Abstract
The kinesin super-family protein (KIF) 4A gene is reported to be overexpressed and associated with poor clinical prognosis in human cancers; however, its clinical significance in prostate cancer (PCa) has not been well studied. The present study performed dataset analyses and revealed that KIF4A expression was significantly increased in castration-resistant PCa patients. Additionally, KIF4A expression was significantly highly expressed in PCa tissues compared with non-cancerous tissues, particularly in advanced PCa pathological stages. Upregulated KIF4A mRNA expression in PCa tissues was significantly correlated with shorter overall survival and prostate-specific antigen failure. Furthermore, both univariate and multivariate analyses revealed that upregulated KIF4A may predict poor biochemical recurrence (BCR)-free survival. The data suggested that KIF4A may play a key role in PCa progression. Notably, increased KIF4A expression may potentially predict poor BCR-free survival in PCa patients.
Keywords: prostate cancer, kinesin super-family, biochemical recurrence-free survival, bioinformatics
Introduction
Prostate cancer (PCa) is the second most common cancer in men with its incidence and mortality increasing in recent years (1–3). As a heterogeneous disease, PCa is influenced by gene aberrations, cellular context and environmental factors (4). Analysis of gene expression profiles with high-throughput platforms are increasingly considered to be effective tools for researching oncology progression mechanisms. Many gene expression databases and PCa profiles have been built using microarray and sequence technology. Accumulating evidence has shown that multiple genes and signaling pathways participate in PCa tumor carcinogenesis, progression and recurrence (5,6); however, the tumor's mechanisms require further study. Therefore, identifying effective biomarkers to better predict the diagnostic and prognostic levels of this malignancy is of vital importance.
Kinesins are a family of molecular motor proteins that travel along microtubule tracks, playing multiple roles in intracellular transport and cell division (7). Previous studies have revealed that kinesins have several biofunctions in tumor malignancies, such as development and progression (8). Multiple studies have shown that kinesin super-family protein 4A (KIF4A) plays critical roles in controlling spindle organization and completing cytokinesis chromosome alignment and chromosome condensation (9–14). Dysregulating KIF4A causes abnormal spindle separation and further induces aneuploidy, which can cause cells to gain or lose genetic material (15). In addition, aneuploidy is highly associated with cancer progression (16). KIF4A is reported to be overexpressed in many cancers, such as liver, lung, oral, and gastric cancers and is amplified in cervical cancer (17–21). KIF4A is considered to be an oncogene that prompts malignancy. Previous studies showed that overexpressed KIF4A inhibits cancer cell growth in the stomach (22); however, its clinical values and roles in PCa are unclear. In the present study, bioinformatic analysis was performed to study KIF4A expression patterns and investigate the relationship between KIF4A and clinical prognosis in PCa patients.
In the present study, we demonstrated that increased KIF4A expression may be a potential clinical and prognostic biomarker in PCa. Cox proportional hazards regression model analysis showed that increased KIF4A expression was correlated with clinicopathological PCa values. This KIF4A expression pattern could be used as a prognostic biomarker in PCa to predict poor patient outcomes.
Materials and methods
Differential gene expression analysis (DGEA)
High-throughput genomic datasets were downloaded from the Gene Expression Omnibus (GEO) database (accession no. GSE32269 and GSE35988). Raw expression data were downloaded as a SOFT formatted family file. GENE-E software (version 3.0.215, Broad Institute, Inc., Cambridge, MA, USA) was used to perform the DGEA. We used a t-test to identify differential expressed genes (DEGs) with a change ≥2-fold and P<0.05 was considered to indicate a statistically significant difference.
Patients samples
Clinical information was obtained from the Taylor dataset (including 150 PCa tissues and 29 adjacent non-cancerous prostate tissues; accession no. GSE21032), TCGA dataset [498 PCa tissues with no normal prostate tissue; Prostate Adenocarcinoma (TCGA, Provisional) http://www.cbioportal.org/study?id=prad_tcga#summary] and Michigan dataset [50 metastatic castration-resistant prostate cancer (CRPC) and 11 high-grade localized PCa tissues; http://www.cbioportal.org/study?id=prad_mich#summary] (23–25). The Taylor and Michigan datasets were used to investigate the KIF4A mRNA expression profile, while the Taylor and TCGA datasets were used to perform further survival analysis.
