Abstract
Thirteen percent of pregnancies result in preterm birth or stillbirth, accounting for fifteen million preterm births and three and a half million deaths annually. A significant cause of these adverse pregnancy outcomes is in utero infection by vaginal microorganisms. To establish an in utero infection, vaginal microbes enter the uterus by ascending infection; however, the mechanisms by which this occurs are unknown. Using both in vitro and murine models of vaginal colonization and ascending infection, we demonstrate how a vaginal microbe, group B streptococcus (GBS), which is frequently associated with adverse pregnancy outcomes, uses vaginal exfoliation for ascending infection. GBS induces vaginal epithelial exfoliation by activation of integrin and β-catenin signaling. However, exfoliation did not diminish GBS vaginal colonization as reported for other vaginal microbes. Rather, vaginal exfoliation increased bacterial dissemination and ascending GBS infection, and abrogation of exfoliation reduced ascending infection and improved pregnancy outcomes. Thus, for some vaginal bacteria, exfoliation promotes ascending infection rather than preventing colonization. Our study provides insight into mechanisms of ascending infection by vaginal microbes.
Keywords: Infectious disease, Reproductive Biology
Keywords: Bacterial infections
Introduction
Adverse pregnancy outcomes, including premature birth and stillbirth, are the leading causes of neonatal morbidity and mortality (1, 2). Fifteen million babies are born prematurely each year (3), and it is estimated that one and a half million pregnancies result in stillbirth (4). A frequent cause of preterm birth and stillbirth is intrauterine infection, which occurs when bacteria ascend from the vagina into the uterus (5, 6) and invade the amniotic cavity, leading to inflammation, tissue damage, and adverse pregnancy outcomes (7). One such bacterium associated with ascending infection and adverse pregnancy outcomes is group B streptococcus (GBS), or Streptococcus agalactiae (8). The principal risk factor for GBS-associated adverse pregnancy outcomes is vaginal colonization (9, 10), and 11%–22% of healthy women are colonized with GBS during pregnancy (11). It is estimated that 3.5 million preterm births are attributable to GBS (8). Despite the substantial impact on pregnancy outcomes, the mechanisms by which bacteria ascend from the vagina to the uterus remain unknown.
A fundamental question that has yet to be answered is why some women experience ascending microbial infections during pregnancy, while others do not. Although the answer to this question is complex, it must be the case that there is a breakdown in host defenses that prevent ascending infection. Epithelial cells are the primary defense against many invading pathogens. Epithelial defense is multifaceted and includes the physical barrier created by the epithelium, the ability of epithelial cells to recognize and respond to pathogenic insult, and communication of epithelial cells with cellular defense mechanisms upon pathogen recognition (12). A key mechanism used by epithelial layers to defend against pathogens is exfoliation (also known as shedding or sloughing) of infected epithelial cells (13, 14). Upon pathogenic challenge, epithelial cells lose their tight junctions and adhesive properties and detach from their basement membrane, thus preventing further pathogen colonization and dissemination (15). For example, vaginal epithelial cell exfoliation prevents colonization by Neisseria gonorrheae deficient for the opacity-associated protein (Opa), but Opa+ strains prevent exfoliation and thus colonize more efficiently (13). Similarly, E. coli strains that induce high levels of bladder epithelial exfoliation through increased α-hemolysin expression have lower bacterial loads in models of urinary tract infection (16). Given that vaginal GBS colonization increases the risk of ascending infection and adverse pregnancy outcomes, we hypothesized that vaginal epithelial exfoliation may regulate GBS colonization.
Here, we describe a mechanism by which GBS utilizes epithelial exfoliation to establish an invasive perinatal infection. We show that GBS stimulates vaginal epithelial exfoliation by activating integrin and β-catenin signaling, leading to the loss of barrier function and epithelial-to-mesenchymal transition (EMT) in vaginal epithelial cells. Although epithelial exfoliation is thought to prevent pathogen colonization of the vaginal tract (12, 17), we found that GBS-induced epithelial exfoliation had no impact on vaginal colonization, but rather permitted bacterial dissemination and ascending infection. Our study shows that preventing GBS-induced exfoliation by blocking upstream integrin signaling decreases ascending infection and improves pregnancy outcomes. This work represents a shift in the current paradigm that vaginal epithelial exfoliation prevents colonization of the vaginal tract.
Results
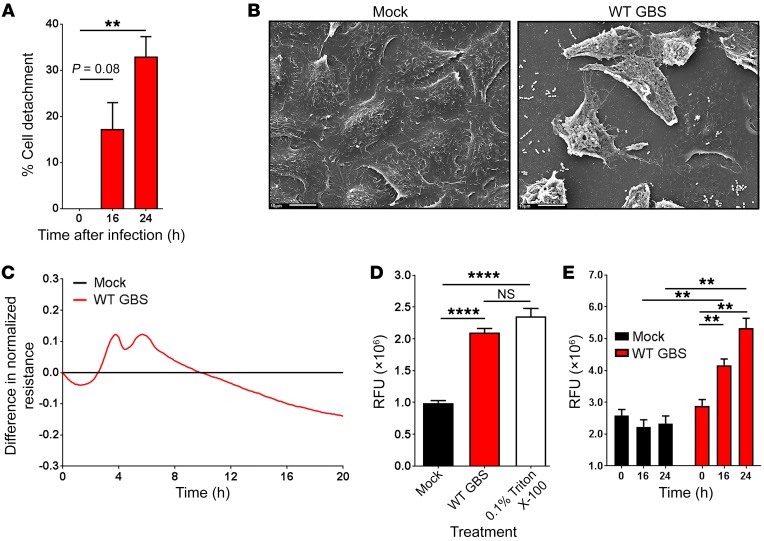
Epithelial exfoliation is a complex process that is characterized by a loss of barrier function and cellular detachment (18). We hypothesized that GBS may trigger the loss of barrier function and cellular detachment of vaginal epithelial cells, leading to exfoliation. To test this hypothesis, we infected immortalized human vaginal epithelial cells (hVECs) with the GBS WT strain COH1 (19). Interestingly, we observed a significant amount of cellular detachment (Figure 1A) and differences in cell shape and structure (Figure 1B) in response to GBS infection. To determine whether cellular detachment was due to a loss of barrier function, we used electric cell-substrate impedance sensing (ECIS) (20) to measure transepithelial resistance. Indeed, we observed a loss of barrier function in WT GBS–infected hVECs (Figure 1C). Additionally, GBS induced a significant increase in transepithelial permeability as measured by fluorescein migration across GBS-infected hVEC monolayers on Transwells (Figure 1D). These data indicate that GBS indeed stimulates vaginal epithelial cell exfoliation; however, we wanted to explore the physiological consequences of GBS-induced exfoliation. To this end, we measured the ability of GBS to migrate across hVEC monolayers in the Transwell assay. A significant amount of GBS crossed the epithelial barrier at time points associated with epithelial exfoliation (Figure 1E). Taken together, these findings suggest that GBS induces vaginal epithelial exfoliation, allowing for increased GBS migration across the vaginal epithelia.
Figure 1. GBS stimulates exfoliation and disrupts vaginal epithelial cell barrier function in vitro.
(A) hVECs were infected with WT GBS for 0, 16, or 24 hours and stained with 10% crystal violet for 30 minutes. Loosely adherent cells were removed by centrifugation, and crystal violet staining intensity was measured. Data were normalized to mock-infected controls (n = 3; **P < 0.005 and P = 0.08, by ANOVA followed by Sidak’s multiple comparisons test; data represent the mean ± SEM). (B) hVECs were infected with WT GBS for 24 hours and analyzed by scanning electron microscopy. Images show hVEC detachment 24 hours after infection with control PBS (mock) or WT GBS. Scale bars: 10 μm. (C) The barrier function of hVEC monolayers was monitored in real time using ECIS. Infection with WT GBS led to a disruption in barrier function as determined by the decrease in resistance of the infected monolayers compared with the uninfected control (n = 3; data represent the mean). (D) hVECs grown on Transwells were infected with WT GBS for 24 hours or treated with 0.1% Triton X-100, which was used as a positive control. After 24 hours, fluorescein dye was added to the apical compartment of the Transwell, and migration of the dye to the basal compartment was measured after 1 hour (n = 3; ****P < 0.00005, by ANOVA followed by Sidak’s multiple comparisons test; data represent the mean ± SEM). (E) hVECs grown on Transwells were either mock infected or infected with WT GBS for 24 hours. At 0, 16, and 24 hours after infection, the basal compartment of each well was sampled and stained with SYTO9 nucleic acid to enumerate bacteria (n = 3; **P < 0.005, by ANOVA followed by Sidak’s multiple comparisons test; data represent the mean ± SEM). RFU, relative fluorescence units.
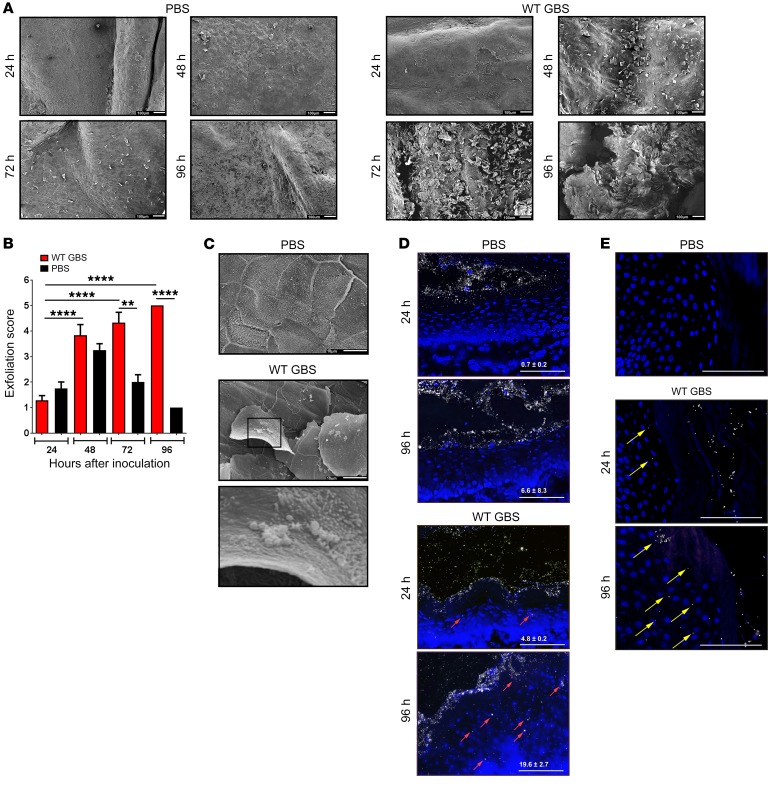
To corroborate these findings in vivo, female mice were vaginally inoculated with WT GBS or saline, and scanning electron microscopy was used to observe epithelial exfoliation as described previously (13, 17). Interestingly, we observed exfoliation in response to GBS inoculation, but not in response to control saline (Figure 2A). Blinded quantification and scoring of these images revealed significantly higher levels of vaginal exfoliation in GBS-treated animals compared with levels in saline-treated controls 72 and 96 hours after inoculation (Figure 2B). The slight increase in epithelial exfoliation observed in saline controls 48 to 72 hours after inoculation (Figure 2, A and B) is consistent with the natural estrus cycle (21). Exfoliated cells appeared to be associated with bacteria reminiscent of GBS, which were not observed in the saline-treated control animals (Figure 2C). To confirm that GBS-induced exfoliation leads to a loss of barrier function, we used fluorescent nanoparticles to probe vaginal epithelial barrier function in vivo. We found that more nanoparticles penetrated the epithelial barrier in GBS-inoculated mice over time (Figure 2D), but did not observe intraepithelial nanoparticles in the control animals (Figure 2D). This increase in epithelial permeability suggested that GBS may also be able to penetrate the vaginal epithelial barrier. Indeed, intraepithelial GBS were observed in conjunction with epithelial disruption (Figure 2E), indicating that GBS are able to disseminate into epithelial tissues. Collectively, these data indicate that GBS stimulates vaginal epithelial exfoliation in vivo and that disruption of epithelial barrier function is associated with increased bacterial dissemination.
