Abstract
Altered epigenetic reprogramming contributes to breast cancer progression and metastasis. How the epigenetic reader mediates breast cancer progression remains poorly understood. Here, we showed that the epigenetic reader zinc finger MYND-type containing 8 (ZMYND8) is induced by HIF-1 and HIF-2 in breast cancer cells and also upregulated in human breast tumors, and is correlated with poor survival of patients with breast cancer. Genetic deletion of ZMYND8 decreases breast cancer cell colony formation, migration, and invasion in vitro, and inhibits breast tumor growth and metastasis to the lungs in mice. The ZMYND8’s oncogenic effect in breast cancer requires HIF-1 and HIF-2. We further showed that ZMYND8 interacts with HIF-1α and HIF-2α and enhances elongation of the global HIF-induced oncogenic genes by increasing recruitment of BRD4 and subsequent release of paused RNA polymerase II in breast cancer cells. ZMYND8 acetylation at lysines 1007 and 1034 by p300 is required for HIF activation and breast cancer progression and metastasis. These findings uncover a primary epigenetic mechanism of HIF activation and HIF-mediated breast cancer progression, and discover a possible molecular target for the diagnosis and treatment of breast cancer.
Keywords: Oncology
Keywords: Breast cancer, Epigenetics, hypoxia
Introduction
Metastasis is a major clinical obstacle to curative cancer therapy and is responsible for about 90% of breast cancer–related deaths in women. The development of a benign and nonmalignant ductal carcinoma in situ to the advanced and metastatic breast cancer requires many dysregulated oncogenes and tumor suppressor genes, each of which provides a unique growth advantage to breast cancer cells and controls a specific phenotypic trait of tumor progression and metastasis (1, 2). Epigenetic reprogramming plays a crucial role in dysregulation of these genes and represents an important mechanism of breast tumor progression and metastasis (2). Several epigenetic writers and erasers, including DOT1L, G9a, histone deacetylase 3 (HDAC3), WDR5, and Jumonji domain–containing protein 3 (JMJD3), have been shown to induce repression of the CDH1 gene (encoding E-cadherin) in breast cancer cells and modulate the epithelial-mesenchymal transition, a key cellular program in the initiation of metastasis, thereby triggering breast tumor metastasis to distant organs (3–6). Our previous work showed that JMJD2C promotes triple-negative breast tumor growth and metastasis to the lungs in mice through inducing glycolytic and metastasis genes (7). Similarly, EZH2, JMJD2B, MLL4, and UTX also regulate invasiveness of breast tumors (8–10). Recent studies have uncovered that the epigenetic readers also emerge to influence breast tumor growth. BRD4 inhibition by its shRNA or a pharmacological inhibitor JQ1 dramatically blocks triple-negative breast tumor growth in xenograft mice (11). Conversely, another epigenetic reader, zinc finger MYND-type containing 11 (ZMYND11), suppresses triple-negative breast tumorigenesis (12). However, how the epigenetic readers control breast tumor progression and metastasis remains poorly understood.
The tumor microenvironment is increasingly recognized as a critical factor that regulates epigenetic reprogramming. A notable feature of the microenvironment of human breast tumors is reduced O2 availability (hypoxia) with median partial pressure of oxygen (PO2) values of 10 mmHg, which is markedly lower than 65 mmHg in normal breast tissues (13). The HIFs are the master transcriptional regulators mediating the adaptive responses to intratumoral hypoxia to drive breast tumor progression (14). HIFs have 3 family members, HIF-1, HIF-2, and HIF-3, each of which consists of an O2-regulated α subunit and a constitutively expressed β subunit (15–17). In well-oxygenated cells, HIF-α protein is subjected to proteasomal degradation, which is mediated by the von Hippel-Lindau protein–dependent ubiquitin system, after it is hydroxylated by prolyl hydroxylases (18). Under hypoxia, HIF-α escapes from proteasomal degradation and is translocated into the nucleus, where it dimerizes with HIF-1β (16). The heterodimer binds to the hypoxia response elements (HREs; 5′-A/GCGTG-3′) in the genome, leading to transcriptional activation of hundreds of oncogenic genes (19), whose protein products regulate angiogenesis, epigenetic reprogramming, metabolism, cell migration and invasion, cell survival, and stem cell maintenance, leading to tumor growth and metastasis (14). For example, HIF-1 and HIF-2 directly activate the transcription of the proangiogenesis factor VEGFA to increase tumor angiogenesis (20). Other HIF-1 target genes — ANGPTL4, AQP1, and AGR2 — are also known to induce angiogenesis and cell migration (21–23). Lysyl oxidase (LOX) regulates collagen crosslinking and is essential for premetastatic niche formation. HIF-1 and HIF-2 are required for this important premetastatic phenotype in breast cancer by inducing expression of the members of the LOX family, including LOX, LOXL2, and LOXL4 (24, 25). Therefore, these phenotypic traits controlled by the specific genes mediate hypoxia-driven breast tumor growth and metastasis.
Epigenetic regulators are essential for HIF-mediated transactivation (26). The histone acetyltransferases p300, CBP, and TIP60 induce acetylation of histones H3 and H4 to increase transcription of a subset of HIF-1 target genes (27, 28). HDACs 1–7 are also known to enhance or suppress HIF-1 transcriptional activity via the different mechanisms (26). We have demonstrated that JMJD2C demethylates trimethyl lysine 9 of histone H3 at the HREs to increase HIF-1–mediated transactivation in human cancer cells (7). The role of chromatin remodelers in HIF-1–mediated transactivation has been also reported (29, 30). Overall, the diverse epigenetic regulators, including writers and erasers, have been functionally linked to HIF activation. However, how the epigenetic reader modulates hypoxia-induced genes to promote breast cancer progression is unknown.
In the present study, we identified a hypoxia-induced epigenetic reader, ZMYND8, in breast cancer cells. ZMYND8 interacts with HIF-1α and HIF-2α, and coactivates HIF-1– and HIF-2–induced oncogenes by recruiting BRD4 and subsequently increasing RNA polymerase II phosphorylation, thereby increasing angiogenesis and cell motility and decreasing cancer cell death to promote breast tumor growth and metastasis to the lungs. ZMYND8 is acetylated by p300 and acetylated ZMYND8 is necessary for HIF activation and breast tumor progression and metastasis. ZMYND8 is highly expressed in human breast tumors, and thus may be a potential biomarker and therapeutic target for the diagnosis and treatment of breast cancer.
Results
ZMYND8 is a novel direct HIF-1 and HIF-2 target gene in breast cancer cells.
To survey the new hypoxia-induced epigenetic regulators in breast cancer, we analyzed mRNA expression changes of 720 epigenetic genes in a microarray gene expression data set of breast cancer MCF-7 cells exposed to 20% or 1% O2 for 18 hours from the Gene Expression Omnibus (GEO) database, and found KDM3A, a known HIF-1 and HIF-2 target gene (31), as the top hit (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI95089DS1), which validated our analysis approach. Of 3 ZMYND8 cDNA oligonucleotides, 2 were ranked as the second and third highest hypoxia-induced hits in this microarray data set, respectively (Supplemental Figure 1A). Similar results were also observed in another microarray data set of MCF-7 cells (Supplemental Figure 1B). ZMYND8 was initially identified in a cDNA library screen as a member of the receptors for activated C-kinase family (32). Recent studies demonstrated that ZMYND8 functions as an epigenetic reader that contains a plant homeodomain (PHD), a bromodomain (BRD), and a Pro-Trp-Trp-Pro (PWWP) domain at its N-terminus, which recognizes modified histones including di- and trimethyl lysine 36 of histone H3 (H3K36me2/3), acetyl lysine 14 of histone H3 (H3K14ac), and acetyl lysine 16 of histone H4 (H4K16ac) as well as DNA (33–35). These data identified ZMYND8 as a new hypoxia-induced epigenetic reader.
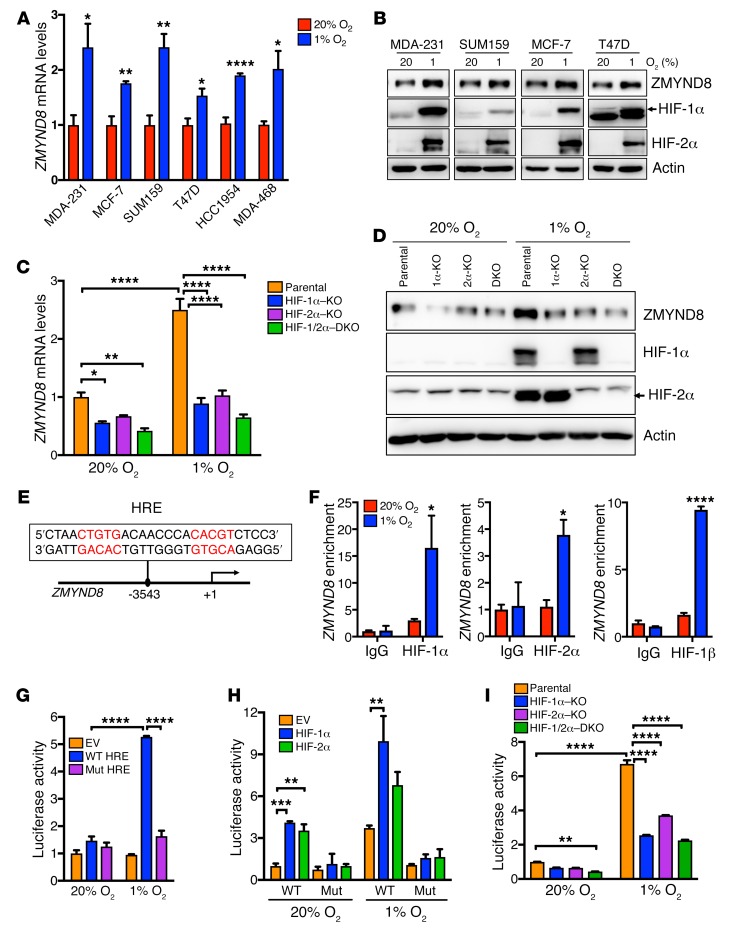
To validate the microarray data, a panel of human breast cancer cell lines, including MCF-7, MDA-MB-231, SUM159, T47D, HCC1954, and MDA-MB-468, was exposed to 20% or 1% O2 for 24 hours. Quantitative reverse transcription–polymerase chain reaction (RT-qPCR) assays showed that ZMYND8 mRNA was significantly upregulated by hypoxia in all breast cancer cell lines we tested (Figure 1A). ZMYND8 protein levels were also elevated, along with induced HIF-1α and HIF-2α proteins, in these cell lines under hypoxia (Figure 1B). Knockout (KO) of HIF-1α, HIF-2α, or both by the CRISPR/Cas9 technique significantly decreased levels of ZMYND8 mRNA and protein in nonhypoxic and hypoxic MDA-MB-231 cells (Figure 1, C and D). These data indicate that ZMYND8 is induced by hypoxia in a HIF-1– and HIF-2–dependent manner in breast cancer cells.
Figure 1. ZMYND8 is a direct HIF-1 and HIF-2 target gene.
(A) RT-qPCR analysis of ZMYND8 mRNA levels in breast cancer cells exposed to 20% or 1% O2 for 24 hours (mean ± SEM, n = 3). *P < 0.05, **P < 0.01, ****P < 0.0001 versus 20% O2, by 2-tailed Student’s t test. MDA-231, MDA-MB-231; MDA-468, MDA-MB-468. (B) Immunoblot assays of indicated proteins in breast cancer cells exposed to 20% or 1% O2 for 24 hours (n = 3). (C) RT-qPCR analysis of ZMYND8 mRNA levels in parental, HIF-1α–KO, HIF-2α–KO, and HIF-1/2α–DKO MDA-MB-231 cells exposed to 20% or 1% O2 for 24 hours (mean ± SEM, n = 3). *P < 0.05, **P < 0.01, ****P < 0.0001, by 2-way ANOVA with Tukey’s t test. DKO, double KO. (D) Immunoblot assays of indicated proteins in parental, HIF-1α–KO, HIF-2α–KO, or HIF-1/2α–DKO MDA-MB-231 cells exposed to 20% or 1% O2 for 24 hours (n = 3). (E) Nucleotide sequence of the HRE (in red) at the promoter of the ZMYND8 gene. (F) ChIP-qPCR assays in T47D cells exposed to 20% or 1% O2 for 24 hours (mean ± SEM, n = 3). *P < 0.05, ****P < 0.0001 versus 20% O2, by 2-way ANOVA with Sidak’s t test. (G–I) Luciferase reporter assays in HEK293T (G and H) and HeLa (I) cells transfected with the indicated plasmids and exposed to 20% or 1% O2 for 24 hours. The FLuc/RLuc activity was determined (mean ± SEM, n = 3). **P < 0.01, ***P < 0.001, ****P < 0.0001, by 2-way ANOVA with Tukey’s t test. Mut, mutant.
