Abstract
Malignant gliomas are treated with temozolomide (TMZ) at present, but often exhibit resistance to this agent. Cancer-initiating cells (CICs) have been suggested to lead to TMZ resistance. The mechanisms underlying CICs-based TMZ resistance are not fully understood. MicroRNAs (miRNAs) have been demonstrated to serve important roles in tumorigenesis and TMZ resistance. In the present study, a sphere forming assay and western blot analysis were performed to detect the formation of CICs and fibroblast activation protein α (FAP-α) protein expression. It was revealed that TMZ resistance promoted the formation of CICs and upregulated FAP-α expression in glioblastoma cells. Over-expressing FAP-α was also demonstrated to promote TMZ resistance and induce the formation of CICs in U251MG cells. In addition, using a reverse transcription-quantitative polymerase chain reaction, it was observed that miR-204 was downregulated in U251MG-resistant (-R) cells. miR-204 expression negatively correlated with the FAP-α levels in human glioblastoma tissues, and it may inhibit the formation of CICs and reverse TMZ resistance in U251MG-R cells. Therefore, it was concluded that miR-204 reversed temozolomide resistance and inhibited CICs phenotypes by degrading FAP-α in glioblastoma.
Keywords: microRNA-204, fibroblast activation protein α, temozolomide resistance, cancer-initiating cells
Introduction
Glioblastoma is the most frequently occurring primary malignant brain tumor in adults and exhibits a spectrum of aberrantly aggressive phenotypes in the USA (1,2). The median survival time is generally <1 year from the time of diagnosis, and even in the most favorable situations, the majority of patients succumb within 2 years (3–5). At present, treatment options include surgery, radiation and chemotherapy with methylating agents and nitrosoureas (6). Unfortunately, the treatment options often do not result in a cure, subsequently leading to tumor progression and mortality of the patient. One of the reasons for failure of treatment is due to de novo or acquired resistance to chemotherapeutic agents. However, the molecular mechanisms underlying chemotherapy resistance of glioblastoma cells remain unknown.
Fibroblast activation protein α (FAP-α) is a member of the serine integral membrane peptidases (SIMPs) family, which also includes prolyl endopeptidase, dipeptidyl peptidase IV and dipeptidyl peptidase IIX. These peptidases are inducible, specific for proline-containing peptides and are active on the cell surface (7–9). FAP-α is significantly associated with poor outcome in patients with breast cancer (10). In vitro, it may promote proliferation and inhibit migration of breast cancer cells, potentially by regulating the focal adhesion kinase pathway, and its overexpression is associated with neoplastic progression of esophageal lesions (10). FAP-α is highly expressed on the surface of glioma cells and contributes to diffuse glioma invasion through extracellular matrix components (11). However, to the best of our knowledge, its role has not been studied in cancer-initiating cells (CICs) or chemotherapy resistance of glioblastoma.
MicroRNAs (miRNAs/miRs) are a class of small noncoding RNAs (~22 nucleotides) and negatively regulate protein-coding gene expression by targeting mRNA degradation or translation inhibition (12–14). Deregulation of miRNAs has been implicated in the development and progression of glioblastoma, and they serve pivotal roles in development, particularly in modulating stem cell-specific pathways (15–17). Previously, it was demonstrated that miR-204 may be a useful drug target in the treatment and diagnosis of glioblastoma multiforme (GBM) (9,18–20). In the present study, it was identified that miR-204 may reverse temozolomide resistance and inhibit CICs phenotypes by degrading FAP-α in glioblastoma.
Materials and methods
Patients
Between November 2013 and October 2015, 18 patients (10 male, 8 female) with glioblastoma were enrolled at the Department of Neurosurgery, Beijing Chaoyang Hospital. The mean age was 57 years (range, 35–78 years). All tissues were examined histologically, and pathologists confirmed the diagnosis. The present study was approved by the ethics committee of Beijing Chaoyang Hospital, and each patient signed an informed consent form at the time of enrollment.
Human glioblastoma cell line
U251MG cells were purchased from the Institute of Biochemistry and Cell Biology Institute of Shanghai, Chinese Academy of Sciences (Shanghai, China) within 3 months of experiments. To obtain temozolomide (TMZ)-resistant U251MG cells (U251MG-R cells), U251MG cells were treated with increasing concentrations of TMZ (10−7, 10−6 and 10−5 M). The U251MG-R cells were considered to be established when colonies grew at similar rate in the presence or absence of 10−5 M TMZ for 3 days (data not shown). The half maximal inhibitory concentration (IC50) of U251MG-R cells increased by 12-fold, as compared with the U251MG cells (data not shown). Cells were cultured in normal culture medium which was composed of Dulbecco modified Eagle medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and antibiotics (100 mg/ml penicillin; 100 U/ml streptomycin) in a 5% CO2 incubator at 37°C.
FAP-α expressing plasmid/empty vector, pre-miR-204/control miR and transfection experiments
FAP-α expressing plasmids and empty vectors were donated by Dr. Chao Wang (Cardiff University-Peking University Cancer Institute, Cardiff University School of Medicine, Cardiff, Wales, UK) and produced as described previously (10). Pre-miR-204 (sequence: 5′-UUCCCUUUGUCAUCCUAUGCCU-3′)/control miR (sequence: 5′-UACCGUAUCUCUUCGUAAGCGU-3′) were purchased from Ambion (Ambion; Thermo Fisher Scientific, Inc.). For transfection experiments, U251MG-R cells were cultured in serum-free medium (DMEM) without antibiotics at 60% confluence for 24 h in a 5% CO2 incubator at 37°C, and then transfected with 50 nM pre-miR-204 or 50 nM control miR using a transfection reagent (Lipofectamine 2000; Thermo Fisher Scientific, Inc.) at room temperature according to the manufacturer's protocol. Following incubation for 6 h at 37°C, the medium was removed and replaced with normal culture medium for 48 h, unless otherwise specified. Subsequent experiments were performed 48 h after transfection.
