Abstract
Hepatitis C virus (HCV)-induced human hepatocellular carcinoma (HCC) progression may be due to a complex multi-step processes. The developmental mechanism of these processes is worth investigating for the prevention, diagnosis and therapy of HCC. The aim of the present study was to investigate the molecular mechanism underlying the progression of HCV-induced hepatocarcinogenesis. First, the dynamic gene module, consisting of key genes associated with progression between the normal stage and HCC, was identified using the Weighted Gene Co-expression Network Analysis tool from R language. By defining those genes in the module as seeds, the change of co-expression in differentially expressed gene sets in two consecutive stages of pathological progression was examined. Finally, interaction pairs of HCV viral proteins and their directly targeted proteins in the identified module were extracted from the literature and a comprehensive interaction dataset from yeast two-hybrid experiments. By combining the interactions between HCV and their targets, and protein-protein interactions in the Search Tool for the Retrieval of Interacting Genes database (STRING), the HCV-key genes interaction network was constructed and visualized using Cytoscape software 3.2. As a result, a module containing 44 key genes was identified to be associated with HCC progression, due to the dynamic features and functions of those genes in the module. Several important differentially co-expressed gene pairs were identified between non-HCC and HCC stages. In the key genes, cyclin dependent kinase 1 (CDK1), NDC80, cyclin A2 (CCNA2) and rac GTPase activating protein 1 (RACGAP1) were shown to be targeted by the HCV nonstructural proteins NS5A, NS3 and NS5B, respectively. The four genes perform an intermediary role between the HCV viral proteins and the dysfunctional module in the HCV key genes interaction network. These findings provided valuable information for understanding the mechanism of HCV-induced HCC progression and for seeking drug targets for the therapy and prevention of HCC.
Keywords: hepatitis C virus, hepatocellular carcinoma, co-expression, differential co-expression, interaction network
Introduction
Hepatocellular carcinoma (HCC) is one of the most common types of cancer in the world (1), and it was the third leading cause of cancer-associated mortality worldwide in 2013 (2). A number of effective measures, including percutaneous ablation, surgical resection and liver transplantation, have been developed for HCC therapy (3). Although the short-term prognosis of patients with HCC has improved due to advances in early diagnosis and treatments, the long-term prognosis remains poor due to frequent recurrence or metastasis (4).
HCC is a heterogeneous cancer that usually develops in patients with chronic liver disease, particularly those with cirrhosis. The progression between chronic liver disease and HCC is a complex and multistep process. Chronic viral hepatitis caused by hepatitis C virus (HCV) infection has become one of the main causes of HCC. A typical HCV-induced HCC progression may undergo the following five successive stages: Normal; cirrhosis; dysplasia; early HCC; and advanced HCC (5).
In the development from hepatic lesions to HCC, not only does the expression level of numerous genes change, but dynamic changes may also appear in the regulation of different genes and protein-protein interaction (6). Classical approaches, including identification of differentially expressed genes, may not reveal the complex interactions and functional association among genes in biological processes. Alternatively, gene co-expressed network analysis may provide a powerful approach for elucidating the co-regulation and interaction between proteins in biological processes (7). Comparing with identification of differentially expressed genes network-based analysis may provide valuable information for understanding complex interaction among genes in biological processes (7,8). At present, a number of studies have identified the altered gene co-expression associated with tumors (9–11).
HCV is a type of single-stranded RNA virus that replicates in the cytoplasm of host hepatocytes. HCV viral proteins are mainly divided into two classes: Structural proteins (CORE, envelope proteins 1 and 2 and p7) and non-structural (NS) proteins (NS2, NS3, NS4B, NS5A and NS5B) (12). Multiple important HCV viral proteins, including CORE, NS3, NS4B and NS5A, were identified to target important cancer-associated proteins. These proteins can promote cell growth and cell cycle deregulation and destroy the stable structure of the host cell genome. These viral proteins were revealed to potentiate oncogenic transformation (12) and perform crucial roles in HCC (13).
However, the exact role of HCV in HCC progression remains to be determined. Therefore, studying the association between the deregulated biological networks and HCV may help to understand the molecular mechanisms of HCC development.
Materials and methods
Microarray data and differentially expressed genes
The HCC gene expression dataset GSE6764 (CEL file) was downloaded (14) from the Gene Expression Omnibus database (15). GSE6764 was based on the GPL570 platform: Affymetrix Human Genome U133 Plus 2.0 Array (Affymetrix; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The dataset contained 75 tissue samples representing the stepwise carcinogenic process from pre-neoplastic lesions (cirrhosis and dysplasia) to HCC. The raw sample groups were categorized into five groups: Normal; cirrhosis; dysplasia, including low-grade dysplastic nodules and high-grade dysplastic nodules; early HCC group, including early HCC and extremely early HCC; and advanced HCC group, including advanced HCC and extremely advanced HCC. The numbers of samples contained in these groups were 10, 13, 17, 18 and 17, respectively.
The Robust Multichip Analysis (RMA) algorithm in the ‘affy’ package (16) (http://www.bioconductor.org/packages/3.6/bioc/html/affy.html) in Bioconductor (17) (http://www.bioconductor.org/) was used to perform background correction, quantile normalization and summarization. For multiple probe sets matching to the same gene, the probes with the largest variation were selected. The linear models for microarray and RNA-sequencing data (LIMMA) software package (https://bioconductor.org/packages/release/bioc/html/limma.html, The Walter and Eliza Hall Institute of Medical Research, Melbourne, Australia, Version 3.24.15) (18) in Bioconductor was utilized for linear models and testing for differential expression, and to adjust for multiple testing. t-tests and F-tests were performed on the matrix. The genes with 2-fold expression change (increase or decrease) (P<0.001) and a false discovery rate (FDR) <0.05, compared with the normal group, were selected as differential expression genes (DEGs) in each disease stage. To reveal the expression dynamic of DEGs across all stages, the DEGs in each stage were merged into a DEG set for subsequent analysis.
Analysis of the modules significantly associated with disease progression
In order to identify the modules of highly correlated genes involved in disease progression, Weighted Gene Co-expression Network Analysis (WGCNA) software (https://labs.genetics.ucla.edu/horvath/CoexpressionNetwork/Rpackages/WGCNA/) (19) was implemented in R language (https://cran.r-project.org/, version 3.2.2). First, the WGCNA package computed co-expressed association among genes in the DEG set and inferred the co-expressed networks in 75 samples. Based on a scale-free topology criterion (20), β=12 was selected as the soft threshold power in the present study. Deep split was set as 2, cut height was set as 0.975 and ‘minModuleSize’ was set as 10, and other parameters were set at default levels.
