Abstract
Epigenetic transgenerational inheritance of disease and phenotypic variation can be induced by several toxicants, such as vinclozolin. This phenomenon can involve DNA methylation, non-coding RNA (ncRNA) and histone retention, and/or modification in the germline (e.g. sperm). These different epigenetic marks are called epimutations and can transmit in part the transgenerational phenotypes. This study was designed to investigate the vinclozolin-induced concurrent alterations of a number of different epigenetic factors, including DNA methylation, ncRNA, and histone retention in rat sperm. Gestating females (F0 generation) were exposed transiently to vinclozolin during fetal gonadal development. The directly exposed F1 generation fetus, the directly exposed germline within the fetus that will generate the F2 generation, and the transgenerational F3 generation sperm were studied. DNA methylation and ncRNA were altered in each generation rat sperm with the direct exposure F1 and F2 generations being distinct from the F3 generation epimutations. Interestingly, an increased number of differential histone retention sites were found in the F3 generation vinclozolin sperm, but not in the F1 or F2 generations. All three different epimutation types were affected in the vinclozolin lineage transgenerational sperm (F3 generation). The direct exposure generations (F1 and F2) epigenetic alterations were distinct from the transgenerational sperm epimutations. The genomic features and gene pathways associated with the epimutations were investigated to help elucidate the integration of these different epigenetic processes. Our results show that the three different types of epimutations are involved and integrated in the mediation of the epigenetic transgenerational inheritance phenomenon.
Keywords: epigenetics, sperm, spermatogenesis, transgenerational, inheritance, DNA methylation
Introduction
Environmentally induced epigenetic transgenerational inheritance is defined as “germline (sperm or egg) transmission of epigenetic information between generations in the absence of any continued exposure or genetic manipulation.” In 1987 the word epimutation was introduced to describe heritable changes in genes which were not due to changes in DNA sequence [1]. A number of different epigenetic factors have been described and are critical in regulating gene expression including chromatin structure and non-coding RNA (ncRNA) methylation which have become an intensively active field of research. The modification of histones, particularly acetylation and methylation, play a crucial role in regulating genome activity [2]. DNA methylation can provide a primary switch where methylation triggers the changes that lead to a closed (not allowing gene transcription) or opened chromatin configuration (allowing gene transcription) [3]. The role of ncRNA in epigenetic events has become increasingly important [4–6]. All these epigenetic processes can be altered by numerous environmental exposures [7]. These environmental exposures can alter the early development of various organ systems that result in later life adult individuals with greater susceptibilities to develop disease when they encounter a second disease-promoting stimulus [8].
Epigenetic alterations in the germline mediate the heritable molecular relationship between the environment and gene expression [9, 10]. Studies have observed epigenetic transgenerational inheritance effects in different species, including plants [11], flies [12], worms [13], fish [14], birds [15] rodents [16–18], pigs [19], and humans [20]. This phenomenon has been observed in a large number of organisms showing that epigenetic transgenerational mechanisms are conserved and induced by environmental factors [21]. Many studies on environmental toxicants have been conducted, such as 2, 3, 7, 8-tetrachlorodibenzo [p]dioxin [22], permethrin and N, N-diethyl-meta-toluamide mixture [23], jet fuel JP-8 [24], plastics mixture [bisphenol-A, bis (2-ethylhexyl)phthalate, dibutyl phthalate] [25], dichlorodiphenyltrichloroethane (DDT) [26], methoxychlor [27], and atrazine [28]. For each exposure, alterations in DNA methylation have been observed in F3 generation rat sperm.
The first compound showing a transgenerational effect was vinclozolin. Vinclozolin [3-(3, 5-dichlorophenyl)-5methyl-5-vinyl-1, 3-oxazolidine-2, 4-dione] is a fungicide used on fruits and vegetables crops. This chemical was found to exert significant antiandrogenic effects [29, 30]. Vinclozolin administration to pregnant female mice during the gestation period of embryonic days E13–E17 (which corresponds to fetal gonadal sex determination) was found to affect the offspring [16, 31]. This chemical also demonstrated transgenerational (F1 through F4 generations) effects on male fertility and altered the DNA methylation patterns in the F2 and F3 generation sperm [16]. This suggested that environment can induce epigenetic changes in the germline and be inherited to contribute to disease. Other exposures since then have been shown to promote epigenetic transgenerational phenotypes [21]. One of the proposed mechanisms is that “epimutations” induced in the germline become “imprinted-like” and therefore escape the post-fertilization DNA methylation erasure. The altered epigenome leads to modified transcriptomes in somatic cells resulting in adult-onset disease susceptibility [27, 32, 33]. Anway and collaborators revealed that following transient gestational exposure to F0 generation females between E8 and E15 (corresponding to the epigenetic reprogramming of germ cells and gonadal sex determination) the F1, F2, F3, and F4 generation adult males showed increased spermatogenic apoptosis and a decreased sperm number and motility [16]. Altered DNA methylation patterns in F2 and F3 generation vinclozolin lineages were also observed [34]. Prostate abnormalities were noted in F1–F4 generation males [35], severe anemia during pregnancy in F1–F3 generation females [36] and also sexually dimorphic changes in anxiety behaviors [37]. The mRNA expression of 196 genes was found to be consistently different in testis among the F1–F3 generation vinclozolin animals compared to the F1–F3 generation controls [35]. Transcription alterations in prostate from F3 males [35], hippocampus and amygdala in F3 males and females [37], and Sertoli cells [38] were also reported. These studies demonstrate the effects of vinclozolin transgenerationally on somatic gene expression.
The involvement of sperm small non-coding RNA (sncRNA) in the impact of traumatic stress in early life across generations was also investigated. Sperm RNAs from traumatized males were injected into fertilized wild-type oocytes which reproduced the behavioral and metabolic alterations in their offspring [39]. This suggests that sncRNAs are sensitive to environmental factors in early life which can be passed to the next generation and contribute to the inheritance of trauma-induced phenotypes [39]. Subsequently vinclozolin has also been shown to alter transgenerationally the sncRNA in sperm [40]. Previous studies have revealed that ncRNA can modulate gene expressions and many of them have been identified to participate in the epigenetic control by affecting DNA methylation [41, 42] or by interacting with various types of proteins involved in histone modification or chromatin remodeling [43–46]. Therefore, long ncRNA (lncRNA) is an ideal mediator to regulate local and sequence-specific DNA methylation or demethylation which may also trigger epigenetic disorders and transgenerational effects [43].
The lncRNAs have been hypothesized to maintain epigenetic memory and a recent study has correlated lncRNA differential expression to DDT-induced epigenetic transgenerational inheritance [7]. The sncRNAs were also implicated when the effects of starvation were found to induce the production of sncRNAs that persisted for multiple generations and resulted in a longer lifespan for the F3 generation [47]. Total estimates for both lncRNA and sncRNA in a single spermatozoa have exceeded 20 000 [48, 49]. As such, there are various different kinds of small RNAs found in spermatozoa including: micro-RNA (miRNAs) and siRNAs involved in regulating spermatogenesis via translation; piwi-interacting RNAs (piRNAs) believed to regulate transcript stability in the germline; and many smaller classes of sncRNA such as tRNA-derived sncRNAs and mitosRNAs have also been discovered in spermatozoa [50, 51]. The expression of ncRNAs throughout spermatogenesis has been shown to be differential [51], suggesting an environmentally induced disruption of ncRNA expression in spermatogenic cells can have lasting effects for generations. Both DNA methylation and ncRNA transgenerational alterations have been reported to be altered in the epigenetic transgenerational inheritance phenomenon, but to the best of our knowledge no studies have examined histone modifications until recently [52]. Several studies have observed conserved genes associated with histone retention sites and modifications [53] indicating modifications of histones may have an impact transgenerationally.
This study was designed to investigate concomitantly the epigenetic modifications of DNA methylation, ncRNA, and histones retention in sperm. The transgenerational model used for this study involves epigenetic transgenerational inheritance of sperm epigenetic alterations and disease triggered by vinclozolin exposure. This study will use this vinclozolin-induced epigenetic transgenerational inheritance of disease in rats to investigate the epigenetic changes in the F1, F2, and F3 generation sperm. This information combined with associated genes will be useful in the elucidation of the molecular processes implicated in the epigenetic transgenerational inheritance phenomenon. Observations demonstrate all three epigenetic processes are potentially involved and reflect transgenerational epimutations, including a novel role of new induced transgenerational differential histone retention sites (DHRs), and distinct transgenerational DNA methylation and ncRNA epimutations.
Results
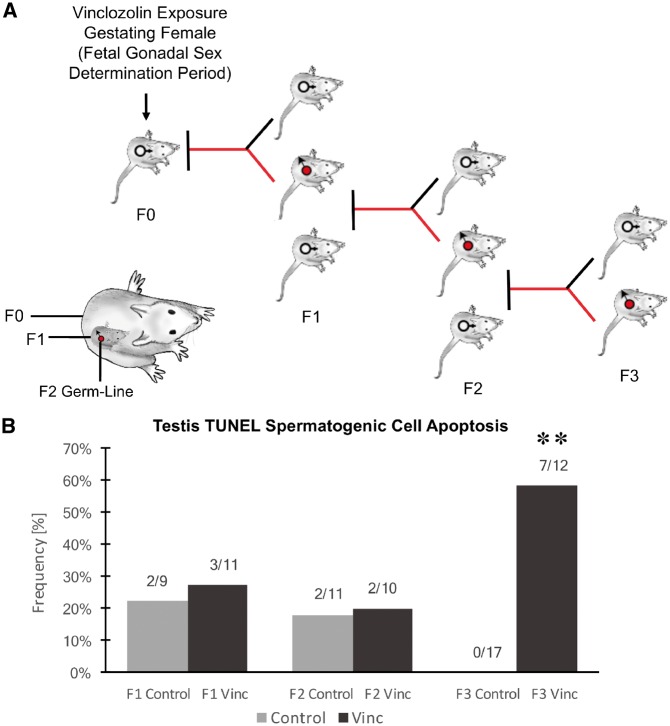
The experimental design (Fig. 1A) involved a daily transient exposure of gestating female F0 generation rats during embryonic days E8–E14 to 100 mg/kg per day of vinclozolin in dimethylsulfoxide (DMSO) using intraperitoneal injection as previously described [16]. A separate control generation lineage was exposed during Days E8–E14 of gestation to vehicle DMSO alone. Six different gestating females from different litters for each control and vinclozolin lineage were used. The F1 generation offspring were obtained and aged to 90-day postnatal age and selected males and females bred within the control or vinclozolin lineage. The F2 generation offspring were obtained and aged to 90 days and selected males and females from different litters bred to generate the F3 generation. No sibling or cousin breeding was used to avoid any inbreeding artifacts. All the males were sacrificed at 120 days of age for epididymal sperm collection. In previous studies, onset of disease was primarily observed between 6 and 12 months of age [54], hence the postnatal 120-day (P120) males were used to avoid any disease artifacts. The only disease detectable at P120 is testis spermatogenic cell apoptosis [16]. Selected male testis at each generation for both the control and vinclozolin lineages was used for spermatogenic cell apoptosis analysis. A significant level of apoptosis was observed in the F3 generation vinclozolin lineage (Fig. 1B) supporting the transgenerational phenotype of the vinclozolin model used. The sperm collected was then used for epigenetic analysis.
