at 15° in the dark for 1 d. Sterilized seeds were imbibed at 30° in the dark for 1.5 d. Uniform seedlings were selected and transferred onto a plastic net supported by a polystyrene plate floating on a hydroponic solution in a plastic tub (L × W × H = 54.4 cm × 34.4 cm × 26.0 cm). Each polystyrene floater included 12 rows with 10 or 12 seeds per row; each tub contained up to four floaters. Seeds were spaced 1.0 cm within and between rows. Tubs were wrapped in foil and filled with 43 liters of 1/4-strength Kimura B solution [91.25 µM (NH4)2SO4, 22.75 µM K2SO4, 136.75 µM MgSO4, 45.75 µM KNO3, 91.25 µM Ca(NO3)2, 45.5 µM KH2PO4, 4.45 µM FeC6H5O7·H2O] with 5 mM MES (C6H13NO4S·H2O) in a greenhouse. The final pH of hydroponic solution was adjusted to 5.5. We renewed the nutrient solution every 2 d, at which time we rerandomized tubs in the greenhouse and floaters in the tubs. The average air temperature was 30° during the day and 26° at night; the average relative humidity was 50%; plants were illuminated during the daytime from 6:00 to 18:00 by 400 W metal-halide lamps (M400DL/BUDP; Iwasaki Electric Co., Ltd., Tokyo, Japan). At approximately the two-leaf-stage of IR64 used as a control (7–9 d after sowing), the plants were removed from the plastic net, and the lengths of roots and shoots were measured with a ruler. The difference in the day of measurement was due to differences in plant growth rate between different cultivation periods. A randomized block design with two replications (12 seedlings per replicate) was used for evaluation of maximal root length in the IK-CSSLs.
DNA marker analysis
The genotypes of BC4F2 and BC4F3 plants were determined by using simple sequence repeat (SSR) markers selected from the International Rice Genome Sequencing Project (2005) data and sequence-tagged site (STS) markers selected from Uga et al. (2008). After measurement of root and shoot lengths, a leaf was collected from each plant, and total leaf genomic DNA was extracted in 300 μl extraction buffer containing 1 M KCl, 100 mM Tris-HCl (pH 8.0), and 10 mM ethylenediamine tetraacetic acid (EDTA; pH 8.0). DNA was precipitated with 125 μl isopropanol, washed with 200 μl 70% ethanol, and dissolved in 100 μl 1/10 TE buffer containing 1.0 mM Tris-HCl (pH 8.0) and 0.1 mM EDTA (pH 8.0). Polymerase chain reaction (PCR) amplification for SSR analysis was performed in 5-µl reaction mixtures containing 1.0 µl DNA, 2.5 µl of KAPA2G Fast Ready Mix with dye (Kapa Biosystems, Boston, MA), 0.125 µl of a mixture of forward and reverse primers (20 µM each), and 1.375 μl H2O. The PCR program consisted of initial denaturation for 1 min at 95°; 35 cycles of 10 sec at 95°, 10 sec at 55°, and 1 sec at 72°; and a final extension for 30 sec at 72°. PCR products were separated by electrophoresis in 3% agarose gels (Type I-A, low EEO; Sigma-Aldrich, St. Louis, MO) at 200 V for 80 min.
Statistical and QTLs analyses
To identify chromosomal regions responsible for maximal root length in IK-CSSLs, we used Dunnett’s test in JMP v. 11.2 software (SAS Institute, Cary, NC). All lines were compared with IR64 as the reference; 24 plants per line were assessed (12 plants per row × 2 replicates).
Construction of a linkage map and QTLs analysis for the five BC4F2 populations were performed in R/qtl software (http://www.rqtl.org/; Broman et al. 2003). The markers were ordered based on their physical map positions in the latest version of the Rice Annotation Project (RAP) database (IRGSP-1.0; http://rapdb.dna.affrc.go.jp/). Genetic distances were estimated using the software’s Kosambi map function (Kosambi 1944). Putative QTLs were detected using the composite interval mapping (CIM) function. The CIM threshold was based on the results of 1000 permutations at the 5% significance level (Churchill and Doerge 1994).
For fine mapping, we genotyped the same SSR and STS markers used in the QTLs analyses, and used additional SSR markers to saturate the regions around the putative QTLs detected in QTLs analyses. We compared the phenotypic values between two BC4F4 homozygous recombinant lines having homozygous alleles of IR64 or Kinandang Patong. The genotype of each line was estimated from the results of a student’s t-test at the 0.1% significance level in JMP v. 11.2. The sample size per BC4F4 line was 40 plants (10 plants per row × 4 replicates).
