Abstract
The present study aimed to investigate the expression level of DNA mismatch repair gene (MMR) in in sporadic colorectal cancer (SCRC) in eastern China, and to investigate the association between MMR status and prognosis of patients with SCRC. Patient archives from the Department of Gastrointestinal Surgery of Weihai Municipal Hospital (Weihai, China) were retrospectively collected between January 2011 and January 2012. Of the 221 consecutive patients identified, 192 patients who met the criterion were deemed eligible for inclusion. Immunohistochemistry (IHC) was conducted to detect the expression of MMR proteins MutL homolog 1 (MLH1), MutS homolog 2 (MSH2), MSH6 and PMS1 homolog 2, mismatch repair system component (PMS2) expression and mutation in sporadic colorectal cancer (SCRC). Kaplan-Meier plots and log-rank tests were performed to conduct survival analysis and Cox proportional hazard regression models were conducted to determine independent prognostic factors. The total rate of deficient MMR (dMMR) was 14.58% (28/192): MSH6, 0.52% (1/192); PMS2, 4.17% (8/192); MSH2/MSH6, 3.65% (7/192); and MLH1/PMS2, 6.25% (12/192). The dMMR group had a significantly longer overall survival time compared with proficient MMR (pMMR) group (P=0.017). Disease-free survival time of dMMR group was also longer than pMMR group (P=0.027). Multivariate analysis using the Cox regression model confirmed that MMR status was an independent prognostic factor for SCRC. Loss of MMR expression was indicative of a favorable outcome for patients with SCRC, and MMR status could be viewed as an independent prognostic factor.
Keywords: mismatch repair, sporadic colorectal cancer, prognosis, microsatellite instability, immunohistochemistry
Introduction
Colorectal cancer (CRC) is one of the most common types of cancer worldwide and, according to cancer statistics in China, CRC incidence and mortality rates were fifth of all cancer in 2015 (1). In Eastern China, age-standardized (Segi standard population) incidence and mortality rates (2) for CRC remain high. The occurrence and development of colorectal cancer is a process that involves multiple factors, steps and genes, which results from a complex combination of internal and external factors. CRC may be divided into two groups: Inherited colorectal cancer and sporadic colorectal cancer (SCRC), the latter of which accounts for ~85% of all colorectal cancer cases (3). In addition, it was reported that ~2/3 of all CRC tumors with deficient DNA mismatch repair (dMMR) were SCRC (4), and dMMR was associated to a favorable prognosis for CRC (5).
The process of CRC can involve three types of genetic alterations: Activation or upregulation of oncogenes, inactivation or loss of function of tumor suppressor genes, and abnormalities or functional decline in DNA repair gene structure (6). MMR aids the maintenance of genome stability by correcting base-base mismatches and insertion/deletion mis-pairing generated during DNA replication and recombination (7). Deficiencies in MMR are recognized through the presence of microsatellite instability (MSI), which can be identified using polymerase chain reaction amplification of specific tumor microsatellite foci, or by a deficiency in expression of any of the MMR proteins, including MutL homolog 1 (MLH1), MutS homolog 2 (MSH2), MSH6, and PMS1 homolog 2, mismatch repair system component (PMS2), detected by immunohistochemistry (IHC) (8). IHC analysis of expression of MMR proteins is frequently used as it does not require a laboratory and the ability to identify the affected gene by detecting loss of its protein product. Furthermore, the IHC-detected loss of MMR protein expression was highly concordant with DNA-based MSI testing (9). Notably, to the best of our knowledge, only the loss of MLH1 protein expression, observed by IHC, has been described in SCRCs (10).
Alterations in both the epigenome and genome occur commonly in colorectal cancer and likely drive the tumorigenesis process through activation of oncogenes and inactivation of tumor suppressor genes (11). The present study aimed to investigate the association between dMMR expression and prognosis in SCRC with long-term individual survey. To the best of our knowledge, the present study is the first to demonstrate an association between MMR-deficiency and the prognosis of an Eastern Chinese population of patients with sporadic CRC.
