Abstract
Germ–soma differentiation is a hallmark of complex multicellular organisms, yet its origins are not well understood. Volvox carteri is a simple multicellular green alga that has recently evolved a simple germ–soma dichotomy with only two cell-types: large germ cells called gonidia and small terminally differentiated somatic cells. Here, we provide a comprehensive characterization of the gonidial and somatic transcriptomes of V. carteri to uncover fundamental differences between the molecular and metabolic programming of these cell-types. We found extensive transcriptome differentiation between cell-types, with somatic cells expressing a more specialized program overrepresented in younger, lineage-specific genes, and gonidial cells expressing a more generalist program overrepresented in more ancient genes that shared striking overlap with stem cell-specific genes from animals and land plants. Directed analyses of different pathways revealed a strong dichotomy between cell-types with gonidial cells expressing growth-related genes and somatic cells expressing an altruistic metabolic program geared toward the assembly of flagella, which support organismal motility, and the conversion of storage carbon to sugars, which act as donors for production of extracellular matrix (ECM) glycoproteins whose secretion enables massive organismal expansion. V. carteri orthologs of diurnally controlled genes from C. reinhardtii, a single-celled relative, were analyzed for cell-type distribution and found to be strongly partitioned, with expression of dark-phase genes overrepresented in somatic cells and light-phase genes overrepresented in gonidial cells- a result that is consistent with cell-type programs in V. carteri arising by cooption of temporal regulons in a unicellular ancestor. Together, our findings reveal fundamental molecular, metabolic, and evolutionary mechanisms that underlie the origins of germ–soma differentiation in V. carteri and provide a template for understanding the acquisition of germ–soma differentiation in other multicellular lineages.
Keywords: cell differentiation, multicellularity, green algae, evolution, carbon metabolism, stem cells, cooption, photosynthesis, flagella, basal bodies, senescence, extracellular matrix, secretory pathway
Germ–soma or reproductive division of labor is a hallmark of complex multicellular organisms such as plants and animals. In organisms with germ–soma division of labor, somatic cells forgo reproduction and become specialized for support functions, while germ cells or stem cells retain reproductive potential (Buss 1983, 1987; Denis and Lacroix 1993; Agata et al. 2006; Seydoux and Braun 2006; Johnson et al. 2011; Solana 2013). Although multicellularity without germ–soma division of labor has arisen repeatedly (e.g., syncytial fungi and simple colonial algae), some degree of segregated reproductive potential arose at least once independently within multicellular taxa belonging to five major eukaryotic groups: plants (red algae, green algae, and land plants), Amoebozoa (cellular slime molds), Opisthokonts (animals and fungi), Alveolates (ciliates), and Heterokonts (brown algae) (Bonner 1998; Grosberg and Strathmann 2007; Herron et al. 2013). At least two selective advantages are thought to be associated with germ–soma separation. The first is conflict mitigation, which reduces intercellular competition for resources by restricting reproduction to a limited number of germ cells (Buss 1983, 1987; Michod 1997; Kerszberg and Wolpert 1998; Wolpert and Szathmáry 2002). The second advantage is the potential for increased functional specialization of somatic cells whose size, shape, organelle contents, and other attributes can be released from the constraints of undergoing periodic mitosis and cytokinesis (Wolpert 1990; Koufopanou and Bell 1993; Koufopanou 1994; Nedelcu and Michod 2004; Ispolatov et al. 2012; Woodland 2016). Indeed, the most complex multicellular taxa, including plants and animals, possess somatic cell-types that are terminally differentiated and, in some cases, completely incapable of further proliferation (e.g., anucleate mammalian red blood cells and plant vascular cells). The genetic and developmental mechanisms controlling germ–soma differentiation in animals and land plants have been extensively investigated (Strome and Lehmann 2007; Dickinson and Grant-Downton 2009; Lesch and Page 2012; Schmidt et al. 2012, 2015; Strome and Updike 2015; Swartz and Wessel 2015), but the highly-derived body plans and ancient origins of these taxa make it challenging to infer the early evolutionary steps that generated their germ–soma dichotomies.
The multicellular green alga Volvox carteri is a member of a monophyletic group called the volvocine green algae, which includes multicellular species with complete germ–soma differentiation (e.g., V. carteri), multicellular species lacking germ–soma differentiation (e.g., Gonium pectorale), and unicellular species (e.g., Chlamydomonas reinhardtii) (Nozaki et al. 2000; Kirk 2005; Nishii and Miller 2010; Herron 2016). Importantly, multicellularity and germ–soma differentiation arose relatively recently in volvocine algae (∼200 MYA) (Herron et al. 2009), making them attractive models for elucidating the origins of multicellular innovations (Kirk 1998, 2005; Nishii and Miller 2010; Umen and Olson 2012).
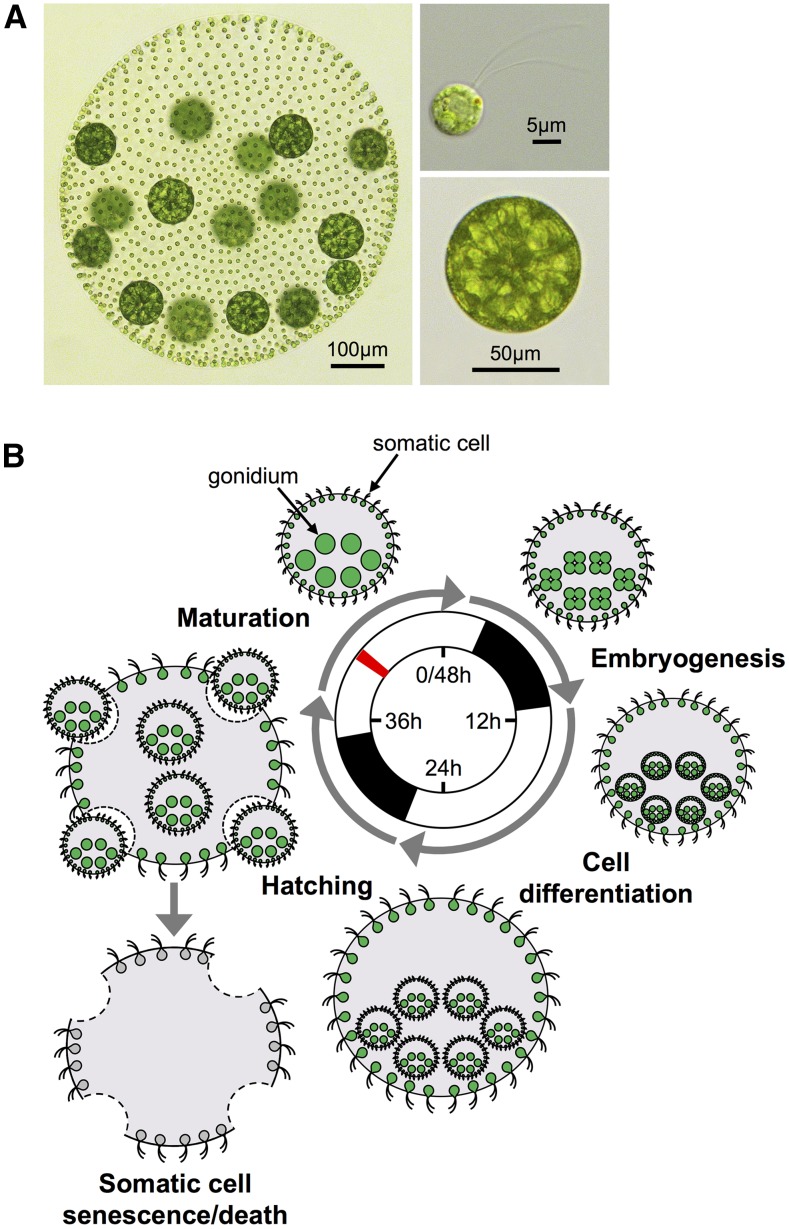
In its asexual phase, V. carteri possesses a simple spheroidal body plan with only two cell-types: ∼16 large aflagellate germ cells called gonidia that are positioned within the spheroid interior and ∼2000 small terminally differentiated somatic cells spaced evenly around the surface layer of the spheroid with flagella projecting outwards (Figure 1A). The majority of the adult spheroid volume is composed of clear secreted glycoprotein extracellular matrix (ECM) that maintains relative cell positioning and spheroid integrity (Hoops 1993; Hallmann and Kirk 2000; Kirk and Nishii 2001). Somatic cells provide phototactic motility to the spheroid through the coordinated beating of their flagella, and they secrete ECM that drives spheroid enlargement; however, somatic cells are terminally differentiated and eventually senesce and die. Gonidial cells serve a reproductive role by undergoing a period of cell growth followed by embryonic cleavage divisions and morphogenesis to produce a new generation of spheroids. Under optimal conditions, the entire vegetative life cycle of V. carteri can be synchronized under a 48 hr diurnal cycle (Kirk 1998, 2001; Kirk and Nishii 2001; Matt and Umen 2016) (Figure 1B).
Figure 1.
V. carteri cell-types and vegetative life cycle. (A) Micrographs of an intact adult spheroid with fully mature somatic and gonidial cells (left), isolated somatic cell (top right), and isolated gonidial cell (bottom right). (B) Diagram of the V. carteri vegetative (asexual) life cycle modified from Matt and Umen (2016). Center, 2 day diurnal regime used to synchronize V. carteri development with alternating 16 hr light (open bars) and 8 hr dark (closed bars) periods. Stages of the asexual life cycle are depicted proceeding clockwise from upper left (mature adult stage, black arrows indicate gonidial and somatic cell-types). Large green circles represent gonidial cells and small green circles with flagella represent somatic cells. Extracellular matrix is shaded gray. Gonidia within adult spheroids undergo cleavage and embryogenesis to form embryos with large gonidial precursors and small somatic precursors. Embryos undergo cytodifferentiation, where somatic precursors elongate their flagella and gonidial precursors grow in size, giving rise to juvenile spheroids. Juvenile spheroids eventually hatch from their mother spheroid whose somatic cells undergo senescence and death. Mature adult stage spheroids were used for cell-type separation and RNA extraction. Red line indicates the period in the diurnal cycle when spheroids were harvested.
Germ–soma differentiation is under genetic control in V. carteri , as evidenced by developmental mutants such as regA−, whose somatic cells are formed initially but then redifferentiate into gonidial cells (Huskey and Griffin 1979; Kirk et al. 1999), and lag− mutants, whose presumptive gonidial cells transiently differentiate as somatic cells but then acquire a gonidial identity (Kirk 1988, 1998, 2001). regA has been cloned and encodes a nuclear-localized putative transcription factor that is expressed only in somatic cells, where it represses gonidial cell fate (Kirk et al. 1999). It has been hypothesized that cell-type-specific genes in V. carteri, including targets of RegA, may have arisen through cooption of temporally or environmentally controlled genes from a unicellular ancestor that came under cell-type control in V. carteri (Nedelcu and Michod 2004, 2006; Nedelcu 2009), but this idea has not been tested on a genomic scale.
Previous studies have identified a few dozen cell-type-specific genes in V. carteri using either differential hybridization or quantitative RT-PCR (Tam and Kirk 1991; Nematollahi et al. 2006; Kianianmomeni and Hallmann 2015). Notably, Tam and Kirk (1991) identified 19 gonidial-specific genes, and subsequent annotation of these genes revealed that 13/19 of the encoded proteins were involved in photosynthesis and other chloroplast-related functions (Choi et al. 1996; Meissner et al. 1999). This result led to the proposal that germ–soma dichotomy in V. carteri is achieved by targeted suppssion of chloroplast biogenesis and photosynthesis in somatic cells (Meissner et al. 1999). However, the suppression of photosynthetic-related gene expression as the possible basis for germ–soma differentiation has not been explored further.
Here, we used deep transcriptome sequencing of highly purified gonidial and somatic cells of V. carteri to characterize their respective differentiation programs. Using intra- and interspecific genomic comparisons we found widespread differences in the compositions and phylogenetic origins of the two transcriptomes, which highlight fundamental differences between germ and somatic cell-types. Moreover, the cell-type transcriptomes in V. carteri could be correlated with temporally regulated diurnal regulons in C. reinhardtii, lending support to the temporal–spatial cooption hypothesis for the origins of V. carteri cell-types. Although our data did not support the photosynthetic suppression hypothesis for somatic differentiation, we did find strong evidence for extensive coordinated cellular and metabolic differentiation between cell-types, with anabolic pathways expressed preferentially in gonidial cells and a catabolic program in somatic cells that appeared specialized for the conversion of carbon stores into sugars that are the major constituent of the ECM glycoproteins.
Materials and Methods
Strains and culture conditions
V. carteri f. nagariensis female strain Eve (HK10), which has been maintained by subculturing since first isolated (Starr 1970), was obtained from Stephen Miller (University of Maryland, Baltimore County) and used for all experiments. Cultures were maintained in Standard Volvox Medium (SVM) in 30 ml glass tubes under fluorescent white light and synchronized under a 16 hr light:8 hr dark cycle at 32° (Tam and Kirk 1991). For cell-type separation and RNA preparation, cultures were grown in 500 ml flasks at 32° in temperature-controlled water baths aerated by bubbling air, and synchronized under a 16 hr light:8 hr dark cycle with illumination from LED lights [250 µEm−2s−1 total, 1:1 fluence ratio of red (625 nm) and blue (465 nm)].
Separation of cell-types
Separation of cell-types was based on a protocol from Tam and Kirk (1991), but modified to reduce processing time and to obtain purer cell-type preparations. Replicate cultures were used to prepare each cell-type. To avoid delays in sample processing only one cell-type was prepared from a given culture. For each of four cell-type preparations (two gonidial and two somatic), 10 flasks containing 350 ml SVM were inoculated with 15–20 manually selected cleavage-stage spheroids and cultures were grown for two generations (90 hr) to a density of 3000–4000 spheroids per flask. Hatched adult spheroids were harvested prior to the initiation of gonidial cleavage (hour 42 in the 48 hr life cycle, which is 6 hr into the second light period and ∼6 hr prior to the initiation of embryonic cleavage, Figure 1B) by collection onto a 30 µM nylon mesh filter (catalog# 03-30/18; SEFAR), and then transferred to a 40 ml Kimble Kontes Dounce Tissue Grinder and suspended in 40 ml SVM.
For gonidial cell purification, spheroids were disrupted by three to four passages of a loose-fitting (A) pestle, and the disrupted spheroid mixture was split into two 50 ml conical tubes and diluted 1:1 with SVM. Percoll (Sigma) was added to each tube to a final concentration of 7% (v/v), and tubes were centrifuged at 100 × g for 2 min in an Eppendorf 5810 centrifuge at room temperature. Supernatants from each tube were discarded and gonidial cell pellets were resuspended in 20 ml SVM, after which cell suspensions were combined and centrifuged at 100 × g for 1 min. Gonidial cells were washed again with SVM using the same centrifugation conditions, resuspended in 10 ml SVM, and then examined visually for purity using a dissecting microscope. Purified cells were transferred to a 15 ml tube and then immediately processed for RNA isolation (see below).
For somatic cell purification, harvested spheroids were disrupted by five passages of a tight-fitting (B) pestle, and the disrupted spheroid mixture was split into two 50 ml conical tubes and diluted 1:1 with SVM. Percoll was added to each tube to a final concentration of 7% (v/v) and tubes were centrifuged at 200 × g for 5 min. The somatic cell sheets at the top of the gradient in each tube were pipetted into 200 ml SVM and then split into four 50 ml conical tubes. Somatic cell sheets were pelleted at 3220 × g for 3 min and resuspended in 5 ml of SVM, after which they were combined into a single 50 ml conical tube and diluted 1:1 with fresh SVM. Cells were recentrifuged using the above conditions, resuspended in 10 ml SVM, and then examined visually for purity using a dissecting microscope. Purified cells were transferred to a 15 ml tube and then immediately processed for RNA isolation.