Western blotting
U251MG and U251MG-R cells and glioblastoma tissues were washed once with PBS and lysed using a radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology, Haimen, China) supplemented with phenylmethanesulfonyl fluoride (Beyotime Institute of Biotechnology) for 30 min at 0°C. Protein samples were measured using Bradford Protein Assay kit (Beyotime Institute of Biotechnology) for 10 min at room temperature and boiled for 10 min in SDS sample buffer (Beyotime Institute of Biotechnology). Then protein extracts (10 µg per lane) were resolved through 8% SDS-PAGE, transferred to polyvinylidene difluoride membranes (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and blocked for 60 min at room temperature in 5% skim milk powder (w/v) in NaCl/Pi at room temperature. Afterwards, the blots were incubated with antibodies including rabbit anti-FAP-α (cat no. ab173904; 1:500; Abcam, Cambridge, MA, USA), rabbit anti-signal transducer and activator of transcription 3 (STAT3; cat no. ab68153; 1:500; Abcam), rabbit anti-mouse double minute 2 homolog (MDM2; cat no. ab38618; 1:500; Abcam), rabbit anti-prominin-1 (CD133; cat no. ab19898; 1:500; Abcam), rabbit anti-tyrosine-protein kinase met (MET; cat no. ab51067; 1:500; Abcam), rabbit anti-ras-related protein rab-22A (RAB22A; cat no. ab137093; 1:500; Abcam), rabbit anti-EZRIN (cat no. ab4069; 1:500; Abcam), rabbit anti-SRY-box 4 (SOX4; cat no. ab80261; 1:500; Abcam), rabbit anti-activating transcription factor 2 (ATF2; cat no. ab47476; 1:500; Abcam), rabbit anti-erythropoietin-producing human hepatocellular receptors (EphB; cat no. ab196793; 1:500; Abcam) or β-actin (cat no. ab5694; 1:500; Abcam) at 4°C, and then with IRDye™-800 conjugated goat anti-rabbit secondary antibody (cat no. ab191866; 1:10,000; Abcam) were used for 30 min at room temperature. The specific proteins were visualized by Odyssey™ Infrared Imaging system using Odyssey Image Studio Software (Version 2009; Gene Company, Ltd., Hong Kong, China).
MTT assay
To monitor resistance to TMZ, U251MG and U251MG-R cells were treated with 10 µM TMZ for 24 h. An MTT assay was performed as described previously (19). Data were analyzed with Origin 7.5 software (OriginLab, Northampton, MA, USA) to fit a sigmoidal curve. IC50 is the TMZ concentration that reduces proliferating cells by 50%.
Sphere formation assay
U251MG and U251MG-R cells (103/ml) in serum-free RPMI-1640/1 mM Na-pyruvate (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) were seeded on 0.5% agar pre-coated 6-well plates. After 10 days, half the medium was exchanged every third day. Single spheres were selected and measured using a phase contrast microscope (magnification, ×100) with scales (Eclipse TS-100; Nikon Corporation, Tokyo, Japan).
Immunofluorescence staining
This protocol was performed as described previously (9). Following transfection, the U251 MG-R cells were fixed in 4% paraformaldehyde for 15 min at room temperature, and then blocked with goat serum blocking solution (Tiangen Biotech Co., Ltd., Beijing, China) for 20 min at room temperature. Then, a rabbit anti-FAP-α antibody (1:200 dilution; Abcam) was added, and the mixtures were incubated in a humid chamber overnight in a 5% CO2 incubator at 37°C. Subsequent to washing 3 times with NaCl/Pi, cells were incubated with goat anti-rabbit secondary antibody (conjugated with Alexa Fluor® 488; cat no. ab150077; 1:10,000; Abcam) for 30 min at 37°C. Following washing with NaCl/Pi, the samples were observed under a laser scanning confocal microscope (magnification, ×100; Olympus Corporation, Tokyo, Japan). The cells nuclei were stained with DAPI (1 µg/ml) for 20 min at 37°C.
Bioinformatics
The analysis of potential microRNA target sites was performed using the prediction algorithm miRanda (http://www.microrna.org/). Target mRNA Search was used (date of access: 2010-11-01) using the following search terms: Target mRNA=FAP (FAP-α) and species=Homo sapiens.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RT-qPCR was performed to determine the expression of miR-204 and FAP-α. Total RNA was extracted from glioblastoma tissues and U251 MG-R cells using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Total RNA (500 ng) was quantitated at 260 nm and reverse-transcribed into cDNA using the PrimeScript RT reagent kit (Takara Biotechnology, Co., Ltd., Dalian, China) according to the manufacturer's protocol, at 37°C for 15 min and 85°C for 30 sec. qPCR was performed using the SYBR Premix Ex Taq™ kit (Takara Biotechnology, Co., Ltd.) according to the manufacturer's protocol in the ABI PRISM 7900HT system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling conditions were as follows: 50°C for 2 min, 95°C for 10 min followed by 40 cycles with each cycle consisting of 30 sec at 95°C, and 1 min at 60°C. Cycle threshold (Cq) values were determined using SDS version 2.4 software (Applied Biosystems; Thermo Fisher Scientific, Inc.). FAP-α expression levels were normalized to GAPDH expression using the 2−ΔΔCq method (21). The primer sequences were as follows: FAP-α forward, 5′-TTAGTCTGACAAAGAGAAACACTG-3′ and reverse, 5′-ATGAAGACTTGGGTAAAAATCG-3′; GAPDH forward, 5′-CGAAGTCAACGGATTTGGTCGTAT-3′ and reverse 5′-AGCCTTCTCGGTGGTGAAGAC-3′.