Figure 2. GBS stimulates exfoliation and disrupts vaginal epithelial cell barrier function in vivo.
(A) Female WT C57BL6/J mice were vaginally inoculated with approximately 108 CFU of either WT GBS or an equal volume of control PBS (n = 3/group). Vaginal tissues were analyzed by scanning electron microscopy, and bacterial burden was assessed in vaginal and uterine tissues. Images show significant vaginal epithelial exfoliation 72 and 96 hours after inoculation with WT GBS and not with control saline. Scale bars: 100 μm. (B) Exfoliated vaginal epithelial cells 24, 48, 72, and 96 hours after inoculation were quantified in a blinded fashion (n = 3 images/2 tissues/group; **P < 0.005 and ****P < 0.00005, by ANOVA followed by Sidak’s multiple comparisons test; data represent the mean ± SEM). (C) High-magnification images of the vaginal epithelium show that GBS was associated with exfoliated epithelial cells. Scale bars: 10 μm; Original magnification, ×1900 (enlarged inset). Images are from 1 of at least 3 experiments. (D) Nanoparticle penetration into mouse vaginal epithelia in control saline– or WT GBS–treated animals 24 and 96 hours after vaginal inoculation, respectively. Nuclei were stained with DAPI and are shown in blue, PEGylated nanoparticles (120 nm diameter) are shown in white, and red arrows indicate intraepithelial nanoparticles. Images are from 1 of at least 2 experiments. Scale bars: 100 μm. Quantitative measurements were calculated using the equation: ([mean area of intraepithelial nanoparticle coverage]/[mean epithelial area]) × 10,000 ± SD. (E) GBS penetration into mouse vaginal epithelia in control saline– or WT GBS–treated animals 24 and 96 hours after vaginal inoculation. Nuclei were stained with DAPI and are shown in blue, GBS are shown in white, and yellow arrows indicate intraepithelial GBS. Images are from 1 of at least 4 experiments. Scale bars: 10 μm.
We next aimed to define the mechanistic underpinnings of GBS-induced vaginal epithelial exfoliation. We first sought to determine whether stimulation of epithelial exfoliation differed by GBS capsular serotypes or is dependent on known virulence factors. To this end, we compared WT GBS strains such as COH1 (capsular serotype III), A909 (capsular serotype Ia), NEM316 (capsular serotype III), and NCTC10/84 (capsular serotype V) for their ability to stimulate exfoliation and isogenic mutants deficient for hemolytic pigment (ΔcylE), capsule biosynthesis (ΔcpsK), pilus function (ΔpilB), or glycosyltransferase, which is known to play a role in GBS virulence (ΔiagA). We observed no significant differences in the levels of exfoliation among different GBS capsular serotypes (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/JCI97043DS1) or among GBS mutants lacking hemolytic pigment (Supplemental Figure 1, A and B), capsule (Supplemental Figure 1, A and B), or pilus function (Supplemental Figure 1, A and E). Although not significant, the slight reduction in uterine bacterial load for the pigment-deficient strain is probably attributable to attenuated virulence in the uterus, as noted in other studies (22–24), rather than a difference in ascending infection. Interestingly, deletion of iagA resulted in significantly less vaginal epithelial exfoliation (Supplemental Figure 1, A and H). These data suggest that the exfoliation is independent of the capsular serotype, hemolytic pigment, capsule, or pilus and is instead dependent on expression of the glycosyltransferase IagA.
GBS lacking iagA shed lipoteichoic acid (LTA) as a result of the loss of a diglucosyl-diacylglycerol membrane anchor, leading to reduced virulence (25). Given that GBS lacking IagA were shown to shed LTA, a TLR2 agonist that is an important regulator of the host response to GBS (26) and a driver for loss of barrier function in response to S. pneumoniae (27), we first examined the role of TLR2 in stimulating epithelial exfoliation by using TLR2-KO mice. We found that vaginal epithelial exfoliation and ascending GBS infection were independent of TLR2, as we observed no differences in either vaginal exfoliation or ascending infection (Supplemental Figure 2, A and B) between WT mice and TLR2-KO mice. These data are consistent with previous findings that attenuated virulence of GBSΔiagA is independent of TLR2 (25). Also, mice deficient for the critical TLR2 adaptor molecule MyD88 showed increased levels of vaginal epithelial exfoliation in response to GBS colonization (Supplemental Figure 2, A and E). These data indicate that GBS induction of epithelial exfoliation is independent of TLR2 signaling.
We next explored cell death as a mechanism of GBS stimulation of epithelial exfoliation, as induction of cell death is a critical component of the host response to genitourinary GBS (22) as well as an important aspect of E. coli–induced bladder epithelial exfoliation (16). We observed no differences in vaginal exfoliation between WT mice and caspase 1–KO mice (Supplemental Figure 2, A and B), indicating that caspase 1 does not control epithelial exfoliation. Also, vaginal exfoliation was independent of cell death, as GBS infection of hVECs did not significantly increase cytotoxicity (Supplemental Figure 3A). Exfoliated murine vaginal epithelial cells (mVECs) (cells expressing CD326 or EpCAM, an epithelial cell marker) collected from GBS- and saline-inoculated mice did not show any differences in propidium iodide uptake (Supplemental Figure 3B). Moreover, exfoliated CD326+ mVECs did not have external phosphatidylserine, a marker of apoptosis, as determined by annexin V staining (Supplemental Figure 3C). Together, these data indicate that GBS induction of epithelial exfoliation is independent of induction of cytotoxicity or cell death. It is likely that the exfoliated cells eventually die, but cell death does not appear to be the cause of exfoliation.
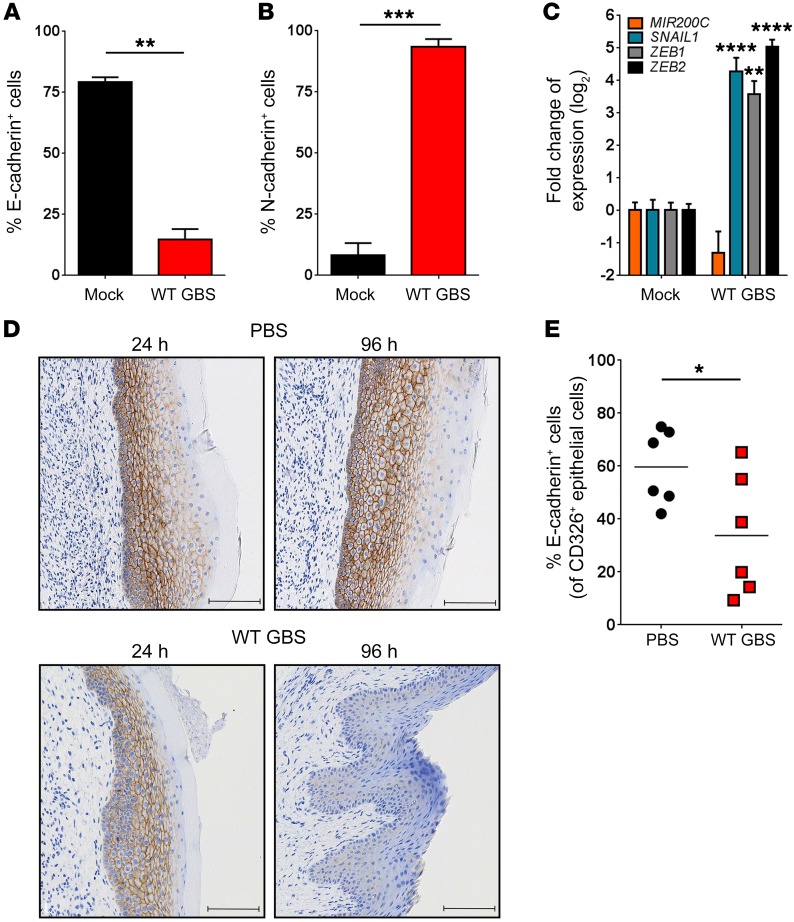
Given that neither immune signaling nor cell death pathways were involved in the stimulation of vaginal epithelial exfoliation, we examined whether EMT is the mechanism by which GBS triggers epithelial exfoliation. Loss of barrier function and increased mobility of epithelial cells are frequently linked to EMT (28), the process whereby nonmotile epithelial cells convert to mobile metastatic cells. EMT is characterized by a loss of the epithelial marker E-cadherin and a gain in expression of the mesenchymal marker N-cadherin (28). To determine whether GBS-infected hVECs display this characteristic cadherin switch, we used flow cytometry to measure surface marker expression (see the gating strategy in Supplemental Figure 4A). Consistent with this hypothesis, WT GBS–infected hVECs had decreased surface expression of E-cadherin (Figure 3A and Supplemental Figure 4B) and increased surface expression of N-cadherin (Figure 3B and Supplemental Figure 4C). These effects were found to be independent of capsular serotype (Supplemental Figure 4, B and C) or bacterial invasion (Supplemental Figure 5). Additionally, hVECs infected with WT GBS showed changes in the transcription of genes such as SNAIL1, ZEB1, and ZEB2 that are well-characterized markers of EMT (28). We detected decreased MIR200C expression and increased SNAIL1, ZEB1, and ZEB2 expression in GBS-infected hVECs compared with expression in controls (Figure 3C). To validate these findings in vivo, we analyzed E-cadherin levels in murine vaginal tracts by immunohistochemistry. We detected decreased levels of E-cadherin in WT GBS–inoculated mice, but not in control animals (Figure 3D). Flow cytometric analysis of primary vaginal epithelial cells isolated from GBS-infected mice (see Supplemental Figure 6 for cellular isolation and gating strategy) had significantly lower E-cadherin expression levels compared with levels in those isolated from control animals (Figure 3E). Interestingly, we did not observe N-cadherin staining in murine vaginal tissue. This finding is not without precedence, as other studies have reported characteristic hallmarks of EMT, specifically the loss of E-cadherin without a gain of N-cadherin (29), due to the variable expression of N-cadherin in different tissues (30, 31). Finally, infection of hVECs with the common vaginal commensal Lactobacillus crispatus failed to induce barrier disruption or EMT (Supplemental Figure 7), suggesting that EMT induction does not occur in response to common vaginal commensals. Together, these data show that GBS are able to induce EMT in vaginal epithelial cells, which is the likely cause of epithelial exfoliation.
Figure 3. GBS induces EMT.
Flow cytometric analysis of surface E-cadherin (A) or N-cadherin (B) expression on GBS-infected hVECs compared with the mock (PBS) control (n = 3; **P < 0.005 and ***P < 0.0005, by 2-sided, unpaired t test; data represent the mean ± SEM). (C) Quantitative reverse transcription PCR analysis of EMT markers in GBS-infected hVECs compared with mock-treated controls (n = 3; **P < 0.005 and ****P < 0.00005, by 1-way ANOVA followed by Sidak’s multiple comparisons test; data represent the mean ± SEM). (D) E-cadherin immunostaining in murine vaginal tracts 24 and 96 hours after vaginal inoculation with PBS or WT GBS, respectively. Images are from 1 of 3 experiments. Scale bars: 100 μm. (E) Flow cytometric analysis of surface E-cadherin on CD326+ mVECs 96 hours after vaginal inoculation with WT GBS or control PBS (n = 6 mice/group; *P < 0.05, by 2-sided, unpaired t test; data represent the mean).