To determine whether ZMYND8 is a direct HIF target gene in breast cancer cells, we analyzed the HIF-1α and HIF-1β chromatin immunoprecipitation–sequencing (ChIP-seq) data sets from T47D cells (36), and found a robust HIF-1α– and HIF-1β–binding peak cluster at the promoter of the ZMYND8 gene (Supplemental Figure 1C), which annotated an HRE containing a consensus core HIF binding sequence (5′-ACGTG-3′) followed by a 5′-CACAG-3′ sequence (Figure 1E). To validate the ChIP-seq data, T47D cells were exposed to 20% or 1% O2 for 24 hours. ChIP-qPCR assays showed that occupancy of HIF-1α or HIF-2α on this HRE was significantly increased by hypoxia in T47D cells (Figure 1F, left and middle), similar to HIF-1α or HIF-2α occupancy on the HRE of a known HIF-1 and HIF-2 target gene VEGFA (Supplemental Figure 1D). Increased HIF-1β enrichment was also detected on the HRE of the ZMYND8 gene in hypoxic T47D cells (Figure 1F, right). These data indicate that HIF-1 and HIF-2 directly bind to the promoter of the ZMYND8 gene.
Next, we studied whether the HRE at the promoter of the ZMYND8 gene is able to enhance gene transcription by a dual-luciferase reporter assay. A 54-bp oligonucleotide sequence containing the ZMYND8 HRE was inserted upstream of the firefly luciferase (FLuc) coding sequence in a pGL2-promoter reporter plasmid. HEK293T cells were cotransfected with WT ZMYND8 HRE reporter plasmid or empty vector (EV) and a control reporter pSV40-Renilla (in which Renilla luciferase [RLuc] is constitutively expressed), and exposed to 20% or 1% O2 for 24 hours. The WT ZMYND8 HRE significantly increased the FLuc activity in hypoxic HEK293T cells (Figure 1G). Mutation of the HIF binding sequence 5′-CGT-3′ to 5′-AAA-3′ within the HRE completely abolished hypoxia-induced FLuc activity (Figure 1G). Overexpression of HIF-1α or HIF-2α significantly enhanced WT but not mutant ZMYND8 HRE-driven FLuc activity in nonhypoxic and hypoxic HEK293T cells (Figure 1H). In contrast, CRISPR/Cas9-based KO of HIF-1α, HIF-2α, or both significantly decreased hypoxia-induced ZMYND8 HRE activity in another reporter cell line, HeLa (Figure 1I). Taken together, these data indicate that HIF-1 and HIF-2 directly bind to the HRE at the ZMYND8 gene promoter to activate its transcription in breast cancer cells.
ZMYND8 is highly expressed in human breast tumors and is associated with poor clinical outcomes in patients.
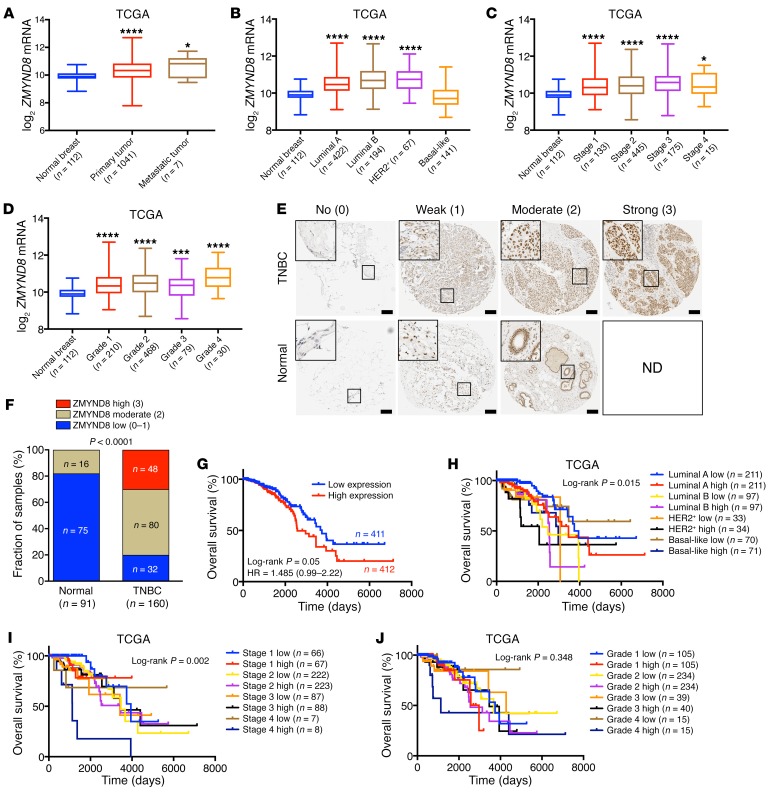
Hypoxia is a hallmark of the microenvironment of breast tumors (13). Thus, we studied whether ZMYND8 is also induced in human breast tumors. Analysis of The Cancer Genome Atlas (TCGA) breast invasive carcinoma data set revealed that ZMYND8 mRNA was significantly elevated in primary human breast tumors as compared with adjacent normal breast tissues (Figure 2A). The highest levels of ZMYND8 mRNA were found in metastatic breast tumors (Figure 2A). Similar findings were also observed in another GEO data set of human breast tumors (Supplemental Figure 2A). ZMYND8 mRNA was upregulated in luminal A, luminal B, and HER2+ subtypes, and stages 1–4 and histological grades 1–4 of human breast tumors (Figure 2, B–D). Next, we performed immunohistochemistry assays to confirm ZMYND8 induction at protein levels in human breast tumors. The specificity of ZMYND8 antibody was validated in control and ZMYND8-KO MDA-MB-231 tumors harvested from an orthotopic human breast cancer xenograft model in SCID mice (Supplemental Figure 2B). ZMYND8 protein was highly expressed in estrogen receptor–positive (ER+) breast tumors as compared with adjacent nontumor cells (Supplemental Figure 2C). Notably, much higher levels of ZMYND8 protein were observed in triple-negative breast cancer (TNBC) (Supplemental Figure 2D). Thus, we focused on TNBC and further analyzed ZMYND8 protein levels in a TNBC tissue microarray (TMA) containing 160 human TNBC specimens and 91 paired adjacent normal breast tissues. Of 160 TNBC specimens, 50% and 30% of tumors expressed moderate and high levels of ZMYND8 protein, respectively (Figure 2, E and F). In contrast, moderate ZMYND8 expression was found in only a small population of adjacent normal breast tissues (18%) and no high ZMYND8 protein levels were detected in normal breast tissues (Figure 2, E and F). Negative and weak expression of ZMYND8 was predominant in normal breast tissues (Figure 2, E and F). Kaplan-Meier analysis of the TCGA data set revealed that high levels of ZMYND8 mRNA were associated with poor overall survival of patients with breast cancer (Figure 2G). A similar negative correlation was found in the gene expression–based outcome (GOBO) data set (Supplemental Figure 2E). ZMYND8 was also inversely correlated with metastasis-free survival of patients with breast cancer (Supplemental Figure 2F). Moreover, ZMYND8 was significantly associated with high mortality of patients with different subtypes and stages of breast cancer, but not different tumor grades (Figure 2, H–J). Together, these data indicate that ZMYND8 is highly expressed in breast tumors and is associated with poor clinical outcomes in patients with breast cancer.
Figure 2. ZMYND8 is highly expressed in breast tumors and predicts poor clinical outcomes in patients with breast cancer.
(A–D) Analysis of ZMYND8 mRNA levels in human breast tumors and normal breast tissues. *P < 0.05, ***P < 0.001, ****P < 0.0001 versus normal breast, by 1-way ANOVA with Tukey’s t test. (E) Representative ZMYND8 immunohistochemical staining for each score (0–3) in a human TNBC tissue microarray. Scale bar, 200 μm. ND, not detected. (F) Quantification of low, moderate, and high ZMYND8 expression in normal breast and TNBC by χ2 test. (G–J) Kaplan-Meier survival analysis for patients with breast cancer by log-rank test. Patients were divided by median expression levels of ZMYND8 mRNA. HR, hazard ratio.
ZMYND8 increases colony formation, migration, and invasion of breast cancer cells in vitro.
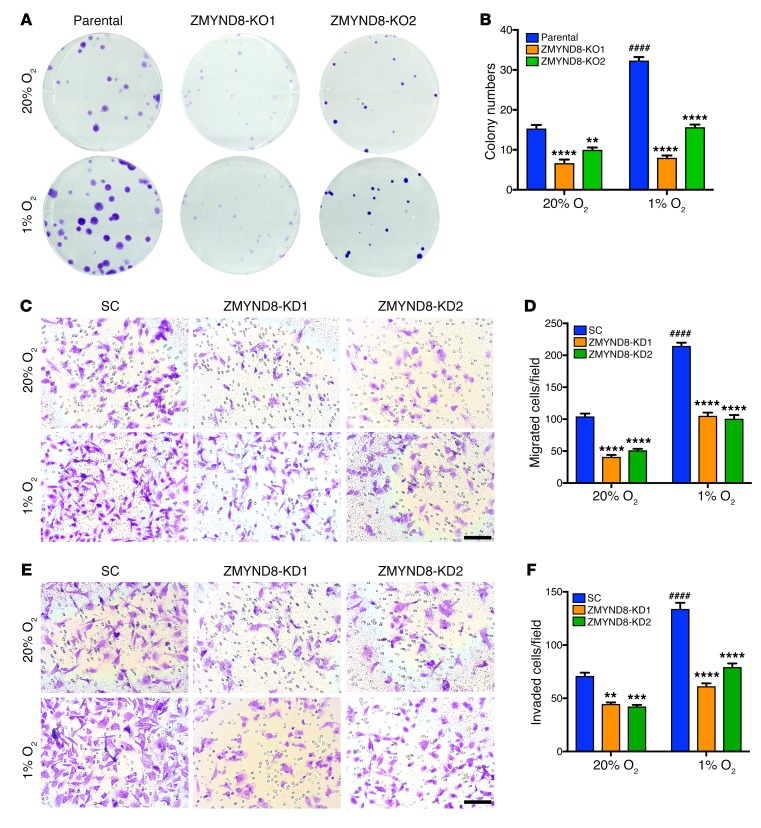
To determine an oncogenic role of ZMYND8 in breast tumors, we performed clonogenic assays in vitro. Two independent ZMYND8-KO MDA-MB-231 cell lines were generated by the CRISPR/Cas9 technique (Supplemental Figure 3A). Single-cell suspensions of parental MDA-MB-231 cells and ZMYND8-KO subclones were seeded on a 6-well plate and exposed to 20% or 1% O2 for 12 days. Hypoxia significantly increased the colony numbers of MDA-MB-231 cells by 2-fold, whereas colony numbers were dramatically diminished by ZMYND8-KO1 or -KO2 (Figure 3, A and B). Similarly, ZMYND8 knockdown (KD) by its shRNA also decreased the colony numbers of MCF-7 cells (Supplemental Figure 3, B–D). These data indicate that ZMYND8 increases breast cancer cell survival and colony formation in vitro.
Figure 3. ZMYND8 depletion suppresses tumorigenic potential of breast cancer cells in vitro.
(A and B) Colony formation of parental and ZMYND8-KO1 or -KO2 MDA-MB-231 cells cultured under 20% or 1% O2 for 12 days. Representative images from 3 experiments are shown in A. Quantification of colony numbers is shown in B (mean ± SEM, n = 3). ####P < 0.0001 versus parental at 20% O2, **P < 0.01, ****P < 0.0001 versus parental, by 2-way ANOVA with Tukey’s t test. (C–F) Migration (C and D) and invasion (E and F) of SC and ZMYND8-KD1 or -KD2 MDA-MB-231 cells under 20% or 1% O2. Representative images from 3 experiments are shown in C and E. Migrated and invaded cell numbers are quantified in D and F, respectively (mean ± SEM, n = 3). ####P < 0.0001 versus SC at 20% O2, **P < 0.01, ***P < 0.001, ****P < 0.0001 versus SC, by 2-way ANOVA with Tukey’s t test. Scale bar, 100 μm.
Next, we studied the effect of ZMYND8 on breast cancer cell migration and invasion in vitro. The Boyden chamber assays demonstrated that hypoxia increased migration of scrambled control (SC) MDA-MB-231 cells across the transwell membrane by 2-fold, whereas migration was significantly decreased by ZMYND8-KD1 or -KD2 (Figure 3, C and D). Similarly, the number of MDA-MB-231 cells invaded through Matrigel in a transwell insert was also significantly decreased by ZMYND8-KD1 or -KD2 under 20% or 1% O2 (Figure 3, E and F). ZMYND8-KD did not alter the rate of in vitro cell proliferation (Supplemental Figure 3, E and F), which rules out the possibility that reduced cell migration and invasion by ZMYND8-KD is due to decreased cell proliferation. Together, these data indicate that ZMYND8 increases breast cancer cell motility in vitro.
ZMYND8 promotes breast tumor growth and metastasis to the lungs in mice in a HIF-1– and HIF-2–dependent manner.