RNA isolation
Cells were pelleted and then resuspended in 5 ml Buffer C (2% SDS in Tris-Buffered Saline) and 5 ml of Trizol (Invitrogen), and then immediately flash frozen in liquid nitrogen and stored at −80°. Lysates were thawed, transferred to a 16 × 100 mm Covaris glass tube (SKU: 500012), and homogenized in a Covaris S220 ultrasonicator at 4° with the following settings: first treatment: Peak Power = 300, Duty Factor = 20, Cycles/Burst = 500, Temperature = 5°, and Time = 200 sec; and second treatment: Peak Power = 250, Duty Factor = 20, Cycles/Burst = 500, Temperature = 5°, and Time = 200 sec. Tubes with lysates were inverted to mix in between the first and second treatments. Lysates were centrifuged at 3000 × g for 10 min at room temperature and supernatants were extracted with 3 ml chloroform in a MaXtract High Density tube (QIAGEN). The aqueous phase was transferred to a 15 ml conical tube, and RNA was precipitated by adding 5 ml of isopropanol and incubating at room temperature for 10 min. RNA was pelleted by centrifugation at 3000 × g for 10 min and then washed with 75% ethanol. DNase digestion and purification of RNA were done using an RNeasy Mini Kit (QIAGEN) according to the manufacturer’s instructions. RNA concentration was determined by measuring absorbance at 260 nm using a nanophotometer (Implen). RNA quality was evaluated by agarose gel electrophoresis and using an Agilent 2100 Bioanalyzer.
Library preparation and deep sequencing
Two biological replicate samples of gonidial and somatic RNA were submitted to the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for Illumina-based polyA library preparation and sequencing. oligo-dT-selected RNA was fragmented to an average size of ∼200 nt and reverse transcribed to produce double-stranded cDNA using random hexamers. cDNA ends were end repaired, 3′ dA-tailed, and then ligated to Illumina sequencing adapters. Adaptor-ligated fragments were amplified for 12 cycles using indexed primers and sequenced on an Illumina HiSeq-2500 using single end reads extending 50 bases.
Mapping sequencing reads and classifying cell-type-specific gene expression
Quality of gonidial and somatic reads was evaluated using FastQC (Andrews 2010). Reads were processed using the Trimmomatic tool (Bolger et al. 2014) by removing adaptor sequences, low-quality bases (quality score < 3) from the 5′ and 3′ ends of reads, and the 3′ ends of reads after the average quality score in a sliding four-base window dropped below 15. Trimmed reads shorter than 25 nt were discarded. Processed reads were aligned to the V. carteri V2 genome assembly available on Phytozome (https://phytozome.jgi.doe.gov) (Prochnik et al. 2010; Goodstein et al. 2012) using STAR (Dobin et al. 2013), with the maximum number of mapped loci set to 10 and a maximum mismatch ratio of 0.06 (Supplemental Material, Table S1 in File S1). For assigning mapped reads to gene models, a merged set of protein-coding gene models was generated by combining all 14,247 V. carteri version 2.1 gene models currently available on Phytozome (Prochnik et al. 2010; Goodstein et al. 2012) with 1761 nonoverlapping (<20% sequence overlap) version 2.0 Phytozome protein-coding gene models (Prochnik et al. 2010; Goodstein et al. 2012) to generate a final set of 16,008 unique protein-coding gene models. Uniquely mapped reads were assigned to the merged set of gene models using HTSeq (Anders et al. 2014). Genes with no expression in any sample were removed from the analysis. Raw read counts for each gene in each sample were used as input for differential expression analysis using DESeq2 (Love et al. 2014). Following the removal of the upper 0.3% quantile of read counts in each sample, the total read count in each sample was divided by 10 million to yield size factors that were used to normalize expression estimates in each sample in DESeq2 (Bullard et al. 2010). Finally, genes in the bottom 10th quantile of normalized expression were removed from the analysis, as this cutoff was shown to maximize the number of significantly differentially expressed genes by minimizing the impact of multiple testing. These filtering steps yielded a set of expressed genes in the transcriptome that were then classified based on their cell-type expression patterns. Genes were considered to be gonidial or somatic genes if they exhibited at least a twofold difference in expression between cell-types with a false discovery rate (FDR) < 0.05. Gonidial and somatic genes were further subclassified based on expression ratio, with those exhibiting a two- to fivefold cell-type expression ratio designated as cell-type-biased, and those having greater than a fivefold expression ratio designated as cell-type-specific (Figure 2B). The remaining genes, all of which had an FDR > 0.05, were classified as “constitutive” if their expression ratio was <2 or “low confidence” if their expression ratio was >2. The low-confidence genes generally had cell-type expression ratios clustered around the twofold threshold but lacked statistical support for classification as cell-type-biased (Figure 2B and Figure S2 in File S1). We also calculated RPKM (reads per kilobase per million mapped reads) normalized expression values (Mortazavi et al. 2008) for all V. carteri genes after adding 0.1 read counts to each gene in order to eliminate zeros in expression ratio calculations. Expression ratios derived from DESeq2 and RPKM normalization were highly correlated (r2 = 0.995) (Figure S9 in File S1), and RPKM expression values were used for all further analyses and reported in Datasets S1, S2, S3, S4, S5, S6, S7, S8, S9 and S10.
Figure 2.
Cell-type sample preparation and gene expression classification. (A) Precleavage spheroids (top micrograph) were disrupted to separate cells types, and then subjected to differential density centrifugation (middle micrograph) to produce separate pure populations of somatic cell sheets on top of the gradient and gonidia in the pellet. Micrographs of separated cell-types at the bottom show sample purity (scale bars = 100 µm). (B) Bioinformatic workflow for classifying cell-type gene expression based on gonidial/somatic expression ratios (see Materials and Methods for details). Numbers in parentheses indicate number of genes in each expression category. G, gonidial expression; S, somatic expression.
Identifying V. carteri orthologs and coorthologs of C. reinhardtii genes
BLASTP (Altschul et al. 1990), with a BLOSUM80 scoring matrix and masking of low complexity and simple repeat sequences, was used to query V. carteri predicted proteins (derived from the merged set of gene models described above; 16,008 proteins) against all C. reinhardtii v5.5-predicted proteins available on Phytozome (17,741 proteins) (Merchant et al. 2007; Goodstein et al. 2012). Alignments were processed using the Smith–Waterman algorithm (Smith and Waterman 1981). Based on manual inspection of alignments, an E-value threshold of 1e−10 and a query coverage of >50% was chosen as a cutoff for homology assignment. V. carteri orthologs of C. reinhardtii proteins were assigned based on a mutual best hit criterion (Moreno-Hagelsieb and Latimer 2008): protein A in V. carteri (VcaA) was considered to be an ortholog of protein A in C. reinhardtii (CreA) if the alignment score between VcaA and CreA was higher than the alignment score between any another V. carteri protein and CreA. V. carteri coorthologs (or in-paralogs) were defined as follows: if VcaA and CreA are orthologs, then VcaB is a coortholog of CreA if CreA is the best hit for VcaB and if the VcaB-VcaA alignment score is higher than the VcaA-CreA alignment score. Using these criteria, we identified 9184 mutual best hit relationships between V. carteri and C. reinhardtii proteins, and 340 additional V. carteri proteins that were assigned as co-orthologs. We used these orthology relationships to annotate V. carteri orthologs and coorthologs of C. reinhardtii genes related to flagella, basal bodies, photosynthesis, central carbon metabolism, glycosylation, and autophagy, which are reported in Datasets S4, S5, S6, S8, and S10.
Functional classification of protein-coding genes
To assign MapMan terms to V. carteri protein-coding genes, we utilized classification terms already assigned to C. reinhardtii version 5.3.1 genes (http://mapman.gabipd.org/) (Thimm et al. 2004). v5.3.1 genes were first converted to version 5.5 C. reinhardtii IDs using a conversion table available on Phytozome (C. reinhardtiiTranscriptNameConversionBetweenReleases.Mch12b.txt.gz) (Merchant et al. 2007; Goodstein et al. 2012). C. reinhardtii annotation terms were then assigned to V. carteri orthologs as defined above. Statistical enrichment tests for MapMan terms in V. carteri cell-types were performed using all expressed V. carteri genes (defined above in Mapping sequencing reads and classifying cell-type-specific gene expression) as a background model. For each MapMan annotation level, we used the two-sided Fisher’s exact test with an odds ratio > 1 and an FDR < 0.05 as criteria for cell-type enrichment. Gene ontology (GO) terms were assigned using Blast2GO (Götz et al. 2008) with default settings, and with input derived from a BLASTP search of the predicted V. carteri proteome queried against the NCBI nonredundant database with a significance threshold of 1e−3. Additional GO terms were identified in Blast2GO using InterProScan (Quevillon et al. 2005), ANNEX (Myhre et al. 2006), and GO-slim (Camon et al. 2004) options. PFAM domain assignments to V. carteri proteins were made using the InterProScan output from Blast2GO.
Phylostratigraphic analysis of cell-type-specific genes
The phylostratigraphy approach was based on a previously reported method (Domazet-Loso et al. 2007). We first assembled a database of proteomes from species representing different phylostrata or taxonomic categories (V. carteri , Volvocaceae, Volvocine algae, Chlorophyte green algae, Viridiplantae, Eukaryotes, and Cellular organisms) (Table S5). The V. carteri proteome was compared to all proteomes using BLASTP with a BLOSUM45 substitution-scoring matrix designed for comparing distantly related protein sequences (Henikoff and Henikoff 1992), with filtering of low complexity and simple repeat sequences. Based on manual inspection of alignments, an E-value threshold of <1e−20 and a query coverage >50% were chosen as similarity cutoffs for assigning homology. A phylostratum assignment for each V. carteri protein was made based on the most distant phylostratum in which at least one homolog was found. Enrichment for cell-type expression was tested using the two-sided Fisher’s exact test with all expressed V. carteri genes as a background, with significance at an odds ratio > 1 and an FDR < 0.05.
Orthology group analysis of V. carteri genes, animal (metazoan) stem cell genes, and land plant (embryophyte) stem cell genes
Metazoan pluripotent stem cell genes have previously been classified into 180 OrthoMCL orthology groups (IDs) (Alié et al. 2015). To define a core set of land plant pluripotent stem cell genes, we used a published dataset that identified genes upregulated in the shoot apical meristem of the angiosperm Zea mays and genes upregulated in the gametophore apical cells of the moss Physcomitrella patens (Frank and Scanlon 2015). Protein sequences of the genes upregulated in the shoot apical meristem of Z. mays were acquired from a set of Z. mays version 2 predicted protein sequences (https://www.maizegdb.org/gene_center/gene#gm_downloads) (Andorf et al. 2016), and protein sequences of the genes upregulated in the gametophore apical cells of P. patens were acquired from a set of P. patens version 1.6 predicted protein sequences (https://www.cosmoss.org/physcome_project/wiki/Genome_Annotation/V1.6#GFF3) (Zimmer et al. 2013). We assigned these protein sequences to OrthoMCL database orthology groups (http://www.orthomcl.org) (Chen et al. 2006; Fischer et al. 2011), and then defined the intersection between the orthology groups containing maize meristem genes and the orthology groups containing Physcomitrella apical cell genes to be the set of land plant (embryophyte) stem cell orthology groups. All V. carteri proteins in the merged proteome were queried against the OrthoMCL database in a similar fashion as above, which allowed us to assign 11,636 V. carteri genes to 8356 unique orthology groups (IDs). Genes from each V. carteri orthology group were then queried for their expression classifications [gonidial, somatic, and not cell-type-regulated (constitutive + low confidence)]. If all V. carteri genes in an orthology group had the same expression classification, then the orthology group ID was assigned to that V. carteri expression classification. Orthology groups containing only genes with no or low expression in the transcriptome (defined in Mapping sequencing reads and classifying cell-type-specific gene expression), or orthology groups containing genes belonging to >1 expression classification, were removed from the analysis. The merged set of V. carteri OrthoMCL IDs associated with each V. carteri expression classification was used as a background for testing enrichment/deenrichment of different subsets using a two-sided Fisher’s exact test. Enrichment was defined as an odds ratio > 1 and an FDR < 0.05, while deenrichment was defined as an odds ratio < 1 and an FDR < 0.05.
Phylogenetic analysis and expression quantification of the light harvesting complex (LHC) genes
V. carteri LHC proteins were identified using BLASTP searches (Altschul et al. 1990) with annotated LHCII, LHCI, and LHCSR proteins from C. reinhardtii (Zones et al. 2015). All alignments and tree building were done using MEGA7 (Kumar et al. 2016). Protein alignments were done using MUSCLE (Edgar 2004) within MEGA. A Jones–Taylor–Thornton amino acid substitution model with a γ parameter of two was used to estimate a neighbor joining tree with 1000 bootstrap replications. Due to the high sequence similarity between paralogs belonging to LHCII clade 1 (19 members) and LHCII clade 4 (seven members), expression of individual members within these two clades could not be assigned. Instead, each clade was treated as a single gene by remapping sequence reads to the V. carteri genome, with up to 19 matches allowed for clade 1 and up to seven matches allowed for clade 4, and allowing for no base mismatches. Mapped reads that intersected with clade 1 genes and clade 4 genes were identified using SAMtools (Li et al. 2009) and BEDTools (Quinlan and Hall 2010). The total read counts were calculated by counting each read mapping to any of the clade 1 genes or to any of the clade 4 genes as one read to the respective clade. The mean transcript length for LHCII genes from clade 1 or from clade 4 was used as a basis to estimate RPKMs for each clade.
Annotation of central carbon metabolism in V. carteri
Metabolic pathways shown in Figure 7 were based on the following references: Calvin cycle (http://www.biocyc.org/CHLAMY/; Serrato et al. 2009), tricarboxylic acid (TCA) cycle (http://www.biocyc.org/CHLAMY/), amino acid metabolism (Vallon and Spalding 2009; Fontaine et al. 2012; De La Torre et al. 2014; Plancke et al. 2014), starch metabolism (Dauvillée et al. 2006; Ball and Deschamps 2009; Johnson and Alric 2013), fatty acid metabolism (Goepfert and Poirier 2007; Li-Beisson et al. 2015), glycolysis/gluconeogenesis (Klein 1986; Johnson and Alric 2013), glyoxylate cycle (Spalding 2009), and nucleotide sugar metabolism (http://www.genome.jp/kegg/pathway.html; Seifert 2004).
Figure 7.
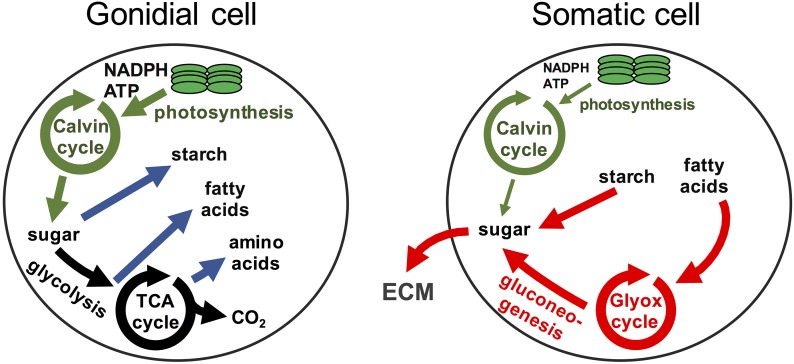
Cell-type expression patterns of carbon metabolism-related genes in V. carteri. Major pathways of central carbon metabolism are depicted as flow diagrams. Large shapes with colored shading and colored text labels are used to indicate subcellular compartments where indicated metabolic steps occur (chloroplast, mitochondrion, peroxisome, and cytosol). Major carbon sinks of starch and free fatty acids are also cartooned. Metabolites are in black lettering, chemical reactions are represented by solid arrows, and metabolite shuttling between subcellular compartments is represented by dashed arrows. Red arrows indicate a putative pathway for sugar biosynthesis in somatic cells. Genes encoding enzymes for indicated steps are in white lettering and surrounded by boxes that are color-coded according to their germ–soma expression patterns (color key in the upper left). Only genes with differential germ–soma gene expression are shown. Genes encoding subunits of a protein complex are grouped together in larger boxes. Paralogs inferred to have similar functions are grouped together in single boxes and separated by commas. Cell-type expression patterns of grouped paralogs were determined by computing the ratios of summed expression levels of paralogs in each cell-type (see Materials and Methods; Datasets S6 and S7). ECM, extracellular matrix; TCA, tricarboxylic acid.