Module eigengene (ME) is the first principle component and explains the majority of information of the module genes. To identify modules that were significantly associated with disease progression, Spearman's rank correlation coefficient was used to analyze the association between the ME of each module and disease stages. Modules that were significantly correlated with disease progression were labeled as candidate modules (absolute correlation coefficient |r|>0.8; P<0.01).
Screening for the key genes significantly associated with disease progression
The present study aimed to identify key genes with functional similarity and high co-expression over all disease developmental stages that were significantly associated with disease progression. The expression pattern of these genes may reflect the trajectory of disease progression. In candidate modules, the genes with absolute gene significance value (GS) >0.75 and modular membership (MM) >0.8 were selected as genes that were highly associated with disease stages. To exclude the gene pairs with lower correlation in all disease stages, the correlation coefficient between all reserved gene pairs were examined. A threshold of |r|=0.85 (P<0.01) was set as the screening criterion. The gene pairs with an absolute correlation coefficient higher than this threshold were retained. The Database for Annotation, Visualization and Integrated Discovery 6.7 (DAVID, https://david-d.ncifcrf.gov/) (21) was utilized to assess the function of these genes. By Gene Ontology (GO) enrichment analysis, the genes with functions associated with disease progression were retained. The heatmap and self-organizing tree algorithm (SOTA) (22) in cogena package (http://www.bioconductor.org/packages/3.2/bioc/html/cogena.html) was implemented for clustering and visualization of expression patterns of the key genes.
Analysis of the interactions among the key genes
GeneMANIA (http://genemania.org/) (23,24) is a web-based tool for the construction of the composite gene-gene functional interaction network and the prediction of protein function, based on multiple networks derived from different databases. The interactions among key genes, including physical and genetic interactions, pathways, co-expression, co-localization, shared protein domain and predicted interactions were analyzed using GeneMANIA.
Differential co-expressed analysis of the key genes
The genes retained as seeds were used to search for highly co-expressed genes (absolute correlation coefficient |r|>0.85, P<0.01) in the DEG set in which seed genes were ruled out in each stage. The software package DiffCorr (https://sourceforge.net/projects/diffcorr/, Version 0.4.1, Slashdot media and dice, Inc., San Jose, CA, USA) (25) in R language was used to search for the differential correlation gene pairs between adjacent stages. Fisher's z-test was used to identify significant differences between adjacent stages. First, the correlation coefficients for each of the two adjacent stages were transformed into z by Fisher's transformation, separately as follows:
Then, differences between the two correlation coefficients were tested using the equation:
na and nb represent the sample size for each of the disease stages for each gene pair. The gene pairs with FDR<0.05 and P<0.001 of differential correlations were considered as differential coexpression between adjacent phases.
Cytoscape software (http://www.cytoscape.org/, version 3.2; Cytoscape Consortium, La Jolla, CA, USA) (26) was used for the construction of a differentially co-expressed network of the key genes.
Construction of interaction networks between HCV viral proteins and products of key genes
Search Tool for the Retrieval of Interacting Genes (STRING) (http://version10.string-db.org/, version 10.0) is an online protein-protein interaction association database curated from experimental interactions of proteins and comprehensive information (27). First, the key genes were directly mapped to the STRING database to get pairwise interactions (combined scores >0.4). Next, the HCV viral proteins and their directly targeted proteins in the key genes set were extracted from the literature and a comprehensive interaction dataset containing 481 interaction pairs between HCV viral proteins and human proteins from yeast two-hybrid experiments (28). Furthermore, the interaction network of the key genes and the interaction network between HCV and their targeted proteins were combined to make an interaction network between HCV viral proteins and key genes, by anchoring the overlapped proteins of these two networks. The interaction network between HCV viral proteins and key genes was visualized using Cytoscape software (26).
Results
DEGs between disease groups and normal group
Subsequent to performing the RMA algorithm for data preprocessing, the LIMMA software package (18) was run to obtain 509, 156, 802 and 1,501 DEGs from the cirrhosis, dysplasia, early HCC and advanced HCC groups, respectively. The DEGs from each disease group were merged into a gene set, termed the DEG set, which contained a total of 1,973 genes.
Identified modules associated with the disease progression. It was hypothesized that there are core gene modules that can exhibit the disease progression dynamic. It is thought that genes with similar expression patterns and similar functions may be regulated by the same mechanisms (29). Therefore, the modules consisting of genes with similar expression patterns and similar functions were expected to be identified via WGCNA (19). The expression profile of the DEG set was used as the input of WGCNA.
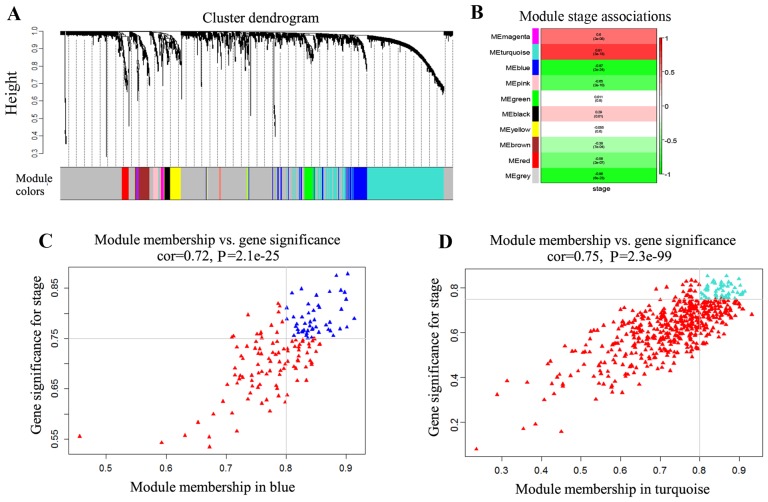
Based on the dissimilarity corresponding to the Topological Overlap Matrix, hierarchical average linkage clustering produced a hierarchical clustering tree (dendrogram) of genes. A total of 10 modules corresponding to the 10 branches of the dendrogram were obtained (Fig. 1A), based on provided parameters. The modules were labeled by 10 colors. The sizes of the 10 modules were ranked between 11 and 1,018 genes. The grey module contained the genes with dissimilar expression patterns.
