Abstract
LN follicles constitute major reservoir sites for HIV/SIV persistence. Cure strategies could benefit from the characterization of CD8+ T cells able to access and eliminate HIV-infected cells from these areas. In this study, we provide a comprehensive analysis of the phenotype, frequency, localization, and functionality of follicular CD8+ T cells (fCD8+) in SIV-infected nonhuman primates. Although disorganization of follicles was a major factor, significant accumulation of fCD8+ cells during chronic SIV infection was also observed in intact follicles, but only in pathogenic SIV infection. In line with this, tissue inflammatory mediators were strongly associated with the accumulation of fCD8+ cells, pointing to tissue inflammation as a major factor in this process. These fCD8+ cells have cytolytic potential and can be redirected to target and kill HIV-infected cells using bispecific antibodies. Altogether, our data support the use of SIV infection to better understand the dynamics of fCD8+ cells and to develop bispecific antibodies as a strategy for virus eradication.
Keywords: AIDS/HIV, Immunology
Keywords: AIDS vaccine, Lymph, T cells
Introduction
The ability of CD8+ T cells to clear virus-infected, including HIV-infected, cells depends on the ability of CD8+ T cells to migrate to sites of active virus replication (1–3). In chronic HIV/SIV infection, follicular helper CD4+ T cells (Tfh) within the lymph nodes (LNs) represent a site for virus persistence and replication, even under suppressive antiretroviral therapy (ART) (4–6). Moreover, replication-competent SIV in chronically infected animals with spontaneous control of viremia is restricted to the Tfh cell population (7). Of particular relevance for HIV/SIV pathogenesis, virus replication in secondary lymphoid organs further impairs the development of adaptive immune responses (8, 9).
Despite the accumulation of CD8+ T cells within the B cell follicles during chronic HIV infection (10–12), the presence of HIV-specific cytotoxic T lymphocytes (CTLs) within germinal centers (GCs) is controversial. While early studies reported the presence of HIV-specific effector CD8+ T cells within GCs (13, 14), in situ tissue staining of HIV-infected LNs revealed that those CTLs are scarce in the vicinity of HIV-infected Tfh cells (12, 15, 16). Despite the low frequency of HIV-specific CTLs within the GC, we have shown that bulk CCR7loCXCR5hi follicular CD8+ (fCD8+) T cells can mediate in vitro killing of HIV-infected cells (11) in the presence of engineered bispecific antibodies designed to bind envelope-expressing HIV-infected cells (17). The use of bispecific antibodies circumvents the relatively low frequency of virus-specific CD8+ T cells and CTL escape variants associated with the latent reservoir (18). Therefore, understanding CD8+ T cell dynamics within the LNs will aid in the development of novel approaches for virus eradication. The use of SIV-infected nonhuman primates (NHPs), a model that recapitulates many aspects of human HIV infection (19), could help delineate the mechanisms of fCD8+ T cell dynamics and aid in the development of cure strategies aimed at bolstering cytolytic activity within B cell follicles and GCs.
Here, we provide a comprehensive analysis of the phenotype, location, and killing activity of LN CD8+ T cells during acute and chronic SIV infection. As with HIV infection, we found accumulation of fCD8+ T cells in chronic infection. These cells are characterized by a potent in vitro bispecific antibody–mediated killing of HIV-infected targets. Our data reveal the role of immune activation–related factors as mediators for the migration of cytolytic fCD8+ T cells into the GC.
Results
Accumulation of fCD8+ T cells in chronic SIV infection.
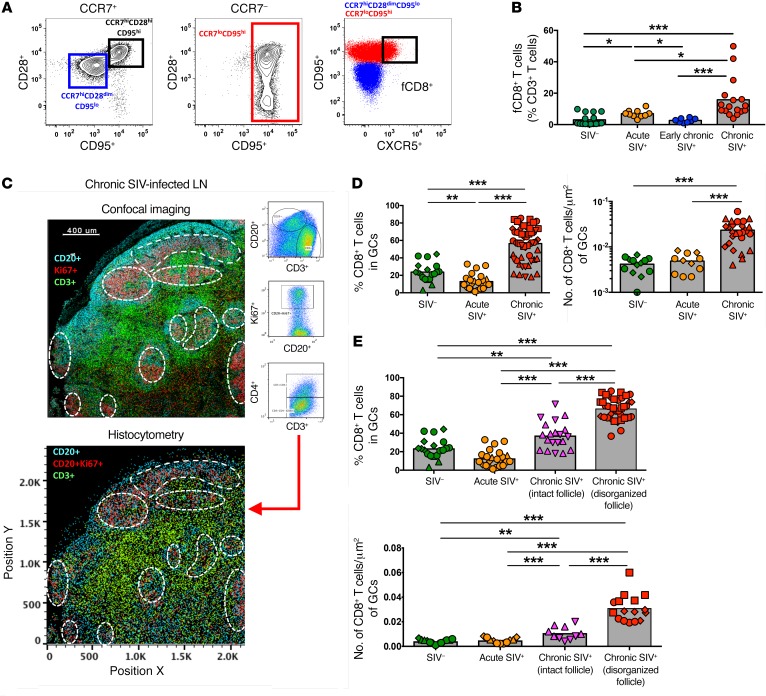
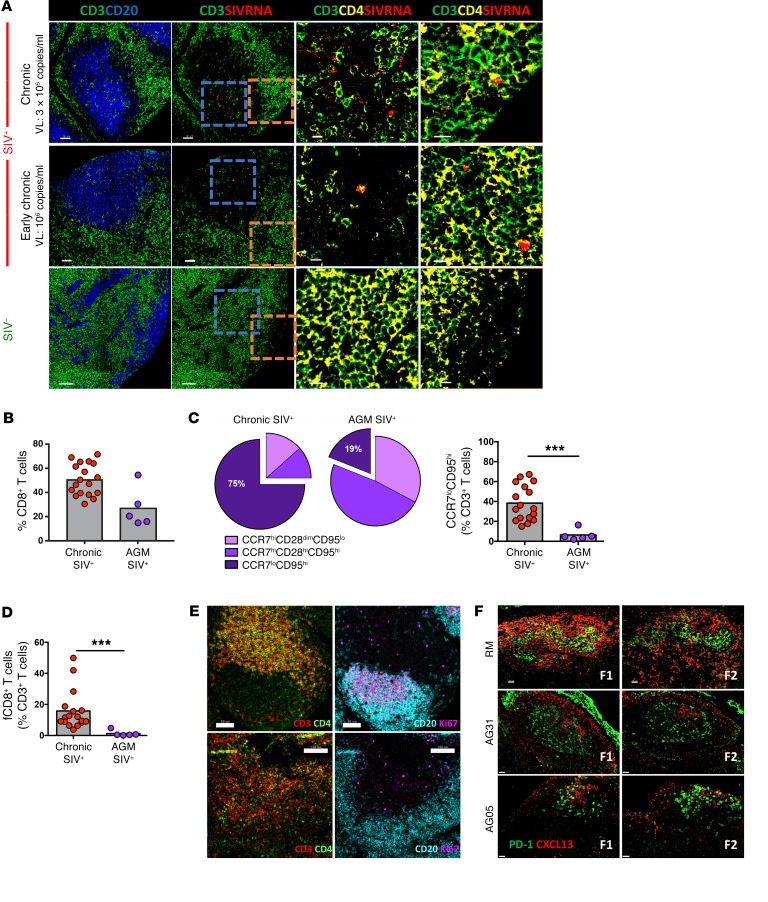
The relative frequency and phenotype of LN-derived CD8+ T cells from rhesus macaques (RMs) in different stages of infection were analyzed by polychromatic flow cytometry (Figure 1A and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI96207DS1). We found a substantial reduction in CD4+ T cells accompanied by a substantial increase in total CD8+ T cells in early chronic (day 45 post infection [p.i.]) infection (Supplemental Figure 1B). Progression was associated with a significant increase of CCR7loCD95hi CD8+ T cells representing, on average, 75% of the total CD8+ T cell pool in chronic (>6 months) infection (Supplemental Figure 1C). As was done for human tissues (11), low expression of CCR7 and high expression of CXCR5 were used to define NHP fCD8+ T cells (Figure 1A and Supplemental Figure 1B). Importantly, the expression levels of CXCR5 were lower on fCD8+ T cells compared with expression on Tfh cells (Supplemental Figure 1D). Despite this lower expression, we observed rapid mobilization of F-actin in fCD8+ T cells in response to CXCL13 (Supplemental Figure 1E). Chronic SIV infection was associated with a clear increase in fCD8+ T cells (Figure 1B).
Figure 1. fCD8+ T cells accumulate in LNs during chronic SIV infection.
(A) Gating strategy for CCR7loCD95hiCXCR5hi fCD8+ cells. (B) Flow cytometric pooled data showing the relative frequency of fCD8+ T cells in noninfected (n = 16 SIV–), acute SIV+ (day 14, n = 10), early chronic SIV+ (day 45, n = 8), and chronic SIV+ (>6 months, n = 17) RMs. *P < 0.05 and ***P < 0.0001, by Mann-Whitney U test. (C) Representative example of histocytometric analysis of follicular cells from 1 chronically SIV-infected animal (7 different samples were analyzed using this method). GCs were defined by CD20+Ki67+ coexpression, and CD4+ (CD3+CD4+) and CD8+ (CD3+CD4–) T cells were quantified within each GC. A representative confocal image and its reconstruction using histocytometry are shown. Scale bar: 400 μm. (D) Histocytometric pooled data showing the relative frequency and actual numbers (per μm2) of CD8+ T cells within GCs. Each point represents an individual GC. Different symbols represent different samples (n = 2 SIV–; n = 2 acute SIV+, n = 3 chronic SIV+). **P < 0.001 and ***P < 0.0001, by Mann-Whitney U test. (E) Pooled data showing the relative frequency and actual numbers (per μm2) of CD8+ T cells within intact and disorganized GCs from chronically SIV-infected animals (n = 5). Data from SIV– (n = 2) and acute SIV-infected animals (n = 2) are also shown. Each point represents an individual GC, and different symbols represent different LN samples. **P < 0.001 and ***P < 0.0001, by Mann-Whitney U test.
