Abstract
Aspergillus fumigatus and multiple other Aspergillus species cause a wide range of lung infections, collectively termed aspergillosis. Aspergilli are ubiquitous in environment with healthy immune systems routinely eliminating inhaled conidia, however, Aspergilli can become an opportunistic pathogen in immune-compromised patients. The aspergillosis mortality rate and emergence of drug-resistance reveals an urgent need to identify novel targets. Secreted and cell membrane proteins play a critical role in fungal-host interactions and pathogenesis. Using a computational pipeline integrating data from high-throughput experiments and bioinformatic predictions, we have identified secreted and cell membrane proteins in ten Aspergillus species known to cause aspergillosis. Small secreted and effector-like proteins similar to agents of fungal-plant pathogenesis were also identified within each secretome. A comparison with humans revealed that at least 70% of Aspergillus secretomes have no sequence similarity with the human proteome. An analysis of antigenic qualities of Aspergillus proteins revealed that the secretome is significantly more antigenic than cell membrane proteins or the complete proteome. Finally, overlaying an expression dataset, four A. fumigatus proteins upregulated during infection and with available structures, were found to be structurally similar to known drug target proteins in other organisms, and were able to dock in silico with the respective drug.
Introduction
Aspergillosis is an umbrella term for a wide array of infections caused by multiple Aspergillus species. The majority of reported aspergillosis cases originate from ten species: Aspergillus fumigatus, Aspergillus flavus, Aspergillus niger, Aspergillus terreus, Aspergillus versicolor, Aspergillus lentulus, Aspergillus nidulans, Aspergillus glaucus, Aspergillus oryzae, and Aspergillus ustus1–3. A. fumigatus alone is responsible for over 90% of the reported cases, followed by A. flavus, A. niger, A. terreus and A. versicolor2,4,5. The disease is an increasing concern for immune-compromised individuals, putting at risk patients with neutropenia, allogenic stem cell transplantation, organ transplantation, and acquired immunodeficiency syndrome (AIDS), among others6. High-risk patients can see mortality rates between 40–90%7. Compounding the issue is the limited number of drugs available to treat aspergillosis, and their widespread use has led to the emergence of drug-resistant strains, creating the pressing need for novel drugs, biomarkers and preventive therapies such as vaccines8.
Aspergillosis is caused by an opportunistic pathogen. Under normal conditions, Aspergilli are saprotrophic fungi which are present in soil and decaying organic matter, thereby, playing a fundamental role in nitrogen and carbon recycling. They have a broad geographical range with colonies typically spread through microscopic airborne conidia. As Aspergillus conidia are ubiquitous in the atmosphere, humans often inhale several hundred spores daily. While these spores are quickly eliminated by a healthy immune system9, in an immune-compromised host, the fungus becomes an opportunistic pathogen and overwhelms the weakened defences. The underlying mechanisms behind the successful initiation of pathogenesis by Aspergilli remain unclear. However, studies have shown that in immune-compromised hosts, A. fumigatus can reach the respiratory epithelia upon inhalation where it secretes proteases and secondary metabolites, specifically gliotoxin, which aids in colonizing healthy lung tissue10,11. Proteases secreted by A. fumigatus have also been implicated in establishing infection by morphologically altering respiratory epithelial cells11,12. In addition, A. fumigatus secretes a diverse array of catabolic enzymes that enable degradation of macromolecular biopolymers like elastin and collagen, which are present in large quantities in lung tissue, for uptake of nutrients13. Apart from secreted proteins, cell membrane and cell wall proteins also play a crucial role in interactions with the host immune system and establishing infection10. Thus, the diverse set of secreted and cell membrane proteins of A. fumigatus and other Aspergillus species play a vital role in pathogenesis.
Given the central role played by the secretome and cell membrane proteins of Aspergillus species during human pathogenesis, they are potential candidates for identifying new biomarkers, vaccines and druggable targets. This is especially urgent as prophylactic use of antifungal therapies has led to the emergence of resistance in many Aspergillus species to antifungal drugs8,14,15. Prospecting the secretome and cell membrane proteins of Aspergillus species for druggable targets is advantageous, since any identified target will be exposed to the extracellular space. In addition, targeting extracellular rather than intracellular proteins will provide fewer mechanisms for resistance to develop in the pathogen.
This study aims to identify and analyze the secretome and cell membrane proteins of A. fumigatus and nine other Aspergillus species known to cause aspergillosis, namely, A. flavus, A. niger, A. terreus, A. versicolor, A. lentulus, A. nidulans, A. glaucus, A. oryzae, and A. ustus. To our knowledge, this is the first dedicated effort to characterize the secretome and cell membrane proteins of Aspergillus species with an aim to understand their role in pathogenesis and to analyze them as potential druggable proteins. While previous studies such as FSD16, FunSecKB217 and SECRETOOL18 have developed computational pipelines for fungal secretome prediction, they rely solely on bioinformatic predictions even though many experimental high-throughput proteomic datasets in Aspergillus species are available. Thus, we have designed a computational pipeline which systematically integrates experimental high-throughput proteomic datasets, UniProt19 annotations with experimental evidence, and predictions from bioinformatic tools to identify a comprehensive set of secreted extracellular proteins and cell membrane proteins in Aspergillus species. Furthermore, small secreted and effector-like proteins20–23 were identified in Aspergillus species, and the set of secreted and cell membrane proteins were analyzed for the abundance of antigenic regions (AAR)24 and similarity to known drug target proteins from DrugBank25. Finally, analysis of a published gene expression dataset26 for A. fumigatus, led to identification of secreted and cell membrane proteins which are upregulated under pathogenesis and have no human homologs, and such candidates were taken for further druggability analysis by computational docking experiments, thereby identifying pre-existing drugs used against other pathogens as candidates for repurposing against aspergillosis.
Results and Discussion
Identification of the secretome from high-throughput experimental studies
An extensive literature search was undertaken to compile high-throughput experimental studies that have characterized the secretome of Aspergillus species. A systematic search led to a comprehensive list of 46 high-throughput proteomic studies27–73 on the secretome of 6 Aspergillus species analyzed here. Note that many high-throughput studies report proteins with transmembrane (TM) domains as part of the secretome but a few studies have specifically identified cell membrane and cell wall associated proteins along with the secretome30,36. Our compilation of high-throughput proteomic studies led to experimentally identified lists of 437, 364, 454, 96, 469 and 171 secreted proteins in A. fumigatus27–41, A. flavus30,42–45, A. niger30,46–58, A. terreus30,59,60, A. nidulans58,61–65 and A. oryzae57,66–73, respectively (Methods; Supplementary Table S1).
Computational pipeline for fungal secretome prediction
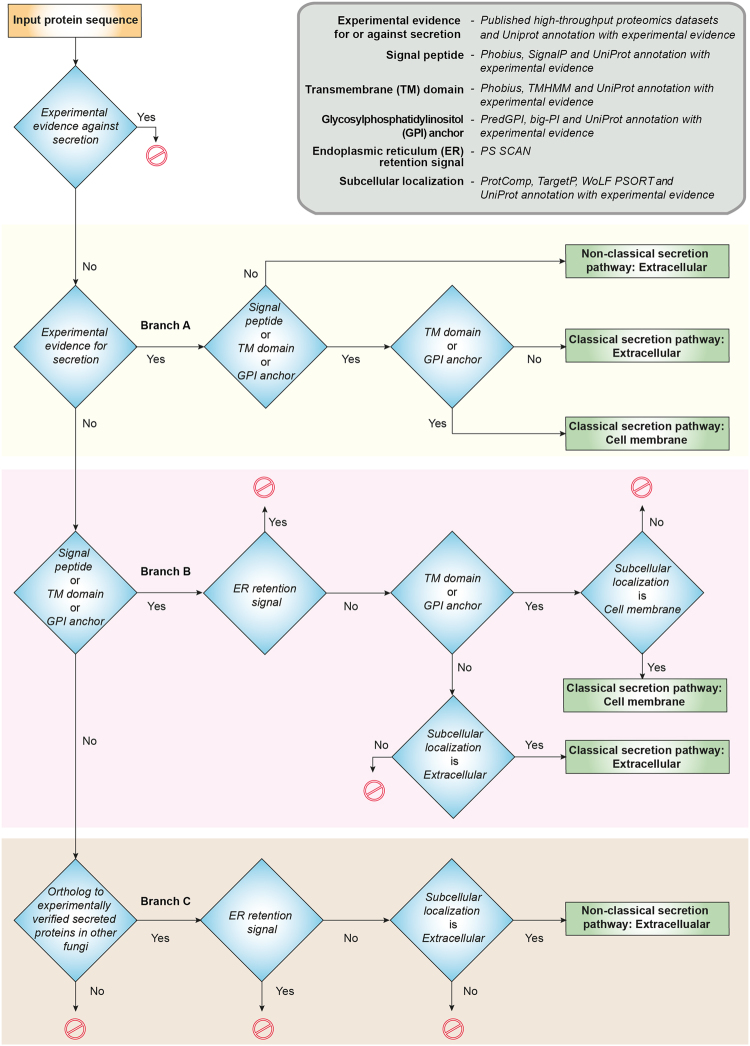
High-throughput experimental studies that identify secretomes can be limited by the detection technology used or the small set of experimental conditions assessed. Such secretomic studies mainly focus on identifying the proteins secreted to the extracellular matrix and not on proteins incorporated into the cell membrane by the eukaryotic secretion pathway. Since cell membrane proteins, including integral proteins, are localized on the cell surface, they can serve as excellent targets for drugs or vaccines. Here we have designed a computational pipeline to predict both secreted extracellular and cell membrane proteins in fungi. Importantly, our pipeline also incorporates and prioritizes available information on the experimentally identified secreted and cell membrane proteins. Furthermore, our pipeline subdivides the secretome into two subsets, proteins that follow the classical secretion pathway through the endoplasmic reticulum (ER) and those that exit the cell boundary through a non-classical secretion pathway. Figure 1 contains a flowchart of our prediction pipeline.
Figure 1.
Flowchart of the computational prediction pipeline for identifying secreted extracellular proteins (secretome) and cell membrane proteins in Aspergillus species. Note that this prediction pipeline can be employed to identify secretome and cell membrane proteins in any fungi with sequenced genome.
Our pipeline starts from the complete proteome of a fungus. Initially, intracellular proteins based on UniProt19 annotation with experimental evidence were removed from later analysis. Subsequently, the remaining proteins without experimental evidence for intracellular localization were classified into two mutually exclusive categories. The first category contained secreted or cell membrane proteins with experimental evidence from a compiled list of high-throughput proteomic studies or UniProt, and the second category contained proteins without experimental evidence of secretion to the extracellular matrix or localization to the cell membrane.
The first category of proteins with experimental evidence were assessed for signal peptide, Glycosylphosphatidylinositol (GPI) anchor or TM domain, confirming passage through the classical pathway (Branch A in Fig. 1; Methods). Next, the presence of GPI anchor or TM domain in proteins sorted by classical pathway is used to separate the cell membrane from extracellular proteins. Next, the proteins without a signal peptide, GPI anchor and TM domain but with experimental evidence of secretion to extracellular matrix were assigned to non-classical pathway (Branch A in Fig. 1).