Annotation of nucleotide sugar metabolism enzymes in V. carteri
We annotated biosynthetic pathways for nucleotide sugars that are the major glycosylation donors for cell wall/ECM glycoproteins in volvocine algae: UDP-arabinose, GDP-mannose, UDP-galactose/GDP-galactose, UDP-xylose, and UDP-glucose (Sumper and Hallmann 1998; Hallmann 2003; Harris 2009). V. carteri and C. reinhardtii genes predicted to encode enzymes involved in the biosynthesis of these nucleotide sugars were obtained from the KEGG website (http://www.genome.jp/kegg-bin/show_pathway?map=map00520&show_description=show) (Kanehisa et al. 2016). Downloaded V. carteri v1 gene models from KEGG were converted to version 2.1/2.0 gene models using BLASTN searching for correspondence (Altschul et al. 1990). C. reinhardtii v3 gene models from KEGG were converted to version 5.5. gene models using the conversion table described above followed by manual curation. V. carteri orthologs of C. reinhardtii genes were identified as described above. Putative UDP-galactopyranose mutases were identified in V. carteri by identifying orthologs of a C. reinhardtii UDP-galactopyranose mutase that was previously identified (Beverley et al. 2005) and identifying other V. carteri genes with the same EC annotation (5.4.99.9, UDP-galactopyranose mutase) in Phytozome. The full set of V. carteri genes predicted to be involved in nucleotide sugar metabolism is reported in Dataset S7.
Annotation of glycosyltransferases in V. carteri
We identified glycosyltransferases that are involved in either O-glycosylation or modification of O-linked glycans, and whose substrates are the sugars found on V. carteri ECM glycoproteins (see above). Putative V. carteri glycosyltransferases were identified through a mutual best-hit approach as follows. Protein sequences of arabinosyltransferases and galactosyltransferases from Arabidopsis thaliana (Showalter and Basu 2016), mannosyltransferases from Saccharomyces cerevisiae (PMT1, AAA02928; KRE2, CAA44516; KTR6/MNN6, NP_015272; and MNN1, AAA53676), xylosyltransferases from Homo sapiens and Phaseolus vulgaris (GXYLT1, NP_001093120; C3ORF21, NP_001294998; TMEM5, NP_001265166; ZOX1, AAD51778; and XYLT1, NP_071449), and glucosyltransferases from Drosophila melanogaster and Ph. lunatus (Rumi, NP_001262849 and ZOG1, AAD04166) were queried against the V. carteri merged proteome using BLASTP (Altschul et al. 1990) (E-value cutoff = 1e−3) to identify candidate V. carteri homologs. Using this approach, we identified candidate V. carteri homologs of A. thaliana arabinosyltransferases and galactosyltransferases, S. cerevisiae mannosyltransferases, and D. melanogaster glucosyltransferases. The protein sequences of these candidate V. carteri homologs were then reciprocally queried against the predicted proteomes of the originally queried species [A. thaliana TAIR10 (https://phytozome.jgi.doe.gov) (Goodstein et al. 2012; Lamesch et al. 2012); S. cerevisiae S288C (http://www.yeastgenome.org/blast-sgd, last updated January 13, 2015) (Engel et al. 2014); and D. melanogaster (http://flybase.org/blast/, DmeI release 6.15) (Gramates et al. 2017)], using BLASTP to verify their similarity to the original enzyme classification and rule out better-scoring classifications. Candidate paralogs of the InvC protein in V. carteri (BAH03159), which encodes a LARGE glycosyltransferase that is required for extracellular vesicle expansion (Ueki and Nishii 2008), were also identified using BLASTP. Protein sequences of candidate InvC paralogs were then queried against the predicted proteome of Mus musculus GRCm38.p5 (https://www.ncbi.nlm.nih.gov/genome?term=mus%20musculus) (Church et al. 2009), using BLASTP to validate their homology to LARGE glycosyltransferases. The full set of V. carteri predicted glycosyltransferases is reported in Dataset S8.
Cell-type expression patterns of paralogous genes encoding central carbon metabolism enzymes
A number of paralogous gene groups encoding isoforms of the same central carbon metabolism enzyme (e.g., Rubisco small subunit) were considered functionally similar, unless there was evidence available about differential isoform localization or differential function in algae or land plants. Expression estimates of functionally similar paralogs were treated additively, based on the rationale that summed expression provides a better estimate of relative activity in each cell-type, especially in cases where one isoform is clearly dominant (Figure 7 and Datasets S6 and S7). These summed cell-type RPKM levels were then used to calculate cell-type expression ratios for each gene group. Assignment of each gene group into a cell-type expression classification was based on the same criteria described for single genes in Mapping sequencing reads and classifying cell-type-specific gene expression, but no confidence statistics were calculated for the expression ratios of summed paralogs.
Predicting subcellular localizations of V. carteri proteins
Predictions for subcellular localizations of all V. carteri predicted proteins from the merged set of gene models described above were done using PredAlgo (https://giavap-genomes.ibpc.fr/cgi-bin/predalgodb.perl?page=main) (Tardif et al. 2012). Enrichment testing (Figure S6 in File S1) was done using a two-sided Fisher’s exact test with FDR < 0.05, with the subcellular localization distribution for all V. carteri proteins in the expressed transcriptome used as a background model.
Correspondence between V. carteri cell-type-regulated genes and C. reinhardtii diurnal clusters
We identified V. carteri orthologs of C. reinhardtii genes found in each of 18 diurnal expression clusters and an unclustered group (Zones et al. 2015) (coorthologs were excluded from the analysis). V. carteri orthologs with either low expression or those within the low-confidence expression classification were removed from the analysis. The cell-type expression pattern classifications for V. carteri orthologs derived from each C. reinhardtii diurnal cluster were then tested for relative enrichment using a two-sided Fisher’s exact test with an FDR < 0.05, with the distribution of cell-type expression pattern classifications for all V. carteri orthologs in the analysis used as a background distribution. A reciprocal analysis was also performed by first merging the C. reinhardtii diurnal clusters into four major superclusters: light-phase genes from clusters 1–8, light–dark transition-phase genes from diurnal clusters 9–11, dark-phase genes from diurnal clusters 12–18, and a group of unclustered genes. The supercluster classifications for C. reinhardtii orthologs of V. carteri gonidial genes, somatic genes, and constitutive genes were then tested for relative enrichment using a two-sided Fisher’s exact test with an FDR < 0.05, with the distribution of supercluster classifications for all C. reinhardtii orthologs of all expressed V. carteri genes used as a background distribution. When testing the effect of flagella genes on the distributions of supercluster classifications, flagella genes (Dataset S4) were removed from the analysis and significance testing was redone.
Data availability
Wild-type V. carteri f. nagariensis strain Eve (HK10) can be obtained from UTEX (http://utex.org/) or NIES (http://mcc.nies.go.jp/), and our current subclone is available upon request. Names and descriptions of supplemental tables and datasets can be found in the supplemental material. Primary data are available at the NCBI Gene Expression Omnibus repository under accession number GSE104835. V. carteri locus ID numbers for genes described in this study are listed in the appropriate supplemental datasets and can be accessed from Phytozome (http://phytozome.jgi.doe.gov).
Results
Identification of germ- and somatic-specific genes
To characterize the germ and somatic transcriptomes of V. carteri, we purified gonidial and somatic cells from hatched, precleavage adult spheroids (Figure 1B and Figure 2A). This life cycle stage was chosen for three reasons: (1) adult spheroids have fully differentiated germ and somatic cell-types; (2) since gonidia are not dividing the cell-type comparisons are not complicated by gene expression programs associated with cell division and embryogenesis; and (3) the two cell-types have the largest difference in size and are easiest to separate into pure populations (Figure 2A).
RNA was prepared from two biological replicates of each cell-type and processed for high-throughput Illumina-based sequencing. First, 15–17 million single-end reads were generated from each RNA-seq library, with 80–90% of reads mapping uniquely to version 2 of the V. carteri genome assembly (Prochnik et al. 2010) (Table S1 in File S1). A nonoverlapping set of 16,008 gene models comprised of version 2.0 and version 2.1 predictions from Phytozome was used for quantitative analysis of gene expression (Materials and Methods). After filtering out poorly expressed genes we obtained a set of 13,350 expressed genes (∼83% of predicted genes), which were subjected to analyses for differential expression between cell-types. Replicate samples for each cell-type had highly correlated expression profiles, while comparisons between cell-types indicated a large number of differentially expressed genes (Figures S1 and S2 in File S1). We classified genes into different categories based on their cell-type expression patterns (Materials and Methods) (Figure 2B). Genes that had at least a twofold difference in expression between cell-types were classified as either gonidial or somatic genes based on whether they had higher expression levels in gonidial cells or somatic cells, respectively. Gonidial genes that had between a two- and fivefold gonidial/somatic expression ratio were considered gonidial-biased (688 genes, ∼5% of the transcriptome), and those with greater than a fivefold gonidial/somatic expression ratio were considered gonidial-specific (3410 genes, ∼25%). Somatic genes were classified in a similar manner into somatic-biased (378 genes, ∼3%) and somatic-specific subsets (2054 genes, ∼15%). Genes that had less than a twofold difference in expression between cell-types were classified as constitutive (3428 genes, ∼25%). An additional 3392 genes (∼25%) had cell-type expression ratios close to the twofold cutoff for classification as cell-type genes, but lacked statistical support for differential expression and were classified as low confidence (Figure 2B and Figure S2 in File S1).
The purity of cell-type-specific RNA preparations was validated by examining the expression pattern of a known somatic-specific gene, regA (Kirk et al. 1999), whose average transcript level in somatic cells (6.594 RPKM) was nearly 1000-fold higher than in gonidial cells, where it was at or near the detection limit (0.007 RPKM) (Datasets S1 and S2). Moreover, we identified numerous other genes whose expression was highly specific (>1000-fold expression ratio) for one or the other cell-type (Table S2 in File S1), a finding that rules out the presence of significant cross-contamination between cell-type preparations. Our data are also generally in agreement with those from previous studies that examined cell-type expression of a limited number of genes (Dataset S3).
Asymmetric expression profiles in gonidial vs. somatic cells
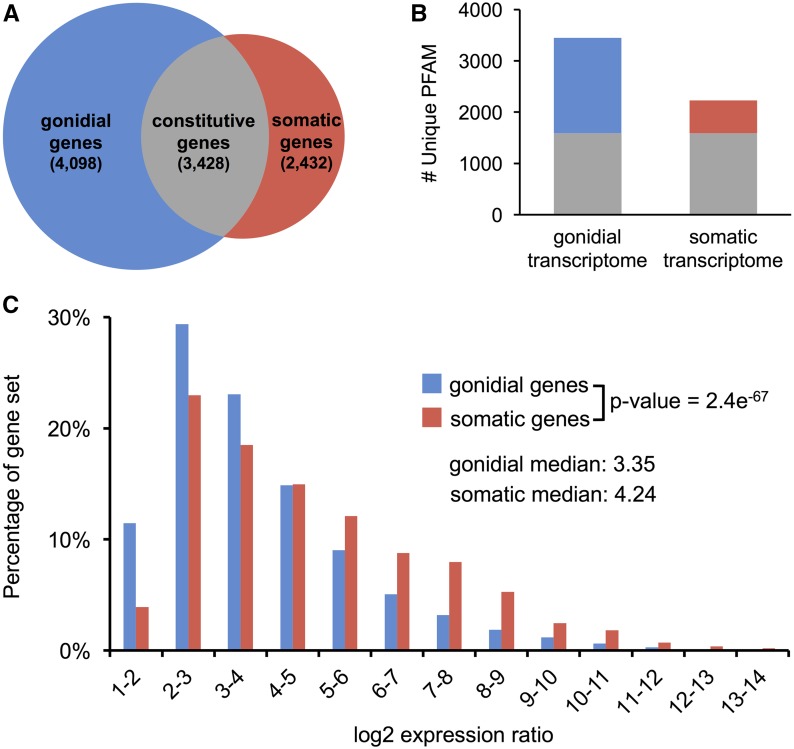
We found that over 6500 genes in V. carteri (∼50% of the expressed transcriptome) had significantly different expression levels between cell-types (Figure 3A, and Figure S2 and Dataset S1). Comparative metrics of cell-type expression patterns further revealed unanticipated asymmetry between the two cell-type transcriptomes. First, there were almost 1.7 times more gonidial genes (4098) than somatic genes (2432) (Figure 3A), with significantly more PFAM domains encoded by the gonidial transcriptome, both of which indicate greater gonidial vs. somatic transcriptome diversity (Figure 3B). On the other hand, somatic transcripts had a significantly higher degree of cell-type specificity (i.e., magnitude of expression bias) than gonidial transcripts (Figure 3C), indicating a greater degree of somatic cell-type specialization. Taken together, these findings demonstrate that the somatic transcriptome is more specialized than the gonidial transcriptome: somatic cells express fewer genes than gonidial cells, and the genes that are expressed in somatic cells are expressed in a more cell-type-specific manner. This somatic cell specialization pattern was retained even when genes related to flagella (Dataset S4) were removed from the analysis (Figure S3 in File S1), suggesting that specialization of the somatic transcriptome extends beyond flagella gene expression, which was examined in more detail below.
Figure 3.
Characterization of V. carteri germ and somatic transcriptomes. (A) Venn diagram showing the numbers of gonidial genes (G/S > 2) in blue, constitutively expressed genes in gray, and somatic genes (S/G > 2) in red. (B) Numbers of different PFAM domain identifiers associated with the gonidial and somatic transcriptomes are shown as stacked bar plots, and color coded as in (A). (C) Histograms of expression ratios (G/S or S/G) of gonidial vs. somatic genes. p-value of Wilcoxon rank sum test for the two distributions is indicated. The median log2 expression ratio for each gene set is also indicated. G, gonidial expression; S, somatic expression.
Functional classification of gonidial and somatic genes
We used MapMan annotations (Thimm et al. 2004), which were derived from C. reinhardtii genes and assigned to V. carteri genes based on orthology, to search for functional enrichment in the proteins encoded by the V. carteri gonidial and somatic transcriptomes (Materials and Methods). From 8763 functional assignments in V. carteri we identified 12 categories that were significantly enriched among gonidial genes, most of which were associated with biosynthetic processes including DNA synthesis, RNA processing, amino acid synthesis, protein assembly/cofactor ligation, and tetrapyrrole synthesis (Table S3 in File S1). In contrast, only three categories were enriched in somatic genes: motility (related to flagella genes), cell organization (also found among flagella/cytoskeletal related genes), and post-translational modification (mostly protein kinases and phosphatases). GO terms enriched in each gene set were consistent with MapMan enrichments, with signal transduction-associated functions also found to be enriched in the somatic gene set (Table S4 in File S1). Together, these classification data validate the known properties of the two cell-types, with gonidia enriched for expression of biosynthetic functions related to cell growth and somatic cells enriched for expression of motility functions.
