Abstract
The mass accuracy and peak intensity of ions detected by mass spectrometry (MS) measurements are essential to facilitate compound identification and quantitation. However, high concentration species can yield erroneous results if their ion intensities reach beyond the limits of the detection system, leading to distorted and non-ideal detector response (e.g. saturation), and largely precluding the calculation of accurate m/z and intensity values. Here we present an open source computational method to correct peaks above a defined intensity (saturated) threshold determined by the MS instrumentation such as the analog-to-digital converters or time-to-digital converters used in conjunction with time-of-flight MS. In this method, the isotopic envelope for each observed ion above the saturation threshold is compared to its expected theoretical isotopic distribution. The most intense isotopic peak for which saturation does not occur is then utilized to re-calculate the precursor m/z and correct the intensity, resulting in both higher mass accuracy and greater dynamic range. The benefits of this approach were evaluated with proteomic and lipidomic datasets of varying complexities. After correcting the high concentration species, reduced mass errors and enhanced dynamic range were observed for both simple and complex omic samples. Specifically, the mass error dropped by more than 50% in most cases for highly saturated species and dynamic range increased by 1–2 orders of magnitude for peptides in a blood serum sample.
Keywords: Mass spectrometry, Detector saturation, Analog-to-digital converter saturation, Saturation correction, Isotopic envelope, Isotopic ratios, Quantitation
1. Introduction
Current mass spectrometers provide molecular measurements (i.e. m/z) with both high mass accuracy and resolving power for concentrations typically ranging from 3 to 4 orders of magnitude in a single mass spectrum depending upon instrumental details. These measurements are extremely important for identifying molecules occurring in complex samples and determining how they change under varying biological and environmental conditions. Two important characteristics for confident molecular identifications from mass spectrometry (MS) are the isotopic peaks (isotopic envelope) for the ion observed and tandem (MS/MS) measurements of fragment ion species. Together these observations yield information about the molecular mass, observed charge state, elemental composition and structural arrangement for the potential molecule. However, both mass and quantitation errors occur in MS measurements for numerous reasons and understanding their source is necessary to ensure that incorrect conclusions are not made. Mass error has been discussed in detail in several publications [1–5] and is associated with all MS measurements for reasons related to both the MS platform type and non-ideal performance through the instrument (e.g. slight deviations in voltage and pressure). Fundamental limitations also influence mass error when insufficient ion statistics result in the inability to accurately define a peak. Quantitation errors are associated with ion statistics and limits of the MS detector dynamic range, which impair accurate measurements of both the high and low concentration species in a sample. If the concentration of a molecule is too low, ions can be lost while traveling through the instrument, resulting in no detection or not enough ion accumulation to create a well-defined peak. High concentration species also cause issues if their signal intensity exceeds the detector capacity saturating one or more of the isotopic peaks. Furthermore, if the isotopic envelope contains one or more saturated peaks, the natural distribution is distorted and results in incorrect quantitative readings [6]. These incorrect intensity values returned by the deisotoping algorithms are not accurate representations of the actual ion abundances regardless of whether the maximum intensity, area or volume is used. Detector saturation is commonly observed in analog-to-digital converters (ADC) or time-to-digital converters used in conjunction with time-of-flight (TOF) mass spectrometers [2,6–8]. However, saturation has also been observed in other MS instrumentation such as triple quadrupoles [9,10] and trap based [11] instruments, and its extent usually depends on the detector design. In addition, the incorporation of on-line separations, such as liquid chromatography (LC) and ion mobility spectrometry (IMS) also result in saturation due to the concentrating effect of the separations.
Practical approaches for avoiding saturation include optimizing sample concentrations and instrument operating settings in order to keep the abundances below the known saturation levels of the platform [8,12]. However, this precludes covering the large dynamic range needed in many applications such as that occurring in blood plasma and environmental samples. Over the last decade, several other approaches have been implemented to extend dynamic range and reduce saturation effects. A majority of these advances have been based on hardware or instrumental techniques applied during MS data acquisition to avoid or minimize saturating the detection systems [9,13,14]. However, as hardware has improved, a handful of post-acquisition software approaches have also been utilized or suggested to correct the saturated MS data. For example, in targeted MS analysis with selected reaction monitoring, an algorithm called SignalFinder was integrated into the SCIEX proprietary software MultiQuant and utilized for saturation correction in data from triple quadrupole MS systems [10]. In the case of untargeted MS-studies, to our knowledge, no proprietary or open source software is available to perform saturation correction post-acquisition. Nevertheless, strategies to correct the saturated peaks using isotopic distributions have been suggested in the literature. For instance, theoretical isotopic distributions can be compared against the most intense observed isotopic envelopes in the data to detect saturation [7]. Furthermore, the intensity and mass shift errors of a saturated ion peak can be corrected by using information from the adjacent (e.g. second or third C13) isotopic peak that is not saturated [2]. Several methods addressing the computationally demanding calculations for theoretical isotopic distributions have been reported [15–22] and reviewed in particular for large biomolecules [23]. Among the cited examples, methods based on utilizing the molecular formula of a known molecular species are preferred and known to be most accurate. However, the elemental composition of ionized molecular species are not always known and, indeed, establishing their identity is often a key aspect of the measurements. In proteomic analyses, methods utilizing known elemental compositions to determine the monoisotopic mass based on the makeup of amino acids provide a good estimation [21,23].
Considering these initial ideas, we implemented a computational method to first detect all possible saturated peaks in a spectrum by flagging those with an intensity above a defined ADC threshold (e.g., 70%), which is close to the ADC capacity of the utilized instrument. The saturated peaks were then corrected using the unsaturated peaks in the isotopic envelope and theoretical isotopic models based upon assumed elemental compositions of the analytes (e.g. peptides). The utility of this approach was then evaluated by analyzing datasets of varying complexity and determining their dynamic range, mass error and quantitation accuracy before and after correction.
2. Materials and methods
2.1. Software algorithm
The software algorithm described in this manuscript first identifies saturated peaks in the mass spectrum. Since a 256 channel 8-bit ADC digitizer was utilized in the MS studies, the maximum signal possible per pixel was 256. The total number of pixels per accumulated scan was then considered, and a peak at or above a threshold of 70% of this number was determined to be saturated. Each flagged peak was then further investigated by utilizing the unsaturated peaks from the ion’s isotopic envelope detected in a mass spectrum. To correct the saturated peaks, the intensity values of the peaks of the isotopic envelope {A1, A2 … An} and the intensity values of the peaks of a theoretical isotopic envelope {T1, T2 … Tn} were required, where n is the number of isotopic peaks.
