Abstract
The aim of this study was to investigate the function of microRNA (miR)-193-3p in human intrahepatic cholangiocarcinoma (ICC) tissues and cells. To evaluate whether miR-193-3p was aberrantly upregulated, we used reverse transcription-quantitative polymerase chain reaction to detect the level of miR-193-3p in ICC tissues and ICC-9810 cells. The effects of miR-193-3p downregulation on ICC cell proliferation, migration and invasion were also measured by MTT, wound-healing and Transwell assays. Additionally, transforming growth factor-β receptor type 3 (TGFBR3) was investigated as a direct target of miR-193-3p by dual-luciferase reporter assays and western blot analyses. The results demonstrated that miR-193-3p was aberrantly upregulated in ICC tissues and cell lines. Furthermore, TGFBR3 was confirmed to be a target of miR-193-3p in ICC and was notably upregulated by miR-193-3p knockdown in the ICC-9810 cells. The knockdown of miR-193-3p also exerted direct inhibitory effects on the proliferation, migration and invasion of the ICC-9810 cells. Therefore, we present evidence that miR-193-3p plays a key role in promoting human ICC by regulating TGFBR3.
Keywords: intrahepatic cholangiocarcinoma, microRNA-193-3p, transforming growth factor-β receptor III, migration, invasion
Introduction
Intrahepatic cholangiocarcinoma (ICC) represents the second most common primary hepatic malignancy, accounting for 5–10% of liver cancers (1). The incidence rate of ICC is relatively low in most populations, including those of the the USA and Europe, although is considerably higher in China.
MicroRNAs (miRNA/miRs), a class of non-protein-coding RNAs, are short ribonucleic acid molecules of approximately 17–25 nucleotides in length that typically bind to the 3′ untranslated regions (3′UTRs) of target mRNAs to regulate their expression levels (2). miRNA are important regulators of gene expression, and the expression levels of miRNAs have been closely associated with cell growth, proliferation, and development, as well as the occurrence and progression of several diseases (3). miRNAs also regulate cell apoptosis, migration, invasion, proliferation, and tumorigenesis by regulating target genes (4). Human miR-193-3p has been demonstrated to be upregulated in ICC cells; however, the detailed functional mechanisms of miR-193-3p in ICC remain unclear.
The transforming growth factor-β (TGF-β) receptor type 3 (TGFBR3) serves as an important tumor-suppressive factor in many cancer types. For instance, TGFBR3 has been reported to suppresse breast cancer progression by inhibiting cell migration and invasion, as well as angiogenesis. Additionally, loss or reduced levels of TGFBR3 expression have been identified in many types of human cancer, including non-small cell lung cancer (NSCLC), ovarian, prostate, pancreatic, breast and renal cell carcinoma, and overexpression of TGFBR3 can lead to inhibited cancer cell migration and invasion. Therefore, it has been conclusded that TGFBR3 might play a suppressive role in the progression of several cancers (5–10). However, Gatza et al (11) reported that TGFBR3 was markedly upregulated in human colon cancer and may advance the development of colon cancer to a certain degree. These findings indicated that TGFBR3 might play a dual role in the formation of tumors.
TGFBR3 is an accessory receptor that binds and modulates the activity of TGF-β, and these two members of the TGF-β growth factor superfamily regulate many of the functions in reproductive biology (12). Furthermore, a recent study showed that downregulation of TGF-β signaling in cholangiocytes promoted cholangiocarcinoma by increasing cholangiocyte proliferation, which suggests the role of TGF-β signaling in cholangiocarcinoma (13).
In the present study, we explored the expression of miR-193-3p in human ICC tissues and cells and determined whether downregulation of miR-193-3p could exert inhibitory effects on ICC cell proliferation, migration, and invasion. In addition, miR-193-3p was upregulated to investigate its direct effect on TGFBR3 levels in ICC. Thus, this study aimed to investigate the roles of miR-193-3p in ICC, and to verify whether TGFBR3 is a downstream target gene of miR-193-3p in ICC.
Materials and methods
ICC clinical samples and grouping
Paired ICC tissues and adjacent normal tissues were surgically obtained from 17 ICC patients who underwent surgery at the Department of Hepatobilliary Laparoscopic Surgery Ward Affilated Zhongshan Hospital of Dalian University (Dalian, China) from May 2014 to December 2016. All tissues were obtained from patients who did not receive adjuvant treatments in order to avoid treatment-induced changes in gene expression. The normal tissues were sampled from ≥3 cm away from the visible edge of the tumor tissues. After extraction, all tissue samples were immediately frozen in liquid nitrogen and stored at −80°C until subsequent RNA isolation. 17 cases of ICC patients with paraffin specimens, including 10 males and 7 females, aged 41 to 73 years, the median age of onset of 61 years old. In accordance with the 2014 American Hepatobiliary and Pancreatic Society consensus statement: ICC management (14), clinical staging criteria are divided into: stage II 4 cases, III stage 7 cases, IV stage 6 cases. In our study, all experimental protocols were carried out in accordance to the principles of the Declaration of Helsinki as well as approved by the Ethics Committee of Affilated Zhongshan Hospital of Dalian University. The informed consent of the ICC samples was obtained from all patients of the study. The follow-up ranged from 1 to 28 months with a median of 15 months.
