Abstract
Following a pathogenic attack, plants are able to mount a defense response with the coordinated activation of a battery of defense-related genes. In this study we have characterized the mode of expression of the P69B and P69C genes from tomato (Lycopersicon esculentum Mill.), which encodes two closely related subtilisin-like proteases associated with the defense response. We have compared the mode of gene regulation in heterologous transgenic Arabidopsis plants harboring promoter-β-glucuronidase (GUS) and promoter-luciferase (LUC) gene fusions for these two genes. These studies revealed that the P69B and P69C promoters are induced by salicylic acid as well as during the course of both a compatible and an incompatible interaction with Pseudomonas syringae. Furthermore, P69B and P69C expression takes place in both the local and the distal (noninoculated) leaves upon inoculation with bacteria but following different and unique tissue-specific patterns of expression that are also different to that described for most other classical PR genes. Also, we report that luciferin, the substrate for the reporter luciferase (LUC) gene, is able to activate expression of PR genes, and this may pose a problem when using this gene reporter system in studies related to plant defense.
Plants are equipped with an array of defense responses to prevent their invasion by pathogens. Some of these defensive tools are already established in the plant, whereas others are inducible upon perception of the pathogen. The most common feature of disease resistance in incompatible plant-pathogen interactions is the rapid development of a cell death process at the infection sites (the so-called hypersensitive response (HR) (Agrios, 1988). The onset of HR in turn activates a signaling process throughout the plant that makes it more refractory to subsequent infections by a broad spectrum of pathogens. This latter response is coined systemic acquired resistance (SAR) (Ross, 1961; Hunt and Ryals, 1996; Ryals et al., 1996).
Salicylic acid (SA) is a master regulatory molecule that accumulates to considerable levels following pathogen recognition, is implicated in the promotion of HR, and is also necessary for SAR induction (Yalpani et al., 1991; Shirasu et al., 1997; for review, see Hammond-Kosack and Jones, 1996;). An elevation in the endogenous level of SA or exogenous application of SA or its synthetic analogs results in a selective and concerted activation of a plethora of genes (SAR genes) encoding proteins related to defense (Ward et al., 1991; Lawton et al., 1993). A major subset of these proteins is known as pathogenesis-related (PR) proteins, which comprises several families of proteins (Cutt and Klessig, 1992). The initial expression of PR genes takes place in dying tissues that are in direct contact with the pathogen and are thus developing HR. Later on, expression of PR genes is induced in the distal tissues during the course of SAR induction. This expression profile along with the demonstrated antimicrobial activity of some PRs (Mauch et al., 1988; Broglie et al., 1991; Zhu et al., 1994) lead to the suggestion that PRs are at least partially responsible for maintaining the disease-resistant state of the plant (Lawton et al., 1993).
A novel and interesting PR gene subfamily is that encoding members of the subtilisin-like Ser proteinases (EC 3.4.21.14) that represent an ancient family of evolutionarily conserved proteins (Siezen and Leunissen, 1997). In tomato plants, recent sequence comparison revealed that the subtilisin-like genes fall into five distinct subfamilies (Meichtry et al., 1999) with the P69 subfamily members being the best characterized so far. The P69 subtilisin-like proteases belong to a multigene family of high complexity that encodes protein isoforms of approximately 69 kD that accumulate extracellularly (Tornero et al., 1996, 1997a). Genomic clusters comprising a tandem array of four genes (P69A, P69B, P69C, and P69D) and a tandem array of two genes (P69E and P69F) encoding closely related P69 proteases were identified recently in tomato plants (Jordá et al., 1999, 2000). Detailed expression analysis of each of these genes revealed that they are tightly regulated by developmental and environmental cues in tomato plants (Jordá et al., 1999, 2000). The P69A gene was shown to be expressed constitutively, whereas the P69D gene showed transient expression in rapidly expanding leaves. At variance, the P69E gene is only expressed in root tissues, whereas the P69F gene is specifically expressed in hydathodes. Although P69A, P69D, P69E, and P69F gene expression is not induced over basal levels during pathogenesis, it cannot be ruled out that these genes are implicated in pathogenesis by acting as an early line of defense, as proposed for other constitutively expressed PR genes (Samac and Shah, 1991; Tornero et al., 1997b and references therein).
In marked contrast, two other gene members of the P69 family, namely P69B and P69C, do not show constitutive expression but are notably induced in tomato plants by infection with Pseudomonas syringae or upon treatment with SA (Jordá et al., 1999). This suggests that both, P69B and P69C, may play roles as active defense weapons against the attacking pathogens.
In the present paper we show a comparative study of the mode of gene regulation of the P69B and P69C genes in transgenic Arabidopsis plants harboring promoter-β-glucuronidase (GUS) and promoter-luciferase (LUC) gene fusions. Also, we show that luciferin, the substrate for the reporter luciferase (LUC) gene is able to activate expression of defenses in the plant, and this may pose a problem when using this reporter system in defense-related studies.