We further analyzed the location of CD8+ T cells using multicolor confocal imaging (Supplemental Figure 2A). In preliminary experiments, we were not able to find a reliable anti-CD8 antibody for paraffin-embedded tissues. Since we detected a very low frequency (<5%) of CD4–CD8– double-negative T cells within the CD3+ population (Supplemental Figure 2B), we are confident that the CD3+CD4– phenotype accurately defines CD8+ T cells. In agreement with the flow cytometric data (Supplemental Figure 1C), we detected a higher frequency of CD8+ T cells in the T cell area in early chronically SIV-infected LNs (Supplemental Figure 2C). However, in chronic SIV infection, we observed an accumulation of CD8+ T cells around and within B cell follicles and GCs (Supplemental Figure 2C). We performed histocytometry to quantify relevant cell populations (11, 20). We quantified CD8+ T cells for each individual GC (Figure 1C and Supplemental Figure 3A) and confirmed the accumulation of fCD8+ T cells during chronic SIV infection (Figure 1D). Follicular disorganization (Supplemental Figure 3B), a marker of disease progression (21), was observed in 3 of the 5 chronically infected RMs that we studied, but not in acute or early infected animals. Although we observed the highest accumulation of fCD8+ T cells in disorganized follicles during chronic SIV infection, intact follicles also contained a significantly higher percentage of fCD8+ T cells compared with follicles from uninfected and acutely infected LNs (Figure 1E). Therefore, as with chronic HIV infection (11), we found CD8+ T cell accumulation within B cell follicles and GCs during chronic SIV infection.
No preferential accumulation of SIV-specific fCD8+ T cells in chronic infection.
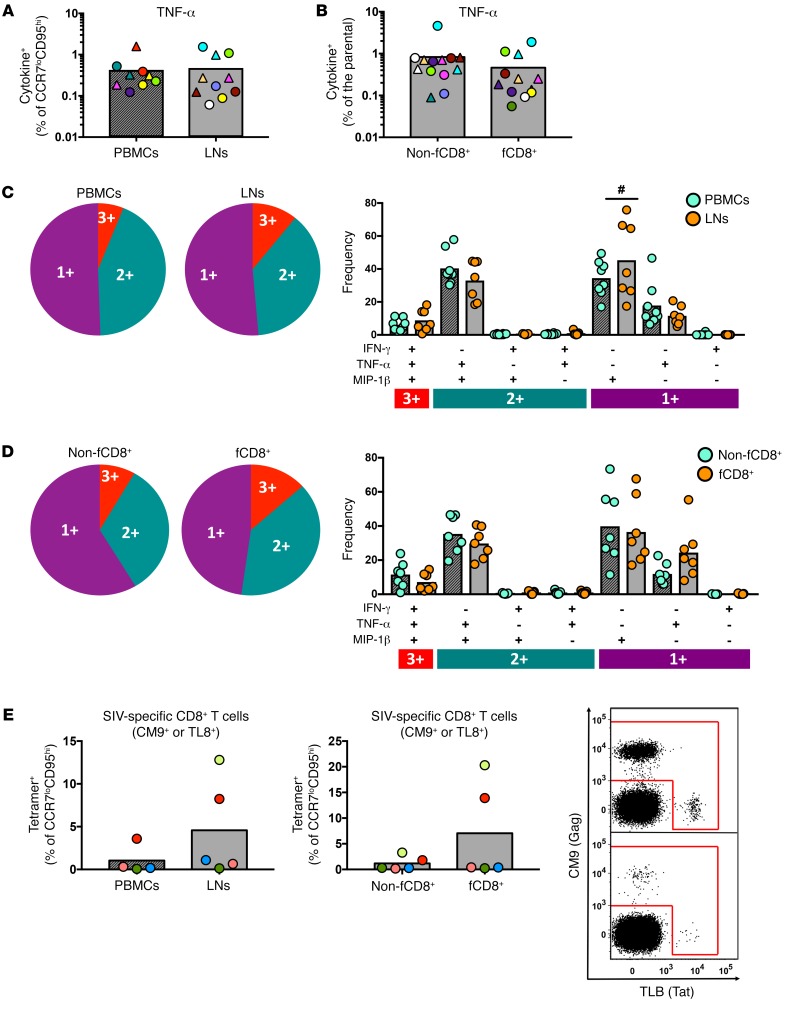
Bulk and SIV-specific responses were determined by cytokine production after stimulation with either anti-CD3 beads or SIV-Gag and SIV-Env peptide pools (Supplemental Figure 4A). We found that responses to CD3 and T cell receptor (TCR) stimulation were similar between LNs and peripheral blood mononuclear cells (PBMCs) (Supplemental Figure 4B). Although fewer LN samples compared with PBMC samples responded to in vitro stimulation with SIV peptide pools (Supplemental Figure 4C), we detected a similar distribution of virus-specific CD8+ T cell responses between PBMCs and LNs among the responders (Figure 2A). Furthermore, we found a similar frequency of SIV-specific CD8+ T cells in non-fCD8+ and fCD8+ T cell subsets (Figure 2B). Polyfunctionality was also similar between LN and PBMC samples (Figure 2C), but LN samples had a higher frequency of MIP-1β–expressing, SIV-specific CD8+ T cells (Figure 2C). Further analysis revealed that, in chronic SIV infection, non-fCD8+ and fCD8+ T cell subsets had similar polyfunctionality (Figure 2D). Since LN SIV-specific CD8+ T cells express a PD-1hi phenotype that could compromise their cytokine response (22), we further quantified virus-specific CD8+ T cells using a tetramer-based approach. The results confirmed that there was no preferential sequestration of SIV-specific CD8+ T cells in the follicular areas (Figure 2E).
Figure 2. SIV-specific CD8+ T cells with a follicular phenotype do not preferentially accumulate in LNs during chronic SIV infection.
(A) Pooled data showing the frequency of TNF-α+ CCR7loCD95hi CD8+ T cells in PBMC (n = 6 responders) and LN (n = 8 responders) samples after short ex vivo stimulation with Gag (circles) or Env (triangles) peptide pools. Matching samples are highlighted with paired colors. Mann-Whitney U test. (B) Frequency of TNF-α+ CD8+ T cells in non-fCD8+ (n = 9 responders) and fCD8+ (n = 9 responders) compartments. Matching samples are highlighted with paired colors. Mann-Whitney U test. The polyfunctionality of virus-specific CD8+ T cell responses after short ex vivo stimulation (Gag or Env peptide pools) in paired samples of (C) LN cells (n = 12 responders) and PBMCs (n = 9 responders) and (D) paired LN non-fCD8+ and fCD8+ T cells. #P < 0.05, by Wilcoxon signed-rank test. (E) Relative frequency of tetramer+, SIV-specific CD8+ T cells in PBMC (n = 5) and LN (n = 5) samples and LN non-fCD8+ and fCD8+ T cell compartments. Representative flow cytometric plots for CM9+ (Gag) and TL8+ (Tat) fCD8+ T cells from 2 animals (LNs) are also shown. All results are from chronically SIV-infected animals. A Mann-Whitney U test was used for unpaired comparisons and a Wilcoxon test for paired comparisons.
fCD8+ T cells are characterized by increased cytolytic and killing potential.
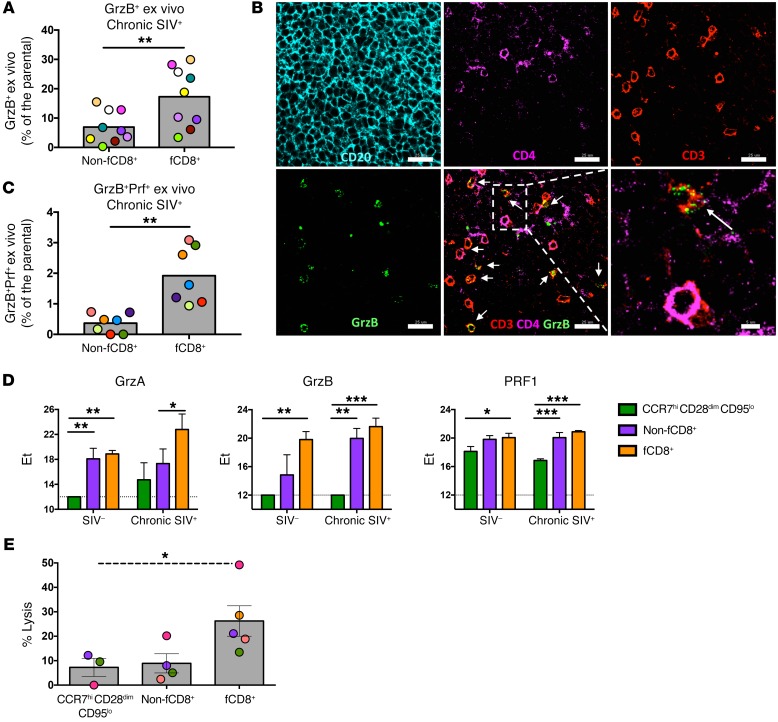
We detected ex vivo granzyme B (GrzB) expression almost exclusively in the CCR7loCD95hi CD8+ T cell subset, the frequency of which was significantly higher in PBMCs than in LNs (Supplemental Figure 5, A and B). In LNs from chronically infected RMs, fCD8+ T cells had higher ex vivo expression of GrzB than did non-fCD8+ T cells (Figure 3A and Supplemental Figure 5C). Image analysis (representative example is shown in Figure 3B) confirmed the presence of CD8+ T cells expressing a GrzBhi phenotype, especially within the GC, during chronic infection. The ex vivo expression of GrzB and perforin (Prf) (Supplemental Figure 5D) was significantly lower in LNs, independent of the infectious status (Supplemental Figure 5, E and F). Within the LNs, fCD8+ T cells had the highest coexpression of GrzB and Prf (Figure 3C). Fluidigm analysis of sorted LN cell populations showed that, while naive and non-fCD8+ T cells had characteristic profiles independent of the infectious status, the fCD8+ T cell expression pattern was strongly affected by SIV infection (Supplemental Figure 5G). Individual gene expression analysis confirmed higher GrzB and Prf expression among the follicular cell subsets (Figure 3D). In line with their cytolytic potential, fCD8+ T cells were the most potent mediators of bispecific antibody–redirected killing of HIV-infected CD4+ T cell targets (Figure 3E). Collectively, these data show that, like HIV, chronic SIV infection is associated with the accumulation of cytolytic fCD8+ T cells able to mediate the redirected killing of infected cells.
Figure 3. A high cytolytic profile of fCD8+ T cells is associated with potent in vitro bispecific antibody–mediated killing of infected cells.