Figure 1:
animal breeding scheme and disease. (A) Experimental design of F0 generation gestating female exposure then F1, F2, and F3 generations being generated for sperm collection. The direct exposure of the F0 generation female, F1 generation fetus, and F2 generation germline is also shown. (B) Testis spermatogenic cell apoptosis as determined with TUNEL analysis of testis histology sections with frequency (%) of apoptosis for each generation and lineage shown. The (**) indicates statistical significance with control with a P < 0.01 with a Fishers exact T-test
Sperm DNA Methylation Alterations
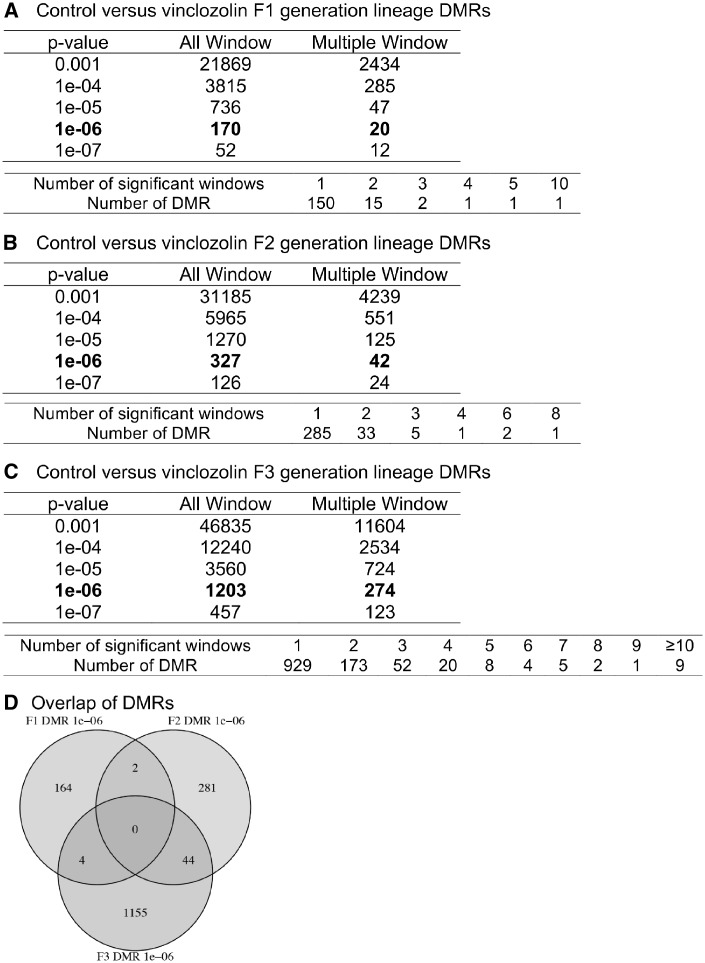
Methylated DNA immunoprecipitation (MeDIP) followed by DNA next-generation sequencing (NGS) for a MeDIP-Seq analysis was used to identify the differential DNA-methylated regions (DMRs) between the control versus vinclozolin lineage male sperm. Initially the sperm DNA was sonicated to produce DNA fragments of 200–500 bp. A methylcytosine antibody was used to immunoprecipitate the methylated DNA fragments. Libraries were generated for sequencing 50-bp paired-end reads to assess differential levels (read depths) of DNA methylation. The DMRs for the F1, F2, and F3 generation sperm were determined and different threshold p values are shown in Fig. 2. A P-value of P < 1e-06 was chosen for comparison of all single 100-bp windows, as well as DMRs with multiple (≥2) adjacent windows. The direct exposure F1 generation sperm had the lowest number of DMRs (Fig. 2A), the F2 generation an increased number of DMRs (Fig. 2B), and the F3 generation the highest number of DMRs (Fig. 2C). The DMR lists are presented in the Supplementary Tables S1–S3. A comparison of the DMRs at P < 1e-06 demonstrated minimal overlap with a higher level of overlap between the F2 and F3 generations (Fig. 2D). The majority of the alterations in sperm DNA methylation was unique between the generations. A small set of 44 overlapping DMRs between the F2 and F3 generation sperm are presented in the Supplementary Table S4.
Figure 2:
DMR analysis. (A) F1 generation control versus vinclozolin lineage DMRs. (B) F2 generation control versus vinclozolin lineage DMRs. (C) F3 generation control versus vinclozolin lineage DMRs. The number of DMRs found using different P-value cutoff thresholds is presented. The All Window column shows all DMRs. The Multiple Window column shows the number of DMRs containing at least two adjacent significant windows. (D) The DMR overlap (P ≤ 1e-06) for the F1, F2, and F3 generation sperm
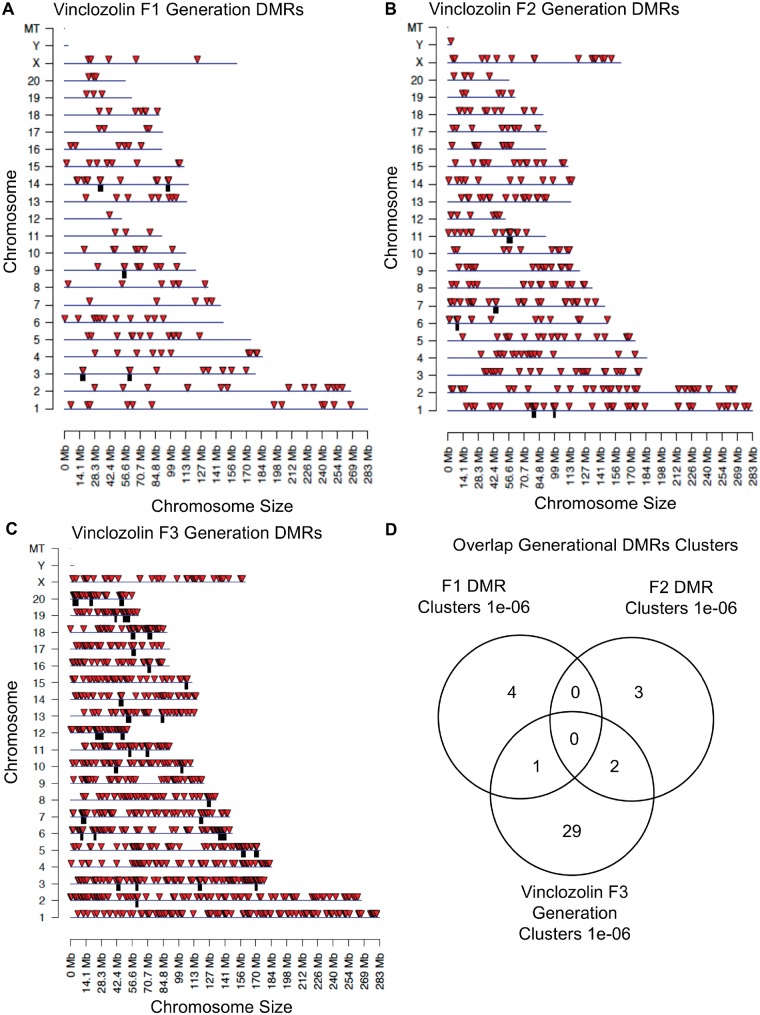
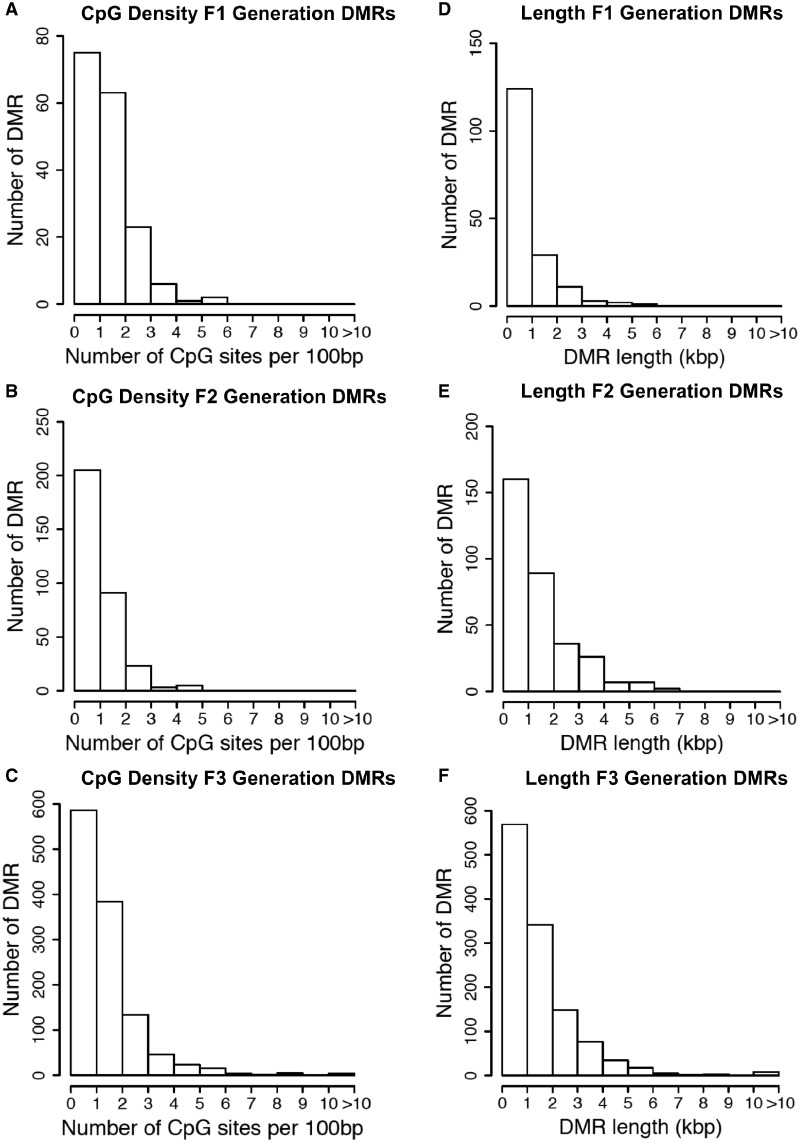
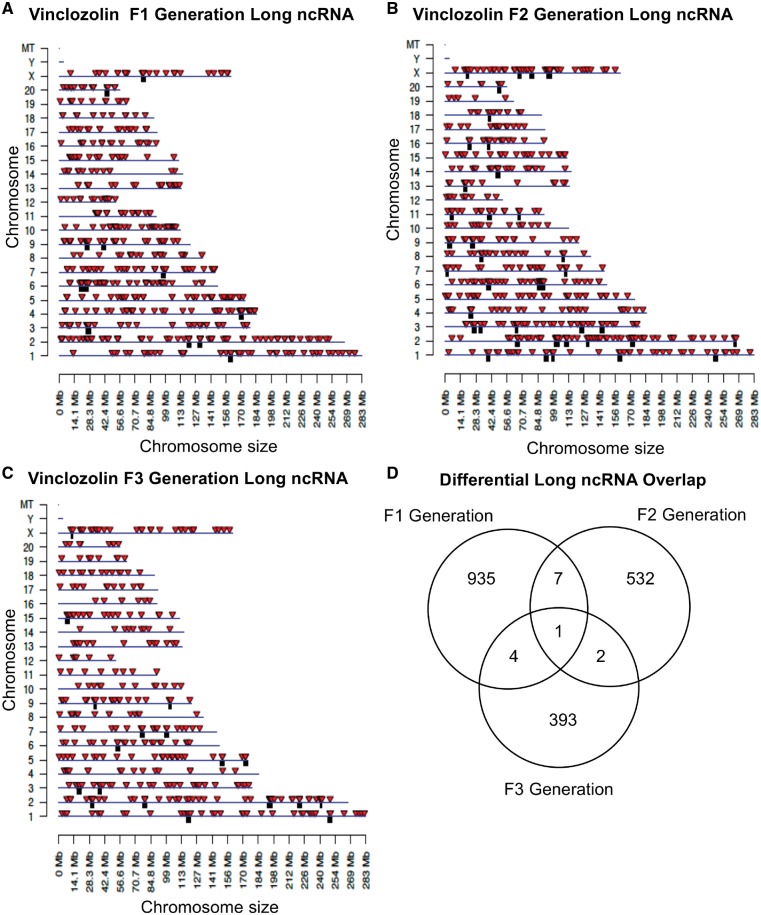
The chromosomal locations of the DMRs for each generation are shown in Fig. 3. Nearly all the chromosomes display DMRs as indicated by the red arrowheads and some present clusters of DMRs indicated by black boxes. These different generation DMR clusters are also primarily distinct between the generations with little overlap (Fig. 3D). The CpG density of the DMRs for the F1, F2, and F3 generation sperm is shown in Fig. 4A–C. The CpG density of the DMRs at P < 1e-06 is shown and reveals that the predominant density is one CpG per 100 bp with a range between 1 and 4 CpG. The lengths of the DMRs are presented in Fig. 4D–F and shows that their predominant length is 1 kb and a range of 1–5 kb for each generation DMRs. Thus, the DMRs are associated with CpG deserts with 10–20 CpG within 1 kb [55]. The lists of DMRs with their chromosomal locations, size, and CpG density are presented in the Supplementary Tables S1–S3 for the F1, F2, and F3 generation DMRs, respectively.