Cell lines and culture
The human ICC-9810 cell line and normal human intrahepatic biliary epithelial cell (HIBEC) line were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA) and were maintained in 25 cm2 flasks containing DMEM (ATCC® 30–2006™; ATCC, Shanghai, China) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in an incubator at 5% CO2 and 37°C.
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from the tissue samples and cell lines with an RNeasy mini kit (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's instructions, and cDNA synthesis was performed with a reverse transcription kit (Promega Corporation, Madison, WI, USA). The mRNA expression levels of miR-193-3p, TGFBR3, TGF-β and GAPDH (internal control) were determined with a Power SYBR-Green Real-Time PCR Master Mix Kit (Takara Bio, Inc., Otsu, Japan). The following primers were used for PCR: For miR-193-3p, 5′-AACTGGCCTACAAAGTCCCAGT-3′; for TGFBR3 forward, 5′-CCTTCCGTTTCCTTTCCCAGA-3′ and reverse, 5′-CACATTTGACAGACAGGGCAAT-3′ (product size, 170 bp); for TGF-β forward, 5′-CAATTCCTGGCGATACCTCAG-3′ and reverse, 5′-GCACAACTCCGGTGACATCAA-3′ (product size, 86 bp); GAPDH forward, 5′-ACAACTTTGGTATCGTGGAAGG-3′ and reverse, 5′-GCCATCACGCCACAGTTTC-3′ (product size, 101 bp). All RT-qPCR reactions were performed in a BIO-RAD CFX96™ system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The relative expression levels were quantified using the 2−ΔΔCq method (15). All RT-qPCR reactions were repeated three times.
Cell transfection and grouping
The function of miR-193-3p in ICC-9810 cells was assessed to determine its role in the development of ICC. The cells were transfected with miR-193-3p inhibitor or control-miR-193-3p inhibitor (Shanghai GenePharma Co., Ltd.,. Shanghai, China) using 30 µl Lipofectamine® 2000 transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturers' instructions. Following incubation for 48 h, the transfected ICC-9810 cells were subjected to RT-qPCR as above to verify the miR-193-3p knockdown, and then used in subsequent assays. The groups were designed as follows: HIBEC group, ICC-9810 group, negative control (NC) group, miR-193-3p inhibitor group and miR-193-3p inhibitor control group.
MTT cell proliferation assay
The viability of ICC-9810 cells was measured with an MTT assay. After the transfection was stabilized, ICC-9810 cells were plated in 96-well plates at a density of 1×105 cells/well. Every 24 h between days 1 and day 3 of culture, 20 µl 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution (5 mg/Ml; Invitrogen; Thermo Fisher Scientific, Inc.) was added to each experimental well, after which the plates were returned to the incubator and maintainedat 37°C in a humidified atmosphere of 5% CO2 for 4 h. Following aspiration of the supernatant, 200 µl dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was added to each well to dissolve the formazan crystals. Cell viability was subsequently analyzed at a wavelength of 570 nm using a micro plate reader.
Dual-luciferase reporter gene assay
For the luciferase assays, a pGL3 firefly luciferase reporter gene vector (30 ng/Ml; Promega Corporation) with the 3′UTR-WT or 3′UTR-MUT fragment of human TGFBR3 Cdna, containing the putative target site for miR-193-3p, was co-transfected with 50 nM miR-193-3p or miR-NC into ICC-9810 cells. A Renilla luciferase construct was also co-transfected to normalize the transfection efficiency. After 24 h, a Dual-Luciferase Reporter Assay System (Promega Corporation) was used to detect the relative luciferase activities, according to the manufacturer's instructions, under a FL500 microplate fluorescence reader (Omega Bio-Tek, Inc., Norcross, GA, USA). More than three independent experiments were performed.