The second category of proteins without experimental evidence were screened based on predictions from computational tools as follows. Firstly, the second category of proteins were screened for signal peptide, GPI anchor or TM domain, suggesting translocation into the ER and passage through classical pathway. Next, the proteins predicted to have a signal peptide, GPI anchor or TM domain but also with an ER retention signal were removed from later analysis (Branch B in Fig. 1; Methods). Next, the proteins predicted to have GPI anchor or TM domain along with subcellular localization as cell membrane were classified as cell membrane proteins, and proteins without GPI anchor and TM domain along with subcellular localization as extracellular were classified as extracellular proteins sorted by classical pathway (Branch B in Fig. 1).
Lastly, the subset of the second category of proteins which lack signal peptide, GPI anchor and TM domain, were assessed for presence of orthologs in the list of secreted proteins with experimental evidence in other fungi (Methods; Branch C in Fig. 1). Next, those proteins in the subset which are orthologs of experimentally identified secreted proteins in other fungi were assessed for an ER retention signal and their predicted subcellular localization, and those without an ER retention signal and predicted subcellular localization as extracellular were classified as extracellular proteins secreted through a non-classical secretion pathway (Branch C in Fig. 1). To our knowledge, SecretomeP74,75 is the only prediction tool for non-classical secretion pathway, however, SecretomeP74,75 is designed for bacteria and mammals and not for fungi. Still FSD16 has employed SecretomeP74,75 to predict approximately 40% of the proteome in several fungi as secreted via a non-classical secretion pathway which clearly is an overestimation. For example, FSD predicts 3546 A. fumigatus proteins, which is 35% of the proteome, to be secreted by a non-classical pathway. Thus, we employ here an alternate ortholog-based method to predict proteins secreted via a non-classical pathway.
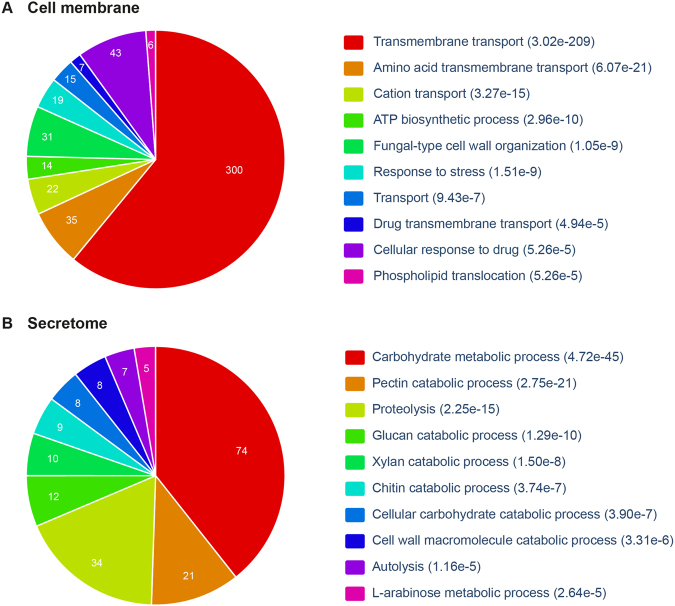
Using this pipeline, we identified comprehensive sets of secreted and cell membrane proteins in ten Aspergillus species causing aspergillosis; Figure 2 displays the size of each set, Supplementary Table S2 lists the proteins in each set, and Supplementary Table S3 gives detailed annotations such as protein family, conserved domain, carbohydrate-binding modules and gene ontology (GO) terms for proteins in each set. Additionally, our pipeline provides a refined view of the protein sorting mechanisms in Aspergillus species by classifying secreted proteins into classical and non-classical pathway. In A. fumigatus, the predicted set of secreted and cell membrane proteins contained 662 and 1129 proteins, respectively, representing 6.7% and 11.5% of the proteome. Among the 662 proteins in A. fumigatus secretome, 64 were predicted to be secreted by a non-classical pathway. Figure 3 shows the significantly enriched GO biological processes in the predicted secretome and cell membrane proteins of A. fumigatus. Within the A. fumigatus secretome, 598 proteins sorted by classical pathway have GO annotations related to carbohydrate metabolism, proteolysis and cell wall modification, and 64 proteins secreted by a non-classical pathway have GO annotations related to carbohydrate metabolism, response to reactive oxygen species (ROS), alcohol degradation and gliotoxin metabolism. Many of the GO processes annotated with A. fumigatus proteins secreted by non-classical pathway are associated with virulence. Especially, gliotoxin has immunosuppressive properties and is suspected to be an important virulence factor in Aspergillus pathogenesis76–79. This suggests that non-classical pathways may be important for the secretion of virulence factors in A. fumigatus.
Figure 2.

Number of proteins in the complete proteome, set of cell membrane proteins, set of secreted extracellular proteins, set of small secreted proteins (SSPs) and set of effector-like SSPs identified by our computational pipeline in Aspergillus species considered here.
Figure 3.
Gene ontology (GO) enrichment analysis for (A) cell membrane proteins and (B) secreted extracellular proteins in A. fumigatus Af293. In each case, the top 10 significantly enriched biological processes are shown along with p-value computed using Benjamini-Hochberg procedure. The GO enrichment analysis was performed using FungiFun2147 webserver (Methods).
Finally, we also performed an extensive search of the published literature in an attempt to locate experimental evidence from detailed low-throughput studies that may validate our predictions on cell membrane proteins in Aspergillus species from Branch B of our computational pipeline shown in Fig. 1. Through this exercise, we were able to find direct experimental evidence, typically, Green fluorescent protein (GFP) tagged experiments, for 17 predicted cell membrane proteins in Aspergillus species of which 3 are in A. fumigatus80,81, 13 in A. nidulans82–94 and 1 in A. oryzae95 (Supplementary Table S4). These findings support predictions from the computational pipeline.
Small secreted and effector-like proteins
Within the fungal secretome, small secreted proteins (SSPs) with sequence length less than 300 amino acids have been widely studied for their role in fungal-plant pathogenesis20,21. A few of the SSPs have been found to act as effectors that play a central role in establishing plant infection22,23. Typically, fungal effector proteins do not share conserved domains which renders effector prediction a challenge96,97. Still, fungal effectors often share certain sequence characteristics such as size, cysteine content, or small motifs identified in fungi and oomycetes96,97. A recent study98 compared the SSPs across eight Aspergillus species focusing on plant biomass degradation. However, the role of SSPs and effector-like proteins in fungal-human pathogenesis, including aspergillosis, remains largely unanswered99. Thus, we have identified SSPs and effector-like proteins within the secretomes of Aspergillus species to enable discovery of potential virulence proteins (Supplementary Table S5). Note that effector-like proteins of Aspergillus species were predicted based on EffectorP97 predictions or cysteine content of SSPs (Methods). We found that SSPs and effector-like proteins account for more than 30% and 10%, respectively, of the secretomes in Aspergillus species (Fig. 2). Specifically, 96 effector-like proteins were predicted among 210 SSPs in the A. fumigatus secretome, of which, 44 do not have conserved Pfam domains and 4 contain one of the known fungal or oomycete effector motifs96, making them intriguing candidates for future virulence experiments.
Upregulated secretome and cell membrane proteins in A. fumigatus during pathogenesis
To place the identified secretome and cell membrane proteins of A. fumigatus within the context of aspergillosis, we overlaid a previously published gene expression dataset26. Significantly differentially expressed and upregulated genes coding for secreted and cell membrane proteins may suggest a vital path or key towards pathogenesis. Note that several gene expression studies in A. fumigatus have been published on various cell cultures. However, due to the difficulty in obtaining sufficient high-quality RNA directly from sites of infection, gene expression studies of A. fumigatus in live animal models are limited. Thus, we selected for our analysis a published microarray dataset26 from a murine lung model which provided one of the first transcriptional snapshots of A. fumigatus during initiation of mammalian infection.
In this dataset26, A. fumigatus genes significantly upregulated over 2-fold during pathogenesis when compared to control conditions were selected for further analysis, totaling, 1264 proteins (12.9%) of the proteome. Within the predicted A. fumigatus secretome, 121 out of 662 proteins (18.3%) were upregulated over 2-fold during pathogenesis, indicating that a larger fraction of secretome is employed during pathogenesis. Moreover, 113 out of the 121 upregulated proteins in the A. fumigatus secretome are secreted via classical pathway and their GO analysis revealed involvement in carbohydrate metabolic processes and regulation of host immune response, while the remaining 8 are secreted via a non-classical pathway and their GO analysis revealed involvement in response to mycotoxin, synthesis of mycotoxin and gliotoxin, and response to oxidative stress apart from carbohydrate metabolic processes. Within the cell membrane proteins of A. fumigatus, 151 out of 1129 proteins (13.3%) were upregulated during pathogenesis, and GO analysis revealed that many of them are involved in transmembrane transport processes. Moreover, 45 SSPs in the A. fumigatus secretome, of which 21 have effector-like features, were upregulated over 2-fold during pathogenesis. For example, Afu3g14940 a predicted peptidase inhibitor I78 family protein which has a known conserved effector motif ([KRHQSA][DENQ]EL) and Afu1g01210 with the adhesion fasciclin domain, are among the 21 effector-like SSPs with over 2-fold upregulation during pathogenesis.
Conservation of secretome across Aspergillus species
The ability of multiple Aspergilli to switch into an opportunistic pathogen may be derived from conserved proteins. Conversely, species-specific proteins may explain the observed differences in virulence between Aspergilli. To determine the unique and conserved proteins across Aspergillus species, OrthoMCL100 was used to identify orthologs between the proteins of nine Aspergillus species. As A. ustus has an incomplete genome, it was omitted from this comparative analysis. Figure 4A–C display for each species the fraction of unique and conserved proteins within the complete proteome, cell membrane proteins and secretomes, respectively, across the nine Aspergillus species. In comparison to the cell membrane proteins, the secretomes of Aspergillus species have a larger set of unique proteins and a smaller set of conserved proteins across the nine Aspergillus species. Interestingly, 3.6% of the secretome of A. fumigatus is unique, while 27.3% and 26.9%, respectively, of the secretomes of A. niger and A. glaucus is unique, when compared across the nine Aspergillus species. Thus, the secretome of A. fumigatus has among the smallest fraction of unique proteins which is in contrast to its position as the most prevalent agent of aspergillosis. The unique and conserved proteins identified here across the secretomes of Aspergillus species could lead to a better understanding of the important players for virulence and provide potential targets for the development of broad spectrum antifungals.
Figure 4.
