Abstract
MicroRNAs (miRNAs) perform important roles in the development and progression of human malignancy. miR-874, as a tumor suppressor, is decreased in malignant tumors, including osteosarcoma. However, the potential mechanism of miR-874 in osteosarcoma remains largely unknown. In the present study, a dual-luciferase reporter system supported cyclin-dependent kinase 9 (CDK9) as a direct target gene of miR-874. miR-874 mimics evidently repressed CDK9 expression in osteosarcoma. Furthermore, osteosarcoma tissues and cell lines exhibited increased expression of CDK9, and CDK9 expression levels were inversely correlated (R=−0.725) with that of miR-874 in osteosarcoma tissues. Finally, CDK9 overexpression reversed the potential effect of miR-874 on the proliferation and invasion of osteosarcoma cells. Overall, the results revealed that miR-874, as a tumor suppressor, is involved in osteosarcoma progression and metastasis by targeting CDK9, indicating that the miR-874/CDK9 pathway may serve as a potential therapeutic target for the treatment of osteosarcoma.
Keywords: microRNA-874, osteosarcoma, cyclin-dependent kinase 9, proliferation, invasion
Introduction
Osteosarcoma is the most common bone malignancy encountered in childhood and adolescence (1). Although conventional chemotherapy and new agents were employed to overcome osteosarcoma, the clinical diagnosis and prognosis was always poor (2). Accumulating studies have demonstrated that miRNAs and their target genes, which perform essential roles in the formation and progression of osteosarcoma, are a promising therapeutic strategy (3). However, the molecular mechanisms of osteosarcoma pathogenesis remain poorly understood. Therefore, it is critical to identify novel diagnostic and prognostic biomarkers for improving the clinical outcome of patients with osteosarcoma.
MicroRNAs (miRNAs), a class of small non-coding RNA species, regulate or degrade target genes by binding to the 3′ untranslated regions (3′UTR) (4). miRNAs perform critical roles in the regulation of diverse biological processes, including embryogenesis, development, cell maintenance, proliferation and apoptosis. In previous years, accumulating studies have reported that miRNAs are involved in the development and progression of various human cancers, including osteosarcoma (3,5,6). Studies have revealed that miRNAs are dysregulated and act as prognostic biomarkers in osteosarcoma (6,7). Furthermore, identifying the mechanism of miRNAs involved in osteosarcoma progression may aid the development of strategies for the future diagnosis, treatment and prognosis of osteosarcoma.
miR-874 has been found to be downregulated and serve as a potential cancer suppressor in various types of cancer, including breast cancer (8), gastric cancer (9), maxillary sinus squamous cell carcinoma (10), non-small cell lung cancer (11) and head and neck squamous cell carcinoma (12). It has been reported that miR-874 can suppress cell migration and invasion by targeting its target genes (13). Previously, miR-874 was reported to be significantly downregulated in osteosarcoma cell lines and clinical specimens, and the decreased miR-874 expression was significantly associated with large tumor size, distant metastasis and advanced clinical stage, and was an independent predictor of poor survival (14). However, little is known about the mechanism of miR-874 in osteosarcoma proliferation and invasion.
In the present study, cyclin-dependent kinase 9 (CDK9) was identified as a direct target of miR-874. CDK9 expression was upregulated in osteosarcoma and inversely correlated with that of miR-874. In addition, miR-874 suppressed osteosarcoma proliferation and invasion via the downregulation of CDK9. The present results demonstrated the mechanism of miR-874 in regulating the proliferation and invasion of the osteosarcoma cells, and indicated a potential therapeutic target for the treatment of osteosarcoma.
Materials and methods
Clinical tissues samples
A total of 30 pairs of human osteosarcoma tissues and matched adjacent noncancerous bone tissues were obtained from patients (age, 41–65; 14 female and 16 male) at the Department of Orthopedics, Yantaishan Hospital (Shandong, China) from January 2013 to December 2014. The present study was approved by the Research Ethics Committee of Yantaishan Hospital and all patients provided written informed consent.