(A) Pooled data showing the frequency of ex vivo GrzB expression in non-fCD8+ and fCD8+ T cell cells from chronically SIV-infected animals (n = 9). **P < 0.001, by Mann-Whitney U test. (B) Representative confocal image showing GrzB expression in CD8+ T cells (CD3+CD4–) within the GCs of a chronically SIV-infected LN. Individual staining and merged images (CD3/CD4/GrzB) are shown. Arrows highlight CD8+ T cells (defined as CD3+CD4-) positive for GrzB staining. Original magnification, ×63; scale bars: 25 μm and 5 μm (enlarged inset of white boxed area). (C) Pooled data showing expression of the GrzB+Prf+ cell population in non-fCD8+ and fCD8+ T cells from chronically SIV-infected animals (n = 7). Matching samples are highlighted with paired colors. **P < 0.001, by Wilcoxon paired test. (D) Fluidigm analysis of mRNA expression levels of granzyme A (GrzA), GrzB, and PRF1 among different CD8+ T cell subsets from chronically SIV-infected LNs (n =3). *P < 0.05, **P < 0.001, and ***P < 0.0001, by Wilcoxon paired test. (E) Level of in vitro bispecific antibody–mediated killing of infected cells in the presence of relevant sorted CD8+ T cells from chronically SIV-infected LNs (n = 5). Matching samples are highlighted with paired colors. *P < 0.05, by Kruskal-Wallis test.
Combination ART therapy leads to a modest reduction of fCD8+ T cells.
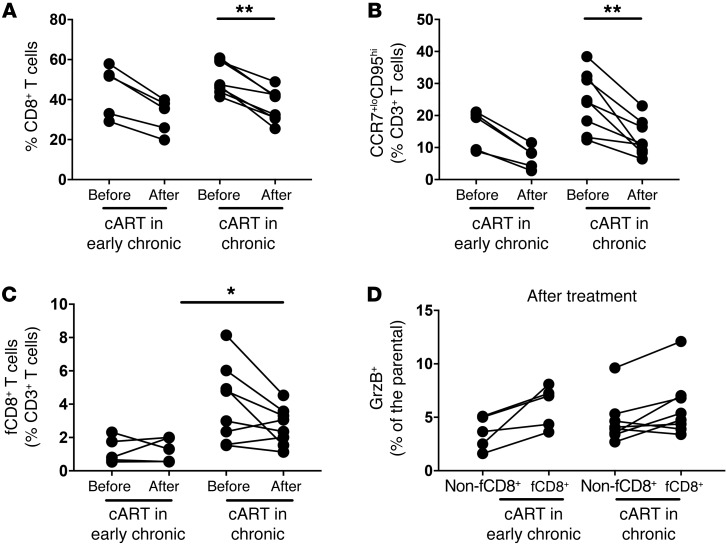
Next, we studied the dynamics of fCD8+ T cells in viremic NHPs that received combination ART (cART). We analyzed animals that began treatment in the early chronic stage of infection (7–8 weeks p.i.) and were treated for 27 weeks as well as animals that were treated during the chronic stage of infection (50 weeks p.i.) and treated for 7 weeks (Table 1). The viral load (VL) was successfully suppressed (VL, mean ± SEM: 264,788 ± 93,207 vs. 217.5 ± 77.75, before and after cART, respectively) in all animals analyzed (Table 1). Our flow cytometric analysis showed no difference in either group in the relative frequencies of total LN CD3+ T cells before or after cART (Supplemental Figure 6A). An increased frequency of total LN CD4+ T cells (Supplemental Figure 6) was associated with a significant reduction in the frequency of total and bulk CCR7loCD95hi memory LN CD8+ T cells, but only in the animals treated during chronic infection (Figure 4, A and B). However, cART had a less profound effect on fCD8+ T cell frequencies (Figure 4C). We observed no accumulation of fCD8+ T cells in animals treated during the early infection stage (Figure 4C). Consistent with what we observed in the nontreated animals (Figure 3A), fCD8+ T cells in the treated animals more frequently expressed GrzB, although not significantly, compared with non-fCD8+ T cells after cART (Figure 4D). Collectively, our data show that cART, at least during the first 2 months of treatment, does not significantly affect fCD8+ T cell dynamics.
Table 1. VL before and after cART for each RM analyzed.
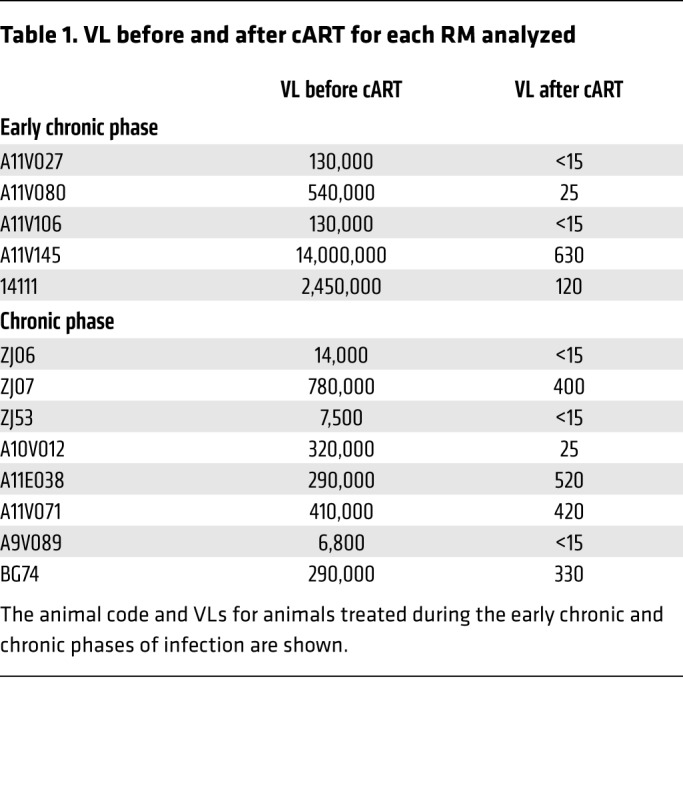
Figure 4. cART has a modest effect on the frequency of fCD8+ T cells.
(A) Pooled data showing the relative frequency of total LN CD8+ T cells before and after cART from RMs treated during early (n = 5) or late (n = 8) chronic SIV infection. The relative frequency of bulk memory LN CCR7loCD95hi CD8+ T cells (B) and fCD8+ T cells (C) is also shown. *P < 0.05 and **P < 0.001, by Mann-Whitney U test for unpaired comparisons and Wilcoxon test for paired comparisons. (D) Pooled data showing the relative frequency of GrzB+ non-fCD8+ and fCD8+ T cells from RMs treated during early (n = 5) or late (n = 8) chronic SIV infection. Mann-Whitney U test for unpaired comparisons and Wilcoxon test for paired comparisons.
Immune activation is associated with the accumulation of fCD8+ T cells in chronic SIV infection.
We used RNA confocal imaging that allowed for the localization of SIV mRNA in tissue samples in order to study the role of active viral transcription within the LNs in the dynamics of fCD8+ T cells. We detected SIV RNA during all stages of SIV infection in both follicular and extrafollicular areas (Supplemental Figure 7). In chronic SIV–infected samples, in addition to the intracellular RNA signal found in CD4+ T cells, we could detect, within the GC, extracellular viral RNA not associated with CD3+CD4+ T cells (Supplemental Figure 7), possibly representing virions captured by B cells and follicular DCs (23). We detected intracellular SIV mRNA in B cell follicles of animals with different levels of viremia (Supplemental Figure 8). Image analysis, however, did not show a preferential accumulation of fCD8+ T cells in the vicinity of actively transcribed virus (Figure 5A), suggesting that locally transcribed virus may not represent a major driving force for fCD8+ T cell recruitment.
Figure 5. Immune activation is associated with the accumulation of fCD8+ T cells during chronic SIV infection.
(A) Representative confocal images showing CD20+, CD3+, and CD4+ staining with SIV RNA ISH (RNAscope). Follicular areas from a noninfected, an early chronic, and a late chronic SIV-infected LN are shown. SIV mRNA is shown in red. Scale bars: 50 μm and 10 μm (enlarged images of boxed areas). (B) Pooled data showing the relative frequency of total LN CD8+ T cells in chronically SIV-infected RMs (n = 17) and AGMs (n = 5), expressed as the frequency of total CD3+ T cells. Mann-Whitney U test. (C) Relative appearance of LN CD8+ T cell subsets (top) and frequency of LN CCR7loCD95hi CD8+ T cells (bottom) in chronically SIV-infected RMs (n = 17) and AGMs (n = 5). ***P < 0.0001, by Mann-Whitney U test. (D) Relative frequency of fCD8+ T cells. ***P < 0.0001, by Mann-Whitney U test. (E) Representative confocal images showing B cell follicles and T cell distribution in 2 SIV-infected AGMs. Individual staining and merged images (CD3/CD4 and CD20/Ki67) are shown. Scale bars: 50 μm (top) and 100 μm (bottom). Original magnification, ×20. (F) Confocal images showing the distribution of PD-1hi cells and the presence of CXCL13 in follicular areas from chronically infected RMs (n = 1) and AGMs (n = 2). Two follicles (F1, F2) from each animal are shown. Original magnification, ×40. Scale bars: 20 μm.
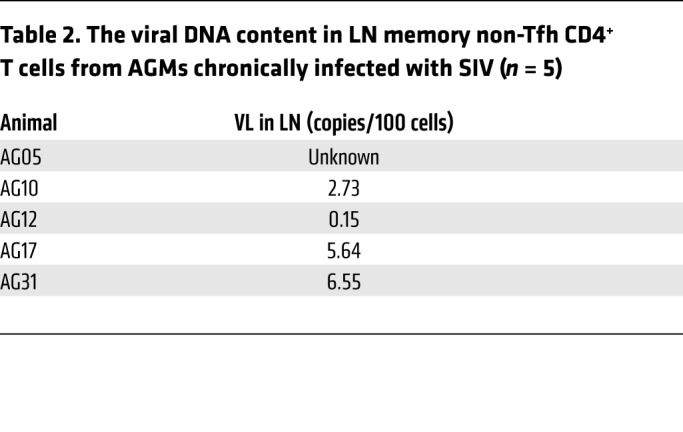
To further assess the role of immune activation in LN CD8+ T cell dynamics, we analyzed 5 African green monkeys (AGMs) with chronic SIV infection and detectable viral DNA in their LNs (Table 2). We detected negligible levels of immune activation, as judged by circulating levels of soluble CD14 (sCD14) (24) (data not shown). Flow cytometric analysis revealed a lower, but nonsignificant, frequency of total LN CD8+ T cells in AGMs compared with frequencies detected in RMs (Figure 5B). Chronic SIV infection in AGMs failed to result in the accumulation of either differentiated CCR7loCD95hi CD8+ T cells (Figure 5C) or fCD8+ T cells (Figure 5D) within LNs. In line with this profile, we found no accumulation of Tfh CD4+ T cells in LNs from AGMs compared with LNs from RMs (Supplemental Figure 9A). In contrast to chronically infected RMs (Supplemental Figure 2 and Supplemental Figure 3B), we observed no signs of follicular disorganization in the AGMs, not even in the 1 animal (AG31) that had ongoing GC reactivity (as determined by the presence of polarized Ki67hiCD20hi B cells) (Supplemental Figure 9B). Furthermore, image analysis confirmed the lack of fCD8+ T cells in chronically infected AGMs (Figure 5E). In line with the low frequency of Tfh cells in AGMs, we detected low expression of CXCL13, the ligand of CXCR5, in the LNs of SIV-infected AGMs (Figure 5F). NK cells were also more frequent in some follicular areas of AGMs compared with those of RMs (Supplemental Figure 9C), a finding that is in line with the results of a recent study (25). Although our study does not directly address the role of specific inflammatory mediators in the described dynamics, our data suggest that tissue inflammation plays a crucial role in the dynamics of LN-derived CD8+ T cells, particularly fCD8+ T cells, during SIV infection.
