Abstract
The object of this study was to analyze the expression of miR-194 and miR-29 in gastric cancer and their roles in the regulation of malignant phenotype of gastric cancer cells, and to explore the application value of miR-194 and miR-29 in diagnosis and prognosis of gastric cancer. Tumor tissue and adjacent healthy tissue of 165 gastric cancer patients diagnosed by pathologic examinations were collected. Expression of miR-194 and miR-29 in the tissues was detected by RT-PCR. The relationship between miR-194 and miR-29 expression and clinical data was analyzed. SGC7901 cells were treated with miR-194 and miR-29 mimics, respectively. Effects of miR-194 and miR-29 on proliferation and invasion of SGC7901 cells were investigated. Expression levels of miR-194 and miR-29 in tumor tissue were lower than those in adjacent tissues (P<0.001). There was no significant difference in expression level of miR-194 and miR-29 in cancer tissues derived from gastric cancer patients in different age and gender groups (P>0.05). Expression of miR-194 and miR-29 in tumor tissue was closely related to TNM stage, differentiation degree of cancer cells and lymph node metastasis (P<0.05). Proliferation and migration of SGC7901 cells were significantly inhibited by miR-194 mimic and miR-29 mimic transfection (P<0.05). miR-194 and miR-29 are downregulated in gastric cancer, and the expression levels of miR-194 and miR-29 were closely related to tumor differentiation and metastasis. Overexpression of miR-194 and miR-29 significantly inhibited the proliferation and migration of gastric cancer. The detection of the expression of miR-194 and miR-29 can provide basis for the diagnosis and prognosis of gastric cancer.
Keywords: miR-194, miR-29, RT-PCR, gastric cancer
Introduction
MicroRNAs (miRNAs) are a class of single-stranded, non-coding small RNAs that are widely present in eukaryotic cells and are highly conserved during evolution. The length of miRNA is generally 19–24 nt. miRNA is the largest class of gene expression transcriptional regulatory factors that are involved in the regulation of a variety of biological activities in human body. The disorders of miRNA expression can lead to the occurrence of diabetes, hypertension and other diseases (1,2). miRNA expression is also related to the onset, development and metastasis of breast cancer (3), lung cancer (4), osteosarcoma (5), liver cancer (6) and other malignant tumors.
Gastric cancer is one of the common malignancies in the digestive system, and the incidence of gastric cancer is high in many countries, especially in China. Gastric cancer is the fifth most common cancer among all the malignant tumors and the third major cause of cancer-related deaths. Due to the lack of indicators for early diagnosis, gastric cancer cannot be easily detected. Most patients are diagnosed at advanced stages with metastasis, and the prognosis is usually poor, seriously threatening the health of people. It is an urgent task to study the molecular mechanism of gastric cancer and to find indicators for early diagnosis and treatment targets. In recent years, abnormal miRNA expression was found in gastric cancer, indicating that miRNA is also involved in the occurrence of gastric cancer (7).
Materials and methods
Specimen collection
Specimens were collected from 165 cases of gastric cancer patients who received surgical resection in Jinan Central Hospital Affiliated to Shandong University from January 2011 to January 2017. All patients were diagnosed and treated for the first time, and none of them received radiotherapy or chemotherapy before this study. The patients included 102 males and 63 females, with an average age of 56±11.5 years. All patients were diagnosed as gastric cancer by biopsy, and the cancer tissues and adjacent tissues (>5 cm away from the cancer tissue) were collected. All samples were frozen in the liquid nitrogen and stored at −80°C. According to the TNM classification criteria and WHO histological grading method, all patients were subjected to clinical staging and classification. Specific clinical data are shown in Table I. This study was approved by the Ethics Committee of Jinan Central Hospital Affiliated to Shandong University (Jinan, China). Signed written informed consents were obtained from the patients and/or guardians.
Table I.