Materials and methods
Samples
A total of 221 patients with colorectal cancer underwent radical surgery treatments with open surgery or laparoscopic surgery in the Department of Gastrointestinal Surgery of Weihai Municipal Hospital (Shandong, China) between January 2011 and January 2012. All patients had been pathologically confirmed to have colorectal adenocarcinoma. Pathological stage was determined according to tumor-node-metastasis staging system of American Joint Committee on Cancer (AJCC) (12). The tumor tissues were collected from 192 patients and additional non-tumoral normal epithelial tissue samples (~5 cm from the border of the main tumor lesion) were collected from 138 of these patients. A total of 16 cases were excluded with strict exclusion criteria; including, familial adenomatous polyposis (FAP), hereditary non-polyposis colorectal cancer (HNPCC) based on Bethesda guidelines (13), mortality within 1 month of surgery, preoperative adjuvant therapy, and non-colorectal adenocarcinoma (including neuroendocrine neoplasm and lymphoma) (14,15). In order to exclude genetic effects, patients with a known family history or those suspected to have hereditary or familial CRC, FAP or HNPCC were excluded. In order to exclude the effects of treatment, patients who succumbed within one month following operation and those who underwent preoperative adjuvant therapy were also excluded. One sample was diagnosed as neuroendocrine neoplasm.
A total of 16 cases were excluded, which included FAP (n=1), HNPCC (n=4), death from cardiopulmonary complication (n=1), neuroendocrine neoplasm (n=1), recurrence of colorectal carcinoma (n=1) and neo-adjuvant chemotherapy (n=8). A further 13 of 205 cases refused to participate in the present study; therefore 192 cases were enrolled. The age range of the patients was 33–89 years (mean, 63.2±10.8 years). A total of 112 of the patients were male (mean age, 63.5±10.8 years) and 80 of the patients were female (mean age, 62.9±11.0 years). The median follow-up time was 43.5 months. At the last follow-up (January 2016), 107 patients were alive and 85 patients had succumbed to disease. An estimated 4-year survival time for the entire population was 55.73%. The present study was approved by the Ethics and Scientific Committees of Weihai Municipal Hospital. Written informed consent was obtained from all participants in the study.
Immunohistochemistry staining
Primary antibodies, including rabbit polyclonal antibodies against human MSH2 (cat. no. AP08394PU-N), MSH6 (cat. no. TA326879) and PMS2 (cat. no. AP00189PU-N) were purchased from OriGene Technologies, Inc., Beijing, China. Primary mouse antibody against human MLH1 (cat. no. sc-56161) was purchased from Santa Cruz Biotechnology, Inc., Dallas, TX, USA. All specimens were fixed with 10% formalin and embedded in paraffin at room temperature for 24 h, and each block was sectioned at 4 µm. All sections were deparaffinized and rehydrated in a descending alcohol series. Slides were heated (96–98°C) in 1 mmol/l EDTA buffer for 20 min for antigen retrieval. Slides were incubated with 0.3% hydrogen peroxide to quench endogenous peroxidase activity at room temperature for 30 min, and non-specific binding was blocked in 10% goat serum (DAB Detection kit (Streptavidin-Biotin) cat. no. SP-9000; OriGene Technologies, Inc., Beijing, China) at room temperature for 1 h. Slides were incubated at 4°C, overnight with primary antibodies against MLH1, MSH2, MSH6 and PMS2 (dilution 1:500). The subsequent steps were performed according to streptavidin-peroxidase method protocols (16). Then each slide was incubated with 30 µl of goat anti-rabbit or anti-mouse biotin-conjugated secondary antibody for 30 min at 37°C (cat. no. SP-9000; ZSGB-BIO; Beijing, China). A volume of 100 µl HRP conjugates were applied to the sections, and incubated in a humidified chamber at room temperature for 30 min. The primary antibody was replaced with normal goat serum (OriGene Technologies, Inc.) or PBS for negative controls, and nuclear staining of MMR proteins in normal colonic epithelium cells and lymphocytes served as positive controls.
Staining evaluation
The samples with >10% of tumor cells stained for any MMR protein were considered to be MMR-positive. The criteria used for semi-quantification of immunohistochemical staining included the staining intensity and the percentage of positively stained cells. A range of 0–3 was defined for classifying the intensity of staining: 0, Absence of staining; 1, weak staining; 2, moderate staining; and 3, intense staining. Furthermore, extent of staining was scored as 0 (<10%), 1 (11–25%), 2 (26–50%), 3 (51–75%), and 4 (76–100%) for evaluation. The final scores were calculated by multiplying the staining intensity by the extension (17). In the present study, all the final scores were stratified as negative MMR expression (0 score) or positive MMR expression (>0). The MMR-positive group included low expression (1–4 score), moderate expression (5–8 score) and high expression (9–12 score). All pathological sections were reviewed by at least two experienced pathologists affiliated to the Department of Pathology of Weihai Municipal Hospital.