Table 2. The viral DNA content in LN memory non-Tfh CD4+ T cells from AGMs chronically infected with SIV (n = 5).
Functional monocytes accumulate within the LNs during chronic SIV.
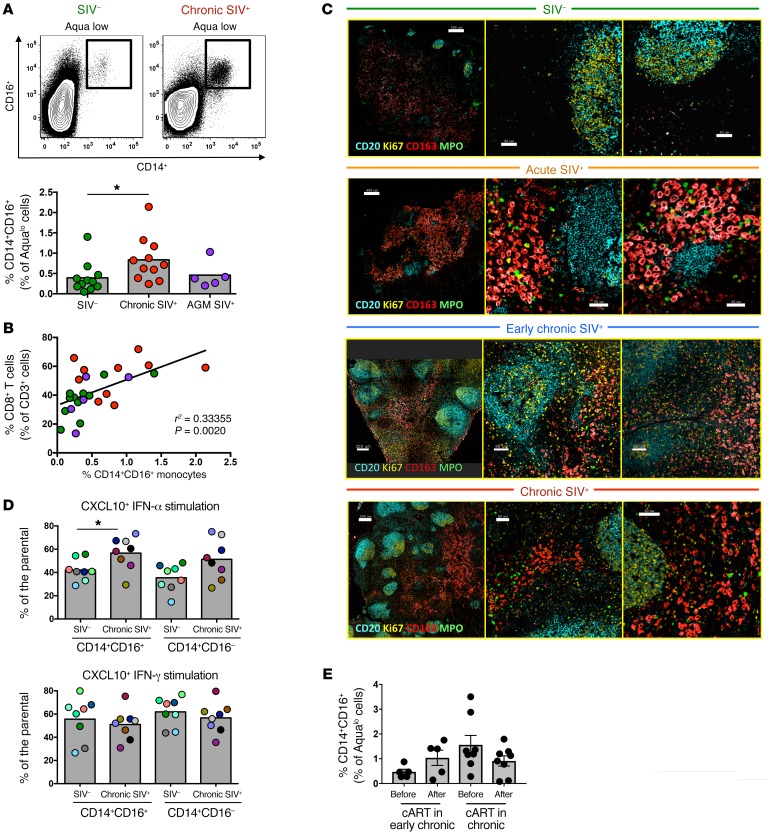
Given the possible role of tissue inflammation and immune activation in the movement and localization of LN CD8+ T cells, we sought to investigate the role of monocytes, an inflammatory cell population that accumulates in tissues during SIV infection (26), in CD8+ T cell dynamics within the LNs. We detected a significant accumulation of CD14hiCD16hi monocytes in chronically SIV-infected RMs, but not in AGMs (Figure 6A). Within the LNs, the frequency of monocytes was significantly associated with the frequency of CD8+ T cells (Figure 6B). Confocal imaging of CD20, Ki67, CD163, and myeloperoxidase (MPO) confirmed an increased presence of activated monocytes surrounding the B cell follicles during acute and chronic infection (Figure 6C). In vitro stimulation with LPS revealed similar monocyte functionality, as assessed by IL-1β production, between noninfected and chronically SIV-infected LNs (Supplemental Figure 10A). Monocytes were also stimulated in vitro with either IFN-γ or IFN-α to determine their capacity to produce CXCL10 (Supplemental Figure 10B), a ligand of the CXCR3 receptor implicated in the trafficking of effector T cells to secondary lymphoid organs (27). Both cytokines induced high levels of CXCL10, but, upon IFN-α stimulation, monocytes from chronically infected RMs had significantly higher CXCL10 production when compared with noninfected samples (Figure 6D). Furthermore, both CD14hiCD16hi and CD14hiCD16lo monocytes were able to produce CXCL10 upon stimulation (Figure 6D), despite a significantly different level of receptor expression (Supplemental Figure 10C). Plasma levels of IFN-γ and CXCL10 were similar, regardless of infection status (Supplemental Figure 10D). As seen for fCD8+ T cells (Figure 4B), cART also resulted in a moderate reduction of CD14hiCD16hi monocytes (Figure 6E). Therefore, infiltration of monocytes into the LNs could be a critical mechanism that triggers LN CD8+ T cell recruitment in SIV infection.
Figure 6. Accumulation of functional monocytes in proximity to follicular areas during SIV infection.
(A) Representative flow cytometric plots showing the gating scheme for identification of monocyte subsets and pooled data showing the relative frequency of CD14hiCD16hi monocytes in LNs from noninfected RMs (n = 11), chronically infected RMs (n = 11), and chronically infected AGMs (n = 5). *P < 0.05, by Mann-Whitney U test. (B) Linear regression analysis showing the association between the frequency of LN CD14hiCD16hi monocytes and LN total CD8+ T cells. (C) Representative confocal images showing the distribution of monocytes (CD163hi, in red) and granulocytes (MPOhi, in green) in LN tissues from noninfected and acutely and chronically SIV-infected RMs. Two zoomed areas close to the B cell follicle (defined by CD20 and Ki67 expression) from each animal are also shown. Scale bars: 400μm (top two), 200 μm (third row), and 300 μm (lower); enlarged 50 μm; 40 μm (second row, right). Original magnification, ×20. (D) Pooled data showing CXCL10 production by CD14hiCD16hi and CD14hiCD16lo monocytes (flow cytometric intracellular staining analysis) after short in vitro stimulation with either IFN-α or IFN-γ. Cells from noninfected (n = 8) and chronically SIV-infected (n = 8) RMs were analyzed. *P < 0.05, by Mann-Whitney U test. (E) Relative frequency of LN CD14hiCD16hi monocytes before and after cART from RMs treated during early (n = 5) or late (n = 8) SIV infection. Mann-Whitney U test for unpaired comparisons and Wilcoxon test for paired comparisons.
The majority of LN CD8+ T cells express a functional CXCR3 receptor.
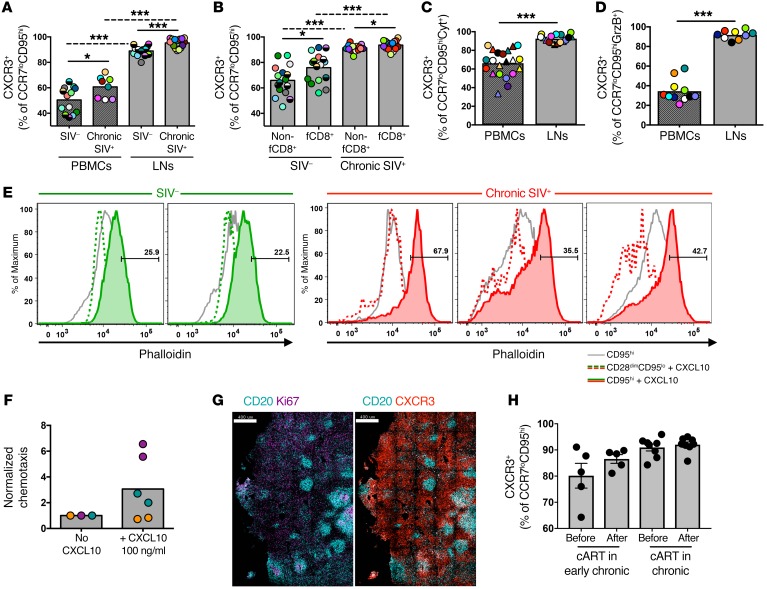
Previous studies have shown that CXCR3 can serve as an alternative trafficking mediator of CCR7lo CD8+ T cells into the LNs (28–30). Therefore, we analyzed the relative expression CXCR3 on circulating and LN CD8+ T cells (Supplemental Figure 11A). In contrast to circulating cells, LN CCR7loCD95hi CD8+ T cells consistently expressed a CXCR3hi phenotype (Figure 7A and Supplemental Figure11B). fCD8+ T cells had higher CXCR3 expression than did non-fCD8+ T cells (Figure 7B), and CXCR3 was expressed in the majority of SIV-specific (Figure 7C) and GrzB+ (Figure 7D) LN CCR7loCD95hi CD8+ T cells. Bulk memory CD8+ T cells, especially from SIV-infected LNs, showed a strong, rapid mobilization of F-actin in response to CXCL10 (Figure 7E), and chemotaxis of SIV-infected LN CD8+ T cells was evident in response to CXCL10 stimuli (Figure 7F). Therefore, the CXCR3 receptor on CCR7loCD95hi CD8+ T cells from SIV-infected LNs is functional and highly responsive to CXCL10-mediated signaling. Our image analysis confirmed the accumulation of CXCR3hi cells around and within the B cell follicle (Figure 7G). Finally, cART had no effect on the expression of CXCR3 by memory LN CD8+ T cells (Figure 7H). Our data indicate that CXCR3 could play an important role in CD8+ T cell trafficking into LNs and GCs during SIV infection.
Figure 7. The majority of LN CD8+ T cells express a functional CXCR3 receptor.