Correlations between the expression of miR-194 and miR-29 in cancer tissue and the clinicopathological features.
| Clinicopathological parameters | Cases | Relative content of miR-194 | P-value | Relative content of miR-29 | P-value |
|---|---|---|---|---|---|
| Age (years) | 0.318 | 0.478 | |||
| <60 | 86 | 3.11±1.18 | 4.02±1.23 | ||
| ≥60 | 79 | 3.44±1.25 | 3.59±1.05 | ||
| Sex | 0.347 | 0.236 | |||
| Male | 102 | 2.78±1.09 | 4.25±0.98 | ||
| Female | 63 | 3.21±1.10 | 3.43±0.89 | ||
| TNM staging | 0.029 | 0.009 | |||
| I + II | 95 | 2.62±0.96 | 4.59±1.43 | ||
| III + IV | 70 | 3.56±1.22 | 3.36±1.37 | ||
| Differentiation | 0.017 | 0.024 | |||
| Low | 93 | 2.61±1.03 | 3.36±0.97 | ||
| Medium/High | 72 | 3.52±1.62 | 4.34±1.13 | ||
| Lymph node metastasis | 0.009 | 0.010 | |||
| Yes | 76 | 2.56±1.14 | 3.18±0.87 | ||
| No | 89 | 3.45±1.45 | 4.27±1.28 |
Materials and reagents
TRIzol, reverse transcription kit and real-time quantitative PCR (qRT-PCR) kit were from Promega (Madison, WI, USA); specific primers of miR-194, miR-29 and U6, miR-194 mimic, miR-29 mimic, and scramble mimic were from Sangon (Shanghai, China); RIMP-1640 and fetal calf serum were from HyClone (Thermo Fisher Scientific, Waltham, MA, USA); MTT and dimethyl sulfoxide (DMSO) were from Sigma (St. Louis, MO, USA); Lipofectamine™ 2000 was provided by Invitrogen (Thermo Fisher Scientific), human gastric cancer cell line SGC7901 was purchased from the cell bank of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences; 7500 Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific).
Cell culture and transfection
Human gastric cancer cell line SGC7901 was cultured in RPMI-1640 medium containing 10% fetal bovine serum (FBS) in an incubator (37°C, 5% CO2). Cells were transferred to 6-well plate during subculture, and transfection was performed when cells reached 80% confluent. All operations were performed according to the instructions of Lipofectamine 2000 kit. Liposomes were mixed with mimics, the mixture was added into medium without serum to culture the cells for 8 h, then the medium was replaced by complete medium.
qRT-PCR
Total RNA was extracted from cells or tissues using TRIzol reagent. RNA was used as template to synthesize cDNA through reverse transcription. PCR reaction was performed for 40 cycles according to the instructions of qRT-PCR kit with U6 as endogenous control. Relative expression of miRNA is expressed as 2−∆∆Ct. miR-194 primers are 5′-CACGCATGTAACAGCAAC (forward) and 5′-CCAGTGCAGGGTCCGAGGTA (reverse); miR-29 primers are 5′-GGTACCGGTTGTCTTGGGTTTATTGs' (forward) and 5′-GAATTCAAATACTTCAGAGCTG-3′ (reverse).
MTT assay
Cells were inoculated into a 96-well plate with 5000 cells in each well. Cells were transfected with miR-194 mimic, miR-29 mimic and scramble mimic, 6 wells were set for each transfection. After incubation for 48 h, 10 µl of MTT (5 mg/l) was added, followed by incubation at 37°C for 4 h. The medium was removed, and 100 µl of DMSO was added, followed by shaking for 10 min. OD values at 490 nm were measured.
Transwell assay
Matrigel was kept at 4°C overnight. Matrigel (50 µl) (1:10 dilution) was added to the upper chamber, followed by incubation at 37°C for 4 h. The lower chamber was filled with 500 µl of culture medium containing 10% FBS. Cells transfected with miR-194 mimic, miR-29 mimic and scramble mimic, and control cells were digested, and were resuspended in RPMI-1640 medium containing 0.2% bovine serum albumin to adjust the cell density to 2×105/ml. Then 200 µl of cell suspension was added to the upper chamber. After incubation for 24 h, Matrigel and the cells that failed to invade were removed. The membrane was fixed for 30 min, and staining with 0.1% crystal violet was performed for 10 min. The results were observed and counted under an inverted microscope. The experiment was repeated three times.
