Abstract
Tissues contain distinct stem cell niches, but whether cell turnover is coordinated between niches during growth is unknown. Here, we report that in mouse skin, hair growth is accompanied by sebaceous gland and interfollicular epidermis expansion. During hair growth, cells in the bulge and outer root sheath temporarily upregulate the glutamate transporter SLC1A3, and the number of SLC1A3+ basal cells in interfollicular epidermis and sebaceous gland increases. Fate mapping of SLC1A3+ cells in mice revealed transient expression in proliferating stem/progenitor cells in all three niches. Deletion of slc1a3 delays hair follicle anagen entry, uncouples interfollicular epidermis and sebaceous gland expansion from the hair cycle, and leads to reduced fur density in aged mice, indicating a role of SLC1A3 in stem/progenitor cell activation. Modulation of metabotropic glutamate receptor 5 activity mimics the effects of SLC1A3 deletion or inhibition. These data reveal that stem/progenitor cell activation is synchronized over distinct niches during growth and identify SLC1A3 as a general marker and effector of activated epithelial stem/progenitor cells throughout the skin.
Keywords: epithelial stem cells, glutamate signaling, skin, Slc1a3, stem cell activation
Subject Categories: Development & Differentiation, Signal Transduction, Stem Cells
Introduction
Proliferation of tissue‐specific stem and progenitor cells for production of differentiated progeny and stem cell renewal is the basis of organ growth and homeostasis. However, whether or how cell turnover is coordinated between distinct stem cell niches during growth is unknown. Skin contains distinct epithelial stem cell niches (Fig 1A). Hair follicles cycle through synchronized phases of rest (telogen), growth (anagen), and regression (catagen; Paus & Cotsarelis, 1999), corresponding to activation of hair follicle stem cells. In contrast, in sebaceous gland (SG) and interfollicular epidermis (IFE), stem and progenitor cells continuously replace lost cells. Whether cell turnover in SG and IFE is influenced by the hair cycle and whether there is a common activity marker and effector is unclear.
Figure 1. Hair follicle, SG, and IFE stem cell niches simultaneously increase Slc1a3 expression and proliferation during hair growth.
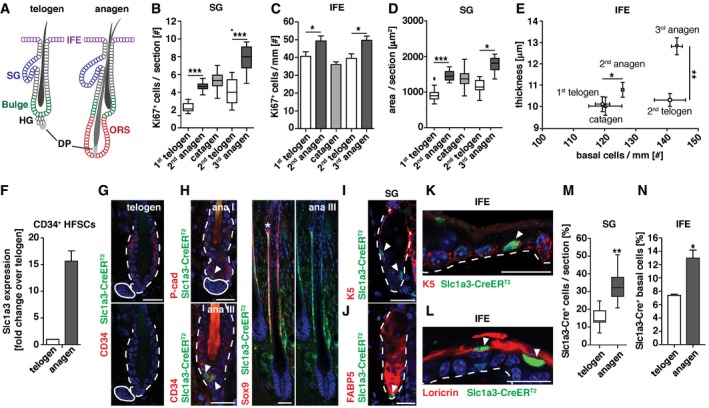
-
ASchematic depiction of the telogen and anagen hair follicle including bulge, hair germ (HG), dermal papilla (DP), and outer root sheath (ORS). Adjacent to the hair follicle (HF) are sebaceous gland (SG) and interfollicular epidermis (IFE).
-
BProliferation of SG in relation to hair cycle (1st telogen, n = 5, 95 HFs; 2nd anagen, n = 7, 140 HFs, catagen n = 3, 60 HFs; 2nd telogen, n = 4, 79 HFs; 3rd anagen, n = 3, 60 HFs).
-
CProliferation of IFE in relation to hair cycle (1st telogen, n = 7, 30 mm; 2nd anagen, n = 6, 26 mm; catagen n = 7, 54 mm; 2nd telogen, n = 7, 49 mm; 3rd anagen, n = 3, 7 mm).
-
DGrowth of SG in relation to hair cycle (1st telogen n = 5, 60 SGs; 2nd anagen, n = 8, 131 SGs; catagen n = 3, 73 SGs; 2nd telogen, n = 6, 60 SGs; 3rd anagen, n = 3, 60 SGs).
-
EGrowth of IFE in relation to hair cycle (1st telogen, n = 5, 9 mm; 2nd anagen, n = 5, 8 mm; catagen n = 7, 54 mm; 2nd telogen, n = 7, 48 mm; 3rd anagen, n = 3, 5 mm).
-
FSlc1a3 mRNA expression in hair follicle stem cells (HFSCs) during growth (anagen) compared to rest (telogen; two technical replicates; anagen, n = 4; telogen, n = 7).
-
G, HSlc1a3‐CreERT2 is absent in telogen hair follicles (G) but expressed in HG, bulge, and ORS during anagen (H).
-
I–LSlc1a3‐CreERT2 is expressed in SG basal cells (I) but not in sebocytes (J) and in IFE scattered basal (K) and most suprabasal cells (L).
-
MPercentage of Slc1a3‐CreERT2+ basal cells in SG during telogen (n = 3, 63 SGs) compared to anagen (n = 3, 59 SGs).
-
NPercentage of Slc1a3‐CreERT2+ basal cells in IFE during telogen (n = 4, 9 mm) compared to anagen (n = 3, 6 mm).
Hair follicle stem cells in the bulge are quiescent in telogen but active during anagen (Fuchs & Nowak, 2008). Anagen can be divided into six stages (I–VI; Muller‐Rover et al, 2001), with an initial expansion of the hair germ (HG), a brief activation of bulge CD34+ hair follicle stem cells and sustained proliferation of the outer root sheath (ORS) in which most actively cycling hair follicle stem cells are located (Hsu et al, 2011; Rompolas & Greco, 2014). In the SG, proliferating basal cells continuously replace sebocytes, which produce sebum. There are several contradicting reports regarding the origin and self‐renewing capacity of SG basal cells with the latest report pointing toward a self‐sustained progenitor pool within the SG (Horsley et al, 2006; Snippert et al, 2010a; Page et al, 2013; Fullgrabe et al, 2015). The IFE is sustained through stem and progenitor cells, located in the basal layer, which constantly replace lost keratinocytes. The degree of heterogeneity within the IFE stem and progenitor cell population is still under investigation (Schepeler et al, 2014).
Stem cell activation has been shown to involve chromatin remodeling and gene expression changes (Ezhkova et al, 2011; Lien et al, 2011), and several transcription factors that govern niche‐specific stem cell behavior have been identified (Horsley et al, 2008; Osorio et al, 2008; Genander et al, 2014). The expression of these factors is controlled by extracellular signals. BMP signaling has been demonstrated to maintain quiescence, while Wnt signaling in the absence of BMP promotes activation of HF stem cells (Kobielak et al, 2007; Greco et al, 2009; Kandyba et al, 2013).
Glutamate signaling is common to several stem cell niches (Julio‐Pieper et al, 2011). In Drosophila, dietary l‐glutamate stimulates intestinal stem cell division and gut growth via the metabotropic glutamate receptor (Deng et al, 2015). Neural stem cells express the metabotropic glutamate receptor 5 (mGluR5), the activity of which can impact proliferation and differentiation (Nochi et al, 2012; Xiao et al, 2013). Glutamate receptor‐mediated synaptic transmission is dependent on extracellular glutamate clearance, effectuated by glutamate transporters. The removal of glutamate in the proximity of glutamate receptors enables fast receptor deactivation, which determines the frequency in which new signaling events can be registered.
Here, we identify the glial high‐affinity glutamate transporter Slc1a3, also known as EAAT1 or Glast, as a unifying marker of activated stem and progenitor cells in the hair follicle, IFE and SG. Using in vivo lineage tracing, we show that Slc1a3‐expressing cells sustain all three epithelial compartments long‐term, identifying them as stem or progenitor cells. All three epithelial compartments synchronize growth during anagen, temporarily increasing stem and progenitor cell activation and Slc1a3 expression. Deletion of Slc1a3 delays the onset of the growth phase, uncouples IFE and SG expansion from the hair cycle, and leads to reduced fur density over time. Slc1a3 acts in conjunction with mGluR5 and inhibition of Slc1a3 or mGluR5 delays growth phase onset and uncouples IFE and SG expansion from the hair cycle. These data reveal that stem/progenitor cell activation is synchronized over distinct niches during growth and identify Slc1a3 as a general marker and effector of activated epithelial stem/progenitor cells throughout the skin.
Results
Differential expression of Slc1a3 during growth and rest
To understand whether growth is coordinated between adjacent epithelial stem cell niches in skin, we quantified cell proliferation in SG and IFE during distinct phases of the hair cycle. Interestingly, we found elevated numbers of Ki67+ proliferating cells in SG and IFE in 2nd anagen compared to 1st telogen (growing mice), and also in 3rd anagen compared to 2nd telogen (adult mice; Fig 1B and C), corresponding to growth of SG and IFE (Fig 1D and E). This suggests that independent of overall growth of the animal, SG and IFE proliferation is correlated to the hair cycle.
Comparing mRNA expression of CD34+ hair follicle stem cells in telogen and anagen, we found increased expression of the glutamate transporter Slc1a3 during anagen (Fig 1F). Immunohistochemistry failed to detect Slc1a3 in the hair follicle during telogen (Fig EV1A), confirming low Slc1a3 expression in quiescent hair follicle stem cells, but revealed expression in the ORS during anagen (Fig EV1B). Employing Slc1a3‐CreER T2 transgenic mice (Slezak et al, 2007), in which CreERT2 is expressed under the gene regulatory elements of Slc1a3, we further investigated Slc1a3 expression throughout the hair cycle. CreERT2 was absent during telogen (Fig 1G) and first detectable in single, P‐cadherin+ cells in the expanding hair germ during anagen I (Fig 1H). In anagen III, CreERT2 could be found in a fraction of CD34+ hair follicle stem cells (Fig 1H), validating Slc1a3 mRNA expression. CreERT2 was continuously present in ORS cells during anagen III‐V, overlapping with Sox9 and Slc1a3 expression (Figs 1H and EV1B and C). CreERT2 was not found in the matrix and inner layers of the growing hair follicle (Figs 1H and EV1C). In addition to the hair follicle, Slc1a3 and CreERT2 were robustly expressed in a fraction of K5+/K5lowFABP5− basal cells in the SG, whereas a few K5+FABP5low SG cells were weakly Slc1a3+ (Figs 1I and J, and EV1D and E). In the IFE, Slc1a3 and CreERT2 marked a subset of K5+ basal and most suprabasal cells (Figs 1K and L, and EV1F and G). Importantly, during anagen‐associated growth, the percentage of CreERT2+ SG and IFE basal cells increased compared to telogen (Fig 1M and N).
Figure EV1. Slc1a3 is expressed in ORS, SG, and IFE stem cell niches but not in the telogen hair follicle.