Statistical analysis
Statistical analyses were performed using SPSS version 17.0 software for Windows (SPSS Inc., Chicago, IL, USA). We used an independent Student's t-test to analyze the results and data expressed as the mean ± standard deviation (SD). The Kaplan-Meier curve method was conducted for survival analysis. Cox proportional hazard regression models were constructed to determine the prognostic value of KIF4A expression for biochemical recurrence-free survival. First, we analyzed connections between biochemical recurrence-free survival and potential prognostic factors, including Gleason score, prostate-specific antigen (PSA), pathological stage, age and clinical stage, considering one factor at a time and all factors as continuous parameters. Second, multivariate Cox analysis was applied using backward stepwise procedures that always forced KIF4A expression into the model. P<0.05 was considered to indicate a statistically significant difference.
Results
Identifying upregulated genes in CRPC
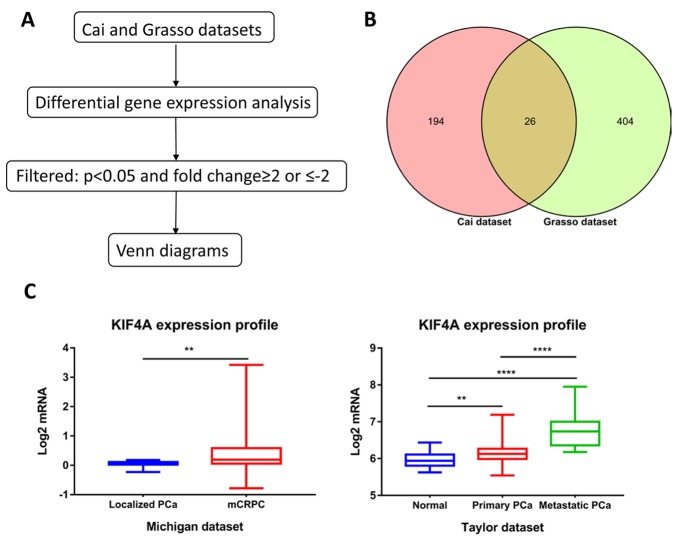
DGEA was analyzed from two datasets (GSE32269 and GSE35988). In the Cai dataset (GSE32269), 22 primary hormone-dependent PCa samples and 29 metastatic CRPC samples were analyzed using GENE-E software, and DEGs lists were identified. A total of 279 genes were identified to be differentially expressed, with thresholds of P<0.05 and fold changes ≥2.0 or ≤-2, which contained 220 upregulated and 59 downregulated genes. In the Grasso dataset (GSE35988), we analyzed differential gene expression between 59 localized PCa samples and 35 metastatic CRPC samples. In total, 430 upregulated and 1,001 downregulated genes were chosen (Fig. 1A). Next, we calculated the intersection of two datasets via Venn diagrams, and 26 genes were commonly upregulated in these datasets (Fig. 1B; Table I).
Figure 1.
Identification commonly upregulated genes. (A) Flowchart for identifying dysregulated genes through DGEA. (B) Venn diagram depicting the commonly upregulated genes in Cai and Grasso datasets. (C) Upregulated genes expression profiles in Michigan and Taylor datasets. (**P<0.001; ****P<0.0001). KIF, kinesin super-family protein; PCa, prostate cancer;
Table I.
Commonly upregulated genes list.