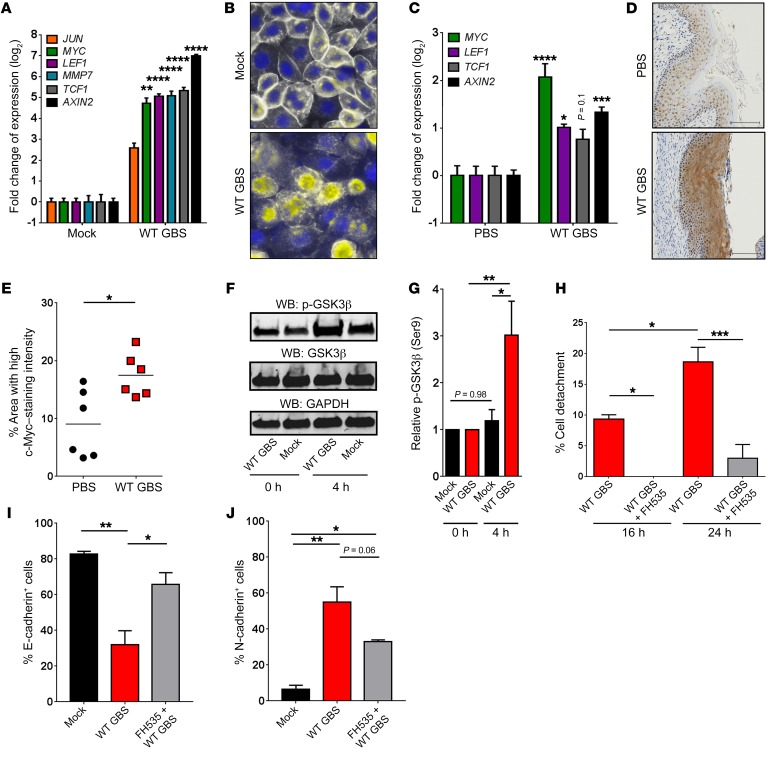
We next sought to determine whether GBS can trigger the β-catenin signaling pathway, which can regulate EMT (28). β-Catenin is sequestered at the membrane by E-cadherin and, when released, is either degraded or translocated to the nucleus. There, it binds the transcription factors T cell factor 1 (TCF1) and lymphoid enhancer factor 1 (LEF1), resulting in the transcription of genes associated with cell-cycle progression, EMT, and oncogenesis (32). Intriguingly, β-catenin–regulated genes were significantly upregulated in WT GBS–infected hVECs (Figure 4A). Visualization of β-catenin in GBS-infected hVECs showed relocalization to the cytoplasm and nucleus (Figure 4B). Additionally, many β-catenin–regulated genes were significantly upregulated in vaginal tissues from GBS-inoculated mice compared with that seen in tissues from controls (Figure 4C), and we observed increased levels of the β-catenin target c-Myc in GBS-inoculated vaginal tissues compared with levels in control animal tissues (Figure 4D). Quantification of these images revealed a significant increase in the amount of c-Myc expression across GBS-inoculated vaginal tissues (Figure 4E). β-Catenin signaling is facilitated by deactivation of glycogen synthase kinase-3 β (GSK3β), which occurs when GSK3β is phosphorylated at Ser9 (33). Deactivation of GSK3β prevents cytoplasmic degradation of β-catenin, leading to its nuclear translocation (34). Consistent with the above, GBS infection of hVECs induced the phosphorylation of GSK3β (p-GSK3β) (Figure 4, F and G). Finally, inhibition of β-catenin signaling by the chemical inhibitor FH535 (35) blocked cell detachment (Figure 4H) and loss of the EMT phenotype in response to WT GBS infection (Figure 4, I and J). These data show that β-catenin signaling controls EMT in vaginal epithelial cells and that GBS exploits this signaling mechanism to induce epithelial exfoliation.
Figure 4. GBS induces β-catenin signaling.
(A) Expression of β-catenin target genes in GBS-infected hVECs compared with expression in mock-treated controls 24 hours after infection (n = 3; **P < 0.005 and ****P < 0.00005, by 1-way ANOVA followed by Sidak’s multiple comparisons test; data represent the mean ± SEM). (B) Localization of β-catenin (white) in GBS-infected hVECs compared with mock-treated controls 24 hours after infection (nuclei are stained with DAPI [blue]; overlap is shown in yellow). Images are from 1 of 3 experiments. Original magnification, ×100. (C) Expression of β-catenin target genes in murine vaginal tissues 96 hours after vaginal inoculation with WT GBS compared with expression in control PBS–treated tissues (n = 4/group; *P < 0.05, ***P < 0.0005, ****P < 0.00005, and P = 0.1, by 1-way ANOVA followed by Sidak’s multiple comparisons test; data represent the mean ± SEM). (D) c-Myc immunostaining in murine vaginal tracts 96 hours after vaginal inoculation with PBS or WT GBS. Images are from 1 of 3 experiments. Scale bars: 100 μm. (E) Quantification of c-Myc immunostaining in murine vaginal tracts 96 hours after vaginal inoculation with WT GBS or control PBS (6 mice/group; *P < 0.05, by 2-sided, unpaired t test; data represent the mean). (F) Western blot (WB) for p-GSK3β in GBS-infected hVECs compared with mock-treated controls, 0 and 4 hours after infection. GAPDH was used as a loading control. Blots are from 1 of 4 experiments. (G) Quantification of p-GSK3β band intensity. The band intensity was first normalized to GAPDH and then to t0 of the corresponding treatment (n = 4/group; *P < 0.05, **P < 0.005, and P = 0.98, by 1-way ANOVA followed by Sidak’s multiple comparisons test; data represent the mean ± SEM). (H) hVECs were left untreated or were treated for 16 or 24 hours with the β-catenin signaling inhibitor FH535 (15 μM) prior to WT GBS infection, and cell detachment was measured. Data were normalized to the uninfected controls (n = 3; *P < 0.05 and ***P < 0.0005, by ANOVA followed by Sidak’s multiple comparisons test; data represent the mean ± SEM). (I and J) Flow cytometric analysis of surface E-cadherin (I) and N-cadherin (J) on mock-treated or WT GBS–infected hVECs, with or without FH535 pretreatment (n = 3; *P < 0.05, **P < 0.005, and P = 0.06; data represent the mean ± SEM by ANOVA followed by Sidak’s multiple comparisons test.
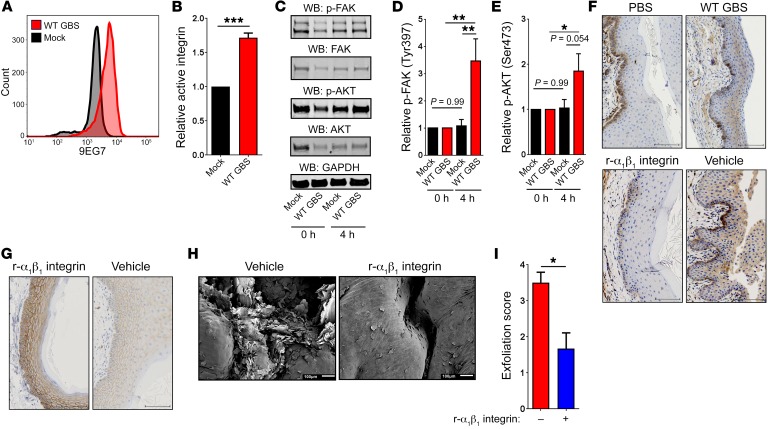
We next sought to identify the extracellular signal that GBS uses to stimulate β-catenin signaling. To this end, we explored the integrin signaling pathway. Integrins are a widely expressed family of proteins that control various cellular processes, ranging from proliferation to migration to organogenesis (36). Activation of integrins through clustering at the cell surface stimulates a signaling cascade that begins with focal adhesion kinase (FAK), which is autophosphorylated at Tyr397 (37). FAK activates protein kinase B (also known as AKT) by phosphorylation at Ser473 (38), which proceeds to phosphorylate GSK3β at Ser9, leading to its deactivation (Figure 4, F and G). To test whether integrin signaling is induced by GBS in vaginal epithelial cells, we used the 9EG7 β1 integrin antibody, which specifically recognizes the active conformation of β1 integrin (39). Interestingly, GBS-infected vaginal epithelial cells had significantly more active integrin than did mock-infected cells (Figure 5, A and B). Also, GBS infection induced an increase in phosphorylation of FAK and AKT (Figure 5, C–E). We did not observe this trend of increased phosphorylation of FAK and AKT in GBS-treated cells in the mock-infected controls (Figure 5, C–E). Furthermore, vaginal tissues from GBS-inoculated animals had substantially higher levels of active integrin at the epithelial layer near the vaginal lumen in contrast to control tissues, in which active integrin staining was localized to the basement membrane (Figure 5F). These findings are similar to those observed in carcinoma biology, in which active integrins are normally expressed at the basement membrane of healthy stratified squamous epithelia; in contrast, expression above the basement membrane is typical in cancerous tissues or tissues undergoing EMT (40). To fully define the mechanistic connection between integrin signaling activation and epithelial exfoliation, we used recombinant α1β1 integrin as a competitive inhibitor of GBS integrin activation. To this end, recombinant murine α1β1 integrin was administered intravaginally 24 hours prior to vaginal GBS inoculation, in conjunction with GBS inoculation, and then every 24 hours following inoculation. Intravaginal administration of α1β1 integrin during GBS colonization reduced active integrin staining, whereas treatment with a vehicle control had no effect (Figure 5F). To mechanistically link integrin signaling and EMT, vaginal tissues from GBS-inoculated animals administered intravaginal α1β1 integrin or vehicle control were stained for E-cadherin. Animals treated with α1β1 integrin showed high levels of E-cadherin, whereas the vehicle controls did not (Figure 5G), indicating that the blockade of integrin signaling also prevents EMT progression. Next, we observed that intravaginal administration of α1β1 integrin significantly diminished epithelial exfoliation (Figure 5, H and I). Together, these data indicate that GBS are able to stimulate integrin signaling, which permits nuclear translocation of β-catenin, potentially leading to EMT, barrier disruption, and vaginal epithelial exfoliation.
Figure 5. GBS activates integrin signaling.
(A) Flow cytometric plot of active β1 integrin (9EG7 antibody) on the surface of GBS-infected hVECs compared with mock-infected controls after 24 hours. Data are from 1 of 3 experiments. (B) Colorimetric assay of active integrin on the surface of mock-infected or WT GBS–infected hVECs (n = 3/group; ***P < 0.0005, by 2-sided, unpaired t test; data represent the mean ± SEM). (C) Western blots for p-FAK and p-AKT in GBS-infected hVECs compared with mock-treated controls, 0 and 4 hours after infection. GAPDH was used as a loading control. Blots are from 1 of 4 experiments. (D and E) Band intensity quantification for p-FAK (D) and p-AKT (E). Band intensity was first normalized to GAPDH as a loading control and then to t0 of the corresponding treatment (n = 4/group; *P < 0.05 and **P < 0.005, by 1-way ANOVA followed by Sidak’s multiple comparisons test; data represent the mean ± SEM). (F) Active β1 integrin immunostaining in murine vaginal tracts 96 hours after vaginal inoculation with PBS, WT GBS, WT GBS with recombinant murine α1β1 integrin, or WT GBS with vehicle control. Images are from 1 of 3 experiments. Scale bars: 100 μm. (G) E-cadherin immunostaining in murine vaginal tracts 96 hours after vaginal inoculation of GBS with recombinant murine α1β1 integrin or vehicle control. Images are from 1 of 3 experiments. Scale bars: 100 μm. (H) Vaginal epithelial exfoliation in mice 96 hours after vaginal inoculation with WT GBS and treatment with vehicle control or recombinant murine α1β1 integrin. Scale bars: 100 μm. (I) Blinded quantification of exfoliated vaginal epithelial cells 96 hours after inoculation (n = 3 tissues/group; *P < 0.05, by 2-sided, unpaired t test; data represent the mean ± SEM).