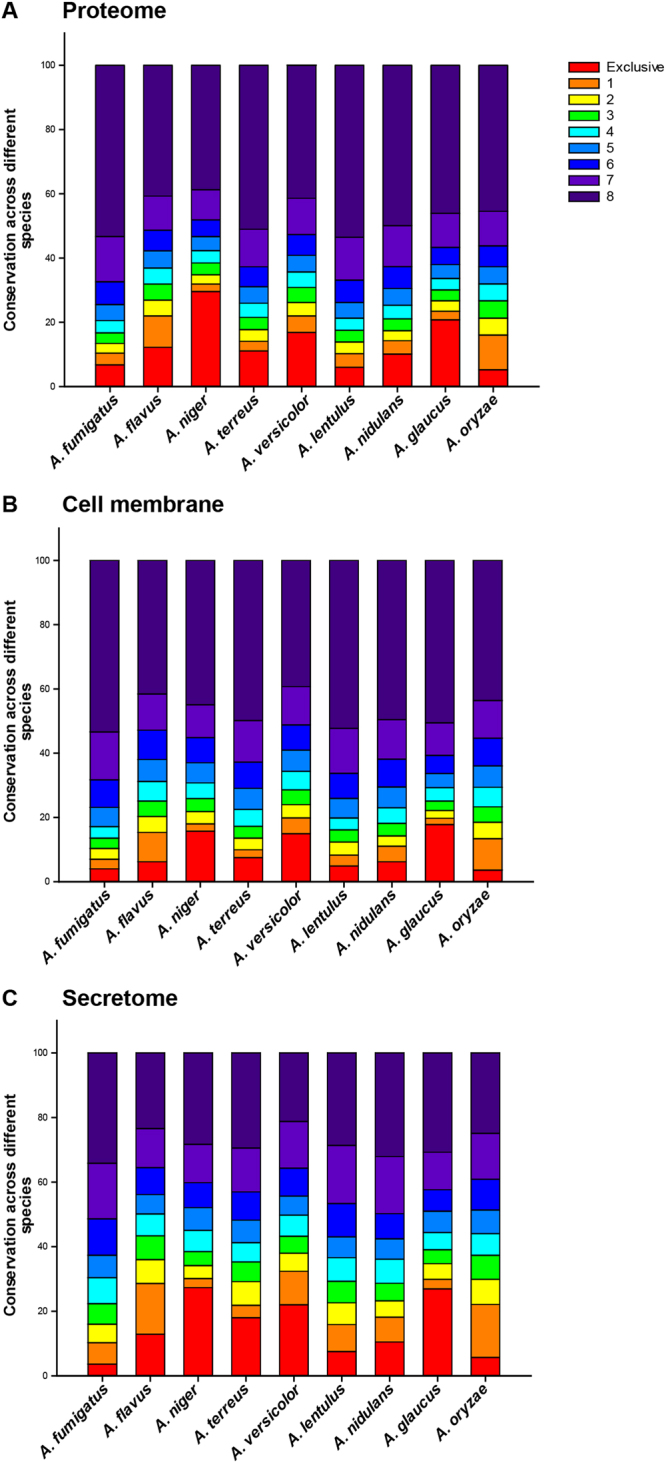
Conservation of (A) complete proteome, (B) the set of cell membrane proteins, and (C) the set of secreted extracellular proteins across the nine Aspergillus species considered here. The fraction of proteins that are unique to a specific Aspergillus species is shaded in orange at the bottom of the stacked bar chart while the fraction of proteins conserved across all nine Aspergillus species are shaded in dark violet at the top of the stacked bar chart.
Comparison with the human proteome
Ideally, antifungals should target fungi-specific proteins to prevent cross-toxicity with human proteins. In order to identify fungi-specific druggable targets, BLASTP with an E-value cut-off of ≤1e-3 was used to identify Aspergillus proteins without a close homolog in the human proteome. We find that more than 70% of the secretomes in Aspergillus species is unique, without a homolog in humans (Fig. 5). In contrast to the secretomes, the cell membrane fraction or whole proteome of Aspergillus species have a much lower fraction of unique proteins without a homolog in humans (Fig. 5). Specifically, A. fumigatus has 71% of its secretome and 52% of its cell membrane proteins without a homolog to humans, and this offers a significant candidate set that may be screened for druggability or biomarker design. The striking difference between conservation of whole proteome and secretome of Aspergillus species in humans can likely be attributed to the large presence of proteins associated with plant cell wall degradation101 in the Aspergillus secretomes.
Figure 5.
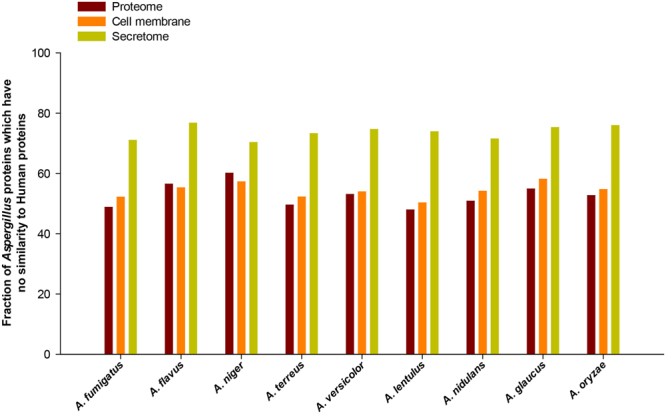
Fraction of the complete proteome, set of cell membrane proteins and set of secreted extracellular proteins in Aspergillus species without a close sequence homolog in human proteome.
Antigenicity of the secretome
Protein vaccines have been proven successful for several invasive fungi102–104. For example, vaccination with the A. fumigatus allergen Aspf3 protected immunosuppressed mice from developing aspergillosis105. One measure of clinical importance is the number of potential antigenic regions on a protein. The more antigenic a protein the more likely it can be used as a biomarker, targeted for immunotherapy, or used in vaccinations. To help prioritize Aspergillus proteins, the Abundance of Antigenic Regions (AAR)24 value of proteins was calculated using two methods, Kolaskar-Tongaonkar106 and BepiPred 2.0107 (Methods). The lower the AAR value the more antigenic the protein24. Interestingly, in each Aspergillus species, the average AAR of the secretome is always significantly lower than cell membrane proteins (Fig. 6; Methods). Furthermore, the average AAR of the secretome and cell membrane proteins was also found to be significantly lower (p < 0.001) and significantly higher (p < 0.001), respectively, in comparison to the average AAR of randomly chosen, equally sized sets of proteins from the whole proteome (Methods). Note that similar observations on relative AAR of secretome and cell membrane proteins were also recently made in tapeworms, Taenia solium24 and Echinococcus multilocularis108, and in bacterium Mycobacterium tuberculosis109. Thus, secreted proteins are likely to be more antigenic than cell membrane proteins, and while choosing candidates for vaccine development, the secretome may provide a more antigenic landscape than cell membrane proteins. While a comparison of the AAR of secretome and cell membrane proteins in Aspergillus species provides insight on relative virulence within a proteome, no significant difference in the average AAR could be detected between A. fumigatus and the Aspergillus cohort.
Figure 6.
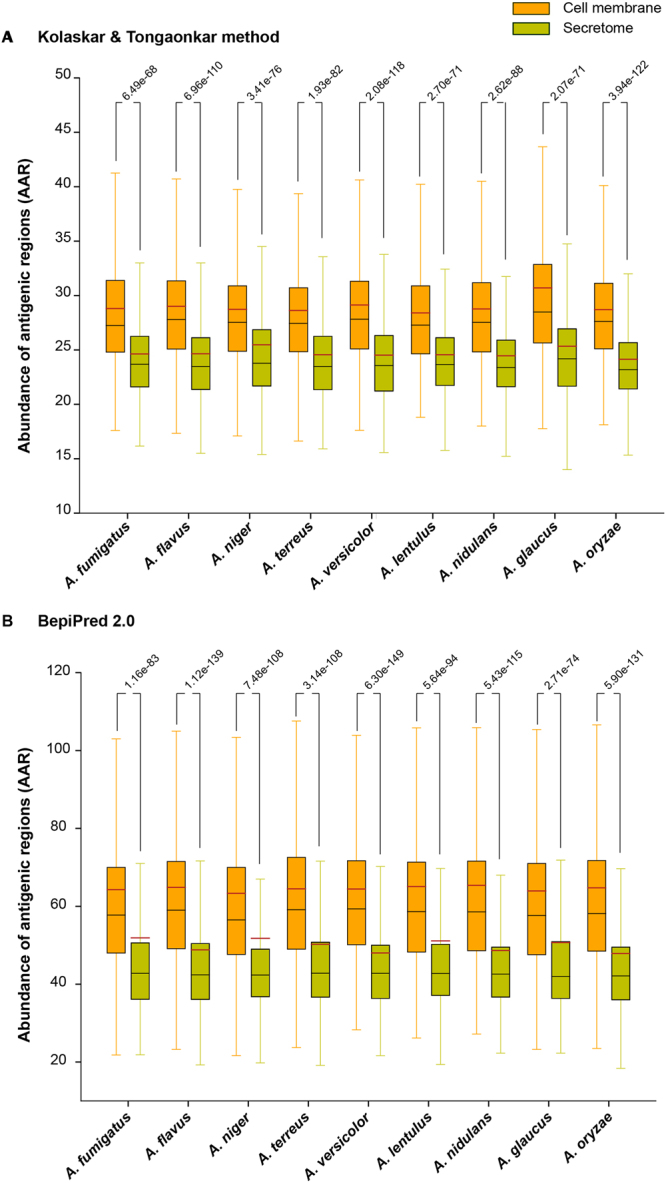
Distribution of Abundance of Antigenic Regions (AAR) values for the set of cell membrane proteins and secreted extracellular proteins in Aspergillus species considered here. AAR values were computed using two different methods: (A) Kolaskar-Tongaonkar106 method and (B) BepiPred 2.0107. In each box plot, the lower end of the box represents the first quartile, black line inside the box is the median, brown line is the mean and the upper end of the box represents the third quartile of the distribution. In this figure, we also report the p-value from the comparison of the distributions of AAR values in the set of cell membrane proteins and in the set of secreted proteins for each Aspergillus species performed using Wilcoxon rank-sum test.
Druggability analysis of A. fumigatus secretome and cell membrane proteins upregulated during pathogenesis
Beyond immune-therapy, biomarker, and vaccine development, drugs are often the main line of defence against aspergillosis. However, the current arsenal to combat aspergillosis is limited to 7 approved drugs. Furthermore, resistance to these few drugs is an ever-emerging threat14. Particularly, fungi are adept at developing resistance to drugs that act on intracellular proteins by pumping them back out into the extracellular matrix through efflux pumps. However, targeting of secreted proteins and cell membrane proteins could circumvent this specific defence mechanism altogether. To expedite the drug discovery pipeline, DrugBank25 provides a database of known drugs and their targets, many of which have been successfully repurposed against similar protein targets in different pathogens110.
Firstly, the list of 4063 known drug target proteins was compiled from DrugBank25. Secondly, the secreted and cell membrane proteins in Aspergillus species with no close human homologs based on BLASTP with an E-value cut-off of ≤1e-3 were determined. Thereafter, secreted and cell membrane proteins in Aspergillus species with no close human homologs but with sequence similarity to known drug target proteins based on BLASTP E-value cut-off of ≤1e-5 were determined (Supplementary Table S6). In A. fumigatus, 50 secreted and 16 cell membrane proteins were found to have sequence similarity with known drug target proteins. Subsequently, we focused on upregulated genes in a transcriptome dataset26 for A. fumigatus during pathogenesis to identify potential target proteins for drug repurposing.