RESULTS
Local and Systemic Induction of Reporter Gene Expression Driven by P69B and P69C Promoters
The P69B and P69C genes were shown to be induced following infection of tomato plants with P. syringae (Jordá et al., 1999). To understand how this induction is regulated and where does it take place in the infected plant, we constructed different chimeric promoter fusions with the bacterial β-glucuronidase (GUS) and the firefly luciferase (LUC) reporter genes. These were introduction into Arabidopsis plants and used to study the transcriptional regulation of these genes by histochemical analysis of GUS activity or by capturing the luminescence emission derived from LUC expression in intact plants.
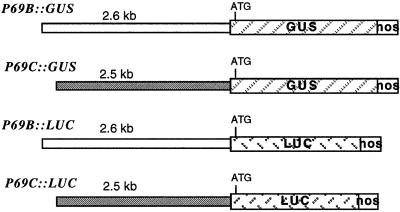
The chimeric P69B::GUS gene was constructed by fusion of the 2.6-kb region of the P69B 5′-promoter region (Jordá et al., 1999) to the ATG initiation codon of the GUS coding sequence and the 3′-untranslated region of the nopaline synthase (NOS) gene (Fig. 1) in the binary vector pBI101 (Jefferson et al., 1987). The chimeric P69C::GUS gene was constructed similarly by fusion of the 2.5-kb region of the P69C 5′ promoter (Jordá et al., 1999) to the ATG initiation codon of the GUS coding sequence in pBI101.
Figure 1.
Schematic representation of the different P69::GUS and P69::LUC gene fusions. The diagonally striped boxes represent the GUS or LUC genes. The white box at the right represents the 3′-region of the nopaline synthase gene. The length of each of the promoter regions is shown above each construct in kilobase pairs. The ATG codon represents the first translation initiation codon that resides in the reporter gene.
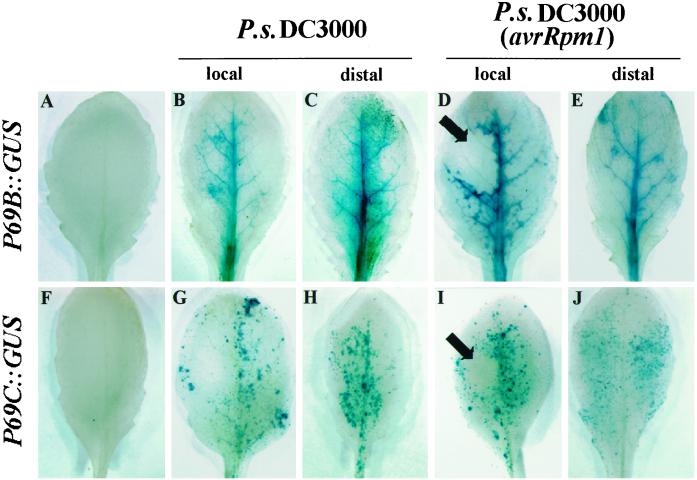
Preliminary studies for the spatial distribution of GUS activity driven by these two P69 promoters during different developmental stages of growth revealed no constitutive expression for either two genes (data not shown). To study the mode of gene induction following pathogen infection, we inoculated leaves of transgenic Arabidopsis plants with the bacterial pathogen P. syringae DC3000, carrying or not carrying the avirulence gene Rpm1, and the extent of GUS induction determined directly in leaf tissues by histochemical staining with 5-bromo-4-chloro-3-indolyl-β-glucuronic acid (X-gluc). These studies revealed that both P69B and P69C promoters are induced in both the local inoculated leaves as well as in distal (noninoculated) leaves from the same plant (Fig. 2). P69B::GUS expression follows a similar tissue pattern of induction upon inoculation with either the virulent or the avirulent bacteria. For both types of bacteria, the induced expression of GUS activity takes place in the inoculated leaves as well as in distal (noninoculated) leaves of the same plant (Fig. 2, A–E). This induction is always delimited to primary and secondary veins of the afflicted plant and with no preferential expression around the tissue zone showing the characteristic HR response (marked with arrow in Fig. 2D) when the plant was inoculated with the incompatible bacteria. Likewise, P69C::GUS expression was also induced by both bacterial strains and also was activated in local as well as in distal leaves (Fig. 2, F–J). However, at variance with the P69B promoter, the P69C promoter is activated with different tissue specificity. Its expression takes place in the form of spots scattered all along the leaf lamina. In the case of the incompatible interaction, neither of these promoters showed preferential expression in the region encompassing the HR lesion (marked with arrow in Fig. 2I).
Figure 2.