Data availability
Supplemental Material, File S1 contains genotypes and phenotypes of the IK-CSSLs and BC4F2 populations used in this study.
Results
Maximal root length in IK-CSSLs grown under hydroponic conditions
Kinandang Patong had longer roots than IR64 in two trials (Figure 1). We also confirmed that this phenotypic difference persisted from the early seedling stage until 6 wk after sowing (Figure S1). These results indicate that the floating hydroponic system allows evaluation of the natural variation in rice root length. In the first trial, six CSSLs (SL1006, SL1011, SL1012, SL1015, SL1016, and SL1019) had significantly longer roots than IR64, whereas three CSSLs (SL1008–SL1010) had significantly shorter roots (Figure 1A). In the second trial, six CSSLs (SL1006, SL1007, SL1011, SL1015, SL1016, and SL1019) had significantly longer roots than IR64, whereas two CSSLs (SL1008 and SL1010) had significantly shorter roots (Figure 1B). These trials indicated that five CSSLs (SL1006, SL1011, SL1015, SL1016, and SL1019) carried robust QTL(s) whose Kinandang Patong alleles increase maximal root length. We also measured shoot length to clarify its association with maximal root length in IK-CSSLs. None of the CSSLs with longer roots than IR64 had significantly longer shoots, although SL1007, SL1012, and SL1015 had shorter shoots than IR64 (Figure S2).
Figure 1.
Maximal root lengths of 26 IK-CSSLs, IR64, and Kinandang Patong (KP) grown in hydroponic conditions. Values are given as mean ± SD (n = 24). Black and gray bars indicate lines with longer and shorter roots, respectively, than those of IR64 (P < 0.001, Dunnett’s test). Dashed lines show the mean values of IR64. Substituted chromosome in each line is indicated at the bottom. (A) First trial (data at 9 d after germination). (B) Second trial (data at 8 d after germination).
Detection of QTLs for maximal root length
To validate the presence of QTLs associated with maximal root length in the five CSSLs selected, we conducted QTLs analyses in five BC4F2 mapping populations segregating for target chromosomal regions. We detected a QTLs for maximal root length on each of chromosomes 2, 6, and 8 (Figure 2), but not on chromosomes 5 or 7 (data not shown), at the logarithm of the odds (LOD) threshold based on the results of 1000 permutations at the 5% significance level.