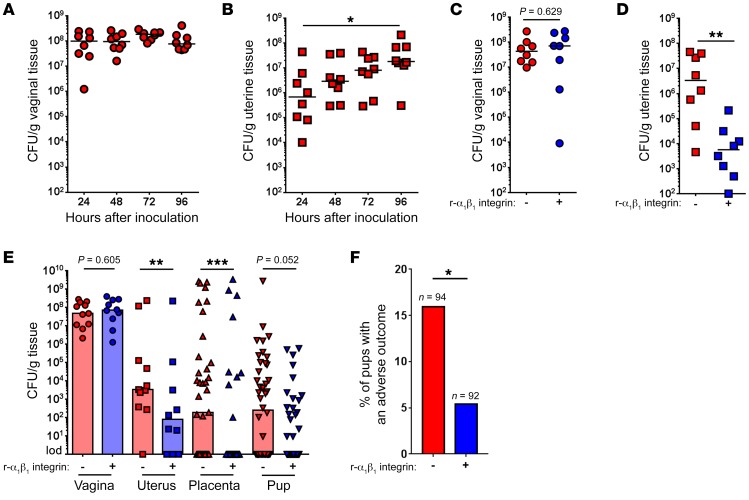
Next, we attempted to determine the role of vaginal epithelial exfoliation in GBS pathogenesis. Heavy vaginal colonization with GBS is the primary risk factor for GBS-associated adverse pregnancy outcomes (9, 10, 41). Prior studies with E. coli and N. gonorrhoeae have demonstrated a role for epithelial exfoliation in decreasing bacterial colonization of the urinary tract (16, 17) and vagina (13, 15). Given that GBS stimulate epithelial exfoliation and are associated with exfoliating cells (Figure 2C), we hypothesized that this response would diminish GBS vaginal colonization and reduce adverse pregnancy outcomes. Despite GBS being present on exfoliating vaginal epithelial cells, we found that epithelial exfoliation did not correlate with significantly decreased bacterial burden in the vagina (Figure 6A). Rather, we observed a significant increase in GBS CFU in the uterus (Figure 6B), suggesting a potential association between increased epithelial exfoliation and GBS ascending infection. Notably, GBS strains with similar levels of epithelial exfoliation (Supplemental Figure 1, A, B, and E) had similar bacterial loads in the vagina and uterus (Supplemental Figure 1, C, D, F, and G), whereas iagA-deficient GBS strains that displayed reduced exfoliation (Supplemental Figure 1, A and H) had reduced bacterial loads in the uterus (Supplemental Figure 1J) but not in the vagina (Supplemental Figure 1I). Also, TLR2-KO, MyD88-KO, and caspase 1–KO mice, which showed levels of exfoliation similar to those seen in WT mice (Supplemental Figure 2, A, B, and E), showed no significant differences in uterine bacterial CFU (Supplemental Figure 2, D and G; vaginal loads are shown in Supplemental Figure 2, C and F). To determine the statistical relationship between epithelial exfoliation and uterine bacterial load, we performed linear regression analysis on bacterial loads and exfoliation scores for all WT GBS–infected animals. Indeed, we found a positive correlation between uterine bacterial loads and the exfoliation score (Supplemental Figure 8), indicating that epithelial exfoliation and GBS ascending infection may be mechanistically linked.
Figure 6. Integrin-mediated epithelial exfoliation promotes ascending GBS infection.
(A) Bacterial burden in the vagina (n = 8; data represent the median). (B) Bacterial burden in the uterus (n = 8; *P < 0.05, by ANOVA followed by Sidak’s multiple comparisons test; data represent the median). (C) Bacterial burden in vaginal tissue from mice that were intravaginally treated with recombinant murine α1β1 integrin or control vehicle 96 hours after vaginal inoculation with WT GBS (n = 7–8; P = 0.629, by Mann-Whitney U test; data represent the median). (D) Bacterial burden in uterine tissue from mice that were intravaginally treated with recombinant murine α1β1 integrin or control vehicle 96 hours after vaginal inoculation with WT GBS (n = 8; **P < 0.05, by Mann-Whitney U test; data represent the median). (E) Pregnant female mice were vaginally inoculated with approximately 108 CFU of WT GBS and intravaginally treated with recombinant murine α1β1 integrin or vehicle control. Seventy-two hours after inoculation or at the first sign of preterm birth (vaginal bleeding and/or pups in cage), mice were euthanized, and the bacterial burden in vaginal tissue, uterine tissue, placental tissue, or fetal tissue was enumerated (n = 10–11; **P < 0.005, ***P < 0.0005, P = 0.605, and P = 0.052, by Mann-Whitney U test; data represent the median). (F) Pups with adverse birth outcomes (either in utero fetal demise or premature birth) from pregnant female mice that were vaginally inoculated with WT GBS and either intravaginally treated with recombinant murine α1β1 integrin (5 of 92 pups) or not (15 of 92 pups). Data represent the percentage of pups that had an adverse birth outcome (*P < 0.05, by 2-sided Fisher’s exact test. Bars represent % of pups with an adverse outcome out of total pups (indicated by n).
To determine whether bacteria in the uterus primarily represented those ascending from the vagina or those arising by replication of the initially ascended bacteria, we vaginally inoculated mice with WT GBS expressing a plasmid-encoded GFP (42). Retention of this plasmid requires the presence of erythromycin, and in the absence of antibiotic pressure, daughter cells do not inherit the plasmid (Supplemental Figure 9A), and GFP+ (mother) and GFP– (daughter) cell populations are generated (Supplemental Figure 9B). We confirmed GFP fluorescence under microaerobic and anaerobic conditions that represented genitourinary oxygen levels (43) (Supplemental Figure 9C). These data indicate that any potential differences in GFP expression observed in vivo are not solely due to differences in environmental conditions. Flow cytometric analysis of GFP+ GBS in vaginal and uterine tissues (Supplemental Figure 9, D and E) revealed a modest decrease in GFP+ GBS in the vagina (Supplemental Figure 9F) over a 96-hour period; in contrast, we observed a significant increase in GFP+ GBS in the uterus (Supplemental Figure 9G). As the plasmid (GFP signal) is lost during replication in the absence of antibiotic selection pressure (Supplemental Figure 9, A and B), it is plausible that any GFP+ GBS in the uterus originated in the vagina rather than through bacterial replication in the uterus. Although the growth dynamics that occur in vivo are difficult to recapitulate in vitro, our results suggest that GBS in the uterus probably ascended from the vagina and indicate an association between epithelial exfoliation and GBS trafficking.
Finally, we hypothesized that abrogation of epithelial exfoliation by blocking integrin signaling may lead to decreased ascending infection. To this end, recombinant murine α1β1 integrin was administered intravaginally before and after GBS infection (see Methods for details). As expected, we noted that intravaginal administration of recombinant murine α1β1 integrin significantly diminished ascending infection without affecting vaginal colonization (Figure 6, C and D). Given the importance of ascending GBS infection for preterm birth (7) and to further verify the effects of decreased GBS-mediated integrin signaling and vaginal epithelial exfoliation, we used a pregnant model of GBS ascending infection (44). Like the observations in nonpregnant mice (Figure 6, C and D), administration of recombinant murine α1β1 integrin diminished GBS ascension, leading to decreased CFU in the uterus, placenta, and fetal tissues (Figure 6E; see Supplemental Figure 10 for a diagram of the murine female reproductive tract). Finally, the number of pups affected by an adverse birth outcome (premature birth or death utero) was significantly reduced for mice that were administered recombinant murine α1β1 integrin (Figure 6F). We also explored the role of iagA in ascending infection, as this GBS strain showed a diminished ability to induce exfoliation (Supplemental Figure 1, A and H) as well as reduced ascending infection (Supplemental Figure 1J). Although GBS deficient for iagA (GBSΔiagA) are less able to invade human brain microvascular endothelial cells and are less virulent than WT GBS (25), we confirmed that both WT GBS and GBSΔiagA invaded vaginal epithelial cells to a similar degree both in vitro (Supplemental Figure 11A) and in vivo (Supplemental Figure 11, B and C). Interestingly, hVECs infected with GBSΔiagA showed less active β1 integrin (Supplemental Figure 12A), had reduced β-catenin signaling (Supplemental Figure 12B), and did not exhibit EMT (Supplemental Figure 12, C and D), or displayed loss of barrier function (Supplemental Figure 12E). Finally, pregnant mice inoculated with GBSΔiagA had substantially fewer CFU in the uterus, placenta, and fetal tissues (Supplemental Figure 12F) and fewer adverse birth outcomes (premature birth or death in utero) (Supplemental Figure 12G), similar to what was observed in mice administered recombinant murine α1β1 integrin (Figure 6, E and F). Together, these data are the first to our knowledge to indicate that GBS stimulate epithelial exfoliation via integrin signaling and EMT and, as a result, are able to cause ascending infection and adverse pregnancy outcomes.
Discussion
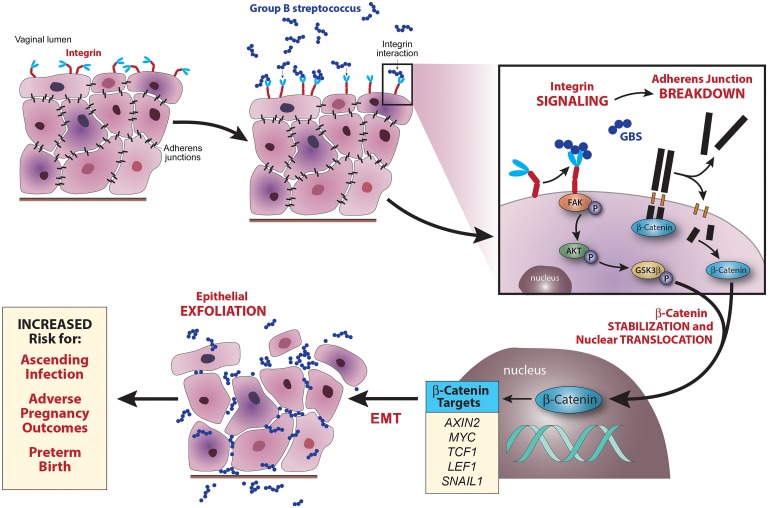
The research presented here identifies a mechanism of ascending bacterial infection. We show that GBS induces epithelial exfoliation, which is associated with bacterial ascension from a commensal to an invasive niche. Epithelial exfoliation is driven by a loss of barrier function and cellular detachment through EMT, which is stimulated by integrin signaling and controlled by β-catenin signaling. Finally, we observed that interruption of these signaling processes decreases epithelial exfoliation, ascending infection, and adverse pregnancy outcomes (Figure 7). This work suggests a shift in the current paradigm of vaginal epithelial exfoliation in response to pathogen colonization. Previous studies on pathogenic Neisseria have revealed a role for exfoliation in the prevention of bacterial colonization (13, 15), as have some studies on uropathogenic E. coli (14, 16, 17), whereas, we have found the opposite to be true for GBS. While some studies have shown beneficial effects of E. coli–induced bladder epithelial exfoliation for urinary tract infections (45–47), we believe ours is the first to show that vaginal epithelial exfoliation can confer a benefit to some invading pathogens that lead to adverse birth outcomes.