For miR-204 detection, 1 µg total RNA extracted from clinical samples was converted to cDNA using the TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The resulting cDNA was diluted at a ratio of 1:40 and mixed with 1 µl miR-204 or U6 TaqMan primers in triplicate wells using TaqMan Universal Master Mix II without Uracil DNA glycosylase (Applied Biosystems; Thermo Fisher Scientific, Inc.). The primer sequences were as follows: miR-204 forward, 5′-CTGTCACTCGAGCTGCTGGAATG-3′ and reverse, 5′-ACCGTGTCGTGGAGTCGGCAATT-3′; U6 forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′, reverse 5′-CGCTTCACGAATTTGCGTGTCAT-3′. The thermocycling conditions were as follows: Denaturation at 95°C for 30 sec, followed by 40 cycles at 95°C for 5 sec and 60°C for 30 sec, and extension at 95°C for 15 sec. The plates were read using the ABI PRISM 7900HT system (Applied Biosystems; Thermo Fisher Scientific, Inc.). Cq values were calculated using SDS version 2.4 software (Applied Biosystems; Thermo Fisher Scientifc, Inc.). miR-204 expression level was normalized to that of U6 using the 2−ΔΔCq method (21). The TaqMan probes for miR-204 and U6 were purchased from Applied Biosystems (Thermo Fisher Scientific, Inc.). The assay was performed in triplicate.
Statistical analysis
Statistical analysis was performed using SAS statistical software (version 9.4; SAS Institute, Cary, NC). Data are presented as the mean ± the standard error of the mean. Student's t test (two-tailed) was used to compare between two groups. The correlation between the expression levels of miR-204 and FAP-α were analyzed using Spearman correlation analysis (22). P<0.05 was considered to indicate a statistically significant difference.
Results
TMZ resistance promotes formation of CICs and upregulates FAP-α expression in glioblastoma
To identify U251MG-R cells that were resistant to TMZ, an MTT assay using TMZ was performed using the U251MG and U251MG-R cells. The results indicated that the U251MG-R cells were resistant to TMZ (Fig. 1A). In order to identify whether TMZ resistance affected CICs traits in U251MG-R cells, sphere forming assays were performed to assess the capacity of CIC or CIC-like cell self-renewal in U251MG, and U251MG-R cells. The sphere forming assay indicated that U251MG-R cells formed larger spheres after 14 days of culture compared with U251MG cells (Fig. 1B). STAT3 and MDM2 may promote TMZ-resistance (22–24). CD133 and MET is essential for glioblastoma stem cell maintenance (25,26). Western blotting identified that STAT3, MDM2, CD133 and MET were markedly upregulated in U251MG-R cells compared with U251MG cells (Fig. 1C). In order to detect whether TMZ-resistance is associated with FAP-α protein expression, FAP-α protein expression was analyzed in U251MG and U251MG-R cells. The results demonstrated that the level of FAP-α protein was markedly upregulated in U251MG-R cells (Fig. 1D).
Figure 1.
TMZ resistance promotes the formation of cancer-initiating cells and upregulates FAP-α expression in glioblastoma (n=3). (A) MTT for cell viability in U251MG and U251MG-R cells. Cells were untreated or treated with TMZ. (B) Sphere growth for U251MG and U251MG-R cells. Scale bar, 200 µm. (C) Western blotting for STAT3, MDM2, CD133 and MET in U251MG and U251MG-R cells. β-actin was the loading control. (D) Western blotting for FAP-α in U251MG and U251MG-R cells. β-actin was the loading control. R, resistant; TMZ, temozolomide; FAP-α, Fibroblast activation protein α; STAT3, Signal transducer and activator of transcription 3; MDM2, Mouse double minute 2 homolog; CD133, Prominin-1; MET, Tyrosine-protein kinase Met.