Figure 3:
chromosomal locations and overlaps of DMRs. (A) Vinclozolin F1 generation DMRs. (B) Vinclozolin F2 generation DMRs. (C) Vinclozolin F3 generation DMRs. The DMR locations on the individual chromosomes are shown with red arrowheads and clusters of DMRs with black boxes. All window DMRs at a P-value threshold of 1e-06 are shown. (D) Overlap of the different generations DMR clusters
Figure 4:
DMR CpG density. (A) F1 generation, (B) F2 generation, and (C) F3 generation. Histograms of the number of DMRs at different CpG densities (CpG/100 bp). All DMRs at a P-value threshold of 1e-06 are shown. DMR length (kb) for (D) F1 generation, (E) F2 generation, and (F) F3 generation. Histograms of the number of DMR at different length (kb) are shown
Sperm ncRNA Alterations
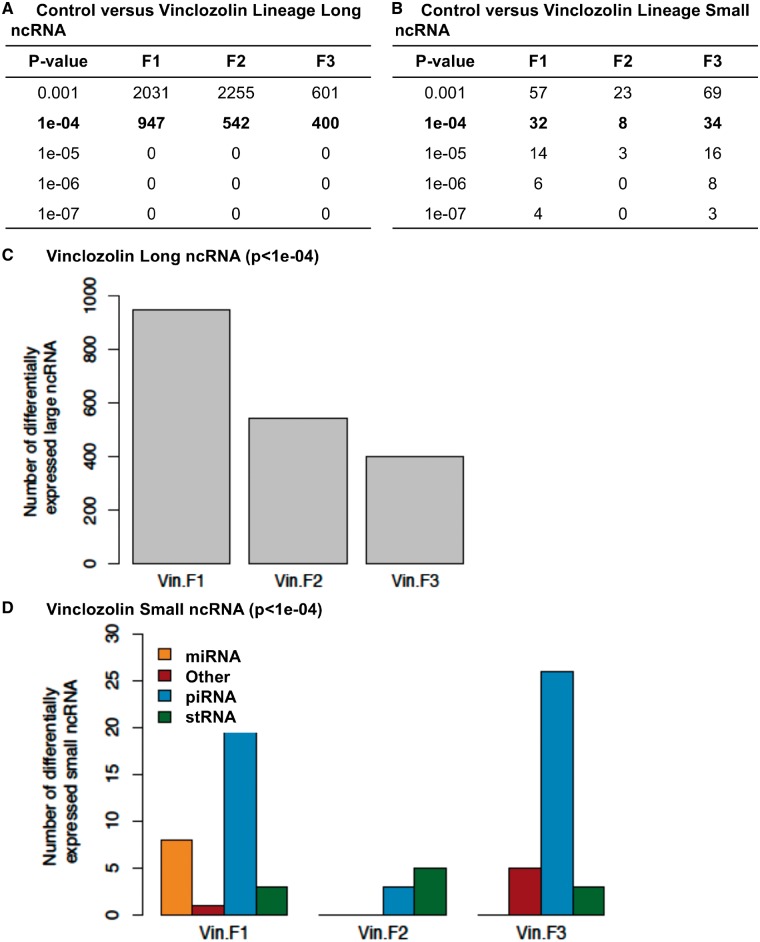
The differential ncRNA present in sperm between the control versus vinclozolin lineages was determined with NGS using RNA-Seq analysis previously described [40]. The sperm total ncRNA was collected and then sncRNA and lncRNA prepared with appropriate ncRNA extraction and sequencing library procedures described in the Methods section. The read depths of differential ncRNA levels were determined for the F1, F2, and F3 generation sperm with a comparison of the control versus vinclozolin lineage sperm. The lncRNA for each generation with different P-value thresholds is compiled in Fig. 5A. The P-value used for subsequent analysis selected was P < 1e-04. The sncRNA for each generation with several P-value thresholds is shown in Fig. 5B. The P-value used for sncRNA analysis selected was P < 1e-04. The lncRNA is higher in number than the sncRNA. For the lncRNA, the F1 generation has a higher number than the F3 generation, and the F3 generation has a lower number than the F2 generation (Fig. 5C). The sncRNA was divided into categories with the piRNA, the miRNA, and the small tRNA being present, as well as a mixture of uncategorized sncRNA (other) (Fig. 5D). Therefore, each generation had differential ncRNA present and they were different between each generation.
Figure 5:
ncRNA differentially regulated in control versus vinclozolin lineage F1, F2, and F3 generation sperm. (A) lncRNA and (B) sncRNA numbers at different P-value thresholds. (C) The number of lncRNA (P < 1e-04) for the F1, F2, and F3 generation correlated to the number of differential lncRNA. (D) The number of sncRNA (P < 1e-04) for the F1, F2, and F3 generation correlated to the number of miRNA, piRNA, stRNA, and other sncRNA
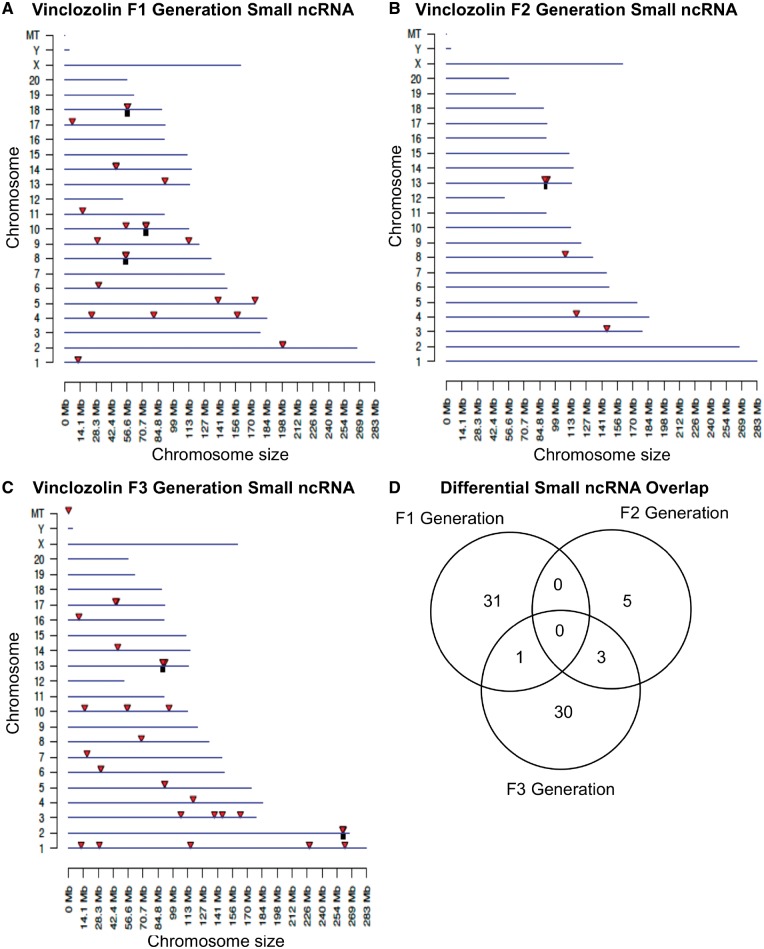
The chromosomal locations of the differential ncRNA are presented for lncRNA in Fig. 6. The red arrowheads identify individual lncRNA and regional clusters of lncRNA are shown in black boxes. The F1, F2, and F3 generation lncRNAs are present on all chromosomes (Fig. 6A–C). The analysis of the lncRNA overlap between the generations indicated that the vast majority of differential lncRNA was distinct for each generation (Fig. 6D). The chromosomal locations of the differential ncRNA are shown for sncRNA in Fig. 7A–C. The overlap analysis between the three generations is shown in Fig. 7D. It reveals that the majority of differential sncRNA was unique for each generation. Both the long and small differential ncRNAs were predominantly distinct from the transgenerational F3 generation ncRNA. The lists of differential ncRNA separated by category for identification, chromosomal location, size, statistics and gene associations are presented in the Supplementary Tables S5–S7 for lncRNA and Supplementary Tables S8–S10 for sncRNA for the F1, F2, and F3 generations, respectively.