| Cai dataset | Grasso dataset | |||
|---|---|---|---|---|
| Gene | logFC | P | logFC | P-value |
| AR | 2.17 | 1.19×10−14 | 2.1 | 2.67×10−08 |
| ASPM | 2.15 | 1.24×10−12 | 3.99 | 7.95×10−19 |
| BIRC5 | 1.75 | 6.06×10−11 | 2.53 | 6.12×10−14 |
| BUB1 | 1.85 | 5.21×10−15 | 2.3 | 6.28×10−11 |
| CENPA | 1.53 | 1.52×10−12 | 3.06 | 1.34×10−11 |
| CENPF | 1.55 | 9.40×10−14 | 3.5 | 6.98×10−09 |
| DDIT4 | 1.96 | 1.43×10−10 | 2.06 | 1.77×10−13 |
| ESPL1 | 1.50 | 2.38×10−10 | 2.39 | 4.72×10−19 |
| EZH2 | 1.79 | 2.81×10−13 | 2.05 | 6.60×10−15 |
| FOXM1 | 1.53 | 3.08×10−11 | 2.63 | 3.15×10−18 |
| GTSE1 | 1.27 | 3.08×10−09 | 2.47 | 4.20×10−11 |
| HBG1 | 3.39 | 6.40×10−14 | 2.03 | 6.23×10−05 |
| IBSP | 3.69 | 1.92×10−12 | 2.61 | 8.29×10−04 |
| KIAA0101 | 1.53 | 1.04×10−10 | 4.20 | 3.11×10−03 |
| KIF11 | 1.30 | 5.87×10−10 | 2.83 | 5.82×10−15 |
| KIF14 | 1.48 | 9.75×10−15 | 2.29 | 3.40×10−04 |
| KIF18B | 1.54 | 1.27×10−09 | 2.69 | 2.09×10−11 |
| KIF20A | 2.22 | 7.21×10−13 | 2.33 | 5.10×10−12 |
| KIF2C | 1.17 | 2.90×10−12 | 2.52 | 1.90×10−19 |
| KIF4A | 2.15 | 4.84×10−17 | 2.06 | 5.01×10−13 |
| MELK | 1.77 | 1.03×10−10 | 2.26 | 9.06×10−11 |
| NUSAP1 | 1.90 | 2.22×10−11 | 2.08 | 5.21×10−10 |
| POLQ | 1.38 | 3.71×10−11 | 2.67 | 1.22×10−14 |
| PTTG1 | 1.50 | 3.43×10−13 | 2.24 | 3.63×10−16 |
| SPAG5 | 1.26 | 8.53×10−10 | 2.71 | 1.09×10−22 |
| TPX2 | 1.94 | 1.01×10−10 | 2.45 | 3.08×10−15 |
EZH2, polycomb protein enhancer of zeste homolog 2.
Increased KIF4A expression in human PCa and CRPC tissues
Among the 26 commonly upregulated genes, androgen receptor (AR) was broadly considered to contribute to castration-resistant disease progression via multiple mechanisms including AR overexpression. Polycomb protein enhancer of zeste homolog 2 (EZH2) as a methyltransferase in the PRC2 complex (Polycomb Repressive Complex 2) is regularly overexpressed in several human cancers, particularly CRPC. KIF4A (abnormal spindle microtubule assembly) is associated with poor clinical prognosis in most tumors and may be a biomarker for predicting poor BCR-free survival in PCa patients. Interestingly, 6 genes (KIF11, KIF18B, KIF14, KIF4A, KIF2C, and KIF20A) from the kinesin superfamily are motor proteins that convert chemical energy into mechanical force (21). Previous studies reported that the KIF4 subfamily was vitally important for tumor development and progression (26–28). We performed KIF4A gene expression profiles on the Taylor and Michigan datasets to further evaluate the dysregulation (Fig. 1C). In the Taylor dataset, we confirmed that KIF4A expression increased significantly with tumor progression (P<0.001), and KIF4A was significantly upregulated in metastatic CRPC compared to localized PCa in the Michigan dataset (P<0.003). Moreover, we examined the connection of KIF4A mRNA expression with different clinic-pathological characteristics according to Taylor and TCGA dataset (Table II). The results showed that patients with high Gleason score (≥8), short overall survival time, positive PSA failure had upregulated expression levels than those with low Gleason score (<8), long over survival time, negative PSA failure (P<0.001, in both datasets).
Table II.
