Abstract
Recent evidence indicates that the long non-coding RNA gastric carcinoma highly expressed transcript 1 (GHET1) is involved in the development and carcinogenesis of several tumor types; however, the exact roles of GHET1 and its underlying mechanisms in head and neck cancer (HNC) remain largely unknown. In the present study, the expression patterns of GHET1 in HNC were determined and its clinical significance was assessed. The expression level of GHET1 was significantly increased in HNC tissues, compared with paired adjacent normal tissues. High GHET1 expression was significantly associated with advanced Tumor-Node-Metastasis stages and poor prognosis. Furthermore, inhibition of GHET1 suppressed cell proliferation, induced cell apoptosis and caused cell cycle arrest in vitro. In addition, GHET1 silencing inhibited cell migration and invasion. Taken together, the results of the present study indicated that GHET1 acts as an oncogene in HNC and may represent a novel therapeutic target.
Keywords: gastric carcinoma highly expressed transcript 1, head and neck cancer, prognosis, proliferation, invasion
Introduction
Head and neck cancer (HNC) is the seventh most common malignancy worldwide, and remains a serious public health problem (1,2). Previous studies have demonstrated that heredity, Epstein-Barr virus infection and environmental factors serve notable roles in HNC development (1,3,4). Although advances in combination treatment strategies involving surgery, radiotherapy and chemotherapy for advanced HNC have been made, patient prognosis remains poor owing to an advanced local regional disease at diagnosis and a high incidence of distant metastasis (2,5,6). Therefore, understanding the molecular mechanisms that underlie tumor progression may lead to the identification of novel molecular predictors to improve the prognosis of HNC.
Long non-coding RNAs (lncRNAs) are a class of transcripts that are >200 nucleotides in size and lack the ability to encode proteins (7). Evidence indicates that aberrant lncRNA expression can contribute to the pathogenesis of major diseases, including HNC (8–10). Recent studies have indicated that lncRNAs could influence various cellular processes, including cell growth, cell apoptosis, cell cycle progression, cell invasion and metastasis (11–14). For instance, it has been reported that HOX antisense intergenic RNA (HOTAIR) was overexpressed in head and neck squamous cell carcinoma and HOTAIR silencing inhibited cell proliferation by inducing cell apoptosis (15). Aberrant H19 expression has also been observed in HNC and was significantly associated with tumor grade, differentiation, neck nodal metastasis and clinical stage (16). H19 promoted cellular proliferation, migration and invasion by competitively binding to microRNA (miR)-148a-3p (16). Expression of maternally expressed gene 3 (MEG3) was significantly reduced in tongue squamous cell carcinoma tissues, and low MEG3 expression was significantly associated with tumor size and lymph node metastasis (17). Overexpression of MEG3 in HNC cells could inhibit cellular proliferation, promote cell apoptosis and arrest cell cycle (17). Gastric carcinoma highly expressed transcript 1 (GHET1), a recently identified lncRNA, has been determined to be unregulated in several cancer types, including gastric, bladder and colorectal cancer (18–20). However, the expression and role of GHET1 in HNC remains largely unknown.
In the present study, the expression of GHET1 was detected and the association between GHET1 and clinicopathological factors of patients with HNC was analyzed. Furthermore, the results indicated that knockdown of GHET1 led to cell proliferation repression, triggering of late apoptosis, decreases in the proportion of cells in S phase and inhibition of cell migration and invasion. These results provide a novel potential biomarker for HNC diagnosis and therapy.
Materials and methods
Tissue samples
A total of 86 pairs of HNC and adjacent normal tissues were collected from patients [26 females and 51 male patients (age range, 37–67 years; mean age, 53 years)] with primary with esophageal squamous cell carcinoma who received treatment in Fujian Cancer Hospital (Fuzhou, China) from January 2007 to December 2009. None of the patients have received any chemotherapy or radiation prior to surgery. All patients were classified according to the sixth edition of the American Joint Committee on Cancer system for esophageal cancer (21). Written informed consent was obtained from all participants and the study was approved by the Board and Ethics Committee of Fujian Cancer University (Fuzhou, China). The tissues were immediately frozen in liquid nitrogen following surgery and stored at −80°C until use.