Figure 7. Model of GBS-induced epithelial exfoliation and ascending infection.
Upon GBS colonization, integrins are either directly or indirectly activated by GBS. Integrin activation induces phosphorylation of FAK, which in turn phosphorylates AKT, which phosphorylates GSK3β. As adherens junctions break down, β-catenin is released into the cytoplasm. In its dephosphorylated form, GSK3β marks β-catenin for degradation, thus preventing β-catenin stabilization, nuclear translocation, and signaling; however, when it is phosphorylated, it cannot mark β-catenin for degradation, leading to β-catenin stabilization and nuclear translocation. Once in the nucleus, β-catenin stimulates the expression of a variety of genes, including those that drive EMT and epithelial exfoliation. Rather than eliminating colonized GBS, epithelial exfoliation permits bacterial dissemination through the loss of barrier function. This leads to increased ascending infection, which increases the rates of adverse pregnancy outcomes and preterm birth.
One potential explanation for the difference between GBS and other bacteria that prevent vaginal epithelial exfoliation lies at the host-pathogen interface. Both N. gonorrhoeae and uropathogenic E. coli express proteins that target a family of host epithelial cell glycoproteins known as CEACAMs, and for these bacteria, CEACAM engagement prevents vaginal epithelial exfoliation (15, 17). We have not found CEACAM-binding proteins in GBS, indicating differences in the interactions between vaginal microbes and host epithelial cells. These differences in host-microbe interactions explain the functional consequences for epithelial exfoliation. It is intriguing that GBS and bacteria that prevent epithelial exfoliation such as N. gonorrhoeae and E. coli all target integrin signaling (13, 17). It is logical that pathogens would target these proteins as a means of controlling cellular behaviors and circumventing host defenses and that different pathogens would subvert integrin signaling in different ways for their benefit. CEACAM binding leads to integrin activation and increased epithelial adhesion (13, 17), whereas integrin activation without CEACAM binding leads to decreased epithelial adhesion. This suggests that activation of integrin signaling is a conserved strategy for pathogen interaction with vaginal epithelial cells, but that the host target is an important determinant of cell behavior. Although N. gonorrhoeae and E. coli target integrin signaling to prevent exfoliation (13, 17), the downstream signaling components of this process are currently unknown and may explain the divergent strategies used by these bacteria and GBS. Modification of integrin signaling may be a common mechanism for pathogenic manipulation of host cells, as it has been shown to play a role in viral infection (48), fungal infection (49), Gram-negative bacterial infection (13, 17, 50), and now, Gram-positive bacterial infection. Interestingly, N. gonorrhoeae also targets α1β1 integrins during urethral mucosa infection (51). The ubiquitous occurrence of integrin signaling during host-pathogen interactions, as well as the increasingly limited availability of antibiotics, makes integrin signaling an attractive target for therapeutic intervention and thus warrants further research.
GBS induction of epithelial exfoliation and loss of barrier function are a result of integrin activation and induction of β-catenin signaling and EMT. While the present study identifies the role of EMT, β-catenin signaling, and integrin activation in response to GBS, their role during infection by other bacterial pathogens still remains poorly understood. Bacterial induction of EMT is largely thought to be TLR mediated (52) rather than integrin mediated; however, we have found EMT to be TLR2 independent for GBS. Recent studies have focused on the induction of EMT by bacterial pathogens that are either associated with or causal agents of cancer (53–55); however, EMT induction by bacterial pathogens not associated with cancer has received little attention. β-Catenin is a master regulator of many different processes, including organogenesis, cell adhesion, and cancer development (56). As with EMT, the role of β-catenin in bacterial infection is not known. Interestingly, direct and indirect inhibition of the Wnt/β-catenin signaling pathway is an active area of research in cancer progression and metastasis (57–59). Insights from this field may reveal novel mechanisms that are applicable to bacterial disruption of the epithelial barrier and useful for the development of therapeutic compounds that will prevent bacterial epithelial colonization and dissemination. Given the importance of EMT and β-catenin signaling in maintaining the vaginal epithelial barrier, which is the primary defense against many pathogens, further insight is necessary to fully understand how bacterial pathogens colonize and disrupt epithelial surfaces.
Additionally, the mechanisms by which bacteria ascend from the vagina to the uterus remain a black box (7). This is especially true for bacteria that cause ascending infection despite being nonmotile organisms (e.g., GBS). Previous studies have suggested that factors not specific to the pathogen, such as viral coinfection (60) or changes in microbial ecology (61), are at the root of ascending infection. While these factors may play a role, they do not solely explain why ascending infection occurs in some cases and not in others. Previously, we have identified multiple bacterial virulence factors that promote ascending infection and adverse pregnancy outcomes (22, 44), and in this study, we have identified a host mechanism that inadvertently promotes ascending GBS infection. GBS strains that are more proficient in their ability to induce integrin signaling, EMT, and epithelial exfoliation are better at establishing ascending infection. In conjunction with host factors, there may be a subset of bacteria that are more likely to establish an ascending infection in some pregnant women. Identifying both the pathogen and host factors that predispose certain individuals to ascending infection and preterm birth is critical for the development of effective interventions and merits further study.
Despite our findings, the present study has some limitations. First, our findings stem from experiments performed with immortalized cells and animal models. As performing these studies in humans is not possible, these models are extensively used to provide insight into mechanisms of disease processes (13, 15, 17). However, there are significant biological differences between humans and animal and cell culture models. To fully confirm the importance of EMT and epithelial exfoliation during ascending bacterial infection, these processes should be explored in humans. Second, we were unable to attribute vaginal epithelial exfoliation to a single microbial factor, as we aimed to understand how the host vaginal epithelium responds to GBS colonization. In this study, GBS lacking iagA led to reduced epithelial exfoliation in ascending infection; however, IagA is a cytoplasmic glycosyltransferase (25) that probably does not directly interact with β1 integrin. It is possible that multiple GBS factors are directly or indirectly involved in these processes, and this merits further exploration. The identification of a microbial factor responsible for integrin activation warrants further investigation, as the virulence factor that controls GBS induction of integrin signaling and EMT may be an attractive vaccine target for the prevention of GBS-associated preterm birth. It is also possible that multiple GBS factors are directly or indirectly involved in these processes. Finally, this study shows a correlation between a cellular process occurring in the vagina (epithelial exfoliation) and a phenotypic result of this process (ascending bacterial infection), but does not comprehensively identify the mechanism of bacterial trafficking. We speculate that (a) the loss of epithelial barrier function may permit pathogen entry into the microvasculature and dissemination into uterine tissues; (b) increases in cellular mobility may inadvertently enable bacterial trafficking to new locations; and/or (c) changes in the cellular architecture may prevent the clearance of disseminated bacteria and thereby permit persistence, resulting in increased bacterial trafficking. While these findings represent a significant advancement in our understanding of ascending infection, they are not exhaustive. It is our expectation that more nuanced mechanisms of ascending bacterial infection will be revealed as the importance of this field is fully realized.
The importance of preterm birth and stillbirth as leading causes of neonatal morbidity and mortality cannot be overstated. Premature neonates are at higher risk for many severe disorders and death (62), and both stillbirth and preterm birth can have severe health consequences for the mother (7, 63). Unfortunately, our knowledge of the causes of preterm birth and stillbirth is incomplete, and available interventions are therefore limited. A significant portion of this disease burden is attributable to ascending bacterial infection (7). For successful infection, pathogen-specific factors must be involved in circumventing host defenses. Multiple bacterial virulence factors that promote ascending infection and adverse pregnancy outcomes have been previously identified (22, 23, 44), and in this study, we have suggested a host mechanism that inadvertently promotes entry of GBS into the uterus. To fully confirm the importance of EMT and vaginal epithelial exfoliation during infections occurring in pregnancy, clinical studies should be designed to explore these processes in humans, with the ultimate aim of developing interventions that reduce the global burden of preterm birth.
Methods
Materials, bacterial strains, and cell lines
All chemicals were purchased from Sigma-Aldrich, unless stated otherwise. WT GBS COH1 is a capsular serotype III, hypervirulent ST-17 clone obtained from an infected newborn (64) and is referred to herein as WT GBS. The following other WT GBS strains were also used: A909 (capsular serotype Ia), NEM316 (capsular serotype III), and NCTC10/84 (capsular serotype V). The ΔcylE, ΔcpsK, and ΔiagA mutants were derived from COH1, and the ΔpilB mutant was derived from NCTC10/84 (25, 65). GBS were grown in tryptic soy broth or agar (TSB/TSA; Difco Laboratories) at 30°C or 37°C in 5% CO2. Erythromycin (5 μg/ml) was used to retain the plasmid expressing GFP. L. crispatus was a gift of Michael Fischbach (UCSF, San Francisco, California, USA) and was cultured in De Man, Rogosa, and Sharpe (MRS) broth at 37°C under anaerobic conditions. hVECs were obtained from the American Type Culture Collection (ATCC) (CRL-2616). Cells were maintained in keratinocyte serum-free medium (KSFM) (Invitrogen, Thermo Fisher Scientific), supplemented with 65 μg/ml bovine pituitary extract (Invitrogen, Thermo Fisher Scientific), 67.419 pg/ml human recombinant EGF (Invitrogen, Thermo Fisher Scientific), and 50–100 IU/ml penicillin and 50–100 μg/ml streptomycin (Corning). Cells were split every 3 to 4 days and passaged at a 1:10 dilution. All assays were performed at passages 14 through 30, and prior to infection, antibiotic-containing media were aspirated, cells were washed with sterile PBS, and media were replaced with antibiotic-free supplemented KSFM. Cells were determined to be mycoplasma free using the Universal Mycoplasma Detection Kit (ATCC).
Cell detachment assay
hVEC monolayers were infected with GBS at an MOI of approximately 1.0. At 0, 16, or 24 hours, 100% crystal violet was added to each well to a concentration of 10% and incubated at 37° C for 30 minutes. Excess crystal violet and loosely adherent cells were removed by centrifuge at 1,000 g for 10 minutes. Following centrifugation, the remaining crystal violet was measured with a Spectramax i3x Plate Reader (Molecular Devices) at 585 nm. For FH535 experiments, 1 hour before infection, hVEC culture media were replaced with FH535-containing media (1.5 μl of 50 mM FH535 in DMSO in 5 ml KFSM; Tocris Biosciences). Following preincubation, GBS infection and cell detachment assay were performed as described above. Data are represented as the percentage of cell detachment, which is calculated as follows: (OD585 mock-infected cells – OD585 GBS-infected cells)/(OD585 mock-infected cells).
Scanning electron microscopy
Vaginal GBS colonization was performed as described below. Excised vaginal tissues were split vertically to expose the entire vaginal lumen. Tissues were pinned onto 3.5% agarose pads, immediately fixed with approximately 3 ml half-strength Karnovsky’s fixative for 5 to 10 minutes, transferred to approximately 10 ml fixative, and stored at 4°C until imaging was performed. For hVEC imaging, hVEC monolayers on glass coverslips were infected with GBS at an MOI of approximately 1.0. At 24 hours, media were replaced with 1 ml of half-strength Karnovsky’s fixative and stored at 4°C until imaging was performed. Samples were prepared for scanning electron microscopy by the Electron Microscopy Core of the Fred Hutch Cancer Research Center. Fixed tissues were washed in 0.1 M cacodylate buffer and dehydrated by an ethanol wash (50%, 70%, 95%, 2 × 100%; 30 min each). Samples were dried using a Critical Point Dryer (Tousimis), mounted, and sputter coated with Au/Pd. Images were captured using a JEOL 5800 Scanning Electron Microscope equipped with a JEOL Orion Digital Acquisition System. For quantification, at least 3 images of equal surface area from at least 2 tissues were deidentified, and individual exfoliated cells were counted in a blinded manner. An exfoliation score was assigned to each image on the basis of the number of exfoliated cells per image (1: ≥0 and <100, 1.5: ≥100 and <200, 2: ≥200 and <300, 2.5: ≥300 and <400, 3: ≥400 and <500, 3.5: ≥500 and <600, 4: ≥600 and <700, 4.5: ≥700 and <800, and 5: ≥800).