Figure 1.
Weighted gene co-expression network analysis. (A) A hierarchical clustering tree of the differential expression genes set was constructed by average linkage of hierarchical clusters. Gene set is grouped into 10 co-expression modules. (B) Heatmap of correlation between MEs and disease stages. The first value of each rectangle represents Spearman's correlation coefficient between the MEs and stages. The second value in each rectangle is the P-value. (C) Blue spots are genes with |GS|>0.75 and |MM|>0.8 in the blue module. (D) Turquoise spots are genes with |GS|>0.75 and |MM|>0.8 in the turquoise module. In (C) and (D), red spots represent the genes with |GS|<0.75 or |MM|<0.8 in the modules. ME, module eigengene; GS, gene significance; MM, module membership.
In order to identify the association between the modules and disease progression, MEs were utilized, instead of single genes, as the overall expression levels of the module. The association between MEs and disease progression obtained was presented as a heatmap (Fig. 1B). In the 10 modules, the most strongly associated with the disease stage modules (|r|>0.8; P<0.01), the turquoise and blue modules, were selected as the candidate modules. The turquoise module was positively associated with disease stages, while the blue module was negatively associated with disease stages.
Key genes associated with disease development and progression
Since the genes in modules may not have equivalent contribution to the association of the module with disease stages, GS and MM were used to identify the genes highly associated with disease stages in the two candidate modules. The numbers of retained genes (|GS|>0.75 and |MM|>0.8) were 53 in the turquoise module and 48 in the blue module (Fig. 1C and D). To yield the consistent co-expression of genes in modules across all stages, the pairwise correlation coefficient among genes in the two candidate modules was examined. The genes that weakly correlated (|r|<0.85 or P>0.01) with any gene in the same module were removed. Finally, 44 genes in the turquoise module and 31 genes in the blue module were obtained. The significantly enriched GO terms are shown in Tables I and II.
Table I.
Significant functions of genes retained in the blue module.
| Term | Name | P-value |
|---|---|---|
| GO:0030246 | Carbohydrate binding | 1.78×10−6 |
| GO:0005529 | Sugar binding | 1.96×10−5 |
| GO:0016485 | Protein processing | 7.59×10−4 |
| GO:0051604 | Protein maturation | 9.72×10−4 |
| GO:0006956 | Complement activation | 2.09×10−3 |
| GO:0002541 | Activation of plasma proteins involved in acute inflammatory response | 2.19×10−3 |
| GO:0005509 | Calcium ion binding | 4.40×10−3 |
| GO:0006959 | Humoral immune response | 7.21×10−3 |
| GO:0044421 | Extracellular region part | 7.35×10−3 |
| GO:0005615 | Extracellular space | 7.91×10−3 |
| GO:0051605 | Protein maturation by peptide bond cleavage | 8.50×10−3 |
| GO:0009611 | Response to wounding | 9.71×10−3 |
| GO:0001867 | Complement activation, lectin pathway | 9.71×10−3 |
GO, Gene Ontology.
Table II.
Top 30 significant functions of genes retained in the turquoise module.
| Term | Name | P-value |
|---|---|---|
| GO:0000278 | Mitotic cell cycle | 1.58×10−23 |
| GO:0000279 | M phase | 6.52×10−23 |
| GO:0022403 | Cell cycle phase | 1.68×10−22 |
| GO:0022402 | Cell cycle process | 3.53×10−21 |
| GO:0007067 | Mitosis | 4.37×10−21 |
| GO:0000280 | Nuclear division | 4.37×10−21 |
| GO:0000087 | M phase of mitotic cell cycle | 5.96×10−21 |
| GO:0048285 | Organelle fission | 8.73×10−21 |
| GO:0007049 | Cell cycle | 1.35×10−19 |
| GO:0051301 | Cell division | 6.69×10−19 |
| GO:0015630 | Microtubule cytoskeleton | 1.60×10−15 |
| GO:0005819 | Spindle | 4.93×10−15 |
| GO:0044430 | Cytoskeletal part | 1.94×10−11 |
| GO:0005856 | Cytoskeleton | 1.07×10−10 |
| GO:0007346 | Regulation of mitotic cell cycle | 2.78×10−10 |
| GO:0043228 | Non-membrane-bounded organelle | 6.31×10−10 |
| GO:0043232 | Intracellular non-membrane-bounded organelle | 6.31×10−10 |
| GO:0051726 | Regulation of cell cycle | 1.06×10−9 |
| GO:0007093 | Mitotic cell cycle checkpoint | 1.84×10−9 |
| GO:0007059 | Chromosome segregation | 2.30×10−9 |
| GO:0000075 | Cell cycle checkpoint | 1.82×10−7 |
| GO:0007017 | Microtubule-based process | 4.03×10−7 |
| GO:0000922 | Spindle pole | 2.59×10−6 |
| GO:0000070 | Mitotic sister chromatid segregation | 2.92×10−6 |
| GO:0000819 | Sister chromatid segregation | 3.27×10−6 |
| GO:0005694 | Chromosome | 4.74×10−6 |
| GO:0051276 | Chromosome organization | 5.86×10−6 |
| GO:0007051 | Spindle organization | 7.26×10−6 |
| GO:0007052 | Mitotic spindle organization | 9.09×10−6 |
GO, Gene Ontology.
For those retained genes in the blue module, the GO terms that were significantly enriched were associated with carbohydrate binding (GO:0030246), sugar binding (GO:0005529), protein processing (GO:0016485), protein maturation (GO:0051604), complement activation (GO:0006956) and activation of plasma proteins involved in the acute inflammatory response (GO:0002541). These genes may be involved in energy metabolism and protein synthesis.
The GO terms that the genes retained in the turquoise module significantly enriched included mitotic cell cycle (GO:0000278), M phase (GO:0000279), cell cycle phase (GO:0022403), cell cycle process (GO:0022402), mitosis (GO:0007067), nuclear division (GO:0000280) and cell division (GO:0051301). In previous studies the process of tumorigenesis, cell cycle regulation was significantly altered and mitosis was significantly faster (30). The protein associated with cell cycle and mitosis notably changed along with tumorigenesis (31). In the turquoise module, the functions of these genes involved in the process of mitotic cell cycle and cell division were associated with tumorigenesis.
