Abstract
Long non-coding RNAs (lncRNAs) have been investigated as a novel class of regulators of cellular processes, including cell growth, apoptosis and carcinogenesis. lncRNA BRAF-activated non-protein coding RNA (BANCR) has recently been revealed to be involved in tumorigenesis of numerous types of cancer, including papillary thyroid carcinoma, melanoma, non-small cell lung cancer and colorectal cancer. However, the expression profiles and biological relevance of lncRNA BANCR in hepatocellular carcinoma (HCC) has not yet been reported. In the present study, the expression level of BANCR in tumor tissues and para-cancerous tissues was determined by reverse transcription-quantitative polymerase chain reaction in patients with hepatitis B virus (HBV)-associated HCC, and its association with clinicopathological characteristics of patients was analyzed. The results demonstrated that the expression level of BANCR was significantly reduced in tumor tissues in comparison with in para-cancerous tissues (P<0.001). Furthermore, the present study demonstrated that BANCR expression level was closely associated with serum α-fetoprotein levels (P<0.01) and HCC tumor number (P<0.05). To the best of our knowledge, these results revealed for the first time that BANCR downregulated in patients with HBV-associated HCC and BANCR expression level may be a potential valuable diagnosis and therapeutic biomarker in HCC.
Keywords: BRAF-activated non-protein coding RNA, hepatocellular carcinoma, reverse transcription-quantitative polymerase chain reaction
Introduction
Hepatocellular carcinoma (HCC) is one of the most common and aggressive human malignancies worldwide (1). The majority of the cancer burden (85%) is present in developing countries, with a particularly high prevalence in regions where hepatitis B virus (HBV) infection is endemic, including South-East Asia and Sub-Saharan Africa (2). The majority of HCC remains incurable once it has become metastatic and has a poor prognosis, although there have previously been advances in cancer treatment with respect to surgery, chemotherapy and biological therapy (3). The major causes of HCC are viral infections, alcohol and tobacco use and HBV infection (4). Despite efforts made to identify appropriate prognostic markers for HCC, including primary tumor size, elevated α-fetoprotein (AFP) levels and gene expression markers in the primary tumor, these methods have not proven adequate to predict the prognosis of all patients with HCC (5,6). Therefore, a reliable clinical biomarker for HCC diagnosis and prediction of clinical outcome is urgently required.
Long non-coding RNAs (lncRNAs) are >200 nucleotides in length and do not code for proteins but interact with them (7). Although lncRNAs are not as well characterized as small non-coding microRNAs, lncRNAs are critical in the regulation of mechanisms underlying cellular processes, including stem cell pluripotency, cell growth, cell cycle, apoptosis, metabolism and cancer migration (8–13). Functional lncRNAs can be used for cancer diagnosis and prognosis, in addition to serving as potential therapeutic targets. Tang et al (14) identified three lncRNAs, RP11-160H22.5, XLOC_014172 and LOC149086, which were upregulated in HCC tissues in comparison with in the cancer-free controls of their study. Furthermore, XLOC_014172 and LOC149086 were confirmed to be highly expressed in patients with metastatic HCC (14). The majority of patients demonstrated a decreased expression level of the three lncRNAs following surgery. In 2013, Xu et al (15) identified a lncRNA, lncRNA-LALR1, which participated in liver regeneration. In addition, in 2014, Yuan et al (16) observed a lncRNA, lncRNA-ATB, that was upregulated in HCC tissues and modulated tumorigenesis and tumor progression. In 2014, Wang et al (17) revealed that the oncofetal long noncoding RNA PVT1 promoted proliferation and stem cell-like properties of HCC cells.