Cell culture and transfection
Human osteosarcoma MG-63, U2OS and HOS and human normal osteoblast NHOst cell lines were obtained from American Type Culture Collection (Manassas, VA, USA) and cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% (v/v) fetal bovine serum (HyClone; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 mg/ml streptomycin. Cultures were maintained at 37°C in a humidified atmosphere with 5% CO2.
MG-63 cells were seeded on 12-well plates with 40% confluence prior to transfection and incubated at 37°C in a humidified atmosphere with 5% CO2 overnight. Cells were transiently transfected with miR-874 mimics and miR-874 inhibitor sequences using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The full-length CDK9 cDNA was amplified by polymerase chain reaction (PCR) and cloned into the pcDNA3.1 vector to generate the pcDNA-CDK9 constructs, which were used in the rescue assays. MG-63 cells were co-transfected with miR-874 mimic and the pcDNA-CDK9.
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from cells and tissues using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The first strand of cDNA was synthesized by RT (Takara Biotechnology Co., Ltd., Dalian, China), and the expression of miRNA was detected by RT-qPCR analysis using the SYBR-Green detection system (Roche Applied Science, Penzberg, Germany). U6 and GAPDH mRNA were used as the internal controls. All tests were run in triplicate. The levels of mRNA expression were normalized to the level of the β-actin mRNA expression using the 2−ΔΔCq cycle threshold method (15). CDK9 primers (forward, 5′-ATGGCAAAGCAGTACGACTCG3′ and reverse, 5′-GCAAGGCTGTAATGGGGAAC-3′; GAPDH primers (forward, 5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse, 5′-GGCTGTTGTCATACTTCTCATGG-3′, miR-874 primers (forward, 5′-TGCGGCTGCCCTGGCCCGAGGGAC-3′ and reverse, 5′-CCAGTGCAGGGTCCGAGGT-3′) and U6 primers (forward, 5′-TGCGGGTGCTCGCTTCGGCAGC-3′ and reverse, 5′-CCAGTGCAGGGTCCGAGGT-3′) were purchased from Shanghai Genepharma Co., Ltd. (Shanghai, China).
Luciferase reporter assay
The 3′UTR of the human CDK9 with the predicted miR-874 binding site was amplified from a cDNA library of MG-63 cells and cloned to a pGL3-control vector. The binding site mutant of CDK9/pGL3 was also generated using a Muta-direct™ site-directed mutagenesis kit (SBS Genetech Co., Ltd., Beijing, China). The cells were seeded on 24-well plates. MG-63 cells were co-transfected with the CDK9/pGL3 vectors and miR-874 or the miRNA control. Luciferase activity values were determined using the Dual-Luciferase reporter assay system (Promega Corporation, Madison, WI, USA).
Cell proliferation assay
Cell proliferation was measured using the MTT assay using a Cell Proliferation kit I (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). In brief, the cells were plated on 96-well plates at a density of 5×103 cells per well following transfection. A MTT assay was conducted. Dimethyl sulfoxide, an acidified ethanol solution, was added to dissolve the insoluble purple formazan product into a colored solution. Finally, the optical density was determined at 570 nm using the ELISA plate reader (Model 550; Bio-Rad Laboratories, Inc., Hercules, CA, USA). The MG-63 cells were used as the control group and compared with transfected MG-63 cells.
Cell invasion assay
Invasion assays were performed in triplicate using Transwell invasion chambers (Costar 3422; Corning Incorporated, Corning, NY, USA) coated with Matrigel (50 µl per filter; BD Biosciences, Franklin Lakes, NJ, USA), according to the manufacturer's protocol. The MG-63 cells were transferred to the top of the Matrigel-coated invasion chambers in 1% fetal calf serum DMEM/F12 (2×104 cells/well). To the wells of the lower chamber, 1.5 ml of DMEM supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) was added as an attractant. Following an overnight incubation at 37°C in an atmosphere containing 5% CO2, the cells in the lower side of the insert membrane were fixed with 5% glutaraldehyde 37°C for 10 min, followed by staining with 1% crystal violet in 2% ethanol 37°C for an additional 20 min. Living cells (with diameters ranging from 7–14 µm) that passed through the membrane were collected from the lower well and counted using a cell Coulter Counter Channelizer 256 (Beckman Coulter, Inc., Brea, CA, USA). The values for invasion were obtained by counting 8 fields per membrane and represented the average of three independent experiments.