GUS staining patterns in rosette leaves of transgenic Arabidopsis (Col-0) plants carrying the P69B::GUS and P69C::GUS transgenes. Top, GUS staining pattern in leaves from P69B::GUS transgenic plants. Bottom, GUS staining pattern in leaves from P69C::GUS. A and F, GUS staining pattern in leaves from noninoculated plants. B and G, GUS staining pattern in leaves inoculated with P.s. DC3000. C and H, GUS staining pattern in distal (noninoculated) leaves from plants inoculated with P.s. DC3000. D and I, GUS staining pattern in leaves inoculated with P.s. DC3000 carrying the avirulent Rpm1 gene. E and J, GUS staining pattern in distal (noninoculated) leaves from plants inoculated with P.s. DC3000 carrying the avirulent Rpm1 gene. Leaves were analyzed 72 h after inoculation. The characteristic HR responses elicited in the inoculated leaves with the incompatible bacteria are indicated with arrows. The experiments were repeated with plants from at least three different transgenic lines for each construct and in all cases render similar results.
Analysis of P69B and P69C Gene Expression in Transgenic Intact Plants
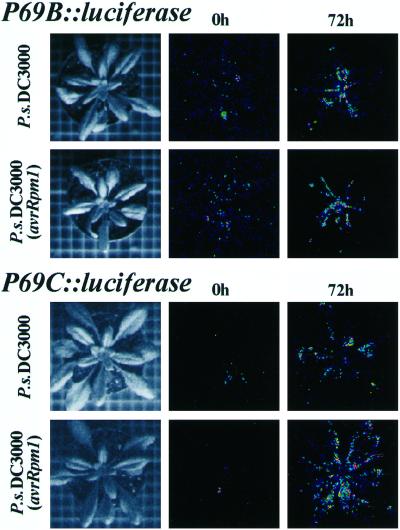
To study the expression pattern of the P69B and P69C genes in the entire plant in a non-destructive fashion, new gene constructs were generated in which the GUS reporter gene was replaced by the luciferase (LUC) reporter gene to render P69B::LUC and P69C::LUC gene constructs (Fig. 1). These new constructs were used to generate new Arabidopsis transgenic plants. The extent of LUC gene expression driven by each of these two promoters was followed in intact plants by capturing the luminescence emission in a CDC camera upon spraying plants with luciferin, the substrate of the LUC protein. The comparison of luminescence images revealed that these transgenic plants showed induced expression of the reporter LUC gene at 72 h postinoculation with both types of bacteria and with nearly undetectable expression in noninoculated plants (Fig. 3). In the case of P69B::LUC plants, image analysis of luminescence showed a preferential emission in the central veins of the rosette leaves (Fig. 3). In the case of P69C::LUC plants, luminescence emission is less concentrated and extends along the entire plant with preferential emission in leaf blades and not around veins (Fig. 3). Each of the two patterns of LUC gene expression was reproducibly generated when the plants where inoculated with either the compatible (P.s. DC3000) or with the incompatible (P.s. DC3000 avrRpm1) bacteria (Fig. 3).
Figure 3.
Monitoring of light emissions in entire P69B::LUC and P69C::LUC transgenic Arabidopsis plants by low-light video image analysis following inoculation with compatible and incompatible bacteria. Left, Standard photographs of the type of plants used in these experiments. Middle, Luminescence from plants at the time of inoculation (0 h) with either P.s. DC3000 or P.s. DC3000 (avrRpm1). Right, Luminescence from plants at 72-h postinoculation with either P.s. DC3000 or P.s. DC3000 (avrRpm1). At the times indicated, plants were sprayed once with 1 mm luciferin, and images were obtained immediately after 10 min of photon collection. The plants shown in each of the panels are different plants but derived from the same homozygous transgenic lines. The experiments were repeated with plants from at least three different transgenic lines for each construct and in all cases render similar results.
Thus, from these experiments it can be concluded that the P69B and P69C genes are induced locally and systemically but with different tissue specificity. In neither case is the expression delimited to the necrotic zone where the majority of the PR genes have been shown to be expressed (Tornero et al., 1997b and references therein).