Functional lncRNAs are considered to be promising candidates for future cancer diagnosis and therapeutic strategies (18). BRAF-activated non-protein coding RNA (BANCR) is a recurrently overexpressed, previously unannotated 693-baspair transcript on chromosome 9 with a potential functional role in melanoma cell migration (19,20). BANCR is strongly associated with V600EBRAF, the most frequent mutation type of the BRAF gene. Furthermore, high frequencies of V600EBRAF mutations are detected in malignant melanoma (70%), papillary thyroid cancer (36–53%) and colorectal cancer (CRC; 5–22%) (21). In 2014, Li et al (22) demonstrated that BANCR promoted proliferation in malignant melanoma by regulating the mitogen-activated protein kinase signaling pathway activation. Wang et al (23) revealed that BANCR contributed to cell proliferation and activated autophagy in papillary thyroid carcinoma cells. In a study investigating CRC, Guo et al (24) demonstrated that BANCR contributed to colorectal cancer migration by inducing epithelial-mesenchyme transition and the results suggested the potential application of BANCR in the therapeutic treatment of CRC; however, the expression pattern and biological functions of BANCR in HCC remain unknown. In the present study, we investigated the expression level of BANCR in HCC tumor tissues by performing reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The results revealed that BANCR expression level is significantly lower in HCC tumors compared with in para-cancerous tissues. In the tumors tissues samples, lower BANCR expression level was closely associated with serum AFP levels and HCC tumor number.
Materials and methods
Patients and specimens
Patient's data were accessed from the databank of the Department of Hepatobiliary Surgery of Beijing 302 Hospital (Beijing, China) of 46 consecutive patients (39 male and 7 female; mean age, 52.2±10.1 years) with HBV-associated HCC. The patients enrolled in the present study underwent surgery at Beijing 302 Hospital between October 2013 and March 2015. None of the patients had received preoperative chemotherapy or radiation therapy prior to surgery. All the diagnoses of HCC were histopathologically confirmed. The present study was approved by the Ethics Committee of Beijing 302 Hospital. Written informed consent was obtained from all patients prior to collection of tumor tissues and para-cancerous liver tissues samples. The resected tumor tissues and para-cancerous tissue samples were immediately snap-frozen in RNA ladder following resection and stored in the tissue bank until analysis. All the clinical data were collected by doctors, and the experimental operators were blinded to the clinical data.
RNA preparation and RT-qPCR
Total RNA from frozen HCC tissues and para-cancerous tissues samples (n=46) was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer's protocol. The RNA integrity was evaluated using a NanoDrop 1000 spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc.). cDNA from all samples was synthesized from 50 ng total RNA from each sample. BANCR expression levels were quantified by RT-qPCR, which was performed using the ABI7500 system (Applied Biosystems; Thermo Fisher Scientific, Inc.) and Thermo Fisher Scientific Maxima SYBR® Green qPCR Master kit (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Briefly, reactions were performed in a 20 µl mixture containing 100 ng cDNA template, 10 µl 2X PCR Mix and 0.5 µl each of forward and reverse primers. The thermocycling conditions used were as follows: 95°C for 10 min, followed by 25 cycles of 95°C for 10 sec, 60°C for 30 sec and 72°C for 30 sec. RT-PCR was performed in duplicate for each sample. The expression level of GAPDH was also detected as the endogenous control, and all the samples were normalized to human GAPDH. The primer sequences used in the present study were as follows: BANCR forward, 5′-ACAGGACTCCATGGCAAACG-3′ and reverse, 5′-ATGAAGAAAGCCTGGTGCAGT-3′; and GAPDH forward, 5′-CAGCCTCAAGATCATCAGCA-3′ and reverse, 5′-TGTGGTCATGAGTCCTTCCA-3′. The median from experiments performed in triplicate was used to determine relative lncRNA concentrations (ΔCq=Cq median lncRNAs-Cq median GAPDH). Expression fold changes were calculated using the 2−ΔΔCq method (25).