Cell lines and cultures
A total of five human HNC cell lines (FaDu, OECM1, SCC25, SAS and Cal-27) were purchased from the Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). Normal human oral keratinocytes (HOK) were obtained from ScienCell Research Laboratories, Inc., (San Diego, CA, USA) and grown in keratinocyte growth medium (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). All cells were cultured in RPMI-1640 (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc.), 2 mM glutamine, 0.4 µg/ml of hydrocortisone, 100 µg/ml penicillin and 100 U/ml penicillin G (Hyclone; GE Healthcare Life Sciences) at 37°C in a humidified incubator containing 5% CO2.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from HNC tumor samples and cell lines using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. A total of 500 ng RNA was reverse transcribed to cDNA using PrimeScript™ RT Master Mix (Takara Biotechnology Co., Ltd., Dalian, China). qPCR was performed in a total reaction volume of 20 ml using SYBR-Green qPCR Master Mix (Takara Biotechnology Co., Ltd.) in the ABI PRISM 7900HT Sequence Detection system (Applied Biosystems; Thermo Fisher Scientific, Inc.) with the following primer sequences: GAPDH forward, 5′-GTCAACGGATTTGGTCTGTATT-3′ and reverse, 5′-AGTCTTCTGGGTGGCAGTGAT-3′; and GHET1 forward, 5′-CCCCACAAATGAAGACACT-3′ and reverse, 5′-TTCCCAACACCCTATAAGAT-3′. Amplification conditions were set as follows: 95°C for 30 sec, followed by 95°C for 10 sec and 60°C for 30 sec for 40 cycles. The expression of GHET1 was normalized to GADPH and data was analyzed using the 2−∆∆Cq method (22).
GHET1 siRNA transfection
siRNAs specifically targeting GHET1 were synthesized by Invitrogen (Thermo Fisher Scientific, Inc.) as previously described (19). FaDu and Cal-27 cells were transfected with Silencer™ Negative Control No. 2 siRNA (cat. no. AM4637) or GHET1 siRNA (cat. no. n321955) at a final concentration of 25 nM using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. The silencing efficiency of GHET1 was verified by RT-qPCR, as aforementioned, 48 h following transfection.
Cell proliferation assay
The proliferation of FaDu and Cal-27 cells was determined using the Cell Counting kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan), according to the manufacturer's protocol. Cells were seeded at a density of 3×103 cells/well in 96-well plates. Next, 10 µl CCK-8 reagent was added to each well at 24, 48, 72 and 96 h, which were incubated for 2 h at 37°C. The absorbance was measured at a wavelength of 450 nm using a Model 680 microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA). All experiments were performed in triplicate in three in independent times.
Flow cytometry analysis
To analyze cell apoptosis, HNC cells were harvested and resuspended in staining buffer (BD Biosciences, San Jose, CA, USA) at a concentration of 1×106 cells/ml. Following this, cells were stained with Annexin V and propidium iodide (PI) using the Annexin V-Fluorescein Isothiocyanate Apoptosis Detection kit (BD Biosciences), according to the manufacturer's instructions. Apoptotic cells were measured using a BD FACSCanto™ Flow Cytometer (BD Biosciences) and analyzed with Summit v5.0 software (BD Biosciences).
For cell cycle analysis, cells were harvested by trypsin in the logarithmic growth phase and fixed in 75% ethanol at 4°C overnight. Next, cells were re-suspended in PBS and incubated with BD Pharmingen™ PI/RNase staining buffer (cat. no. 550825; BD Biosciences) at room temperature for 30 min in the dark. Finally, cell cycle distribution and DNA content were analyzed using a BD FACSCanto™ Flow Cytometer (version 5.0; BD Biosciences).