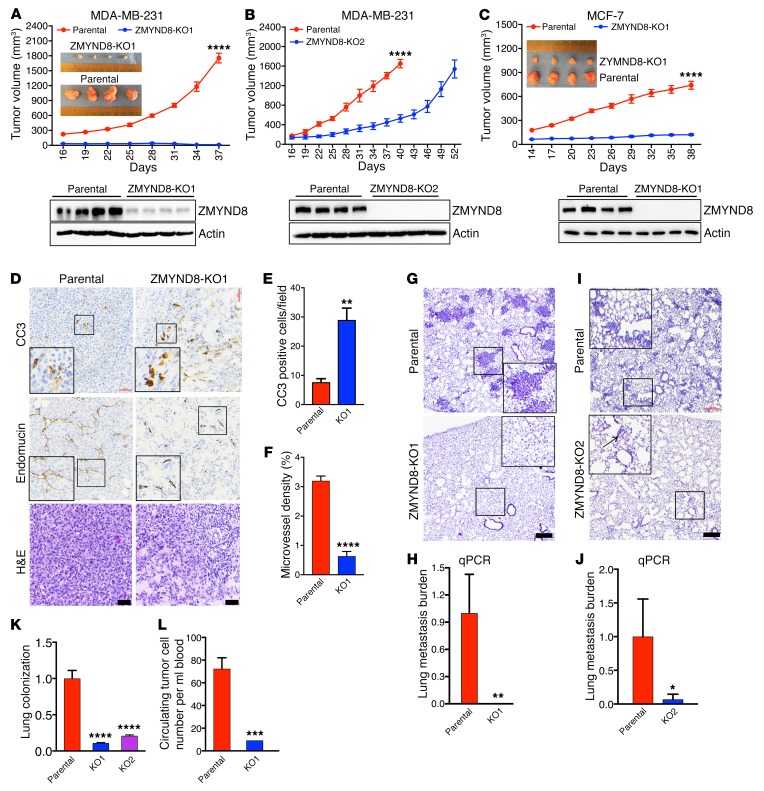
To determine whether ZMYND8 regulates breast tumor growth in vivo, we performed the orthotopic implantation of parental or ZMYND8-KO1 MDA-MB-231 cells into the mammary fat pad of SCID mice, and found that genetic deletion of ZMYND8 gene robustly blocked spontaneous breast tumor growth in mice (Figure 4A). Immunoblot assays confirmed reduced ZMYND8 protein levels in ZMYND8-KO1 tumors as compared with parental control tumors (Figure 4A). ZMYND8 protein weakly expressed in ZMYND8-KO1 tumors may be the murine form in tumor stromal cells. Similarly, ZMYND8-KO2 or -KD2 also significantly impaired MDA-MB-231 tumor growth in xenograft mice (Figure 4B and Supplemental Figure 4A). We further found that ZMYND8-KO1 or -KD2 significantly inhibited MCF-7 tumor growth in xenograft SCID mice subcutaneously administered slow-release 17β-estradiol pellets (Figure 4C, and Supplemental Figure 4, B and C). Immunohistochemistry assays showed that the number of cleaved caspase 3–positive (CC3-positive) cells was significantly increased in ZMYND8-KO1 and -KD2 MDA-MB-231 tumors and ZMYND8-KO1 MCF-7 tumors as compared with their respective control tumors (Figure 4, D and E, and Supplemental Figure 4, D, E, G, and H), suggesting increased cell death in tumors by ZMYND8-KO or -KD. Conversely, the microvessel density shown by endomucin immunohistochemical staining was significantly decreased in these tumors (Figure 4, D and F, and Supplemental Figure 4, D, F, G, and I), suggesting decreased tumor angiogenesis by ZMYND8-KO or -KD. However, expression of Ki-67, a cell proliferation marker, was not significantly different between SC and ZMYND8-KD2 MDA-MB-231 tumors (Supplemental Figure 4D), consistent with our in vitro cell growth data above (Supplemental Figure 3F). These data indicate that ZMYND8 promotes breast tumor growth in mice by increasing tumor angiogenesis and cell survival.
Figure 4. Loss of ZMYND8 suppresses breast tumor growth and metastasis to the lungs in mice.
(A–C) Growth of parental, ZMYND8- KO1 (A) and -KO2 (B) MDA-MB-231, and ZMYND8-KO1 MCF-7 (C) tumors in mice (mean ± SEM, n = 4). ****P < 0.0001 versus parental by 2-way ANOVA with Sidak’s t test. The image of parental and ZMYND8-KO1 tumors harvested at the end time point is shown in the insets. ZMYND8-KO in tumors was confirmed by immunoblot assays (bottom). (D–F) Representative H&E and immunohistochemical staining of CC3 and endomucin in parental or ZMYND8-KO1 MDA-MB-231 tumors (D). Magnified images of the boxed area are shown in the insets. Scale bar, 200 μm. CC3-positive cell numbers (E) and endomucin-positive areas (F) in tumors are quantified (mean ± SEM, n = 4). **P < 0.01, ****P < 0.0001 versus parental by 2-tailed Student’s t test. (G–J) Lung metastasis in mice bearing parental or ZMYND8-KO1 or -KO2 MDA-MB-231 tumors by H&E staining (G and I) and qPCR assays (H and J, mean ± SEM, n = 4). Magnified images of the boxed area are shown in the insets (G and I). *P < 0.05, **P < 0.01 versus parental by 2-tailed Student’s t test. Scale bar, 200 μm. (K) Lung colonization of parental and ZMYND8-KO MDA-MB-231 cells by qPCR assays (mean ± SEM, n = 11). ****P < 0.0001 versus parental by 1-way ANOVA with Dunnett’s test. (L) Circulating tumor cells in blood from mice bearing parental or ZMYND8-KO1 MDA-MB-231 tumors by qPCR assays (mean ± SEM, n = 4). ***P < 0.001 versus parental by 2-tailed Student’s t test.
Next, we studied the effect of ZMYND8 on spontaneous breast cancer metastasis to the lungs. H&E staining detected extensive metastatic burden in the lungs of mice 37 days after MDA-MB-231 cell implantation (Figure 4G). In contrast, no lung metastases were found in mice bearing ZMYND8-KO1 or -KD2 tumors (Figure 4G and Supplemental Figure 4J). Analysis of human genomic DNA by qPCR further confirmed that no human cancer cells were detected in the lungs of these mice (Figure 4H and Supplemental Figure 4K). To rule out the possibility that the loss of metastatic ability in mice bearing ZMYND8-KO1 or -KD2 tumors is due to the small primary tumors, mice orthotopically implanted with ZMYND8-KO2 MDA-MB-231 cells were euthanized 12 days later than control mice when the volume of ZMYND8-KO2 tumors matched that of control MDA-MB-231 tumors at day 40 (Figure 4B). Again, little lung metastasis burden was detected in mice bearing ZMYND8-KO2 tumors as compared with mice bearing control tumors (Figure 4, I and J), even though the tumor volume was comparable in these 2 groups of mice (Figure 4B). These data indicate that ZMYND8 mediates spontaneous metastasis of breast cancer cells to the lungs in mice.
Cancer cell extravasation and colonization at distant organs is critical for metastatic tumor outgrowth. To determine whether ZMYND8 regulates breast cancer cell extravasation and colonization in the lungs, we injected parental, or ZMYND8-KO1 or -KO2 MDA-MB-231 cells into the tail vein of female SCID mice. Three weeks after injection, mouse lungs were harvested for detection of human genomic DNA by qPCR. We found that less human genomic DNA was present in the lungs of mice injected with ZMYND8-KO1 or -KO2 cells as compared with mice bearing parental MDA-MB-231 cells (Figure 4K). We also studied the effect of ZMYND8-KO on circulating tumor cells in the orthotopic xenograft mouse model, and detected a fair number of human tumor cells in the circulating blood of mice bearing parental MDA-MB-231 tumors (Figure 4L). However, the number of circulating tumor cells was significantly decreased in mice bearing ZMYND8-KO1 tumors (Figure 4L). The reduced number of circulating tumor cells was even found in mice bearing ZMYND8-KO2 tumors (Supplemental Figure 4L) whose volume was comparable to that of control tumors (Figure 4B), indicating that reduction in circulating tumor cells by ZMYND8-KO is not due to its inhibitory effect on tumor growth. Taken together, these data indicate that ZMYND8 increases circulating tumor cell populations to promote their extravasation and colonization, leading to metastasis of breast cancer cells to the lungs.
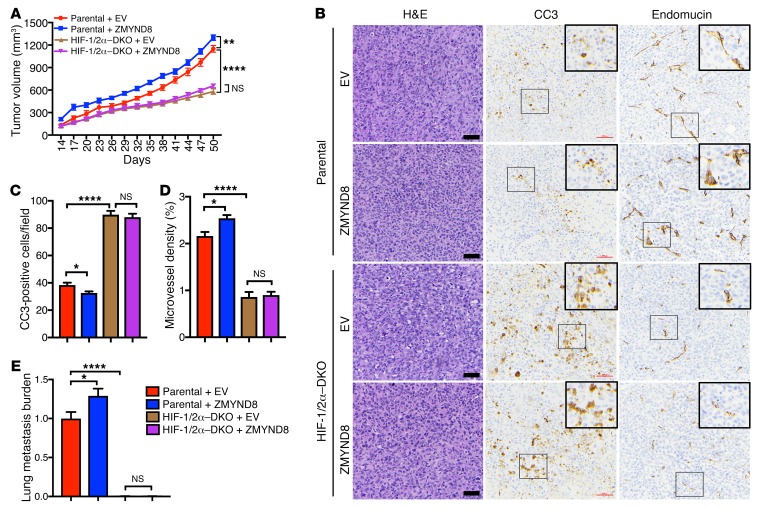
Last, we performed gain-of-function studies to complement loss-of-function effects of ZMYND8 on breast tumor progression and metastasis. MDA-MB-231 cells were transduced with lentivirus carrying FLAG-ZMYND8 or EV. These stable cell lines were implanted into the mammary fat pads of female SCID mice. Overexpression of FLAG-ZMYND8 significantly increased MDA-MB-231 tumor growth in mice (Figure 5A). HIF-1 and HIF-2 are the key drivers of invasive breast tumor growth (14). HIF-1/2α–DKO abolished ZMYND8-induced MDA-MB-231 tumor growth in mice (Figure 5A). Immunohistochemistry assays showed FLAG-ZMYND8 overexpression significantly decreased CC3-positive cell numbers but increased microvessel density in parental tumors (Figure 5, B–D). However, increased CC3-positive cell numbers and decreased microvessel density were found in HIF-1/2α–DKO tumors, which were not affected by FLAG-ZMYND8 overexpression (Figure 5, B–D). Lung metastasis was also significantly increased in mice bearing parental and FLAG-ZMYND8 tumors, which were eliminated by HIF-1/2α–DKO (Figure 5E). These data indicate that HIF-1 and HIF-2 are required for ZMYND8-mediated breast tumor progression and metastasis to the lungs.
Figure 5. HIF-1 and HIF-2 are required for ZMYND8-mediated breast tumor growth and metastasis in mice.
(A) Growth of Parental+EV, Parental+ZMYND8, HIF-1/2α–DKO+EV, and HIF-1/2α–DKO+ZMYND8 MDA-MB-231 tumors in mice (mean ± SEM, n = 5). **P < 0.01, ****P < 0.0001 by 2-way ANOVA with Tukey’s t test. NS, not significant. (B–D) Representative H&E and immunohistochemical staining of CC3 and endomucin in primary tumors. Magnified images of the boxed area are shown in the insets. Scale bar, 200 μm. CC3-positive cell numbers (C) and endomucin-positive areas (D) in tumors are quantified (mean ± SEM, n = 5). *P < 0.05, ****P < 0.0001 by 1-way ANOVA with Sidak’s t test. (E) Lung metastasis are quantified by qPCR (mean ± SEM, n = 5). *P < 0.05, ****P < 0.0001 by 1-way ANOVA with Dunnett’s test.
ZMYND8 interacts with HIF-1α and HIF-2α in breast cancer cells.
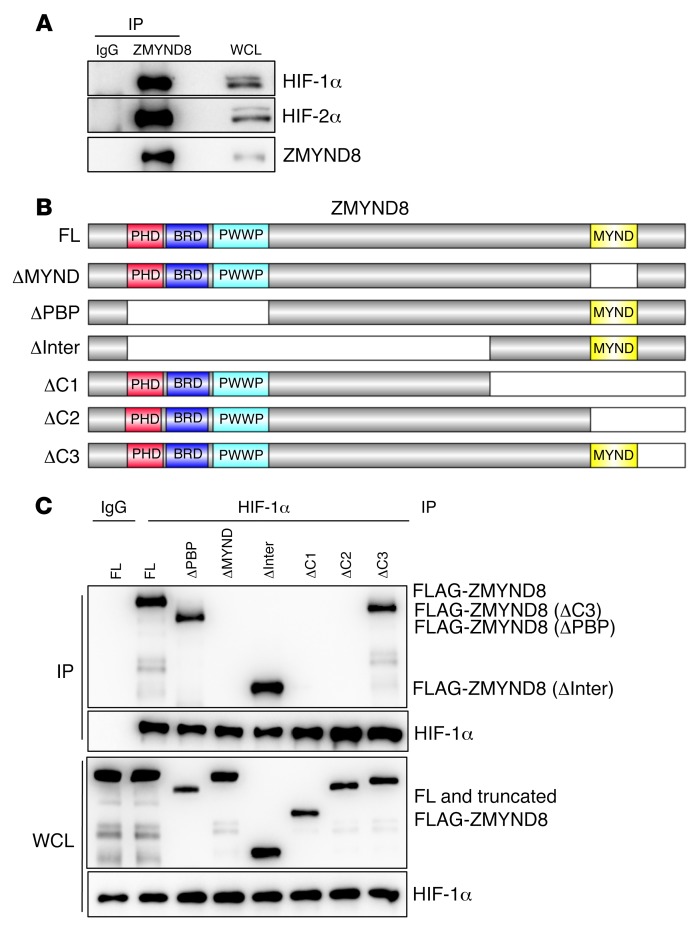
The data described above (Figure 5) suggest that ZMYND8 may be also an upstream regulator of HIF signaling in breast cancer cells. Thus, we studied whether ZMYND8 physically interacts with HIF-1α or HIF-2α. HEK293T cells were cotransfected with V5-tagged ZMYND8 vector and vector encoding FLAG-tagged HIF-1α, FLAG-tagged HIF-2α, or EV, and exposed to 1% O2 for 6 hours. Coimmunoprecipitation (co-IP) assays using anti–FLAG antibody showed that FLAG–HIF-1α or FLAG–HIF-2α, but not EV, pulled down ZMYND8-V5 in HEK293T cells (Supplemental Figure 5A). We also performed a reciprocal co-IP assay using anti–V5 antibody and found that ZMYND8-V5 precipitated FLAG–HIF-1α or FLAG–HIF-2α in hypoxic HEK293T cells (Supplemental Figure 5, B and C). Further, anti–ZMYND8 antibody coprecipitated endogenous ZMYND8, HIF-1α, and HIF-2α in MDA-MB-231 cells exposed to 1% O2 for 6 hours (Figure 6A). These data indicate that ZMYND8 physically interacts with HIF-1α and HIF-2α in human breast cancer cells.