Connection of KIF4A expression with clinicopathological characteristics of PCa in Taylor dataset and TCGA dataset.
| Taylor dataset (23) | TCGA dataset (25) | |||||
|---|---|---|---|---|---|---|
| Variable | n | Mean ± SD | P | n | Mean ± SD | P-value |
| KIF4A expression | ||||||
| Benign | 29 | 5.97±0.23 | – | |||
| PCa | 150 | 6.23±0.35 | <0.001 | 498 | 0.121±1.569 | – |
| Serum PSA | <0.001 | |||||
| <4 (ng/ml) | 24 | 6.21±0.34 | 411 | 0.011±1.000 | ||
| ≥4 (ng/ml) | 123 | 6.21±0.32 | 0.977 | 27 | 1.517±5.048 | |
| Age, years | 0.306 | |||||
| <66 | 124 | 6.21±0.33 | 352 | 0.075±1.664 | ||
| ≥66 | 26 | 6.31±0.41 | 0.201 | 143 | 0.235±1.300 | |
| Pathological stage | <0.001 | |||||
| <T3A | 86 | 6.16±0.24 | 186 | −0.300±0.480 | ||
| ≥T3A | 55 | 6.27±0.42 | 0.056 | 302 | 0.307±1.245 | |
| Gleason score | <0.001 | |||||
| <8 | 117 | 6.14±0.24 | 290 | −0.255±0.528 | ||
| ≥8 | 22 | 6.50±0.47 | <0.001 | 205 | 0.653±2.252 | |
| Overall survival | <0.001 | |||||
| Alive | 131 | 6.18±0.29 | 485 | 0.069±1.068 | ||
| Decease | 19 | 6.57±0.51 | <0.001 | 10 | 2.653±7.752 | |
| PSA failure | <0.001 | |||||
| Negative | 104 | 6.13±0.24 | 398 | −0.032±0.904 | ||
| Positive | 36 | 6.39±0.43 | <0.001 | 91 | 0.524±1.508 | |
-, lack of relative information. KIF, kinesin super-family protein; PCa, prostate cancer; PSA, prostate-specific antigen.
High KIF4A expression as a prognostic factor of human PCa
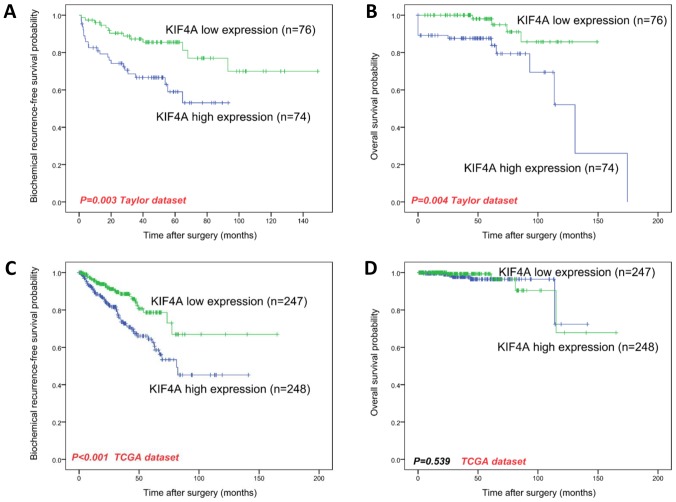
Biochemical recurrence (BCR) is defined as a surrogate endpoint following radical prostatectomy. As with overall survival, BCR-free survival is vital to PCa patients, and it is clinically used to identify those who would benefit from early initiation of salvage treatment for additional therapy. We used the Kaplan-Meier curve method to evaluate the relationship between KIF4A expression and both overall and BCR-free survival in PCa datasets. We used the median KIF4A mRNA expression level as the cutoff point to split all samples into KIF4A-high (n=248, in the TCGA dataset; n=74, in the Taylor dataset) and KIF4A-low (n=247, in the TCGA dataset; n=76, in the Taylor dataset) groups. We confirmed that the BCR-free survival between KIF4A-high and KIF4A-low group samples was statistically significant (P<0.001, in the TCGA dataset; P=0.003, in the Taylor dataset, Fig. 2 A, C), and overall survival in the Taylor dataset was statistically significant (P=0.004, Fig. 2B); however, overall survival in the TCGA dataset did not significantly differ (P=0.539, Fig. 2D).
Figure 2.
Kaplan-Meier survival analysis of BCR-free survival (A and C) and overall survival (B and D) for KIF4A expression in PCa. KIF4A mRNA expression level showed a prognostic value in BCR-free survival (A, and C; both P<0.05) in two PCa datasets and in overall survival (C) in Taylor dataset, but not in overall survival (D) in TCGA dataset. KIF, kinesin super-family protein; PCa, prostate cancer; BCR, biochemical recurrence.