Barrier function analysis
ECIS was used to measure changes in hVEC barrier function in real time. hVEC monolayers were grown to confluence on polyethylene 8W10E+ arrays (Applied BioPhysics). Cells were infected with approximately 105 CFU GBS or L. crispatus and monitored for changes in resistance at 1,000 Hz using an ECIS Zθ Instrument (Applied BioPhysics). Data were normalized to resistance values at the point of infection and subtracted from the mock-infected resistance values. For permeability assays, hVECs were grown to confluence on tissue culture–treated polycarbonate 0.4-μm or 8.0-μm Transwells (Corning). For fluorescein permeability assays, cells on 0.4-μm Transwells were infected with 5 × 105 CFU GBS or mock controls for 24 hours. One hour before analysis, an untreated well was treated with 0.1% (v/v) Triton X-100, which served as a positive control. After 24 hours, the media from both the apical and basal compartments were replaced with either HBSS (Corning) alone (basal) or HBSS containing 50 μg/ml FITC (apical). After 1 hour, 300 μl media from each basal compartment was removed, and fluorescence was measured in triplicate with a Spectramax i3x Plate Reader with excitation at 485 nm and emission at 535 nm. For bacterial penetration assays, cells on 8.0-μm Transwells were infected with 5 × 105 CFU GBS or mock controls for 24 hours. At specified times, 100 μl basal compartment contents was removed, stained with 50 μM SYTO 9 Dye (Molecular Probes) for 15 minutes at room temperature (RT), and fluorescence was measured with a Spectramax i3x Plate Reader with excitation at 485 nm and emission at 535 nm.
Murine model of GBS vaginal colonization and ascending infection
To determine sample size, power calculations (α level of 0.05) were performed to detect a predetermined effect size for adverse birth outcomes or changes in bacterial load in genital tissue to obtain 80% power.
Vaginal colonization.
Six- to eight-week-old female C57BL/6J, B6.129-Tlr2tm1Kir/J (TLR2-KO), B6.129P2(SJL)-Myd88tm1.1Defr/J (MyD88-KO), and B6N.129S2-Casp1tm1Flv/J (caspase 1–KO) mice obtained from The Jackson Laboratory were used for colonization studies at eight to sixteen weeks as described previously (66). One day before GBS colonization, mice were i.p. injected with 500 μg 17β-estradiol in 100 μl sterile canola oil as described previously (67) and randomly assigned to an experimental group. No blinding was performed during group assignment. Mice were anesthetized using 4% isoflurane, and 10 μl (~108 CFU) GBS was administered into the vaginal tract using a micropipette. Mice were left inverted for 5 minutes under anesthesia and then returned to their cages and monitored until ambulatory. After inoculation, mice were monitored daily for signs of distress and euthanized at specified times, and vaginal and uterine tissues were excised for further analysis. For inhibitor studies, 300 ng recombinant murine α1β1 integrin (R&D Systems) resuspended in 10 μl PBS or PBS alone was administered 1 day prior to GBS inoculation, 2 hours prior to inoculation, and every 24 hours following inoculation. To obtain lavage fluids, mice were euthanized, and the vaginal lumen of each mouse was lavaged twice with 20 μl sterile PBS. For fluorescent nanoparticle experiments, following euthanasia, fluorescent nanoparticles (10 μl in PBS) were administered intravaginally by micropipette, and then vaginal tracts were excised, embedded and frozen in OCT (Tissue-Tek), and stored at –80° C until sectioning.
Ascending infection.
Six-to-eight week old female C57BL/6J mice were mated, randomly assigned to an experimental group, and inoculated with GBS as described previously (44). No blinding was performed for the group assignments. GBS was inoculated as described above, and mice were monitored twice a day for signs of preterm labor (vaginal bleeding and/or pups in the cage). Seventy-two hours after infection, or earlier if preterm labor was observed, mice were euthanized, and a midline laparotomy was performed to identify fetal injury or loss of pregnancy. For CFU enumeration, maternal (vagina, uterus), placenta (2 proximal, 2 distal), and fetal tissues (2 proximal, 2 distal pups; see Supplemental Figure 10 for the diagram) were excised, homogenized in 1 ml PBS, serially diluted, and plated on TSA and confirmed as GBS using Granada media (Hardy Diagnostics) or CHROMAgar StrepB (CHROMagar). For inhibitor studies, either 300 ng recombinant murine α1β1 integrin dissolved in 10 μl PBS or PBS alone was administered as described above. Pregnant mice infected with WT GBS that were corresponding controls for ΔiagA (Supplemental Figure 12) were described in a previous publication (44). All other mouse work was conducted as part of this study and was not previously reported.
Histology
Vaginal GBS colonization was performed as described above. At 24 or 96 hours, mice were euthanized, and vaginal tracts were excised, mounted onto cardboard, and immediately fixed in formalin overnight at 4°C or embedded in OCT. Following fixation, formalin was replaced with 70% ethanol and stored at 4°C. Tissues were sectioned, baked, and deparaffinized for 30 minutes at 60°C. Antigens were revived in EDTA buffer (pH 9.0; Leica Bond Epitope Retrieval Solution 2) at 100°C for 20 minutes, blocked for 20 minutes in 10% normal goat serum at RT, and stained with one of the following primary antibodies: anti–E-cadherin (1:400; clone 24E10; Cell Signaling Technology); anti–c-Myc (1:2,500; clone D84C12; Cell Signaling Technology); anti–β1 integrin (1:250; clone 9EG7; BD Biosciences); or anti–rabbit IgG (1:1,000) for 30 minutes at RT. Subsequently, slides were treated with Bond Polymer Refine DAB Reagent (Leica) for 8 minutes at RT, blocked with peroxide block (Leica) for 5 minutes at RT, treated twice with Bond Mixed Refine DAB Detection Reagent for 10 minutes at RT, and counterstained with hematoxylin (Leica) for 4 minutes. For OCT-embedded tissues, slides were stained with 2 μg/ml DAPI (Life Technologies, Thermo Fisher Scientific) for 10 minutes, washed with PBS, and stored in SlowFade Gold Anti-fade Reagent (Life Technologies, Thermo Fisher Scientific) until imaging. Following antigen revival, slides were blocked with 5% BSA (Sigma-Aldrich) in PBS for 1 hour at RT and then stained with anti-GBS antibody (1:1,000, generated in-house; ref. 68) in 5% BSA in PBS for 2 hours at RT. Slides were washed in PBS plus 0.025% Tween-80 (PBST) (Thermo Fisher Scientific) and then stained with Cy3-conjugated secondary antibody (1:5,000; affinity-purified polyclonal, catalog ab6939; Abcam) and 2 μg/ml DAPI in PBS for 30 minutes at RT. Slides were washed in PBST and PBS and then stored in SlowFade Gold Anti-fade Reagent until imaging. Images were collected with a Leica DMI6000B inverted microscope equipped with a Leica DFC310FX camera (Leica Application Suite, version 4.0.0, was used for image acquisition) or a Keyence BZ-X710 fluorescence microscope. Images were quantified using ImageJ, version 1.6.0_24 (NIH).
Flow cytometric analysis
hVEC monolayers were infected with GBS at an MOI of approximately 1.0. After 24 hours, K was replaced with 1 ml FACS buffer (1 mM EDTA, 25 mM HEPES, and 0.1% BSA [w/v] in PBS). Cells were collected by cell scraper, pelleted at 300 g for 5 minutes, washed in FACS buffer, resuspended in FACS buffer containing human Fc block (1:200; catalog 564219; BD Biosciences), and incubated at RT for 15 minutes. Approximately 106 cells were then pelleted, washed in FACS buffer, and stained with either PE-conjugated anti–E-cadherin (1:20; clone 36; BD Biosciences) or Alexa Fluor 488–conjugated anti–N-cadherin (1:20; catalog FAB6426G; R&D Systems) for 30 minutes at RT, and then washed and resuspended in FACS buffer. For FH535 experiments, 1 hour prior to infection, culture media were replaced with FH535-containing media (1.5 μl of 50 mM FH535 in DMSO in 5 ml KFSM). Following preincubation with FH535, GBS infection and flow cytometry were performed as described above. For β1 integrin staining, cells were prepared as above and stained with anti–β1 integrin primary antibody (1:200; clone 9EG7; BD Biosciences) for 1 hour at 4°C. Cells were then pelleted, washed in FACS buffer, and stained with Cy3-conjugated secondary antibody (1:200; affinity-purified polyclonal, catalog ab6953; Abcam). For flow cytometric analysis of isolated murine epithelial cells, cells were stained with PE-conjugated anti–E-cadherin (1:200; clone DECMA-1; BioLegend) and APC-conjugated anti-CD326 (1:100; clone G8.8; BioLegend) as described above. PE- and APC-stained beads (BD Biosciences) were used for flow cytometric compensation. All data were collected using an LSR II Instrument (BD Biosciences). Surface marker expression was analyzed using FlowJo software, version 10 (TreeStar).
Quantitative reverse transcription PCR analysis
hVEC monolayers were infected with GBS at an MOI of approximately 1.0. After 24 hours, KFSM was aspirated, and total RNA was isolated using the RNeasy Miniprep Kit (QIAGEN) according to the manufacture’s protocol. cDNA was generated from whole RNA using the iScript cDNA Synthesis Kit (Bio-Rad). Equal amounts of cDNA were used to determine gene expression as previously described (69). The primers are listed in Supplemental Table 1.
Immunofluorescence
hVEC monolayers on glass coverslips were infected with GBS at an MOI of approximately 1.0. After 24 hours, media were replaced with 1 ml 10% formalin and fixed at 4°C overnight. Following fixation, cells were washed in PBS and permeabilized for 20 minutes in 1.0% Triton X-100. Coverslips were washed with PBS and then blocked for at least 1 hour in 1 ml SuperBlock PBS Blocking Buffer (Life Technologies, Thermo Fisher Scientific). Next, coverslips were incubated overnight with anti–β catenin antibody (1 μg/ml; catalog ab16051; Abcam), washed in PBS, and then incubated with Alexa Fluor 594–conjugated anti–rabbit IgG antibody (1:1,000; catalog A-11037; Invitrogen, Thermo Fisher Scientific) for 2 hours. Coverslips were washed in PBS and mounted in DAPI-containing VectaShield Anti-fade Mounting Media (Vector Laboratories) and imaged using a Leica DMI6000B inverted microscope equipped with a Leica DFC310FX camera. Leica Application Suite, version 4.0.0, was used for image acquisition.