In the transcriptome dataset26 for A. fumigatus, we found 97 secreted and 101 cell membrane proteins with no close human homologs to be significantly upregulated over 2-fold during pathogenesis. These 97 secreted and 101 cell membrane proteins in A. fumigatus were searched against known target protein sequences for sequence similarity to known drug targets. Seven secreted proteins, Afu7g06140, Afu8g01670, Afu2g15160, Afu5g14380, Afu6g09740, Afu3g00610 and Afu1g09900, of which three proteins, Afu8g01670, Afu6g09740 and Afu1g09900, are secreted via non-classical pathways, and one membrane protein, Afu6g03570, had a significant hit to known drug target proteins. After establishing the sequence similarity between A. fumigatus proteins and known drug target proteins, structural similarity was probed (Methods). Experimentally identified protein structure was available only for Afu6g09740, and it was downloaded from the protein data bank111 (PDB). Since a good-quality structure model was also unavailable for Afu2g15160, it was omitted from later analysis. For the remaining five secreted proteins and one cell membrane protein, structure models were obtained from ModBase112 and SWISS-MODEL repository113 and were subsequently checked for their quality (Methods). Thereafter, the compiled protein structures were compared to their DrugBank counterparts for structural similarity. Four secreted proteins, Afu5g14380, Afu8g01670, Afu1g09900 and Afu6g09740, had significant structural similarity with TM scores greater than 0.8 and root-mean-square deviation (RMSD) values lower than 3Å2 (Table 1; Methods). One of the proteins, Afu8g01670, is a putative bifunctional catalase-peroxidase which has been shown to be involved in virulence114. Afu5g14380 is an α-glucuronidase involved in the hydrolysis of xylan. Afu1g09900 is involved in degradation of arabinoxylan. Afu6g09740 is a thioredoxin reductase which is a part of the gene cluster involved in biosynthesis of gliotoxin in A. fumigatus, and its knockout has been shown to affect the oxidation of gliotoxin and render the fungi hypersensitive to exogenous gliotoxin115. Afu6g09740 was also found to be immunogenic in humans and a potential biomarker of aspergillosis116.
Table 1.
Evaluation of the structural similarity of secreted and cell membrane proteins upregulated in the transcriptome of Aspergillus fumigatus Af293 with their close sequence homologs among known drug target proteins in DrugBank25.
| Protein identifier | Structure Source | PDB identifier of template used to create protein structure models | Log2 fold change in transcriptome | UniProt identifier of drug target protein | PDB identifier of drug target protein | TM score | RMSD (Å2) |
|---|---|---|---|---|---|---|---|
| Afu5g14380 | SWISS-MODEL | 1MQQ A | 1.42 | Q8VVD2 | 1L8N | 0.98 | 0.19 |
| Afu8g01670 | SWISS-MODEL | 5KSN A | 2.34 | Q939D2 | 5L05 | 0.99 | 0.28 |
| P9WIE5 | 1SJ2 | 0.97 | 0.72 | ||||
| Afu1g09900 | SWISS-MODEL | 3S2C A | 1.10 | Q9XBQ3 | 1PZ3 | 0.91 | 0.99 |
| Afu6g09740$ | PDB (4NTC) | NA | 1.27 | P66010 | 4GCM | 0.86 | 2.68 |
| Afu7g06140 # | SWISS-MODEL | 3ZYZ A | 7.16 | P40406 | 3BMX | 0.73 | 4.06 |
| Afu6g03570 # | SWISS-MODEL | 5FJJ A | 1.30 | P40406 | 3BMX | 0.72 | 3.29 |
| Afu3g00610 # | ModBase | 1GAI A | 1.19 | P05618 | 1PAM | 0.26 | 15.24 |
| P26827 | 1CIU | 0.25 | 19.25 |
Note that the last three A. fumigatus proteins italicized in this table are not structurally similar to their close sequence homologs among known drug target proteins. $An experimentally solved crystal structure was available from PDB for this protein.
#These three A. fumigatus proteins are not structurally similar to their close sequence homologs among known drug target proteins.
Given the sequence and structural similarity of these four A. fumigatus proteins, Afu5g14380, Afu8g01670, Afu1g09900 and Afu6g09740, to known drug targets, the proteins were subsequently tested whether they could bind to their respective drugs (Table 2; Methods). The four proteins were analyzed for the presence of ligand binding pockets using metaPocket 2.0117. For each of the four proteins, the top three ligand binding pockets were tested for ligand binding affinity using AutoDock Vina118. Table 2 lists the four Aspergillus proteins, the corresponding target proteins in DrugBank with both sequence and structural similarity, their reported approved or experimental drugs, and the binding affinity of each drug to the ligand pockets. Using AutoDock Vina118, it was found that most of the identified drug candidates have an affinity value of ≤−5.0 kcal/mol for their respective Aspergillus proteins which signifies good binding and suggests that the drugs may be able to bind to the upregulated and secreted A. fumigatus proteins (Table 2; Methods). Further experimental validation of these hit molecules is needed to verify whether the drug will act upon the A. fumigatus protein in the same fashion and whether the drug will have an impact upon the ability of A. fumigatus to cause aspergillosis.
Table 2.
Binding affinity of potential drugs against predicted pockets of the four candidate proteins in Aspergillus fumigatus.
| Protein identifier | Protein identifier of drug target protein | Drug identifier in DrugBank | Drug name | Drug class | Mean binding affinity (kcal/mol) | ||
|---|---|---|---|---|---|---|---|
| First pocket | Second pocket | Third pocket | |||||
| Afu8g01670 | P9WIE5 | DB00609 | Ethionamide | Approved | −5.9 ± 0.0 | −5.9 ± 0.0 | −5 ± 0.0 |
| DB00951 | Isoniazid | Approved | −5.53 ± 0.015 | −5.91 ± 0.01 | −5.3 ± 0.0 | ||
| Q939D2 | DB08638 | 2-Amino-3-(1-hydroperoxy-1H-indol-3-yl)propan-1-ol | Experimental | −7.6 ± 0.033 | −7.6 ± 0.0 | −5.87 ± 0.015 | |
| Afu5g14380 | Q8VVD2 | DB03156 | D-Glucuronic Acid | Experimental | −5.1 ± 0.0 | −6.5 ± 0.0 | −5.6 ± 0.0 |
| DB04303 | 4-O-methyl-alpha-D-glucuronic acid | Experimental | −5 ± 0.0 | −6.3 ± 0.0 | −5.3 ± 0.0 | ||
| DB02722 | 4-O-methyl-beta-D-glucuronic acid | Experimental | −5 ± 0.0 | −5.8 ± 0.0 | −5.11 ± 0.01 | ||
| Afu6g09740 | P66010 | DB00548 | Azelaic Acid | Approved | −5.53 ± 0.026 | −5.52 ± 0.065 | −5.21 ± 0.031 |
| Afu1g09900 | Q9XBQ3 | DB03196 | 4-Nitrophenyl-Ara | Experimental | −6.3 ± 0.0 | −6 ± 0.0 | −5.54 ± 0.031 |
| DB03870 | Ara-Alpha(1,3)-Xyl | Experimental | −5.51 ± 0.023 | −5.54 ± 0.016 | −5.62 ± 0.029 | ||
Note that the drug candidates were identified based on sequence and structural similarity of the four upregulated proteins in A. fumigatus with known drug target proteins in DrugBank25 database. The ligand binding pockets were predicted using metaPocket 2.0117 webserver and binding affinities of drugs to proteins were computed using AutoDock Vina118 (Methods).
Comparison of our prediction pipeline with earlier work
We have designed here a computational prediction pipeline to identify the secretome and cell membrane proteins in Aspergillus species, and the pipeline can be used for any fungi. To our knowledge, FSD16 and FunSecKB217 are the only databases on pan-fungi secretome prediction. While FSD and FunSecKB2 contain pre-computed secretome predictions for several fungal genomes, SECRETOOL18 provides an online sever that enables implementation of a computational pipeline to predict secreted proteins among user-input protein sequences. In Supplementary Text and Supplementary Table S7, we report a detailed comparison of the secretome predictions from our pipeline with those from FSD, FunSecKB2 and SECRETOOL.
Based on this comparative analysis with FSD, FunSecKB2 and SECRETOOL, our pipeline has following additional features. Firstly, we incorporate available experimental information from high-throughput studies and UniProt on secreted proteins (Branch A in Fig. 1). Secondly, we incorporate and prioritize UniProt annotations with experimental evidence for presence of signal peptide, GPI anchor or TM domain in a protein sequence over bioinformatic prediction tools (Branch B in Fig. 1). Thirdly, we provide a sub-classification of the secreted extracellular proteins into those sorted by classical pathway and those transported via non-classical pathways. Fourthly, we have used an alternate approach whereby orthologs to secreted proteins with experimental evidence in other fungi is used to predict secretion via non-classical pathways (Branch C in Fig. 1). We remark that our pipeline predicts a much smaller set of secreted proteins via non-classical pathways (albeit with much higher confidence) in comparison to FSD which uses SecretomeP74,75, a tool designed for bacteria and mammals rather than fungi. Lastly, to our knowledge, this is the first study to perform a comparative analysis of both secreted and cell membrane proteins across Aspergillus species causing aspergillosis.
Conclusions
Aspergillosis is a serious concern among immune-compromised patients worldwide. Fungal secretome and cell membrane proteins are often the first point of contact between fungal-host interactions, and thus are key factors in initiating infection or to mounting any defence against the disease. Thus, here, we report a comprehensive set of secreted and cell membrane proteins in Aspergillus fumigatus and nine other Aspergillus species that will serve as a valuable resource for understanding the pathogenesis of aspergillosis. We also identified protein cohorts that could be further screened and targeted for development of novel treatments. To summarize, we have firstly implemented a computational pipeline that integrates data from experiments and bioinformatic-based predictions to identify a comprehensive set of secreted and cell membrane proteins. Secondly, the identified secretome was analyzed for SSPs and effector-like SSPs for correlating their role during pathogenesis. Thirdly, the antigenicity of the secretome and cell membrane proteins were computed to aid in understanding their role in inducing immune responses. Fourthly, the secreted and cell membrane proteins without homologs in humans were scanned for similarity with known drug target proteins in DrugBank to identify new druggable proteins. Lastly, a systematic representative drug repurposing analysis was performed in A. fumigatus responsible for overwhelming majority of the aspergillosis cases. Contextualization of an expression dataset for A. fumigatus within its secretome and cell membrane proteins led to identification of potential targets with over 2-fold upregulation during pathogenesis, and similar to DrugBank target proteins based on both sequence-wise and structure-wise comparison, and able to dock with their respective drug. This led to identification of new potential drug candidates against aspergillosis from existing drugs. The identified secretome and cell membrane proteins in the ten Aspergillus species along with their sub-classification and functional annotations is also hosted at https://cb.imsc.res.in/aspertome for convenient access and retrieval.
Methods
Protein sequences and associated annotations for Aspergillus species
The proteomes of A. fumigatus Af293, A. niger CBS 513.88, A. terreus NIH2624, A. versicolor CBS 583.65, A. lentulus IFM 54703 T, A. nidulans FGSC A4, A. glaucus CBS 516.65 and A. oryzae RIB40 were retrieved from the Aspergillus genome database (AspGD119; http://www.aspgd.org/). The proteomes of A. flavus NRRL3357 and A. ustus 3.3904 were retrieved from FungiDB120 (http://fungidb.org/fungidb/) and Ensembl Genomes121 (http://ensemblgenomes.org/), respectively. Note that the A. ustus 3.3904 genome sequence is still incomplete, and thus, its incomplete proteome was used here for predictions.
Experimentally identified secreted proteins
We performed an extensive literature search to compile 46 published high-throughput experimental studies27–73 which have characterized the secretome of 6 Aspergillus species studied here (Supplementary Table S1). Note that protein identifiers in these experimental studies for the reference strain or other strains of the same Aspergillus species were mapped to standard identifiers in the reference sequence using OrthoMCL100 (http://orthomcl.org/orthomcl/) with inflation option set at 1.5. In addition to high-throughput studies, experimentally verified secreted or cell membrane proteins in Aspergillus species were retrieved from UniProt19 (http://www.uniprot.org/) by filtering proteins with subcellular localization as extracellular or cell membrane, respectively, with evidence code of ECO:0000269. Note that ECO:0000269 corresponds to manually curated information with published experimental evidence. Similarly, experimentally verified secreted proteins in all other fungal species were retrieved from UniProt19 by filtering proteins with subcellular localization as extracellular with evidence code of ECO:0000269. OrthoMCL100 with inflation option set at 1.5 was used to identify orthologs of experimentally verified secreted proteins in other fungi in Aspergillus species.