Cell migration and invasion assay
A cell migration assay was performed using transwell insert chambers (8-µm pore size; Corning Incorporated, Corning, NY, USA). A total of ~5×104 FaDu and Cal-27 cells in 200 µl serum-free RPMI-1640 medium were seeded into the upper chamber of the transwell chamber. Following this, 600 µl RPMI-1640 medium containing 10% FBS, as a chemoattractant, was added to the lower chamber. After incubation for 36 h, non-migrating cells were removed with a cotton swab, and the migrating cells on the lower side of the insert filter were fixed using 100% methanol and stained with 0.1% crystal violet for 20 min at room temperature. The number of migrated cells was counted in five random visual fields and imaged using a light microscope (×100, DP73; Olympus Corporation, Tokyo, Japan). The invasion assay was performed according to the same method of migration, but the transwell insert chambers were coated with Matrigel (BD Biosciences).
Statistical analysis
The data are expressed as mean ± standard deviation and were analyzed using SPSS 14.0 (SPSS, Inc., Chicago, IL, USA). Overall survival (OS) time was defined from the date of the surgical treatment to the date of mortality or the most recent follow-up. Survival curves were assessed through the Kaplan-Meier method, and statistical analysis was performed using the log-rank test. The difference between two groups was evaluated using a two-tailed Student's t-test. Differences between multiple groups were assessed using one-way analysis of variance with Dunnett's post hoc test. The association between GHET1 and clinicopathological characteristics was analyzed using a χ2 test. P<0.05 was considered to indicate a statistically significant difference.
Results
GHET1 was upregulated in HNC tissues
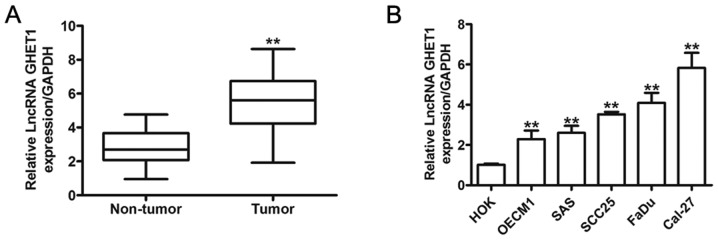
To determine whether GHET1 was dysregulated in HNC, GHET1 expression in HNC tissues and pair-matched adjacent normal tissues was analyzed using RT-qPCR. As depicted in Fig. 1A, expression of GHET1 was significantly increased in HNC tissues, compared with the adjacent normal tissues (P<0.01). The aberrant expression level of GHET1 in the cancer tissues indicated that GHET1 may serve a notable role in the development and progress of HNC. Furthermore, the expression of GHET1 was assessed in HNC cell lines (FaDu, OECM1, SAS and Cal-27) and normal HOK. These results demonstrated that the expression of GHET1 was significantly higher in HNC cells than that in HOK cells (P<0.01; Fig. 1B). These results indicated that GHET1 contributed to the development and progression of HNC.
Figure 1.
Expression of lncRNA GHET1 is upregulated in HNC tissues and cell lines. (A) The relative expression of GHET1 was increased in HNC tissues, compared with the adjacent non-cancer tissues, as determined by reverse transcription-quantitative polymerase chain reaction. (B) Analysis of lncRNA GHET1 expression in FaDu, OECM1, SCC25, SAS and Cal-27 cells, and normal HOK cells. **P<0.01 vs. non-tumor or HOK cells. HOK, human oral keratinocytes; lncRNA, long non-coding RNA; GHET1, gastric carcinoma highly expressed transcript 1; HNC, head and neck cancer.