To further discuss KIF4A's prognostic value in PCa, we conducted univariate and multivariate Cox proportional hazards regression to verify KIF4A's clinical prognostic value in PCa datasets (Tables III and IV). We confirmed that KIF4A (P<0.001 in the Taylor dataset), the Gleason score (P<0.001 in both datasets), PSA (P=0.009 in the Taylor dataset) and pathological stage (P<0.001 in the Taylor dataset) were appropriate for being considered as prognosis factors for BCR-free survival via univariate analysis. Next, we performed multivariate analysis and found that high KIF4A expression levels (P=0.005 in the Taylor datasets), Gleason scores (P=0.003 in the Taylor dataset, P<0.001 in TCGA dataset), PSA (P=0.010 in the Taylor dataset) and pathological stage (P=0.004 in the Taylor dataset) are potentially independent factors for predicting shorter BCR-free survival. Due to the number of deceased patients in TCGA dataset (10 deaths in 499 samples), KIF4A's clinical prognostic value in overall survival still need to be studied in the future.
Table III.
Prognostic value of KIF4A mRNA expression level for the BCR-free survival via Cox proportional hazards model.
| Taylor dataset (23) | TCGA dataset (25) | |||
|---|---|---|---|---|
| Variable | Hazard ratio (95% CI) | P | Hazard ratio (95% CI) | P-value |
| Univariate analysis | ||||
| KIF4A mRNA | 3.76 (2.086–6.779) | <0.001 | 1.200 (0.996–1.445) | 0.055 |
| Gleason score | 2.665 (1.879–3.779) | <0.001 | 1.730 (1.300–2.302) | <0.001 |
| PSA | 1.005 (1.001–1.008) | 0.009 | – | |
| Pathological stage | 3.143 (1.961–5.038) | <0.001 | 1.190 (0.681–2.080) | 0.541 |
| Age | 1.024 (0.973–1.076) | 0.363 | 0.992 (0.960–1.026) | 0.651 |
| Clinical stage | 1.429 (0.808–2.525) | 0.219 | – | |
| Multivariate analysis | ||||
| KIF4A mRNA | 4.591 (1.584–13.309) | 0.005 | 1.436 (1.271–1.623) | 0.051 |
| Gleason score | 1.844 (1.240–2.741) | 0.003 | 2.202 (1.769–2.741) | <0.001 |
| PSA | 1.005 (1.001–1.009) | 0.010 | – | |
| Pathological stage | 2.198 (1.281–3.770) | 0.004 | 2.590 (1.717–3.908) | 0.067 |
| Age | 1.024 (0.974–1.077) | 0.837 | 1.027 (0.925–1.060) | 0.589 |
| Clinical stage | 1.415 (0.799–2.505) | 0.514 | – | |
-, lack of relative information. KIF, kinesin super-family protein; BCR, biochemical recurrence; PSA, prostate-specific antigen.
Table IV.
Prognostic value of KIF4A mRNA expression level for the overall survival via Cox proportional hazards model.
| Taylor dataset (23) | TCGA dataset (25) | |||
|---|---|---|---|---|
| Variable | Hazard ratio (95% CI) | P | Hazard ratio (95% CI) | P-value |
| Univariate analysis | ||||
| KIF4A mRNA | 38.090 (7.044–205.973) | <0.001 | 0.610 (0.174–2.134) | 0.439 |
| Gleason score | 1.127 (0.743–1.710) | 0.574 | 1.730 (1.300–2.302) | 0.074 |
| PSA | 1.005 (1.000–1.010) | 0.067 | – | |
| Pathological stage | 1.206 (0.454–3.205) | 0.707 | 0.129 (0.015–1.095) | 0.061 |
| Age | 1.048 (0.960–1.144) | 0.294 | 0.993 (0.882–1.118) | 0.911 |
| Clinical stage | 1.955 (0.757–5.065) | 0.168 | – | |
| Multivariate analysis | ||||
| KIF4A mRNA | 41.798 (9.665–151.730) | <0.001 | – | – |
| Gleason score | 1.844 (1.240–2.741) | 0.003 | – | – |
| PSA | 1.006 (1.001–1.010) | 0.013 | – | – |
| Pathological stage | – | – | – | – |
| Age | – | – | – | – |
| Clinical stage | – | – | – | – |
-, lack of relative information. KIF, kinesin super-family protein; BCR, biochemical recurrence; PSA, prostate-specific antigen.