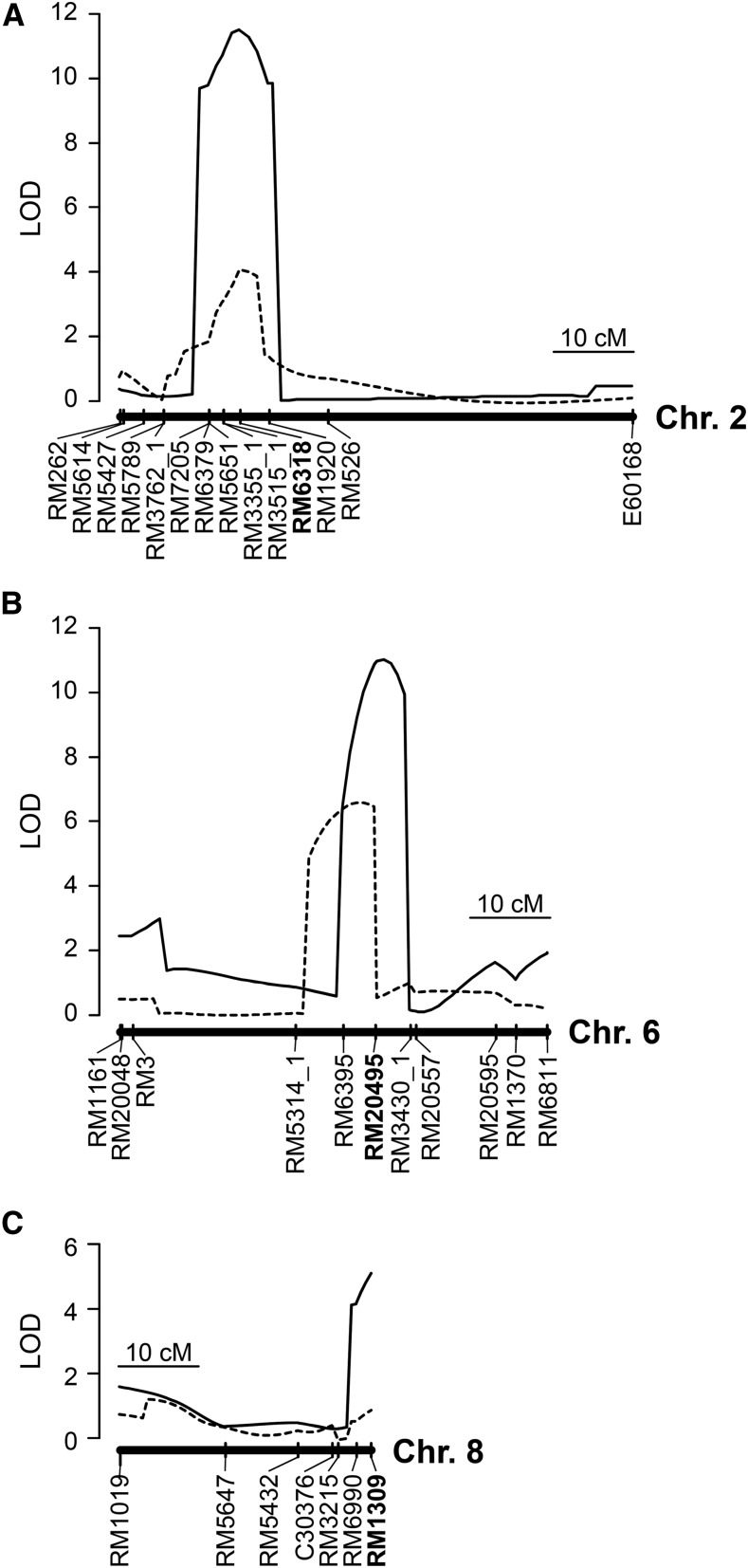
Figure 2.
LOD score curves of QTLs for root and shoot lengths in the three BC4F2 mapping populations. Peaks indicate the putative QTLs positions. Solid lines indicate maximal root length; dotted lines indicate shoot length. Vertical ticks indicate the genetic positions (cM) of DNA markers. DNA markers closest to LOD peaks are shown in bold. (A) Population #09-6642-34 segregating for the long arm of chromosome 2; (B) population #15-6456-7 segregating for the long arm of chromosome 6; (C) population #09-S19-1 segregating for chromosome 8.
One significant LOD peak (11.51) for maximal root length was detected at RM6318 on chromosome 2 (Figure 2A). This QTLs explained 30.7% of the total phenotypic variance (Table 1). The additive effect (AE) of the homozygous Kinandang Patong allele at this QTLs on maximal root length was 2.06 cm (i.e., 2× the AE for a single copy of the allele). The mean maximal root lengths of lines homozygous for the Kinandang Patong allele at RM6318 were significantly larger than those of lines homozygous for the IR64 allele (Figure 3A). We also found a QTLs for shoot length at RM6318, although the shoot length of SL1006 was not significantly different from that of IR64 (Figure 2A and Table 1). Lines homozygous for the Kinandang Patong allele at RM6318 had significantly longer shoots than lines homozygous for the IR64 allele (Figure 3A).
Table 1. Putative QTLs for root and shoot length detected in BC4F2 populations.
| Population | Trait | Chr. | Nearest Marker | Mba | cMb | LOD | Shc | AEd | DEe | R2f |
|---|---|---|---|---|---|---|---|---|---|---|
| 09-6642-34 | Root length | 2 | RM6318 | 24.43 | 0.0 | 11.51 | 2.64 | 1.03 | −0.13 | 30.7 |
| Shoot length | 2 | RM6318 | 24.43 | 0.0 | 4.07 | 2.95 | 0.50 | 0.44 | 16.5 | |
| 15-6456-7 | Root length | 6 | RM20495 | 26.55 | 1.4 | 11.04 | 2.66 | 0.56 | −0.17 | 30.7 |
| Shoot length | 6 | RM6395 | 26.00 | 2.6 | 6.56 | 2.73 | −0.36 | 0.60 | 26.7 | |
| 09-S19-1 | Root length | 8 | RM1309 | 19.18 | 0.0 | 4.85 | 2.75 | 0.46 | 0.24 | 21.4 |
Physical map position of each marker based on the latest version of the RAP database (http://rapdb.dna.affrc.go.jp/).