Association between GHET1 expression and clinical characteristics and prognosis
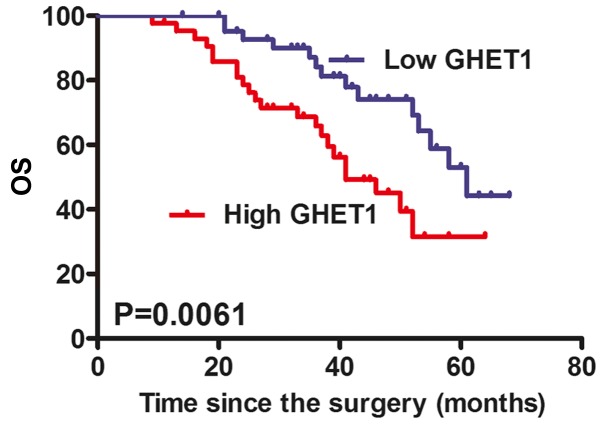
To assess whether GHET1 expression was associated with clinical pathological parameters and prognosis of HNC, the 86 patients with HNC were classified into two groups according to the median of GHET1 in tumor tissues: The high GHET1 group (n=43, >median value); and the low GHET1 group (n=43, ≤ median value). Notably, high expression is significantly associated with lymph node metastasis and advanced Tumor-Node-Metastasis stage (TNM; Table I). Other clinical parameters, including sex, age, smoking status, tumor size and differentiation, were determined not to be significantly associated with GHET1. Furthermore, it was determined that patients with high GHET1 expression exhibited reduced OS time (median survival, 41 months), compared with those with low GHET1 expression (median survival, 61 months) (Fig. 2).
Table I.
Association between clinicopathological characteristics and GHET1 expression.
| GHET1 expression, n | |||
|---|---|---|---|
| Variables | High | Low | P-value |
| Total | 43 | 43 | |
| Age, years | 0.6610 | ||
| <60 | 19 | 16 | |
| ≥0 | 24 | 27 | |
| Sex | 0.4838 | ||
| Male | 29 | 32 | |
| Female | 14 | 11 | |
| Smoker | 0.5169 | ||
| Yes | 22 | 18 | |
| No | 21 | 25 | |
| Differentiation | 0.1280 | ||
| Well or moderate | 20 | 28 | |
| Poor | 23 | 15 | |
| Tumor size, cm | 0.1297 | ||
| <5 | 16 | 24 | |
| ≥5 | 27 | 19 | |
| Lymph node metastasis | 0.0019 | ||
| No | 18 | 33 | |
| Yes | 25 | 10 | |
| TNM stage | 0.0171 | ||
| I–II | 16 | 28 | |
| III | 27 | 15 | |
TNM, Tumor-Node-Metastasis; GHET1, gastric carcinoma highly expressed transcript 1.
Figure 2.
High lncRNA GHET1 expression predicts poor survival in patients with head and neck cancer. Kaplan-Meier OS curves depicted patients with high lncRNA GHET1 expression have reduced OS times compared with those with low lncRNA GHET1 expression. OS, overall survival; lncRNA, long non-coding RNA; GHET1, gastric carcinoma highly expressed transcript 1.