Evolutionary origins of germ and somatic genes
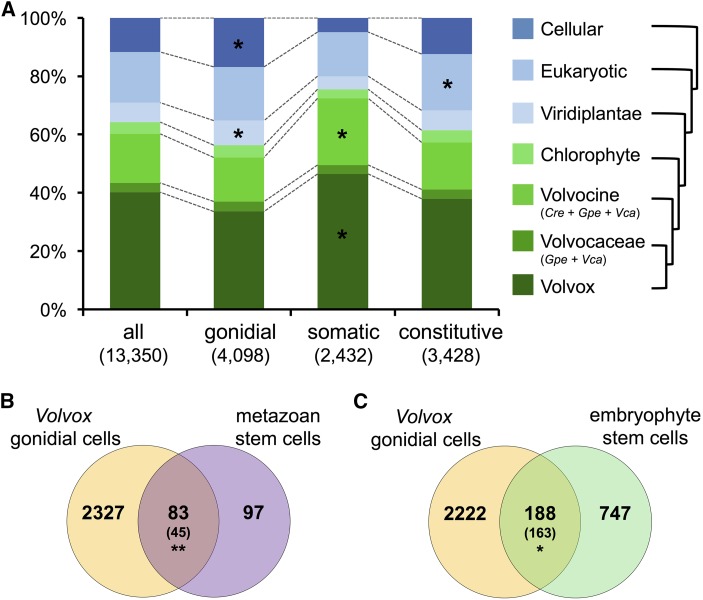
We next investigated the evolutionary origins of cell-type specialization in V. carteri using phylostratigraphy (Domazet-Loso et al. 2007; Domazet-Loso and Tautz 2010; Lopez et al. 2015; Hanschen et al. 2016). Briefly, well-annotated proteomes were collected from species with increasing divergence times from V. carteri and were grouped into a nested set of taxonomic categories or phylostrata: V. carteri (V. carteri only), Volvocaceae (multicellular volvocine algae: G. pectorale + V. carteri), all volvocine algae (C. reinhardtii + G. pectorale + V. carteri), chlorophyte green algae, viridiplantae (chlorophyte algae + streptophytes), all eukaryotes, and all cellular organisms (eukaryotes + prokaryotes + archaea) (Table S5 in File S1). The phylogenetic origin of each V. carteri protein-coding gene was then determined by identifying the most distantly related phylostratum that contained a homolog based on BLASTP searches that were customized for long-distance comparisons (Materials and Methods). Compared to the entire transcriptome, gonidial genes were enriched for those with ancient origins (genes conserved across all cellular organisms) and those originating within green eukaryotes (Viridiplantae). In contrast, somatic genes showed a reciprocal phylostratigraphic pattern, with a strong enrichment for lineage-specific genes that were found only in V. carteri or only in the volvocine algae (Figure 4A). Constitutively- expressed genes had a phylostratigraphic distribution similar to the background transcriptome, though they showed a modest enrichment for genes conserved across eukaryotes. Interestingly, no enrichments were observed for genes found only among the multicellular Volvocine algae (G. pectorale + V. carteri). Together, our comparative phylostratigraphy data show that somatic cells preferentially express a more derived set of lineage-specific genes, many of which arose in the most recent unicellular ancestor of V. carteri, while gonidial genes encode more deeply ancestral and conserved functions.
Figure 4.
Evolutionary genomics analyses of gonidial and somatic genes. (A) Phylostratigraphic profiling. Bar graphs show the fraction of genes from each V. carteri cell-type expression classification group in each phylostratum. Color key on right shows nested phylostrata progressing from the most recent phylostratum of V. carteri-specific genes on the bottom to the most ancient phylostratum of genes found across all cellular organisms at the top. Asterisk indicates significant enrichment compared to all expressed genes (* FDR < 0.01). (B and C) Venn diagrams showing overlap between OrthoMCL IDs associated with V. carteri gonidial cells and animal (metazoan) stem cells (B) or land plant (embryophyte) stem cells (C). Expected values are in parentheses. Asterisks indicates significant enrichment (** FDR < 0.01 and * FDR < 0.05). Cre, C. reinhardtii; FDR, false discovery rate; Gpe, Gonium pectorale; IDs, identifiers; Vca, V. carteri.
The older and more conserved origins of gonidial genes prompted us to compare the genetic program of V. carteri gonidia, a pluripotent cell-type, with the genetic programs of pluripotent cells from metazoans and land plants (embryophytes), both of which have independently evolved multicellularity. A previous study identified a core set of metazoan pluripotent stem cell genes based on shared OrthoMCL classifications among early-diverging animal taxa (Chen et al. 2006; Alié et al. 2015). We extended this analysis to include V. carteri cell-type genes and embryophyte apical stem cell genes that were previously found to be shared between maize shoot apical meristems and moss (P. patens) gametophore apical cells (Frank and Scanlon 2015) (Materials and Methods) (Figure 4, B and C). In this analysis we found a significant nearly twofold enrichment of OrthoMCL IDs shared between V. carteri gonidia and metazoan stem cells (83 observed vs. 45 expected) (Figure 4B), suggesting that these two lineages converged on similar networks of pluripotency-associated genes. There was also a modest but significant enrichment of OrthoMCL IDs shared between V. carteri gonidia and embryophyte stem cells (188 observed vs. 163 expected), whose total overlap was larger than with metazoan stem cells (188 vs. 83 IDs) but whose relative enrichment was less pronounced (Figure 4C). Interestingly, the greatest enrichment between V. carteri gonidia and embryophyte stem cells was in the subset of OrthoMCL IDs that were also shared with metazoan stem cells (Figure S4A in File S1). In contrast, most of the V. carteri–metazoan overlap for stem cell IDs was not shared with embryophytes. Additional two-way comparisons between metazoan or embryophyte stem cells with different V. carteri expression groups (somatic and constitutive + low-confidence) yielded complementary results, where we found strong deenrichment for overlap of V. carteri somatic cell OrthoMCL IDs and metazoan or embryophyte stem cell OrthoMCL IDs (Figure S4, B and C in File S1). V. carteri genes with little or no expression bias (constitutive + low-confidence genes) had modest deviations from expected OrthoMCL ID overlaps, showing deenrichment with metazoan stem cells and enrichment in embryophyte stem cells (Figure S4, D and E in File S1). Taken together, our data suggest commonality and convergence between stem cell genetic networks across the three multicellular lineages, with V. carteri gonidia and metazoan stem cells having retained a more significant proportion of overlap despite their greater divergence time, and with a larger magnitude of overlap between V. carteri gonidia and embryophyte apical stem cells. We also note that the metazoan stem cell data set of 180 OrthoMCL IDs (Alié et al. 2015) was more stringently derived than the embryophyte stem cell data set (935 OrthoMCL IDs) (Frank and Scanlon 2015), making it difficult to draw firm conclusions about the relative similarities among the three stem cell comparison groups. Nonetheless, our study shows that a comparative genomics approach to stem cell evolution may help refine our understanding of the origins and maintenance of pluripotency in different multicellular lineages.
In the following sections, we describe cellular and metabolic processes or pathways that are differentially expressed between gonidial and somatic cells, and which highlight the functional specializations of each cell-type. We then test a more general model of cooption based on conversion of temporally segregated gene expression patterns in a unicellular ancestor to cell-type segregated expression patterns in V. carteri.
Distinct gene expression patterns of flagellar, basal body, and transition zone (TZ) genes
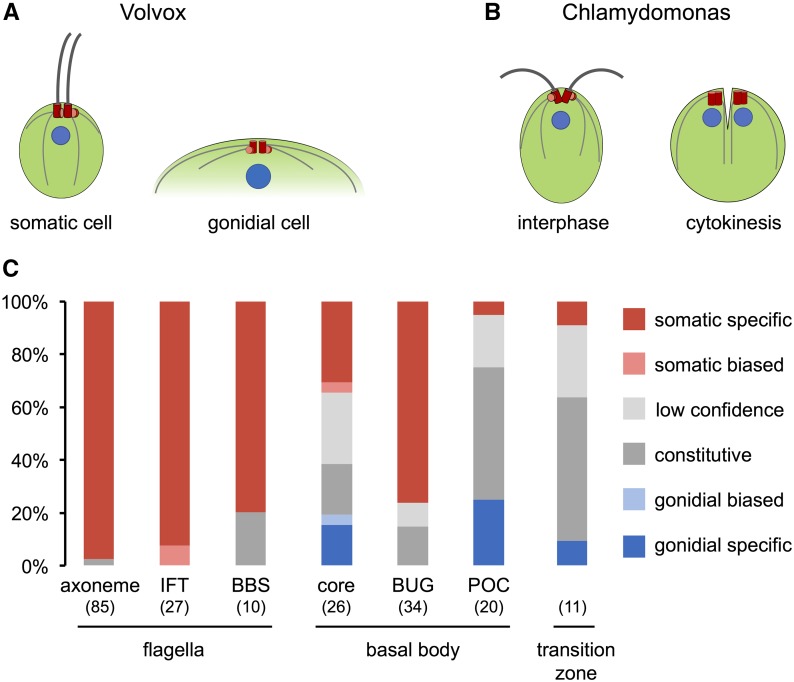
Besides their vastly different sizes, a distinctive derived feature of V. carteri cell-types is the absence and presence of flagella on gonidia and somatic cells, respectively (Figure 5A). In C. reinhardtii, flagella genes are coexpressed as a tightly coordinated regulon that is activated after flagella loss or during the cell cycle after flagella resorption and cell division (Stolc et al. 2005; Chamberlain et al. 2008; Albee et al. 2013; Zones et al. 2015). It seemed possible that the entire flagella regulon of C. reinhardtii might be conserved in V. carteri and was coopted to be active only in somatic cells. Alternatively, suppressed expression of one or a few key flagella biogenesis genes in gonidia could also completely block flagella formation (McVittie 1972; Huang et al. 1977; Huskey et al. 1979; Huskey 1979; Adams et al. 1982; Tam and Lefebvre 1993; Rosenbaum and Witman 2002). A third possibility is that flagella biogenesis is controlled post-transcriptionally in V. carteri, in which case no significant differences in flagella-related mRNA abundance would be observed between cell-types. As shown in Figure 5C, nearly all flagella genes in V. carteri were somatic-specific, a result that is consistent with the flagella regulon cooption model (Figure 5C and Dataset S4).
Figure 5.
Expression patterns of flagella, basal body, and transition zone genes. (A) Cartoon diagrams of the cytoskeletal apparatus from a V. carteri somatic cell (left) and the apical portion of a V. carteri gonidial cell (right). Flagella and microtubule rootlets are shaded dark and light gray, respectively, nuclei are shaded blue, mature basal bodies are shaded red, and probasal bodies shaded pink/red. (B) Cartoon diagrams of interphase (left) and dividing (right) C. reinhardtii cell shaded as described in (A). Interphase basal bodies in C. reinhardtii are arranged with 180° symmetry, causing the flagella to beat in opposite directions, while basal bodies in a V. carteri somatic cell are each rotated 90° relative to those in C. reinhardtii, causing the flagella to beat in parallel. During cytokinesis, C. reinhardtii cells resorb flagella and basal bodies function as centrioles to coordinate positioning of the cleavage furrow between daughter nuclei. (C) Stacked bar graph shows the fraction of flagella-related genes [axonemal genes, intraflagellar transport (IFT) genes, and Bardet–Biedel Syndrome (BBS) genes], basal body genes (core genes with validated function/localization, BUG genes, and POC genes), and transition zone genes (see Dataset S4) in each cell-type expression classification shown in the color key on the right. Number of genes for each group is in parentheses.
Flagella biogenesis also requires basal bodies that template axoneme assembly, position the flagella at the cell’s anterior pole, and organize the internal microtubule cytoskeleton (Dutcher 2003; Marshall 2008). In ciliated or flagellated cells, including C. reinhardtii, basal bodies have dual functions that are mutually exclusive: during interphase they template the assembly of cilia/flagella, while during cell division they replicate and help spatially coordinate mitosis and cytokinesis (Figure 5B) (Coss 1974; Ehler et al. 1995; Marshall and Rosenbaum 2000; Simons and Walz 2006; Dawe et al. 2007; Parker et al. 2010). Since gonidia are flagella-less, yet undergo cell division, and somatic cells are motile, yet do not divide, expression of different basal body gene subsets corresponding to cell division or flagella biogenesis functions might also be segregated between the cell-types. Most of the core basal body genes (those with validated basal body function) did not show a clear pattern of preferential cell-type expression, with many being constitutive or low confidence (Figure 5C). We also searched for cell-type expression biases among V. carteri orthologs of less well-characterized candidate basal body genes identified through two proteomics surveys in C. reinhardtii (Keller et al. 2005, 2009). Candidate basal body proteins from these studies were split into two sets based on validation methods: BUG genes were upregulated during flagellar assembly and may be related to flagella function, while POC genes were conserved in species with centrioles and are expected to be related to centriolar and cell division functions (Keller et al. 2005, 2009). Consistent with these validation methods, most BUG genes in V. carteri were preferentially expressed in somatic cells where flagella functions dominate, while most POC genes in V. carteri were either constitutively expressed or preferentially expressed in gonidia where core replication and mitotic functions of basal bodies are required (Figure 5C). These data suggest that cell-type expression patterns in V. carteri could be useful as a functional filter for distinguishing flagella/motility-related functions and cell division/replication-related functions of basal body proteins.
Lastly, we examined the cell-type expression patterns of genes encoding proteins of the TZ, a distinct region near the base of each flagellum that is connected to the plasma membrane by transition fibers and which is thought to serve as a protein diffusion barrier between the flagellum and cytosol (Craige et al. 2010; Awata et al. 2014; Dutcher and O’Toole 2016). In C. reinhardtii, the TZ protein-encoding genes were expressed with high periodicity just prior to or during cell division (Zones et al. 2015). Interestingly, V. carteri orthologs of most C. reinhardtii TZ genes were expressed at similar levels in somatic and gonidial cells, suggesting a requirement for TZ protein function in both cell-types (Figure 5C). The expression of TZ genes in aflagellate gonidia may reflect a cryptic requirement for a TZ or related structure in this cell-type, a need to stockpile TZ proteins prior to their deployment in the next generation of somatic cells, or additional nonflagella-specific roles for TZ proteins (e.g., vesicle trafficking), as was predicted in C. reinhardtii (Diener et al. 2015).
Differential photosynthetic gene expression in gonidial and somatic cells
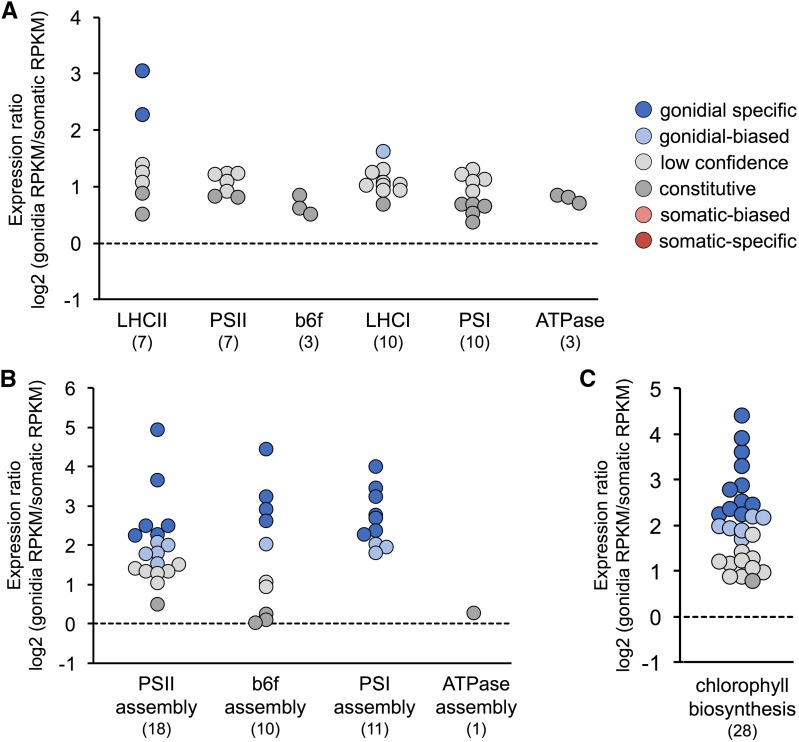
Previous studies of cell-type gene expression in V. carteri led to a hypothesis that photosynthetic gene expression is suppressed in somatic cells, which in turn blocks or slows cell growth and thereby ensures terminal differentiation (Tam and Kirk 1991; Choi et al. 1996; Meissner et al. 1999). We revisited this hypothesis by examining the cell-type expression patterns of genes in V. carteri that are predicted to encode photosynthesis-related proteins. In our transcriptome data, nearly all core photosynthetic complex genes corresponding to subunits of LHCII, photosystem II (PSII), cytochrome b6f complex (b6f), LHCI, photosystem I (PSI), and ATP synthase (ATPase) were expressed at high levels in both cell-types, and were classified as constitutive or low confidence, demonstrating that they did not show strong cell-type expression biases (Figure 6A and Dataset S5). Nonetheless, collectively they exhibited a clear pattern of 1.5–2.5-fold higher expression in gonidia than in somatic cells, suggesting that photosynthesis genes are expressed with a modest gonidial preference, but are not strongly cell-type regulated as suggested by earlier studies. Exceptions to this general finding included one major LHCII gene family (subclade I containing 19 paralogs) and a minor LHCII gene (LHCB7) having gonidial-specific expression patterns, and a variant LHCI paralog, LHCA3.2, which is found in V. carteri but not C. reinhardtii, having gonidial-biased expression (Figure S5 and Dataset S5). These exceptions might indicate differential fine-tuning of photosynthetic antenna function between V. carteri cell-types. In contrast to the genes encoding core subunits of photosynthetic complexes, many genes encoding photosynthetic complex assembly factors and chlorophyll biosynthesis enzymes were gonidial-biased or gonidial-specific in expression (Figure 6, B and C), suggesting a greater requirement for photosynthetic complex assembly and/or repair in gonidial cells.