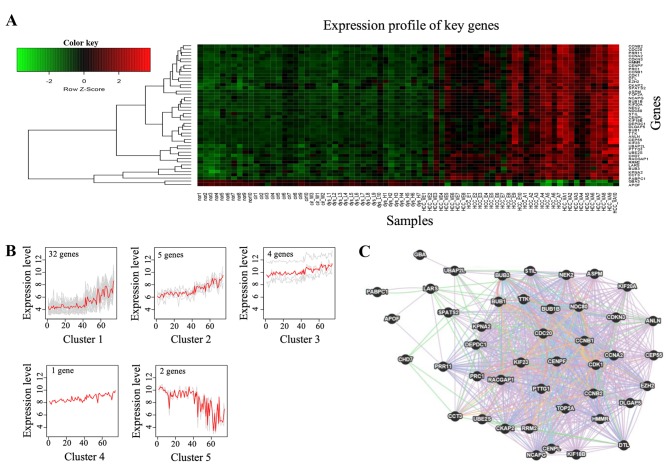
The heatmap showed that the expression of certain genes changed gradually between the normal stage and HCC; specifically, between the dysplasia stage and advanced HCC (Fig. 2A). Furthermore, the expression intensity of those genes in early HCC was in a transitional state between the dysplasia stage and advanced HCC. The expression patterns and clustering of these genes in the turquoise module are shown in Fig. 2B. A total of 44 genes were grouped into five clusters, with an evident trend along with the disease progression between the normal stage and advanced HCC. As the majority of the 44 genes showed gradually increased expression, they were selected as key genes of the core module. The interactions between the 44 key genes were analyzed using GeneMANIA (23,24). The co-localization, co-expression, pathway, shared protein domains, genetic, physical and predictive interactions of the 44 key genes are shown in Fig. 2C.
Figure 2.
Expression patterns of the 44 key genes. (A) Heatmap shows the gradual change in the expression level of key genes between the normal stage and hepatocellular carcinoma. (B) The 44 key genes are grouped into five clusters by self-organizing tree algorithm, and the genes show gradual expression trends along with the disease stages (|r|>0.8, P<0.01). (C) Interaction network of the 44 genes, based on the GeneMANIA online tool.
Altered association of co-expression with key genes in disease progression
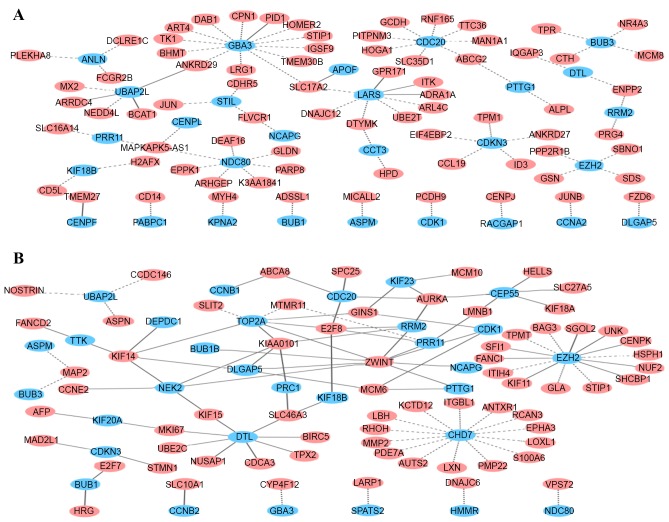
As aforementioned, the changes in expression of the 44 key genes reflected the progression dynamic between the normal stage and advanced HCC. Therefore, it was inferred that co-expressed associations among these genes and others may be altered correspondingly in this process. The differential co-expression genes of 44 key genes in disease progression were investigated. The results showed that the association of co-expression of 9 gene pairs was enhanced (P<0.01, FDR<0.05), while for 76 gene pairs it was reduced (P<0.01, FDR<0.05), between the normal and cirrhosis stages (Fig. 3A). In the cirrhosis-dysplasia stage, there were only 4 pairs of differentially co-expressed genes. In total, 103 pairs of genes were differentially co-expressed in dysplasia-early HCC stages. Among those, the correlations of the 32 gene pairs demonstrated either significant decrease, including the changes from strong correlation to weak correlation or to no correlation, or a transformation from positive correlation to negative correlation (the difference of the correlation coefficients in the adjacent phases: r2-r1<0; P<0.01; FDR<0.05). By contrast, the 71 gene pairs exhibited either significant increase in correlation, including from weak correlation or no correlation to strong correlation, or the transformation from negative correlation to positive correlation (r2-r1>0; P<0.01; FDR<0.05; Fig. 3B). However, between the early HCC stage and the advanced HCC stage, no differentially co-expressed gene pairs were identified. This analysis showed that the association of co-expression among key genes and other genes had maximum ratio of change in normal-cirrhosis stages and dysplasia-early HCC stages.
Figure 3.
Differentially co-expressed network in normal-cirrhosis and dysplasia-early HCC stages. (A) Differentially co-expressed network in normal-cirrhosis stages. (B) Differentially co-expressed network in dysplasia-early HCC stages. Turquoise nodes represent key genes; pink nodes represent differential co-expression genes with key genes. Edges represent the association between genes. Solid lines and dashed lines represent enhanced and weakened association between consecutive stages, respectively. HCC, hepatocellular carcinoma.
In normal-cirrhosis stages, genes that were differentially co-expressed with key genes significantly enriched the GO terms: Organic acid biosynthetic process (GO:0016053) and carboxylic acid biosynthetic process (GO:0046394) (P<0.01). According to the functional annotation of the genes, these genes are involved in the biological synthesis of the liver. The differential co-expression indicated the disorder in some of biosynthesis functions of the liver during the transition from normal to cirrhosis. In dysplasia-early HCC stages, in cases of weakened correlation, no GO terms were significantly enriched in those genes (P<0.01). The genes that enhanced correlation with key genes, including aurora kinase A (AURKA), marker of proliferation Ki-67 (MKI67), baculoviral IAP repeat containing 5 (BIRC5), ZW10 interacting kinetochore protein (ZWINT), cell division cycle associated 3 (CDCA3), cyclin (CCN) E2, kinesin family member 18A (KIF18A), NUF2 and nucleolar and spindle-associated protein 1 (NUSAP1), were involved in cell cycle, nuclear division, mitosis, spindle, microtubule-based process and spindle (Table III), which were closely associated with cancer. The differential co-expression reflected the dysfunction of cell cycle, mitosis and other associated processes during the development from dysplasia to HCC.