Genetic distance from the QTLs LOD peak to the nearest marker.
Threshold of LOD value based on 1000 permutation tests at the 5% level.
Additive effect of the allele from Kinandang Patong compared with that from IR64.
Dominance effect of the allele from Kinandang Patong compared with that from IR64.
Percentage of the phenotypic variance explained by each QTLs.
Figure 3.
Frequency distributions of root and shoot lengths in the three BC4F2 mapping populations, showing three genotype classes of the DNA markers closest to the detected QTLs. For each allele, an inverted triangle indicates the mean, and a horizontal bar indicates SD. The same shading is used for triangles and corresponding bars. The means labeled with different letters differ significantly (P < 0.05, Tukey’s multiple comparison test). (A) Population #09-6642-34 segregating for the long arm of chromosome 2; (B) population #15-6456-7 segregating for the long arm of chromosome 6; (C) population #09-S19-1 segregating for chromosome 8.
One significant LOD peak (11.04) for maximal root length was detected near RM20495 on chromosome 6 (Figure 2B). This QTLs also explained 30.7% of the total phenotypic variance (Table 1). The AE of the homozygous Kinandang Patong allele at this QTLs on maximal root length was 1.12 cm. The mean maximal root lengths of lines homozygous for the Kinandang Patong allele at RM20495 were significantly larger than those of lines homozygous for the IR64 allele (Figure 3B). We also found a QTLs for shoot length near RM6395 (Figure 2B and Table 1). Lines homozygous for the Kinandang Patong allele at RM6395 had significantly shorter shoots than lines homozygous for the IR64 allele (Figure 3B), suggesting that this QTLs may be associated with the shorter shoots of SL1015 than of IR64.
One significant QTLs for maximal root length was detected at RM1309 on chromosome 8, but no significant QTLs for shoot length was found in the same region (Figure 2C). This QTLs explained 21.4% of the total phenotypic variance (Table 1). The AE of the homozygous Kinandang Patong allele at this QTLs on maximal root length was 0.92 cm. The mean maximal root lengths of lines homozygous for the Kinandang Patong allele at RM1309 were significantly greater than those of lines homozygous for the IR64 allele (Figure 3C).
Fine mapping of maximal root length QTLs
To map the three QTLs for maximal root length detected on chromosomes 2, 6, and 8, we used three BC4F3 populations in which recombination occurred within the target region of each chromosome.
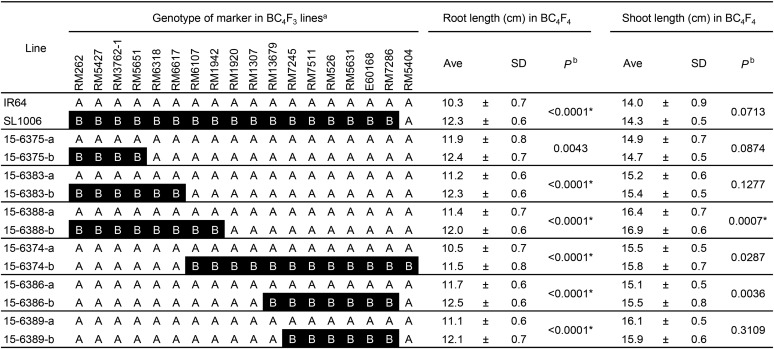
For the QTLs on chromosome 2, significant differences in maximal root length, ranging from 0.6 to 1.1 cm, between lines were found in all line pairs except #15-6375 (Figure 4), whereas the roots of SL1006 were 2.0 cm longer than those of IR64. These results suggest that this QTLs alone does not fully explain the total phenotypic variance in SL1006, and that two or more QTLs in this region are involved. On the basis of this hypothesis, we compared the genotypes of BC4F3 lines and the phenotypes of their BC4F4 progeny and identified two chromosomal regions related to maximal root length: one between RM5651 (23.57 Mb) and RM6107 (25.28 Mb), and the other between RM13679 (26.11 Mb) and RM5404 (33.68 Mb) (Figure 5). We designated the maximal root length QTLs in the former region QUICK ROOTING 1 (QRO1). On the other hand, we could not narrow down the latter region sufficiently as a single locus. A significant difference in shoot length was found only in one pair of lines, #15-6388, demonstrating that the shoot length QTLs on chromosome 2 was located between RM6617 (24.77 Mb) and RM6107 (25.28 Mb), i.e., within the candidate region of QRO1.