GHET1 regulates HNC cell proliferation by affecting the cell apoptosis and cycle
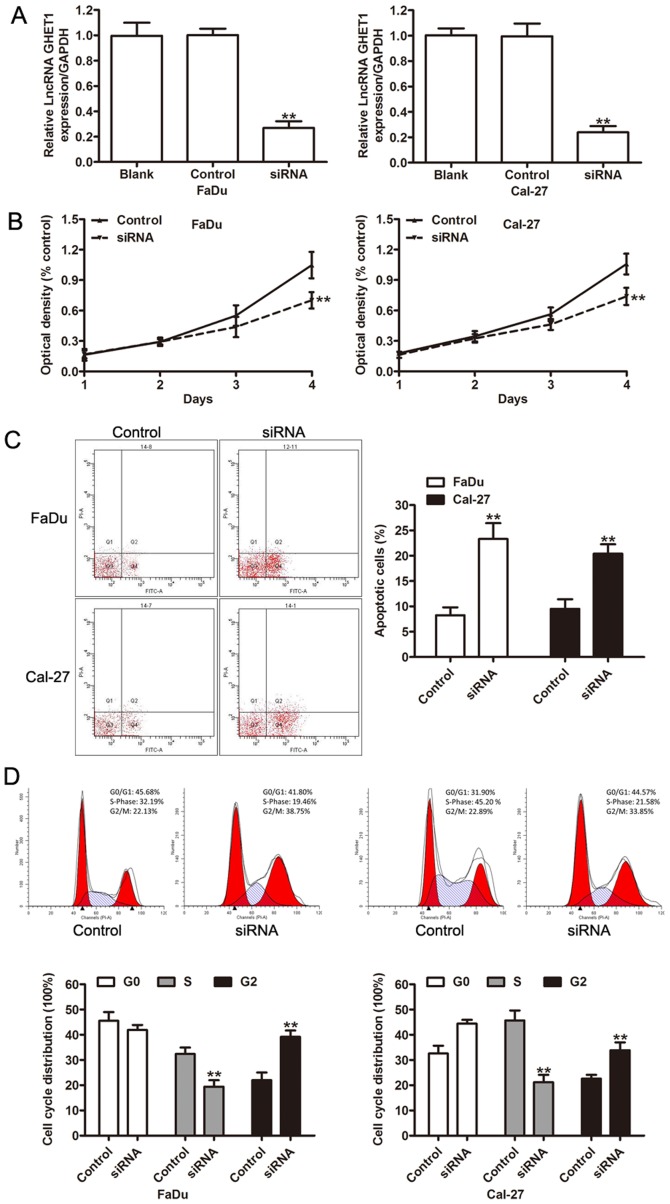
Since GHET1 may act as an oncogene in HNC, the cellular functions of GHET1 was investigated by employing specific siRNA oligomers. GHET1-specific siRNAs were transfected into FaDu and Cal-27 cells knockdown efficiency was confirmed using RT-qPCR (Fig. 3A). To investigate whether GHET1 affected the growth of HNC cells, cell proliferation was assessed using a CCK-8 assay. The results of this analysis demonstrated that downregulation of GHET1 significantly decreased the growth of HNC cells, compared with control cells (P<0.01; Fig. 3B). To investigate whether cell apoptosis and cell cycle progression contributed to the proliferation of HNC cells in vitro, the effects of GHET1 on cell apoptosis and the cell cycle distribution of FaDu and Cal-27 cells were analyzed. A flow cytometry assay revealed that the apoptosis of FaDu and Cal-27 cells was significantly enhanced following knockdown of GHET1 (P<0.01; Fig. 3C). Furthermore, the percentage of cells in S phase significantly decreased upon GHET1 silencing in the two cell lines, whereas the percentage of cells in G2 phase significantly increased (P<0.01; Fig. 3D). These results indicated that GHET1 regulated cell proliferation by mediating cell apoptosis and the cell cycle.
Figure 3.
lncRNA GHET1 regulates the proliferation of head and neck cancer cells in vitro. (A) Reverse transcription-quantitative polymerase chain reaction analysis was performed to detect the expression of lncRNA GHET1 in FaDu and Cal-27 cells transfected with negative control or GHET1 siRNA. (B) Cell proliferation was measured using a Cell Counting kit-8 assay in FaDu and Cal-27 cells transfected with negative control or GHET1 siRNA at an indicated time. (C) Representative flow cytometry analysis depicting that knockdown of lncRNA GHET1 induces apoptosis using annexin V-FITC and PI staining. (D) Flow cytometry analysis demonstrated that lncRNA GHET1 depletion significantly decreased the proportion of cancer cells in S phase. **P<0.01 vs. control. lncRNA, long non-coding RNA; GHET1, gastric carcinoma highly expressed transcript 1; siRNA, small interfering RNA; OD, optical density; FITC, fluorescein isothiocyanate; PI, propidium iodide.