Figure 6:
chromosomal locations of lncRNA. (A) F1 generation, (B) F2 generation, and (C) F3 generation sperm. The lncRNA locations on the individual chromosomes are indicated with red arrowheads and clusters with black boxes. The lncRNA at a P-value threshold of 10−4 is shown. (D) The overlap between the lncRNA (P < 1e-04) in the three vinclozolin generations. Overlaps were determined based on common ncRNA names
Figure 7:
chromosomal locations of small ncRNA. (A) F1 generation, (B) F2 generation, and (C) F3 generation. The sncRNA locations on the individual chromosomes are indicated with red arrowheads and clusters with black boxes. The sncRNA at an FDR-adjusted P-value threshold of P < 1e-04 is shown. (D) The overlap between the sncRNAs (P < 1e-04) in the three vinclozolin generations. Overlaps were determined based on common RNA names
Sperm Histones Alterations
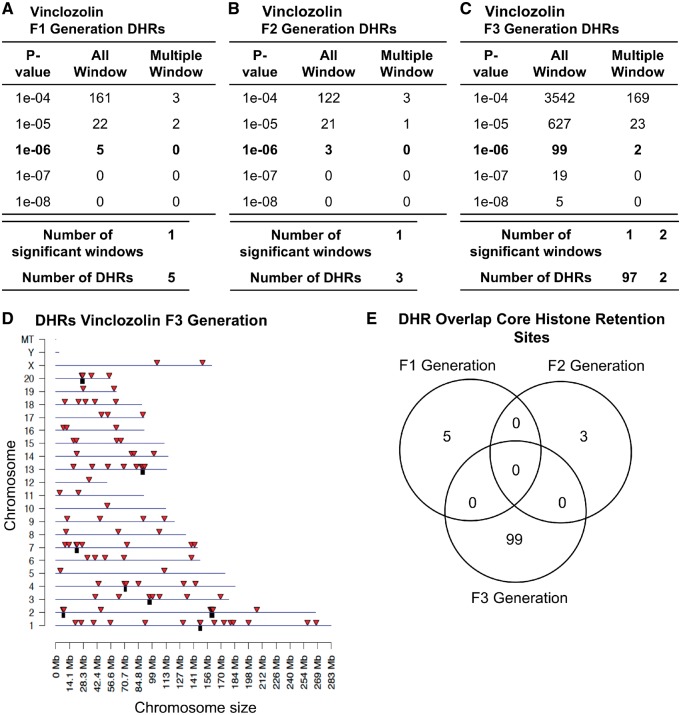
A core of histone retention sites have been previously shown in control and vinclozolin lineage rat sperm that are not altered following exposure, but the F3 generation vinclozolin lineage sperm had an increase in the number of histone retention sites [52]. This study examined the sites (DHRs for the F1, F2, and F3 generation control versus vinclozolin lineage sperm with a procedure similar to the DMR analysis. Interestingly, the F1 and F2 generations had negligible DHRs showing no major difference between control and vinclozolin lineages (Fig. 8A and B). However, the F3 generation comparison of the control versus vinclozolin lineage sperm identified an increased number of DHRs (Fig. 8C). The chromosomal locations of these DHRs are represented in Fig. 8D. An overlap analysis of these DHRs between F1, F2, and F3 revealed that each of them was unique (Fig. 8E). DHRs were induced in the F3 generation lineage sperm thus displaying a transgenerational effect whereas a direct exposure did not trigger major alterations. The DHRs are listed with identification, location, size and associated genes in the Supplementary Tables S11–S13 for the F1, F2, and F3 generations, respectively.
Figure 8:
vinclozolin DHRs. (A) F1 generation, (B) F2 generation, and (C) F3 generation. The number of DHRs found using different P-value cutoff thresholds. The All Window column shows all DHRs. The Multiple Window column shows the number of DHRs containing at least two significant windows. (D) Chromosomal locations of the DHRs in the F3 generation on individual chromosomes are indicated by red arrowheads and DHR clusters with black boxes. All DHRs at a P-value threshold of 1e-06 are shown. (E) The DHR overlap (P < 1e-06) for the F1, F2, and F3 generation sperm
Epimutation Gene Associations
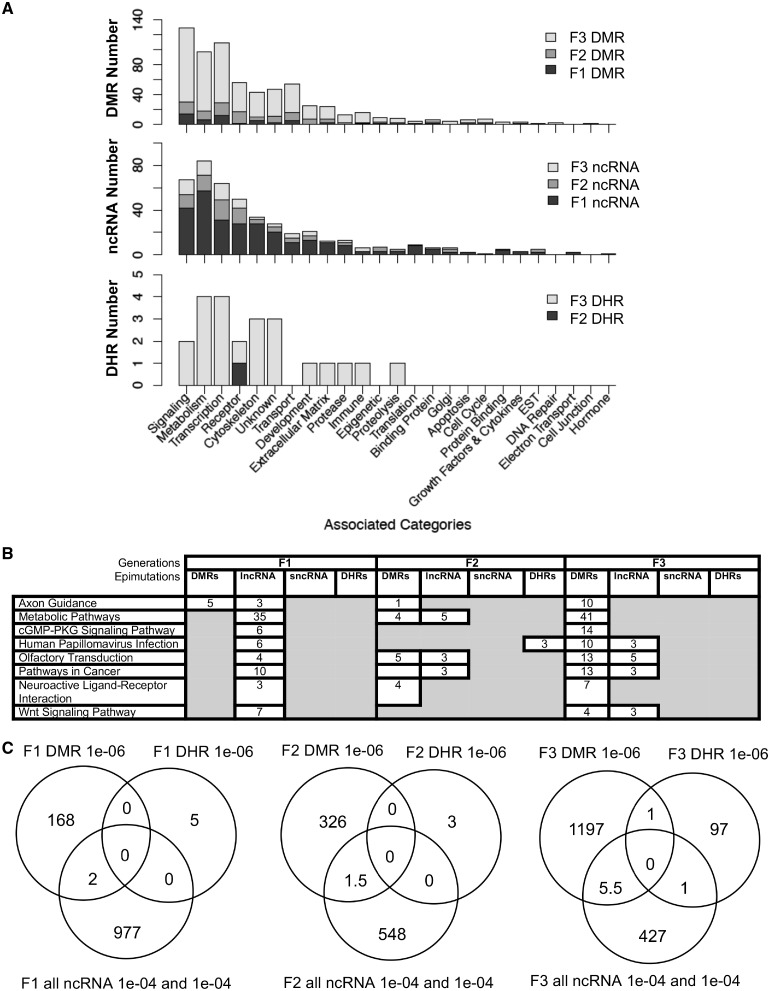
The Supplementary Tables S1–S9 provide the lists of DMRs, ncRNAs, and DHRs for all the epigenetic alterations (i.e. epimutations) identified. The known gene associations for these epimutations were determined using genomic location and are also listed. All the associated genes were classified by relevant functions and the functional categories are presented for each generation in Fig. 9A. For this analysis, sncRNA and lncRNA were combined. The top ten gene categories containing multiple genes for F1, F2, and F3 generations are presented for DMRs, ncRNA, and DHRs separately. Epimutations were found predominantly in the metabolism, signaling, and transcription categories. The predominant gene categories for the F1 generation were mostly found with the lncRNA (Fig. 9A). The predominant gene categories for the F3 generation were similar as the F1 generation but found in the DMRs. The ratio of epimutations between the generations was highest for DMRs in the F3 generation, highest for ncRNA in the F1 generation, and negligible DHRs are present in the F1 and F2 generations, but present in the F3 generation (Fig. 10A).
Figure 9:
epimutation overlaps and gene associations. (A) The number of DMR, ncRNA, and DHR epimutation-associated genes in each gene category for the F1, F2, and F3 generations. (B) Gene pathways with epimutation-associated genes for the DMRs, DHRs and ncRNAs in the F1, F2 and F3 generations. The pathways with greater than three associated genes for each epimutation type are listed that were in general common between generations. (C) Overlaps of the F1, F2, and F3 generation epimutations between the DMRs, DHRs, and ncRNAs
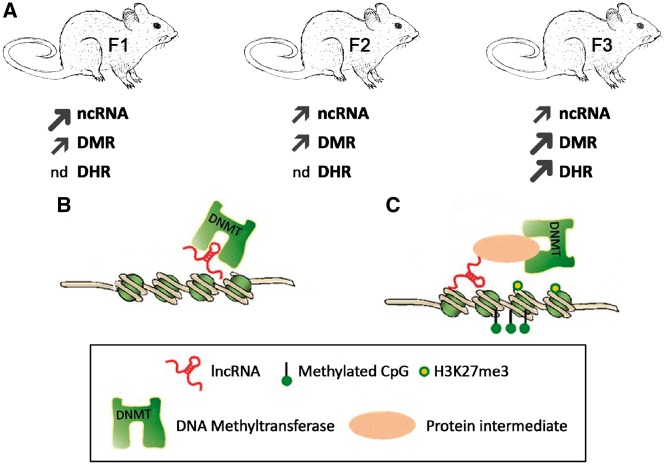
Figure 10:
The regulatory mechanisms of DNA methylation by lncRNAs. (A) Summary of the different epimutations in the F1, F2, and F3 generation exposed rat sperm with larger/thicker arrows reflecting larger numbers of epimutations than thinner arrows and nd (negligible detected). (B) lncRNA/DNMT interaction prevents locus-specific DNA methylation locally in cis.(C) lncRNA interacts with DNMT1 (methyltransferase 1) indirectly through a protein intermediate. Figure modified from [43]
The integration of the different epigenetic alterations is suggested in Fig. 9A and B with epimutation-associated genes in specific pathways and gene categories. The majority of the gene categories and pathways observed in the F1 generation was associated with the lncRNAs. The same phenomenon was found in the F3 generation but associated with the DMRs (Fig. 9A and B). Two pathways, axon guidance and pathways in cancer, were selected to investigate the potential overlap with genes involving the lncRNA- and DMR-associated genes (Supplementary Figs. S1 and S2). For the F1 and F3 generation lncRNA and DMRs the pathways were represented by both types of epimutations at independent genes, and few had the same gene involving a lncRNA in the F1 generation and DMR in the F3 generation. In both generations the pathways were affected, but with different types of epimutations.
The Venn diagrams of DMRs, DHRs, and ncRNA suggest negligible overlap between the different types of epimutations within the same generation comparison (Fig. 9C). So, the different epigenetic alterations have different genomic locations and minimal overlap between each other in the same generation. The total list of associated genes with the DMRs, ncRNAs, and DHRs are presented in the Supplementary Tables S1–S13.
Discussion
This study was designed to help understand how three different epigenetic processes in sperm are correlated with vinclozolin-induced epigenetic transgenerational inheritance of disease. A number of transgenerational diseases including testis disease, ovarian disease, kidney pathology, prostate abnormalities, and anxiety behaviors have been shown to be induced by vinclozolin [37, 56]. Analyzing in parallel DNA methylation, ncRNA content, and histone retention in the same sperm samples from F1, F2, and F3 generation control versus vinclozolin lineage rats provides a broader comprehensive analysis of epigenetic alterations linked with environmentally induced germline epimutations. Classic toxicology studies for risk assessment investigate the direct exposure of the individual only, disregarding the impacts on future generations. However, transgenerational studies on environmental toxicants have shown that even if a compound seems safe or has negligible risk from direct exposure mechanisms, it can affect the offspring of future generations [28]. Treatment of a gestating F0 generation female exposes the F1 generation fetus and the germline that will generate the F2 generation (Fig. 1A). Thus, the F1 generation represents a direct exposure, the F2 generation is a mix of direct and generational exposure, and the F3 generation and subsequent generations are considered purely transgenerational. This study investigates and compares the sperm epimutations in each of these generations.