FAP-α promotes TMZ resistance and induces formation of CICs in U251MG cells
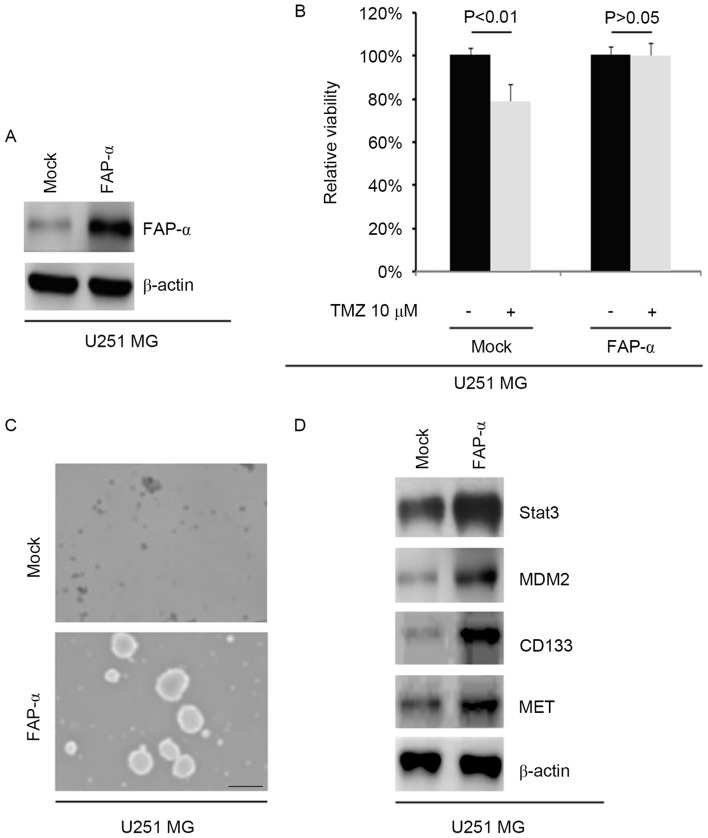
To identify the role of FAP-α, whether FAP-α expressing plasmids were able to stably express FAP-α protein in U251MG cells was tested. The results indicated that the levels of FAP-α protein were markedly increased by FAP-α-expressing plasmids in the cells (Fig. 2A). To additionally identify whether FAP-α affected TMZ efficacy in glioblastoma cells, U251MG cells were transfected with FAP-α-expressing plasmids. Then an MTT assay was performed in the cells transfected with FAP-α expressing plasmids. The results indicated that overexpressing FAP-α transformed U251MG cells into U251MG-R cells (Fig. 2B), suggesting that its overexpression promotes TMZ resistance.
Figure 2.
FAP-α promotes TMZ resistance and induces the formation of cancer-initiating cells in U251MG cells (n=3). (A) Western blotting for FAP-α in U251MG cells transfected with FAP-α expressing plasmids. β-actin was the loading control. (B) MTT for cell viability in U251MG cells. U251MG cells transfected with FAP-α expressing plasmids and empty vectors (mock) were untreated or treated with TMZ. (C) Sphere growth for U251MG cells transfected with FAP-α expressing plasmids and empty vectors (mock). Scale bar, 200 µm. (D) Western blotting for STAT3, MDM2, CD133 and MET in U251MG cells transfected with FAP-α expressing plasmids and empty vectors (mock). β-actin was the loading control. TMZ, temozolomide; FAP-α, Fibroblast activation protein α; STAT3, Signal transducer and activator of transcription 3; MDM2, Mouse double minute 2 homolog; CD133, Prominin-1; MET, Tyrosine-protein kinase Met.
In order to identify if FAP-α affected the CIC traits of U251MG cells, a sphere forming assay was performed to assess the capacity of CIC or CIC-like cell self-renewal in U251MG cells. The sphere forming assay demonstrated that FAP-α overexpressing cells formed larger spheres after 14 days of culture, thus expressing more CIC-like traits compared with the mock group (Fig. 2C). To identify whether FAP-α affects STAT3, MDM2, CD133 and MET protein expression, western blotting was performed to detect their expression in U251MG cells transfected with FAP-α expressing plasmids. All four protein levels were markedly increased following FAP-α overexpression compared with the mock control group (Fig. 2D).
miR-204 inhibits FAP-α protein expression in U251MG-R cells
Having demonstrated that FAP-α expression was specifically upregulated in U251MG-R cells and promoted the formation of CICs in U251MG cells, the mechanisms that induced FAP-α expression in U251MG-R cells were then investigated. miRNAs are a class of small (~22 nucleotide) noncoding RNAs that negatively regulate protein-coding gene expression by targeted mRNA degradation or translation inhibition (23). The downregulation of specific miRNA may contribute to the upregulation of oncogenes and TMZ resistance (24,25). Therefore, the present study hypothesized whether FAP-α was upregulated by the downregulation of specific miRNA in U251MG-R cells.
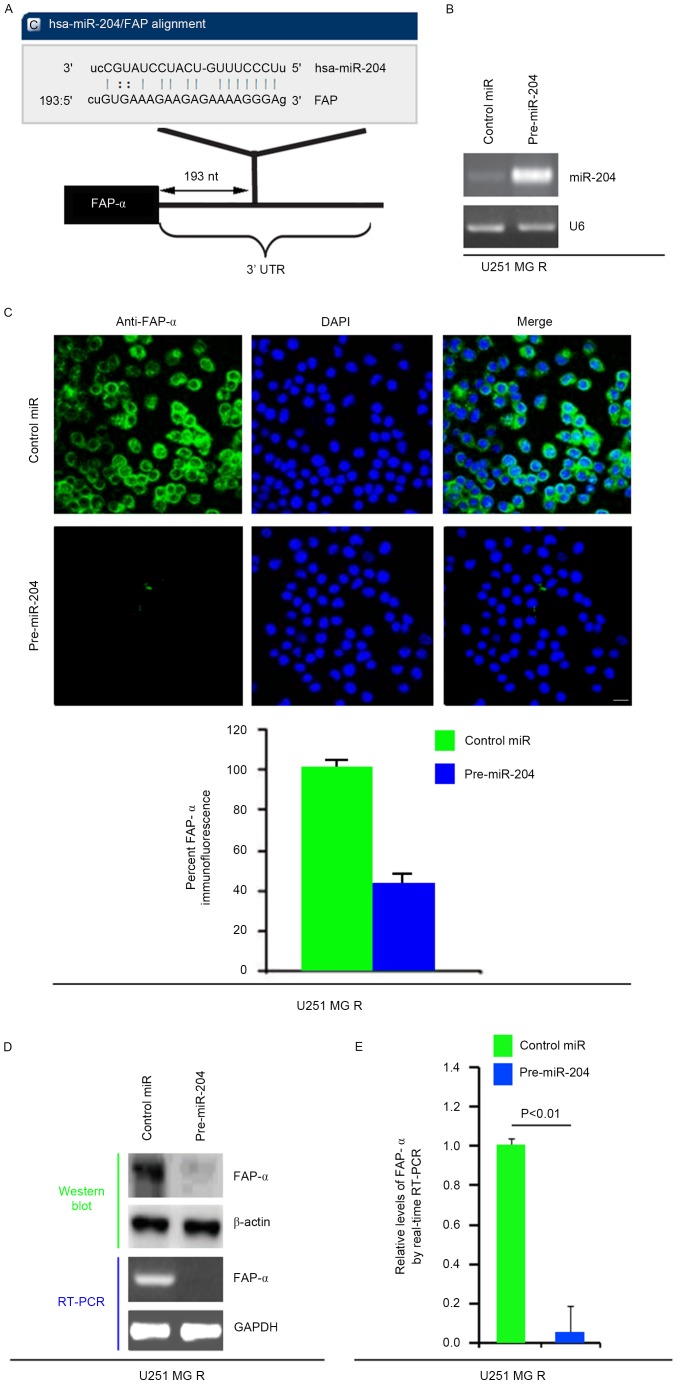
To confirm this hypothesis, a commonly used prediction algorithm, miRanda (http://www.microrna.org/), was used to analyze the 3′ untranslated region (UTR) of FAP-α. The algorithm predicted that miR-204 targeted the 3′UTR of FAP-α (Fig. 3A).