Discussion
PCa has imposed a considerable economic burden on our society (29). Although many PCa patients have benefitted from improved techniques such as PSA screening, the time between tumor progression onset and BCR varies (30). Most castration-resistant PCa patients eventually relapse, resulting in metastasis and invasion, and because no effective treatments are currently available, their prognosis declines and eventually results in death. Therefore, identifying reliable biomarkers is vitally important for predicting recurrence and prognosis. To our knowledge, abnormally increased KIF4A expression has been reported in lung, oral, cervical, gastric and liver cancers (17–21); however, no research has been conducted on KIF4A in prostate neoplasms.
KIF4A mRNA and protein were reported to be significantly upregulated in vitro, and KIF4A protein expression was found to be significantly overexpressed in primary oral squamous cell carcinomas, which was closely correlated with tumor size (17). Further research demonstrated that KIF4A was a diagnostic/prognostic biomarker that was highly transactivated in many lung cancers by cDNA microarray, tumor tissue microarray, and immunohistochemical staining and was associated with male sex, non-adenocarcinoma histology, and shorter survival for the lung cancer patients (18). KIF4A was also reported to be upregulated in cervical cancer by cDNA array comparative genomic hybridization (aCGH) analysis (19). In addition, Hou et al demonstrated that KIF4A was overexpressed in hepatocellular carcinoma (HCC) tissue, which was significantly associated with survival time and clinical information (stage, metastasis and tumor dimension). Further functional analysis of siRNA-mediated silencing and KEGG pathway enrichment analysis showed that KIF4A promoted HCC cell growth and metastasis by mediating cell cycle-related and p53 signaling pathways (20). In the present study, using DGEA we found that KIF4A was highly expressed in CRPC, and further survival analysis showed that KIF4A may be an important oncogene. We investigated KIF4A gene expression profiles in different PCa datasets and confirmed that the KIF4A expression level was significantly increased in PCa samples, especially those in advanced pathological stages. To further validate the representativeness of its prognostic value in PCa, we conducted Kaplan-Meier survival analysis and Cox proportional hazards regression. The results showed that increased KIF4A expression was correlated with adverse BCR-free survival.
Furthermore, KIF4A overexpression enhanced migration and proliferation in lung cancer and HCC (18,20), and KIF4A was found to control cellular proliferation via spindle assembly checkpoint (SAC) activation (17–20). Recent studies have revealed that the KIF4 subfamily is important in tumor development and progression (9–14). KIF4A plays critical roles in controlling spindle organization and completing cytokinesis chromosome alignment and chromosome condensation. Dysregulated KIF4A may induce cellular aneuploidy, causing cells to gain or lose genetic material (15). KIF4A is reported to be overexpressed in many cancers, and the present study showed that in PCa, KIF4A was overexpressed and may indicate poor prognosis. We innovatively used bioinformatic analysis in different datasets to cross-validate and screen possible genes that may potentially function as novel prognostic biomarkers. In addition, studies have demonstrated that several KIF family proteins are involved in developing drug resistance in cancer cells (31). Liu et al showed silencing MPHOSPH1 (also referred to as KIF20B) plus chemotherapy in HCC greatly improved antitumor treatment (32). Therefore, a combined strategy that targets KIF4A during PCa treatment may provide great efficacy. KIF4A is considered to be an oncogene that prompts malignancy; however, the mechanisms of KIF4A influencing PCa progression and prognosis still need further study.
In summary, our data showed for the first time that KIF4A was overexpressed in PCa, especially in CRPC. Increased KIF4A expression significantly predicted worse BCR-free survival and has the potential to be a prognostic PCa biomarker for predicting poor patient outcomes. Although the mechanism of increased KIF4A expression in PCa requires further study, KIF4A may serve as a therapeutic target for PCa.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant nos. 81472682 and 81772756), and Natural Science Foundation of Tianjin (grant nos. 17JCZDJC35300, 15JCZDJC35400 and 15JCYBJC27200).