Luciferin and SA Are Potent Inducers of P69B and P69C Gene Expression
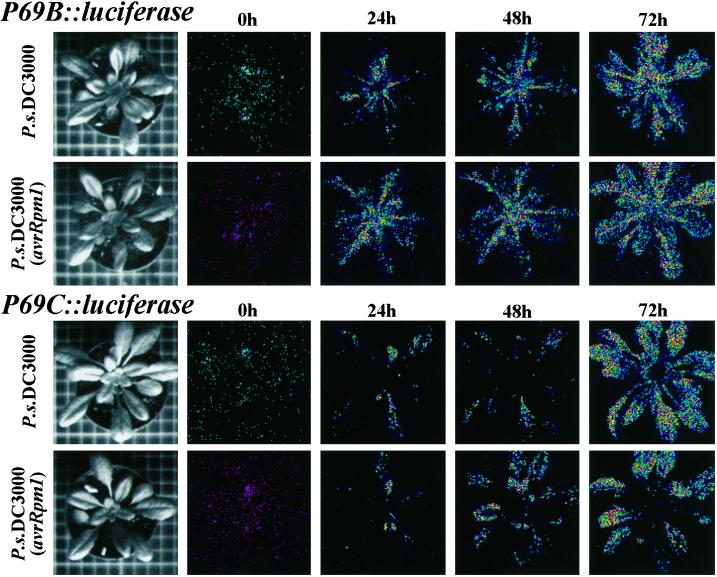
In contrast to the assay of GUS activity, the advantage of the LUC assay is that it is not destructive and thus allows studies of gene expression to be performed in the same plant at different times or following different treatments. In attempts to perform this type of study with the LUC gene driven by the P69B or the P69C promoters, transgenic plant harboring P69B::LUC or P69C::LUC constructs were inoculated with either virulent or avirulent bacteria, and the extent of luminescence emission due to LUC expression was recorded in the same plant at 24-h intervals. At each time point in the study the plant was sprayed with a 1-mm solution of luciferin to provide fresh substrate to detect newly induced LUC enzyme. This type of experiment, summarized in Figure 4, shows that for both gene constructs the activation of LUC expression following pathogen inoculation increased continuously along the time. However, a comparison of luminescence images among the many different experiments performed revealed that LUC induction was reproducibly higher than that observed previously (Fig. 3) in experiments in which LUC expression was determined in plants that received only a single treatment with luciferin. These dramatic differences in the extent of LUC induction suggested that presumably luciferin was acting as a potent inducer of P69B and P69C gene expression. To test this possibility, noninfected transgenic plants were sprayed every 24 h with a solution containing 1 mm luciferin, and the extent of LUC induction was recorded at 72 h. For comparison, other plants from the same transgenic lines were sprayed with a solution containing 0.5 mm SA, which is a potent inducer of defense-related genes. The experiment in Figure 5 shows that the sole application of luciferin is sufficient to bring activation of the P69B and P69C promoters in a manner similar to that provoked by an application of SA alone.
Figure 4.
Monitoring of light emissions by low-light video image analysis in a single transgenic Arabidopsis plant during the course of a compatible or an incompatible interaction. Two leaves from each plant were inoculated with either P.s. DC3000 or P.s. DC3000 (AvrRpm1) and at each time point the plant was taken from the growth chamber, sprayed with 1 mm luciferin, and immediately the photon collection was performed for 10 min. This process was repeated in the same plant at 0, 24, 48, and 72 h postinoculation with the bacteria. The left column of pictures shows a standard photograph of the single plant used in each experiment. Top, Two plants derived from the same P69B::LUC transgenic line. Bottom, Two plants derived from the same P69C::LUC transgenic line. The experiments were repeated with plants from at least three different transgenic lines for each construct and in all cases render similar results.
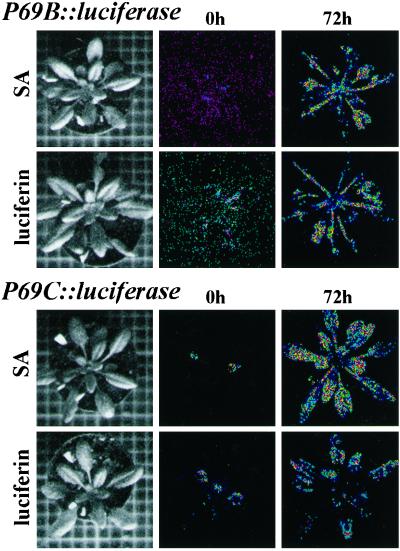
Figure 5.
Effect of exogenous application of SA and luciferin on the activation of P69B::LUC and P69C::LUC gene expression. P69B::LUC transgenic Arabidopsis plants (top) and P69C::LUC transgenic Arabidopsis plants (bottom) were sprayed three times (at 24-h intervals) with a solution containing 0.5 mm SA or 1 mm luciferin. The same plants were monitored for light emissions by low-light video image at 0 or 72 h after receiving the first chemical treatment. Images were obtained after 10 min of photon collection.
To verify if the real inducer of gene expression was luciferin or a side product of its ATP-dependent metabolic conversion (Karl and Holm-Hansen, 1976) in the living cells by the action of trace amounts of pre-existing LUC, the same experiment shown in Figure 5 was performed in transgenic plants containing P69B::GUS or P69C::GUS. Figure 6 shows that luciferin alone was able to induce GUS activity driven by the P69B and P69C promoters (Fig. 6, C and F), and these induction patterns were similar to those achieved upon treatment with SA alone (Fig. 6, B and E). It is interesting that both luciferin and SA activate the expression of these two genes in a tissue-specific pattern that coincides with that observed when expression was induced by bacterial infection. This suggests that these two molecules operate following a mechanism similar to that of the endogenous plant signal produced during pathogenesis.
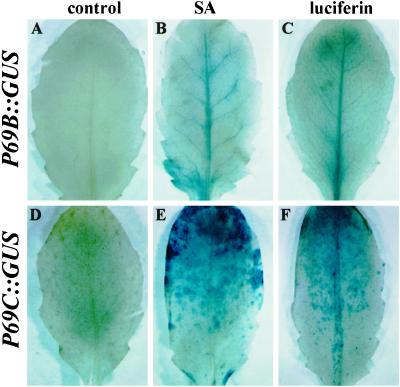
Figure 6.