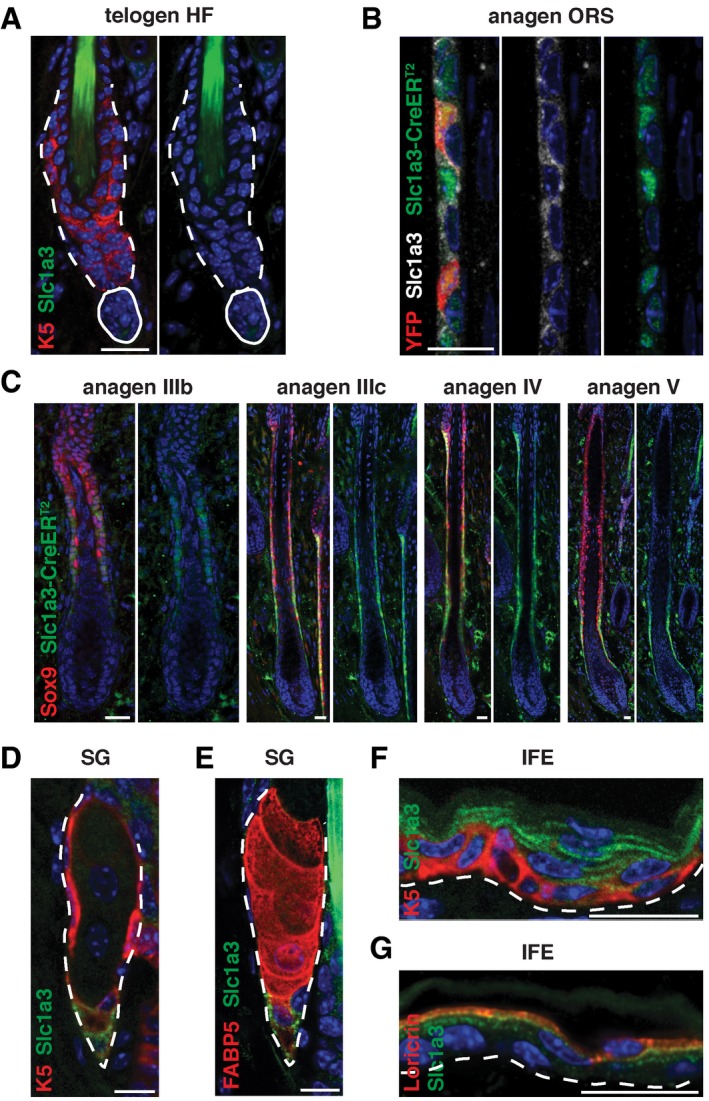
-
ASlc1a3 is not expressed in hair follicle stem cells in telogen.
-
BSlc1a3 and Slc1a3‐CreERT2 expression overlap. YFP expression is restricted to Slc1a3‐CreERT2+ cells immediately after recombination.
-
CSlc1a3‐CreERT2 is expressed in the Sox9+ ORS and absent from the matrix and inner layers of the growing hair follicle.
-
D, ESlc1a3 is robustly expressed in a fraction of K5+/K5lowFABP5− SG basal cells and a few K5+FABP5low SG cells.
-
F, GIn the IFE, Slc1a3 is expressed in a subset of K5+ basal and all Loricrin+ suprabasal cells.
Deletion of Slc1a3 delays anagen entry and uncouples SG and IFE growth from the hair cycle
To investigate the functional role of Slc1a3 in hair follicle, SG, and IFE stem cell compartments, we compared Slc1a3 +/− and Slc1a3 −/− siblings (Watase et al, 1998) during the first postnatal hair cycle. Slc1a3 −/− hair follicles did not show differences in either density or hair follicle stem cell marker expression when compared to Slc1a3 +/− littermates (Fig EV2A–C), excluding major developmental effects on hair follicle stem cell establishment due to abrogation of Slc1a3. However, we found that anagen initiation was significantly delayed in Slc1a3 −/− animals (Fig 2A and B). At P24, the number of telogen hair follicles was increased whereas the number of hair follicles in anagen II to IV was reduced in Slc1a3 −/− compared to Slc1a3 +/− littermates (Fig 2A and B), corresponding to an overall shortened hair follicle outgrowth with less Ki67+ cells (Fig 2C and D). Furthermore, quantification of anagen I hair follicles showed that hair follicle stem cells in Slc1a3 −/− mice proliferated less compared to Slc1a3 +/− animals (Fig 2E). At P28, 82% of the hair follicles in Slc1a3 −/− mice had entered 2nd anagen (99% in Slc1a3 +/− mice), but were significantly delayed in stage progression (Fig 2F and G). From the 3rd anagen phase on, hair cycle phases are no longer synchronized. Instead, anagen is randomly initiated at different sites. We found that at 3 months of age in average 32.5 ± 7.2% of the back‐skin area of Slc1a3 +/+ mice was in 3rd anagen, compared to 6.6 ± 4.9% in Slc1a3 −/− animals, measured by the area of pigmented back‐skin characteristic for anagen follicles (Figs 2H and EV2D). The number of club hairs per hair follicle cluster was similar between Slc1a3 +/+ and Slc1a3 −/− mice (Fig EV2E) suggesting that club hairs are not selectively lost in Slc1a3 −/− hair follicles. Furthermore, Slc1a3 +/+, but not Slc1a3 −/−, hair follicles contained one pigmented anagen hair shaft consistent with them having entered growth phase (Fig 2I), indicating that Slc1a3 −/− mice fail to efficiently initiate anagen.
Figure EV2. Slc1a3 deletion does not alter the number of hair follicles and hair anchoring.
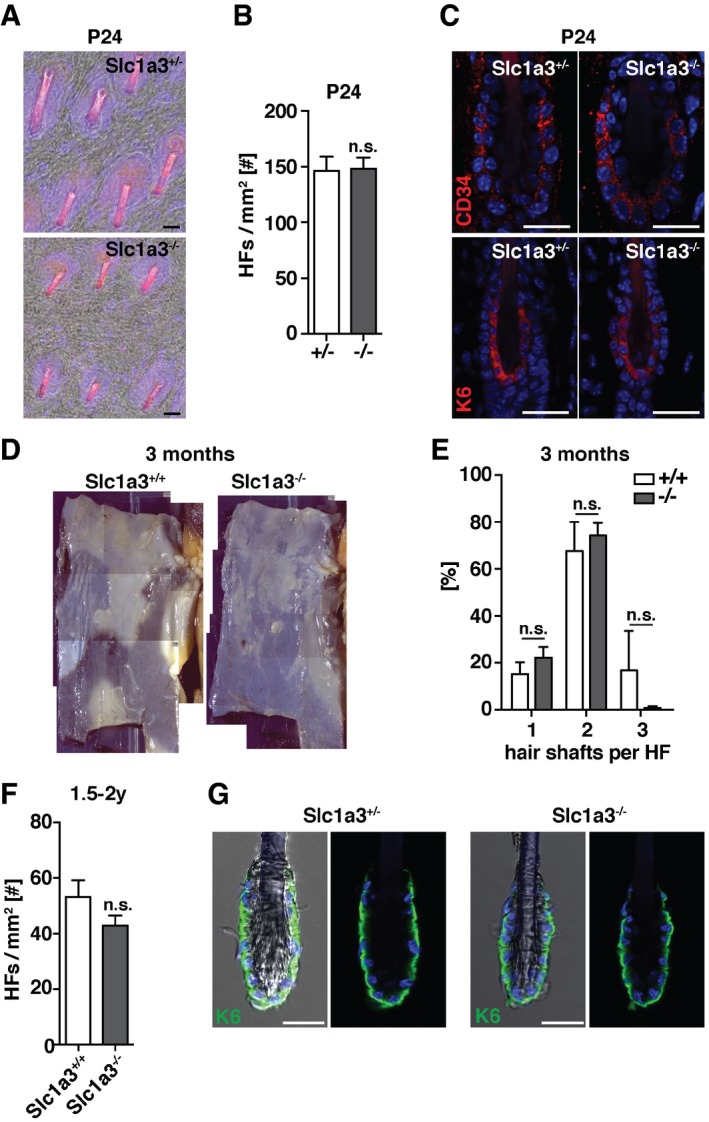
-
A–CHair follicle density (A, B) and CD34 and K6 stem cell niche marker expression (C) of P24 Slc1a3+/− animals (n = 4, 1,6 mm2 skin per animal) compared to Slc1a3−/− littermates (n = 4, 1,6 mm2 skin per animal).
-
DTile scan micrographs of back skin indicating pigmented regions in which hair follicles are in anagen of 3‐month‐old Slc1a3+/+ and Slc1a3−/− animals.
-
EHair shaft density of 3‐month‐old Slc1a3+/+ (n = 4, 355 HFs) compared to Slc1a3−/− (n = 4, 391 HFs) animals.
-
FComparison of hair follicle density of aged Slc1a3+/+ (n = 4, 1.6 mm2 skin per animal) and Slc1a3−/− animals (n = 4, 1.6 mm2 skin per animal).
-
GPlucked hair shafts of Slc1a3+/− and Slc1a3−/− mice show no difference in the amount of attached K6+ inner bulge cells.
Figure 2. Deletion of Slc1a3 impairs anagen initiation, uncouples SG and IFE growth from the hair cycle and leads to reduced hair density.
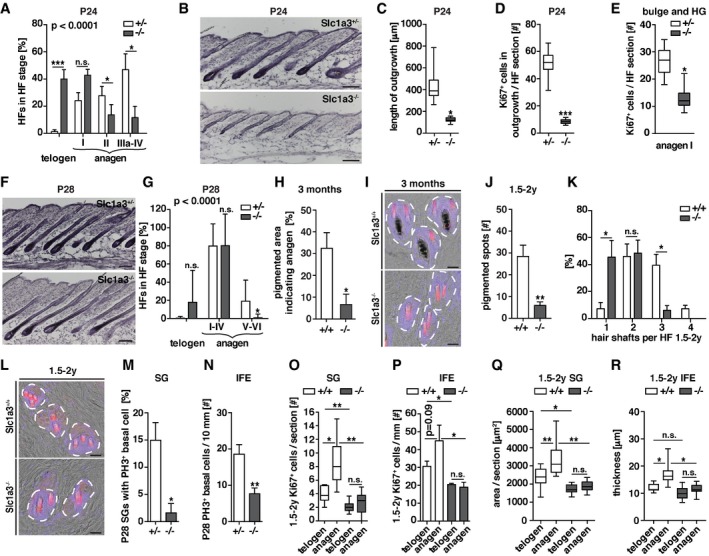
-
AHair follicle stage distribution at P24 in Slc1a3+/− mice (n = 11, 220 HFs) compared to Slc1a3−/− littermates (n = 12, 240 HFs). P < 0.0001 derived from grouped comparison of ranked HF stages in Slc1a3+/− and Slc1a3−/− mice.
-
BH&E staining of dorsal skin sections of Slc1a3+/− compared to Slc1a3−/− littermates at P24.
-
C, DHair follicle outgrowth length (C) and number of Ki67+ cells (D) of Slc1a3+/− animals at P24 (n = 4, 80 HFs) compared to Slc1a3−/− littermates (n = 4, 80 HFs).
-
EProliferation of anagen I hair follicles in Slc1a3+/− (n = 6, 40 HFs) compared to Slc1a3−/− (n = 5, 45 HFs) mice.
-
FH&E staining of dorsal skin sections of Slc1a3+/− compared to Slc1a3−/− littermates at P28.
-
GHair follicle stage analysis of Slc1a3+/− mice at P28 (n = 9, 180 HFs) compared to Slc1a3−/− (n = 8, 160 HFs) littermates. P < 0.0001 derived from grouped comparison of ranked HF stages in Slc1a3+/− and Slc1a3−/− mice.
-
HQuantification of melanin‐enriched (pigmented) areas, indicating regions in which hair follicles are in anagen, of 3‐month‐old Slc1a3+/+ (n = 4, 5,361 mm2) compared to Slc1a3−/− (n = 4, 5,048 mm2) animals.
-
IRepresentative top‐view micrographs of back skin showing hair shafts per follicle in 3‐month‐old Slc1a3+/+ compared to Slc1a3−/− animals.
-
JQuantification of melanin‐enriched (pigmented) spots in aged (1.5–2 years old) Slc1a3+/+ (n = 4, 7,671 mm2) compared to Slc1a3−/− (n = 4, 6,511 mm2) animals.