Figure 6.
Cell-type expression patterns of photosynthesis-related genes. Each dot represents a single gene (see Dataset S5) whose expression category is color-coded according to the key at the top right. y-axis is log2-transformed gonidial/somatic expression ratio. Dashed line demarcates a 1:1 gonidial/somatic expression ratio. Numbers of genes for each subgroup are in parentheses. (A) Subunits of indicated complexes carrying out photosynthetic light reactions. (B) Assembly factors for indicated photosynthetic complexes. (C) Chlorophyll biosynthesis enzymes. RPKM, reads per kilobase per million mapped reads.
Most genes encoding enzymes of the photosynthetic dark reactions [also known as the Calvin–Benson–Bassham (CBB) cycle] did not exhibit strong cell-type expression patterns, though the small subunits of the Rubisco enzyme complex (RBCS1–RBCS7), collectively showed a modest twofold gonidial bias in expression (Figure 7 and Dataset S6). Two other genes encoding CBB enzymes, RPI1 and TPIC1, were also found to be preferentially expressed in gonidial cells. In summary, our data for photosynthetic gene expression do not support the hypothesis that photosynthesis is strongly suppressed in somatic cells, but do suggest a greater requirement for photosynthetic complex assembly and/or repair in gonidia.
Carbon metabolism gene expression in germ and somatic cells
To gain further insight into the metabolic bases of germ–soma differentiation, we investigated the cell-type expression patterns of genes encoding enzymes involved in carbon metabolism (Materials and Methods; Dataset S6). Here, we found a clear dichotomy between carbon storage anabolism and catabolism in the two cell-types (Figure 7). Two major sinks for fixed carbon in green algae are fatty acids and starch. Nearly all genes encoding enzymes for fatty acid biosynthesis, starting with those for the plastidic pyruvate dehydrogenase complex that generates acetyl-CoA, were gonidial-specific. Conversely, genes encoding the fatty acid breakdown enzymes acyl-CoA oxidase (ACX1-ACX4) and enoyl-CoA hydratase (ECH1) were either somatic-specific or somatic-biased. A similar anabolic/catabolic dichotomy was observed for starch metabolism with preferential expression of genes for starch biosynthetic enzymes in gonidial cells—most notably STA1 that encodes ADP-glucose pyrophosphorylase, which catalyzes the rate-limiting step for starch accumulation (Ball et al. 1991; Zabawinski et al. 2001). Conversely, genes encoding amylase enzymes (AMB1-AMB3) that catalyze the hydrolysis of starch were preferentially expressed in somatic cells.
Glycolysis and gluconeogenesis share most enzymes, but differ in two key steps: interconversion of fructose-6-phosphate and fructose-1,6-bisphosphate, which is catalyzed by either phosphofructokinase (PFK, glycolysis) or fructose-1,6-bisphosphatase (FBP, gluconeogenesis); and synthesis/breakdown of phosphoenolpyruvate, which is catalyzed by pyruvate kinase (PYK, glycolytic breakdown into pyruvate) or phosphoenolpyruvate carboxykinase (PCK, gluconeogenic synthesis from oxaloacetate). PYK and PCK genes showed opposing cell-type expression patterns, with PYK genes being gonidial-biased and PCK genes being somatic-specific. PFK did not show cell-type preference, but FBP1 was somatic-specific (Figure 7). Taken together, these profiles suggest opposing directions of net carbon flow through glycolysis in gonidia and through gluconeogenesis in somatic cells.
Consistent with the glycolysis/gluconeogenesis gene expression dichotomy, we found that expression of TCA cycle and glyoxylate cycle genes was also cell-type-biased. Genes encoding enzymes or subunits of complexes that catalyze the first four out five reactions specific to the TCA cycle—pyruvate dehydrogenase (PDH1 and DLD1), citrate synthase (CIS1), isocitrate dehydrogenase (IDH1–IDH3), and oxoglutarate dehydrogenase (DLD1–DLD2)—were expressed preferentially in gonidia (Figure 7 and Dataset S6). TCA cycle intermediates are also used for amino acid biosynthesis, and genes encoding enzymes that catalyze two major amino acid biosynthetic entry points, glutamate synthase (GSN1) and aspartate aminotransferase (AST1–AST3 and AST5), were also gonidial-specific. Conversely, glutamate dehydrogenase paralogs (GDH1 and GDH2), which function in amino acid catabolism, had somatic-specific expression. The glyoxylate cycle makes use of some TCA cycle enzymes but bypasses the decarboxylation steps, and is often coupled to fatty acid catabolism and gluconeogenesis to produce sugars (Eastmond and Graham 2001; Graham 2008). Genes encoding the two dedicated enzymes of the glyoxylate cycle, isocitrate lyase (ICL1) and malate synthase (MAS1), were expressed specifically in somatic cells, suggesting preferential use of the glyoxylate cycle over the TCA cycle. Our finding that genes encoding enzymes involved in fatty acid breakdown, the glyoxylate cycle, and gluconeogenesis were all preferentially expressed in somatic cells suggests that these pathways function coordinately to promote sugar biosynthesis in somatic cells (Figure 7, red arrows; see Discussion).
Together, our analyses of central carbon metabolism gene expression are consistent with a well-orchestrated dichotomy in metabolism between V. carteri cell-types, with gonidial cell metabolism geared towards cell growth and anabolism, and somatic cell metabolism geared towards starch breakdown and conversion of fatty acids into sugar through the glyoxylate cycle and gluconeogenesis.
The sink for somatic cell sugar biosynthesis may be the extracellular matrix
Because somatic cells showed relatively lower expression of starch biosynthetic enzymes and higher expression of starch hydrolysis enzymes compared with gonidial cells (Figure 7 and Dataset S6), it seemed unlikely that sugars derived from gluconeogenesis in somatic cells would be used for starch biosynthesis. An alternative sink for sugars in somatic cells is the extracellular matrix (ECM), which comprises the vast majority of the spheroid volume (>99%) and which is composed primarily of glycoproteins (Kirk et al. 1986; Sumper and Hallmann 1998; Hallmann 2003). If the ECM is the sugar sink of somatic cells, then we might expect to see genes involved in protein glycosylation and genes encoding ECM glycoproteins to be expressed preferentially in somatic cells compared to gonidial cells. We first examined genes encoding enzymes involved in the biosynthesis of nucleotide sugars (UDP-xylose, UDP-arabinose, UDP-galactose, UDP-glucose, and GDP-mannose) that serve as sugar donors for cell wall/ECM glycosylation in volvocine algae (Sumper and Hallmann 1998; Hallmann 2003; Seifert 2004; Harris 2009). Genes encoding five nucleotide sugar biosynthesis enzymes—phosphomannose isomerase (PMI1), GDP-D-mannose 3′, 5′-epimerase (GME1), UDP-glucose dehydrogenase (UGD1-UGD3), UDP-xylose synthase (UXS1), and UDP-arabinopyranose mutase (RGP1-RGP3)—were expressed preferentially in somatic cells (Figure 7 and Dataset S7). The only genes involved in nucleotide sugar biosynthesis that were expressed preferentially in gonidia—phosphoglucose isomerase (PGI1) and phosphoglucomutase (GPM1 and GPM2)—also play a central role in starch biosynthesis, which is likely to be preferentially expressed in gonidia, as described above (Figure 7 and Dataset S6).
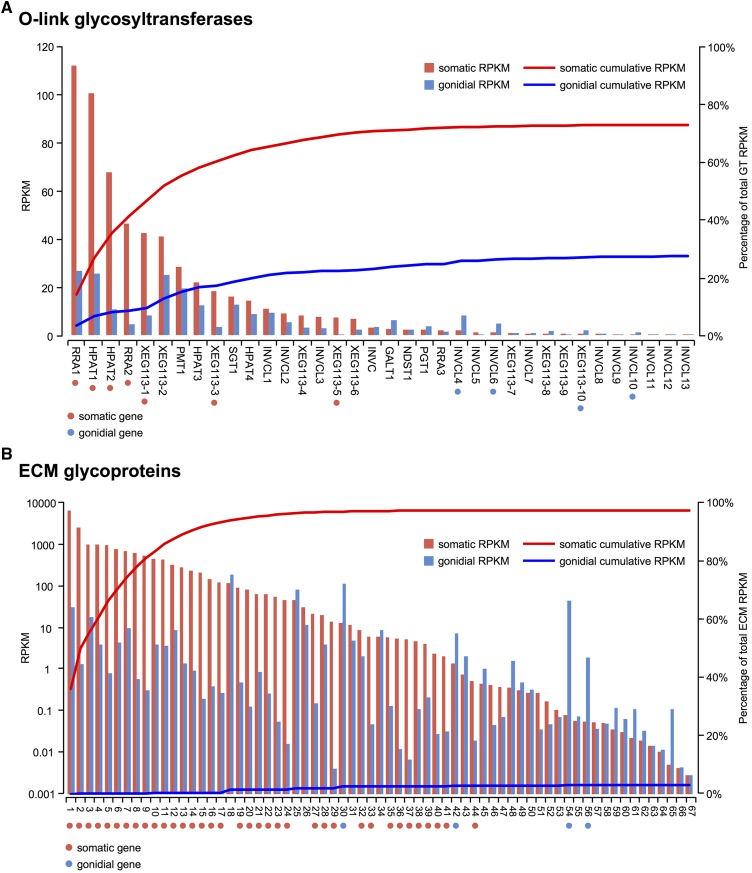
We also assessed expression patterns of genes encoding glycosyltransferases that are involved in O-linked glycosylation and are predicted to glycosylate the hydroxyproline-rich glycoproteins (HRGPs) that comprise most of the V. carteri ECM (Materials and Methods) (Sommer-Knudsen et al. 1998; Sumper and Hallmann 1998; Hallmann 2003; Showalter and Basu 2016). Out of the 36 predicted glycosyltransferase-encoding genes in V. carteri, seven were somatic and four were gonidial. However, >73% of total glycosyltransferase gene expression summed over all 36 genes was found in somatic cells, suggesting a higher demand for this activity in soma (Figure 8A and Dataset S8).
Figure 8.
V. carteri ECM-related gene expression. Combined bar and cumulative expression plots of somatic (red) and gonidial (blue) expression for genes encoding predicted glycosyltransferases (GTs) involved in O-linked glycosylation (A) (Dataset S8) or ECM-targeted glycoproteins (B) (Dataset S9). Bars indicate expression levels of genes in RPKM (y-axis, left side) in each cell-type and lines indicate cumulative expression in each cell-type, summed across all genes (left to right) as percentages of total expression (y-axis, right side). Note that the y-axis for RPKMs in (B) is log-scaled. Red and blue dots mark somatic and gonidial genes, respectively. ECM, extracellular matrix; RPKM, reads per kilobase per million mapped reads.
Finally, we assessed cell-type expression patterns of predicted ECM glycoprotein genes and found that 36/67 genes were somatic, while only 4/67 were gonidial. Moreover, >95% of total ECM gene expression (summed expression across all ECM genes) was in somatic cells (Figure 8B and Dataset S9).
To test more generally whether somatic cells might be specialized for secretion, we predicted the subcellular localization of all V. carteri proteins (Materials and Methods) and then grouped them with respect to cell-type expression pattern. In doing so, we found a significant enrichment of predicted secreted or endomembrane-targeted proteins among somatic genes, while gonidia were enriched for expression of predicted chloroplast- and mitochondrial-targeted proteins (Figure S6 in File S1). Together, our findings suggest that somatic cells are specialized for the production and secretion of ECM glycoproteins, and that a major sink for somatic cell sugars is the ECM.
Testing the temporal–spatial cooption hypothesis for the origin of cell-type gene expression in V. carteri
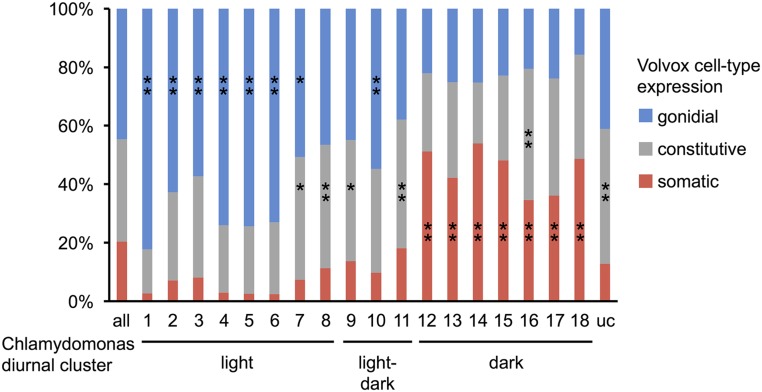
As detailed above, V. carteri genes related to flagellar motility, photosynthesis, central carbon metabolism, and ECM secretion exhibited coherent expression patterns, with many showing a clear germ–soma dichotomy. Interestingly, C. reinhardtii orthologs for most of these genes showed strong and coordinate periodic expression during a diurnally synchronized cell cycle (Zones et al. 2015). This correlation between cell-type expression patterns in V. carteri and diurnal expression patterns in C. reinhardtii suggests that cell-type gene expression programs in V. carteri might have originated through cooption of preexisting temporal expression programs in a C. reinhardtii-like unicellular ancestor. To more directly test for temporal–spatial cooption, we investigated the relationship between diurnal gene expression in C. reinhardtii and cell-type gene expression in V. carteri. We identified V. carteri orthologs of C. reinhardtii genes from previously characterized diurnal gene expression clusters (Zones et al. 2015) and assessed their cell-type expression patterns. If temporal clustering in C. reinhardtii were unrelated to cell-type expression in V. carteri, then we would expect to see a similar representation of V. carteri expression patterns within each C. reinhardtii temporal cluster. On the contrary, we found a strong relationship between diurnal gene expression in C. reinhardtii and cell-type gene expression in V. carteri: C. reinhardtii genes belonging to light-phase clusters (c1–c8) were enriched for genes whose V. carteri orthologs were gonidial, while C. reinhardtii genes belonging to dark-phase clusters (c12–c18) were enriched for genes whose V. carteri orthologs were somatic (Figure 9). We performed a reciprocal version of this enrichment test by first grouping C. reinhardtii diurnal genes into four major superclusters (light-phase, light/dark transition, dark-phase, and unclustered) (Materials and Methods), and then determined the composition of each supercluster based on orthology to V. carteri cell-type genes. Again, we found significantly skewed distributions where C. reinhardtii orthologs of V. carteri gonidial genes were enriched for light-phase supercluster assignments, while C. reinhardtii orthologs of V. carteri somatic genes were enriched for dark-phase supercluster assignments (Figure S7A in File S1). These results were essentially unchanged when we removed flagella-associated genes from the analysis, indicating that the diurnal and cell-type expression patterns of the ∼120 flagella-related genes in the two species (Dataset S4) were not solely responsible for the results we observed supporting cooption (Dataset S4) (Figure S7B in File S1). Together, our results are consistent with large-scale cooption, where regulons that exhibited diurnal regulatory patterns in a unicellular ancestor came under cell-type control in V. carteri.
