GmPIB1, previously reported to be up-regulated in Rps near-isogenic lines, enhances resistance to Phytophthora sojae in soybean by repressing expression of GmSPOD1, a key enzyme for producing reactive oxygen species.
Keywords: bHLH transcription factor, Glycine max, Phytophthora sojae, root, ROS
Abstract
Phytophthora sojae Kaufmann and Gerdemann causes Phytophthora root rot, a destructive soybean disease worldwide. A basic helix–loop–helix (bHLH) transcription factor is thought to be involved in the response to P. sojae infection in soybean, as revealed by RNA sequencing (RNA-seq). However, the molecular mechanism underlying this response is currently unclear. Here, we explored the function and underlying mechanisms of a bHLH transcription factor in soybean, designated GmPIB1 (P. sojae-inducible bHLH transcription factor), during host responses to P. sojae. GmPIB1 was significantly induced by P. sojae in the resistant soybean cultivar ‘L77-1863’. Analysis of transgenic soybean hairy roots with elevated or reduced expression of GmPIB1 demonstrated that GmPIB1 enhances resistance to P. sojae and reduces reactive oxygen species (ROS) accumulation. Quantitative reverse transcription PCR and chromatin immunoprecipitation–quantitative PCR assays revealed that GmPIB1 binds directly to the promoter of GmSPOD1 and represses its expression; this gene encodes a key enzyme in ROS production. Moreover, transgenic soybean hairy roots with GmSPOD1 silencing through RNA interference exhibited improved resistance to P. sojae and reduced ROS generation. These findings suggest that GmPIB1 enhances resistance to P. sojae by repressing the expression of GmSPOD1.
Introduction
Phytophthora root and stem rot caused by Phytophthora sojae is one of the most destructive soybean diseases worldwide, resulting in annual losses of $1–2 billion globally (Tyler, 2007). The most economical and effective way to protect soybeans against P. sojae infection is by breeding for dominant resistance to P. sojae (Rps) genes (Sugimoto et al., 2012). However, the continuous utilization of a single Rps gene can result in selective pressure that promotes the evolution of more pathogenic races of P. sojae. Thus, a particular Rps gene is effective for only 8–15 years (Walker and Schmitthenner, 1984; Tooley and Grau, 1984; Sugimoto et al., 2012). Moreover, some genes encode proteins that most likely function in direct protection, such as key enzymes for osmolyte biosynthesis, antioxidant and reactive oxygen species (ROS) scavengers, and enzymes involved in many metabolic processes (Yan et al., 2014; Cheng et al., 2015; Wang et al., 2015a; Yan et al., 2016). The products of regulatory genes, including membrane-localized receptors, calcium sensors, kinases, and transcription factors (TFs), participate in further signal transduction and the regulation of gene expression (Wang et al., 2015a). Several TF families play important roles in plant stress tolerance, such as basic helix–loop–helix (bHLH), DREB, ERF, WRKY, MYB, bZIP, and NAC TFs (Tran et al., 2004; Hu et al., 2006; Kim and Kim, 2006; Liao et al., 2008a,b; Zhou et al., 2008; Seo et al., 2010; Hao et al., 2011; Niu et al., 2012; Liu et al., 2014; Dong et al., 2015). These TFs separately or cooperatively affect the expression of various downstream genes and constitute gene networks for stress adaptation (Wang et al., 2015a).
Members of the bHLH family, which are distinguished by the bHLH domain, are universally found in eukaryotes (Duek and Fankhauser, 2005; Liu et al., 2014). The bHLH domain consists of 50–60 amino acids with two functionally distinct regions: the basic region (containing 13–17 primarily basic amino acids for DNA binding) and the HLH region (which enables the formation of homodimers or heterodimers with one or several different partners) (Toledo-Ortiz et al., 2003; Feller et al., 2011). The bHLH TFs are involved in essential plant physiological and developmental processes by binding to E-box (CANNTG)/G-box (CACGTG) sequences in the promoters of stress-response genes (Kim and Kim, 2006; Liu et al., 2013; Liu et al., 2014). For instance, CIB1 is a bHLH TF that binds to the G-box DNA motif in vitro but heterodimerizes with other CIB1-related proteins that in turn bind to E-box sequences to regulate transcription in vivo (Liu et al., 2013). bHLH122 binds directly to the G-box/E-box cis-elements in the CYP707A3 promoter and represses its expression, and bHLH122 is strongly induced by drought, NaCl, and osmotic stress in Arabidopsis (Liu et al., 2014). Increasing evidence indicates that bHLHs regulate plant responses to biotic and abiotic stresses (Zhang et al., 2011; Liu et al., 2014; Wang et al., 2015b; Turnbull et al., 2017). For example, phytochrome-interacting factor 4 (PIF4), a nucleus-localized bHLH protein, interacts directly with brassinazole-resistant 1 (BZR1) and forms a module that integrates steroid and environmental signaling (Oh et al., 2012). Abscisic acid (ABA)-inducible bHLH TF/jasmonic acid (JA)-associated MYC2-like 1 (JAM1), a repressor of JA signaling, plays a pivotal role in the fine-tuning of JA-mediated stress responses and plant growth (Nakata et al., 2013). ABA-inducible gene (AtAIG1), encoding a bHLH-type TF in Arabidopsis, is up-regulated after exposure to ABA but not to cold or NaCl, suggesting that AtAIG1 might be involved in ABA-mediated responses (Kim and Kim, 2006). ICE1, which is constitutively expressed in Arabidopsis, encodes a bHLH TF that regulates the expression of CBF genes in response to cold stress (Chinnusamy et al., 2003; Lee et al., 2005). Overexpressing OrbHLH001 improves freezing and salt tolerance in Arabidopsis. Moreover, the Arabidopsis bHLH TF HBI1 is a negative regulator of the basal defense response. Loss-of-function of HBI1 increases resistance to bacterial infection, and constitutive overexpression of HBI1 reduces pathogen-associated molecular pattern (PAMP)-induced immune responses (Fan et al., 2014). The transient overexpression of StCHL1 significantly increases leaf colonization of Nicotiana benthamiana by P. infestans, which is consistent with the finding that its homologs, HBI1 and CIB1, are negative regulators of immunity responses (Turnbull et al., 2017). However, the potential functions of most bHLH family members in soybean are still unclear.
A bHLH TF gene was shown to be up-regulated in all 10 near-isogenic lines (NILs) examined, each with a unique Rps gene/allele, based on sequencing and comparative transcriptome analysis of the NILs and the susceptible parent ‘Williams’ pre- and post-inoculation with P. sojae (Lin et al., 2014). Therefore, in the current study, we isolated this bHLH TF gene from P. sojae-resistant soybean cultivar ‘L77-1863’, which we designated GmPIB1 (P. sojae-inducible bHLH transcription factor; Glyma.01g129700). Overexpressing GmPIB1 in transgenic soybean hairy roots increased resistance to P. sojae, whereas RNA interference (RNAi) of this gene in transgenic soybean hairy roots increased susceptibility to this pathogen. GmPIB1 bound directly to the promoter of GmSPOD1 and inhibited its expression, leading to improve resistance to P. sojae. Taken together, these results indicate that GmPIB1 facilitates the resistance response of soybean to P. sojae infection by repressing the expression of GmSPOD1.
Materials and methods
Plant material, treatments, and primers
The P. sojae-susceptible soybean cultivar ‘Williams’ (rps1b) and the resistant cultivar ‘L77-1863’ (Rps1b) (Shan et al., 2004) were used in this study. The seeds were sown in pots in a growth chamber maintained at 25 °C and 70% relative humidity with a 16 h light/8 h dark cycle. Fourteen days after planting, seedlings at the first-node stage (V1; Fehr et al., 1971) were subjected to various treatments.
For abiotic treatments, ‘L77-1863’ plants were exposed to one of three different hormones, namely, methyl jasmonate (MeJA), ethylene (ET), or salicylic acid (SA). SA (2 mM) and MeJA (100 µM) were dissolved in 0.01% Tween 20 and sprayed onto young leaves for 0, 1, 3, 6, 9, 12, or 24 h. Ethylene treatment was performed by injecting gaseous ethylene at a concentration of 200 µl l−1 into a sealed Plexiglas chamber for 0, 1, 3, 6, 9, 12, or 24 h. The control leaves were sprayed with an equal volume of 0.01% (v/v) Tween 20.
For P. sojae treatment, plants of the susceptible cultivar ‘Williams’ and the resistant cultivar ‘L77-1863’ were inoculated with P. sojae race 1 (Zhang et al., 2010) zoospores as described by Ward et al. (1979). Unifoliate leaves were treated for 0, 6, 9, 12, 24, 36, 48, or 72 h. The susceptible soybean cultivar ‘Williams’ and resistant cultivar ‘L77-1863’ were obtained from the Key Laboratory of Soybean Biology at the Chinese Ministry of Education, Harbin, and used for the gene transformation experiments. All primers used for vector construction, PCR, and quantitative reverse transcription (qRT)-PCR assays for all target genes are listed in Supplementary Table S1 at JXB online.
RT-PCR and qRT-PCR analysis
Total RNA was isolated from ‘Williams’ and ‘L77-1863’ soybean leaves using Trizol reagent (Invitrogen, Shanghai, China). cDNA synthesis was conducted using an M-MLV reverse transcriptase kit (Takara, Dalian, China) according to the manufacturer’s instructions. RT-PCR was performed to analyse GmPIB1 transcript levels in ‘Williams’ and ‘L77-1863’ plants according to Zhang et al. (2012). The soybean housekeeping gene GmEF1β (GenBank accession no. NM_001248778) was used as the internal control. qRT-PCR analysis was performed to measure GmPIB1 transcript levels on a CFX96 Touch™ Real-Time PCR machine (Bio-Rad, USA) using a real-time PCR kit (Toyobo, Japan). The soybean housekeeping gene GmEF1β was used as an internal reference to normalize all data. The relative transcript level of the target gene was calculated using the 2−ΔΔCT method. Three biological replications per line were performed in each test.
Subcellular localization of GmPIB1 fusion protein
The coding sequence of GmPIB1 was amplified by RT-PCR using primers GmPIB1GF and GmPIB1GR. The coding sequence was fused to the N-terminus of green fluorescent protein (GFP) under the control of the constitutive CaMV35S promoter. The resulting expression vector, p35S:GmPIB1-GFP, was transformed into Arabidopsis protoplasts via polyethylene glycol (PEG)-mediated transfection as described by Yoo et al. (2007). Fluorescence signals were imaged using a TCS SP2 spectral confocal microscope imaging system (Leica, Germany). The p35S:GFP vector was used as a control.