Wound-healing migration assay
The migratory ability of ICC-9810 cells was measured with a Wound healing assay. The ICC-9810 cells were plated into 6-well plates (25,000 cells/well) with 2 ml of cell suspension in each well. When cell confluence reached 80%, a sterile Eppendorf pipette tip was used to scratch the cell surface to create a wound area. Following removal of the debris, the culture was replenished with fresh medium and incubated under 5% CO2 at 37°C for 24 h. The migration of the ICC-9810 cells into the wound area was detected under phase contrast objectives (×10) on a CK2 inverted microscope (DM16000; Leica Microsystems GmbH, Wetzlar, Germany) (16).
Transwell assay
The invasion of ICC-9810 cells was measured using a Transwell assay. The upper membrane surface was coated with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) and 500 µl RPMI-1640 medium containing 10% FBS was added into the lower chambers. Serum-free DMEM and ICC-9810 cell suspension (1×105 cells) were placed in the upper chambers. After culturing for 48 h, 4% paraformaldehyde and 0.05% crystal violet were used to fix and stain the invaded cells, respectively, and a cotton swab was used to remove the non-invaded cells in the upper chamber. A microscope (×100) was subsequently used to count the numbers of cells in 10 random fields in order to estimate the relative invasiveness of cells in the different groups (16).
Western blot assay
The relative protein expression levels of TGFBR3 and TGF-β in the tissue samples and cell lines were detected by western blotting. The total protein content of the tissues and cells was harvested using radio-immunoprecipitation assay buffer (RIPA Buffer; BioVision, Inc., Milpitas, CA, USA), and protein concentration was determined by the BCA method. Subsequently, total protein (40 µg/sample) was separated by 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes (Thermo Fisher Scientific, Inc.). The membranes were blocked with 10% nonfat milk in Tris-buffered saline with Tween (TBST; Invitrogen; Thermo Fisher Scientific, Inc.) for 2 h at room temperature, and then incubated with primary antibodies against TGFBR3 (ab78421), TGF-β (ab220084) and GAPDH (ab8245; all from Abcam Cambridge, UK) overnight at 4°C. Subsenquently, the membranes were washed with TBST and incubated with horseradish peroxidase-coujugated secondary antibodies for 2 h at room temperature. The blots were visualized using an enhanced chemiluminescence film kit (New England Nuclear, Boston, MA, USA) according to the manufacturer's instructions, and Image J software was used to analyse protein density.
Statistical analysis
All data were expressed as the mean ± standard deviation of at least three independent experiments. All statistical analyses was performed using SPSS version 19.0 statistical software (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism 6.0 software (GraphPad Software, Inc., La Jolla, CA, USA). A Student's t-test was used to evaluate the statistical significance of difference between two groups, whiledifferences between multiple groups were assessed by one-way analysis of variance followed by the Dunnett's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of miR-193-3p, TGFBR3 and TGF-β in the tissues of ICC patients
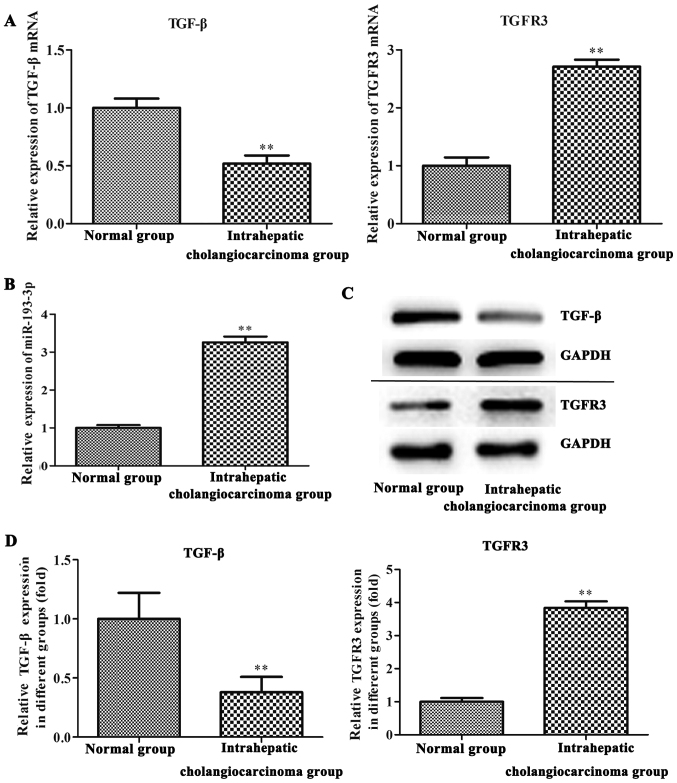
On RT-qPCR and western blot analysis, the expression levels of miR-193-3p and TGFBR3 were found to be increased in tumor tissue samples of ICC patients when compared with adjacent normal tissues (P<0.01; Fig. 1). By contrast, TGF-β was significantly downregulated in the ICC tissues of the patients (P<0.01; Fig. 1).