-
KHair shaft density of aged Slc1a3+/+ mice (n = 4, 1.6 mm2 per animal) compared to Slc1a3−/− mice (n = 4, 1.6 mm2 per animal).
-
LRepresentative top‐view micrographs of back skin showing hair shafts per follicle in aged Slc1a3+/+ compared to Slc1a3−/− animals.
-
MBasal cell proliferation of the SG in Slc1a3+/− (n = 4, 60 SGs) compared to Slc1a3−/− mice during anagen (n = 4, 60 SGs).
-
NBasal cell proliferation of the IFE in Slc1a3+/− (n = 4, 41 mm) compared to Slc1a3−/− mice during anagen (n = 4, 41 mm).
-
OBasal cell proliferation of the SG in anagen and telogen in aged Slc1a3+/+ (n = 3, 58 SGs) compared to Slc1a3−/− (n = 4, 41 SGs) mice.
-
PBasal cell proliferation of the IFE in anagen and telogen in aged Slc1a3+/+ (n = 4, 11 mm for telogen, 8 mm for anagen) compared to Slc1a3−/− (n = 3, 12 mm for telogen, 10 mm for anagen) mice.
-
QComparison of SG area between anagen and telogen regions of aged Slc1a3+/+ (n = 4, 28 SGs for telogen, 25 for anagen) and Slc1a3−/− (n = 4, 29 SGs for telogen, 28 for anagen) mice.
-
RComparison of IFE thickness between anagen and telogen regions of aged Slc1a3+/+ (n = 6) and Slc1a3−/− (n = 4) mice.
The deficiency in anagen initiation was also pronounced in aged (1.5–2 years old) animals. Aged Slc1a3 +/+ mice presented with more pigmented spots than aged Slc1a3 −/− mice, suggesting that in absence of Slc1a3 normal anagen initiation is disturbed (Fig 2J). Although the number of hair follicles was maintained (Fig EV2F) and hair anchoring was not altered in Slc1a3 −/− mice (Fig EV2G), abrogating Slc1a3 long‐term resulted in reduced fur density. Whereas more than 45% of Slc1a3 +/+ hair follicles contained three to four hair shafts, < 10% of Slc1a3 −/− hair follicles consisted of groups of more than two hair shafts (Fig 2K and L). Together, these data suggest that genetic ablation of Slc1a3 leads to reduced hair follicle stem cell activation and proliferation, consequently resulting in disturbed anagen initiation, impaired hair follicle cycling, and, over time, reduced fur density.
Deletion of Slc1a3 also affected SG and IFE growth. The number of dividing basal cells in SG and IFE at P28 was reduced in Slc1a3 −/− animals compared to Slc1a3 +/− littermates although most hair follicles in Slc1a3 −/− mice were in anagen (Fig 2M and N). Comparing anagen and telogen regions within each aged animal, we found increased proliferation of SG and IFE basal cells in Slc1a3 +/+ mice during anagen. Intriguingly however, in telogen, SG and IFE proliferation was reduced by 50 and 40%, respectively, in Slc1a3 −/− animals and failed to adjust to the hair cycle (Fig 2O and P). In line with these observations, SG size and IFE thickness was increased in anagen regions in aged Slc1a3 +/+ mice, but were similar to telogen levels in Slc1a3 −/− mice (Fig 2Q and R). Together, this shows that deletion of Slc1a3 not only delays hair follicles anagen entry and leads to a diminution of SG and IFE proliferation, but also uncouples SG and IFE proliferation from the hair cycle, resulting in an overall failure of SG and IFE to adjust to the tissue remodeling associated with hair follicle growth.
Slc1a3 is expressed in hair follicle, SG, and IFE stem/progenitor cells
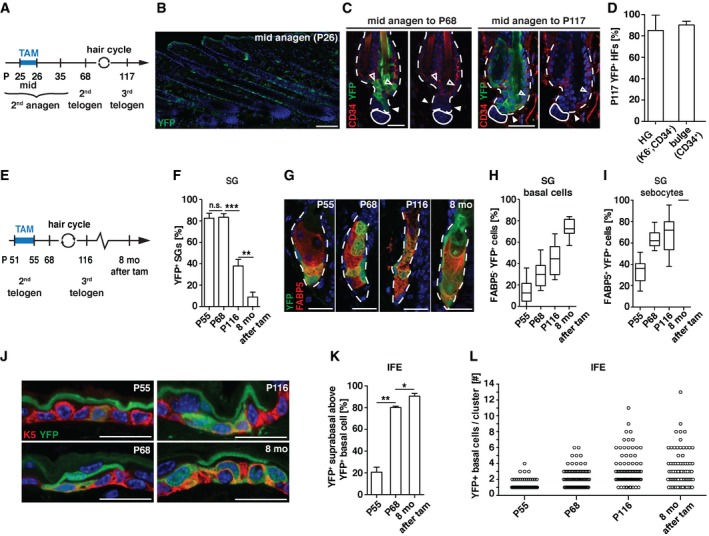
Proliferation in hair follicles, SGs and IFE is driven by stem and progenitor cells. To examine whether Slc1a3 is indeed expressed by stem/progenitor cells, we performed lineage tracing using Slc1a3‐CreER T2 transgenic mice (Slezak et al, 2007) carrying a Rosa26‐YFP reporter allele (Srinivas et al, 2001). We fate mapped Slc1a3+ hair follicle cells from mid 2nd anagen (P25/26) and assessed the contribution to telogen follicles after completing one (2nd telogen, P68) or two hair cycles (3rd telogen, P117; Fig 3A and B). YFP‐labeled hair follicle cells survived catagen and contributed to the new bulge and HG at P68 and P117. Strikingly, the vast majority of HGs and bulges at P117 were YFP+, indicating that Slc1a3‐expressing cells labeled at P25/26 include long‐term hair follicle stem cells (Fig 3C and D).
Figure 3. Slc1a3 is expressed in hair follicle, SG, and IFE stem/progenitor cells.
-
AExperimental timeline for hair follicle lineage tracing.
-
B–DAfter tamoxifen (TAM)‐mediated recombination (P26), YFP+ cells reside in the ORS (B) and contribute to YFP+ hair follicle stem cells at P68 and P117 (C, D; n = 5, 64 HFs).
-
EExperimental layout for SG and IFE lineage tracing.
-
FPercentage of YFP+ SGs over time (P55, n = 3, 150 SGs; P68, n = 4, 200 SGs; P116 n = 4, 200 SGs, 8 months, n = 4, 200 SGs).
-
GRepresentative micrographs of lineage‐traced SGs at P55, P68, P116 and 8 months after recombination induction.
-
H, IPercentage of YFP+ FABP5− basal cells (H) and YFP+ FABP5+ sebocytes (I) in YFP+ SGs over time (P55, n = 3, 60 SGs; P68, n = 4, 80 SGs; P116 n = 4, 80 SGs, 8 months, n = 4, 40 SGs).
-
JRepresentative micrographs of lineage‐traced IFE at P55, P68, P116, and 8 months after recombination induction.
-
KPercentage of YFP+ suprabasal cells above YFP+ basal cells over time (P55 n = 3, 122 YFP+ clusters; P68 n = 4, 78 YFP+ clusters; 8 months n = 4, 171 YFP+ clusters).
-
LNumber of YFP+ basal cells directly neighboring each other over time (P55, n = 3, 60 YFP+ clusters; P68, n = 4, 80 YFP+ clusters; P116 n = 4, 80 YFP+ clusters; 8 months, n = 4, 40 YFP+ clusters).
To explore the stem/progenitor potential of Slc1a3+ basal cells in the SG we initiated fate mapping during 2nd telogen (P51–P55) to exclude contribution of hair follicle stem cells (Figs 1G and EV3A; Petersson et al, 2011). CreERT2 and YFP were not detected in cells around the SG orifice or in the isthmus (Fig EV3B and C), excluding contributing cells from these sources (Horsley et al, 2006; Snippert et al, 2010a; Page et al, 2013). Skin was analyzed in 2nd telogen (P55 and P68), in 3rd telogen (P116) and 8 months after labeling (Fig 3E). At P55, 83% of SGs were recombined and contained on average 14 ± 2% YFP+ basal cells, corresponding to 34 ± 3% YFP+FABP5+ sebocytes. Over time, the number of recombined SGs gradually decreased (Figs 3F and EV3D). However, SGs that did maintain recombined cells showed a steady increase in the number of YFP+ basal cells and differentiated sebocytes until at 8 months after recombination, 73 ± 4% of basal cells and all sebocytes were YFP+ (Fig 3G and I). These data show that some Slc1a3+ basal cells are able to renew the entire SG over an extended period of time and support previous reports of the SG as a self‐maintained stem cell niche (Fullgrabe et al, 2015).
Figure EV3. Slc1a3 is transiently expressed in SG basal cells, which can self‐maintain SGs for at least 8 months.

-
ARecombination induction during telogen (P51‐55) labels cells in the sebaceous gland (SG) but not in the hair follicle.
-
B, CThe Slc1a3‐CreERT2 mouse line does not target cells in the isthmus or around the SG orifice and these regions neither contain YFP+ cells 5 days after recombination induction (B) nor express Slc1a3‐CreERT2 (C), excluding contribution of YFP+ SG cells from these regions.
-
DSections of lineage‐traced dorsal back skin directly (P55) or 8 months (8 mo) after 5 consecutive days of tamoxifen injection. Over time, the number of YFP+ cells in YFP+ SGs increases while the number of YFP+ SGs decreases. FABP5+ sebocytes in red mark SGs, and asterisks indicate SGs containing YFP+ cells.
-
EFollowing lineage tracing, YFP− SGs contain CreERT2+Slc1a3+ basal cells.
To assess self‐renewal potential and lineage contribution of Slc1a3+ IFE basal cells, we analyzed the same time points as described for the SG. At P55, most suprabasal and a minor fraction of scattered basal cells were YFP+ (Fig 3J), reflecting CreERT2 and Slc1a3 expression (Figs 1K and L, and EV1F and G). At this time point, 21% of the YFP+ suprabasal cells were neighboring a YFP+ basal cell (Fig 3K). At P68, the predominantly uniform suprabasal YFP expression seen at P55 was replaced by single or clusters of YFP+ suprabasal cells of which 80% were positioned directly above YFP+ basal cells, suggesting that the originally labeled suprabasal cells had shed off and were replaced by YFP+ suprabasal cells derived from underlying recombined basal cells. YFP+ basal cells were still present at 8 months after recombination and continuously produced YFP+ suprabasal cells (Fig 3J and K). Over time, the number of YFP+ basal cells increased and could be found in clusters of 1‐13 YFP+ cells (Fig 3L). Together, these data demonstrate that Slc1a3 labels a population of long‐term stem/progenitor cells, corroborating recent work identifying Slc1a3 as a marker for IFE stem cells (Sada et al, 2016).