Western blot analysis
All primary antibodies used for Western blotting were obtained from Cell Signaling Technology (1:1,000; anti–p-AKT, catalog 9271; anti–p-FAK, clone D20B1; anti–p-GSK3β, clone D3A4; anti-AKT, catalog 9272; anti-FAK, catalog 3285; anti-GSK3β, clone D5C5Z), except anti-GAPDH (1:40; catalog sc20357; Santa Cruz Biotechnology). hVEC monolayers were infected with GBS at an MOI of approximately 100 for 4 hours. Cells were lysed in 500 μl lysis buffer (150 mM NaCl, 15 mM Tris, 1 mM MgCl2, 1 mM CaCl2, 1% Triton X-100) with protease inhibitor plus phosphatase inhibitor (1 tablet per 5 ml; PhosphoSTOP; Roche). The cell lysate protein concentration was measured by Bradford assay. Equal amounts of protein were added to Laemmli buffer (6×), boiled at 95°C for 5 minutes, loaded onto precast 4%–20% acrylamide Mini-PROTEAN TGX Protein Gels (Bio-Rad), and run at 200 V for 40 minutes. Proteins were transferred onto PVDF membranes at 100 V for 75 minutes or at 30 V overnight. Membranes were blocked for at least 1 hour in 1:1 Odyssey Blocking Buffer (Li-Cor Biosciences) in TBS and then incubated with a primary antibody. Membranes were washed in TBS with 1.0% Tween-20 (Thermo Fisher Scientific) and incubated with Alexa Fluor 680–conjugated anti–rabbit IgG antibody (1:5,000; catalog A-21076; Invitrogen, Thermo Fisher Scientific) or Alexa Fluor 680–conjugated anti–goat IgG antibody (1:5,000; catalog A-21084; Invitrogen, Thermo Fisher Scientific). Membranes were washed in TBS with 1.0% Tween-20, rinsed several times in TBS, and visualized with the Odyssey Li-Cor Infrared Imager (Li-Cor Biosciences). Western blot images were quantified using ImageJ, version 1.6.0_24.
Integrin activity assay
For integrin activity assays, hVEC monolayers were infected with GBS at an MOI of approximately 100. After 1 hour, cells were scraped, counted, washed with PBS, centrifuged at 300 g for 5 minutes, and fixed with 4% paraformaldehyde. Cells (2.5 × 105) from each group were plated onto a 96-well plate, washed once with PBS, and blocked in 2% BSA-PBS for 30 minutes prior to staining with a primary antibody (1:1,000; clone 9EG7; BD Biosciences). After 1 hour, cells were washed with PBS and incubated for 20 minutes with a secondary reagent (1:20,000; recombinant protein A/G, peroxidase conjugated; Thermo Fisher Scientific). Cells were washed with PBS and developed with the TMB Microwell Peroxidase Substrate Kit (KPL). Samples were read at 620 nm before the reaction was stopped with 2 M H2SO4 and then read at 450 nm.
Statistics
All experimental replicates represent biological replicates. A 2-sided Student’s t test, 2-sided Mann-Whitney U test, 2-sided Fisher’s exact test, or ANOVA followed by Sidak’s multiple comparisons test was used to estimate differences as appropriate. A P value of less than 0.05 was considered statistically significant. These tests were performed using GraphPad Prism, version 5.0, for Windows, (GraphPad Software).
Study approval
All animal experiments were approved by the Seattle Children’s Research Institutional Animal Care and Use Committee (protocol 13907) and performed in strict accordance with the recommendations set forth in the NIH Guide for the Care and Use of Laboratory Animals (National Academies Press, 8th ed., 2011).
Author contributions
JV, BA, CG, SM, MC, CLM, EN, and LR designed the experiments. JV, BA, VSU, CG, SM, MC, PQ, EB, VA, LYN, and CW performed the experiments. CC prepared the nanoparticles. KSD provided reagents, and KMAW provided the model figures. JV, EN, KMAW, and LR analyzed the results and wrote the manuscript. All authors reviewed the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Bobbie Schneider, Sharmon Knecht, and Steve MacFarlane (all from Fred Hutchinson Cancer Research Center, Seattle, WA, USA) for their invaluable assistance with the scanning electron microscopy. We also thank Adam Lacy-Hulbert, Caroline Stefani (both from Benaroya Research Institute, Seattle, WA, USA), Barry Gumbiner, Melanie Gasper (both from Seattle Children’s Research Institute, Seattle, WA, USA), and Bill Carter (Fred Hutchinson Cancer Research Center, Seattle, WA, USA) for insightful discussions. We thank Jan Hamanishi (University of Washington, Seattle, WA, USA) for assistance with graphic design, members of the Seattle Children’s Research Institute Vivarium staff for their assistance, and Connie Hughes (Seattle Children’s Research Institute, Seattle, WA, USA) for administrative assistance. This work was supported by funding from NIH grants R01AI100989 and R01AI33976 (to LR and KMAW), and R01AI112619, R21AI109222, and R21 AI125907 (to LR). The NIH training grants T32 AI07509 (PI: Lee Ann Campbell) supported JV, and T32 HD007233 (PI: Lisa Frenkel) supported EB. EN was supported by a Burroughs Wellcome Fund Career Award and the University of Washington. KSD acknowledges grant R01NS051247. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Version 1. 04/09/2018
Electronic publication
Version 2. 05/01/2018
Print issue publication
Footnotes
JV’s present address is: University of Michigan, Ann Arbor, Michigan, USA.
EB’s present address is: Calvin College, Grand Rapids, Michigan, USA.
CW’s present address is: Pacific Northwest National Laboratory, Richland, Washington, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(5):1985–1999.https://doi.org/10.1172/JCI97043.
Contributor Information
Jay Vornhagen, Email: jayvorn@umich.edu.
Blair Armistead, Email: Blair.Armistead@seattlechildrens.org.
Claire Gendrin, Email: Claire.Gendrin@seattlechildrens.org.
Sean Merillat, Email: seanmerillat@gmail.com.
Michelle Coleman, Email: michelle.coleman@seattlechildrens.org.
Phoenicia Quach, Email: pjfquach@gmail.com.
Erica Boldenow, Email: erica.boldenow@gmail.com.
Varchita Alishetti, Email: Varchita@uw.edu.
Christina Leonhard-Melief, Email: christinam@miltenyibiotec.com.
Christopher Whidbey, Email: christopher.whidbey@pnnl.gov.
Chad Curtis, Email: ccurtis7@uw.edu.
Elizabeth Nance, Email: eanance@uw.edu.
Lakshmi Rajagopal, Email: lakshmi.rajagopal@seattlechildrens.org.
References
- 1.Lawn JE, Gravett MG, Nunes TM, Rubens CE, Stanton C, GAPPS Review Group Global report on preterm birth and stillbirth (1 of 7): definitions, description of the burden and opportunities to improve data. BMC Pregnancy Childbirth. 2010;10 Suppl 1:S1. doi: 10.1186/1471-2393-10-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu L, et al. Global, regional, and national causes of under-5 mortality in 2000-15: an updated systematic analysis with implications for the sustainable development goals. Lancet. 2016;388(10063):3027–3035. doi: 10.1016/S0140-6736(16)31593-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blencowe H, et al. Born too soon: the global epidemiology of 15 million preterm births. Reprod Health. 2013;10 Suppl 1:S2. doi: 10.1186/1742-4755-10-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lawn JE, et al. Stillbirths: rates, risk factors, and acceleration towards 2030. Lancet. 2016;387(10018):587–603. doi: 10.1016/S0140-6736(15)00837-5. [DOI] [PubMed] [Google Scholar]
- 5.Romero R, Gómez R, Chaiworapongsa T, Conoscenti G, Kim JC, Kim YM. The role of infection in preterm labour and delivery. Paediatr Perinat Epidemiol. 2001;15 Suppl 2:41–56. doi: 10.1046/j.1365-3016.2001.00007.x. [DOI] [PubMed] [Google Scholar]
- 6.Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. Lancet. 2008;371(9606):75–84. doi: 10.1016/S0140-6736(08)60074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romero R, Dey SK, Fisher SJ. Preterm labor: one syndrome, many causes. Science. 2014;345(6198):760–765. doi: 10.1126/science.1251816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seale AC, et al. Estimates of the burden of group B streptococcal disease worldwide for pregnant women, stillbirths, and children. Clin Infect Dis. 2017;65(suppl_2):S200–S219. doi: 10.1093/cid/cix664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen U, Nimrod C, Macdonald N, Toye B, Stephens D, Marchessault V. Relationship between antenatal group B streptococcal vaginal colonization and premature labour. Paediatr Child Health. 1999;4(7):465–469. doi: 10.1093/pch/4.7.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seale AC, et al. Maternal colonization with Streptococcus agalactiae and associated stillbirth and neonatal disease in coastal Kenya. Nat Microbiol. 2016;1(7):16067. doi: 10.1038/nmicrobiol.2016.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwatra G, et al. Prevalence of maternal colonisation with group B streptococcus: a systematic review and meta-analysis. Lancet Infect Dis. 2016;16(9):1076–1084. doi: 10.1016/S1473-3099(16)30055-X. [DOI] [PubMed] [Google Scholar]
- 12.Schleimer RP, Kato A, Kern R, Kuperman D, Avila PC. Epithelium: at the interface of innate and adaptive immune responses. J Allergy Clin Immunol. 2007;120(6):1279–1284. doi: 10.1016/j.jaci.2007.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muenzner P, Bachmann V, Zimmermann W, Hentschel J, Hauck CR. Human-restricted bacterial pathogens block shedding of epithelial cells by stimulating integrin activation. Science. 2010;329(5996):1197–1201. doi: 10.1126/science.1190892. [DOI] [PubMed] [Google Scholar]
- 14.Mulvey MA, et al. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science. 1998;282(5393):1494–1497. doi: 10.1126/science.282.5393.1494. [DOI] [PubMed] [Google Scholar]
- 15.Muenzner P, Rohde M, Kneitz S, Hauck CR. CEACAM engagement by human pathogens enhances cell adhesion and counteracts bacteria-induced detachment of epithelial cells. J Cell Biol. 2005;170(5):825–836. doi: 10.1083/jcb.200412151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagamatsu K, et al. Dysregulation of Escherichia coli α-hemolysin expression alters the course of acute and persistent urinary tract infection. Proc Natl Acad Sci U S A. 2015;112(8):E871–E880. doi: 10.1073/pnas.1500374112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muenzner P, et al. Uropathogenic E. coli exploit CEA to promote colonization of the urogenital tract mucosa. PLoS Pathog. 2016;12(5):e1005608. doi: 10.1371/journal.ppat.1005608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams JM, Duckworth CA, Burkitt MD, Watson AJ, Campbell BJ, Pritchard DM. Epithelial cell shedding and barrier function: a matter of life and death at the small intestinal villus tip. Vet Pathol. 2015;52(3):445–455. doi: 10.1177/0300985814559404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Musser JM, Mattingly SJ, Quentin R, Goudeau A, Selander RK. Identification of a high-virulence clone of type III Streptococcus agalactiae (group B Streptococcus) causing invasive neonatal disease. Proc Natl Acad Sci U S A. 1989;86(12):4731–4735. doi: 10.1073/pnas.86.12.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giaever I, Keese CR. A morphological biosensor for mammalian cells. Nature. 1993;366(6455):591–592. doi: 10.1038/366591a0. [DOI] [PubMed] [Google Scholar]