Knockdown of GHET1 inhibits cell migration and invasion
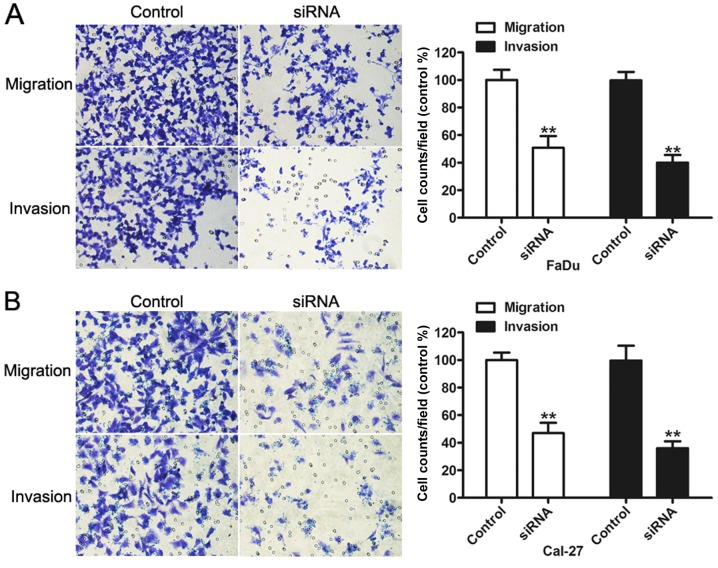
A crucial process of cancer progression is the degradation of the extracellular matrix and penetration of the basement membrane (22). To assess whether GHET1 regulated the migratory and invasive ability of HNC cells, migration and invasion assays were performed. Migration and invasion assays revealed that the ablation of endogenous GHET1 significantly inhibited HNC cell migration and invasiveness, compared with the control (P<0.01; Fig. 4A and B). These data indicated that GHET1 knockdown may decrease malignant gastric cancer cell mobility.
Figure 4.
Knockdown of lncRNA GHET1 obstructs the migration of head and neck cancer cells. (A) Migration and invasion assays revealing that lncRNA GHET1 silencing significantly decreased the number of migrated and invaded FaDu cells. (B) Migration and invasion assays demonstrating that lncRNA GHET1 silencing significantly decreased the number of migrated and invaded Cal-27 cells. **P<0.01 vs. control. lncRNA, long non-coding RNA; GHET1, gastric carcinoma highly expressed transcript 1; siRNA, small interfering RNA.
Discussion
Previous studies demonstrated that lncRNAs serve critical roles in the development and progression of HNC; aberrant lncRNA expression is associated with metastasis, recurrence and poor prognosis in patients with HNC (6,13,17). The results of recent studies have indicated that GHET1 was overexpressed in several human cancer types, and dysregulation of GHET1 was associated with tumor size, metastasis and poor prognosis in gastric carcinoma (18–20). In addition, GHET1 has also been demonstrated to have an increased expression in bladder cancer tissues, which is associated with tumor size, TNM status and poor survival rates (19). In the present study, the expression of GHET1 in HNC tissues was measured and its clinical implications were investigated. In accordance with the aforementioned data, GHET1 was determined to be significantly upregulated in HNC tissues, as well as in the HNC cell lines. In addition, patients with high GHET1 expression levels exhibited an advanced TNM stage, more lymph node metastasis and reduced OS times, compared with those with low GHET1 expression; however, tumor size was not observed to be associated with the expression of GHET1. These data indicated that GHET1 may promote the malignant progression of HNC.