Figure 4.
Genotypes of 18 DNA markers on chromosome 2 in BC4F3 lines and phenotypes of their progeny (BC4F4). aA (white background), IR64 homozygous; B (black background), Kinandang Patong homozygous. bProbability of no significant difference between lines in a pair (student’s t-test). *Significance at the 0.1% level.
Figure 5.
Location of QRO1 on chromosome 2. (A) Graphical genotype of SL1006. White and gray boxes represent regions homozygous for the IR64 and Kinandang Patong alleles, respectively. S indicates short arm; L indicates long arm. (B) Physical map position of QRO1. Double-headed arrows indicate candidate regions of QRO1 and other putative QTLs for maximal root length. DNA markers are shown on the left side of each map; numbers in parentheses indicate physical map positions of the markers in the latest version of the RAP database. Underlines indicate no recombination with the predicted genotype of QRO1.
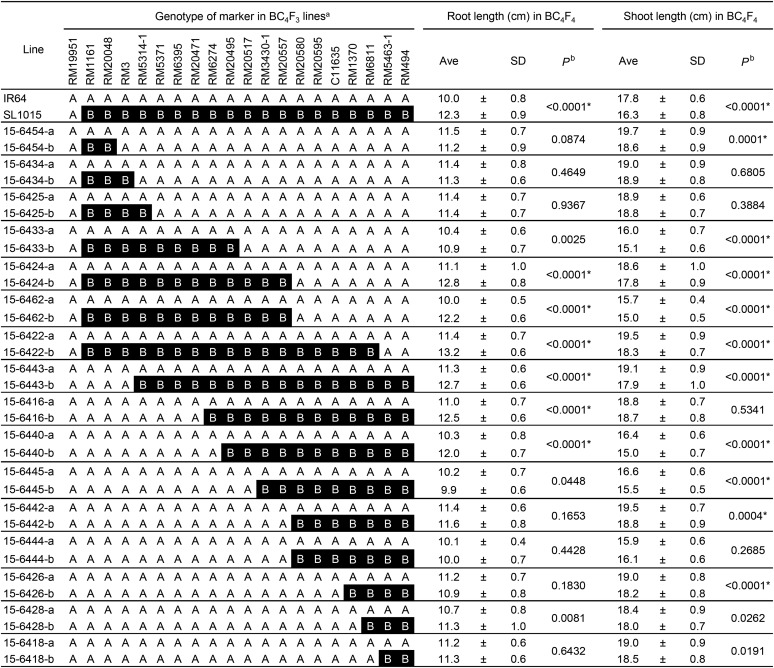
For the QTLs on chromosome 6, significant differences in maximal root length were found in the line pairs #15-6424, #15-6462, #15-6422, #15-6443, #15-6416, and #15-6440 (Figure 6), indicating that the QTLs is located between RM20495 (26.55 Mb) and RM3430-1 (27.43 Mb) (Figure 7). We designated this QTLs as QUICK ROOTING 2 (QRO2). We could not identify the specific region of the QTLs for shoot length by comparing the genotypes of BC4F3 lines and BC4F4 phenotypes.
Figure 6.
Genotypes of 20 DNA markers on chromosome 6 in BC4F3 lines and phenotypes of their progeny (BC4F4). aA (white background), IR64 homozygous; B (black background), Kinandang Patong homozygous. bProbability of no significant difference between lines in a pair (student’s t-test). *Significance at the 0.1% level.
Figure 7.
Location of QRO2 on chromosome 6. (A) Graphical genotype of SL1015. White and gray boxes represent regions homozygous for the IR64 and Kinandang Patong alleles, respectively. S indicates short arm; L indicates long arm. (B) Physical map position of QRO2. A short double-headed arrow indicates the candidate region of QRO2. DNA markers are shown on the left side of each map; numbers in parentheses indicate physical map positions of the markers in the latest version of the RAP database. Underline indicates no recombination with the predicted genotype of QRO2.
For the QTLs on chromosome 8, significant differences in maximal root length were detected in all line pairs except #15-6518 and #15-6519 (Figure S3). Comparison between the genotype and phenotype data suggested that SL1019 might have two QTLs for maximal root length in different regions: between C30376 and RM6215 and between RM5351-1 and RM23229. We were unable to narrow down the candidate region of the target QTLs in this mapping population, as both intervals were very wide.