For the peptide analyses, the software implementation reported here used the so-called averagine method to create an approximate peptide molecular formula from the given monoisotopic mass [21]. Since the purpose of the algorithm is to report an accurate intensity value for each observed isotopic envelope instead of attempting to correct every saturated value in the profile mass spectrum or ion distribution, a peak centroiding method was applied to the isotopic peaks for both the observed and theoretical isotopic envelopes prior to comparison. Intensity values of saturated isotopic peaks were corrected by assuming that, when unsaturated, the relative intensities of individual peaks of the observed isotopic envelope will match the relative intensity of the isotopic peaks of the theoretical isotopic envelope. In other words, the intensity ratio for each isotope in the form observed/theoretical, is constant for all isotopic peaks. The correction of each saturated peak was accomplished using the formula:
where A is the group of observed isotopic peaks, T is the group of theoretical isotopic peaks, x is the index of the isotopic peak being corrected, and u is the index of the most intense observed isotopic peak that is not saturated. The algorithm then iterates over the observed isotopic peaks to find the most intense isotopic peak (Au) that is not saturated since it has both an accurate abundance and the best signal quality. The current version of the algorithm requires defining the saturation level of the detector, which is characteristic for a given mass spectrometer. Using the index of the selected peak, the relative intensity of the corresponding peak (Tu) of the theoretical isotopic envelope is determined. For each saturated isotopic peak (Ax), the new intensity value is computed by multiplying the relative intensity of the corresponding theoretical peak (Tx) by the defined ratio (Au ÷ Tu). Similarly, the new m/z value of the saturated isotopic peak is computed by back calculating from the m/z of the unsaturated isotopic peak.
The described method for saturation correction was implemented as a command-line application in C# and it has been integrated into the data processing pipeline for LC-IMS–MS datasets at the Pacific Northwest National Laboratory. Raw MS files in the Unified Ion Mobility Frame (UIMF) file format [24] were processed by DeconTools [25,26], where the saturation correction was integrated. R software (v3.3.1 × 64) was then used for visualization and comparison of the results from the DeconTools output. This software is freely available and can be downloaded from http://omics.pnl.gov/software/decontools-decon2ls. The source code is available at https://github.com/PNNL-Comp-Mass-Spec/DeconTools. Saturation correction was enabled in DeconTools by setting the “ScanBasedWorkflowType” parameter to uimf_saturation_repair. See, for example, the parameter file SampleParameterFileIMS.xml in the repository on GitHub.
2.2. Instrumentation
All data files were acquired with an IMS-MS platform previously described [27,28]. Briefly, the IMS instrument couples a 1-m IM drift cell with an Agilent 6224 TOF MS (Agilent Technologies, Santa Clara, CA) that was upgraded to a 1.5-m flight tube. The signal from the TOF detector was routed to a 8-bit ADC (AP240, Acqiris, Switzerland) and processed using a custom control-software written in C#. The software saves all experimental parameters as well as data into a UIMF file. All experiments described were collected from the 100 to 3200 m/z range.
LC separations were utilized for the more complex proteomic and lipidomic studies. The LC system operated in the proteomic experiments was custom built using two Agilent 1200 nanoflow pumps and one Agilent 1200 cap pump (Agilent Technologies, Santa Clara, CA), various Valco valves (Valco Instruments Co., Houston, TX), and a PAL autosampler (Leap Technologies, Carrboro, NC). Full automation was made possible by custom software that allows for parallel event coordination and nearly 100% MS duty cycle through use of two analytical columns. Reversed phase columns were prepared in-house by slurry packing 3 µm Jupiter C18 (Phenomenex, Torrence, CA) into fused silica columns (Polymicro Technologies Inc., Phoenix, AZ) with a 1-cm sol-gel frit at the end for media retention. Both a 60-min LC gradient (30-cm long columns × 360 µm o.d. × 75 µm i.d.) and 100-min LC gradient (60-cm long columns × 360 µm o.d. × 75 µm i.d.) were performed in this manuscript. Mobile phases consisted of 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B) and the gradient profiles were as follows: 60-min gradient profile, (min:%B); 0:0, 1.2:8, 12:12, 51:35, 58.2:60, 60:95, 64:95, 65:0 and 100-min profile, (min:%B); 0:5, 2:8, 20:12, 75:35, 97:60, 100:75, 103:5; and each was followed by a column wash, (min:%B); 0:5, 1.25:35, 2.5:5, 3.75:35, 5:5, 6:90, 14:90, 16:5, 16.25:35, 17.5:5, 18.75:35, 20:5. Five micro-liters of sample was injected for both analyses and the HPLC was operated under a constant flow rate of 0.3 µL/min for the 100-min gradient and 1 µL/min for the 60-min gradient.
For the lipidomic LC analyses, a Waters Aquity UPLC H class system was used. Lipid extracts were dried in vacuo and reconstituted in 200 µL methanol. 10 µL was then injected onto a reversed phase Waters CSH column (3.0 mm × 150 mm × 1.7 µm particle size). The lipids were then separated over a 34-min gradient, (min:%B); 0:40, 2:50, 3:60, 12:70, 15:75, 17:78, 19:85, 22:92, 25:99, 34:99, where mobile phase A consisted of ACN/H2O (40:60) containing 10 mM ammonium acetate and mobile phase B was ACN/IPA (10:90) containing 10 mM ammonium acetate. The HPLC which was operated under a constant flow rate of 250 µL/min during the 34-min gradient.
2.3. Standards and sample preparation
Peptide standards (angiotensin I, melittin, and leucin-enkephalin (leu-enkephalin)) and reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA). The tryptically digested bovine serum albumin (BSA) and yeast were purchased from Promega (Madison, WI, USA) and the porcine brain total lipid extract (BTLE) was acquired from Avanti Polar Lipids (Alabaster, AL, USA). Human serum datasets from the study detailed in (Nielson et al. [29]) were also used for the complex proteomic analyses. In this study, the serum was prepared by unthawing 150 µL and depleting it of the 14 high abundance proteins using IgY14 immunoaffinity depletion columns (Sigma-Aldrich). The serum was subsequently digested with trypsin and prepared for instrumental analysis with LC-IMS–MS.