Availability of data and materials
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YN and ZS supervised the whole study and participated in study design and coordination, analysis and interpretation of data, material support for obtained funding. HG and XC performed most of the experiments and data statistical analysis and were major contributors in writing the manuscript. QC carried out the data collection. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Chang CC, Lee YC, Tsai HW, Yii SC, Yen TH, Chu FY. Diagnostic role of serum free-to-total prostate specific antigen (PSA) ratio in prostate cancer with serum total concentration of PSA below 4 ng/ml. Asian Pac J Cancer Prev. 2015;16:5261–5264. doi: 10.7314/APJCP.2015.16.13.5261. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Koo KC, Park SU, Kim KH, Rha KH, Hong SJ, Yang SC, Chung BH. Predictors of survival in prostate cancer patients with bone metastasis and extremely high prostate-specific antigen levels. Prostate Int. 2015;3:10–15. doi: 10.1016/j.prnil.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, Tamayo P, Renshaw AA, D'Amico AV, Richie JP, et al. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1:203–209. doi: 10.1016/S1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 5.Heestand GM, Kurzrock R. Molecular landscape of pancreatic cancer: Implications for current clinical trials. Oncotarget. 2015;6:4553–4561. doi: 10.18632/oncotarget.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khemlina G, Ikeda S, Kurzrock R. Molecular landscape of prostate cancer: Implications for current clinical trials. Cancer Treat Rev. 2015;41:761–766. doi: 10.1016/j.ctrv.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 7.Myers SM, Collins I. Recent findings and future directions for interpolar mitotic kinesin inhibitors in cancer therapy. Future Med Chem. 2016;8:463–489. doi: 10.4155/fmc.16.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu X, Gong H, Huang K. Oncogenic role of kinesin proteins and targeting kinesin therapy. Cancer Sci. 2013;104:651–656. doi: 10.1111/cas.12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rath O, Kozielski F. Kinesins and cancer. Nat Rev Cancer. 2012;12:527–539. doi: 10.1038/nrc3310. [DOI] [PubMed] [Google Scholar]
- 10.Bernasconi P, Cappelletti C, Navone F, Nessi V, Baggi F, Vernos I, Romaggi S, Confalonieri P, Mora M, Morandi L, Mantegazza R. The kinesin superfamily motor protein KIF4 is associated with immune cell activation in idiopathic inflammatory myopathies. J Neuropathol Exp Neurol. 2008;67:624–632. doi: 10.1097/NEN.0b013e318177e5fd. [DOI] [PubMed] [Google Scholar]
- 11.Hu CK, Coughlin M, Field CM, Mitchison TJ. KIF4 regulates midzone length during cytokinesis. Curr Biol. 2011;21:815–824. doi: 10.1016/j.cub.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samejima K, Samejima I, Vagnarelli P, Ogawa H, Vargiu G, Kelly DA, de Lima Alves F, Kerr A, Green LC, Hudson DF, et al. Mitotic chromosomes are compacted laterally by KIF4 and condensin and axially by topoisomerase IIα. J Cell Biol. 2012;199:755–770. doi: 10.1083/jcb.201202155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurasawa Y, Earnshaw WC, Mochizuki Y, Dohmae N, Todokoro K. Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J. 2004;23:3237–3248. doi: 10.1038/sj.emboj.7600347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mazumdar M, Sundareshan S, Misteli T. Human chromokinesin KIF4A functions in chromosome condensation and segregation. J Cell Biol. 2004;166:613–620. doi: 10.1083/jcb.200401142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wordeman L. How kinesin motor proteins drive mitotic spindle function: Lessons from molecular assays. Semin Cell Dev Biol. 2010;21:260–268. doi: 10.1016/j.semcdb.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oki E, Hisamatsu Y, Ando K, Saeki H, Kakeji Y, Maehara Y. Clinical aspect and molecular mechanism of DNA aneuploidy in gastric cancers. J Gastroenterol. 2012;47:351–358. doi: 10.1007/s00535-012-0565-4. [DOI] [PubMed] [Google Scholar]