Statistical analysis
Statistical significances between groups were determined by two-tailed Student's t-test. A receiver operating characteristics (ROC) curve was plotted to determine how well the expression level of BANCR discriminated between HBV HCC tumor tissues and para-cancerous tissues. The association between BANCR expression level and clinicopathological characteristics was analyzed using one-way analysis of variance with Bonferroni correction. All statistical analyses were performed using SPSS version 13.0 (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
lncRNA BANCR is downregulated in HBV-associated HCC tissues compared with in para-cancerous tissues
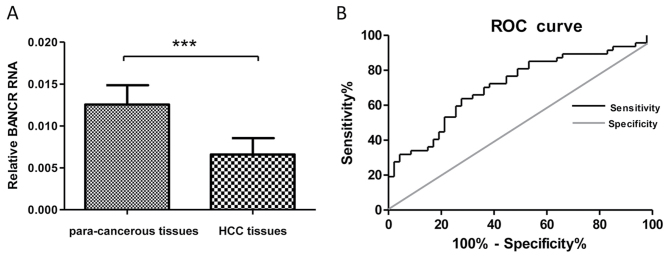
To assess the potential clinical significance of BANCR, its expression levels in HCC tissues and para-cancerous tissue samples were analyzed. Firstly, the expression level of BANCR in HBV HCC tissues samples and para-cancerous tissues samples were determined by RT-qPCR. The results demonstrated that the expression levels of BANCR were markedly decreased in HCC tissues compared with in para-cancerous tissues (P<0.001; Fig. 1A). Subsequently, ROC analysis was used to evaluate the suitability of BANCR expression level to discriminate between the tumor and control tissue samples. Total area under the curve (AUC) for BANCR was 0.76 (Fig. 1B), which suggested that BANCR has adequate sensitivity and specificity to discriminate between HCC and para-cancerous tissue samples. These results revealed that the expression levels of BANCR significantly decreased in HBV-associated HCC tissues compared with in para-cancerous tissue samples.
Figure 1.
Clinical significance of the expression level of lncRNA BANCR in patients with HBV-associated HCC. (A) Lower relative BANCR lncRNA expression levels were detected in HCC tissues compared with in para-cancerous tissues in patients with HBV HCC. (B) The area under the ROC curve was 0.76 in distinguishing hepatocellular carcinoma vs. para-cancerous tissues. ***P<0.001. lncRNA, long non-coding RNA; BANCR, BRAF-activated non-protein coding RNA; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; ROC, receiver operating characteristics.
lncRNA BANCR expression level is associated with the AFP levels and tumor numbers in patients with HBV-associated HCC
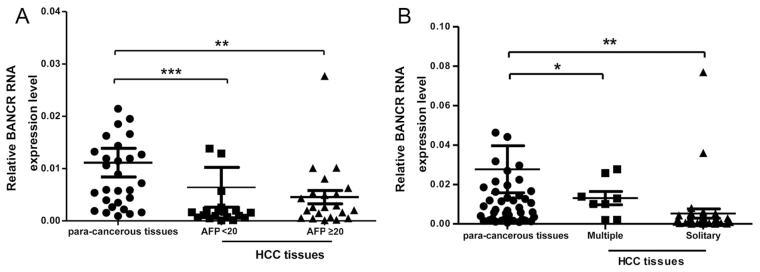
To investigate whether the expression levels of lncRNA BANCR in the HBV-associated HCC tissues are associated with the disease clinicopathological parameters, the present study analyzed the expression profiles of BANCR in tumor tissue samples according to AFP levels and tumors numbers. Since serum AFP level is a critical tumor biomarker for patients with HCC, the evidence resulted in the investigation of whether there is any association between BANCR expression level and serum AFP concentration status in patients with HBV-associated HCCs. As presented in Fig. 2A, the BANCR expression level was lower in the tissue samples with serum AFP ≥20 ng/ml compared with in tissue samples that had serum AFP <20 ng/ml from patients with HBV HCC. In addition, the present study compared the BANCR expression level and solitary or multiple tumor numbers in patients with HBV-associated HCC (Fig. 2B); these results revealed that the BANCR expression level and tumor numbers were positively associated in these patients.
Figure 2.
lncRNA BANCR expression level is associated with serum AFP value and number of tumors. (A) Comparison of the relative expression levels of BANCR with serum AFP value <20 ng/ml and serum AFP value ≥20 ng/ml groups in patients with HBV-associated HCC. (B) Comparison of the relative expression levels of BANCR from solitary and multiple groups in patients with HBV-associated HCC. ***P<0.001, **P<0.01, *P<0.05. lncRNA, long non-coding RNA; BANCR, BRAF-activated non-protein coding RNA; AFP, α-fetoprotein; HBV, hepatitis B virus; HCC, hepatocellular carcinoma.
Association between BANCR expression level and the clinicopathological parameters of patients with HBV-associated HCC
To further analyze the association between lncRNA expression level and various clinicopathological parameters of patients with HBV HCC, 46 patients were divided into high (n=23) and low (n=23) BANCR expression level groups, according to the mean value of the expression levels of BANCR in tumor tissue samples. As presented in Table I, clinicopathological factors were investigated between the two groups and the low BANCR expression level group demonstrated significantly higher serum AFP levels (P=0.008) and more advanced tumor numbers (P=0.047) compared with the high BANCR expression level group. However, no significant association was revealed between BANCR expression level and other clinicopathological features, including age, gender, tumor size, clinical stage and liver cirrhosis (P>0.05).