Table III.
Top 30 significant functions of genes showed enhanced association with key genes in dysplasia-early hepatocellular carcinoma stages.
| Term | Name | P-value |
|---|---|---|
| GO:0007049 | Cell cycle | 4.43×10−21 |
| GO:0000279 | M phase | 4.62×10−20 |
| GO:0022403 | Cell cycle phase | 2.29×10−18 |
| GO:0022402 | Cell cycle process | 4.21×10−16 |
| GO:0000280 | Nuclear division | 6.09×10−16 |
| GO:0007067 | Mitosis | 6.09×10−16 |
| GO:0000087 | M phase of mitotic cell cycle | 7.71×10−16 |
| GO:0048285 | Organelle fission | 1.03×10−15 |
| GO:0000278 | Mitotic cell cycle | 1.79×10−14 |
| GO:0051301 | Cell division | 2.53×10−11 |
| GO:0007059 | Chromosome segregation | 4.96×10−10 |
| GO:0000775 | Chromosome, centromeric region | 9.18×10−10 |
| GO:0007017 | Microtubule-based process | 3.42×10−9 |
| GO:0005819 | Spindle | 3.56×10−9 |
| GO:0000226 | Microtubule cytoskeleton organization | 3.28×10−8 |
| GO:0000793 | Condensed chromosome | 3.88×10−8 |
| GO:0007051 | Spindle organization | 5.16×10−8 |
| GO:0005694 | Chromosome | 1.99×10−7 |
| GO:0000779 | Condensed chromosome, centromeric region | 7.80×10−7 |
| GO:0043228 | Non-membrane-bound organelle | 9.53×10−7 |
| GO:0043232 | Intracellular non-membrane-bound organelle | 9.53×10−7 |
| GO:0015630 | Microtubule cytoskeleton | 1.02×10−6 |
| GO:0044427 | Chromosomal part | 5.86×10−6 |
| GO:0000777 | Condensed chromosome kinetochore | 1.60×10−5 |
| GO:0044430 | Cytoskeletal part | 2.14×10−5 |
| GO:0007010 | Cytoskeleton organization | 4.63×10−5 |
| GO:0000776 | Kinetochore | 4.92×10−5 |
| GO:0005876 | Spindle microtubule | 5.99×10−5 |
| GO:0005524 | ATP binding | 6.05×10−5 |
| GO:0032559 | Adenyl ribonucleotide binding | 6.85×10−5 |
GO, Gene Ontology.
Interaction networks between HCV viral proteins and their targeted proteins in the key genes set
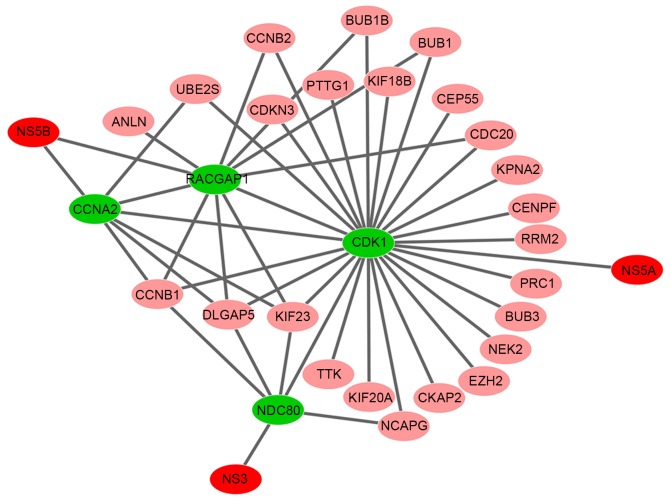
In the HCV-human interactome from Chassey's yeast two-hybrid experiments (28), HCV viral proteins NS3 and NS5A were revealed to target the proteins of NDC80 and cyclin-dependent kinase 1 (CDK1) separately, which were present in the key gene set. In addition, HCV NS5B was reported to specifically interact with CCNA2 (32) and Rac GTPase-activating protein 1 (RACGAP1) (33). The interaction networks between HCV viral proteins (NS3, NS5A and NS5B) and their targeted proteins in the key genes set are shown in Fig. 4.
Figure 4.
The interaction networks between HCV viral proteins and their targets in the key genes set. The red nodes in the network are the HCV viral proteins. The green nodes are the proteins targeted by HCV viral proteins. The pink nodes are the other members in the key genes set. The grey edges represent interactions between proteins. HCV, hepatitis C virus.
Discussion
HCV infection has become one of the predominant causes of HCC in patients with chronic viral hepatitis (34). The exploration of HCV-induced HCC progression may help to understand the common developmental mechanism of HCC induced by other risk factors, including HBV and alcoholic liver diseases. In the present study, the WGCNA tool was performed for screening of the module consisting of the key genes that exhibited HCC progression. The 44 genes identified as key genes were associated with disease progression. The majority of these genes had similar expression patterns and were associated and interconnected with each other. The enriched GO terms were mainly involved in the cell cycle process, mitotic cell cycle, M phase, cell cycle phase, mitosis, nuclear division, cell division and phosphorylation of proteins. This indicated that the dynamic of the module consisting of these genes may contribute to or be driven by disease progression. As cell cycle deregulation is one of the common hallmark traits of cancer (35), dynamic features of this module may also be associated with numerous other cancer progressions.
In the key genes set, genes including CDK1, cell-division cycle protein 20 (CDC20), CCNB2, NIMA related kinase 2 (NEK2) and CCNB1 were known to be involved in common regulatory processes of the cell cycle, mitosis and cell division. Additionally, a number of the 44 genes were closely associated with HCC. DLG associated protein 5 (DLGAP5), also termed HURP (hepatoma upregulated protein), is overexpressed in HCC (36,37). DLGAP5 activates p38/nuclear factor κ-light chain-enhancer of activated B cells (NF-κB), and combines NF-κB into the HURP/NF-κB complex to regulate CCNE1 expression (38). Previous studies have revealed that silencing or knockdown of DLGAP5 significantly inhibited the proliferation and invasion of HCC cells (36,37). The TTK gene encodes a dual specificity protein kinase with the ability to phosphorylate tyrosine, serine and threonine. TKK is essential for chromosome alignment at the centromere during mitosis and is required for centrosome duplication. TTK is upregulated in the majority of HCC specimens and its overexpression can promote cell proliferation, anchor-dependent colony formation and resistance to sorafenib of HCC cells (39). CDKN3 is frequently upregulated in HCC and is associated with poor prognosis of HCC. Overexpression of CDKN3 can stimulate the proliferation of HCC cells by promoting G1/S phase transition (40). In addition, certain genes are associated with multiple cancers. For example, enhancer of zeste homolog 2 (EZH2) promotes lung cancer progression via the vascular endothelial growth factor-A/AKT signaling pathway in non-small cell lung cancer (41), and it is also involved in aggressive breast cancer (42,43).