To understand the biological function of GHET1 in HNC progression, a range of in vitro assays were performed. A previous study demonstrated that GHET1 promoted cellular proliferation by increasing c-Myc mRNA stability and expression (18). Inhibition of GHET1 impaired proliferation of bladder cancer cells by inducing G0/G1 arrest and bladder cell invasion by reversing the epithelial-mesenchymal-transition (EMT) (19). Results of a recent study revealed that GHET1 expression was significantly upregulated in colorectal cancer, and the downregulation of GHET1 suppressed malignant phenotypes by reversing EMT (20). The in vitro data of the present study demonstrated that siRNA-mediated knockdown of GHET1 resulted in significant decrease in cell viability, the proportion of the cells in S phase and a significant increase in the percentage of apoptotic HNC cells.
Evidence has demonstrated that metastasis is a key biological process that contributes to disease relapse and the progression of tumors (20,23). Since high GHET1 expression was positively associated with lymph node metastasis, the effects of GHET1 on HNC cells in vitro were evaluated. The experiments demonstrated that knockdown of GHET1 notably inhibited cell migration and invasion in HNC cells.
In summary, the data presented in the present study provide evidence that upregulated GHET1 promoted the progression of HNC by mediating the proliferation and migration of HNC cells. Analysis indicated that high GHET1 expression was significantly associated with lymph node metastasis and advanced TNM stage; however, further studies are required to confirm these data and reveal the potential molecular mechanism underlying GHET1 in regulating HNC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
HL and YW performed the experiments and contributed to the writing of the manuscript. YW conceived of the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Mehanna H, Paleri V, West CM, Nutting C. Head and neck cancer-Part 1: Epidemiology, presentation, and prevention. Clin Otolaryngol. 2011;36:65–68. doi: 10.1111/j.1749-4486.2010.02231.x. [DOI] [PubMed] [Google Scholar]
- 2.Mehanna H, West CM, Nutting C, Paleri V. Head and neck cancer-Part 2: Treatment and prognostic factors. BMJ. 2010;341:c4690. doi: 10.1136/bmj.c4690. [DOI] [PubMed] [Google Scholar]
- 3.Xiong W, Zeng ZY, Xia JH, Xia K, Shen SR, Li XL, Hu DX, Tan C, Xiang JJ, Zhou J, et al. A susceptibility locus at chromosome 3p21 linked to familial nasopharyngeal carcinoma. Cancer Res. 2004;64:1972–1924. doi: 10.1158/0008-5472.CAN-03-3253. [DOI] [PubMed] [Google Scholar]
- 4.Song Y, Li X, Zeng Z, Li Q, Gong Z, Liao Q, Li X, Chen P, Xiang B, Zhang W, et al. Epstein-Barr virus encoded miR-BART11 promotes inflammation-induced carcinogenesis by targeting FOXP1. Oncotarget. 2016;7:36783–36799. doi: 10.18632/oncotarget.9170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmitz S, Machiels JP. Molecular biology of squamous cell carcinoma of the head and neck: Relevance and therapeutic implications. Expert Rev Anticancer Ther. 2010;10:1471–1484. doi: 10.1586/era.10.115. [DOI] [PubMed] [Google Scholar]
- 6.Yang L, Tang Y, He Y, Wang Y, Lian Y, Xiong F, Shi L, Zhang S, Gong Z, Zhou Y, et al. High Expression of LINC01420 indicates an unfavorable prognosis and modulates cell migration and invasion in nasopharyngeal carcinoma. J Cancer. 2017;8:97–103. doi: 10.7150/jca.16819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gutschner T, Diederichs S. The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pickard MR, Williams GT. The hormone response element mimic sequence of GAS5 lncRNA is sufficient to induce apoptosis in breast cancer cells. Oncotarget. 