Discussion
We analyzed the phenotypes of 26 CSSLs and identified five CSSLs that had longer roots at the seedling stage because of the presence of chromosome segments from Kinandang Patong. These results indicate that several regions, including those on chromosomes 2, 5, 6, 7, and 8, are associated with the difference in maximal root length between IR64 and Kinandang Patong. Furthermore, the possible presence of multiple QTLs on chromosomes 2 and 8 suggests that maximal root length is controlled by multiple genes on the same chromosome. For such a trait, the development and use of mapping populations with a simple genetic background such as CSSLs may become a powerful tool to reliably detect QTLs.
We narrowed down the candidate regions of two QTLs for maximal root length on chromosomes 2 (1.7 Mb) and 6 (884 kb) by fine mapping, and designated these QTLs as QRO1 and QRO2, respectively (Figure 5 and Figure 7). Next, we surveyed the presence of genes and QTLs related to root length reported by previous studies in the QRO1 and QRO2 candidate regions in three online databases: Overview of Functionally Characterized Genes in Rice Online (OGRO, http://qtaro.abr.affrc.go.jp/ogro/table; Yamamoto et al. 2012), QTLs Annotation Rice Online (Q-TARO, http://qtaro.abr.affrc.go.jp/qtab/table, Yonemaru et al. 2010), and RAP-DB (http://rapdb.dna.affrc.go.jp; Sakai et al. 2013). No genes or QTLs related to root length were found in OGRO or Q-TARO in the 1.7-Mb QRO1 candidate region. In RAP-DB, among ∼200 genes predicted in this region, we found cell division-related genes such as those encoding cyclins and transcription factors, including MYBs. In the 884-kb QRO2 candidate region, we found no genes or QTLs in OGRO or Q-TARO. In RAP-DB, ∼120 genes, including those encoding an auxin efflux carrier and several transcription factors, are predicted in this region. These genes might be candidates for QRO1 and QRO2. To identify the causative genes of QRO1 and QRO2, we are now performing high-resolution mapping. Comparison of the regions of two QTLs on chromosome 2 detected in this study and meta-QTL for root length reported by Courtois et al. (2009) showed that both QTLs regions included meta-QTL, suggesting that two regions may have a robust effect on root length in different genetic backgrounds if they are identical to these meta-QTL. On the other hand, Courtois et al. (2009) reported no hot spot of QTLs for root length on chromosome 6. Obara et al. (2010) fine-mapped such a QTLs (qRL6.1) on the long arm of chromosome 6, but its location is distinct from that of QRO2. These comparisons indicate that QRO2 is likely a novel QTLs for root length.
Seminal root elongation immediately after germination is a valuable trait for any crop, because it accelerates access to water and nutrients, resulting in the early establishment of vigorous seedlings (Rogers and Benfey 2015). QRO1 and QRO2 certainly promote quick seminal root elongation for a given length of time after germination. In Arabidopsis, mutants with a larger root meristem tend to have longer roots, and root meristem size is regulated by the balance between the plant hormones auxin and cytokinin (Dello Ioio et al. 2007, 2008). However, these studies measured root growth rate but not the maximal root length. Maximal root length may depend on cell division activity in the root apical meristem, but no report has identified genes contributing to the maximal root growth in any plant species. Therefore, whether maximal root length correlates with root meristem size or root growth rate remains unknown. To assess the durability of root growth promotion by QRO1 and QRO2, we will investigate their effects on maximal root length using QRO1-NIL and QRO2-NIL in greenhouse and field trials.
In the field, lower soil layers contain more water owing to gravity. The grain yield of Dro1-NIL, which carries a functional allele of DRO1 derived from Kinandang Patong in the genetic background of IR64, was higher than that of IR64 under drought conditions, because Dro1-NIL had deeper rooting and could obtain more water from lower soil layers than IR64 (Uga et al. 2013a). Dro1-NIL also had a higher grain yield than IR64 in paddy fields, because deeper rooting enhances nitrogen uptake from lower soil layers (Arai-Sanoh et al. 2014). On the other hand, the maximal root length of Dro1-NIL was identical to that of IR64, because DRO1 controls mainly root growth angle (Uga et al. 2013a). If QRO1 and/or QRO2 increase maximal root length, rice plants possessing their functional allele(s) could obtain water and nutrients under a wide range of field conditions. Crossing Dro1-NIL with QRO1-NIL and/or QRO2-NIL may enable development of rice plants with a greater capacity to obtain water and nutrients from deeper soil layers than that of Dro1-NIL. To develop QRO2-NIL, we have to separate QRO2 from the QTLs for short shoots on chromosome 6 to ensure vigorous early growth, if these QTLs are derived from respective genes (Figure 2B). We expect that the use of these root-related genes may allow us to improve the rice root system, resulting in high yields.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.300147/-/DC1.