Bioinformatic-based predictions
The following computational tools were employed to predict protein-based features in our secretome prediction pipeline. The presence of a signal peptide in the N-terminus was predicted using SignalP 4.1122 (http://www.cbs.dtu.dk/services/SignalP/) and Phobius123 (http://phobius.sbc.su.se/). The presence of GPI anchor was predicted using PredGPI124 (http://gpcr.biocomp.unibo.it/predgpi/) and big-PI125 (http://mendel.imp.ac.at/gpi/fungi_server.html). The presence of TM domain was predicted using TMHMM 2.0126 (http://www.cbs.dtu.dk/services/TMHMM/) and Phobius123. Note that TM domain predictions by TMHMM within the last 70 amino acid residues of the N-terminus of a protein sequence were not considered as the tool can sometimes predict signal peptides as false-positive TM domains. The presence of ER retention signal was predicted using PS SCAN127 with PROSITE (https://prosite.expasy.org/) pattern PS00014128. The subcellular localization of proteins was predicted using WoLF PSORT 0.2129 (https://psort.hgc.jp/), TargetP 1.1130 (http://www.cbs.dtu.dk/services/TargetP/) and ProtComp 9131 (http://www.softberry.com/berry.phtml?topic=protcomp&group=help&subgroup=proloc). Importantly, UniProt19 annotations with published experimental evidence (ECO:0000269) for the protein-based features were also retrieved for Aspergillus proteins. UniProt identifiers for Aspergillus proteins were mapped to reference sequence identifiers using pre-existing maps from AspGD119, UniProt19, and in-house python scripts.
While combining information from different sources to decide on the presence of signal peptide or GPI anchor or TM domain, a decision is made based on tool predictions using an inclusive rule if UniProt annotation is not available, else decision is made only on UniProt annotation by overriding tool predictions. While combining information from different sources to decide on subcellular localization, the decision is made based on tool predictions using a majority rule if UniProt annotation is not available, else decision is made only on UniProt annotation by overriding tool predictions. Thus, our pipeline gives precedence to published experimental evidence over bioinformatic-based predictions. Supplementary Table S2 contains tool-based predictions and UniProt annotations for secreted and cell membrane proteins in Aspergillus species.
Note that the computational tools used in our secretome prediction pipeline have been widely-used in the scientific literature. Sperschneider et al.132 have evaluated five bioinformatic tools employed here for predictions in fungi, and have found the sensitivity and specificity of SignalP 4.1122 to be 94.9% and 99.7%, respectively, of Phobius123 to be 95.4% and 98.7%, respectively, of WoLF PSORT 0.2129 to be 88% and 99.8%, respectively, of TargetP 1.1130 to be 95.1% and 98.3%, respectively, and of ProtComp 9131 to be 63.3% and 97.2%, respectively. In addition, Pierleoni et al.124 found the sensitivity and specificity of PredGPI124 to be 77.2% and 99.9%, respectively, and of big-PI125 to be 54.5% and 99.7%, respectively. Note that sensitivity measures the fraction of true positive predictions while specificity measures the fraction of true negative predictions.
Functional annotation of secreted proteins
The predicted secretomes in Aspergillus species were annotated with information on protein family from Pfam133 database (http://pfam.xfam.org/) and carbohydrate-binding modules from CAZy134 (http://www.cazy.org/)134 and dbCAN v5.0135 (http://csbl.bmb.uga.edu/dbCAN/) databases using hmmpress and hmmscan utilities in HMMER3 (http://hmmer.org/) with profile-specific GA (gathering) thresholds. Furthermore, the predicted secretomes in Aspergillus species were annotated with protein family and domain information from TIGRFAM136 (http://www.jcvi.org/cgi-bin/tigrfams/index.cgi), SFLD137 (http://sfld.rbvi.ucsf.edu/django/), SMART138 (http://smart.embl-heidelberg.de/), CDD139(https://www.ncbi.nlm.nih.gov/cdd), PROSITE128, SUPERFAMILY140 (http://supfam.org/SUPERFAMILY/), PRINTS141 (http://130.88.97.239/PRINTS/index.php), PANTHER142 (http://www.pantherdb.org/), COILS143 (https://embnet.vital-it.ch/software/COILS_form.html) and MobiDB-lite144 (http://protein.bio.unipd.it/mobidblite/) accessed through InterPro version 64.0 (https://www.ebi.ac.uk/interpro/) using InterProScan 5145,146. In addition, the secreted proteins in Aspergillus species were annotated with GO terms using FungiFun2147 (https://elbe.hki-jena.de/fungifun/fungifun.php). Supplementary Table S3 contains detailed annotations for secreted and cell membrane proteins in Aspergillus species.
Identification of small secreted and effector-like proteins
In the predicted secretomes of Aspergillus species, SSPs were defined as those with sequence length less than or equal to 300 amino acid residues21,98,148. The SSPs were then evaluated for effector-like properties based on their EffectorP97 (http://effectorp.csiro.au/) predictions or cysteine-richness. For identifying effector-like SSPs, cysteine-rich sequences are those which contain at least 4 cysteine residues and have greater than 5% of their total amino acid residues as cysteines20,149. The predicted SSPs in the secretomes of Aspergillus species were also scanned for known fungal or oomycetes effector motifs96 (DEER, RXLR, RXLX[EDQ], [KRHQSA][DENQ]EL, [YW]XC and RSIVEQD) using the FIMO package in MEME program suite150 with E-value cut-off of ≤1e-4 (Supplementary Table S5).
Antigenicity of secreted proteins
Antigenic regions in Aspergillus proteins were predicted using Kolaskar-Tongaonkar106 method implemented in EMBOSS package151 with a threshold of 1.0 and BepiPred 2.0107 (http://www.cbs.dtu.dk/services/BepiPred/index.php) with the default threshold of 0.5. Note that only predicted antigenic regions with a length ≥6 amino acids were accounted in the later analysis. For each protein in Aspergillus species, the AAR value was computed following the method proposed by Gomez et al.24. The AAR value of a given protein is computed by dividing the length of its amino acid sequence by the number of predicted antigenic regions. The average AAR value was computed for the set of secreted and cell membrane proteins, respectively, in each of the Aspergillus species considered here (Fig. 6). To test whether the computed average AAR values for the set of secreted and cell membrane proteins, respectively, were significantly different from the average AAR value for the whole proteome of the same species, a p-value was computed by comparing against the average AAR values for 1000 equally-sized sets of randomly drawn proteins from the complete proteome of the organism. To test whether the average AAR value for the set of secreted proteins is significantly different from that of the set of cell membrane proteins of an Aspergillus species, Wilcoxon rank-sum test was performed to compare the two sets of different sizes (Fig. 6).
Identification of candidate drug targets in secreted proteins
A microarray dataset for A. fumigatus Af293 from infected murine lungs26 was obtained from Array Express (Accession number E-TABM-327). The microarray dataset from infected murine lungs26 gave a reliable signal for 9121 genes in A. fumigatus Af293 which covers more than 90% of the secretome and 88% of the cell membrane proteins in the fungus. Gene expression data26 for A. fumigatus Af293 was next analyzed within the context of the predicted secreted and cell membrane proteins to identify differentially expressed proteins with over 2-fold upregulation in each fraction during pathogenesis for further analysis. Thereafter, secreted and cell membrane proteins in A. fumigatus Af293 with over 2-fold upregulation during pathogenesis and sequence similarity with known drug target proteins from DrugBank25 database were identified using BLASTP (https://blast.ncbi.nlm.nih.gov/Blast.cgi) with E-value cut-off of ≤1e-5 (as specified by DrugBank database).
To further ascertain the similarity between the filtered set of A. fumigatus proteins in secretome and cell membrane fraction with over 2-fold upregulation during pathogenesis and with sequence similarity with known drug target proteins from DrugBank25, the structural similarity between filtered proteins and their sequence-based homologs among known drug targets was assessed. Structure of proteins from x-ray diffraction experiments were retrieved from PDB111 (https://www.rcsb.org/pdb/home/home.do). However, experimentally identified protein structures are not available for most of the filtered set of secreted and cell membrane proteins in A. fumigatus Af293, and in such cases, the protein structure models from ModBase112 (https://modbase.compbio.ucsf.edu/modbase-cgi/index.cgi) and SWISS-MODEL repository113 (https://swissmodel.expasy.org/repository) were used to evaluate structural similarity. Note that only protein structure models which have more than 40% sequence identity with the template used to generate the corresponding structure models were selected for subsequent analysis. Further the selected protein structure models were checked for their quality using Verify3D152 (http://servicesn.mbi.ucla.edu/Verify3D/) and PROCHECK153 (http://servicesn.mbi.ucla.edu/PROCHECK/) (Supplementary Table S8). Thus, good quality structure models of filtered proteins in A. fumigatus Af293 were compared to the structure of their corresponding sequence-based homolog among the known drug target proteins in DrugBank25 based on TM score computed using TM Align154 (https://zhanglab.ccmb.med.umich.edu/TM-align/) and root-mean-square deviation (RMSD) value between locally aligned atoms computed using PyMOL (The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC). Filtered proteins in A. fumigatus Af293 with significant structural similarity to known drug target proteins were subsequently tested for binding by the respective drug using molecular protein-ligand docking. Molecular docking is a widely used computational approach in structure-based drug design which predicts with substantial accuracy the plausible binding poses and binding affinity of a ligand to the binding site in the target protein of interest155. Here we employ AutoDock Vina to perform rigid docking wherein the target protein of interest is kept rigid while the ligand is flexible. The flexibility of the ligand is determined by torsion tree built while the ligand is prepared for molecular docking using the prepare_ligand4.py script from AutoDockTools156.
Firstly, three ligand binding pockets for each filtered protein in A. fumigatus Af293 were predicted using metaPocket 2.0117 (http://projects.biotec.tu-dresden.de/metapocket/). Secondly, for docking drugs to the predicted protein pockets, both protein and drug molecule were prepared by adding explicit hydrogen atoms and cleaned up using python scripts from AutoDockTools156, prepare_receptor4.py and prepare_ligand4.py respectively, with the default options. Finally, AutoDock Vina118 (http://vina.scripps.edu/) was used for docking the drug against the predicted pockets in proteins with the option for exhaustiveness set at 24 to obtain consistent results157. The binding affinity results obtained by docking drugs to the protein pockets from AutoDock Vina118 were tabulated (Table 2). We would like to note that the docking protocol implemented here is capable of predicting with substantial accuracy the plausible docked poses with their binding affinities reported against the ligand. However, the docking protocol and chosen software have their limitations such as the target protein is kept rigid during our computations and protein structure models are used whenever experimental structures for target proteins are unavailable.
Data availability
All data generated or analyzed during this study is included in this article and its supplementary information.
Electronic supplementary material
Acknowledgements
We thank S. Krishnaswamy and Sankar Basu for discussions, Aashish Shivkumar for help in data collection, and B. Raveendra Reddy and P. Mangalapandi for help with webserver for hosting our dataset. AS would like to acknowledge financial support from the Department of Science and Technology (DST) India through the award of a start-up grant (YSS/2015/000060) and Ramanujan fellowship (SB/S2/RJN-006/2014), and Max Planck Society Germany through the award of a Max Planck Partner Group on Mathematical Biology.