Western blot analysis
Total protein was extracted from H1299 and A549 cells with a radioimmunoprecipitation assay buffer with 0.5% SDS and 3% proteinase inhibitor cocktail (Sigma-Aldrich; Merck KGaA) for 30 min on ice. The concentration of protein was determined using the bicinchoninic acid protein assay kit (Santa Cruz Biotechnology, Inc., Dallas, TX, USA). For western blotting, proteins were separated by 10% SDS-PAGE and transferred to polyvinylidene fluoride membranes (Invitrogen; Thermo Fisher Scientific, Inc.). Subsequent to blocking with 5% bovine serum albumin (Gibco; Thermo Fisher Scientific, Inc.) at room temperature for 1 h, membranes were incubated with primary antibody against CDK9 (cat no. ab76320; dilution, 1:200; Abcam, Shanghai, China) at 4°C overnight. Membranes were then incubated with secondary antibody goat anti-rabbit IgG (cat no. sc-2007; dilution, 1:2,500; Santa Cruz Biotechnology, Inc.) for 2 h at room temperature. The signals were detected using a RapidStep™ ECL Reagent (EMD Millipore, Billerica, MA, USA).
Statistical analysis
All experiments were repeated at least three times with similar results. Representative data are shown. The Student's t-test was used for comparisons between two groups. One-way analysis of variance was used for comparisons between multiple groups followed by Turkey multiple comparison post-hoc analysis. Pearson's correlation coefficient was used to evaluate the relationships among the relative expression levels of miR-874 and CDK9 in osteosarcoma tissues. P<0.05 was considered to indicate a statistically significant difference.
Results
CDK9 is a direct target of miR-874 in osteosarcoma
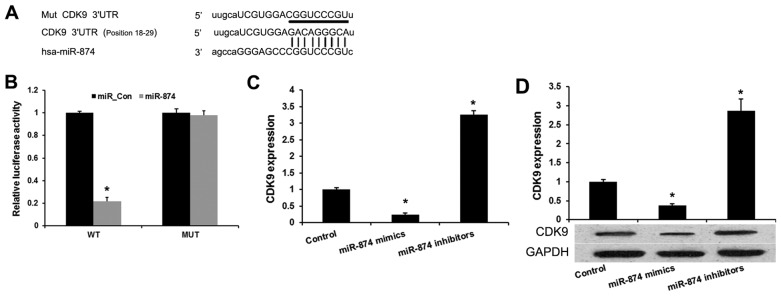
A previous study demonstrated that miR-874 was downregulated in osteosarcoma and inhibits osteosarcoma cell proliferation and metastasis (14). Therefore, it was speculated that the specific genes suppressed by miR-874 may predominantly function in this process. In the present study, miRanda (http://www.microrna.org/microrna/home.do) and Targetscan (http://www.targetscan.org/) were used to predict that miR-874 directly binds to the 3′UTR of CDK9 mRNA (Fig. 1A). To verify the prediction, wild-type CDK9 3′UTR and mutated luciferase reporter plasmids were constructed (Fig. 1B). The luciferase activity assays indicated that miR-874 mimics evidently inhibited the reporter activity of the wild type, but not the mutant, CDK9 3′UTR. It was then investigated whether miR-874 affects endogenous CDK9 expression. Enforced expression of miR-874 induced a reduction, while silencing of miR-874 induced an upregulation of endogenous CDK9 mRNA in MG-63 cells (Fig. 1C). miR-874 was also found to inhibit the CDK9 protein level in MG-63 cells (Fig. 1D). These results indicated that miR-874 directly targets and modulates the expression of CDK9 in osteosarcoma cells.