Figure 6. ZMYND8 interacts with HIF-1α and HIF-2α.
(A) Co-IP assays of endogenous ZMYND8 and HIF-1α or HIF-2α in MDA-MB-231 cells exposed to 1% O2 for 6 hours (n = 3). WCL, whole-cell lysate. (B and C) Mapping of the ZMYND8 domain binding to HIF-1α. Schematic depiction of FL and domain-deleted ZMYND8 (B). Co-IP assays of endogenous HIF-1α and FL or truncated FLAG-ZMYND8 in transfected HEK293T cells exposed to 1% O2 for 6 hours (n = 3) (C).
Next, we mapped the ZMYND8 domain binding to HIF-1α. HEK293T cells were transfected with vector encoding full-length (FL) or domain-deleted FLAG-ZMYND8 (Figure 6B), and exposed to 1% O2 for 6 hours. Co-IP assays using anti–HIF-1α antibody showed that FL and PBP (ΔPBP), inter (ΔInter), or C3 (ΔC3) fragment–deleted FLAG-ZMYND8 interacted with HIF-1α (Figure 6C). In contrast, deletion of the MYND domain (ΔMYND) or 2 C-terminal fragments (ΔC1 and ΔC2) abolished ZMYND8 binding to HIF-1α in hypoxic HEK293T cells (Figure 6C). These data indicate that the MYND domain of ZMYND8 is required and sufficient for interaction with HIF-1α.
ZMYND8 activates the global HIF target genes in breast cancer cells.
To determine whether ZMYND8 regulates HIF transcriptional activity, we first performed HIF luciferase reporter assays. HeLa cells were cotransfected with a HIF reporter (containing 2 copies of HREs from the human VEGFA gene upstream of SV40 promoter and FLuc coding sequence), pSV-Renilla, and ZMYND8-V5 vector or EV, and exposed to 20% or 1% O2 for 24 hours. Overexpression of ZMYND8-V5 significantly increased HIF transcriptional activity in HeLa cells, as compared with EV (Supplemental Figure 6A).
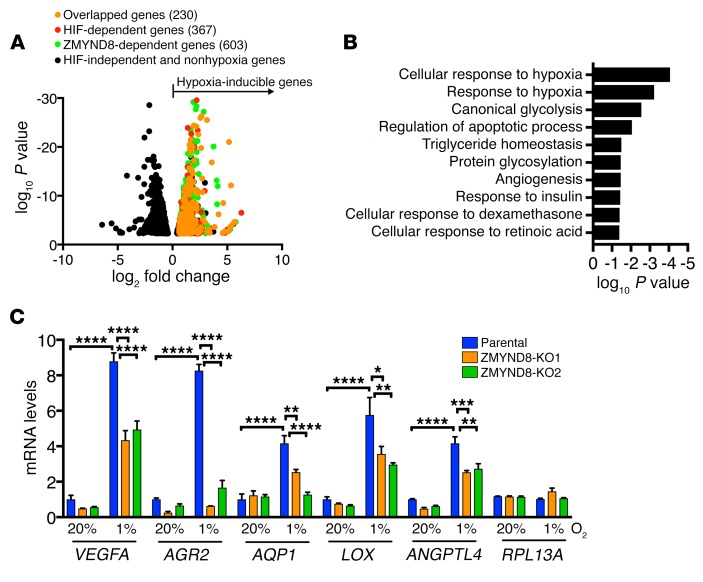
We next set out to analyze HIF transcriptome profiling controlled by ZMYND8 in breast cancer cells. To this end, parental, HIF-1/2α–DKO, and ZMYND8-KO1 MDA-MB-231 cells were exposed to 20% or 1% O2 for 24 hours and subjected to RNA sequencing (RNA-seq). RNA-seq analysis identified 1,124 hypoxia-inducible genes in MDA-MB-231 cells, of which about one-third (367, 32.7%) were induced by HIF-1 and HIF-2 and about half (603, 53.6%) were induced by ZMYND8 (Figure 7A). Notably, 230 genes were overlapped between HIF-upregulated and ZMYND8-upregulated hypoxia-inducible genes, which represented 62.7% of HIF target genes (Figure 7A). Those genes were involved in classic HIF-dependent biological functions, including glycolysis, apoptosis, and angiogenesis (Figure 7B). Consistent with the RNA-seq data, RT-qPCR assays confirmed that hypoxia induced transcription of the HIF target genes VEGFA, ANGPTL4, AGR2, AQP1, and LOX in MDA-MB-231 cells, and induction of these genes was significantly attenuated by ZMYND8-KO1 or -KO2 (Figure 7C). However, mRNA transcription of non–HIF target gene RPL13A was not altered by ZMYND8-KO1 or -KO2 in MDA-MB-231 cells (Figure 7C). Similar results were also found in ZMYND8-KD2 SUM159 cells (Supplemental Figure 6, B and C). ZMYND8-KO or -KD did not affect protein levels of HIF-1α and HIF-2α in breast cancer cells (Supplemental Figure 3A and Supplemental Figure 6B), which rules out regulation of HIF-1α and HIF-2α protein stability as the cause of ZMYND8-enhanced HIF transcriptional activity.
Figure 7. ZMYND8 activates the global HIF target genes in breast cancer cells.
(A) Volcano plot of ZMYND8 and HIF target genes in MDA-MB-231 cells exposed to 20% or 1% O2 for 24 hours (n = 2). (B) Gene ontology analysis of ZMYND8-dependent HIF target genes (n = 2). (C) RT-qPCR analysis of indicated mRNAs in parental and ZMYND8-KO1 or -KO2 MDA-MB-231 cells exposed to 20% or 1% O2 for 24 hours (mean ± SEM, n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, by 2-way ANOVA with Tukey’s t test.
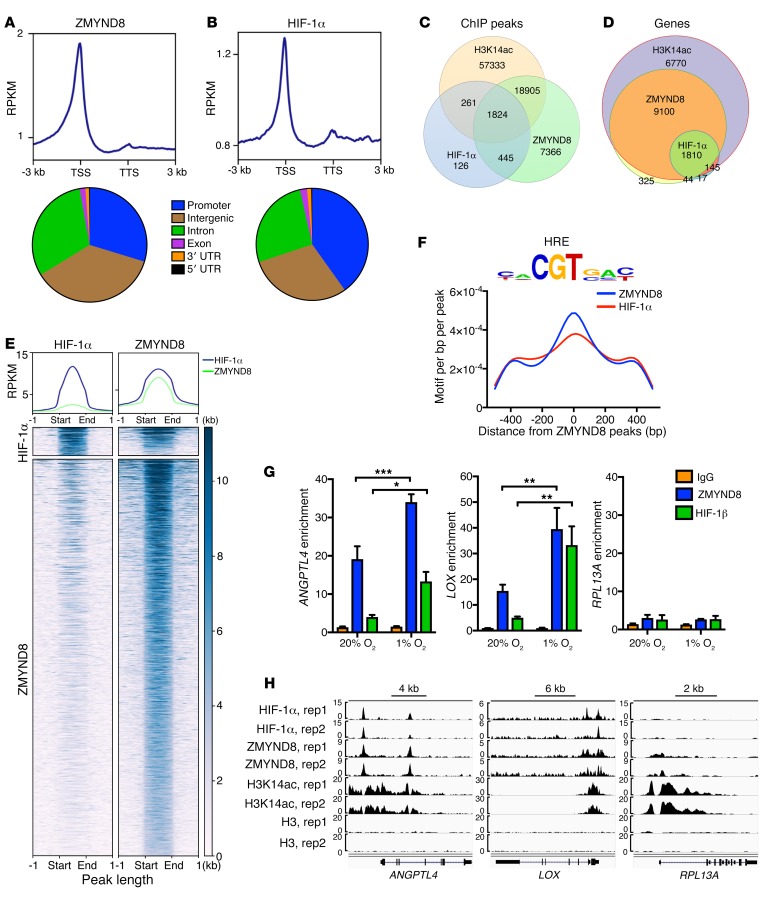
We further performed ChIP-seq in hypoxic MDA-MB-231 cells to determine whether ZMYND8 colocalizes with HIF-1α at the HREs. MDA-MB-231 cells were exposed to 1% O2 for 24 hours. ZMYND8 was highly enriched near the transcription start sites, similar to HIF-1α (Figure 8, A and B). Its ChIP-seq peaks were mainly located at the promoter (29.7%), intergenic (36.5%), and intron (30.9%) regions (Figure 8A). Strikingly, about 85.4% of HIF-1α peaks overlapped with ZMYND8 peaks (Figure 8C), and 92% of HIF-1 target genes were co-occupied by HIF-1α and ZMYND8 (Figure 8D). The metagene analysis confirmed strong colocalization of HIF-1α and ZMYND8 (Figure 8E). The motif analysis of ZMYND8 peaks further revealed marked enrichment of the HRE (Figure 8F). These data indicate genome-wide colocalization of HIF-1α and ZMYND8 at the HREs.
Figure 8. ZMYND8 globally colocalizes with HIF-1α at the HREs in breast cancer cells.
(A and B) Metagene analysis of the genomic distribution of ZMYND8 (A) and HIF-1α (B) in MDA-MB-231 cells exposed to 1% O2 for 24 hours (n = 2). RPKM, reads per kilobase per million mapped reads; TSS, transcription start site; TTS, transcription termination site. (C and D) Venn diagram of the overlapped ChIP-seq peaks (C) and co-occupied genes (D) by ZMYND8, HIF-1α, and H3K14ac (n = 2). (E) Co-occupancy analysis of HIF-1α and ZMYND8 ChIP-seq peaks (n = 2). (F) Motif density analysis of ZMYND8 ChIP-seq peaks (n = 2). HRE is shown in top panel. (G) ZMYND8 and HIF-1β ChIP-qPCR assays in MDA-MB-231 cells exposed to 20% or 1% O2 for 24 hours (mean ± SEM, n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, by 2-way ANOVA with Sidak’s t test. (H) Genome browser snapshots of HIF-1α, ZMYND8, H3K14ac, and H3 ChIP-seq peaks. rep, replicate.
To validate the ChIP-seq data, ChIP-qPCR assays were performed in MDA-MB-231 cells exposed to 20% or 1% O2 for 24 hours. ZMYND8 was highly enriched at the HREs of LOX and ANGPTL4, but weakly enriched on RPL13A, as compared with control IgG in nonhypoxic MDA-MB-231 cells. ZMYND8 occupancy on the LOX and ANGPTL4 HREs was significantly increased 2.6- and 1.8-fold, respectively, by hypoxia (Figure 8, G and H). HIF-1β occupancy was similarly detected on the HREs of LOX and ANGPTL4, but not RPL13A (Figure 8, G and H). Taken together, the mRNA-seq and ChIP-seq data indicate that ZMYND8 colocalizes with HIF-1 and activates its global target genes in breast cancer cells.
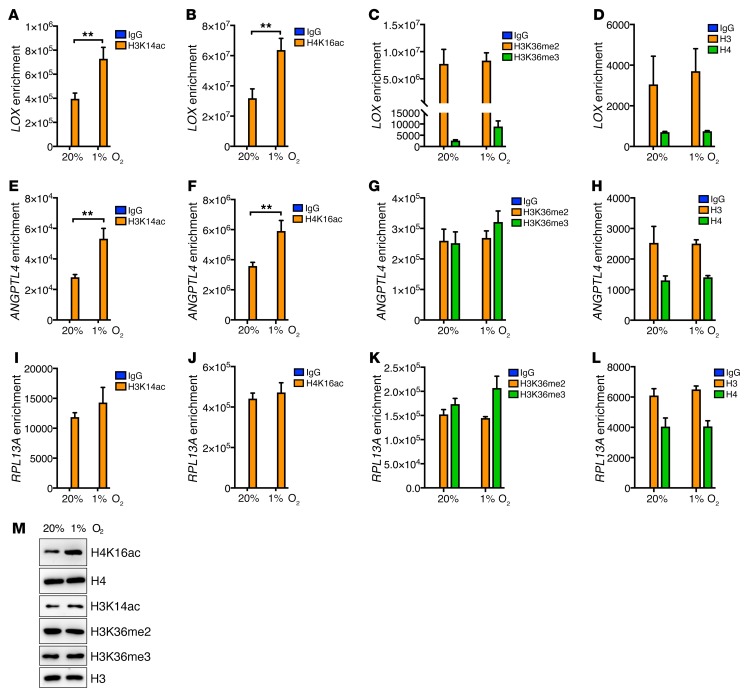
ZMYND8 is associated with H3K14ac and H4K16ac at the HREs and induces release of paused RNA polymerase II in breast cancer cells.