Effect of SA and luciferin on the activation of P69B::GUS and P69C::GUS gene expression. P69B::GUS transgenic Arabidopsis plants (top) and P69C::GUS transgenic Arabidopsis plants (bottom) were sprayed three times (at 24-h intervals) with a solution containing 0.5 mm SA or 1 mm luciferin, and GUS expression was detected by histochemical staining of leaves with X-gluc. A and D, GUS staining pattern before treatment. B and E, GUS staining pattern at 72 h after SA treatments. C and F, GUS staining pattern at 72 h after luciferin treatments.
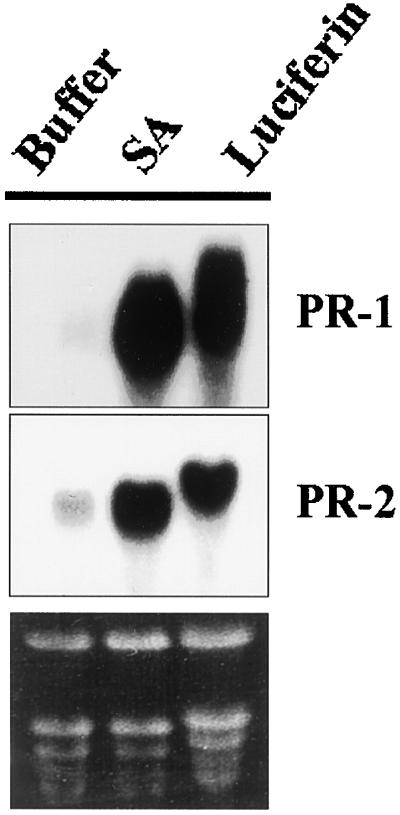
To extend this observation to other defense-related genes, wild-type Arabidopsis plants were treated similarly with luciferin or SA, and total mRNA was extracted from these plants. Northern blots shown in Figure 7 demonstrate that PR-1 and PR-2 mRNAs accumulate in response to luciferin to an extent comparable with that achieved by SA. This observation thus reinforces the consideration that luciferin, as is true for SA, is a potent inducer of defenses in the plant. However, at this stage we cannot disregard the possibility that the true inducer of PR gene expression is a breakdown product of luciferine.
Figure 7.
Northern-blot analyses of endogenous Arabidopsis PR-1 and PR-2 gene expression at 48 h following treatment of plants with SA and luciferin.
DISCUSSION
Here we describe the mode of expression of two closely related genes (P69B and P69C) from tomato plants encoding members of the PR family of P69 subtilisin-like proteases. Comparison of the P69B and P69C nucleotide and deduced amino acid sequences reveal remarkable similarity all along the open reading frames (87.3% identity) (Jordá et al., 1999). The observation that these two genes are tightly linked within an approximately 30-kb genomic cluster that comprise a total of four P69-like genes leads to the suggestion that this multigene family might have derived from a common ancestral gene by recent gene duplication events (Jordá et al., 2000). However, the high-sequence identity existing within the coding regions for these genes does not extend outside of the open reading frames, thus suggesting that the genes may have acquired a different transcriptional regulation during evolution.
To understand how the P69B and P69C genes, previously shown to be up-regulated in diseased tomato plants, might be transcriptionally regulated during pathogenesis, transgenic Arabidopsis plants harboring different promoter-reporter gene constructs were generated. Analysis of transgenic plants containing the GUS or the LUC reporter genes under the control of the 5′-promoter region of the P69B or P69C genes did not reveal any detectable constitutive expression for any of them in healthy plants. Conversely, both promoters are able to drive expression of either GUS or LUC when transgenic plants are inoculated with the compatible P.s. DC3000 or the incompatible P.s. DC3000 AvrRpm1. This induced expression is observable in both the local inoculated leaves as well as in distal noninoculated leaves of the same plant. This type of gene expression pattern coincides with that reported for other defense-related genes (referred to as SAR genes, Ryals et al., 1996), which are concomitantly induced in inoculated, as well as in upper noninoculated leaves (Brederode et al., 1991; Ward et al., 1991; Lawton et al., 1993). However, when the infected transgenic plants are analyzed by histochemical staining to detect expression of GUS activity driven by the P69 promoters, differences with the mode of expression of other inducible PR genes are observed. Our studies reveal that while the induced expression driven by the P69B promoter is restricted to the veins of the inoculated plant, the expression pattern derived from the P69C promoter takes place in group of cells that are distributed all along the leaf blade and not in the veins. Furthermore, both P69B and P69C gene expression takes place far away from the necrotic lesion that is derived from the HR during the incompatible interaction. These two tissue patterns of expression are highly different to that observed for most classical PR genes, which are highly expressed in the tissues that surround the HR lesions (Ohshima et al., 1990; Van der Rhee et al., 1990; Eyal et al., 1993; Meller et al., 1993; Uknes et al., 1993; Alonso et al., 1995; Tornero et al., 1997). Thus the confined transcriptional activation observed for the P69B and P69C genes constitutes examples, to some extent unexpected, of novel and precise mechanisms of expression of PR genes.