Table I.
Association between BANCR expression and clinicopathological characteristics of patients with HBV-associated HCC.
| BANCR expression level, n | ||||
|---|---|---|---|---|
| Parameters | Total, n | Low | High | P-value |
| Gender | 0.665 | |||
| Male | 40 | 19 | 21 | |
| Female | 6 | 4 | 2 | |
| Age | 0.475 | |||
| <60 years | 36 | 17 | 19 | |
| ≥60 years | 10 | 6 | 4 | |
| Tumor size | 0.767 | |||
| <5 cm | 25 | 12 | 13 | |
| ≥5 cm | 21 | 11 | 10 | |
| AFP | 0.008b | |||
| <20 ng/ml | 21 | 15 | 6 | |
| ≥20 ng/ml | 25 | 8 | 17 | |
| Histological grade | 0.349 | |||
| Well | 41 | 20 | 21 | |
| Moderately-poorly | 5 | 3 | 2 | |
| TNM stage | 0.491 | |||
| I–II | 35 | 19 | 16 | |
| III | 11 | 4 | 7 | |
| Liver cirrhosis | 0.326 | |||
| Absence | 33 | 15 | 18 | |
| Presence | 13 | 8 | 5 | |
| Tumor number | 0.047a | |||
| Solitary | 38 | 22 | 16 | |
| Multiple | 8 | 1 | 7 | |
summarizes the association between BANCR expression level and clinicopathological features in patients with HBV HCC.
P<0.05
P<0.01. BANCR, BRAF-activated non-protein coding RNA; AFP, α-fetoprotein; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; TNM, tumor-node-metastasis.
Discussion
With advancements in genome wide sequencing and high-resolution microarray technology, more attention has been received by lncRNAs (26–28). A previous study revealed that dysexpression of lncRNAs is associated with numerous types of disease (29). Furthermore, lncRNAs have been identified to perform an important function in the progression of various types of cancer. Numerous well-studied lncRNAs, including Hox transcription antisense RNA (30–32), maternally expressed 3 (33) and LOC285194 (34) have been reported to be strongly associated with survival of patients with cancer; thus, they have been determined to be prognostic factors for specific types of cancer.
LncRNA BANCR was first revealed to have a potential functional role in melanoma cell migration (22). Other previous studies also investigated the expression levels and functions of BANCR in various malignancies. BANCR has been demonstrated to be upregulated in numerous types of solid tumors, including papillary thyroid cancer (23) and CRC (21). Therefore, the present study hypothesized that BANCR also had similar effects on the tumorigenesis, development and progression of HCC. To confirm these hypotheses, the present study firstly determined the BANCR expression levels in 46 pairs of HBV-associated HCC tissues and their para-cancerous tissues by RT-qPCR. HCC para-cancerous tissues always demonstrate liver cirrhosis or chronic inflammations and fibrosis (35); thus, para-cancerous tissues were used as the control in the present study. It was revealed that BANCR was significantly downregulated in HBV-associated HCC and this differs from the overexpression of BANCR observed in melanoma tissue samples. The results of the present study may provide evidence for the tissue specificity of lncRNA. However, the underlying molecular mechanisms of lncRNA BANCR require further investigation. Only HBV-associated HCC patients were included in the present study and it was demonstrated that the expression level of BANCR in HBV-associated HCC tissues were lower compared with in para-cancerous tissues. Furthermore, the ROC curve analysis was performed in order to identify the diagnostic significance of BANCR expression level in HBV-associated HCC and the AUC area was 0.76, revealing a possible diagnostic value of BANCR. However, a considerably larger cohort of patients with HBV-associated HCC is required to better support these findings.