Statistical analysis
All data are presented as the mean ± standard deviation (SD) or the median SD. For example, data for OS or PFS were median ± SD, and data for age were presented as mean ± SD. SPSS 18.0 software (SPSS Inc., Chicago, IL, USA) was used for statistical analysis. Fisher's exact test and χ2 test were used to evaluate clinicopathological significance of enrolled patients' characteristics in SCRC. The Kaplan-Meier method and log rank test were used to calculate survival data. The Cox regression tests were used for independent prognosis factor analysis. Two-sided P-values were calculated, and P<0.05 was considered to indicate a statistically significant difference.
Results
Patient characteristics
Samples from 192 patients with sporadic colorectal cancer who had undergone complete surgical resection were obtained in the present study (Table I). The age distribution of cases was between 33 and 89 years old and the mean age was 63.2±10.8 years. A total of 112 of 192 cases were males and the mean age was 63.5±10.8 years; 80 of 192 patients were female and the mean age was 62.9±11.0 years.
Table I.
Clinical characteristics of 192 patients with sporadic colorectal cancer.
| Variables | Patients, n (%) |
|---|---|
| Sex | |
| Male | 112 (58.3) |
| Female | 80 (41.7) |
| Age, years | |
| ≥60 | 124 (64.6) |
| <60 | 68 (35.4) |
| Location | |
| Right colon | 40 (20.8) |
| Left colon | 44 (22.9) |
| Rectum | 108 (56.3) |
| Differentiation | |
| Well/moderate | 139 (72.4) |
| Poor | 53 (27.6) |
| Tumor stage | |
| T1+T2 | 42 (21.9) |
| T3+T4 | 150 (78.1) |
| Lymph node status | |
| pN0 | 121 (63.0) |
| pN1 | 45 (23.4) |
| pN2 | 26 (13.6) |
| Metastasis status | |
| Negative | 187 (97.4) |
| Positive | 5 (2.6) |
| Invasion | |
| Negative | 129 (67.2) |
| Positive | 63 (32.8) |
| MMR status | |
| dMMR | 28 (14.6) |
| pMMR | 164 (85.4) |
dMMR, deficient DNA mismatch repair; pMMR, proficient MMR.
Expression of MMR in sporadic colorectal cancer
To investigate the status of the MMR proteins MLH1, MSH2, MSH6, PMS2 in SCRC, an IHC assay was performed to evaluate the expression of MMR. Fig. 1 demonstrates representative typical IHC staining images of positive and negative nuclear expression of MLH1, MSH2, MSH6 and PMS2 in different patients with SCRC. Fig. 1A depicts tumor cells with retained MLH1, MSH2, MSH6 and PMS2 expression, which were regarded as MMR proficient, while Fig. 1B depicts cells lacking MLH1, MSH2, MSH6 and PMS2, which were regarded as deficient MMR. Stromal cells and lymphocytes served as internal positive controls and non-tumoral normal epithelial tissue as normal controls (data not shown). In the present study, the total rate of deficient MMR (dMMR) was 14.58% (28/192): MSH6, 0.52% (1/192); PMS2, 4.17% (8/192); MSH2/MSH6, 3.65% (7/192); and MLH1/PMS2, 6.25% (12/192). These differences in expression may be due to race, sample size, test methods and result evaluation. Distant metastasis was not observed in the dMMR group on count of sample number probably, while there were 5 cases of distant metastasis in the pMMR group.
Figure 1.

Representative typical immunohistochemical staining images of positive and negative nuclear expression of MLH1, MSH2, MSH6 and PMS2 in different patients with sporadic colorectal cancer. Tumor cells with (A) retained MLH1, MSH2, MSH6 and PMS2 expression, which were regarded as MMR proficient, and with (B) absent MLH1, MSH2, MSH6 and PMS2 expression, which were regarded as MMR deficient. MLH1, MutL homolog 1; MSH2, MutS homolog 2; PMS2, PMS1 homolog 2, mismatch repair system component; MMR, mismatch repair gene.