Alterations in DNA methylation patterns were the first epigenetic process shown in F3 generation sperm [16, 57]. Therefore, many studies have focused on DNA methylation alterations in the germline and since then numerous chemicals, nutrition or stress exposures have been shown to induced transgenerational phenotypes [21]. A previous study identified a distinct set of DMRs in vinclozolin lineage rat sperm DNA methylation between the F1 and the F3 generation [58]. Most DMRs identified in this study are unique between the F1, F2, and F3 generations. This shows that the direct exposure F1 generation sperm and transgenerational F3 generation sperm DMRs are specific to each generation. Interestingly, we observed that the F2 generation seems to have fewer epimutations and has minimal overlap with the F1 and F3 generations. A small set of overlapping DMRs between the F2 and F3 generation sperm are presented in the Supplementary Fig. S4. These represent the potential transgenerational DMR that appear first in the F2 generation. In addition, no known imprinted genes correlated to the DMRs identified. As classic imprinted genes are resistant to environmental alterations in regards to DNA methylation [59], they do not appear in the differential DNA methylation regions observed.
Environmental conditions such as traumatic stress in early life in mice altered miRNA expression and behavioral and metabolic responses in the progeny. Several miRNAs were affected in the serum and brain of the traumatized animals and their progeny when adult. Sperm of traumatized males was also affected. Injection of sperm ncRNAs from these males into fertilized wild-type oocytes reproduced the behavioral and metabolic changes in the resulting offspring. These results suggest that sncRNAs are sensitive to environmental factors in early life and contribute to the inheritance of trauma-induced phenotypes across generations [39]. Vinclozolin-induced epigenetic transgenerational inheritance of disease has also been shown to involve alterations in sperm sncRNA [40]. Transgenerational lncRNA and sncRNA have been proposed to have diverse functions [5]. The sncRNA often targets specific genes through alterations in mRNA metabolism or binding protein [60], whereas the lncRNA regulates the transcriptional machinery and targets multiple distal genes. In this study, the number of lncRNA was much higher than the number of sncRNA. The overlap between each generation was very low or nonexistent. The piRNA was the most common class of sncRNA for the F1 and F3 generations, stRNAs (small tRNA fragments) were the most common for the F2 generation (Fig. 5D). Our results show that concurrent DNA methylation and ncRNA alterations in sperm may be associated with the epigenetic transgenerational inheritance phenomenon.
Sperm chromatin not only has a unique structure to condense and protect the paternal DNA, but it also provides epigenetic information supporting early embryonic development. The histones and other chromatin proteins remaining are located in structurally and transcriptionally relevant positions in the genome and carry diverse posttranslational modifications relevant to the control of embryonic gene expression. In Caenorhabditiselegans and Drosophila, inheritance of phenotypes has been associated with histone modifications [13, 61] which has not been reported in mammals. Histone modifications and retentions have been shown to be implicated in male fertility [53, 62]. However, until recently the role of histone retention has not been implicated in transgenerational inheritance studies [52]. At the end of spermatogenesis the histones are removed in many species and the DNA is condensed by protamines forming highly dense nucleoprotamine complexes [63]. Generally, 5–10% of histones are retained and are thought to play a role in the zygotic and early embryonic gene expression [53, 64–67]. In this study, the hypothesis that vinclozolin could alter histone retentions in a transgenerational manner was investigated. In a previous study, control sperm rats were shown to have a highly reproducible core set of histones [52]. This core set of histones generally remained the same in the control animals and the animals exposed to vinclozolin between the F1, F2, and F3 generations. Because of its high reproducibility and consistency between each generation, and despite the treatment with DDT or vinclozolin, these core histone sites are highly conserved and speculated to be critical for early development [52]. The analysis of DHRs revealed that the F1 and the F2 generation control versus vinclozolin lineage sperm had negligible DHRs (Fig. 8). This observation suggests that the direct vinclozolin exposure does not alter histone retention or trigger any changes. However, the F3 generation control versus vinclozolin lineage sperm DHRs increased considerably in comparison to the F1 and F2 generation DHRs. In addition to the DMRs and ncRNAs, alterations in histone retention sites are also associated with the vinclozolin-induced transgenerational F3 generation sperm. Unlike the ncRNAs and DMRs, the F1 and F2 generation histone retention sites were not affected in the exposed animals. The DHR epimutations are all present in the transgenerational F3 generation sperm and therefore might also play a role in mediating the epigenetic transgenerational inheritance phenomenon.
In order to investigate the integration of DMRs, ncRNA, and DHR, the overlap of the different genomic sites between the different epimutations was analyzed. The chromosomal locations of the DMR, ncRNA, and DHR epimutations were distinct with a low number of overlaps. These transgenerational epimutations have chromosomal localizations that are distinct between each other, but generally they are present in regions with similar genomic features (e.g. CpG desert and size). The genome evolving over time develops regions that have very low cytocine (C) levels due to the ability of DNA methylation of the CpG site to promote susceptibility for a C to T conversion point mutation. Over 90% of the known point mutations in the genome are C to T conversions. Therefore, if a cluster of CpG sites with DNA methylation are conserved evolutionarily in these deserts of CpG, they likely are functionally important [55].
The gene associations of the transgenerational epimutations identified two similar categories of genes for the ncRNA and the DMRs involving metabolism, signaling, and transcription. The analysis comparing the F1, F2, and F3 generation epimutation-associated genes showed minimal overlap between the different types of epimutations (Fig. 9C). Each one of them (DMRs, ncRNA, DHRs) appears to associate with different pathways and may be integrated on a functional level. Although the epimutations in sperm can have associated genes, the sperm are transcriptionally silenced due to the compaction of DNA with protamines and complete loss of transcriptional machinery. The epimutations observed will primarily function in the early embryo to impact gene expression and epigenetic programming to alter the embryonic stem cell epigenome and transcriptome. Therefore all somatic cells derived from this stem cell population will have altered epigenomes and transcriptomes, and those sensitive to that will have a susceptibility to develop disease later in life. Therefore, the epimutation gene associations may be important for later somatic cell development and differentiation, but not relevant to sperm function.
Interestingly, the comparisons in the F1, F2, and F3 generations provide insights into the direct exposure effects to vinclozolin and transgenerational impacts of the exposure. The F1 and F2 generations are exposed directly to vinclozolin (Fig. 1B), whereas the F3 generation is the first transgenerational generation. The effects observed in the F1 and F2 generations were different from the ones observed in the F3 generation. Predominantly, ncRNAs were altered by the direct exposure to vinclozolin in the F1 generation whereas the DNA methylation and histone retention sites were more predominant in the F3 generation. Histone retention did not seem to be affected by a direct exposure of vinclozolin. Therefore, the number of ncRNA was dramatically altered in the F1 generation, but to a lesser extent in the F2 and F3 generations. The altered ncRNA was also distinct between each generation (Fig. 9B). The transgenerational F3 generation alterations in DNA methylation, ncRNAs, and histone retention are distinct from the direct exposure in the F1 and F2 generations.
The analysis of the different epimutations associated gene pathways revealed that the F1 generation ncRNA seemed to be targeted by the direct vinclozolin exposure. However, these ncRNA epimutations (predominantly lncRNA) associated gene pathways disappeared in the F3 generation and similar associated gene pathways were found within the DMRs. The hypothesis is that the ncRNA could be more sensitive to the direct exposure to vinclozolin, then mediate the formation of the other epimutations in the next generation with DNA methylation and histone retention becoming permanent and mediating the epigenetic transgenerational inheritance in the future generations. An emerging hypothesis is the capacity of ncRNAs to modulate gene expression and many of them have been identified to participate in epigenetic control by interacting with various types of proteins involved in histone modification or chromatin remodeling [43]. Although these ncRNA regulations have not been observed in transgenerational animals, lncRNAs appear to be able to modulate DNA and/or histone methylation [43, 68]. Therefore, ncRNA may be the epimutation that helps mediate the formation and actions of the other types of epimutations. Based on Zhao et al., two hypotheses are proposed to explain how lncRNA can interact directly or indirectly with diverse DNMT members (Fig. 10B) [43]. Through recruitment or eviction the lncRNA can either promote or repress DNA methylation in cis or in trans. The dynamic of the lncRNA repertoire and the ncRNA plasticity to interact with DNA, ncRNA, and protein makes lncRNA a potential mediator to regulate local and sequence-specific DNA methylation or demethylation. Although lncRNA are not as well annotated or functionally linked as sncRNA, as seen in Supplementary Tables S5–S7 nearly a third of those observed have associated genes. These modifications can result in global changes in DNA methylation profile, thus changing how cells can respond to diverse stimuli (Fig. 10). Even though it has not been shown yet, an interplay between lncRNA and histones is likely. Clearly an integration and interplay between the different epigenetic processes will be critical in the epigenetic transgenerational phenomenon and regulation of genome activity.
Studies are in progress to explore the integration of these transgenerational epimutations. One of the possibilities is that the transgenerational epimutations developed in the F1 and F2 generations alter the developmental epigenetic programming of the primordial germ cells (PGCs) which are the stem cells for the germline. This epigenetic alteration in the PGCs development and become permanently established and is transmitted through the germline to the next generation. This germline reprogramming will promote the epigenetic transgenerational inheritance phenomenon. Epigenetic alterations in the transgenerational PGCs have already been observed [32, 69], but further studies are required to better understand the PGCs’ transgenerational mechanisms. This study provides new insights into the distinct epigenetic alterations between a direct exposure phenomenon and the transgenerational impact of the exposure. Additional support that vinclozolin is concurrently altering these different types of epimutations comes from similar observations of concurrent alterations following DDT exposure [7].
Further investigation is needed to better understand the integrated mechanisms of the unique transgenerational F3 generation germline epigenetic alterations. It appears that the phenomenon is more complex than just a direct exposure triggering the formation of epimutations that are then simply maintained in the subsequent generations. This study finds concurrent alterations of DNA methylation, ncRNA, and histone retention in the sperm. Observations indicate that all three types of epimutations are involved in the epigenetic transgenerational inheritance phenomenon. The ncRNA have already been shown to act on DNA methylation or histone methylation [43, 68]; so it is not surprising to discover that DNA methylation, ncRNA, and histone retention appear to be integrated to facilitate the epigenetic transgenerational inheritance. The potential impact of these epimutations on disease etiology and other areas in biology requires further investigation and clarification of the mechanisms involved.
Methods
Animal Studies and Breeding
Female and male rats of an outbred strain Hsd: Sprague Dawley SD®™ (Harlan) at about 70 and 100 days of age were fed ad lib with a standard rat diet and received ad lib tap water for drinking. To obtain time-pregnant females, the female rats in proestrus were pair-mated with male rats. The sperm-positive (Day 0) rats were monitored for diestrus and body weight. On Days 8 through 14 of gestation [36], the females received daily intraperitoneal injections of vinclozolin (100 mg/kg BW/day) or DMSO. The vinclozolin was obtained from Chem Service Inc. (West Chester, PA) and was injected in a 20-μl DMSO vehicle as previously described [70]. Treatment lineages are designated “control” or “vinclozolin” lineages. The treated gestating female rats were designated as the F0 generation. The offspring of the F0 generation rats were the F1 generation. Non-littermate females and males aged 70–90 days from the F1 generation of control or vinclozolin lineages were bred to obtain F2 generation offspring. The F2 generation rats were bred to obtain F3 generation offspring. Individuals were maintained for 12 months and euthanized for sperm collection. The F1–F3 generation offspring were not themselves treated directly with vinclozolin. The control and vinclozolin lineages were housed in the same room and racks with lighting, food, and water as previously described [54, 70, 71]. All experimental protocols for the procedures with rats were pre-approved by the Washington State University Animal Care and Use Committee (IACUC approval # 02568-49).