Figure 3.
miR-204 inhibits FAP-α protein expression in U251MG-R cells. (A) Schematic of predicted miR-204 binding sites in the 3′UTR of FAP-α mRNA by miRanda. (B) qPCR for miR-204 in U251MG-R cells. U251MG-R cells were infected with pre-miR-204 or control miR (mock). U6 was the loading control. (n=3). (C) Immunofluorescence analyses for U251MG-R cells transfected with pre-miR-204 and control miR (mock). Upper panel demonstrates microscopy images of immunofluorescence staining of one representative experiment (magnification, ×100). Bottom panel indicates graphic representation of mean fluorescence intensities. (n=3). Scale bar, 20 µm. (D) Western blotting for and RT-PCR for FAP-α protein and FAP-α mRNA, respectively, in U251MG-R cells infected as indicated. β-actin and GAPDH were the loading controls for the western blotting and RT-PCR (n=3). (E) qPCR for FAP-α in U251MG-R cells transfected with pre-miR-204 or control miR (mock). GAPDH was the loading control (n=3). UTR, untranslated region; miR, microRNA; R, resistant; PCR, polymerase chain reaction; RT, reverse transcription; q, quantitative; FAP-α, Fibroblast activation protein α.
Therefore, it was reasoned that miR-204 downregulated FAP-α expression by targeting its 3′UTR in U251MG-R cells, and that FAP-α was increased in U251MG-R cells due to the downregulation of miR-204 (26). In an attempt to identify the role of miR-204 in regulating FAP-α expression in U251MG-R cells, cells were transfected with pre-miR-204 and control miR. Following transfection, miR-204 expression was detected by qPCR and the results demonstrated that miR-204 was markedly increased following pre-miR-204 transfection in the cells (Fig. 3B). To confirm this observation, immunofluorescence analyses were performed in U251MG-R cells transfected with pre-miR-204 or control miR. The results indicated that FAP-α protein was suppressed in the cells transfected with pre-miR-204 (Fig. 3C). RT-PCR and western blotting was next performed to detect FAP-α expression in U251MG-R cells transfected with pre-miR-204 or control miR. The results demonstrated that FAP-α protein and mRNA were markedly downregulated in the cells transfected with pre-miR-204 (Fig. 3D). Consistent with the results of the RT-PCR, the qPCR data demonstrated that FAP-α mRNA was significantly reduced in U251MG-R cells transfected with pre-miR-204 compared with control miR-transfected group (Fig. 3E). All the data demonstrated that miR-204 may suppress FAP-α mRNA and protein expression in U251MG-R cells.
miR-204 inhibits the formation of CICs and reverses TMZ resistance in U251MG-R cells
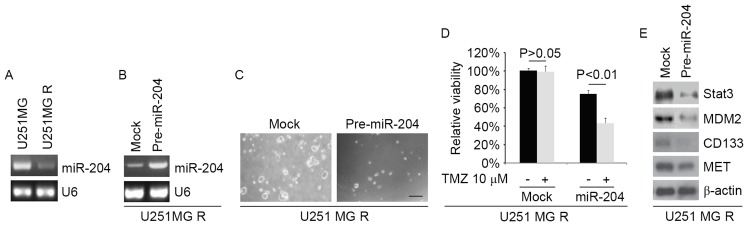
In order to detect whether TMZ resistance is associated with miR-204 expression, miR-204 expression was analyzed in U251MG and U251MG-R cells. The results suggested that miR-204 expression was markedly downregulated in U251MG-R cells (Fig. 4A). To identify the role of miR-204 in CIC formation, the ability of pre-miR-204 to stably express miR-204 in U251MG-R cells was examined. The results indicated that the levels of miR-204 were increased by pre-miR-204 in the cells (Fig. 4B). To additionally identify whether miR-204 affected the formation of CICs in U251MG-R cells, U251MG-R cells were transfected with pre-miR-204 and then a sphere formation assay was performed. The results demonstrated that miR-204 inhibited the formation of CICs in U251MG-R cells (Fig. 4C). To additionally identify whether miR-204 was able to affect TMZ efficacy in U251MG-R cells, U251MG-R cells were transfected with pre-miR-204. Then, an MTT assay in U251MG-R cells transfected with pre-miR-204 was performed. The results demonstrated that miR-204 was able to transform U251MG-R to U251MG cells (Fig. 4D), suggesting that miR-204 restoration reversed TMZ resistance. To identify whether miR-137 affected STAT3, MDM2, CD133 and MET expression, western blotting was performed to detect their expression in Huh7-R cells. The results suggested that STAT3, MDM2, CD133 and MET expression levels were attenuated by miR-204 (Fig. 4E).