A simple 3-peptide sample with large dynamic range was prepared for the initial studies by spiking the three peptide standards at 100 pM (angiotensin I), 1 µM (melittin) and 100 µM (leu-enkephalin) in a (49.75/49.75/0.5) water/methanol/acetic acid buffer to cover a dynamic range of 106. This sample was directly injected in the IMS-MS at a flow rate of 200 nL/min. A more complex sample was then prepared by spiking four different concentrations of a BSA digest (1 nM, 10 nM, 100 nM and 1 µM) into a 0.1 µg/uL yeast digest. Each BSA/yeast mixture was analyzed with the 100 min LC-IMS–MS platform. The most complex proteomic sample was the human serum which was analyzed with the 60 min LC-IMS–MS platform [29] and had numerous species that co-eluted. Finally, to investigate how the algorithm worked in lipidomic studies, the BTLE sample was analyzed using the 34-min lipid LC method coupled to the IMS-MS platform.
3. Results and discussion
Quantitation and mass error resulting from peak saturation can be a major limitation in studies where samples have a large dynamic range (i.e. plasma, tissue, etc.). While saturated peaks have a detrimental impact on the dynamic range and mass error observed, these can be mitigated using data processing software to correct both values. Our program specifically looks for a defined saturation level according to the MS instrument used. Basically, the effects of excessive ion current are manifested differently and vary depending upon details of the detector design. For instance, the Agilent TOF system with an ADC displays a discrete saturation level where highly saturated peaks are often observed with a “flat top” at the apex. Other MS instruments, such as those using a TDC, will have more gradual onset and simply result in a less linear gain. Moreover, some MS systems may “lock up” detection, recording no signal at all. Defining the saturation threshold on any MS instrumentation should be possible by identifying the transition from unsaturated to saturated peak based on the hardware present. In our setup, the first saturation point in the hardware is the ADC. Thus, we defined this level and flagged all peaks within 70% of this value for further investigation. The isotopic peaks from the flagged ions were then grouped and the expected isotopic distribution was computed for comparison after determining the first unsaturated peak. This is extremely important because if any of the isotopic peaks of a particular envelope are saturated, then the reported intensity for the ion will be too low if they are used. Thus, this method corrects all saturated peaks of the isotopic envelope as described in the methods section, allowing for an accurate intensity value to be reported. In the studies described below, we analyzed the performance of the saturation correction algorithm on samples with different complexities. To understand the full effect of the saturation correction method, the complete spectra from each sample was processed two times, one time with the correction set-ting disabled (before correction) and a second time with this setting enabled (after correction).
3.1. Increased dynamic range and reduced mass error
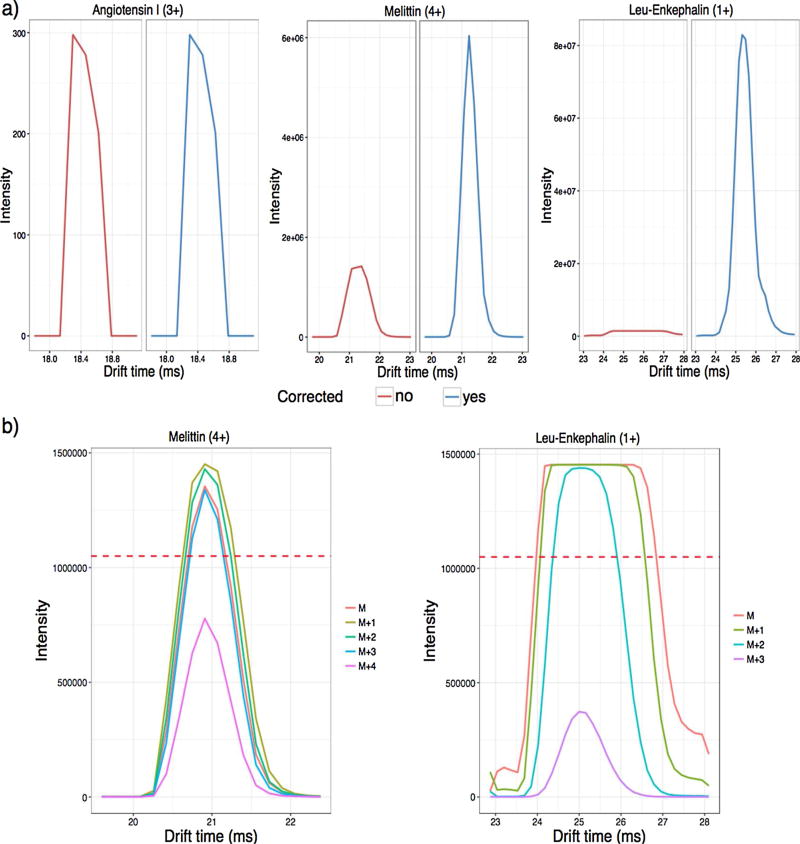
We first evaluated our saturation correction software by processing the dataset acquired from the 3-peptide dynamic range sample which was directly infused into the IMS-MS platform. Since the peptides were spiked at specific concentrations, we could compare the expected dynamic ranges to the results reported by the data processing software before and after correction. Initially, we examined the IMS arrival time distributions (ATDs) for the monoisotopic ion of each peptide (Fig. 1a). Because angiotensin I was only injected at 100 pM, it was not near the saturation level and therefore no correction was applied. However, both melittin and leu-enkephalin, which were several orders of magnitude more concentrated, were significantly saturated with several isotopic peaks reaching intensities above the 70% saturation threshold (shown with a dotted line in Fig. 1b), illustrating that correction was needed. When the isotopic envelopes were extracted, the most intense isotopic peak chosen by the algorithm to apply the correction corresponded to the (M + 4) peak for melittin (or 4th C13 peak) and the (M + 3) peak for leu-enkephalin (or 3rd C13 peak), due to the differences in the envelopes for a 4+ peptide and 1+ peptide. Each of these was below the 70% saturation threshold as shown in Fig. 1b. Even though our method performs saturation correction for each mass spectrum individually, the improvements can be observed across the whole IMS ATD profile. As shown in Fig. 1a, the ATDs of the corrected peptides preserved the base width, however, the intensity was corrected around the apex, where the values exceeded the saturation threshold.