Acknowledgments
We thank S. Tatsumi, J. Nakatsuji, and the staff of the technical support section of the Institute of Crop Science for plant management and measurement. This work was supported by grants of Science and Technology Research Partnership for Sustainable Development (SATREPS) from the Japan Science and Technology Agency (JST) and Japan International Collaboration Agency (JICA).
Footnotes
Communicating editor: J. Faris
Literature Cited
- Abe J., Morita S., 1994. Growth direction of nodal roots in rice: its variation and contribution to root system formation. Plant Soil 165: 333–337. [Google Scholar]
- Arai-Sanoh Y., Takai T., Yoshinaga S., Nakano H., Kojima M., et al. , 2014. Deep rooting conferred by DEEPER ROOTING 1 enhances rice yield in paddy fields. Sci. Rep. 4: 5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki H., Morita S., Tatsumi J., Iijima M., 2002. Physio-morphological analysis on axile root growth in upland rice. Plant Prod. Sci. 5: 286–293. [Google Scholar]
- Broman K. W., Wu H., Sen S., 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. [DOI] [PubMed] [Google Scholar]
- Canadell J., Jackson R. B., Ehleringer J. B., Mooney H. A., Sala O. E., et al. , 1996. Maximum rooting depth of vegetation types at the global scale. Oecologia 108: 583–595. [DOI] [PubMed] [Google Scholar]
- Churchill G. A., Doerge R. W., 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois B., Ahmadi N., Khowaja F., Price A. H., Rami J. F., et al. , 2009. Rice root genetic architecture: meta-analysis from a drought QTL database. Rice 2: 115–128. [Google Scholar]
- de Dorlodot S., Forster B., Pagés L., Price A., Tuberosa R., et al. , 2007. Root system architecture: opportunities and constraints for genetic improvement of crops. Trends Plant Sci. 12: 474–481. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R., Linhares F. S., Scacchi E., Casamitjana-Martinez E., Heidstra R., et al. , 2007. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 17: 678–682. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R., Nakamura K., Moubayidin L., Perilli S., Taniguchi M., et al. , 2008. A genetic framework for the control of cell division and differentiation in the root meristem. Science 322: 1380–1384. [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project , 2005. The map-based sequence of the rice genome. Nature 436: 793–800. [DOI] [PubMed] [Google Scholar]
- Inukai Y., Sakamoto T., Morinaka Y., Miwa M., Kojima M., et al. , 2012. ROOT GROWTH INHIBITING, a rice endo-1,4-β-D-glucanase regulates cell wall loosening and is essential for root elongation. J. Plant Growth Regul. 31: 373–381. [Google Scholar]
- Kawata S., Soejima M., Yamazaki K., 1978. The superficial root function and yield of hulled rice. Jpn. J. Crop. Sci. 47: 617–628. [Google Scholar]
- Kitomi Y., Kanno N., Kawai S., Mizubayashi T., Fukuoka S., et al. , 2015. QTLs underlying natural variation of root growth angle among rice cultivars with the same functional allele of DEEPER ROOTING 1. Rice 8: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosambi D. D., 1944. The estimation of map distance from recombination values. Ann. Eugen. 12: 172–175. [Google Scholar]
- Lou Q., Chen L., Mei H., Wei H., Feng F., et al. , 2015. Quantitative trait locus mapping of deep rooting by linkage and association analysis in rice. J. Exp. Bot. 66: 4749–4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J., 2013. Steep, cheap and deep: an ideotype to optimize water and N acquisition by maize root systems. Ann. Bot. 112: 347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S., Iwabuchi A., Yamazaki K., 1986. Relationships between the growth direction of primary roots and yield in rice plants. Jpn. J. Crop. Sci. 56: 530–535. [Google Scholar]
- Morita S., Suga T., Yamazaki K., 1988. The relationship between root length density and yield in rice plants. Jpn. J. Crop. Sci. 57: 438–443. [Google Scholar]