To analyse the expression of GmPIB1 fusion protein in plants, membrane, nuclear, and cytoplasmic proteins were extracted using a Cytoplasmic, Nuclear, and Membrane Protein Extraction Kit (Sangon Biotech, Shanghai, China, C510002). The supernatants of extracts were separated by SDS-PAGE. After electrophoresis, the proteins were transferred to polyvinylidene difluoride membranes (Millipore) and probed using anti-GFP antibodies (Abmart, M2004).
Expression and purification of fusion protein
The open reading frame of GmPIB1 was fused to the N-terminus of the 6×His-tag at the EcoRI and XhoI restriction sites of the vector pET29b(+) (Novagen, Germany). The recombinant fusion plasmid was expressed in Transetta (DE3) E. coli cells (TransGen Biotech, China). His-tagged protein production was induced with 0.5 mM isopropyl-β-D-thiogalactoside (IPTG) at 37 °C for 4 h. The fusion protein was purified at 4 °C according to the pET System Manual (Novagen). The GmPIB1–His fusion protein was subsequently analysed by SDS-PAGE and immunoblotting using an anti-His antibody.
Electrophoretic mobility shift assay
The DNA-binding activity of GmPIB1 was examined using a digoxigenin-ddUTP-labeled double-stranded oligonucleotide E-box probe as described previously (Meng et al., 2013). The sequence of the probe for the E-box was 5′-AGGAGAGTGGGCCANNT GCGCTCTTTTGCATTC-3′ and that of the mutant E-box (mE-box) was 5′-AGGAGAGTGGGCCCNN CGCGCTCTTTTGCATTC-3′. The electrophoretic mobility shift assay (EMSA) was performed as described by Kass et al. (2000).
Transactivation assay
For the transactivation assay, the β-glucuronidase (GUS) gene in pCAMBIA3301 was replaced by GmPIB1 as the effector plasmid. The E-box was multimerized four times and placed upstream of the cauliflower mosaic virus (CaMV) 35S promoter (–42 to +8) containing a TATA box. This construct was inserted into pXGUS-P (Chen et al., 2009) and fused to the GUS gene as the reporter plasmid. The transactivation assay was performed by PEG transfection of Arabidopsis protoplasts as described by Yoo et al. (2007). Twenty micrograms of reporter plasmid and 20 µg of effector plasmid or control plasmid (pXGUS-P-35Smini) were co-transfected into 4 × 104 protoplasts. The transfected cells were incubated at 22 °C in the light for 18–20 h. GUS activity was determined as described (Lu et al., 1998).
Agrobacterium rhizogenes-mediated transformation of soybean hairy roots
To construct the p35S:GmPIB1-Myc overexpression vector, the coding sequence of GmPIB1 with a C-terminal 4×Myc fusion sequence was cloned into plant expression vector pCAMBIA3301 with gene-specific primers. To construct the GmPIB1 RNAi vector, the cDNA fragment of GmPIB1 was amplified using the primer set PIB1RNAi-F/R and inserted into vector pFGC5941 (Kerschen et al., 2004). Transgenic soybean hairy roots were generated by A. rhizogenes-mediated transformation as described by Graham et al. (2007) and Kereszt et al. (2007) with some modifications. The cotyledons were cut into rough triangles and immediately placed in Petri dishes containing 0.6% agar medium to keep them moist. The cut surface was treated with 20 µl A. rhizogenes suspension. The dishes were sealed with Parafilm and placed in an incubator at 25 °C. Transformed hairy roots were abundant along a callus ridge on the inoculated cotyledons after approximately 3 weeks. Overexpression of the target gene in transgenic hairy roots was tested via quantitative PCR (qPCR) and immunoblotting, and RNAi transgenic hairy roots were verified by qPCR and Southern blot analysis.
Promoter–GUS analysis
The 1494 bp promoter sequence of GmPIB1 was amplified using gene-specific primers GmPIB1PF and GmPIB1PR and cloned into the pBI121 expression vector. The GmPIB1 promoter–GUS construct was transformed into the hairy roots of ‘L77-1863’ soybean plants by A. rhizogenes-mediated transformation. When the hairy roots generated at the infection site were approximately 8 cm long, the original main roots were treated with P. sojae zoospores for 48 h, or MeJA, ET, or SA for 6 h. Soybean hairy roots transformed with empty vector (EV) were used as controls. Histochemical GUS staining was performed 3 h after treatment using GUS staining buffer (1 mM 5-bromo-4-chloro-3-indolyl-b-D-GlcA solution in 100 mm sodium phosphate pH 7.0, 0.1 mM EDTA, 0.5 mM ferrocyanide, 0.5 mm ferricyanide, and 0.1% Triton X-100) at 37 °C overnight. GUS activity was measured as described by Jefferson et al. (1987).
Pathogen response assays of transgenic soybean hairy roots
To investigate whether GmPIB1-transformed hairy roots were resistant to pathogen infection, artificial inoculation procedures were performed as described by Ward et al. (1979). When the hairy roots generated at the infection site were approximately 8 cm long, the original main roots were incubated with P. sojae zoospores in a mist chamber at 25 °C with 100% relative humidity for 2 d. EV soybean hairy roots were used as controls. Disease symptoms on each root were observed after inoculation and photographed with a Nikon B7000 camera.
In situ ROS detection
To investigate whether the GmPIB1-transformed soybean hairy roots would respond to oxidative stress, GmPIB1 transgenic and EV (control) hairy roots were treated with P. sojae zoospores for 48 h as described by Ward et al. (1979). In situ H2O2 and O2− detection were performed using diaminobenzidine (DAB) or Nitro blue tetrazolium (NBT) as described by Lu et al. (2011). Total ROS levels were measured according to the instructions supplied with the Reactive Oxygen Species Assay Kit (Beyotime Institute of Biotechnology, Haimen, China). Fluorescence was detected at 485 nm for excitation and 530 nm for emission with a fluorescence microplate reader (Bio-TEK, USA; Qian et al., 2009). Relative ROS levels, i.e. the ratio of total ROS levels in hairy roots under P. sojae zoospore versus water treatment (mock) at the same time point were measured.
Yeast two-hybrid assays
For interaction studies, full-length GmPIB1 was amplified using gene-specific primers GmPIB1YF and GmPIB1YR and cloned in the pGBKT7 vector and pGADT7 vector. Fusion plasmids pGADT7-GmPIB1 and pGBKT7-GmPIB1 were transformed into yeast strain Y2HGold (Clontech). After selection on SD (−Trp, −Leu) medium, the transformants were transferred to SD (−Trp, −His, −Trp, −Ade) medium to identify protein–protein interactions.
Bimolecular fluorescence complementation assays
The coding sequence of GmPIB1 was cloned into serial pSAT6 vectors encoding either N- and C-terminal-enhanced yellow fluorescent protein fragments. The resulting constructs were used for transient assays via PEG transfection of Arabidopsis protoplasts as described by Yoo et al. (2007). Transfected cells were imaged using a TCS SP2 confocal spectral microscope imaging system (Leica).
Chromatin immunoprecipitation–qPCR assays
For chromatin immunoprecipitation (ChIP)–qPCR assays, EV and p35S:PIB1-Myc transgenic lines were subjected to chromatin extraction and immunoprecipitation as described by Saleh et al. (2008). Briefly, soybean hairy roots were harvested for fixation. Chromatin was isolated and sonicated to generate DNA fragments with an average size of 500 bp. The soluble chromatin fragments were isolated and pre-absorbed with 30 µl Protein G Plus/Protein A Agarose Suspension (Merck Millipore Biotechnology) to eliminate non-specific binding and immunoprecipitated by 30 µl Protein G Plus/Protein A Agarose Suspension with anti-Myc (Santa Cruz Biotechnology). The precipitated DNA was recovered and analysed by qRT-PCR with SYBR Premix ExTaq Mix (Takara Bio). The precipitated and input DNA samples were analysed by qPCR with the gene-specific primers. The data were normalized to input transcript levels and represent the means from three biological replicates.
Transient expression assay
A transient dual-luciferase assay was performed as previously described (Shang et al., 2010; Song et al., 2013). Briefly, the 1.761 kb promoter sequence of pGmSPOD1 was cloned using gene-specific primers GmSPOD1P-F/R and inserted into the ScaI and XbaI sites of the pBI121 vector (Clontech, CA, USA) after its GUS gene had been replaced with the firefly luciferase gene. The reporter construct pGmSPOD1:GUS and the effector construct p35S:GmPIB1-Myc were transformed into A. rhizogenes strain K599 and transfected into soybean hairy roots by A. rhizogenes-mediated transformation. When the hairy roots generated at the infection site were approximately 8 cm long, the original main roots were stained for GUS. The reporter construct pGmSPOD1:LUC and the effector construct p35S:GmPIB1-Myc were transformed into Agrobacterium tumefaciens strain GV3101 and transfected into healthy 21-day-old N. benthamiana tobacco leaves by agroinfiltration as described previously (Liu et al., 2012, Meng et al., 2013). The plants were incubated 3 d after infiltration, sprayed with luciferin (1 mM), and photographed with a CCD camera (Berthold Technologies) at 72 h after infiltration.
Protein extraction, immunoblotting, and Southern blotting
To analyse protein expression in transgenic plants, total proteins were extracted with protein extraction buffer (50 mM Tris–HCl at pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.1% Triton X-100, and protease inhibitor cocktail (Roche)). Total proteins (200 mg) were separated by SDS-PAGE. After electrophoresis, the proteins were transferred to polyvinylidene difluoride membranes (Millipore) and probed using anti-Myc antibodies (Santa Cruz Biotechnology).
Southern blotting was conducted according to the modified protocol of Zhang et al. (2012), in which 20 μg of genomic DNA digested with the restriction enzyme HindIII was hybridized to a probe derived from the bar-specific fragment (354 bp).