Fig. 1.
IMS arrival time distributions (ATDs) for the 3 peptides in the dynamic range sample. a) The IMS ATDs for the monoisotopic ions before (red) and after (blue) saturation correction, showing no change for angiotensin I, but huge modifications in the abundances of melittin and leu-enkephalin. b) The IMS ATDs of the precursor ion and following isotopic peaks extracted from the raw data for melittin and leu-enkephalin (the two saturated peptides). The dashed line shows the 70% saturation threshold and illustrates why the algorithm picked the (M + 4) peak for melittin and the (M + 3) peak for leu-enkephalin for correction. Angiotensin I is not shown in these plots due to the large difference between the observed intensity and 70% saturation threshold used for correction. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The abundance and ppm error of the three peptides were further evaluated in Table 1. Even after processing the complete dataset with the saturation correction enabled, no changes were observed in abundance and ppm error for angiotensin I, since it was below the 70% saturation threshold. However, abundances increased for the other two peptides and their mass variability was reduced. This was most notable for leu-enkephalin, which was the most concentrated peptide at 100 µM and thus most prone to saturation. After applying saturation correction, the resulting dynamic ranges were also much closer to the known dynamic range of the sample. Small deviations from the actual dynamic ranges were still observed and we attribute these variations mainly to three different factors: multimerization, ionization efficiency, and the presence of multiple charge states for melittin. Previously, we have observed that at high concentrations peptides can form multimers, and hence, obtaining accurate abundances may not be possible without more advanced data analysis methods to include all abundances of present dimers, trimers, etc. Leu-enkephalin was observed as a monomer, dimer, trimer and tetramer in the spectra. However, we only compared the monomer to the other ions in Table 1, since multimer post-processing is not currently performed in the algorithm. In addition, the key point of our saturation correction software is to establish more precise relative abundances. Therefore, even though the leu-enkephalin abundance is lower than the known sample concentration, we preferred the table to reflect the actual output achieved by the algorithm. We did add the dimer and trimer onto the abundance manually, but this still was not sufficient to obtain the values equal to the actual dynamic range of the solution. This suggests that other issues related to detector recovery, such as a reduced response for the isotopic peaks that follow a saturated peak (i.e. close in m/z), may occur after a high concentration ion. Ionization efficiency can also have an effect on the intensity of each peptide and we know that melittin does not ionize as well as the other two peptides due to the higher charge state needed. While this may be one of the reasons it shows a lower dynamic range than expected, melittin was also observed as a 3+, 4+ and 5+ ion. Both the 3+ and 5+ charge states were very low in abundance though, so Table 1 only reflects the dynamic range for the dominant 4+ charge state. Despite these limitations, saturation correction enhanced the dynamic range observed between leu-enkephalin/melittin and leu-enkephalin/angiotensin I by an order of magnitude in both cases. These improvements in abundances and mass errors for the saturated ions illustrate the utility of this algorithm, even in simple samples.
Table 1.
Abundances and mass errors before and after saturation correction for the 3 peptide dynamic range sample.
| Analyte | Concentration (µM) | Abundance | Mass Error (ppm) | Dynamic Range (vs. Leu-Enkephalin) | ||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||
| Before | After | Before | After | Before | After | Actual | ||
| Angiotensin I (3+) | 1.00E − 04 | 7.77E + 02 | 7.77E + 02 | 0.56 | 0.56 | 4.09E + 04 | 7.27E + 05 | 1.00E + 06 |
| Melittin (4+) | 1.00E + 00 | 6.64E + 06 | 2.22E + 07 | 0.94 | 0.80 | 4.79E + 00 | 2.55E + 01 | 1.00E + 02 |
| Leu-Enkephalin (1+) | 1.00E + 02 | 3.18E + 07 | 5.65E + 08 | 6.33 | 0.97 | – | – | – |
Note: Abundance is the area of the monoisotopic peak across the IM ATD and mass error was computed from the standard deviation of the monoisotopic ion across the IM ATD.
3.2. Improved quantitation linearity
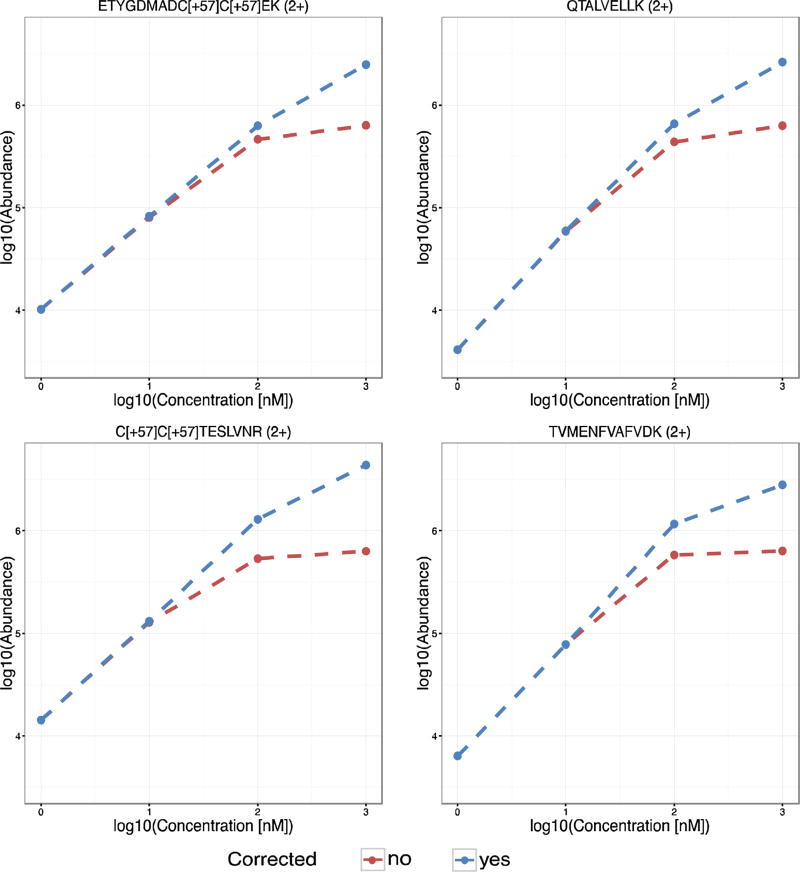
To further validate the saturation correction software, we examined calibration curves acquired by spiking different concentrations of a BSA tryptic digest (1 nM, 10 nM, 100 nM and 1 µM) into a tryptic digest of yeast (0.1 µg/uL). This evaluation was crucial to assess the quantitation accuracy of our method and applicability to conditions closer to real proteomics studies, where an LC gradient is used in most cases because of sample complexity. Each data file corresponded to a single spiked concentration was individually processed, allowing calibration curves to be constructed. Saturation was observed at 100 nM and 1 µM for several peptides in the BSA digest and correlates to when the analyte response flattens as the concentration increases, as seen in Fig. 2 for four example BSA digest peptides. Saturation correction significantly improved the quantitation linearity for all four peptides with the best results illustrated for ETYGDMADC[+57]C[+57]EK (2+). Although linearity was not fully recovered for any of the four peptides, saturation correction increased the experimental abundances at 100 nm and 1 µM for all peptides to values closer to those expected based on extrapolation from the lower concentrations. As previously mentioned, at such high concentrations the peptides can form multimers and, although saturation correction significantly improves the quantitative measurement, complete quantitation accuracy may not be possible without more complex data reconstruction. Nevertheless, caution should be taken to avoid over processing the spectra and introducing errors.