Figure 9.
Cell-type expression patterns of V. carteri orthologs of diurnally expressed C. reinhardtii genes. Stacked bar plots show fractions of V. carteri genes from each cell-type expression classification whose corresponding orthologs in C. reinhardtii belong to the indicated diurnal cluster [all genes, c1–c18, or unclustered (uc) (Zones et al. 2015)]. Clusters are arranged in temporal order with peak expression within the light-, light–dark transition, or dark- phase of the diurnal cycle indicated below groups of clusters. Color key is shown to the right. Significant enrichment of V. carteri cell-type expression within each C. reinhardtii diurnal cluster is indicated as follows: ** FDR < 0.01 and * FDR < 0.05.
Discussion
Although germ–soma differentiation has been studied extensively in animals and plants, very little is known about the early evolution of germ–soma specialization, especially the nature of genetic programming required to initially achieve this fundamental cell-type dichotomy. As discussed below, our transcriptome data from V. carteri has yielded insights about this key innovation that may apply more broadly to other multicellular lineages outside of volvocine algae.
An asymmetric germ–soma dichotomy may be a convergent trait
Our data revealed not only extensive differential expression between V. carteri cell-types (50% of expressed genes were cell-type-regulated), but also an unanticipated asymmetry in expression patterns. V. carteri germ cells, though specialized for reproduction, are more generalist in their overall genetic programming as they express a significantly larger transcriptome than somatic cells, but with less overall expression bias (i.e., degree of cell-type specificity) (Figure 3). We also found that gonidia express a more conserved and ancient set of genes than do somatic cells (Figure 4A). This generalist and ancestral nature of the gonidial transcriptome may be a convergent trait in the transcriptomes of pluripotent cell-types. Similar to V. carteri, pluripotent embryonic stem cells in mammals and pluripotent adult stem cells in early diverging metazoan taxa express more genes than their differentiated daughter cells (Brandenberger et al. 2004; Miura et al. 2004; Denis et al. 2011; Hemmrich et al. 2012; Gan et al. 2014; Zhao et al. 2014; Alié et al. 2015), and preferentially express genes with phylogenetically older origins (Hemmrich et al. 2012; Alié et al. 2015). Though transcriptomic analyses have been conducted on the pluripotent stem cell populations of land plants (Nawy et al. 2005; Yadav et al. 2009, 2014; Frank and Scanlon 2015), comparable pairwise analyses of stem cells and specific differentiated daughter lineages have not been reported for land plants. For example, the land plant stem cell gene datasets used for our analyses (Figure 4, B and C and Figure S4 in File S1) were derived from overlapping genes upregulated in the stem cell populations of Z. mays or P. patens relative to whole-plant samples (Frank and Scanlon 2015). Our finding that a significant fraction of pluripotency-associated genes is conserved between V. carteri, animals, and land plants (Figure 4, B and C and Figure S4A in File S1) suggests potential convergent evolution of pluripotency, and may indicate a common underlying set of constraints that shaped stem cell evolution in these divergent lineages of multicellular eukaryotes.
Lineage-specific genes and the early evolution of differentiated cell-types
All multicellular taxa trace their roots back to unicellular ancestors whose genetic toolkits were exploited and modified to solve a set of problems associated with multicellular organization, such as cell adhesion, morphogenesis, and cell-type specialization (Cock et al. 2010; Richter and King 2013; Umen 2014). While genes whose origins are restricted to a multicellular clade are obvious candidates for encoding multicellular innovations, genes shared by a multicellular clade and its closest unicellular relatives (but not found outside this grouping) may also have special significance for understanding the transition to multicellularity, as they represent clade-specific adaptations that arose in a common ancestor and persisted in both unicellular and multicellular descendants.
Our germ–soma transcriptome data suggests that lineage-specific genes did play a large role in the evolution of cell-type specialization in V. carteri where two categories of such genes were disproportionately expressed in somatic cells: (1) those that were found only in V. carteri, meaning that they arose in the V. carteri clade after the transition to simple multicellularity; and (2) those shared with C. reinhardtii but not elsewhere, meaning that they arose in a recent common unicellular ancestor of all volvocine algae (Figure 4A). Absent from somatic enrichment were genes found exclusively in multicellular volvocine algae (V. carteri + G. pectorale), suggesting that the program of somatic cell differentiation did not strongly rely on genes that originated during the transition to simple multicellularity (i.e., typified by G. pectorale colonies that have undifferentiated groups of cells).
Category 1 somatic cell genes (those exclusive to V. carteri) evolved during the evolution of germ–soma differentiation, a trait found in two volvocine genera, Volvox and Pleodorina. While complete germ–soma differentiation is a synapomorphy in the clade containing V. carteri (three V. carteri forma plus V. obversus), partial germ–soma differentiation may have evolved more than once in the volvocine tree and was also likely to have been lost in some sublineages (Herron and Michod 2008). Future studies of cell-type gene expression may shed light on whether lineage-specific genes were recruited for somatic differentiation in other volvocine species where germ–soma division of labor was independently gained or lost.
Category 2 somatic cell genes (those that are pan-volvocine) presumably arose as genetic specializations in a unicellular ancestor, and were subsequently coopted or redeployed for somatic cell differentiation. Our finding that a large fraction of cell differentiation genes in V. carteri originated in a close unicellular ancestor parallels findings in metazoans, where much of the apparently specialized genetic toolkit associated with animal multicellularity also first evolved in unicellular ancestors that were similar to present-day choanoflagellates (Rokas 2008; Ruiz-Trillo et al. 2008; Richter and King 2013). Our data not only help to generalize the relationship between unicellular clade-specific innovations and multicellular innovation, but further suggest that the earliest stages of cell-type specialization may be strongly dependent on lineage-specific genes that provide a functional connection between the unicellular and multicellular lifestyles (Figure 4A). For example, V. carteri somatic cells resemble free-living, nonmitotic, C. reinhardtii cells just as choanocytes—a specialized cell-type in multicellular sponges—resemble free-living extant choanoflagellates (Kochert and Olson 1970; Coleman 2012; Mah et al. 2014). Fewer data are available for land plants and their closest unicellular relatives, the charophyte algae, but several studies indicate that the genetic bases for important land plant multicellular/developmental pathways, such as those for phytohormone biosynthesis and signaling, were present in early charophytes (Becker and Marin 2009; Hori et al. 2014; Umen 2014; Bowman et al. 2017).
Cooption of temporal gene expression programs for cell-type differentiation
It has been previously proposed that the evolution of cell-type differentiation in multicellular organisms involved the cooption of different life cycle stages of a unicellular ancestor (Wolpert 1990, 1992; Nedelcu and Michod 2004; Mikhailov et al. 2009; Nedelcu 2012). The C. reinhardtii vegetative life cycle has strong temporal programming, with transitions between a motility/cell growth phase and an immotile cell division phase (Cross and Umen 2015). Koufopanou (1994) proposed a flagellar constraint hypothesis to explain the evolution of germ–soma segregation in the largest volvocine genera, where the evolution of permanently motile somatic cells allowed germ cells to escape the presumed loss-of-fitness associated with long periods of immotility during division (Koufopanou 1994). A similar trade-off between reproduction and motility has been proposed as a selective pressure for the evolution of cell-type differentiation in animals (Buss 1987; King 2004).
In our study, genes encoding flagella proteins were a clear example supporting temporal–spatial cooption, as they showed strong diurnal regulation in C. reinhardtii (Wood et al. 2012; Zones et al. 2015) and strong differential expression between V. carteri cell-types (Figure 5). More broadly, we found extensive evidence for a general pattern of temporal–spatial expression cooption based on comparisons of C. reinhardtii cell cycle/diurnal cycle genes (likely similar to an ancestral unicellular program) and V. carteri cell-type genes (Figure 9 and Figure S7 in File S1). Importantly, the V. carteri gonidial program was most closely associated with light-phase regulons in C. reinhardtii, while the V. carteri somatic program was most closely associated with dark-phase regulons in C. reinhardtii, even though our cell-type transcriptome samples were taken during the light-phase of a diurnal cycle. These differences are unlikely to reflect a complete blockage of light-based gene expression programs in somatic cells that retained relatively high levels of photosynthetic complex gene expression (Figure 6 and Dataset S5), but instead indicate a rewiring of metabolism to promote catabolic processes that typically occur in the dark phase for C. reinhardtii and which might also reduce or prevent net cell growth and proliferation of somatic cells. While our results support temporal–spatial cooption as a model for the origin of cell-type expression programs, this cooption could occur by a number of different mechanisms. One possibility is that temporal programs in the unicellular ancestor were converted into fixed spatial programs in V. carteri, such that the temporal regulatory cues of these programs were completely replaced by developmental cues. A second possibility is that the temporal programs were converted into spatial programs through phase-shifting of ancestral temporal patterns between V. carteri cell-types. However, phase-shifting seems like a less plausible scenario, as there are many functions that are either adaptively tied to a specific diurnal cue (e.g., photosynthetic machinery expression in light) or would never be appropriate to be expressed in one cell-type vs. the other, regardless of relative timing (e.g., cell division genes in somatic cells). A more extensive time course will be required to conclusively distinguish phase shifting vs. fixed expression differences between cell-types. Overall, very little is known about how diurnal and other gene expression programs are regulated in volvocine algae, though many transcription factor genes have diurnally controlled expression profiles in C. reinhardtii (Zones et al. 2015), including RLS1 (Nedelcu 2009) whose V. carteri homolog, regA, is a master regulator of somatic cell fate (Kirk et al. 1999; Duncan et al. 2006, 2007). Our results suggest widespread cooption of temporal gene expression programs in V. carteri for cell-type gene expression, and further predict that some form of cell-type control has evolved for V. carteri orthologs of transcription factors that govern diurnal regulons in C. reinhardtii.
A model for somatic cell metabolic specialization in V. carteri
Previous studies of differential gene expression in V. carteri led to a model for somatic cell differentiation based on suppression of photosynthetic gene expression (Tam and Kirk 1991; Choi et al. 1996; Meissner et al. 1999). Our data indicate that this model must be revised. While we did see an approximately twofold higher expression of core photosynthetic complex genes in gonidia vs. somatic cells (Figure 6A), photosynthetic gene transcripts were still among the most abundant in somatic cells (Dataset S5). Our results and those of a previous study (Pommerville and Kochert 1982) suggest that both V. carteri cell-types actively support photosynthesis, with a higher capacity in gonidial cells to support active cell growth. The elevated expression of photosynthetic complex assembly factors and chlorophyll biosynthetic enzymes in gonidia is consistent with their faster rates of growth and rates of chloroplast DNA synthesis compared to somatic cells (Coleman and Maguire 1982; Kirk 1998) (Figure 6, B and C).
While our data did not support the photosynthesis suppression hypothesis, they did suggest that gonidial and somatic cells have very different metabolic programs. Based on expression profiles, gonidial metabolism was geared toward anabolic processes such as starch, lipid, and amino acid biosynthesis, while somatic cell metabolism was programmed to couple starch and fatty acid breakdown to the glyoxylate cycle and gluconeogenesis to facilitate sugar biosynthesis (Figure 7, Figure 10, and Table S3 in File S1). The predicted somatic cell metabolic profile we inferred is similar to that of germinating oil seeds in land plants where fatty acid oxidation, the glyoxylate cycle, and gluconeogenesis are coordinately upregulated to convert stored lipids into sugars (Canvin and Beevers 1961; Runquist and Kruger 1999; Eastmond and Graham 2001; Rylott et al. 2001; Graham 2008). Somatic cell metabolism also resembles that of senescing leaves, where fatty acid breakdown is coupled to the glyoxylate cycle or to the TCA cycle to promote the biosynthesis and export of sugars or amino acids (Troncoso-Ponce et al. 2013). Autophagy is known to be induced during leaf senescence, where it is involved in breaking down chloroplasts to recycle nutrients (Schippers et al. 2015). An examination of V. carteri homologs of autophagy (ATG) genes in our study did not reveal a coherent bias toward expression in either cell-type (Figure S8 and Dataset S10), though we did see significantly higher expression of the autophagy marker gene ATG8 in somatic cells (72 RPKM) vs. gonidia (9 RPKM). Because the absolute somatic expression level of V. carteri ATG8 is within the range observed for rapidly proliferating C. reinhardtii cells (Zones et al. 2015), the significance of its higher expression in somatic cells vs. gonidia is unclear. It may be that autophagy plays a clearer or more prominent role in somatic cell biology later in the somatic life cycle, when cells are older and have exhausted nutrient stores.
Figure 10.
Model for gonidial and somatic cell metabolic specialization. Gonidia (left) have active photosynthesis as well as production of new photosynthetic machinery to support growth metabolism with net production of starch, lipids, and amino acids. Glycolysis and the TCA cycle are the preferred route for carbon flow in gonidial cells. Somatic cells (right) maintain active photosynthesis at somewhat reduced levels compared to gonidia, and with reduced production of new photosynthetic machinery. Lipids are broken down and metabolized through the glyoxylate cycle and gluconeogenesis to make sugars. Starch is also catabolized into sugar monomers which are used as substrates to produce excreted ECM glycoproteins. ECM, extracellular matrix; TCA, tricarboxylic acid.
Our data not only suggest a coordinated program of catabolism in somatic cells to produce sugars, but also a potential sink for those sugars as ECM glycoproteins. This idea is supported by expression data for nucleotide sugar biosynthetic genes, glycosyltransferase genes, and ECM glycoprotein genes, many of which were strongly expressed in somatic cells (Figure 7, Figure 8, and Dataset S7, S8 and S9). Thus, we propose that somatic cells—which are terminally differentiated and destined to senesce and die—slowly cannibalize themselves to generate an extensive ECM, while at the same time maintain moderately active photosynthesis to power basal metabolism, flagellar motility, and secretory activity (Figure 10). This model for germ–soma metabolic specialization in V. carteri provides some insight into the metabolic basis for how reproductive altruism might have evolved in the volvocine algae and possibly in other multicellular lineages. During the evolution of germ–soma differentiation, the metabolism of putative somatic cells was shifted away from cell growth-related processes and instead toward secretion that supports spheroid expansion. Secretory functions of somatic cells may also serve to support growth of gonidial cells, as previous work has shown that gonidia within intact spheroids grow faster than isolated gonidia (Koufopanou and Bell 1993). The metabolic model we propose for V. carteri cell-type specialization provides a framework for further investigation of how reproductive division of labor evolved within a simple multicellular organism.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.300253/-/DC1.
Acknowledgments
We thank David Kirk for advice on V. carteri cytology, Sa Geng for advice and help with culturing V. carteri, James Matt Zones for advice on bioinformatics, Takashi Hamaji for advice on phylostratigraphy, Margaret Frank for advice on stem cell comparative genomics, Jie Li for laboratory support, and the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for help with genomic analysis. The Center is partially supported by National Cancer Institute Cancer Center Support grant #P30 CA91842 to the Siteman Cancer Center and by an Institute for Clinical and Translational Science/Clinical and Translational Sciences Award grant #UL1TR000448 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and the NIH Roadmap for Medical Research. This publication is solely the responsibility of the authors and does not necessarily represent the official view of NCRR or NIH. This work was supported by NIH grant R01 GM-078376 to J.G.U. and a William H. Danforth fellowship to G.Y.M.