Results
GmPIB1 expression is induced upon P. sojae infection
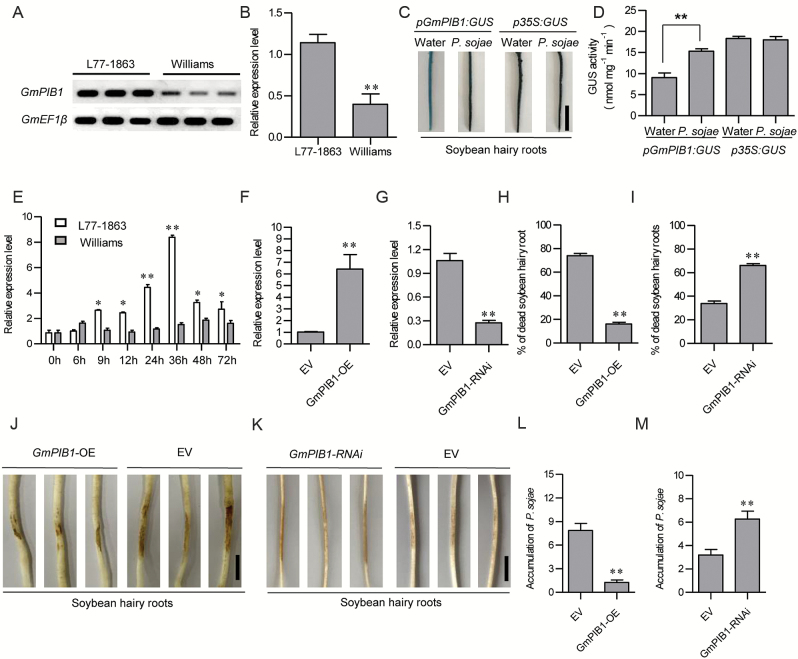
To evaluate whether GmPIB1 is involved in the response of soybean to P. sojae infection, we performed RT-PCR and qRT-PCR to examine the transcript levels of this gene in the susceptible soybean cultivar ‘Williams’ and the resistant cultivar ‘L77-1863’. As shown in Fig. 1A, B, the expression level of GmPIB1 was much higher in the resistant cultivar ‘L77-1863’ than in the susceptible cultivar ‘Williams’. qRT-PCR assays showed that GmPIB1 transcript levels were significantly elevated and reached a maximum level at 36 h after P. sojae treatment in ‘L77-1863’ (Fig. 1D). However, in ‘Williams’, GmPIB1 transcript levels did not increase under P. sojae treatment (Fig. 1E).
Fig. 1.
Transcriptional analysis of GmPIB1. (A) Expression patterns of GmPIB1 in susceptible soybean cultivar ‘Williams’ and resistant cultivar ‘L77-1863’, as assessed by RT-PCR. (B) Expression patterns of GmPIB1 in susceptible cultivar ‘Williams’ and resistant cultivar ‘L77-1863’, as assessed by qPCR. (C) GmPIB1 promoter-driven GUS expression in transgenic soybean hairy roots treated with P. sojae or water for 48 h. Bars, 1 cm. (D) GUS activity analysis of GmPIB1 promoter expression. GUS activity was measured using a 4-methylumbelliferyl-D-glucuronide assay. The data represent the means ±SD of three independent experiments. (E) Relative expression of GmPIB1 in soybean cultivars ‘Williams’ and ‘L77-1863’ upon P. sojae infection. The infected samples were collected at 0, 6, 9, 12, 24, 36, 48, and 72 h after inoculation with P. sojae (race 1). Relative GmPIB1 transcript levels were compared with mock-treated plants at the same time point. (F, G) qRT-PCR analysis of relative GmPIB1 expression in transgenic soybean hairy roots. Empty vector (EV) transgenic soybean hairy roots were used as controls. (H, I) Percentages of dead EV, GmPIB1-OE, and GmPIB1-RNAi roots after 5 d of P. sojae infection. Each experiment contained at least 50 roots per line, and roots were scored as dead when they were completely rotten. (J, K) Typical infection phenotypes of GmPIB1-OE, GmPIB1-RNAi, and EV soybean hairy roots after 2 d of P. sojae inoculation. Bars, 1 cm. (L, M) Accumulation of P. sojae biomass in transgenic soybean hairy roots and EV. Phytophthora sojae TEF1 (EU079791) transcript levels in infected soybean hairy roots (2 d) were plotted relative to soybean GmEF1β (NM_001248778) expression levels, as determined by qRT-PCR. The amplification of soybean GmEF1β was used as an internal control to normalize all data. The experiment was performed using three biological replicates, each with three technical replicates, and differences were statistically analysed using Student’s t-test (*P<0.05, **P<0.01). Bars indicate standard error of the mean. (This figure is available in color at JXB online.)
We used the 1494 bp promoter region of GmPIB1 to drive the expression of the GUS reporter gene in the pBI121 expression vector, which we transformed into ‘L77-1863’ soybean hairy roots via high-efficiency A. rhizogenes-mediated transformation as described by Graham et al. (2007) and Kereszt et al. (2007). When the hairy roots generated at the infection site were approximately 8 cm long, we subjected the original main roots to gene expression analysis and P. sojae treatment. Compared with control roots (treated with water), GmPIB1 promoter activity was highly induced in roots subjected to P. sojae treatment (Fig. 1C, D). Together, these results suggest that GmPIB1 is involved in the defense response of soybean to P. sojae.
Cloning full-length GmPIB1 cDNA
We then examined whether the GmPIB1 gene and promoter sequences differ between ‘Williams’ and ‘L77-1863’. We cloned and sequenced the cDNA and promoter of GmPIB1 in ‘Williams’ and ‘L77-1863’ and found no difference in sequence between the two cultivars (data not shown). GmPIB1 encodes a deduced 151 amino acid polypeptide with a bHLH domain at amino acid positions 9–63 (see Supplementary Fig. S1A). The predicted three-dimensional model of GmPIB1 consists of two α-helices (Supplementary Fig. S1C). To further explore the evolutionary relationship among plant bHLH proteins, we constructed a phylogenetic tree using MEGA4.0 (Tamura et al., 2007) based on amino acid sequences. Sequence alignment and phylogenetic tree analysis revealed that GmPIB1 shares 65.5–95.2% identity in overall amino acid sequence with bHLH TFs from Glycine max (XP_003551597), Arachis ipaensis (XP_016186634), Theobroma cacao (XP_017974773), Vigna radiata var. radiata (XP_014491943), Vitis vinifera (XP_002268100), Gossypium arboretum (XP_017609785), and Cicer arietinum bHLH (XP_004492536) (Supplementary Fig. S1B, D).
GmPIB1 enhances resistance to P. sojae in transgenic soybean hairy roots
To examine the effect of the loss and overexpression of GmPIB1 on resistance to P. sojae in soybean, we generated GmPIB1-overexpressing (GmPIB1-OE) and GmPIB1-RNA interference (GmPIB1-RNAi) transgenic soybean hairy roots by high-efficiency A. rhizogenes-mediated transformation (Graham et al., 2007; Kereszt et al., 2007) in susceptible cultivar ‘Williams’ and resistant cultivar ‘L77-1863’. We examined the GmPIB1-OE transgenic hairy roots by immunoblotting (see Supplementary Fig. S2A) and qRT-PCR (Fig. 1F) and the GmPIB1-RNAi transgenic hairy roots by Southern blot analysis (Supplementary Fig. S2B) and qRT-PCR (Fig. 1G). As shown in Fig. 1H, ~75% of EV (vector control) transgenic hairy roots of the susceptible cultivar ‘Williams’ inoculated with P. sojae were completely dead at 5 d of treatment, whereas only ~18% of inoculated GmPIB1-OE transgenic hairy roots were completely dead. However, ~35% of inoculated EV transgenic hairy roots of resistant cultivar ‘L77-1863’ and ~95% of inoculated GmPIB1-RNAi transgenic hairy roots were completely dead at 5 d of inoculation with P. sojae (Fig. 1I). After 2 d of incubation with P. sojae zoospores, the three GmPIB1-OE lines displayed almost no visible lesions compared with EV control roots in susceptible cultivar ‘Williams’ (Fig. 1J). By contrast, the three GmPIB1-RNAi transgenic hairy root lines exhibits enhanced wilting symptoms and chlorosis compared with EV hairy roots in resistant cultivar ‘L77-1863’ (Fig. 1K).
We also analysed the relative biomass of P. sojae in infected soybean hairy roots after 2 d of incubation with P. sojae zoospores. The biomass of P. sojae (based on the transcript level of P. sojae TEF1; GenBank accession no. EU079791) (Blair et al., 2008) was significantly (P<0.01) lower in the GmPIB1-OE lines than in EV hairy roots (Fig. 1L). However, the biomass of P. sojae was significantly (P<0.01) higher in the GmPIB1-RNAi lines than in EV hairy roots (Fig. 1M). These results indicate that overexpressing GmPIB1 in soybean hairy roots improves resistance to P. sojae and that silencing this gene increases susceptibility to P. sojae.
GmPIB1 transcript levels under different hormone treatments
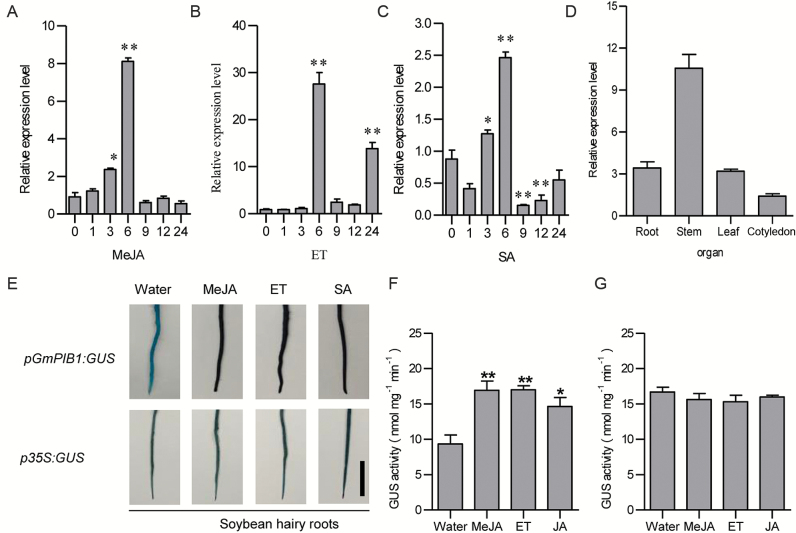
To investigate the expression pattern of GmPIB1 in response to phytohormone treatment, we performed qRT-PCR to examine GmPIB1 transcript levels in ‘L77-1863’ soybean plants. GmPIB1 expression was responsive to MeJA, ET, and SA treatment. GmPIB1 mRNA levels rapidly increased under these treatments, reaching a maximum level at 6 h after treatment, followed by a rapid decline (Fig. 2A–C). In ‘L77-1863’ plants, GmPIB1 was constitutively and highly expressed in stems, follow by roots and leaves (Fig. 2D). To elucidate the regulatory mechanism of GmPIB1 under MeJA, ET, and SA treatment, we measured GmPIB1 promoter activity in hairy roots at 6 h after treatment. GUS activity driven by the GmPIB1 promoter (pGmPIB1) was weak under control (water) conditions, but it increased approximately 8- and 2.5-fold compared with the control under MeJA and SA treatment, respectively (Fig. 2E, F). These results suggest that GmPIB1 is primarily involved in the response to MeJA treatment.