Testis sections from the males were obtained, fixed for histology, and examined by Terminal Deoxynucleotidyl Transferase-mediated dUTP Nick End Labeling (TUNEL) assay (in situ cell death detection kit, Fluorescein, Sigma, St. Louis, MO) to assess spermatogenic cell apoptosis.
Epididymal Sperm Collection and DNA
The epididymis was dissected free of connective tissue, a small cut made to the cauda and tissue placed in 5 ml of 1× PBS solution for up to 24 h at 4°C. The epididymal tissue was minced and the released sperm was centrifuged at 6000 g, then the supernant removed, and the pellet resuspended in NIM buffer, to be stored at −80°C until further use. One hundred microliters of sperm suspension was sonicated to destroy somatic cells and tissue, spun down at 6000 g, the sperm pellet washed with 1× PBS once, and then combined with 820 µl DNA extraction buffer and 80 µl 0.1 M DTT. The sample was incubated at 65°C for 15 min. Following this incubation 80 µl proteinase K (20 mg/ml) was added and the sample incubated at 55°C for at least 2 h under constant rotation. Then 300 µl of protein precipitation solution (Promega, A7953) was added, the sample mixed thoroughly and incubated for 15 min on ice. The sample was centrifuged at 12 500 g for 30 min at 4°C. One milliliter of the supernatant was transferred to a 2-ml tube and 2 µl of glycoblue and 1 ml of cold 100% isopropanol were added. The sample was mixed well by inverting the tube several times, then left in −20°C freezer for at least 1 h. After precipitation the sample was centrifuged at 12 500 g for 20 min at 4°C. The supernatant was taken off and discarded without disturbing the (blue) pellet. The pellet was washed with 70% cold ethanol by adding 500 µl of 70% ethanol to the pellet and returning the tube to the freezer for 20 min. After the incubation the tube was centrifuged for 10 min at 4°C at 12 500 g and the supernatant discarded. The tube was spun again briefly to collect residual ethanol to the bottom of the tube and then as much liquid as possible was removed with gel loading tip. Pellet was air-dried at RT until it looked dry (about 5 min). Pellet was then resuspended in 100 µl of nuclease free water.
Equal amounts of DNA or ncRNA from each individuals sperm from 3–4 different individuals for the F1 and F2 generations and 4–6 different individuals for the F3 generation were pooled and three different pools generation for each F1, F2, and F3 generation control and vinclozolin lineage males. Therefore, each pool had different individuals and there were three pools for each group. A summary of the pools for the DNA and ncRNA pools is shown in Supplementary Table S14. Two analyses of the same pool were taken, one for DNA and one for ncRNA. Therefore, between 10 and 17 individuals were present in the three pools for the subsequent analysis.
RNA Isolation
The F1–F3 generation vinclozolin and control lineage male epididymal sperm were collected and processed as previously described and stored at −80°C until use [57]. The total RNA (messenger RNA, lncRNA, ribosomal RNA, sncRNA) was isolated using the mirVana miRNA Isolation Kit (Life Technologies) following the manufacturer’s instructions with modifications at the lysis stage. In brief, after addition of lysis buffer, the sperm pellets were manually homogenized, followed by a 20-min incubation at 65°C. Samples were then placed on ice, and the default protocol was resumed. For quality control, RNA integrity numbers (RIN) were obtained by RNA 6000 Pico chips run on an Agilent 2100 Bioanalyzer (Agilent). A RIN of 2–4 indicates good sperm RNA quality. Concentration was determined using the Qubit RNA HS Assay Kit (ThermoFisher). Biological replicates of sperm were pooled by equal ncRNA content and were concentrated using Agencourt AMPure XP beads (Beckman Coulter). Some pools had underrepresented replicates due to low concentration. In this case, the maximum RNA content from the replicate was used in the pool and the pool was concentrated. The pools and samples that were underrepresented are as follows: F1 control pool 3, samples 1 and 2; F2 control pool 2, sample 3; and F2 vinclozolin pool 2, sample 3. Equal amounts of each pool were used in the final analysis.
Methylated DNA Immunoprecipitation
MeDIP with genomic DNA was performed as follows: rat sperm DNA pools were generated using the appropriate amount of genomic DNA from each individual for three pools each of control and vinclozolin lineage animals. Genomic DNA was sonicated using the Covaris M220 the following way: the pooled genomic DNA was diluted to 130 μl with TE buffer into the appropriate Covaris tube. Covaris was set to 300-bp program and the program was run for each tube in the experiment. Each sonicated DNA of 10 μl was run on 1.5% agarose gel to verify fragment size. The sonicated DNA was transferred from the Covaris tube to a 1.7-ml microfuge tube and the volume measured. The sonicated DNA was then diluted with TE buffer (10 mM Tris HCl, pH7.5; 1 mM EDTA) to 400 μl, heat-denatured for 10 min at 95°C, then immediately cooled on ice for 10 min. Then 100 μl of 5× IP buffer and 5 μg of antibody (monoclonal mouse anti-5-methyl cytidine; Diagenode #C15200006) were added to the denatured sonicated DNA. The DNA–antibody mixture was incubated overnight on a rotator at 4°C.
The following day magnetic beads (Dynabeads M-280 Sheep anti-Mouse IgG; 11201 D) were pre-washed as follows: the beads were resuspended in the vial, then the appropriate volume (50 μl per sample) was transferred to a microfuge tube. The same volume of Washing Buffer (at least 1 ml PBS with 0.1% BSA and 2 mM EDTA) was added and the bead sample was resuspended. Tube was then placed into a magnetic rack for 1–2 min and the supernatant was discarded. The tube was removed from the magnetic rack and the beads were washed once. The washed beads were resuspended in the same volume of 1× IP buffer (50 mM sodium phosphate ph7.0, 700 mM NaCl, 0.25% TritonX-100) as the initial volume of beads. Fifty microliters of beads was added to the 500 μl of DNA–antibody mixture from the overnight incubation, then incubated for 2 h on a rotator at 4°C.
After the incubation the bead–antibody–DNA complex was washed three times with 1× IP buffer as follows: the tube was placed into magnetic rack for 1–2 min and the supernatant discarded, then washed with 1× IP buffer three times. The washed bead–DNA solution is then resuspended in 250 μl digestion buffer with 3.5 μl Proteinase K (20 mg/ml). The sample was then incubated for 2–3 h on a rotator at 55°C and then 250 μl of buffered phenol–chloroform–isoamylalcohol solution was added to the supe and the tube vortexed for 30 s then centrifuged at 12 500 g for 5 min at room temperature. The aqueous supernatant was carefully removed and transferred to a fresh microfuge tube. Then 250 μl chloroform was added to the supernatant from the previous step, vortexed for 30 s and centrifuged at 12 500 g for 5 min at room temperature. The aqueous supernatant was removed and transferred to a fresh microfuge tube. To the supernatant 2 μl of glycoblue (20 mg/ml), 20 μl of 5 M NaCl and 500 μl ethanol were added and mixed well, then precipitated in −20°C freezer for 1 h to overnight.
The precipitate was centrifuged at 12 500 g for 20 min at 4°C and the supernatant removed, while not disturbing the pellet. The pellet was washed with 500 μl cold 70% ethanol in −20°C freezer for 15 min, then centrifuged again at 12 500 g for 5 min at 4°C and the supernatant was discarded. The tube was spun again briefly to collect residual ethanol to the bottom of the tube and as much liquid as possible was removed with gel loading tip. Pellet was air-dried at RT until it looked dry (about 5 min) then resuspended in 20 μl H2O or TE. DNA concentration was measured in Qubit (Life Technologies) with ssDNA kit (Molecular Probes Q10212).
ncRNA Sequencing Analysis
LRNA and noncoding RNA were constructed using total RNA with the KAPA Stranded RNA-Seq Library Preparation kit with RiboErase, according to the manufacturer’s instructions, with some modifications. The adaptor and barcodes used were from NEBNext Multiplex Oligos for Illumina. Prior to PCR amplification, libraries were incubated at 37°C for 15 min with the USER enzyme (NEB). PCR cycle number was determined using qPCR with the KAPA RealTime Library Amplification kit before final amplification. Size selection (300–700 bp) was performed using Agencourt AMPure XP beads (Beckman Coulter). Quality control was performed using Agilent DNA High Sensitivity chips (Agilent) and Qubit dsDNA high sensitivity assay (ThermoFisher). Libraries were pooled and loaded onto an Illumina NextSeq High Output v2 1x75 chip, and sequenced on an Illumina NextSeq 500 sequencer. Bioinformatics analysis was used to separate mRNA libraries from ncRNA libraries (see ncRNA Bioinformatics section).
Prior to sncRNA library preparation, total sperm RNA samples were enriched for sncRNAs using the miRNA enrichment protocol (Beckman Coulter), and libraries were constructed using the NEBNext Multiplex Small RNA Library Prep Set for Illumina, and barcoded with NEBNext Multiplex Oligos for Illumina. Size selection (135–170 bp) was performed using the Pippin Prep (Sage Science). Quality control was performed using Agilent DNA High Sensitivity chips (Agilent) and Qubit dsDNA high sensitivity assay (ThermoFisher). Libraries were pooled and loaded onto an Illumina NextSeq High Output v2 1x75 chip, and sequenced on an Illumina NextSeq 500 sequencer.
Histone Chromatin Immunoprecipitation ChIP-Seq
Histone chromatin immunoprecipitation with genomic DNA was performed as follows (adapted from [72]): rat sperm pools were generated using a total of 8 million sperm per pool for 3 pools of control and vinclozolin lineage animals. The control pools contained equal amounts of sperm for each of 3–4 individuals for a total of n = 11 rats and the vinclozolin pools contained equal amounts of sperm for each of 3 individuals for a total of n = 9 rats per exposure group. To remove any somatic cell contamination, sperm samples from each animal were sonicated 10 s using a Sonic Dismembrator Model 300 (Thermo Scientific Fisher, USA) then centrifuged 4000 g for 5 min at 4°C. The supernatant was discarded and the pellet was resuspended and counted individually on a Neubauer counting chamber (Propper Manufacturing Co., Inc., New York, USA) prior to pooling. The sperm pools were reconstituted up to 1 ml with 1× PBS. To reduce disulfide bonds, 50 μl of 1 M DTT was added to each pool and the pools were then incubated for 2 h at room temperature under constant rotation. To quench any residual DTT in the reaction, 120 μl of fresh 1 M NEM (N-ethylmaleimide, Thermo Scientific, Rockford, USA) was then added and the samples were incubated for 30 min at room temperature under constant rotation. The sperm cells were pelleted at 2000 g for 5 min at room temperature and the supernatant was discarded. Pellets were resuspended in 1× PBS and then spun again at 2000 g for 5 min at room temperature. The supernatant was discarded. The sperm cell DNA was divided into aliquots of 4 μg of DNA and was then suspended in “complete buffer” [final concentration: 15 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 0.1 mM EGTA; all filtered through a 0.22-μm filter and stored at room temperature; 0.5% tergitol (vol/vol) and 1% DOC (wt/vol), sodium deoxycholate]. A 130 μl of this complete buffer was added to each DNA aliquot. The samples were mixed and incubated for 20 min on ice. These aliquots were sonicated using the Covaris M220 the following way: 4 μg of genomic DNA was resuspended in 130 μl of complete buffer supplemented with tergitol 0.5% and DOC 1%. Covaris was set to a 10-min “Chromatin shearing” program and the program was run for each tube in the experiment.