Association between MMR expression and overall survival (OS) or disease-free survival (DFS) in sporadic colorectal cancer
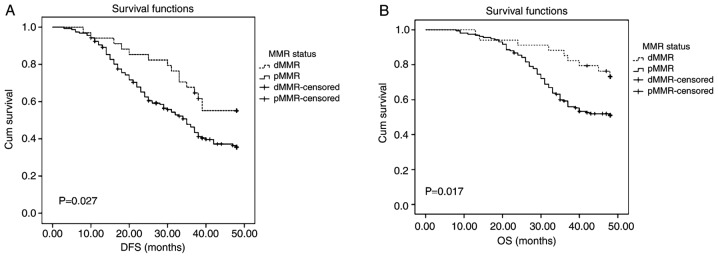
Patients were divided into pMMR and dMMR expression groups. Kaplan-Meier curves and log-rank test results for DFS and OS were demonstrated in Fig. 2. Patients with dMMR presented with longer DFS and OS times (median survival ± SD, 40±10.83 and 49±8.52, respectively) compared with those with pMMR (median survival ± SD, 28±12.11 and 39±10.02, respectively) (Fig. 2; P=0.027 and P=0.017, respectively).
Figure 2.
Survival analyses of the dMMR and pMMR groups. (A) The DFS of patients with SCRC with dMMR expression status was higher compared with that with pMMR expression status, P=0.036. (B) The OS of patients with SCRC with dMMR expression status was higher than that with pMMR expression status, P=0.035. dMMR, deficient DNA mismatch repair; pMMR, proficient MMR; DFS, disease-free survival; SCRC, sporadic colorectal cancer; OS, overall survival.
Univariate and multivariate analysis of prognostic factors in sporadic colorectal cancer
To determine the association between MMR status and prognostic factors in SCRC, Cox proportional hazard model was used, and univariate analysis revealed that patient sex, age, tumor location, tumor differentiation, tumor stage, lymph node status, metastasis status, invasion and MMR status were significantly associated with DFS and OS. Differentiation, tumor stage, lymph node status, metastasis status, invasion and MMR status were identified to be significant prognostic factors for DFS by univariate analysis. For multivariate analysis, differentiation, tumor stage, lymph node status and MMR status were independent significant prognostic factors for DFS (P=0.006, P<0.001, P<0.001, P=0.003, respectively; Table II). Age, differentiation, tumor stage, lymph node status, metastasis status, invasion and MMR status were identified to be significant prognostic factors for OS by univariate analysis. For multivariate analysis, age, differentiation, tumor stage, lymph node status, metastasis status, invasion and MMR status were identified to be independent prognostic factors for OS (Table III; P=0.003, P=0.047, P<0.001, P<0.001, P=0.027, P=0.004, P<0.001, respectively).
Table II.
Univariate and multivariate analysis of prognostic factors for disease-free survival in sporadic colorectal cancer.
| 95% CI for HR | |||||||
|---|---|---|---|---|---|---|---|
| Factors | B | SE | Wald | P-value | HR | Lower | Upper |
| Univariate analysis | |||||||
| Age (years) | −0.358 | 0.194 | 3.398 | 0.065 | 0.699 | 0.478 | 1.023 |
| Sex | 0.164 | 0.192 | 0.732 | 0.392 | 1.179 | 0.809 | 1.717 |
| Location | −0.044 | 0.059 | 0.555 | 0.456 | 0.957 | 0.852 | 1.075 |
| Differentiation | −0.862 | 0.188 | 21.044 | 0.001a | 0.422 | 0.292 | 0.610 |
| Tumor stage | 1.151 | 0.226 | 25.987 | 0.001a | 3.162 | 2.031 | 4.923 |
| Lymph node status | 1.239 | 0.123 | 101.937 | 0.001a | 3.453 | 2.714 | 4.391 |
| Metastasis status | 2.471 | 0.471 | 27.541 | 0.001a | 11.834 | 4.703 | 29.779 |
| Invasion | −1.033 | 0.194 | 28.264 | 0.001a | 0.356 | 0.243 | 0.521 |
| MMR status | 0.599 | 0.278 | 4.623 | 0.032a | 1.819 | 1.054 | 3.140 |
| Multivariate analysis | |||||||
| Differentiation | −0.574 | 0.208 | 7.623 | 0.006a | 0.563 | 0.375 | 0.847 |
| Tumor stage | 1.283 | 0.232 | 30.586 | 0.001a | 3.606 | 2.289 | 5.680 |
| Lymph node status | 1.333 | 0.149 | 79.598 | 0.001a | 3.793 | 2.830 | 5.083 |
| Metastasis status | 0.593 | 0.503 | 1.391 | 0.238 | 1.809 | 0.676 | 4.845 |
| Invasion | 0.324 | 0.112 | 1.892 | 0.329 | 0.672 | 0.492 | 1.168 |
| MMR status | 0.863 | 0.288 | 8.985 | 0.003a | 2.369 | 1.348 | 4.165 |
P<0.05. B, partial regression coefficient; SE, standard error of partial regression coefficient; CI, confidence interval; HR, hazard ratio; MMR, DNA mismatch repair. Wald was used to compare if there was difference between total partial regression coefficient and 0.