Footnotes
Communicating editor: K. Gunsalus
Literature Cited
- Adams G. M., Huang B., Luck D. J., 1982. Temperature-sensitive, assembly-defective flagella mutants of Chlamydomonas reinhardtii. Genetics 100: 579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agata K., Nakajima E., Funayama N., Shibata N., Saito Y., et al. , 2006. Two different evolutionary origins of stem cell systems and their molecular basis. Semin. Cell Dev. Biol. 17: 503–509. [DOI] [PubMed] [Google Scholar]
- Albee A. J., Kwan A. L., Lin H., Granas D., Stormo G. D., et al. , 2013. Identification of cilia genes that affect cell-cycle progression using whole-genome transcriptome analysis in Chlamydomonas reinhardtti. G3 3: 979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alié A., Hayashi T., Sugimura I., Manuel M., Sugano W., et al. , 2015. The ancestral gene repertoire of animal stem cells. Proc. Natl. Acad. Sci. USA 112: E7093–E7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., Huber W., 2014. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andorf C. M., Cannon E. K., Portwood J. L., Gardiner J. M., Harper L. C., et al. , 2016. MaizeGDB update: new tools, data and interface for the maize model organism database. Nucleic Acids Res. 44: D1195–D1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, S., 2010 FastQC: a quality control tool for high throughput sequence data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed: November 8, 2013.
- Awata J., Takada S., Standley C., Lechtreck K. F., Bellve K. D., et al. , 2014. NPHP4 controls ciliary trafficking of membrane proteins and large soluble proteins at the transition zone. J. Cell Sci. 127: 4714–4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball S., Marianne T., Dirick L., Fresnoy M., Delrue B., et al. , 1991. A Chlamydomonas reinhardtii low-starch mutant is defective for 3-phosphoglycerate activation and orthophosphate inhibition of ADP-glucose pyrophosphorylase. Planta 185: 17–26. [DOI] [PubMed] [Google Scholar]
- Ball S. G., Deschamps P., 2009. Starch metabolism, in The Chlamydomonas Sourcebook, Vol. 2, chap 1, edited by Stern D. B., Harris E. H. Academic Press, Inc., San Diego. [Google Scholar]
- Becker B., Marin B., 2009. Streptophyte algae and the origin of embryophytes. Ann. Bot. 103: 999–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beverley S. M., Owens K. L., Showalter M., Griffith C. L., Doering T. L., et al. , 2005. Eukaryotic UDP-galactopyranose mutase (GLF gene) in microbial and metazoal pathogens. Eukaryot. Cell 4: 1147–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner J. T., 1998. The origins of multicellularity. Integr. Biol. Issues News Rev. 1: 27–36. [Google Scholar]
- Bowman J. L., Kohchi T., Yamato K. T., Jenkins J., Shu S., et al. , 2017. Insights into Land Plant Evolution Garnered from the Marchantia polymorpha Genome. Cell 171: 287–304.e15. [DOI] [PubMed] [Google Scholar]
- Brandenberger R., Wei H., Zhang S., Lei S., Murage J., et al. , 2004. Transcriptome characterization elucidates signaling networks that control human ES cell growth and differentiation. Nat. Biotechnol. 22: 707–716. [DOI] [PubMed] [Google Scholar]
- Bullard J. H., Purdom E., Hansen K. D., Dudoit S., 2010. Evaluation of statistical methods for normalization and differential expression in mRNA-seq experiments. BMC Bioinformatics 11: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss L. W., 1983. Evolution, development, and the units of selection. Proc. Natl. Acad. Sci. USA 80: 1387–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss L. W., 1987. The Evolution of Individuality. Princeton University Press, Priceton, NJ. [Google Scholar]
- Camon E., Magrane M., Barrell D., Lee V., Dimmer E., et al. , 2004. The Gene Ontology Annotation (GOA) database: sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res. 32: D262–D266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canvin D. T., Beevers H., 1961. Sucrose synthesis from acetate in the germinating castor bean: kinetics and pathway. J. Biol. Chem. 236: 988–995. [PubMed] [Google Scholar]
- Chamberlain K. L., Miller S. H., Keller L. R., 2008. Gene expression profiling of flagellar disassembly in Chlamydomonas reinhardtii. Genetics 179: 7–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Mackey A. J., Stoeckert C. J., Roos D. S., 2006. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 34: D363–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi G., Przybylska M., Straus D., 1996. Three abundant germ line-specific transcripts in Volvox carteri encode photosynthetic proteins. Curr. Genet. 30: 347–355. [DOI] [PubMed] [Google Scholar]
- Church D. M., Goodstadt L., Hillier L. W., Zody M. C., Goldstein S., et al. , 2009. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol. 7: e1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock J. M., Sterck L., Rouzé P., Scornet D., Allen A. E., et al. , 2010. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 465: 617–621. [DOI] [PubMed] [Google Scholar]
- Coleman A. W., 2012. A comparative analysis of the Volvocaceae (Chlorophyta). J. Phycol. 48: 491–513. [DOI] [PubMed] [Google Scholar]
- Coleman A. W., Maguire M. J., 1982. A microspectrofluorometric analysis of nuclear and chloroplast DNA in Volvox. Dev. Biol. 94: 441–450. [DOI] [PubMed] [Google Scholar]
- Coss R. A., 1974. Mitosis in Chlamydomonas reinhardtii basal bodies and the mitotic apparatus. J. Cell Biol. 63: 325–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craige B., Tsao C. C., Diener D. R., Hou Y., Lechtreck K. F., et al. , 2010. CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J. Cell Biol. 190: 927–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross F. R., Umen J. G., 2015. The Chlamydomonas cell cycle. Plant J. 82: 370–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauvillée D., Chochois V., Steup M., Haebel S., Eckermann N., et al. , 2006. Plastidial phosphorylase is required for normal starch synthesis in Chlamydomonas reinhardtii. Plant J. 48: 274–285. [DOI] [PubMed] [Google Scholar]
- Dawe H. R., Farr H., Gull K., 2007. Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. J. Cell Sci. 120: 7–15. [DOI] [PubMed] [Google Scholar]
- De La Torre F., Cañas R. A., Pascual M. B., Avila C., Cánovas F. M., 2014. Plastidic aspartate aminotransferases and the biosynthesis of essential amino acids in plants. J. Exp. Bot. 65: 5527–5534. [DOI] [PubMed] [Google Scholar]
- Denis H., Lacroix J. C., 1993. The dichotomy between germ line and somatic line, and the origin of cell mortality. Trends Genet. 9: 7–11. [DOI] [PubMed] [Google Scholar]
- Denis J. A., Rochon-Beaucourt C., Champon B., Pietu G., 2011. Global transcriptional profiling of neural and mesenchymal progenitors derived from human embryonic stem cells reveals alternative developmental signaling pathways. Stem Cells Dev. 20: 1395–1409. [DOI] [PubMed] [Google Scholar]
- Dickinson H. G., Grant-Downton R., 2009. Bridging the generation gap: flowering plant gametophytes and animal germlines reveal unexpected similarities. Biol. Rev. Camb. Philos. Soc. 84: 589–615. [DOI] [PubMed] [Google Scholar]
- Diener D. R., Lupetti P., Rosenbaum J. L., 2015. Proteomic analysis of isolated ciliary transition zones reveals the presence of ESCRT proteins. Curr. Biol. 25: 379–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., et al. , 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazet-Loso T., Tautz D., 2010. Phylostratigraphic tracking of cancer genes suggests a link to the emergence of multicellularity in metazoa. BMC Biol. 8: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazet-Loso T., Brajković J., Tautz D., 2007. A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet. 23: 533–539. [DOI] [PubMed] [Google Scholar]
- Duncan L., Nishii I., Howard A., Kirk D., Miller S. M., 2006. Orthologs and paralogs of regA, a master cell-type regulatory gene in Volvox carteri. Curr. Genet. 50: 61–72. [DOI] [PubMed] [Google Scholar]
- Duncan L., Nishii I., Harryman A., Buckley S., Howard A., et al. , 2007. The VARL gene family and the evolutionary origins of the master cell-type regulatory gene, regA, in Volvox carteri. J. Mol. Evol. 65: 1–11. [DOI] [PubMed] [Google Scholar]
- Dutcher S. K., 2003. Elucidation of basal body and centriole functions in Chlamydomonas reinhardtii. Traffic 4: 443–451. [DOI] [PubMed] [Google Scholar]
- Dutcher S. K., O’Toole E. T., 2016. The basal bodies of Chlamydomonas reinhardtii. Cilia 5: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond P. J., Graham I. A., 2001. Re-examining the role of the glyoxylate cycle in oilseeds. Trends Plant Sci. 6: 72–78. [DOI] [PubMed] [Google Scholar]
- Edgar R. C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehler L. L., Holmes J. A., Dutcher S. K., 1995. Loss of spatial control of the mitotic spindle apparatus in a Chlamydomonas reinhardtii mutant strain lacking basal bodies. Genetics 141: 945–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel S. R., Dietrich F. S., Fisk D. G., Binkley G., Balakrishnan R., et al. , 2014. The reference genome sequence of Saccharomyces cerevisiae: then and now. G3 4: 389–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer S., Brunk B. P., Chen F., Gao X., Harb O. S., et al. , 2011. Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr. Protoc. Bioinformatics Chapter 6: Unit 6.12.1–6.12.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine J.-X., Terce-Laforgue T., Armengaud P., Clement G., Renou J.-P., et al. , 2012. Characterization of a NADH-dependent glutamate dehydrogenase mutant of Arabidopsis demonstrates the key role of this enzyme in root carbon and nitrogen metabolism. Plant Cell 24: 4044–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M. H., Scanlon M. J., 2015. Transcriptomic evidence for the evolution of shoot meristem function in sporophyte-dominant land plants through concerted selection of ancestral gametophytic and sporophytic genetic programs. Mol. Biol. Evol. 32: 355–367. [DOI] [PubMed] [Google Scholar]
- Gan L., Schwengberg S., Denecke B., 2014. Transcriptome analysis in cardiomyocyte-specific differentiation of murine embryonic stem cells reveals transcriptional regulation network. Gene Expr. Patterns 16: 8–22. [DOI] [PubMed] [Google Scholar]
- Goepfert S., Poirier Y., 2007. Beta-oxidation in fatty acid degradation and beyond. Curr. Opin. Plant Biol. 10: 245–251. [DOI] [PubMed] [Google Scholar]
- Goodstein D. M., Shu S., Howson R., Neupane R., Hayes R. D., et al. , 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40: 1178–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz S., García-Gómez J. M., Terol J., Williams T. D., Nagaraj S. H., et al. , 2008. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36: 3420–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham I. A., 2008. Seed storage oil mobilization. Annu. Rev. Plant Biol. 59: 115–142. [DOI] [PubMed] [Google Scholar]
- Gramates L. S., Marygold S. J., Dos Santos G., Urbano J.-M., Antonazzo G., et al. , 2017. FlyBase at 25: looking to the future. Nucleic Acids Res. 45: D663–D671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosberg R. K., Strathmann R. R., 2007. The evolution of multicellularity: a minor major transition? Annu. Rev. Ecol. Evol. Syst. 38: 621–654. [Google Scholar]
- Hallmann A., 2003. Extracellular matrix and sex-inducing pheromone in Volvox. Int. Rev. Cytol. 227: 131–182. [DOI] [PubMed] [Google Scholar]
- Hallmann A., Kirk D. L., 2000. The developmentally regulated ECM glycoprotein ISG plays an essential role in organizing the ECM and orienting the cells of Volvox. J. Cell Sci. 113: 4605–4617. [DOI] [PubMed] [Google Scholar]
- Hanschen E. R., Marriage T. N., Ferris P. J., Hamaji T., Toyoda A., et al. , 2016. The Gonium pectorale genome demonstrates co-option of cell cycle regulation during the evolution of multicellularity. Nat. Commun. 7: 11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E., 2009. Cell architecture, in The Chlamydomonas Sourcebook, Vol. 1, chap 2. Academic Press, Inc., San Diego. [Google Scholar]
- Hemmrich G., Khalturin K., Boehm A. M., Puchert M., Anton-Erxleben F., et al. , 2012. Molecular signatures of the three stem cell lineages in hydra and the emergence of stem cell function at the base of multicellularity. Mol. Biol. Evol. 29: 3267–3280. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Henikoff J. G., 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89: 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron M. D., 2016. Origins of multicellular complexity: Volvox and the volvocine algae. Mol. Ecol. 25: 1213–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron M. D., Michod R. E., 2008. Evolution of complexity in the volvocine algae: transitions in individuality through Darwin’s eye. Evolution 62: 436–451. [DOI] [PubMed] [Google Scholar]
- Herron M. D., Hackett J. D., Aylward F. O., Michod R. E., 2009. Triassic origin and early radiation of multicellular volvocine algae. Proc. Natl. Acad. Sci. USA 106: 3254–3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron M. D., Rashidi A., Shelton D. E., Driscoll W. W., 2013. Cellular differentiation and individuality in the “minor” multicellular taxa. Biol. Rev. Camb. Philos. Soc. 88: 844–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoops H. J., 1993. Flagellar, cellular and organismal polarity in Volvox carteri. J. Cell Sci. 117: 105–117. [Google Scholar]
- Hori K., Maruyama F., Fujisawa T., Togashi T., Yamamoto N., et al. , 2014. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat. Commun. 5: 3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Rifkin M. R., Luck D. J., 1977. Temperature-sensitive mutations affecting flagellar assembly and function in Chlamydomonas reinhardtii. J. Cell Biol. 72: 67–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huskey R. J., 1979. Mutants affecting vegetative cell orientation in Volvox carteri. Dev. Biol. 72: 236–243. [DOI] [PubMed] [Google Scholar]
- Huskey R. J., Griffin B. E., 1979. Genetic control of somatic cell differentiation in Volvox: analysis of somatic regenerator mutants. Dev. Biol. 72: 226–235. [DOI] [PubMed] [Google Scholar]
- Huskey R. J., Griffin B. E., Cecil P. O., Callahan A. M., 1979. A preliminary genetic investigation of Volvox carteri. Genetics 91: 229–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ispolatov I., Ackermann M., Doebeli M., 2012. Division of labour and the evolution of multicellularity. Proc. Biol. Sci. 279: 1768–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. D., Richardson E., Bachvarova R. F., Crother B. I., 2011. Evolution of the germ line-soma relationship in vertebrate embryos. Reproduction 141: 291–300. [DOI] [PubMed] [Google Scholar]
- Johnson X., Alric J., 2013. Central carbon metabolism and electron transport in Chlamydomonas reinhardtii: metabolic constraints for carbon partitioning between oil and starch. Eukaryot. Cell 12: 776–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M., 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44: D457–D462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller L. C., Romijn E. P., Zamora I., Yates J. R., Marshall W. F., 2005. Proteomic analysis of isolated chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr. Biol. 15: 1090–1098. [DOI] [PubMed] [Google Scholar]
- Keller L. C., Geimer S., Romijn E., Yates J., Zamora I., et al. , 2009. Molecular architecture of the centriole proteome: the conserved WD40 domain protein POC1 is required for centriole duplication and length control. Mol. Biol. Cell 20: 1150–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerszberg M., Wolpert L., 1998. The origin of metazoa and the egg: a role for cell death. J. Theor. Biol. 193: 535–537. [DOI] [PubMed] [Google Scholar]
- Kianianmomeni A., Hallmann A., 2015. Transcriptional analysis of Volvox photoreceptors suggests the existence of different cell-type specific light-signaling pathways. Curr. Genet. 61: 3–18. [DOI] [PubMed] [Google Scholar]
- King N., 2004. The unicellular ancestry of animal development. Dev. Cell 7: 313–325. [DOI] [PubMed] [Google Scholar]
- Kirk D. L., 1988. The ontogeny and phylogeny of cellular differentiation in Volvox. Trends Genet. 4: 32–36. [DOI] [PubMed] [Google Scholar]