To determine whether ZMYND8 binds to the HREs through modified histones, we studied occupancy of H3K14ac, H3K36me2, H3K36me3, and H4K16ac on the HREs in MDA-MB-231 cells exposed to 20% or 1% O2 for 24 hours. ChIP-seq showed that H3K14ac peaks were markedly overlapped with ZMYND8 peaks and co-occupied at the HREs (Figure 8, C and H). H3K14ac occupancy at the LOX and ANGPTL4 HREs was significantly increased by hypoxia in MDA-MB-231 cells (Figure 9, A and E). Likewise, H4K16ac was strongly enriched on the HREs of LOX and ANGPTL4 as compared with control IgG in nonhypoxic MDA-MB-231 cells, and hypoxia increased its occupancy on both HREs by 2-fold and 1.7-fold, respectively (Figure 9, B and F). In contrast, enrichment of H3K14ac and H4K16ac on RPL13A was not regulated by hypoxia in MDA-MB-231 cells (Figure 9, I and J). Occupancy of H3K36me2, H3K36me3, histone H3, or histone H4 on LOX, ANGPTL4, and RPL13A was also not altered by hypoxia in MDA-MB-231 cells (Figure 9, C, D, G, H, K, and L). The global levels of H4K16ac, but not H3K14ac, H3K36me2, H3K36me3, histone H3, and histone H4, were increased by hypoxia in MDA-MB-231 cells (Figure 9M). These data indicate that H3K14ac and H4K16ac, but not H3K36me2 and H3K36me3, mediate ZMYND8 binding to the HREs of the HIF target genes in breast cancer cells.
Figure 9. Hypoxia increases H3K14ac and H4K16ac at the HREs in breast cancer cells.
(A–L) ChIP-qPCR assays in MDA-MB-231 cells exposed to 20% or 1% O2 for 24 hours (mean ± SEM, n = 3). **P < 0.01 by 2-way ANOVA with Sidak’s t test. (M) Immunoblot assays of indicated proteins in MDA-MB-231 cells exposed to 20% or 1% O2 for 24 hours (n = 3).
Previous studies have shown that RNA polymerase II is preloaded and paused at the promoter of the HIF target genes and that paused RNA polymerase II release is essential for HIF-mediated transactivation (37). Paused RNA polymerase II is phosphorylated at serine 5 (S5P), whereas serine 2 phosphorylation (S2P) of RNA polymerase II controls release of paused RNA polymerase II (38). To determine whether ZMYND8 regulates paused RNA polymerase II release to promote HIF-mediated transactivation, parental and ZMYND8-KO2 MDA-MB-231 cells were exposed to 20% or 1% O2 for 24 hours and subjected to ChIP-qPCR assays. Hypoxia significantly increased RNA polymerase II–S2P occupancy on the LOX and ANGPTL4 genes but did not affect enrichment of RNA polymerase II–S5P in MDA-MB-231 cells (Figure 10, A and B), confirming the previous findings (37). ZMYND8-KO2 significantly decreased enrichment of RNA polymerase II–S2P, but not RNA polymerase II–S5P and total RNA polymerase II, on the LOX and ANGPTL4 genes in hypoxic MDA-MB-231 cells (Figure 10, A and B). ZMYND8-KO2 had no effects on occupancy of RNA polymerase II–S2P, RNA polymerase II–S5P, or total RNA polymerase II on the RPL13A gene in MDA-MB-231 cells (Figure 10C). Together, these findings indicate that ZMYND8 induces release of paused RNA polymerase II to promote transcriptional elongation of the HIF target genes in breast cancer cells.
Figure 10. ZMYND8 mediates hypoxia-induced RNA polymerase II–S2P occupancy on the HIF target genes in breast cancer cells.
(A–C) RNA polymerase II ChIP-qPCR assays in parental and ZMYND8-KO2 MDA-MB-231 cells exposed to 20% or 1% O2 for 24 hours (mean ± SEM, n = 3). *P < 0.05, **P < 0.01, by 2-way ANOVA with Sidak’s t test.
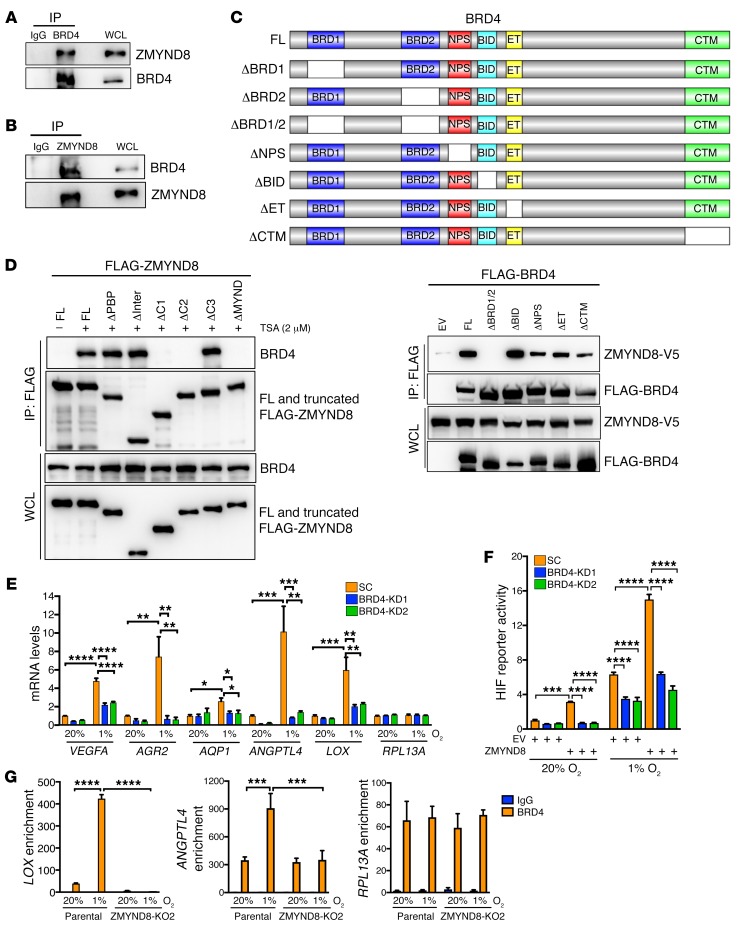
BRD4 recruited by ZMYND8 is required for ZMYND8-mediated HIF activation in breast cancer cells.
A recent study showed that ZMYND8 binds to RNA polymerase II–S5P, but not RNA polymerase II–S2P (34), suggesting that a ZMYND8-interacting protein may mediate ZMYND8 signaling to RNA polymerase II–S2P, leading to elongation of the HIF target genes. BRD4 is a known transcriptional regulator to induce RNA polymerase II–S2P (39). To determine whether BRD4 is involved in ZMYND8-mediated HIF activation, we first studied ZMYND8 interaction with BRD4 by co-IP assays. HEK293T cells were cotransfected with ZMYND8-V5 vector and FLAG-BRD4 vector. Anti–FLAG antibody, but not IgG, precipitated both FLAG-BRD4 and ZMYND8-V5 (Supplemental Figure 7A). Similarly, anti–BRD4 antibody coprecipitated endogenous BRD4 and ZMYND8 in MDA-MB-231 cells, but control IgG failed to do so (Figure 11A). We further performed a reciprocal co-IP assay and found that ZMYND8-V5 pulled down FLAG-BRD4 in HEK293T cells treated with HDAC inhibitor trichostatin A (TSA) (Supplemental Figure 7B). Precipitation of endogenous ZMYND8 by anti–ZMYND8 antibody also pulled down endogenous BRD4 in MDA-MB-231 cells treated with TSA (Figure 11B). Hypoxia did not influence physical interaction of ZMYND8 with BRD4 in MDA-MB-231 cells (Supplemental Figure 7C). These data indicate that ZMYND8 physically interacts with BRD4 in breast cancer cells, and that ZMYND8-BRD4 interaction is enhanced upon inhibition of HDAC but independent of hypoxia.
Figure 11. BRD4 binds to ZMYND8 and is required for ZMYND8-mediated HIF activation in breast cancer cells.
(A and B) Co-IP assays of endogenous BRD4 and ZMYND8 in MDA-MB-231 cells (n = 3). (C) Mapping the BRD4 domain binding to ZMYND8. Schematic depiction of FL and domain-deleted BRD4 (top). Co-IP assays of ZMYND8-V5 and FL or truncated FLAG-BRD4 in transfected HEK293T cells (bottom, n = 3). (D) Mapping the ZMYND8 domain binding to BRD4. Co-IP assays of endogenous BRD4 and FL or truncated FLAG-ZMYND8 in HEK293T cells treated with TSA or DMSO (–) for 6 hours (n = 3). (E) RT-qPCR analysis of indicated mRNAs in SC and BRD4-KD1 or -KD2 MDA-MB-231 cells exposed to 20% or 1% O2 for 24 hours in the presence of doxycycline (mean ± SEM, n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, by 2-way ANOVA with Tukey’s t test. (F) HIF luciferase reporter assays in HeLa cells transfected with indicated plasmids and exposed to 20% or 1% O2 for 24 hours in the presence of doxycycline. The FLuc/RLuc activity was determined (mean ± SEM, n = 3). ***P < 0.001, ****P < 0.0001, by 2-way ANOVA with Tukey’s t test. (G) BRD4 ChIP-qPCR assays in parental and ZMYND8-KO2 MDA-MB-231 cells exposed to 20% or 1% O2 for 24 hours (mean ± SEM, n = 3). ***P < 0.001, ****P < 0.0001 by 2-way ANOVA with Sidak’s t test.
Next, we mapped the ZMYND8-BRD4 binding domains in HEK293T cells by co-IP assays. HEK293T cells were transiently cotransfected with ZMYND8-V5 vector and FL or domain-deleted FLAG-BRD4 vector or EV. Analysis of FLAG immunoprecipitates revealed that ZMYND8-V5 was precipitated by FL and BID (ΔBID), NPS (ΔNPS), ET (ΔET), or CTM (ΔCTM) domain–deleted FLAG-BRD4 (Figure 11C). However, deletion of the BRD1 and BRD2 (ΔBRD1/2) of FLAG-BRD4 or EV failed to pull down ZMYND8-V5 in HEK293T cells (Figure 11C). Interestingly, deletion of the individual BRD1 (ΔBRD1) or BRD2 (ΔBRD2) of FLAG-BRD4 had little effect on its interaction with ZMYND8-V5 in HEK293T cells (Supplemental Figure 7D), indicating that ZMYND8 binds to both BRD1 and BRD2 of BRD4. JQ1 competitively binds to BRD1/2 to block BRD4 functions (40). Treatment of JQ1 remarkably decreased FL, ΔBRD1, or ΔBRD2 of FLAG-BRD4 binding to ZMYND8-V5 in HEK293T cells (Supplemental Figure 7D), further confirming that both BRD1 and BRD2 of BRD4 are required for ZMYND8 binding. On the other side, ΔPBP, ΔInter, or ΔC3 FLAG-ZMYND8 bound to BRD4 in HEK293T cells treated with TSA, similar to FL FLAG-ZMYND8 (Figure 11D). However, ΔC1, ΔC2, or ΔMYND of FLAG-ZMYND8 lost the ability to bind to BRD4 (Figure 11D), indicating that the MYND domain is necessary and sufficient for BRD4 interaction. Taken together, these findings indicate that the MYND domain of ZMYND8 interacts with BRD1/2 of BRD4 in cells.
To determine whether BRD4 regulates HIF-mediated transactivation, we established 2 independent Tet-inducible BRD4-KD MDA-MB-231 cell lines (Supplemental Figure 7E). SC and BRD4-KD1 or -KD2 MDA-MB-231 cells were exposed to 20% or 1% O2 for 24 hours in the presence of doxycycline. RT-qPCR assays showed that BRD4-KD1 or -KD2 significantly decreased hypoxia-induced transcription of the HIF target genes VEGFA, LOX, AGR2, AQP1, and ANGPTL4, but not RPL13A, in MDA-MB-231 cells (Figure 11E). This effect was not due to the protein stability of HIF-1α and HIF-2α, as their protein levels were not affected by BRD4-KD1 or -KD2 (Supplemental Figure 7E). These data indicate that BRD4 enhances HIF-mediated transactivation in breast cancer cells.
To determine whether BRD4 is required for ZMYND8-mediated HIF activation, we performed the HIF luciferase reporter assays. HeLa cells were transfected with HIF reporter plasmid, pSV-Renilla, SC or BRD4-KD1 or -KD2 vector, and ZMYND8-V5 vector or EV, and exposed to 20% or 1% O2 for 24 hours in the presence of doxycycline. Consistent with HIF target gene expression (Figure 11E), BRD4-KD1 or -KD2 significantly decreased HIF transcriptional activity in hypoxic HeLa cells (Figure 11F). Moreover, BRD4-KD1 or -KD2 dramatically blocked ZMYND8-induced HIF activation in nonhypoxic and hypoxic HeLa cells (Figure 11F). These data indicate that BRD4 promotes HIF transcriptional activity, and that it is required for ZMYND8-mediated HIF activation.