Fig. 2.
Calibration curves for BSA peptides spiked in a yeast background at 1 nM, 10 nM, 100 nM and 1 µM. The calibration curves show the linearity improvements before (red) and after (blue) saturation correction for four example saturated BSA peptides. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Saturation correction is also relevant to data-independent acquisition (DIA) MS analyses, which are continuous MS/MS maps for all eluting LC peaks [30,31]. The datasets of the BSA tryptic digest spiked at different concentration in the yeast background were acquired in DIA mode by alternating low and high collision energy to generate MS and MS/MS spectra. While the total number of saturated MS/MS features in the high collision energy spectra were less than those from the precursor MS spectra, we observed that some fragment ions did reach saturation levels, especially at the 1 µM spiked concentration (Supplementary Fig. 1). Our algorithm was also able to correct the fragment ion values, allowing MS/MS quantitation to be preserved. To enable saturation correction on the DIA data, the same “ScanBasedWorkflowType” parameter was set to uimf_saturation_repair so that correction was performed for each spectrum individually. The MS features in the output CSV file can then be split according to the used MS acquisition settings for alternating low and high collision energy, for example, using the frameNum column with even number values corresponding to MS features and odd number values corresponding to fragment ion MS/MS features as long as this alternating order was maintained across the complete acquisition. Correcting saturated MS/MS features is very important to obtain consistent and improved results, since saturation also affects the DIA computational processing, where the similarity of the elution profiles and comparison of the peptide fragmentation pattern against library spectra are used for scoring peptide identification. For example, in approaches like SWATH [32,33], the apex of the extracted ion chromatogram (XIC) for saturated fragment ions is obscured by a flat line and is not similar to the unsaturated fragment ions, which impacts the final peptide score. Likewise, in untargeted data processing such as DIA-Umpire [34,35], the similarity of the saturated precursor LC peak and unsaturated fragment LC peaks will be reduced, potentially affecting the precursor/fragment groupings which generate the pseudo-DDA spectra used for identification. Saturation correction is thus pivotal to enhance the grouping and identification steps in DIA.
3.3. Correction of saturated features detected in blood serum
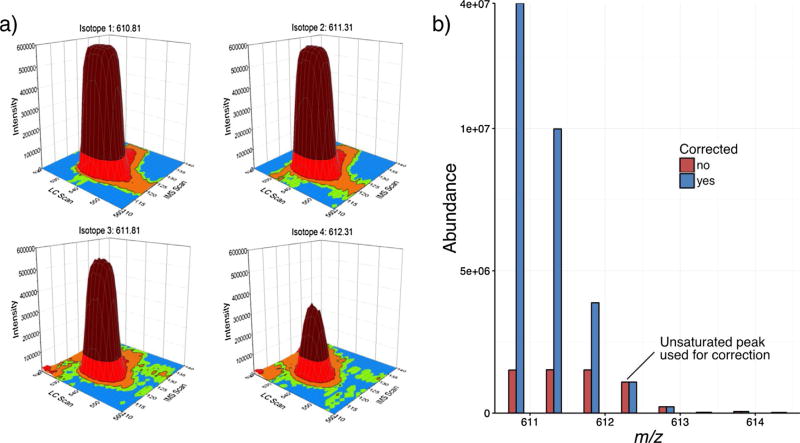
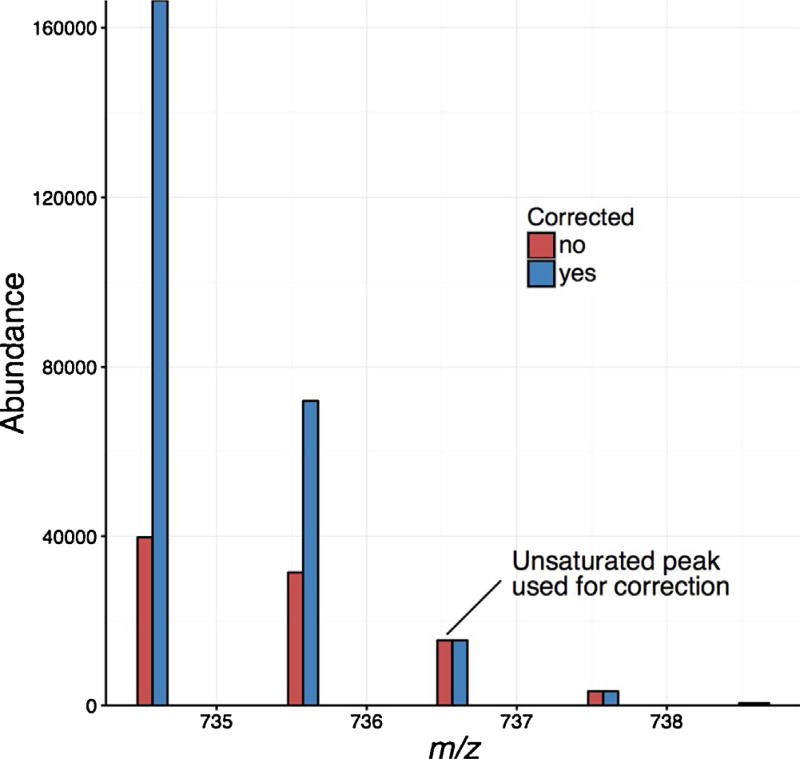
To implement saturation correction on a very complex sample, depleted human serum datasets acquired from a previous proteomics study [29] were also evaluated. Serum datasets are particularly useful for exploring the benefits of our saturation correction method since they have an extended dynamic range and numerous species with saturated isotopic profiles. Since the serum datasets were acquired using LC and IMS separations, 2D-XICs were reconstructed from the raw data. Fig. 3a shows an example set of isotopic peaks for one of the saturated peptides. The characteristic flat profile can be observed at the apex of the isotopic peaks affected by saturation (m/z values: 610.81, 611.31 and 611.81), and is most notable for the monoisotopic peak. When examining the isotopic envelope, the first unsaturated peak was (M + 3). Upon using it for saturation correction, a nine-fold increase was observed for the monoisotopic peak of this peptide (Fig. 3b), further extending the observed sample dynamic range.