Aberrant expression level of BANCR was previously reported to be involved in the progression of numerous tumors and can be used as a prognostic indicator (36). Subsequently, the present study described the association between BANCR expression level and various clinicopathological parameters. The results also demonstrated positive associations between the BANCR expression level and tumor number and serum AFP value. As a diagnostic indicator, AFP serves an essential role in the diagnosis of HCC and there was a significant association between BANCR expression level and serum AFP in the present study. Thus, BANCR may be a biomarker for the occurrence of HBV-associated HCC. However, no significant association was identified between the BANCR expression level and other clinic pathological features, including age, gender, tumor size, clinical stage and liver cirrhosis. Further in vitro and in vivo experiments are presently being performed to investigate the biological function and molecular basis of lncRNA BANCR expression in patients with HCC.
In conclusion, the results of the present study demonstrated, to the best of our knowledge, for the first time that lncRNA BANCR expression level was significantly lower in HBV-associated HCC tissues, and downregulation of lncRNA BANCR was positively associated with serum AFP level and tumor number in patients with HBV-associated HCC. These results suggested that lncRNA BANCR may be a potential novel diagnosis biomarker for HBV-associated HCC.
Acknowledgements
This work was supported by the funding from Natural Science Foundation of Beijing Municipality (7162185).
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Venook AP, Papandreou C, Furuse J, de Guevara LL. The incidence and epidemiology of hepatocellular carcinoma: A global and regional perspective. Oncologist. 2010;15(Suppl 4):5–13. doi: 10.1634/theoncologist.2010-S4-05. [DOI] [PubMed] [Google Scholar]
- 3.Blum HE. Hepatocellular carcinoma: Therapy and prevention. World J Gastroenterol. 2005;11:7391–7400. doi: 10.3748/wjg.v11.i47.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu YR, Tang RX, Huang WT, Ren FH, He RQ, Yang LH, Luo DZ, Dang YW, Chen G. Long noncoding RNAs in hepatocellular carcinoma: Novel insights into their mechanism. World J Hepatol. 2015;7:2781–2791. doi: 10.4254/wjh.v7.i28.2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan K, Liang XT, Zhang HK, Zhao JJ, Wang DD, Li JJ, Lian Q, Chang AE, Li Q, Xia JC. Characterization of bridging integrator 1 (BIN1) as a potential tumor suppressor and prognostic marker in hepatocellular carcinoma. Mol Med. 2012;18:507–518. doi: 10.2119/molmed.2011.00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tu ZQ, Li RJ, Mei JZ, Li XH. Down-regulation of long non-coding RNA GAS5 is associated with the prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol. 2014;7:4303–4309. [PMC free article] [PubMed] [Google Scholar]
- 7.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 8.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: Past, present and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Xie G, Singh M, Ghanbarian AT, Raskó T, Szvetnik A, Cai H, Besser D, Prigione A, Fuchs NV, et al. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature. 2014;516:405–409. doi: 10.1038/nature13804. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Wang Y, Li J, Zhang Y, Yin H, Han B. CRNDE, a long-noncoding RNA, promotes glioma cell growth and invasion through mTOR signaling. Cancer Lett. 2015;367:122–128. doi: 10.1016/j.canlet.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 11.Leveille N, Melo CA, Rooijers K, Díaz-Lagares A, Melo SA, Korkmaz G, Lopes R, Moqadam Akbari F, Maia AR, Wijchers PJ, et al. Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nat Commun. 2015;6:6520. doi: 10.1038/ncomms7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qiu JJ, Wang Y, Ding JX, Jin HY, Yang G, Hua KQ. The long non-coding RNA HOTAIR promotes the proliferation of serous ovarian cancer cells through the regulation of cell cycle arrest and apoptosis. Exp Cell Res. 2015;333:238–248. doi: 10.1016/j.yexcr.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 13.Cui M, Xiao Z, Wang Y, Zheng M, Song T, Cai X, Sun B, Ye L, Zhang X. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. 2015;75:846–857. doi: 10.1158/0008-5472.CAN-14-1192. [DOI] [PubMed] [Google Scholar]