- Kirk D. L., 1998. Volvox: Molecular-Genetic Origins of Multicellularity and Cellular Differentiation. Cambridge University Press, Cambridge, UK. [Google Scholar]
- Kirk D. L., 2001. Germ-soma differentiation in Volvox. Dev. Biol. 238: 213–223. [DOI] [PubMed] [Google Scholar]
- Kirk D. L., 2005. A twelve-step program for evolving multicellularity and a division of labor. Bioessays 27: 299–310. [DOI] [PubMed] [Google Scholar]
- Kirk D. L., Nishii I., 2001. Volvox carteri as a model for studying the genetic and cytological control of morphogenesis. Dev. Growth Differ. 43: 621–631. [DOI] [PubMed] [Google Scholar]
- Kirk D. L., Birchem R., King N., 1986. The extracellular matrix of Volvox: a comparative study and proposed system of nomenclature. J. Cell Sci. 80: 207–231. [DOI] [PubMed] [Google Scholar]
- Kirk M. M., Stark K., Miller S. M., Müller W., Taillon B. E., et al. , 1999. regA, a Volvox gene that plays a central role in germ-soma differentiation, encodes a novel regulatory protein. Development 126: 639–647. [DOI] [PubMed] [Google Scholar]
- Klein U., 1986. Compartmentation of glycolysis and of the oxidative pentose-phosphate pathway in Chlamydomonas reinhardtii. Planta 167: 81–86. [DOI] [PubMed] [Google Scholar]
- Kochert G., Olson L. W., 1970. Ultrastructure of Volvox carteri. Arch. Mikrobiol. 74: 19–30. [Google Scholar]
- Koufopanou V., 1994. The evolution of soma in the volvocales. Am. Nat. 143: 907–931. [Google Scholar]
- Koufopanou V., Bell G., 1993. Soma and germ: an experimental approach through Volvox. Proc. Biol. Sci. 254: 107–113. [Google Scholar]
- Kumar S., Stecher G., Tamura K., 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33: msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P., Berardini T. Z., Li D., Swarbreck D., Wilks C., et al. , 2012. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40: D1202–D1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch B. J., Page D. C., 2012. Genetics of germ cell development. Nat. Rev. Genet. 13: 781–794. [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y., Beisson F., Riekhof W., 2015. Metabolism of acyl-lipids in Chlamydomonas reinhardtii. Plant J. 82: 504–522. [DOI] [PubMed] [Google Scholar]
- Lopez D., Hamaji T., Kropat J., De Hoff P., Morselli M., et al. , 2015. Dynamic changes in the transcriptome and methylome of Chlamydomonas reinhardtii throughout its life cycle. Plant Physiol. 169: 2730–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S., 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah J. L., Christensen-Dalsgaard K. K., Leys S. P., 2014. Choanoflagellate and choanocyte collar-flagellar systems and the assumption of homology. Evol. Dev. 16: 25–37. [DOI] [PubMed] [Google Scholar]
- Marshall W. F., 2008. Basal bodies: platforms for building cilia. Curr. Top. Dev. Biol. 85: 1–22. [DOI] [PubMed] [Google Scholar]
- Marshall W. F., Rosenbaum J. L., 2000. How centrioles work: lessons from green yeast. Curr. Opin. Cell Biol. 12: 119–125. [DOI] [PubMed] [Google Scholar]
- Matt G., Umen J., 2016. Volvox: a simple algal model for embryogenesis, morphogenesis and cellular differentiation. Dev. Biol. 419: 99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVittie A., 1972. Flagellum mutants of Chlamydomonas reinhardii. J. Gen. Microbiol. 71: 525–540. [DOI] [PubMed] [Google Scholar]
- Meissner M., Stark K., Cresnar B., Kirk D. L., Schmitt R., 1999. Volvox germline-specific genes that are putative targets of RegA repression encode chloroplast proteins. Curr. Genet. 36: 363–370. [DOI] [PubMed] [Google Scholar]
- Merchant S. S., Prochnik S. E., Vallon O., Harris E. H., Karpowicz S. J., et al. , 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michod R. E., 1997. Evolution of the individual. Am. Nat. 150: S5–S21. [DOI] [PubMed] [Google Scholar]
- Mikhailov K. V., Konstantinova A. V., Nikitin M. A., Troshin P. V., Rusin L. Y., et al. , 2009. The origin of Metazoa: a transition from temporal to spatial cell differentiation. Bioessays 31: 758–768. [DOI] [PubMed] [Google Scholar]
- Miura T., Luo Y., Khrebtukova I., Brandenberger R., Zhou D., et al. , 2004. Monitoring early differentiation events in human embryonic stem cells by massively parallel signature sequencing and expressed sequence tag scan. Stem Cells Dev. 13: 694–715. [DOI] [PubMed] [Google Scholar]
- Moreno-Hagelsieb G., Latimer K., 2008. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics 24: 319–324. [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B., 2008. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Myhre S., Tveit H., Mollestad T., Lægreid A., 2006. Additional Gene Ontology structure for improved biological reasoning. Bioinformatics 22: 2020–2027. [DOI] [PubMed] [Google Scholar]
- Nawy T., Lee J.-Y., Colinas J., Wang J. Y., Thongrod S. C., et al. , 2005. Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell 17: 1908–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelcu A. M., 2009. Environmentally induced responses co-opted for reproductive altruism. Biol. Lett. 5: 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelcu A. M., 2012. The evolution of self during the transition to multicellularity. Adv. Exp. Med. Biol. 738: 14–30. [DOI] [PubMed] [Google Scholar]
- Nedelcu A. M., Michod R. E., 2004. Evolvability, modularity, and individuality during the transition to multicellularity in volvocalean green algae, pp. 466–489 in Modularity in Development and Evolution, edited by Schlosser G., Wagner G. University of Chicago Press, Chicago, London. [Google Scholar]
- Nedelcu A. M., Michod R. E., 2006. The evolutionary origin of an altruistic gene. Mol. Biol. Evol. 23: 1460–1464. [DOI] [PubMed] [Google Scholar]
- Nematollahi G., Kianianmomeni A., Hallmann A., 2006. Quantitative analysis of cell-type specific gene expression in the green alga Volvox carteri. BMC Genomics 7: 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishii I., Miller S. M., 2010. Volvox: simple steps to developmental complexity? Curr. Opin. Plant Biol. 13: 646–653. [DOI] [PubMed] [Google Scholar]
- Nozaki H., Misawa K., Kajita T., Kato M., Nohara S., et al. , 2000. Origin and evolution of the colonial volvocales (Chlorophyceae) as inferred from multiple, chloroplast gene sequences. Mol. Phylogenet. Evol. 17: 256–268. [DOI] [PubMed] [Google Scholar]
- Parker J. D. K., Hilton L. K., Diener D. R., Rasi M. Q., Mahjoub M. R., et al. , 2010. Centrioles are freed from cilia by severing prior to mitosis. Cytoskeleton (Hoboken) 67: 425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plancke C., Vigeolas H., Höhner R., Roberty S., Emonds-Alt B., et al. , 2014. Lack of isocitrate lyase in Chlamydomonas leads to changes in carbon metabolism and in the response to oxidative stress under mixotrophic growth. Plant J. 77: 404–417. [DOI] [PubMed] [Google Scholar]
- Pommerville J., Kochert G., 1982. Effects of senescence on somatic cell physiology in the green alga Volvox carteri. Exp. Cell Res. 140: 39–45. [DOI] [PubMed] [Google Scholar]
- Prochnik S. E., Umen J., Nedelcu A. M., Hallmann A., Miller S. M., et al. , 2010. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science 329: 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon E., Silventoinen V., Pillai S., Harte N., Mulder N., et al. , 2005. InterProScan: protein domains identifier. Nucleic Acids Res. 33: W116–W120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M., 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter D. J., King N., 2013. The genomic and cellular foundations of animal origins. Annu. Rev. Genet. 47: 509–537. [DOI] [PubMed] [Google Scholar]
- Rokas A., 2008. The origins of multicellularity and the early history of the genetic toolkit for animal development. Annu. Rev. Genet. 42: 235–251. [DOI] [PubMed] [Google Scholar]
- Rosenbaum J. L., Witman G. B., 2002. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 3: 813–825. [DOI] [PubMed] [Google Scholar]
- Ruiz-Trillo I., Roger A. J., Burger G., Gray M. W., Lang B. F., 2008. A phylogenomic investigation into the origin of Metazoa. Mol. Biol. Evol. 25: 664–672. [DOI] [PubMed] [Google Scholar]
- Runquist M., Kruger N., 1999. Control of gluconeogenesis by isocitrate lyase in endosperm of germinating castor bean seedlings. Plant J. 19: 423–431. [DOI] [PubMed] [Google Scholar]
- Rylott E. L., Hooks M. A., Graham I. A., 2001. Co-ordinate regulation of genes involved in storage lipid mobilization in Arabidopsis thaliana. Biochem. Soc. Trans. 29: 283–287. [DOI] [PubMed] [Google Scholar]
- Schippers J. H. M., Schmidt R., Wagstaff C., Jing H.-C., 2015. Living to die and dying to live: the survival strategy behind leaf senescence. Plant Physiol. 169: 914–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A., Schmid M. W., Grossniklaus U., 2012. Analysis of plant germline development by high-throughput RNA profiling: technical advances and new insights. Plant J. 70: 18–29. [DOI] [PubMed] [Google Scholar]
- Schmidt A., Schmid M. W., Grossniklaus U., 2015. Plant germline formation: common concepts and developmental flexibility in sexual and asexual reproduction. Development 142: 229–241. [DOI] [PubMed] [Google Scholar]
- Seifert G. J., 2004. Nucleotide sugar interconversions and cell wall biosynthesis: how to bring the inside to the outside. Curr. Opin. Plant Biol. 7: 277–284. [DOI] [PubMed] [Google Scholar]
- Serrato A. J., de Dios Barajas-López J., Chueca A., Sahrawy M., 2009. Changing sugar partitioning in FBPase-manipulated plants. J. Exp. Bot. 60: 2923–2931. [DOI] [PubMed] [Google Scholar]
- Seydoux G., Braun R. E., 2006. Pathway to totipotency: lessons from germ cells. Cell 127: 891–904. [DOI] [PubMed] [Google Scholar]
- Showalter A. M., Basu D., 2016. Extensin and arabinogalactan-protein biosynthesis: glycosyltransferases, research challenges, and biosensors. Front. Plant Sci. 7: 814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M., Walz G., 2006. Polycystic kidney disease: cell division without a c(l)ue? Kidney Int. 70: 854–864. [DOI] [PubMed] [Google Scholar]
- Smith T. F., Waterman M. S., 1981. Identification of common molecular subsequences. J. Mol. Biol. 147: 195–197. [DOI] [PubMed] [Google Scholar]
- Solana J., 2013. Closing the circle of germline and stem cells: the primordial stem cell hypothesis. Evodevo 4: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer-Knudsen J., Bacic A., Clarke A. E., 1998. Hydroxyproline-rich plant glycoproteins. Phytochemistry 47: 483–497. [Google Scholar]
- Spalding M. H., 2009. The CO2-concentrating mechanism and carbon assimilation, in The Chlamydomonas Sourcebook, Vol. 2, chap 8, edited by Stern D. B., Harris E. H. Academic Press, Inc., San Diego. [Google Scholar]
- Starr R. C., 1970. Control of differentiation in Volvox. Symp. Soc. Dev. Biol. 29: 59–100. [DOI] [PubMed] [Google Scholar]
- Stolc V., Samanta M. P., Tongprasit W., Marshall W. F., 2005. Genome-wide transcriptional analysis of flagellar regeneration in Chlamydomonas reinhardtii identifies orthologs of ciliary disease genes. Proc. Natl. Acad. Sci. USA 102: 3703–3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S., Lehmann R., 2007. Germ vs. soma decisions: lessons from flies and worms. Science 316: 392–393. [DOI] [PubMed] [Google Scholar]
- Strome S., Updike D., 2015. Specifying and protecting germ cell fate. Nat. Rev. Mol. Cell Biol. 16: 406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumper M., Hallmann A., 1998. Biochemistry of the extracellular matrix of Volvox. Int. Rev. Cytol. 180: 51–85. [DOI] [PubMed] [Google Scholar]
- Swartz S. Z., Wessel G. M., 2015. Germ line vs. soma in the transition from egg to embryo. Curr. Top. Dev. Biol. 113: 149–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam L. W., Kirk D. L., 1991. Identification of cell-type-specific genes of Volvox carteri and characterization of their expression during the asexual life cycle. Dev. Biol. 145: 51–66. [DOI] [PubMed] [Google Scholar]
- Tam L. W., Lefebvre P. A., 1993. Cloning of flagellar genes in Chlamydomonas reinhardtii by DNA insertional mutagenesis. Genetics 135: 375–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif M., Atteia A., Specht M., Cogne G., Rolland N., et al. , 2012. Predalgo: a new subcellular localization prediction tool dedicated to green algae. Mol. Biol. Evol. 29: 3625–3639. [DOI] [PubMed] [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., et al. , 2004. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37: 914–939. [DOI] [PubMed] [Google Scholar]
- Troncoso-Ponce M. A., Cao X., Yang Z., Ohlrogge J. B., 2013. Lipid turnover during senescence. Plant Sci. 205–206: 13–19. [DOI] [PubMed] [Google Scholar]
- Ueki N., Nishii I., 2008. Idaten is a new cold-inducible transposon of Volvox carteri that can be used for tagging developmentally important genes. Genetics 180: 1343–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen J. G., 2014. Green algae and the origins of multicellularity in the plant kingdom. Cold Spring Harb. Perspect. Biol. 6: a016170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen J. G., Olson B. J., 2012. Genomics of volvocine algae. Adv. Bot. Res. 64: 185–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallon O., Spalding M. H., 2009. Amino acid metabolism, in The Chlamydomonas Sourcebook, Vol. 2, Chap 4, edited by Stern D. B., Harris E. H. Academic Press, Inc, San Diego. [Google Scholar]
- Wolpert L., 1990. The evolution of development. Biol. J. Linn. Soc. Lond. 39: 109–124. [Google Scholar]
- Wolpert L., 1992. Gastrulation and the evolution of development. Dev. Suppl. 116: 7–13. [PubMed] [Google Scholar]
- Wolpert L., Szathmáry E., 2002. Evolution and the egg. Nature 420: 745. [DOI] [PubMed] [Google Scholar]
- Wood C. R., Wang Z., Diener D., Zones J. M., Rosenbaum J., et al. , 2012. IFT proteins accumulate during cell division and localize to the cleavage furrow in Chlamydomonas. PLoS One 7: e30729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodland H. R., 2016. The birth of animal development: multicellularity and the germline. Curr. Top. Dev. Biol. 117: 609–630. [DOI] [PubMed] [Google Scholar]
- Yadav R. K., Girke T., Pasala S., Xie M., Reddy G. V., 2009. Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc. Natl. Acad. Sci. USA 106: 4941–4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R. K., Tavakkoli M., Xie M., Girke T., Reddy G. V., 2014. A high-resolution gene expression map of the Arabidopsis shoot meristem stem cell niche. Development 141: 2735–2744. [DOI] [PubMed] [Google Scholar]
- Zabawinski C., Koornhuyse N., D’Hulst C., Schlichting R., Giersch C., et al. , 2001. Starchless mutants of Chlamydomonas reinhardtii lack the small subunit of a heterotetrameric ADP-glucose pyrophosphorylase. J. Bacteriol. 183: 1069–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Ji S., Wang J., Huang J., Zheng P., 2014. mRNA-seq and microRNA-seq whole-transcriptome analyses of rhesus monkey embryonic stem cell neural differentiation revealed the potential regulators of rosette neural stem cells. DNA Res. 21: 541–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer A. D., Lang D., Buchta K., Rombauts S., Nishiyama T., et al. , 2013. Reannotation and extended community resources for the genome of the non-seed plant Physcomitrella patens provide insights into the evolution of plant gene structures and functions. BMC Genomics 14: 498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zones J. M., Blaby I. K., Merchant S. S., Umen J. G., 2015. High-resolution profiling of a synchronized diurnal transcriptome from Chlamydomonas reinhardtii reveals continuous cell and metabolic differentiation. Plant Cell 27: 2743–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Wild-type V. carteri f. nagariensis strain Eve (HK10) can be obtained from UTEX (http://utex.org/) or NIES (http://mcc.nies.go.jp/), and our current subclone is available upon request. Names and descriptions of supplemental tables and datasets can be found in the supplemental material. Primary data are available at the NCBI Gene Expression Omnibus repository under accession number GSE104835. V. carteri locus ID numbers for genes described in this study are listed in the appropriate supplemental datasets and can be accessed from Phytozome (http://phytozome.jgi.doe.gov).