Fig. 3.
An example saturated peptide from human serum. a) The 2D XICs for the LC and IMS profiles of the saturated (m/z values: 610.81, 611.31 and 611.81) and unsaturated (m/z value 612.31) peaks from the raw data. The colors in the plots correspond to the intensity of each peak. b) The mass spectrum for this peptide before (red) and after (blue) saturation correction using the most abundant unsaturated peak (M + 3). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
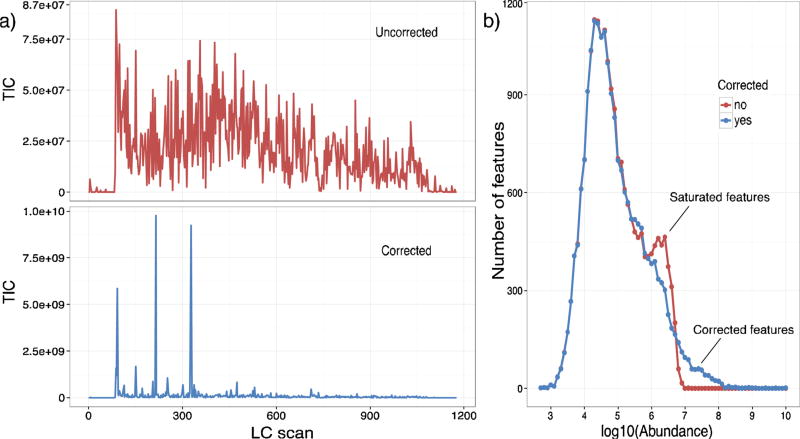
Next, we investigated the overall outcome of saturation correction on all the peptide LC-IMS features detected in the serum datasets. The LC-IMS-MS FeatureFinder [36], available at https://github.com/PNNL-Comp-Mass-Spec/LC-IMS-MS-Feature-Finder, was used to group the MS features as LC-IMS features. As Fig. 4a illustrates, the maximum abundances of the total ion chromatograms (TICs) greatly increased upon saturation correction and reconstruction of the feature abundances. Specifically, the observed total ion abundance increased by two orders of magnitude (from 8.75E + 07 to ~1E + 10), indicating a much greater dynamic range for the serum sample. To further investigate the intensity change, all features were plotted against their log10 abundances in Fig. 4b. This distribution confirmed that the low abundance features were unchanged by the correction method, while the saturated features increased after correction from a log10 abundance of ~6.5 to >8. Since the lowest concentration species had a log10 abundance of ~3, utilization of the saturation algorithm increased the dynamic range from 103.5 to 105. Thus, saturation correction should be beneficial for increasing the dynamic range in all complex biological and environmental samples.
Fig. 4.
Dynamic range increases in serum samples. The abundance changes in all human serum peptide features were analyzed before and after saturation correction. a) The abundance increase in the TICs (reconstructed from feature abundances) for an example serum dataset before (top) and after (bottom) saturation correction. b) The distribution of feature abundances in that same dataset before (red) and after correction (blue) illustrates a drop off of features at the saturation level before correction and an increase in dynamic range following. Each set of isotopic peaks in the LC and IMS dimensions were grouped and counted as a single feature. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Correcting other omic data
Currently, our open source saturation software only supports proteomic data as it utilizes the averagine model to create the theoretical isotopic pattern. However, the same algorithm can be applied to other types of analytes such as metabolites and lipids by adapting software components that generate the theoretical isotopic envelope. To demonstrate this capability, we analyzed a brain total lipid extract sample with LC-IMS–MS and flagged lipid profiles that were above the ADC saturation threshold. One of the phosphatidylcholine lipids, PC(16:0/16:0), was found to be greatly saturated as shown in Fig. 5. To correct its theoretical isotopic distribution, we utilized www.chemcalc.org [37] to generate the theoretical distribution based on the molecular formula of the lipid. A comparison of the experimental data to the theoretical spectrum showed that the highest unsaturated peak was (M + 2), so it was utilized in the abundance correction. Correction of lipidomic data is very useful since lipid classes vary greatly in intensity. Some researchers even perform two analyses per sample, one at higher concentration and one at lower concentration to detect the diverse classes in the higher concentration sample and avoid saturation in the lower concentration version. By applying saturation correction to the lipidomic data, this type of double analysis could be eliminated.
Fig. 5.
The lipid isotopic profile for PC(16:0/16:0) before (red) and after (blue) saturation correction. The theoretical isotopic distribution for PC(16:0/16:0) was computed with www.chemcalc.org, which illustrated that the (M + 2) peak was the first unsaturated peak in this profile, thus it was used for correction. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Implementing automatic saturation correction for small molecule studies such as in lipidomic and metabolomic measurements is challenging since specific formulas must be known to correct the isotopic profiles. We are currently examining tools that perform this function and hope to add this capability in future versions. For instance, our software FormulaFinder (https://github.com/PNNL-Comp-Mass-Spec/Molecular-Weight-Calculator-DLL/blob/master/clsFormulaFinder.vb) can be used to first generate the molecular formula from accurate masses and then produce theoretical isotopic distributions. To this aim, we are working to integrate all “seven golden rules” [38] into an improved version of the tool, which will be essential to automatically constrain the thousands of possible candidate structures while selecting the most likely and chemically correct molecular formula. We expect these additions will make the saturation correction algorithm even more valuable to the MS community.