(A) Pooled data showing the relative frequency of CXCR3hiCCR7loCD95hi CD8+ T cells in samples of PBMCs (n = 13 SIV–; n = 8 chronic SIV) and LNs (n = 16 SIV–; n = 16 chronic SIV). *P < 0.05 and ***P < 0.0001, by Mann-Whitney U test for unpaired comparisons (solid lines) and Wilcoxon test for paired comparisons (dotted lines). (B) Pooled data showing the relative frequency of CXCR3hi non-fCD8+ and fCD8+ T cells from noninfected (n = 16) and chronically SIV-infected RMs (n = 16). *P < 0.05 and ***P < 0.0001, by Mann-Whitney U test. Dotted lines show paired analysis (Wilcoxon test). (C) Pooled data showing the frequency of CXCR3hi SIV-specific CCR7loCD95hi CD8+ T cells identified after short ex vivo stimulation with Gag (circles) or Env (triangles) peptide pools (Cyt+, IFN-γ, and/or TNF-α and/or MIP-1β). ***P < 0.0001, by Wilcoxon paired test. (D) Frequency of CXCR3hiCCR7loCD95hi CD8+ T cells that expressed an ex vivo GrzBhi phenotype in PBMC (n = 11) and LN (n = 9) samples from chronically SIV-infected RMs. Matching samples are highlighted with paired colors. ***P < 0.0001, by Mann-Whitney U test. (E) Histograms depicting F-actin mobilization, judged by phalloidin increase, in naive (CD28dimCD95lo) and memory CD95hi CD8+ T cells after short (5 s) ex vivo stimulation with CXCL10. Data from 2 noninfected and 3 chronically SIV-infected animals are shown. (F) Normalized chemotaxis levels of CD8+ T cells from SIV-infected LNs (n = 3) after a 3-hour stimulation with CXCL10. Matching samples are highlighted with paired colors. No statistically significant difference was found by Wilcoxon paired test. (G) Representative confocal images showing CXCR3 expression within LNs of a chronically SIV-infected RM. Scale bars: 400 μm. Original magnification, ×20. (H) Pooled data showing the relative frequency of CXCR3hiCCR7loCD95hi CD8+ T cells before and after cART from RMs treated during early (n = 5) or late (n = 8) SIV infection. Mann-Whitney U test for unpaired comparisons and Wilcoxon test for paired comparisons.
Discussion
We provide a comprehensive analysis of CD8+ T cell dynamics in LNs from noninfected; SIV-infected; and cART-treated, SIV-infected NHPs. Our data, which are in line with the fCD8+ T cell dynamics in LNs from HIV-infected individuals reported in another study (11), further validate the use of the SIV NHP model for the study of such dynamics. We demonstrate that fCD8+ accumulation (a) occurs during chronic SIV infection, (b) is present in both intact and disorganized follicles, and (c ) is associated with immune activation in and around B cell follicles. Furthermore, we provide evidence of possible inflammatory signals that could play an important role in this process.
Similar to HIV infection (11), chronic SIV infection is associated with the accumulation of CD8+ T cells expressing a CCR7loCXCR5hi phenotype, consistent with their follicular localization. While CXCR5 expression is lower on fCD8+ T cells than it is on Tfh cells, cross-linking of CXCR5 still initiates cellular signaling (judged by actin mobilization) in fCD8+ T cells compared with other LN CD8+ T cells. The use of the SIV model further allowed for the analysis of fCD8+ T cell dynamics at different stages of infection. Despite the accumulation of total CD8+ T cells in early chronic infection (day 45 p.i.) compared with the acute phase (day 14 p.i.), we did not observe fCD8+ T cell accumulation until late chronic infection (>6 months p.i.). Therefore, non-fCD8+ and fCD8+ T cells follow different kinetics during SIV infection. Whether this pattern reflects differential trafficking or delayed differentiation of relevant cell populations needs to be determined. We observed an initial accumulation of CXCR5loCCR7lo CD8+ T cells during early chronic infection (data not shown), possibly preceding the appearance of CXCR5hi fCD8+ T cells. However, CXCR5lo CD8+ T cells are, after TCR-driven stimulation, incapable of generating CXCR5hi T cells (31). Further investigation of these cell populations could inform the differentiation pathways of LN CD8+ T cells. Our histocytometric analysis confirmed the increased frequency of CD8+ T cells within the follicular and GC areas. It is well known that chronic HIV/SIV infection is characterized by major changes of LN tissue structure (32–35). However, although the sequestration of fCD8+ T cells was higher in disorganized follicles, we also observed fCD8+ T cell accumulation in intact follicles from chronically infected LNs. Our data indicate that both passive diffusion after barrier disruption in disorganized follicles and active trafficking/infiltration mechanisms may be involved in directing local fCD8+ T cell dynamics. We hypothesize that altered local cytokine/chemokine gradients could provide at least one mechanism for the in vivo movement of fCD8+ T cells. Therefore, chronic HIV/SIV infection creates a local environment in which CD8+ T cells are able to penetrate immunologically privileged areas like the GC.
As with previous reports in humans (11) and NHPs (36), we found that chronic SIV infection was characterized by a significantly increased frequency of fCD8+ T cells expressing GrzB ex vivo. Image analysis confirmed the presence of GrzBhi CD8+ T cells within B cell follicles and GCs. This cytolytic phenotype was associated with a higher capacity to mediate in vitro bispecific antibody–redirected killing of infected targets. Previous studies using in situ tetramer staining have shown that bulk and SIV-specific CD8+ T cells are less represented in the follicular areas (16, 36). Using a flow cytometric–based approach, we also did not find a preferential accumulation of SIV-specific CTLs (identified by either cytokine production or tetramer binding) in follicles in the small group of chronically infected animals tested. We should emphasize that our assay measures the relative frequencies of SIV-specific CD8+ T cells, a measurement affected by the overall presence of bulk CD8+ T cells in both the extrafollicular and follicular areas. Thus, our results are in line with those of previous studies showing that higher numbers of SIV-specific CTLs in extrafollicular areas are strongly associated with higher numbers of SIV-specific CTLs within the GC, suggesting that frequencies remain constant between these 2 compartments (37). Despite the lack of preferential recruitment of SIV-specific CD8+ T cells within the GC, we provide evidence that fCD8+ T cells can target and kill infected cells if redirected with bispecific antibodies, consistent with what has been shown in humans (11). Although fCD8+ T cells do not accumulate in acute infection, this type of immunotherapy could operate in extrafollicular areas where active viral replication predominates during acute infection. Our data provide an immunological basis for the evaluation of such immunotherapies in the SIV NHP model (38, 39).
Unraveling the dynamics of fCD8+ T cells will provide clues for the development of novel curative strategies targeting the potential killing activity of these T cells. To this end, investigation of LN CD8+ T cell dynamics in cART-treated subjects is of great importance. We were able to show that suppressive cART had an overall modest effect on the frequency of NHP fCD8+ T cells, which is in line with the data described for human fCD8+ T cells (11). However, a significant drop in fCD8+ cells was found in animals with a higher baseline frequency of fCD8+ cells. Although cART effectively controls viral replication, the immune activation is not reduced back to the levels found in healthy individuals (40). We hypothesize that the higher the accumulation of fCD8+ cells, which is associated with higher levels of tissue inflammation and immune activation, the more impactful the cART will be in correcting the local activation and thereby affecting fCD8+ T cell accumulation. Furthermore, cART had no effect on the expression of GrzB by fCD8+ cells. Unfortunately, we were not able to find samples from NHPs treated for long periods of time. We should emphasize, however, that the short treatment duration (2 months) successfully suppressed the VL, a profile associated with a significant effect on the frequency of total and bulk memory LN CD8+ T cells. Our data reveal differential kinetics of fCD8+ T cells compared with other LN CD8+ T cells in cART-treated NHPs, a finding that could be extended to cART treatment of HIV-infected individuals.
The mechanisms regulating the dynamics of CD8+ T cells within the LNs during HIV/SIV infection are largely unknown. We have shown that local active replication is not the primary driver for fCD8+ T cell dynamics in HIV infection (11). Here, we performed ISH and confocal imaging to detect SIV mRNA within the LN areas. Again, we did not observe a preferential accumulation of fCD8+ T cells adjacent to SIV RNA+ cells, even in animals with high viremia. Therefore, SIV replication, while a necessary component for the accumulation of fCD8+ T cells within the LNs (31), may not be sufficient to induce fCD8+ T cell trafficking. Thus, we analyzed the role of local immune activation in this process. We took advantage of the AGM SIV model, in which immune activation in the LNs is very low, despite active viral replication (41, 42). The frequency of fCD8+ T cells in chronically infected AGMs was negligible. Furthermore, AGM follicles showed canonical LN and GC morphology, even in 1 animal that had active GC formation. The low frequency of fCD8+ T cells was accompanied by a low frequency of Tfh cells and low expression of CXCL13, the ligand for CXCR5 and a major regulator of the follicular trafficking of CD4+ and CD8+ T cells, in the follicular areas in AGMs compared with those in RMs, indicating an interplay between local immune activation and adaptive T cell dynamics. Therefore, immune activation could provide critical signals for the observed fCD8+ T cell dynamics in chronic SIV and HIV infection.
During acute viral infection, monocytes are recruited to the site of infection and then migrate to the LNs to exert inflammatory signals involved in T cell priming (43). Accordingly, monocytes accumulate in SIV-infected LNs, which are inflammatory sites where active SIV replication occurs. We found that monocytes and macrophages, surrogates of local inflammation and immune activation, accumulated around the B cell follicles during acute infection and persisted during chronic infection, while fCD8+ accumulation was not observed until the chronic phase. Therefore, tissue sequestration of inflammatory cells could represent an important factor for the recruitment of fCD8+ T cells during SIV infection. We found a strong correlation between the frequencies of LN CD8+ T cells and monocytes in chronic SIV, suggesting a mechanistic link. In line with the possible role of tissue immune activation and inflammation in this process is the reduced frequency of monocytes found in the AGM LNs. Furthermore, a comparison of tissue dynamics before and after cART revealed a modest reduction in tissue monocytes, which was associated with a similarly overall modest reduction in the frequency of fCD8+ T cells. To dissect specific inflammatory pathways in this process, we investigated the possible role of CCXL10 (IP10), a strong chemoattractant for CD8+ T cells (44). In particular, CXCL10 has a dominant role in the recruitment, through CXCR3, of effector CD8+ T cells that lack CCR7 expression (27–29). We found that monocytes from SIV-infected, as compared with uninfected, RMs secreted significantly higher amounts of CXCL10 in response to IFN-α stimulation. Recent studies have shown that in vivo stimulation of type I IFN pathways results in decreased virus replication (45) and slows disease progression (46). Our data indicate that recruitment of fCD8+ T cells in the LNs could, at least in part, be responsible for this effect. As with HIV infection (47), we found that the frequency of CD8+ T cells expressing CXCR3 was markedly increased in chronically SIV-infected LNs. Despite the accumulation of total LN CD8+ T cells, we detected similar frequencies of bulk LN CXCR3hi memory CD8+ T cells between noninfected and early infected animals. This could be due to a low frequency of circulating CXCR3hi effector CD8+ T cells at this phase of infection, limited recruitment of these cells to the LNs, or a combination of these factors. Interestingly, cART had no effect on the expression of CXCR3 by LN CD8+ T cells. Our in vitro studies show that CXCR3 expressed on CD8+ T cells from chronically infected LNs is functional. Therefore, our results suggest that the accumulation of monocytes within the LNs in chronic SIV infection could facilitate a positive feedback loop that leads, through the CXCR3/CXCL10 axis, to the recruitment of differentiated CD8+ T cells to B cell follicles. Overall, we identify a model in which the availability of relevant cells in circulation, accompanied by dramatic changes in LN architecture and local inflammation signals, facilitates the recruitment and accumulation of potential cytolytic CD8+ T cells within follicular areas. However, further study is needed to fully understand the mechanisms regulating CD8+ T cell trafficking to GCs.