Fig. 2.
Expression patterns of GmPIB1 in soybean. (A–C) GmPIB1 expression in soybean leaves in response to exogenous hormones: 100 μM MeJA, 2 mM SA, and ET treatment for 0, 1, 3, 6, 9, 12, and 24 h. Fourteen-day-old plants were used for treatments and analyses. Relative GmPIB1 transcript levels were compared with mock-treated plants at the same time point. Soybean GmEF1β was used as an internal control to normalize all data. Three biological replicates were averaged and statistically analysed using Student’s t-test (*P<0.05, **P<0.01). Bars indicate standard error of the mean. (D) GmPIB1 mRNA levels in various soybean plant tissues. Leaves, roots, and stems were harvested from 14-day-old plants. The experiment was performed on three biological replicates, each with three technical replicates. Bars indicate standard error of the mean. (E) GUS histochemical staining analysis of pGmPIB1:GUS. pGmPIB1:GUS and p35S:GUS transgenic soybean hairy roots were produced by A. tumefaciens-mediated transformation and treated with 100 μM MeJA, 2 mM SA, or ET for 6 h. GUS histochemical staining results 3 h after treatment are shown compared with roots treated with water. Bars, 1 cm. (F) GUS activity analysis of GmSPOD1 promoter expression. GUS activity was measured using a 4-methylumbelliferyl-D-glucuronide assay. The data represent the means ±SD of three independent experiments. (This figure is available in color at JXB online.)
GmPIB1 is a transcriptional repressor that binds to the E-box sequence
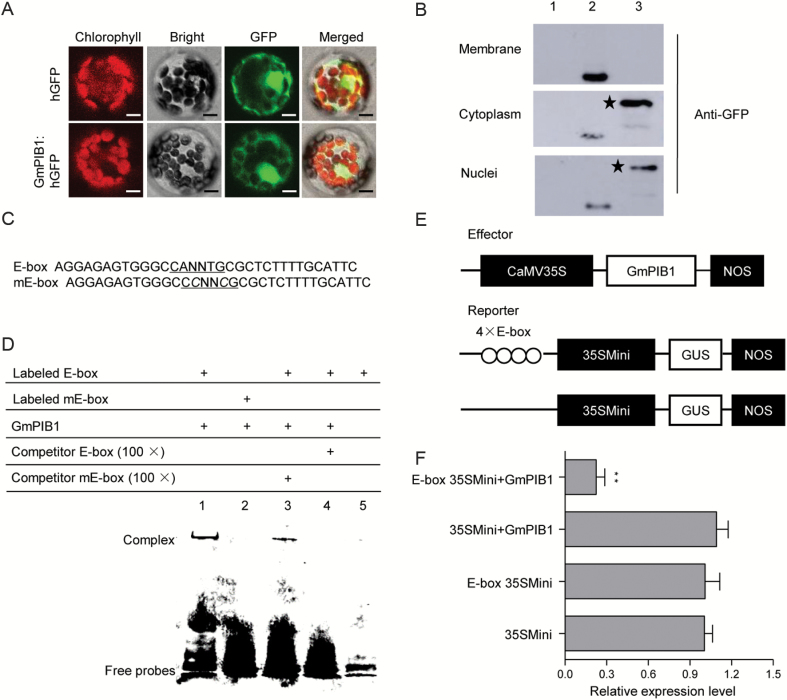
To investigate the subcellular localization of GmPIB1, we expressed a gene construct encoding GmPIB1–humanized GFP (hGFP) fusion protein under the control of the 35S promoter in Arabidopsis protoplasts. Confocal immunofluorescence and immunoblot analysis showed that hGFP alone was uniformly distributed throughout the cell, whereas transformed cells carrying GmPIB1–hGFP fusion protein localized to the cytoplasm and nuclei (Fig. 3A, B).
Fig. 3.
Sequence-specific binding activity of GmPIB1 to the E-box element. (A) Subcellular localization of GmPIB1–hGFP fusion protein. Subcellular localization was investigated in Arabidopsis protoplasts by confocal microscopy. The fluorescence from humanized GFP (hGFP) and the fusion protein GmPIB1–hGFP was observed under white light, UV light, and red light separately. Bars, 10 μm. (B) Immunoblot analysis detecting GmPIB1–hGFP fusion protein in the cytoplasm and nuclei. Line 1, Arabidopsis protoplasts (negative control); line 2, hGFP; line 3, GmPIB1–hGFP fusion protein. Anti-GFP was used to detect GmPIB1–GFP fusion protein in Arabidopsis cells. An asterisk denotes the specific band of the fusion protein GmPIB1–hGFP. (C) Nucleotide sequences of the E-box and mE-box probes. (D) EMSA showing sequence-specific binding of the recombinant GmPIB1 protein to the E-box. Lane 1, labeled E-box probe and GmPIB1 protein; lane 2, labeled mE-box probe and GmPIB1 protein; lane 3, titration using a cold mE-box sequence as a competitor; lane 4, titration using a cold E-box sequence as a competitor; lane 5, EMSA performed with only the free E-box probe. (E) Schematic diagram of the reporter and effector constructs. The reporter plasmids contained four repeats of the E-box sequence and 35Smini, and the effector plasmids encoded GmPIB1 under the control of the CaMV 35S promoter. (F) Relative GUS activity in transactivation assays. The effector and reporter plasmids were co-transfected into Arabidopsis protoplasts. The numbers show the fold increase in GUS activity compared with the vector E-box/35Smini promoter (E-box 35SMini) alone. The experiments were performed on three biological replicates and statistically analysed using Student’s t-test (**P<0.01). Bars indicate standard error of the mean. (This figure is available in color at JXB online.)
To express GmPIB1 in Transetta (DE3) E. coli cells, we cloned the coding sequence of GmPIB1 into pET-29b, an expression vector with a His-tag. Upon induction by IPTG, GmPIB1 was expressed as a major soluble protein product at 1, 2, and 4 h (Supplementary Fig. S3, lanes 2, 3, and 4). The molecular mass of the purified protein was approximately 21 kDa, as revealed by SDS-PAGE (Supplementary Fig. S3, lane 5), which is consistent with its calculated molecular mass (21.33 kDa). Immunoblotting of purified recombinant GmPIB1 protein confirmed its specific immune reactivity to anti-His antibodies (Supplementary Fig. S3, lane 6).
To determine whether GmPIB1 binds to the cis-acting element of the E-box in its target promoters in vitro, we subjected purified His-tagged GmPIB1 to an EMSA with a digoxigenin-ddUTP-labeled double-stranded oligonucleotide E-box probe. The sequences of the E-box and mutated E-box (mE-box) are shown in Fig. 3C. When the E-box was used as a probe, GmPIB1 caused a mobility shift in labeled E-box probe (Fig. 3D, lane 1), which migrated more slowly than the free probe (Fig. 3D, lane 5). Furthermore, when mE-box was used in the assay, this mobility shift was not observed (Fig. 3D, lane 2). We conducted competition experiments to examine the specificity of the mobility shift. When the ratio of unlabeled-to-labeled E-box probe was 100:1, almost no labeled probe was bound (Fig. 3D, lane 4), and when 100-fold unlabeled mE-box probe was used as the competitor, no binding competition was observed (Fig. 3D, lane 3).
To investigate whether GmPIB1 is a transcriptional repressor, we performed a transactivation assay in Arabidopsis protoplasts using a reporter gene with four tandem copies of the E-box and effector plasmids with GmPIB1 (Fig. 3E). As shown in Fig. 3F, GmPIB1 appeared to repress reporter gene expression, since GUS expression was reduced to 71% of control levels in the presence of this protein. Overall, these results suggest that GmPIB1 is an E-box-specific DNA binding protein that acts as a transcriptional repressor in plant cells.
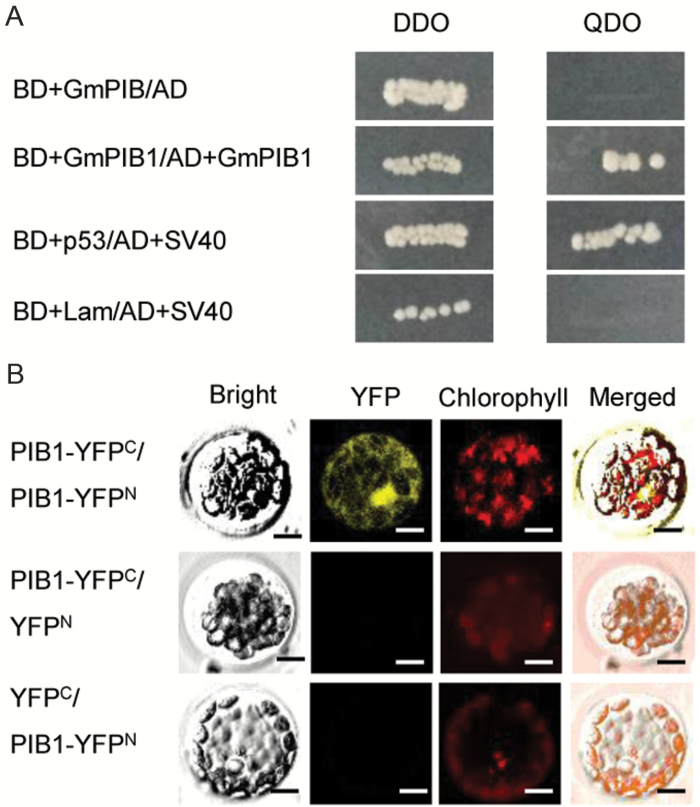
GmPIB1 can form homodimers
The bHLH TFs form homodimers or heterodimers, which is a prerequisite for DNA binding, and each partner binds to half of the DNA recognition sequence (Ma et al., 1994; Shimizu et al., 1997; Feller et al., 2011). To determine whether GmPIB1 forms homodimers in yeast cells, we fused full-length GmPIB1 to the DNA-binding domain of GAL4 (BD) (Clontech, Palo Alto, CA, USA) and subjected it to a transcriptional activation activity by growing the yeast cells on SD/–Leu/–Trp (DDO) and SD/–Ade/–His/–Leu/–Trp (QDO) media. Together with the GAL4 activation domain (AD), yeast cells carrying full-length GmPIB1 fused to the GAL4 DNA binding domain grew on DDO, but not on QDO medium (Fig. 4A). Further analysis suggested that in yeast cells carrying BD-GmPIB1 and AD-GmPIB1, the transcription of downstream reporter genes was activated, and the cells grew on QDO medium (Fig. 4A).
Fig. 4.