2016;7:10104–10106. doi: 10.18632/oncotarget.7173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu X, Zhou C, Li R, Liang Z, Zhai W, Zhao L, Zhang S. Critical role for the long non-coding RNA AFAP1-AS1 in the proliferation and metastasis of hepatocellular carcinoma. Tumour Biol. 2016;37:9699–9707. doi: 10.1007/s13277-016-4858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng X, Hu H, Li S. High expression of lncRNA PVT1 promotes invasion by inducing epithelial-to-mesenchymal transition in esophageal cancer. Oncol Lett. 2016;12:2357–2362. doi: 10.3892/ol.2016.5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin Y, Cui Z, Li X, Jin X, Peng J. Upregulation of long non-coding RNA PlncRNA-1 promotes proliferation and induces epithelial-mesenchymal transition in prostate cancer. Oncotarget. 2017;8:26090–26099. doi: 10.18632/oncotarget.15318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu J, Liu Y, Gong Z, Zhang S, Guo C, Li X, Tang Y, Yang L, He Y, Wei F, et al. Overexpression long non-coding RNA LINC00673 is associated with poor prognosis and promotes invasion and metastasis in tongue squamous cell carcinoma. Oncotarget. 2017;8:16621–16632. doi: 10.18632/oncotarget.14200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fayda M, Isin M, Tambas M, Guveli M, Meral R, Altun M, Sahin D, Ozkan G, Sanli Y, Isin H, et al. Do circulating long non-coding RNAs (lncRNAs) (LincRNA-p21, GAS 5, HOTAIR) predict the treatment response in patients with head and neck cancer treated with chemoradiotherapy? Tumour Biol. 2016;37:3969–3978. doi: 10.1007/s13277-015-4189-1. [DOI] [PubMed] [Google Scholar]
- 14.Li X, Cao Y, Gong X, Li H. Long noncoding RNAs in head and neck cancer. Oncotarget. 2017;8:10726–10740. doi: 10.18632/oncotarget.12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong L, Zhou X, Wu Y, Wang Y, Chen L, Li P, Liu S, Sun S, Ren Y, Mei M, et al. Targeting HOTAIR induces mitochondria related apoptosis and inhibits tumor growth in head and neck squamous cell carcinoma in vitro and in vivo. Curr Mol Med. 2015;15:952–960. doi: 10.2174/1566524016666151123112716. [DOI] [PubMed] [Google Scholar]
- 16.Wu T, Qu L, He G, Tian L, Li L, Zhou H, Jin Q, Ren J, Wang Y, Wang J, et al. Regulation of laryngeal squamous cell cancer progression by the lncRNA H19/miR-148a-3p/DNMT1 axis. Oncotarget. 2016;7:11553–11566. doi: 10.18632/oncotarget.7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia LF, Wei SB, Gan YH, Guo Y, Gong K, Mitchelson K, Cheng J, Yu GY. Expression, regulation and roles of miR-26a and MEG3 in tongue squamous cell carcinoma. Int J Cancer. 2014;135:2282–2293. doi: 10.1002/ijc.28667. [DOI] [PubMed] [Google Scholar]
- 18.Yang F, Xue X, Zheng L, Bi J, Zhou Y, Zhi K, Gu Y, Fang G. Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability. FEBS J. 2014;281:802–813. doi: 10.1111/febs.12625. [DOI] [PubMed] [Google Scholar]
- 19.Li LJ, Zhu JL, Bao WS, Chen DK, Huang WW, Weng ZL. Long noncoding RNA GHET1 promotes the development of bladder cancer. Int J Clin Exp Pathol. 2014;7:7196–7205. [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou J, Li X, Wu M, Lin C, Guo Y, Tian B. Knockdown of Long Noncoding RNA GHET1 Inhibits Cell Proliferation and Invasion of Colorectal Cancer. Oncol Res. 2016;23:303–309. doi: 10.3727/096504016X14567549091305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson SK, Ruszkiewicz AR, Jamieson GG, Esterman A, Watson DI, Wijnhoven BP, Lamb PJ, Devitt PG. Improving the accuracy of TNM staging in esophageal cancer: A pathological review of resected specimens. Ann Surg Oncol. 2008;15:3447–3458. doi: 10.1245/s10434-008-0155-0. [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen DX, Bos PD, Massague J. Metastasis: From dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9:274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.