The analysis indicated that the changes in expression levels of key genes were associated with transition between chronic liver disease and HCC. These changes may contribute to or be driven by the dysregulation of the interaction or regulated associations between certain genes and these key genes in the disease progression.
Differentially co-expressed analysis identified several genes that showed enhanced association of co-expression with key genes in dysplastic-early HCC stages, including AURKA, MKI67, BIRC5, cell division cycle associated 3 (CDCA3), ZWINT, NUSAP1, ubiquitin conjugating enzyme E2 C (UBE2C), CCNE2, KIF18A and NUF2, which were highly associated with cancer. In other enhanced co-expression genes, the α-fetoprotein (AFP) gene, is a well-known marker for primary HCC.KIAA0101 (alternative symbol NS5ATP9) is highly co-expressed with several key genes. A previous study revealed that KIAA0101 may be involved in the pathogenesis of HCV-associated HCC and is upregulated by HCV NS5A protein (44). Furthermore, KIAA0101 potentially has an important role in NS5A-induced hepatocyte autophagy (45). Notably, the correlation coefficient between ZWINT and NEK2 in the early HCC stage (r=0.904; P=2.69×10−7) is significantly increased (P=1.19×10−5, FDR=0.022) compared with the dysplastic stage (r=−0.134). A previous study reported that the functions of the two genes were associated with chromosomal instability, which is a major characteristic of numerous cancers (46). Similarly, pituitary tumor transforming gene 1 (PTTG1), differentially co-expressed with ZWINT, is also associated with chromosomal instability (46). Therefore, these findings indicated that chromosomal instability of HCC may be associated with not only the increasing expression intensity of these genes, but also the altered interaction among them. Centrosomal protein of 55 kDa (CEP55) and a DNA helicase/putative stem cell marker HELLS (gene encoding lymphoid-specific helicase) are downstream targets of the key oncogene forkhead box M1 (FOXM1). Waseem et al suggested the proliferation-associated gene set consisting of FOXM1, CEP55 and HELLS shared a progressive expression pattern during the progression of head and neck squamous cell carcinoma (47). In this analysis, CEP55 and HELLS showed enhanced correlation in the early HCC stage, compared with the dysplastic stage. Whether these genes possess a similar function in HCC, as demonstrated in a previous study (47), is worth additional investigation. These altered interactive associations among important genes may be associated with tumorigenesis.
To study the interaction association between the key genes identified and HCV, an interaction network between HCV viral proteins and their targeted proteins, which were contained in the key genes set, was constructed by mining the HCV-human database and published literature. In the present study, CDK1, NDC80, CCNA2 and RACGAP1 were revealed to be targets of HCV NS5A, NS3 and NS5B, respectively. CDK1 is a key regulatory kinase of the cell cycle in the CDK family. A previous study demonstrated that CDK1 is upregulated in HCC (48), and serves a crucial role of the G2/M modulators in the cell cycle and cell proliferation of HCC (49). As CDK1 activation is required for mitosis, the increase of CDK1 activation in HCC may indicate the increase of cancer cell proliferation. NDC80 is a coiled-coil protein critical for cell mitosis, and performs essential roles in chromosome segregation by interacting with several proteins that modulate the G2/M phase through its coiled-coil domains (50). NDC80 is overexpressed in a variety of human cancers, such as gastric and breast cancer (51,52), and is associated with multiple cancers (53,54).
In the key genes set, CCNA2 is a cell cycle regulatory protein that can bind to and activate CDK1 or CDK2 kinases, and thus promote cell cycle G1/S and G2/M transitions. NS5B was reported to specifically interact with CCNA2 in vitro and in vivo (32). However, unlike NDC80 and CDK1, CCNA2 has dual regulatory roles in cell cycle and viral propagation. CCNA2 is required for HCV replication. Small interfering RNA-mediated depletion of CCNA2 may significantly inhibit HCV replication in HCV subgenomic replicon cells and HCVcc-infected cells (32). In addition, RACGAP1 belongs to the GTPase-activating protein family (55). A previous study suggested that the level of RACGAP1 is upregulated by HCV infection in human hepatoma cells to enhance HCV replication by binding to and affecting NS5B polymerase activity (33). The interaction network between the key genes and HCV showed that a number of important proteins directly interacted with CDK1, NDC80, CCNA2 and RACGAP1. Therefore, these proteins that are targeted by HCV may perform an intermediary role between the HCV viral proteins and the dysfunctional module. The results suggested that CDK1, NDC80, RACGAP1 and CCNA2 inhibition or interference by drugs may have potential as an effective method of therapy for HCC and prevention of the HCC progression.
Overall, in the present study, using the WGCNA tool, a core gene module was identified, in which the dynamic characteristics of the genes were associated with HCV-induced HCC progression. The marked changes in expression levels and association of co-expression of the key genes may contribute to or be driven by progression of HCV-induced HCC. The changes may be markers of transition from precancerous to HCC states. Particularly, in the identified core module, CDK1, NDC80, CCNA2 and RACGAP1 were revealed to be targeted by HCV viral proteins. These findings confirmed the close link between the identified module and HCV-induced HCC. The present study may be helpful for understanding the molecular mechanism that underlies the progression of HCV-induced HCC and provides valuable information for seeking the drug targets for the therapy of HCC and prevention of HCC development.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets analyzed during the current study are available in the Gene Expression Omnibus (GEO) repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE6764).