- 14.Tang J, Jiang R, Deng L, Zhang X, Wang K, Sun B. Circulation long non-coding RNAs act as biomarkers for predicting tumorigenesis and metastasis in hepatocellular carcinoma. Oncotarget. 2015;6:4505–4515. doi: 10.18632/oncotarget.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu D, Yang F, Yuan JH, Zhang L, Bi HS, Zhou CC, Liu F, Wang F, Sun SH. Long noncoding RNAs associated with liver regeneration 1 accelerates hepatocyte proliferation during liver regeneration by activating Wnt/b-catenin signaling. Hepatology. 2013;58:739–751. doi: 10.1002/hep.26361. [DOI] [PubMed] [Google Scholar]
- 16.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al. A long noncoding RNA activated by TGF-b promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 17.Wang F, Yuan JH, Wang SB, Yang F, Yuan SX, Ye C, Yang N, Zhou WP, Li WL, Li W, Sun SH. Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology. 2014;60:1278–1290. doi: 10.1002/hep.27239. [DOI] [PubMed] [Google Scholar]
- 18.Takahashi K, Yan I, Haga H, Patel T. Long noncoding RNA in liver diseases. Hepatology. 2014;60:744–753. doi: 10.1002/hep.27043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flockhart RJ, Webster DE, Qu K, Mascarenhas N, Kovalski J, Kretz M, Khavari PA. BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Res. 2012;22:1006–1014. doi: 10.1101/gr.140061.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCarthy N. Epigenetics. Going places with BANCR. Nat Rev Cancer. 2012;12:451. doi: 10.1038/nrc3307. [DOI] [PubMed] [Google Scholar]
- 21.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 22.Li R, Zhang L, Jia L, Duan Y, Li Y, Bao L, Sha N. Long non-coding RNA BANCR promotes proliferation in malignant melanoma by regulating MAPK pathway activation. PLoS One. 2014;9:e100893. doi: 10.1371/journal.pone.0100893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Guo Q, Zhao Y, Chen J, Wang S, Hu J, Sun Y. BRAF-activated long non-coding RNA contributes to cell proliferation and activates autophagy in papillary thyroid carcinoma. Oncol Lett. 2014;8:1947–1952. doi: 10.3892/ol.2014.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo Q, Zhao Y, Chen J, Hu J, Wang S, Zhang D, Sun Y. BRAF-activated long non-coding RNA contributes to colorectal cancer migration by inducing epithelial-mesenchymal transition. Oncol Lett. 2014;8:869–875. doi: 10.3892/ol.2014.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Ma H, Hao Y, Dong X, Gong Q, Chen J, Zhang J, Tian W. Molecular mechanisms and function prediction of long noncoding RNA. ScientificWorldJournal. 2012;2012:541786. doi: 10.1100/2012/541786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang JY, Lee JC, Chang YT, Hou MF, Huang HW, Liaw CC, Chang HW. Long noncoding RNAs-related diseases, cancers, and drugs. ScientificWorldJournal. 2013;2013:943539. doi: 10.1155/2013/943539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu MX, Chen X, Chen G, Cui QH, Yan GY. A computational framework to infer human disease-associated long noncoding RNAs. PLoS One. 2014;9:e84408. doi: 10.1371/journal.pone.0084408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakagawa T, Endo H, Yokoyama M, Abe J, Tamai K, Tanaka N, Sato I, Takahashi S, Kondo T, Satoh K. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem Biophys Res Commun. 2013;436:319–324. doi: 10.1016/j.bbrc.2013.05.101. [DOI] [PubMed] [Google Scholar]
- 33.Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu WQ, Xie WP, Hou YY. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer. 2013;13:461. doi: 10.1186/1471-2407-13-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qi P, Xu MD, Ni SJ, Huang D, Wei P, Tan C, Zhou XY, Du X. Low expression of LOC285194 is associated with poor prognosis in colorectal cancer. J Transl Med. 2013;11:122. doi: 10.1186/1479-5876-11-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zou H, Shao CX, Zhou QY, Zhu GQ, Shi KQ, Braddock M, Huang DS, Zheng MH. The role of lncRNAs in hepatocellular carcinoma: Opportunities as novel targets for pharmacological intervention. Expert Rev Gastroenterol Hepatol. 2016;10:331–340. doi: 10.1586/17474124.2016.1116382. [DOI] [PubMed] [Google Scholar]
- 36.Fan YH, Ye MH, Wu L, Wu MJ, Lu SG, Zhu XG. BRAF-activated lncRNA predicts gastrointestinal cancer patient prognosis: A meta-analysis. Oncotarget. 2017;24:6295–6303. doi: 10.18632/oncotarget.14061. [DOI] [PMC free article] [PubMed] [Google Scholar]