GmPIB1 forms a homodimer in yeast cells and in planta. (A) Yeast cells of strain Y2H harboring pGBKT7-GmPIB1 and pGADT7-GmPIB1 plasmid combinations were grown on either SD/−Trp/−Leu or SD/−Trp/−Leu/−His/−Ade medium. Yeast cells carrying the pGBKT7-53 and pGADT7-SV40 plasmids were used as the positive control; yeast cells harboring the pGBKT7-Lam and pGADT7-SV40 plasmids were used as the negative control. (B) BiFC analysis of the interaction of GmPIB1 with itself. GmPIB1–YFPN and GmPIB1–YFPC were co-transfected into Arabidopsis protoplasts. The bright-field, YFP fluorescence (yellow), chlorophyll autofluorescence (red), and combined images were visualized under a confocal microscope 16 h after transfection. Bars, 10 μm. (This figure is available in color at JXB online.)
To further confirm the occurrence of these interactions in planta, we performed a bimolecular fluorescence complementation (BiFC) assay involving transient expression in Arabidopsis protoplasts. Co-expression of both N-terminal yellow fluorescent protein (YFPN)-tagged GmPIB1 and C-terminal YFP (YFPC)-tagged GmPIB1 resulted in significant fluorescence in the chloroplasts of Arabidopsis protoplasts (Fig. 4B). However, no fluorescence was detected in Arabidopsis protoplasts co-transformed with YFPN–GmPIB1 and YFPC or YFPC–GmPIB1 and YFPN. These results suggest that GmPIB1 interacts with itself in planta.
Expression of GmPIB1 in soybean hairy root affects ROS levels
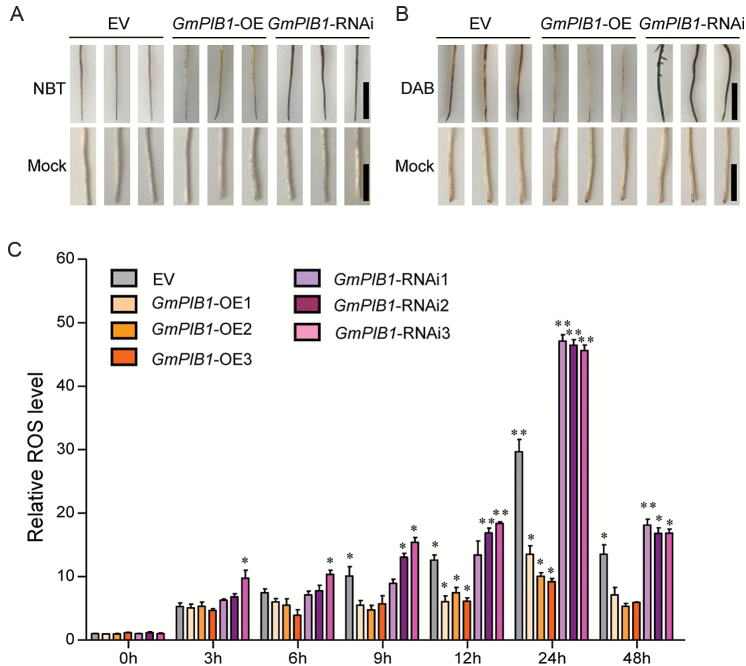
ROS are key signaling molecules that are produced in response to biotic and abiotic stress and trigger a variety of plant defense responses (Hückelhoven and Kogel, 2003; Soosaar et al., 2005; Takabatake et al., 2007; Shetty et al., 2008; Perez and Brown, 2014). H2O2 and superoxide (O2−) are the primary ROS components (Mittler et al., 2004; Foyer and Shigeoka 2011). We therefore compared ROS production in EV, GmPIB1-OE, and GmPIB1-RNAi hairy roots after P. sojae zoospore inoculation by in situ NBT staining of superoxide anions and DAB staining of H2O2. Upon infection with P. sojae zoospores, we observed a dramatic increase in superoxide anion and H2O2 contents in EV hairy roots at 48 h after inoculation (Fig. 5A, B). Compared with EV hairy roots, lower levels of superoxide anion and H2O2 were detected in GmPIB1-OE roots, whereas higher levels were detected in GmPIB1-RNAi roots (Fig. 5A, B). We also measured relative ROS levels in EV, GmPIB1-OE, and GmPIB1-RNAi transgenic hairy roots at 0, 3, 6, 12, 24, and 48 h after incubation with P. sojae. The relative ROS levels gradually increased in EV, GmPIB1-OE, and GmPIB1-RNAi with increasing incubation time (Fig. 5C) and were significantly lower in the GmPIB1-OE lines and significantly higher in the GmPIB1-RNAi lines compared with EV hairy roots at the same time point (Fig. 5C). These results suggest that overexpressing GmPIB1 efficiently reduces ROS accumulation in soybean.
Fig. 5.
Analysis of ROS levels in GmPIB1-OE, GmPIB1-RNAi, and EV transgenic soybean hairy roots. (A) NBT staining of O2− in 20-day-old EV, GmPIB1-OE, and GmPIB1-RNAi soybean hairy roots after P. sojae zoospore treatment for 48 h. Bars, 1 cm. (B) DAB staining of H2O2 in 20-day-old EV, GmPIB1-OE, and GmPIB1-RNAi soybean hairy roots under P. sojae zoospore treatment for 48 h. Bars, 1 cm. (C) Relative ROS levels in EV, GmPIB1-OE1, GmPIB1-OE2, GmPIB1-OE3, GmPIB1-RNAi1, GmPIB1-RNAi2, and GmPIB1-RNAi3 soybean hairy roots at 0, 3, 6, 12, 24, and 48 h after P. sojae infection. Relative ROS levels were measured, i.e. the ratio of total ROS levels in soybean hairy roots treated with P. sojae zoospores versus that in hairy roots treated with equal amounts of sterile water (mock) at the same time point. Three biological replicates, each with three technical replicates, were averaged and statistically analysed using Student’s t-test (*P<0.05, **P<0.01). Bars indicate standard error of the mean. (This figure is available in color at JXB online.)
GmPIB1 represses the expression of GmSPOD1 in transgenic soybean hairy roots
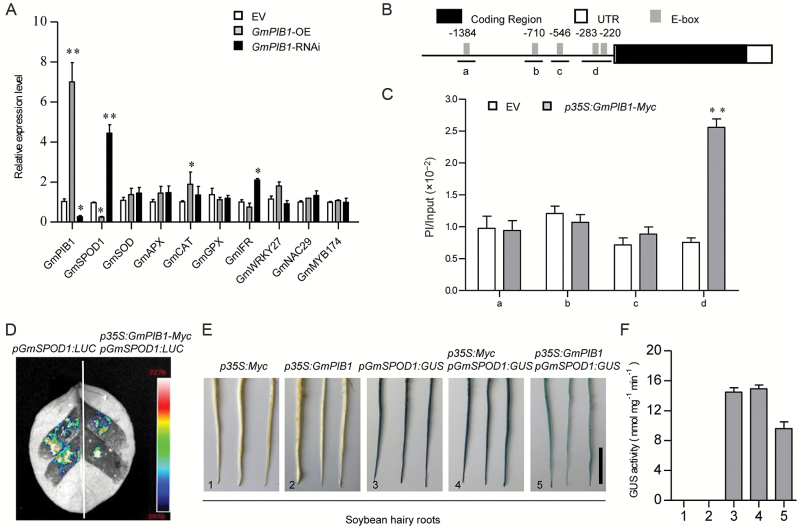
To address how GmPIB1 affects ROS generation, we performed qRT-PCR in EV, GmPIB1-OE, and GmPIB1-RNAi hairy roots to measure the relative expression of genes that are known to take part in ROS production, such as the peroxidase gene GmSPOD1 (NM_001252802); the ascorbate peroxidase gene GmAPX (L10292.1); the catalase gene GmCAT (AK286272.1); the superoxide dismutase gene GmSOD (XM_003526765.3); the glutathione peroxidase gene GmGPX (XM_006600055.2); the TF genes GmNAC29 (XM_003556741), GmWRKY27 (DQ322695), and GmMYB174 (DQ822939); and the isoflavone reductase gene GmIFR (NM_001254100). SPOD1 was significantly down-regulated in GmPIB1-OE hairy roots but markedly up-regulated in the GmPIB1-RNAi lines compared with EV (Fig. 6A). GmCAT was up-regulated in GmPIB1-OE hairy roots, and GmIFR was up-regulated in GmPIB1-RNAi lines, compared with the EV control. However, the expression of the other genes was not affected in GmPIB1-OE or GmPIB1-RNAi hairy roots versus the control (Fig. 5A).
Fig. 6.
Analysis of ROS-induced gene expression in GmPIB1 transgenic and EV soybean hairy roots. (A) GmPIB1-modulated gene expression in GmPIB1-OE and GmPIB1-RNAi hairy roots compared with EV, as revealed by qRT-PCR. Soybean GmEF1β was used as an internal control to normalize all data. (B, C) ChIP analysis of GmPIB1 binding to the GmSPOD1 promoter in GmPIB1-Myc transgenic soybean hairy roots and EV. Chromatin from GmPIB1-Myc transgenic and EV hairy roots was immunoprecipitated with anti-Myc antibody and treated without antibodies. The precipitated chromatin fragments were analysed by qPCR using four primer sets amplifying four regions upstream of GmSPOD1 (GmSPOD1a, GmSPOD1b, GmSPOD1c, GmSPOD1d), as indicated. One-tenth of the input (without antibody precipitation) of chromatin was analysed and used as a control. Three biological replicates, each with three technical replicates, were averaged and statistically analysed using Student’s t-test (*P<0.05, **P<0.01). Bars indicate standard error of the mean. (D) GmPIB1 represses GmSPOD1 promoter activity in N. benthamiana leaves. Agrobacterium tumefaciens GV3101 strains harboring pGmSPOD1:LUC and p35S: GmPIB1 were transfected into N. benthamiana leaves. Luciferase imaging was performed 72 h after injection. (E) GmPIB1 represses GmSPOD1 promoter activity in soybean hairy roots. Agrobacterium rhizogenes K599 strains harboring p35S: GmPIB1, and pGmSPOD1:GUS were transfected into soybean hairy roots. Line 1, pGmSPOD1:GUS; line 2, p35:Myc; line 3, p35S: GmPIB1-Myc; line 4, p35:Myc and pGmSPOD1:GUS; line 5, p35S:GmPIB1-Myc and pGmSPOD1:GUS. (F) GUS activity analysis of GmSPOD1 promoter expression. GUS activity was measured using a 4-methylumbelliferyl-D-glucuronide assay. The x-axis numbers correspond to the numbers 1–5 in (E). The data represent the means ±SD of three independent experiments. (This figure is available in color at JXB online.)