Figure 4.
miR-204 inhibits formation of cancer-initiating cells and reverses TMZ resistance in U251MG-R cells. (A) qPCR for miR-204 in U251MG and U251MG-R cells. U6 was the loading control (n=3). (B) qPCR for miR-204 in U251MG-R cells transfected with pre-miR-204 and control miR (mock). β-actin was the loading control (n=3). (C) Sphere growth for U251MG-R cells transfected with pre-miR-204 and control miR (mock). Scale bar, 200 µm (n=3). (D) MTT assay for cell viability in U251MG-R cells. U251MG-R cells transfected with pre-miR-204 and control miR (mock) were untreated or treated with TMZ. (E) Western blotting for STAT3, MDM2, CD133 and MET in U251MG-R cells transfected with pre-miR-204 and control miR (mock). β-actin was the loading control (n=3). TMZ, temozolomide; STAT3, Signal transducer and activator of transcription 3; MDM2, Mouse double minute 2 homolog; CD133, Prominin-1; MET, Tyrosine-protein kinase Met; miR, microRNA; R, resistant; PCR, polymerase chain reaction; RT, reverse transcription; q, quantitative.
miR-204 expression is negatively associated with FAP-α levels in human glioblastoma tissues
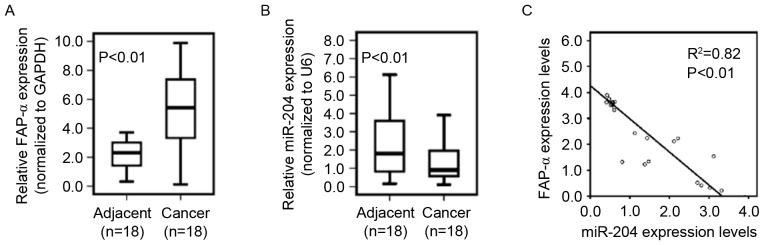
To determine whether reduced miR-204 expression correlates with increased levels of FAP-α in glioblastoma tissues, 18 pairs of primary glioblastoma tissues and adjacent normal tissues were used to determine FAP-α, and miR-204 expression using qPCR. The results indicated that the FAP-α mRNA level in glioblastoma tissues was significantly increased compared with that of adjacent normal tissues (Fig. 5A) and that the miR-204 level in glioblastoma tissues was significantly decreased compared with that in adjacent normal tissues (Fig. 5B). As demonstrated in Fig. 5C, linear correlation analysis suggested a significant inverse correlation between miR-204 and FAP-α mRNA expression in glioblastoma tissues (P<0.01).
Figure 5.
miR-204 expression is negatively correlated with FAP-α levels in human glioblastoma tissues. (A) qPCR for FAP-α in glioblastoma tissues and adjacent normal tissues. GAPDH was the loading control. (n=18). (B) qPCR for miR-204 in glioblastoma tissues and adjacent normal tissues. U6 was the loading control (n=18). (C) Pearson correlation for miR-204 and FAP-α of glioblastoma tissues and adjacent normal tissues (n=18). FAP-α, Fibroblast activation protein α; miR, microRNA; PCR, polymerase chain reaction; q, quantitative.
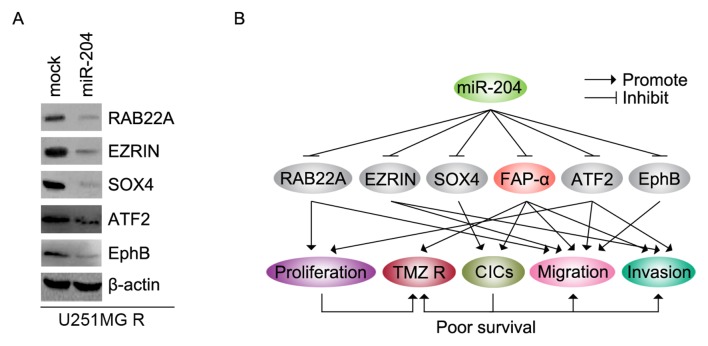
miR-204 inhibits RAB22A, EZRIN, SOX4, ATF2 and EphB protein in U251MG-R cells
In order to detect whether miR-204 inhibits RAB22A, EZRIN, SOX4, ATF2 and EphB protein expression, U251MG-R cells were transfected with pre-miR-204. Then, western blotting was performed to detect the protein levels. The results demonstrated that the levels of RAB22A, EZRIN, SOX4, ATF2 and EphB proteins were downregulated in U251MG-R cells transfected with pre-miR-204 (Fig. 6A).
Figure 6.
miR-204 inhibits RAB22A, EZRIN, SOX4, ATF2 and EphB protein in U251MG-R cells. (A) Western blotting for RAB22A, EZRIN, SOX4, ATF2 and EphB protein in U251MG-R cells transfected with pre-miR-204 and control miR (mock). (B) miR-204 regulates proliferation, TMZ resistance, CICs, migration and invasion by targeting RAB22A, EZRIN, SOX4, ATF2 and EphB protein in glioblastoma. miR, microRNA; R, resistant; TMZ, temozolomide; RAB22A, Ras-related protein Rab-22A; SOX4, SRY-box 4; ATF2, Activating transcription factor 2; EphB, erythropoietin-producing human hepatocellular receptors.