Author's contributions
GB, WZ and WM conceived and designed the experiments, GB performed the experiments and analyzed the data. GB, WZ and WM drafted the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Global Burden of Disease Cancer Collaboration, corp-author. Fitzmaurice C, Dicker D, Pain A, Hamavid H, Moradi-Lakeh M, MacIntyre MF, Allen C, Hansen G, Woodbrook R, et al. The global burden of cancer 2013. JAMA Oncol. 2015;1:505–527. doi: 10.1001/jamaoncol.2015.0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamane B, Weber S. Liver-directed treatment modalities for primary and secondary hepatic tumors. Surg Clin North Am. 2009;89:97–113. doi: 10.1016/j.suc.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Ishikawa T. Strategy for improving survival and reducing recurrence of HCV-related hepatocellular carcinoma. World J Gastroenterol. 2013;19:6127–6130. doi: 10.3748/wjg.v19.i47.8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu H, Lin CC, Li YY, Zhao Z. Dynamic protein interaction modules in human hepatocellular carcinoma progression. BMC Syst Biol. 2013;5(7 Suppl):S2. doi: 10.1186/1752-0509-7-S5-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi JK, Yu U, Yoo OJ, Kim S. Differential coexpression analysis using microarray data and its application to human cancer. Bioinformatics. 2005;21:4348–4355. doi: 10.1093/bioinformatics/bti722. [DOI] [PubMed] [Google Scholar]
- 7.Lee HK, Hsu AK, Sajdak J, Qin J, Pavlidis P. Coexpression analysis of human genes across many microarray data sets. Genome Res. 2004;14:1085–1094. doi: 10.1101/gr.1910904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He B, Zhang H, Shi T. A comprehensive analysis of the dynamic biological networks in HCV induced hepatocarcinogenesis. PLoS One. 2011;6:e18516. doi: 10.1371/journal.pone.0018516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varelas X, Bouchie MP, Kukuruzinska MA. Protein N-glycosylation in oral cancer: Dysregulated cellular networks among DPAGT1, E-cadherin adhesion and canonical Wnt signaling. Glycobiology. 2014;24:579–591. doi: 10.1093/glycob/cwu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mentzen WI, Floris M, de la Fuente A. Dissecting the dynamics of dysregulation of cellular processes in mouse mammary gland tumor. BMC Genomics. 2009;10:601. doi: 10.1186/1471-2164-10-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Southworth LK, Owen AB, Kim SK. Aging mice show a decreasing correlation of gene expression within genetic modules. PLoS Genet. 2009;5:e1000776. doi: 10.1371/journal.pgen.1000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banerjee A, Ray RB, Ray R. Oncogenic potential of hepatitis C virus proteins. Viruses. 2010;2:2108–2133. doi: 10.3390/v2092108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGivern DR, Lemon SM. Virus-specific mechanisms of carcinogenesis in hepatitis C virus associated liver cancer. Oncogene. 2011;30:1969–1983. doi: 10.1038/onc.2010.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wurmbach E, Chen YB, Khitrov G, Zhang W, Roayaie S, Schwartz M, Fiel I, Thung S, Mazzaferro V, Bruix J, et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45:938–947. doi: 10.1002/hep.21622. [DOI] [PubMed] [Google Scholar]
- 15.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, et al. NCBI GEO: Archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 17.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 19.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 21.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 22.Herrero J, Valencia A, Dopazo J. A hierarchical unsupervised growing neural network for clustering gene expression patterns. Bioinformatics. 2001;17:126–136. doi: 10.1093/bioinformatics/17.2.126. [DOI] [PubMed] [Google Scholar]
- 23.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zuberi K, Franz M, Rodriguez H, Montojo J, Lopes CT, Bader GD, Morris Q. GeneMANIA prediction server 2013 update. Nucleic Acids Res. 2013;41:W115–W122. doi: 10.1093/nar/gkt533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukushima A. DiffCorr: An R package to analyze and visualize differential correlations in biological networks. Gene. 2013;518:209–214. doi: 10.1016/j.gene.2012.11.028. [DOI] [PubMed] [Google Scholar]
- 26.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Chassey B, Navratil V, Tafforeau L, Hiet MS, Aublin-Gex A, Agaugué S, Meiffren G, Pradezynski F, Faria BF, Chantier T, et al. Hepatitis C virus infection protein network. Mol Syst Biol. 2008;4:230. doi: 10.1038/msb.2008.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allocco DJ, Kohane IS, Butte AJ. Quantifying the relationship between co-expression, co-regulation and gene function. BMC Bioinformatics. 2004;5:18. doi: 10.1186/1471-2105-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mas VR, Maluf DG, Archer KJ, Yanek K, Kong X, Kulik L, Freise CE, Olthoff KM, Ghobrial RM, McIver P, Fisher R. Genes involved in viral carcinogenesis and tumor initiation in hepatitis C virus-induced hepatocellular carcinoma. Mol Med. 2009;15:85–94. doi: 10.2119/molmed.2008.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Giorgi V, Buonaguro L, Worschech A, Tornesello ML, Izzo F, Marincola FM, Wang E, Buonaguro FM. Molecular signatures associated with HCV-induced hepatocellular carcinoma and liver metastasis. PLoS One. 2013;8:e56153. doi: 10.1371/journal.pone.0056153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pham LV, Ngo HT, Lim YS, Hwang SB. Hepatitis C virus non-structural 5B protein interacts with cyclin A2 and regulates viral propagation. J Hepatol. 2012;57:960–966. doi: 10.1016/j.jhep.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 33.Wu MJ, Ke PY, Horng JT. RacGTPase-activating protein 1 interacts with hepatitis C virus polymerase NS5B to regulate viral replication. Biochem Biophys Res Commun. 2014;454:19–24. doi: 10.1016/j.bbrc.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 34.Koike K, Moriya K, Kimura S. Role of hepatitis C virus in the development of hepatocellular carcinoma: Transgenic approach to viral hepatocarcinogenesis. J Gastroenterol Hepatol. 2002;17:394–400. doi: 10.1046/j.1440-1746.2002.02763.x. [DOI] [PubMed] [Google Scholar]