Using the PLACE program (Higo et al., 1999), we detected five E-box cis-elements in the 1.761-kb region upstream of the GmSPOD1 promoter (Fig. 6B). To further determine the binding capacity of GmPIB1 to the promoter of GmSPOD1, we performed a ChIP-qPCR assay to compare the relative enrichment of specific GmSPOD1 sequences in GmPIB1-OE and EV hairy roots using anti-Myc antibodies. GmPIB1 protein was highly enriched in the GmSPOD1 promoter d site in the GmPIB1-OE lines, whereas it was present at extremely low levels in the EV control (Fig. 6C).
To further examine the regulatory effect of GmPIB1 on the expression of its target gene, we performed transient expression assays using 1.761 kb of the GmSPOD1 promoter fused to GUS or LUC as a reporter (pGmSPOD1:GUS or pGmSPOD1:LUC). The effector construct harbored GmPIB1 expressed under the control of the 35S promoter (p35S:GmPIB1-Myc). We transformed the reporter construct (pGmSPOD1:LUC) and the effector construct (p35S:GmPIB1-Myc) into healthy N. benthamiana leaves, finding that GmPIB1 significantly repressed the expression of GmSPOD1 (Fig. 6D). When we transformed the reporter construct (pGmSPOD1:GUS) and the effector construct (p35S:GmPIB1-Myc) into soybean hairy roots, we detected GUS activity driven by the GmSPOD1 promoter (Fig. 6Ea, F), but not by p35S:Myc (Fig. 6Eb, F) or p35S:GmPIB1-Myc (Fig. 6Ec, F). GmPIB1 significantly repressed the expression of GmSPOD1 (Fig. 6Ee, F), whereas there was no change in expression when pGmSPOD1:GUS and p35S:Myc were co-transformed into hairy roots (Fig. 6Ed, F). Taken together, these findings strongly support the idea that GmPIB1 directly inhibits the expression of the downstream GmSPOD1 gene.
GmSPOD1 also functions in responses to P. sojae infection
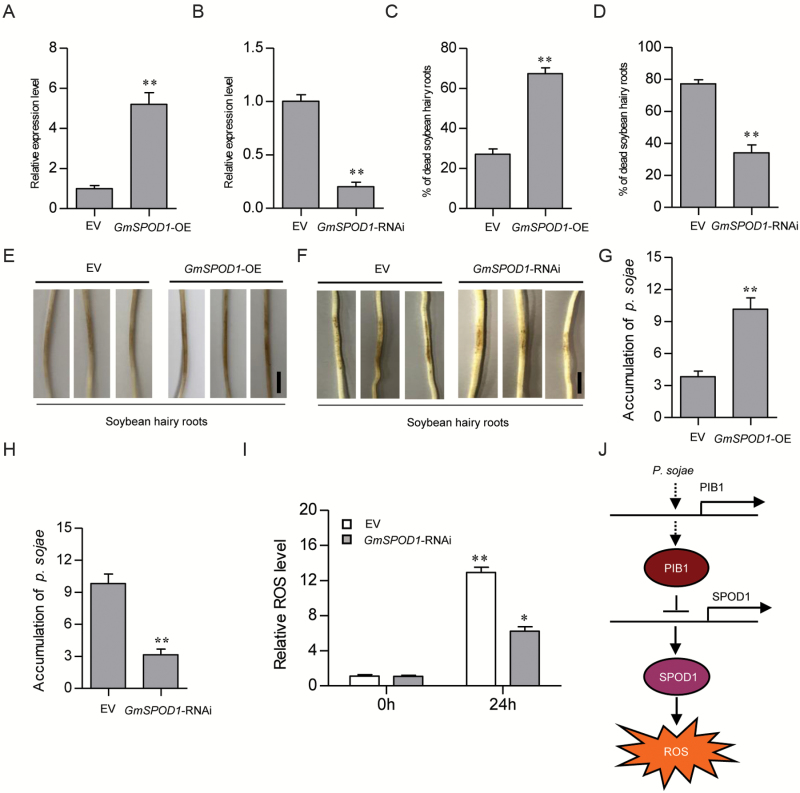
We then explored the possible role of GmSPOD1 in the response to P. sojae infection by analysing the phenotypes of EV, GmSPOD1-RNAi, and GmSPOD1-OE hairy roots after incubation with P. sojae zoospores. First, we tested GmSPOD1-OE and GmSPOD1-RNAi transgenic hairy roots using qRT-PCR (Fig. 7A, B). We then selected transgenic hairy roots and investigated their resistance to P. sojae. As shown in Fig. 7C, ~27% of inoculated EV hairy roots were completely dead and only ~67% of inoculated GmSPOD1-OE transgenic soybean hairy roots were completely dead at 5 d of incubation in resistant cultivar ‘L77-1863’. However, ~77% of inoculated EV hairy roots were completely dead and only ~35% of inoculated GmSPOD1-RNAi transgenic hairy roots were completely dead at 5 d of incubation in susceptible cultivar ‘Williams’ (Fig. 7D). After 2 d of incubation with P. sojae zoospores, all three GmSPOD1-OE transgenic hairy root lines exhibited enhanced wilting symptoms and chlorosis (Fig. 7E), whereas the GmSPOD1-RNAi lines displayed almost no visible lesions compared with the EV control (Fig. 7F).
Fig. 7.
Knockdown of GmSPOD1 increases resistance to P. sojae. (A, B) qRT-PCR analysis of GmSPOD1 expression in EV, GmSPOD1-OE, and GmSPOD1-RNAi transgenic lines. (C, D) Percentage of dead hairy roots in EV, GmSPOD1-OE, and GmSPOD1-RNAi lines after P. sojae infection for 5 d. Each experiment contained at least 50 roots per line, and hairy roots were scored as dead when they were completely rotten. (E, F) Infection phenotypes of GmSPOD1-OE, GmSPOD1-RNAi, and EV soybean hairy roots after P. sojae inoculation for 2 d. (G, H) qRT-PCR analysis of relative P. sojae biomass based on the transcript level of P. sojae TEF1. (I) Relative ROS levels in EV versus GmSPOD1-RNAi lines at 0 and 24 h after P. sojae infection. Three biological replicates, each with three technical replicates, were averaged and statistically analysed using Student’s t-test (**P<0.01). Bars indicate standard error of the mean. (J) Model of the GmPIB1-mediated response to P. sojae. GmPIB1 expression is induced by P. sojae. GmPIB1 inhibits GmSPOD1 transcription by binding to the E-box element in its promoter. The suppression of GmSPOD1 expression leads to decreased intracellular ROS levels. (This figure is available in color at JXB online.)
We also analysed the relative biomass of P. sojae in infected hairy roots after 2 d of incubation with P. sojae zoospores. The biomass of P. sojae (based on P. sojae TEF1 (GenBank accession no. EU079791) transcript levels) was significantly (P<0.01) higher in the roots of GmSPOD1-OE plants versus the EV control (Fig. 7G). The biomass of P. sojae was significantly (P<0.01) lower in the roots of GmSPOD1-RNAi plants compared with EV (Fig. 7H). Finally, we measured relative ROS levels in EV and GmSPOD1-RNAi transgenic hairy roots at 0 and 24 h after incubation with P. sojae. Relative ROS levels gradually increased with increasing inoculation time in both EV and GmSPOD1-RNAi plants (Fig. 7I). However, the relative ROS levels were significantly lower in GmSPOD1-RNAi roots than in EV roots at the same time point (Fig. 7I). These results indicate that repressing GmSPOD1 expression in soybean hairy roots improves resistance to P. sojae.
Discussion
A bHLH TF gene was previously found to be up-regulated in all 10 Rps NILs examined under P. sojae treatment, as revealed by RNA-seq (Lin et al., 2014). In this study, we determined that the bHLH TF designated GmPIB1 plays a crucial role in the response of soybean to P. sojae infection. Consistent with this finding, we found that GmPIB1 transcript levels were much higher in the P. sojae-resistant soybean cultivar ‘L77-1863’ than in the susceptible cultivar ‘Williams’ (Fig. 1A, B). Under P. sojae treatment, GmPIB1 was significantly up-regulated in ‘L77-1863’ but not in ‘Williams’ (Fig. 1E). We also compared the gene and promoter sequences of GmPIB1 between ‘Williams’ and ‘L77-1863’, finding no difference. Perhaps the difference in GmPIB1 expression levels between the two cultivars is due to differences in Rps-mediated defense pathways. To date, a number of genes involved in P. sojae infection have been identified in soybean (Xu et al., 2014; Cheng et al., 2015; Dong et al., 2015; Fan et al., 2015, 2017; Jiang et al., 2015; Yan et al., 2016; Jing et al., 2016; Zhao et al., 2017). For example, in GmERF5-overexpressing soybean plants, PR10, PR1-1, and PR10-1 are up-regulated and P. sojae resistance is significantly enhanced compared with wild type (Dong et al., 2015). GmIFR encodes a NAD(P)H-dependent oxidoreductase and enhances resistance to P. sojae when overexpressed in soybean plants (Cheng et al., 2015). Moreover, GmBips, which are targets of the P. sojae RxLR effector, negatively regulate plant defense responses against P. sojae infection (Jing et al., 2016). Although some genes were shown to be involved in P. sojae responses, little is known about the biological functions of bHLH family members in soybean. To explore the molecular function of GmPIB1 in the response to P. sojae, we overexpressed GmPIB1 in transgenic soybean hairy roots. These hairy roots exhibited significantly increased resistance to P. sojae, whereas resistance to P. sojae was compromised in GmPIB1-RNAi transgenic hairy roots compared with the control (Fig. 1H–M). These results indicate that GmPIB1 plays an important role in defense responses to P. sojae in soybean.
Plants encounter many environmental stresses in their natural environments and have evolved a wide range of mechanisms to cope with these stresses (Dixon and Paiva, 1995; Zhang et al., 2008). When plants are overcome by certain pathogens, they recruit an inducible defense system to limit further pathogen ingression. The phytohormones SA, JA, and ET play central roles in biotic stress signaling following pathogen infection (Pieterse et al., 2009; Robert-Seilaniantz et al., 2011; Sugano et al., 2013). The transcriptional cofactor NPR1 plays a key role in the SA-signaling pathway in several plant species (Vlot et al., 2009). ERF1 plays a crucial role in ET-mediated disease resistance (Berrocal-Lobo et al., 2002). ERF1 also regulates other hormone responses, particularly the JA-mediated defense response (Lorenzo et al., 2003). ET and JA mediate defense responses against pathogen attack (partly) by inducing the expression of defense genes such as PLANT DEFENSIN1.2 (PDF1.2). In the current study, we analysed the expression of GmPIB1 following various hormone treatments (Fig. 2A–C) and determined that GmPIB1 might be primarily involved in responses to MeJA treatment.