Discussion
TMZ has improved the prognosis of patients with GBM (27,28); however, almost all patients succumb to recurrence following chemoradiotherapy. miRNAs are involved in the acquisition of TMZ resistance in GBM (29). The expression level of miR-204 was significantly reduced in clinical glioma tissues compared with normal brain tissues (20). The introduction of miR-204 dramatically suppressed glioma cell growth, migration and invasion by targeting RAB22A (a member of the RAS oncogene family) (20), and miR-204 may suppress cell proliferation, migration and invasion through inhibiting ATF2 and EZRIN expression (18,19). miR-204 suppresses self-renewal, the stem cell-associated phenotype of glioma cells by targeting the stemness-governing transcriptional factor SOX4, and the migration-promoting receptor EphB2 (30). The results of the present study confirmed that RAB22A, EZRIN, SOX4, ATF2 and EphB were inhibited by miR-204 (Fig. 6A). Consistent with a previous study (30), the present study demonstrated that miR-204 significantly inhibited the formation of glioma initiating cells (GICs) in glioblastoma, and that SOX4 and EphB2 may be significantly downregulated by miR-204. GICs, also termed glioma stem cells, are responsible for tumor initiation, relapse and therapeutic resistance (26). The results of the present study demonstrated that the formation of CICs induced by the decrease in miR-204 levels may serve an important role in promoting TMZ resistance during the progression of glioblastoma.
FAP-α is highly expressed on the surface of glioma cells and contributes to diffuse glioma invasion through extracellular matrix components (11). However, its role in regulating the formation of GICs and TMZ resistance has not been identified at present. The present study demonstrated that it serves a crucial role in regulating TMZ resistance and GIC formation. In addition, consistent with a previous study, it was identified that FAP-α may promote proliferation, migration and invasion in glioma cells (11). The results suggest that FAP-α may serve an important role in the prognosis of glioblastoma and is a potential target for therapy.
In conclusion, the present study demonstrated that miR-204 serves a crucial role in the extensive network of cellular pathways underlying glioblastoma carcinogenesis, and identified a novel target gene, FAP-α. Thus, the restoration of miR-204's levels to physiological levels observed in non-tumor tissues may represent a novel therapeutic strategy for glioblastoma.
Acknowledgements
Not applicable.
Funding
The present study was supported by Beijing Chaoyang Hospital, Capital Medical University (Beijing, China).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
YNY and ZG performed the majority of the experimental work, initially conceived the study and wrote a draft of the manuscript. XHZ, YMW and XZ performed the remainder of the experimental work and helped during the preparation of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the ethics committee of Beijing Chaoyang Hospital, and each patient signed an informed consent form at the time of enrollment.
Consent for publication
Consent for publication was obtained from each patient.
Competing interests
All authors declare that there are no competing interests.
References
- 1.Schwartzbaum JA, Fisher JL, Aldape KD, Wrensch M. Epidemiology and molecular pathology of glioma. Nat Clin Pract Neurol. 2006;2:494–503. doi: 10.1038/ncpneuro0289. [DOI] [PubMed] [Google Scholar]
- 2.Ostrom QT, Gittleman H, Farah P, Ondracek A, Chen Y, Wolinsky Y, Stroup NE, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neuro Oncol. 2013;15(Suppl 2):ii1–ii56. doi: 10.1093/neuonc/not151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buckner JC. Factors influencing survival in high-grade gliomas. Seminars in oncology Elsevier. 2003:10–14. doi: 10.1053/j.seminoncol.2003.11.031. [DOI] [PubMed] [Google Scholar]
- 4.Curran WJ, Jr, Scott CB, Horton J, Nelson JS, Weinstein AS, Fischbach AJ, Chang CH, Rotman M, Asbell SO, Krisch RE, et al. Recursive partitioning analysis of prognostic factors in three Radiation Therapy Oncology Group malignant glioma trials. J Natl Cancer Inst. 1993;85:704–710. doi: 10.1093/jnci/85.9.704. [DOI] [PubMed] [Google Scholar]
- 5.DeAngelis LM. Brain tumors. N Engl J Med. 2001;344:114–123. doi: 10.1056/NEJM200101113440207. [DOI] [PubMed] [Google Scholar]
- 6.Fine HA. The basis for current treatment recommendations for malignant gliomas. J Neurooncol. 1994;20:111–120. doi: 10.1007/BF01052722. [DOI] [PubMed] [Google Scholar]
- 7.Chen WT, Kelly T. Seprase complexes in cellular invasiveness. Cancer Metastasis Rev. 2003;22:259–269. doi: 10.1023/A:1023055600919. [DOI] [PubMed] [Google Scholar]
- 8.Rosenblum JS, Kozarich JW. Prolyl peptidases: A serine protease subfamily with high potential for drug discovery. Curr Opin Chem Biol. 2003;7:496–504. doi: 10.1016/S1367-5931(03)00084-X. [DOI] [PubMed] [Google Scholar]