Slc1a3 is transiently expressed in active hair follicle stem cells
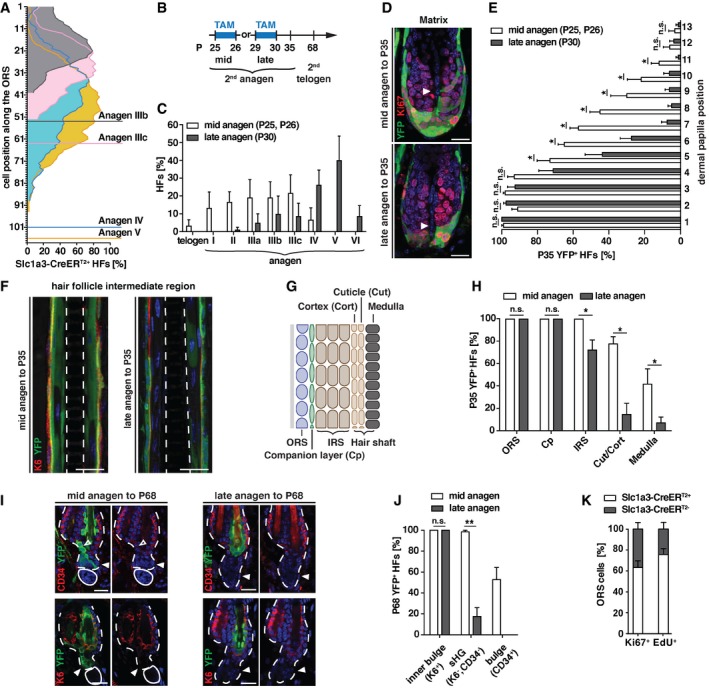
Considering the relatively small number of Slc1a3+ stem/progenitor cells labeled in hair follicles, SG, and IFE, we were intrigued by the significant effects genetic abrogation of Slc1a3 had on growth of these stem cell niches. To determine whether Slc1a3 expression is temporally regulated or continuously expressed in individual stem cells, we turned to the ORS, in which most cycling hair follicle stem cells are located, and mapped CreERT2 expression to cell position along the ORS (as quantified from the bulge to the hair follicle base). Comparing anagen stages IIIb, IIIc, IV, and V, we found that the number of CreERT2 expressing cells increased together with the continuous growth of the follicle. While CreERT2+ cells could be localized starting from the first position below the bulge (+1), the density was highest in the middle of the ORS at all four stages (Fig 4A). This dynamic distribution of Slc1a3+ cells along the ORS offered a temporal resolution that we exploited. We fate mapped Slc1a3+ hair follicle cells from mid‐ (P25/26) or late (P29/30) anagen and assessed the contribution to full‐grown follicles at P35 and to telogen follicles at P68 (Fig 4B). Hair follicles of P30 mice were more progressed in anagen compared to hair follicles of P25/P26 mice (Fig 4C). Fate mapping to full‐grown follicles (P35) revealed contribution of Slc1a3+ cells to matrix and inner differentiated layers. Progeny of Slc1a3+ cells labeled at mid‐anagen contributed significantly more often to higher matrix positions and to the inner root sheath and hair shaft lineages than progeny of late anagen Slc1a3‐targeted cells (Fig 4D–H). While Slc1a3+ ORS cells traced from mid‐anagen contributed to new bulge and HG hair follicle stem cells in 2nd telogen, Slc1a3+ cells in late anagen failed to do so efficiently (Fig 4I and J). In contrast, Slc1a3+ cells in mid‐ and late anagen both contributed to the K6+ inner bulge layer (Fig 4I and J). These results show that stem cells, which form the new outer CD34+ bulge, terminate Slc1a3 expression first, followed by HG‐forming cells and K6+ inner bulge‐forming cells, and that Slc1a3 is transiently expressed in stem/progenitor cells.
Figure 4. Slc1a3 is transiently expressed in active hair follicle stem cells.
-
ADistribution of Slc1a3‐CreERT2 expressing cells along the ORS during anagen (IIIb, n = 3, 20 HFs; IIIc, n = 3, 20 HFs; IV, n = 4, 23 HFs; V, n = 3, 28 HFs). Position +1 is the ORS cell next to the bulge. Horizontal lines indicate the base of the HF for each anagen stage.
-
BExperimental timeline for hair follicle stem cell fate mapping.
-
CHair follicle stage distribution at the onset of lineage tracing at mid‐ (n = 6, 120 HFs) and late anagen (n = 4, 80 HFs), determined in littermates of traced animals.
-
D, ERepresentative micrographs (D) and quantification (E) of matrix contribution (YFP) of lineage‐traced Slc1a3+ cells at mid‐ (n = 5, 64 HFs) and late anagen (n = 4, 40 HFs).
-
FRepresentative micrographs showing inner layer contribution (YFP) of lineage‐traced Slc1a3+ cells at mid‐ and late anagen.
-
GSchematic depiction of the different hair follicle layers; outer root sheath (ORS), companion layer (Cp), inner root sheath (IRS), Cuticle/Cortex (Cut/Cort).
-
HQuantification of hair follicle layer contribution (YFP) of lineage‐traced Slc1a3+ cells at mid‐ (n = 5, 70 HFs) and late anagen (n = 4, 40 HFs).
-
I, JBulge and HG contribution to the following telogen hair follicle of lineage‐traced Slc1a3+ ORS cells from mid‐ (n = 7, 70 HFs) compared to late anagen (n = 4, 40 HFs).
-
KQuantification of co‐expression of Slc1a3‐CreERT2 and proliferation markers in ORS cells (n = 6, 61 HFs for Ki67, 72 HFs for EdU).
New bulge‐contributing ORS cells stop proliferating before HG and inner bulge‐forming ORS cells (Hsu et al, 2011). To correlate the transient Slc1a3 expression to stem cell activity, we assessed the number of cycling ORS cells that co‐expressed CreERT2. The majority (64 ± 6%) of Ki67+ cells co‐expressed CreERT2. Similarly, the majority of cells in S‐phase (76 ± 6%; labeled by injection of the nucleotide analog EdU 2 h before sacrifice) were CreERT2+ (Fig 4K). Together, these results show that Slc1a3‐CreERT2 expression is associated with proliferation.
Slc1a3 is transiently expressed in active SG and IFE stem and progenitor cells
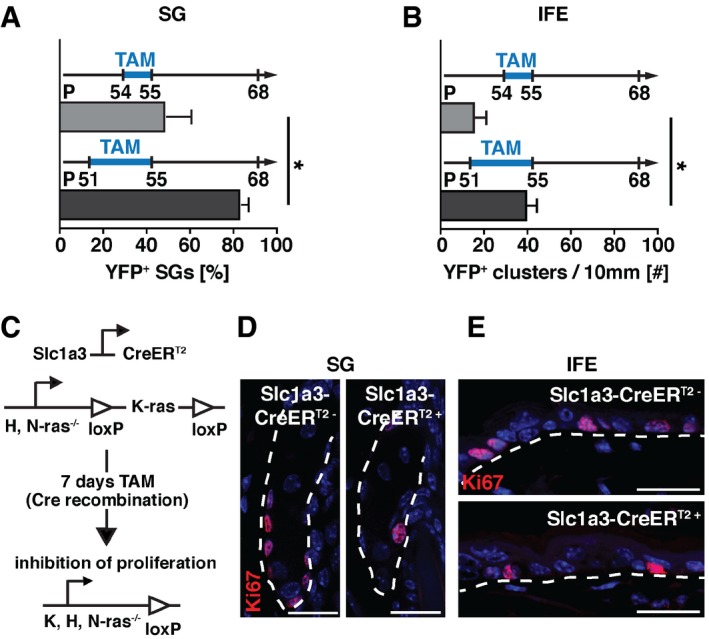
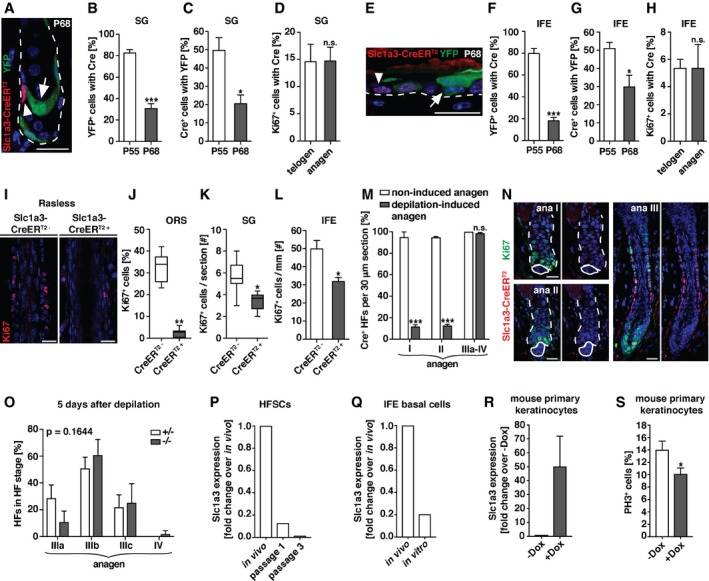
In SGs, a small fraction of basal cells expressed Slc1a3 and CreERT2 at all times (Figs 1I and J, EV1D and E, and EV3E), while a large proportion of lineage‐traced SGs lost YFP expression over time (Figs 3F and EV3D), suggesting that Slc1a3 expression is transient. To address this question, we reduced tamoxifen administration to 2 days (P54‐55) instead of five (P51‐55; Fig 3E–I) and compared the YFP expression. When analyzed at P68, we detected a significant reduction in the percentage of YFP+ SGs from 83 ± 3% to 49 ± 12%. Also, the number of YFP+ clusters/10 mm of IFE reduced from 40 ± 5 to 16 ± 6 (Fig EV4A and B). These results suggest that Slc1a3 expression in SG and IFE is changed within days and verifies the 5‐days tamoxifen regime. To test this further, we examined skin sections from mice that had received tamoxifen between P51‐55 and were fate mapped until P55 or P68 for YFP and CreERT2 expression. At P55, 83 ± 3% of all YFP+ basal cells in recombined SGs co‐expressed CreERT2+ and 50 ± 7% of all CreERT2+ basal cells co‐expressed YFP. At P68, only 31 ± 4% of all YFP+ cells co‐expressed CreERT2+ and 20 ± 5% of the CreERT2 expressing basal cells were YFP+ (Fig 5A–C), showing that previously Slc1a3‐expressing basal cells had lost Slc1a3 expression, whereas others upregulated Slc1a3. Importantly, the ratio of proliferating Ki67+ cells that co‐expressed CreERT2 remained constant in telogen and anagen (Fig 5D).
Figure EV4. Slc1a3 is transiently expressed in active cycling SG and IFE cells.
-
APercentage of YFP+ SGs after 2 (n = 4, 200 SGs) compared to 5 (n = 4, 200 SGs) consecutive days of tamoxifen injection, assessed at the same time during telogen.
-
BNumber of YFP+ cell clusters in 10 mm IFE after 2 (n = 4, 40 mm) compared to 5 (n = 3, 30 mm) consecutive days of tamoxifen injection, assessed at the same time during telogen.
-
CSchematic depiction of the genetic strategy to block proliferation of Slc1a3+ cells, by tamoxifen inducible cell‐specific deletion of all Ras genes.
-
D, ERepresentative micrographs showing proliferating (Ki67+) cells in SG (D) and IFE (E) after recombination‐mediated block of proliferation in Slc1a3+ cells (Slc1a3‐CreERT2+) compared to control (Slc1a3‐CreERT2−) animals.
Figure 5. Slc1a3 is transiently expressed in actively cycling hair follicle, SG, and IFE stem/progenitor cells but not in response to injury.
-
A–CRepresentative micrograph (A) and quantification of YFP+ SG basal cells expressing Slc1a3‐CreERT2 (B) and Slc1a3‐CreERT2+ cells expressing YFP (C) following recombination induction (P51‐55) at P55 (n = 3, 72 SGs) and P68 (n = 3, 64 SGs).
-
DPercentage of Ki67+ SG cells that co‐express Slc1a3‐CreERT2 in telogen (n = 3, 60 SGs) compared to anagen (n = 4, 80 SGs).