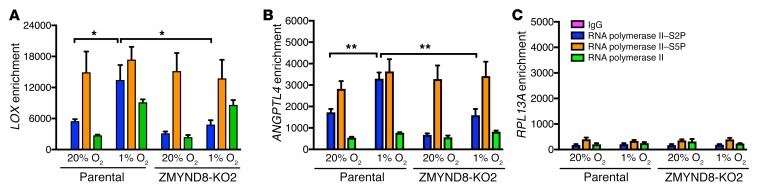
To determine whether ZMYND8 recruits BRD4 to the HREs of the HIF target genes, we performed ChIP-qPCR assays in parental and ZMYND8-KO2 MDA-MB-231 cells exposed to 20% or 1% O2 for 24 hours, and found that BRD4 occupied on the HREs of LOX and ANGPTL4 genes in nonhypoxic MDA-MB-231 cells, and hypoxia significantly increased its occupancy (Figure 11G, left and middle). ZMYND8-KO2 abolished hypoxia-induced BRD4 enrichment on the HREs of the LOX and ANGPTL4 genes but not the RPL13A gene in MDA-MB-231 cells (Figure 11G). Conversely, we found that BRD4-KD2 did not regulate ZMYND8 binding to the HREs of LOX, ANGPTL4, and RPL13A genes in nonhypoxic and hypoxic MDA-MB-231 cells (Supplemental Figure 7F). These data indicate that ZMYND8 recruits BRD4 to the HREs to induce HIF-mediated transactivation in breast cancer cells.
Previous studies showed that ZMYND8 recruits CHD4, a core helicase in the nucleosome remodeling and deacetylase complex, to the chromatin (33, 41). To determine whether CHD4 is involved in ZMYND8-mediated HIF activation, we performed co-IP assays and found that CHD4 physically interacted with HIF-1α and HIF-2α in MDA-MB-231 cells under hypoxia (Supplemental Figure 8A). However, ZMYND8-KO did not influence CHD4 binding to HIF-1α or HIF-2α in hypoxic MDA-MB-231 cells (Supplemental Figure 8A), suggesting that ZMYND8 does not mediate CHD4 recruitment to the HIF transactivation complex. Further, we found that CHD4 knockdown significantly inhibited expression of the HIF target genes VEGFA, LOX, and AGR2, but not AQP1, in MDA-MB-231 cells under hypoxia (Supplemental Figure 8, B and C), indicating that CHD4 does not fully phenocopy ZMYND8’s effect on HIF activation in breast cancer cells. These data indicate that ZMYND8 and CHD4 independently increase HIF-mediated transactivation in breast cancer cells.
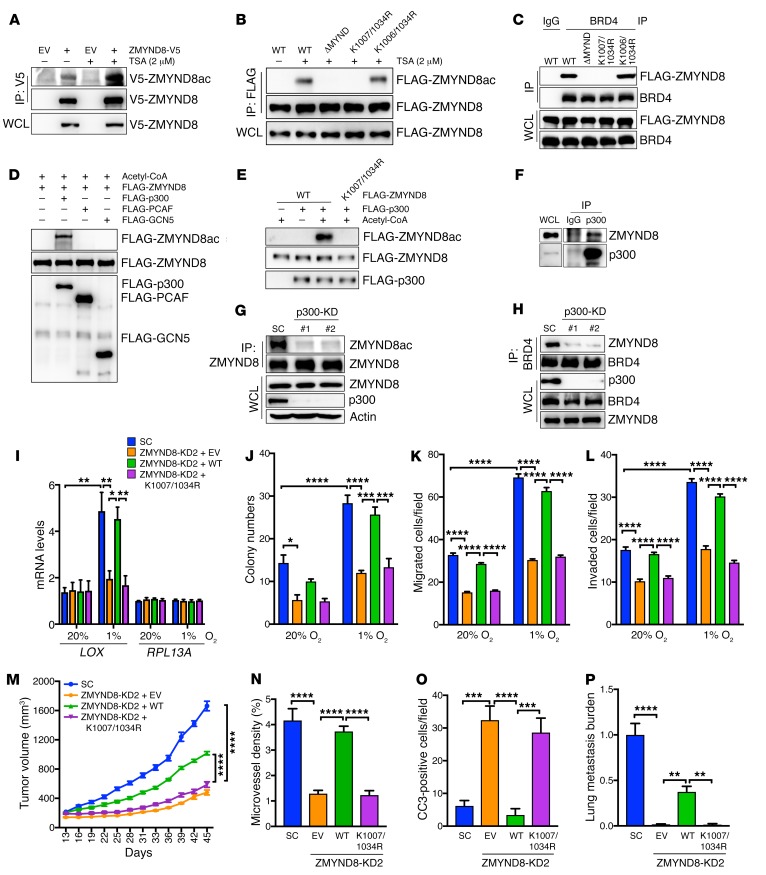
ZMYND8 acetylation at lysines 1007 and 1034 by p300 is required for interaction with BRD4 and HIF activation in breast cancer cells.
The BRD1/2 of BRD4 specifically recognizes acetylated lysine residues of its interacting protein (40). Our binding domain mapping studies (Figure 11, C and D) suggest that ZMYND8 may be acetylated at lysine residue(s) within the MYND domain. To test this hypothesis, HEK293T cells were transfected with vector encoding ZMYND8-V5 or EV, and treated with TSA or DMSO for 6 hours. Immunoblot analysis of V5 immunoprecipitates by anti–acetyl lysine antibody showed that ZMYND8-V5 is acetylated in cells, and that treatment of TSA dramatically increased lysine acetylation of ZMYND8-V5 (Figure 12A). Hypoxia had no effect on ZMYND8-V5 acetylation in HEK293T cells (Supplemental Figure 9A). Acetylation of endogenous ZMYND8 was also detected in MCF-7 cells treated with TSA (Figure 12G). Furthermore, we found that ΔMYND abolished lysine acetylation of FLAG-ZMYND8 (Figure 12B), indicating acetylation of the lysine residue(s) within the MYND domain. The MYND domain of ZMYND8 contains 3 lysine residues, and lysine 1007 and 1034 residues are highly conserved across species of vertebrates (Supplemental Figure 9B). Individual mutation of lysine into arginine did not reduce FLAG-ZMYND8 acetylation (Supplemental Figure 9C). Double mutation of lysines 1007 and 1034 to arginine (K1007/1034R) recapitulated the loss of acetylation of ΔMYND (Figure 12B), whereas double mutation of lysines 1006 and 1034 to arginine (K1006/1034R) did not. Consistently, K1007/1034R, but not K1006/1034R or individual lysine to arginine mutation, completely blocked ZMYND8 binding to BRD4 in HEK293T cells (Figure 12C, and Supplemental Figure 9D). None of these lysine mutations altered ZMYND8 binding to HIF-1α (Supplemental Figure 9E). These data indicate that ZMYND8 is acetylated at lysines 1007 and 1034, and that acetylated ZMYND8 is required for interaction with BRD4 in cells.
Figure 12. Acetylation of ZMYND8 by p300 is necessary for HIF activation and breast tumor progression.
(A and B) Acetylation of ZMYND8-V5 (A) and WT or mutant FLAG-ZMYND8 (B) in HEK293T cells treated with TSA or DMSO (–) for 6 hours (n = 3). (C) Co-IP assays of BRD4 and WT or mutant FLAG-ZMYND8 in transfected HEK293T cells (n = 3). (D and E) In vitro acetylation assays of WT or K1007/1034R FLAG-ZMYND8 by purified FLAG-p300, FLAG-PCAF, or FLAG-GCN5 (n = 3). (F) Co-IP assays of endogenous ZMYND8 and p300 in MCF-7 cells (n = 3). (G) Acetylation of endogenous ZMYND8 in SC and p300-KD MCF-7 cells (n = 3). (H) Co-IP assays of endogenous ZMYND8 and BRD4 in SC and p300-KD MCF-7 cells (n = 3). (I) RT-qPCR analysis of indicated mRNAs in ZMYND8-rescued MDA-MB-231 cells exposed to 20% or 1% O2 for 24 hours (mean ± SEM, n = 3). *P < 0.05, **P < 0.01, by 2-way ANOVA with Tukey’s t test. (J–L) Clonogenic assays (J), migration assays (K), and invasion assays (L) in ZMYND8-rescued MDA-MB-231 cells exposed to 20% or 1% O2 for 12 days (J), 16 hours (K), or 24 hours (L) (mean ± SEM, n = 3). *P < 0.05, ***P < 0.001, ****P < 0.0001, by 2-way ANOVA with Tukey’s t test. (M–P) Growth of ZMYND8-rescued MDA-MB-231 tumors in mice (M, mean ± SEM, n = 5). Endomucin-positive areas (N) and CC3-positive cell numbers (O) in tumors and lung metastasis (P) were quantified (mean ± SEM, n = 5). **P < 0.01, ***P < 0.001, ****P < 0.0001, by 2-way ANOVA with Tukey’s t test.
To identify the lysine acetyltransferase that acetylates ZMYND8, we screened acetyltransferases by in vitro acetylation assays. FLAG-ZMYND8 was expressed in HEK293T cells and purified by anti–FLAG antibody. Purified FLAG-ZMYND8 was then incubated in the presence of acetyl-CoA with the acetyltransferase FLAG-p300, FLAG-PCAF, or FLAG-GCN5 that was expressed and purified from Sf9 cells (42), and analyzed by immunoblot assays with anti–acetyl lysine antibody. FLAG-p300 strongly induced acetylation of FLAG-ZMYND8 in vitro (Figure 12D). In contrast, no detectable acetylation of FLAG-ZMYND8 was found after incubation with FLAG-PCAF or FLAG-GCN5 (Figure 12D). Moreover, K1007/1034R abolished FLAG-p300–induced acetylation of FLAG-ZMYND8 (Figure 12E). We next studied whether p300 catalyzes ZMYND8 acetylation in vivo. Co-IP assays showed that endogenous p300 interacted with endogenous ZMYND8 in MCF-7 cells (Figure 12F), and their interaction was not regulated by hypoxia (Supplemental Figure 9F). p300-KD by 2 independent shRNAs completely abolished lysine acetylation of endogenous ZMYND8 in MCF-7 cells (Figure 12G). Pharmacological inhibition of p300 by its inhibitor L002 also reduced acetylation of ZMYND8 in MCF-7 cells (Supplemental Figure 9G). Further, we found that p300-KD blocked ZMYND8-BRD4 interaction in MCF-7 cells (Figure 12H). Together, these findings indicate that p300 is a lysine acetyltransferase for ZMYND8.
To determine whether ZMYND8 acetylation is required for HIF-mediated transactivation, we generated the rescued cell lines by transducing ZMYND8-KD2 MDA-MB-231 cells with lentivirus encoding WT or K1007/1034R FLAG-ZMYND8 or EV. WT or K1007/1034R FLAG-ZMYND8 protein was expressed at comparable levels to endogenous ZMYND8 protein in SC MDA-MB-231 cells, and did not affect the levels of HIF-1α and HIF-2α proteins (Supplemental Figure 10A). These rescued cell lines and SC MDA-MB-231 cells were exposed to 20% or 1% O2 for 24 hours. RT-qPCR assays showed that the reduced transcription of LOX mRNA in ZMYND8-KD2 MDA-MB-231 cells was restored by overexpression of WT FLAG-ZMYND8 to that in SC MDA-MB-231 cells under hypoxia (Figure 12I). In contrast, overexpression of FLAG-ZMYND8 (K1007/1034R) failed to restore reduced transcription of LOX mRNA in ZMYND8-KD2 MDA-MB-231 cells (Figure 12I). Neither WT FLAG-ZMYND8 nor FLAG-ZMYND8 (K1007/1034R) influenced the RPL13A mRNA levels in nonhypoxic and hypoxic MDA-MB-231 cells (Figure 12I). These data indicate that acetylation of lysines 1007 and 1034 is necessary for ZMYND8-mediated HIF activation in breast cancer cells.
Acetylated ZMYND8 mediates breast tumor growth and lung metastasis.
Next, we studied whether ZMYND8 acetylation mediates the oncogenic effects in vitro and in vivo. Overexpression of WT FLAG-ZMYND8, but not FLAG-ZMYND8 (K1007/1034R), restored the colony formation, migration, and invasion abilities of ZMYND8-KD2 MDA-MB-231 cells (Figure 12, J–L, and Supplemental Figure 10, B and C). Reduced MDA-MB-231 tumor growth conferred by ZMYND8-KD2 was partially rescued by WT but not K1007/1034R FLAG-ZMYND8 in xenograft mice (Figure 12M, and Supplemental Figure 10D). Notably, overexpression of WT but not K1007/1034R FLAG-ZMYND8 completely restored microvessel density and CC3-positive cell numbers in ZMYND8-KD2 tumors (Figure 12, N and O, and Supplemental Figure 10E). Further, lung metastasis burden shown by H&E staining and qPCR assays was also partially but significantly rescued by overexpression of WT, but not K1007/1034R, FLAG-ZMYND8 in xenograft mice (Figure 12P and Supplemental Figure 10E). These data indicate that ZMYND8 acetylation is required for breast cancer progression and metastasis to the lungs.