Author Contributions
A.S. conceived the project. A.S., R.P.V., J.P.C. designed research including the computational prediction pipeline. R.P.V., K.M., M.V. compiled and curated data from various sources. R.P.V. implemented the computational prediction pipeline. R.P.V., A.J., J.P.C., A.S. analyzed data. R.P.V., J.P.C., A.S. wrote the manuscript. All authors have read and approved the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-25016-4.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Patterson, T. F. In Essentials of Clinical Mycology (eds Carol A. Kauffman, Peter G. Pappas, Jack D. Sobel, & William E. Dismukes) 243–263 (Springer New York, 2011).
- 2.Paulussen C, et al. Ecology of aspergillosis: insights into the pathogenic potency of Aspergillus fumigatus and some other Aspergillus species. Microbial Biotechnology. 2017;10:296–322. doi: 10.1111/1751-7915.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baddley JW, et al. Patterns of susceptibility of Aspergillus isolates recovered from patients enrolled in the Transplant-Associated Infection Surveillance Network. Journal of clinical microbiology. 2009;47:3271–3275. doi: 10.1128/JCM.00854-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lass‐Flörl C, et al. Epidemiology and outcome of infections due to Aspergillus terreus: 10‐year single centre experience. British journal of haematology. 2005;131:201–207. doi: 10.1111/j.1365-2141.2005.05763.x. [DOI] [PubMed] [Google Scholar]
- 5.Balajee SA, et al. Aspergillus alabamensis, a new clinically relevant species in the section Terrei. Eukaryotic Cell. 2009;8:713–722. doi: 10.1128/EC.00272-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patterson TF, et al. Practice guidelines for the diagnosis and management of aspergillosis: 2016 update by the Infectious Diseases Society of America. Clinical Infectious Diseases. 2016;63:e1–e60. doi: 10.1093/cid/ciw326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin SJ, Schranz J, Teutsch SM. Aspergillosis case-fatality rate: systematic review of the literature. Clin Infect Dis. 2001;32:358–366. doi: 10.1086/318483. [DOI] [PubMed] [Google Scholar]
- 8.Perfect, J. R. & Casadevall, A. In Molecular principles of fungal pathogenesis 3–11 (American Society of Microbiology, 2006).
- 9.Segal BH. Aspergillosis. New England Journal of Medicine. 2009;360:1870–1884. doi: 10.1056/NEJMra0808853. [DOI] [PubMed] [Google Scholar]
- 10.Abad A, et al. What makes Aspergillus fumigatus a successful pathogen? Genes and molecules involved in invasive aspergillosis. Rev Iberoam Micol. 2010;27:155–182. doi: 10.1016/j.riam.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Dagenais TR, Keller NP. Pathogenesis of Aspergillus fumigatus in invasive aspergillosis. Clinical microbiology reviews. 2009;22:447–465. doi: 10.1128/CMR.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kauffman HF, Tomee JF, van de Riet MA, Timmerman AJ, Borger P. Protease-dependent activation of epithelial cells by fungal allergens leads to morphologic changes and cytokine production. J Allergy Clin Immunol. 2000;105:1185–1193. doi: 10.1067/mai.2000.106210. [DOI] [PubMed] [Google Scholar]
- 13.Hohl TM, Feldmesser M. Aspergillus fumigatus: principles of pathogenesis and host defense. Eukaryot Cell. 2007;6:1953–1963. doi: 10.1128/EC.00274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howard SJ, Arendrup MC. Acquired antifungal drug resistance in Aspergillus fumigatus: epidemiology and detection. Med Mycol. 2011;49(Suppl 1):S90–95. doi: 10.3109/13693786.2010.508469. [DOI] [PubMed] [Google Scholar]
- 15.Chowdhary A, Sharma C, Hagen F, Meis JF. Exploring azole antifungal drug resistance in Aspergillus fumigatus with special reference to resistance mechanisms. Future Microbiol. 2014;9:697–711. doi: 10.2217/fmb.14.27. [DOI] [PubMed] [Google Scholar]
- 16.Choi J, et al. Fungal secretome database: integrated platform for annotation of fungal secretomes. BMC Genomics. 2010;11:105. doi: 10.1186/1471-2164-11-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meinken, J. et al. FunSecKB2: a fungal protein subcellular location knowledgebase. Computational Molecular Biology4 (2014).
- 18.Cortázar AR, Aransay AM, Alfaro M, Oguiza JA, Lavín JL. SECRETOOL: integrated secretome analysis tool for fungi. Amino Acids. 2014;46:471–473. doi: 10.1007/s00726-013-1649-z. [DOI] [PubMed] [Google Scholar]
- 19.The UniProt C. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim KT, et al. Kingdom-Wide Analysis of Fungal Small Secreted Proteins (SSPs) Reveals their Potential Role in Host Association. Front Plant Sci. 2016;7:186. doi: 10.3389/fpls.2016.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hacquard S, et al. A comprehensive analysis of genes encoding small secreted proteins identifies candidate effectors in Melampsora larici-populina (poplar leaf rust) Mol Plant Microbe Interact. 2012;25:279–293. doi: 10.1094/MPMI-09-11-0238. [DOI] [PubMed] [Google Scholar]
- 22.Panstruga R, Dodds PN. Terrific protein traffic: the mystery of effector protein delivery by filamentous plant pathogens. Science. 2009;324:748–750. doi: 10.1126/science.1171652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stergiopoulos I, de Wit PJGM. Fungal Effector Proteins. Annual Review of Phytopathology. 2009;47:233–263. doi: 10.1146/annurev.phyto.112408.132637. [DOI] [PubMed] [Google Scholar]
- 24.Gomez S, et al. Genome analysis of Excretory/Secretory proteins in Taenia solium reveals their Abundance of Antigenic Regions (AAR) Sci Rep. 2015;5:9683. doi: 10.1038/srep09683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Law V, et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 2014;42:D1091–1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDonagh A, et al. Sub-telomere directed gene expression during initiation of invasive aspergillosis. PLoS Pathog. 2008;4:e1000154. doi: 10.1371/journal.ppat.1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adav SS, Ravindran A, Sze SK. Proteomic analysis of temperature dependent extracellular proteins from Aspergillus fumigatus grown under solid-state culture condition. J Proteome Res. 2013;12:2715–2731. doi: 10.1021/pr4000762. [DOI] [PubMed] [Google Scholar]
- 28.Adav SS, Ravindran A, Sze SK. Quantitative proteomic study of Aspergillus Fumigatus secretome revealed deamidation of secretory enzymes. J Proteomics. 2015;119:154–168. doi: 10.1016/j.jprot.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 29.Asif AR, et al. Proteome of conidial surface associated proteins of Aspergillus fumigatus reflecting potential vaccine candidates and allergens. J Proteome Res. 2006;5:954–962. doi: 10.1021/pr0504586. [DOI] [PubMed] [Google Scholar]
- 30.Champer, J., Ito, J. I., Clemons, K. V., Stevens, D. A. & Kalkum, M. Proteomic Analysis of Pathogenic Fungi Reveals Highly Expressed Conserved Cell Wall Proteins. J Fungi (Basel) 2, 10.3390/jof2010006 (2016). [DOI] [PMC free article] [PubMed]
- 31.Farnell E, Rousseau K, Thornton DJ, Bowyer P, Herrick SE. Expression and secretion of Aspergillus fumigatus proteases are regulated in response to different protein substrates. Fungal Biol. 2012;116:1003–1012. doi: 10.1016/j.funbio.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim KH, Brown KM, Harris PV, Langston JA, Cherry JR. A proteomics strategy to discover beta-glucosidases from Aspergillus fumigatus with two-dimensional page in-gel activity assay and tandem mass spectrometry. J Proteome Res. 2007;6:4749–4757. doi: 10.1021/pr070355i. [DOI] [PubMed] [Google Scholar]
- 33.Kumar A, Ahmed R, Singh PK, Shukla PK. Identification of virulence factors and diagnostic markers using immunosecretome of Aspergillus fumigatus. J Proteomics. 2011;74:1104–1112. doi: 10.1016/j.jprot.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 34.Liu D, et al. Secretome diversity and quantitative analysis of cellulolytic Aspergillus fumigatus Z5 in the presence of different carbon sources. Biotechnol Biofuels. 2013;6:149. doi: 10.1186/1754-6834-6-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neustadt M, et al. Characterization and identification of proteases secreted by Aspergillus fumigatus using free flow electrophoresis and MS. Electrophoresis. 2009;30:2142–2150. doi: 10.1002/elps.200800700. [DOI] [PubMed] [Google Scholar]
- 36.Ouyang H, Luo Y, Zhang L, Li Y, Jin C. Proteome analysis of Aspergillus fumigatus total membrane proteins identifies proteins associated with the glycoconjugates and cell wall biosynthesis using 2D LC-MS/MS. Mol Biotechnol. 2010;44:177–189. doi: 10.1007/s12033-009-9224-2. [DOI] [PubMed] [Google Scholar]
- 37.Sharma Ghimire P, et al. Insight into Enzymatic Degradation of Corn, Wheat, and Soybean Cell Wall Cellulose Using Quantitative Secretome Analysis of Aspergillus fumigatus. J Proteome Res. 2016;15:4387–4402. doi: 10.1021/acs.jproteome.6b00465. [DOI] [PubMed] [Google Scholar]
- 38.Sharma M, Soni R, Nazir A, Oberoi HS, Chadha BS. Evaluation of glycosyl hydrolases in the secretome of Aspergillus fumigatus and saccharification of alkali-treated rice straw. Appl Biochem Biotechnol. 2011;163:577–591. doi: 10.1007/s12010-010-9064-3. [DOI] [PubMed] [Google Scholar]
- 39.Singh B, et al. Immuno-reactive molecules identified from the secreted proteome of Aspergillus fumigatus. J Proteome Res. 2010;9:5517–5529. doi: 10.1021/pr100604x. [DOI] [PubMed] [Google Scholar]
- 40.Sriranganadane D, et al. Aspergillus protein degradation pathways with different secreted protease sets at neutral and acidic pH. J Proteome Res. 2010;9:3511–3519. doi: 10.1021/pr901202z. [DOI] [PubMed] [Google Scholar]
- 41.Wartenberg D, et al. Secretome analysis of Aspergillus fumigatus reveals Asp-hemolysin as a major secreted protein. Int J Med Microbiol. 2011;301:602–611. doi: 10.1016/j.ijmm.2011.04.016. [DOI] [PubMed] [Google Scholar]
- 42.Mellon JE, Mattison CP, Grimm CC. Aspergillus flavus-secreted proteins during an in vivo fungal-cotton carpel tissue interaction. Physiological and Molecular Plant Pathology. 2015;92:38–41. [Google Scholar]
- 43.Mellon JE, Mattison CP, Grimm CC. Identification of hydrolytic activities expressed by Aspergillus flavus grown on cotton carpel tissue. Physiological and Molecular Plant Pathology. 2016;94:67–74. [Google Scholar]
- 44.Muthu Selvam R, et al. Data set for the mass spectrometry based exoproteome analysis of Aspergillus flavus isolates. Data Brief. 2015;2:42–47. doi: 10.1016/j.dib.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Selvam RM, et al. Exoproteome of Aspergillus flavus corneal isolates and saprophytes: identification of proteoforms of an oversecreted alkaline protease. J Proteomics. 2015;115:23–35. doi: 10.1016/j.jprot.2014.11.017. [DOI] [PubMed] [Google Scholar]
- 46.de Oliveira JM, van Passel MW, Schaap PJ, de Graaff LH. Proteomic analysis of the secretory response of Aspergillus niger to D-maltose and D-xylose. PLoS One. 2011;6:e20865. doi: 10.1371/journal.pone.0020865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Souza WR, et al. Transcriptome analysis of Aspergillus niger grown on sugarcane bagasse. Biotechnol Biofuels. 2011;4:40. doi: 10.1186/1754-6834-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Braaksma M, Martens-Uzunova ES, Punt PJ, Schaap PJ. An inventory of the Aspergillus niger secretome by combining in silico predictions with shotgun proteomics data. BMC Genomics. 2010;11:584. doi: 10.1186/1471-2164-11-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu X, et al. The intra- and extracellular proteome of Aspergillus niger growing on defined medium with xylose or maltose as carbon substrate. Microb Cell Fact. 2010;9:23. doi: 10.1186/1475-2859-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borin GP, et al. Comparative Secretome Analysis of Trichoderma reesei and Aspergillus niger during Growth on Sugarcane Biomass. PLoS One. 2015;10:e0129275. doi: 10.1371/journal.pone.0129275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Florencio C, et al. Secretome data from Trichoderma reesei and Aspergillus niger cultivated in submerged and sequential fermentation methods. Data Brief. 2016;8:588–598. doi: 10.1016/j.dib.2016.05.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krijgsheld P, et al. Spatially resolving the secretome within the mycelium of the cell factory Aspergillus niger. J Proteome Res. 2012;11:2807–2818. doi: 10.1021/pr201157b. [DOI] [PubMed] [Google Scholar]