Our data are in line with the intra-LN CD8+ T cell dynamics recently described in HIV-infected individuals (11) and support the use of SIV infection of NHPs to delineate the mechanisms that regulate LN CD8+ T cell dynamics. Our data suggest that tissue inflammation and immune activation are at least one of the driving forces in fCD8+ T cell dynamics and provide the basis for future studies aimed at manipulating CD8+ T cell trafficking in the LNs. Importantly, inflammation also affects viral and Tfh CD4+ T cell dynamics and, as a result, affects this important viral reservoir. Therefore, targeting inflammatory molecules and pathways that could specifically affect CD8+ T cell dynamics could represent a strategy to augment cytotoxic responses in LNs. Furthermore, fCD8+ T cells are characterized by potent ex vivo killing of infected cells via bispecific antibodies, justifying the further exploration of such tools in combination with therapies that increase fCD8+ T cell trafficking to GCs as potential curative strategies.
Methods
Animals
LN samples and PBMCs from Indian RMs (Macaca mulatta) and AGMs (Chlorocebus pygerythrus) were analyzed in this study. We included non–SIV-infected RMs (n = 15), acute SIV-infected RMs (14 days, n = 10), early chronically SIV-infected RMs (45 days, n = 8), chronically SIV-infected (>6 months) RMs (n = 20), cART-treated RMs (n = 13) and AGMs (n = 5). Animals were treated for 7 weeks (for animals treated during late chronic infection) or 27 weeks (for animals treated during early chronic infection) with a combination of 5 drugs given daily: s.c. injections of 20 mg/kg/day tenofovir and 30 mg/kg/day emtricitabine (Gilead Sciences) and 100 mg b.i.d. raltegravir (Merck), 800 mg b.i.d. darunavir (Janssen Pharmaceuticals), and 100 mg b.i.d. ritonavir (Abbvie Inc.), all mixed with food. MamuA01 chronically SIV-infected animals (n = 5) were used for experiments including tetramer staining.
Tissue processing
Biopsied LNs were washed with cold R-10 medium (RPMI 1640, 10% FBS, 2 mM L-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin). Fat and connective tissue encircling the LN was completely removed. A fraction of each tissue was fixed in 10% neutral buffered formalin (Sigma-Aldrich) overnight at room temperature (RT) and embedded facedown in a paraffin block. The remainder of the tissue was cut into small pieces, transferred into C tubes (Miltenyi Biotec) containing 5 ml R-10 medium, and processed to cell suspensions with a GentleMACS Cell Dissociator (Miltenyi Biotec). Cells were frozen (10% DMSO in FBS) and stored in liquid nitrogen until further use. PBMCs were isolated by Ficoll gradient density (Sigma-Aldrich), frozen (10% DMSO in FBS), and stored in liquid nitrogen until further use. Monocyte stimulations and receptor analyses were performed on fresh PBMCs.
Antibodies
Flow cytometry.
The following directly conjugated monoclonal antibodies were used: (BD Biosciences) CD3–APC Cy7 (catalog 557757, clone SP34-2); CCR7–Alexa Fluor 700 (catalog 561143, clone 150503) or CCR7-BV421 (catalog 562555, clone 150503), CD8–Pacific blue (catalog 558207, clone RPA-T8), IFN-γ–PE (catalog 554701, clone B27), MIP-1b–Cy7PE (catalog 560687, clone D21-1351), GrzB-PE (catalog 561142, clone GB11), CD14-Cy7PE (catalog 561385, clone M5E2), CD16–Pacific blue (catalog 558122, clone 3G8), IL-6–FITC (catalog 554696, clone MQ2-6A3), and CD95-Cy5PE (catalog 561977, clone DX2); (Beckman Coulter) CD28-ECD (catalog 6607111, clone Tp44); (BioLegend) PD-1–BV711 (catalog 329928, clone EH12.2H7) or PD-1–BV421 (catalog 329920, clone EH12.2H7), CD20-BV570 (catalog 302336, clone 2H7), IFN-γR–PE (catalog 308504, clone 2HUB-159), and CXCL10-PE (catalog 519504, clone J034D6); (Life Technologies, Thermo Fisher Scientific) CD4-QD605 (catalog Q10008, clone S3.5) and CD8-QD655 (catalog Q10055, clone 3B5); (eBioscience) CXCR5-Cy7PE (catalog 25-9185-41, clone MU5UBEE) or CXCR5–PerCP eFluor 710 (catalog 46-9185-42, clone MU5UBEE), CXCR3-FITC (catalog 11-1831-82, clone CXCR3-173), and IL-1β–PE (catalog 12-7018-82, clone CRM56); and (Mabtech) Prf-FITC (catalog 3465-7, clone pf-344). Purified anti–IFN-αRII antibody was obtained from Fitzgerald (catalog 10-I64A, clone M99012521). TNF-α–Alexa Fluor 594 (clone MAb11) was conjugated in-house. Aqua amine viability dye was obtained from Invitrogen (Thermo Fisher Scientific).
Confocal microscopy.
We used directly conjugated antibodies against Ki67-Ax700 (catalog 561277) or Ki67-Ax488 (catalog 561165) (BD Biosciences, clone B56); CXCR3-FITC (eBioscience, catalog 11-1831-82, clone CXCR3-173); CD20–eFluor 615 (eBiosciences, catalog 42-0202-82, clone L26); and CD4-Ax488 (R&D Systems, catalog FAB8165G, goat polyclonal); and nonconjugated antibodies against CD3 (Dako, catalog M725401, clone F7.2.38); CD163 (Thermo Fisher Scientific, catalog MA5-11458, clone 10D6); MPO (Dako, catalog A0398, rabbit polyclonal); GrzB (Dako, catalog M723501-2, clone GrB-7); NKG2A (Beckman Coulter, catalog COIM2750, clone Z199); and CXCL13 (Thermo Fisher Scientific, catalog PA5-47035, goat polyclonal). The following nonconjugated antibodies were used in combination with secondary antibodies obtained from Life Technologies (Thermo Fisher Scientific): goat anti–mouse IgG2a-Ax488 or IgG2a-Ax680 for GrzB detection and goat anti–mouse IgG1-Ax647 for CD3 and CD163 detection; or from BioLegend: donkey anti–rabbit IgG-BV421 for MPO detection. Jojo-1 (Life Technologies, Thermo Fisher Scientific) was used for nuclear staining.
Peptide–MHC class I tetramers
Soluble fluorochrome–labeled peptides (pMamu-A*01) specific for SIV Tat TL8 (TTPESANL; Tat, residues 28–35) and SIV Gag CM9 (CTPYDINQM; Gag, residues 181–189) were provided by David Watkins of the Department of Pathology, Miller School of Medicine, University of Miami (Miami, Florida, USA) and used as previously described (48, 49).
Polychromatic flow cytometry
Phenotypic analysis.
Cells were thawed and rested for 2 hours before further use. Cells were stained with Aqua amine viability dye and then with titrated amounts of antibodies against surface markers. For T cell analysis, anti-CD3, anti-CD4, anti-CD8, anti-CD28, anti-CD95, anti-CXCR5, anti-CCR7, anti-CXCR3, anti–PD-1, and anti-CD20 were used. PE- and APC-conjugated CM9 and TL8 in-house–produced tetramers were used to quantify SIV-specific CD8+ T cells in MamuA01 samples. For monocyte analysis, anti-CD3, anti-CD20, anti-CD14, anti-CD16, and, for some experiments, anti–IFN-αR and anti–IFN-γR were used. After a final washing step, cells were fixed with 1% paraformaldehyde.
Functional analysis.
For ex vivo detection of GrzB and Perf, thawed cells were rested for 2 hours and then incubated with Aqua amine viability dye and surface stained with titrated amounts of anti-CD3, anti-CD4, anti-CD8, anti-CD95, anti-CD28, anti-CXCR3, anti-CCR7, and anti-CXCR5 antibodies, and then washed, permeabilized (Cytofix/Cytoperm, BD Biosciences), and stained with anti-GrzB and anti-Prf antibodies. Cells were washed and fixed with 1% paraformaldehyde. For de novo production of cytokines, thawed cells were rested for 2 hours and stimulated (2 × 106 cells/ml in the presence of 10 μg/ml brefeldin A) with either anti-CD3, anti-CD2, and anti-CD28 beads (Miltenyi Biotec) or peptide pools containing 400 μg SIV Gag or Env peptides. Peptides in incubation mixtures were each present at 2 μg/ml and were at least 70% pure. After 6 hours of stimulation, samples were surface stained with Aqua amine viability dye and titrated amounts of anti-CD4, anti-CD8, anti-CD28, anti-CD95, anti-CCR7, anti-CXCR5, and anti-CXCR3. Cells were washed, fixed, and permeabilized (Cytofix/Cytoperm, BD Biosciences) and intracellularly stained with anti-CD3, anti–IFN-γ, anti–MIP-1b, and anti–TNF-α. Cells were washed and fixed with 1% paraformaldehyde. At least 5 × 105 events were acquired for each sample.
Monocyte stimulation.
Fresh PBMCs were stimulated with IFN-γ (20 ng/ml), IFN-α (1041 U/ml), or LPS (1 ng/ml) for 6 hours in the presence of 10 μg/ml brefeldin A and 0.7 μg/ml monensin. After stimulation, cells were stained with Aqua amine viability dye and titrated amounts of anti-CD3, anti-CD20, anti-CD14, and anti-CD16. Cells were washed, fixed, and permeabilized (Cytofix/Cytoperm, BD Biosciences), and intracellular staining was performed with either anti-CXCL10 (for IFN-γ and IFN-α stimulation) or anti–IL-1β (LPS stimulation). A minimum of 1 × 106 events were acquired for each sample.