- 17.Minakawa Y, Kasamatsu A, Koike H, Higo M, Nakashima D, Kouzu Y, Sakamoto Y, Ogawara K, Shiiba M, Tanzawa H, Uzawa K. Kinesin Family member 4A: A potential predictor for progression of human oral cancer. PLoS One. 2013;8:e85951. doi: 10.1371/journal.pone.0085951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taniwaki M, Takano A, Ishikawa N, Yasui W, Inai K, Nishimura H, Tsuchiya E, Kohno N, Nakamura Y, Daigo Y. Activation of KIF4A as a prognostic biomarker and therapeutic target for lung cancer. Clin Cancer Res. 2007;13:6624–6631. doi: 10.1158/1078-0432.CCR-07-1328. [DOI] [PubMed] [Google Scholar]
- 19.Narayan G, Bourdon V, Chaganti S, Arias-Pulido H, Nandula SV, Rao PH, Gissmann L, Dürst M, Schneider A, Pothuri B, et al. Gene dosage alterations revealed by cDNA microarray analysis in cervical cancer: Identification of candidate amplified and overexpressed genes. Genes Chromosomes Cancer. 2007;46:373–384. doi: 10.1002/gcc.20418. [DOI] [PubMed] [Google Scholar]
- 20.Hou G, Dong C, Dong Z, Liu G, Xu H, Chen L, Liu L, Wang H, Zhou W. Upregulate KIF4A enhances proliferation, invasion of hepatocellular carcinoma and indicates poor prognosis across human cancer types. Sci Rep. 2017;7:4148. doi: 10.1038/s41598-017-04176-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imai T, Oue N, Nishioka M, Mukai S, Oshima T, Sakamoto N, Sentani K, Matsusaki K, Yoshida K, Yasui W. Overexpression of KIF11 in gastric cancer with intestinal mucin phenotype. Pathobiology. 2017;84:16–24. doi: 10.1159/000447303. [DOI] [PubMed] [Google Scholar]
- 22.Gao J, Sai N, Wang C, Sheng X, Shao Q, Zhou C, Shi Y, Sun S, Qu X, Zhu C. Overexpression of chromokinesin KIF4 inhibits proliferation of human gastric carcinoma cells both in vitro and in vivo. Tumour Biol. 2011;32:53–61. doi: 10.1007/s13277-010-0090-0. [DOI] [PubMed] [Google Scholar]
- 23.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research Network: The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu C, Jiang W. Cell cycle-dependent translocation of PRC1 on the spindle by Kif4 is essential for midzone formation and cytokinesis. Proc Natl Acad Sci USA. 2005;102:343–348. doi: 10.1073/pnas.0408438102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miki H, Okada Y, Hirokawa N. Analysis of the kinesin superfamily: Insights into structure and function. Trends Cell Biol. 2005;15:467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 28.Miki H, Setou M, Kaneshiro K, Hirokawa N. All Kinesin Superfamily Protein, KIF, Genes in Mouse and Human. Proc Natl Acad Sci USA. 2001;98:7004–7011. doi: 10.1073/pnas.111145398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skolarus TA, Zhang Y, Miller DC, Wei JT, Hollenbeck BK. The economic burden of prostate cancer survivorship care. J Urol. 2010;184:532–538. doi: 10.1016/j.juro.2010.03.136. [DOI] [PubMed] [Google Scholar]
- 30.Esfahani M, Ataei N, Panjehpour M. Biomarkers for evaluation of prostate cancer prognosis. Asian Pac J Cancer Prev. 2015;16:2601–2611. doi: 10.7314/APJCP.2015.16.7.2601. [DOI] [PubMed] [Google Scholar]
- 31.Liu J, Wang H, Huang C, Qian H. Subcellular localization of MTA proteins in normal and cancer cells. Cancer Metastasis Rev. 2014;33:843–856. doi: 10.1007/s10555-014-9511-7. [DOI] [PubMed] [Google Scholar]
- 32.Liu X, Zhou Y, Liu X, Peng A, Gong H, Huang L, Ji K, Petersen RB, Zheng L, Huang K. MPHOSPH1: A potential therapeutic target for hepatocellular carcinoma. Cancer Res. 2014;74:6623–6634. doi: 10.1158/0008-5472.CAN-14-1279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request.