4. Conclusions
We developed and evaluated a computational method to correct saturated ion abundances in MS data. This algorithm flags all peaks at a defined instrumental threshold, which can be based to on ADC or detector saturation level. The isotopic envelopes for all flagged peaks are then compared to the theoretical distributions and the most intense unsaturated peak is used for mass and abundance correction. The algorithm was evaluated using proteomic datasets of varying complexities from a 3 peptide sample to human serum, and illustrated enhancements in quantitation, mass accuracy and dynamic range for all complexity ranges. While we primarily demonstrated the saturation correction method for IMS-MS and LC-IMS–MS data, the method performs the correction at the mass spectrum level, therefore the software can be applied to MS, IMS-MS, LC–MS, LC-IMS–MS data, etc. In addition, we anticipate the algorithm will work for correcting saturated ions in data from other detection systems, such as the ones using TDC. By defining an appropriate saturation threshold, according to the hardware response, the same comparison principle of observed and theoretical isotopic envelops should be applicable.
Our approach best applies where a theoretical isotopic distribution can be assumed with some level of confidence. We are working on further improving the software to automatically process other omic data, although this requires formulas and theoretical isotopic envelopes to be generated by a model other than averagine, which is substantially more challenging and is still under development. As currently implemented, we believe that our saturation correction software provides significant advantages for large scale MS-based studies. While the fraction of saturated analytes in a sample might be low compared to all detected features, the accurate quantitation of those features could be critical in revealing changes related to an environmental perturbation or understanding the development of a disease.
Supplementary Material
Acknowledgments
The authors would like to acknowledge John Fjeldsted for insightful discussions about instrumentation and peak saturation. Portions of this research were supported by grants from the National Institute of Environmental Health Sciences of the NIH (R01ES022190), National Institute of General Medical Sciences (P41 GM103493), the Laboratory Directed Research and Development Program at Pacific Northwest National Laboratory, and the U.S. Department of Energy Office of Biological and Environmental Research Genome Sciences Program under the Pan-omics program. This work was performed in the W. R. Wiley Environmental Molecular Sciences Laboratory (EMSL), a DOE national scientific user facility at the Pacific Northwest National Laboratory (PNNL). PNNL is operated by Battelle for the DOE under contract DE-AC05-76RL0 1830.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.ijms.2017.11.003.
References
- 1.Blom KF. Estimating the precision of exact mass measurements on an orthogonal time-of-flight mass spectrometer. Anal. Chem. 2001;73:715–719. doi: 10.1021/ac001064v. [DOI] [PubMed] [Google Scholar]
- 2.Chernushevich IV, Loboda AV, Thomson BA. An introduction to quadrupole-time-of-flight mass spectrometry. J. Mass Spectrom. 2001;36:849–865. doi: 10.1002/jms.207. [DOI] [PubMed] [Google Scholar]
- 3.Colombo M, Sirtori FR, Rizzo V. A fully automated method for accurate mass determination using high-performance liquid chromatography with a quadrupole/orthogonal acceleration time-of-flight mass spectrometer. Rapid Commun. Mass Spectrom. 2004;18:511–517. doi: 10.1002/rcm.1368. [DOI] [PubMed] [Google Scholar]
- 4.Gibbons BC, Chambers MC, Monroe ME, Tabb DL, Payne SH. Correcting systematic bias and instrument measurement drift with mzRefinery. Bioinformatics. 2015;31:3838–3840. doi: 10.1093/bioinformatics/btv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu J, McAllister H. Exact mass measurement on an electrospray ionization time-of-flight mass spectrometer: error distribution and selective averaging. J. Mass Spectrom. 2003;38:1043–1053. doi: 10.1002/jms.516. [DOI] [PubMed] [Google Scholar]
- 6.Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 2007;389:1017–1031. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- 7.Cappadona S, Baker PR, Cutillas PR, Heck AJ, van Breukelen B. Current challenges in software solutions for mass spectrometry-based quantitative proteomics. Amino Acids. 2012;43:1087–1108. doi: 10.1007/s00726-012-1289-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schriemer DC, Li L. Mass discrimination in the analysis of polydisperse polymers by MALDI time-of-flight mass spectrometry. 2. Instrumental issues. Anal. Chem. 1997;69:4176–4183. [Google Scholar]
- 9.Moulds R, Kenny D, Worthington KR, Pringle SD. Extending the linear dynamic range of quadrupole detectors. 62th ASMS Conference Mass Spectrometry Allied Topics; Baltimore, USA. 2014. [Google Scholar]
- 10.Quinn-Paquet P, Beaudet S, Zhong F, Ivosev G, Denison J. Performance of a novel algorithm for reliable LC peak integration of triple quadrupole MRM data. 58th ASMS Conference Mass Spectrometry Allied Topics; Salt Lake City, USA. 2010. [Google Scholar]
- 11.Makarov A, Denisov E, Lange O, Horning S. Dynamic range of mass accuracy in LTQ Orbitrap hybrid mass spectrometer. J. Am. Soc. Mass Spectrom. 2006;17:977–982. doi: 10.1016/j.jasms.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 12.Gutierrez JA, Dorocke JA, Knierman MD, Gelfanova V, Higgs RE, Koh NL, Hale JE. Quantitative determination of peptides using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Biotechniques. 2005;6:13–18. doi: 10.2144/05386su02. [DOI] [PubMed] [Google Scholar]
- 13.Chernushevich IV, Loboda A. Dynamic range extension for TOF MS with orthogonal injection. 55th ASMS Conference Mass Spectrometry Allied Topics; Indianapolis, USA. 2007. [Google Scholar]
- 14.Hanson CD, Just CL. Selective background suppression in MALDI-TOF mass spectrometry. Anal. Chem. 1994;66:3676–3680. [Google Scholar]