Discussion
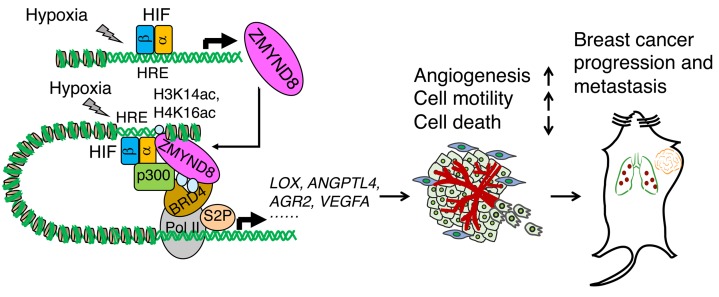
In the present study, we delineate an epigenetic mechanism by which the epigenetic reader ZMYND8 promotes HIF-mediated transactivation and breast cancer progression and metastasis (Figure 13). ZMYND8 physically interacts with HIF-1α and HIF-2α and also binds to the HREs possibly through H3K14ac and H4K16ac, where ZMYND8 is acetylated at lysines 1007 and 1034 by the HIF coactivator p300. Acetylated ZMYND8 recognizes the bromodomains of BRD4 and recruits the latter to the HREs, leading to increased RNA polymerase II phosphorylation at serine 2 and subsequent transcriptional elongation of the HIF target genes in breast cancer cells. Consequently, the ZMYND8/p300/BRD4/HIF axis increases angiogenesis and cell motility and decreases cancer cell death to promote breast tumor progression and metastasis. ZMYND8 itself is a HIF-1 and HIF-2 target gene, and thus provides a positive feedback mechanism that amplifies HIF-mediated transactivation and subsequent breast cancer progression and metastasis (Figure 13).
Figure 13. A proposed model of ZMYND8 in regulation of HIF transcriptional activity and breast cancer progression and metastasis.
ZMYND8 is localized at the HREs through H3K14ac and H4K16ac and acetylated at lysines 1007 and 1034 by p300 in breast cancer cells. Acetylated ZMYND8 recruits BRD4 to the HREs to form a HIF transactivation complex. Upon HIF binding to the HRE under hypoxia, ZMYND8/BRD4 are further enriched at the HREs and enhance RNA polymerase II phosphorylation at serine 2 and subsequent transcriptional elongation of the HIF target genes in breast cancer cells, thereby increasing angiogenesis and cell motility and decreasing cell death to promote breast cancer progression and metastasis. ZMYND8 itself is induced by HIF-1 and HIF-2 and thus amplifies HIF activity and HIF-mediated breast cancer progression and metastasis.
Several epigenetic regulators have been shown to coactivate a subset of HIF target genes in cancer cells (7, 26, 27, 43). Our RNA-seq and ChIP-seq data indicate that ZMYND8 co-occupies more than 85% of the genome-wide HIF-1 binding sites and activates more than 60% of the global HIF-dependent coding genes in breast cancer cells, indicating that ZMYND8 also regulates HIF-dependent long noncoding RNAs as they are the direct HIF targets under hypoxia (37). These findings reveal that ZMYND8 is a primary HIF regulator that plays a dominant role in HIF activation in breast cancer. In line with its dominant role in transcription of the HIF target genes in breast cancer cells, ZMYND8 phenocopies HIF’s oncogenic effects in breast cancer progression and metastasis. Strikingly, ZMYND8 acetylation at lysines 1007 and 1034 by p300 is the key to switch on HIF activation and breast cancer progression and metastasis. Our data indicate that this novel posttranslational modification mediates assembly of the HIF transcription machinery to promote HIF activation in breast cancer cells. ZMYND8 acetylation may be reversible in breast cancer cells, as treatment of an HDAC inhibitor TSA dramatically increases ZMYND8 acetylation. HDAC1 and HDAC2 have been shown to interact with ZMYND8 (33), and are also known to regulate HIF-1 transcriptional activity (26). Further studies are needed to determine which HDAC deacetylates ZMYND8.
Recent studies suggest that RNA polymerase II pausing and release finely switch HIF activation on and off (37, 43). The HIF transcription machinery, including RNA polymerase II, is preassembled and preloaded at the promoter of the target genes under nonhypoxic conditions, and release of paused RNA polymerase II triggers gene elongation once HIF binds to the HREs under hypoxia (37). We found that BRD4 is recruited by ZMYND8 to the HREs of the HIF target genes. Their interaction is constitutive and not regulated by O2 tension. BRD4 is known to interact with positive transcription elongation factor b (P-TEFb) and regulates P-TEFb–induced phosphorylation of RNA polymerase II at serine 2, thereby promoting release of paused RNA polymerase II at the promoter (39). Thus, the ZMYND8/BRD4 axis well supports the current model of HIF activation and represents an important molecular mechanism underlying release of paused RNA polymerase II and elongation of the HIF target genes.
Apart from BRD4, P-TEFb–mediated release of paused RNA polymerase II is also regulated by the super elongation complex (SEC) via a distinct mechanism (44). A previous report demonstrated a role of the SEC in release of paused RNA polymerase II at the promoter of a subset of HIF-1 target genes in HCT116 cells (43). CDK8 controls the recruitment of AFF4, a key component of the SEC, to the HIF-1 target gene ANKRD37, but has no effect on BRD4 recruitment. Notably, ZMYND8-controlled HIF target genes such as AQP1, AGR2, and LOX are not regulated by the CDK8/AFF4 axis (43). Therefore, ZMYND8/BRD4 and CDK8/AFF4 selectively regulate P-TEFb–induced release of paused RNA polymerase II on their respective HIF target genes and subsequent gene elongation.
Our extensive functional studies in multiple ER+ and TNBC cell lines in vitro and breast cancer mouse models indicate that ZMYND8 is an oncoprotein in breast cancer. This concept may be extended to prostate cancer because a previous study showed that ZMYND8 increases VEGFA expression and angiogenesis in prostate DU145 xenograft tumors from zebrafish (45). Previous genetic studies identified ZMYND8 fusion proteins in patients with acute erythroid leukemia and breast cancer and they may be pathogenic for the diseases (46, 47), also supporting an oncogenic role of ZMYND8 in human cancers. However, recent studies showed that ZMYND8 suppresses growth of ZR-75-30 or DU145 xenograft tumors by inducing gene silencing (48, 49). It is unclear whether these discrepancies are due to the experimental context or cell types. Nevertheless, we showed that DKO of HIF-1α and HIF-2α abolished ZMYND8-mediated breast cancer progression and metastasis in mice, suggesting that ZMYND8’s oncogenic functions in breast cancer require HIF-1 and HIF-2. ZMYND8 is known to bind to and enhance ER α transcriptional activity in MCF-7 cells (50), and also regulates the transcription of all-trans retinoic acid–dependent genes in SH-SY5Y cells (34). Whether or not these HIF-independent ZMYND8 functions contribute to breast tumorigenesis needs to be investigated.
In conclusion, our studies in human breast cancer cells, breast cancer mouse models, and human breast cancer patients provide a strong rationale for ZMYND8 as a potential biomarker and therapeutic target for the diagnosis and treatment of breast cancer.
Methods
Cell culture and transfection.
MDA-MB-231 (gift from Rolf Brekken, UT Southwestern, Dallas, Texas, USA), MDA-MB-468 (ATCC), HEK293FT (Invitrogen), HeLa, HEK293T, MCF-7, HCC1954, T47D, and SUM159 (gifts from Gregg L. Semenza, Johns Hopkins, Baltimore, Maryland, USA) cells were cultured in DMEM, RPMI1640, or DMEM/Ham’s F-12 supplemented with 10% heat-inactivated FBS at 37°C in a 5% CO2/95% air incubator. Hypoxic cells were placed in a modular incubator chamber (Billups-Rothenberg) and flushed with a gas mixture of 1% O2, 5% CO2, and balanced N2. Cells were transfected using PolyJet (SignaGen) or FuGENE6 (Promega). All cell lines were mycoplasma-free and were authenticated by short tandem repeat DNA profiling analysis.
Clonogenic assays.
One hundred cells were seeded on a 6-well plate and exposed to 20% or 1% O2 for 12–18 days. Colonies were washed with PBS, fixed with 4% paraformaldehyde, and stained with 0.5% crystal violet (MilliporeSigma). After staining, colonies were gently washed and counted.
Boyden chamber migration and invasion assays.
Cells (5 × 104) were resuspended in serum-free medium, seeded in a transwell insert (for migration) or a Matrigel-coated transwell insert (for invasion) in the presence of cell culture medium with 10% FBS at the bottom chamber, and exposed to 20% or 1% O2 for 16 hours (migration) or 24 hours (invasion). Cells that invaded to the lower side of the transwell insert were fixed with methanol, stained with 0.5% crystal violet, and counted.
IP and immunoblot assays.
Cells were lysed in NETN lysis buffer (150 mM NaCl, 1 mM EDTA, 10 mM Tris-HCl, pH 8.0, 0.5% NP-40, and protease inhibitor cocktail) for 30 minutes on ice and followed by sonication (33). After centrifugation at 13,000 g for 15 minutes, supernatant was collected for IP overnight with the following antibodies: V5, FLAG, HIF-1α, ZMYND8, BRD4, CHD4, or p300 in the presence of protein A/G magnetic beads (Bio-Rad). The next day, proteins bound on the beads were washed 4 times with NETN lysis buffer, boiled in 1× Laemmli buffer, and fractionated by SDS-PAGE, followed by immunoblot assays with the antibodies listed in Supplemental Table 1.
Luciferase reporter assays.
HeLa or HEK293T cells were seeded on a 48-well plate, transfected with the indicated plasmids, and exposed to 20% or 1% O2 for 24 hours. The FLuc/RLuc activities were measured using the Dual Luciferase Reporter Assay System (Promega) (7).
In vitro acetylation assays.
Immunoprecipitated WT or K1007/1034R FLAG-ZMYND8 protein was incubated for one hour at 30°C in the presence or absence of acetyl-CoA and purified FLAG-p300, FLAG-PCAF, or FLAG-GCN5 (gifts from Cheng-Ming Chiang, UT Southwestern, Dallas, Texas, USA) (42), resolved by SDS-PAGE, and analyzed by immunoblot assays using antibodies against acetyl lysine (1: 2,000) or FLAG epitope (1: 5,000).
RT-qPCR assays.
Total RNA was isolated from cultured cells using Trizol reagent (ThermoFisher) and reverse-transcribed. RT-qPCR assays were performed with the primers listed in Supplemental Table 2 (7).
RNA-seq assays.
Total RNA was isolated using the RNeasy mini kit (Qiagen), and treated with DNase (Qiagen). mRNA was used for library preparation with the TruSeq Stranded mRNA Library Prep Kit (Illumina), and sequenced on the Illumina NextSeq 500. See Supplemental Methods for analysis details.
ChIP assays.
Cells were crosslinked with 1% formaldehyde and quenched in 0.125 M glycine. ChIP-qPCR assays were performed with the primers listed in Supplemental Table 3 (7). For ChIP-seq, chromatin was isolated using the SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling Technology), sonicated to 200–300 bp in length, and subjected to IP using antibodies against ZMYND8, HIF-1α, H3K14ac, or histone H3. ChIP DNA libraries were prepared with the KAPA Hyper Prep Kit and sequenced on the Illumina NextSeq 500. See Supplemental Methods for analysis details.
Animal studies.
For the orthotopic breast cancer mouse model (7), 2 × 106 cells in 100 μl PBS/Matrigel (1:1, Corning) were injected into the second left mammary fat pad of female SCID mice (6–8 weeks old, Envigo). Tumor volume was measured with a caliper every 3 days beginning on day 11–16 after cell implantation, and calculated according to the formula: volume = 0.52 × length × height × width. Lungs were perfused with PBS and analyzed by H&E staining and qPCR assays with primers for human HK2 gene and mouse and human 18S rRNA.
For the tail vein injection model, 1 × 106 cells in 100 μl PBS were injected into the tail vein of female SCID mice. Three weeks later, lungs were perfused with PBS and subjected to qPCR assays with primers for human HK2 gene and mouse and human 18S rRNA.
Measurement of circulating tumor cells.
Genomic DNA was extracted from peripheral blood in SCID mice bearing parental or ZMYND8-KO MDA-MB-231 tumors using the QIAamp DNA Blood Mini Kit (Qiagen), and quantified by qPCR assays with primers for the human HK2 gene and mouse and human 18S rRNA. MDA-MB-231 cells were mixed with blood from tumor-free SCID mice to generate a standard curve. The number of circulating tumor cells in mouse blood was calculated according to the standard curve.
Immunohistochemistry assays.
Immunohistochemistry assays were performed by the Dako Autostainer Link 48 system. Briefly, the slides were baked, deparaffinized, and hydrated, followed by antigen retrieval in a Dako PT Link. The tissues were incubated with a peroxidase block and then a following primary antibody: ZMYND8 (1: 1,000), cleaved caspase 3 (1: 1,500), Ki-67 (ready to use), or endomucin (1: 50). The staining was visualized using the EnVision FLEX visualization system (Dako).
Human breast tumor studies.
Human ER+ breast tumor and TNBC tissues and a TNBC TMA were obtained from surgical breast cancer pathologist Yan Peng and UT Southwestern Tissue Resource, and analyzed by immunohistochemistry assays. Each staining was scored by Yan Chen and Yan Peng using 4 grades (range 0–3) according to the percentage of immunopositive cells and immunostaining intensity.
Statistics.