Statistical analysis
Statistical analysis was performed using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA). Measurement data are expressed as mean ± SD, and processed by Student's t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
Comparison of expression levels of miR-194 and miR-29 in gastric cancer tissues and adjacent tissues
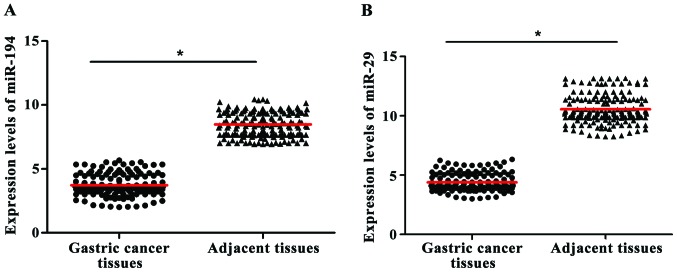
RT-PCR results showed that the relative contents of miR-194 and miR-29 in gastric cancer tissue were 3.72±1.65 and 4.39±1.47, respectively. While the relative contents of miR-194 and miR-29 in normal tissue were 8.47±3.79 and 10.56±3.95, respectively. Expression levels of miR-194 and miR-29 were significant lower in cancer tissue than in normal tissue (t=10.84, P<0.001) (Fig. 1).
Figure 1.
Comparison of miR-194 and miR-29 expression in gastric cancer tissues and adjacent tissues. RNA was extracted from tissue, and RT-PCR was used to detect the expression of miRNAs in different tissues. (A) The expression of miR-194 in tumor tissue was lower than that in adjacent tissues (B). The expression of miR-29 in tumor tissue was lower than that in adjacent tissues (*P<0.05).
Correlations between the expression of miR-194 and miR-29 in cancer tissue and the clinicopathological features
No significant differences in expression levels of miR-194 and miR-29 were found between patients in different gender and age groups. Expression levels of miR-194 and miR-29 in gastric cancer tissue were closely related to TNM stage, differentiation degree of cancer cells and lymph node metastasis. Expression levels of miR-194 and miR-29 in TNM stage III+IV were lower than that in stage I+II. Expression levels of miR-194 and miR-29 in tumors with lower degree of differentiation and with lymph node metastasis were significantly lower than those in tumor with higher degree of differentiation and without lymph node metastasis (P<0.05), indicating that expression of miR-194 and miR-29 is closely related to the degree of malignancy, metastasis and prognosis of gastric cancer (Table I).
Overexpression of miR-194 and miR-29 in gastric cancer SGC7901 cells
miR-194 mimic, miR-29 mimic and scramble mimic were transfected into SGC7901 cells. Expression levels of miR-194 and miR-29 were detected by qRT-PCR. Results showed that, compared with control group and scramble group, expression levels of miR-194 and miR-29 were significantly increased in SGC7901 cells after the transfection of miR-194 and miR-29 mimics (Fig. 2).
Figure 2.