Regulatory elements controlling pathogen-induction of PR genes have been studied in a number of cases; however they are widely different, making it difficult to define a minimal promoter necessary for pathogen induction of different PR genes. Lebel et al. (1998) recently have identified that the CGTCA motif (as-1 element) is important for SA induction of PR-1 gene expression. However, this 5-bp element is not found in any of the two P69 promoters, and this may suggest that the regulation of the latter is in part different to that of PR-1. Also, the conserved GCCGCCTC DNA motif is present in the promoter region of a number of genes that encode “basic” isoforms of PRs and appears to be necessary for induction of these genes around the HR zone in an ethylene-dependent manner (Eyal et al., 1993; Meller et al., 1993; Alonso et al., 1995; Tornero et al., 1997 and references therein). This cis-acting element is not found in any of the two promoters under consideration, and since neither of them respond to exogenous ethylene (data not shown), it is tempting to speculate that the observed induction of these two P69 genes is not controlled by the level of endogenous ethylene produced by infection. Thus this might explain why these two promoters avoid expression around the necrotic lesion during the HR.
SA is a master regulatory molecule that accumulates following pathogen recognition. It has been proposed that SA is a mobile signal that can be transported to distal tissues from the infection site to participate in SAR by activating defenses systemically (for review, see Hammond-Kosack and Jones, 1996). Also, when applied exogenously, SA induces expression of PR genes (Ohshima et al., 1990; Van der Rhee et al., 1990; Ward et al., 1991; Yalpani et al., 1991; Uknes et al., 1993). Since P69B and P69C are responsive to SA in a tissue-specific manner coincident with that achieved in infected plant, this favors the interpretation that SA may be the likely signal that set in motion the transcriptional activation of these two genes in such a precise manner.
How such a variety of different cell-type-specific expression patterns has evolved for a common set of pathogen-inducible PR genes and how are they coordinately activated during disease development either locally or in distant tissues in such a precise fashion remains enigmatic. The availability of such a variety of pathogen-inducible promoters, including the P69B and P69C presently described, will help increase our understanding of the complex biological signaling processes that are set in motion during disease resistance in plants. Also, they may be used as molecular tools in experiments aimed to engineer specific aspects of the resistance of plants to challenging pathogens.
Derived from the present study, additional considerations are the results from the observed transcriptional activation of PR genes when luciferin is used as a substrate to detect the activity of the reporter firefly luciferase (LUC) protein in vivo. Luciferin, a 6-hydroxy-benzothiazol (Karl and Holm-Hansen, 1976; Koncz et al., 1990), or one of its breakdown products, is presumably acting as an analog of SA thus priming the expression of SA-inducible genes in a manner similar to that of other benzoic acid derivatives (e.g. 2,6-dichloroisouicotinic acid). With this observation, we want to bring to the attention of researchers in the field of defense-related genes that the use of the LUC/luciferin system might bring false interpretation of results and that experiments using this gene reporter system (e.g. those directed toward isolation of mutant plants with altered expression patterns of defense-related genes) should be performed with care.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Treatments
Arabidopsis (Col-0) plants were grown at 22°C in growth chambers programmed for a 14-h light and 10-h dark cycle. Rosette leaves were either sprayed with SA (0.5 mm), luciferin (1 mm), or buffer alone (50 mm phosphate buffer, pH 7.2) as described before. Leaves were also inoculated with Pseudomonas syringae DC3000 carrying or not carrying the avirulence Rpm1 gene as previously described (Jordá et al., 1999).
Promoter Constructs, Plant Transformation, and Analysis of Transgenic Plants
Oligonucleotides GEN69b (5′-GCCCGGGGGCTAGCTAATACAACAAGTG-3′) and GEN69c (5′-GCCCGGGGGCTGCAAATACAAGAAG-3′) in combination with the T7 oligonucleotide, served as primers for the incorporation of a synthetic SmaI restriction site in each promoter by site-directed mutagenesis (Kunkel et al., 1987). These primers introduced the SmaI site at positions −1 relative to the translation initiation sites in each gene. SmaI-BamHI fragments encompassing 2.6 kb and 2.5 kb of the promoter regions of P69B and p69C, respectively, were cloned upstream of the uidA gene in pBI101.1 (Jefferson et al., 1987) to generate plasmids pP69E::GUS and pP69F::GUS. For transcriptional fusions with the luciferase (LUC) gene, these constructs were digested with SmaI-SacI to release the GUS coding region that was replaced by the LUC coding region. The resulting transcriptional fusions were verified by nucleotide sequence analysis using specific primers. The constructs were introduced into Arabidopsis (Col-O) by Agrobacterium tumefaciens-mediated transformation (Bechtold et al., 1993). Transformants were selected on Murashige and Skoog agar medium containing kanamycin, transferred to soil, and allowed to self pollinate. The transgenic lines were assayed for GUS activity by a fluorimetric assay or by an in situ assay using the chromogenic substrate X-gluc (Jefferson, 1987) and for luciferase activity as described below. For each construct, expression was determined in at least four independent transgenic lines.