Statistical analysis was performed by 2-tailed Student’s t test between 2 groups, and 1-way or 2-way ANOVA with multiple testing correction within multiple groups. Quantification of ZMYND8 protein levels between normal breast tissues and human TNBC tissues was determined by χ2 test. Kaplan-Meier survival curve was analyzed by log-rank test. RNA-seq and ChIP-seq were repeated twice, and other experiments were repeated at least 3 times. Data were expressed as mean ± SEM. P < 0.05 was considered significant.
Accession number.
The RNA-seq and ChIP-seq data were deposited at the GEO database with accession number GSE108833.
Study approval.
Animal experiments were approved by the Animal Care and Use Committee at UT Southwestern Medical Center. The de-identified human ER+ breast tumor, TNBC, and adjacent normal breast tissues were used and the study was approved by the institutional review board at UT Southwestern Medical Center with informed consent.
Author contributions
WL and YW conceived the study, analyzed the data, and wrote the paper; YC performed most experiments, analyzed the data, and wrote the paper; BZ generated mutant ZMYND8 plasmids; LB generated the HIF reporter plasmid, HIF-1/2α sgRNA plasmids, and HIF-1/2α–KO cell lines; LJ assisted in animal studies; MY performed tail vein injection experiments; YP provided and analyzed TMA; AK and CX analyzed RNA-seq and ChIP-seq data; CW prepared cell lysates; JEW performed mouse breeding and lentivirus production; XZ generated p300-KD cells.
Supplementary Material
Acknowledgments
We thank Gregg L. Semenza (Johns Hopkins) for HeLa, HEK293T, MCF-7, T47D, HCC1954, and SUM159 cells; Rolf Brekken (UT Southwestern) for MDA-MB-231 cells; Cheng-Ming Chiang (UT Southwestern) for BRD4 plasmids and purified p300, PCAF, and GCN5 proteins; Kyle Miller (The University of Texas at Austin)for SFB-ZMYND8 plasmids; and Lei Guo (UT Southwestern) for generation of FLAG–HIF-2α plasmid. We also thank the UTSW Cancer Center Tissue Resource for assistance with immunohistochemistry (supported by NCI Cancer Center grant P30CA142543), the UTSW Next Generation Sequencing Core for assistance with RNA-seq and ChIP-seq, and the UTSW Pathology digital imaging core for TMA slide scanning. We are grateful to Carole Baas (Dallas, Texas, USA) for comments on the manuscript. This work was supported by grants from Susan G. Komen (CCR16376227), NIH (R00CA168746), the Cancer Prevention & Research Institute of Texas (CPRIT, RR140036), and the American Cancer Society/UTSW Simmons Cancer Center (ACS-IRG-02-196) to WL, and grants from NIH (R00NS078049, R35GM124693) to YW. CX was partially supported by NIH grant UL1TR001105. WL is a CPRIT Scholar in Cancer Research.
Version 1. 04/09/2018
Electronic publication
Version 2. 05/01/2018
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(5):1937–1955.https://doi.org/10.1172/JCI95089.
Contributor Information
Yan Chen, Email: Yan.Chen2@UTSouthwestern.edu.
Bo Zhang, Email: bo2.zhang@utsouthwestern.edu.
Lei Bao, Email: lei.bao@utsouthwestern.edu.
Lai Jin, Email: jinlai2007happy@163.com.
Mingming Yang, Email: mingming.yang@utsouthwestern.edu.
Yan Peng, Email: yan.peng@utsouthwestern.edu.
Ashwani Kumar, Email: Ashwani.Kumar@UTSouthwestern.edu.
Jennifer E. Wang, Email: jennifer.wang@utsouthwestern.edu.
Chenliang Wang, Email: chenliang.wang@utsouthwestern.edu.
Xuan Zou, Email: wooair@gmail.com.
Chao Xing, Email: chao.xing@utsouthwestern.edu.
Yingfei Wang, Email: Yingfei.Wang@Utsouthwestern.edu.
Weibo Luo, Email: weibo.luo@utsouthwestern.edu.
References
- 1.Bradner JE, Hnisz D, Young RA. Transcriptional addiction in cancer. Cell. 2017;168(4):629–643. doi: 10.1016/j.cell.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ell B, Kang Y. Transcriptional control of cancer metastasis. Trends Cell Biol. 2013;23(12):603–611. doi: 10.1016/j.tcb.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho MH, et al. DOT1L cooperates with the c-Myc-p300 complex to epigenetically derepress CDH1 transcription factors in breast cancer progression. Nat Commun. 2015;6:7821. doi: 10.1038/ncomms8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong C, et al. G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J Clin Invest. 2012;122(4):1469–1486. doi: 10.1172/JCI57349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu MZ, et al. Interplay between HDAC3 and WDR5 is essential for hypoxia-induced epithelial-mesenchymal transition. Mol Cell. 2011;43(5):811–822. doi: 10.1016/j.molcel.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 6.Ramadoss S, Chen X, Wang CY. Histone demethylase KDM6B promotes epithelial-mesenchymal transition. J Biol Chem. 2012;287(53):44508–44517. doi: 10.1074/jbc.M112.424903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo W, Chang R, Zhong J, Pandey A, Semenza GL. Histone demethylase JMJD2C is a coactivator for hypoxia-inducible factor 1 that is required for breast cancer progression. Proc Natl Acad Sci U S A. 2012;109(49):E3367–E3376. doi: 10.1073/pnas.1217394109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim JH, et al. UTX and MLL4 coordinately regulate transcriptional programs for cell proliferation and invasiveness in breast cancer cells. Cancer Res. 2014;74(6):1705–1717. doi: 10.1158/0008-5472.CAN-13-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang CJ, et al. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-β-catenin signaling. Cancer Cell. 2011;19(1):86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi L, et al. Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis. Proc Natl Acad Sci U S A. 2011;108(18):7541–7546. doi: 10.1073/pnas.1017374108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shu S, et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature. 2016;529(7586):413–417. doi: 10.1038/nature16508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wen H, et al. ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature. 2014;508(7495):263–268. doi: 10.1038/nature13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaupel P, Mayer A, Höckel M. Tumor hypoxia and malignant progression. Meth Enzymol. 2004;381:335–354. doi: 10.1016/S0076-6879(04)81023-1. [DOI] [PubMed] [Google Scholar]
- 14.Semenza GL. Hypoxia-inducible factors: mediators of cancer progression and targets for cancer therapy. Trends Pharmacol Sci. 2012;33(4):207–214. doi: 10.1016/j.tips.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997;11(1):72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 16.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995;92(12):5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu YZ, Moran SM, Hogenesch JB, Wartman L, Bradfield CA. Molecular characterization and chromosomal localization of a third alpha-class hypoxia inducible factor subunit, HIF3alpha. Gene Expr. 1998;7(3):205–213. [PMC free article] [PubMed] [Google Scholar]
- 18.Epstein AC, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107(1):43–54. doi: 10.1016/S0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 19.Semenza GL, et al. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J Biol Chem. 1996;271(51):32529–32537. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- 20.Pugh CW, Ratcliffe PJ. Regulation of angiogenesis by hypoxia: role of the HIF system. Nat Med. 2003;9(6):677–684. doi: 10.1038/nm0603-677. [DOI] [PubMed] [Google Scholar]
- 21.Esteva-Font C, Jin BJ, Verkman AS. Aquaporin-1 gene deletion reduces breast tumor growth and lung metastasis in tumor-producing MMTV-PyVT mice. FASEB J. 2014;28(3):1446–1453. doi: 10.1096/fj.13-245621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong XY, Wang J, Li Z. AGR2 expression is regulated by HIF-1 and contributes to growth and angiogenesis of glioblastoma. Cell Biochem Biophys. 2013;67(3):1487–1495. doi: 10.1007/s12013-013-9650-4. [DOI] [PubMed] [Google Scholar]
- 23.Zhang H, et al. HIF-1-dependent expression of angiopoietin-like 4 and L1CAM mediates vascular metastasis of hypoxic breast cancer cells to the lungs. Oncogene. 2012;31(14):1757–1770. doi: 10.1038/onc.2011.365. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Erler JT, et al. Hypoxia-induced lysyl oxidase is a critical mediator of bone marrow cell recruitment to form the premetastatic niche. Cancer Cell. 2009;15(1):35–44. doi: 10.1016/j.ccr.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong CC, et al. Hypoxia-inducible factor 1 is a master regulator of breast cancer metastatic niche formation. Proc Natl Acad Sci U S A. 2011;108(39):16369–16374. doi: 10.1073/pnas.1113483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo W, Wang Y. Epigenetic regulators: multifunctional proteins modulating hypoxia-inducible factor-α protein stability and activity. Cell Mol Life Sci. 2018;75(6):1043–1056. doi: 10.1007/s00018-017-2684-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perez-Perri JI, et al. The TIP60 complex is a conserved coactivator of HIF1A. Cell Rep. 2016;16(1):37–47. doi: 10.1016/j.celrep.2016.05.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arany Z, et al. An essential role for p300/CBP in the cellular response to hypoxia. Proc Natl Acad Sci U S A. 1996;93(23):12969–12973. doi: 10.1073/pnas.93.23.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JS, et al. Negative regulation of hypoxic responses via induced Reptin methylation. Mol Cell. 2010;39(1):71–85. doi: 10.1016/j.molcel.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JS, et al. Hypoxia-induced methylation of a pontin chromatin remodeling factor. Proc Natl Acad Sci U S A. 2011;108(33):13510–13515. doi: 10.1073/pnas.1106106108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pollard PJ, et al. Regulation of Jumonji-domain-containing histone demethylases by hypoxia-inducible factor (HIF)-1alpha. Biochem J. 2008;416(3):387–394. doi: 10.1042/BJ20081238. [DOI] [PubMed] [Google Scholar]
- 32.Fossey SC, Kuroda S, Price JA, Pendleton JK, Freedman BI, Bowden DW. Identification and characterization of PRKCBP1, a candidate RACK-like protein. Mamm Genome. 2000;11(10):919–925. doi: 10.1007/s003350010174. [DOI] [PubMed] [Google Scholar]
- 33.Gong F, et al. Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription-associated DNA damage that promotes homologous recombination. Genes Dev. 2015;29(2):197–211. doi: 10.1101/gad.252189.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adhikary S, et al. Selective recognition of H3.1K36 dimethylation/H4K16 acetylation facilitates the regulation of all-trans-retinoic acid (ATRA)-responsive genes by putative chromatin reader ZMYND8. J Biol Chem. 2016;291(6):2664–2681. doi: 10.1074/jbc.M115.679985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Savitsky P, et al. Multivalent histone and DNA engagement by a PHD/BRD/PWWP triple reader cassette recruits ZMYND8 to K14ac-rich chromatin. Cell Rep. 2016;17(10):2724–2737. doi: 10.1016/j.celrep.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J, et al. EglN2 associates with the NRF1-PGC1α complex and controls mitochondrial function in breast cancer. EMBO J. 2015;34(23):2953–2970. doi: 10.15252/embj.201591437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choudhry H, et al. Extensive regulation of the non-coding transcriptome by hypoxia: role of HIF in releasing paused RNApol2. EMBO Rep. 2014;15(1):70–76. doi: 10.1002/embr.201337642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu M, et al. RNA polymerase II-associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II. Science. 2015;350(6266):1383–1386. doi: 10.1126/science.aad2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19(4):523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 40.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia L, et al. CHD4 has oncogenic functions in initiating and maintaining epigenetic suppression of multiple tumor suppressor genes. Cancer Cell. 2017;31(5):653–668.e7. doi: 10.1016/j.ccell.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu SY, Lee AY, Lai HT, Zhang H, Chiang CM. Phospho switch triggers Brd4 chromatin binding and activator recruitment for gene-specific targeting. Mol Cell. 2013;49(5):843–857. doi: 10.1016/j.molcel.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galbraith MD, et al. HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response to hypoxia. Cell. 2013;153(6):1327–1339. doi: 10.1016/j.cell.2013.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo Z, Lin C, Shilatifard A. The super elongation complex (SEC) family in transcriptional control. Nat Rev Mol Cell Biol. 2012;13(9):543–547. doi: 10.1038/nrm3417. [DOI] [PubMed] [Google Scholar]
- 45.Kuroyanagi J, et al. Zinc finger MYND-type containing 8 promotes tumour angiogenesis via induction of vascular endothelial growth factor-A expression. FEBS Lett. 2014;588(18):3409–3416. doi: 10.1016/j.febslet.2014.07.033. [DOI] [PubMed] [Google Scholar]
- 46.Panagopoulos I, et al. Fusion of ZMYND8 and RELA genes in acute erythroid leukemia. PLoS One. 2013;8(5):e63663. doi: 10.1371/journal.pone.0063663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wada Y, et al. Development of detection method for novel fusion gene using GeneChip exon array. J Clin Bioinforma. 2014;4(1):3. doi: 10.1186/2043-9113-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen H, et al. Suppression of enhancer overactivation by a RACK7-histone demethylase complex. Cell. 2016;165(2):331–342. doi: 10.1016/j.cell.2016.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li N, et al. ZMYND8 reads the dual histone mark H3K4me1-H3K14ac to antagonize the expression of metastasis-linked genes. Mol Cell. 2016;63(3):470–484. doi: 10.1016/j.molcel.2016.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malovannaya A, et al. Analysis of the human endogenous coregulator complexome. Cell. 2011;145(5):787–799. doi: 10.1016/j.cell.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.