Figure 1.
The expression of miR-193-3p, TGFBR3 and TGF-β in ICC patients tissue. (A) The mRNA expression of TGFBR3 and TGF-β in the tissues of ICC patients was validated by RT-qPCR. **P<0.01, vs. normal group and ICC group. (B) RT-qPCR analysis of miR-193-3p expression in tissues of the ICC patients. **P<0.01, vs. normal group and ICC group. (C) The expression of TGFBR3 and TGF-β at the protein level in the tissues of ICC patients. (D) Levels of TGFBR3 and TGF-β in normal groups and ICC groups. **P<0.01, vs. normal group and ICC group. TGFBR3, transforming growth factor-β receptor III; TGF-β, transforming growth factor-β; ICC, intrahepatic cholangiocarcinoma; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; miR, microRNA.
miR-193-3p expression is upregulated in the ICC-9810 cell line
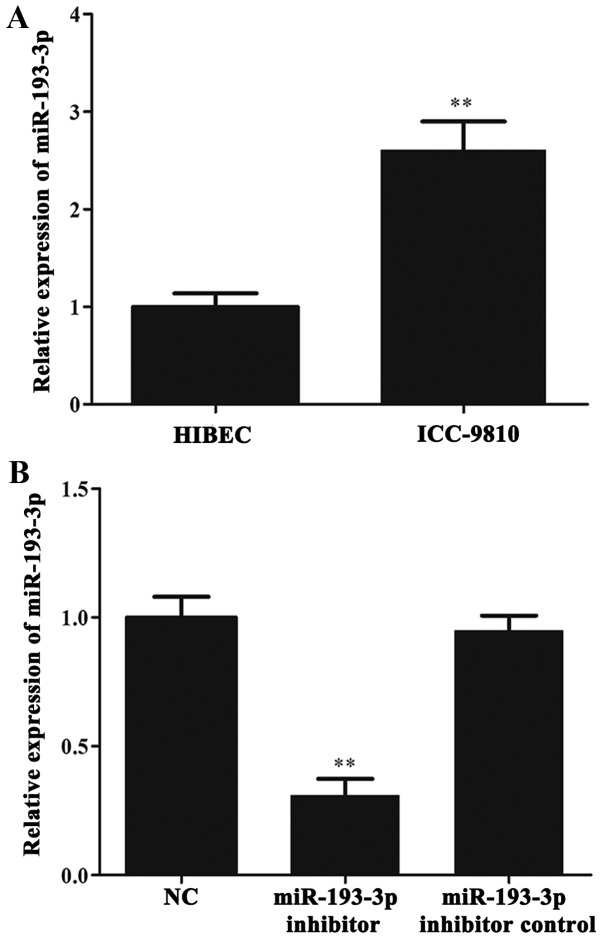
RT-qPCR was used to investigate whether miR-193-3p was aberrantly expressed in ICC cells. First, we determined the level of miR-193-3p in ICC-9810 and HIBEC cells. The results of RT-qPCR showed that miR-193-3p was significantly upregulated in the ICC-9810 cells when compared with the HIBEC cells (P < 0.01; Fig. 2A). Subsequently, the levels of miR-193-3p in the NC group, miR-193-3p inhibitor group and miR-193-3p inhibitor control group were detected by RT-qPCR. The results indicated a marked reduction in miR-193-3p levels in the miR-193-3p inhibitor group when compared with the NC and miR-193-3p inhibitor control groups (P<0.01; Fig. 2B). Meanwhile, there was no significant difference in the level of miR-193-3p between the NC and miR-193-3p inhibitor control groups.
Figure 2.
The expression of miR-193-3p in ICC-9810 cell lines. (A) RT-qPCR was carried out to compare miR-193-3p expression levels between ICC-9810 cell line and HIBEC cell line. **P<0.01, vs. HIBEC group and ICC-9810 group. (B) miR-193-3p expression in NC group, miR-193-3p inhibitor group and miR-193-3p inhibitor control group. **P<0.01, vs. miR-193-3p inhibitor group and NC group; P>0.05, vs. miR-193-3p inhibitor control group and NC group. ICC, intrahepatic cholangiocarcinoma; HIBEC, human intrahepatic biliary epithelial cell; miR, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; NC, negative control.