Figure 1.
miR-874 directly targets CDK9 in osteosarcoma cells. (A) Sequence alignment of miR-874 and 3′UTR of CDK9 using mirco-RNA.org. (B) Luciferase reporter assay. HEK293T cells were transiently co-transfected with Wt/Mut 3′UTR of CDK9 with miRNAs as indicated. (C) Quantitative polymerase chain reaction assay revealed the effects of miR-874 on the expression levels of CDK9 mRNA. (D) Western blot analysis revealed the effects of miR-874 on the expression levels of CDK9 miRNA in cells. Data are presented as the mean ± standard deviation from three independent experiments. *P<0.01 vs. the control group. CDK9, cyclin-dependent kinase 9; UTR, untranslated region; Mut, mutant; Wt, wild-type; miRNA, microRNA; miR-874, microRNA-874.
CDK9 expression levels in osteosarcoma tissues and cell lines
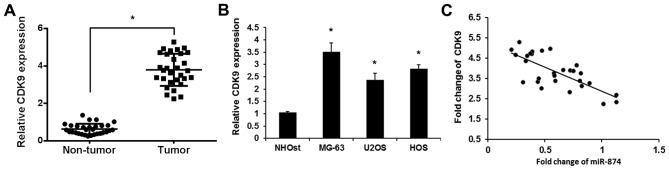
To validate the role of CDK9 in osteosarcoma, the expression levels of CDK9 were detected in osteosarcoma tissues and cell lines using RT-qPCR. As shown in Fig. 2A, increased levels of CDK9 were detected in osteosarcoma tissues compared with adjacent noncancerous tissues. CDK9 expression was also upregulated in three osteosarcoma cell lines (MG-63, U2OS and HOS) compared with the human normal osteoblast cell line (Fig. 2B). The correlation between miR-874 and CDK9 in osteosarcoma was then assessed using Pearson's correlation coefficient. As expected, the levels of miR-874 exhibited a significant negative correlation with the levels of CDK9 mRNA (r=−0.715; P<0.01; Fig. 2C). Overall, the present findings indicated that the expression level of CDK9 was upregulated and negatively associated with those of miR-874 in clinical osteosarcoma tissues.
Figure 2.
CDK9 expression in osteosarcoma tissues and cell lines. (A) The mean level of CDK9 expression in osteosarcoma tissues (n=30) was significantly increased compared with corresponding adjacent normal tissues (n=30). Data are presented as the mean ± SD from three independent experiments. *P<0.01 vs. non-tumor tissues. (B) RT-qPCR revealed the expression levels of CDK9 in 3 osteosarcoma cell lines (MG-63, U2OS and HOS) and the normal osteoblast NHOst cell line. Data are presented as the mean ± SD from three independent experiments. *P<0.01 vs. NHOst. (C) Pearson's correlation coefficient of miR-874 levels with CDK9 mRNA levels was examined by RT-qPCR in 30 cases of osteosarcoma tissues (r=−0.715). CDK9, cyclin-dependent kinase 9; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; SD, standard deviation; miR-874, microRNA-874.
miR-874 inhibits osteosarcoma cell proliferation through CDK9
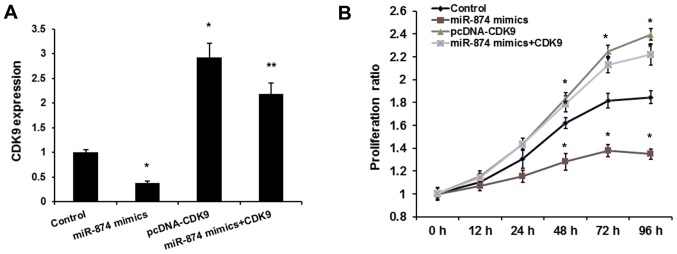
A previous study reported that miR-874 inhibited osteosarcoma progression (14). Therefore, the present study aimed to determine whether miR-874 promotes the proliferation of osteosarcoma cells by targeting CDK9. To determine whether miR-874 exerts its function by targeting CDK9, MG-63 cells were co-transfected with miR-874 mimics and pcDNA-CDK9. As shown in Fig. 3A, transfection of the pcDNA-CDK9 plasmid reversed the inhibitory effects of miR-874 on the expression of CDK9. Consistent with a previous study, the MTT assay revealed that miR-874 decreased osteosarcoma cell proliferation. Overexpression of CDK9 rescued the oncogenic effects of miR-874 on osteosarcoma cell proliferation (Fig. 3B). Therefore, it was proposed that miR-874 promotes the proliferation of osteosarcoma cells by inhibiting CDK9.