- 53.Adav SS, Li AA, Manavalan A, Punt P, Sze SK. Quantitative iTRAQ secretome analysis of Aspergillus niger reveals novel hydrolytic enzymes. J Proteome Res. 2010;9:3932–3940. doi: 10.1021/pr100148j. [DOI] [PubMed] [Google Scholar]
- 54.Tsang A, Butler G, Powlowski J, Panisko EA, Baker SE. Analytical and computational approaches to define the Aspergillus niger secretome. Fungal Genet Biol. 2009;46(Suppl 1):S153–S160. doi: 10.1016/j.fgb.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 55.Wang L, et al. Mapping N-linked glycosylation sites in the secretome and whole cells of Aspergillus niger using hydrazide chemistry and mass spectrometry. J Proteome Res. 2012;11:143–156. doi: 10.1021/pr200916k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nitsche BM, Jorgensen TR, Akeroyd M, Meyer V, Ram AF. The carbon starvation response of Aspergillus niger during submerged cultivation: insights from the transcriptome and secretome. BMC Genomics. 2012;13:380. doi: 10.1186/1471-2164-13-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benoit-Gelber I, et al. Mixed colonies of Aspergillus niger and Aspergillus oryzae cooperatively degrading wheat bran. Fungal Genet Biol. 2017;102:31–37. doi: 10.1016/j.fgb.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 58.Couturier M, et al. Post-genomic analyses of fungal lignocellulosic biomass degradation reveal the unexpected potential of the plant pathogen Ustilago maydis. BMC Genomics. 2012;13:57. doi: 10.1186/1471-2164-13-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han MJ, Kim NJ, Lee SY, Chang HN. Extracellular proteome of Aspergillus terreus grown on different carbon sources. Curr Genet. 2010;56:369–382. doi: 10.1007/s00294-010-0308-0. [DOI] [PubMed] [Google Scholar]
- 60.M S, Singh S, Tiwari R, Goel R, Nain L. Do cultural conditions induce differential protein expression: Profiling of extracellular proteome of Aspergillus terreus CM20. Microbiol Res. 2016;192:73–83. doi: 10.1016/j.micres.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 61.Nekiunaite L, Arntzen MO, Svensson B, Vaaje-Kolstad G, Abou Hachem M. Lytic polysaccharide monooxygenases and other oxidative enzymes are abundantly secreted by Aspergillus nidulans grown on different starches. Biotechnol Biofuels. 2016;9:187. doi: 10.1186/s13068-016-0604-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saykhedkar S, et al. A time course analysis of the extracellular proteome of Aspergillus nidulans growing on sorghum stover. Biotechnol Biofuels. 2012;5:52. doi: 10.1186/1754-6834-5-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martins I, et al. Investigating Aspergillus nidulans secretome during colonisation of cork cell walls. J Proteomics. 2014;98:175–188. doi: 10.1016/j.jprot.2013.11.023. [DOI] [PubMed] [Google Scholar]
- 64.Martins I, et al. Elucidating how the saprophytic fungus Aspergillus nidulans uses the plant polyester suberin as carbon source. BMC Genomics. 2014;15:613. doi: 10.1186/1471-2164-15-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rubio MV, et al. Mapping N-linked glycosylation of carbohydrate-active enzymes in the secretome of Aspergillus nidulans grown on lignocellulose. Biotechnol Biofuels. 2016;9:168. doi: 10.1186/s13068-016-0580-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang B, Guan ZB, Cao Y, Xie GF, Lu J. Secretome of Aspergillus oryzae in Shaoxing rice wine koji. Int J Food Microbiol. 2012;155:113–119. doi: 10.1016/j.ijfoodmicro.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 67.Zhu Y, et al. A comparative secretome analysis of industrial Aspergillus oryzae and its spontaneous mutant ZJGS-LZ-21. Int J Food Microbiol. 2017;248:1–9. doi: 10.1016/j.ijfoodmicro.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 68.Oda K, et al. Proteomic analysis of extracellular proteins from Aspergillus oryzae grown under submerged and solid-state culture conditions. Appl Environ Microbiol. 2006;72:3448–3457. doi: 10.1128/AEM.72.5.3448-3457.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liang Y, Pan L, Lin Y. Analysis of extracellular proteins of Aspergillus oryzae grown on soy sauce koji. Biosci Biotechnol Biochem. 2009;73:192–195. doi: 10.1271/bbb.80500. [DOI] [PubMed] [Google Scholar]
- 70.Zhu LY, Nguyen CH, Sato T, Takeuchi M. Analysis of secreted proteins during conidial germination of Aspergillus oryzae RIB40. Biosci Biotechnol Biochem. 2004;68:2607–2612. doi: 10.1271/bbb.68.2607. [DOI] [PubMed] [Google Scholar]
- 71.Te Biesebeke R, Boussier A, van Biezen N, van den Hondel CA, Punt PJ. Identification of secreted proteins of Aspergillus oryzae associated with growth on solid cereal substrates. J Biotechnol. 2006;121:482–485. doi: 10.1016/j.jbiotec.2005.08.028. [DOI] [PubMed] [Google Scholar]
- 72.Imanaka H, Tanaka S, Feng B, Imamura K, Nakanishi K. Cultivation characteristics and gene expression profiles of Aspergillus oryzae by membrane-surface liquid culture, shaking-flask culture, and agar-plate culture. J Biosci Bioeng. 2010;109:267–273. doi: 10.1016/j.jbiosc.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 73.Eugster PJ, Salamin K, Grouzmann E, Monod M. Production and characterization of two major Aspergillus oryzae secreted prolyl endopeptidases able to efficiently digest proline-rich peptides of gliadin. Microbiology. 2015;161:2277–2288. doi: 10.1099/mic.0.000198. [DOI] [PubMed] [Google Scholar]
- 74.Bendtsen JD, Jensen LJ, Blom N, Von Heijne G, Brunak S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng Des Sel. 2004;17:349–356. doi: 10.1093/protein/gzh037. [DOI] [PubMed] [Google Scholar]
- 75.Bendtsen JD, Kiemer L, Fausboll A, Brunak S. Non-classical protein secretion in bacteria. BMC Microbiol. 2005;5:58. doi: 10.1186/1471-2180-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kwon-Chung KJ, Sugui JA. What do we know about the role of gliotoxin in the pathobiology of Aspergillus fumigatus? Medical Mycology. 2009;47:S97–S103. doi: 10.1080/13693780802056012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Spikes S, et al. Gliotoxin Production in Aspergillus fumigatus Contributes to Host-Specific Differences in Virulence. The Journal of Infectious Diseases. 2008;197:479–486. doi: 10.1086/525044. [DOI] [PubMed] [Google Scholar]
- 78.Lewis RE, et al. Detection of gliotoxin in experimental and human aspergillosis. Infection and Immunity. 2005;73:635–637. doi: 10.1128/IAI.73.1.635-637.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sugui JA, et al. Gliotoxin Is a Virulence Factor of Aspergillus fumigatus: gliP Deletion Attenuates Virulence in Mice Immunosuppressed with Hydrocortisone. Eukaryotic Cell. 2007;6:1562–1569. doi: 10.1128/EC.00141-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dichtl K, Helmschrott C, Dirr F, Wagener J. Deciphering cell wall integrity signalling in Aspergillus fumigatus: identification and functional characterization of cell wall stress sensors and relevant Rho GTPases. Mol Microbiol. 2012;83:506–519. doi: 10.1111/j.1365-2958.2011.07946.x. [DOI] [PubMed] [Google Scholar]
- 81.Malvezzi M, et al. Ca2+-dependent phospholipid scrambling by a reconstituted TMEM16 ion channel. Nat Commun. 2013;4:2367. doi: 10.1038/ncomms3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Apostolaki A, et al. AgtA, the dicarboxylic amino acid transporter of Aspergillus nidulans, is concertedly down-regulated by exquisite sensitivity to nitrogen metabolite repression and ammonium-elicited endocytosis. Eukaryot Cell. 2009;8:339–352. doi: 10.1128/EC.00270-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Athanasopoulos A, Gournas C, Amillis S, Sophianopoulou V. Characterization of AnNce102 and its role in eisosome stability and sphingolipid biosynthesis. Sci Rep. 2015;5:15200. doi: 10.1038/srep15200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Calcagno-Pizarelli AM, et al. Establishment of the ambient pH signaling complex in Aspergillus nidulans: PalI assists plasma membrane localization of PalH. Eukaryot Cell. 2007;6:2365–2375. doi: 10.1128/EC.00275-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Colabardini AC, et al. Functional characterization of a xylose transporter in Aspergillus nidulans. Biotechnol Biofuels. 2014;7:46. doi: 10.1186/1754-6834-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Forment JV, Flipphi M, Ramon D, Ventura L, Maccabe AP. Identification of the mstE gene encoding a glucose-inducible, low affinity glucose transporter in Aspergillus nidulans. J Biol Chem. 2006;281:8339–8346. doi: 10.1074/jbc.M508198200. [DOI] [PubMed] [Google Scholar]
- 87.Soid-Raggi G, Sanchez O, Aguirre J. TmpA, a member of a novel family of putative membrane flavoproteins, regulates asexual development in Aspergillus nidulans. Mol Microbiol. 2006;59:854–869. doi: 10.1111/j.1365-2958.2005.04996.x. [DOI] [PubMed] [Google Scholar]
- 88.Valdez-Taubas J, Diallinas G, Scazzocchio C, Rosa AL. Protein expression and subcellular localization of the general purine transporter UapC from Aspergillus nidulans. Fungal Genet Biol. 2000;30:105–113. doi: 10.1006/fgbi.2000.1197. [DOI] [PubMed] [Google Scholar]
- 89.Zhang S, et al. FigA, a putative homolog of low-affinity calcium system member Fig. 1 in Saccharomyces cerevisiae, is involved in growth and asexual and sexual development in Aspergillus nidulans. Eukaryot Cell. 2014;13:295–303. doi: 10.1128/EC.00257-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Erpapazoglou Z, Kafasla P, Sophianopoulou V. The product of the SHR3 orthologue of Aspergillus nidulans has restricted range of amino acid transporter targets. Fungal Genet Biol. 2006;43:222–233. doi: 10.1016/j.fgb.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 91.Flipphi M, et al. Functional analysis of alcS, a gene of the alc cluster in Aspergillus nidulans. Fungal Genet Biol. 2006;43:247–260. doi: 10.1016/j.fgb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 92.Pantazopoulou A, et al. Differential physiological and developmental expression of the UapA and AzgA purine transporters in Aspergillus nidulans. Fungal Genet Biol. 2007;44:627–640. doi: 10.1016/j.fgb.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 93.Takeshita N, Ohta A, Horiuchi H. CsmA, a class V chitin synthase with a myosin motor-like domain, is localized through direct interaction with the actin cytoskeleton in Aspergillus nidulans. Mol Biol Cell. 2005;16:1961–1970. doi: 10.1091/mbc.E04-09-0761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Takeshita N, Yamashita S, Ohta A, Horiuchi H. Aspergillus nidulans class V and VI chitin synthases CsmA and CsmB, each with a myosin motor-like domain, perform compensatory functions that are essential for hyphal tip growth. Mol Microbiol. 2006;59:1380–1394. doi: 10.1111/j.1365-2958.2006.05030.x. [DOI] [PubMed] [Google Scholar]
- 95.Higuchi Y, Nakahama T, Shoji JY, Arioka M, Kitamoto K. Visualization of the endocytic pathway in the filamentous fungus Aspergillus oryzae using an EGFP-fused plasma membrane protein. Biochem Biophys Res Commun. 2006;340:784–791. doi: 10.1016/j.bbrc.2005.12.077. [DOI] [PubMed] [Google Scholar]
- 96.Sonah H, Deshmukh RK, Belanger RR. Computational Prediction of Effector Proteins in Fungi: Opportunities and Challenges. Front Plant Sci. 2016;7:126. doi: 10.3389/fpls.2016.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sperschneider J, et al. EffectorP: predicting fungal effector proteins from secretomes using machine learning. New Phytol. 2016;210:743–761. doi: 10.1111/nph.13794. [DOI] [PubMed] [Google Scholar]
- 98.Valette N, et al. Secretion of small proteins is species-specific within Aspergillus sp. Microb Biotechnol. 2017;10:323–329. doi: 10.1111/1751-7915.12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lowe RG, Howlett BJ. Indifferent, affectionate, or deceitful: lifestyles and secretomes of fungi. PLoS Pathog. 2012;8:e1002515. doi: 10.1371/journal.ppat.1002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fischer, S. et al. In Current Protocols in Bioinformatics (John Wiley & Sons, Inc., 2002).