Expression of TGFBR3 and TGF-β in the ICC-9810 cell line
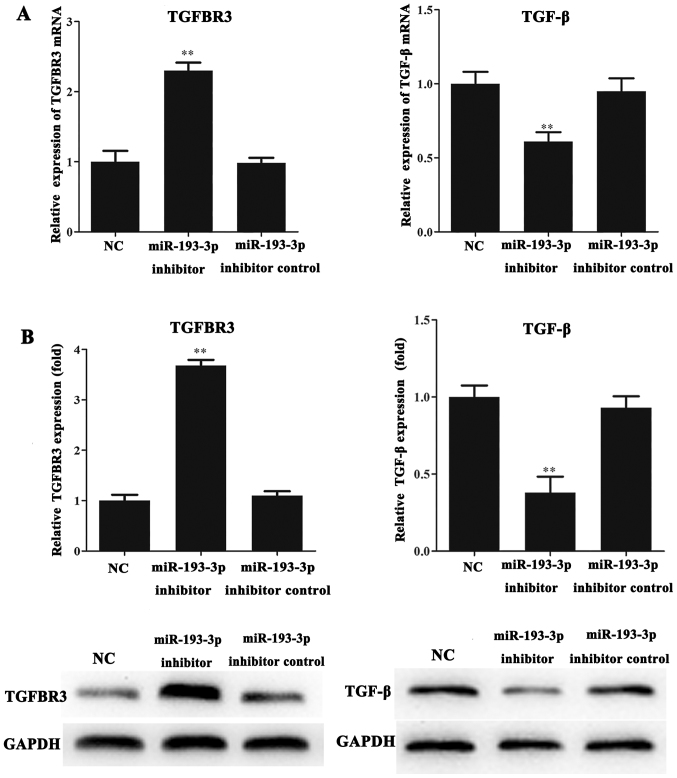
The mRNA and protein levels of TGFBR3 and TGF-β in the ICC-9810 cells were detected by RT-qPCR and western blot analysis. The results indicated that the mRNA and protein levels of TGFBR3 in the miR-193-3p inhibitor group were significantly increased compared with those in the NC and miR-193-3p inhibitor control groups (P<0.01; Fig. 3A and B). Conversely, we found that the mRNA and protein expression levels of TGF-β in the miR-193-3p inhibitor group were lower than that in the NC and miR-193-3p inhibitor control groups (P<0.01; Fig. 3A and B).
Figure 3.
The expression of TGFBR3 and TGF-β in ICC-9810 cell lines. (A) The mRNA expression of TGFBR3 and TGF-β in ICC-9810 cell line. TGFBR3 is upregulated in miR-193-3p inhibitor group (**P<0.01 vs. miR-193-3p inhibitor group and NC group), but TGF-β is downregulated in miR-193-3p inhibitor group (**P < 0.01 vs. miR-193-3p inhibitor group and NC group). (B) Western blottiong was used to compare TGFBR3 and TGF-β protein expression levels in NC group, miR-193-3p inhibitor group and miR-193-3p inhibitor control group, the result is consistent with the RT-qPCR results. **P<0.01, vs. miR-193-3p inhibitor group and NC group. TGFBR3, transforming growth factor-β receptor III; TGF-β, transforming growth factor-β; ICC, intrahepatic cholangiocarcinoma; miR, microRNA; NC, negative control.
ICC-9810 cell proliferation was repressed following miR-193-3p inhibition
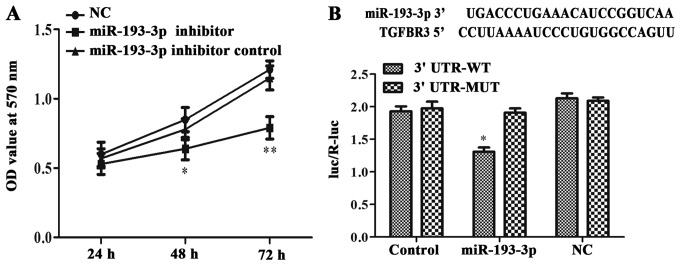
The MTT assays results showed that the viability of ICC-9810 cells transfected with miR-193-3p inhibitor was lower than that of cells in the NC and miR-193-3p inhibitor control groups at 48 h (P<0.05; Fig. 4A) and at 72 h (P<0.01; Fig. 4A). Thus, the inhibition of miR-193-3p was hinder the proliferation of the ICC-9810 cells.
Figure 4.
miR-193-3p affected ICC-9810 cell lines proliferation and directly targeted TGFBR3 and suppressed its expression. (A) In ICC-9810 cells transduced with miR-193-3p inhibitor, a three-day MTT assay was carried out to detect the proliferation rates which has a significant drop (at 48 h, *P < 0.05 vs. miR-193-3p inhibitor group and miR-193-3p inhibitor control group; at 72 h, **P < 0.01 vs. miR-193-3p inhibitor group and miR-193-3p inhibitor control group). (B) The putative binding of human miR-193-3p on human TGFBR3 gene 3′UTR was shown. ICC-9810 cells were co-transfected with luciferase plasmids with TGFBR3 3′UTR-WT or with TGFBR3 3′UTR-MUT. The relative luciferase activities were measured by a dual-luciferase assay in different groups (*P<0.05). TGFBR3, transforming growth factor-β receptor III; TGF-β, transforming growth factor-β; ICC, intrahepatic cholangiocarcinoma; miR, microRNA; UTR, untranslated region; WT, wild type; MUT, mutant; NC, negative control.