- 9.Liao XH, Lu DL, Wang N, Liu LY, Wang Y, Li YQ, Yan TB, Sun XG, Hu P, Zhang TC. Estrogen receptor α mediates proliferation of breast cancer MCF-7 cells via a p21/PCNA/E2F1-dependent pathway. FEBS J. 2014;281:927–942. doi: 10.1111/febs.12658. [DOI] [PubMed] [Google Scholar]
- 10.Jia J, Martin TA, Ye L, Jiang WG. FAP-α (Fibroblast activation protein-α) is involved in the control of human breast cancer cell line growth and motility via the FAK pathway. BMC Cell Biol. 2014;15:16. doi: 10.1186/1471-2121-15-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mentlein R, Hattermann K, Hemion C, Jungbluth AA, Held-Feindt J. Expression and role of the cell surface protease seprase/fibroblast activation protein-α (FAP-α) in astroglial tumors. Biol Chem. 2011;392:199–207. doi: 10.1515/bc.2010.119. [DOI] [PubMed] [Google Scholar]
- 12.Yoshikawa K, Noguchi K, Nakano Y, Yamamura M, Takaoka K, Hashimoto-Tamaoki T, Kishimoto H. The Hippo pathway transcriptional co-activator, YAP, confers resistance to cisplatin in human oral squamous cell carcinoma. Int J Oncol. 2015;46:2364–2370. doi: 10.3892/ijo.2015.2948. [DOI] [PubMed] [Google Scholar]
- 13.Lee RC, Feinbaum RL, Ambros V. The C. Elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 14.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Müller P, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 15.Goscinski MA, Suo Z, Flørenes VA, Vlatkovic L, Nesland JM, Giercksky KE. FAP-alpha and uPA show different expression patterns in premalignant and malignant esophageal lesions. Ultrastruct Pathol. 2008;32:89–96. doi: 10.1080/01913120802034934. [DOI] [PubMed] [Google Scholar]
- 16.Jiang C, Shen F, Du J, Hu Z, Li X, Su J, Wang X, Huang X. MicroRNA-564 is downregulated in glioblastoma and inhibited proliferation and invasion of glioblastoma cells by targeting TGF-β1. Oncotarget. 2016;7:56200–56208. doi: 10.18632/oncotarget.8987. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Gao YT, Chen XB, Liu HL. Up-regulation of miR-370-3p restores glioblastoma multiforme sensitivity to temozolomide by influencing MGMT expression. Sci Rep. 2016;6:32972. doi: 10.1038/srep32972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song S, Fajol A, Tu X, Ren B, Shi S. miR-204 suppresses the development and progression of human glioblastoma by targeting ATF2. Oncotarget. 2016;7:70058–70065. doi: 10.18632/oncotarget.11732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mao J, Zhang M, Zhong M, Zhang Y, Lv K. MicroRNA-204, a direct negative regulator of ezrin gene expression, inhibits glioma cell migration and invasion. Mol Cell Biochem. 2014;396:117–128. doi: 10.1007/s11010-014-2148-6. [DOI] [PubMed] [Google Scholar]
- 20.Xia Z, Liu F, Zhang J, Liu L. Decreased expression of MiRNA-204-5p contributes to glioma progression and promotes glioma cell growth, migration and invasion. PLoS One. 2015;10:e0132399. doi: 10.1371/journal.pone.0132399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Lu DL, Sookthai D, Le Cornet C, Katzke VA, Johnson TS, Kaaks R, Fortner RT. Reproducibility of serum oxysterols and lanosterol among postmenopausal women: Results from EPIC-Heidelberg. Clin Biochem. 2017;52:117–122. doi: 10.1016/j.clinbiochem.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 23.Sato A, Sunayama J, Matsuda K, Seino S, Suzuki K, Watanabe E, Tachibana K, Tomiyama A, Kayama T, Kitanaka C. MEK-ERK Signaling Dictates DNA-Repair Gene MGMT expression and temozolomide resistance of stem-like glioblastoma cells via the MDM2-p53 Axis. Stem Cells. 2011;29:1942–1951. doi: 10.1002/stem.753. [DOI] [PubMed] [Google Scholar]
- 24.Li P, Lu X, Wang Y, Sun L, Qian C, Yan W, Liu N, You Y, Fu Z. MiR-181b suppresses proliferation of and reduces chemoresistance to temozolomide in U87 glioma stem cells. J Biomed Res. 2010;24:436–443. doi: 10.1016/S1674-8301(10)60058-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.She X, Yu Z, Cui Y, Lei Q, Wang Z, Xu G, Luo Z, Li G, Wu M. miR-181 subunits enhance the chemosensitivity of temozolomide by Rap1B-mediated cytoskeleton remodeling in glioblastoma cells. Med Oncol. 2014;31:892. doi: 10.1007/s12032-014-0892-9. [DOI] [PubMed] [Google Scholar]
- 26.Anido J, Sáez-Borderías A, Gonzàlez-Juncà A, Rodón L, Folch G, Carmona MA, Prieto-Sánchez RM, Barba I, Martínez-Sáez E, Prudkin L, et al. TGF-β receptor inhibitors target the CD44(high)/Id1(high) glioma-initiating cell population in human glioblastoma. Cancer Cell. 2010;18:655–668. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 27.Stupp R, Mason WP, Van Den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 28.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 29.Wong ST, Zhang XQ, Zhuang JT, Chan HL, Li CH, Leung GK. MicroRNA-21 inhibition enhances in vitro chemosensitivity of temozolomide-resistant glioblastoma cells. Anticancer Res. 2012;32:2835–2841. [PubMed] [Google Scholar]
- 30.Ying Z, Li Y, Wu J, Zhu X, Yang Y, Tian H, Li W, Hu B, Cheng SY, Li M. Loss of miR-204 expression enhances glioma migration and stem cell-like phenotype. Cancer Res. 2013;73:990–999. doi: 10.1158/0008-5472.CAN-12-2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.