Fluidigm analysis
Fluidigm analysis using 96 primer pools was performed as previously described (50). Briefly, 30 cells from each population were sorted in triplicate and retro-transcribed with a pool of the 59 gene primers to specifically amplify the transcripts of interest. Then, multiplex quantitative PCR (qPCR) was performed using a microfluidic chip from the Biomark system. Et values (Et = 40 – Ct) were calculated by subtracting the Ct value from the total number of cycles performed during the qPCR process.
F-actin mobilization
Cells derived from noninfected (n = 2) and chronically SIV-infected (n = 3) LNs were thawed, rested at 37°C, and surface stained with titrated amounts of anti-CD3, anti-CD4, anti-CD8, anti-CD28, and anti-CD95 antibodies. After a washing step, cells were resuspended in R-10 and rested for 15 minutes at 37°C. Samples were then stimulated or not with 1 μg/ml CXCL10. After 5 seconds, the stimulation was stopped by adding 750 μl Cytofix/Cytoperm buffer (BD Biosciences) for 20 minutes at RT. Cells were then incubated for 20 minutes with 0.8 U/ml Alexa 647–conjugated phalloidin (Thermo Fisher Scientific), a high-affinity probe for F-actin. After a final wash, cells where fixed with 1% PFA and analyzed by flow cytometry.
Chemotaxis assay
Cells (2 × 105) from chronically SIV-infected LNs were located in the upper chamber of a multiwell Boyden chamber. CXCL10 (100 ng/ml) was added to the lower chamber for the positive wells. After a 3-hour incubation at 37°C, Dynosphere beads (15 μm, 104 beads, Thermo Fisher Scientific) were added to the lower chamber, and the contents were recovered and transferred to a FACS tube. A fixed number of events was collected for each tube to quantify normalized chemotaxis levels.
VL measurement
Memory LN non-Tfh CD4+ T cells were sorted, and SIVagm DNA was measured by qPCR as previously described (51).
Imaging studies
Confocal microscopy and histocytometric analysis.
Formalin-fixed, paraffin-embedded sections (6- to 10-μm thick) were warmed for 1 hour at 50°C and then deparaffinized and rehydrated in sequential baths of xylene, ethanol, and water. Antigen retrieval was performed in a decloaking chamber (Biocare Medical) for 15 minutes at 110°C (6 psi) using Borg Decloaker Buffer (Biocare Medical). After a 1-hour incubation with blocking buffer (0.1 M Tris, 0.3% Triton X-100, 1% BSA), the sections were incubated overnight with titrated amounts of purified antibodies. Slides were washed with PBS (3 × 20 min) and incubated with the appropriate secondary antibodies for 2 hours at RT. Slides were washed again with PBS (3 × 20 min) and, after a second blocking step with a 1:10 dilution of normal mouse serum (1 h at RT), incubated with titrated amounts of conjugated antibodies (2 h at RT). After a final washing step (3 × 20 min) and Jojo-1 staining (1:15,000, 15 min at RT), slides were mounted with Fluoromount G (Southern Biotech). Tissues were imaged using a Leica SP8 confocal microscope with ×20 0.75 NA or ×63 1.40 NA objectives with a ×1.5 optical zoom at 512 × 512 pixel density. Multiparameter confocal images analyzed with the histocytometry pipeline (20) were obtained with a ×40 1.30 NA objective at 1,024 × 1,024 pixel density. Compensation of imaging data was carried out by applying a compensation matrix constructed from single-staining tissues. Histocytometry was performed as previously reported, with minor modifications (11). In brief, 3D segmented surfaces (based on the nuclear signal) of spillover corrected images were generated with Imaris (Bitplane) via the Surface Creation module. Average voxel intensities for all channels were exported to Microsoft Excel and combined into a unified spreadsheet. This file, in comma-separated values format, was imported to FlowJo, version X0.7, for further analysis.
SIV-RNA confocal imaging.
Formalin-fixed, paraffin-embedded tissue sections were stained with SIV RNA probes using the RNAscope ISH protocol (ACD) according to the manufacturer’s instructions. Briefly, sections were baked for 1 hour at 60°C and deparaffinized using serial xylene and ethanol baths. This was followed by an antigen retrieval step performed at 100°C for 15 minutes and a proteinase K treatment step for 20 minutes at 40°C. Subsequently, sections were incubated with SIVmac251-specific RNA probes (ACD) at 40°C for 2 hours before being subjected to 4 rounds of signal amplification that were performed using the amplification reagents provided in the RNAscope kit. To visualize the fCD8+, CD4+, and CD20+ cell subsets, sections were stained with antibodies against CD20 (L26, eBioscience), CD3 (F7.2.38, Dako), and CD4 (1F6, Thermo Fisher Scientific) at the end of the RNAscope protocol. Images were acquired on a TCS SP8 Leica confocal microscope using a ×40 objective (NA 1.3) and analyzed using Imaris, version 8.2.0.
Bispecific antibody studies
Construction and purification.
Bispecific antibodies were constructed and used as previously reported (17). In brief, the cDNA for human VRC07-523 Fab was amplified from the IgG vector and assembled to a cross-reactive anti–human CD3 using overlapping PCR. The scFv fragment of a cross-reactive anti–human CD3 monoclonal antibody was linked by a 16-aa GS linker to the light chain of VRC07-523, leaving intact the VH/CH1 domains to form the bispecific antibody. Cross-reactive anti–human CD3 scFv sequences were synthesized using human preferred codons (GenScript). All bispecific antibodies were configured as Fab-scFv. Assembled cDNAs were cloned into a mammalian expression vector for protein production using 293F cells. Large quantities of bispecific antibodies were produced by transfecting 293F cells using 293Fectin according to the manufacturer’s protocol (Life Technologies, Thermo Fisher Scientific). Five days after transfection, cell culture supernatants were harvested and filtered. The proteins were purified using Kappaselect beads (GE Healthcare Biosciences), and only the monomer fractions were collected for further characterization. The endotoxin level of all purified antibodies was measured, and samples were passed through an endotoxin removal column (Hyglos) when necessary. Only samples with endotoxin levels below 1 EU/mg were used in this study.
In vitro killing assay.
LN cells were sorted on the basis of CD8, CD28, CD95, CCR7, and CXCR5 expression. Different populations of sorted CD8+ T cells were cocultured with HIV-infected CEM cells (CEM cells infected with HIV-IIIB virus, infectivity >90%) previously labeled with PKH (Sigma-Aldrich) in the presence of an anti-CD3/VRC07 bispecific antibody (500 ng/ml). After 8 hours, the cells were harvested, washed, and stained with Aqua amine viability dye and annexin V Alexa Fluor 647. The frequency of the lysis was calculated on the basis of the Aqua+annexin V+ and Aqua–annexin V+ cell ratios in the target cell population.
Statistics
Experimental variables were analyzed using the nonparametric Mann-Whitney U and Wilcoxon tests for unpaired or paired variables, respectively. P values of less than 0.05 were considered statistically significant. GraphPad Prism Version 7.0a (GraphPad Software) was used to create the graphs and perform the statistical analysis. The data are shown as individual points, with mean values highlighted as bars with SD. Analysis and graphical representation of cytokine production (polyfunctionality assays) were done using Simple Presentation of Incredibly Complex Evaluations (SPICE)software (version 5.05013) provided by M. Roederer (NIH).
Study approval
All studies were reviewed and approved by the IRBs of the VRC, NIAID, and NCI, NIH; the Yerkes National Primate Research Center at Emory University; and the Wisconsin National Primate Research Center at the University of Wisconsin (Madison, Wisconsin, USA). All animals were handled in accordance with the standards of the American Association of Laboratory Animal Care.
Author contributions
RAK, SFM, and CP conceived the study and designed the experiments. SFM, EM, SA, and DA performed and analyzed experiments. MP, GNP, JMB, KNM, DP, and DD provided samples. ABM gave conceptual advice. AP and JRM provided the bispecific antibodies. RAK, SFM, and CP wrote the manuscript.
Supplementary Material
Acknowledgments
Authors would like to thank the staff of the Flow Cytometry Core at the VRC, NIAID and David Watkins at the Miller School of Medicine, University of Miami for providing cells and reagents (tetramers). This research was supported by the Intramural Research Program of the VRC, NIAID and NCI, NIH, and a Collaboration for AIDS Vaccine Discovery (CAVD) grant (OP1032325) from the Bill and Melinda Gates Foundation (to RAK). This project was supported in part by a Yerkes National Primate Research Center Base Grant (ORIP/OD P51OD011132).
Version 1. 04/16/2018
Electronic publication
Version 2. 05/01/2018
Print issue publication
Footnotes
Phone: 301.761.6993; Email: petrovasc@mail.nih.gov.
SFM’s present address is: Microbial Sciences, MedImmune LLC, Gaithersburg, Maryland, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(5):2089–2103.https://doi.org/10.1172/JCI96207.
Contributor Information
Eirini Moysi, Email: eirini.moysi@gmail.com.
Amarendra Pegu, Email: pegua@niaid.nih.gov.
Sarah Andrews, Email: sarah.andrews2@nih.gov.
Adrian B. McDermott, Email: adrian.mcdermott@nih.gov.
David Palesch, Email: david.johannes.palesch@emory.edu.
Mirko Paiardini, Email: mirko.paiardini@emory.edu.
Jason M. Brenchley, Email: jbrenchl@mail.nih.gov.
John R. Mascola, Email: jmascola@nih.gov.
Constantinos Petrovas, Email: petrovasc@mail.nih.gov.
Richard A. Koup, Email: rkoup@mail.nih.gov.