Validation of TGFBR3 as a direct target of miR-193-3p
A dual-luciferase reporter gene assay was used to test the binding relationship between miR-193-3p and TGFBR3. The complimentary sequences for miR-193-3p in the 3′UTR of TGFBR3 were obtained from the TargetScan database (Fig. 4B). Cells co-transfected with miR-193-3p and the TGFBR3 3′UTR-WT exhibited lower intracellular luciferase activity than those transfected TGFBR3 3′UTR-WT alone (P<0.05; Fig. 4B). Meanwhile, the luciferase activity of cells co-transfection with miR-193-3p and mutant TGFBR3 3′UTR-MUT did not differ from that of the NC + TGFBR3 3′UTR-MUT or the control + TGFBR3 3′UTR-MUT groups. This date indicated that TGFBR3 was negatively regulated by miR-193-3p, consistently with the western blotting results, which indicated that the inhibition of miR-193-3p could increase the expression of TGFBR3 (Fig. 2A). Thus, we concluded that miR-193-3p could directly target TGFBR3 and inhibit its expression at the mRNA and protein levels.
miR-193-3p downregulation or TGFBR3 overexpression suppresses the migration and invasion ability of ICC-9810 cells
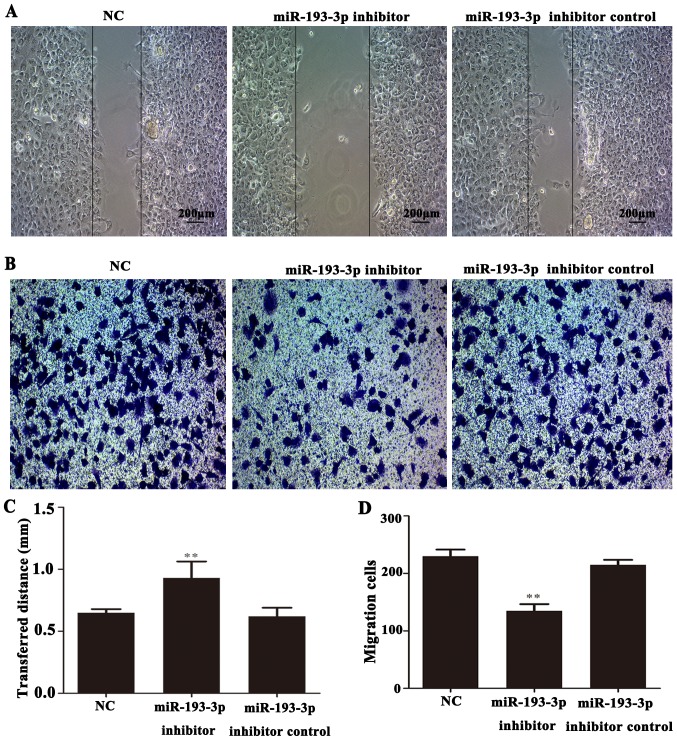
The migratory ability of ICC-9810 cells was measured using a wound healing assay. The results showed that the migration distance of ICC-9810 cells in the miR-193-3p inhibitor group was markedly lower than that NC and miR-193-3p inhibitor control groups (P<0.01; Fig. 5A and C). Similarly, to investigate the invasive ability of ICC-9810 cells, we measured the numbers of invaded in a Transwell assay. In the microscope images (magnification, ×200), significantly fewer invaded cells were observed in the miR-193-3p inhibitor group than in the NC and miR-193-3p inhibitor control groups (P<0.01; Fig. 5B and D). In summary, these results indicated that the downregulation of miR-193-3p leading to TGFBR3 overexpression could inhibit the invasion and migration of ICC-9810 cells.
Figure 5.
miR-193-3p affected the migratory and invasion ability of ICC-9810 cells. (A) A wound-healing assay was carried out to compare the migrating capabilities in miR-193-3p inhibitor group, the pictures indicated that the transferred distance in the miR-193-3p inhibitor group was higher than it in NC group (magnification, ×200). (B) Transwell assay results demonstrated the effects of ICC-9810 cells invasion (magnification, ×200). (C) The result showed that miR-193-3p suppressed cell migration. (D) The data indicated that miR-193-3p could promote cell invasion. **P<0.01 vs. miR-193-3p inhibitor group and NC group. miR, microRNA; ICC, intrahepatic cholangiocarcinoma; NC, negative control.