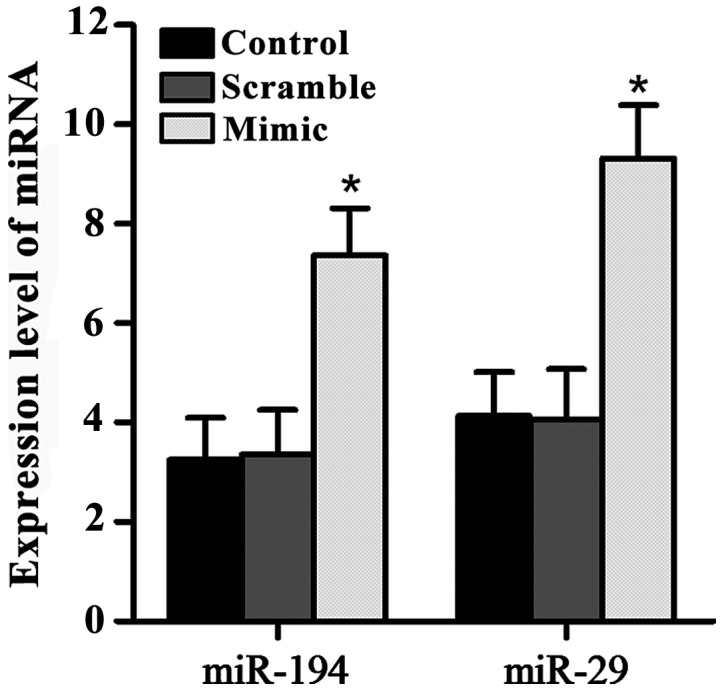
Overexpression of miR-194 and miR-29 in gastric cancer SGC7901 cells. SGC7901 cells were transfected with miR-194 mimic, miR-29 mimic and scramble mimic. Significantly increased expression levels of miR-194 and miR-29 were observed after the transfection with miR-194 mimic and miR-29 mimic (*P<0.05).
Effects of miR-194 and miR-29 overexpression on the proliferation of SGC7901 cells
MTT assay showed that the absorbance values of SGC7901 cells with miR-194 mimic and miR-29 mimic transfection were 0.49±0.11 and 0.51±0.10, respectively, after incubation for 48 h. While the absorbance value of control cells was 0.91±0.08 and the cells transfected with scramble mimics was 0.88±0.09. Significant differences were found between them (P<0.05) (Fig. 3).
Figure 3.
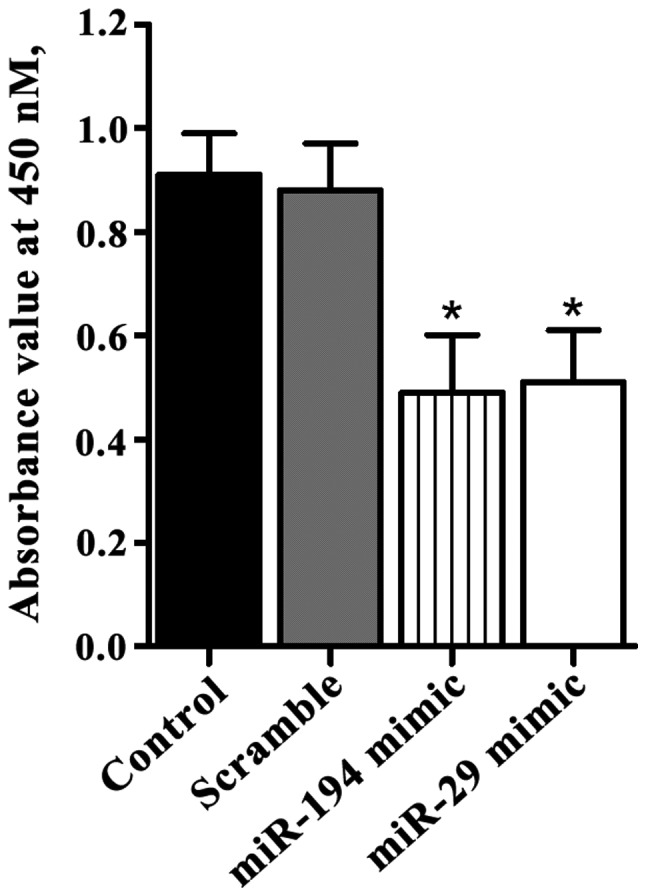
miR-194 and miR-29 overexpression inhibited the proliferation of SGC7901 cells. MTT assay was used to detect cell proliferation at 48 h after transfection. miR-194 and miR-29 overexpression significantly inhibited the proliferation of SGC7901 cells (*P<0.05).