For each sample 10 μl was run on a 1.5% agarose gel to verify fragment size. Aliquoted samples were pooled back together and centrifuged at 12 500 g for 10 min at room temperature. The supernatant was transferred to a fresh microfuge tube. Sixty-five microliters of protease inhibitor cocktail (1 tablet dissolved in 500 μl, 20× concentrated) (Roche, cat. no. 11 873 580 001) was added to each sample as well as 3 μl of antibody (monoclonal rabbit antihistone H3, Millipore 05-928). The DNA–antibody mixture was incubated overnight on a rotator at 4°C. The following day, magnetic beads (ChIP-Grade protein G magnetic beads, Cell Signaling 9006) were pre-washed as follows: the beads were resuspended in the vial, then 30 μl per sample was transferred to a microfuge tube. The same volume of Washing Buffer (at least 1 ml) was added and the bead sample was resuspended. Tube was then placed into a magnetic rack for 1–2 min and the supernatant was discarded. The tube was removed from the magnetic rack and the beads were washed once. The washed beads were resuspended in the same volume of 1× IP buffer as the initial volume of beads. Thirty microliters of beads was added to each DNA–antibody mixture from the overnight incubation, then incubated for 2 h on a rotator at 4°C. After the incubation, the bead–antibody–DNA complex was washed three times with 1× IP buffer as follows: the tube was placed into a magnetic rack for 1–2 min and the supernatant was discarded, then washed with 1× IP buffer 3 times. The washed bead–antibody–DNA solution was then resuspended in 300 μl of digestion buffer and 3 μl proteinase K (20 mg/ml). The sample was incubated for 3 h on a rotator at 56°C. After incubation the samples were extracted with phenol–chloroform–isoamylalcohol and precipitated with 2 μl of glycoblue (20 mg/ml), a one-tenth volume of 3 M sodium acetate and two volumes of ethanol overnight at −20°C.
The precipitate was centrifuged at 14 000 g for 30 min at 4°C and the supernatant was removed, while not disturbing the pellet. The pellet was washed with 500 μl cold 70% ethanol, then centrifuged again at 14 000 g for 10 min at 4°C and the supernatant was discarded. The tube was spun briefly to collect residual ethanol and as much liquid as possible was removed with a gel loading tip. Pellet was air-dried at RT until it looked dry (about 5 min) then resuspended in 20 μl H20. DNA concentration was measured in the Qubit (Life Technologies) with the BR dsDNA kit (Molecular Probes Q32853).
MeDIP-Seq Analysis
The MeDIP pools were used to create libraries for NGS using the NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, San Diego, CA) starting at step 1.4 of the manufacturer’s protocol to generate double-stranded DNA. After this step the manufacturer’s protocol was followed. Each pool received a separate index primer. NGS was performed at WSU Spokane Genomics Core using the Illumina HiSeq 2500 with a PE50 application, with a read size of ∼50 bp and ∼100 million reads per pool. Two to three libraries were run in one lane.
Histone ChIP-Seq Analysis
The ChIP pools were used to create libraries for NGS using the NEBNext® UltraTM II DNA Library Prep Kit for Illumina® (NEB, San Diego, CA). The manufacturer’s protocol was followed. Each pool received a separate index primer. NGS was performed at WSU Spokane Genomics Core using Ilumina HiSeq 2500 with a PE50 application, with a read size of ∼50 bp and ∼35 million reads per pool. Six libraries were run in one lane.
ncRNA Bioinformatics
The sncRNA-Seq data were annotated as follows: reads shorter than 15 nt were discarded by Cutadapt [73]. The remaining reads were matched to known rat sncRNA, consisting of mature miRNA (miRBase, release 21), tRNA (Genomic tRNA Database, rn5), piRNA (piRNABank), rRNA (ENSEMBL, release 76), and mitochondrial RNA (ENSEMBL, release 76) using AASRA pipeline with default parameters [74]. Read counts generated by AASRA were statistically normalized by DESeq2 [75].
The lncRNA data were annotated as follows: trimmomatic was used to remove adaptor sequences and the low-quality reads from the RNA sequencing data [76]. To identify all the transcripts, we used Tophat2 and Cufflinks to assemble the sequencing reads based on the Ensembl_Rnor_6.0 [77]. The differential expression analyses were performed by Cuffdiff. The coding and the noncoding genes were primarily annotated through rat CDS data ensembl_Rnor_6.0. The non-annotated genes were extracted through our in-house script, then analyzed by CPAT, indicating the true ncRNAs [78].
Statistics and Bioinformatics
For the DMR and DHR analyses, the basic read quality was verified using summaries produced by the FastQC program (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The raw reads were trimmed and filtered using Trimmomatic. The reads for each MeDIP sample were mapped to the Rnor 6.0 rat genome using Bowtie2 [76] with default parameter options. The mapped read files were then converted to sorted BAM files using SAMtools [79]. To identify DMRs and DHRs, the reference genome was broken into 100-bp windows. The MEDIPS R package [80] was used to calculate differential coverage between control and exposure sample groups. The edgeR P-value [81] was used to determine the relative difference between the two groups for each genomic window. Windows with an edgeR P-value <10−6 were considered DMRs or DHRs. The DMR or DHR edges were extended until no genomic window with a P-value <0.1 remained within 1000 bp of the DMR or DHR. CpG density and other information were then calculated for the DMR based on the reference genome.
DMRs, DHRs, and ncRNA were annotated using the biomaRt R package [82] to access the Ensembl database [83]. The genes that fell within 10 kb of the DMR, DHR, or ncRNA edges were then input into the KEGG pathway search [84, 85] to identify associated pathways. The associated genes were then sorted into functional groups by consulting information provided by the DAVID [86], Panther [87], and Uniprot databases incorporated into an internal curated database (www.skinner.wsu.edu under genomic data).
Supplementary Material
Acknowledgments
We acknowledge Ms Deepika Kubsad and Ms Jayleana Barton for technical assistance and Ms Heather Johnson for assistance in preparation of the manuscript. We thank the Genomics Core Laboratories at WSU and UNR. This study was supported by a John Templeton Foundation (50183) grant to M.K.S. and NIH (ES012974) grant to M.K.S. and NIH grants (HD085506 and GM110767) to W.Y.
Supplementary data
Supplementary data are available at EnvEpig online.
Conflict of interest statement. None declared.
References
- 1. Holliday R. The inheritance of epigenetic defects. Science 1987; 238:163–70. [DOI] [PubMed] [Google Scholar]
- 2. Cosgrove MS, Wolberger C.. How does the histone code work? Biochem Cell Biol 2005; 83:468–76. [DOI] [PubMed] [Google Scholar]
- 3. Gray SG, Teh BT.. Histone acetylation/deacetylation and cancer: an “open” and “shut” case? Curr Mol Med 2001; 1:401–29. [DOI] [PubMed] [Google Scholar]
- 4. Cao J. The functional role of long non-coding RNAs and epigenetics. Biol Proced Online 2014; 16:11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yan W. Potential roles of noncoding RNAs in environmental epigenetic transgenerational inheritance. Mol Cell Endocrinol 2014; 398:24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mohammad F, Mondal T, Kanduri C.. Epigenetics of imprinted long noncoding RNAs. Epigenetics 2009; 4:277–86. [PubMed] [Google Scholar]
- 7. Skinner Mk, Ben Maamar M, Sadler-Riggleman I, Beck D, Nilsson E, McBirney M, Klukovich R, Xie Y, Tang C, Yan W, et al. Alterations in sperm DNA methylation, non-coding RNA and histone retention associate with DDT induced epigenetic transgenerational inheritance of disease. Epigenetics Chromatin 2018; 11:8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burdge GC, Lillycrop KA, Jackson AA.. Nutrition in early life, and risk of cancer and metabolic disease: alternative endings in an epigenetic tale? Br J Nutr 2009; 101:619–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jirtle RL, Skinner MK.. Environmental epigenomics and disease susceptibility. Nat Rev Genet 2007; 8:253–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tang WY, Ho SM.. Epigenetic reprogramming and imprinting in origins of disease. Rev Endocr Metab Disord 2007; 8:173–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Quadrana L, Colot V.. Plant transgenerational epigenetics. Annu Rev Genet 2016; 50:467–91. [DOI] [PubMed] [Google Scholar]
- 12. Brookheart RT, Duncan JG.. Drosophila melanogaster: an emerging model of transgenerational effects of maternal obesity. Mol Cell Endocrinol 2016;435:20–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kelly WG. Transgenerational epigenetics in the germline cycle of Caenorhabditis elegans. Epigenetics Chromatin 2014;7:6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carvan MJ, Kalluvila TA, Klingler RH, Larson JK, Pickens M, Mora-Zamorano FX, Connaughton VP, Sadler-Riggleman I, Beck D, Skinner MK, et al. Mercury-induced epigenetic transgenerational inheritance of abnormal neurobehavior is correlated with sperm epimutations in zebrafish. PLoS One 2017; 12:e0176155.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leroux S, Gourichon D, Leterrier C, Labrune Y, Coustham V, Rivière S, Zerjal T, Coville J-L, Morisson M, Minvielle F, et al. Embryonic environment and transgenerational effects in quail. Genet Sel Evol 2017; 49:14.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anway MD, Cupp AS, Uzumcu M, Skinner MK.. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 2005; 308:1466–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Padmanabhan N, Jia D, Geary-Joo C, Wu X, Ferguson-Smith AC, Fung E, Bieda MC, Snyder FF, Gravel RA, Cross JC, et al. Mutation in folate metabolism causes epigenetic instability and transgenerational effects on development. Cell 2013; 155:81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F.. RNA-mediated non-Mendelian inheritance of an epigenetic change in the mouse. Nature 2006; 441:469–74. [DOI] [PubMed] [Google Scholar]
- 19. Braunschweig M, Jagannathan V, Gutzwiller A, Bee G.. Investigations on transgenerational epigenetic response down the male line in F2 pigs. PLoS One 2012; 7:e30583.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Northstone K, Golding J, Davey Smith G, Miller LL, Pembrey M.. Prepubertal start of father's smoking and increased body fat in his sons: further characterisation of paternal transgenerational responses. Eur J Hum Genet 2014; 22:1382–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Skinner MK. Endocrine disruptor induction of epigenetic transgenerational inheritance of disease. Mol Cell Endocrinol 2014; 398:4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Manikkam M, Tracey R, Guerrero-Bosagna C, Skinner MK.. Dioxin (TCDD) induces epigenetic transgenerational inheritance of adult onset disease and sperm epimutations. PLoS One 2012; 7:e46249.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Manikkam M, Tracey R, Guerrero-Bosagna C, Skinner M.. Pesticide and insect repellent mixture (permethrin and DEET) induces epigenetic transgenerational inheritance of disease and sperm epimutations. Reprod Toxicol 2012; 34:708–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tracey R, Manikkam M, Guerrero-Bosagna C, Skinner M.. Hydrocarbons (jet fuel JP-8) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. Reprod Toxicol 2013; 36:104–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Manikkam M, Tracey R, Guerrero-Bosagna C, Skinner M.. Plastics derived endocrine disruptors (BPA, DEHP and DBP) induce epigenetic transgenerational inheritance of adult-onset disease and sperm epimutations. PLoS One 2013; 8:e55387.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Skinner MK, Manikkam M, Tracey R, Guerrero-Bosagna C, Haque MM, Nilsson E.. Ancestral dichlorodiphenyltrichloroethane (DDT) exposure promotes epigenetic transgenerational inheritance of obesity. BMC Med 2013; 11:228.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Manikkam M, Haque MM, Guerrero-Bosagna C, Nilsson E, Skinner MK.. Pesticide methoxychlor promotes the epigenetic transgenerational inheritance of adult onset disease through the female germline. PLoS One 2014; 9:e102091.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McBirney M, King SE, Pappalardo M, Houser E, Unkefer M, Nilsson E, Sadler-Riggleman I, Beck D, Winchester P, Skinner MK, et al. Atrazine induced epigenetic transgenerational inheritance of disease, lean phenotype and sperm epimutation pathology biomarkers. PLoS One 2017; 12:e0184306.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bayley M, Larsen PF, Baekgaard H, Baatrup E.. The effects of vinclozolin, an anti-androgenic fungicide, on male guppy secondary sex characters and reproductive success. Biol Reprod 2003; 69:1951–6. [DOI] [PubMed] [Google Scholar]