- Obara M., Tamura W., Ebitani T., Yano M., Sato T., et al. , 2010. Fine-mapping of qRL6.1, a major QTL for root length of rice seedlings grown under a wide range of NH4+ concentrations in hydroponic conditions. Theor. Appl. Genet. 121: 535–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., Wang S., Shen C., Zhang S., Chen Y., et al. , 2012. OsARF12, a transcription activator on auxin response gene, regulates root elongation and affects iron accumulation in rice (Oryza sativa). New Phytol. 193: 109–120. [DOI] [PubMed] [Google Scholar]
- Qin C., Li Y., Gan J., Wang W., Zhang H., et al. , 2013. OsDGL1, a homolog of an oligosaccharyltransferase complex subunit, is involved in N-glycosylation and root development in rice. Plant Cell Physiol. 54: 129–137. [DOI] [PubMed] [Google Scholar]
- Rogers E. D., Benfey P. N., 2015. Regulation of plant root system architecture: implications for crop advancement. Curr. Opin. Biotechnol. 32: 93–98. [DOI] [PubMed] [Google Scholar]
- Saengwilai P., Tian X., Lynch J. P., 2014. Low crown root number enhances nitrogen acquisition from low-nitrogen soils in maize. Plant Physiol. 166: 581–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H., Lee S. S., Tanaka T., Numa H., Kim J., et al. , 2013. Rice Annotation Project Database (RAP-DB): an integrative and interactive database for rice genomics. Plant Cell Physiol. 54: e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uga Y., Okuno K., Yano M., 2008. QTL underlying natural variation in stele and xylem structures of rice root. Breed. Sci. 58: 7–14. [Google Scholar]
- Uga Y., Okuno K., Yano M., 2011. Dro1, a major QTL involved in deep rooting of rice under upland field conditions. J. Exp. Bot. 62: 2485–2494. [DOI] [PubMed] [Google Scholar]
- Uga Y., Hanzawa E., Nagai S., Sasaki K., Yano M., et al. , 2012. Identification of qSOR1, a major rice QTL involved in soil-surface rooting in paddy fields. Theor. Appl. Genet. 124: 75–86. [DOI] [PubMed] [Google Scholar]
- Uga Y., Sugimoto K., Ogawa S., Rane J., Ishitani M., et al. , 2013a Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 45: 1097–1102. [DOI] [PubMed] [Google Scholar]
- Uga Y., Yamamoto N., Kanno S., Kawai T., Mizubayashi T., et al. , 2013b A major QTL controlling deep rooting on rice chromosome 4. Sci. Rep. 3: 3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uga Y., Kitomi Y., Ishikawa S., Yano M., 2015a Genetic improvement for root growth angle to enhance crop production. Breed. Sci. 65: 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uga Y., Kitomi Y., Yamamoto E., Kanno N., Kawai S., et al. , 2015b A QTL for root growth angle on rice chromosome 7 is involved in the genetic pathway of DEEPER ROOTING 1. Rice 8: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Xu X., Zhan X., Zhai R., Wu W., et al. , 2013. Identification of qRL7, a major quantitative trait locus associated with rice root length in hydroponic conditions. Breed. Sci. 63: 267–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Xu Y., Li Z., Zhang S., Lim J. M., et al. , 2014. OsMOGS is required for N-glycan formation and auxin-mediated root development in rice (Oryza sativa L.). Plant J. 78: 632–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto E., Yonemaru J., Yamamoto T., Yano M., 2012. OGRO: the overview of functionally characterized genes in rice online database. Rice 5: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Uga Y., Yano M., 2014. Genomics-assisted allele mining and its integration into rice breeding, pp. 251–265 in Genomics of Plant Genetic Resources, edited by Tuberosa R., Graner A., Frison E. Springer, Heidelberg. [Google Scholar]
- Yonemaru J. I., Yamamoto T., Fukuoka S., Uga Y., Hori K., et al. , 2010. Q-TARO: QTL Annotation Rice Online database. Rice 3: 194–203. [Google Scholar]
- Zhang J. W., Xu L., Wu Y. R., Chen X. A., Liu Y., et al. , 2012. OsGLU3, a putative membrane-bound endo-1,4-beta-glucanase, is required for root cell elongation and division in rice (Oryza sativa L.). Mol. Plant 5: 176–186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplemental Material, File S1 contains genotypes and phenotypes of the IK-CSSLs and BC4F2 populations used in this study.