-
E–GRepresentative micrograph (E) and quantification of YFP+ IFE basal cells expressing Slc1a3‐CreERT2 (F) and Slc1a3‐CreERT2+ basal cells expressing YFP (G) following recombination induction (P51‐55) at P55 (n = 3, 150 YFP+ and 234 CreERT2+ basal cells, respectively) and P68 (n = 3, 209 YFP+ and 123 CreERT2+ basal cells, respectively).
-
HPercentage of Ki67+ IFE cells that co‐express Slc1a3‐CreERT2 in telogen (n = 3, 150 Ki67+ cells) compared to anagen (n = 3, 150 Ki67+ cells).
-
I–LRepresentative micrographs (I) and quantification of proliferating cells in ORS (J), SG (K) and IFE (L) of Rasless mice (n = 3, 22 HFs, 52 SGs and 7 mm IFE, respectively), in which mitosis was inhibited in Slc1a3‐CreERT2+ cells, compared to control (ORS n = 3, 18 HFs; SG and IFE n = 4, 65 SGs and 10 mm IFE, respectively).
-
M, NQuantification of percentage of Slc1a3‐CreERT2+ HFs in 30 μm sections in non‐induced anagen (anagen I, n = 2, 18 HFs; anagen II, n = 2, 39 HFs; anagen III‐IV, n = 3, 60 HFs) and depilation (anagen I, n = 3, 60 HFs; anagen II, n = 3, 56 HFs; anagen III‐IV, n = 4, 119 HFs; M) and representative micrographs of Slc1a3‐CreERT2 and Ki67 in depilated HFs (N).
-
OHair follicle stage distribution 5 days after depilation in Slc1a3+/− (n = 3, 45 HFs) compared to Slc1a3−/− (n = 3, 60 HFs) mice. P = 0.1644 derived from grouped comparison of ranked HF stages in Slc1a3+/− and Slc1a3−/− mice.
-
PSlc1a3 mRNA expression in hair follicle stem cells (HFSCs) after passage 1 and passage 3 compared to expression in vivo.
-
QSlc1a3 mRNA expression in basal IFE cells in culture compared to expression in vivo.
-
RSlc1a3 mRNA expression in mouse primary keratinocytes after overexpression of hSlc1a3 (+Dox; n = 3 repeats) compared to control (−Dox; n = 3 repeats).
-
SPercentage of proliferating PH3+ mouse primary keratinocytes expressing hSlc1a3 (+Dox; n = 18 technical replicates) compared to control (−Dox; n = 18 technical replicates).
Lineage tracing of IFE Slc1a3+ cells confirmed their progenitor cell identity. After tracing, however, many YFP+ basal cells were CreERT2−, and instead, neighboring YFP− basal cells were found to be CreERT2+ (Fig 5E), suggesting that Slc1a3 is transiently expressed in basal cells. We confirmed this finding by analyzing skin sections from mice that had received tamoxifen P51‐55 and were fate mapped until P55 and P68 for YFP and CreERT2 expression. At P55, 80 ± 5% of all YFP+ basal cells co‐expressed CreERT2 and 51 ± 3% of all CreERT2+ basal cells co‐expressed YFP. At P68, only 17 ± 3% of all YFP+ basal cells co‐expressed CreERT2 and 30 ± 6% of the CreERT2 expressing basal cells were YFP+ (Fig 5E–G). As for the SG, the ratio of Ki67+ cells with CreERT2 expression was constant comparing telogen and anagen (Fig 5H). Together, these results show that Slc1a3 is transiently expressed in actively cycling SG and IFE stem and progenitor cells.
Accumulative reduction in stem/progenitor cell division due to genetic cell cycle arrest in Slc1a3‐expressing cells
To further test the association of Slc1a3 expression with stem cell activity, we specifically blocked proliferation in Slc1a3+ cells using Slc1a3‐Rasless mice, which, in addition to carrying the Slc1a3‐CreERT2 allele, are homozygous for H‐Ras and N‐Ras null alleles and for floxed K‐Ras alleles. Thus, Slc1a3+ cells lack all Ras genes after recombination (Fig EV4C). Ras genes are obligatory for cell cycle progression and mitosis, so deleting all Ras genes in Slc1a3+ cells abrogates their proliferation (Göritz et al, 2011). Tamoxifen‐induced recombination of Slc1a3‐Rasless mice during anagen (P23‐29) almost completely abrogated ORS proliferation (Fig 5I and J) and significantly reduced the number of Ki67+ cells in SG and IFE (Figs 5K and L, and EV4D and E), analyzed at P30. In all three niches, proliferation was reduced to a much larger extent than the co‐expression of Ki67 and CreERT2 at a specific time point would have suggested, validating the transient Slc1a3 expression in actively cycling stem/progenitor cells in hair follicle, SG, and IFE.
Injury‐induced stem cell activation is Slc1a3 independent
Depilation is considered as a mild injury, leading to hair follicle stem cell proliferation and hair growth. We investigated Slc1a3‐CreERT2 expression after depilation‐induced injury and discovered that in contrast to growth‐induced stem cell activation, hair follicle stem cells do not upregulate Slc1a3 after injury. After depilation, hair follicle stem cells in anagen I and II show significantly reduced CreERT2 expression when compared to non‐induced anagen hair follicles. Interestingly, during anagen III‐IV a normal morphology and proliferation pattern is reestablished and CreERT2 is expressed in the ORS of depilated and non‐induced anagen hair follicles alike (Fig 5M and N). To address the functional role of Slc1a3 in depilation‐induced anagen, we analyzed Slc1a3 +/− and Slc1a3 −/− hair follicles 5 days after depilation. In contrast to non‐induced anagen (Fig 2A–G), the absence of Slc1a3 did not affect the growth of the depilation‐induced new hair follicle (Fig 5O). To address the potential loss of Slc1a3 in a different injury model, we sorted CD34+ anagen hair follicle stem cells as well as IFE basal cells (Sca1+/Itga6+) and cultured them under proliferative conditions. While we consistently detected Slc1a3 mRNA in in vivo isolated cells, Slc1a3 expression was downregulated in culture (Fig 5P and Q). Re‐expression of Slc1a3 in cultured primary mouse keratinocytes (Fig 5R) resulted in reduced proliferation (Fig 5S). These results suggest Slc1a3 acts as a cell‐autonomous modulator of epidermal stem cell activation during growth and homeostasis, but not after wounding.
Slc1a3 acts in conjunction with mGluR5
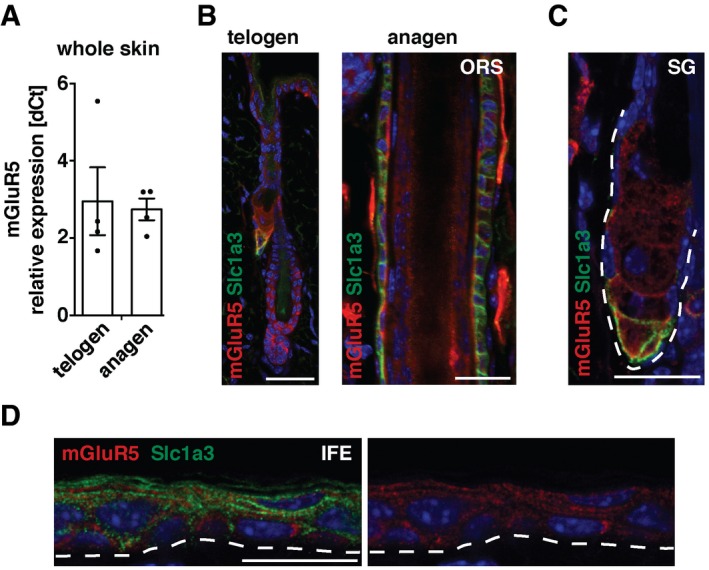
The function of Slc1a3 in the nervous system is to buffer and clear extracellular glutamate in the proximity of glutamate receptors to regulate local glutamate abundance and receptor excitability (Huang et al, 2004; Tzingounis & Wadiche, 2007; Divito & Underhill, 2014). We found mRNA expression of the metabotropic glutamate receptor mGluR5 both during anagen and telogen (Fig 6A). Immunohistochemistry localized mGluR5 expression to telogen and anagen hair follicle, HG, SG, and IFE as well as to the ORS during anagen, partly co‐localizing with Slc1a3 in all three niches (Fig 6B–D).
Figure 6. MGluR5 expression partly co‐localizes with Slc1a3 in hair follicle, SG, and IFE stem cell niches.
-
ARelative mGluR5 mRNA expression in whole telogen and anagen skin (dCt, n = 4 for each group). Data are mean ± SEM.
-
B–DMGluR5 is expressed in the hair follicle (B), SG (C) and IFE (D), partially co‐localizing with Slc1a3. Dashed line outlines the SG (C) and indicates the epidermal–dermal border (D). Scale bar = 20 μm.
Source data are available online for this figure.
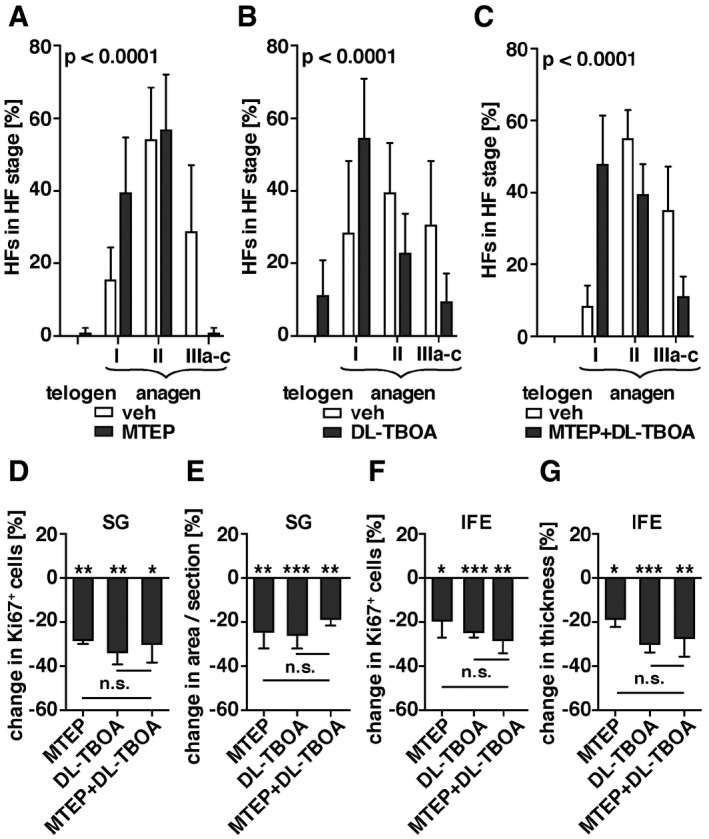
To test for a co‐dependence of Slc1a3 and mGluR5, we topically treated mice with the mGluR5 allosteric antagonist MTEP at the beginning of anagen (P22–P25) and staged hair follicles directly after (P26). MTEP treatment delayed anagen stage progression compared to vehicle‐treated animals (Fig 7A). Topical treatment with the competitive Slc1a3 inhibitor DL‐TBOA caused a similar delay in anagen progression (Fig 7B), which was comparable to the phenotype observed in Slc1a3 −/− mice (Fig 2A and G). Combined application of MTEP and DL‐TBOA did not exceed the effect of single compounds, suggesting that Slc1a3 and mGluR5 act on the same pathway (Fig 7C). Furthermore, treatment of Slc1a3 −/− mice with MTEP had no effect on anagen progression beyond the Slc1a3 −/− phenotype alone (Fig EV5A), further strengthening the conclusion of Slc1a3 and mGluR5 cooperation. Treatment with l‐glutamate only slightly expedited anagen entry compared to vehicle‐treated animals (Fig EV5B). Moreover, we found that blocking mGluR5 or Slc1a3 significantly reduced anagen‐induced growth of SG and IFE. Again, there was no additive effect of the combined antagonist treatment (Fig 7D–G) and no effect of MTEP on Slc1a3 −/− mice (Fig EV5C–F). Application of l‐glutamate did not significantly influence SG and IFE growth (Fig EV5G–J). These data imply that Slc1a3 acts in conjunction with mGluR5.