The bHLH TFs play important roles in stress responses, which they mediate by binding to the E- and G-boxes present in the promoters of stress-related genes (Qian et al., 2007; Liu et al., 2014). AtbHLH122 specifically binds the E-box of the promoter regions of CYP707A3 and represses its expression, thereby increasing ABA content to positively regulate drought, salt, and osmotic stress signaling in Arabidopsis (Liu et al., 2014). PsGBF (a bHLH-type G-box binding factor) binds to the PsCHS1 promoter and activates its expression to regulate the phenylpropanoid biosynthesis pathway in pea (Qian et al., 2007). bHLH TFs also bind to the G- or E-box DNA motif to regulate plant development (Meng et al., 2013; Liu et al., 2013). For example, GmCIB1 (for cryptochrome-interacting bHLH1) interacts with the E-box-containing promoter sequence of WRKY53b to mediate light-induced regulation of leaf senescence in soybean (Meng et al., 2013). In the current study, we demonstrated that GmPIB1 is localized to the nucleus and cytoplasm and specifically binds to the E-box in vitro (Fig. 3A–D). We also found that GmPIB1 suppressed the basal transcription levels of a reporter gene in Arabidopsis protoplasts (Fig. 3E, F). These findings suggest that GmPIB1 acts as an E-box-mediated transcriptional repressor.
ROS such as H2O2 and O2− act as signaling molecules to regulate plant responses to biotic stress (Mittler et al., 2004; Foyer and Shigeoka 2011; Shigeoka and Maruta 2014). Therefore, we measured ROS levels in GmPIB1-OE, GmPIB1-RNAi, and EV soybean transgenic hairy roots. ROS levels were reduced in GmPIB1-overexpressing transgenic soybean hairy roots (Fig. 5C), suggesting that GmPIB1 improves resistance to P. sojae, possibly by affecting ROS levels. ROS are produced not only as by-products of primary metabolism but also by plasma membrane- or apoplast-localized oxidases, peroxidases, and some TFs (Suzuki et al., 2011; Cheng et al., 2015; Wang et al., 2105a; Zhang et al., 2015, 2016; Noshi et al., 2016). We therefore performed qRT-PCR in EV, GmPIB1-OE, and GmPIB1-RNAi soybean hairy roots to measure the relative expression of genes known to be responsible for ROS production. Among GmPIB1-modulated genes, the expression of GmSPOD1, encoding a key enzyme for ROS production, was down-regulated in hairy roots overexpressing GmPIB1 and up-regulated in GmPIB1 RNA interference lines (Fig. 6A). Using ChIP-qPCR analysis, we also demonstrated that GmPIB1 directly binds to the E-box within the d site region of the GmSPOD1 promoter (Fig. 6B, C). These results suggest that GmPIB1 directly represses GmSPOD1 expression by binding to the E-box in its promoter.
Some enzymes involved in P. sojae infection have been identified in soybean (Subramanian et al., 2005; Graham et al., 2007; Cheng et al., 2015). For example, silencing of either isoflavone synthase or chalcone reductase genes led to the breakdown of resistance to race 1 P. sojae in soybean (Subramanian et al., 2005; Graham et al., 2007). In the current study, GmSPOD1-OE hairy roots showed increased susceptibility to P. sojae, whereas GmSPOD1-RNAi hairy roots showed increased resistance to this pathogen (Fig. 7E, F). These results indicate that the inhibition of GmSPOD1 expression by GmPIB1 enhances resistance to P. sojae in soybean.
Based on our data, we propose a model for the pathway regulating the defense response against P. sojae infection in soybean (Fig. 7J). According to this model, the bHLH TF GmPIB1 is a positive regulator of the response to P. sojae infection. During P. sojae infection, GmPIB1 transcription is activated and this TF binds to the promoter of GmSPOD1, thereby directly inhibiting its expression. Subsequently, the reduced expression of GmSPOD1 leads to decreased intracellular ROS levels and enhanced resistance to P. sojae in soybean plants. Our findings provide important insights into the mechanism underlying the response of soybean to P. sojae infection and offer a strategy for designing and breeding P. sojae-resistant soybean by genetically manipulating a bHLH gene.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Nucleotide and amino acid sequences of GmPIB1 cDNA.
Fig. S2. Resistance analysis of GmPIB1 transgenic soybean hairy roots.
Fig. S3. Expression and purification of fusion protein.
Table S1. List of primers used in this study.
Author contributions
PX, SZ, QC, and LD designed the research. QC, LD, TG, TL, NL, and LW performed the research. XC and JW analysed the data. PX, SZ, QC, and LD wrote the article.
Acknowledgements
This work was supported by NSFC (31171577, 31671719), National Key Research and Development Program of China (2017YFD0101300), Outstanding Talents and Innovative Team of Agricultural Scientific Research, Young and Middle-aged scientific and Technological innovation leader (MOST), Academic backbone of NEAU (17XG21), Natural Science Foundation of Heilongjiang Province (JC201308, C2015010), and Changjiang Scholar Candidates Program for Provincial Universities in Heilongjiang (2013CJHB003).
References
- Berrocal-Lobo M, Molina A, Solano R. 2002. Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. The Plant Journal 29, 23–32. [DOI] [PubMed] [Google Scholar]
- Blair JE, Coffey MD, Park SY, Geiser DM, Kang S. 2008. A multi-locus phylogeny for Phytophthora utilizing markers derived from complete genome sequences. Fungal Genetics and Biology 45, 266–277. [DOI] [PubMed] [Google Scholar]
- Chen S, Songkumarn P, Liu J, Wang GL. 2009. A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiology 150, 1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Li N, Dong L, Zhang D, Fan S, Jiang L, Wang X, Xu P, Zhang S. 2015. Overexpression of soybean isoflavone reductase (GmIFR) enhances resistance to Phytophthora sojae in soybean. Frontiers in Plant Science 6, 1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK. 2003. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes & Development 17, 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Paiva NL. 1995. Stress-induced phenylpropanoid metabolism. The Plant Cell 7, 1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong LD, Cheng YX, Wu JJ, et al. . 2015. Overexpression of GmERF5, a new member of the soybean EAR motif-containing ERF transcription factor, enhances resistance to Phytophthora sojae in soybean. Journal of Experimental Botany 9, 2635–2647. [DOI] [PubMed] [Google Scholar]
- Duek PD, Fankhauser C. 2005. bHLH class transcription factors take centre stage in phytochrome signalling. Trends in Plant Science 10, 51–54. [DOI] [PubMed] [Google Scholar]
- Fan M, Bai MY, Kim JG, et al. . 2014. The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern-triggered immunity in Arabidopsis. The Plant Cell 26, 828–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan SJ, Dong LD, Han D, et al. . 2017. GmWRKY31 and GmHDL56 enhances resistance to Phytophthora sojae by regulating defense-related gene expression in soybean. Frontiers in Plant Science 8, 781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan SJ, Jiang LY, Wu JJ, Dong LD, Cheng Q, Xu PF, Zhang SZ. 2015. A novel pathogenesis-related class 10 protein Gly m 4l, increases resistance upon Phytophthora sojae infection in soybean (Glycine max [L.] Merr.). PLoS One 10, e014036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr WR, Caviness CE, Burmood DT, Pennington J. 1971. Stage of development descriptions for soybeans, Glycine max (L.) Merrill. Crop Science 11, 929–931. [Google Scholar]
- Feller A, Machemer K, Braun EL, Grotewold E. 2011. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. The Plant Journal 66, 94–116. [DOI] [PubMed] [Google Scholar]
- Foyer CH, Shigeoka S. 2011. Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiology 155, 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TL, Graham MY, Subramanian S, Yu O. 2007. RNAi silencing of genes for elicitation or biosynthesis of 5-deoxyisoflavonoids suppresses race-specific resistance and hypersensitive cell death in Phytophthora sojae infected tissues. Plant Physiology 144, 728–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao YJ, Wei W, Song QX, et al. . 2011. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. The Plant Journal 68, 302–313. [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. 1999. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Research 27, 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L. 2006. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proceedings of the National Academy of Sciences, USA 103, 12987–12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hückelhoven R, Kogel KH. 2003. Reactive oxygen intermediates in plant-microbe interactions: who is who in powdery mildew resistance?Planta 216, 891–902. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Wu J, Fan S, Li W, Dong L, Cheng Q, Xu P, Zhang S. 2015. Isolation and characterization of a novel pathogenesis-related protein gene (GmPRP) with induced expression in soybean (Glycine max) during infection with Phytophthora sojae. PLoS One 10, e0129932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing M, Guo B, Li H, et al. . 2016. A Phytophthora sojae effector suppresses endoplasmic reticulum stress-mediated immunity by stabilizing plant binding immunoglobulin proteins. Nature Communications 7, 11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass J, Ruben A, Baylies MK. 2000. Non-radioactive electrophoretic mobility shift assay using digoxigenin-ddUTP labeled probes. Drosophila Information Service 83, 185–188. [Google Scholar]
- Kereszt A, Li D, Indrasumunar A, Nguyen CD, Nontachaiyapoom S, Kinkema M, Gresshoff PM. 2007. Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nature Protocols 2, 948–952. [DOI] [PubMed] [Google Scholar]
- Kerschen A, Napoli CA, Jorgensen RA, Müller AE. 2004. Effectiveness of RNA interference in transgenic plants. FEBS Letters 566, 223–228. [DOI] [PubMed] [Google Scholar]
- Kim J, Kim HY. 2006. Molecular characterization of a bHLH transcription factor involved in Arabidopsis abscisic acid-mediated response. Biochimica et Biophysica Acta 1759, 191–194. [DOI] [PubMed] [Google Scholar]
- Lee BH, Henderson DA, Zhu JK. 2005. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. The Plant Cell 17, 3155–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Zou HF, Wang HW, Zhang WK, Ma B, Zhang JS, Chen SY. 2008a Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants. Cell Research 18, 1047–1060. [DOI] [PubMed] [Google Scholar]
- Liao Y, Zou HF, Wei W, Hao YJ, Tian AG, Huang J, Liu YF, Zhang JS, Chen SY. 2008b Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 228, 225–240. [DOI] [PubMed] [Google Scholar]
- Lin F, Zhao M, Baumann DD, et al. . 2014. Molecular response to the pathogen Phytophthora sojae among ten soybean near isogenic lines revealed by comparative transcriptomics. BMC Genomics 15, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Tai H, Li S, Gao W, Zhao M, Xie C, Li WX. 2014. bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytologist 201, 1192–1204. [DOI] [PubMed] [Google Scholar]