- 101.Samal A, et al. Network reconstruction and systems analysis of plant cell wall deconstruction by Neurospora crassa. Biotechnol Biofuels. 2017;10:225. doi: 10.1186/s13068-017-0901-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kniemeyer O. Proteomics of eukaryotic microorganisms: The medically and biotechnologically important fungal genus Aspergillus. Proteomics. 2011;11:3232–3243. doi: 10.1002/pmic.201100087. [DOI] [PubMed] [Google Scholar]
- 103.Bozza S, et al. Immune sensing of Aspergillus fumigatus proteins, glycolipids, and polysaccharides and the impact on Th immunity and vaccination. The Journal of immunology. 2009;183:2407–2414. doi: 10.4049/jimmunol.0900961. [DOI] [PubMed] [Google Scholar]
- 104.Bozza S, et al. Vaccination of mice against invasive aspergillosis with recombinant Aspergillus proteins and CpG oligodeoxynucleotides as adjuvants. Microbes and infection. 2002;4:1281–1290. doi: 10.1016/s1286-4579(02)00007-2. [DOI] [PubMed] [Google Scholar]
- 105.Ito JI, Lyons JM, Diaz-Arevalo D, Hong TB, Kalkum M. Vaccine progress. Medical mycology. 2009;47:S394–S400. doi: 10.1080/13693780802552614. [DOI] [PubMed] [Google Scholar]
- 106.Kolaskar AS, Tongaonkar PC. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990;276:172–174. doi: 10.1016/0014-5793(90)80535-q. [DOI] [PubMed] [Google Scholar]
- 107.Jespersen, M. C., Peters, B., Nielsen, M. & Marcatili, P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res, 10.1093/nar/gkx346 (2017). [DOI] [PMC free article] [PubMed]
- 108.Wang S, Wei W, Cai X. Genome-wide analysis of excretory/secretory proteins in Echinococcus multilocularis: insights into functional characteristics of the tapeworm secretome. Parasit Vectors. 2015;8:666. doi: 10.1186/s13071-015-1282-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cornejo-Granados F, et al. Secretome Prediction of Two M. tuberculosis Clinical Isolates Reveals Their High Antigenic Density and Potential Drug Targets. Front Microbiol. 2017;8:128. doi: 10.3389/fmicb.2017.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jin G, Wong ST. Toward better drug repositioning: prioritizing and integrating existing methods into efficient pipelines. Drug Discov Today. 2014;19:637–644. doi: 10.1016/j.drudis.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Berman HM, et al. The Protein Data Bank. Nucleic Acids Research. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pieper U, et al. ModBase, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2014;42:D336–346. doi: 10.1093/nar/gkt1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bienert S, et al. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 2017;45:D313–D319. doi: 10.1093/nar/gkw1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lessing F, et al. The Aspergillus fumigatus transcriptional regulator AfYap1 represents the major regulator for defense against reactive oxygen intermediates but is dispensable for pathogenicity in an intranasal mouse infection model. Eukaryot Cell. 2007;6:2290–2302. doi: 10.1128/EC.00267-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Owens RA, et al. Interplay between Gliotoxin Resistance, Secretion, and the Methyl/Methionine Cycle in Aspergillus fumigatus. Eukaryot Cell. 2015;14:941–957. doi: 10.1128/EC.00055-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shi LN, et al. Immunoproteomics based identification of thioredoxin reductase GliT and novel Aspergillus fumigatus antigens for serologic diagnosis of invasive aspergillosis. BMC Microbiol. 2012;12:11. doi: 10.1186/1471-2180-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang Z, Li Y, Lin B, Schroeder M, Huang B. Identification of cavities on protein surface using multiple computational approaches for drug binding site prediction. Bioinformatics. 2011;27:2083–2088. doi: 10.1093/bioinformatics/btr331. [DOI] [PubMed] [Google Scholar]
- 118.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cerqueira GC, et al. The Aspergillus Genome Database: multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic Acids Res. 2014;42:D705–710. doi: 10.1093/nar/gkt1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Stajich JE, et al. FungiDB: an integrated functional genomics database for fungi. Nucleic Acids Res. 2012;40:D675–681. doi: 10.1093/nar/gkr918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kersey, P. J. et al. Ensembl Genomes 2018: an integrated omics infrastructure for non-vertebrate species. Nucleic Acids Research, gkx1011–gkx1011, 10.1093/nar/gkx1011 (2017). [DOI] [PMC free article] [PubMed]
- 122.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 123.Kall L, Krogh A, Sonnhammer EL. Advantages of combined transmembrane topology and signal peptide prediction–the Phobius web server. Nucleic Acids Res. 2007;35:W429–432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pierleoni A, Martelli PL, Casadio R. PredGPI: a GPI-anchor predictor. BMC Bioinformatics. 2008;9:392. doi: 10.1186/1471-2105-9-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Eisenhaber B, Bork P, Eisenhaber F. Prediction of Potential GPI-modification Sites in Proprotein Sequences. Journal of Molecular Biology. 1999;292:741–758. doi: 10.1006/jmbi.1999.3069. [DOI] [PubMed] [Google Scholar]
- 126.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 127.Gattiker A, Gasteiger E, Bairoch A. ScanProsite: a reference implementation of a PROSITE scanning tool. Appl Bioinformatics. 2002;1:107–108. [PubMed] [Google Scholar]
- 128.Sigrist CJA, et al. New and continuing developments at PROSITE. Nucleic Acids Research. 2013;41:D344–D347. doi: 10.1093/nar/gks1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Horton P, et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 131.Klee EW, Ellis LB. Evaluating eukaryotic secreted protein prediction. BMC Bioinformatics. 2005;6:256. doi: 10.1186/1471-2105-6-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sperschneider J, Williams AH, Hane JK, Singh KB, Taylor JM. Evaluation of Secretion Prediction Highlights Differing Approaches Needed for Oomycete and Fungal Effectors. Front Plant Sci. 2015;6:1168. doi: 10.3389/fpls.2015.01168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Finn RD, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Research. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yin Y, et al. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Research. 2012;40:W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Haft DH, et al. TIGRFAMs and Genome Properties in 2013. Nucleic Acids Research. 2013;41:D387–D395. doi: 10.1093/nar/gks1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Akiva E, et al. The Structure–Function Linkage Database. Nucleic Acids Research. 2014;42:D521–D530. doi: 10.1093/nar/gkt1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Letunic, I. & Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Research, gkx922–gkx922 10.1093/nar/gkx922 (2017). [DOI] [PMC free article] [PubMed]
- 139.Marchler-Bauer A, et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45:D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 141.Attwood TK, et al. The PRINTS database: a fine-grained protein sequence annotation and analysis resource–its status in 2012. Database (Oxford) 2012;2012:bas019. doi: 10.1093/database/bas019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mi H, et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 144.Necci, M., Piovesan, D., Dosztanyi, Z. & Tosatto, S. C. MobiDB-lite: Fast and highly specific consensus prediction of intrinsic disorder in proteins. Bioinformatics, 10.1093/bioinformatics/btx015 (2017). [DOI] [PubMed]
- 145.Finn RD, et al. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017;45:D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jones P, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Priebe S, Kreisel C, Horn F, Guthke R, Linde J. FungiFun2: a comprehensive online resource for systematic analysis of gene lists from fungal species. Bioinformatics. 2015;31:445–446. doi: 10.1093/bioinformatics/btu627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Feldman D, Kowbel DJ, Glass NL, Yarden O, Hadar Y. A role for small secreted proteins (SSPs) in a saprophytic fungal lifestyle: Ligninolytic enzyme regulation in Pleurotus ostreatus. Sci Rep. 2017;7:14553. doi: 10.1038/s41598-017-15112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Krijger JJ, Thon MR, Deising HB, Wirsel SG. Compositions of fungal secretomes indicate a greater impact of phylogenetic history than lifestyle adaptation. BMC Genomics. 2014;15:722. doi: 10.1186/1471-2164-15-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 152.Eisenberg, D., Lüthy, R. & Bowie, J. U. In Methods in Enzymology Vol. 277, 396–404 (Academic Press, 1997). [DOI] [PubMed]
- 153.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- 154.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ferreira LG, dos Santos RN, Oliva G, Andricopulo AD. Molecular docking and structure-based drug design strategies. Molecules. 2015;20:13384–13421. doi: 10.3390/molecules200713384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Morris GM, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Forli S, et al. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc. 2016;11:905–919. doi: 10.1038/nprot.2016.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study is included in this article and its supplementary information.