Figure 7. Slc1a3 acts in conjunction with mGluR5.
-
A–CQuantification of hair cycle progression assessed by hair follicle staging following topical treatment with mGluR5 antagonist MTEP (n = 5, 75 HFs; A), Slc1a3‐antagonist DL‐TBOA (n = 4, 60 HFs; B) or combined MTEP and DL‐TBOA treatment (n = 4, 60 HFs; C) in comparison with vehicle‐treated mice (n = 5, 75 HFs; n = 3, 60 HFs; n = 3 60 HFs, respectively). P < 0.0001 derived from grouped comparison of ranked HF stages in mice treated with vehicle and antagonist.
-
D–GQuantification of anagen‐induced proliferation (D, F) and growth (E, G) of SG (D, E) and IFE (F, G) following topical treatment with MTEP (n = 4, 29 SGs and 10 mm, respectively), DL‐TBOA (n = 5, 38 SGs and n = 6, 10 mm), or MTEP + DL‐TBOA (n = 4, 38 SGs and 12 mm, respectively) relative to vehicle (n = 9, 72 SGs and 20 mm, respectively). P‐values derived from comparison of the mean induced proliferation and growth of SG and IFE in mice treated with vehicle and antagonist. Graph shows the percentage change normalized to the vehicle group.
Figure EV5. Slc1a3 and mGluR5 act together.

-
AQuantification of hair cycle progression assessed by hair follicle staging following topical treatment with mGluR5 antagonist MTEP (n = 4, 120 HFs) or vehicle (n = 4, 120 HFs) in Slc1a3−/− mice. P‐value derived from grouped comparison of ranked HF stages in mice treated with vehicle and MTEP.
-
BQuantification of hair cycle progression assessed by hair follicle staging following cutaneous treatment with l‐glutamate (l‐Glu; n = 5, 75 HFs) in comparison with vehicle‐treated mice (n = 3, 48 HFs). P‐value derived from grouped comparison of ranked HF stages in mice treated with vehicle and l‐Glu.
-
C–FQuantification of proliferation (C, E) and growth (D, F) of SG (C, D) and IFE (E, F) following topical treatment with MTEP (n = 4, 79 SGs and 7 mm IFE, respectively) or vehicle (n = 4, 77 SGs and 8 mm IFE, respectively) in Slc1a3−/− mice.
-
G–JQuantification of proliferation (G, I) and size (H, J) of SG (G, H) and IFE (I, J) following cutaneous treatment with l‐glutamate (n = 4, 46 and 48 SGs, 5 mm, respectively) compared to vehicle (n = 3, 32 and 22 SGs, 3 mm, respectively).
Discussion
We report that skin growth is mediated through coordinated stem/progenitor cell proliferation in three distinct epithelial stem cell niches. As hair follicle stem cells transition from quiescence to activation, their proliferative burst is mirrored by the proliferative kinetics of IFE and SG. We identified the glutamate transporter Slc1a3 as a general marker and effector of activated epithelial stem cells throughout the skin. Each niche has a distinct ratio of proliferating to Slc1a3‐expressing cells, which remains constant during growth and homeostasis. Such a system enables each individual niche to maintain its characteristics while expanding in synchrony with surrounding niches. Neural stem cells express Slc1a3 and mGluR5 (Shibata et al, 1997; Mori et al, 2006; Slezak et al, 2007; Gilley & Kernie, 2011; Nochi et al, 2012), and in Drosophila, dietary l‐glutamate stimulates intestinal stem cell division and gut growth via the metabotropic glutamate receptor (Deng et al, 2015), suggesting that niche‐spanning stem cell activation could be a general mechanism. In conclusion, our data reveal new properties of epithelial tissue growth and a role for Slc1a3 in inter‐niche modulation of stem/progenitor cell activation.
Slc1a3 is transiently expressed in activated stem cells
We identified the glutamate transporter Slc1a3 as highly enriched in a subset of activated hair follicle stem cells when compared to their quiescent counterparts. Furthermore, a distinct subset of basal SG and IFE cells expressed Slc1a3 (Figs 1 and EV1).
We discovered that stem cell niches in the skin require Slc1a3 for their coordinated growth. When Slc1a3 was ablated, the hair cycle was delayed and proliferation rates decreased (Fig 2A–G). With time, the absence of Slc1a3 resulted in a thinning of the hair coat, suggesting an accumulative effect of failing to properly initiate hair follicle cycling (Figs 2J–L and EV2F). Additionally, Slc1a3 ablation resulted in reduced SG and IFE proliferation but also, importantly, a failure to respond to the activation signals that normally couple these niche proliferation patterns to the hair cycle. As a consequence, aged mice lacking Slc1a3 developed smaller SGs and thinner IFE (Fig 2M–R). This enhanced aging phenotype is similar to human skin aging, but the opposite of aging phenotypes observed in, for example, circadian clock abrogation (Janich et al, 2011). It will be important to determine whether increased Slc1a3 function would rejuvenate skin.
At first, we were puzzled by the severe consequences of ablating Slc1a3 in a small subset of epithelial cells, but tracing of progeny derived from Slc1a3‐expressing cells showed that Slc1a3 marked a subset of stem/progenitor cells in all compartments (Figs 3 and EV3). During anagen progression, Slc1a3 expression is transiently detected in defined subpopulations of hair follicle stem cells. We detected the same transitory expression of Slc1a3 in both SG and IFE when following recombined YFP+ basal cells over time and determining the fraction also expressing CreERT2, indicative of their Slc1a3 expression (Figs 4A and 5A–C and E–G). In both SG and IFE, we found that the increase in proliferation detected as the skin transitions from telogen to anagen arose from the Slc1a3+ population, suggesting that Slc1a3 is a marker or inducer of an activated stem and progenitor cell state. Interestingly however, the ratio of CreERT2+ cells contained within the cycling Ki67‐expressing population varied between niches (Fig 5D and H), indicating the differential proliferative needs and kinetics of individual stem cell systems. To more directly address the dynamic expression of Slc1a3, we haltered proliferation of Slc1a3+ cells by genetically ablating all Ras genes in Slc1a3‐expressing cells. Seven days of tamoxifen administration during anagen almost completely abrogated proliferation in the ORS and dramatically reduced proliferation in the SG and IFE. The reduction in proliferation exceeds predictions based on Ki67 and static CreER expression (Figs 5I–L and EV4), and further supports the transient nature of Slc1a3‐expression.
Behavior of activated stem cells
In the ORS, Slc1a3+ cells were both spatially and temporally regulated and this correlated with their fate. Slc1a3+ cells in the ORS contributed to the matrix and inner differentiated layers of the hair follicle in mid‐ and late anagen (Fig 4A–H), in contrast to the expansion of the activated HG, which drives matrix formation in early anagen (Rompolas et al, 2013). However, we also found that the ORS fate was correlated with the temporal regulation of Slc1a3 expression along the hair follicle. This finding is intriguing in light of previous work correlating the potential of ORS cells and their cycling behavior (Hsu et al, 2011). It is interesting that we did not detect any Slc1a3 expression in hair bulb transit amplifying cells, perhaps a reflection of their short‐lived nature or their rapid proliferation. Furthermore, Slc1a3‐expression remains low in hair follicle and IFE stem/progenitor cells after wounding such as depilation‐induced hair growth or cell culture (Fig 5M, N, P and Q), and reintroduction of Slc1a3 in cultured keratinocytes negatively affects their proliferation (Fig 5S). It is tempting to speculate that glutamate signaling, through regulation of Slc1a3 expression, modulates the activated stem cell state during tissue growth and homeostasis while hyperproliferation of hair follicle transit amplifying cells or hair follicle stem cell proliferation as a response to injury, is Slc1a3‐independent and regulated by other mechanisms. Using a different Slc1a3‐CreERT2 mouse line, a recent study reported selective targeting of the inner root sheath in the anagen follicle (Sada et al, 2017). We could detect neither Slc1a3 nor CreERT2 protein expression in the inner root sheath but only in the outer root sheath, indicating that the line used by Sada et al does not recapitulate the endogenous expression faithfully.
Slc1a3 labels long‐term contributing stem and progenitor cells in three epithelial compartments
We used lineage tracing to examine stem cell potential in Slc1a3‐expressing cells. We targeted Slc1a3+ cells in the ORS at 2nd anagen (P25/26) and traced them over two consecutive hair cycles to P68 and P117. The vast majority of analyzed hair follicles contained YFP+ daughter cells contributing to the new bulge and HG, demonstrating long‐term stem cell potential of targeted ORS cells (Fig 3A–D).
The identity and location of SG progenitor cells have been much debated (Horsley et al, 2006; Jensen et al, 2009; Kretzschmar et al, 2014; Fullgrabe et al, 2015). Recombined Slc1a3+, YFP+ cells in the SG basal layer generate differentiated progeny and expand the YFP+ basal cell population (Fig 3G–I). Some Slc1a3+ basal cells eventually compete out other basal cells, hence generating a clonal structure, in striking reminiscence of the intestinal stem cell niche, in which single Lgr5+ basal crypt cells divide stochastically to generate a fixed number of stem cells within the crypt, with neutral competition for the restricted niche eventually leading to clonality (Lopez‐Garcia et al, 2010; Snippert et al, 2010b).
Clonal analysis from cells targeted for recombination using Slc1a3‐CreER T2 in the IFE showed that the majority of clusters were maintained over time, with the vast majority of clones producing differentiated progeny 8 months after tamoxifen (Fig 3K and L). However, the clones varied dramatically in size, with some single basal cell clones being maintained for up to 8 months, whereas others readily expanded laterally. These data demonstrate that Slc1a3+ stem cells differ in their spatial restrictions. Furthermore, it unveils a novel feature of IFE stem cells, identifying a subset of Slc1a3+ basal cells that only generate suprabasal progeny without expanding laterally during homeostasis.
Glutamate signaling in stem cell niches
We identified mGluR5 as a constitutively expressed interaction partner for Slc1a3 in the skin epithelia (Fig 6). Strikingly, inhibition of mGluR5 phenocopies the effects seen in Slc1a3 −/− (Figs 2A–G, M, N, and 7A and D–G), while Slc1a3 −/− mice are refractory to mGluR5 inhibition (Fig EV5A and C–F).
In skin, glutamate is released by keratinocytes and is increased in the basal layer by disruption of the outer epithelial layer (Nordlind et al, 1993; Fuziwara et al, 2003). Macrophages located in epidermis and dermis also release glutamate, and this increases in response to inflammation (Nordlind et al, 1993; Zhao et al, 2004). In Drosophila, dietary l‐glutamate stimulates intestinal stem cell division and gut growth via the metabotropic glutamate receptor (Deng et al, 2015), showing that extracellular glutamate can influence stem cell proliferation. However, application of l‐glutamate did not significantly affect anagen onset or SG and IFE proliferation (Fig EV5B and G–J), suggesting that extracellular glutamate is abundant in the skin.