Effects of miR-194 and miR-29 overexpression on migration of SGC7901 cells
Transwell assay showed that 10.6±1.6 cells with miR-194 mimic transfection passed through the membrane, and the number for cells with miR-29 mimic transfection was 9.6±1.5, no significant differences were found between those two types of cells. Significant differences were found between these two types of cells and untreated SGC7901 cells 23.1±2.6 and cells transfected with scramble mimic 20.5±1.9 (P<0.05) (Fig. 4).
Figure 4.
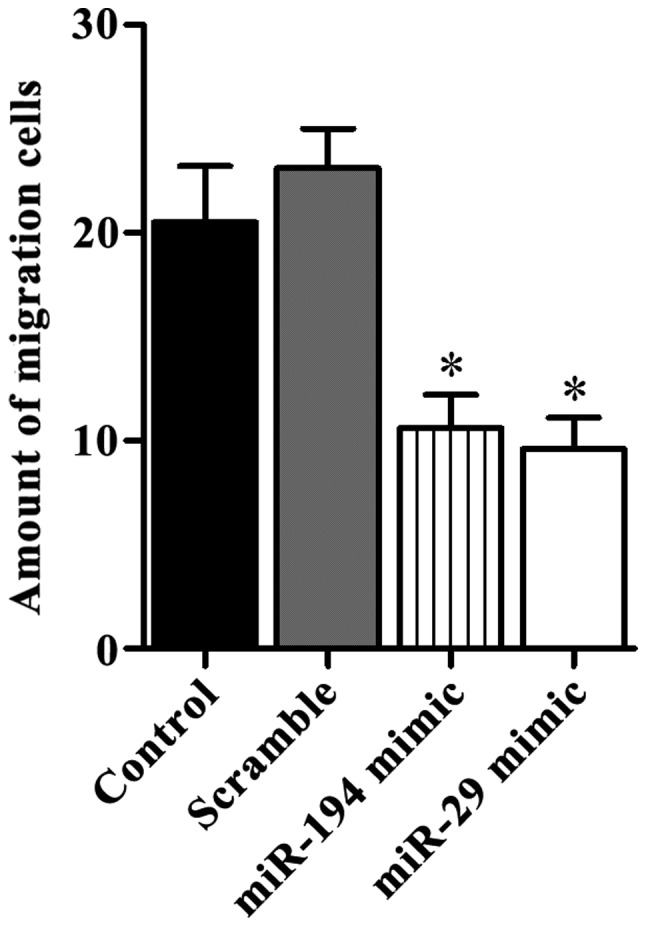
Overexpression of miR-194 and miR-29 inhibit migration of SGC7901 cells. Transwell invasion assay was performed and the results showed that miR-194 and miR-29 overexpression could significantly inhibit the migration of SGC7901 cells (*P<0.05).
Discussion
miRNAs are widely distributed in many species. Each miRNA has approximately 200 potential target genes. miRNAs in the human genome are involved in the regulation of 30–50% gene expression. Any change in the expression of miRNAs may lead to the development of diseases (8). miRNA plays an important role in the development of the tumor. Research on miRNA in tumors is mainly focused on the mechanism of the onset, growth and metastasis of tumor. miRNAs are upregulated or downregulated in many tumors to perform a role as oncogene or tumor suppressor. With the important roles in the diagnosis, treatment and prognosis of tumors, miRNAs have become a hot topic in tumor-related research (9,10).