Table III.
Univariate and multivariate analysis of prognostic factors for overall survival in sporadic colorectal cancer.
| 95.0% CI for HR | |||||||
|---|---|---|---|---|---|---|---|
| Factors | B | SE | Wald | P-value | HR | Lower | Upper |
| Univariate analysis | |||||||
| Age (years) | −0.580 | 0.227 | 6.515 | 0.011a | 0.560 | 0.359 | 0.874 |
| Sex | 0.188 | 0.218 | 0.744 | 0.388 | 1.207 | 0.788 | 1.849 |
| Location | −0.096 | 0.065 | 2.198 | 0.138 | 0.908 | 0.799 | 1.032 |
| Differentiation | −0.790 | 0.212 | 13.849 | 0.001a | 0.454 | 0.299 | 0.688 |
| Tumor stage | 1.036 | 0.248 | 17.420 | 0.001a | 2.819 | 1.733 | 4.585 |
| Lymph node status | 1.297 | 0.134 | 94.256 | 0.001a | 3.658 | 2.815 | 4.752 |
| Metastasis status | 2.832 | 0.482 | 34.472 | 0.001a | 16.978 | 6.597 | 43.696 |
| Invasion | −1.015 | 0.218 | 21.655 | 0.001a | 0.362 | 0.236 | 0.556 |
| MMR status | 0.813 | 0.353 | 5.307 | 0.021a | 2.255 | 1.129 | 4.504 |
| Multivariate analysis | |||||||
| Age | −0.789 | 0.237 | 11.232 | 0.003a | 0.238 | 0.278 | 0.696 |
| Differentiation | −0.459 | 0.231 | 3.957 | 0.047a | 0.632 | 0.402 | 0.993 |
| Tumor stage | 1.070 | 0.251 | 18.115 | 0.001a | 2.917 | 1.782 | 4.775 |
| Lymph node status | 1.270 | 0.154 | 67.866 | 0.001a | 3.559 | 2.631 | 4.814 |
| Metastasis status | 1.126 | 0.509 | 4.892 | 0.027a | 3.084 | 1.137 | 8.367 |
| Invasion | 1.072 | 0.368 | 8.482 | 0.004a | 2.920 | 1.420 | 6.005 |
| MMR status | 1.175 | 0.332 | 12.496 | 0.001a | 3.237 | 1.688 | 6.207 |
P<0.05. B, partial regression coefficient; SE, standard error of partial regression coefficient; CI, confidence interval; HR, hazard ratio. Wald was used to compare if there was difference between total partial regression coefficient and 0, CI.
Discussion
Colorectal cancer is divided into two types, of which one is an inherited disease, including FAP and HNPCC, and the other is a sporadic disease (18). The genetic foundation of HNPCC is intimately associated with MMR, which has been well studied and proven (19). Approximately 15% of sporadic colorectal cancer cases have been confirmed to be attributable to the same processes and mechanisms involved in HNPCC (20); among which dMMR was a major type of genomic instability caused by a failure to correct errors during DNA replication. Mutation or modification of MMR genes (including by methylation) usually causes the absence of MMR protein expression and MSI (3). There are two ways that tumorigenesis arises as a consequence of MMR function. MSI can induce the activation of oncogenes or inhibition of tumor suppressor genes. Alternatively, deficient MMR directly brings about activation of oncogenes or the inhibition of tumor suppressor genes (21).