- 21.Byers SL, Wiles MV, Dunn SL, Taft RA. Mouse estrous cycle identification tool and images. PLoS ONE. 2012;7(4):e35538. doi: 10.1371/journal.pone.0035538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whidbey C, et al. Gundlach JH, Elovitz MA, et al. A streptococcal lipid toxin induces membrane permeabilization and pyroptosis leading to fetal injury. EMBO molecular medicine. 2015;7(4):488–505. doi: 10.15252/emmm.201404883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boldenow E, et al. Group B Streptococcus circumvents neutrophils and neutrophil extracellular traps during amniotic cavity invasion and preterm labor. Sci Immunol. 2016;1(4):eaah4576. doi: 10.1126/sciimmunol.aah4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Randis TM, et al. Group B Streptococcus β-hemolysin/cytolysin breaches maternal-fetal barriers to cause preterm birth and intrauterine fetal demise in vivo. J Infect Dis. 2014;210(2):265–273. doi: 10.1093/infdis/jiu067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doran KS, et al. Blood-brain barrier invasion by group B Streptococcus depends upon proper cell-surface anchoring of lipoteichoic acid. J Clin Invest. 2005;115(9):2499–2507. doi: 10.1172/JCI23829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henneke P, et al. Role of lipoteichoic acid in the phagocyte response to group B streptococcus. J Immunol. 2005;174(10):6449–6455. doi: 10.4049/jimmunol.174.10.6449. [DOI] [PubMed] [Google Scholar]
- 27.Clarke TB, Francella N, Huegel A, Weiser JN. Invasive bacterial pathogens exploit TLR-mediated downregulation of tight junction components to facilitate translocation across the epithelium. Cell Host Microbe. 2011;9(5):404–414. doi: 10.1016/j.chom.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Costa LC, et al. Expression of epithelial-mesenchymal transition markers at the invasive front of oral squamous cell carcinoma. J Appl Oral Sci. 2015;23(2):169–178. doi: 10.1590/1678-775720140187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berx G, van Roy F. Involvement of members of the cadherin superfamily in cancer. Cold Spring Harb Perspect Biol. 2009;1(6):a003129. doi: 10.1101/cshperspect.a003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stemmler MP. Cadherins in development and cancer. Mol Biosyst. 2008;4(8):835–850. doi: 10.1039/b719215k. [DOI] [PubMed] [Google Scholar]
- 32.Tian X, et al. E-cadherin/β-catenin complex and the epithelial barrier. J Biomed Biotechnol. 2011;2011:567305. doi: 10.1155/2011/567305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang X, Yu SX, Lu Y, Bast RC, Woodgett JR, Mills GB. Phosphorylation and inactivation of glycogen synthase kinase 3 by protein kinase A. Proc Natl Acad Sci U S A. 2000;97(22):11960–11965. doi: 10.1073/pnas.220413597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen P, Frame S. The renaissance of GSK3. Nat Rev Mol Cell Biol. 2001;2(10):769–776. doi: 10.1038/35096075. [DOI] [PubMed] [Google Scholar]
- 35.Handeli S, Simon JA. A small-molecule inhibitor of Tcf/beta-catenin signaling down-regulates PPARgamma and PPARdelta activities. Mol Cancer Ther. 2008;7(3):521–529. doi: 10.1158/1535-7163.MCT-07-2063. [DOI] [PubMed] [Google Scholar]
- 36.Streuli CH. Integrins as architects of cell behavior. Mol Biol Cell. 2016;27(19):2885–2888. doi: 10.1091/mbc.E15-06-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sackmann E. How actin/myosin crosstalks guide the adhesion, locomotion and polarization of cells. Biochim Biophys Acta. 2015;1853(11 Pt B):3132–3142. doi: 10.1016/j.bbamcr.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 38.Huang D, Cheung AT, Parsons JT, Bryer-Ash M. Focal adhesion kinase (FAK) regulates insulin-stimulated glycogen synthesis in hepatocytes. J Biol Chem. 2002;277(20):18151–18160. doi: 10.1074/jbc.M104252200. [DOI] [PubMed] [Google Scholar]
- 39.Byron A, Humphries JD, Askari JA, Craig SE, Mould AP, Humphries MJ. Anti-integrin monoclonal antibodies. J Cell Sci. 2009;122(Pt 22):4009–4011. doi: 10.1242/jcs.056770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Janes SM, Watt FM. New roles for integrins in squamous-cell carcinoma. Nat Rev Cancer. 2006;6(3):175–183. doi: 10.1038/nrc1817. [DOI] [PubMed] [Google Scholar]
- 41.Vornhagen J, Adams Waldorf KM, Rajagopal L. Perinatal Group B Streptococcal Infections: Virulence Factors, Immunity, and Prevention Strategies. Trends Microbiol. 2017;25(11):919–931. doi: 10.1016/j.tim.2017.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mu R, Kim BJ, Paco C, Del Rosario Y, Courtney HS, Doran KS. Identification of a group B streptococcal fibronectin binding protein, SfbA, that contributes to invasion of brain endothelium and development of meningitis. Infect Immun. 2014;82(6):2276–2286. doi: 10.1128/IAI.01559-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hill DR, et al. In vivo assessment of human vaginal oxygen and carbon dioxide levels during and post menses. J Appl Physiol. 2005;99(4):1582–1591. doi: 10.1152/japplphysiol.01422.2004. [DOI] [PubMed] [Google Scholar]
- 44.Vornhagen J, et al. Bacterial hyaluronidase promotes ascending GBS infection and preterm birth. MBio. 2016;7(3):e00781. doi: 10.1128/mBio.00781-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eto DS, Sundsbak JL, Mulvey MA. Actin-gated intracellular growth and resurgence of uropathogenic Escherichia coli. Cell Microbiol. 2006;8(4):704–717. doi: 10.1111/j.1462-5822.2006.00691.x. [DOI] [PubMed] [Google Scholar]
- 46.Justice SS, Hunstad DA. UPEC hemolysin: more than just for making holes. Cell Host Microbe. 2012;11(1):4–5. doi: 10.1016/j.chom.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dhakal BK, Mulvey MA. The UPEC pore-forming toxin α-hemolysin triggers proteolysis of host proteins to disrupt cell adhesion, inflammatory, and survival pathways. Cell Host Microbe. 2012;11(1):58–69. doi: 10.1016/j.chom.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hussein HA, Walker LR, Abdel-Raouf UM, Desouky SA, Montasser AK, Akula SM. Beyond RGD: virus interactions with integrins. Arch Virol. 2015;160(11):2669–2681. doi: 10.1007/s00705-015-2579-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Svoboda E, et al. Secreted aspartic protease 2 of Candida albicans inactivates factor H and the macrophage factor H-receptors CR3 (CD11b/CD18) and CR4 (CD11c/CD18) Immunol Lett. 2015;168(1):13–21. doi: 10.1016/j.imlet.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 50.Zhao Y, et al. Endothelial cell proteomic response to Rickettsia conorii infection reveals activation of the janus kinase (JAK)-signal transducer and activator of transcription (STAT)-inferferon stimulated gene (ISG)15 pathway and reprogramming plasma membrane integrin/cadherin signaling. Mol Cell Proteomics. 2016;15(1):289–304. doi: 10.1074/mcp.M115.054361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edwards JL, Butler EK. The pathobiology of neisseria gonorrhoeae lower female genital tract infection. Front Microbiol. 2011;2:102. doi: 10.3389/fmicb.2011.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hofman P, Vouret-Craviari V. Microbes-induced EMT at the crossroad of inflammation and cancer. Gut Microbes. 2012;3(3):176–185. doi: 10.4161/gmic.20288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han IH, Kim JH, Kim SS, Ahn MH, Ryu JS. Signalling pathways associated with IL-6 production and epithelial-mesenchymal transition induction in prostate epithelial cells stimulated with Trichomonas vaginalis. Parasite Immunol. 2016;38(11):678–687. doi: 10.1111/pim.12357. [DOI] [PubMed] [Google Scholar]
- 54.Ha NH, et al. Prolonged and repetitive exposure to Porphyromonas gingivalis increases aggressiveness of oral cancer cells by promoting acquisition of cancer stem cell properties. Tumour Biol. 2015;36(12):9947–9960. doi: 10.1007/s13277-015-3764-9. [DOI] [PubMed] [Google Scholar]
- 55.Sougleri IS, Papadakos KS, Zadik MP, Mavri-Vavagianni M, Mentis AF, Sgouras DN. Helicobacter pylori CagA protein induces factors involved in the epithelial to mesenchymal transition (EMT) in infected gastric epithelial cells in an EPIYA- phosphorylation-dependent manner. FEBS J. 2016;283(2):206–220. doi: 10.1111/febs.13592. [DOI] [PubMed] [Google Scholar]
- 56.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17(1):9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pai P, Rachagani S, Dhawan P, Batra SK. Mucins and Wnt/β-catenin signaling in gastrointestinal cancers: an unholy nexus. Carcinogenesis. 2016;37(3):223–232. doi: 10.1093/carcin/bgw005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pez F, Lopez A, Kim M, Wands JR, Caron de Fromentel C, Merle P. Wnt signaling and hepatocarcinogenesis: molecular targets for the development of innovative anticancer drugs. J Hepatol. 2013;59(5):1107–1117. doi: 10.1016/j.jhep.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 59.Takahashi-Yanaga F, Sasaguri T. Drug development targeting the glycogen synthase kinase-3beta (GSK-3beta)-mediated signal transduction pathway: inhibitors of the Wnt/beta-catenin signaling pathway as novel anticancer drugs. J Pharmacol Sci. 2009;109(2):179–183. doi: 10.1254/jphs.08R28FM. [DOI] [PubMed] [Google Scholar]
- 60.Racicot K, et al. Viral infection of the pregnant cervix predisposes to ascending bacterial infection. J Immunol. 2013;191(2):934–941. doi: 10.4049/jimmunol.1300661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hyman RW, et al. Diversity of the vaginal microbiome correlates with preterm birth. Reprod Sci. 2014;21(1):32–40. doi: 10.1177/1933719113488838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blencowe H, et al. Preterm birth-associated neurodevelopmental impairment estimates at regional and global levels for 2010. Pediatr Res. 2013;74 Suppl 1:17–34. doi: 10.1038/pr.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Bernis L, et al. Stillbirths: ending preventable deaths by 2030. Lancet. 2016;387(10019):703–716. doi: 10.1016/S0140-6736(15)00954-X. [DOI] [PubMed] [Google Scholar]
- 64.Martin TR, Rubens CE, Wilson CB. Lung antibacterial defense mechanisms in infant and adult rats: implications for the pathogenesis of group B streptococcal infections in the neonatal lung. J Infect Dis. 1988;157(1):91–100. doi: 10.1093/infdis/157.1.91. [DOI] [PubMed] [Google Scholar]
- 65.Pritzlaff CA, Chang JC, Kuo SP, Tamura GS, Rubens CE, Nizet V. Genetic basis for the beta-haemolytic/cytolytic activity of group B Streptococcus. Mol Microbiol. 2001;39(2):236–247. doi: 10.1046/j.1365-2958.2001.02211.x. [DOI] [PubMed] [Google Scholar]
- 66.Gendrin C, et al. Mast cell degranulation by a hemolytic lipid toxin decreases GBS colonization and infection. Sci Adv. 2015;1(6):e1400225. doi: 10.1126/sciadv.1400225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sheen TR, Jimenez A, Wang NY, Banerjee A, van Sorge NM, Doran KS. Serine-rich repeat proteins and pili promote Streptococcus agalactiae colonization of the vaginal tract. J Bacteriol. 2011;193(24):6834–6842. doi: 10.1128/JB.00094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chaffin DO, Mentele LM, Rubens CE. Sialylation of group B streptococcal capsular polysaccharide is mediated by cpsK and is required for optimal capsule polymerization and expression. J Bacteriol. 2005;187(13):4615–4626. doi: 10.1128/JB.187.13.4615-4626.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lembo A, et al. Regulation of CovR expression in Group B Streptococcus impacts blood-brain barrier penetration. Mol Microbiol. 2010;77(2):431–443. doi: 10.1111/j.1365-2958.2010.07215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.