Figure 3.
miR-874 regulates osteosarcoma cell proliferation by repressing CDK9 expression. (A) pcDNA-CDK9 rescued the regulation of miR-874 on CDK9 expression. *P<0.01 vs. control group; **P<0.01 vs. miR-874 mimics group. (B) MTT assay was utilized to analyze the effect of miR-874 on cell proliferation of MG-63 cells. The groups were as follows: miR-874 mimics (MG-63 cells transfected with miR-874 mimics); pcDNA-CDK9 (MG-63 cells transfected with pcDNA-CDK9); and miR-874 mimics+CDK9 (MG-63 cells co-transfected with miR-874 mimics and pcDNA-CDK9). Data are presented as the mean ± standard deviation from three independent experiments. *P<0.01 vs. control group; **P<0.01 vs. miR-874 mimics group. CDK9, cyclin-dependent kinase 9; miR-874, microRNA-874.
miR-874 inhibits osteosarcoma cell invasion through CDK9
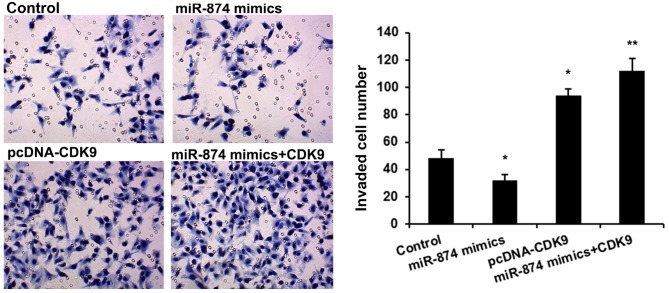
A Transwell invasion assay was then performed to detect the effects of miR-874 and CDK9 on osteosarcoma cell invasion. As shown in Fig. 4, MG-63 cells treated with miR-874 mimics had a significantly reduced cell invasion (P<0.01), and CDK9 upregulation rescued the oncogenic effects of miR-874 on cell invasion (P<0.01).
Figure 4.
miR-874 regulates osteosarcoma cell invasion by repressing CDK9 expression. The Transwell invasion assay was utilized to analyze the effect of miR-874 on cell invasion of MG-63 cells (magnification, ×40). The groups were as follows: miR-874 mimics (MG-63 cells transfected with miR-874 mimics); pcDNA-CDK9 (MG-63 cells transfected with pcDNA-CDK9); and miR-874 mimics+CDK9 (MG-63 cells co-transfected with miR-874 mimics and pcDNA-CDK9). Data are presented as the mean ± standard deviation from three independent experiments. *P<0.01 vs. control group; **P<0.01 vs. miR-874 mimics group. CDK9, cyclin-dependent kinase 9; miR-874, microRNA-874.
Discussion
The underlying mechanism of osteosarcoma carcinogenesis is critical for predicting prognosis and developing a therapeutic strategy. Therefore, it is urgent to investigate the function of deregulated molecules in osteosarcoma progression. In the present study, miR-874, as a tumor suppressor, was found to regulate osteosarcoma cell proliferation and metastasis by directly targeting CDK9.
Downregulated miR-874 has been commonly observed in various types of cancers (10,13). A previous study demonstrated that dysregulation of miR-874 is involved in osteosarcoma carcinogenesis (14). The present study confirmed that miR-874 is downregulated in osteosarcoma cells, and gain- and loss-of-function analysis was performed to reveal the effect of miR-874 on osteosarcoma cells. It was revealed that miR-874 overexpression significantly inhibited tumor cell proliferation (P<0.01) and invasion (P<0.01), indicating that restoration of miR-874 expression may be a novel therapeutic strategy in osteosarcoma treatment.