References
- 1.Koup RA, et al. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68(7):4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmitz JE, et al. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283(5403):857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 3.Lifson JD, et al. Role of CD8(+) lymphocytes in control of simian immunodeficiency virus infection and resistance to rechallenge after transient early antiretroviral treatment. J Virol. 2001;75(21):10187–10199. doi: 10.1128/JVI.75.21.10187-10199.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindqvist M, et al. Expansion of HIV-specific T follicular helper cells in chronic HIV infection. J Clin Invest. 2012;122(9):3271–3280. doi: 10.1172/JCI64314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perreau M, et al. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med. 2013;210(1):143–156. doi: 10.1084/jem.20121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu Y, et al. Simian immunodeficiency virus infects follicular helper CD4 T cells in lymphoid tissues during pathogenic infection of pigtail macaques. J Virol. 2013;87(7):3760–3773. doi: 10.1128/JVI.02497-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukazawa Y, et al. B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers. Nat Med. 2015;21(2):132–139. doi: 10.1038/nm.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iyengar S, Chin B, Margolick JB, Sabundayo BP, Schwartz DH. Anatomical loci of HIV-associated immune activation and association with viraemia. Lancet. 2003;362(9388):945–950. doi: 10.1016/S0140-6736(03)14363-2. [DOI] [PubMed] [Google Scholar]
- 9.Mudd PA, et al. Vaccine-induced CD8+ T cells control AIDS virus replication. Nature. 2012;491(7422):129–133. doi: 10.1038/nature11443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quigley MF, Gonzalez VD, Granath A, Andersson J, Sandberg JK. CXCR5+ CCR7- CD8 T cells are early effector memory cells that infiltrate tonsil B cell follicles. Eur J Immunol. 2007;37(12):3352–3362. doi: 10.1002/eji.200636746. [DOI] [PubMed] [Google Scholar]
- 11.Petrovas C, et al. Follicular CD8 T cells accumulate in HIV infection and can kill infected cells in vitro via bispecific antibodies. Sci Transl Med. 2017;9(373):eaag2285. doi: 10.1126/scitranslmed.aag2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Folkvord JM, Armon C, Connick E. Lymphoid follicles are sites of heightened human immunodeficiency virus type 1 (HIV-1) replication and reduced antiretroviral effector mechanisms. AIDS Res Hum Retroviruses. 2005;21(5):363–370. doi: 10.1089/aid.2005.21.363. [DOI] [PubMed] [Google Scholar]
- 13.Brodie SJ, et al. In vivo migration and function of transferred HIV-1-specific cytotoxic T cells. Nat Med. 1999;5(1):34–41. doi: 10.1038/4716. [DOI] [PubMed] [Google Scholar]
- 14.Hosmalin A, et al. HIV-specific effector cytotoxic T lymphocytes and HIV-producing cells colocalize in white pulps and germinal centers from infected patients. Blood. 2001;97(9):2695–2701. doi: 10.1182/blood.V97.9.2695. [DOI] [PubMed] [Google Scholar]
- 15.Folkvord JM, Anderson DM, Arya J, MaWhinney S, Connick E. Microanatomic relationships between CD8+ cells and HIV-1-producing cells in human lymphoid tissue in vivo. J Acquir Immune Defic Syndr. 2003;32(5):469–476. doi: 10.1097/00126334-200304150-00001. [DOI] [PubMed] [Google Scholar]
- 16.Connick E, et al. CTL fail to accumulate at sites of HIV-1 replication in lymphoid tissue. J Immunol. 2007;178(11):6975–6983. doi: 10.4049/jimmunol.178.11.6975. [DOI] [PubMed] [Google Scholar]
- 17.Pegu A, et al. Activation and lysis of human CD4 cells latently infected with HIV-1. Nat Commun. 2015;6:8447. doi: 10.1038/ncomms9447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marsden MD, Zack JA. Double trouble: HIV latency and CTL escape. Cell Host Microbe. 2015;17(2):141–142. doi: 10.1016/j.chom.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evans DT, Silvestri G. Nonhuman primate models in AIDS research. Curr Opin HIV AIDS. 2013;8(4):255–261. doi: 10.1097/COH.0b013e328361cee8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerner MY, Kastenmuller W, Ifrim I, Kabat J, Germain RN. Histo-cytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity. 2012;37(2):364–376. doi: 10.1016/j.immuni.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pantaleo G, Fauci AS. New concepts in the immunopathogenesis of HIV infection. Annu Rev Immunol. 1995;13:487–512. doi: 10.1146/annurev.iy.13.040195.002415. [DOI] [PubMed] [Google Scholar]
- 22.Petrovas C, et al. SIV-specific CD8+ T cells express high levels of PD1 and cytokines but have impaired proliferative capacity in acute and chronic SIVmac251 infection. Blood. 2007;110(3):928–936. doi: 10.1182/blood-2007-01-069112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heesters BA, et al. Follicular Dendritic Cells Retain Infectious HIV in Cycling Endosomes. PLoS Pathog. 2015;11(12):e1005285. doi: 10.1371/journal.ppat.1005285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sandler NG, et al. Plasma levels of soluble C independently predict mortality in HIV infection. J Infect Dis. 2011;203(6):780–790. doi: 10.1093/infdis/jiq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huot N, et al. Natural killer cells migrate into and control simian immunodeficiency virus replication in lymph node follicles in African green monkeys. Nat Med. 2017;23(11):1277–1286. doi: 10.1038/nm.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hasegawa A, et al. The level of monocyte turnover predicts disease progression in the macaque model of AIDS. Blood. 2009;114(14):2917–2925. doi: 10.1182/blood-2009-02-204263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffith JW, Sokol CL, Luster AD. Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu Rev Immunol. 2014;32:659–702. doi: 10.1146/annurev-immunol-032713-120145. [DOI] [PubMed] [Google Scholar]
- 28.Kastenmüller W, Brandes M, Wang Z, Herz J, Egen JG, Germain RN. Peripheral prepositioning and local CXCL9 chemokine-mediated guidance orchestrate rapid memory CD8+ T cell responses in the lymph node. Immunity. 2013;38(3):502–513. doi: 10.1016/j.immuni.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khan IA, et al. IP-10 is critical for effector T cell trafficking and host survival in Toxoplasma gondii infection. Immunity. 2000;12(5):483–494. doi: 10.1016/S1074-7613(00)80200-9. [DOI] [PubMed] [Google Scholar]
- 30.Guarda G, et al. L-selectin-negative CCR7- effector and memory CD8+ T cells enter reactive lymph nodes and kill dendritic cells. Nat Immunol. 2007;8(7):743–752. doi: 10.1038/ni1469. [DOI] [PubMed] [Google Scholar]
- 31.Mylvaganam GH, et al. Dynamics of SIV-specific CXCR5+ CD8 T cells during chronic SIV infection. Proc Natl Acad Sci U S A. 2017;114(8):1976–1981. doi: 10.1073/pnas.1621418114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schacker TW, et al. Lymphatic tissue fibrosis is associated with reduced numbers of naive CD4+ T cells in human immunodeficiency virus type 1 infection. Clin Vaccine Immunol. 2006;13(5):556–560. doi: 10.1128/CVI.13.5.556-560.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng M, Haase AT, Schacker TW. Lymphoid tissue structure and HIV-1 infection: life or death for T cells. Trends Immunol. 2012;33(6):306–314. doi: 10.1016/j.it.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 34.Estes JD. Pathobiology of HIV/SIV-associated changes in secondary lymphoid tissues. Immunol Rev. 2013;254(1):65–77. doi: 10.1111/imr.12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tabb B, et al. Reduced inflammation and lymphoid tissue immunopathology in rhesus macaques receiving anti-tumor necrosis factor treatment during primary simian immunodeficiency virus infection. J Infect Dis. 2013;207(6):880–892. doi: 10.1093/infdis/jis643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Connick E, et al. Compartmentalization of simian immunodeficiency virus replication within secondary lymphoid tissues of rhesus macaques is linked to disease stage and inversely related to localization of virus-specific CTL. J Immunol. 2014;193(11):5613–5625. doi: 10.4049/jimmunol.1401161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li S, et al. Simian immunodeficiency virus-producing cells in follicles are partially suppressed by CD8+ cells in vivo. J Virol. 2016;90(24):11168–11180. doi: 10.1128/JVI.01332-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang Y, et al. Engineered bispecific antibodies with exquisite HIV-1-neutralizing activity. Cell. 2016;165(7):1621–1631. doi: 10.1016/j.cell.2016.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu L, et al. Trispecific broadly neutralizing HIV antibodies mediate potent SHIV protection in macaques. Science. 2017;358(6359):85–90. doi: 10.1126/science.aan8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klatt NR, Chomont N, Douek DC, Deeks SG. Immune activation and HIV persistence: implications for curative approaches to HIV infection. Immunol Rev. 2013;254(1):326–342. doi: 10.1111/imr.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paiardini M, Pandrea I, Apetrei C, Silvestri G. Lessons learned from the natural hosts of HIV-related viruses. Annu Rev Med. 2009;60:485–495. doi: 10.1146/annurev.med.60.041807.123753. [DOI] [PubMed] [Google Scholar]
- 42.Brenchley JM, Silvestri G, Douek DC. Nonprogressive and progressive primate immunodeficiency lentivirus infections. Immunity. 2010;32(6):737–742. doi: 10.1016/j.immuni.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Randolph GJ, Inaba K, Robbiani DF, Steinman RM, Muller WA. Differentiation of phagocytic monocytes into lymph node dendritic cells in vivo. Immunity. 1999;11(6):753–761. doi: 10.1016/S1074-7613(00)80149-1. [DOI] [PubMed] [Google Scholar]
- 44.Padovan E, Spagnoli GC, Ferrantini M, Heberer M. IFN-alpha2a induces IP-10/CXCL10 and MIG/CXCL9 production in monocyte-derived dendritic cells and enhances their capacity to attract and stimulate CD8+ effector T cells. J Leukoc Biol. 2002;71(4):669–676. [PubMed] [Google Scholar]
- 45.Vanderford TH, et al. Treatment of SIV-infected sooty mangabeys with a type-I IFN agonist results in decreased virus replication without inducing hyperimmune activation. Blood. 2012;119(24):5750–5757. doi: 10.1182/blood-2012-02-411496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sandler NG, et al. Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature. 2014;511(7511):601–605. doi: 10.1038/nature13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brainard DM, et al. Decreased CXCR3+ CD8 T cells in advanced human immunodeficiency virus infection suggest that a homing defect contributes to cytotoxic T-lymphocyte dysfunction. J Virol. 2007;81(16):8439–8450. doi: 10.1128/JVI.00199-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Price DA, et al. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity. 2004;21(6):793–803. doi: 10.1016/j.immuni.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 49.Price DA, et al. Public clonotype usage identifies protective Gag-specific CD8+ T cell responses in SIV infection. J Exp Med. 2009;206(4):923–936. doi: 10.1084/jem.20081127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dominguez MH, et al. Highly multiplexed quantitation of gene expression on single cells. J Immunol Methods. 2013;391(1-2):133–145. doi: 10.1016/j.jim.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beaumier CM, et al. CD4 downregulation by memory CD4+ T cells in vivo renders African green monkeys resistant to progressive SIVagm infection. Nat Med. 2009;15(8):879–885. doi: 10.1038/nm.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.