- 30. Uzumcu M, Suzuki H, Skinner MK.. Effect of the anti-androgenic endocrine disruptor vinclozolin on embryonic testis cord formation and postnatal testis development and function. Reprod Toxicol 2004; 18:765–74. [DOI] [PubMed] [Google Scholar]
- 31. Buckley J, Willingham E, Agras K, Baskin LS.. Embryonic exposure to the fungicide vinclozolin causes virilization of females and alteration of progesterone receptor expression in vivo: an experimental study in mice. Environ Health 2006; 5:4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Skinner M, Guerrero-Bosagna C, Haque MM, Nilsson E, Bhandari R, McCarrey J.. Environmentally induced transgenerational epigenetic reprogramming of primordial germ cells and subsequent germline. PLoS One 2013; 8:e66318.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nilsson EE, Skinner MK.. Environmentally induced epigenetic transgenerational inheritance of disease susceptibility. Transl Res 2015; 165:12–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Anway MD, Memon MA, Uzumcu M, Skinner MK.. Transgenerational effect of the endocrine disruptor vinclozolin on male spermatogenesis. J Androl 2006; 27:868–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Anway MD, Rekow SS, Skinner MK.. Transgenerational epigenetic programming of the embryonic testis transcriptome. Genomics 2008; 91:30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nilsson EE, Anway MD, Stanfield J, Skinner MK.. Transgenerational epigenetic effects of the endocrine disruptor vinclozolin on pregnancies and female adult onset disease. Reproduction 2008; 135:713–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Skinner MK, Anway MD, Savenkova MI, Gore AC, Crews D.. Transgenerational epigenetic programming of the brain transcriptome and anxiety behavior. PLoS One 2008; 3:e3745.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guerrero-Bosagna C, Savenkova M, Haque MM, Nilsson E, Skinner MK.. Environmentally induced epigenetic transgenerational inheritance of altered Sertoli cell transcriptome and epigenome: molecular etiology of male infertility. PLoS One 2013; 8:e59922.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gapp K, Jawaid A, Sarkies P, Bohacek J, Pelczar P, Prados J, Farinelli L, Miska E, Mansuy IM.. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci 2014; 17:667–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schuster A, Skinner MK, Yan W.. Ancestral vinclozolin exposure alters the epigenetic transgenerational inheritance of sperm small noncoding RNAs. Environ Epigenet 2016; 2:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mancini-Dinardo D, Steele SJ, Levorse JM, Ingram RS, Tilghman SM.. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev 2006; 20:1268–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mohammad F, Mondal T, Guseva N, Pandey GK, Kanduri C.. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development 2010; 137:2493–9. [DOI] [PubMed] [Google Scholar]
- 43. Zhao Y, Sun H, Wang H.. Long noncoding RNAs in DNA methylation: new players stepping into the old game. Cell Biosci 2016; 6:45.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT.. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell 2010; 40:939–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cifuentes-Rojas C, Hernandez AJ, Sarma K, Lee JT.. Regulatory interactions between RNA and polycomb repressive complex 2. Mol Cell 2014; 55:171–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA 2009; 106:11667–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rechavi O, Houri-Ze’evi L, Anava S, Goh WSS, Kerk SY, Hannon GJ, Hobert O.. Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell 2014;158:277–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Krawetz SA, Kruger A, Lalancette C, Tagett R, Anton E, Draghici S, Diamond MP.. A survey of small RNAs in human sperm. Hum Reprod 2011; 26:3401–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang X, Gao F, Fu J, Zhang P, Wang Y, Zeng X.. Systematic identification and characterization of long non-coding RNAs in mouse mature sperm. PLoS One 2017; 12:e0173402.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jodar M, Selvaraju S, Sendler E, Diamond MP, Krawetz SA.. The presence, role and clinical use of spermatozoal RNAs. Hum Reprod Update 2013; 19:604–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen Q, Yan W, Duan E.. Epigenetic inheritance of acquired traits through sperm RNAs and sperm RNA modifications. Nat Rev Genet 2016; 17:733–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ben Maamar M, Sadler-Riggleman I, Beck D, Skinner MK. Epigenetic transgenerational inheritance of altered sperm histone retention sites. Sci Rep2018; 8:5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hammoud SS, Nix DA, Hammoud AO, Gibson M, Cairns BR, Carrell DT.. Genome-wide analysis identifies changes in histone retention and epigenetic modifications at developmental and imprinted gene loci in the sperm of infertile men. Hum Reprod 2011; 26:2558–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Anway MD, Leathers C, Skinner MK.. Endocrine disruptor vinclozolin induced epigenetic transgenerational adult-onset disease. Endocrinology 2006; 147:5515–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Skinner MK, Guerrero-Bosagna C.. Role of CpG deserts in the epigenetic transgenerational inheritance of differential DNA methylation regions. BMC Genomics 2014; 15:692.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Anway MD, Skinner MK.. Transgenerational effects of the endocrine disruptor vinclozolin on the prostate transcriptome and adult onset disease. Prostate 2008; 68:517–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Guerrero-Bosagna C, Settles M, Lucker B, Skinner M.. Epigenetic transgenerational actions of vinclozolin on promoter regions of the sperm epigenome. PLoS One 2010; 5:e13100.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Beck D, Sadler-Riggleman I, Skinner MK.. Generational comparisons (F1 versus F3) of vinclozolin induced epigenetic transgenerational inheritance of sperm differential DNA methylation regions (epimutations) using MeDIP-Seq. Environ Epigenet 2017; 3:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tang L, Liu Z, Zhang R, Su C, Yang W, Yao Y, Zhao S.. Imprinting alterations in sperm may not significantly influence ART outcomes and imprinting patterns in the cord blood of offspring. PLoS One 2017; 12:e0187869.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dallaire A, Simard MJ.. The implication of microRNAs and endo-siRNAs in animal germline and early development. Dev Biol 2016; 416:18–25. [DOI] [PubMed] [Google Scholar]
- 61. Ruden DM, Lu X.. Hsp90 affecting chromatin remodeling might explain transgenerational epigenetic inheritance in Drosophila. Curr Genomics 2008;9:500–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Oliva R, Ballesca JL.. Altered histone retention and epigenetic modifications in the sperm of infertile men. Asian J Androl 2012; 14:239–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Balhorn R. The protamine family of sperm nuclear proteins. Genome Biol 2007; 8:227.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jenkins TG, Carrell DT.. The paternal epigenome and embryogenesis: poising mechanisms for development. Asian J Androl 2011; 13:76–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR.. Distinctive chromatin in human sperm packages genes for embryo development. Nature 2009; 460:473–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hammoud SS, Purwar J, Pflueger C, Cairns BR, Carrell DT.. Alterations in sperm DNA methylation patterns at imprinted loci in two classes of infertility. Fertil Steril 2010; 94:1728–33. [DOI] [PubMed] [Google Scholar]
- 67. Zhang X, San Gabriel M, Zini A.. Sperm nuclear histone to protamine ratio in fertile and infertile men: evidence of heterogeneous subpopulations of spermatozoa in the ejaculate. J Androl 2006; 27:414–20. [DOI] [PubMed] [Google Scholar]
- 68. Joh RI, Palmieri CM, Hill IT, Motamedi M.. Regulation of histone methylation by noncoding RNAs. Biochim Biophys Acta 2014; 1839:1385–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Miyoshi N, Stel JM, Shioda K, Qu NA, Odahima J, Mitsunaga S, Zhang X, Nagano M, Hochedlinger K, Isselbacher KJ, et al. Erasure of DNA methylation, genomic imprints, and epimutations in a primordial germ-cell model derived from mouse pluripotent stem cells. Proc Natl Acad Sci USA 2016; 113:9545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Manikkam M, Guerrero-Bosagna C, Tracey R, Haque MM, Skinner MK.. Transgenerational actions of environmental compounds on reproductive disease and identification of epigenetic biomarkers of ancestral exposures. PLoS One 2012; 7:e31901.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Skinner MK, Manikkam M, Guerrero-Bosagna C.. Epigenetic transgenerational actions of environmental factors in disease etiology. Trends Endocrinol Metab 2010; 21:214–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hisano M, Erkek S, Dessus-Babus S, Ramos L, Stadler MB, Peters AH.. Genome-wide chromatin analysis in mature mouse and human spermatozoa. Nat Protoc 2013; 8:2449–70. [DOI] [PubMed] [Google Scholar]
- 73. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 2011; 17:10–2. [Google Scholar]
- 74. Tang C, Xie Y, Yan W. AASRA: an anchor alignment-based small RNA annotation pipeline. bioRxiv. 2017: 132928. doi: https://doi.org/10.1101/132928. [DOI] [PMC free article] [PubMed]
- 75. Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15:550.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bolger AM, Lohse M, Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012; 7:562–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wang L, Park HJ, Dasari S, Wang S, Kocher JP, Li W.. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res 2013; 41:e74.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Langmead B, Salzberg Sl.. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012; 9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lienhard M, Grimm C, Morkel M, Herwig R, Chavez L.. MEDIPS: genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics 2014; 30:284–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Robinson MD, McCarthy DJ, Smyth GK.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Durinck S, Spellman PT, Birney E, Huber W.. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc 2009; 4:1184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, et al. Ensembl 2015. Nucleic Acids Res 2015; 43 (Database issue): D662–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kanehisa M, Goto S.. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000; 28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M.. Data, information, knowledge and principle: back to metabolism in KEGG. Nucl Acids Res 2014; 42:D199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Huang da W, Sherman BT, Lempicki RA.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 87. Mi H, Muruganujan A, Casagrande JT, Thomas PD.. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 2013; 8:1551–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.