Gastric cancer is a common digestive system tumor that seriously affects human health. The pathogenesis of this disease remains to be further studied. miR-194 is specifically expressed in the gastrointestinal tract, and it has been reported that miR-29 expression is downregulated in gastric cancer (11). In order to study the roles of miR-194 and miR-29 in gastric cancer, tumor tissue and adjacent tissue were collected from 165 cases of gastric cancer patients. Results showed that expression levels of miR-194 and miR-29 were significantly lower in gastric cancer tissue than in adjacent healthy tissue, indicating than miR-194 and miR-29 are closely related to the development of tumors. Further studies showed that miR-194 and miR-29 expression levels in TNM stage III+IV were lower than that in stage I+II, and expression levels of those two miRNAs were significantly lower in patients with lymph node metastasis than in patients without. Those results suggest that the low expression levels of miR-194 and miR-29 are closely related to the degree of malignancy. Higher degree of malignancy is accompanied with lower expression levels and worse prognosis. miR-194 and miR-29 are tumor suppressor genes in most tumors. For example, miR-29 can downregulate anti-apoptotic factor Mcl-1 to promote apoptosis of bile duct epithelial cancer cells (12); level of miR-29 expression was significantly lower in breast cancer patients with poor prognosis (13); the low expression of miR-29 is closely related to the metastasis and prognosis of malignant tumors, and miR-29 can be used as a key biomarker for predicting the recurrence and progression of tumors (14).
MYC can reduce the expression level of miR-29 in pancreatic ductal adenocarcinoma (PDAC), increase the degree of malignancy, and MYC knockout can upregulate the level of miR-29 expression in PDAC, to increase sensitivity of the tumor to gemcitabine (15). Low expression level of miR-29 was also detected in other tumors such as head and neck squamous cell carcinoma, and prostate cancer (16,17). Studies have shown that miR-194 is downregulated in non-small cell lung cancer and clear cell renal cell carcinoma (ccRCC), and the reduced expression level is closely related to prognosis (18,19). Low expression levels of miR-194 was detected in ovarian cancer, colorectal cancer, melanoma and other tumors, and miR-194 can participate in the regulation of drug sensitivity and tumor growth through a series of signal pathways (20–22).
To further study the role and possible mechanism of miR-194 and miR-29 in gastric cancer, CGC7901 cells with miR-194 and miR-29 overexpression were established to observe cell proliferation and invasion. After transfection with miR-194 mimic and miR-29 mimic, RT-PCR results showed that expression levels of miR-194 and miR-29 were significantly increased in CGC7901 cells. Further study showed that miR-194 and miR-29 overexpression could inhibit the proliferation and invasion of SGC7901 cells. In laryngeal squamous cell carcinoma and ccRCC, lysyl oxidase 2 (LOXL2) gene can directly interact with miR-29s to inhibit the migration and invasion of cancer cells (23,24). Stable expression of miR-194 can inhibit the proliferation and invasion of non-small cell lung cancer cells and promote drug sensitivity of tumor cells to cisplatin (18). miR-194 can inhibit the proliferation and invasion of ovarian cells through protein tyrosine phosphatase non-receptor type 12 (PTPN12) (25). miR-194 can also inhibit the proliferation and migration of bladder cancer cells (26).
miRNAs can participate in the regulation of tumor-related signaling pathways such as PI3K/KT signaling pathway (27), RAS-MAPK signaling pathway to affect the occurrence and development of tumors (28). Some miRNAs can enter the body fluid, and the detection of miRNAs in serum or body fluids has important roles in early diagnosis of tumors (29,30). In this study, we found that the expression levels of miR-194 and miR-29 were low in gastric cancer tissue, and the low expression level was correlated with poor prognosis. miR-194 and miR-29 are closely related to tumor cell proliferation, migration and invasion.