Subsequently, the present study focused on the underlying mechanism of how miR-874 exerts its effect on cancer cells. CDK9 is a CDC2-associated kinase and the catalytic subunit of positive-transcription elongation factor b and Tat-activating kinase (16). CDK9 regulates androgen receptor transcriptional activity and performs an important role in prostate cancer cell growth (17). miRanda and Targetscan predicted that miR-874 has putative binding CDK9 sites in the 3′UTR. The effects of miR-874 on CDK9 were then validated via a luciferase activity reporter assay and western blot analysis, which indicated that miR-874 may be a novel negative regulator for CDK9. In addition, the upregulated expression level of CDK9 and negative association with the miR-874 expression level were also observed in clinical osteosarcoma tissues. Studies have demonstrated that deregulation of CDK9 has been associated with numerous types of cancer (18–20) and inhibition of CDK9 may represent potential as a cancer therapeutic target (21,22). In the present study, it was revealed that overexpressed CDK9 attenuated the tumor suppressor role of miR-874, indicating the oncogene function of CDK9 in osteosarcoma development. Therefore, it was speculated that CDK9 has important roles in tumorigenesis, and the mechanism of how CDK9 affected cancer development requires additional study.
In conclusion, the present study provides new insights into the mechanism of miR-874 in osteosarcoma proliferation and invasion, and proposes that targeting of the miR-874/CDK9 axis may provide a potential therapeutic strategy for osteosarcoma.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
WT and WGW conceived and designed the study. WT, YCZ and ZYZ performed the experiments. WGW, ZYZ and WT wrote the paper. All authors read and approved the manuscript.
Ethics approval and consent to participate
The present study was approved by the Research Ethics Committee of Yantaishan Hospital and all patients provided written informed consent.
Consent for publication
The study participants provided consent for the data to be published
Competing interests
The authors declare that they have no competing interests.
References
- 1.Sun XH, Geg XL, Zhang J, Zhang C. miRNA-646 suppresses osteosarcoma cell metastasis by downregulating fibroblast growth factor 2 (FGF2) Tumour Biol. 2015;36:2127–2134. doi: 10.1007/s13277-014-2822-z. [DOI] [PubMed] [Google Scholar]
- 2.Yang J, Zhang W. New molecular insights into osteosarcoma targeted therapy. Curr Opin Oncol. 2013;25:398–406. doi: 10.1097/CCO.0b013e3283622c1b. [DOI] [PubMed] [Google Scholar]
- 3.Han K, Zhao T, Chen X, Bian N, Yang T, Ma Q, Cai C, Fan Q, Zhou Y, Ma B. microRNA-194 suppresses osteosarcoma cell proliferation and metastasis in vitro and in vivo by targeting CDH2 and IGF1R. Int J Oncol. 2014;45:1437–1449. doi: 10.3892/ijo.2014.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun K, Lai EC. Adult-specific functions of animal microRNAs. Nat Rev Genet. 2013;14:535–548. doi: 10.1038/nrg3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li X, Yang H, Tian Q, Liu Y, Weng Y. Upregulation of microRNA-17-92 cluster associates with tumor progression and prognosis in osteosarcoma. Neoplasma. 2014;61:453–460. doi: 10.4149/neo_2014_056. [DOI] [PubMed] [Google Scholar]
- 6.Wang G, Shen N, Cheng L, Lin J, Li K. Downregulation of miR-22 acts as an unfavorable prognostic biomarker in osteosarcoma. Tumour Biol. 2015;36:7891–7895. doi: 10.1007/s13277-015-3379-1. [DOI] [PubMed] [Google Scholar]