LUC Imaging
Imaging with the firefly LUC reporter requires application of the exogenous substrate luciferin. Luciferin (Promega, Madison, WI) was dissolved in water and stored frozen as a 1-mm solution. This 1-mm solution was applied uniformly by spraying onto plants. For LUC imaging, the plants were kept for 2 min in the dark after the luciferin application. The imaging system consists of a high-performance CCD camera mounted in a dark chamber, a camera controller, and a computer. Image acquisition and processing were performed with the WinView software provided by the camera manufacturer (Hamamatsu, Japan). Exposure time was 10 min unless stated otherwise.
RNA-Blot Analysis
RNA was purified as described (Jordá et al., 1999), and 15 μg of total RNA were electrophoresed on 1% agarose gels containing formaldehyde and blotted onto Nytran membranes (Schleicher & Schull, Keene, NH). Equal loading of RNA was verified by ethidium bromide staining of the gel before transfer to the membrane. Radiolabeled probes were prepared by random priming using T7 polymerase (Pharmacia Biotech, Piscataway, NJ). Hybridization and washing conditions of filters were done as described (Jordá et al., 1999).
ACKNOWLEDGMENT
We thank Dr. J. Salinas for providing the luciferase gene.
Footnotes
This work was supported by the Spanish Ministry of Science and Education (grant no. DIGICYT PB96–1055 to P.V.) and by a Conselleria de Educación y Ciencia de la Generalitat de Valencia fellowship (to L.J.).
LITERATURE CITED
- Agrios GN. Plant Pathology. London: Academic Press; 1988. [Google Scholar]
- Alonso E, de Carvalho Niebel E, Obregón P, Gheysen G, Inzé D, Van Montagu M, Castresana C. Differential in vitro DNA binding activity to a promoter element of the gn1 β-1,3-glucanase gene in hypersensitively reacting tobacco plants. Plant J. 1995;7:309–320. doi: 10.1046/j.1365-313x.1995.7020309.x. [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR Acad Sci Paris Life Sci. 1993;316:1194–1199. [Google Scholar]
- Brederode FT, Linthorst HJM, Bol JF. Differential induction of acquired resistance and PR gene expression in tobacco by virus infection, ethephon treatment, UV light and wounding. Plant Mol Biol. 1991;17:1117–1125. doi: 10.1007/BF00028729. [DOI] [PubMed] [Google Scholar]
- Broglie K, Chet I, Holliday M, Cressman R, Riddle P, Knowlton S, Mauvais CJ, Broglie R. Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani. Science. 1991;254:1194–1197. doi: 10.1126/science.254.5035.1194. [DOI] [PubMed] [Google Scholar]
- Cutt JR, Klessig DF. Pathogenesis-related proteins. In: Boller T, Meins Jr F, editors. Genes Involved in Plant Defense. Wien, Austria: Springer-Verlag; 1992. pp. 209–243. [Google Scholar]
- Eyal Y, Meller Y, Lev-Yadun S, Fluhr R. A basic-type PR-1 promoter directs ethylene responsiveness, vascular and abscission-specific expression. Plant J. 1993;4:225–234. doi: 10.1046/j.1365-313x.1993.04020225.x. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Jones JJ. Resistance gene-dependent plant defense responses. Plant Cell. 1996;8:1773–1791. doi: 10.1105/tpc.8.10.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt MD, Ryals JA. Systemic acquired resistance signal transduction. Crit Rev Plant Sci. 1996;15:583–606. [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordá L, Coego A, Conejero V, Vera P. A genomic cluster containing four differentially regulated subtilisin-like processing protease genes is in tomato plants. J Biol Chem. 1999;274:2360–2365. doi: 10.1074/jbc.274.4.2360. [DOI] [PubMed] [Google Scholar]
- Jordá L, Conejero V, Vera P. Characterization of two differentially regulated genes (P69E and P69F) encoding new members of the subtilisin-like protease clan from tomato plants. Plant Physiol. 2000;122:67–74. doi: 10.1104/pp.122.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl DM, Holm-Hansen O. Effect of luciferin concentration on the quantitative assay of ATP using crude luciferase preparations. Anal Biochem. 1976;75:100–112. doi: 10.1016/0003-2697(76)90060-9. [DOI] [PubMed] [Google Scholar]
- Koncz S, Langridge WHR, Olsson O, Schell J, Szalay AA. Bacterial and firefly luciferase genes in transgenic plants: advantages and disadvantages of a reporter gene. Dev Genet. 1990;11:224–232. [Google Scholar]
- Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–381. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lawton K, Uknes S, Friedrich L, Gaffney T, Alexander D, Goodman R, Metraux J-P, Kessmann H, Ahl Goy P, Gut Rella M, Ward E, Ryals J. The molecular biology of systemic acquired resistance. In: Fritig B, Legrand M, editors. Mechanisms of Defense Responses in Plants. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1993. pp. 410–420. [Google Scholar]