- Liu Y, Li X, Li K, Liu H, Lin C. 2013. Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis. PLoS Genetics 9, e1003861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZQ, Yan L, Wu Z, et al. . 2012. Cooperation of three WRKY-domain transcription factors WRKY18, WRKY40, and WRKY60 in repressing two ABA-responsive genes ABI4 and ABI5 in Arabidopsis. Journal of Experimental Botany 63, 6371–6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R. 2003. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. The Plant Cell 15, 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CA, Lim EK, Yu SM. 1998. Sugar response sequence in the promoter of a rice α-amylase gene serves as a transcriptional enhancer. The Journal of Biological Chemistry 273, 10120–10131. [DOI] [PubMed] [Google Scholar]
- Lu Y, Hall DA, Last RL. 2011. A small zinc finger thylakoid protein plays a role in maintenance of photosystem II in Arabidopsis thaliana. The Plant Cell 23, 1861–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma PC, Rould MA, Weintraub H, Pabo CO. 1994. Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell 77, 451–459. [DOI] [PubMed] [Google Scholar]
- Meng Y, Li H, Wang Q, Liu B, Lin C. 2013. Blue light-dependent interaction between cryptochrome2 and CIB1 regulates transcription and leaf senescence in soybean. The Plant Cell 25, 4405–4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. 2004. Reactive oxygen gene network of plants. Trends in Plant Science 9, 490–498. [DOI] [PubMed] [Google Scholar]
- Nakata M, Mitsuda N, Herde M, Koo AJ, Moreno JE, Suzuki K, Howe GA, Ohme-Takagi M. 2013. A bHLH-type transcription factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in arabidopsis. The Plant Cell 25, 1641–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu CF, Wei W, Zhou QY, et al. . 2012. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant, Cell & Environment 35, 1156–1170. [DOI] [PubMed] [Google Scholar]
- Noshi M, Mori D, Tanabe N, Maruta T, Shigeoka S. 2016. Arabidopsis clade IV TGA transcription factors, TGA10 and TGA9, are involved in ROS-mediated responses to bacterial PAMP flg22. Plant Science 252, 12–21. [DOI] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY. 2012. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nature Cell Biology 14, 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez IB, Brown PJ. 2014. The role of ROS signaling in cross-tolerance: from model to crop. Frontiers in Plant Science 5, 754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CM, Leon-Reyes A, Van der Ent S, Van Wees SC. 2009. Networking by small-molecule hormones in plant immunity. Nature Chemical Biology 5, 308–316. [DOI] [PubMed] [Google Scholar]
- Qian H, Chen W, Sun L, Jin Y, Liu W, Fu Z. 2009. Inhibitory effects of paraquat on photosynthesis and the response to oxidative stress in Chlorella vulgaris. Ecotoxicology 18, 537–543. [DOI] [PubMed] [Google Scholar]
- Qian W, Tan G, Liu H, He S, Gao Y, An C. 2007. Identification of a bHLH-type G-box binding factor and its regulation activity with G-box and Box I elements of the PsCHS1 promoter. Plant Cell Reports 26, 85–93. [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JD. 2011. Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annual Review of Phytopathology 49, 317–343. [DOI] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova Z. 2008. An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nature Protocols 3, 1018–1025. [DOI] [PubMed] [Google Scholar]
- Seo YJ, Park JB, Cho YJ, Jung C, Seo HS, Park SK, Nahm BH, Song JT. 2010. Overexpression of the ethylene-responsive factor gene BrERF4 from Brassica rapa increases tolerance to salt and drought in Arabidopsis plants. Molecules and Cells 30, 271–277. [DOI] [PubMed] [Google Scholar]
- Shan W, Cao M, Leung D, Tyler BM. 2004. The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b. Molecular Plant-Microbe Interactions 17, 394–403. [DOI] [PubMed] [Google Scholar]
- Shang Y, Yan L, Liu ZQ, et al. . 2010. The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. The Plant Cell 22, 1909–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty NP, Lyngs Jørgensen HJ, Jensen JD, Collinge DB, Shetty HS. 2008. Roles of reactive oxygen species in interactions between plants and pathogens. European Journal Plant Pathology 121, 267–280. [Google Scholar]
- Shigeoka S, Maruta T. 2014. Cellular redox regulation, signaling, and stress response in plants. Bioscience, Biotechnology, and Biochemistry 78, 1457–1470. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Toumoto A, Ihara K, Shimizu M, Kyogoku Y, Ogawa N, Oshima Y, Hakoshima T. 1997. Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition. The EMBO Journal 16, 4689–4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song QX, Li QT, Liu YF, et al. . 2013. Soybean GmbZIP123 gene enhances lipid content in the seeds of transgenic Arabidopsis plants. Journal of Experimental Botany 64, 4329–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soosaar JL, Burch-Smith TM, Dinesh-Kumar SP. 2005. Mechanisms of plant resistance to viruses. Nature Reviews. Microbiology 3, 789–798. [DOI] [PubMed] [Google Scholar]
- Subramanian S, Graham MY, Yu O, Graham TL. 2005. RNA interference of soybean isoflavone synthase genes leads to silencing in tissues distal to the transformation site and to enhanced susceptibility to Phytophthora sojae. Plant Physiology 137, 1345–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano S, Sugimoto T, Takatsuji H, Jiang J. 2013. Induction of resistance to Phytophthora sojae in soybean (Glycine max) by salicylic acid and ethylene. Plant Pathology 62, 1048–1056. [Google Scholar]
- Sugimoto T, Kato M, Yoshida S, et al. . 2012. Pathogenic diversity of Phytophthora sojae and breeding strategies to develop Phytophthora-resistant soybeans. Breeding Science 61, 511–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Miller G, Morales J, Shulaev V, Torres MA, Mittler R. 2011. Respiratory burst oxidases: the engines of ROS signaling. Current Opinion in Plant Biology 14, 691–699. [DOI] [PubMed] [Google Scholar]
- Takabatake R, Ando Y, Seo S, Katou S, Tsuda S, Ohashi Y, Mitsuhara I. 2007. MAP kinases function downstream of HSP90 and upstream of mitochondria in TMV resistance gene N-mediated hypersensitive cell death. Plant & Cell Physiology 48, 498–510. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24, 1596–1599. [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH. 2003. The Arabidopsis basic/helix-loop-helix transcription factor family. The Plant Cell 15, 1749–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooley PW, Grau CR. 1984. The relationship between rate-reducing resistance to Phytophthora megasperma f. sp. glycinea and yield of soybean. Phytopathology 74, 1209–1216. [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, et al. . 2004. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. The Plant Cell 16, 2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull D, Yang L, Naqvi S, et al. . 2017. RXLR Effector AVR2 up-regulates a brassinosteroid-responsive bHLH transcription factor to suppress immunity. Plant Physiology 174, 356–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler BM. 2007. Phytophthora sojae: root rot pathogen of soybean and model oomycete. Molecular Plant Pathology 8, 1–8. [DOI] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF. 2009. Salicylic acid, a multifaceted hormone to combat disease. Annual Review of Phytopathology 47, 177–206. [DOI] [PubMed] [Google Scholar]
- Walker AK, Schmitthenner AF. 1984. Heritability of tolerance to Phytophthora rot in soybean. Crop Science 24, 490–491. [Google Scholar]
- Wang F, Chen HW, Li QT, et al. . 2015. a GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants. The Plant Journal 83, 224–236. [DOI] [PubMed] [Google Scholar]
- Wang JY, Hu ZZ, Zhao T, Yang YW, Chen TZ, Yang M, Yu WG, Zhang BL. 2015b Genome-wide analysis of bHLH transcription factor and involvement in the infection by yellow leaf curl virus in tomato (Solanum lycopersicum). BMC Genomics 16, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward EWB, Lazarovits G, Unwin CH, Buzzell RI. 1979. Hypocotyl reactions and glyceollin in soybeans inoculated with zoospores of Phytophthora megasperma var. sojae. Phytopathology 69, 951–955. [Google Scholar]
- Xu P, Jiang L, Wu J, Li W, Fan S, Zhang S. 2014. Isolation and characterization of a pathogenesis-related protein 10 gene (GmPR10) with induced expression in soybean (Glycine max) during infection with Phytophthora sojae. Molecular Biology Reports 41, 4899–4909. [DOI] [PubMed] [Google Scholar]
- Yan Q, Cui X, Lin S, Gan S, Xing H, Dou D. 2016. GmCYP82A3, a soybean cytochrome P450 family gene involved in the jasmonic acid and ethylene signaling pathway, enhances plant resistance to biotic and abiotic stresses. PLoS One 11, e0162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Cui X, Su L, Xu N, Guo N, Xing H, Dou D. 2014. GmSGT1 is differently required for soybean Rps genes-mediated and basal resistance to Phytophthora sojae. Plant Cell Reports 33, 1275–1288. [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. 2007. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhang G, Chen M, Chen X, et al. . 2008. Phylogeny, gene structures, and expression patterns of the ERF gene family in soybean (Glycine max L.). Journal of Experimental Botany 59, 4095–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Hedhili S, Montiel G, Zhang Y, Chatel G, Pré M, Gantet P, Memelink J. 2011. The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. The Plant Journal 67, 61–71. [DOI] [PubMed] [Google Scholar]
- Zhang H, Hong Y, Huang L, Li D, Song F. 2016. Arabidopsis AtERF014 acts as a dual regulator that differentially modulates immunity against Pseudomonas syringae pv. tomato and Botrytis cinerea. Scientific Reports 6, 30251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Huang L, Dai Y, et al. . 2015. Arabidopsis AtERF15 positively regulates immunity against Pseudomonas syringae pv. tomato DC3000 and Botrytis cinerea. Frontiers in Plant Science 6, 686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SZ, Xu PF, Wu JJ, Xue AG, Zhang JX, Li WB, Chen C, Chen WY, Lv HY. 2010. Races of Phytophthora sojae and their virulences on soybean cultivars in Heilongjiang, China. Plant Disease 94, 87–91. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Liu X, Wang X, Zhou M, Zhou X, Ye X, Wei X. 2012. An R2R3 MYB transcription factor in wheat, TaPIMP1, mediates host resistance to Bipolaris sorokiniana and drought stresses through regulation of defense- and stress-related genes. New Phytologist 196, 1155–1170. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Chang X, Qi D, et al. . 2017. A Novel soybean ERF transcription factor, GmERF113, increases resistance to Phytophthora sojae infection in soybean. Frontiers in Plant Science 8, 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou QY, Tian AG, Zou HF, et al. . 2008. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnology Journal 6, 486–503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.