Discussion
ICC is a common primary liver tumor with increasing incidence worldwide. The clinical outcomes of ICC are typically worse than those for hepatocellular carcinoma due to its non-specific presentation and detection at more advanced stages (17). A previous study suggested that miR-193-3p predominantly exerted an oncogenic effect in gastric cancer (18); however, interestingly, in human NSCLC, miR-193-3p was downregulated in the cancer cells, indicating that miR-193-3p may act as a tumor suppressor to inhibit the proliferation and migration of NSCLC (19). Similarly, a previous miRNA analysis demonstrated that miR-193-3p was significantly downregulated in the lung tissue and serum of PAH patients (4). In the current study, which aimed to explore the functional significance of miR-193-3p in ICC, we demonstrated by RT-qPCR that miR-193-3p was upregulated in ICC-9810 cells when compared with HIBECs, and in ICC tissues compared with adjacent normal tissues. Further data showed that miR-193-3p enhanced the proliferation, invasion and migration of ICC-9810 cells. Collectively these findings indicated that miR-193-3p may act as an oncogene in ICC. Considering the multiple functions of miR-193-3p within complex signaling pathways during the development of different cancers, we conclude that miR-193-3p may act as either an oncogene or a tumor suppressor gene depending on the cancer type.
Previous research has shown that TGFBR3 may play a dichotomous role in human bladder cancer, acting as both a tumor suppressor and tumor promoter (20). Based on these findings, we speculated that there may be a potential relationship between TGFBR3 and miR-193-3p in ICC, and found that TGFBR3 was a direct target gene of miR-193-3p in ICC. Our studies also showed that TGFBR3 expression was upregulated by miR-193-3p downregulation in ICC, thus indicating that TGFBR3 is not only a tumor suppressor, but is also directly regulated by miRNA in ICC.
In conclusion, the downregulation of miR-193-3p directly affected TGFBR3 expression and suppressed the proliferation, migration and invasion of ICC cells. Therefore, our study provides that miR-193-3p may promote the invasion and migration of ICC cells by directly targeting TGFBR3 and reducing its expression.
To the best of our knowledge, this study is the first to elucidate mechanistic functions of miR-193-3pregarding its role in human ICC. Notably, we identified that TGFBR3 was closely related to the regulatory effects of miR-193-3p in ICC. These findings may provide a basis for therapeutic strategies in the treatment of ICC.
References
- 1.Wang LJ, He CC, Sui X, Cai MJ, Zhou CY, Ma JL, Wu L, Wang H, Han SX, Zhu Q. MiR-21 promotes intrahepatic cholangiocarcinoma proliferation and growth in vitro and in vivo by targeting PTPN14 and PTEN. Oncotarget. 2015;6:5932–5946. doi: 10.18632/oncotarget.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Narouie B, Ziaee SAM, Basiri A, Hashemi M. Functional polymorphism at the miR-502-binding site in the 3′untranslated region of the SETD8 gene increased the risk of prostate cancer in a sample of Iranian population. Gene. 2017;626:354–357. doi: 10.1016/j.gene.2017.05.060. [DOI] [PubMed] [Google Scholar]
- 3.Mandujano-Tinoco EA, García-Venzor A, Muñoz-Galindo L, Lizarraga-Sanchez F, Favela-Orozco A, Chavez-Gutierrez E, Krötzsch E, Salgado RM, Melendez-Zajgla J, Maldonado V. miRNA expression profile in multicellular breast cancer spheroids. Biochim Biophys Acta. 2017;1864:1642–1655. doi: 10.1016/j.bbamcr.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 4.Sharma S, Umar S, Potus F, Iorga A, Wong G, Meriwether D, Breuils-Bonnet S, Mai D, Navab K, Ross D, et al. Apolipoprotein A-I mimetic peptide 4F rescues pulmonary hypertension by inducing microRNA-193-3p. Circulation. 2014;130:776–785. doi: 10.1161/CIRCULATIONAHA.114.007405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper SJ, Zou H, Legrand SN, Marlow LA, von Roemeling CA, Radisky DC, Wu KJ, Hempel N, Margulis V, Tun HW, et al. Loss of type III transforming growth factor-beta receptor expression is due to methylation silencing of the transcription factor GATA3 in renal cell carcinoma. Oncogene. 2010;29:2905–2915. doi: 10.1038/onc.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finger EC, Turley RS, Dong M, How T, Fields TA, Blobe GC. TbetaRIII suppresses non-small cell lung cancer invasiveness and tumorigenicity. Carcinogenesis. 2008;29:528–535. doi: 10.1093/carcin/bgm289. [DOI] [PubMed] [Google Scholar]