- Lebel E, Heifetz P, Thorne L, Uknes S, Ryals J, Ward E. Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J. 1998;16:223–234. doi: 10.1046/j.1365-313x.1998.00288.x. [DOI] [PubMed] [Google Scholar]
- Mauch F, Mauch-Mani B, Boller T. Plant Physiol 88: 936–942. 1988. Antifungal hydrolases in pea tissue: II. Inhibition of fungal growth by combinations of chitinase and β-1,3-glucanase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meichtry J, Amrhein N, Schaller A. Characterization of the subtilase gene family in tomato (Lycopersicon esculentum Mill.) Plant Mol Biol. 1999;39:749–760. doi: 10.1023/a:1006193414434. [DOI] [PubMed] [Google Scholar]
- Meller Y, Sessa G, Eyal Y, Fluhr R. DNA-protein interactions on a cis-DNA element essential for ethylene regulation. Plant Mol Biol. 1993;23:453–463. doi: 10.1007/BF00019294. [DOI] [PubMed] [Google Scholar]
- Ohshima M, Itoh H, Matsuoka M, Murakami T, Ohashi Y. Analysis of stress-induced or salicylic acid-induced expression of the pathogenesis-related 1a protein gene in transgenic tobacco. Plant Cell. 1990;2:95–106. doi: 10.1105/tpc.2.2.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AF. Systemic acquired resistance induced by localized virus infections in plants. Virology. 1961;14:340–358. doi: 10.1016/0042-6822(61)90319-1. [DOI] [PubMed] [Google Scholar]
- Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner H-Y, Hunt MD. Systemic acquired resistance. Plant Cell. 1996;8:1809–1819. doi: 10.1105/tpc.8.10.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samac DA, Shah DM. Developmental and pathogen-induced activation of the Arabidopsis acidic chitinnase promoter. Plant Cell. 1991;3:1063–1072. doi: 10.1105/tpc.3.10.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siezen RJ, Leunissen JAM. Subtilases: the subtilisin-like serine proteases. Protein Sci. 1997;6:501–523. doi: 10.1002/pro.5560060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu K, Nakajima K, Rajasekhar VK, Dixon RA, Lamb CJ. Salicylic acid potentiates an agonistic-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms. Plant Cell. 1997;9:261–271. doi: 10.1105/tpc.9.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero P, Conejero V, Vera P. Primary structure and expression of a pathogen-induced protease (PR-P69) in tomato plants: similarity of functional domains to subtilisin-like endoproteases. Proc Natl Acad Sci USA. 1996;93:6332–6337. doi: 10.1073/pnas.93.13.6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero P, Conejero V, Vera P. Identification of a new pathogen-induced member of the subtilisin-like processing protease family from plants. J Biol Chem. 1997a;272:14412–14419. doi: 10.1074/jbc.272.22.14412. [DOI] [PubMed] [Google Scholar]
- Tornero P, Gadea J, Conejero V, Vera P. Two PR-1 genes from tomato are differentially regulated and reveal a novel mode of expression for a pathogenesis-related gene during the hypersensitive response and development. Mol Plant-Microbe Interact. 1997b;10:624–634. doi: 10.1094/MPMI.1997.10.5.624. [DOI] [PubMed] [Google Scholar]
- Uknes S, Dincher S, Friedrich L, Negrotto D, Williams S, Thompson-Taylor H, Potter S, Ward E, Ryals J. Regulation of pathogenesis-related protein-1a gene expression in tobacco. Plant Cell. 1993;5:159–169. doi: 10.1105/tpc.5.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Rhee MD, Van Kan JAL, Gonzalez-Jaen MT, Bol J. Analysis of regulatory elements involved in the induction of two tobacco genes by salicylate treatment and virus infection. Plant Cell. 1990;2:357–366. doi: 10.1105/tpc.2.4.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward ER, Uknes SJ, Williams SC, Dincher SS, Wiederhold DL, Alexander DC, Ahl-Goy P, Métraux JP, Ryals JA. Coordinate gene activation in response to agents that induce systemic acquired resistance. Plant Cell. 1991;3:1085–1094. doi: 10.1105/tpc.3.10.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalpani N, Silverman P, Wilson TMA, Kleier DA, Raskin I. Salicylic acid is a systemic signal and an inducer of pathogenesis-related proteins in virus-infected tobacco. Plant Cell. 1991;3:809–818. doi: 10.1105/tpc.3.8.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QE, Maher S, Masoud R, Dixon R, Lamb CJ. Enhanced protection against fungal attack by constitutive coexpression of chitinase and glucanase genes in transgenic tobacco. Biotechnology. 1994;12:807–812. [Google Scholar]