- 7.Pencheva N, Tavazoie SF. Control of metastatic progression by microRNA regulatory networks. Nat Cell Biol. 2013;15:546–554. doi: 10.1038/ncb2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L, Gao W, Hu F, Xu Z, Wang F. MicroRNA-874 inhibits cell proliferation and induces apoptosis in human breast cancer by targeting CDK9. FEBS Lett. 2014;588:4527–4535. doi: 10.1016/j.febslet.2014.09.035. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X, Tang J, Zhi X, Xie K, Wang W, Li Z, Zhu Y, Yang L, Xu H, Xu Z. miR-874 functions as a tumor suppressor by inhibiting angiogenesis through STAT3/VEGF-A pathway in gastric cancer. Oncotarget. 2015;6:1605–1617. doi: 10.18632/oncotarget.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nohata N, Hanazawa T, Kikkawa N, Sakurai D, Fujimura L, Chiyomaru T, Kawakami K, Yoshino H, Enokida H, Nakagawa M, et al. Tumour suppressive microRNA-874 regulates novel cancer networks in maxillary sinus squamous cell carcinoma. Br J Cancer. 2011;105:833–841. doi: 10.1038/bjc.2011.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kesanakurti D, Maddirela DR, Chittivelu S, Rao JS, Chetty C. Suppression of tumor cell invasiveness and in vivo tumor growth by microRNA-874 in non-small cell lung cancer. Biochem Biophys Res Commun. 2013;434:627–633. doi: 10.1016/j.bbrc.2013.03.132. [DOI] [PubMed] [Google Scholar]
- 12.Nohata N, Hanazawa T, Kinoshita T, Inamine A, Kikkawa N, Itesako T, Yoshino H, Enokida H, Nakagawa M, Okamoto Y, Seki N. Tumour-suppressive microRNA-874 contributes to cell proliferation through targeting of histone deacetylase 1 in head and neck squamous cell carcinoma. Br J Cancer. 2013;108:1648–1658. doi: 10.1038/bjc.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang B, Li Z, Zhang W, Wang H, Zhi X, Feng J, Chen Z, Zhu Y, Yang L, Xu H, Xu Z. miR-874 inhibits cell proliferation, migration and invasion through targeting aquaporin-3 in gastric cancer. J Gastroenterol. 2014;49:1011–1025. doi: 10.1007/s00535-013-0851-9. [DOI] [PubMed] [Google Scholar]
- 14.Zhang LQ, Sun SL, Li WY, Feng Z, Xu XY, Zhuang QS, Fang J. Decreased expression of tumor suppressive miR-874 and its clinical significance in human osteosarcoma. Genet Mol Res. 2015;14:18315–18324. doi: 10.4238/2015.December.23.19. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Garriga J, Bhattacharya S, Calbó J, Marshall RM, Truongcao M, Haines DS, Graña X. CDK9 is constitutively expressed throughout the cell cycle, and its steady-state expression is independent of SKP2. Mol Cell Biol. 2003;23:5165–5173. doi: 10.1128/MCB.23.15.5165-5173.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon V, Bhadel S, Wunderlich W, Zhang J, Ficarro SB, Mollah SA, Shabanowitz J, Hunt DF, Xenarios I, Hahn WC, et al. CDK9 regulates AR promoter selectivity and cell growth through serine 81 phosphorylation. Mol Endocrinol. 2010;24:2267–2280. doi: 10.1210/me.2010-0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krystof V, Baumli S, Fürst R. Perspective of cyclin-dependent kinase 9 (CDK9) as a drug target. Curr Pharm Des. 2012;18:2883–2890. doi: 10.2174/138161212800672750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitra P, Yang RM, Sutton J, Ramsay RG, Gonda TJ. CDK9 inhibitors selectively target estrogen receptor-positive breast cancer cells through combined inhibition of MYB and MCL-1 expression. Oncotarget. 2016;23:9069–9083. doi: 10.18632/oncotarget.6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morales F, Giordano A. Overview of CDK9 as a target in cancer research. Cell Cycle. 2016;15:519–527. doi: 10.1080/15384101.2016.1138186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin T, Lallena MJ, Kreklau EL, Fales KR, Carballares S, Torrres R, Wishart GN, Ajamie RT, Cronier DM, Iversen PW, et al. A novel CDK9 inhibitor shows potent antitumor efficacy in preclinical hematologic tumor models. Mol Cancer Ther. 2014;13:1442–1456. doi: 10.1158/1535-7163.MCT-13-0849. [DOI] [PubMed] [Google Scholar]
- 22.Liu X, Shi S, Lam F, Pepper C, Fischer PM, Wang S. CDKI-71, a novel CDK9 inhibitor, is preferentially cytotoxic to cancer cells compared to flavopiridol. Int J Cancer. 2012;130:1216–1226. doi: 10.1002/ijc.26127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.