Neural stem cells in the developing cerebral cortex and adult hippocampus express Slc1a3 (Mori et al, 2006). While it is known that injury increases Slc1a3 expression in adult hippocampal stem cells (Gilley & Kernie, 2011), the in vivo function of Slc1a3 in neural stem cells is still unknown. However, deletion of Slc1a3 in cultured neural stem cells alters proliferation and neurosphere formation in an mGluR5‐dependent manner (Gilley & Kernie, 2011). Melanocytes express mGluR5 and overexpression induces melanoma formation in vivo (Choi et al, 2011), indicating a role in regulation of proliferation. A role for glutamate signaling in regulating stem cell proliferation is beginning to appear, but it is clear that the cellular outcome is context dependent. It is tempting to speculate that glutamate release from keratinocytes and macrophages could stimulate mGluR5‐dependent stem cell activation; however, our data imply that mGluR5 activity is gated by other factors. Slc1a3 could play such a role by increasing the receptor deactivation rate. Another possible interpretation is that low receptor activity is important for stem cell activation and that no or high activity both could lead to quiescence. By temporally regulating the Slc1a3 expression in a subpopulation of stem and progenitor cells in the constitutive presence of the glutamate receptor, each stem cell niche creates an elegant and highly dynamic signaling mechanism, which allows stem cells to rapidly respond to environmental cues. Here, we propose a modulatory role for Slc1a3 in stem and progenitor cell activation during growth and homeostasis, distinct from the proliferative burst initiated by an acute injury.
Materials and Methods
Mice and lineage‐tracing experiments
Slc1a3‐CreERT2 (Slezak et al, 2007) were crossed to Rosa26‐YFP reporter (Srinivas et al, 2001) or Rasless (Drosten et al, 2010) mice. CreERT2‐mediated recombination was initiated by intraperitoneal injection or oral gavage of 20 mg/ml tamoxifen (Sigma) in corn oil at different time points. For hair follicle stem cell lineage tracing, 50 μl tamoxifen was administered two times per day for 2 days in 2nd anagen at mid‐anagen (P24/25 or P25/P26) or late anagen (P29/P30). For SG and IFE lineage tracing, 100 μl tamoxifen was administered once per day for 5 days in 2nd telogen from P51 to P55. For Rasless experiments, 50 μl tamoxifen was administered by oral gavage two times per day from P23–P29 and mice were perfused at P30. For CreERT2 detection, tamoxifen was administered 2 h before perfusion to mediate nuclear CreERT2 localization. The thymidine analogue 5‐ethynyl‐2′‐deoxyuridine (EdU) was administered by intraperitoneal injection 2 h before perfusion. Slc1a3 −/− mice were described previously (Watase et al, 1998), and Slc1a3tm1.1(KOMP)Mbp was purchased from the KOMP Repository (MGI ID = MGI:5443926). Animal experiments were carried out in accordance to Swedish and European law and approved by Jordbruksverket.
Topical pharmacological treatment
The lower back of female mice was shaved, and 50 μl of the respective compound or vehicle was applied twice per day for 4 days, starting at P22. The following compounds were applied: DL‐TBOA (21 mM in DMSO; Tocris) and MTEP (15 mM in DMSO; Abcam). l‐Glutamate (2.5%; l‐glutamic acid; Sigma) was applied in a cutaneous emulsion (Propyless, containing 20% propylene glycol and 9% fat; Merck Sharp & Dome).
Depilation and hair plucking
Depilation of the lower back of mice was done using Veet Warm Wax. Hair was plucked from the lower back and fixed in 4% formaldehyde solution before immunohistochemistry.
Histology and immunohistochemistry
Mice were perfused and postfixed with 4% formaldehyde solution. Caudal back skin was cryopreserved in 30% sucrose and embedded in OCT for sagittal or horizontal (12 or 30 μm) cryosectioning. For histological analysis, sections were stained with hematoxylin and eosin. For immunohistochemistry, sections were permeabilized and blocked for 45–60 min with 10% normal donkey serum, 0.3% Triton in PBS followed by primary and species‐specific fluorophore‐conjugated secondary antibody incubation. Antibodies were diluted in 10% normal donkey serum in PBS. Biotinylated primary antibodies were detected with a Cy3‐conjugated streptavidin (Jackson laboratories). For Cre and Loricrin co‐labeling, immunostaining for Cre was completed after which slides were blocked with 10% rabbit serum followed by Loricrin labeling using a directly fluorophore‐conjugated anti‐Loricrin antibody (Zenon® antibody labeling kit, Thermo Fisher Scientific). For visualization of EdU, sections were incubated in 4 mM CuSo4, 10 mM ascorbic acid, and 1 μM Azide Fluorophore (Alexa Fluor® 647 Azide, Thermo Fisher) in PBS for 30 min.
Nuclei were stained with DAPI (4′6′‐diamidino‐2‐phenylindole; 1 μg/ml, Sigma). Slides were mounted with Vectashield Mounting Medium (Vector labs) or Mowiol supplemented with DABCO (both Carl Roth).
The following primary antibodies were used: CD34 (rat, 1:50; Abcam #ab8158), Cre (rabbit, 1:3,000; generous gift from Günther Schütz Heidelberg or rabbit, 1:500; Cell Signaling #15036), FABP5 (goat biotinylated, 1:1,000–1:4,000; R&D Systems #BAF1476), Loricrin (rabbit, 1:1,000; Abcam #ab24722), Ki67 (rat, 1:2,000; eBioscience #14‐56898 or rabbit, 1:250–1:500; Thermo Scientific #RM‐9106), K5 (Cytokeratin 5; rabbit, 1:1,000; Abcam #ab52635 or guinea pig, 1:1,000; Acris #BP5006), K6 (Cytokeratin 6; rabbit, 1:2,000; Abcam #ab24646), mGluR5 (rabbit, 1:250; Abcam #ab53090), P‐cadherin (rat, 1:50; Invitrogen #12‐2000Z), PH3 (Histone H3 pS10; rabbit, 1:1,000; Abcam #ab5176), Slc1a3 (Glast; guinea pig, 1:500; Frontier Institute #GLAST‐GP‐Af1000), Sox9 (goat, 1:500; R&D Systems #AF3075), and GFP (chicken, 1:4,000; Aves Lab #GFP‐1020 or goat, 1:1,000; FITC‐labeled; Abcam #ab6662).
Images were acquired with a Leica SP8X or a Zeiss LSM700 confocal microscope. Quantifications were performed on overview and high‐resolution images using Image J/Fiji software.
Hair cycle staging
Hair cycle phases and anagen stages were determined according to morphological criteria previously described (Muller‐Rover et al, 2001). For quantitative purposes, 15–20 vertically sectioned hair follicles from the lower back were staged per animal. Siblings, including both male and female mice, were analyzed, except for topical pharmacological treatments, where only females were treated and quantified.
Quantifications and statistical analysis
All quantifications were performed in mixed groups of male and female mice except for topical pharmacological treatments, where only females were treated and quantified.
Quantifications of 1st telogen were performed at P22–P24. Quantifications of 2nd anagen were performed in hair follicle stages III‐IV (mid‐anagen) at P25–P26. Quantifications of catagen were performed at P40. Quantifications of 2nd telogen were performed at P49, P55, P58, and P68. Quantifications of 3rd anagen were performed at P89. Quantifications of early anagen were performed at P24 or P102. In the IFE, thickness was measured from the basal cell layer to and including Loricrin+ suprabasal cells.
Sample sizes were not predetermined using statistical methods but were based on previous experience. Data are presented as mean ± standard error of mean (SEM) or ± SEM. In some graphs, individual data points are plotted. Sample sizes (n) are indicated in the Figure legends. Data analysis and statistics were performed using GraphPad Prism or Excel. Mann–Whitney U‐tests for general differences of hair follicle stages in Slc1a3 +/− and Slc1a3 −/− animals and upon cutaneous treatment were performed on ranked hair follicle stages in the different animal groups.
Overexpression of hSlc1a3 in keratinocytes
Mouse primary keratinocytes were infected with a lentiviral pCW57.1 vector (Addgene #41393) expressing hSlc1a3 under a doxycycline‐inducible promoter. Infected cells were selected using puromycin for 7 days, after which doxycycline was administered for 3 days.
Skin dissociation and fluorescence‐activated cell sorting
For isolation of cells in telogen, postnatal mouse skin was dissected and adipose tissue was scraped off. Remaining skin was digested in 0.05% trypsin (GIBCO) and incubated for 30 min at 37°C. Cells were scraped off from the epidermal side, filtered through strainers (70, 40 μm) and pelleted. For isolation of cells in anagen, skin was placed in Collagenase (5 mg/ml in HBSS; Sigma) and incubated for 1 h at 37°C. Cells were scraped off from the dermal side and incubated in 0.05% trypsin (GIBCO) for 10–15 min at 37°C. The cell suspension was filtered through strainers (70, 40 μm) and pelleted. The single‐cell suspensions were incubated with the respective antibodies for 30 min on ice. The following antibodies were used: CD34‐A700 (rat, 1:100; eBioscience #56‐0341‐82), CD49f (Integrin alpha 6)‐PE (rat, 1:1,000; eBioscience #12‐0495‐81), Sca1‐PerCP‐Cy5.5 (rat, 1:500; eBioscience #45‐5981). After sorting, the CD34+/CD49f+/Sca1‐ hair follicle bulge cells were purified using a Becton Dickinson FACS Aria flow cytometer with the FACSDiva software (BD Biosciences). Purity of the sorted population was validated with RT–qPCR for CD34 and Sca1 expression.
Quantitative reverse transcription PCR (RT–qPCR)
Total RNA was isolated from FACS‐sorted cells using Trizol‐LS (Life Technologies) followed by RNA extraction: The RNA phase was separated by mixing the isolated cells with chloroform; for RNA precipitation, EtOH was added to the sample before loading it onto RNEasy columns and processing according to the protocol (RNEasy Mini Kit, Qiagen).
For RNA extraction from whole skin, the Direct‐zol™ RNA MiniPrep Kit (Zymo Research) was used. RNA was reverse‐transcribed using the SuperScript® VILO cDNA Synthesis Kit (Invitrogen). RT–qPCR was conducted using a 7500 Fast Real‐time PCR System from Applied Biosystems and Fast CYBR® Green Master Mix (Life Technologies). Mouse Hprt was used as endogenous control. Relative expression was calculated as dCt. Fold change in mRNA levels was calculated as 2ddCt. Primers used are listed below.
The RT–qPCR for Slc1a3 analysis was performed with pooled samples obtained from seven animals in telogen and four animals in anagen. The RT–qPCR was performed using triplicates in two technical replicates. The RT–qPCR for mGluR5 analysis was performed comparing samples from four mice for telogen and anagen each. Each RT–qPCR was performed as triplicates.
Primers (and their sequences) used for RT–qPCR were HPRT‐F (tcagtcaacgggggacataaa), HPRT‐R (ggggctgtactgcttaaccag), HSLC1A3‐F (tacgagtgacagctgcagat), HSLC1A3‐R (tccatggcctcagacacatt), MGLUR5‐F (agaaacccatagtgggagtcat), MGLUR5‐R (atgctagttgcagagtaagcaat), SLC1A3‐F (ccaaaagcaacggagaagag), and SLC1A3‐R (acctcccggtagctcatttt).