The MMR system involves nine proteins: MLH1, MSH2, MSH3, MSH6, MLH3, PMS1, PMS2, MSH4 and MSH5. Between 87 and 90% of all mutated genes associated with colorectal cancer are MLH1 and MSH2 (22). A study undertaken by Herman et al (23) demonstrated that 5′-CpG hypermethylation of MLH1 of SCRC often led to the absence of MLH1 expression (23). In another study, Herman et al (10) proposed that inhibition of hypermethylation in the promoter of MLH1 of tumor cells with demethylating agents would produce the reappearance of MLH1 expression. Another study demonstrated that the loss of MSH2 expression was linked to missense mutations (24). Other previous studies have demonstrated that the mutation rate of MMR gene in SCRC was 10–20% (25,26). Lindor et al (9) revealed that the absence of MLH1 protein expression was present in 20.4% cases and that of MSH2 was absent in in 8.8% cases in a sample of 1,114 patients with SCRC. Other previous reports revealed that tumors in several patients with SCRC with a deficient MMR system were frequently accompanied by poor differentiation, mucinous subtype and occurred in the ascending colon, as in patients with HNPCC (27,28). In the present study, there were no significant differences in age, sex, tumor staging, lymph node metastasis or vascular invasion between the two groups.
Benatti et al (25) reported that mutations in the MMR gene may only affect early-phase tumors, and were not associated with invasion and metastasis. Other studies have demonstrated that the outcome of patients with negative MMR expression with SCRC, concerning overall survival and disease-free survival, were more favorable those for the positive expression group (29). In addition, there was a reduced relapse rate in the negative expression group (28,30). The results from the present study are in accordance with the conclusion above. In the present study, MMR expression status was associated with OS and DFS rates of patients with SCRC, and with one of the independent prognostic factors. The dMMR group had a significantly higher OS rate than the pMMR group (P=0.017). The DFS rate of dMMR group was also higher than those of the pMMR group (P=0.027).
In addition, MMR system detection may have vital predictive and guidance value for colorectal cancer response to chemotherapy. Several studies (31,32) have revealed that a deficiency in MMR protein expression status may be a predictive marker of decreased benefit, and possibly even a detrimental effect, from adjuvant therapy with fluoropyrimidine alone in patients with stage II disease (29). Compared with patients that underwent surgical resection alone, treatment with fluoropyrimidine following surgery, exhibited a lower 5-year survival rate. However, it has been reported that MMR status cannot be recommended to inform adjuvant treatment decisions in patients with stage III CRC (33). National Comprehensive Cancer Network (NCCN) guidelines (34) state that MMR testing should be performed for all patients with colorectal cancer diagnosed at ≤70 years, including patients diagnosed at older ages that meet the Bethesda guidelines, to assess for the possibility of Lynch syndrome. Poorly differentiated histology is not considered to be a high-risk feature for patients with stage II disease whose tumors are dMMR (35).
There were certain limitations in the present study. Although MMR status is associated with MSI level, MSI testing was not performed. Secondly, MMR associated gene mutations were not addressed in the present study, including those to KRAS proto-oncogene, GTPase (KRAS) and B-Raf proto-oncogene, serine/threonine kinase (BRAF). Mutation analysis for KRAS and BRAF, in addition to MMR/MSI testing may be beneficial for patients with CRC, in accordance with the NCCN guidelines (36).
In conclusion, the results of the present study demonstrated that MMR status, as an independent prognostic factor, has critical prognostic value in an Eastern Chinese population. MMR testing may therefore have potential benefits in clinical practice.