- 15.Olson MT, Yergey AL. Calculation of the isotope cluster for polypeptides by probability grouping. J. Am. Soc. Mass Spectrom. 2009;20:295–302. doi: 10.1016/j.jasms.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rockwood AL, Van Orman JR, Dearden DV. Isotopic compositions and accurate masses of single isotopic peaks. J. Am. Soc. Mass Spectrom. 2004;15:12–21. doi: 10.1016/j.jasms.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 17.Valkenborg D, Jansen I, Burzykowski T. A model-based method for the prediction of the isotopic distribution of peptides. J. Am. Soc. Mass Spectrom. 2008;19:703–712. doi: 10.1016/j.jasms.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 18.Datta BP. Polynomial method of molecular isotopic abundance calculations: a computational note. Rapid Commun. Mass Spectrom. 1997;11:1767–1774. [Google Scholar]
- 19.Hsu CS. Diophantine approach to isotopic abundance calculations. Anal. Chem. 1984;56:1356–1361. [Google Scholar]
- 20.Yergey JA. A general approach to calculating isotopic distributions for mass spectrometry. Int. J. Mass Spectrom. Ion Phys. 1983;52:337–349. doi: 10.1002/jms.4498. [DOI] [PubMed] [Google Scholar]
- 21.Senko MW, Beu SC, McLafferty FW. Determination of monoisotopic masses and ion populations for large biomolecules from resolved isotopic distributions. J. Am. Soc. Mass Spectrom. 1995;6:229–233. doi: 10.1016/1044-0305(95)00017-8. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto H, McCloskey JA. Calculations of isotopic distribution in molecules extensively labeled with heavy isotopes. Anal. Chem. 1977;49:281–283. [Google Scholar]
- 23.Valkenborg D, Mertens I, Lemiere F, Witters E, Burzykowski T. The isotopic distribution conundrum. Massspectrom. Rev. 2012;31:96–109. doi: 10.1002/mas.20339. [DOI] [PubMed] [Google Scholar]
- 24.Beagley N, Scherrer C, Shi Y, Clowers BH, Danielson WF, Shah AR. Increasing the efficiency of data storage and analysis using indexed compression; EScience,2009, eScience’09. Fifth IEEE International Conference; 2009. pp. 66–71. [Google Scholar]
- 25.Jaitly N, Mayampurath A, Littlefield K, Adkins JN, Anderson GA, Smith RD. Decon2LS: an open-source software package for automated processing and visualization of high resolution mass spectrometry data. BMC Bioinf. 2009;10:87. doi: 10.1186/1471-2105-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slysz GW, Baker ES, Shah AR, Jaitly N, Anderson GA, Smith RD. The DeconTools framework: an application programming interface enabling flexibility in accurate mass and time tag workflows for proteomics and metabolomics; Proc58th ASMS Conf Mass Spectrom Allied Topics; 2010. [Google Scholar]
- 27.Baker ES, Clowers BH, Li F, Tang K, Tolmachev AV, Prior DC, Belov ME, Smith RD. Ion mobility spectrometry–mass spectrometry performance using electrodynamic ion funnels and elevated drift gas pressures. J. Am. Soc. Mass Spectrom. 2007;18:1176–1187. doi: 10.1016/j.jasms.2007.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ibrahim YM, Baker ES, Danielson WF, Norheim RV, Prior DC, Anderson GA, Belov ME, Smith RD. Development of a new ion mobility-time-of-flight mass spectrometer. Int. J. Mass Spectrom. 2015;377:655–662. doi: 10.1016/j.ijms.2014.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nielson CM, Wiedrick J, Shen J, Jacobs J, Baker ES, Baraff A, Piehowski P, Lee CG, Baratt A, Petyuk V, McWeeney S, Lim JY, Bauer DC, Lane NE, Cawthon PM, Smith RD, Lapidus J, Orwoll ES. Identification of hip BMD loss and fracture risk markers through population-based serum proteomics. J. Bone Miner. Res. 2017;32:1559–1567. doi: 10.1002/jbmr.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bilbao A, Varesio E, Luban J, Strambio-De-Castillia C, Hopfgartner G, Müller M, Lisacek F. Processing strategies and software solutions for data-independent acquisition in mass spectrometry. Proteomics. 2015;15:964–980. doi: 10.1002/pmic.201400323. [DOI] [PubMed] [Google Scholar]
- 31.Bilbao A, Zhang Y, Varesio E, Luban J, Strambio-De-Castilla C, Lisacek F, Hopfgartner G. Ranking fragment ions based on outlier detection for improved label-free quantification in data-independent acquisition LC-MS/MS. J. Proteome Res. 2015;14:4581–4593. doi: 10.1021/acs.jproteome.5b00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gillet LC, Navarro P, Tate S, Röst H, Selevsek N, Reiter L, Bonner R, Aebersold R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteom. 2012;11 doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Röst HL, Rosenberger G, Navarro P, Gillet L, Miladinovi SM, Schubert OT, Wolski W, Collins BC, Malmström J, Malmström L. others OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat. Biotechnol. 2014;32:219–223. doi: 10.1038/nbt.2841. [DOI] [PubMed] [Google Scholar]
- 34.Tsou C-C, Avtonomov D, Larsen B, Tucholska M, Choi H, Gingras A-C, Nesvizhskii AI. DIA-Umpire: comprehensive computational framework for data independent acquisition proteomics. Nat. Methods. 2015;12:258–264. doi: 10.1038/nmeth.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsou C-C, Tsai C-F, Teo G, Chen Y-J, Nesvizhskii AI. Untargeted, spectral library-free analysis of data independent acquisition proteomics data generated using Orbitrap mass spectrometers. Proteomics. 2016;16:2257–2271. doi: 10.1002/pmic.201500526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crowell KL, Slysz GW, Baker ES, LaMarche BL, Monroe ME, Ibrahim YM, Payne SH, Anderson GA, Smith RD. LC-IMS-MS Feature Finder: detecting multidimensional liquid chromatography ion mobility and mass spectrometry features in complex datasets. Bioinformatics. 2013;29:2804–2805. doi: 10.1093/bioinformatics/btt465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patiny L, Borel A. ChemCalc: a building block for tomorrow’s chemical infrastructure. J. Chem. Inf. Model. 2013;53:1223–1228. doi: 10.1021/ci300563h. [DOI] [PubMed] [Google Scholar]
- 38.Kind T, Fiehn O. Seven Golden Rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry. BMC Bioinf. 2007;8:105. doi: 10.1186/1471-2105-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.