Further study of the mechanism and signal regulation of miR-194 and miR-29 in gastric cancer and the detection of miR-194 and miR-29 in serum are of great significance for the early diagnosis and prognosis of gastric cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
XZ contributed to the conception and design of the study. YH drafted this manuscript. ZT collected the data. YH and ZT interpreted the data. FW revised the manuscript critically for important intellectual content. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Jinan Central Hospital Affiliated to Shandong University (Jinan, China). Signed written informed consents were obtained from the patients and/or guardians.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 3.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 4.Rabinowits G, Gerçel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: A diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10:42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 5.Sampson VB, Yoo S, Kumar A, Vetter NS, Kolb EA. MicroRNAs and potential targets in osteosarcoma: Review. Front Pediatr. 2015;3:69. doi: 10.3389/fped.2015.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Callegari E, Gramantieri L, Domenicali M, D'Abundo L, Sabbioni S, Negrini M. MicroRNAs in liver cancer: A model for investigating pathogenesis and novel therapeutic approaches. Cell Death Differ. 2015;22:46–57. doi: 10.1038/cdd.2014.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: A microRNA expression analysis. Lancet Oncol. 2010;11:136–146. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang JB, Song W, Wang YY, Liu MG, Sun MM, Liu H. Study on correlation between PKIB and pAkt expression in breast cancer tissues. Eur Rev Med Pharmacol Sci. 2017;21:1264–1269. [PubMed] [Google Scholar]
- 9.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang D, Fan Z, Liu F, Zuo J. Hsa-miR-21 and Hsa-miR-29 in tissue as potential diagnostic and prognostic biomarkers for gastric cancer. Cell Physiol Biochem. 2015;37:1454–1462. doi: 10.1159/000438514. [DOI] [PubMed] [Google Scholar]
- 12.Mott JL, Kobayashi S, Bronk SF, Gores GJ. miR-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–6140. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shinden Y, Iguchi T, Akiyoshi S, Ueo H, Ueda M, Hirata H, Sakimura S, Uchi R, Takano Y, Eguchi H, et al. miR-29b is an indicator of prognosis in breast cancer patients. Mol Clin Oncol. 2015;3:919–923. doi: 10.3892/mco.2015.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi Y, Huang Y, Pang L, Gu W, Wang N, Hu J, Cui X, Zhang J, Zhao J, Liu C, et al. Prognostic value of the MicroRNA-29 family in multiple human cancers: A meta-analysis and systematic review. Clin Exp Pharmacol Physiol. 2017;44:441–454. doi: 10.1111/1440-1681.12726. [DOI] [PubMed] [Google Scholar]
- 15.Kwon JJ, Quirin K, Aljoufi A, Nafiseh A, Kota J. Restoration of MYC-repressed miR-29 in pancreatic cancer cells leads to increased reactive oxygen species and gemcitabine sensitization. Cancer Res. 2017;77(Suppl 13):LB–327. doi: 10.1158/1538-7445.AM2017-LB-327. [DOI] [Google Scholar]
- 16.Kinoshita T, Nohata N, Hanazawa T, Kikkawa N, Yamamoto N, Yoshino H, Itesako T, Enokida H, Nakagawa M, Okamoto Y, et al. Tumour-suppressive microRNA-29s inhibit cancer cell migration and invasion by targeting laminin-integrin signalling in head and neck squamous cell carcinoma. Br J Cancer. 2013;109:2636–2645. doi: 10.1038/bjc.2013.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishikawa R, Goto Y, Kojima S, Enokida H, Chiyomaru T, Kinoshita T, Sakamoto S, Fuse M, Nakagawa M, Naya Y, et al. Tumor-suppressive microRNA-29s inhibit cancer cell migration and invasion via targeting LAMC1 in prostate cancer. Int J Oncol. 2014;45:401–410. doi: 10.3892/ijo.2014.2437. [DOI] [PubMed] [Google Scholar]