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Zheng R, Zeng H, Zhang S, Chen W. Estimates of cancer incidence and mortality in China, 2013. Chin J Cancer. 2017;36:66. doi: 10.1186/s40880-017-0234-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawakami H, Zaanan A, Sinicrope FA. Implications of mismatch repair-deficient status on management of early stage colorectal cancer. J Gastrointest Oncol. 2015;6:676–684. doi: 10.3978/j.issn.2078-6891.2015.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609–618. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 6.Worthley DL, Whitehall VL, Spring KJ, Leggett BA. Colorectal carcinogenesis: Road maps to cancer. World J Gastroenterol. 2007;13:3784–3791. doi: 10.3748/wjg.v13.i28.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sameer AS, Nissar S, Fatima K. Mismatch repair pathway: Molecules, functions and role in colorectal carcinogenesis. Eur J Cancer Prev. 2014;23:246–257. doi: 10.1097/CEJ.0000000000000019. [DOI] [PubMed] [Google Scholar]
- 8.Korphaisarn K, Pongpaibul A, Limwongse C, Roothumnong E, Klaisuban W, Nimmannit A, Jinawath A, Akewanlop C. Deficient DNA mismatch repair is associated with favorable prognosis in Thai patients with sporadic colorectal cancer. World J Gastroenterol. 2015;21:926–934. doi: 10.3748/wjg.v21.i3.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindor NM, Burgart LJ, Leontovich O, Goldberg RM, Cunningham JM, Sargent DJ, Walsh-Vockley C, Petersen GM, Walsh MD, Leggett BA, et al. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol. 2002;20:1043–1048. doi: 10.1200/JCO.20.4.1043. [DOI] [PubMed] [Google Scholar]
- 10.Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079–1099. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edge SB, Compton CC. The American joint committee on cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 13.Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Rüschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R, et al. Revised bethesda guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Compton CC, Greene FL. The staging of colorectal cancer: 2004 and beyond. CA Cancer J Clin. 2004;54:295–308. doi: 10.3322/canjclin.54.6.295. [DOI] [PubMed] [Google Scholar]
- 15.Compton CC, Fielding LP, Burgart LJ, Conley B, Cooper HS, Hamilton SR, Hammond ME, Henson DE, Hutter RV, Nagle RB, et al. Prognostic factors in colorectal cancer. College of American Pathologists Consensus Statement 1999. Arch Pathol Lab Med. 2000;124:979–994. doi: 10.5858/2000-124-0979-PFICC. [DOI] [PubMed] [Google Scholar]
- 16.Wang LG, Ni Y, Su BH, Mu XR, Shen HC, Du JJ. MicroRNA-34b functions as a tumor suppressor and acts as a nodal point in the feedback loop with Met. Int J Oncol. 2013;42:957–962. doi: 10.3892/ijo.2013.1767. [DOI] [PubMed] [Google Scholar]
- 17.Ma H, Wang L, Zhang T, Shen H, Du J. Loss of β-arrestin1 expression predicts unfavorable prognosis for non-small cell lung cancer patients. Tumour Biol. 2016;37:1341–1347. doi: 10.1007/s13277-015-3886-0. [DOI] [PubMed] [Google Scholar]
- 18.Bhattacharya P, McHugh TW. StatPearls. StatPearls Publishing StatPearls Publishing LLC Treasure Island (FL); 2017. Lynch Syndrome. [Google Scholar]
- 19.Kawakami H, Zaanan A, Sinicrope FA. Microsatellite instability testing and its role in the management of colorectal cancer. Curr Treat Options Oncol. 2015;16:30. doi: 10.1007/s11864-015-0348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Win AK, Young JP, Lindor NM, Tucker KM, Ahnen DJ, Young GP, Buchanan DD, Clendenning M, Giles GG, Winship I, et al. Colorectal and other cancer risks for carriers and noncarriers from families with a DNA mismatch repair gene mutation: A prospective cohort study. J Clin Oncol. 2012;30:958–964. doi: 10.1200/JCO.2011.39.5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sheng JQ, Chan TL, Chan YW, Huang JS, Chen JG, Zhang MZ, Guo XL, Mu H, Chan AS, Li SR, et al. Microsatellite instability and novel mismatch repair gene mutations in northern Chinese population with hereditary non-polyposis colorectal cancer. Chin J Dig Dis. 2006;7:197–205. doi: 10.1111/j.1443-9573.2006.00269.x. [DOI] [PubMed] [Google Scholar]