Author contributions
JC and BR performed experiments and analysis. CG and BR performed lineage‐tracing experiments. TA and KT contributed Slc1a3 k.o. experiments. MG designed experiments. CG designed and supervised the study. JC, BR, MG, and CG wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Acknowledgements
We thank Maria Kasper, Jonas Frisén, Christopher Uhde, Pekka Katajisto, and members of the Göritz laboratory for valuable comments on the manuscript. We thank M. Barbacid for providing Ras mutant mice through a material transfer agreement with the Centro Nacional de Investigaciones Oncológicas (Spanish National Cancer Institute, CNIO), Spain. We thank F. W. Pfrieger for providing GLAST‐CreERT2 transgenic mice through a material transfer agreement with the Institut Génétique Biologie Moléculaire Cellulaire (Strasbourg, France). Requests for mice should be directed to M. Barbacid (CNIO, Spain) and F. W. Pfrieger (CNRS, France), respectively. M.G. is a Söderberg fellow. C.G. is a Hållsten Academy and a Knut och Alice Wallenberg Academy Fellow. Research in the Göritz laboratory was supported by the European Union's Seventh Framework Programme (FP7)/ERC‐2012‐StG 310938 PERICYTESCAR, Swedish Research Council, SFO StratRegen, Hjärnfonden, Ming Wai Lau Centre, Swedish Cancer Foundation and the Åke Wiberg foundation.
The EMBO Journal (2018) 37: e98280
See also: https://doi.org/10.15252/embj.201899393 (May 2018)
References
- Choi KY, Chang K, Pickel JM, Badger JD II, Roche KW (2011) Expression of the metabotropic glutamate receptor 5 (mGluR5) induces melanoma in transgenic mice. Proc Natl Acad Sci USA 108: 15219–15224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Gerencser AA, Jasper H (2015) Signal integration by Ca(2+) regulates intestinal stem‐cell activity. Nature 528: 212–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divito CB, Underhill SM (2014) Excitatory amino acid transporters: roles in glutamatergic neurotransmission. Neurochem Int 73: 172–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten M, Dhawahir A, Sum EY, Urosevic J, Lechuga CG, Esteban LM, Castellano E, Guerra C, Santos E, Barbacid M (2010) Genetic analysis of Ras signalling pathways in cell proliferation, migration and survival. EMBO J 29: 1091–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Lien WH, Stokes N, Pasolli HA, Silva JM, Fuchs E (2011) EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev 25: 485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E, Nowak JA (2008) Building epithelial tissues from skin stem cells. Cold Spring Harb Symp Quant Biol 73: 333–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullgrabe A, Joost S, Are A, Jacob T, Sivan U, Haegebarth A, Linnarsson S, Simons BD, Clevers H, Toftgard R, Kasper M (2015) Dynamics of Lgr6 progenitor cells in the hair follicle, sebaceous gland, and interfollicular epidermis. Stem Cell Rep 5: 843–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuziwara S, Inoue K, Denda M (2003) NMDA‐type glutamate receptor is associated with cutaneous barrier homeostasis. J Invest Dermatol 120: 1023–1029 [DOI] [PubMed] [Google Scholar]
- Genander M, Cook PJ, Ramskold D, Keyes BE, Mertz AF, Sandberg R, Fuchs E (2014) BMP signaling and its pSMAD1/5 target genes differentially regulate hair follicle stem cell lineages. Cell Stem Cell 15: 619–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley JA, Kernie SG (2011) Excitatory amino acid transporter 2 and excitatory amino acid transporter 1 negatively regulate calcium‐dependent proliferation of hippocampal neural progenitor cells and are persistently upregulated after injury. Eur J Neurosci 34: 1712–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göritz C, Dias DO, Tomilin N, Barbacid M, Shupliakov O, Frisen J (2011) A pericyte origin of spinal cord scar tissue. Science 333: 238–242 [DOI] [PubMed] [Google Scholar]
- Greco V, Chen T, Rendl M, Schober M, Pasolli HA, Stokes N, Dela Cruz‐Racelis J, Fuchs E (2009) A two‐step mechanism for stem cell activation during hair regeneration. Cell Stem Cell 4: 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsley V, O'Carroll D, Tooze R, Ohinata Y, Saitou M, Obukhanych T, Nussenzweig M, Tarakhovsky A, Fuchs E (2006) Blimp1 defines a progenitor population that governs cellular input to the sebaceous gland. Cell 126: 597–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsley V, Aliprantis AO, Polak L, Glimcher LH, Fuchs E (2008) NFATc1 balances quiescence and proliferation of skin stem cells. Cell 132: 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YC, Pasolli HA, Fuchs E (2011) Dynamics between stem cells, niche, and progeny in the hair follicle. Cell 144: 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YH, Sinha SR, Tanaka K, Rothstein JD, Bergles DE (2004) Astrocyte glutamate transporters regulate metabotropic glutamate receptor‐mediated excitation of hippocampal interneurons. J Neurosci 24: 4551–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janich P, Pascual G, Merlos‐Suarez A, Batlle E, Ripperger J, Albrecht U, Cheng HY, Obrietan K, Di Croce L, Benitah SA (2011) The circadian molecular clock creates epidermal stem cell heterogeneity. Nature 480: 209–214 [DOI] [PubMed] [Google Scholar]
- Jensen KB, Collins CA, Nascimento E, Tan DW, Frye M, Itami S, Watt FM (2009) Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell Stem Cell 4: 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julio‐Pieper M, Flor PJ, Dinan TG, Cryan JF (2011) Exciting times beyond the brain: metabotropic glutamate receptors in peripheral and non‐neural tissues. Pharmacol Rev 63: 35–58 [DOI] [PubMed] [Google Scholar]
- Kandyba E, Leung Y, Chen YB, Widelitz R, Chuong CM, Kobielak K (2013) Competitive balance of intrabulge BMP/Wnt signaling reveals a robust gene network ruling stem cell homeostasis and cyclic activation. Proc Natl Acad Sci USA 110: 1351–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobielak K, Stokes N, de la Cruz J, Polak L, Fuchs E (2007) Loss of a quiescent niche but not follicle stem cells in the absence of bone morphogenetic protein signaling. Proc Natl Acad Sci USA 104: 10063–10068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar K, Cottle DL, Donati G, Chiang MF, Quist SR, Gollnick HP, Natsuga K, Lin KI, Watt FM (2014) BLIMP1 is required for postnatal epidermal homeostasis but does not define a sebaceous gland progenitor under steady‐state conditions. Stem Cell Rep 3: 620–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien WH, Guo X, Polak L, Lawton LN, Young RA, Zheng D, Fuchs E (2011) Genome‐wide maps of histone modifications unwind in vivo chromatin states of the hair follicle lineage. Cell Stem Cell 9: 219–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Garcia C, Klein AM, Simons BD, Winton DJ (2010) Intestinal stem cell replacement follows a pattern of neutral drift. Science 330: 822–825 [DOI] [PubMed] [Google Scholar]
- Mori T, Tanaka K, Buffo A, Wurst W, Kuhn R, Gotz M (2006) Inducible gene deletion in astroglia and radial glia–a valuable tool for functional and lineage analysis. Glia 54: 21–34 [DOI] [PubMed] [Google Scholar]
- Muller‐Rover S, Handjiski B, van der Veen C, Eichmuller S, Foitzik K, McKay IA, Stenn KS, Paus R (2001) A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J Invest Dermatol 117: 3–15 [DOI] [PubMed] [Google Scholar]
- Nochi R, Kato T, Kaneko J, Itou Y, Kuribayashi H, Fukuda S, Terazono Y, Matani A, Kanatani S, Nakajima K, Hisatsune T (2012) Involvement of metabotropic glutamate receptor 5 signaling in activity‐related proliferation of adult hippocampal neural stem cells. Eur J Neurosci 36: 2273–2283 [DOI] [PubMed] [Google Scholar]
- Nordlind K, Johansson O, Liden S, Hokfelt T (1993) Glutamate‐ and aspartate‐like immunoreactivities in human normal and inflamed skin. Virchows Arch B Cell Pathol Incl Mol Pathol 64: 75–82 [DOI] [PubMed] [Google Scholar]
- Osorio KM, Lee SE, McDermitt DJ, Waghmare SK, Zhang YV, Woo HN, Tumbar T (2008) Runx1 modulates developmental, but not injury‐driven, hair follicle stem cell activation. Development 135: 1059–1068 [DOI] [PubMed] [Google Scholar]
- Page ME, Lombard P, Ng F, Gottgens B, Jensen KB (2013) The epidermis comprises autonomous compartments maintained by distinct stem cell populations. Cell Stem Cell 13: 471–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paus R, Cotsarelis G (1999) The biology of hair follicles. N Engl J Med 341: 491–497 [DOI] [PubMed] [Google Scholar]
- Petersson M, Brylka H, Kraus A, John S, Rappl G, Schettina P, Niemann C (2011) TCF/Lef1 activity controls establishment of diverse stem and progenitor cell compartments in mouse epidermis. EMBO J 30: 3004–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P, Mesa KR, Greco V (2013) Spatial organization within a niche as a determinant of stem‐cell fate. Nature 502: 513–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P, Greco V (2014) Stem cell dynamics in the hair follicle niche. Semin Cell Dev Biol 25–26: 34–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sada A, Jacob F, Leung E, Wang S, White BS, Shalloway D, Tumbar T (2016) Defining the cellular lineage hierarchy in the interfollicular epidermis of adult skin. Nat Cell Biol 18: 619–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sada A, Jain P, Wang S, Leung E, Tumbar T (2017) Slc1a3‐CreER as a targeting tool for the K6+ epithelial stem cell niche and its precursors during mouse hair follicle cycle. J Invest Dermatol 137: 1569–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepeler T, Page ME, Jensen KB (2014) Heterogeneity and plasticity of epidermal stem cells. Development 141: 2559–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, Yamada K, Watanabe M, Ikenaka K, Wada K, Tanaka K, Inoue Y (1997) Glutamate transporter GLAST is expressed in the radial glia‐astrocyte lineage of developing mouse spinal cord. J Neurosci 17: 9212–9219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slezak M, Goritz C, Niemiec A, Frisen J, Chambon P, Metzger D, Pfrieger FW (2007) Transgenic mice for conditional gene manipulation in astroglial cells. Glia 55: 1565–1576 [DOI] [PubMed] [Google Scholar]
- Snippert HJ, Haegebarth A, Kasper M, Jaks V, van Es JH, Barker N, van de Wetering M, van den Born M, Begthel H, Vries RG, Stange DE, Toftgard R, Clevers H (2010a) Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science 327: 1385–1389 [DOI] [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon‐Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, Clevers H (2010b) Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143: 134–144 [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F (2001) Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol 1: 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzingounis AV, Wadiche JI (2007) Glutamate transporters: confining runaway excitation by shaping synaptic transmission. Nat Rev Neurosci 8: 935–947 [DOI] [PubMed] [Google Scholar]
- Watase K, Hashimoto K, Kano M, Yamada K, Watanabe M, Inoue Y, Okuyama S, Sakagawa T, Ogawa S, Kawashima N, Hori S, Takimoto M, Wada K, Tanaka K (1998) Motor discoordination and increased susceptibility to cerebellar injury in GLAST mutant mice. Eur J Neurosci 10: 976–988 [DOI] [PubMed] [Google Scholar]
- Xiao XL, Ma DL, Wu J, Tang FR (2013) Metabotropic glutamate receptor 5 (mGluR5) regulates proliferation and differentiation of neuronal progenitors in the developmental hippocampus. Brain Res 1493: 1–12 [DOI] [PubMed] [Google Scholar]
- Zhao J, Lopez AL, Erichsen D, Herek S, Cotter RL, Curthoys NP, Zheng J (2004) Mitochondrial glutaminase enhances extracellular glutamate production in HIV‐1‐infected macrophages: linkage to HIV‐1 associated dementia. J Neurochem 88: 169–180 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7