- 35.Chan KS, Koh CG, Li HY. Mitosis-targeted anti-cancer therapies: Where they stand. Cell Death Dis. 2012;3:e411. doi: 10.1038/cddis.2012.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuo TC, Chang PY, Huang SF, Chou CK, Chao CC. Knockdown of HURP inhibits the proliferation of hepacellular carcinoma cells via downregulation of gankyrin and accumulation of p53. Biochem Pharmacol. 2012;83:758–768. doi: 10.1016/j.bcp.2011.12.034. [DOI] [PubMed] [Google Scholar]
- 37.Liao W, Liu W, Yuan Q, Liu X, Ou Y, He S, Yuan S, Qin L, Chen Q, Nong K, et al. Silencing of DLGAP5 by siRNA significantly inhibits the proliferation and invasion of hepatocellular carcinoma cells. PLoS One. 2013;8:e80789. doi: 10.1371/journal.pone.0080789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen JM, Chiu SC, Wei TY, Lin SY, Chong CM, Wu CC, Huang JY, Yang ST, Ku CF, Hsia JY, Yu CT. The involvement of nuclear factor-kappaB in the nuclear targeting and cyclin E1 upregulating activities of hepatoma upregulated protein. Cell Signal. 2015;27:26–36. doi: 10.1016/j.cellsig.2014.09.020. [DOI] [PubMed] [Google Scholar]
- 39.Liang XD, Dai YC, Li ZY, Gan MF, Zhang SR, Yin-Pan, Lu HS, Cao XQ, Zheng BJ, Bao LF, et al. Expression and function analysis of mitotic checkpoint genes identifies TTK as a potential therapeutic target for human hepatocellular carcinoma. PLoS One. 2014;9:e97739. doi: 10.1371/journal.pone.0097739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xing C, Xie H, Zhou L, Zhou W, Zhang W, Ding S, Wei B, Yu X, Su R, Zheng S. Cyclin-dependent kinase inhibitor 3 is overexpressed in hepatocellular carcinoma and promotes tumor cell proliferation. Biochem Biophys Res Commun. 2012;420:29–35. doi: 10.1016/j.bbrc.2012.02.107. [DOI] [PubMed] [Google Scholar]
- 41.Geng J, Li X, Zhou Z, Wu CL, Dai M, Bai X. EZH2 promotes tumor progression via regulating VEGF-A/AKT signaling in non-small cell lung cancer. Cancer Lett. 2015;359:275–287. doi: 10.1016/j.canlet.2015.01.031. [DOI] [PubMed] [Google Scholar]
- 42.Collett K, Eide GE, Arnes J, Stefansson IM, Eide J, Braaten A, Aas T, Otte AP, Akslen LA. Expression of enhancer of zeste homologue 2 is significantly associated with increased tumor cell proliferation and is a marker of aggressive breast cancer. Clin Cancer Res. 2006;12:1168–1174. doi: 10.1158/1078-0432.CCR-05-1533. [DOI] [PubMed] [Google Scholar]
- 43.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi L, Zhang SL, Li K, Hong Y, Wang Q, Li Y, Guo J, Fan WH, Zhang L, Cheng J. NS5ATP9, a gene up-regulated by HCV NS5A protein. Cancer Lett. 2008;259:192–197. doi: 10.1016/j.canlet.2007.10.029. [DOI] [PubMed] [Google Scholar]
- 45.Quan M, Liu S, Li G, Wang Q, Zhang J, Zhang M, Li M, Gao P, Feng S, Cheng J. A functional role for NS5ATP9 in the induction of HCV NS5A-mediated autophagy. J Viral Hepat. 2014;21:405–415. doi: 10.1111/jvh.12155. [DOI] [PubMed] [Google Scholar]
- 46.Brendle A, Brandt A, Johansson R, Enquist K, Hallmans G, Hemminki K, Lenner P, Försti A. Single nucleotide polymorphisms in chromosomal instability genes and risk and clinical outcome of breast cancer: A Swedish prospective case-control study. Eur J Cancer. 2009;45:435–442. doi: 10.1016/j.ejca.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 47.Waseem A, Ali M, Odell EW, Fortune F, Teh MT. Downstream targets of FOXM1: CEP55 and HELLS are cancer progression markers of head and neck squamous cell carcinoma. Oral Oncol. 2010;46:536–542. doi: 10.1016/j.oraloncology.2010.03.022. [DOI] [PubMed] [Google Scholar]
- 48.Li KK, Ng IO, Fan ST, Albrecht JH, Yamashita K, Poon RY. Activation of cyclin-dependent kinases CDC2 and CDK2 in hepatocellular carcinoma. Liver. 2002;22:259–268. doi: 10.1046/j.0106-9543.2002.01629.x. [DOI] [PubMed] [Google Scholar]
- 49.Ito Y, Takeda T, Sakon M, Monden M, Tsujimoto M, Matsuura N. Expression and prognostic role of cyclin-dependent kinase 1 (cdc2) in hepatocellular carcinoma. Oncology. 2000;59:68–74. doi: 10.1159/000012140. [DOI] [PubMed] [Google Scholar]
- 50.Chen Y, Riley DJ, Zheng L, Chen PL, Lee WH. Phosphorylation of the mitotic regulator protein Hec1 by Nek2 kinase is essential for faithful chromosome segregation. J Biol Chem. 2002;277:49408–49416. doi: 10.1074/jbc.M207069200. [DOI] [PubMed] [Google Scholar]
- 51.Qu Y, Li J, Cai Q, Liu B. Hec1/Ndc80 is overexpressed in human gastric cancer and regulates cell growth. J Gastroenterol. 2014;49:408–418. doi: 10.1007/s00535-013-0809-y. [DOI] [PubMed] [Google Scholar]
- 52.Bièche I, Vacher S, Lallemand F, Tozlu-Kara S, Bennani H, Beuzelin M, Driouch K, Rouleau E, Lerebours F, Ripoche H, et al. Expression analysis of mitotic spindle checkpoint genes in breast carcinoma: Role of NDC80/HEC1 in early breast tumorigenicity, and a two-gene signature for aneuploidy. Mol Cancer. 2011;10:23. doi: 10.1186/1476-4598-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Y, Riley DJ, Chen PL, Lee WH. HEC, a novel nuclear protein rich in leucine heptad repeats specifically involved in mitosis. Mol Cell Biol. 1997;17:6049–6056. doi: 10.1128/MCB.17.10.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest. 2005;115:1503–1521. doi: 10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liang Y, Liu M, Wang P, Ding X, Cao Y. Analysis of 20 genes at chromosome band 12q13: RACGAP1 and MCRS1 overexpression in nonsmall-cell lung cancer. Genes Chromosomes Cancer. 2013;52:305–315. doi: 10.1002/gcc.22030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed during the current study are available in the Gene Expression Omnibus (GEO) repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE6764).