- 22.Ericson K, Halvarsson B, Nagel J, Rambech E, Planck M, Piotrowska Z, Olsson H, Nilbert M. Defective mismatch-repair in patients with multiple primary tumours including colorectal cancer. Eur J Cancer. 2003;39:240–248. doi: 10.1016/S0959-8049(02)00584-1. [DOI] [PubMed] [Google Scholar]
- 23.Herman JG, Merlo A, Mao L, Lapidus RG, Issa JP, Davidson NE, Sidransky D, Baylin SB. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 24.Belvederesi L, Bianchi F, Galizia E, Loretelli C, Bracci R, Catalani R, Amati M, Cellerino R. MSH2 missense mutations and HNPCC syndrome: Pathogenicity assessment in a human expression system. Hum Mutat. 2008;29:E296–E309. doi: 10.1002/humu.20875. [DOI] [PubMed] [Google Scholar]
- 25.Benatti P, Gafa R, Barana D, Marino M, Scarselli A, Pedroni M, Maestri I, Guerzoni L, Roncucci L, Menigatti M, et al. Microsatellite instability and colorectal cancer prognosis. Clin Cancer Res. 2005;11:8332–8340. doi: 10.1158/1078-0432.CCR-05-1030. [DOI] [PubMed] [Google Scholar]
- 26.Ohrling K, Edler D, Hallström M, Ragnhammar P. Mismatch repair protein expression is an independent prognostic factor in sporadic colorectal cancer. Acta Oncol. 2010;49:797–804. doi: 10.3109/02841861003705786. [DOI] [PubMed] [Google Scholar]
- 27.Greenson JK, Huang SC, Herron C, Moreno V, Bonner JD, Tomsho LP, Ben-Izhak O, Cohen HI, Trougouboff P, Bejhar J, et al. Pathologic predictors of microsatellite instability in colorectal cancer. Am J Surg Pathol. 2009;33:126–133. doi: 10.1097/PAS.0b013e31817ec2b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hutchins G, Southward K, Handley K, Magill L, Beaumont C, Stahlschmidt J, Richman S, Chambers P, Seymour M, Kerr D, et al. Value of mismatch repair, KRAS and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J Clin Oncol. 2011;29:1261–1270. doi: 10.1200/JCO.2010.30.1366. [DOI] [PubMed] [Google Scholar]
- 29.Sargent DJ, Marsoni S, Monges G, Thibodeau SN, Labianca R, Hamilton SR, French AJ, Kabat B, Foster NR, Torri V, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28:3219–3226. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sinicrope FA, Foster NR, Thibodeau SN, Marsoni S, Monges G, Labianca R, Kim GP, Yothers G, Allegra C, Moore MJ, et al. DNA mismatch repair status and colon cancer recurrence and survival in clinical trials of 5-fluorouracil-based adjuvant therapy. J Natl Cancer Inst. 2011;103:863–875. doi: 10.1093/jnci/djr153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hemminki A, Mecklin JP, Järvinen H, Aaltonen LA, Joensuu H. Microsatellite instability is a favorable prognostic indicator in patients with colorectal cancer receiving chemotherapy. Gastroenterology. 2000;119:921–928. doi: 10.1053/gast.2000.18161. [DOI] [PubMed] [Google Scholar]
- 32.Elsaleh H, Joseph D, Grieu F, Zeps N, Spry N, Iacopetta B. Association of tumour site and sex with survival benefit from adjuvant chemotherapy in colorectal cancer. Lancet. 2000;355:1745–1750. doi: 10.1016/S0140-6736(00)02261-3. [DOI] [PubMed] [Google Scholar]
- 33.Sinicrope FA, Yang ZJ. Prognostic and predictive impact of DNA mismatch repair in the management of colorectal cancer. Future Oncol. 2011;7:467–474. doi: 10.2217/fon.11.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carroll PR, Parsons JK, Andriole G, Bahnson RR, Castle EP, Catalona WJ, Dahl DM, Davis JW, Epstein JI, Etzioni RB, et al. NCCN Guidelines Insights: Prostate cancer early detection, version 2.2016. J Natl Compr Canc Netw. 2016;14:509–519. doi: 10.6004/jnccn.2016.0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engstrom PF, Arnoletti JP, Benson AB, III, Chen YJ, Choti MA, Cooper HS, Covey A, Dilawari RA, Early DS, Enzinger PC, et al. NCCN clinical practice guidelines in oncology: Colon cancer. J Natl Compr Canc Netw. 2009;7:778–831. doi: 10.6004/jnccn.2009.0056. [DOI] [PubMed] [Google Scholar]
- 36.Benson AB, III, Venook AP, Bekaii-Saab T, Chan E, Chen YJ, Cooper HS, Engstrom PF, Enzinger PC, Fenton MJ, Fuchs CS, et al. Colon cancer, version 3.2014. J Natl Compr Canc Netw. 2014;12:1028–1059. doi: 10.6004/jnccn.2014.0112. [DOI] [PubMed] [Google Scholar]