- 18.Zhu X, Li D, Yu F, Jia C, Xie J, Ma Y, Fan S, Cai H, Luo Q, Lv Z, et al. miR-194 inhibits the proliferation, invasion, migration, and enhances the chemosensitivity of non-small cell lung cancer cells by targeting forkhead box A1 protein. Oncotarget. 2016;7:13139–13152. doi: 10.18632/oncotarget.7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nofech-Mozes R, Khella HWZ, Scorilas A, Youssef L, Krylov SN, Lianidou E, Sidiropoulos KG, Gabril M, Evans A, Yousef GM. MicroRNA-194 is a marker for good prognosis in clear cell renal cell carcinoma. Cancer Med. 2016;5:656–664. doi: 10.1002/cam4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakamura K, Sawada K, Yoshimura A, Nakatsuka E, Kinose Y, Mabuchi S, Kimura T. MiR-194 modulates paclitaxel resistance in ovarian cancer cells through the regulation of MDM-2 expression. Cancer Res. 2017;77(Suppl 13):3436. doi: 10.1158/1538-7445.AM2017-3436. [DOI] [Google Scholar]
- 21.Wang B, Shen ZL, Gao ZD, Zhao G, Wang CY, Yang Y, Zhang JZ, Yan YC, Shen C, Jiang KW, et al. MiR-194, commonly repressed in colorectal cancer, suppresses tumor growth by regulating the MAP4K4/c-Jun/MDM2 signaling pathway. Cell Cycle. 2015;14:1046–1058. doi: 10.1080/15384101.2015.1007767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo B, Hui Q, Zhang Y, Chang P, Tao K. miR-194 is a negative regulator of GEF-H1 pathway in melanoma. Oncol Rep. 2016;36:2412–2420. doi: 10.3892/or.2016.5020. [DOI] [PubMed] [Google Scholar]
- 23.Mizuno K, Seki N, Mataki H, Matsushita R, Kamikawaji K, Kumamoto T, Takagi K, Goto Y, Nishikawa R, Kato M, et al. Tumor-suppressive microRNA-29 family inhibits cancer cell migration and invasion directly targeting LOXL2 in lung squamous cell carcinoma. Int J Oncol. 2016;48:450–460. doi: 10.3892/ijo.2015.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishikawa R, Chiyomaru T, Enokida H, Inoguchi S, Ishihara T, Matsushita R, Goto Y, Fukumoto I, Nakagawa M, Seki N. Tumour-suppressive microRNA-29s directly regulate LOXL2 expression and inhibit cancer cell migration and invasion in renal cell carcinoma. FEBS Lett. 2015;589:2136–2145. doi: 10.1016/j.febslet.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 25.Liang T, Li L, Cheng Y, Ren C, Zhang G. MicroRNA-194 promotes the growth, migration, and invasion of ovarian carcinoma cells by targeting protein tyrosine phosphatase nonreceptor type 12. Onco Targets Ther. 2016;9:4307–4315. doi: 10.2147/OTT.S90976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang M, Zhuang Q, Cui L. MiR-194 inhibits cell proliferation and invasion via repression of RAP2B in bladder cancer. Biomed Pharmacother. 2016;80:268–275. doi: 10.1016/j.biopha.2016.03.026. [DOI] [PubMed] [Google Scholar]
- 27.Coffre M, Benhamou D, Riess D, Blumenberg L, Snetkova V, Hines MJ, Chakraborty T, Bajwa S, Jensen K, Chong MMW, et al. miRNAs are essential for the regulation of the PI3K/AKT/FOXO pathway and receptor editing during B cell maturation. Cell Rep. 2016;17:2271–2285. doi: 10.1016/j.celrep.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masliah-Planchon J, Garinet S, Pasmant E. RAS-MAPK pathway epigenetic activation in cancer: miRNAs in action. Oncotarget. 2016;7:38892–38907. doi: 10.18632/oncotarget.6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madhavan B, Yue S, Galli U, Rana S, Gross W, Müller M, Giese NA, Kalthoff H, Becker T, Büchler MW, et al. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int J Cancer. 2015;136:2616–2627. doi: 10.1002/ijc.29324. [DOI] [PubMed] [Google Scholar]
- 30.Jiang X, Du L, Wang L, Li J, Liu Y, Zheng G, Qu A, Zhang X, Pan H, Yang Y, et al. Serum microRNA expression signatures identified from genome-wide microRNA profiling serve as novel noninvasive biomarkers for diagnosis and recurrence of bladder cancer. Int J Cancer. 2015;136:854–862. doi: 10.1002/ijc.29041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.