- 7.Gordon KJ, Dong M, Chislock EM, Fields TA, Blobe GC. Loss of type III transforming growth factor beta receptor expression increases motility and invasiveness associated with epithelial to mesenchymal transition during pancreatic cancer progression. Carcinogenesis. 2008;29:252–262. doi: 10.1093/carcin/bgm249. [DOI] [PubMed] [Google Scholar]
- 8.Hempel N, How T, Dong M, Murphy SK, Fields TA, Blobe GC. Loss of betaglycan expression in ovarian cancer: Role in motility and invasion. Cancer Res. 2007;67:5231–5238. doi: 10.1158/0008-5472.CAN-07-0035. [DOI] [PubMed] [Google Scholar]
- 9.Turley RS, Finger EC, Hempel N, How T, Fields TA, Blobe GC. The type III transforming growth factor-beta receptor as a novel tumor suppressor gene in prostate cancer. Cancer Res. 2007;67:1090–1098. doi: 10.1158/0008-5472.CAN-06-3117. [DOI] [PubMed] [Google Scholar]
- 10.Dong M, How T, Kirkbride KC, Gordon KJ, Lee JD, Hempel N, Kelly P, Moeller BJ, Marks JR, Blobe GC. The type III TGF-beta receptor suppresses breast cancer progression. J Clin Invest. 2007;117:206–217. doi: 10.1172/JCI29293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gatza CE, Holtzhausen A, Kirkbride KC, Gordon KJ, Lee JD, Hempel N, Kelly P, Moeller BJ, Marks JR, Blobe GC. Type III TGF-β receptor enhances colon cancer cell migration and anchorage-independent growth. J Clin Invest. 2007;117:206–217. [Google Scholar]
- 12.Sarraj MA, Chua HK, Umbers A, Loveland KL, Findlay JK, Stenvers KL. Differential expression of TGFBR3 (betaglycan) in mouse ovary and testis during gonadogenesis. Growth Factors. 2007;25:334–345. doi: 10.1080/08977190701833619. [DOI] [PubMed] [Google Scholar]
- 13.Mu X, Pradere JP, Affό S, Dapito DH, Friedman R, Lefkovitch JH, Schwabe RF. Epithelial transforming growth factor-β signaling does not contribute to liver fibrosis but protects mice from cholangiocarcinoma. Gastroenterology. 2016;150:720–733. doi: 10.1053/j.gastro.2015.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou X, Zang H, Liu H L. An excerpt of intrahepatic cholangiocarcinoma: expert consensus statement. J Clin Hepatol. 2015;31:1584–1587. (In Chinese) [Google Scholar]
- 15.Deng M, Jing DD, Meng XJ. Effect of MUC1 siRNA on drug resistance of gastric cancer cells to trastuzumab. Asian Pac J Cancer Prev. 2013;14:127–131. doi: 10.7314/APJCP.2013.14.1.127. [DOI] [PubMed] [Google Scholar]
- 16.He B, Lin X, Tian F, Yu W, Qiao B. MiR-133a-3p inhibits oral squamous cell carcinoma (OSCC) proliferation and invasion by suppressing COL1A1. J Cell Biochem. 2017 Jun 1; doi: 10.1002/jcb.26182. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 17.Guro H, Kim JW, Choi Y, Cho JY, Yoon YS, Han HS. Multidisciplinary management of intrahepatic cholangiocarcinoma: Current approaches. Surg Oncol. 2017;26:146–152. doi: 10.1016/j.suronc.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 18.Jian B, Li Z, Xiao D, He G, Bai L, Yang Q. Downregulation of microRNA-193-3p inhibits tumor proliferation migration and chemoresistance in human gastric cancer by regulating PTEN gene. Tumor Biol. 2016;37:8941–8949. doi: 10.1007/s13277-015-4727-x. [DOI] [PubMed] [Google Scholar]
- 19.Yu T, Li J, Yan M, Liu L, Lin H, Zhao F, Sun L, Zhang Y, Cui Y, Zhang F, et al. MicroRNA-193a-3p and −5p suppress the metastasis of human non-small-cell lung cancer by downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway. Oncogene. 2015;34:413–423. doi: 10.1038/onc.2013.574. [DOI] [PubMed] [Google Scholar]
- 20.Liu XL, Xiao K, Xue B, Yang D, Lei Z, Shan Y, Zhang HT. Dual role of TGFBR3 in bladder cancer. Oncol Rep. 2013;30:1301–1308. doi: 10.3892/or.2013.2599. [DOI] [PubMed] [Google Scholar]